Abstract
Objective
Accumulating evidence from microbial studies have highlighted the modulatory roles of intestinal microbes in numerous human diseases, however, the shared microbial signatures across different diseases remain relatively unclear.
Methods
To consolidate existing knowledge across multiple studies, we performed meta-analyses of 17 disease types, covering 34 case–control datasets of 16S rRNA sequencing data, to identify shared alterations among different diseases. Furthermore, the impact of a microbial species, Lactobacillus salivarius, was established in a dextran sodium sulphate–induced colitis model and a collagen type II–induced arthritis mouse model.
Results
Microbial alterations among autoimmune diseases were substantially more consistent compared with that of other diseases (cancer, metabolic disease and nervous system disease), with microbial signatures exhibiting notable discriminative power for disease prediction. Autoimmune diseases were characterized by the enrichment of Enterococcus, Veillonella, Streptococcus and Lactobacillus and the depletion of Ruminococcus, Gemmiger, Oscillibacter, Faecalibacterium, Lachnospiracea incertae sedis, Anaerostipes, Coprococcus, Alistipes, Roseburia, Bilophila, Barnesiella, Dorea, Ruminococcus2, Butyricicoccus, Phascolarctobacterium, Parabacteroides and Odoribacter, among others. Functional investigation of L. salivarius, whose genus was commonly enriched in numerous autoimmune diseases, demonstrated protective roles in two separate inflammatory mouse models.
Conclusion
Our study highlights a strong link between autoimmune diseases and the gut microbiota, with notably consistent microbial alterations compared with that of other diseases, indicating that therapeutic strategies that target the gut microbiome may be transferable across different autoimmune diseases. Functional validation of L. salivarius highlighted that bacterial genera associated with disease may not always be antagonistic, but may represent protective or adaptive responses to disease.
Keywords: microbiome, autoimmune diseases, meta-analysis, machine learning, general pattern
Rheumatology key messages.
Microbial alterations across autoimmune diseases were more consistent than those of other diseases.
The diagnostic power of the microbial signatures from autoimmune diseases was significantly higher than from other diseases.
L. salivarius exhibited protective functions in two mouse inflammatory models, indicating significantly associated bacterial taxa may not be antagonistic.
Introduction
The pathogenesis of various human diseases cannot be fully explained by underlying genetic susceptibilities of the host, which highlights the necessity to explore the contributions by environmental factors. Supported by our understanding of the pathogenic roles of microbes [1, 2], increasing evidence has highlighted the correlation between abnormal alterations of gut microbiota and various disease types, including autoimmune diseases [such as IBD, primary APS (PAPS), RA, SS, SLE and UCTD), metabolic diseases (such as obesity and polycystic ovary syndrome), cancer (such as breast, colorectal and pancreatic) and even nervous system diseases (such as multiple system atrophy and Parkinson’s disease) [3–7]. Disease-specific microbial signatures have been identified for multiple diseases, yet the biological significance of changes in microbial composition has not been thoroughly assessed [8].
Particularly for autoimmune diseases, the underlying genetics of the host forms the basis for susceptibility to disease, yet environmental factors such as microbial dysbiosis have been noted to frequently trigger and drive disease progression, with the severity and clinical heterogeneity of diseases invariably modulated by gut microbiota [9]. A large proportion of genetic risk factors are shared among autoimmune diseases and possibly contribute towards unique alterations in the human gut microbiota [10, 11], thereby potentially driving disease [12].
However, it remains poorly understood whether diverse disease types share common microbial signatures. Prior case–control gut microbiome studies have yielded inconsistent results, which makes comparisons between studies difficult and contextualization of their findings across multiple diseases difficult. Primary issues include insufficient sample size, differences in underlying cohorts and demographics and a lack of data standardization and normalization. In the current study we address this by reprocessing 34 faecal 16S rRNA datasets spanning 17 disease types with a standardized bioinformatics workflow. Meta-analyses of these data revealed bacterial taxa commonly associated with a diverse array of disease types. Moving beyond these correlations, we also examined the causative role of a microbe putatively associated with multiple diseases using mouse models.
Methods
Data acquisition and selection
Data from 41 faecal 16S rRNA sequencing experiments were acquired from the Sequence Read Archive (SRA) of the National Center for Biotechnology Information (NCBI). Studies without sufficient raw sequencing data, meta information or of poor sequencing quality were excluded, resulting in 24 high-quality datasets. Additionally, we acquired 10 datasets from the MicrobiomeHD database [13]. In total, 34 case–control datasets covering 17 disease types were classified into four broad categories (autoimmune disease, metabolic disease, cancer and nervous system disease) according to the International Classification of Diseases, 11th revision (ICD-11).
16S rRNA amplicon data processing
Raw sequencing data were processed through a standardized 16S processing pipeline (https://github.com/thomasgurry/amplicon_sequencing_pipeline) and used to generate the operational taxonomic unit (OTU) tables from the MicrobiomeHD database (Supplementary Table S1, available at Rheumatology online) [13]. In brief, downloaded pre-trimmed and de-multiplexed reads with quality scores <20 were removed. Reads with more than two expected errors were also removed. Reads were trimmed in accordance with the original sequencing length for each data set. OTU clustering was performed at 100% similarity using USEARCH [14] and the RDP classifier was used to obtain taxonomic assignments with a confidence cut-off of 0.5 [15]. OTUs with <10 reads or unannotated at the genus level were discarded. The relative abundance of each OTU was calculated and collapsed to the genus level.
