Significance
Cytomegaloviruses (CMVs) have coevolved with their mammalian hosts throughout mammalian history. Persisting lifelong, they have to counteract host antiviral mechanisms, e.g., presentation of viral antigens by highly polymorphic MHC-I (major histocompatibility class I) molecules to cytotoxic T cells. We analyzed the adaptation of the CMV-encoded MHC-I inhibitor US11 to the MHC-I molecule MHC-A and found that MHC-A and US11 have co-evolved in great apes. One MHC-A lineage, A2, acquired a point mutation to resist an ancestral US11. In response, US11 gained a new independent function, which targets the antigen-binding domain of A2. Next, the human CMV US11 perfected this strategy to block the most prevalent human MHC-A, HLA-A*02. This indicates that a persisting herpesvirus can promote MHC-I diversification.
Keywords: cytomegalovirus, HLA-A, MHC class I, coevolution, tapasin
Abstract
Recurrent, ancient arms races between viruses and hosts have shaped both host immunological defense strategies as well as viral countermeasures. One such battle is waged by the glycoprotein US11 encoded by the persisting human cytomegalovirus. US11 mediates degradation of major histocompatibility class I (MHC-I) molecules to prevent CD8+ T-cell activation. Here, we studied the consequences of the arms race between US11 and primate MHC-A proteins, leading us to uncover a tit-for-tat coevolution and its impact on MHC-A diversification. We found that US11 spurred MHC-A adaptation to evade viral antagonism: In an ancestor of great apes, the MHC-A A2 lineage acquired a Pro184Ala mutation, which confers resistance against the ancestral US11 targeting strategy. In response, US11 deployed a unique low-complexity region (LCR), which exploits the MHC-I peptide loading complex to target the MHC-A2 peptide-binding groove. In addition, the global spread of the human HLA-A*02 allelic family prompted US11 to employ a superior LCR strategy with an optimally fitting peptide mimetic that specifically antagonizes HLA-A*02. Thus, despite cytomegaloviruses low pathogenic potential, the increasing commitment of US11 to MHC-A has significantly promoted diversification of MHC-A in hominids.
CD8+ T cells detect peptide antigens presented by major histocompatibility class I (MHC-I) molecules, leading to the elimination of cancerous and virally infected cells. In the human genome, classical MHC-I is encoded by the most polymorphic human genes, the human leukocyte antigen class I (HLA-I) genes HLA-A, -B, and -C. Among these, HLA-B displays the highest level of diversity likely due to its ability to adapt to emerging infections (1). HLA-A and HLA-C loci are less diverse, with HLA-A having coevolved with DNA viruses, such as herpesviruses (2). The evolution of HLA-A provides a unique opportunity to understand the cadence and mechanisms of immune restriction of DNA viruses, and viral evolution to counteract this restriction.
Cytomegaloviruses (CMVs) belong to the family of Betaherpesviridae. Despite vigorous immune responses, CMV infection is lifelong and characterized by periodic phases of latency, reactivation, and viral dissemination. Both with respect to its genome length and numbers of transcribed open reading frames (3), the human cytomegalovirus (HCMV) is by far the largest of all human herpesviruses. A considerable proportion of its coding capacity is dedicated to evading both innate and adaptive pathways (4). Despite a subclinical course of infection in immunocompetent hosts, HCMV infection has a permanent impact on immune cell subsets, such as CD8+ memory T cells and memory-like NK cells (5, 6). For immunocompromised individuals, HCMV-related complications are a major cause of morbidity and mortality and can result in severe and permanent sequelae to the developing fetus if maternal HCMV is acquired and congenitally transmitted during pregnancy (7).
CMVs have coevolved in a species-specific manner with mammalian hosts throughout mammalian history (8) and impacted immune evolution (9–13). Coevolution is indicated both by the species-specificity of infection and by the genetic distance between cytomegalovirus subfamily members that infect, e.g., mice, rats, and primates such as rhesus macaques and chimpanzees (14). As a consequence, immune evasion strategies by CMVs are often species-specific. One such immune evasion strategy is encoded by the genes of the US2 and US6 gene families that inhibit MHC-I antigen presentation to CD8+ T cells. These gene families are only found in primate CMVs (15–17).
Heterodimeric MHC-I molecules comprise a membrane-attached heavy chain (HC), which forms an α1/α2 peptide–binding domain above the membrane-proximal α3 domain, and the soluble non-covalently bound beta-2-microglobulin (β2m). The majority of MHC-I peptide ligands are generated by the proteasome in the cytosol and transported into the endoplasmic reticulum (ER) by the transporter associated with antigen processing (TAP). In the ER, MHC-I heterodimers are recruited to the peptide loading complex (PLC), consisting of TAP, tapasin, calreticulin, and ERp57. The PLC controls optimal peptide loading and release of MHC-I for trafficking to the cell surface. The HCMV US2 and US6 gene families encode at least five immunoevasins that manipulate MHC-I levels at the cell surface. For example, US3 retains MHC-I in the ER and blocks tapasin function (18, 19), whereas US6 inhibits TAP. (20–22). By exploiting ER-associated protein degradation (ERAD), US2 and US11 initiate MHC-I retro-translocation to the cytosol and proteasomal degradation (23, 24).
Here, we investigated the coevolution between MHC-I and US11, with an ectodomain that comprises an Ig domain and an N-terminal low-complexity region (LCR) (25). LCRs are often involved in protein–protein interactions (26), and we recently demonstrated that the US11 N-terminus is crucial for US11 interaction with β2m-assembled MHC-I and the PLC (25). Furthermore, HLA-B allotypes can escape US11-mediated degradation, whereas HLA-A allotypes are efficiently blocked (25). Hence, this prompted further examination to gain insight into the so far poorly understood specificity of cytomegaloviral MHC-I immunoevasins. By assessing the interplay between primate MHC-A lineages and US11 homologs, we identified two distinct mechanisms, LCR-dependent and -independent, of US11-mediated downregulation of MHC-A, which arose as a result of a coevolutionary arms race. We show that US11’s LCR-independent mechanism is ancestral and targets the α3 domain of MHC-A. However, in an early ancestor of great apes, a Pro184Ala escape mutation emerged with the A2 lineage in the α3 domain. In response, US11 deployed an LCR-dependent mechanism retargeting the α1/α2 antigen-binding groove. Furthermore, US11 specifically targets HLA-A*02:01 using peptide mimicry. Our data provide evidence for a tit-for-tat dynamics (27) that shaped the coevolution of herpesviral US11-like molecules and diversification of MHC-A proteins in hominids.
Results
The HCMV-Encoded Immunoevasin US11 Differentially Regulates the Surface Expression of HLA-A Allotypes.
The US11 N-terminus contains a proline-rich LCR (residues 27 to 40, pink line in SI Appendix, Fig. S1A), which is critical for US11’s conformational specificity. Previous studies have shown that the N-terminus is required for the interaction with β2m-assembled MHC-I but not with free HCs (25). Further, we observed differences in the level of surface expression between HLA-A allomorphs and hypothesized that US11 has a preference for certain HLA-A allomorphs. Therefore, we assessed the importance of the US11 N-terminus for HLA-A regulation.
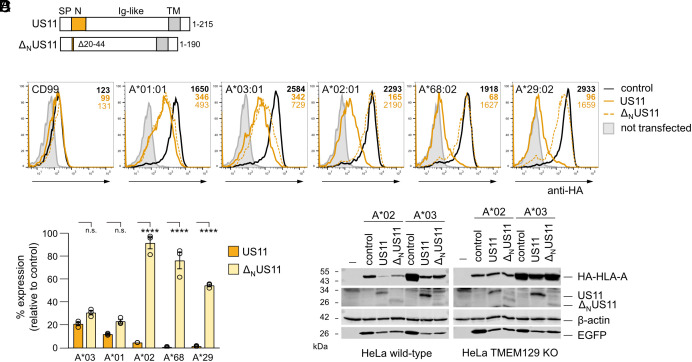
N-terminally hemagglutinin (HA) epitope-tagged HLA-A allotypes were expressed along with either full-length US11 or an N-terminal truncation mutant (ΔNUS11 with deletion of residues 20 to 44; Fig. 1A and SI Appendix, Fig. S1A), and HLA-A cell surface expression was quantified. The N-terminal truncation did not significantly affect the US11-mediated downregulation of HLA-A*03:01 and A*01:01. In contrast, the surface expression of HLA-A*02:01, A*29:02, and A*68:02 was strongly increased (more than 20-fold) in the absence of the US11 N-terminus (Fig. 1 B and C). These findings were confirmed by western blot analyses, which showed higher levels of HLA-A*02:01 when expressed with ΔNUS11 as compared to co-expression with US11 (Fig. 1 D, Left). No difference in total protein level was apparent for HLA-A*03.
Fig. 1.
The HCMV-encoded immunoevasin US11 differentially regulates the surface expression of HLA-A allotypes. (A) Schematic representation of full-length US11 and ΔNUS11. SP, signal peptide, Ig-like, immunoglobulin-like domain; TM, transmembrane domain. (B) HeLa cells were transiently co-transfected with plasmids encoding HA-tagged HLA-I and CD99 and pIRES-EGFP encoding either US11, ΔNUS11, or a control protein. At 20 h post-transfection, surface expression was determined by flow cytometry (anti-HA) on EGFP-positive cells. Representative histograms are shown. Example of gating is shown in SI Appendix, Fig. S1B. (C) The percentage of MFI (mean fluorescence intensity) as compared to control transfected cells was determined from analysis in (B). Dots represent individual values and bars mean values ± SEM from three independent experiments. Significance was analyzed by two-way ANOVA. (D) HeLa wild-type cells and TMEM129 KO cells were transfected as in (B). At 20 h post-transfection, cells were lysed and western blot analysis was performed with antibodies detecting indicated proteins (anti-HA for detection of HA-tagged HLA-A).
