Abstract
Only five of the nine subunits of human eukaryotic translation initiation factor 3 (eIF3) have recognizable homologs encoded in the Saccharomyces cerevisiae genome, and only two of these (Prt1p and Tif34p) were identified previously as subunits of yeast eIF3. We purified a polyhistidine-tagged form of Prt1p (His-Prt1p) by Ni2+ affinity and gel filtration chromatography and obtained a complex of ≈600 kDa composed of six polypeptides whose copurification was completely dependent on the polyhistidine tag on His-Prt1p. All five polypeptides associated with His-Prt1p were identified by mass spectrometry, and four were found to be the other putative homologs of human eIF3 subunits encoded in S. cerevisiae: YBR079c/Tif32p, Nip1p, Tif34p, and YDR429c/Tif35p. The fifth Prt1p-associated protein was eIF5, an initiation factor not previously known to interact with eIF3. The purified complex could rescue Met-tRNAiMet binding to 40S ribosomes in defective extracts from a prt1 mutant or extracts from which Nip1p had been depleted, indicating that it possesses a known biochemical activity of eIF3. These findings suggest that Tif32p, Nip1p, Prt1p, Tif34p, and Tif35p comprise an eIF3 core complex, conserved between yeast and mammals, that stably interacts with eIF5. Nip1p bound to eIF5 in yeast two-hybrid and in vitro protein binding assays. Interestingly, Sui1p also interacts with Nip1p, and both eIF5 and Sui1p have been implicated in accurate recognition of the AUG start codon. Thus, eIF5 and Sui1p may be recruited to the 40S ribosomes through physical interactions with the Nip1p subunit of eIF3.
The initiation of protein synthesis in eukaryotic cells is dependent on multiple initiation factors (eIFs) that stimulate the binding of mRNA and methionyl-initiator tRNA (tRNAiMet) to 40S ribosomes to form the 48S preinitiation complex (39). The Met-tRNAiMet is delivered to 40S ribosomes in a ternary complex with eIF2 and GTP, whereas the binding of mRNA to ribosomes is stimulated by eIF4F, eIF4A, eIF4B (39), and the poly(A)-binding protein Pab1p (54). Joining of the 60S subunit to form an 80S initiation complex requires hydrolysis of the GTP bound to eIF2, dissociation of the ternary complex, and release of the eIF2-GDP binary complex, and eIF5 promotes these events by stimulating GTP hydrolysis on ternary complexes bound to 40S ribosomes (39).
Mammalian eIF3 is a multisubunit complex that has been implicated in several aspects of 48S complex formation. The purified factor promotes dissociation of 80S ribosomes into 40S and 60S subunits, forming a complex with the 40S subunits, and stabilizes binding of the eIF2–GTP–Met-tRNAiMet ternary complex to the 40S ribosome. It also stimulates binding of mRNA to 40S subunits (9, 56), presumably through its interactions with the cap-binding initiation factor eIF4F (36, 38) or eIF4B (41). A mammalian eIF3 complex, purified by its ability to promote methionylpuromycin (Met-puromycin) synthesis by an 80S initiation complex in an assay containing purified eIF1A, eIF2, eIF5, eIF5A, and ribosomal subunits in addition to eIF3 (11), contained nine nonidentical polypeptides, called p170, p116, p110, p66, p48, p47, p44, p40, and p36. All nine polypeptides could be coimmunoprecipitated with affinity-purified antibodies against the p170 subunit (3–5, 40), supporting the idea that they are subunits of the same complex.
With the isolation of cDNAs encoding all nine subunits of mammalian eIF3 (5, 30), it became apparent that only five of these proteins have identifiable homologs encoded in the genome of the yeast Saccharomyces cerevisiae. The yeast proteins encoded by YBR079c, PRT1 (44), NIP1 (25), TIF34 (42), and YDR429c have similar predicted molecular weights and strong sequence similarities to the mammalian eIF3 subunits p170, p116, p110, p36, and p44, respectively. The 90- and 39-kDa products of PRT1 and TIF34, respectively, were shown to copurify through several fractionation steps with a yeast eIF3 complex purified by its ability to substitute for mammalian eIF3 in the Met-puromycin synthesis assay described above (42, 44). Prt1p is an essential protein required in vivo for translation initiation (28), and the prt1-1 mutation was shown previously to impair ternary complex binding to 40S subunits in cell extracts (17, 21). TIF34 is also essential and is required in vivo for normal rates of translation initiation (42, 58). The p33 polypeptide that copurified with yeast eIF3 activity in the Met-puromycin assay is similar in size to the predicted 30.5-kDa product of YDR429c, the putative homolog of human eIF3-p44. Recent evidence that the YDR429c product interacts genetically and physically with Tif34p (2, 58) supports the idea that it represents the yeast counterpart of human eIF3-p44. Based on these findings and their sequence similarity to subunits of human eIF3, the products of YBR079c and YDR429c have been referred to as Tif32p and Tif35p, respectively (30), a nomenclature adopted henceforth in this report.
A 16-kDa polypeptide that copurified with yeast eIF3 activity in the Met-puromycin assay was identified as the product of the SUI1 gene (43), first identified genetically by recessive mutations that permit increased utilization of UUG triplets as translation initiation codons (61). This phenotype suggests that Sui1p is required for accurate recognition of the AUG start codon by initiator tRNAMet in the 43S preinitiation complex. The same phenotype has been observed for mutations in subunits of eIF2 (16, 19, 20, 31) and eIF5 (31). Interestingly, Sui1p shows sequence similarity to mammalian eIF1 (34), a poorly characterized factor not known to interact physically with mammalian eIF3 (8). It appears that only a fraction of Sui1p is associated with eIF3 in yeast (43), and it is unclear whether the postulated function of Sui1p in selection of AUG start codons involves the eIF3-associated or the free form of Sui1p.
The p62 RNA-binding protein present in the yeast eIF3 preparations of Naranda et al. (44) was shown to be encoded by GCD10 (23), a gene first identified by recessive mutations that impaired translational repression of GCN4 mRNA (23). It was originally believed that Gcd10p was the yeast counterpart of the p66 subunit of human eIF3 because of their similar molecular weights, immunological cross-reactivity, and strong in vitro RNA-binding activities (5, 23). However, Gcd10p and human p66 do not have strong sequence similarities, and a human protein homologous to Gcd10p can be predicted from expressed sequence tag sequences (1a), suggesting that a Gcd10p homolog is present in humans but is not an integral subunit of eIF3.
The above comparison of yeast and human eIF3 complexes purified by their activities in the Met-puromycin assay suggests numerous differences in subunit composition between the two species. It is particularly noteworthy that the yeast NIP1 and TIF32 products, homologous to human eIF3 subunits p110 and p170, respectively, apparently did not copurify with yeast eIF3 activity in the experiments of Naranda et al. (44). Danaie et al. (17) described a distinct Prt1p-containing complex that contains four polypeptides with relative molecular weights similar to the products of TIF32, NIP1, TIF34, and TIF35 but did not determine the identities of these proteins. This complex could rescue translation initiation and Met-tRNAiMet binding to 40S ribosomes in cell extracts prepared from a prt1 mutant following incubation at the nonpermissive temperature (17). Thus, it seemed possible that yeast contains a functional eIF3 complex containing the five yeast proteins homologous to subunits of human eIF3.
We have tested this hypothesis by purifying a polyhistidine-tagged version of Prt1p (His-Prt1p) by Ni2+ affinity and gel filtration chromatography and identified the polypeptides specifically associated with the tagged protein by mass spectrometry (MS). The results revealed that Prt1p resides in a stable complex containing the other four predicted yeast homologs of human eIF3 subunits and that this complex could rescue translation and binding of Met-tRNAiMet to 40S ribosomes in both prt1 mutant and Nip1p-depleted extracts. Unexpectedly, a substantial fraction of the eIF5 also was physically associated with the eIF3 complex, and we found that eIF5 can interact directly with the NIP1-encoded eIF3 subunit by in vivo and in vitro protein binding assays.
MATERIALS AND METHODS
Plasmids.
A ClaI DNA fragment containing PRT1 was ligated to plasmid pRS316 (51) to yield plasmid pJA100. To construct pLP102, PRT1 was excised from pJA100 by digesting with ClaI and PstI and ligated to pRS315 (51). Plasmid pLP101, encoding Prt1p tagged with eight histidines at its C terminus, was constructed in the following two steps. PCR fusion (60) was used to mutate nucleotide 2172 (A→C) of PRT1 (numbered relative to the translation start site) to create PRT1-salI, which contains a novel SalI site immediately prior to the stop codon and encodes Asp instead of Glu as the last amino acid. The final 879-bp PCR fusion product and plasmid pJA100 were digested with BamHI and PstI and ligated together to form plasmid pLP100. Two complementary oligonucleotides, PRT1HISA (5′-TCGACCACCACCACCACCACCACCACCACTAAAGATCT-3′) and PRT1HISB (5′-TCGAGATCTTTAGTGGTGGTGGTGGTGGTGGTGGTGG-3′), were annealed to form a DNA duplex containing eight consecutive histidine codons, a BglII restriction site (underlined), and SalI sites at each end. The ends of the duplex were phosphorylated with T4 kinase and ligated to plasmid pLP100 digested with SalI and dephosphorylated with calf intestine phosphatase, producing pLP101. The insert was confirmed by digestion with BglII and XhoI and also by PCR using oligonucleotides PRT1HISB and PRT1-E (5′-CGATATGGACTATCCAGG-3′) as primers, the latter complementary to a sequence 553 bp upstream of the insertion site. Henceforth, the allele on pLP101 will be referred to as PRT1-His.
Plasmid pGAD-TIF5, encoding a fusion between eIF5 and the Gal4p activation domain (GAD) used for two-hybrid analysis, was constructed by synthesizing the complete TIF5 open reading frame (ORF) by PCR amplification from chromosomal DNA, using oligonucleotide primers that introduced BamHI and EcoRI sites upstream and downstream from the ORF, respectively. The 1.2-kb PCR product was digested with BamHI and EcoRI and ligated to plasmid pGAD424 (6) to produce pGAD-TIF5. The derivatives of plasmid pGBT9 listed in Fig. 7A encoding fusions to the Gal4p DNA-binding domain (GBD) were described previously (2). Plasmid pGEX-TIF5, encoding a fusion between glutathione S-transferase (GST) and eIF5, was constructed by digesting pGAD-TIF5 with EcoRI and SalI to yield a 1.3-kb DNA fragment containing the TIF5 ORF, which was inserted into pGEX-4T-1 (Pharmacia). pGEX-4T-1 was described previously (52), as was pT7-NIP1 (2). Plasmid pRG166 encoding a polyadenylated luciferase mRNA under control of the T7 promoter was provided by Simon Green, Ribogene Inc.
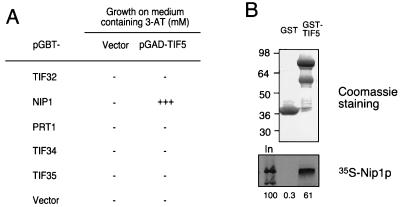
FIG. 7.
