Abstract
The Artemisia genus belongs to the Asteraceae family and is used in the treatment of many different diseases such as hepatitis and cancer. So far, around 500 species of Artemisia have been found in different regions of the world. Artemisinin is one of the medicinal compounds found in Artemisia species. Hence, this medical feature encourages researchers to pay attention to various species of this genus to discover more genetic and phytochemical information. In the present study, five species of Artemisia including A. fragrans, A. annua, A. biennis, A. scoparia, and A. absinthium were compared to each other in terms of the artemisinin content and other phytochemical components. Moreover, the relative expression profiles of eight genes related to the accumulation and synthesis of artemisinin [including 4FPSF, DBR2, HMGR1, HMGR2, WIRKY, ADS, DXS, and SQS] were determined in investigated species. The result of high-performance liquid chromatography (HPLC) analysis showed that the content of artemisinin in various species was in the order of A. fragrans > A. annua > A. biennis > A. scoparia > A. absinthium. Based on the gas chromatography–mass spectrometry (GC-MS) analysis, 34, 26, 26, 24, and 20 phytochemical compounds were identified for A. scoparia, A. biennis, A. fragrans, A. absinthum, and A. annua species, respectively. Moreover, camphor (38.86%), β-thujone (68.42%), spathulenol (48.33%), β-farnesene (48.16%), and camphor (29.04%) were identified as the considerable compounds A. fragrans, A. absinthium, A. scoparia, A. biennis, and A. annua species, respectively. Considering the relative expression of the targeted genes, A. scoparia revealed higher expression for the 4FPSF gene. The highest relative expression of the DBR2, WIRKY, and SQS genes was found in A. absinthium species. Moreover, A. annua showed the highest expression of the ADS and DXS genes than the other species. In conclusion, our findings revealed that various species of Artemisia have interesting breeding potential for further investigation of different aspects such as medicinal properties and molecular studies.
Keywords: Medicinal plants, Artemisia, Artemisinin, Chromatography, Gene expression
1. Introduction
The Artemisia L. genus is one of the important medicinal plants belonging to the Asteraceae family. This genus consists of 200–500 species at the specific or sub-specific level [1], which are mainly distributed in the temperate regions of Australia, North America, Asia, North Africa, and Europe [2]. Among them, Asia has the greatest number of Artemisia species [3]. The secondary metabolites extracted from the Artemisia species have critical roles in the treatment of several diseases such as cancer, malaria, inflammation, hepatitis, and infections by viruses, fungi, and bacteria [4]. It has been reported that Artemisia species have bitter tastes and characteristic strong aromas, which are caused by the presence of terpenes and sesquiterpene lactones [5]. Nevertheless, several phenolic compounds such as phenolic acids, flavonoids, and coumarins have been identified in various species [5]. Moreover, various species of this genus have been served to synthesize essential oil used in food commodities, medicine, and cosmetics, due to their remedial and therapeutic medicinal virtues [6].
Artemisinin is one of the most abundant secondary metabolites in the genus Artemisia [7]. This metabolite and its derivatives are all sesquiterpene lactones containing an unusual peroxide bridge. It has been reported that this endoperoxide 1,2,4-trioxane ring is responsible for its antimalarial properties [8]. Moreover, several studies revealed the anticancer effect of artemisinin on lung, leukemia, breast, prostate, and ovarian cancers [9]. Hence, the medicinal properties of Artemisia species are attributed to as this secondary metabolite. It has been proved that artemisinin is synthesized through two distinct pathways: the non-mevalonate (MEP) in the plastid and the mevalonate (MVA) in the cytosol [10]. The MVA pathway has a critical role in artemisinin biosynthesis. In this pathway, the first specific precursor of artemisinin is formed via the conversion of farnesyl diphosphate (FDP) into amorpha-4,11-diene by using an amorpha-4, 11-diene synthase (ADS) catalyzer [11]. In this conversion pathway, sterol and sesquiterpene production are regulated, simultaneously. The production of sesquiterpene is dependent on sesquiterpene cyclase (SQC), while in the other pathway (sterol production) squalene synthase (SQS) plays a key role. In other words, by decreasing sterol production artemisinin production will increase, and vice versa. The regulation of SQS and SQC genes has been demonstrated by using miconazole to limit SQS in A. annua [12]. All artemisinin pathways are not fully known, however, two commonly known pathways along with the main involved genes in each of them are shown in Fig. 1.
Fig. 1.
Artemisinin biosynthetic pathway. Art acid artemisinic acid, ADS amorpha-4, 11-diene synthase, CYP cytochrome P 450 CYP71AV1, DBR2 double bond reductase 2, Aldh1 aldehyde dehydrogenase 1, DXR 1-deoxyxylulouse 5-phosphate reductoisomerase, RED1 dihydroartemisinic aldehyde reductase 1, DXS 1-deoxyxylulose 5- phosphate synthase, FPS farnesyl diphosphate synthase, MEP nonmevalonate pathway, HMGR 3-hydroxy-3-methylglutaryl-CoA reductase, and HMG-CoA 3-hydroxy-3-methylglutaryl-CoA [13].
It is clear that various factors such as plant and cell organs, phytohormones, signaling molecules and pathways, and environmental conditions affect the production of artemisinin [13]. Moreover, up-or down-regulation of different involved genes in artemisinin production pathways also affected artemisinin production. For instance, Olofsson et al. [14] and Wang et al. [15] reported that various sesquiterpene synthases genes may have a negative effect on the amount of produced artemisinin as a result of competition for the substrate FDP. Hence, an investigation of the expression of genes related to artemisinin biosynthesis can complete our knowledge of increasing artemisinin content.
In general, several studies demonstrated that artemisinin biosynthesis is unique to A. annua species [12,15,16], whereas recent studies [[17], [18], [19], [20]] have revealed that other Artemisia species such as A. indica, A. afftangutica, A. bushriences, A. absinthium, A. parviflora, A. vulgaris, A. sieberi, A. cina, A. dracunculus, A. dubia, A. moorcroftiana, A. roxburghiana, and A. japonica can be used to the extraction of this important secondary metabolite. To our knowledge, there is no enough information on the artemisinin biosynthetic pathways in various Artemisia species. In the present study, we aimed to compare phytochemical profiles and the expression of eight genes involved in artemisinin biosynthesis among five different Artemisia species. We believe that the findings obtained in this study are important for natural products research in the near future.
2. Materials and methods
2.1. Plant materials and growth conditions
The seeds of five Artemisia species, including A. fragrans Will. (Accession No. IBRC P1000099), A. scoparia Waldst & Kit (Accession No. IBRC P1006514), A. biennis Rchb. (Accession No. IBRC P1006581), A. absinthium L. (Accession No. IBRC P1000003), and A. annua L. (Accession No. IBRC P1000008), were collected from the Iranian Biological Resource Center (IBRC). All seeds of five species were sterilized with 20% (v/v) sodium hypochlorite for 20 min and then washed three times using tap water as proposed by Liu et al. [21]. Next, sterilized seeds were planted in plastic experimental pots filled with perlite and peat moss. The experimental trays were transferred into a growth chamber with controlled conditions (photoperiod of 16 h and light intensity of 5000 1X LUX). After 45 days, seedlings were transferred into plastic pots filled with soil and sand (1:1 ratio) and until the flowering stage kept under the same controlled conditions with minor modification (photoperiod of 12 h and light intensity of 7000 1X LUX).
