Cystic fibrosis (CF) is the most common genetic life-threatening disease. It is defined as a mutation in the gene coding for the cystic fibrosis transmembrane conductance regulator (CFTR) membrane channel (1). Despite the monogenic cause of CF, there is a strikingly poor correlation between the CFTR genotype and clinical phenotype. Our incomplete understanding of the disease is linked to the very complex biology that underpins airway epithelial tissues, which are the main location of morbidity and mortality in the disease. CF is at heart a genetic disorder, but this is already not trivially a single gene problem: Previous studies have demonstrated major contributions to clinical outcomes of other genes although these remain poorly understood. As a consequence, therefore, we cannot currently predict from the genetics at an individual patient level any of i) the rate of decline of lung function, ii) response to a specific treatment, or iii) the risk of infection with particular pathogens. Of the nearly 2,000 CFTR mutations that have been identified so far (http://www.genet.sickkids.on.ca) (2), only the most common mutations expressed by large groups of subjects have been targeted for drug screening due to the high cost and time-consuming nature of clinical trials.
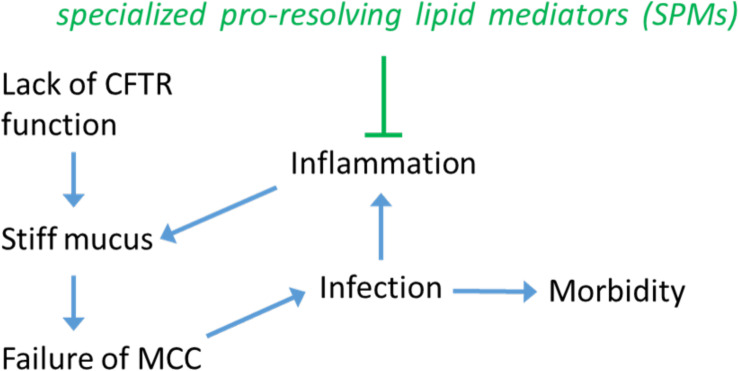
Lack of CFTR function generally leads to a cascade of other effects that mutually reinforce each other; see Fig. 1: a reduction of airway surface liquid and incompletely hydrated mucin layers, resulting in a thick layer of mucus that obstructs the airways and promotes chronic bacterial infections and inflammatory lung damage. Inflammation is of course a complex biochemical network of processes and is the aspect studied in a new Proceedings of National Academic Sciences paper (3). These physiological and biological elements are also in cross-talk with aspects that require an understanding of fluid mechanics and physics. This is because the overarching emerging problem in CF, as a consequence of the altered mucus, is a failure to sustain healthy transport of mucus out of the airways, known as mucociliary clearance (MCC). MCC is itself a multi-scale phenomenon, dependent on quite specific properties of the activity of motile cilia at the level of individual filaments and cells, but also on coordination of ciliary beating across large distances (many cell diameters) which is mediated by the physical parameters of both the liquid surrounding the cilia (the periciliary liquid, PCL) and of the more elastic mucus layer above the cilia (4). In individuals with CF, cilia beating is greatly compromised to the extent that the mucus cannot be properly cleared, and the range of cilia movement is severely restricted (5).
Fig. 1.
Cystic fibrosis is a complex disease. From a genetic cause affecting the CFTR ion transport channel, many other biological and physical factors contribute to the disease outcome. Failure of the MCC is a recognized key node in this network of factors affecting each other. Arrows indicate what we think are the main causal effects. The paper by Briottet et al. (3) studies the effect of SPM factors RvE1 and LXB4 not just as anti-inflammatory agents, but also all the way to affecting the physical properties of the mucus, and restoring a physiological MCC.
A good understanding of how a functioning clearance comes about is required to rationally intervene on the flow properties of the CF mucus. Unfortunately despite its importance, our understanding of cilia beating coordination in the context of human physiology and respiratory disease is incomplete. We think that the key elements are the cilia density, arrangement, orientation, together with their individual beating properties, and the rheological properties of the fluids (4), but we do not have a model that incorporates them all in a predictive way, let alone how they inter-relate to the biochemical level outlined earlier.
Phenotyping ciliary activity has itself been a long-standing challenge. Traditionally, one of three experiments/observations have been attempted (6): A) imaging of cilia beats in “profile”; B) quantification of cilia beat frequency (CBF) in air liquid interface (ALI) cultures; C) assays of clearance. In brief, these are of limited value in understanding CF: A) is powerful for ciliopathies, but cilia beat itself is functional in CF, and not much is learned; B) and C) are relevant in CF, and indeed mucus rheology affects both the CBF and the overall clearance, but these parameters by themselves are not informative of what are the mucus properties nor of what is compromising the clearance, e.g., are the cilia not beating in a stiff environment, or are they beating but failing to coordinate with neighbors? In our recent work on beating cilia in CF conditions, we introduced a novel approach we termed “multiscale differential dynamic microscopy” (multi-DDM) to quantitatively assess collective cilia beating in a nonbiased, automated manner over an entire population of human airway epithelial cells (7, 8). We proposed that measuring the spatial scale of coherent dynamics of cilia would be an informative parameter for assessing physiological cilia beating (8).
