Abstract
Endophytic fungi live inside virtually every plant species, without causing any apparent disease or damage to the host. Nevertheless, under particular conditions, mutualistic lifestyle of endophytes may change to pathogenic. In this study, the biodiversity of Alternaria and Fusarium species, the two most abundant endophytic fungi isolated from healthy potato plants in two climatically different regions of Iran, Ardebil in the north-west and Kerman in the south-east, was investigated. Seventy-five Fusarium strains and 83 Alternaria strains were molecularly characterized by multi-locus gene sequencing. Alternaria strains were characterized by the sequences of gpd and caM gene fragments and the phylogenetic tree was resolved in 3 well-separated clades. Seventy-three strains were included in the clade A, referred as Alternaria section, 6 strains were included in clade B, referred as Ulocladioides section, and 4 strains were included in clade C, referred as Infectoriae section. Fusarium strains, identified by sequencing the translation elongation factor 1α (tef1), β-tubulin (tub2) and internal transcribed spacer (ITS) genomic regions, were assigned to 13 species, viz. F. brachygibosum, F. clavum, F. equiseti, F. flocciferum, F. incarnatum, F. nirenbergiae, F. nygamai, F. oxysporum, F. proliferatum, F. redolens, F. sambucinum, F. solani and F. thapsinum. Twenty-six selected strains, representative of F. equiseti, F. nirenbergiae, F. oxysporum, F. nygamai, F. proliferatum, and F. sambucinum, were also tested for production of the mycotoxins deoxynivalenol (DON), nivalenol (NIV), diacetoxyscirpenol (DAS), T-2 toxin (T-2), beauvericin (BEA), enniatins (ENNs), fumonisins (FBs), fusaric acid (FA) and moniliformin (MON). None of the tested strains produced trichothecene toxins (DON, NIV, DAS and T-2). Two out of 2 F. equiseti isolates, 1/6 F. oxysporum, 1/3 F. proliferatum, and 1/9 F. nygamai did not produce any of the tested toxins; the rest of strains produced one or more BEA, ENNs, FBs, FA and MON toxins. The most toxigenic strain, F. nygamai ITEM-19012, produced the highest quantities of FBs (7946, 4693 and 4333 μg/g of B1, B2, and B3 respectively), along with the highest quantities of both BEA (4190 μg/g) and MON (538 μg/g). These findings suggest that contamination of potato tubers with mycotoxins in the field or at post-harvest, due to a change in lifestyle of endophytic microflora, should be carefully considered and furtherly investigated.
Keywords: Endophytic fungi, Molecular identification, Solanum tuberosum, Fusarium, Alternaria, Mycotoxins
1. Introduction
Under natural conditions plants are not individual entities; they are associated with microorganisms to form the plant holobiont [1]. The nature of the interaction of these microorganisms with plants varies from mutualism to parasitism [2]. Endophytic fungi live inside healthy plant tissues without causing any apparent disease symptoms or damage to their hosts. Endophytic fungi have been recovered in all plant tissues, including leaves, stems, roots, flowers and fruits [3], and every plant examined to date was found to harbour at least one species of endophytic fungi. Research on the ecology of endophytic fungi mostly supports their non-pathogenic nature and their capability to enhance biotic and abiotic stress tolerance [4] and improve mineral nutrition in the host plant [5,6], so that endophytes have been proposed as biocontrol agents or beneficial bioinoculants [7]. Nevertheless, conceptual aspects related to the ecological nature of endophytes are still under debate, because some reports have highlighted that, under specific conditions, the mutualistic relationship of endophytes with plants may change to be detrimental [8].
Potato (Solanum tuberosum L.) is the fourth-largest world crop, surpassed in total production only by wheat, rice and corn. It is a rich source of carbohydrates for human nutrition and an important part of the diet for over 1.5 billion people worldwide. Iran is the fourth-largest potato producer in Asia, where the crop is extensively cultivated in the provinces of Ardebil, Hamedan, Esfahan, East Azarbaijan, and South Kerman, with the Ardebil province being the most important production area because of its favourable climate conditions [9]. During a large-scale investigation on endophytic fungi of potato plants in two major potato-producing areas of Iran, viz. Ardebil area (northwestern Iran) and South Kerman area (southeastern Iran), almost 400 endophytic fungal strains were isolated from above-ground and below-ground organs of potato plants, and 22 fungal genera, and 52 fungal species were identified [10]. In both the regions investigated, Alternaria was the most abundant genus in the above-ground plant parts (stems and leaves) and Fusarium in the below-ground plant parts (roots and tubers). Both Alternaria and Fusarium were previously reported as endophytic in potato. O'Callaghan et al. [11] examined the microbial communities of magainin-producing transgenic lines of S. tuberosum in New Zealand, and identified six species of Fusarium and two species of Alternaria. In Germany, Gotz et al. [12] isolated and identified root endophytic fungi from two different potato lines by traditional techniques and cultivation-independent DNA-based methods. They obtained 67 isolates of Fusarium and 73 isolates of Alternaria. Marak and Kayang [13] isolated and identified endophytic fungi associated with potato plants from South-West Garo Hills, Meghalaya, India. In that survey, the species F. oxysporum, F. redolens, F. semitectum, F. solani, F. sporotrichioides, A. alternata, A. brassicicola, and A. solani were identified.
Alternaria species that cause “early blight” and “brown spot” on potato, as well as on tomato, are A. tomatophila E.G. Simmons, A. solani Sorauer, A. alternata (Fr.) Keissl., A. tenuissima (Kunze) Wiltshire, A. infectoria E.G. Simmons and A. arborescens E.G. Simmons [14]. Alternaria solani is the second most devastating foliar pathogen of potato crops after Phytophthora infestans, worldwide [15]. The common symptoms of Alternaria diseases are necrotic lesions on leaves, which are created by the diffusion of fungal toxins [16].
Fusarium dry rot is a postharvest fungal disease affecting potato tubers, which is a cause of up to 60% loss of stored tubers [17]. The disease is caused by several species of Fusarium, mainy F. solani var. coeruleum, F. sambucinum, F. oxysporum, F. avenaceum, and F. culmorum. Some Fusarium species associated with the disease are known to produce mycotoxins that have been implicated in acute toxicoses of humans and domesticated animals [17,18]. In addition, the prolonged exposure to low sub-lethal doses of mycotoxin may result in chronic toxicoses which often are associated with development of cancer. Several Fusarium mycotoxins are regulated in most countries [19]. In addition, concerns about the “emerging mycotoxins” which may co-occur with regulated ones and contribute to the overall health risk of the contaminated commodities, have been growing [19]. Fusarium graminearum, a major mycotoxigenic species that produces the regulated mycotoxins zearalenone and tricothecenes in corn and small grains, was reported as a causal agent of Fusarium dry rot of potato in USA [20]. It has also been shown that this species is able to produce the trichothecene mycotoxins deoxynivalenon (DON) and nivalenol (NIV) in rotten potato tuber tissue [21]. Potato tubers artificially infected with F. sambucinum, another toxigenic species, contained the trichothecene toxin diacetoxyscirpenol (DAS) in concentrations up to 200 μg/tuber [22]. Also, the emerging mycotoxins enniatins were found in potato tuber tissue infected by a complex of six Fusarium strains from different sources [23].
In this article, we present the molecular characterization of endophytic strains of Alternaria spp. and Fusarium spp., isolated from leaf, stem, root, and tuber of potato plants from two geographically diverse potato-producing areas in Iran. We also investigated the mycotoxigenicity of the Fusarium strains isolated from tubers and roots, to assess their potential risk of mycotoxin production.
2. Materials and methods
2.1. Sample collection, isolation, and preservation of the endophytic Alternaria and Fusaria
Eighty mature and disease symptomless potato plants, thirty-five from the Ardebil area (northwestern Iran) and forty-five from the South Kerman area (southeastern Iran) were collected in August 2018 and February 2019. In each inspected field, two potato plants were collected from the opposite sides of each field, and the points of sampling were geolocalized. The visited fields were at least 5 km apart from each other. Samples were kept in paper bags under refrigeration at +4 °C and immediately transferred to the laboratory for further processing. Plant organs were surface sterilizedwithin 72 h from collection, as described by Alijani Mamaghani et al. [10]. Briefly, samples were immersed in 70% (v/v) ethanol for 2 min, then in 5% (w/v) sodium hypochlorite for 5 min, rinsed 3 times in sterile distilled water for at least 5 min, dabbed between sterile tissues and let dry in sterile conditions for at least 15 min. Isolations of endophytic fungi from stems, leaves, roots and tubers were done on antibiotic-supplemented Potato Dextrose Agar (PDA, Merck, Darmstadt, Germany) followed by single germinated-spore isolation. The cultures were preserved on sterile filter paper pieces stored at −20 °C. Pure cultures of the endophytes were deposited in the Mycology Laboratory of the College of Agriculture and Natural Resources, University of Tehran, Iran; representative isolates that were used for molecular identification and analysis of mycotoxins were also cryopreserved at −80 °C in the Agro-Food Microbial Culture Collection of Institute of Sciences of Food Production, CNR, Italy (http://server.ispa.cnr.it/ITEM/Collection/) under an “ITEM” accession number (Table 1, Table 2).
Table 1.
Endophytic Fusarium strains used for molecular characterization, and accession numbers for their gene sequences in NCBI GeneBank database. The strains used for analysis of mycotoxigenicity are in bold.
| Origin and Strain No.(a) | ITEM No..(b) | Plant part | Species | Gene Bank Accession number |
||
|---|---|---|---|---|---|---|
| tef1 | tub2 | ITS | ||||
| Ardebil Province | ||||||
| FU-41 | 18962 | Root | F. clavum | OQ419383 | OQ419308 | OQ404982 |
| FU-1 | 18964 | Root | F. equiseti | OQ419350 | OQ419275 | OQ404949 |
| FU-3 | 18956 | Stem | F. equiseti | OQ419372 | OQ419297 | OQ404971 |
| FU-73 | 18965 | Root | F. equiseti | OQ419402 | OQ419327 | OQ405001 |
| FU-87 | 18966 | Root | F. equiseti | OQ419414 | OQ419339 | OQ405013 |
| FU-94 | 18967 | Tuber | F. equiseti | OQ419422 | OQ419347 | OQ405021 |
| FU-96 | 18968 | Root | F. equiseti | OQ419423 | OQ419348 | OQ405022 |
| FU-103 | 18969 | Stem | F. equiseti | OQ419352 | OQ419277 | OQ404951 |
| FU-34 | 18960 | Root | F. flocciferum | OQ419376 | OQ419301 | OQ404975 |
| FU-84 | 18972 | Root | F. nirenbergiae | OQ419411 | OQ419336 | OQ405010 |
| FU-86 | 18973 | Root | F. nirenbergiae | OQ419413 | OQ419338 | OQ405012 |
| FU-97 | 18974 | Root | F. nirenbergiae | OQ419424 | OQ419349 | OQ405023 |
| FU-2 | 18941 | Tuber | F. oxysporum | OQ419362 | OQ419287 | OQ404961 |
| FU-35 | 18946 | Tuber | F. oxysporum | OQ419377 | OQ419302 | OQ404976 |
| FU-66 | 18986 | Tuber | F. oxysporum | OQ419398 | OQ419323 | OQ404997 |
| FU-70 | 18987 | Root | F. oxysporum | OQ419399 | OQ419324 | OQ404998 |
| FU-92 | 18994 | Leaf | F. proliferatum | OQ419420 | OQ419345 | OQ405019 |
| FU-36 | 18990 | Root | F. redolens | OQ419378 | OQ419303 | OQ404977 |
| FU-38 | 18991 | Root | F. redolens | OQ419380 | OQ419305 | OQ404979 |
| FU-40 | 18992 | Root | F. redolens | OQ419382 | OQ419307 | OQ404981 |
| FU-59 | 18997 | Tuber | F. solani | OQ419391 | OQ419316 | OQ404990 |
| FU-60 | 18998 | Tuber | F. solani | OQ419393 | OQ419318 | OQ404992 |
| FU-63 | 19000 | Root | F. solani | OQ419396 | OQ419321 | OQ404995 |
| FU-64 | – | Root | F. solani | OQ419397 | OQ419322 | OQ404996 |
| FU-72 | – | Root | F. solani | OQ419401 | OQ419326 | OQ405000 |
| FU-85 | 19003 | Stem | F. solani | OQ419412 | OQ419337 | OQ405011 |
| Kerman Province | ||||||
| FU-14 | 18957 | Tuber | F. brachygibbosum | OQ419356 | OQ419281 | OQ404955 |
| FU-81 | 18961 | Root | F. brachygibbosum | OQ419408 | OQ419333 | OQ405007 |
| FU-30 | 18970 | Root | F. equiseti | OQ419373 | OQ419298 | OQ404972 |
| FU-37 | 18971 | Root | F. equiseti | OQ419379 | OQ419304 | OQ404978 |
| FU-83 | 18963 | Root | F. incarnatum | OQ419410 | OQ419335 | OQ405009 |
| FU-20 | 18975 | Tuber | F. nirenbergiae | OQ419363 | OQ419288 | OQ404962 |
| FU-21 | 18976 | Root | F. nirenbergiae | OQ419364 | OQ419289 | OQ404963 |
| FU-24 | 18977 | Tuber | F. nirenbergiae | OQ419367 | OQ419292 | OQ404966 |
| FU-25 | 18945 | Tuber | F. nirenbergiae | OQ419368 | OQ419293 | OQ404967 |
| FU-29 | 18978 | Tuber | F. nirenbergiae | OQ419371 | OQ419296 | OQ404970 |
| FU-33 | 18979 | Root | F. nirenbergiae | OQ419375 | OQ419300 | OQ404974 |
| FU-78 | 18980 | Root | F. nirenbergiae | OQ419405 | OQ419330 | OQ405004 |
| FU-88 | 18981 | Tuber | F. nirenbergiae | OQ419415 | OQ419340 | OQ405014 |
| FU-89 | 18982 | Root | F. nirenbergiae | OQ419416 | OQ419341 | OQ405015 |
| FU-91 | 18983 | Tuber | F. nirenbergiae | OQ419419 | OQ419344 | OQ405018 |
| FU-4 | 18947 | Tuber | F. nygamai | OQ419381 | OQ419306 | OQ404980 |
| FU-5 | 18948 | Tuber | F. nygamai | OQ419388 | OQ419313 | OQ404987 |
| FU-6 | 18984 | Root | F. nygamai | OQ419392 | OQ419317 | OQ404991 |
| FU-10 | 18949 | Root | F. nygamai | OQ419351 | OQ419276 | OQ404950 |
| FU-11 | 18985 | Tuber | F. nygamai | OQ419353 | OQ419278 | OQ404952 |
| FU-12 | 18950 | Tuber | F. nygamai | OQ419354 | OQ419279 | OQ404953 |
| FU-15 | 18951 | Tuber | F. nygamai | OQ419357 | OQ419282 | OQ404956 |
| FU-23 | 18952 | Tuber | F. nygamai | OQ419366 | OQ419291 | OQ404965 |
| FU-26 | 18953 | Root | F. nygamai | OQ419369 | OQ419294 | OQ404968 |
| FU-27 | 19005 | Tuber | F. nygamai | OQ419370 | OQ419295 | OQ404969 |
| FU-31 | 19006 | Root | F. nygamai | OQ419374 | OQ419299 | OQ404973 |
| FU-43 | 19007 | Root | F. nygamai | OQ419385 | OQ419310 | OQ404984 |
| FU-44 | 19008 | Root | F. nygamai | OQ419386 | OQ419311 | OQ404985 |
| FU-48 | 19009 | Root | F. nygamai | OQ419387 | OQ419312 | OQ404986 |
| FU-54 | – | Tuber | F. nygamai | OQ419389 | OQ419314 | OQ404988 |
| FU-71 | 19011 | Root | F. nygamai | OQ419400 | OQ419325 | OQ404999 |
| FU-77 | 19012 | Root | F. nygamai | OQ419404 | OQ419329 | OQ405003 |
| FU-79 | 19013 | Root | F. nygamai | OQ419406 | OQ419331 | OQ405005 |
| FU-90 | 19014 | Tuber | F. nygamai | OQ419418 | OQ419343 | OQ405017 |
| FU-8 | 18942 | Tuber | F. oxysporum | OQ419407 | OQ419332 | OQ405006 |
| FU-9 | 18943 | Tuber | F. oxysporum | OQ419417 | OQ419342 | OQ405016 |
| FU-18 | 18944 | Stem | F. oxysporum | OQ419360 | OQ419285 | OQ404959 |
| FU-76 | 18988 | Root | F. oxysporum | OQ419403 | OQ419328 | OQ405002 |
| FU-82 | 18989 | Stem | F. oxysporum | OQ419409 | OQ419334 | OQ405008 |
| FU-13 | 18954 | Tuber | F. proliferatum | OQ419355 | OQ419280 | OQ404954 |
| FU-17 | 18955 | Tuber | F. proliferatum | OQ419359 | OQ419284 | OQ404958 |
| FU-62 | 18993 | Tuber | F. proliferatum | OQ419395 | OQ419320 | OQ404994 |
| FU-19 | 18958 | Root | F. sambucinum | OQ419361 | OQ419286 | OQ404960 |
| FU-16 | – | Root | F. solani | OQ419358 | OQ419283 | OQ404957 |
| FU-42 | 18996 | Tuber | F. solani | OQ419384 | OQ419309 | OQ404983 |
| FU-61 | – | Root | F. solani | OQ419394 | OQ419319 | OQ404993 |
| FU-93 | – | Root | F. solani | OQ419421 | OQ419346 | OQ405020 |
| FU-22 | 18959 | Tuber | F. thapsinum | OQ419365 | OQ419290 | OQ404964 |
| FU-55 | 19015 | Tuber | Rectifusarium robinianum | OQ419390 | OQ419315 | OQ404989 |
Department of Plant Protection, Mycology Laboratory of the College of Agriculture and Natural Resources, University of Tehran, Iran.
