Abstract
A screen for suppressors of a U2 snRNA mutation identified CUS2, an atypical member of the RNA recognition motif (RRM) family of RNA binding proteins. CUS2 protein is associated with U2 RNA in splicing extracts and interacts with PRP11, a subunit of the conserved splicing factor SF3a. Absence of CUS2 renders certain U2 RNA folding mutants lethal, arguing that a normal activity of CUS2 is to help refold U2 into a structure favorable for its binding to SF3b and SF3a prior to spliceosome assembly. Both CUS2 function in vivo and the in vitro RNA binding activity of CUS2 are disrupted by mutation of the first RRM, suggesting that rescue of misfolded U2 involves the direct binding of CUS2. Human Tat-SF1, reported to stimulate Tat-specific, transactivating region-dependent human immunodeficiency virus transcription in vitro, is structurally similar to CUS2. Anti-Tat-SF1 antibodies coimmunoprecipitate SF3a66 (SAP62), the human homolog of PRP11, suggesting that Tat-SF1 has a parallel function in splicing in human cells.
In eukaryotes, the removal of introns from nuclear transcripts requires two transesterification reactions carried out by spliceosomes. Five small nuclear ribonucleoprotein particles (snRNPs), U1, U2, U5, U4, and U6, and many extrinsic protein factors act in concert to build a spliceosome and execute the splicing reactions (38, 39, 49, 50, 53). The spliceosome is clearly the most dynamic of the RNP enzymes. Major changes in the secondary structure of U4, U6, and U2 snRNAs during the spliceosome cycle are inferred from genetic and cross-linking studies (reviewed in reference 63). The snRNAs arrive at the assembling spliceosome in a form unlike that necessary for the catalytic activity of splicing and must be rearranged before splicing can proceed (7, 45). A less studied corollary of this finding is that these rearrangements must be undone during spliceosome disassembly, and snRNA structure must be regenerated in an appropriate form for another round of spliceosome assembly and splicing (56). Although individual proteins are clearly linked to changes in the composition and organization of splicing complexes at distinct points in the splicing pathway (for a review, see reference 63), it has been difficult to assign responsibility for a specific RNA rearrangement event to any single protein.
The first ATP-dependent step during in vitro spliceosome assembly is the stable binding of the U2 snRNP to the branch point region of the intron, an event normally dependent on formation of ATP-independent complexes between the pre-mRNA and other proteins, as well as the U1 snRNP (51). These ATP-independent complexes (called the E complex in mammalian studies and commitment complexes in yeast studies) contain pre-mRNA that has been recognized both at the 5′ splice site by the U1 snRNP and at the branch point by the homologous mammalian (SF1) or yeast (BBP) branch point-interacting proteins (2, 13, 29, 38, 48). Formation of this complex is expected to specify an exon joining event because the 5′ splice site and branch point are selected, and in most cases the 3′ splice site used will be the first YAG downstream from the branch point (54, 62). The ATP-dependent binding of U2 snRNP to the complex presumably seals its fate. This reaction forms the prespliceosome (called the A complex in mammals and the B complex or III in yeast) and the subsequent steps of spliceosome assembly and splicing proceed.
Prior to its stable docking with the commitment complex to form the prespliceosome, an assembly-competent U2 snRNP must be formed. A number of requirements for U2 participation in prespliceosome formation have been identified through both yeast genetics and biochemical approaches in the mammalian system. In yeast, stem-loop IIa of U2 RNA contributes to this binding through a network of functional interactions with the products of the PRP5, PRP9, PRP11, PRP21, and CUS1 genes (1, 54, 58, 62, 66, 67, 70). By several criteria, the PRP9, PRP11, and PRP21 proteins are homologs of the subunits of mammalian SF3a, also called SAP61, -62, and -114 or SF3a60, -66, and -120, respectively (for reviews, see references 32 and 38). The CUS1 and HSH49 proteins are yeast splicing factors homologous to two mammalian SF3b subunits, SAP145 and SAP49 (21, 29, 35, 67). SF3a and SF3b are distinct multimeric protein complexes that bind to the 12S form of the mammalian U2 snRNP to create the 17S form that is recruited to the spliceosome (for a review, see reference 39). SF3a binding to U2 is dependent on binding of SF3b (18), and additional proteins are found associated with the 17S U2 snRNP (10). Once assembled into the spliceosome, this set of proteins and U2 RNA remain through both steps of splicing (11, 21, 23, 39).
Despite the enumeration of these components, none have been shown to interact directly with U2 RNA and little is known about how their assembly with the core U2 snRNP is regulated. In this report, we describe CUS2, a novel gene whose mutation improves the performance of a cold-sensitive stem IIa mutant U2 RNA (5). The CUS2 protein is an atypical member of the RNA recognition motif (RRM) family of RNA binding proteins (16, 37, 53) closely related to human Tat-SF1, identified as a host cofactor important for Tat-dependent, transactivating region (TAR)-dependent human immunodeficiency virus (HIV) transcription (72). We show that Tat-SF1 and CUS2 have parallel associations with the splicing protein homologs SAP62 and PRP11. The data provide evidence that CUS2, and by inference Tat-SF1, is a splicing factor that aids assembly of the splicing-competent U2 snRNP in vivo.
MATERIALS AND METHODS
Cloning of CUS2.
All yeast and Escherichia coli techniques (growth, transformation, sporulation, and tetrad dissection) were performed by using standard procedures (8, 59, 60). A library of fragments from the Saccharomyces cerevisiae CUS2-9 strain was constructed and screened as described for the isolation of CUS1-54 (67). DNA spanning open reading frame YNL286w (accession no. Z71562) from CUS2-9 suppresses the U2 RNA mutation. A single base change was observed between the cloned sequence and the genome project sequence. Genetic linkage between the cloned YNL286w open reading frame and CUS2-9 was demonstrated by inserting the LEU2 marker next to the YNL286w open reading frame in a haploid wild-type CUS2+ (nonsuppressing) strain of yeast carrying the cold-sensitive U2 mutation. This strain was mated to a leu2− CUS2-9 suppressor strain carrying the cold-sensitive U2 mutation, and tetrads were dissected. Of 42 complete tetrads, 40 displayed 2:2 segregation of parental, nonrecombinant spores: two cold resistant (CUS2-9) and Leu−, and two cold sensitive (CUS2+) and Leu+. The remaining two produced 3:1 segregation of the same two classes, most likely by gene conversion. Since no spores to indicate recombination between the LEU2 marker near YNL286w and the CUS2-9 suppressor were observed, CUS2-9 is tightly linked to YNL286w.
Yeast strains and growth.
Description of yeast strains carrying U2 mutations and plasmids can be found elsewhere (30, 71). BJ81 is MATa leu2-3,112 ura3-52 trp1 pep4-3 prb1 prc1 and carries a chromosomal glucose-repressible GAL-U2 gene (49). HI227 is MATa leu2-3,112 ura3-52 trp1 pep4-3 prb1 prc1 his3Δ lys2Δ. D2 is MATa U2Δ ade2 ade3 his3 lys2 leu2 can and carries a U2 gene on pCH1122 (YCp50 carrying ADE3; gift of C. Holm). Strains HI227CUS2KO and D2CUS2KO were made from strains HI227 and D2, respectively, by replacing the chromosomal copy of CUS2 with the cus2::HIS3 allele (see below). RP01 was made from HI227CUS2KO by replacing the chromosomal copy of U2 with the same glucose-repressible GAL-U2 gene as for BJ81. Diploid strain SS330/SS328 (gift of S. Ruby) is MATa/MATa ade2-101/ade2-101 his3-d200/his3-d200 ura3-52/ura3-52 LYS2/lys2-801 tyr1/TYR. MA80 is MATa his4-619 leu2-3,112 ura3-52 lys2. The U2 allele specificity of CUS2 suppression was tested in strains BJ81 or RP01 by cotransforming mutant U2 genes on a LEU2 plasmid together with various CUS2 genes on a TRP1 plasmid. Transformants were selected on synthetic complete (SC) medium containing galactose to maintain expression of the GAL-controlled wild-type U2 gene on the chromosome. Transformants were then streaked on glucose to test suppression in the absence of wild-type U2.
Two disruptions of CUS2 were made. One has an insertion of the 1.8-kb BamHI fragment of the HIS3 gene in the BclI site of the PstI fragment of CUS2 in pTZ19R. The second is a deletion that replaces the PstI fragment spanning the entire CUS2 gene with HIS3. Both give identical results; however, the deletion disruption is more difficult to integrate due to the presence of repetitive sequences downstream of CUS2. The insertional disruption, cus2::HIS3, was released from vector sequences by digestion with SphI and SalI. Transformation with this linear DNA resulted in replacement of one wild-type CUS2 gene in the SS328/SS330 diploid with the cus2::HIS3 allele as determined by blots of genomic DNA (57). Haploid strains carrying the disruption were transformed directly. Enhancement of U2 mutations by cus2::HIS3 was tested in strain D2CUS2KO. The strain was transformed with mutant U2 genes carried on a LEU2 plasmid. Transformants were streaked on plates containing 5-fluoroorotic acid (17) to select for cells that had lost the URA3 plasmid carrying the wild-type U2 gene. Growth defects revealed by loss of the wild-type U2 gene indicate enhancement.
