Abstract
Telomeres in the budding yeast Kluyveromyces lactis consist of perfectly repeated 25-bp units, unlike the imprecise repeats at Saccharomyces cerevisiae telomeres and the short (6- to 8-bp) telomeric repeats found in many other eukaryotes. Telomeric DNA is synthesized by the ribonucleoprotein telomerase, which uses a portion of its RNA moiety as a template. K. lactis telomerase RNA, encoded by the TER1 gene, is ∼1.3 kb long and contains a 30-nucleotide templating domain, the largest ever examined. To examine the mechanism of polymerization by this enzyme, we identified and analyzed telomerase activity from K. lactis whole-cell extracts. In this study, we exploited the length of the template and the precision of copying by K. lactis telomerase to examine primer elongation within one round of repeat synthesis. Under all in vitro conditions tested, K. lactis telomerase catalyzed only one round of repeat synthesis and remained bound to reaction products. We demonstrate that K. lactis telomerase polymerizes along the template in a discontinuous manner and stalls at two specific regions in the template. Increasing the amount of primer DNA-template RNA complementarity results in stalling, suggesting that the RNA-DNA hybrid is not unpaired during elongation in vitro and that lengthy duplexes hinder polymerization through particular regions of the template. We suggest that these observations provide an insight into the mechanism of telomerase and its regulation.
Telomeres, the essential protein-DNA elements at the ends of most eukaryotic chromosomes, confer chromosome stability and constitute protective terminal caps for the genetic material of the cell (for a review, see reference 45). Telomeric DNA typically consists of tandem arrays of a precisely repeated 5- to 8-bp sequence (for a review, see reference 17). However, the telomeric repeat units of yeast species have greatly diverged in precision and length (4, 26). The budding yeast Kluyveromyces lactis, with a perfectly repeated 25-bp telomeric repeat unit (27), is one of several budding yeasts with exceptionally large telomeric repeats (26). In Saccharomyces cerevisiae and the fission yeast Schizosaccharomyces pombe, the telomeric sequences are shorter, imprecisely repeated units (5′-TG1–3-3′ and primarily 5′-GGTTACA-3′, respectively [18, 36]). Despite their variability between species, all of these telomeric sequences are specified by the enzyme telomerase.
Telomerase, a ribonucleoprotein (RNP) reverse transcriptase, facilitates the complete biosynthesis and maintenance of telomeres. Telomerase activity, as shown initially for the ciliate Tetrahymena thermophila (12) and subsequently for many other eukaryotes (3, 24, 25, 31, 35, 37, 44), is dependent on an integral telomerase RNA component (13, 14) which contains a sequence that serves as a template for telomeric DNA synthesis (14, 43). While in T. thermophila the templating domain is 5′-CAACCCCAA-3′, complementary to one and a half telomeric repeats (14), in K. lactis it is a 30-nucleotide (nt) RNA sequence complementary to one and a fifth telomere repeat units (27). Specific mutations within the templating domain of both T. thermophila and S. cerevisiae telomerase RNA drastically reduce or alter telomerase activity in vitro and in vivo, suggesting that bases in the template are not simply copied but play crucial roles in active-site functions (8, 9, 33). Active-site functions are also carried by the reverse transcriptase protein component of telomerase (the hTERT gene product), which has been identified in Euplotes aediculatus, yeasts, and human cells (16, 23, 30, 32), confirming that telomerase requires both reverse transcriptase protein and RNA components.
Unlike the extensive RNA genome copying carried out by the more typical reverse transcriptases of viruses and retroposons, the polymerization activity of telomerase is restricted to copying the discrete template portion of the telomerase RNA. In vitro, telomerase elongates a telomeric DNA primer substrate, which aligns within the templating domain via Watson-Crick base pairing (14). The sequence of the oligonucleotide primer determines the positioning of the 3′ end and therefore the site of initiation. Telomerase then extends the primer by polymerization of one nucleotide at a time along the RNA template to the 5′-end boundary. In T. thermophila and S. cerevisiae, mutating the RNA sequence adjacent to the templating domain allows polymerization to proceed beyond the normal template. Hence, telomerase RNA structures or interactions outside of the template also appear to prevent polymerization beyond the template boundary (1, 33). It is not known whether during primer elongation a constant length of template RNA-product DNA hybrid is maintained (monotonic polymerization) or if the RNA-DNA duplex builds up, although it has been proposed that T. thermophila telomerase maintains a minimal 3- to 4-bp hybrid during elongation (21). During in vitro reactions, telomerases from most organisms catalyze multiple rounds of telomere repeat synthesis, and two modes of synthesis, distributive and translocative, have been distinguished. In the distributive synthesis mode, T. thermophila telomerase dissociates from its DNA product and then binds a new primer to repeat the cycle (5, 21). Translocative synthesis, catalyzed by telomerase from T. thermophila, E. aediculatus, Saccharomyces castellii, and human cells, involves repositioning of the 3′ end of the newly elongated primer at the beginning of the template without release of the product (for a review, see reference 11). Such translocation and the resulting processive synthesis of multiple repeats on a single primer in vitro are influenced by interactions of the 5′ end of the primer with an anchor site within a protein and/or RNA component of telomerase (5, 15, 20).
Synthesis of small repeats (such as the 6-nt repeat of T. thermophila) requires minimal relative movement within the telomerase RNP of the built-in RNA template as it crosses the catalytic site of the TERT protein (41). However, the templating domains of the yeast telomerase RNAs that have been identified, TLC1 RNA from S. cerevisiae and TER1 RNA from K. lactis, are considerably larger, posing interesting mechanistic challenges to telomere repeat synthesis. TLC1 RNA contains a templating domain maximally 17 nt long (39). An 11-nt portion of this domain has been shown to be copied (33), although it is rarely copied in its entirety in vitro, resulting in a series of incomplete single-round extension products (3, 23, 33, 34). Since the frequent stalling exhibited in vitro produces variable 3′-end sequences, alignment of the partially redundant template sequence at telomeric ends can presumably take place in multiple registers and may underlie the degeneracy of the telomeric repeat sequences in vivo (3, 33). The templating domain of K. lactis TER1 RNA is theoretically 30 nt, longer than any examined to date, and in contrast to the irregular repeats of S. cerevisiae telomeres, the telomeric repeat units in K. lactis are tandem arrays of perfect copies of a 25-bp sequence (27). These features suggest that K. lactis telomerase faithfully copies its entire template.
We predicted that the properties that enable K. lactis telomerase to combine a precise mode of copying with an exceptionally long template would be important for understanding general mechanistic features of telomerase action. In particular, we anticipated that analysis of telomerase polymerization along a lengthy template would allow in-depth dissection of the steps in primer elongation occurring within a round of repeat synthesis. In addition, K. lactis has already been established as a highly informative and experimentally advantageous model system for studies on telomere maintenance and length regulation (19, 27, 28).
Here we report the identification and characterization of telomerase activity from K. lactis. We show that in vitro, K. lactis telomerase catalyzes no more than a single round of repeat synthesis, remains bound to its elongated DNA products, and stalls at specific positions along the template. Stalled complexes result from position-specific arrest and pausing; both are exacerbated by increased complementarity between DNA product and the RNA template. These observations provide new insights into the mechanism of polymerization by telomerase and have implications for the in vivo functioning of the enzyme.
MATERIALS AND METHODS
Strain construction.
The mutant telomerase RNA yeast strains used in this study (29) were constructed in a K. lactis 7B520 background, using procedures described previously (27).
DNA oligonucleotides.
All oligonucleotides (Cruachem) were purified on denaturing polyacrylamide gels (15% polyacrylamide, 8 M urea) run in 1× Tris-borate-EDTA (TBE). DNA was eluted from excised gel fragments by overnight incubation at 30°C in distilled water with nutation. Purified oligonucleotides were desalted on Sep-Pak C18 columns (Waters). The concentration of purified oligonucleotides was calculated based on (i) taking 1 OD260 (unit of optical density at 260 nm) as representing 30 μg of DNA and (ii) the molecular weight of the individual oligonucleotide.
