Abstract
mRNA in the yeast Saccharomyces cerevisiae is primarily degraded through a pathway that is stimulated by removal of the mRNA cap structure. Here we report that a mutation in the SPB8 (YJL124c) gene, initially identified as a suppressor mutation of a poly(A)-binding protein (PAB1) gene deletion, stabilizes the mRNA cap structure. Specifically, we find that the spb8-2 mutation results in the accumulation of capped, poly(A)-deficient mRNAs. The presence of this mutation also allows for the detection of mRNA species trimmed from the 3′ end. These data show that this Sm-like protein family member is involved in the process of mRNA decapping, and they provide an example of 3′-5′ mRNA degradation intermediates in yeast.
The function of the poly(A) tail on mRNA in eucaryotes is the subject of much research. It has been shown that in the yeast Saccharomyces cerevisiae, the poly(A) tail acts to enhance the translation of the mRNA (reviewed in reference 36). This activity requires the poly(A)-binding protein Pab1p, which, through an interaction with a protein complex recognizing the cap structure, is thought to stimulate the binding of ribosomes to the 5′ end of the mRNA (23, 39–41). Another potential function of the poly(A) tail is to stabilize mRNA. Several independent observations support this hypothesis. A series of detailed studies of both yeast and mammalian cells has documented that mRNA deadenylation usually precedes mRNA degradation (reviewed in reference 8). More specifically, it has been shown that in yeast, mRNA decapping, the initiating event in the degradation of the majority of mRNAs, occurs after deadenylation (9, 26, 27). Following decapping, these mRNAs are destroyed by the 5′-3′ exoribonuclease Xrn1p (20, 26). It has also been shown that decreasing the rate of poly(A) tail removal by mutagenizing mRNA (for example, see reference 28) results in lower rates of mRNA degradation.
Degradation through the pathway of decapping and then digestion by the 5′-3′ exonuclease Xrn1p is not the sole means by which mRNA is degraded in yeast. Three key observations form the basis for this conclusion. First, targeted disruption of either the decapping enzyme gene DCP1 or the exonuclease gene XRN1 does not lead to cell inviability or greater than four- to fivefold stabilization of mRNAs that are normally unstable (4, 20). Second, the disruption of these genes does not change the stability of the most stable yeast mRNAs, such as PGK1 or ACT1, by more than a factor of 2 (4, 27). Finally, for yeast strains containing a disruption of the XRN1 gene or a mutation in the Dcp1 protein, the existence of mRNA species trimmed from the 3′ end has been reported (27).
A common feature of both the mRNA translation and degradation reactions is that the cap structure and the poly(A) tail appear to be involved in their regulation (reviewed in reference 42). The possibility that the roles of these two structures in the degradation reaction are functionally linked was supported by the recent report that mutations in the yeast decapping enzyme Dcp1p and the unidentified gene products of the MRT1 and MRT3 genes, which also appear to regulate mRNA decapping rates, can allow yeast cells to survive in the absence of Pab1p (17). These mutations were therefore suggested to allow for cell viability in the absence of Pab1p by stabilizing the mRNA to an extent that allowed for sufficient expression in the absence of Pab1p.
We originally chose to further explore the essential roles of Pab1p in yeast in order to define the mechanistic roles of Pab1p and poly(A) in mRNA metabolism (33, 34). A series of genetic suppression experiments identified mutations in other yeast genes which allowed for cell growth in the absence of Pab1p. Most of these mutations resulted in aberrant 60S ribosomal subunit production. Other laboratories have also identified similar types of pab1Δ bypass suppressors (43). To identify proteins involved in mRNA degradation, including those that functionally interact with Pab1p and the poly(A) tail during this process, we chose to focus our newer studies on those pab1Δ bypass suppressor mutations that did not alter the levels of the ribosomal subunits. Based on published data (17), we reasoned that such mutations could lead to viability in the absence of Pab1p by stabilizing the mRNA. The current lack of sequence information on genes other than DCP1 and XRN1 that are known to alter the pathway of 5′-3′ mRNA degradation in yeast provided us with further incentive to pursue this avenue of investigation.
Here we report that a null mutation in the nonessential yeast SPB8 (YJL124c) gene leads to bypass suppression of a PAB1 deletion without altering ribosomal subunit levels. Spb8p was found to contain an Sm-like domain. Mutations within Spb8p led to the accumulation of capped, deadenylated degradation intermediates and in some cases the stabilization of mRNA. Cap stabilization was so complete in the spb8-2 strain that 3′-5′ mRNA degradation intermediates also became readily detectable. These data support the hypotheses that Spb8p is needed for normal rates of mRNA decapping in yeast and that Spb8p allows for rates of decapping and 5′-3′ degradation that preclude the detection of 3′-5′ degradative intermediates.
MATERIALS AND METHODS
Yeast methods.
All yeast strains and their relevant genotypes are listed in Table 1. The parent strain used in this study is a derivative of W303a, YAS306 (MATa ade2-1 his3-11 leu2-3,112 trp1-1 ura3-1 can 1-100). All yeast cultures were grown in standard media (16). Yeast cells were transformed with DNA by the lithium acetate method (14).
TABLE 1.
Yeast strains and plasmids used in this study
| Strain | Relevant genotypea | Plasmid description (name; source or reference) |
|---|---|---|
| YAS1070 | MATa TRP1 | pPAB1 URA3 LEU2 CEN (pAS580; this study) |
| YAS1071 | MATα lys2 | pPAB1 URA3 LEU2 CEN (pAS580; this study) |
| YAS1915 | MATa | pPAB1 TRP1 CEN (pAS80) |
| YAS2266 | MATa | pPAB1 URA3 CEN (pAS77; 35) |
| pTRP1 CEN (pUN15; 11) | ||
| YAS2267 | MATa | pPAB1 URA3 CEN (pAS77) |
| pSPB8 TRP1 CEN (pAS579; this study) | ||
| YAS2268 | MATa spb8-1 | pPAB1 URA3 CEN (pAS77) |
| pTRP1 CEN (pUN15) | ||
| YAS2269 | MATa spb8-1 | pPAB1 URA3 CEN (pAS77) |
| pSPB8 TRP1 CEN (pAS579) | ||
| YAS2270 | MATα spb8-2 lys2 | pPAB1 URA3 CEN (pAS77) |
| pTRP1 CEN (pUN15) | ||
| YAS2271 | MATα spb8-2 lys2 | pPAB1 URA3 CEN (pAS77) |
| pSPB8 TRP1 CEN (pAS579) | ||
| YAS2272 | MATa | pPAB1 TRP1 CEN (pAS80; 35) |
| pGAL1 MFA2pG URA3 CEN (pRP485; 9) | ||
| YAS2273 | MATα spb8-2 lys2 | pPAB1 TRP1 CEN (pAS80) |
| pGAL1 MFA2pG URA3 CEN (pRP485) | ||
| YAS2274 | MATa | pPAB1 TRP1 CEN (pAS80) |
| pGAL1 B55TPGK1pG URA3 CEN (pRP602; 27) | ||
| YAS2275 | MATα spb8-2 lys2 | pPAB1 TRP1 CEN (pAS80) |
| pGAL1 B55TPGK1pG URA3 CEN (pRP602) | ||
| YAS2276 | MATa | pPAB1 TRP1 CEN (pAS80) |
| pGAL1 B55TPGK1NSpG URA3 CEN (pRP611; 26) | ||
| YAS2277 | MATα spb8-2 lys2 | pPAB1 TRP1 CEN (pAS80) |
| pGAL1 B55TPGK1NSpG URA3 CEN (pRP611) | ||
| YAS2278 | MATα spb8-2 lys2 | pPAB1 TRP1 CEN (pAS80) |
| YAS2279 | MATα spb8-2 lys2 | pGAL1 MFA2pG URA3 CEN (pRP485) |
| YAS2280 | MATα ADE2 lys2 | pPAB1 URA3 TRP1 CEN (pAS521; this study) |
| YAS2281 | MATa | pPAB1 URA3 TRP1 CEN (pAS521; this study) |
All strains are PAB1::HIS3 ade2-1 his3-11 leu2-3,112 trp1-1 ura3-1 can 1-100 unless otherwise stated.
