Abstract
MyoD is a basic helix-loop-helix transcription factor involved in the activation of genes encoding skeletal muscle-specific proteins. Independent of its ability to transactivate muscle-specific genes, MyoD can also act as a cell cycle inhibitor. MyoD activity is regulated by transcriptional and posttranscriptional mechanisms. While MyoD can be found phosphorylated, the functional significance of this posttranslation modification has not been established. MyoD contains several consensus cyclin-dependent kinase (CDK) phosphorylation sites. In these studies, we examined whether a link could be established between MyoD activity and phosphorylation at putative CDK sites. Site-directed mutagenesis of potential CDK phosphorylation sites in MyoD revealed that S200 is required for MyoD hyperphosphorylation as well as the normally short half-life of the MyoD protein. Additionally, we determined that turnover of the MyoD protein requires the proteasome and Cdc34 ubiquitin-conjugating enzyme activity. Results of these studies demonstrate that hyperphosphorylated MyoD is targeted for rapid degradation by the ubiquitin pathway. The targeted degradation of MyoD following CDK phosphorylation identifies a mechanism through which MyoD activity can be regulated coordinately with the cell cycle machinery (CDK2 and CDK4) and/or coordinately with the cellular transcriptional machinery (CDK7, CDK8, and CDK9).
In animals, skeletal muscle is a critical organ, as it is responsible for carrying out coordinated and directed movements. The developmental events that ultimately give rise to mature skeletal muscle can be divided into two major steps (reviewed in references 27, 47, 48, and 50). The first step is determination of which multipotential mesodermal cells become restricted to the myogenic lineage. The second step is commitment of the myogenically determined cells (myoblasts) to terminal differentiation. Terminal differentiation involves cell cycle arrest, fusion of myoblasts leading to myotube formation, and the production of skeletal muscle-specific structural proteins. Cellular restriction to the myogenic lineage is in large part under the control of determination genes which encode a family of skeletal muscle-specific basic helix-loop-helix transcription factors, including MyoD, Myf-5, myogenin, and MRF4. The first determination gene, MyoD, was found by its ability to initiate the conversion of fibroblasts into myoblasts (6), a property subsequently found to be true for all four myogenic determination genes. While mice containing a homozygous deletion of MyoD develop normally, mice lacking both MyoD and Myf-5 do not form skeletal muscle (36, 37). These results indicate that MyoD and Myf-5 are critical for the determination step in skeletal muscle development. Once MyoD is activated, it continues to be expressed throughout development, presumably through the ability of the MyoD protein to activate MyoD transcription (46).
In addition to activating skeletal muscle-specific genes, MyoD expression can also lead to cell cycle arrest, even in the absence of terminal myogenic differentiation (5, 26, 45, 47). The importance of MyoD as an inhibitor of cell cycle progression can also be seen during wound repair (29). When muscle from mice lacking MyoD is injured, the animals are deficient in the regeneration of muscle tissue. The defect in regeneration appears to be the result of the increased potential of satellite cells for self-renewal rather than fusion and myotube formation. One potential mechanism by which MyoD can arrest cell cycle progression is through the transcriptional activation of the CIP1 gene, which encodes the cyclin-dependent kinase (CDK)-inhibitory protein p21 (14, 33). Consistent with a role for MyoD in negatively regulating cell cycle progression during G1, overexpression of cyclin D1 results in an inhibition of MyoD-dependent transcription and might lead to a decrease in p21 production (35, 42). The cyclin D1-mediated decrease in MyoD transactivation activity correlates with a concomitant increase in the abundance of a phosphorylated form of the MyoD protein, although MyoD has not been shown to be a substrate of cyclin D complexes (35, 42, 46). Alternatively, MyoD has been proposed to inhibit cell cycle progression by binding the Rb protein and interfering with E2F-dependent transcription events, which include the activation of a number of genes required for exit from G1 or DNA replication (3, 13, 44).
These results suggest that a CDK controls MyoD protein activity, possibly through direct phosphorylation of the MyoD protein. The CDKs have been termed proline-directed protein kinases, as their substrates are phosphorylated on serines or threonines that are preceded by a proline (30). MyoD protein has seven putative CDK phosphorylation sites. We demonstrate here that the MyoD protein is likely to be phosphorylated on a serine residue within one of these CDK consensus sites and that the prevention of phosphorylation at this site leads to the stabilization of the MyoD protein. Furthermore, agents that inhibit the ubiquitin-dependent pathway of protein degradation, a pathway intimately linked to the turnover of several key regulators of cell cycle progression, also cause the stabilization of MyoD. Based on these results, we propose that the functional significance of MyoD phosphorylation is to control the level of MyoD protein rather than to alter the transactivating activity of the MyoD protein.
MATERIALS AND METHODS
Cell culture.
Cell culture and transient transfections were performed as described previously (15). Nuclear extracts were prepared as described elsewhere (1). Where indicated, extracts were treated with 1 U of calf intestine alkaline phosphatase (Gibco BRL Co.) for 30 min at 37°C prior to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
Plasmids and DNA manipulations.
Plasmid 4TRTK-CAT (kindly provided by S. Konieczny, Purdue University) contains four copies in tandem of the E-box sequence present in the muscle creatine kinase gene enhancer. The E-box sequences are upstream of the thymidine kinase (TK) gene basal promoter that controls the expression of the bacterial chloramphenicol acetyltransferase (CAT) gene.
