Abstract
Ebola virus (EBOV) causes severe hemorrhagic fever in humans and is lethal in a large percentage of those infected. The EBOV matrix protein viral protein 40 kDa (VP40) is a peripheral binding protein that forms a shell beneath the lipid bilayer in virions and virus-like particles (VLPs). VP40 is required for virus assembly and budding from the host cell plasma membrane. VP40 is a dimer that can rearrange into oligomers at the plasma membrane interface, but it is unclear how these structures form and how they are stabilized. We therefore investigated the ability of VP40 to form stable oligomers using in vitro and cellular assays. We characterized two lysine-rich regions in the VP40 C-terminal domain (CTD) that bind phosphatidylinositol-4,5-bisphosphate (PI(4,5)P2) and play distinct roles in lipid binding and the assembly of the EBOV matrix layer. The extensive analysis of VP40 with and without lipids by hydrogen deuterium exchange mass spectrometry revealed that VP40 oligomers become extremely stable when VP40 binds PI(4,5)P2. The PI(4,5)P2-induced stability of VP40 dimers and oligomers is a critical factor in VP40 oligomerization and release of VLPs from the plasma membrane. The two lysine-rich regions of the VP40 CTD have different roles with respect to interactions with plasma membrane phosphatidylserine (PS) and PI(4,5)P2. CTD region 1 (Lys221, Lys224, and Lys225) interacts with PI(4,5)P2 more favorably than PS and is important for VP40 extent of oligomerization. In contrast, region 2 (Lys270, Lys274, Lys275, and Lys279) mediates VP40 oligomer stability via lipid interactions and has a more prominent role in release of VLPs.
Supplementary key words: Ebola virus; lipid-protein interaction; phosphatidylserine; PI(4,5)P2; plasma membrane; protein oligomerization; virus assembly; VP40
Ebola virus (EBOV) is a collective name for six species of filovirus, four of which are known to cause a severe and often fatal hemorrhagic fever in humans (1). The FDA has approved a vaccine and antibody cocktail focused on the EBOV glycoprotein (2, 3), which studs the exterior of the virus particle during infection. Despite these efforts, there are currently no approved small molecule therapies for EBOV. Accordingly, other targets are being explored in filovirus infection such as host-virus interactions (4, 5) and the matrix protein viral protein 40 kDa (VP40) (6, 7, 8).
EBOV VP40 is a 326-residue protein required for virus assembly, budding, and escape from the host cell plasma membrane (9, 10). VP40 in the absence of other EBOV proteins is sufficient to produce virus-like particles (VLPs) from the host cell plasma membrane that resemble authentic virions (11, 12, 13, 14). VP40 therefore fulfils several distinct and essential tasks, which is facilitated by its ability to exist as a dimer, filament of dimers, and RNA-binding octameric ring (9, 15, 16). Following viral infection and uncoating, VP40 enters the host cell nucleus (17) and is also thought to bind RNA as an octamer to regulate viral transcription in the cytoplasm (9, 15). Subsequently, VP40 dimers interact with host lipids such as phosphatidylserine (PS) (18, 19) in order to localize at the inner leaflet of the plasma membrane (9, 18, 19, 20, 21) and form stable dimer-dimer interactions (9, 16, 18, 19, 20, 21, 22, 23). If the inner leaflet of the host plasma membrane contains phosphatidylinositol 4,5-bisphosphate (PI(4,5)P2), this allows the formation of large VP40 oligomers (23) from PS-induced VP40 hexamers (9, 18, 19, 20, 24). Lipid-induced VP40 oligomerization is supported by a recent cryoelectron tomography study showing the VP40 matrix layer forms from a patchwork of VP40 assemblies initiated at different points at the plasma membrane interface (16).
Although PS and PI(4,5)P2 play important roles in the EBOV life cycle, the molecular basis and structural consequences of VP40–PI(4,5)P2 interactions are mostly unknown. VP40 has two cationic loop regions in its C-terminal domain (CTD region 1: Lys221, Lys224, Lys225, CTD region 2: Lys270, Lys274, Lys275, Lys279), and molecular dynamics (MD) studies have shown that PS interacts predominantly with CTD region 1 in the absence of PI(4,5)P2 (19). Notably, the VP40 dimer is necessary for localization to the plasma membrane, which does not occur for the VP40 monomer (9, 19). Dimer interactions with plasma membrane lipids may induce the structural changes needed to form VP40 dimer-dimer interactions (16) and larger oligomers as the basis of the viral matrix layer (9, 16, 22, 23). The VP40 octamer that regulates viral transcription has not been detected at the plasma membrane, nor in virions or VLPs, and has a much lower affinity for PS than the VP40 dimer (19).
We previously reported that VP40 requires the presence of PI(4,5)P2 in the plasma membrane to form large oligomers, which are in turn needed for VLP formation and budding (23). A MD model indicated that VP40 could cause the clustering of PI(4,5)P2 via C-terminal lysine residues (25). We therefore investigated the mechanism of VP40–lipid binding at the plasma membrane in more detail and the structural consequences of VP40 interactions with PS and PI(4,5)P2.
Materials and Methods
Molecular biology
Primers were designed for single and double VP40 mutants and synthesized by Integrated DNA Technologies (Coralville, Iowa). The following primers were used: K104A: Forward 5′-CCTCTAGGTGTCGCTGATCAAGCGACCTACAGCTTTGACTC-3′, Reverse 5′-GAGTCAAAGCTGTAGGTCGCTTGATCAGCGACACCTAGAGG-3′. K221A: Forward 5′-GCCCCATTCTTTTACCCAACGCAAGTGGGAAGAAGGGGAAC-3′, Reverse 5′-GTTCCCCTTCTTCCCACTTGCGTTGGGTAAAAGAATGGGGC-3′. K224A: Forward 5′-CCCAACAAAAGTGGGGCGAAGGGGAACAGTGCCGATCTAACATCTCCGG-3′, Reverse 5′-CCGGAGATGTTAGATCGGCACTGTTCCCCTTCGCCCCACTTTTGTTGGG-3′. K225A: Forward 5′-CCCAACAAAAGTGGGAAGGCGGGGAACAGTGCCGATCTAACATCTCCGG-3′, Reverse 5′-CCGGAGATGTTAGATCGGCACTGTTCCCCGCCT TCCCACTTTTGTTGGG-3′. K236A: Forward 5′-GCCGATCTAACATCTCCGGAGGCA ATCCAAGCAATAATGACTTCAC-3′, Reverse 5′-GTGAAGTCATTATTGCTTGGATTG CCTCCGGAGATGTTAGATCGGC-3′. H269A: Forward 5′-GCCAGAAACTCTGGTCGCCAAGCTGACCGGTAAGAAGG-3′, Reverse 5′-CCTTCTTACCGGTCAGCTTGGCGACCAGAGTTTCTGGC-3′. K270A: Forward 5′-GCCAGAAACTCTGGTCCACGCGCTGACCGGTAAGAAGG-3′, Reverse 5′-CCTTCTTACCGGTCAGCGCGTGGACCAGAGTTTCTGGC-3′. K274A: Forward 5′-GGTCCACAAGCTGACCGGTGCGAAGGTGACTTCTAAAAATGG-3′, Reverse 5′-CCATTTTTAGAAGTCACCTTCGCACCGGTCAGCTTGTGGACC-3′. K275A: Forward 5′-GGTCCACAAGCTGACCGGTAAGGCGGTGACTTCTAAAAATGG-3′, Reverse 5′-CCATTTTTAGAAGTCACCGCCTTACCGGTCAGCTTGTGGACC-3′. K279A: Forward 5′-CCGGTAAGAAGGTGACTTCTGCAAATGGACAACCAATCATCCC-3′, Reverse 5′-GGGATGATTGGTTGTCCATTTGCAGAAGTCACCTTCTTACCGG-3′. K291A: Forward 5′-CCCTGTTCTTTTGCCAGCGTACATTGGGTTGGACCCG-3′, Reverse 5′-CGGGTCCAACCCAATGTACGCTGGCAAAAGAACAGGG-3′. K224A/K225A: Forward 5′-CCCAACAAAAGTGGGGCGGCGGGGAACAGTGCCGATCTAACATCTCCGG-3′, Reverse 5′-CCGGAGATGTTAGATCGGCACTGTTCCCCGCCGCCCCACTTTTGTTGGG-3′. K274/K275A (made from K274A template): Forward 5′-GGTCCACAAGCTGACCGGTGCGGCGGTGACTTCTAAAAATGG-3′, Reverse 5′-CCATTTTTAGAAGTCACCGCCGCACCGGTCAGCTTGTGGACC-3′. Site-directed mutagenesis was performed using a Quikchange XL cloning kit as described by the manufacturer (Agilent Technologies, Santa Clara, CA). Sanger sequencing was used to verify mutagenesis (University of Notre Dame Genomics Facility).
Protein expression and purification
VP40-6xHis-pET46 Ek/LIC was expressed in Rosetta BL21 DE3 cells (EMD Millipore Corp, Billerica, MA) as previously described (19, 26). Briefly, cells were grown to A600 of 0.6 then induced with 1 mM IPTG for 5 h at room temperature. Protein was purified with a Ni-NTA affinity column followed by size-exclusion chromatography (HiLoad Superdex 200 pg). Protein concentration was determined by BCA assay (Thermo Fisher Scientific, Waltham, MA), and protein was stored at 4°C for up to 2 weeks.