Spontaneous bypass suppressors of a pab1Δ were directly selected for by plating out more than 500 independent single colonies of yeast strain YAS1070 (MATa PAB1::HIS3 ade2 his3 leu2 ura3 pPAB1 URA3 LEU2 CEN) or YAS1071 (MATα PAB1::HIS3 ade2 his3 leu2 trp1 ura3 lys2 pPAB1 URA3 LEU2 CEN) grown on YPD medium onto YM medium containing 1 mg of 5-fluoro-orotic acid (5-FOA) per ml (16). A single 5-FOAr Leu− colony derived from each of the independent colonies was then tested for mating and sporulation competency. Crude extracts from strains exhibiting good mating and sporulation, as well as a recessive Spb phenotype, were prepared and analyzed by sucrose gradient sedimentation (33) for alterations in ribosome content. Mutants displaying normal ribosome content were placed into different complementation groups by standard techniques. One such mutant, the spb8-1 strain, was identified as having no observable effect on ribosome biogenesis and containing a single mutation responsible for the pab1Δ bypass suppression phenotype. Following several backcrosses to the parental strains, we were unable to identify other independent growth phenotypes associated with the spb8-1 mutation that would allow for complementation-based cloning of the SPB8 gene. We were also unable to identify yeast genomic DNA fragments on either single- or multiple-copy plasmids that led to a 5-FOAs phenotype in an spb8-1 strain containing a pPAB1 URA3 CEN plasmid and a PAB1 deletion.
Transposon-based mutagenesis of yeast strains YAS2280 (MATα PAB1::HIS3 his3 leu2 lys2 trp1 ura3 pPAB1 URA3 TRP1 CEN) and YAS2281 (MATa PAB1::HIS3 ade2 his3 leu2 trp1 ura3 pPAB1 URA3 TRP1 CEN), using 15 independent mini-Tn3 genomic insertion libraries digested with NotI prior to yeast transformation, was done essentially as described previously (6). Approximately 5 × 104 Leu+ transformants were replica plated onto YM medium containing 5-FOA in order to identify the 5-FOAr colonies. 5-FOAr colonies that were also Trp− were analyzed further. The spb8-2 mutant was one of several isolates that did show linkage with the inserted transposon and which did not exhibit ribosome biogenesis defects. The yeast genomic DNA flanking the Tn3 insertion site was rescued in two steps as previously described (6). First, the spb8-2 strain was transformed with plasmid pRSQ2 (31), which recombines with the transposon inserted in the genome and brings a procaryotic origin of replication into the vicinity of the transposon. Genomic DNA was then prepared, digested with EcoRI, and circularized with DNA ligase. DNA containing the bacterial origin of replication, the Ampr gene, plus the flanking region at the site of the transposon insertion was rescued by transforming Escherichia coli. The isolated DNA was sequenced with an oligonucleotide primer specific for the end of the mini-Tn3 sequence. A full description of this screen for pab1Δ bypass suppressors will appear elsewhere (24a).
DNA methods.
Plasmid pAS521 (pPAB1 TRP1 URA3 CEN) was constructed by inserting the URA3 gene into pPAB1 TRP1 CEN (pAS80) (35). The URA3 gene was amplified from YIplac211 (15) by using oligonucleotides oAS322 and oAS323 (see below). PCR was performed on 10 ng of YIplac211 plasmid DNA in 100 μl of buffer supplied by the manufacturer (30 cycles of 1-min denaturation at 94°C, 2-min annealing at 45°C, and 2-min extension at 75°C), using 2 U of Vent DNA polymerase (New England Biolabs). The amplified fragment was digested with EcoRI, purified on an agarose gel, and inserted into pAS80 previously digested with EcoRI. Plasmid pAS579 (pSPB8 TRP1 CEN) was constructed by inserting the SPB8 gene into the pUN15 vector (pTRP1 CEN) (11). SPB8 was amplified from yeast genomic DNA with oAS319 and oAS320, which are located 450 nucleotides (nt) upstream of the initiation codon and 155 nt downstream of the stop codon, respectively. PCR was performed on 10 ng of wild-type yeast genomic DNA in 100 μl of buffer supplied by the manufacturer (30 cycles of 1-min denaturation at 94°C, 2-min annealing at 45°C, and 2-min extension at 75°C), using 2 U of Vent DNA polymerase (New England Biolabs). The amplified 1,237-bp fragment was digested with KpnI and XbaI, purified on an agarose gel, and inserted into the pUN15 vector previously digested with KpnI and SpeI. Plasmid pAS580 (pPAB1 URA3 LEU2 CEN) was constructed by inserting the LEU2 gene into pPAB1 URA3 CEN (pAS77) (35). The LEU2 gene was isolated from YEp13 (5) as a SalI/XhoI fragment, purified on an agarose gel, and inserted into pAS77 previously digested with XhoI.
All other plasmids have been described previously and are listed in Table 1.
The indicated oligonucleotides (10 ng of each) were 3′-end labeled with 2 U of recombinant terminal deoxynucleotidyltransferase (GibcoBRL), using 50 μCi of [α-32P]dCTP as recommended by the supplier. Fifty nanograms of DNA fragment, derived either from a PCR or from a plasmid, was labeled with 50 μCi of [α-32P]dCTP, using random hexamer primers and the Klenow fragment of DNA polymerase.
Oligonucleotides used were oRP70 (5′-CGGATAAGAAAGCAACACCTGG-3′ [9]), oRP121 (5′-AATTCCCCCCCCCCCCCCCCCCA-3′ [26]), oAS22 (5′-TTAAGCGATAACACAGGCGGG-3′), oAS318 (5′-GCCAGCAACACGTAATAAATGAAAGGGTAG-3′), oAS319 (5′-GGGGTACCGTCGACTGAATGGGTAAAGGAATGGAT-3′), oAS320 (5′-GCTCTAGAAACGAAGTGTAAGAGGAAAAAGAAT-3′), oAS321 (5′-GGCCAGCAATTTCAAGTTAACTCC-3′), oAS322 (5′-GAAGATCTGAATTCCTGACGTCTAAGAAACCATT-3′), and oAS323 (5′-GAAGATCTGAATTCGGTTTTCACCGTCATCACC-3′).
RNA methods.
For the pGAL1:MFA2/PGK1 transcriptional repression experiments, yeast cells were grown at 25°C to mid-log phase (optical density at 600 nm = 0.4 to 0.5) in 400 ml of galactose-containing YM supplemented with the appropriate nutrients. Cells were collected by centrifugation, washed once in 50 ml of sterile H2O, and resuspended in 20 ml of YM containing 4% dextrose. When required, cycloheximide (Sigma) was added at this point to a final concentration of 100 μg/ml. Cells from 1.5-ml aliquots were collected at fixed time intervals, quick-frozen in liquid nitrogen, and stored at −80°C.
For the CUP1 induction experiments, yeast cells were grown in 400 ml of YMD supplemented with the appropriate nutrients at 25°C to mid-log phase (optical density at 600 nm = 0.4 to 0.5). Cells were concentrated by centrifugation and resuspended in 20 ml of YMD. CuSO4 was added to a final concentration of 0.5 mM. Cells were incubated at 25°C, and cells from 1.5-ml aliquots were collected at defined times, quick-frozen in liquid nitrogen, and stored at −80°C.
RNA was extracted from frozen cell pellets by the hot-phenol procedure. Cell pellets were resuspended in 600 μl of lysis buffer (300 mM NaCl, 20 mM Tris-HCl [pH 7.4], 10 mM EDTA, 1% sodium dodecyl sulfate [SDS]), and 600 μl of preheated (65°C) phenol was added. The tubes were vortexed for 30 s on a multitube vortexer, followed by a 4-min incubation at 65°C and a 3-min incubation on ice. Phases were separated by a 4-min microcentrifugation, and 600 μl of the aqueous phase was reextracted with 65°C preheated phenol as described above. Finally, the aqueous phase was extracted once in phenol-chloroform-isoamyl alcohol (25:24:1) and once in chloroform-isoamyl alcohol (24:1). RNA was precipitated with 2 volumes of ethanol, resuspended in 50 μl of diethyl pyrocarbonate-treated H2O, and stored at −80°C.
RNase H cleavage analysis was performed on 5 μg of total RNA, which was incubated for 1 h at 30°C with 300 ng of oligonucleotide complementary to the cleavage site, and 0.25 U of RNase H (GibcoBRL) in RNase H buffer (20 mM Tris-HCl [pH 7.4], 10 mM MgCl2, 0.5 mM EDTA, 50 mM NaCl, 1 mM dithiothreitol, 30 μg of bovine serum albumin per ml). Reactions were stopped by the addition of 1 volume of RNA loading buffer (90% formamide, 0.05% bromophenol blue, 0.05% xylene cyanol FF).
RNA was separated either by polyacrylamide gel electrophoresis (PAGE) or by agarose gel electrophoresis. Prior to loading, all RNA samples were denatured for 1 min in RNA loading buffer at 94°C. For PAGE, 5-μg aliquots of total RNA were loaded onto 0.75-mm-thick, 24-cm-long 6% polyacrylamide–8.3 M urea–0.5× Tris-borate-EDTA gels, which were run for 2,500 to 4,000 V · h, depending on the size of the RNA analyzed. RNA was transferred to a Zetaprobe membrane (Bio-Rad) by electroblotting. For agarose gel electrophoresis, 10 μg of total RNA was loaded onto 1.2% agarose–6% formaldehyde–1× morpholinepropanesulfonic acid (MOPS) buffer (20 mM MOPS, 8 mM sodium acetate, 1 mM EDTA). RNA was transferred in 10× SSC (1.5 M NaCl, 0.15 M Na3C6H5O7 · 2H2O) to a Zetaprobe membrane (Bio-Rad) by using a TransVac vacuum blotter (Hoefer).