All DNA manipulations were performed by using standard techniques (38). The MyoD gene was liberated from plasmid pVZC11b (6) (kindly provided by A. Lassar) by cleavage with EcoRI/HindIII and ligated into the EcoRI/HindIII fragment of pALTER-1 (Promega, Madison, Wis.). Site-directed mutagenesis was performed by using the Altered Sites Mutagenesis System II (Promega), with oligonucleotides described in the legend to Fig. 2, according to the manufacturer’s instructions. The presence of each mutation was verified by DNA sequencing using Sequenase (U.S. Biochemicals, Cleveland, Ohio) according to the manufacturer’s instructions. An EcoRI/HindIII DNA fragment containing MyoD was ligated into the EcoRI/HindIII fragment of pCB6 (28) (kindly provided by F. Rauscher). Cloning into pCB6 allows for expression under the control of the cytomegalovirus immediate-early gene promoter in mammalian cells (15).
FIG. 2.
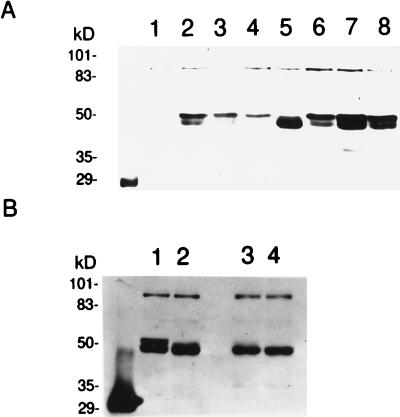
Hyperphosphorylation of MyoD requires S200. Thirty micrograms of pCB6+ vector or pCB6+ containing wild-type or mutated MyoD was transfected into C3H10T1/2 fibroblasts as described in Materials and Methods. At 36 h after transfection, cells were harvested; nuclear extracts were prepared and separated by SDS-PAGE. Separated proteins were subjected to Western analysis using anti-MyoD antibodies. (A) Lanes (oligonucleotide sequences in brackets): 1, vector alone; 2, MyoD (wild-type MyoD); 3, SP1-MyoD (MyoD S5A [ELLSPPLR ELLAPPLR]); 4, SP2-MyoD (MyoD S37A [CFDSPDLR CFDAPDLR]); 5, SP3-MyoD (MyoD S200A [DASSPRSN DASAPRSN]); 6, SP4-MyoD (MyoD S262A [STDSPAAP STDAPAAP]); 7, SP5-MyoD (MyoD S277A [PPESPPGP PPEAPPGP]); 8, SP6-MyoD (MyoD T296A/S298A [GTQTPSPDA GTQA PAPDA]). (B) Lanes: 1, wild-type MyoD-containing nuclear extracts; 2, wild-type MyoD-containing nuclear extracts pretreated with calf intestine alkaline phosphatase; 3, MyoD S200A-containing nuclear extracts; 4, MyoD S200A nuclear extracts pretreated with calf intestine alkaline phosphatase.
Plasmid pADANS-tx61 (kindly provided by S. Plon), which contains the human CDC34 gene (9, 34), was used to generate a PCR product consisting of the coding region of CDC34 with an EcoRI site at the 5′ end and a HindIII site at the 3′ end of the DNA fragment, respectively, using Taq DNA polymerase with primer 1 (5′ GTGGTCGAATTCCCCCGCGCTGCTCCGACC 3′) and primer 2 (5′ TTTATTCTGAAGCTTTCAGGACTCCTCCGTGCC 3′) according to the manufacturer’s instructions (Boehringer Mannheim Corp.). The PCR product was then ligated into the EcoRI/HindIII fragment of the mammalian expression vector pCB6 (15) to generate pCB6-CDC34. pCB6-dnCDC34 was generated as follows. Plasmid pADANS-tx61 was used as template for PCR using primers 1 and 4 (5′ ACGGGGGACGTGTCTATCTCCATCTCCCACCCGCCGGTG 3′) as well as primers 2 and 3 (5′ CACCGGCGGGTGGGAGATGGAGATAGACACGTCCCCCGT 3′) in two separate reactions as described above. These two PCR products were then placed together with primers 1 and 2 for several further rounds of amplification. The resulting PCR product was placed into pCB6 as described above.
Half-life determination.
To determine the MyoD protein half-life, cell cultures were incubated for 16 h following transfection and then harvested by trypsinization. Equal numbers of cells were replated into 60-mm-diameter dishes; 24 h after replating, one set of cultures was harvested and nuclear extracts were prepared. The remaining cultures were treated with cycloheximide (final concentration, 35.5 μM). At the indicated time points, one set of cultures was harvested and nuclear extracts were prepared. Equal volumes of nuclear extracts from each sample were separated by SDS-PAGE (10% gel) and analyzed by Western blotting with affinity-purified anti-MyoD. The relative amounts of MyoD protein were quantitated with a Bio-Rad GS-250 Molecular Imager. The half-lives were calculated as described previously (40). Where noted, identical blots were analyzed by Western analysis using anti-Cdc2 antibodies as instructed by the manufacturer (Santa Cruz, Inc.).
Antibody generation and Western blot analysis.
Plasmid pGEX MyoD (23) (kindly provided by the late H. Weintraub) encodes a glutathione S-transferase (GST)–MyoD fusion protein that can be produced in bacteria. Recombinant GST-MyoD was prepared as described previously (23). Purified recombinant GST-MyoD fusion protein was used to prepare rabbit polyclonal antisera (performed by HRP Inc., Denver, Colo.). Antibodies were affinity purified as described previously (9), using the recombinant GST-MyoD fusion protein. SDS-PAGE and Western blotting were performed as described previously (9).
