Abstract
Both of the major immediate-early (IE) proteins IE1 and IE2 of human cytomegalovirus (HCMV) as well as input viral DNA and sites of viral IE transcription colocalize with or adjacent to punctate PML domains (PML oncogenic domains [PODs] or nuclear domain 10) in the nucleus within the first few hours after infection of permissive human fibroblasts. However, colocalization of IE1 and PML in PODs is only transient, with both proteins subsequently redistributing into a nuclear diffuse form. These processes are believed to promote efficient viral IE transcription and initiation of DNA synthesis especially at low multiplicities of infection. To examine the mechanism of PML displacement by IE1, we carried out indirect immunofluorescence assay experiments with plasmids expressing intact or deleted forms of PML and IE1 in DNA-transfected cells. The results demonstrated that deletion of the C-terminal acidic region of IE1 uncouples the requirements for displacement of both endogenous and coexpressed PML from those needed to target to the PODs. Mutant PML proteins containing either a Cys point mutation within the N-terminal RING finger domain or a small deletion (of positions 281 to 304) within the coiled-coil region did not localize to the PODs but instead gave a nuclear diffuse distribution, similar to that produced by intact PML in the presence of IE1. Endogenous PML also colocalized with IE1 in metaphase chromosomes in HCMV or recombinant adenovirus type 5-IE1-infected HF cells undergoing mitosis, implying that there may be a direct physical interaction between IE1 and PML. Indeed, a specific interaction between IE1 and PML was observed in a yeast two-hybrid assay, and the strength of this interaction was comparable to that of IE2 with the retinoblastoma protein. The RING finger mutant form of PML showed a threefold-lower interaction with IE1 in the yeast system, and deletion of the N-terminal RING finger domain of PML abolished the interaction. Consistent with the IFA results, a mutant IE1 protein that lacks the C-terminal acidic region was sufficient for interaction with PML in the yeast system. The two-hybrid interaction assay also showed that both the N-terminal RING finger domain and the intact coiled-coil region of PML are required cooperatively for efficient self-interactions involving dimerization or oligomerization. Furthermore, truncated or deleted GAL4/PML fusion proteins that retained the RING finger domain but lacked the intact coiled-coil region displayed an unmasked cryptic transactivator function in both yeast and mammalian cells, and the RING finger mutation abolished this transactivation property of PML. Therefore, we suggest that a direct interaction between IE1 and the N-terminal RING finger domain of PML may inhibit oligomerization and protein-protein complex formation by PML, leading to displacement of PML and IE1 from the PODs, and that this interaction may also modulate a putative conditional transactivator function of PML.
Several herpesvirus nuclear regulatory proteins expressed at immediate-early (IE) times after infection target to specific punctate subdomains in the nucleus of host cells, where they appear to exert as yet unknown roles in facilitating initial events in viral mRNA transcription and DNA replication. Interactions within the first few hours of infection between these viral regulatory proteins and appropriate cellular proteins present at these punctate loci, such as the PML tumor suppressor, may play a key enhancing (although not necessarily essential) role for increasing the efficiency of the productive lytic cycle processes. In this study, we evaluated the possibility of a direct interaction between the cytomegalovirus (CMV) IE1 protein and the cellular PML protein that might be responsible for both the targeting to and disruption of the PML oncogenic domains (PODs).
Human CMV (HCMV) typically causes asymptomatic infection in immunocompetent individuals. However, infection of newborns and of immunocompromised individuals, as well as reactivation from latent infection, can lead to severe disease complications and pathogenesis (7, 55). During permissive lytic HCMV infection, viral gene expression occurs in a three-step sequential fashion (IE, early, and late), in which both the IE proteins and virion factors are required for the subsequent efficient induction of the early and late genes (48, 63). Two IE nuclear phosphoproteins, IE1 (UL123, IE72 [72-kDa IE protein]) and IE2 (UL122, IE86), which are expressed from differentially spliced mRNA species generated from the major IE (MIE) locus (65, 66), are the first and most abundantly expressed HCMV IE gene products and are also the only viral proteins synthesized in some nonpermissive cell types (43).
The role of the IE2 (IE86) protein as an essential transcriptional transactivator and DNA binding repressor has been partially defined (11, 12, 30, 45, 49, 58–60). However, the role of IE1, which is important for efficient lytic cycle infection at low multiplicity of infection (MOI) but is not essential in cell culture at high MOI (26, 54), is poorly understood. One study has suggested that IE1 can transactivate the MIE promoter through upstream NFκB sites (10), but this has not been confirmed. The 491-amino-acid (aa) IE1 protein shares 87 aa at the N terminus with IE2 but is otherwise largely hydrophobic, except for a highly acidic Glu-rich C-terminal region. Unlike IE2 or the IE175 (ICP4), IE110 (ICP0), IE63 (ICP27), and IE68 (ICP22) nuclear regulatory proteins of herpes simplex virus (HSV), stable constitutive expression of IE1 in cell lines is fully compatible with long-term cell survival. Because the IE1 coding region displays selective CpG suppression and the IE1 protein preferentially associates with metaphase chromosomes, it has been suggested that like EBNA-1 of Epstein-Barr virus (EBV), it may potentially play a role in maintenance of the latent state of HCMV DNA (31, 44).
Recently, considerable attention has been focused on the fact that several key regulatory proteins encoded by DNA viruses target to a set of punctate subdomains within the nucleus at very early times after infection (reviewed in reference 16). These PML-containing subdomains (PODs or nuclear domain 10) are spherical structures with a size of 0.3 to 0.5 μm in which the PML RING finger protein surrounds an electron-dense core associated with the nuclear matrix (19, 39, 70). They appear to be dynamic structures that form between 10 and 20 distinct bodies distributed throughout the nucleus in most cell types (3, 53). Several cellular proteins including PML (19, 39, 70), SP100 (68), NDP55 (27), PIC 1 (5), and PLZF (38) have all been reported to be present in the PODs. Among them, the PML proto-oncogene was first identified as part of a fusion protein with the alpha retinoic acid receptor (RARα) that resulted from the t(15;17) translocation in acute promyelocytic leukemia (APL) (14, 25, 35, 36, 57). In the NB4 cell line derived from an APL tumor, the localization of PML and PML/RARα is changed from the normal punctate nuclear bodies to a nuclear and cytoplasmic micropunctate pattern. However, when NB4 cells are treated with retinoic acid (RA), the normal punctate PML distribution pattern is restored and the cells regain both normal cell growth controls and the ability to differentiate (19, 27, 39).
The crucial role of the dominant negative PML/RARα fusion protein in the pathogenesis of APL has been confirmed by the observation that PML/RARα transgenic mice develop a form of acute leukemia with a differentiation block at the promyelocyte stage (29). The PML punctate pattern is also regulated by cell cycle progression (19). Although little is known about the function of proteins present in the PODs, they appear to be involved in cell proliferation processes because PML acts as a growth and tumor suppressor when overexpressed (46, 50, 56), and its gene expression is up-regulated by alpha and gamma interferons (64). Also, PML has been suggested to have a transcriptional modulator function because the PML/RARα fusion protein shows altered transactivator or repressor properties (depending on the target gene tested) compared to RARα (14, 35).
In human HSV type 1 (HSV-1) infection, the viral regulatory protein IE110 (or ICP0), which is also a member of the RING finger protein family, transiently colocalizes with PML in the PODs and then apparently displaces PML, leading to complete loss of detectable PML indirect immunofluorescence assay (IFA) signals in the cell (20, 52, 53). Adenovirus type 5 (Ad5) infection causes a morphological change in the PODs from the spherical punctate structures to fibrous “track” structures, and some POD proteins relocalize into viral replication compartments (8, 15, 61). Furthermore, during Ad5 infection, at least three early proteins (E1A, E1b 55-kDa protein, and E4-ORF3 11-kDa protein) associate with the PML bodies, although the viral E4-ORF3 11-kDa protein alone is sufficient to induce reorganization of the PODs (8, 15). The nuclear antigen EBNA-5 of EBV stably colocalizes with the PODs in both EBV-infected B cells and cell lines (67), and large tumor antigen of simian virus 40 (SV40) is distributed adjacent to the PODs in some transfected cell types (8).
Several groups have shown that HCMV infection in permissive human fibroblasts in culture also causes displacement of PML from the PODs into a nuclear diffuse form (1, 37, 41). We also showed previously that the PODs are targeted by both the isolated IE1 and IE2 proteins of HCMV (1). However, HCMV IE1 only transiently colocalizes with PML in the PODs, and subsequently both IE1 and PML become distributed as nuclear diffuse forms. This contrasts with IE110 itself becoming punctate but with an accompanying complete loss of the PML signal as in HSV-1 infection, or the reorganization of the spherical PODs structures into fibrous track structures as seen in Ad5 infection, and suggests that each virus has developed different ways to interact with and disrupt or reorganize the PODs. The fact that the HCMV IE1 protein alone is sufficient for displacement and redistribution of PML in transiently transfected cells, as well as in cell lines stably expressing IE1 and in cells infected with a recombinant defective Ad5 vector expressing IE1 (Ad5-IE1) (1), led us to investigate the mechanism for targeting to and disruption of the PODs by IE1.
In this study, we used double-label IFA, cotransfection experiments, and yeast two-hybrid interaction assays to show that targeting to and disruption of the PODs by HCMV may be mediated through direct interaction of the IE1 protein with the N-terminal RING finger domain of PML. We also demonstrate that removal of the intact α-helical coiled-coil (dimerization) region of PML uncovers a cryptic activation domain and that this unmasked transactivator property of PML requires the N-terminal RING finger domain.
MATERIALS AND METHODS
Mammalian cell cultures and virus infection.
Permissive human fibroblast (HF) cells and Vero cells were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum. The HCMV(Towne) virus stock used was prepared as described by LaFemina et al. (44). Preparation of Ad5-IE1, referred to previously as RAd31 (71), was described previously (1). For infection, HF cells were seeded into four-well chamber slides (0.6 × 105/well), and the subconfluent cells were infected with the HCMV or Ad5-IE1 at an MOI of <1.0 PFU per cell. Input supernatant virus (25 μl) was adsorbed for 1.5 h at 37°C, and then the inoculum was replaced with 500 μl of fresh warmed medium at time zero (1). For the experiment using IE1 (exon 4)-deleted HCMV (26), samples of the defective mutant virus CR208 and its parent wild-type Towne virus were provided by Edward S. Mocarski (Stanford University, Stanford, Calif.). HF cells were seeded into four-well chamber slides and were infected with either CR208 or its parent Towne virus at an MOI of either 0.1 or 10 PFU per cell.
Mammalian expression plasmids.
Genomic versions of the HCMV(Towne) IE1 and IE2 coding regions were all derived from Escherichia coli plasmid pRL103, which contains the 20.8-kb HindIII C fragment encompassing the entire leftward-oriented MIE gene transcription unit (42). The parent effector plasmid pRL45 contains a 6.6-kb EcoRI-SalI subfragment expressing both IE1 and IE2 under the control of their natural transcriptional and splicing signals, whereas pMP17 and pMP18 express either IE1 or IE2 only in the same background. Further derivatives in plasmids pMP10, pMP11, and pRL55, expressing both mutant IE1 and wild-type IE2, and in plasmids pRL60, pRL61, and pRL74, expressing mutant IE1 only, were described previously (44, 60).
Plasmid pCMX-PML (35), expressing the intact human PML protein (560 aa) was provided by Ronald M. Evans (The Salk Institute, San Diego, Calif.); plasmid pGH623-5, encoding a version of PML containing point mutations within the RING finger domain (C88P89→S88R89), was described previously (13). To generate plasmids expressing PML(1-447) (PML containing aa 1 to 447) in pJHA286, PML(1-267) in pJHA287, PML(1-267, C88P89→S88R89) in pJHA288, PML(224-560) in pJHA289, PML(1-560, Δ281-304) in pJHA290, and PML(1-560, C88P89→S88R89 Δ281-304) in pJHA291, the NcoI-BamHI fragment containing the wild-type PML of pCMX-PML was replaced by the NcoI-BamHI fragments containing mutant PML from the yeast versions expressing GAL4-DB (GAL4 DNA-binding domain; aa 1 to 147)/PML fusion proteins in pJHA252, pJHA253, pJHA273, pJHA280, pEB1, and pEB2 (see below), respectively.
To construct a set of mammalian expression plasmids for GAL4-DB/PML fusion proteins, XhoI-BamHI fragments containing truncated GAL4-DB/PML fusions from the yeast versions in pJHA238, pJHA247, pJHA252, pJHA253, pJHA273, pJHA277, and pJHA250 (see below) were moved between the XhoI-BglII sites of a parent modified pSV2-GAL4-DB vector (pGH250) to generate pSV2-GAL4-DB/PML(1-560) in pJHA258, pSV2-GAL4-DB/PML(1-560, C88P89→S88R89) in pJHA259, pSV2-GAL4-DB/PML(1-447) in pJHA260, pSV2-GAL4-DB/PML(1-267) in pJHA261, pSV2-GAL4-DB/PML(1-267, C88P89→S88R89) in pJHA275, pSV2-GAL4-DB/PML(97-267) in pJHA278, and pSV2-GAL4-DB/PML(447-560) in pJHA263, respectively.
Transient DNA transfection and CAT assays.
