Highlights
-
•
This study reports for the first time the value of genes that regulate tumor cell sensitivity to T cell-mediated cytotoxicity (GSTTKs) in the prognosis of COAD patients.
-
•
This study constructs a new gene signature based on GSTTKs. The risk score calculated by gene signature is significantly correlated with immune cell infiltration, immune therapy response, and sensitivity to chemotherapy. Gene signature helps to accurately stratify the risk of COAD patients and improve the current status of personalized treatment.
-
•
This study explores the mechanism of GSTTKs in colon cancer and provides new therapeutic targets for colon cancer patients.
-
•
Single cell analysis revealed the correlation between the risk genes involved in constructing gene signatures and the tumor microenvironment.
Keywords: Colon adenocarcinoma, Gene signature, T cell-mediated tumor killing, Single cell data
Abstract
Background
Colorectal cancer (CRC) is a prevalent malignancy of the digestive tract. A new prognostic scoring model for colon adenocarcinoma (COAD) is developed in this study based on the genes involved in tumor cell-mediated killing of T cells (GSTTKs), accurately stratifying COAD patients, thus improving the current status of personalized treatment.
Method
The GEO and TCGA databases served as the sources of the data for the COAD cohort. This study identified GSTTKs-related genes in COAD through single-factor Cox analysis. These genes were used to categorize COAD patients into several subtypes via unsupervised clustering analysis. The biological pathways and tumor microenvironments of different subgroups were compared. We performed intersection analysis between different subtypes to obtain intersection genes. Single-factor Cox regression analysis and Lasso-Cox analysis were conducted to establish clinical prognostic models. Two methods are used to assess the accuracy of model predictions: ROC and Kaplan-Meier analysis. Next, the prediction model was further validated in the validation cohort. Differential immune cell infiltration between various risk categories was identified via single sample gene set enrichment analysis (ssGSEA). The COAD model's gene expression was validated via single-cell data analysis and experiments.
Result
We established two distinct GSTTKs-related subtypes. Biological processes and immune cell tumor invasion differed significantly between various subtypes. Clinical prognostic models were created using five GSTTKs-related genes. The model's risk score independently served as a prognostic factor. COAD patients were classified as low- or high-risk depending on their risk scores. Patients in the low-risk category recorded a greater chance of surviving. The outcomes from the validation cohort match those from the training set. Risk scores and several tumor-infiltrating immune cells were strongly correlated, according to ssGSEA. Single-cell data illustrated that the model's genes were linked to several immune cells. The experimental results demonstrated a significant increase in the expression of HOXC6 in colon cancer tissue.
Conclusion
Our research findings established a new gene signature for COAD. This gene signature helps to accurately stratify the risk of COAD patients and improve the current status of individualized care.
Introduction
Colorectal cancer (CRC) is a prevalent cancerous growth of the gastrointestinal system. In a 2020 research report, the incidence rate of CRC was estimated to reach the third-highest and the second-highest fatality rate [1]. A prevalent subtype of CRC is colon adenocarcinoma (COAD). Each year, COAD is attributed to around 10 % of all cancer cases and fatalities caused by cancer globally [2]. In both women and men, COAD is the third most prevalent type of malignancy, with cancer mortality ranking second globally [3,4]. Although the diagnosis and treatment of COAD have developed rapidly recently, its annual incidence rate is still steadily rising [5]. Radiation therapy, chemotherapy, and surgical treatment are the traditional treatment methods for COAD. However, surgery-based comprehensive treatment can achieve good therapeutic results in the early management stage of COAD [6]. At present, the use of novel therapies like biologically targeted therapy, precision therapy, and immunotherapy for colon cancer has significantly expanded. However, the therapeutic efficacy of these treatments is limited by the development of immune escape and drug resistance [7], [8], [9]. On the other hand, the late metastasis of COAD also makes cancer treatment more challenging, resulting in unsatisfactory 5-year survival rates [10]. The poor treatment status of COAD indicates that better and deeper research on the potential mechanisms that affect tumor occurrence and progression is necessary. Finding promising treatment targets and reliable prognostic biological markers for COAD is essential, which may help improve the treatment status of COAD patients.
The emergence of immunotherapy has changed the current status of cancer treatment. The working principle of immunotherapy is to get rid of cancerous cells by boosting the body's defensive mechanisms. Even though immunotherapy is rapidly becoming the preferred treatment option for cancer, the overall poor response rate is a significant roadblock [11]. An important mechanism of tumor escape is the upregulation of the expression of immune regulatory ligands. In CRC, the expression of programmed death - 1 (PD-L1) on tumor cells and tumor-infiltrating lymphocytes (TILs) is particularly important for this immune escape phenotype [12,13]. A tumor cell's reaction to endogenous anti-tumor activity is characterized by the PD-1/PD-L1 pathway, which is the process behind adaptive immune resistance. Even though the PD-1/PD-L1 signal axis is crucial in CRC, anti-PD-1 medication is not often successful in this scenario, and a proportion of patients may still experience tumor progression after treatment [14]. The tumor killing mediated by T cells is among the key determinants of immunotherapy response [15,16]. Recent emerging studies have found that inhibition of Prbm1, Arid2, and Brd7 enhances T cell-mediated cytotoxicity against tumor cells [17]. The study by Ru et al. illustrated that CD47 and PTPN2 genes are linked to the sensitivity of T-cell-mediated tumor killing (TTK) [18]. However, the mechanism of action of genes that regulate tumor cell sensitivity to T cell-mediated cytotoxicity (GSTTKs) in COAD remains a blank field.
Therefore, this study integrated GSTTKs and identified GSTTKs-related subtypes from the COAD cohort integrated by the Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) databases. Notably, the features of immune cell infiltration vary across GSTTK subtypes. Our goal is to design a new prognostic scoring model for COAD using GSTTKs and evaluate its clinical utility in the prognosis of CPAD patients. This model may characterize the immune microenvironment, predict COAD patient prognosis with high accuracy, and effectively stratify COAD patients, thus improving the current status of precise treatment for these patients.
Methods
Raw data download
A total of two COAD datasets (TCGA-PAAD, GSE39582) were acquired from TCGA and GEO databases. Firstly, we pre-processed the raw datasets, including background correction and quantile normalization. The TCGA-COAD and GEO datasets were merged into one COAD cohort, and the ComBat function of the "SVA" package in R was employed to eliminate batch processing effects in different datasets. A TCGA database was also used to obtain data on somatic mutations and copy number variations (CNVs) of COAD patients. Thereafter, the TISIDB database (http://cis.hku.hk/TISIDB/) was searched for genes that are sensitive to T cell-mediated tumor killing in cancer immunotherapy [18].
Pattern recognition of different GSTTKs based on unsupervised clustering in COAD
By means of univariate Cox regression analysis, we identified genes in the TCGA-COAD cohort that were substantially linked to overall survival (OS). We generated two clusters using the R software "ConsensusClusterPlus" for consensus clustering based on the GSTTKs associated genes linked to prognosis. The heat map displays the association of various clusters with clinical characteristics [19]. Additionally, to identify differentially expressed genes (DEGs) among different clusters, the "limma" R program was utilized, with logFC=1 and P < 0.05 as the threshold.
Functional enrichment analysis and single sample gene set enrichment analysis algorithm (ssGSEA)
The MSigDB database was searched to retrieve the Kyoto Encyclopedia of Genes and Genomes (KEGG) gene set (C2. cp. kegg. V7.4). The biological functional variations between two distinct subgroups were identified via gene set variation analysis (GSVA) [20]. The potential molecular functions (MFs), cellular components (CCs), and biological processes (BPs) KEGG pathways of DEGs amongst various clusters are examined using the 'clusterProfiler' in R. The biological pathways between various risk categories were investigated via GSEA. Significant enrichment was determined as per the criteria of log 2-fold change (FC) | ≥ 1 and p-value<0.05. Furthermore, based on the ssGSEA algorithm, the association between BM-related subtypes and the proportion of 23 infiltrating immune cells was examined using a Normalized Enrichment Score (NES). In the COAD cohort, ssGSEA was conducted to estimate the relative level of infiltration of 22 immune cells in the tumor microenvironment of COAD. Enrichment fraction is used to express each immune cell type's relative abundance. Wilcoxon rank sum test was completed to examine the variations in immune cell abundance between various clusters and the association between distinct clusters and infiltrating immune cells in COAD.
Development and validation of a clinical prognostic model using TTK-sensitive genes
For the prognostic evaluation of DEGs, we implemented a univariate Cox regression model, followed by the use of the Least Absolute Shrinkage and Selection Operator (LASSO) and 10-fold cross-validation to further narrow the scope of screening for prognostic genes. Finally, a clinical prognosis model was developed with the Cox proportional risk regression algorithm. The formula is as indicated: riskscore= Σ (Expressioni * Coefi), whereby, Coefi and Expressioni represent the risk factors and each gene's expression, respectively.
The training and validation groups were generated by randomly classifying the COAD dataset. Patients were classified into low- and high-risk categories in the training and validation groups as per the median risk score. To contrast the survival rates of high- and low-risk populations, the Kaplan-Meier (KM) curve was employed. For both low- and high-risk groupings, the expression patterns of the genes used to develop clinical prognostic models were shown using heat maps. For both low- and high-risk groupings, the expression profiles of the genes used to build clinical prognostic models were shown using heat maps. Risk scores are examined via univariate and multivariate analyses to ascertain whether they function as independent prognostic indicators.