Normalization of the variation between datasets for meta-analyses
Since each dataset differs by the genetic background of the participants, as well as technical differences in data acquisition, we normalized the data to remove batch effects prior to analysis. In brief, we transformed discrete taxonomic count data to approximately normally distributed log-count per million (log-CPM) data to remove heteroscedasticity. We then performed supervised normalization (SNM) on the data to remove significant batch effects while preserving biological effects [16].
Divergence of metagenomic approaches and study design, such as differences in population, sample collection, preservation, preparation, amplification and sequencing platform, constitute heterogeneities across different datasets [17, 18]. As each study integrally would be considered as a confounding factor, a confounder analysis was therefore performed by analysis of variance (ANOVA) to quantify the effect of confounders combined relative to that of each one alone. Variance calculations were performed to rank the microbiome abundance following a non-Gaussian distribution.
Univariate analysis of disease-associated microbial species
Since microbiome data are characterized by non-Gaussian distributions with excessive dispersion, non-parametric significance testing using Wilcoxon rank-sum testing was implemented in the R coin package (R Foundation for Statistical Computing, Vienna, Austria) [19]. Informed by the results of the preceding confounder analysis, the ‘study’ was blocked for all disease categories in meta-analysis. The generalized fold change was calculated as the logarithmic mean fold change in a set of predefined quantiles of the case and control distributions, which extended the mean-based fold change to provide higher resolution in the sparse microbiome data [20]. Quantiles ranging from 0.1 to 0.9 by an increment of 0.05 were used and false discovery rate (FDR)-adjusted P-values were used to adjust for multiple hypothesis testing [21].
α and β diversity
The α diversity measurements were calculated at the OTU level using vegan in R [22]. To calculate the effect of a disease on the composition of the gut microbiota, we assessed the proportion of explained variance [R2 from permutational ANOVA (PERMANOVA)] between samples based on the Bray–Curtis distance by the adonis function from the R package vegan [22].
Correlation of bacterial signatures with diseases
The cor function in R was used to calculate the Spearman correlation coefficients of fold change data for each pair of datasets. The genera were clustered using the Ward algorithm implemented in the R function hclust. The cor and hclust functions were part of the R STATS package.
Random forest–based model for regression analysis of bacterial signatures for disease diagnosis
Random forest–based models were established to discriminate between cases and controls using Python’s scikit-learn [23]. In a cross-validation, this model was trained and tested by 5-fold cross-validation, which divided the relative abundance values randomly into five sets (one set for testing and four sets for training), for five times.
Animal and disease models
C57BL/6J littermates were used for dextran sodium sulphate (DSS)-induced colitis model. Colitis was induced by exposure to 2% DSS (MP Biomedicals, Irvine, CA, USA) in the drinking water for 7 days. The clinical manifestations of the colitis were monitored daily by the changes in body weight and stool starting from day 1 of DSS treatment for 9 days. At the end of the study, the serum and colon tissues were collected after the mice had been sacrificed. DBA/1J littermates were used for a collagen type II (CII)-induced arthritis model. Mice were immunized with 0.25 mg bovine type II collagen (Chondrex, Woodinville, WA, USA) in 100 µL complete Freund’s adjuvant (Difco Laboratories, Franklin Lakes, NJ, USA) containing 5 mg/ml killed Mycobacterium tuberculosis by s.c. injection at the base of the tail. Mice were challenged again with the same product in incomplete Freund’s adjuvant 21 days after the initial immunization. The animal protocol was approved by the Tsinghua University Institutional Animal Care and Use Committee (no. 21-XHJ1).
Antibiotic preconditioning and bacterial inoculation
Mice were cohoused for 3–4 weeks before antibiotic treatment to allow for equilibration of the microbiota. Broad-spectrum antibiotic consisted of metronidazole (1 g/l), neomycin (1 g/l), ampicillin (1 g/l), vancomycin (0.5 g/l) and streptomycin (1 g/l) (all from Solarbio, Beijing, China) was administered in the drinking water for 1 week and then was replaced with regular water for the rest of the experiment.
Mice were orally inoculated 24 h later with 1 × 109 CFUs Lactobacillus salivarius. L. salivarius was grown at 37°C in MRS broth (Solarbio) and were washed twice with PBS prior to use.
Histology
Samples were obtained at autopsy and were fixed in 4% paraformaldehyde. Histological sections were stained with haematoxylin and eosin (H&E) and scored in a blinded fashion. Colonic epithelial damage was scored as follows: 0 = normal; 1 = hyperproliferation, irregular crypts and goblet cell loss; 2 = mild–moderate crypt loss (10–50%); 3 = severe crypt loss (50–90%); 4 = complete crypt loss, surface epithelium intact; 5 = small to medium-sized ulcer (<10 crypt widths); 6 = large ulcer (≥10 crypt widths). Infiltration with inflammatory cells was assigned scores as follows: 0 = normal; 1 = mucosa; 2 = mucosa and submucosa; 3 = mucosa, submucosa and transmural.
Statistical analysis for animal experiments
Statistical significance was determined by a Student’s t-test using Prism version 6.03 (GraphPad Software, San Diego, CA, USA). Statistical significance was represented by P < 0.05, P < 0.01, P < 0.001 and P < 0.0001 in the figures. Data variances were represented as mean (s.e.m.) as indicated in the text.
Results
Collected datasets and normalization
The meta-analysis was performed on 34 case–control datasets of faecal 16S rRNA sequencing data, with sample sizes ranging from 23 to 644. These 34 datasets were grouped into four disease categories according to the ICD-11:
Autoimmune disease, comprised of 13 datasets covering eight diseases, including Crohn’s disease (CD), PsA, PAPS, RA, SS, SLE, ulcerative colitis (UC) and UCTD
Cancers, comprised of 8 datasets from three cancers, including breast (BC), colorectal (CRC) and pancreatic ductal adenocarcinoma (PDAC)
Metabolic disease, comprised of 8 datasets from three diseases, including obesity (OB, BMI <25 as control and >30 as case), polycystic ovary syndrome (POS) and type 1 diabetes (T1D)
Nervous system disease, comprised of 6 datasets from three diseases, including multiple sclerosis (MS), multiple system atrophy (MSA) and Parkinson’s disease (PD).
