HIGHLIGHTS
-
•
Inflammatory Breast Cancer is a rare but highly aggressive variant responsible for 10 % of breast cancer-related mortality.
-
•
XIAP is involved in the regulation of programmed cell death.
-
•
In IBC, XIAP is associated with more tumor associated macrophages and an immunosuppressive tumor microenvironment.
-
•
The TNF-a signaling pathway is a likely candidate governing macrophage-induced XIAP overexpression in IBC.
-
•
Birinapant, a pan IAP antagonist, induced apoptotic signals by decreasing XIAP and has potential as therapy to overcome an immunosuppressive tumor microenvironment.
Keywords: PD-L1; XIAP; Inflammatory breast cancer (IBC); Tumor immune-microenvironment (TiME); Tumor-associated macrophages (TAM); Tumor necrosis factor - alpha (TNF-α); Birinapant, IAP-antagonist
Abstract
Objective
To assess the expression pattern of X-linked inhibitor of apoptosis protein (XIAP), a cellular stress sensor, and delineate the associated changes in the tumor immune microenvironment (TiME) for prognostic value and new therapeutic targets in inflammatory breast cancer (IBC).
Methods
Immunohistochemistry was conducted to assess the spatial localization of immune subsets, XIAP, and PDL1 expression in IBC and non-inflammatory breast cancer (nIBC) pretreatment tumors (n = 142). Validation and further exploration were performed by gene expression analysis of patient tumors along with signaling studies in a co-culture model.
Results
High XIAP in 37/81 IBC patients correlated significantly with high PD-L1, increased infiltration of FOXP3+ Tregs, CD163+ tumor-associated macrophages (TAMs), low CD8/CD163 ratio in both tumor stroma (TS) and invasive margins (IM), and higher CD8+ T cells and CD79α+ B cells in the IM. Gene set enrichment analysis identified cellular stress response- and inflammation-related genes along with tumor necrosis factor receptor 1 (TNFR1) expression in high-XIAP IBC tumors. Induction of TNFR1 and XIAP was observed when patient-derived SUM149 IBC cells were co-cultured with human macrophage-conditioned media simulating TAMs, further demonstrating that the TNF-α signaling pathway is a likely candidate governing TAM-induced XIAP overexpression in IBC cells. Finally, addition of Birinapant, a pan IAP antagonist, induced cell death in the pro-survival cytokine-enriched conditions.
Conclusion
Using immunophenotyping and gene expression analysis in patient biospecimens along with in silico modeling and a preclinical model with a pan-IAP antagonist, this study revealed an interplay between increased TAMs, TNF-α signaling, and XIAP activation during (immune) stress in IBC. These data demonstrate the potential of IAP antagonists as immunomodulators for improving IBC therapeutic regimens.
Introduction
Although breast cancer has historically not been considered among the malignancies with a highly active immune microenvironment, studies have clearly demonstrated that the presence of tumor-infiltrating lymphocytes (TIL) prior to treatment can predict the response to therapy and improve prognosis in breast cancer patients [1] including those diagnosed with inflammatory breast cancer (IBC) [2], a rare but highly aggressive and lethal variant [3]. Apoptosis, a type of programmed cell death [4] plays a crucial role in modulating both immune- and non-immune-mediated responses in the tumor microenvironment (TiME) [5,6]. Induction of tumor cell apoptosis by CD4+/CD8+ cytotoxic T cells and natural killer cells (NK) occurs via the intrinsic cell death pathway with the release of cytotoxic granules (perforin, granzyme) and/or the extrinsic death pathway via engagement of death receptors FAS, TNFR, and TRAILR. These multi-step apoptotic cascades eventually lead to the activation of effector caspases-3/−7 [7,8] and are regulated by X-linked inhibitor of apoptosis protein (XIAP), a member of the IAP family, and are considered the most potent inhibitors of mitochondrial/intrinsic and extrinsic death receptor-mediated apoptotic signaling. XIAP suppresses granzyme and caspase activation when overexpressed [9], [10], [11] and plays a substantial role in the crosstalk between the NFκB and MAPK pro-inflammatory pathways, which are generally induced during the activation of immune cells. Characterization of XIAP knock-out mice [12] shows the requirement of XIAP in regulatory T- cell (Treg) function, as Tregs isolated from these mice had reduced suppressive capacity without a decrease in total number [13].
Interestingly, XIAP is one of a handful of eukaryotic proteins that have an internal ribosomal entry sequence (IRES). This sequence allows for the initiation of the cap-independent protein translation during cellular stress when the normal protein synthesis that occurs via cap-dependent pathway is suppressed to allow for repair and elimination of mutated cells [14]. Furthermore, XIAP is known to play a key role in postpartum breast remodeling [15], which requires a balance of cell death, immune, and inflammatory factors in the breast [16]. This is particularly relevant in IBC, wherein recent epidemiological studies have identified reproductive factors, including multiparity, as potential risk factors [17]. Previous studies, including ours characterizing XIAP function in IBC patient-derived cell lines exposed to therapeutic and oxidative stressors, evaluated stress factors in IBC [18]. These studies showed increased XIAP expression due to its IRES activity and suppressed cell death in response to these stress stimuli [19,20]. Interestingly, in murine xenograft models, XIAP knockdown suppresses tumor growth and migration of IBC cells [21]. In addition, when IBC cell lines were characterized by immune assays with XIAP overexpression, XIAP knockdown, or selected for drug resistance, it was found that IBC cells exhibiting XIAP overexpression were associated with resistance to both antibody-dependent cellular cytotoxicity (ADCC) mediated by monoclonal antibodies cetuximab (anti-EGFR) and trastuzumab (anti-HER2), as well as T-cell-mediated lysis [22]. Transcriptome analysis also identified differential expression of genes involved in immunosuppression and oxidative stress response in XIAP-overexpressing cells [23]. These observations, coupled with the paucity of data on XIAP and immune subsets in breast cancer, in particular IBC, provided the rationale for investigating the impact of differential XIAP expression on the type and spatial distribution of immune cells and PDL1 expression in IBC and subtype-matched nIBC patients.
Materials and methods
Patients
This study was conducted in accordance with the guidelines of the Declaration of Helsinki and was approved by the ethical committee of the Antwerp University Hospital. Written informed consent was obtained from each patient in the study and by the Duke University Institutional Review Board. Patient tumors pathologically confirmed as invasive breast carcinoma and diagnosed with IBC using the clinical definition agreed upon by international experts were selected (n = 81) from a retrospective cohort collected [2,24] from patients at diagnosis (open or core biopsy) between June 1, 1996, and December 31, 2017, at GZA Hospital Sint-Augustinus (Antwerp, Belgium), Antwerp University Hospital (Edegem, Belgium), or Institut Paoli-Calmettes (Marseille, France). These samples had complete hospital records (including pathologic response data) along with estrogen (ER) and progesterone receptor (PR) expression scores defined as positive if Allred score ≥ 3/8 and designated HER2-positive when a fluorescence in situ hybridization (FISH) test documented amplification. Pathological complete response (pCR) was defined as the absence of residual invasive carcinoma in the resected breast specimen (mastectomy) and in all sampled regional lymph nodes after completion of anthracycline/taxane-based neoadjuvant chemotherapy (NACT), with most HER2+ patients receiving targeted therapy with Trastuzumab (n = 17/30). Pretreatment nIBC biopsy samples (n = 61) were randomly selected from a previously reported [2] retrospective cohort sampled in 2006 to match the same period in which most IBC cases were diagnosed. Exclusion criteria included IBC disease, previous breast cancer treatment, diagnosis of ductal carcinoma in situ, or loss of follow-up, and all these nIBC patients had received adequate local and systemic treatment after a pathologically confirmed diagnosis. We decided to subtype-match, rather than stage-match the nIBC and the IBC cohort because Van Laere et al. compared differentially expressed genes in IBC to alternatively composed nIBC control groups (i.e. non-stage matched, advanced stage (III-IV) only, or low stage (I-II) only) and demonstrated a very high concordance of 87 % between the lists of differentially expressed genes between IBC versus the 3 nIBC groups. They concluded that stage matching had limited influence [25]. Furthermore, IBC can be considered as an early but rapid evolving disease, while stage III nIBC cases might often reflect later stage disease. The time between the oncogenic insult, the elicited immune-response and the symptoms might therefore be better comparable with early nIBC cases.
Stromal tumor-infiltrating lymphocytes (sTIL) scoring
TIL scoring was performed on hematoxylin and eosin (H&E)-stained 5-μm sections of formalin-fixed paraffin-embedded (FFPE) pretreatment tumor tissue by two different researchers and independently scored by JG, a board-certified pathologist with expertise in breast cancer biomarker analysis, according to the recommendations of the International TILs Working Group [2]. Given the specific pathology of IBC with often small and dispersed tumor cell nests, TILs were reported for the stromal compartment (% stromal TILs, sTIL) in all areas containing invasive tumor cells on the H&E slide containing the most invasive tumor.
Immunohistochemistry
Consecutive (5 µm thick) FFPE slides were stained for the following validated antibodies: CD79α for activated B cells and plasma cells, CD8 for cytotoxic T-cells, FOXP3 for Tregs, and CD163 for tumor-associated macrophages (TAM). Staining was performed using Bond/Leica or Benchmark/Ventana Autostainers. Antibodies were visualized using HRP 3,3′-diaminobenzidine () and the slides were counterstained with hematoxylin. All stained slides were digitized using a digital slide scanner (3DHISTECH, Hungary). PD-L1 expression was assessed (clone SP142, Ventana Benchmark) in the tumor (TC) and infiltrating immune cells (IC), as previously described [26]. Scoring was performed by two different researchers based on the percentage of the tumor area that was occupied by PD-L1+ immune cells or the percentage of PD-L1+ tumor cells per tumor area, with consensus scoring followed in cases of any differences between the researchers. For XIAP staining, the slides were incubated in a 1:60 dilution of mouse anti-human XIAP (BD Biosciences) overnight at 4 °C, washed, and incubated with anti-mouse secondary antibody (Dako anti-mouse Envision kit) for 30 min at room temperature. Imaging was performed on a Zeiss Axio Observer A1 microscope, and images were analyzed using MetaMorph. The slides were manually scored in a blinded manner by a board-certified pathologist (JG) as previously reported [27]. Staining intensity was graded on a qualitative scale (0, 0.5, 1, 2, 3) and for statistical analysis, tumors with a score > 1 were considered “XIAP high.”
