Abstract
The highly conserved SR family contains a growing number of phosphoproteins acting as both essential and alternative splicing factors. In this study, we have cloned human genomic and cDNA sequences encoding a novel SR protein designated SRp46. Nucleotide sequence analyses have revealed that the SRp46 gene corresponds to an expressed PR264/SC35 retropseudogene. As a result of mutations and amplifications, the SRp46 protein significantly differs from the PR264/SC35 factor, mainly at the level of its RS domain. Northern and Western blot analyses have established that SRp46 sequences are expressed at different levels in several human cell lines and normal tissues, as well as in simian cells. In contrast, sequences homologous to SRp46 are not present in mice. In vitro splicing studies indicate that the human SRp46 recombinant protein functions as an essential splicing factor in complementing a HeLa cell S100 extract deficient in SR proteins. In addition, complementation analyses performed with β-globin or adenovirus E1A transcripts and different splicing-deficient extracts have revealed that SRp46 does not display the same activity as PR264/SC35. These results demonstrate, for the first time, that an SR splicing factor, which represents a novel member of the SR family, is encoded by a functional retropseudogene.
Pre-mRNA splicing is a fundamental process in the expression of most eukaryotic genes. The spliceosome, which catalyzes the precise removal of intronic sequences from primary mRNA transcripts, consists of several small nuclear ribonucleoprotein particles (snRNPs) and numerous non-snRNP proteins playing an essential role in pre-mRNA splicing. Several of these non-snRNP factors belong to a remarkably conserved family of structurally and functionally highly related phosphoproteins called SR proteins (20, 40, 75). The SR protein family contains at least nine members mostly designated according to their apparent molecular weights: SRp75 (76), SRp55 (B52 in Drosophila melanogaster) (13, 52, 55), p54 (79), SRp40 (HRS in rats) (6, 16), SRp30a (ASF/SF2) (25, 33), SRp30b (PR264/SC35) (22, 67), SRp30c (55), 9G8 (11), and SRp20 (X16 in mice and RBP1 in Drosophila) (3, 30, 75). These SR proteins contain RNA binding domains constituted by one or two copies of the RNA recognition motif, also called the RNP-type RNA binding domain (RBD) (6, 29), and a C-terminal region of variable length, rich in Arg and Ser residues (RS domain).
SR proteins act as essential splicing factors in that they are individually able to complement S100 extracts (11, 23, 32, 43, 55, 75, 79). These factors are involved in the early steps of spliceosome assembly and pre-mRNA commitment. Several studies have reported that SR proteins promote the binding of the U1 snRNP to the 5′ splice site (18, 31) and stimulate the binding of the U2AF and U2 snRNPs to the 3′ splice site region (22, 57). Based on results showing that the RS domains of SR factors interact with themselves and with each other, as well as with the U1-70K and U2AF35 proteins (31, 74), it has been proposed that a role of SR proteins in splicing consists in bringing the 5′ and 3′ splice sites together via a bridge of protein contacts.
SR proteins are also involved in the control of alternative splicing. These factors have been shown to influence the selection of alternative splice sites in a concentration-dependent manner both in vitro (23, 25, 33, 77) and in transfected cells (10, 55, 69). For some, if not all SR proteins, this activity can be antagonized by members of the hnRNP A/B family (23, 41, 42) as well as by other SR factors (24, 28). This indicates that accurate alternative splice site selection depends on the respective concentrations of activating SR proteins and antagonist hnRNP or SR factors.
Specific binding of SR proteins to pre-mRNA has been shown to rely on exonic or intronic sequences, designated splicing enhancer elements (24, 35, 39, 50, 59), which may be involved in the control of stage-, sex-, or tissue-specific splicing events. Despite the considerable overlap in SR protein activity, it appears that individual members of the family are not functionally redundant. Indeed, different specific activities for constitutive and alternative splicing with various pre-mRNA substrates have been described (21, 24, 55, 59, 62, 69, 77–79). Moreover, SRp55/B52 and ASF/SF2 have been reported to be essential genes for Drosophila development (34, 51) and chicken B-cell viability (70), respectively. This suggests unique cellular functions for individual members of the SR protein family.
In the present study, we have isolated and characterized human PR264/SC35-related sequences corresponding to a processed pseudogene. We show that this pseudogene, termed H430, is differentially expressed at the RNA level in several human cell lines and normal tissues. The H430 translation product, which we designate SRp46 because of its apparent molecular mass of 46 kDa, shows significant modifications compared to the PR264/SC35 splicing factor. Consistent with Northern blot analyses, we have observed that the SRp46 protein is expressed at different levels in various human cell lines as well as in simian cells. The results of in vitro splicing experiments demonstrate that recombinant human SRp46 is able to fully complement S100 extracts and exhibits the general characteristics of SR factors. Furthermore, we provide evidence that SRp46 activity differs from that of PR264/SC35.
MATERIALS AND METHODS
Cell cultures and tissues.
With the exception of HeLa cells, all human (293, CCRF-CEM, HL60, KATO III, MCF7, SVK14), simian (CV-1, COS-1), and murine [NIH/3T3, AT20, L-M (TK−)] cell lines were obtained from the American Type Culture Collection (Manassas, Va.) and cultured as recommended. The human thymus used in these studies was a surgery sample from a 1-month-old girl.
Probes, library screening, nucleotidic sequencing, and Southern and Northern blot analyses.
The RR200 probe was obtained following RsaI digestion of the H230 clone (66). The SS800 and HB1600 probes, subcloned into the pBluescript KS vector (Stratagene), correspond to a 0.8-kb SstI-SstI fragment containing the RR200 homologous sequences and to a 1.6-kb HindIII-BamHI fragment located upstream from the H430 pseudogene sequences, respectively. The ES165 probe, specific for the H430 locus, was amplified by PCR with a pair of mutated oligodeoxynucleotides (5′-GTGAGACCGCGGGCTGTGAT-3′ and 5′-ATAGGAATTCTCGCTATAGCC-3′) in order to generate the SstII and EcoRI restriction sites used for subsequent cloning into the pBluescript KS plasmid. The human placenta genomic library (Clontech) was screened with the RR200 probe. The human thymus oligo(dT)-primed cDNA library, constructed from 5 μg of polyadenylated RNA as recommended by the manufacturer (Amersham), was screened with the ES165 probe. The nucleotide sequence of the 2.4-kb EcoRI-HindIII fragment containing most of the H430 sequences (see Fig. 1) was determined by automated procedures. Additional sequences presented in this study were obtained from dideoxynucleotide sequencing reactions performed with α35-S-dATP and the Pharmacia T7 sequencing kit. Southern blot analyses, RNA purification, selection of polyadenylated species, and Northern blot hybridizations were performed as previously described (64). Human multiple-tissue Northern blots were purchased from Clontech. The human glyceraldehyde-3-phosphate dehydrogenase (GAPDH)-specific and β-actin probes (Clontech) were used as internal controls to normalize for RNA amounts.
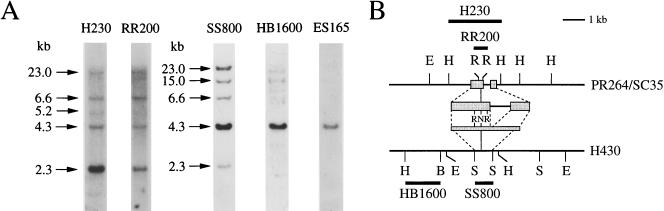
FIG. 1.
Characterization of human PR264-related sequences (A) Southern blot analysis of HindIII-digested human thymus DNA (15 μg per lane) hybridized with the various probes as indicated above the lanes. Sizes are in kilobases. (B) Organization and structure of human PR264/SC35 and H430 sequences. The partial restriction maps of both loci are represented. Restriction sites: B, BamHI; E, EcoRI; H, HindIII; N, NcoI; R, RsaI; S, SstI. The probes used for Southern blot analyses and their correspondence with both loci are indicated. Shaded boxes correspond to PR264/SC35 coding exons 1 and 2 and to their related sequences in H430.
Fluorescence in situ hybridization analysis.
Chromosomes were prepared from human normal peripheral blood lymphocyte cultures, after bromodeoxyuridine incorporation during the last 7 h before harvesting. The 1.6-kb HB1600 probe was nick translated with biotin-14-dATP according to the Gibco-BRL protocol. Classical hybridization was performed as described previously (19). The probe was used at a concentration of 20 ng/μl in 15 μl of hybridization buffer. Immunochemical detection of the hybridized probe was performed with goat anti-biotin antibodies (Vector Laboratories, Burlingame, Calif.) diluted to 1:100 and fluorescein-conjugated anti-goat antibodies (Biosys, Compiègne, France) diluted to 1:200. Chromosomes were stained with propidium iodide, mounted in PPD11 (36), and observed with a fluorescence microscope without amplification. Metaphases were photographed on Ektachrome ASA 400 film (Kodak, Rochester, N.Y.).
RNase protection analyses.
In vitro transcription of the ES165 template was performed with T7 RNA polymerase (Promega) following linearization at the EcoRI restriction site. Samples of polyadenylated RNA (5 μg) were hybridized overnight at 55°C with 3 ng of [α32-P]UTP-labeled probe. RNA-RNA hybrids were then digested with RNase A (40 μg/ml) and RNase T1 (1,000 U/ml) (Boehringer Mannheim) and analyzed in a 6% sequencing gel.
