Abstract
The adenovirus E1A protein both activates and represses gene expression to promote cellular proliferation and inhibit differentiation. Here we report the identification and characterization of a cellular protein that antagonizes transcriptional activation and cellular transformation by E1A. This protein, termed CREG for cellular repressor of E1A-stimulated genes, shares limited sequence similarity with E1A and binds both the general transcription factor TBP and the tumor suppressor pRb in vitro. In transfection assays, CREG represses transcription and antagonizes 12SE1A-mediated activation of both the adenovirus E2 and cellular hsp70 promoters. CREG also antagonizes E1A-mediated transformation, as expression of CREG reduces the efficiency with which E1A and the oncogene ras cooperate to transform primary cells. Binding sites for E2F, a key transcriptional regulator of cell cycle progression, were found to be required for repression of the adenovirus E2 promoter by CREG, and CREG was shown to inhibit activation by E2F. Since both the adenovirus E1A protein and transcriptional activation by E2F function to promote cellular proliferation, the results presented here suggest that CREG activity may contribute to the transcriptional control of cell growth and differentiation.
Studies of the transforming proteins of small DNA tumor viruses, such as adenovirus E1A, simian virus 40 large tumor antigen, or human papillomavirus E7, have revealed a great deal about the proteins and pathways that regulate cellular proliferation. In normal cells, the transition from G1 to S phase and the start of DNA synthesis is tightly controlled by mechanisms that include transcriptional regulation of genes encoding proteins required in the S phase. In many cell types, the adenovirus E1A protein dramatically alters the transcriptional program of the host cell to stimulate cell division and inhibit differentiation. The ability of E1A to reprogram cellular gene expression to promote entry into S phase correlates with the ability of E1A to cooperate with oncogenes, such as ras, to transform primary cells (38, 62).
The protein products of both the 12S and 13S mRNA forms of E1A (12SE1A and 13SE1A, respectively) regulate the expression of a number of viral and cellular genes. Although 13SEIA has a unique transcriptional activation domain encoded by CR3, the sequences present in 12SE1A are sufficient to mediate cellular transformation. Investigations into the mechanisms by which E1A activates and represses expression of particular genes have revealed that 12SE1A interacts with several transcriptional regulators of cell proliferation, including the retinoblastoma tumor suppressor protein, pRb, and the coactivators p300 and CBP. Two conserved regions of E1A, CR1 and CR2, have been shown to mediate binding to pRb, and CR1 also participates in binding to p300 (14, 71). The functional importance of these interactions is supported by the observation that mutations in CR1 and CR2 result in E1A proteins defective in transcriptional regulation and cellular transformation (62, 71). These same regions of E1A have also been implicated in interactions with other cellular transcription factors, such as TATA-binding protein (TBP), raising the possibility that transcriptional regulation and cellular transformation by E1A may involve additional mechanisms (22, 61).
12SE1A has been observed to activate transcription through several different response elements, including both sequences in the core promoter and binding sites for specific regulatory proteins. The binding site for the cell cycle-regulated transcription factor E2F is an E1A response element that was first identified in the adenoviral E2 (AdE2) promoter (49). A variety of other E1A response elements have been identified in the E1A-stimulated hsp70, PCNA, and the adeno-associated virus P5 promoters (32, 36, 50, 54). Multiple sequence elements in the hsp70 promoter have been implicated in the response to E1A, including the TATA box and the CAAT box (43, 55, 72). Transcriptional stimulation of the AdE2 and hsp70 promoters by 12SE1A has therefore been thought to involve distinct mechanisms and, in fact, these promoters have been shown to respond differently to some E1A mutants (35). It is clear, however, that E1A employs multiple mechanisms to regulate gene expression, and it remains possible that some common mechanisms may be involved in the activation of these disparate promoters.
Although initially identified as an E1A response element, binding sites for E2F have been shown to be important for the regulated transcription of many genes whose products contribute to cell cycle progression or DNA synthesis. In mammalian cells, E2F activity is composed of at least five E2F family proteins and two DP subunits that form E2F-DP heterodimers, whose activity is highly regulated during the cell cycle (reviewed in reference 56). Overexpression of E2F in cell culture leads to increased cell proliferation, often accompanied by apoptosis, which is dependent on the transcriptional activation function (for a review, see reference 1 and references therein). These observations have been confirmed in animal studies demonstrating that regulated E2F activity is critical for normal cell cycle progression, cell survival, and possibly differentiation in vivo (5, 8, 12, 13, 17, 73). The transcriptional activity of the E2F proteins is regulated by association with the retinoblastoma protein, pRb, and the related p107 and p130 proteins. pRb not only inhibits activation by E2F, but the E2F-pRb complex also functions to repress transcription from other activators bound at the promoter (70). E2F activity is also regulated at other levels, including expression, nuclear localization, DNA binding, and protein stability (3, 21, 27, 39, 47, 56, 68). The molecular mechanisms that ensure proper regulation of the different E2F family proteins during cell proliferation and differentiation are complex and not fully understood.
We have identified a human cellular repressor of E1A-stimulated genes, designated hCREG, that shares limited sequence similarity with E1A and binds both the general transcription factor TBP and the tumor suppressor pRb in vitro. When tethered to a promoter by fusion to a heterologous DNA binding domain, hCREG represses transcription. In transient-transfection assays, hCREG was found to repress transcription and antagonize the ability of adenovirus E1A to stimulate the AdE2 and hsp70 promoters. Expression of hCREG also reduces the ability of E1A to cooperate with the oncogene ras in the transformation of primary cells. Analysis of mutant derivatives of the AdE2 promoter revealed that binding sites for the cell cycle regulator E2F constitute one target of hCREG-mediated repression. Analysis of CREG activity on several different E2F-regulated promoters and Gal4E2F fusion proteins indicates that hCREG functions by inhibiting the transcriptional activation function of E2F. The results presented here suggest that hCREG may contribute to the transcriptional control of cell growth by repression of specific activators such as E2F.
MATERIALS AND METHODS
Plasmids.
Fragments of 0.7 kb containing the hCREG open reading frame were inserted into the HindIII and XbaI sites of Rc/CMV (Invitrogen) to give CMVhCREG, the EcoRI and XbaI sites of pGEX4T-1 (Pharmacia Biotech) to give pGEX+hCREG, into the EcoRI and XbaI sites of pSG424 (53) to give pSG424+hCREG, and into pTβSTOP for in vitro transcription and translation. All other Gal4(1–147) fusion proteins have been previously described: pSG147 and pSGVP (53); Gal4+E2F1 expresses a Gal4(1–147)+E2F1(aa284–437) fusion protein; Gal4+E2F1(Δ417–437) expresses a Gal4(1–147)+E2F1(aa284–417) fusion protein, and Gal4+E2F1(Y411C) expresses a Gal4(1–147)+E2F1(aa284–437) fusion protein with a single amino acid change from tyrosine to cystine at position 411 (24); pJ3-Gal4-E2F4 and pJ3-Gal4-E2F5 express Gal4+E2F4(aa276–412) and Gal4+E2F5(aa222–346) fusion proteins (25).
The reporters used in transfection assays have all been previously described: Gal4TkCAT (54); pBLCAT2 (42); G5luc, which contains the luciferase gene under the control of the minimal E1B TATA with five Gal4 binding sites upstream (23); pE2w.t.CAT and the mutants (−80−70)E2CAT and (−64−60)(−45−36)E2CAT (41); pMaeWTDHFR contains the wild-type dihydrofolate reductase (DHFR) promoter, and pMaeNWDHFR contains a mutant DHFR promoter in which the E2F sites have been disrupted (45); pGL2-(−536) contains the wild-type b-myb promoter, and pGL2-(−536)mut contains a mutant b-myb promoter in which the E2F site has been disrupted (37); pGL2AN contains the E2F1 promoter and the pGL2AN 5′-3′ mutant contains an E2F1 promoter mutant in which the E2F sites have been disrupted (48); and pHC1170 contains the hsp70 promoter (55).
GST-Rb, GST-Rb(379–792), pRb, and pRbΔ22 expression constructs were provided by Bill Kaelin (Dana Farber Cancer Institute). CMV12SE1A and GST-12SE1A plasmids were provided by Yang Shi (Harvard Medical School).
Expression plasmids used in the baby rat kidney (BRK) assay were 13S-SVE expressing the adenovirus type 5 13S cDNA from the simian virus 40 early promoter and pucEJRas containing an oncogenic allele of Ha-ras under the control of the EJ promoter (26).
Analysis of protein interactions in vitro.