Extract preparation and fractionation.
K. lactis cells were harvested at an OD595 of 1.2 to 1.5 and resuspended in TMG buffer (10 mM Tris-HCl [pH 8], 1.2 mM MgCl2, 10% glycerol, 0.1 mM EDTA, 0.1 mM EGTA, 1.5 mM dithiothreitol [DTT], 1 μM pepstatin A, 1× EDTA-free COMPLETE protease inhibitor tablet [Boehringer Mannheim]). Whole-cell extracts and S-100 supernatants were prepared essentially as described previously (3) except for extract used in the experiment shown in Fig. 1A, lanes 1 to 5, which was prepared by disrupting cells with a mortar and pestle under liquid nitrogen (7). For partial purification of telomerase, S-100 supernatants at protein concentrations of 10 to 15 mg/ml were adjusted to 0.5 M sodium acetate (pH 8.0) and loaded onto 5-ml disposable columns (Iso-Lab) packed with 2 ml of DEAE-agarose (Bio-Rad) equilibrated in the same buffer. Columns were washed with TMG containing 0.5 M sodium acetate, and telomerase eluted with 3 ml of TMG containing 0.7 M sodium acetate. Fractions were desalted by dialysis (Dispo-dialysers; Spectra/Por) against TMG for 2 h and then aliquoted and stored at −80°C. Mutant cell extracts were prepared alongside wild-type cell extracts, and equivalent amounts of protein (by Bradford assay, 10 to 15 mg/ml) were loaded onto DEAE-agarose columns. TER1 RNA levels were compared by dot blot and hybridization with a labeled TER1 probe to ensure that extracts contained similar telomerase RNA concentrations.
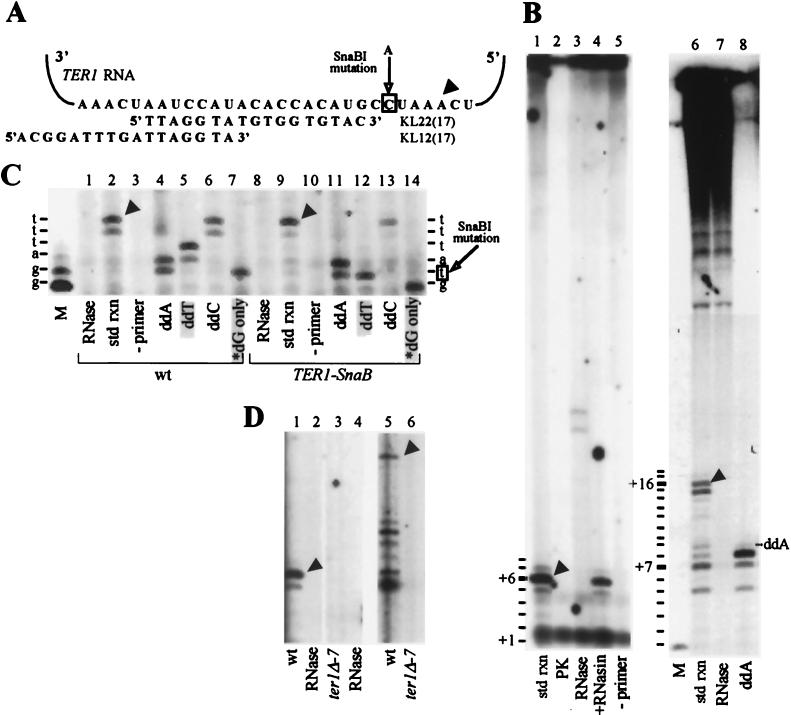
FIG. 1.
Identification of K. lactis telomerase activity in vitro. (A) Schematic of K. lactis telomerase RNA (TER1) template region and the expected alignment of the primers KL22(17) and KL12(17). Arrowhead indicates the 3′ end position of correspondingly marked products in panels B and C. The boxed template residue corresponds to the site of the C-to-A mutation in the Ter1-SnaBI strain examined in panel C. (B) Telomerase reactions were carried out with primers KL22(17) (lanes 1 to 5) and KL12(17) (lanes 6 to 8) as described in Materials and Methods, with the following changes: lane 2, pretreatment of extract with proteinase K (PK; 0.5 mg/ml) for 5 min at 25°C; lanes 3 and 7, pretreatment of extract with RNase A (10 μg/ml) at 25°C for 5 min (lane 3), followed by an additional 5-min incubation with 50 U of RNasin and 1 mM DTT; lane 4, pretreatment of extract with 50 U of RNasin and 1 mM DTT at 25°C for 5 min, followed by RNase A (10 μg/ml) for an additional 5 min; lane 5, no input primer; lane 8, ddATP substituted for dATP, with chain termination product marked. Reactions in lanes 6 to 8 were performed with 7.5 μM [α-32P]dTTP (400 Ci/mmol) as the radioactive label. The primer +1 position for terminal transferase-labeled KL22(17) is shown on the lower left side of lane 1. Product positions up to +8 are marked, but +8 products were visible only on longer gel exposures. Terminal transferase-labeled KL12(17) primer is in the lane marked M. Product positions up to +18 are marked, but +18 products were visible only on longer gel exposures. The high-molecular-weight products at the top of the gel were, as described previously for comparable S. cerevisiae extracts (3, 33), independent of RNase pretreatment or telomerase RNA as described below and were therefore not attributable to telomerase activity. Also, the diffuse band between +1 and +2 in lanes 1 to 5 was not produced by telomerase, as it formed independent of primer and was insensitive to RNase A and proteinase K. Std rxn, standard reaction. (C) Reactions with DEAE-fractionated extracts from wild-type (wt; lanes 1 to 7) and Ter1-SnaBI (lanes 8 to 14) strains were carried out with primer KL22(17) as described in Materials and Methods, with the following changes: lanes 1 and 8, pretreatment with RNase A as described above; lanes 3 and 10, no input primer; lanes 4 to 6 and 11 to 13, substitution of each indicated ddNTP for its partner dNTP; lanes 7 and 14, [α-32P]dGTP as the sole dNTP substrate. Shaded lanes highlight nucleotide incorporation differences between the wild-type and mutant extracts. Terminal transferase-labeled KL22(17) primer (lane M) marks the primer +1 position. The nucleotides predicted to be incorporated by wild-type and Ter1-SnaBI telomerase are marked on the left and right, respectively. (D) Reactions with DEAE-fractionated extracts from wild-type (lanes 1, 2, and 5) and ter1-Δ7 (lanes 3, 4, and 6) were carried out with primer KL22(17) (lanes 1 to 4) and primer KL12(12) (lanes 5 and 6). Extract in lanes 2 and 4 was pretreated with RNase A as described above.
In vitro telomerase reactions.
Unless otherwise indicated, standard 20-μl telomerase reaction mixtures contained 50% (vol/vol) DEAE fraction, 50 mM Tris-HCl (pH 8), 1 mM spermidine, 1 mM DTT, 50 μM dTTP, 50 μM dATP, 50 μM dCTP, 3.75 μM [α-32P]dGTP (800 Ci/mmol), and 1 μM gel-purified oligonucleotide. Reactions were incubated at 30°C for 20 min, stopped with the addition of 2.5 μl of stop buffer (2% sodium dodecyl sulfate [SDS], 250 mM Tris-HCl [pH 8], 250 mM EDTA) and 2.5 μl of proteinase K (20 mg/ml), and incubated at 65°C for 25 min. An equal amount of terminal transferase-labeled 10-mer was added to each reaction after the stop to monitor the recovery of the products. Reaction products were extracted with phenol-chloroform-isoamyl alcohol (25:24:1) and precipitated with 1/10 volume 3 M sodium acetate (pH 5.2), 2.5 volumes of ethanol, and 10 μg of tRNA or glycogen as a carrier. For the pulse-chase and time course reactions, a nucleotide mix and a mix of the other reaction components were prewarmed separately at 30°C for 5 min before being mixed together, aliquots were removed at various time points, and reactions were stopped with the addition of stop buffer and proteinase K. Reaction products were resolved on denaturing 15% acrylamide (20:1 acrylamide/bisacrylamide)–8 M urea gels in 1× TBE and visualized by autoradiography. Reaction products were quantified either by using a Molecular Dynamics PhosphorImager and ImageQuant or by scanning and using the NIH Image program.