RACE-PCR.
Rapid amplification of cDNA ends (RACE)-PCR experiments were performed with human normal thymus polyadenylated mRNA species and the Marathon cDNA amplification kit (Clontech) according to the manufacturer’s instructions. Double-stranded cDNA was amplified in the presence of a 0.8 μM concentration of each amplimer (H430-1507 [5′-TAGCGAGAGTTACTGTAACCA-3′] and AP1 adapter primer)–500 μM dNTP–50 mM Tris-HCl (pH 9.2)–14 mM (NH4)SO4–3 mM MgCl2–2% dimethyl sulfoxide–0.1% Tween 20–2.5 U of Expand enzyme mixture (Boehringer Mannheim).
Construction of the H430 expression vectors.
The transcription plasmid used for in vitro translation of the wild-type H430 protein was constructed by insertion of an ApaI-HindIII fragment containing the entire H430 coding sequences into the NotI-HindIII site of the pTL2(glo) vector (49). In vitro translation of the H430 protein was performed with reticulocyte lysate extract in the presence of [35S]methionine, according to the manufacturer’s instructions (Promega). The H430 baculoviral vector was constructed by inserting a NotI-BamHI fragment containing the coding sequence of H430 deprived of the first three codons into the pAc SG His NT-B vector (PharMingen).
Antibody production.
H430 polyclonal antibodies were obtained by immunizing rabbits against three peptides, P213 (SRYRESRYGGSHYSS), P214 (RYRGSRYSRSPYSRS), and P215 (SGYSNSRYSRYHSSRSH), from the H430 repeated region coupled to ovalbumin. Antisera were tested 4, 6, and 8 weeks after multiple intradermal injections (64). Only sera directed against peptides P213 and P214 were positive in Western blotting analyses. Their specific antibodies were further affinity purified on Sulfolink agarose beads to which the corresponding peptide was bound, according to the instructions of the manufacturer (Pierce). The monoclonal antibody (MAb) directed against the 15 C-terminal amino acids of SC35 (24) and MAb 104, which recognizes a phosphorylated epitope of the SR domain (75), were used as hybridoma culture supernatants. MAb 9G8, which recognizes primarily splicing factor 9G8 and which cross-reacts with SC35 (11), was used as ascites fluid. It was coupled to cyanogen bromide-activated Sepharose (30 mg of antibodies per ml of swollen matrix) to perform immunodepletion.
Immunoblot analysis.
Protein samples were separated by sodium dodecyl sulfate–10% polyacrylamide gel electrophoresis (SDS–10% PAGE) and electroblotted onto nitrocellulose membranes in 25 mM Tris-HCl–192 mM glycine buffer (pH 8.3) containing 20% methanol. The blots were saturated in phosphate-buffered saline–0.5% Tween 20 containing 10% nonfat milk powder (blocking buffer) and then probed overnight at 4°C with primary antibodies diluted in the same buffer. The detection was performed with a peroxidase-conjugated secondary antibody, donkey anti-rabbit immunoglobulin G (IgG) or goat anti-mouse IgG plus IgM (Jackson Immunoresearch), and the ECL system (Amersham).
Recombinant proteins.
Recombinant baculovirus expressing the H430 protein was prepared with the PharMingen Baculogold system and used at a multiplicity of infection of 2 PFU per cell. Sf9 cells were harvested 48 h postinfection and lysed by mild sonication in buffer E (20 mM Tris-HCl [pH 8.0], 100 mM NaCl, 1 mM dithiothreitol, 5 mM β-glycerophosphate, 5 mM KF, and a protease inhibitor mixture). Protein extraction was continued for 30 min at 0°C in the presence of 0.33 M NaCl, and cellular debris were removed by ultracentrifugation. The histidine-tagged H430 protein was purified by metal affinity chromatography using a Co2+-IDA chelated resin (Talon; Clontech). The recombinant protein was stepwise eluted with binding buffer containing 100 to 500 mM imidazole. The fractions containing the recombinant H430 protein were dialyzed against dialysis buffer D, which was described by Dignam et al. (17). The same mock fractions, originating from Sf9 cells infected with baculovirus expressing transcriptional mediator/intermediary factor 2 (TIF2), characterized by Voegel et al. (68) were used as a control. Recombinant SRp30 factors, all histidine tagged at their N termini, were purified on a Hitrap chelating column (Pharmacia) as previously described for SC35 and ASF/SF2 (24) or on a Talon column as described above for 9G8. Proteins were quantified by Coomassie blue staining after SDS-PAGE, with bovine serum albumin as a standard.
Preparation of splicing-competent and splicing-deficient extracts.
Nuclear extracts competent for splicing and cytoplasmic S100 fractions were prepared from HeLa or 293 cells, grown in suspension culture, by the protocol of Dignam et al. (17). Nuclear extracts deficient in SR splicing factors were obtained by immunodepletion with the 9G8 antibody linked to activated Sepharose, as previously described (11). This treatment results in a complete depletion of 9G8 factor and in a significant depletion (>50%) of the other SR factors. A nuclear extract fraction deficient in splicing was also prepared by precipitation to 55% saturation with (NH4)2SO4 solution. After being stirred for 30 min on ice, the precipitated material was pelleted and redissolved in buffer D containing only 5% glycerol and having 70% of the initial extract volume. This fraction (NE-55), which contained detectable levels of SRp40 and 9G8 factor but no other SR factors, was deficient in splicing but could be rescued by the addition of certain SR factors (24).
In vitro splicing assays.
Splicing assays were carried out in 25-μl reaction mixtures in the presence of 3.2 mM MgCl2 and 60 mM KCl as described previously (54). Twenty femtomoles of capped, 32P-labeled RNA substrates, corresponding to the adenoviral E1A pre-mRNA (Sp4 transcript) or to the 5′ half of β-globin pre-mRNA (β-glo transcript), was used for each assay (11). The standard reaction mixtures contained 10 μl of HeLa nuclear extract. The amounts of other nuclear fractions, S100 extract, and recombinant proteins are indicated in the legends of the figures. Following incubation at 30°C for 2 h, the RNA was extracted and analyzed by urea-PAGE and autoradiography.
Nucleotide sequence accession numbers.
GenBank accession numbers for the human H430 genomic and cDNA sequences are AF031165 and AF031166, respectively.
RESULTS
Characterization of a PR264-related pseudogene.
Southern blot analysis of HindIII-digested human genomic DNA with the H230 probe derived from the PR264 gene revealed a 2.3-kb fragment specific for the PR264/SC35 locus as well as the existence of PR264/SC35-related sequences in three fragments of 4.3, 6.6, and 23.0 kb (Fig. 1A) (66). As a first step in the characterization of these related sequences, we screened a human placenta genomic library with an internal 200-bp probe (RR200), which allows better detection of the PR264-related sequences (Fig. 1A). Two overlapping recombinant phages (λ22 and λ23) were shown, by restriction mapping and Southern blot analyses, to contain sequences corresponding to the same PR264-related locus. Following hybridization of human genomic DNA with an 800-bp SacI-SacI probe (SS800) derived from the λ23 phage, a 4.3-kb HindIII fragment was predominantly detected (Fig. 1A). The corresponding PR264-related locus was designated H430. Under the same conditions, hybridization signals obtained for the 23.0- and 6.6-kb fragments as well as for an additional 15.0-kb fragment were stronger than that of the PR264/SC35-specific 2.3-kb fragment, indicating that the corresponding sequences are more closely related to the H430 than to the H230 (PR264/SC35) locus.
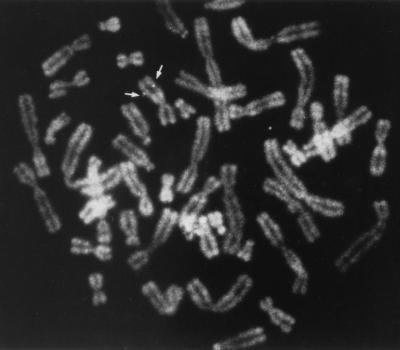
An analysis of human-rodent hybrid cell lines had previously allowed us to establish a positive correlation between chromosome 11 and the PR264-related H430 locus (66). In order to determine more precisely the chromosomal localization of this locus, in situ hybridization was performed with the HB1600 probe (depicted in Fig. 1B), which essentially detected H430 sequences in Southern blot hybridization experiments (Fig. 1A). As shown in Fig. 2, fluorescence in situ hybridization on human metaphase chromosomes revealed recurrent spots on the long arm of chromosome 11, with a very low background level. Among the 25 metaphases which were analyzed, 80% exhibited at least one fluorescent spot on chromosome 11 and 25% had signal on both chromosome 11 homologs. Of all fluorescent spots, 73% were located on chromosome 11 and 25% of the specific spots were double spots. Direct R-banding induced by an alkaline solution of phenylenediamine (PPD11) when chromosomes were stained with propidium iodide and observed under a fluorescence microscope (data not shown) allowed us to precisely localize the H430 sequences on band 11q22-2. These observations indicate that the chromosomal localization of H430 differs from that of the genuine PR264/SC35 gene, which resides, like the ASF/SF2 locus, on human chromosome 17 (5, 66).