Glutathione S-transferase (GST) and GST fusion proteins were purified from DH5 cells with glutathione-Sepharose 4B (Pharmacia) beads. 35S-methionine-labeled proteins were generated by in vitro transcription and translation with the Promega TNT reticulocyte lysate kit and then diluted with NETN (0.5% Nonidet P-40, 20 mM Tris-HCl [pH 8.0], 100 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol [DTT]). A 20-μl portion of diluted in vitro-translated protein was reserved as input, and 200 μl was combined with 20 μl of GST slurry (1:1). Binding reactions were carried out with gentle rotation at 4°C for 1 h, after which the beads were pelleted. The beads were washed four or five times with NETN, and bound protein was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and visualized by autoradiography. Whole-cell lysate (1 mg) prepared in ELB (250 mM NaCl, 50 mM HEPES, 0.1% Nonidet P-40, 1 mM DTT, 1 mM EDTA, 0.4 mM phenylmethylsulfonyl fluoride [PMSF]) from a p107-overexpressing stable cell line, U2OS-p107 (74), was used in binding reactions as described above. Bound proteins were analyzed by Western blotting with mouse anti-p107 (a gift from N. Dyson).
Transfections and reporter assays.
CV-1 monkey kidney cells were seeded onto 10- or 6-cm plates 24 to 30 h before transfection. Medium was replaced 1 to 3 h prior to transfection. DNA (10 μg/10-cm plate or 5 μg/6-cm plate) was precipitated by the calcium phosphate method and spread over the cells. At 16 to 20 h after transfection, the medium was removed, and the plates were washed once with phosphate-buffered saline (PBS); then fresh medium was added to each plate. For transcription assays, cells were harvested 36 to 44 h after transfection, by which time the plates were up to 95% confluent. For luciferase assays, cells were washed twice with PBS and then harvested into 200 μl of lysis buffer (25 mM Tris-PO4, 15% [vol/vol] glycerol, 2% [wt/vol] CHAPS (3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate), 1% [wt/vol] lecithin, 1% [wt/vol] bovine serum albumin, 4 mM EGTA, 8 mM MgCl2, 1 mM DTT, 0.4 mM PMSF). A luminometer was used to inject 1 mM d-luciferin into 300 μl of luciferase assay buffer (25 mM glycylglycine, 15 mM MgSO4, 15 mM potassium phosphate [pH 7.8], 4 mM EGTA, 1 mM DTT, 1 mM ATP), to which 20 μl of cell lysate had just been added. The luminescence over 20 s was then recorded as the luciferase activity. Chloramphenicol acetyltransferase (CAT) assays were carried out as previously described (58).
Preparation of BRK cells and BRK transformation assay.
BRK cells were prepared by dissociation of 5- to 6-day-old Sprague-Dawley rat kidneys with trypsin and plated out at 4 × 105 to 6 × 105 cells per 6-cm plate in Dulbecco modified Eagle medium that included 5 mM penicillin-streptomycin and 10% (vol/vol) fetal calf serum.
At 2 to 3 days after preparation, cells were transfected with a total of 10 μg of DNA per plate containing 3 μg of SV13SE1A, 2 μg of EJ-Ras and either 5 μg of CMVhCREG or 5 μg of carrier DNA. At 16 to 20 h after transfection, the medium was discarded and each plate was rinsed four times with 2 ml of PBS before the addition of fresh medium. At 48 h after transfection the medium was removed and replaced with fresh medium containing 5% (vol/vol) fetal calf serum, and at 14 days after transfection the foci were counted on each plate.
RESULTS
Cloning of hCREG.
In an attempt to identify novel transcriptional regulators, a Drosophila cDNA library was screened by the yeast two-hybrid method for proteins that interact with the Drosophila TBP. A novel protein, dCREG, has been identified in this screen; the identification and characterization of dCREG will be described elsewhere. Sequence analysis of dCREG revealed it to share amino acid sequence similarity with the regions of the adenovirus E1A protein, CR1 and CR2, that have been shown to be important for the transcriptional and transforming functions of this viral oncoprotein (Fig. 1B). Sequences in CR1 mediate binding to both pRb and p300 family proteins, and the pRb binding domain in CR2 contains an LXCXE motif found in many pRb-binding proteins (64, 71). A partial cDNA capable of encoding a human homolog was identified in the GenBank human EST sequence database. The cloning and characterization of this protein, human CREG (hCREG), are described here.
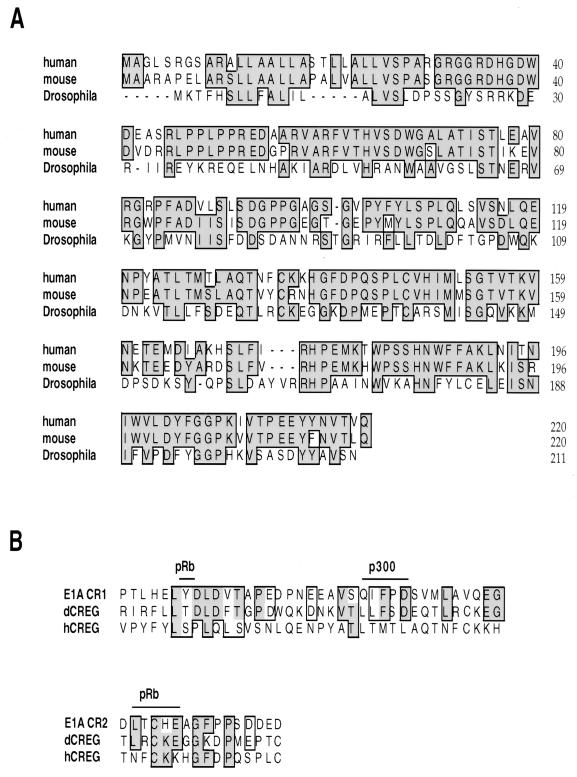
FIG. 1.
Amino acid sequence of human CREG. (A) The amino acid sequence of human CREG aligned with the sequences of the mouse and Drosophila CREG proteins. The human and Drosophila proteins are 31% identical; the human and mouse CREG sequences are 77% identical. (B) Alignment of human and Drosophila CREG sequences with conserved regions of the adenovirus E1A protein. CR1 E1A(41–77) and CR2 E1A(121–136) are shown. Positions at which mutations in E1A disrupt binding to pRb and p300 are indicated. Identical amino acids are shaded and boxed, and similar amino acids are boxed.
A 2.0-kb hCREG cDNA was cloned from a HeLa cDNA library. This cDNA contained a 660-base open reading frame followed by approximately 1.2 kb of 3′ untranslated region (UTR). Northern blot analysis revealed that hCREG mRNA is widely expressed in adult human tissues (data not shown). As shown in Fig. 1A, the CREG protein sequence is well conserved between species; the predicted human CREG protein is 31% identical and 55% similar to the Drosophila CREG. We have also identified a murine CREG homolog that is 77% identical to the human protein (Fig. 1A). Although CREG is well conserved across species, the human and mouse CREG homologs are less similar to E1A. Human CREG shows some similarities with E1A CR1, particularly in the region implicated in pRb binding; however, this protein lacks the LXCXE motif that is critical for CR2 function (11). Despite this limited sequence similarity, we have found that hCREG regulates expression from several E1A-responsive genes (see below).
hCREG binds TBP, pRb, p107, and p130 in vitro.
Since Drosophila CREG was cloned as a TBP-binding protein, the ability of the human homolog to interact with TBP was investigated. As shown in Fig. 2A, in vitro-translated TBP bound to a GST-hCREG fusion protein but not to GST alone. The hCREG interaction with TBP was not reduced by the presence of ethidium bromide in the reaction, indicating that this interaction is not mediated by DNA (data not shown).
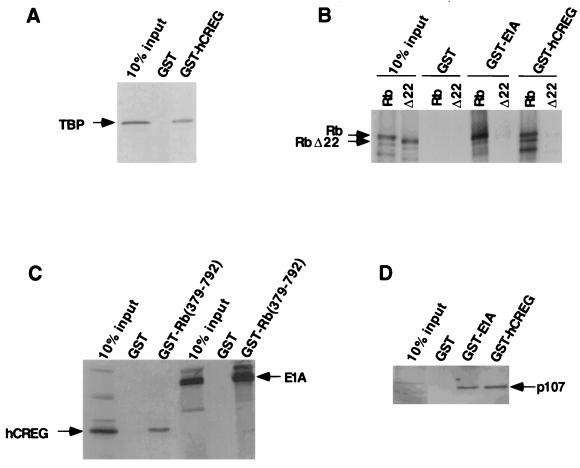
FIG. 2.
hCREG interacts with the transcriptional regulators TBP, pRb, and p107. (A) hCREG binds TBP in vitro. In vitro-translated, 35S-methionine-labeled, full-length human TBP bound to GST-hCREG but not to GST alone. (B and C) Like E1A, hCREG binds pRb in vitro, and the pocket domain of pRb is necessary for this interaction. (B) Binding reactions were carried out between GST-hCREG or GST-12SE1A and in vitro-translated, 35S-methionine-labeled, full-length human pRb or the mutant pRbΔ22. (C) GST-pRb(379–792) bound in vitro-translated hCREG and E1A. Binding reactions were carried out between GST-Rb(379–792) and in vitro-translated, 35S-methionine-labeled, hCREG or 12SE1A. (D) GST-hCREG binds p107 in an extract from p107-overexpressing U2OS cells.