In transfection experiments for IFA, Vero cells were seeded into two-well chamber slides (0.4 × 105/well) and DNA was introduced into the subconfluent cells for 48 h using the HEPES-buffered saline version of the calcium phosphate procedure described previously (58). In transfection experiments for chloramphenicol acetyltransferase (CAT) assays, the target control adenovirus E1b promoter (E1b-CAT) and the test reporter gene GAL45/E1b-CAT with five tandomly repeated 17-bp GAL4 binding sites added upstream were used (47). Vero cells were seeded into six-well plates (2 × 105/well), and DNA transfection, harvesting at 48 h, and the CAT assay were carried out as described previously (58). Percent conversion of [14C]chloramphenicol to acetylated forms was measured with an Instant Imager (Packard Instrument Company, Downer Grove, Ill.).
Antibodies and IFA.
Mouse monoclonal antibodies (MAbs) 6E1 and 12E2 against the IE1 (exon 4) and IE2 (exon 5), respectively, were obtained from Vancouver Biotech (Vancouver, B.C., Canada), and MAb CH810, which detects epitopes present in both IE1 and IE2 (exons 2 and 3), was purchased from Chemicon (Temecula, Calif.). Rabbit antipeptide polyclonal antibodies (PAbs) directed against amino acids at positions 484 to 498 (PML-C) or at positions 1 to 17 (PML-N) of the human PML oncoprotein were described elsewhere (1, 13).
For IFA, both virus-infected and DNA-transfected cells were fixed by either the methanol or the paraformaldehyde procedure. For the methanol procedure (used for metaphase chromosome analysis), the cells were washed in Tris-buffered saline (TBS), then permeabilized with absolute methanol at 20° for 10 min, and rehydrated in ice-cold TBS for 5 min. For the more usual standard paraformaldehyde procedure, the cells were washed in phosphate-buffered saline (PBS), fixed with 1% paraformaldehyde solution in PBS at 20°C for 5 min, and then permeabilized in ice-cold 0.2% Triton X-100 solution in PBS for 20 min. The cells were incubated with mouse MAbs at dilutions of 1:200-fold for 6E1, 12E2, and CH810, 1:1,000-fold for the PML-C antibody, and 1:500-fold for the PML-N antibody. The antibody incubations were carried out in TBS at 30°C for 1 h, followed by incubation with fluorescein isothiocyanate (FITC)-labeled goat anti-mouse immunoglobulin G (IgG) or by rhodamine-coupled goat anti-rabbit IgG antibody at 1:100-fold dilution at 37°C for 45 min. For double labeling, monoclonal and polyclonal antibodies were incubated together. Slides were screened and photographed with a 40× oil immersion objective on a Leitz Dialux 20EB epifluorescence microscope using Kodak T-Max P3200 film. For confocal microscopy, Noran OZ CLSM confocal microscope system with intervision software (Noran Inc., Madison, Wis.) was used.
In vitro transcription and translation.
Plasmid pCMX-PML was linearized downstream of the coding region by BamHI and in vitro transcribed by using the T7 polymerase plus mRNA capping kit from Stratagene. In vitro translation was carried out with rabbit reticulocyte lysates as specified by the manufacturer (Promega) and as described elsewhere (12).
Western blot assay.
Vero cells were transfected for 48 h with 3 μg of pCMX-PML plasmid DNA per well in a six-well plate. HF cells were infected with HCMV(Towne) at an MOI of 1.0 for 6 h in a 100-mm-diameter dish. Untransfected or transfected Vero cells and mock-infected or HCMV-infected HF cells were washed twice with cold PBS and lysed in ice-cold lysis buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1.0% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS]). Cell extracts were subjected to SDS-polyacrylamide gel electrophoresis through a 10% acrylamide gel followed by electroblotting onto nitrocellulose. The blots were blocked by incubation for 1 h at 20°C in 1× PBS containing 0.1% Tween 20 and 5% nonfat dry milk. The blots were then washed three more times for 10 min and incubated for 1.5 h at 20°C with either 1:1,000-diluted mouse MAb 5E10 against PML (provided by K. van der Krann; 66a), rabbit PAb PML-C diluted 1:2,000, or mouse MAb 6E1 directed against IE1 diluted 1:3,000. After three 10-min washes with PBS-Tween 20, the blots were incubated with horseradish peroxidase-conjugated goat anti-mouse or anti-rabbit IgG (Bio-Rad) for 1 h at 20°C. The blots were washed three times, and reacting protein bands were detected with an enhanced chemiluminescence system (Amersham RP2106) using Kodak XAR film.
Construction of plasmids for yeast analyses. (i) GAL4-DB fusions.
All GAL4-DB fusions for expression in yeast were generated in pAS1-CYH2 (2, 18). Plasmid pCJC442 expressing the GAL4-DB/IE1(1-491) fusion protein was generated by placing the BamHI fragment containing the entire IE1 cDNA from pCJC180 into pAS1-CYH2. Plasmid pJHA238 expressing the GAL4-DB/PML(1-560) fusion protein was constructed by subcloning of the NcoI-BamHI fragment containing the entire PML cDNA from pCMX-PML into pAS1-CYH2. Plasmids pYW18 containing the GAL4-DB/EBNA-1(1-641, Δ102-325) fusion protein (69) and pCJC420 containing GAL4-DB/IE2(290-579) (2) were described elsewhere. Control plasmids pRb2 expressing GAL4-DB/Rb(301-928), pSE1112 expressing GAL4-DB/SNF1, and pSE1111 containing GAL4-A/SNF4 (18) were provided by Stephen J. Elledge.
To generate plasmids expressing the GAL4-DB/mutant PML fusions, an NcoI-SmaI fragment (codons 1 to 447 in pJHA252), an NcoI-PvuII fragment (codons 1 to 267 in pJHA253), an NcoI-AvrII fragment (codons 1 to 96 in pJHA254), and an AvrII-PvuII fragment (codons 97 to 267 in pJHA277) from the parent plasmid pJHA238 were placed in frame behind the GAL4-DB of pAS1-CYH2. Plasmid pJHA250 expressing GAL4-DB/PML(448-560) was generated by an in-frame deletion between the NcoI and SmaI sites from pJHA238. To generate plasmid pJHA280 expressing GAL4-DB/PML(224-560), the NcoI-BamHI fragment containing PML codons 224 to 560 was PCR amplified (5′ primer [LGH3013], TAGGATCCATGGAGCTCAAGTGC GACATC; 3′ primer [LGH1737], CACGATGCACAGTTGAAG) from pJHA238 and placed into the NcoI and BamHI sites of pAS1-CYH2. In addition, plasmid pJHA247 expressing GAL4-DB/PML(1-560, C88P89→S88R89) was constructed by placing the NcoI-BamHI fragment from pGH623-5 (13) into pAS1-CYH2. Subsequently, plasmids pJHA273 expressing GAL4-DB/PML(1-267, C88P89→S88R89) and pJHA274 expressing GAL4-DB/PML(1-96, C88P89→S88R89) were generated from plasmid pJHA247 by replacing a restriction fragment containing the point mutations into pAS1-CYH2. To generate plasmids pEB1 expressing GAL4-DB/PML(1-560, Δ281-304) and pEB2 expressing GAL4-DB/PML(1-560, C88P89→S88R89 Δ281-304), the NcoI-KpnI fragment containing PML(1-304) from pJHA238 was replaced by the NcoI-KpnI fragments containing PML(1-280) PCR amplified from pJHA238 or containing PML(1-280, C88P89→S88R89) PCR amplified from pJHA247 (5′ primer [LGH 3026], GAAGATCTTCC ATGGAGCCTGCAC; 3′ primer [LGH 3027], GGGGTACCCGCGGATCAGCTCCT). Plasmid pJHA294 expressing GAL4-DB/PML(96-560, Δ281-304) was generated by deleting an NcoI-AvrII fragment from pEB1. Plasmids pLZ59 expressing GAL4-DB/110(104-240) and pLZ60 expressing GAL4-DB/110(104-240, C152P153→S152R153) were described elsewhere (72).
(ii) GAL4-A fusions.
All GAL4-A (GAL4 activation domain; aa 768 to 881) fusions were generated in pACTII (2, 18). Plasmids pJHA140 expressing GAL4-A/IE2(1-579) and pJHA239 expressing GAL4-A/IE1(1-491) were generated by placing the BglII fragment containing the entire IE2 cDNA from pJHA122 (2) and the BamHI fragment containing the entire IE1 cDNA from pCJC442, respectively, into pACTII.
To generate plasmids expressing the GAL4-A/mutant IE1 fusion proteins, an NcoI fragment (codons 1 to 231 in pJHA255), an NcoI-BamHI fragment (codons 232 to 491 in pJHA249), an NcoI-BamHI fragment containing a deletion between two EcoRV sites (codons 1 to 491 Δ132 to 274 in pJHA251), and an EcoRV fragment (codons 132 to 274 in pJHA257) from the parent plasmid pJHA239 were placed in frame behind the GAL4-A of pACTII. Plasmid pJHA300 encoding GAL4-A/IE1(1-346) was constructed by deleting a BglII fragment (codons 347 to 491) from the parent plasmid pJHA239.
For plasmids expressing GAL4-A/PML fusions, the NcoI-SalI fragments containing PML fragments from yeast version expressing GAL4-DB/PML fusions were moved into the NcoI-XhoI sites of pACTII to generate plasmids expressing GAL4-A/PML(1-560) in pJHA266, GAL4-A/PML(1-560, C88P89→S88R89) in pJHA267, GAL4-A/PML(1-447) in pJHA268, GAL4-A/PML(1-267) in pJHA269, GAL4-A/PML(1-96) in pJHA270, GAL4-A/PML(224-560) in pJHA281, GAL4-A/PML(447-560) in pJHA271, GAL4-A/PML(1-560, Δ281-304) in pEB5, and GAL4-A/PML(1-560, C88P89→S88R89 Δ281-304) in pEB6.
Yeast two-hybrid interaction assays.
Saccharomyces cerevisiae Y190 (MATa gal4Δ gal80Δ his3-Δ200 trp1-901 ade2-101 ura3-52 leu2-3,-112 URA3::GAL1-lacZ LYS2::GAL-HIS3 cyhR), a derivative of Y153 (18), was used in a two-hybrid system. Complete and synthetic media for yeast growth and the yeast transformation method were described elsewhere (62). Both a plasmid encoding the GAL4-DB fusion (Trp+) and a plasmid encoding the GAL4-A fusion (Leu+) were introduced into Y190 cells. The double transformants were selected in the plates lacking both Trp and Leu, and the production of β-galactosidase was assayed by both a 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) filter assay and a quantitative assay using o-nitrophenyl-β-d-galactopyranoside (ONPG) as described previously (2). For rapid in situ assays of lacZ expression from yeast colonies, an X-Gal filter assay was used and between 6 and 10 independent colonies of each type were screened simultaneously. The nitrocellulose filters were laid onto the plate and allowed to wet completely, then lifted off of the plate carefully to avoid smearing the colonies, and placed into liquid nitrogen to permeabilize the cells. After 10 s, the filters were removed from the liquid nitrogen and placed cell side up in a petri dish containing 3MM paper soaked with Z buffer (60 mM Na2HPO4, 40 mM Na2H2PO4 · 2H2O, 10 mM KCl, 1 mM MgSO4 · 7H2O, 50 mM β-mercaptoethanol) plus 1 mg of X-Gal per ml. The filters were then incubated at 30°C for appropriate times for development of a positive blue color. For quantitation of the β-galactosidase activity in yeast, 2-ml cultures were grown in the appropriate synthetic medium to an optical density at 600 nm of 2.0, then 0.4 ml of the culture was harvested, and the β-galactosidase activity within the cells was assayed by the standard method using ONPG after permeabilizing the cells with chloroform and SDS (27a). The unit of β-galactosidase was defined as 1,000(A420 − 1.75A550)/(A600 × t × v) (t, reaction time [minutes]; v, reaction volume [milliliters]).
RESULTS
An IE1-defective mutant virus does not disrupt PODs.
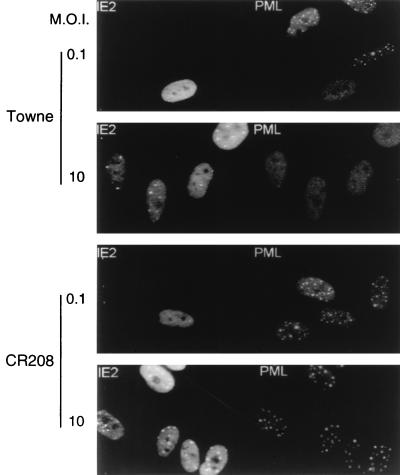
In permissive HF cells infected with wild-type HCMV(Towne), IE1 transiently targets to PODs and subsequently displaces the cellular PML protein from the PODs, with both proteins redistributing into a nuclear diffuse form (1). The IE1 protein alone is sufficient for this process in Vero cells transiently transfected with a plasmid expressing IE1, as well as in permissive U373 cell lines constitutively expressing IE1 and in HF cells infected with Ad5-IE1 (1). The specific requirement for IE1 in this process in HCMV-infected cells was further investigated by using a recently described IE1-defective mutant (26). The CR208 virus contains a deletion of the exon 4 segment of IE1 within the HCMV(Towne) background and fails to give efficient lytic cycle infection at low MOI, although it is able to grow relatively normally in cell culture at high MOI (26). HF cells were infected with either the parent wild-type Towne or CR208 virus for 6 h at both low and high MOI (i.e., 0.1 or 10 PFU/cell). The staining patterns of both IE2 and PML in infected cells were analyzed by double-label IFA with both mouse MAb 12E2 against IE2 and rabbit PAb PML-C. Cells infected with the Towne virus parent showed the typical mixture of diffuse plus punctate IE2 staining (1), together with the uniform dispersed nuclear pattern for PML at both low- and high-MOI infection (Fig. 1, upper two panels). However, in cells infected with the CR208 virus, an unaltered nuclear punctate staining pattern of PML was observed in all infected cells under both low- and high-MOI conditions and the IE2 punctate patterns were unaffected (Fig. 1, lower two panels). This result clearly demonstrates that displacement of PML from the PODs in HCMV-infected cells depends on expression of IE1 and that even at high MOI, the IE1-defective mutant virus does not affect the distribution pattern of PML.