FIG. 2.
Human metaphase chromosomes showing the specific hybridization of the HB1600 probe to chromosome 11q22-2.
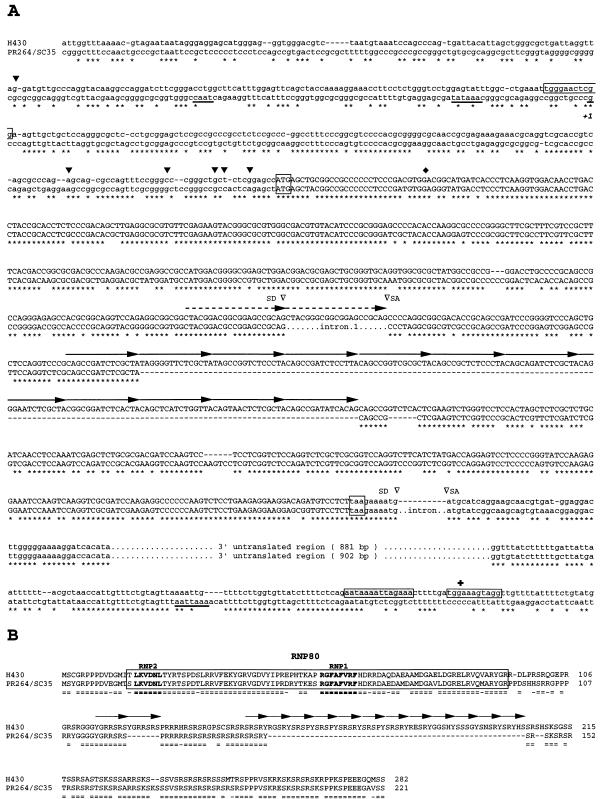
The nucleotide sequence corresponding to the first half of a 5.5-kb EcoRI fragment derived from the λ22 recombinant phage was then determined and compared to that previously established for the PR264/SC35 gene (60, 66, 67). As shown in Fig. 3A, exon 1 sequences encoding the RBD of the PR264/SC35 splicing factor are well conserved in the H430 locus. The highest level of homology (85%) is observed between the ATG translation initiation codon and the first splice donor site identified in the PR264 gene (67). This homology decreases to 70% between the ATG codon and the PR264 transcriptional start site (+1) (54) and abruptly falls to 35% upstream from this start site. Exon 2 sequences encoding the PR264/SC35 RS-rich domain are also well conserved in H430 (80%), but these sequences are split in two parts as a result of a 165-bp insertion. An analysis of this sequence indicated that it likely arose from amplification and subsequent mutation of a 15-bp nucleotide motif (5′-CAGCCGA/GTCTCG/ACTA-3′) which is represented immediately upstream from the insertion site in both the PR264 and H430 sequences. A striking feature of the H430 genomic structure is the lack of identified intronic sequences (334 bp) between the two coding exons in the PR264 gene (67). In H430, the sequences homologous to PR264 exons 1 and 2 are separated by a 21-bp motif which likely results from duplication of the first exon 3′-proximal sequences. The 3′ noncoding region represented in the major PR264 mRNA species (60) is also conserved (81% identity) in the H430 sequences. Again, the 986-bp intron separating PR264 exon 2 from these noncoding sequences is not present in the H430 gene.
FIG. 3.
Nucleotide and deduced amino acid sequences of the human H430 and PR264/SC35 genes. (A) Comparison of H430 and PR264/SC35 nucleotide sequences. Conserved nucleotides are indicated by stars, and gaps have been introduced in order to maximize alignment. The 21-bp duplication found at the junction of the homologous sequences of exons 1 and 2 and the 15-bp degenerate repeats constituting the H430 165-bp insertion are indicated by dashed and solid arrows, respectively. Splice donors (SD), splice acceptors (SA), and introns represented in PR264/SC35 sequences are indicated. The PR264/SC35 CAAT box, the TATA box, the transcription start site (+1), and the polyadenylation signal are underlined. Conserved translation initiation and stop codons are boxed. H430 and PR264/SC35 ORFs are in capital letters. The H430 poly(A) tail remnant and the imperfect repeats flanking the H430 retropseudogene are indicated by shaded and open boxes, respectively. H430 transcript 5′ ends mapped by RACE-PCR experiments are represented by solid arrowheads above the sequence. The H430 cDNA 5′ end (⧫) and polyadenylation site (✚) are indicated. (B) Comparison of H430 and PR264/SC35 amino-acid sequences. Identical amino acids (=) and semiconservative substitutions (-) are indicated. Gaps have been introduced in order to maximize alignment. The RBD is boxed, and the RNP1 and RNP2 motifs are indicated in boldface. YGRRSRS and degenerate SRSRY repeats resulting from the 21-bp duplication and the 165-bp insertion, respectively, are represented by arrows.
Taken together, these observations indicate that H430 likely represents a pseudogene which arose by the reverse transcription and integration of a processed PR264 mRNA. On the basis of the genomic organization of the H430 sequences, it is likely that this pseudogene originates from reverse transcription of the major 2.0-kb PR264/SC35 mRNA species (HPR4) that we previously characterized (60). This hypothesis is supported by the fact that the homology between the H430 and PR264 3′-proximal sequences ends abruptly at the level of an A-rich region (AATAAAATTAGAAA in Fig. 3A) which could constitute the remnant of a poly(A) tail. Furthermore, H430 sequences are flanked by short imperfect repeats (Fig. 3A) reminiscent of the target site duplications which are a hallmark of integrated transposons (see reference 72 for a review).
DNA sequence analysis of the H430 gene also revealed the existence of an uninterrupted open reading frame (ORF) largely colinear with those of PR264 cDNAs. This ORF encodes a potential 282-amino-acid protein homologous to the PR264/SC35 splicing factor. Both the RBD and RS domains of the H430 potential product exhibit 80% homology with the corresponding regions of the PR264/SC35 protein (Fig. 3B). In the putative H430 translation product, the 21-bp duplication results in a tandemly repeated YGRRSRS sequence, while most of the 165-bp insertion within homologous sequences of PR264 exon 2 encodes eleven degenerate repeats of the initial SRSRY motif. The existence of an ORF in the H430 gene was confirmed by translation experiments performed in a cell-free system with in vitro-transcribed H430 RNA species (data not shown). The apparent molecular mass of the resulting product was 46 kDa, whereas its theoretical molecular mass was estimated to be 34.5 kDa. Such a difference indicates that the in vitro-translated H430 protein exhibits, like the other members of the SR splicing factor family (76), an abnormal gel mobility.
H430 is an expressed retropseudogene.
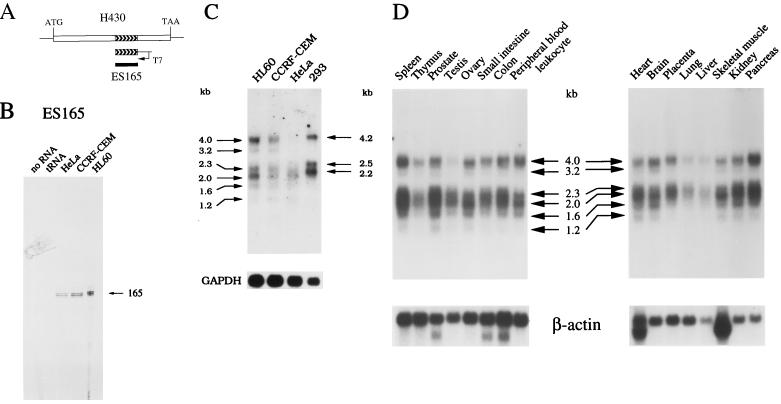
Whether the PR264-related H430 retropseudogene is expressed was first assessed by RNase protection experiments. These studies were performed with a riboprobe corresponding to the 165-bp insertion which was identified in the H430 sequences (Fig. 4A) and which is specific for this locus, as revealed by Southern blot analysis (see Fig. 1A). As shown in Fig. 4B, this riboprobe was fully and specifically protected by polyadenylated RNA species purified from the human epithelial HeLa, T-lymphoma CCRF-CEM, and myelomonocytic HL60 cell lines.
FIG. 4.
Expression pattern of the human H430 retropseudogene. (A) Correspondence of the DNA and RNA probes with the human H430 sequences. (B) An RNase protection analysis of H430 transcripts was performed with the ES165 riboprobe and 7 μg of the indicated polyadenylated RNA. The sizes of the specifically protected fragments are in nucleotides. (C and D) Northern blot analyses of H430 mRNA species. Polyadenylated RNA samples from the indicated human cells lines (5 μg per lane) and normal tissues (2 μg per lane) were hybridized with the 32P-labeled ES165 DNA probe. Transcripts detected in the different samples are indicated by arrows. Sizes are in kilobases. Human GAPDH and β-actin were used as the internal controls to normalize for variations in RNA amounts.