Membranes were hybridized in 7% SDS–250 mM NaPO4 (pH 7.4)–2 mM EDTA either at 60°C [when oligo(C) or hexamer-labeled DNA was used as a probe] or at 40°C (when oAS22 or oAS318 was used). After overnight hybridization, membranes were washed at the hybridization temperature twice in 5% SDS–20 mM NaPO4 (pH 7.4)–2 mM EDTA and once in 1% SDS–20 mM NaPO4 (pH 7.4)–2 mM EDTA.
Membranes were exposed to a Phosphorscreen, scanned on a PhosphorImager (Molecular Dynamics), and quantitated with the ImageQuant software (Molecular Dynamics). The RNA half-lives were calculated by plotting the natural log (ln) of each band’s intensity against time, and the half-life was determined as −ln(2)/slope.
Xrn1p assay.
Five micrograms of total RNA was incubated with 400 ng of purified Xrn1p (kind gift of N. Cozzarelli, University of California, Berkeley) in a final volume of 10 μl of 33 mM Tris-HCl (pH 8.0)–50 mM NaCl–2.5 mM MgCl2–0.2 mM dithiothreitol in the presence or absence of 5 mM EDTA. Reactions were incubated for 30 min at 37°C and stopped by the addition of 1 volume of RNA loading buffer.
RESULTS
Transposon insertion mutagenesis identifies new pab1Δ bypass suppressor mutations.
Yeast genomic mutations that lead to suppression of a PAB1 deletion were initially identified by plating yeast cells containing a PAB1 genomic deletion and the PAB1 gene on a URA3 plasmid onto 5-FOA medium and selecting for viable cells. Following the identification of the recessive bypass suppressor mutations (spb [bypass suppressor of PAB]) which segregated as single genes and did not affect the relative amounts of ribosomal subunits (see Materials and Methods), we identified several yeast mutants belonging to different complementation groups. However, none of these mutants had an independent scorable growth phenotype that could be used for cloning purposes. Furthermore, despite repeated attempts, none of the wild-type counterparts of the mutated genes in these strains could be cloned by screening for a yeast genomic DNA fragment that would prevent the spb mutant from growing in the absence of Pab1p.
As a result of these technical difficulties, we chose to mutagenize the yeast genome by homologous recombination with a randomly mutagenized yeast genomic library. This library was mutagenized in E. coli with a mini-Tn3 transposon containing a LEU2 gene, a lacZ gene, and an Ampr gene (6). The mutagenized library was transformed into a yeast strain containing a genomic PAB1 deletion and PAB1 on a URA3 TRP1 plasmid. Following selection for Leu+ cells, we isolated cells which could grow on 5-FOA and screened for the ones that also became Trp− to confirm that the 5-FOAr phenotype was due to a loss of the URA3 TRP1 plasmid and not to a mutation in the URA3 gene (see Materials and Methods). The Leu+ 5-FOAr mutants which could be mated to form diploids were then sporulated. Spores exhibiting linkage of the Leu+ phenotype with the bypass suppression phenotype, and which did not exhibit alterations in the relative amounts of ribosomal subunits within the cell, were then subjected to further analysis. A complete description of this screen and the results from it will be presented elsewhere (24a). One of the mutants identified in this way (spb8-2) was found to be in the same complementation group as one of the mutants (spb8-1) identified in the first selection procedure. Experiments designed to study the phenotypes associated with an SPB8 mutation were then undertaken.
Cloning and partial characterization of SPB8.
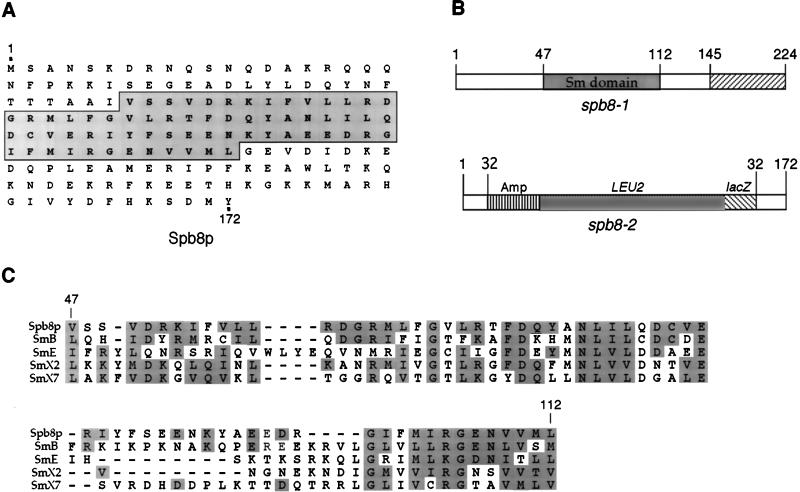
A small fragment of yeast genomic DNA flanking the site of the transposon insertion in the spb8-2 mutant was isolated by standard protocols (6). Sequencing of the yeast genomic DNA immediately flanking the insertion site of the transposon revealed that spb8-2 contained an insertion within the yeast YJL124c protein, a 172-amino-acid polypeptide containing a putative Sm domain (see below and Fig. 1A). The insertion site of the transposon occurred within amino acid 32, and subsequent sequencing of the spb8-1 allele isolated during the first screen revealed a frameshift mutation at codon 145 (Fig. 1B). A complete deletion of SPB8 by directed gene replacement further showed that SPB8 was not essential for yeast cell viability (data not shown). A BLAST search for proteins homologous to Spb8p revealed extensive similarity between Spb8p and the large family of Sm proteins (19, 38) (Fig. 1C). A similar homology involving Spb8p has been previously reported (13). These predominantly nuclear proteins have been shown to be associated with small RNAs within the cell and are involved in various types of RNA processing reactions. Furthermore, overexpression of a mammalian protein exhibiting the greatest homology to Spb8p (CaSm; 30% identity and 67.7% similarity) has recently been found to be associated with maintenance of the transformed state in several types of cancers (37).
FIG. 1.
Identification of YJL124c as the SPB8 gene. (A) Predicted open reading frame of Spb8p. The Sm-like region of the protein that is homologous to the Sm proteins is boxed. (B) Diagram of the YJL124c gene and the sites of mutations in the spb8-1 and spb8-2 alleles. spb8-1 contains a frameshift mutation at codon 145; spb8-2 contains a mini-Tn3 insertion within codon 32. (C) Sequence alignment of Spb8p with other Sm proteins based on an Sm domain alignment previously described (38). Database accession numbers for protein sequences: SmB (human), S10594; SmE (human), P08578; and SmX2 (alfalfa), P24715. SmX7, Brassica campestris pekinensis sequence derived by translation from nucleic acid sequence L33514.
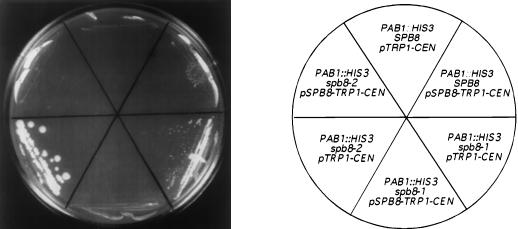
We confirmed that the mutations within the SPB8 genes of the spb8-1 and spb8-2 mutants were responsible for the Spb phenotype (growth in the absence of Pab1p) by showing that the entire SPB8 gene on a plasmid complemented this phenotype (Fig. 2). Given the Spb phenotype associated with both the mrt1-1 and mrt3-1 mutations (17), we decided to investigate whether any of these mutations was associated with the SPB8 gene. We found by DNA sequencing that the SPB8 open reading frames in the mrt1-1 and mrt3-1 mutants were wild type in sequence and that the level of SPB8 mRNA in these two mutants was near that of wild-type cells (data not shown). These data strongly suggest that SPB8 is not allelic to either MRT1 or MRT3.
FIG. 2.
Complementation of the spb8 phenotype by wild-type SPB8. Yeast cells (YAS2266 to YAS2271) containing a deletion of PAB1 and the indicated allele of SPB8 in the genome, PAB1 on a URA3 CEN plasmid, and a TRP1 CEN plasmid with either SPB8 or no insert were streaked onto minimal medium lacking tryptophan and containing 1 mg of 5-FOA per ml (16) and grown at 30°C for 8 days. A photograph of the plate is shown.
Novel mRNA species are found in the spb8-2 mutant.
