Abstract
The chemokine 20 (CCL20) is a member of the CC chemokine family and plays a role in tumor immunity and autoimmune disease. This work investigated the value of CCL20 as a serum diagnostic marker for primary hepatocellular carcinoma (HCC). Based on the data of hepatocellular carcinoma patients in the TCGA database, the up-regulated genes encoding secretory proteins were analyzed in each pathological stage, and the candidate marker CCL20 gene was selected. Serum concentrations of CCL20 in patients with primary HCC, benign liver disease, and healthy subjects were analyzed by enzyme-linked immunosorbent assay (ELISA). The ROC curve evaluated the efficacy of CCL20 alone or in combination with AFP in the diagnosis of HCC. It was found the expression of CCL20 in HCC patients was significantly higher than that in the benign liver disease group and healthy controls (P < 0.05); The AUC of ROC curve to distinguish HCC patients from healthy controls was 0.859, the sensitivity was 73.42%, and the specificity was 86.84%. After combination with AFP, the AUC increased to 0.968, the sensitivity was 88.16%, and the specificity was 97.37%. Although CCL20 was increased in the serum of patients with benign liver diseases, combined with AFP, the AUC to distinguish HCC patients from non-HCC cohorts (benign liver disease group and healthy control group) was 0.902, with a sensitivity of 91.67% and a specificity of 75.26%. Collectively, serum CCL20 is closely related to the occurrence of HCC, and detection of serum CCL20 can assist AFP in improving the diagnostic sensitivity of HCC.
Keywords: Hepatocellular carcinoma, Serum biomarker, Diagnosis, CCL20, AFP
1. Introduction
Liver cancer (LC) is one of the most common malignant tumors in the world, and HCC is the most common type of primary LC [1]. Medical statistics show that liver cancer has the third highest mortality rate for malignant tumors and is the sixth most commonly diagnosed cancer worldwide in 2020 [2]. Hepatitis B virus (HBV) infection is a common cause of hepatocellular carcinoma in Asia, and cirrhosis is the leading risk factor for HCC [3]. Early diagnosis and treatment are essential for a good prognosis of liver cancer patients [4]. A large number of studies have confirmed the effectiveness of blood markers in tumor diagnosis and monitoring [5,6]. Compared with invasive puncture pathology, its non-inventiveness makes it more acceptable and easier to apply. Alpha-fetoprotein AFP, a blood marker commonly used in the diagnosis of hepatocellular carcinoma, is not sufficient to be used as a single diagnostic tool for monitoring and diagnosing hepatocellular carcinoma due to its insufficient diagnostic sensitivity [7]. Therefore, it is necessary to find new sensitive and specific blood markers for HCC diagnosis.
Many chemokines, such as CXCL1, CXCL13, CCL5 and CCL4, have been well studied for their diagnostic value in HCC [[8], [9], [10]]. Chemokine ligand (CCL20), also known as macrophage inflammatory protein (MIP) -3α or liver Activation regulatory chemokine (LARC), is a member of the CC subfamily of chemokines [11]. CCL20 is generally produced by epithelial cells and its expression is increased under inflammatory conditions [9]. Studies have shown that it plays a variety of biological functions in tumorigenesis, such as invasion, migration, immunosuppression, angiogenesis, and epithelial-mesenchymal transformation [[12], [13], [14], [15]]. It has been reported that the ccl20 expression is up-regulated in tissues and body fluids of patients with multiple tumor types, including colorectal cancer [16], oral squamous cell carcinoma [17], gastric cancer [18], pancreatic carcinoma [19], indicating that serum CCL20 may have the potential as a tumor marker. Several works have also explored the value of CCL20 as a diagnostic marker for HCC [20,21], but these studies either only looked at HCV associated hepatocellular carcinoma or lacked comparisons in patients with hepatitis. Considering that the major viral type of hepatitis in China is hepatitis B virus. Differences between different viral phenotypes may exist. By studying the expression of related markers in hepatitis B patients and combining with clinically available markers, it may be more meaningful for the diagnosis of hepatitis B-related liver cancer. In this study, the secreted proteins differentially expressed in each pathological stage of primary HCC were analyzed through the TCGA database, and CCL20 was selected as a candidate protein for study to evaluate its diagnostic value as a serum marker for HCC patients.