Accession number of the strain in the Agro-Food Microbial Culture Collection of Institute of Sciences of Food Production, CNR, Italy (http://server.ispa.cnr.it/ITEM/Collection/).
Table 2.
Endophytic Alternaria strains used for molecular characterization, and accession numbers for their gene sequences in NCBI GeneBank database.
| Origin and Strain No.(a) | Item No..(b) | Plant part | Section | Gene Bank Accession number |
|
|---|---|---|---|---|---|
| Gpd | caM | ||||
| Ardebil Province | |||||
| AL-1 | 19017 | Leaf | Alternaria | OQ419109 | OQ419192 |
| AL-2 | 19018 | Stem | Alternaria | OQ419119 | OQ419202 |
| AL-3 | 19019 | Stem | Alternaria | OQ419129 | OQ419212 |
| AL-4 | – | Leaf | Alternaria | OQ419138 | OQ419221 |
| AL-11 | – | Stem | Alternaria | OQ419111 | OQ419194 |
| AL-13 | – | Stem | Alternaria | OQ419113 | OQ419196 |
| AL-15 | – | Stem | Alternaria | OQ419114 | OQ419197 |
| AL-17 | 19025 | Stem | Alternaria | OQ419116 | OQ419199 |
| AL-18 | – | Stem | Alternaria | OQ419117 | OQ419200 |
| AL-21 | – | Stem | Alternaria | OQ419120 | OQ419203 |
| AL-24 | – | Leaf | Alternaria | OQ419123 | OQ419206 |
| AL-25 | – | Stem | Alternaria | OQ419124 | OQ419207 |
| AL-26 | 19031 | Stem | Alternaria | OQ419125 | OQ419208 |
| AL-27 | – | Leaf | Alternaria | OQ419126 | OQ419209 |
| AL-28 | – | Leaf | Alternaria | OQ419127 | OQ419210 |
| AL-30 | 19035 | Stem | Alternaria | OQ419130 | OQ419213 |
| AL-33 | – | Stem | Alternaria | OQ419132 | OQ419215 |
| AL-34 | – | Stem | Alternaria | OQ419133 | OQ419216 |
| AL-37 | 19040 | Leaf | Alternaria | OQ419136 | OQ419219 |
| AL-57 | – | Leaf | Alternaria | OQ419157 | OQ419240 |
| AL-59 | – | Stem | Alternaria | OQ419159 | OQ419242 |
| AL-67 | – | Leaf | Alternaria | OQ419162 | OQ419245 |
| AL-70 | – | Stem | Alternaria | OQ419165 | OQ419248 |
| AL-73 | 19033 | Stem | Alternaria | OQ419168 | OQ419251 |
| AL-74 | – | Stem | Alternaria | OQ419169 | OQ419252 |
| AL-80 | 19037 | Stem | Alternaria | OQ419175 | OQ419258 |
| AL-85 | – | Leaf | Alternaria | OQ419180 | OQ419263 |
| AL-95 | 19038 | Stem | Alternaria | OQ419190 | OQ419273 |
| AL-96 | 19039 | Leaf | Alternaria | OQ419191 | OQ419274 |
| AL-16 | – | Leaf | Infectoria | OQ419115 | OQ419198 |
| AL-22 | 19042 | Root | Infectoria | OQ419121 | OQ419204 |
| AL-23 | – | Leaf | Infectoria | OQ419122 | OQ419205 |
| AL-51 | – | Leaf | Infectoria | OQ419151 | OQ419234 |
| AL-12 | 19043 | Stem | Ulocladioides | OQ419112 | OQ419195 |
| Kerman Province | |||||
| AL-5 | – | Leaf | Alternaria | OQ419149 | OQ419232 |
| AL-10 | 19022 | Leaf | Alternaria | OQ419110 | OQ419193 |
| AL-19 | 19027 | Stem | Alternaria | OQ419118 | OQ419201 |
| AL-29 | 19034 | Stem | Alternaria | OQ419128 | OQ419211 |
| AL-32 | 19036 | Leaf | Alternaria | OQ419131 | OQ419214 |
| AL-35 | – | Leaf | Alternaria | OQ419134 | OQ419217 |
| AL-36 | – | Stem | Alternaria | OQ419135 | OQ419218 |
| AL-39 | – | Leaf | Alternaria | OQ419137 | OQ419220 |
| AL-40 | – | Leaf | Alternaria | OQ419139 | OQ419222 |
| AL-41 | – | Stem | Alternaria | OQ419140 | OQ419223 |
| AL-42 | 19017 | Leaf | Alternaria | OQ419141 | OQ419224 |
| AL-43 | 19020 | Leaf | Alternaria | OQ419142 | OQ419225 |
| AL-44 | 19021 | Leaf | Alternaria | OQ419143 | OQ419226 |
| AL-45 | – | Root | Alternaria | OQ419144 | OQ419227 |
| AL-46 | 19023 | Stem | Alternaria | OQ419145 | OQ419228 |
| AL-47 | – | Stem | Alternaria | OQ419146 | OQ419229 |
| AL-50 | 19024 | Leaf | Alternaria | OQ419150 | OQ419233 |
| AL-52 | 19026 | Leaf | Alternaria | OQ419152 | OQ419235 |
| AL-53 | 19028 | Leaf | Alternaria | OQ419153 | OQ419236 |
| AL-55 | 19029 | Stem | Alternaria | OQ419155 | OQ419238 |
| AL-56 | 19030 | Leaf | Alternaria | OQ419156 | OQ419239 |
| AL-61 | 19032 | Leaf | Alternaria | OQ419160 | OQ419243 |
| AL-63 | – | Stem | Alternaria | OQ419161 | OQ419244 |
| AL-68 | – | Leaf | Alternaria | OQ419163 | OQ419246 |
| AL-69 | 19044 | Leaf | Alternaria | OQ419164 | OQ419247 |
| AL-71 | - | Leaf | Alternaria | OQ419166 | OQ419249 |
| AL-72 | 19045 | Root | Alternaria | OQ419167 | OQ419250 |
| AL-75 | - | Leaf | Alternaria | OQ419170 | OQ419253 |
| AL-76 | - | Leaf | Alternaria | OQ419171 | OQ419254 |
| AL-77 | 19046 | Leaf | Alternaria | OQ419172 | OQ419255 |
| AL-78 | 19047 | Leaf | Alternaria | OQ419173 | OQ419256 |
| AL-79 | - | Root | Alternaria | OQ419174 | OQ419257 |
| AL-81 | 19048 | Leaf | Alternaria | OQ419176 | OQ419259 |
| AL-82 | 19049 | Stem | Alternaria | OQ419177 | OQ419260 |
| AL-83 | - | Root | Alternaria | OQ419178 | OQ419261 |
| AL-84 | - | Leaf | Alternaria | OQ419179 | OQ419262 |
| AL-86 | - | Leaf | Alternaria | OQ419181 | OQ419264 |
| AL-87 | 19050 | Leaf | Alternaria | OQ419182 | OQ419265 |
| AL-88 | - | Leaf | Alternaria | OQ419183 | OQ419266 |
| AL-89 | - | Leaf | Alternaria | OQ419184 | OQ419267 |
| AL-91 | 19051 | Leaf | Alternaria | OQ419186 | OQ419269 |
| AL-92 | 19052 | Leaf | Alternaria | OQ419187 | OQ419270 |
| AL-93 | - | Leaf | Alternaria | OQ419188 | OQ419271 |
| AL-94 | 19053 | Leaf | Alternaria | OQ419189 | OQ419272 |
| AL-48 | 19054 | Stem | Ulocladioides | OQ419147 | OQ419230 |
| AL-49 | 19055 | Stem | Ulocladioides | OQ419148 | OQ419231 |
| AL-54 | 19056 | Leaf | Ulocladioides | OQ419154 | OQ419237 |
| AL-58 | – | Stem | Ulocladioides | OQ419158 | OQ419241 |
| AL-90 | 19057 | Stem | Ulocladioides | OQ419185 | OQ419268 |
Department of Plant Protection, Mycology Laboratory of the College of Agriculture and Natural Resources, University of Tehran, Iran.
Accession number of the strain in the Agro-Food Microbial Culture Collection of Institute of Sciences of Food Production, CNR, Italy (http://server.ispa.cnr.it/ITEM/Collection/).
2.2. Morphological identification of endophytic Fusarium and Alternaria strains
Isolates of endophytes putatively assigned to Fusarium and Alternaria genera based on conidial morphology, were re-isolated from single spores and transferred on specific media to examine their macroscopic and microscopic features for the identification to the species level. Fusarium strains were incubated on PDA at 25 °C for 5–7 days with a 12/12 h day/night photoperiod to examine growth rate, colony features and production of chlamydospores; Carnation Leaf Agar (CLA) and Synthetic Nutrient Agar (SNA) were used for examination of macroconidia, chlamydospores, and phialides, according to Leslie and Summerell [24]. Morphological identification of Alternaria strains was carried out according to Simmons [25]. Small plugs of pure isolates were cultured on Potato Carrot Agar -PCA [26] and kept under 8/16 h of fluorescent light/dark cycle at 22 °C for 5–7 days. Then, colony features, sporulation patterns, conidial chains, shape, size and septa of conidia and primary and secondary conidiophores were examined.
2.3. Molecular characterization of Fusarium and Alternaria endophytic strains
Seventy-five (26 from Ardebil and 49 from Kerman) Fusarium strains (Table 1) and 83 Alternaria strains (34 from Ardebil and 49 from Kerman) (Table 2) were selected as representative of the Fusarium and Alternaria populations from different parts of potato plants, viz. leaves, stems, tubers, and roots. Selection of the strains was based on morphological features, sampling location (geographical area and farm), and potato plant tissue type.
For genomic DNA extraction and molecular analyses, the cryopreserved strains at ISPA-CNR were refreshed on PDA and then cultured on cellophane disks overlaid on PDA Petri dishes. After 3 days of growth at 25 °C, mycelia were scraped, transferred to 2 ml microtubes, frozen and lyophilized. Ten to fifteen mg of powdered lyophilized mycelium were used for DNA extraction by using the “Wizard Magnetic DNA Purification System for food” kit (Promega Corporation, Madison, WI), based on the manufacturer's protocol. The quantity and quality of extracted DNA were examined with Thermo-Scientific Nanodrop (LabX, Midland, ON, Canada), and by 0.8 % agarose gel electrophoresis, in comparison with a standard DNA (1 kb DNA Ladder, Fermentas GmbH).
2.3.1. Polymerase chain reaction (PCR) and sequencing
To molecularly identify Fusarium and Alternaria strains to the species level, and to evaluate the phylogenetic relationships within the two genera, a multi-locus sequencing approach was used. For the Fusarium strains, internal transcribed spacer regions (ITS), translation elongation factor 1α (tef1) and β-tubulin (tub2) genes were selected among the most informative genomic regions. For Alternaria strains, glyceraldephyde-3-phosphate dehydrogenase (gpd) and calmodulin (caM) gene fragments were chosen for the molecular analyses. The sequences of the primers used for PCR and the relevant references are summarised in Table 3.
Table 3.