The absence of detectable differences in the ribosomal subunit profiles of the spb8-2 strain versus a wild-type strain suggested to us that some other aspect of cellular metabolism was altered. We found that the spb8-2 mutant did not accumulate unspliced U3 RNA (data not shown), a phenotype previously associated with yeast splicing mutants (2, 30). This result indicates that despite the extensive homology of Spb8p to Sm proteins, it is probably not directly involved in mRNA splicing. We also found that translation extracts prepared from the spb8-1 strain did not exhibit any significant differences in translation of capped polyadenylated luciferase mRNA compared with wild-type extracts (data not shown). This finding indicates that Spb8p does not play an essential role in the in vitro translation activity of yeast extracts. Based on the report that mutations in yeast which stabilize the cap structure also allow for cell viability in the absence of Pab1p (17), we decided to examine the degradation patterns of several mRNAs in the spb8-2 strain.
The MFA2pG mRNA molecule is degraded rapidly in wild-type yeast (9, 26). A short fragment corresponding to the trapped 5′-3′ degradation intermediate is detectable upon insertion of an oligo(G) tract in the 3′ untranslated region (3′UTR) of the mRNA. This oligo(G) tract will form a stable secondary structure and impede passage of the Xrn1p exonuclease (9). We will refer to this fragment as the oligo(G) degradation intermediate. This modified transcript has been used to show that yeast mutants exhibiting delayed rates of decapping accumulate full-length capped and deadenylated mRNA (4, 17). Mutants defective in Xrn1p exoribonuclease activity accumulate uncapped, deadenylated mRNA and little oligo(G) degradation intermediate (26, 27), while pab1 mutants defective in deadenylation accumulate polyadenylated oligo(G) degradation intermediates (7).
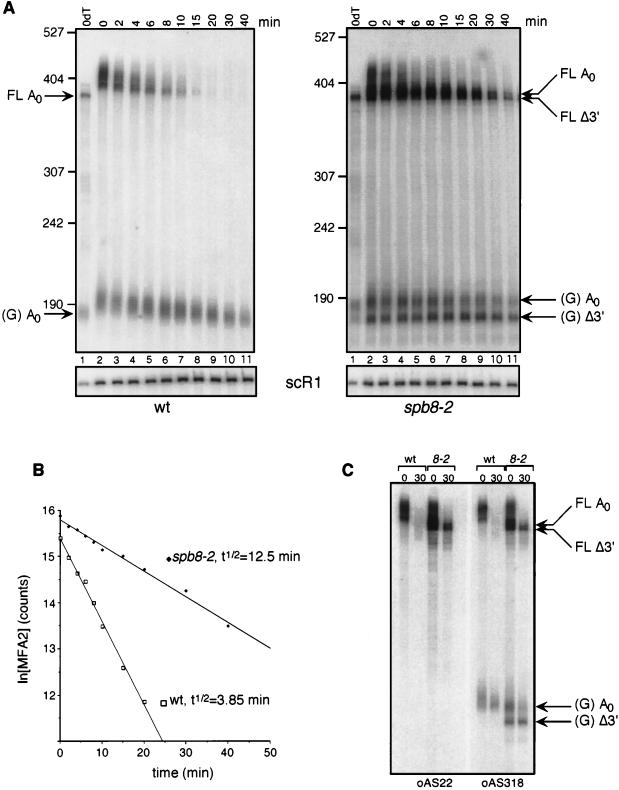
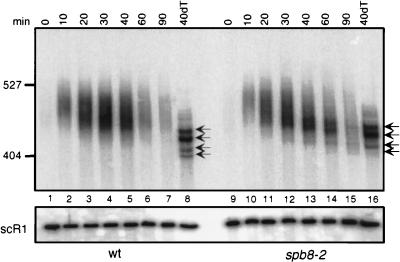
The MFA2pG mRNA degradation rate and pattern in the spb8-2 strain exhibited several significant differences from those of wild-type strains (Fig. 3). First, the overall stability of the mRNA was increased 3.2-fold (Fig. 3A and B). Second, fragments equal in length to and shorter than the deadenylated full-length mRNA were also apparent (Fig. 3A, lanes 7 to 11). Finally, the presence of shorter fragments in the population of oligo(G) degradation intermediates was also observed. We confirmed that the shorter oligo(G) degradation intermediates did not represent some aberrant RNA fragment containing the oligo(G) tract at its 3′ end by showing specific hybridization of this fragment to an oligonucleotide homologous to sequences 3′ but not 5′ to the site of the oligo(G) insertion (Fig. 3C). Analysis of endogenous MFA2 mRNA [lacking the poly(G) tract] in an spb8-1 strain also revealed the accumulation of fragments equal in length to and shorter than the deadenylated full-length mRNA (data not shown). This observation argues against the possibility that these shorter fragments are a consequence of the poly(G) tract inserted into the 3′UTR.
FIG. 3.
Degradation of MFA2pG mRNA in the spb8-2 strain. (A) Accumulation of MFA2pG mRNA degradation intermediates. Yeast strains YAS2272 (SPB8) and YAS2273 (spb8-2) carrying the GAL1:MFA2pG reporter (pRP485) (9) were grown in galactose-containing minimal medium. Glucose was then added to repress transcription of the reporter. RNA was recovered from cells harvested at the indicated times after repression, separated on a 6% polyacrylamide gel, and detected by hybridization with oligo(C) probe oRP121 (26). The length and mobility of size standards are indicated in nucleotides to the left of each gel. The lower panel shows the hybridization signal with the SCR1 probe, which recognizes a polymerase III transcript (12) that serves as an internal loading standard for these experiments (9). The lane containing the RNA sample treated with RNase H-oligo(dT) (0dT) serves to provide size markers for the deadenylated mRNA species. Positions of the full-length and oligo(G) deadenylated mRNAs [FL A0 and (G) A0] and the shorter fragments [FL Δ3′ and (G) Δ3′] are indicated. wt, wild type. (B) MFA2 mRNA is stabilized in an spb8-2 strain. The level of full-length MFA2 mRNA in each lane shown in panel A was quantified by phosphorimaging and then plotted as a function of time. The plotted values have been normalized to an SCR1 loading control. t1/2, half-life. (C) The shortened oligo(G) degradation intermediates in the spb8-2 strain lack mRNA sequences 5′ to the oligo(G) tract. mRNA samples derived from the 0- and 30-min time points in panel A were resolved on a 6% polyacrylamide gel and detected by Northern blot analysis with end-labeled oligonucleotides complementary to a region ending 19 nt 5′ to the oligo(G) tract (oAS22) or starting 13 nt 3′ to the oligo(G) tract (oAS318). Positions of the full-length and oligo(G) deadenylated mRNAs [FL A0 and (G) A0] and the shorter fragments [FL Δ3′ and (G) Δ3′] are indicated.
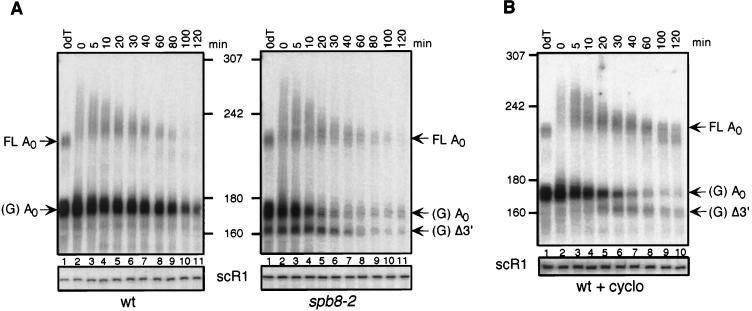
We also examined the degradation patterns of the PGK1pG mRNA in the spb8-2 strain in order to confirm that the alterations we observed were not unique to MFA2pG mRNA. Like the MFA2pG mRNA, degradation of this molecule involves decapping and the action of 5′-3′ exoribonucleases, with oligo(G) degradation intermediates accumulating during the period of degradation (27). Unlike MFA2, however, the degradation rate of PGK1pG mRNA is not as sensitive to mutations in genes involved in either the decapping or the 5′-3′ exonuclease reaction (4, 17, 27). As shown in Fig. 4, the pattern of PGK1pG mRNA degradation was again very different in the spb8-2 strain than in the wild-type strain. Although we observed that this mRNA was not significantly stabilized by this mutation (ca. 15 to 30% stabilization), accumulation of full-length deadenylated species could be detected, and mRNA species shorter than the oligo(G) degradation intermediate were again observable.
FIG. 4.