CUS2 expression in yeast.
The coding sequence of CUS2 was amplified by PCR using primers that contained NotI restriction sites at either end of the PCR product. The PCR product was cloned into pTAG (67) at the NotI site. This produced a GAL-regulated gene encoding a tagged CUS2 protein containing three alanines, the nine-amino-acid hemagglutinin (HA) epitope, and six histidines at the carboxyl terminus (pTAGCUS2). To replace the GAL1 promoter with the native CUS2 promoter, pTAGCUS2 was digested with BamHI and SspI, and the fragment containing the GAL1 promoter was replaced with a BamHI-SspI fragment containing the native CUS2 promoter. This plasmid (pTAGnCUS2) was transformed into strain HI227CUS2KO to express tagged CUS2 protein under CUS2 control. The tagged gene complements the defects of the cus2::HIS3 knockout allele but is not a dominant suppressor of U2 G53A. The primers used were CUS2NOT5 (5′AATATGCGGCCGCCATGGATGCTGATGAATTGGAATT) and CUS2NOT3′ (5′AATATGCGGCCGCTATAAGGTCATCTTCCACTTCGCT).
To generate CUS2 mutant proteins, the promoter and coding sequence of CUS2 was amplified by PCR with primers designed to engineer BamHI restriction sites at both ends of the products (BAM5 [CGGGATCCGC ATGCAAATAA GGGAATGAAC AG] and BAM3 [CGGGATCCGT ACTGGGAAGT CATAAAGCCC TA]). The PCR product was subcloned into pRS314 (30) to create p314CUS2. The carboxyl terminus was tagged with the nine-amino-acid HA epitope and six histidines from pTAGCUS2 by subcloning a DraI/blunt MluI fragment into DraI/blunt SacI-digested p314CUS2 to create p314CUS2TAG. Y48D and C-terminal CUS2 truncation mutations were made in p314CUS2TAG by standard site-directed mutagenesis using oligonucleotides RNP-2Y (AATACTTCAATAVATATTTC TGGTCTT) and CUS2trunc (ATATGGGTAT GCGGCCGCCT CGAGAATAAT GAAAGCAAGC AA), respectively (42). The Y48D mutation substitutes tyrosine 48 with an aspartic acid, and the C-terminal truncation deletes to amino acid 264 on the CUS2 protein. The Y48DCUS2-9 mutation was made by site-directed mutagenesis of pY48DCUS2TAG, using the oligonucleotide 2-9TAG (GTATGCGGCC GCTATAAAGT CATCTTCCA).
Yeast splicing extracts and immunoprecipitations.
Splicing extracts were made from strain HI227 and strain HI227CUS2KO carrying pTAGnCUS2, using a method described previously (65). Immunoprecipitation experiments were performed essentially as described elsewhere (66). Protein A-Sepharose CL-4B (PAS)-conjugated beads were swollen overnight in NET-150 (50 mM Tris-Cl [pH 7.5], 0.01% Nonidet P-40, 150 mM NaCl) at 100 mg/4 ml at 4°C with slow rotation and washed three times with 4 ml of NET-150. The amount of prewashed PAS beads equivalent to 20 μl of packed volume was suspended in 0.4 ml of NET-150 and incubated with 5 μl of anti-HA antibody 12CA5 with slow rotation for 4 h at 4°C. PAS bead-antibody complexes were washed three times with NET-X (X equal to 50, 100, 150, or 200 mM NaCl). Complexes were suspended in 0.4 ml of NET-X before incubation with 20 μl of the splicing extracts at 4°C with slow rotation for 1 h. Immune complexes were collected and washed three times on ice with the same salt concentration of NET as in the binding reactions, each for 5 min with occasional mixing. To extract RNA, the beads were resuspended in 100 μl of 0.3 mM sodium acetate (pH 5.2)–0.2% sodium dodecyl sulfate (SDS)–1 mM EDTA–10 μg of proteinase K per ml and incubated for 10 min at 65°C. Samples were phenol (pH 4.0) extracted and ethanol precipitated in the presence of 0.5 μg of linear acrylamide as carrier. The recovered RNA was subjected to reverse transcription as previously described (5).
RNA mobility shift assay.
His6-tagged CUS2 or mutant Y48D-CUS2 protein was expressed in E. coli and purified by using Ni-nitrilotriacetic acid beads as specified by the manufacturer (Qiagen). RNA mobility shift assays were performed essentially as described previously (35). Twenty femtomoles of 32P-labeled RNA probe corresponding to T7-U2Δ107 (61) was incubated with increasing amounts of purified CUS2 or Y48D-CUS2 protein for 10 min on ice in the presence of assay mixture (20 mM HEPES [pH 7.9], 125 mM KCl, 1 mM dithiothreitol, 0.1% Triton X-100, 5% glycerol, 1 mM EDTA, 0.3 U of RNasin) and 2 pmol of tRNA as competitor in 10 μl. CUS2-RNA complexes were resolved on 5% native polyacrylamide gel (60 acrylamide:1 bisacrylamide) supplemented with 10% glycerol and 0.5× Tris-borate-EDTA running buffer. The gel was prerun for 20 min at 4°C, and then samples were loaded and run for 3 h at 120 V and complexes were visualized with a phosphorimager.
Two-hybrid assay.
A plasmid based on pAS2 containing the CUS2 coding region (or as a negative control, part of the URA3 coding region) fused to the GAL4 DNA binding domain was made and tested in combination with pACT plasmids expressing the GAL4 activation domain fusions with one of the following: PRP9, PRP11, PRP21 (generous gifts of Pierre Legrain), MUD2 (generous gift of M. Rosbash), CUS1, HSH49, or (as a negative control) a fragment of the URA3 gene. All of the activation domain constructs (with the exception of URA3) were previously shown to function in the two-hybrid system in combination with other DNA binding domain constructs (43, 44). Plasmids were cotransformed into yeast Y187 and then selected and arrayed on Trp- and Leu-deficient SCD agar medium in 90-mm-diameter petri dishes. After growth (usually 2 to 3 days), the plates were overlaid with 10 ml of 0.5% agarose–0.5 M sodium phosphate (pH 7.0)–0.1% SDS–2% (vol/vol) dimethylformamide–0.2% 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) and incubated at 37°C overnight.
Sequence comparison between CUS2 and Tat-SF1.
The first 420 amino acids of Tat-SF1 were used to search the complete yeast DNA sequence, using TBLASTN at http://genome-www2.stanford.edu/cgi-bin/nph-blastsgd. The complete CUS2 protein sequence was used to search the dbest database, using TBLASTN at http://www.ncbi.nlm.nih.gov/cgi-bin/BLAST/nph-blast?Jform=0. Ungapped segments of homology were joined by visual inspection and the conservative introduction of gaps. Match to the RRM fold was judged according to published criteria (16, 37, 53). A potential casein kinase II (CKII) phosphorylation site was identified as described previously (46).
Immunoprecipitation from HeLa nuclear extract.
Immunoprecipitation experiments were done as described previously (66). PAS beads were preswollen in NET-150 (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 0.05% Nonidet P-40) at 4°C overnight. Then a 20-μl bed volume of beads was incubated in NET-150 with various antibodies or control serum by slow rotation for 4 h at 4°C. Beads were washed three times with 5-min incubation on ice between washes; 20-μl aliquots of HeLa cell nuclear extract were then incubated with PAS beads conjugated with rabbit preimmune serum or polyclonal anti-Tat-SF1 (gift of Q. Zhou and P. Sharp) (72) or anti-SF3a60 (gift of A. Kramer) (40) antibody in 0.4 ml of NET-150 at 4°C for 1 h with slow rotation. Beads were collected and washed three times as described above. Proteins eluted from the beads were suspended in 10 μl of SDS loading dye, boiled, and separated on SDS–10% polyacrylamide gels. Proteins were electrotransferred to nitrocellulose in 0.24% Tris base–1.13% glycine–20% methanol. Blots were blocked in 3% bovine serum albumin in TBST (15 mM NaCl, 100 mM Tris [pH 7.5], 0.1% Tween) for 30 min at room temperature. Mouse anti-SF3a66 monoclonal antibody (MAb; gift of A. Kramer) (18) was added at 1:5,000 dilution in TBST for 2 h at room temperature. Blots were washed in TBST, and immunoglobulin G goat anti-mouse secondary antibody was added at 1:1,000 dilution for 1 h at room temperature. Blots were washed in TBST, and products were visualized with conjugated alkaline phosphatase and 5-bromo-4-chloro-3-indolyl phosphate–nitroblue tetrazolium.
RESULTS
The CUS2 suppressor corresponds to yeast open reading frame YNL286w.