Primers used for the molecular analysis of endophytic Fusarium spp. and Alternaria spp.
| Gene | Primer | Sequence (5'-3') | Reference |
|---|---|---|---|
| ITS | ITS5 | GGA AGT AAA AGT CGT AAC AAG G | [118] |
| ITS4 | TCC GCT TAT TGA TAT GC | [118] | |
| tef1 | EF1 | ATG GGT AAG GAR GAC AAG AC | [119] |
| EF2 | GGA RGT ACC AGT SAT CAT GTT | [119] | |
| tub2 | Bt2a | GGT AAC CAA ATC GGT GCT TTC | [120] |
| Bt2b | GGT AAC CAA ATC GGT GCT TTC | [120] | |
| gpd | Gpd1 | CAA CGG CTT CGG TCG CAT TG | [121] |
| Gpd2 | GCC AAG CAG TTG GTT GTG C- | [121] | |
| CaM | CALDF1 | AGC AAG TCT CCG AGT TCA AGG | [122] |
| CALDR2 | CTT CTG CAT CAY CTG GAC G | [122] |
Each PCR reaction (total volume of 15 μl), containing 15 ng of genomic DNA, 300 nM each primer, 0.8 mM dNTPs, 1x PCR buffer and 0.6 U of Hot Start Taq DNA polymerase (Fisher Molecular Biology, Roma, Italy), was performed in the Mastercycler epgradient thermocycler (Eppendorf). The following PCR conditions were used: 95 °C for 2 min; 35 cycles for ITS, gpd, and caM, and 40 cycles for tef1 and tub2 of denaturation at 95 °C for 30 s, annealing at 52 °C for 40 s for ITS, 58 °C for 40 s for tef1, 58 °C for 30 s for tub2, gpd, and caM, extension step at 72 °C for 50 s, and final extension at 72 °C for 7 min. The quality and quantity of PCR products were examined and visualized by UV light after electrophoresis separation in 1 × TAE buffer, on 1.5 % agarose gel, in comparison with 100 bp DNA ladder (Invitrogen, Thermo Fisher Scientific). The PCR products were purified with enzymatic mixture of Exonuclease I/FastAP thermosensitive alkaline phosphatase (Thermo Fisher Scientific, Vilnius, Lithuania). Subsequent sequencing was performed with the Big Dye Terminator v3.1 Cycle Sequencing Ready Reaction Kit (Applied Biosystems, Foster City, CA, USA), according to the manufacturer's instructions. Each labelled product was purified by filtration through Sephadex G-50 (5%) (Sigma-Aldrich, Saint Louis, MO, USA) and sequenced in “ABI PRISM 3730 Genetic Analyzer” (Applied Biosystems, Foster City, CA, USA).
2.3.2. Phylogenetic analysis
The raw DNA sequences were edited, cleaned and assembled using the BioNumerics v. 5.1 software (Applied Maths, Kortrijk, Belgium). For each considered locus, partial FASTA sequences of endophytic Fusarium or Alternaria strains, and sequences of reference strains, were aligned using the ClustalW algorithm [27]. The sequences of the strains used as reference for Fusarium and Alternaria specieswere downloaded through the Fusarioid-ID database (http://www.fusarium.org) and the National Center for Biotechnology Information (NCBI).
For each fungal genus, the combined phylogenetic trees were built using the maximum likelihood method with the MEGA software version 7 [28]. The bootstrap analysis [29] was conducted to determine the confidence of internal nodes using a heuristic search with 1000 replicates, removing gaps. In the phylogenetic analyses of Fusarium, the strain Atractium crassum CBS 180.31 was used as outgroup taxon. Whereas, for the phylogenetic analyses of Alternaria, A. malorum CBS 135.31, belonging to the Chalastospora section, was utilized. Sequences obtained in this study were deposited in the GenBank database, with the accession numbers listed in Table 1, Table 2
2.4. Production of mycotoxins by the endophytic fusarium isolates
Twenty-six Fusarium isolates belonging to 6 species, viz. F. equiseti (2 isolates), F. nirenbergiae (5 isolates), F. oxysporum (6 isolates), F. nygamai (9), F. proliferatum (3 strains), and F. sambucinum (1 strain), mostly isolated from tubers, were tested for mycotoxin production in duplicate (Table 1). For production of mycotoxins, isolates were grown on 50 g of rice in 250 mL Erlenmeyer Flasks left to imbibe overnight with 30 mL (approx. 60% v/w) distilled water and then sterilized at 121 °C for 30 min. Each flask was inoculated with five pieces of a fresh fungal culture on PDA and incubated for 21 days at 25 °C, shaking every day. Then, the inoculated kernels were dried at 55 °C for 48 h and finely milled.
2.4.1. Extraction of mycotoxins from rice cultures
Based on the mycotoxigenicity of the Fusarium species considered, as reported by Munkvold [30], cultures were analyzed for the presence of beauvericin (BEA), enniatins (ENNs, namely ENN A, ENN A1, ENN B, ENN B1), fumonisins B (FBs, namely FB1, FB2 and FB3), fusaric acid (FA), moniliformin (MON), the Type A trichothecenes diacetoxyscirpenol (DAS) and T-2 toxin (T-2), and the type B trichothecenes deoxynivalenol (DON), nivalenol (NIV) and their respective acetylates 3-AcDON, 15-AcDON and 4-AcNIV.
Two different extraction protocols were used. A multi-toxin extraction procedure was used for the extraction of BEA, ENNs, FBs, FA, and MON. One gram of ground rice culture was extracted with 5 mL of methanol/water (70:30, v/v) in an orbital shaker at 250 rpm for 60 min. The extract was filtered using Whatman No. 4 filters (Waters, Milford, MA, USA). One milliliter was diluted with 1 mL of water and then was filtered through 0.20 μm regenerated cellulose filter (Phenomenex, Torrance, CA, USA). The diluted culture extracts were used for the HPLC analyses.
The second procedure was used for the extraction of DON, as described in Quarta et al. [31]. One gram of ground rice culture was extracted in orbital shaker with 5 mL of acetonitrile/water (84:16, v/v) and 1% of acetic acid for 2 h. After filtration through filter paper (Whatman No. 4), 100 μL were diluted with 900 μL ultrapure water. The residue was filtered through 0.20 μm regenerated cellulose filter and analyzed by UHPLC/DAD.
2.4.2. Analyses of beauvericin and enniatins
Analyses of the cyclic hexadepsipeptides BEA and ENNs (ENN A, ENNA1, ENN B, ENN B1) were carried out by HPLC as described by Prosperini et al. [32] with minor modifications. One hundred microliters of the filtered extract of the culture (first extraction protocol) were injected into an HPLC apparatus (Agilent 1100 Series, Agilent Technology, Santa Clara, CA, USA) equipped with a binary solvent manager, a column heater set at 40 °C and a diode array (DAD) detector that was set at 205 nm wavelength. The analytical column was a Gemini (150 × 4.6 mm, 5 μm; Phenomenex) preceded by a SecurityGuard™ cartridge Gemini (4 × 3 mm, Phenomenex). The mobile phase was water as solvent A and acetonitrile as solvent B, eluted at a flow rate of 1 mL/min. A gradient elution was performed by changing the mobile phase composition. After 5 min at 70% eluent B, the proportion was set at 90% in 10 min, then kept constant for 3 min. The column was re-equilibrated with 70% eluent B for 5min. The retention times were: 11.3 min for BEA; 13.2 min for ENN A; 11.9 min for ENN A1; 10.5 min for ENN B; 9.1 min for ENN B1. The detection limits (LOD) based on signal-to-noise ratio of 3:1, were as follows: BEA = 0.01 μg/g, ENN A = 0.2 μg/g, ENN A1 = 0.5 μg/g, ENN B = 0.06 μg/g, ENN B1 = 0.07 μg/g. The mycotoxins were quantified by comparing peak areas with the calibration curves obtained with standard solutions.
2.4.3. Analysis of fumonisins
Analyses of FBs (FB1, FB2 and FB3) were carried out by HPLC/FLD according to the procedure described by Haidukowski et al. [33] with minor modifications. The culture extract (first procedure) diluted and filtered (50 μL) was derivatized with o-phtaldialdehyde (50 μL) using the HPLC autosampler Agilent 1100 and injected after 3 min. The analytical column was a SymmetryShield RP18 (15 cm x 4,6 mm, 5 μm; Waters) set at 30 °C. The mobile phase consisted of a binary gradient applied as follows: the initial composition of the mobile phase 60% of (A) acetonitrile-water-acetic acid (B) acetonitrile-water-acetic acid (60/39/1, v/v/v), was kept constant for 5 min, then B solvent was linearly increased to 88% in 21 min, and kept constant for 4 min. The flow rate of the mobile phase was 1 mL/min. The fluorometric detector was set at wavelengths, ex = 335 nm, em = 440 nm. Retention time was about 16.4 min for FB1, 25.4 min for FB2 and 26.6 min for FB3. Fumonisins were quantified by measuring peak areas and comparing them with the calibration curves obtained with standard solutions. LOD was 0.02 μg/g based on a signal-to-noise ratio of 3:1 for FB1, FB2 and FB3.
2.4.4. Analysis of fusaric acid
FA was determined by HPLC using a 1100 Agilent instrument. The analytical column was a Gemini (150 × 4.6 mm, 5 μm; Phenomenex). The temperature of the column was maintained at 40 °C. Constant flow was set at 1.0 mL/min and mobile phase was 1 % formic acid as solvent A and methanol with 1% of formic acid as solvent B. The starting gradient ratio was 80% of solvent A and the final ratio was set at 30% of solvent A in 15 min, then kept constant for 2 min. FA was detected at 272 nm. Retention time was about 7.9 min, and the LOD was 0.25 μg/g.
2.4.5. Analysis of moniliformin
Analysis of MON was carried out according to the procedure described by Parich et al. [34] with minor modifications. Fifty microliters of the extract obtained with the first procedure were injected into the HPLC apparatus with a column thermostat set at 30 °C and a DAD detector set at 229 nm. The analytical column was a Symmetry C18 (150 × 4.6 mm, 5 μm; Waters). The mobile phase was a mixture of water/formic acid (99:1, v/v, solvent A) and methanol/formic acid (99:1, v/v, solvent B) eluted at a flow rate of 1 mL/min. A gradient elution was performed as follows: 50% B solvent for 5 min; then was linearly increased to 70% in 2 min, 50% at 12 min and kept constant for 5 min. The retention time of mycotoxin was about 3.9 min, and the LOD was 0.1 μg/g.
2.4.6. Analyses of trichothecenes type B (DON, NIV, 3-AcDON and 4-AcNIV)
Analysis of trichothecenes group B was carried out according to the procedure described by Pascale et al. [35]. Seven point 5 μL of extract were injected into a Waters Acquity UPLC/PDA system. The analytical column was Aquity UPLC BEH RP-C18 (2.1 × 100 mm, 1.7 μm; Waters) with an Acquity UPLC column in-line filter (0.2 μm). The column heater was set at 50 °C and the detector at 220 nm. The isocratic flow was set at 0.350 mL/min and the mobile phase was water/methanol (85:15 v/v). Under these analytical conditions the retention times of target toxins were NIV 1.3 min, DON 2.1 min, 4-AcNIV 4.3 min, 3-AcDON 9.9 min and 15-AcDON 10.5 min. The detection limits (LOD) were 0.02 μg/g for NIV, DON and 4-AcNIV, 0.05 μg/g for 3-AcDON and 15-AcDON. DON production was confirmed using DONTest™ immunoaffinity column (VICAM, Watertown, MA, USA) method. One gram of rice culture was extracted with 5 mL of water in an orbital shaker at 250 rpm for 60 min. The extract was filtered, and 2 mL was passed through immunoaffinity column. After washing with 5 mL of water, DON was eluted with 1.5 mL of methanol. Then, the extract was dried and solubilized with 250 μL of acetonitrile/water (10:90, v/v). Seven point five μL of extract was injected into to UPLC apparatus as described above. The detection limit (LOD) of DON based on signal-to-noise ratio of 3:1 was 0.02 μg/g. DON was quantified by comparing peak areas with a calibration curve obtained with standard solutions.
2.4.7. Analyses of trichothecenes type A (DAS and T-2 toxin)
Analysis of trichothecenes group A was carried out according to the procedure described by Pascale et al. [36] with minor modifications. Seven point 5 μL of extract (second protocol) were injected into a Waters Acquity UPLC system. The analytical column was Aquity UPLC BEH RP-C18 (2.1 × 100 mm, 1.7 μm; Waters) with an Acquity UPLC column in-line filter (0.2 μm). The column heater was set at 50 °C and the detector at 202 nm. The chromatographic separation was performed by a gradient elution water as solvent A and acetonitrile as solvent B. The initial composition of the mobile phase. (80 % solvent A, 20% solvent B) was kept constant for 2 min, then solvent B was linearly increased to 50% in 3 min, and kept constant for 1 min. The flow rate of the mobile phase was 0.7 mL/min. Retention times were 1.7 min for DAS and 4.2 min for T-2 toxin. LOD for DAS and T-2 toxin were 4 μg/kg and 8 μg/kg, respectively.
3. Results and discussion
3.1. Endophytic fusarium species
The anamorphic genus Fusarium Link is arguably the most agronomically important fungal genus associated to potato plants, since members of the genus are well-known for their phytopathogenicity and mycotoxin production. Out of almost 400 total fungal endophytic isolates from potato, 75 were belonging to the genus Fusarium, accounting for 18.75% of total isolates [10]. Fusaria were mostly isolated from the underground parts of potato plants (roots and tubers), suggesting some tissue specificity.
Seventy-five isolates were molecularly identified to species level based on the multi-locus sequencing of the genes tef1, tub2 and ITS, and assigned to 13 species, viz. F. brachygibosum, F. clavum, F. equiseti, F. flocciferum, F. incarnatum, F. nirenbergiae, F. nygamai, F. oxysporum, F. proliferatum, F. redolens, F. sambucinum, F. solani and F. thapsinum (Table 1). Five Fusarium species, namely F. oxysporum, F. redolens, F. semitectum, F. solani, and F. sporotrichioides were previously reported as endophytic in potato [13]. In our survey, we did not find any endophytic strains of either F. semitectum or F. sporotrichioides.
