Abstract
Background
Helicobacter pylori infection remains a major public health threat leading to gastrointestinal illness and increased risk of gastric cancer. Mostly affecting populations in developing countries no vaccines are yet available and the disease is controlled by antimicrobials which, in turn, are driving the emergence of AMR.
Materials and Methods
We have engineered spores of Bacillus subtilis to display putative H. pylori protective antigens, urease subunit A (UreA) and subunit B (UreB) on the spore surface. Following oral dosing of mice with these spores, we evaluated immunity and colonization in animals challenged with H. pylori.
Results
Oral immunization with spores expressing either UreA or UreB showed antigen‐specific mucosal responses (fecal sIgA) including seroconversion and hyperimmunity. Following challenge, colonization by H. pylori was significantly reduced by up to 1‐log.
Conclusions
This study demonstrates the utility of bacterial spores for mucosal vaccination to H. pylori infection. The heat stability and robustness of Bacillus spores coupled with their existing use as probiotics make them an attractive solution for either protection against H. pylori infection or potentially for therapy and control of active infection.
Keywords: Helicobacter pylori infection, oral and parenteral immunization, vaccine
1. INTRODUCTION
Helicobacter pylori (HP) is a Gram‐negative human pathogenic bacterium believed to be carried by an estimated 50% of the world's population. 1 Infection typically occurs in childhood and if untreated an individual will likely remain colonized for life. 2 Of those infected about 15% may go on to develop pathological symptoms, typically chronic gastritis and peptic ulcer disease (gastric and duodenal ulceration) with the potential for gastric cancer (adenocarcinoma and lymphoma). H. pylori infection is thought to be linked with more than 90% of gastric cancers and normally results if the infection is left untreated. 3 Gastric adenocarcinoma is now the 5th most common cancer and the 3rd in terms of cancer‐related deaths. Unfortunately, by the time gastric cancer has developed it is nearly impossible to eradicate the underlying infection. Humans appear to acquire H. pylori in two ways, oral‐oral being the most common route of transmission and vertical transmission from mother to child. 2 H. pylori is unusual in that it is able to colonize the stomach where this highly motile bacterium can penetrate mucus, cause inflammation, and degrade the stomach lining. Incidence rates appear higher in developing countries where there is poor sanitation, particularly Asia/SE Asia and in some countries such as Vietnam >70% of the population are carriers with up to 10%–25% exhibiting symptoms. 4 , 5 , 6
Infections are treated using a combination of antibiotics 2 , 7 but often multidrug regimens are required often in combination with proton pump inhibitors lasting for up to 14 days. This approach has often discouraged patient compliance. However, the prevalence of AMR (antimicrobial resistance; notably to clarithromycin and metronidazole 8 ) is so high that many infected patients are now considered as having fully resistant infections 9 and in some cases are unable to access antibiotic therapy. 10 Clarithromycin has been used around the world in standard triple therapy but clarithromycin‐resistant H. pylori isolates have been rapidly increasing worldwide, for example, in China from ~15% in 2000 to ~53% in 2014. 11 In 2017, clarithromycin‐resistant H. pylori was included in the WHO's list of 12 antibiotic‐resistant “priority pathogens” that pose the greatest threat to human health. 12 Importantly, antimicrobial therapy cannot protect against reinfection, and the rate of reinfection is as high as 15%–30% per year. 13
The greatest burden of H. pylori infection is in developing countries (notably China and SE Asia) where there is a clear link to an increased risk of gastric cancer. It has been suggested that a 10‐year vaccination program might significantly reduce the impact of H. pylori infection both with regard to symptoms, gastric cancer, and the associated economic burden of disease management. 14
Conceptually, a vaccine would best be administered orally to enable the production of secretory IgA (sIgA) in the stomach mucosa preventing colonization. 15 , 16 , 17 However, other mucosal delivery routes (intranasal, rectal) have been successfully used. 15 , 16 , 17 Based on the pathogenesis of H. pylori, a number of putative protective antigens have been evaluated including urease (subunits UreA and UreB), flagellar antigens (FlaA and FlaB), cytotoxin‐associated gene A (CagA), vacuolating toxin (VacA), and others. 15 , 16 , 17 Vaccine formulations including subunit vaccines, live vector vaccines, DNA vaccines, and other delivery systems have been evaluated. 15 , 16 , 17 One of major problems with oral immunization is that resulting immunity is weak. Accordingly, adjuvants such as cholera toxin (CT), the closely related heat‐labile toxin (LT) of E. coli or the B subunit of CT (CTB) have been extensively evaluated. 18 , 19
Although there has been considerable effort in vaccine development, few human studies have demonstrated convincing levels of protective immunity. 16 , 17 The one promising exception being a recently described vaccine consisting of an orally‐administered protein formulation comprised of UreB fused to LT. 20 Despite this there is a case for vaccination where even reduced efficacy might shorten existing treatment regimens and help protect against reinfection. 17 Here, we have evaluated bacterial spore vaccines using UreA and UreB as putative protective antigens. Using oral delivery spore vaccines induced antigen‐specific mucosal IgA and in a mouse colonization model a 1‐log reduction in stomach colonization was observed. Taken together, the use of spore vaccines could have utility for a prophylactic and potentially therapeutic strategy for reducing the impact of H. pylori infection.
2. MATERIALS AND METHODS
2.1. Strains
Bacillus subtilis strain PY79 is a prototrophic laboratory strain. A clinical strain of H. pylori, strain HP34, was obtained from the Hospital of the University of Medicine and Pharmacy, Hue University, Vietnam. HP34 was isolated (May 5, 2020) from a 54‐year‐old female patient, peptic ulcer patient, with endoscopy displaying superficial duodenal ulceration, inflammation in the fundus, and antral erosions. The virulence genotypes were shown to be positive to ure, ureB, cagA, and vacA (Genbank accession number CP122516). Identity was confirmed by whole genome sequencing. The strain was resistant to clarithromycin but sensitive to tetracycline, metronidazole, amoxicillin, and levofloxacin. H. pylori was cultured using either selective Horse Blood Agar (HBA) which was prepared using 4% (w/v) Blood Agar Base No. 2 (Oxoid), supplemented with 8% (w/v) defibrinated horse blood (IVAC, Vietnam), 0.2% (v/v) Skirrow's antibiotic selective supplement (consisting of vancomycin [Sigma], 10 μg/mL; polymyxin B [Sigma], 25 ng/mL; trimethoprim [Sigma], 5 μg/mL; amphotericin B [Sigma], 2.5 μg/mL) and 1% (v/v) sodium lactate (Sigma), or Brain Heart Infusion (BHI) medium (Oxoid) containing 5% (v/v) fetal bovine serum (FBS, Thermofisher Scientific). Incubation was made in a microaerophilic chamber using an Oxoid CampyGen 2.5 L Sachet (5%–7% O2, 5%–10% CO2, and 85% N2) at 37°C, with passaging every 48 h. The strain was preserved in BHI supplemented with 15% (v/v) glycerol at −80°C.