The expression pattern of H430 sequences was then analyzed by Northern blot hybridizations performed with the 165-bp DNA probe (Fig. 4C). Three major polyadenylated transcripts (4.0, 2.3, and 2.0 kb) and three minor polyadenylated species (3.2, 1.6, and 1.2 kb) were detected, although at different levels, in HL60, CCRF-CEM, and HeLa cells. In human epithelial 293 cells, three major species (4.2, 2.5, and 2.2 kb) were three to five times more abundantly expressed than in HL60 cells, as determined following GAPDH normalization. The differential expression of H430 sequences was also observed following hybridization of human multiple-tissue Northern blots with the ES165 probe (Fig. 4D). Large amounts of H430 polyadenylated transcripts were detected in pancreas, spleen, and prostate, while H430 sequences were weakly expressed in lung, liver, and thymus. In addition, we observed variations between the expression patterns of the different H430 mRNA species in several tissues.
Screening a human thymic cDNA library with the ES165 probe allowed us to isolate a 1.95-kb cDNA clone whose nucleotide sequence was determined and compared to that of the H430 retropseudogene. The cDNA clone corresponds to an H430 mRNA species which is polyadenylated at the level of the 3′ direct repeat identified in the H430 sequences, 18 nucleotides downstream from an AATAAA motif (Fig. 3). However, this cDNA is likely incomplete in the 5′ part since genomic and cDNA sequences were found to diverge 28 nucleotides downstream from the potential translation initiation codon.
The characterization of the 5′ ends of H430 transcripts was performed by RACE-PCR experiments performed with human thymic polyadenylated RNAs. Sequencing the resulting products revealed that the potential transcription start sites (Fig. 3) of the H430 gene are spread over a region of 260 nucleotides located within and upstream from the 5′ noncoding sequences homologous to PR264. Taken together, these observations indicate that the H430 retropseudogene is expressed through different polyadenylated mRNA species resulting, in part, from heterogeneous transcription start sites. Our results also suggest that H430 expression is regulated in a tissue-specific way.
In vivo expression of the H430 protein.
In order to determine whether H430 mRNA species are translated in vivo, polyclonal antibodies were raised against three peptides represented in the H430-specific region containing the degenerate SRSRY repeats. To avoid a cross-reaction of the polyclonal antibodies with the other SR factors, these three peptides (P213, P214, and P215; see Materials and Methods) were defined so that no consecutive SR dipeptides were present in their sequences. Following rabbit immunization, we obtained two positive sera (S213 and S214) which recognized the H430 recombinant protein expressed in a baculoviral system and which exclusively bound to their own peptides. For immunoblot analyses, we also used MAb 104 (75) and an SC35 MAb raised against the 15 carboxy-terminal residues of the PR264/SC35 protein (24). Indeed, a cross-reaction of MAb SC35 with the H430 translation product was anticipated since the corresponding H430 sequence only contains three amino acid changes, preserving a nine-amino-acid homologous peptide (Fig. 3B).
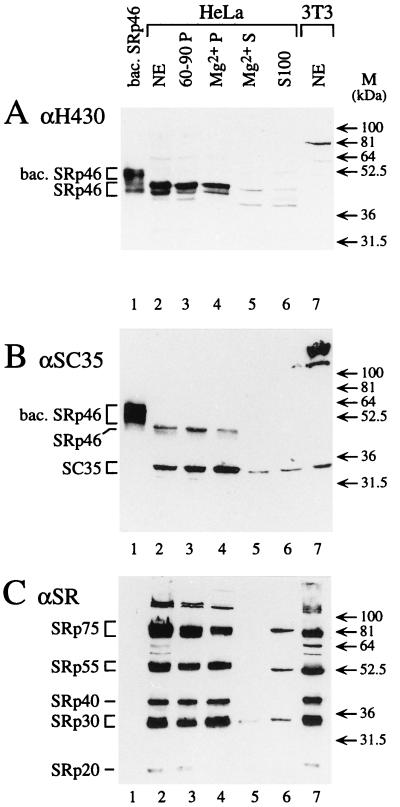
The expression and cellular localization of the H430 protein were analyzed with the different antibodies and various subcellular fractions of HeLa cells. As shown in Fig. 5A, the H430 antibody directed against the P214 peptide and revealing the baculoviral H430 protein as a diffuse band (lane 1) led to the detection of a major band and a minor band migrating as a doublet in the HeLa nuclear extract (lane 2). The differences between the apparent molecular masses of the baculoviral H430 (51 kDa) and H430 antigen in the nuclear extract (46 kDa) resulted from the presence of 40 additional amino acids (leading to a 4,780-Da excess) in the N-terminal part of the recombinant protein. As previously mentioned, the apparent molecular mass of the H430 antigen (46 kDa) was significantly higher than that deduced from the gene (34.5 kDa). However, this phenomenon is systematically observed with SR proteins, mainly as a result of extensive phosphorylation of serine residues. The minor band detected under the major one (lane 2) likely resulted from a degradative process because its intensity was found to increase following repeated freezing and thawing of the nuclear extract. Significantly, exactly the same major and minor bands were detected with the P213-specific H430 antibody (data not shown), indicating that the antigenic protein of the nuclear extract is indeed the genuine H430 product, which we designated SRp46.
FIG. 5.
Identification of the H430 gene product expressed in vivo. Immunoblot analyses of various extracts or fractions performed with the H430 antibody directed against the P214 peptide (A), the SC35 antibody (B), and MAb 104 (C). Nuclear extracts of HeLa cells (lanes 2) and derived fractions including an ammonium sulfate pellet (60-90P), an Mg2+ pellet (Mg2+P), and supernatant (Mg2+S) were analyzed in parallel with S100 cytoplasmic extract of HeLa cells (S100), nuclear extract of NIH/3T3 cells (NE), and baculovirus-expressed SRp46 (bac. SRp46). The apparent molecular masses of markers are indicated. Note that no signal was obtained in panel A when the H430 antibody was preincubated with an excess of P214 peptide coupled to ovalbumin.
Immunoblotting experiments performed with MAb SC35 indicated that this antibody also recognizes the baculoviral SRp46 protein (Fig. 5B, lane 1). In the HeLa nuclear extract (lane 2), MAb SC35 detected an intense band of 35 kDa corresponding to the PR264/SC35 factor, as well as a less-intense 46-kDa band which corresponds to the major SRp46 species. An analysis of the cytoplasmic S100 fraction with the P214 antibody (Fig. 5A, lane 6) only revealed a faint band of 46 kDa. This indicates that the SRp46 protein, like other SR factors, is localized in the nucleus and is not significantly released during isolation of the nuclei.
In contrast to MAb SC35, MAb 104 did not efficiently recognize the baculoviral SRp46 protein (Fig. 5C, lane 1). When a nuclear extract was tested (lane 2), a classical pattern of SR species was obtained but no bands were detected in the 42- to 50-kDa region, even following overexposure of the blot. An examination of the PR264/SC35 and SRp46 RS domains revealed that the two longest perfect repeats of seven and eight SR dipeptides represented in PR264/SC35 are reduced, in SRp46, to repetitions of only four SR dipeptides which might be too short to constitute an efficient epitope for MAb 104. In agreement with this, we noted that the SRp20 factor, which contains only two perfect repeats of four SR dipeptides, is also poorly revealed when the immunoblot analysis is performed under similar conditions (Fig. 5C, lanes 2 to 4).
Whether the SRp46 protein fully behaves as do the other SR factors was assessed by analyzing the different fractions obtained through a purification of SR factors performed by the procedure of Zahler et al. (75). Following immunodetection with MAb 104 antiserum (Fig. 5C), we observed that the SR proteins are concentrated in the 60- to 90% ammonium sulfate pellet (lane 3) and in the 20 mM MgCl2 precipitate (compare lanes 4 and 5). Interestingly, the SRp46 protein, detected with the P214 antibodies or MAb SC35, behaves as the other SR factors do because it is also concentrated in the ammonium sulfate and MgCl2 pellets (lanes 3 to 5 in Fig. 5A and B, respectively). Due to the specificity of the Mg2+ precipitation, these results indicate that the SRp46 protein is either phosphorylated itself, or may interact strongly with other SR factors and be trapped with these proteins during the precipitation. Following alkaline phosphatase treatment of the baculoviral SRp46 recombinant protein, we have observed a 5- to 7-kDa reduction of its apparent molecular mass, thus implying that the SRp46 product is indeed phosphorylated in vivo (data not shown).
To determine whether SRp46 is expressed in various cell types, aliquots of nuclear extracts purified from different cell lines and containing equivalent amounts of SR factors were analyzed. Consistent with the results of Northern blot hybridizations (Fig. 4C), we observed that 293 cells contain larger amounts of SRp46 protein than HeLa cells (Fig. 6, lanes 2 and 3). Other human cells, such as KATO III cells (lane 4) and MCF7 and SVK14 cells (data not shown) were also found to contain significant amounts of SRp46, indicating that expression of this protein is not restricted to a few cell types. Finally, we searched for H430 expression in other species. The SRp46 protein was not detected in either NIH/3T3 (Fig. 5 and 6, lanes 7), AT20, or L-M (TK−) murine cells (data not shown). Since no hybridization signal was observed following Northern and Southern blot analyses of murine RNA and genomic DNA performed with the ES165 probe (data not shown), these results indicate that the H430 retropseudogene has no homolog in mice. In contrast, two bands corresponding to proteins of 48 and 43 kDa were detected in CV-1 and COS-1 simian cells (Fig. 6, lanes 5 and 6). The same bands were also revealed with the P213 antibody and MAb SC35, indicating that they correspond to genuine SRp46 products and that the lower band does not result from a C-terminal cleavage of this protein (data not shown). In addition, the results obtained with MAb SC35 indicate that the intensity of the signal corresponding to the SRp46 products is five times higher than that of the PR264/SC35 factor in both CV-1 and COS-1 cells.
