Abstract
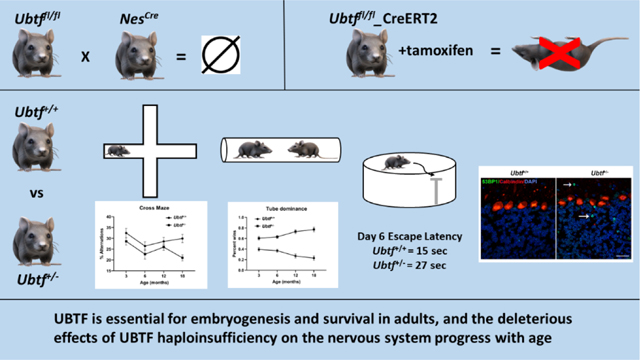
The UBTF E210K neuroregression syndrome is caused by de novo dominant mutations in UBTF (NM_014233.3:c.628G>A, p.Glu210Lys). In humans, onset is typically at 2.5 to 3 years and characterized by slow progression of global motor, cognitive and behavioral dysfunction. Other potentially pathogenic UBTF variants have been reported in humans with severe neurological disease and it remains undetermined if the UBTF E210K mutation operates via gain- and/or loss-of-function. Here we examine the behavioral, cognitive, motor, and molecular effects of Ubtf knockout and knockdown in mice as a means of gauging the role of loss-of-function in humans. Ubtf+/− mice show progression of behavioral (dominance tube), cognitive (cross maze), and mild motor abnormalities from 3 to 18 months. At 18 months, Ubtf+/− mice had more slips on a raised 9-mm round beam task, shorter latencies to fall on the accelerated rotarod, reduced open field vertical and jump counts, and significant deficits in spatial learning and memory. Via crosses to Nestin-Cre (NesCre) mice we found that homozygous Ubtf deletion limited to the central nervous system was embryonic lethal. Tamoxifen-induced homozygous knockdown of Ubtf in adult mice with the Cre-ERT2 system was associated with precipitous deterioration in neurological functioning. At the molecular level, 18-month-old Ubtf+/− mice showed mild increases in cerebellar 53BP1 immunoreactivity. These findings show that UBTF is essential for embryogenesis and survival in adults, and the deleterious effects of UBTF haploinsufficiency progress with age. Loss-of-function mechanisms may contribute, in part, to the human UBTF E210K neuroregression syndrome.
Keywords: neuroregression, nucleolus, rRNA, UBTF, mouse
Graphical abstract
1. Introduction
De novo dominant mutations in UBTF [NM_014233.3:c.628G>A, p.Glu210Lys] cause a distinct, predominantly neurological disorder, dubbed the UBTF E210K neuroregression syndrome (ClinVar SCV000598648.1)(Edvardson et al., 2017; Toro et al., 2018). Other deleterious UBTF variants including likely pathogenic nonsense variants have also been reported in human clinical databases (ClinVar: Q635*, Q672*, Y271*, Y308*, E56*). UBTF (Upstream Binding Transcription Factor) is a multi-HMGB (High Mobility Group B)-box architectural DNA binding protein essential for ribosomal RNA (rRNA) transcription by RNA polymerase 1 (Pol I) and for ribosome biogenesis in the nucleolus (Hamdane et al., 2014; Herdman et al., 2017). Initial molecular studies suggested that the E210K variant may function through a gain-of-function mechanism with increased binding to rDNA and increased expression of 18S rRNA (Edvardson et al., 2017; Toro et al., 2018). Recent work suggests that the UBTF E210K variant causes a mild reduction in the cooperative interaction between UBTF and Selectivity Factor (SL1) leading to reduced rDNA transcription but an increase in active rDNA copies (Tremblay et al., 2022). Here we describe a range of targeted studies using murine model systems to understand the contribution of loss-of-function mechanisms to the pathobiology of UBTF mutations.
UBTF exists as two isoforms, UBTF1 (CCDS42346.1) and UBTF2 (CCDS1140.1), that are expressed at similar levels in differentiated cell types. These replace histone chromatin across active copies of the several hundred rDNA genes present in the genome (Tchelidze et al., 2019), the UBTF1/UBTF2 ratio corresponding closely with the fraction of active rRNA genes. The UBTF c.628G>A mutation results in the same amino acid change in both UBTF1 and UBTF2 (E210K). Disease onset is at 2.5 to 3 years and characterized by slow progression of global motor, cognitive and behavioral dysfunction. UBTF E210K shows modified binding to the rDNA promoter that affects formation of the preinitiation complex by the Pol I-specific TBP-complex SL1 (Tremblay et al., 2022). UBTF E210K fibroblasts show increased levels of pre-rRNA and 18S rRNA, nucleolar abnormalities, markedly increased numbers of DNA double-strand breaks (DSBs), defective cell-cycle progression, and apoptosis (Toro et al., 2018).
In previously published work from our laboratories, adult (3 month-old) Ubtf+/− mice and sex-matched Ubtf+/+ littermates were subjected to a battery of motor and behavioral examinations including open-field activity, rotarod, vertical rope climbing, raised-beam task, grip strength, gait analysis (DigiGait™), dominance tube, and cross-maze test (Toro et al., 2018). No evidence of involuntary movements or seizures was noted. Ubtf+/− mice tended to fall off the rotarod at shorter latencies than their Ubtf+/+ littermates. Ubtf+/− mice also took more time to traverse raised beams in comparison to Ubtf+/+ littermates. Quantitative measures of gait exposed minor differences between Ubtf+/− mice and their Ubtf+/+ littermates. Ubtf−/− is early embryonic lethal in mice (Hamdane et al., 2014). Quantitative real-time PCR (QRT-PCR) showed significant reductions in total brain Ubtf transcript in Ubtf+/− mice with only minor, statistically insignificant, reductions in Ubft1 transcript (Toro et al., 2018). These findings suggest that Ubtf gene dosage is important in establishing and maintaining normal neural function, but since UBTF1 is essential for rDNA function, the maintenance of UBTF1 levels may provide partial compensation for the loss of one allele.
Here we extend previous work and examine mice through 18 months of age to determine if neuroregression is rapidly progressive as is the case for the human UBTF E210K neuroregression syndrome. In the gnomAD database (gnomad.broadinstitute.org) there are no high-confidence UBTF putative LoF (loss-of-function) variants. In contrast, ClinVar (www.ncbi.nlm.nih.gov/clinvar) reports several LoF variants that are likely pathogenic, pathogenic, or of uncertain significance. As such, study of knockdown and knockout of Ubtf in murine models is relevant to clinical genetics diagnostics in humans and understanding the biology of UBTF.
2. Results
2.1. Homozygous knockout of Ubtf in neural tissues is embryonic lethal
B6.Cg-Tg(Nes-cre)1Kln/J mice, commonly known as nestin-Cre mice, express Cre recombinase in the central and peripheral nervous systems by embryonic day 11 (Tronche et al., 1999). Using crosses between Ubtffl/fl and nestin-Cre mice we wanted to determine if UBTF is vital for late embryonic development of the nervous system. We did not identify surviving Postnatal Day 0 (P0) pups with elimination of Ubtf expression in the nervous system. Therefore, UBTF must be essential for late embryonic development. However, pregnant dams were not euthanized for assessment of embryos before or after embryonic day 11 to establish a temporal profile of embryonic death. To assess the effects of heterozygous UBTF loss, we euthanized Ubtf+/fl_nestin-Cre mice at 3 months-of-age and harvested RNA from liver and hippocampus. Ubtf+/+ mice were used as controls. In comparison to Ubtf+/+ mice, Ubtf+/fl_nestin-Cre mice showed less than 50% reductions in hippocampal total Ubtf (37%) and Ubtf1 (38%). Similar reductions were seen in 18S, 28S and 45S rRNA (Table S1). For unclear reasons, there were modest increases in liver expression of total Ubtf, 28S rRNA and 45S rRNA (Table S1).
2.2. Ubtf Knockdown in Adult Mice
To bypass the potential effects of developmental compensation and determine the subacute effects of haploinsufficiency in a mammalian system in vivo, Ubtffl/fl_CreERT2 mice were treated with tamoxifen to knockdown Ubtf expression in 3-month-old mice. Ubtf knockdown was associated with rapid weight loss and marked deterioration in sensorimotor functioning over the course of 8 days. Open-field activity measures including distance traveled, ambulatory counts, stereotypic counts, vertical counts, and jump counts were notably reduced in comparison to groups of control mice (Supplementary Material, Table S2). Gait analyses (DigiGait™) showed reduced stride lengths and paw areas in the tamoxifen treated Ubtffl/fl_CreERT2 mice (Table S2). Mice were euthanized 10 days after initial treatment with tamoxifen and RNA was extracted from fresh postmortem brain. Relative quantitative real-time PCR (QRT-PCR) showed an approximate 90% reduction in total Ubtf expression in cerebral cortex, cerebellum, striatum, and liver (Table S2).
