Abstract
Previous work has demonstrated that two key melanocyte-specific elements termed the MSEu and MSEi play critical roles in the expression of the melanocyte-specific tyrosinase-related protein 1 (TRP-1) promoter. Both the MSEu and MSEi, located at position −237 and at the initiator, respectively, bind a melanocyte-specific factor termed MSF but are also recognized by a previously uncharacterized repressor, since mutations affecting either of these elements result in strong up-regulation of TRP-1 promoter activity in melanoma cells. Here we demonstrate that repression mediated by the MSEu and MSEi also operates in melanocytes. We also report that both the MSEu and MSEi are recognized by the brachyury-related transcription factor Tbx2, a member of the recently described T-box family, and that Tbx2 is expressed in melanocyte and melanoblast cell lines but not in melanoblast precursor cells. Although Tbx2 and MSF each recognize the TRP-1 MSEu and MSEi motifs, it is binding by Tbx-2, not binding by MSF, that correlates with repression. Several lines of evidence tend to point to the brachyury-related transcription factor Tbx2 as being the repressor of TRP-1 expression: both the MSEu and MSEi bind Tbx2, and mutations in either element that result in derepression of the TRP-1 promoter diminish binding by Tbx2; the TRP-1 promoter, but not the tyrosinase, microphthalmia, or glyceraldehyde-3-phosphate dehydrogenase (G3PDH) promoter, is repressed by Tbx2 in cotransfection assays; a high-affinity consensus brachyury/Tbx2-binding site is able to constitutively repress expression of the heterologous IE110 promoter; and a low-affinity brachyury/Tbx2 binding site is able to mediate Tbx2-dependent repression of the G3PDH promoter. Although we cannot rule out the presence of an additional, as yet unidentified factor playing a role in the negative regulation of TRP-1 in vivo, the evidence presented here suggests that Tbx2 most likely is the previously unidentified repressor of TRP-1 expression and as such is likely to represent the first example of transcriptional repression by a T-box family member.
In attempting to understand how the precise temporal and spatial pattern of gene expression necessary for the development of an organism is achieved, consideration should be given not only to the question of why a specific gene is expressed in a particular cell type at any given time, but also to why it is not expressed elsewhere or at other times. Analysis of tissue-specific promoters in many cell types has revealed that they often contain binding sites for widely expressed transcription factors which may act together with factors with a more restricted tissue distribution. However, while these tissue-specific factors may be present in only a very limited number of cell types, they frequently fall into transcription factor families, members of which have identical or highly similar DNA-binding properties. For example, the CANNTG E-box motif is recognized by members of the basic helix-loop-helix (bHLH) class of transcription factors, and while some bHLH factors are clearly expressed in a highly tissue restricted fashion, all cells contain bHLH factors capable of recognizing the E-box motif. To enable the expression of a promoter containing an E-box element to be limited to a specific cell type, some mechanism must operate to restrict its expression in other cells. It seems likely that in addition to the positive regulators of transcription which have received much attention over recent years, sequence-specific repressors acting to restrict expression to specific cell types may be equally important. Indeed, lessons learned from genetic studies of Drosophila suggest that transcriptional repressors may be as common as activators (37).
Melanocytes afford a particularly attractive system for understanding the molecular mechanisms operating to achieve the commitment and differentiation of a cell lineage. Melanocytes originate in the neural crest as a dispersed population of melanoblasts which migrate primarily to the hair follicles and epidermis before differentiating into mature, pigmented cells. In addition to being responsible for skin, hair, and eye color (41), melanocytes perform an essential function in the generation of action potentials in the inner ear (43). Moreover, in response to UV irradiation, skin melanocytes increase the production of the pigment melanin, which is then transferred to the surrounding keratinocytes as protection from UV-induced DNA damage (15). Since melanocytes are not essential for viability, and because pigmentation is an obvious phenotype, around 70 genes which affect the melanocyte lineage have been identified by genetic analysis; of these 70, more than 20 have now been cloned. The cloned genes include those whose products play crucial roles in melanocyte commitment, survival, or differentiation, such as the c-Kit (31) and the endothelin B receptors (4, 21, 36) and the transcription factors microphthalmia (20, 29, 46), Pax3 (2, 11), and Sox10 (34, 43), as well as genes encoding melanogenic enzymes, such as tyrosinase and tyrosinase-related protein 1 (TRP-1), which map to the albino and brown loci, respectively (22, 27, 38). The isolation and analysis of the promoters which control expression of the tyrosinase and TRP-1 genes (7, 8, 9, 16, 17, 23, 26, 28, 35, 40, 49) have provided an insight into how melanocyte-specific gene expression is achieved. Although it was anticipated that because tyrosinase and TRP-1 are both melanogenic enzymes, they would be subject to coordinate regulation by a shared set of transcription factors, it is now clear that their proximal promoters contain only a single common element, the M box (28). The 11-bp M box element contains at its core a CATGTG motif recognized by the bHLH-leucine zipper (LZ) transcription activator microphthalmia (7, 17, 18, 48, 50), which plays an essential role in melanocyte development. The proximal tyrosinase promoter contains both the M box and a second essential CATGTG motif located at its initiator, as well as a conserved Sp1 element (7). By contrast, the TRP-1 promoter is considerably more complex. In addition to the M box located at position −100 with respect to the transcription initiation site, two key melanocyte-specific elements termed the MSEu and MSEi play critical roles in the regulation of TRP-1 expression (49). Both the MSEu and the MSEi, located at position −237 and at the initiator, respectively, bind a melanocyte-specific factor termed MSF. Mutational analysis of each element has indicated that MSF plays a positive role in regulation of TRP-1 promoter activity. In contrast, both the MSEu and MSEi are also recognized by a repressor, since mutations affecting either of these elements result in strong up-regulation of TRP-1 promoter activity. Although repressor DNA-binding activity was not detected in cell extracts, point mutation of the MSEu and MSEi coupled to functional analysis suggests that its DNA-binding specificity is related to but different from that of MSF. Clearly, establishing the identity of both MSF and the repressor which play key roles in regulation of melanocyte-specific gene expression is a major goal.
In this paper, we report that melanocyte and melanoblast cell lines, but not melanoblast precursor cells, express the brachyury-related transcription factor Tbx2. Although Tbx2 and MSF recognize both the TRP-1 MSEu and MSEi motifs, it is binding by Tbx-2, not binding by MSF, that correlates with repression.
MATERIALS AND METHODS
Cell lines and transfection assays.
The mouse melanocyte cell line melan-c was grown in RPMI 1640 with 10% fetal calf serum and 200 ng of phorbol myristyl acetate per ml. Transfections were performed by using Lipofectamine reagent (Life Technologies). Cells were plated at 5 × 104/ml in 5-cm-diameter dishes 1 day before transfection. After two washes with serum-free medium, 1.7 ml of serum-free medium was added. A total of 1 μg of DNA in 100 μl of serum-free medium was mixed with an appropriate amount of Lipofectamine in 200 μl of serum-free medium, left for 30 min at room temperature, and then added to the cells. After 5 h at 37°C, the medium was removed and the cells were washed once in serum-free medium before the addition of 4 ml of medium with serum. Cells were harvested 2 days later and processed. The optimal amount of Lipofectamine used per microgram of DNA was determined empirically for each batch of Lipofectamine and was found to vary between 1 and 14 μl. All transfections were repeated with different preparations of DNA, and pCH110 containing the simian virus 40 promoter driving expression of a LacZ reporter was used as an internal control for transfection efficiency (1 μg per transfection). Chloramphenicol acetyltransferase (CAT) assays were performed as described previously (49) and were quantitated by excising the spots following thin-layer chromatography and scintillation counting. Luciferase assays were carried out as instructed by the manufacturer of the luciferase assay reagent (Promega) and were quantitated with a Bertholdt Microlumat LB 96Vplate luminometer. The origin and culture of the melanocyte, melanoblast, and melanoblast precursor cell lines used for the Northern blots have been described previously (3, 5, 6, 45).
Band shift assays.
The band shift assays were performed in a final volume of 20 μl containing 20 mM HEPES (pH 7.9), 10% glycerol, and 112 mM KCl; nuclear extracts were prepared as described previously (49). In vitro-transcribed/translated (ITT) protein was made by using a TNT T7 Quick Coupled Transcription kit as instructed by the manufacturer (Promega). Nuclear extracts or ITT Tbx2 were preincubated at 0°C with 1 μg of poly(dIdC-dIdC) for 10 min before the addition of 10, 50, or 250 ng of cold competitor DNA. After a further incubation for 10 min, approximately 0.5 ng of oligonucleotide probe, labeled at each end by filling in 5′ overhangs with a Klenow fragment and the appropriate α-32P-deoxynucleoside triphosphate, was added to the reaction for a further 20 min before loading onto a 8% polyacrylamide gel (44:1 acrylamide/bisacrylamide ratio) and electrophoresis at 200 V for 1.5 h.
