Summary
Background
Altered lipid metabolism is a hallmark of cancer development. However, the role of specific lipid metabolites in colorectal cancer development is uncertain.
Methods
In a case–control study nested within the European Prospective Investigation into Cancer and Nutrition (EPIC), we examined associations between pre-diagnostic circulating concentrations of 97 lipid metabolites (acylcarnitines, glycerophospholipids and sphingolipids) and colorectal cancer risk. Circulating lipids were measured using targeted mass spectrometry in 1591 incident colorectal cancer cases (55% women) and 1591 matched controls. Multivariable conditional logistic regression was used to estimate odds ratios (ORs) and 95% confidence intervals (CIs) for associations between concentrations of individual lipid metabolites and metabolite patterns with colorectal cancer risk.
Findings
Of the 97 assayed lipids, 24 were inversely associated (nominally p < 0.05) with colorectal cancer risk. Hydroxysphingomyelin (SM (OH)) C22:2 (ORper doubling 0.60, 95% CI 0.47–0.77) and acylakyl-phosphatidylcholine (PC ae) C34:3 (ORper doubling 0.71, 95% CI 0.59–0.87) remained associated after multiple comparisons correction. These associations were unaltered after excluding the first 5 years of follow-up after blood collection and were consistent according to sex, age at diagnosis, BMI, and colorectal subsite. Two lipid patterns, one including 26 phosphatidylcholines and all sphingolipids, and another 30 phosphatidylcholines, were weakly inversely associated with colorectal cancer.
Interpretation
Elevated pre-diagnostic circulating levels of SM (OH) C22:2 and PC ae C34:3 and lipid patterns including phosphatidylcholines and sphingolipids were associated with lower colorectal cancer risk. This study may provide insight into potential links between specific lipids and colorectal cancer development. Additional prospective studies are needed to validate the observed associations.
Funding
World Cancer Research Fund (reference: 2013/1002); European Commission (FP7: BBMRI-LPC; reference: 313010).
Keywords: Colorectal cancer, Metabolomics, Lipids, Glycerophospholipids, Sphingolipids, Acylcarnitines
Research in context.
Evidence before this study
The role of specific lipid metabolites on colorectal cancer risk is unclear. Prospective studies have reported both positive and inverse associations with colorectal cancer for glycerophospholipids following multiple comparison correction. However, these studies have been smaller in scale or focused on lipid clusters rather than specific lipid metabolites.
Added value of this study
This large-scale (n = 1591 cases) nested case–control study investigated the association between both pre-diagnostic circulating lipid metabolites and metabolite patterns with colorectal cancer. Comprehensive covariate data on participants were also used to identify potential fatty acid side chains and lipid determinants. We found inverse associations between concentrations of hydroxysphingomyelin (SM (OH)) C22:2 and acylakyl-phosphatidylcholine (PC ae) C34:3 with colorectal cancer risk. Crucially, these associations were consistent following lag analyses excluding early years of follow-up and according to subgroups of age, sex, BMI, and colorectal subsite.
Implications of all the available evidence
This study identified two lipid metabolites (SM (OH) C22:2 and PC ae C34:3) associated with colorectal cancer risk. These findings may provide insight into the biological mechanisms underlying the potential role of lipid metabolism in colorectal cancer development.
Introduction
Colorectal cancer was estimated to be the third most common cancer and second most common cause of cancer death globally in 2020.1 There is evidence that diet, lifestyle, and metabolic dysfunction play a role in colorectal cancer development, however, the underlying mechanisms are unclear.2, 3, 4
Metabolomic profiling enables the investigation of associations between small molecules involved in metabolic processes and disease outcomes. In targeted metabolomics, a defined set of metabolites can be identified, allowing for a better understanding of potential biological pathways of disease development and for comparison of effect estimates across studies. The role of lipid metabolites in metabolic disease outcomes is a growing area of scientific interest. Lipids play key roles in biological processes such as energy metabolism, cell membrane structure, cell signalling and proliferation,5, 6, 7, 8 and the dysregulation of lipid metabolism is often one of the first stages of tumorigenesis and is considered an emerging hallmark of cancer.7 A potential relationship between circulating lipid metabolites and colorectal cancer has also been supported by epidemiological studies linking several lipids to known colorectal cancer risk factors such as diet, cholesterol levels, adiposity, alcohol consumption, smoking, diabetes, and other lifestyle measures.9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20
Previous studies investigating the association between individual circulating lipid metabolites–specifically glycerophospholipids, sphingolipids, and acylcarnitines–and colorectal cancer risk have reported inconsistent results.21, 22, 23, 24, 25, 26 However, some of these studies collected blood samples after cancer diagnosis which may represent disease rather than aetiological risk biomarkers given the high likelihood for reverse causation.21,22,25
Few prospective studies, including three nested case–control studies, have investigated the associations between individual circulating pre-diagnostic lipid metabolites and colorectal cancer.23,24,26 A study in the European Prospective Investigation into Cancer and Nutrition (EPIC)-Heidelberg cohort (n = 163 colorectal cancer cases),24 found that circulating lysophosphatidylcholine (lysoPC) a 18:0 levels were inversely associated with colorectal cancer risk, but not after correction for multiple comparisons.24 Two studies used an untargeted metabolomics approach.23,26 One using data from the Shanghai Women's Health Study (SWHS) and the Shanghai Men's Health Study (SMHS) (n = 250 colorectal cancer cases) reported inverse associations for nine, and positive associations for three, glycerophospholipids with colorectal cancer risk.23 Another conducted on data from the Cancer Prevention Study (CPS) II Nutrition cohort (n = 517 colorectal cancer cases) investigated associations between individual lipids including acylcarnitines, glycerophosphospholipids and sphingolipids.26 However, as for the Heidelberg study,24 no associations were reported following multiple comparisons correction. There is a need for larger-scale studies to clarify the role of lipid metabolites in colorectal cancer development.
Here, we used a targeted metabolomics approach to examine the association between pre-diagnostic blood concentrations of 97 lipids and colorectal cancer risk within a large case–control study nested within the EPIC cohort (n = 1591 colorectal cancer cases; 1591 matched controls). In addition, to provide insights into fatty acid side chain composition and possible determinants of lipid concentrations, we conducted a cross-sectional analysis examining associations between circulating lipid metabolites and dietary, lifestyle and anthropometric factors.
Methods
Study population
The EPIC study is a cohort of approximately 520,000 men and women, mostly between the ages of 35–70 years at recruitment, who were enrolled between 1992 and 2000 from 23 centres across 10 European countries (Denmark, France, Germany, Greece, Italy, Netherlands, Norway, Spain, Sweden, United Kingdom [UK]).27 Participants were volunteers, largely selected from the general population, or in some instances, from breast screening (Italy [Florence], Netherlands [Utrecht]) or health insurance programmes (France), blood donors (Spain, Italy [Ragusa, Turin]) or vegetarian and vegan societies (UK [Oxford]). Blood plasma and serum were also collected from over 70% of EPIC participants and were stored at the International Agency for Research on Cancer (IARC) in liquid nitrogen at −196 °C, or for samples collected in Denmark, were stored locally in nitrogen vapour at −150 °C.27
Data for the current analysis were obtained from an established colorectal cancer case–control study nested within the cohort including participants from all countries except Sweden, Norway, and Greece for which biosamples and/or data were unavailable.
Cancer incidence data were mainly obtained from population-based cancer registries or in France and Germany through health insurance records, cancer and pathology registries or active follow-up.27 Vital statistics data were obtained from mortality registries or through active follow-up (Germany).27,28
Nested case–control study
Data for this current study were available for 3194 participants from a colorectal cancer case–control study nested within the EPIC cohort. Eligible participants were those without a prior diagnosis of invasive cancer (except for non-melanoma skin cancer) at the time of blood collection. Colorectal cancer cases were randomly selected from among invasive first primary cancers diagnosed during follow-up and defined according to International Classification of Diseases for Oncology third edition (ICD-O-3) codes C18–C20 corresponding to cancers occurring between the caecum and rectum. Controls were free of cancer (except non-melanoma skin cancer) at the time of diagnosis of cases and were selected by 1:1 incidence density sampling and matched to cases in several rounds on study centre, sex, age at blood sampling (±3–±5 years in subsequent rounds), follow-up time from blood sampling to the date of colorectal cancer case diagnosis, date (±1–±6 months) and time of day of blood sampling (±1–±3 h) and fasting status at blood collection. Women were also matched on menopausal status, phase of menstrual cycle (pre-menopausal women only) and current use of exogenous hormones, all at baseline. When more than one control was eligible for matching to a case, one was chosen at random.
Lipid metabolite data
A targeted metabolomics approach was used to analyse circulating lipid metabolites. Samples were analysed at IARC or at Helmholtz Zentrum München, Germany using a Biocrates AbsoluteIDQ™ p180 or p150 assay (Biocrates Life Science AG, Innsbruck Austria) respectively, according to manufacturer specifications.29 These assays allowed for the identification of three lipid classes: acylcarnitines (denoted as C), glycerophospholipids (lysophosphatidylcholines [denoted as lysoPC], diacylphosphatidylcholines [denoted as PC aa] and acylalkylphosphatidylcholines [denoted as PC ae]), and sphingolipids (sphingomyelins [denoted as SM] and hydroxysphingomyelins [denoted as SM (OH)]). Measurements were performed at IARC using an Agilent 1290 liquid chromatography system and a SCIEX Triple Quad 4500 mass spectrometer, whereas in Germany an Agilent 1200 liquid chromatography system and a SCIEX API 4000 mass spectrometer was used. Each case–control pair was assayed in the same laboratory and batch, and laboratory staff were blinded to case–control status.30, 31, 32
Among the 3194 participants, metabolite data were available for 113 lipids in blood samples from IARC (plasma, n = 925) and 118 lipids in blood samples from Germany (serum, n = 2269). These were lipids which could be assayed using the Biocrates kits and for which there was a detectable lipid metabolite concentration. Lipid metabolites not common to both datasets (n = 19) or lipid metabolites within each dataset with >20% of sample concentrations below the batch-specific limit of detection (LOD) were excluded from analyses (n = 9). After exclusions, metabolite data were available for 97 lipids (10 acylcarnitines, 75 glycerophospholipids and 12 sphingolipids). For the remainder of metabolites with ≤20% of sample concentrations below the batch specific LOD (n = 12), the value was imputed with half the LOD. Participants in incomplete case–control sets after data pooling were excluded (n = 12) leaving 3182 (IARC n = 922; Germany n = 2260) participants for analysis.