Evidence that eIF5 interacts with Nip1p. (A) Analysis of interactions between full-length eIF5 and the five yeast proteins homologous to subunits of eIF3 in the yeast two-hybrid assay. Fusions between GBD and full-length Tif32p, Nip1p, Prt1p, Tif34p, and Tif35p encoded by the plasmids listed in the first column were tested for interactions with a fusion between full-length eIF5 and GAD encoded by pGAD-TIF5 or with GAD alone (Vector). The pGBT9 derivatives and pGBT9 alone were introduced into yeast strain Y190, and the resulting Trp+ Leu− transformants were mated to Trp− Leu+ transformants of strain Y187 containing pGAD-TIF5 or pGAD424 alone. The resulting Trp+ Leu+ diploids were tested for growth on SC medium lacking leucine, tryptophan, and histidine and containing different concentrations of 3-AT. The extent of two-hybrid interaction is indicated by the degree of 3-AT resistance (22, 27). −, no growth at 5 mM 3-AT; +++, growth at 30 mM 3-AT. (B) The GST-eIF5 fusion protein or GST alone was immobilized on glutathione-Sepharose beads and incubated with 35S-labeled Nip1p synthesized by in vitro translation. After extensive washing, proteins bound to the beads were eluted and separated by SDS-PAGE. The gel was stained with Coomassie blue to visualize the eluted GST proteins (top), followed by autoradiography to detect the [35S]Nip1p (bottom). The species of greatest apparent molecular weight (molecular weights are indicated in thousands on the left) visible in the GST-TIF5 lane (top) migrated with the mobility expected for the full-length GST-TIF5 fusion. The less abundant species migrating more rapidly are presumed to be degradation products of the full-length fusion. Lane In in the bottom panel contains 100% of the input amount of 35S-labeled Nip1p used in the binding reactions. Phosphorimaging analysis of the bottom panel showed that 61% of the input amount of 35S-labeled full-length Nip1p bound to GST-TIF5, whereas only 0.3% bound to GST alone.
Yeast strains.
The prt1-1 strain H1676 was constructed by tetrad analysis of a cross involving strain TP11B-4-1 (provided by G. Johnston) and transformed with plasmid pJA100 or pLP101 to yield strain LPY100 or LPY101, respectively. PCR-based gene disruption (59) was used to replace chromosomal PRT1 in LPY101 with the kanMX module encoding kanamycin resistance. Plasmid pFA6 (59) was used as the template for amplifying the KanMX module by PCR using oligonucleotides containing 19 to 22 nucleotides corresponding to the multiple cloning sequences flanking the module and 35 nucleotides corresponding to sequences either immediately upstream of the start codon or downstream of the stop codon of PRT1. The 1.3-kb PCR product was used to transform LPY101 by the lithium acetate method (33) to resistance to G418 (Gibco BRL) at a concentration of 200 μg/ml on YPD (50) plates. The G418-resistant clones were screened for disruption of chromosomal PRT1 by determining their ability to lose the plasmid-borne copy of PRT1 on pLP101 after replica plating colonies to synthetic complete (SC) (50) plates supplemented with 5-fluoro-orotic acid (5-FOA) (1 μg/ml) (10). To eliminate false positives, we showed that the G418-resistant, 5-FOA-sensitive strains thus identified could be cured of plasmid pLP101 on medium containing 5-FOA after introduction of the LEU2 plasmid pLP102 bearing PRT1. One such Ura− Leu+ strain (LPY199) was selected to produce isogenic strains LPY200 and LPY201 bearing PRT1-salI on pLP100 and PRT1-His on pLP101, respectively. Following introduction of pLP100 or pLP101, the resulting Ura+ Leu+ transformants were cultured on minimal medium supplemented with leucine to promote loss of the LEU2 PRT1 plasmid pLP102.
Strains KAY1 (MATα his1-29 gcn2-508 ura3-52 leu2-3,112 tif34Δ-1 <HIS4-lacZ ura3-52> YCpL-TIF34 [TIF34 LEU2]) and KAY8 (MATα his1-29 gcn2-508 ura3-52 leu2-3,112 tif34Δ-1 <HIS4-lacZ ura3-52> YCpL-TIF34 [TIF34-HA LEU2]) were described previously (2), as were Y190 (MATa leu2-3,112 ura3-52 trp1-901 his3-Δ200 ade2-101 gal4Δ gal80Δ URA3::GAL-lacZ LYS::GAL-HIS3) and Y187 (MATα gal4 gal80 his3 trp1-901 ade2-101 ura3-52 leu2-3,112 met URA3::GAL-lacZ) (27). Strains YJA146 (MATα gcd10Δ::hisG ura3-52 trp1 leu2Δ1 his3Δ200 pep::HIS4 prb1Δ1.6 can1 [p1775:IMT4]) and YJA158 (MATα GCD10 ura3-52 trp1 leu2Δ1 his3Δ200 pep::HIS4 prb1Δ1.6 can1 [p1775:IMT4]) were described previously (1), except that YJA158 was listed as a transformant of BJ5464 containing high-copy-number plasmid p1775 bearing the IMT4 gene which encodes tRNAiMet. The construction of strain NIP1KR4R1, containing a fusion between ubiquitin and Nip1p expressed under the control of the GAL10 promoter, from strain Ad (MATa NIP1 ade2 his3 leu2 trp1 ura3 can1 rho+ L-0 M-0) is described elsewhere (24).
Preparation of RSW fractions.
Ribosomal salt wash (RSW) fractions were prepared from strains LPY200 and LPY201 essentially as described previously (17), with some modifications. Cells were grown in 10 liters of YPD medium to an optical density at 600 nm (OD600) of 7.5 to 8.0, harvested by centrifugation at 7,000 × g for 15 min, and washed with ice-cold water. All subsequent steps were performed at 4°C. About 80 g of cells was resuspended in 160 ml of buffer A (20 mM Tris-HCl [pH 7.5], 100 mM KCl, 5 mM MgCl2, 0.1 mM EDTA, 7 mM β-mercaptoethanol, 1 mM phenylmethylsulfonyl fluoride [PMSF], 1× Complete protease inhibitor cocktail [Boehringer Mannheim]) and homogenized in a bead beater with 2 cell volumes of glass beads. The cells were homogenized eight times for 30 s and cooled on ice for 30 s between cycles. The pooled homogenate was clarified by centrifugation at 17,000 × g and then at 25,000 × g for 15 min, and ribosomes were pelleted at 200,000 × g for 2 h. The ribosomal pellet was resuspended in 25 ml of buffer B (buffer A containing 350 mM KCl) and centrifuged at 200,000 × g for 2 h.
Purification of His-Prt1p/eIF5 complex from the RSW fraction by Ni2+-NTA agarose and Superose-6 FPLC chromatography.
The supernatant containing the RSW was dialyzed against 1 liter of binding buffer (20 mM Tris-HCl [pH 7.5], 350 mM KCl, 5 mM MgCl2, 7 mM β-mercaptoethanol, 10% glycerol, 1 mM PMSF, 20 mM imidazole) for 1 h and bound in batch format to Ni2+-nitrilotriacetic acid (NTA) agarose (Qiagen) (0.2 ml of 50% slurry per liter of cells) in a 15-ml conical tube, rocking on a Nutator for 1 h. The Ni2+-NTA agarose was pelleted at 1,000 × g for 5 min and washed with 10 ml of binding buffer three times. After the last wash, the bound protein was eluted in elution buffer (binding buffer containing 250 mM imidazole). The fractions containing Prt1p-His, as determined by immunoblot analysis, were pooled, dialyzed against GF buffer (20 mM Tris-Cl [pH 7.0], 75 mM KCl, 1 mM PMSF, 10% glycerol) and concentrated in a Centricon-10 spin column (Amicon) to a volume of 300 μl and a final protein concentration of 1 to 1.5 μg/μl. An aliquot (200 μl) of the concentrated eluate was injected onto a Superose-6 sizing column of a fast-phase liquid chromatography (FPLC) system (Pharmacia Biotech) and chromatographed in GF buffer at a flow rate of 0.3 ml/min, collecting 0.9-ml fractions. This column separates proteins in the molecular mass range of 5 × 103 to 1 × 106 kDa. Fractions were analyzed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) followed by silver staining using a SilverXpress kit (Novex) or by immunoblot analysis (described below).
MS.
Following SDS-PAGE separation of Superose-6 column fraction 18 (Fig. 1A), the gel was stained with copper stain (Bio-Rad), and protein bands (containing <100 ng of each protein) were excised, destained, and ground to a fine powder. The in-gel digestion with trypsin and extraction of peptides was performed essentially as described previously (48). The dried peptides were redissolved in 10 μl of 50% acetonitrile for MS. A 1.2-m matrix-assisted laser desorption–ionization–time-of-flight mass spectrometer with delayed extraction (Voyager-DE; PerSeptive Biosystems, Framingham, Mass.) was used for mass analysis of the peptides. Data were collected with an internal 500-MHz digital board. The working matrix solution was a twofold dilution of a saturated solution of 2,5-dihydroxybenzoic acid in acetonitrile-water (1:1). Aliquots of 0.5 μl of the peptide mixture and 0.5 μl of the working matrix solution were mixed on the sample plate and air dried prior to MS analysis. Masses were calibrated internally with two peptides derived from trypsin autolysis (m/z 2164.3 and 2274.6). Only the average mass could be measured with better than 1-Da mass accuracy. For oxidation of methionines in the peptides prior to MS, 1 μl of the extracted peptide mixture was mixed with 0.5 μl of 30% H2O2 and dried instantly in a SpeedVac concentrator. The dried sample was redissolved in 1.5 μl of the working matrix solution and subjected to MS again.
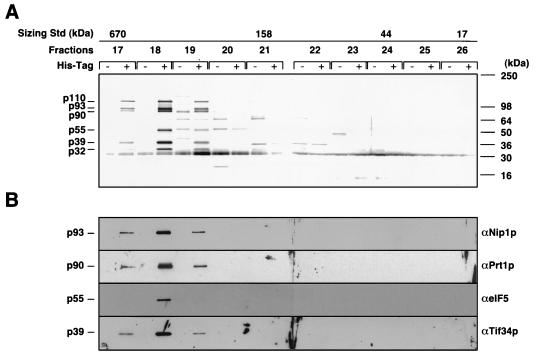
FIG. 1.
Affinity purification of a high-molecular-weight complex containing yeast homologs of mammalian eIF3 subunits and eIF5. Equal amounts of RSW fractions prepared from strains LPY200 and LPY201 containing wild-type Prt1p or His-Prt1p, respectively, were bound to Ni2+-NTA agarose, eluted, and separated on a Pharmacia Superose-6 FPLC column precalibrated with known size standards (Std) (4 to 670 kDa; Bio-Rad). The masses of the standards are shown above the fractions in which they eluted. (A) Aliquots (20 μl) of column fractions (numbered across the top) were separated by SDS-PAGE using 4 to 20% gradient gels, and the proteins were visualized by silver staining. Identical fractions derived from LPY200 and LPY201 were loaded in adjacent lanes (His-Tag − and +, respectively). The molecular masses of SDS-PAGE size standards (Novex) are shown on the right. The six major polypeptides whose amounts peaked in fraction 18 from LPY201 and were absent in the corresponding fraction from LPY200 are indicated on the left. (B) A gel identical to that in panel A was subjected to immunoblot analysis with antibodies against the proteins listed on the right, used at the following dilutions: Nip1p, 1:1,000; Prt1p, 1:1,000; eIF5, 1:2,500; and Tif34p, 1:500.
To identify each protein, the measured masses of tryptic peptides were compared to the calculated tryptic peptide maps of all yeast proteins in the OWL database with a Bayesian algorithm (29), using the ProFound program available on the World Wide Web (http://prowl.rockefeller.edu). Up to three undigested trypsin recognition sites were allowed, and a mass tolerance of ±1 Da was used for the searches. An identification was considered positive only if the top candidate had a probability score of 1 and the second-best candidate had a much lower probability (of 10−4 or less). See the footnotes to Table 1 for additional details.
