Abstract
Sensorineural hearing loss is irreversible and can be caused by loss of auditory neurons. Regeneration of neural cells from endogenous cells may offer a future tool to restore the auditory circuit and to enhance the performance of implantable hearing devices. Neurons and glial cells in the peripheral nervous system are closely related and originate from a common progenitor. Prior work in our lab indicated that in the early postnatal mouse inner ear, proteolipid protein 1 (Plp1) expressing glial cells could act as progenitor cells for neurons in vitro. Here, we used a transgenic mouse model to transiently overexpress Lin28, a neural stem cell regulator, in Plp1-positive glial cells. Lin28 promoted proliferation and conversion of auditory glial cells into neurons in vitro. To study the effects of Lin28 on endogenous glial cells after loss of auditory neurons in vivo, we produced a model of auditory neuropathy by selectively damaging auditory neurons with ouabain. After neural damage was confirmed by the auditory brainstem response, we briefly upregulated the Lin28 in Plp1-expressing inner ear glial cells. One month later, we analyzed the cochlea for neural marker expression by quantitative RT-PCR and immunohistochemistry. We found that transient Lin28 overexpression in Plp1-expressing glial cells induced expression of neural stem cell markers and subsequent conversion into neurons. This suggests the potential for inner ear glia to be converted into neurons as a regeneration therapy for neural replacement in auditory neuropathy.
Keywords: glial conversion, inner ear, mouse models, neural stem cells, regeneration
1 |. INTRODUCTION
Sensorineural hearing loss (SNHL) is generally thought of as loss of primary sensory cells (hair cells), and/or spiral ganglion neurons (SGNs). Auditory neuropathy describes the sole loss or functional impairment of SGNs, which may be inherited or acquired.
In cases of partial hair cell loss, hearing can be aided with amplifying devices, for example, hearing aids. In cases of major hair cell loss, implantable devices such as cochlear implants are able to replace hair cell function and transmit the signal to the brain. In auditory neuropathy, however, these devices have little success, as they rely on intact auditory (cochlear) nerves for signal transmission.1–6 Regeneration strategies for auditory neurons would offer a treatment to restore biological hearing.
Postmitotic cells that have assumed their mature, differentiated fate can, undergo adaptive cellular reprogramming driven by overexpression of transcription factors in vitro6–8 and in vivo.9,10 Recent work suggests that reprogramming of cells in the same lineage may be expedited: neuronal and glial lineages are developmentally related through common progenitors,11–13 and glial cells of the central nervous system (CNS) can become neurons upon forced overexpression of several proneural genes.11,14–16 We and others found that myelin proteolipid protein 1 (Plp1)-positive glial cells in the cochlea spontaneously differentiated into neurons and grew neurites and formed synapses.17–25 The glial cells of the spiral ganglion are derived from neural crest and continue to express Plp1 postnatally in Schwann cells of the osseous spiral lamina (OSL) along the peripheral SGN axons, as well as along the central axons, and in the satellite cells surrounding SGN cell bodies.20,26,27 During development, Plp1 is present in early, undifferentiated precursors and migratory neural crest cells, and Plp1 expressing cochlear glial cells may represent a dormant population of early neural crest progenitors that is responsive to Lin28 signaling.
In the adult inner ear in vivo, reactive gliosis after damage may create an environment more permissive for transdifferentiation and neural reprogramming, similar to the CNS,28,29 but to date, the contribution of Plp1-positive cells to postnatal gliogenesis and neurogenesis remains unclear.30,31
The RNA binding protein, Lin28, together with Sox2, Oct4, and NANOG, can reinduce pluripotency in murine cells32,33; Lin28 promotes proliferation of neural precursors.34 Overexpression of Lin28 during early development causes proliferation, whereas at later time points, Lin28 interacts with Let-7 to promote differentiation.35–37 Cell cycle regulator high mobility group AT-hook 2 (Hmga2) is another downstream target of Lin28.38 In combination with Sox2, Hmga2 facilitates conversion of human adult fibroblasts and umbilical cord cells into induced neural stem cells.8,39,40 Decreasing expression of Hmga2 with maturation is believed to be due to Let-7 microRNA regulation.41 A recently described Sox2-Lin28/Let-7 axis maintains neural progenitors and activates neuronal differentiation34,42–44 via interaction with bHLH transcription factors.34,45
Lin28 is widely expressed during early cochlear development, but is downregulated before the onset of hearing46,47; expression of Hmga2 mirrors that of Lin28, but narrows to hair cells and supporting cells postnatally.48 Prolonged expression of Lin28 delays cell cycle exit and maintains an undifferentiated state in inner ear sensory cells.49 Proneural bHLH transcription factors are upregulated in reactive glia after damage to adult auditory neurons,31 and forced overexpression of Ascl1 or Neurod1 in non-neural inner ear cells in vitro can generate neuron-like cells.50,51
We show here that Lin28 initiates proliferation of Pl p1-positive cochlear glial cells isolated and grown as neurospheres in vitro and promotes neurogenesis by activating proneural bHLH transcription factors through inhibition of Let-7. We further show that transient Lin28 overexpression reprograms inner ear glial cells to neurons in an in vivo model of auditory neuropathy.
2 |. MATERIALS AND METHODS
All animal studies were approved by our Institutional Animal Care and Use Committee (IACUC) prior to conduction of experiments.
2.1 |. Isolation of spiral ganglion cells for neurosphere culture
For each experiment, ears of neonatal Plp1-CreER;rtTA;Lin28a-TetO; or Plp1-CreER;Lin28a-fl;tdTm postnatal day P3-P4 pups (generous gift from George Q. Daley) were used. Plp1-CreER;rtTA;Lin28a-TetO;tdTm mice express tdTomato, a red fluorescent reporter, rtTA, and Lin28-TetO in Plp1-positive glial cells. After treatment with tamoxifen (Tam), rtTA, and tdTomato are expressed. We use tdTomato to permanently label and trace Plp1-positive glial cells. In the presence of doxycycline (Dox), rtTA interacts with the Tet-responsive element (TetO) to transiently upregulate Lin28 in a concentration dependent manner. Plp1-CreER;Lin28a-fl;tdTm mice allow for timed deletion of Lin28a in Plp1 glia which are also labeled with the tdTomato fluorescent reporter after treatment with Tam. Plp1-CreER;tdTm mice were used as controls. These mice have Plp1 positive glial cells labeled with red fluorescent reporter to allow for tracing after treatment with Tam, but are otherwise unchanged (Figure 1A).
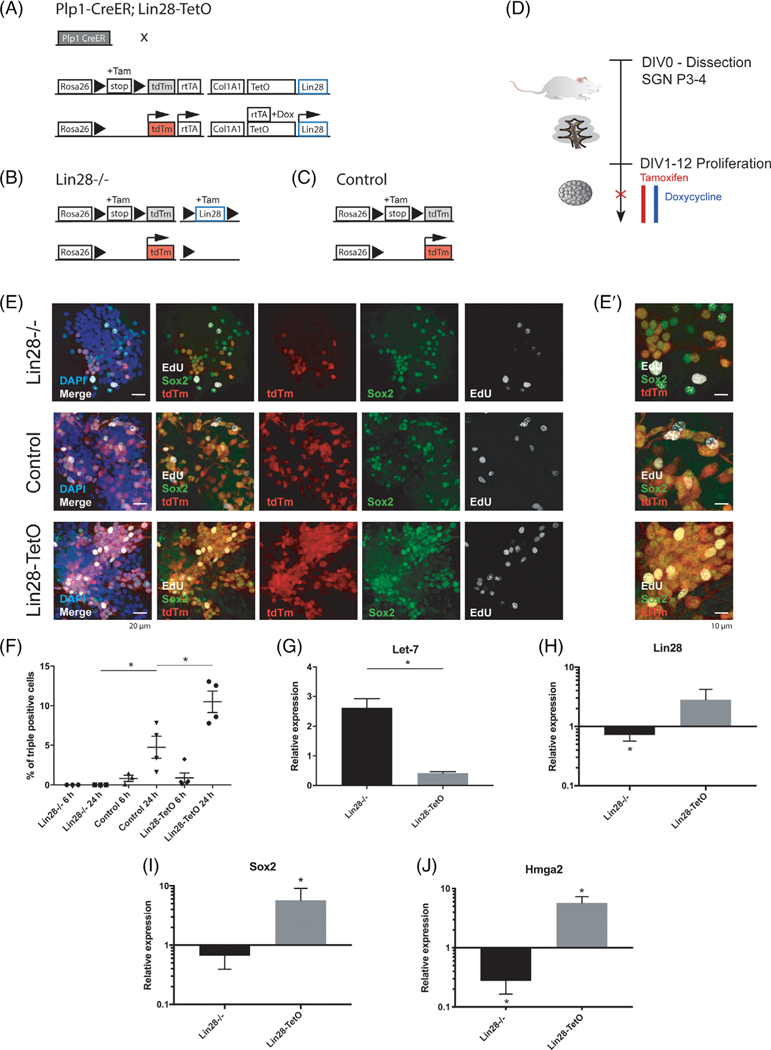
FIGURE 1.