Tests for normality of the distribution of metabolite concentrations were conducted using the Shapiro–Francia test for normality. Lipid metabolite concentrations were shown to be non-normally distributed and therefore for statistical analyses, lipid metabolite concentrations were log2-transformed.
Covariate data
At recruitment, data on participant demographics including self-reported age, sex and education were collected via questionnaires. Dietary data were collected using self-administered (Denmark, France, Germany, Italy [Florence, Turin and Varese], Netherlands, UK) or face-to-face (Italy [Ragusa, Naples], Spain) quantitative (France, Germany, Italy, Netherlands, Spain), or semi-quantitative food-frequency questionnaires (Denmark, Italy [Naples], Spain, UK).27 Macronutrient intakes were estimated by matching dietary data to the EPIC nutrient database (ENDB).33 Lifestyle variables on physical activity, smoking, and alcohol consumption were collected using questionnaires.27 Anthropometric measurements of weight, height, waist and hip circumference were taken objectively by trained staff using standardized protocols (or self-reported in France and UK [Oxford]).27 Data on plasma phospholipid fatty acid concentrations (expressed as a percentage of total fatty acids and available for a subset of those participants whose circulating lipid metabolites were assayed at IARC; n = 922). For subsets of the sample of 3182 participants levels of serum total cholesterol (mmol/l), low-density lipoprotein (LDL) cholesterol (mmol/l), high-density lipoprotein (HDL) cholesterol (mmol/l), triglycerides (mmol/l), C-reactive protein (CRP) (ng/ml), C-peptide (ng/ml), and insulin-like growth factor-1 (IGF-1) (nmol/l) were available from previous analyses.34, 35, 36, 37, 38
Statistical analyses
Spearman pairwise correlations were estimated for log2-transformed lipid metabolite concentrations among controls, due to evidence of non-normality for some lipid metabolite concentrations, and presented with lipids ordered by hierarchical clustering using the complete method (Appendix Figure 1). Multivariable conditional logistic regression models were used to estimate odds ratios (ORs) and 95% confidence intervals (CIs) for associations of a doubling in lipid metabolite concentration and colorectal cancer risk. These estimates represent differences of metabolite levels between cases and controls within each case–control pair. Due to the incidence density sampling, the ORs provide an unbiased estimate of the hazard ratio, which would have been obtained from a cohort study in the source population (i.e., EPIC participants who provided a blood sample).39,40
Models were conditioned on matching variables and adjusted for potential confounders using the modified disjunctive cause criterion as clear knowledge for a causal diagram of the association between all lipids under study and CRC was not known.41 Confounders chosen were known causes of CRC and we had no prior knowledge that any included covariate was a known instrumental variable for the association between lipid levels and CRC.41 Potential confounders included baseline observed measurements of education level, Cambridge physical activity index, smoking status, height, body mass index (BMI), alcohol intake, energy intake, red and processed meat intake, fibre intake and dairy product intake. Additional adjustment for diabetes status did not materially alter our observed results and therefore the more parsimonious model was used for analyses. A Bonferroni-corrected p-value was estimated as p = 0.05 ∗ the effective number of independent tests (ENT).42 ENT was defined as the number of principal components which explained more than an a priori defined proportion (95% for this study) of the variance of metabolite matrices. In this study the ENT was 28.
Analyses were also conducted including metabolite concentrations categorised into quartiles based on distributions among controls (concentrations among cases below or above the range for controls were included in the lowest or highest quartiles respectively). To assess for trend across categories, quartile medians were additionally modelled as continuous variables. Tests for non-linearity were performed using restricted cubic splines. Due to the large number of lipids under study, the number and placement of knots were determined a priori, with five knots placed at equally spaced percentiles of the variable's marginal distribution as recommended by Harrell's (i.e., 5, 27.5, 50, 72.5 and 95).43 Departures from linearity were evaluated using likelihood ratio tests, comparing the model with restricted cubic splines to linear models.
To evaluate the possible influence of reverse causation, lag analyses were performed excluding the first 2 or 5 years of follow-up after blood collection (in this nested case-control design, this was the equivalent of excluding pairs for which the case was diagnosed within the first 2 years [n = 162 cases, 162 controls] or 5 years [n = 501 cases, 501 controls] after blood collection). Subgroup analyses for circulating lipids associated with colorectal cancer risk were conducted according to sex (men, women), age at diagnosis (<55, ≥55 years), and BMI (<25, ≥25 kg/m2). Age and BMI were dichotomised based on their distribution among participants and to ensure adequate case/control numbers in each subgroup and, for age specifically, based on evidence of differing colorectal cancer incidence trends among individuals <55 years compared to those ≥55 years.44 Associations between these lipid metabolites and colorectal cancer subsites (colon and rectum) were also examined. Tests for heterogeneity in the associations between subgroups and colorectal cancer risk were performed using likelihood ratio tests comparing a model with and without an interaction term between the lipid concentration and subgroup variable.
Due to the correlated nature of individual lipid metabolites, dimension reduction was used to convert these data into orthogonal components. Dimension reduction was performed using an approach called treelet transformation which was developed by Gorst-Rasmussen et al.45,46 Unlike principal component analysis (PCA) which results in uncorrelated components within which all original variables contribute a numerical value (or loading), treelet transform combines methodology from PCA and hierarchical clustering to reduce the loadings of variables which contribute minimally to zero thus improving the interpretability of the resultant components.45,46 The treelet transform works by joining the two metabolites with the highest correlation by PCA methods and replacing them by their principal component.45,46 For a set of n variables, this joining process is then repeated n-1 times until a cluster tree or dendrogram is created.45,46 Treelet components (TC) are determined by choosing a cut-level which balances the trade-off between greater explained variation and sparsity of components (i.e., improved interpretability).
Among controls, treelet transform was performed on log2 transformed lipid metabolite data. The optimal cut-level for a choice of between 1 and 10 TCs was obtained using ten-fold cross-validation and was observed to be between 91 and 97.47,48 Following inspection of scree plots, pattern interpretability, and cross-validation results, five TCs were retained and the lowest cut-level of 91 was selected. Component scores for each of the five TCs (i.e., TC1–TC5) were calculated for all participants. Conditional logistic regression models were run to estimate associations between each component and colorectal cancer risk, matching and adjusting for the same covariates as for analyses with individual metabolites.
Sensitivity analyses were performed to evaluate the impact of using ±3 cut levels of the chosen cut point. The stability of the treelet transform components was assessed by subsampling.45,46 A random sample of 80% of the data was selected 100 times and the sign patterns (+for positive loadings and—for negative loadings) of the highest-variance components were evaluated. The stability of each treelet component was assessed based on the frequency of its corresponding sign pattern among the 100 subsamples. See Supplementary information for more details on sensitivity and stability evaluations.
Using data from control participants, we examined associations between circulating lipid metabolites and plasma fatty acid levels (n = 461 controls), diet (n = 1591 controls), lifestyle (n = 1591 controls), anthropometric (n = 1591 controls) and serum biomarkers (range n = 503 to n = 619). Associations were evaluated using multivariable linear regression models with the lipid metabolite as the dependent variable, the exposure as the independent variable and adjusting for a priori confounders (See Supplementary methods). Data were missing for at least one covariate for <5% of study controls, with ≤3% of participants missing data for time of day at blood collection, education level, physical activity index and smoking status. Due to these small proportions of missingness, data were imputed using the median value for continuous variables and the mode for categorical variables. See Supplementary methods for more information on these analyses.
Analyses were performed using Stata version 17.0 (StataCorp Inc).
Ethics
Written informed consent was obtained from all study participants and ethical approval was obtained from the ethical review committee of the IARC (No. 14-08) and participating study centres. The study was conducted in compliance with ethics approval.
Role of the funding source
The funders played no role in the study design, collection, analysis, interpretation of data, writing of the report, or the decision to submit the paper for publication.
Results
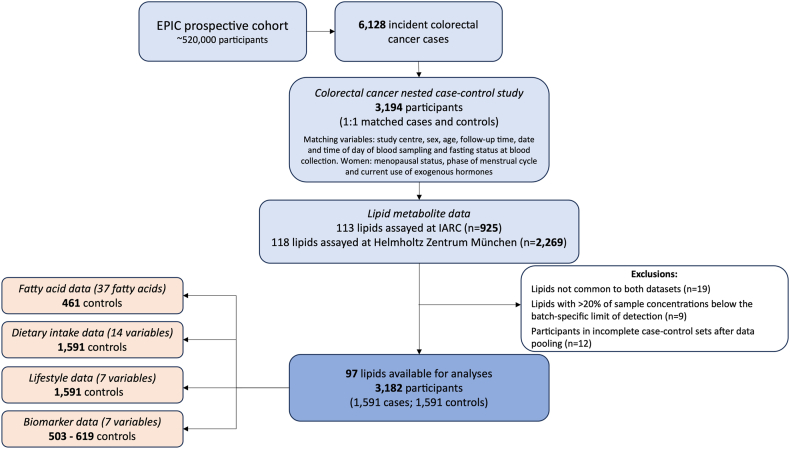
A data flow diagram showing the study design, exclusions and data availability is presented in Fig. 1.