TABLE 1.
Identification of proteins physically associated with His-Prt1p by mass measurements of tryptic peptides by using MSa
| Gene product identified | Molecular mass (kDa)
|
No. of peptides observed | Probability of correct identificationb
|
Frequency (no. observed/no. predicted) of peptides thatc:
|
Human homolog (% identity with yeast protein)d | |||
|---|---|---|---|---|---|---|---|---|
| Apparente | Calculatedf | Gene product | Next closest candidate gene product | Contain Met | Do not contain Met | |||
| Tif32p | 110 | 110.3 | 32 | 1 | 2.5 × 10−22 | 14/14 | 18/18 | eIF3-p170 (29) |
| Nip1p | 93 | 93.2 | 16 | 1 | 1.0 × 10−4 | 2/2 | 14/14 | eIF3-p110 (31) |
| Prt1p | 90 | 88.1 | 28 | 1 | 3.7 × 10−8 | 9/9 | 19/19 | eIF3-p116 (31) |
| eIF5 | 55 | 45.3 | 28 | 1 | 3.1 × 10−5 | 9/9 | 19/19 | eIF5 (40) |
| Tif34p | 39 | 38.8 | 24 | 1 | 3.7 × 10−8 | 1/1 | 23/23 | eIF3-p36 (46) |
| Tif35p | 30 | 30.5 | 15 | 1 | 1.4 × 10−6 | 1/1 | 14/14 | eIF3-p44 (33) |
A sample of Superose-6 column fraction 18 from strain LPY201 (∼1 μg) and an equivalent sample from LPY200 (Fig. 1A) were separated by SDS-PAGE, and the gel was stained with copper stain (Bio-Rad). Six protein bands (numbered 1 to 6 from the top) were found exclusively in the LPY201-derived sample, and these were excised and digested in gel with trypsin. The tryptic peptides were eluted, and their masses were determined by MS (see Materials and Methods). Gel slices were removed from the equivalent positions in the LPY200 lane and treated identically. The masses of all tryptic peptides identified for each band in the LPY201 lane which were absent in the corresponding LPY200-derived sample were compared to the calculated tryptic peptide maps of all yeast proteins in the OWL database, using the ProFound program (see Materials and Methods). The proteins in gel bands 1 and 3 to 6 were identified as Tif32p, Prt1p, eIF5, Tif34p, and Tif35p; band 2 was found to contain Prt1p and Nip1p, which are similar in molecular weight. The search program identified Prt1p as the top candidate, with a probability score of 1, and Nip1p as the second candidate, with a much lower probability score. After assigning all of the measured peptides derived from Prt1p, we used the masses of the remaining peptides to search the database again, and Nip1p was identified as the top candidate, with a probability score of 1.0.
Calculated by the ProFound program.
To confirm the identifications listed in column 1, oxidation of methionine residues in the extracted peptide mixtures was carried out. The masses of peptides shifted by 16 Da following oxidation were presumed to contain methionine; those peptides whose masses did not shift following oxidation were presumed to lack a methionine residue. On average, 30% of the tryptic peptides contained one or more Met residues. For a set of 20 measured tryptic peptides, if 5 contain methionine and 15 do not, the probability of an incorrect identification is further decreased by a factor of (0.3)5 × (1 − 0.3)15 = 1.1 × 10−5 below that achieved with database searching with the peptide masses alone. Therefore, the chance of false identification for the six polypeptides listed here is negligible.
The percentages of identity between the yeast polypeptides and their presumed homologs in mammalian eIF3 were calculated by using the Bestfit program in the Genetics Computer Group software package (18). Accession numbers (in parentheses) for the mammalian protein sequences are as follows: eIF3-p170 (D50929), eIF3-p110 (U91326), eIF3-p116 (U62583), eIF5 (P55010), and eIF3-p36 (S60335). The human eIF3-p44 sequence was made available by Hershey (29a).
Based on mobility in SDS-PAGE analysis.
Isolation of His-Prt1p and associated proteins from whole-cell extracts by Ni2+ affinity chromatography.
For small-scale purification of His-Prt1p, yeast strains were grown in 50 ml of YPD, and cells were pelleted by centrifugation at 7,000 × g for 15 min, washed once with ice-cold distilled water, and resuspended in 2 cell volumes of breaking buffer (above-described binding buffer with 150 mM KCl). Extract was prepared by adding 2 cell volumes of acid-washed glass beads and breaking cells by vortexing at high speed for 30 s, followed by 30 s on ice, a total of eight times. Extract was clarified by two successive centrifugations at 17,000 × g for 15 min. Total extract was bound in batch format to 75 μl of Ni2+-silica (Qiagen) (30% slurry) for 1 h at 4°C. Protein bound to Ni2+-silica was collected by centrifugation at 6,000 rpm for 2 min and washed four times with 300 μl of breaking buffer containing 20 mM imidazole. Bound protein was eluted in 300 μl of breaking buffer containing 250 mM imidazole. Fractions were collected and analyzed by SDS-PAGE and immunoblotting as described for Fig. 2.
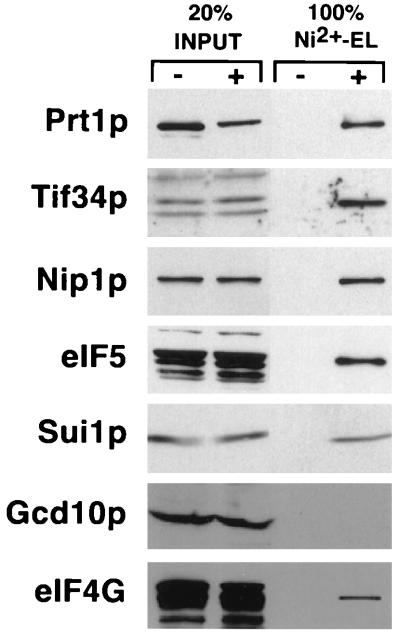
FIG. 2.
Identification of proteins specifically associated with His-Prt1p in whole-cell extracts. Equal amounts of total protein in whole-cell extracts prepared from strains LPY200 and LPY201 containing wild-type Prt1p or His-Prt1p, respectively, were bound to Ni2+-silica, eluted, resolved by SDS-PAGE using 4 to 20% gradient gels, and subjected to immunoblot analysis using antibodies against the proteins listed on the left of each panel, used at the following dilutions: Prt1p, 1:1,000; Tif34p, 1:500; Nip1p, 1:1,000; eIF5, 1:2,500; Sui1p, 1:1,000; Gcd10p, 1:500; eIF4G, 1:1,000; and Gcd6p, 1:1,000. Lanes 1 and 2 contain 20% of the input amounts of whole-cell extract from LPY200 (His-Tag −) and LPY201 (His-Tag +), respectively; lanes 3 and 4 contain the entire Ni2+-silica eluates (Ni2+-EL) from LPY200 and LPY201, respectively.
Coimmunoprecipitation of proteins with HA-Tif34p from whole-cell extracts.
Yeast strains KAY1 and KAY8 were grown in 50 ml of SC medium and harvested at early log phase and whole-cell extracts were prepared and immunoprecipitated with hemagglutinin (HA)-specific mouse monoclonal antibody 12CA5 (Boehringer Mannheim) as previously described (2). Immunoprecipitated complexes were separated on SDS–8 to 16% gradient polyacrylamide gels, transferred to a polyvinylidene membrane, and probed with the antibodies indicated in Fig. 3. Antibodies against eIF4G, Sui1p, eIF5, eIF4E, and Gcd11p were provided by Michael Altmann, Thomas Donahue, Umadas Maitra, John McCarthy, and Ernest Hannig, respectively. Antibodies against Prt1p (15), Gcd6p (12), Gcd10p (23), Tif34p (2), and Nip1p (24) were described previously.
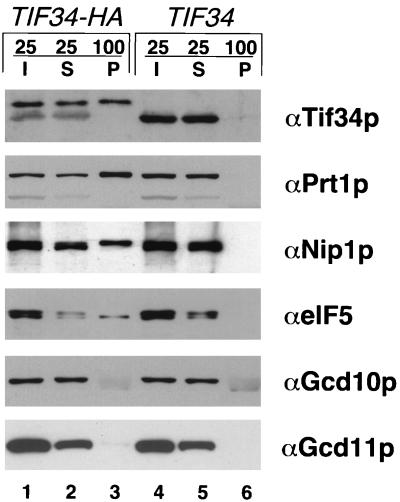
FIG. 3.
Coimmunoprecipitation of Prt1p, Nip1p, and eIF5 with HA-tagged Tif34p. Whole-cell extracts from strains KAY8 (TIF34-HA) and KAY1 (TIF34) were immunoprecipitated with monoclonal antibody 12CA5 against the HA epitope. Aliquots containing 25% of the input whole-cell extracts (I), 25% of the supernatant fractions (S), and the entire immunoprecipitated pellets (P) were separated by SDS-PAGE using 12% gels and subjected to immunoblot analysis using antibodies against the proteins listed to the right of each panel.
In vitro assays of eIF3 activity.
The stimulation of Met-puromycin synthesis was assayed as described (44) except that the empirically determined optimized amounts of HeLa initiation factors used here were somewhat different from those described previously. Whole-cell extracts for assaying in vitro translation of LUC mRNA and 40S binding of Met-tRNAiMet to 40S subunits were prepared from yeast strains grown in YPD medium (50) as previously described (32). For preparing extracts from strains NIP1KR4R1 and Ad, cells were grown in 5 liters of YPG medium (50) (containing galactose as carbon source) to an OD600 of 0.6 to 1.0, harvested by centrifugation, washed once with sterile distilled water, reinoculated into 5 liters of YPD medium (containing dextrose as the carbon source) at an OD600 of 0.2 and grown to an OD600 of 1.5 to 2.0. The final extracts (OD260 = 100) were stored as 200-μl aliquots in liquid nitrogen and thawed on ice before use. Capped luciferase mRNA was transcribed in vitro from plasmid pRG166, after linearization by digestion with SmaI, using a mMessage mMachine kit (Ambion) according to the manufacturer’s instructions. The mRNA was purified by using an RNAeasy spin column (Qiagen), eluted with 30 μl of diethyl pyrocarbonate-treated water, and stored at −70°C. In vitro translation reactions with luciferase mRNA were carried out as described for Fig. 4 by mixing equal volumes of translation extract and 2× translation buffer (40 mM HEPES [pH 7.4], 60 mM potassium acetate, 4 mM magnesium acetate, 1.5 mM ATP, 3 mM dithiothreitol [DTT], 50 mg of creatine phosphate per ml, 333 μg of creatine phosphate kinase per ml [Sigma]) containing 1 mM GTP, 0.1 mM amino acid mix (Promega), 4 μg of luciferase mRNA, and 10 U of RNAsin (Promega) RNase inhibitor. To measure the amounts of luciferase produced, 8-μl aliquots were withdrawn at the times indicated in Fig. 4, diluted with 22 μl of distilled water, and frozen in a dry ice-ethanol bath. Subsequently, the diluted reaction samples were thawed on ice, and 20 μl was mixed with 100 μl of premixed luciferase assay reagents (Promega) and assayed for light production over a 10-s interval, using an automated injection luminometer (Analytical Luminescence Laboratory).
FIG. 4.