2. Materials and methods
2.1. Screening of candidate serum biomarkers
Transcriptome RNA-seq and clinical data for HCC were collected from the TCGA database (https://portal.gdc.cancer.gov/) [22]. Differentially expressed gene (DEGs) analysis is achieved by the “limma” R package. The screening criteria for DEGs were set as, logFC>1.0, adjusted p-Value<0.05, and the mean expression value in adjacent normal tissue (conMean) < 15, We used the DAVID (https://david.ncifcrf.gov/summary.jsp) [23] online tool to analyze the up-regulated DEGs for screening secretory proteins. GEPIA2 (http://gepia2.cancer-pku.cn/) [24] online tool was used to verify the differential expression of candidate DEGs. The research process is shown in Supplementary Fig. 1.
2.1.1. Patient and specimen collection
A total of 176 serum samples were collected from Hefei Cancer Hospital, Chinese Academy of Sciences, of which 79 were from HCC patients (15 patients with stage I, 17 patients with stage II, 27 patients with stage III, and 15 patients with stage IV), details are in Supplementary Table 1. 59 samples were from patients with benign liver diseases (29 samples from patients with chronic hepatitis B and 30 samples from patients with cirrhosis) and 38 samples were from healthy subjects with no history of liver disease. All HCC patients were primary cases. The clinical staging of HCC was based on the TNM classification criteria of the American Joint Committee on Cancer. This study was reviewed and approved by the Ethics Committee of the Hefei Institutes of Physical Science, Chinese Academy of Sciences (Approval No. Y-2018-19).
2.1.2. Serum ELISA assay
Venous blood was collected, hemolysis and lipid samples were removed, and serum was collected after centrifugation. CCL20 concentration was detected using an ELISA kit (R&D Systems, Minneapolis, MN, USA) according to the manufacturer's instructions. Briefly, a 100-μL detection diluent and a 100-μL standard or serum sample were each added to a 96-well microwell coated with CCL20 antibody and incubated at room temperature for 2 h before cleaning. Then the HRP-labeled secondary antibody was added to the micropores and incubated for 2 h at room temperature, cleaned again, and then TMB solution was added. The reaction time was 30 min at room temperature, and the termination solution was added to measure the absorbance at 450 nm within 30 min. In addition, the concentration of AFP in serum was detected by Beckman DXI800 automatic chemiluminescence analyzer (kit purchased from Beckman).
2.2. Statistical analysis
All statistical analyses were performed using GraphPad Prism 9.0 software (GraphPad Software Inc., CA, USA). The nonparametric Mann-Whitney test was used to determine the difference between two independent sets of data with non-normal distribution. The combined diagnosis model was established by binary logistic regression. The diagnostic efficiency of single and combined tests was analyzed using receiver operating characteristic (ROC) curves. The AUC (area under the curve) and 95% CI (confidence interval), sensitivity, specificity and accuracy were calculated from the ROC curve. The largest Youden index is used as the best cutoff value for the test item. P value < 0.05 was considered statistically significant (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001).