Image analysis
All stained slides were digitized for further evaluation using VISIOPHARM® software. The first quality check was performed after the staining and scanning. During quality control, we checked for out-of-focus scanning, staining artifacts, and disconnected tissue. If possible, the slides were scanned again; otherwise, the patients were excluded. All slides were evaluated to quantify the number of DAB+ immune cells and the relative marker area (RMA: DAB+ area/total area of interest) in the tumor stroma (TS) and invasive margin (IM). Subsequently, we scored the slides for immune cell density (number of DAB+ immune cells/total area of interest). Immune cell positivity was defined using specific image analysis algorithms for every staining, and each image analysis algorithm was validated using 10 images in which the positive immune cells were manually marked by a trained pathologist.
Gene expression analysis
Gene expression data from patients with and without IBC were generated and used as described before [25,28]. Briefly, both mRNA and XIAP protein expression data were available for 30 IBC and 18 nIBC patients. For these patients, XIAP mRNA was evaluated using five informative probe sets and XIAP activation scores were calculated using single-sample gene set enrichment analysis (ssGSEA; GSVA package) based on sets of genes repressed and overexpressed upon XIAP activation [21]. The final XIAP activation score was determined by subtracting the ssGSEA scores generated using the repressed gene set from those generated using the overexpressed gene set. To identify biological processes associated with XIAP protein expression, generalized linear regression models were set up, comparing the XIAP protein expression to mRNA expression levels for all genes in the data set. The gene wise regression coefficients were then subjected to GSEA (fgsea package) for the hallmark gene sets of the Molecular Signatures Database (Broad Institute). In addition, regression modeling was repeated by incorporating molecular subtypes as blocking variables to account for confounding effects. Finally, CIBERSORT [29] was run in absolute mode using the immunedeconv package, and the resulting quantifications of various immune sets were compared with XIAP protein expression using correlation analysis.
To delineate IBC cell-macrophage interactions that govern XIAP overexpression and activation, expression data of tumor biopsies from 68 and 132 patients with respectively IBC and nIBC were selected [26]. To mitigate the influence of stromal expression on the results, selection was based on a minimal cancer cell fraction of 80 %, scored using the algorithm provided by the R-package estimate. For these samples, the XIAP activation score was calculated as described above and dichotomized according to the median. Next, differences in gene expression were calculated using generalized linear models (BioC-package limma) and analyzed using the BioC-package VIPER to identify master regulators of XIAP activation. The VIPER algorithm was run in multi-sample mode without a null model, and protein activity levels were virtually inferred using a regulon of breast cancer specific protein-target gene interactions obtained through the BioC-package aracne.networks. In addition, no pleiotropic interactions were considered. Master regulators of XIAP were defined as proteins with significantly increased activity level in XIAP active samples (i.e. normalized enrichment score > 0 and false discovery corrected p-value inferior to 10 %) that additionally directly interact with XIAP in the STRING network (https://string-db.org) of physical protein-protein interactions (PPIs). Next, identified master regulators of XIAP activation were mapped onto the same PPI network and shortest paths were calculated linking any of the master regulators with any receptor of a ligand secreted by M2 macrophages. Only receptors overexpressed in IBC or nIBC tissue samples (Z-transformed mean expression value > 1.96) were included in this analysis. The resulting subgraph was subjected to louvain clustering and normalized node degree statistics were calculated. All network analyses were done using the R-package igraph package and network visualization was performed using R-packages tidygraph, ggraph, and ggpubr. Finally, a heat diffusion algorithm (R-package diffusr) was used on the resulting networks to identify the most likely receptor that regulates XIAP expression and activation. The heat diffusion algorithm was initiated using the scaled virtually inferred protein activities of the XIAP master regulators as initial weights. Heat accumulation in each node of the network was recorded over 41 time points ranging from 0 to 20 by steps of 0.5 and visualized.
Cell lines and reagents
SUM149 cells were obtained from Asterand Inc. (Detroit, MI) and routinely cultured in Ham's F12 medium (Mediatech Inc., Manassas, VA) supplemented with 5 % FBS (Atlanta Biologicals, Lawrenceville, GA), 1 % penicillin/streptomycin, 1 % antibiotic/antimycotic, hydrocortisone (Invitrogen, Carlsbad, CA), and an insulin/transferrin/selenium cocktail (Gibco, Carlsbad, CA).
Co-Culture of tumor cells with macrophage conditioned media
To generate macrophage-conditioned media, peripheral blood mononuclear cells isolated from leukopak material (Stemcell Technologies) from three independent de-identified donors were cultured in 6 well plates (one per donor) for 1 hour in Aim-V (Gibco) media to allow attachment of monocytes. Non-adherent cells were removed and macrophage differentiation media containing DMEM, 10 % FBS, and 50 ng/mL M-CSF (Stemcell Technologies) was added. Media was also added to a plate without monocytes to serve as mock media. The plates were incubated for 7 days to generate monocyte-derived macrophages (MDMs), after which MDM-conditioned media (MDM-CM) or mock media were pooled and frozen at −80 °C for downstream studies. TNF-α levels in the mock media and MDM-CM were measured using the human anti-virus legendplex kit (BioLegend), with data collected on a BD FACs Canto analyzer (Duke Cancer Institute Flow Cytometry Core) and analyzed using the manufacturer (BioLegend). Minimum threshold values were used if cytokine concentrations were below the detection sensitivity (automatically calculated by the analysis software). SUM149 cells were plated at a density of 7.5 × 104 cells per well in 6-well plates in 1.8 mL of SUM149 growth medium and allowed to adhere overnight. Cells were treated with control media (SUM149 growth media or mock media) or MDM-CM as treatment alone or supplemented with 1000 nM Birinapant for 24 and 96 h.
Trypan blue exclusion assay
After treatment, the top medium of each well with non-adherent cells was collected and spun down. The cells were then resuspended in media, and 10uL of cell suspension was added to 10uL of 0.4 % trypan blue (Thermo Fisher, Waltham, MA). 10uL of the mixture was loaded into a counting chamber slide that was read by the Countess II Cell Counter (Invitrogen, Waltham, MA). Viability of Birinapant-treated cells was calculated as the fold change of non-adherent cells compared to control media.
Western immunoblot
Cells from each treatment group were pooled from their respective triplicate wells and lysed using 150 mM sodium chloride, 1 % Triton X-100, 50 mM Tris pH 8.0, 100X Halt Protease Inhibitor Cocktail (Thermo Fisher, Waltham, MA) and 1 M DTT (ThermoFisher, Waltham, MA). Protein concentration was determined using Pierce 660 nm Protein Assay Reagent (ThermoFisher, Waltham, MA) and read at 660 nm with a Nanodrop 2000 (ThermoFisher, Waltham, MA). The cell lysates were boiled for 6 min and immediately cooled on ice. The lysates were then subjected to gel electrophoresis on NuPage 4–12 % Bis-Tris gels (Invitrogen, Waltham, MA) with MES SDS Page running buffer (Invitrogen, Waltham, MA). The protein was transferred onto a nitrocellulose blotting membrane (Amersham, St. Louis, MO) previously soaked in transfer buffer by the XCell II Blot Module transfer cell (Life Technologies, Carlsbad, CA). After the transfer, the membranes were incubated with blocking buffer (5 % BSA in 1X TBS-0.1 % Tween 20) for 1 hr in room temperature. Membranes were incubated with primary antibodies for XIAP, cIAP1, SOD2, TNFR1 (all 1:1000, Cell Signaling Technology), or GAPDH (1:5000, Santa Cruz Biotechnology) overnight at 4 °C. Membranes were washed and incubated with anti-mouse or anti-rabbit HRP-conjugated antibodies (Santa Cruz Biotechnology, Dallas, TX) for 1 hour at room temperature. Chemiluminescent reagent (Thermo Scientific, Waltham, MA) and LI-COR Odyssey FC imager with Image Studio software (LI-COR) was used for detection. Densitometric analysis was performed using the Image Studio Lite software.
Statistical analysis
Statistical analysis of preclinical data was performed using GraphPad Prism Version 9 (GraphPad Software, Inc.) using Student's two-tailed t-test. Clinical data were analyzed using R studio (version 1.1.463, using the following packages: dplyr, tidyr, survival, survminer, and ggplot2). To assess the differences between the clinicopathological, XIAP, and immune parameters in the nIBC and IBC cohorts, we used the Pearson Chi2 test (categorical variables) and Kruskal-Wallis (continuous variables) tests. The same tests were used to compare the XIAP high versus the XIAP low group and a multivariate logistic regression model included all significant parameters. Survival data were updated by December 31, 2021, and patients were censored at the date of their last follow-up visit or event. The evaluated survival endpoints were disease-free survival (DFS), defined as the period from the date of diagnosis to the date of cancer recurrence; metastasis-free survival (MFS), defined as the interval between the date of diagnosis and distant relapse; and breast cancer-specific survival (BCSS), defined as the interval between pathological diagnosis and death. Survival curves were calculated using Kaplan-Meier estimates. The log-rank test was used for comparison. P-values (2-sided) were considered statistically significant at P< 0.05.
Results
Clinicopathological characteristics
We investigated expression of XIAP and a panel of immune subsets in pretreatment invasive breast cancer samples by comparing IBC with subtype-matched nIBC cases (Table 1 summarizes the patient clinicopathological parameters and Table S1 details the M0-stage IBC and nIBC cases). As we selected the nIBC cases to subtype match the IBC cohort, there were no differences in hormone receptor (HR) and HER2 expression. Compared to nIBC cases, IBC presented, more often, with high grade disease (P = 0.039).
Table 1.