Development and verification of column chart predictions
The 1-, 3-, and 5-year OS rates of COAD patients were predicted using the "rms" and "survival" packages, which were also utilized to create a column chart model as per risk score and several clinical characteristics. The discrimination and accuracy of the column chart were estimated using the calibration curve.
Correlation between clinical prognosis model related to GSTTKs, TME invasion, and somatic mutation
The CIBERSORT method evaluates the proportion of 22 types of immune cells from a vast volume of expression data from tumor samples using a set of reference gene expression matrices and is centered on the theory of linear support vector regression [21]. Using the CIBERSORT method, we investigated how risk scores and infiltrating immune cells in clinical prognostic models correlate. The 'maftools' R package was employed to extract mutation annotation formats (MAFs) from the TCGA database and detect mutations in COAD patients across different risk groups.
Exploring the dry characteristics and drug sensitivity analysis of gene signature in clinical prognosis
From the Cancer Drug Sensitivity Genomics (GDSC) database, we evaluated drug response predictions using the "pRRophic" R-package, where Ridge regression was utilized to measure the median maximum inhibitory concentration (IC50) for each patient. 10-fold cross-validation was utilized to measure the prediction ability.
Tumor immune single cell hub (TISCH) database
The comprehensive single-cell RNA sequencing database known as TISCH was specifically designed for TME, targeting tumor immune single-cell centers (http://tisch.comp-genomics.org) [22]. From this platform, an extensive investigation was conducted into the TME heterogeneity across distinct datasets and cell types.
Cell culture and transient transfection
Human colon cancer cell lines SW480, SW620 and Human normal colon epithelial cells NCM460 were obtained from BNCC (Beijing, China). SW480, SW620 and NCM460 cells were cultured in DMEM F12 with 10 % FBS (Gibco, Thermo Fisher, USA). Cells were grown at 37 °C in a humidified environment containing 5 % CO2. The target sequences of NPM1 siRNA were shown in Table 1.
Table 1.
The target sequences for NPM1 siRNA.
| Gene | target sequence (5′−3′) |
|---|---|
| si HOXC6#1 | CCGTATGACTATGGATCTAATTC |
| si HOXC6#2 | GACTATGGATCTAATTCCTTTTA |
Quantitative reverse transcription-polymerase chain reaction (qRT-PCR)
TRIzol (Thermo Fisher, USA) reagent was used to extract total RNA from the SW480, SW620 and NCM460 cell lines. Using FastStart Universal SYBR Green Master, quantitative reverse transcription-polymerase chain reaction (qRT-PCR) was performed on the RNA extracted from each sample (2 μg) on a LightCycler 480 PCR System (Roche, USA). The cDNA was utilized as a template with a reaction volume of 20 μl (2 μl of cDNA template, 10 μl of PCR mixture, 0.5 μl of forward and reverse primers, and an appropriate water volume). The following procedures were utilized for the PCR reactions: Cycling conditions started with an initial DNA denaturation phase at 95 °C for 30 s, followed by 45 cycles at 94 °C for 15 s, 56 °C for 30 s, and 72 °C for 20 s. Three separate analyses were performed on each sample. Based on the 2-ΔΔCT method, data from the threshold cycle (CT) were obtained and standardized to the levels of GAPDH in each sample. The expression levels of mRNA were compared to controls obtained from normal tissues. The sequence table of primer pairs for the target genes was shown in Table 2
Table 2.
A list of the sequences of primer pairs for target genes.
| Gene | Forward primer sequence (5′−3′) | Reverse primer sequence (5′−3′) |
|---|---|---|
| C11orf96 | TCACGCCAACACTCTCGCTGAA | CAATCCTCCAGACGCAGTAGCA |
| CXCL9 | CTGTTCCTGCATCAGCACCAAC | TGAACTCCATTCTTCAGTGTAGCA |
| HOXC6 | TTACCCCTGGATGCAGCGAATG | CCGCGTTAGGTAGCGATTGAAG |
| VSIG4 | GATGGCAACCAAGTCGTGAGAG | CCTGGCATTGAAGGCTAATCCTC |
| CXCL13 | TATCCCTAGACGCTTCATTGATCG | CCATTCAGCTTGAGGGTCCACA |
| CXCL1 | AGCTTGCCTCAATCCTGCATCC | TCCTTCAGGAACAGCCACCAGT |
| CXCL8 | GAGAGTGATTGAGAGTGGACCAC | GAGAGTGATTGAGAGTGGACCAC |
| IL1B | CCACAGACCTTCCAGGAGAATG | GTGCAGTTCAGTGATCGTACAGG |
| IL4 | CCGTAACAGACATCTTTGCTGCC | GAGTGTCCTTCTCATGGTGGCT |
| IL10 | TCTCCGAGATGCCTTCAGCAGA | TCAGACAAGGCTTGGCAACCCA |
| GAPDH | GTCTCCTCTGACTTCAACAGCG | ACCACCCTGTTGCTGTAGCCAA |
Immunohistochemistry (IHC) staining
The tissue sections through deparaffinization and dehydration were incubated with polyclonal rabbit anti-human HOXC6 antibodies (1:500, Abcam, ab151575) overnight at 4 °C after epitope retrieval, H2O2 treatment, and non-specific antigens blocking. Next, sections were incubated with secondary antibodies (1:1000, Proteintech, SA00001–2) for two hours at room temperature, and then the signal was detected with an enhanced DAB staining kit (Proteintech, China).
Cell viability
Cell viability was detected using the Cell Counting Kit-8 assay (Beyotime, China). Cells from different treatments were cultured in 96-well plates at a density of 1 × 103 cells per well. CCK-8 solution was applied at the indicated time points. After incubation at 37 °C for 2 h, the O.D 450 values of each well were detected using a microplate reader (Thermo Fisher, USA).
Transwell assay
Transwell assays for migration and invasion of SW480 and SW620 cell lines were performed. Briefly, cells (5 × 104) were inoculated into chambers coated (for invasion) or uncoated with Matrigel (BD Biosciences, USA) (for migration). Serum-free medium was added to the upper layer and a complete DMEM medium was added to the lower layer. After 24 h of incubation, migrating or invading cells were fixed with 4 % paraformaldehyde and stained with 0.1 % crystalline violet.
Statistical analysis
R version 4.01 was applied to conduct all analyses of statistical data. For a comparison between the two groups, we employed the Wilcoxon test. The Kruskal-Wallis test and one-way ANOVA were employed to examine variations across two or more groups. For survival analysis, the KM method was employed, and Pearson correlation analysis was executed to assess correlations. Limma was applied to construct principal component analysis (PCA) between different clusters. ROC curve analysis was conducted based on the "survival" and "timeROC" R packages. P < 0.05 denotes the significance criterion.
