Abstract
HOX proteins and some orphan homeodomain proteins form complexes with either PBX or MEIS subclasses of homeodomain proteins. This interaction can increase the binding specificity and transcriptional effectiveness of the HOX partner. Here we show that specific members of both PBX and MEIS subclasses form a multimeric complex with the pancreatic homeodomain protein PDX1 and switch the nature of its transcriptional activity. The two activities of PDX1 are exhibited through the 10-bp B element of the transcriptional enhancer of the pancreatic elastase I gene (ELA1). In pancreatic acinar cells the activity of the B element requires other elements of the ELA1 enhancer; in β-cells the B element can activate a promoter in the absence of other enhancer elements. In acinar cell lines the activity is mediated by a complex comprising PDX1, PBX1b, and MRG1 (MEIS2). In contrast, β-cell lines are devoid of PBX1b and MRG1, so that a trimeric complex does not form, and the β-cell-type activity is mediated by PDX1 without PBX1b and MRG1. The presence of specific nuclear isoforms of PBX and MEIS is precisely regulated in a cell-type-specific manner. The β-cell-type activity can be detected in acinar cells if the B element is altered to retain binding of PDX1 but prevent binding of the PDX1-PBX1b-MRG1 complex. These observations suggest that association with PBX and MEIS partners controls the nature of the transcriptional activity of the organ-specific PDX1 transcription factor in exocrine versus endocrine cells.
The mammalian pancreas is a compound gland of endocrine and exocrine tissues derived from the embryonic endoderm (62). Approximately 90% of the pancreas is exocrine tissue, comprising acinar cells that synthesize and secrete digestive enzymes and ductal cells that secrete and channel the fluid that transports the acinar enzymes to the duodenum. About 1% of the pancreas is endocrine tissue, comprising four principal cell types synthesizing insulin (β-cells), glucagon (α-cells), somatostatin (δ-cells), and pancreatic polypeptide (PP cells) organized into islets scattered throughout the exocrine pancreas. The endocrine and exocrine compartments are structurally and functionally integrated through an islet-acinar portal blood system that facilitates the regulation of acinar cell functions directly by islet peptide hormones (81).
The exocrine-endocrine relationship begins at the inception of pancreogenesis: both tissues are derived from a common endodermal cell lineage (for reviews, see references 34 and 73). This shared lineage may be expected to result in shared strategies and effectors for gene transcription, but with the important requirement that neither endocrine hormones or exocrine digestive enzymes be synthesized in the wrong compartment. Divergence of islet and acinar cell lineages may occur by modification of a pancreatic regulatory network through the differential expression of regulatory molecules such as transcription factors (or possibly by the differential use of a shared factor) to activate exocrine or endocrine-specific gene sets.
Several homeodomain proteins are important components of the transcriptional network that controls pancreogenesis (1, 22, 50, 69, 74). The homeodomain transcription factors encoded by the HOX gene clusters are key mediators in setting up the body plan during animal development (29, 33). Different members of this large gene family are essential for correct specification of cell identity, despite having very similar DNA binding sites in vitro. The exquisite specificity of many of these proteins in vivo depends in part on interaction with the PBC family of homeodomain proteins (for reviews, see references 39 and 82). Other homeodomain proteins whose genes are not part of the HOX gene clusters, such as engrailed and the pancreas- and duodenum-specific factor PDX1, also interact with PBC family members (57, 58).
PDX1 (previously termed IPF1 [51], STF1 [35], X1Hbox8 [84], IDX1 [41], or β-TF1 [32]) is critical to pancreogenesis. PDX1-deficient mice are born without endocrine and exocrine pancreatic tissues (22, 50). An apancreatic human who is also PDX1 deficient due to a homozygous point mutation in the PDX1 gene has been identified (76). In adult mice PDX1 has a selective endodermal expression pattern limited to the epithelial cells of the pancreas and the rostral duodenum (50, 84). PDX1 expression is first detected in the primitive endodermal gut tube at sites that give rise to dorsal and ventral pancreatic buds about one-half day later (19, 22). Subsequently during normal development, PDX1 is present in the epithelial cells of the bud that give rise to pancreatic islet, acinar, and ductal cells (19). Pancreogenesis in PDX1-deficient mice is arrested at this stage, prior to the appearance of differentiated islet and acinar cells. As development progresses, PDX1 is present in amylase-expressing precursors of acinar cells, as well as in islet cell precursors expressing insulin, somatostatin, or glucagon (19). In the pancreas of adult mice, PDX1 is present at the highest levels in β-cells of the islets and at much lower levels in other islet cell types and in acinar cells (85). In mature β-cells, PDX1 participates in the activation of several β-cell-specific promoters, including those of the insulin, Glut2 transporter, and glucokinase genes (51, 56, 79, 80).
In this report we show that PDX1 also participates in the activation of the transcriptional enhancer of at least one acinar cell-specific gene, that for elastase I (ELA1), and therefore plays a role in acinar cell function as well. The rat ELA1 enhancer consists of three transcription factor binding sites: elements A, B, and C. The A element is the primary determinant of acinar cell-specific transcription; a homomultimeric repeat of a 26-bp A element directs pancreatic acinar cell expression in transgenic mice (66). The activity of the A element is mediated by the acinar cell-specific factor PTF1 (14, 66), which is proposed to be a key regulator that defines the acinar phenotype (14, 27, 59). The C element is bound by tissue-nonspecific factors that augment the activity of PTF1 (32). The third element of the enhancer, the B element, is active in β-cells as well as acinar cells (31, 32). However, its roles in these two cell types are different. In acinar cells of mice, the B element acts only in concert with the other enhancer elements; in this context it plays a secondary role by augmenting the activity of the acinar cell-specific A element. In islets of transgenic mice, the B element directs expression selectively to β-cells. This β-cell-specific activation property of the B element is manifested both within the context of the enhancer and as a homomultimeric repeat. The repeated form of the B element is inactive in acinar cells. Thus, the B element alone can activate a simple promoter in β-cells, whereas in acinar cells it requires the participation of other enhancer elements (31).
We show that the distinct acinar cell and β-cell activities of the B element are mediated by complexed and uncomplexed forms of PDX1, respectively. In acinar cell lines, a PDX1 complex containing two additional homeodomain proteins, PBX1b and MRG1 (MEIS2), participates in the enhancer. In β-cell lines, this complex does not form and PDX1 in the absence of these two cofactors is capable of activating a reporter gene containing a repeat of the B element. The trimeric complex, which acts within the three-element enhancer, cannot activate the B-element repeat. Conversely, PDX1 without PBX1b and MRG1 can activate the B repeat but is not functional within the context of the enhancer. Therefore, the different activities of PDX1 are dependent on whether it is complexed with PBX1b and MRG1. We suggest that the use of this shared pancreatic transcription factor in two different ways contributes to the activation of different gene sets that distinguish the exocrine and endocrine lineages.
MATERIALS AND METHODS
Gene construction.
The ELA1 enhancer-promoter construct contains ELA1 gene sequences from nucleotide −205 to +8 fused to a human growth hormone (hGH) reporter gene as described previously (54). In the various ELA1 B-element multimer constructs, five or six copies of double-stranded oligonucleotides (described below) were multimerized and placed in front of the ELA1 minimal promoter from nucleotide −92 to +8, linked to an hGH reporter gene, as described previously (32). To express PDX1 in transfected cells, the coding sequence of mouse PDX1, with 5′ and 3′ untranslated regions from the Xenopus laevis β-globin gene (28), was cloned into the pBK-CMV expression plasmid (Stratagene, La Jolla, Calif.) (details are available upon request). The lacZ region of the plasmid, between the cytomegalovirus (CMV) promoter and the cloning cassette, was removed to enhance eukaryotic expression. Full-length clones of PBX1a and PBX1b were in the pBK-RSV plasmid (Stratagene) (23).
Cell lines.
The endocrine lines were βTC-3, derived from an insulinoma of a mouse bearing a simian virus 40 T-antigen gene directed by insulin gene regulatory elements (16), and Ins1 and RIN1046-38 (RIN38), rat insulinoma cell lines induced by radiation (2, 61). The acinar cell lines were 266-6 (ATCC CRL-2151), derived from an acinar pancreatic tumor of a mouse bearing a T-antigen transgene directed by ELA1 regulatory sequences (53); C5-2E, derived from an acinar pancreatic tumor of a mouse bearing a T-antigen transgene directed by rat trypsin 1 gene regulatory sequences (32a); and AR42J (ATCC CRL-1492), derived from an azaserine-induced acinar pancreatic carcinoma of a rat (21). AR42J is a widely used model of pancreatic acinar cells with some amphicrine properties (68). An additional exocrine cell line, ARIP (ATCC CRL-1674), was also derived from an azaserine-induced pancreatic carcinoma of a rat (21). The cell lines HeLa (ATCC CCL-2), Rat2 (ATCC CRL-1764), NIH 3T3 (ATCC CRL-1658), and Jurkat (ATCC TIB-152) were utilized as nonpancreatic controls. All key results were confirmed with multiple endocrine, exocrine, and nonpancreatic cell lines because of the limitations of any single cell line as a model of the tissue from which it was derived.
Transfections.
266-6, AR42J, and HeLa cells were transfected by the calcium phosphate technique (18). RIN38 and βTC-3 cells were transfected by electroporation (83). Expression of the transfected genes was monitored by determination of hGH accumulation in the medium (70) by using a radioimmunoassay (Nichols Institute, San Juan Capistrano, Calif.). All transfection results were corrected for varying transfection efficiencies, based on the activity of a cotransfected reporter gene control. Internal control plasmids contained either a Rous sarcoma virus-chloramphenicol acetyltransferase fusion gene (30) or a CMV–β-galactosidase fusion gene (pCMVβ-2; Clontech, San Francisco, Calif.). Chloramphenicol acetyltransferase enzyme activity was assayed as described by Nielsen et al. (48), and β-galactosidase activity was assayed according to directions provided by Tropix, Inc., Bedford, Mass. The activities of all constructs were assayed in duplicate in two or more independent experiments.
EMSA and antibody supershift assay.
Nuclear extracts were prepared by a slight modification of the procedure of Dignam et al. (15) as described by Kruse et al. (32). Mobility shift binding reactions and electrophoresis were as described by Swift et al. (78). In some of the electrophoretic mobility shift assays (EMSAs) of 266-6 nuclear extracts, the GATA4–B-oligonucleotide complex was competed with a GATA binding site oligonucleotide, βG1 (78). In EMSA with added antibodies, nuclear extract was incubated for 10 min at room temperature with 1 to 3 μl of rabbit serum or purified immunoglobulin G fractions prior to incubation with oligonucleotides under standard EMSA conditions. EMSA with the rabbit reticulocyte lysate containing in vitro-translated (IVT) factors was done similarly.
Oligonucleotides.
The unmodified (wild-type) B-element oligonucleotides and the transversion mutants B through H have been described previously (78). The wild-type B element had the coding-strand sequence CTTATCAGATAAATGAGTTGA and noncoding-strand sequence AGTCAACTCATTTATCTGATA, creating a double-stranded oligonucleotide encompassing nucleotides −162 to −144 of the rat ELA1 gene. The bases mutated by transversion were changed from A to C, C to A, G to T, and T to G. The PBX-PDX1 consensus binding site oligonucleotide from the TseII site of the somatostatin gene (35, 57) had the top-strand sequence GTACTGCATGATTAATTACTGA and bottom-strand sequence GATCTCAGTAATTAATCATGCA. The TseII site is identical at 12 of 13 nucleotides to the selected binding site of PBX1 and HOXB-7 (46).