FIG. 1.
Cells infected with an HCMV mutant that does not express IE1 fail to disperse the PML protein from punctate domains (PODs) into a nuclear diffuse pattern. The photographs show a comparison of PML staining patterns in HF cells infected with either HCMV(Towne) (upper two panels) or the CR208(ΔIE1) virus (lower two panels) at a low MOI of 0.1 and a high MOI of 10. At 6 h after infection, the cells were fixed with paraformaldehyde followed by double-label IFA. IE2 was detected with mouse MAb 12E2 and FITC-labeled anti-mouse IgG (left). PML was detected in the same fields with rabbit anti-PML PML-C PAb and rhodamine-coupled anti-rabbit IgG (right). Note the typical punctate POD patterns of PML in adjacent uninfected cells at low MOI.
PML protein levels in HF cells are not affected by HCMV infection.
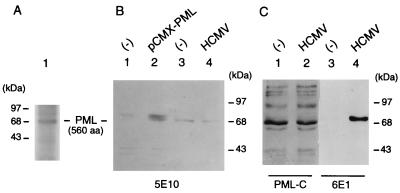
Because several variant PML proteins are expressed by C-terminal alternative splicing (25), we investigated whether either the total PML protein levels or the ratio of the different forms of PML were changed in HCMV-infected HF cells after being displaced from the PODs. To confirm the validity of our assay for PML, the protein produced by human cDNA plasmid pCMX-PML in total-cell extracts from DNA-transfected Vero cells was subjected to Western blot analysis using MAb 5E10 as a probe (66a). This antibody is expected to detect only the predominant nuclear isoforms of endogenous PML as well as the protein produced from pCMX-PML. The result revealed that indeed a novel PML protein with a molecular mass of 68 kDa was enriched in pCMX-PML-transfected cells (Fig. 2B, lane 2) compared to those in untransfected cells (Fig. 2B, lane 1). As a control, the 35S-labeled PML protein that was produced from pCMX-PML by in vitro transcription and translation also proved to migrate at 68 kDa (Fig. 2A). When total extracts prepared from mock-infected HF cells or cells infected with HCMV for 6 h were used, very similar levels of a predominant 68-kDa form of the protein were also detected from both extracts with MAb 5E10 (Fig. 2B, lanes 3 and 4). Furthermore, Western blot analysis with PAb PML-C directed against the C terminus of PML showed that the entire complex patterns of multiple isoforms of the PML protein detectable with this antibody were very similar in mock-infected (Fig. 2C, lane 1) and HCMV-infected (lane 2) HF cells, whereas with MAb 6E1 against IE1, the IE1 (72-kDa) protein was detected only in extracts from the HCMV-infected HF cells (lane 4). Overall, these results suggest that the total levels of PML protein in HCMV-infected cells remain unaffected after displacement from the PODs compared to those in uninfected cells, and that there are also no obvious modifications in the sizes or ratios of the different isoforms of PML present.
FIG. 2.
Comparison of the PML protein levels between mock-infected, transfected, and HCMV-infected cells. (A) Detection of the in vitro-translated pCMX-PML cDNA protein product (560 aa). [35S]Met-labeled PML proteins were synthesized in reticulocyte extracts from template RNA transcribed in vitro with T7 polymerase from BamHI-linearized plasmid pCMX-PML and analyzed on SDS–10% polyacrylamide gels. (B) Detection of nuclear forms of the PML protein by mouse MAb 5E10, using extracts prepared from pCMX-PML-transfected Vero cells and HCMV-infected HF cells. Total extracts (35 μg of protein) prepared from untransfected (lane 1) or pCMX-PML-transfected Vero cells (lane 2) and from mock-infected (lane 3) or HCMV-infected HF cells (lane 4) were electrophoretically fractionated on SDS–10% polyacrylamide gels, and Western blot analysis was performed by incubating the membrane with MAb 5E10 directed against PML. (C) Comparison of the levels of all forms of the PML protein detectable in mock-infected and HCMV-infected HF cells. Gel fractionated extracts from mock-infected (30 μg for lane 2 and 8 μg for lane 3) and HCMV-infected HF cells (30 μg for lane 2 and 8 μg for lane 4) were subjected to Western blot analysis using either rabbit PAb PML-C directed against the C terminus of PML (lanes 1 and 2) or mouse MAb 6E1 against IE1 (lanes 3 and 4).
Evaluation of the IE1 protein domains required for PML displacement and the nuclear diffuse distribution pattern.
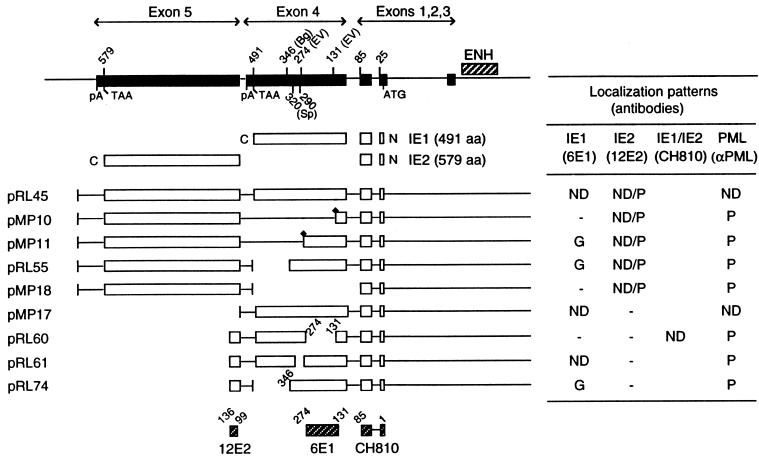
To determine whether specific regions of the IE1 protein are required for PML displacement, the PML staining patterns of Vero cells transfected with plasmids encoding mutant IE1 proteins were investigated by double-label IFA. A schematic representation describing the proteins encoded by each plasmid and a summary of the localization patterns of IE1 and IE2 in transfected Vero cells are presented in Fig. 3. In the initial experiments, we first mapped the epitope for mouse MAb 6E1 to a region from codons 131 to 274 in IE1 exon 4 and that for MAb 12E2 to a region from codons 99 to 136 in IE2 exon 5 (data not shown) (Fig. 3, bottom). The epitope for MAb CH810 has been previously mapped to within the exon 2 and 3 region common to both IE1 and IE2 (44).
FIG. 3.
Summary of the localization patterns of IE1, IE2, and PML in Vero cells transiently transfected with genomic plasmids expressing deleted versions of IE1. At the top is an illustration of the overlapping five exon structure (solid bar) of the MIE gene transcription unit in the inverted (i.e., viral) genomic orientation. The positions of key restriction sites used to generate the deleted or truncated versions of IE1 are indicated above the diagram. Bg, BglII; EV, EcoRV; Sp, SpeI. The enhancer/promoter region of the MIE locus (ENH; hatched bar) and the translation start (ATG) and termination (TAA) sites as well as polyadenylation sites (pA) are also indicated. Below is a comparison of the structures of the proteins encoded by the variant MIE expression gene plasmids used. Open bars represent coding regions, with gaps denoting in-frame deletions; diamonds indicate inserted triple-terminator oligonucleotides. The estimated map locations for the epitopes recognized by MAbs 6E1, 12E2, and CH810 are shown at the bottom (hatched bars). To detect IE1, IE2, and PML, FITC-labeled MAb 6E1 (for IE1), 12E2 (for IE2), and CH810 (for both IE1 and IE2) were used in double-label IFA experiments together with rhodamine-coupled rabbit PAb against PML. IFA patterns; ND, nuclear diffuse; P, punctate; ND/P, mixture of nuclear diffuse and punctate; G, nuclear granular structures.
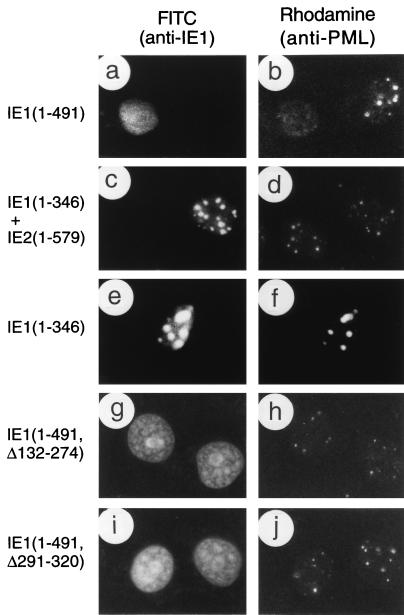
We have previously shown (1, 44) that a mutant IE1 protein truncated in the C-terminal region at codon 347 (encoded by plasmid pRL74) gave mostly large spherical globular or ring structures within the nucleus, rather than the normal nuclear diffuse distribution pattern seen with the transfected parent wild-type IE1 protein (pMP17). This aberrant distribution pattern of the C-terminally truncated mutant IE1 protein was observed even in the presence of IE2 (both are encoded by plasmid pRL55 [Fig. 4c]) and with another similar IE1 deletion protein truncated at codon 274 (pMP11 [not shown]). In contrast, two mutant IE1 proteins containing internal in-frame deletions between codons 132 and 274 (pRL60 [Fig. 4g]) or between codons 291 and 320 (pRL61 [Fig. 4i]) displayed a typical nuclear diffuse pattern. Since the 48-h transient expression assay does not detect the intermediate stage of transient targeting to PODs seen at the earliest times in infection, the latter result does not address whether these two deleted forms of IE1 have lost or retain the ability to target to the PODs.
FIG. 4.
Effects of wild-type or mutant IE1 expression on distribution of the endogenous PML proteins in transient expression assays. Vero cells were transfected with plasmids encoding various mutant IE1 proteins and fixed with paraformaldehyde followed by double-label IFA at 48 h after transfection. (a and b) Paired photographs of cells receiving plasmid pMP17 encoding wild-type IE1(1-491); (c and d) pRL55 encoding both IE1(1-346) and wild-type IE2; (e and f) pRL74 encoding IE1(1-346); (g and h) pRL60 encoding IE1(Δ132-274); (i and j) pRL61 encoding IE1(Δ291-320). (a, c, e, g, and i) Detection of IE1 with mouse MAb 6E1 (a, c, e, and i) or MAb CH810 (g) and FITC-labeled anti-mouse IgG. (b, d, f, h, and j) Detection of PML in the same fields with rabbit anti-PML-C PAb and rhodamine-coupled anti-rabbit IgG.
The patterns of displacement of PML from the PODs by mutant IE1 proteins was also examined by double-label IFA approaches using rhodamine-coupled rabbit anti-PML (PML-C) and FITC-labeled mouse anti-IE1 (6E1) antibodies. The results for nuclear diffuse or punctate PML signals are summarized in Fig. 3. As a positive control, the transfected wild-type IE1 protein completely displaced endogenous PML from the PODs into a nuclear diffuse form in all cells expressing IE1 in the presence or absence of IE2 (Fig. 4a and b). However, truncated IE1 proteins that lack the C-terminal regions between 346 to 491 (pRL55 and pRL74) (Fig. 4c to f) as well as between 131 to 491 (pMP10) and 274 to 491 (pMP11) (not shown) all failed to displace PML from the punctate bodies. Similarly, the internally deleted IE1 proteins that lacked only the regions from codons 132 to 274 (pRL60) or codons 290 to 320 (pRL61) also failed to displace PML from the punctate bodies, although in this case the two mutant IE1 proteins themselves showed a typical nuclear diffuse pattern (Fig. 4g to j). Importantly, the large aberrant IE1 globules seen with IE1(1-346) still touched or encompassed all of the much smaller endogenous PML punctate domains.
We conclude that parts of at least three segments of IE1 exon 4 including codons 132 to 274, 291 to 320, and beyond 346 are all required for normal PML displacement, whereas the region between codons 1 to 346 encompasses all domains necessary for targeting to the PODs. Since the internally deleted IE1(Δ132-274) and IE1(Δ291-320) versions neither form stable punctate structures nor displace PML, they appear likely to have lost the ability to target to the PODs. Unlike the RING finger mutants of HSV IE110, none of the HCMV IE1 mutants tested resulted in direct stable colocalization with PML in the PODs, although those lacking the C-terminal acidic domain did produce a stable interaction in larger aberrant structures that either surround or lie adjacent to endogenous PODs. Therefore, the latter mutant appears to uncouple the requirements for targeting to POD-related structures from actual displacement of PML.
Evaluation of the PML protein domains required for POD localization.