The sequences of double-stranded oligonucleotides used as probes and competitors in Fig. 1, 3, and 5 are as follows: MSEi, 5′-ctagaGAATTCACTGGTGTGAGAAGGGATTAGTt-3′; MSEu, 5′-ctagaAAAGCTAACAGAAAATACAAGTGTGACATTt-3′; LS-MSEi, 5′-ctagaGAATTCACTGGTCGACGAAGGGATTAGTt-3′; and brachyury, 5′-ctagaGGGAATTTCACACCTAGGTGTGAAATTCCCT-3′. Lowercase letters indicate bases added to facilitate cloning.
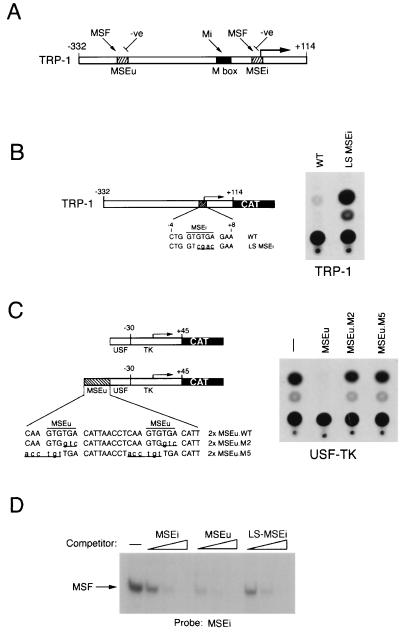
FIG. 1.
Repression of TRP-1 in melanocytes is not mediated by MSF. (A) Schematic showing the elements required for regulation of the TRP-1 promoter. The M box is recognized by the transcription factor microphthalmia, while the MSEu and MSEi are targets for MSF and for negative regulation. (B) The LS-MSEi mutation relieves repression of TRP-1 in melanocytes. The melan-c melanocyte cell line was transfected with the indicated WT or LS-MSEi mutant TRP-1–CAT reporters, and CAT activity was determined. (C) Repression mediated by the MSEu in melanocytes. Melan-c cells were transfected with the USF-TK-CAT reporter or the indicated derivatives containing WT or mutant MSEu elements, and CAT activity was determined 48 h posttransfection. (D) MSF binds the WT and mutants LS-MSEi elements. A radioactive oligonucleotide probe containing the MSEu was used in a band shift assay with B16 melanoma cell nuclear extract, and MSF binding was competed with 10, 50, and 250 ng of unlabeled oligonucleotides containing the MSE, MSEu, or LS-MSEi. Only the bound DNA is shown. The sequence of each oligonucleotide probe and competitor is shown in Fig. 6.
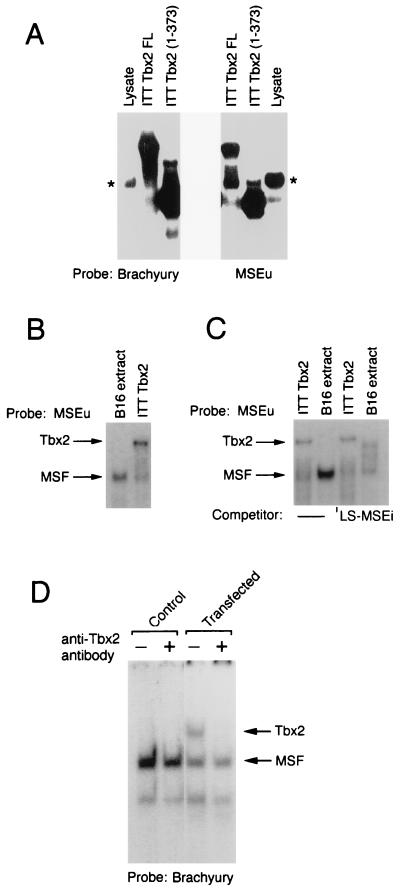
FIG. 3.
Tbx2 can bind the MSEu and MSEi and is distinct from MSF. (A) Band shift assay using a radiolabeled brachyury-binding site or the MSEu as the probe together with either unprogrammed reticulocyte lysate, ITT Tbx2FL, or the C-terminal truncation Tbx2(1-373). Only the bound DNA is shown. The asterisk indicates the position of a complex originating in the unprogrammed reticulocyte lysate and which has a mobility similar to that of MSF. (B) Band shift assay as in panel A, using the MSEu probe and either B16 nuclear extract or ITT Tbx2. The relative positions of the MSF- and Tbx2-containing complexes are indicated. (C) As for panel B but with 50 ng of the indicated LS-MSEi competitor where indicated. (D) Band shift assay using B16 extract derived from untransfected cells (control) or cells transfected with a Tbx2 expression vector, together with a brachyury consensus binding site as a probe. The DNA-binding reaction was performed in the presence or absence of a polyclonal anti-Tbx2 antiserum as indicated.
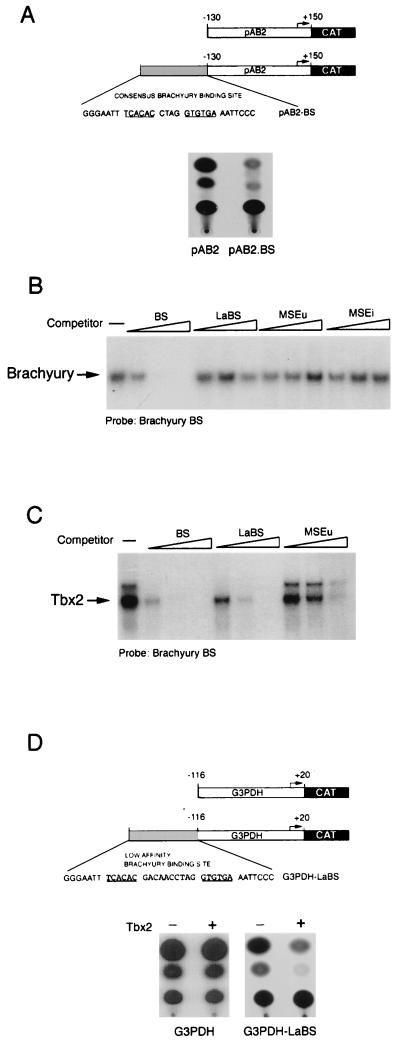
FIG. 5.
Transcriptional repression by Tbx2 in melanocytes. (A) Schematic showing the structure of the herpes simplex virus IE110 promoter (in pAB2) and the consensus brachyury site (BS). (B) Brachyury does not bind the LaBS, the MSEu, or the MSEi, as determined by band shift assay using ITT brachyury together with the consensus brachyury probe and 10, 50, or 250 ng of the indicated consensus BS, LaBS, and MSEu, and MSEi competitors. (C) Tbx2 binds the consensus brachyury site around 5-fold better than the LaBS and around 15-fold better than the MSEu, as determined by band shift assay using ITT Tbx2 together with the consensus brachyury probe and 10, 50, or 250 ng of the consensus (BS) or low-affinity (LaBS) brachyury site or MSEu as the competitor. Only the bound DNA is shown. The full sequences of the BS and LaBS sites are shown in panels A and D, respectively. (D) Transcriptional repression by Tbx2. The indicated G3PDH-CAT or G3PDH.LaBS-CAT reporters were transfected into the melanocyte cell line melan-c either alone or together with a Tbx2 expression vector, and CAT activity was determined 48 h posttransfection.
Plasmids and DNA constructs.
The TRP-1 and thymidine kinase (TK) promoter reporter plasmids used for Fig. 1 have been described previously (49). pAB2 contains the herpes simplex virus IE110 promoter upstream from a CAT reporter and has also been described previously (32). Plasmid pAB2-BS was constructed by insertion into the unique XbaI site upstream from the IE110 promoter of a double-stranded oligonucleotide containing the brachyury consensus binding site (24): 5′-ctagaGGGAATTTCACACCTAGGTGTGAAATTCCCt-3′. The reporter G3PDH (glyceraldehyde-3-phosphate dehydrogenase)-CAT has been described previously (1). Plasmid G3PDH-LaBS-CAT was made by inserting in the unique HindIII site upstream from the G3PDH promoter a double-stranded oligonucleotide containing a low-affinity brachyury-binding site (LaBS): agcttGGGAATTTCACACgacaacctagGTGTGAAATTCCCa.