Antibodies.
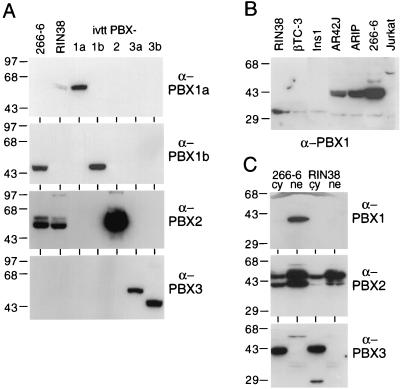
Polyclonal rabbit antibodies used in this study included anti-PDX1 (anti-STF1 [a generous gift of Marc Montminy] or anti-X1Hbox8 [19]); anti-PBX (raised against PBX1b; it detects all members of the PBX family [5]); antibody specific for PBX1, PBX2, or PBX3 (Santa Cruz Biotechnology, Inc.); or antibody specific for PBX1a or PBX1b. The antibody specific for PBX1a was raised against a peptide corresponding to amino acids 383 to 400 of the PBX1a protein, and the antibody specific for PBX1b was raised against a peptide corresponding to amino acids 331 to 347 of the PBX1b protein. The specificity and suitability of each PBX antibody in both Western blotting (shown in Fig. 6A) and EMSA conditions (data not shown) were tested with the IVT PBX proteins (see below). Polyclonal rabbit antibodies specific for MEIS1 and MRG1 were raised against peptides containing the first 16 amino acids of each protein.
FIG. 6.
Nuclear and cytoplasmic distribution of PBX species for endocrine and exocrine pancreatic cells. (A) 266-6 nuclear extract contains PBX1b and PBX2, whereas RIN38 nuclear extract contains only PBX2. Western analyses of nuclear extracts (30 μg of total protein/lane) from 266-6 or RIN38 cells were probed with antibodies specific to PBX1a, PBX1b, PBX2, and PBX3. IVT PBX species (0.5 μl) present in each panel demonstrate the specificity and reactivity of each antibody. (B) PBX1b is present in three different exocrine cell lines but is absent in three endocrine cell lines. Western analyses of nuclear extracts (30 μg of total protein/lane) from the acinar cell lines AR42J, ARIP, and 266-6 and the β-cell lines RIN38, βTC-3, and Ins1, as well as a control (Jurkat pre-B) cell line, were probed with antibodies specific for PBX1. (C) Cytoplasmic versus nuclear localization of PBX species in 266-6 acinar and RIN38 endocrine cells. Cytoplasmic and nuclear extracts (30 μg of total protein/lane) probed with antibodies specific to PBX1, PBX2, and PBX3 detected PBX1b only in the 266-6 nuclear compartment, PBX2 in both the nuclei and cytoplasm of both the endocrine and acinar cell lines, and PBX3b only in the cytoplasm of the two cell lines. Numbers on the left in each panel are molecular masses in kilodaltons.
Northern blots.
RNAs from cell lines were isolated with RNA STAT 60 (Tel-Test B, Inc., Friendswood, Tex.). Pancreatic RNA was isolated by the guanidine thiocyanate procedure (37). Poly(A)-enriched RNA was obtained by binding to oligo(dT) columns. RNA was assayed by Northern blotting of RNA resolved by electrophoresis in methyl mercury hydroxide (77) and transferred to Zeta-probe membranes (Bio-Rad).
Cloning of mouse PBX cDNAs.
A cDNA library from the mouse acinar pancreatic cell line 266-6 was created in LambdaZap. Of a total library of 1.2 × 106 clones, 3.5 × 105 clones were screened with a mixture of a full-length human PBX1a clone and partial clones of human PBX2 and PBX3. Seventy-three clones were identified as potential members of the mouse PBX family. These clones were further identified by hybridization with short fragments specific for the N-terminal coding sequence of each class of human PBX. The complete sequence of a full-length clone of each mouse PBX species was determined with an ABI 377 sequencer.
IVT.
Constructs for in vitro translation (IVT) were prepared as follows. Mouse PDX1-coding sequence with the Xenopus β-globin 5′ untranslated region was cloned into the HindIII and SalI sites of the pSP64 vector (Promega, Madison, Wis.) so that the PDX1-coding sequence is under the control of SP6 promoter. Human PBX1a- and PBX1b-coding sequences with the Xenopus β-globin 5′ untranslated region were cloned into the BglII and PstI sites of the pSP73 vector (Promega). To construct the PBX2 expression plasmid, mouse PBX2-coding sequence was amplified by PCR and cloned into the NcoI and XhoI sites of the pSP73 vector that contains the Xenopus β-globin 5′ untranslated region. Mouse PBX3a- and PBX3b-coding sequences were also amplified by PCR and cloned into the NcoI and SphI sites of the same vector. The PBX-coding sequences were all under the control of SP6 promoter. MRG1a cDNA was under the control of the T7 promoter in the vector pET-28b (Novagen, Madison, Wis.).
Coupled in vitro transcription and translation were performed with the Promega TNT coupled reticulocyte lysate system according to the manufacturer’s instructions. Relative molar amounts of IVT proteins were estimated from the [35S]methionine incorporation and the molar fraction of methionine in each protein.
Western blots.
Thirty micrograms of cytoplasmic or nuclear extracts was resolved by sodium dodecyl sulfate–10% polyacrylamide gel electrophoresis and electrotransferred to Immobilon polyvinylidene difluoride membranes (Millipore, Bedford, Mass.). Filters were then blocked overnight with a solution of 5% nonfat dry milk and 0.5% Tween 20 in phosphate-buffered saline. After blocking, filters were incubated with antibody for 1 h. The bound primary antibody was detected by incubation with horseradish peroxidase-conjugated donkey anti-rabbit immunoglobulins (Amersham, Arlington Heights, Ill.) and reaction with an enhanced chemiluminescence detection kit (Pierce, Inc., Rockford, Ill.).
RESULTS
Activity of the B element in cultured pancreatic cells.
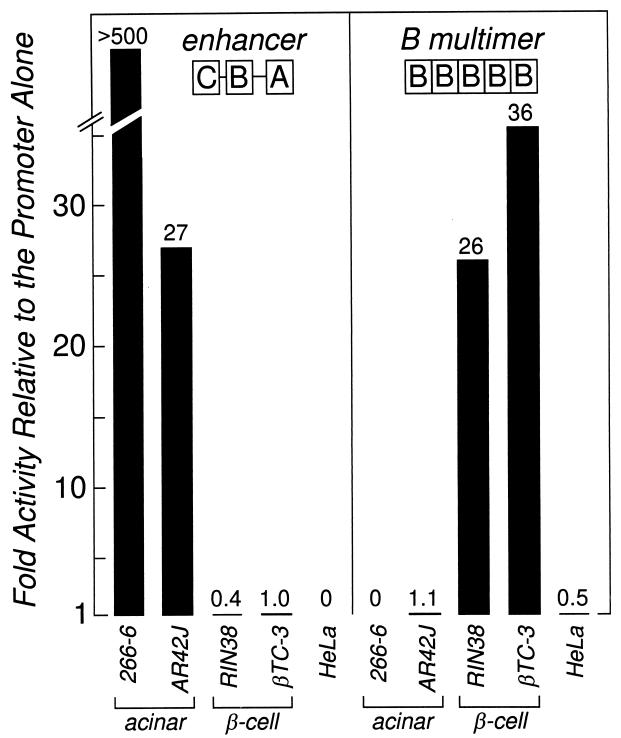
To determine the basis for the two different activities of the ELA1 B element in the acinar cells and β-cells of the pancreas, we analyzed its activity both as a homomultimeric repeat and as part of the ELA1 enhancer in various pancreatic acinar cell and β-cell lines. The B-element repeat activated expression only in the β-cell lines (Fig. 1), consistent with its restricted activity only in β-cells of transgenic mice. The complete three-element enhancer is active only in the acinar cell lines (Fig. 1), consistent with its activity in acinar cells of mice. However, the single B element residing within the enhancer is not active in the β-cell lines, in contrast to its action in β-cells of mice (32). Both the repeat and enhancer forms of the B element are inactive in nonpancreatic cell lines (i.e., HeLa [Fig. 1] and NIH 3T3 and Rat2 [data not shown]). Thus, the two different activities of the B element in mice are largely reproduced in acinar cell and β-cell lines.
FIG. 1.
Activity of the ELA1 enhancer and the ELA1 B-element repeat in acinar cell, β-cell, and nonpancreatic cell lines. The enhancer (nucleotides −205 to −93 of the ELA1 gene) contains three elements: A, B, and C. The B-element repeat has five tandem copies of the B element. The reporter gene in each case was the hGH gene under the direction of the rat ELA1 minimal promoter (nucleotides −92 to +8 in the ELA1 gene). Construct details are in Materials and Methods. Fold activity is expressed as the ratio of the activity of the test construct to the activity of the minimal promoter. The activity of the enhancer relative to that of the promoter is much greater in 266-6 than in the acinar AR42J cells because the activity of the minimal promoter in the 266-6 cells is extremely low and the activity of the enhancer is high. The differences in levels of reporter gene activity between the expressing and nonexpressing cell lines for both the ELA1 enhancer and the ELA1 B-element repeat are statistically significant (P < 0.001) in all instances.
Different regions of the B element mediate the endocrine and acinar cell activities.
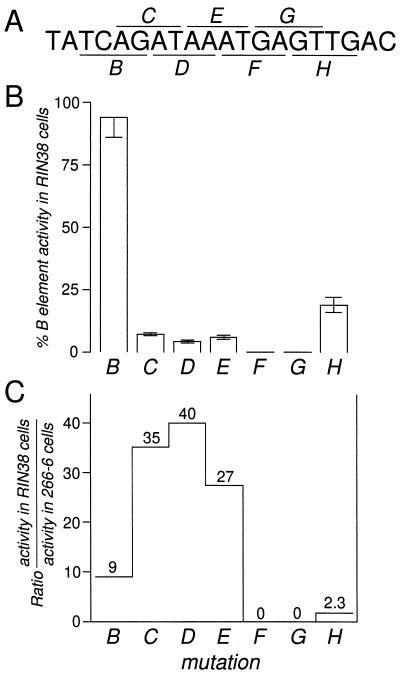
Previous results suggested that two different regions of the initially defined B element may be active in acinar cells versus β-cells (32, 78). To define the minimal region required for activity of the B element in β-cells, overlapping 4-bp transversion mutations of the element (Fig. 2A) were tested as homomultimeric repeats by transfection into the RIN38 β-cell line. The mutant repeats were linked to the minimal promoter of the ELA1 gene (nucleotides −92 to +8). The results are not dependent on the ELA1 promoter specifically, because the B-element multimer (31, 32b) and the ELA1 enhancer (52) can also activate other minimal promoters, resulting in the same specificity of expression in transgenic mice and transfected cells. Mutations within an 8-bp region, ATAAATGA, effectively inactivated the B-element multimer in RIN38 cells (Fig. 2B).
FIG. 2.