Guided by the sequence similarity with E1A, we had previously shown that dCREG interacts with RBF, the Drosophila homolog of the retinoblastoma protein, both in vitro and in vivo (to be described elsewhere). Therefore, in vitro binding assays were carried out to determine whether hCREG was able to interact with the human retinoblastoma protein pRb. As shown in Fig. 2B, pRb bound GST-hCREG in vitro. The central domain of pRb, often called the “pocket”, is required for binding viral oncoproteins, such as E1A, and is also necessary for Rb-mediated growth arrest (64). The pocket of Rb is necessary and sufficient for hCREG binding, since hCREG binds Rb(379–792), which contains only the pocket, and does not bind Δ22, a tumor-derived Rb mutant from which the pocket is absent (Fig. 2B and C) (28). The pattern and extent of binding observed with hCREG are similar to those observed with E1A in these experiments. The GST-hCREG fusion was also used in affinity chromatography experiments with mammalian cell lysates. Western blotting analysis revealed that hCREG also bound the pRb-related p107 and p130 proteins from cell lysates (Fig. 2D and data not shown). Consistent with the observation that hCREG binds pRb and related proteins in vitro, we have shown that hCREG is able to regulate transcription from some pRb-responsive promoters (see below).
hCREG represses transcription when tethered to the promoter.
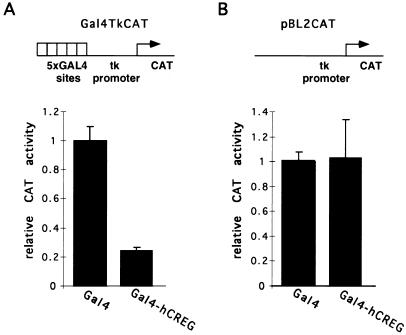
Analysis of the amino acid sequence of hCREG did not reveal the presence of any sequences characteristic of known DNA-binding domains. In order to determine whether hCREG affects transcriptional activity when tethered to the promoter, the entire hCREG open reading frame was fused in frame to the cDNA encoding the DNA binding domain of GAL4, Gal4(1–147). CV-1 cells were cotransfected with a plasmid expressing the Gal4-hCREG fusion and a reporter plasmid, Gal4TkCAT, that contains five Gal4 binding sites 105 bases upstream of the herpes simplex virus thymidine kinase (tk) promoter. As shown in Fig. 3, the Gal4-hCREG fusion lowered expression from this promoter by fivefold relative to Gal4(1–147). Similar results were obtained when this experiment was carried out in HeLa or U2OS cells (data not shown). The observed fivefold repression is comparable to the level seen when other repressors, e.g., pRb, are tethered to this promoter (2, 7). Gal4-hCREG had no effect on expression from the TkCAT reporter, which lacks Gal4 binding sites (Fig. 3B). It is therefore unlikely that Gal4-hCREG reduces CAT activity via a nonspecific, global effect on transcription, translation, or cell viability. Instead, these data support the conclusion that, when tethered to the promoter, hCREG functions as a transcriptional repressor.
FIG. 3.
hCREG represses transcription when tethered to a promoter by a heterologous DNA binding domain. (A) A Gal4-hCREG fusion represses transcription from a promoter bearing Gal4 binding sites. CV-1 cells on 10-cm plates were transfected with 5 μg of Gal4TkCAT, a reporter plasmid bearing Gal4 binding sites upstream of the HSVtk promoter; 1 μg of pSG147 expressing Gal4(1–147); or 1 μg of pSG424+hCREG expressing Gal4-hCREG. Each group of CV-1 cells was also transfected with 4 μg of carrier DNA. The CAT activity observed with Gal4-hCREG is shown relative to the CAT activity with Gal4(1–147), which was taken as 100%. (B) Repression by Gal4-hCREG is dependent on the presence of Gal4 binding sites in the reporter. CV-1 cells were transiently transfected as described above but with pBL2CAT, a reporter plasmid expressing CAT under the control of the tk promoter. The experiment was performed in triplicate more than three times; results from a representative experiment are shown. Error bars represent the standard deviation of the mean.
CREG functions antagonistically to E1A to repress transcription from the AdE2 promoter.
Having established that hCREG can repress transcription when tethered to the promoter, we considered the possibility that hCREG may repress particular target promoters. In vitro binding studies demonstrated that hCREG can bind the E1A-binding proteins TBP and pRb. We therefore investigated whether CREG was able to regulate the expression of any E1A-responsive promoters. E1A stimulates transcription of the other early adenoviral genes including AdE2. 12SE1A activates the AdE2 promoter through a direct interaction with pRb (and the related p107 and p130 proteins), thereby relieving pRb-mediated repression of E2F (reviewed in reference 49).
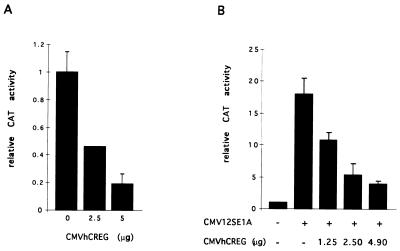
To investigate whether hCREG can regulate the AdE2 promoter, CV-1 cells were cotransfected with a CAT reporter plasmid containing the AdE2 promoter and an expression plasmid containing hCREG under the control of the CMV promoter. A four- to sixfold repression of the AdE2 promoter was observed in cells transfected with CMVhCREG compared with cells transfected with a control CMV plasmid. Repression of the AdE2 promoter showed a dose-dependent response to the amount of CMVhCREG DNA added (Fig. 4A). Repression by hCREG depends on specific sequences in the AdE2 promoter (see below).
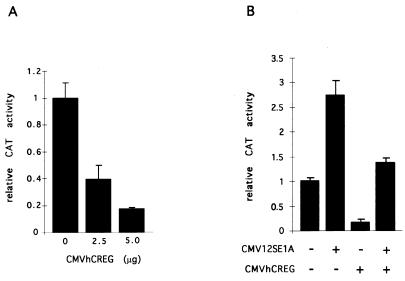
FIG. 4.
hCREG and E1A have opposing activities on the AdE2 promoter. (A) hCREG represses expression from the AdE2 promoter. CV-1 cells on 10-cm plates were transfected with 10 μg of DNA, of which 5 μg was the pE2w.t.CAT reporter plasmid. Cells were also transfected with increasing amounts of CMVhCREG DNA, as indicated, which was made up to a total of 5 μg of expression plasmid with CMVβgal. (B) hCREG abrogates E1A-mediated activation of AdE2. CV-1 cells on 10-cm plates were transfected with 10 μg of DNA, of which 5 μg was the pE2w.t.CAT reporter plasmid. Cells were also transfected with 50 ng of the E1A expression vector CMV12SE1A, 4.95 μg of CMVhCREG, or the control plasmid (CMVβgal), as indicated in the figure. The experiment was carried out in triplicate and repeated at least three times. Data are from a representative experiment, and the error bars indicate the standard deviation.
Since hCREG was found to repress the E1A-stimulated AdE2 promoter, the effects of cotransfecting hCREG and E1A on AdE2 CAT expression were determined. As shown in Fig. 4B, hCREG and E1A have mutually opposing effects on expression from the AdE2 promoter; as expected, 12SE1A stimulated expression, but this increase was not observed in the presence of CMVhCREG. Similarly, E1A relieves CREG-mediated repression. The ability of hCREG to inhibit the activation of the AdE2 promoter by E1A was not due to reduced expression of E1A, as cotransfection of CREG did not significantly reduce the level of E1A protein detected on a Western blot of transfected cells (data not shown). The abrogation of E1A activation of AdE2 by hCREG is consistent with the hypothesis that CREG and E1A have opposing effects on the expression of a common set of target genes.
hCREG abrogates E1A-mediated activation of the hsp70 promoter.