Antibodies against human Cdc34 were generated as follows. PCR was performed on pADANS-TX61 with the primers TCCGCCGCCCATATGGCTCGGCCGCTAGTGCCC and TTTATTCTGCATATGTCAGGACTCCTCCGTGCC to generate a DNA fragment containing the human CDC34 gene with an NdeI site at both the 5′ and 3′ ends. This PCR product was ligated into the NdeI site of pET28a (Novagen, Inc.), generating a fusion gene encoding a Cdc34 protein tagged with six histidine residues at the amino terminus. The His-tagged Cdc34 was produced in bacteria as previously described (9), purified by using Ni2+-nitrilotriacetic acid agarose (Qiagen Inc.) according to the manufacturer’s instructions, and used for the production of antisera (performed by HRP Inc.). Antibodies against human Cdc34 were affinity purified as described previously, using His-tagged Cdc34 (9).
RESULTS
Serine 200 is required for the hyperphosphorylation of MyoD.
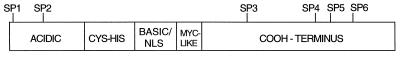
Overexpression of cyclin D1 leads to hyperphosphorylation of MyoD protein and inhibition of MyoD-dependent transcription (35, 42). To explore the possibility that MyoD is a CDK substrate, five of the seven potential CDK phosphorylation sites were individually changed to alanines by site-directed mutagenesis. At the sixth and seventh potential CDK sites (GTQSPTPDAA), S296 and T298 were changed to alanines simultaneously (Fig. 1). The MyoD genes encoding the six mutant proteins or the wild-type protein were subcloned into a mammalian expression vector that places the cDNAs under the control of the cytomegalovirus immediate-early gene promoter and simian virus 40 polyadenylation signals (see Materials and Methods). The expression constructs were transfected into C3H10T1/2 fibroblasts, and nuclear extracts were prepared 36 h later. MyoD proteins were then visualized by Western analysis of the separated nuclear proteins with anti-MyoD antibodies (Fig. 2A). Nuclear extracts were used, as preliminary experiments revealed that the wild-type MyoD protein was difficult to visualize consistently in whole-cell lysates (unpublished observations). Western analysis indicated that cells expressing wild-type MyoD or all but one of the serine-to-alanine mutations produced a 45-kDa form of MyoD as well as a slower-migrating form (Fig. 2A, lanes 2 to 4 and 6 to 8). However, cells expressing SP3-MyoD, encoding the MyoD S200A protein, lacked the slower-migrating form (Fig. 2A, lane 5). Treatment of nuclear extracts with alkaline phosphatase prior to SDS-PAGE also caused a loss of the slower-migrating form of MyoD expressed from the wild-type gene (Fig. 2B; compare lanes 1 and 2); however, alkaline phosphatase had no effect on the mobility of the MyoD S200A protein (Fig. 2B, lanes 3 and 4). These results are consistent with experiments on endogenous MyoD, demonstrating that phosphorylation of MyoD retards its migration during SDS-PAGE (46). These results indicate that the slower-migrating form of MyoD is phosphorylated and that this phosphorylation event requires a serine residue at position 200 in the MyoD protein. However, from these experiments we cannot determine whether S200 is the site of phosphorylation or whether phosphorylation of S200 is required for phosphorylation at a different position.
FIG. 1.
Schematic diagram of the MyoD protein indicating functional domains and putative CDK phosphorylation sites. NLS, nuclear localization signal.
Mutation of serine 200 to alanine increases the ability of MyoD to transactivate a muscle-specific promoter.
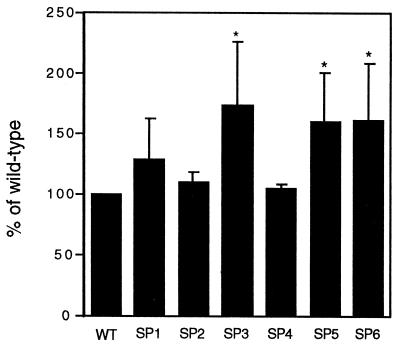
Since SP3-MyoD, encoding the MyoD S200A protein, prevented formation of the slower-migrating MyoD species, we set out to determine whether this mutation would also affect MyoD function. One consequence of preventing the formation of the hyperphosphorylated form of MyoD might be an increase or decrease in the ability of the MyoD protein to transactivate muscle-specific genes. To determine the importance of phosphorylation for MyoD transactivation activity, the ability of the mutant proteins to transactivate expression of a reporter construct under the control of the muscle creatine kinase promoter/enhancer E box was examined (see Materials and Methods). In transiently transfected C3H10T1/2 fibroblasts, MyoD S200A exhibited a modest but significant increase in reporter transactivation compared to wild-type MyoD (Fig. 3). All six mutant MyoD proteins were at least as effective as wild-type MyoD at activating transcription from this promoter. These results indicate that the phosphorylation event abolished by the MyoD S200A mutant protein increases MyoD transactivating activity. Interestingly, SP5-MyoD and SP6-MyoD also led to increased 4TRTK-CAT activity, although these mutants had no obvious effect on the abundance of the hyperphosphorylated form of MyoD.
FIG. 3.
Increased 4TRTK-CAT activity in C3H10T1/2 fibroblasts expressing MyoD proteins containing mutations of CDK consensus phosphorylation sites. Wild-type (WT) or mutant MyoD expression vectors, 4TRTK-CAT, and a TK-luciferase vector were cotransfected into C3H10T1/2 fibroblasts. Cultures were harvested 36 h posttransfection, and luciferase and CAT activities were determined. The results are presented as percentages of the transactivating ability of wild-type MyoD, which was arbitrarily set to 100%. Each experiment was done in duplicate, and each CAT assay contained equivalent amounts of luciferase activity. The results presented are means ± standard deviations of three separate experiments.
Mutation of serine 200 to alanine causes an increase in the half-life of MyoD.