Sephacryl S-300 gel filtration, native gel electrophoresis, and Northern analysis.
A scaled-up standard 260-μl telomerase reaction mixture with primer KL13(12) was incubated at 30°C for 7 min, mixed with a terminal transferase-labeled 30-mer (TGGGTGTGGTGTGTGGGTGTGGTGTGTGGG), and then separated on Sephacryl S-300 (Pharmacia) essentially as described previously (34). The reaction mixture was loaded onto a 2-ml column and eluted with TMG buffer in 150-μl fractions. A 15-μl portion of each fraction was Cerenkov counted. A 30-μl portion was further processed as described for the standard telomerase reaction. Two 30-μl aliquots from each fraction were loaded onto separate native gels composed of 3% acrylamide (80:1 acrylamide/bisacrylamide) and 0.6% agarose and electrophoresed in 50 mM Tris-acetate at 200 V for 3 h. One gel was exposed to film immediately for 2 h. The other gel was soaked in 50% (wt/vol) urea for 20 min and transferred to a Hybond Plus membrane (Amersham) in 0.5× TBE. Since the transferred labeled products generated considerable background signal, the membrane was set aside for approximately 2 months to allow the radioisotope to decay to a level that would not interfere with hybridization. To ensure low background levels, the membrane was exposed to film for 2 h. The membrane was then hybridized to a mixture of two 32P-labeled probes prepared by random-prime labeling (Amersham) of PCR fragments amplified from the TER1 gene. The fragments span nt 26 to 277 and 701 to 1273 of the TER1 gene, which exclude the template region. The membrane was then reexposed to film for 2 h.
RESULTS
Identification of telomerase activity from K. lactis.
To identify K. lactis telomerase in vitro, we fractionated S-100 extracts prepared from wild-type cells by ion-exchange chromatography on a DEAE-agarose column (see Materials and Methods) and assayed for telomerase activity. In these experiments, DEAE fractions were incubated with one α-32P-radiolabeled deoxynucleoside triphosphate (dNTP), three unlabeled dNTPs, and an oligonucleotide complementary to part of the previously identified K. lactis TER1 RNA templating domain (27). After incubation of the reaction mixture for 20 min at 30°C, reactions were terminated and products were purified and separated on 15% denaturing polyacrylamide gels. Initially, a 17-nt-long oligonucleotide (17-mer) [KL22(17) (Fig. 1A)] was used as the DNA primer to detect polymerization activity in extracts. 32P-labeled products longer than the input primer by up to 8 nt (+1 to +8 products) were expected if the primer was elongated all the way along the template. Such products were revealed, with the majority of the labeled products appearing at +6 (Fig. 1B, lane 1, arrowhead), corresponding to a position near the presumed end of the template (Fig. 1A, arrowhead). Through the remainder of this report, we refer to these prominent products as near-terminal products. The high-molecular-weight products seen at the top of the gel were, as described previously for comparable S. cerevisiae extracts (3, 33), independent of RNase pretreatment or telomerase RNA as described below and were therefore not attributable to telomerase activity.
Telomerase activity requires input of a DNA primer and is dependent on both protein and RNA components. K. lactis polymerization activity fulfilled these essential criteria, as no products were detected in the absence of a primer (Fig. 1B, lane 5), and the activity was abolished by preincubation of the extract with either proteinase K or RNase A (Fig. 1B, lanes 2 and 3). RNase-pretreated extracts sometimes produced bands (Fig. 1B, lane 3), but these were often variable in appearance and have never been attributable to telomerase activity. Specific polymerization still occurred when the telomerase preparation was incubated with RNase inhibitor (RNasin) before pretreatment with RNase A (Fig. 1B, lane 4). Hence, RNase inhibition of the activity was due to RNA digestion rather than other aspects of the preincubation conditions.
Further evidence that this activity was telomerase came from testing its ability to prime synthesis from another telomeric oligonucleotide, KL12(17). This 17-mer was designed to align on the template 10 nt 3′ of KL22(17) (Fig. 1A). The longest major near-terminal product produced in assays with KL12(17) (Fig. 1B, lane 6, +16 product, arrowhead) was 10 nt longer than the major product from KL22(17) (Fig. 1B, lane 1, +6 product). This result is in agreement with the predicted alignment of the primers on the TER1 RNA template. The RNase-sensitive products seen with KL12(17) included both the near-terminal products and a set of shorter products (Fig. 1B, lane 7). The shorter products, which we will refer to as mid-template products, are further characterized below.
To test the prediction for TER1 RNA template-directed synthesis, we performed assays in which each unlabeled dNTP was substituted by its ddNTP analog. With both primers, this resulted in premature chain termination at positions predicted from the template sequence (Fig. 1B, lane 8; Fig. 1C, lanes 4 to 6). In addition, omission of unlabeled dNTPs from the KL22(17)-primed reaction resulted in the expected strong stop at +2, after the addition of two [32P]dG residues to the input primer (Fig. 1C, lane 7).
As a direct test for dependence on the telomerase RNA template, extracts were prepared from strains in which the telomerase RNA gene was mutated. First, extracts were assayed from the ter1-Δ7 strain (27), in which a 300-bp fragment of the TER1 gene, including the template, is deleted. As predicted, these extracts had no RNase-sensitive polymerization activity with either primer, demonstrating that the template region is required for synthesis of both mid-template and near-terminal products (Fig. 1D, lanes 3 and 6, and data not shown). Confirmation of template-directed synthesis came from mutations introduced in the template region. The Ter1-SnaBI strain (29) was engineered to contain a C-to-A mutation within the TER1 RNA template (boxed residue in Fig. 1A). Fractionated extracts prepared from this strain produced the predicted patterns of incorporation: specifically, in assays with KL22(17), chain termination due to incorporation of ddTTP occurred at +2 instead of +4 (Fig. 1C; compare lanes 5 and 12), and polymerization ended at +1 instead of +2 in [32P]dGTP-only reactions (Fig. 1C; compare lanes 7 and 14). Thus, the SnaBI mutation, which is incorporated into telomeres in vivo, produced the expected changes in dNTP incorporation in vitro. In addition, polymerization was specifically altered in extracts prepared from both the Ter1-SnaBI mutant and another template mutant, Ter1-TaqI (data not shown) (29). Together, these results showed that telomerase is responsible for the K. lactis polymerization activity.