To reduce batch effects in the data from different studies (as described in the methods), we converted discrete taxonomic counts into log-CPM per sample and then performed SNM [24]. Although the batch effect was greatly reduced (Supplementary Fig. S1, available at Rheumatology online), the factor ‘study’ in all four disease categories still had a predominant impact on species composition (Supplementary Fig. S2, available at Rheumatology online). Thus we assessed the differential abundance in microbiome by Wilcoxon tests, while accounting for ‘study’ as a confounding effect that was treated as a blocking factor to avoid bias.
Microbial alterations in autoimmune disease were more consistent than for other disease
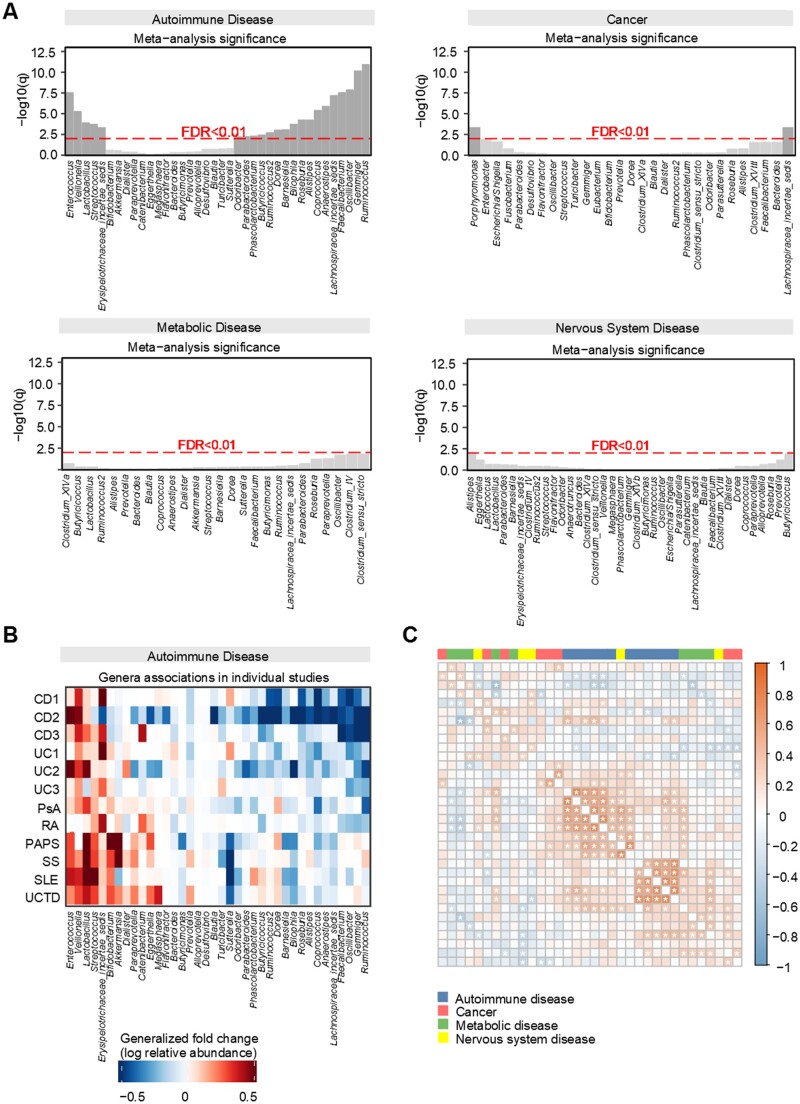
For each disease category, we performed a meta-analysis to identify microbial signatures shared across disease types. By comparing the microbial composition between cases and healthy controls, we revealed that autoimmune diseases had more consistent alteration patterns than the other disease categories (Fig. 1A and B). For autoimmune diseases, significantly enriched (FDR <0.01) genera included Enterococcus, Veillonella, Streptococcus, Lactobacillus and Erysipelotrichaceae incertae sedis, whereas Ruminococcus, Gemmiger, Oscillibacter, Faecalibacterium, Lachnospiracea incertae sedis, Anaerostipes, Coprococcus, Alistipes, Roseburia, Bilophila, Barnesiella, Dorea, Ruminococcus2, Butyricicoccus, Phascolarctobacterium, Parabacteroides and Odoribacter were characteristically depleted. In contrast, only two genera were found to be commonly enriched in cancers (BC, CRC and PDAC) (Fig. 1A, Supplementary Fig. S3A, available at Rheumatology online), and no genera were identified for metabolic diseases (OB, POS, T1D and MS, PD) (Fig. 1A, Supplementary Fig. S3A, available at Rheumatology online).
Figure 1.
Microbial alterations in autoimmune diseases were more consistent than in other diseases. (A) Meta-analysis identified a core set of gut microbes associated with diseases in each of the four broad categorizations. Species are ordered by significance and direction of change (the left side for enrichment in patients and the right side for depletion in patients). (B) Genera-level generalized fold change within individual studies. (C) Microbial correlations between diseases. Matrix of pairwise microbial correlations are coloured by correlation coefficient. Cells with asterisks indicate pairwise interactions that remained significant (P < 0.05)
To further investigate the common microbial signatures within each disease category, we analysed microbial correlations across all pairs of these 34 datasets. On average, autoimmune disease typically demonstrated stronger correlations between microbial signatures and disease subtypes than the other three disease categories (Fig. 1C).