All-atom simulations of VP40 dimer and plasma membrane
The crystal structure of VP40 dimer was obtained from the Protein Data Bank (PDB ID: 4LDB). The protein and plasma membrane complex was set up (27) using the Charmm-Gui web server (28). The plasma membrane systems consisted of different types of lipids: 1-palmitoyl-2-oleoyl-sn-phosphatidylcholine (POPC), 1-palmitoyl-2-oleoyl-sn-phosphatidyl-ethanolamine (POPE), 1-palmitoyl-2-oleoyl-sn-phosphatidyl-serine (POPS), palmitoylsphingomyelin (PSM), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoinositol (POPI), palmitoyl-oleoyl-phosphatidylinositol-(4,5)-bisphosphate (PIP2), and cholesterol (CHOL). Two different systems were constructed: one with PIP2 and another with POPI. The lipid composition for the PIP2 containing membrane was in the ratio (21:11:32:17:9:10) (CHOL:POPC:POPE:POPS:PIP2:PSM) for the inner leaflet of the plasma membrane, and the outer leaflet had the ratio (19:33:8:3:4:33). There were 250 lipids in the outer leaflet and 244 in the inner leaflet. A similar membrane was constructed containing POPI with a lipid composition ratio of (21:11:32:17:9:10) (CHOL: POPC: POPE: POPS: POPI: PSM) for the inner leaflet and (19:32:8:3:4:34) for the outer leaflet. There were 244 lipids in outer leaflet and 247 in the inner leaflet for this system. Lipids were placed randomly in the bilayer, while VP40 was placed in close proximity to the bilayer to optimize computational efficiency, but there were only a few protein atoms in contact within 2 Å of any lipid atoms.
Both dimer-membrane systems were solvated using TIP3 water molecules. Both systems (PIP2 setup and POPI set up) consisted of a total of ∼210,000 atoms. We performed all-atom MD simulations using NAMD 3.0 (29) with the CHARMM36m force field for both systems. The particle mesh Ewald method was used to treat long-range electrostatic interactions, and the SHAKE algorithm was employed to constrain hydrogen-containing covalent bonds. The system was minimized for 10,000 steps followed by a six-step equilibration process. Pressure was controlled using a Nose-Hoover Langevin-piston method. Langevin temperature coupling with a friction coefficient of 1 ps-1 was used to control the temperature, and a 2 fs time step was used for production runs. Each system was simulated for 500 ns.
Lipid vesicle pelleting assay
All lipids were purchased from Avanti Polar Lipids, Inc. (Alabaster, AL). The liposome pelleting assay was adapted from Julkowska et al. (30). Lipid films were hydrated with fresh raffinose buffer (250 mM Raffinose pentahydrate, 150 mM NaCl, 10 mM Tris, pH 8.0) by thorough heating to 37°C and vortexing. Resuspended lipids were then extruded through a 200 nm filter with an Avanti Polar Lipids lipid extruder. Dynamic light scattering (Delsa Nano S particle size analyzer, Brea, CA) was used to confirm liposome size. Liposomes were then diluted and added to the reaction at a final concentration of 1.6 mM lipid and 3.3 μM protein. Reaction mixtures were incubated at room temperature for 30 min then centrifuged at 75,000 g for 30 min at 22°C. Three independent experiments were performed for each VP40 protein tested.
Surface plasmon resonance analysis
Surface plasmon resonance measurements were made using a Biacore X (Cytiva, Marlborough, MA) with an L1 sensor Chip (Cytiva). Liposomes were prepared by hydrating lipid films in 10 mM Tris, pH 7.4, containing 150 mM NaCl followed by extrusion through a 100 nm filter. All experiments were performed at room temperature in 150 mM NaCl, 10 mM Tris, pH 7.4, using the method previously described in detail (18). The lipid composition used was POPC:DOPE (80:20) on flow cell 1 and POPC:DOPE:PI(4,5)P2 75:20:5 on flow cell 2. The apparent affinity (Kd) was determined in Kaleidagraph (version 4.1) with the following equation: Req= Rmax/(1+Kd/[P]), where [P] is protein concentration, and R is response units.
Cell culture and transfection
COS-7 cells were maintained in a humidified incubator at 37°C and 5% CO2. The cells were kept in DMEM with L-glutamine, D-glucose, and sodium pyruvate (Life Technologies, Carlsbad, CA) with 10% FBS (Sigma, St. Louis, MO) and 1% penicillin-streptomycin (Life Technologies) at 37°C, 5% CO2. For imaging, cells were seeded in 8-well imaging plates (MatTek, Ashland, MA) and transfected with Lipofectamine 2000 or Lipofectamine LTX™ with Plus reagent (Life Technologies) and 0.4 μg DNA in optiMEM (Life Technologies) for 12–14 h. Transfection reactions were scaled up proportionally for scanning electron microscopy and VLP experiments.
Confocal imaging
A Zeiss LSM710 laser scanning microscope was used to image the enhanced green fluorescent protein (EGFP)-VP40 phenotype in COS-7 cells. A 488 nm laser was used to excite EGFP. All EGFP-VP40 expressing cells as either presenting VLPs or not were counted. EGFP-VP40 mutants were compared to WT as previously described (23). Each construct was counted and imaged over at least three independent experiments (at least 100 cells counted in each experiment). For number and brightness (N&B) experiments, an Olympus FV1000 instrument was used according to the procedure explained in depth previously (31). Data analysis was performed in SimFCS software (Globals Software, Irvine, CA) as previously described in detail (32, 33), and three independent experiments were performed for each VP40 construct.
Scanning electron microscopy analysis
Transfected cells were scraped from plates 12–14 h post transection as described previously in detail (23). Images were acquired on a Field Emission Scanning Electron Microscope Magellan 400 (FEI, Hillsboro, OR) at the University of Notre Dame Integrated Imaging Facility. Five to seven cells were imaged for each experiment.
VLP collection and Western blot analysis
VLPs were harvested 24 h post transfection as described previously in detail (23). Briefly, VLPs were washed from cells with 1X PBS and applied to a sucrose cushion. Samples were centrifuged at 100,000 g, at 4°C for 2 h. During the centrifugation, cells were scraped from plates and lysed with RIPA buffer. Soluble protein was isolated from cell lysate via centrifugation at 17,000 g at 4°C for 15 min, followed by removal of the soluble fraction of samples. Samples were then stored at −80°C. A BCA assay was used to determine protein concentration of all samples, 15 μg of each cell lysate was loaded for SDS PAGE and Western Blot. A proportional volume of the VLP sample was loaded for SDS PAGE and Western blot. For VP40 detection, mouse anti-EGFP (F56-6A1.2.3, ThermoFisher Scientific) primary antibody and sheep anti-mouse HRP (AB 6808, Abcam, Cambridge, United Kingdom) secondary antibody was used. For GAPDH detection, an anti-GAPDH antibody was used (AB 8245, Abcam, Cambridge, MA) and sheep anti mouse HRP (AB 6808, Abcam) secondary antibody was used. ECL blotting substrate was used according to the manufactures instructions (Thermo Scientific). Three independent experiments were performed for each construct. Blots were imaged on a GeneGnome (Syngene, Frederick, MD). ImageJ was used to quantify band density and determine budding efficiency.
Hydrogen deuterium exchange analysis
Hydrogen deuterium exchange mass spectrometry (HDX-MS) experiments were performed as described in detail previously with Marburg virus VP40 (32, 34). For EBOV VP40, multilamellar vesicles (MLVs) of the following compositions were used: phosphatidylcholine (PC) (50% POPC, 50% DOPE), 12.5% PI(4,5)P2 (47.5% POPC, 40% DOPE, 12.5% PI(4,5)P2), and 60% POPS (30% POPC, 10% DOPE, 60% POPS). 3 μg protein and 2.8 mM MLV were incubated at room temperature for 30 min prior to the HDX experiments, and technical replicates were performed for each condition. HDX data were analyzed as previously described in detail (32, 34) and mapped to VP40 dimer (PDB ID: 4LDB) with modeled CTDs (25, 27).
Reproducibility and statistics
At least three experimental replicates were performed for all experiments unless otherwise noted. A two-tailed students t test was used to determine statistical differences. A P value of 0.05 or less was considered statistically significant. Error bars are shown as the standard error of the mean in all graphs unless otherwise noted.
Results
VP40 features two CTD cationic regions that are hypothesized to bind PI(4,5)P2
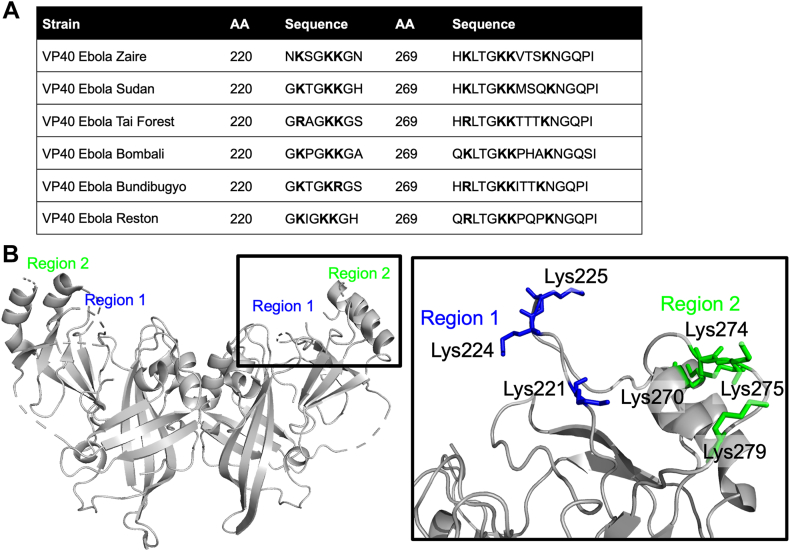
VP40 sequences from Zaire, Sudan, Taï Forest, Bundibugyo, Bombali, and Reston EBOV were aligned using UniProt to reveal sequence conservation among the C-terminal lysine residues that may bind PI(4,5)P2. We found that Lys224, Lys225, Lys274, and Lys279 were completely conserved, whereas Lys221 had mutated to Arg in the Taï Forest strain, Lys225 had mutated to Arg in the Bundibugyo strain, and residue 270 can be Arg or Lys among the six strains (Fig. 1A). Both Arg and Lys are commonly found in protein domains that bind PI(4,5)P2 (35), so we hypothesized a similar role in VP40. We therefore used scanning mutagenesis in the conserved CTD of Zaire VP40 to convert Lys104, Lys221, Lys224, Lys225, Lys236, His269, Lys270, Lys274, Lys275, Lys279, and Lys291 into alanine residues. When mapped onto the VP40 structure, the lysine residues in the 220–230 region (blue) and those in the 270–280 region (green) formed distinct, adjacent domains, which we named region 1 and region 2, respectively (Fig. 1B and zoomed inset).