2.2. High-performance liquid chromatography (HPLC)
To prepare of plant extracts, 2 g of dried leaf samples were powdered and added to 50 mL of 60% acetonitrile, then kept in an ultrasonic water bath for 10 min (Elmasonic E30H, 60Hz, Germany). In the next step, extracts were filtered using Whatman filter paper 0.45 μm. The HPLC analysis was performed using a Knauer HPLC-DAD system (DAD detector, Azura, Germany) with an Eclipse-XBD-C18 column (4.6 mm ID × 250 mm (5 μm 80A, USA) at room temperature. The mobile phase was acetonitrile:water (60–40) at a flow rate of 1 mL min−1. The injected volume was 20 μL. The detection wavelength for artemisinin (Sigma-Aldrich, USA) was set at 260 nm [22]. The injection volume of the sample was 25 μL. The artemisinin accumulation was estimated from the standard curve of concentration versus the peak area.
2.3. Extract preparation and GC-MS analysis
0.5 g of dried and powdered leaves were extracted in 20 mL of methanol and maintained in an incubator shaker at 25 °C for 2 days. The extract was filtered using Whatman filter papers No. 1 and stored at 4 °C for further use. The phytochemical compounds of Artemisia species were identified using GC-MS (TRACE MS., TermoQuest-Finnigan) coupled to a 5973 MSD operated in electron impact mode at 70 eV ion source energy. The gas chromatograph was fitted with a DB-5 GC column (30 m length, 0.25 mm inner diameter, and 0.25 μm film thickness). The oven temperature was programmed initially at 60 °C and raised to 250 °C at a rate of 5 °C min−1. The injector and detector temperatures were set to 250 °C. The total run time of the sample was 40 min. Helium gas was used as the carrier with a flow rate of 1.1 mL min−1. The detection of phytochemical compounds in the extract was achieved using the matches percentages and commercial libraries of the National Institute of Standards and Technology (WILEY 9th edition, NIST-08 MS library, Gaithersburg, MD, USA).
2.4. RNA extraction, cDNA synthesis and real-time PCR
To investigate eight genes related to artemisinin accumulation (4FPSF, DBR2, HMGR1, HMGR2, WIRKY, ADS, DXS, and SQS) (Table 1), total RNA was extracted from young leaves of the investigated Artemisia species using DENAZIST ASIA kit (Tehran, Iran) according to the manufacturer's instructions. The concentration of the isolated RNA was determined using a Nano-Drop Spectrophotometers device (Thermo Scientific-2000C, USA). Then, cDNAs were synthesized using EasyTM cDNA Synthesis Kit (Parstos, Tehran, Iran) according to the manufacturer's instructions. Afterward, RT-qPCR analysis was carried out in a 12 μL volume containing 3.4 μL of RNAse-free water, 2 μL of cDNA (50 ng μL−1), 6 μL of 2 × RealQ Plus 2 × Master Mix Green (Ampliqon), and 0.3 μL of (0.3 l M) forward and reverse primers. Amplifications were run in a MiniOpticon™ Real-Time PCR device (Bio-Rad, USA) under the following steps: 95 °C for 10 min, 40 cycles of 95 °C for 10 s, 53–60 °C for 20 s, 72 °C for 30 s, and finally the temperature was increased from 65 °C to 95 °C by one degree per second. Normalization of the relative expression of examined genes was done using the Actin gene. Based on the CT values for each reaction, the relative expression for each investigated gene was calculated as suggested by Pfaffl [23].
Table 1.
The sequences of the selected genes related to Artemisinin accumulation.
| Gene | Forward/Reverse | Sequence | Reference |
|---|---|---|---|
| ADS | F | GTCGAATGGGCTGTCTCTGC | [24] |
| R | CCATCAATAACGGCCTTGGA | ||
| DBR2 | F | CATCAACAAGCAAGCCCATTTC | [24] |
| R | GCGATAGTCTTCAACCACCTC | ||
| HMGR-1 | F | GGTCAGGATCCGGCCCAAAACATT | [25] |
| R | CCAGCCAACACCGAACCAGCAACT | ||
| HMGR-2 | F | TGCTGGTTCTCTTGGTGGAT | [22] |
| R | CTCCAACTGTGCCAACCTCT | ||
| WIRKY | F | CAAGAACTACCAAGACCGAATCC | [26] |
| R | GGAGATAACAGGTGGCGAATAGAC | ||
| DXS | F | ATGGGTTGGCGGGATTCAC | [25] |
| R | CCGTCAAGATTGGCAGTAGGTAAA | ||
| FPS | F | GTATGATTGCTGCGAACGATGGA | [25] |
| R | CGGCGGTGAATAGACAATGAATAC | ||
| SQS | F | TTTGAAAGCAGTATTGAAACAC | [27] |
| R | CAGACAGCATCACGAAGC | ||
| Actin | F | AGTGCTCCTGGTTAGTTGTC | [27] |
| R | CTTGTTGCCTCGTAATCTTCG |
2.5. Statistical analysis
Based on the artemisinin content detected by HPLC, the differences among the studied species were investigated. An analysis of variance (ANOVA) was performed for the relative expression data based on a completely randomized design with three replications. Mean values were compared by using Duncan's multiple range test at the significant probability level for each experimental treatment. Data were analyzed by IBM SPSS Statistics software ver. 26 (Armonk, NY, USA). To investigate the association between the relative expression of studied genes and artemisinin content in each Artemisia species, Pearson's correlation coefficients were determined using IBM SPSS software ver. 26.
3. Results
3.1. Artemisinin content
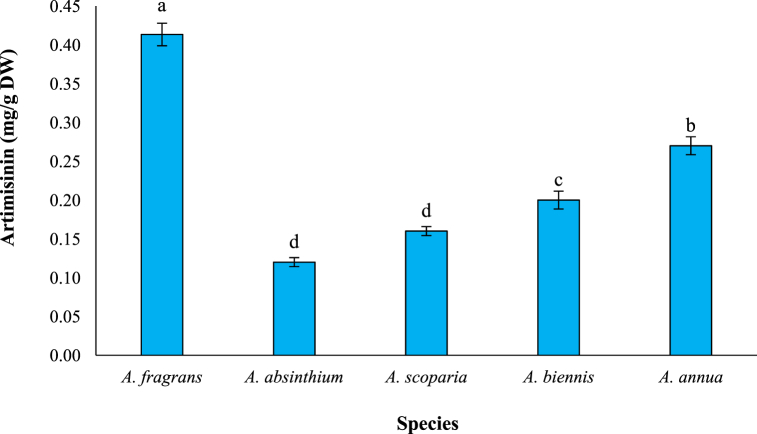
In the present study, artemisinin content was estimated using the HPLC with a specific standard (Fig. 2A). As shown in Fig. 2B, there was significant difference among investigated species in term of artemisinin content (P ≤ 0.01). The results demonstrated that A. fragrans has a significant amount of this compound (0.41 mg/g DW), followed by A. annua (0.27 mg/g DW), A. biennis (0.20 mg/g DW), A. scoparia (0.16 mg/g DW), and A. absinthium (0.12 mg/g DW). Indeed, the amount of artemisinin in A. fragrans was 1.52, 2.05, 2.56, and 3.42 times higher than in A. annua, A. biennis, A. scoparia, and A. absinthium, respectively (Fig. 3).
Fig. 2.
The HPLC chromatogram for the standard (A) and extracted samples for Artemisia species (B).
Fig. 3.
