Abstract
Transcriptional coactivators play a crucial role in gene expression by communicating between regulatory factors and the basal transcription machinery. The coactivator multiprotein bridging factor 1 (MBF1) was originally identified as a bridging molecule that connects the Drosophila nuclear receptor FTZ-F1 and TATA-binding protein (TBP). The MBF1 sequence is highly conserved across species from Saccharomyces cerevisiae to human. Here we provide evidence acquired in vitro and in vivo that yeast MBF1 mediates GCN4-dependent transcriptional activation by bridging the DNA-binding region of GCN4 and TBP. These findings indicate that the coactivator MBF1 functions by recruiting TBP to promoters where DNA-binding regulators are bound.
Studies on transcriptional control in eukaryotes have revealed many regulatory proteins that bind to control elements on DNA (17, 25). Typical regulators such as GAL4 and GCN4 consist of two domains (29), a DNA-binding domain that binds to a control element in a sequence-specific fashion and a transcriptional activation domain that somehow stimulates the basal transcription machinery. A variety of observations have led to the proposal that some transactivation domains may facilitate binding of TATA-binding protein (TBP) to a promoter (32, 37 see reference 35 for a review). In many cases, a member of another class of transcription factors, termed coactivators, adapters, or mediators, is necessary to connect a regulatory protein and a component of the basal transcription machinery (see references 23 and 30 for reviews). Recent studies demonstrated the importance of these non-DNA-binding transcription factors in gene expression (1, 4, 5, 11, 14, 19, 20, 27, 31).
In in vitro transcription studies, it was found that an insect coactivator, multiprotein bridging factor 1 (MBF1), can recruit TBP to a promoter carrying the FTZ-F1-binding site by interconnecting FTZ-F1 and TBP (24, 34). A homology search of the databases showed that the MBF1 sequence is highly conserved across species from Saccharomyces cerevisiae to human (34). A key question concerning MBF1, and its close relatives, is its in vivo function. To address this issue, we have started genetic studies of MBF1. We report here that MBF1 serves as a coactivator of GCN4 in the yeast S. cerevisiae.
MATERIALS AND METHODS
Yeast strains and plasmids.
The GCN4 disruptant used in this study was KY803 (trp1-Δ1 ura3-52 leu2-P1 gcn4-Δ1) (13). Wild-type strain KT130 (trp1-Δ1 ura3-52 leu2-P1) was constructed by homologous recombination after introduction of GCN4 genomic DNA into KY803. KT130 was used to generate the Δmbf1 strain by replacement of the sequence encoding amino acid residues 64 to 131 of yeast MBF1 (yMBF1) (34) with a LEU2 selectable marker.
To construct pMBF1, the 3-kb EcoRI genomic fragment encompassing the entire MBF1 regulatory and coding regions was subcloned into YCplac33 (9). YCp88 (13), a centromeric vector containing the constitutive DED1 promoter, was used to express GCN4 or its mutants in KY803. pYCGCN4, pYCGCN4ΔbZIP, and pYCGCN4bZIP carry the entire coding region of GCN4, the coding region excluding bZIP (amino acid positions 1 to 220), and the coding region including bZIP (amino acid positions 221 to 281), respectively. pYCGCN4ΔAD lacks the coding region of the major activation domain (amino acid positions 88 to 147) of GCN4. To express yMBF1, its point mutant carrying D112A, and yMBF1ΔNT in the Δmbf1 mutant, each coding region tagged with the GAPDH promoter was inserted into YCplac22 (9) to yield pYCMBF1, pD112A, and pMBF1ΔNT. For overexpression of yMBF1 and the D112A mutant under the control of GAPDH promoter, each coding region tagged with the GAPDH promoter was cloned into YEplac112 (9) to yield pYEMBF1 and pYED112A. yTBP was overexpressed from pyTBP under the control of the ADH promoter in pDB20 (3). Point mutations of GCN4 and MBF1 were introduced by site-directed PCR mutagenesis.
Expression of proteins in bacteria.
To produce yMBF1, GCN4, or its mutant derivatives in Escherichia coli, each NdeI-BamHI tagged open reading frame was subcloned into 6HisT-pET11d (Novagen). GCN4ΔbZIP and GCN4bZIP consist of amino acid positions 1 to 221 and 222 to 281 of GCN4, respectively. For GCN4 and its derivatives, the sequence encoding hemagglutinin (HA) epitope (8) was inserted in frame at the NdeI site to generate HA epitope-tagged protein. Histidine-tagged yTBP was expressed as described previously (12). These histidine-tagged proteins were recovered in a soluble fraction and purified with Ni-nitrilotriacetic acid resin (Novagen) followed by Mono S (Pharmacia) chromatography. To produce glutathione S-transferase–yMBF1 (GST-yMBF1) and its mutant derivatives in E. coli, each BclI tagged coding region was subcloned into the BamHI site of pGEX-2T (Pharmacia). GST-yMBF1ΔNT and GST–yMBF1Δ41-119 lack the N-terminal 40 amino acids and the amino acid residues from 41 to 119, respectively, of yMBF1. GST and GST fusion proteins were purified by glutathione-Sepharose 4B (Pharmacia) chromatography.
GST pull-down assays.