Clinicopathological parameters of the study cohort. Comparison between nIBC and IBC was done using a Chi-square test. nIBC: non-inflammatory breast cancer, IBC: inflammatory breast cancer, HR: Hormone receptor, sTIL: stromal tumor infiltrating lymphocytes and NACT: neoadjuvant chemotherapy.
| Parameter | IBC (n = 81) | nIBC (n = 61) | p-value | |
|---|---|---|---|---|
| Age | 61.8 [32.9 - 91.0] | 59.1 [30 - 86.1] | 0.67 | |
| Menopausal status | No | 23 | 18 | 0.99 |
| Yes | 58 | 43 | ||
| APO | Ductal | 76 | 54 | 0.53 |
| Lobular | 3 | 4 | ||
| Mixed | 2 | 3 | ||
| Differentation | Grade 1 | 1 | 4 | 0.039 |
| Grade 2 | 19 | 24 | ||
| Grade 3 | 55 | 33 | ||
| cT | 1 | 0 | 18 | < 0.001 |
| 2 | 0 | 33 | ||
| 3 | 0 | 7 | ||
| 4 | 81 | 3 | ||
| cN | 0 | 3 | 29 | < 0.001 |
| 1 | 29 | 17 | ||
| 2 | 32 | 11 | ||
| 3 | 17 | 4 | ||
| cM | 0 | 57 | 61 | < 0.001 |
| 1 | 24 | 0 | ||
| HR | 0 | 34 | 25 | 0.99 |
| 1 | 47 | 36 | ||
| HER2+ | 0 | 50 | 37 | 0.96 |
| 1 | 30 | 24 | ||
| Subtype | HER2+ | 30 | 24 | 0.96 |
| HR-HER2- | 18 | 14 | ||
| HR+ | 32 | 23 | ||
| PD-L1 Positivity | Negative | 46 | 42 | 0.15 |
| Positive | 35 | 18 | ||
| sTIL Infiltration | < 10% | 30 | 15 | 0.29 |
| 10 - 40 % | 39 | 35 | ||
| > 40 % | 12 | 11 | ||
| XIAP overexpression | No | 44 | 49 | 0.002 |
| Yes | 37 | 12 |
XIAP expression associated with sTIL infiltration and PD-L1 expression on the immune cells in invasive breast tumors
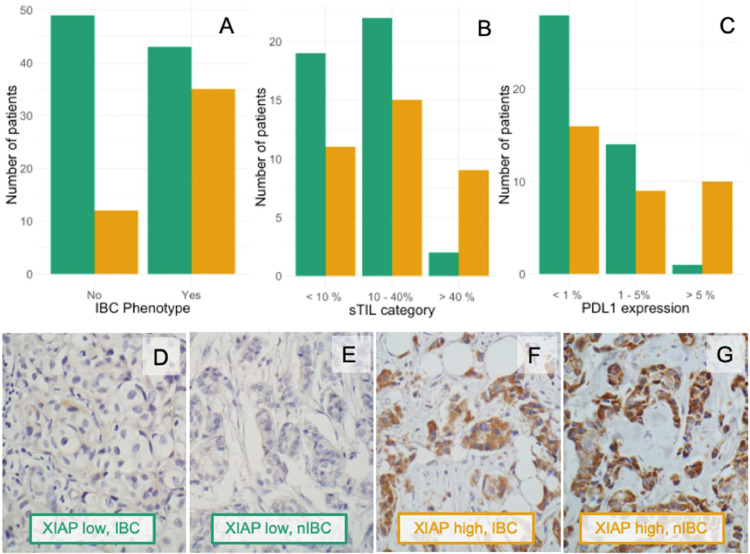
Immunohistochemical analysis of XIAP showed granular cytoplasmic staining in positive tumor cells. Average expression in a tumor was qualitatively graded from 0 (negative) to 3 (strong) and tumors with a score of >1 were considered high. Based on this, almost half of the IBC patients (n = 37/81, 45.6 %) had high XIAP expression. This was significantly more than in the nIBC cohort (P = 0.002), in which 19.6 % of patients (n = 12/61) had high XIAP levels (Fig. 1A). sTIL infiltration was observed in both IBC and nIBC samples, with no significant differences (Table 1). XIAP expression was associated with sTIL infiltration (Fig. 1B) and PD-L1 positivity (Fig. 1C) in the total cohort [PD-L1 expression (P = 0.004) and sTIL infiltration (P = 0.028)] and in the IBC cohort [PD-L1 expression (P< 0.001) and sTIL infiltration (P = 0.05)] (Table 2, Table S2A). In a multivariate model including all significant parameters, only high PD-L1 expression on the immune cells (≥ 5 %, P = 0.0089) and the IBC phenotype (P = 0.0023) remained significant for XIAP high tumors. Within the 61 nIBC samples, we did not observe any significant associations (Table S2B).
Fig. 1.
(A) Barplot of high XIAP associated with the IBC phenotype (P = 0.002) (B) Barplot of high XIAP associated with high sTIL infiltration in IBC (P = 0.028). (C) Barplot showing high XIAP in high PDL1 expressing IBC tumors (P = 0.004). (D-G) Representative slides of patients with low (green, D. IBC & E. nIBC) and high (orange, F. IBC & G. nIBC) XIAP expression. Original magnifications 400x. IBC: inflammatory breast cancer, nIBC: non-inflammatory breast cancer; sTIL: stromal tumor infiltrating lymphocytes. Green bars: XIAP low, Orange bars: XIAP high.
Table 2.
Chi-square (univariate) analysis and multivariate analysis with logistic regression of the clinicopathological parameters associated with XIAP overexpression in the total cohort. nIBC: non-inflammatory breast cancer, IBC: inflammatory breast cancer, HR: hormone receptor status, sTIL: stromal tumor infiltrating lymphocytes, OR: odds ratio, CI: Confidence Interval.
| Parameter |
P-value Univariate |
OR (95 % CI), P-value Multivariate |
|---|---|---|
| Phenotype: IBC vs. nIBC | 0.0033 | 3.86 (1.67 – 9.62), 0.0023 |
| Menopausal state: pre vs. postmenopausal | 0.52 | / |
| APO: Ductal vs. Lobular vs. mixed | 0.50 | / |
| Differentation: Grade 1 vs.Grade 2 vs. Grade 3 | 0.06 | / |
| Nodal disease: Negative vs. Positive | 0.92 | / |
| Distant disease: Negative vs. Positive | 0.29 | / |
| HR: Negative vs. Positive | 0.96 | / |
| HER2+: Negative vs. Positive | 0.92 | / |
| PD-L1 Positivity: <1% vs. 1 – 5 % < 1% vs. ≥ 5 % |
< 0.001 | 0.73 (0.27 – 1.89), 0.53 6.04 (1.67 – 26.07), 0.0089 |
| Stromal TILs: < 10% vs. 10 - 40 % < 10% vs. ≥ 40 % |
0.052 | 1.26 (0.52 – 3.19), 0.61 2.55 (0.68 – 9.59), 0.16 |
XIAP overexpression and differential pattern of immune infiltrates in tumor stroma and invasive margins of IBC vs nIBC
Based on the abovementioned data identifying a strong association between XIAP expression and infiltration with sTIL, we investigated the composition of the immune microenvironment in the tumor stroma and if there were any differences between low and high XIAP in nIBC (Table S3) and IBC (Table S4) tumors. To address this, we conducted an immunohistochemical analysis for CD79α for activated B-cells and plasma cells, CD8 for cytotoxic T-cells, FOXP3 for Tregs, and CD163 for TAMs in consecutive sections and assessed these immune infiltrates by looking at the number of immune cells and the ratio of immune cells in the IM compared to the number of immune cells in the TS in relation to XIAP expression.
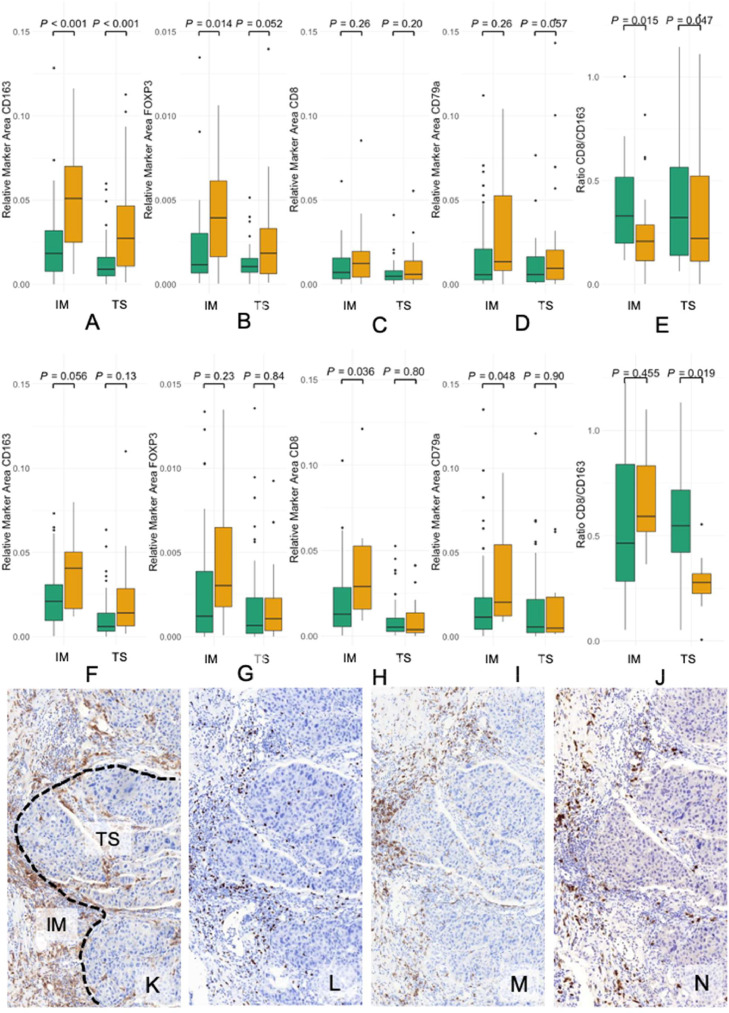
In the nIBC cohort (n = 61, Table S3), high XIAP expressing tumors had higher numbers of CD8+ cytotoxic T-cells [XIAP+: 2.91 % (0.9 – 12.11) vs. XIAP-: 1.29 % (0.03 – 10.26), P = 0.036] and CD79α+ B-cells [XIAP+: 2.86 % (0.88 – 30.29) vs. XIAP-: 1.24 % (0.03 – 27.89), P = 0.048] in the IM (Fig. 2F-I) compared to relative lower levels of CD8 [XIAP+: 0.22 (0.1 – 0.55) vs. XIAP-: 0.44 (0.06 – 5.28), P = 0.013] and CD79α+ [XIAP+: 0.25 (0.15 – 0.66) vs. XIAP-: 0.53 (0.1 – 19.01), P = 0.019] in the TS (Fig. S1A-B). This reduced infiltration of CD8+ T-cells in XIAP high tumors also corresponded with relatively higher CD163+ macrophages in the TS [XIAP+: 0.29 (0.01 – 3.66) vs. XIAP-: 0.72 (0.05 – 22.07), P = 0.019] (Fig. 2J).