As US11 and ΔNUS11 were expressed at different levels, this could affect the ability to counteract HLA-A expression. Therefore, HLA-A surface expression was re-examined after transfection with a lower concentration of US11- and ΔNUS11-encoding plasmids. Still, ΔNUS11 downregulated HLA-A*03:01, whereas regulation of HLA-A*02:01 required the LCR regardless of the US11 concentration (SI Appendix, Fig. S1C), showing a qualitative difference between the full-length US11 and the truncation mutant. Previous studies demonstrated that US11 exploits the E3 ligase TMEM129 for HLA-A*02:01 degradation (28, 29). Accordingly, we observed no degradation HLA-A*02:01 and A*03:01 in TMEM129 knock-out HeLa cells (Fig. 1 D, Right). Therefore, despite apparent differences in HLA-A targeting, both US11 and ΔNUS11 utilize the same ERAD pathway to degrade HLA-A. These results suggest that US11 uses the LCR to initiate degradation of a specific subset of HLA-A allotypes, including HLA-A*02:01.
The US11 LCR Protects HCMV from HLA-A*02:01-Restricted CD8+ T Cells.
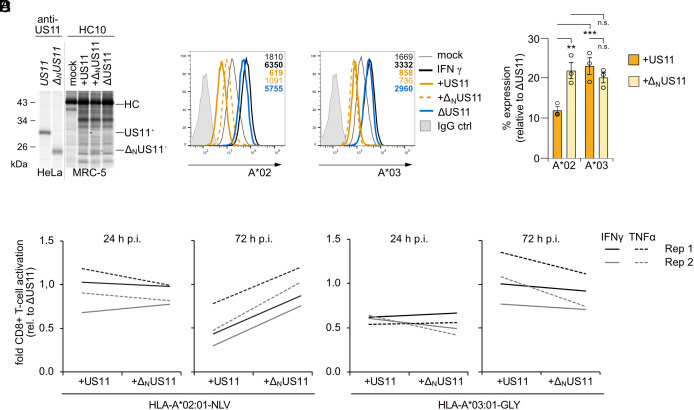
We extended our analyses of HLA-A antagonism from transient transfection of US11 to HCMV infection. On the basis of the HCMV AD169VarL strain–derived BAC virus (30) that lacks the genes US2-US6 we deleted the US11-coding sequence (ΔUS11) and replaced it with either wild-type US11 (+US11), or with ΔNUS11 (+ΔNUS11). Similar levels of expression and binding of US11 and ΔNUS11 to non-assembled HLA-I were controlled for by immunoprecipitation with the HC10 mAb (Fig. 2A). Next, human foreskin fibroblasts (HF-α) endogenously expressing both HLA-A*02:01 and A*03:01 were infected with the HCMV mutants. Previous analyses showed that US11-mediated regulation of HLA-A reaches a maximum at 48 to 72 h post-infection (p.i.) (31). Therefore, HLA-A surface expression was monitored at 48 h p.i. by flow cytometry. To estimate the maximum of HLA-A expression, control cells were treated with saturating concentration of IFNγ. Both HLA-A*02:01 and A*03:01 were markedly reduced on +US11-infected cells as compared to ΔUS11-infected cells. Similar to our results in Fig. 1B, HLA-A*02:01 was more efficiently downregulated by US11 than HLA-A*03:01. Cells infected with +US11 and +ΔNUS11 HCMV mutants showed a similar level of HLA-A*03:01 expression, whereas the HLA-A*02:01 level was twofold higher on +ΔNUS11-infected cells than on +US11-infected cells (Fig. 2 B and C). Hence, this verified that also in HCMV-infected cells the US11 LCR is important to control HLA-A*02:01, but not HLA-A*03:01.
Fig. 2.
The US11 LCR protects HCMV from HLA-A*02:01-restricted CD8+ T-cells. (A) US11 or ΔNUS11 in pIRES-EGFP were transiently transfected into HeLa cells. MRC-5 fibroblast were mock treated or infected with an MOI of 7 with indicated HCMV mutants. At 48 h p.i., cells were metabolically labeled for 2 h. The anti-US11 antiserum or the mAb HC10 were applied for immunoprecipitation. Retrieved proteins were separated on sodium dodecyl sulfate – polyacrylamid gel electrophoresis (SDS-PAGE) and detected by autoradiography. (B) HF-α fibroblasts were treated as indicated and infected with an MOI of 8. At 48 h p.i., HLA-A*02:01 and A*03:01 cell surface expression was determined by flow cytometry. (C) The percentage of MFI as compared to ΔUS11-infected cells was determined from the analysis in (A). Dots represent individual values and bars mean values ± SEM from three independent experiments. Significance was determined using one-way ANOVA. (D) The fold change in the percentage of activated CD8+ T cells as compared to CD8+ T cells co-cultured with ΔUS11-infected cells is depicted. HF-α fibroblasts were mock treated or infected as indicated with an MOI of 8. At 24 and 72 h p.i., the cells were co-cultured for 5 h with HLA-A*02:01NLV or HLA-A*03:01GLY specific polyclonal ex vivo expanded CD8+ T cells. Activation of CD8 and tetramer-positive T cells was determined by staining of intracellular IFNγ and TNFα.
To determine the impact of the US11 N-terminus for antigen presentation, we used ex vivo expanded polyclonal CD8+ T cells, which had been cultured for 14 d with either an HLA-A*02:01-restricted, pp65-derived peptide (NLVPMVATV, NLV) or with an HLA-A*03:01-restricted, UL77-derived peptide (GLYTQPRWK, GLY) (32). The CD8+ T cells were co-cultured for 5 h with HCMV-infected HF-α fibroblasts starting at 24 or 72 h p.i., representing early and late time-points of the protracted HCMV replication cycle. Epitope-specific activation of CD8+ T cells was determined by intracellular staining of IFNγ and TNFα (SI Appendix, Fig. S2). At 24 h p.i., no differences in activation were observed between HLA-A*02:01NLV-specific CD8+ T cells that had been co-cultured with +US11, +ΔNUS11, or ΔUS11-infected cells (Fig. 2D). This was not unexpected, since the highly abundant tegument protein pp65 (33) has previously been shown to efficiently activate CD8+ T cells at early time points of infection despite the presence of immunoevasins (34, 35). At 72 h p.i., activation of HLA-A*02:01NLV-specific CD8+ T cells was strongly blocked by +US11-infected cells (twofold decreased IFNγ expression), and to a lesser extent by +ΔNUS11, in comparison to ΔUS11-infected cells. This pattern clearly contrasted with the US11-mediated block of activation of HLA-A*03:01GLY-specific CD8+ T cells, which remained largely unaffected by the deletion of the US11 N-terminus. This result further supports that US11-mediated inhibition of HLA-A*02:01 antigen presentation is dependent on the LCR.
Peptide-Loaded HLA-A*02:01 Confers Resistance to US11.
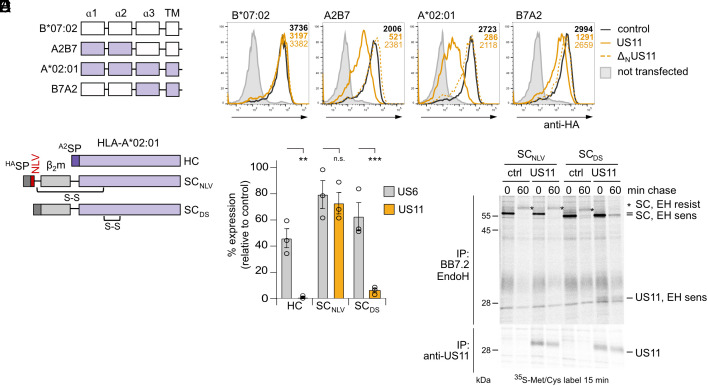
We next investigated which domain of HLA-A*02:01 is targeted by the LCR. For this purpose, HA-tagged HLA-I chimeras with segments from HLA-A*02:01 swapped into the US11-resistant HLA-B*07:02 were generated (Fig. 3A). In accordance with previous studies (25), HLA-B*07:02 cell surface levels were minimally affected by US11 and ΔNUS11. However, when the α1-2 domains of HLA-A*02:01 were transferred into HLA-B*07:02 (A2B7), US11 reduced the expression to ca 20% as compared to control cells, while ΔNUS11 did not affect expression (Fig. 3B and SI Appendix, Fig. S3A). Hence, the α1-2 domains of HLA-A*02:01 are sufficient for LCR-dependent targeting. Expression of the reciprocal B7A2 chimera, containing the α1-2 domains of HLA-B*07:02 in the context of HLA-A*02:01, was reduced by US11 to ca 40%, yet moderately by ΔNUS11, suggesting that the HLA-B*07:02 α1-2 domains are not resistant to US11.
Fig. 3.
Peptide-loaded HLA-A*02:01 confers resistance to US11. (A) Schematic representation of HA-HLA-A*02:01 and B*07:02 chimeras. (B) HA-HLA-I surface expression was determined as in Fig. 1B. Representative histograms are shown. Statistical analysis is depicted in SI Appendix, Fig. S3A. (C) Schematic representation of HLA-A*02:01 SC constructs. HASP, HA SP; A2SP, HLA-A*02 SP; S-S, disulfide stabilization. (D) HLA-A*02:01 HC or SC constructs encoded in pcDNA3.1 were transiently co-transfected with pIRES-EGFP encoding a control protein, US6 or US11 into HeLa cells. At 20 h post-transfection HLA-A*02:01 cell surface expression was analyzed by flow cytometry using the mAb BB7.2. The percentage of MFI as compared to control transfected cells was determined. Dots represent individual values and bars mean values ± SEM from three independent experiments. Significance was analyzed by two-way ANOVA. (E) Cells were transfected as in (D) with indicated constructs. At 20 h post-transfection, cells were metabolically labeled for 15 min and a pulse-chase experiment was performed. The mAb BB7.2 and anti-US11 were applied for immunoprecipitation. Retrieved proteins were EndoH (EH) digested as indicated prior to separation on SDS-PAGE and detection by autoradiography.