Autoimmune disease exhibits a stronger association with gut microbiome than other diseases
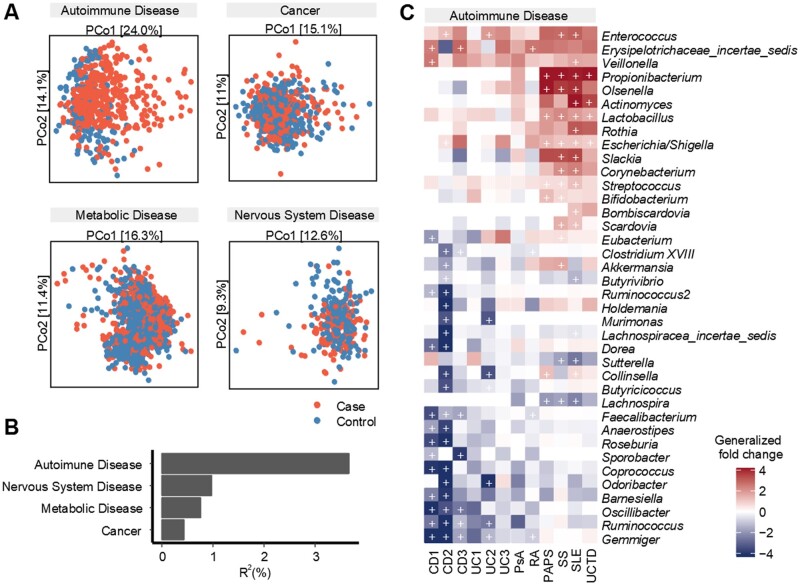
Analysis of β diversity via PERMANOVA and principal coordinates analysis (PCoA) revealed that autoimmune disease had the strongest association with variations in the gut microbiota (R2 = 3.67%, P < 0.001, using Bray–Curtis distances) compared with cancer, metabolic disease and nervous system disease (Fig. 2A and B). Investigation of specific genera associated with autoimmune disease revealed 79 significant associations (FDR <0.05), with 30 microbes shared by at least two subcategories of autoimmune disease (Fig. 2C). In particular, Enterococcus, Veillonella, Propionibacterium and Rothia were enriched in patients across all datasets; Erysipelotrichaceae incertae sedis, Olsenella, Lactobacillus, Escherichia/Shigella and Streptococcus were also enriched in patients in an overwhelming majority of datasets. Many of these microbes, such as Propionibacterium, Escherichia/Shigella and Streptococcus are well-known pathogenic bacteria [25]. For cancer (BC, CRC, PDAC), metabolic disease (OB, POS, T1D) and nervous system disease (MS, PD), the disease-associated markers appeared to be inconsistent, with only two commonly shared markers identified for cancer (Supplementary Fig. S4, available at Rheumatology online) and no shared marker for the other two disease categories. In addition, we did not observe consistently significant changes in α diversity for either of the disease categories (Supplementary Fig. S5, available at Rheumatology online).
Figure 2.
Interdisease comparisons of microbial composition associated with broad disease category. (A) Case–control PCoA, based on Bray–Curtis dissimilarity. (B) The microbial variation explained by disease category, represented by the PERMANOVA R2 value, was significant across four cross-sectional PERMANOVA tests. (C) Commonly shared microbial markers for autoimmune disease. Each row includes genera that were significant in at least two datasets within each disease category (two-sided Wilcoxon test, FDR-corrected P-value <0.05); columns represent datasets. Generalized fold change is coloured by direction of the effect, where red indicates enriched abundance in cases and blue indicates depletion. White indicates that the genus was not present in that data set. +: P < 0.05
Diagnostic microbial signatures across disease types
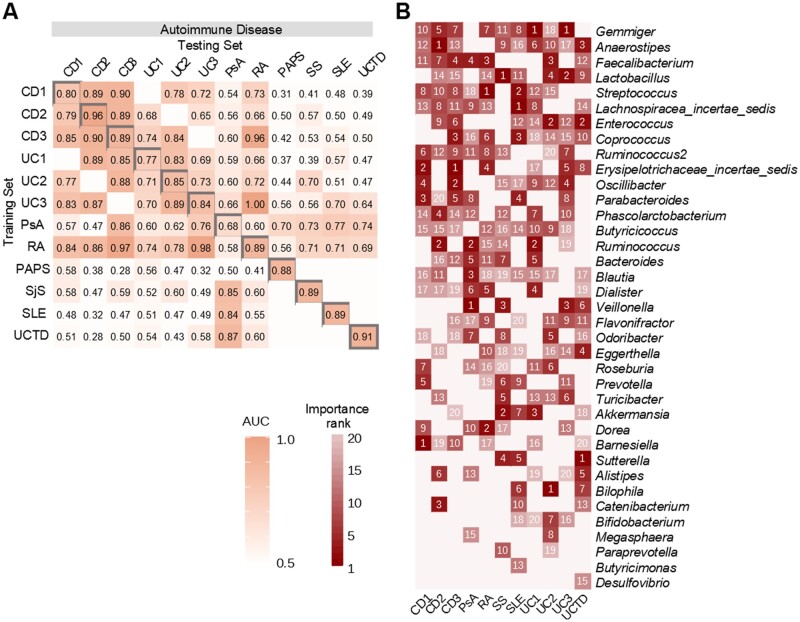
To determine the fitness of our diagnostic model, we performed a 5-fold cross-validation on all 34 datasets (Supplementary Fig. S6, available at Rheumatology online). The areas under the curve (AUCs) for most of the datasets (27/34) were >0.7, indicating the model had fair diagnostic power. Next, we evaluated the accuracy of the diagnostic model to differentiate diseases of the same category via a study-to-study transfer validation method (Fig. 3A, Supplementary Fig. S7, available at Rheumatology online). Fitting of datasets of the same type of autoimmune disease (Fig. 3A) demonstrated consistent AUCs compared with other disease categories (Supplementary Fig. S7, available at Rheumatology online), indicating autoimmune diseases may share more similarities in bacterial composition than the other diseases. Taken together, these results imply that autoimmune diseases have higher marker homogeneity than cancers, metabolic diseases and nervous system diseases.