To study the mechanism of PML displacement to a nuclear diffuse form by HCMV IE1, we also needed to examine the domain requirements within PML for POD localization. PML contains an N-terminal RING finger domain, two adjacent Cys/His-rich B-box regions, a coiled-coil probable dimerization region, and a C-terminal Ser-rich domain with a possible nuclear localization signal (NLS) (see Fig. 6). Therefore, we obtained a mammalian cDNA expression plasmid encoding the intact human PML protein (in pCMX-PML) under the control of the HCMV MIE enhancer/promoter region (35) and generated several variants expressing mutant deleted or truncated PML proteins. Initially, we investigated the localization patterns of overexpressed PML proteins by IFA in transiently transfected Vero cells by using two antipeptide PAbs directed against the N (PML-N) and C (PML-C) termini of PML, although preliminary tests showed that unlike the PML-C antibody, the PML-N antibody did not detect the endogenous PML signals in Vero cells (data not shown). The exogenous wild-type PML protein alone formed five to six large globular bodies, which appeared to have coalesced with or incorporated all of the normal smaller endogenous PML containing PODs (Fig. 5a).
FIG. 6.
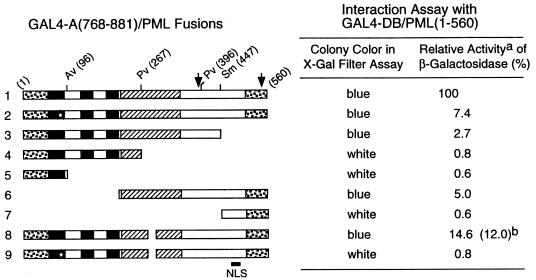
Effects of mutant PML proteins on self-interaction measured with a yeast two-hybrid assay. (Left) Diagram illustrating the structure of the GAL4-A/PML fusion proteins used. The two major translocation fusion points occurring within the PML protein (at positions 552 and 955) in PML/RARα fusions in APL are indicated by arrows. The proposed NLS (at positions 467 to 490) is indicated (36). The location of the paired RING finger point mutations (C88P89→S88R89) is indicated by a star. The amino acid positions of the restriction enzyme cleavage sites used to generate the GAL4-A/mutant PML fusion are indicated. Dotted bars, N-terminal Pro-rich domain and C-terminal Ser-rich domain; black bars, RING finger domain (left) and two adjacent Cys/His-rich domains (right); hatched bars, putative α-helical region. Av, AvrII; Pv, PvuII; Sm, SmaI. (Right) Qualitative and quantitative results of the yeast self-interaction assay. No detectable β-galactosidase activity was measured in Y190 cells transformed with the plasmid encoding the GAL4-DB/PML(1-560) fusion protein alone. Plasmids encoding a variety of GAL4-A/PML fusion proteins were then introduced together with GAL4-DB/PML(1-560) into Y190 cells. Transformants were selected on plates lacking Trp and Leu, and β-galactosidase activity of the transformants was measured as described in Materials and Methods. Lines: 1, GAL4-A/PML(1-560) in pJHA266; 2, GAL4-A/PML(1-560, C88P89→S88R89) (pJHA267); 3, GAL4-A/PML(1-447) (pJHA268); 4, GAL4-A/PML(1-267) (pJHA269); 5, GAL4-A/PML(1-96) (pJHA270); 6, GAL4-A/PML(224-560) (pJHA281); 7, GAL4-A/PML(447-560) (pJHA271); 8, GAL4-A/PML(1-560, Δ281-304) (pEB5); 9, GAL4-A/PML(1-560, C88P89→S88R89 Δ281-304) (pEB6). aMean values for β-galactosidase units for interaction between GAL4-DB/PML(1-560) and GAL4-A/PML(1-560) in duplicated assays are indicated as 100%. The relative activity of control SNF1/SNF4 interaction in the same assay was 148%. bSelf-interaction with the GAL4-DB/PML(96-560, Δ281-304) fusion protein encoded by plasmid pJHA294.
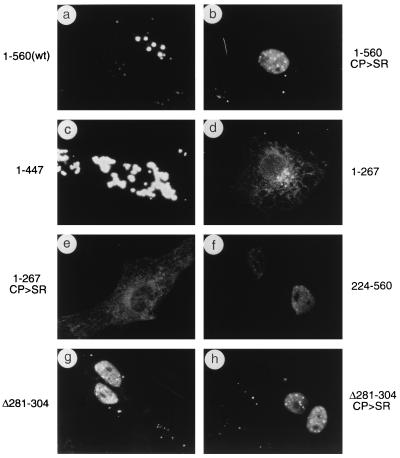
FIG. 5.
Localization patterns of wild-type and mutant PML in transfected Vero cells. Vero cells were transfected with plasmids expressing the wild-type (wt) PML(1-560) protein (pCMX-PML) (a), PML(1-560, C88P89→S88R89) (pGH623-5) (b), PML(1-447) (pJHA286) (c), PML(1-267) (pJHA287) (d), PML(1-267, C88P89→S88R89) (pJHA288) (e), PML(224-560) (pJHA289) (f), PML(1-560, Δ282-304) (pJHA290) (g), and PML(1-560, C88P89→S88R89 Δ282-304) (pJHA291) (h). The cells were fixed with paraformaldehyde at 48 h after transfection followed by IFA with anti-PML-C PAb (a, b, g, and h) or anti-PML-N PAb (c, d, e, and f) and rhodamine-coupled anti-rabbit IgG. Phase-contrast images confirming the nuclear plus cytoplasmic locations of PML(1-447) are available upon request.
Unlike the transfected wild-type PML protein, four of the five mutant PML proteins tested all gave nuclear diffuse distribution patterns of PML detectable by the PML-C antibody. These four mutants contained either point mutations (C88P89→S88R89) within the RING finger domain (Fig. 5b), a deletion of 25 aa (Δ281-304) within the coiled-coil region (Fig. 5g), both of the above-mentioned mutations (C88P89→S88R89 Δ281-304) (Fig. 5h), or an N-terminal truncation (Δ1-223) (Fig. 5f). Furthermore, none of these four mutant PML proteins interfered with endogenous PML, since normal-sized PODs were still detected within the nuclear diffuse background in those cells expressing mutant PML proteins. However, a C-terminal truncated PML(1-447) protein that lacked the proposed NLS formed large aggregated bodies that were detectable with the PML-N antibody and were predominantly cytoplasmic (Fig. 5c). Two smaller PML proteins, which were truncated at codon 267 with or without the RING finger mutation and lacked both the coiled-coil region and the NLS (Fig. 5d and e), were both distributed as diffuse forms throughout the cell as detected with the PML-N antibody. Therefore, these IFA results showed that the N-terminal RING finger domain and the intact coiled-coil region of PML are both required for punctate body formation (presumably through dimerization or oligomerization and other protein-protein interactions) and that the putative NLS or some other C-terminal feature is also needed for efficient nuclear POD localization.
Evaluation of the PML protein domains required for self-interaction.
Because both the RING finger motif and the coiled-coil motif of PML are believed to be involved in protein-protein interactions including self-interaction, we investigated the ability of the mutant PML proteins that were used for IFA experiments to self-interact in a yeast two-hybrid genetic assay. Various plasmids encoding GAL4-A/PML fusion proteins were introduced into yeast Y190 cells together with a plasmid encoding GAL4-DB/PML(1-560), and the ability to self-interact was tested in both an initial X-Gal filter color assay and a quantitative ONPG assay. In control experiments, yeast cells that received only a single plasmid encoding each of the GAL4-DB/PML fusion proteins alone or each of the GAL4-A/PML fusion proteins alone did not activate the target UASGal/GAL1-lacZ reporter gene (data not shown), whereas cells that received both wild-type plasmids expressing GAL4-DB/PML(1-560) and GAL4-A/PML(1-560) produced very strong β-galactosidase activity (Fig. 6, line 1). Three deleted PML fusion proteins, GAL4-A/PML(1-267), GAL4-A/PML(1-96), and GAL4-A/PML(447-560), that each lacked the entire coiled-coil region did not gave any significant interaction (less than 1% of the wild-type level) with the intact GAL4-DB/PML(1-560) fusion protein (lines 4, 5, and 7), suggesting that the coiled-coil region is probably essential for self-interactions. However, three mutant PML fusion proteins, one containing only a C88P89→S88R89 change within the RING finger domain (line 2), another representing the C terminus and coiled-coil region only (codons 224 to 560) (line 6), and a third containing a 25-amino-acid deletion within the coiled-coil region (Δ281-304) (line 8), retained only 7.4, 5, and 14.6%, respectively, of wild-type interaction activity with the GAL4-DB/PML(1-560) fusion protein. Furthermore, a double-mutant PML fusion protein containing both the C88P89→S88R89 change and the coiled-coil domain deletion (Δ281-304) completely abolished the ability to interact with the wild-type protein (line 9). These results demonstrate that both the N-terminal RING finger domain and the coiled-coil region of PML are required to be intact for efficient self-interaction.
Interestingly, when we characterized the coiled-coil deletion PML(Δ281-304) protein in a homodimerization assay with itself, the GAL4-A/PML(1-560, Δ281-304) fusion protein was still able to interact (at 12% efficiency) with the GAL4-DB/PML(96-560, Δ281-304) fusion protein (line 8), suggesting that the small coiled-coil region deletion between codons 281 and 304 does not abolish all self-interaction functions of the protein. Nevertheless, this mutant PML(Δ281-304) protein does not appear to retain the ability to assemble normal protein-protein complexes because of its failure to be incorporated into PODs (Fig. 5g). Taken together with the IFA results, the lack of efficient self-interaction (or oligomerization) by several distinct mutant PML proteins appears to correlate with the failure of POD localization (seen in Fig. 5). Furthermore, the C-terminal truncated PML fusion protein GAL4-A/PML(1-447), although it retained both an intact N-terminal RING finger domain and the coiled-coil region, also showed only 2.7% self-interaction activity (Fig. 6, line 3). Perhaps the property of strong self-interaction (and aggregation) in the cytoplasm observed with the nonfusion version (Fig. 5c) also prevents this version of the GAL4-fusion PML protein from undergoing efficient nuclear transport in yeast.
Coexpression of IE1 and PML also leads to an altered diffuse distribution of exogenous PML.
We have shown above that expression of wild-type IE1 alone causes displacement of endogenous PML from the PODs and leads to the subsequence redistribution of PML together with IE1 in a nuclear diffuse form. This process occurs in DNA-transfected HF or Vero cells, in U373 astrocytoma or Vero cell lines constitutively expressing IE1, and in HF cells infected with recombinant adenovirus (Ad5-IE1) (1). Furthermore, the two events correlate to such a high degree that in our previous studies of HCMV-infected HF cells at different times between 2 and 12 h after infection, all 110 cells scored that displayed punctate IE1 also had fully colocalized punctate PML, whereas all 390 cells scored that displayed diffuse IE1 also had fully diffuse PML patterns. This was the case irrespective of whether rabbit PAb PML-C or mouse MAb 5E10 was used to detect PML and whether rabbit anti-IE1 PAb or mouse MAb CH810 or 6E1 was used to detect IE1. To examine this phenomenon further, we cotransfected Vero cells with the two plasmids encoding intact PML and wild-type IE1 together. Nevertheless, despite the large globular pattern of overexpressed PML, the results again showed that the presence of IE1 changed the PML distribution to a uniform nuclear diffuse pattern in all coexpressing cells (Fig. 7a and b).
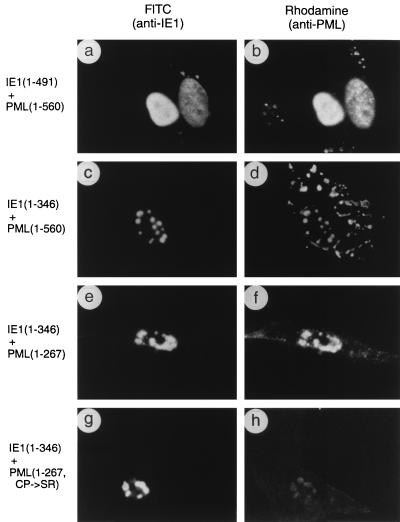
FIG. 7.
Effects of wild-type or mutant IE1 expression on distribution of the PML proteins in cotransfection assays. Vero cells were cotransfected with the following plasmid pairs: (a and b) pMP18 encoding wild-type IE1(1-491) and pCMX-PML encoding intact PML(1-560); (c and d) pJHA300 encoding IE1(1-346) and pCMX-PML; (e and f) pJHA300 and pJHA261 encoding mutant PML(1-267); (g and h) pJHA300 and pJHA275 encoding mutant PML(1-267, C88P89→S88R89). At 48 h after transfection, cells were fixed with paraformaldehyde followed by double-label IFA. (a, c, e, and g) Detection of IE1 with mouse MAb 6E1 and FITC-labeled anti-mouse IgG. (b, d, f, and h) Detection of PML in the same fields with rabbit anti-PML PML-C (b and d) or PML-N (f and h) PAb and rhodamine-coupled anti-rabbit IgG. Phase-contrast images confirming the nuclear plus cytoplasmic locations of PML(1-560) in the presence of IE1(1-346) are available upon request.
The targeting of the mutant protein IE1(1-346) to POD-related structures without displacement of PML seen in Fig. 4 was also further investigated in cotransfection assays. The IFA result in cells expressing both mutant IE1(1-346) and intact PML showed that IE1(1-346) colocalized with overexpressed PML in large nuclear POD-like structures, although PML signals were also detected as cytoplasmic bodies (Fig. 7c and d). This result reinforces our observation that the mutant IE1 protein lacking the C-terminal acidic region still targets to PODs but fails to displace PML from the PODs.