The artemisinin content in different Artemisia species. Different letters show significant differences among species at 0.01 the probability level.
3.2. GC-MS analysis
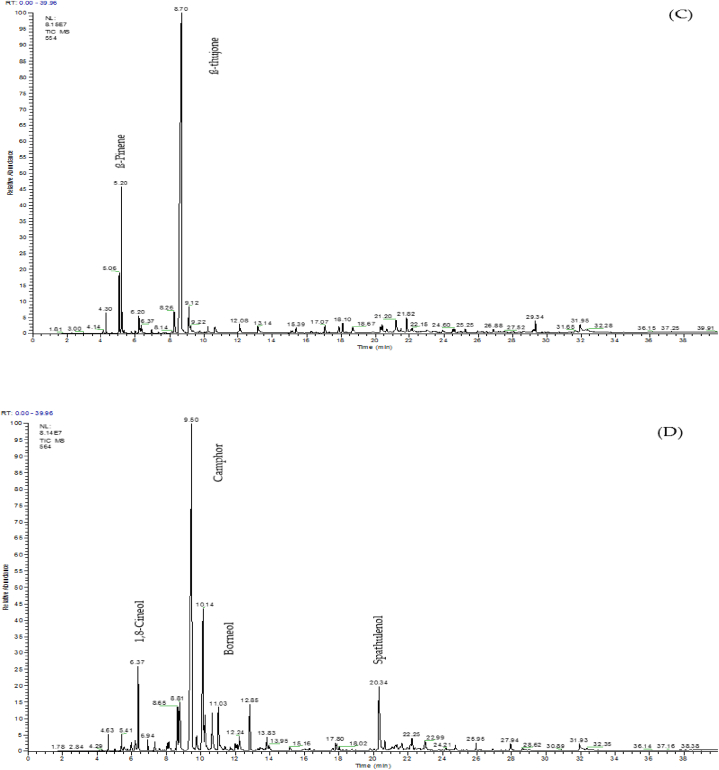
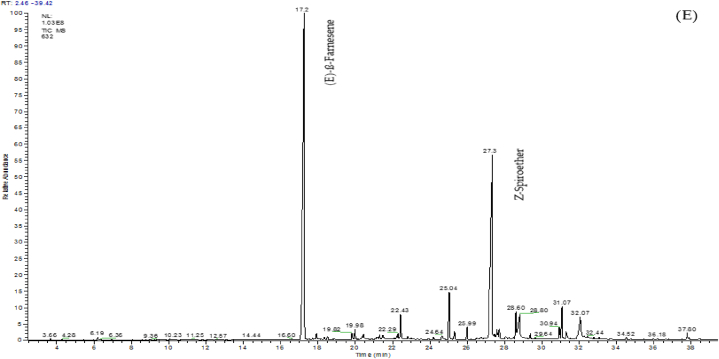
A GC-MS analysis was performed to identify different phytochemical compounds in five investigated Artemisia species. Fig. 4 shows the rendered chromatograms for each species. Moreover, details of identified phytochemical compounds in each species are presented in Table 2. According to results, there was found many different phytochemical compounds in the investigated species. As a result, 34, 26, 26, 24, and 20 phytochemical compounds were identified for the species A. scoparia, A. biennis, A. fragrans, A. absinthum, and A. annua, respectively. In A. fragrans, the most compounds were related to camphor (38.86%), followed by borneol (14.28%) and β-terpineol (8.17%). The most identified compounds in A. absinthium were β-thujone (68.42%), followed by β-pinene (8.61%), and sabinene (3.83%). In A. scoparia species, spathulenol (48.33%), palmitic acid (4.47%), and oleic acid (4.31%) were identified as the most phytochemical compounds; whereas, the most compounds in A. biennis were β-farnesene (48.16%), spiroether (28.53%), and Octadecenoic acid (3.63%). In A. annua species, camphor (29.04%), β-selenin (27.87%), and trans-pinocarveol (7.54%) were highlighted as the dominant compounds. Among the identified compounds, p-Cymene and 1, 8-Cineol were common compounds in all investigated species. Species A. fragrans, A. absinthium, and A. scoparia were common in terms of 4-Terpineol and Spathulenol compounds. Moreover, the common compounds in A. scoparia, A. biennis, and A. annua were Hexahydrofarnesyl acetone and Palmitic acid. The Limonene and (E)-β-Farnesense were common compounds in A. fragrans, A. absinthium, and A. scoparia species. Other common compounds in various species are presented in Table 2.
Fig. 4.
Chromatograms of five different Artemisia species. (A) A. annua, (B) A. scoparia, (C) A. absinthium, (D) A. fragrans, and (E) A. biennis.
Table 2.
Phytochemical compounds in the five Iranian Artemisia species detected using GC-MS analysis.
| No. | RT (min) | Compounds name | Area (%) |
||||
|---|---|---|---|---|---|---|---|
| A. fragrans | A. absinthium | A. scoparia | A. biennis | A. annua | |||
| 1 | 4.14 | α-thujene | – | 0.15 | – | – | – |
| 2 | 4.31 | α-Pinene | – | 1.02 | 0.56 | 0.07 | 5.26 |
| 3 | 4.63 | Camphene | 0.67 | – | – | – | 2.28 |
| 4 | 4.69 | Thuja-2,4(10)-diene | – | – | – | – | 0.12 |
| 5 | 5.06 | Sabinene | – | 3.83 | – | – | – |
| 6 | 5.17 | β-Pinene | – | 8.61 | 2.12 | – | 0.44 |
| 7 | 5.3 | β-Myrcene | – | – | 0.1 | – | – |
| 8 | 5.41 | dehydro-1,8-Cineole | 0.71 | – | – | – | – |
| 9 | 5.98 | α-Terpinene | 0.33 | – | – | – | – |
| 10 | 6.2 | p-Cymene | 0.57 | 1.12 | 0.62 | 0.15 | 0.66 |
| 11 | 6.25 | Limonene | – | 0.33 | 1.18 | 0.04 | – |
| 12 | 6.38 | 1,8-Cineol | 4.05 | 0.5 | 0.31 | 0.04 | 6.66 |
| 13 | 6.95 | γ-Terpinene | 0.56 | 0.18 | – | 0.05 | – |
| 14 | 7.34 | Cis-Sabinene hydrate | 0.54 | – | – | – | – |
| 15 | 8.05 | Filifolone | 0.4 | – | – | – | – |
| 16 | 8.16 | Trans-Sabinene hydrate | 0.8 | – | – | – | – |
| 17 | 8.22 | cis-Thujone | – | – | 0.11 | – | – |
| 18 | 8.26 | α-thujone | – | 2.21 | – | – | – |
| 19 | 8.51 | trans-Thujone | – | – | 0.53 | – | – |
| 20 | 8.63 | Chrysanthenone | 2.83 | – | 0.19 | – | – |
| 21 | 8.7 | β-thujone | – | 68.42 | – | – | – |
| 22 | 8.72 | 1-Terpineol | 4.59 | – | 0.14 | – | – |
| 23 | 8.78 | α-Campholenal | – | – | – | – | 0.24 |
| 24 | 9.12 | Thujanol <neo-3-> | – | 2.24 | – | – | – |
| 25 | 9.35 | trans-Pinocarveol | – | 0.87 | 0.32 | – | 7.54 |
| 26 | 9.39 | β-Terpineol | 8.17 | – | – | – | – |
| 27 | 9.49 | Camphor | 38.86 | – | 4.25 | – | 29.04 |
| 28 | 9.79 | Pinocarvone | 1.1 | – | 0.12 | – | 4.81 |
| 29 | 10.03 | Borneol | 14.28 | – | 0.31 | – | – |
| 30 | 10.06 | 4-Terpineol | 1.73 | 0.59 | 0.18 | – | 1.54 |
| 31 | 10.64 | Myrtenol | – | 0.52 | 0.59 | – | – |
| 32 | 10.65 | Myrtenal | – | – | – | – | 1.21 |
| 33 | 10.67 | α-Terpineol | 2.91 | – | – | – | – |
| 34 | 11.03 | Trans-Piperitol | 3.12 | – | – | – | – |
| 35 | 12.08 | Cis-Chrysanthenyl acetate | – | – | 0.19 | – | – |
| 36 | 12.08 | Carvotanacetone | – | 0.83 | – | – | – |
| 37 | 12.24 | Piperitone | 1.07 | – | – | – | – |
| 38 | 12.83 | Bornyl acetate | 2.67 | – | 0.28 | – | – |