Fig. 1.
VP40 sequence and structure revels two potential PI(4,5)P2 binding sites. A: Consensus sequence of Ebola VP40 strains highlighting several lysine residues in the 220 and 269 amino acid regions of the protein and positioned at the surface of the C-terminal domain. B: VP40 dimer structure (PDB ID: 4LDB with missing Lys loop residues modeled in) with region 1 lysine residues colored in blue, and region 2 lysine residues colored in green.
Molecular dynamics simulations of VP40 and membrane bilayers
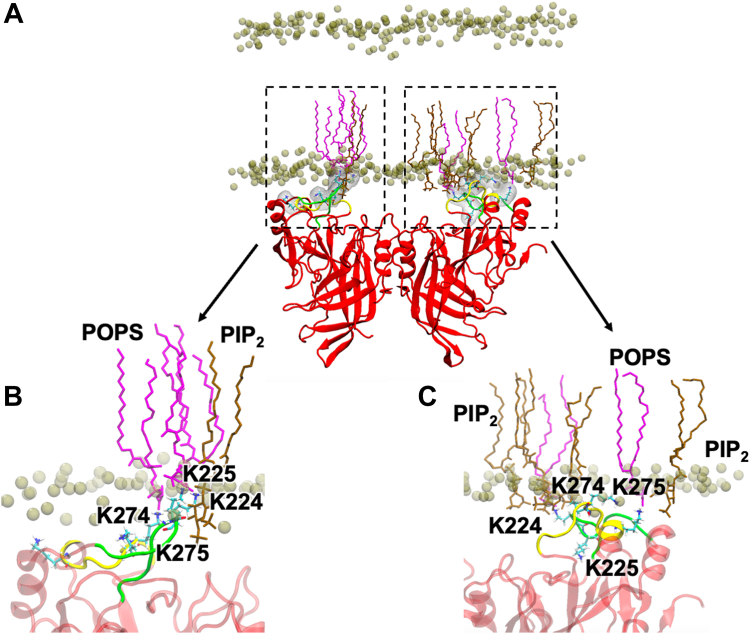
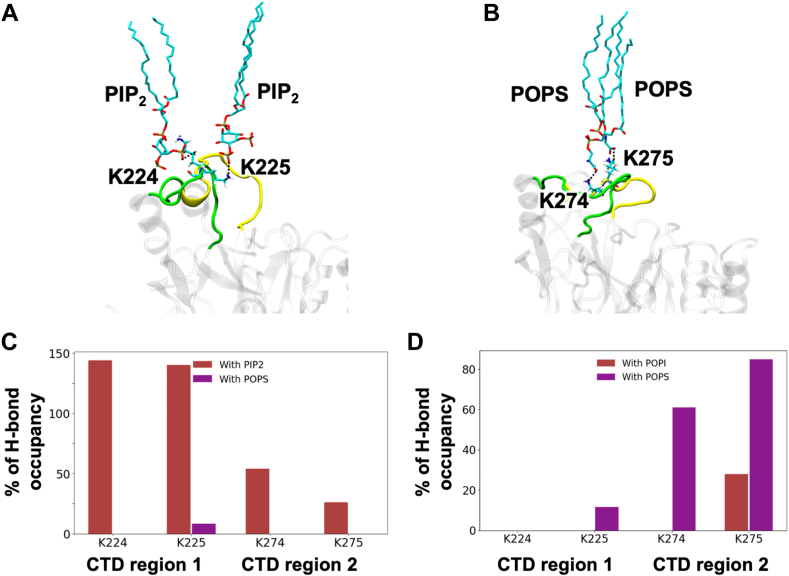
To gain insight into the molecular basis of PS and PI(4,5)P2 coordination by VP40, we performed MD simulations of VP40 dimers (Fig. 2) with bilayer systems containing PI(4,5)P2 or POPI for 500 ns each. Both membrane systems were asymmetric bilayers to partially mimic the cellular plasma membrane. For the PI(4,5)P2 system, CHOL:POPC:POPE:POPS:PIP2:PSM was used with different ratios for the inner (21:11:32:17:9:10) and outer (19:33:8:3:4:33) leaflets. For the POPI system, CHOL:POPC:POPE:POPS:POPI:PSM was used with inner (21:11:32:17:9:10) and outer (19:32:8:3:4:34) membrane ratios differing. H-bonding was calculated from the last 100 ns of each simulation. VP40 forms key H-bonds with the anionic lipids by basic residues Lys224, Lys225, Lys274, and Lys275 (Figs. 2 and 3). The PI(4,5)P2/PS membrane system showed that PI(4,5)P2 dominated PS in protein binding (Fig. 3C) with appreciable H-bonding of PI(4,5)P2 by Lys224, Lys225, Lys274, and Lys275. Whereas in the POPI/PS system lacking PI(4,5)P2, PS H-bonds predominantly with Lys225, Lys274, and Lys275, a result supported by previous identification of VP40 residues that modulate PS-binding (19).
Fig. 2.
Molecular dynamics simulation of a VP40 dimer. A: interacting with an asymmetric membrane containing CHOL:POPC:POPE:POPS:PIP2:PSM (21:11:32:17:9:10) on the bottom leaflet. POPS lipids are shown in purple and PI(4,5)P2 lipids in light brown. B: Close up of the CTD of the VP40 protomer on the left highlighting CTD region 1 residue interactions with POPS and CTD region 2 interactions with PI(4,5)P2. C: Close up of the VP40 protomer on the right demonstrating abundant PI(4,5)P2 and POPS interactions with CTD regions 1 and 2.
Fig. 3.
Molecular dynamics simulations of VP40 dimer. A: Snapshot from MD simulations in Figure 2 demonstrates interaction of region 1 (Lys224) with PS. B: MD simulations from Figure 2 demonstrated predominant PI(4,5)P2 interactions with region 2 (Lys270 and Lys274 here) and a lack of PS interactions for this region. C: Percentage of hydrogen bond occupancy for region 1 and region 2 lysine residues over the time course of MD simulations (from Fig. 2). Region 1 residues display interactions with PS and PI(4,5)P2, whereas region 2 residues interact predominantly with PI(4,5)P2. D: Percentage of hydrogen bond occupancy for regions 1 and 2 lysine residues over the time course of MD simulations with membranes containing POPI (in place of PI(4,5)P2) and PS. Region 2 residues interact with PS as does region 1 Lys225.
HDX-MS differentiates the mode of PS and PI(4,5)P2 interactions by VP40
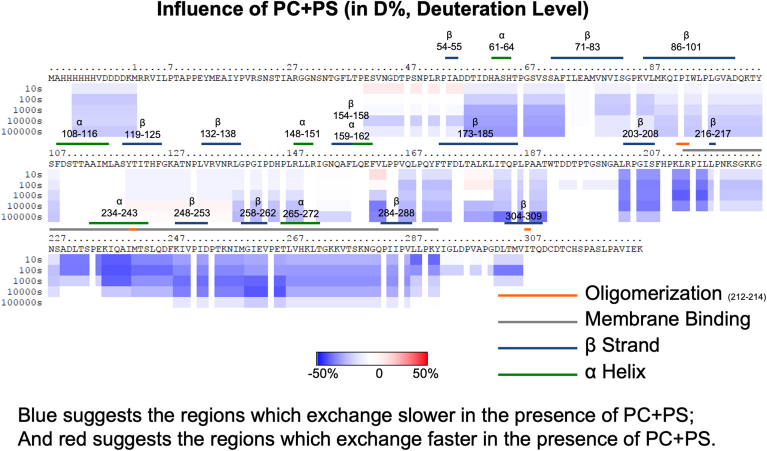
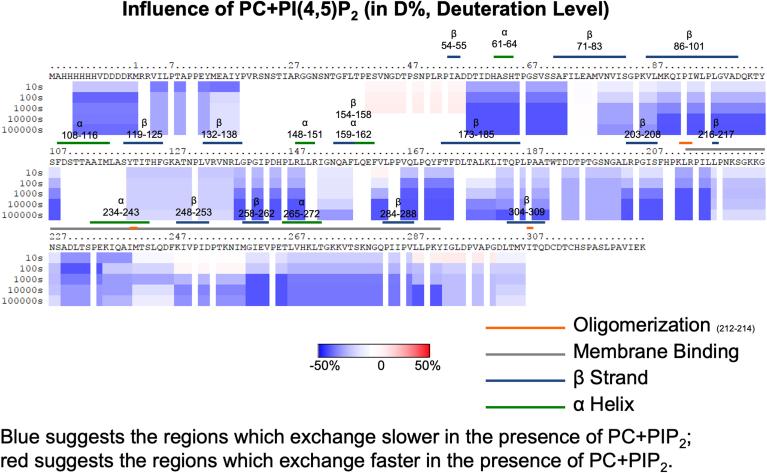
HDX-MS was used to investigate the structural changes that occur when VP40 binds PS and PI(4,5)P2. Complete coverage of the VP40 protein was achieved with many overlapping peptides (supplemental Fig S1) as we previously determined with MARV VP40 (34). To ensure lipid samples that allow for saturation of EBOV VP40 membrane binding, we prepared MLVs with elevated levels of PI(4,5)P2 or PS (supplemental Fig S2). We chose this MLV method as it previously allowed for saturation of MARV VP40 lipid binding to compare different lipid binding conditions (34). Samples were incubated for 10, 100, 1000, 10,000, or 100,000 s. Changes in deuterium incorporation during these time periods are shown in supplemental Fig S3 for VP40 alone and VP40 with PC MLVs and supplemental Fig S4 for VP40 with PS- and PI(4,5)P2-containing MLVs, respectively. Figure 4 shows ribbon maps with the level of deuterium incorporation mapped to the protein sequence, whereas Fig. 4, Fig. 5, Fig. 6 show the influence of PC, PS, and PI(4,5)P2, respectively.