Fig. 1.
Overview of the study design. EPIC, European Prospective Investigation into Cancer and Nutrition; IARC, International Agency for Research on Cancer; Serum fatty acid data (expressed as a percentage of total fatty acids)—industrial trans fatty acid, monounsaturated fatty acid, natural trans fatty acid, polyunsaturated fatty acid; Dietary intake data (g/day)—mono-unsaturated fatty acid, polyunsaturated fatty acid, saturated fatty acid, total cholesterol, fat, animal fat, carbohydrate, starch, sugar, protein, animal protein, red and processed meat, dairy, fibre; Lifestyle data—alcohol consumption at recruitment (g/day), Cambridge physical activity index (inactive, moderately inactive, moderately active, active), smoking status (never, former, current), body mass index (kg/m2), height (cm), waist circumference (cm), waist-to-hip ratio; Biomarker data—cholesterol (mmol/l), low-density lipoprotein (LDL) cholesterol (mmol/l), high-density lipoprotein (HDL) cholesterol (mmol/l), triglycerides (mmol/l), C-peptide [ng/ml], C-reactive protein [ng/ml], insulin-like growth factor-1 (IGF-1) [nmol/l]).
The mean age of participants at blood collection was 56.8 years (56.8 years among cases). Median time to diagnosis of cases was 7.3 years following blood collection. The distributions of covariates among cases and controls are described in Table 1 and for men and women Appendix Table 1. The median and interquartile range (IQR) of lipid metabolite concentrations among cases and controls are presented in Appendix Table 2. Strong positive correlations (r > 0.5) were observed between individual lipids within each lipid grouping (i.e., acylcarnitines, glycerophospholipids, and sphingolipids) and strong inverse correlations (r > −0.5) between some glycerophospholipids and sphingolipids (Appendix Figure 1).
Table 1.
Characteristics of study population.
| Total | Controls | Cases | |
|---|---|---|---|
| All sexes | |||
| N = 3182 | N = 1591 | N = 1591 | |
| Age at recruitment (years) [mean, sd] | 56.8 (7.5) | 56.7 (7.5) | 56.8 (7.5) |
| Age at recruitment (years) | |||
| <55 | 1218 (38%) | 608 (38%) | 610 (38%) |
| ≥55 | 1964 (62%) | 983 (62%) | 981 (62%) |
| Age at blood collection (years) [mean, sd] | 56.9 (7.5) | 56.9 (7.5) | 57.0 (7.5) |
| Follow-up time after blood collection (years) [median, IQR] | – | – | 7.3 (4.2–11.3) |
| Colorectal cancer subsite | |||
| Colon | – | – | 1362 (86%) |
| Rectal | – | – | 229 (14%) |
| Sex | |||
| Men | 1442 (45%) | 721 (45%) | 721 (45%) |
| Women | 1740 (55%) | 870 (55%) | 870 (55%) |
| Highest education level | |||
| None/primary school | 1400 (44%) | 721 (45%) | 679 (43%) |
| Technical/professional/secondary school | 1163 (37%) | 559 (35%) | 604 (38%) |
| Longer education including university | 511 (16%) | 264 (17%) | 247 (16%) |
| Not specified/missing | 108 (3%) | 47 (3%) | 61 (4%) |
| Cambridge physical activity index | |||
| Inactive | 858 (27%) | 402 (25%) | 456 (29%) |
| Moderately inactive | 1076 (34%) | 541 (34%) | 535 (34%) |
| Moderately active | 618 (19%) | 310 (19%) | 308 (19%) |
| Active | 584 (18%) | 315 (20%) | 269 (17%) |
| Missing | 46 (1%) | 23 (1%) | 23 (1%) |
| Smoking status | |||
| Never | 1428 (45%) | 753 (47%) | 675 (42%) |
| Former | 991 (31%) | 476 (30%) | 515 (32%) |
| Current smoker | 731 (23%) | 346 (22%) | 385 (24%) |
| Unknown | 32 (1%) | 16 (1%) | 16 (1%) |
| Height (cm) [mean, sd] | 165.9 (9.3) | 165.6 (9.3) | 166.2 (9.3) |
| Body Mass Index (kg/m2) [mean, sd] | 26.7 (4.1) | 26.4 (3.9) | 27.0 (4.4) |
| Body Mass Index (kg/m2) | |||
| <25 | 1161 (36%) | 608 (38%) | 553 (35%) |
| ≥25 | 2021 (64%) | 983 (62%) | 1038 (65%) |
| Alcohol intake at recruitment (g/day) [median, IQR] | 8.0 (1.0–23.9) | 8.1 (1.1–23.0) | 7.8 (0.8–24.5) |
| Total energy intake (kcal/day) [mean, sd] | 2168.1 (674.4) | 2174.6 (643.5) | 2161.7 (704.3) |
| Meat intake (red and processed) (g/day) [median, IQR] | 77.2 (49.4–110.7) | 78.8 (50.4–110.7) | 74.9 (48.7–110.8) |
| Dietary fibre intake (g/day) [mean, sd] | 23.4 (8.1) | 23.7 (8.2) | 23.2 (8.0) |
| Dairy intake (g/day) [median, IQR] | 270.6 (150.4–432.1) | 281.3 (160.6–444.7) | 256.1 (138.2–420.0) |
| Women only | |||
| N = 1740 | N = 870 | N = 870 | |
| Menopausal status | |||
| Premenopausal | 291 (17%) | 148 (17%) | 143 (16%) |
| Postmenopausal | 1168 (67%) | 586 (67%) | 582 (67%) |
| Perimenopausal/unknown menopausal status | 181 (10%) | 90 (10%) | 91 (10%) |
| Surgical postmenopausal | 100 (6%) | 46 (5%) | 54 (6%) |
| Ever use of hormones for menopause | |||
| Never | 1297 (75%) | 646 (74%) | 651 (75%) |
| Yes | 387 (22%) | 194 (22%) | 193 (22%) |
| Missing | 56 (3%) | 30 (3%) | 26 (3%) |
| Ever use of oral contraceptives | |||
| Never | 1018 (59%) | 498 (57%) | 520 (60%) |
| Yes | 701 (40%) | 365 (42%) | 336 (39%) |
| Missing | 21 (1%) | 7 (1%) | 14 (2%) |
Number and percentage reported unless stated otherwise; IQR, interquartile range; sd, standard deviation.
Associations between metabolites and colorectal cancer risk
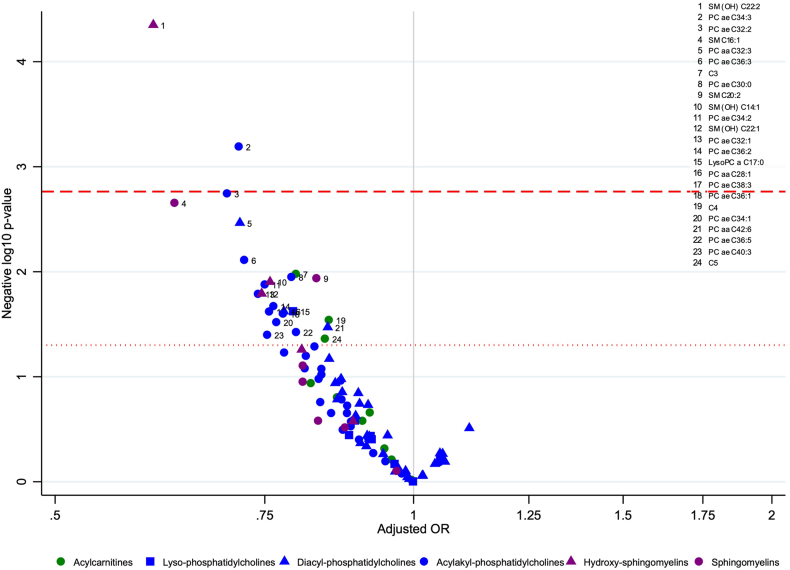
For 24 of the 97 lipid metabolites, a doubling in their concentration was inversely associated with colorectal cancer risk at a nominal p-value threshold (p < 0.05). These included 3 acylcarnitines, 16 glycerophospholipids, and 5 sphingolipids (Fig. 2). After correcting for multiple comparisons, SM (OH) C22:2 (ORper doubling 0.60, 95% CI 0.47–0.77, pent = 0.0012) and PC ae C34:3 (ORper doubling 0.71, 95% CI 0.59–0.87, pent = 0.018) were inversely associated with colorectal cancer risk (Fig. 2, Appendix Table 3). These inverse associations were also observed for men and women separately but did not meet the multiple comparisons threshold for women (Appendix Table 3).
Fig. 2.
Volcano plot for the association between lipid metabolites and colorectal cancer risk in the EPIC cohort. Negative log10 p-values (y-axis) and adjusted odds ratios (x-axis) obtained from conditional logistic regression models per doubling of lipid metabolite concentration, conditioned on matching variables (study centre, sex, age at blood collection, follow-up time since blood collection, time of day and fasting status at blood collection, menopausal status [women], phase of menstrual cycle [pre-menopausal women only] and current use of exogenous hormones [women] and adjusted for baseline measurements of education level (no schooling/primary, secondary/professional/technical, university/higher, not specified/missing), Cambridge physical activity index (inactive, moderately inactive, moderately active, active, unknown), smoking status (never, former, current, unknown), height (cm, continuous), Body mass index (kg/m2, continuous), alcohol use at recruitment (g/day, continuous), energy intake (kcal/day, continuous), red and processed meat consumption (g/day, continuous), dairy intake (g/day, continuous) and fibre intake (g/day, continuous). Each coloured shape represents one lipid metabolite with acylcarnitines in green, phosphatidylcholines in blue and sphingomyelins in purple. The vertical grey line represents an adjusted odds ratio of 1.00. Circles above the horizontal red dotted line or red dashed line represent lipid metabolites inversely (left of grey line) associated with colorectal cancer risk at a significance level of p-value <0.05 or after correction for multiple comparisons based on the effective number of independent tests (ENT) pENT <0.05, respectively. C, acylcarnitine; OR, odds ratio; PC aa, diacyl-phosphatidylcholine; PC ae, acylakyl-phosphatidylcholine; SM, sphingomyelin, SM (OH), Hydroxysphingomyelin.