| 39 | 13.14 | E-Anethole | – | 1.06 | – | – | – |
| 40 | 15.39 | β-Bourbonene | – | 0.3 | – | – | – |
| 41 | 16.02 | Methyl eugenol | – | – | 0.73 | – | – |
| 42 | 16.33 | trans- Caryophyllene | – | – | 1.95 | – | – |
| 43 | 17.06 | (E)-β-Farnesene | – | 0.45 | 0.5 | 48.16 | – |
| 44 | 17.86 | Ar-Curcumene | – | – | 2.04 | 0.5 | – |
| 45 | 17.88 | Germacrene D | – | 0.48 | – | – | – |
| 46 | 18.2 | β-Selinene | – | 0.73 | – | – | 27.87 |
| 47 | 18.34 | Ledene | – | – | – | 0.13 | – |
| 48 | 18.48 | β-Bisabolene | – | – | 0.41 | 0.16 | – |
| 49 | 18.75 | δ-Cadinene | – | – | 0.74 | – | – |
| 50 | 19.82 | (E)-Nerolidol | – | – | – | 0.48 | – |
| 51 | 20.08 | Citronellyl butanoate | – | – | 0.97 | – | – |
| 52 | 20.44 | Caryophyllene oxide | 0.61 | 0.71 | – | 0.78 | 4.99 |
| 53 | 20.46 | Globulol | – | – | – | 0.42 | – |
| 54 | 20.5 | Spathulenol | 4.74 | 0.4 | 48.33 | – | – |
| 55 | 20.66 | Salvial-4(14)-en-1-one | 0.49 | – | – | – | – |
| 56 | 20.95 | Bornyl angelate | – | – | – | – | 0.5 |
| 57 | 20.99 | Ledol | – | – | 0.59 | – | – |
| 58 | 21.11 | Humulene epoxide II | – | – | 0.81 | – | – |
| 59 | 21.48 | Ledeneoxide II | – | – | – | 0.3 | – |
| 60 | 21.65 | Cadin-4-en-7-ol | – | – | – | – | 0.74 |
| 61 | 21.77 | Caryophylla-4(12),8(13)-dien-5α-ol | – | – | – | – | 0.73 |
| 62 | 21.82 | α-epi-Cadinol | – | 1.29 | – | – | – |
| 63 | 22.15 | α-Cadinol | – | 0.49 | – | – | – |
| 64 | 22.25 | neo-Intermedeol | 0.82 | – | – | 0.38 | – |
| 65 | 22.43 | Eudesm-7(11)-en-4-ol | – | – | – | 1.81 | – |
| 66 | 22.57 | epoxide Aromadendrene | – | – | 0.73 | – | – |
| 67 | 22.59 | Iso-Aromadendrene epoxide | – | – | – | – | 1.11 |
| 68 | 22.63 | Isospathulenol | – | – | 1.39 | – | – |
| 69 | 24.98 | Diazinone | – | – | 0.65 | 3.11 | – |
| 70 | 25.31 | (2Z,6E)-Farnesyl acetate | – | – | – | 0.43 | – |
| 71 | 25.95 | Hexahydrofarnesyl acetone | 0.4 | – | 0.67 | 0.75 | 0.65 |
| 72 | 27.35 | Z-Spiroether | – | – | – | 28.53 | – |
| 73 | 27.58 | E-Spiroether | – | – | – | 0.86 | – |
| 74 | 27.72 | Methyl palmitate | – | – | – | 0.57 | – |
| 75 | 28.63 | Palmitic acid | – | – | 4.47 | 3.43 | 0.58 |
| 76 | 30.94 | Linoleic acid, methyl ester | – | – | – | 0.72 | – |
| 77 | 31.07 | Methyl linolenate | – | – | – | 1.94 | – |
| 78 | 31.29 | Phytol | – | – | – | 0.77 | – |
| 79 | 32.05 | oleic acid | 0.87 | – | 4.31 | – | – |
| 80 | 32.07 | 9-Octadecenoic acid, (E) | – | – | – | 3.63 | – |
3.3. Expression of the genes involved in the metabolite biosynthesis pathway
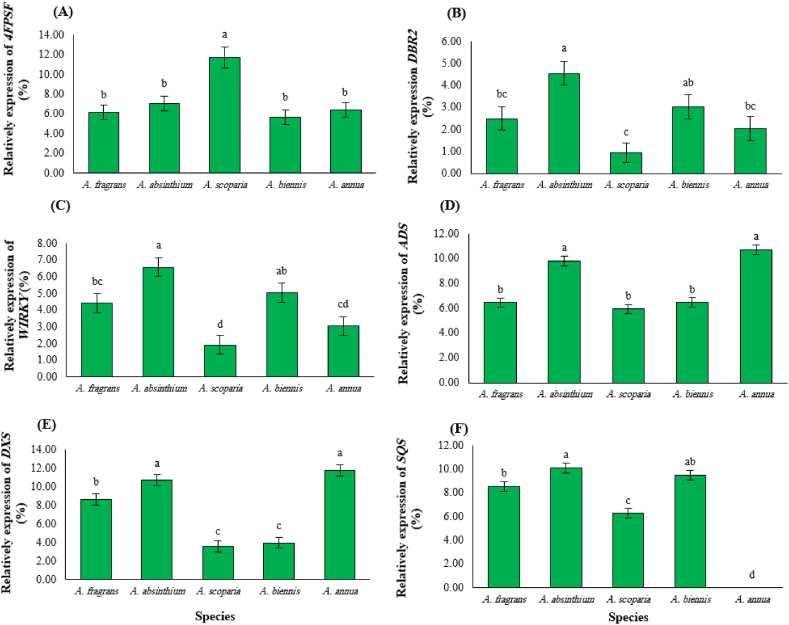
The relative expression of eight genes including 4FPSF, DBR2 (artemisinic aldehyde Delta (11(13)) reductase), HMGR1 (3-hydroxy-3-methyl-glutaryl-CoA reductase 1), HMGR2 (3-hydroxy-3-methyl-glutaryl-CoA reductase 2), WIRKY, ADS (amorpha-4, 11-diene synthase), DXS (1-deoxy-d-xylulose 5-phosphate synthase), and SQS (squalene synthase) were investigated using qPCR technique. The result of ANOVA showed significant differences among investigated species in terms of the relative expression of 4FPSF, DBR2, WIRKY, ADS, and SQS genes (Table 3). A. scoparia had a higher relative expression of 4FPSF (11.75%), which was 1.89, 1.66, 2.06, and 1.83 times higher than A. fragrans (6.20%), A. absinthium (7.08%), A. biennis (5.71%), and A. annua (6.42%), respectively (Fig. 5A). The expression of DBR2, HMGR1, HMGR2, WIRKY, and SQS in A. absinthium was higher than other species. In A. absinthium, the expression of DBR2 (4.56%) was 1.84-, 4.80-, 1.50-, and 2.23-fold higher than A. fragrans, (2.50%), A. scoparia (0.95%), A. biennis (3.04%), and A. annua (2.04%), respectively (Fig. 5B). Moreover, the expression of WIRKY (6.58%) was 1.48-, 3.41-, 1.30-, and 2.16-fold higher than A. fragrans, (4.44%), A. scoparia (1.93%), A. biennis (5.06%), and A. annua (3.05%), respectively (Fig. 5C); and the expression of SQS (10.11%) was 1.18-, 1.61-, and 1.06-fold higher than A. fragrans, (8.54%), A. scoparia (6.26%), and A. biennis (9.49%), respectively (Fig. 5F). The maximum number of transcripts for ADS and DXS genes were found in A. annua. Accordingly, the expression of ADS (10.71%) was 1.66-, 1.09-, 1.81-, and 1.65-fold higher than A. fragrans, (2.50%), A. absinthium (7.08%), A. scoparia (1.93%), and A. biennis (5.06%), respectively (Fig. 5D). Moreover, the expression of DXS (11.78%) was 1.36-, 1.10-, 3.25-, and 2.96-fold higher than A. fragrans, (8.63%), A. absinthium (10.74%), A. scoparia (3.62%), and A. biennis (3.98%), respectively (Fig. 5E).