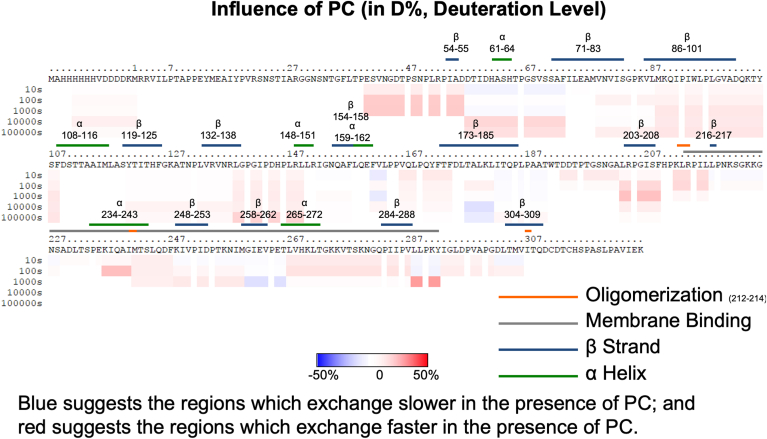
Fig. 4.
Influence of phosphatidylcholine vesicles on the HDX-MS profile of VP40. VP40 was incubated with MLVs containing POPC, and the reaction stopped at 10 s, 100 s, 1000 s, 10,000 s, and 100,000 s. The complete amino acid sequence is shown for VP40 including the N-terminal His tag. Red (see scale bar) indicates regions with faster exchange of deuterium in the presence of PC compared to VP40 in solution. Blue shows regions that exchange slower in the presence of PC. Three technical replicates were performed for each experiment.
Fig. 5.
Influence of phosphatidylserine containing vesicles on the HDX-MS profile of VP40. VP40 was incubated with MLVs containing POPC, and the reaction stopped at 10 s, 100 s, 1000 s, 10,000 s, and 100,000 s. The complete amino acid sequence is shown for VP40 including the N-terminal His tag. Red (see scale bar) indicates regions with faster exchange of deuterium in the presence of PC compared to VP40 in solution. Blue shows regions that exchange slower in the presence of PC. Three technical replicates were performed for each experiment.
Fig. 6.
Influence of PI(4,5)P2 containing vesicles on the HDX-MS profile of VP40. VP40 was incubated with MLVs containing POPC, and the reaction stopped at 10 s, 100 s, 1000 s, 10,000 s, and 100,000 s. The complete amino acid sequence is shown for VP40 including the N-terminal His tag. Red (see scale bar) indicates regions with faster exchange of deuterium in the presence of PC compared to VP40 in solution. Blue shows regions that exchange slower in the presence of PC. Three technical replicates were performed for each experiment.
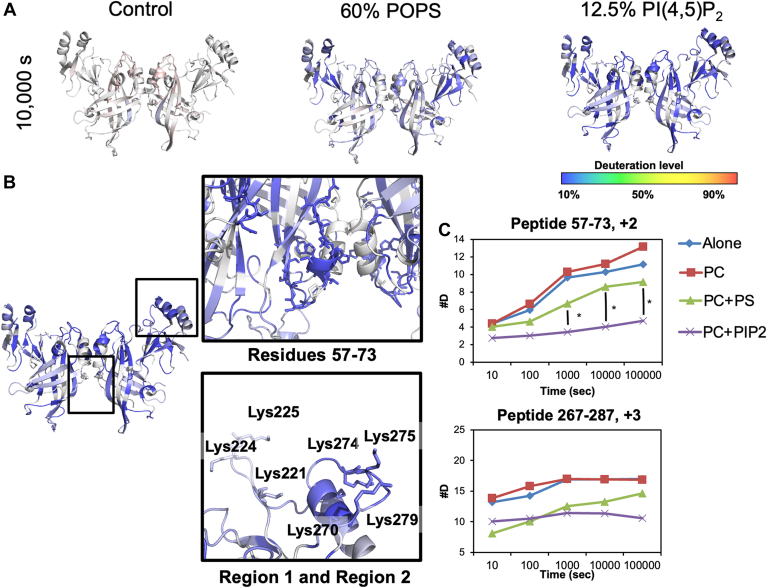
The deuterium incorporations for each time point and lipid were also mapped to the VP40 dimer structure (Fig. 7), the relevant structure for VP40 matrix assembly (16). Figure 7A shows the percent deuterium incorporation mapped to the dimer structure, whereas Figure 7B shows the influence of PI(4,5)P2 focused on an area (residues 57–73) just below the α-helical dimer interface and CTD regions 1 and 2. In the presence of PC, VP40 deuterium exchange is slightly increased compared to samples without the lipid as shown by the light red regions in the N- and CTDs (Figs. 4 and 7A). The structural changes deduced from the less extensive incorporation of deuterium after VP40 binds PI(4,5)P2 were highly stable, with less than a 20% change in deuterium incorporation between the 10 s and 100,000 s time points (Fig. 6). Most of the decrease in deuterium incorporation was abolished in the PS samples after the 10,000 s time point (Fig. 5). These data strongly support a model in which PS is required for VP40’s initial association and conformational change, whereas PI(4,5)P2 plays a more important role in stabilizing the resulting VP40 oligomers.
Fig. 7.
HDX-MS reveals changes in VP40 solvent accessibility when coincubated with lipid vesicles of different compositions. A: Deuteration level changes of VP40 at 10,000 s time point after a 30-min incubation with control liposomes, PS containing liposomes, or PI(4,5)P2 containing liposomes. B: The VP40 dimer structure is shown with deuteration profile changes highlighted (as in A) with influence of PC+PI(4,5)P2 vesicles at the 100,000 s time points. The black boxes around the NTD dimer interface and CTD lipid-binding regions are further highlighted with two insets (residues 57–73 and regions 1 and 2). C: The influence of PC, PC+PS, or PC+PI(4,5)P2 containing vesicles on deuterium incorporation in the two regions highlighted in B. The y-axis is the number of deuterium incorporated into the peptide fragment (peptide 57–73 or peptide 267–287) with respect to time on the x-axis. Three technical replicates were performed for each experiment, ∗ P value < 0.05.
Deuterium incorporation was reduced at all time points in the regions containing lysine residues proposed to bind PI(4,5)P2. Notably, CTD region 1 (Lys221, Lys224, and Lys225) and CTD region 2 (Lys270, Lys274, Lys275, and Lys279) yielded different deuteration profiles, with region 1 registering a smaller decrease in deuterium incorporation (Figs. 6 and 7C). The residues between positions 221 and 229 and 270 and 275 have previously been shown to interact with PS, and the HDX-MS data support these observations (Fig. 5). PI(4,5)P2-binding residues in CTD region 2 (Lys270, Lys274, Lys275, and Lys279) had a slower rate of deuterium exchange with both PS and PI(4,5)P2 vesicles (Figs. 5, 6, and 7C), although a more substantial reduction was observed with PI(4,5)P2 compared to PS over the time course of the interaction.
The most dramatic changes were observed in the N-terminal domain (NTD) residues 57–73 (Figs. 6 and 7), which appear to be important for VP40 dimer and perhaps dimer-dimer interaction stability. Residues 57–73 pack against the N–N terminal α-helical dimer interface (Fig. 7B), and residues in this region (residues 52–61) were recently shown to play a critical role in VP40 dimer stability (36). HDX-MS studies are highly sensitive to secondary structural formation and although we did not observe secondary structure changes in VP40 residues 57–73 in MD simulations, we cannot rule out that such changes occur. The α-helical dimer interface structure (residues 109–119) is extremely stable, given that no deuterium is incorporated even after 100,000 s under all conditions (supplemental Figs S3 and S4). PI(4,5)P2 and PS both significantly reduced deuterium incorporation between residues 57 and 73, with PI(4,5)P2 providing greater stability over the experimental time frame, highlighting the importance of PI(4,5)P2 in VP40 dimer stability and potentially dimer-dimer interactions in the membrane bound state.