FIG. 6.
Expression of SRp46 in various cell lines. Aliquots of nuclear extracts, prepared from various human (HeLa, 293, KATO III), simian (CV, COS), and mouse (3T3) cell lines and containing comparable amounts of SR factors as detected with MAb 104 were analyzed by SDS–10% PAGE. SRp46 protein was detected by immunoblot analysis with the P214 antibody. Comparable SRp46 expression levels were observed with the P213 and SC35 antibodies.
The SRp46 protein acts as an SR splicing factor.
The potential activity of the SRp46 factor in the splicing reaction was analyzed by using purified baculoviral SRp46 recombinant protein (see Materials and Methods). The absence of contaminating SR factors from the infected Sf9 cells was tested by Western blot analyses with the MAb 104 antiserum. Specifically, we have determined that, for the largest amount of recombinant SRp46 protein tested, only trace amounts of insect SR factors were present (data not shown). These trace amounts were much less than the very small amounts of SR proteins which persist in all the splicing-deficient extracts used below. To assess in detail the SRp46 activity, we performed complementation analyses with the E1A and β-globin splicing substrates and with three different extracts deficient in splicing: (i) cytoplasmic S100 extracts which contain only limited amounts of SR species (Fig. 5C, lane 6), (ii) a nuclear extract immunodepleted with the 9G8 MAb (11), and (iii) a nuclear extract fraction obtained by precipitation with 55% ammonium sulfate (55P), which contains detectable levels of only SRp40 and 9G8 factors.
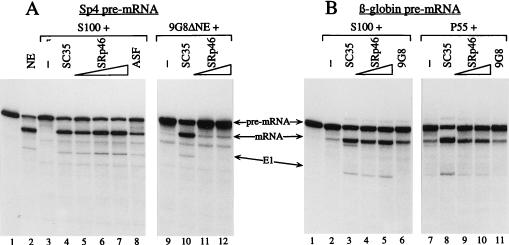
As shown in Fig. 7A, the E1A transcript was spliced efficiently in 13S mRNA in the presence of the nuclear extract (lane 2) while no splicing occurred in the presence of S100 (lane 3). The S100 fraction was efficiently complemented by SC35 and ASF/SF2 (lanes 4 and 8, respectively) as well as by increasing amounts of SRp46 (lanes 5 to 7). Comparable results were obtained with the β-globin transcript (Fig. 7B, lanes 2 to 6), showing that H430 is practically as efficient as SC35, ASF/SF2, and 9G8 factors for the complementation of S100 extracts.
FIG. 7.
In vitro analyses of SRp46 protein splicing activity. Complementing properties of SRp46 and other SR factors were analyzed in in vitro splicing assays using an E1A (A) or a β-globin (B) pre-mRNA substrate. (A) In vitro splicing assays were performed with 10 μl of standard nuclear extracts (lane 2), 9G8-depleted nuclear extract (lanes 9 to 12), or S100 extract (lanes 3 to 8). (B) Splicing was performed with 10 μl of S100 extract (lanes 2 to 6) or the P55 nuclear fraction (lanes 7 to 11). The various deficient extracts were complemented with 120 ng of recombinant SC35 or 9G8 protein, 150 ng of recombinant ASF, and increasing amounts of recombinant SRp46 protein (panel A: 70 to 210 ng in lanes 5 to 7 and 70 to 140 ng in lanes 11 to 12; panel B: 100 to 200 ng in lanes 4, 5, 9, and 10). We have verified that an aliquot of a mock fraction, originating from Sf9 cells infected with baculovirus expressing a nonrelated protein (TIF2) and corresponding to the largest amount of SRp46, was unable to complement the S100 fraction (data not shown).
To determine whether SRp46 and SC35 exhibit distinguishable splicing activities, we then tested other deficient extracts. A 9G8-depleted nuclear extract was deficient in splicing (Fig. 7A, lane 9) but was complemented by SC35 (lane 10). In contrast, we observed that this extract was only poorly rescued by SRp46 (lanes 11 and 12). A nuclear extract 55P fraction, inefficient in splicing with the β-globin substrate (Fig. 7B, lane 7), was strongly stimulated by SC35 (lane 8) but not by 9G8, which is already present in this fraction (lane 11). Complementation with SRp46 was found to stimulate splicing (lanes 9 and 10), but less efficiently than SC35.
Taken together, these results demonstrated that SRp46, which is able to fully complement S100 extracts, exhibits the general characteristics of SR factors. However, it does not display exactly the same splicing activity as SC35. This is consistent with the fact that mutations are present all along the protein sequence and that important insertions interrupt the SR domain.
DISCUSSION
H430 is a PR264/SC35 retropseudogene.
The presence of PR264/SC35-related sequences in humans has raised questions as to their origin and possible functions. The results presented in this study indicate that the PR264/SC35-related H430 sequences exhibit the three hallmarks of nonretroviral retroposons (reviewed in references 65 and 72). Indeed, striking features of the H430 gene are the absence of intronic sequences, the acquisition of a 3′ poly(A) tail, and the presence of flanking direct repeats at the points of divergence with the PR264 genomic sequences. As already mentioned for other mammalian retropseudogenes such as the human phosphoglycerate kinase-2 gene (7, 44), the apparent degeneration of the poly(A) tail remnant in the processed H430 gene from a presumed original status of 100% adenines to its current status of 71% adenines is a predictable consequence of evolutionary divergence. Consistent with this, the two imperfect repeats flanking the H430 pseudogene were found to be 72% homologous.
Another striking feature of the H430 gene is the presence of an ORF, largely colinear with those of PR264/SC35 cDNAs (60), whose coding capacity is significantly greater than that of the progenitor PR264/SC35 gene, mainly due to a multimerization of preexisting sequences in exon 2. In contrast, most of the processed pseudogenes characterized thus far exhibit, with respect to their founder gene, either an unrelated (46) or a shorter ORF arising from frameshift and stop mutations (4, 71).
H430 is an expressed retroposon.
The identification, in H430 sequences, of a unique 165-bp region has allowed us to establish that this pseudogene is transcribed at different levels in several human cell lines and normal tissues. It is currently admitted that, aside from those with deleterious mutations, most retropseudogenes are nonfunctional because they correspond to cDNA copies lacking promoter sequences (8, 47). Most retroposons that are expressed have been shown to originate from aberrantly transcribed mRNA molecules that included promoter sequences (12, 56, 65). But occasionally, a retropseudogene is expressed by association with foreign regulatory sequences, either preexisting at the site of insertion or generated by mutation (8, 58). This is likely the case for the H430 pseudogene since we observed an abrupt reduction in homology between H430 and PR264/SC35 genomic sequences upstream from the PR264/SC35 transcriptional start site.
Northern blot analyses have revealed that the H430 gene is expressed through several polyadenylated mRNA species ranging from 4.2 to 1.2 kb in size. The sizes of two of the major transcripts (2.3 and 2.0 kb) nearly correspond to that of the H430 pseudogene, suggesting that the transcription start and polyadenylation sites of these mRNAs are close to the H430 boundaries. Consistent with this, a polyadenylation signal (AATAAA) is present in the H430 poly(A) tail remnant and most of the transcriptional start sites mapped by RACE-PCR experiments are located immediately upstream from the H430 translation initiation codon. The large H430 transcripts (4.2, 4.0, and 3.2 kb) could correspond to readthrough species and/or contain additional 5′-proximal sequences. In agreement with the latter hypothesis, RACE-PCR and preliminary RNase protection experiments (data not shown) have revealed the existence of potential transcription start sites located upstream from the H430 5′ flanking repeat.
The existence of heterogenous H430 transcription start sites accounting for the fuzzy appearance of the mRNAs detected by Northern blot analyses is reminiscent of GC box-containing promoters (45). An examination of the sequences surrounding the H430 potential start sites indeed revealed the presence of GC-rich motifs located in the region homologous to the PR264/SC35 5′-untranslated sequences. Since most of these motifs are interrupted by A/T insertions in the corresponding region of PR264/SC35, one can hypothesize that mutations occurring after the retroposition event have revealed a promoting activity. Further studies will be needed to precisely characterize the promoter regions involved in the expression of the different H430 mRNA species.
Evolutionary divergence of H430 sequences.
The time of origin of the H430 pseudogene was estimated from the extent of divergence of the pseudogene noncoding sequences from those of the functional human (60) and mouse PR264/SC35 genes (23a) (GenBank accession no., X98511). In this analysis, only the 5′-untranslated sequences represented in human PR264/SC35 mRNA species were compared to those of the corresponding regions of H430 and murine PR264/SC35 because the high level of conservation of the 3′ noncoding sequences (81% identity between H430 and PR264/SC35 sequences) likely reflects functional constraints that reduced the rate of divergence. We observed the substitution of 46 of 166 nucleotides (27.7% divergence) between the H430 and human PR264/SC35 sequences, 61 of 156 nucleotides (39.1% divergence) between the H430 and murine PR264/SC35 sequences, and 65 of 162 nucleotides (40.1% divergence) between the human and murine PR264/SC35 sequences. The method of Fitch and Margoliash as detailed by Wu et al. (73) was used to calculate the distribution of divergence among the three sequences. Of the 27.7% divergence between the human PR264/SC35 and H430 genes, 13.35% was due to base substitutions within the pseudogene since its origin and 14.35% was due to base substitutions in the PR264/SC35 gene, indicating that both the PR264/SC35 and the H430 genes have evolved at the same rate. If the average rate for divergence of ancient primate pseudogenes is 1.5 × 10−9 substitutions per site per year (37), then the calculated 0.1335 substitutions per site in the pseudogene would require 89 million years (Myr) to accumulate. A comparison of this value with the date of divergence of primates and rodents, which has been recently reestimated to about 118 Myr (1), allows an explanation of why H430 sequences are not represented in the murine genome.