2.3. Motor, behavioral, and cognitive testing of Ubtf+/−mice
2.3.1. Vertical rope climbing
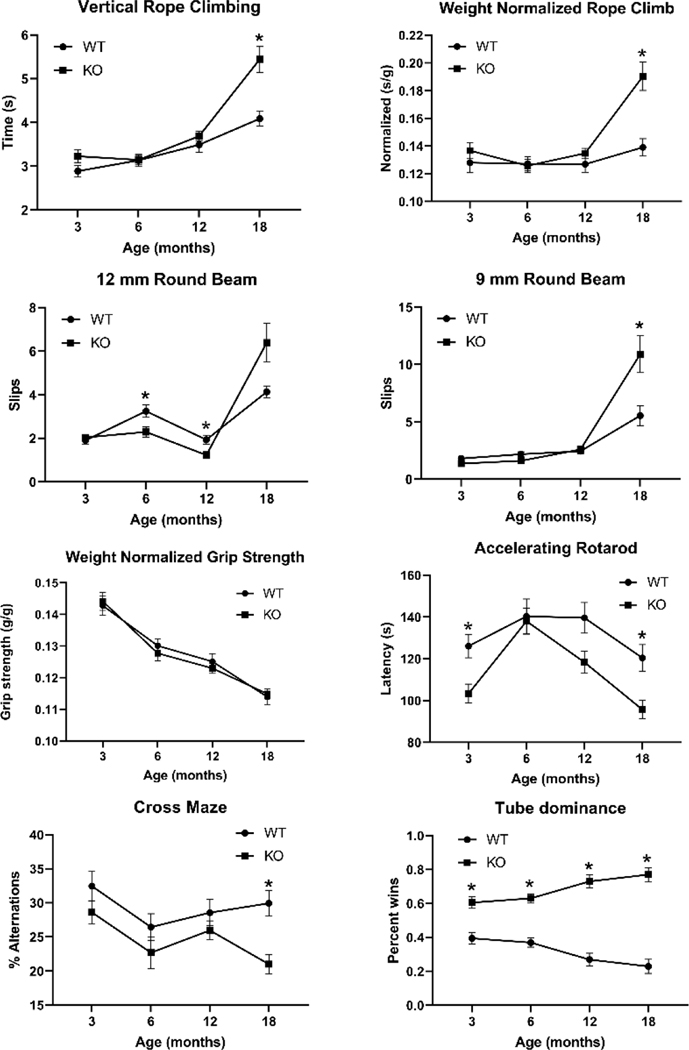
At 18 months, there were significant effects of genotype (F1,69 = 18.47, P < 0.0001) and the sex*genotype interaction (F1,69 = 4.45, P = 0.038) on vertical rope climbing times (Table 1). Ubtf+/+ males but not females were faster than sex-matched Ubtf+/− mice. There were similar effects of genotype (F1,69 = 20.32, P < 0.0001) and the sex*genotype interaction (F1,69 = 11.44, P = 0.15) on weight-normalized vertical rope climbing (Table 1).
Table 1.
Phenotypes of Ubtf +/− Mice at 18 mo
| Ubtf+/+ Male | Ubtf+/− Male | Ubtf+/+ Female | Ubtf+/− Female | |
|---|---|---|---|---|
| Body weight (g) | 33.4±0.9 (N=18) | 30.5±0.4* (N=20) | 26.6±0.7 (N=18) | 28.0±0.7 (N=19) |
| Brain weight (g) | 0.460±0.006 (N=7) | 0.440±0.005 (N=7) | 0.456±0.005 (N=6) | 0.446±0.007 (N=6) |
| Rope climb (s) | 4.32±0.23 (N=17) | 6.34±0.44* (N=19) | 3.86±0.24 (N=18) | 4.55±0.28 (N=19) |
| Normalized rope climb (s/g) | 0.13±0.01 (N=17) | 0.22±0.02* (N=19) | 0.15±0.01 (N=18) | 0.16±0.01 (N=19) |
| Grip strength (g) | 350.6±5.2 (N=18) | 338.3±5.8 (N=19) | 313.0±5.0 (N=18) | 320.0±4.4 (N=19) |
| Normalized grip strength (g/g) | 10.49±0.21 (N=18) | 11.28±0.21 (N=19) | 12.30±0.34 (N=18) | 11.71±0.24 (N=19) |
| Rotarod Day 5 (s) | 118.3±9.6 (N=15) | 96.3±5.0* (N=19) | 112.3±8.4 (N=17) | 95.0±7.1* (N=17) |
| Raised Beam slips | ||||
| 12 mm square | 1.50±0.21 (N=18) | 1.48±0.22 (N=18) | 1.28±0.16 (N=18) | 1.16±0.15 (N=18) |
| 9 mm square | 2.56±0.36 (N=18) | 3.78±0.79 (N=18) | 2.12±0.24 (N=18) | 2.39±0.34 (N=18) |
| 12 mm round | 4.48±0.32 (N=18) | 8.23±1.45* (N=18) | 3.79±0.38 (N=18) | 4.52±0.68 (N=18) |
| 9 mm round | 3.39±0.70 (N=18) | 13.3±2.15* (N=18) | 7.52±1.46 (N=18) | 8.47±2.19 (N=18) |
| Open field activity | ||||
| Distance traveled (cm) | 1081.1±103.5 (N=20) | 1393.8±76.8* (N=20) | 1470.4±111.5 (N=18) | 1433.4±52.7 (N=19) |
| Ambulatory count | 583.4±53.5 (N=20) | 717.9±44.7 (N=20) | 765.7±66.0 (N=18) | 758.5±31.5 (N=19) |
| Stereotypic count | 1403.2±69.5 (N=20) | 1645.2±47.6* (N=20) | 1608.8±66.5 (N=18) | 1491±52.9 (N=19) |
| Vertical count | 59.6±7.5 (N=20) | 48.3±4.2 (N=20) | 48.9±5.1 (N=18) | 38.7±3.7 (N=19) |
| Jump count | 13.5±2.8 (N=20) | 10.2±1.4 (N=20) | 21.5±2.5 (N=18) | 14.4±2.3 (N=19) |
| Average velocity (cm/s) | 31.5±0.9 (N=20) | 32.6±0.6 (N=20) | 32.2±1.3 (N=18) | 30.6±0.7 (N=19) |
| Ambulatory episodes | 48.5±3.9 (N=20) | 57.7±3.4 (N=20) | 61.4±4.0 (N=18) | 59.4±2.3 (N=19) |
| DigiGait™ | ||||
| Propel (s) Forelimb | 0.120±0.004 (N=11) | 0.111±0.006 (N =12) | 0.121±0.005 (N=15) | 0.122±0.004 (N=15) |
| Propel (s) Hindlimb | 0.185±0.005 (N=11) | 0.187±0.007 (N=12) | 0.191±0.004 (N=15) | 0.204±0.015 (N=15) |
| Stride length (cm) Forelimb | 6.65±0.13 (N=11) | 6.61±0.16 (N=12) | 7.06±0.14 (N=15) | 7.18±0.13 (N=15) |
| Stride length (cm) Hindlimb | 6.71±0.12 (N=11) | 6.70±0.14 (N=12) | 7.12±0.15 (N=15) | 7.68±0.46 (N=15) |
| Stride frequency (steps/s) Forelimb | 3.04±0.07 (N=11) | 3.02±0.08 (N=12) | 2.83±0.06 (N=15) | 2.77±0.06 (N=15) |
| Stride frequency (steps/s) Hindlimb | 3.03±0.06 (N=11) | 2.99±0.07 (N=12) | 2.82±0.07 (N=15) | 2.96±0.21 (N=15) |
| Stance width (cm) Forelimb | 1.84±0.06 (N=11) | 1.84±0.02 (N=12) | 1.88±0.04 (N=15) | 1.88±0.04 (N=15) |
| Stance width (cm) Hindlimb | 3.05±0.07 (N=11) | 3.20±0.06 (N=12) | 3.01±0.06 (N=15) | 3.10±0.08 (N=15) |
| Step angle (deg) Forelimb | 66.27±1.41 (N=11) | 63.61±1.63 (N=12) | 64.50±1.27(N=15) | 61.44±1.42 (N=15) |
| Step angle (deg) Hindlimb | 52.62±1.45 (N=11) | 51.40±1.69 (N=12) | 53.13±1.14 (N=15) | 55.30±0.96 (N=15) |
| Paw area (cm2) Forelimb | 0.448±0.014 (N=11) | 0.438±0.013 (N=12) | 0.362±0.012 (N=15) | 0.390±0.010 (N=15) |
| Paw area (cm2) Hindlimb | 0.764±0.031 (N=11) | 0.740±0.015 (N=12) | 0.674±0.022 (N=15) | 0.731±0.045 (N=15) |
| Tube dominance (%) | 28.8±6.2 (N=20) | 71.2±6.2* (N=20) | 16.8±0.5 (N=19) | 83.2±0.5* (N=19) |
| Cross maze (%) | 27.7±2.4 (N=17) | 21.0±1.7 (N=18) | 32.0±2.8 (N=18) | 27.5±2.4 (N=18) |
| Morris water maze | ||||
| Day 6 escape latency(s) | 12.68±1.43 (N=15) | 24.89±2.51* (N=15) | 17.05±1.39 (N=15) | 29.57±4.03* (N=15) |
| Probe - time in target quadrant (s) | 25.13±2.73 (N=15) | 19.76±1.60 (N=15) | 22.69±1.41 (N=15) | 18.84±2.32 (N=15) |
| Nociception latency (s) | 10.04±0.44 (N=18) | 12.16±0.86* (N=19) | 10.09±0.47 (N=18) | 12.92±0.62* (N=19) |
Values ± standard error of the mean (SEM).