GST or GST fusion proteins (200 ng) were incubated with bacterially expressed and purified target protein (100 ng) at 4°C for 1 h in 20 μl of protein-binding buffer (PBB) (20 mM HEPES-KOH [pH 7.9], 20% glycerol, 0.5 mM EDTA, 60 mM KCl, 6 mM MgCl2, 0.1% Nonidet P-40, 5 mM 2-mercaptoethanol, 1 mM phenylmethylsulfonyl fluoride). After incubation, 1 μl (packed volume) of glutathione-Sepharose 4B (Pharmacia) in 20 μl of PBB was added, and the mixture was rolled at 4°C for 1 h. The beads were collected and washed twice, with 1.5 ml of PBB with rotation at 4°C for 20 min each time, and then bound proteins were eluted for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). HA epitope-tagged GCN4 and its derivatives and yTBP were detected on immunoblots by an anti-HA antibody (Boehringer GmbH, Mannheim, Germany) at 5 μg/ml and an anti-yTBP antibody (Upstate Biotechnology) at a 1:50,000 dilution, respectively. To examine the recovery of the GST and GST fusion proteins, all pull-down blots were probed with an antibody against GST. More than 95% of the input proteins were recovered.
Electrophoresis mobility shift assays.
DNA-binding assays for GCN4 and its derivatives were carried out in reaction mixtures (10 μl) containing 12 mM HEPES-KOH (pH 7.9), 5 mM MgCl2, 70 mM KCl, 1 mM dithiothreitol, 6% glycerol, 1 mg of poly(dI-dC) · poly(dI-dC) (Pharmacia) per ml, 5 mg of bovine serum albumin (BSA) per ml, 1 ng of bacterially expressed GCN4 or its derivative, and 10 fmol of the probe. The samples were incubated at 30°C for 30 min and then electrophoresed at room temperature and 100 V for 2.5 h on a 0.8% agarose gel in Tris-borate-EDTA (TBE). The sequence of the GCN4-binding site probe was 5′-TCCACCTAGCGGATGACTCTTTTTTTTTCTTAG-3′. The binding of yTBP to the TATA element was carried out in reaction mixtures (10 μl) containing 12 mM HEPES-KOH (pH 7.9), 4 mM MgCl2, 60 mM KCl, 12% glycerol, 2% polyvinyl alcohol (average molecular weight, 10,000; Sigma), 0.5 mg of poly(dG-dC) · poly(dG-dC) (Pharmacia) per ml, 5 mg of BSA per ml, 10 ng of bacterially expressed yTBP, and 5 fmol of the probe. After incubation at 30°C for 30 min, the samples were electrophoresed at 22°C and 100 V for 2.5 h on a 1% agarose gel in 0.5× TBE containing 2 mM MgCl2. The sequence of the TATA element probe was 5′-GAATTATACATTATATAAAGTAATGTGATTTC-3′.
S1 nuclease analyses.
Total RNA was isolated by a hot-phenol method (22) from yeast cells grown in synthetic complete medium lacking histidine and then treated with 30 mM aminotriazole (AT) for 6 h. Portions of the RNA (40 μg) were subjected to S1 nuclease analyses with 32P-labeled oligonucleotide probes for HIS3 and DED1 genes as described previously (15).
Western analyses.
Yeast protein extracts were prepared from the histidine-starved cells grown as described above, according to the method of C. Kaiser et al. (18). Exactly 20 μg of proteins in the extract was resolved by SDS–10% PAGE for detection of GCN4 or SDS–15% PAGE for detection of MBF1 and then subjected to Western blot analyses as described previously (26). The bound antibodies were visualized by using a Super Signal CL-HRP substrate system (Pierce). Antisera against GCN4 and MBF1 were gifts from A. G. Hinnebusch and M. Jindra, respectively. The antiserum against GCN4 was used after 250-fold dilution followed by incubation with a membrane that had been blotted with proteins in the extract from the Δgcn4 strain. The antiserum against MBF1 was used after 10,000-fold dilution.
Nucleotide sequence accession number.
The nucleotide sequence encoding a part of yMBF1 has been deposited in the GenBank/EMBL/DDBJ databases as an expressed sequence tag of unknown function with accession no. T38224.
RESULTS AND DISCUSSION
Yeast MBF1 is essential for transcriptional activation of HIS3.
Curiously, the partial yMBF1 sequence was not recognized as an open reading frame in the yeast genome sequence (10). The nucleotide sequence of the complete open reading frame has been determined, and the deduced amino acid sequence has been compared with MBF1 sequences of other organisms (34). yMBF1 has 43% amino acid identity with the Drosophila counterpart. To investigate the role of yMBF1 in vivo, we inactivated the MBF1 locus by one-step gene replacement. The disruptant (the Δmbf1 strain) was viable (Fig. 1A) and able to grow on galactose, sucrose, or inositol-free media (data not shown), indicating that yMBF1 is not a general transcription factor. But the Δmbf1 strain was sensitive to AT, an inhibitor of the HIS3 gene product (Fig. 1A). This sensitivity was suppressed by introducing a yMBF1 expression plasmid, pMBF1. We also examined the level of HIS3 mRNA by quantitative S1 nuclease mapping (Fig. 1B). Upon histidine starvation, no activation of HIS3 mRNA synthesis was detected in the Δmbf1 strain, and this phenotype was the same as that observed for a GCN4 disruptant (Δgcn4 strain). Activation of HIS3 transcription was restored when pMBF1 was introduced into the Δmbf1 strain. These results clearly show that yMBF1 is essential for transcriptional activation of the HIS3 gene. The Δmbf1 strain was also defective in transcriptional activation of the HIS5 gene upon histidine starvation (data not shown), suggesting that yMBF1 plays a role in GCN4-dependent activation.