Fig. 2.
Boxplots of the different immune cell types and XIAP in IBC (first row, A - E) and nIBC (second row, F-J) cohorts. (A) Boxplot of the RMA of CD163 showing significantly more infiltration in the high XIAP cohort in both the IM (P< 0.001) and TS (P< 0.001). (B) Boxplot of the RMA of FOXP3 showing significantly more infiltration in the high XIAP cohort in the IM (P = 0.014), but not the TS (P = 0.052). (C) Boxplot of the RMA of CD8 showing no significant differences. (D) Boxplot of the RMA of CD79α showing no significant differences. (E) Boxplot of the ratio of CD8/CD163 infiltration. In both the IM (P = 0.015) and the TS (P = 0.047) this ratio was lower in the high XIAP tumors. (F) Boxplot of the RMA of CD163 showing no significant differences. (G) Boxplot of the RMA of FOXP3 showing no significant differences. (H) Boxplot of the RMA of CD8 showing significantly more infiltration in the IM of high XIAP cohort (P = 0.036), but not in the TS (P = 0.80) in nIBC. (I) Boxplot of the RMA of CD79α showing significantly more infiltration in the IM of the high XIAP nIBC tumors (P = 0.048), but not the TS (P = 0.90). (J) Boxplot of the ratio of CD8/CD163 infiltration. In the TS (P = 0.019) this ratio was lower in the high XIAP nIBC. (K-N) Immunostainings of one patient with high XIAP expression for (K) CD163 (L) FOXP3 (M) CD8 and (N) CD79α. RMA = Relative Marker Area, IM= invasive margin, TS= tumor stroma. Green bars: XIAP low, Orange bars: XIAP high.
Compared to the nIBC samples, in the IBC cohort (n = 81, Table S4) XIAP high tumors had increased infiltration of CD163+ TAM in both TS [XIAP+: 2.73 % (0.11 – 11.26) vs. XIAP-: 0.89 % (0.0 – 5.97), P< 0.001] and in the IM [XIAP+: 5.23 % (0.62 – 18.57) vs. XIAP-: 1.84 % (0.0 – 12.84), P< 0.001] (Fig. 2A-D). Furthermore, the ratio of cytotoxic T-cells compared to TAM was lower in both the IM [XIAP+: 0.22 (0.0 – 3.29) vs. XIAP-: 0.43 (0.11 - NA), P = 0.015] and the TS [XIAP+: 0.26 (0.0 – 3.45) vs. XIAP-: 0.46 (0.06 – NA), P = 0.047] in XIAP high samples (Fig. 2E). Finally, FOXP3+ Tregs were higher in high XIAP tumors, especially in the IM [XIAP+: 0.39 % (0.0 – 1.06) vs. XIAP-: 0.12 % (0.01 – 1.35), P = 0.014] (Fig. 2A-D). Overall, in a multivariate setting, high PD-L1 expression, CD163+ TAM infiltration, and a low CD8/CD163 ratio remained significantly associated with high XIAP expression in IBC (Table 3). Collectively, these findings suggest that XIAP expression is associated with a lower level of cytotoxic immune cell infiltration in nIBC and a CD163-dominated immunosuppressive tumor microenvironment in IBC.
Table 3.
Univariate, Kruskal wallis analysis and multivariate analysis with logistic regression of the immune parameters significantly associated with XIAP overexpression in the IBC cohort. *The median value was used for dichotomization. RMA: Relative Marker Area IBC: inflammatory breast cancer, HR: hormone receptor status, sTIL: stromal tumor infiltrating lymphocytes, OR: odds ratio, CI: Confidence Interval.
| Parameter | Median (Range) | Pval | OR (95 % CI) | Pval |
|---|---|---|---|---|
| RMA CD163 IM* < 3%vs. ≥ 3 % |
XIAP+: 5.23 % (0.62 – 18.57) XIAP-: 1.84 % (0.0 – 12.84) |
<0.001 |
0.86 (0.08 – 7.97) |
0.90 |
| RMA CD163 Tumor* < 1.35%vs. ≥ 1.35 % |
XIAP+: 2.73 % (0.11 – 11.26) XIAP-: 0.89 % (0.0 – 5.97) |
<0.001 |
12.19 (1.65 – 145.76) |
0.023 |
| RMA FOXP3 IM* < 0.24%vs. ≥ 0.24 % |
XIAP+: 0.39 % (0.0 – 1.06) XIAP-: 0.12 % (0.01 – 1.35) |
0.014 |
4.29 (0.72 – 34.18) |
0.13 |
| Ratio CD8/CD163 IM* < 0.41vs. ≥ 0.41 |
XIAP+: 0.22 (0.0 – 3.29) XIAP-: 0.43 (0.11 – NA) |
0.015 |
7.22 (1.04 – 79.15) |
0.07 |
| Ratio CD8/CD163 Tumor* < 0.29vs. ≥ 0.29 |
XIAP+: 0.26 (0.0 – 3.45) XIAP-: 0.46 (0.06 – NA) |
0.047 |
0.087 (0.009 – 0.54) |
0.017 |
| PD-L1 Positivity <1% vs. 1 – 5 % < 1% vs. ≥ 5 % |
<0.001 |
0.87 (0.18 – 4.22) 15.75 (1.35 – 464.89) |
0.53 0.05 |
|
| Stromal TILs < 10% vs. 10 - 40 % < 10% vs. ≥ 40 % |
0.052 |
0.65 (0.086 – 4.13) 1.93 (0.069 – 83.52) |
0.66 0.70 |
To validate this with the IHC data, we examined the number of patients that had respectively high XIAP expression and more influx of TAMs (HH), high XIAP expression and less influx of TAMs (HL), low XIAP expression and more influx of TAMs (LH) and low XIAP expression and less influx of TAMs (LL). This was significantly different between IBC and nIBC (P = 0.014). In both IBC and nIBC, XIAP overexpression was uncommon when TAM infiltration was low (HL) (n = 8/78, 10.2% vs. 3/54, 5.6 %). The biggest difference between IBC and nIBC was seen in the HH (n = 27/78, 34.6% vs. n = 7/54, 13.0 %, P = 0.021) and LL (n = 23/78, 29.5% vs. n = 27/54, 50.0 %, P = 0.068) group. Seemingly demonstrating that lower XIAP expression in nIBC could be caused by lesser infiltration with TAMs, although TAM infiltration did not always cause XIAP overexpression (LH, n = 20/78, 25.6% vs. n = 17/54, 31.5 %, P= NS) (Fig. S1C).
Relationship between XIAP, immune subsets, and clinical outcome in IBC
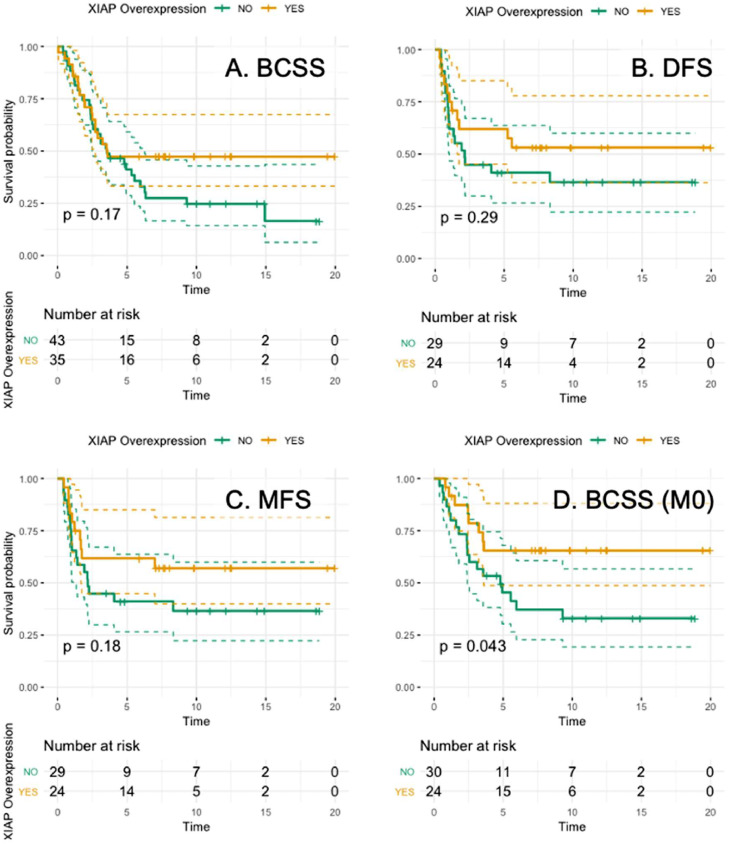
Next, we assessed how the expression levels of the immune subsets and XIAP expression in pretreatment samples of IBC patients who underwent mastectomy after NACT correlated with pCR. PD-L1 expression (P = 0.05) and infiltration of both CD8+ T cells (P = 0.02) and CD163+ TAM (P = 0.004) were associated with pCR, but high XIAP was not (P = 0.12). The median follow-up was 10.4 years (95 % CI:7.32 – 14.12) with a median BCSS of 3.55 years (95 % CI:2.71 – 6.34). No significant relationship was found between high XIAP levels and BCSS (P = 0.17), DFS (P = 0.29), or MFS (P = 0.18) (Fig. 3A-C). However, interestingly, in the group of non-metastatic IBC patients, we observed a correlation between high XIAP and longer BCSS (HR:0.44, 95 %CI:0.20 – 0.99, P = 0.04) (Fig. 3D).