Degradation of PGK1pG mRNA in the spb8-2 strain. (A) Accumulation of shortened PGK1pG mRNA species in an spb8-2 strain. Yeast strains YAS2274 (SPB8) and YAS2275 (spb8-2) carrying the GAL1:PGK1pG reporter (pRP602) (27) were grown in galactose-containing medium. RNA transcriptional shutoff and subsequent analysis were as for Fig. 3A except that all RNA samples were treated with RNase H-oRP70 in order to resolve the full-length PGK1pG molecules. Positions of the full-length and oligo(G) deadenylated mRNAs [FL A0 and (G) A0] and the oligo(G) shorter fragment [(G) Δ3′] are indicated. wt, wild type. (B) Cycloheximide induces the appearance of shortened PGK1pG mRNA species in a wild-type strain. Yeast strain YAS2274 (SPB8) carrying the GAL1:PGK1pG reporter was grown in galactose-containing medium and shifted to glucose medium containing 100 μg of cycloheximide (cyclo) per ml. RNA samples were analyzed as for Fig. 3A. Positions of the full-length and oligo(G) deadenylated mRNAs [FL A0 and (G) A0] and the oligo(G) shorter fragment [(G) Δ3′] are indicated. Sizes are indicated in nucleotides.
The last mRNA to be examined in detail in the spb8-2 strain was CUP1. In these experiments, we induced CUP1 synthesis by the addition of copper to the growth medium and examined by high-resolution Northern analysis the amount and size distribution of CUP1 mRNA as a function of time. As shown in Fig. 5, the rates of shortening of the poly(A) tail on the CUP1 mRNA in the wild-type and spb8-2 strains were not significantly different. Similar conclusions can be drawn from experiments studying the MFA2pG and PGK1pG mRNAs (Fig. 3 and 4). This result suggests that the spb8-2 mutation does not lead to alterations in deadenylation rates. The degradation pattern of the CUP1 mRNA in the wild-type strain did not include a stable deadenylated intermediate, since transcripts equal in size to the poly(A)− transcripts were not observed. The four different forms of CUP1 mRNA that are observed in the RNase H-oligo(dT) samples (Fig. 5, lanes 8 and 16) result from the creation of alternative 5′ ends on this molecule (22). In contrast to the wild-type strain, however, the spb8-2 strain accumulated CUP1 mRNA which appeared to be completely deadenylated (Fig. 5, lanes 13 and 14). However, and in contrast to the MFA2pG mRNA, no shorter forms of mRNA were observed. In combination with the above studies on the degradation of the MFA2pG and PGK1pG mRNAs, these data on CUP1 mRNA support the conclusions that the spb8-2 mutation leads to an increase in abundance of mRNAs whose lengths are equal to that of a deadenylated mRNA and that it can lead to the appearance of novel mRNA species shorter than deadenylated mRNA from wild-type cells.
FIG. 5.
Accumulation of deadenylated CUP1 mRNA in spb8-2 strains. Yeast strains YAS1915 (SPB8) (wild type [wt]) and YAS2278 (spb8-2) were grown to mid-log phase in YM medium and then induced for CUP1 synthesis by the addition of 0.5 mM CuSO4 to the medium. Aliquots of cells were taken at the indicated time points after induction; the mRNA within them was extracted, resolved on 6% polyacrylamide gel, and analyzed by Northern blot analysis with a CUP1 probe. Positions of the four deadenylated CUP1 transcripts containing alternative 5′ ends are indicated with arrows. Sizes are indicated in nucleotides. 40dT, 40-min time point for RNase H-oligo(dT) treatment.
Stabilization of capped, deadenylated mRNA in the spb8-2 mutant.
Our observation that each of the three mRNAs examined in our studies accumulated mRNA species that were equal to or less than the size expected for a deadenylated mRNA was similar to those previously reported for mRNAs in yeast strains lacking either decapping activity (dcp1 mutants), mrt1 or mrt3 activity, or 5′-3′ exoribonuclease activity (xrn1 mutants) (4, 17, 26, 27). We therefore tested whether the deadenylated MFA2pG mRNA in the spb8-2 mutant accumulated as a capped or uncapped species. This information was needed in order to determine whether decapping or 5′-3′ exonucleolytic degradation was delayed in the spb8-2 strain.
Methods to examine the relative amount of capped versus uncapped deadenylated mRNA in RNA preparations through the use of 7-methyl-cap-specific antibody immunoprecipitation (for instance, see reference 26) or the use of immobilized eIF4E and affinity chromatography (10) have been described. However, we found that these methods did not efficiently deplete the RNA preparations of capped mRNA and as a result did not provide us with an absolute measure of the percentage of capped versus uncapped mRNA in each preparation. We therefore chose to take advantage of the availability of purified Xrn1p (21, 24) and its property of selective degradation of uncapped RNA.
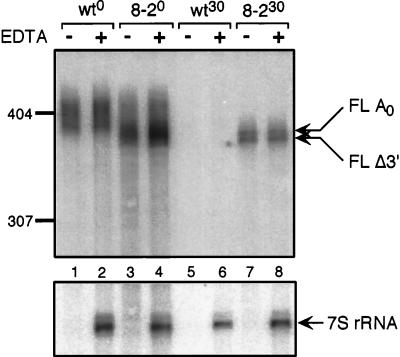
In these experiments, RNA preparations from either the wild type or spb8-2 mutants expressing the MFA2pG mRNA at the steady-state level and 30 min after transcriptional shutoff were incubated with high concentrations of purified Xrn1p in the presence or absence of magnesium, an essential cofactor for the enzyme. Following resolution of the digested RNA on polyacrylamide gels, the extent of RNA degradation by Xrn1p was first determined by examining the degradation of the uncapped 7S rRNA precursor by Northern analysis. As seen in Fig. 6, this species was completely destroyed by Xrn1p treatment.
FIG. 6.
Stabilization of capped, deadenylated mRNA in the wild-type (wt) and spb8-2 (8-2) mutant strains. RNA samples derived from the zero (wt0 or 8-20) or 30-min (wt30 or 8-230) time points of the experiments shown in Fig. 2 were subject to treatment with 400 ng of purified Xrn1p in the presence (+) or absence (−) of neutralizing EDTA. Following resolution of the RNA samples on a 6% polyacrylamide gel, the MFA2pG mRNA was detected with an oligo(C) probe (oRP121). The 7S pre-rRNA was detected with an end-labeled oligonucleotide probe (oAS321) that specifically recognizes the 7S rRNA precursor. Positions of the full-length deadenylated mRNA (FL A0) and the shorter fragments (FL Δ3′) are indicated. Sizes are indicated in nucleotides.
The degradation of the MFA2pG mRNA in both the wild-type and spb8-2 RNA preparations was then examined by Northern analysis. As shown in Fig. 6, Xrn1p treatment results in very little degradation of MFA2pG mRNA prepared from wild-type and spb8-2 strains which are continually expressing the transcript (Fig. 6, lanes 1 and 3). This result is consistent with previous work which showed that full-length uncapped MFA2 mRNA is undetectable in these preparations (26). Importantly, MFA2pG mRNA that accumulates in the spb8-2 strain 30 min after transcriptional shutoff was also resistant to Xrn1p (Fig. 6, lane 7). The resistance of the deadenylated full-length and the shortened MFA2pG mRNA species to Xrn1p digestion provides strong evidence that these mRNAs contain a cap structure. Based on these data, we conclude that the spb8-2 mutation leads to a stabilization of capped, deadenylated mRNA molecules. These data also lend support to our hypothesis (see below) that the shortened MFA2pG mRNAs arise from 3′-to-5′ exonuclease shortening.
Shortened mRNAs similar to those in the spb8-2 mutant arise in wild-type strains treated with cycloheximide.
The appearance of mRNA species in the spb8-2 mutant that were shorter than their deadenylated mRNA counterparts from wild-type cells raised the possibility that they were 3′-5′ exonucleolytic degradation intermediates. Alternatively, it was possible that the spb8-2 mutation led to aberrant 3′-end site selection during the cleavage and polyadenylation reaction. Two different experiments were performed to examine these possibilities.
In the first experiment, we hypothesized that the spb8-2 mutation was allowing for the appearance of 3′-5′ degradation intermediates because of its inhibition of decapping and the 5′-3′ exonucleolytic pathway. This hypothesis was based on the observation that PGK1 3′-5′ degradation intermediates are detectable in xrn1Δ yeast strains (27). Cycloheximide has also been shown to inhibit 5′-3′ mRNA degradation, leading to the accumulation of deadenylated capped mRNAs, and has therefore also been used to reveal these same PGK1 3′-5′ intermediates (27). Based on these data, we anticipated that treatment of our wild-type cells with cycloheximide would lead to the appearance of degradation intermediates identical to those found in the spb8-2 strain.