FIG. 1.
Phenotype of the Δmbf1 strain. (A) AT sensitivity. Cells of the wild type (WT), the Δgcn4 strain, the Δmbf1 strain, and the Δmbf1 strain transformed with pMBF1 were grown on plates in the presence or absence of 10 mM AT for 3 days at 30°C. (B) Effect on HIS3 transcription. Total RNA from the four strains grown under the histidine-starved conditions was used for S1 nuclease analyses of HIS3 and DED1 transcripts. Constitutive HIS3 transcripts initiate from the +1, +13, and +22 sites, whereas GCN4-activated transcripts initiate preferentially from the +13 sites; DED1 is not affected by GCN4 and serves as an internal control (33). (C) Expression of GCN4 in the Δmbf1 strain. The levels of GCN4 in the four strains were estimated by Western analysis with an anti-GCN4 antiserum.
GCN4-MBF1 interaction is necessary for GCN4-dependent activation of HIS3.
Because GCN4 is required for activation of the HIS3 gene (33), we examined expression of GCN4 in the Δmbf1 strain. Virtually the same levels of GCN4 were observed in the wild type, the Δmbf1 strain, and the Δmbf1 strain harboring pMBF1 when Western blots of 20 μg of proteins in each yeast extract were probed with the antiserum against GCN4 (Fig. 1C). Under these conditions, signal intensity increased with protein loading up to at least 20 μg (data not shown). No GCN4 protein was detected with the antiserum in the Δgcn4 strain, which was used as a control.
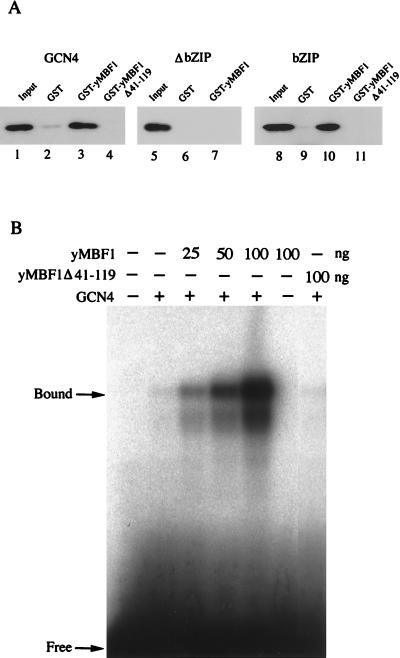
We next analyzed interaction between yMBF1 and GCN4. GST pull-down assays of bacterially expressed and purified proteins showed that GCN4 binds directly to yMBF1 (Fig. 2A; compare lanes 2 and 3). In this and all subsequent GST pull-down assays, addition of 50 μg of ethidium bromide per ml to the binding mixture did not affect the interactions, suggesting that the observed interactions were not due to bridging through contaminating nucleic acid (data not shown). The DNA-binding domain of GCN4 (bZIP; amino acid positions 222 to 281) is necessary and sufficient for the binding (Fig. 2A; compare lanes 6 and 7 with lanes 9 and 10). No binding occurred in the absence of Mg2+, and the complex that formed in the presence of Mg2+ dissociated immediately upon removal of Mg2+ (data not shown). Deletion of a central region (amino acid positions 41 to 119) of yMBF1 (yMBF1Δ41-119) abolished the binding of GCN4 (Fig. 2A, lanes 4 and 11). The corresponding region of insect MBF1 has been shown to be essential for the binding of FTZ-F1 (34). Furthermore, we observed a significant increase in the DNA binding of GCN4 upon addition of purified yMBF1 to gel mobility shift assay mixtures (Fig. 2B). Competition with unlabeled oligonucleotide carrying the wild-type or mutant GCN4-binding site revealed that the shifted bands represent the GCN4-DNA complex (data not shown). When we used yMBF1Δ41-119, which does not interact with GCN4, no increase in the GCN4 binding to DNA was detectable (Fig. 2B). Essentially the same results were obtained when we used GCN4bZIP in place of intact GCN4 (data not shown). The effect appears to be specific to GCN4, as yMBF1 did not increase the DNA binding of other sequence-specific factors (for example, see Fig. 6 for TBP binding). Because the mixture contained 5 mg of BSA per ml, it is highly unlikely that the yMBF1 simply prevented either GCN4 denaturation or sticking of GCN4 to the test tube. Accumulation, but not supershift, of the GCN4-DNA complex was detected upon addition of MBF1, as observed for the FTZ-F1–DNA complex (34). This is due to immediate dissociation of the GCN4-MBF1 complex in the electrophoresis buffer without Mg2+. No qualitative changes were observed in the DNase I footprinting patterns of GCN4 upon addition of yMBF1 (data not shown). Therefore, we surmise that the contact of yMBF1 with the DNA-binding domain of GCN4 induces some conformational change in GCN4, allowing increased binding to DNA.