P < 0.05, within sex.
Repeated measures analysis (Fig. 1A) found significant effects of age (F2.19,157.7 = 60.42, P < 0.001), genotype (F1,72 = 8.68, P = 0.0043), and the age*genotype interaction (F3,216 = 8.80, P < 0.0001) on vertical rope climbing times. The effect of genotype at the four postnatal ages was only significant at 18 months (adjusted P = 0.0007, post-hoc Šídák’s multiple comparisons test). There were also significant effects of age (F2.35,169.2 = 20.88, P <0.001), genotype (F1,72 = 7.524, P = 0.0077), and the age*genotype interaction (F3,216 = 9.339, P < 0.0001) on weight-normalized vertical rope climbing times (Fig. 1B). The effect of genotype on weight-normalized vertical rope climbing times at the four postnatal ages was only significant at 18 months (adjusted P = 0.0003, post-hoc Šídák’s multiple comparisons test).
Fig. 1.
Repeated measures analysis of vertical rope climbing in 74 mice (A), weight normalized vertical rope climbing in 74 mice (B), 12 mm raised round beam task in 72 mice (C), 9 mm raised round beam task in 72 mice (D), weight normalized grip strength in 74 mice (E), accelerating rotarod in 68 mice (F), cross maze analysis in 71 mice (G), and tube dominance in 78 mice (H). *, adjusted P < 0.05 Šídák’s multiple comparisons test
2.3.2. Raised-beam task
Dystonia and Parkinsonism have been reported in humans with the UBTF-associated neuroregression (Ikeda et al., 2021; Toro et al., 2018). The raised-beam task is a sensitive measure of motor dysfunction in mouse models of dystonia and Parkinson’s disease (Khan et al., 2018; Xiao et al., 2016; Zhao et al., 2008). At 18 months there were no significant differences between Ubtf+/− mice and their Ubtf+/+ littermates in the number of slips on the 12 mm and 9 mm square beams. On the 12 mm round beam there were significant effects of genotype (F1, 69 = 6.88, P = 0.011) and sex (F1, 69 = 6.66, P = 0.012) but no effect of the sex*genotype interaction (Table 1). Post-hoc analysis identified a significant difference between male Ubtf+/− and Ubtf+/+ mice (adjusted P = 0.017) but no difference between female Ubtf+/− and Ubtf+/+ mice (Table 1). On the 9 mm round beam there were significant effects of genotype (F1, 69 = 8.62, P = 0.005) and the sex*genotype interaction (F1, 69 = 5.74, P = 0.019) but no effect of sex (Table 1). Post-hoc analysis identified a significant difference between male Ubtf+/− and Ubtf+/+ mice (adjusted P = 0.002) but no difference between female Ubtf+/− and Ubtf+/+ mice (Table 1).
Repeated measures analyses of mean slips at 3, 6, 12, and 18 months on the 12 mm round beam showed significant effects of age (F1.6, 111.1 = 44.1, P < 0.0001) and the age*genotype interaction (F3, 210 = 8.6, P < 0.001), but no effect of genotype (F1, 70 = 0.39, P = 0.54). Post-hoc analyses showed that Ubtf+/− mice had fewer slips than Ubtf+/+ littermates at 6 and 12 months (adjusted P < 0.05, for both). Variance increased at 18 months, particularly in the Ubtf+/− mice (Fig. 1C). On the 9 mm round beam (Fig. 1D) there were effects of age (F1.135, 79.43 = 44.72, P < 0.0001), genotype (F1, 70 = 5.21, P = 0.026), and the age*genotype interaction (F3, 210 = 9.16, P < 0.0001). Post-hoc analyses showed that the differences between Ubtf+/+ and Ubtf+/− mice were only significant at 18 months (adjusted P = 0.020).
2.3.3. Grip strength analysis
Based on repeated measures analysis, there were no significant effects of genotype or age on grip strength. Similarly, there were no effects of genotype or sex on grip strength at 18 months. However, there was a significant effect of age on weight normalized grip strength (F2.47,178.0 = 97.99, P < 0.0001). As seen in Fig. 1E, weight normalized grip strength deteriorated with increasing age in both Ubtf+/− mice and their Ubtf+/+ littermates.
2.3.4. Accelerating rotarod
The accelerating rotarod is used to access general motor coordination. At 18 months, the was an effect of genotype (F1,64 = 10.12, P = 0.002) but no effect of sex or the sex*genotype interaction on Day 5 rotarod latencies (Table 1). Repeated measures analysis using rotarod Day 5 latencies at 3, 6, 12, and 18 months, showed significant effects of age (F2.800, 184.8 = 18.07, P < 0.001) and genotype (F1, 66 = 7.86, P = 0.0066), but no overall effect of the age*genotype interaction (F3, 198 = 2.47, P = 0.063). Šídák’s multiple comparisons test identified significant effects of genotype at 3 and 18 months but not at 6 or 12 months (Fig. 1F).
2.3.5. Open-field activity
Measures included distance traveled (cm), ambulatory count, stereotypic count, vertical count, jump count, average velocity (cm/s), and ambulatory episodes. At 18 months (Table 1), there were overall effects of genotype on vertical (F1, 73 = 4.00, P = 0.049) and jump (F1, 73 = 5.25, P = 0.025) counts, and sex on distance traveled (F1, 73 = 5.83, P = 0.018), ambulatory count (F1, 73 = 5.83, P = 0.018), jump counts (F1, 73 = 7.2, P = 0.0094), and ambulatory episodes (F1, 73 = 4.43, P = 0.039). Post-hoc Šídák’s multiple comparisons tests showed differences between male Ubtf+/− and Ubtf+/+ mice on distance traveled (adjusted P = 0.026) and stereotypic counts (adjusted P = 0.027). There were no significant effects of genotype on repeated measures analyses (3, 6, 12, and 18 months) of distance traveled, ambulatory count, stereotypic count, vertical count, jump count, average velocity (cm/s), or ambulatory episodes.
2.3.6. Gait analysis
There were no effects of genotype on any forelimb or hindlimb gait parameter (propel, stride length, stride frequency, stance width, step angle or paw area) at 18 months (Table 1). In addition, there were no significant effects of genotype on repeated measures analyses (3, 6, 12, and 18 months) of gait parameters.
2.3.7. Cross-maze analysis
The cross-maze test was used to evaluate spatial working memory and exploratory behavior. At 18 months, there were effects of genotype (F1, 67 = 5.3, P = 0.024) and sex (F1, 67 = 4.9, P = 0.030), but no interaction effect (Table 1). Overall, females outperformed males and Ubtf+/− mice outperformed their Ubtf+/+ littermates. There were effects of genotype (F1, 69 = 12.1, P = 0.0009) and age (F2.624, 181.1 = 4.11, P = 0.010) but no interaction effects (Fig. 1G) on repeated measures analysis. Post-hoc analyses were only significant for group differences at 18 months (adjusted P = 0.0012).