SRp46 as a novel member of the SR family.
The results reported in this study demonstrate that the H430 gene has remained functional despite the long period of evolution of primates. Remarkably, the multiple mutations, amplifications, and deletions have not affected the global ORF of the H430 gene, suggesting that a strong selection pressure has preserved its functionality and that its expression resulted in a general benefit for the primate ancestors. In agreement with this, we present several pieces of evidence indicating that the H430 gene product may be considered a novel member of the SR family. First, it exhibits all the general characteristics of the SR factors. The only exception, likely explaining why SRp46 was not identified previously, is that it is not detected by MAb 104. The most important characteristic of SRp46 is that it complements a splicing-deficient S100 extract. In addition, we have shown that SRp46 is localized in nuclear fractions. An immunofluorescence microscopy analysis has allowed us to detect a speckled distribution of SRp46 in the nucleus (data not shown), reminiscent of that of other SR factors and indicating that the targeting signals to the speckles have been preserved in SRp46 (9). Second, SRp46 differs significantly from the PR264/SC35 factor in several respects. (i) Despite the limited number of nonconservative amino acid replacements in the RBD, four of seven affect loop 3, which is positioned between the β2 and β3 sheets. Thus, the RYTKESR peptide is changed to a PHTKAPR peptide, resulting in the loss of two charged residues. Since it has been shown previously for snRNP proteins U1A and U2B′ that the loop 3 sequence is primarily involved in the RNA recognition specificity of the RBD (53), it is reasonable to assume that SRp46 exhibits an RNA substrate specificity different from that of PR264/SC35. (ii) In the RS domain of SRp46, the 11-fold repetition of the highly degenerate SRSRY motif results in an RS divergent region, which contains only two SRSR peptides and which exhibits a very regular tyrosine repetition. Such important alterations might modify significantly the interaction properties of the RS domain. Indeed, the well-conserved RS domains of ASF/SF2 and SC35 are interchangeable (14), in agreement with the fact that they develop similar protein-protein interactions with other factors containing RS domains or RS motifs (74). However, a comparable analysis including SR factor p54, which exhibits the less-conserved RS domain of the SR family, indicates that a divergence of the RS domain induces some modifications in the protein-protein interactions (79). Therefore, it is possible that, like p54, SRp46 plays different roles in the processes involving protein-protein interactions. Consistent with this, we have shown that SRp46 exhibits complementing splicing activities different from those of PR264/SC35. (iii) A last feature which differentiates SRp46 and PR264/SC35 concerns their respective levels of expression in different tissues. For instance, we observed that SRp46 transcripts are as well expressed in spleen as those of PR264/SC35 but are weakly expressed in thymus, in contrast to those of PR264/SC35 (data not shown) (79). Therefore, because the respective activities of SR factors in alternative splicing may depend on their relative expression levels in cells (20, 40), the tissue-specific expression of SRp46 may be of biological significance. Taken together, our results strongly suggest that the H430 retropseudogene has acquired the status of a novel functional gene.
Among the numerous retropseudogenes characterized thus far in mammals (65, 72), only a few are functionally expressed (47). This is the case for the mouse phosphoglycerate kinase-2 (Pgk-2) (7), pyruvate dehydrogenase-2 (Pdha-2) (15), Zfa (2), and glucose-6-phosphate dehydrogenase-2 (G6pd-2) (26) genes, which are expressed in spermatogenic cells where they likely compensate for the loss of expression of the X chromosome genes Pgk-1, Pdha-1, Zfx, and G6pd-1 encoding the corresponding isotypic proteins (2, 7, 44, 63). In contrast, the SRp46 protein encoded by the H430 retropseudogene is detected in cells which also express the PR264/SC35 splicing factor, and SRp46 appears to exhibit a splicing activity different from that of its founder gene product. Thus, the H430 gene represents, to our knowledge, the first clear example of a retropseudogene which encodes a novel trans-acting factor. Inasmuch as the existence of retropseudogenes related to the SRp20 gene has recently been suggested (27), it will be interesting to determine whether other retroposition events generating functional SR-processed pseudogenes contribute to increase the diversity of the SR splicing factor family.
ACKNOWLEDGMENTS
We thank G. Hildwein for excellent technical assistance, Y. Lutz for the immunochemistry analysis, and J. Voegel and H. Gronemeyer for the baculovirus construct expressing TIF2. We also thank the IGBMC services for cell culture, baculovirus expression, antibody preparation, DNA sequencing, and oligonucleotide and peptide synthesis. D. Lawrence is acknowledged for the critical reading of the manuscript.
C.G., M.P., and E.L.R. were supported by a fellowship from the Ministère de la Recherche et de la Technologie (MRT). This work was supported by funds from the Institut National de la Santé et de la Recherche Médicale, the Centre National de la Recherche Scientifique, the Centre Hospitalier Universitaire Régional (Strasbourg), the Ligue Nationale Contre le Cancer (Comité National et Comité du Cher), the Fondation de France, and the Association pour la Recherche sur le Cancer. Part of this work was performed at and funded by the Institut Curie when the Laboratoire d’Oncologie Virale et Moléculaire was affiliated to the CNRS-UMR 146.
REFERENCES
- 1.Arnason U, Gullberg A, Janke A, Xu X. Pattern and timing of evolutionary divergences among hominoids based on analyses of complete mtDNAs. J Mol Evol. 1996;43:650–661. doi: 10.1007/BF02202113. [DOI] [PubMed] [Google Scholar]
- 2.Ashworth A, Skene B, Swift S, Lovell-Badge R. Zfa is an expressed retroposon derived from an alternative transcript of the Zfx gene. EMBO J. 1990;9:1529–1534. doi: 10.1002/j.1460-2075.1990.tb08271.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ayane M, Preuss U, Kohler G, Nielsen P J. A differentially expressed murine RNA encoding a protein with similarities to two types of nucleic acid binding motifs. Nucleic Acids Res. 1991;19:1273–1278. doi: 10.1093/nar/19.6.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bard J A, Nawoschik S P, O’Dowd B F, George S R, Branchek T A, Weinshank R L. The human serotonin 5-hydroxytryptamine 1D receptor pseudogene is transcribed. Gene. 1995;153:295–296. doi: 10.1016/0378-1119(94)00693-m. [DOI] [PubMed] [Google Scholar]
- 5.Bermingham J R, Arden K C, Naumova A K, Sapienza C, Viars C S, Fu X D, Khotz J, Manley J L, Rosenfeld M G. Chromosomal localization of mouse and human genes encoding the splicing factors ASF/SF2 (SFRS1) and SC-35 (SFRS2) Genomics. 1995;29:70–79. doi: 10.1006/geno.1995.1216. [DOI] [PubMed] [Google Scholar]
- 6.Birney E, Kumar S, Krainer A R. Analysis of the RNA-recognition motif and RS and RGG domains: conservation in metazoan pre-mRNA splicing factors. Nucleic Acids Res. 1993;21:5803–5816. doi: 10.1093/nar/21.25.5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boer P H, Adra C N, Lau Y-F, McBurney M W. The testis-specific phosphoglycerate kinase gene pgk-2 is a recruited retroposon. Mol Cell Biol. 1987;7:3107–3112. doi: 10.1128/mcb.7.9.3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brosius J. Retroposons: seeds of evolution. Science. 1991;251:753. doi: 10.1126/science.1990437. [DOI] [PubMed] [Google Scholar]
- 9.Caceres J F, Misteli T, Screaton G R, Spector D L, Krainer A R. Role of the modular domains of SR proteins in subnuclear localization and alternative splicing specificity. J Cell Biol. 1997;138:225–238. doi: 10.1083/jcb.138.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caceres J F, Stamm S, Helfman D M, Krainer A R. Regulation of alternative splicing in vivo by overexpression of antagonistic splicing factors. Science. 1994;265:1706–1709. doi: 10.1126/science.8085156. [DOI] [PubMed] [Google Scholar]