The phylogenetic analysis of the concatenated sequences of 1337 sites resulted in a phylogenetic combining dataset comprising 92 taxa, including 75 Fusarium field strains, 16 Fusarium reference sequences and the strain Atractium crassum CBS 180.31 as outgroup taxon. Four out of the 79 endophytic Fusarium strains isolated (FU-57, -28, −56, −80) were not included in phylogenetic analyses since they did not give PCR products of tef1. However, the comparison of tub and ITS sequences through the Blast N program (http://www.ncbi.nlm.nih.gov/) allowed us to assign these strains to the genus Plectospharella. A high homology (more than 99%) was found with the deposited sequences of P. cucumerina (syn. Fusarium tabacinum) strains.
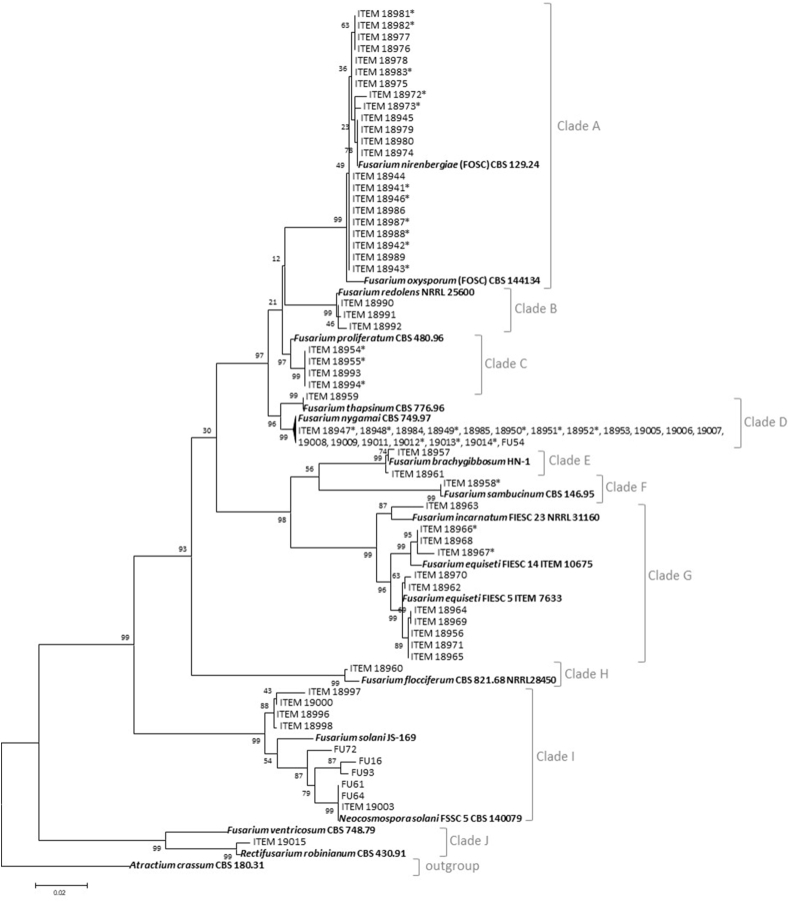
The phylogenetic tree, rooted to outgroup taxon, was resolved in 10 well-separated clades, supported by high significance (bootstrap values more than 96), as reported in Fig. 1. Twenty-two out of 75 strains and F. oxysporum CBS 144,134 and F. nirembergiae CBS 129.24 reference strains grouped together in a well-supported clade corresponding to the Fusarium oxysporum species complex (FOSC, clade A). In this clade, a very low genetic variability was observed. Indeed, all the strains showed high level of similarity among them and with the two reference strains included in the analyses. In particular, the strains ITEM-18945, -18979, −18980, −18974 showed 100% homology with the F. nirembergiae reference strain. Clade B, referred to as Fusarium redolens species complex (FRSC), grouped the reference strain F. redolens NRRL 25600 together with the strains ITEM-18990, -18991, and −18992 with high level of homology. Twenty-four endophytic Fusarium strains, identified as belonging to Fusarium fujikuroi species complex (FFSC), were grouped in clades C and D. Four Fusarium strains, showing 100% of homology among them, clustered with F. proliferatum reference strain (clade C). In clade D, two clusters were distinguished: the first cluster contained a single strain (ITEM-18959) that showed high similarity with F. thapsinum CBS 776.96 and the second cluster contained majority of the strains (19 out of 20) along with F. nygamai CBS 749.97 reference strain.Only two strains (ITEM-18957 and -18961) clustered with F. brachygibbosum NH-1 reference strains (clade E) and one single strain with F. sambucinum CBS 146.95 strain (Clade F). Eleven endophytic Fusarium strains included in the clade G, shared high genetic diversity. This clade included strains assigned to different species of Fusarium incarnatum-equiseti species complex (FIESC). One single strain (ITEM-18963) was genetically closely related to the reference strain F. incarnatum FIESC 23. Three strains (ITEM-18966, -18968, and −18967) grouped with the reference strains F. equiseti FIESC 14. Finally, in a well-supported group, 7 Fusarium strains clustered with the reference strain F. equiseti FIESC 5. Likewise, large diversity was observed in the clade I, in which 10 endophytic F. solani strains clustered with the two F. solani strains used as references of the Fusarium solani species complex (FSSC). On the other hand, two strains (ITEM-18960 and -19015) showed a high similarity with F. flocciferum CBS 821.68, member of Fusarium tricinctum species complex, and F. robinianum (syn. Rectifusarium robinianum), respectively (clades H and J).
Fig. 1.
Phylogenetic tree generated by Maximum Likelihood method (bootstrap 1000 replicates) from combined DNA sequences of tub, tef1, and ITS fragments of 75 endophytic Fusarium strains isolated from different organs of potato plants, in Iran. Atractium crassum strain CBS 180.31 was used as an outgroup. Isolates with an asterisk were used for mycotoxin analysis.
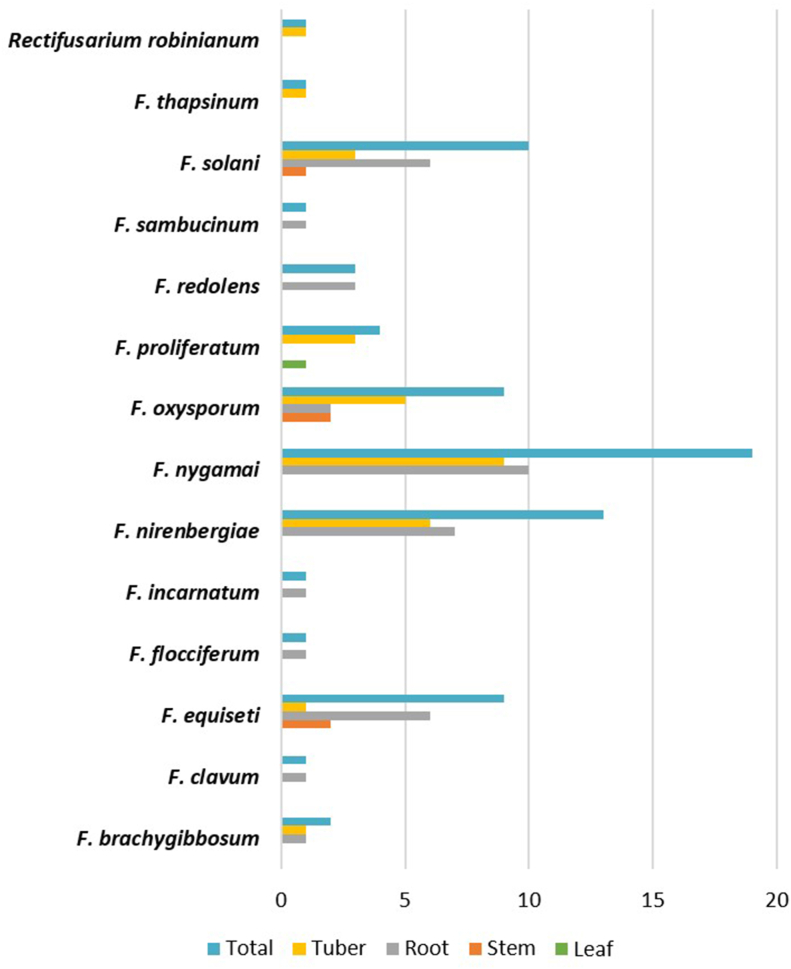
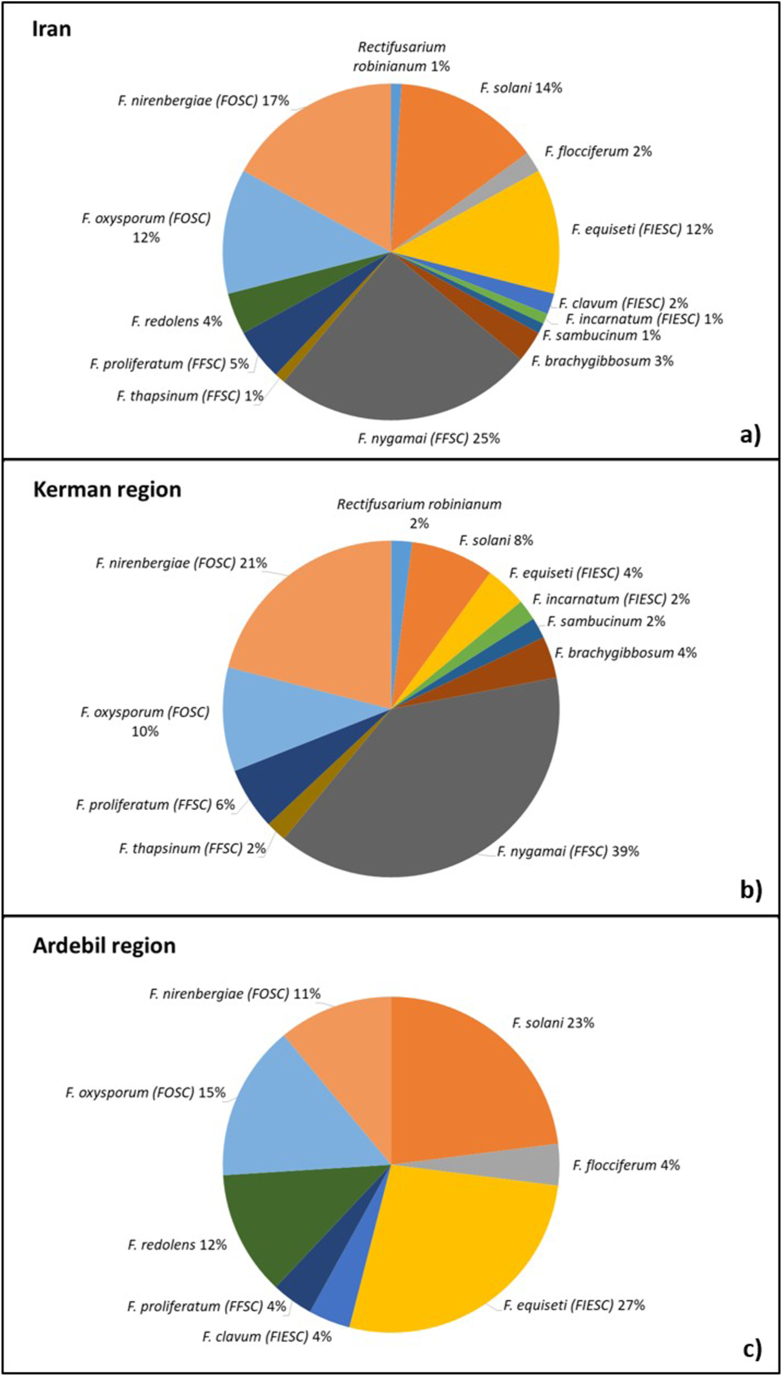
Ninety percent of endophytic Fusaria strains were isolated from roots and tubers. Only four species, viz. F. nygamai, F. oxysporum and F. solani, were isolated from stems and only one isolate of F. proliferatum was obtained from potato leaves (Fig. 2). At a whole, F. nygamai, member of FFSC, F. oxysporum and F. nirembergiae, both members of FOSC, F. solani and F. equiseti (FIESC), were the most abundant species (Fig. 3a). In particular, F. nygamai was predominant in south Kerman (Fig. 3b), and F. equiseti, F. oxysporum and F. solani were more frequently isolated in Ardebil (Fig. 3c). Therefore, F. nygamai seems to be more adapted to dry and hot climate conditions, while F. oxysporum and F. solani can apparently colonize potato plants under diverse climate conditions.
Fig. 2.
Occurrence and distribution of endophytic Fusarium species in different organs of potato plants.
Fig. 3.
Frequency of endophytic Fusarium species in potato plants from Iran (a), detailed for South Kerman (b) and Ardebil (c) regions. The species complexes are reported in brackets: FFSC: Fusarium fujikuroi species complex, FOSC: F. oxysporum species complex, FIESC: F. incarnatum-equiseti species complex.
The species F. brachygibbosum, F. clavum, F. flocciferum and F. nirenbergiae are herein reported as endophytic for the first time. So far, F. brachygibbosum has been reported as a plant pathogen of different crops, including wheat [37], maize [38], date palm [39,40], Cannabis sativa [41], 2019) and several medicinal plants [42,43], but not potato. Fusarium clavum was previously isolated from crop plants cultivated under both conventional and organic farming [44]. The species causes post-harvest contamination of wheat, barely, and maize [45] and associated to leaf spot disease in vegetable plants [46] and leaf wilt in date palm [47]. Fusarium flocciferum is a common species in temperate regions, soil, roots, fruits, stems and twigs of various plants [48]. Fusarium nirenbergiae is a plant pathogenic species and an agent of vascular diseases such as wilting of common bean [49], passion fruit (Passiflora edulis Sims) [50] and ornamental plants [51], but also reported as the agent of saffron corm rot disease [52].
The species F. equiseti, F. incarnatum, F. proliferatum, F. nygamai, F. sambucinum and F. thapsinum were reported to be endophytes in different plants, but not in potato. Particularly, F. equiseti was isolated from leaf of the medicinal plant Sophora tonkinensis [53], barley roots [54], and Salicornia bigelovii [55]. Fusarium incarnatum is the causal agent of postharvest fruit rot in muskmelon (Cucumis melo) [56], and of a fruit disease of bell peppers [57], but the species is reportedly endophytic in the mangrove plant Aegiceras corniculatum [58]. Fusarium proliferatum is a widespread phytopathogen in a number of major crops, including rice, wheat, maize, garlic, asparagus, date palm, and Chinese chive [59], and an endophyte in wheat [60]. On the other hand, few studies report the endophytic associations of F. nygamai with medicinal plants, namely Alhagi graecorum, Cressa cretica, Citrullus colocynthis, Tamarix nilotica, Achillea fragrantissima, Artemisia sieberi, and Neurospora retusa [42], and rice root [61]. Fusarium sambucinum, which is recognized as a major agent of dry rot of potato tubers [24] has also been reported as an endophyte from Nicotiana tabacum [62](Zhang et al., 2019). Fusarium thapsinum causes stalk rot and grain mold of sorghum [24], but it was also found to be endophytic in the same plant [63].