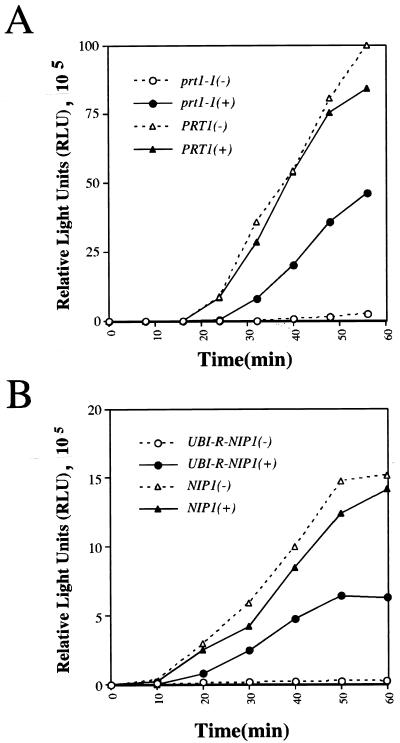
Complementation of a heat-treated prt1-1 mutant or Ubi-Nip1p-depleted extract for translation of exogenous luciferase mRNA. Whole-cell extracts were tested for the ability to translate capped luciferase mRNA. (A) Extracts prepared from isogenic strains H1616 (prt1-1) or LPY200 (PRT1) were incubated at 37°C for 5 min prior to performance of the assay; 35-μl aliquots of extract were mixed with an equal volume of 2× translation buffer containing 1 mM GTP, 4 μg of mRNA, 0.1 mM complete amino acid mix (Promega), and 10 U of RNasin (Promega) RNase inhibitor and then incubated at 26°C. Reactions designated (+) were performed in the presence of ∼4 pmol of purified His-Prt1p complex from Superose-6 column fraction 18 (shown in Fig. 1A) from strain LPY201; reactions designated (−) received an equivalent proportion of column fraction 18 lacking the Prt1p complex from strain LPY200. At the indicated times, 8-μl aliquots were withdrawn and diluted with 22 μl of distilled H2O and frozen in a dry ice-ethanol bath. The amount of luciferase produced (in relative light units [RLU]) in each sample was measured subsequently as described in Materials and Methods. (B) NIP1KR4R1 (UBI-R-NIP1) and its isogenic parent Ad (NIP1) were grown in galactose-containing medium for 17 h and shifted to glucose-containing medium and allowed to double three to four times before harvesting. Extracts were prepared and tested for the ability to translate capped luciferase mRNA as described for panel A.
To measure binding of [3H]Met-tRNAiMet to 40S ribosomes in extracts, 15 μl of translation extract was mixed with 20 μl of 2× translation buffer containing 1.2 mM nonhydrolyzable GTP analog guanylyl-(β,γ-imido)diphosphonate (GMPPNP; Boehringer Mannheim) and 5 μl of [3H]Met-tRNAiMet (0.154 μCi/1.95 pmol/μl), prepared as described previously (19), and incubated at 26°C for 20 min. The reaction was stopped and fixed by adding 4 μl of 3% formaldehyde on ice and resolved by velocity sedimentation through a 12.5-ml 10 to 30% sucrose gradient in a Beckman SW41 rotor at 41,000 rpm for 5 h at 4°C. Fractions of 0.7 ml were collected and filtered through nitrocellulose disks. The disks were washed extensively with wash buffer (20 mM Tris-HCl [pH 7.5], 100 mM KCl, 2 mM MgCl2, 1 mM DTT) to remove unincorporated [3H]Met-tRNAiMet, dried, and counted by liquid scintillation.
Nip1p-eIF5 interaction assays.
The yeast two-hybrid analysis and GST pulldown assays shown in Fig. 7 were carried out as described previously (2). Briefly, for the latter, the GST-TIF5 or GST fusion encoded by pGEX-TIF5 or pGEX-4T-1, respectively, was induced in Escherichia coli, purified from cell extracts by binding to glutathione-Sepharose 4B beads (Pharmacia), and resuspended in 0.75 ml of binding buffer (20 mM HEPES [pH 7.5], 75 mM KCl, 0.1 mM EDTA, 2.5 mM MgCl2, 1% skim milk, 1 mM DTT, 0.05% Nonidet P-40), to which 10 μl of rabbit reticulocyte lysate containing 35S-labeled Nip1p was added. The latter was synthesized by in vitro translation using [35S]methionine and plasmid pT7-NIP1 as the template, using a TnT transcription/translation kit (Promega). Binding was conducted at 4°C for 2 h, followed by washing of the beads five times with 1 ml of phosphate-buffered saline (140 mM NaCl, 2.7 mM KCl, 10.1 mM Na2HPO4, 1.8 mM KH2PO4 [pH 7.3]). Bound proteins were eluted in Laemmli buffer (35) at 95°C for 2 min and separated by SDS-PAGE. Gels were stained with Coomassie blue to visualize the GST fusion proteins, followed by autoradiography or fluorography. The amounts of bound 35S-labeled proteins were quantitated by phosphorimaging analysis using a STORM model 860 (Molecular Dynamics).
RESULTS
Purification of a high-molecular-weight complex containing polyhistidine-tagged Prt1p.
We set out to isolate the S. cerevisiae eIF3 complex by purifying a histidine-tagged form of Prt1p and any proteins stably associated with it, using Ni2+ affinity chromatography. By carrying out the same purification scheme in parallel with extracts from strains expressing His-Prt1p or untagged Prt1p, we could distinguish between proteins specifically associated with His-Prt1p and those that bound nonspecifically to the Ni2+ affinity resin. We found that a prt1Δ strain containing the PRT1-His allele (encoding His-Prt1p) on a single-copy plasmid (LPY201) had a growth rate indistinguishable from that of an isogenic strain containing wild-type PRT1 (LPY200) (data not shown). Therefore, we concluded that addition of the histidine tag did not significantly affect PRT1 function. His-Prt1p and associated proteins were purified from the RSW fraction from strain LPY201 by chromatography on Ni2+-NTA agarose followed by gel filtration chromatography on a Superose-6 column. The same procedure was carried out in parallel for an equivalent amount of RSW prepared from strain LPY200 containing untagged Prt1p. SDS-PAGE and immunoblot analysis of the Superose-6 column fractions showed that Prt1p was detected only in fractions 17 to 19, corresponding to apparent molecular weights of 400,000 to 600,000, and only for the RSW preparation from LPY201 (Fig. 1B, panel αPrt1p, + lanes). These results indicated that a high-molecular-weight complex of ca. 600 kDa containing His-Prt1p could be isolated on Ni2+-NTA resin in a manner completely dependent on the presence of the histidine tag on His-Prt1p.
Silver staining of the Superose-6 peak fractions containing His-Prt1p (fractions 17 to 19) also contained several major polypeptides with apparent molecular masses of 110, 93, 55, 39, and 32 kDa (Fig. 1A, fractions 17 to 19, + lanes). Because these polypeptides were completely absent from the corresponding fractions from LPY200 (− lanes), they were judged to be physically associated with His-Prt1p. In accordance with this conclusion, the abundances of all six proteins varied in similar fashion across fractions 17 to 19, all peaking in fraction 18 (Fig. 1A). There were other minor polypeptides (of 89, 75, 57, 44, 30, 25, and 20 kDa) evident in the silver-stained gel; however, these were judged to be contaminating proteins that bound nonspecifically to the Ni2+ resin because they were present at equal abundance in the LPY201 and LPY200 preparations (Fig. 1A).
Identification of subunits of the His-Prt1p complex by MS.
To identify the five polypeptides that copurified with His-Prt1p, samples of Superose-6 fraction 18 from the LPY201 (tagged) and LPY200 (untagged) preparations were resolved by SDS-PAGE (as shown in Fig. 1A) and each polypeptide in the LPY201 fraction was subjected to in-gel trypsin digestion and MS to determine the mass spectrum of its tryptic peptides. As a negative control, we analyzed gel slices from the LPY200 fraction at positions in the gel corresponding to the six specific protein bands present in the LPY201 lane. By comparing the observed mass spectra of tryptic peptides for each band to the predicted spectra of all proteins encoded in the yeast genome, we identified the p110, p93, p90, p39, and p32 polypeptides in the LPY201 fraction as the products of TIF32, NIP1, PRT1, TIF34, and TIF35, the five yeast homologs of human eIF3 subunits (Table 1). Interestingly, the p55 subunit was identified as eIF5, encoded by TIF5. To confirm these assignments, the tryptic digests were treated with hydrogen peroxide to oxidize the methionine side chains and analyzed again by MS. As shown in Table 1, all of the tryptic peptides assigned to each of the six identified proteins showed the predicted change in mass expected from oxidation of methionine side chains.
We also confirmed the identification of Nip1p, Tif34p, and eIF5 as proteins specifically associated with His-Prt1p by immunoblot analysis of the column fractions shown in Fig. 1A with polyclonal antibodies against these three proteins. As shown in Fig. 1B, antibodies against these proteins reacted with polypeptides of the expected molecular weights that were distributed across fractions 17 to 19 in a pattern identical to that seen for His-Prt1p and for the corresponding polypeptides visualized by silver staining (Fig. 1A). (Antibodies were not available for the products of TIF32 and TIF35.) From the findings in Fig. 1 and Table 1, we concluded that Tif32p, Nip1p, Prt1p, Tif34p, Tif35p, and eIF5 are components of a high-molecular-mass complex of ca. 600 kDa.
Based on the intensity of silver staining of the polypeptides in fraction 18 of Fig. 1A, we estimated that Tif32p, Nip1p, Prt1p, eIF5, and Tif35p were recovered in approximately equimolar amounts. Tif34p appeared to be present at relatively higher levels, suggesting that two molecules of Tif34p may be present in each molecule of the His-Prt1p complex. It should be noted that in some preparations, the relative amount of eIF5 in the complex was lower than that shown in Fig. 1A, perhaps indicating that eIF5 is less tightly bound to the complex than are the other five polypeptides.
Substoichiometric interactions of Sui1p and eIF4G with the Prt1p complex.
We did not detect Sui1p or Gcd10p in the complex of proteins isolated in association with His-Prt1p (Fig. 1A and Table 1), whereas both proteins were present in the eIF3 complex purified by stimulation of Met-puromycin synthesis (23, 43). It is possible that these two proteins are physically associated with the His-Prt1p complex in cell extracts but dissociated from it during preparation of the RSW, which involves a high-ionic-strength buffer, or during subsequent purification steps. We wished to address this possibility and also to investigate whether the eIF4G component of eIF4F was physically associated with the His-Prt1p complex, as mammalian eIF4G was shown to bind eIF3 (36, 38). Toward these ends, we incubated whole-cell extracts from strain LPY201 (PRT1-His) or LPY200 (PRT1) directly with Ni2+-silica resin, washed the resin with buffer containing 20 mM imidazole, and eluted the proteins in buffer containing 250 mM imidazole. With this minimum purification procedure, we hoped to minimize loss of proteins associated with His-Prt1p due to high-ionic-strength buffers or proteolysis. Analysis of the eluates by SDS-PAGE and immunoblotting showed that the LPY201 eluate contained proportions of the input amounts of Nip1p, eIF5, and Tif34p that were comparable to the proportion of His-Prt1p recovered in the eluate (ca. 25%), whereas none of these proteins was detected in the eluate from LPY200 (Fig. 2). These results indicate that the association between the Prt1p complex and eIF5 shown in Fig. 1 was not an artifact of the high-salt buffer used to prepare the RSW. A considerable fraction of Sui1p and a small fraction of eIF4G were also recovered in association with His-Prt1p (Fig. 2). By comparing the recoveries of Sui1p and eIF4G with that of His-Prt1p itself (see the legend to Fig. 2), we calculated that 20% of the Sui1p in the cell extracts was recovered with the His-Prt1p complex, similar to the estimate of 30% of Sui1p that copurified with yeast eIF3 activity (43). However, only 5% of the total eIF4G was recovered with the His-Prt1p complex. We conclude that a considerable fraction of Sui1p is physically associated with the His-Prt1p complex in whole-cell extracts but dissociates from the complex during the purification scheme used in Fig. 1.