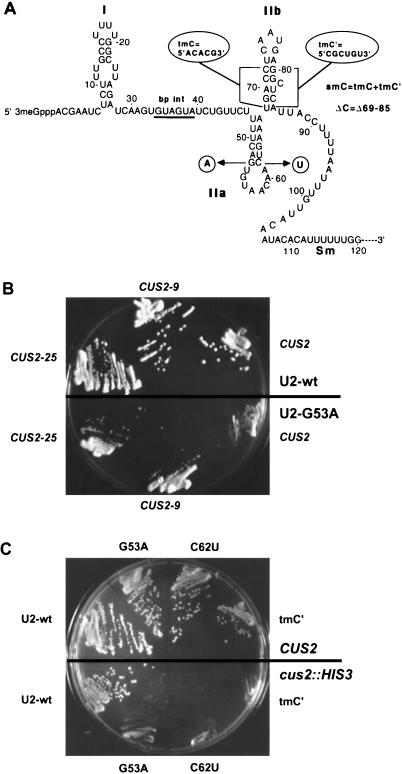
Three independent CUS2 mutations, CUS2-9, CUS2-25, and CUS2-62, were isolated in a genetic screen for suppressors of the cold-sensitive G53A mutation in stem IIa of U2 snRNA (67). All of the alleles are dominant to wild-type CUS2+, and none has a detectable growth defect in an otherwise wild-type haploid cell. We made a yeast genomic library from the CUS2-9 strain and selected for plasmids able to mediate dominant suppression of U2 G53A cold sensitivity. One plasmid uniformly reproduced suppression upon retransformation. Analysis of this plasmid revealed that DNA spanning open reading frame YNL286w could suppress the U2 RNA mutation. A single base change was observed between the cloned sequence from the CUS2-9 library plasmid and the yeast genome sequence (provided prior to release by Bart Scherens at the Munich Information Center for Protein Sequences [MIPS]).
To prove that the cloned YNL286w open reading frame corresponds to the genetically identified CUS2-9 suppressor, we tested for genetic linkage between CUS2-9 and cloned sequences near YNL286w (see Materials and Methods). No recombination was observed between YNL286w sequences and CUS2-9, indicating that they are tightly linked. Amplification and sequencing of YNL286w from our wild-type strain and from strains carrying the independently isolated CUS2 alleles showed that CUS2-9 and CUS2-62 carry a substitution of leucine 284 with phenylalanine (L284F) and that CUS2-25 carries a nearby substitution of aspartic acid 282 with asparagine (D282N). We conclude that YNL286w is the same as CUS2 and that the substitutions account for suppression of the U2 mutation. The CUS2 protein contains two unusual RRMs (16, 37, 53), but neither of the suppressor substitutions is within a conserved RRM structure (see below).
CUS2 suppression is restricted to mutations in U2 stem-loop IIa.
We tested the ability of CUS2-9 and CUS2-25 to suppress U2 mutations with lethal and conditional lethal phenotypes (Fig. 1A and B). In addition to U2 G53A, CUS2-9 and CUS2-25 partially suppress G53C, as well as a C-to-U mutation at position 62, the base-pairing partner of G53 in stem IIa (5) (Fig. 1A and data not shown). None of the cold-sensitive single mutations (U44A or U44G), lethal double mutations (U40A G43A, U40A G43C, U40G G43A, or U40G G43C), or a lethal triple mutation (43A 44C 45C) in the conserved nucleotides immediately downstream of the U2 intron branch point recognition region is suppressed by either CUS2 allele (data not shown), despite the rescue of these U2 mutations by other suppressors (67, 68). CUS2 suppressor alleles do not rescue the lethal tetraloop replacement of loop IIa (BTL) or a lethal triple mutation in stem IIa (tmA [6]) (data not shown), indicating that the CUS2 suppressors do not bypass the function of stem-loop IIa. Thus, the U2 allele specificity of CUS2 suppression is restricted to mutations in the G53-C62 base pair of stem IIa. This specificity is higher than that observed with the SF3b subunit suppressor CUS1-54 (67), suggesting that the action of CUS2 is directly related to U2 stem IIa function.
FIG. 1.
(A) Secondary structure model of the 5′ end of yeast U2 snRNA. Mutations G53A, C62U, A61Δ, tmC (68ACACG72), tmC′ (80CGCUGU85), smC (tmC plus tmC′), ΔC (deletion of nucleotides 69 to 85), and SMB (58AAC60 plus 99UUG101) discussed in the text are indicated. (B) Suppression of G53A and C62U by different CUS2 alleles. Strain BJ81 was transformed with either wild-type (wt) U2 (top half of plate) or the cold-sensitive G53A mutation (bottom half) and the indicated CUS2 allele on a plasmid. Strains were streaked onto glucose-containing medium and incubated at 18°C for 5 days. (C) Enhancement of U2 mutations by the cus2::HIS3 disruption. Strain D2 (top half of plate) or D2CUS2KO (bottom half) was transformed with a plasmid carrying the indicated U2 allele and streaked on 5-fluoroorotic acid at 26°C to shuffle out the wild-type U2 gene on pCH1122. Growth was for 4 days.
Loss of CUS2 enhances the phenotype of U2 RNA folding mutations.
To determine the phenotype of CUS2 loss of function, we inserted the HIS3 gene within one of the CUS2+ alleles in a diploid strain. No loss of spore viability and normal 2:2 segregation of His+:His− spores (and thus of the disrupted CUS2 gene) were observed upon tetrad dissection, indicating that the CUS2 gene is not essential for growth. We next tested whether the loss of CUS2 might enhance the growth defects caused by mutant U2 snRNA. Five of the many U2 mutations tested show enhanced growth defects when combined with the cus2::HIS3 allele (three examples are shown in Fig. 1C). Strikingly, two of the single mutations enhanced by the cus2::HIS3 disruption (G53A and C62U) are the same as those suppressed by CUS2-9 and CUS2-25. Since the CUS2 loss-of-function allele enhances the defects of U2 mutants and the CUS2-9 and CUS2-25 suppressor alleles are dominant, the suppressor alleles must be hyperactive, and the mechanism of suppression is related to the normal wild-type function of CUS2. A third single mutation, A61Δ (68), and two multiple U2 mutants, one which disrupts stem IIb (tmC′ [5]) and another which hyperstabilizes an alternative base-pairing interaction between loop IIa and the downstream region (smB [5]), also display enhanced growth defects in the absence of CUS2 (Fig. 1C and data not shown).
Both the G53A and C62U U2 mutations enhanced by the cus2::HIS3 knockout have demonstrated U2 RNA folding defects (70, 71). Although each of the component single mutations is enhanced (Fig. 1), the G53A C62U compensatory double mutant is not enhanced by the disruption of CUS2 (data not shown), indicating that restoration of the 53-62 base pair compensates for the loss of CUS2. Given that the primary defect in these mutations is RNA folding, this result argues that CUS2 is required when this region of U2 RNA cannot fold itself correctly. A similar result is obtained with the triple mutation tmC′ in the adjacent stem IIb (Fig. 1A). The tmC′ mutation causes slight cold sensitivity in a wild-type background, but the cold sensitivity is greatly enhanced in the absence of CUS2 (Fig. 1). The growth defect of U2-tmC′ in the absence of CUS2 must be due to deleterious folding by the tmC′ mutant RNA, because CUS2 is not required (i) if tmC′ is combined with its compensatory mutation tmC (smC mutant [6]) so that stem IIb is restored or (ii) if the nonessential stem IIb is deleted (as in the ΔC mutation) (reference 5, data not shown, and Fig. 1A). Because folding-defective mutations in stem-loops IIa and IIb are enhanced by the absence of CUS2, we suggest that the normal wild-type function of CUS2 is to help wild-type U2 RNA that is accidentally misfolded or to cycle U2 from an alternative conformation that it acquires during splicing to a form necessary for a new round of spliceosome assembly.
Mutations tested that do not show enhanced phenotypes with the cus2::HIS3 allele include those 3′ of the branch point interaction region (G32A; U40A or -G; C41G; U42A, -C, or -G; G43A; U44C; U45A, -C, or -G; C46A or -U; U47A; U28A and U44A; U28C and U44A [68]), single mutations in stem IIa (U49A; U54A or -G; A65G; A66U [5]) and in loop IIa (U56C; U56G; A57C; A57G; C59G [5]), multiple mutations in the stem-loop IIa and IIb regions (U49A and A66U; G53A and G100A; G53C and C62G; G53A and C62U; tmB and 58GUU60; tmC, smC, and ΔC [5]), as well as deletion of nucleotides 123 to 1081 (34).
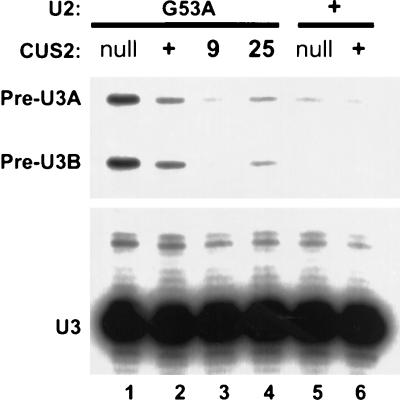
To determine whether CUS2 contributes to splicing efficiency, we examined levels of unspliced U3A and U3B RNA in strains carrying different combinations of U2 and CUS2 alleles (Fig. 2). Unspliced U3 levels are low in wild-type cells (lane 6) or in cells lacking CUS2 but containing wild-type U2 (lane 5). The U2 G53A mutation causes accumulation of unspliced U3A and U3B (lane 2) (70), and this accumulation is increased by the CUS2 disruption (lane 1). The CUS2 suppressor alleles (lanes 3 and 4) reduce the accumulation of unspliced U3A and U3B transcripts caused by the U2 mutation. These data indicate that CUS2 supports the function of the U2 snRNP in splicing and suppresses the growth defects caused by the U2 mutations through rescue of their splicing defects.