3.2. Endophytic Alternaria species
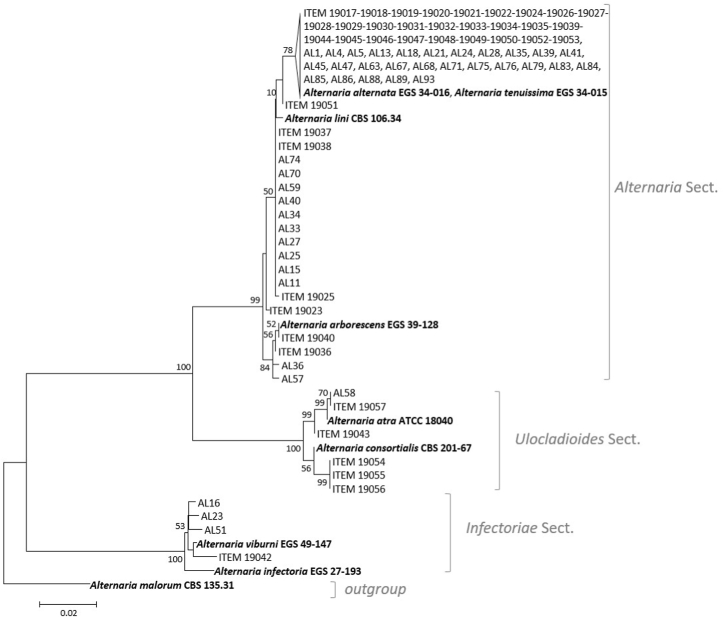
The phylogenetic combining dataset comprised 82 taxa, including 83 Alternaria field strains, and 9 additional Alternaria reference sequences, among which A. malorum CBS 135.31 which was used as outgroup taxon. The Alternaria isolates were molecularly identified at the species level based on the multi-locus sequencing of the genes gpd and CaM (Table 3).
The phylogenetic tree, rooted to outgroup taxon, was resolved in 3 well-separated clades, supported by high bootstrap values (Fig. 4). Most of the strains (73 out of 83) were included in the clade A, referred as Alternaria section. In this clade, high genetic homology was observed among the strains. Indeed, except for two strains (ITEM-19051 and -19023), 54 Alternaria strains clustered with A. alternata EGS34-016 and A. tenuissima EGS34-015 reference strains; 13 strains were highly similar to A. lini CBS 106.34 reference strain, and four strains clustered, in a well-separated group (bootstrap value 84) with A. arborescens 39–128 reference strain. In clade B, referred as Ulocladioides section, 6 strains grouped with two members of this section, A. consortialis and A. utra. The strains ITEM-9043, AL-58, and ITEM-19057 were very close to A. utra ATCC18040 reference strain; whereas ITEM-19054, -19055 and −19057, showing 100% similarity among them, were very close to A. consortialis CBS 201.67 reference strain. Finally, four strains (ITEM-19042, AL-51, -16, and −23) grouped with A. infectoria EGS27-193 and A. viburni EGS 49–147 reference strains in the Infectoriae section (clade C).
Fig. 4.
Phylogenetic tree generated by Maximum Likelihood method (bootstrap 1000 replicates) from combined DNA sequences of gpd and caM fragments of 83 endophytic Alternaria strains isolated from different organs of potato plants, in Iran. Alternaria malorum strain CBS 135.31 was used as an the outgroup.
Alternaria spp. are ubiquitous fungi that include endophytic, saprophytic, and pathogenic species commonly found in soil, air, food commodities, and on decaying plant tissues [64]. The pathogenic species of Alternaria cause major losses on a wide range of crops such as cereals, oil crops, ornamentals, vegetables and fruits [65]. In our survey, out of almost 400 total fungal endophytic isolates, 96 were identified as Alternaria spp. [10], that is 24.06 % of total isolates. Ninety-five percent of endophytic Alternaria strains were isolated from the above-ground plant parts (stems and leaves). Only five isolates were originated from roots, four of which belonged to the Alternaria section and one to the Infectoriae section (Fig. 4). It appears that the different climatic conditions in the two Iranian regions of Ardebil and Kerman do not affect significantly the distribution of endophytic Alternaria; in both the regions the Alternaria section was predominant, supposedly due to greater tolerance to diverse climate conditions.
The species A. alternata, A. tenuissima, and A. lini were the most abundantly occurring species in this study. However, in the last decade Alternaria taxonomy has been deeply revised, based on genetic and genomic investigations. Sequencing analyses of the most informative genomic regions led to the conclusion that species that were morphologically different could not be distinguished genetically [66,67]. Therefore, around 35 morpho-species, including A. alternata, A. tenuissima and A. lini, should be considered one single species and synonymized as A. alternata [66]. Recently, this finding has been supported also in a genome-wide study based on phylogenomic analyses of Alternaria species belonging to Alternaria Section [68]. Anyway, morpho-species names continue to be reported in several studies. Previously, only three Alternaria species, namely A. alternata, A. solani, and A. brassicola were reported as endophytic in potato [11,13]. Alternaria alternata is a cosmopolitan species with a wide host range worldwide, including cereals [69,70], vegetables [64,71] and halophyte plants [72]. It is pathogenic in many important crops and also lives in asymptomatic symbiosis as an endophyte of many plants [73], including potato, Nicotiana spp. [74], forest tree species [75], and Juncus spp [76]. This is the first report of A. lini as an endophytic species. Alternaria tenuissima has been widely reported as endophyte from different plants [77,78], but it is herein reported as an endophyte of potato for the first time. Alternaria arborescens was isolated as endophyte from root tissue of Combretum latifolium [79], and is also reported as a phytopathogen of potato and tomato [64,71,80,81], and a human pathogen [82]. Alternaria atra (previously known as Ulocladium atra [65] was reported as an endophyte of common yew (Taxus baccata L.) in Iran [83]. Alternaria consortialis (previuosly knwon as U. consortilis [65], was reported as endophyte of several plants in Iran, including Prunus trees [84], and spinach [85], and as a phytopathogen of declined Persian oak trees in Iran [86] and on date palm showing leaf spot in Tunisia [87]. Alternaria viburni, previously known as Lewia viburni [65], is reported as endophyte from timothy (Phleum pratense) and perennial ryegrass [88]. This species belongs to the Infectoriae Section, which is phylogenetically divergent from the Alternaria Section and characterized by a great genetic variability within and among its species members [67].
While Alternaria spp. are mainly regarded as plant pathogenic fungi that cause diseases of aerial parts of plants and are also capable of producing potentially harmful metabolites [89], beneficial effects of endophytic Alternaria on plant growth have also been reported [90]. Whether the endophytic Alternaria strains associated with potato plants may turn to be pathogenic and under which conditions this may occur, or conversely may have beneficial effect on potato plants, remains to be clarified and is worth of further investigation.
3.3. Mycotoxin production by potato endophytic fusarium strains
The toxigenicity of twenty-six Fusarium strains from potato root and tubers was assessed. The strains tested belonged to 6 species, viz. F. equiseti (2 strains), F. nirenbergiae (5 strains), F. oxysporum (6 strains), F. nygamai (9 strains), F. proliferatum (3 strains), and F. sambucinum (1 strain). Twenty-one out of 26 strainswere able to produce one or more mycotoxin (Table 4).
Table 4.
Production of mycotoxins by representative isolates of major endophytic Fusarium species from potato root and tuber. None of the isolates tested produced either the type B trichothecenes deoxynivalenol, nivalenol or their respective acetylates or the type A trichothecenes diascetoxyscirpenol and T-2 toxin.
| Species |
Strain |
Mycotoxin production (μg/g)a |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Enniatins |
BEA |
Fumonisins |
MON |
FA |
|||||||
| B | B1 | A | A1 | B1 | B2 | B3 | |||||
| F. nirenbergiae | ITEM 18972 | n.d. | n.d. | n.d. | n.d. | 396 | n.d. | n.d. | n.d. | 82 | n.d. |
| ITEM 18973 | n.d. | n.d. | n.d. | n.d. | 101 | n.d. | n.d. | n.d. | 63 | n.d. | |
| ITEM 18983 | n.d. | n.d. | n.d. | n.d. | 1382 | n.d. | n.d. | n.d. | 66 | n.d. | |
| ITEM 18981 | n.d. | n.d. | n.d. | n.d. | 952 | n.d. | n.d. | n.d. | 31 | n.d. | |
| ITEM 18982 | n.d. | n.d. | n.d. | n.d. | 332 | n.d. | n.d. | n.d. | 27 | n.d. | |
| F. oxysporum | ITEM 18988 | n.d. | n.d. | n.d. | n.d. | 46 | n.d. | n.d. | n.d. | 21 | n.d. |
| ITEM 18941 | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | 105 | n.d. | |
| ITEM 18946 | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | |
| ITEM 18987 | n.d. | n.d. | n.d. | n.d. | 525 | n.d. | n.d. | n.d. | n.d. | n.d. | |
| ITEM 18942 | n.d. | n.d. | n.d. | 648 | n.d. | n.d. | n.d. | n.d. | n.d. | 156 | |
| ITEM 18952 | n.d. | n.d. | n.d. | 446 | 7 | n.d. | n.d. | n.d. | n.d. | n.d. | |
| F. proliferatum | ITEM 18954 | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. |
| ITEM 18955 | n.d. | n.d. | n.d. | n.d. | 235 | 62 | 23 | 11 | n.d. | n.d. | |
| ITEM 18994 | n.d. | n.d. | n.d. | n.d. | 58 | 3 | 2 | 2 | n.d. | n.d. | |
| F. nygamai | ITEM 18952 | n.d. | n.d. | n.d. | n.d. | n.d. | 5 | 2 | 2 | n.d. | n.d. |
| ITEM 18947 | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | |
| ITEM 18948 | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | 26 | n.d. | |
| ITEM 18951 | n.d. | n.d. | n.d. | n.d. | n.d. | 105 | 29 | 24 | 67 | n.d. | |
| ITEM 19012 | n.d. | n.d. | n.d. | n.d. | 4190 | 7946 | 4693 | 4333 | 538 | n.d. | |
| ITEM 19013 | n.d. | n.d. | n.d. | n.d. | 41 | 49 | 18 | 13 | 4 | n.d. | |
| ITEM 19014 | n.d. | n.d. | n.d. | n.d. | n.d. | 126 | 5 | 4 | 26 | n.d. | |
| ITEM 18949 | n.d. | n.d. | n.d. | n.d. | 458 | n.d. | n.d. | n.d. | 75 | n.d. | |
| ITEM 18950 | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | 4 | n.d. | |
| F. equiseti | ITEM 18966 | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. |
| ITEM 18967 | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | |
| F. sambucinum | ITEM 18958 | n.d. | n.d. | n.d. | n.d. | 21 | n.d. | n.d. | n.d. | n.d. | n.d. |
amounts are the mean of two replicated cultures on sterile rice; BEA = beauvericin; MON = moniliformin; FA = fusaric acid; n.d. = not detected.
None of the tested strains produced either type A or type B trichothecenes. Two out of two F. equiseti strains, 1/6 F. oxysporum, 1/3 F. proliferatum, and 1/9 F. nygamai did not produce any of the other tested toxins. All the 5 strains of F. nirenbergiae produced both BEA (from moderate to high quantity, 101.0–1382.5 μg/g) and MON (2.9–82.1 μg/g). Fusarium oxysporum showed a more variable mycotoxin profile: ENN A1 was produced by 2/6 strains, BEA by 3/6, MON by 2/6 and FA by 1/6. BEA was found in combination with MON in a single strain; ENN A1 was found in combination with either FA (1 strain) or BEA (1 strain). Among the F. proliferatum strains, 1/3 did not produce any of the mycotoxins analyzed, while the other 2 strains produced both BEA and FBs (B1, B2, and B3). In F. nygamai, which was the most numerous group, 3/9 strains produced BEA, 4/9 produced FBs (B1, B2, and B3), and 7/9 produced MON. All the fumonisins-producing strains also produced MON and 2 of them produced BEA, as well. One isolate produced BEA in combination with MON. The most toxigenic strain was ITEM-19012, which produced the highest quantities of FBs (7946.5, 4693.0 and 4333.0 μg/g of B1, B2, and B3 respectively), along with the highest quantities of both BEA (4190.0 μg/g) and MON (538.4 μg/g). The only toxin produced by F. sambucinum was BEA.
Amongst the regulated mycotoxins, only FBs were produced by potato endophytic strains of Fusarium spp., while trichothecenes were not found in cultures of two representative isolates of F. equiseti and one F. sambucinum isolate, reportedly the only two trichothecene-producing species among the ones analyzed [30]. Fumonisins were produced by strains of F. proliferatum and F. nygamai, in all of the three most important forms B1, B2, and B3. Particularly, the ITEM 19012 F. nygamai strainproduced significantly more FBs than the other strains. This particular strain also produced the highest levels of both BEA and MON. Indeed, members of F. fujikuroi complex are known to produce multiple mycotoxins, including BEA, ENNs, FUMs, fusaproliferin, FA, fusarins, and MON [18]. Co-occurrence of MON with the cyclodepsipeptides BEA and ENNs has been also reported in F. sporotrichioides, F. equiseti, F. avenaceum and F. oxysporum [18].