No detectable Gcd10p was recovered with His-Prt1p (Fig. 2), suggesting that Gcd10p does not stably interact with the His-Prt1p complex. As expected, the ɛ subunit of eIF2B (Gcd6p) did not bind to the Ni2+-silica resin (Fig. 2), providing evidence for the specificity of the protein-protein interactions detected by this assay. Because eIF4E and Pab1p are believed to interact with eIF4G (38, 53), we also probed the Ni2+-NTA eluates depicted in Fig. 2 for these two proteins. No Pab1p was detected in either LPY200 or LPY201 eluates, whereas the same amounts of eIF4E were found in both eluates (data not shown). The latter presumably results from nonspecific binding of eIF4E to the Ni2+ resin, making it impossible to determine whether the small proportion of eIF4G associated with the His-Prt1p complex is bound to eIF4E.
Coimmunoprecipitation of Prt1p, Nip1p, and eIF5 with epitope-tagged Tif34p.
We carried out coimmunoprecipitation experiments to examine physical interactions between the subunits of the Prt1p complex by an independent approach. For these studies, we used a pair of isogenic strains in which the TIF34 chromosomal gene had been deleted and replaced with plasmid-borne wild-type TIF34 (strain KAY1) or a TIF34-HA allele encoding an HA epitope-tagged form of Tif34p (KAY8). We showed previously that the TIF34-HA and TIF34 alleles were indistinguishable in complementing the lethality of a chromosomal tif34Δ mutation (2). Whole-cell extracts from the two strains were immunoprecipitated with monoclonal anti-HA antibodies, and the resulting immune complexes were resolved by SDS-PAGE and probed by immunoblot analysis. We found that Prt1p, HA-Tif34p, Nip1p, and eIF5 were coimmunoprecipitated with anti-HA antibodies from the TIF34HA extract but not from the wild-type TIF34 extract (Fig. 3, TIF34-HA versus TIF34, P lanes), confirming that these proteins were stably associated with one another in whole-cell extracts. By comparing the relative amounts of these proteins in the pellet and supernatant fractions, it appeared that the efficiency of coimmunoprecipitation with Tif34p was somewhat higher for Prt1p than for Nip1p and eIF5. (A considerable fraction of the eIF5 seems to have been degraded during incubation with anti-HA antibody.) In contrast to these results, we did not detect any Gcd10p coimmunoprecipitating with HA-Tif34p, providing further evidence that Gcd10p is not stably associated with subunits of eIF3. The γ subunit of eIF2 (Gcd11p) also did not coimmunoprecipitate with HA-Tif34p (Fig. 3), establishing the specificity of the interaction between Prt1p, Nip1p, and eIF5 with HA-Tif34p in the cell extracts.
Biochemical activities of the purified Prt1p complex.
The His-Prt1p complex that we isolated (Fig. 1) appeared to have only two or three polypeptides with apparent molecular masses in common with subunits of the yeast complex purified using the Met-puromycin synthesis assay (44), namely, Prt1p, Tif34p, and possibly the 30-kDa Tif35p protein. Therefore, it was of interest to determine whether our complex could substitute for human eIF3 in this assay, stimulating Met-puromycin synthesis by an 80S initiation complex in the presence of human eIF1A, eIF2, eIF5, eIF5A, and ribosomal subunits. Under conditions where purified human eIF3 stimulated Met-puromycin synthesis 3.0- to 3.5-fold, we consistently found that the Superose-6 fraction 18 from strain LPY201 (Fig. 1) stimulated synthesis by a factor of 1.5 to 2.0. Although this stimulation was modest, none whatsoever was given by the corresponding fraction derived from strain LPY200 (PRT1) that was assayed as a negative control. Thus, the stimulatory activity appeared to be specific for the His-Prt1p complex.
To provide an alternative demonstration of the biochemical activity of the purified His-Prt1p complex, we examined whether it could complement a prt1-1 mutant extract for defects in translation initiation. In the first experiment, we assayed translation of a recombinant mRNA encoding luciferase that was synthesized in vitro and added to whole-cell extracts competent for in vitro translation. In agreement with previous observations (17, 21), brief incubation of the prt1-1 mutant extract at 37°C abolished translation of the luciferase mRNA. Addition of an aliquot of purified His-Prt1p complex from LPY201 restored high-level translation in the heated extract, whereas addition of the corresponding control sample from strain LPY200 had no stimulatory effect (Fig. 4A). In contrast, preincubation at 37°C of an extract prepared from an isogenic PRT1 strain did not reduce translation of the luciferase mRNA, and addition of the purified His-Prt1p complex had no stimulatory effect on the translational activity of this extract (Fig. 4A). Thus, the purified His-Prt1p complex rescued the translation of luciferase mRNA in a heat-inactivated prt1-1 extract.
To provide evidence that Nip1p is required for translation, we also analyzed luciferase mRNA translation in extracts depleted of Nip1p. For this we used strain NIP1KR4R1, which lacks chromosomal NIP1 and expresses a ubiquitin-Nip1p (Ubi-Nip1p) fusion under the control of a galactose-inducible promoter. Removal of the ubiquitin moiety at the N terminus of the fusion protein by a deubiquitinating enzyme is expected to generate a Nip1p polypeptide containing Arg at the N terminus, making it susceptible to ubiquitin-dependent degradation through the N-terminal recognition pathway (45). Under inducing growth conditions with galactose as the carbon source, strain NIP1KR4R1 containing the UBI-NIP1 allele had a growth rate equivalent to that of NIP1 parental strain Ad, showing that the essential function of Nip1p was supplied by the Ubi-Nip1p fusion. When shifted to noninducing medium containing glucose as the carbon source, the cells grew slowly over a period of ca. 20 h with a doubling time fourfold greater than that of NIP1 strain Ad. Immunoblot analysis of an extract from strain NIP1KR4R1 after 17 h in glucose-containing medium revealed no detectable Nip1p, whereas Nip1p was readily detected in a comparable extract prepared from strain Ad (24). Thus, expression of the Ubi-Nip1p fusion was repressed and the preexisting fusion protein was degraded in cells grown on glucose-containing medium. In contrast, the steady-state levels of eIF5, the eIF3 subunits of Prt1p and Tif34p, and Sui1p were all essentially unaffected by depletion of Nip1p in strain NIP1KR4R1 (24). As shown in Fig. 4B, an extract from NIP1KR4R1 depleted of Ubi-Nip1p was completely defective for translation of luciferase mRNA in vitro, and translation was stimulated more than 30-fold by addition of the purified His-Prt1 complex containing wild-type Nip1p. Addition of the His-Prt1 complex had no stimulatory effect on translation in the wild-type NIP1 extract. Thus, the NIP1-encoded subunit of the His-Prt1p complex is required for translation in vitro. These data support recent findings that depletion of Ubi-Nip1p leads to an inhibition of translation in vivo at the initiation step (24).
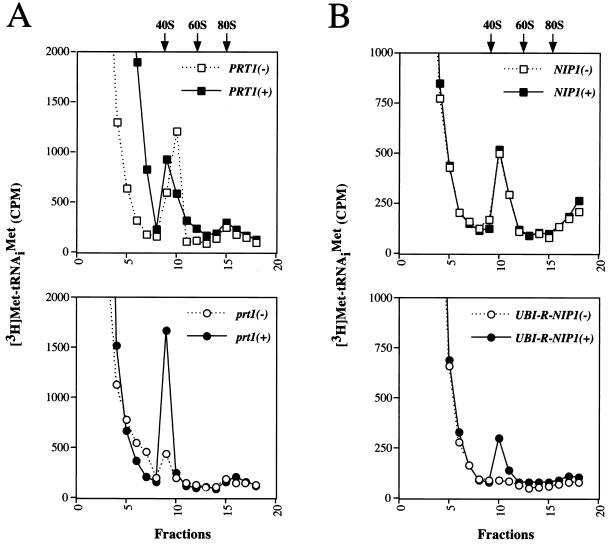
It was shown previously that prt1-1 extracts are temperature sensitive for binding of Met-tRNAiMet to 40S ribosomes (17, 21), a step in translation initiation known to be stimulated by eIF3. We sought to determine whether the purified His-Prt1p complex could rescue this defect in the prt1-1 extract described above. tRNAiMet was acylated in vitro with [3H]methionine and added to the heat-treated prt1-1 and PRT1 extracts, with or without an aliquot of purified His-Prt1p complex. A nonhydrolyzable GTP analog (GMPPNP) was included to inhibit the conversion of 48S to 80S initiation complexes by preventing eIF5-catalyzed GTP hydrolysis on eIF2–GTP–Met-tRNAiMet ternary complexes bound to 40S ribosomes (54). As shown in Fig. 5A, the amount of [3H]Met-tRNAiMet that cosedimented with 40S ribosomes was consistently higher in the reaction containing prt1-1 extract supplemented with purified His-Prt1p complex compared to the control supplement. In contrast, addition of the purified His-Prt1p complex to the PRT1 extract had no stimulatory effect on the relatively high level of [3H]Met-tRNAiMet binding to 40S ribosomes in these extracts. We conclude that the purified His-Prt1p complex restored binding of tRNAiMet to 40S ribosomes in the heat-inactivated prt1-1 mutant extract. The degree of stimulation of 40S ribosome binding by tRNAiMet seen in these experiments is comparable to that described for the Prt1p complex purified previously from yeast (17) and by eIF3 isolated from both mammalian (46, 57) and plant (14) sources.
FIG. 5.
Complementation of a heat-treated prt1-1 mutant and Ubi-Nip1p-depleted extracts for [3H]Met-tRNAiMet binding to 40S ribosomal subunits. The binding of [3H]Met-tRNAiMet to 40S ribosomal subunits was assayed by mixing 15 μl of extract with an equal volume of 2× translation buffer containing ∼10 pmol of [3H]Met-tRNAiMet (0.77 μCi) and 2.4 mM nonhydrolyzable GTP analog GMPPNP. As for Fig. 4, duplicate reactions were carried out for each extract containing an aliquot of fraction 18 (Fig. 1A) from strain LPY201 containing ∼2 pmol of purified His-Prt1p complex (+) or an equivalent proportion of fraction 18 from LPY200 lacking Prt1p complex (−). The reactions were incubated at 26°C for 20 min, fixed by addition of formaldehyde to 0.3%, and resolved by velocity sedimentation on 10 to 30% sucrose gradients by centrifugation at 41,000 rpm for 5 h in an SW41 rotor. Fractions (0.7 ml) were collected starting at the top of the gradient and analyzed by filter assay and counted for [3H]Met-tRNAiMet by liquid scintillation (dashed line). The positions of the 40S and 60S ribosomal subunits and 80S ribosomes were determined by monitoring the OD254 while collecting fractions from the gradient. (A) Extracts prepared from isogenic strains H1616 (prt1-1) (bottom) and LPY200 (PRT1) (top) were incubated at 37°C for 5 min prior to performance of the assay. (B) Extracts prepared from isogenic strains Ad (NIP1) (top panel) and NIP1KR4R1 (UBI-RNIP1) (bottom) grown under conditions described for Fig. 4B to deplete Ubi-Nip1p from the NIP1KR4R1 extract.