In lag analyses excluding the first 2 or 5 years of follow-up, estimates per doubling in lipid concentration for the association between SM (OH) C22:2 and PC ae C34:3 with colorectal cancer were similar in direction and magnitude to those reported in main analyses (Appendix Table 4).
In models mutually adjusting for both lipid metabolites associated with colorectal cancer, despite slight attenuation of the strength of associations, both lipids remained inversely associated with colorectal cancer (SM (OH) C22:2 (ORper doubling 0.67, 95% CI 0.51–0.89, p = 0.0046) and PC ae C34:3 (ORper doubling 0.83, 95% CI 0.66–1.03, p = 0.087).
In tests for non-linearity, despite no evidence after adjusting for multiple comparisons (Appendix Table 3), there was suggestive evidence of departures from linearity for the association between LysoPC a C20:4, PC aa C32:2, PC aa C36:4, PC aa C38:0, PC ae C40:2, PC ae C40:3 and PC ae C40:6 prior to this adjustment at a nominal p-value of 0.05, however, in quartile models there was little evidence of clear associations with colorectal cancer and with no trends found across quartiles or associations found when comparing extreme quartiles (Q4 vs Q1) (Appendix Table 5).
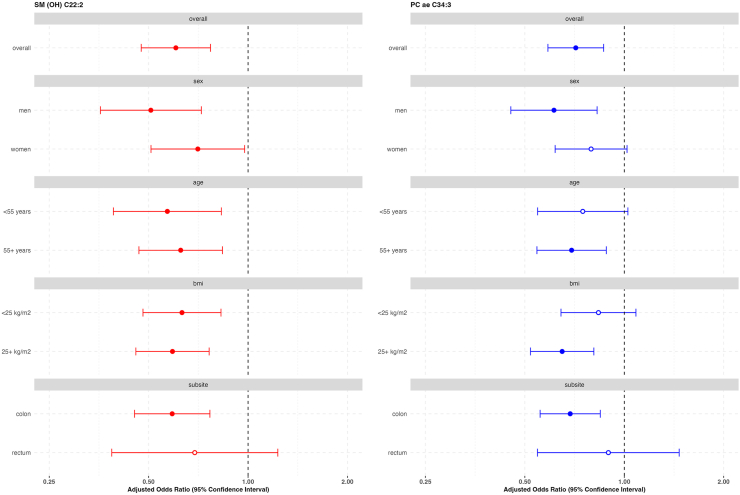
In subgroup analyses, similar magnitude inverse associations between SM (OH) C22:2 and PC ae C34:3 with colorectal cancer were observed according to sex, age, BMI, and anatomical subsite (Fig. 3).
Fig. 3.
Risk of colorectal cancer for concentrations of SM (OH) C22:2 and PC ae C34:3 by subgroups in the EPIC cohort. PC ae, acylakyl-phosphatidylcholine; SM (OH), Hydroxysphingomyelin. Estimates obtained from multivariable conditional logistic regression models for the association between each lipid metabolite (per doubling in concentration) and colorectal cancer risk, conditioned on matching variables (study centre, sex, age at blood collection, follow-up time since blood collection, time of day and fasting status at blood collection, menopausal status [women], phase of menstrual cycle [pre-menopausal women only] and current use of exogenous hormones [women] and adjusted for baseline measurements of education level (no schooling/primary, secondary/professional/technical, university/higher, not specified/missing), Cambridge physical activity index (inactive, moderately inactive, moderately active, active, unknown), smoking status (never, former, current, unknown), height (cm, continuous), Body mass index (kg/m2, continuous), alcohol use at recruitment (g/day, continuous), energy intake (kcal/day, continuous), red and processed meat consumption (g/day, continuous), dairy intake (g/day, continuous) and fibre intake (g/day, continuous). Solid circles represent an association between the specified lipid metabolite and exposure variable at a significance level of p-value <0.05. p-het, p-value for heterogeneity for the multivariable conditional logistic regression model including an interaction term between the lipid metabolite concentration and sex, age, body mass index or colorectal subsite.
Following treelet transform, five treelet components were retained which accounted for 63.1% of metabolite variation among all participants. The screeplot and dendrogram are presented in Appendix Figures 2 and 3. All components comprised only positive loadings. TC1 included 11 diacyl-, 15 acylakyl-phosphatidylcholines and all hydroxy-sphingomyelins and sphingomyelins (including SM (OH) C22:2), TC2 included 15 diacyl- and 15 acylakyl-phosphatidylcholines (including PC ae C34:3), TC3 included 7 diacyl- and 4 acylakyl-phosphatidylcholines, TC4 included 6 acylcarnitines and TC5 included 8 lyso-phosphatidylcholines (Appendix Figure 4) Associations between metabolite patterns, and colorectal cancer risk yielded a similar pattern of results with inverse associations observed for TC1, ORper doubling 0.93, 95% CI 0.88–0.99, p = 0.016, and weaker evidence of an inverse association with TC2, ORper doubling 0.95, 95% CI 0.90–1.00, p = 0.053 (Table 2).
Table 2.
Associations between treelet component scores and colorectal cancer in the EPIC cohort.
| Treelet components | OR | 95% CI | p-value |
|---|---|---|---|
| TC1 | 0.93 | (0.88–0.99) | 0.016 |
| TC2 | 0.95 | (0.90–1.00) | 0.053 |
| TC3 | 0.97 | (0.91–1.03) | 0.28 |
| TC4 | 0.96 | (0.87–1.04) | 0.32 |
| TC5 | 0.96 | (0.88–1.05) | 0.35 |
CI, confidence interval; OR, odds ratio; TC, treelet component.
Estimates obtained from multivariable conditional logistic regression models for the association between each treelet component and colorectal cancer risk, conditioned on matching variables (study centre, sex, age at blood collection, follow-up time since blood collection, time of day and fasting status at blood collection, menopausal status [women], phase of menstrual cycle [pre-menopausal women only] and current use of exogenous hormones [women] and adjusted for baseline measurements of education level (no schooling/primary, secondary/professional/technical, university/higher, not specified/missing), Cambridge physical activity index (inactive, moderately inactive, moderately active, active, unknown), smoking status (never, former, current, unknown), height (cm, continuous), body mass index (kg/m2, continuous), alcohol use at recruitment (g/day, continuous), energy intake (kcal/day, continuous), red and processed meat consumption (g/day, continuous), dairy intake (g/day, continuous) and fibre intake (g/day, continuous).
Associations between individual lipids and baseline plasma fatty acid levels, serum biomarkers, dietary, lifestyle and anthropometric factors
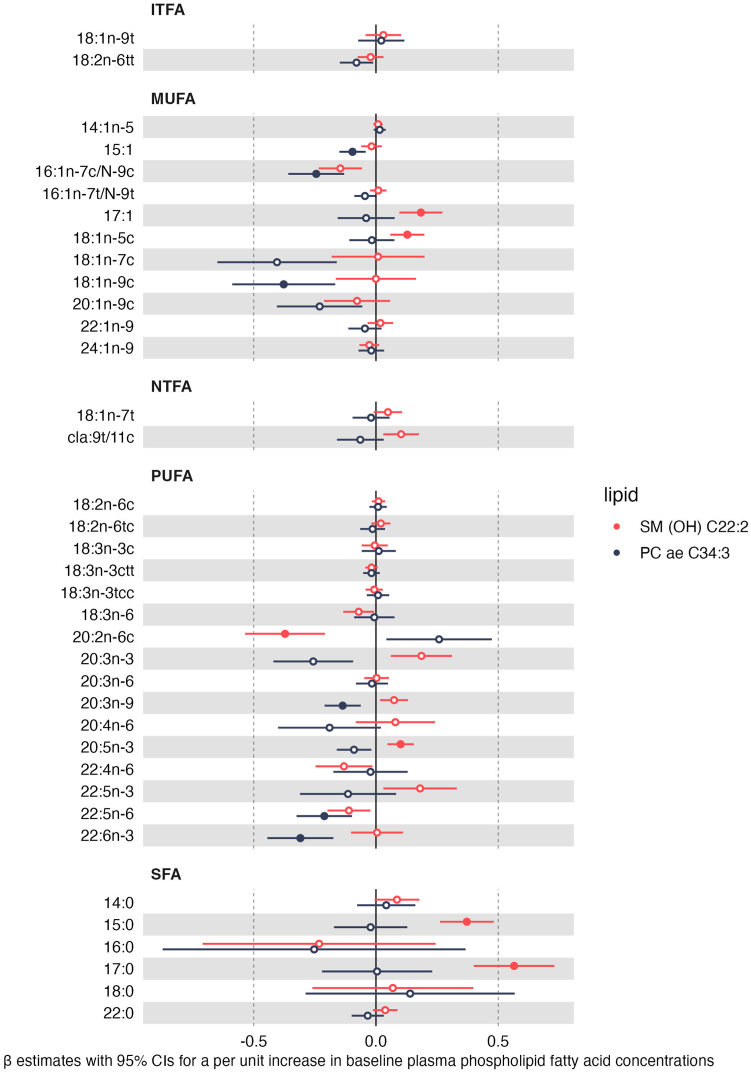
Among controls whose samples were analysed at IARC (n = 461), circulating levels of SM (OH) C22:2 were positively associated with two saturated fatty acids (SFAs) (15:0 and 17:0), two monounsaturated fatty acids (MUFAs) (17:1 [heptadecenoic acid] and 18:1n-5c) and an omega-3 polyunsaturated fatty acid (PUFA) (20:5n-3 [eicosapentaenoic acid (EPA]) whereas it was inversely associated with PUFA 20:2n-6c (eicosadienoic acid), an omega-6 fatty acid. Inverse associations were also observed between PC ae C34:3 and three MUFAs (15:1 [pentadecanoic acid], 16:1n-7cN-9c [palmitoleic acid], 18:1n-9c [oleic acid]) and three PUFAs (one omega-3 (22:6n-3 [docosahexaenoic acid (DHA)]), one omega-6 (22:5n-6 [docosapentaenoic acid (DPA)]) and one omega-9 PUFA (20:3n-9 [mead acid])) (Fig. 4). There was evidence of non-linearity for the relationship between 18:2n-6c and 20:3n-6 with PC ae C34:3 and for 20:5n-3 with SM (OH) C22:2. In models comparing quartiles of fatty acid concentrations, there was an apparent increasing trend across quartiles for PC ae C34:3 and 18:2n-6c and a decreasing trend across quartiles for PC ae C34:3 and 20:3n-6. An increase in SM (OH) C22:2 was observed for Q2 vs Q1 of 20:5n-3 which plateaued across quartiles (Appendix Table 6).