Results
Genetic characteristics and transcriptional changes of GSTTKs in COAD
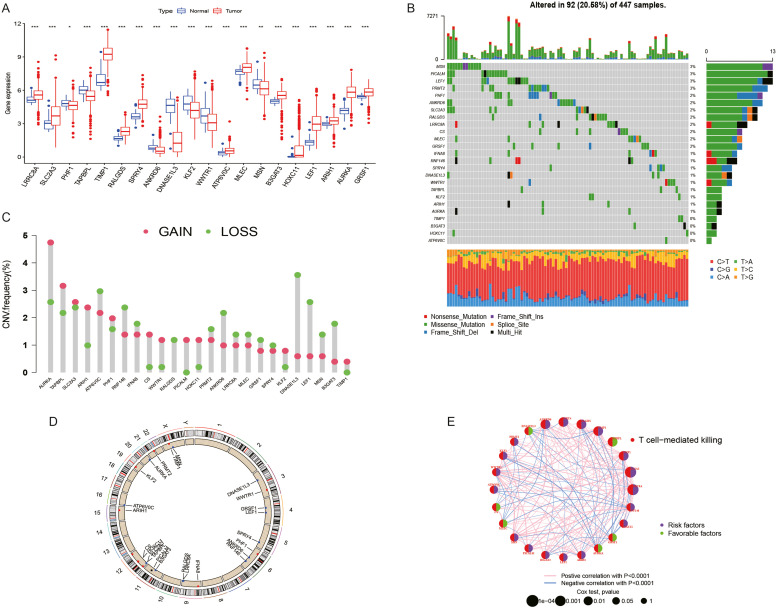
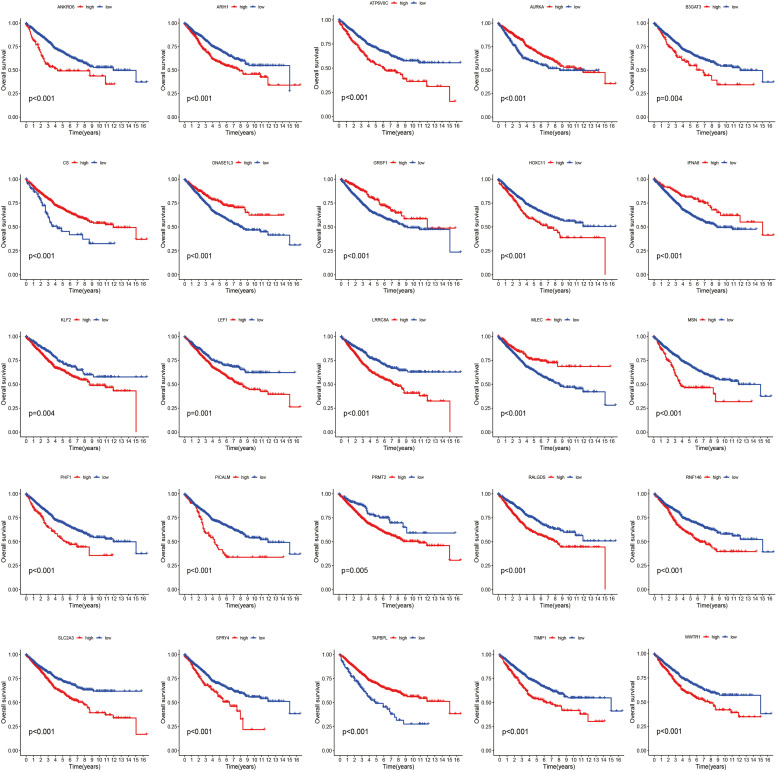
In the TCGA-COAD cohort, univariate analysis was conducted to identify potential prognosis-related genes. The threshold was set at P < 0.01. An aggregate of 25 prognosis-related genes was discovered. The differential expression of these 25 prognosis-related genes in the TCGA-COAD cohort is depicted in Fig. 1A. More specifically, we determined the prevalence of somatic mutation and CNV in 25 GSTTKs in COAD. 98 out of 447 COAD samples (20.58 %) had mutations. We then examined the copy number alterations of these 25 GSTTKs. Among them, the CNV of ATP6V0C, RNF146, IFNA8, RALGDS, PRMT2, ANKRD6, LRRC8A, MLEC, GRSF1, SPRY4, DNASE1L3, LEF1, MSN, and B3GAT3 significantly decreased. The copy number of other genes significantly increased. (Fig. 1C) The placement of these GSTTKs' CNVs on the chromosome is depicted in Fig. 1D. The molecular interactions between 25 GSTTKs are depicted in Fig. 1E. In COAD patients, upregulation of IFNA8, CS, GRSF1, TAPBPL, MLEC, AURKA, and DNASE1L3 was linked to a better OS. The upregulation of other genes (HOXC11, WWTR1, LEF1, MSN, SLC2A3, KLF2, TIMP1, RNF146, PICALM, ANKRD6, SPRY4, ARIH1, B3GAT3, RALGDS, ATP6V0C, PHF1, PRMT2, and LRRC8A) was associated with poor OS in PAAD patients. (Fig. 2)
Fig. 1.
Genetic characteristics of GSTTKs in COAD. (A) The differential expression of 25 genes linked to prognosis between tumors and normal samples in the TCGA-COAD cohort. (B) The waterfall plot shows the mutation frequency of 25 GSTTKs in 447 TCGA-COAD samples. The list shows a single patient, TMB appears on the upper bar chart, the percentage of each variant type is depicted in the right Bar Chart, and each patient's conversion score is displayed in the lower stacked bar chart. (C) The CNV variation frequency of 25 GSTTKs in TCGA-COAD. Green: deletion frequency; Red dot: Amplify the frequency. (D) The position of 25 GSTTKs on chromosomes. (E) The interaction network between 25 GSTTKs.
Fig. 2.
Survival curve of 25 GSTTKs in the TCGA-COAD cohort.
Determination of COAD subtypes associated with GSTTKs
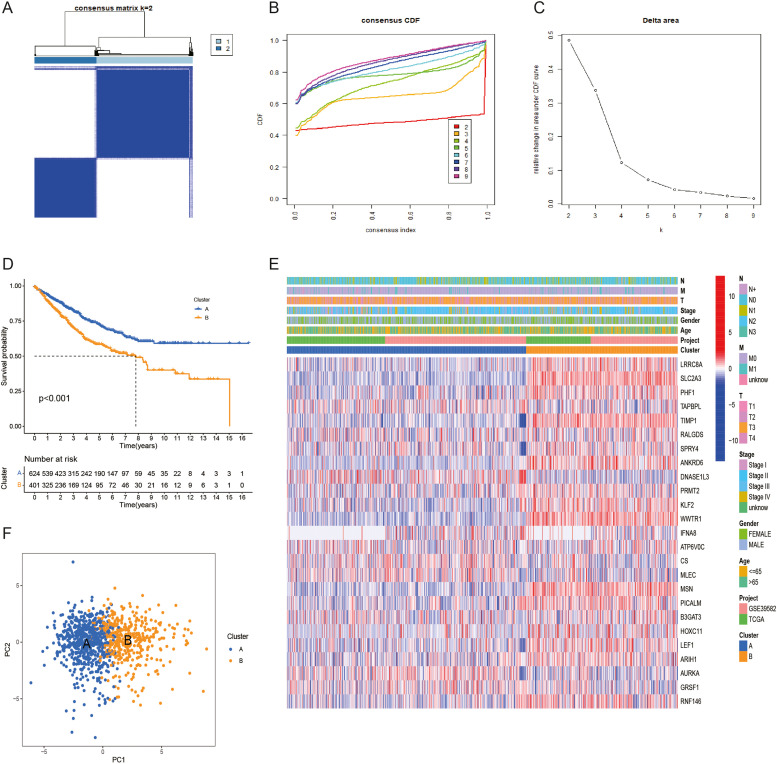
The TCGA-COAD dataset and the GEO dataset were merged into a single COAD cohort. A consensus clustering approach was applied to cluster and evaluate COAD samples in the PAAD cohort to explore the expression features and probable biological properties of GSTTKs in COAD, yielding two subtypes (cluster A and cluster B) (Fig. 3A-C). The survival curve results show that cluster A's OS is superior to cluster B's OS. The heat map (Fig. 3D) exhibits the correlation between two different subtypes of GSTTKs and clinical characteristics. The PCA diagram (Fig. 3E) shows a significantly different distribution between clusters A and B (Fig. 3F).
Fig. 3.
Identification of GSTTKs-related subtypes in COAD. (A) When k = 2, merge the heatmap of the cohort cluster. (B) The Cumulative Distribution Function (CDF) curve's distribution. (C) The area under the CDF curve. (D) The variations in survival between the two subtypes are shown by the Kaplan-Meier survival curve. (E) The link between the subtypes of GSTTKs and the clinical pathological parameters is displayed in the heat map. (F) Two GSTTK subtypes were compared by principal component analysis (PCA).
Characteristics of biological pathways and TME between subtypes of GSTTKs in COAD
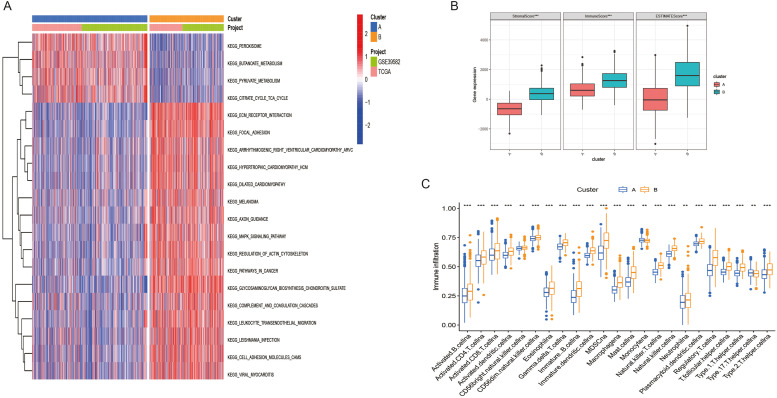
The results of GSVA showed a significant correlation between cluster A and metabolic pathways such as butyric acid metabolism and pyruvate metabolism. Cluster B is significantly correlated with ECM receptor interactions, MAPK signaling pathways, and cancer pathways. (Fig. 4A) Using the CIBERSORT and ssGSEA methods, we evaluated the variations in immune cell infiltration levels between Clusters A and B. Cluster B's immune score is substantially greater than Cluster B's (Fig. 4B). Additionally, Cluster B has a greater proportion of infiltrating immune cells than Cluster A. The infiltration level of immune cells such as Monocytena and Type.17.T.helper.cellna in Cluster A is higher than that in Cluster A and Cluster B. (Fig. 4C)
Fig. 4.
Characteristics of biological pathways and tumor microenvironment between GSTTKs related subtypes in COAD. (A) the biological pathway between Clusters A and B is in an activated state. (B) Variations between Clusters A and B in terms of estimated scores, immune scores, and matrix scores. (D) Clusters A and B have varying degrees of immune cell infiltration.
