Summary
Chronic kidney disease (CKD) is a major public health burden, with dietary acid load (DAL) and gut microbiota playing crucial roles. As DAL can affect the host metabolome, potentially via the gut microbiota, we cross-sectionally investigated the interplay between DAL, host metabolome, gut microbiota, and early-stage CKD (TwinsUK, n = 1,453). DAL was positively associated with CKD stage G1-G2 (Beta (95% confidence interval) = 0.34 (0.007; 0.7), p = 0.046). After adjusting for covariates and multiple testing, we identified 15 serum, 14 urine, 8 stool, and 7 saliva metabolites, primarily lipids and amino acids, associated with both DAL and CKD progression. Of these, 8 serum, 2 urine, and one stool metabolites were found to mediate the DAL-CKD association. Furthermore, the stool metabolite 5-methylhexanoate (i7:0) correlated with 26 gut microbial species. Our findings emphasize the gut microbiota’s therapeutic potential in countering DAL’s impact on CKD through the host metabolome. Interventional and longitudinal studies are needed to establish causality.
Subject areas: Health sciences, Medicine, Medical specialty, Internal medicine, Nephrology, Natural sciences, Biological sciences, Systems biology, Metabolomics
Graphical abstract
Highlights
-
•
Multi-biological sample metabolites associate with dietary acid load and kidney function
-
•
Multiple associated metabolites mediated between DAL and mild kidney function decline
-
•
The stool metabolite, 5-methylhexanoate(i7:0), correlated with several gut microbial species
Health sciences; Medicine; Medical specialty; Internal medicine; Nephrology; Natural sciences; Biological sciences; Systems biology; Metabolomics
Introduction
Chronic kidney disease (CKD) is a major cause of morbidity and mortality, with incident rates increasing globally.1 Multiple modifiable risk factors for CKD development have been identified, including diet and gut microbiota composition and function.2,3 Recently, there has been an evidence-based shift toward manipulating dietary acid load (DAL), when managing CKD progression.4 DAL is defined as the difference (in mEq H+/day) between endogenously produced acid and base, originating from diet.4 Modern Western diets generally contain large amounts of acid-forming protein, processed foods, and limited amounts of base-producing fruits and vegetables, thereby inducing a state of chronic metabolic acidosis.5 An almost linear relationship exists between acidosis and poorer clinical outcomes in individuals with CKD, including CKD progression and mortality.6
The mechanisms by which DAL contributes to CKD are complex and poorly understood but have been reported to lead to vascular dysfunction and increased peripheral insulin resistance.7,8,9
Indeed, a systematic review and meta-analysis of 31 observational studies reported that a higher DAL is associated with increased systolic and diastolic blood pressure.10 Increased DAL calculated via potential renal acid load (PRAL) methods was also associated with impaired fasting glucose and increased levels of HbA1c in this study.
However, the underlying metabolic pathways resulting in this dysregulation remain unclear. Metabolomics, a high-throughput technology that provides a snapshot of an individual’s metabolic profile at a particular time point, can be used to provide insights into these pathways and has been used successfully to identify novel markers of CKD risk.11,12 Recently, Tariq and colleagues (2022) identified circulating levels of the amino acid N-methylproline to be inversely associated with both DAL and CKD (stage G3-G4), suggesting a protective effect of diet.13
Nevertheless, the effect of DAL on the host metabolome of individuals with early-stage CKD remains largely unexplored. Moreover, the role of the gut microbiome in this relationship is unknown. It is paramount to identify molecular pathways and gut microbiota signatures regulating the DAL-CKD association, as this could lead to effective strategies for mitigating kidney function decline.
Therefore, the aims of this large population-based study were to (i) explore the relationship between DAL and early-stage CKD (stage G1-G2), (ii) investigate the biological pathways underlying this association using metabolomics profiling from four biological samples (serum, urine, stool, and saliva) with mediation analyses to estimate effects, and (iii) assess the interaction between DAL and gut microbiota composition in kidney function decline.
Results
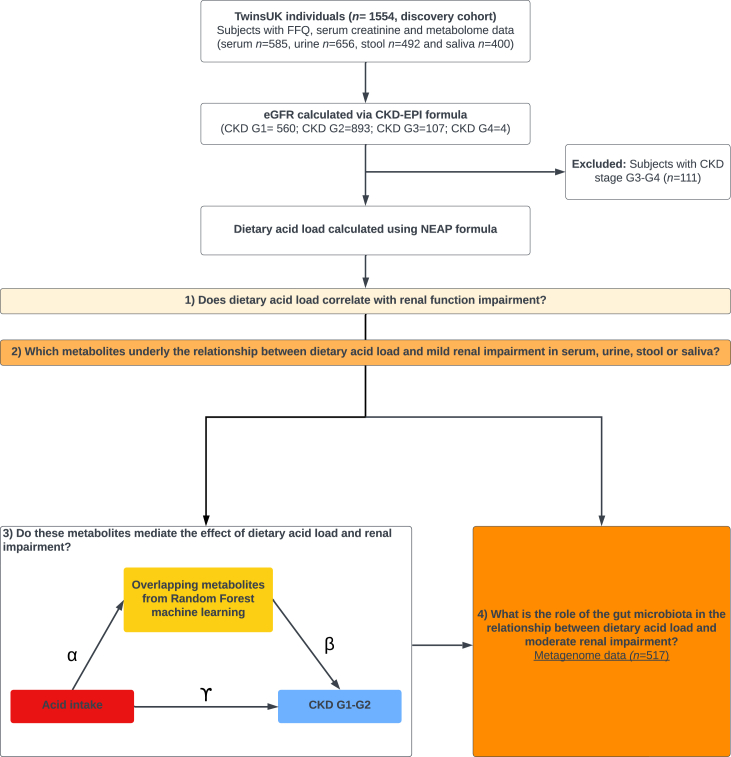
The descriptive characteristics of the study population are presented in Table 1. This study included 1,453 individuals from TwinsUK with metabolomics profiling available in serum (fasting), urine (spot), stool, and saliva, as well as estimated glomerular filtration rate (eGFR) and dietary data available. Briefly, the predominantly female (89.7%) sample included 560 individuals with CKD stage G1 and 893 CKD stage G2, and on average they were 61.3 (±12) years of age with an average body mass index (BMI) of 25.9 (±4.7) kg/m2. A flowchart of the study design is presented in Figure 1.
Table 1.