2.2. General methods
Methods for B. subtilis including preparation of spores and extraction of spore coat proteins are described elsewhere. 21
2.3. Construction of B. subtilis spores expressing urease antigens
A cloning method referred to as THY‐X‐CISE® 22 was used to introduce heterologous genes into the chromosome of B. subtilis. First, chimeric genes were synthesized (Azenta Life Sci.) carrying the 5′ segment (including promoter) of the cotB gene of B. subtilis fused at their 3′‐end to DNA encoding either the full‐length ureA gene (encoding UreA, amino acids 1–237; MW 26.4 kDa) or a segment of ureB termed ureB CT (encoding the non‐enzymatic carboxy‐terminus of UreB, 23 , 24 amino acids 365–568, MW 22.9 kDa; Figure 1). The ureA and ureB sequences were those from H. pylori strain 26,695. 25 The resulting CotB‐UreA and CotB‐UreBCT chimeric polypeptides have predicted MWs of 63 and 61 kDa, respectively. Chimeras were then subcloned into the plasmid pThyA. 22 This places the chimeric gene between the left (proximal) and right (distal) arms of the thyA gene that encodes thymidine synthetase A (Figure 1). Linearization of the resulting plasmid and introduction into the B. subtilis chromosome (strain PY79) by a double crossover recombination generates trimethoprim‐resistant transformants that will starve in the absence of thymine (thymine‐dependent). The resulting thyA::insertion strain was then used as a recipient in a second transformation where an empty pThyB vector is introduced thus disrupting the thyB locus (thyB::Δ). The resulting strain (thyA::insertion thyB::Δ) is thymine‐dependent but resistant to a higher concentration of trimethoprim. Strains constructed were PK78 thyA::cotB‐ureB CT thyB::Δ and PK82 thyA::cotB‐ureA thyB::Δ referred to henceforth as PK78 (cotB‐ureB CT ) and PK82 (cotB‐ureA), respectively. An isogenic strain, PK118 (thyA::Δ thyB::Δ), that carries no gene insertions was made using the same procedure with empty pThyA and pThyB vectors that carries no gene insertions and referred to henceforth as PK118 (WT). Strains were confirmed by preparing spores, extracting spore coat proteins, and probing western blots with polyclonal (rabbit) antibodies recognizing UreA (Fisher Sci. Cat No. 17240004) and UreB (Fisher Sci. Cat. No. 17250004).
FIGURE 1.
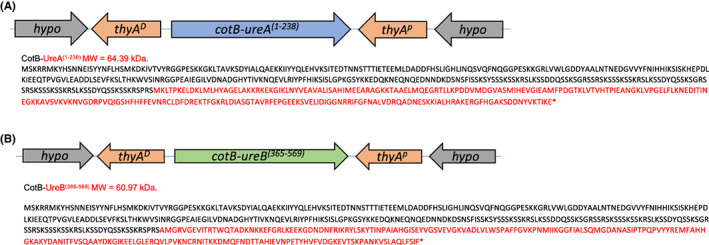
Amino acid sequences of the fusion genes. Chimeric genes inserted at the thyA loci (proximal (thyA P ) and distal (thyB D ) segments are indicated) of B. subtilis are shown together with a schematic of the chromosomal region (hypo = hypothetical gene). Urease A (ureA) and urease B (ureB, C‐terminal region) were fused in frame to the 3′‐end of the spore coat protein gene. The MW of the chimeric proteins are shown.
2.4. Detection of surface display of antigens using enzyme‐linked immunosorbent assay
The whole‐spore enzyme‐linked immunosorbent assay (ELISA) protocol was followed as described elsewhere. 26 Briefly, spores were diluted to 2 × 108 spores/mL in PBS, and 50 μL of suspension was used to coat microplate wells (Greiner, high binding) overnight at 4°C. This was followed by blocking for 1 h at 37°C with PBS containing 0.05% (v/v) Tween 20 and 2% (w/v) bovine serum albumin (BSA). Primary antibodies to the relevant H. pylori domains: anti‐UreA raised against the entire UreA polypeptide (Thermo Fisher Sci., PA5‐117505) and anti‐UreB raised against the entire UreB polypeptide (Thermo Fisher Sci., PA5‐32168) were diluted in conjugate buffer (1:2000 in 0.01 M PBS, 1% [w/v] BSA, 1% [v/v] and 0.05% [v/v] Tween 20) and incubated for 2 h at 30°C. The appropriate horseradish peroxidase‐conjugated anti‐rabbit IgG (Sigma Cat No. 12‐348) or anti‐mouse IgG (Dako Cat No. P0447) was diluted in conjugate buffer (1:2000 in 0.01 M PBS, 1% [w/v] BSA, 1% [v/v] and 0.05% [v/v] Tween 20) and used as a secondary antibody. Plates were incubated for 1 h at RT and then developed using tetramethyl benzidine (TMB) substrate (0.1 mg/mL 3.3′,5.5′‐tetramethylbenzidine in 0.1 M sodium acetate buffer [pH 5.5]). Reactions were stopped using 2 M H2SO4, and ODs read at 450 nm.