Differential expression analysis and functional annotation between subtypes of GSTTKs
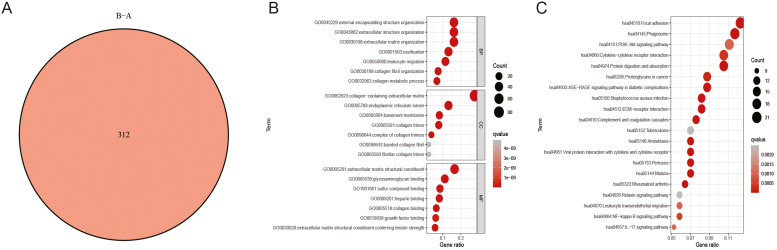
We identified 312 DEGs by performing a differential expression analysis between clusters A and B using the "limma" program. We subsequently employed these DEGs to perform functional enrichment analysis (Fig. 5A). The GO findings illustrated that these DEGs were significantly correlated with BPs including extracellular matrix structural components, extracellular matrix tissue, and basement membrane. (Fig. 5B) KEGG results illustrate that the ECM receptor interaction, PI3K Akt signaling pathway, and NF-κB pathways including the B signaling pathway are enriched. (Fig. 5C)
Fig. 5.
Analysis of differential expression and functional annotation of GSTTK-related subtypes. (A) The differentially expressed genes (DEGs) at the intersection between Clusters A and B. (B) GO enrichment analysis between these DEGs. (C) The potential KEGG pathway of these DEGs.
Development and verification of a clinical prognostic model incorporating GSTTKs
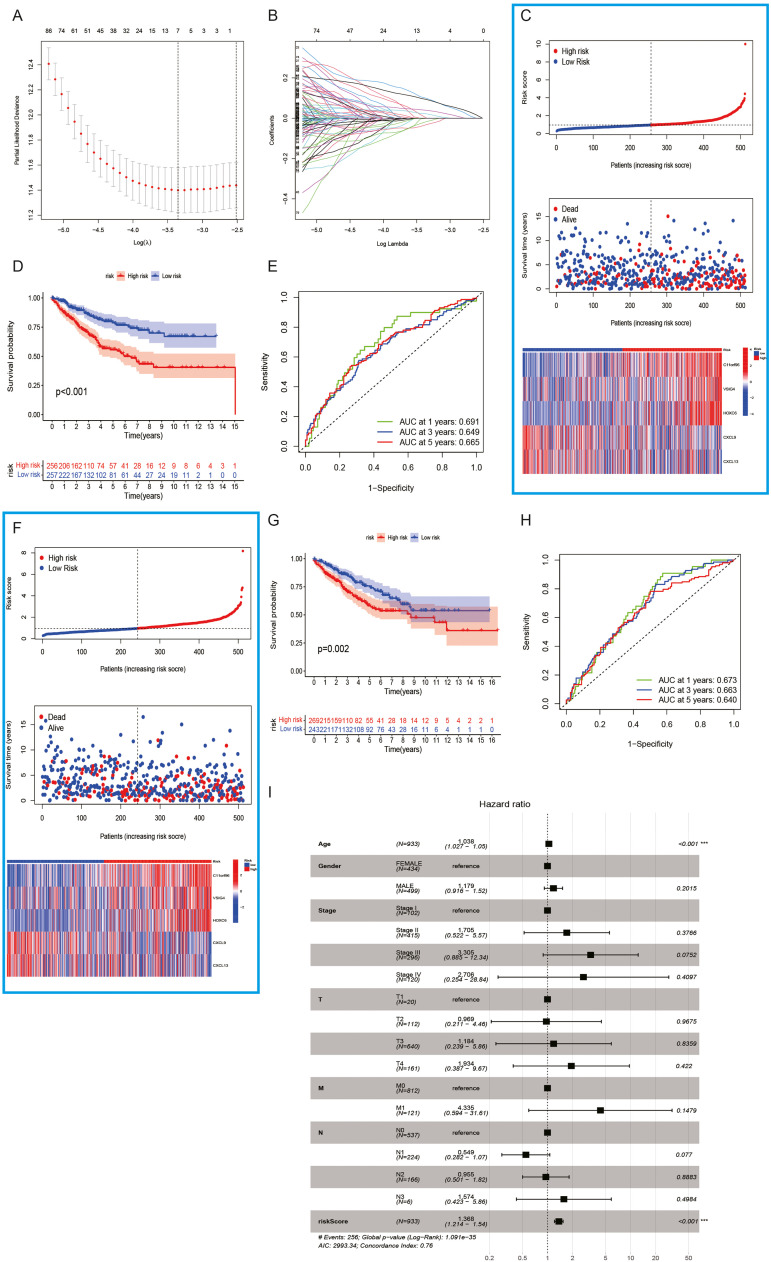
We used 312 DEGs between clusters A and B for conducting univariate analysis and obtained 163 GSTTKs related to prognosis for constructing clinical prognosis models. Finally, we constructed a clinical prognosis model containing 5 characteristic genes (C11orf96, VSIG4, HOXC6, CXCL9, and CXCL13) using the LASSO—COX algorithm. The formula (Fig. 5A and B) is as follows: riskscore=(0.2100) * C11orf96+(0.2018) * VSIG4+(0.1650) * HOXC6+(−0.1489) * CXCL9+(−0.1115) * CXCL13
In the training group, COAD patients were divided into low-risk and high-risk groups based on the risk score of the model. More fatalities and shorter OS periods were linked to higher risk scores. The expression levels of five characteristic genes (C11orf96, VSIG4, HOXC6, CXCL9, and CXCL13) were substantially varied across high- and low-risk subgroups (Fig. 6C) As opposed to the high-risk patients, those at low-risk exhibited a superior OS, according to a KM analysis (P < 0.001). (Fig. 6D) AUC values serve as a representation of the risk score's 1-, 3-, and 5-year survival probabilities, which were 0.691, 0.649, and 0.665, respectively. More importantly, the model was well-validated in the validation group (Fig. 6E). As per the model's risk score, the COAD patients in the validation group were classified into low- and high-risk categories. More fatalities and shorter OS periods are linked to higher risk scores. Furthermore, the expression patterns of the five characteristic genes varied significantly between high- and low-risk groupings (Fig. 6F) When contrasted with the high-risk patients, those with lower risk recorded increased OS, according to a KM analysis (P < 0.001). (Fig. 6G) The 1-year, 3-year, and 5-year survival probabilities of the risk score are represented by AUC values, which are 0.673, 0.663, and 0.740, respectively. The results of the forest plot illustrated that the risk score of the clinical prognosis model is a potential risk factor. (Fig. 6I).
Fig. 6.
Construction and validation of clinical prognostic models related to GSTTKs. (A, B) Regression screening of characteristic genes based on the minimal criterion for least absolute shrinkage and selection operator (LASSO). (C) COAD patients with various risk scores, survival statuses, and GSTTK expression profiles were shown in the training set. (D) High- and low-risk patients in the training set were analyzed utilizing the Kaplan-Meier method. (E) The clinical prognostic model's ability in predicting patient prognosis in the training cohort is demonstrated by the receiver operating characteristic (ROC) curve. (F) Patients with COAD were shown in the validation cohort along with their risk scores, survival statuses, and GSTTKs. (G) Patients in the high-risk category had considerably worse prognoses in the validation cohort. (H) The clinical prognostic model's prognostic effectiveness in the validation cohort is displayed by the ROC curve. (I) Multivariate COX regression analysis indicates that risk scores can serve as independent prognostic factors.
Development of a column chart to predict the COAD patients' survival rate
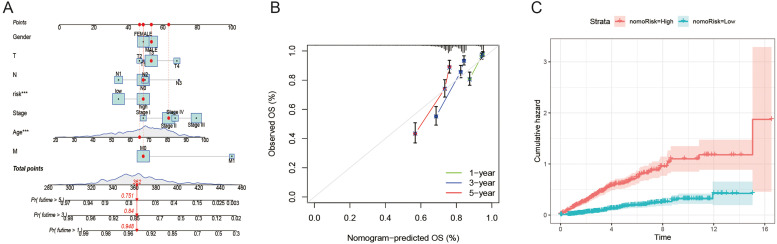
Depending on the data collected, we used the rms program to generate a column chart that predicted the COAD patients' life expectancy at 1-, 3-, and 5-year periods (Fig. 7A). The nomogram outperforms the ideal model, as shown by the calibration diagram (Fig. 7B). In addition, the cumulative risk of high-risk patients is significantly higher than that of low-risk patients over time. (Fig. 7C).
Fig. 7.
Generating and evaluating prognostic column charts. (A) A column chart was used to evaluate 1-, 3-, and 5-year survival rates. (B) The calibration curve of the column chart. (C) Cumulative incidence rate by risk stratification.
Correlation between clinical prognosis model and TME
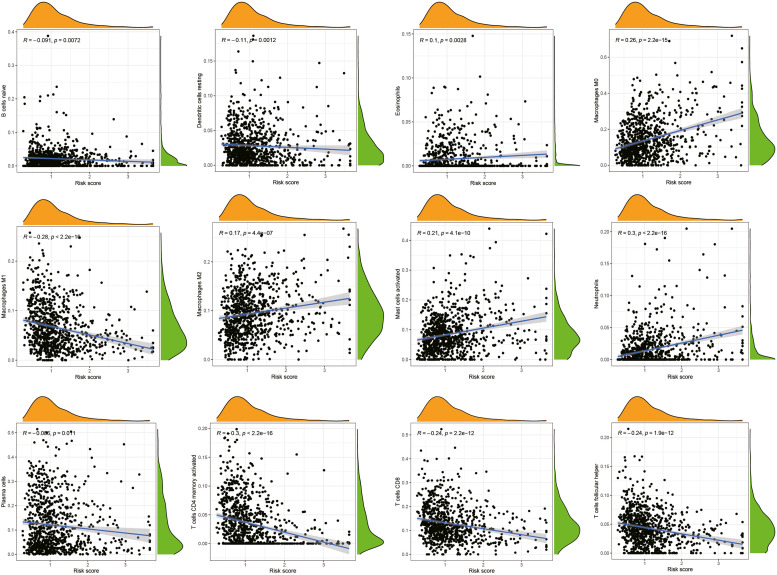
To assess the association of risk score with infiltrating immune cells, the CIBERSORT method was applied. Neutrophils, activated mast cells, macrophage M0, and macrophage M2 all had positive associations with the model's risk score. Conversely, the T cells follicular helper, T cells CD4 memory activated, CD8T cells, Macrophages M1, Dendritic cells resting, B cells naive, and Plasma cells exhibited an inverse correlation with the risk score of the model (Fig. 8).