Figure 3.
Disease diagnostic models. Classification accuracy via cross-validation within each study (grey boxes along the diagonal) and study-to-study model transfer (external validations off the diagonal) as measured by the AUC. The blank box indicates the two datasets share the same healthy controls and was excluded in the cross-study validation. Datasets with cross-validation <0.65 are not shown here
The function of L. salivarius in the DSS-induced IBD model
Abnormalities of the gut microbiome are thought to cause inflammation in response to innate and adaptive immune responses, particularly for diseases such as IBD [26, 27]. RA is an autoimmune disease with unknown aetiology, and although joint inflammation is a typical feature of RA, inflammation may develop in other parts of body, such as the gut, years prior [28]. In the current study, we chose IBD and RA as representative autoimmune diseases. To date, very few cohort studies have implicated specific bacterial species in RA [29, 30]. In one such study, L. salivarius was found to be more abundant in very active cases of RA [28-joint DAS (DAS28) >5.1] compared with mild to moderately active (DAS28 ≤5.1) cases [29]. In the current meta-analysis, Lactobacillus was also noted as the third-ranked bacterial genera that was enriched in patients with autoimmune disease. Thus L. salivarius was selected as a candidate bacterial species for further functional interrogation.
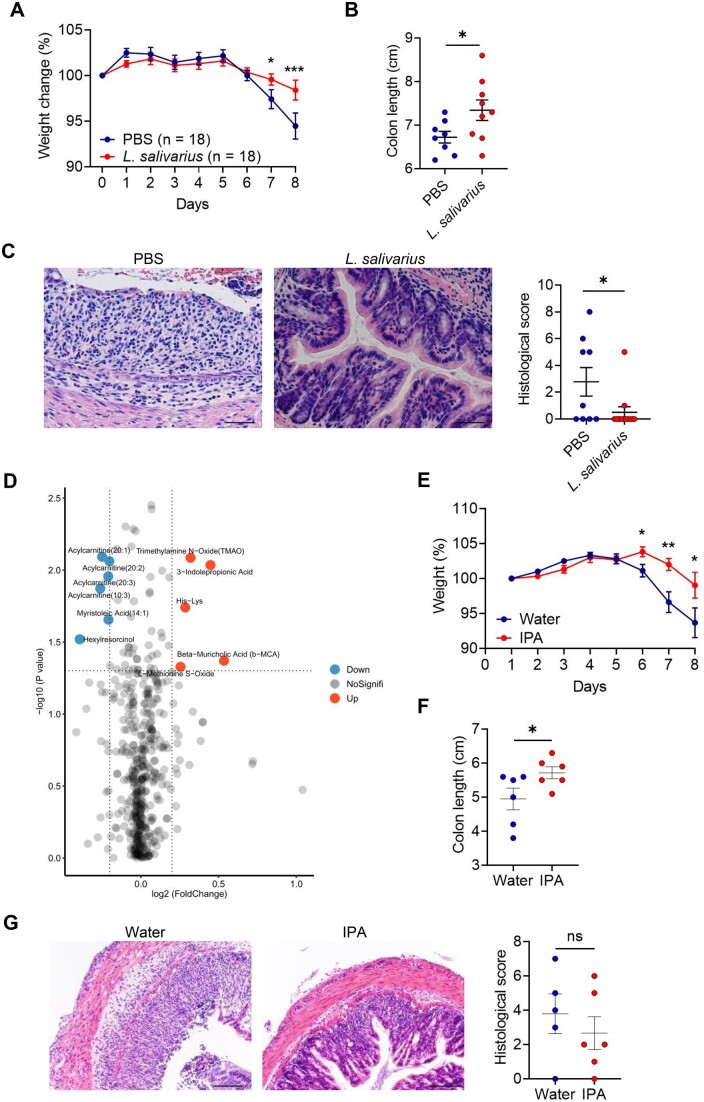
To delineate the role of L. salivarius in intestinal inflammation, we gavaged the antibiotic-pretreated mice with PBS or L. salivarius and then treated the mice with 2% DSS for 7 days. Mice with PBS developed severe clinical symptoms of IBD, including rapid weight loss, severe diarrhoea and bloody stools, compared with mice with L. salivarius (Fig. 4A). Mice with PBS also had a shorter colon than mice with L. salivarius (Fig. 4B). Histological analysis of the colon showed increased severity of disease, including massive inflammatory infiltrates, crypt loss and disruption of mucosal structures in mice with PBS compared with mice with L. salivarius (Fig. 4C). Thus L. salivarius colonization in the gut appeared to relieve colonic inflammation in DSS-treated mice.
Figure 4.