We showed above that the N-terminal segment of PML(1-267) was distributed as a diffuse form throughout both the nucleus and cytoplasm (Fig. 5d). However, when we investigated the effect of IE1(1-346) on the distribution pattern of PML(1-267), a strong association of PML(1-267) with IE1(1-346) was found in the nucleus in cotransfected cells (Fig. 7e and f). In contrast, when cells were cotransfected with both IE1(1-346) and PML(1-267, C88P89→S88R89), only a very weak association of both proteins was detected (Fig. 7g and h). These results again suggest that IE1 might interact directly with PML, that the intact RING finger domain of PML is required for this interaction, and that the Glu-rich acidic terminus of IE1 is necessary or responsible for displacement from the PODs.
PML associates together with IE1 in metaphase chromosomes in some infected HF cells.
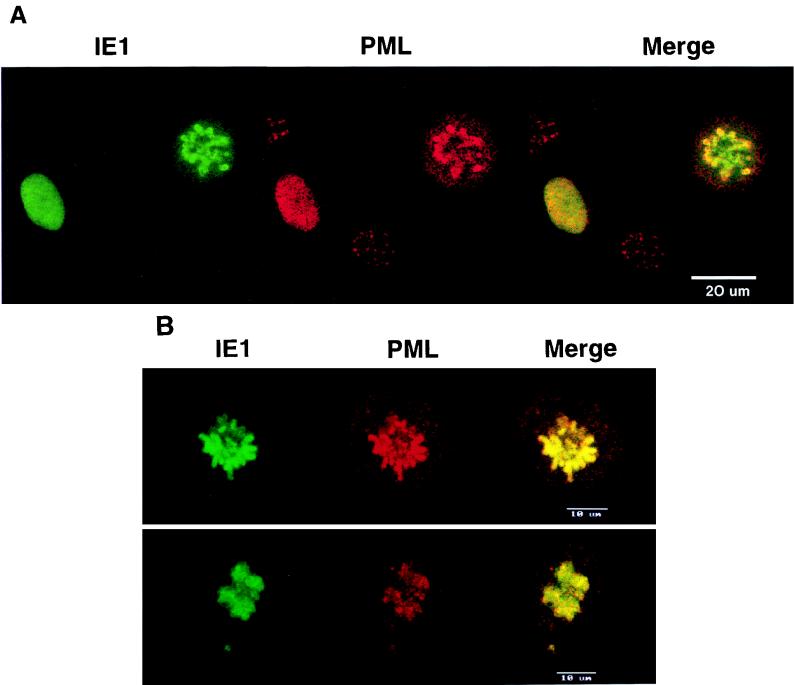
The PML staining pattern alters with cell cycle progression in uninfected cells, displaying the typical nuclear body staining in G1 phase but changing to multiple smaller dots in late S phase and almost disappearing briefly in M phase (16, 40). We have previously shown that IE1 associates with metaphase chromosomes after a methanol permeabilization procedure in those 1 to 2% of transiently DNA-transfected cells, or of constitutively IE1-expressing cells in stable cell lines, that are undergoing mitosis (44). This is also the case in the occasional mitotic cells found during HCMV infection in both permissive and nonpermissive cell types. Because of the precise correlation between displacement of PML from the PODs into a nuclear diffuse form and the apparently simultaneous change in IE1 distribution from PODs to a nuclear diffuse pattern in individual HCMV-infected cells (1), we also investigated whether the distribution pattern of the two proteins again correlated in those cells showing an association of IE1 with metaphase chromosomes. When HF cells were infected with HCMV(Towne) at an MOI of 0.5 and stained at 72 h after infection, nearly 4% of the infected cells showed a metaphase chromosome-associated IFA pattern of IE1 after methanol treatment, whereas the nonmitotic cells gave the typical uniform nuclear diffuse pattern (Fig. 8A, green fluorescence). Double-label IFA of these cells showed that PML also associated with (or precipitated onto) metaphase chromosomes together with IE1 in all of these same cells, whereas infected cells with a nuclear diffuse IE1 staining pattern always gave a nuclear diffuse PML staining pattern (Fig. 8A). Similar results were obtained for HF cells infected with Ad5-IE1 that were undergoing mitosis (Fig. 8B). Importantly, PML was never seen to be associated with metaphase chromosomes after methanol treatment in mitotic cells from uninfected cultures or in those infected with Ad5 expressing β-galactosidase. These results strongly imply that PML may be physically associated with IE1 after displacement from the PODs during HCMV infection.
FIG. 8.
Confocal microscopy images demonstrating colocalization between PML and IE1 in metaphase chromosomes in HCMV- or Ad5-IE1-infected cells undergoing mitosis. HF cells were infected with either HCMV(Towne) (A) or Ad5-IE1 (B) at an MOI of 0.5. At 72 h after infection, the cells were permeabilized in absolute methanol at −20°C followed by double-label IFA. Left-hand panels (green fluorescence), detection of IE1 with mouse MAb 6E1 and FITC-labeled anti-mouse IgG; center panels (red fluorescence), detection of PML in the same fields with rabbit anti-PML-C PAb and rhodamine-coupled anti-rabbit IgG; Right-hand panels (yellow merge fluorescence), confocal images from each fluorochrome were recorded and superimposed to demonstrate colocalization. Two representative cells are shown in the upper and lower sections of panel B.
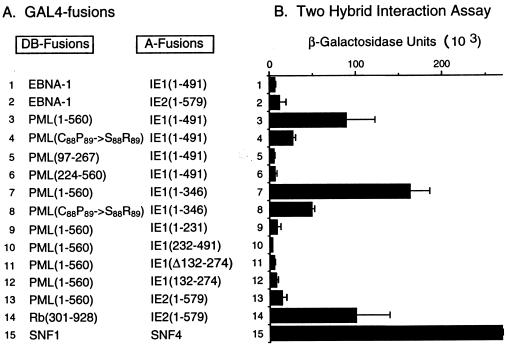
Specific interactions between IE1 with PML in two-hybrid assays in yeast.
The observation that the distribution of PML in infected cells precisely correlated with that of IE1, when the latter was either transiently associated with PODs or present as nuclear diffuse forms in most cells after displacement from the PODs, as well as in a form associated with metaphase chromosomes in the small percentage of mitotic cells, led us to investigate the possibility of direct protein-protein interactions between IE1 and PML. To do so, we used a standard yeast two-hybrid interaction assay (23). The results of representative experiments are shown in Fig. 9. As negative controls, when yeast cells were transformed only with the single plasmids encoding each of the GAL4-A/IE1 fusions alone, or with the GAL4-DB fusions with intact PML(1-560) or deleted PML(97-267) or PML(224-560) alone, none of these proteins activated the target UASGal/GAL1-lacZ reporter gene (data not shown). However, when plasmids containing the intact GAL4-DB fusion and the intact GAL4-A fusion proteins were present together in the same yeast cells, they gave blue color development in an X-Gal filter assay (data not shown) and produced moderate levels of β-galactosidase activity that were 20-fold higher than in negative interaction controls (e.g., the combination of EBV EBNA-1 with IE1 or IE2) (Fig. 9; compare line 3 with lines 1 and 2). Interestingly, the version of PML containing point mutations within the RING finger domain (C88P89→S88R89) exhibited an interaction affinity 3 fold lower than that of the wild-type protein (line 4). When we investigated the ability of PML fusion proteins that either retained just the Cys/His-rich domain (codons 97 to 267) or lacked the entire N-terminal domain (Δ1-223) to interact with IE1, neither was able to do so (lines 5 and 6), indicating that probably either the RING finger domain alone or both the RING finger and adjacent Cys/His-rich domains may be essential for the interaction with IE1. Note that truncated GAL4-DB domain fusions containing the intact N terminus of PML could not be evaluated in this assay because of their constitutive activator properties (see below). However, experiments set up in the reverse two-hybrid format eliminated this complication and showed that the GAL4-DB/IE1(1-491) fusion protein was able to interact equally efficiently with both GAL4-A/PML(1-560) and GAL-4-A/PML(1-267) in X-Gal filter assays (not shown), although the level of interactions with this version of the assay were weaker than with the original version of the assay. As a negative control, the GAL4-DB/IE1(1-491) fusion protein alone did not activate the target reporter gene. Therefore, the N-terminal region of PML containing the RING finger domain may be sufficient for interaction with IE1.
FIG. 9.
Yeast two-hybrid assays demonstrating specific interactions between IE1 and PML. The GAL4-DB fusion proteins used were GAL4-DB/EBNA-1(1-641, Δ102-325) (encoded by plasmid pYW18), GAL4-DB/PML(1-560) (pJHA238), GAL4-DB/PML(1-560, C88P89→S88R89) (pJHA247), GAL4-DB/PML(97-267) (pJHA277), GAL4-DB/PML(224-560) (pJHA280), GAL4-DB/SNF1 (pSE1112), and GAL4-DB/Rb(301-928) (pRb2). The GAL4-A fusion proteins used were GAL4-A/IE1(1-491) (in pJHA239), GAL4-A/IE2(1-579) (pJHA140), GAL4-A/IE1(1-346) (pJHA300), GAL4-A/IE1(1-231) (pJHA255), GAL4-A/IE1(232-491) (pJHA249), GAL4-A/IE1(Δ132-274) (pJHA251), GAL4-A/IE1(132-274) (pJHA257), and GAL4-A/SNF4 (pSE1111). Paired plasmids encoding the GAL4-DB and GAL4-A fusion proteins were introduced together into Y190 cells. Transformants were selected on plates lacking Trp and Leu, and β-galactosidase activity of the transformants were measured. Mean values for β-galactosidase units in duplicated assays are denoted by the black bars with error range indicated.
To determine whether any specific isolated subfragments of IE1 might be capable of interaction with PML, we generated a series of yeast expression plasmids encoding truncated GAL4-A/IE1 fusions and investigated the abilities of these mutant IE1 proteins to bind to the intact GAL4-DB/PML(1-560) fusion protein. The results showed that the GAL4-A/IE1(1-346) fusion protein lacking the acidic C-terminal region of IE1 was able to interact strongly with the intact GAL4-DB/PML(1-560) protein (Fig. 9, line 7) but did so with threefold-reduced efficiency with the GAL4-DB/PML(C88P89→S88R89) mutant (line 8). However, the other fusion proteins used in this experiment, GAL4-A/IE1(1-231), GAL4-A/IE1(232-491), GAL4-A/IE1 (Δ132-274), and GAL4-A/IE1(132-274), all failed to interact with GAL4-DB/PML(1-560) (Fig. 9, lines 9 to 12). This set of fusion proteins contained a hemagglutinin epitope tag (inserted behind the GAL4-A on plasmid pACTII), which allowed us to confirm by using Western blot analysis with anti-hemagglutinin antibody (data not shown) that these GAL4-A/mutant IE1 fusion proteins were of the expected sizes and all gave comparable levels of expression relative to the wild-type fusion protein in the yeast cells. Therefore, the isolated segment of IE1(1-346) but not smaller fragments is sufficient for interaction with PML in yeast, which is fully consistent with the results obtained above with similar mutant IE1 proteins by IFA studies in transfected Vero cells (Fig. 4 and 7).
Importantly, PML did not demonstrate any significant interaction with IE2 in the same assay (Fig. 9, line 13), and IE1 does not interact with itself in such assays (data not shown). Compared to other protein-protein interactions that were used as positive controls, the strength of the interaction of the intact IE1 and PML fusion proteins (90 × 10−3 U) was 20% to 40% as efficient as either the self-dimerization interaction of the intact PML protein in the same yeast cell background (516 × 10−3 U [data not shown]) or the SNF1 interaction with SNF4 (line 15), but was comparable to the affinity of the IE2 interaction with Rb (100 × 10−3 U) (line 14) (27b). Therefore, consistent with our IFA data, these genetic assays using a yeast two-hybrid system identified a specific protein-protein interaction between IE1 and PML that appears to be mediated through the RING finger domain of PML and does not require the C-terminal acidic domain of IE1.
Unmasking of a cryptic transactivator domain in PML in yeast GAL4 fusion assay.
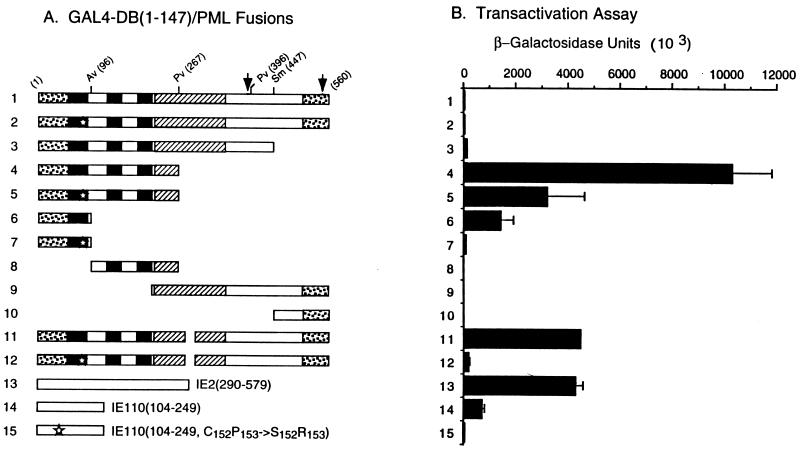
In APL cells, the t(15;17) chromosome translocation produces C-terminally truncated aberrant PML proteins as well as the PML/RARα fusion proteins (57). The N-terminal portion of PML has been suggested to have some form of transcriptional modulator function because the PML/RARα fusion causes an alteration in the RA-dependent transactivation function of the RARα (14, 35). In our initial study to investigate possible biological functions of the interaction of IE1 with the RING finger domain of PML (in addition to the disruption of PODs), we generated a set of yeast expression plasmids encoding C-terminal truncated GAL4-DB/PML fusion proteins (illustrated in Fig. 10A) but discovered that several of these forms gave a direct transactivator function in yeast (Fig. 10B). Neither the intact GAL4-DB/PML(1-560) protein nor GAL4-DB/PML(1-447) containing both the N-terminal Cys-rich domains and the intact coiled-coil region of PML had any activity in this assay (lines 1 and 3). However, the shorter form GAL4-DB/PML(1-267), which contains both the intact RING finger domain and the adjacent Cys/His-rich domains but lacks the coiled-coil region, showed positive transactivation on the target UASGal/GAL1-lacZ reporter gene (line 4). Similarly, a GAL4-DB/PML(1-96) protein containing the intact RING finger domain only (line 6) also showed transactivation properties, although with a sixfold reduction in activity compared to GAL4-DB/PML(1-267). In contrast, the GAL4-DB/PML(96-267) (line 8), GAL4-DB/PML(224-560) (line 9), and GAL4-DB/PML(447-560) (line 10) fusion proteins did not show any transactivator properties.