2.3.8. Dominance tube
This test was used for identifying a general abnormality in social interaction through the measurement of aggression. Overall, Ubtf+/− mice were more aggressive than their Ubtf+/+ littermates. At 18 months, there were significant effects of genotype (F1, 74 = 88.3, P < 0.0001) and the sex*genotype interaction (F1, 74 = 4.246, P = 0.043) on % tube dominance (Table 1). Differences between Ubtf+/− mice and their Ubtf+/+ littermates were most pronounced for females. Repeated measures analysis (3 mo., 6 mo., 12 mo., and 18 mo.), exposed a significant effect of genotype (F1, 76 = 150.9, P < 0.0001) and the age*genotype interaction (F3, 228 = 11.6, P < 0.0001 but no effect of age on % tube dominance. The effect of genotype became more pronounced with increasing age (Fig. 1H).
2.4.9. Morris water maze
The Morris water maze is used to access spatial learning and memory. At 18 months, there was a large effect of genotype but no effect of sex or the genotype*sex interaction on Day 6 escape latencies (F1, 56 = 23.03, P < 0.0001). A smaller effect of genotype was found on the probe trial which measured time in the target quadrant (F1, 56 = 4.57, P = 0.037). There was no effect of sex or the genotype*sex interaction on the probe trial.
2.4.10. Nociception
Some patients with the UBTF E210K neuroregression syndrome exhibit increased tolerance to noxious stimuli (Toro et al., 2018). This is not surprising given the protean neurological features of this syndrome and ubiquitous expression of UBTF. Findings in mice support this clinical observation. In comparison to Ubtf+/+ littermates (10.06 ± 0.32 s), Ubtf+/− mice (12.54 ± 0.54 s) showed prolonged latencies on the nociception assay (F1,69 = 14.75, P = 0.003). There were no significant effects of sex or the sex*genotype interaction on nociception latency (Table 1).
2.5. QRT-PCR
At 18 months, reductions of total Ubtf were larger than reductions of Ubtf1 but notably less than 50% in both Ubtf+/− cerebral cortex and hippocampus (Table 2). Reductions in Ubtf1 were significant in cerebral cortex but did not reach statistical significance in hippocampus. There were no significant changes in brain 18S, 28S or 45S rRNA (Table 2).
Table 2.
Expression of Ubtf and rRNA at 18 months
| Cerebral Cortex | Hippocampus | |||
|---|---|---|---|---|
| Ubtf +/+ | Ubtf +/− | Ubtf +/+ | Ubtf +/− | |
| Ubtf Total | 1.00 ± 0.03 | 0.58 ± 0.01* | 0.68 ± 0.05 | 0.45 ± 0.01* |
| Ubtf1 | 1.00 ± 0.03 | 0.77 ± 0.03* | 0.65 ± 0.02 | 0.58 ±0.02 |
| 18S rRNA | 1.00 ± 0.02 | 0.83 ± 0.05 | 0.97 ± 0.07 | 0.72 ± 0.04 |
| 28S rRNA | 1.00 ± 0.04 | 0.84 ± 0.08 | 0.69 ± 0.02 | 0.66 ±0.02 |
| 45S rRNA | 1.00 ± 0.05 | 1.12 ± 0.04 | 0.73 ± 0.02 | 0.83 ±0.03 |
P < 0.05
2.6. Comet Assay
The comet assay, or single cell gel electrophoresis, is a sensitive method for detection of DNA strand breaks in individual cells. The assay is based on denatured, cleaved DNA fragments migrating out of cells under an electric field and forming “tails”. The distribution of DNA between the head and the tail is used to evaluate the degree of DNA damage. In cerebellum, percent tail DNA did not differ between Ubtf+/−mice (7.0%) and their Ubtf+/+ littermates (6.4%). Similarly, in striatum, percent tail DNA did not differ between Ubtf+/−mice (6.3%) and their Ubtf+/+ littermates (5.7%).
2.7. γ-H2A.X ELISA
Histone H2A.X is required for DNA repair following DSBs. DNA damage triggers rapid phosphorylation of H2A.X at Ser139 (γ-H2A.X). Detection of γ-H2A.X is widely used as a surrogate marker of DNA damage. Using γ-H2A.X ELISA we did not detect differences in DNA damage between 18-month-old Ubtf+/− mice and Ubtf+/+ littermates (N = 6/group) using fresh post-mortem cerebellar and striatal tissues (P = NS, for both).
2.8. Di-methyl histone H3K4 quantification
H3K4me2/3 and H2A.Zac are chromatin marks that along with CCCTC-Binding Factor (CTCF) form the so-called “Enhancer Boundary Complex,” upstream boundary of rDNA, and are augmented after loss of UBTF (Herdman et al., 2017). In general, transcriptionally active genes correlate with H3K4me2/3 (di- and trimethyl). ELISA was used to access the overall effects of UBTF haploinsufficiency on global changes in histone H3K4 di-methylation. No differences were detected in the cerebella and striata of 18-month-old Ubtf+/− mice in comparison to Ubtf+/+ littermates (P = NS, for both).
2.9. 53BP1 immunoreactivity
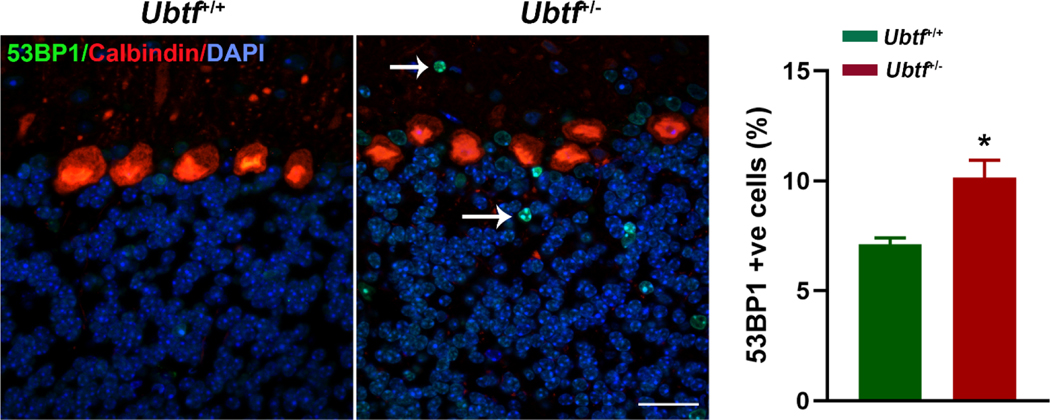
53BP1 is a sensor protein of DNA damage and plays an important role in the non-homologous end-joining DNA repair pathway. More 53BP1 immunoreactive foci were found in Ubtf+/− cerebellum in comparison to Ubtf+/+ cerebellum at 18 months (Welch-corrected t = 3.65, P = 0.048). Most commonly, multiple 53BP1-immunoreactive foci were found in individual neurons (Fig. 2).
Fig. 2.
Cerebellar 53BP1 immunoreactivity (green) in cerebellar cortex from Ubtf+/+ (N = 3) and Ubtf+/− (N = 3) mice. Calbindin immunoreactivity (red) is used to label Purkinje cells and define the layers of cerebellar cortex. Nuclei are labeled with DAPI. Graph shows percentages of 53BP1-positive (+ve) cells. Scale bar, 100 μm.
2.10. Routine histochemistry
We used the Luxol fast blue stain to access myelin in the central nervous system of Ubtf+/− mice and Ubtf+/+ littermates at 18 months. Based on qualitative assessments, there was no evidence of demyelination or dysmyelination (Supplementary Material, Figure S1). There were no neuroanatomical differences between Ubtf+/− mice and Ubtf+/+ littermates at the level of light microscopy on qualitative analysis of cresyl violet and hematoxylin and eosin (H & E) stained sections (Figure S1). In addition, there were no abnormal neuronal patterns, abnormal neuronal populations, or microstructural abnormalities in Ubtf+/− mouse brain in comparison to Ubtf+/+ brains at 18 months.
2.11. Body and brain weights
Ubtf+/− mice (28.7 ± 3.3 g) were not significantly smaller than Ubtf+/+ littermates (29.8 ± 4.9) at 18 months of age (F1,70 = 2.12, P = 0.15). However, there was a significant sex*genotype interaction (F1, 70 = 19.49, P < 0.0001) and post-hoc Šídák’s multiple comparisons test showed a significant difference in the weights of male mice (adjusted P = 0.0006, Table 1). Overall, Ubtf+/− mice had smaller brain weights (mean ± SEM = 0.444 ± 0.123 g) than Ubtf+/+ littermates (mean ± SEM = 0.458 ± 0.127 g) at 18 months (F1,22 = 5.00, P = 0.036; Table 1).