2.5. rUreA and rUreBCT polypeptides
Recombinant versions of UreA and UreBCT intended for ELISA assays were expressed and purified from E. coli. Essentially, PCR was used to amplify DNA fragments encoding open reading frames for UreA (residues 1–237) and UreBCT (residues 365–568) using primer pairs 3080 and 3081 and 3082 and 3083, respectively. The pET‐3d expression vector backbone was amplified using primer pair 522 and 1497. Primer sequences are available upon request. Purified PCR amplicons were assembled using the Klenow Assembly Method and then used to transform E. coli Turbo to produce plasmids P90‐pET‐3d‐UreA and P91‐pET‐3d‐ureB CT . Protein expression was conducted using E. coli Rosetta cells freshly transformed with either plasmid. Cells were cultured in 400 mL LB medium at 37°C until an A600 of approx. 0.8 was attained, and then, the temperature reduced to 16°C for 20 min before inducing protein expression by adding IPTG to 0.2 mM. Expression continued overnight (~18 h) before the cells were harvested, lysed by sonication, and then clarified by centrifugation. Recombinant UreA and UreBCT, both of which were designed to be expressed with C‐terminal hexa‐histidine tags, were purified from cell lysates using Ni‐NTA agarose resin (Qiagen) and then buffer exchanged using Amicon Ultra‐4 Centrifugal filters (10 kDa MWCO) into 50 mM Tris–HCl, pH 7.5, containing 50 mM NaCl.
2.6. Animal studies
2.6.1. Oral immunizations
Inbred mice (C57 BL/6, females, 9 weeks of age) were used for immunity studies and were housed in groups (n = 6). The dosing intra‐gastric (i.g., 0.2 mL) regimen is shown Figure S1A. Groups were Gp1 (naive), dosed with PBS; Gp2, dosed with PK118 (WT) spores; Gp3, dosed with PK78 (cotB‐ureB CT ) spores and Gp4, dosed with PK82 (cotB‐ureA) spores. The dosing regimen consisted of four doses with each dose corresponding to three daily administrations (i.g.) of 0.2 mL (PBS or spore vaccine). For Gps 2–4 each i.g. administration consisted of 1 × 1010 spore CFU and daily administrations were used to reduce viscosity of the i.g. inoculation. Samples of feces were taken on Days−1, 15, 31, 46, and 61, and serum was taken on Day 62.
2.6.2. Challenge studies
Mice (Mlac:ICR, males, 5–6 weeks of age, 18–20 g) were used for this study. Dosing schedules are shown in Figure S1B and consisted of four oral (i.g.) doses on Days 0, 14, 28, and 53. Four groups (Gp; n = 6) were used; Gp1, naive receiving sterile PBS, Gp2, PK118 (WT) spores, Gp3, PK82 (cotB‐ureA), and Gp4, PK78 (cotB‐ureB CT ). i.g. dosing consisted of 0.2 mL of either PBS (Gp 1) or spores (1 × 1010 CFU; Gps 2–4). On Days 7–9 following the last dose animals were challenged daily with 0.2 mL/day of freshly grown H. pylori HP34 culture qualified by OD600 measurements to contain ~108 H. pylori CFU. Samples of stomach were taken on Day 83 to enumerate H. pylori CFU by plating on HBA.
2.7. Determination of mucosal titers by indirect enzyme‐linked immunosorbent assay
For analysis of immunological responses, fecal samples were collected on Days−1, 15, 31, 46, and 61 and serum on Day 62 and stored at −80°C. For feces, sample extractions were made at a one‐fifth (w/v) dilution in extraction buffer (2% [v/v] fetal calf serum) containing protease inhibitors, EDTA (0.05 mg/mL), as previously described. 27 Samples were gently shaken for 2 h at 4°C to disrupt solid material, centrifuged (8000 g, 15 min) and the supernatant used for analysis. Antibody levels in feces (IgA) and serum (IgG) were quantified by indirect enzyme‐linked immunosorbent assay (ELISA). Greiner 96‐well plates (MaxiSorp) were coated with 8 μg/mL of rUreA or 4 μg/mL of rUreBCT (50 μL/well) in PBS overnight at 4°C, followed by blocking for 1 h at RT with PBS containing 2% (w/v) bovine serum albumin (BSA). Fecal samples were diluted 1:20 in PBS. Serum samples were diluted 1:10, in diluent buffer (0.01 M PBS, 1% [w/v] BSA, 2% [v/v] FBS, 0.1% [v/v] Triton X‐100, 0.05% [v/v] Tween 20). Samples were added to plates and twofold serially diluted. Plates containing fecal samples were incubated for 2 h at 30°C and those containing serum samples incubated for 2 h at RT. Levels of IgA and IgG were detected using the appropriate horseradish peroxidase‐conjugated anti‐mouse IgA (Sigma Cat No. A4789‐1) or anti‐mouse IgG (Dako Cat No. P0447) in conjugate buffer (2% [v/v] FBS, 1% [v/v] BSA, 0.05% [v/v] Tween 20 in 0.01 PBS). Plates were incubated for 1 h at RT and then developed using tetramethyl benzidine (TMB) substrate (0.1 mg/mL 3.3′,5.5′‐tetramethylbenzidine in 0.1 M sodium acetate buffer [pH 5.5]). Reactions were stopped using 2 M H2SO4, and ODs read at 450 nm. Dilution curves were created for each sample and endpoint titers estimated as the maximum dilution that gave an absorbance reading above the average naive sample.
2.8. Ethics approval
Murine studies were conducted with approval from Royal Holloway University of London Ethics Committee under and an approved UK Home Office animal project license PB9FA6ABB. Challenge studies were conducted with approval from the Research and Ethics Committee of the Institute of Vaccines and Biological Medicals (IVAC; decision no. 241/QD‐VXSPYT 29/07/2022).
2.9. Statistical analysis
Statistical significance was assessed by the Mann–Whitney U‐test or the Dunnett's test using Prism (GraphPad, Dotmatics).
3. RESULTS
3.1. Display of urease antigens on the spore coat of B. subtilis
The complete urease A protein and the carboxy‐terminus of urease B of H. pylori were expressed on the surface of B. subtilis spores by in‐frame fusion of the relevant ureA and ureB coding ORFs to the B. subtilis cotB gene (Figure 1; n.b., we were unable to express the complete UreB protein on the spore surface). Both UreA and UreB have been shown to confer protection when delivered orally. 28 , 29 , 30 , 31 Here, however, we used a truncated urease B that lacked the amino‐terminal enzymatic domain. 23 , 24 The cotB gene has been used repeatedly for expression of heterologous antigens on B. subtilis spores and enables stable presentation of chimeric polypeptides. Our method for cloning used the THY‐X‐CISE® system that places the chimeric genes at the thyA (thymidylate synthetase A) gene of the prototrophic B. subtilis strain PY79. 22
This was followed by insertional disruption of the thyB locus resulting in strains (PK82 cotB‐ureA and PK78 cotB‐ureB CT ) that are unable to grow in the absence of thymine (or thymidine). Using a congenic strain (PK118) that carried insertional disruption of both thyA and thyB but carried no chimeric genes two immunological methods were used to verify surface display. First, western blotting using PAbs that recognize UreA and UreB and second, whole spore ELISA (Figure 2).