Protection from DSS-induced colon inflammation via the colonization of L. salivarius. To induce colitis, mice with PBS or L. salivarius were administered 2% DSS in drinking water for 7 days. (A) Weight loss of mice with PBS or L. salivarius. (B) Colon length of mice with PBS or L. salivarius. (C) Representative H&E staining of distal colon sections and histological score of mice with PBS or L. salivarius. (D) Differential analysis for serum metabonomics of mice with PBS or L. salivarius at homeostasis. (E) Weight loss of mice with water or IPA. (F) Representative H&E staining of distal colon sections and histological score of mice with water or IPA. Data are represented as mean (s.e.m.). *P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001 by unpaired Student’s t-test. Scale bar = 40 μm
In order to explore how the colonization of L. salivarius protected mice from DSS-induced colon inflammation, we administered PBS or L. salivarius to mice at homeostasis and collected the serum of mice on day 7 for metabolomics analysis. Overall, there were six metabolites significantly depleted and five metabolites significantly enriched in the serum of mice with L. salivarius, suggesting that the colonization of L. salivarius influenced the composition of the serum metabolites. Through metabolite screening, we found that indolepropionic acid (IPA) can alleviate DSS-induced intestinal inflammation (Fig. 4D). Compared with normal water supply mice, IPA supplementation significantly protected mice against weight loss (Fig. 4E), increased colon length (Fig. 4F) and decreased intestinal histological score (Fig. 4G), suggesting that IPA can alleviate the colitis phenotype in mice.
The function of L. salivarius in CII-induced RA model
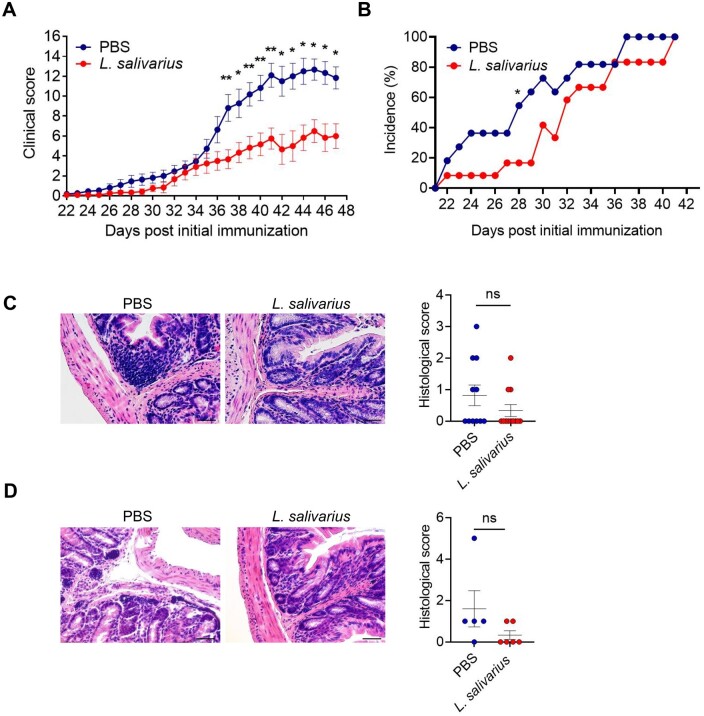
To investigate the role of L. salivarius in RA, we orally administrated the antibiotic-pretreated mice with PBS or L. salivarius and treated the mice with bovine type II collagen immunization at day 0 and day 21. Mice with PBS developed more severe clinical symptoms and a high incidence of RA (Fig. 5A and B). The infiltration of immune cells in the colon of the mice colonized with L. salivarius was reduced (Fig. 5C). Previous studies have shown that intestinal inflammation accompanied by arthritis occurs before the onset of arthritic symptoms [31]. In the current study, the colon histology revealed the onset of arthritis on day 14, and it was found that the colonization of L. salivarius weakened the infiltration of colon immune cells (Fig. 5D). Therefore we hypothesized that the colonization of L. salivarius improved the colon microenvironment in the initial stage of arthritis and alleviated the severity of arthritis in the later stage.
Figure 5.
Colonization of L. salivarius alleviated joint inflammation. To induce arthritis, mice were immunized with CII on day 0 and day 21. (A) Articular scores of mice with PBS or L. salivarius. The paws were evaluated for joint swelling and grip strength three times per week. Each paw was individually scored using a 4-point scale: 0 = normal paw, 1 = minimal swelling or redness, 2 = redness and swelling involving the entire forepaw, 3 = redness and swelling involving the entire limp and 4 = joint deformity or ankylosis or both. (B) The incidence of arthritis of mice with PBS or L. salivarius. (C) Representative H&E staining of distal colon sections and histological score of mice with PBS or L. salivarius in late stage of disease (day 40+). (D) Representative H&E staining of distal colon sections and histological score of mice with PBS or L. salivarius in initiation of disease (day 14). Scale bar = 40 μm
Discussion
This study presents the first meta-analysis of gut microbiota across a broad array of diseases, revealing distinct interdisease association patterns. Overall, 34 publicly available case–control 16S rRNA sequencing studies were collected. These studies encompassed 17 diseases that were broadly grouped into four main categories: autoimmune disease, cancer, metabolic disease and nervous system disease. By reprocessing these data through a standardized bioinformatics pipeline, numerous shared microbial signatures were noted across specific disease subtypes within the same broader disease categorization. In particular, our analysis revealed that compared with other disease categories, signals associated with autoimmune diseases were surprisingly strong and consistent, typically characterized by the enrichment of Enterococcus, Veillonella, Streptococcus, Lactobacillus and Erysipelotrichaceae incertae sedis and the depletion of Ruminococcus, Gemmiger, Oscillibacter, Faecalibacterium, Lachnospiracea incertae sedis, Anaerostipes, Coprococcus, Alistipes, Roseburia, Bilophila, Barnesiella, Dorea, Ruminococcus2, Butyricicoccus, Phascolarctobacterium, Parabacteroides and Odoribacter, among others (Fig. 1). We further validated the function of a bacterial species, L. salivarius, that has been previously reported to be enriched in RA and is also a member of one of the genera noted to be commonly enriched in autoimmune diseases in our study. The functional validation of L. salivarius showed that the colonization of L. salivarius alleviated the intestinal inflammation and arthritic symptoms. For the other broad disease types (cancer, metabolic disease and nervous system disease), the lack of consistent microbial alterations and the lower degree of significant associations overall may be because the microbiomes associated with these broad diseases are more heterogeneous in nature, but also may be due to the heterogeneity of the diseases themselves.