Demographic characteristics of the study population overall and by CKD status
| Overall (n = 1453) |
CKD G1 (n = 560) |
CKD G2 (n = 893) |
||||
|---|---|---|---|---|---|---|
| n | % | n | % | n | % | |
| Females/males | 1,303/150 | 503/57 | 800/93 | |||
| CKD stage G1 | 560 | 38.5 | 560 | 100 | ||
| CKD stage G2 | 893 | 61.5 | 893 | 100 | ||
| Mean | SD | Mean | SD | Mean | SD | |
|---|---|---|---|---|---|---|
| Age (years) | 61.3 | 12.0 | 55.8 | 12.7 | 64.7 | 10.2 |
| Energy (kcal/day) | 1,864 | 556.2 | 1,842 | 554.7 | 1,878 | 557.0 |
| BMI (kg/m2) | 25.9 | 4.74 | 24.93 | 4.49 | 26.5 | 4.79 |
| Dietary acid (mEq H+/day) | 51.7 | 9.5 | 51.8 | 9.8 | 51.7 | 9.3 |
Figure 1.
Flow chart of the study design
High DAL is associated with renal function decline
Linear mixed models adjusting for age, sex, BMI, and family relatedness identified a positive association between DAL and CKD (β = 0.337 (confidence interval [CI] 0.007; 0.7), p = 0.046). Results were consistent when exploring eGFR as a continuous variable (β = −1.45 (CI −2.89; −0.0047), p = 0.049).
DAL and renal function exhibit a coordinated signature on the serum, urine, and stool metabolome
To explore the serum, urine, stool, and saliva metabolite profiles of individuals in CKD stage G1 and G2 in relation to their DAL, we performed principal-component analysis and Permutational multivariate analysis of variance (PERMANOVA). We found that the metabolome profile of all biological samples, except for saliva, clustered differently according to their DAL on a global scale (p < 0.05; Figure S1).
We then performed random forest (RF) models on the residuals-adjusted metabolites and identified 41 metabolites in serum that overlap between CKD and DAL, of which 15 passed FDR correction, 41 in urine, of which 14 passed FDR correction, 27 in stool, of which 8 passed FDR correction, and 22 in saliva, of which 7 passed FDR correction (Figure 2; Table S6).
Figure 2.
Metabolites associated with CKD and acid intake
Metabolites identified through random forest machine learning that pass multivariable regression analysis (p < 0.05, FDR corrected [Benjamini & Hochberg]) and are associated with CKD (red) and acid intake (blue) in (A) serum, (B) urine, (C) stool, and (D) saliva. All analyses were corrected for age, sex, and BMI.
Effect of DAL on renal function is mediated through serum, urine, and stool metabolites
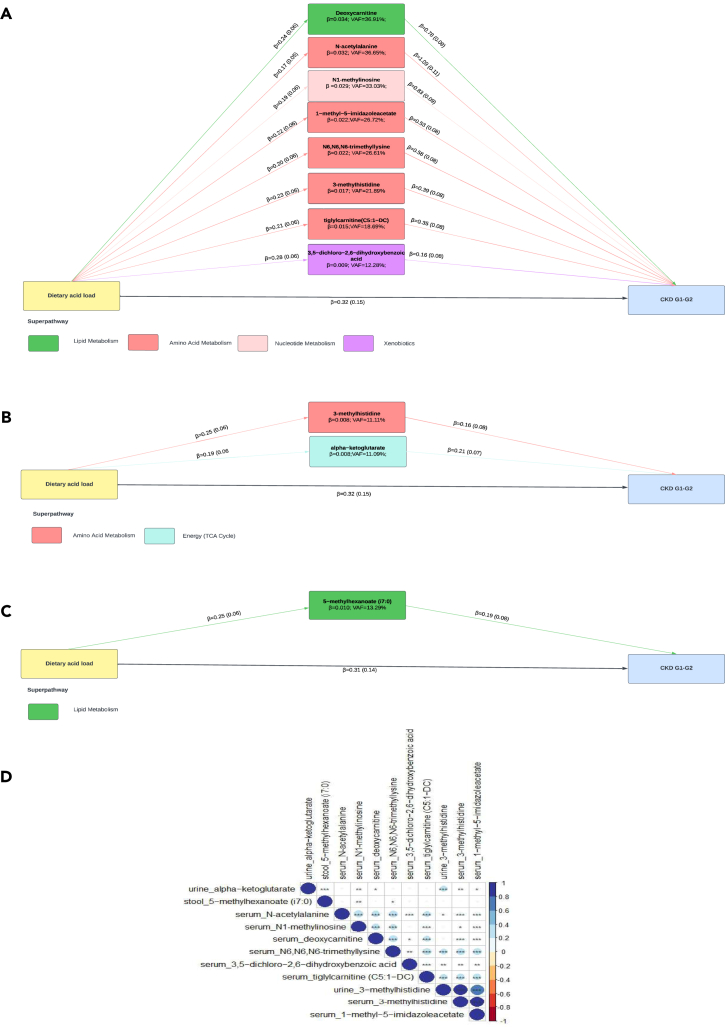
We next tested if the overlapping metabolites associated with both DAL and early-stage CKD potentially mediated the association between DAL and CKD stage G1-G2 progression through mediation analysis, correcting for confounding factors such as age, sex, and BMI. We tested both the DAL→metabolite of interest→CKD stage G1-G2 as well as the DAL→CKD stage G1-G2→ metabolite of interest order (Figures 3 and S2).
Figure 3.
Mediation analysis between DAL and CKD stage G1-G2
Mediation analysis of the association between DAL and CKD stage G1-G2, using metabolite of interest as potential mediator in (A) serum,(B) urine, and (C) stool. Path coefficients are illustrated beside each path and variance accounted for (VAF) score is denoted below the mediator. All associations are statistically significant (p < 0.05).
(D) Spearman correlation between mediating metabolites across biological samples (∗p < 0.05; ∗∗p < 0.01 and ∗∗∗p < 0.001). All analyses are corrected for age, sex, and BMI. Abbreviations: DAL, dietary acid load; CKD, chronic kidney disease; VAF, variance accounted for.
All 8 positively associated metabolites in serum mediated the effect of DAL and stage G1-G2 progression, and the variance accounted for (VAF) ranged between 12.28% (3,5-dichloro-2,6-dihydrpxybenzoic acid) to 36.91% (deoxycarnitine). Interestingly, 3-methlyhistidine was the only mediating metabolite which was found in both serum (VAF 21.89%) and urine (VAF 11.11% [Figure 3A]).
In urine, only 2/7 positively associated metabolites mediated the effect with VAF ranging from 11.09% (alpha-ketoglutorate) to 11.11% (3-methylhistidine [3-MH]) (Figure 3B). In stool, only 1/3 metabolite, 5-methylhexanoate (i7:0), mediated the association between DAL and CKD stage G1-G2 progression with a VAF of 13.29% (Figure 3C). We did not observe evidence for mediation of the 3 saliva metabolites, which corresponds with the results of the principal-component analysis (Figure S1).