Having established that hCREG abrogates E1A-mediated activation of the AdE2 promoter, we investigated whether hCREG also repressed the expression of an E1A-stimulated promoter lacking E2F sites. For this purpose, we chose the hsp70 promoter, a cellular gene whose expression is stimulated by E1A. Although many sequence elements in the hsp70 promoter have been implicated in the response to E1A, including the CAAT box and the TATA box, they are distinct from the E1A response elements in AdE2 (43, 55, 72). In order to examine the effect of hCREG on expression from the hsp70 promoter, CV-1 cells were cotransfected with a reporter expressing CAT under the control of the hsp70 promoter and CMVhCREG or a control CMV plasmid. In these experiments, hCREG was observed to repress the activity of the hsp70 promoter four- to sixfold (Fig. 5A). Thus, the hsp70 promoter is also a target for hCREG-mediated repression. In an experiment similar to that described above, the ability of E1A to stimulate expression from the hsp70 promoter was severely impaired, in a dose-dependent manner, by cotransfection with hCREG (Fig. 5B). This experiment also revealed the ability of E1A to relieve CREG-mediated repression of the hsp70 promoter. Thus, hCREG and E1A have opposing effects on transcription from two dissimilar promoters.
FIG. 5.
hCREG represses the E1A-stimulated hsp70 promoter. (A) hCREG represses the hsp70 promoter. CV-1 cells on 10-cm plates were transfected with a total of 10 μg of DNA, of which 5 μg was pHC1170, a CAT reporter plasmid containing the hsp70 promoter (55). Cells were also transfected with increasing amounts of CMVhCREG DNA as indicated. (B) hCREG antagonizes activation by E1A. Cells were transfected as described above with the addition of 100 ng of CMV12SE1A where indicated (+). The total amount of expression plasmid was brought to 5 μg per plate with CMVβgal. These experiments were performed in triplicate and repeated at least three times. Representative experiments are shown, and the error bars indicate the standard deviation.
hCREG interferes with the ability of E1A and ras to transform BRK cells.
E1A, together with a cooperating oncogene such as activated ras, will transform primary BRK cells, giving rise to foci of proliferating cells that are no longer contact inhibited (52). Since hCREG was found to antagonize E1A-mediated activation of both the AdE2 and hsp70 promoters, we tested whether hCREG would also antagonize the transforming activity of E1A. BRK cells were prepared and transfected with vectors expressing 13SE1A and an oncogenic allele of Ha-Ras (hereafter referred to as Ras) or 13SE1A, Ras, and hCREG. Foci of rapidly dividing cells were counted 14 days after transfection. Although the average number of foci per plate varied between experiments, cotransfection with CMVhCREG reproducibly reduced the number of foci per plate (Table 1). Overall, expression of hCREG was observed to lower the transformation efficiency by approximately threefold. A threefold reduction in the number of foci is similar to the level of inhibition observed upon cotransfection of CREB binding protein (CBP) with E1A and E1B (59). In a separate experiment, cotransfection of hCREG was not found to detectably alter the levels of E1A and Ras expression as determined by Western blot analysis (data not shown). No foci were ever observed on plates transfected with E1A, Ras, or hCREG alone or on plates cotransfected with hCREG and Ras, indicating that hCREG displays no oncogenic activity in this assay. hCREG’s ability to interfere with the combined oncogenic activity of E1A and Ras suggests that CREG inhibits cell proliferation, presumably through its ability to repress the expression of cellular genes required for immortalization.
TABLE 1.
hCREG inhibits the transformation of BRK cells by E1A and Rasa
| Expt | No. of foci/plate with:
|
|
|---|---|---|
| E1A + Ras | E1A + Ras + hCREG | |
| 1 | 20 | 6.0 |
| 2 | 21 | 3.0 |
| 3 | 4.0 | 1.3 |
| 4 | 3.0 | 1.5 |
| 5 | 3.5 | 1.0 |
| 6 | 4.5 | 1.3 |
| 7 | 3.4 | 4.4 |
| 8 | 7.8 | 4.5 |
| 9 | 5.3 | 1.0 |
BRK cells were prepared and transfected as described in the Materials and Methods. Foci on each plate were counted macroscopically and confirmed under the microscope 14 days after transfection. The table summarizes the data from nine independent transfections carried out with BRK cells prepared on six separate days.
CREG repression of the AdE2 promoter is E2F site dependent.
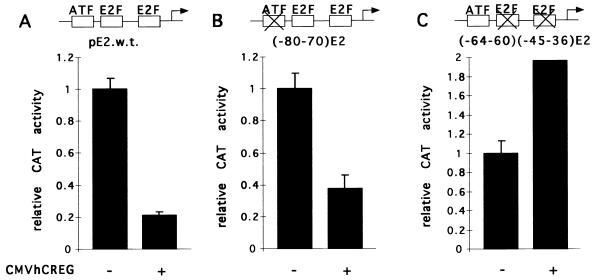
To address the mechanisms by which hCREG acts to antagonize some of the transcriptional and transforming activities of E1A, we wished to determine if hCREG mediates repression through specific promoter elements and, if so, if these correspond to E1A response elements. The AdE2 promoter is somewhat simpler than the large hsp70 promoter fragment used in these studies, and the E1A response elements of the AdE2 promoter have been well defined. This promoter contains two E2F binding sites and an ATF site through which E1A is able to stimulate expression (Fig. 6A) (41). Activation through the ATF site requires E1A CR3, which is unique to the 13SE1A product (40). Sequences in CR1 and CR2 of E1A, present in both the 12S and 13S gene products, participate in activation through the E2F sites. By using AdE2 promoters with mutations in the known E1A response elements, the ATF or E2F sites, the contribution of these sequence elements to repression by hCREG was determined. The ATF and E2F sites both contribute to the expression of AdE2; however, these sites respond differently to transfection with CREG. The mutant promoter with a disruption of the ATF site [pE2(−80/−70)CAT] was still repressed by cotransfection with CMVhCREG (Fig. 6B). In contrast, hCREG was unable to repress the activity of the AdE2 mutant reporter in which both E2F sites have been disrupted, i.e., the pE2(−64/−60, −45/−36)CAT reporter (Fig. 6C). Curiously, the activity of this mutant reporter, lacking E2F sites, was slightly elevated in cells cotransfected with CMVhCREG. Thus, hCREG repression of the AdE2 promoter is mediated through specific upstream promoter elements, the E2F sites, that also support activation by E1A.
FIG. 6.
hCREG repression of the AdE2 promoter is E2F site dependent. (A) The wild-type AdE2 promoter contains three E1A-responsive sites: an ATF site and two E2F sites. Two previously characterized mutant promoters—E2(−80−70), in which the ATF site (B) had been disrupted, and E2(−64−60)(−45−36), in which the E2F sites (C) had been disrupted—were also used (41). CV-1 cells on 10-cm plates were cotransfected with either 5 μg of wild-type or mutant AdE2 CAT reporter and either 5 μg of CMVhCREG(+) or 5 μg of control plasmid CMVβgal(−) DNA. (A) hCREG represses the wild-type AdE2 promoter. (B) hCREG represses the (−80−70)AdE2 promoter. (C) hCREG does not repress the (−64−60)(−45−36)AdE2 promoter. Relative CAT activities are shown with the activity of each reporter with the control effector plasmid (CMVβgal) taken to be 1. Experiments were performed at least three times. The data shown in all three panels are from a single representative experiment, and the error bars indicate the standard deviation.
E2F site-dependent repression by CREG is context dependent.
Since hCREG-mediated repression of AdE2 is E2F site dependent, the effect of hCREG on the expression from cellular E2F-regulated promoters was examined. E2F sites have been shown to be important for the cell cycle-regulated expression of many cellular genes whose products are involved in cell cycle progression or DNA synthesis, including the DHFR, b-myb, and E2F1 genes (6, 29, 31, 37, 48, 57). Transcriptional regulation by E2F appears to involve both repression by complexes between E2F and members of the pRb protein family during G0-early G1 phase and activation by E2F at G1-S. The E2F sites in the DHFR promoter, for example, contribute to activation at the G1-S phase (6, 57). In the b-myb and E2F1 promoters, however, the E2F sites have been implicated in transcriptional repression during G0-early G1 (29, 31, 37, 48). We have examined the ability of CREG to regulate the expression of promoters subject to both E2F site-dependent activation and repression.