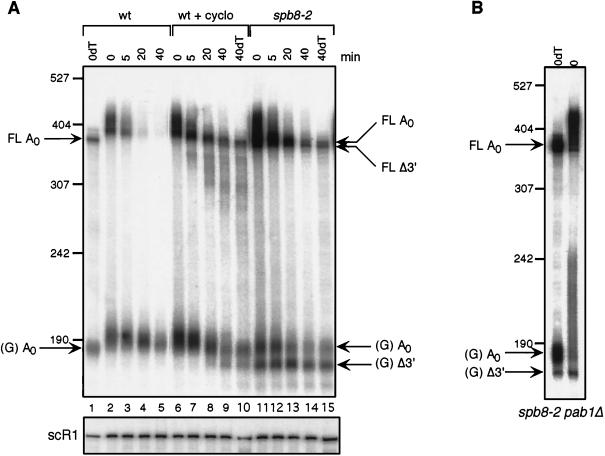
Accordingly, wild-type yeast cells containing either the MFA2pG or PGK1pG mRNA were treated with cycloheximide just prior to shutting off the transcription of these genes, and mRNA from these cells was then analyzed at different times after this shutoff. Cycloheximide treatment resulted in the accumulation of shortened MFA2pG mRNA species in wild-type cells, with mobilities identical to those observed in the spb8-2 mutant (Fig. 7A; compare lanes 8 and 9 with lanes 13 and 14). These included both the shortened full-length molecules and the oligo(G) degradation intermediates. We do not yet understand why our wild-type yeast strain accumulates larger amounts of these products in the presence of cycloheximide than does the strain used in previous studies (3). Degradation of PGK1pG mRNA in a wild-type strain in the presence of cycloheximide also resulted in the appearance of a shorter oligo(G) degradation intermediate (Fig. 4B). The absence of detectable shorter full-length PGK1pG mRNA forms in the spb8-2 mutant could be attributed to a lack of resolution of these gels as well as to the weak signal obtained in this type of analysis, which requires RNase H digestion in order to resolve the full-length PGK1pG molecules. However, the fact that mRNA species in cycloheximide-treated wild-type cells are identical in mobility to those in the spb8-2 cells supports the hypothesis that these species are 3′-5′ mRNA degradation intermediates whose abundance is increased as a result of stabilization of the cap structure. The greater abundance of mRNA degradation intermediates in cycloheximide-treated wild-type cells (Fig. 4B, lanes 9 and 10; Fig. 7A, lanes 7 to 9) also indicates that cycloheximide inhibits 5′-3′ degradation of mRNA to a greater extent than the spb8-2 mutation.
FIG. 7.
The shortened mRNA species in the spb8-2 mutant arise in wild-type yeast cells treated with cycloheximide and are not polyadenylated in pab1Δ strains. (A) Accumulation of shortened MFA2pG mRNA in a wild-type (wt) strain treated with cycloheximide (cyclo). Yeast strains YAS2272 (SPB8) and YAS2273 (spb8-2) carrying the GAL1:MFA2pG reporter were grown in galactose-containing medium and shifted to glucose medium to shut off transcription of the MFA2 mRNA. When indicated, cycloheximide was added at time zero of transcriptional shutoff. RNA samples derived from cells harvested at the indicated times after glucose addition were resolved on 6% polyacrylamide gels, and the MFA2 mRNA was detected by hybridization to end-labeled oligo(C). The length and mobility of size standards are indicated in nucleotides at the left. The lower panel shows the hybridization signal with the SCR1 probe. The lanes containing the RNA samples treated with RNase H-oligo(dT) (0dT and 40dT) provide size markers for the deadenylated mRNA species. The position of the full-length and oligo(G) deadenylated mRNAs [FL A0 and (G) A0] and the shorter fragments [FL Δ3′ and (G) Δ3′] are indicated. (B) The shortened MFA2pG mRNA intermediate is not derived from a polyadenylated precursor. Yeast strain YAS2279 (spb8-2 pab1Δ) carrying the GAL1:MFA2pG reporter and growing in galactose was harvested, and its MFA2pG mRNA was detected by Northern blot analysis with either prior treatment (0dT) or no treatment (0) with RNase H-oligo(dT) to remove the poly(A) tail. Positions of the full-length and oligo(G) deadenylated mRNAs [FL A0 and (G) A0] and the shorter oligo(G) fragment [(G) Δ3′] are indicated. The MFA2 mRNA was detected by hybridization to end-labeled oligo(C).
In the second experiment to determine the origin of the shortened mRNA species, we investigated whether they resulted from aberrant 3′-end formation. For these experiments, we took advantage of the observation that yeast strains lacking PAB1 accumulate oligo(G) degradation intermediates that are polyadenylated (7). We reasoned that if the shorter versions of these degradation intermediates were polyadenylated in the spb8-2 pab1Δ RNA preparations (i.e., represented natural 3′ ends), then their relative abundance would be increased in an mRNA sample which had been deadenylated by treatment with RNase H-oligo(dT), since the smear observed on a gel and resulting from poly(A) tails of different lengths would be resolved into a single band. As shown in Fig. 7B, the abundance of the longer oligo(G) intermediate was greatly increased upon RNase H-oligo(dT) treatment relative to the untreated sample. In contrast, the abundance of the shorter oligo(G) fragment in an RNA sample from an spb8-2 pab1Δ double mutant that had been subject to deadenylation by RNase H treatment was not significantly different from that of the untreated RNA sample. These data show that very little, if any, of the oligo(G) degradation intermediate is polyadenylated. Based on these and the above data, we tentatively conclude that the shortened mRNA fragments in the spb8-2 mutant result from 3′-5′ exonucleolytic degradation of mRNA containing a stabilized cap structure.
The spb8-2 mutation does not lead to stabilization of mRNA degraded via the Upf pathway.
The degradation of mRNA molecules containing a premature nonsense codon depends on the group of Upf proteins that allow for the stimulation of mRNA decapping once the nonsense codon and RNA sequences downstream of it are recognized (reviewed in reference 32). That decapping is the rate-limiting step of this degradation pathway is supported by the observation that mutations in either Dcp1p or Xrn1p lead to stabilization of nonsense codon-containing mRNAs (4, 29). In contrast, the mrt1 and mrt3 mutations, which lead to stabilization of cap structures on mRNAs degraded via the normal pathway, do not stabilize the cap structures on nonsense codon-containing mRNAs (17). These data have resulted in the creation of a model which suggests that Dcp1p activity is separately regulated via the Upf proteins for nonsense codon stimulated mRNA decay and by the MRT gene products for regular mRNA decay (4). We examined whether the spb8-2 mutation, which stabilizes cap structures on mRNAs undergoing the normal pathway of degradation, would stabilize cap structures on mRNAs containing a premature stop codon.
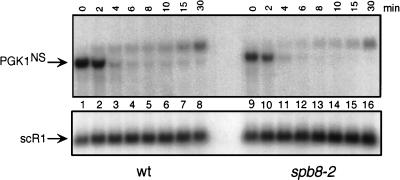
The mRNA substrate used in this experiment (PGK1NSpG) contains a destabilizing nonsense codon in the yeast PGK1pG mRNA (4). It has previously been demonstrated that the rapid degradation of this mRNA in yeast results from the activity of the Upf proteins (18) and that this degradation can be inhibited by mutations in Dcp1p or Xrn1p (4, 29). In our experiments, the transcription of this mRNA, which is driven by the yeast GAL1 promoter, was stimulated by growth of the cells in galactose medium and then repressed by transferring them to glucose medium. As shown in Fig. 8, the degradation rates of this mRNA were nearly identical in both wild-type and spb8-2 strains. The origin of the hybridizing species above the PGK1pG in these experiments is currently unknown. These data lead us to conclude that while the spb8-2 mutation results in stabilization of cap structures on wild-type mRNA, it does not lead to stabilization of cap structures on mRNAs subject to the nonsense codon-stimulated degradation pathway. We therefore place the Spb8 protein into the same category as the MRT1 and MRT3 gene products with respect to the ability to stabilize selectively a subclass of mRNA molecules.
FIG. 8.
The spb8-2 mutation does not alter the stability of a nonsense codon containing mRNA. mRNA samples were prepared from either the wild-type (wt; YAS2276) or spb8-2 (YAS2277) strain carrying the GAL1:PGK1NSpG reporter mRNA (pRP611) (29). Yeast cells were grown in galactose minimal medium and then shifted to glucose for the indicated times. PGK1NSpG mRNAs were detected by Northern blot analysis with an end-labeled oligo(C) probe. Positions of the PGK1NSpG mRNA and the SCR1 RNA loading control are indicated.
DISCUSSION
We have identified a bypass suppressor mutation of a pab1Δ mutant that lies within the SPB8 (YJL124c) gene. Mutations in this gene also lead to the accumulation of capped mRNA degradation intermediates, the appearance of degradation intermediates arising from 3′-5′ nucleolytic attack, and in some cases the stabilization of mRNA. They do not, however, lead to stabilization of mRNA containing a premature nonsense codon. Based on these data, we conclude that the Spb8 protein either directly or indirectly controls the rate of yeast mRNA decapping. We presume that the mRNA stabilization resulting from loss-of-function mutations in SPB8 allows for cell growth in the absence of Pab1p by allowing for sufficient translation of key mRNAs required for cell viability.