3. Results
3.1. Screening of candidate diagnostic markers for HCC
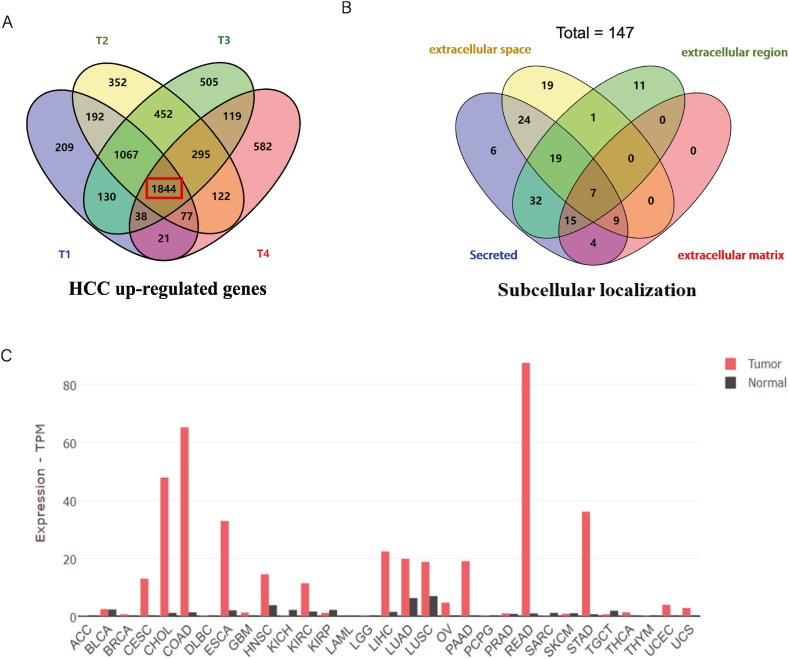
To identify potential HCC diagnostic markers, we downloaded transcriptomic RNA-seq data from the TCGA database for primary hepatocellular carcinoma (TCGA-LIHC), including 50 normal liver tissue samples and 371 liver cancer tissue samples, as well as patient clinical information, and analyzed the genes that were significantly upregulated at each HCC pathological stage compared to normal liver tissue (screening criteria: The average expression value in normal tissues conMean<15, logFC>1.0, adjust p-Value<0.05) (Fig. 1A). Then, the subcellular localization of 1844 co-upregulated genes was analyzed through the DAVID database to screen out genes encoding secretory proteins. Considering that secreted proteins are located in extracellular space, only those localized in extracellular space (79 genes), extracellular domain (85 genes), extracellular matrix (35 genes), and clearly defined as secreted proteins (116 genes) were selected. After removing the genes with the same name in each group, 147 candidate secreted proteins were finally obtained (Fig. 1B), the heatmaps of 147 genes were shown in Supplementary Fig. 2. Theoretically, a good serum marker should be expressed at low levels in all normal tissues. This will result in a lower background amount being secreted into the blood. So that the tumor secretes this marker and it will be sensitively reflected in the blood. We validated the expression of these 147 genes using the GEPIA2 online tool [25] and selected genes with low expression in all paracancerous tissues as the dominant candidate markers. CCL20 meets the above screening criteria and is a potential ideal serum marker (Fig. 1C).
Fig. 1.
Screening of candidate diagnostic markers for HCC. (A) Genes were commonly up-regulated in all pathological stages of HCC in the TCGA-LIHC data set. (B) 147 genes encoding potential secretory proteins were identified by subcellular localization analysis. (C) Expression levels of CCL20 in tumors and corresponding paracancer tissues in 33 tumor types.
3.1.1. Comparison of serum CCL20 levels
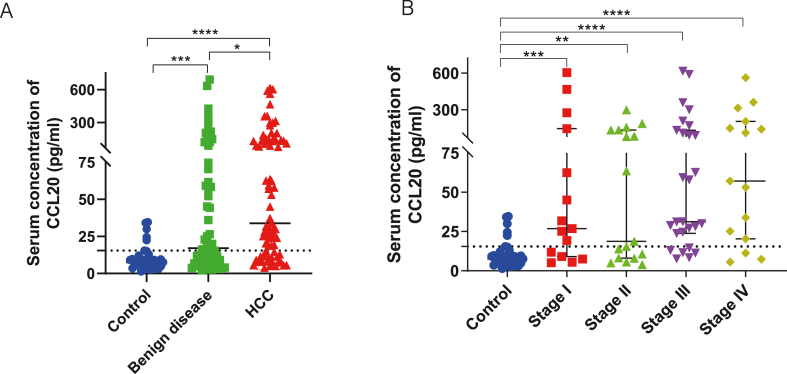
The levels of serum CCL20 in the HCC group, benign liver disease group, and 38 healthy subjects were shown in Table 1. The median concentration of serum CCL20 in the HCC group was significantly higher than that in the healthy group (p < 0.0001) and benign disease group (p = 0.0152), and the liver benign disease group was significantly higher than that in the healthy group (p = 0.0001) (Fig. 2A). The median concentration of serum CCL20 in HCC patients at each TNM stage was higher than that in the healthy group (Fig. 2B). In addition, liver cirrhosis, HBV infection, medication, and HBV viral load and ALT levels in patients with hepatitis did not affect serum CCL20 levels (Supplementary Fig. 3). Therefore, serum CCL20 concentration was significantly up-regulated in HCC, indicating its value as a serum diagnostic marker.
Table 1.