VP40 oligomerization
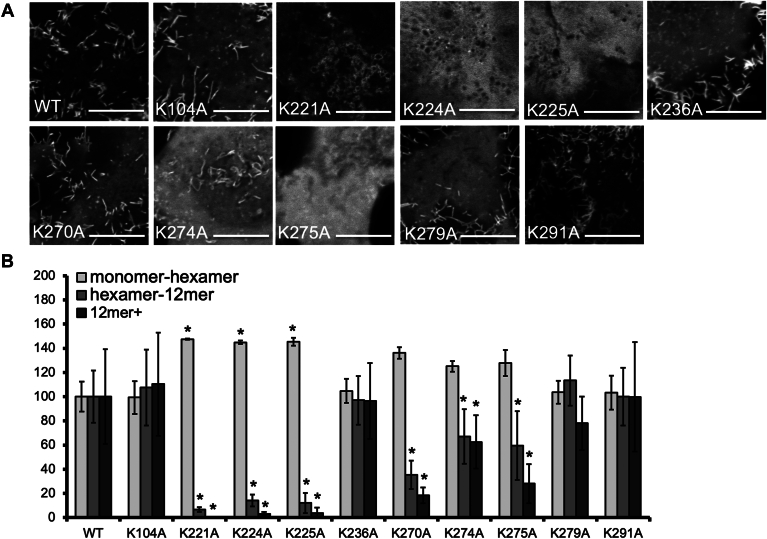
The depletion of PI(4,5)P2 in the plasma membrane by Myc5-PtaseIV was previously shown to inhibit the formation of large VP40 oligomers (23). Given that VP40 oligomer formation depends on the presence of PS in the plasma membrane (20, 33), these data suggest that PS and PI(4,5)P2 play different roles in the structure and assembly of the viral matrix layer. We therefore used a VP40 oligomerization assay in live cells (20, 21, 22, 23, 32, 33, 37) to compare the oligomerization abilities of mutants deficient in PI(4,5)P2 binding compared to wild-type VP40. N&B analysis was used as previously described (20, 21, 22, 23, 31, 32, 33, 37). As VP40 assembles via dimer-dimer interaction at the plasma membrane inner leaflet in patches (16, 37), N&B analysis allows for assessment of VP40 oligomerization when fluorescently tagged (37). Since VP40 builds filaments via addition of dimers, we defined VP40 size as small (monomer-hexamer), medium (hexamer-12mer), and large VP40 oligomers (12mer and greater (12mer+)), for the purpose of analysis and the resolution of the assay (33). We found that the region 1 mutants K221A, K224A, and K225A strongly inhibited the assembly of oligomers larger than a hexamer and significantly increased the monomer-hexamer population compared to wild-type VP40 (Fig. 8). In contrast, some large oligomers were detected for the region 2 mutants K270A, K274A and K275A, although still significantly less than the amount formed by wild-type VP40 (Fig. 8). There was also a small drop in the number of oligomers formed by mutant K279A. These data confirm the previous results with Myc5-PtaseIV (23) but suggest the two cationic regions may contribute differently to the oligomerization and stabilization of VP40 filaments where region 1 had more prominent effects on oligomerization compared to region 2.
Fig. 8.
Number and Brightness analysis to determine VP40 oligomerization in live cells. A: Representative image frame from the 100 images acquired for each imaging experiment. All images were collected at the same zoom (16.4). Scale bar = 10 μm. B: Normalized VP40 oligomer formation. To compare changes between WT VP40 and mutations, the average EGFP-VP40 population determined for monomer-hexamer, hexamer-12mer, and 12mer+ was normalized to 100% for each condition. WT and mutant EGFP-VP40 oligomer population values are shown ± the SEM, ∗ P value < 0.05.
Assessment of potential PI(4,5)P2 binding residues for PI(4,5)P2-binding, plasma membrane localization, and budding
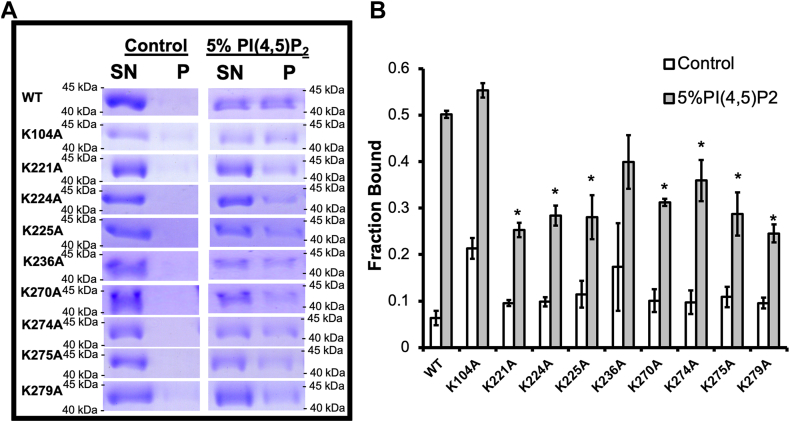
To determine which VP40 residues are responsible for PI(4,5)P2 interactions, a LUV assay was employed to monitor the binding of WT VP40 and VP40 lysine mutants to PI(4,5)P2 containing vesicles. There was no significant difference in binding when comparing wild-type VP40 and the mutants K104A and K236A, whereas mutants K221A, K224A, K225A, K270A, K274A, K275A, and K279A showed a significant loss in binding to LUVs containing PI(4,5)P2 (Fig. 9).
Fig. 9.
VP40 C-terminal domain lysine residues are important for PI(4,5)P2 binding. A: Representative SDS PAGE of the LUV pelleting assay to measure VP40 binding to control (1,2-dipalmitoyl-sn-glycero-3-phosphocholine and cholesterol (50:50)) or vesicles containing 5% PI(4,5)P2. Both vesicle mixtures had a trace amount of dansylPE for visualization of the lipid vesicle pellet. Molecular weight is labeled for each VP40 protein gel panel above and below the VP40 band in kDa. B: Average fraction bound to LUVs shown ± the SEM, ∗ P value < 0.05. Three independent replicates were performed for each protein construct.
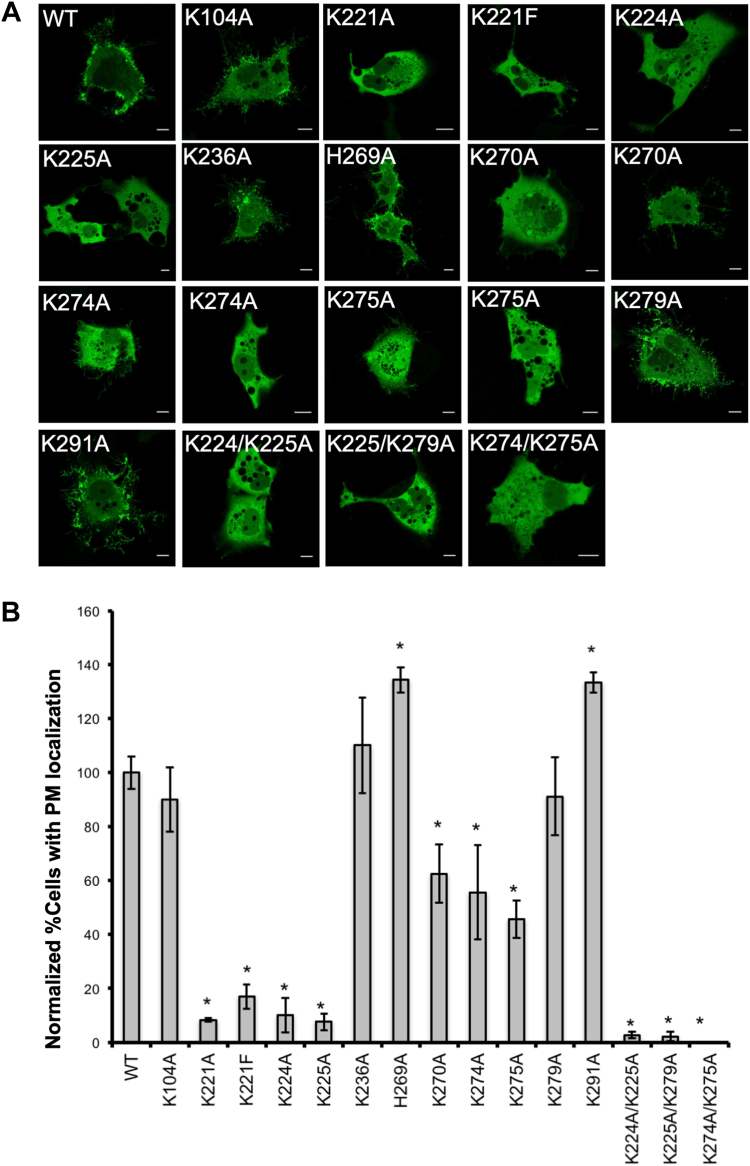
The transfection of COS-7 cells with a construct encoding wild-type VP40 fused to the EGFP produced an abundance of fluorescent VLPs at the plasma membrane 12–14 h posttransfection (Fig. 10). The WT signal for plasma membrane localization was normalized to 100% to compare a series of EGFP-VP40 constructs based on the mutant VP40 sequences and the proportion of transfected COS-7 cells producing VP40 plasma membrane localization to those expressing the wild-type EGFP-VP40 control (Fig. 10). Mutants K221A and K225A showed the most dramatic reduction in plasma membrane localization, with <10% of the transfected cells producing detectable plasma membrane localization. Furthermore, <20% of cells expressing K221F and K224A produced detectable plasma membrane localization, and ∼50% of cells expressing K270A, K274A, and K275A produced some level of plasma membrane localization. Also, note in Figure 10 additional images for K270A, K274A, and K275A indicating some pre-VLP structures detectable in some of the cells with plasma membrane localization. The remaining mutants (K104A, K236A, H269A, K279A, and K291A) had a phenotype-like wild-type VP40. The limited PI(4,5)P2 binding by K221A may reflect a loss of PS or nonspecific electrostatic interactions as MD simulations indicated little detectable H-bonding by Lys221 with PI(4,5)P2, while the LUV binding assay indicated K221A had reduced binding to vesicles containing PI(4,5)P2.