For the luciferase assays, the TRP-1 promoter extending from −310 to +114 was cloned as an XbaI-HindIII fragment between the NheI and HindIII sites of the pGL3 basic luciferase reporter vector (Promega). The microphthalmia promoter between −387 and +97 was cloned by PCR from genomic DNA by using primers which placed SstI and HindIII sites at the 5′ and 3′ ends, respectively, between the SstI and HindIII sites of the pGL3 basic luciferase reporter vector. The tyrosinase promoter extending from −300 to +80 was cloned as an XbaI-XhoI fragment between the NheI and XhoI sites of the same reporter vector. The tyrosinase and TRP-1 promoter fragments used have been described previously (7, 28).
Antibody production.
A partial Tbx2 cDNA encoding amino acids 361 to 701 was cloned into a glutathione S-transferase expression vector, and the resulting fusion protein was expressed in Escherichia coli prior to purification and injection into rabbits. This region of Tbx2 was chosen because it lies outside the conserved T-box DNA-binding domain and consequently the resulting anti-Tbx2 antiserum was unlikely to cross-react with other members of the T-box family. The anti-Tbx2 antibody was affinity purified before use and could immunoprecipitate Tbx2 but not brachyury.
RNA extraction, RT-PCR, and Northern blot analysis.
Isolation of RNA and blotting procedures were as described (14). For the reverse transcription (RT)-PCR isolation of the Tbx2 cDNA, total melan-a RNA was subjected to RT with avian myeloblastosis virus reverse transcriptase (Boehringer) followed by a first-strand cDNA synthesis (Amersham First Strand cDNA synthesis kit). The initial identification of T-box protein was performed by PCR amplification from melan-a cDNA with two degenerate primers, 5′-agacagatctAGATA{TC} AT{TCA}CA{IC}CCIGA{TC}{AT}{GC}ICC-3′ and 5′-agacagatctAGATT{TC}TG{AG}TAIGCIGTIACIGC-3′, which place BglII sites at each end of the PCR product. PCR was performed with a reaction mixture containing 250 μM deoxynucleotides, 2 μg of primers, 5% dimethyl sulfoxide, and 0.5 U of Taq polymerase (Life Technologies). Cycling parameters were 1 min at 94°C, 2 min at 40°C, and 2 min at 65°C for 36 cycles. Amplified fragments were digested with BglII and cloned into the BamHI site of pBluescript II KS (Stratagene). After the clones obtained by using the degenerate primers were identified by sequencing (Amersham T7 sequencing kit), a full-length mouse Tbx2 (Tbx2FL) cDNA was cloned as two separate pieces by PCR using the following two pairs of primers: 5′-agacggattcGGATCCATGGCTTACCACCCGTTCC-3′ plus 5′-agacgaattcAGATCTCTCCTCCCCCGTCCGCTC-3′, and 5′-GGAGATCTGCAGCGCCCCTGTGCCGCAG-3′ plus 5′-agactctagaggatccTCACTTGGGCGACTCCC-3′. The underlined sequences correspond to a silent mutation allowing the creation of a BglII site in the Tbx2 cDNA. The reaction amplifications were as follows: 32 cycles comprising 1 min at 94°C, 1 min at 50°C, and 2 min at 72°C. The 5′ Tbx2 PCR product was cloned into pGEX-2TK digested with BamHI and EcoRI, followed by the cloning of the 3′ Tbx2 PCR product into the pGEX vector containing the previously cloned 5′ Tbx2 cDNA fragment digested with BglII-XbaI. The Tbx2FL clone was then excised as a BamHI fragment and cloned into pT7plink (39), an ITT vector, and into pCMV19a (50) for expression in mammalian cells.
RESULTS
Repression of the TRP-1 promoter in melanocytes.
Previous work (49) using transfection assays of melanoma cells has established that the melanocyte-specific TRP-1 promoter is primarily regulated by three specific elements: the M box, the MSEi, and the MSEu, the latter two located at the initiator and at position −237, respectively (Fig. 1A). While the M box is regulated positively by the product of the microphthalmia gene, the MSEi and MSEu, which each contain a conserved GTGTGA motif, are recognized by both MSF and an uncharacterized repressor. Mutational analysis of the MSEu and MSEi provided convincing evidence that the repressor was responsible for the extremely low levels of TRP-1 promoter activity observed in melanoma cell lines. Whether the repressor was also active in nontransformed melanocytes or whether its activity was peculiar to melanomas was not established. Therefore, before attempting to characterize further the repressor, we wished to determine whether the TRP-1 promoter was also subject to repression mediated by the MSEu and MSEi in melanocytes.
To determine whether the repression acting through the MSEi was operating in untransformed melanocytes, we transfected the melanocyte cell line melan-c with a TRP-1 promoter–CAT reporter or an identical construct (LS-MSEi) in which four residues within the core MSEi motif had been mutated. The LS-MSEi mutation has been shown previously to result in around an 80-fold increase in TRP-1 promoter activity in transfected melanomas (49). The result (Fig. 1B) demonstrates that the activity of the TRP-1 promoter is significantly (around 15-fold) derepressed by the LS-MSEi mutation, indicating that repression through this element occurs in melanocytes as well as in melanomas. To verify that repression also was mediated in melanocytes by the related MSEu, we made use of a sensitive assay for MSEu function in which a double-MSEu element is placed upstream from a minimal TK promoter fused to a USF-binding site. We have used this assay previously to demonstrate the repression activity mediated via the MSEu (49). The USF-TK reporter was therefore transfected into the melan-c melanocyte cell line either in the presence or in the absence of wild-type (WT) MSEu or MSEu mutants (M2 and M5) which we have previously shown to be defective for transcriptional repression in melanomas. The results (Fig. 1C) reveal that as in melanoma cells, WT MSEu, but not the M2 or M5 mutant, confers efficient repression on the USF-TK minimal promoter. Thus, the activity of the MSEu and MSEi repressor is not restricted to transformed cells.
The MSEu and MSEi are bound by MSF, and previous work using point mutations in the MSEu suggested that MSF and the repressor bound related sequences (49). It was therefore possible that at the MSEi, MSF participates in repression of the TRP-1 promoter, particularly since no factor other than MSF had been found to bind to this element. Since individual point mutations in the MSEu GTGTGA motif severely diminished MSF binding, it was anticipated that MSF would not recognize the LS-MSEi mutant in which four of the six bases in the conserved GTGTGA sequence had been mutated. However, this had never previously been tested. Demonstration that MSF could recognize the LS-MSEi would provide convincing evidence that the repressor and MSF were distinct. To test this possibility, we performed band shift assays using a radiolabeled MSEi probe together with B16 melanoma cell nuclear extract and tested it for MSF binding activity in competition with WT MSEi, MSEu, and LS-MSEi oligonucleotides (see Fig. 6 or Materials and Methods for full sequences of the oligonucleotides used). The result shown in Fig. 1D demonstrates that, surprisingly, MSF binding to the MSEi element was competed not only by the WT MSEi and MSEu but also by the LS-MSEi mutant. Thus, since the LS-MSEi cannot repress transcription yet retains the ability to bind MSF, we conclude that MSF does not act as the TRP-1 repressor. Moreover, the fact that MSF can bind the LS-MSEi sequence suggests that at the initiator, sequences outside the GTGTGA motif play a significant role in recruiting MSF to the TRP-1 promoter. Indeed, we have recently identified residues some 8 to 10 bp 3′ to the MSEi which, in addition to the first two bases of the GTGTGA sequence are essential for MSF binding (reference 49 and unpublished observations).
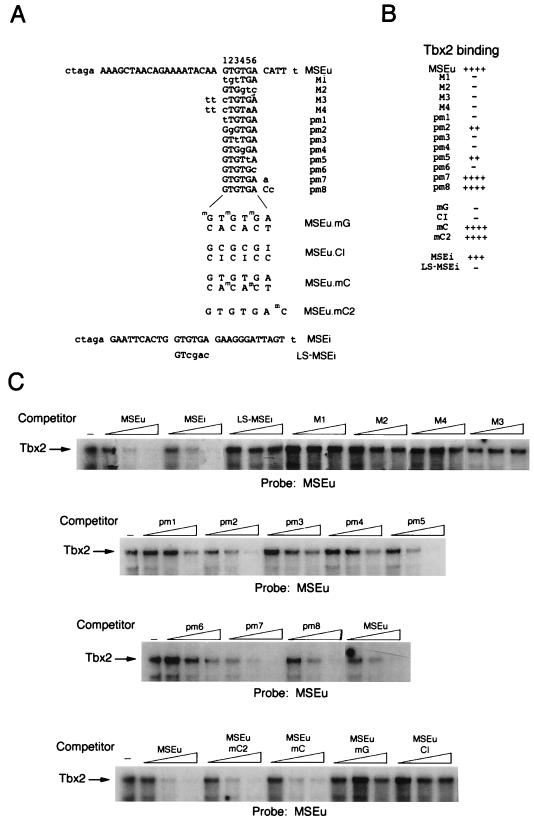
FIG. 6.