When considering CKD stage G1-G2 as a potential mediator in the relationship between DAL→ metabolite of interest, we found that 7/8 serum metabolites, 1 urine metabolite, and 1 stool metabolite were mediated by CKD stage (Figure S2). The VAF was smaller in this model compared to the model using metabolites of interest as potential mediators (Figure 3). The VAF had a range from 8.38% (tiglylcarnitine (C5:1−DC)) to 18.88% (N-acetylalanine) in serum metabolites, 5.94% in urine alpha-ketoglutorate, and 4.14% in stool 5-methylhexanoate (i7:0). CKD stage G1-G2 did not affect saliva metabolites. A step-by-step report of the mediation models used is reported in Tables S2–S5.
The major super pathways involved with metabolites that mediated the association between DAL→CKD stage G1-G2 were mainly amino acid (n = 6) and lipid metabolism (n = 2) (Table S7) across all biological samples. Spearman’s correlation showed that the metabolites strongly correlate with each other, even across biological fluids (Figure 3D).
DAL and the gut microbiota
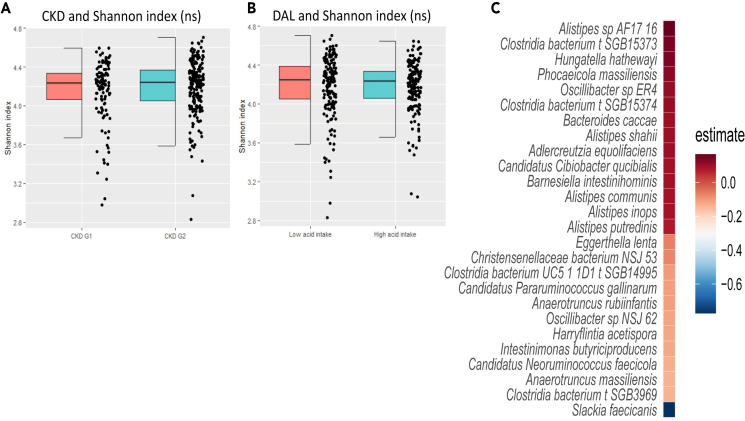
Next, we explored if the metabolites mediating the DAL-CKD relation were associated with gut microbiota composition (alpha diversity, represented as Shannon index) and microbial species (Figure 4). We found that the gut microbiota could predict 13% of the variance in stool abundances of the metabolite, 5-methylhexanoate (i7:0), and we detected 26 bacterial species that were significantly associated with it. Out of these 3 were belonging to different Clostridia bacterium species genomic bins (SGBs).
Figure 4.
Gut microbiota, CKD and DAL
Effect of (A) CKD and (B) DAL on Shannon alpha diversity (ns = non-significant) and (C) gut microbiota species identified to significantly (p < 0.05; FDR corrected [Benjamini & Hochberg]) associate with the stool metabolite 5-methylhexanoate (i7:0) in the discovery cohort. All analyses are corrected for age, sex, and BMI.
Furthermore, we found no association between the mediating serum and urine metabolites and gut microbiota composition.
Discussion
In this large cross-sectional study integrating serum, urine, stool, and saliva metabolites, we report, for the first time, that the effect of early-stage kidney function declines (CKD stage G1-G2) due to DAL being possibly mediated by serum, urine, and stool, but not by saliva metabolites. The metabolites mediating the association are predominantly related to amino acid (n = 6) and lipid (n = 2) metabolism and strongly correlate with each other, even across biological sample types. Moreover, RF machine learning models identified that 13% of the variance of the stool metabolite, 5-methylhexanoate (i7:0) that potentially mediates the DAL-CKD association, could be explained via the gut microbiota composition. However, it is important to note that longitudinal, as well as interventional studies, are warranted to further confirm the potential mediating effects of the found metabolites.
This study also revealed multiple microbial species linked with the mediating stool metabolite, thereby providing potential for novel treatment options.
Our finding that DAL contributes to CKD progression supports previous reports and meta-analysis.14,15 The underlying pathophysiology of this relationship is complex and poorly understood, with the metabolome likely to play an important role.11
To date, only one study has investigated the role of the serum metabolome in the DAL-CKD relationship.13 In this study performed by Tariq et al. (2022), the authors identified 12 metabolites to be inversely associated with DAL, of which only one (N-methylproline) was also inversely associated with incident CKD. Our study identified, through machine learning models, 3 serum metabolites to be inversely associated with both DAL and CKD, and 8 positively associated. Our study confirms that indolepropionate and tartronate (hydroxymalonate) are negatively associated with DAL, as previously reported,16 highlighting the robustness of our result. However, we also found these metabolites to be negatively associated with CKD. The contrast in our findings likely originates from the fact that Tariq et al. (2022) studied these associations using a population with overt CKD (stage 2–4) that also used medications known to affect the serum metabolome, such as antihypertensive drugs,17 as well as different metabolomic platforms to those used in our study.
Moreover, our study identified 8 serum, 2 urine, and 1 stool metabolite to potentially mediate the positive association between DAL and CKD stage G1-G2. The majority of the mediating metabolites identified play a role in amino acid metabolism, which is to be expected as DAL is largely driven by protein intake.4,18 Interestingly, one metabolite, 3-MH, was identified as a mediating metabolite in both serum and urine. 3-MH is a product of histidine metabolism and produced after actin and myosin degradation.19 3-MH has been proposed as a marker for protein turnover but is influenced by meat intake.20 3-MH, therefore, has gained attention as a metabolite that can potentially serve as a positive biomarker for meat intake, especially poultry.21,22 However, the effects of 3-MH on kidney function remain inconclusive. One study using rat models showed that serum 3-MH is a candidate biomarker for acute renal injury, as its levels increased when drug-induced nephrotoxicity was induced.23 Another human observational study showed higher levels of 3-MH in subjects with moderate kidney failure compared to healthy individuals.24 However, another study in individuals on hemodialysis identified low levels of 3-MH to be predictive of cardiovascular events.25
This study also identified the stool metabolite 5-methylhexanoate (i7:0) to potentially mediate the effects of DAL on CKD and identified several gut microbiota species that correlate with this metabolite. 5-methylhexanoate belongs to the group of medium-chain fatty acids (MCFAs) but is not well investigated as an individual metabolite. However, the effect of MCFA in general on human health has been extensively reviewed elsewhere,26 and was generally found to be positive for human health, but deleterious effects have also been reported.27 DAL itself did not associate with gut microbiota composition, which aligns with previous studies reporting that protein intake, a major source of DAL, does not affect gut microbiota diversity.28,29
As kidney function can also affect host metabolome,11,30 we also performed mediation analyses by using CKD stage as potential mediator, i.e., DAL→ CKD stage G1-G2 (potential mediator)→ metabolite of interest. Though we found that early-stage CKD partially mediates the DAL→ metabolome relationship in selected metabolites, the VAF was lower when using metabolites as the mediator. This could be explained by the fact that we only used early-stage CKD and the association may be stronger as kidney function declines toward end-stage CKD.30
Multiple studies have reported an intricate relationship between the gut microbiota and kidney disease,31,32,33,34 with the gut microbiota correlating with end-stage CKD34 In our data, we did not find an association between gut microbiota composition and CKD; however, our study participants were early-stage CKD (stage G1-G2).34 We did however find 26 microbial species to be associated with fecal abundances of the stool metabolite 5-methylhexanoate (i7:0) that mediated the association between DAL and CKD, thus linking the gut microbiome’s involvement in CKD.