FIGURE 2.
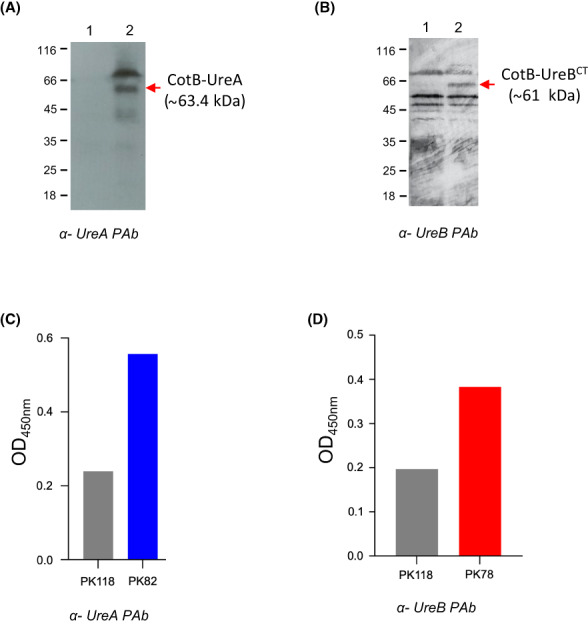
Spore coat expression of urease proteins. B. subtilis vaccine strains carrying insertions at the thyA loci were examined by western blotting of SDS‐PAGE (12% w/v) size‐fractionated spore coat proteins extracted from preparations of pure spores (approx. 2 × 109 spores/extraction). Blots were probed with PAbs as shown (panels A, B). Surface expression was also determined by whole spore enzyme‐linked immunosorbent assay (ELISA) using microtiter plates coated with spores at 1 × 108 CFU/well (panels C, D). Panel A shows blots of coat proteins extracted from PK82 (cotB‐ureA) and the isogenic parent strain PK118 (WT). Lane 1, spores of the PK118; lane 2, PK82 probed with anti‐UreA PAbs. Bands corresponding to CotB‐UreA (~63.4 kDa) are shown. Panel B shows blots of coat proteins extracted from PK118 (WT) and PK78 (cotB‐ureBCT). Lane 1, PK118 spore coat extracts; lane 2, PK78 spore coat extracts. Blots were probed with anti‐UreB PAbs. A band corresponding in size to CotB‐UreBCT and of the correct size (~61 kDa) of the CotB‐UreBCT chimera is indicated. Panel C, PK118 (WT) and PK82 (cotB‐ureA) spores used in ELISA and labeled with anti‐UreA PAbs (1:1000) followed by anti‐rabbit IgG‐HRP secondary antibody (1:3000). Panel D PK118 (WT) and PK78 (cotB‐ureBCT) spores used in ELISA and labeled with anti‐UreB PAbs (1:2000) followed by anti‐rabbit IgG‐HRP secondary antibody (1:3000).
Whole spore ELISA clearly demonstrated recognition of UreA and UreB on spores with some cross‐reaction to PK118 spores (Figure 2C,D). It should be noted that a tricistronic urease operon (ureABC) is present in most strains of B. subtilis with ureC corresponding to the enzymatic subunit that in H. pylori is named ureB. 32 B. subtilis UreA shares some homology with H. pylori UreA (~31%) and with UreC about 75% homology with H. pylori UreB. The operon is transcribed during ordinary vegetative cell growth but only at high levels during nitrogen‐limited growth. 32 , 33 Using a standard agar‐based biochemical method (Christensen's slant agar 34 ), we have confirmed that all three strains (PK118, PK78, and PK82) do not produce functional urease (not shown). Although we produced crops of spores, we cannot, however, rule out the possibility of the presence of low levels of B. subtilis‐produced urease being present (possibly adsorbed to spores) and possibly accounting for this cross‐reaction.
Blotting of size‐fractionated spore coat extracts for PK82 (cotB‐ureA) revealed three bands (~40, [diffuse], 64 and 70 kDa.) that were absent in PK118 spores (Figure 2A,B). One of these bands was in close agreement with the predicted size (63.4 kDa) of the CotB‐UreA chimera (Figure 2A; the other bands most likely being multimeric or breakdown species). Western blotting of PK78 (cotB‐ureB CT ) was less clean with cross‐reacting bands in PK118 spores. However, one abundant band of the correct size for CotB‐UreB (~61 kDa) was clearly present in PK78 and absent in PK118 spores (Figure 2B). Since the cross‐reacting bands are associated with the spore coat the most likely explanation is that of cross‐recognition with B. subtilis spore coat proteins. However, we were unable to identify any spore coat protein that showed significant levels of amino acid homology with either urease A or B.
3.2. Immune responses in mice dosed with spore vaccines
Mice were dosed orally (i.g.) four times with spores of PK82 (CotB‐UreA), PK78 (CotB‐UreBCT) and the isogenic control PK118 strain that expressed no heterologous polypeptides. To enable a dose of 3 × 1010 spore CFU three daily administrations of 1 × 1010 spores were required since high concentrations of spores in suspension are typically overly viscous. A naive group receiving only PBS provided a baseline.
Measurement of antigen‐specific sIgA in fecal samples showed seroconversion to both UreA (PK82‐dosed) and UreB (PK78‐dosed; Figure 3). Maximal antibody responses were observed at Day 61. Responses for both PK78 and PK82‐dosed animals, at maximum, were significantly (p = 0.0001) greater than in mice dosed with PK118 spores or the naive group. Very low levels of UreA‐specific sIgA were observed in PK118‐dosed mice but these were not statistically significant.
FIGURE 3.