Table 3.
Analysis of variance was conducted for the relative expression data of eight investigated genes in the five Iranian Artemisia species.
| Gene | Source of variation |
Gene | Source of variation |
||
|---|---|---|---|---|---|
| Species (df = 4) | Error (df = 10) | Species (df = 4) | Error (df = 10) | ||
| 4FPSF | 18.207 ** | 1.973 | WIRKY | 9.702 ** | 0.989 |
| DBR2 | 5.318 ** | 0.822 | ADS | 14.627 ** | 0.419 |
| HMGR1 | 4.919 ns | 2.173 | DXS | 42.919 ** | 1.068 |
| HMGR2 | 4.406 ns | 1.169 | SQS | 50.790 ** | 0.391 |
ns, *, and **: Non-significant, and significant at P < 0.05 and P < 0.01, respectively.
Fig. 5.
The relative expression of genes including 4FPSF, DBR2, WIRKY, ADS, DXS, and SQS in different Artemisia species. Different letters show significant differences among species at 0.01 the probability level.
3.4. Correlation analysis
To investigate relationships between artemisinin content and the relative expression of studied genes, Pearson's correlation analysis was performed based on the obtained data for each species, and the results are presented in Table 4. In A. fragrans and A. absinthium species, there was no significant correlation between artemisinin content and expression of studied genes. However, the relative expression of HMGR1 with DRB2 and SQS with ADS showed a positive and significant correlation. In A. scoparia specie, artemisinin content positively and significantly correlated with the relative expression of DBR2 and HMGR1 genes. Moreover, a significant and positive correlation was found between expression of HNGR1 with DBR2, and SQS with ADS genes. Similar to A. fragrans and A. absinthium species, there was found a positive and significant correlation between the relative expression of HMGR1 with DRB2 and SQS with ADS genes. In A. annua species, a positive and significant correlation was only found between expression of HMGR1 and DBR2 genes.
Table 4.
Correlation coefficients among artemisinin content and the relative expression of studied genes in different Artemisia species.
| Species | Variables | Artemisinin | 4FPSF | DBR2 | WIRKY | HMGR1 | HMGR2 | ADS | DXS | SQS |