Upon cotransfection into CV-1 cells, hCREG was found to repress the activity of the mouse DHFR promoter 4.4-fold (Table 2). In the absence of functional E2F sites in the DHFR promoter, cotransfection with CMVhCREG resulted in a twofold reduction of DHFR promoter-driven luciferase activity. Thus, maximal hCREG-mediated repression of DHFR, like AdE2, requires the E2F site(s). Additional elements in the DHFR promoter appear to contribute to the hCREG response, however, since twofold repression of the DHFR promoter was observed in the absence of E2F sites in marked contrast to the AdE2 promoter (Fig. 6C). When CMVhCREG was cotransfected into CV-1 cells with either a luciferase reporter containing the b-myb or the E2F1 promoters only a 2- to 2.5-fold decrease in luciferase activity was observed (Table 2). This moderate repression was also observed in the absence of functional E2F sites, suggesting that in the b-myb and E2F1 promoters the E2F sites are not sufficient to confer a response to hCREG. The sequence elements in these promoters that confer the twofold response to hCREG have not been determined. Thus, although maximal hCREG-mediated repression of the AdE2 and DHFR promoters is E2F site dependent, these experiments suggest that the ability of an E2F site to support hCREG-mediated repression is context dependent.
TABLE 2.
hCREG-mediated repression of E2F sites is context dependenta
| Promoter | Fold repression (± SEM) by hCREG in:
|
|
|---|---|---|
| Wild type | Mutant | |
| DHFR | 4.4 ± 0.1 | 1.9 ± 0.1 |
| E2F1 | 2.5 ± 0.1 | 2.0 ± 0.3 |
| b-myb | 2.2 ± 0.1 | 1.9 ± 0.1 |
The table presents the fold repression observed upon cotransfection with CMVhCREG relative to transfection with a control plasmid. For each promoter, “mutant” refers to the disruption of the E2F site(s). CV-1 cells on 6-cm plates were cotransfected with 2.5 μg of luciferase reporter and 2.5 μg of CMVhCREG, and the fold repression by hCREG was measured relative to the luciferase activity of the reporter with 2.5 μg of control DNA (CMVβgal). The activity of the DHFR mutant promoter was threefold lower than that of the wild type. The activity of the mutant b-myb promoter was twofold higher than that of the wild type, and the E2F1 mutant and wild-type promoters had comparable activities, a finding consistent with previous reports (37, 48). The values are the average from at least three experiments.
CREG is able to repress activation by Gal4E2F1, Gal4E2F4, or Gal4E2F5.
The E2F site-dependent repression of transcription by hCREG observed for the AdE2 and DHFR promoters suggests that hCREG is able to specifically repress transcriptional activation by E2F. E2F activity in mammalian cells results from heterodimers of at least five different E2F family members with two DP proteins. The E2F proteins share a similar overall structure, with domains for sequence-specific DNA binding and dimerization located towards the amino-terminal half and sequences required for transcriptional activation and binding to the pRb family of repressors at the C terminus (56). Although the different E2F family proteins have overlapping activities in many in vitro and overexpression assays, specific E2F family members are subject to differential regulation of expression, subcellular localization, and complex formation (10, 39, 47, 56, 68).
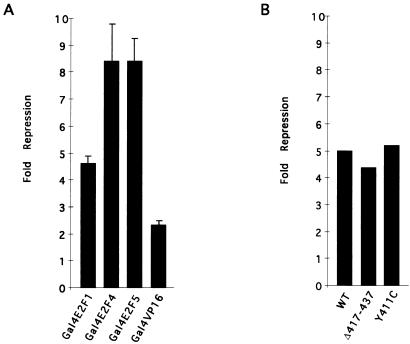
In order to shed light upon the mechanism of E2F site-dependent repression by hCREG, experiments were performed to investigate whether hCREG repressed stimulation by hybrid activators consisting of the C-terminal activation domain of E2F1, E2F4, or E2F5 fused to the Gal4(1–147) DNA-binding domain. Gal4E2F1, Gal4E2F4, or Gal4E2F5 each strongly activated transcription of the G5luc reporter (at least 100-fold; data not shown). Cotransfection of CMVhCREG strongly repressed activation by both the Gal4E2F4 and Gal4E2F5 fusions as shown in Fig. 7A; these activators were repressed eight- to ninefold by hCREG. Cotransfection of CMVhCREG with Gal4E2F1 resulted in four- to fivefold repression. This is comparable to the magnitude of repression observed when Gal4E2F1 was cotransfected with Rb in Rb-deficient cells (24). In contrast, activation by Gal4VP16 was only reduced twofold, indicating that hCREG acts preferentially to inhibit the activity of E2F. Expression of the Gal4+E2F fusions was not reduced by cotransfection with CMV-hCREG, as revealed by immunoblot analysis (data not shown). Since the Gal4E2F fusion proteins lack the E2F DNA binding and dimerization domains, these data indicate that hCREG does not repress E2F site-dependent transcription by interfering with the ability of E2F to bind its cognate binding site.
FIG. 7.
hCREG represses the activation function of E2F. (A) hCREG represses activation by E2F1, E2F4, and E2F5. CV-1 cells on 6-cm plates were cotransfected with 2.5 μg of G5 luciferase, a reporter plasmid containing a minimal promoter (E1B TATA) and five binding sites for the yeast transactivator Gal4; 10 ng of expression vector for the indicated Gal4(1–147) fusion protein; and 2.5 μg of CMVhCREG or control plasmid (CMVβgal). Each Gal4-activation domain fusion protein activated transcription by more than 100-fold. The figure shows the average fold repression from at least three independent experiments performed in triplicate, and the error bars indicate the standard error of the mean. (B) hCREG represses activation by Gal4E2F1 mutants that are unable to bind to Rb. Experiments were carried out as in panel A with the indicated Gal4E2F1 fusions. WT contains the wild-type E2F1 activation domain; Δ417–437 contains a deletion and Y411C contains a point mutation in the activation domain, both of which eliminate Rb binding in vitro (24). The experiment was performed in triplicate and repeated twice. The figure shows the fold repression from a single representative experiment.
The retinoblastoma protein, which interacts with hCREG in in vitro binding assays (Fig. 2), represses E2F-dependent transcription through binding to specific sequences in the activation domain. We have been unable to detect an interaction in in vitro binding assays between E2F1 and hCREG (15). Mutants of E2F1 that fail to bind pRb but retain the activation function have been described (24). As shown in Fig. 7B, cotransfection with hCREG represses these Gal4+E2F1 mutants as efficiently as did the wild type. These data show that pRb binding by E2F is not required for repression by CREG and thus rule out the simple model whereby pRb serves as a bridge for recruiting the hCREG repressor to E2F. Consistent with the context dependence of E2F site-dependent repression, these data suggest that, rather than augmenting the repression activity associated with E2F-pRb complexes, CREG acts to inhibit the activation function of E2F.
DISCUSSION
We report here the identification of a novel cellular protein, hCREG, that antagonizes the transcriptional activation and cellular transformation activities of the adenovirus E1A oncoprotein. These studies suggest that, in addition to the other known activities of E1A, its ability to functionally antagonize hCREG-mediated repression may contribute to the transcriptional and transforming properties of this viral oncoprotein. Cotransfection of hCREG was found to inhibit the ability of E1A and Ras to cooperate in the oncogenic transformation of primary cells. Other cellular proteins such as p300, CBP, and p53, that have been shown to inhibit transformation in this assay have clear and dramatic effects on cell growth and/or differentiation (18, 59). Transcriptional activation by E2F, an important regulator of cell cycle progression, is specifically repressed by hCREG. The complete inability of hCREG to repress transcription from a mutant AdE2 promoter lacking functional E2F sites indicates that repression is specific for certain target promoters and not due to global inhibitory effects on transcription, translation, or cell viability. Although understanding the full biological activity of hCREG will require additional studies, the data presented here suggest that the normal role of CREG may be to inhibit proliferation and/or promote differentiation.
The adenovirus E1A protein utilizes multiple mechanisms to regulate gene expression. The ability of E1A to repress transcription of many genes implicated in terminal differentiation has largely been correlated with binding to p300 and CBP, although interactions with additional proteins, including TBP and promoter-specific activators, may also be involved (60–63, 69). Interestingly, CREG shares sequence similarity with E1A CR1, which is required for E1A-mediated repression. We have so far failed to observe an interaction between hCREG and p300 or CBP, and the ability of hCREG to regulate transcription of promoters repressed by E1A has not been extensively studied to date (16). The best-understood mechanism of transcriptional activation by 12SE1A is through a direct interaction with the tumor suppressor pRb and the related proteins p107 and p130; for example, E1A binding to pRb relieves pRb-mediated repression of the E2F sites in the AdE2 promoter (49). In vitro, hCREG interacts with the E1A-binding proteins TBP, pRb, p107, and p130, raising the possibility that CREG may antagonize E1A by direct binding to the same protein targets. Alternatively, CREG may antagonize E1A by acting at a different step in an E1A-regulated pathway.