DNA-binding specificity of Tbx2. A series of WT and mutant oligonucleotides based on the MSEu or MSEi was used in band shift assays (C) together with ITT Tbx2. The probes and competitors are indicated. Competitors were used at 10, 50, and 250 ng. The sequences or the probes and competitors are shown in panel A along with the full sequence of the parental MSEu or MSEi oligonucleotide. The derivatives used in the competition assays are identical except for the indicated residues shown in lowercase. mG, methylated G residue; mC, methylated C residue; I, inosine. For ease of reference, the bases within the GTGTGA motif are numbered 1 to 6. (B) Summary of the binding assays shown in panel C. Only the bound DNA is shown.
The brachyury-related factor Tbx2 is expressed in the melanocyte lineage.
The results outlined above indicate clearly that the repression of TRP-1 is not mediated by MSF. Given that we were unable to detect repressor DNA-binding activity in vitro, it would not be possible to isolate the repressor by standard biochemical means. However, one clue to the nature of the repressor was afforded by the fact that repression at both the MSEu and MSEi was absolutely dependent on the conserved GTGTGA motifs. One protein known to recognize a GTGTGA sequence is the transcription factor brachyury (19, 24, 25), which plays an essential role in mesoderm induction during development (47). However, from what is known of the pattern of brachyury expression during development, it would be surprising if it were expressed in the melanocyte lineage. Indeed, the absence of brachyury expression in melanocytes and melanomas was confirmed by using both RT-PCR and a specific antibrachyury antibody (not shown). Despite this result, it was nevertheless possible that a factor sharing brachyury DNA-binding specificity was present in melanocytes. In this respect, recent work from Bollag et al. (10) revealed that brachyury is encoded by a gene that belongs to a gene family whose members possess a highly conserved DNA-binding domain, the T box. It was therefore possible that the TRP-1 repressor, or possibly MSF, was a member of this transcription factor family which plays a major role in development (for reviews of the T-box family, see references 33 and 42). To explore this possibility, we designed degenerate PCR primers corresponding to highly conserved regions of the T-box DNA-binding domain. These primers were used to amplify cDNA prepared from the melan-a melanocyte cell line. Analysis of the PCR products by agarose gel electrophoresis revealed a single band of around 250 bp, consistent with the size expected to result from the amplification of a T-box-encoding sequence. The PCR products were cloned, and 20 independent clones were sequenced. All 20 clones analyzed contained the same sequence, encoding the T box of the previously identified Tbx2 factor (10). Using 5′ and 3′ RACE (rapid amplification of cDNA ends), we isolated a full-length cDNA which when sequenced corresponded exactly to that reported for mouse Tbx2 with the exception of two amino acid substitutions, Cys476 to Gly and Arg679 to Ser, which are found in the human protein (12). Despite repeated attempts using different primer pairs corresponding to sequences derived from T-box factors for which sequence information was available (Tbx-1, -3, -5, and -6 and Tbr1), no other T-box mRNAs were detected by RT-PCR using mRNA derived from one melanoma and two different melanocyte cell lines. Although we cannot absolutely rule out the presence of an unidentified family member with a sequence not compatible with any of the degenerate PCR primers tested, it seems likely that Tbx-2 is the only member of the T-box family present in the melanocyte lineage.
Previously, Tbx2 message has been detected by Northern blotting in lung and kidney and at lower levels in the heart and ovary (10). Using whole-mount hybridization in mouse embryos, Chapman et al. (13) were also able to detect Tbx2 expression in a number of other tissues during development, including the central and peripheral nervous system and the neural retina and myotome, though expression remained restricted to a subset of cell types. No expression was found in melanocytes or melanoblasts, possibly because these cells are present only as a dispersed population and are consequently difficult to detect.
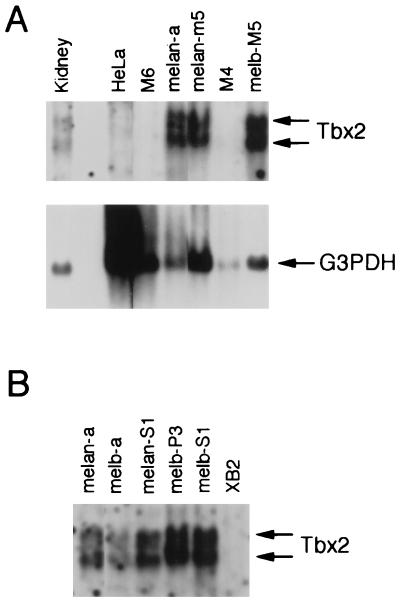
The presence of Tbx2 mRNA in melan-a cells raised the possibility that Tbx2 can recognize the MSEu and MSEi. However, we were also aware that RT-PCR was an extremely sensitive tool for the extraction of specific cDNAs and may sometimes result in the detection of rare messages. Therefore, we wished to confirm the expression of Tbx2 in a range of cell types derived from the melanocyte lineage. RNA was therefore prepared from cell lines corresponding to different stages of melanocyte differentiation, including melanoblast precursors, melanoblasts, and melanocytes, and analyzed by Northern blotting using a nonconserved region of the Tbx2 cDNA as probe. The results (Fig. 2) revealed that Tbx2 mRNA was present as two species, of 2.6 and 3.6 kb, which were expressed at similar levels in three melanocyte cell lines (melan-a, melan-m5, and melan-S1) and in four melanoblast cell lines (melb-M5, melb-a, melb-P3, and melb-S1). Interestingly, no expression was detected in two premelanoblast cell lines, M6 and M4. This pattern of expression is reminiscent of that of the microphthalmia and c-kit genes, both of which play an essential role in melanocyte development and which, like Tbx2, are expressed in melanocytes and melanoblasts but not in premelanoblasts. Tbx2 was also expressed in kidney, consistent with previous observations (10), but not in the mouse keratinocyte cell line XB2 or in HeLa cells.
FIG. 2.
The T-box transcription factor Tbx-2 is expressed in melanocytes and melanoblasts. (A) Northern blot of 5 μg of poly(A)+ mRNA derived from the indicated cell lines or from mouse kidney probed with either Tbx-2 or G3PDH cDNA. Tbx2 mRNA is present as two species of approximately 2.6 and 3.6 kb. The same blot was also probed for G3PDH message as a loading control. Note that the HeLa lane was deliberately overloaded to illustrate that these cells do not express Tbx2 message. M6 and M4 are premelanoblast cell lines; melan-a and melan-M5 are mouse melanocyte cell lines; melb-M5 is a mouse melanoblast cell line. (B) Northern blot of mRNA derived from the indicated cell lines and probed with radiolabeled Tbx-2 cDNA. Each lane was loaded with the same amount of RNA, as judged by staining with ethidium bromide and visualization under UV. melan-a and melan-S1 are mouse melanocyte cell lines; melb-a, melb-P3, and melb-S1 are mouse melanoblast cell lines; XB2 is a mouse keratinocyte cell line.
Tbx2 binds the MSEu and MSEi.
As members of the T-box family have similar DNA-binding domains, Tbx2 might be expected to recognize a sequence similar to that bound by the transcription factor brachyury. The presence of a T-box family member in melanocytes raised the possibility that Tbx2 either was the repressor of TRP-1 expression or was MSF. Apart from brachyury, which preferentially binds a palindromic GTGTGA sequence separated by 4 bp (24), no information was available as to the DNA-binding requirements of other members of the T-box family. To determine whether Tbx2 was competent to bind the MSEu sequence, ITT Tbx2 was used in a band shift assay together with either an MSEu probe or, as a control, a consensus palindromic brachyury binding site (Fig. 3A). Using either Tbx2FL or the C-terminally truncated form Tbx(1-373) Tbx2 together with the brachyury probe, we detected a specific complex with a mobility reflecting the size of the translated protein and which was distinct from a low level of nonspecific DNA-binding activity present in unprogrammed reticulocyte lysate. A similar pattern of DNA-binding activity was observed with the MSEu probe, although in this case significantly more nonspecific binding arising from the reticulocyte lysate was apparent. Nevertheless the results obtained from these assays demonstrate that Tbx2 can recognize both the palindromic, consensus brachyury-binding site as well as the MSEu which comprises a single GTGTGA motif.