K. lactis telomerase catalyzes a single round of telomere repeat synthesis in vitro.
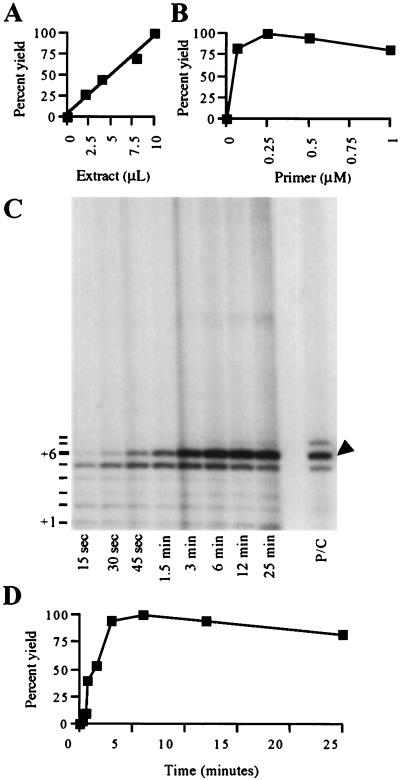
Under a variety of in vitro conditions, S. cerevisiae telomerase remains bound to its DNA reaction products in a manner that prevents further elongation (34). With many different primers, polymerization by K. lactis telomerase in vitro was also found to be confined to one round of synthesis across the template, which was often incomplete (Fig. 1B and reference 7). To address whether these properties of the K. lactis telomerase reaction have a basis similar to that of S. cerevisiae telomerase, we first characterized the kinetics of K. lactis telomerase by varying the amounts of extract and primer [KL22(17)] under standard assay conditions. Product yield increased linearly with increasing enzyme concentration, indicating that telomerase was limiting (Fig. 2A). Reactions with various primer concentrations demonstrated that the 1 μM concentration used in standard assays was well above the apparent Km for the reaction (Fig. 2B). In fact, maximal telomerase activity was observed at primer concentrations as low as 0.04 μM, with no change in product distribution (data not shown). At 1 μM primer, despite the large excess of primer and dNTP substrates used in all of the experiments described (see below), product yield reached a plateau by ∼3 min (Fig. 2C; quantitation shown in Fig. 2D), which would not be expected if repeated cycles of extension occurred. For example, in similar experiments with T. thermophila telomerase, total product yield increases over a period of up to 60 min (10, 20). In a pulse-chase experiment, excess unlabeled dGTP was added to a standard reaction after 45 s and incubated for a total of 25 min. The shorter (+1 to +5) products were chased into longer (+6 and +7) products (Fig. 2C; compare 45 sec lane to P/C lane), consistent with telomerase remaining bound to the +1 to +5 products during elongation. Notably, the chase did not stimulate synthesis of multiple consecutive 25-bp repeats (Fig. 2C, in which second-round products would have been visible in the top quarter of the autoradiogram portion shown). These results suggested that K. lactis telomerase consistently catalyzes only a single round of synthesis.
FIG. 2.
Kinetics of K. lactis telomerase activity. Reactions were performed with wild-type DEAE-fractionated extract and primer KL22(17). (A) Plot of product yield from reactions containing 0, 2, 4, 8, or 10 μl of wild-type DEAE-fractionated cell extract. The sums of the +1 to +7 products for each reaction were quantified and are presented as percentages of the yield with 10 μl of extract. (B) Plot of product yield from reactions containing 0, 0.125, 0.25, 0.5, and 1 μM primer. Product yield was quantified as in panel A. (C) Reaction with 10 μl of extract and 1 μM primer incubated for various time periods. Products from a 45-s reaction followed by a 24-min chase with 100 μM unlabeled dGTP are shown in lane P/C. Any products resulting from translocative synthesis would have appeared in the top quarter of the gel region shown. (D) Plot of product yield from reactions shown in panel C. The sums of the +5 and +6 (marked by arrowhead) products were quantified from a scanned autoradiogram, using the program NIH Image, and the results are presented as percentages of the yield at 3 min.
K. lactis telomerase remains bound to reaction products.
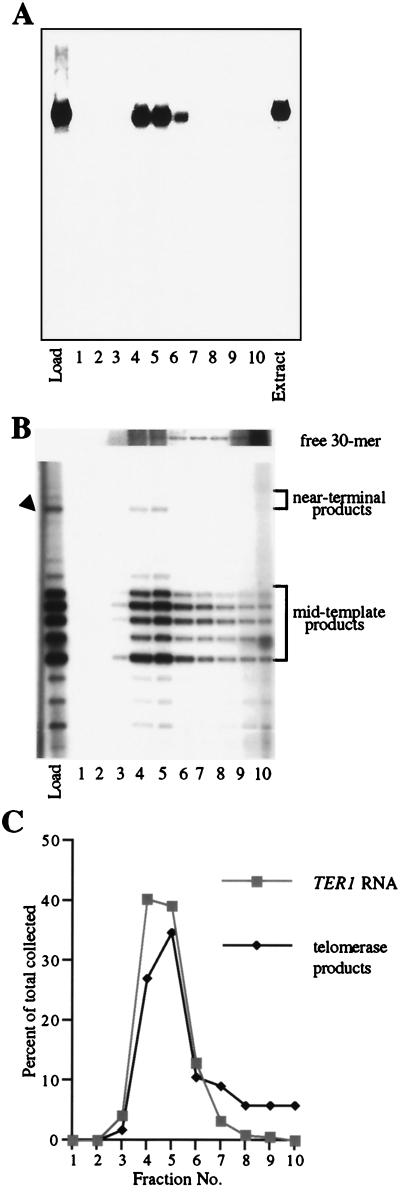
The early plateau in product yield seen in Fig. 2 was similar to that previously observed with S. cerevisiae telomerase (34) and could have been attributed to (i) dissociation and inactivation of telomerase at the completion of the reaction or (ii) telomerase remaining bound to its reaction products, thus precluding binding and extension of the excess free primer present in the reaction. To test directly whether telomerase enzyme remains bound to its products, we used size-exclusion chromatography to determine whether K. lactis telomerase products coeluted with the large telomerase RNP complex, as has been shown for S. cerevisiae telomerase (34). Reactions were primed with the 12-mer oligonucleotide KL13(12), which produces a product profile similar to KL12(17), including both mid-template and near-terminal products. After a 7-min incubation, the reaction was size fractionated on a Sephacryl S-300 column. A portion of each fraction was electrophoresed on a native gel, transferred to a membrane, and hybridized with a radiolabeled probe for the TER1 RNA subunit of telomerase to determine the elution profile of the telomerase complex (Fig. 3A). The hybridization was performed after the 32P label incorporated into the products had decayed for 2 months, to prevent interference from any signal resulting from products of the telomerase reaction bound to telomerase complexes. The 32P-labeled DNA products of the telomerase reaction, purified from a second aliquot of each fraction from the sizing column (see Materials and Methods), were separated by denaturing polyacrylamide gel electrophoresis (Fig. 3B). The remainder of each fraction was also electrophoresed on a separate native gel and exposed to film directly, to visualize large complexes carrying 32P label after the telomerase reaction. Although a large 32P-labeled complex in this directly exposed native gel cofractionated and comigrated with the TER1 RNA (data not shown), it was partially obscured by a background of the high-molecular-weight, non-telomerase-generated, 32P-labeled products shown in Fig. 1B. However, quantitation of the results shown in Fig. 3A and B clearly showed that after Sephacryl S-300 chromatography, both mid-template and near-terminal products cofractionated with the TER1 RNA, peaking in fractions 4 and 5 (Fig. 3C). Reaction products that had dissociated from the telomerase RNP would have eluted in a peak following the TER1 RNA-telomerase product peak. This was demonstrated by loading a marker 30-mer (previously 32P labeled by terminal transferase) together with the telomerase reaction mix onto the Sephacryl S-300 column; the 30-mer eluted after the telomerase product-TER1 RNA peak (Fig. 3B, top panel). A small population of telomerase products eluted after the peak (Fig. 3B and C), which may be attributable to the stability of the enzyme-product complex. These results indicated that K. lactis telomerase remained bound to the majority of both near-terminal and mid-template products of the extension reaction and suggested that each telomerase-active site carries out only one round of extension on a given primer molecule.
FIG. 3.