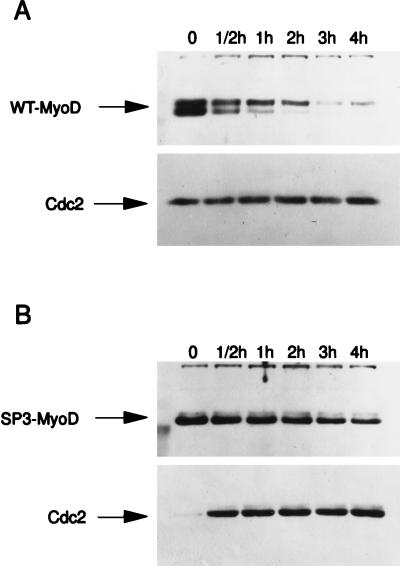
CDK-dependent phosphorylation events have been proposed to control protein stability (22, 25). To determine if the SP3-MyoD mutation affected MyoD protein stability, the half-lives of the mutant MyoD proteins were determined as described in Materials and Methods. The half-life of wild-type MyoD in nuclear extracts was found to be about 30 min (Fig. 4 and Table 1), which compares well with the previously determined value of 30 to 60 min (46). The half-life of each of the mutant MyoD proteins described above was also determined (see Materials and Methods). The MyoD S200A mutant was found to have a half-life of about 147 min, approximately fivefold longer than that of the wild-type protein (Fig. 4 and Table 1). These results indicate that one role of MyoD phosphorylation is to destabilize the MyoD protein. Additionally, they suggest that the increased 4TRTK-CAT transactivation in response to SP3-MyoD may be due to increased steady-state levels of the mutant MyoD protein resulting from decreased protein turnover. Interestingly, the SP4-MyoD and SP5-MyoD proteins were also slightly more stable than wild-type MyoD protein (Table 1).
FIG. 4.
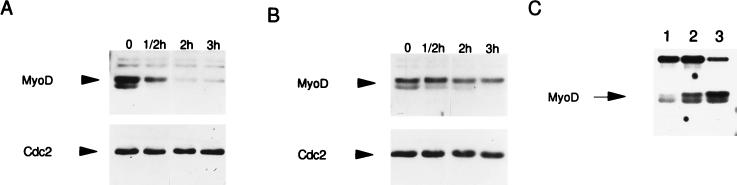
Increased stability of MyoD S200A. The half-lives of nuclear wild-type MyoD and MyoD S200A were determined as described in Materials and Methods. C3H10T1/2 fibroblasts transiently transfected with either wild-type MyoD (WT-MyoD) (A) or MyoD S200A (B) were treated with cycloheximide for the times indicated; nuclear extracts were prepared and subjected to Western analysis. Identical blots were subjected to Western analysis with anti-Cdc2 antibodies to verify sample loading.
TABLE 1.
Half-lives of MyoD proteins
| MyoD protein | Mean half-life (min) ± SD |
|---|---|
| Wild type | 30 ± 3 |
| S5A | 33 ± 8 |
| S37A | 35 ± 1 |
| S200A | 147 ± 3 |
| S262A | 55 ± 10 |
| S277A | 46 ± 4 |
| T296A/S298A | 31 ± 6 |
Requirement for components of the ubiquitin pathway in MyoD degradation.
Short-lived proteins are often degraded by a large, complex protease known as the 26S proteasome (11, 17). The relatively short half-life of MyoD led us to determine whether the degradation of MyoD might be dependent on 26S proteasome activity. Leucyl-leucyl-norleucinal (LLnL) has been described as a potent inhibitor of the 26S proteasome (39), and therefore the effect of LLnL on the stability of the MyoD protein was determined. In the presence of LLnL, wild-type MyoD protein becomes more stable (compare Fig. 5A and B). This result indicates that the turnover of MyoD is dependent on the function of the 26S proteasome. Further, in the presence of LLnL, there is a preferential accumulation of the phosphorylated form of the MyoD protein under steady-state conditions (Fig. 5C; compare lanes 2 and 3 to lane 1). A preferential accumulation of phosphorylated MyoD would be expected if phosphorylated MyoD were the preferred substrate of the proteasome.
FIG. 5.
The proteasome inhibitor LLnL stabilizes the hyperphosphorylated form of MyoD. (A and B) C3H10T1/2 fibroblasts transiently transfected with wild-type MyoD were treated with cycloheximide for the times indicated in the absence (A) or presence (B) of LLnL; nuclear extracts were prepared and subjected to Western analysis. The same blots were subjected to Western analysis with anti-Cdc2 antibodies to verify sample loading. (C) C3H10T1/2 fibroblasts transiently transfected with wild-type MyoD were not treated (lane 1) or treated with LLnL for 1 h (lane 2) or 2 h (lane 3) prior to harvest; nuclear extracts were prepared and subjected to Western analysis.