Of the 26 microbial species, Alistipes spp. had the strongest positive association and has been consistently reported to be increased in CKD, and to be associated with an increased intake of a Western diet, high in animal protein and fat but low in fiber.2,35 Our study also identified Intestimonas butyriciproducens, a known producer of the beneficial metabolite butyrate, a short-chain fatty acid (SCFA),36 to be negatively associated with 5-methylhexanoate (i7:0). This finding further emphasizes a role for SCFA in line with a previous study reporting that supplementation with the probiotic L. casei Zhang increased SCFA levels and reduced CKD progression in mice.31
In conclusion, we identified serum, stool, and urinary metabolites to possibly mediate the positive relationship between DAL and CKD stage G1-G2. Moreover, this study identified several gut microbiota species, such as Alistipes spp and Intestimonas butyriciproducens, that can be used as novel targets to mitigate this relationship, suggesting that the gut microbiota may be a therapeutic option to combat the effects of DAL on kidney function decline by altering host metabolome. Longitudinal, as well as interventional, studies are warranted to investigate the exact relationship of the identified metabolites and microbial species and to elucidate potential causality in the relationship between DAL and kidney function decline.
Limitations of the study
This study has limitations and strengths. Strengths of this study include the use of a large population-based cohort with accurate phenotyping, dietary information, and concurrent metabolomics in multiple biological fluids. Another strength of our study is that we explored the host metabolome, using the Metabolon, Inc, platform, that covers a wide range of metabolites from multiple biological pathways leading to a more detailed understanding of the underlying biology.
However, our findings should also be appreciated in the context of some limitations. First, DAL was determined using self-reported dietary intake. Although a previously validated formula was used to calculate DAL,4,37 this approach is prone to several biases, including misreporting bias, which may cause misclassification bias. However, there is no gold standard to measure DAL and each method has its own drawbacks.4
Second, our study sample was limited to individuals with normal-to-mild renal impairment and was predominantly females of Caucasian ancestry. Accordingly, our results cannot be generalized to those with moderate-to-end-stage renal disease, and future research should explore the relationships identified here in those with later-stage CKD.
Third, the mediation analysis has some limitations. Although our study identified multiple metabolites that potentially mediate the relationship between DAL and CKD progression, our data do not permit comments on causality due to the lack of a temporal relationship. Moreover, our study cannot rule out a different directionality between exposure, mediator, and outcomes. Hence, we tested both directions. These analyses indicate a potential mediating effect of CKD stage on the metabolite of interest and, to a larger extent, of the metabolite of interest on CKD.
Finally, we were unable to replicate our results in an independent sample as, to our knowledge, there are currently no cohorts with such a comprehensive assessment of serum, urine, stool, and saliva metabolomics as well as clinical creatinine, dietary data, and shotgun metagenome profiling of the gut microbiome.
Longitudinal metabolomics data, as well as dietary and kidney function, are needed to confirm causality and understand directionality.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological samples | ||
| Human plasma/urine/fecal/saliva metabolomics data | TwinsUK; measured via Metabolon,Inc, Durham, North Carolina, USA | N/A |
| Human fecal metagenomics data | TwinsUK | EBI (https://www.ebi.ac.uk/) accession number PRJEB32731 |
| Critical commercial assays | ||
| DNA extraction kit | QIAamp DNA Mini kit | N/A |
| Deposited data | ||
| Dietary data | TwinsUK | twinsuk.ac.uk/resources-for-researchers/access-our-data |
| Anthropometric data | TwinsUK | twinsuk.ac.uk/resources-for-researchers/access-our-data |
| Software and algorithms | ||
| open software program R version 4.2.1 | ( R Core Team (2022). R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. URL https://Www.R-Project.Org/., n.d.) | https://www.R-project.org/ |
| FactoMineR package in R (version 2.7) | Comprehensive R Archive Network | http://factominer.free.fr/ |
| Vegan package in R (version 2.6-4) | Comprehensive R Archive Network | https://github.com/vegandevs/vegan |
| randomForest R package (version 4.7.1.1) | Comprehensive R Archive Network | cran.r-project.org/web/packages/randomForest/randomForest.pdf |
Resource availability
Lead contact
Further information should be directed to and will be fulfilled by the lead contact Dr. Cristina Menni (cristina.menni@kcl.ac.uk).
Materials availability
This study did not generate new unique reagents or materials.
Data and code availability
-
•
The data used in this study are held by the Department of Twin Research at King’s College London. The data can be released to bona fide researchers using our normal procedures overseen by the Wellcome Trust and its guidelines as part of our core funding (https://twinsuk.ac.uk/resources-for-researchers/access-our-data/). The gut microbiome data is available on EBI (https://www.ebi.ac.uk/) under accession PRJEB32731 (TwinsUK).
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon reasonable request.
Experimental model and study participant details
Study population and ethics approval
We included 1,453 individuals from TwinsUK38 with good estimated kidney function (CKD stage G1), or moderately impaired (CKD stage G2) as defined by the 2021 CKD-EPI formula (based on serum creatinine).39 Included participants also had data on dietary intake, and metabolites measured from multiple biological samples (serum, urine, stool and saliva). TwinsUK is the largest adult twin registry in the world, recruited as research volunteers, without selecting for any particular disease or trait, and has been shown to be comparable to the general British population for lifestyle characteristics.40 The baseline characteristics of participants are described in Table 1.