Association of K. lactis telomerase with elongation products. An in vitro telomerase reaction with primer KL13(12) was performed, and then the mixture was size fractionated on Sephacryl S-300 as described in Materials and Methods. (A) Aliquots of each fraction were separated on a native gel composed of 3% polyacrylamide (80:1 acrylamide/bisacrylamide) and 0.6% agarose (lanes 1 to 10; numbers correspond to fraction numbers), along with a portion of the nonfractionated telomerase reaction (Load lane) and 10 μl of the partially purified K. lactis extract that was used in the reaction (Extract lane). The gel was transferred to a Hybond Plus membrane (Amersham) and hybridized to a mixture of two 32P-labeled fragments of the TER1 gene that exclude the template region. (B) Aliquots of each fraction were separated on a denaturing 15% polyacrylamide (20:1 acrylamide/bisacrylamide)–8 M urea gel (lanes 1 to 10), along with a portion of the nonfractionated telomerase reaction (Load lane). Arrowhead corresponds to products described in Fig. 1B, and positions of mid-template and near-terminal products are bracketed. The upper portion of the panel shows the fractionation pattern of a telomeric 30-mer, 32P labeled by terminal transferase and loaded on the S-300 column with the completed telomerase reaction. This portion of the panel is from an exposure of the gel 30-fold longer than that used to show telomerase products. (C) Plot of the relative amounts of TER1 RNA (A) and telomerase products (B) recovered in each fraction. Products in each fraction were quantified and are represented as a percentage of the total collected.
Telomerase stalls at specific template regions in arrested and paused conformations.
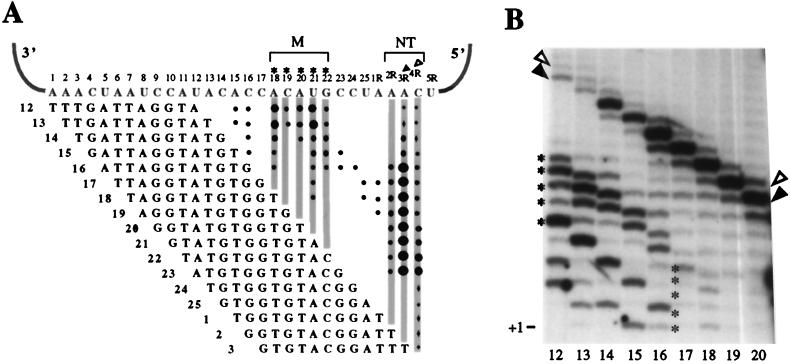
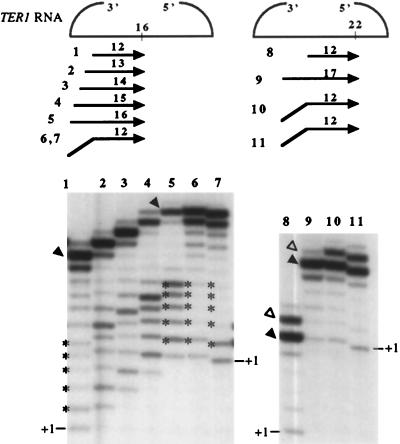
As described above, the majority of the products of K. lactis telomerase were shorter than would be expected if polymerization reached the end of the template (Fig. 1 to 3). To investigate the influence of the primer on the progression of polymerization along the template, we primed telomerase reactions with a series of template-complementary 12-mers which align stepwise across the template (Fig. 4A). Each primer was designed to anneal completely within the templating domain, with 3′ ends at template positions 12 through 3R (numbering as in Fig. 4A). Two distinct regions of product accumulation emerged from this data set: region M (mid-template) at positions 18 through 22, and region NT (near-terminal) at positions 2R through 4R (Fig. 4). A somewhat altered set of mid-template products was also seen with Ter1-SnaBI mutant extracts (data not shown), again confirming that the mid-template products as well as the near-terminal products were generated by telomerase activity. As shown in Fig. 4B for the wild-type enzyme, as primers aligned closer to the 3′ end of the template, the amount of products ending in the M region increased (compare bands in Fig. 4B marked with asterisks in lanes 12 and 17), while the amount of products ending in the NT region decreased proportionally (Fig. 4B; compare bands marked with arrowheads in lanes 12 and 20). Quantitation of the individual products and correction for the number of labeled incorporated nucleotides substantiated this trend and also confirmed that the amounts of total products formed were similar for all primers. These results suggest that mid-template products accumulate at the expense of near-terminal products. Because, as shown above, K. lactis telomerase remains bound to the majority of its extension products (Fig. 3), we hypothesized that product accumulation in the M and NT regions represents stalled telomerase-DNA complexes. Interestingly, stalling was not uniform along the template but instead occurred specifically at these two preferred regions.
FIG. 4.
Primer-dependent stalling by K. lactis telomerase. (A) Schematic of results from assays using 12-mer oligonucleotides aligning at different positions along the template. Numbers next to primers reflect the positions of the primers’ 3′ end on the template, as indicated by numbers above the template. Product bands are denoted at their appropriate template positions with black dots. The size of each dot represents the relative intensity of signal from products stalled at each position, and shaded columns of dots indicate the two preferred regions of stalling. Asterisks above positions 18 through 22 and the arrowheads above positions 3R and 4R mark 3′-end positions of products shown in panel B. (B) Reactions using primers 12 to 20, as depicted in panel A. Since all primers are the same length but align stepwise across the template, elongation of primers to the same position resulted in products varied by 1 nt in length (compare positions of arrowheads and asterisks).
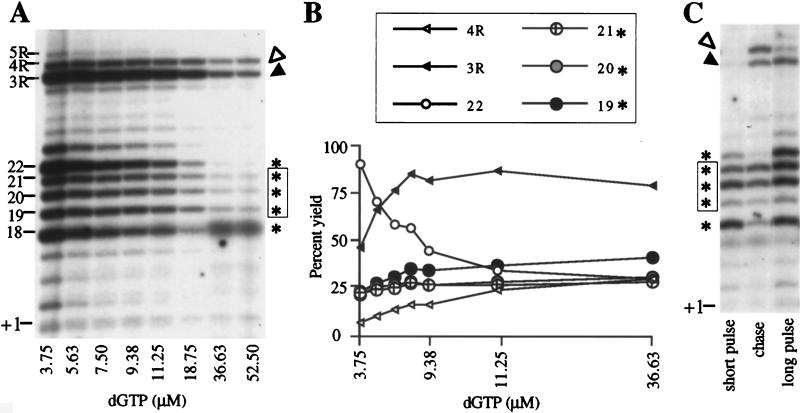
At limiting dNTP concentrations, telomerases from other species pause at template positions just prior to incorporation of that particular dNTP (20, 31, 38). Similarly, a subset of the stalls in the K. lactis telomerase reactions resulted from the low [32P]dGTP concentration used in standard in vitro reactions, as shown by titrating unlabeled dGTP into telomerase reactions with primer KL12(12) and monitoring product formation (Fig. 5A and B). Along with the expected quantitative decrease in total incorporated 32P label as the unlabeled dGTP concentration was increased, the distribution of products also changed, suggesting that stalling was overcome at some positions along the template. However, strikingly, products at template positions 19, 20, and 21 (M stalls) remained visible even at 50 μM dGTP (Fig. 5A, boxed asterisks). This result is in agreement with the accumulation of products at these same template positions in the experiment shown in Fig. 1B, in which dTTP was the limiting radiolabeled nucleotide and dGTP was not limiting (Fig. 1B, lane 6, +7 to +9 products). After correction for both the decreasing specific activity and the number of radiolabeled dG residues incorporated into each product, total product formation was similar at all dGTP concentrations, confirming that higher dGTP does not stimulate turnover. Product yields at 18 (data not shown) and 22 (Fig. 5B, small open circles) dropped as the dGTP concentration was increased, as expected for positions preceding incorporation of dG (pre-G positions). This finding suggests that telomerase stalled in the M region at positions 18 and 22 was paused, awaiting incorporation of a dG residue. Yields of the longer products at 3R and 4R in the NT region increased as the dGTP concentration was raised (Fig. 5B, filled and open triangles, respectively). However, high dGTP did not promote their extension to the extreme terminus of the template (Fig. 5A, 5R position). Strikingly, the high dGTP concentrations only slightly affected the product yields for positions 19, 20, and 21 (Fig. 5B, large circles), with the observed minor increase being attributable to depletion of smaller intermediates. Hence we conclude that the strong stalls at positions 19 through 21 in the M region and 3R and 4R in the NT region are functionally distinct from the pauses at pre-G positions 18 and 22. As we have shown that K. lactis telomerase remains associated with its products, we will refer to the stalls which are not elongated in response to high dGTP concentrations (at positions 19, 20, 21, 3R, and 4R) as arrested telomerase-product complexes.