Serum concentrations of CCL20 and AFP.
| Markers | Control group |
Benign disease group |
HCC group |
Stage I |
Stage II |
Stage III |
Stage IV |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Median | IQR | Median | IQR | Median | IQR | Median | IQR | Median | IQR | Median | IQR | Median | IQR | |
| CCL20 | 7.57 | 1.45–34.67 | 17.10 | 7.71–70.38 | 33.80 | 3.86–615.68 | 26.87 | 10.47–103.50 | 18.73 | 8.15–132.17 | 31.3 | 24.24–119.10 | 57.15 | 22.74–174.63 |
| AFP | 2.50 | 1.98–3 | 2.70 | 2.10–5.70 | 352.15 | 13.18–3000 | 49.70 | 10.05–3000 | 629.00 | 12.8–3000 | 1168.8 | 44.68–3000 | 98.5 | 8.61–1563.6 |
Fig. 2.
Detection of CCL20 in the serum of HCC patients. (A) Serum levels of CCL20 were determined by ELISA in 79 patients with hepatocellular carcinoma, 59 patients with benign liver disease, and 38 healthy subjects. (B) Expression of CCL20 in the serum of HCC patients was compared in different pathological stages.
3.1.2. Diagnostic efficacy of serum CCL20 in differentiating HCC patients from healthy subjects
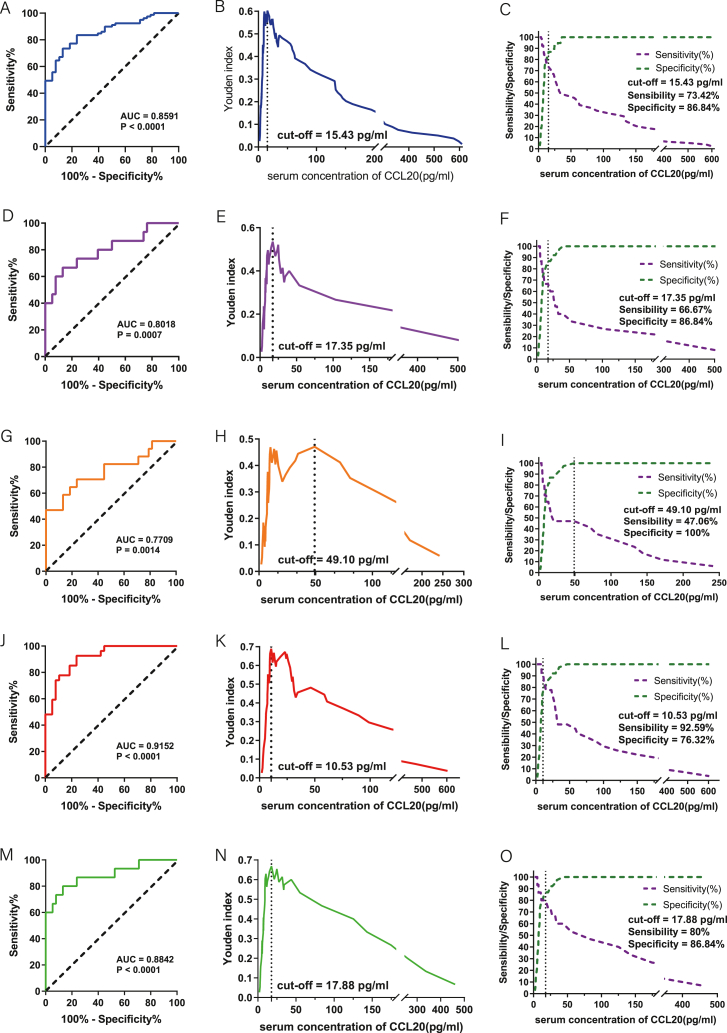
Univariate logistic regression analysis showed that serum CCL20 was a risk factor for HCC compared with the healthy group (OR:1.096; 95% CI: 1.041 1.154; p < 0.0001); The AUC under ROC curve of serum CCL20 for the diagnosis of HCC was 0.8591 (95%CI:0.7932–0.9250; P < 0.0001) (Fig. 3A). With the maximum Youden index corresponding to 15.43 pg/ml as the optimal diagnostic threshold (Fig. 3B), the corresponding diagnostic sensitivity was 73.42% and the specificity was 86.84% (Fig. 3C). For stage I-IV of HCC, the diagnostic AUC values based on serum CCL20 were 0.8018 (95%CI:0.6577–0.9458; P = 0.0007), 0.7709 (95%CI:0.6222–0.9196; P = 0.0014), 0.9152 (95%CI: 0.85–0.9804; P < 0.0001), 0.8842 (95%CI:0.7709–0.9975; P < 0.0001) (Fig. 3D–G,J,M). There was no significant difference among all stages (Stage I vs II: P = 0.7078; Stage I vs III: P = 0.1020; Stage I vs IV: P = 0.1747; Stage II vs III: P = 0.0845; Stage II vs IV: P = 0.2323; Stage III vs IV: P = 0.5032). The cut-off values of CCL20 level for diagnosis of stage I-IV tumor are shown in Fig. 3E–H,K,N. The corresponding sensitivity and specificity are shown in Fig. 3F–I,L,O.