We next sought to delineate the mechanism by which the US11 N-terminus targeted the HLA-A*02:01 α1-2 domains. In particular, we aimed to determine whether the US11 N-terminus bound the peptide-bound or peptide-free HLA-A*02:01. For this, we generated two single-chain (SC) HLA-A*02:01 constructs. First, the HC with mutation Tyr84Cys, β2m, and the disulfide-trapped NLV peptide were covalently connected by flexible linker sequences (SCNLV), thereby enforcing stable peptide loading (36). In a second SC construct without a ligand, a disulfide bridge was introduced (SCDS; Tyr84Cys/Ala139Cys), stabilizing the peptide-binding domain in the absence of peptides (37, 38) (Fig. 3C). We expressed the SC constructs and the wild-type HLA-A*02:01 HC in HeLa cells in the presence of US11, or the TAP inhibitor US6. Consistent with previous studies (39, 40), we found that TAP inhibition reduced HLA-A*02:01 surface expression by about 50% on HLA-A*02:01 HC transfected cells, but this effect was less pronounced for both SC constructs (Fig. 3D), demonstrating less TAP dependency for these constructs. Intact US11 almost completely abolished surface expression of HLA-A*02:01 HC transfected cells and significantly reduced surface expression of SCDS to ca. 10% as compared to the control. However, US11 only modestly affected surface expression of SCNLV (ca. 70% remaining). A metabolic pulse-chase co-immunoprecipitation experiment confirmed that SCNLV was able to gain Endoglycosidase H (EndoH)–resistant glycans in the presence of US11, indicating its escape from US11 regulation and transit through the Golgi complex. This was not the case for SCDS, which remained EndoH sensitive in the presence of US11 (Fig. 3E). As a final confirmation, we found that US11 robustly co-immunoprecipitated with SCDS, but not with SCNLV. To analyze whether SCNLV escaped US11 regulation due to enhanced ER–Golgi trafficking, we trapped proteins in the ER with brefeldin A (BFA). Still, no interaction between SCNLV and US11 was observed, implying that SCNLV escaped US11 due to lack of recognition rather than through enhanced ER–Golgi trafficking (SI Appendix, Fig. S3B). Altogether, our results indicate that the N-terminus of US11 targets peptide-free HLA-A*02:01 and that occupation of the HLA-A*02:01 peptide-binding groove protects against US11.
The US11 LCR Mimics Peptide Binding to HLA-A*02:01.
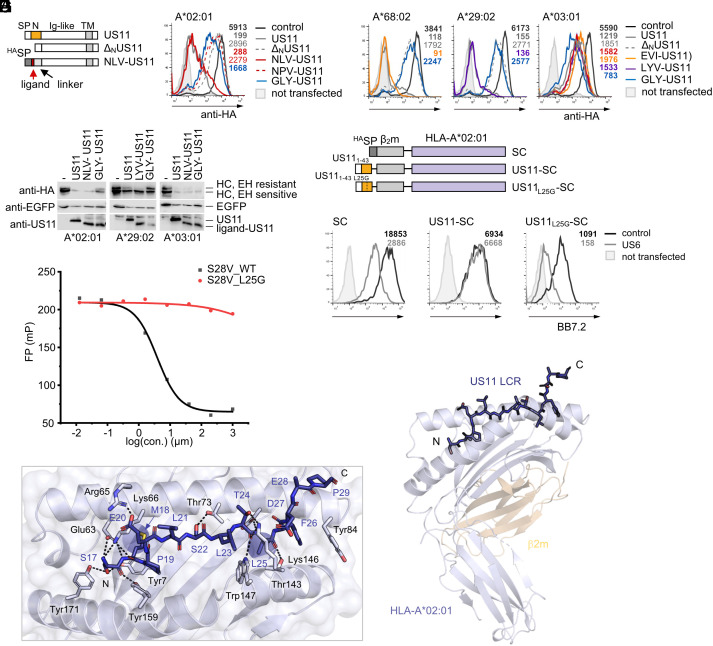
Following from our finding that peptide-bound HLA-A*02:01 escapes US11 antagonism, we investigated whether US11 targets the HLA-A peptide-binding groove by mimicking a peptide ligand. To test this, we replaced the US11 N-terminus with either the high-affinity HLA-A*02:01 ligand NLV (NLV-US11) or the mutated NPV (NPV-US11; Fig. 4A), predicted to ablate binding to HLA-A*02:01 (NetMHC4.1 (41) was applied for peptide-binding predictions). Indeed, NLV-US11 robustly downregulated HLA-A*02:01, similarly to full-length US11, while the mutated NPV-US11 did not (Fig. 4B). Prompted by these findings, we next tested ligand sequences of HLA-A*68:02, A*29:02, and A*03:01 (SI Appendix, Fig. S4A) as replacements of the US11 N-terminus. Both HLA-A*68:02 and A*29:02 were strongly downregulated by their ligand-US11 counterparts, but not by the unmatched HLA-A*03:01 ligand (GLY-US11; Fig. 4C). Unmatched ligand-chimeras maintained the level of regulation of HLA-A*03:01 relative to ΔNUS11 (Fig. 4C). Although fusion with the GLY peptide also endowed ΔNUS11 with the ability to downregulate HLA-A*03:01, its efficacy was less powerful compared to other matching ligand-US11 HLA-A combinations. In addition, western blot analysis demonstrated that the ligand-US11 constructs facilitated degradation of HLA-A target molecules (Fig. 4D).
Fig. 4.
The US11 LCR mimics peptide binding to HLA-A*02:01. (A) Schematic representation of chimeric HLA-A ligand-US11 constructs. (B and C) Panels show flow cytometry histograms of HA-tagged HLA-A expressed by transient co-transfection with pIRES-EGFP encoding ligand-US11 variants or a control. At 20 h post-transfection, surface expression was determined by flow cytometry (anti-HA) on EGFP-positive cells. Representative histograms are shown. (D) Cells were transfected as in (B and C). At 20 h post-transfection, cells were lysed and lysates were treated with EndoH. Proteins were detected by western blot analysis with indicated antibodies. (E) Schematic representation of HLA-A*02:01 SC constructs. (F) HLA-A*02:01 SC constructs encoded in pcDNA3.1 were transiently co-transfected with pIRES-EGFP encoding a control protein or US6 into HeLa cells. At 20 h post-transfection, SC cell surface expression was analyzed by flow cytometry using the mAb BB7.2. Representative histograms are shown. Statistical analysis is depicted in SI Appendix, Fig. S4B. (G) Effect of the US11 peptide S28V_WT (amino acids 17 to 44) and the mutated peptide S28V_L25G on binding of the fluorescein isothiocyanate (FITC)-labeled GV9 peptide to HLA-A*02:01 in an MHC-I fluorescence polarization assay. A representative experiment out of three independent experiments to determine the IC50 value is shown. Primary data are shown in SI Appendix, Fig. S4C. (H) X-ray crystal structure of a US1117-43-HLA-A*02:01 SC. Cartoon representation of HLA-A*02:01 and β2m, colored in lavender and wheat, respectively. The US11 LCR (blue) is shown in stick format with the N and C termini marked (PDB ID: 8FRT). (I) Close-up of the peptide-binding groove of H with US11 LCR residues indicated in single-letter code, and key HLA-A*02:01 residues in stick format and in three-letter code. Hydrogen bonds are displayed as dashed lines.
The ability of the HLA-A ligand sequences to compensate for the N-terminus suggested that this part of US11 may itself act as a peptide ligand mimic. To further explore this, we replaced the signal peptide and the NLV peptide sequence of the SC construct SCNLV with the US11 residues 1 to 43 (US11-SC; Fig. 4E). In addition, we constructed a control molecule with a signal peptide placed directly in front of β2m without stabilizing the peptide-binding groove (SC). As expected, the TAP inhibitor US6 significantly reduced surface expression of HLA-A*02:01 SC (Fig. 4F and SI Appendix, Fig. S4B) but not of US11-SC, demonstrating a stabilizing effect of the US11 LCR in a peptide-deprived situation.
Two potential HLA-A*02:01 high-affinity ligand sequences were predicted in the US11 N-terminus: SMPELSLTL (SMP) (17 to 25, 22 nM) and TLFDEPPPL (TLF) (24 to 32, 5 nM). In both sequences, Leu25 occupies an anchor residue position (underlined letter). Hence, accordingly, a leucine to glycine mutation at position 25 should disrupt binding to HLA-A*02:01. As predicted, the mutated US11L25G-SC was expressed at a lower level compared to US11-SC and was significantly blocked by US6 (Fig. 4F and SI Appendix, Fig. S4B).
Finally, using a fluorescence polarization-based binding competition assay, we found that the US11 LCR can compete with ligand (GILGKFITCVFTV, GV9) binding to HLA-A*02:01. The wild-type US11 peptide S28V_WT (residues 17 to 44) clearly reduced GV9 loading of HLA-A*02:01 with an IC50 of 3.83 μM, while the mutated S28V_L25G peptide did not show any effect on GV9 binding (Fig. 4G and SI Appendix, Fig. S4C).
To understand the molecular basis for the US11 peptide mimicry, we determined the X-ray crystal structure of US11-SC at 1.8 Å resolution (Fig. 4 H and I and SI Appendix, Table S1). The US11-SC fusion did not significantly affect the HLA-A*02:01 fold, with a root mean squared deviation of 0.5 Å between the HLA-A*02:01 of US11-SC and a previously determined binary X-ray crystal structure (42). Clear unambiguous electron density was observed for 13 residues of US11 (S17MPELSLTLFDEP29) with the N-terminus captured within the peptide-binding pocket. The structure revealed that US11 adopted a binding mode characteristic of HLA-A*02:01 peptide docking (43) with Ser17 stabilized by a network of hydrogen bonds with Tyr7, Glu63, Lys66, Tyr159, and Tyr171 of HLA-A*02:01 (Fig. 4I). Met18 penetrated deeply within the B pocket and the US11 Glu20 carboxyl formed salt bridge interactions with Arg65 on the α-chain of HLA-A*02:01. Centrally, Leu21-Thr24 adopted a linear epitope and via this linearity, Leu25 buried into the F pocket acting as the PΩ anchor. To accommodate the C-terminal extension and US11 Phe26, HLA-A*02:01 Tyr84 rotated away from the peptide, as previously observed for HLA-A*02:01 presenting binders with extended C termini (44, 45). Overall, US11 docked within HLA-A*02:01 reminiscent of binding modes previously observed for C-terminally extended peptides.