Fig. 10.
VP40 and VP40 mutant phenotypes in COS-7 cells. A: Representative images of WT-EGFP-VP40 and EGFP-VP40 mutants 14 h posttransfection in COS-7 cells. Scale bars are 10 μm. B: Percentage of the cell population expressing detectable plasma membrane localization 14 h posttransfection, normalized to WT EGFP-VP40 plasma membrane localization. Three independent experiments were performed with at least 100 cells counted for each experiment. WT VP40 pre-VLPs detectable in cells were normalized to 100% to compare changes in pre-VLP detection for mutant constructs. Average values are shown ± the SEM, ∗ P value < 0.05.
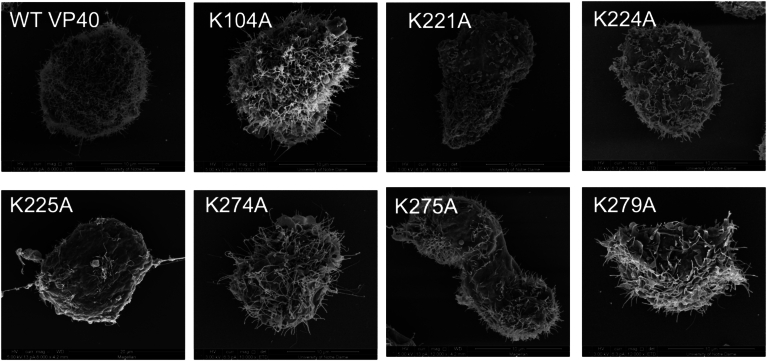
Scanning electron microscopy (SEM) was used to determine the morphological phenotype of COS-7 cells transfected with wild-type VP40 or mutations. For SEM analysis, we selected three mutants from region 1 and three from region 2, representing mutants that affected PI(4,5)P2 binding and plasma membrane localization, and one (K104A) that had no effect on either of these processes (Fig. 11). In cells transfected with K221A, K224A, or K225A, the filopodia were sparser compared to WT or K104A (Fig. 11). In contrast to region 1 mutations, region 2 mutations (K274A, K275A, K279A) were less abundant than WT but more prominent than region 1 mutations. This showed that K104A formed VLPs similar in length and density to wild-type VP40, whereas region 1 mutations greatly reduced VLP density, and region 2 mutations reduced VLP density slightly.
Fig. 11.
Scanning electron microscopy of VP40 and VP40 mutant expressing cells to assess virus-like particle production prior to release (pre-VLPs). Representative SEM images of COS-7 cells (5–7 cells imaged per experiment) expressing WT and mutant VP40 constructs at 12–14 h posttransfection. Scale bar is as shown in each figure.
PI(4,5)P2-binding residues are needed for efficient VLP formation
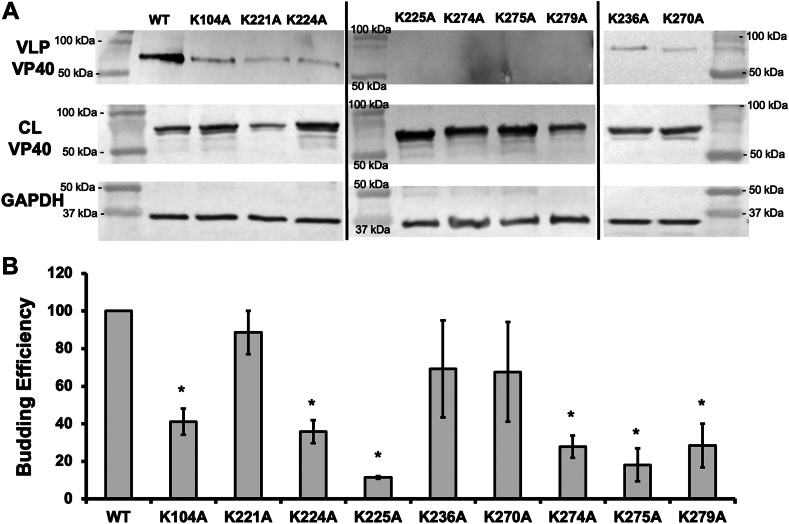
We used lysates and VLPs prepared from COS-7 cells 20–24 h post transfection to determine the budding efficiency of wild-type VP40 and corresponding mutants in Western blot experiments. The region 1 K225A mutation made 85% less VLPs than WT (Fig. 12) consistent with a low level of plasma membrane localization and pre-VLP formation as detected using SEM (Fig. 11). K224A, also in region 1, reduced VLP formation by 60%. Given that oligomer formation and plasma membrane localization were inhibited by more than 90% in mutant K221A, we were surprised to observe little difference in budding efficiency compared to WT (Fig. 12). Region 2 mutations K274A and K275A did not produce detectable VLPs despite some detectable plasma membrane localization and pre-VLPs in SEM. K279A also showed a significant decrease in budding efficiency, which is consistent with the limited binding to PI(4,5)P2 in vitro. However, this mutant produced detectable quantities of pre-VLPs and showed only a small decrease in the formation of large oligomers. Because Lys279 is important for lipid binding in vitro, we hypothesize that even a small decrease in PI(4,5)P2 binding and VP40 dimer-dimer interactions may have dramatic effects on the late stages of budding required for high efficiency of VLP production. It should be noted in the context of interpreating VP40 cellular data that WT VP40 and some mutations did not always have equal expression in cell lysates (e.g., K221A and K279A). Different expression of VP40 mutants has been previously observed (14, 19, 20, 26, 32) and may be attributed to flux of VP40 going out of the cell in VLP formation or interactions with host proteins that sequester or alter VP40 stability.
Fig. 12.
VLP budding efficiency of WT and mutant VP40. A: Western blots of WT and mutant VP40 detect VP40 in the VLP and the cell lysate (CL) fractions. GAPDH (CL) was used as a loading control for these experiments. Molecular weight markers (in kDa) are shown for all blots to verify band size for EGFP-VP40 and GAPDH. B: Quantified budding efficiency normalized to VP40-WT. Average values are shown of three independent replicates ± the SEM, ∗P < 0.05.
K104A reduced the efficiency of budding even though it did not affect the efficiency of pre-VLP formation and did not block interactions with PI(4,5)P2 in vitro. Although this mutation is found in the NTD, it folds against the CTD adjacent to Asp193 in the dimer. Its interaction with lipids is therefore unlikely to be required for PI(4,5)P2-dependent assembly, but we cannot rule out a role in interdomain interactions or host protein interactions necessary for VLP release.
Discussion
Binding of VP40 to PI(4,5)P2 is mediated in part by lysine residues in region 1 (Lys221, Lys224, and Lys225) and region 2 (Lys270, Lys274, Lys275, and Lys279), which contributed to in vitro PI(4,5)P2 binding, plasma membrane localization, VP40 oligomerization, and/or budding. In addition, these two binding regions have distinct VP40 phenotypes. For example, region 1 lysine residues are required for the formation of oligomers larger than a hexamer, whereas region 2 lysine residues harbor more significantly reduced VLP formation but did not have as a profound of an effect on the formation of large VP40 oligomers. These data agree with the confocal imaging and assessment of cell populations with SEM imaging (Fig. 11) and VLP formation (Fig. 12).
The HDX-MS data confirmed these VP40 binding sites are distinct as they have different deuterium incorporations when bound to PI(4,5)P2 containing vesicles. HDX-MS was recently used to study Marburg virus VP40 oligomerization and lipid binding (32, 34). MARV VP40 (mVP40) interacts with anionic membranes through electrostatic interactions (37) and HDX-MS confirmed that these interactions occur via CTD residues and that mVP40 undergoes oligomer formation when bound to anionic membranes (32, 34). In contrast to EBOV VP40, the lifetime of the mVP40 interactions with anionic membranes was relatively short (34). In region 1 of the VP40 CTD, PS contributed to a slower exchange rate with deuterium than that of PI(4,5)P2, and this region was more important for formation of VP40 oligomers larger than a hexamer. Previously, region 1 was shown to interact with PS (19), and PS was necessary for formation of VP40 oligomers larger than hexamers (18, 33). Thus, region 1 may be able to bind PS or PI(4,5)P2 with PS providing an interaction that is able to induce VP40 dimer conformational change to a greater extent. Lys225 may be a key residue in this regard as the current and previous study (19) suggest Lys225 interacts with PS more significantly than other CTD region 1 residues (Fig. 3). More significant exploration of dynamic and structural consequences of lipid head group binding by VP40 will be necessary to delineate the molecular basis of these conformational transitions.