FIG. 2.
Direct interaction between yMBF1 and GCN4. (A) GST pull-down assay for interaction of yMBF1 with GCN4. Bacterially expressed and purified HA epitope-tagged GCN4 (lanes 2 to 4), GCN4ΔbZIP (lanes 6 and 7), or GCN4bZIP (lanes 9 to 11) (100 ng each) was incubated with 200 ng of either GST (lanes 2, 6, and 9), GST-yMBF1 (lanes 3, 7, and 10), or GST–yMBF1Δ41-119 (lanes 4 and 11). The bound GCN4 was electrophoresed by SDS–10% PAGE, and its immunoblot was probed with an anti-HA antibody. Lanes 1, 5, and 8: 1/10 the input GCN4 or its derivatives. (B) yMBF1 increases GCN4 binding to DNA. The binding of bacterially expressed and purified GCN4 to a 32P-labeled oligonucleotide carrying the GCN4 binding site of HIS3 was analyzed by an electrophoresis mobility shift assay in the presence or absence of the indicated amounts of purified His-tagged yMBF1 or yMBF1Δ41-119.
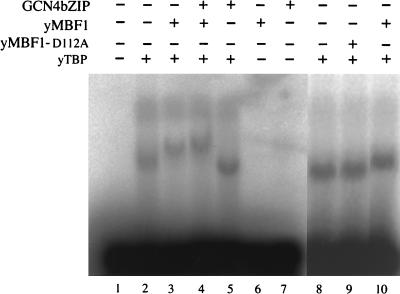
FIG. 6.
yTBP, yMBF1, and GCN4bZIP form a complex on a TATA element. 32P-labeled oligonucleotide bearing the TATA element of the HIS3 gene was incubated with various combinations of bacterially expressed and purified proteins. When indicated, the incubation mixture contained 10 ng of yTBP, 100 ng of yMBF1, 100 ng of yMBF1-D112A, and/or 100 ng of GCN4bZIP. The samples in lanes 1 to 7 were run on the same gel; those in lanes 8 to 10 were run on another gel. A faint band above the yTBP-TATA element complex appears to be a nonspecific complex, as it was competed out with a mutant TATA element carrying GCGC sequence in place of TATA, and hence we ignored it.
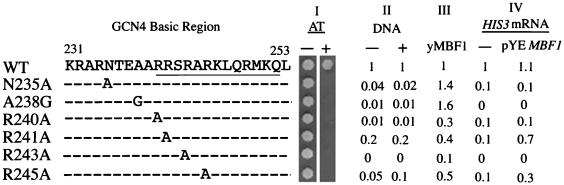
Interestingly, a part of the basic region in the GCN4 bZIP motif, RRSRARKLQRMKQ (underlined residues in Fig. 3), has homology with the C-terminal half of the FTZ-F1 box KRDRARKLQVMRQ (36) to which insect MBF1 binds (34) (identity of 9 amino acids and similarity of 11 amino acids of 13 residues). Therefore, we prepared four mutant forms of GCN4 in which each of the four arginine residues within the homologous region was replaced with alanine (R240A, R241A, R243A, and R245A). We also prepared two point mutants carrying replacements outside the homologous region (N235A and A238G). Bacterially expressed and purified mutant proteins migrated during SDS-PAGE with essentially the same mobility as the wild-type GCN4 (data not shown). GST interaction assays with these purified proteins showed that all four mutations within the homologous region reduced the binding to yMBF1 while the two point mutations outside the homologous region did not diminish but rather enhanced the binding (Fig. 3, column III). Gel mobility shift assays with purified proteins revealed that all six mutations reduced the DNA binding to various degrees (Fig. 3, column II). In the presence of 200 ng of yMBF1, the relative DNA-binding activities of the mutants carrying R241A and R245A were higher than those of the other mutants (Fig. 3, column II).
FIG. 3.
Genetic interaction between yMBF1 and the basic region of GCN4. Amino acid sequences of the basic region of wild-type GCN4 (WT) and its point mutants are shown. (Column I) AT sensitivity of GCN4 mutants. Indicated GCN4 derivatives were expressed in the Δgcn4 strain. Approximately 105 cells were spotted onto plates in the presence or absence of 20 mM AT and incubated for 3 days at 30°C. (Column II) DNA-binding activities of bacterially expressed and purified GCN4 mutants in the presence or absence of 200 ng of purified yMBF1 as determined by electrophoresis mobility shift assays. Band intensities of complexes of GCN4 mutant forms with DNA were quantitated with a Fuji BAS-2000II bioimage analyzer, and the levels relative to that of the WT are shown. (Column III) yMBF1-binding activities of GCN4 mutants analyzed by GST assays. Band intensities of GCN4 mutant forms on Western blots were quantitated with a Bio Image advanced quantifier, and the levels relative to that of the WT are shown. (Column IV) Suppression of gcn4 mutations by overexpression of yMBF1. Strains expressing the indicated GCN4 derivatives with or without pYEMBF1 were assessed for HIS3 and DED1 transcripts by S1 nuclease analyses. Relative levels of the HIS3 + 13 transcript normalized to the levels of the DED1 internal control after subtraction of the basal level in the Δgcn4 strain are shown.