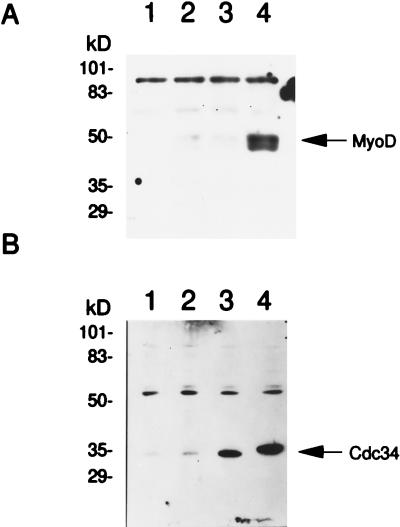
Most proteins that are degraded by the 26S proteasome are first targeted to the proteasome by the attachment of ubiquitin (17, 39). Consistent with this observation, MyoD protein is ubiquitinated in vitro following the addition of purified ubiquitin machinery components (12). In yeast, several cell cycle ubiquitination events have been shown to be under the control of the Cdc34p (Ubc3p) ubiquitin-conjugating enzyme (7, 10, 34). Furthermore, Cdc34p has also been shown to control the function of the transcription factors Gcn4p and Rgt1p (21, 24). In mammalian cells, human Cdc34 has been proposed to serve a function analogous to that of its yeast counterpart (32, 34). The prolonged half-life of MyoD in the presence of LLnL as well as the effects of phosphorylation on MyoD stability led us to consider a role for the Cdc34 enzyme in controlling the degradation of MyoD. This possibility was tested directly by examining the effect of expressing wild-type and a dominant-negative allele of human CDC34 (dnCDC34) (2) on the stability of MyoD. C3H10T1/2 fibroblasts were transiently transfected with wild-type MyoD and either wild-type CDC34 or dnCDC34. Thirty-six hours later, nuclear extracts were prepared and the steady-state levels of the MyoD protein were compared by Western analysis. The steady-state level of MyoD protein was dramatically elevated by the presence of the dnCdc34 protein, whereas expression of wild-type CDC34 had no obvious effect on MyoD levels (Fig. 6A; compare lanes 3 and 4). The effect of dnCdc34 protein was not due to differential expression of the dominant-negative and wild-type Cdc34 proteins, as both forms of Cdc34 were efficiently produced in the C3H10T1/2 fibroblasts (Fig. 6). Interestingly, the levels of Cdc34 were high in the nuclear extracts (Fig. 6) and low in the cytoplasm (data not shown), indicating that the subcellular location of Cdc34 is also consistent with a role for Cdc34 in MyoD degradation. These results indicate that the rapid degradation of nuclear MyoD is controlled by Cdc34 ubiquitin-conjugating activity.
FIG. 6.
Expression of a dnCDC34 stabilizes MyoD. C3H10T1/2 fibroblasts were transiently transfected with MyoD, CDC34, or both; nuclear extracts were prepared and subjected to Western analysis as described for Fig. 5. (A) Western analysis of nuclear extracts prepared from cells transfected with the indicated plasmids and incubated in the presence of anti-MyoD. Lanes: 1, pCB6+; 2, pCB6+ and wild-type MyoD; 3, wild-type MyoD and wild-type CDC34; 4, wild-type MyoD and dnCDC34. (B) Western analysis of the samples described for panel A, incubated with anti-Cdc34.
DISCUSSION
We demonstrate here that serine residue 200, located within a CDK consensus phosphorylation site in the MyoD protein, is required for the formation of a specific phosphorylated form of MyoD, often referred to as hyperphosphorylated MyoD. When serine 200 is changed to an alanine, the hyperphosphorylated form of MyoD is absent. Mutations that eliminate other potential CDK phosphorylation sites do not affect the presence of hyperphosphorylated MyoD. While we do not know the nature of the protein kinase responsible for MyoD phosphorylation, we can propose several possible candidates based on the function of MyoD as a transcription factor with roles in differentiation and cell cycle progression. One possibility is that MyoD is a substrate of Cdc2, Cdk2, or Cdk4 complexes that control progression through the cell cycle. Such a phosphorylation event might regulate the cell cycle-inhibitory effects of MyoD. In fact, increasing the levels of cyclin D1 resulted in an elevation of the hyperphosphorylated form of MyoD as well as inhibition of terminal myogenic differentiation (35, 42). However, Cdk4-cyclin D complexes have not been shown to phosphorylate MyoD. An alternative model is that CDKs associated with the transcriptional machinery, Cdk7, Cdk8, and Cdk9, modify MyoD. These kinases and their associated cyclin subunits have been proposed to regulate transcriptional elongation (for a review, see reference 19). In this instance, transcriptional elongation may be regulated by a process that requires the modification and subsequent elimination of transcription factors like MyoD. Under these conditions, MyoD would be required for initiation of transcription, and its targeted degradation would be required for processivity of the transcriptional machinery along the transcribed gene (elongation).
When the half-lives of wild-type and the various mutant MyoD proteins were determined, the S200A change was found to dramatically alter the half-life of MyoD. MyoD S200A has a half-life (147 min) about five times longer than that of wild-type MyoD (30 min). The increased stability of MyoD S200A is likely to be responsible for the increased 4TRTK-CAT activity detected in C3H10T1/2 fibroblasts overexpressing MyoD S200A compared to wild-type MyoD. In contrast, removal of the other potential CDK sites had a much more modest effect on the MyoD half-life. The 30-min half-life was determined for nuclear MyoD. Whether the MyoD present in the cytoplasm has the same half-life is not clear. These results suggest that phosphorylation of MyoD at serine 200 is a prerequisite for MyoD degradation.
Most short-lived proteins in the cell are targeted for degradation by ubiquitin modification (11, 17). Ubiquitination of many proteins leads to their destruction via the 26S proteasome. Indeed, wild-type MyoD protein is a short-lived protein. The steady-state level of wild-type MyoD protein is increased in the presence of the LLnL, a proteasome inhibitor (39), which suggests that inhibition of proteasome activity stabilizes MyoD. Interestingly, in the presence of LLnL, there is a preferential increase in the amount of hyperphosphorylated MyoD, as would be expected if the hyperphosphorylated form of MyoD is the actual target of the proteasome. Recently, Horwitz (18) described a myoblast cell line in which nuclear MyoD had become unstable and the cells had lost the ability to differentiate into myotubes. These studies point to a relationship between MyoD activity and the stability of the MyoD protein.