Fig. 3.
Diagnostic efficacy of serum CCL20 in differentiating patients with HCC from healthy subjects. (A) ROC diagnostic curve analysis of all HCC patients in different pathological stages and healthy subjects. (B) Relationship between serum CCL20 concentration and Yoden index in all patients with HCC and healthy subjects. (C) Sensitivity and specificity of ROC diagnostic curves for serum CCL20 concentrations in all patients with HCC and healthy subjects. (D–F) ROC diagnostic curve, Yoden index analysis, diagnostic sensitivity, and specificity based on CCL20 serum concentrations in stage I HCC patients and healthy subjects. (G–I) ROC diagnostic curve, Yoden index analysis, diagnostic sensitivity, and specificity based on CCL20 serum concentrations in Stage II HCC patients and healthy subjects. (J–L) ROC diagnostic curve, Yoden index analysis, diagnostic sensitivity, and specificity based on CCL20 serum concentrations in Stage III of HCC patients and healthy subjects. (M − O) ROC diagnostic curve, Yoden index analysis, diagnostic sensitivity, and specificity based on CCL20 serum concentrations in stage IV of HCC patients and healthy subjects.
3.1.3. Diagnostic efficacy of serum CCL20 in differentiating HCC patients from non-HCC patients
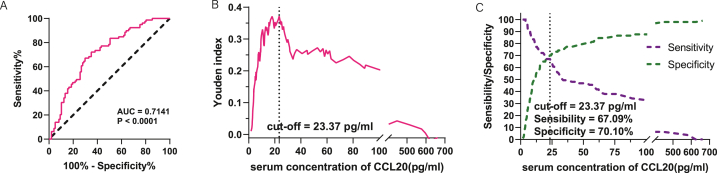
Univariate logistic regression analysis showed that serum CCL20 was a risk factor for HCC compared with non-HCC (healthy group and benign liver disease group) (OR:1.003; 95% CI: 1.001 1.006; p = 0.013), the ROC diagnosis AUC was 0.7141(95%CI:0.6385–0.7897; P < 0.0001) (Fig. 4A). If 23.37 pg/ml corresponding to the maximum Youden index was taken as the threshold value (Fig. 4B), the sensitivity and specificity of HCC diagnosis were 67.09% and 70.10% respectively (Fig. 4C). These data indicate that CCL20 has broad potential as a novel biomarker for hepatocellular carcinoma.
Fig. 4.
Evaluation of the efficacy of serum CCL20 in differentiating HCC patients from non-HCC cohorts (healthy subjects and those with benign liver disease). (A) ROC diagnostic curve of serum CCL20 in HCC patients and non-HCC population. (B) The relationship between serum CCL20 concentration and Yoden index. (C) Sensitivity and specificity of ROC diagnostic curve based on serum CCL20 concentration.