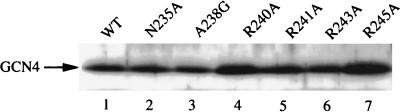
Δgcn4 cells expressing any of these mutant constructs were sensitive to AT (Fig. 3, column I) and defective in transcriptional activation of the HIS3 gene upon histidine starvation (Fig. 3, column IV). The mutant proteins were as stable as the wild-type GCN4 in vivo (Fig. 4). Overexpression of yMBF1 enhanced the transcription of HIS3 mRNA in mutants carrying R241A and R245A but not in those carrying R240A and R243A (Fig. 3, column IV). The sensitivities to 20 mM AT of the R241A and R245A mutants but not those of the R240A and R243A mutants were suppressed by overexpression of yMBF1, albeit partially in the mutant carrying R245A (data not shown). These in vivo data are consistent with the relatively high levels of DNA binding of R241A and R245A in the presence of excess yMBF1 (Fig. 3 column II). It is likely that arginine residues at 241 and 245 constitute a binding surface for yMBF1, whereas arginine residues at 240 and 243, on the opposite surface of the α helix, are known to face DNA (7). These data suggest that the binding of yMBF1 to the basic region of GCN4 is necessary for GCN4-dependent activation of the HIS3 gene.
FIG. 4.
Stability of GCN4 mutant proteins in yeast. The levels of wild-type GCN4 (WT) and the mutant forms in 20 μg of proteins of each yeast cell extract were estimated by Western analysis with the anti-GCN4 antiserum.
MBF1-TBP interaction is required for transcriptional activation of HIS3.
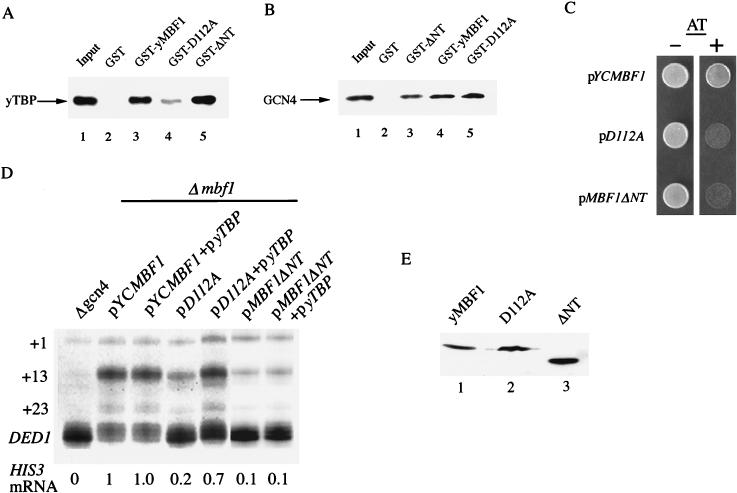
Next, we analyzed interaction between yMBF1 and yTBP. GST pull-down assays with bacterially expressed and purified proteins showed that yMBF1 comes into direct contact with yTBP (Fig. 5A; compare lanes 2 and 3). Like that of insect MBF1 (34), the central region of yMBF1 (amino acid positions 41 to 119) is essential for the TBP binding (data not shown). The yMBF1-yTBP interaction was confirmed by retardation of a yTBP-TATA element complex upon addition of purified yMBF1 in gel mobility shift assays (Fig. 6; compare lanes 2 and 3). We also found that substitution of alanine for aspartic acid at position 112, within the highly conserved domain of yMBF1 (34) (D112A), reduced the binding of yTBP (Fig. 5A; compare lanes 3 and 4), whereas the binding of yTBP to a truncated version of yMBF1 lacking its N-terminal 40 amino acids (yMBF1ΔNT) remained unaffected (Fig. 5A; compare lanes 3 and 5). The binding of GCN4 was not altered by the D112A mutation (Fig. 5B; compare lanes 4 and 5) but was slightly reduced by the N-terminal deletion of yMBF1 (Fig. 5B; compare lanes 3 and 4). Both the D112A and mbf1ΔNT mutants are sensitive to AT (Fig. 5C) and express significantly lower levels of HIS3 mRNA than the wild type (Fig. 5D). Overexpression of yTBP enhanced the HIS3 mRNA level in the mbf1D112A strain (Fig. 5D) and suppressed the AT sensitivity (data not shown). Neither the HIS3 mRNA level (Fig. 5D) nor the AT sensitivity (data not shown) of the mbf1ΔNT strain, used as a negative control, was affected by overexpression of yTBP. These mutant MBF1 proteins were as stable as the wild-type yMBF1 in vivo (Fig. 5E). These results indicate that the yMBF1-yTBP interaction is required for transcriptional activation of the HIS3 gene. Expression of GAL4-MBF1 or LexA-MBF1 fusion protein caused severe growth inhibition in yeast. This inhibition was alleviated by overexpression of TBP (34a). These results also suggest that MBF1 and TBP interact in yeast.
FIG. 5.