3. Discussion
Our results verify the importance of normal UBTF expression to embryogenesis, survival, and neural functioning. The relatively minimal effects of UBTF1 haploinsufficiency on several measures of motor functioning previously described at 3 months (Toro et al., 2018) suggests the presence of minor neurodevelopmental defects in Ubtf+/− mice. Comparably, subtle to mild neurodevelopmental abnormalities have been reported in children with the UBTF E210K mutation (Edvardson et al., 2017). The more pronounced and progressive abnormalities noted after 12 months of age in mice indicate the presence of superimposed neurodegeneration. In this regard, neurodegeneration in childhood is commonly referred to as developmental regression, neurodevelopmental regression or developmental neuroregression. It is quite possible that the behavioral, motor, cognitive, and molecular abnormalities reported herein in 18-month-old Ubtf+/− mice would have been more prominent if mice lived longer given that neuroregression in humans with the UBTF E210K syndrome does not become clinically manifest until 2.5 to 3 years of age. This age constraint can be a limitation of mouse models used to study some neurodegenerative disorders.
Comparison of behavioral (dominance tube), cognitive (Morris water maze, cross maze), motor (rope climbing, raised beam, rotarod), and sensory testing results permits cross-correlation with clinical findings in humans with UBTF mutations (Bastos et al., 2020; Edvardson et al., 2017; Ikeda et al., 2021; Sedlackova et al., 2019; Toro et al., 2018). Ubtf+/− mice exhibited abnormal social interactions on tube dominance testing at 3 months with notable progression from 12 to 18 months (Fig. 1H). Autistic behavior patterns, impulsivity, and hyperactive behaviors are well described in some patients with UBTF mutations and, correspondingly, abnormal tube dominance testing has been reported in several murine models of autism (Han et al., 2020; Jiang-Xie et al., 2014). Cognitive regression is perhaps the earliest and most overt manifestation of the UBTF E210K neuroregression syndrome. The impairments on cross maze and Morris water maze testing were marked at 18 months. In combination, the results of behavioral and cognitive testing indicate the presence of both impaired learning and memory in Ubtf+/− mice. Similarly, behavioral, and cognitive deficits typically precede overt motor abnormalities in humans with UBTF mutations.
Relative to the notable behavioral and cognitive impairments in Ubtf+/− mice, the abnormalities on vertical rope climbing, the raised beam task, and open field activity were modest and mostly limited to 18-month-old animals, and there were no significant differences in gait parameters between Ubtf+/− mice and their isogenic Ubtf+/+ littermates. Overall, Ubtf+/− mice were less active in the vertical axis with fewer jump and vertical counts during open-field activity. There were also differences on the accelerating rotarod at both 3 and 18 months with a quadratic effect of age on both Ubtf+/− and Ubtf+/+ mice (Fig. 1F). Variance increased at 18 months on most motor assessments in Ubtf+/− mice suggesting that some animals were nearing the precipice of neuroregression at 12 months of age. In aggregate, these data from Ubtf+/− mice are compatible with clinical data from humans with the UBTF E210K neuroregression syndrome. In humans, postural instability and axial hypotonia tend to precede the appearance of spasticity, dystonia, ataxia, and Parkinsonism by several years (Bastos et al., 2020; Ikeda et al., 2021; Sedlackova et al., 2019; Toro et al., 2018). In our study, Ubtf+/− mice did not exhibit overt spasticity, dystonia, ataxia, or Parkinsonism on open field behavior. Furthermore, these motor signs would likely manifest as abnormalities on gait analysis.
The sensory abnormality detected at 18 months in Ubtf+/− mice is not surprising given the ubiquitous expression of UBTF in neural tissues. Sensory abnormalities, particularly impaired responses to noxious stimuli, has been reported in patients with the UBTF E210K neuroregression syndrome (Toro et al., 2018). The integrity of other sensory systems (vision, hearing, taste, and smell) was not assessed in our mouse model and have not been detailed in clinical reports of the UBTF E210K neuroregression syndrome.
Relative to Ubtf+/+ littermates, brain weights were reduced in Ubtf+/− mice at 18 months. Magnetic resonance imaging in humans with the UBTF E210K neuroregression syndrome shows progressive loss of brain volume, and, in patients older than 10 years, marked ex vacuo ventriculomegaly (Toro et al., 2018). In humans, supratentorial atrophy is greater than infratentorial atrophy which is compatible with the appearance of overt cognitive and behavioral deficits prior to motor abnormalities seen herein in mice.
UBTF1 and UBTF2 are found on active rDNA and at GC-rich nucleosome-free regions (NFRs) genome-wide as heterodimers or homodimers. UBTF regulates rRNA transcription by RNA Pol I employing several mechanisms that involve assembly of transcription complexes (Beckmann et al., 1995; Jantzen et al., 1992; Watt et al., 2022), regulation of chromatin state (Herdman et al., 2017; Stefanovsky et al., 2006), modulating promoter escape (Panov et al., 2006) and transcriptional elongation (Stefanovsky et al., 1996; Stefanovsky et al., 2006). UBTF1 but not UBTF2 cooperates with SL1 to form the preinitiation complex at the Pol I promoter, explaining why UBTF2 cannot replace UBTF1 for rRNA gene expression (Tremblay et al., 2022). However, UBTF2 may be sufficient to regulate mRNA transcription by Pol II (Grueneberg et al., 2003; Sanij et al., 2015). In some respects, UBTF2 functionally resembles an ERK phosphorylated form of UBTF1 and is more permissive to Pol I transcription elongation (Stefanovsky et al., 2001; Stefanovsky and Moss, 2008), suggesting that it is a modulator/diluter of UBTF1 activity. Although the expression of total Ubtf was reduced by approximately half in cerebral cortex, cerebellum, and liver, Ubtf1 levels were not significantly reduced in 3-month-old Ubtf+/− mice (Toro et al., 2018) suggesting compensatory regulation of Ubtf primary transcript splicing via feedback mechanisms. At 18 months, the upregulation was less robust with modest but significant reduction in Ubtf1 in cerebral cortex of Ubtf+/− mice, although there were no significant effects on levels of 18S, 28S, and 45S rRNA (Table 2). These findings suggest that Ubtf gene dosage is important in establishing and maintaining normal neural function, but since UBTF1 is essential for rDNA function, the maintenance of UBTF1 levels may provide partial compensation for the loss of one allele. However, compensatory mechanisms could begin to fail in older animals thereby contributing to small reductions in UBTF1 with aging. In addition, UBTF2 deficiency may also contribute to late-onset neurological deterioration in mice.
Depletion of UBTF has been shown to cause abnormalities of cell-cycle progression, the DNA damage response, and genome instability (Hamdane et al., 2015; Sanij et al., 2015). This may be related to the essential role this factor plays in rDNA transcription and/or its ability to replace nucleosomes and stabilize NFRs. Defects in ribosome biogenesis lead to nucleolar stress, the stabilization of p53, cell cycle arrest and apoptosis (Calo et al., 2018; Nicolas et al., 2016; Orsolic et al., 2016; Zhou et al., 2015), and loss of UBTF has been shown to have these effects (Hamdane et al., 2015; Sanij et al., 2015). Loss or mutation of UBTF could affect the formation of both rDNA-specific and genome-wide NFRs, possibly leaving the underlying DNA poorly protected and explaining the enhanced damage that has been observed. Fibroblasts from patients with the UBTF E210K neuroregression syndrome show nucleolar abnormalities, markedly increased numbers of DSBs, defective cell-cycle progression, and apoptosis (Toro et al., 2018). Using postmortem neural tissue from 18-month-old Ubtf+/− mice, we did not detect statistically significant evidence of abnormal DNA damage with the comet assay or γ-H2A.X ELISA. However, increased cerebellar 53BP1-IR was detected in the Ubtf+/− mice suggesting that morphological assessment of DNA damage may be more sensitive than whole tissue ELISA and comet assays. In future studies, more mechanistic approaches will be required to link specific molecular and cellular consequences of Ubtf1 and/or Ubtf2 deficiency to the development of late-onset cognitive, behavioral, and motor dysfunction in Ubtf+/− mice.