This study was carried out under TwinsUK BioBank ethics, approved by North West – Liverpool Central Research Ethics Committee (REC reference 19/NW/0187), IRAS ID 258513. This approval supersedes earlier approvals granted to TwinsUK by the St Thomas’ Hospital Research Ethics Committee, later London – Westminster Research Ethics Committee (REC reference EC04/015), which have now been subsumed within the TwinsUK BioBank.
Method details
Study design
A flowchart of the study design is presented in Figure 1. Our primary objective is to investigate the relationship between DAL and kidney function decline (CKD stage G1-G2) and to understand the metabolites underlying the relationship. Our secondary aims were (i) to explore the mediating effects of the identified metabolites per biological sample; and (ii) to identify underlying gut microbial species that drive the mediating metabolites per biological sample.
Metabolite profiling
Metabolite concentrations were measured at fasting from serum, urine, stool and saliva samples by Metabolon Inc. (Durham, USA) using an untargeted Liquid chromatography–mass spectrometry (LC-MS) platform as previously described.41 This platform was used for both the discovery, and the replication cohort. Metabolites with more than 20% missingness were excluded, and the remaining metabolites were day median normalized, imputed to the day minimum, and inverse normalized.42 The Metabolon platform measured 585 serum, 656 urine, 492 stool and 400 saliva metabolites post-quality check from different metabolic pathways including amino acid, lipid, carbohydrate and vitamin metabolism.
Metabolite measurement and standardization
To quantify metabolites, the area-under-the-curve method was employed. Each metabolite’s raw area count in every sample underwent standardization to mitigate variations caused by differences in daily instrument tuning. This was achieved by adjusting the median values for each day of the run to 1.0. Such normalization allowed for maintaining sample-to-sample variations while enabling the comparison of metabolites with significantly differing raw peak areas on a uniform scale.
Quality control
In addressing batch effects in metabolite measurements, the experimental sample values for each metabolite were normalized against the median value of those in the same batch. This process standardized each batch, setting the median for each metabolite at one. For missing values, imputation was done using the lowest value from all the batches in the median-adjusted data.
Dietary intake
Dietary intake was estimated using a modified European Prospective Investigation into Cancer and Nutrition food frequency questionnaire (FFQ).43 FFQs were excluded if more than 10 food items were left unanswered or if the total energy intake estimate derived from FFQ as a ratio of the subject’s estimated basal metabolic rate (determined by the Harris–Benedict equation)44 was more than 2 standard deviations outside the mean of this ratio (<0.52 or >2.58), as previously described.45
DAL was calculated as the net endogenous acid production from FFQ data using a validated formula4,37: Net Endogenous Acid Production (mEq H+/day) = 54.5 × [protein intake (g/day)/potassium intake (mEq/day)) − 10.2.
Gut microbiota fecal collection, DNA extraction and metagenome profiling
Fecal sample collection, DNA extraction and metagenome profiling were performed as previously described.46 Briefly, participants collected stool samples at home in pre-labelled kits (containing 2 x 25ml tube or 1 x 25ml tube and 1 x 10ml Zymo buffer) posted to them before their clinic visit date and brought with them to the visit. Alternatively, samples could be posted to the clinic using blue Royal Mail safe boxes, ensuring correct cooled temperature. In the laboratory, samples were homogenised, aliquoted into 4 bijou tubes, and stored at −80°C, within 2 hours of receipt. TwinsUK sequenced metagenomes were processed using the YAMP pipeline (v. 0.9.5.3)31.46 The metagenomic analysis was conducted following the general guidelines47 and based on the bioBakery computational environment.48 High-resolution taxonomic profiling of the metagenomes was performed using MetaPhlAn 4.beta.2 with the Jan21 database that comprises 26,970 species-level genome bins, with default parameters.49
Quantification and statistical analysis
Statistical analyses were performed using open software program R (version 4.2.1).50
DAL was adjusted for energy intake using the residual method.51 DAL was then converted to tertiles and the top and bottom tertiles were used for analyses.
Linear and logistic mixed-effect models were used to investigate high/low DAL - CKD/eGFR associations, correcting for age, sex, body mass index (BMI) and family relatedness (random effect). CKD and DAL were dichotomous, whereas eGFR was a continuous variable.
Principal component analysis and PERMANOVA were performed using the FactoMineR (version 1.34)52 and vegan packages (version 2.6)50 to explore the serum, urine, stool, and saliva metabolite profiles of individuals in CKD stage G1 and G2 in relation to their DAL.
For each biological tissue (urine, stool, saliva, serum) two random forest (RF) models were employed to identify the most influential metabolites (from multiple biological samples) affecting (1) DAL, and (2) CKD stage G1-G2. Models were adjusted for traditional risk factors (age, sex, and BMI), by calculating the residuals per metabolite, using the risk factors as independent variables. For each RF, the dataset was split (80:20), with 80% going into a training set and 20% into the test set. Hyperparameters were tuned using the adaptive resampling search, and the optimum number of features was calculated using five-fold cross-validation and selected by node purity. Using the optimum number of metabolites from the RF model, we generated a panel of metabolites (per biological sample) that overlapped between the DAL model and the CKD model.
To identify a direction of effect for the metabolite panels we ran regression models adjusting for covariates. The Benjamini-Hochberg method was used to correct for multiple testing (false discovery rate (FDR)<0.05).53
Metabolites that passed the multiple testing threshold (FDR; p<0.05), were tested for a potential mediating effect in the relationship between DAL and CKD stage G1-G2. Mediation analysis was performed using the ‘mediate’ function in the R package ‘mediation’ (version 4.5.0).54 The variance accounted for (VAF) was determined as the ratio of indirect-to-total effect and distinguishes the proportion of the variance explained by the mediation process (the proportion of the effect of DAL on CKD status that goes through the metabolite). Mediation analyses was conducted in accordance with the AGReMA guidelines for conducting and reporting mediation analyses (Table S1).55
The main mediation model was constructed as DAL (independent variable)→ Metabolite of interest (potential mediator) → CKD stage G1-G2 (dependent variable).
However, as kidney function is also associated with changes in metabolites30 we also tested the model DAL(independent variable)→ CKD stage G1-G2 (potential mediator)→ metabolite of interest (dependent variable). All available data was used for the mediation models and no sample size calculation was conducted prior to the analyses, as is often the case in mediation analyses.55 All models and corresponding output are reported in Tables S2–S5.
The relationship between the mediatory metabolites and the gut microbiota was then tested using multivariable regression models with FDR correction. Explained variance of the gut microbiota with mediating metabolites was determined using RF models as described above. Alpha diversity (Shannon index) was calculated using the “vegan” package (version 2.6).56
Acknowledgments
We express our gratitude to all of the participants and staff at TwinsUK.