Fig. 8.
The link between the tumor microenvironment and the risk scores of the clinical prognostic models.
Association between risk score and mutation in clinical prognosis models
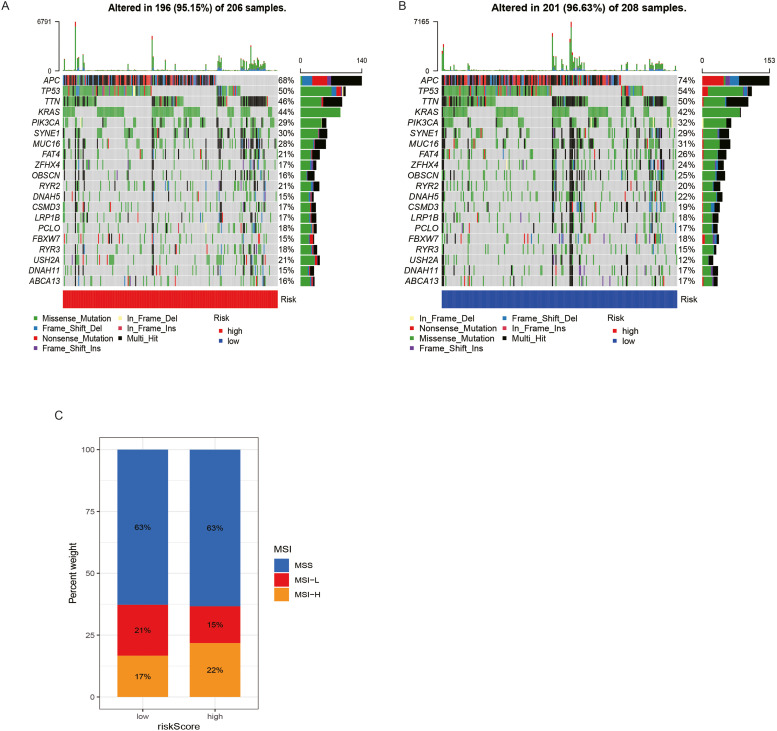
We examined the somatic mutation distribution differences between the low- and high-risk categories utilizing the maftools program in the TCGA-PAAD cohort. Additionally, the findings demonstrated that the low-risk patients exhibited a higher prevalence of mutations. The most prominent somatic mutations were APC (74 %) and TP53 (54 %) in the low-risk population and APC (68 %) and TP53 (50 %) in the high-risk population. Nevertheless, when contrasted with the low-risk population, the high-risk patients recorded a larger percentage of KRAS mutations (44 % vs 42 %) (Fig. 9A and B). According to Fig. 9C, there are more MSI-L cases in the low-risk category than in the high-risk category. In comparison to the high-risk patients, those at low risk exhibited a lower proportion of MSI-H.
Fig. 9.
The association of risk scores with mutations in clinical prognostic models. (A, B) Waterfall plots of tumor body mutations established in high- and low-risk patients. (C) The association of MSI with the risk scores of the clinical prognostic models.
Drug sensitivity analysis
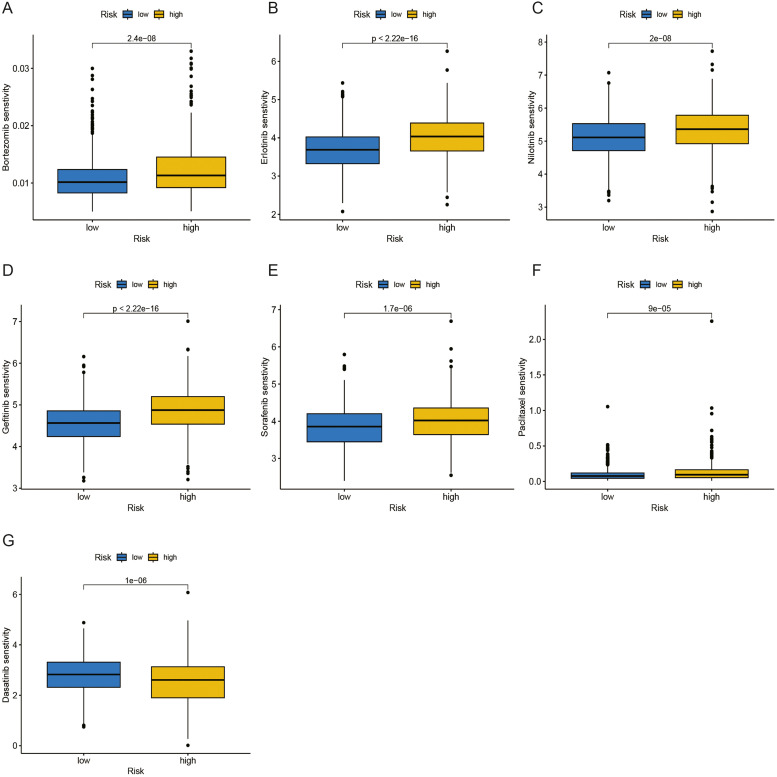
The pRRophytic algorithm was used to estimate the responsiveness to two commonly used chemotherapeutic medications (Fig. 7A). The sensitivity of many chemotherapeutic agents varied substantially between high- and low-risk categories. The estimated IC50 values for Bortezomib, Erlotinib, Nilotinib, Gefitinib, Sorafenib, and Paclitaxel in the high-risk patients were greater compared to those in the low-risk category (Fig. 10A-F). This implies that low-risk patients have a better chance of gaining benefits from these chemotherapy medications. Interestingly, the low-risk group's IC50 values for dasatinib were elevated relative to those for the high-risk patients (Fig. 10G).
Fig. 10.
Drug sensitivity analysis. (A-F) Bortezomib, Erlotinib, Nilotinib, Gefitinib, Sorafenib, and Paclitaxel have higher IC50 values in the high-risk group. A: Bortezomib; B: Erlotinib; C: Nilotinib; D: Gefitinib; E: Sorafenib; F: Paclitaxel (G) Dasatinib has a higher IC50 value in the low-risk patients.
Analysis of the correlation between characteristic genes of the model and TME
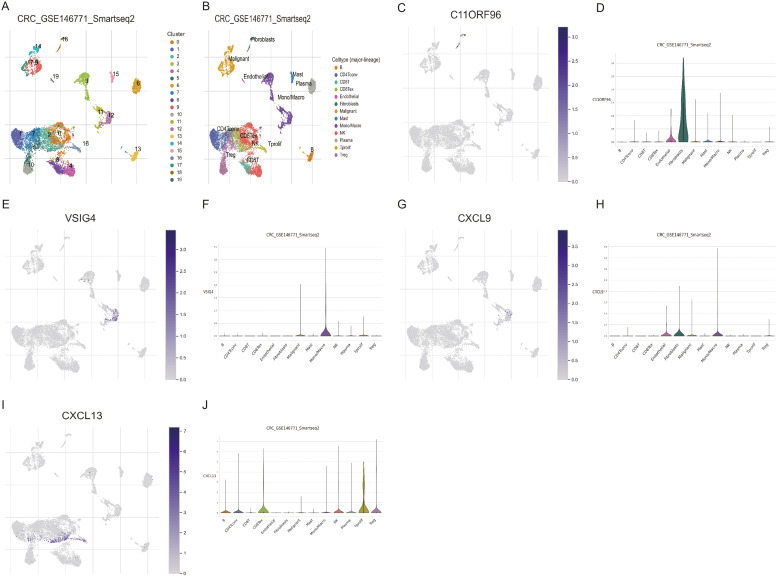
We applied the single-cell dataset GSE146771 from the TISCH database for CRC to determine the expression of five characteristic genes (C11orf96, VSIG4, HOXC6, CXCL9, and CXCL13) in TME. There are 20 cell populations and 13 intermediate cell types in the GSE146771 dataset, and the images present their distribution and quantity (Fig. 11A and B). C11orf96 is mainly expressed in fibroblasts. (Figs. 11C and 10D) VSIG4 is predominantly expressed in monocytes or macrophages. (Fig. 11E and F) CXCL9 is mainly expressed in endothelial cells, fibroblasts, monocytes, and macrophages. (Fig. 11G and H) CXCL13 is mainly expressed in tprolif cells. (Fig. 11I and J) However, no expression of HOXC6 was detected in this dataset.
Fig. 11.