In region 2, PI(4,5)P2 slowed exchange more significantly than PS, indicating a more stable VP40 complex. Residues in region 2 had differential effects on VP40 dependent properties depending upon the assay. Lys274 seems to be a key PI(4,5)P2 interacting residue, which was supported by the MD simulations. Lys275 also made important contributions to PI(4,5)P2 binding in in vitro and in silico assays, and like K274A, K275A reduced VP40 oligomerization and VLP formation. Lys270 and Lys279 in region 2 also play a significant role in aspects of PI(4,5)P2-mediated budding as both K270A and K279A mutants show reduced binding to LUVs and K279A reduced pre-VLP formation. In contrast, K279A large oligomers formed like WT (Fig. 8B), suggesting the contacts K279A facilitates with the membrane or among VP40 oligomers is critical to budding. Despite a reduction in PI(4,5)P2 binding and VP40 oligomerization, K270A was sufficient for approximately the same budding efficiency as WT. K104A is a thought-provoking mutation as it sits at the N-C terminal domain interface and may be an important residue involved in the unlatching in larger oligomers (as determined with N&B analysis, Fig. 8), indicating it may promote larger oligomer formation.
The HDX-MS studies also revealed PI(4,5)P2 induced structural changes in residues 57–73 as evidenced by dramatic reduction in deuterium exchange in this region. It should be noted that recent computational and experimental studies demonstrated mutation of residues at or near this region (R52A and H61A) abrogated VP40 dimer formation (36). Mutation of R52A and H61A abrogated VP40 dimer formation in vitro (36) and computational studies predicted other amino acids in this region are critical for VP40 dimer stability and interactions across the two protomers. Thus, PI(4,5)P2 may stabilize interactions across the dimer interface in the membrane-bound form facilitating more effective dimer-dimer interactions or VP40 oligomer stability. However, it should be noted that HDX-MS analysis is highly sensitive to secondary structural changes in proteins, and structural changes in this region cannot be ruled out. Although our MD analysis could predict and validate lipid-protein interactions for the VP40 CTD, we did not observe structural changes in residues 57–73 throughout the simulation time. Thus, more detailed in vitro and computational studies will be required to rule out secondary structural changes in these residues. An alternative explanation could be that residues in this region are interacting with the membrane. This hypothesis is supported by recent MD simulations of VP40 interactions with the membrane of VLPs, which demonstrated residues 58 to 67 contacted the membrane (38).
Late-stage budding and scission of EBOV or EBOV VLPs is still not well understood. Several host proteins have been deemed critical for effective budding of EBOV or VP40-based VLPs (39, 40, 41, 42, 43, 44) through direct or indirect VP40 interactions, but the mechanisms by which VP40-lipid interactions facilitates interaction or dissociation of VP40 and host components is still unknown. VP40 also contains two Late domains, a PTAP and PPxY motif (14, 45), that can mediate interactions with E3 ubiquitin ligases and components of the ESCRT pathway (46, 47). While the VP40 based VLPs can still be released from the plasma membrane when the Late domains are deleted (48), VP40-lipid interactions could facilitate conformational changes of the late domain regions facilitating or disassembling VP40-host interactions. Notably, some VP40 host interactions have been deemed antiviral by causing sequestration or degradation of VP40 (49, 50). VP40-mediated lipid interactions could mediate VP40 stability and weaken interactions with host proteins. While the unveiling of the mechanisms by which VP40 protein-protein and lipid-protein interactions facilitate release of virus particles from the plasma membrane is still emerging, this study has opened new horizons in determining how PS and PI(4,5)P2 differentially effect these processes. Region 1 lysine residues interactions with PS or PI(4,5)P2 most prominently reduced budding by limiting large VP40 oligomers from forming and reducing pre-VLPs when mutated to alanine. Region 2 lysine residues more readily stabilized the VP40 structure when bound to PI(4,5)P2 compared to PS strengthening the stability of VP40 oligomers with a more notable effect on VLP release.
Data availability
The data generated in this study are included in the article and supplemental data. Further inquiries can be directed to rstaheli@purdue.edu.
Supplemental data
This article contains supplemental data.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of the article.
Acknowledgments
Author contributions
K. A. J. and R. V. S. conceptualization; K. A. J., M. R. B., N. B., T. S., S. U., P. P. C., and S. L. data curation; K. A. J., M. R. B., N. B., T. S., S. U., P. P. C., and S. L. formal analysis; B. S. G., P. P. C., S. L., and R. V. S. funding acquisition; K. A. J., M. R. B., N. B., T. S., S. U., P. P. C., and S. L. investigation; K. A. J., M. R. B., N. B., T. S., S. U., B. S. G., P. P. C., S. L., and R. V. S. methodology; B. S. G., P. P. C., S. L., and R. V. S. project administration; B. S. G., P. P. C., S. L., and R. V. S. resources; B. S. G., P. P. C., S. L., and R. V. S. supervision; K. A. J. and R. V. S. writing–original draft; K. A. J., M. R. B., N. B., T. S., S. U., B. S. G., P. P. C., S. L., and R. V. S. writing–review & editing.
Funding and additional information
This work was supported by the National Institutes of Health grant numbers: AI081077 and AI158220 (R. V. S.) and AI117905, GM020501, NS070899, and GM121964 (S. L.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Current address for Kristen A. Johnson: Fred Hutch Cancer Center, Seattle, WA, USA.
Current address for Melissa R. Budicini: Department of Microbiology, University of Texas Southwestern, Medical Center, Dallas, TX, USA.
Current address for Nisha Bhattarai: Department of Physiology and Biophysics, Case Western Reserve University, Cleveland, OH, USA.
Supplementary Data
References
- 1.Feldmann H., Sprecher A., Geisbert T.W. Ebola. N. Engl. J. Med. 2020;382:1832–1842. doi: 10.1056/NEJMra1901594. [DOI] [PubMed] [Google Scholar]
- 2.Ollmann Saphire E. A vaccine against Ebola virus. Cell. 2020;181:6. doi: 10.1016/j.cell.2020.03.011. [DOI] [PubMed] [Google Scholar]
- 3.Taki E., Ghanavati R., Navidifar T., Dashtbin S., Heidary M., Moghadamnia M. EbangaTM: the most recent FDA-approved drug for treating Ebola. Front. Pharmacol. 2023;14 doi: 10.3389/fphar.2023.1083429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Han Z., Lu J., Liu Y., Davis B., Lee M.S., Olson M.A., et al. Small-molecule probes targeting the viral PPxY-host Nedd4 interface block egress of a broad range of RNA viruses. J. Virol. 2014;88:7294–7306. doi: 10.1128/JVI.00591-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Loughran H.M., Han Z., Wrobel J.E., Decker S.E., Ruthel G., Freedman B.D., et al. Quinoxaline-based inhibitors of ebola and Marburg VP40 egress. Bioorg. Med. Chem. Lett. 2016;26:3429–3435. doi: 10.1016/j.bmcl.2016.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harty R.N. No exit: targeting the budding process to inhibit filovirus replication. Antivir. Res. 2009;81:189–197. doi: 10.1016/j.antiviral.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stahelin R.V. Could the ebola virus matrix protein VP40 be a drug target? Expert Opin. Ther. Targets. 2013;18:115–120. doi: 10.1517/14728222.2014.863877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Madara J., Han Z., Ruthel G., Freedman B.D., Harty R.N. The multifunctional Ebola virus VP40 matrix protein is a promising therapeutic target. Futur. Virol. 2015;10:537–546. doi: 10.2217/fvl.15.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bornholdt Z.A., Noda T., Abelson D.M., Halfmann P., Wood M.R., Kawaoka Y., et al. Structural rearrangement of ebola virus VP40 begets multiple functions in the virus life cycle. Cell. 2013;154:763–774. doi: 10.1016/j.cell.2013.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stahelin R.V. Membrane binding and bending in Ebola VP40 assembly and egress. Front. Microbiol. 2014;5:300. doi: 10.3389/fmicb.2014.00300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jasenosky L.D., Neumann G., Lukashevich I., Kawaoka Y. Ebola virus VP40-induced particle formation and association with the lipid bilayer. J. Virol. 2001;75:5205–5214. doi: 10.1128/JVI.75.11.5205-5214.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Timmins J., Scianimanico S., Schoehn G., Weissenhorn W. Vesicular release of ebola virus matrix protein VP40. Virology. 2001;283:1–6. doi: 10.1006/viro.2001.0860. [DOI] [PubMed] [Google Scholar]