We have shown that hCREG and E1A have opposing effects on transcription from both the AdE2 and hsp70 promoters. Several models have been put forth to explain the mechanism by which E1A activates the hsp70 promoter, including the activation of CAAT box-dependent transcription and the relief of Dr1-mediated repression (34, 43). Our results suggest an additional mechanism for E1A-mediated activation that is common to the hsp70 and AdE2 genes: the relief of hCREG-mediated repression. Since repression of the AdE2 promoter by CREG is dependent on specific promoter elements, it is likely that repression of the hsp70 promoter by hCREG, as well as the modest twofold repression seen with several other promoters, is also mediated through specific upstream or core promoter elements. Interestingly, expression of the hsp70 promoter has been found to be regulated during the cell cycle (33, 46). It is not known at present if the different CREG responsive promoters are downstream of a common pathway or if CREG, like E1A, affects the activity of several pathways important for regulation of gene expression.
Although the E2F sites in both the AdE2 and DHFR promoters were required for full repression by hCREG, we did not observe E2F site-dependent repression of the E2F1 and b-myb promoters. Mutation of the E2F sites in the AdE2 and DHFR promoters causes a decrease in promoter activity, indicating that E2F contributes to activation of these promoters (6, 41, 57). In contrast, mutation of the E2F sites in the E2F1 and b-myb promoters results in an elevated level of expression, indicating that E2F-containing complexes contribute predominantly to repression (29, 31, 37, 48). Consistent with this view, in vivo footprinting of the b-myb promoter has shown that the E2F sites are only occupied during G0-G1, when the gene is not expressed (75). Although the underlying mechanism remains obscure, whether a given E2F binding site contributes to positive or negative regulation depends on the promoter context (19, 66). Similarly, in our assays, the ability of an E2F site to support repression by hCREG is context dependent, and we have only observed E2F site-dependent repression by hCREG on positively acting E2F sites. Although it remains possible that the failure of the E2F sites in the b-myb and E2F-1 promoters to support repression by hCREG is due to other sequence elements present in, or absent from, these promoters, these observations suggest that it is the activation function of E2F that is inhibited by CREG.
We have shown that hCREG preferentially represses activation by Gal4 fusion proteins bearing E2F activation domains. Thus, in contrast to other factors that repress E2F activity, such as p202 and PPARγ, hCREG does not act predominantly through inhibition of the DNA binding activity of E2F (4, 9). Interestingly, hCREG showed twofold-greater repression of Gal4E2F4 and Gal4E2F5 than Gal4E2F1, suggesting that hCREG-mediated repression may contribute to differential regulation of these highly related proteins. Since in in vitro binding assays we have been unable to detect any interaction between E2F1 and hCREG (15), we consider it unlikely that the repression of E2F activity by hCREG is via a direct interaction. hCREG interacts in vitro with the pRb, p107, and p130 repressor proteins, raising the possibility that these interactions may contribute to specific repression by hCREG in vivo. The simple model whereby pRb serves as a bridge to recruit the hCREG repressor to E2F is not supported by our observation that hCREG efficiently inhibited activation by E2F1 mutants defective in binding pRb. The results with Gal4+E2F fusions are consistent with the context-dependent effects of CREG on E2F sites and further support the hypothesis that CREG represses E2F site-dependent transcription by inhibiting the activation by “free” E2F and not by augmenting repression by E2F-pRb complexes.
The mechanism of transcriptional activation by E2F has not been determined. Fry et al. (19) have reported that the N-terminal VP16 activation domain will not substitute for E2F in stimulation of the DHFR promoter, indicating that there is some unique aspect to the E2F activation function. Although E2F4 and E2F5 activation domains have not been extensively characterized, the activation domain of E2F1 has been shown to interact with TBP, TFIIH, MDM2, p300, and CBP (20, 44, 51, 65), any or all of which may function as coactivators for E2F-dependent activation and are therefore potential targets for hCREG-mediated repression. The observation that hCREG represses transcription when tethered to the promoter suggests that hCREG has a repression domain that inhibits some aspect of the transcription process common to many genes. Many transcriptional repressors, such as Dr1, interfere with the function of the general transcription factor TBP (30). Future studies should reveal whether hCREG-mediated repression results from inhibition of the activity of general transcription factors such as TBP, coactivators, alterations in chromatin structure, or other mechanisms.
The results reported here demonstrate that the hCREG protein represses E2F-dependent activation and antagonizes the ability of the adenovirus E1A oncoprotein to stimulate the expression of several genes and to cooperate with oncogenic ras in the transformation of primary cells. Both the adenovirus E1A protein and transcriptional activation by E2F function to promote cellular proliferation. hCREG activity is therefore likely to play a role in inhibiting cell growth and/or promoting differentiation. Although Northern analysis revealed that hCREG mRNA is widely expressed in human tissues, we have found that hCREG mRNA levels increase during the terminal differentiation of several cell lines (67). Future studies will examine how changes in hCREG activity affect cell proliferation and differentiation or influence the cellular response to infection by DNA tumor viruses.
ACKNOWLEDGMENTS
We thank Amelia Tung for technical assistance. Studies of Drosophila CREG were initiated in the laboratory of Robert Tjian at the University of California, Berkeley. We are grateful to Jennifer Dowhanick for help in setting up the BRK transformation assays. Reagents used in this study were kindly provided by Rene Bernards, Nicholas Dyson, Bill Kaelin, Karl Munger, Joseph Nevins, Bill Sellers, and Yang Shi. We also thank Keith Blackwell, Mike Carey, Phil Hinds, Karl Munger, and Yang Shi for helpful comments on the manuscript.
This work was supported in part by a grant from The Jessie B. Cox Charitable Trust and The Medical Foundation to G.G.
REFERENCES
- 1.Adams P D, Kaelin W G. The cellular effects of E2F overexpression. Curr Opin Microbiol Immunol. 1996;208:79–93. doi: 10.1007/978-3-642-79910-5_4. [DOI] [PubMed] [Google Scholar]
- 2.Adnane J, Shao Z, Robbins P D. The retinoblastoma susceptibility gene product represses transcription when directly bound to the promoter. J Biol Chem. 1995;270:8837–8843. doi: 10.1074/jbc.270.15.8837. [DOI] [PubMed] [Google Scholar]
- 3.Allen K, Luna S, Kerkhoven R, Bernards R, Thangue N. Distinct mechanisms of nuclear accumulation regulate the functional consequence of E2F transcription factors. J Cell Sci. 1997;110:2819–2831. doi: 10.1242/jcs.110.22.2819. [DOI] [PubMed] [Google Scholar]
- 4.Altiok S, Xu M, Spiegelman B M. PPARgamma induces cell cycle withdrawal: inhibition of E2F/DP DNA-binding activity via down-regulation of PP2A. Genes Dev. 1997;11:1987–1998. doi: 10.1101/gad.11.15.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Asano M, Nevins J R, Wharton R P. Ectopic E2F expression induces S phase and apoptosis in Drosophila imaginal discs. Genes Dev. 1996;10:1422–1432. doi: 10.1101/gad.10.11.1422. [DOI] [PubMed] [Google Scholar]
- 6.Blake M C, Azizkhan J C. Transcription factor E2F is required for efficient expression of the hamster dihydrofolate reductase gene in vitro and in vivo. Mol Cell Biol. 1989;9:4994–5002. doi: 10.1128/mcb.9.11.4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bremner R, Cohen B L, Sopta M, Hamel P A, Ingles C J, Gallie B L, Phillips R A. Direct transcriptional repression by pRB and its reversal by specific cyclins. Mol Cell Biol. 1995;15:3256–3265. doi: 10.1128/mcb.15.6.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brook A, Xie J-E, Du W, Dyson N. Requirements for dE2F function in proliferating cells and in post-mitotic differentiating cells. EMBO J. 1996;15:3676–3683. [PMC free article] [PubMed] [Google Scholar]
- 9.Choubey D, Li S-J, Datta B, Gutterman J U, Lengyel P. Inhibition of E2F-mediated transcription by p202. EMBO J. 1996;15:5668–5678. [PMC free article] [PubMed] [Google Scholar]
- 10.Cobrinik D. Regulatory interactions among E2Fs and cell cycle control proteins. Curr Top Microbiol Immunol. 1996;208:32–61. doi: 10.1007/978-3-642-79910-5_2. [DOI] [PubMed] [Google Scholar]
- 11.Corbeil H B, Branton P E. Functional importance of complex formation between the retinoblastoma tumor suppressor family and adenovirus E1A proteins as determined by mutational analysis of E1A conserved region 2. J Virol. 1994;68:6697–6709. doi: 10.1128/jvi.68.10.6697-6709.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Du W, Xie J-E, Dyson N. Ectopic expression of dE2F and dDP induces cell proliferation and death in the Drosophila eye. EMBO J. 1996;15:3684–3692. [PMC free article] [PubMed] [Google Scholar]
- 13.Duronio R J, O’Farrell P H, Xie J-E, Brook A, Dyson N. The transcription factor E2F is required for S phase during Drosophila embryogenesis. Genes Dev. 1995;9:1445–1455. doi: 10.1101/gad.9.12.1445. [DOI] [PubMed] [Google Scholar]
- 14.Dyson N, Guida P, McCall C, Harlow E. Adenovirus E1A makes two distinct contacts with the retinoblastoma protein. J Virol. 1992;66:4606–4611. doi: 10.1128/jvi.66.7.4606-4611.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eisenstein, M., and G. Gill. Unpublished data.