One side of the B element is more important in acinar cells than in islet cells. (A) Sequence of the B element (rat ELA1 nucleotides −162 to −143) with the region of each 4-bp transversion mutation indicated by underlining or overlining. (B) Percent activity of repeats of each mutant in RIN38 cells compared to the activity of the repeat of the unmodified element. Error bars represent standard errors of the mean. (C) Comparison of the relative activity of each mutant B element in RIN38 and 266-6 cells revealed that the 5′ side of the element is more critical to activity in 266-6 acinar cells than in RIN38 β-cells. The data for each mutant B element within the context of the enhancer in acinar 266-6 cells was taken from reference 78.
The same series of mutations introduced into the B element within the context of the −205/−93 ELA1 enhancer previously delineated a longer sequence of 10 bp, AGATAAATGA, required in 266-6 acinar cells (78). In addition to the differences in the length of the active element in the two cell types and in the context in which it is active (as a repeat in β-cells and as part of the three-element enhancer in acinar cells), the relative importance of particular nucleotide positions within the element differed. We compared the relative effects of the mutations in the B-element repeats in RIN38 cells and of the same mutations in the B element within the ELA1 enhancer in 266-6 cells (Fig. 2C). The bases at the 5′ side of the B element are critical to its activity in the 266-6 acinar cells and are of less importance in the β-cell line. This result implies that the acinar cell and β-cell activities are mediated by different factor complexes with different sequence requirements and is consistent with the different complexes that form on this element in nuclear extracts from each cell type (see below).
Different complexes containing PDX1 are active in β-cells and acinar cells.
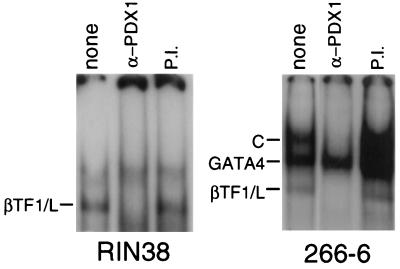
To understand the basis of the two activities of the B element in the β-cells and acinar cells, we identified the transcription factors that are responsible for its activity in each cell type. Previous results for RIN38 β-cells identified a binding activity, termed βTF-1, with nucleotide sequence requirements coincident with the 8-bp region essential to activity in RIN38 cell transfections (32). A different binding activity, termed the C complex (Fig. 3), was identified in nuclear extracts of 266-6 acinar cells (78). This complex is not present in RIN38 nuclear extracts. The binding specificity of the C complex matched the B-element region critical to the activity of the enhancer in the acinar cell line (78). The C complex is formed by the interaction of a factor (termed L) with migration and binding specificity similar to those of βTF-1 and another factor(s) which did not appear to bind to the B element on its own. The results presented below demonstrate that βTF-1 and L are identical and are contained in the C complex.
FIG. 3.
The active complexes in RIN38 β-cells and in 266-6 acinar cells contain PDX1. Anti-PDX1 serum (α-PDX1) or preimmune serum (P.I.) was added to EMSA mixtures with the B-element oligonucleotide (see Materials and Methods). The identity of each major gel shift band is indicated at the left of each panel. Antibody against PDX1 eliminated the complex previously termed βTF-1 in RIN38 cells (32) and L in 266-6 cells (78). Antibody against PDX1 also eliminated the slow-mobility complex in 266-6 cells previously termed the C complex (78). The EMSA of 266-6 nuclear extract without added antibody (lane none) is shown as a shorter exposure than for the other lanes in the panel in order to better visualize the three binding activities. GATA4 is present in 266-6 nuclear extracts and bound to the B oligonucleotide in EMSAs but is not responsible for the activity of the B element in these cells (78).
The high AT content of the B element suggested that the active factors in each cell type might be homeodomain proteins. An appropriate candidate was the pancreas- and duodenum-specific homeodomain protein PDX1, which binds to an element in the insulin gene promoter (51, 56) and is critical to pancreatic development (22, 50). Antibody specific for either rat or Xenopus PDX1 specifically eliminated the βTF-1 band from RIN38 nuclear extracts (Fig. 3, left panel, and data not shown). Thus, the βTF-1 binding activity from RIN38 β-cells contains PDX1. In nuclear extracts from 266-6 acinar cells, anti-PDX1 antibody eliminated both the L complex, with mobility similar to that of βTF-1, and the acinar cell-specific C complex (Fig. 3, right panel). Therefore, the 266-6 acinar cell nuclear extract has two B-element binding activities containing PDX1. The slower-migrating form (the C complex) corresponds to the active species in these cells (78).
PDX1 can activate the B element in nonpancreatic cells.
To confirm the ability of PDX1 to activate transcription through the B element, we tested the effect of PDX1 on the expression of the B-element repeat reporter gene in HeLa cells (Table 1). The B repeat does not increase the activity of a minimal promoter in HeLa cells (Fig. 1), consistent with the absence of the pancreatic transcription factor PDX1. When exogenous PDX1 was added by cotransfection, the activity directed by the B repeat increased 100-fold (Table 1), whereas the activity of the minimal promoter without the repeat was not affected (data not shown). In contrast, increasing the level of PDX1 in 266-6 cells did not activate the B-repeat reporter gene, consistent with the inability of the acinar form of PDX1 (the C complex) to activate the B element in the absence of the A and C elements of the ELA1 enhancer.
TABLE 1.
Activation of B element activity by PDX1
| PDX1 | Activitya of the B element in transfected:
|
||
|---|---|---|---|
| HeLa cells | 266-6 cells
|
||
| B | B-P2b | ||
| Absent | 0.12 | 0.04 | 20 |
| Presentc | 12 | 0.02 | 180 |
| Fold increase | 100 | 0.5 | 9 |
Amount of expression (nanograms of hGH) of the B-element-driven hGH reporter gene (5B.EIp.hGH).
This B-element mutant has two base pair changes in the PBX binding motif that abolish PBX binding (Fig. 8).
Cotransfection with the PDX1 expression plasmid. Transfections without the PDX1 expression plasmid contained an equal amount of the pBKCMV expression vector without insert.
PDX1 mRNA is present in exocrine as well as endocrine cell lines.
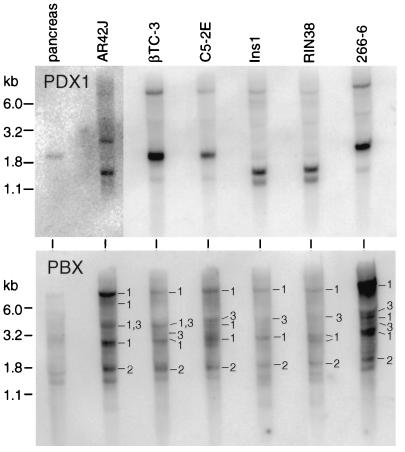
Although it is not strongly expressed throughout the acinar tissue of the adult mouse pancreas, PDX1 is detectable in amylase-positive exocrine cells in the developing mouse pancreas at least as late as 16.5 days postcoitum (19, 50) and in the acinar cells of the frog throughout life (84). Moreover, lacZ reporter genes under the direction of the PDX1 transcriptional control region express β-galactosidase in the acinar cells of the adult mouse, although at a substantially lower level than in the islets (85). Therefore, the presence of PDX1 in the acinar cell line 266-6 is not surprising. Indeed, PDX1 mRNA was present in all pancreatic cell lines tested, including three insulinoma lines (RIN38, βTC-3, and Ins1) and three acinar cell lines (266-6, AR42J, and C5-2E) of widely divergent origins, as well as mouse pancreas (Fig. 4, top panel) and rat pancreas (data not shown). The levels of PDX1 mRNA in the cell lines are similar, except for smaller amounts in the rat AR42J acinar cell line. PDX1 protein levels, as assayed by Western blot analysis, parallel the mRNA levels in the various pancreatic cell lines (data not shown). PDX1 mRNA was not detected in Northern blots of poly(A)-enriched RNA from HeLa, NIH 3T3, or Rat2 fibroblast cells (data not shown). Therefore, the presence of PDX1 is a general and selective property of pancreatic acinar cell as well as β-cell lines.
FIG. 4.
PDX1 mRNA and multiple species of PBX mRNA are present in both acinar and endocrine cell lines. Each lane contained 5 μg of poly(A) RNA from mouse pancreas or the cell line indicated at the top. (Top panel) Hybridization with a PDX1 probe revealed PDX1 message in both exocrine and endocrine cell lines. As previously observed (41, 51), a 2.3-kb mouse PDX1 mRNA was detected in the mouse cells, whereas shorter mRNAs of 1.5 and 1.2 kb were found in rat cells. (Bottom panel) The same Northern blot membrane as in the top panel was hybridized to a full-length PBX1 cDNA probe, which hybridizes with all three known PBX family members. The panel was then hybridized with short probes specific for either PBX1, PBX2, or PBX3. Bands specific for PBX1, PBX2, and PBX3 are indicated by 1, 2, and 3, respectively. The three major PBX1 RNAs from the murine cell lines were 7.7, 3.8, and 2.8 kb, whereas human transcripts of 7.6 and 2.2 kb have been reported (42). The major PBX2 transcript in these cell lines was 1.7 kb, smaller than the 3.2-kb PBX2 RNA of human cells (42). The PBX3 RNA present in each cell line is approximately 4 kb.
PDX1 forms a complex with PBX in acinar cell but not β-cell lines.
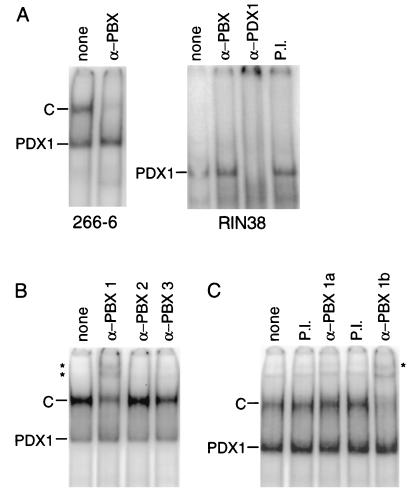
PDX1 is one of a group of homeodomain proteins that interacts with members of the PBC family of homeodomain proteins (13, 47, 57, 60). In humans, this family includes PBX1a and PBX1b (splice variants differing at the carboxyl terminus), PBX2, and PBX3a and PBX3b (also carboxyl-terminal splice variants) (24, 42, 49). In Drosophila the PBX homolog EXD is an essential partner to many of the HOX proteins controlling cell specification (38, 82). A complex containing PDX1 and a PBX can bind to the TseII element of the rat somatostatin gene (57). To determine whether a PBX protein is part of the C complex, a polyclonal antibody that recognizes all members of the PBX family was added to 266-6 nuclear extract. This addition eliminated the C complex (Fig. 5A). Therefore, the complex responsible for activity of the B element in 266-6 acinar cells contains both PDX1 and PBX. Anti-PBX antibodies had no effect on the B-element binding activities in RIN38 nuclear extract (Fig. 5A). Thus, we infer that PDX1 is bound to the B element without a PBX partner in RIN38 β-cells and in this uncomplexed form can activate a linked minimal promoter. In contrast, a complex containing both PDX1 and PBX binds the B element in 266-6 cells, but this complex is not sufficient to activate transcription, requiring instead the cooperation of the factors bound to the other enhancer elements.
FIG. 5.