The expression of characteristic genes for constructing models in the COAD single-cell dataset. (A) There are 20 cell populations in the GSE146771 dataset. (B) 20 cell populations were annotated as 13 cell subpopulations. The levels of C11orf96, VSIG4, CXCL9, and CXCL13 expressed in COAD samples. C. D: C11orf96; E. F: VSIG4; G. H: CXCL9; I. J: CXCL13.
Experimental validation of bioinformatics models
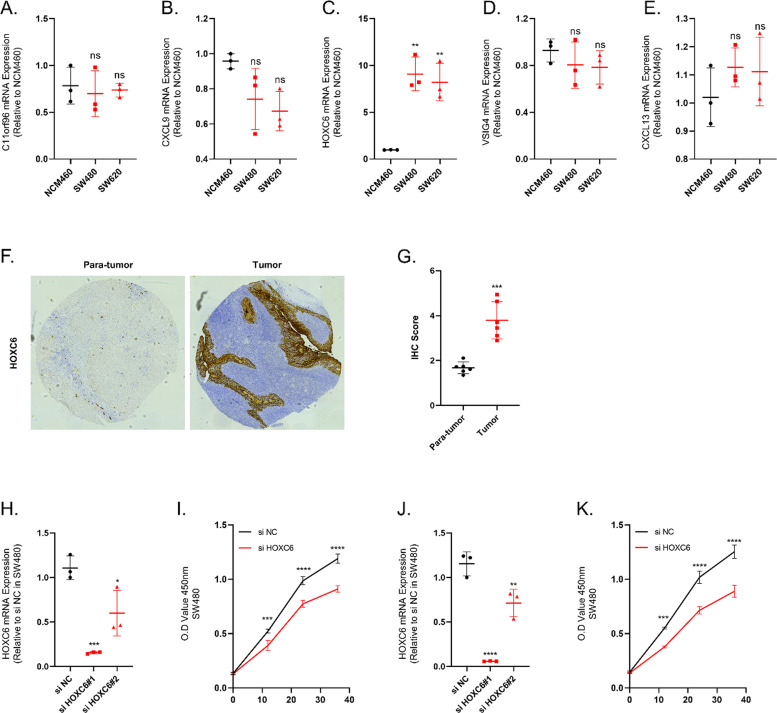
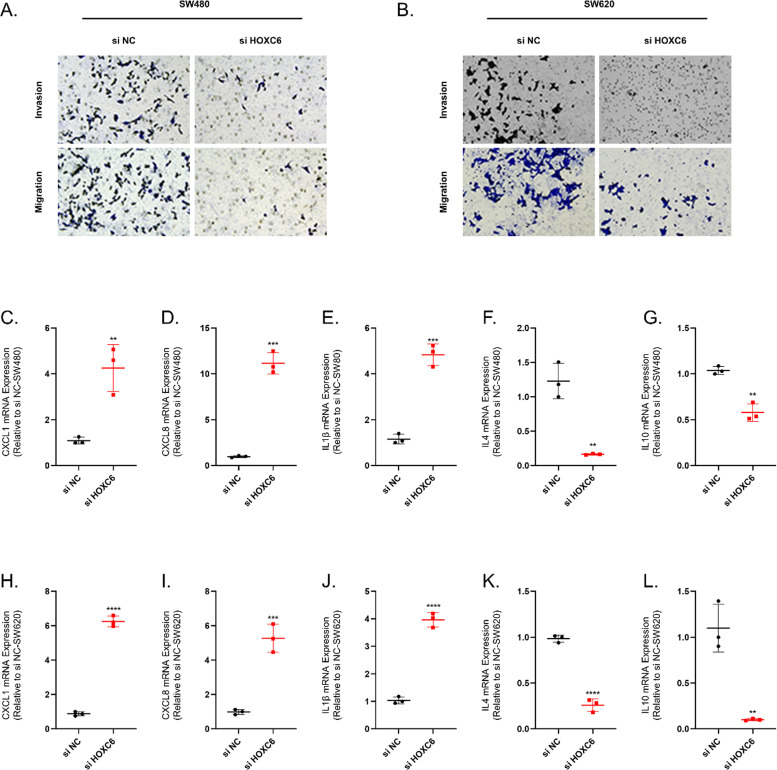
We examined the expression of C11orf96, CXCL9, HOXC6, VISG4 and CXCL13 by qRT-PCR in human colon cancer cell lines SW480, SW620 and human normal colon epithelial cells NCM460. (Fig. 12A-E) The qRT-PCR results indicated that HOXC6 expression was elevated in SW480 and SW620 relative to NCM460. We then examined the expression of HOXC6 in cancer and paracancer tissues in a tissue microarray of colon cancer. The results of IHC showed that the expression of HOXC6 was significantly elevated in colon cancer tissues. (Fig. 12F and G) We then tested the efficiency of using small interfering RNAs to inhibit HOXC6 expression in SW460 and SW480, and the results showed that si HOXC6#1 exhibited good inhibition efficiency. (Fig. 12H-K) Meanwhile, the cell viability of SW480 and SW620 cell lines was significantly decreased after the inhibition of HOXC6 expression. The results of Transwell experiments showed that the invasion and migration abilities of SW480 and SW620 cell lines were significantly decreased after the inhibition of HOXC6 expression.(Fig. 13A and B) Finally, we examined the expression of immune-related cytokines in SW480 and SW620 cell lines before and after HOXC6 inhibition, and we could observe a rise in the expression of pro-inflammatory cytokines CXCL1, CXCL8 and IL1β after inhibition of HOXC6 expression. Whereas the expression of anti-inflammatory cytokines such as IL4 and IL10 was suppressed upon inhibition of HOXC6 expression. (Fig. 13C-L)
Fig. 12.
HOXC6 is elevated in colon cancer tissues. (A-E) The expression of C11orf96, CXCL9, HOXC6, VISG4 and CXCL13 in NCM460, SW480 and SW620 was detected by qRT-PCR and analyzed for relative quantification. (F-G) The expression of HOXC6 in cancer and paracancer tissues was detected and quantified using IHC. (H) The inhibition efficiency of small interfering RNAs was detected and analyzed for relative quantification in SW480 cell line. (I) Cell viability of SW480 was assayed after inhibition of HOXC6 expression in SW480 cell line. (J) The inhibition efficiency of small interfering RNAs was detected and analyzed for relative quantification in SW620 cell line. (K) Cell viability of SW480 was assayed after inhibition of HOXC6 expression in SW620 cell line. N = 3. *≤0.05, **≤0.01, ***≤0.001, ****≤0.0001. N = 6/3, The results are presented as mean ± SD.
Fig. 13.
HOXC6 promotes invasion and immunosuppression of colon cancer cell lines. (A-B) Alterations in cell migration as well as invasive capacity after inhibition of HOXC6 expression in SW480 and SW620 cell lines. (C-G) The expression levels of CXCL1, CXCL8, IL1β, IL4 and IL10 in SW480 cell line were detected by qRT-PCR before and after HOXC6 inhibition. (H-L) The expression levels of CXCL1, CXCL8, IL1β, IL4 and IL10 in SW620 cell line were detected by qRT-PCR before and after HOXC6 inhibition. N = 3. *≤0.05, **≤0.01, ***≤0.001, ****≤0.0001. N = 6/3, The results are presented as mean ± SD.
Discussion
Colon cancer has the highest prevalence among primary malignant tumors. The current treatment status of COAD is still inadequate because of its high rates of recurrence and metastasis, as well as its growth resistance [23,24]. Consequently, there is a pressing need to investigate novel characteristics for COAD patients to assess prognosis, detect high-risk populations, and provide individualized care. We comprehensively evaluated the expression and prognosis of GSTTKs in this study and divided COAD patients into 2 categories (clusters A and B) as per the expression patterns of GSTTKs. Between clusters A and B, there were considerable variations in OS, the proportion of immune cells that infiltrate tumors, and immune scores. This implies that these GSTTKs-related genes are implicated in the COAD TME. Cluster A was significantly correlated with metabolic pathways such as butyric acid and Pyruvic acid metabolism. Cluster B was significantly correlated with ECM receptor interactions, MAPK signaling pathways, and cancer pathways. To additionally examine the mechanistic basis of GSTTKs-related genes in COAD, we analyzed the DEGs between two subtypes and conducted LASSO and multiple Cox regression analysis on DEGs to establish a gene signature based on GSTTKs improve patient risk stratification for COAD and facilitate individualized screening.