Interaction between yMBF1 and yTBP in vitro and in vivo. (A) GST pull-down assays for binding of yMBF1, the D112A protein, and yMBF1ΔNT to yTBP. Bacterially expressed and purified yTBP was incubated with either GST (lane 2), GST-yMBF1 (lane 3), GST-D112A protein (lane 4), or GST-yMBF1ΔNT (lane 5). The bound yTBP was separated by SDS–12.5% PAGE and, after transfer, probed with an anti-yTBP antibody. Lane 1, 1/10 the input yTBP. (B) GST pull-down assays for binding of the yMBF1 D112A protein and yMBF1ΔNT to GCN4. Purified HA epitope-tagged GCN4 was incubated with GST or indicated GST fusion proteins (lanes 2–5), and the bound GCN4 was detected as described in the legend for Fig. 2. Lane 1, 1/10 the input GCN4. (C) yMBF1 D112A and ΔNT mutants show AT sensitivity. The Δmbf1 strain transformed with pYCMBF1, pD112A, or pMBF1ΔNT was tested for growth as described in the legend for Fig. 3. (D) Suppression of mbf1 D112A by overexpression of yTBP. Total RNA from the Δgcn4 strain or the Δmbf1 stain harboring the indicated plasmids was subjected to S1 nuclease analysis. Relative levels of the HIS3 + 13 transcript are expressed as in Fig. 3. (E) Stability of yMBF1 mutant proteins in yeast. The levels of wild-type yMBF1 (lane 1) and its mutant forms, the D112A protein (lane 2) and yMBF1ΔNT (lane 3), in 20 μg of proteins of each yeast cell extract were estimated by Western analysis with an anti-MBF1 antiserum.
Formation of a TBP-MBF1-GCN4 ternary complex in vitro.
As described above, when yMBF1 was added to gel mobility shift assay mixtures consisting of yTBP and the TATA element of the HIS3 gene, we observed a supershift of the yTBP-TATA element complex (Fig. 6; compare lanes 2 and 3). Such supershift was not detectable upon addition of yMBF1-D112A (Fig. 6; compare lanes 8 to 10). This result confirms the weak nature of the interaction of yMBF1-D112A and TBP. Upon further addition of the DNA-binding domain of GCN4 (GCN4bZIP), further retardation of the supershifted complex was detected (Fig. 6; compare lanes 3 and 4). The retardation was observed only in the presence of yMBF1 and not in its absence (Fig. 6; compare lanes 4 and 5). Similar results were obtained when we used intact GCN4 instead of GCN4bZIP (data not shown). These results suggest formation of a yTBP-yMBF1-GCN4 ternary complex on the TATA element. We were unable to use the anti-GCN4 antiserum or the anti-HA antibody for detection of GCN4 in the ternary complex by supershift, because neither of these antibodies supershifted any complex, even the GCN4-DNA complex (data not shown).
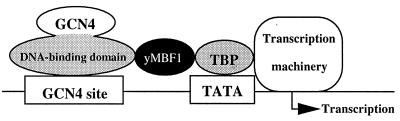
Based on these biochemical and genetic data, we conclude that yMBF1 acts as a coactivator of GCN4 by tethering TBP to the DNA-binding region of GCN4 (Fig. 7).
FIG. 7.
Model for GCN4-dependent activation through yMBF1. yMBF1 serves as a coactivator of GCN4 by tethering TBP to the DNA-binding domain of GCN4.
Recruitment of TBP through MBF1 is rate limiting in GCN4-dependent activation.
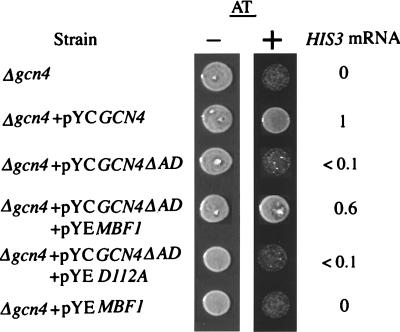
When TBP is artificially recruited by covalent attachment to a promoter-bound protein, an activation domain is no longer required for transcription (6, 21, 38). It is possible that the recruitment of TBP by yMBF1 requires the aid of the activation domain because the protein-protein interaction for tethering of TBP is not as firm as the covalent linkage of TBP. In agreement with this idea, transcription of the HIS3 gene in the absence of the major activation domain of GCN4 was partially activated by overexpression of yMBF1 but not by overexpression of D112A protein (Fig. 8). GCN4ΔAD retains a residual activation domain near its N terminus (16). It is possible that the observed suppression may be due to enhanced expression of GCN4ΔAD by overexpression of yMBF1. However, Western analysis of yeast cell extracts showed that the expression of GCN4ΔAD was not altered by overexpression of yMBF1 (data not shown). These results suggest that the recruitment of TBP through MBF1 is a rate-limiting step in GCN4-dependent activation and hence is a target of regulation.
FIG. 8.
Overexpression of yMBF1 restores activation of HIS3 to a GCN4 mutant lacking the activation domain. For evaluation of AT sensitivity, approximately 105 cells of the Δgcn4 strain or the Δgcn4 strain transformed with the indicated plasmid were grown on plates in the presence or absence of 2.5 mM AT for 3 days at 30°C. For evaluation of HIS3 mRNA, total RNA from the Δgcn4 strain harboring the indicated plasmids was subjected to S1 nuclease analysis; relative levels of the HIS3 +13 transcript are expressed as in Fig. 3.