FIG. 2.
Splicing defects caused by U2-G53A are suppressed or enhanced by different CUS2 alleles. Yeast strains carrying the indicated alleles of U2 and CUS2 were grown at 30°C and shifted to 18°C for 4 h before extraction of RNA and primer extension with a labeled oligonucleotide complementary to U3 snRNA. Samples were normalized to contain the same amount of spliced U3 so that the level of unspliced U3 indicates splicing inhibition. Lane 1, U2-G53A with cus2::HIS3; lane 2, U2-G53A with CUS2+; lane 3, U2-G53A with CUS2-9; lane 4, U2-G53A with CUS2-25; lane 5, wild-type U2 with cus2::HIS3; lane 6, wild-type U2 with CUS2+.
CUS2 is associated with U2 and the PRP11 subunit of yeast SF3a.
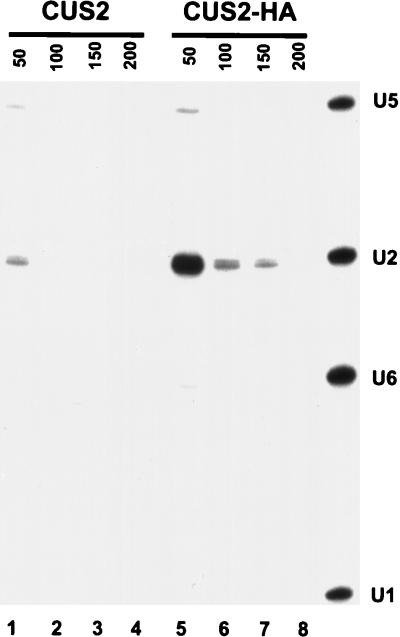
To determine whether the CUS2 protein is physically associated with snRNPs, we immunoprecipitated epitope-tagged CUS2 protein from splicing extracts and examined the immunoprecipitate for the presence of snRNAs (Fig. 3). After immunoprecipitation with the anti-HA antibody 12CA5, RNA was extracted and used for primer extension with a mixture of labeled oligonucleotides complementary to yeast U1, U2, U5, and U6 snRNAs. Of these, only U2 was detectably associated with the tagged CUS2 protein (lanes 5 to 7). The signal-to-noise ratio (as measured by the ratio of phosphorimager counts in cDNA bands derived from tagged versus untagged extracts) is highest for U2 at 50 mM NaCl (compare lanes 1 and 5). Salt concentrations above 100 mM reduce the yield of U2 RNA in the immunoprecipitate, but the signal-to-noise ratio remains high. In all cases, only a small fraction of the total U2 snRNA in the splicing extract is immunoprecipitated with epitope-tagged CUS2. This physical association argues that the function of CUS2 and the mechanism of CUS2 suppression are mediated through the binding of CUS2 to the U2 snRNP.
FIG. 3.
Coimmunoprecipitation of snRNA with tagged-CUS2. Extracts prepared from strains carrying untagged (lanes 1 to 4) or tagged (lanes 5 to 8) CUS2 protein were incubated with anti-HA antibody 12CA5 bound to PAS and washed with NET buffer containing 50 mM (lanes 1 and 5), 100 mM (lanes 2 and 6), 150 mM (lanes 3 and 7), and 200 mM (lanes 4 and 8) NaCl. RNA was extracted from the immunoprecipitate and used as the template for a reverse transcription reaction using a mixture of 5′-end-labeled oligonucleotides complementary to U1, U2, U5, and U6 snRNAs. The right-hand lane is a sample from a reverse transcription reaction using total yeast RNA as the template.
To test for associations between CUS2 and other proteins known to interact with U2, we used a two-hybrid system (28, 32). Of the splicing factors tested, only the combination of CUS2 and PRP11 generates a signal consistent with in vivo protein-protein interaction (Fig. 4). This signal is also observed with both CUS2-9 and CUS2-25 suppressor proteins (data not shown), indicating that the suppressor mutations do not greatly affect the interaction with PRP11 observed in this assay. These results indicate that CUS2 interacts directly or indirectly (via other yeast proteins) with the essential U2-associated splicing factor PRP11 (22), the yeast homolog of SF3a subunit SF3a66/SAP62 (12, 33, 39).
FIG. 4.
CUS2 interacts with PRP11 in vivo. Plasmids designed to express fusion proteins of URA3 with the GAL4 DNA binding domain (left) or CUS2 with the GAL4 DNA binding domain (right) were cotransformed into yeast Y190 with plasmids designed to express fusion proteins of the GAL4 activation domain with the proteins indicated at the left. After selection for both plasmids, independent transformants were arrayed on the plate and assayed for β-galactosidase by an X-Gal overlay method. Strains that express β-galactosidase turn blue (shown as black in this photograph).
CUS2 is the yeast protein most related to human Tat-SF1.
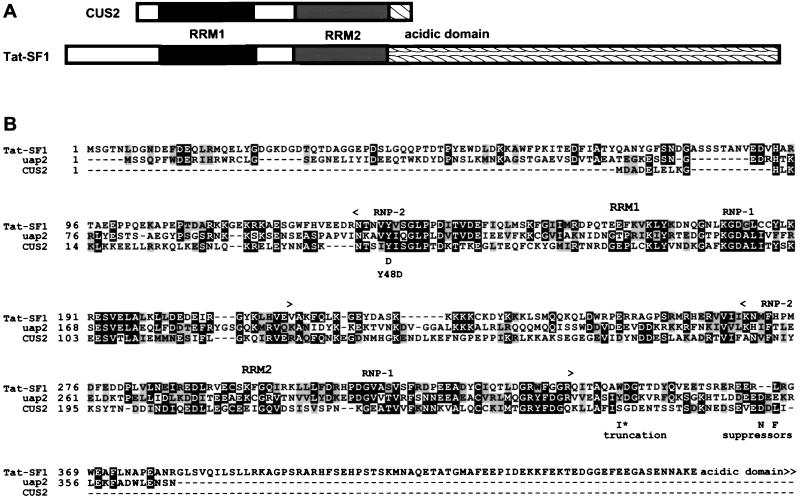
Sequence comparisons using the BLAST program (3, 4) indicate that CUS2 is a member of the RRM group of RNA binding proteins (16, 37, 53). CUS2 contains 42 amino-terminal residues followed by two copies of the RRM (Fig. 5), which are separated by a 46-amino-acid region rich in charged and polar residues. The protein terminates 25 amino acids beyond the second RRM, and it is within this acidic C-terminal domain (9 of 25 residues are D or E) that the suppressor proteins differ from wild-type CUS2. Although both CUS2 RRMs lack the solvent-exposed aromatic residues commonly found in the RNP-1 sequence of RRMs, they are true family members, as indicated by similarity elsewhere in the conserved RNP-1 and RNP-2 elements, conservation of a pattern of hydrophobic core residues, and potential to adopt the structural fold of the RRM domain (16, 37, 53).
FIG. 5.
CUS2 is structurally homologous to Tat-SF1. (A) Schematic comparison of CUS2 and Tat-SF1. Tat-SF1 is more than twice as large as CUS2, but most of this difference resides in the C-terminal acidic domain that is greatly shortened in CUS2. The unfilled regions share a charged nature but little sequence identity. (B) “Stitched” BLAST results generated by comparison of CUS2 with UAP2 and the first 420 residues of Tat-SF1. Each RRM is designated between angle brackets with RNP-1 and RNP-2 within each RRM indicated. Identical and similar amino acids are boxed in black and gray, respectively. The positions of the Y48D mutation, the C-terminal truncation (I∗), and CUS2-9 (L284F) and CUS2-25 (D282N) suppressor substitutions are shown.
The known human protein most related to CUS2 is Tat-SF1 (Fig. 5), a phosphoprotein present in fractions required for efficient Tat-specific, TAR-dependent in vitro transcription from the HIV promoter (72). In addition to Tat-SF1, a Schizosaccharomyces pombe protein called UAP2 was isolated through a two-hybrid screen interaction with the large subunit of S. pombe, U2AF (47). UAP2 has many of the same features as CUS2 and Tat-SF1, including the unusual RNP-1 sequences and the acidic C-terminal tail (47) (Fig. 5).