Besides the regulated mycotoxins, we also investigated production of some “emerging” mycotoxins, which are neither routinely determined, nor legislatively regulated because of their lesser occurrence or uncertain toxicity in vivo. ENNs and BEA are toxic compounds similar for both structure and mode of action. They are cyclic hexadepsipeptides consisting of three d-α-hydroxy-isovaleryl-(2-hydroxy-3-methylbutanoic acid) alternating with three amino acid units and are biosynthesized by the non-ribosomal multifunctional enzymes enniatin and beauvericin synthetases. They both have ionophoric properties; they can form stable lipophilic complexes with cations and transport them into cell membrane and form cation-selective channels in membranes, thus impairing the membrane functions [91,92]. In potato, ENNs are virulence factors within the plant-pathogen interaction [93]. They have phytotoxic effects that include wilting and necrosis of tissues and are produced in vivo in potato tuber tissue infected by Fusariun spp [23]. However, their function as virulence factor seems to be host dependent [94]. Likewise, BEA showed strong phytotoxicity to tomato protoplasts, conceivably by inducing release of ascorbate from the cytosol to the apoplast of plant cell, with a consequent increase in cell wall plasticity that facilitates pathogen penetration [95]. BEA, along with FA, were contaminants of banana plants and fruits infected by F. oxysporum f. sp. cubense and proved to have phytotoxic effects on banana protoplasts [96]. In that study, virulence of F. oxysporum f. sp. cubense isolates correlated well with toxin accumulation. BEA was produced, in combination with unusual forms of ENNs, by fifteen F. oxysporum and two not-identified Fusarium sp. strains, out of twenty-eight Fusarium isolates from potato samples in Korea [97]. ENNs and BEA exhibit different toxic effects in vitro, including cytotoxicity and necrotic and pro-apoptotic effects to animal cell lines, but toxicity in vivo is generally low. This is thought to be due to their rapid metabolization rather than low bioavailability. MON is a phytotoxin, first isolated by Cole et al. [98]. Despite its name, nowadays MON is thought to be produced by a few Fusariun species, including F. oxysporum, F. proliferatum, F. nygamai, F. equiseti and F. sambucinum, but not by F. moniliforme [30]. MON is extremely soluble in water, due to its polarity, thus it is easily translocated through the plant. MON causes growth inhibition, necrosis, and chlorosis in many plants [99]. MON has a selective cytotoxicity in vitro, but shows severe effects in vivo, with symptoms of acute intoxication in test animals that include muscular weakness, respiratory stress, myocardial degeneration, and histopathological changes in kidneys, lungs, and pancreas and ultimately coma and death. FA has long been known as a wilt toxin [100] and is regarded as a virulence factor in plant tracheo-fusariosis caused by F. oxysporum ff. spp. in different crops [[101], [102], [103]], including potato [104]. FA is moderately toxic to animals, but it may potentiate the effect of other Fusarium toxins.
Although the above toxins are mainly regarded as phytotoxins that participate in the pathogenicity process, because of their occurrence in vivo and toxicity in animal models, they are emerging as possible concern for food safety. A recent study showed that BEA was the emergent mycotoxin with the highest prevalence in feed and feed ingredients, followed by ENNs [105]. Naturally, MON often co-occurs with ENNs and BEA, but also with trichothecenes, since several trichothecene-producing species produce MON, including F. acuminatum, F. culmorum, F. equiseti, and F. sporotrichioides [106]. FA is widespread on corn and corn-based food and feeds, frequently in association to other mycotoxigenic Fusarium species [107]. These toxins may contribute to the overall health risk of contaminated foods or feeds, either because of their direct toxic effects or because of additive or synergistic interaction with other co-occurring mycotoxins.
All the isolates we analyzed for mycotoxin production were endophytes of potato plants, recovered from healthy tubers or roots. Endophytic vs. pathogenic behavior of fungi is the outcome of a finely tuned balance, which depends on a molecular cross-talking between plant and symbiont [108]. Even if the molecular and physiological aspect of this interaction and its determinants are not completely clear, it is widely accepted that endophytism is a continuum which results from an equilibrium between fungal virulence and plant defense, and that endophytic behavior might revert to pathogenic, if one or more factors change [109]. In this sense, the chance that endophytic Fusaria become pathogenic and produce mycotoxins in tubers cannot be ruled out. Logrieco et al. [110] examined the production of zearalenone, zearalenols, trichothecenes and MON in cultures of some Fusarium species isolated from rotted potato tubers. They found that strains able to produce mycotoxins on autoclaved rice and maize kernels were not capable to produce toxins on fresh potato under any of the conditions tested. Conversely, Ellner [22] reported that potato tubers artificially infected with F. sambucinum were contaminated with the trichothecene toxin diacetoxyscirpenol in concentrations up to 200 μg/tuber, depending on the susceptibility of the cultivar tested. The toxin was also detected in tubers with no apparent disease symptoms. Likewise, Delgado et al. [21]studied the accumulation and diffusion of trichothecenes in potato tubers affected with dry rot caused by F. graminearum during storage. They found accumulation of either DON or NIV, depending on the F. graminearum genotype, in rotten tissue but not in the surrounding tissue. These findings suggest that contamination of potato tubers with mycotoxins in the field or at post-harvest, due to a change in lifestyle of endophytic microflora, is a potential risk that should be carefully considered and that is worth of further studies, along with possible means of control.
4. Conclusions
Plant-microbe endophytic association may bring a vast range of beneficial effects to the host plants. Endophytes produce biologically active metabolites with diverse activities, which interact with their host's physiology and ecology in different ways. These compounds can benefit host plants by protecting them against insect herbivores [111,112], by inhibiting plant pathogen growth and by eliciting host immune system against pathogens attacks [113]. On the other hand, some endophytes may actually become detrimental to their hosts, especially under stress conditions [109], or they may become pathogenic to crop plants other than their original host [114]. Thus, some endophytic fungi can be actually regarded as latent pathogens [8]. The chance that under certain conditions the endophytes of potato may change their lifestyle and become pathogenic to potato or other crop plants, should be considered.
Members of Fusarium are found worldwide, predominantly as soil inhabitants and plant pathogens, with the existence of nonpathogenic isolates [115,116]. In addition, some members of Fusarium, including some herein found to live endophytically in potato, are known to produce numerous toxins, some of which are regarded as virulence factors [93] or emerging mycotoxins (viz. ENNs, BEA, MON and FA) and others are regulated mycotoxins (viz. fumonisins), which may pose safety concerns for consumers. It is not yet well-established if, in certain conditions, the endophytic Fusaria may switch to a parasitic lifestyle and, in that case, if accumulation of toxins in the edible tubers can occur in the field or during storage. The findings of this and previous work [10], also open the prospect to conceivable strategies aimed at controlling pathogens of potato and, particularly, for biological control of mycotoxigenic Fusaria. Classical biological control is founded on the concept that most effective biocontrol agents should be sought amongst the coevolved antagonists of the target pathogen, which are supposed to have the highest specificity and biocontrol activity. In this regard, the report of the occurrence of fungi belonging to Trichoderma and Clonostachys, two well-known genera of biocontrol agents [117], as endophytes of potato in Iran [10] paves the way for studies on their use for sustainable control of potentially pathogenic endophytic fungi and for prevention of mycotoxin contamination of potatoes.
Funding
This work was supported by the Iran National Science Foundation (INSF) [grant number 98015699].
Additional information
No additional information is available for this paper.
Data availability statement
Data will be made available on request.
The data and supportive information are available within the article. Data associated with the study have been deposited into publicly available repositories. Sequences were deposited in the GenBank database; strains were deposited in the microbial ITEM collection (http://server.ispa.cnr.it/ITEM/Collection/).
CRediT authorship contribution statement
Nasim Alijani Mamaghani: Writing – original draft, Visualization, Resources, Investigation, Formal analysis, Data curation. Mario Masiello: Writing – original draft, Visualization, Validation, Methodology, Investigation, Formal analysis, Data curation. Stefania Somma: Writing – review & editing, Visualization, Validation, Methodology, Investigation, Formal analysis, Data curation. Antonio Moretti: Writing – review & editing, Supervision, Resources, Methodology, Conceptualization. Hossein Saremi: Writing – original draft, Supervision, Resources, Project administration, Funding acquisition, Conceptualization. Miriam Haidukowski: Writing – review & editing, Validation, Resources, Methodology, Investigation, Formal analysis, Data curation. Claudio Altomare: Writing – review & editing, Writing – original draft, Visualization, Supervision, Funding acquisition, Conceptualization.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Miriam Haidukowski reports travel was provided by Iran National Science Foundation. Miriam Haidukowski reports financial support, administrative support, article publishing charges, and equipment, drugs, or supplies were provided by Institute of Sciences of Food Production, National Research Council, Italy. If there are other authors, they declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
Nasim Aljani Mamaghani thanks Prof. Orlando Petrini, Prof. Mohammad Javan-Nikkhah, Prof. Mauro Tonolla and Dr. Sophie De Respinis for all their support and kindness during her PhD, the “University of Tehran”, Tehran, Iran, “University of Applied Sciences and Arts of Southern Switzerland (SUPSI)”, and the “Institute of Sciences of Food Production, National Research Council” (ISPA-CNR), Bari, Italy, for providing scientific guidance and training during her research work for doctorate.
Contributor Information
Stefania Somma, Email: stefania.somma@ispa.cnr.it.
Miriam Haidukowski, Email: miriam.haidukowski@ispa.cnr.it.
References
- 1.Mishra S., Bhattacharjee A., Sharma S. An ecological insight into the multifaceted world of plant-endophyte association. Crit. Rev. Plant Sci. 2021;40(2):127–146. doi: 10.1080/07352689.2021.1901044. [DOI] [Google Scholar]
- 2.Verma S.K., Sahu P.K., Kumar K., Pal G., Gond S.K., Kharwar R.N., et al. Endophyte roles in nutrient acquisition, root system architecture development and oxidative stress tolerance. J. Appl. Microbiol. 2021;131(5):2161–2177. doi: 10.1111/jam.15111. [DOI] [PubMed] [Google Scholar]
- 3.Dey P., Datta D., Saha D., Parida S., Panda D. Plant-endophyte interaction and its application to abiotic stress management of crop plants. Int J Curr Microbiol Appl Sci. 2019;8(7):2708–2716. doi: 10.20546/ijcmas.2019.807.332. [DOI] [Google Scholar]
- 4.Rodriguez R.J., White J.F., Arnold A.E., Redman R.S. Fungal endophytes: diversity and functional roles. New Phytol. 2009;182(2):314–330. doi: 10.1111/j.1469-8137.2009.02773.x. [DOI] [PubMed] [Google Scholar]
- 5.White J.F., Kingsley K.L., Zhang Q., Verma R., Obi N., Dvinskikh S., et al. Review: endophytic microbes and their potential applications in crop management. Pest Manag. Sci. 2019;75(10):2558–2565. doi: 10.1002/ps.5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xia Y., Sahib M.R., Amna A., Opiyo S.O., Zhao Z., Gao Y.G. Culturable endophytic fungal communities associated with plants in organic and conventional farming systems and their effects on plant growth. Sci. Rep. 2019;9(1):1669. doi: 10.1038/s41598-018-38230-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chitnis V.R., Suryanarayanan T.S., Nataraja K.N., Prasad S.R., Oelmüller R., Shaanker R.U. Fungal endophyte-mediated crop improvement: the way ahead. Front. Plant Sci. 2020;11 doi: 10.3389/fpls.2020.561007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hardoim P.R., van Overbeek L.S., Berg G., Pirttilä A.M., Compant S., Campisano A., et al. The hidden world within plants: ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol. Mol. Biol. Rev. 2015;79(3):293–320. doi: 10.1128/MMBR.00050-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Imani B., Allahyari M.S., Bondori A., Emami N., El Bilali H. Adoption of organic potato production in Ardabil plain, Iran: an application of the extended theory of planned behaviour. Potato Res. 2020:177–195. doi: 10.1007/s11540-020-09471-z. [DOI] [Google Scholar]
- 10.Alijani Mamaghani N., Saremi H., Javan-Nikkhah M., Respinis S.D., Pianta E., Tonolla M. Endophytic Cephalotrichum spp. from Solanum tuberosum (potato) in Iran – a polyphasic analysis. Sydowia. 2022;74:287–301. [Google Scholar]