Proliferation of Plp1-expressing glial cells after knockout or overexpression of Lin28 in vitro. A, For Lin28 overexpression in Plp1-positive cells, Plp1-CreER;STOP fl; rtTA mice and Lin28a/b-TetO, Lafayette, Colorado; STOP fl; tdTm (tdTm) mice were crossed to obtain Plp1-CreER;rtTA;Lin28a/b-TetO;tdTm (Lin28-TetO) transgenic mice. Tamoxifen application initiated Cre recombinase dependent tdTm and tetracycline-controlled transactivator (rtTA) expression. In the presence of doxycycline, the tetracycline operator (TetO) was transiently activated and led to overexpression of Lin28. B, For the Lin28 knockout experiments, Plp1-CreER mice were crossed with Lin28a-fl;tdTm mice to obtain Plp1-CreER;Lin28a-fl;tdTm (Lin28−/−) transgenic mice. Application of tamoxifen led to conditional knockout of Lin28 in tdTm expressing Plp1 positive cells. C, Plp1-CreER;tdTm mice were used as controls for Lin28 overexpression and knockout experiments (control). D, Timeline for neurosphere assay from early postnatal spiral ganglion cells. Pups were dissected at P3-P4 and kept under proliferating conditions for 12 days, undergoing two passages; DIV, days in vitro. Spheres were treated after the second passage and analyzed by immunohistochemistry (red X). E, Second-generation neurospheres were treated with ethynyl-desoxyuridine (EdU) for either 6 or 24 hours and harvested for immunohistochemistry. Neurospheres of Lin28−/− (first row), control (second row), and Lin28-TetO (third row) mice were stained for EdU (white) and Sox2 (green), nuclei were labeled with DAPI (blue). Plp1-positive cells expressed tdTm (red). Merged images in the first two columns show co-localization. Scale bar = 20 μm. E′, High-magnification image of EdU-Sox2-tdTm colocalization. Scale bar = 10 μm. F, Quantification of tdTm, Sox2, and EdU-triple-positive (+) cells as percentage of the total number of tdTm+ cells after 6 or 24 hours of EdU treatment (*P < .05, mean ± SEM). G, Relative expression of Let-7 in proliferating Lin28−/− and Lin28-TetO neurospheres (mean ± SEM, *P < .05, n = 3). H-J, Quantitative RT-PCR of Lin28 (n = 7), Sox2 (n = 5), and Hmga2 (n = 6) in proliferating Lin28−/− and Lin28-TetO neurospheres compared to control (mean ± SEM, *P < .05, control set as 1)
Ears were dissected in Hank’s Balanced Salt Solution (HBSS) and the modiolus containing spiral ganglion tissue with Plp1 positive glial cells was separated from the organ of Corti and stria vascularis. Given that only few pups per litter produced the desired genotype (quadruple transgenic), we modified our assay to allow reliable sphere formation from two ears (one mouse, roughly 50 000 cells per spiral ganglion). This was technically more challenging due to lower cell numbers, but allowed for good control of clonal expansion of spheres.21 The tissues were incubated in TrypLE for 20 minutes at 37°C and were manually dissociated to a single cell solution (P100 pipette). The triturated cells were passed through a 70 μm cell strainer (BD Labware Franklin Lakes, New Jersey) to remove larger tissue debris.
For cell culture, basic medium (DMEM/F12 [1:1] containing GlutaMAX) was supplemented with N2 (100X, Invitrogen Waltham, Massachusetts) and B27 (50X, Invitrogen, Waltham, Massachusetts), amphotericin B (0.83 μg/mL, Gibco, Waltham, Massachusetts), ampicillin (50 ng/mL), and Hepes (10 mM, Gibco). Single cells (2 spiral ganglia per well) were maintained in ultra-low attachment surface 24-well plates (Costar, Corning, Tewksbury, Massachusetts). To obtain first-generation spheres, the medium was supplemented with EGF (20 ng/mL; Chemicon, Temecula, California), bFGF (10 ng/mL; Chemicon), IGF-1 (50 ng/mL; Chemicon), and heparan sulfate (50 ng/mL; Sigma, St. Louis, Missouri).
First-generation spheres are highly heterogenous and contain primary, nonproliferating cells. Two additional passages were therefore performed, after 4 and 8 DIV (days in vitro), to purify neurosphere populations and improve clonal expansion of Plp1-positive spheres. Passaging was performed by incubating first- or second-generation spheres in TrypLE for 20 minutes at 37°C, followed by dissociation with a P100 pipette 20 times. Single cell suspensions were cultured in fresh medium to form second- and third-generation spheres, respectively. Roughly 100 spheres can be obtained per animal, and number of spheres roughly doubles with each passage.
2.2 |. Treatment of neurospheres during proliferation
Single cells were cultured in the proliferation medium described above with supplemented 4-hydroxytamoxifen (200 nm/mL; Sigma) or 4-hydroxytamoxifen and doxycycline (20 ng/mL, Sigma), starting at 0 DIV. Fresh supplemented medium with growth factors and drugs were used at 4 and 8 DIV after splitting.
2.3 |. Treatment of neurospheres during differentiation
Third-generation neurospheres were plated in basic culture medium without growth factors in 4-well plates (Greiner, Kremsmünster, Austria) on round 10 mm glass coverslips coated with 10% Matrigel (Corning) in basic medium. Cells designated for RNA extraction and quantitative reverse transcription polymerase chain reaction (RT-PCR) were cultured in treated 24-well plates (CytoOne, USA Scientific, Ocala, Florida). Neurospheres were differentiated for 9 to 12 days. For treatment, 4-hydroxytamoxifen or 4-hydroxytamoxifen and doxycycline were added. Cells were harvested and further analyzed by immunohistochemistry or RT-PCR as described for cultured neurospheres.
2.4 |. SiRNA (small interfering RNA) silencing
Cochlear neurospheres were transfected with siRNA (100 nM ON-TARGET plus, Smart Pool for Sox2, Hmga2, or non-targeting siRNA from Dharmacon), using Gene Silencer (Genlantis, San Diego, California) for 24 hours according to the manufacturer’s instructions. Transfection efficiency was monitored using siGlo Red Transfection Indicator (Dharmacon). After transfection, the cells were washed and cultured in DMEM/F12 and N2/B27. For immunohistochemistry and quantification after siRNA treatment, neurospheres were differentiated for 24 hours after transfection and were cultured for up to 6 days after siRNA treatment.
2.5 |. Immunohistochemistry of neurospheres
Differentiated or proliferating neurospheres were fixed at room temperature in 4% paraformaldehyde/phosphate buffered saline (PBS) for 10 minutes and then washed in PBS. Permeabilization and blocking was performed for spheres or sections with blocking solution (0.3% Triton X-100, 15% heat-inactivated goat or donkey serum in PBS) for 1 hour. Diluted primary antibody (0.1% Triton X-100, and 10% heat inactivated goat or donkey serum in PBS) was applied overnight at 4°C. Secondary antibodies were applied for 2 hours at room temperature. Nuclei were visualized with 4,6-diamidino-2-phenylindole (DAPI, Vector Laboratories, Burlingame, California). EdU (5-ethynyl-2′-deoxyuridine) was added to the culture medium for 6 or 24 hours and staining was performed according to the manufacturer’s instructions (Click-iT EdU Imaging Kit 647, Invitrogen). Immunohistochemical staining was analyzed by confocal microscopy (SP8, Leica, Wetzlar, Germany). The LAS X software was used for data acquisition for qualitative and quantitative analysis. The number of cells was manually quantified with the ImageJ-Fiji software. Cell counts were expressed as mean ± SE of the mean. The Graphpad Prism software was used for statistical evaluation.
2.6 |. Measurement of neurosphere diameter and proliferation
Fifty first-generation neurospheres were picked per condition (three replicates) and passaged. Right after passage, neurospheres were treated with siRNA as described above, and each well of each replicate was analyzed with bright field (inverted microscope, Leica DMi 8). After the second passage, EdU was added to each well for 12 hours. Third-generation neurospheres were then fixed and stained with DAPI, mounted and analyzed with confocal microscopy for EdU and DAPI (SP8, Leica). Cell diameter was measured with the ImageJ-Fiji software. Differentiating spheres were fixed after 7 days and stained and quantified for TuJ and DAPI.
2.7 |. Antibodies
The antibodies used were polyclonal goat antibody against Sox2 (used at 1:300; Santa Cruz), monoclonal mouse antibody against Tubulin beta 3, TuJ (used at 1:250–1:500; Bio Legend, San Diego, California), polyclonal rabbit antibody against Lin28 (used at 1:500, Abcam, Cambridge, UK), polyclonal rabbit antibody against doublecortin, DCX (used at 1:1000, Abcam). Secondary antibodies for detection of primary antibodies: Alexa Fluor 488, 568, and 647-conjugated (1:500; Invitrogen). EdU staining was performed according to the manufacturer’s instructions of the Click-iT EdU Alexa Fluor 647 Imaging Kit (Invitrogen).
2.8 |. Quantitative RT-PCR
Proliferated or differentiated spheres were harvested and stored in RLT lysis buffer at −80°C according to the manufacturer’s instructions. The same storage conditions were applied to embryonic and postnatal tissue for RNA extraction. For adult RT-PCR analysis after treatment in vivo, the cochlea was collected and dissected. Bone fragments covering the modiolus were carefully removed as much as possible to collect the modiolar tissue.
Total RNA was extracted from spheres or embryonic tissue with the RNeasy Micro Kit (Qiagen, Hilden, Germany) according to manufacturer’s instructions. RNA was denatured at 65°C for 5 minutes. For reverse transcription, ImProm II (Promega) was used with random hexamers. The reverse transcription conditions were 25°C for 5 minutes followed by 42°C for 60 minutes. The reaction was terminated at 70°C for 15 minutes. The cDNAs were mixed with Platinum quantitative PCR Mastermix ROX with UDG (Invitrogen) and primers for Gapdh (Mm99999915_g1), Lin28 (Mm00524077_m1), Neurod1 (Mm01946604_s1), Neurog1 (Mm00440466_s1), Ascl1 (Mm03058063_m1), Hmga2 (Mm04183367_g1), or Sox2 (Mm03053810_s1). All probes are TaqMan FAM-MDG inventoried probes (Applied Biosystems, Foster City, California/Thermo Fisher Scientific, Waltham, Massachusetts).