The first RRMs of CUS2 and Tat-SF1 are 37% identical and 59% similar, and this similarity extends beyond the conserved elements of the RRMs to include putative loop residues in which functional specificity elements may reside (16, 37, 53). The comparison between the second RRMs of each protein is less striking (30% identity and 56% similarity) but still significantly better than with other RRM proteins in the database. UAP2 is slightly more related to Tat-SF1 than to CUS2. The extensive acidic C-terminal domain containing multiple potential CKII phosphorylation sites in Tat-SF1 is reduced in CUS2 to a 25-amino-acid segment containing a single strong potential CKII site at serine 278 (S278). This residue is near the sites of the suppressor substitutions, suggesting a possible role for phosphorylation by CKII in regulation of CUS2 activity. CUS2 is by far the best match to Tat-SF1 encoded by the yeast genome (data not shown). Searches of the EST (expressed sequence tag) database with CUS2 reveal no vertebrate proteins more similar to CUS2 than Tat-SF1 (data not shown). Homology between CUS2 and Tat-SF1 is greater than that between the homologous pair of human and yeast U2-associated splicing factors PRP11 and SF3a66/SAP62 (12) or PRP21 and SF3a120/SAP114 (41).
Tat-SF1 interacts with SF3a66/SAP62, the human homolog of yeast PRP11.
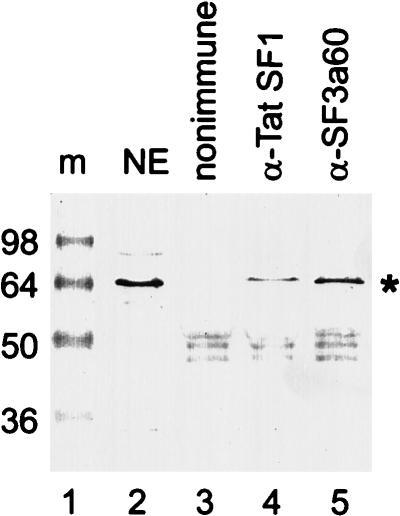
The two-hybrid interaction between CUS2 and PRP11 (Fig. 4) prompted us to test whether Tat-SF1 interacts with the human homolog of PRP11, SF3a66/SAP62 (12, 33, 39). SF3a66/SAP62 is a subunit of the heterotrimeric splicing factor SF3a, along with SF3a120/SAP114 and SF3a60/SAP61 (reviewed in reference 39). We obtained rabbit polyclonal anti-Tat-SF1 antibodies (gift of Q. Zhou and P. Sharp) (72), a polyclonal rabbit serum against the SF3a60/SAP61 subunit of SF3a, and MAb 66, against the SF3a66/SAP62 subunit of SF3a (both gifts of A. Kramer) (19, 40). Immunoprecipitates from HeLa cell nuclear extract were prepared by using either a nonimmune control rabbit serum (as a negative control) or rabbit anti-Tat-SF1 or rabbit anti-SF3a60/SAP61 (as a positive control) antibody bound to PAS. The presence of SF3a66/SAP62 in the different immunoprecipitates was monitored by Western blotting using MAb 66 (Fig. 6). As expected for the SF3a complex (19), anti-SF3a60 coimmunoprecipitates SF3a66/SAP62 (lane 5). Anti-Tat-SF1 also coimmunoprecipitates SF3a66/SAP62 (lane 4), indicating that a fraction of Tat-SF1 in HeLa cell nuclear extracts is associated with a fraction of splicing factor SF3a. This interaction is stable in 150 mM NaCl but is disrupted by 250 mM NaCl (data not shown), a salt concentration which disrupts the 17S U2 snRNP (10). The coimmunoprecipitation of Tat-SF1 with SF3a66/SAP62 parallels the two-hybrid interaction between CUS2 and PRP11 and suggests that Tat-SF1 is a functional as well as structural homolog of CUS2.
FIG. 6.
Coimmunoprecipitation of SF3a66 with anti-Tat-SF1 antibody; Western blot of protein preparations from HeLa nuclear extract probed with anti-SF3a66 MAb. NaCl washes (150 mM) of nuclear extract were incubated with PAS bound with preimmune serum (lane 3), anti-Tat-SF1 antibody (lane 4), or anti-SF3a60 antibody (lane 5). Samples (with Sepharose) were boiled and separated on an SDS–10% polyacrylamide gel. Lane 2 is total nuclear extract (NE) not purified via PAS. Positions of molecular mass standards (m) are indicated in kilodaltons.
CUS2 rescue of U2 G53A is mediated by both the C-terminal tail containing the suppressor mutations and an RNA binding activity supported by the first RRM.
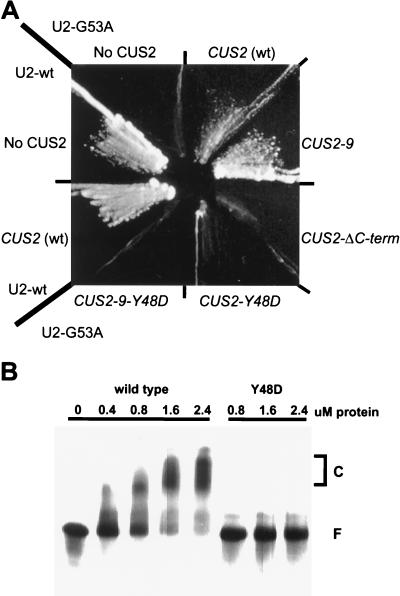
To test the functional roles of RRM1 as well as other parts of CUS2 in vivo, we assessed the ability of CUS2 mutants to rescue the enhancement of U2 G53A caused by the cus2::HIS3 disruption. An HA-His6-tagged wild-type CUS2-encoding gene and one encoding a CUS2 derivative in which a tyrosine at a conserved position of the β1 strand in RNP-2 of the first RRM is changed to aspartate (CUS2-Y48D) were introduced into a strain carrying the U2 G53A mutation and the cus2::HIS3 disruption (Fig. 7A). Although the strain with the wild-type CUS2 gene grows well, the CUS2-Y48D mutation fails to rescue the U2 mutation. Similarly, when the Y48D mutation is introduced into the CUS2-9 (CUS2-9-Y48D) gene, CUS2-9 protein can no longer suppress the U2 G53A mutation. These results indicate that RRM1 is required for normal CUS2 function as well as for suppression in vivo. Western blots of extracts from these yeast strains using the antibody against the tag reveal that the mutant and wild-type proteins are present in the same amounts (data not shown).
FIG. 7.
CUS2 rescue of U2 G53A requires the C-terminal tail where the suppressor mutations are located and an RNA binding activity supported by RRM1. (A) Enhancement of U2 G53A mutation by mutant CUS2 alleles. Strain RP01 was transformed with plasmids expressing U2 G53A and the indicated CUS2 mutant alleles. Strains were streaked onto glucose-containing medium and incubated at 18°C for 5 days. wt, wild type; term, terminus. (B) His6-CUS2 binds RNA in vitro. U2 RNA (20 fmol) and CUS2 (wild type) or Y48D protein were mixed, incubated in 10 μl, and then run on a native polyacrylamide gel. Lanes 1 to 5, U2 plus CUS2; lanes 6 to 8, U2 plus Y48D. C and F, RNA-protein complex and free U2 RNA, respectively.
To obtain evidence for RNA binding by CUS2 protein, we produced and partially purified a tagged CUS2 protein from E. coli and tested it for the ability to bind different RNAs, including U2. We found that CUS2 protein can bind numerous RNAs of different sequences with an apparent Kd of about 800 nM (Fig. 7B and data not shown). CUS2-Y48D protein purified in the same way no longer binds RNA (apparent Kd of >2.4 μM), suggesting that RRM1 is required for in vitro RNA binding activity (Fig. 7B). Because the Y48D substitution blocks both CUS2 activity in vivo (Fig. 7A) and RNA binding in vitro (Fig. 7B), we suggest that CUS2 function in vivo requires the RNA binding activity of RRM1.
We also tested the C-terminal acidic region containing the suppressor substitutions and the potential CKII phosphorylation site (Fig. 5 and 7A). Expression of a C-terminal deletion of CUS2 also fails to rescue the enhancement of U2 G53A cold sensitivity caused by the CUS2 disruption, indicating that this part of the protein is also important for CUS2 function. Taken together, these results suggest that for CUS2 protein to assist U2 G53A snRNA in splicing, both the RRM1-mediated RNA binding activity and the C-terminal tail containing the potential CKII phosphorylation site must be intact.
DISCUSSION
Specific functional (Fig. 1, 2, and 7A) and physical (Fig. 3 and 7B) interactions between CUS2 and mutant or wild-type U2 RNAs, and a two-hybrid interaction between CUS2 and PRP11 (Fig. 4), provide strong evidence that CUS2 is a splicing factor. CUS2 is similar to the yeast U2AF homolog MUD2 in that neither is essential in S. cerevisiae (reference 1 and Fig. 1). In contrast, the S. pombe homolog of CUS2, UAP2, is essential (47). In this respect, CUS2 is also similar to MUD2 in that U2AF homologs in S. pombe and other species are essential (36, 55). The human structural homolog of CUS2, Tat-SF1 (72), shares a parallel function with its yeast homolog: association with the conserved U2-associated splicing factor SF3a66 (Fig. 6). These findings suggest that Tat-SF1 and CUS2 may have parallel roles in splicing and that the role of Tat-SF1 in splicing may also be essential.
How does CUS2 function in splicing?