Interestingly, the Ubi-Nip1p-depleted extract prepared from strain NIP1KR4R1 also was defective for [3H]Met-tRNAiMet binding to 40S ribosomes, and this activity was stimulated threefold by addition of the purified His-Prt1p complex (Fig. 5B). In the experiments shown in Fig. 4 and 5, the samples of purified His-Prt1p complex used to restore translation and ribosome binding by Met-tRNAiMet contained quantities of Prt1p and the other subunits of the complex which were comparable to the amounts of these proteins present in the extracts. Thus, the fact that purified His-Prt1p complex did not restore translation and Met-tRNAiMet binding in the Ubi-Nip1p-depleted extract to wild-type levels is probably not due to insufficient amounts of the His-Prt1p complex in the reconstituted extract. Rather, there may be reduced amounts or activities of one or more additional factors in the Ubi-Nip1p-depleted extract which cannot be complemented by addition of His-Prt1p complex. Alternatively, the function of the purified His-Prt1p complex in the Ubi-Nip1p-depleted extract could be hindered by the binding to 40S subunits of inactive eIF3 complexes lacking Nip1p. Nonetheless, the strong stimulation given by the His-Prt1p complex indicates a direct role for Nip1p in translation initiation at the step of Met-tRNAiMet binding to 40S ribosomes.
Gcd10p is not required for Met-tRNAiMet binding to 40S ribosomes in vitro.
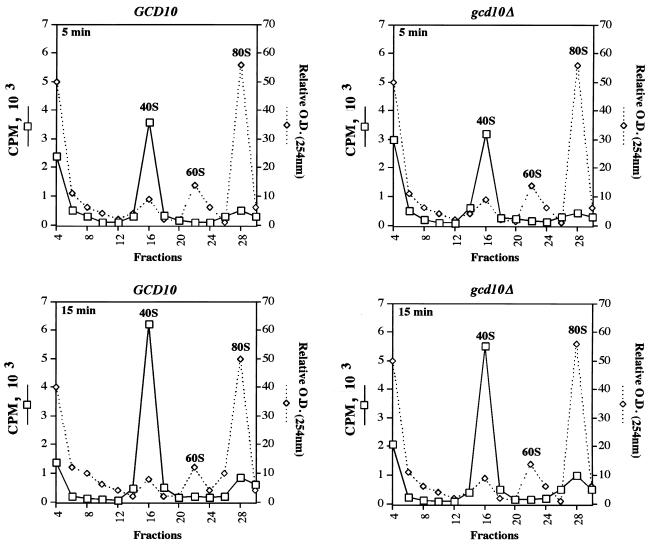
We previously reported that Gcd10p is physically associated with purified eIF3 (23) and suggested that gcd10 mutations affect GCN4 translation by impairing a function of eIF3 involved in the binding of eIF2–GTP–Met- tRNAiMet ternary complexes to 40S ribosomes. To test this possibility, we measured Met-tRNAiMet binding to 40S ribosomes in extracts lacking Gcd10p by exploiting our recent finding that GCD10 can be deleted from strains overexpressing tRNAiMet (1). As shown in Fig. 6, the amount of [3H]Met-tRNAiMetadded to the extracts that sedimented with 40S subunits in the GCD10 extract increased by 60% between 5 and 15 min of incubation, indicating that binding of [3H]Met- tRNAiMet to 40S ribosomes increased at a nearly constant rate over this time course. The results in Fig. 6 also revealed that [3H]Met-tRNAiMet binding to 40S ribosomes was only slightly less in the gcd10Δ extract than in the GCD10 extract, suggesting that Gcd10p does not play a critical role in this step of translation initiation.
FIG. 6.
Efficient Met-tRNAiMet binding to 40S ribosomal subunits in the absence of Gcd10p. Whole-cell extracts were prepared from isogenic strains YJA146 (gcd10Δ) and YJA158 (GCD10), and binding of [3H]Met-tRNAiMet to 40S ribosomal subunits was assayed by mixing 100 μl of extract with an equal volume of 2× translation buffer containing ∼10 pmol of [3H]Met-tRNAiMet (80 pCi/pmol) and 2.4 mM nonhydrolyzable GTP analog GMPPNP and then incubating the mixture for 5 or 15 min at 26°C. Reactions were stopped and resolved by velocity sedimentation essentially as described for Fig. 5 except that 0.4-ml fractions were collected from the gradients.
Interactions between eIF5 and the NIP1-encoded subunit of eIF3.
The results presented above indicated that a substantial fraction of the eIF5 in cell extracts was specifically associated with the His-Prt1p complex. To confirm this finding and identify a specific subunit of the Prt1p complex that interacts with eIF5, we tested each of the five identified components of the complex for interactions with eIF5, using the yeast two-hybrid system. Fusions between GBD and full-length Tif32p, Nip1p, Prt1p, Tif34p, or Tif35p were coexpressed with a fusion between full-length eIF5 and GAD in a yeast strain containing multiple copies of the Gal4p binding site located upstream from the HIS3 gene. Interaction between GBD and GAD fusions in this strain stimulates HIS3 transcription and confers resistance to 3-aminotriazole (3-AT), an inhibitor of His3p. As shown in Fig. 7A, among the five GBD fusions tested, only the GBD-Nip1p fusion (encoded by pGBT9-NIP1) interacted with the GAD-eIF5 fusion (encoded by pGAD-TIF5). (At least for GBD-Tif34p and GBD-Tif35p, we observed two-hybrid interactions between these fusions and GAD fusions to other subunits of the Prt1p complex [2].)
We verified that Nip1p and eIF5 can form a stable complex in vitro in the absence of other yeast proteins by examining interactions between a fusion of eIF5 to bacterial GST protein (GST-TIF5) expressed in E. coli and 35S-labeled Nip1p protein synthesized by in vitro translation. As shown in Fig. 7B, ≈60% of the input amount of [35S]Nip1p was recovered with GST-TIF5 on glutathione-agarose beads, whereas less than 1% of [35S]Nip1p was recovered with GST alone. (In similar experiments, we found that in vitro-translated [35S]Prt1p, [35S]Tif34p, and [35S]Tif35p did not bind to GST-TIF5 above the background level of binding to GST alone, whereas these in vitro-translated proteins showed high-level binding to GST-Prt1p, GST-Tif34p, or GST-Tif35p fusion protein [2].) These results provide evidence that the Nip1p subunit provides a binding site for eIF5 in the Prt1p complex.
DISCUSSION
Affinity purification of an eIF3 core complex conserved between yeast and mammals.
We have purified a high-molecular-weight complex by using Ni2+-NTA affinity chromatography directed against a histidine-tagged form of Prt1p and identified five polypeptides that are physically associated with His-Prt1p. Four of these, together with Prt1p, encompass the five polypeptides encoded in S. cerevisiae with strong sequence similarities to mammalian eIF3 subunits; the fifth was eIF5. All six subunits were identified by MS of tryptic digests of the individual polypeptides resolved by SDS-PAGE. In the case of Nip1p, Tif34p, and eIF5, these assignments were confirmed by immunoblot analysis using the antibodies available against these three proteins. The same conclusion was reached by coimmunoprecipitation experiments for Nip1p, Tif34p, Prt1p, and eIF5, using a yeast strain expressing an HA epitope-tagged form of Tif34p (Fig. 3). We recently found that epitope-tagged Tif35p and a protein with the molecular weight of Tif32p also could be coimmunoprecipitated specifically with HA-Tif34p (2). Pairwise interactions among Prt1p, Tif34p, and Tif35p (2, 58), between Prt1p and Tif32p (2), and between Nip1p and Tif32p (2) have been demonstrated by using the two-hybrid assay or in vitro binding experiments with recombinant proteins. Moreover, we and others (58) found that temperature-sensitive mutations in TIF34 can be suppressed in vivo by multiple copies of TIF35, and we showed that the suppressible tif34 mutations weaken the interaction between Tif34p and Tif35p in vitro (2). The results in Fig. 7 indicate that eIF5 can interact with Nip1p in two-hybrid and in vitro binding assays. Thus, by several independent approaches, we demonstrated that the five yeast homologs of human eIF3 subunits, plus eIF5, comprise a heteromeric complex that can be detected in crude extracts and withstands the high-salt conditions (0.35 M KCl) used to prepare the RSW.
The results of experiments shown in Fig. 2 and 3, in which the eIF3 complex was isolated from whole-cell extracts by affinity chromatography (directed against His-Prt1p) or immunoprecipitation (directed against HA-Tif34p), suggested that the proportion of total eIF5 in the extract recovered with the tagged eIF3 subunit was somewhat smaller than that seen for the other subunits of the complex. This may indicate that a significant fraction of eIF5 is not associated with eIF3 in vivo, existing as an individual polypeptide. It also appeared that eIF5 was present in substoichiometric amounts in most preparations of the highly purified eIF3 complex (e.g., in Fig. 1A, p55). In addition, we found recently that eIF5 dissociated from the core eIF3 subunits in 0.45 M KCl, whereas Nip1p, Prt1p, and Tif34p (the other three subunits that we could detect by immunoblotting) remained associated in up to 1.0 M KCl (data not shown). These results suggest that eIF5 is less stably associated with the eIF3 complex than are the other five subunits identified here. Accordingly, we propose that Tif32p, Nip1p, Prt1p, Tif34p, and Tif35p comprise a highly stable core eIF3 complex which exhibits a strong, but salt-sensitive, interaction with eIF5. It remains to be seen whether eIF5 is physically associated with eIF3 in mammalian systems. In view of the salt sensitivity of the yeast eIF3-eIF5 interaction, detecting an interaction with eIF5 may require purification of mammalian eIF3 by affinity and size exclusion chromatography in relatively low salt buffers. As the concentration of eIF5 appears to be much lower than that of eIF3 in mammalian cells (8), it is possible that the almost stoichiometric interaction we detected between eIF3 and eIF5 is unique to the yeast system.
When we tested our purified complex for the ability to substitute for human eIF3 in the Met-puromycin synthesis assay, we consistently observed a 1.5- to 2.0-fold stimulation over the eIF3-independent level of Met-puromycin synthesis. This stimulation was smaller than the threefold reported for the eight-subunit yeast complex purified by Naranda et al. (44) using similar amounts of the two complexes. It is possible that the Sui1p (p16), Gcd10p (p62), p135, or p21 polypeptide present in the eIF3 preparation of Naranda et al. was responsible for its greater activity in this assay. An important role for Sui1p has been suggested from the observation that eIF3 activity in the RSW fraction of a temperature-sensitive sui1 mutant grown at the nonpermissive temperature did not stimulate Met-puromycin synthesis (43), although an indirect effect of the mutation on one of the eIF3 subunits was not ruled out. More experimentation is required to resolve this question, particularly since the mammalian counterpart of Sui1p, eIF1, does not seem to be required for eIF3 activity in the Met-puromycin assay (5, 11).