Gene expression was measured relative to Gapdh. Samples were analyzed in 96-well plates in triplicate by PCR (Applied Biosystems 7900HT) using the following conditions: initial denaturation at 95°C for 2 minutes, denaturation at 95°C for 40 seconds, and annealing/extension at 60°C for 35 seconds for 45 cycles. To detect expression levels of miRNA, miRNA Let-7 was extracted with the miRNeasy Mini Kit (Qiagen) from proliferating or differentiating neurospheres obtained from 2 to 4 ears. The reverse transcription conditions were 16°C for 30 minutes followed by 42°C for 30 minutes. The reaction was terminated at 85°C for 5 minutes. The cDNAs were mixed with platinum quantitative PCR Mastermix ROX with UDG (Invitrogen) and primers for Let-7 (ABI, Applied Biosystems). The amount of cDNA was normalized and measured relative to other samples on the plate.52 Samples were analyzed in 96-well plates in triplicate by PCR (Applied Biosystems 7900HT) using the same conditions described above. All statistical analyses were performed with the software Graphpad Prism.
2.9 |. Statistics
Statistical analysis of immunostained proliferating spheres was performed by a two-tailed t test. Statistical analysis of immunostained differentiated spheres was performed by one-way analysis of variance with Tukey correction. Quantitative RT-PCR of proliferating and differentiating spheres and analysis of Let-7 expression was performed by unpaired Kolmogorov-Smirnov t test. Analysis of immunostained adult SGN and quantitative RT-PCR data from adult SGN after ouabain was performed by unpaired Kolmogorov-Smirnov t test. All experiments were repeated at least three times. All statistics were analyzed using the Graphpad Prism software.
2.10 |. Tissue collection of embryos
Tissue was collected from embryos of wildtype mice at E9, E14, and E18, or from pups at postnatal day 4. In E9 embryos, the invaginating otic placode was identified microscopically and sharply dissected with two #55 forceps (FST, Fine Science Tools, Foster City, California) under high magnification. At E14, the developing otic capsule was identified after removal of the brain, and the cochlear portion was removed. At E18 and P4, the modiolus containing the spiral ganglion tissue was removed from the cochlea. The tissue was then stored in RLT buffer at −80°C and processed as described above for RT-PCR.
2.11 |. Immunohistochemistry of cryosections
Cryosections were dried and rehydrated in PBS. Antigen retrieval was performed with Antigen Unmasking Solution (Vector) diluted in deionized water for 5 minutes. After washing in PBS, cryosections were permeabilized, blocked, and stained as described above for neurospheres.
2.12 |. Mice for in vivo experiments
Plp1-CreER;rtTA/rtTA were mated with Lin28a-TetO/+;tdTm/tdTm or Lin28b-TetO/+;tdTm/tdTm (Lin28a-TetO and Lin28b-TetO) mice, a generous gift from George Q. Daley, Harvard University Boston to obtain heterozygous Plp1-CreER/+;rtTA/+;Lin28a/b-TetO/+;tdTm/+ mice to overexpress Lin28 in Plp1-expressing glial cells after tamoxifen and doxycycline treatment. For Plp1-CreER;tdTm and Plp1-CreER/+;rtTA/+;Lin28a/b-TetO/+;tdTm/+ mice, the right ear was treated with ouabain and the left ear served as control.
2.13 |. Neuropathy model
Neurons of the right ear were ablated by ouabain application to the round window of 6-week-old male and female Lin28-TetO mice or Plp1-CreER;tdTm mice, as described.53,54 Mice were anesthetized with ketamine (100 mg/kg, intraperitoneal, i.p.) and xylazine (10 mg/kg, i.p.) and injected with a half-dose every 30 minutes to maintain anesthesia during surgery. Underlying soft tissues including muscle, nerve, and vessels were bluntly separated through a retroauricular incision to expose the auditory bulla. Using a 30 G insulin syringe, the bulla was perforated above the round window niche without damaging the stapedial artery. Ouabain (1–2 μL, 1 mM in distilled water) was applied to the round window membrane using a 10 μL NanoFil syringe (34–36 G), exchanging the solution every 10 minutes for 60 minutes. The opening of the bulla was then closed with muscle and fascia and the incision was closed with non-absorbable 7–0 nylon suture and bacitracin ointment. If bleeding occurred during surgery, mice were injected with 100 μL saline directly after and 24 hours after surgery. For postsurgical pain control, mice were injected with buprenex (0.05 mg/kg). For recovery, mice were kept in a heated room or on a homeothermic blanket at 39.8°C under supervision. At day 3 to day 5 after surgery, 100 μL tamoxifen (50 mg/mL, Sigma, in corn oil, i.p.) was injected. A 100 μL doxycycline (100 μg in saline, i.p.) was injected on 3 days subsequently, starting at day 4 after surgery. In addition, 1 mg/mL doxycycline was administered to the drinking water. One or 4 weeks after injection, mice were sacrificed via cardiac perfusion with 4% paraformaldehyde. After removing the skullcap, mice heads were prepared for immunohistochemistry as described above. For analysis, the left ear served as an ouabain-untreated but tamoxifen-doxycycline-injected control. Plp1-CreER;tdTm mice served as tamoxifen-doxycycline-injected, ouabain-treated control without Lin28 upregulation.
3 |. RESULTS
3.1 |. Proliferation of Plp1-expressing glial cells in the presence of Lin28
Lin28 promotes proliferation at early developmental stages and cell fate specification at later developmental time points.55–58 To ask whether Lin28 could influence the fate of glial cells in the cochlea, we employed a Cre-recombinase-dependent, doxycycline-inducible Lin28a and Lin28b transgenic mouse model, containing a red reporter tag (tdTomato, tdTm), preceded by a loxP flanked STOP cassette (Plp1-CreER;rtTA;Lin28-TetO;tdTm) to transiently overexpress Lin28 in postnatal Plp1-positive cochlear glial cells grown as neurospheres in vitro (Figure 1A,D). This allowed us to activate Lin28 in Plp1-expressing cells and to perform lineage tracing at specific time points after tamoxifen treatment. Inner ear neurospheres demonstrate a low level of endogenous Lin28 expression when kept in proliferating conditions. We used a Cre-loxP model to knock out Lin28 in Plp1-positive glial cells (Plp1-CreER;Lin28a-fl;tdTm; Figure 1B). We used Plp1-CreER-tdTm reporter mice as controls (Figure 1C). We obtained third-generation neurospheres from early postnatal spiral ganglion tissue of mice overexpressing Lin 28 (Plp1-CreER;rtTA; Lin28a/b-TetO;tdTm, referred to as Lin28-TetO), knockout mice (Plp1-CreER;Lin28a-fl;tdTm, referred to as Lin28−/−), and controls (Plp1-CreER;tdTm) after treatment with tamoxifen or a combination of tamoxifen and doxycycline (Figure 1D). Immunohistochemical analysis demonstrated that tdTm-positive Plp1-positive glial cells coexpressed progenitor marker Sox2 (control, Figure 1E,E′). When we included EdU in the cultures, triple-positive glial cells, coexpressing tdTm, Sox2, and EdU in Plp1 positive cells, were found in the Lin28-TetO and control but not the Lin28−/− neurospheres (Figure 1E, E′) after 6 or 24 hours of EdU treatment (Figure 1F). No significant difference was found in the number of triple-positive proliferating progenitors between control (0.8%) and Lin28 overexpressing (0.9%) spheres after 6 hours of EdU treatment (Figure 1F). In contrast, at 24 hours of EdU exposure, we found a significant difference between control (4.8%) and Lin28 overexpressing (10.5%) neurospheres (Figure 1F). These data demonstrate a pro-proliferative role of Lin28 in Plp1-expressing progenitors. Although we initially tested both the Lin28a and Lin28b Cre-recombinase-dependent, doxycycline-inducible Lin28 transgenic mouse models, we did not see any significant difference between them. We therefore continued with the Lin28a model and refer to it as the Lin28 model throughout the paper.
Lin28 regulates its downstream target, Hmga2, by inhibiting Let-7 microRNA that otherwise inhibits Hmga2 expression59 (Figure 1G,H, J). Let-7 microRNA levels were significantly increased after Lin28 deletion, and decreased after Lin28 overexpression (Figure 1G,H). Lin28 increased the expression of Sox2 and Hmga2 (Figure 1H-J), whereas Lin28 knockout reduced the level of the two transcription factors (Figure 1H-J). Since Lin28 upregulated Hmga2 in the neurospheres, we further investigated its role. Hmga2 or Sox2 downregulation in proliferating neurospheres with siRNA significantly inhibited new sphere formation after the second passage compared to control siRNA, as demonstrated by reduced size of spheres. From passage 1 (p1) to passage 2 (p2), the number of spheres with diameter larger than 50 μm had roughly doubled in spheres treated with control siRNA (p1: 50.34 ± 1.20 to p2: 97.34 ± 1.85; mean ± SEM). The number of large spheres had significantly decreased in spheres treated with Hmga2 siRNA (p1: 49.67 ± 0.88 to p2: 63.67 ± 1.86; mean ± SEM, P = .0012, n = 3 per passage) or Sox2 siRNA (p1: 50.67 ± 0.88 to p2: 66.334 ± 2.19; mean ± SEM, P = .0004, n = 3 per passage) after passage compared to control. To further assess proliferation, EdU was added for 24 hours in third-generation (after 2 passages) proliferating spheres. Immunohistochemistry for EdU revealed a significantly reduced incorporation of EdU in spiral ganglion neurospheres treated with Hmga2 siRNA (15.37 ± 3.77; mean% ± SEM, P = .033, n = 5) or Sox2 siRNA (19.77 ± 4.30; mean% ± SEM, P = .041, n = 4) compared to control siRNA (31.92 ± 5.71; mean% ± SEM), and differentiating neurospheres had a roughly threefold reduced percentage of neurons per DAPI positive cells after Hmga2 siRNA treatment (1.27 ± 0.34; mean% ± SEM) compared to control siRNA (4.52 ± 1.29; mean% ± SEM, P = .027, n = 9). These data show that the Sox2-Lin28/Let-7-Hmga2 axis in spiral ganglion progenitors maintains an undifferentiated state and that high levels of Lin28 and Sox2 favor proliferation of these cells.