This research was funded in whole, or in part, by the Wellcome Trust (WT212904/Z/18/Z). For the purpose of open access, the authors have applied a CC BY public copyright to any Author Accepted Manuscript version arising from this submission. The Department of Twin Research receives support from grants from the Wellcome Trust (212904/Z/18/Z) and the Medical Research Council (MRC)/British Heart Foundation (BHF) Ancestry and Biological Informative Markers for Stratification of Hypertension (AIM-HY; MR/M016560/1), European Union, Chronic Disease Research Foundation (CDRF), Zoe Global Ltd., and the NIHR Clinical Research Facility and Biomedical Research Centre (based at Guy’s and St Thomas’ NHS Foundation Trust in partnership with King’s College London). C.M. is funded by the Chronic Disease Research Foundation (CDRF), the UKRI grant (MR/W026813/1), and the MRC/BHF (AIM-HY; MR/M016560/1). I.A. is funded by Amsterdam Cardiovascular Sciences Post-Doctoral grant. B.B.-C. is funded by a CRN West Midlands Personal Development Award and the Centre for Care Excellence at UHCW NHS Trust. P.L. is supported by the CDRF (CDRF–15/2018). J.T.B. was supported by JPI ERA-HDHL-funded DIMENSION project through BBSRC (BB/S020845/1 to J.T.B.). I.A. was funded through an Amsterdam Cardiovascular Sciences (ACS) post-doctoral grant (2022).
Author contributions
I.A. ran the analyses and verified the underlying data; I.A., B.B.-C., and C.M. wrote the original manuscript. P.L., A.N., F.T., A.V., K.W., G.M., M.F., and T.D.S. contributed methods/materials/analysis tools. All authors have read and approved the final version of the manuscript.
Declaration of interests
T.D.S. is a co-founder and shareholder of ZOE Ltd.
Published: February 5, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2024.109132.
Contributor Information
Ilias Attaye, Email: ilias.attaye@kcl.ac.uk.
Cristina Menni, Email: cristina.menni@kcl.ac.uk.
Supplemental information
References
- 1.Kovesdy C.P. Epidemiology of chronic kidney disease: an update 2022. Kidney Int. Suppl. 2022;12:7–11. doi: 10.1016/j.kisu.2021.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang X., Yang S., Li S., Zhao L., Hao Y., Qin J., Zhang L., Zhang C., Bian W., Zuo L., et al. Aberrant gut microbiota alters host metabolome and impacts renal failure in humans and rodents. Gut. 2020;69:2131–2142. doi: 10.1136/gutjnl-2019-319766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hobby G.P., Karaduta O., Dusio G.F., Singh M., Zybailov B.L., Arthur J.M. Chronic kidney disease and the gut microbiome. Am. J. Physiol. Renal Physiol. 2019;316:F1211–F1217. doi: 10.1152/ajprenal.00298.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scialla J.J., Anderson C.A.M. Dietary acid load: a novel nutritional target in chronic kidney disease? Adv. Chronic Kidney Dis. 2013;20:141–149. doi: 10.1053/j.ackd.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berkemeyer S. Acid-base balance and weight gain: are there crucial links via protein and organic acids in understanding obesity? Med. Hypotheses. 2009;73:347–356. doi: 10.1016/j.mehy.2008.09.059. [DOI] [PubMed] [Google Scholar]
- 6.Han E., Kim G., Hong N., Lee Y.-H., Kim D.W., Shin H.J., Lee B.-W., Kang E.S., Lee I.-K., Cha B.-S. Association between dietary acid load and the risk of cardiovascular disease: nationwide surveys (KNHANES 2008-2011) Cardiovasc. Diabetol. 2016;15:122. doi: 10.1186/s12933-016-0436-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parohan M., Sadeghi A., Nasiri M., Maleki V., Khodadost M., Pirouzi A., Sadeghi O. Dietary acid load and risk of hypertension: A systematic review and dose-response meta-analysis of observational studies. Nutr. Metab. Cardiovasc. Dis. 2019;29:665–675. doi: 10.1016/j.numecd.2019.03.009. [DOI] [PubMed] [Google Scholar]
- 8.Dehghan P., Abbasalizad Farhangi M. Dietary acid load, blood pressure, fasting blood sugar and biomarkers of insulin resistance among adults: Findings from an updated systematic review and meta-analysis. Int. J. Clin. Pract. 2020;74 doi: 10.1111/ijcp.13471. [DOI] [PubMed] [Google Scholar]
- 9.Hayata H., Miyazaki H., Niisato N., Yokoyama N., Marunaka Y. Lowered extracellular pH is involved in the pathogenesis of skeletal muscle insulin resistance. Biochem. Biophys. Res. Commun. 2014;445:170–174. doi: 10.1016/j.bbrc.2014.01.162. [DOI] [PubMed] [Google Scholar]
- 10.Daneshzad E., Haghighatdoost F., Azadbakht L. Dietary acid load and cardiometabolic risk factors: a systematic review and meta-analysis of observational studies. Public Health Nutr. 2019;22:2823–2834. doi: 10.1017/S1368980019001125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao Y.-Y. Metabolomics in chronic kidney disease. Clin. Chim. Acta. 2013;422:59–69. doi: 10.1016/j.cca.2013.03.033. [DOI] [PubMed] [Google Scholar]
- 12.Sekula P., Goek O.-N., Quaye L., Barrios C., Levey A.S., Römisch-Margl W., Menni C., Yet I., Gieger C., Inker L.A., et al. A Metabolome-Wide Association Study of Kidney Function and Disease in the General Population. J. Am. Soc. Nephrol. 2016;27:1175–1188. doi: 10.1681/ASN.2014111099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tariq A., Chen J., Yu B., Boerwinkle E., Coresh J., Grams M.E., Rebholz C.M. Metabolomics of Dietary Acid Load and Incident Chronic Kidney Disease. J. Ren. Nutr. 2022;32:292–300. doi: 10.1053/j.jrn.2021.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mofrad M.D., Daneshzad E., Azadbakht L. Dietary acid load, kidney function and risk of chronic kidney disease: A systematic review and meta-analysis of observational studies. Int. J. Vitam. Nutr. Res. 2021;91:343–355. doi: 10.1024/0300-9831/a000584. [DOI] [PubMed] [Google Scholar]