Fig. 4.
Relationship between concentrations of serum fatty acid levels and lipid metabolites shown to be associated with colorectal cancer risk in the EPIC cohort (n = 461 controls). ITFA, industrial trans fatty acid; MUFA, monounsaturated fatty acid; NTFA, natural trans fatty acid; PC ae, acylakyl-phosphatidylcholine; PUFA, polyunsaturated fatty acid; SFA, saturated fatty acid; SM (OH), Hydroxysphingomyelin. Beta difference and 95% confidence intervals obtained from multivariable linear regression models for the association between a per unit increase in log-transformed serum fatty acid concentration levels (recorded as a percentage of total fatty acid levels) and lipid metabolite concentration adjusted for sex, batch, energy intake (kcal/day, continuous) (models including dietary variables only), age at blood collection, time of day at blood collection, fasting status at blood collection (yes, no, in between), education level (no schooling/primary, secondary/professional/technical, university/higher), Cambridge physical activity index (inactive, moderately inactive, moderately active, active), smoking status (never, former, current), body mass index (kg/m2, continuous), alcohol use at recruitment (g/day, continuous), dietary intakes (g/day) of red and processed meat, fibre intake and dairy. Solid circles represent an association between the specified lipid metabolite and fatty acid after correction for multiple comparisons based on the effective number of independent tests (ENT) (i.e., pENT <0.05 (ENT = 26; pENT = 0.05∗[26∗2 lipid metabolites]).
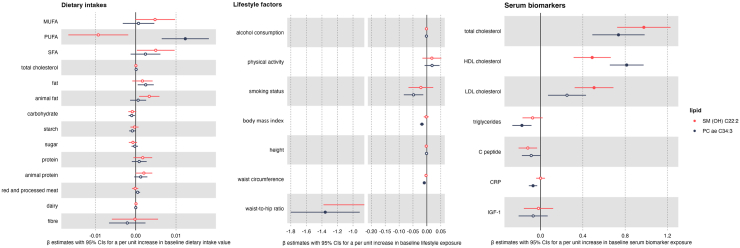
PC ae C34:3 was positively associated and with dietary intakes of PUFA (Fig. 5) and inversely associated with several measures of body size (i.e., BMI, waist circumference and waist-to-hip ratio). In a subset of participants (range n = 503–619) PC ae C34:3 was inversely associated with serum levels of CRP and triglycerides and positively associated with serum levels of total and HDL cholesterol, and SM (OH) C22:2 was positively associated with serum levels of total, HDL, and LDL cholesterol (Fig. 5).
Fig. 5.
Relationships between baseline exposures and lipid metabolites shown to be associated with colorectal cancer risk in the EPIC cohort. CRP, C reactive protein; IGF-1, insulin growth factor 1; MUFA, mono-unsaturated fatty acid; PUFA, poly-unsaturated fatty acid; SFA, saturated fatty acid; PC ae, acylakyl-phosphatidylcholine; SM (OH), Hydroxysphingomyelin. Beta difference and 95% confidence intervals obtained from multivariable linear regression models for the association between a per unit increase of each baseline exposure i.e., log-transformed biomarker concentration (n = 503–619) (total cholesterol (mmol/l), low-density lipoprotein (LDL) cholesterol (mmol/l), high-density lipoprotein (HDL) cholesterol (mmol/l), triglycerides (mmol/l), C-peptide [ng/ml], C-reactive protein [ng/ml], IGF-1 [nmol/l]), dietary intake (g/day) (n = 1591) or lifestyle variable (n = 1591) (alcohol consumption at recruitment [g/day], Cambridge physical activity index [inactive, moderately inactive, moderately active, active], smoking status [never, former, current], body mass index [kg/m2], height [cm], waist circumference [cm], waist-to-hip ratio) and lipid metabolite concentration. Models were adjusted for sex, centre, energy intake (kcal/day) (for dietary and biomarker variables only), age at blood collection, time of day at blood collection, fasting status at blood collection (yes, no, in between), education level (no schooling/primary, secondary/professional/technical, university/higher), Cambridge physical activity index (inactive, moderately inactive, moderately active, active), smoking status (never, former, current), height (cm), alcohol use at recruitment (g/day, continuous), body mass index (kg/m2, continuous) for all models except those with waist circumference and waist-to-hip ratio as the exposure of interest, and for non-dietary exposure variables, dietary intakes (g/day) of red and processed meat, fibre intake and dairy. Solid circles represent an association between the specified lipid metabolite and baseline exposure after correction for multiple comparisons based on the effective number of independent tests (ENT) (i.e., pENT <0.05 (ENT = 17; pENT = 0.05∗[17∗2 lipid metabolites]).
Discussion
We conducted the largest and most comprehensive study to date examining the associations between individual pre-diagnostic circulating levels of 97 lipids and colorectal cancer risk. Of these, 24 circulating lipid metabolites were nominally associated, all inversely, with colorectal cancer risk. After controlling for multiple comparisons, inverse associations remained between SM (OH) C22:2 and PC ae C34:3, and colorectal cancer risk, which persisted after mutual adjustment. These robust inverse associations were unaltered after excluding the first 5 years of follow-up after blood collection, and were consistent according to sex, age at diagnosis, BMI, and colorectal subsite.
Few studies have investigated associations between pre-diagnostic levels of circulating lipid metabolites and colorectal cancer risk, each using different methods of metabolite identification (targeted vs untargeted), assays and analytical methodology (NMR vs mass-spectrometry).23,24,26,49 A previous study using data from the EPIC-Heidelberg cohort (also included in this current analysis) and the Biocrates AbsoluteIDQ™ assay did not observe associations between SM (OH) C22:2 nor PC ae C34:3 and colorectal cancer neither prior to or following FDR correction.24 The smaller sample size (n = 163 cases) of the Heidelberg cohort analysis may explain these divergent findings. A prior nested case–control study (n = 250 cases) was conducted including data from two Chinese cohorts. Using an untargeted metabolomics approach, this study found associations between 3 phosphatidylcholines PC (16:0/16:0) (positive), PC (18:3/16:0) (positive) and PC 22:6/18:0 (inverse) and colorectal cancer risk.23 Different naming conventions made direct comparisons difficult but the corresponding lipid synonyms were identified as PC aa C32:0, PC aa C34:3 and PC aa C40:6 respectively in the Human Metabolome Database (HMDB).50 None of these lipid metabolites were associated with colorectal cancer risk in the current analyses, however. In another untargeted nested case–control study on a US population no lipid metabolites were associated with colorectal cancer risk after correction for multiple comparisons, but in contrast to our study all but four lipids (not quantified in this current study) were found to have positive associations with colorectal cancer risk prior to multiple comparisons correction.26 The different metabolomics methodology and study population (Asian/American compared to European) may have contributed to observed differences.
A large-scale pan-cancer study (n = 1500 colorectal cancer cases) within EPIC examined shared associations between metabolites and overall cancer leveraging data included in our current analysis. That study reported inverse associations between a phosphatidylcholine metabolite cluster including PC ae C34:3 along with PC ae C34:2, PC ae C36:3, PC ae C36:2 and PC aa C32:3 and colorectal cancer risk but not with the cluster including SM (OH) C22:2 in subsite-specific analyses.49 As analyses by subsite were examined for lipid metabolite clusters this may have masked important associations by specific individual metabolites.
Interestingly, most observed lipid and colorectal cancer risk associations were inverse (both for individual circulating lipids and lipid metabolite components), despite only reaching statistical significance for SM (OH) C22:2 and PC ae C34:3. As fatty acid dysregulation is a key early stage in cancer tumorigenesis,7,51 our findings may reflect some underlying process that perturbs lipid metabolism which is relevant for colorectal cancer development. This is supported by evidence from Mendelian randomisation studies of a likely causal pathway between specific fatty acids and lipids and colorectal cancer development.52
The biological mechanisms underlying the association between SM (OH) C22:2 and colorectal cancer development are unclear. Ceramides, precursors to sphingomyelins,53 can be produced through the hydrolysis of sphingomyelins51,53,54 and evidence from experimental studies suggests that ceramides can have pro- and anti-tumorigenic roles dependent on factors such as the length of their associated side-chain fatty acids.51,55,56 The observed association with SM (OH) C22:2 may suggest that some aspect of its structure could be contributing to associations with this, over other sphingomyelin metabolites.