- 11.O'Callaghan M., Gerard E.M., Waipara N.W., Young S.D., Glare T.R., Barrell P.J., et al. Microbial communities of Solanum tuberosum and magainin-producing transgenic lines. Plant Soil. 2005;266(1–2):47–56. doi: 10.1007/s11104-005-3714-1. [DOI] [Google Scholar]
- 12.Gotz M., Nirenberg H., Krause S., Wolters H., Draeger S., Buchner A., et al. Fungal endophytes in potato roots studied by traditional isolation and cultivation-independent DNA-based methods: fungal endophytes in potato roots. FEMS Microbiol. Ecol. 2006;58(3):404–413. doi: 10.1111/j.1574-6941.2006.00169.x. [DOI] [PubMed] [Google Scholar]
- 13.Marak M.C.N., Kayang H. Isolation and identification of endophytic fungi associated with Solanum tuberosum L. Of south-west Garo Hills, Meghalaya. IJAAST. 2018;5(1):58–65. [Google Scholar]
- 14.Kokaeva L.Y., Belosokhov A.F., Doeva L.Y., Skolotneva E.S., Elansky S.N. Distribution of Alternaria species on blighted potato and tomato leaves in Russia. JPDP. 2018;125(2):205–212. doi: 10.1007/s41348-017-0135-3. [DOI] [Google Scholar]
- 15.Van De Vijver R., Mertens K., Heungens K., Somers B., Nuyttens D., et al. In-field detection of Alternaria solani in potato crops using hyperspectral imaging. Comput. Electron. Agric. 2020;168 doi: 10.1016/j.compag.2019.105106. [DOI] [Google Scholar]
- 16.Saharan G.S., Mehta N., Meena P.D. Springer Singapore; 2016. Alternaria Diseases of Crucifers: Biology, Ecology and Disease Management. [DOI] [Google Scholar]
- 17.Bojanowski A., Avis T.J., Pelletier S., Tweddell R.J. Management of potato dry rot. Postharvest Biol. Technol. 2013;84:99–109. doi: 10.1016/j.postharvbio.2013.04.008. [DOI] [Google Scholar]
- 18.Munkvold G.P., Proctor R.H., Moretti A. Mycotoxin production in Fusarium according to contemporary species concepts. Annu. Rev. Phytopathol. 2021;59(1) doi: 10.1146/annurev-phyto-020620-102825. annurev-phyto-020620-102825. [DOI] [PubMed] [Google Scholar]
- 19.Altomare C., Logrieco A.F., Gallo A. Mycotoxins and mycotoxigenic fungi: risk and management. A challenge for future global food safety and security. Encycl. Mycol. 2021;1:64–93. doi: 10.1016/B978-0-12-819990-9.00032-9. [DOI] [Google Scholar]
- 20.Ali S., Rivera V.V., Secor G.A. First report of Fusarium graminearum causing dry rot of potato in North Dakota. Plant Dis. 2005;89:105. doi: 10.1094/PD-89-0105B. [DOI] [PubMed] [Google Scholar]
- 21.Delgado J.A., Schwarz P.B., Gillespie J., Rivera-Varas V.V., Secor G.A. Trichothecene mycotoxins associated with potato dry rot caused by Fusarium graminearum. Phytopathol. 2010;100:290–296. doi: 10.1094/phyto-100-3-0290. [DOI] [PubMed] [Google Scholar]
- 22.Ellner F.M. Mycotoxins in potato tubers infected by Fusarium sambucinum. Mycotoxin Res. 2002;18:57–61. doi: 10.1007/bf02946697. [DOI] [PubMed] [Google Scholar]
- 23.Herrmann M., Zocher R., Haese A. Enniatin production by Fusarium strains and its effect on potato tuber tissue. AEM. 1996;62:393–398. doi: 10.1128/aem.62.2.393-398.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leslie J.F., Summerell B.A. first ed. Blackwell Pub.; Ames: 2006. The Fusarium Laboratory Manual. [Google Scholar]
- 25.Simmons E.G. sixth ed. CBS Fungal Biodiversity Centre; Utrecht, The Netherlands: 2007. Alternaria: an Identification Manual. [Google Scholar]
- 26.Simmons E.G., Roberts R. Alternaria themes and variations (73) Mycotaxon. 1993;48:109–140. [Google Scholar]
- 27.Thompson J.D., Higgins D.G., Gibson T.J., Clustal W. Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kumar S., Stecher G., Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 30.Munkvold G.P. In: Moretti A., Susca A., editors. vol. 1542. Springer; New York: 2017. Fusarium species and their associated mycotoxins; pp. 51–106. (Mycotoxigenic Fungi). [DOI] [PubMed] [Google Scholar]
- 31.Quarta A., Mita G., Haidukowski M., Santino A., Mulè G., Visconti A. Assessment of trichothecene chemotypes of Fusarium culmorum occurring in Europe. Food Addit. Contam. 2005;22(4):309–315. doi: 10.1080/02652030500058361. [DOI] [PubMed] [Google Scholar]
- 32.Prosperini A., Meca G., Font G., Ruiz M.J. Study of the cytotoxic activity of beauvericin and fusaproliferin and bioavailability in vitro on Caco-2 cells. FCT. 2012;50(7):2356–2361. doi: 10.1016/j.fct.2012.04.030. [DOI] [PubMed] [Google Scholar]
- 33.Haidukowski M., Cozzi G., Dipierro N., Bavaro S., Logrieco A., Paciolla C. Decontamination of Fumonisin B1 in maize grain by Pleurotus eryngii and antioxidant enzymes. Phytopathol. Mediterr. 2017;56(1):134–145. doi: 10.14601/Phytopathol_Mediterr-20358. [DOI] [Google Scholar]
- 34.Parich A., Boeira L.S., Castro S.P., Krska R. Determination of moniliformin using SAX column clean-up and HPLC/DAD-detection. Mycotoxin Res. 2003;19:203–206. doi: 10.1016/j.ab.2019.113530. [DOI] [PubMed] [Google Scholar]
- 35.Pascale M., Panzarini G., Powers S., Visconti A. Determination of deoxynivalenol and nivalenol in wheat by ultra-performance liquid chromatography/photodiode-array detector and immunoaffinity column cleanup. Food Anal. Methods. 2014;7:555–562. doi: 10.1007/s12161-013-9653-1. [DOI] [Google Scholar]
- 36.Pascale M., Panzarini G., Visconti A. Determination of HT-2 and T-2 toxins in oats and wheat by ultra-performance liquid chromatography with photodiode array detection. Talanta. 2012;89:231–236. doi: 10.1016/j.talanta.2011.12.017. [DOI] [PubMed] [Google Scholar]
- 37.Ali H.Z., Hameed M.S., Abdulrahman A.A., Saood H.M. First report on Fusarium brachygibbosum Isolate FIR 16_ITS isolated from Iraqi wheat plant. Journal of Ecological Engineering. 2020;21(3):81–86. doi: 10.12911/22998993/118295. [DOI] [Google Scholar]
- 38.Fallahi M., Saremi H., Javan-Nikkhah M., Somma S., Haidukowski M., Logrieco A.F., et al. Isolation, molecular identification and mycotoxin profile of Fusarium species isolated from maize kernels in Iran. Toxins. 2019;11:297. doi: 10.3390/toxins11050297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Namsi A., Rabaoui A., Masiello M., Moretti A., Othmani A., Gargouri S., et al. First report of leaf wilt caused by Fusarium proliferatum and F. brachygibbosum on date palm (Phoenix dactylifera) in Tunisia. Plant Dis. 2021;105:1217. doi: 10.1094/pdis-08-20-1791-pdn. [DOI] [Google Scholar]
- 40.Nishad R., Ahmed T.A. Survey and identification of date palm pathogens and indigenous biocontrol agents. Plant Dis. 2020;104(9):2498–2508. doi: 10.1094/PDIS-12-19-2556-RE. [DOI] [PubMed] [Google Scholar]
- 41.Punja Z.K., Collyer D., Scott C., Lung S., Holmes J., Sutton D. Pathogens and molds affecting production and quality of Cannabis sativa L. Front. Plant Sci. 2019;10:1120. doi: 10.3389/fpls.2019.01120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gashgari R., Gherbawy Y., Ameen F., Alsharari S. Molecular characterization and analysis of antimicrobial activity of endophytic fungi from medicinal plants in Saudi Arabia. Jundishapur J. Microbiol. 2016;9(1) doi: 10.5812/jjm.26157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.EFSA Panel on Plant Health Pest categorisation of Fusarium brachygibbosum. EFSA J. 2021;19(11) doi: 10.2903/j.efsa.2021.6887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xia J.W., Sandoval-Denis M., Crous P.W., Zhang X.G., Lombard L. Numbers to names - restyling the Fusarium incarnatum-equiseti species complex. Persoonia. 2019;43:186–221. doi: 10.3767/persoonia.2019.43.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Villani A., Moretti A., De Saeger S., Han Z., Di Mavungu J.D., Soares C.M.G., et al. A polyphasic approach for characterization of a collection of cereal isolates of the Fusarium incarnatum-equiseti species complex. Int. J. Food Microbiol. 2016;234:24–35. doi: 10.1016/j.ijfoodmicro.2016.06.023. [DOI] [PubMed] [Google Scholar]
- 46.Matić S., Tabone G., Gullino M.L., Garibaldi A., Guarnaccia V. Emerging leafy vegetable crop diseases caused by the Fusarium incarnatum-equiseti species complex. Phytopathol. Mediterr. 2020;59(2):2. doi: 10.14601/Phyto-10883. [DOI] [Google Scholar]
- 47.Rabaaoui A., Dall'Asta C., Righetti L., Susca A., Logrieco A.F., Namsi A., et al. Phylogeny and mycotoxin profile of pathogenic Fusarium species isolated from sudden decline syndrome and leaf wilt symptoms on date palms (Phoenix dactylifera) in Tunisia. Toxins. 2021;13:463. doi: 10.3390/toxins13070463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Crous P.W., Hernández-Restrepo M., van Iperen A.L., Starink-Willemse M., Sandoval-Denis M., Groenewald J.Z. Citizen science project reveals novel fusarioid fungi (Nectriaceae, Sordariomycetes) from urban soils. Fungal Syst Evol. 2021;8(1):101–127. doi: 10.3114/fuse.2021.08.09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.de Carvalho Paulino J.F., de Almeida C.P., Barbosa C.C.F., de Moraes G., Gonçalves C., Bueno C.J., et al. Molecular and pathogenicity characterization of Fusarium oxysporum species complex associated with Fusarium wilt of common bean in Brazil. Trop Plant Pathol. 2022;47:485–494. doi: 10.1007/s40858-022-00502-3. [DOI] [Google Scholar]
- 50.Aiello D., Fiorenza A., Leonardi G.R., Vitale A., Polizzi G. Fusarium nirenbergiae (Fusarium oxysporum Species Complex) causing the wilting of passion fruit in Italy. Plants. 2021;10(10):10. doi: 10.3390/plants10102011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aiello D., Gusella G., Vitale A., Polizzi G. Characterization of Fusarium nirenbergiae and F. elaeidis causing diseases on Dipladenia and Grevillea plants. Eur. J. Plant Pathol. 2022;162(4):885–896. doi: 10.1007/s10658-021-02444-z. [DOI] [Google Scholar]
- 52.Mirghasempour S.A., Studholme D.J., Chen W., Cui D., Mao B. Identification and characterization of Fusarium nirenbergiae associated with saffron corm rot disease. Plant Dis. 2022;106(2):486–495. doi: 10.1094/PDIS-04-21-0871-RE. [DOI] [PubMed] [Google Scholar]
- 53.Liu X.B., Zheng N., Liang L.Q., Zhao D.M., Qin Y.Y., Li J., et al. Yang Secondary metabolites from the endophytic fungus Fusarium equiseti and their antibacterial activities. Chem. Nat. Compd. 2019;55(6):1141–1144. doi: 10.1007/s10600-019-02915-0. [DOI] [Google Scholar]
- 54.Maciá-Vicente J.G., Rosso L.C., Ciancio A., Jansson H.B., Lopez-Llorca L. Colonisation of barley roots by endophytic Fusarium equiseti and Pochonia chlamydosporia: effects on plant growth and disease. Ann. Appl. Biol. 2009;155(3):391–401. doi: 10.1111/j.1744-7348.2009.00352.x. [DOI] [Google Scholar]
- 55.Wang H., Liu T., Xin Z. A new glucitol from an endophytic fungus Fusarium equiseti Salicorn 8. Eur Food Res Technol. 2014;239(3):365–376. doi: 10.1007/s00217-014-2230-z. [DOI] [Google Scholar]
- 56.Intana W., Kheawleng S., Sunpapao A. Trichoderma asperellum T76-14 released volatile organic compounds against postharvest fruit rot in muskmelons (Cucumis melo) caused by Fusarium incarnatum. J Fungi. 2021;7(1):1. doi: 10.3390/jof7010046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ramdial H., Hosein F., Rampersad S.N. First report of Fusarium incarnatum associated with fruit disease of bell peppers in Trinidad. Plant Dis. 2016;100(2) doi: 10.1094/PDIS-05-15-0550-PDN. 526–526. [DOI] [Google Scholar]
- 58.Ding L., Dahse H.M., Hertweck C. Cytotoxic alkaloids from Fusarium incarnatum associated with the mangrove tree Aegiceras corniculatum. J Nat Prod. 2012;75(4):617–621. doi: 10.1021/np2008544. [DOI] [PubMed] [Google Scholar]
- 59.Proctor R.H., Desjardins A.E., Moretti A. In: The Role Of Plant Pathology In Food Safety And Food Security, 97–111. Strange R.N., Gullino M.L., editors. Springer Netherlands; 2010. Biological and chemical complexity of Fusarium proliferatum. [DOI] [Google Scholar]
- 60.Yap L.S., Lee W.L., Ting A.S.Y. Optimization of L-asparaginase production from endophytic Fusarium proliferatum using OFAT and RSM and its cytotoxic evaluation. J. Microbiol. Methods. 2021;191 doi: 10.1016/j.mimet.2021.106358. [DOI] [PubMed] [Google Scholar]
- 61.Pili N.N., França S.C., Kyndt T., Makumba B.A., Skilton R., Höfte M., et al. Analysis of fungal endophytes associated with rice roots from irrigated and upland ecosystems in Kenya. Plant Soil. 2016;405(1):371–380. doi: 10.1007/s11104-015-2590-6. [DOI] [Google Scholar]
- 62.Zhang P., Yuan X.L., Du Y.M., Zhang H.B., Shen G.M., Zhang Z.F., et al. Angularly prenylated indole alkaloids with antimicrobial and insecticidal activities from an endophytic fungus Fusarium sambucinum TE-6L. J. Agric. Food Chem. 2019;67(43):11994–12001. doi: 10.1021/acs.jafc.9b05827. [DOI] [PubMed] [Google Scholar]
- 63.Thio G.I., Zida E.P., Neya J.B., Wulff E.G., Lund O.S., Boelt B. Genetic diversity of Fusarium endophytes strains from sorghum (Sorghum bicolor L.) tissues in Burkina Faso. Int. J. Biotechnol. Mol. Biol. Res. 2021;11(1):1–9. doi: 10.5897/IJBMBR2021.0316. [DOI] [Google Scholar]
- 64.Habib W., Masiello M., El Ghorayeb R., Gerges E., Susca A., Meca G., et al. Mycotoxin profile and phylogeny of pathogenic Alternaria species isolated from symptomatic tomato plants in Lebanon. Toxins. 2021;13:513. doi: 10.3390/toxins13080513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Woudenberg J.H.C., Groenewald J.Z., Binder M., Crous P.W. Alternaria redefined. Stud. Mycol. 2013;75:171–212. doi: 10.3114/sim0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Woudenberg J., Seidl M.F., Groenewald J., de Vries M., Stielow J., Thomma B., et al. Alternaria section Alternaria: species, formae speciales or pathotypes? Stud. Mycol. 2015;82:1–21. doi: 10.1016/j.simyco.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Somma S., Amatulli M.T., Masiello M., Moretti A., Logrieco A.F. Alternaria species associated to wheat black point identified through a multilocus sequence approach. Int. J. Food Microbiol. 2019;293:34–43. doi: 10.1016/j.ijfoodmicro.2019.01.001. [DOI] [PubMed] [Google Scholar]