This recent work studies for the first time the SPM factors RvE1 (Resolvin E1) and LXB4 (lipoxin B4) in the context of exploring their effects on the PCL and mucus layers.
In this context, the recent work by Briottet et al. (3) is a step precisely in the right direction. One of the problems to fix in the CF disease, and opportunities for intervention, is the misregulation of inflammation. Previous work by that group and others had already shown that in CF tissues that there was a reduced expression of anti-inflammatory factors. Inflammation is normally followed by an active resolution phase requiring specialized pro-resolving lipid mediators (SPMs) to return to homeostasis. This recent work studies for the first time the SPM factors RvE1 (Resolvin E1) and LXB4 (lipoxin B4) in the context of exploring their effects on the PCL and mucus layers. Working on cell culture models, their results show that the liquid layer returns to a more physiological state, and that the MCC is restored. This work builds on the previous video analysis tools (7, 8) to add a new metric of the degree of alignment in the flow. This gives a multi-parametric video analysis technique that seems very effective in characterizing the cilia beating in CF nasal epithelial cells. This novel use of video microscopy, including the new way to quantify alignment in collective cilia beating, allows the authors to distinguish CF from non-CF by PCA analysis, and to quantify the importance of RvE1 in restoring a non-CF ciliary dynamics phenotype. In the cell models, specialized pro-resolving mediator ResolvinThis recent work studies for the first time the SPM factors RvE1 (Resolvin E1) and LXB4 (lipoxin B4) in the context of exploring their effects on the PCL and mucus layers. E1 corrects the altered cystic fibrosis nasal epithelium cilia beating dynamics.
Future work in CF will need to be bold and ambitious, aiming to define the genetic determinants for different clinical trajectories, to reveal underlying biology and new potential therapeutic targets and how the physical aspects regulate and affect all this. For example, a recent study showed that select CFTR-modulating drugs used to treat patients with the common F508del-CFTR mutation can in some cases be effective in treating patients with rare, uncharacterized CFTR mutations that are currently not registered for treatment, if only cost-effective assays could be used to screen such samples (9). Given the number of patient specific factors in CF disease, it is likely in our opinion that future advances in CF treatment will require the development of further such personalized medicine approaches, where robust and affordable functional phenotyping is a crucial element.
Acknowledgments
P.C. thanks Erika Causa, Viridiana Carmona Sosa and Jurij Kotar for their recent research contributions on this topic and many conversations. The project was funded by the Cystic fibrosis Trust SRC 016.
Author contributions
P.C. wrote the paper.
Competing interests
The author declares no competing interest.
Footnotes
See companion article, “Specialized proresolving mediator Resolvin E1 corrects the altered cystic fibrosis nasal epithelium cilia beating dynamics,” 10.1073/pnas.2313089121.
References
- 1.Ratjen F., et al. , Cystic fibrosis. Nat. Rev. Dis. Primers. 14, 15010 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lukacs G., Verkman A., CFTR: Folding, misfolding and correcting the F508 conformational defect. Trends. Mol. Med. 18, 81–91 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Briottet M., et al., Specialized proresolving mediator resolvin E1 corrects the altered cystic fibrosis nasal epithelium cilia beating dynamics. Proc. Natl. Acad. Sci. U.S.A. 121, e2313089121 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cicuta P., The use of biophysical approaches to understand ciliary beating. Biochem. Soc. Trans. 48, 221–229 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cutting G., Cystic fibrosis genetics: From molecular understanding to clinical application. Nat. Rev. Genet. 16, 45–56 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.E. Causa, R. Fradique, P. Cicuta, “Measuring biophysical properties of cilia motility from mammalian tissues via quantitative video analysis methods” in Cilia: Methods and Protocols - Methods in Molecular Biology, V. Mennella, Ed. (Springer, NL, 2023), pp. 251–262. [DOI] [PubMed]
- 7.Feriani L., et al. , Assessing the collective dynamics of motile cilia in cultures of human airway cells by multiscale DDM. Biophys. J. 113, 109–119 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chioccioli M., Feriani L., Kotar J., Bratcher P., Cicuta P., Phenotyping ciliary dynamics and coordination in response to CFTR-modulators and thymosin-alpha1 in cystic fibrosis respiratory epithelial cells. Nat. Commun. 10, 1763 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dekkers J., et al. , Characterizing responses to CFTR-modulating drugs using rectal organoids derived from subjects with cystic fibrosis. Sci. Transl. Med. 8, 344ra84 (2016). [DOI] [PubMed] [Google Scholar]