Fig. 3.
Kaplan-Meier curves for (A) BCSS (P = 0.17), (B) DFS (P = 0.29), (C) MFS (P = 0.18) and (D) BCSS in the non-metastatic setting (P = 0.04) in IBC.
Cellular stress response- and inflammation-related gene set enrichment in XIAP expressing IBC tumors
To validate the immunohistochemistry findings, gene expression data from 30 IBC and 18 nIBC were analyzed. In this series and in line with previous results, XIAP protein expression was significantly lower in nIBC than in the IBC series (i.e. 0.528 unit difference; P< 0.001) and this observation was not biased by alternative subtype distributions (i.e. 0.362 unit difference after correction for PAM50 subtypes – P = 0.047). In addition, XIAP protein expression was not significantly different between IBC samples stratified according to the PAM50 molecular subtypes (Kruskal-Wallis test, P = 0.314), demonstrating the minimal impact of the molecular subtypes as potential confounding variables.
Although gene expression data analysis did not reveal any correlation between protein and mRNA expression of XIAP for any of the five probes assessed, there was a significant correlation between an XIAP activation signature identified in the well characterized pretreatment patient-derived SUM149 IBC cell line [27] and the protein expression in the IBC patient tumors (Rs=0.460; P = 0.010) (Fig. S2A). Furthermore, we discovered a strong association between XIAP protein expression and overexpression of genes related to cellular stress response- & inflammation-related gene sets (Fig. S2B). Interestingly, genes overexpressed in IBC tumor samples with low XIAP protein expression are associated with processes related to cell proliferation (i.e. DNA Repair: NES=−1.590; E2F target genes: NES=−1.506; early estrogen receptor response: NES=−1.351; MYC target genes: NES=−1.393 and oxidative phosphorylation: NES=−1.249; All FDR<10 %). Within the nIBC series, no correlation was observed with the XIAP activity score as described above for IBC (Fig. S2C). Gene set enrichment analysis (GSEA) of XIAP associated genes identified a distinct set of metabolic hallmark processes (mTORC1 signaling, PI3K-AKT-mTOR signaling, ROS signaling, glycolysis, protein secretion) for high XIAP nIBC (Fig. S2D).
Finally, XIAP protein expression in IBC revealed strong correlations with infiltration scores for M2 macrophages, neutrophils & activated dendritic cells (Fig. S3A). In contrast, in nIBC only activated NK cells amongst all the immune cell types quantified by means of CIBERSORT correlated with XIAP expression (Fig. S3B).
XIAP-overexpressing tumor cells express high levels of TNFR1
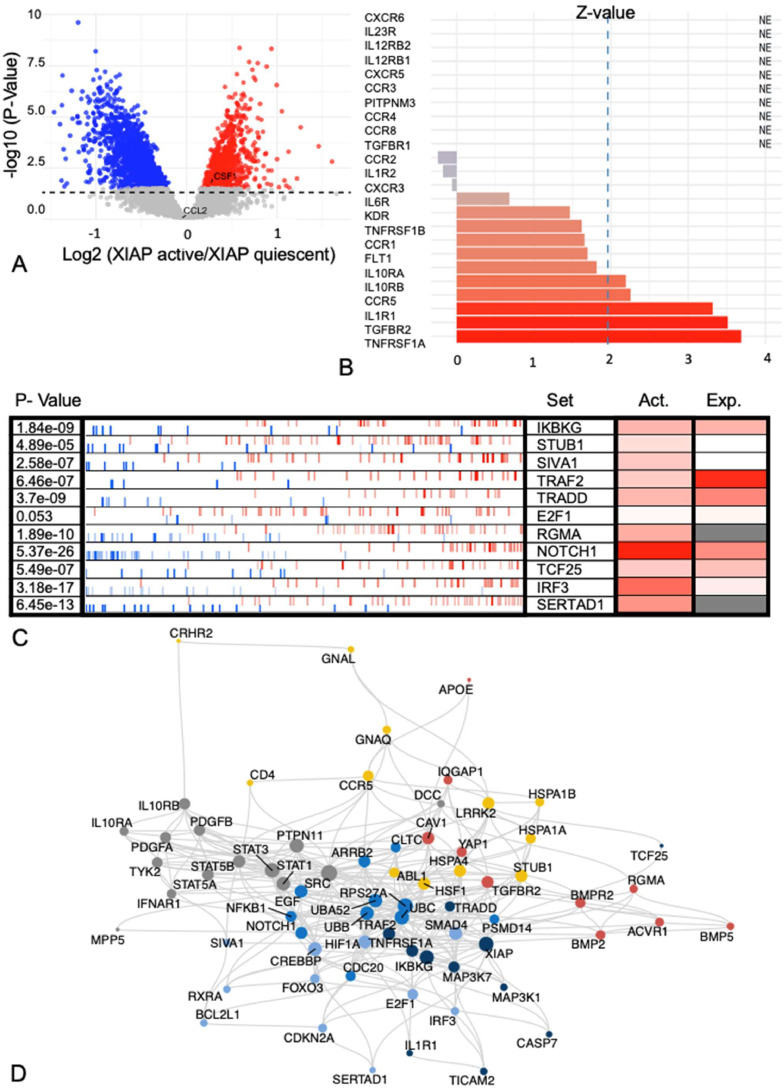
To evaluate paracrine interactions between cancer cells and M2 macrophages that govern XIAP overexpression in IBC, we first evaluated the gene expression data of tissue biopsies from patients with IBC described above. To mitigate the influence of stromal expression on the results, this analysis was restricted to 68 IBC samples having cancer cell fractions exceeding 80 %. For these samples, the XIAP activation score calculated earlier [27] was used to classify the IBC biospecimens as XIAP active or quiescent by dichotomizing the XIAP score vector to the median XIAP activity score. Using generalized linear models, 2605 differentially expressed genes were identified of which 818 (31.40 %) were overexpressed in XIAP activated IBC samples. When evaluating ligands that either engage macrophage chemotaxis (i.e. CCL1, CCL2, CSF1, or CSF2) or induce macrophage M2 polarization (i.e. IL4, IL10, or IL13), only CSF1 and CCL2 exhibit detectable mRNA levels but expression differences between XIAP active and quiescent IBC samples were only noted for CSF1 (Fig. 4A).
Fig. 4.
(A) Volcano plot looking at mRNA expression differences between XIAP active and quiescent IBC samples shows that only CSF-1was differentially expressed. (B) Expression levels of receptors for 14 ligands expressed by macrophages. Significant overexpression (i.e. Z-value>1.96) was seen for TNFRSF1A, TGFBR2, IL1R1 and CCR5. IL10RA tended to be overexpressed in IBC (Z-value=1.81). (C) Master regulators of XIAP activation in IBC were determined using the VIPER algorithm. (D) Shortest paths between any receptor/master regulator pair were calculated on the PPI network STRING, identifying a fully connected network containing 69 nodes and 329 edges.
Next, we investigated the expression of receptors for factors specific to M2 macrophages in the same series. For this analysis, we selected (based on available literature) 14 ligands secreted by different subtypes of M2 macrophages: CCL1, CCL17, CCL18, CCL22, CCL24, CXCL13, IL1, IL6, IL10, IL12, IL23, TGF-β1, TNF-ɑ, and VEGFA and corresponding receptors for each of these ligands (i.e. CCR1, CCR2, CCR3, CCR4, CCR5, CCR8, CXCR3, CXCR5, CXCR6, FLT1, IL1R1, IL1R2, IL6R, IL10RA, IL10RB, IL12RB1, IL12RB2, IL23R, KDR, PITPNM3, TGFBR1, TGFBR2, TNFRSF1A, and TNFRSF1B). Based on the vector of Z-transformed mean expression values calculated across all genes in the expression series, IL10RA overexpression (Z-value=1.81) along with significant overexpression (i.e. Z-value>1.96) of TNFRSF1A, TGFBR2, IL1R1, CCR5 and IL10RB was identified (Fig. 4B).
Thirdly, master regulators of XIAP activation in IBC were determined using the VIPER algorithm, wherein differences in gene expression between XIAP active and quiescent IBC samples were translated into virtually inferred protein activity scores using breast cancer specific regulons. This analysis revealed 587 regulators of XIAP activity in IBC cells (false discovery rate inferior to 10 % and NES>0). This list was prioritized by considering only proteins that directly interact with XIAP in the protein-protein interaction (PPI) network STRING, resulting in the identification of 11 potential XIAP master regulators: E2F1, IKBKG, IRF3, NOTCH1, RGMA, SERTAD1, SIVA1, STUB1, TCF25, TRADD, and TRAF2 (Fig. 4C). To delineate potential signal transduction mechanisms linking one or more receptors of ligands secreted by M2 macrophages to any of the 11 XIAP master regulators identified, shortest paths between any receptor/master regulator pair were calculated on the PPI network STRING. This analysis identified a fully connected network containing 69 nodes and 329 edges (Fig. 4D). The network was clustered, and 6 identified communities were largely centered around the different cytokine receptors (Fig. S4A).