4. Materials and methods
4.1. Heterozygous germline deletion of Ubtf
All mouse experiments were performed in accordance with the National Institutes of Health’s Guidelines for the Care and Use of Laboratory Animals and approved by our Institutional Animal Care and Use Committee. Experiments utilized Ubtf+/− mice along with sex-matched isogenic Ubtf+/+ littermates. Derivation and genotyping of Ubtf+/− mice was detailed in previous work (Hamdane et al., 2014; Toro et al., 2018). Serial and quantitative behavioral, cognitive, and sensorimotor testing (cross maze, grip strength, dominance tube, rotarod, open-field activity, raised-beam task, vertical rope climbing, DigiGait™ gait analysis) were completed from the early postnatal period through late adulthood (18 mo). Hot-plate analgesia testing and Morris water maze were only completed at 18 months. At each age (3, 6, 12, and 18 months), testing occurred over a time window of 14 days. Data from mice that died or were euthanized for other studies prior to 18 months were not included in repeated measures analyses. Moreover, some mice did not reliably participate in certain tests, particularly gait analyses such that group sizes differ among specific tests. Upon completion of behavioral, cognitive, and sensorimotor testing, mice were euthanized (pentobarbital overdose, > 100 mg/kg) for molecular and morphological studies. Behavioral testing and post-mortem analyses were blinded to sex and genotype.
4.2. Conditional knockout of Ubtf in the central nervous system
Hemizygous B6.Cg-Tg(Nes-cre)1Kln/J (nestin-Cre) male and female mice were bred to male and female Ubtffl/fl mice through two generations. F1 mice were genotyped for the cre and floxed Ubtf alleles. Ubtffl/fl mice were bred to Ubtf+/fl_nestin-Cre mice. F2 Postnatal Day 0 pups were decapitated, and RNA was extracted from brain. A total of 6 litters were assessed.
4.3. Tamoxifen-induced Ubtf Knockdown in Adult Mice
Gt(ROSA)26Sor<tm1(cre/ERT2)Tyj> mice [Gt(ROSA)26Sortm1(cre/ERT2)Tyj], The Jackson Laboratory, Bar Harbor, ME, USA] were crossed to Ubtffl/fl floxed mice. Three-month-old F2 generation mice of the desired genotype (Gt(ROSA)26Sortm1(cre/ERT2)Tyj_ Ubtffl/fl) were treated with tamoxifen (75 mg/kg, i.p., q.d.) for 5 consecutive days and then monitored for an additional 5 days prior to euthanasia. Tamoxifen (Sigma-Aldrich, St. Louis, MO, USA) was solubilized at a concentration of 10mg/ml in corn oil (Sigma-Aldrich) containing 5% ethanol. Ubtf+/+ and Ubtf+/− mice were also injected with tamoxifen and served as controls. Mice underwent behavioral testing 8 days after the initial injection of tamoxifen and then euthanized 2 days later for RNA extraction from brain.
4.4. Behavioral testing
4.4.1. Vertical rope climbing
Mice were acclimated to a vertical 40-cm long, 10-mm thick rope prior to testing. The bottom of the rope was suspended 15 cm above a padded base and the top entered an escape box. Three trials with at least 5-min inter-trial intervals were completed for each mouse.
4.4.2. Raised-beam task
Mice were acclimated to an 80-cm long, 26-mm wide beam elevated 50 cm above a padded foundation. A 60W lamp at the start served as an aversive stimulus whereas the opposite end of the beam entered an escape box. After initial runs, trials were completed with 12-mm and 9-mm beams, and 12-mm and 9-mm rods. Inter-trial intervals were at least 5 min.
4.4.3. Grip strength analysis
Mice were held by the scruff of the neck with one hand and the base of the tail with the other hand. Mice were free to grasp a metal grid attached to a grip strength meter (Columbus Instruments, Columbus, OH, USA) as they are moved along the axis of the grid. Maximal grip strength (g) was measured in triplicate trials with a minimal inter-trial interval of 5 min.
4.4.4. Rotarod
Mice were acclimated to a Rotamex-5 rotarod (Columbus Instruments) rotating at 5 revolutions per minute (rpm) for 5 min on the day prior to data acquisition. On the following day, mice were exposed to a 30 s acclimation period at 4 rpm followed by an acceleration of 4 rpm every 30 s to a target of 40 rpm at 5 min. Mice were given 3 trials at the same time each day for 5 consecutive days. Mean values were used for statistical comparisons.
4.4.5. Morris water maze
The test was performed in a circular pool, approximately 1.5 m in diameter, filled with opaque water (25 °C) using non-toxic white tempera paint (Crayola), as previously described (Khan et al., 2018). The depth of the water was 30 cm, and the walls were sufficiently high so that mice could see external visuospatial cues but not escape from the pool. An escape platform measuring approximately 10 × 10 cm was submerged 1 cm below the surface of the water. Mice were trained for three days using a visible platform. Mice were allowed to swim for 60 s until they located the platform. Latency times for mice to reach the platform were recorded for each trial. On the final day a probe trial was given, in which the platform was removed from the pool, all mice swam for 60 s, and the percentage of time spent in the target quadrant was calculated. During the entire duration of the task, an experimenter was present and observing the mice. If there was any indication that the mouse was struggling or in danger of drowning, it was removed from the pool immediately and excluded from the study. The entire test was recorded with a video camera mounted on the ceiling. Data was analyzed with ANY-maze software (Stoelting Co., Wood Dale, IL, USA). Mice were tested once for 6 days with the probe trial on Day 7.
4.4.6. Gait analysis
The DigiGait™ imaging system consists of a motorized treadmill with a digital camera positioned below a transparent belt (Mouse Specifics Inc., Boston, MA). For each mouse, the location and timing of each paw contact on the belt was automatically recorded at a belt speed of 20 cm/s for 5 s. A minimum of 800 video frames collected at 160 frames/s was digitized and gait parameters were calculated for both the forelimbs and hindlimbs by the accompanying software. Three trials with 30 min inter-trial intervals were completed for each mouse at 3, 6, 12 and 18 months. Median values at each age were used for statistical analyses.
4.4.7. Cross-maze analysis
The testing apparatus consisted of 4 radial arms, each 25 cm-long, and a camera along with ANY-maze software. Mice were placed into the apparatus for 5 min and entries into each arm were recorded sequentially using an ABCD format with each letter representing an individual arm of the maze. Alternation was counted if mice went into radial arms sequentially (% alternations = 100 × total alternations / [total entries − 3]). Each mouse was tested once at 3, 6, 12 and 18 months.
4.4.8. Open-field activity assay
Mice were placed in activity monitors (MED Associates, Inc., Georgia, VT, USA) for 10-min sessions. The activity monitors measure 27 × 27 cm, with 16 infrared photocell beams equally spaced in the x and y axes of the horizontal plane, 1 cm from the floor of the chambers. An additional array of 16 photocells is situated 5 cm above the floor to track rearing. All tests were conducted in the dark. One hour prior to testing, cages were moved from the housing racks to a quiet anteroom adjacent to the testing room. Following this period of habituation, animals were removed from their home cage, immediately placed in the center of the open field, and allowed to freely explore the apparatus for a test interval of 10 min. Animals were scored for total distance traveled, ambulatory count, stereotypic count, vertical count, jump count, average velocity, and ambulatory episodes. Ambulatory count: the total number of X + Y photo beam breaks while in ambulatory movement status. Stereotypic count: any partial-body movements that occur within the ambulatory box such as grooming, head-weaving, or scratching. Vertical count: number of periods of continuous Z photo beam breaks. Jump count: the number of times that the mouse leaves the photo beam array for a period of time. Ambulatory episodes: the number of times the mouse has started moving after the resting delay has expired. The data recorded during testing was scored in post-session analyses using Activity Monitor 5.1 (MED Associates). The testing apparatus was cleaned with a 70% ethanol solution and allowed to air dry between mice. Mice were given 1 trial at the same time daily for 3 consecutive days at 3 mo., 6 mo., 12 mo., and 18 mo. Median values of activity parameters were used for statistical comparisons.
4.4.9. Dominance tube
Ubtf+/+ and Ubtf+/− mice were released into opposite ends of a dark, narrow tube (30-cm long). Ubtf+/+ and Ubtf+/− mice interacted in the tube and the more dominant or aggressive mouse forcing its opponent out of the tube. When one mouse had all four paws out of the tube, it was declared the loser while the mouse remaining inside the tube was the winner, ending the match. The number of wins was reported as a percentage of the total number of matches. The match was deemed a draw when both mice remained in the tube greater than 120 s (Khan et al., 2018; Xiao et al., 2016; Xiao et al., 2017).