The active complex in 266-6 cells contains PBX1b as well as PDX1. (A) An antibody specific for the PBX family (α-PBX) eliminated the C complex from 266-6 nuclear extract (left panel); neither an antibody specific for the PBX family nor preimmune serum (P.I.) eliminated the PDX1 complex in RIN38 nuclear extract, although this complex was completely eliminated by anti-PDX1 antibody (α-PDX1) (right panel). (B) An antibody specific for PBX1 (α-PBX1), but not an antibody specific for PBX2 (α-PBX2) or PBX3 (α-PBX3), supershifted the PDX1-PBX-containing C complex. Asterisks mark the positions of the supershifted complexes. (C) An antibody specific for PBX1b (α-PBX1b), but neither an antibody specific for PBX1a (α-PBX1a) nor preimmune serum, also supershifts the C complex. An asterisk marks the position of the supershifted complex. To simplify the binding pattern by eliminating the binding of GATA4, the EMSA mixtures include a competitor oligonucleotide bearing a GATA binding site (78) (see Materials and Methods). The B-element oligonucleotide (see Materials and Methods) was the 32P-labeled probe throughout. Each panel shows lanes from a single gel.
mRNAs for PBX1, PBX2, and PBX3 are in acinar and endocrine cell lines.
To determine whether the presence of PBX in a PDX1-PBX complex in pancreatic cells was correlated simply with the presence of a Pbx mRNA, we examined the distribution of the mRNAs for PBX1, PBX2, and PBX3 in the pancreatic acinar cell and β-cell lines (Fig. 4, bottom panel). Multiple species of Pbx1 RNA are present in each cell line, regardless of their endocrine or exocrine origins. Transcripts of Pbx2 and Pbx3 are also present in each cell line. The presence of Pbx mRNAs in both cell types cannot explain the selective formation of the PDX1-PBX complex in acinar cells but is consistent with previous reports that mRNAs for all three Pbx genes are widespread (42, 65). The acinar cell specificity of the complex must therefore be controlled after transcription of the Pbx genes.
To further examine the PBX mRNAs present in the 266-6 acinar cell line, we screened a 266-6 cDNA library and identified cDNAs encoding PBX1a and -1b, PBX2, and PBX3a and -3b (Table 2). The clones for the mouse PBX1 splice variants, PBX1a and PBX1b, encode amino acid sequences identical to those of the human proteins. The mouse forms of PBX2 and the PBX3 splice variants, PBX3a and PBX3b, are extremely similar to the human proteins, with only nine amino acid residues different for PBX2, one for PBX3a, and two for PBX3b. The presence of cDNA clones representing all three classes of PBX is consistent with the detection of mRNAs for all three classes by Northern hybridization.
TABLE 2.
Homology between human and mouse PBX proteinsa
| PBX protein | No. of identical amino acids/total | % Amino acid identity |
|---|---|---|
| 1a | 430/430 | 100 |
| 1b | 347/347 | 100 |
| 2 | 421/430 | 97.9 |
| 3a | 433/434 | 99.8 |
| 3b | 349/351 | 99.4 |
PBX1b is a component of the active PDX1-PBX complex of 266-6 acinar cells.
To identify the PBX protein(s) present in the C complex from 266-6 nuclear extracts, we examined the effect of adding antibodies specific for PBX1, -2, or -3 on the gel mobility complexes formed on the B element with nuclear extract from 266-6 acinar cells. The C complex was diminished by the antibody specific for PBX1 but was unaffected by antibodies specific for PBX2 or PBX3 (Fig. 5B). Furthermore, an antibody specific for PBX1b eliminated the C complex, whereas complex formation was unaffected by an antibody specific for PBX1a (Fig. 5C). Therefore, although mRNAs for all five PBXs are present in 266-6 cells, only a single PBX protein species, PBX1b, is present in the C complex with PDX1.
Of the five PBX isoforms assayed by Western analysis, only PBX1b and PBX2 were detected in nuclear extracts of 266-6 cells (Fig. 6A). Therefore, the selective presence of the 1b isoform of PBX in the C complex is explained in part by the absence of all other PBX proteins except PBX2, and PBX2 must be excluded from the complex by some other process.
PBX1b was present in nuclear extracts from all three exocrine pancreatic cell lines examined but was absent from the nuclear extracts of all three β-cell lines (Fig. 6B). Therefore, the presence of nuclear PBX1b may be a distinguishing trait of exocrine pancreatic cell lines. Because PBX1 transcripts were detectable in the β-cell lines (Fig. 4B) and the activity of the Drosophila PBX homolog EXD is regulated by cytoplasmic-to-nuclear translocation (3, 40), we compared cytoplasmic and nuclear fractions from RIN38 and 266-6 cells (Fig. 6C). Neither cytoplasmic fraction contained either PBX1 isoform, demonstrating that the endocrine cells are devoid of PBX1 protein and not simply blocked in its nuclear import. These results show that the C complex containing PBX1b cannot form in the β-cell lines because PBX1b protein is absent.
It is noteworthy that each PBX isoform has a different pattern of subcellular and cell type distribution (Fig. 6C). PBX1a protein is not detected in either 266-6 or RIN38 cells. PBX1b protein is absent from RIN38 cells and is present in 266-6 cells almost exclusively in the nucleus. PBX2 protein is in both the nuclear and cytoplasmic fractions of both cell lines. Whereas PBX3a is not detected, PBX3b is in the cytoplasmic fractions, but not the nuclear fractions, of both the endocrine and acinar cells.
PBX1, PBX2, and PBX3 can form complexes with PDX1 in vitro.
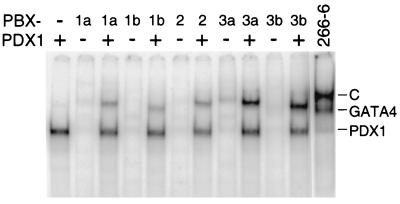
To determine whether each of the PBX isoforms is capable of forming a complex with PDX1 on the B element, in vitro-translated mouse PDX1 and each species of IVT PBX were tested for their abilities to bind separately and as a PDX1-PBX complex. IVT PDX1 bound to the B element had a mobility identical to that of L in the 266-6 nuclear extract (Fig. 7). In contrast, among the IVT PBX species little if any complex formed in the absence of PDX1. The faint but detectable complexes formed by PBX3a are consistent with previous findings that only PBX3 can bind a PBX consensus site in the absence of a homeodomain protein partner (46). All of the IVT PBX species formed complexes with PDX1 on the B element. However, the electrophoretic mobility of each heterodimeric complex, including that of PDX1-PBX1b, was greater than the mobility of the native C complex (Fig. 7). The slower electrophoretic mobility of the native C complex than that of the IVT PDX1-PBX1b suggested the presence of an additional factor(s) in the native complex.
FIG. 7.
IVT PDX1 and PBX can form complexes on the B element. cDNA clones for PDX1 and each of the species of PBX were transcribed and translated in vitro (see Materials and Methods). An equal molar amount of each IVT PBX species was added to an EMSA mixture containing 32P-labeled B oligonucleotide in the presence or absence of the same amount of IVT PDX1. The IVT PDX1 was capable of forming a complex on the B oligonucleotide (first lane), but each PBX isoform was largely unable to bind the oligonucleotide in the absence of PDX1. PDX1 and each of the PBX species were able to form a heteromeric complex on the B element. All of the PDX1-PBX complexes had a greater mobility than the PDX1- and PBX1b-containing C complex from 266-6 nuclear extract (rightmost lane).
The third protein of the complex is MRG1.
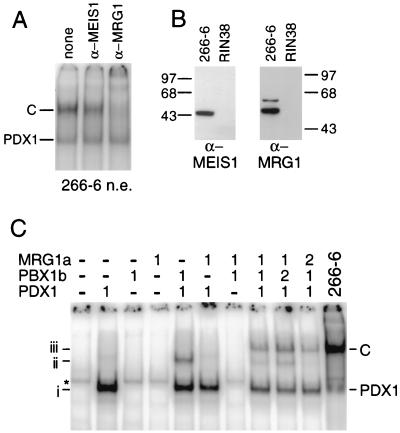
Because the MEIS family of homeodomain cofactors can interact with members of both the PBX and HOX protein families (12, 64, 72), we tested whether a MEIS family protein is present in the C complex, although a trimeric complex containing both MEIS and PBX had not yet been described. Antibodies against MRG1 (MEIS2), decreased the intensity of the C complex by more than half (Fig. 8A). Therefore, the acinar cell-specific C complex contains three different homeodomain proteins: PDX1, PBX1b, and MRG1. The slight decrease in intensity of the C complex when anti-MEIS1 is added may be due to the presence of MEIS1 in a small fraction of the complexes.
FIG. 8.
The C complex contains MRG1 (MEIS2). (A) An antibody specific for MRG1 (α-MRG1) largely eliminates the C complex bound to the B element. The binding of GATA4 present in the 266-6 nuclear extract was eliminated by inclusion of a competitor oligonucleotide bearing a GATA binding site (see Materials and Methods). (B) Western analyses of nuclear extracts (30 μg of total protein/lane) from the 266-6 or RIN38 cell line probed with antibodies specific for MEIS1 or MRG1 revealed that MEIS1 and MRG1 are present only in the acinar cell line. Numbers on the left and right indicate molecular masses in kilodaltons. (C) A complex containing IVT PDX1, IVT PBX1b, and IVT MRG1a has a mobility identical to that of the C complex. Each of the three IVT proteins was incubated alone or in combinations, as indicated at the top, with an oligonucleotide containing a consensus PBX-PDX1 binding site (see Materials and Methods). The addition of equal molar amounts of the IVT proteins or of twofold molar amounts is indicated above the lanes. The rightmost lane shows the PDX1 and C complex formed on this oligonucleotide when incubated with 266-6 extract. The mobilities of IVT PDX1 (i), IVT PDX1-PBX1b (ii), and IVT PDX1-PBX1b-MRG1a (iii) complexes are indicated at the left.
Consistent with the presence of MRG1 in the exocrine specific C complex, MRG1 is found in 266-6, but not RIN38, nuclear extracts (Fig. 8B). MRG1 is also present in the other two exocrine pancreatic lines examined, AR42J and ARIP, and is absent from the two other insulinoma cell lines, Ins1 and βTC-3 (data not shown). Therefore, the presence of both MRG1 and PBX1b is a distinguishing characteristic of exocrine versus endocrine pancreatic cell lines. MEIS1 is also found in 266-6, but not RIN38, nuclear extracts (Fig. 8B).
IVT PDX1, PBX1b, and MRG1a form a complex with a mobility identical to that of the C complex on a consensus PDX1-PBX binding site (Fig. 8C). MRG1a cannot bind alone or in pairwise combination with either PDX1 or PBX1b but requires both of the other factors to be present in order to participate in the complex. Doubling the amount of PBX1b increased the levels of the PDX1-PBX1b and PDX1-PBX1b-MRG1a complexes. Doubling the amount of MRG1a led to a disappearance of the PDX1-PBX1b complex.
PBX binding alters the activity of PDX1 in acinar cells.