3.2. Serum CCL20 combined with AFP can improve the diagnostic efficacy of HCC
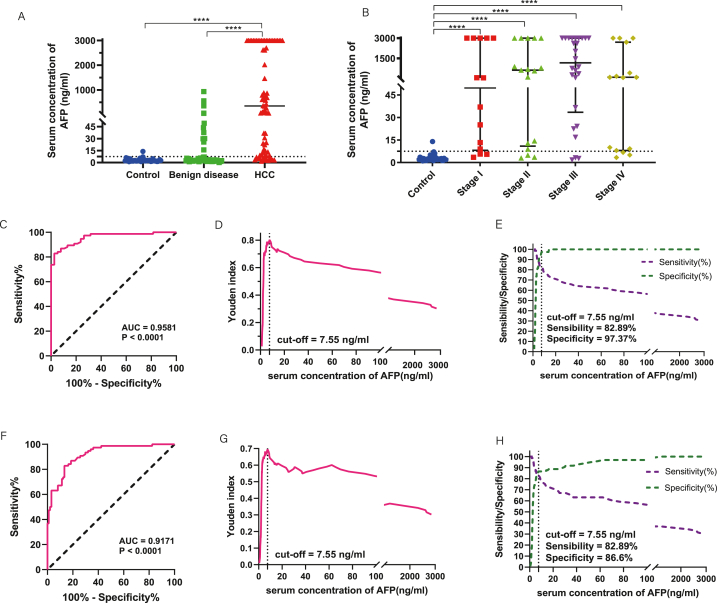
First, we analyzed the diagnostic efficacy of serum AFP in the above samples. The median concentration of AFP in HCC serum was significantly higher than that in the healthy group and benign liver disease group (P < 0.0001) (Fig. 5A). There was no difference between the benign liver disease group and the healthy group (P = 0.0967). The median concentration of serum AFP in stage I-IV patients with HCC was higher than that in the control group (Fig. 5B). The AUC for AFP to distinguish between healthy and HCC patients was 0.9581(95%CI:0.9254–0.9908; P < 0.0001) (Fig. 5C). Taking the maximum Yoden index as the threshold (7.55 ng/ml) (Fig. 5D), the diagnostic sensitivity was 82.89% and the specificity was 97.37% (Fig. 5E).
Fig. 5.
Analysis of AFP expression in the serum of HCC patients. (A) Serum CCL20 expression was compared in 76 patients with hepatocellular carcinoma, 59 patients with benign liver disease, and 38 healthy subjects. (B) Comparison of serum AFP expression in different pathological stages. (C) ROC diagnostic curve of serum AFP in patients with HCC and healthy subjects. (D) Relationship between serum AFP concentration and Yoden index in patients with HCC and healthy subjects. (E) Diagnostic sensitivity and specificity of ROC curves for serum AFP concentrations in patients with HCC and healthy subjects. (F) ROC diagnostic curve of serum AFP in HCC patients and non-HCC (healthy subjects and people with benign liver disease). (G) Relationship between serum AFP concentration and Yoden index in HCC patients and non-HCC population. (H) Sensitivity and specificity of ROC diagnostic curves for serum AFP concentrations in HCC patients and non-HCC populations.
Further, we analyzed the ability of serum AFP to differentiate HCC from non-HCC (healthy subjects and benign liver disease group) using ROC curves. The results showed that the diagnostic AUC was 0.9171(95%CI:0.8769–0.9573; P < 0.0001) (Fig. 5F). When the Yoden index reached its maximum, the threshold value was 7.55 ng/ml (Fig. 5G), the diagnostic sensitivity was 82.89% and the specificity was 86.60% (Fig. 5H). For HCC patients at different pathological stages (stage I-IV), the diagnostic AUC values were 0.9146 (95%CI:0.8553–0.9739; P < 0.0001), 0.9203 (95%CI:0.8598–0.9808; P < 0.0001), 0.9192 (95%CI:0.8473–0.9911; P < 0.0001), 0.9182 (95%CI:0.8605–0.9759; P < 0.0001) (Supplementary Fig. 4). There was no significant difference in diagnostic AUC among different pathological stages (Stage I vs II: P = 0.8877; Stage I vs III: P = 0.5714; Stage I vs IV: P = 0.8917; Stage II vs III: P = 0.4956; Stage II vs IV: P = 0.9930; Stage III vs IV: P = 0.5029).