Inverse associations between phosphatidylcholines and other cancer sites have been reported.30,48,49,57 Although unclear, the suggested inverse association of PC ae C34:3 with colorectal cancer in our analysis could be due to a potential anti-inflammatory role in the colorectum.58 Additional experimental studies are needed to further elucidate specific pathways through which this phosphatidylcholine may be involved in colorectal cancer development.
This study found that metabolite patterns including both sphingomyelins and phosphatidylcholines were inversely associated with colorectal cancer. Common factors between the two lipid classes may affect colorectal cancer development. For example, choline, present in both sphingomyelin and phosphatidylcholine, has been investigated for its potential role in colorectal cancer.59,60
In this analysis it was not possible to annotate the fatty acid side chains that are components of the lipid metabolites under study. A strength of this study was the availability of fatty acid data in the same population which allowed for inferences to be made on potential lipid metabolite side-chain components. Strong positive associations were observed between SM (OH) C22:2 with two odd-chain SFAs (15:0 and 17:0). Higher levels of these odd-chain SFAs 15:0 and 17:0 have been shown to be associated with higher dietary dairy fat61,62 and fibre intake,63 lower levels of serum CRP64 and to be linked to lower risks of chronic inflammation, type 2 diabetes, obesity and metabolic syndrome,65 all of which have been implicated in colorectal cancer risk. The generally positive associations observed between SM (OH) C22:2 with SFAs and MUFAs and inverse associations between PC ae C34:3 with MUFAs and PUFAs may highlight some particularity of the fine structure of their fatty acid side chains which underly associations between these specific phosphatidylcholine and sphingomyelin metabolites and colorectal cancer.
Linear regression analyses also allowed for an investigation of potential determinants of SM (OH) C22:2 and PC ae C34:3. SM (OH) C22:2 was found to the positively associated with serum cholesterol levels (total, HDL, and LDL). PC ae C34:3 was positively associated with serum total and HDL cholesterol but inversely related to body size measures and levels of serum CRP and triglycerides. Previous studies have reported inverse associations between PC ae C34:3 with obesity14,66 and body size measures (within the EPIC cohort).15 Observed positive associations between PC ae C34:3 and dietary PUFA intake and inverse associations between serum triglycerides and CRP align with evidence of inverse associations between dietary PUFA, and positive associations between triglycerides and CRP, with colorectal cancer risk in meta-analyses.67, 68, 69, 70, 71, 72, 73 Findings from two meta-analyses of a possible inverse association between HDL cholesterol and colorectal cancer risk also support our findings,69,71 however, reported positive associations between total cholesterol and null associations between LDL cholesterol and colorectal cancer risk did not.69, 70, 71
This was the largest and most comprehensive prospective study to date to investigate associations between lipid metabolites and colorectal cancer risk. Detailed covariate data allowed us to control for other colorectal cancer risk factors and findings were consistent following exclusion of up to five years of follow-up after blood draw. Limitations include the measurement of pre-diagnostic circulating lipid metabolite concentrations at baseline only. Previous studies have reported high reliability of metabolites (measured using the Biocrates AbsoluteIDQ™ assay) over time with relatively good reproducibility reported within 2 years following initial blood draw which was lower for non-fasting individuals and more pronounced for acylcarnitines, phosphatidylcholines and sphingomyelins (median intraclass correlation coefficients for lipid metabolites between 0.62 and 0.74 for fasting, and 0.34 and 0.62 for non-fasting individuals).74 Poorer reproducibility has been reported in the longer term (median Spearman's correlation coefficient (ρ) 0.13–0.42) for lipid metabolites.75 Changes in metabolite measurements over time would likely have led to an attenuation of results.74 In this study, non-fasting participants were also included in analyses which may have affected the levels of detected metabolites; however, we matched cases and controls by fasting status and prior data suggests that only a small proportion of the variation in metabolite concentrations can be attributed to fasting status.75 In the Biocrates AbsoluteIDQ™ kit, the flow injection analysis with tandem mass spectrometry (FIA-MS/MS) analytical method for lipid identification does not allow for specification of the positions, locations of double bonds, or chain lengths of associated fatty acids, meaning metabolite signals could correspond to several different lipid isobars or isomers.76,77 However, we examined correlations between circulating lipid metabolites that were associated with colorectal cancer and plasma fatty acid levels to supplement the interpretation of the potential lipid compounds identified. A limitation of the cross-sectional analyses was the lack of availability of data on serum fatty acid levels and biomarker levels for all 1591 controls included in the main analyses. If some characteristics of those participants with these data missing were associated with fatty acid or biomarker levels and/or lipid concentrations this may have affected the observed results. Estimates may also have been affected by unmeasured confounding (e.g., no data were available on colorectal cancer screening) or residual confounding as a result of measurement error in covariate data collected via questionnaires (e.g., smoking status and alcohol consumption). In addition, this study matched cases and cotrols based on a range of values for each matching variable which may have led to bias when there was an under sampling of controls at the extremes of the distributions of the matching variable.78 Even though this bias is thought to be small, this may have had an effect on observed estimates.78 Finally, this study was conducted in a European population therefore findings may not be generalisable to other settings.
In summary, our study found evidence of inverse associations between several lipid metabolites and colorectal cancer risk with SM (OH) C22:2 and PC ae C34:3 remaining associated after correction for multiple comparisons. Additional prospective studies are needed to confirm these associations alongside experimental studies to evaluate the role of dysregulation of lipid metabolism in colorectal carcinogenesis. Future studies could also utilise data on genetic instruments, such as in Mendelian randomisation, to allow for examination of a causal role of lipids in colorectal cancer development. Elucidating any potential roles of specific lipid metabolites in colorectal cancer development could be important to inform targeted prevention strategies including metabolic targets for drug or lifestyle intervention.
Contributors
RH—methodology, formal analysis, visualisation, writing—original draft; MJG, NM—conceptualization, methodology, supervision; MJG—funding acquisition, investigation; AT, GS, RK, MBS, GM, SP, CS, MJS, MDC, BVG—investigation; AG, CP, DA and JA—data curation and verification; JS—methodology; all authors–writing—review and editing. All authors read and approved the final version of the manuscript.
Data sharing statement
Data from EPIC are not publicly available, but access requests can be made to the EPIC Steering Committee (https://epic.iarc.fr/access/submit_appl_access.php).
Disclaimer
Where authors are identified as personnel of the International Agency for Research on Cancer/World Health Organization, the authors alone are responsible for the views expressed in this article and they do not necessarily represent the decisions, policy, or views of the International Agency for Research on Cancer/World Health Organization.
Declaration of interests
None declared.
Acknowledgements
The authors thank all EPIC centers for provision of biological samples and participant data. The coordination of EPIC is financially supported by the International Agency for Research on Cancer (IARC) and by the Department of Epidemiology and Biostatistics, School of Public Health, Imperial College London, with infrastructure support provided by the NIHR Imperial Biomedical Research Centre (BRC).
The national cohorts are supported by Danish Cancer Society (Denmark); Ligue Contre le Cancer, Institut Gustave-Roussy, Mutuelle Générale de l’Education Nationale, Institut National de la Santé et de la Recherche Médicale (INSERM) (France); German Cancer Aid, German Cancer Research Center (DKFZ), German Institute of Human Nutrition Potsdam-Rehbruecke (DIfE), Federal Ministry of Education and Research (BMBF) (Germany); Associazione Italiana per la Ricerca sul Cancro-AIRC-Italy, Compagnia di SanPaolo and National Research Council (Italy); The National Institute for Public Health and the Environment (RIVM), Bilthoven, Dutch Ministry of Public Health, Welfare and Sports (VWS), Netherlands Cancer Registry (NKR), LK Research Funds, Dutch Prevention Funds, Dutch ZON (Zorg Onderzoek Nederland), World Cancer Research Fund (WCRF), Statistics Netherlands (The Netherlands); Health Research Fund (FIS)–Instituto de Salud Carlos III (ISCIII), Regional Governments of Andalucía, Asturias, Basque Country, Murcia and Navarra, and the Catalan Institute of Oncology–ICO (Spain); Cancer Research UK (14136 to EPIC-Norfolk; C8221/A29017 to EPIC-Oxford), Medical Research Council (1000143 to EPIC-Norfolk; MR/M012190/1 to EPIC-Oxford) (UK). IDIBELL acknowledges support from the Generalitat de Catalunya through the CERCA Program. The nested colorectal cancer study was funded by World Cancer Research Fund (reference: 2013/1002; www.wcrf.org/) and the European Commission (FP7: BBMRI-LPC; reference: 313010; https://ec.europa.eu/).
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2024.105024.