The function of CUS2 is especially critical under conditions where correct U2 RNA folding may be limiting due to U2 mutations (Fig. 1 and 2) (70, 71). Although we do not observe any splicing defects in the absence of CUS2 when wild-type U2 is present, it is conceivable that under environmental conditions beyond those tested in the current study, a requirement for CUS2 may become apparent, if the rate of wild-type U2 refolding becomes limiting. CUS2 function places it with the set of proteins that include the subunits of the conserved multimeric splicing factors SF3a and SF3b, which must associate with the U2 snRNP prior to its recruitment into splicing complexes (18, 29). A single mutation in RRM1 of CUS2 abolishes RNA binding activity in vitro (Fig. 7B) and function in vivo (Fig. 7A). This finding suggests that direct interaction with RNA is an integral part of CUS2 function in vivo. The U2 allele specificity displayed by CUS2 and its derivatives suggests that this interaction is likely to be in the region around stem IIa (Fig. 1), perhaps helping form and maintain this essential region of U2 snRNA during the activation of U2 snRNP for spliceosome assembly (70). Deleting the C-terminal domain containing the potential CKII phosphorylation site also reduces CUS2 function with U2 G53A (Fig. 7A). The importance of the C-terminal region is underscored by the location of both the suppressor mutations, which appear to increase the activity of the protein. The proximity of the suppressor mutations to the phosphorylation site may mean that these amino acid substitutions affect protein activity by influencing the phosphorylation state or function of one of the phosphorylated forms of the molecule.
CUS2 protein interacts with SF3a subunit PRP11 (Fig. 4); thus, CUS2 might also be involved in recruiting SF3a to the U2 snRNP. The CUS2 suppressor proteins also associate with PRP11 in the two-hybrid system, suggesting that suppression does not involve an alteration in the interaction of CUS2 with PRP11. Preliminary results (54a) indicate that CUS2 is required for suppression of certain U2 alleles by CUS1-54, a suppressor allele of the yeast homolog of the SF3b subunit SF3b145/SAP145 (29, 67). Taken together with order-of-assembly data from mammalian studies (18), these data suggest a working model (Fig. 8) in which CUS2 aids in the formation of both correct U2 snRNA secondary structure and recruitment of SF3b and then SF3a protein components to the U2 snRNP for spliceosome assembly (18). Alternatively, CUS2 may recruit U2 snRNP already containing SF3a to the assembling spliceosome, or to some step after the binding of the U2 snRNP to the spliceosome. Although we did not observe a two-hybrid interaction between the yeast U2AF homolog MUD2 and CUS2, a parallel interaction is observed between U2AF and the CUS2 homolog UAP2 in S. pombe (47) and could mean that CUS2 helps mediate U2AF-dependent association of the U2 snRNP with the pre-mRNA. Further experiments will be necessary to test this hypothesis.
FIG. 8.
Model for CUS2 action in preparing U2 snRNP for recruitment to the spliceosome. The U2 allele specificity of CUS2 suppression and enhancement suggests a close interaction with stem IIa, as shown. Newly synthesized U2, U2 snRNA emerging from the splicing pathway, or otherwise misfolded U2 interacts with CUS2 and is refolded. ySF3b and ySF3a are the S. cerevisiae protein complexes homologous to the human splicing factors SF3b and SF3a (39). Protein-protein interactions (35, 39, 43) (Fig. 4) are indicated by small gray boxes. The interaction between CUS2 and PRP11 may assist the recruitment of ySF3a to the U2 snRNP as indicated. The ySF3b subunit labeled “155?” is a yeast open reading frame homologous to a vertebrate SF3b subunit suggested to be SF3b155/SAP155 (58a). Once completely assembled, the U2 snRNP can bind to the commitment complex. Given the interaction between the S. pombe homologs of CUS2 and U2AF (47), CUS2 could have a role in U2 snRNP binding to the commitment complex through MUD2 (1).
The observation that the absence of CUS2 enhances U2-RNA misfolding mutations implies that CUS2 may help overcome the effects of RNA misfolding (Fig. 1). Misfolding mutants are defined as those that stabilize an alternative folding of the RNA and can be suppressed by second-base changes that destabilize this alternate structure (5, 70, 71). We found CUS2 to be active on stem IIa and stem IIb misfolding mutants (Fig. 1) but observed no effect on other misfolding mutants elsewhere in U2 snRNA (data not shown). This suggests that CUS2 is specifically required to help fold stems IIa and IIb of U2 (Fig. 8). CUS2 may be involved in helping fold both new and recycled U2 RNA. Here, CUS2 would interact with newly transcribed U2 as well as U2 snRNPs which have been released from the spliceosome to effect the intramolecular rearrangements required for a new round of splicing (63). Several folded forms of wild-type U2 snRNA appear to exist in the cell (71). Chemical cross-links can be formed in human U2, which suggests the existence of base pairing between the loop of stem IIa and the downstream conserved complementarity (26). Oligonucleotides directed against stem-loop IIa allow spliceosome assembly and the first step of splicing but block the second step of splicing (9), suggesting that U2 undergoes intramolecular changes between the first and second transesterifications. CUS2 may be involved in the refolding of several or all of these U2 snRNA forms.
General RNA binding proteins of a variety of types have been shown to accelerate the reaction rates of different ribozymes (15, 24, 64). In these cases, it is thought that general strand displacement or strand exchange activities of these proteins assist the RNA in adopting a functional structure. If CUS2 acts in a similar way, it must have some specificity for a part of U2 RNA, and this activity must require RRM1 (Fig. 7). Activity could be due to enhanced rates of folding or stabilization of a final folded RNA form. Thus far, our RNA binding studies do not show that CUS2 binds tightly to U2 with high specificity (Fig. 7b and data not shown); however, a specific RNA rearrangement activity involving a high off rate would not have been detected by these experiments. An experiment to investigate the folding of the U2 G53A mutant in the presence of CUS2-9 did not detect a change in the steady-state levels of the different folded forms in vivo (data not shown); thus, we have no evidence that CUS2 protein greatly stabilizes a particular folded form of U2 by complexing with it.
What is the functional relationship between CUS2 and Tat-SF1?
A human homolog of CUS2, Tat-SF1 (Fig. 5), has been identified as a host factor that assists Tat-TAR-dependent transcription enhancement from the HIV promoter in vitro (72). In addition, UAP2, the S. pombe homolog of CUS2/Tat-SF1, interacts with both subunits of the splicing factor U2AF (47). We show that Tat-SF1 associates with SF3a66/SAP62 (Fig. 6), a subunit of SF3a and an integral component of the spliceosome (39), suggesting that Tat-SF1 also has a role in splicing. We do not yet have evidence that Tat-SF1 or UAP2 can replace CUS2 in S. cerevisiae.
Like CUS2, both Tat-SF1 and UAP2 lack the characteristic aromatic amino acid residues at positions 3 and 5 in the RNP-1 sequences of both RRMs (Fig. 5). Atypical RNP-1 sequences of this type have also been found in several other RNA binding proteins which otherwise fit the RNP consensus motif (16). Those containing acidic substitutions at the normally aromatic position 3 of RNP-1 include EWS (Ewing’s sarcoma [27]), FUS/TLS (25) (also known as hnRNP P2 [20]), and hTAFII68 (14). Both FUS/TLS and EWS are preoncoproteins and exhibit extensive sequence and functional homology with hTAFII68, which is part of the multiprotein transcription complex TFIID (14). The functional significance of these amino acid substitutions is not clear, but it has been suggested that they represent a new subclass of the RRM family (14, 25). Interestingly, another spliceosomal protein, human 69KD, which predominantly associates with the U1 snRNP, is also a member of this family, suggesting that these types of proteins may be required in a wide range of RNA-protein binding activities (31).
Could CUS2 be active in the elongation phase of transcription? CUS2 does not influence U2 RNA levels (data not shown) and thus does not suppress U2 mutations by acting as a U2 transcription factor. CUS2 function in splicing could stem from a general role in determining the fate of nascent pre-mRNA transcripts with respect to their association with U2 snRNP. In this model, CUS2 would act during cotranscriptional spliceosome assembly (52, 69). The weight of our data clearly supports a direct role for CUS2 in splicing. It seems possible that Tat-SF1’s role in the uninfected human cell is like that of CUS2: to help fold U2 snRNA and assemble SF3a and SF3b into the 17S U2 snRNP. It may be that upon HIV infection, some Tat-SF1 activity in U2 snRNP assembly is diverted to Tat-TAR complex assembly for stimulating elongation of viral transcripts.
ACKNOWLEDGMENTS
We are grateful to Bart Scherens, Laboratoire de Microbiologie, Universite Libre de Bruxelles, and his colleagues at MIPS for providing the sequence of the region of the yeast genome containing CUS2 prior to its release. Thanks also go to Michelle Haynes Pauling for tetrad dissection, Pierre Legrain and Michael Rosbash and their colleagues for providing plasmids and yeast strains, Al Zahler for HeLa nuclear extract, and Angela Kramer, Qiang Zhou, and Phil Sharp for antibodies. We are grateful to Roland Nagel, Marc Spingola, and Michelle Haynes Pauling for critical reading of the manuscript.
This work was supported by grant GM47408 from the National Institutes of Health to M.A. R.P. was supported by an American Cancer Society postdoctoral fellowship.