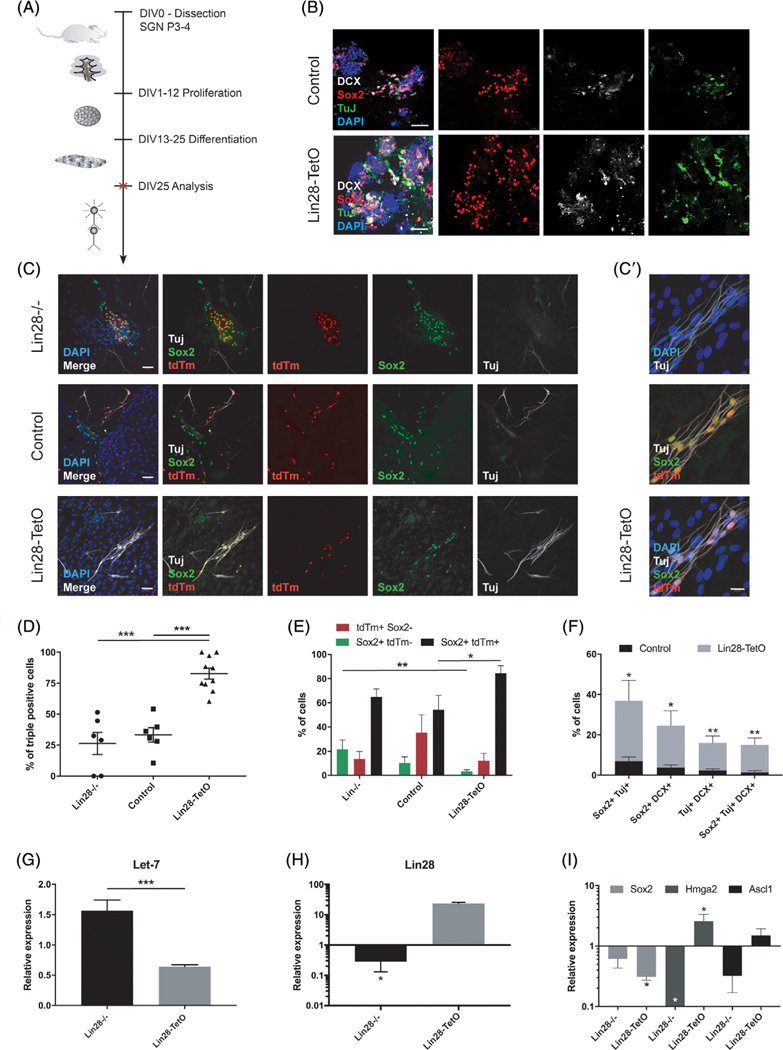
3.2 |. Effect of Lin28 on neural differentiation of Plp1-expressing glial cells
Our data on proliferating neurospheres indicated that Lin28 and Sox2 maintained and expanded Plp1-positive spiral ganglion glial cells. To assess whether Lin28 could influence cell fate specification as well as progenitor maintenance, as previously shown for Sox2 in the cochlea, retina, and CNS,60–64 we used third-generation neurospheres from the same mouse models as described above under differentiation-promoting conditions (Figure 2A). After 12 days of differentiation in vitro, immunohistochemical analysis revealed that in the absence of manipulation of Lin28 expression, differentiating Plp1-positive glial cells continued to coexpress Sox2 (control, Figure 2B,C), and exhibited spontaneous differentiation into neurons, albeit at a low yield,21 based on coexpression of neuron-specific marker class III ß tubulin (TuJ, Figure 2C) and proneural marker doublecortin (DCX, Figure 2B). Deletion of Lin28 did not significantly change the number of triple positive tdTm-Sox2-TuJ cells (Figure 2C,D), indicating that Lin28 was not required for neural differentiation after initiation of Sox2 expression in Plp1-positive glial cells. Lin28 overexpression however increased the number of Plp1-Sox2 positive progenitors that coexpressed TuJ (Figure 2C,C′,D,E), and number of immature neurons expressing TuJ and DCX (Figure 2B,F). Quantitative RT-PCR at day 12 of differentiation demonstrated a significant reduction in Hmga2 expression after Lin28 deletion, and no significant effects on Sox2 or Ascl1 expression (Figure 2I). Overall lower levels of Lin28, Sox2, Hmga2, and Ascl1 are expected at this time in differentiation, as downregulation of neural stem cell markers and early proneural genes is required for maturation of neurons.65,66 In contrast, upregulation of Lin28 led to an increased number of Plp1-positive, proliferating progenitors (Figure 1A,E,H) that differentiated into new neurons based on expression of doublecortin (Figure 2B,F) and TuJ (Figure 2C,C′,D,F). After 12 days of differentiation, Sox2 was markedly downregulated (Figure 2I), whereas Hmga2 and Ascl1 remained slightly increased (Figure 2I), which may indicate maturation of Plp1-positive progenitors into neurons. Similar to proliferating Plp1 glial cells, expression of Let-7 in differentiating progenitors was reduced after forced Lin28 upregulation, and increased after Lin28 deletion (Figure 2G), suggesting that the Sox2-Lin28/Let-7-Hmga2 axis continued to be active in differentiating progenitors. Quantification revealed that Lin28 overexpression increased the neuronal differentiation of Plp1-positive glial cells, whereas loss of Lin28 did not decrease the number of neurons (Figure 2D-F).
FIGURE 2.
Differentiation of Plp1-expressing glial cells after knockout or overexpression of Lin28 in vitro. A, Timeline for in vitro neurosphere assay from early postnatal spiral ganglion cells. Pups were dissected at P3-P4 and kept under proliferating conditions for 12 days, undergoing two passages; DIV, days in vitro. Spheres were treated after the second passage and analyzed by immunohistochemistry (red cross). B, Immunohistochemical analysis of third-generation spiral ganglion neurospheres after 12 days of differentiation. Differentiating neurospheres of Lin28-TetO and control mice were stained for DCX (white), Tuj (green), and Sox2 (red). Nuclei were labeled with DAPI (blue). Scale bar = 50 μm. C, Immunohistochemical analysis of third-generation spiral ganglion neurospheres after 12 days of differentiation. Differentiating neurospheres of Lin28−/− (first row), control (second row), and Lin28-TetO mice (third row) were stained for TuJ (white) and Sox2 (green), and colocalized with tdTm (red). Nuclei were labeled with DAPI (blue). Scale bar = 50 μm. C′, High-magnification of Lin28-TetO, Plp1-positive glial cells (tdTm) coexpressing Sox2 (green) and TuJ (white). Nuclei were labeled with DAPI (blue). Scale bar = 20 μm. D, Quantification of tdTm, Sox2, and TuJ positive (+) cells in differentiating neurospheres of Lin28−/−, control, and Lin28-TetO mice as percentage of the total number of tdTm+ Plp1 progenitors after 12 days of differentiation (***P < .001, mean ± SEM). E, Quantification of tdTm, Sox2 double positive, tdTm single positive, and Sox2 single positive cells in spheres on day 1 of differentiation (**P < .01, *P < .05, mean ± SEM, n = 3). F, Number of cells coexpressing Sox2, TuJ, and/or DCX per total cell number in differentiating neurospheres from control or Lin28-TetO mice at 12 days of differentiation (**P < .01, *P < .05, mean ± SEM, n = 3). G, Quantitative expression of Let-7 in differentiating spiral ganglion neurospheres after Lin28 deletion (Lin28−/−) or Lin28 overexpression (Lin28-TetO) (***P < .001, mean ± SEM, n = 3). H-I, Quantitative RT-PCR of Lin28, Sox2, Hmga2, and Ascl1 in differentiating Lin28−/− and Lin28-TetO neurospheres compared to control (*P < .05, mean ± SEM, n = 3, control set as 1)
3.3 |. Endogenous expression of Lin28 and Hmga2
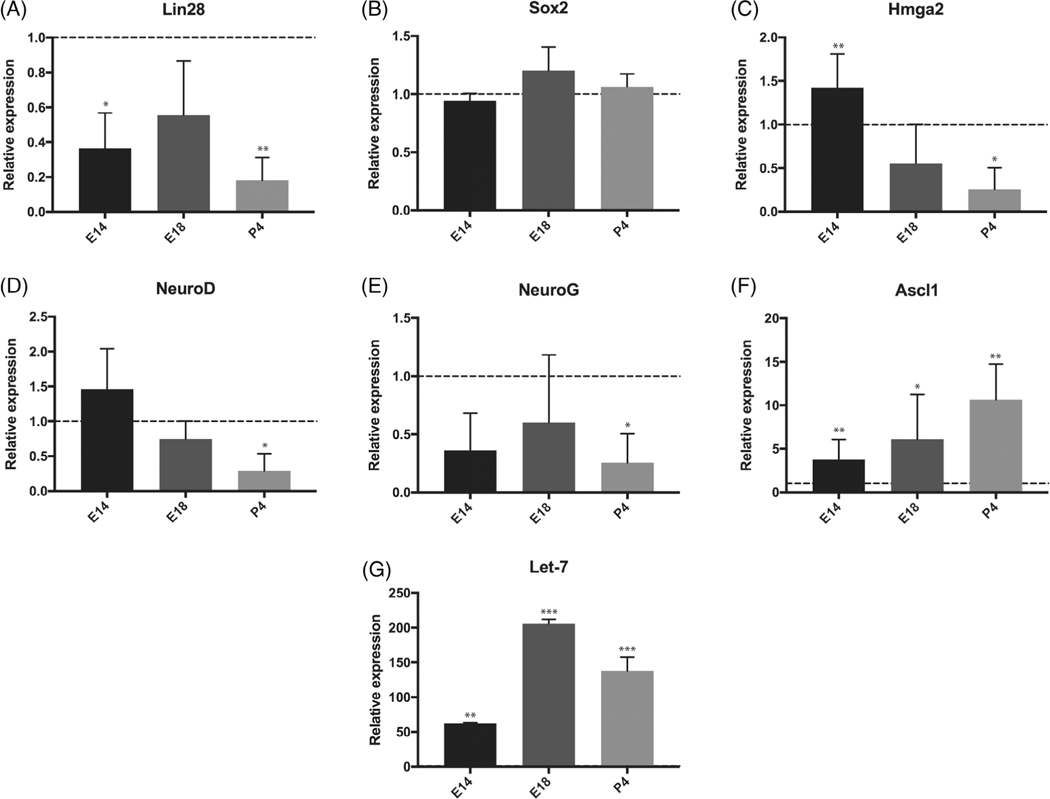
To better understand the balance of Lin28, Let-7, and proneural gene expression during cochlear maturation, we analyzed expression at E9 (otocyst stage), E14 (developing spiral ganglion), E18 (postmitotic stage of spiral ganglion), and P4 (early mature spiral ganglion).