|---|---|---|---|---|---|---|---|---|---|---|
| A. fragrans | Artemisinin | |||||||||
| 4FPSF | 0.23 | |||||||||
| DBR2 | 0.97 | 0.46 | ||||||||
| WIRKY | −0.06 | 0.95 | 0.18 | |||||||
| HMGR1 | 0.96 | 0.48 | 1** | 0.21 | ||||||
| HMGR2 | 0.57 | 0.93 | 0.75 | 0.78 | 0.77 | |||||
| ADS | 0.87 | 0.67 | 0.97 | 0.42 | 0.97 | 0.89 | ||||
| DXS | 0.75 | 0.81 | 0.89 | 0.60 | 0.91 | 0.96 | 0.98 | |||
| SQS | 0.91 | 0.62 | 0.98 | 0.36 | 0.99 | 0.86 | 0.99 | 0.96 | ||
| A. absinthium | Artemisinin | |||||||||
| 4FPSF | 0.34 | |||||||||
| DBR2 | 0.99 | 0.46 | ||||||||
| WIRKY | 0.05 | 0.96 | 0.18 | |||||||
| HMGR1 | 0.98 | 0.48 | 1** | 0.21 | ||||||
| HMGR2 | 0.66 | 0.93 | 0.75 | 0.78 | 0.77 | |||||
| ADS | 0.93 | 0.67 | 0.97 | 0.42 | 0.97 | 0.89 | ||||
| DXS | 0.83 | 0.81 | 0.89 | 0.60 | 0.91 | 0.97 | 0.97 | |||
| SQS | 0.95 | 0.62 | 0.98 | 0.36 | 0.98 | 0.86 | 0.99** | 0.96 | ||
| A. scoparia | Artemisinin | |||||||||
| 4FPSF | 0.59 | |||||||||
| DBR2 | 0.99** | 0.63 | ||||||||
| WIRKY | 0.05 | 0.83 | 0.08 | |||||||
| HMGR1 | 1** | 0.61 | 1** | 0.06 | ||||||
| HMGR2 | 0.66 | 0.99 | 0.68 | 0.78 | 0.67 | |||||
| ADS | 0.93 | 0.85 | 0.94 | 0.42 | 0.93 | 0.89 | ||||
| DXS | 0.83 | 0.94 | 0.85 | 0.60 | 0.83 | 0.97 | 0.98 | |||
| SQS | 0.95 | 0.82 | 0.96 | 0.36 | 0.95 | 0.86 | 0.99** | 0.96 | ||
| A. biennis | Artemisinin | |||||||||
| 4FPSF | −0.34 | |||||||||
| DBR2 | −0.99 | 0.46 | ||||||||
| WIRKY | −0.05 | 0.95 | 0.17 | |||||||
| HMGR1 | −0.98 | 0.48 | 1** | 0.21 | ||||||
| HMGR2 | −0.66 | 0.93 | 0.75 | 0.78 | 0.77 | |||||
| ADS | −0.92 | 0.67 | 0.96 | 0.42 | 0.97 | 0.89 | ||||
| DXS | −0.82 | 0.81 | 0.89 | 0.60 | 0.91 | 0.97 | 0.98 | |||
| SQS | −0.95 | 0.62 | 0.98 | 0.36 | 0.98 | 0.86 | 0.99** | 0.96 | ||
| A. anuua | Artemisinin | |||||||||
| 4FPSF | 0.34 | |||||||||
| DBR2 | 0.99 | 0.46 | ||||||||
| WIRKY | 0.05 | 0.96 | 0.18 | |||||||
| HMGR1 | 0.98 | 0.49 | 1** | 0.21 | ||||||
| HMGR2 | 0.66 | 0.93 | 0.75 | 0.78 | 0.77 | |||||
| ADS | 0.93 | 0.67 | 0.97 | 0.42 | 0.97 | 0.89 | ||||
| DXS | 0.83 | 0.81 | 0.89 | 0.60 | 0.91 | 0.97 | 0.98 | |||
| SQS† |
** significant at P < 0.01.
4. Discussion
There are more than 300 species of Artemisia in the world [28]. Artemisia species differ from each other in terms of chemical composition, but some compounds are present in all of them. The common characteristic of these species is sesquiterpen lactones. Artemisinin is a known sesquiterpen lactone that exists in A. annua, A. abrotanum, and A. vulgaris [29]. Similar to sesquiterpen lactones, the flavonoid composition is different in different species. The most common flavonoids of the Artemisia are artemetin and casticin, which are identified in the extracts of A. abrotanum, A. absinthium, and A. annua [30]. In addition, another common group of metabolites in Artemisia is coumarins [31]. In the present study, artemisinin content varied between 0.12 and 0.41 mg/g DW. The species A. fragrans revealed the highest amount of artemisinin, followed by A. annua, A. biennis, A. scoparia, and A. absinthium. Similarly, Salehi et al. [32] reported a wide range of variability in artemisinin content across different species of Artemisia. Various environmental and genetic factors affect the amount of artemisinin in Artemisia species [20]. Many genes are involved in artemisinin synthesis, and changes in their expression increase or decrease the amount of artemisinin. However, the extent of this change in gene expression and artemisinin biosynthesis also varies depending on the species and growth stages [33]. Artemisinin has been found in about 40 different species of Artemisia, varying from 0.0005% to 38.1% depending on the growth stage and plant organ [34]. In different studies, the highest amount of artemisinin was obtained in the A. annua species followed by the A. deserti, A. marschalliana, and A. absinthium [[35], [36], [37]]. Nomonov et al. [33] studied seven species of Artemisia in Tajikistan and found that the amount of artemisinin is between 0.07% and 0.45% of the dry mass. Salehi et al. [22] reported that A. deserti had 5.13 mg/g DW artemisinin. Salih et al. [32] showed that the leaf extract of A. sieberi, A. Judaica, and A. monosperma had about 3.01, 2.5, and 1.9 mg/g DW of artemisinin, respectively.
Plants use secondary metabolites to deal with both biotic and abiotic stresses. These compounds have medicinal properties and are used in the pharmaceutical industry [38]. So far, many of these medicinal compounds in nature have not been identified and many studies should be done for this purpose [39]. Different species of Artemisia are among the most important plants that have medicinal compounds and play an important role in traditional medicine [28]. Based on the result of GC-MS analysis, many bioactive compounds were found in different investigated species. Indeed, the high rate of variability in identified phytochemical compounds may be caused by the nature and physicochemical response of different species to environmental conditions. In addition, genetic, environmental, and seasonal factors can also be effective [40,41]. In the present study, the investigated species also differed in terms of their compositions. For example, monoterpenoids are abundant in the essential oils of A. abrotanum, A. absinthium, A. annua, and A. vulgaris, while phenylpropanoids are predominant in the essential oil of A. dracunculus [42]. However, our results indicated that alcohol esters of thujyl, α-thujone, β-thujone, camphene, (Z)-epoxycymene, trans-sabinyl acetate, and chrysanthyl acetate are common components in A. absinthium, which is in accordance with the findings reported by Kazemi et al. [43]. The most common monoterpenoids found in A. abrotanum are 1-terpineol, trans-piperitol, 1,8-cineole, and camphor [42]. While, camphene, camphor, β-pinene, borneol, and cuminal are commonly in A. annua [44]. Sabinene, terpinen-4-ol, β-osimene, cis-osimene, α-trans-osimene, limonene, α-flandrin, β-flandrin, (Z)-artemidin, and capylene have been identified in A. dracunculus [45]; and 1,8-cineole, sabinene, camphor, camphene, caryophyllene oxide, α-thugone, and β-thugone were founded in A. vulgaris [28]. From the geographical viewpoint, there are significant difference in phytochemical compositions. For instance, A. vulgaris sampled from the Republic of Bashkortostan have large amounts of Cpinene, trans chrysanthenol, 5-pinene, C-myrcene, and [46]. Mucciarelli et al. [47] emphasized that camphene and camphor are two main compounds in Italian A. vulgaris, and Pino et al. [48] reported caryophyllene oxide as the main compound of this plant from Cuba.
Genes play an important role in the synthesis of secondary metabolites, and increasing or decreasing their expression has a great impact on the production of secondary metabolites. For example, in a study, SQS gene silencing increased artemisinin synthesis in A. annua [26]. In addition, it has been reported that blocking the synthesis pathways of other secondary metabolites increases the synthesis of artemisinin [49]. In our study, the highest expression of the SQS gene was observed in the A. absinthium species, while A. fragrans, which had the highest content of artemisinin, showed the lowest relative expression of the studied genes. These results further confirmed the HPCL analysis (Fig. 3, Fig. 5F). Therefore, this gene has a negative effect on artemisinin biosynthesis. ADS and DBR2 genes play an important role in the production of artemisinin, and reducing their expression levels will reduce its production. The possible reason for the decrease in the expression of some studied genes in the mentioned species can be due to the decrease in the precursor material required for the activity of enzymes and the biosynthesis of artemisinin. Indeed, if the precursor material for artemisinin biosynthesis is consumed and becomes the final product, the expression of genes involved in this pathway will decrease. These results are further supported by the correlation analysis, where there was no significant association between artemisinin content and the relative expression of ADS and DBR2 genes (Table 4). In previous studies, it has been reported that the A. annua species has a lower amount of artemisinin than other species, which is due to the low expression of these two genes [22,26]. Yuan et al. [50] reported that overexpression of DBR2 gene in transgenic A. annua resulted in increasing the artemisinin concentration. Indeed, this finding was not in accordance with our results. As shown in Fig. 3, Fig. 5B, there was no association between artemisinin content and the relative expression of DBR2 gene, especially in A. annua species. Transcription factors regulate the synthesis of secondary metabolites by binding to cis-acting regulatory elements in promoters. The WIRKY transcription factor is one of the important transcription factors in the production of secondary metabolites, which affects the synthesis of artemisinin through binding to the W-box in the ADS promoter and activating its expression [51]. In another study, the overexpression of the WIRKY gene caused an increase in the expression of the CYP71AV gene, but it did not have a significant effect on the transcription of the ADS and DBR2 genes [52]. However, we found a positive association between expression of WIRKY and ASD genes across all investigated species (Fig. 5C and D). In general, our findings indicated that a large class of phytochemical compounds are available in various Artemisia species, which may be responsible for many pharmacological activities. Hence, further work to discover pharmacological activities in various species is required.
5. Conclusions
According to the present study, the results conclude that artemisinin content depends on genetic background. In contrast to numerous studies that reported A. annua has a considerable artemisinin content, A. fragrans showed the highest amount of artemisinin than other species. Moreover, we found a direct association between artemisinin accumulations in different species with the relative expression of artemisinin accumulation-related genes in them. The result of GC-MS analysis identified 80 phytochemical compounds in five investigated species, which in turn emphasized more attention to these species in search of novel medicinal properties.