REFERENCES
- 1.Abovich N, Liao X C, Rosbash M. The yeast MUD2 protein: an interaction with PRP11 defines a bridge between commitment complexes and U2 snRNP addition. Genes Dev. 1994;8:843–854. doi: 10.1101/gad.8.7.843. [DOI] [PubMed] [Google Scholar]
- 2.Abovich N, Rosbash M. Cross-intron bridging interactions in the yeast commitment complex are conserved in mammals. Cell. 1997;89:403–412. doi: 10.1016/s0092-8674(00)80221-4. [DOI] [PubMed] [Google Scholar]
- 3.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 4.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ares M, Jr, Igel A H. Lethal and temperature-sensitive mutations and their suppressors identify an essential structural element in U2 small nuclear RNA. Genes Dev. 1990;4:2132–2145. doi: 10.1101/gad.4.12a.2132. [DOI] [PubMed] [Google Scholar]
- 6.Ares M, Jr, Igel A H. Mutations define essential and nonessential U2 RNA structures. Mol Biol Rep. 1990;14:131–132. doi: 10.1007/BF00360444. [DOI] [PubMed] [Google Scholar]
- 7.Ares M, Jr, Weiser B. Rearrangement of snRNA structure during assembly and function of the spliceosome. Prog Nucleic Acid Res Mol Biol. 1995;50:131–159. doi: 10.1016/s0079-6603(08)60813-2. [DOI] [PubMed] [Google Scholar]
- 8.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Greene/John Wiley & Sons; 1987. [Google Scholar]
- 9.Barabino S M, Sproat B S, Lamond A I. Antisense probes targeted to an internal domain in U2 snRNP specifically inhibit the second step of pre-mRNA splicing. Nucleic Acids Res. 1992;20:4457–4464. doi: 10.1093/nar/20.17.4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Behrens S E, Galisson F, Legrain P, Luhrmann R. Evidence that the 60-kDa protein of 17S U2 small nuclear ribonucleoprotein is immunologically and functionally related to the yeast PRP9 splicing factor and is required for the efficient formation of prespliceosomes. Proc Natl Acad Sci USA. 1993;90:8229–8233. doi: 10.1073/pnas.90.17.8229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bennett M, Michaud S, Kingston J, Reed R. Protein components specifically associated with prespliceosome and spliceosome complexes. Genes Dev. 1992;6:1986–2000. doi: 10.1101/gad.6.10.1986. [DOI] [PubMed] [Google Scholar]
- 12.Bennett M, Reed R. Correspondence between a mammalian spliceosome component and an essential yeast splicing factor. Science. 1993;262:105–108. doi: 10.1126/science.8211113. [DOI] [PubMed] [Google Scholar]
- 13.Berglund J A, Chua K, Abovich N, Reed R, Rosbash M. The splicing factor BBP interacts specifically with the pre-mRNA branchpoint sequence UACUAAC. Cell. 1997;89:781–787. doi: 10.1016/s0092-8674(00)80261-5. [DOI] [PubMed] [Google Scholar]
- 14.Bertolotti A, Lutz Y, Heard D J, Chambon P, Tora L. hTAF(II)68, a novel RNA/ssDNA-binding protein with homology to the pro-oncoproteins TLS/FUS and EWS is associated with both TFIID and RNA polymerase II. EMBO J. 1996;15:5022–5031. [PMC free article] [PubMed] [Google Scholar]
- 15.Bertrand E L, Rossi J J. Facilitation of hammerhead ribozyme catalysis by the nucleocapsid protein of HIV-1 and the heterogeneous nuclear ribonucleoprotein A1. EMBO J. 1994;13:2904–2912. doi: 10.1002/j.1460-2075.1994.tb06585.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Birney E, Kumar S, Krainer A R. Analysis of the RNA-recognition motif and RS and RGG domains: conservation in metazoan pre-mRNA splicing factors. Nucleic Acids Res. 1993;21:5803–5816. doi: 10.1093/nar/21.25.5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boeke J D, Trueheart J, Natsoulis G, Fink G R. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- 18.Brosi R, Groning K, Behrens S E, Luhrmann R, Kramer A. Interaction of mammalian splicing factor SF3a with U2 snRNP and relation of its 60-kD subunit to yeast PRP9. Science. 1993;262:102–105. doi: 10.1126/science.8211112. [DOI] [PubMed] [Google Scholar]
- 19.Brosi R, Hauri H P, Kramer A. Separation of splicing factor SF3 into two components and purification of SF3a activity. J Biol Chem. 1993;268:17640–17646. [PubMed] [Google Scholar]
- 20.Calvio C, Neubauer G, Mann M, Lamond A I. Identification of hnRNP P2 as TLS/FUS using electrospray mass spectrometry. RNA. 1995;1:724–733. [PMC free article] [PubMed] [Google Scholar]
- 21.Champion-Arnaud P, Reed R. The prespliceosome components SAP 49 and SAP 145 interact in a complex implicated in tethering U2 snRNP to the branch site. Genes Dev. 1994;8:1974–1983. doi: 10.1101/gad.8.16.1974. [DOI] [PubMed] [Google Scholar]
- 22.Chang T H, Clark M W, Lustig A J, Cusick M E, Abelson J. RNA11 protein is associated with the yeast spliceosome and is localized in the periphery of the cell nucleus. Mol Cell Biol. 1988;8:2379–2393. doi: 10.1128/mcb.8.6.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chiara M D, Gozani O, Bennett M, Champion-Arnaud P, Palandjian L, Reed R. Identification of proteins that interact with exon sequences, splice sites, and the branchpoint sequence during each stage of spliceosome assembly. Mol Cell Biol. 1996;16:3317–3326. doi: 10.1128/mcb.16.7.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coetzee T, Herschlag D, Belfort M. Escherichia coli proteins, including ribosomal protein S12, facilitate in vitro splicing of phage T4 introns by acting as RNA chaperones. Genes Dev. 1994;8:1575–1588. doi: 10.1101/gad.8.13.1575. [DOI] [PubMed] [Google Scholar]
- 25.Crozat A, Aman P, Mandahl N, Ron D. Fusion of CHOP to a novel RNA-binding protein in human myxoid liposarcoma. Nature. 1993;363:640–646. doi: 10.1038/363640a0. [DOI] [PubMed] [Google Scholar]
- 26.Datta B, Weiner A M. Cross-linking of U2 snRNA using nitrogen mustard. Evidence for higher order structure. J Biol Chem. 1992;267:4497–4502. [PubMed] [Google Scholar]
- 27.Delattre O, Zucman J, Plougastel B, Desmaze C, Melot T, Peter M, Kovar H, Joubert I, de Jong P, Rouleau G, et al. Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature. 1992;359:162–165. doi: 10.1038/359162a0. [DOI] [PubMed] [Google Scholar]
- 28.Durfee T, Becherer K, Chen P L, Yeh S H, Yang Y, Kilburn A E, Lee W H, Elledge S J. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- 29.Gozani O, Feld R, Reed R. Evidence that sequence-independent binding of highly conserved U2 snRNP proteins upstream of the branch site is required for assembly of spliceosomal complex A. Genes Dev. 1996;10:233–243. doi: 10.1101/gad.10.2.233. [DOI] [PubMed] [Google Scholar]
- 30.Guthrie C, Fink G. Guide to yeast genetics and molecular biology. San Diego, Calif: Academic Press; 1991. [Google Scholar]
- 31.Hackl W, Luhrmann R. Molecular cloning and subcellular localisation of the snRNP-associated protein 69KD, a structural homologue of the proto-oncoproteins TLS and EWS with RNA and DNA-binding properties. J Mol Biol. 1996;264:843–851. doi: 10.1006/jmbi.1996.0681. [DOI] [PubMed] [Google Scholar]
- 32.Harper J W, Adami G R, Wei N, Keyomarsi K, Elledge S J. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 33.Hodges P E, Beggs J D. RNA splicing. U2 fulfils a commitment. Curr Biol. 1994;4:264–267. doi: 10.1016/s0960-9822(00)00061-0. [DOI] [PubMed] [Google Scholar]
- 34.Igel A H, Ares M., Jr Internal sequences that distinguish yeast from metazoan U2 snRNA are unnecessary for pre-mRNA splicing. Nature. 1988;334:450–453. doi: 10.1038/334450a0. [DOI] [PubMed] [Google Scholar]
- 35.Igel H, Wells S, Perriman R, Ares M., Jr Conservation of structure and subunit interactions in yeast homologues of splicing factor 3b (SF3b) subunits. RNA. 1998;4:1–10. [PMC free article] [PubMed] [Google Scholar]
- 36.Kanaar R, Roche S E, Beall E L, Green M R, Rio D C. The conserved pre-mRNA splicing factor U2AF from Drosophila: requirement for viability. Science. 1993;262:569–573. doi: 10.1126/science.7692602. [DOI] [PubMed] [Google Scholar]
- 37.Kenan D J, Query C C, Keene J D. RNA recognition: towards identifying determinants of specificity. Trends Biochem Sci. 1991;16:214–220. doi: 10.1016/0968-0004(91)90088-d. [DOI] [PubMed] [Google Scholar]