Expression of Lin28 and Hmga2 in the developing cochlea decreased sharply from embryonic day 9 to postnatal day 4 (Figure 3A,C). Proneural transcription factors Neurog1 and Neurod1, which play major roles in the acquisition of neural cell fate in the ear,50,67 showed a similar pattern (Figure 3D,E). The more mature neural precursor marker Ascl1 increased from E9 to P4 (Figure 3F). Expression of Sox2 remained relatively stable across embryonic development in the spiral ganglion, consistent with continued expression in postnatal glia (Figure 3B).64,68,69 In contrast to Lin28, Let-7 expression increased and remained high until postnatal day 4 (Figure 3G), indicative of a Lin28-Let-7 interaction.
FIGURE 3.
Expression of Lin28, Let-7, and neural stem cell markers in the developing ear. A-F, RT-PCR in wild-type embryos from embryonic day E9 to P4. Expression level at E9 is set as 1 and serves as a control. Experimental values at E14, E18, and P4 are compared with E9 for each gene. Neural stem cell genes Lin28 and Hmga2 were downregulated between E9 and P4. The expression of early neural progenitor genes, Neurog1 (NeuroG) and Neurod1 (NeuroD), was decreased during maturation of SGNs, while expression of more mature neural precursor marker, Ascl1, was increased between E9 and P4. Sox2 expression was unchanged (*P < .05; **P < .01, mean ± SEM, n = 3). G, Let-7 expression was increased from E9 to P4 (**P < .01; ***P < .001, mean ± SEM, n = 3)
3.4 |. Effect of Lin28 on the adult cochlea
Given that Lin28 expression is lost in the adult inner ear, we studied effects of transient, forced overexpression of Lin28 in a murine model of auditory nerve damage in vivo. Ouabain, a sodium-potassium-ATPase inhibitor, selectively destroys type I SGNs to induce auditory neuropathy, without direct damage to glia or the sensory epithelium.54 Ouabain treatment of adult ears induced increased numbers of Plp1-tdTm positive glial cells (Figure 4A,B), consistent with prior findings of glial cell proliferation,31 and many of the Plp1-tdTm positive cells coexpressed progenitor marker Sox2. Upregulation of Lin28 on day 7 after ouabain treatment in Plp1-TetO mice led to an increase in expression of Sox2, as well as early proneural markers Ascl1, Neurog1, and Neurod1 (Figure 4A,C-E). These genes were significantly more upregulated in damaged ears with Lin28 upregulation, compared to Lin28 upregulation alone (Figure 4E).
FIGURE 4.
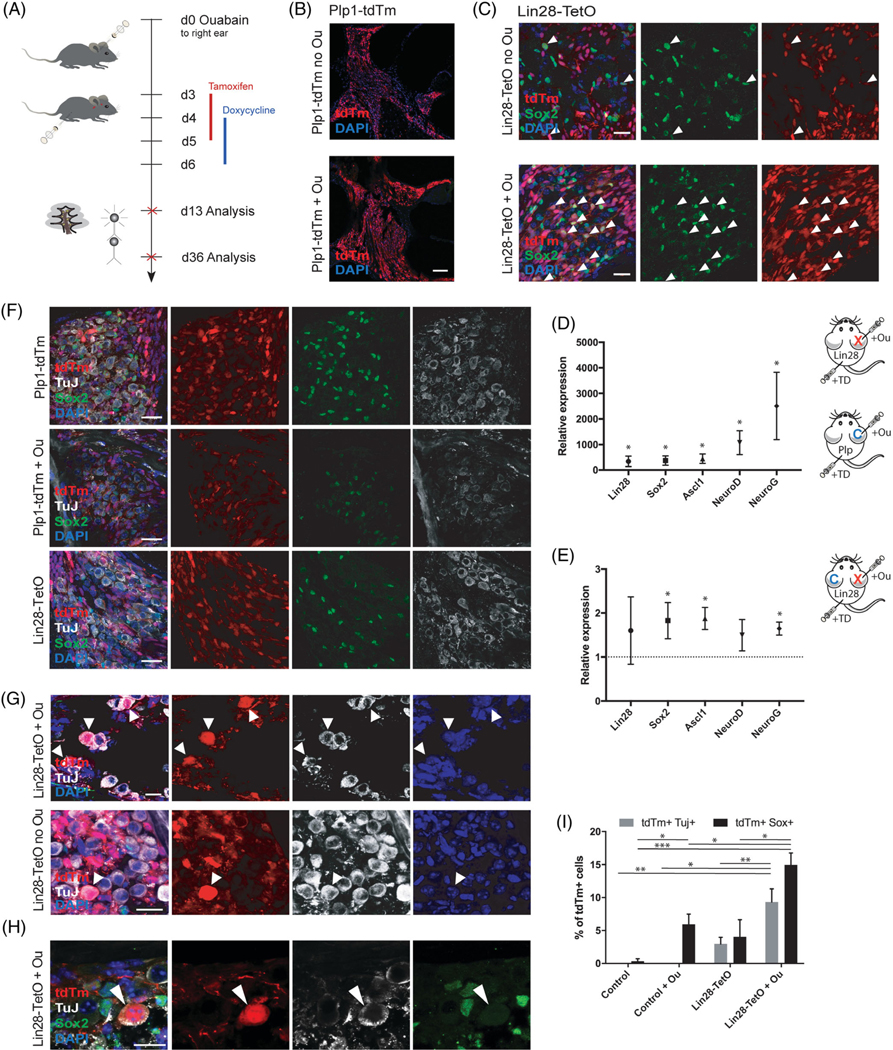
Neural conversion of adult spiral ganglion glia after damage and transient Lin28 overexpression. A, Timeline for in vivo experiment. Ouabain surgery was performed on 6-week-old Lin28-TetO mice or Plp1-tdTm mice at experimental time point day 0 (d0), followed by tamoxifen injections from day 3 to day 5 and doxycycline injections from day 4 to day 6 after surgery. Animals were analyzed 7 days after injection at experimental day 13 or 30 days after injections at experimental day 36. B, Low-magnification view of proliferating Plp1-positive glia in the modiolus of the cochlea without (no Ou) or with (+Ou) ouabain treatment. Plp1-positive glia are shown in red (tdTm reporter). Nuclei were labeled with DAPI (blue). Scale bar = 100 μm. C Coexpression (arrowheads) of tdTm reporter (red) and Sox2 (green) was increased after ouabain treatment and Lin28 overexpression (Lin28-TetO + Ou) compared to Lin28 overexpression alone (Lin28-TetO no Ou). Scale bar = 20 μm. D, Seven days post Lin28 upregulation after tamoxifen/doxycycline injection (+TD) and ouabain treatment (+Ou). RT-PCR showed increased expression of Lin28, Sox2, Ascl1, Neurod1 (NeuroD), and Neurog1 (NeuroG) in Lin28-TetO (Lin28, X) mice compared to tamoxifen/doxycycline injected Plp1-tdTm control (Plp, C) mice after ouabain treatment (*P < .05, mean ± SEM, n = 5). E, Seven days post Lin28 upregulation after tamoxifen/doxycycline injection (+TD) and ouabain treatment (+Ou), RT-PCR showed higher expression of Sox2, Ascl1, Neurod1 (NeuroD), and Neurog1 (NeuroG) in ouabain damaged right ears (Lin28, X) of Lin28-TetO mice compared to undamaged left ears (Lin28, C, set at 1) (*P < .05, mean ± SEM, n = 4). F, Low-magnification of Plp1-tdTm mice after tamoxifen injections with or without ouabain, showing no coexpression of Plp1-expressing glial cells (tdTm in red) and Tuj (white). Ouabain-treated and tamoxifen injected Plp1-tdTm mice (+ Ou) showed diminished Tuj expression. Plp1 glia were found with Sox2 coexpression (green), but not with Tuj (white). Uninjected Lin28-TetO mice without ouabain never demonstrated Tuj-expressing Plp1-glia. Nuclei were labeled with DAPI (blue). Scale bar = 25 μm. G, Multiple lineage-traced (white arrowheads, upper row) Plp1-positive glial cells (tdTm, red) 4 weeks after ouabain-induced neural damage (+Ou) and Lin28 upregulation via tamoxifen/doxycycline injections expressed TuJ (white). Lin28 upregulation without previous ouabain treatment (no Ou) resulted in less Plp1-positive cells that expressed TuJ (single white arrowhead, bottom row). Scale bar = 10 μm. H, Plp1-positive glial cell (tdTm, in red) 4 weeks after ouabain-induced neural damage and Lin28 upregulation via tamoxifen/doxycycline injections (white arrowhead) co-expressed TuJ (white) but had lost Sox2 (green) expression. Nuclei were labeled with DAPI (blue). Scale bar = 10 μm. I, Quantification of surviving lineage-traced Plp1-positive glial cells (tdTm) co-expressing TuJ after ouabain and Lin28 upregulation via tamoxifen/doxycycline injections, compared to Lin28 upregulation only without ouabain. Lin28 overexpression after ouabain led to higher numbers of converted glial cells than Lin28 upregulation without previous ouabain treatment (**P < .01, mean ± SEM, n = 7)
Immunohistochemical analysis of the cochlea 4 weeks after Lin28 upregulation indicated neuronal differentiation of Plp1-positive glial cells based on co-labeling of tdTm and TuJ and loss of Sox2 expression (Figure 4G-I). This was never seen in Plp1-positive control mice or Plp1-TetO mice without activation of Lin28 (Figure 4F). A 3× increase in reprogramming was apparent in ears where Lin28 was upregulated after damage, compared to Lin28 upregulation in undamaged ears (Figure 4G,I). Neural stem cell marker Sox2 was not observed in reprogrammed cells, consistent with the observation of a negative feedback loop among more mature proneural Sox2 downstream targets such as Neurog1, Neurod1, and Ascl1 (Figure 4H).