Consistent with previous studies, the strongest microbial link to autoimmune disease was an enrichment of Enterococcus (Fig. 1). Enterococcus gallinarum has been reported to translocate from the small intestine to the liver, driving organ-specific and systemic autoimmunity both in mice and humans [4]. Our results were also consistent with another recent study that revealed an enrichment of Lactobacillus potentially contributes to susceptibility to autoimmune disease. Specifically, this study demonstrated that Lactobacillus reuteri was a commensal species that was unexpectedly linked to exacerbation of CNS autoimmunity [32]. Although the roles of Enterococcus and Lactobacillus in autoimmune disease require further elucidation, and different species within these genera can have either a protective or antagonistic effect, these observations in combination with our meta-analysis support a role for these genera across multiple disease aetiologies (Figs 1 and 2B). Further fine-grained investigations utilizing whole-genome metagenomics are therefore essential for this research to progress. It follows that the broad causal implications of intestinal microbiota such as Enterococcus and Lactobacillus, but also Veillonella and Streptococcus, may be missed in single-disease studies. By combining multiple small cohorts of potentially low generalizability, it is possible to obtain better representation of the spectrum of cases and controls, therefore our meta-analysis highlights numerous bacterial taxa that may previously have been overlooked. Moreover, the general pattern identified in our study should be indicative of a shared response to autoimmune disease, which should be interpreted carefully for future microbial research.
Human diseases are increasingly noted as exhibiting associations with a dysbiotic gut microbiome. However, whether such alterations are causal, consequential or coincidental to disease is unresolved for the most part, and the majority of case–control studies also typically do not attempt to identify the causal microbes. As previously discussed, the current study demonstrated the functional potential of L. salivarius, as the colonization of L. salivarius may alleviate intestinal inflammation by enriching the amount of IPA in the gut. Previous studies have shown that IPA was significantly depleted in the metabolic profile of an IBD cohort, indicating that IPA may also play an important role in human enteritis [33]. Interestingly, L. salivarius was noted to be enriched in patients with arthritic disease and also alleviates joint inflammation in mice. These results indicate that the microbiota associated with disease may not always be pathogenic bacteria, but may be protective, thus highlighting the importance of functional validation in case–control microbiome studies.
In summary, we identified numerous common microbial signatures associated with autoimmune disease, implying a potential common microbial function. The function of L. salivarius contributes key evidence that microbiota associated with autoimmune disease may not always be antagonistic microbes, but may be microbes eliciting a protective or adaptive response to disease.
Supplementary Material
Acknowledgements
We thank the members of C.X. and H.Z.’s group for their inspiring discussions. We also appreciate C. Duvallet’s group for their 16S bioinformatic pipeline.
Contributor Information
Tianjiao Wang, School of Medicine, Tsinghua University, Beijing, China.
Peter R Sternes, Centre for Microbiome Research, School of Biomedical Sciences, Queensland University of Technology, Brisbane, QLD, Australia.
Xue-Kun Guo, School of Medicine, Tsinghua University, Beijing, China.
Huiying Zhao, Sun Yat-sen Memorial Hospital, Guangzhou, China; Guangdong Provincial Key Laboratory of Malignant Tumor Epigenetics and Gene Regulation, Guangzhou, China.
Congmin Xu, Biomap (Beijing) Intelligence Technology Ltd., Beijing, China.
Huji Xu, School of Medicine, Tsinghua University, Beijing, China; Peking-Tsinghua Center for Life Sciences, Tsinghua University, Beijing, China; Department of Rheumatology and Immunology, Changzheng Hospital, Naval Medical University, Shanghai, China.
Supplementary material
Supplementary material is available at Rheumatology online.
Data availability
The data that support the findings of this study are available from the corresponding author (H.X.) upon reasonable request.
Authors’ contributions
T.W., H.X. and C.X. conceived the study. T.W. performed the data collection, taxonomic profiling, machine learning, statistical analyses, bacterial cultures, animal experiments and molecular experiments. X.K.G. provided conceptual advice. T.W. wrote the manuscript with contributions from X.K.G., P.R.S., C.X. and H.X. All authors discussed and approved the manuscript.
Funding
H.X. was supported by the National Natural Science Foundation of China (31821003) and the China Ministry of Science and Technology (2018AAA0100302). H.Z. was supported by Natural Science Foundation of China (81801132 and 81971190) and Guangdong Province Key Laboratory of Malignant Tumor Epigenetics and Gene Regulation (2017B030314026).
Disclosure statement: The authors have declared no conflicts of interests.