Appendix A. Supplementary data
References
- 1.Sung H., Ferlay J., Siegel R.L., et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Vieira A.R., Abar L., Chan D.S.M., et al. Foods and beverages and colorectal cancer risk: a systematic review and meta-analysis of cohort studies, an update of the evidence of the WCRF-AICR continuous update project. Ann Oncol. 2017;28(8):1788–1802. doi: 10.1093/annonc/mdx171. [DOI] [PubMed] [Google Scholar]
- 3.Dekker E., Tanis P.J., Vleugels J.L.A., Kasi P.M., Wallace M.B. Colorectal cancer. Lancet. 2019;394(10207):1467–1480. doi: 10.1016/S0140-6736(19)32319-0. [DOI] [PubMed] [Google Scholar]
- 4.Keum N., Giovannucci E. Global burden of colorectal cancer: emerging trends, risk factors and prevention strategies. Nat Rev Gastro Hepatol. 2019;16(12):713–732. doi: 10.1038/s41575-019-0189-8. [DOI] [PubMed] [Google Scholar]
- 5.Wenk M.R. Lipidomics: new tools and applications. Cell. 2010;143(6):888–895. doi: 10.1016/j.cell.2010.11.033. [DOI] [PubMed] [Google Scholar]
- 6.Yan G.F., Li L.Q., Zhu B., Li Y.S. Lipidome in colorectal cancer. Oncotarget. 2016;7(22):33429–33439. doi: 10.18632/oncotarget.7960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu R., Huang Y. Lipid signaling in tumorigenesis. Mol Cell Pharmacol. 2014;6(1):1–9. [PMC free article] [PubMed] [Google Scholar]
- 8.Rohrig F., Schulze A. The multifaceted roles of fatty acid synthesis in cancer. Nat Rev Cancer. 2016;16(11):732–749. doi: 10.1038/nrc.2016.89. [DOI] [PubMed] [Google Scholar]
- 9.Grenville Z.S., Noor U., His M., et al. Diet and BMI correlate with metabolite patterns associated with aggressive prostate cancer. Nutrients. 2022;14(16):3306. doi: 10.3390/nu14163306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.His M., Viallon V., Dossus L., et al. Lifestyle correlates of eight breast cancer-related metabolites: a cross-sectional study within the EPIC cohort. BMC Med. 2021;19(1):312. doi: 10.1186/s12916-021-02183-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang S.J., Kwak S.Y., Jo G., Song T.J., Shin M.J. Serum metabolite profile associated with incident type 2 diabetes in Koreans: findings from the Korean Genome and Epidemiology Study. Sci Rep. 2018;8(1):8207. doi: 10.1038/s41598-018-26320-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu T., Holzapfel C., Dong X., et al. Effects of smoking and smoking cessation on human serum metabolite profile: results from the KORA cohort study. BMC Med. 2013;11(1):60. doi: 10.1186/1741-7015-11-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lacruz M.E., Kluttig A., Tiller D., et al. Cardiovascular risk factors associated with blood metabolite concentrations and their alterations during a 4-year period in a population-based cohort. Circ Cardiovasc Gene. 2016;9(6):487–494. doi: 10.1161/CIRCGENETICS.116.001444. [DOI] [PubMed] [Google Scholar]
- 14.Frigerio G., Favero C., Savino D., et al. Plasma metabolomic profiling in 1391 subjects with overweight and obesity from the SPHERE study. Metabolites. 2021;11(4):194. doi: 10.3390/metabo11040194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kliemann N., Viallon V., Murphy N., et al. Metabolic signatures of greater body size and their associations with risk of colorectal and endometrial cancers in the European Prospective Investigation into Cancer and Nutrition. BMC Med. 2021;19(1):101. doi: 10.1186/s12916-021-01970-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Merz B., Nothlings U., Wahl S., et al. Specific metabolic markers are associated with future waist-gaining phenotype in women. PLoS One. 2016;11(6) doi: 10.1371/journal.pone.0157733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rothwell J.A., Murphy N., Besevic J., et al. Metabolic signatures of healthy lifestyle patterns and colorectal cancer risk in a European cohort. Clin Gastroenterol Hepatol. 2022;20(5):E1061–E1082. doi: 10.1016/j.cgh.2020.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Assi N., Gunter M.J., Thomas D.C., et al. Metabolic signature of healthy lifestyle and its relation with risk of hepatocellular carcinoma in a large European cohort. Am J Clin Nutr. 2018;108(1):117–126. doi: 10.1093/ajcn/nqy074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cornish A.J., Law P.J., Timofeeva M., et al. Modifiable pathways for colorectal cancer: a mendelian randomisation analysis. Lancet Gastroenterol. 2020;5(1):55–62. doi: 10.1016/S2468-1253(19)30294-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rodriguez-Broadbent H., Law P.J., Sud A., et al. Mendelian randomisation implicates hyperlipidaemia as a risk factor for colorectal cancer. Int J Cancer. 2017;140(12):2701–2708. doi: 10.1002/ijc.30709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Farshidfar F., Kopciuk K.A., Hilsden R., et al. A quantitative multimodal metabolomic assay for colorectal cancer. BMC Cancer. 2018;18(1):26. doi: 10.1186/s12885-017-3923-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Geijsen A.J.M.R., Brezina S., Keski-Rahkonen P., et al. Plasma metabolites associated with colorectal cancer: a discovery-replication strategy. Int J Cancer. 2019;145(5):1221–1231. doi: 10.1002/ijc.32146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shu X., Xiang Y.B., Rothman N., et al. Prospective study of blood metabolites associated with colorectal cancer risk. Int J Cancer. 2018;143(3):527–534. doi: 10.1002/ijc.31341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuhn T., Floegel A., Sookthai D., et al. Higher plasma levels of lysophosphatidylcholine 18:0 are related to a lower risk of common cancers in a prospective metabolomics study. BMC Med. 2016;14(1):13. doi: 10.1186/s12916-016-0552-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tan B.B., Qiu Y.P., Zou X., et al. Metabonomics identifies serum metabolite markers of colorectal cancer. J Proteome Res. 2013;12(6):3000–3009. doi: 10.1021/pr400337b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCullough M.L., Hodge R.A., Campbell P.T., Stevens V.L., Wang Y. Pre-diagnostic circulating metabolites and colorectal cancer risk in the cancer prevention study-II nutrition cohort. Metabolites. 2021;11(3):156. doi: 10.3390/metabo11030156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Riboli E., Hunt K.J., Slimani N., et al. European prospective investigation into cancer and nutrition (EPIC): study populations and data collection. Public Health Nutr. 2002;5(6b):1113–1124. doi: 10.1079/PHN2002394. [DOI] [PubMed] [Google Scholar]
- 28.Jenab M., Bueno-De-Mesquita H.B., Ferrari P., et al. Association between pre-diagnostic circulating vitamin D concentration and risk of colorectal cancer in European populations: a nested case-control study. Brit Med J. 2010;340:b5500. doi: 10.1136/bmj.b5500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Romisch-Margl W., Prehn C., Bogumil R., Rohring C., Suhre K., Adamski J. Procedure for tissue sample preparation and metabolite extraction for high-throughput targeted metabolomics. Metabolomics. 2012;8(1):133–142. [Google Scholar]
- 30.His M., Viallon V., Dossus L., et al. Prospective analysis of circulating metabolites and breast cancer in EPIC. BMC Med. 2019;17(1):178. doi: 10.1186/s12916-019-1408-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmidt J.A., Fensom G.K., Rinaldi S., et al. Pre-diagnostic metabolite concentrations and prostate cancer risk in 1077 cases and 1077 matched controls in the European Prospective Investigation into Cancer and Nutrition. BMC Med. 2017;15(1):122. doi: 10.1186/s12916-017-0885-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rothwell J.A., Besevic J., Dimou N., et al. Circulating amino acid levels and colorectal cancer risk in the European Prospective Investigation into Cancer and Nutrition and UK Biobank cohorts. BMC Med. 2023;21(1):80. doi: 10.1186/s12916-023-02739-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Slimani N., Deharveng G., Unwin I., et al. The EPIC nutrient database project (ENDB): a first attempt to standardize nutrient databases across the 10 European countries participating in the EPIC study. Eur J Clin Nutr. 2007;61(9):1037–1056. doi: 10.1038/sj.ejcn.1602679. [DOI] [PubMed] [Google Scholar]
- 34.Aglago E.K., Murphy N., Huybrechts I., et al. Dietary intake and plasma phospholipid concentrations of saturated, monounsaturated and trans fatty acids and colorectal cancer risk in the European Prospective Investigation into Cancer and Nutrition cohort. Int J Cancer. 2021;149(4):865–882. doi: 10.1002/ijc.33615. [DOI] [PubMed] [Google Scholar]
- 35.Aleksandrova K., Jenab M., Boeing H., et al. Circulating C-reactive protein concentrations and risks of colon and rectal cancer: a nested case-control study within the European prospective investigation into cancer and nutrition. Am J Epidemiol. 2010;172(4):407–418. doi: 10.1093/aje/kwq135. [DOI] [PubMed] [Google Scholar]
- 36.Jenab M., Riboli E., Cleveland R.J., et al. Serum C-peptide, IGFBP-1 and IGFBP-2 and risk of colon and rectal cancers in the European prospective investigation into cancer and nutrition. Int J Cancer. 2007;121(2):368–376. doi: 10.1002/ijc.22697. [DOI] [PubMed] [Google Scholar]
- 37.Rinaldi S., Cleveland R., Norat T., et al. Serum levels of IGF-I, IGFBP-3 and colorectal cancer risk: results from the EPIC cohort, plus a meta-analysis of prospective studies. Int J Cancer. 2010;126(7):1702–1715. doi: 10.1002/ijc.24927. [DOI] [PubMed] [Google Scholar]
- 38.van Duijnhoven F.J.B., Bueno-De-Mesquita H.B., Calligaro M., et al. Blood lipid and lipoprotein concentrations and colorectal cancer risk in the European Prospective Investigation into Cancer and Nutrition. Gut. 2011;60(8):1094–1102. doi: 10.1136/gut.2010.225011. [DOI] [PubMed] [Google Scholar]