4 |. DISCUSSION
We have shown here that transient overexpression of Lin28 in Plp1-positive inner ear glia leads to increased proliferation and conversion of these cells to neurons. Loss of Lin28 reduces their proliferation and differentiation. The ability of Lin28 to induce neural conversion in inner ear glia in vitro led us to investigate its effect after neural damage in the inner ear in vivo. We demonstrated that upregulation of Lin28 after auditory nerve damage induced reprogramming of Plp1-positive glial cells into TuJ-positive cells.
Lin28 is well established as a gene that maintains pluripotency in ES cells, and several studies have suggested a role in reprogramming. Loss of Lin28 is associated with reduced proliferation of CNS progenitors and overall reduction in brain size, whereas overexpression of Lin28 increases brain volume and progenitor proliferation.70 Our in vitro data suggest similar effects on progenitors of the inner ear: knockout of Lin28 reduced proliferation of Plp1-positive glial cells and their differentiation to neurons, while transient Lin28 overexpression augmented the number of Plp1-positive cells and the Plp1-derived neurons. We found that Sox2 was upregulated in Plp1-positive cells after Lin28 overexpression. Sox2 is known to control both glial and neural fate in the CNS and PNS in a dose- and time-dependent manner.60,61,63 In the inner ear, Sox2-positive supporting cells act as progenitors for sensory hair cells.61,71–73 In the retina, Lin28 interacts with Sox2 to promote glial proliferation, subsequent commitment to neural cell fate, and conversion of Müller glia into sensory photoreceptors after upregulation of Ascl1.74 In the brain, while Ascl1 alone was not sufficient to drive neural reprogramming of astrocytes, Sox2 was sufficient to convert astrocytes into Ascl1-positive neural progenitors and adult neuroblasts.75 In addition, Sox2 and Ascl1 reprogram human somatic stem cells into functional neurons.76 In our study, Lin28 may exert these effects through interaction with Sox2 to induce and maintain proneural gene expression and induce neural cell fate in inner ear glia. Alternatively, Lin28 may directly control expression of Ascl1 in parallel to Sox2. Plp1-positive cells maintained Sox2 expression in the absence of Lin28, raising the possibility that Sox2 is not a direct downstream target of Lin28; it may be controlled by additional upstream regulators that stimulate Sox2-dependent pathways to promote differentiation. However, the lack of Lin28 did not promote neural differentiation of Plp1/Sox2 positive cells; rather, most Lin28-negative cells retained a glial phenotype, indicating that glial cell fate is favored in the absence of Lin28.77
Hmga2, a direct downstream target of Lin28, interacts with Sox2 and promotes conversion into neural progenitor cells.40,41 We found sharp upregulation of Hmga2 after transient Lin28 activation in proliferating glial cells in vitro, and loss of Hmga2 had a similar effect as loss of Sox2 in inner ear neurospheres, confirming that Hmga2 is a direct downstream target of Lin28. Our data suggest that Lin28 and Sox2 reprogram inner ear glial cells into neural progenitors via the Lin28/Sox2/Hmga2 axis, which promotes a neural stem cell fate and ensures dedifferentiation and proliferation of glial cells. Thus, the temporal interplay of several downstream targets of Lin28 and Sox2 may initiate proliferation of Plp1-positive glial cells as well as oscillatory expression of proneural downstream targets for initiation of neural cell fate and neural differentiation.78
Decreasing expression of Hmga2 is believed to be due to direct influence of Let-7 miRNA regulation.41 Our developmental data confirm downregulation of Lin28 and increased Let-7 early during inner ear development. Let-7 miRNA families are directly involved in competitive inhibition of Lin28 and Hmga2 to create a delicate balance between stem cell maintenance and neural differentiation.39 In retinal progenitors in the PNS, Lin28 expression maintained low Let-7 levels, which upregulated Hmga2 levels and promoted stemness. Higher levels of Let-7 inhibited Hmga2 and promoted neurogliogenesis.41,79–81 Dose-dependent Let-7 inhibition by Lin28 and Ascl1 was required for direct reprogramming of Müller glia into retinal neural progenitors.82,83 We found a similar reciprocal relationship between Lin28 and Let-7 miRNA families in Plp1-positive inner ear glial cells, suggesting an active Lin28-Let-7 pathway in inner ear glial cells.84 High levels of Lin28 in Lin28-overexpressing Plp1 glial cells competitively suppressed Let-7 miRNA levels, while Lin28 knockout increased Let-7 levels. An increase in Let-7 levels is generally believed to facilitate neural differentiation and cell cycle exit.84–86 However, we did not find an increase in neural differentiation after Lin28 knockout with Let-7 upregulation in Plp1-positive glial cells. This could be explained by a cell fate switch in the absence of Lin28, whereby a Let-7 mediated Notch activation in Plp1-positive glial cells favors gliogenesis over neurogenesis and increases glial cell differentiation.87 This would be consistent with findings in the adult inner ear, where Lin28 expression is decreased or absent, but a subset of inner ear glia continues to express Sox2. Sox2 is upregulated sharply in inner ear glial cells after damage.31,53 These cells undergo an activation that includes proliferation and upregulation of proneural genes. However, they fail to undergo reprogramming into neurons, and instead proceed to form a glial scar.53
We describe here for the first time a dormant progenitor cell population with neurogenic potential in the adult inner ear. Plp1-positive cochlear glial cells originate from neural crest stem cells that migrate to the inner ear early in development. Neural crest stem cells display multipotency during embryonic stages and can migrate and adopt new cell fates, including neurons, glial cells, melanocytes, chondrocytes, and myofibroblasts.88–90
Neural crest stem cell and progenitor niches with neurogenic potential have been identified in adult human and mouse skin, heart, carotid body, and cornea.91–93 Cochlear Plp1-positive glial cells may represent such a niche of neural crest progenitors in the inner ear.
More work is needed to clarify the extent of neural differentiation that can be achieved after the upregulation of proneural genes such as Ascl1, NeuroD, and Neurog1 in response to Lin28 and to dissect the interactions of reprogrammed inner ear neurons with their environment. CNS astroglial cells have been shown to convert into neurons after upregulation of proneural genes such as Ascl1, Neurog2, or NeuroD.94–95 It has been suggested that while reprogrammed astroglial cells may predominantly favor a GABAergic or glutamatergic phenotype in vitro, they can, upon transplantation in vivo, adapt various different neuronal fates based on neurotrophic cues from their new environment.94,96 Taken together, our data suggest that neural crest-derived, Plp1-positive glial progenitors harbor intrinsic neurogenic potential, and might adopt peripheral sensory neural fate in response to specific neurotrophic cues in the cochlear environment.
In summary, we have shown that Lin28 induces neural reprogramming of inner ear glial cells in vitro and in vivo. Transient postnatal upregulation of Lin28 after neural damage induced proneural gene expression and reprogramming into TuJ-positive cells lacking Sox2 expression. Prior damage and activation of glia created a more permissive environment for reprogramming and increased the level of reprogramming.31 This effect may be useful in future studies aimed at inducing neuronal regeneration in the ear.
5 |. CONCLUSION
Transient Lin28 overexpression in Plp1 expressing inner ear glial cells induced early expression of neural stem cell markers and subsequent conversion into neurons in vivo. This suggests that inner ear glia could be an endogenous source of reprogrammable cells for regenerative therapies.
Significance statement.
Loss or damage of auditory neurons is associated with sensorineural hearing loss and deafness. To date, no cure is available and amplification, as well as cochlear implants, relies on surviving neurons to convey the auditory signal to the brain. Regeneration strategies focusing on endogenous cell therapy may offer a future treatment option for the replacement of lost neurons to restore the auditory circuit. In a transgenic mouse model, Plp1-positive glial cells of the inner ear have a capacity for regeneration and differentiate into neurons after transient activation of neural stem cell regulator Lin28 in vitro and in vivo. The study presents evidence that Lin28 acts through stem cell regulatory genes, Sox2 and Hmga2, to stimulate proliferation and reprogramming of inner ear glia to neurons, increasing the possibility of a new avenue for regeneration that could replace dying neurons in auditory neuropathy.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grant DC007174. We would like to thank George Q. Daley (Harvard Medical School, Boston) for generously providing us with the Lin28 mouse strains used in this paper.
Funding information
National Institutes of Health, Grant/Award Number: DC007174
Footnotes
CONFLICT OF INTEREST
The authors declared no potential conflicts of interest.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available on request from the corresponding author.