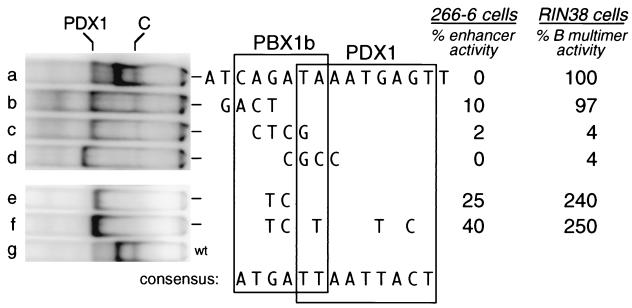
Our results indicate that binding of the C complex (PDX1-PBX1b-MRG1) to the B element is required to complement the activity of the A and C enhancer elements in acinar cells. In contrast, a repeat of the B element is inactive in these cells, although it is active in β-cells in which both PBX1b and MRG1 are absent. Therefore, the presence of PBX1b and/or MRG1 in the acinar binding complex appears to suppress the ability of PDX1 to activate the B-element repeat. The different transcriptional activities of PDX1 and the PDX1-PBX1b-MRG1 complex are summarized in Fig. 9. To test whether eliminating binding of the C complex while retaining binding of PDX1 would create elements that are active as repeats in the acinar 266-6 cell line, we tested several of the 4-bp transversion mutants shown in Fig. 2. Mutant B, which reduces C-complex binding without affecting PDX1 binding, was indeed active as a repeat in 266-6 cells, to a level 10% that of the intact enhancer (Figure 10, row b). Mutant C, which strongly decreases C-complex formation but also decreased PDX1 binding, was less active (Figure 10, row c). Mutant D eliminates both C-complex and PDX1 binding and, as predicted, was inactive (Figure 10, row d). In contrast, in the RIN38 β-cell line, mutant B was as active as the unmodified B element (Figure 10, row b), consistent with the activity of the B repeat requiring only PDX1 binding in these cells. Both mutants C and D, which interfered with PDX1 binding, had greatly reduced activity in the β-cell line (Figure 10, rows c and d).
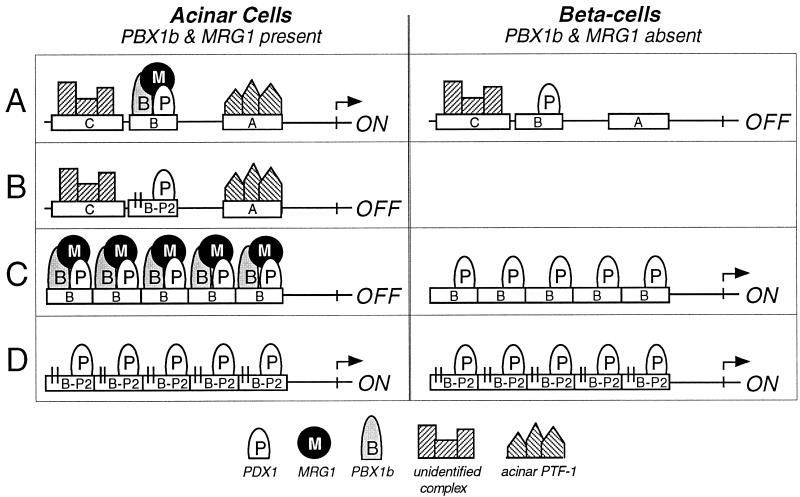
FIG. 9.
Summary of the activities of the complexed and uncomplexed forms of PDX1. (A) The minimal elastase I enhancer comprises three short sequence elements, all of which are required for enhancer activity in acinar cell lines. The pancreatic acinar cell-specific factor PTF1 binds the A element, an unidentified complex binds the C element, and the PDX1-PBX1b-MRG1 complex binds the B element. In β-cell lines, PTF1 is absent and PDX1 with the C-element binding factors is not sufficient for enhancer activity. (B) A 2-bp mutation (indicated by the pair of short vertical lines) in the PBX half site prevents PBX binding and inactivates the enhancer in acinar cell lines. (C) In acinar cells the PDX1-PBX1b-MRG1 complex binds and the repeat is inactive. In β-cells the B element repeat binds PDX1 without PBX1b-MRG1 and is active. (D) Mutation of the PBX half site in the B element (B-P2) has no effect on the activity of the repeat in β-cells but activates the repeat in acinar cells by preventing PBX1b-MRG1 binding.
FIG. 10.
Activity of a mutant B-element repeat in acinar 266-6 cells requires the absence of a PBX binding site and an intact PDX1 binding site, whereas activity in RIN38 cells is largely unaffected by the presence or absence of a PBX binding site. The sequence of the ELA1 B element is shown in row a, and the changes in each modified element tested are shown below the wild-type sequence. Rows: a, wild-type B element; b, mutant B; c, mutant C; d, mutant D; e, mutant B-P2; f, B-P2I+; g, wild type (wt). Mutants B, C, and D correspond to those shown in Fig. 2. Boxes around the sequences indicate the binding sites for PBX and PDX1 based on the effects of mutations on PDX1 and C-complex binding and the model of Knoepfler et al. (26). The panel at the left shows the EMSA binding patterns with oligonucleotides of each element in 266-6 nuclear extract. The positions of PDX1 and the C complex are marked at the top of the panel. The top of the gel is to the right. The GC-rich mutant D created a binding site for an unknown factor that migrated faster than the PDX1 complex and was incapable of activating the repeat. The band between the PDX1 and the C complex is the GATA4 complex. At the right is the activity of each repeat construct in transfections of 266-6 or RIN38 cells. The activities of the mutant B-element repeats in RIN38 cells are expressed as percentages of the activity of the unmodified B repeat. The activities of the mutant B repeats in 266-6 cells are expressed as percentages of the activity of the ELA1 enhancer construct (nucleotides −205 to −93), because the unmodified B repeat is inactive in 266-6 cells.
The PBX binding site of the B element contains a central AGAT, in contrast to the TGAT of the PBX consensus binding site (11, 26, 46). To more clearly establish the dependence of the activity of a homomultimeric element on the absence of PBX1b binding, we created a 2-base mutation of this core PBX binding site (AGAT to ATCT). This mutation completely eliminated C-complex binding retained PDX1 binding (B-P2; Figure 10, row e). As predicted for selectively eliminating PBX binding, this mutant was highly active as a repeat in 266-6 cells as well as RIN38 cells (Fig. 10, row e, and 9D). A further change (Fig. 10, row f) which strengthens PDX1 binding by making the element better match the PDX1 site of the strong PBX-PDX1 site found in the somatostatin gene (57) further increased the activity of the repeat in the acinar cell line. In contrast to the activity of the B-P2 repeat, the same 2-bp mutation in the context of the three-element enhancer reduced the activity of the enhancer 10-fold in 266-6 acinar cells (data not shown), consistent with the acinar cell requirement for PBX binding for enhancer activity (Fig. 9B). These results indicate that the association of PBX1b-MRG1 with PDX1 acts as a switch to change the nature of PDX1 activity.
In 266-6 cells the B-element multimer was not activated by adding exogenous PDX1 (Table 1), presumably because there is sufficient PBX1b and MRG1 to form the C complex with the additional PDX1. Because the B-P2 multimer construct (Fig. 10, row e) (which binds PDX1 but not the C complex) was active in 266-6 cells, we tested whether it would respond to increased levels of PDX1. Increasing PDX1 led to an ninefold increase in the activity of the B-P2 mutation (Table 1). This induction confirms that PDX1 can activate transcription even in acinar cells, provided that binding by the complete C complex is prevented.
DISCUSSION
PDX1 is active alone and as part of a PDX1-PBX1b-MRG1 complex.
PBX and MEIS proteins are generally thought to affect HOX factor activity by providing additional binding site specificity and affinity that are important for HOX function in vivo (39, 72). More recently, the Drosophila MEIS homolog homothorax (HTH) has been implicated in controlling HOX function by regulating the nuclear import of the HOX cofactor and PBX homolog EXD (64). Here we provide evidence that PBX and MEIS in combination can change the qualitative nature of the transcriptional activity of an organ-specific homeodomain factor in a cell-specific manner. Our results show that two different pancreatic transcription factor complexes can bind to and activate transcription through the B element of the ELA1 enhancer and that the nature of the transcriptional activation differs for the two complexes. The pancreatic homeodomain factor PDX1 is common to both complexes. In pancreatic acinar cell lines a complex containing PDX1, PBX1b, and MRG1 (MEIS2) binds the B element. In β-cell lines PDX1 binds alone.
PBX1b is a member of the PBC family of homeodomain proteins (8, 9), which includes EXD of Drosophila, Ceh-20 of Caenorhabditis, and PBX1, PBX2, and PBX3 of vertebrates. In addition to containing a highly conserved homeodomain, the PBC proteins are characterized by an extra three amino acid residues between helices 1 and 2 of the homeodomain, a feature of the TALE superclass of homeodomain proteins (7). The PBC proteins form heterodimers with members of the Antennepedia class of homeodomain proteins, which share similar homeodomain sequences and a conserved pentapeptide motif (consensus, YPWMR) required for the HOX-PBC interaction near and amino terminal to the homeodomain (13, 47, 60). Compared to either HOX or PBC proteins alone, HOX-PBC heterodimers have increased binding affinities, extended DNA recognition sites of 10 to 13 rather than 6 bp (11, 46), and in some instances a recognition sequence different from the simple composite of the HOX and PBC consensus half sites (reviewed in reference 39). Thus, the association of a PBC protein with a HOX protein is thought to confer the affinity and binding site recognition specificity required for the in vivo functions of HOX proteins.
PDX1 is a member of the Antennepedia class of homeodomain proteins (35) and is part of the recently discovered ParaHox cluster expressed in the endoderm (6). PDX1 expression is restricted to the pancreas and rostral duodenum (50). PDX1 binds its recognition site effectively without a PBX partner (reference 57 and this study), although a PBX partner does stabilize binding in vitro (57). PDX1 is inherently capable of interacting with all five PBX family members tested (Fig. 7). Our results show that (depending on the context) PDX1 can be transcriptionally effective without a PBX partner: it is active in β-cell lines, which do not form a PDX1-PBX complex, and its action through the B element (outside the context of the elastase enhancer) does not require an intact PBX half site.
A third homeodomain protein, MRG1 (MEIS2), is also in the acinar PDX1-containing complex. Mrg1 is one of the three known members of the Meis family of homeobox genes (Meis1, Mrg1, and Mrg2) (44, 75). Each member encodes multiple isoforms (43, 55). The prototypic Meis family gene, Meis1, was identified as a common site for retroviral integration in myeloid leukemias of BXH-2 mice (43). The MEIS family homeodomain is closely related to the PBC homeodomain, including the three-amino-acid insertion characteristic of the TALE superclass (7). The relatedness of the MEIS and PBC homeodomains suggested that MEIS proteins may have functions similar to those of the PBC proteins, such as binding cooperatively with other homeodomain proteins (43). Indeed, the frequent coactivation of Meis1 with Hoxa7 or Hoxa9 in myeloid leukemias suggested that MEIS proteins may work in concert with some HOX proteins (45). Shen et al. (72) have shown that MEIS proteins form cooperative DNA binding complexes with the ABD-B-like subset of HOX proteins (HOX paralog groups 9 through 13). This paralog subgroup complements the subgroup (HOX1 through -10) which form complexes with PBX (71). MEIS1 can also bind cooperatively with PBX1b to a combined PBX-MEIS consensus site (TGAT/TGACAG) (12). Aside from contributing binding stability, a transcriptional role for MEIS in these complexes has not been shown. However, a natural cyclic AMP-responsive element in the bovine CYP17 gene has been shown to bind a PBX1-MEIS complex (5). Our results add to the understanding of the role of MEIS proteins by demonstrating that MRG1 forms a multimeric complex containing both a PBX and an organ-specific homeodomain protein.