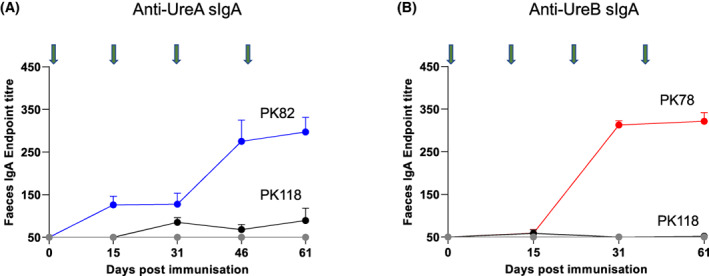
Mucosal responses following oral administration of spore vaccines expressing H. pylori antigens. Mice (C57 BL/6) were dosed (i.g.) with spores of PK118 (WT), PK82 (CotB‐UreA), or PK78 (CotB‐UreBCT) four times (green arrows). Each dose comprised three separate administrations (1 × 1010 CFU/administration); dose 1 (Days 1–3), dose 2 (Days 16–18), dose 3 (Days 32–34), and dose 4 (Days 47–49). rUreA‐specific (panel A) and rUreB‐specific sIgA (panel B) in longitudinal fecal samples in fecal samples from PK82, PK78, or PK118 dosed mice are shown.
Serum IgG responses measured at Day 61 also showed that both PK78 and PK82 were able to induce systemic immunity (Figure 4). Taken together, oral administration of spores expressing either UreA or UreBCT on the spore surface can elicit both mucosal and systemic responses.
FIGURE 4.
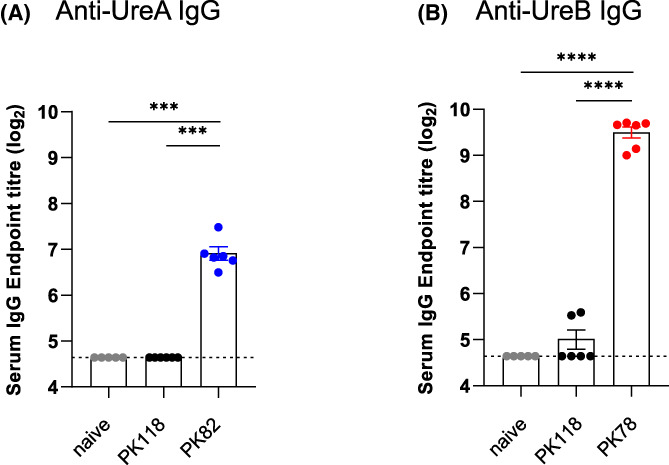
Systemic responses following oral administration of spore vaccines expressing H. pylori antigens. Mice (C57 BL/6) were dosed (i.g.) with spores of PK118 (WT), PK82 (CotB‐UreA), or PK78 (CotB‐UreB CT ) four times. Each dose comprised three separate administrations (1 × 1010 CFU/administration); dose 1 (Days 1–3), dose 2 (Days 16–18), dose 3 (Days 32–34), and dose 4 (Days 47–49). rUreA‐specific (panel A) and rUreB‐specific (panel B) IgG in serum samples taken at Day 61 are shown. Mann–Whitney, ***p = 0.001, ****p = 0.0001.
3.3. Protection in a murine colonization model
Mice were given four oral (i.g.) doses of spores (1 × 1010/dose) of either PK118 (WT), PK78 (CotB‐UreBCT) or PK82 (CotB‐UreA), as well as a naive group, and then challenged with H. pylori (i.g.) using a challenge dose of ~108 CFU. Stomach samples were taken 21 days post‐challenge for enumeration of H. pylori CFU. The study was repeated, and combined CFU data are shown in Figure 5.
FIGURE 5.
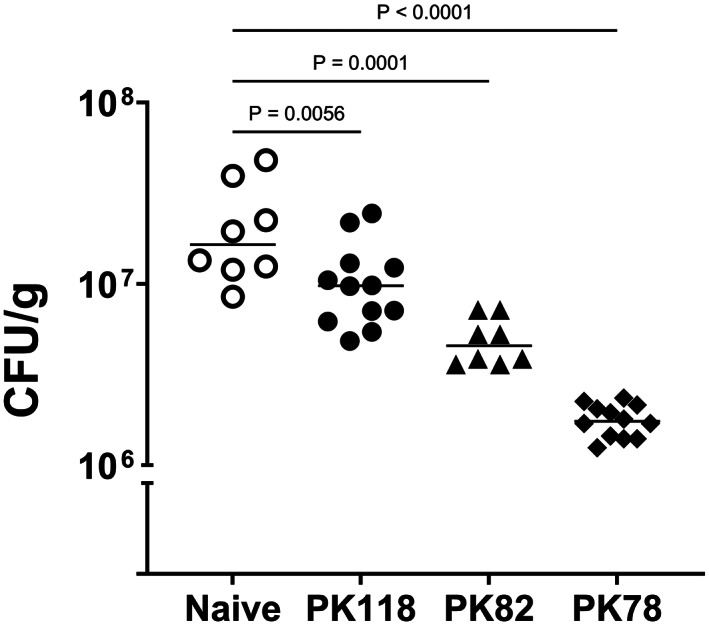
H. pylori colonization in immunized mice. Bacterial loads of H. pylori HP34 in stomach samples 21‐day post‐challenge. Animals had been orally (i.g.) dosed four times with spores (1 × 1010 CFU/dose) of PK118 (WT), PK82 (CotB‐UreA), or PK78 (CotB‐UreB CT ) and challenged 7–9 days after the last immunization. Naive mice received PBS. The data combine samples from two independent repeat studies with p values as shown.
PK78 (CotB‐UreBCT) immunized animals showed the greatest reduction in H. pylori CFU of about 1‐log. PK82 (CotB‐UreA) dosed animals also showed a significant reduction (~72%, median values; p = 0.0001) in CFU but less so than PK78 dosed animals (89%, median values, p < 0.0001). Interestingly, animals dosed with “naked” spores (PK118), that is, spores displaying no H. pylori antigens, also showed a reduction (~40%, median, p < 0.01) in H. pylori CFU compared to naive animals. In conclusion, both spore vaccines expressing either UreA or UreBCT were able to confer protective immunity sufficient to reduce H. pylori colonization in mice.