REFERENCES
- 1.Nadol JB Jr, Young YS, Glynn RJ. Survival of spiral ganglion cells in profound sensorineural hearing loss: implications for cochlear implantation. Ann Otol Rhinol Laryngol. 1989;98(6):411–416. [DOI] [PubMed] [Google Scholar]
- 2.Nadol JB Jr. Patterns of neural degeneration in the human cochlea and auditory nerve: implications for cochlear implantation. Otolaryngol Head Neck Surg. 1997;117(3 Pt 1):220–228. [DOI] [PubMed] [Google Scholar]
- 3.Seyyedi M, Viana LM, Nadol JB Jr. Within-subject comparison of word recognition and spiral ganglion cell count in bilateral cochlear implant recipients. Otol Neurotol. 2014;35(8):1446–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moller AR. History of cochlear implants and auditory brainstem implants. Adv Otorhinolaryngol. 2006;64:1–10. [DOI] [PubMed] [Google Scholar]
- 5.Lustig LR, Yeagle J, Driscoll CL, Blevins N, Francis H, Niparko JK. Cochlear implantation in patients with neurofibromatosis type 2 and bilateral vestibular schwannoma. Otol Neurotol. 2006;27(4):512–518. [DOI] [PubMed] [Google Scholar]
- 6.Yamanaka S, Blau HM. Nuclear reprogramming to a pluripotent state by three approaches. Nature. 2010;465(7299):704–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sisakhtnezhad S, Matin MM. Transdifferentiation: a cell and molecular reprogramming process. Cell Tissue Res. 2012;348(3):379–396. [DOI] [PubMed] [Google Scholar]
- 8.Ma W, Yan RT, Li X, Wang SZ. Reprogramming retinal pigment epithelium to differentiate toward retinal neurons with Sox2. STEM CELLS. 2009;27(6):1376–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsonis PA, Madhavan M, Tancous EE, del Rio-Tsonis K. A newt’s eye view of lens regeneration. Int J Dev Biol. 2004;48(8–9):975–980. [DOI] [PubMed] [Google Scholar]
- 10.Jessen KR, Mirsky R, Arthur-Farraj P. The role of cell plasticity in tissue repair: adaptive cellular reprogramming. Dev Cell. 2015;34(6): 613–620. [DOI] [PubMed] [Google Scholar]
- 11.Delaunay D, Heydon K, Cumano A, et al. Early neuronal and glial fate restriction of embryonic neural stem cells. J Neurosci. 2008;28(10):2551–2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doetsch F. The glial identity of neural stem cells. Nat Neurosci. 2003; 6(11):1127–1134. [DOI] [PubMed] [Google Scholar]
- 13.Kriegstein A, Alvarez-Buylla A. The glial nature of embryonic and adult neural stem cells. Annu Rev Neurosci. 2009;32:149–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jessen KR, Mirsky R. The repair Schwann cell and its function in regenerating nerves. J Physiol. 2016;594(13):3521–3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davis AA, Temple S. A self-renewing multipotential stem cell in embryonic rat cerebral cortex. Nature. 1994;372(6503):263–266. [DOI] [PubMed] [Google Scholar]
- 16.Wu SX, Goebbels S, Nakamura K, et al. Pyramidal neurons of upper cortical layers generated by NEX-positive progenitor cells in the subventricular zone. Proc Natl Acad Sci USA. 2005;102(47):17172–17177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martinez-Monedero R, Yi E, Oshima K, Glowatzki E, Edge ASB. Differentiation of inner ear stem cells to functional sensory neurons. Dev Neurobiol. 2008;68(5):669–684. [DOI] [PubMed] [Google Scholar]
- 18.Guo F, Ma J, McCauley E, Bannerman P, Pleasure D. Early postnatal proteolipid promoter-expressing progenitors produce multilineage cells in vivo. J Neurosci. 2009;29(22):7256–7270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Le Bras B, Chatzopoulou E, Heydon K, et al. Oligodendrocyte development in the embryonic brain: the contribution of the plp lineage. Int J Dev Biol. 2005;49(2–3):209–220. [DOI] [PubMed] [Google Scholar]
- 20.Harlow DE, Saul KE, Culp CM, Vesely EM, Macklin WB. Expression of proteolipid protein gene in spinal cord stem cells and early oligodendrocyte progenitor cells is dispensable for normal cell migration and myelination. J Neurosci. 2014;34(4):1333–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McLean WJ, McLean DT, Eatock RA, et al. Distinct capacity for differentiation to inner ear cell types by progenitor cells of the cochlea and vestibular organs. Development. 2016;143(23):4381–4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McLean WJ, Yin X, Lu L, et al. Clonal expansion of Lgr5-positive cells from mammalian cochlea and high-purity generation of sensory hair cells. Cell Rep. 2017;18(8):1917–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Diensthuber M, Zecha V, Wagenblast J, Arnhold S, Edge ASB, Stöver T. Spiral ganglion stem cells can be propagated and differentiated into neurons and glia. Biores Open Access. 2014;3(3):88–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shi F, Edge AS. Prospects for replacement of auditory neurons by stem cells. Hear Res. 2013;297:106–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li X, Aleardi A, Wang J, Zhou Y, Andrade R, Hu Z. Differentiation of spiral ganglion-derived neural stem cells into functional synaptogenetic neurons. Stem Cells Dev. 2016;25(10):803–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Breuskin I, Bodson M, Thelen N, et al. Glial but not neuronal development in the cochleo-vestibular ganglion requires Sox10. J Neurochem. 2010;114(6):1827–1839. [DOI] [PubMed] [Google Scholar]
- 27.Wan G, Corfas G. Transient auditory nerve demyelination as a new mechanism for hidden hearing loss. Nat Commun. 2017;8:14487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robel S, Berninger B, Gotz M. The stem cell potential of glia: lessons from reactive gliosis. Nat Rev Neurosci. 2011;12(2):88–104. [DOI] [PubMed] [Google Scholar]
- 29.Meas SJ, Zhang CL, Dabdoub A. Reprogramming glia into neurons in the peripheral auditory system as a solution for sensorineural hearing loss: lessons from the central nervous system. Front Mol Neurosci. 2018;11:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gomez-Casati ME, Murtie J, Taylor B, et al. Cell-specific inducible gene recombination in postnatal inner ear supporting cells and glia. J Assoc Res Otolaryngol. 2010;11(1):19–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lang H, Xing Y, Brown LN, et al. Neural stem/progenitor cell properties of glial cells in the adult mouse auditory nerve. Sci Rep. 2015;5: 13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang J, Ratanasirintrawoot S, Chandrasekaran S, et al. LIN28 regulates stem cell metabolism and conversion to primed pluripotency. Cell Stem Cell. 2016;19(1):66–80. [DOI] [PubMed] [Google Scholar]
- 33.Chien CS, Wang ML, Chu PY, et al. Lin28B/Let-7 regulates expression of Oct4 and Sox2 and reprograms oral squamous cell carcinoma cells to a stem-like state. Cancer Res. 2015;75(12):2553–2565. [DOI] [PubMed] [Google Scholar]
- 34.Cimadamore F, Amador-Arjona A, Chen C, Huang CT, Terskikh AV. SOX2-LIN28/let-7 pathway regulates proliferation and neurogenesis in neural precursors. Proc Natl Acad Sci USA. 2013;110(32):E3017E3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsialikas J, Romer-Seibert J. LIN28: roles and regulation in development and beyond. Development. 2015;142(14):2397–2404. [DOI] [PubMed] [Google Scholar]
- 36.Faas L, Warrander FC, Maguire R, et al. Lin28 proteins are required for germ layer specification in Xenopus. Development. 2013;140(5): 976–986. [DOI] [PubMed] [Google Scholar]
- 37.Polesskaya A, Cuvellier S, Naguibneva I, Duquet A, Moss EG, Harel-Bellan A. Lin-28 binds IGF-2 mRNA and participates in skeletal myogenesis by increasing translation efficiency. Genes Dev. 2007;21 (9):1125–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nishino J, Kim S, Zhu Y, Zhu H, Morrison SJ. A network of heterochronic genes including Imp1 regulates temporal changes in stem cell properties. Elife. 2013;2:e00924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu KR, Shin JH, Kim JJ, et al. Rapid and efficient direct conversion of human adult somatic cells into neural stem cells by HMGA2/let-7b.Cell Rep. 2015;10:441–452. [DOI] [PubMed] [Google Scholar]
- 40.Kim JJ, Shin JH, Yu KR, et al. Direct conversion of human umbilical cord blood into induced neural stem cells with SOX2 and HMGA2. Int J Stem Cells. 2017;10(2):227–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nishino J, Kim I, Chada K, Morrison SJ. Hmga2 promotes neural stem cell self-renewal in young but not old mice by reducing p16Ink4a and p19Arf expression. Cell. 2008;135(2):227–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Newman MA, Thomson JM, Hammond SM. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing.RNA. 2008;14(8):1539–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Viswanathan SR, Daley GQ. Lin28: a microRNA regulator with a macro role. Cell. 2010;140(4):445–449. [DOI] [PubMed] [Google Scholar]
- 44.Heo I, Joo C, Cho J, Ha M, Han J, Kim VN. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol Cell. 2008;32(2): 276–284. [DOI] [PubMed] [Google Scholar]