Data availability statement
Data will be made available on request.
CRediT authorship contribution statement
Bita Jamshidi: Writing – original draft, Software, Investigation. Alireza Etminan: Methodology, Conceptualization. Alimehras Mehrabi: Methodology, Conceptualization. Lia Shooshtari: Investigation, Conceptualization. Alireza Pour-Aboughadareh: Writing – review & editing, Software.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Contributor Information
Bita Jamshidi, Email: bita.alimer@epu.edu.iq.
Alireza Etminan, Email: alietminan55@yahoo.com.
Alimehras Mehrabi, Email: alimehrasmehrabi@yahoo.com.
Lia Shooshtari, Email: Lia.shooshtari@iau.ac.ir.
Alireza Pour-Aboughadareh, Email: a.poraboghadareh@edu.ikiu.ac.ir.
References
- 1.Badr A., El-Shazly H.H., Helail N.S., Ghanim W. Genetic diversity of Artemisia populations in central and north Saudi Arabia based on morphological variation and RAPD polymorphism. Plant Systemat. Evol. 2012;298:871–886. [Google Scholar]
- 2.Abad M.J., Bedoya L.M., Apaza L., Bermejo P. The Artemisia L. genus: a review of bioactive essential oils. Molecules. 2012;17:2542–2566. doi: 10.3390/molecules17032542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hamidi F., Karimzadeh G., Rashidi Monfared G., Salehi M. Assessment of Iranian endemic Artemisia khorassanica: karyological, genome size, and gene expressions involved in artemisinin production. Turk. J. Biol. 2018;42:322–333. doi: 10.3906/biy-1802-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Willcox M. Artemisia species: from traditional medicines to modern antimalarials-and back again. J. Alternative Compl. Med. 2009;15:101–109. doi: 10.1089/acm.2008.0327. [DOI] [PubMed] [Google Scholar]
- 5.Trifan A., Zengin G., Sinan K.I., Sieniawska E., Sawicki R., Maciejewska-Turska M., Skalikca-Wozniak K., Luca S.V. Unveiling the phytochemical profile and biological potential of five Artemisia species. Antioxidants. 2022;11:1017. doi: 10.3390/antiox11051017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bisht D., Kumar D., Kumar D., Dua K., Chellappan D.K. Phytochemistry and pharmacological activity of the genus Artemisia. Arch Pharm. Res. (Seoul) 2021;44:439–474. doi: 10.1007/s12272-021-01328-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arsenault P.R., Wobbe K.K., Weathers P.J. Recent advances in artemisinin production through heterologous expression. Curr. Med. Chem. 2008;15:2886–2896. doi: 10.2174/092986708786242813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown G.D., Sy L.K. In vivo transformations of artemisinic acid in Artemisia annua plants. Tetrahedron. 2007;63:9548–9566. [Google Scholar]
- 9.Liu S., Tian N., Li J., Huang J., Liu Z. Isolation and identification of novel genes involved in artemisinin production from flowers of Artemisia annua using suppression subtractive hybridization and metabolite analysis. Planta Med. 2009;74:1542–1547. doi: 10.1055/s-0029-1185809. [DOI] [PubMed] [Google Scholar]
- 10.Mohadjerani M., Rasolizadeh A. Evaluation of kinetic parameters of acetylcholinesterase from the bovine brain and its inhibition with Mentha pulegium L. extract. J. Ilam Univ. 2020;27:74–87. [Google Scholar]
- 11.Kim S.H., Chang Y.J., Kim S.U. Tissue specificity and developmental pattern of amorpha-4, 11-diene synthase (ADS) proved by promoter-driven GUS expression in the heterologous plant Arabidposis thaliana. Planta Med. 2008;74:188–193. doi: 10.1055/s-2008-1034276. [DOI] [PubMed] [Google Scholar]
- 12.Weathers P.J., Elkholy S., Wobbe K.K. Artemisinin: the biosynthetic pathway and its regulation in Artemisia annua, a terpenoid-rich species, in Vitro Cell. Dev. Pol. 2006;42:309–317. [Google Scholar]
- 13.Nguyen K.T., Arsenault P.R., Weathers P.J. Trichomes + roots + ROS = artemisinin: regulating artemisinin biosynthesis in Artemisia annua L., in Vitro Cell. Dev. Biol-Plant. 2011;47:329–338. doi: 10.1007/s11627-011-9343-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Olofsson L., Engström A., Lundgren A., Brodelius P.E. Relative expression of genes of terpene metabolism in different tissues of Artemisia annua L. BMC Plant Biol. 2011;11:45–57. doi: 10.1186/1471-2229-11-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang H., Ma C., Li Z., Ma L., Wang H., Ye H., Xu G., Liu B. Effects of exogenous methyl jasmonate on artemisinin biosynthesis and secondary metabolites in Artemisia annua L. Ind. Crops Prod. 2009;31:214–218. [Google Scholar]
- 16.Wang H., Liu Y., Chong K., Liu B.Y., Ye H.C., Li Z.Q., Yan F., Li G.F. Earlier flowering induced by over-expression of CO gene does not accompany increase of artemisinin biosynthesis in Artemisia annua, Plant. Biol. 2007;9:442–446. doi: 10.1055/s-2006-924634. [DOI] [PubMed] [Google Scholar]
- 17.Arab H.A., Rahbari S., Rassouli A., Moslemi M.H., Khosravirad F. Determination of artemisinin in Artemisia sieberi and anticoccidial effects of the plant extract in broiler chickens. Trop. Anim. Health Prod. 2006;38:497–503. doi: 10.1007/s11250-006-4390-8. [DOI] [PubMed] [Google Scholar]
- 18.Hsu E. The history of qing hao in the Chinese Materia medica. Trans. R. Soc. Trop. Med. Hyg. 2006;100:505–508. doi: 10.1016/j.trstmh.2005.09.020. [DOI] [PubMed] [Google Scholar]
- 19.Zia M., Mannan A., Chaudhary M.F. Effect of growth regulators and amino acids on artemisinin production in the callus of Artemisia absinthium, Pakistan. J. Bot., Le. 2007;39:799–805. [Google Scholar]
- 20.Mannan A., Ahmed I., Arshad W., Asim M.F., Qureshi R.A., Hussain I., Bushra M. Survey of artemisinin production by diverse Artemisia species in northern Pakistan. Malar. J. 2010;9:310. doi: 10.1186/1475-2875-9-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu H., Qiu N., Ding H., Yao R. Polyphenols contents and antioxidant capacity of 68 Chinese herbals suitable for medical or food uses. Food Res. Int. 2008;41:363–370. [Google Scholar]
- 22.Salehi M., Karimzadeh G., Naghavi M.R., Naghdi Badi H., Rashidi Monfared S. Expression of key genes affecting artemisinin content in five Artemisia species. Sci. Rep. 2018;8 doi: 10.1038/s41598-018-31079-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pfaffl M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ghassemi B., Dehghan Nayeri F., Hosseini R. The effects of chitosan nanoparticles on genes expression of artemisinin synthase in suspension culture of Artemisia annua L: a comparative study. Int. J. Adv. Biol. Biomed. Res. 2021;2:214–227. [Google Scholar]