- 15.Rebholz C.M., Coresh J., Grams M.E., Steffen L.M., Anderson C.A.M., Appel L.J., Crews D.C. Dietary Acid Load and Incident Chronic Kidney Disease: Results from the ARIC Study. Am. J. Nephrol. 2015;42:427–435. doi: 10.1159/000443746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rebholz C.M., Surapaneni A., Levey A.S., Sarnak M.J., Inker L.A., Appel L.J., Coresh J., Grams M.E. The Serum Metabolome Identifies Biomarkers of Dietary Acid Load in 2 Studies of Adults with Chronic Kidney Disease. J. Nutr. 2019;149:578–585. doi: 10.1093/jn/nxy311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Altmaier E., Fobo G., Heier M., Thorand B., Meisinger C., Römisch-Margl W., Waldenberger M., Gieger C., Illig T., Adamski J., et al. Metabolomics approach reveals effects of antihypertensives and lipid-lowering drugs on the human metabolism. Eur. J. Epidemiol. 2014;29:325–336. doi: 10.1007/s10654-014-9910-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lennon E.J., Lemann J., Jr., Litzow J.R. The effects of diet and stool composition on the net external acid balance of normal subjects. J. Clin. Invest. 1966;45:1601–1607. doi: 10.1172/JCI105466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Young V.R., Munro H.N. Ntau-methylhistidine (3-methylhistidine) and muscle protein turnover: an overview. Fed. Proc. 1978;37:2291–2300. [PubMed] [Google Scholar]
- 20.Kochlik B., Gerbracht C., Grune T., Weber D. The Influence of Dietary Habits and Meat Consumption on Plasma 3-Methylhistidine-A Potential Marker for Muscle Protein Turnover. Mol. Nutr. Food Res. 2018;62 doi: 10.1002/mnfr.201701062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cuparencu C., Praticó G., Hemeryck L.Y., Sri Harsha P.S.C., Noerman S., Rombouts C., Xi M., Vanhaecke L., Hanhineva K., Brennan L., Dragsted L.O. Biomarkers of meat and seafood intake: an extensive literature review. Genes Nutr. 2019;14:35. doi: 10.1186/s12263-019-0656-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Myint T., Fraser G.E., Lindsted K.D., Knutsen S.F., Hubbard R.W., Bennett H.W. Urinary 1-methylhistidine is a marker of meat consumption in Black and in White California Seventh-day Adventists. Am. J. Epidemiol. 2000;152:752–755. doi: 10.1093/aje/152.8.752. [DOI] [PubMed] [Google Scholar]
- 23.Uehara T., Horinouchi A., Morikawa Y., Tonomura Y., Minami K., Ono A., Yamate J., Yamada H., Ohno Y., Urushidani T. Identification of metabolomic biomarkers for drug-induced acute kidney injury in rats. J. Appl. Toxicol. 2014;34:1087–1095. doi: 10.1002/jat.2933. [DOI] [PubMed] [Google Scholar]
- 24.Ceballos I., Chauveau P., Guerin V., Bardet J., Parvy P., Kamoun P., Jungers P. Early alterations of plasma free amino acids in chronic renal failure. Clin. Chim. Acta. 1990;188:101–108. doi: 10.1016/0009-8981(90)90154-k. [DOI] [PubMed] [Google Scholar]
- 25.Bres E., Pagan C., Bouchara A., Pastural M., Granjon S., Laville M., Fouque D., Soulage C.O., Koppe L. 3-methylhistidine and clinical outcomes in maintenance haemodialysis patients. Nephrol. Dial. Transplant. 2022;37:1951–1961. doi: 10.1093/ndt/gfac050. [DOI] [PubMed] [Google Scholar]
- 26.Roopashree P.G., Shetty S.S., Suchetha Kumari N. Effect of medium chain fatty acid in human health and disease. J. Funct. Foods. 2021;87 [Google Scholar]
- 27.Temme E.H., Mensink R.P., Hornstra G. Effects of medium chain fatty acids (MCFA), myristic acid, and oleic acid on serum lipoproteins in healthy subjects. J. Lipid Res. 1997;38:1746–1754. [PubMed] [Google Scholar]
- 28.Bel Lassen P., Attaye I., Adriouch S., Nicolaou M., Aron-Wisnewsky J., Nielsen T., Chakaroun R., Le Chatelier E., Forslund S., Belda E., et al. Protein Intake, Metabolic Status and the Gut Microbiota in Different Ethnicities: Results from Two Independent Cohorts. Nutrients. 2021;13 doi: 10.3390/nu13093159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bartlett A., Kleiner M. Dietary protein and the intestinal microbiota: An understudied relationship. iScience. 2022;25 doi: 10.1016/j.isci.2022.105313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Y.-N., Ma S.-X., Chen Y.-Y., Chen L., Liu B.-L., Liu Q.-Q., Zhao Y.-Y. Chronic kidney disease: Biomarker diagnosis to therapeutic targets. Clin. Chim. Acta. 2019;499:54–63. doi: 10.1016/j.cca.2019.08.030. [DOI] [PubMed] [Google Scholar]
- 31.Zhu H., Cao C., Wu Z., Zhang H., Sun Z., Wang M., Xu H., Zhao Z., Wang Y., Pei G., et al. The probiotic L. casei Zhang slows the progression of acute and chronic kidney disease. Cell Metab. 2021;33:2091–2093. doi: 10.1016/j.cmet.2021.08.015. [DOI] [PubMed] [Google Scholar]
- 32.Miao H., Wang Y.-N., Yu X.-Y., Zou L., Guo Y., Su W., Liu F., Cao G., Zhao Y.-Y. Lactobacillus species ameliorate membranous nephropathy through inhibiting the aryl hydrocarbon receptor pathway via tryptophan-produced indole metabolites. Br. J. Pharmacol. 2024;181:162–179. doi: 10.1111/bph.16219. [DOI] [PubMed] [Google Scholar]
- 33.Shi X., Li Z., Lin W., Shi W., Hu R., Chen G., Li X., Li X., Zhang S. Altered intestinal microbial flora and metabolism in patients with idiopathic membranous nephropathy. Am. J. Nephrol. 2023;54:451–470. doi: 10.1159/000533537. [DOI] [PubMed] [Google Scholar]
- 34.Simões-Silva L., Araujo R., Pestana M., Soares-Silva I., Sampaio-Maia B. The microbiome in chronic kidney disease patients undergoing hemodialysis and peritoneal dialysis. Pharmacol. Res. 2018;130:143–151. doi: 10.1016/j.phrs.2018.02.011. [DOI] [PubMed] [Google Scholar]
- 35.Beam A., Clinger E., Hao L. Effect of diet and dietary components on the composition of the gut Microbiota. Nutrients. 2021;13:2795. doi: 10.3390/nu13082795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kläring K., Hanske L., Bui N., Charrier C., Blaut M., Haller D., Plugge C.M., Clavel T. Intestinimonas butyriciproducens gen. nov., sp. nov., a butyrate-producing bacterium from the mouse intestine. Int. J. Syst. Evol. Microbiol. 2013;63:4606–4612. doi: 10.1099/ijs.0.051441-0. [DOI] [PubMed] [Google Scholar]