- 45.Kumar RM, Cahan P, Shalek AK, et al. Deconstructing transcriptional heterogeneity in pluripotent stem cells. Nature. 2014;516(7529): 56–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sajan SA, Warchol ME, Lovett M. Toward a systems biology of mouse inner ear organogenesis: gene expression pathways, patterns and network analysis. Genetics. 2007;177(1):631–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Doetzlhofer A, Avraham KB. Insights into inner ear-specific gene regulation: epigenetics and non-coding RNAs in inner ear development and regeneration. Semin Cell Dev Biol. 2017;65:69–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smeti I, Watabe I, Savary E, Fontbonne A, Zine A. HMGA2, the architectural transcription factor high mobility group, is expressed in the developing and mature mouse cochlea. PLoS One. 2014;9(2):e88757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Golden EJ, Benito-Gonzalez A, Doetzlhofer A. The RNA-binding protein LIN28B regulates developmental timing in the mammalian cochlea. Proc Natl Acad Sci USA. 2015;112(29):E3864–E3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Noda T, Meas SJ, Nogami J, et al. Direct reprogramming of spiral ganglion non-neuronal cells into neurons: toward ameliorating sensorineural hearing loss by gene therapy. Front Cell Dev Biol. 2018;6:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meas SJ, Nishimura K, Scheibinger M, Dabdoub A. In vitro methods to cultivate spiral ganglion cells, and purification of cellular subtypes for induced neuronal reprogramming. Front Neurosci. 2018;12:822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schwarzenbach H, da Silva AM, Calin G, Pantel K. Data normalization strategies for microRNA quantification. Clin Chem. 2015;61(11):1333–1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lang H, Li M, Kilpatrick LA, et al. Sox2 up-regulation and glial cell proliferation following degeneration of spiral ganglion neurons in the adult mouse inner ear. J Assoc Res Otolaryngol. 2011;12(2):151–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yuan Y, Shi F, Yin Y, et al. Ouabain-induced cochlear nerve degeneration: synaptic loss and plasticity in a mouse model of auditory neuropathy. J Assoc Res Otolaryngol. 2014;15(1):31–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vadla B, Kemper K, Alaimo J, et al. Lin-28 controls the succession of cell fate choices via two distinct activities. PLoS Genet. 2012;8(3): e1002588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ambros V. A hierarchy of regulatory genes controls a larva-to-adult developmental switch in C. elegans. Cell. 1989;57(1):49–57. [DOI] [PubMed] [Google Scholar]
- 57.Ambros V, Horvitz HR. Heterochronic mutants of the nematode Caenorhabditis elegans. Science. 1984;226(4673):409–416. [DOI] [PubMed] [Google Scholar]
- 58.Moss EG, Lee RC, Ambros V. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell. 1997;88(5):637–646. [DOI] [PubMed] [Google Scholar]
- 59.Copley MR, Babovic S, Benz C, et al. The Lin28b-let-7-Hmga2 axis determines the higher self-renewal potential of fetal haematopoietic stem cells. Nat Cell Biol. 2013;15(8):916–925. [DOI] [PubMed] [Google Scholar]
- 60.Taranova OV, Magness ST, Fagan BM, et al. SOX2 is a dose-dependent regulator of retinal neural progenitor competence. Genes Dev. 2006;20(9):1187–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kempfle JS, Turban JL, Edge AS. Sox2 in the differentiation of cochlear progenitor cells. Sci Rep. 2016;6:23293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cavallaro M, Mariani J, Lancini C, et al. Impaired generation of mature neurons by neural stem cells from hypomorphic Sox2 mutants. Development. 2008;135(3):541–557. [DOI] [PubMed] [Google Scholar]
- 63.Favaro R, Valotta M, Ferri AL, et al. Hippocampal development and neural stem cell maintenance require Sox2-dependent regulation of Shh. Nat Neurosci. 2009;12(10):1248–1256. [DOI] [PubMed] [Google Scholar]
- 64.Puligilla C, Dabdoub A, Brenowitz SD, Kelley MW. Sox2 induces neuronal formation in the developing mammalian cochlea. J Neurosci. 2010;30(2):714–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hagey DW, Muhr J. Sox2 acts in a dose-dependent fashion to regulate proliferation of cortical progenitors. Cell Rep. 2014;9(5):1908–1920. [DOI] [PubMed] [Google Scholar]
- 66.Sanchez-Calderon H, Rodriguez-de la Rosa L, Milo M, et al. RNA microarray analysis in prenatal mouse cochlea reveals novel IGF-I target genes: implication of MEF2 and FOXM1 transcription factors. PLoS One. 2010;5(1):e8699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Evsen L, Sugahara S, Uchikawa M, Kondoh H, Wu DK. Progression of neurogenesis in the inner ear requires inhibition of Sox2 transcription by neurogenin1 and neurod1. J Neurosci. 2013;33(9):3879–3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Steevens AR, Sookiasian DL, Glatzer JC, Kiernan AE. SOX2 is required for inner ear neurogenesis. Sci Rep. 2017;7(1):4086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mak AC, Szeto IY, Fritzsch B, et al. Differential and overlapping expression pattern of SOX2 and SOX9 in inner ear development. Gene Expr Patterns. 2009;9(6):444–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang M, Yang SL, Herrlinger S, et al. Lin28 promotes the proliferative capacity of neural progenitor cells in brain development. Development. 2015;142(9):1616–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bramhall NF, Shi F, Arnold K, Hochedlinger K, Edge ASB. Lgr5-positive supporting cells generate new hair cells in the postnatal cochlea. Stem Cell Rep. 2014;2(3):311–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shi F, Hu L, Edge AS. Generation of hair cells in neonatal mice by beta-catenin overexpression in Lgr5-positive cochlear progenitors. Proc Natl Acad Sci USA. 2013;110(34):13851–13856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shi F, Kempfle JS, Edge AS. Wnt-responsive Lgr5-expressing stem cells are hair cell progenitors in the cochlea. J Neurosci. 2012;32(28): 9639–9648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gorsuch RA, Lahne M, Yarka CE, Petravick ME, Li J, Hyde DR. Sox2 regulates Muller glia reprogramming and proliferation in the regenerating zebrafish retina via Lin28 and Ascl1a. Exp Eye Res. 2017; 161:174–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Niu W, Zang T, Smith DK, et al. SOX2 reprograms resident astrocytes into neural progenitors in the adult brain. Stem Cell Rep. 2015;4(5): 780–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Araujo JAM, Hilscher MM, Marques-Coelho D, et al. Direct reprogramming of adult human somatic stem cells into functional neurons using Sox2, Ascl1, and Neurog2. Front Cell Neurosci. 2018;12:155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Johnston AP, Naska S, Jones K, et al. Sox2-mediated regulation of adult neural crest precursors and skin repair. Stem Cell Rep. 2013;1(1):38–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kageyama R, Shimojo H, Ohtsuka T. Dynamic control of neural stem cells by bHLH factors. Neurosci Res. 2018;138:12–18. [DOI] [PubMed] [Google Scholar]
- 79.Lee YS, Dutta A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 2007;21(9):1025–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xia X, Teotia P, Ahmad I. Lin28a regulates neurogliogenesis in mammalian retina through the Igf signaling. Dev Biol. 2018;440(2):113–128. [DOI] [PubMed] [Google Scholar]
- 81.Parameswaran S, Xia X, Hegde G, Ahmad I. Hmga2 regulates self-renewal of retinal progenitors. Development. 2014;141(21):4087–4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ramachandran R, Fausett BV, Goldman D. Ascl1a regulates Muller glia dedifferentiation and retinal regeneration through a Lin-28-dependent, let-7 microRNA signalling pathway. Nat Cell Biol. 2010;12(11):1101–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nelson CM, Gorsuch RA, Bailey TJ, Ackerman KM, Kassen SC, Hyde DR. Stat3 defines three populations of Muller glia and is required for initiating maximal muller glia proliferation in the regenerating zebrafish retina. J Comp Neurol. 2012;520(18):4294–4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Morgado AL, Rodrigues CM, Sola S. MicroRNA-145 regulates neural stem cell differentiation through the Sox2-Lin28/let-7 signaling pathway. STEM CELLS. 2016;34(5):1386–1395. [DOI] [PubMed] [Google Scholar]
- 85.Xia X, Ahmad I. Let-7 microRNA regulates neurogliogenesis in the mammalian retina through Hmga2. Dev Biol. 2016;410(1):70–85. [DOI] [PubMed] [Google Scholar]
- 86.Rehfeld F, Rohde AM, Nguyen DT, et al. Lin28 and let-7: ancient milestones on the road from pluripotency to neurogenesis. Cell Tissue Res. 2015;359(1):145–160. [DOI] [PubMed] [Google Scholar]
- 87.Patterson M, Gaeta X, Loo K, et al. Let-7 miRNAs can act through notch to regulate human gliogenesis. Stem Cell Rep. 2014;3(5): 758–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Achilleos A, Trainor PA. Neural crest stem cells: discovery, properties and potential for therapy. Cell Res. 2012;22(2):288–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Motohashi T, Yamanaka K, Chiba K, et al. Neural crest cells retain their capability for multipotential differentiation even after lineagerestricted stages. Dev Dyn. 2011;240(7):1681–1693. [DOI] [PubMed] [Google Scholar]
- 90.Motohashi T, Yamanaka K, Chiba K, Aoki H, Kunisada T. Unexpected multipotency of melanoblasts isolated from murine skin. STEM CELLS. 2009;27(4):888–897. [DOI] [PubMed] [Google Scholar]
- 91.Pardal R, Ortega-Saenz P, Duran R, et al. Glia-like stem cells sustain physiologic neurogenesis in the adult mammalian carotid body. Cell. 2007;131(2):364–377. [DOI] [PubMed] [Google Scholar]
- 92.Sieber-Blum M, Grim M, Hu YF, Szeder V. Pluripotent neural crest stem cells in the adult hair follicle. Dev Dyn. 2004;231(2):258–269. [DOI] [PubMed] [Google Scholar]
- 93.Fernandes KJ, McKenzie IA, Mill P, et al. A dermal niche for multipotent adult skin-derived precursor cells. Nat Cell Biol. 2004;6 (11):1082–1093. [DOI] [PubMed] [Google Scholar]
- 94.Chouchane M, Melo de Farias AR, DMS M, et al. Lineage reprogramming of Astroglial cells from different origins into distinct neuronal subtypes. Stem Cell Rep. 2017;9(1):162–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Guo Z, Zhang L, Wu Z, Chen Y, Wang F, Chen G. In vivo direct reprogramming of reactive glial cells into functional neurons after brain injury and in an Alzheimer’s disease model. Cell Stem Cell. 2014;14(2): 188–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Adameyko I, Lallemend F, Aquino JB, et al. Schwann cell precursors from nerve innervation are a cellular origin of melanocytes in skin. Cell. 2009;139(2):366–379. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available on request from the corresponding author.