In this study, we have demonstrated that expression of a dominant-negative allele of human CDC34 leads to the accumulation of the MyoD protein. While expression of a dnCdc34 protein may indirectly perturb the ubiquitination machinery, we favor the possibility that the Cdc34 protein is directly involved in the destruction of MyoD. The Cdc34 protein was first identified in yeast as a regulator of the G1-to-S phase transition (10). It is a member of the family of ubiquitin-conjugating enzymes that can catalyze the addition of ubiquitin to other protein substrates. A characteristic of Cdc34 substrates in yeast is that they must first be phosphorylated by Cdc28-cyclin complexes (8, 16, 22, 43, 51). In mammals, Cdc34 has been invoked to destabilize p27Kip1 and therefore is a likely regulator of the G1-to-S phase transition (32). Furthermore, human Cdc34 is a predominantly nuclear protein (our results and reference 26). MyoD can be ubiquitinated in vitro (12), and its half-life is controlled by phosphorylation through a potential CDK site. Therefore, we propose that phosphorylation of nuclear MyoD leads to its Cdc34-dependent ubiquitination, thus targeting MyoD for destruction via the 26S proteasome.
The potential control of MyoD levels through CDK phosphorylation-dependent ubiquitination suggests a mechanism for coordinating transcription events during the G1-to-S phase progression in myoblasts. At least two other proteins associated with the transcriptional machinery, p53 and Rb, are substrates of CDKs as well as potential substrates of Cdc34. The stability of the p53 protein also appears to be controlled by CDK-dependent phosphorylation at a site, SSSPQPKKK, very similar to that found in MyoD, ASSPRSN (25). Recently, Connell-Crowley et al. (4) reported that phosphorylation of a similar site in Rb, PSSPLRI, is critical to the inactivation of the Rb protein (see also reference 20). Therefore, depending on cellular growth status, or potentially as a mechanism to enhance transcriptional elongation, the activities of MyoD, Rb, and p53 may be coordinately activated or alternatively eliminated by CDK-dependent phosphorylation. A role for phosphorylation in targeting a regulatory protein for degradation has been postulated in a number of cases, including p27Kip1 and B-lymphocyte-specific Rag2 (25, 31, 41, 49). In fact, one could speculate that the activities of unrelated cellular events are coordinated by the regulated degradation of several proteins simultaneously.
ACKNOWLEDGMENTS
A.S. and Q.W. contributed equally to this work.
We acknowledge the National Institutes of Health (grants GM43792 and GM45460), the Indiana Affiliate of the American Heart Association, and the Diabetes Research and Training Center for financial support. M.A.H. is a Scholar of the Leukemia Society of America.
We thank Peter A. Jones and Sharon Plon for their insightful comments.
REFERENCES
- 1.Andrews N C, Faller D V. A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 1991;19:2499. doi: 10.1093/nar/19.9.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banerjee A, Deshaies R J, Chau V. Characterization of a dominant negative mutant of the cell cycle ubiquitin-conjugating enzyme Cdc34. J Biol Chem. 1995;270:26209–26215. doi: 10.1074/jbc.270.44.26209. [DOI] [PubMed] [Google Scholar]
- 3.Cobrinik D. Regulatory interactions among E2Fs and cell cycle control proteins. Curr Top Microbiol Immunol. 1996;208:32–59. doi: 10.1007/978-3-642-79910-5_2. [DOI] [PubMed] [Google Scholar]
- 4.Connell-Crowley L, Harper J W, Goodrich D W. Cyclin D1/Cdk4 regulates retinoblastoma protein-mediated cell cycle arrest by site-specific phosphorylation. Mol Biol Cell. 1997;8:287–301. doi: 10.1091/mbc.8.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crescenzi M, Fleming T P, Lassar A B, Weintraub H, Aaronson S A. MyoD induces growth arrest independent of differentiation in normal and transformed cells. Proc Natl Acad Sci USA. 1990;87:8442–8446. doi: 10.1073/pnas.87.21.8442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davis R L, Weintraub H, Lassar A. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51:987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- 7.Deshaies R J, Chau V, Kirschner M. Ubiquitination of the G1 cyclin Cln2p by a Cdc34p-dependent pathway. EMBO J. 1995;14:303–312. doi: 10.1002/j.1460-2075.1995.tb07004.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feldman R M R, Correll C C, Kaplan K B, Deshaies R J. A complex of Cdc4p, Skp1p, and Cdc53p/Cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell. 1997;91:221–230. doi: 10.1016/s0092-8674(00)80404-3. [DOI] [PubMed] [Google Scholar]
- 9.Goebl M G, Goetsch L, Byers B. The Ubc3 (Cdc34) ubiquitin-conjugating enzyme is ubiquitinated and phosphorylated in vivo. Mol Cell Biol. 1994;14:3022–3029. doi: 10.1128/mcb.14.5.3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goebl M G, Yochem J, Jentsch S, McGrath J P, Varshavsky A, Byers B. The yeast cell cycle gene CDC34 encodes a ubiquitin-conjugating enzyme. Science. 1988;241:1331–1335. doi: 10.1126/science.2842867. [DOI] [PubMed] [Google Scholar]