The gene signature of the GSTTKs-related genes constructed in this study includes five genes (C11orf96, VSIG4, HOXC6, CXCL9, and CXCL13). HOXC6 belongs to the family of homeobox genes and is involved in the embryonic development of vertebrates and performs crucial functions in breast cancer, lung cancer, prostate cancer, leukemia, and other cancer types [25], [26], [27], [28], [29]. The study by Lina Qi et al. discovered that HOXC6 is overexpressed in right colon cancer and is linked to a dismal prognosis. Overexpression of HOXC6 can be achieved by activating the Wnt/β-catenin signaling pathway and inhibiting DKK1 production to induce EMT, thereby promoting the motility and invasiveness of colon cancer cells [30]. In addition, our experiment also demonstrated a significant increase in the expression of HOXC6 in colon cancer tissue. After inhibiting the expression of HOXC6, the expression of pro-inflammatory cytokines CXCL1, CXCL8, and IL1β increases. The expression of anti-inflammatory cytokines such as IL4 and IL10 is inhibited when inhibiting HOXC6 expression. CXCL13 belongs to the family of CXC chemokines [31]. CXCL13 is an exclusively selective ligand of CXCR5, a member of the G protein-coupled receptor (GPCR) family [32]. According to Zhenyu Zhu et al., CXCL13 can stimulate colon cancer cells to proliferate, migrate, and invade matrix gel. The CXCL13-CXCR5 axis can boost colon cancer cell proliferation, motility, and invasive potential through the PI3K/AKT pathway [33]. V-set and immunoglobulin domain containing 4 (VSIG4) is a Complement receptor of the immunoglobulin superfamily [34]. According to earlier research, VSIG4 binding to the macrophages' immunosuppressive function in the microenvironment can accelerate cancer progression [35], [36], [37]. CXCL9 is also known as γ Interferon-induced single factor and primarily mediates lymphocyte infiltration into lesions and inhibits tumor growth [38,39]. However, there is still limited research on the mechanisms of action of C11orf96, VSIG4, and CXCL9 in COAD. Therefore, our future research needs to additionally investigate the exact action mechanisms of C11orf96, VSIG4, and CXCL9 in COAD.
Subsequently, using risk scores, we classified COAD patients into high- and low-risk categories. In the training and validation cohorts, patients' OS in the low-risk subgroup was considerably higher than in the high-risk category, demonstrating that risk scores can be employed as a distinguishing factor in the COAD survival rate. Risk score could be applied independently as a predictor of COAD prognosis, according to multivariate analysis. The 1-, 3-, and 5-year OS of CRC patients were better quantified using a column chart that included these independent prognostic variables. The nomogram's prognostic prediction ability was significantly good, as evidenced by the calibration curve and cumulative incidence curve results. This study concludes that this quantitative characteristic can act as a supplemental tool to facilitate prognosis assessment and individualized COAD treatment.
We also found a strong link between the gene signature of COAD patients and tumor-infiltrating immune cells. Neutrophils, Mast cells activated, Macrophages M2, and Macrophages M0 were significantly positively linked to risk scores. On the other hand, the association of risk score with T cells follicular helper, CD8 T cells, memory-activated CD4 T cells, and Macrophages M1 was substantially negative. Among the inflammatory cells in the TME, mast cell and their mediators not only participate in tumor progression but also play an anti-tumor role. [40]. By secreting growth factors and cytokines, mast cells can accelerate tumor development. More importantly, Mast cells could also suppress tumor development by secreting IL-1, IL-6, Monocyte chemoattractant protein-3 and −4, etc [41]. At present, more and more studies have proved that Mast cells perform an integral function in CRC [42], [43], [44]. Jochen Wedemeyer et al. illustrate that B6Kit (W)/Kit (W-v) mice lacking Mast cells were less susceptible to chemically induced intestinal tumors [45]. Ji Hyun Lee et al. found that the lack of Mast cells can inhibit the progress of colitis and colon cancer by minimizing inflammation and regulating different inflammatory markers [46]. However, there is also some evidence that Mast cells seem to have a protective function in the development of CRC. Feifei Song et al. found that Mast cells are capable of selectively inducing Endoplasmic Reticulum Stress and triggering the Unfolded Protein Response in CRC cells, thereby inhibiting the development of CRC [47]. Consequently, more investigations are required to validate the apparent Mast cell mechanism in COAD.
Currently, chemotherapy remains an important strategy for treating COAD patients [48]. Therefore, We evaluated how two risk categories of patients responded to immunotherapy and chemotherapy. Bortezomib, Erlotinib, Gefitinib, Nilotinib, Sorafenib, and Paclitaxel exhibited higher sensitivity among low-risk patients. Furthermore, high-risk patients were more responsive to Dasatinib. This indicates that the clinical model of GSTTKs-related genes can serve as a tool for screening COAD patients suitable for chemotherapy and immunotherapy.
Several limitations remain in this study. Firstly, the sample size included in the research is limited. In order to expand the sample size, increase the statistical power and improve the reliability of the results, we combined the TCGA dataset and the GEO dataset. Although the "sva" package can eliminate the batch effect between different datasets, there may still be some risks. Therefore, the clinical model still needs further validation in multicenter trials and larger patient cohorts. Secondly, due to technical constraints, we are unable to verify the possible mechanisms of unreported genes. In the future, additional research will be required to validate the findings of our analysis.
Conclusion
In summary, this study innovatively investigated a gene signature based on genes associated with GSTTKs, offering a fresh approach to risk assessment and possible biological markers for COAD patients. This clinical model feature can help with stratified screening and individualized COAD patient treatment by assessing the prognosis, immune infiltration, and chemotherapy responsiveness among COAD patients.
CRediT authorship contribution statement
Jing Chen: Writing – original draft, Conceptualization. Zhengfang Wang: Writing – original draft, Formal analysis. Qin Zhu: Writing – original draft, Formal analysis. Shiqi Ren: Software, Methodology. Yanhua Xu: Validation, Supervision. Guangzhou Wang: Writing – review & editing, Data curation. Lin Zhou: Writing – review & editing, Writing – original draft, Data curation, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgments
Not applicable
Data Availability Statement
All data comes from TCGA database (https://portal.gdc.cancer.gov) and GEO database (https://www.ncbi.nlm.nih.gov/geo/).
Funding
This project was supported by Project Fund of Northern Jiangsu People's Hospital Affiliated to Yangzhou University (SBKY21011).
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Contributor Information
Guangzhou Wang, Email: 18051060706@yzu.edu.cn.
Lin Zhou, Email: zhou-lin2001@163.com.
References
- 1.Sung H., Ferlay J., Siegel R.L., et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.El Kinany K., Mint Sidi Deoula M., Hatime Z., et al. Consumption of modern and traditional Moroccan dairy products and colorectal cancer risk: a large case control study. Eur. J. Nutr. 2020;59(3):953–963. doi: 10.1007/s00394-019-01954-1. [DOI] [PubMed] [Google Scholar]
- 3.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2020. CA Cancer J. Clin. 2020;70(1):7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 4.Siegel R.L., Miller K.D., Goding Sauer A., et al. Colorectal cancer statistics, 2020. CA Cancer J. Clin. 2020;70(3):145–164. doi: 10.3322/caac.21601. [DOI] [PubMed] [Google Scholar]
- 5.Roslan N.H., Makpol S., Mohd Yusof Y.A. A Review on dietary intervention in obesity associated colon cancer. Asian Pacific J. Cancer Prev.: APJCP. 2019;20(5):1309–1319. doi: 10.31557/APJCP.2019.20.5.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang H., Li Q., Wu Y., et al. Long non‑coding RNA RP11‑400N13.3 promotes the progression of colorectal cancer by regulating the miR‑4722‑3p/P2RY8 axis. Oncol. Rep. 2020;44(5):2045–2055. doi: 10.3892/or.2020.7755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sandhu J., Lavingia V., Fakih M. Systemic treatment for metastatic colorectal cancer in the era of precision medicine. J. Surg. Oncol. 2019;119(5):564–582. doi: 10.1002/jso.25421. [DOI] [PubMed] [Google Scholar]