- 11.Cavaloc Y, Popielarz M, Fuchs J P, Gattoni R, Stevenin J. Characterization and cloning of the human splicing factor 9G8: a novel 35 kDa factor of the serine/arginine protein family. EMBO J. 1994;13:2639–2649. doi: 10.1002/j.1460-2075.1994.tb06554.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chakrabarti R, McCracken J B, Jr, Chakrabarti D, Souba W W. Detection of a functional promoter/enhancer in an intron-less human gene encoding a glutamine synthetase-like enzyme. Gene. 1995;153:163–199. doi: 10.1016/0378-1119(94)00751-d. [DOI] [PubMed] [Google Scholar]
- 13.Champlin D T, Frasch M, Saumweber H, Lis J T. Characterization of a Drosophila protein associated with boundaries of transcriptionally active chromatin. Genes Dev. 1991;5:1611–1621. doi: 10.1101/gad.5.9.1611. [DOI] [PubMed] [Google Scholar]
- 14.Chandler S D, Mayeda A, Yeakley J M, Krainer A R, Fu X D. RNA splicing specificity determined by the coordinated action of RNA recognition motifs in SR proteins. Proc Natl Acad Sci USA. 1997;94:3596–3601. doi: 10.1073/pnas.94.8.3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dahl H H, Brown R M, Hutchison W M, Maragos C, Brown G K. A testis-specific form of the human pyruvate dehydrogenase E1 alpha subunit is coded for by an intronless gene on chromosome 4. Genomics. 1990;8:225–232. doi: 10.1016/0888-7543(90)90275-y. [DOI] [PubMed] [Google Scholar]
- 16.Diamond R H, Du K, Lee V M, Mohn K L, Haber B A, Tewari D S, Taub R. Novel delayed-early and highly insulin-induced growth response genes. Identification of HRS, a potential regulator of alternative pre-mRNA splicing. J Biol Chem. 1993;268:15185–15192. [PubMed] [Google Scholar]
- 17.Dignam J D, Lebovitz R M, Roeder R G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eperon I C, Ireland D C, Smith R A, Mayeda A, Krainer A R. Pathways for selection of 5′ splice sites by U1 snRNPs and SF2/ASF. EMBO J. 1993;12:3607–3617. doi: 10.1002/j.1460-2075.1993.tb06034.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eychene A, Vianney-Barnier J, Apiou F, Dutrillaux B, Calothy G. Chromosomal assignment of two human B-raf (Rmil) proto-oncogene loci: B-raf-1 encoding the p94Braf/Rmil and B-raf-2, a processed pseudogene. Oncogene. 1992;7:1657–1660. [PubMed] [Google Scholar]
- 20.Fu X D. The superfamily of arginine/serine-rich splicing factors. RNA. 1995;1:663–680. [PMC free article] [PubMed] [Google Scholar]
- 21.Fu X D. Specific commitment of different pre-mRNAs to splicing by single SR proteins. Nature. 1993;365:82–85. doi: 10.1038/365082a0. [DOI] [PubMed] [Google Scholar]
- 22.Fu X D, Maniatis T. Isolation of a complementary DNA that encodes the mammalian splicing factor SC35. Science. 1992;256:535–538. doi: 10.1126/science.1373910. [DOI] [PubMed] [Google Scholar]
- 23.Fu X D, Mayeda A, Maniatis T, Krainer A R. General splicing factors SF2 and SC35 have equivalent activities in vitro, and both affect alternative 5′ and 3′ splice site selection. Proc Natl Acad Sci USA. 1992;89:11224–11228. doi: 10.1073/pnas.89.23.11224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23a.Gaillard, C., and B. Perbal. Unpublished results.
- 24.Gallego M E, Gattoni R, Stevenin J, Marie J, Expert-Bezancon A. The SR splicing factors ASF/SF2 and SC35 have antagonistic effects on intronic enhancer-dependent splicing of the beta-tropomyosin alternative exon 6A. EMBO J. 1997;16:1772–1784. doi: 10.1093/emboj/16.7.1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ge H, Zuo P, Manley J L. Primary structure of the human splicing factor ASF reveals similarities with Drosophila regulators. Cell. 1991;66:373–382. doi: 10.1016/0092-8674(91)90626-a. [DOI] [PubMed] [Google Scholar]
- 26.Hendriksen P J, Hoogerbrugge J W, Baarends W M, De Boer P, Vreeburg J T, Vos E A, Van Der Lende T, Grootegoed J A. Testis-specific expression of a functional retroposon encoding glucose-6-phosphate dehydrogenase in the mouse. Genomics. 1997;41:350–359. doi: 10.1006/geno.1997.4673. [DOI] [PubMed] [Google Scholar]
- 27.Jumaa H, Guenet J L, Nielsen P J. Regulated expression and RNA processing of transcripts from the SRp20 splicing factor gene during the cell cycle. Mol Cell Biol. 1997;17:3116–3124. doi: 10.1128/mcb.17.6.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jumaa H, Nielsen P J. The splicing factor SRp20 modifies splicing of its own mRNA and ASF/SF2 antagonizes this regulation. EMBO J. 1997;16:5077–5085. doi: 10.1093/emboj/16.16.5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kenan D J, Query C C, Keene J D. RNA recognition: towards identifying determinants of specificity. Trends Biochem Sci. 1991;16:214–220. doi: 10.1016/0968-0004(91)90088-d. [DOI] [PubMed] [Google Scholar]
- 30.Kim Y J, Zuo P, Manley J L, Baker B S. The Drosophila RNA-binding protein RBP1 is localized to transcriptionally active sites of chromosomes and shows a functional similarity to human splicing factor ASF/SF2. Genes Dev. 1992;6:2569–2579. doi: 10.1101/gad.6.12b.2569. [DOI] [PubMed] [Google Scholar]
- 31.Kohtz J D, Jamison S F, Will C L, Zuo P, Luhrmann R, Garcia-Blanco M A, Manley J L. Protein-protein interactions and 5′-splice-site recognition in mammalian mRNA precursors. Nature. 1994;368:119–124. doi: 10.1038/368119a0. [DOI] [PubMed] [Google Scholar]
- 32.Krainer A R, Conway G C, Kozak D. Purification and characterization of pre-mRNA splicing factor SF2 from HeLa cells. Genes Dev. 1990;4:1158–1171. doi: 10.1101/gad.4.7.1158. [DOI] [PubMed] [Google Scholar]
- 33.Krainer A R, Mayeda A, Kozak D, Binns G. Functional expression of cloned human splicing factor SF2: homology to RNA-binding proteins, U1 70K, and Drosophila splicing regulators. Cell. 1991;66:383–394. doi: 10.1016/0092-8674(91)90627-b. [DOI] [PubMed] [Google Scholar]
- 34.Kraus M E, Lis J T. The concentration of B52, an essential splicing factor and regulator of splice site choice in vitro, is critical for Drosophila development. Mol Cell Biol. 1994;14:5360–5370. doi: 10.1128/mcb.14.8.5360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lavigueur A, La Branche H, Kornblihtt A R, Chabot B. A splicing enhancer in the human fibronectin alternate ED1 exon interacts with SR proteins and stimulates U2 snRNP binding. Genes Dev. 1993;7:2405–2417. doi: 10.1101/gad.7.12a.2405. [DOI] [PubMed] [Google Scholar]
- 36.Lemieux N, Dutrillaux B, Viegas-Péquignot E. A simple method for simultaneous R- or G-banding and fluorescence in situ hybridization of a small single copy gene. Cytogenet Cell Genet. 1992;59:311–312. doi: 10.1159/000133277. [DOI] [PubMed] [Google Scholar]
- 37.Li W H, Tanimura M, Sharp P M. An evaluation of the molecular clock hypothesis using mammalian DNA sequences. J Mol Evol. 1987;25:330–342. doi: 10.1007/BF02603118. [DOI] [PubMed] [Google Scholar]
- 38.Linnenbach A J, Seng B A, Wu S, Robbins S, Scollon M, Pyrc J J, Druck T, Huebner K. Retroposition in a family of carcinoma-associated antigen genes. Mol Cell Biol. 1993;13:1507–1515. doi: 10.1128/mcb.13.3.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lynch K W, Maniatis T. Assembly of specific SR protein complexes on distinct regulatory elements of the Drosophila doublesex splicing enhancer. Genes Dev. 1996;10:2089–2101. doi: 10.1101/gad.10.16.2089. [DOI] [PubMed] [Google Scholar]
- 40.Manley J L, Tacke R. SR proteins and splicing control. Genes Dev. 1996;10:1569–1579. doi: 10.1101/gad.10.13.1569. [DOI] [PubMed] [Google Scholar]
- 41.Mayeda A, Krainer A R. Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. Cell. 1992;68:365–375. doi: 10.1016/0092-8674(92)90477-t. [DOI] [PubMed] [Google Scholar]
- 42.Mayeda A, Munroe S H, Caceres J F, Krainer A R. Function of conserved domains of hnRNP A1 and other hnRNP A/B proteins. EMBO J. 1994;13:5483–5495. doi: 10.1002/j.1460-2075.1994.tb06883.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mayeda A, Zahler A M, Krainer A R, Roth M B. Two members of a conserved family of nuclear phosphoproteins are involved in pre-mRNA splicing. Proc Natl Acad Sci USA. 1992;89:1301–1304. doi: 10.1073/pnas.89.4.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McCarrey J R, Thomas K. Human testis-specific PGK gene lacks introns and possesses characteristics of a processed gene. Nature. 1987;326:501–505. doi: 10.1038/326501a0. [DOI] [PubMed] [Google Scholar]