The origin of the mRNA degradation intermediates that appeared in the spb8-2 mutant can be inferred from the knowledge that this mutation leads to stabilization of the mRNA cap structure. The slowing of mRNA decapping in the mutant strain probably allows for complete deadenylation of the mRNA through the activity of the normal poly(A) tail degradation machinery. Then, because of the stability of the cap and the resulting resistance of the mRNA to the normally highly active 5′-3′ exonuclease Xrn1p, endogenous 3′-5′ exonuclease(s) chews into the mRNA from the 3′ end. Once the cap structure is removed from either the deadenylated or 3′-end trimmed mRNA species, the activity of Xrn1p results in the appearance of oligo(G) degradation intermediates of different lengths. In this model, therefore, the shorter oligo(G) intermediates can arise from the 3′-end trimmed capped mRNA species. This aspect of our model is most consistent with our observation that shortened oligo(G) degradation intermediates do not appear to accumulate in wild-type cells as a function of time following transcriptional shutoff, even though the longer oligo(G) intermediates continue to be slowly degraded by presumably other 3′-5′ exoribonucleases. The alternative model, that both cycloheximide and the spb8-2 mutation lead to the activation of a 3′-5′ exonuclease activity which shortens both the full-length mRNA and the oligo(G) intermediate, could be supported by the data presented in Fig. 4. As seen in Fig. 4A, the degradation rate of the oligo(G) intermediate seems to be higher in the presence of the spb8-2 mutation, and the shortened oligo(G) fragment would appear to be derived from the deadenylated oligo(G) fragment in a wild-type cell treated with cycloheximide (Fig. 4B). A more thorough analysis of the degradation rates of the oligo(G) intermediates for both the MFA2pG and PGK1pG mRNAs should help to distinguish between each of these models.
The nucleases trimming the mRNA in the spb8-2 strain appear to be stalled at each of several unique positions within the MFA2 and PGK1 3′UTRs. In the case of the CUP1 mRNA, a major stall site would appear to be at or very near the site of poly(A) tail addition since shortened degradation intermediates are not detected in our experiments. We assume that these 3′-5′ exoribonucleases can move past their stall sites and eventually degrade the mRNA. Although we have not looked directly for shorter degradation intermediates that would arise from this process, shorter 3′-5′ exonuclease products have been previously found upon analysis of the degradation of PGK1pG mRNA in a wild-type strain treated with cycloheximide (27). The stall sites within the 3′UTR may represent RNA sequences that are particularly resistant to degradation by the nuclease(s), or they may result from the presence of proteins bound to the mRNA 3′UTR that are involved in some other aspect of mRNA metabolism. Future work on identifying and characterizing the locations and sequences of these sites could help to distinguish between these possibilities.
The detection of 3′-5′ degradation intermediates of the normally unstable MFA2 mRNA and the stable PGK1 mRNA in the spb8-2 strain indicates that mRNAs of all stability classes could be subject to 3′-5′ degradation when the decapping reaction is slowed. This mode of degradation could explain why the stable PGK1 mRNA is not significantly stabilized when either the decapping or 5′-3′ exonucleolytic activities of yeast are compromised (4, 27). Similar conclusions have been reached by other laboratories studying 3′-5′ degradation intermediates stalled at an oligo(G) tract in the 3′UTR (27). The existence of a reasonably active 3′-5′ exonucleolytic activity in yeast may also explain why the stabilities of most yeast mRNAs are enhanced only severalfold when either DCP1 or XRN1 is deleted (4, 20, 27). A protein complex consisting of five essential proteins, and required for the maturation of the 5.8S rRNA, has recently been identified in yeast and termed the exosome (25). This complex displays 3′-5′ exonuclease activity, and two of its components, Ski6p/Rrp41p and Rrp4p, have been shown to be required for 3′-to-5′ decay of mRNA in yeast (1). It will be interesting to determine whether loss of this enzymatic activity in a spb8-2 strain prevents the appearance of the 3′-5′ degradation intermediates.
Previous isolation and characterization of many bypass suppressor mutations of pab1Δ in yeast have led to the model that these mutations act by altering the ratio of ribosomal subunits (33, 34, 40). A smaller number of pab1Δ bypass suppressor mutations appear to act by making mRNA more stable than it is normally in a wild-type cell. These include bypass suppressor mutations in the genes encoding Spb8p, as well as those encoding Dcp1p, Mrt1p, and Mrt3p (17). Based on the hypothesis that the yeast translational system is compromised in the absence of Pab1p, we assume that this class of suppressor acts by maintaining higher than normal levels of mRNA so as to compensate for their decreased translational rates. This would result in increased levels of total protein production per newly synthesized transcript, which could then allow for cell viability in the absence of Pab1p. A third group of pab1Δ bypass suppressor mutations, of which we (24a) and others (24b) have identified one member, the SPB9/PBP1 gene, appears to exert its effects through neither of these two mechanisms. Understanding the mechanism of suppression of this class should shed even more light on how mRNA translation can be enhanced in a Pab1p-deficient yeast cell.
In the absence of more detailed information about the normal functions of Spb8p, we can only hypothesize how mutations in it lead to stabilization of the cap structure. We envision that these mutations could result in any one of several changes which could delay decapping of mRNA. These include, but are not limited to, a direct inhibition of the Dcp1 enzyme, an inhibition of an activator of Dcp1, an inhibition of some aspect of the translation cycle that indirectly leads to slowed decapping, and even possibly a modification of part of the ribosome that both leads to enhanced binding of the 40S subunit in the absence of Pab1p and, as an indirect effect, stabilization of the mRNA cap structure.
The identification of Spb8p as a factor involved in controlling mRNA decapping in vivo adds another player to the roster of yeast proteins now known to be involved in this step. As with the MRT1 and MRT3 gene products, Spb8p appears to exert its activity specifically during the normal mRNA degradation process. The presence of an Sm-like domain in Spb8p does suggest the interesting possibility that it could be part of an RNA-protein complex in the cell and that it could be localized within the nucleus. Future work aimed at examining each of these facets of Spb8p should help to define further its function within the yeast cell.
ACKNOWLEDGMENTS
We thank members of our laboratory for advice during the course of this work and for critical reading of the manuscript. We thank M. Snyder (Yale University) for the mini-Tn3 mutagenized yeast genomic library, R. Parker (University of Arizona, Tucson) for many of the plasmids used in this study, and N. Cozzarelli (University of California, Berkeley) for the purified Xrn1p.
R.B. is a recipient of a Human Frontier Science Program postdoctoral fellowship. This work was supported by grant NP944 to A.B.S. from the American Cancer Society.