- 8.Bao X., Zhang H., Wu W., et al. Analysis of the molecular nature associated with microsatellite status in colon cancer identifies clinical implications for immunotherapy. J. ImmunOther Cancer. 2020;8(2) doi: 10.1136/jitc-2020-001437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fidelle M., Yonekura S., Picard M., et al. Resolving the paradox of colon cancer through the integration of genetics, immunology, and the microbiota. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.600886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuipers E.J., Grady W.M., Lieberman D., et al. Colorectal cancer. Nature Rev. Dis. Primers. 2015;1:15065. doi: 10.1038/nrdp.2015.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Varadé J., Magadán S., González-Fernández Á. Human immunology and immunotherapy: main achievements and challenges. Cell. Mol. Immunol. 2021;18(4):805–828. doi: 10.1038/s41423-020-00530-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Y., Liang L., Dai W., et al. Prognostic impact of programed cell death-1 (PD-1) and PD-ligand 1 (PD-L1) expression in cancer cells and tumor infiltrating lymphocytes in colorectal cancer. Mol. Cancer. 2016;15(1):55. doi: 10.1186/s12943-016-0539-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosenbaum M.W., Bledsoe J.R., Morales-Oyarvide V., Huynh T.G., Mino-Kenudson M. PD-L1 expression in colorectal cancer is associated with microsatellite instability, BRAF mutation, medullary morphology and cytotoxic tumor-infiltrating lymphocytes. Modern Pathol. 2016;29(9):1104–1112. doi: 10.1038/modpathol.2016.95. [DOI] [PubMed] [Google Scholar]
- 14.Dunne P.D., McArt D.G., O'Reilly P.G., et al. Immune-derived PD-L1 gene expression defines a subgroup of stage II/III colorectal cancer patients with favorable prognosis who may be harmed by adjuvant chemotherapy. Cancer Immunol. Res. 2016;4(7):582–591. doi: 10.1158/2326-6066.CIR-15-0302. [DOI] [PubMed] [Google Scholar]
- 15.Li Z.L., Zhang H.L., Huang Y., et al. Autophagy deficiency promotes triple-negative breast cancer resistance to T cell-mediated cytotoxicity by blocking tenascin-C degradation. Nat. Commun. 2020;11(1):3806. doi: 10.1038/s41467-020-17395-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carpenter K.J., Valfort A.C., Steinauer N., et al. LXR-inverse agonism stimulates immune-mediated tumor destruction by enhancing CD8 T-cell activity in triple negative breast cancer. Sci. Rep. 2019;9(1):19530. doi: 10.1038/s41598-019-56038-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pan D., Kobayashi A., Jiang P., et al. A major chromatin regulator determines resistance of tumor cells to T cell-mediated killing. Science (New York, NY) 2018;359(6377):770–775. doi: 10.1126/science.aao1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ru B., Wong C.N., Tong Y., et al. TISIDB: an integrated repository portal for tumor-immune system interactions. Bioinformatics. 2019;35(20):4200–4202. doi: 10.1093/bioinformatics/btz210. [DOI] [PubMed] [Google Scholar]
- 19.Wilkerson M.D., Hayes D.N. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26(12):1572–1573. doi: 10.1093/bioinformatics/btq170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hnzelmann S., Castelo R., Guinney J. GSVA: gene set variation analysis for microarray and RNA-Seq data. BMC Bioinformatics. 2013;14(1):7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Newman A.M., Liu C.L., Green M.R., et al. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods. 2015;12(5):453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun D., Wang J., Han Y., et al. TISCH: a comprehensive web resource enabling interactive single-cell transcriptome visualization of tumor microenvironment. Nucleic Acids Res. 2021;49(D1):D1420–d1430. doi: 10.1093/nar/gkaa1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dekker E., Tanis P.J., Vleugels J.L.A., Kasi P.M., Wallace M.B. Colorectal cancer. Lancet. 2019;394(10207):1467–1480. doi: 10.1016/S0140-6736(19)32319-0. [DOI] [PubMed] [Google Scholar]
- 24.Biller L.H., Schrag D. Diagnosis and treatment of metastatic colorectal cancer: a review. JAMa. 2021;325(7):669–685. doi: 10.1001/jama.2021.0106. [DOI] [PubMed] [Google Scholar]
- 25.Castronovo V., Kusaka M., Chariot A., Gielen J., Sobel M. Homeobox genes: potential candidates for the transcriptional control of the transformed and invasive phenotype. Biochem. Pharmacol. 1994;47(1):137–143. doi: 10.1016/0006-2952(94)90447-2. [DOI] [PubMed] [Google Scholar]
- 26.Ansari K.I., Hussain I., Shrestha B., Kasiri S., Mandal S.S. HOXC6 Is transcriptionally regulated via coordination of MLL histone methylase and estrogen receptor in an estrogen environment. J. Mol. Biol. 2011;411(2):334–349. doi: 10.1016/j.jmb.2011.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bodey B., Bodey B., Gröger A.M., Siegel S.E., Kaiser H.E. Immunocytochemical detection of homeobox B3, B4, and C6 gene product expression in lung carcinomas. Anticancer Res. 2000;20(4):2711–2716. Jr. [PubMed] [Google Scholar]
- 28.Hamid A.R., Hoogland A.M., Smit F., et al. The role of HOXC6 in prostate cancer development. Prostate. 2015;75(16):1868–1876. doi: 10.1002/pros.23065. [DOI] [PubMed] [Google Scholar]
- 29.Bijl J., van Oostveen J.W., Kreike M., et al. Expression of HOXC4, HOXC5, and HOXC6 in human lymphoid cell lines, leukemias, and benign and malignant lymphoid tissue. Blood. 1996;87(5):1737–1745. [PubMed] [Google Scholar]
- 30.Qi L., Chen J., Zhou B., et al. HomeoboxC6 promotes metastasis by orchestrating the DKK1/Wnt/β-catenin axis in right-sided colon cancer. Cell Death. Dis. 2021;12(4):337. doi: 10.1038/s41419-021-03630-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gunn M.D., Ngo V.N., Ansel K.M., Ekland E.H., Cyster J.G., Williams L.T. A B-cell-homing chemokine made in lymphoid follicles activates Burkitt's lymphoma receptor-1. Nature. 1998;391(6669):799–803. doi: 10.1038/35876. [DOI] [PubMed] [Google Scholar]
- 32.Pevzner V., Wolf I., Burgstahler R., Förster R., Lipp M. Regulation of expression of chemokine receptor BLR1/CXCR5 during B cell maturation. Curr. Top. Microbiol. Immunol. 1999;246:79–84. doi: 10.1007/978-3-642-60162-0_10. discussion 85. [DOI] [PubMed] [Google Scholar]
- 33.Zhu Z., Zhang X., Guo H., Fu L., Pan G., Sun Y. CXCL13-CXCR5 axis promotes the growth and invasion of colon cancer cells via PI3K/AKT pathway. Mol. Cell. Biochem. 2015;400(1–2):287–295. doi: 10.1007/s11010-014-2285-y. [DOI] [PubMed] [Google Scholar]
- 34.Helmy K.Y., Katschke K.J., Gorgani N.N., et al. CRIg: a macrophage complement receptor required for phagocytosis of circulating pathogens. Cell. 2006;124(5):915–927. doi: 10.1016/j.cell.2005.12.039. Jr. [DOI] [PubMed] [Google Scholar]
- 35.Small A.G., Al-Baghdadi M., Quach A., Hii C., Ferrante A. Complement receptor immunoglobulin: a control point in infection and immunity, inflammation and cancer. Swiss. Med. Wkly. 2016;146:w14301. doi: 10.4414/smw.2016.14301. [DOI] [PubMed] [Google Scholar]
- 36.Chow A., Schad S., Green M.D., et al. Tim-4(+) cavity-resident macrophages impair anti-tumor CD8(+) T cell immunity. Cancer Cell. 2021;39(7):973–988. doi: 10.1016/j.ccell.2021.05.006. e979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xia H., Li S., Li X., et al. Autophagic adaptation to oxidative stress alters peritoneal residential macrophage survival and ovarian cancer metastasis. JCI Insight. 2020;5(18) doi: 10.1172/jci.insight.141115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Farber J.M. Mig and IP-10: CXC chemokines that target lymphocytes. J. Leukoc. Biol. 1997;61(3):246–257. [PubMed] [Google Scholar]
- 39.Gorbachev A.V., Kobayashi H., Kudo D., et al. CXC chemokine ligand 9/monokine induced by IFN-gamma production by tumor cells is critical for T cell-mediated suppression of cutaneous tumors. J. Immunol. (Baltimore, Md: 1950) 2007;178(4):2278–2286. doi: 10.4049/jimmunol.178.4.2278. [DOI] [PubMed] [Google Scholar]
- 40.Ribatti D. A double-edged sword in tumor angiogenesis and progression. Dual roles of mast cells, macrophages, and neutrophils. Pathol. Res. Pract. 2022;240 doi: 10.1016/j.prp.2022.154167. [DOI] [PubMed] [Google Scholar]
- 41.Ribatti D., Crivellato E. The controversial role of mast cells in tumor growth. Int. Rev. Cell Mol. Biol. 2009;275:89–131. doi: 10.1016/S1937-6448(09)75004-X. [DOI] [PubMed] [Google Scholar]
- 42.Heijmans J., Büller N.V., Muncan V., van den Brink G.R. Role of mast cells in colorectal cancer development, the jury is still out. Biochim. Biophys. Acta. 2012;1822(1):9–13. doi: 10.1016/j.bbadis.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 43.Varricchi G., Galdiero M.R., Loffredo S., et al. Are mast cells MASTers in cancer? Front. Immunol. 2017;8:424. doi: 10.3389/fimmu.2017.00424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rigoni A., Colombo M.P., Pucillo C. Mast cells, basophils and eosinophils: from allergy to cancer. Semin. Immunol. 2018;35:29–34. doi: 10.1016/j.smim.2018.02.001. [DOI] [PubMed] [Google Scholar]
- 45.Wedemeyer J., Galli S.J. Decreased susceptibility of mast cell-deficient Kit(W)/Kit(W-v) mice to the development of 1, 2-dimethylhydrazine-induced intestinal tumors. Lab. Invest. 2005;85(3):388–396. doi: 10.1038/labinvest.3700232. [DOI] [PubMed] [Google Scholar]
- 46.Lee J.H., Jeon Y.D., Xin M., Lim J.Y., Lee Y.M., Kim D.K. Mast cell modulates tumorigenesis caused by repeated bowel inflammation condition in azoxymethane/dextran sodium sulfate-induced colon cancer mouse model. Biochem. Biophys. Rep. 2022;30 doi: 10.1016/j.bbrep.2022.101253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Song F., Zhang Y., Chen Q., et al. Mast cells inhibit colorectal cancer development by inducing ER stress through secreting Cystatin C. Oncogene. 2023;42(3):209–223. doi: 10.1038/s41388-022-02543-z. [DOI] [PubMed] [Google Scholar]
- 48.Johdi N.A., Sukor N.F. Colorectal Cancer Immunotherapy: options and Strategies. Front. Immunol. 2020;11:1624. doi: 10.3389/fimmu.2020.01624. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data comes from TCGA database (https://portal.gdc.cancer.gov) and GEO database (https://www.ncbi.nlm.nih.gov/geo/).