- 16.Eisenstein, M., E. Veal, and G. Gill. Unpublished data.
- 17.Field S J, Tsai F-Y, Kuo F, Zubiaga A M, Kaelin W G, Livingston D M, Orkin S H, Greenberg M E. E2F-1 functions in mice to promote apoptosis and suppress proliferation. Cell. 1996;85:549–562. doi: 10.1016/s0092-8674(00)81255-6. [DOI] [PubMed] [Google Scholar]
- 18.Finlay C A, Hinds P W, Levine A J. The p53 proto-oncogene can act as a suppressor of transformation. Cell. 1989;57:1083–1093. doi: 10.1016/0092-8674(89)90045-7. [DOI] [PubMed] [Google Scholar]
- 19.Fry C J, Slansky J E, Farnam P J. Position-dependent transcriptional regulation of the murine dihydrofolate reductase promoter by the E2F transactivation domain. Mol Cell Biol. 1997;17:1966–1976. doi: 10.1128/mcb.17.4.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hagemeier C, Cook A, Kouzarides T. The retinoblastoma protein binds E2F residues required for activation in vivo and TBP binding in vitro. Nucleic Acids Res. 1993;21:4998–5004. doi: 10.1093/nar/21.22.4998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hateboer G, Kerkhoven R M, Shvarts A, Bernards R, Beijersbergen R L. Degradation of E2F by the ubiquitin-proteasome pathway: regulation by retinoblastoma family proteins and adenovirus transforming proteins. Genes Dev. 1996;10:2960–2970. doi: 10.1101/gad.10.23.2960. [DOI] [PubMed] [Google Scholar]
- 22.Hateboer G, Timmers H T M, Rustgi A K, Billaud M, Van’t Veer L J, Bernards R. TATA-binding protein and the retinoblastoma gene product bind to overlapping epitopes on c-Myc and adenovirus E1A protein. Proc Natl Acad Sci USA. 1993;90:8489–8493. doi: 10.1073/pnas.90.18.8489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haviv I, Vaizel D, Shaul Y. The X protein of hepatitis B virus coactivates potent activation domains. Mol Cell Biol. 1995;15:1079–1085. doi: 10.1128/mcb.15.2.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Helin K, Harlow E, Fattaey A. Inhibition of E2F-1 transactivation by direct binding of the retinoblastoma protein. Mol Cell Biol. 1993;13:6501–6508. doi: 10.1128/mcb.13.10.6501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hijmans E M, Voorhoeve P M, Beijersbergen R L, van’t Veer L J, Bernards R. E2F-5, a new E2F family member that interacts with p130 in vivo. Mol Cell Biol. 1995;15:3082–3089. doi: 10.1128/mcb.15.6.3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hinds P W, Dowdy S F, Eaton E N, Arnold A, Weinberg R A. Function of a human cyclin as an oncogene. Proc Natl Acad Sci USA. 1994;91:709–713. doi: 10.1073/pnas.91.2.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hofmann F, Martelli F, Livingston D, Wang Z. The retinoblastoma gene product protects E2F-1 from degradation by the ubiquitin-proteasome pathway. Genes Dev. 1996;10:2949–2959. doi: 10.1101/gad.10.23.2949. [DOI] [PubMed] [Google Scholar]
- 28.Horowitz J M, Yandell D W, Park S-H, Canning S, Whyte P, Buchkovich K, Harlow E, Weinberg R A, Dryja T P. Point mutational inactivation of the retinoblastoma antioncogene. Science. 1989;243:937–940. doi: 10.1126/science.2521957. [DOI] [PubMed] [Google Scholar]
- 29.Hsiao K M, McMahon S L, Farnham P J. Multiple DNA elements are required for the growth regulation of the mouse E2F1 promoter. Genes Dev. 1994;8:1526–1537. doi: 10.1101/gad.8.13.1526. [DOI] [PubMed] [Google Scholar]
- 30.Inostroza J A, Mermelstein F H, Ha I, Lane W S, Reinberg D. Dr1, a TATA-binding protein-associated phosphoprotein and inhibitor of class II gene transcription. Cell. 1992;70:477–489. doi: 10.1016/0092-8674(92)90172-9. [DOI] [PubMed] [Google Scholar]
- 31.Johnson D G, Ohtani K, Nevins J R. Autoregulatory control of E2F1 expression in response to positive and negative regulators of cell cycle progression. Genes Dev. 1994;8:1514–1525. doi: 10.1101/gad.8.13.1514. [DOI] [PubMed] [Google Scholar]
- 32.Kao H, Nevins J R. Transcriptional activation and subsequent control of the human heat shock gene during adenovirus infection. Mol Cell Biol. 1983;3:2058–2065. doi: 10.1128/mcb.3.11.2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kao H T, Capasso O, Heintz N, Nevins J R. Cell cycle control of the human HSP70 gene: implications for the role of a cellular E1A-like function. Mol Cell Biol. 1985;5:628–633. doi: 10.1128/mcb.5.4.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kraus V B, Inostroza J A, Yeung K, Reinberg D, Nevins J R. Interaction of the Dr1 inhibitory factor with the TATA binding protein is disrupted by adenovirus E1A. Proc Natl Acad Sci USA. 1994;91:6279–6282. doi: 10.1073/pnas.91.14.6279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kraus V B, Moran E, Nevins J R. Promoter-specific trans-activation by the adenovirus E1A12S product involves separate E1A domains. Mol Cell Biol. 1992;12:4391–4399. doi: 10.1128/mcb.12.10.4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Labrie C, Morris G F, Mathews M B. A complex promoter element mediates transactivation of the human proliferating cell nuclear antigen promoter by the 243-residue adenovirus E1A oncoprotein. Mol Cell Biol. 1993;13:1697–1707. doi: 10.1128/mcb.13.3.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lam E W-F, Watson R J. An E2F-binding site mediates cell-cycle regulated repression of mouse B-myb transcription. EMBO J. 1993;12:2705–2713. doi: 10.1002/j.1460-2075.1993.tb05932.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lillie J W, Green M, Green M R. An adenovirus E1A protein region required for transformation and transcriptional repression. Cell. 1986;46:1043–1051. doi: 10.1016/0092-8674(86)90704-x. [DOI] [PubMed] [Google Scholar]
- 39.Lindeman G J, Gaubatz S, Livingston D M, Ginsberg D. The subcellular localization of E2F-4 is cell-cycle dependent. Proc Natl Acad Sci USA. 1997;94:5095–5100. doi: 10.1073/pnas.94.10.5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu F, Green M R. A specific member of the ATF transcription factor family can mediate transcription activation by the adenovirus E1A protein. Cell. 1990;61:1217–1224. doi: 10.1016/0092-8674(90)90686-9. [DOI] [PubMed] [Google Scholar]
- 41.Loeken M R, Brady J. The adenovirus EIIA enhancer. Analysis of regulatory sequences and changes in binding activity of ATF and EIIF following adenovirus infection. J Biol Chem. 1989;264:6572–6579. [PubMed] [Google Scholar]
- 42.Luckow B, Schutz G. CAT constructs with multiple restriction sites for the functional analysis of eukaryotic promoters and regulatory elements. Nucleic Acids Res. 1987;15:5490. doi: 10.1093/nar/15.13.5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lum L S Y, Hsu S, Vaewhongs M, Wu B. The hsp70 gene CAAT-binding factor mediates transcriptional activation by the adenovirus E1A protein. Mol Cell Biol. 1992;12:2599–2605. doi: 10.1128/mcb.12.6.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martin K, Trouche D, Hagemeier C, Sorensen T S, La Thangue N B, Kouzarides T. Stimulation of E2F1/DP1 transcriptional activity by MDM2 oncoprotein. Nature. 1995;375:691–694. doi: 10.1038/375691a0. [DOI] [PubMed] [Google Scholar]