FIG. 10.
Unmasking of a cryptic transactivator property within the N-terminal domain of PML proteins in yeast cells. (A) Diagram illustrating the structure of the GAL4-DB/PML (lines 1 to 12) and other control GAL4-DB/IE2 (line 13) or GAL4-DB/IE110 (lines 14 and 15) fusion proteins used. See Fig. 6 for details of PML protein features. (B) β-Galactosidase activity measured in Y190 cells transformed with plasmids encoding the GAL4-DB/PML or other control fusion proteins. Single-transformants were selected on plates lacking Trp, and β-galactosidase activity of the transformants was measured. Lines: 1, GAL4-DB/PML(1-560) in pJHA238; 2, GAL4-DB/PML(1-560, C88P89→S88R89) (pJHA247); 3, GAL4-DB/PML(1-447) (pJHA252); 4, GAL4-DB/PML(1-267) (pJHA253); 5, GAL4-DB/PML(1-267, C88P89→S88R89) (pJHA273); 6, GAL4-DB/PML(1-96) (pJHA254); 7, GAL4-DB/PML(1-96, C88P89→S88R89) (pJHA274); 8, GAL4-DB/PML96-267) (pJHA277); 9, GAL4-DB/PML(224-560) (pJHA280); 10, GAL4-DB/PML(447-560) (pJHA250); 11, GAL4-DB/PML(1-560, Δ281-304) (pEB1); 12, GAL4-DB/PML(1-560, C88P89→S88R89 Δ281-304) (pEB2); 13, GAL4-DB/IE2(290-579) (pCJC420); 14, GAL4-DB/IE110(104-240) (pLZ59); 15, GAL4-DB/IE110(104-240, C152P153→S152R153) (pLZ60).
Quantitatively, the level of transactivation by GAL4-DB/PML(1-267) on the yeast target reporter gene was twofold higher than that of GAL4-DB/IE2(290-579) (Fig. 10, line 13), which contains the C-terminal activator domain of HCMV IE2 (2, 59). Since the GAL4-DB/PML(1-267) protein lacks most of the coiled-coil region of PML that is required for dimerization and POD localization, we also examined whether a nearly intact GAL4-DB/PML(1-560, Δ281-304) fusion protein containing just a small 25-aa deletion within the coiled-coil region might also unmask the cryptic transactivator properties in the same yeast one-hybrid assay. Indeed, this small deletion in the PML dimerization region also revealed transactivator properties comparable to those of the isolated N terminus (line 11). Overall these results demonstrate that N-terminal PML fragments containing both the RING finger and the adjacent Cys/His-rich domains, or less efficiently just the RING finger domain alone, display cryptic transactivator functions that are not evident in the intact PML protein. Furthermore, the coiled-coil domain may be involved in blocking this function of the N-terminal RING finger domain region of PML.
The RING finger domain of PML plays a critical role in the unmasked transactivator property of PML.
To investigate the role of the RING finger domain itself in transactivation by PML, a mutation of a Cys residue (C88P89→S88R89) within the RING finger domain was generated in each of the GAL4-DB/ PML(1-267), GAL4-DB/PML(1-96), and GAL4-DB/PML (1-560, Δ281-304) backgrounds. Interestingly, GAL4-DB/PML(1-267, C88P89→S88R89) (Fig. 10, line 5) showed a 3-fold lower level of transactivation compared to the parent GAL4-DB/PML(1-267) protein, whereas both GAL4-DB/PML(1-96, C88P89→S88R89) (line 7) and GAL4-DB/PML(1-560, C88P89→S88R89 Δ281-304) (line 12) displayed 100- and 60-fold reductions, respectively, and were almost devoid of any remaining transactivator function. A similar pattern of both a significant transactivation function with an isolated intact RING finger domain fusion protein from HSV IE110 [in GAL4-DB/IE110(104-240) (line 14)] but an almost complete loss of this activity by mutation of a Cys residue within the RING finger domain [in GAL4-DB/IE110(104-240), C152P153→S152R153) (line 15)] was observed in control experiments. These results demonstrate that the RING finger domain itself plays a major role in the unmasked transactivation activity of the C-terminal truncated PML proteins and that such properties may be a common feature of RING finger domains.
Masking of the cryptic N-terminal transactivator activity of PML is also evident in mammalian cells.
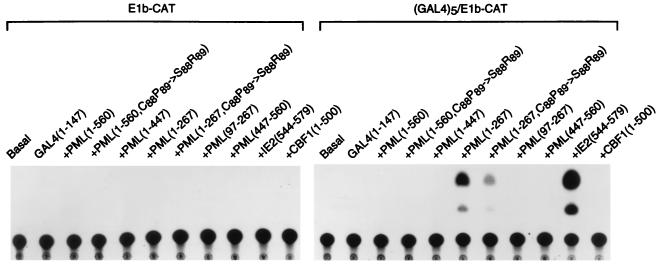
To confirm that unmasking of a transactivator function within the N terminus of the truncated PML protein also occurred in mammalian cells, the GAL4-DB/mutant PML proteins were also placed into an SV40 enhancer-derived vector and expressed in Vero cells by transient DNA transfection together with the E1b-CAT or GAL45/E1b-CAT target reporter plasmids. The results of the CAT assay revealed that GAL4-DB/PML(1-267) was able to activate the GAL45/E1b-CAT target reporter gene 22-fold but failed to do so on the E1b-CAT control (Fig. 11, lanes 17 and 6). Consistent with the results obtained with the yeast system, the RING finger domain mutant GAL4-DB/PML(1-267, C88P89→S88R89) produced a threefold-lower level of transactivation compared to its parent version, and none of the other GAL4-DB domain fusion proteins containing segments of PML, including a version of GAL4-DB/PML(97-267) with the RING finger domain omitted (lane 19), had any activity. Quantitatively, the level of transactivation by GAL4-DB/PML(1-267) in this assay was at least 20-fold greater than that obtained with the intact PML fusion protein [GAL4-DB/PML(1-560)], although it was 4-fold less than that obtained in parallel assays with the HCMV IE2 C-terminal activator domain (codons 544 to 579) as a GAL4-DB fusion protein (59) (lane 21). Importantly, the masking effect of using the intact PML protein in GAL4-DB/PML(1-560) or of just adding the adjacent coiled-coil domain to the N-terminal activator domain in GAL4-DB/PML(1-447) again blocked all transactivator activity (lanes 14 and 16). These results demonstrate that the 267-amino-acid N-terminal PML protein fragment containing just the RING finger and adjacent Cys/His-rich motifs behaves as a functional transactivator in domain swap assays in both yeast and mammalian cells, but that addition of the adjacent α-helical coiled-coil region that is thought to be required for dimerization masks this activity within the wild-type PML protein.
FIG. 11.
Assays for activator domains in PML using GAL4 fusion proteins expressed in transient assays in mammalian cells. Vero cells were cotransfected with plasmids containing either the parent E1b-CAT or GAL45/E1b-CAT reporter target gene together with plasmids encoding the GAL4-DB (pGH250) or GAL4-DB/PML fusion proteins. A representative autoradiograph of a transient CAT assay is shown. The basal samples show the levels of E1b-CAT or GAL45/E1b-CAT expression in the presence of vector plasmid DNA only. GAL4/IE2(544-579) in plasmid pMP54a containing the C-terminal IE2 transactivator domain described previously (59) and GAL4/CBF1(1-500) in plasmid pJH93 were used as positive and negative controls, respectively (32). GAL4-DB/PML fusion proteins tested were GAL4-DB/PML(1-560) in pJHA258, GAL4-DB/PML(1-560, C88P89→S88R89) in pJHA259, GAL4-DB/PML(1-447) in pJHA260, GAL4-DB/PML(1-267) in pJHA261, GAL4-DB/PML(1-267, C88P89→S88R89) in pJHA275, GAL4-DB/PML(97-267) in pJHA278, and GAL4-DB/PML(447-560) in pJHA263.
DISCUSSION
The role of the PML-containing nuclear bodies (PODs or nuclear domain 10) in cell growth control and in very early events in DNA virus infection appears likely to prove to be very important. Maul and colleagues have demonstrated that input herpesvirus, adenovirus, and SV40 DNA genomes, HCMV IE transcription, and even initial HSV and adenovirus DNA synthesis are all localized in close proximity to POD structures (33, 34, 51). Furthermore, they have also pointed out that PODs and HCMV IE transcription domains often also lie directly adjacent to or touching the RNP containing SC35-positive spliceosome domains (3). Herpesvirus IE transcription processes are very similar to those of rapidly induced or activated cellular genes, including being under the control of strong inducible enhancers and often (unlike delayed-early or late genes) yielding spliced mRNA products. Therefore, it would not be surprising if at IE stages of infection the DNA viruses all utilize preexisting cellular machinery that might be localized to a limited number of highly active chromosomal sites. At least some PODs represent the cellular dense bodies detectable by electron microscopy in which a central matrix-associated core, including chromatin-like nucleofibrils, is decorated by a layer of PML and other POD proteins (19, 39, 70).
The apparent replacement or displacement of PML and its associated proteins from the PODs by the HSV IE110 (ICP0) protein or the CMV IE1 (IE72) protein could represent a way to increase the efficiency of IE transcription; alternatively, at later times it could either be part of the process of disrupting cellular transcription controls or represent a way to recruit POD factors to other more dispersed sites for delayed-early phase transcription events, or both. However, although the suspicion is increasing that these phenomena involve transcriptional events and factors, there is no direct evidence as yet to pinpoint such a mechanism or to exclude other processes as well. Because of the RA-reversible correlation between disruption of PODs and the transformed or myeloid cell phenotype of NB4 cells (19, 27, 39), the dispersion or loss of PODs in M-phase cells (16, 19), and the disruption of PODs within 2 h of adenovirus, HSV, or HCMV infection, it seems clear that protein-protein interactions and protein complex aggregation and disaggregation are central events in these processes.
Our results confirm and extend previous suggestions that although both IE1 and IE2 independently target initially to the PODs, expression of the HCMV IE1 protein only and not IE2 leads to displacement of the POD proteins (1, 41). This is also unlike the IE110 and IE68 proteins of HSV, which both also very efficiently displace PML from PODs but themselves end up stably-associated with either of two distinct types of nuclear punctate bodies (20, 73). Instead, the HCMV IE1 protein is subsequently found distributed uniformly throughout the nucleoplasm, and this correlates with the timing of a parallel change in the location of PML into a uniform diffuse distribution also. Furthermore, endogenous PML also colocalized with IE1 in condensed metaphase chromosomes in those IE1-expressing transfected or infected cells that were undergoing mitosis, but PML was never detected there in mitotic cells in the absence of IE1.
Our observations also confirmed previous findings that a transiently expressed version of PML that is mutated in one of the Cys residues of the RING finger domain fails to target to or colocalize with endogenous PML despite presumably retaining the ability to heterodimerize with it (6, 36, 46). Curiously, this finding is different than that for the HSV IE110 protein, which still targets to PODs when its RING finger is mutated or deleted but now is unable to displace PML, leading to a stable colocalization of the two proteins in the PODs (13, 20). Although neither IE1 nor IE2 or any of the other cellular proteins found in PODs are RING finger proteins, these results clearly implicate the RING finger domains of PML as contributing in some way to protein aggregation and complex formation in the PODs. The RING class zinc finger domains found in PML, IE110 and a number of other cellular proteins (including BRCA-1 and RAG-1) are believed to be protein-protein interaction motifs with similar cross-braced zinc-coordinated core structures but with distinct targeting specificities (4, 6). For example, Everett et al. (21) have shown that replacement of the IE110 RING finger domain within IE110 by that of PML in an HSV genomic background fails to restore an IE110-positive phenotype.
There is no evidence for a direct interaction between IE110 or its RING finger domain with PML, but our data from yeast two-hybrid assays strongly reinforces the concept that HCMV IE1 does interact directly with PML. Furthermore, the RING finger point mutant form of PML was severely compromised in its ability to interact with IE1 in yeast, and deletion of the N-terminal 97 amino acids of PML including the RING finger domain abolished the interaction. In both mammalian cell cotransfection and yeast interaction studies we showed that the truncated IE1 (1-346) protein is sufficient for interaction with PML, although the ability of IE1 to target to the PODs and to displace PML can apparently be dissociated by removal of the highly acidic C terminus. The large central hydrophobic segment of IE1 is required to be intact for both activities. The isolated N-terminal PML(1-267) protein fragment encompassing the RING finger domain was also sufficient for interaction with IE1 in yeast two-hybrid assays, and overexpressed PML(1-267) was capable of being targeted to POD-like structures in mammalian cells in the presence of IE1(1-346), although it did not do so on its own. Despite previous statements (24, 28) about the presence of both zinc finger and leucine zipper motifs in HCMV IE1 (which might be obvious candidates for PML interaction domains), we do not consider that this is the case, because neither of these proposed motifs fits with standard consensus patterns nor are they conserved in the simian or rodent CMV homologs of the human CMV IE1 protein (9).