- 11.Goldberg A L. Functions of the proteasome: the lysis at the end of the tunnel. Science. 1995;268:522–523. doi: 10.1126/science.7725095. [DOI] [PubMed] [Google Scholar]
- 12.Gonen H, Stancovski I, Shkedy D, Hadari T, Bercovich B, Bengal E, Mesilat S, Abu-Hatoum O, Schwartz A L, Ciechanover A. Isolation, characterization, and partial purification of a novel ubiquitin-protein ligase, E3. J Biol Chem. 1996;271:302–310. doi: 10.1074/jbc.271.1.302. [DOI] [PubMed] [Google Scholar]
- 13.Gu W, Schneider J W, Condorelli G, Kaushal S, Mahdavi V, Nadal-Ginard B. Interaction of myogenic factors and the retinoblastoma protein mediates muscle cell commitment and differentiation. Cell. 1993;72:309–324. doi: 10.1016/0092-8674(93)90110-c. [DOI] [PubMed] [Google Scholar]
- 14.Halevy O, Novitch B G, Spicer D B, Skapek S X, Rhee J, Hannon G J, Beach D, Lassar A B. Correlation of terminal cell cycle arrest of skeletal muscle with induction of p21 by MyoD. Science. 1995;267:1018–1021. doi: 10.1126/science.7863327. [DOI] [PubMed] [Google Scholar]
- 15.Harrington M A, Konicek B, Song A, Xia X L, Fredericks W J, Rauscher F J., III Inhibition of colony-stimulating factor-1 promoter activity by the product of the Wilms’ tumor locus. J Biol Chem. 1993;268:21271–21275. [PubMed] [Google Scholar]
- 16.Henchoz S, Chi Y, Catarin B, Herskowitz I, Deshaies R J, Peter M. Phosphorylation- and ubiquitin-dependent degradation of the cyclin-dependent kinase inhibitor Far1p in budding yeast. Genes Dev. 1997;11:3046–3060. doi: 10.1101/gad.11.22.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hochstrasser M. Ubiquitin, proteasomes, and the regulation of intracellular protein degradation. Curr Opin Cell Biol. 1995;7:215–223. doi: 10.1016/0955-0674(95)80031-x. [DOI] [PubMed] [Google Scholar]
- 18.Horwitz M. Hypermethylated myoblasts specifically deficient in MyoD autoactivation as a consequence of instability of MyoD. Exp Cell Res. 1996;226:170–182. doi: 10.1006/excr.1996.0216. [DOI] [PubMed] [Google Scholar]
- 19.Jones K A. Taking a new TAK on Tat transactivation. Genes Dev. 1997;11:2593–2599. doi: 10.1101/gad.11.20.2593. [DOI] [PubMed] [Google Scholar]
- 20.Kato J, Matsushime H, Hiebert S W, Ewen M E, Sherr C J. Direct binding of cyclin D to the retinoblastoma gene product (pRb) and pRb phosphorylation by the cyclin D-dependent kinase CDK4. Genes Dev. 1993;7:331–342. doi: 10.1101/gad.7.3.331. [DOI] [PubMed] [Google Scholar]
- 21.Kornitzer D, Raboy B, Kulka R G, Fink G R. Regulated degradation of the transcription factor Gcn4. EMBO J. 1994;13:6021–6030. doi: 10.1002/j.1460-2075.1994.tb06948.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lanker S, Valdivieso M H, Wittenberg C. Rapid degradation of the G1 cyclin Cln2 induced by CDK-dependent phosphorylation. Science. 1996;271:1597–1601. doi: 10.1126/science.271.5255.1597. [DOI] [PubMed] [Google Scholar]
- 23.Lassar A B, Buskin J N, Lockshon D, Apone S, Hauschka S D, Weintraub H. MyoD is a sequence-specific DNA binding protein requiring a region of myc homology to bind to the muscle creatine kinase promoter. Cell. 1989;58:823–831. doi: 10.1016/0092-8674(89)90935-5. [DOI] [PubMed] [Google Scholar]
- 24.Li F N, Johnston M. Grr1 of Saccharomyces cerevisiae is connected to the ubiquitin proteolysis machinery through Skp1: coupling glucose sensing to gene expression and the cell cycle. EMBO J. 1997;16:5629–5638. doi: 10.1093/emboj/16.18.5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin W-C, Desiderio S. Regulation of V(D)J recombination activator protein RAG-2 by phosphorylation. Science. 1993;260:953–959. doi: 10.1126/science.8493533. [DOI] [PubMed] [Google Scholar]
- 26.Lisztwan J, Marti A, Sutterluty H, Gstaiger M, Wirbelauer C, Krek W. Association of human CUL-1 and ubiquitin-conjugating enzyme CDC34 with the F-box protein p45SKP2: evidence for evolutionary conservation in the subunit composition of the CDC34-SCF pathway. EMBO J. 1998;17:368–383. doi: 10.1093/emboj/17.2.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ludolph D C, Konieczny S F. Transcription factor families: muscling in on the myogenic program. FASEB J. 1995;9:1595–1604. doi: 10.1096/fasebj.9.15.8529839. [DOI] [PubMed] [Google Scholar]
- 28.Madden S L, Cook D M, Morris J F, Gashler A, Sukhatme V P, Rauscher F J., III Transcriptional repression mediated by the WT1 Wilm’s tumor gene product. Science. 1991;253:1550–1553. doi: 10.1126/science.1654597. [DOI] [PubMed] [Google Scholar]
- 29.Megeney L A, Kablar B, Garrett K, Anderson J E, Rudnicki M A. MyoD is required for myogenic stem cell function in adult skeletal muscle. Genes Dev. 1996;10:1173–1183. doi: 10.1101/gad.10.10.1173. [DOI] [PubMed] [Google Scholar]