The presence of PDX1, PBX1b, and MRG1 in a single mobility shift complex indicates that the active form of PDX1 in acinar cells is part of a novel trimeric complex of an organ-specific homeodomain protein, a PBC family member, and a MEIS protein. As the MEIS1 and HOX interaction domains map to different regions of the PBX1 protein (25), it is likely that the complex forms through the simultaneous interaction of MRG1 and PDX1 with PBX1b. The short, 10- to 12-bp binding site for the C complex on the B element is consistent with DNA contacts by both components of a PDX1-PBX1 dimer but is inconsistent with contacts by all three members of the PDX1-PBX1b-MRG1 trimer. The ability of MEIS to interact with both PBX and a subset of HOX proteins in the absence of DNA (12, 72), coupled with the ability of the B element to bind IVT PDX1-PBX, but not IVT PBX1b-MRG1a or IVT MRG1a-PDX1, implies that the MRG1 interaction in the trimeric complex does not require a specific DNA recognition site. Berthelsen et al. (4) have recently described a trimeric complex of HOXB1, PBX1, and PREP1 (pKNOX1) that binds a HOXB1-PBX site. PREP1 (pKNOX1) is a member of the TALE homeodomain family with a homeodomain most closely related to the MEIS subfamily. In the trimeric complex, the PREP1 homeodomain is not required for DNA binding of the complex, similar to the model we propose for PDX1, PBX1b, and MRG1.
PDX1 and the PDX1-PBX1b-MRG1 complex have different transcriptional activities.
The activity of the ELA1 transcriptional enhancer in the acinar 266-6 cell line requires the binding of the PDX1-PBX1b-MRG1 trimer to the B element, the acinar cell-specific factor PTF1 to the A element (66, 67), and an as-yet-unidentified factor to the C element. The PDX1-PBX1b-MRG1 complex apparently acts in concert with the other enhancer-bound transcription factors, and in this context, PDX1 alone bound to the B element is not sufficient. Therefore, mutations in the PBX binding site within the B element that eliminate binding of the PDX1-PBX1b-MRG1 trimer, while still permitting PDX1 binding, inactivate the enhancer in the acinar cell line.
Although the PDX1-PBX1b-MRG1 complex is required for the activity of the three-element elastase enhancer, PDX1 is sufficient for activation of an artificial enhancer constructed by multimerizing the B element. Moreover, formation of the PDX1-PBX1b-MRG1 complex prevents transcriptional activation from the binding site repeat. Therefore, in this context the association with PBX1b-MRG1 represses transcriptional activation by PDX1, whereas in the context of the enhancer the trimeric complex is required and PDX1 alone is insufficient.
In the 266-6 acinar cell line, all three proteins of the PDX1-PBX1b-MRG1 complex are present, and the different activities of the complex and of PDX1 can be elicited under appropriate conditions. Thus, B elements with PBX half-site mutations that eliminate trimer binding are inactive in the enhancer. However, the same mutation activates a repeat construct in these acinar cells, because it allows PDX1 binding without associated PBX1b and MRG1.
RIN38 β-cells cannot form a PDX1-PBX1b-MRG1 complex because they contain neither PBX1b nor MRG1. Therefore, the activity of a B-element repeat is dependent only on the presence of a PDX1 binding site, and the presence or absence of a PBX half-site has little, if any, effect. Although nuclei of the β-cell line contain PBX2, this factor does not form a complex with PDX1 on the B element in nuclear extracts, and the lack of importance of the PBX binding site within the B element in these cells implies that it also does not form a complex with PDX1 on the B element in living cells.
It is not clear whether PBX1b or MRG1 (or both) alters the transcriptional activity of PDX1. The interaction with PBX1b may confer this change; in this instance MRG1 may contribute properties not detected in our analyses. PBX1a and -1b proteins, for example, have a curious repressing activity associated with a region N terminal to the homeodomain (36). This activity is detected in transfected cells, is not dependent on DNA binding, and can repress the effect of some but not all transcription factors (e.g., Sp1 but not p53 or VP16). However, inhibitory effects on homeodomain partners have not been reported. Alternatively, because the PDX1-PBX1b-MRG1 complex does not form in the absence of the PBX half site, PBX1b may serve to recruit MRG1, which then would provide the functions required within the context of the elastase enhancer and coincidentally interfere with the PDX1 activation of the B-element repeat.
The disparate regulation of Pbx family members implies the existence of distinct functions for each isoform.
We have shown that Pbx family members are regulated at multiple posttranscriptional points in both acinar cell and β-cell lines, including control of protein synthesis or stability, nuclear localization, and formation of multimeric complexes with other homeodomain proteins. For example, although mRNAs for PBX1a and -1b, PBX2, and PBX3a and -3b are all present in the acinar 266-6 cell line (Table 2), PBX1a and PBX3a are not found in these cells, indicating regulation of either the synthesis or stability of these PBX species. Regulation in RIN38 cells is similarly complex: PBX1b as well as PBX1a and PBX3a are absent from these cells, although transcripts for PBX1, as well as PBX2 and PBX3, are present (Fig. 4).
The nuclear import of the PBX proteins is also differentially controlled. PBX1b is detectable only in the nuclei of the 266-6 cells. In both the acinar cell and β-cell lines, PBX3b is detectable only in the cytoplasm, whereas PBX2 is distributed between the two compartments.
A final level of control of nuclear PBX species must occur to regulate the formation of the complex. PDX1 in acinar cell nuclear extracts is complexed only with PBX1b and MRG1, even though other PBX and MEIS isoforms are present. Thus, although PBX2 is present in both RIN38 and 266-6 nuclei and IVT PBX2 and PDX1 can form a complex on the B element, no PDX1-PBX2-containing complex can be detected in nuclear extracts of either cell type.
The result of this complex regulatory scheme is the presence of only the PBX1b isoform in the C complex of acinar cells, despite the presence in these cells of the mRNAs for all five isoforms and the proteins of three isoforms. This implies that the other PBX isoforms might have different activities that would be inappropriate for the C complex.
MEIS and the nuclear localization of PBX isoforms.
The translocation of EXD from cytoplasm to nucleus is dependent on the presence of HTH, the Drosophila MEIS homolog (64). Because MEIS1 can substitute for HTH to induce this translocation in Drosophila S2 cells, the nuclear localization of mammalian PBX species is also likely to depend on the presence of an MEIS isoform. We have shown, however, that MRG1 and MEIS1 (which are present in 266-6 cells but absent in RIN38 cells) are not sufficient for the nuclear localization of all PBX isoforms, as PBX3b is found only in the cytoplasm of both cell types. Similarly, PBX2, which is partially nuclear in both cell lines, must require the presence of an unidentified MEIS species (e.g., MRG2) in both cell lines or not require an MEIS partner to be imported into the nucleus. It remains to be determined whether MEIS1 or MRG1 mediates the nuclear import of PBX1.
Recently, Casares and Mann (10) have shown that HTH is required for proper development of the Drosophila head. Because nuclear EXD is not sufficient for head development, HTH appears to have a required function in addition to inducing the nuclear translocation of EXD. Its participation in multisubunit homeoprotein complexes of the sort described in this report may be one such function.
The existence of multiple MEIS genes (44, 75), as well as multiple splice variants of MEIS transcripts (43, 55), potentially adds complexity both to the regulation of PBX localization and to the variety of dimer and trimer complexes of PBX, MEIS, and other homeodomain proteins which may control developmental pathways. Also yet to be determined is whether posttranscriptional regulation of MEIS expression occurs in various cells and tissues. Labial, a HOX partner of EXD (63), is also regulated during Drosophila development by translocation from the cytoplasm to the nucleus (20), raising the possibility that control of nuclear import may be a widespread mode of regulating homeodomain protein activity.
The endocrine-exocrine dichotomy.
The presence of both PBX1b and MRG1 in all three exocrine cell lines and their absence in all three β-cell lines tested suggest that the PDX1-PBX1b-MRG1 complex and the uncomplexed PDX1 perform important cell-type-specific functions in the exocrine and endocrine tissue compartments, respectively. The insulin gene promoter is the best studied of the regulatory regions of β-cell genes known to be regulated by PDX1. The rat insulin gene promoter elements A1 and A3/A4 (17) are activated by PDX1 (51), do not bind PDX1-PBX complexes (57), and do not contain PBX or MEIS binding sites. The activity of PDX1 on the insulin promoter without participation of a PBX partner is as would be expected if the absence of PBX1b and MRG1 in β-cell lines extends to the β-cells of islets. The genes for glucokinase and GLUT2, which also are expressed selectively in β-cells, have been shown to have functional PDX1 binding sites in their promoters (79, 80). We predict that these sites as well will be found not to depend on PBX or MEIS binding. If a PDX1-PBX1b-MRG1 complex is the active form of PDX1 in the mature acinar cells of animals, whereas PDX1 unfettered by other homeodomain partners is active in β-cells, a switch in PDX1 activity may be an important control point in distinguishing the acinar cell and β-cell lineages during pancreogenesis. It will be important to discover how and when PBX and MEIS appear during pancreogenesis and how this is related to the adoption of acinar cell and β-cell fates.
ACKNOWLEDGMENTS
We thank Fred Kruse for early contributions to this work, Jeana Buxton for excellent technical assistance, Mike Waterman for helpful discussions and critique of the manuscript, Marc Montminy for providing anti-STF1 antiserum, and Ira O. Daar for providing MEIS1 and MRG1 antisera.
This work was supported by grant DK-27430 from the National Institutes of Health (NIH) and a grant from the Juvenile Diabetes Foundation International to R.J.M. and by NIH grants CA-21124 to A.M.B., DK-42502 to C.V.E.W., and DK-28350 to M. R. Waterman.
Galvin H. Swift and Ying Liu contributed equally to this work.