The present study illustrates the importance of the basic region within the bZIP motif of GCN4 for the interaction with yMBF1. The basic amino acids relevant to genetic interaction with yMBF1, R241 and R245, are conserved at corresponding positions in a subgroup of bZIP proteins. This, together with the evolutionary conservation of MBF1, raises the possibility that MBF1 serves as a coactivator for a wider spectrum of bZIP proteins. The human T-cell leukemia virus transactivator Tax has been shown to interact with the basic region of certain bZIP proteins and to increase the DNA binding of these proteins (2, 28). Although MBF1 does not have apparent sequence homology with Tax, the two may carry out their functions through a common mechanism.
ACKNOWLEDGMENTS
We are grateful to K. Struhl for yeast strain KY803 and YCp88-GCN4, M. Horikoshi for yTBP DNA, R. D. Gietz for yeast vectors, A. G. Hinnebusch for anti-GCN4 antiserum, and M. Jindra for anti-MBF1 antiserum. We also thank I. Herskowitz, M. Jindra, J.-I. Tomizawa, and A. Ishihama for critical reading of the manuscript, H. Mitsuzawa for helpful discussions, and M. Fujino for advice on RNA isolation.
This work was supported by grants-in-aid for scientific research from the Ministry of Education, Science, Sports and Culture of Japan. K.T. was supported by a C.O.E. program.
REFERENCES
- 1.Akimaru H, Chen Y, Dai P, Hou D-X, Nonaka M, Smolik S M, Armstrong S, Goodman R H, Ishii S. Drosophila CBP is a co-activator of cubitus interruptus in hedgehog signalling. Nature. 1997;386:735–738. doi: 10.1038/386735a0. [DOI] [PubMed] [Google Scholar]
- 2.Baranger A M, Palmer C R, Hamm M K, Giebler H A, Brauweiler A, Nyborg J K, Schepartz A. Mechanism of DNA-binding enhancement by the human T-cell leukaemia virus transactivator Tax. Nature. 1995;376:606–608. doi: 10.1038/376606a0. [DOI] [PubMed] [Google Scholar]
- 3.Becker D M, Fikes J D, Guarente L. A cDNA encoding a human CCAAT-binding protein cloned by functional complementation in yeast. Proc Natl Acad Sci USA. 1991;88:1968–1972. doi: 10.1073/pnas.88.5.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brownell J E, Zhou J, Ranalli T, Kobayashi R, Edmondson R, Roth S Y, Allis C D. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 5.Chakravarti D, LaMorte V J, Nelson M C, Nakajima T, Schulman I G, Juguilon H, Montiminy M, Evans R M. Role of CBP/p300 in nuclear receptor signaling. Nature. 1996;383:99–103. doi: 10.1038/383099a0. [DOI] [PubMed] [Google Scholar]
- 6.Chatterjee S, Struhl K. Connecting a promoter-bound protein to TBP bypasses the need for a transcriptional activation domain. Nature. 1995;374:820–822. doi: 10.1038/374820a0. [DOI] [PubMed] [Google Scholar]
- 7.Ellenberger T E, Brand C J, Struhl K, Harrison S C. The GCN4 basic region leucine zipper binds DNA as a dimer of uninterrupted α helices: crystal structure of the protein-DNA complex. Cell. 1992;71:1223–1237. doi: 10.1016/s0092-8674(05)80070-4. [DOI] [PubMed] [Google Scholar]
- 8.Field J, Nikawa J, Broek D, MacDonald B, Rodgers L, Wilson I A, Lerner R A, Wigler M. Purification of a RAS-responsive adenylyl cyclase complex from Saccharomyces cerevisiae by use of an epitope addition method. Mol Cell Biol. 1988;8:2159–2165. doi: 10.1128/mcb.8.5.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gietz R D, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 10.Goffeau A, Barrell B G, Bussey H, Davis R W, Dujon B, Feldmann H, Galibert F, Hoheisel J D, Jacq C, Johnston M, et al. Life with 6000 genes. Science. 1996;274:546–567. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- 11.Grant P A, Duggan L, Côtté J, Roberts S M, Brownell J E, Candau R, Ohba R, Owen-Hughes T, Allis C D, Winston F, Berger S L, Workman J L. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- 12.Hoffmann A, Roeder R G. Purification of his-tagged proteins in non-denaturing conditions suggests a convenient method for protein interaction studies. Nucleic Acids Res. 1991;19:177–188. doi: 10.1093/nar/19.22.6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hope I A, Struhl K. Functional dissection of a eukaryotic transcriptional activator protein, GCN4 of yeast. Cell. 1986;46:885–894. doi: 10.1016/0092-8674(86)90070-x. [DOI] [PubMed] [Google Scholar]
- 14.Horiuchi J, Silverman N, Marcus G A, Guarente L. ADA3, a putative transcriptional adaptor [sic], consists of two separable domains and interacts with ADA2 and GCN5 in a trimeric complex. Mol Cell Biol. 1995;15:1203–1209. doi: 10.1128/mcb.15.3.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iyer V, Struhl K. Absolute mRNA levels and transcriptional initiation rates in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1996;93:5208–5212. doi: 10.1073/pnas.93.11.5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jackson B M, Drysdale C M, Natarajan K, Hinnebusch A G. Identification of seven hydrophobic clusters in GCN4 making redundant contributions to transcriptional activation. Mol Cell Biol. 1996;16:5557–5571. doi: 10.1128/mcb.16.10.5557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson P, McKnight S. Eukaryotic transcriptional regulatory proteins. Annu Rev Biochem. 1989;58:799–839. doi: 10.1146/annurev.bi.58.070189.004055. [DOI] [PubMed] [Google Scholar]