- 30.Moreno S, Nurse P. Substrates for p34cdc2: in vivo veritas? Cell. 1990;61:549–551. doi: 10.1016/0092-8674(90)90463-o. [DOI] [PubMed] [Google Scholar]
- 31.Morisaki H, Fujimoto A, Ando A, Nagata Y, Ikeda K, Nakanishi M. Cell cycle-dependent phosphorylation of p27 cyclin-dependent kinase inhibitor by cyclin E/CDK2. Biochem Biophys Res Commun. 1997;240:386–390. doi: 10.1006/bbrc.1997.7590. [DOI] [PubMed] [Google Scholar]
- 32.Pagano M, Tam S W, Theodoras A M, Beer-Romero P, Dell Sal G, Chau V, Yew P R, Draetta G R, Rolfe M. Role of the ubiquitin-proteasome pathway in regulating abundance of the cyclin-dependent kinase inhibitor p27. Science. 1995;269:682–685. doi: 10.1126/science.7624798. [DOI] [PubMed] [Google Scholar]
- 33.Parker S B, Eichele G, Zhang P, Rawls A, Sands A T, Bradley A, Olson E N, Harper J W, Elledge S J. p53-independent expression of p21Cip1 in muscle and other terminally differentiating cells. Science. 1995;267:1024–1027. doi: 10.1126/science.7863329. [DOI] [PubMed] [Google Scholar]
- 34.Plon S E, Leppig K A, Do H-N, Groudine M. Cloning of the human homolog of the CDC34 cell cycle gene by complementation in yeast. Proc Natl Acad Sci USA. 1993;90:10484–10488. doi: 10.1073/pnas.90.22.10484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rao S S, Chu C, Kohtz D S. Ectopic expression of cyclin D1 prevents activation of gene transcription by myogenic basic helix-loop-helix regulators. Mol Cell Biol. 1994;14:5259–5267. doi: 10.1128/mcb.14.8.5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rudnicki M A, Braun T, Hinuma S, Jaenisch R. Inactivation of MyoD in mice leads to up-regulation of the myogenic HLH gene Myf-5 and results in apparently normal muscle development. Cell. 1992;71:383–390. doi: 10.1016/0092-8674(92)90508-a. [DOI] [PubMed] [Google Scholar]
- 37.Rudnicki M A, Schnegelsberg P N J, Stead R H, Braun T, Arnold H-H, Jaenisch R. MyoD or Myf-5 is required for the formation of skeletal muscle. Cell. 1993;75:1351–1359. doi: 10.1016/0092-8674(93)90621-v. [DOI] [PubMed] [Google Scholar]
- 38.Sambrook J E, Fritsch F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 39.Seemüller E, Lupas A, Stock D, Löwe J, Huber R, Baumeister W. Proteasome from Thermoplasma acidophilum: a threonine protease. Science. 1995;268:579–582. doi: 10.1126/science.7725107. [DOI] [PubMed] [Google Scholar]
- 40.Segel I H. p376. Biochemical calculations. 2nd ed. New York, N.Y: John Wiley & Sons; 1976. [Google Scholar]
- 41.Sheaff R J, Groudine M, Gordon M, Roberts J M, Clurman B E. Cyclin E-CDK2 is a regulator of p27Kip1. Genes Dev. 1997;11:1464–1478. doi: 10.1101/gad.11.11.1464. [DOI] [PubMed] [Google Scholar]
- 42.Skapek S X, Rhee J, Spicer D B, Lassar A B. Inhibition of myogenic differentiation in proliferating myoblasts by cyclin D1-dependent kinase. Science. 1995;267:1022–1024. doi: 10.1126/science.7863328. [DOI] [PubMed] [Google Scholar]
- 43.Skowyra D, Craig K L, Tyers M, Elledge S J, Harper J W. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell. 1997;91:209–219. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- 44.Slansky J E, Farnham P J. Introduction to the E2F family: protein structure and gene regulation. Curr Top Microbiol Immunol. 1996;208:1–31. doi: 10.1007/978-3-642-79910-5_1. [DOI] [PubMed] [Google Scholar]
- 45.Sorrentino V, Pepperkok R, Davis R L, Ansorge W, Philipson L. Cell proliferation inhibited by MyoD independently of myogenic differentiation. Nature (London) 1990;345:813–815. doi: 10.1038/345813a0. [DOI] [PubMed] [Google Scholar]
- 46.Thayer M J, Tapscott S J, Davis R L, Wright W E, Lassar A B, Weintraub H. Positive autoregulation of the myogenic determination gene MyoD. Cell. 1989;58:241–248. doi: 10.1016/0092-8674(89)90838-6. [DOI] [PubMed] [Google Scholar]
- 47.Thorburn A M, Walton P A, Feramisco J R. MyoD induced cell cycle arrest is associated with increased nuclear affinity of the Rb protein. Mol Biol Cell. 1993;4:705–713. doi: 10.1091/mbc.4.7.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Venuti J M, Cserjesi P. Molecular embryology of skeletal myogenesis. Curr Top Dev Biol. 1996;34:169–206. doi: 10.1016/s0070-2153(08)60711-5. [DOI] [PubMed] [Google Scholar]
- 49.Vlach J, Hennecke S, Amati B. Phosphorylation-dependent degradation of the cyclin-dependent kinase inhibitor p27Kip1. EMBO J. 1997;16:5334–5344. doi: 10.1093/emboj/16.17.5334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weintraub H, Davis R, Tapscott S, Thayer M, Krause M, Benezra R, Blackwell T K, Turner D, Hollenberg S, Zhuang Y, Lassar A. The MyoD family: nodal point during specification of the muscle cell lineage. Science. 1991;251:761–766. doi: 10.1126/science.1846704. [DOI] [PubMed] [Google Scholar]
- 51.Willems A R, Lanker S, Patton E E, Craig K L, Nason T F, Mathias N, Kobayashi R, Wittenberg C, Tyers M. Cdc53 targets phosphorylated G1 cyclins for degradation by the ubiquitin proteolytic pathway. Cell. 1996;62:453–463. doi: 10.1016/s0092-8674(00)80118-x. [DOI] [PubMed] [Google Scholar]