4. DISCUSSION
Spores of B. subtilis have been used extensively as mucosal vaccine vectors where oral (intra‐gastric or sublingual) or nasal administration efficiently induces mucosal immunity (typically sIgA) as well as a Th1 bias. 35 , 36 , 37 Antigens are displayed on the spore surface (diameter ~ 1 μm) typically fused (as chimeric fusions) to proteins associated with the outermost layers of the spore coat. Bacillus spores are dormant yet able to germinate and outgrow under favorable conditions. They are also particularly robust being able to survive exposure to extremes of heat, desiccation as well as noxious compounds including gastric fluids. 38 Remarkably, and as a rule, chimeric spore expression does not normally lead to significant degradation of the exposed heterologous protein. Spores are members of the aerobiome and also found in soil and vegetation. 39 , 40 As such animals and humans are exposed to a low level of Bacillus on a daily basis. 41 , 42 Considered together, the use of bacterial spores as oral vaccine vehicles is compelling.
For H. pylori vaccination, we evaluated two “classical” antigens, UreA (urease A) and UreB (urease B) since these have been used extensively in vaccine formulations and in animal studies show evidence of protection. 29 , 31 , 43 , 44 , 45 Our data show firstly that 12 oral administrations of spores expressing either UreA or UreBCT evoked mucosal immunity evident from seroconversion of antigen‐specific sIgA in fecal samples although it should be noted that IgG is also present in mucosal samples 46 and we did not assess levels of this immunoglobulin. UreB has previously been used for H. pylori vaccination utilizing spores for oral delivery but this has incorporated the entire UreB polypeptide fused to the CotC spore coat anchor. 28 , 47 Here, we chose a truncated UreB domain, UreBCT, to optimize spore expression since the use of the CotB anchor partner significantly increases the size of the resulting hybrid protein. Secondly, systemic antigen‐specific IgG responses were also induced. Finally, when mice were dosed with UreA or UreBCT spores and then challenged with H. pylori the resulting counts of H. pylori CFU in the stomach were reduced by about 1‐log (for CotB‐UreBCT). These data are broadly similar to those obtained by Zhou et al. 28 using spores expressing the full‐length UreB protein (CotC‐UreB) although there were some differences that need discussion. First, Zhou et al. 28 evaluated, in parallel, spores of an isogenic control strain that did not express any H. pylori antigens and in protection studies mice showed no reduction in counts of H. pylori. This is in marked contrast to our work here which showed that spores alone (i.e., PK118 spores) conferred a low level of protection (40% reduction in gastric CFU). These spores do not evoke antigen‐specific sIgA so the most probable explanation is that of innate immunity. Bacillus spores have been well documented as being able to evoke innate immunity and for some pathogens such as influenza this can be protective. 48 , 49 , 50 This has included murine studies showing reduced colonization by Clostridium difficile following oral dosing with “naked” spores. 35 We suspect that repeat dosing with B. subtilis spores may trigger an innate immune response sufficient to exert some level of protection. A second point is that Zhou et al. 28 also evaluated a trimeric fusion protein comprising a CotC anchor fused to CTB (cholera toxin subunit B) and UreB. This vaccine provided the highest reduction in gastric CFU of ~90% and was thus similar to our data found here for CotB‐UreBCT. 28 CTB was employed as a mucosal adjuvant but we suspect that the natural and well documented microparticulate adjuvant properties of spores are sufficient to provide adjuvancy dispensing with the need for an auxiliary mucosal adjuvant. 50 , 51
Here, we evaluated two different antigens and neither have been previously evaluated using spores (note that Zhou et al. used complete UreB 28 ). Examination of other in vivo studies on H. pylori live‐vectored vaccines reveals that 90% reduction in H. pylori CFU is close to the maximum that can be achieved. That both UreA and UreB delivered on spores confers some level of protection reinforces the general observation that a variety of H. pylori antigens can be used for vaccination. 17 It also supports the notion that other elements of the immune system may be required to achieve full sterilizing immunity. 15 , 52 It is well documented that cell‐mediated immunity plays an important role in protection against H. pylori infection. 15 , 16 Gastric biopsy samples from infected patients display an increase in CD4+ T cells, 52 and a bias of Th1 cells has been considered necessary for protection. 53 In addition, UreB has been shown to induce Th17 cells that, in turn, are responsible for the production of the proinflammatory cytokines, IL‐17, IL‐17F, and IL‐22. 54 Oral administration of Bacillus spores has been shown in mice to interact with components of the cellular immune system, notably toll‐like receptors (TLRs), with in vivo induction of proinflammatory cytokines (TNF‐α and IL‐6). 55 Potentially, these phenotypes may be linked with the abovementioned innate response but we suspect that the immunostimulatory properties of spores alone may also be contributing to the inhibition of H. pylori colonization. Lastly, as humans are exposed to low levels of Bacillus on a daily basis future development of the spore platform must consider and address the issue of tolerance and suppression of the immune response. 56
5. CONCLUSION
This work has shown the potential utility of spores for prophylactic vaccination to H. pylori infection. The use of a system that ensures containment of genetically modified probiotic spores is a further advantage primarily because the use of GMOs in humans remains contentious and biological containment using the approach reported here is assured. A therapeutic application of a H. pylori spore vaccine is also worthy of consideration and is under current investigation. Finally, the spore platform enables other potential H. pylori antigens to be evaluated and potentially a multivalent vaccine to be formulated. Such an approach might further boost levels of protection.
AUTHOR CONTRIBUTIONS
PMK, TLPN, TKCN, GM, and DMDB conducted experimental studies. VDN, GC, HAH, and SMC designed studies. SMC wrote the manuscript.
Supporting information
Figure S1.
ACKNOWLEDGMENTS
This work was supported by the Newton Fund program on Infectious Diseases and funded by the UK Medical Research Council (MRC) under grant number MR/R026262/1 to SMC and from the Vietnam Ministry of Science & Technology (MOST) under grant number NDT.79.GB/20 to VDN.