The behavior of SP100 in NB4 cells, where it is released from PODs when PML/RAR is dispersed but returns when PML/RAR is restored to the PODs in the presence of RA, suggests that PML may be a key aggregation factor that holds the POD multiprotein complexes together. The targeting of both HCMV IE1 and HSV IE110 to PODs leading to the subsequent release of both PML and SP100 would appear to be consistent with this simplistic view. However, unlike the apparent direct interaction of IE1 with PML, one would have to argue that IE110 and PML compete for interaction with some other POD component that is not itself displaced when PML is lost. Interestingly PML is known to interact with a protein called PIC 1 that is related to ubiquitin (5) and the C terminus of IE110 is known to bind to a ubiquitin pathway regulating protein (22), which suggests that some of these events might involve protein degradation and stabilization, or even that the function of IE110 and PML might be mediated by regulating the stability of ubiquitin-dependent cell cycle or growth control proteins.
Recently, Le et al. (46) have also reported that deletions of the coiled-coil domain or of the RING finger domains of the PML protein resulted in its inability to assemble into PODs leading to a nuclear diffuse distribution in transfected cells. Our results with mutant PML proteins containing either point mutations within the RING finger domain or a small deletion within the coiled-coil region corroborate and extend their findings. Furthermore, our self-interaction assay of mutant PML proteins using the yeast two-hybrid assay demonstrated that the inability of mutant PML to localize in PODs may result from a failure to either dimerize or to oligomerize efficiently. Therefore, we suggest that the N-terminal RING finger domain of PML may normally be involved in assembly of protein complexes in the PODs by homo- and hetero-oligomerization. The interaction of the HCMV IE1 protein with the RING finger domain of PML would then disrupt this process, leading to displacement of PML into a nuclear diffuse form in association with IE1.
Although the PML/RAR fusion protein apparently produces a RA-dependent stimulation of AP-1 activity (17), we have been unable to detect any direct or specific effects of wild-type PML, IE1, or IE110 on AP-1-containing target reporter genes in cotransfection experiments in Vero cells (13a). The HSV IE110/PML RING finger replacement protein also does not restore the transcriptional transactivation properties of the intact IE110 protein (21). Nevertheless, while investigating the properties of PML variants in GAL4 domain swap assays in transfected mammalian cells, we discovered that removal of the C terminus or even a small deletion within the coiled-coil domain of PML uncovered a latent or cryptic transactivator function that encompassed the RING finger and adjacent Cys/His-rich domains. Furthermore, this function was inactivated by mutation of a Cys residue within the RING finger, and the isolated RING finger domains of PML or IE110 as GAL4 fusion proteins were both also active in yeast one-hybrid transactivator domain assays in a totally RING finger-dependent manner. These results suggest that the PML/RAR fusion protein may generate a novel constitutive transactivator function that is probably targeted to RAR- and RXR-containing promoter motifs and that the wild-type PML protein itself has conditional cryptic transactivator characteristics. This transactivator domain appears to be normally masked by the coiled-coil C-terminal domain of the protein but has the potential to be uncovered either by appropriate protein modifications or by interactions with other unknown proteins. Whether the masking involves subunit homodimerization interactions directly or alternatively involves either conformational changes or an intramolecular folding event is unresolved at present, although we have been unable to detect any interaction between the isolated N-terminal and C-terminal halves of PML in yeast two-hybrid assays (2a). Therefore, it seems likely that interaction of IE1 with the RING finger region of PML not only leads to disaggregation of PML-containing POD protein complexes but also either blocks or unmasks the ability of the N terminus of PML to function as a conditional transactivator domain. Considering that IE1 also has a cryptic transactivator domain, which lies within the N-terminal 87 aa that are shared with IE2 (59), the possible functional outcomes of IE1-PML interactions appear to be rather complex.
ACKNOWLEDGMENTS
This study was funded by Public Health Service research grant RO1 AI24576 to G.S.H. from the National Institute for Allergy and Infectious Diseases.
We thank Peter O’Hare, James J.-D. Hsieh, and Yilong Wang for gifts of plasmids. We also thank Dolores Ciufo for plasmids and rabbit PAbs against PML. Generous gifts of yeast strains and plasmids from Stephen J. Elledge (Baylor College of Medicine, Waco, Tex.), plasmids for PML from Ronald M. Evans (The Salk Institute, San Diego, Calif.), the Ad-IE1 vector from Gavin Wilkinson (University of Wales, College of Medicine, Cardiff, United Kingdom), and MAb 5E10 from K. van der Krann (Universiteit van Amsterdam) are greatly acknowledged. We are also grateful to Edward S. Mocarski (Standford University, Calif.) for a gift of samples of the pair of IE1-deleted CR208 virus and its parent HCMV(Towne) virus. We also thank Mike Delannoy (Department of Cell Biology, Johns Hopkins School of Medicine) for assistance with the confocal microscopy analysis and Sarah Heaggans for help in preparation of the manuscript.
REFERENCES
- 1.Ahn J-H, Hayward G S. The major immediate-early proteins IE1 and IE2 of human cytomegalovirus colocalize with and disrupt PML-associated nuclear bodies at very early times in infected permissive cells. J Virol. 1997;71:4599–4613. doi: 10.1128/jvi.71.6.4599-4613.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahn J-H, Chiou C-J, Hayward G S. Evaluation and mapping of the DNA binding and oligomerization domains of the IE2 regulatory protein of human cytomegalovirus using yeast one and two hybrid interaction assays. Gene. 1998;210:25–36. doi: 10.1016/s0378-1119(98)00056-0. [DOI] [PubMed] [Google Scholar]
- 2a.Ahn, J.-H. Unpublished data.
- 3.Ascoli C A, Maul G G. Identification of a novel nuclear domain. J Cell Biol. 1991;112:785–795. doi: 10.1083/jcb.112.5.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barlow P N, Luisi B, Milner A, Elliott M, Everett R D. Structure of the C3HC4 domain by 1H-nuclear magnetic resonance spectroscopy. J Mol Biol. 1994;237:201–211. doi: 10.1006/jmbi.1994.1222. [DOI] [PubMed] [Google Scholar]
- 5.Boddy M N, Howe K, Etkin L D, Solomon E, Freemont P S. PIC 1, a novel ubiquitin-like protein which interacts with the PML component of a multiprotein complex that is disrupted in acute promyelocytic leukemia. Oncogene. 1996;13:971–982. [PubMed] [Google Scholar]
- 6.Borden K L B, Boddy M N, Lally J, O’Reilly N J, Martin S, Howe K, Solomon E, Freemont P S. The solution structure of the RING finger domain from the acute promyelocytic leukaemia proto-oncogene PML. EMBO J. 1995;14:1532–1541. doi: 10.1002/j.1460-2075.1995.tb07139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Britt W J, Alford C A. Cytomegalovirus. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2493–2523. [Google Scholar]
- 8.Carvalho T, Seeler J-S, Ohman K, Jordan P, Pettersson U, Akusjarvi G, Carmo-Fonseca M, Dejean A. Targeting of adenovirus E1A and E4-ORF3 proteins to nuclear matrix-associated PML bodies. J Cell Biol. 1995;131:45–56. doi: 10.1083/jcb.131.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang Y-N, Jeang K-T, Lietman T, Hayward G S. Structural organization of the spliced immediate-early gene complex that encodes the major acidic nuclear (IE1) and trans-activator (IE2) proteins of African green monkey cytomegalovirus. J Biomed Sci. 1995;2:105–130. doi: 10.1007/BF02253062. [DOI] [PubMed] [Google Scholar]
- 10.Cherrington J M, Mocarski E S. Human cytomegalovirus IE1 transactivates the promoter-enhancer via an 18-base-pair repeat element. J Virol. 1989;63:1435–1440. doi: 10.1128/jvi.63.3.1435-1440.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cherrington J M, Khoury E L, Mocarski E S. Human cytomegalovirus ie2 negatively regulates alpha gene expression via a short target sequence near the transcription start site. J Virol. 1991;65:887–896. doi: 10.1128/jvi.65.2.887-896.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chiou C-J, Zong J, Waheed I, Hayward G S. Identification and mapping of dimerization and DNA-binding domains in the C terminus of the IE2 regulatory protein of human cytomegalovirus. J Virol. 1993;67:6201–6214. doi: 10.1128/jvi.67.10.6201-6214.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ciufo, D., and G. S. Hayward. Evaluation of colocalization interactions between the herpes simplex virus IE110 nuclear regulatory protein and the cellular PML and SP100 proteins in infected and DNA-transfected cells. Submitted for publication.
- 13a.Ciufo, D., and G. S. Hayward. Unpublished data.
- 14.de The H, Lavau C, Marchio A, Chomienne C, Degos L, Dejean A. The PML-RARα fusion mRNA generated by the t(15;17) translocation of acute promyelocytic leukemia encodes a functionally altered RAR. Cell. 1991;66:675–684. doi: 10.1016/0092-8674(91)90113-d. [DOI] [PubMed] [Google Scholar]
- 15.Doucas V, Ishov A M, Romo A, Juguilon H, Weitzman M D, Evans R M, Maul G G. Adenovirus replication is coupled with the dynamic properties of the PML nuclear structure. Genes Dev. 1996;10:196–207. doi: 10.1101/gad.10.2.196. [DOI] [PubMed] [Google Scholar]
- 16.Doucas V, Evans R M. The PML nuclear compartment and cancer. Biochem Biophys Acta. 1996;1288:M25–M29. doi: 10.1016/s0304-419x(96)00028-5. [DOI] [PubMed] [Google Scholar]
- 17.Doucas V, Brockes J P, Yaniv M, de The H, Dejean A. The PML-retinoic acid receptor α translocation converts the receptor from an inhibitor to a retinoic acid-dependent activator of transcription factor AP-1. Proc Natl Acad Sci USA. 1993;90:9345–9349. doi: 10.1073/pnas.90.20.9345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Durfee T, Becherer K, Chen P L, Yeh S H, Yang Y, Kilburn A, Lee W H, Elledge S J. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- 19.Dyck J A, Maul G G, Miller W H, Chen J D, Kakizuka A, Evans R M. A novel macromolecular structure is a target of the promyelocyte-retinoic acid receptor oncoprotein. Cell. 1994;76:333–343. doi: 10.1016/0092-8674(94)90340-9. [DOI] [PubMed] [Google Scholar]
- 20.Everett R D, Maul G G. HSV-1 IE protein Vmw110 causes redistribution of PML. EMBO J. 1994;13:5062–5069. doi: 10.1002/j.1460-2075.1994.tb06835.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Everett R D, Maul G G, Orr A, Elliott M. The cellular RING finger protein PML is not a functional counterpart of the herpes simplex virus type 1 RING finger protein Vmw110. J Gen Virol. 1995;76:791–798. doi: 10.1099/0022-1317-76-4-791. [DOI] [PubMed] [Google Scholar]
- 22.Everett R D, Meredith M, Orr A, Cross A, Kathoria M, Parkinson J. A novel ubiquitin-specific protease is dynamically associated with PML nuclear domain and binds to a herpesvirus regulatory protein. EMBO J. 1997;16:556–577. doi: 10.1093/emboj/16.7.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fields S, Song O-K. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 24.Ghazal P, Nelson J A. Transcription factors and viral regulatory proteins as potential mediators of human cytomegalovirus pathogenesis. Berlin, Germany: Springer-Verlag; 1993. [Google Scholar]
- 25.Goddard A D, Borrow J, Freemont P S, Solomon E. Characterization of a zinc finger gene disrupted by the t(15:17) in acute promyelocytic leukemia. Science. 1991;254:1371–1374. doi: 10.1126/science.1720570. [DOI] [PubMed] [Google Scholar]
- 26.Greaves R F, Mocarski E S. Defective growth correlates with reduced accumulation of a viral DNA replication protein after low-multiplicity infection by a human cytomegalovirus ie1 mutant. J Virol. 1998;72:366–379. doi: 10.1128/jvi.72.1.366-379.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grignani F, Fagioli M, Ferrucci P F, Alcalay M, Pelicci P G. The molecular genetics of acute promyelocytic leukaemia. Blood Rev. 1993;7:87–93. doi: 10.1016/s0268-960x(05)80018-9. [DOI] [PubMed] [Google Scholar]