4.4.10. Nociception assay
Mice were placed on a Hot-Plate Analgesia Meter which consists of a hot plate with builtin digital thermometer confined by a clear acrylic enclosure (Columbus Instruments, Columbus, OH, USA). The stage of the hot plate was set to 55 °C. Individual mice were placed on the stage as the timer was started using a foot pedal. The time to the first sign of nociception including paw licking, flinching, or jumping was recorded as the latency and the mouse was immediately removed from the hot plate. A cut-off period of 20 s was maintained to avoid damage to the paws. The assay was performed for three days. Three trials were conducted each day with a minimum of 15 min between each trial. Median values from each day were used to calculate means for each mouse.
4.5. QRT-PCR
Relative levels of mouse Ubtf mRNA, 18S, 28S and 45S were determined in the cerebral cortex and hippocampus of 18 mo. Ubtf+/− and Ubtf+/− mice (n= 3/genotype) using primer pairs targeting different regions of Ubtf for Isoform 1 or all isoforms (Supplementary Material, Table S3). TaqMan-based QRT-PCR was performed with a LightCycler® 480 System (Roche, Indianapolis, IN, USA). Mouse β-actin was used as an endogenous control. Detailed methods are provided in previous work from our laboratories (Toro et al., 2018; Xiao et al., 2016).
4.6. Comet assay
Alkaline comet assays were performed using cerebellar cortex and striatum from 18-month-old Ubtf+/− and Ubtf+/+ (N = 3 males/genotype) using Trevigen’s CometAssay kit (Trevigen, Gaithersburg, MD, USA). Tissues were homogenized with glass homogenizer and washed in ice-cold Ca2+/Mg2+-free 1X phosphate buffered saline (PBS). Approximately 1 × 105 cells were mixed with 500 μl of pre-warmed low melting agarose (1:10, v/v) and 50 μl of the mixture was plated onto a slide (CometSlide™, Trevigen). After agarose solidified and attached to the slides, the slides were immersed in pre-chilled lysis solution overnight at 4 °C, then in alkaline unwinding solution (200 mM NaOH and 1 mM EDTA) for one hour at 4 °C. Electrophoresis was performed in pre-chilled alkaline electrophoresis solution (200 mM NaOH and 1 mM EDTA) at 4 °C for 30 min at 21 V. The slides were then washed twice with distilled water for 5 min, followed by a 5 min incubation with 70% ethanol. Slides were air-dried at 37 °C for 15 min in the dark and the agarose gels were stained with SYBR™ Gold for 30 min at room temperature. After SYBR™ Gold was removed and slides were air-dried, images were acquired with fluorescence microscopy. For each sample, 100 cells were selected randomly selected to quantify the percentage of tail DNA using automated OpenComet v1.3.1 (Gyori et al., 2014). Assays were carried out in triplicate and analyzed by an investigator blinded to genotype.
4.7. γ-H2A.X ELISA
A PathScan® Phospho-Histone H2A.X (Ser139) Sandwich ELISA Kit was used to quantify endogenous levels of γ-H2A.X (Ser139) in the brains (cerebellum and striatum) of 18-month-old Ubtf+/− mice and Ubtf+/+ littermates. Brain tissues were lysed in buffer containing Halt™ protease and phosphatase inhibitor cocktail and pipetted into the wells of a coated plate. The plate was incubated overnight at 4 °C. After incubation and washing, a phospho-histone H2A.X (Ser139) mouse antibody was added into each well to detect the captured phospho-histone H2A.X protein. Horseradish peroxidase (HRP)-conjugated affinity-purified anti-mouse antibody was added to recognize the bound detection antibody. Following a wash to remove any unbound antibody–enzyme reagent, a substrate solution was added to the wells and color developed in proportion to the amount of bound protein. The reaction was terminated by the addition of stop solution. The absorbance was measured at 450 ηm with a microtiter plate reader (SpectraMax M2e, Molecular Devices, San Jose, CA, USA).
4.8. Di-methyl histone H3K4 quantification
The global changes in histone H3K4 di-methylation were determined using an EpiQuik™ Global Di-Methyl Histone H3-K4 Quantification Kit (Cat. P-3022, EpiGentek Group Inc., NY, USA). Total histone proteins from the cerebellae and striata of 18-month-old Ubtf+/− mice and Ubtf+/+ littermates were extracted by treatment with pre-lysis, lysis, and balance buffers according to the manufacturer’s instructions. Absorbance was measured at a 450 ηm with a SpectraMax M2e microtiter plate reader.
4.9. 53BP1 immunohistochemistry
Ubtf+/− mice and Ubtf−/− littermates were overdosed with pentobarbital (> 100 mg/kg) and transcardially perfused, first with ice cold saline and then 4% paraformaldehyde in 0.1M phosphate buffer (pH 7.4). Brains were postfixed with 4% paraformaldehyde, and cryoprotected with 30% sucrose in 0.1M phosphate buffered saline (PBS). Brains were sectioned on a cryostat at 20 μm. Sections through the cerebellum were rinsed with PBS followed by blocking with 2.5% bovine serum albumin (BSA), 1% goat serum, and 0.3% Triton X-100. Sections were incubated overnight with primary antibodies for 53BP1 (ab175933; abcam, Boston, MA, USA), and calbindin D-28K (Sigma-Aldrich). The following day, slides were rinsed in PBS and incubated in secondary antibody (1:500 dilutions of goat anti-rabbit IgG conjugated to AlexaFluor488, or goat anti-mouse IgG conjugated to AlexaFluor555, Thermo-Fischer) and finally stained with DAPI (Thermo-Fischer) to label nuclei. Slides were then washed in PBS and mounted using ProLong™ Diamond antifade mountant (Thermo-Fischer). Three non-contiguous sections from the cerebellum of each mouse were analyzed for 53BP1 immunoreactive neurons. Approximately 100 DAPI+ neurons per section were assessed for 53BP1 immunoreactivity and the percentage of 53BP1 immunoreactive neurons was calculated for each mouse.
4.10. Routine histochemistry
Perfusion-fixed (4% paraformaldehyde) brains from 18-month-old Ubtf+/− mice (2 male, 2 female) and matched controls were sectioned on a cryostat in the frontal plane and processed with Luxol fast blue, cresyl violet, and H & E stains. Slides were examined for by two investigators blinded to genotype.
4.11. Brain weights
After pentobarbital overdose, mice were perfused with cold saline and then decapitated. Brains were removed and section rostrally at the frontal pole and caudally at the pyramidal decussation and weighted immediately.
5. Conclusions
Mice are not men and the study of neurodegenerative disorders including neuroregression syndromes is, at least in part, limited by the lifespan of mice. Here we have detailed cognitive, behavioral, and motor functioning during the lifespan of Ubtf+/− mice and identified marked behavioral and cognitive impairments and modest motor abnormalities, predominantly at 18 months of age. When integrated with previous molecular and cellular studies using tissues from humans with the UBTF E210K syndrome and engineered mice harboring the E210K mutation (Edvardson et al., 2017; Toro et al., 2018; Tremblay et al., 2022), our findings suggest that UBTF loss-of-function mutations are likely deleterious in humans and the human E210K variant may cause neural dysfunction through both loss- and gain-of-function mechanisms at the molecular level. Our findings and conclusions should inform clinical geneticists and provide a platform for the development of targeted therapeutics for patients the E210K and other mutations in UBTF.
Supplementary Material
Highlights.
UBTF is essential for embryogenesis and survival in adults
Ubtf+/− mice show significant deficits in spatial learning and memory
The deleterious effects of UBTF haploinsufficiency progress with age
Loss-of-function mechanisms may contribute to the UBTF E210K neuroregression syndrome
Acknowledgements
The work was supported by grants from the National Institutes of Health: R56NS123059 (Mark LeDoux), R21GM118962 (Roderick Hori and Mark LeDoux), and R03NS114616 (Mohammad Khan).