As mentioned, also an additional high-affinity ligand for HLA-A*02:01 was predicted reflecting the central region of the US11 N-terminus (TLF; T27LFDEPPPL32). We refolded recombinant HLA-A*02:01 in the presence of the 9-mer peptide and determined the X-ray crystal structure to 1.6 Å resolution (SI Appendix, Fig. S4D and Table S1). The peptide bound conventionally within the binding groove, with Leu25 and Leu32 forming the canonical anchor residues, docking within the B pocket and F pocket, respectively. Without US11 C-terminal extension, Tyr84 adopted a conventional position, effectively closing off the binding pocket.
Collectively, our results strongly support that the US11 LCR serves as a peptide ligand mimic to anchor itself in the peptide-binding groove and restrict surface levels of HLA-A*02:01. Our findings further suggest that the US11 N-terminus may provide multiple HLA-A*02:01 binding interfaces to mimic ligand binding.
Optimal US11 LCR Targeting of HLA-A*29:02 and A*68:02 Is Tapasin Dependent.
Although peptide mimicry provides a satisfying explanation of US11 antagonism of HLA-A*02:01, it cannot explain US11 regulation of HLA-A*29:02 and A*68:02 allotypes. Unlike HLA-A*02:01, no optimal sequences for binding to the peptide-binding grooves of HLA-A*29:02 and A*68:02 were identified within the US11 N-terminal sequence. Consistent with our predictions, the US11 N-terminus did not robustly stabilize HLA-A*29:02 in a SC construct (SI Appendix, Fig. S5). Thus, how US11 regulates these HLA-A allotypes remained elusive.
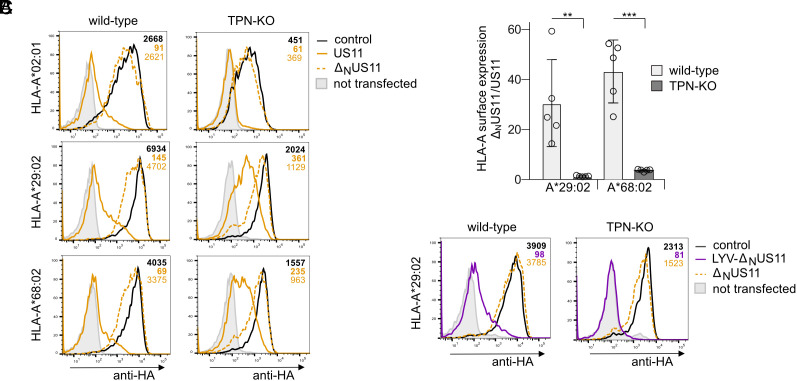
We postulated that other proteins could increase the avidity of US11 for slightly mismatched HLA-A allotypes. One candidate protein that could accomplish this compensation is tapasin, which plays an important role in stabilizing the peptide-binding pocket (46). To test this, we determined the ratio of HLA-A*68:02 and A*29:02 surface expression between ΔNUS11- and US11-expressing cells in the presence and absence of tapasin. In wild-type HeLa cells, HLA-A*29:02 and A*68:02 were 30- to 40-fold higher expressed when US11 lacked the N-terminus (Fig. 5 A and B). However, in tapasin knock-out cells (TPN-KO), this difference was reduced to twofold to fourfold, due to loss of efficacy of the full-length US11 in TPN-KO cells. We validated the effect of LYV-ΔNUS11 (N-terminal HLA-A*29:02 ligand) on HLA-A*29:02 and observed a complete loss of HLA-A*29:02 expression in TPN-KO cells (Fig. 5C). These findings support our hypothesis that tapasin is able to compensate for the weaker avidity of the US11-LCR for some HLA-allotypes, whereas tapasin is not necessary when the N-terminus is well matched, e.g., to HLA-A*02:01. Thus, the better the LCR matches the peptide-binding groove, the less LCR targeting is tapasin dependent. These results point at a key role for tapasin in HLA-A regulation by US11 and underlines the importance of a previous finding, demonstrating an interaction between US11 and the PLC in HCMV-infected cells (25).
Fig. 5.
Optimal US11 LCR targeting of HLA-A*29:02 and A*68:02 is tapasin dependent. (A) Wild-type and TPN-KO HeLa cells were transiently co-transfected with plasmids encoding HA-HLA-I and pIRES-EGFP encoding US11, ΔNUS11, or a control protein. At 20 h post-transfection, HA-HLA-I surface expression was determined by flow cytometry (anti-HA) on EGFP-positive cells. Representative histograms are shown. (B) The ratio of MFI determined in ΔNUS11 and US11-expressing cells in A. Dots represent individual values and bars mean values ± SEM from five independent experiments. Significance was calculated using two-way ANOVA. (C) Flow cytometry analysis as described in A with US11 variants as indicated. Representative histograms are shown.
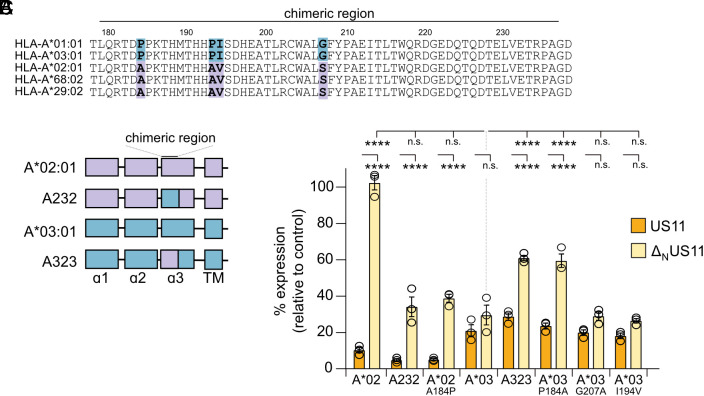
A Single Residue in the α3 Domain Dictates N-terminus–Independent Regulation of HLA-A by US11.
Although targeting of HLA-A*02:01, A*68:02, and A*29:02 requires the US11 N-terminal LCR, this region of US11 is dispensable for efficient inhibition of HLA-A*03:01 and A*01:01. We next investigated the mechanism of this LCR-independent regulation, which might also reveal insights into which mechanism of regulation evolved earlier. Alignment of HLA-A sequences identified four variable residues in the α3 domain of HLA-A: 184, 193, 194, and 207 (Fig. 6A and SI Appendix, Fig. S6A). To delineate their impact on US11 susceptibility, HA-tagged HLA-A chimeras were generated by swapping a segment (residues 179-236, marked with a black line in Fig. 6A), comprising all four varying residues: HLA-A*02:01 with the α3 segment from A*03:01 (A232) and HLA-A*03:01 with the α3 segment from A*02:01 (A323) (Fig. 6B). When co-expressed with US11, no major changes of the surface level of the chimeras were observed as compared to their wild-type counterparts (HLA-A*02:01 compared to A232, and HLA-A*03:01 to A323; Fig. 6C and SI Appendix, Fig. S6B). However, co-expression of ΔNUS11 revealed striking differences. First, compared to HLA-A*03:01, A323 surface expression was significantly enhanced. Second, the A232 chimera was more efficiently downregulated than HLA-A*02:01. Finally, we found that the A232 chimera was regulated comparably to HLA-A*03:01. Thus, the insertion of the α3 fragment from HLA-A*03:01 into HLA-A*02:01 resulted in a full gain of sensitivity toward an LCR-independent inhibition. To identify the specific residues required for this targeting, we introduced single point mutations into HLA-A*02:01 and A*03:01. No effects were observed when amino acids 194 and 207 were changed (Fig. 6C). In contrast, we found that a single Pro184Ala mutation in HLA-A*03:01 and Ala184Pro in HLA-A*02:01 recapitulated the effects of swapping the entire α3 fragment. This suggests that Pro at position 184 in the α3 domain is critical and Ala at position 184 of HLA-A*02:01 confers resistance against ΔNUS11 regulation.
Fig. 6.
A single residue in the α3 domain dictates N-terminus–independent regulation of HLA-A by US11. (A) HLA-A protein sequence alignment. (B) Schematic representation of HA-HLA-A*02:01 (lavender) and A*03:01 (blue) chimeras with a swapped α3 segment (residues 179 to 236). (C) HeLa cells were transiently co-transfected with plasmids encoding HA-HLA-A and pIRES-EGFP encoding US11 versions as indicated or a control protein. At 20 h post-transfection, HA-HLA-A surface expression was determined by flow cytometry (anti-HA) on EGFP-positive cells. The percentage of MFI as compared to control transfected cells was determined. Dots represent individual values and bars mean values ± SEM from three independent experiments. Significance was calculated using two-way ANOVA.
To allow simultaneous engagement with a surface comprising position 184 on the HLA-A*02:01 α3 domain, it would require that the LCR reaches the peptide-binding groove from its C-terminal end. To investigate how US11 interacts at both sites, we used the AlphaFold Multimer structural prediction program to generate a model of β2m-assembled HLA-A*02:01 in complex with full-length US11. The fold of HLA-A*02:01 closely agreed with experimentally determined X-ray crystal structures, with a root mean squared deviation of 0.5 Å with HLA-A*02:01 from our US11-SC X-ray crystal structure (SI Appendix, Fig. S6C). In further agreement with our US11-SC X-ray crystal structure, the US11 LCR was captured within the HLA-A*02:01 peptide–binding pocket with Met18 buried in the B pocket (SI Appendix, Fig. S6D). Of interest, the Ig domain of US11 docked adjacent to Ala184 on the α3 domain, illustrating that US11 can both engage this part of the molecule and hook the LCR to occupy the peptide-binding pocket in the correct orientation to mimic peptide binding.