Finally, to identify the receptor from Fig. 4B that correlates with high XIAP in IBC tumor cells, a heat diffusion analysis was performed on the network using the scaled virtually inferred protein activity scores of the XIAP master regulators as starting temperatures. Heat accumulation by each of the receptors for M2 secreted cytokines, representing the information flow from the XIAP master regulators through the network to the receptors, was measured over 41 time points ranging from 0 to 20 by steps of 0.5 (Fig. S4B). Heat accumulation was the strongest for TNFRSF1A, revealing it as the most likely candidate receptor wherein ligand TNF-α binding and downstream signaling could mediate paracrine interactions between high XIAP expressing IBC tumor cells and M2 macrophages (known to secrete TNF-α [30]) in the TiME. Furthermore, we correlated the VIPER activity scores of the master modulators also to the XIAP protein expression. Positive correlations were found for SERTAD1, TCF25, IKBKG, TRAF2, and IRF3. Interestingly 3 out of 5 master modulators with positive correlations belonged to the network cluster that involves TNFRSF1A (Fig S4, cluster 6) corroborating the importance of this cluster and the associated signaling pathway (TNF-α) in steering XIAP activity in IBC. The fact that not all master modulators exhibit a positively correlated activity with XIAP protein expression can be explained by the fact that the master modulator analysis was done using the XIAP activity score and the correlation between the XIAP activity score and the protein expression was not perfect. Interestingly, we identified the same mechanism in which M2 macrophages can lead to XIAP overexpression via TNF-α also in nIBC (Fig. S5 & Fig. S6).
XIAP and TNFR1 expression induced in IBC cells in a TAM simulated co-culture
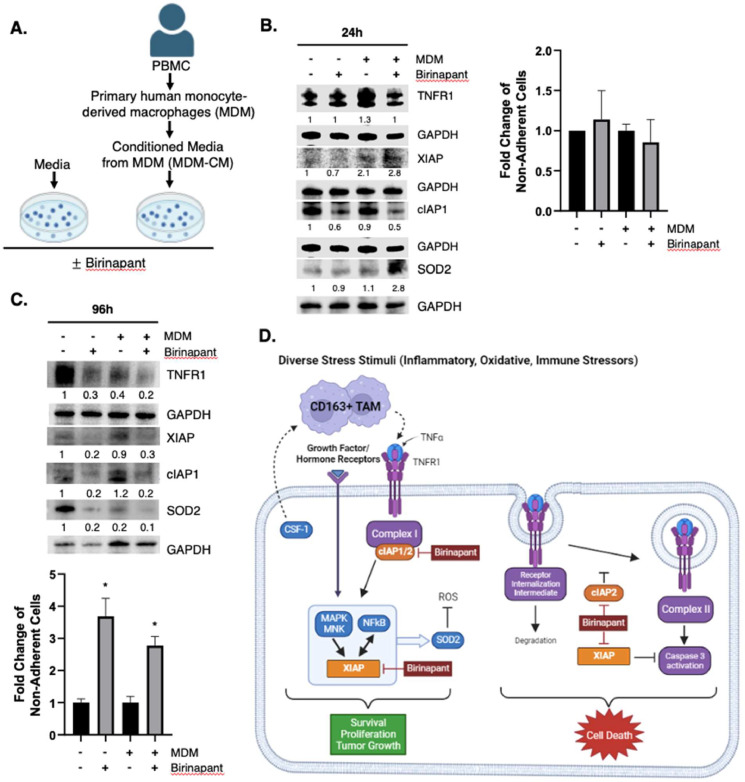
Based on the aforementioned data that XIAP-overexpressing IBC patient tumors have high levels of TNFR1 and increased infiltration of TAMs, we investigated their expression pattern in a simulated assay (Fig. 5A) comprising of IBC patient-derived, treatment naïve SUM149 cells cultured in primary human monocyte-derived macrophage conditioned media (MDM-CM). TNF-α is routinely detected in MDM-CM [31], and we confirmed that the pooled MDM-CM from 3 donors used in these experiments contained 3747.42 pg/mL TNF-α which was undetected in control media. Immunoblot analysis of the lysates at 24 h from SUM149 cells treated with MDM-CM compared to cells treated with control media reveals increased TNFR1 and XIAP levels (Fig. 5B; lanes 1, 3) with no change in cell viability of tumor cells growing in a TNF-α-enriched environment provided by the MDM-CM (Fig. 5B, graph). Overall, these results reveal that XIAP acts as a stress sensor contributed by factors like TNF-α in the tumor microenvironment and its upregulation fosters pro-survival and anti-apoptotic signaling.
Fig. 5.
Birinapant inhibits TNFR1 signaling, XIAP and SOD2 expression in a TAM-simulated co-culture model. (A) Conditioned media from primary human monocyte-derived macrophages (MDM-CM) was collected. MDM-CM and control media was prepared with or without 1000 nM Birinapant and used to treat SUM149 cells for 24 and 96 h. Western immunoblot analysis of indicated targets in SUM149 cells treated with control media or with MDM-CM (± 1000 nM Birinapant) for 24hr. (B) or 96 h (C). Densitometry values for each target signal normalized to loading control GAPDH in the Western blot images in B, C. Graphs represent mean fold change of non-adherent cells ± SEM (3); *P < 0.05) as determined by trypan blue exclusion for each indicated condition. (D) Schema of pan-IAP antagonism in TNFR1 pathway. The current data shows that patient tumor samples with high levels of XIAP are associated with increased infiltration of TAMs, high expression of TNFR1 and CSF1. Macrophages secrete TNF-α, which upon binding to TNFR1 leads to the recruitment of pro-survival factors including cIAP1 that form Complex I, which activates MAPK signaling that promotes XIAP expression and downstream NFκB target genes like SOD2 that have the ability to clear reactive oxygen species (ROS) induced during immune stress and protect against cell death. Birinapant degrades cIAP1/2 and inhibits XIAP and its caspase, which collectively fosters signaling toward Complex II formation and caspase mediated cell death.
IAP antagonist Birinapant inhibits XIAP and TNFR1 to induce cell death in IBC cells under TAM simulated conditions
Binding of TNF-α to TNFR1 can potentiate tumor cell survival signaling (MAPK and NFκB activation via Complex I), which is largely dependent on the expression of IAP family members. Therefore, we investigated whether pan-IAP antagonism can modulate this signaling toward cell death. Toward this, Birinapant, currently in clinical trials, was used in a TAM simulated co-culture assay. Immunoblot analysis was carried out with cell lysates from Birinapant- treated SUM149 in control media and when cultured in the MDM-CM media for 24 h (Fig. 5B; lanes 2, 4) and 96 h (Fig. 5C; lanes 2, 4). Although there was a decrease in target IAP1 levels in Birinapant-treated cells at the 24 h time point, an increase in XIAP as well as SOD2, an XIAP-activated NFκB target gene involved in suppression of reactive oxygen species (ROS) mediated apoptosis during cellular stress, was also observed. Additionally, this corresponded with no change in tumor cell viability (Fig. 5B graph). In contrast, Birinapant treatment for a longer time period (96 h) caused inhibition of IAP1; and TNFR1, XIAP and SOD2 that were induced in the presence of TNF-α enriched conditions (Fig. 5C; lanes 2, 4). This corresponded with a significant decrease in tumor cell viability in control media (P = 0.0342) and MDM-CM (P = 0.0123) (Fig. 5C graph). Collectively, these datasets demonstrate that the Birinapant-mediated inhibition of IAP1 (necessary for complex I formation) and XIAP, which fosters MAPK and NFκB survival signaling and antioxidant expression, while also inhibiting caspase activation and apoptosis, causes increased cell death in the tumor cells growing in the cytokine enriched MDM-CM culture conditions (Fig. 5D).
Discussion
To the best of our knowledge, this is the first study to describe the spatial distribution of immune subsets in relation to XIAP expression patterns. Our findings showed that IBC tumors with high XIAP protein expression are characterized by an immunosuppressive tumor microenvironment comprising increased levels of CD163+ TAMs, a low CD8/CD163 ratio, and higher PD-L1 expression. This was confirmed by deconvolution analysis, which revealed strong correlations between XIAP and M2 macrophages and activated dendritic cells in IBC. This is significant as the immune effectors like T and NK cells kill tumor cells by activating extrinsic death receptor pathway (engagement of death receptors FAS, TNFR and TRAILR) and/or the intrinsic granule exocytosis (perforin, granzyme) and macrophages and neutrophils predominantly activate oxidative stress (reactive oxygen species/ROS) mediated cell death. Expression of high levels of anti-apoptotic factors like XIAP by the tumor cells, as seen in IBC, can lead to evasion of immune-mediated cell death by suppressing granzymes and caspase activation [9], [10], [11]. Furthermore, this study demonstrates the immunomodulatory benefit of targeting TNFR1 signaling to induce cell death in IBC cells using Birinapant, a pan-IAP antagonist in clinical trials.
Emerging evidence in the past five years has reported the importance of tumor stroma and infiltrating immune cells in IBC tumors [2,32,33] including higher mutational burden [34,35] along with the expression of signatures associated with tertiary lymphoid structures and T-cell-inflamed signature [26]. Importantly, higher levels of sTIL are not only associated with a better outcome; sTIL in IBC also seem to function and behave differently, as shown by their different responses to NACT, compared to nIBC [36]. We demonstrated that XIAP is associated with a specific TiME in both IBC and nIBC. In nIBC, XIAP expression was associated with a lower influx of cytotoxic immune cells, whereas in IBC, a CD163-dominated immunosuppressive TiME was observed. A major question that remains not fully answered is whether a pre-existing immune microenvironment with more TAMs in IBC is responsible for higher XIAP protein expression or vice versa. However, based on gene expression data of the IBC-IC dataset in which we selected 68 samples with cancer cell fractions exceeding 80 %, it seems that XIAP overexpression in IBC cells does not induce a tumor-suppressive phenotype in adjacent macrophages via paracrine interactions (Fig. 4A); rather, TAMs can induce XIAP overexpression in IBC cells (Fig. 4B-4D). This hypothesis is further corroborated by the observation that tumor-infiltrating CD163+ macrophages are present in IBC tumors with low XIAP expression. Using in silico modeling and cell line experiments, we further demonstrated that the TNF-α-signaling pathway is a likely candidate that governs M2 macrophage-induced overexpression of XIAP in IBC cells.