The other assays of yeast eIF3 activity described in the literature involve complementing a heat-inactivated prt1-1 mutant extract for defects in translation of exogenously added mRNA (which requires initiation) and efficient binding of Met- tRNAiMet to 40S ribosomes (17, 21). Our purified His-Prt1p complex was active in both assays. All of the His-Prt1p protein was found in the high-molecular-weight Prt1p-eIF5 complex (Fig. 1), and there is no evidence that Prt1p functions independently of the eIF3 complex. In fact, it was reported that depletion of the Tif34p subunit in vivo led to the disappearance of Prt1p, suggesting that Prt1p is unstable outside of the eIF3 complex (42). Consequently, rescue of translation and Met-tRNAiMet binding to 40S ribosomes in the heat-inactivated prt1-1 extract most likely resulted from addition of a functional Prt1p complex and not the Prt1p subunit alone. Supporting this conclusion, we recently found that monomeric His-At1p purified from a strain overexpressing only this protein failed to rescue translation of luciferase mRNA in the prt1-1 extract (data not shown). We also showed that Ubi-Nip1p-depleted extracts were defective for translation and Met-tRNAiMet binding to 40S ribosomes and that both activities were partially restored by addition of purified His-Prt1p complex (Fig. 4 and 5). The latter results provided biochemical evidence that the NIP1-encoded subunit of the His-Prt1p complex is required for efficient Met-tRNAiMet binding to 40S ribosomes. It remains to be determined whether Prt1p or Nip1p functions directly, or whether their inactivation or removal from the complex alters the conformation of another subunit which acts more directly in this reaction. As the stimulation of Met-tRNAiMet binding to 40S ribosomes is a well-established activity of mammalian eIF3, our findings suggest that Tif32p, Nip1p, Prt1p, Tif34p, and Tif35p comprise an eIF3 core complex conserved between yeast and mammals.
The SDS-PAGE profile of the complex shown in Fig. 1A qualitatively resembles that of the Prt1p-containing complex purified previously (17) and also that reported for eIF3 purified from wheat germ (37). Summing the molecular masses of the six polypeptides in the His-Prt1p complex (assuming two molecules of Tif34p per complex) yields a molecular mass of ∼450 kDa. Although the molecular mass estimate obtained from gel filtration chromatography is about 600 kDa (Fig. 1), electron microscopic analysis of mammalian eIF3 revealed a flat triangular prism shape (7), which would increase its apparent molecular weight in gel filtration experiments.
We found that Sui1p was associated with His-Prt1p when it was affinity purified on Ni2+-silica resin from whole-cell extracts (Fig. 2) but was absent when the His-Prt1p/eIF5 complex was purified from the RSW fraction by Ni2+-NTA agarose and Superose-6 chromatography. This finding suggests that Sui1p may be associated with the His-Prt1p/eIF5 complex less tightly than are the five core eIF3 subunits and eIF5. We could not detect an association between Gcd10p and the His-Prt1p/eIF5 complex in whole-cell extracts by binding to Ni2+-silica (Fig. 2) or by coimmunoprecipitation with HA-tagged Tif34p (Fig. 3). The effect of gcd10 mutations on GCN4 mRNA translation suggests that Gcd10p is required for ternary complex formation or for binding of the ternary complex to 40S subunits, both of which involve eIF3 (9, 26, 56). We detected no requirement for Gcd10p in either reaction by measuring the rate at which exogenously labeled Met-tRNAiMet bound to 40S ribosomes in extracts containing or lacking Gcd10p. Recent genetic and biochemical experiments indicate that Gcd10p is required for maturation and accumulation of tRNAiMet (1). As this function would be required for ternary complex formation in vivo (but not in our in vitro assays), these findings provide an alternative explanation for the known phenotypes of gcd10 mutants. Combining these results with the absence of the putative Gcd10p homolog in purified human eIF3 leads us to suggest that Gcd10p either is not a subunit of eIF3 or readily dissociates from the rest of the complex in cell extracts.
The six-subunit eIF3/eIF5 complex purified here is functional for stimulating Met-tRNAiMet binding to 40S ribosomes in cell extracts and in stimulating Met-puromycin synthesis in conjunction with mammalian initiation factors. Determining whether it can perform the other known functions ascribed to mammalian eIF3, including 40S-60S subunit dissociation and stimulating mRNA binding to 40S ribosomes, must await the development of protocols for assaying these individual reactions with purified yeast ribosomes and initiation factors. Sui1p, p135, p21, or Gcd10p might contribute to one of these other proposed functions of yeast eIF3. It is also conceivable that Sui1p, p135, or p21 would stimulate the Met-tRNAiMet binding activity of the core eIF3 complex defined here. Until these questions can be addressed experimentally, we cannot state the complete subunit composition of yeast eIF3.
Possible role of the Nip1p subunit in binding of eIF5 and Sui1p to eIF3.
The identification of yeast Nip1p and its human homolog as subunits of eIF3 complexes in these organisms was unexpected since yeast Nip1p was first identified genetically as a factor involved in nuclear import (25) and was not detected in previous yeast eIF3 preparations (44). However, we and others (24) have obtained evidence that Nip1p is associated with 40S subunits in cell extracts and is required in translation initiation at the step of Met-tRNAiMet binding to 40S ribosomes, a demonstrated activity of eIF3. We recently demonstrated that Nip1p interacts with Sui1p in yeast two-hybrid and in vitro binding assays with recombinant proteins (2). This observation suggests that Nip1p could mediate physical interaction between the eIF3 complex and Sui1p, observed previously (43) and confirmed here (Fig. 2). Considering that Nip1p also interacted with eIF5 in vitro and in the two-hybrid assay (Fig. 7), it could be proposed that Nip1p functions in stabilizing physical association between eIF3 and both eIF5 and Sui1p.
eIF5 catalyzes the hydrolysis of GTP in the eIF2–GTP–Met-tRNAiMet ternary complex, causing release of eIF2-GDP and eIF3 from the 40S ribosome, followed by joining of the 60S subunit to form an 80S initiation complex (9, 13, 47, 49, 55). Mutations in eIF5 which allow increased utilization of UUG triplets as the initiation codon (Sui− phenotype) increase the specific activity of eIF5 in stimulating GTP hydrolysis on eIF2. Sui− alleles have also been identified in SUI1 (61) and the genes encoding subunits of eIF2 (16, 19, 20, 31). A Sui− mutation in eIF2γ appeared to cause increased dissociation of tRNAiMet from ternary complexes independently of GTP hydrolysis, whereas Sui− mutations in eIF2β conferred a higher rate of GTP hydrolysis on eIF2 independent of eIF5 (31). These findings have prompted the idea that accurate recognition of AUG as the start codon is dependent on the functions of eIF2, eIF5, and Sui1p and is coupled to the rate of GTP hydrolysis on eIF2 in ternary complexes bound to the 43S preinitiation complex (31).
The biochemical role of Sui1p in determining the fidelity of AUG recognition is unknown; however, the fact that eIF5 and Sui1p have been linked at this step in initiation is intriguing in view of our finding that eIF5 and a fraction of Sui1p are associated with eIF3 and that both proteins interact directly with Nip1p. Binding of eIF3 to the 40S ribosome is thought to precede the binding of the ternary complex or eIF5 (39). Thus, an interesting possibility is that eIF5 and Sui1p are recruited to the 43S preinitiation complex by docking with the Nip1p subunit of eIF3 bound to the 40S ribosome. This physical interaction might also be involved in release of eIF3 from the 40S ribosome subunit upon hydrolysis of GTP and dissociation of the ternary complex catalyzed by eIF5.
ACKNOWLEDGMENTS
We are indebted to Scot Kimball for the gift of labeled initiator tRNA, Tom Donahue for antibodies against Sui1p, Umadas Maitra for antibodies against eIF5, Michael Altmann for antibodies against eIF4G, John McCarthy for antibodies against eIF4E, Simon Green for the luciferase construct, John Hershey for communicating results prior to publication, and Bobbie Felix for help in preparation of the manuscript.
This study was supported in part by grant BE-104 from the American Cancer Society to David Goldfarb.
REFERENCES
- 1.Anderson, J., M. Pak, L. Phan, R. Cuesta, K. Asano, M. Tamame, and A. G. Hinnebusch. A nuclear complex containing Gcd10p and Gcd14p controls translation by promoting maturation of initiator methionyl-tRNA. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 1a.Asano, K., J. Anderson, and A. G. Hinnebusch. Unpublished data.