- 45.Means A L, Slansky J E, McMahon S L, Knuth M W, Farnham P J. The HIP1 binding site is required for growth regulation of the dihydrofolate reductase gene promoter. Mol Cell Biol. 1992;14:6607–6615. doi: 10.1128/mcb.12.3.1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Milarski J L, Morimoto R I. Expression of human HSP70 during the synthetic phase of the cell cycle. Proc Natl Acad Sci USA. 1986;83:9517–9521. doi: 10.1073/pnas.83.24.9517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Muller H, Moroni M C, Vigo E, Petersen B O, Bartek J, Helin K. Induction of S-phase entry by E2F transcription factors depends on their nuclear localization. Mol Cell Biol. 1997;17:5508–5520. doi: 10.1128/mcb.17.9.5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Neuman E, Flemington E K, Sellers W R, Kaelin W G. Transcription of the E2F-1 gene is rendered cell cycle dependent by E2F DNA-binding sites within its promoter. Mol Cell Biol. 1994;14:6607–6615. doi: 10.1128/mcb.14.10.6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nevins J R. E2F: a link between the Rb tumor suppressor protein and viral oncoproteins. Science. 1992;258:424–429. doi: 10.1126/science.1411535. [DOI] [PubMed] [Google Scholar]
- 50.Nevins J R. Induction of the synthesis of a 70,000 dalton mammalian heat shock protein by the adenovirus E1A gene product. Cell. 1982;29:913–919. doi: 10.1016/0092-8674(82)90453-6. [DOI] [PubMed] [Google Scholar]
- 51.Pearson A, Greenblatt J. Modular organization of the E2F1 activation domain and its interaction with general transcription factors TBP and TFIIH. Oncogene. 1997;15:2643–2658. doi: 10.1038/sj.onc.1201451. [DOI] [PubMed] [Google Scholar]
- 52.Ruley H E. Adenovirus early region 1A enables viral and cellular transforming genes to transform primary cells in culture. Nature. 1983;304:602–606. doi: 10.1038/304602a0. [DOI] [PubMed] [Google Scholar]
- 53.Sadowski I, Ma J, Ptashne P. GAL4-VP16 is an unusually potent transcriptional activator. Nature. 1988;335:563–564. doi: 10.1038/335563a0. [DOI] [PubMed] [Google Scholar]
- 54.Shi Y, Seto E, Chang L-S, Shenk T. Transcriptional repression by YY1, a human GL1-Kruppel-related protein, and relief of repression by adenovirus E1A protein. Cell. 1991;67:377–388. doi: 10.1016/0092-8674(91)90189-6. [DOI] [PubMed] [Google Scholar]
- 55.Simon M C, Fisch T M, Benecke B J, Nevins J R, Heintz N. Definition of multiple, functionally distinct TATA elements, one of which is a target in the hsp70 promoter for E1A regulation. Cell. 1988;52:723–729. doi: 10.1016/0092-8674(88)90410-2. [DOI] [PubMed] [Google Scholar]
- 56.Slansky J E, Farnham P J. Introduction to the E2F family: protein structure and gene regulation. Curr Top Microbiol Immunol. 1996;208:1–30. doi: 10.1007/978-3-642-79910-5_1. [DOI] [PubMed] [Google Scholar]
- 57.Slansky J E, Li Y, Kaelin W G, Farnham P J. A protein synthesis-dependent increase in E2F1 mRNA correlates with growth regulation of the dihydrofolate reductase promoter. Mol Cell Biol. 1993;13:1610–1618. doi: 10.1128/mcb.13.3.1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sleigh M J. A nonchromatographic assay for expression of the chloramphenicol acetyltransferase gene in eucaryotic cells. Anal Biochem. 1986;156:251–256. doi: 10.1016/0003-2697(86)90180-6. [DOI] [PubMed] [Google Scholar]
- 59.Smits P H, de Wit L, van der Eb A J, Zantema A. The adenovirus E1A-associated 300 kDa adaptor protein counteracts the inhibition of collagenase promoter by E1A and represses transformation. Oncogene. 1996;12:1529–1535. [PubMed] [Google Scholar]
- 60.Somasundaram K, Jayaraman G, Williams T, Moran E, Frisch S, Thimmapaya B. Repression of a matrix metalloprotease gene by E1A correlates with its ability to bind to cell type-specific transcription factor AP-2. Proc Natl Acad Sci USA. 1996;93:3088–3093. doi: 10.1073/pnas.93.7.3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Song C-Z, Loewenstein P M, Toth K, Green M. Transcription factor TFIID is a direct functional target of the E1A transcription-repression domain. Proc Natl Acad Sci USA. 1995;92:10330–10333. doi: 10.1073/pnas.92.22.10330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stein R W, Corrigan M, Yaciuk P, Whelan P, Moran E. Analysis of E1A-mediated growth regulation functions: binding of the 300-kilodalton cellular product correlates with E1A enhancer repression function and DNBA synthesis-inducing activity. J Virol. 1990;64:4421–4427. doi: 10.1128/jvi.64.9.4421-4427.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Taylor D A, Kraus V B, Schwarz J J, Olson E N, Kraus W E. E1A-mediated inhibition of myogenesis correlates with a direct physical interaction of E1A12S and basic helix-loop-helix proteins. Mol Cell Biol. 1993;13:4714–4727. doi: 10.1128/mcb.13.8.4714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tiemann F, Musunuru K, Hinds P W. The retinoblastoma protein and cancer. In: Swallow D M, Edwards Y H, editors. Protein dysfunction in human genetic disease. Oxford, England: Bios Scientific Publishers; 1997. pp. 163–185. [Google Scholar]
- 65.Trouche D, Cook A, Kouzarides T. The CBP co-activator stimulates E2F1/DP1 activity. Nucleic Acids Res. 1996;24:4139–4145. doi: 10.1093/nar/24.21.4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.van Ginkel P R, Hsiao K-M, Schjerven H, Farnham P J. E2F-mediated growth regulation requires transcription factor cooperation. J Biol Chem. 1997;272:18367–18374. doi: 10.1074/jbc.272.29.18367. [DOI] [PubMed] [Google Scholar]
- 67.Veal, E., and G. Gill. Unpublished data.
- 68.Verona R, Moberg K, Estes S, Starz M, Vernon J P, Lees J A. E2F activity is regulated by cell cycle-dependent changes in subcellular localization. Mol Cell Biol. 1997;17:7268–7282. doi: 10.1128/mcb.17.12.7268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang H-G H, Rikitake Y, Carter M C, Yaciuk P, Abraham S E, Zerler B, Moran E. Identification of specific adenovirus E1A N-terminal residues critical to the binding of cellular proteins and to their control of cell growth. J Virol. 1993;67:476–488. doi: 10.1128/jvi.67.1.476-488.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Weintraub S J, Chow K N B, Luo R X, Zhang S H, He S, Dean D C. Mechanism of active transcriptional repression by the retinoblastoma protein. Nature. 1995;375:812–815. doi: 10.1038/375812a0. [DOI] [PubMed] [Google Scholar]
- 71.Whyte P, Williamson N M, Harlow E. Cellular targets for transformation by the adenovirus E1A proteins. Cell. 1989;56:67–75. doi: 10.1016/0092-8674(89)90984-7. [DOI] [PubMed] [Google Scholar]
- 72.Williams G T, McClanahan T K, Morimoto R I. E1A transactivation of the human HSP70 promoter is mediated through the basal transcription complex. Mol Cell Biol. 1989;9:2574–2587. doi: 10.1128/mcb.9.6.2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yamasaki L, Jacks T, Bronson R, Goillot E, Harlow E, Dyson N. Tumor induction and tissue atrophy in mice lacking E2F-1. Cell. 1996;85:537–548. doi: 10.1016/s0092-8674(00)81254-4. [DOI] [PubMed] [Google Scholar]
- 74.Zhu L, van den Heuvel S, Helin K, Fattaey A, Ewen M, Livingston D, Dyson N, Harlow E. Inhibition of cell proliferation by p107, a relative of the retinoblastoma protein. Genes Dev. 1993;7:1111–1125. doi: 10.1101/gad.7.7a.1111. [DOI] [PubMed] [Google Scholar]
- 75.Zwicker J, Liu N, Engeland K, Lucibello F C, Muller R. Cell cycle regulation of E2F site occupation in vivo. Science. 1996;271:1595–1597. doi: 10.1126/science.271.5255.1595. [DOI] [PubMed] [Google Scholar]