The ability of Tbx2 to recognize the MSEu element was consistent with the possibility that Tbx2 was either MSF or the repressor of TRP-1. To distinguish between these possibilities, we compared the mobility of MSF derived from B16 melanoma cell nuclear extracts bound to an MSEu probe to the mobility of ITT Tbx2 bound to the same probe. The result (Fig. 3B) demonstrates unequivocally that the mobilities of the two complexes are different, strongly suggesting that Tbx2 is not MSF. This conclusion was further substantiated by comparing the abilities of Tbx2 and MSF to bind the LS-MSEi mutant. We have already shown (Fig. 1) that the LS-MSEi mutation abolishes repression at the TRP-1 promoter but fails to prevent binding of MSF. In assays using an MSEu probe and either MSF derived from B16 cells or ITT Tbx2, it is evident that an LS-MSEi competitor binds MSF but not Tbx2 (Fig. 3C; see also Fig. 6). Thus, binding by Tbx2, but not MSF, correlates with repression of TRP-1 at the initiator.
Although we were able to demonstrate binding to the MSEu and MSEi elements by using Tbx2 protein generated by ITT, we also wished to show that Tbx2 present in cell extracts was similarly able to bind DNA. As mentioned above, we were consistently unable to detect any Tbx2 DNA-binding activity in extracts from Tbx2-expressing cells. Although other explanations are possible, this may be a result of a low level of Tbx2 in the melanocyte lineage or the fact that Tbx2 is poorly extractable from cells. However, by transfecting B16 melanoma cells with a Tbx2 expression vector, a specific Tbx2 DNA-binding activity could readily be detected with a consensus brachyury-binding site as the probe (Fig. 3D). The identity of the Tbx2-DNA complex, which was apparent only in extracts from the transfected cells, was confirmed by the addition to the DNA-binding assay of a specific anti-Tbx2 antiserum which abolished formation of the complex.
Transcription repression by Tbx2.
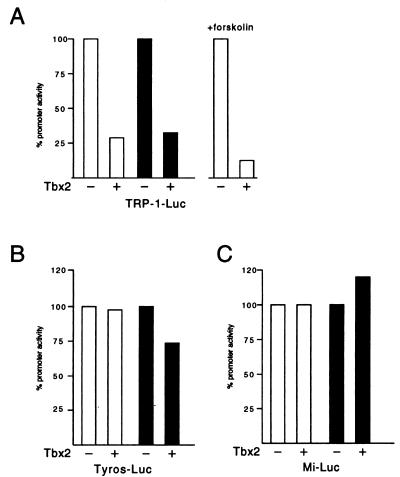
The results described above implicate Tbx2 in repression of the TRP-1 promoter. To assess the ability of Tbx2 to act as a repressor in a more direct assay, we examined whether Tbx2 was able to repress the TRP-1 promoter in cotransfection assays. Given that the TRP-1 promoter was already strongly repressed in melanocytes or melanomas and is not expressed in other cell types, any additional effect of a transfected Tbx2 expression vector might be expected to be minimal. However, when B16 melanoma cells were transfected with a TRP-1 promoter-luciferase reporter in the presence or absence of a Tbx2 expression vector, we were able to demonstrate up to 20-fold repression by Tbx2 in some experiments, although the more usual level of repression was in the region of 3- to 4-fold, as shown for two independent transfection experiments in Fig. 4A. The degree of repression obtained could be increased in either CAT (not shown) or luciferase (Fig. 4A) assays if the basal TRP-1 promoter activity was elevated by stimulating cells with forskolin, which activates TRP-1 promoter indirectly via transiently increasing intracellular levels of the transcription factor microphthalmia (9).
FIG. 4.
Specific repression of the TRP-1 promoter by Tbx2. B16 melanoma cells were transfected with the indicated TRP-1 (A), tyrosinase (Tyros) (B), or microphthalmia (Mi) (C) promoter-luciferase (Luc) reporters either alone or together with the Tbx2 expression vector. The open and filled columns represent the results from two independent transfection experiments. Forskolin was added at 20 μM to the indicated transfection assay. Luciferase activity was determined 48 h posttransfection.
As a control for specificity, we also assayed the effects of Tbx-2 on the promoters driving expression of the melanocyte-specific tyrosinase and microphthalmia genes. Neither promoter contains a Tbx2 binding site and as such would not expect to be repressed by expression of Tbx2 in a cotransfection assay. Consistent with this, no repression of either promoter was observed in the presence of a Tbx2 expression vector (Fig. 4B and C). Thus, repression of TRP-1 by Tbx2 is promoter specific.
While it was evident that Tbx2 could specifically repress the TRP-1 promoter, it was also possible that repression was dependent on the specific arrangement of elements within the TRP-1 promoter. To demonstrate that Tbx2 could also act independently of promoter context, we also examined whether Tbx2 could repress a non-melanocyte-specific promoter when linked to a consensus brachyury-binding site. We initially tried the herpes simplex virus IE110 promoter, which was strongly expressed following transfection of melan-c cells. However, even in the absence of any cotransfected Tbx2 expression vector, the presence of a brachyury/Tbx2 consensus binding site upstream from the IE110 promoter resulted in efficient repression (Fig. 5A). This result illustrated that in melanocytes, the brachyury/Tbx2 consensus binding sequence could act as a strong negative regulatory element and that repression was not peculiar to the MSEu and MSEi elements; in addition, we demonstrated for the first time that the repression mediated by a Tbx-2-binding site was not confined to melanocyte-specific promoters. Nevertheless, the fact that the IE110-CAT reporter was so severely repressed by the brachyury/Tbx2-binding sequence meant that this particular reporter could not readily be used to determine whether Tbx2 expression directed repression in a cotransfection assay. In an attempt to circumvent this problem, we considered using a low-affinity brachyury-binding site. Previously, Kispert et al. (25) demonstrated that compared to the high-affinity consensus sequence, brachyury was able to activate transcription around fivefold less well from an element in which the two half sites are separated by 9 bp. We therefore designed a similar site (LaBS; Fig. 5D) in which the spacing between the two half sites was increased to 10 bp. Using ITT brachyury in a DNA-binding band shift assay together with the high-affinity consensus site as a probe, we competed brachyury for binding with either the high-affinity site or the altered spacing mutant. The result (Fig. 5B) revealed that as expected, brachyury could not recognize this altered spacing mutant. Similarly, brachyury was unable to recognize either the MSEu or the MSEi from the TRP-1 promoter, consistent with previous reports that DNA binding by brachyury requires two half sites (24).
Since we already knew that Tbx2 was able to bind the single half sites present in the MSEu and MSEi, we anticipated that it would also recognize the low-affinity altered spacing mutant, though less well than the full consensus sequence. To verify that Tbx2 would indeed recognize the low-affinity binding site less well than the consensus sequence, we used the consensus site as a probe in a band shift assay together with ITT Tbx2 and performed competition for Tbx2 binding, using either the high-affinity consensus sequence, the LaBS, in which the spacing between the two half sites had been extended to 10 bp, or the MSEu. The result (Fig. 5C) showed that as expected, Tbx2 bound the LaBS around fivefold less well than the consensus sequence. Nevertheless Tbx2 binding to the LaBS was still more efficient than to the MSEu, illustrating that while Tbx2 can bind the single half sites present in the MSEu and MSEi, binding is significantly enhanced by the presence of two half sites.
On the basis of these results, we constructed a CAT reporter comprising the highly active G3PDH promoter linked to the brachyury/Tbx2 LaBS in which the spacing between the two half sites had been increased from 4 to 10 bp. In transfected melan-c cells, expression of the G3PDH promoter was repressed no more than 20% in the presence of the modified brachyury/Tbx2-binding site (Fig. 5D). In contrast, expression of the G3PDH promoter linked to the modified brachyury/Tbx2-binding site (G3PDH.LaBS) was repressed around eightfold by Tbx2 in this assay, while the G3PDH promoter itself was barely affected. We conclude from these experiments that expression of Tbx2 results in transcriptional repression from promoters containing appropriate Tbx2 recognition sequences.
DNA-binding specificity of Tbx2.