Footnotes
Declarations of interest: none
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Bastos F, Quinodoz M, Addor MC, Royer-Bertrand B, Fodstad H, Rivolta C, Poloni C, Superti-Furga A, Roulet-Perez E, Lebon S, 2020. Childhood neurodegeneration associated with a specific UBTF variant: a new case report and review of the literature. BMC Neurol. 20, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann H, Chen JL, O’Brien T, Tjian R, 1995. Coactivator and promoter-selective properties of RNA polymerase I TAFs. Science. 270, 1506–9. [DOI] [PubMed] [Google Scholar]
- Calo E, Gu B, Bowen ME, Aryan F, Zalc A, Liang J, Flynn RA, Swigut T, Chang HY, Attardi LD, Wysocka J, 2018. Tissue-selective effects of nucleolar stress and rDNA damage in developmental disorders. Nature. 554, 112–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edvardson S, Nicolae CM, Agrawal PB, Mignot C, Payne K, Prasad AN, Prasad C, Sadler L, Nava C, Mullen TE, Begtrup A, Baskin B, Powis Z, Shaag A, Keren B, Moldovan GL, Elpeleg O, 2017. Heterozygous De Novo UBTF Gain-of-Function Variant Is Associated with Neurodegeneration in Childhood. Am J Hum Genet. 101, 267–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grueneberg DA, Pablo L, Hu KQ, August P, Weng Z, Papkoff J, 2003. A functional screen in human cells identifies UBF2 as an RNA polymerase II transcription factor that enhances the beta-catenin signaling pathway. Mol Cell Biol. 23, 3936–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyori BM, Venkatachalam G, Thiagarajan PS, Hsu D, Clement MV, 2014. OpenComet: an automated tool for comet assay image analysis. Redox Biol. 2, 457–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamdane N, Stefanovsky VY, Tremblay MG, Nemeth A, Paquet E, Lessard F, Sanij E, Hannan R, Moss T, 2014. Conditional inactivation of Upstream Binding Factor reveals its epigenetic functions and the existence of a somatic nucleolar precursor body. PLoS Genetics. 10, e1004505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamdane N, Herdman C, Mars JC, Stefanovsky V, Tremblay MG, Moss T, 2015. Depletion of the cisplatin targeted HMGB-box factor UBF selectively induces p53-independent apoptotic death in transformed cells. Oncotarget. 6, 27519–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han KA, Yoon TH, Shin J, Um JW, Ko J, 2020. Differentially altered social dominance- and cooperative-like behaviors in Shank2- and Shank3-mutant mice. Mol Autism. 11, 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herdman C, Mars JC, Stefanovsky VY, Tremblay MG, Sabourin-Felix M, Lindsay H, Robinson MD, Moss T, 2017. A unique enhancer boundary complex on the mouse ribosomal RNA genes persists after loss of Rrn3 or UBF and the inactivation of RNA polymerase I transcription. PLoS Genet. 13, e1006899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda C, Kawarai T, Setoyama C, Orlacchio A, Imamura H, 2021. Recurrent de novo missense variant E210K in UBTF causes juvenile dystonia-parkinsonism. Neurol Sci. 42, 1217–1219. [DOI] [PubMed] [Google Scholar]
- Jantzen HM, Chow AM, King DS, Tjian R, 1992. Multiple domains of the RNA polymerase I activator hUBF interact with the TATA-binding protein complex hSL1 to mediate transcription. Genes Dev. 6, 1950–63. [DOI] [PubMed] [Google Scholar]
- Jiang-Xie LF, Liao HM, Chen CH, Chen YT, Ho SY, Lu DH, Lee LJ, Liou HH, Fu WM, Gau SS, 2014. Autism-associated gene Dlgap2 mutant mice demonstrate exacerbated aggressive behaviors and orbitofrontal cortex deficits. Mol Autism. 5, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan MM, Xiao J, Patel D, LeDoux MS, 2018. DNA damage and neurodegenerative phenotypes in aged Ciz1 null mice. Neurobiol Aging. 62, 180–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas E, Parisot P, Pinto-Monteiro C, de Walque R, De Vleeschouwer C, Lafontaine DL, 2016. Involvement of human ribosomal proteins in nucleolar structure and p53-dependent nucleolar stress. Nat Commun. 7, 11390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsolic I, Jurada D, Pullen N, Oren M, Eliopoulos AG, Volarevic S, 2016. The relationship between the nucleolus and cancer: Current evidence and emerging paradigms. Semin Cancer Biol. 37–38, 36–50. [DOI] [PubMed] [Google Scholar]
- Panov KI, Friedrich JK, Russell J, Zomerdijk JC, 2006. UBF activates RNA polymerase I transcription by stimulating promoter escape. Embo J. 25, 3310–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanij E, Diesch J, Lesmana A, Poortinga G, Hein N, Lidgerwood G, Cameron DP, Ellul J, Goodall GJ, Wong LH, Dhillon AS, Hamdane N, Rothblum LI, Pearson RB, Haviv I, Moss T, Hannan RD, 2015. A novel role for the Pol I transcription factor UBTF in maintaining genome stability through the regulation of highly transcribed Pol II genes. Genome Res. 25, 201–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedlackova L, Lassuthova P, Sterbova K, Haberlova J, Vyhnalkova E, Neupauerova J, Stanek D, Sediva M, Krsek P, Seeman P, 2019. UBTF Mutation Causes Complex Phenotype of Neurodegeneration and Severe Epilepsy in Childhood. Neuropediatrics. 50, 57–60. [DOI] [PubMed] [Google Scholar]
- Stefanovsky VY, Bazett-Jones DP, Pelletier G, Moss T, 1996. The DNA supercoiling architecture induced by the transcription factor xUBF requires three of its five HMG-boxes. Nucleic Acids Research. 24, 3208–3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanovsky VY, Pelletier G, Hannan R, Gagnon-Kugler T, Rothblum LI, Moss T, 2001. An immediate response of ribosomal transcription to growth factor stimulation in mammals is mediated by ERK phosphorylation of UBF. Mol Cell. 8, 1063–73. [DOI] [PubMed] [Google Scholar]
- Stefanovsky VY, Langlois F, Bazett-Jones D, Pelletier G, Moss T, 2006. ERK modulates DNA bending and enhancesome structure by phosphorylating HMG1-boxes 1 and 2 of the RNA polymerase I transcription factor UBF. Biochemistry. 45, 3626–34. [DOI] [PubMed] [Google Scholar]
- Stefanovsky VY, Moss T, 2008. The splice variants of UBF differentially regulate RNA polymerase I transcription elongation in response to ERK phosphorylation. Nucleic Acids Res. 36, 5093–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchelidze P, Kaplan H, Terryn C, Lalun N, Ploton D, Thiry M, 2019. Electron tomography reveals changes in spatial distribution of UBTF1 and UBTF2 isoforms within nucleolar components during rRNA synthesis inhibition. J Struct Biol. 208, 191–204. [DOI] [PubMed] [Google Scholar]
- Toro C, Hori RT, Malicdan MCV, Tifft CJ, Goldstein A, Gahl WA, Adams DR, Harper F, Wolfe LA, Xiao J, Khan MM, Tian J, Hope KA, Reiter LT, Tremblay MG, Moss T, Franks AL, Balak C, Group CRR, LeDoux MS, 2018. A recurrent de novo missense mutation in UBTF causes developmental neuroregression. Hum Mol Genet. 27, 691–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay MG, Sibai DS, Valere M, Mars JC, Lessard F, Hori RT, Khan MM, Stefanovsky VY, LeDoux MS, Moss T, 2022. Ribosomal DNA promoter recognition is determined in vivo by cooperation between UBTF1 and SL1 and is compromised in the UBTF-E210K neuroregression syndrome. PLoS Genet. 18, e1009644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tronche F, Kellendonk C, Kretz O, Gass P, Anlag K, Orban PC, Bock R, Klein R, Schutz G, 1999. Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nat Genet. 23, 99–103. [DOI] [PubMed] [Google Scholar]
- Watt KE, Macintosh J, Bernard G, Trainor PA, 2022. RNA Polymerases I and III in development and disease. Semin Cell Dev Biol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J, Vemula SR, Xue Y, Khan MM, Kuruvilla KP, Marquez-Lona EM, Cobb MR, LeDoux MS, 2016. Motor phenotypes and molecular networks associated with germline deficiency of Ciz1. Exp Neurol. 283, 110–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J, Vemula SR, Xue Y, Khan MM, Carlisle FA, Waite AJ, Blake DJ, Dragatsis I, Zhao Y, LeDoux MS, 2017. Role of major and brain-specific Sgce isoforms in the pathogenesis of myoclonus-dystonia syndrome. Neurobiol Dis. 98, 52–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, DeCuypere M, LeDoux MS, 2008. Abnormal motor function and dopamine neurotransmission in DYT1 DeltaGAG transgenic mice. Exp Neurol. 210, 719–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Liao WJ, Liao JM, Liao P, Lu H, 2015. Ribosomal proteins: functions beyond the ribosome. J Mol Cell Biol. 7, 92–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.