- 38.Kramer A. Purification of splicing factor SF1, a heat-stable protein that functions in the assembly of a presplicing complex. Mol Cell Biol. 1992;12:4545–4552. doi: 10.1128/mcb.12.10.4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kramer A. The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu Rev Biochem. 1996;65:367–409. doi: 10.1146/annurev.bi.65.070196.002055. [DOI] [PubMed] [Google Scholar]
- 40.Kramer A, Legrain P, Mulhauser F, Groning K, Brosi R, Bilbe G. Splicing factor SF3a60 is the mammalian homologue of PRP9 of S. cerevisiae: the conserved zinc finger-like motif is functionally exchangeable in vivo. Nucleic Acids Res. 1994;22:5223–5228. doi: 10.1093/nar/22.24.5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kramer A, Mulhauser F, Wersig C, Groning K, Bilbe G. Mammalian splicing factor SF3a120 represents a new member of the SURP family of proteins and is homologous to the essential splicing factor PRP21p of Saccharomyces cerevisiae. RNA. 1995;1:260–272. [PMC free article] [PubMed] [Google Scholar]
- 42.Kunkel T A, Roberts J D, Zakour R A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 43.Legrain P, Chapon C. Interaction between PRP11 and SPP91 yeast splicing factors and characterization of a PRP9-PRP11-SPP91 complex. Science. 1993;262:108–110. doi: 10.1126/science.8211114. [DOI] [PubMed] [Google Scholar]
- 44.Legrain P, Dokhelar M C, Transy C. Detection of protein-protein interactions using different vectors in the two-hybrid system. Nucleic Acids Res. 1994;22:3241–3242. doi: 10.1093/nar/22.15.3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Madhani H D, Guthrie C. Dynamic RNA-RNA interactions in the spliceosome. Annu Rev Genet. 1994;28:1–26. doi: 10.1146/annurev.ge.28.120194.000245. [DOI] [PubMed] [Google Scholar]
- 46.Marshak D R, Carroll D. Synthetic peptide substrates for casein kinase II. Methods Enzymol. 1991;200:134–156. doi: 10.1016/0076-6879(91)00135-j. [DOI] [PubMed] [Google Scholar]
- 47.McKinney R, Wentz-Hunter K, Schmidt H, Potashkin J. Molecular characterization of a novel fission yeast gene spUAP2 that interacts with the splicing factor spU2AF59. Curr Genet. 1997;32:323–330. doi: 10.1007/s002940050284. [DOI] [PubMed] [Google Scholar]
- 48.Michaud S, Reed R. An ATP-independent complex commits pre-mRNA to the mammalian spliceosome assembly pathway. Genes Dev. 1991;5:2534–2546. doi: 10.1101/gad.5.12b.2534. [DOI] [PubMed] [Google Scholar]
- 49.Miraglia L, Seiwert S, Igel A H, Ares M., Jr Limited functional equivalence of phylogenetic variation in small nuclear RNA: yeast U2 RNA with altered branchpoint complementarity inhibits splicing and produces a dominant lethal phenotype. Proc Natl Acad Sci USA. 1991;88:7061–7065. doi: 10.1073/pnas.88.16.7061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moore M J, Query C C, Sharp P A. Splicing of precursors to mRNA in the spliceosome. In: Gestland R, Atkins J, editors. The RNA world. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1993. pp. 303–357. [Google Scholar]
- 51.Moore M J, Sharp P A. Evidence for two active sites in the spliceosome provided by stereochemistry of pre-mRNA splicing. Nature. 1993;365:364–368. doi: 10.1038/365364a0. [DOI] [PubMed] [Google Scholar]
- 52.Mortillaro M J, Blencowe B J, Wei X, Nakayasu H, Du L, Warren S L, Sharp P A, Berezney R. A hyperphosphorylated form of the large subunit of RNA polymerase II is associated with splicing complexes and the nuclear matrix. Proc Natl Acad Sci USA. 1996;93:8253–8257. doi: 10.1073/pnas.93.16.8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nagai K, Oubridge C, Ito N, Avis J, Evans P. The RNP domain: a sequence-specific RNA-binding domain involved in processing and transport of RNA. Trends Biochem Sci. 1995;20:235–240. doi: 10.1016/s0968-0004(00)89024-6. [DOI] [PubMed] [Google Scholar]
- 54.Patterson B, Guthrie C. A U-rich tract enhances usage of an alternative 3′ splice site in yeast. Cell. 1991;64:181–187. doi: 10.1016/0092-8674(91)90219-o. [DOI] [PubMed] [Google Scholar]
- 54a.Perriman, R., T. Hoblitzell, and M. Ares. Unpublished data.
- 55.Potashkin J, Naik K, Wentz-Hunter K. U2AF homolog required for splicing in vivo. Science. 1993;262:573–575. doi: 10.1126/science.8211184. [DOI] [PubMed] [Google Scholar]
- 56.Raghunathan P L, Guthrie C. A spliceosome recycling factor that reanneals U4 and U6 small nuclear ribonucleoprotein particles. Science. 1998;279:857–860. doi: 10.1126/science.279.5352.857. [DOI] [PubMed] [Google Scholar]
- 57.Rothstein R. Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol. 1991;194:281–301. doi: 10.1016/0076-6879(91)94022-5. [DOI] [PubMed] [Google Scholar]
- 58.Ruby S W, Chang T H, Abelson J. Four yeast spliceosomal proteins (PRP5, PRP9, PRP11, and PRP21) interact to promote U2 snRNP binding to pre-mRNA. Genes Dev. 1993;7:1909–1925. doi: 10.1101/gad.7.10.1909. [DOI] [PubMed] [Google Scholar]
- 58a.Schmidt-Zachmann M, Knecht S, Kramer A. Molecular characterization of a novel, widespread nuclear protein that colocalizes with spliceosome components. Mol Biol Cell. 1998;9:143–160. doi: 10.1091/mbc.9.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- 60.Sherman F, Hicks J. Micromanipulation and dissection of asci. Methods Enzymol. 1991;194:21–37. doi: 10.1016/0076-6879(91)94005-w. [DOI] [PubMed] [Google Scholar]
- 61.Shuster E O, Guthrie C. Two conserved domains of yeast U2 snRNA are separated by 945 nonessential nucleotides. Cell. 1988;55:41–48. doi: 10.1016/0092-8674(88)90007-4. [DOI] [PubMed] [Google Scholar]
- 62.Smith C W, Chu T T, Nadal-Ginard B. Scanning and competition between AGs are involved in 3′ splice site selection in mammalian introns. Mol Cell Biol. 1993;13:4939–4952. doi: 10.1128/mcb.13.8.4939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Staley J P, Guthrie C. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell. 1998;92:315–326. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- 64.Tsuchihashi Z, Khosla M, Herschlag D. Protein enhancement of hammerhead ribozyme catalysis. Science. 1993;262:99–102. doi: 10.1126/science.7692597. [DOI] [PubMed] [Google Scholar]
- 65.Umen J G, Guthrie C. The second catalytic step of pre-mRNA splicing. RNA. 1995;1:869–885. [PMC free article] [PubMed] [Google Scholar]
- 66.Wells S E, Ares M., Jr Interactions between highly conserved U2 small nuclear RNA structures and Prp5p, Prp9p, Prp11p, and Prp21p proteins are required to ensure integrity of the U2 small nuclear ribonucleoprotein in Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:6337–6349. doi: 10.1128/mcb.14.9.6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wells S E, Neville M, Haynes M, Wang J, Igel H, Ares M., Jr CUS1, a suppressor of cold-sensitive U2 snRNA mutations, is a novel yeast splicing factor homologous to human SAP 145. Genes Dev. 1996;10:220–232. doi: 10.1101/gad.10.2.220. [DOI] [PubMed] [Google Scholar]
- 68.Yan D, Ares M., Jr Invariant U2 RNA sequences bordering the branchpoint recognition region are essential for interaction with yeast SF3a and SF3b subunits. Mol Cell Biol. 1996;16:818–828. doi: 10.1128/mcb.16.3.818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yuryev A, Patturajan M, Litingtung Y, Joshi R V, Gentile C, Gebara M, Corden J L. The C-terminal domain of the largest subunit of RNA polymerase II interacts with a novel set of serine/arginine-rich proteins. Proc Natl Acad Sci USA. 1996;93:6975–6980. doi: 10.1073/pnas.93.14.6975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zavanelli M I, Ares M., Jr Efficient association of U2 snRNPs with pre-mRNA requires an essential U2 RNA structural element. Genes Dev. 1991;5:2521–2533. doi: 10.1101/gad.5.12b.2521. [DOI] [PubMed] [Google Scholar]
- 71.Zavanelli M I, Britton J S, Igel A H, Ares M., Jr Mutations in an essential U2 small nuclear RNA structure cause cold-sensitive U2 small nuclear ribonucleoprotein function by favoring competing alternative U2 RNA structures. Mol Cell Biol. 1994;14:1689–1697. doi: 10.1128/mcb.14.3.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhou Q, Sharp P A. Tat-SF1: cofactor for stimulation of transcriptional elongation by HIV-1 Tat. Science. 1996;274:605–610. doi: 10.1126/science.274.5287.605. [DOI] [PubMed] [Google Scholar]