We previously reported XIAP overexpression in invasive breast tumors (including IBC) compared to normal, benign and DCIS breast samples [22,37]. We identified that XIAP acts as an oncogenic signaling intermediate that links MNK (ser/thr kinase in the growth factor receptor/MAPK pathway) with NFκB signaling in cellular (immune) stress [27] and showed that XIAP gene expression in breast cancer is associated with shorter survival and resistance to chemotherapy [38]. Furthermore, a XIAP-driven adaptive stress response signature with gene sets was associated with poor survival outcomes [39]. In the current study, we observed that high XIAP expression in non-metastatic IBC patients was associated with a better overall survival outcome. Although unexpected, this observation is supported by the data in this study wherein, compared to high XIAP-metastatic IBC TiME that have increased presence of immunosuppressive TAM and Tregs, the non-metastatic high-XIAP IBC TiME show increased cytotoxic CD8+ T-cell infiltrates along with lower ratio of CD8/CD163 in the IM and TS (Fig. S7). This highlights the role of XIAP as a stress sensor and immunomodulator such that initially XIAP protein expression is associated with increased infiltration of all types of immune cells. However, the abundance of immunosuppressive polarized TAMS [40] that produce TNF-α can act via NFκB and other pathways to cause chronic inflammation. Chronic activation of NFκB can result in the production of inflammatory cytokines, leading to chronic inflammation and eventually promoting tumor progression via proliferation, stemness, angiogenesis, invasion, and metastasis [41,42]. Therefore, we hypothesized that XIAP protein expression may function as a marker of the initial immune cell response. The more immunogenic environment of IBC might induce cellular stress that is countered by upregulation of XIAP as shown in previous studies [18,19] such that XIAP upregulation is an immediate-early response to cellular stress caused by immune cell activation, but the chronic exposure to TNF-α will lead to further XIAP upregulation and subsequent evasion of immune-mediated cell death. Consequently, in metastatic IBC, which may be considered later stage disease in which the tumor seems to have evaded the immune response, there was a borderline significant association with increased TAMs (Fig. S7), and increased infiltration of Tregs. The prognostic benefit of XIAP has been completely lost. Furthermore, our observations are based on XIAP protein levels, which are along the lines of evidence for translational control of XIAP through the IRES during cellular stress [43].
Overall, it seems that IBC tumor cells are addicted to TNFR1 and NFκB signaling pathways that are at least partially induced by the higher infiltration with CD163+ TAMs that produce TNF-α in the TME. Subsequent XIAP activation will put a forward loop in motion that will lead to lead to evasion of immune-mediated cell death and chronic activation of NFκB (Fig. 5D). This aligns with our previous findings demonstrating the higher levels of XIAP in IBC cells during cellular stress [22,44,45]. Interestingly, also in nIBC, M2 macrophages could lead to XIAP overexpression via TNF-α. Collectively, these results reveal that chronic cellular and immune stress (example provided by TNF-α) in the tumor microenvironment can exacerbate resistance to therapeutic agents. Therefore, we tested whether Birinapant, a biindole-based bivalent mimetic of naturally occurring Smac protein that we and others [46], [47], [48] have shown can induce potent tumor cell death in breast cancer cells, albeit with different potency, which depends on the amount of birinapant- induced autocrine secretion of TNF-α by the tumor cells. Our results showing the efficacy of Birinapant in inducing cell death in IBC tumor cells growing in an enriched cytokine environment in the TAM simulated cultures supports the significance of antagonizing the IAP pathway to enhance immunotherapy like with TAM antagonists [49].
This study has a few limitations. Although we characterized the immune subsets in subtype-matched nIBC patient samples, the sample size within each PAM50 subtype was not large enough for robust molecular subtype-specific analysis. Systemic treatment strategies for breast cancer [50] have also evolved during the study period. However, there is still no specific treatment approved for IBC patients, and they are treated with similar regimens approved for nIBC patients, wherein they receive anthracycline/taxane-based neoadjuvant therapy (including HER2 targeting kinase inhibitors if HER2 positive) followed by surgery and radiation. One of the strengths of this study is that gene expression data validated our findings from the immunohistochemical analysis. Furthermore, the gene expression data showed a strong association between XIAP and inflammation-related gene sets in IBC. Additionally, although we mostly reported RMA as a marker for immune cell infiltration in this study, we also looked at our results using density as a marker and observed similar results, providing further validation.
Conclusion
In this study we demonstrated that IBC is characterized by XIAP overexpression. Furthermore, high XIAP-expressing IBC tumors show increased infiltration of immunosuppressive TAMs and are associated with inflammation-related and cellular stress response gene signatures. Immune stress caused by TAMs seems to induce XIAP overexpression in IBC cells via the TNF-α-signaling pathway. Therefore, overcoming the observed immunosuppressive tumor microenvironment in IBC by combining XIAP antagonists with immunotherapeutics, including therapies targeting TAMs, has the potential to improve clinical outcomes in IBC.
Declarations
Ethics approval and consent to participate
The study was conducted in accordance with the guidelines of the Declaration of Helsinki and ethical standards of the University of Antwerp. The study was approved by the Ethics Committee of Antwerp University Hospital (File number:16/33/338) and Duke University (Pro00087933 and Pro00078454).
Availability of data and material
The datasets used and/or analyzed during the current study are available from the corresponding authors on reasonable request.
Competing interests
The authors declare no potential conflicts of interest.
Funding
This work was supported by a PhD grant for CVB from the Research Foundation—Flanders (FWO) [grant number 1189617N], by the Ligue Nationale Contre le Cancer & Ruban Rose for FB and by the Department of Defense awards W81XWH-17–10297, W81XWH-20–10153 and NIH award from NCI RO1CA264529–02 for GRD.
CRediT authorship contribution statement
Christophe Van Berckelaer: Writing – review & editing, Writing – original draft, Visualization, Project administration, Methodology, Investigation, Funding acquisition, Formal analysis, Data curation, Conceptualization. Steven Van Laere: Writing – review & editing, Writing – original draft, Supervision, Methodology, Formal analysis, Data curation, Conceptualization. Seayoung Lee: Writing – review & editing, Writing – original draft, Methodology, Investigation. Michael A Morse: Methodology, Investigation. Joseph Geradts: Methodology, Investigation. Luc Dirix: Resources, Funding acquisition, Data curation. Mark Kockx: Visualization, Software, Resources, Project administration. François Bertucci: Resources, Investigation, Funding acquisition, Data curation. Peter Van Dam: Writing – review & editing, Writing – original draft, Resources, Project administration, Investigation, Data curation, Conceptualization. Gayathri R Devi: Writing – review & editing, Writing – original draft, Supervision, Methodology, Investigation, Funding acquisition, Formal analysis, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors thank Dr. Shannon McCall at the Duke BioRepository & Precision Pathology Center (Duke BRPC), a shared resource of the School of Medicine and the Duke Cancer Institute and Stacy Murray, regulatory coordinator at the Duke Surgery Office of Clinical Research for their assistance.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2024.101907.
Contributor Information
Peter Van Dam, Email: Peter.vandam@uza.be.
Gayathri R Devi, Email: gayathri.devi@duke.edu.
Appendix. Supplementary materials
References
- 1.Loi S., et al. The journey of tumor-infiltrating lymphocytes as a biomarker in breast cancer: clinical utility in an era of checkpoint inhibition. Ann. Oncol. 2021;32(10):1236–1244. doi: 10.1016/j.annonc.2021.07.007. [DOI] [PubMed] [Google Scholar]
- 2.Van Berckelaer C., et al. Infiltrating stromal immune cells in inflammatory breast cancer are associated with an improved outcome and increased PD-L1 expression. Breast Cancer Res. 2019;21(1):28. doi: 10.1186/s13058-019-1108-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fouad T.M., T K., Liu D.D., Shen Y., Masuda H., El-Zein R., Woodward W.A., Chavez-MacGregor M., Alvarez R.H., Arun B., Lucci A., Krishnamurthy S., Babiera G., Buchholz T.A., Valero V., Ueno N.T. Overall survival differences between patients with inflammatory and noninflammatory breast cancerpresenting with distant metastasis at diagnosis. Breast Cancer Res. Treat. 2015;152(2):407–416. doi: 10.1007/s10549-015-3436-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reed J.C. Dysregulation of apoptosis in cancer. J. Clin. Oncol. 1999;17(9) doi: 10.1200/JCO.1999.17.9.2941. 2941-2941. [DOI] [PubMed] [Google Scholar]
- 5.Ravi R., et al. Resistance of cancers to immunologic cytotoxicity and adoptive immunotherapy via X-Linked inhibitor of apoptosis protein expression and coexisting defects in mitochondrial death signaling. Cancer Res. 2006;66(3):1730. doi: 10.1158/0008-5472.CAN-05-3377. [DOI] [PubMed] [Google Scholar]
- 6.Gadiyar V, et al. Cell death in the tumor microenvironment: implications for cancer immunotherapy. Cells. 2020;9(10):2207. doi: 10.3390/cells9102207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lalaoui N., Vaux D.L. Recent advances in understanding inhibitor of apoptosis proteins. F1000Research, 2018;7 doi: 10.12688/f1000research.16439.1. p. F1000 Faculty Rev-1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dougan S.K., Dougan M. Regulation of innate and adaptive antitumor immunity by IAP antagonists. Immunotherapy. 2018;10(9):787–796. doi: 10.2217/imt-2017-0185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lewis J., et al. Uncoupling of the signaling and caspase-inhibitory properties of X-linked inhibitor of apoptosis. J. Biol. Chem. 2004;279(10):9023–9029. doi: 10.1074/jbc.M312891200. [DOI] [PubMed] [Google Scholar]