Although Tbx2 was able to bind the MSEu and MSEi from the TRP-1 promoter, and each of these elements contained a conserved GTGTGA motif, the precise requirements for DNA binding by Tbx2 were not apparent. Understanding how Tbx2 recognized DNA would be of use in identifying alternative binding sites for this factor in other promoters and would in addition yield insight into DNA recognition by the T-box family in general. DNA binding by Tbx2 was therefore assessed by using the MSEu probe and an extensive series of competitors bearing modifications to the MSEu or MSEi sequence. The sequences of the probes and competitors used are shown in Fig. 6A; results of the DNA-binding assays with the indicated competitors are summarized in Fig. 6B and displayed in Fig. 6C. We initially concentrated on introducing mutations in the conserved GTGTGA motifs. Thus, as shown above, Tbx2 was able to bind both the MSEu and MSEi with around the same efficiency but was not able to bind the LS-MSEi mutant. Tbx2 also failed to bind an MSEu in which either the left (mutant M1) or right (M2) half site of the GTGTGA motif was mutated. Similarly, no competition was observed in assays using either the M3 or M4 mutant in which the two 5′ flanking bases in addition to residues 1 or 1 and 5, respectively, within the GTGTGA sequence were affected. Since the M1 to M4 mutations affected multiple base pairs, we also wished to assess the impact of single base changes within the conserved sequence. The results showed that G-to-T or A-to-C transversions at positions 1, 3, 4, and 6 (pm1, pm3, pm4, and pm6) all severely inhibit binding by Tbx2. In contrast, transversion mutations at positions 2 and 5 (pm2 and pm5) do not affect binding more than two- to threefold. Thus, Tbx2 may be able to recognize not only a GTGTGA motif but also the sequence GGGTGA or GTGTTA.
We next examined whether bases flanking the 3′ side of the core GTGTGA sequence were important for recognition by Tbx2 by using mutants pm7 and pm8 as competitors. We observed no effect of pm7 and only a marginal effect of pm8. Thus, bases to the 3′ side of the MSEu appear to play little role in binding Tbx2.
In addition to examining the sequence requirements for DNA binding by Tbx2, we wondered whether the specific contacts between Tbx2 and its target sequence occurred in the major or minor groove. To address this question, we made use of oligonucleotide competitors bearing modifications which affected specific aspects of the major groove. Thus, the three G residues at positions 1, 3, and 5 within the GTGTGA motif of the MSEu.mG competitor were modified by the addition of a methyl group which would extend into the major groove and inhibit DNA binding by any protein making intimate major groove contacts. Consistent with Tbx2 binding in the major groove, the MSEu.mG competitor failed to bind Tbx2. However, surprisingly, the MSEu.mC oligonucleotide, in which the major groove is modified by the presence of a methyl group on the C residues at positions 3 and 5 on the bottom strand, bound Tbx2 as well as the WT MSEu. These data suggest that in binding the MSEu, Tbx2 makes asymmetric contacts within the major groove since methylation of G residues on the top strand inhibits binding whereas methylation of the C residues on the bottom strand does not. The importance of the major groove contacts was confirmed by using the MSEu.CI oligonucleotide, in which each T residue within the GTGTGA motif is substituted by C and each A is changed to inosine. In this mutant, the minor groove would provide an identical interface to the WT MSEu, while the major groove would be significantly different. Consistent with Tbx2 making major groove contacts, the MSEu.CI mutant failed to bind Tbx2. Finally, an additional competitor (MSEu.mC2) in which the 3′ C residue on the top strand was methylated, bound Tbx2 efficiently, confirming the results using the pm7 and pm8 mutants which showed little effect of mutating bases 3′ to the conserved GTGTGA element.
DISCUSSION
Previously we proposed a model in which the TRP-1 promoter was regulated positively by microphthalmia and MSF and negatively by an unidentified repressor (49). We suggested further that MSF and the repressor might act antagonistically through the MSEu and MSEi elements and that the repression observed in melanoma cells might reflect the fact that these cells were transformed. In this report, we have demonstrated that repression of the TRP-1 promoter occurs not only in melanoma cells but also in the melanocyte cell line melan-c. The repression of the TRP-1 promoter can be overcome by mutation of the MSEu and MSEi, and although both elements can bind MSF, it is clear, most convincingly from assays using the LS-MSEi mutation, that MSF is not the repressor of TRP-1 expression. In contrast, several lines of evidence tend to point to the brachyury-related transcription factor Tbx2 as being the repressor of TRP-1 expression: both the MSEu and the MSEi bind Tbx2, and mutations in either element that result in derepression of the TRP-1 promoter diminish binding by Tbx2; the TRP-1 promoter, but not the tyrosinase, microphthalmia, or G3PDH promoter, was repressed by Tbx2 in cotransfection assays; a high-affinity consensus brachyury/Tbx2-binding site was able to repress expression of the herpes simplex virus IE110 promoter; and a low-affinity brachyury/Tbx2-binding site was able to mediate Tbx2-dependent repression of the strong G3PDH promoter. Although we cannot absolutely rule out the presence of an additional, as yet unidentified factor playing a role in the negative regulation of TRP-1 in vivo, the evidence presented here suggests that Tbx2 is the previously unidentified repressor of TRP-1 expression and as such is likely to represent the first example of transcriptional repression by a T-box family member.
In the melanocyte lineage, Tbx2 mRNA is expressed in both melanoblast and melanocyte cell lines but not in melanoblast precursors. What controls the expression pattern of Tbx2 in development is not known, though it is intriguing that the Tbx2 promoter contains a potential microphthalmia-binding site (our unpublished observations). In light of this observation, it is possible that the onset of microphthalmia expression upon commitment to the melanocyte lineage in turn results in activation of Tbx2 expression, while in other cell types Tbx2 would be regulated by other tissue-specific bHLH or bHLH-LZ factors. While this is an attractive possibility, it is at present no more than speculation, as is the reason why the TRP-1 promoter may be controlled by Tbx2. Although melanoblasts are nonpigmented, they nevertheless express TRP-1 mRNA (4a). Since Tbx2 is expressed in both melanoblast and melanocyte cell lines, it might be expected that TRP-1 would be repressed in both cell types. However, the presence of Tbx2 in a cell does not necessarily mean that Tbx2 will act constitutively as a repressor of transcription. Indeed, we have obtained preliminary evidence that Tbx2 expression is regulated by cellular stress and that Tbx2 may be regulated by a variety of signal transduction pathways which may act to modulate its function (our unpublished observations). Thus, the regulation of TRP-1 expression by Tbx2 may be dependent on environmental cues. This may be especially important in vivo during the migration of the melanoblast from the neural crest and its subsequent differentiation in the hair follicles and epidermis. The constitutive repression of TRP-1 by Tbx2 observed in tissue culture should therefore be taken only as indicative that at some stage in the development or differentiation of the melanocyte lineage, Tbx2 is likely to modulate the activity of the TRP-1 promoter in vivo.
Although here we have provided evidence for Tbx2-mediated repression of the TRP-1 promoter, given that Tbx2 expression appears in melanoblasts, Tbx2 may also play a role in the commitment to or maintenance of the melanocyte lineage. The identification of additional Tbx2 target genes will be essential if the function of Tbx2 expression is to be understood. Our analysis of the DNA-binding specificity of Tbx2 suggests that it may be able to recognize not only the GTGTGA motifs present in the MSEu and MSEi but also GGGTGA or GTGTTA. Analysis of the binding specificity of brachyury by using binding site selection (24) revealed that like Tbx2, brachyury can recognize the GTGTTA sequence in addition to GTGTGA. However, in the selection assay, no GGGTGA sequence bound by brachyury was selected, which may mean that this sequence represents a relatively low affinity binding site for brachyury or possibly that Tbx2 and Brachyury have related but distinct DNA-binding specificities. The fact that brachyury and Tbx2 bind DNA in distinct fashions is highlighted by the fact that while brachyury binding requires two half sites, Tbx2 can bind, although with reduced affinity, to the single half sites present in the MSEu and MSEi as well as the LaBS created by increasing the spacing between the two half sites to 10 bp.
Recently, the crystal structure of the brachyury DNA-binding domain bound to DNA was determined (30). Given that the Tbx2 DNA-binding domain has a high degree of homology with that of brachyury, it is likely that the structure of the Tbx2 DNA-binding domain is similar to that of Brachyury. The brachyury T-box–DNA cocrystal structure revealed three major features of interest. First, the sequence used, an inverted repeat of an AGGTGTGA motif, exhibits a substantially widened minor groove but no DNA bending; second, bp 3 and 5 within the half sites used for the crystallization are important specificity determinants, while bp 6 and 7 are less important; and third, the contacts made with the DNA occur in both the minor and major grooves. Our analysis of Tbx2-binding specificity is broadly in agreement with this. Thus, mutations such as pm1 and pm3 (Fig. 6) which affect the equivalent of bp 3 and 5 of the DNA in the crystal structure abolish Tbx2 binding, while mutant pm5, which affects the equivalent of bp 7, has a relatively minor impact. Moreover, the main contact point for brachyury in the major groove is a single G residue (bp 5 in the crystal structure). Thus, methylation of the G residues within the binding site should severely affect binding of Tbx2, while methylation of the C residues on the opposite strand should have little effect, exactly as observed (Fig. 6). Thus, the data from our DNA-binding studies and the structure of the brachyury T-box–DNA complex are also in agreement on this point. Nonetheless, some differences are apparent. Notably, the results from the crystal structure suggest that the close contact between Phe215 of brachyury and the T/A base pair at position 4 will not tolerate its substitution. Yet while that specific Phe residue is conserved in Tbx2, mutation of that T/A base pair to G/C (pm2) fails to abolish binding by Tbx2. It is therefore possible that the protein-DNA contacts for the T-box family are different on different target sites or that residues which are not conserved between the brachyury and Tbx2 DNA-binding domains influence DNA-binding specificity.