- 18.Kaiser C, Michaelis S, Mitchell A, editors. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1994. pp. 149–150. [Google Scholar]
- 19.Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, Lin S-C, Heyman R A, Rose D W, Glass C K, Rosenfeld M G. A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- 20.Kim U, Qin X-F, Gong S, Stevens S, Luo Y, Nussenzweig M, Roeder R G. The B-cell-specific transcription coactivator OCA-B/OBF-1/Bob-1 is essential for normal production of immunoglobulin isotypes. Nature. 1996;383:542–547. doi: 10.1038/383542a0. [DOI] [PubMed] [Google Scholar]
- 21.Klages N, Strubin M. Stimulation of RNA polymerase II transcription initiation by recruitment of TBP in vivo. Nature. 1995;374:822–823. doi: 10.1038/374822a0. [DOI] [PubMed] [Google Scholar]
- 22.Köhrer K, Domdey H. Preparation of high molecular weight RNA. Methods Enzymol. 1991;194:398–405. doi: 10.1016/0076-6879(91)94030-g. [DOI] [PubMed] [Google Scholar]
- 23.Lewin B. Commitment and activation at pol II promoters: a tail of protein-protein interactions. Cell. 1990;61:1161–1164. doi: 10.1016/0092-8674(90)90675-5. [DOI] [PubMed] [Google Scholar]
- 24.Li F-Q, Ueda H, Hirose S. Mediators of activation of fushi tarazu gene transcription by BmFTZ-F1. Mol Cell Biol. 1994;14:3013–3021. doi: 10.1128/mcb.14.5.3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mitchell P J, Tjian R. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science. 1989;245:371–378. doi: 10.1126/science.2667136. [DOI] [PubMed] [Google Scholar]
- 26.Murata T, Kageyama Y, Hirose S, Ueda H. Regulation of the EDG84A gene by FTZ-F1 during metamorphosis in Drosophila melanogaster. Mol Cell Biol. 1996;16:6509–6515. doi: 10.1128/mcb.16.11.6509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ogryzko V V, Schiltz R L, Russanova V, Howard B H, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 28.Perini G, Wagner S, Green M R. Recognition of bZIP proteins by the human T-cell leukaemia virus transactivator Tax. Nature. 1995;376:602–605. doi: 10.1038/376602a0. [DOI] [PubMed] [Google Scholar]
- 29.Ptashne M, Gann A. Transcriptional activation by recruitment. Nature. 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- 30.Roeder R G. The complexities of eukaryotic transcription initiation: regulation of preinitiation complex assembly. Trends Biochem Sci. 1991;16:402–408. doi: 10.1016/0968-0004(91)90164-q. [DOI] [PubMed] [Google Scholar]
- 31.Schubart D B, Rolink A, Kosco-Vilbois M H, Botteri F, Matthias P. B-cell-specific coactivator OBF-1/OCA-B/Bob1 required for immune response and germinal centre formation. Nature. 1996;383:538–542. doi: 10.1038/383538a0. [DOI] [PubMed] [Google Scholar]
- 32.Stringer K F, Ingles C J, Greenblatt F. Direct and selective binding of an acidic transcriptional activation domain to the TATA-box factor TFIID. Nature. 1990;345:783–786. doi: 10.1038/345783a0. [DOI] [PubMed] [Google Scholar]
- 33.Struhl K. Constitutive and inducible Saccharomyces cerevisiae promoters: evidence for two distinct molecular mechanisms. Mol Cell Biol. 1986;6:3847–3853. doi: 10.1128/mcb.6.11.3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takemaru K, Li F-Q, Ueda H, Hirose S. Multiprotein bridging factor 1 (MBF1) is an evolutionarily conserved transcriptional coactivator that connects a regulatory factor and TATA element-binding protein. Proc Natl Acad Sci USA. 1997;94:7251–7256. doi: 10.1073/pnas.94.14.7251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34a.Takemaru, K., and S. Hirose. Unpublished observation.
- 35.Treizenberg S J. Structure and function of transcriptional activation domains. Curr Opin Genet Dev. 1995;5:190–196. doi: 10.1016/0959-437x(95)80007-7. [DOI] [PubMed] [Google Scholar]
- 36.Ueda H, Sun G-C, Murata T, Hirose S. A novel DNA-binding motif abuts the zinc finger domain of insect nuclear hormone receptor FTZ-F1 and mouse embryonal long terminal repeat-binding protein. Mol Cell Biol. 1992;12:5667–5672. doi: 10.1128/mcb.12.12.5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu Y, Reece R J, Ptashne M. Quantitation of putative activator-target affinities predicts transcriptional activating potentials. EMBO J. 1996;15:3951–3963. [PMC free article] [PubMed] [Google Scholar]
- 38.Xiao H, Friesen J D, Lis J T. Recruiting TATA-binding protein to a promoter: transcriptional activation without an upstream activator. Mol Cell Biol. 1995;15:5757–5761. doi: 10.1128/mcb.15.10.5757. [DOI] [PMC free article] [PubMed] [Google Scholar]