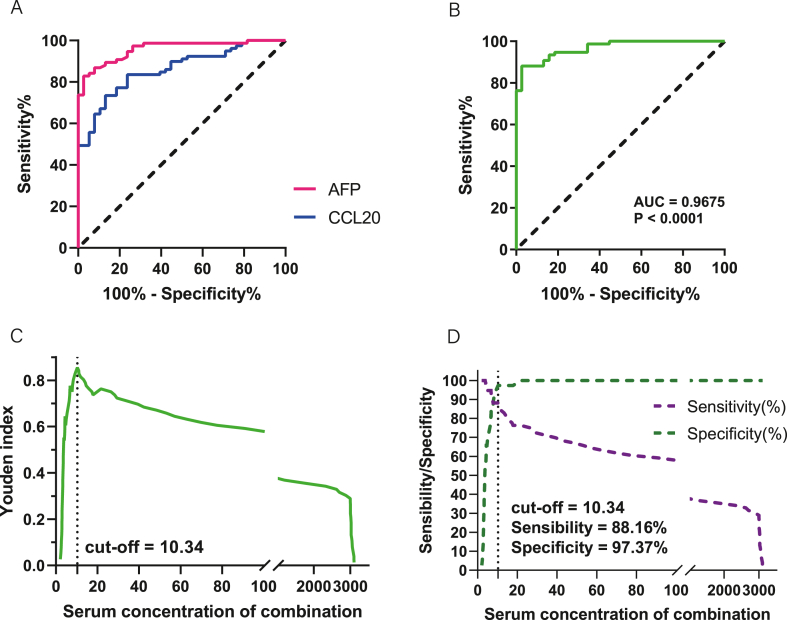
The above experiments showed that compared with the traditional diagnostic marker AFP, CCL20 can also be used effectively in the diagnosis of HCC, but the efficacy is slightly lower (Fig. 6A). Therefore, we considered combining the two to improve the efficacy of HCC diagnosis. The results showed that the AUC, sensitivity, specificity, PPV, PPV, and NPV were increased to 0.9675 (95%CI:0.9411–0.9938), 88.16%, 97.37%, 98.53%, and 80.43%, respectively. Corresponding to stage I-IV HCC patients, AUC values based on the combination of the two markers were 0.9586 (95%CI:0.9032–1.000; P < 0.0001), 0.9396 (95%CI:0.8712–1.000; P < 0.0001), 0.9808 (95%CI:0.9515–1.000; P < 0.0001), 0.9842 (95%CI:0.9578–1.000; P < 0.0001) (Supplementary Fig. 5). There was no significant difference in diagnostic AUC between stages (Stage I vs II: P = 0.7092; Stage I vs III: P = 0.8714; Stage I vs IV: P = 0.4198; Stage II vs III: P = 0.6163; Stage II vs IV: P = 0.3133; Stage III vs IV: P = 0.5285). In addition, the diagnostic AUC (95%CI:0.8581–0.9460), sensitivity (91.76%), specificity (75.26%), PPV (73.33%), and NPV (92.40%) for the differentiation between HCC and non-HCC populations were obtained by the combination of the two markers (Table 2). Correspondingly, the diagnostic AUC values for stage I-IV of HCC patients were 0.8476 (95%CI:0.7524–0.9428; P < 0.0001), 0.8708 (95%CI: 0.7712–0.9704; P < 0.0001), 0.9243 (95%CI:0.8685–0.9801; P < 0.0001), 0.8969 (95%CI:0.8315–0.9624; P < 0.0001) (Supplementary Fig. 6), there was no significant difference in the diagnostic AUC among different stages (Stage I vs II: P = 0.7726; Stage I vs III: P = 0.3670; Stage I vs IV: P = 0.4869; Stage II vs III: P = 0.7440; Stage II vs IV: P = 0.7907; Stage III vs IV: P = 0.9120). In addition, our data showed that serum CCL20 levels were effective in differentiating benign liver disease from healthy people. Although the distinction between benign liver disease and HCC is not effective, it can be achieved in combination with AFP. (Supplementary Fig. 7).Therefore, the combination of CCL20 and AFP can greatly improve the diagnostic efficacy of HCC (Fig. 6B,C,D).
Fig. 6.
Combinination diagnosis of HCC with AFP and CCL20. (A) Comparison of the diagnostic efficacy of AFP and CCL20 in HCC patients. (B) ROC diagnostic curve of a combination of two biomarkers. (D) Yoden index analysis of combination diagnosis. (C) Sensitivity and specificity of combination diagnosis.
Table 2.