US11 Evolved an LCR-Dependent Strategy Specifically to Counteract A2 Lineage Escape from N-terminus–Independent Antagonism.
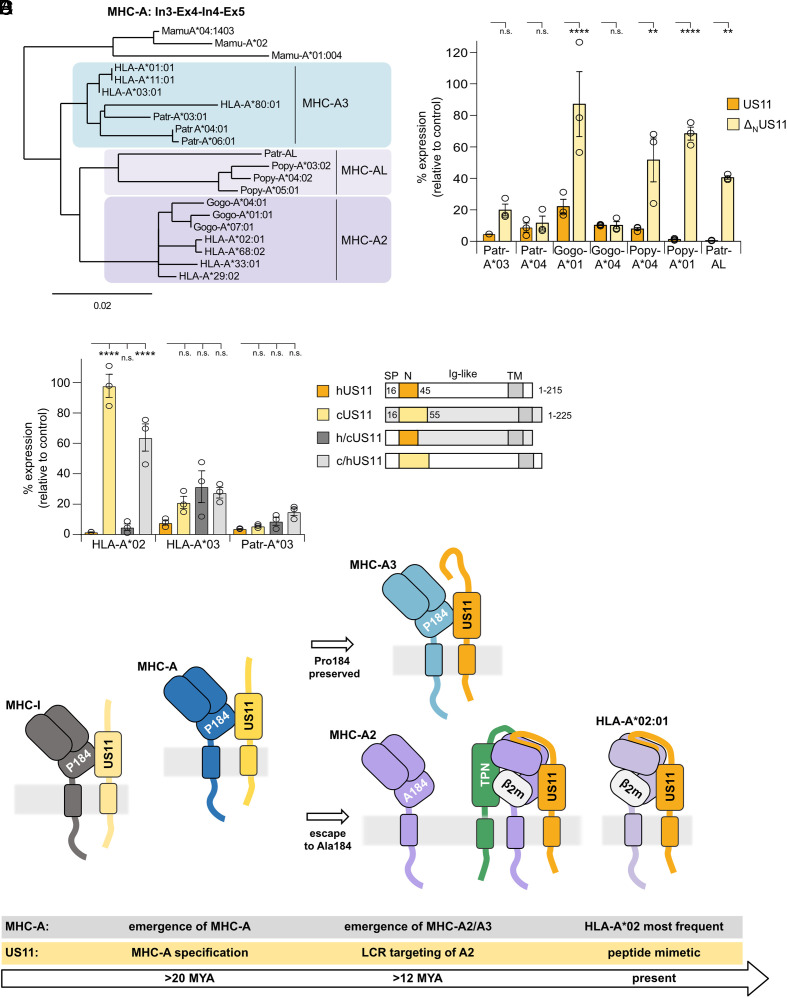
Position 184 is a key determinant for LCR-independent US11 regulation. This residue differs between the two major polymorphic MHC-A lineages in great apes, A2 and A3 (47). While the A3 lineage has a highly prevalent Pro at position 184 (SI Appendix, Fig. S6A), found in both classical and non-classical primate MHC-I, this position was mutated to an Ala during evolution of A2. Gorillas only have alleles of the A2 lineage, whereas chimpanzees only have polymorphic alleles of the A3 lineage (47, 48). Humans have both A2 and A3 HLA-A alleles. Intriguingly, the LCR-sensitive allotypes HLA-A*02:01, A*29:02, and A*68:02 belong to the A2 lineage, while HLA-A*03:01 and A*01:01 belong to the A3 lineage (49), visualized by aligning genomic sequences comprising intron 3, exon 4, intron 4, and exon 5 (Fig. 7A).
Fig. 7.
US11 evolved an LCR-dependent strategy specifically to counteract A2 lineage escape from N-terminus–independent antagonism. (A) Alignment of MHC-A genomic sequences from intron 3, exon 4, intron 4, and exon 5. Phylogeny.fr was used to build the phylogenetic tree [84]). (B and C) HA-MHC-A surface expression was determined as in Fig. 1B. (C) In the Right panel, a schematic representation of hUS11, cUS11, h/cUS11, and c/hUS11 is depicted. (D) Model of US11 and MHC-A coevolution. After the split of New World monkeys and Old World monkeys, the MHC-A locus emerged, which might have resulted in better control of CMV, spurring dedication of US11 to regulate MHC-A. The lineages A2 and A3 evolved in an ancestor of great apes. The A2 lineage is characterized by Ala at position 184, resisting control by the concurrent US11. US11 gained a new function at the N-terminus; in a tapasin-dependent manner, this sequence is able to interact with the peptide-binding domain of assembled A2 molecules and redirects the target molecule for degradation. In the human population, the HLA-A*02 allelic family became highly prevalent prompting US11 to target this allotype specifically by a peptide mimetic in the LCR.
As a result of a gene duplication, an additional A locus, MHC-A like (MHC-AL), has been maintained as an active gene in some species (50). In orangutans, it evolved into the major antigen-presenting A locus (Popy-A), which displays a high level of polymorphism. About 50% of chimpanzees have a haplotype comprising the MHC-AL locus as an active but non-polymorphic gene (Patr-AL) (50). Parts of this locus could be more related to A2 than to A3 (Fig. 7A), as it lacks the A3-specific deletion of 17 nucleotides in intron 3 (51), and, also, similarly to A2 proteins, it encodes an alanine at position 184.
To test the conservation of US11 specificity for MHC-A in great apes, we assessed US11 regulation of MHC-A from chimpanzee (Pan troglodytes; Patr-A*03:01, A*04:01, and AL), gorilla (Gorilla gorilla; Gogo-A*01:01 and A*04:01), and orangutan (Pongo pygmaeus; Popy-A*01:01 and A*04:01). Full-length US11 strongly downregulated all MHC-A (Fig. 7B). In contrast, ΔNUS11 did not inhibit Gogo-A*01, Popy-A*01, Popy-A*04, and Patr-AL to the same extent as wild-type US11, supporting conservation of the N-terminus dependent targeting of A2 lineage molecules. Interestingly, ΔNUS11 efficiently downregulated Gogo-A*04 despite an Ala at position 184. This suggests that even the Ala “resistance” variant could be overcome by other changes during the US11-MHC-A coevolution, which may be revealed by analysis of US11 and MHC-A sequences from further CMV and host pairs.
These results suggest that US11 evolved distinct targeting strategies in a stepwise manner to cope with MHC-A lineages. Since chimpanzees have lost the polymorphic A2 lineage, we hypothesized that the chimpanzee CMV US11 homolog (cUS11) should not depend upon an LCR-dependent mechanism to control this lineage. Consistent with this hypothesis, there is a modest 45% sequence identity between hUS11 (HCMV) and cUS11, which is reduced to only 26% identity in the N-terminus alone (alignment in SI Appendix, Fig. S1A). To test cUS11 specificity, we compared surface expression levels of MHC-A molecules co-expressed with either hUS11 or cUS11. As hypothesized, cUS11 downregulated HLA-A*03:01 but not HLA-A*02:01 (Fig. 7 C, Left). However, when the N-terminus of cUS11 was substituted with the corresponding hUS11 region (residues 1 to 44; schematic representation Fig. 7 C, Right), the chimeric h/cUS11 gained the ability to downregulate HLA-A*02:01. In contrast, a near-reciprocal chimera, with the cUS11 N-terminus replacing residues 20 to 44 in US11, did not efficiently regulate HLA-A*02:01. All four native and chimeric US11 variants strongly reduced surface expression of HLA-A*03:01 and the chimpanzee Patr-A*03. This finding implies that the US11 LCR evolved to target the MHC-A A2 lineage. In conclusion, our data support the overarching model that the viral immunoevasin US11 specifically developed an additional MHC-A targeting mechanism using its N-terminal region early during evolution of the great apes, prior to the gene duplication of MHC-A2 and MHC-AL.
Discussion
US2 and US11 were the first viral proteins shown to exploit the ERAD pathway for elimination of host proteins (23, 24). While the contact sites between US2 and HLA-A*02:01 were resolved in a X-ray crystal structure (52), the basis of binding specificity of US11 has remained unclear due to lack of structural data. Here, we have characterized the specification of US11 toward HLA-A locus products and demonstrated that the A2 and A3 subfamilies are targeted by US11 through two independent spatially separated subdomains, with the variability centered on the use of a LCR at the N-terminus of US11. The presence of the LCR is decisive for control of HLA-A*02:01-restricted CD8+ T cells in HCMV infection.
In agreement with earlier studies (53), our data demonstrate the importance of the HLA-A*02:01 α1-2 domains for US11-mediated regulation. However, this mechanism applies only to the A2 and not to the A3 lineage HLA-A. Our analysis further extends this insight, demonstrating that it is the N-terminal LCR that targets the peptide-binding groove.
As proof-of-principle, the US11 N-terminus could be functionally substituted by HLA-A peptide ligand sequences, which led to degradation of target molecules. However, in the natural US11 N-terminus stabilizing ligand sequences were not identified for HLA-A*29:02 and A*68:02. Instead, US11 takes advantage of the endogenous MHC-I quality control and overcomes the lack of specificity by exploiting the function of tapasin, which prevents loading of low-affinity peptides and maintains an empty peptide-binding groove in the PLC (54). Further studies are required to better understand how US11 utilizes this and which part of the peptide-binding groove US11 targets. Importantly, we have demonstrated that US11 interacts with the PLC in HCMV-infected cells (25), underlining the relevance of these interactions for US11 function.