- 2.Asano, K., L. Phan, J. Anderson, and A. G. Hinnebusch. Complex formation by all five homologues of mammalian translation initiation factor 3 subunits from yeast Saccharomyces cerevisiae. J. Biol. Chem., in press. [DOI] [PubMed]
- 3.Asano K, Kinzy T G, Merrick W C, Hershey J W B. Conservation and diversity of eukaryotic translation initiation factor eIF3. J Biol Chem. 1997;272:1101–1109. doi: 10.1074/jbc.272.2.1101. [DOI] [PubMed] [Google Scholar]
- 4.Asano K, Merrick W C, Hershey J W B. The translation initiation factor eIF3-p48 subunit is encoded by int-6, a site of frequent integration by the mouse mammary tumor virus genome. J Biol Chem. 1997;272:23477–23480. doi: 10.1074/jbc.272.38.23477. [DOI] [PubMed] [Google Scholar]
- 5.Asano K, Vornlocher H-P, Richter-Cook N J, Merrick W C, Hinnebusch A G, Hershey J W B. Structure of cDNAs encoding human eukaryotic initiation factor 3 subunits: possible roles in RNA binding and macromolecular assembly. J Biol Chem. 1997;272:27042–27052. doi: 10.1074/jbc.272.43.27042. [DOI] [PubMed] [Google Scholar]
- 6.Bartel P L, Chien C T, Stemglanz R, Fields S. Using the two-hybrid system to detect protein-protein interactions. In: Hartley D A, editor. Cellular interactions in development: a practical approach. Oxford, England: Oxford University Press; 1993. pp. 153–179. [Google Scholar]
- 7.Behlke J, Bommer U A, Lutsch G, Henske A, Bielka H. Structure of initiation factor eIF-3 from rat liver. Hydrodynamic and electron microscopic investigations. Eur J Biochem. 1986;157:523–530. doi: 10.1111/j.1432-1033.1986.tb09698.x. [DOI] [PubMed] [Google Scholar]
- 8.Benne R, Brown-Luedi M L, Hershey J W B. Protein synthesis initiation factors from rabbit reticulocytes: purification, characterization, and radiochemical labeling. Methods Enzymol. 1979;60:15–35. doi: 10.1016/s0076-6879(79)60005-8. [DOI] [PubMed] [Google Scholar]
- 9.Benne R, Hershey J W B. The mechanism of action of protein synthesis initiation factors from rabbit reticulocytes. J Biol Chem. 1978;253:3078–3087. [PubMed] [Google Scholar]
- 10.Boeke J D, LaCroute F, Fink G R. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197:345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- 11.Brown-Luedi M L, Meyer L J, Milburn S C, Yau P M P, Corbett S, Hershey J W B. Protein synthesis initiation factors from human HeLa cells and rabbit reticulocytes are similar: comparison of protein structure, activities, and immunochemical properties. Biochemistry. 1982;21:4202–4206. doi: 10.1021/bi00261a002. [DOI] [PubMed] [Google Scholar]
- 12.Bushman J L, Foiani M, Cigan A M, Paddon C J, Hinnebusch A G. Guanine nucleotide exchange factor for eIF-2 in yeast: genetic and biochemical analysis of interactions between essential subunits GCD2, GCD6, and GCD7 and regulatory subunit GCN3. Mol Cell Biol. 1993;13:4618–4631. doi: 10.1128/mcb.13.8.4618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chakrabarti A, Maitra U. Function of eukaryotic initiation factor 5 in the formation of an 80 S ribosomal polypeptide chain initiation complex. J Biol Chem. 1991;21:14039–14045. [PubMed] [Google Scholar]
- 14.Checkley J W, Cooley L, Ravel J M. Characterization of initiation factor eIF-3 from wheat germ. J Biol Chem. 1981;256:1582–1586. [PubMed] [Google Scholar]
- 15.Cigan A M, Foiani M, Hannig E M, Hinnebusch A G. Complex formation by positive and negative translational regulators of GCN4. Mol Cell Biol. 1991;11:3217–3228. doi: 10.1128/mcb.11.6.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cigan A M, Pabich E K, Feng L, Donahue T F. Yeast translation initiation suppressor sui2 encodes the alpha subunit of eukaryotic initiation factor 2 and shares identity with the human alpha subunit. Proc Natl Acad Sci USA. 1989;86:2784–2788. doi: 10.1073/pnas.86.8.2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Danaie P, Wittmer B, Altmann M, Trachsel H. Isolation of a protein complex containing translation initiation factor Prt1 from Saccharomyces cerevisiae. J Biol Chem. 1995;270:4288–4292. doi: 10.1074/jbc.270.9.4288. [DOI] [PubMed] [Google Scholar]
- 18.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Donahue T F, Cigan A M, Pabich E K, Castilho-Valavicius B. Mutations at a Zn(II) finger motif in the yeast eIF-2α gene alter ribosomal start-site selection during the scanning process. Cell. 1988;54:621–632. doi: 10.1016/s0092-8674(88)80006-0. [DOI] [PubMed] [Google Scholar]
- 20.Dorris D R, Erickson F L, Hannig E M. Mutations in GCD11, the structural gene for eIF-2γ in yeast, alter translational regulation of GCN4 and the selection of the start site for protein synthesis. EMBO J. 1995;14:2239–2249. doi: 10.1002/j.1460-2075.1995.tb07218.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feinberg B, McLaughlin C S, Moldave K. Analysis of temperature-sensitive mutant ts187 of Saccharomyces cerevisiae altered in a component required for the initiation of protein synthesis. J Biol Chem. 1982;257:10846–10851. [PubMed] [Google Scholar]
- 22.Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 23.Garcia-Barrio M T, Naranda T, Cuesta R, Hinnebusch A G, Hershey J W B, Tamame M. GCD10, a translational repressor of GCN4, is the RNA-binding subunit of eukaryotic translation initiation factor-3. Genes Dev. 1995;9:1781–1796. doi: 10.1101/gad.9.14.1781. [DOI] [PubMed] [Google Scholar]
- 24.Greenberg, J. R., L. Phan, Z. Gu, A. deSilva, C. Apolito, F. Sherman, A. G. Hinnebusch, and D. S. Goldfarb. Nip1p associates with 40S ribosomes and the Prt1p subunit of eIF3 and is required for efficient translation initiation. J. Biol. Chem., in press. [DOI] [PubMed]
- 25.Gu Z, Moerschell R P, Sherman F, Goldfarb D S. NIP1, a gene required for nuclear transport in yeast. Proc Natl Acad Sci USA. 1992;89:10355–10359. doi: 10.1073/pnas.89.21.10355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gupta N K, Roy A L, Nag M K, Kinzy T G, MacMillan S, Hileman R F, Dever T E, Wu S, Merrick W C, Hershey J W B. New insights into an old problem: ternary complex (Met-tRNAieIF-2-GTP) formation in animal cells. In: McCarthy J E G, Tuite M F, editors. Post-transcriptional control of gene expression. H49. Berlin, Germany: Springer-Verlag; 1990. pp. 521–526. [Google Scholar]
- 27.Harper J W, Adami G R, Wei N, Keyomarsi K, Elledge S J. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 28.Hartwell L H, McLaughlin C S. A mutant of yeast apparently defective in the initiation of protein synthesis. Proc Natl Acad Sci USA. 1969;62:468–474. doi: 10.1073/pnas.62.2.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Henzel W J, Billeci T M, Stults J T, Wong S C, Grimley C, Watanabe C. Identifying proteins from two-dimensional gels by molecular mass searching of peptide fragments in protein sequence databases. Proc Natl Acad Sci USA. 1993;90:5011–5015. doi: 10.1073/pnas.90.11.5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29a.Hershey, J. W. B. Personal communication.
- 30.Hershey J W B, Asano K, Naranda T, Vornlocher H P, Hanachi P, Merrick W C. Conservation and diversity in the structure of translation initiation factor eIF3 from humans and yeast. Biochimie. 1996;78:903–907. doi: 10.1016/s0300-9084(97)86711-9. [DOI] [PubMed] [Google Scholar]
- 31.Huang H, Yoon H, Hannig E M, Donahue T F. GTP hydrolysis controls stringent selection of the AUG start codon during translation initiation in Saccharomyces cerevisiae. Genes Dev. 1997;11:2396–2413. doi: 10.1101/gad.11.18.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hussain I, Leibowitz M J. Translation of homologous and heterologous messenger RNAs in a yeast cell-free system. Gene. 1986;46:13–23. doi: 10.1016/0378-1119(86)90162-9. [DOI] [PubMed] [Google Scholar]
- 33.Ito H, Fukada Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kasperaitis M A, Voorma H O, Thomas A A. The amino acid sequence of eukaryotic translation initiation factor 1 and its similarity to yeast initiation factor SUI1. FEBS Lett. 1995;365:47–50. doi: 10.1016/0014-5793(95)00427-b. [DOI] [PubMed] [Google Scholar]
- 35.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 36.Lamphear B J, Kirchweger R, Skern T, Rhoads R E. Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornaviral proteases. J Biol Chem. 1995;270:21975–21983. doi: 10.1074/jbc.270.37.21975. [DOI] [PubMed] [Google Scholar]
- 37.Lauer S J, Burks E A, Ravel J M. Characterization of initiation factor 3 from wheat germ. 1. Effects of proteolysis on activity and subunit composition. Biochemistry. 1985;24:2924–2928. doi: 10.1021/bi00333a016. [DOI] [PubMed] [Google Scholar]
- 38.Mader S, Lee H, Pause A, Sonenberg N. The translation initiation factor eIF-4E binds to a common motif shared by the translation factor eIF-4γ and the translational repressors 4E-binding proteins. Mol Cell Biol. 1995;15:4990–4997. doi: 10.1128/mcb.15.9.4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Merrick W C, Hershey J W B. The pathway and mechanism of eukaryotic protein synthesis. In: Hershey J W B, Matthews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 31–69. [Google Scholar]
- 40.Methot N, Rom E, Olsen H, Sonenberg N. The human homologue of the yeast Prt1 protein is an integral part of the eukaryotic initiation factor 3 complex and interacts with p170. J Biol Chem. 1997;272:1110–1116. doi: 10.1074/jbc.272.2.1110. [DOI] [PubMed] [Google Scholar]
- 41.Methot N, Song M S, Sonenberg N. A region rich in aspartic acid, arginine, tyrosine, and glycine (DRYFG) mediates eukaryotic initiation factor 4B (eIF4B) self-association and interaction with eIF3. Mol Cell Biol. 1996;16:5328–5334. doi: 10.1128/mcb.16.10.5328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Naranda T, Kainuma M, McMillan S E, Hershey J W B. The 39-kilodalton subunit of eukaryotic translation initiation factor 3 is essential for the complex’s integrity and for cell viability in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:145–153. doi: 10.1128/mcb.17.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Naranda T, MacMillan S E, Donahue T F, Hershey J W. SUI1/p16 is required for the activity of eukaryotic translation initiation factor 3 in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:2307–2313. doi: 10.1128/mcb.16.5.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Naranda T, MacMillan S E, Hershey J W B. Purified yeast translational initiation factor eIF-3 is an RNA-binding protein complex that contains the PRT1 protein. J Biol Chem. 1994;269:32286–32292. [PubMed] [Google Scholar]
- 45.Park E-C, Finley D, Szostak J W. A strategy for the generation of conditional mutations by protein destabilization. Proc Natl Acad Sci USA. 1992;89:1249–1252. doi: 10.1073/pnas.89.4.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peterson D T, Merrick W C, Safer B. Binding and release of radiolabeled eukaryotic initiation factors 2 and 3 during 80 S initiation complex formation. J Biol Chem. 1979;254:2509–2519. [PubMed] [Google Scholar]
- 47.Peterson D T, Safer B, Merrick W C. Role of eukaryotic initiation factor 5 in the formation of 80S initiation complexes. J Biol Chem. 1979;254:7730–7735. [PubMed] [Google Scholar]
- 48.Qin J, Fenyo D, Zhao Y, Hall W W, Chao D M, Wilson C J, Young R A, Chait B T. A strategy for rapid, high-confidence protein identification. Anal Chem. 1997;69:3995–4001. doi: 10.1021/ac970488v. [DOI] [PubMed] [Google Scholar]
- 49.Raychaudhuri P, Chaudhuri A, Maitra U. Eukaryotic initiation factor 5 from calf liver is a single polypeptide chain protein of Mr = 62,000. J Biol Chem. 1985;260:2132–2139. [PubMed] [Google Scholar]
- 50.Sherman F, Fink G R, Lawrence C W. Methods of yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1974. [Google Scholar]
- 51.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith D B, Johnson K S. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- 53.Tarun S Z, Sachs A B. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J. 1996;15:7168–7177. [PMC free article] [PubMed] [Google Scholar]
- 54.Tarun S Z, Sachs A B. A common function for mRNA 5′ and 3′ ends in translation initiation in yeast. Genes Dev. 1995;9:2997–3007. doi: 10.1101/gad.9.23.2997. [DOI] [PubMed] [Google Scholar]
- 55.Trachsel H, Staehelin T. Binding and release of eukaryotic initiation factor eIF-2 and GTP during protein synthesis initiation. Proc Natl Acad Sci USA. 1978;75:204–208. doi: 10.1073/pnas.75.1.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Trachsel H, Staehelin T. Initiation of mammalian protein synthesis. The multiple functions of the initiation factor eIF-3. Biochim Biophys Acta. 1979;565:305–315. doi: 10.1016/0005-2787(79)90207-7. [DOI] [PubMed] [Google Scholar]
- 57.Trachsel H, Staehelin T. Initiation of mammalian protein synthesis: the multiple functions of the initiation factor eIF-3. Biochim Biophys Acta. 1979;565:305–314. doi: 10.1016/0005-2787(79)90207-7. [DOI] [PubMed] [Google Scholar]
- 58.Verlhac M-H, Chen R-H, Hanachi P, Hershey J W B, Derynck R. Identification of partners of TIF34, a component of the yeast eIF3 complex, required for cell proliferation and translation initiation. EMBO J. 1997;16:6812–6822. doi: 10.1093/emboj/16.22.6812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 60.Yon J, Fried M. Precise gene fusion by PCR. Nucleic Acids Res. 1988;17:4895. doi: 10.1093/nar/17.12.4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yoon H J, Donahue T F. The sui1 suppressor locus in Saccharomyces cerevisiae encodes a translation factor that functions during tRNAiMet recognition of the start codon. Mol Cell Biol. 1992;12:248–260. doi: 10.1128/mcb.12.1.248. [DOI] [PMC free article] [PubMed] [Google Scholar]