- 13.Noda T., Sagara H., Suzuki E., Takada A., Kida H., Kawaoka Y. Ebola virus VP40 drives the formation of virus-like filamentous particles along with GP. J. Virol. 2002;76:4855–4865. doi: 10.1128/JVI.76.10.4855-4865.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Licata J.M., Simpson-Holley M., Wright N.T., Han Z., Paragas J., Harty R.N. Overlapping motifs (PTAP and PPEY) within the ebola virus VP40 protein function independently as late budding domains: involvement of host proteins TSG101 and VPS-4. J. Virol. 2003;77:1812–1819. doi: 10.1128/JVI.77.3.1812-1819.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Landeras-Bueno S., Wasserman H., Oliveira G., VanAernum Z.L., Busch F., Salie Z.L., et al. Cellular mRNA triggers structural transformation of Ebola virus matrix protein VP40 to its essential regulatory form. Cell Rep. 2021;35 doi: 10.1016/j.celrep.2021.108986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wan W., Clarke M., Norris M.J., Kolesnikova L., Koehler A., Bornholdt Z.A., et al. Ebola and Marburg virus matrix layers are locally ordered assemblies of VP40 dimers. eLife. 2020;9 doi: 10.7554/eLife.59225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nanbo A., Watanabe S., Halfmann P., Kawaoka Y. The spatio-temporal distribution dynamics of Ebola virus proteins and RNA in infected cells. Sci. Rep. 2013;3:1206. doi: 10.1038/srep01206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adu-Gyamfi E., Johnson K., Fraser M.E., Scot J.L., Soni S.P., Jones K.R., et al. Host cell plasma membrane phosphatidylserine regulates the assembly and budding of ebola virus. J. Virol. 2015;89:9440–9453. doi: 10.1128/JVI.01087-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Del Vecchio K., Frick C.T., Gc J B., Oda S.-I., Gerstman B.S., Saphire E.O., et al. A cationic, C-terminal patch and structural rearrangements in Ebola virus matrix VP40 protein control its interactions with phosphatidylserine. J. Biol. Chem. 2018;293:3335–3349. doi: 10.1074/jbc.M117.816280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adu-Gyamfi E., Soni S.P., Xue Y., Digman M.A., Gratton E., Stahelin R. The Ebola virus matrix protein penetrates into the plasma membrane: a key step in viral protein 40 (VP40) oligomerization and viral egress. J. Biol. Chem. 2013;288:5779–5789. doi: 10.1074/jbc.M112.443960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Amiar S., Stahelin R.V. The Ebola virus matrix protein VP40 hijacks the host plasma membrane to form virus envelope. J. Lipid Res. 2020;61:971. doi: 10.1194/jlr.ILR120000753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adu-Gyamfi E., Digman M.A., Gratton E., Stahelin R.V. Investigation of ebola VP40 assembly and oligomerization in live cells using number and brightness analysis. Biophys. J. 2012;102:2517–2525. doi: 10.1016/j.bpj.2012.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson K.A., Taghon G., Scott J.L., Stahelin R.V. The Ebola Virus matrix protein, VP40, requires phosphatidylinositol 4,5-bisphosphate (PI(4,5)P2) for extensive oligomerization at the plasma membrane and viral egress. Sci. Rep. 2016;6 doi: 10.1038/srep19125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scianimanico S., Schoehn G., Timmins J., Ruigrok R.H., Klenk H.D., Weissenhorn W. Membrane association induces a conformational change in the Ebola virus matrix protein. EMBO J. 2000;19:6732–6741. doi: 10.1093/emboj/19.24.6732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gc J.B., Gerstman B.S., Stahelin R.V., Chapagain P.P. The Ebola virus protein VP40 hexamer enhances the clustering of PI(4,5)P2 lipids in the plasma membrane. Phys. Chem. Chem. Phys. 2016;18:28409–28417. doi: 10.1039/c6cp03776c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson K.A., Bhattarai N., Budicini M.R., LaBonia C.M., Baker S.C.B., Gerstman B.S., et al. Cysteine mutations in the Ebolavirus matrix protein VP40 promote phosphatidylserine binding by increasing the flexibility of a lipid-binding loop. Viruses. 2021;13:1375. doi: 10.3390/v13071375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gc J B., Gerstman B.S., Chapagain P.P. Membrane association and localization dynamics of the Ebola virus matrix protein VP40. Biochim. Biophys. Acta. 2017;1859:2012–2020. doi: 10.1016/j.bbamem.2017.07.007. [DOI] [PubMed] [Google Scholar]
- 28.Jo S., Kim T., Iyer V.G., Im W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 2008;29:1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- 29.Phillips J.C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., et al. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Julkowska M.M., Rankenberg J.M., Testerink C. Liposome-binding assays to assess specificity and affinity of phospholipid-protein interactions. Methods Mol. Biol. 2013;1009:261–271. doi: 10.1007/978-1-62703-401-2_24. [DOI] [PubMed] [Google Scholar]
- 31.Digman M.A., Dalal R., Horwitz A.F., Gratton E. Mapping the number of molecules and brightness in the laser scanning microscope. Biophys. J. 2008;94:2320–2332. doi: 10.1529/biophysj.107.114645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Amiar S., Husby M.L., Wijesinghe K.J., Angel S., Bhattarai N., Gerstman B.S., et al. Lipid-specific oligomerization of the Marburg virus matrix protein VP40 is regulated by two distinct interfaces for virion assembly. J. Biol. Chem. 2021;296 doi: 10.1016/j.jbc.2021.100796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Husby M.L., Amiar S., Prugar L.I., David E.A., Plescia C.B., Huie K.E., et al. Phosphatidylserine clustering by the Ebola virus matrix protein is a critical step in viral budding. EMBO Rep. 2022;23 doi: 10.15252/embr.202051709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wijesinghe K.J., Urata S., Bhattarai N., Kooijman E.E., Gerstman B.S., Chapagain P.P., et al. Detection of lipid-induced structural changes of the Marburg virus matrix protein VP40 using hydrogen/deuterium exchange-mass spectrometry. J. Biol. Chem. 2017;292:6108–6122. doi: 10.1074/jbc.M116.758300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stahelin R.V., Scott J.L., Frick C.T. Cellular and molecular interactions of phosphoinositdes and peripheral proteins. Chem. Phys. Lipids. 2013;182:3–18. doi: 10.1016/j.chemphyslip.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Narkhede Y.B., Bhardwaj A., Motsa B.B., Saxena R., Sharma T., Chapagain P.P., et al. Elucidating residue level determinants affecting dimerization of Ebola virus matrix protein using high-throughput site saturation mutagenesis and biophysical approaches. J. Phys. Chem. B. 2023;127:6449–6461. doi: 10.1021/acs.jpcb.3c01759. [DOI] [PubMed] [Google Scholar]
- 37.Wijesinghe K.J., Stahelin R.V. Investigation of the lipid binding properties of the Marburg virus matrix protein VP40. J. Virol. 2015;90:3074–3085. doi: 10.1128/JVI.02607-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Winter S.L., Golani G., Lolicato F., Vallbracht M., Thiyagarajah K., Ahmed S.S., et al. The Ebola virus VP40 matrix layer undergoes endosomal disassembly essential for membrane fusion. EMBO J. 2023;42 doi: 10.15252/embj.2023113578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Han Z., Ruthel G., Dash S., Berry C.T., Freedman B.D., Harty R.N., et al. Angiomotin regulates budding and spread of Ebola virus. J. Biol. Chem. 2020;295:8596–8601. doi: 10.1074/jbc.AC120.013171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shepley-McTaggart A., Schwoerer M.P., Sagum C.A., Bedford M.T., Jaladanki C.K., Fan H., et al. Ubiquitin ligase SMURF2 interacts with filovirus VP40 and promotes egress of VP40 VLPs. Viruses. 2021;13:288. doi: 10.3390/v13020288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Han Z., Sagum C.A., Bedford M.T., Sidhu S.S., Sudol M., Harty R.N. ITCH E3 ubiquitin ligase interacts with Ebola virus VP40 to regulate budding. J. Virol. 2016;90:9163–9171. doi: 10.1128/JVI.01078-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mori H., Connell J.P., Donahue C.J., Boytz R.M., Nguyen Y.T.K., Leung D., et al. CAPG is required for Ebola virus infection by controlling virus egress from infected cells. Viruses. 2022;14:1903. doi: 10.3390/v14091903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bhattarai N., Pavadai E., Pokhrel R., Baral P., Hossen M.L., Stahelin R.V., et al. Ebola virus protein VP40 binding to Sec24c for transport to the plasma membrane. Proteins. 2022;90:340–350. doi: 10.1002/prot.26221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamayoshi S., Noda T., Ebihara H., Goto H., Morikawa Y., Lukashevich I.S., et al. Ebola virus matrix protein VP40 uses the COPII transport system for its intracellular transport. Cell Host Microbe. 2008;3:168–177. doi: 10.1016/j.chom.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Han Z., Madara J.J., Liu Y., Ruthel G., Freedman B.D., Harty R.N. ALIX rescues budding of a double PTAP/PPEY L-domain deletion mutant of Ebola VP40: a role for ALIX in Ebola virus egress. J. Infect. Dis. 2015;212:S138–S145. doi: 10.1093/infdis/jiu838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Silvestri L.S., Ruthel G., Kallstrom G., Warfield K.L., Swenson D.L., Nelle T., et al. Involvement of vacuolar protein sorting pathway in Ebola virus release independent of TSG101 interaction. J. Infect. Dis. 2007;196:S264–S270. doi: 10.1086/520610. [DOI] [PubMed] [Google Scholar]
- 47.Martin-Serrano J., Eastman S.W., Chung W., Bieniasz P.D. HECT ubiquitin ligases link viral and cellular PPXY motifs to the vacuolar protein-sorting pathway. J. Cell Biol. 2005;168:89–110. doi: 10.1083/jcb.200408155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yamayoshi S., Kawaoka Y. Mapping a region of Ebola virus VP40 that is important in the production of virus-like particles. J. Inf. Dis. 2007;196:S291–S295. doi: 10.1086/520595. [DOI] [PubMed] [Google Scholar]
- 49.Liang J., Djurkovic M.A., Shtanko O., Harty R.N. Chaperone-assisted selective autophagy targets filovirus VP40 as a client and restricts egress of virus particles. Proc. Natl. Acad. Sci. U. S. A. 2023;120 doi: 10.1073/pnas.2210690120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liang J., Sagum C.A., Bedford M.T., Sidhu S.S., Sudol M., Han Z., et al. Chaperone-mediated autophagy protein BAG3 negatively regulates Ebola and Marburg VP40-mediated egress. PLoS Pathog. 2017;13 doi: 10.1371/journal.ppat.1006132. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated in this study are included in the article and supplemental data. Further inquiries can be directed to rstaheli@purdue.edu.