- 39.Pearce N. What does the odds ratio estimate in a case-control study. Int J Epidemiol. 1993;22(6):1189–1192. doi: 10.1093/ije/22.6.1189. [DOI] [PubMed] [Google Scholar]
- 40.Rothman K. Oxford University Press; Oxford, U.K.: 2012. Epidemiology: an introduction. [Google Scholar]
- 41.VanderWeele T.J. Principles of confounder selection. Eur J Epidemiol. 2019;34(3):Cp3–219. doi: 10.1007/s10654-019-00494-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chadeau-Hyam M., Campanella G., Jombart T., et al. Deciphering the complex: methodological overview of statistical models to derive OMICS-based biomarkers. Environ Mol Mutagen. 2013;54(7):542–557. doi: 10.1002/em.21797. [DOI] [PubMed] [Google Scholar]
- 43.Harrell F.E. Springer; 2001. Regression modeling strategies: with applications to linear models, logistic regression, and survival analysis. [Google Scholar]
- 44.Zaki T.A., Singal A.G., May F.P., Murphy C.C. Increasing incidence rates of colorectal cancer at ages 50-54 years. Gastroenterology. 2022;162(3):964–965.e3. doi: 10.1053/j.gastro.2021.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gorst-Rasmussen A. tt: treelet transform with Stata. Stata J. 2012;12(1):130–146. [Google Scholar]
- 46.Gorst-Rasmussen A., Dahm C.C., Dethlefsen C., Scheike T., Overvad K. Exploring dietary patterns by using the treelet transform. Am J Epidemiol. 2011;173(10):1097–1104. doi: 10.1093/aje/kwr060. [DOI] [PubMed] [Google Scholar]
- 47.Assi N., Moskal A., Slimani N., et al. A treelet transform analysis to relate nutrient patterns to the risk of hormonal receptor-defined breast cancer in the European Prospective Investigation into Cancer and Nutrition (EPIC) Public Health Nutr. 2016;19(2):242–254. doi: 10.1017/S1368980015000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schmidt J.A., Fensom G.K., Rinaldi S., et al. Patterns in metabolite profile are associated with risk of more aggressive prostate cancer: a prospective study of 3,057 matched case-control sets from EPIC. Int J Cancer. 2020;146(3):720–730. doi: 10.1002/ijc.32314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Breeur M., Ferrari P., Dossus L., et al. Pan-cancer analysis of pre-diagnostic blood metabolite concentrations in the European Prospective Investigation into Cancer and Nutrition. BMC Med. 2022;20(1):351. doi: 10.1186/s12916-022-02553-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wishart D.S., Guo A.C., Oler E., et al. Hmdb 5.0: the human metabolome database for 2022. Nucleic Acids Res. 2022;50(D1):D622–D631. doi: 10.1093/nar/gkab1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li Y., Nicholson R.J., Summers S.A. Ceramide signaling in the gut. Mol Cell Endocrinol. 2022;544 doi: 10.1016/j.mce.2022.111554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shu X., Chen Z.S., Long J.R., et al. Large-scale integrated analysis of genetics and metabolomic data reveals potential links between lipids and colorectal cancer risk. Cancer Epidemiol Biomarkers Prev. 2022;31(6):1216–1226. doi: 10.1158/1055-9965.EPI-21-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chaurasia B., Summers S.A. Ceramides - lipotoxic inducers of metabolic disorders. Trends Endocrinol Metab. 2015;26(10):538–550. doi: 10.1016/j.tem.2015.07.006. [DOI] [PubMed] [Google Scholar]
- 54.Ogretmen B. Sphingolipid metabolism in cancer signalling and therapy. Nat Rev Cancer. 2018;18(1):33–50. doi: 10.1038/nrc.2017.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Molendijk J., Robinson H., Djuric Z., Hill M.M. Lipid mechanisms in hallmarks of cancer. Mol Omics. 2020;16(1):6–18. doi: 10.1039/c9mo00128j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Garcia-Barros M., Coant N., Truman J.P., Snider A.J., Hannun Y.A. Sphingolipids in colon cancer. Biochim Biophys Acta. 2014;1841(5):773–782. doi: 10.1016/j.bbalip.2013.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guida F., Tan V.Y., Corbin L.J., et al. The blood metabolome of incident kidney cancer: a case-control study nested within the MetKid consortium. PLoS Med. 2021;18(9) doi: 10.1371/journal.pmed.1003786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Treede I., Braun A., Sparla R., et al. Anti-inflammatory effects of phosphatidylcholine. J Biol Chem. 2007;282(37):27155–27164. doi: 10.1074/jbc.M704408200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bae S., Ulrich C.M., Neuhouser M.L., et al. Plasma choline metabolites and colorectal cancer risk in the women's health initiative observational study. Cancer Res. 2014;74(24):7442–7452. doi: 10.1158/0008-5472.CAN-14-1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nitter M., Norgard B., de Vogel S., et al. Plasma methionine, choline, betaine, and dimethylglycine in relation to colorectal cancer risk in the European Prospective Investigation into Cancer and Nutrition (EPIC) Ann Oncol. 2014;25(8):1609–1615. doi: 10.1093/annonc/mdu185. [DOI] [PubMed] [Google Scholar]
- 61.Brevik A., Veierod M.B., Drevon C.A., Andersen L.F. Evaluation of the odd fatty acids 15: 0 and 17: 0 in serum and adipose tissue as markers of intake of milk and dairy fat. Eur J Clin Nutr. 2005;59(12):1417–1422. doi: 10.1038/sj.ejcn.1602256. [DOI] [PubMed] [Google Scholar]
- 62.Otto M.C.D., Nettleton J.A., Lemaitre R.N., et al. Biomarkers of dairy fatty acids and risk of cardiovascular disease in the multi-ethnic study of atherosclerosis. J Am Heart Assoc. 2013;2(4) doi: 10.1161/JAHA.113.000092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Weitkunat K., Schumann S., Nickel D., et al. Odd-chain fatty acids as a biomarker for dietary fiber intake: a novel pathway for endogenous production from propionate. Am J Clin Nutr. 2017;105(6):1544–1551. doi: 10.3945/ajcn.117.152702. [DOI] [PubMed] [Google Scholar]
- 64.Zheng J.S., Sharp S.J., Imamura F., et al. Association between plasma phospholipid saturated fatty acids and metabolic markers of lipid, hepatic, inflammation and glycaemic pathways in eight European countries: a cross-sectional analysis in the EPIC-InterAct study. BMC Med. 2017;15(1):203. doi: 10.1186/s12916-017-0968-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Venn-Watson S., Lumpkin R., Dennis E.A. Efficacy of dietary odd-chain saturated fatty acid pentadecanoic acid parallels broad associated health benefits in humans: could it be essential? Sci Rep. 2020;10(1):8161. doi: 10.1038/s41598-020-64960-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bagheri M., Djazayery A., Farzadfar F., et al. Plasma metabolomic profiling of amino acids and polar lipids in Iranian obese adults. Lipids Health Dis. 2019;18(1):94. doi: 10.1186/s12944-019-1037-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Aglago E.K., Huybrechts I., Murphy N., et al. Consumption of fish and long-chain n-3 polyunsaturated fatty acids is associated with reduced risk of colorectal cancer in a large European cohort. Clin Gastroenterol Hepatol. 2020;18(3):654–666.e6. doi: 10.1016/j.cgh.2019.06.031. [DOI] [PubMed] [Google Scholar]
- 68.Lu Y., Li D.D., Wang L.J., et al. Comprehensive investigation on associations between dietary intake and blood levels of fatty acids and colorectal cancer risk. Nutrients. 2023;15(3):730. doi: 10.3390/nu15030730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yang Z.P., Tang H.Z., Lu S., Sun X.B., Rao B.Q. Relationship between serum lipid level and colorectal cancer: a systemic review and meta-analysis. BMJ Open. 2022;12(6) doi: 10.1136/bmjopen-2021-052373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang C., Cheng Y.J., Luo D.L., et al. Association between cardiovascular risk factors and colorectal cancer: a systematic review and meta-analysis of prospective cohort studies. eClinicalMedicine. 2021;34 doi: 10.1016/j.eclinm.2021.100794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yao X., Tian Z. Dyslipidemia and colorectal cancer risk: a meta-analysis of prospective studies. Cancer Cause Control. 2015;26(2):257–268. doi: 10.1007/s10552-014-0507-y. [DOI] [PubMed] [Google Scholar]
- 72.Tsilidis K.K., Branchini C., Guallar E., Helzlsouer K.J., Erlinger T.P., Platz E.A. C-reactive protein and colorectal cancer risk: a systematic review of prospective studies. Int J Cancer. 2008;123(5):1133–1140. doi: 10.1002/ijc.23606. [DOI] [PubMed] [Google Scholar]
- 73.Zhou B., Shu B., Yang J., Liu J., Xi T., Xing Y.Y. C-reactive protein, interleukin-6 and the risk of colorectal cancer: a meta-analysis. Cancer Cause Control. 2014;25(10):1397–1405. doi: 10.1007/s10552-014-0445-8. [DOI] [PubMed] [Google Scholar]
- 74.Carayol M., Licaj I., Achaintre D., et al. Reliability of serum metabolites over a two-year period: a targeted metabolomic approach in fasting and non-fasting samples from EPIC. PLoS One. 2015;10(8) doi: 10.1371/journal.pone.0135437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kuhn T., Sookthai D., Rolle-Kampczyk U., et al. Mid- and long-term correlations of plasma metabolite concentrations measured by a targeted metabolomics approach. Metabolomics. 2016;12(12):184. [Google Scholar]
- 76.Kofeler H.C., Ahrends R., Baker E.S., et al. Recommendations for good practice in MS-based lipidomics. J Lipid Res. 2021;62 doi: 10.1016/j.jlr.2021.100138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Siskos A.P., Jain P., Romisch-Margl W., et al. Interlaboratory reproducibility of a targeted metabolomics platform for analysis of human serum and plasma. Anal Chem. 2017;89(1):656–665. doi: 10.1021/acs.analchem.6b02930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Austin H., Flanders W.D., Rothman K.J. Bias arising in case-control studies from selection of controls from overlapping groups. Int J Epidemiol. 1989;18(3):713–716. doi: 10.1093/ije/18.3.713. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.