Not the entirety of US11 regulation is tapasin dependent however. We found that a well-matched sequence at the N-terminus of US11 rendered US11 tapasin independent, possibly by its ability to outcompete peptide binding. Strikingly, in its natural form, the US11 LCR exhibits a specificity that might be unique for the peptide-binding groove of HLA-A*02 alleles. Two distinct US11 sites are capable of docking within the HLA-A*02:01 binding groove, SMP and TLF. Interestingly, TLF was found also by others to stabilize HLA-A*02:01 (55).
The SMP peptide is predicted to bind to the most prevalent HLA-A*02 allelic variants (A*02:02, A*02:03, A*02:05, A*02:06, and A*02:07) with high affinity. Although the highest sequence variability is found in the N-terminal part of US11 (SI Appendix, Fig. S7 A and B), of 231 HCMV-US11 sequences analyzed, only three showed variation in the SMP sequence (Leu23Met; SI Appendix, Fig. S7A), which is predicted not to significantly affect binding to HLA-A*02:01. We speculate that the extraordinary immunodominant HLA-A*02:01NLV-specific CD8+ T-cell responses (56) may have spurred this immune evasion strategy focused against HLA-A*02. Consistent with this hypothesis, a lack of the US11 N-terminus in HCMV infection resulted in a strong activation of HLA-A*02:01NLV-specific CD8+ T cells, almost to the same extent as a complete deletion of US11 (Fig. 2D).
A3 lineage HLA-A allotypes are controlled by US11 independently of the N-terminus. To maintain this independence, a Pro at position 184 in the α3 domain of HLA-A is critical. The interface at the α2/α3 junction is also targeted by the Ig domain of US2 (52) and by the adenovirus immunoevasin E3/19k (57), suggesting this interface may be pre-disposed to be targeted by immunoevasins. Our experimental data and the AlphaFold prediction suggest that the Ig domain of US11 may also dock at this site. It is currently unclear why Ala at position 184 makes HLA-A*02:01 more resistant to ΔNUS11 regulation. However, both LCR-dependent and -independent targeting mechanisms of US11 require TMEM129 for HLA-A degradation, indicating a convergent downstream degradation pathway. In addition, we have shown that the HLA-B HC bears an intrinsic resistance to degradation by US11 (25). Since HLA-B allotypes have an invariable Pro at position 184, additional positions must be decisive for US11 sensitivity.
Our data display compelling evidence for a distinct spatial and temporal shaping of US11 subdomains to both the A2 and A3 lineages of MHC-A. On the basis of hominid MHC-A evolution and the functional as well as structural analysis of US11 homologs, we propose a model for the parallel evolution of US11 as depicted in Fig. 7D. To date, no clear US11 sequence homolog has been found in CMV species infecting New World monkeys (SI Appendix, Fig. S7C). Thus, US11 is likely to have evolved after the split of New World and Old World monkeys, which took place approximately 40 Mya (58), coincident with the emergence of the MHC-A locus in an ancestor of Old World monkeys (59). This locus might have controlled CMV replication better, forcing US11 to focus on this locus. Early during the evolution of great apes, the MHC-A locus segregated into the A2 and A3 lineages. The Pro184Ala mutation in the A2 lineage might have conferred resistance against the ancestral US11. Strikingly, the A2 lineage in great apes appears to be the only primate MHC-I with an Ala at position 184. This unique feature prompts the speculation that US11 induced a balanced evolution with variability at this position. In response, US11 evolved a new function in the N-terminal subdomain targeting the MHC-A2 α1-2 domains. Chimpanzee CMV US11 lacks this function, in accordance with complete loss of the polymorphic MHC-A2 lineage, presumably caused by a simian immunodeficiency virus induced selective sweep (60). As a limitation to our study, the genomes of gorilla and orangutan CMV are still awaiting sequencing and the effects of US11 from these CMV species could not be analyzed.
Last, since a possible introgression into modern human populations from Neandertals, alleles of the HLA-A*02 allelic family have become the most prevalent in the human population (61, 62). In a relatively recent counterattack, the US11 LCR might have gained the ability to act as an HLA-A*02 peptide mimetic to protect HCMV-infected cells from the exceptionally strong response of HLA-A*02-restricted HCMV-specific CD8+ T cells.
Polymorphic MHC-I allele diversification is considered to be permanently driven by the need to control emerging infections and pathogen escape variants (63). This study demonstrates the impact of a viral immunoevasin as an independent driving force for MHC-I allele variability. We speculate that future studies will reveal further DNA viruses expressing tailored immunoevasins targeting antigen-presenting MHC-I.
Materials and Methods
Cell Lines and Transfection.
HeLa cells (ATCC CCL-2), FO-1 (64), MRC-5 fibroblasts (ECACC 05090501), and HF-α fibroblasts (a kind gift of Dieter Neumann-Haefelin and Valeria Kapper-Falcone, University Medical Center Freiburg) were grown in Dulbecco's Modified Eagle Medium supplemented with 10% fetal calf serum (FCS) and penicillin/streptomycin at 37 °C and 5% CO2. A TMEM129 HeLa knock-out clone was generated by nucleofection of a multi-guide sgRNA and Cas9 (Gene Knockout Kit v2, Synthego). A tapasin knock-out HeLa cell line was generated by transient expression of Cas9 and a gRNA targeting the sequence GCCCTATACGCCAGGCCTGG (65).
Molecular Cloning.
HLA-I molecules tagged with an N-terminal HA epitope were constructed as previously described (25). More detailed information about molecular cloning of HLA-I molecules and chimeric constructs can be found in SI Appendix.
RL8 (used as a negative control), US11, ΔNUS11, and US6 in pIRES-EGFP have been described previously (25, 66). The cloning of US11 variants is described in SI Appendix.
Viruses.
Generation, reconstitution, and propagation of the recombinant HCMV viruses have been previously described, as the ΔUS11 deletion mutant (25). The construction of mutants is described in SI Appendix.
Antibodies and Reagents.
Antibodies and reagents used for this study can be found in SI Appendix.
HLA-I Cell Surface Analysis.
HeLa cells (ca. 2 × 105) were transiently transfected with an HA-HLA-I-encoding puc2CL6IP vector (500 ng), pIRES-EGFP encoding an immunoevasin or control protein (150 ng), and a non-coding plasmid (350 ng) using SuperFect (Qiagen). When analyzing HLA-A*02:01 SC constructs, 200 ng expression plasmid (pcDNA3.1) was transfected. Cells were harvested by trypsin, washed, and stained with antibodies diluted in phosphate-buffered saline with 3% FCS.
HF-α fibroblasts were infected with an MOI of 8 and centrifugal enhancement (800 × g, 2× 15 min). Cells were harvested using accutase, washed, and treated with FcR blocking reagent. Antibodies used for staining were diluted in phosphate-buffered saline with 3% FCS. Cells were fixed in 4% paraformaldehyde prior to flow cytometry analysis (FACS Canto II, BD Biosciences).
Western Blot Analysis.
Western blot analysis was performed as described previously (25). Briefly, proteins were separated by SDS-PAGE and transferred to a nitrocellulose membrane. After incubation with specific antibodies, peroxidase-conjugated secondary antibodies were used. Proteins were detected by an ECL reagent and an Odyssey XF Imager (Licor).
HLA-A-Restricted HCMV-Specific CD8+ T-Cell Activation.
PBMCs from four healthy HCMV seropositive donors (two positive for HLA-A*02:01 and two for HLA-A*03) were isolated from EDTA blood by density separation. The PBMCs were cultured in Roswell Park Memorial Institute 1640 medium supplemented with 10% FCS, penicillin/streptomycin, 1.5% HEPES, 0.5 µg/mL anti-CD28, 5 µM peptide, (NLV or GLY), and 20 IU/mL IL-2. After 14 d, the PBMCs were co-cultured for 5 h with HF-α fibroblasts (E:T ratio: 5:1, in the presence of BFA), mock treated, or infected with HCMV BAC2 mutants at an MOI of 8 for 24 or 72 h. Activation was determined by staining with anti-CD8 and peptide-specific tetramers generated from HLA-I easYmers (immunAware) and streptavidin-R-PE. Subsequently, fixed and permeabilized cells were stained with anti-IFNγ and anti-TNFα. As positive controls, PBMCs were cultured with 2 µg/mL PMA and 40 µg/mL ionomycin or with 15 µM peptide (NLV or GLY) for 5 h. The project was examined and approved by the ethical committee at the Albert-Ludwigs-University Freiburg (22 to 1,196), and informed consent was obtained from all participants.
Immunoprecipitation.
Immunoprecipitations were performed as previously described (67). Briefly, cells were metabolically labeled with 35S-Met-Cys. Cells were treated with lysis buffer [140 mM NaCl, 20 mM Tris (pH 7.6), and 5 mM MgCl2] containing 1 % digitonin (Calbiochem) and protease inhibitors. Lysates were incubated with antibodies and protein A Sepharose (Merck) for 1 h at 4 °C. The beads were washed, and, where indicated, an EndoH digestion was performed according to the manufacturer’s instructions (New England Biolabs). Samples were separated by a gradient SDS-PAGE. Gels were fixed, dried, and exposed to phosphor screens, which were scanned by a Typhoon FLA 7000 phosphorimager (GE Healthcare). For better illustration, contrast and light settings were adjusted in the figures.
MHC-I Fluorescence Polarization Assay.
Fluorescence polarization assays were performed with HLA-A*02:01 refolded with photocleavable peptide, photoKV9 (KILGFVFJ*V; J*: 3-amino-3-(2-nitro) phenyl-propionic acid). HLA-A*02/photoKV9 was purified by gel filtration in TBS buffer (20 mM Tris, 150 NaCl, pH 7.23) and protein quality was confirmed by Tm measurement. Exchange to the FITC-labeled high affinity peptide FITC-GV9 (GILGKFITCVFTV) was determined in the presence of US11 peptides (WT, amino acids 17 to 44, or the mutated L25G peptide) after UV-exposure.