- 45.Melton D W. Strategies and mechanisms for the control of transcriptional initiation of mammalian protein-coding genes. J Cell Sci. 1987;88:267–270. doi: 10.1242/jcs.88.3.267. [DOI] [PubMed] [Google Scholar]
- 46.Nelissen R L, Gunnewiek J M, Lambermon M H, Van Venrooij W J. Cloning and characterization of two processed pseudogenes and the cDNA for the murine U1 snRNP-specific protein C. Gene. 1997;184:273–278. doi: 10.1016/s0378-1119(96)00612-9. [DOI] [PubMed] [Google Scholar]
- 47.Nouvel P. The mammalian genome shaping activity of reverse transcriptase. Genetica. 1994;93:191–201. doi: 10.1007/BF01435251. [DOI] [PubMed] [Google Scholar]
- 48.Perbal B. A practical guide to molecular cloning. 2nd ed. New York, N.Y: Wiley; 1988. [Google Scholar]
- 49.Perez A, Kastner P, Sethi S, Lutz Y, Reibel C, Chambon P. PMLRAR homodimers: distinct DNA binding properties and heterodimeric interactions with RXR. EMBO J. 1993;12:3171–3182. doi: 10.1002/j.1460-2075.1993.tb05986.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ramchatesingh J, Zahler A M, Neugebauer K M, Roth M B, Cooper T A. A subset of SR proteins activates splicing of the cardiac troponin T alternative exon by direct interactions with an exonic enhancer. Mol Cell Biol. 1995;15:4898–4907. doi: 10.1128/mcb.15.9.4898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ring H Z, Lis J T. The SR protein B52/SRp55 is essential for Drosophila development. Mol Cell Biol. 1994;14:7499–7506. doi: 10.1128/mcb.14.11.7499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roth M B, Zahler A M, Stolk J A. A conserved family of nuclear phosphoproteins localized to sites of polymerase II transcription. J Cell Biol. 1991;115:587–596. doi: 10.1083/jcb.115.3.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Scherly D, Boelens W, Dathan N A, van Venrooij W J, Mattaj I W. Major determinants of the specificity of interaction between small nuclear ribonucleoproteins U1A and U2B" and their cognate RNAs. Nature. 1990;345:502–506. doi: 10.1038/345502a0. [DOI] [PubMed] [Google Scholar]
- 54.Schmitt P, Gattoni R, Keohavong P, Stevenin J. Alternative splicing of E1A transcripts of adenovirus requires appropriate ionic conditions in vitro. Cell. 1987;50:31–39. doi: 10.1016/0092-8674(87)90659-3. [DOI] [PubMed] [Google Scholar]
- 55.Screaton G R, Caceres J F, Mayeda A, Bell M V, Plebanski M, Jackson D G, Bell J I, Krainer A R. Identification and characterization of three members of the human SR family of pre-mRNA splicing factors. EMBO J. 1995;14:4336–4349. doi: 10.1002/j.1460-2075.1995.tb00108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Soares M B, Schon E, Henderson A, Karathanasis S K, Cate R, Zeitlin S, Chirgwin J, Efstratiadis A. RNA-mediated gene duplication: the rat preproinsulin I gene is a functional retroposon. Mol Cell Biol. 1985;5:2090–2103. doi: 10.1128/mcb.5.8.2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Staknis D, Reed R. SR proteins promote the first specific recognition of pre-mRNA and are present together with the U1 small nuclear ribonucleoprotein particle in a general splicing enhancer complex. Mol Cell Biol. 1994;14:7670–7682. doi: 10.1128/mcb.14.11.7670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stein J P, Munjaal R P, Lagace L, Lai E C, O’Malley B W, Means A R. Tissue-specific expression of a chicken calmodulin pseudogene lacking intervening sequences. Proc Natl Acad Sci USA. 1983;80:6485–6489. doi: 10.1073/pnas.80.21.6485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sun Q, Mayeda A, Hampson R K, Krainer A R, Rottman F M. General splicing factor SF2/ASF promotes alternative splicing by binding to an exonic splicing enhancer. Genes Dev. 1993;7:2598–2608. doi: 10.1101/gad.7.12b.2598. [DOI] [PubMed] [Google Scholar]
- 60.Sureau A, Perbal B. Several mRNAs with variable 3′ untranslated regions and different stability encode the human PR264/SC35 splicing factor. Proc Natl Acad Sci USA. 1994;91:932–936. doi: 10.1073/pnas.91.3.932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sureau A, Soret J, Vellard M, Crochet J, Perbal B. The PR264/c-myb connection: expression of a splicing factor modulated by a nuclear protooncogene. Proc Natl Acad Sci USA. 1992;89:11683–11687. doi: 10.1073/pnas.89.24.11683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tacke R, Manley J L. The human splicing factors ASF/SF2 and SC35 possess distinct, functionally significant RNA binding specificities. EMBO J. 1995;14:3540–3551. doi: 10.1002/j.1460-2075.1995.tb07360.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Takakubo F, Dahl H H. The expression pattern of the pyruvate dehydrogenase E1 alpha subunit genes during spermatogenesis in adult mouse. Exp Cell Res. 1992;199:39–49. doi: 10.1016/0014-4827(92)90459-l. [DOI] [PubMed] [Google Scholar]
- 64.Vaitukaitis J L. Production of antisera with small doses of immunogen: multiple intradermal injections. Methods Enzymol. 1981;73:46–52. doi: 10.1016/0076-6879(81)73055-6. [DOI] [PubMed] [Google Scholar]
- 65.Vanin E F. Processed pseudogenes: characteristics and evolution. Annu Rev Genet. 1985;19:253–272. doi: 10.1146/annurev.ge.19.120185.001345. [DOI] [PubMed] [Google Scholar]
- 66.Vellard M, Soret J, Viegas-Pequignot E, Galibert F, Nguyen V C, Dutrillaux B, Perbal B. C-myb proto-oncogene: evidence for intermolecular recombination of coding sequences. Oncogene. 1991;6:505–514. [PubMed] [Google Scholar]
- 67.Vellard M, Sureau A, Soret J, Martinerie C, Perbal B. A potential splicing factor is encoded by the opposite strand of the trans-spliced c-myb exon. Proc Natl Acad Sci USA. 1992;89:2511–2515. doi: 10.1073/pnas.89.7.2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Voegel J J, Heine M J S, Zechel C, Chambon P, Gronemeyer H. TIF2, a 160 kDa transcriptional mediator for the ligand-dependent activation function AF-2 of nuclear receptors. EMBO J. 1996;15:3667–3675. [PMC free article] [PubMed] [Google Scholar]
- 69.Wang J, Manley J L. Overexpression of the SR proteins ASF/SF2 and SC35 influences alternative splicing in vivo in diverse ways. RNA. 1995;1:335–346. [PMC free article] [PubMed] [Google Scholar]
- 70.Wang J, Takagaki Y, Manley J L. Targeted disruption of an essential vertebrate gene: ASF/SF2 is required for cell viability. Genes Dev. 1996;10:2588–2599. doi: 10.1101/gad.10.20.2588. [DOI] [PubMed] [Google Scholar]
- 71.Weil D, Power M A, Webb G C, Li C L. Antisense transcription of a murine FGFR-3 psuedogene during fetal developement. Gene. 1997;187:115–122. doi: 10.1016/s0378-1119(96)00733-0. [DOI] [PubMed] [Google Scholar]
- 72.Weiner A M, Deininger P L, Efstratiadis A. Nonviral retroposons: genes, pseudogenes, and transposable elements generated by the reverse flow of genetic information. Annu Rev Biochem. 1986;55:631–661. doi: 10.1146/annurev.bi.55.070186.003215. [DOI] [PubMed] [Google Scholar]
- 73.Wu C I, Li W H, Shen J J, Scarpulla R C, Limbach K J, Wu R. Evolution of cytochrome c genes and pseudogenes. J Mol Evol. 1986;23:61–75. doi: 10.1007/BF02100999. [DOI] [PubMed] [Google Scholar]
- 74.Wu J Y, Maniatis T. Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell. 1993;75:1061–1070. doi: 10.1016/0092-8674(93)90316-i. [DOI] [PubMed] [Google Scholar]
- 75.Zahler A M, Lane W S, Stolk J A, Roth M B. SR proteins: a conserved family of pre-mRNA splicing factors. Genes Dev. 1992;6:837–847. doi: 10.1101/gad.6.5.837. [DOI] [PubMed] [Google Scholar]
- 76.Zahler A M, Neugebauer K M, Lane W S, Roth M B. Distinct functions of SR proteins in alternative pre-mRNA splicing. Science. 1993;260:219–222. doi: 10.1126/science.8385799. [DOI] [PubMed] [Google Scholar]
- 77.Zahler A M, Neugebauer K M, Stolk J A, Roth M B. Human SR proteins and isolation of a cDNA encoding SRp75. Mol Cell Biol. 1993;13:4023–4028. doi: 10.1128/mcb.13.7.4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zahler A M, Roth M B. Distinct functions of SR proteins in recruitment of U1 small nuclear ribonucleoprotein to alternative 5′ splice sites. Proc Natl Acad Sci USA. 1995;92:2642–2646. doi: 10.1073/pnas.92.7.2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang W J, Wu J Y. Functional properties of p54, a novel SR protein active in constitutive and alternative splicing. Mol Cell Biol. 1996;16:5400–5408. doi: 10.1128/mcb.16.10.5400. [DOI] [PMC free article] [PubMed] [Google Scholar]