Katsande PM, Nguyen VD, Nguyen TLP, et al. Prophylactic immunization to Helicobacter pylori infection using spore vectored vaccines. Helicobacter. 2023;28:e12997. doi: 10.1111/hel.12997
Paidamoyo M. Katsande and Van Duy Nguyen contributed equally to the project.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Parsonnet J. The incidence of Helicobacter pylori infection. Aliment Pharmacol Ther. 1995;9(Suppl 2):45‐51. [PubMed] [Google Scholar]
- 2. Kusters JG, van Vliet AH, Kuipers EJ. Pathogenesis of Helicobacter pylori infection. Clin Microbiol Rev. 2006;19(3):449‐490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Polk DB, Peek RM Jr. Helicobacter pylori: gastric cancer and beyond. Nat Rev Cancer. 2010;10(6):403‐414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nagy P, Johansson S, Molloy‐Bland M. Systematic review of time trends in the prevalence of Helicobacter pylori infection in China and the USA. Gut Pathog. 2016;8:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Binh TT, Shiota S, Nguyen LT, et al. The incidence of primary antibiotic resistance of Helicobacter pylori in Vietnam. J Clin Gastroenterol. 2013;47(3):233‐238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nguyen TL, Uchida T, Tsukamoto Y, et al. Helicobacter pylori infection and gastroduodenal diseases in Vietnam: a cross‐sectional, hospital‐based study. BMC Gastroenterol. 2010;10:114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Goderska K, Agudo Pena S, Alarcon T. Helicobacter pylori treatment: antibiotics or probiotics. Appl Microbiol Biotechnol. 2018;102(1):1‐7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Frenck RW, Clemens J. Helicobacter in the developing world. Microbes Infect. 2003;8:705‐713. [DOI] [PubMed] [Google Scholar]
- 9. Graham DY, Shiotani A. New concepts of resistance in the treatment of Helicobacter pylori infections. Nat Clin Pract Gastroenterol Hepatol. 2008;5(6):321‐331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yoon K, Kim N, Nam RH, et al. Ultimate eradication rate of Helicobacter pylori after first, second, or third‐line therapy in Korea. J Gastroenterol Hepatol. 2015;30(3):490‐495. [DOI] [PubMed] [Google Scholar]
- 11. Thung I, Aramin H, Vavinskaya V, et al. Review article: the global emergence of Helicobacter pylori antibiotic resistance. Aliment Pharmacol Ther. 2016;43(4):514‐533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. WHO . Prioritization of pathogens to guide discovery, research and development of new antibiotics for drug‐resistant bacterial infections, including tuberculosis . Geneva. WHO/EMP/IAU/2017.12; 2017.
- 13. Parsonnet J. What is the Helicobacter pylori global reinfection rate? Can J Gastroenterol. 2003;17(Suppl B):46B‐48B. [DOI] [PubMed] [Google Scholar]
- 14. Rupnow MF, Shachter RD, Owens DK, Parsonnet J. Quantifying the population impact of a prophylactic Helicobacter pylori vaccine. Vaccine. 2001;20(5–6):879‐885. [DOI] [PubMed] [Google Scholar]
- 15. Agarwal K, Agarwal S. Helicobacter pylori vaccine: from past to future. Mayo Clin Proc. 2008;83(2):169‐175. [DOI] [PubMed] [Google Scholar]
- 16. Czinn SJ, Blanchard T. Vaccinating against Helicobacter pylori infection. Nat Rev Gastroenterol Hepatol. 2011;8(3):133‐140. [DOI] [PubMed] [Google Scholar]
- 17. Svennerholm AM, Lundgren A. Progress in vaccine development against Helicobacter pylori . FEMS Immunol Med Microbiol. 2007;50(2):146‐156. [DOI] [PubMed] [Google Scholar]
- 18. Raghavan S, Svennerholm AM, Holmgren J. Effects of oral vaccination and immunomodulation by cholera toxin on experimental Helicobacter pylori infection, reinfection, and gastritis. Infect Immun. 2002;70(8):4621‐4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lee CK. Vaccination against Helicobacter pylori in non‐human primate models and humans. Scand J Immunol. 2001;53(5):437‐442. [DOI] [PubMed] [Google Scholar]
- 20. Zeng M, Mao XH, Li JX, et al. Efficacy, safety, and immunogenicity of an oral recombinant Helicobacter pylori vaccine in children in China: a randomised, double‐blind, placebo‐controlled, phase 3 trial. Lancet. 2015;386(10002):1457‐1464. [DOI] [PubMed] [Google Scholar]
- 21. Harwood CR, Cutting SM, eds. Molecular Biological Methods for Bacillus. John Wiley & Sons Ltd; 1990. Goodfellow M, ed. Modern microbiological methods. [Google Scholar]
- 22. Hosseini S, Curilovs A, Cutting SM. Biological containment of genetically modified Bacillus subtilis . Appl Environ Microbiol. 2018;84(3):e02334‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jabri E, Carr MB, Hausinger RP, Karplus PA. The crystal structure of urease from Klebsiella aerogenes . Science. 1995;268(5213):998‐1004. [PubMed] [Google Scholar]
- 24. Shin M‐K, Jun J‐S, Kwon S‐W, et al. Characterization of specific IgA response to antigenic determinants of Helicobacter pylori rrease encoded by ureA and ureB in children. J Bacteriol Virol. 2018;48:14‐22. [Google Scholar]
- 25. Tomb JF, White O, Kerlavage AR, et al. The complete genome sequence of the gastric pathogen Helicobacter pylori . Nature. 1997;388(6642):539‐547. [DOI] [PubMed] [Google Scholar]
- 26. Permpoonpattana P, Phetcharaburanin J, Mikelsone A, et al. Functional characterization of Clostridium difficile spore coat proteins. J Bacteriol. 2013;195(7):1492‐1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Smeekens JM, Johnson‐Weaver BT, Hinton AL, et al. Fecal IgA, antigen absorption, and gut microbiome composition are associated with food antigen sensitization in genetically susceptible mice. Front Immunol. 2020;11:599637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhou Z, Dong H, Huang Y, et al. Recombinant Bacillus subtilis spores expressing cholera toxin B subunit and Helicobacter pylori urease B confer protection against H. pylori in mice. J Med Microbiol. 2017;66(1):83‐89. [DOI] [PubMed] [Google Scholar]
- 29. Smythies LE, Novak MJ, Waites KB, Lindsey JR, Morrow CD, Smith PD. Poliovirus replicons encoding the B subunit of Helicobacter pylori urease protect mice against H. pylori infection. Vaccine. 2005;23(7):901‐909. [DOI] [PubMed] [Google Scholar]