In brief, 20 μL of 1 μM purified A*02/photoKV9 was UV-exposed on ice for 5 min and added to the 384-well plate (Corning® 384-well Low Flange Black Flat Bottom Polystyrene NBS Microplate 3575). Subsequently, 40nM FITC-GV9 was added to TBS buffer only or to US11 peptides at different concentrations (in 0.1M Tris pH 8.0). Each condition was measured in three replicates. FITC-peptide was set to baseline at a final concentration of 10 nM. FP signals were recorded on a Tecan Spark reader (λex = 485 nm; λem = 535 nm). For kinetic measurements, the FP signals were monitored for 3 h. Static measurements were performed overnight at room temperature with FITC-GV9 and different concentrations of US11 peptides. Experimental data analysis was processed with Origin 2019, and IC50 was fitted by the mode of dose–response.
Protein Expression and Purification.
A single chain construct (US11SC) encoding the US11 N-terminus, β2m, and HLA-A*02:01 was codon optimized for expression in Escherichia coli. US11SC was expressed as inclusion bodies in Ton A- BL21 (DE3) E. coli, solubilized in 6 M guanidine-HCl and subsequently refolded. Following dialysis, refolded US11SC was purified by a combination of anion exchange (DEAE and Hitrap Q, Cytiva) and size exclusion (Superdex 200, Cytiva) chromatography systems. Purified US11SC was concentrated to 5.2 mg/mL for crystallization experiments. Isolated β2m and the HLA-A*02:01 were also expressed as inclusion bodies in Ton A- BL21 (DE3) E. coli, before being refolded with the TLF peptide in the ratio 3.2 μM HC, 2.6 μM β2m, and 11 μM peptide, and purified as described above. Detailed information is given in SI Appendix.
Crystallization, Data Collection, Structure Determination, and Structure Prediction.
All crystallization conditions that were used are described in SI Appendix. Diffractions images were collected by an EIGER detector on the MX2 beamline at the Australian Synchrotron, part of ANSTO (68). Diffraction data were processed using XDS (69) and merged using AIMLESS (70) within the CCP4 package (Winn 2010). Initial phases were generated by molecular replacement, using PHASER of the CCP4 package, with HLA-A*02:01 stripped of peptide, water molecules and ions (PDB ID: 1S9W) as the search model. One solution was identified, and sequential rounds of model refinement and model building were performed in PHENIX (71) and COOT (72), respectively, until convergence was reached. Atomic contacts were identified using CONTACT within the CCP4 suite, and figures were generated using the PyMOL Molecular Graphics System (Version 2.3.2; Schrödinger, LLC).
The structure of full-length US11 in complex with HLA-A*02:01 was predicted using AlphaFold Multimer v2.3.2 run on a local server (73, 74). Figures were generated using the PyMOL Molecular Graphics System (Version 2.5.5; Schrödinger, LLC).
Statistics.
Calculations based on flow cytometric results were performed using the median fluorescence intensity and shown with the SEM. The applied statistical analysis is mentioned for each experiment in the figure legend and was determined using GraphPad Prism software (v8). A P-value < 0.05 was considered significant (*P < 0.05; **P < 0.005; ***P < 0.0005).
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We are greatly indebted to Harmit Malik (Fred Hutchinson Cancer Research Center), Mirko Trilling (University Hospital Essen), and Hans-Gerhard Burgert and Zsolt Ruzsics (University Medical Center Freiburg) for inspiring discussions and critical reading of the manuscript. We are grateful for technical support by Tim Birnesser and Zhihui Fu. We thank Geoffrey Kong from the Monash Macromolecular Crystallization Facility for assistance with crystallization screens, and Dene Litter for assistance with diffraction data collection. This research was undertaken in part using the MX2 beamline at the Australian Synchrotron, part of ANSTO, and made use of the Australian Cancer Research Foundation detector. The work was funded by the Deutsche Forschungsgemeinschaft: grant FOR2830, project HA 6035/2-2 (A.H.) and HE 2526/9-1 (H.H.), grant TRR 186, project 278001972 (C.F.), and the project FR1325/20-1 (C.F.).
Author contributions
G.M.W., R.B., J.F., M.H., C.F., J.R., F.M., and A.H. designed research; C.Z., G.M.W., L.B., B.C., H.L., C.G., V.O., J.F., and A.H. performed research; M.H., and F.M. contributed new reagents/analytic tools; C.Z., G.M.W., L.B., R.B., B.C., H.L., V.O., M.H., C.F., J.R., F.M., H.H., and A.H. analyzed data; and G.M.W. and A.H. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
Protein structure data have been deposited in RCSB Protein Data Bank (8FRT (75) and 8FU4 (76)).
Supporting Information
References
- 1.Dos Santos Francisco R., et al. , HLA supertype variation across populations: New insights into the role of natural selection in the evolution of HLA-A and HLA-B polymorphisms. Immunogenetics 67, 651–663 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hertz T., et al. , Mapping the landscape of host-pathogen coevolution: HLA class I binding and its relationship with evolutionary conservation in human and viral proteins. J. Virol. 85, 1310–1321 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stern-Ginossar N., et al. , Decoding human cytomegalovirus. Science 338, 1088–1093 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berry R., Watson G. M., Jonjic S., Degli-Esposti M. A., Rossjohn J., Modulation of innate and adaptive immunity by cytomegaloviruses. Nat. Rev. Immunol. 20, 113–127 (2020). [DOI] [PubMed] [Google Scholar]
- 5.Guma M., et al. , Imprint of human cytomegalovirus infection on the NK cell receptor repertoire. Blood 104, 3664–3671 (2004). [DOI] [PubMed] [Google Scholar]
- 6.Sylwester A. W., et al. , Broadly targeted human cytomegalovirus-specific CD4+ and CD8+ T cells dominate the memory compartments of exposed subjects. J. Exp. Med. 202, 673–685 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crough T., Khanna R., Immunobiology of human cytomegalovirus: From bench to bedside. Clin. Microbiol. Rev. 22, 76–98 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McGeoch D. J., Cook S., Dolan A., Jamieson F. E., Telford E. A., Molecular phylogeny and evolutionary timescale for the family of mammalian herpesviruses. J. Mol. Biol. 247, 443–458 (1995). [DOI] [PubMed] [Google Scholar]
- 9.Ashiru O., et al. , NKG2D ligand MICA is retained in the cis-Golgi apparatus by human cytomegalovirus protein UL142. J. Virol. 83, 12345–12354 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arase H., Mocarski E. S., Campbell A. E., Hill A. B., Lanier L. L., Direct recognition of cytomegalovirus by activating and inhibitory NK cell receptors. Science 296, 1323–1326 (2002). [DOI] [PubMed] [Google Scholar]
- 11.Smith H. R., et al. , Recognition of a virus-encoded ligand by a natural killer cell activation receptor. Proc. Natl. Acad. Sci. U.S.A. 99, 8826–8831 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aguilar O. A., et al. , A viral immunoevasin controls innate immunity by targeting the prototypical natural killer cell receptor family. Cell 169, 58–71.e14 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Zeleznjak J., et al. , The complex of MCMV proteins and MHC class I evades NK cell control and drives the evolution of virus-specific activating Ly49 receptors. J. Exp. Med. 216, 1809–1827 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davison A. J., Evolution of the herpesviruses. Vet. Microbiol. 86, 69–88 (2002). [DOI] [PubMed] [Google Scholar]
- 15.Becker S., Fink A., Podlech J., Reddehase M. J., Lemmermann N. A., Host-adapted gene families involved in murine cytomegalovirus immune evasion. Viruses 14, 128 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Verweij M. C., et al. , Viral inhibition of the transporter associated with antigen processing (TAP): A striking example of functional convergent evolution. PLoS Pathog. 11, e1004743 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pande N. T., Powers C., Ahn K., Fruh K., Rhesus cytomegalovirus contains functional homologues of US2, US3, US6, and US11. J. Virol. 79, 5786–5798 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones T. R., et al. , Human cytomegalovirus US3 impairs transport and maturation of major histocompatibility complex class I heavy chains. Proc. Natl. Acad. Sci. U.S.A. 93, 11327–11333 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park B., et al. , Human cytomegalovirus inhibits tapasin-dependent peptide loading and optimization of the MHC class I peptide cargo for immune evasion. Immunity 20, 71–85 (2004). [DOI] [PubMed] [Google Scholar]
- 20.Lehner P. J., Karttunen J. T., Wilkinson G. W., Cresswell P., The human cytomegalovirus US6 glycoprotein inhibits transporter associated with antigen processing-dependent peptide translocation. Proc. Natl. Acad. Sci. U.S.A. 94, 6904–6909 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ahn K., et al. , The ER-luminal domain of the HCMV glycoprotein US6 inhibits peptide translocation by TAP. Immunity 6, 613–621 (1997). [DOI] [PubMed] [Google Scholar]
- 22.Hengel H., et al. , A viral ER-resident glycoprotein inactivates the MHC-encoded peptide transporter. Immunity 6, 623–632 (1997). [DOI] [PubMed] [Google Scholar]
- 23.Wiertz E. J., et al. , The human cytomegalovirus US11 gene product dislocates MHC class I heavy chains from the endoplasmic reticulum to the cytosol. Cell 84, 769–779 (1996). [DOI] [PubMed] [Google Scholar]
- 24.Wiertz E. J., et al. , Sec61-mediated transfer of a membrane protein from the endoplasmic reticulum to the proteasome for destruction. Nature 384, 432–438 (1996). [DOI] [PubMed] [Google Scholar]
- 25.Zimmermann C., et al. , HLA-B locus products resist degradation by the human cytomegalovirus immunoevasin US11. PLoS Pathog. 15, e1008040 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coletta A., et al. , Low-complexity regions within protein sequences have position-dependent roles. BMC Syst. Biol. 4, 43 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Axelrod R., Hamilton W. D., The evolution of cooperation. Science 211, 1390–1396 (1981). [DOI] [PubMed] [Google Scholar]