REFERENCES
- 1.Ahlgren U, Pfaff S L, Jessell T M, Edlund T, Edlund H. Independent requirement for ISL1 in formation of pancreatic mesenchyme and islet cells. Nature. 1997;385:257–260. doi: 10.1038/385257a0. [DOI] [PubMed] [Google Scholar]
- 2.Asfari M, Janjic D, Meda P, Li G, Halban P A, Wollheim C B. Establishment of 2-mercaptoethanol-dependent differentiated insulin-secreting cell lines. Endocrinology. 1992;130:167–178. doi: 10.1210/endo.130.1.1370150. [DOI] [PubMed] [Google Scholar]
- 3.Aspland S E, White R A. Nucleocytoplasmic localisation of extradenticle protein is spatially regulated throughout development in Drosophila. Development. 1997;124:741–747. doi: 10.1242/dev.124.3.741. [DOI] [PubMed] [Google Scholar]
- 4.Berthelsen J, Zappavigna V, Ferretti E, Mavilio F, Blasi F. The novel homeoprotein Prep1 modulates Pbx-Hox protein cooperativity. EMBO J. 1998;8:1434–1445. doi: 10.1093/emboj/17.5.1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bischof L J, Kagawa N, Moskow J J, Takahashi Y, Iwamatsu A, Buchberg A M, Waterman M R. Members of the Meis1 and Pbx homeodomain protein families cooperatively bind a cAMP-responsive sequence (CRS1) from CYP17. J Biol Chem. 1998;273:7941–7948. doi: 10.1074/jbc.273.14.7941. [DOI] [PubMed] [Google Scholar]
- 6.Brooke N M, Garcia-Fernandez J, Holland P W H. The ParaHox cluster is an evolutionary sister of the Hox gene cluster. Nature. 1998;392:920–923. doi: 10.1038/31933. [DOI] [PubMed] [Google Scholar]
- 7.Burglin T R. Analysis of TALE superclass homeobox genes (MEIS, PBC, KNOX, Iroquois, TGIF) reveals a novel domain conserved between plants and animals. Nucleic Acids Res. 1997;25:4173–4180. doi: 10.1093/nar/25.21.4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burglin T R. A comprehensive classification of homeobox genes. In: Duboule D, editor. Guidebook to the homeobox genes. Oxford, United Kingdom: Oxford University Press; 1994. pp. 27–71. [Google Scholar]
- 9.Burglin T R, Ruvkun G. New motif in PBX genes. Nat Genet. 1992;1:319–320. doi: 10.1038/ng0892-319. [DOI] [PubMed] [Google Scholar]
- 10.Casares F, Mann R S. Control of antennal versus leg development in Drosophila. Nature. 1998;392:723–726. doi: 10.1038/33706. [DOI] [PubMed] [Google Scholar]
- 11.Chang C-P, Brocchieri L, Shen W-F, Largman C, Cleary M L. Pbx modulation of Hox homeodomain amino-terminal arms establishes different DNA-binding specificities across the Hox locus. Mol Cell Biol. 1996;16:1734–1745. doi: 10.1128/mcb.16.4.1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang C-P, Jacobs Y, Nakamura T, Jenkins N A, Copeland N G, Cleary M L. Meis proteins are major in vivo DNA binding partners for wild-type but not chimeric Pbx proteins. Mol Cell Biol. 1997;17:5679–5687. doi: 10.1128/mcb.17.10.5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang C-P, Shen W-F, Rozenfeld S, Lawrence H J, Largman C, Cleary M L. Pbx proteins display hexapeptide-dependent cooperative DNA binding with a subset of Hox proteins. Genes Dev. 1995;9:663–674. doi: 10.1101/gad.9.6.663. [DOI] [PubMed] [Google Scholar]
- 14.Cockell M, Stevenson B J, Strubin M, Hagenbuchle O, Wellauer P K. Identification of a cell-specific DNA-binding activity that interacts with a transcriptional activator of genes expressed in the acinar pancreas. Mol Cell Biol. 1989;9:2464–2476. doi: 10.1128/mcb.9.6.2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dignam J D, Lebovitz R M, Roeder R G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Efrat S, Linde S, Kofod H, Spector D, Delannoy M, Grant S, Hanahan D, Baekkeskov S. Beta-cell lines derived from transgenic mice expressing a hybrid insulin gene-oncogene. Proc Natl Acad Sci USA. 1988;85:9037–9041. doi: 10.1073/pnas.85.23.9037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.German M, Ashcroft S, Docherty K, Edlund H, Edlund T, Goodison S, Imura H, Kennedy G, Madsen O, Melloul D, Moss L, Olson K, Permutt M A, Philippe J, Robertson R P, Rutter W J, Serup P, Stein R, Steiner D, Tsai M-J, Walker M D. The insulin gene promoter: a simplified nomenclature. Diabetes. 1995;44:1002–1004. doi: 10.2337/diab.44.8.1002. [DOI] [PubMed] [Google Scholar]
- 18.Graham F L, van der Eb A J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973;52:456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- 19.Guz Y, Montminy M R, Stein R, Leonard J, Gamer L W, Wright C V E, Teitelman G. Expression of murine STF-1, a putative insulin gene transcription factor, in β cells of pancreas, duodenal epithelium and pancreatic exocrine and endocrine progenitors during ontogeny. Development. 1995;121:11–18. doi: 10.1242/dev.121.1.11. [DOI] [PubMed] [Google Scholar]
- 20.Immergluck K, Lawrence M, Bienz M. Induction across germ layers in Drosophila mediated by a genetic cascade. Cell. 1990;62:261–268. doi: 10.1016/0092-8674(90)90364-k. [DOI] [PubMed] [Google Scholar]
- 21.Jessop N W, Hay R J. Characteristics of two rat pancreatic exocrine cell lines derived from transplantable tumors. In Vitro. 1980;16:212. [Google Scholar]
- 22.Jonsson J, Carlsson L, Edlund T, Edlund H. Insulin-promoter-factor 1 is required for pancreas development in mice. Nature. 1994;371:606–609. doi: 10.1038/371606a0. [DOI] [PubMed] [Google Scholar]
- 23.Kagawa N, Ogo A, Takahashi Y, Iwamatsu A, Waterman M R. A cAMP-regulatory sequence of CYP17 is a cellular target for the homeodomain protein PBX1. J Biol Chem. 1994;269:18716–18719. [PubMed] [Google Scholar]
- 24.Kamps M P, Look A T, Baltimore D. A new homeobox gene contributes the DNA binding domain of the t(1:19) translocation in pre-B ALL. Cell. 1991;60:547–555. doi: 10.1016/0092-8674(90)90658-2. [DOI] [PubMed] [Google Scholar]
- 25.Knoepfler P S, Calvo K R, Chen H, Antonarakis S E, Kamps M P. Meis and pKnox1 bind DNA cooperatively with Pbx1 utilizing an interaction surface disrupted in oncoprotein E2a-Pbx1. Proc Natl Acad Sci USA. 1997;94:14553–14558. doi: 10.1073/pnas.94.26.14553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knoepfler P S, Lu Q, Kamps M P. Pbx1-Hox heterodimers bind DNA on inseparable half-sites that permit intrinsic DNA binding specificity of the Hox partner at nucleotides 3′ to a TAAT motif. Nucleic Acids Res. 1996;24:2288–2294. doi: 10.1093/nar/24.12.2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krapp A, Knofler M, Frutiger F, Hughes G J, Hagenbuchle O, Wellauer P K. The p48 DNA-binding subunit of transcription factor PTF1 is a new exocrine pancreas-specific basic helix-loop-helix protein. EMBO J. 1996;15:4317–4329. [PMC free article] [PubMed] [Google Scholar]
- 28.Kreig P A, Melton D A. Functional messenger RNAs are produced by SP6 in vitro transcription of cloned cDNAs. Nucleic Acids Res. 1984;12:7057–7070. doi: 10.1093/nar/12.18.7057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krumlauf R. Hox genes in vertebrate development. Cell. 1994;78:191–201. doi: 10.1016/0092-8674(94)90290-9. [DOI] [PubMed] [Google Scholar]
- 30.Kruse F, Komro C T, Michnoff C H, MacDonald R J. The cell-specific elastase I enhancer comprises two domains. Mol Cell Biol. 1988;8:893–902. doi: 10.1128/mcb.8.2.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kruse F, Rose S D, Swift G H, Hammer R E, MacDonald R J. Cooperation between elements of an organ-specific transcriptional enhancer in animals. Mol Cell Biol. 1995;15:4385–4394. doi: 10.1128/mcb.15.8.4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kruse F, Rose S D, Swift G H, Hammer R E, MacDonald R J. An endocrine-specific element is an integral component of an exocrine-specific pancreatic enhancer. Genes Dev. 1993;7:774–786. doi: 10.1101/gad.7.5.774. [DOI] [PubMed] [Google Scholar]
- 32a.Kruse, F., and R. J. MacDonald. Unpublished data.
- 32b.Lalgudi, R., S. D. Rose, and R. J. MacDonald. Unpublished data.
- 33.Lawrence P A, Morata G. Homeobox genes: their function in Drosophila segmentation and pattern formation. Cell. 1994;78:181–189. doi: 10.1016/0092-8674(94)90289-5. [DOI] [PubMed] [Google Scholar]
- 34.Le Douarin N M. On the origin of pancreatic endocrine cells. Cell. 1988;53:169–171. doi: 10.1016/0092-8674(88)90375-3. [DOI] [PubMed] [Google Scholar]
- 35.Leonard J, Peers B, Johnson T, Ferreri K, Lee S, Montminy M R. Characterization of somatostatin transactivating factor-1, a novel homeobox factor that stimulates somatostatin expression in pancreatic islet cells. Mol Endocrinol. 1993;7:1275–1283. doi: 10.1210/mend.7.10.7505393. [DOI] [PubMed] [Google Scholar]
- 36.Lu Q, Kamps M P. Selective repression by Pbx1 does not require the homeodomain. Proc Natl Acad Sci USA. 1996;93:470–474. doi: 10.1073/pnas.93.1.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.MacDonald R J, Swift G H, Przybyla A E, Chirgwin J M. Isolation of RNA using guanidinium salts. Methods Enzymol. 1987;152:219–227. doi: 10.1016/0076-6879(87)52023-7. [DOI] [PubMed] [Google Scholar]
- 38.Mann R S. The specificity of homeotic gene function. Bioessays. 1995;17:855–863. doi: 10.1002/bies.950171007. [DOI] [PubMed] [Google Scholar]
- 39.Mann R S, Chan S-K. Extra specificity from extradenticle: the partnership between HOX and PBX/EXD homeodomain proteins. Trends Genet. 1996;12:258–262. doi: 10.1016/0168-9525(96)10026-3. [DOI] [PubMed] [Google Scholar]
- 40.Mann R S, Shaar A-S. Nuclear import of the homeodomain protein extradenticle in response to Wg and Dpp signalling. Nature. 1996;383:630–633. doi: 10.1038/383630a0. [DOI] [PubMed] [Google Scholar]
- 41.Miller C P, McGehee R E, Habener J F. IDX-1: a new homeodomain transcription factor expressed in rat pancreatic islets and duodenum that transactivates the somatostatin gene. EMBO J. 1994;13:1145–1156. doi: 10.1002/j.1460-2075.1994.tb06363.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Monica K, Galili N, Nourse J, Saltman D, Cleary M L. PBX2 and PBX3, new homeobox genes with extensive homology to the human proto-oncogene PBX1. Mol Cell Biol. 1991;11:6149–6157. doi: 10.1128/mcb.11.12.6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moskow J J, Bullrich F, Huebner K, Daar I O, Buchberg A M. Meis1, a PBX1-related homeobox gene involved in myeloid leukemia in BXH-2 mice. Mol Cell Biol. 1995;15:5434–5443. doi: 10.1128/mcb.15.10.5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nakamura T, Jenkins N A, Copeland N G. Identification of a new family of Pbx-related homeobox genes. Oncogene. 1996;13:2235–2242. [PubMed] [Google Scholar]