- 30. Gomez‐Duarte OG, Lucas B, Yan ZX, Panthel K, Haas R, Meyer TF. Protection of mice against gastric colonization by Helicobacter pylori by single oral dose immunization with attenuated Salmonella typhimurium producing urease subunits A and B. Vaccine. 1998;16(5):460‐471. [DOI] [PubMed] [Google Scholar]
- 31. Corthesy‐Theulaz IE, Hopkins S, Bachmann D, et al. Mice are protected from Helicobacter pylori infection by nasal immunization with attenuated Salmonella typhimurium phoPc expressing urease A and B subunits. Infect Immun. 1998;66(2):581‐586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cruz‐Ramos H, Glaser P, Wray LV Jr, Fisher SH. The Bacillus subtilis ureABC operon. J Bacteriol. 1997;179(10):3371‐3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wray LV Jr, Ferson AE, Fisher SH. Expression of the Bacillus subtilis ureABC operon is controlled by multiple regulatory factors including CodY, GlnR, TnrA, and Spo0H. J Bacteriol. 1997;179(17):5494‐5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Christensen WB. Urea decomposition as a means of differentiating Proteus and Paracolon cultures from each other and from Salmonella and Shigella types. J Bacteriol. 1946;52(4):461‐466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Permpoonpattana P, Hong HA, Phetcharaburanin J, et al. Immunization with Bacillus spores expressing toxin A peptide repeats protects against infection with Clostridium difficile strains producing toxins A and B. Infect Immun. 2011;79(6):2295‐2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hoang TH, Hong HA, Clark GC, Titball RW, Cutting SM. Recombinant Bacillus subtilis expressing the Clostridium perfringens alpha toxoid is a candidate orally delivered vaccine against necrotic enteritis. Infect Immun. 2008;76(11):5257‐5265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Duc le H, Hong HA, Fairweather N, Ricca E, Cutting SM. Bacterial spores as vaccine vehicles. Infect Immun. 2003;71(5):2810‐2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nicholson WL, Munakata N, Horneck G, Melosh HJ, Setlow P. Resistance of Bacillus endospores to extreme terrestrial and extraterrestrial environments. Microbiol Mol Biol Rev. 2000;64(3):548‐572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nicholson WL. Roles of Bacillus endospores in the environment. Cell Mol Life Sci. 2002;59(3):410‐416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shaffer BT, Lighthart B. Survey of culturable airborne bacteria at four diverse locations in Oregon: urban, rural, forest, and coastal. Microb Ecol. 1997;34(3):167‐177. [DOI] [PubMed] [Google Scholar]
- 41. Hong HA, To E, Fakhry S, Baccigalupi L, Ricca E, Cutting SM. Defining the natural habitat of Bacillus spore‐formers. Res Microbiol. 2009;160(6):375‐379. [DOI] [PubMed] [Google Scholar]
- 42. Ferreira WT, Hong HA, Adams JRG, et al. Environmentally acquired Bacillus and their role in C. difficile colonization resistance. Biomedicine. 2022;10(5):930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bumann D, Metzger WG, Mansouri E, et al. Safety and immunogenicity of live recombinant Salmonella enterica serovar typhi Ty21a expressing urease A and B from Helicobacter pylori in human volunteers. Vaccine. 2001;20(5–6):845‐852. [DOI] [PubMed] [Google Scholar]
- 44. Zhou WY, Shi Y, Wu C, et al. Therapeutic efficacy of a multi‐epitope vaccine against Helicobacter pylori infection in BALB/c mice model. Vaccine. 2009;27(36):5013‐5019. [DOI] [PubMed] [Google Scholar]
- 45. Yang J, Dai LX, Pan X, et al. Protection against Helicobacter pylori infection in BALB/c mice by oral administration of multi‐epitope vaccine of CTB‐UreI‐UreB. Pathog Dis. 2015;73(5):ftv026. [DOI] [PubMed] [Google Scholar]
- 46. Sano K, Bhavsar D, Singh G, et al. SARS‐CoV‐2 vaccination induces mucosal antibody responses in previously infected individuals. Nat Commun. 2022;13(1):5135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhou Z, Gong S, Li XM, et al. Expression of Helicobacter pylori urease B on the surface of Bacillus subtilis spores. J Med Microbiol. 2015;64(Pt 1):104‐110. [DOI] [PubMed] [Google Scholar]
- 48. Song M, Hong HA, Huang JM, et al. Killed Bacillus subtilis spores as a mucosal adjuvant for an H5N1 vaccine. Vaccine. 2012;30(22):3266‐3277. [DOI] [PubMed] [Google Scholar]
- 49. James J, Meyer SM, Hong HA, et al. Intranasal treatment of ferrets with inert bacterial spores reduces disease caused by a challenging H7N9 avian influenza virus. Vaccines. 2022;10(9):1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. de Souza RD, Batista MT, Luiz WB, et al. Bacillus subtilis spores as vaccine adjuvants: further insights into the mechanisms of action. PLoS One. 2014;9(1):e87454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Barnes AG, Cerovic V, Hobson PS, Klavinskis LS. Bacillus subtilis spores: a novel microparticle adjuvant which can instruct a balanced Th1 and Th2 immune response to specific antigen. Eur J Immunol. 2007;37(6):1538‐1547. [DOI] [PubMed] [Google Scholar]
- 52. Chmiela M, Michetti P. Inflammation, immunity, vaccines for Helicobacter infection. Helicobacter. 2006;11(Suppl 1):21‐26. [DOI] [PubMed] [Google Scholar]
- 53. Eaton KA, Mefford M, Thevenot T. The role of T cell subsets and cytokines in the pathogenesis of Helicobacter pylori gastritis in mice. J Immunol. 2001;166(12):7456‐7461. [DOI] [PubMed] [Google Scholar]
- 54. Zhang JY, Liu T, Guo H, et al. Induction of a Th17 cell response by Helicobacter pylori urease subunit B. Immunobiology. 2011;216(7):803‐810. [DOI] [PubMed] [Google Scholar]
- 55. Huang JM, La Ragione RM, Nunez A, Cutting SM. Immunostimulatory activity of Bacillus spores. FEMS Immunol Med Microbiol. 2008;53(2):195‐203. [DOI] [PubMed] [Google Scholar]
- 56. Homayun B, Lin X, Choi HJ. Challenges and recent progress in oral drug delivery systems for biopharmaceuticals. Pharmaceutics. 2019;11(3):129. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