- 68.Dettman J.R., Eggertson Q. Phylogenomic analyses of Alternaria section Alternaria: a high-resolution, genome-wide study of lineage sorting and gene tree discordance. Mycologia. 2021;113(6):1218–1232. doi: 10.1080/00275514.2021.1950456. [DOI] [PubMed] [Google Scholar]
- 69.Ramires F.A., Masiello M., Somma S., Villani A., Susca A., Logrieco A.F., et al. Phylogeny and mycotoxin characterization of Alternaria species isolated from wheat grown in tuscany. Italy, Toxins. 2018;10:472. doi: 10.3390/toxins10110472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Masiello M., El Ghorayeb R., Somma S., Saab C., Meca G., Logrieco A.F., et al. Alternaria species and related mycotoxin detection in Lebanese durum wheat grain. Phytopathol. Mediterr. 2022;61:383–393. doi: 10.36253/phyto-13396. [DOI] [Google Scholar]
- 71.Landschoot S., Vandecasteele M., De Baets B., Höfte M., Audenaert K., Haesaert G. Identification of A. arborescens, A. grandis, and A. protenta as new members of the European Alternaria population on potato. Fungal Biol. 2017;121(2):172–188. doi: 10.1016/j.funbio.2016.11.005. [DOI] [PubMed] [Google Scholar]
- 72.Chalbi A., Sghaier-Hammami B., Meca G., Quiles J.M., Abdelly C., Marangi C., et al. Characterization of mycotoxigenic Alternaria species isolated from the Tunisian halophyte Cakile maritima. Phytopathol. Mediterr. 2020;59(1):107–108. doi: 10.36253/phyto-10720. [DOI] [Google Scholar]
- 73.Lawrence D.P., Rotondo F., Gannibal P.B. Biodiversity and taxonomy of the pleomorphic genus Alternaria. Mycol. Prog. 2015;15(1):3. doi: 10.1007/s11557-015-1144-x. [DOI] [Google Scholar]
- 74.Cabral D., Stone J.K., Carroll G.C. The internal mycobiota of Juncus spp.: microscopic and cultural observations of infection patterns. Mycol. Res. 1993;97(3):367–376. doi: 10.1016/S0953-7562(09)81140-4. [DOI] [Google Scholar]
- 75.Kowalski T., Kehr R. Endophytic fungal colonization of branch bases in several forest tree species. Sydowia. 1992;44:137–168. [Google Scholar]
- 76.Spurr H.W.J., Welty R.E. Characterization of endophytic fungi in healthy leaves of Nicotiana spp. Phytopathol. 1975;65(4):417–422. doi: 10.1094/Phyto-65-417. [DOI] [Google Scholar]
- 77.Sun H., Gao S., Li X., Li C., Wang B. Chemical constituents of marine mangrove-derived endophytic fungus Alternaria tenuissima EN-192. Chin. J. Oceanol. Limnol. 2013;31(2):464–470. doi: 10.1007/s00343-013-2106-2. [DOI] [Google Scholar]
- 78.Ismaiel A.A., Ahmed A.S., Hassan I.A., El-Sayed E.S.R., Karam El-Din A.Z.A. Production of paclitaxel with anticancer activity by two local fungal endophytes, Aspergillus fumigatus and Alternaria tenuissima. Appl. Microbiol. Biotechnol. 2017;101(14):5831–5846. doi: 10.1007/s00253-017-8354-x. [DOI] [PubMed] [Google Scholar]
- 79.Rao H.C.Y., Rakshith D., Harini B.P., Satish S. Antimicrobial profiling and molecular identification of Alternaria arborescens CLB12, A myco-endosymbiont inhabiting Combretum latifolium Blume. JBAPN. 2017;7(1):1–9. doi: 10.1080/22311866.2017.1287592. [DOI] [Google Scholar]
- 80.Komatsu K., Katayama Y., Omatsu T., Mizutani T., Fukuhara T., Kodama M., et al. Genome sequence of a novel mitovirus identified in the phytopathogenic fungus Alternaria arborescens. Arch. Virol. 2016;161(9):2627–2631. doi: 10.1007/s00705-016-2953-1. [DOI] [PubMed] [Google Scholar]
- 81.Somma S., Pose G., Pardo A., Mulè G., Pinto V.F., Moretti A., et al. AFLP variability, toxin production, and pathogenicity of Alternaria species from Argentinean tomato fruits and puree. Int. J. Food Microbiol. 2011;145:414–419. doi: 10.1016/j.ijfoodmicro.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 82.Hu W., Ran Y., Zhuang K., Lama J., Zhang C. Alternaria arborescens infection in a healthy individual and literature review of Cutaneous alternariosis. Mycopathologia. 2015;179(1):147–152. doi: 10.1007/s11046-014-9822-9. [DOI] [PubMed] [Google Scholar]
- 83.Jam Ashkezari S., Fotouhifar K.B. Diversity of endophytic fungi of common yew (Taxus baccata L.) in Iran. Mycol. Prog. 2017;16(3):247–256. doi: 10.1007/s11557-017-1274-4. [DOI] [Google Scholar]
- 84.Hashemloo E., Jamali Zavareh A.H., Ghosta Y. Taxonomic study of endophytic species of Alternaria from Prunus trees in urmia and miandoab in W azarbaijan province (NW Iran) Rostaniha. 2015;16(1):88–100. doi: 10.22092/botany.2015.102002. [DOI] [Google Scholar]
- 85.Ahadi N., Safari Sinegani A.A., Aletaha R.S. Evaluation of capability of fifteen isolates of mycorrhiza-like endophytic fungi on release of phosphorous from phosphorite mineral in the aquatic culture medium. Appl Soil Res. 2021;9(2):87–101. [Google Scholar]
- 86.Alidadi A., Javan-Nikkhah M., Kowsari M., KaramiM S., Rastaghi Ebrahimi. Some species of fungi associated with declined Persian oak trees in Ilam province with emphasis on new records to mycobiota of Iran. Rostaniha. 2018;19(2):75–91. doi: 10.22092/botany.2019.122177.1105. [DOI] [Google Scholar]
- 87.Rabaaoui A., Masiello M., Somma S., Crudo F., Dall'Asta C., Righetti L., et al. Phylogeny and mycotoxin profiles of pathogenic Alternaria and Curvularia species isolated from date palm in southern Tunisia. Front. Microbiol. 2022;13 doi: 10.3389/fmicb.2022.1034658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Przemieniecki S.W., Damszel M., Kurowski T.P., Mastalerz J., Kotlarz K. Identification, ecological evaluation and phylogenetic analysis of non-symbiotic endophytic fungi colonizing timothy grass and perennial ryegrass grown in adjacent plots. Grass Forage Sci. 2019;74(1):42–52. doi: 10.1111/gfs.12404. [DOI] [Google Scholar]
- 89.Pinto V.E.F., Patriarca A. In: Moretti A., Susca A., editors. vol. 1542. Humana Press; New York, NY: 2017. Alternaria species and their associated mycotoxins. (Mycotoxigenic Fungi. Methods in Molecular Biology). [DOI] [PubMed] [Google Scholar]
- 90.Zhou Z., Zhang C., Zhou W., Li W., Chu L., Yan J., et al. Diversity and plant growth-promoting ability of endophytic fungi from the five flower plant species collected from Yunnan, Southwest China. J. Plant Interact. 2014;9(1):585–591. doi: 10.1080/17429145.2013.873959. [DOI] [Google Scholar]
- 91.Kouri K., Lemmens M., Lemmens-Gruber R. Beauvericin-induced channels in ventricular myocytes and liposomes. Biochim. Biophys. Acta Biomembr. 2003;1609(2):203–210. doi: 10.1016/s0005-2736(02)00689-2. [DOI] [PubMed] [Google Scholar]
- 92.Kamyar M., Rawnduzi P., Studenik C.R., Kouri K., Lemmens-Gruber R. Investigation of the electrophysiological properties of enniatins. Arch. Biochem. Biophys. 2004;429:215–223. doi: 10.1016/j.abb.2004.06.013. [DOI] [PubMed] [Google Scholar]
- 93.Herrmann M., Zocher R., Haese A. Effect of disruption of the enniatin synthetase gene on the virulence of. Fusarium avenaceum MPMI. 1996;9(4):226–232. doi: 10.1094/mpmi-9-0226. [DOI] [PubMed] [Google Scholar]
- 94.Eranthodi A., Schneiderman D., Harris L.J., Witte T.E., Sproule A., Hermans A., et al. Enniatin production influences Fusarium avenaceum virulence on potato tubers, but not on durum wheat or peas. Pathogens. 2020;9:75. doi: 10.3390/pathogens9020075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Paciolla C., Dipierro N., Mule G., Logrieco A.F., Dipierro S. The mycotoxins beauvericin and T-2 induce cell death and alteration to the ascorbate metabolism in tomato protoplasts. Physiol. Mol. Plant Pathol. 2004;65:49–56. doi: 10.1016/j.pmpp.2004.07.006. [DOI] [Google Scholar]
- 96.Li c., Zuo C., Deng G., Kuang R., Yang Q., Hu C., et al. Contamination of bananas with beauvericin and fusaric acid produced by Fusarium oxysporum f. sp. cubense. PLoS One. 2013;8(7) doi: 10.1371/journal.pone.0070226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Song H.H., Lee H.S., Jeong J.H., Park H., Lee C. Diversity in beauvericin and enniatins H, I, and MK1688 by Fusarium oxysporum isolated from potato. Int. J. Food Microbiol. 2008;122(3):296–301. doi: 10.1016/j.ijfoodmicro.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 98.Cole R.J., Kirksey J.W., Cutler H.G., Doupink B.L., Peckham J.C. Toxin from Fusarium moniliforme: effects on plants and animals. Science. 1973;179:1324–1326. doi: 10.1126/science.179.4080.1324. [DOI] [PubMed] [Google Scholar]
- 99.Duke S.O. In: The Science of Allelopathy. Putnam A.R., Tang C.S., editors. John Wiley & Sons Inc.; 1986. Microbially produced phytotoxins as herbicides-A perspective; pp. 15–44. [Google Scholar]
- 100.Gäumann E. Fusaric acid as a wilt toxin. Phytopathology. 1957;47:342–357. [Google Scholar]
- 101.Toyoda H., Hashimoto H., Utsumi R., Kobayashi H., Ouchi S. Detoxification of fusaric acid by a fusaric acid-resistant mutant of Pseudomonas solanacearum and its application to biological control of Fusarium wilt of tomato. Phytopathol. 1988;78:1307–1311. doi: 10.1094/phyto-78-1307. [DOI] [Google Scholar]
- 102.Singh V.K., Singh H.B., Upadhyay R.S. Role of fusaric acid in the development of Fusarium wilt' symptoms in tomato: physiological, biochemical and proteomic perspectives. Plant Physiol Biochem. 2017;118:320–332. doi: 10.1016/j.plaphy.2017.06.028. [DOI] [PubMed] [Google Scholar]
- 103.Liu S., Li J., Zhang Y., Liu N., Viljoen A., Mostert D., et al. Fusaric acid instigates the invasion of banana by Fusarium oxysporum f. sp. cubense TR4. New Phytol. 2020;225:913–929. doi: 10.1111/nph.16193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Venter S.L., Steyn P.J. Correlation between fusaric acid production and virulence of isolates of Fusarium oxysporum that causes potato dry rot in South Africa. Potato Res. 1998;41:289–294. doi: 10.1007/BF02358198. [DOI] [Google Scholar]
- 105.Streit E., Schwab C., Sulyok M., Naehrer K., Krska R., Schatzmayr G. Multi-mycotoxin screening reveals the occurrence of 139 different secondary metabolites in feed and feed ingredients. Toxins. 2013;5:504–523. doi: 10.3390/toxins5030504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Jestoi M. Emerging Fusarium -mycotoxins fusaproliferin, beauvericin, enniatins, and moniliformin—a review. Crit. Rev. Food Sci. Nutr. 2008;48(1):21–49. doi: 10.1080/10408390601062021. [DOI] [PubMed] [Google Scholar]
- 107.Bacon C.W., Porter J.K., Norred W.P., Leslie J.F. Production of fusaric acid by Fusarium species. AEM. 1996;62(11):4039–4043. doi: 10.1128/aem.62.11.4039-4043.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kogel K.H., Franken P., Hückelhoven R. Endophyte or parasite--what decides? Curr. Opin. Plant Biol. 2006;9(4):358–363. doi: 10.1016/j.pbi.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 109.Schulz B., Boyle C. The endophytic continuum. Mycol. Res. 2005;109(6):661–686. doi: 10.1017/S095375620500273X. [DOI] [PubMed] [Google Scholar]
- 110.Logrieco A., Bottalico A., Solfrizzo M. Toxigenic Fusarium species isolated from rotted potato tubers. Mycotoxin Res. 1987;3(1):105–110. doi: 10.1007/BF03192040. [DOI] [PubMed] [Google Scholar]
- 111.Barbosa P., Krischik V.A., Jones C.G., editors. Microbial Mediation of Plant-Herbivore Interactions. first ed. John Wiley & Sons, Inc.; New York: 1991. [Google Scholar]
- 112.Carroll G. Fungal endophytes in stems and leaves: from latent pathogen to mutualistic symbiont. Ecol. 1988;69(1):2–9. doi: 10.2307/1943154. [DOI] [Google Scholar]
- 113.Cheplick G.P., Clay K. Acquired chemical defences in grasses: the role of fungal endophytes. Oikos. 1988;52(3):309–318. doi: 10.2307/3565204. [DOI] [Google Scholar]
- 114.Schulz B., Guske S., Dammann U., Boyle C. Endophyte-host interactions. II. Defining symbiosis of the endophyte-host interaction. Symbiosis. 1998;25:213–227. [Google Scholar]
- 115.Gordon T.R., Martyn R.D. The evolutionary biology of Fusarium oxysporum. Annu. Rev. Phytopathol. 1997;35(1):111–128. doi: 10.1146/annurev.phyto.35.1.111. [DOI] [PubMed] [Google Scholar]
- 116.Nelson P.E., Toussoun T.A., Cook R.J. Pennsylvania State University Press; 1981. Fusarium: Diseases, Biology, and Taxonomy. [DOI] [Google Scholar]
- 117.Metz N., Hausladen H. Trichoderma spp. as potential biological control agent against Alternaria solani in potato. Biol. Control. 2022;166 doi: 10.1016/j.biocontrol.2021.104820. [DOI] [Google Scholar]
- 118.White T., Bruns T., Lee S., Taylor J., Innis M., Gelfand D., et al. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protocols: A Guide to Methods and Applications. 1990;31:315–322. [Google Scholar]
- 119.O'Donnell K., Kistler H.C., Cigelnik E., Ploetz R.C. Multiple evolutionary origins of the fungus causing Panama disease of banana: concordant evidence from nuclear and mitochondrial gene genealogies. Proc. Natl. Acad. Sci. U.S.A. 1998;95:2044–2049. doi: 10.1073/pnas.95.5.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Glass N.L., Donaldson G.C. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microbiol. 1995;61(4):1323–1330. doi: 10.1128/aem.61.4.1323-1330.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Berbee M.L., Pirseyedi M., Hubbard S. Cochliobolus phylogenetics and the origin of known, highly virulent pathogens, inferred from ITS and glyceraldehyde-3-phosphate dehydrogenase gene sequences. Mycologia. 1999;91(6):964–977. doi: 10.1080/00275514.1999.12061106. [DOI] [Google Scholar]
- 122.Lawrence D.P., Gannibal P.B., Peever T.L., Pryor B.M. The sections of Alternaria: formalizing species-group concepts. Mycologia. 2013;105(3):530–546. doi: 10.3852/12-249. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.
The data and supportive information are available within the article. Data associated with the study have been deposited into publicly available repositories. Sequences were deposited in the GenBank database; strains were deposited in the microbial ITEM collection (http://server.ispa.cnr.it/ITEM/Collection/).