- 37.Sebastian A., Frassetto L.A., Sellmeyer D.E., Merriam R.L., Morris R.C., Jr. Estimation of the net acid load of the diet of ancestral preagricultural Homo sapiens and their hominid ancestors. Am. J. Clin. Nutr. 2002;76:1308–1316. doi: 10.1093/ajcn/76.6.1308. [DOI] [PubMed] [Google Scholar]
- 38.Verdi S., Abbasian G., Bowyer R.C.E., Lachance G., Yarand D., Christofidou P., Mangino M., Menni C., Bell J.T., Falchi M., et al. TwinsUK: The UK Adult Twin Registry Update. Twin Res. Hum. Genet. 2019;22:523–529. doi: 10.1017/thg.2019.65. [DOI] [PubMed] [Google Scholar]
- 39.Inker L.A., Eneanya N.D., Coresh J., Tighiouart H., Wang D., Sang Y., Crews D.C., Doria A., Estrella M.M., Froissart M., et al. New creatinine- and cystatin C-based equations to estimate GFR without race. N. Engl. J. Med. 2021;385:1737–1749. doi: 10.1056/NEJMoa2102953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moayyeri A., Hammond C.J., Valdes A.M., Spector T.D. Cohort Profile: TwinsUK and healthy ageing twin study. Int. J. Epidemiol. 2013;42:76–85. doi: 10.1093/ije/dyr207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Louca P., Meijnikman A.S., Nogal A., Asnicar F., Attaye I., Vijay A., Kouraki A., Visconti A., Wong K., Berry S.E., et al. The secondary bile acid isoursodeoxycholate correlates with post-prandial lipemia, inflammation, and appetite and changes post-bariatric surgery. Cell Rep. Med. 2023;4 doi: 10.1016/j.xcrm.2023.100993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Louca P., Nogal A., Moskal A., Goulding N.J., Shipley M.J. Cross-sectional Blood Metabolite Markers of Hypertension: A Multicohort Analysis of 44,306 Individuals from the COnsortium of METabolomics Studies. Metabolites. 2022;12:601. doi: 10.3390/metabo12070601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bingham S.A., Welch A.A., McTaggart A., Mulligan A.A., Runswick S.A., Luben R., Oakes S., Khaw K.T., Wareham N., Day N.E. Nutritional methods in the European Prospective Investigation of Cancer in Norfolk. Public Health Nutr. 2001;4:847–858. doi: 10.1079/phn2000102. [DOI] [PubMed] [Google Scholar]
- 44.Frankenfield D.C., Muth E.R., Rowe W.A. The Harris-Benedict studies of human basal metabolism: history and limitations. J. Am. Diet. Assoc. 1998;98:439–445. doi: 10.1016/S0002-8223(98)00100-X. [DOI] [PubMed] [Google Scholar]
- 45.Le Roy C.I., Kurilshikov A., Leeming E.R., Visconti A., Bowyer R.C.E., Menni C., Falchi M., Koutnikova H., Veiga P., Zhernakova A., et al. Yoghurt consumption is associated with changes in the composition of the human gut microbiome and metabolome. BMC Microbiol. 2022;22:39. doi: 10.1186/s12866-021-02364-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Visconti A., Le Roy C.I., Rosa F., Rossi N., Martin T.C., Mohney R.P., Li W., de Rinaldis E., Bell J.T., Venter J.C., et al. Interplay between the human gut microbiome and host metabolism. Nat. Commun. 2019;10:4505. doi: 10.1038/s41467-019-12476-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Quince C., Walker A.W., Simpson J.T., Loman N.J., Segata N. Shotgun metagenomics, from sampling to analysis. Nat. Biotechnol. 2017;35:833–844. doi: 10.1038/nbt.3935. [DOI] [PubMed] [Google Scholar]
- 48.Stoltenberg R.L., Madsen J.A., Schlack S.C., Harms B.A., Jacoby R.F. Neoplasia in ileal pouch mucosa after total proctocolectomy for juvenile polyposis: report of a case. Dis. Colon Rectum. 1997;40:726–730. doi: 10.1007/BF02140904. [DOI] [PubMed] [Google Scholar]
- 49.Truong D.T., Franzosa E.A., Tickle T.L., Scholz M., Weingart G., Pasolli E., Tett A., Huttenhower C., Segata N. MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat. Methods. 2015;12:902–903. doi: 10.1038/nmeth.3589. [DOI] [PubMed] [Google Scholar]
- 50.R Core Team . R Foundation for Statistical Computing; 2022. R: A Language and Environment for Statistical Computing. [Google Scholar]
- 51.McCullough L.E., Byrd D.A. Total Energy Intake: Implications for Epidemiologic Analyses. Am. J. Epidemiol. 2023;192:1801–1805. doi: 10.1093/aje/kwac071. [DOI] [PubMed] [Google Scholar]
- 52.Lê S., Josse J., Husson F. FactoMineR: AnRPackage for Multivariate Analysis. J. Stat. Softw. 2008;25 doi: 10.18637/jss.v025.i01. [DOI] [Google Scholar]
- 53.Benjamini Y., Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. 1995;57:289–300. [Google Scholar]
- 54.Tingley D., Yamamoto T., Hirose K., Keele L., Imai K. mediation:RPackage for Causal Mediation Analysis. J. Stat. Softw. 2014;59 doi: 10.18637/jss.v059.i05. [DOI] [Google Scholar]
- 55.Lee H., Cashin A.G., Lamb S.E., Hopewell S., Vansteelandt S., VanderWeele T.J., MacKinnon D.P., Mansell G., Collins G.S., Golub R.M., et al. A Guideline for Reporting Mediation Analyses of Randomized Trials and Observational Studies: The AGReMA Statement. JAMA. 2021;326:1045–1056. doi: 10.1001/jama.2021.14075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vegan:Community Ecology Package. 2022. https://github.com/vegandevs/vegan
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The data used in this study are held by the Department of Twin Research at King’s College London. The data can be released to bona fide researchers using our normal procedures overseen by the Wellcome Trust and its guidelines as part of our core funding (https://twinsuk.ac.uk/resources-for-researchers/access-our-data/). The gut microbiome data is available on EBI (https://www.ebi.ac.uk/) under accession PRJEB32731 (TwinsUK).
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon reasonable request.