FIG. 5.
Dependence of K. lactis telomerase stalling on nucleotide concentration. (A) Reactions using the primer KL13(12) and dGTP concentrations equaling 3.75, 5.63, 7.50, 9.38, 11.25, 18.75, 35.63, and 52.50 μM (each reaction mixture contains 50 μCi of [α-32P]dGTP and a variable amount of unlabeled dGTP). Products of particular interest are marked on the left side by their 3′-end positions. Asterisks and arrowheads mark products corresponding to those shown in Fig. 4. (B) Plot of telomerase products at various dGTP concentrations. At each concentration, products stalled at template positions 18, 19, 20, 21, 22, 3R, and 4R were quantified, corrected for specific activity differences, and divided by the number of dG residues incorporated into each product. Results for position 18 were omitted due to interference of the diffuse background band visible in the last two lanes, which corresponds to the non-telomerase-generated product seen in Fig. 1B, lanes 1 to 5. (C) A scaled-up reaction mixture with 3.75 μM [α-32P]dGTP was incubated for 1.5 min and then divided into three aliquots; one part was stopped (short pulse lane) with proteinase K and SDS (see Materials and Methods), the second aliquot was mixed with 100 μM unlabeled dGTP and incubated for an additional 23.5 min (long chase lane), and the third was incubated for a total of 25 min with no chase (long pulse lane). Asterisks and arrowheads mark mid-template and near-terminal products, respectively, and boxed asterisks show unchaseable products.
The assignment of stalled complexes as either paused or arrested was confirmed by a pulse-chase experiment with the same primer. Excess unlabeled dGTP was added to a reaction mixture at 45 s, and then the incubation continued for a total of 25 min (Fig. 5C). Consistent with the findings from the dGTP titration, mid-template pre-G intermediates at positions 18 and 22 that had accumulated by 45 s (Fig. 5C, nonboxed asterisks) were chased into 3R and 4R (NT region) products (Fig. 5C, arrowheads). In contrast, product yield at positions 19 through 21 was unaffected by the unlabeled dGTP chase (Fig. 5C, bands marked with asterisks), confirming a steady-state level of arrested complexes. The lack of elongation to 5R during the chase provided further evidence that most 3R and 4R products were also arrested.
Increased primer-template complementarity exacerbates stalling.
The frequency of telomerase stalling in the M region increased as primers annealed further toward the 3′ end of the template (Fig. 4A and B). This could have resulted from the increased length of (i) just the DNA added to the primer by the enzyme, (ii) total potential RNA-DNA duplex created during polymerization, or (iii) the DNA product itself (primer plus added nucleotides) when polymerization reaches the stall region, regardless of its degree of pairing to the template. To test which of these parameters influenced stalling, we performed telomerase reactions using two sets of primers, each set having a common 3′ end but variable 5′ ends. The 3′ end of the first set of primers was at template position 16, and with such a 12-mer primer little stalling occurred in the M region (Fig. 4B, lane 16; Fig. 6, lane 1). However, extending the amount of primer-template complementarity with extra nucleotides on the 5′ end of the primer dramatically increased the ratio of mid-template products (Fig. 6, bands marked with asterisks) to near-terminal products (lanes 1 to 5, arrowheads). In contrast, extra noncomplementary 5′ nucleotides did not exacerbate stalling (lanes 6 and 7). In additional experiments, M region stalling was also alleviated when the length of potential RNA-DNA duplex of a 12-mer ending at position 15 was reduced to 11 nt, by changing the 5′ nucleotide from the complementary dG to a dA, dT, or dC residue (data not shown). Comparable results were seen with a second set of primers with a common 3′ end at position 22, designed to examine stalling in the NT region. Stalling at position 3R relative to 4R was exacerbated when the 5′ end of a 12-mer was extended with five template-complementary nucleotides (lanes 8 and 9) and alleviated when the 5′ end was made noncomplementary (lanes 10 and 11).
FIG. 6.
Dependence of telomerase stalling on primer-template complementarity. Lanes 1 to 7, products from reactions with primers 1 to 7 as illustrated above (lane numbers correspond to the numbers at the left of the primers). All primers have 3′ ends aligning at template position 16. Primers’ amounts of template complementarity increase by 1 nt at a time from 12 to 16 nt (number above each primer). Note that the overall length of the primers also increases, and therefore the +1 position changes on the gel correspondingly. Primers numbered 6 and 7 are both 16 nt long, with 12 nt of complementarity to the template. The 4 nt at the 5′ end of primer 6 are the same as those of primer 5 but scrambled so that none of them are complementary to the template. The nucleotide composition of the 5′ end of primer 7 is nontelomeric (ATAT). Lanes 8 to 11, products from reactions with primers, as illustrated. Primers in this set share 3′ ends that align at position 22. Again, the numbers above each primer reflect their degree of complementarity to the template. Primers 10 and 11 are both 17 nt long, and both have 12 nt of complementarity to the template. The 5 nt at the 5′ end of primer 10 are as in primer 9 but scrambled (as with primer 6). The 5′ end of primer 11 is nontelomeric (ATATA). Asterisks and arrowheads correspond to products previously discussed.
These results demonstrated that stalling by K. lactis telomerase in vitro is not determined by the total length per se of the product or by the length of only the newly synthesized stretch of DNA. Instead, in the M and NT regions of the template, stalling is exacerbated by increasing the length of the potential DNA-RNA template duplex. Interestingly, the class of stalled complexes influenced by limiting dGTP concentrations (at positions 18 and 22; top and bottom bands marked with asterisks) also accumulated in response to increased primer-template complementarity (Fig. 6), suggesting an interplay between dGTP concentration and complementarity in contributing to stalling.
DISCUSSION
The telomerases described heretofore use a short template to synthesize either short, precise repeats or irregular repeats (3, 24, 25, 31, 35, 37, 44). However, K. lactis telomerase synthesizes 25-bp repeats from a 30-nt templating domain, the largest described to date (27). Here we have reported the identification and characterization of in vitro telomerase activity from this budding yeast. This study reveals intriguing properties of K. lactis telomerase, including stalling at specific regions of the template and the dependence of stalling on increased primer-template complementarity. The establishment of an in vitro K. lactis telomerase system will allow other questions about telomere biology to be addressed. For example, certain previously characterized template mutations in K. lactis cause major effects on telomere length and maintenance in vivo (19, 27). It will now be possible to distinguish whether these effects result from altered binding of telomere-associated proteins (for example, Rap1p) or altered telomerase activity.
We found that K. lactis telomerase catalyzes only a single round of primer elongation in vitro, like telomerase from S. cerevisiae (3, 23, 33, 34). In K. lactis and S. cerevisiae cells lacking intact telomerase RNA, telomeres shorten by an average of only 5 bp per cell division (27, 39). Hence in wild-type K. lactis cells, the copying of even one-fifth of a telomeric repeat per cell division on average would be sufficient to maintain telomere length equilibrium. Thus, single-round repeat synthesis by K. lactis telomerase could maintain telomeres in vivo. The in vitro behavior of K. lactis telomerase reported here is consistent with a model in which telomerase acts in each cell cycle to synthesize a small amount of telomeric DNA. Synthesis of only small increments per cell cycle was proposed previously for S. cerevisiae, based on analysis of the observed small-length fluctuations of its telomeres in vivo (36). Another property of K. lactis telomerase activity reported here, its nondissociativity from reaction products, may indicate a structural role for telomerase in a telomere-capping complex, as suggested previously for S. cerevisiae telomerase (34).