Whatever the precise nature of the relative DNA-binding abilities of Tbx2 and brachyury, the analysis of the Tbx2-binding specificity and the fact that the consensus brachyury-binding site used for the band shift assays binds Tbx2 efficiently strongly suggest that at least some brachyury target genes may be regulated by Tbx2. What these targets may be is currently unclear, though embryonic fibroblast growth factor is one possibility. Moreover, brachyury has been reported to be a transcription activator, while in melanocytes a brachyury-binding site acts as a strong repression element and Tbx2 appears to act as a repressor. Thus, different members of the T-box family may have opposing roles in transcription regulation which may be particularly significant in cells where more than one T-box family member is present. However, while brachyury can activate transcription and Tbx2 can repress, does this inevitably mean that these factors have immutably defined but opposing roles in transcription regulation? Not necessarily. Deletion analysis of brachyury suggested that it may contain both activation and repression domains (25). Although we have yet to examine the requirements within Tbx2 for transcription regulation, it is possible that brachyury and Tbx2 each play roles as both activators and repressors, depending on their responsiveness to different signal transduction pathways or the context and arrangement of their binding sites within a target promoter. Since the repertoire of signaling pathways which may be active is likely to differ in different cell types, the role of individual members of the T-box family may vary between tissues or during development. Future work will be directed toward exploring these issues.
ACKNOWLEDGMENTS
We thank Peter O’Hare for plasmid pAB2, Maria Alexander-Bridges for providing the G3PDH-CAT reporter vector, Bernard Herrmann for the antibrachyury antibody, and Dot Bennett for discussions and encouragement.
This work was supported by the Association for International Cancer Research, the Medical Research Council, and Marie Curie Cancer Care.
REFERENCES
- 1.Alexander M C, Lomanto M, Nasrin N, Ramaika C. Insulin stimulates glyceraldehyde-3-phosphate dehydrogenase gene expression through cis-acting DNA sequences. Proc Natl Acad Sci USA. 1988;85:5092–5096. doi: 10.1073/pnas.85.14.5092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baldwin C T, Lipsky N R, Hoth C F, Cohen T, Mamuya W, Milunsky A. Mutations in PAX3 associated with Waardenburg syndrome type I. Hum Mutat. 1994;3:205–211. doi: 10.1002/humu.1380030306. [DOI] [PubMed] [Google Scholar]
- 3.Bassi M T, Incerti B, Easty D J, Sviderskaya E V, Ballabio A. Cloning of the murine homolog of the ocular albinism type 1 (OA1) gene: sequence, genomic structure, and expression analysis in pigment cells. Genome Res. 1996;6:880–885. doi: 10.1101/gr.6.9.880. [DOI] [PubMed] [Google Scholar]
- 4.Baynash A G, Hosoda K, Giaid A, Richardson J A, Emoto N, Hammer R E, Yanagisawa M. Interaction of endothelin-3 with endothelin-B receptor is essential for development of epidermal melanocytes and enteric neurons. Cell. 1994;79:1277–1285. doi: 10.1016/0092-8674(94)90018-3. [DOI] [PubMed] [Google Scholar]
- 4a.Bennett, D. Personal communication.
- 5.Bennett D C, Cooper P J, Hart I R. A line of non-tumorigenic mouse melanocytes, syngeneic with the B16 melanoma and requiring a tumour promoter for growth. Int J Cancer. 1987;39:414–418. doi: 10.1002/ijc.2910390324. [DOI] [PubMed] [Google Scholar]
- 6.Bennett D C, Svedeskaya E V. Advances in immortalization of cultured melanocytes and melanoblasts. In: Nordlund J J, et al., editors. The pigmentary system and its disorders. New York, N.Y: Oxford University Press; 1998. pp. 165–174. [Google Scholar]
- 7.Bentley N J, Eisen T, Goding C R. Melanocyte-specific expression of the human tyrosinase promoter: activation by the microphthalmia gene product and role of the initiator. Mol Cell Biol. 1994;14:7996–8006. doi: 10.1128/mcb.14.12.7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bertolotto C, Busca R, Abbe P, Bille K, Aberdam E, Ortonne J-P, Ballotti R. Different cis-acting elements are involved in the regulation of TRP1 and TRP2 promoter activities by cyclic AMP: pivotal role of M boxes (GTCATGTGCT) and of microphthalmia. Mol Cell Biol. 1998;18:694–702. doi: 10.1128/mcb.18.2.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bertolotto C, K. B, Ortonne J-P, Ballotti R. Regulation of tyrosinase gene expression by cAMP in B16 melanoma cells involves two CATGTG motifs surrounding the TATA box: implication of the microphthalmia gene product. J Cell Sci. 1996;134:747–755. doi: 10.1083/jcb.134.3.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bollag R J, Siegried Z, Cebra-Thomas J A, Garvay N, Davison E M, Silver L M. An ancient family of embryonically expressed mouse genes sharing a conserved protein motif with the T-locus. Nat Genet. 1994;7:383–389. doi: 10.1038/ng0794-383. [DOI] [PubMed] [Google Scholar]
- 11.Butt J, Greenberg J, Winship I, Sellars S, Beighton P, Ramesar R. A splice junction mutation in PAX3 causes Waardenburg syndrome in a South African family. Hum Mol Genet. 1994;3:197–198. doi: 10.1093/hmg/3.1.197. [DOI] [PubMed] [Google Scholar]
- 12.Campbell C, Goodrich K, Casey G, Beatty B. Cloning and mapping of a human gene (TBX2) sharing a highly conserved protein motif with the Drosophila omb gene. Genomics. 1995;28:255–260. doi: 10.1006/geno.1995.1139. [DOI] [PubMed] [Google Scholar]
- 13.Chapman D L, Garvay N, Hancock S, Alexiou M, Agulnik S I, Gibson-Brown J J, Cebra-Thomas J, Bollag R J, Silver L M, Papaioannou V E. Expression of the T-box family genes, Tbx1-Tbx5, during early mouse development. Dev Dyn. 1996;206:379–390. doi: 10.1002/(SICI)1097-0177(199608)206:4<379::AID-AJA4>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 14.Easty D J, Guthrie B A, Maung K, Farr C J, Lindberg R A, Toso R J, Herlyn M, Bennett D C. Protein B61 as a new growth factor: expression of B61 and up-regulation of its receptor epithelial cell kinase during melanoma progression. Cancer Res. 1995;55:2528–2532. [PubMed] [Google Scholar]
- 15.Fitzpatrick T B, Szabo G, Seiji M, Quevado W C. Biology of the melanin pigmentation system. In: Fitzpatrick T B, Eisen A Z, Wolff K, Freedberg I M, Austin K F, editors. Dermatology in general medicine. 2nd ed. New York, N.Y: McGraw-Hill; 1979. pp. 131–163. [Google Scholar]
- 16.Ganss R, Montoliu L, Monaghan A P, Schutz G. A cell-specific enhancer far upstream of the mouse tyrosinase gene confers high level and copy number-related expression in transgenic mice. EMBO J. 1994;13:3083–3093. doi: 10.1002/j.1460-2075.1994.tb06607.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ganss R, Schutz G, Beermann F. The mouse tyrosinase gene. Promoter modulation by positive and negative regulatory elements. J Biol Chem. 1994;269:29808–29816. [PubMed] [Google Scholar]
- 18.Hemesath T J, Steingrimsson E, McGill G, Hansen M J, Vaught J, Hodgkinson C A, Arnheiter H, Copeland N G, Jenkins N A, Fisher D E. Microphthalmia, a critical factor in melanocyte development, defines a discrete transcription factor family. Genes Dev. 1994;8:2770–2780. doi: 10.1101/gad.8.22.2770. [DOI] [PubMed] [Google Scholar]
- 19.Herrmann B G, Labeit S, Poustka A, King T R, Lehrach H. Cloning the T-gene required in mesoderm formation in the mouse. Nature (London) 1990;343:617–622. doi: 10.1038/343617a0. [DOI] [PubMed] [Google Scholar]