- 25.Arsenault P., Vail D.R., Wobbe K.K., Weathers P.J. Effect of sugars on artemisinin production in Artemisia annua L.: transcription and metabolite measurements. Molecules. 2010;15:2302–2318. doi: 10.3390/molecules15042302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salehi M., Karimzadeh G., Naghavi M.R., Badi H.N., Monfared S.R. Expression of artemisinin biosynthesis and trichome formation genes in five Artemisia species. Ind. Crop. Prod. 2018;112:130–140. [Google Scholar]
- 27.Ghasemi B., Hosseini R., Dehghan Nayeri F. Effects of cobalt nanoparticles on artemisinin production and gene expression in Artemisia annua. Turk. J. Bot. 2015;39:769–777. [Google Scholar]
- 28.Ekiert H., Klimek-Szczykutowicz M., Rzepiela A., Klin P., A. Szopa A. Artemisia species with high biological values as a potential source of medicinal and cosmetic raw materials. Molecules. 2022;27:6427. doi: 10.3390/molecules27196427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Efferth T., Zacchino S., Georgiev M.I., Liu L., Wagner H., Panossian A. Nobel Prize for artemisinin brings phytotherapy into the spotlight. Phytomedicine. 2015;22:A1–A3. doi: 10.1016/j.phymed.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 30.Weathers P.J., Towler M.J. The flavonoids casticin and artemetin are poorly extracted and are unstable in an Artemisia annua tea infusion. Planta Med. 2012;78:1024–1026. doi: 10.1055/s-0032-1314949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aydin T., Akincioglu H., Gumustas M., Gulcin I., Kazaz C., Cakir A. Human monoamine oxidase (hMAO) A and hMAO B inhibitors from Artemisia dracunculus L. herniarin and skimmin: human mononamine oxidase A and B inhibitors from A. dracunculus L. Z. Naturforsch. 2020;75:459–466. doi: 10.1515/znc-2019-0227. [DOI] [PubMed] [Google Scholar]
- 32.Salih A.M., Qahtan A.A., Al-Qurainy F. Phytochemicals identification and bioactive compounds estimation of Artemisia species grown in Saudia Arabia. Metabolites. 2023;13:443. doi: 10.3390/metabo13030443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Numonov S., Sharopov F., Salimov A., Sukhrobov P., Atolikshoeva S., Safarzoda R., Aisa H.A. Assessment of artemisinin contents in selected Artemisia species from Tajikistan (central Asia) Medicines. 2019;6:23. doi: 10.3390/medicines6010023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pellicer J., Saslis-Lagoudakis C.H., Carrio E., Ernst M., Garnatje T., Grace O.M., Ronsted N.A. Phylogenetic road map to antimalarial Artemisia species. J. Ethnopharmacol. 225. 2018;225:1–9. doi: 10.1016/j.jep.2018.06.030. [DOI] [PubMed] [Google Scholar]
- 35.Czechowski T., Larson T.R., Catania T.M., Harvey D., Wei C., Essome M., Graham I.A. Detailed phytochemical analysis of high-and low artemisinin-producing chemotypes of Artemisia annua. Front. Plant Sci. 2018;9:641. doi: 10.3389/fpls.2018.00641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zia M., Mannan A., Chaudhary M.F. Effect of growth regulators and amino acids on artemisinin production in the callus of Artemisia absinthium. Pakistan J. Bot. 2007;39:799–805. [Google Scholar]
- 37.Benyagoub E., Nabbou N., Dine A. Antimicrobial effect of Quercus robur L. Leaves selective extracts from the Mezi mountain of Djeniene Bourezg (West of Algeria) Curr. Bioact. Compd. 2020;16:1181–1190. [Google Scholar]
- 38.Pagare S., Bhatia M., Tripathi N., Pagare S., Bansal Y.K. Secondary metabolites of plants and their role: overview. Curr. Trends Biotechnol. Pharm. 2015;9:293–304. [Google Scholar]
- 39.Yang L., Wen K.S., Ruan X., Zhao Y.X., Wei F., Wang Q. Response of plant secondary metabolites to environmental factors. Molecules. 2018;23:762. doi: 10.3390/molecules23040762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Skrovankova S., Misurcova L., Machu L. Antioxidant activity and protecting health effects of common medicinal plants. Adv. Food. Nutr. 2012;67:75–139. doi: 10.1016/B978-0-12-394598-3.00003-4. [DOI] [PubMed] [Google Scholar]
- 41.Kunal M. Antioxidant analysis of essential oils and methanolic extracts of Artemisia vulgaris. Int. J. Agric. Sci. 2018;10:5710–5713. [Google Scholar]
- 42.Beigh Y.A., Ganai A.M. Potential of wormwood (Artemisia absinthium Linn.) herb for use as additive in livestock feeding: a review, Pharma. Innov. 2017;6:176. [Google Scholar]
- 43.Kazemi M. Essential oil of the aerial parts of Artemisia annua (Asteraceae) from Iran. J. Essen. Oil. Bear. 2015;18:1003–1005. [Google Scholar]
- 44.Joshi R.K., Satyal P., Setzer W.N. Himalayan aromatic medicinal plants: a review of their ethnopharmacology, volatile phytochemistry, and biological activities. Medicines. 2016;3:6. doi: 10.3390/medicines3010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lv Z., Zhang F., Pan Q., Fu X., Jiang W., Shen Q., Tang K. Branch pathway blocking in Artemisia annua is a useful method for obtaining high yield artemisinin. Plant Cell Physiol. 2016;57:588–602. doi: 10.1093/pcp/pcw014. [DOI] [PubMed] [Google Scholar]
- 46.Khalilov L., Paramonov E.A., Khalilova A.Z., Odinokov V.N., Muldashev A.A., Baltaev U.A., Dzehemilev U.M. Identification and biological activity of volatile organic compounds emitted by plants and insects. IV. Composition of vapor isolated from certain species of Artemisia plants. Chem. Nat. Compd. 2001;37:339. [Google Scholar]
- 47.Mucciarelli M., Caramiello R., Maffei M., Chialva F. Essential oils from some Artemisia species growing spontaneously in North-West Italy. Flavour Fragrance J. 1995;10:25. [Google Scholar]
- 48.Pino J.A., Rosado A., Fuentes V. Composition of the essential oil of Artemisia vulgaris L., Herb from Cuba. J. Essent. Oil Res. 1999;11:477. [Google Scholar]
- 49.Ranjbar M., Naghavi M.R., Alizadeh H., Soltanloo H. Expression of artemisinin biosynthesis genes in eight Artemisia species at three developmental stages. Ind. Crop. Prod. 2015;76:836–843. [Google Scholar]
- 50.Czechowski T., Larson T.R., Catania T.M., Harvey D., Wei C., Essome M., Brown G.D., Graham I.A. Detailed phytochemical analysis of high- and low artemisinin-producing chemotypes of Artemisia annua. Front. Plant Sci. 2018;9:641. doi: 10.3389/fpls.2018.00641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ma D., Pu G., Lei C., Ma L., Wang H., Guo Y., Liu B. Isolation and characterization of AaWRKY1, an Artemisia annua transcription factor that regulates the amorpha-4, 11-diene synthase gene, a key gene of artemisinin biosynthesis. Plant Cell Physiol. 2009;50:2146–2161. doi: 10.1093/pcp/pcp149. [DOI] [PubMed] [Google Scholar]
- 52.Han J., Wang H., Lundgren A., Brodelius P.E. Effects of overexpression of AaWRKY1 on artemisinin biosynthesis in transgenic Artemisia annua plants. Phytochemistry (Elsevier) 2014;102:89–96. doi: 10.1016/j.phytochem.2014.02.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.