- 28.van de Weijer M. L., et al. , A high-coverage shRNA screen identifies TMEM129 as an E3 ligase involved in ER-associated protein degradation. Nat. Commun. 5, 3832 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van den Boomen D. J., et al. , TMEM129 is a Derlin-1 associated ERAD E3 ligase essential for virus-induced degradation of MHC-I. Proc. Natl. Acad. Sci. U.S.A. 111, 11425–11430 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Le V. T., Trilling M., Hengel H., The cytomegaloviral protein pUL138 acts as potentiator of tumor necrosis factor (TNF) receptor 1 surface density to enhance ULb’-encoded modulation of TNF-alpha signaling. J. Virol. 85, 13260–13270 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ameres S., Besold K., Plachter B., Moosmann A., CD8 T cell-evasive functions of human cytomegalovirus display pervasive MHC allele specificity, complementarity, and cooperativity. J. Immunol. 192, 5894–5905 (2014). [DOI] [PubMed] [Google Scholar]
- 32.Borst E. M., et al. , The essential human cytomegalovirus proteins pUL77 and pUL93 are structural components necessary for viral genome encapsidation. J. Virol. 90, 5860–5875 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Varnum S. M., et al. , Identification of proteins in human cytomegalovirus (HCMV) particles: The HCMV proteome. J. Virol. 78, 10960–10966 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Besold K., Wills M., Plachter B., Immune evasion proteins gpUS2 and gpUS11 of human cytomegalovirus incompletely protect infected cells from CD8 T cell recognition. Virology 391, 5–19 (2009). [DOI] [PubMed] [Google Scholar]
- 35.Besold K., et al. , Processing and MHC class I presentation of human cytomegalovirus pp65-derived peptides persist despite gpUS2-11-mediated immune evasion. J. Gen. Virol. 88, 1429–1439 (2007). [DOI] [PubMed] [Google Scholar]
- 36.Hansen T. H., Connolly J. M., Gould K. G., Fremont D. H., Basic and translational applications of engineered MHC class I proteins. Trends Immunol. 31, 363–369 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Anjanappa R., et al. , Structures of peptide-free and partially loaded MHC class I molecules reveal mechanisms of peptide selection. Nat. Commun. 11, 1314 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hein Z., et al. , Peptide-independent stabilization of MHC class I molecules breaches cellular quality control. J. Cell Sci. 127, 2885–2897 (2014). [DOI] [PubMed] [Google Scholar]
- 39.Wei M. L., Cresswell P., HLA-A2 molecules in an antigen-processing mutant cell contain signal sequence-derived peptides. Nature 356, 443–446 (1992). [DOI] [PubMed] [Google Scholar]
- 40.Weinzierl A. O., et al. , Features of TAP-independent MHC class I ligands revealed by quantitative mass spectrometry. Eur. J. Immunol. 38, 1503–1510 (2008). [DOI] [PubMed] [Google Scholar]
- 41.Nielsen M., Andreatta M., NetMHCpan-3.0; improved prediction of binding to MHC class I molecules integrating information from multiple receptor and peptide length datasets. Genome Med. 8, 33 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Webb A. I., et al. , Functional and structural characteristics of NY-ESO-1-related HLA A2-restricted epitopes and the design of a novel immunogenic analogue. J. Biol. Chem. 279, 23438–23446 (2004). [DOI] [PubMed] [Google Scholar]
- 43.Nguyen A. T., Szeto C., Gras S., The pockets guide to HLA class I molecules. Biochem. Soc. Trans. 49, 2319–2331 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Collins E. J., Garboczi D. N., Wiley D. C., Three-dimensional structure of a peptide extending from one end of a class I MHC binding site. Nature 371, 626–629 (1994). [DOI] [PubMed] [Google Scholar]
- 45.McMurtrey C., et al. , Toxoplasma gondii peptide ligands open the gate of the HLA class I binding groove. eLife 5, e12556 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wearsch P. A., Cresswell P., Selective loading of high-affinity peptides onto major histocompatibility complex class I molecules by the tapasin-ERp57 heterodimer. Nat. Immunol. 8, 873–881 (2007). [DOI] [PubMed] [Google Scholar]
- 47.Lawlor D. A., Warren E., Taylor P., Parham P., Gorilla class I major histocompatibility complex alleles: Comparison to human and chimpanzee class I. J. Exp. Med. 174, 1491–1509 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mayer W. E., et al. , Nucleotide sequences of chimpanzee MHC class I alleles: Evidence for trans-species mode of evolution. EMBO J. 7, 2765–2774 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lienert K., Parham P., Evolution of MHC class I genes in higher primates. Immunol. Cell Biol. 74, 349–356 (1996). [DOI] [PubMed] [Google Scholar]
- 50.Gleimer M., et al. , Although divergent in residues of the peptide binding site, conserved chimpanzee Patr-AL and polymorphic human HLA-A*02 have overlapping peptide-binding repertoires. J. Immunol. 186, 1575–1588 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Firgaira F. A., Male D. A., Morley A. A., The ancestral HLA-A lineage split is delineated by an intron 3 insertion/deletion polymorphism. Immunogenetics 40, 445–448 (1994). [DOI] [PubMed] [Google Scholar]
- 52.Gewurz B. E., et al. , Antigen presentation subverted: Structure of the human cytomegalovirus protein US2 bound to the class I molecule HLA-A2. Proc. Natl. Acad. Sci. U.S.A. 98, 6794–6799 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Barel M. T., et al. , Amino acid composition of alpha1/alpha2 domains and cytoplasmic tail of MHC class I molecules determine their susceptibility to human cytomegalovirus US11-mediated down-regulation. Eur. J. Immunol. 33, 1707–1716 (2003). [DOI] [PubMed] [Google Scholar]
- 54.Domnick A., et al. , Molecular basis of MHC I quality control in the peptide loading complex. Nat. Commun. 13, 4701 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Elkington R., et al. , Ex vivo profiling of CD8+-T-cell responses to human cytomegalovirus reveals broad and multispecific reactivities in healthy virus carriers. J. Virol. 77, 5226–5240 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wills M. R., et al. , The human cytotoxic T-lymphocyte (CTL) response to cytomegalovirus is dominated by structural protein pp65: Frequency, specificity, and T-cell receptor usage of pp65-specific CTL. J. Virol. 70, 7569–7579 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li L., Santarsiero B. D., Bouvier M., Structure of the AdeNovirus type 4 (species E) E3–19K/HLA-A2 complex reveals species-specific features in MHC class I recognition. J. Immunol. 197, 1399–1407 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Perelman P., et al. , A molecular phylogeny of living primates. PLoS Genet. 7, e1001342 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Adams E. J., Parham P., Species-specific evolution of MHC class I genes in the higher primates. Immunol. Rev. 183, 41–64 (2001). [DOI] [PubMed] [Google Scholar]
- 60.de Groot N. G., et al. , Evidence for an ancient selective sweep in the MHC class I gene repertoire of chimpanzees. Proc. Natl. Acad. Sci. U.S.A. 99, 11748–11753 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ellis J. M., et al. , Frequencies of HLA-A2 alleles in five U.S. population groups. Predominance Of A*02011 and identification of HLA-A*0231. Hum. Immunol. 61, 334–340 (2000). [DOI] [PubMed] [Google Scholar]
- 62.Abi-Rached L., et al. , The shaping of modern human immune systems by multiregional admixture with archaic humans. Science 334, 89–94 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Borghans J. A., Beltman J. B., De Boer R. J., MHC polymorphism under host-pathogen coevolution. Immunogenetics 55, 732–739 (2004). [DOI] [PubMed] [Google Scholar]
- 64.D’Urso C. M., et al. , Lack of HLA class I antigen expression by cultured melanoma cells FO-1 due to a defect in B2m gene expression. J. Clin. Invest. 87, 284–292 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gerke C., et al. , Multimodal HLA-I genotype regulation by human cytomegalovirus US10 and resulting surface patterning. bioRxiv [Preprint] (2023). 10.1101/2023.01.10.523457 (Accessed 13 January 2023). [DOI] [PMC free article] [PubMed]
- 66.Matschulla T., et al. , A highly conserved sequence of the viral TAP inhibitor ICP47 is required for freezing of the peptide transport cycle. Sci. Rep. 7, 2933 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Halenius A., et al. , Human cytomegalovirus disrupts the major histocompatibility complex class I peptide-loading complex and inhibits tapasin gene transcription. J. Virol. 85, 3473–3485 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McPhillips T. M., et al. , Blu-ice and the distributed control system: Software for data acquisition and instrument control at macromolecular crystallography beamlines. J. Synchrotron Radiat. 9, 401–406 (2002). [DOI] [PubMed] [Google Scholar]
- 69.Kabsch W., Xds. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Evans P. R., Murshudov G. N., How good are my data and what is the resolution? Acta Crystallogr. D Biol. Crystallogr. 69, 1204–1214 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Afonine P. V., et al. , Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 68, 352–367 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Emsley P., Cowtan K., Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 73.Jumper J., et al. , Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Evans R., et al. , Protein complex prediction with AlphaFold-Multimer. bioRxiv [Preprint] (2021). 10.1101/2021.10.04.463034 (Accessed 10 March 2022). [DOI]
- 75.Watson G. M., Berry R., Littler D. R., Rossjohn J., HCMV US11 peptide binding to HLA-A*02:01. RCSB Protein Data Bank. https://www.rcsb.org/structure/8FU4. Deposited 16 January 20232.
- 76.Watson G. M., Berry R., Rossjohn J., X-ray crystal structure of the N-terminal region from HCMV US11 binding to HLA-A*02:01. RCSB Protein Data Bank. https://www.rcsb.org/structure/8FRT. Deposited 8 January 2023.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
Protein structure data have been deposited in RCSB Protein Data Bank (8FRT (75) and 8FU4 (76)).