An unusual property of the K. lactis telomerase activity is that it stalls during the synthesis of a telomeric repeat in at least two specific regions of the template. The mechanistic basis for this stalling is unknown, but it may be influenced by the constrained nature of the template combined with the potential to create a long RNA-DNA hybrid during polymerization. In Tetrahymena telomerase, the regions of telomerase RNA outside the template region are apparently buried in the RNP (14). Therefore, the movement of the templating domain across the catalytic center of telomerase at each polymerization step is necessarily constrained by the anchoring of the template in the RNP by RNA-protein associations. The exacerbation of stalling that we observed upon increasing the DNA-template RNA complementarity supports the idea that base pairing between template RNA and DNA contributes to stalling. Building up a long, presumably rigid RNA-DNA hybrid duplex during elongation in vitro is likely to interfere with the constrained movement of the template through the catalytic center. Furthermore, such a duplex may also prevent the active site of the RNP from assuming the correct conformation necessary for polymerization. Hence, as the hybrid lengthens, steric restriction may become sufficient to prevent further polymerization.
The formation of lengthy template RNA-primer or product DNA hybrids during polymerization in vitro would imply that telomerase lacks the ability to unpair the duplex, which, as discussed above, may contribute to stalling. Other telomerases analyzed to date, including those from ciliated protozoa, human cells, and the budding yeast S. castellii, catalyze multiple cycles of synthesis in vitro (3, 10, 15, 31), which requires unpairing of the product DNA-RNA template hybrid. Telomerase activity reconstituted from a human telomerase protein (hTERT) and the telomerase RNA component (hTR), as well as extensively purified Tetrahymena and Euplotes telomerase preparations are able to carry out translocative synthesis, indicating that such unpairing ability is intrinsic to these telomerase enzymes and does not require other factors in vitro (6, 22, 42). It has been proposed that Tetrahymena telomerase unpairs the recently made DNA from the template through the energy of binding to another site in the RNP and that this site is analogous to that shown to bind the nascent transcript of RNA polymerase (21). The K. lactis telomerase preparations used in this study may lack telomeric binding proteins, other components of the telomerase complex, or accessory factors such as a helicase that could alleviate stalling in vitro. Alternatively, the buildup of an RNA-DNA hybrid and stalling may be inherent to the in vivo action of telomerase.
The available information is compatible with some occurrence of stalling in vivo. S. cerevisiae telomerase has been suggested to stall frequently in vitro as copying proceeds along its template, and in this yeast, degenerate repeats are found in vivo in a pattern consistent with the in vitro properties of this enzyme (3, 33). Stalling by other polymerases appears to be exploited in vivo. For example, pausing by RNA polymerase in bacterial cells coordinates transcription and other processes including translation, possibly the loading of regulatory proteins important for antitermination and the formation of appropriate RNA structure during rRNA transcription (for a review, see reference 40). The stalling that we have reported here for K. lactis telomerase, which often results in only partial synthesis of a repeat in vitro, is compatible with synthesis of the perfectly repeated telomeric sequence of this species in vivo as long as primer alignment on the template is correct before the next round of synthesis. Since the template lacks internal redundancy, primers with 3′ ends in the M or NT region can align in a unique register, and thus incomplete extension could be tolerated in vivo. Hence stalling by telomerase is a potential step at which telomerase action could be regulated in vivo.
The diversity of telomeric repeat lengths and sequences in budding yeasts suggests differences among the polymerization properties and, potentially, the evolutionary stages of their telomerases. Long-template telomerases may have been derived sporadically in budding yeasts, arising from short template telomerases copying beyond their normal template boundaries. In this case, it is conceivable that the K. lactis telomerase template positions where stalling occurs represent the ancient boundaries of a shorter template. It has been suggested that the ancestor to telomerase was a catalytic self-replicating RNA. This ancestor is proposed to have acquired a reverse transcriptase protein component but to have retained RNA, with at least a portion still functioning as a template (2). By this model, the long-template telomerases of budding yeasts may represent an intermediate stage in the evolution from a ribozyme that completely copied itself to the typical short-template telomerases which copy a small discrete region of RNA. Indeed, stalling by the more primitive telomerases is one possible means that may have helped initiate the eventual evolution into copying progressively shorter regions of telomerase RNA. Further analysis of telomerases from budding yeasts will be valuable for our understanding the origins of their diverse telomere repeats and the general mechanism of telomerase.
ACKNOWLEDGMENTS
We are grateful to M. McEachern for the strains used in this study. We thank Y. Tzfati, T. Ware, A. Krauskopf, and C. Gross for critical reading of the manuscript, and we thank members of the Blackburn laboratory for helpful discussions and support.
This work was supported by NIH grants GM26259 and DE11356 to E.H.B. and NIH training grant T32 CA09270 to T.B.F.
REFERENCES
- 1.Autexier C, Greider C W. Boundary elements of the Tetrahymena telomerase RNA template and alignment domains. Genes Dev. 1995;9:2227–2239. doi: 10.1101/gad.9.18.2227. [DOI] [PubMed] [Google Scholar]
- 2.Blackburn, E. H. Telomerase. In R. F. Gesteland and J. F. Atkins (ed.), The RNA world II, in press. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 3.Cohn M, Blackburn E H. Telomerase in yeast. Science. 1995;269:396–400. doi: 10.1126/science.7618104. [DOI] [PubMed] [Google Scholar]
- 4.Cohn M, McEachern M J, Blackburn E H. Telomeric sequence diversity within the genus Saccharomyces. Curr Genet. 1998;33:83–91. doi: 10.1007/s002940050312. [DOI] [PubMed] [Google Scholar]
- 5.Collins K, Greider C W. Tetrahymena telomerase catalyzes nucleolytic cleavage and nonprocessive elongation. Genes Dev. 1993;7:1364–76. doi: 10.1101/gad.7.7b.1364. [DOI] [PubMed] [Google Scholar]
- 6.Collins K, Kobayashi R, Greider C W. Purification of Tetrahymena telomerase and cloning of genes encoding the two protein components of the enzyme. Cell. 1995;81:677–686. doi: 10.1016/0092-8674(95)90529-4. [DOI] [PubMed] [Google Scholar]
- 7.Fulton, T. B., and E. H. Blackburn. Unpublished data.