References
- 1. Cougnoux A, Dalmasso G, Martinez R. et al. Bacterial genotoxin colibactin promotes colon tumour growth by inducing a senescence-associated secretory phenotype. Gut 2014;63:1932–42. [DOI] [PubMed] [Google Scholar]
- 2. Chung L, Thiele Orberg E, Geis AL. et al. Bacteroides fragilis toxin coordinates a pro-carcinogenic inflammatory cascade via targeting of colonic epithelial cells. Cell Host Microbe 2018;23:203–14.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yin J, Sternes PR, Wang M. et al. Shotgun metagenomics reveals an enrichment of potentially cross-reactive bacterial epitopes in ankylosing spondylitis patients, as well as the effects of TNFi therapy upon microbiome composition. Ann Rheum Dis 2020;79:132–40. [DOI] [PubMed] [Google Scholar]
- 4. Manfredo Vieira S, Hiltensperger M, Kumar V. et al. Translocation of a gut pathobiont drives autoimmunity in mice and humans. Science 2018;359:1156–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wirbel J, Pyl PT, Kartal E. et al. Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. Nat Med 2019;25:679–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Thomas AM, Manghi P, Asnicar F. et al. Metagenomic analysis of colorectal cancer datasets identifies cross-cohort microbial diagnostic signatures and a link with choline degradation. Nat Med 2019;25:667–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Xu H, Yin J.. HLA risk alleles and gut microbiome in ankylosing spondylitis and rheumatoid arthritis. Best Pract Res Clin Rheumatol 2019;33:101499. [DOI] [PubMed] [Google Scholar]
- 8. Fan Y, Pedersen O.. Gut microbiota in human metabolic health and disease. Nat Rev Microbiol 2021;19:55–71. [DOI] [PubMed] [Google Scholar]
- 9. Ruff WE, Greiling TM, Kriegel MA.. Host-microbiota interactions in immune-mediated diseases. Nat Rev Microbiol 2020;18:521–38. [DOI] [PubMed] [Google Scholar]
- 10. Zhernakova A, Withoff S, Wijmenga C.. Clinical implications of shared genetics and pathogenesis in autoimmune diseases. Nat Rev Endocrinol 2013;9:646–59. [DOI] [PubMed] [Google Scholar]
- 11. Russell JT, Roesch LFW, Ördberg M. et al. Genetic risk for autoimmunity is associated with distinct changes in the human gut microbiome. Nat Commun 2019;10:3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Asquith M, Sternes PR, Costello ME. et al. HLA alleles associated with risk of ankylosing spondylitis and rheumatoid arthritis influence the gut microbiome. Arthritis Rheumatol 2019;71:1642–50. [DOI] [PubMed] [Google Scholar]
- 13. Duvallet C, Gibbons SM, Gurry T, Irizarry RA, Alm EJ.. Meta-analysis of gut microbiome studies identifies disease-specific and shared responses. Nat Commun 2017;8:1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010;26:2460–1. [DOI] [PubMed] [Google Scholar]
- 15. Wang Q, Garrity GM, Tiedje JM, Cole JR.. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 2007;73:5261–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mecham BH, Nelson PS, Storey JD.. Supervised normalization of microarrays. Bioinformatics 2010;26:1308–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lawson CE, Harcombe WR, Hatzenpichler R. et al. Common principles and best practices for engineering microbiomes. Nat Rev Microbiol 2019;17:725–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Quince C, Walker AW, Simpson JT, Loman NJ, Segata N.. Shotgun metagenomics, from sampling to analysis. Nat Biotechnol 2017;35:833–44. [DOI] [PubMed] [Google Scholar]
- 19. Hothorn T, Hornik K, van de Wiel MA, Zeileis A.. A Lego system for conditional inference. Am Stat 2006;60:257–63. [Google Scholar]
- 20. Feng J, Meyer CA, Wang Q. et al. GFOLD: a generalized fold change for ranking differentially expressed genes from RNA-seq data. Bioinformatics 2012;28:2782–8. [DOI] [PubMed] [Google Scholar]
- 21. Benjamini Y, Hochberg Y.. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc 1995;57:289–300. [Google Scholar]
- 22. Oksanen J, Simpson GL, Blanchet FG. et al. vegan: Community Ecology Package. Version 1.15. Vienna, Austria: R Project for Statistical Computing, 2009.
- 23. Swami A, Jain R.. Scikit-learn: machine learning in Python. J Mach Learn Res 2012;12:2825–30. [Google Scholar]
- 24. Qin J, Li Y, Cai Z. et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 2012;490:55–60. [DOI] [PubMed] [Google Scholar]
- 25. Ryan FJ, Ahern AM, Fitzgerald RS. et al. Colonic microbiota is associated with inflammation and host epigenomic alterations in inflammatory bowel disease. Nat Commun 2020;11:1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hall AB, Yassour M, Sauk J. et al. A novel Ruminococcus gnavus clade enriched in inflammatory bowel disease patients. Genome Med 2017;9:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lewis JD, Chen EZ, Baldassano RN. et al. Inflammation, antibiotics, and diet as environmental stressors of the gut microbiome in pediatric Crohn’s disease. Cell Host Microbe 2015;18:489–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. McInnes IB, Schett G.. The pathogenesis of rheumatoid arthritis. N Engl J Med 2011;365:2205–19. [DOI] [PubMed] [Google Scholar]
- 29. Zhang X, Zhang D, Jia H. et al. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat Med 2015;21:895–905. [DOI] [PubMed] [Google Scholar]
- 30. He J, Chu Y, Li J. et al. Intestinal butyrate-metabolizing species contribute to autoantibody production and bone erosion in rheumatoid arthritis. Sci Adv 2022;8:eabm1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zaneveld JR, McMinds R, Vega Thurber R.. Stress and stability: applying the Anna Karenina principle to animal microbiomes. Nat Microbiol 2017;2:17121. [DOI] [PubMed] [Google Scholar]
- 32. Montgomery TL, Künstner A, Kennedy JJ. et al. Interactions between host genetics and gut microbiota determine susceptibility to CNS autoimmunity. Proc Natl Acad Sci USA 2020;117:27516–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lloyd-Price J, Arze C, Ananthakrishnan AN. et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 2019;569:655–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author (H.X.) upon reasonable request.