- 27a.Guarente L, Ptashne M. Fusion of Escherichia coli lacZ to the cytochrome c gene of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1981;78:2199–2203. doi: 10.1073/pnas.78.4.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27b.Hagemeier C, Caswell R, Hayhurst G, Sinclair J, Kouzarides T. Functional interaction between the HCMV IE2 transactivator and the retinoblastoma protein. EMBO J. 1994;13:2897–2903. doi: 10.1002/j.1460-2075.1994.tb06584.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hayhurst G P, Bryant L A, Caswell R C, Walker S M, Sinclair J H. CCAAT box-dependent activation of the TATA-less human DNA polymerase α promoter by the human cytomegalovirus 72-kilodalton major immediate-early protein. J Virol. 1995;69:182–188. doi: 10.1128/jvi.69.1.182-188.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He L-Z, Tribioli C, Rivi R, Peruzzi D, Pelicci P G, Soares V, Cattoretti G, Pandolfi P P. Acute leukemia with promyelocytic features in PML/RARα transgenic mice. Proc Natl Acad Sci USA. 1997;94:5302–5307. doi: 10.1073/pnas.94.10.5302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hermiston T W, Malone C L, Witte P R, Stinski M F. Identification and characterization of the human cytomegalovirus immediate-early region 2 gene that stimulates gene expression from an inducible promoter. J Virol. 1987;61:3214–3221. doi: 10.1128/jvi.61.10.3214-3221.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Honess R W, Gompels U A, Craxton M, Cameron K R, Staden R, Barrell B G, Chang Y-N, Hayward G S. Global and local deviation from expected frequencies of CpG-dinucleotides in herpes virus DNAs may be diagnostic of differences in the states of their latent genomes. J Gen Virol. 1989;70:837–855. doi: 10.1099/0022-1317-70-4-837. [DOI] [PubMed] [Google Scholar]
- 32.Hsieh J J-D, Hayward S D. Masking of the CBF1/RBPJκ transcriptional repressor domain by Epstein-Barr virus EBNA2. Science. 1995;268:560–563. doi: 10.1126/science.7725102. [DOI] [PubMed] [Google Scholar]
- 33.Ishov A M, Stenberg R M, Maul G G. Human cytomegalovirus immediate early interaction with host nuclear structures: Definition of an immediate transcription environment. J Cell Biol. 1997;138:5–16. doi: 10.1083/jcb.138.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ishov A M, Maul G G. The periphery of nuclear domain 10 (ND10) as site of DNA virus deposition. J Cell Biol. 1996;134:815–826. doi: 10.1083/jcb.134.4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kakizuka A, Miller W H, Jr, Umesono K, Warrell R P, Jr, Frankel S R, Murty V V V S, Dmitrovsky E, Evans R M. Chromosomal translocation t(15;17) in human acute promyelocytic leukemia fuses RARα with a novel putative transcription factor. Cell. 1991;66:663–674. doi: 10.1016/0092-8674(91)90112-c. [DOI] [PubMed] [Google Scholar]
- 36.Kastner P, Pevez A, Lute Y, Rochette-Egly C, Gaub M P, Durand B, Lanotte M, Berger R, Chambon P. Structure, localization and transcriptional properties of two classes of retinoic acid receptor α fusion proteins in acute promyleocytic leukemia (APL); structural similarities with a new family of oncoproteins. EMBO J. 1992;11:629–642. doi: 10.1002/j.1460-2075.1992.tb05095.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kelly C, Driel R V, Wilkinson G W G. Disruption of PML-associated nuclear bodies during human cytomegalovirus infection. J Gen Virol. 1995;76:2887–2893. doi: 10.1099/0022-1317-76-11-2887. [DOI] [PubMed] [Google Scholar]
- 38.Koken M H, Reid A, Quignon F, Chelbi-Alix M K, Davies J M, Kabarowski J H S, Zhu J, Dong S, Chen S-J, Chen Z, Tan C C, Licht J, Waxman S, de The H, Zelent A. Leukemia-associated retinoic acid receptor α fusion partner, PML and PLZF, heterodimerize and colocalize to nuclear bodies. Proc Natl Acad Sci USA. 1997;94:10255–10260. doi: 10.1073/pnas.94.19.10255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koken M H, Puvion-Dutilleul F, Guillemin M C, Viron A, Linares-Cruz G, Stuurman N, De Jong L, Szostechi C, Calvo F, Chomienne C, Degos L, Puvion E, de The H. The t(15:17) translocation alters a nuclear body in a retinoic acid-reversible fashion. EMBO J. 1994;13:1073–1083. doi: 10.1002/j.1460-2075.1994.tb06356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koken M H, Linares-Cruz G, Quignon F, Viron A, Chelbi-Alix M K, Sobczak-Thepot J, Juhlin L, Degos L, Calvo F, de The H. The PML growth suppressor has an altered expression in human oncogenesis. Oncogene. 1995;10:1315–1324. [PubMed] [Google Scholar]
- 41.Korioth F, Maul G G, Plachter B, Stamminger T, Frey J. The nuclear domain 10 (ND10) is disrupted by the human cytomegalovirus gene product IE1. Exp Cell Res. 1996;229:155–158. doi: 10.1006/excr.1996.0353. [DOI] [PubMed] [Google Scholar]
- 42.LaFemina R L, Hayward G S. Structural organization of the DNA molecules from human cytomegalovirus. ICN-UCLA Symp Mol Biol. 1980;18:39–55. [Google Scholar]
- 43.LaFemina R L, Hayward G S. Differences in cell type-specific blocks to immediate early gene expression and DNA replication of human, simian and murine cytomegalovirus. J Gen Virol. 1988;69:355–374. doi: 10.1099/0022-1317-69-2-355. [DOI] [PubMed] [Google Scholar]
- 44.LaFemina R L, Pizzorno M C, Mosca J D, Hayward G S. Expression of the acidic nuclear immediate-early protein (IE1) of human cytomegalovirus in stable cell lines and its preferential association with metaphase chromosomes. Virology. 1989;172:584–600. doi: 10.1016/0042-6822(89)90201-8. [DOI] [PubMed] [Google Scholar]
- 45.Lang D, Stamminger T. The 86-kilodalton IE-2 protein of human cytomegalovirus is a sequence-specific DNA-binding protein that interacts directly with the negative autoregulatory response element located near the cap site of the IE-1/2 enhancer-promoter. J Virol. 1993;67:323–331. doi: 10.1128/jvi.67.1.323-331.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Le X-F, Yang P, Chang K-S. Analysis of the growth and transformation suppressor domains of promyelocytic leukemia gene, PML. J Biol Chem. 1996;271:130–135. doi: 10.1074/jbc.271.1.130. [DOI] [PubMed] [Google Scholar]
- 47.Lillie J W, Green M R. Transcription activation by the adenovirus E1A protein. Nature. 1989;338:39–44. doi: 10.1038/338039a0. [DOI] [PubMed] [Google Scholar]
- 48.Liu B, Stinski M F. Human cytomegalovirus contains a tegument protein that enhances transcription from promoters with upstream ATF and AP-1 cis-acting elements. J Virol. 1992;66:4434–4444. doi: 10.1128/jvi.66.7.4434-4444.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu B, Hermiston T W, Stinski M F. A “cis”-acting element in the major immediate-early (IE) promoter of human cytomegalovirus is required for negative regulation by IE2. J Virol. 1991;65:897–903. doi: 10.1128/jvi.65.2.897-903.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu J-H, Mu Z-M, Chang K-S. PML suppresses oncogenic transformation of NIH/3T3 cells by activated neu. J Exp Med. 1995;181:1965–1973. doi: 10.1084/jem.181.6.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maul G G, Ishov A M, Everett R D. Nuclear domain 10 as preexisting potential replication start sites of herpes simplex virus type-1. Virology. 1996;217:67–75. doi: 10.1006/viro.1996.0094. [DOI] [PubMed] [Google Scholar]
- 52.Maul G G, Everett R D. The nuclear location of PML, a cellular member of the C3HC4 zinc-binding domain protein family, is rearranged during herpes simplex virus infection by the C3HC4 viral protein ICP0. J Gen Virol. 1994;75:1223–1233. doi: 10.1099/0022-1317-75-6-1223. [DOI] [PubMed] [Google Scholar]
- 53.Maul G G, Guldner H H, Spivack J G. Modification of discrete nuclear domains induced by herpes simplex virus type 1 immediate early gene 1 product (ICP0) J Gen Virol. 1993;74:2679–2690. doi: 10.1099/0022-1317-74-12-2679. [DOI] [PubMed] [Google Scholar]
- 54.Mocarski E S, Kemble G W, Lyle J M, Greaves R F. A deletion mutant in the human cytomegalovirus gene encoding IE1491aa is replication defective due to a failure in autoregulation. Proc Natl Acad Sci USA. 1996;93:11321–11326. doi: 10.1073/pnas.93.21.11321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mocarski E S., Jr . Cytomegalovirus and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2447–2492. [Google Scholar]
- 56.Mu Z M, Chin K V, Liu J H, Lozano G, Chang K-S. PML, a growth suppressor disrupted in acute promyelocytic leukemia. Mol Cell Biol. 1994;14:6858–6867. doi: 10.1128/mcb.14.10.6858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pandolfi P P, Grignani F, Alcalay M, Mencarelli A, Biondi A, LoCoco F, Grignani F, Pellicci P G. Structure and origin of the acute promyelocytic leukemia myl/RARα cDNA and characterization of its retinoid-binding and transactivation properties. Oncogene. 1991;6:1285–1292. [PubMed] [Google Scholar]
- 58.Pizzorno M C, Hayward G S. The IE2 gene products of human cytomegalovirus specifically down-regulate expression from the major immediate-early promoter through a target sequence located near the cap site. J Virol. 1990;64:6154–6165. doi: 10.1128/jvi.64.12.6154-6165.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pizzorno M C, Mullen M-A, Chang Y-N, Hayward G S. The functionally active IE2 immediate-early regulatory protein of human cytomegalovirus is an 80-kilodalton polypeptide that contains two distinct activator domains and a duplicated nuclear localization signal. J Virol. 1991;65:3839–3852. doi: 10.1128/jvi.65.7.3839-3852.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pizzorno M C, O’Hare P, Sha L, LaFemina R L, Hayward G S. trans-activation and autoregulation of gene expression by the immediate-early region 2 gene products of human cytomegalovirus. J Virol. 1988;62:1167–1179. doi: 10.1128/jvi.62.4.1167-1179.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Puvion-Dutilleul F, Chelbi-Alix M K, Koken M, Quignon F, Puvion E, de The H. Adenovirus infection induces rearrangements in the intranuclear distribution of the nuclear body-associated PML protein. Exp Cell Res. 1995;218:9–16. doi: 10.1006/excr.1995.1125. [DOI] [PubMed] [Google Scholar]
- 62.Rose M D, Winston F, Hieter P. Methods in yeast genetics. A laboratory course manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- 63.Spaete R R, Mocarski E S. Insertion and deletion mutagenesis of the human cytomegalovirus genome. Proc Natl Acad Sci USA. 1987;84:7213–7217. doi: 10.1073/pnas.84.20.7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stadler M, Chelbi-Alix M K, Koken M H, Venturini L, Lee C, Saib A, Quignon F, Pelicano L, Guillemin M C, Schindler C. Transcriptional induction of the PML growth suppressor gene by interferons is mediated through an ISRE and a GAS element. Oncogene. 1995;11:2565–2573. [PubMed] [Google Scholar]
- 65.Stenberg R M, Witte P R, Stinski M F. Multiple spliced and unspliced transcripts from human cytomegalovirus immediate-early region 2 and evidence for a common initiation site within immediate-early region 1. J Virol. 1985;56:665–675. doi: 10.1128/jvi.56.3.665-675.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stinski M F, Thomsen D R, Stenberg R M, Goldstein L C. Organization and expression of the immediate early genes of human cytomegalovirus. J Virol. 1983;46:1–14. doi: 10.1128/jvi.46.1.1-14.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66a.Stuurman N, De Graaf A, Floore A, Josso A, Humbel B, De Jong L, Van Driel R. A monoclonal antibody recognizing nuclear matrix associated nuclear bodies. J Cell Sci. 1992;101:773–784. doi: 10.1242/jcs.101.4.773. [DOI] [PubMed] [Google Scholar]
- 67.Szekely L, Pokrovskaja K, Jiang W-Q, de The H, Ringertz N, Klein G. The Epstein-Barr virus-encoded nuclear antigen EBNA-5 accumulates in PML-containing bodies. J Virol. 1996;70:2562–2568. doi: 10.1128/jvi.70.4.2562-2568.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Szostecki C, Guldner H H, Netter H T, Will H. Isolation and characterization of cDNA encoding a human nuclear antigen predominantly recognized by autoantibodies from patients with cirrhosis. J Immunol. 1990;145:4338–4347. [PubMed] [Google Scholar]
- 69.Wang Y, Finan J E, Middeldrop J M, Hayward S D. P32/TAP, a cellular protein that interacts with EBNA-1 of Epstein-Barr virus. Virology. 1997;236:18–29. doi: 10.1006/viro.1997.8739. [DOI] [PubMed] [Google Scholar]
- 70.Weis K, Rambaud S, Lavau C, Janseu J, Carvalho T, Carmo-Fonseca M, Lamond A, Dejean A. Retinoic acid regulates aberrant nuclear localization of PML-RAR alpha in acute promyelocytic leukemia cells. Cell. 1994;76:345–356. doi: 10.1016/0092-8674(94)90341-7. [DOI] [PubMed] [Google Scholar]
- 71.Wilkinson G W G, Akrigg A. Constitutive and enhanced expression from the CMV major IE promoter in a defective adenovirus vector. Nucleic Acids Res. 1992;20:2233–2239. doi: 10.1093/nar/20.9.2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhong L. Role of herpes simplex virus type-1 (HSV-1) immediate early proteins in intranuclear compartmentalization during HSV-1 infection. Ph.D. thesis. Baltimore, Md: Johns Hopkins University; 1997. [Google Scholar]
- 73.Zhong, L., and G. S. Hayward. The herpes simplex virus IE68(ICP22) immediate-early protein is associated with novel nuclear punctate structures and causes the disruption of both cellular PML oncogenic domains and spliceosome domains. Submitted for publication.