- 10.Holcik M., et al. A new internal-ribosome-entry-site motif potentiates XIAP-mediated cytoprotection. Nat. Cell Biol. 1999;1(3):190–192. doi: 10.1038/11109. [DOI] [PubMed] [Google Scholar]
- 11.Kashkar H. X-linked inhibitor of apoptosis: a chemoresistance factor or a hollow promise. Clin. Cancer Res. 2010;16(18):4496–4502. doi: 10.1158/1078-0432.CCR-10-1664. [DOI] [PubMed] [Google Scholar]
- 12.Harlin H., et al. Characterization of XIAP-deficient mice. Mol. Cell. Biol. 2001;21(10):3604–3608. doi: 10.1128/MCB.21.10.3604-3608.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hsieh W.C., et al. IL-6 receptor blockade corrects defects of XIAP-deficient regulatory T cells. Nat. Commun. 2018;9(1):463. doi: 10.1038/s41467-018-02862-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holcik M., et al. A new internal-ribosome-entry-site motif potentiates XIAP- mediated cytoprotection. Nat. Cell Biol. 1999;1(3):190–192. doi: 10.1038/11109. [DOI] [PubMed] [Google Scholar]
- 15.Owens T.W., et al. Analysis of inhibitor of apoptosis protein family expression during mammary gland development. BMC Dev. Biol. 2010;10:71. doi: 10.1186/1471-213X-10-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Faupel-Badger J.M., Arcaro K.F., Balkam J.J., Eliassen A.H., Hassiotou F., Lebrilla C.B., Michels K.B., Palmer J.R., Schedin P., Stuebe A.M., Watson C.J. Postpartum remodeling, lactation, and breast cancer risk: summary of a national cancer institute–sponsored workshop. J. Natl. Cancer Inst. 2012;105(3):166–174. doi: 10.1093/jnci/djs505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mejri N., et al. Reproductive risk factors of inflammatory breast cancer according to luminal, HER2-overexpressing, and triple-negative subtypes: a case comparison study. Oncol. Res. Treat. 2020;43(5):204–210. doi: 10.1159/000506691. [DOI] [PubMed] [Google Scholar]
- 18.Silvera D., Schneider R.J. Inflammatory breast cancer cells are constitutively adapted to hypoxia. Cell Cycle. 2009;8(19):3091–3096. doi: 10.4161/cc.8.19.9637. [DOI] [PubMed] [Google Scholar]
- 19.Aird K.M., et al. X-linked inhibitor of apoptosis protein inhibits apoptosis in inflammatory breast cancer cells with acquired resistance to an ErbB1/2 tyrosine kinase inhibitor. Mol. Cancer Ther. 2010;9(5):1432–1442. doi: 10.1158/1535-7163.MCT-10-0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Allensworth J.L., et al. XIAP inhibition and generation of reactive oxygen species enhances TRAIL sensitivity in inflammatory breast cancer cells. Mol. Cancer Ther. 2012;11(7):1518–1527. doi: 10.1158/1535-7163.MCT-11-0787. [DOI] [PubMed] [Google Scholar]
- 21.Evans M.K., Brown M.C., Geradts J., Bao X., Robinson T.J., Jolly M.K., Vermeulen P.B., Palmer G.M., Gromeier M., Levine H., Morse M.A. XIAP regulation by MNK links MAPK and NFκB signaling to determine an aggressive breast cancer phenotype. Cancer Res. 2018;78(7):1726–1738. doi: 10.1158/0008-5472.CAN-17-1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Evans M.K., et al. X-linked inhibitor of apoptosis protein mediates tumor cell resistance to antibody-dependent cellular cytotoxicity. Cell Death Dis. 2016;7(1) doi: 10.1038/cddis.2015.412. e2073-e2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nair S., et al. Immunologic targeting of FOXP3 in inflammatory breast cancer cells. PLoS ONE. 2013;8(1):e53150. doi: 10.1371/journal.pone.0053150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van Berckelaer C., et al. A high neutrophil-lymphocyte ratio and platelet-lymphocyte ratio are associated with a worse outcome in inflammatory breast cancer. Breast. 2020;53:212–220. doi: 10.1016/j.breast.2020.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van Laere S.J., et al. Uncovering the molecular secrets of inflammatory breast cancer biology: an integrated analysis of three distinct affymetrix gene expression datasets. Clin. Cancer Res. 2013;19(17):4685–4696. doi: 10.1158/1078-0432.CCR-12-2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bertucci F., et al. Immune landscape of inflammatory breast cancer suggests vulnerability to immune checkpoint inhibitors. Oncoimmunology. 2021;10(1) doi: 10.1080/2162402X.2021.1929724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Evans M.K., et al. XIAP regulation by MNK Links MAPK and NFkappaB signaling to determine an aggressive breast cancer phenotype. Cancer Res. 2018;78(7):1726–1738. doi: 10.1158/0008-5472.CAN-17-1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bertucci F., et al. Gene expression profiles of inflammatory breast cancer: correlation with response to neoadjuvant chemotherapy and metastasis-free survival. Ann. Oncol. 2014;25(2):358–365. doi: 10.1093/annonc/mdt496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen B., et al. Profiling Tumor infiltrating immune cells with CIBERSORT. Methods Mol. Biol. 2018;1711:243–259. doi: 10.1007/978-1-4939-7493-1_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Micheau O., Tschopp J. Induction of TNF receptor I-mediated apoptosis via two sequential signaling complexes. Cell. 2003;114(2):181–190. doi: 10.1016/s0092-8674(03)00521-x. [DOI] [PubMed] [Google Scholar]
- 31.Brown M.C., et al. Viral infection of cells within the tumor microenvironment mediates antitumor immunotherapy via selective TBK1-IRF3 signaling. Nat. Commun. 2021;12(1):1858. doi: 10.1038/s41467-021-22088-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lim B., et al. Inflammatory breast cancer biology: the tumour microenvironment is key. Nature Rev. Cancer. 2018;18(8):485–499. doi: 10.1038/s41568-018-0010-y. [DOI] [PubMed] [Google Scholar]
- 33.Valenza C., et al. The immunogram of inflammatory breast cancer. Cancer Treat. Rev. 2023;119 doi: 10.1016/j.ctrv.2023.102598. [DOI] [PubMed] [Google Scholar]
- 34.Bertucci F., et al. NOTCH and DNA repair pathways are more frequently targeted by genomic alterations in inflammatory than in non-inflammatory breast cancers. Mol. Oncol. 2020;14(3):504–519. doi: 10.1002/1878-0261.12621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liang X., et al. Targeted next-generation sequencing identifies clinically relevant somatic mutations in a large cohort of inflammatory breast cancer. Breast Cancer Res. 2018;20(1):88. doi: 10.1186/s13058-018-1007-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van Berckelaer C., et al. The evolution and prognostic role of tumour-infiltrating lymphocytes and peripheral blood-based biomarkers in inflammatory breast cancer patients treated with Neoadjuvant chemotherapy. Cancers (Basel) 2021;13(18) doi: 10.3390/cancers13184656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arora J., et al. Inflammatory breast cancer tumor emboli express high levels of anti-apoptotic proteins: use of a quantitative high content and high-throughput 3D IBC spheroid assay to identify targeting strategies. Oncotarget. 2017;8(16):25848–25863. doi: 10.18632/oncotarget.15667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Devi G.R., et al. Expression of X-linked inhibitor of apoptosis protein (XIAP) in breast cancer is associated with shorter survival and resistance to chemotherapy. Cancers (Basel) 2021;13(11) doi: 10.3390/cancers13112807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Al Abo M, et al. Adaptive stress response genes associated with breast cancer subtypes and survival outcomes reveal race-related differences. NPJ Breast Cancer. 2022;8(1):73. doi: 10.1038/s41523-022-00431-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Valeta-Magara A., et al. Inflammatory breast cancer promotes development of M2 tumor-associated macrophages and cancer Mesenchymal cells through a complex chemokine network. Cancer Res. 2019;79(13):3360–3371. doi: 10.1158/0008-5472.CAN-17-2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Diep S., et al. Interleukin-1 and nuclear factor Kappa B signaling promote breast cancer progression and treatment resistance. Cells. 2022;11(10) doi: 10.3390/cells11101673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.DiDonato J.A., Mercurio F., Karin M. NF-kappaB and the link between inflammation and cancer. Immunol. Rev. 2012;246(1):379–400. doi: 10.1111/j.1600-065X.2012.01099.x. [DOI] [PubMed] [Google Scholar]
- 43.Lewis S.M., Holcik M. IRES in distress: translational regulation of the inhibitor of apoptosis proteins XIAP and HIAP2 during cell stress. Cell Death Differ. 2005;12(6):547–553. doi: 10.1038/sj.cdd.4401602. [DOI] [PubMed] [Google Scholar]
- 44.Aird K.M., et al. Trastuzumab signaling in ErbB2-overexpressing inflammatory breast cancer correlates with X-linked inhibitor of apoptosis protein expression. Mol. Cancer Ther. 2008;7(1):38–47. doi: 10.1158/1535-7163.MCT-07-0370. [DOI] [PubMed] [Google Scholar]
- 45.Aird K.M. Duke University: Ann Arbor; 2010. Role of X-Linked Inhibitor of Apoptosis Protein in Therapeutic Resistance of Inflammatory Breast Cancer Cells; p. 259. [Google Scholar]
- 46.Xie X., et al. Birinapant Enhances Gemcitabine's Antitumor Efficacy in Triple-Negative Breast Cancer by Inducing Intrinsic Pathway–Dependent Apoptosis. Mol. Cancer Ther. 2021;20(2):296. doi: 10.1158/1535-7163.MCT-19-1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Allensworth J.L. Duke University; Ann Arbor: 2013. Identification and Targeting of Therapeutic Resistance Mechanisms in Inflammatory Breast Cancer; p. 286. [Google Scholar]
- 48.Lalaoui N., et al. Targeting triple-negative breast cancers with the Smac-mimetic birinapant. Cell Death Differ. 2020;27(10):2768–2780. doi: 10.1038/s41418-020-0541-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Singh S., et al. Chemotherapy coupled to macrophage inhibition induces T-cell and B-cell infiltration and durable regression in triple-negative breast cancer. Cancer Res. 2022;82(12):2281–2297. doi: 10.1158/0008-5472.CAN-21-3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rizzo A., et al. KEYNOTE-522, IMpassion031 and GeparNUEVO: changing the paradigm of neoadjuvant immune checkpoint inhibitors in early triple-negative breast cancer. Future Oncol. 2022;18(18):2301–2309. doi: 10.2217/fon-2021-1647. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding authors on reasonable request.