REFERENCES
- 1.Anderson J S, Parker R P. The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J. 1998;17:1497–1506. doi: 10.1093/emboj/17.5.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ares M, Jr, Igel A H. Lethal and temperature-sensitive mutations and their suppressors identify an essential structural element in U2 small nuclear RNA. Genes Dev. 1990;4:2132–2145. doi: 10.1101/gad.4.12a.2132. [DOI] [PubMed] [Google Scholar]
- 3.Beelman C A, Parker R. Differential effects of translational inhibition in cis and in trans on the decay of the unstable yeast MFA2 mRNA. J Biol Chem. 1994;269:9687–9692. [PubMed] [Google Scholar]
- 4.Beelman C A, Stevens A, Caponigro G, LaGrandeur T E, Hatfield L, Fortner D M, Parker R. An essential component of the decapping enzyme required for normal rates of mRNA turnover. Nature. 1996;382:642–646. doi: 10.1038/382642a0. [DOI] [PubMed] [Google Scholar]
- 5.Broach J R, Strathern J N, Hicks J B. Transformation in yeast: development of a hybrid cloning vector and isolation of the CAN1 gene. Gene. 1979;8:121–133. doi: 10.1016/0378-1119(79)90012-x. [DOI] [PubMed] [Google Scholar]
- 6.Burns N, Grimwade B, Ross-Macdonald P B, Choi E Y, Finberg K, Roeder G S, Snyder M. Large-scale analysis of gene expression, protein localization, and gene disruption in Saccharomyces cerevisiae. Genes Dev. 1994;8:1087–1105. doi: 10.1101/gad.8.9.1087. [DOI] [PubMed] [Google Scholar]
- 7.Caponigro G, Parker R. Multiple functions for the poly(A)-binding protein in mRNA decapping and deadenylation in yeast. Genes Dev. 1995;9:2421–2432. doi: 10.1101/gad.9.19.2421. [DOI] [PubMed] [Google Scholar]
- 8.Decker C J, Parker R. Mechanisms of mRNA degradation in eukaryotes. Trends Biochem Sci. 1994;19:336–340. doi: 10.1016/0968-0004(94)90073-6. [DOI] [PubMed] [Google Scholar]
- 9.Decker C J, Parker R. A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes Dev. 1993;7:1632–1643. doi: 10.1101/gad.7.8.1632. [DOI] [PubMed] [Google Scholar]
- 10.Edery I, Chu L L, Sonenberg N, Pelletier J. An efficient strategy to isolate full-length cDNAs based on an mRNA cap retention procedure (CAPture) Mol Cell Biol. 1995;15:3363–3371. doi: 10.1128/mcb.15.6.3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elledge S J, Davis R W. A family of versatile centromeric vectors designed for use in the sectoring-shuffle mutagenesis assay in Saccharomyces cerevisiae. Gene. 1988;70:303–312. doi: 10.1016/0378-1119(88)90202-8. [DOI] [PubMed] [Google Scholar]
- 12.Felici F, Cesareni G, Hughes J M. The most abundant small cytoplasmic RNA of Saccharomyces cerevisiae has an important function required for normal cell growth. Mol Cell Biol. 1989;9:3260–3268. doi: 10.1128/mcb.9.8.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fromont-Racine M, Rain J C, Legrain P. Toward a functional analysis of the yeast genome through exhaustive two-hybrid screens. Nat Genet. 1997;16:277–282. doi: 10.1038/ng0797-277. [DOI] [PubMed] [Google Scholar]
- 14.Gietz D, St. Jean A, Woods R A, Schiestl R H. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gietz R D, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 16.Guthrie C, Fink G R. Guide to yeast genetics and molecular biology. Methods Enzymol. 1991;194:1–933. [PubMed] [Google Scholar]
- 17.Hatfield L, Beelman C A, Stevens A, Parker R. Mutations in trans-acting factors affecting mRNA decapping in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:5830–5838. doi: 10.1128/mcb.16.10.5830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He F, Jacobson A. Identification of a novel component of the nonsense-mediated mRNA decay pathway by use of an interacting protein screen. Genes Dev. 1995;9:437–454. doi: 10.1101/gad.9.4.437. [DOI] [PubMed] [Google Scholar]
- 19.Hermann H, Fabrizio P, Raker V A, Foulaki K, Hornig H, Brahms H, Luhrmann R. snRNP Sm proteins share two evolutionarily conserved sequence motifs which are involved in Sm protein-protein interactions. EMBO J. 1995;14:2076–2088. doi: 10.1002/j.1460-2075.1995.tb07199.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hsu C L, Stevens A. Yeast cells lacking 5′→3′ exoribonuclease 1 contain mRNA species that are poly(A) deficient and partially lack the 5′ cap structure. Mol Cell Biol. 1993;13:4826–4835. doi: 10.1128/mcb.13.8.4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson A W, Kolodner R D. Strand exchange protein 1 from Saccharomyces cerevisiae. A novel multifunctional protein that contains DNA strand exchange and exonuclease activities. J Biol Chem. 1991;266:14046–14054. [PubMed] [Google Scholar]
- 22.Karin M, Najarian R, Haslinger A, Valenzuela P, Welch J, Fogel S. Primary structure and transcription of an amplified genetic locus: the CUP1 locus of yeast. Proc Natl Acad Sci USA. 1984;81:337–341. doi: 10.1073/pnas.81.2.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kessler S H, Sachs A B. RNA recognition motif 2 of yeast Pab1p is required for its functional interaction with eukaryotic translation initiation factor 4G. Mol Cell Biol. 1998;18:51–57. doi: 10.1128/mcb.18.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kolodner R, Evans D H, Morrison P T. Purification and characterization of an activity from Saccharomyces cerevisiae that catalyzes homologous pairing and strand exchange. Proc Natl Acad Sci USA. 1987;84:5560–5564. doi: 10.1073/pnas.84.16.5560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24a.Lapeyre, B. Unpublished data.
- 24b.Mangus, D., and A. Jacobson. Personal communication.
- 25.Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D. The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′→5′ exoribonucleases. Cell. 1997;91:457–466. doi: 10.1016/s0092-8674(00)80432-8. [DOI] [PubMed] [Google Scholar]
- 26.Muhlrad D, Decker C J, Parker R. Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5′→3′ digestion of the transcript. Genes Dev. 1994;8:855–866. doi: 10.1101/gad.8.7.855. [DOI] [PubMed] [Google Scholar]
- 27.Muhlrad D, Decker C J, Parker R. Turnover mechanisms of the stable yeast PGK1 mRNA. Mol Cell Biol. 1995;15:2145–2156. doi: 10.1128/mcb.15.4.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muhlrad D, Parker R. Mutations affecting stability and deadenylation of the yeast MFA2 transcript. Genes Dev. 1992;6:2100–2111. doi: 10.1101/gad.6.11.2100. [DOI] [PubMed] [Google Scholar]
- 29.Muhlrad D, Parker R. Premature translational termination triggers mRNA decapping. Nature. 1994;370:578–581. doi: 10.1038/370578a0. [DOI] [PubMed] [Google Scholar]
- 30.Myslinski E, Segault V, Branlant C. An intron in the genes for U3 small nucleolar RNAs of the yeast Saccharomyces cerevisiae. Science. 1990;247:1213–1216. doi: 10.1126/science.1690452. [DOI] [PubMed] [Google Scholar]
- 31.Ross-Macdonald P B, Burns N, Malczynski M, Sheehan A, Roeder G S, Snyder M. Methods for large-scale analysis of gene expression, protein localization, and disruption phenotypes in Saccharomyces cerevisiae. Methods Mol Cell Biol. 1995;5:298–308. [Google Scholar]
- 32.Ruiz E M, Czaplinski K, Peltz S W. Making sense of nonsense in yeast. Trends Biochem Sci. 1996;21:433–438. doi: 10.1016/s0968-0004(96)10055-4. [DOI] [PubMed] [Google Scholar]
- 33.Sachs A B, Davis R W. The poly(A) binding protein is required for poly(A) shortening and 60S ribosomal subunit-dependent translation initiation. Cell. 1989;58:857–867. doi: 10.1016/0092-8674(89)90938-0. [DOI] [PubMed] [Google Scholar]
- 34.Sachs A B, Davis R W. Translation initiation and ribosomal biogenesis: involvement of a putative rRNA helicase and RPL46. Science. 1990;247:1077–1079. doi: 10.1126/science.2408148. [DOI] [PubMed] [Google Scholar]
- 35.Sachs A B, Davis R W, Kornberg R D. A single domain of yeast poly(A)-binding protein is necessary and sufficient for RNA binding and cell viability. Mol Cell Biol. 1987;7:3268–3276. doi: 10.1128/mcb.7.9.3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sachs A B, Sarnow P, Hentze M W. Starting at the beginning, middle, and end: translation initiation in eukaryotes. Cell. 1997;89:831–838. doi: 10.1016/s0092-8674(00)80268-8. [DOI] [PubMed] [Google Scholar]
- 37.Schweinfest C W, Graber M W, Chapman J M, Papas T S, Baron P L, Watson D K. CaSm: an Sm-like protein that contributes to the transformed state in cancer cells. Cancer Res. 1997;57:2961–2965. [PubMed] [Google Scholar]
- 38.Séraphin B. Sm and Sm-like proteins belong to a large family: identification of proteins of the U6 as well as the U1, U2, U4 and U5 snRNPs. EMBO J. 1995;14:2089–2098. doi: 10.1002/j.1460-2075.1995.tb07200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tarun S Z, Jr, Sachs A B. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J. 1996;15:7168–7177. [PMC free article] [PubMed] [Google Scholar]
- 40.Tarun S Z, Jr, Sachs A B. A common function for mRNA 5′ and 3′ ends in translation initiation in yeast. Genes Dev. 1995;9:2997–3007. doi: 10.1101/gad.9.23.2997. [DOI] [PubMed] [Google Scholar]
- 41.Tarun S Z, Jr, Wells S E, Deardorff J A, Sachs A B. Translation initiation factor eIF4G mediates in vitro poly(A) tail-dependent translation. Proc Natl Acad Sci USA. 1997;94:9046–9051. doi: 10.1073/pnas.94.17.9046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wickens M, Anderson P, Jackson R J. Life and death in the cytoplasm: messages from the 3′ end. Curr Opin Genet Dev. 1997;7:220–232. doi: 10.1016/s0959-437x(97)80132-3. [DOI] [PubMed] [Google Scholar]
- 43.Zhong T, Arndt K T. The yeast SIS1 protein, a DnaJ homolog, is required for the initiation of translation. Cell. 1993;73:1175–1186. doi: 10.1016/0092-8674(93)90646-8. [DOI] [PubMed] [Google Scholar]