- 45.Nakamura T, Largaespada D A, Shaughnessy J D J, Jenkins N A, Copeland N G. Cooperative activation of Hoxa and Pbx-1-related genes in murine myeloid leukemias. Nat Genet. 1996;12:149–153. doi: 10.1038/ng0296-149. [DOI] [PubMed] [Google Scholar]
- 46.Neuteboom S T C, Murre C. Pbx raises the DNA binding specificity but not the selectivity of Antennapedia Hox proteins. Mol Cell Biol. 1997;17:4696–4706. doi: 10.1128/mcb.17.8.4696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Neuteboom S T C, Peltenburg L T C, van Dijk M A, Murre C. The hexapeptide LFPWMR in HoxB-8 is required for cooperative DNA binding with PBX1 and PBX2 proteins. Proc Natl Acad Sci USA. 1995;92:9166–9170. doi: 10.1073/pnas.92.20.9166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nielsen D A, Chang T C, Shapiro D J. A highly sensitive, mixed-phase assay for chloramphenicol acetyltransferase activity in transfected cells. Anal Biochem. 1989;179:19–23. doi: 10.1016/0003-2697(89)90193-0. [DOI] [PubMed] [Google Scholar]
- 49.Nourse J, Melletin J D, Galili N, Walkinson J, Stanbridge E, Smith S D, Cleary M L. Chromosomal translocation t(1;19) results in the synthesis of a homeobox fusion mRNA that codes for a potential chimeric transcription factor. Cell. 1991;60:535–545. doi: 10.1016/0092-8674(90)90657-z. [DOI] [PubMed] [Google Scholar]
- 50.Offield M F, Jetton J L, Labosky P A, Ray M, Stein R W, Magnuson M A, Hogan B L M, Wright C V E. PDX-1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development. 1996;122:983–995. doi: 10.1242/dev.122.3.983. [DOI] [PubMed] [Google Scholar]
- 51.Ohlsson H, Karlsson K, Edlund T. IPF1, a homeodomain-containing transactivator of the insulin gene. EMBO J. 1993;12:4251–4259. doi: 10.1002/j.1460-2075.1993.tb06109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ornitz D M, Hammer R E, Davison B L, Brinster R L, Palmiter R D. Promoter and enhancer elements from the rat elastase I gene function independently of each other and of heterologous enhancers. Mol Cell Biol. 1987;7:3466–3472. doi: 10.1128/mcb.7.10.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ornitz D M, Hammer R E, Messing A, Palmiter R D, Brinster R L. Pancreatic neoplasia induced by SV40 T-antigen expression in acinar cells of transgenic mice. Science. 1987;238:188–193. doi: 10.1126/science.2821617. [DOI] [PubMed] [Google Scholar]
- 54.Ornitz D M, Palmiter R D, Hammer R E, Brinster R L, Swift G H, MacDonald R J. Specific expression of an elastase-human growth hormone fusion gene in pancreatic acinar cells of transgenic mice. Nature. 1985;313:600–603. doi: 10.1038/313600a0. [DOI] [PubMed] [Google Scholar]
- 55.Oulad-Abdelghani M, Chazaud C, Bouillet P, Sapin V, Chambon P, Dolle P. Meis2, a novel mouse Pbx-related homeobox gene induced by retinoic acid during differentiation of P19 embryonal carcinoma cells. Dev Dyn. 1997;210:173–183. doi: 10.1002/(SICI)1097-0177(199710)210:2<173::AID-AJA9>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 56.Peers B, Leonard J, Sharma S, Teitelman G, Montminy M R. Insulin expression in pancreatic islet cells relies on cooperative interactions between the helix loop helix factor E47 and the homeobox factor STF-1. Mol Endocrinol. 1994;8:1798–1806. doi: 10.1210/mend.8.12.7708065. [DOI] [PubMed] [Google Scholar]
- 57.Peers B, Sharma S, Johnson T, Kamps M, Montminy M. The pancreatic islet factor STF-1 binds cooperatively with Pbx to a regulatory element in the somatostatin promoter: importance of the FPWMK motif and of the homeodomain. Mol Cell Biol. 1995;15:7091–7097. doi: 10.1128/mcb.15.12.7091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Peltenburg L T C, Murre C. Engrailed and Hox homeodomain proteins contain a related Pbx interaction motif that recognizes a common structure present in Pbx. EMBO J. 1996;15:3385–3393. [PMC free article] [PubMed] [Google Scholar]
- 59.Petrucco S, Wellauer P K, Hagenbüchle O. The DNA-binding activity of transcription factor PTF1 parallels the synthesis of pancreas-specific mRNAs during mouse development. Mol Cell Biol. 1990;10:254–264. doi: 10.1128/mcb.10.1.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Phelan M L, Rambaldi I, Featherstone M S. Cooperative interactions between HOX and PBX proteins mediated by a conserved peptide motif. Mol Cell Biol. 1995;15:3989–3997. doi: 10.1128/mcb.15.8.3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Phillipe J, Chick W L, Habener J F. Multipotential phenotypic expression of genes encoding peptide hormones in rat insulinoma cell lines. J Clin Invest. 1987;79:351–358. doi: 10.1172/JCI112819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pictet R, Rutter W J. Development of the embryonic endocrine pancreas. In: Steiner D F, Freinkel N, editors. Handbook of physiology: the endocrine pancreas. Vol. 1. Baltimore, Md: Williams and Wilkins; 1972. pp. 25–66. [Google Scholar]
- 63.Popperl H, Bienz M, Studer M, Chan S-K, Aparicio S, Brenner S, Mann R S, Krumlauf R. Segmental expression of Hoxb-1 is controlled by a highly conserved autoregulatory loop dependent upon exd/pbx. Cell. 1995;81:1031–1042. doi: 10.1016/s0092-8674(05)80008-x. [DOI] [PubMed] [Google Scholar]
- 64.Rieckhof G E, Casares F, Ryoo H D, Abu-Sharr M, Mann R S. Nuclear translocation of extradenticle requires homothorax, which encodes an extradenticle-related homeodomain protein. Cell. 1997;91:171–183. doi: 10.1016/s0092-8674(00)80400-6. [DOI] [PubMed] [Google Scholar]
- 65.Roberts V J, van Dijk M A, Murre C. Localization of PBX1 transcripts in developing rat embryos. Mech Dev. 1995;51:193–198. doi: 10.1016/0925-4773(95)00364-9. [DOI] [PubMed] [Google Scholar]
- 66.Rose S D, Kruse F, Swift G H, MacDonald R J, Hammer R E. A single element of the elastase I enhancer is sufficient to direct transcription selectively to the pancreas and gut. Mol Cell Biol. 1994;14:2048–2057. doi: 10.1128/mcb.14.3.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rose S D, MacDonald R J. Evolutionary silencing of the human elastase I gene (ELA1) Hum Mol Genet. 1997;6:897–903. doi: 10.1093/hmg/6.6.897. [DOI] [PubMed] [Google Scholar]
- 68.Rosewicz S, Vogt D, Harth N, Grund C, Franke W W, Ruppert S, Schweitzer E, Riecken E-O, Wiedenmann B. An amphicrine pancreatic cell line: AR42J cells combine exocrine and neuroendocrine properties. Eur J Cell Biol. 1992;59:80–91. [PubMed] [Google Scholar]
- 69.Sander M, Neubuser A, Kalamaras J, Ee H C, Martin G R, German M S. Genetic analysis reveals that PAX6 is required for normal transcription of pancreatic hormone genes and islet development. Genes Dev. 1997;11:1662–1673. doi: 10.1101/gad.11.13.1662. [DOI] [PubMed] [Google Scholar]
- 70.Seldon R F, Burke H K, Rowe M E, Goodman H M, Moore D D. Human growth hormone as a reporter gene in regulation studies employing transient gene expression. Mol Cell Biol. 1986;6:3173–3179. doi: 10.1128/mcb.6.9.3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shen W-F, Rozenfeld S, Lawrence H J, Largman C. The Abd-B-like Hox homeodomain proteins can be subdivided by the ability to form complexes with PBX1a on a novel DNA target. J Biol Chem. 1997;272:8198–8206. doi: 10.1074/jbc.272.13.8198. [DOI] [PubMed] [Google Scholar]
- 72.Shen W F, Montgomery J C, Rozenfeld S, Moskow J J, Lawrence H J, Buchberg A M, Largman C. AbdB-like Hox proteins stabilize DNA binding by the Meis1 homeodomain proteins. Mol Cell Biol. 1997;17:6448–6458. doi: 10.1128/mcb.17.11.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Slack J M W. Developmental biology of the pancreas. Development. 1995;121:1569–1580. doi: 10.1242/dev.121.6.1569. [DOI] [PubMed] [Google Scholar]
- 74.Sosa-Pineda B, Chowdhury K, Torres M, Oliver G, Gruss P. The PAX4 gene is essential for differentiation of insulin-producing beta cells in the mammalian pancreas. Nature. 1997;386:399–402. doi: 10.1038/386399a0. [DOI] [PubMed] [Google Scholar]
- 75.Steelman S, Moskow J J, Muzynski K, North C, Druck T, Montgomery J C, Huebner K, Daar I O, Buchberg A M. Identification of a conserved family of Meis1-related homeobox genes. Genome Res. 1997;7:142–156. doi: 10.1101/gr.7.2.142. [DOI] [PubMed] [Google Scholar]
- 76.Stoffers D A, Zinkin N T, Stanojevic V, Clarke W L, Haener J F. Pancreatic agenesis attributable to a single nucleotide deletion in the human IPF1 gene coding sequence. Nat Genet. 1997;15:106–110. doi: 10.1038/ng0197-106. [DOI] [PubMed] [Google Scholar]
- 77.Swift G H, Hammer R E, MacDonald R J, Brinster R L. Tissue-specific expression of the rat pancreatic elastase I gene in transgenic mice. Cell. 1984;38:639–646. doi: 10.1016/0092-8674(84)90258-7. [DOI] [PubMed] [Google Scholar]
- 78.Swift G H, Rose S D, MacDonald R J. An element of the elastase I enhancer is an overlapping bipartite binding site activated by a heteromeric factor. J Biol Chem. 1994;269:12809–12815. [PubMed] [Google Scholar]
- 79.Waeber G, Thompson N, Nicod P, Bonny C. Transcriptional activation of the GLUT2 gene by the IPF-1/STF-1/IDX-1 homeobox factor. Mol Endocrinol. 1996;10:1327–1334. doi: 10.1210/mend.10.11.8923459. [DOI] [PubMed] [Google Scholar]
- 80.Watada H, Kajimoto Y, Umayahara Y, Matsuoka T, Kaneto H, Fujitani Y, Kamada T, Kawamori R, Yamasaki Y. The human glucokinase gene beta-cell-type promoter: an essential role of insulin promoter factor 1/PDX-1 in its activation in HIT-T15 cells. Diabetes. 1996;45:1478–1488. doi: 10.2337/diab.45.11.1478. [DOI] [PubMed] [Google Scholar]
- 81.Williams J A, Goldfine I D. The insulin-acinar relationship. In: Go V L W, editor. The pancreas: biology, pathobiology and disease. 2nd ed. New York, N.Y: Raven Press; 1993. pp. 789–802. [Google Scholar]
- 82.Wilson D S, Desplan C. Cooperating to be different. Curr Biol. 1995;5:32–34. doi: 10.1016/s0960-9822(95)00010-8. [DOI] [PubMed] [Google Scholar]
- 83.Wong T-K, Neumann E. Electric field mediated gene transfer. Biochem Biophys Res Commun. 1982;107:584–587. doi: 10.1016/0006-291x(82)91531-5. [DOI] [PubMed] [Google Scholar]
- 84.Wright C V E, Schnegelsberg P, De Robertis E M. X1Hbox 8: a novel Xenopus homeoprotein restricted to a narrow band of endoderm. Development. 1988;104:787–794. doi: 10.1242/dev.105.4.787. [DOI] [PubMed] [Google Scholar]
- 85.Wu K-L, Gannon M, Peshavaria M, Offield M F, Henderson E, Ray M, Marks A, Gamer L W, Wright C V E, Stein R. Hepatocyte nuclear factor 3β is involved in pancreatic β-cell-specific transcription of the pdx-1 gene. Mol Cell Biol. 1997;17:6002–6013. doi: 10.1128/mcb.17.10.6002. [DOI] [PMC free article] [PubMed] [Google Scholar]