- 20.Hodgkinson C A, Moore K J, Nakayama A, Steingrimsson E, Copeland N G, Jenkins N A, Arnheiter H. Mutations at the mouse microphthalmia locus are associated with defects in a gene encoding a novel basic-helix-loop-helix-zipper protein. Cell. 1993;74:395–404. doi: 10.1016/0092-8674(93)90429-t. [DOI] [PubMed] [Google Scholar]
- 21.Hosoda K, Hammer R E, Richardson J A, Baynash A G, Cheung J C, Giaid A, Yanagisawa M. Targeted and natural (piebald-lethal) mutations of endothelin-B receptor gene produce megacolon associated with spotted coat color in mice. Cell. 1994;79:1267–1276. doi: 10.1016/0092-8674(94)90017-5. [DOI] [PubMed] [Google Scholar]
- 22.Jackson I J. A cDNA encoding tyrosinase-related protein maps to the brown locus in mouse. Proc Natl Acad Sci USA. 1988;85:4392–4396. doi: 10.1073/pnas.85.12.4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jackson I J, Chambers D M, Budd P S, Johnson R. The tyrosinase-related protein-1 gene has a structure and promoter sequence very different from tyrosinase. Nucleic Acids Res. 1991;19:3799–3804. doi: 10.1093/nar/19.14.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kispert A, Herrmann B G. The Brachyury gene encodes a novel DNA-binding protein. EMBO J. 1993;12:3211–3220. doi: 10.1002/j.1460-2075.1993.tb05990.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kispert A, Koschorz B, Herrmann B G. The T protein encoded by Brachyury is a tissue-specific transcription factor. EMBO J. 1995;14:4763–4772. doi: 10.1002/j.1460-2075.1995.tb00158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kluppel M, Beermann F, Ruppert S, Schmid E, Hummler E, Schutz G. The mouse tyrosinase promoter is sufficient for expression in melanocytes and in the pigmented epithelium of the retina. Proc Natl Acad Sci USA. 1991;88:3777–3781. doi: 10.1073/pnas.88.9.3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kwon B S, Haq A K, Pomerantz S H, Halaban R. Isolation and sequence of a cDNA clone for human tyrosinase that maps at the mouse c-albino locus. Proc Natl Acad Sci USA. 1987;84:7473–7477. doi: 10.1073/pnas.84.21.7473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lowings P, Yavuzer U, Goding C R. Positive and negative elements regulate a melanocyte-specific promoter. Mol Cell Biol. 1992;12:3653–3662. doi: 10.1128/mcb.12.8.3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moore K J. Insight into the microphthalmia gene. Trends Genet. 1995;11:442–448. doi: 10.1016/s0168-9525(00)89143-x. [DOI] [PubMed] [Google Scholar]
- 30.Müller C W, Herrmann B G. Crystallographic structure of the T domain-DNA complex of the Brachyury transcription factor. Nature (London) 1997;389:884–889. doi: 10.1038/39929. [DOI] [PubMed] [Google Scholar]
- 31.Nocka K, Majumder S, Chabot B, Ray P, Cervone M, Bernstein A, Besmer P. Expression of c-kit gene products in known cellular targets of W mutations in normal and W mutant mice. Genes Dev. 1989;3:816–826. doi: 10.1101/gad.3.6.816. [DOI] [PubMed] [Google Scholar]
- 32.O’Rourke D, O’Hare P. Mutually exclusive binding of two cellular factors within a critical promoter region of the gene for the IE110k protein of herpes simplex virus. J Virol. 1993;67:7201–7214. doi: 10.1128/jvi.67.12.7201-7214.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Papaioannou V E, Silver L M. The T-box gene family. Bioessays. 1998;20:9–19. doi: 10.1002/(SICI)1521-1878(199801)20:1<9::AID-BIES4>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 34.Pingault V, Bondurand N, Kuhlbrodt K, Goerich D E, Préhu M-O, Puliti A, Herbarth B, Hermans-Borgmeyer I, Legius E, Matthijs G, Amiel J, Lyonnet S, Ceccherini I, Romeo G, Clayton-Smith J, Read A P, Wegner M, Goossens M. SOX10 mutations in patients with Waardenburgs-Hirschprung disease. Nat Genet. 1998;18:171–173. doi: 10.1038/ng0298-171. [DOI] [PubMed] [Google Scholar]
- 35.Porter S D, Meyer C J. A distal tyrosinase upstream element stimulates gene expression in neural crest-derived melanocytes of transgenic mice: position-independent and mosaic expression. Development. 1994;120:2103–2111. doi: 10.1242/dev.120.8.2103. [DOI] [PubMed] [Google Scholar]
- 36.Puffenberger E G, Hosoda K, Washington S S, Nakao K, deWit D, Yanagisawa M, Chakravart A. A missense mutation of the endothelin-B receptor gene in multigenic Hirschsprung’s disease. Cell. 1994;79:1257–1266. doi: 10.1016/0092-8674(94)90016-7. [DOI] [PubMed] [Google Scholar]
- 37.Rivera-Pomar R, Jackle H. From gradients to stripes in Drosophila embryogenesis: filling in the gaps. Trends Genet. 1996;12:478–483. doi: 10.1016/0168-9525(96)10044-5. [DOI] [PubMed] [Google Scholar]
- 38.Ruppert S, Muller G, Kwon B, Schutz G. Multiple transcripts of the mouse tyrosinase gene are generated by alternative splicing. EMBO J. 1988;7:2715–2722. doi: 10.1002/j.1460-2075.1988.tb03125.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sato S, Roberts K, Gambino G, Cook A, Kouzarides T, Goding C R. CBP/p300 as a co-factor for the microphthalmia transcription factor. Oncogene. 1997;14:3083–3092. doi: 10.1038/sj.onc.1201298. [DOI] [PubMed] [Google Scholar]
- 40.Shibata K, Muraosa Y, Tomita Y, Tagami H, Shibahara S. Identification of a cis-acting element that enhances the pigment cell-specific expression of the human tyrosinase gene. J Biol Chem. 1992;267:20584–20588. [PubMed] [Google Scholar]
- 41.Silvers W K. The coat colors of mice. New York, N.Y: Springer-Verlag; 1979. [Google Scholar]
- 42.Smith J. Brachyury and the T-box genes. Curr Opin Genet Dev. 1997;7:474–480. doi: 10.1016/s0959-437x(97)80073-1. [DOI] [PubMed] [Google Scholar]
- 43.Southard-Smith E M, Kos L, Pavan W. Sox10 mutation disrupts neural crest development in Dom Hirschprung mouse model. Nat Genet. 1998;18:60–64. doi: 10.1038/ng0198-60. [DOI] [PubMed] [Google Scholar]
- 44.Steel K P, Barkway C. Another role for melanocytes: their importance for stria vascularis development in the mammalian inner ear. Development. 1989;107:453–463. doi: 10.1242/dev.107.3.453. [DOI] [PubMed] [Google Scholar]
- 45.Sviderskaya E V, Easty D J, Hill S P, Bennett D C. Differential gene expression in immortal melanocytes, melanoblasts and melanoblast precursors. Pigment Cell Res Suppl. 1996;5:36–37. [Google Scholar]
- 46.Tassabehji M, Newton V E, Read A P. Waardenburg syndrome type 2 caused by mutations in the human microphthalmia (MITF) gene. Nat Genet. 1994;8:251–255. doi: 10.1038/ng1194-251. [DOI] [PubMed] [Google Scholar]
- 47.Wilkinson D G, Bhatt S, Herrmann B G. Expression of the mouse T gene and its role in mesoderm formation. Nature (London) 1990;343:657–659. doi: 10.1038/343657a0. [DOI] [PubMed] [Google Scholar]
- 48.Yasumoto K, Yokoyama K, Shibata K, Tomita Y, Shibahara S. Microphthalmia-associated transcription factor as a regulator for melanocyte-specific transcription of the human tyrosinase gene. Mol Cell Biol. 1994;14:8058–8070. doi: 10.1128/mcb.14.12.8058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yavuzer U, Goding C R. Melanocyte-specific gene expression: role of repression and identification of a melanocyte-specific factor, MSF. Mol Cell Biol. 1994;14:3494–3503. doi: 10.1128/mcb.14.5.3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yavuzer U, Keenan E, Lowings P, Vachtenhein J, Currie G, Goding C R. The microphthalmia gene product interacts with the retinoblastoma protein in vitro and is a target for deregulation of melanocyte-specific transcription. Oncogene. 1995;10:123–134. [PubMed] [Google Scholar]