- 8.Gilley D, Blackburn E H. Specific RNA residue interactions required for enzymatic functions of Tetrahymena telomerase. Mol Cell Biol. 1996;16:66–75. doi: 10.1128/mcb.16.1.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gilley D, Lee M S, Blackburn E H. Altering specific telomerase RNA template residues affects active site function. Genes Dev. 1995;9:2214–2226. doi: 10.1101/gad.9.18.2214. [DOI] [PubMed] [Google Scholar]
- 10.Greider C W. Telomerase is processive. Mol Cell Biol. 1991;11:4572–4580. doi: 10.1128/mcb.11.9.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Greider C W. Telomerase biochemistry and regulation. In: Blackburn E H, Greider C W, editors. Telomeres. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1995. pp. 35–68. [Google Scholar]
- 12.Greider C W, Blackburn E H. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985;43:405–413. doi: 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- 13.Greider C W, Blackburn E H. The telomere terminal transferase of Tetrahymena is a ribonucleoprotein enzyme with two kinds of primer specificity. Cell. 1987;51:887–898. doi: 10.1016/0092-8674(87)90576-9. [DOI] [PubMed] [Google Scholar]
- 14.Greider C W, Blackburn E H. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature. 1989;337:331–337. doi: 10.1038/337331a0. [DOI] [PubMed] [Google Scholar]
- 15.Hammond P W, Lively T N, Cech T R. The anchor site of telomerase from Euplotes aediculatus revealed by photo-cross-linking to single- and double-stranded DNA primers. Mol Cell Biol. 1997;17:296–308. doi: 10.1128/mcb.17.1.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harrington L, Zhou W, McPhail T, Oulton R, Yeung D S, Mar V, Bass M B, Robinson M O. Human telomerase contains evolutionarily conserved catalytic and structural subunits. Genes Dev. 1997;11:3109–3115. doi: 10.1101/gad.11.23.3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henderson E. Telomere DNA structure. In: Blackburn E, Greider C W, editors. Telomeres. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1995. pp. 11–34. [Google Scholar]
- 18.Hiraoka Y, Henderson E, Blackburn E H. Not so peculiar: fission yeast telomere repeats. Trends Biochem Sci. 1998;23:126. doi: 10.1016/s0968-0004(98)01176-1. [DOI] [PubMed] [Google Scholar]
- 19.Krauskopf A, Blackburn E H. Control of telomere growth by interactions of RAP1 with the most distal telomeric repeats. Nature. 1996;383:354–357. doi: 10.1038/383354a0. [DOI] [PubMed] [Google Scholar]
- 20.Lee M S, Blackburn E H. Sequence-specific DNA primer effects on telomerase polymerization activity. Mol Cell Biol. 1993;13:6586–6599. doi: 10.1128/mcb.13.10.6586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee M S, Gallagher R C, Bradley J, Blackburn E H. In vivo and in vitro studies of telomeres and telomerase. Cold Spring Harbor Symp Quant Biol. 1993;58:707–718. doi: 10.1101/sqb.1993.058.01.078. [DOI] [PubMed] [Google Scholar]
- 22.Lingner J, Cech T R. Purification of telomerase from Euplotes aediculatus: requirement of a primer 3′ overhang. Proc Natl Acad Sci USA. 1996;93:10712–10717. doi: 10.1073/pnas.93.20.10712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lingner J, Hughes T R, Shevchenko A, Mann M, Lundblad V, Cech T R. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science. 1997;276:561–567. doi: 10.1126/science.276.5312.561. [DOI] [PubMed] [Google Scholar]
- 24.Mantell L L, Greider C W. Telomerase activity in germline and embryonic cells of Xenopus. EMBO J. 1994;13:3211–3217. doi: 10.1002/j.1460-2075.1994.tb06620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCormick-Graham M, Romero D P. A single telomerase RNA is sufficient for the synthesis of variable telomeric DNA repeats in ciliates of the genus Paramecium. Mol Cell Biol. 1996;16:1871–1879. doi: 10.1128/mcb.16.4.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McEachern M J, Blackburn E H. A conserved sequence motif within the exceptionally diverse telomeric sequences of budding yeasts. Proc Natl Acad Sci USA. 1994;91:3453–3457. doi: 10.1073/pnas.91.8.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McEachern M J, Blackburn E H. Runaway telomere elongation caused by telomerase RNA gene mutations. Nature. 1995;376:403–409. doi: 10.1038/376403a0. [DOI] [PubMed] [Google Scholar]
- 28.McEachern M J, Blackburn E H. Cap-prevented recombination between terminal telomeric repeat arrays (telomere CPR) maintains telomeres in Kluyveromyces lactis lacking telomerase. Genes Dev. 1996;10:1822–1834. doi: 10.1101/gad.10.14.1822. [DOI] [PubMed] [Google Scholar]
- 29.McEachern, M. J., and E. H. Blackburn. Unpublished data.
- 30.Meyerson M, Counter C M, Eaton E N, Ellisen L W, Steiner P, Caddle S D, Ziaugra L, Beijersbergen R L, Davidoff M J, Liu Q, Bacchetti S, Haber D A, Weinberg R A. hEST2, the putative human telomerase catalytic subunit gene, is up-regulated in tumor cells and during immortalization. Cell. 1997;90:785–795. doi: 10.1016/s0092-8674(00)80538-3. [DOI] [PubMed] [Google Scholar]
- 31.Morin G B. The human telomere terminal transferase enzyme is a ribonucleoprotein that synthesizes TTAGGG repeats. Cell. 1989;59:521–529. doi: 10.1016/0092-8674(89)90035-4. [DOI] [PubMed] [Google Scholar]
- 32.Nakamura T M, Morin G B, Chapman K B, Weinrich S L, Andrews W H, Lingner J, Harley C B, Cech T R. Telomerase catalytic subunit homologs from fission yeast and human. Science. 1997;277:955–959. doi: 10.1126/science.277.5328.955. [DOI] [PubMed] [Google Scholar]
- 33.Prescott J, Blackburn E H. Telomerase RNA mutations in Saccharomyces cerevisiae alter telomerase action and reveal nonprocessivity in vivo and in vitro. Genes Dev. 1997;11:528–540. doi: 10.1101/gad.11.4.528. [DOI] [PubMed] [Google Scholar]
- 34.Prescott J, Blackburn E H. Functionally interacting telomerase RNAs in the yeast telomerase complex. Genes Dev. 1997;11:2790–2800. doi: 10.1101/gad.11.21.2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prowse K R, Avilion A A, Greider C W. Identification of a nonprocessive telomerase activity from mouse cells. Proc Natl Acad Sci USA. 1993;90:1493–1497. doi: 10.1073/pnas.90.4.1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shampay J, Blackburn E H. Generation of telomere-length heterogeneity in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1988;85:534–538. doi: 10.1073/pnas.85.2.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shippen-Lentz D, Blackburn E H. Telomere terminal transferase activity from Euplotes crassus adds large numbers of TTTTGGGG repeats onto telomeric primers. Mol Cell Biol. 1989;9:2761–2764. doi: 10.1128/mcb.9.6.2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shippen-Lentz D, Blackburn E H. Functional evidence for an RNA template in telomerase. Science. 1990;247:546–552. doi: 10.1126/science.1689074. [DOI] [PubMed] [Google Scholar]
- 39.Singer M S, Gottschling D E. TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science. 1994;266:404–409. doi: 10.1126/science.7545955. [DOI] [PubMed] [Google Scholar]
- 40.Uptain S M, Kane C M, Chamberlin M J. Basic mechanisms of transcript elongation and its regulation. Annu Rev Biochem. 1997;66:117–172. doi: 10.1146/annurev.biochem.66.1.117. [DOI] [PubMed] [Google Scholar]
- 41.Wang H, Gilley D, Blackburn E H. A novel specificity for the primer-template pairing requirement in Tetrahymena telomerase. EMBO J. 1998;17:1152–1160. doi: 10.1093/emboj/17.4.1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weinrich S L, Pruzan R, Ma L, Ouellette M, Tesmer V M, Holt S E, Bodnar A G, Lichtsteiner S, Kim N W, Trager J B, Taylor R D, Carlos R, Andrews W H, Wright W E, Shay J W, Harley C B, Morin G B. Reconstitution of human telomerase with the template RNA component hTR and the catalytic protein subunit hTRT. Nat Genet. 1997;17:498–502. doi: 10.1038/ng1297-498. [DOI] [PubMed] [Google Scholar]
- 43.Yu G L, Bradley J D, Attardi L D, Blackburn E H. In vivo alteration of telomere sequences and senescence caused by mutated Tetrahymena telomerase RNAs. Nature. 1990;344:126–132. doi: 10.1038/344126a0. [DOI] [PubMed] [Google Scholar]
- 44.Zahler A M, Prescott D M. Telomere terminal transferase activity in the hypotrichous ciliate Oxytricha nova and a model for replication of the ends of linear DNA molecules. Nucleic Acids Res. 1988;16:6953–6972. doi: 10.1093/nar/16.14.6953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zakian V. Saccharomyces telomeres: function, structure, and replication. In: Blackburn E, Greider C, editors. Telomeres. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1995. pp. 107–138. [Google Scholar]