The diagnosis values of CCL20 and AFP.
| Biomarkers | Cut-off | AUC (95%CI) | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) |
|---|---|---|---|---|---|---|
| HCC versus healthy control | ||||||
| AFP | 7.55 | 0.9581(0.9254–0.9908) | 82.89 | 97.37 | 98.44 | 74 |
| CCL20 | 15.43 | 0.8591(0.7932–0.9250) | 73.42 | 86.84 | 92.06 | 61.11 |
| Combination | 10.34 | 0.9675(0.9411–0.9938) | 88.16 | 97.37 | 98.53 | 80.43 |
| HCC versus non-HCC | ||||||
| AFP | 7.55 | 0.9171(0.8769–0.9573) | 82.89 | 86.6 | 82.89 | 86.6 |
| CCL20 | 23.37 | 0.7141(0.6385–0.7897) | 67.09 | 70.1 | 65.43 | 72.34 |
| Combination | 56.18 | 0.9021(0.8581–0.9460) | 91.67 | 75.26 | 73.33 | 92.4 |
4. Discussion
The incidence of hepatocellular carcinoma is increasing worldwide, with more than half of the deaths occurring in China [26]. HBV and aflatoxin have been identified as major pathogenic factors [27]. The survival and prognosis of patients with liver cancer are still clinical challenges [[28], [29], [30]]. Ultrasound-bound serum alpha-fetoprotein (AFP) is one of the widely used combinations for HCC screening [31,32]. However, there are many controversies regarding the diagnostic efficacy of AFP, such as abnormal AFP serum levels in only a small percentage of early-stage tumors (10–20%) and false positive AFP results in patients with active liver inflammation [33,34]. Some newly discovered markers lack reproducibility and are difficult to be further applied in the clinic [35]. Biomarker combinations with higher specificity and sensitivity than single biomarkers are important for earlier detection of cancer and thus timely intervention and prevention of HCC [36].
CCL20 has been reported to play a key role in the development of a variety of tumors [37,38]. The CCL20-CCR6 axis promotes cancer progression by enhancing the migration and proliferation of cancer cells and indirectly reshaping the tumor microenvironment by controlling immune cells [39]. Altered CCL20 expression has been found in most cancer types [[16], [17], [18], [19]]. Consistent with these results, this study found that increased serum CCL20 concentration was closely associated with HCC. We detected the serum CCL20 concentration in healthy subjects, patients with benign liver disease, and patients with HCC, and found that the serum CCL20 concentration was significantly increased in HCC patients and slightly increased in patients with benign liver disease. The use of serum CCL20 in the diagnosis of HCC also showed good efficacy. We also compared CCL20 with the traditional HCC serum marker AFP and found that the diagnosis of AUC by AFP was still higher than CCL20, but the combination of CCL20 and AFP could significantly improve the sensitivity of HCC diagnosis and reduce the false negative rate. When a combined diagnosis was made between HCC and healthy people, the negative predictive value was 6.43% higher than that of AFP. When a combined diagnosis was made between HCC and non-HCC (including healthy people and patients with benign liver disease), the negative predictive value was increased by 5.8% compared with AFP. Therefore, the combined diagnosis of CCL20 and AFP has the potential to be used in HCC screening.
Several studies have identified CCL20 as a potential biomarker for HCC [20,21], but there are some differences with the results of our study.Firstly, the concentrations of serum CCL20 detected were different. A reported serum concentration of 100 pg/ml was measured in healthy controls [21], whereas our test results were 7.57 pg/ml. In addition, we found a significant difference in serum CCL20 concentration between patients with benign liver disease and healthy controls, but others reported that the results were not different [21]. Finally, our study focused on hepatitis B-related HCC, but another study focused on hepatitis C-related HCC [20]. It's clear that our work could help to complement and correct the shortcomings of previous work.
It is noteworthy that previous studies have shown that the AUC area of AFP in the diagnosis of hepatocellular carcinoma is between 0.80 and 0.857,34,35 with a sensitivity of 41%–65% [40,41]. However, the area under the curve calculated in this study was 0.917–0.9581, with a sensitivity of 82.89%, which may be due to regional differences in the source of enrolled objects and different pathological stages. In addition, this study found that CCL20 increased significantly in the serum of the benign liver disease group, especially for cirrhosis cases, which would interfere with the use of CCL20 in HCC diagnosis. However, cirrhosis is also widely considered a risk factor for HCC, and its identification is of certain clinical significance. On the other hand, although the diagnosis of HCC by CCL20 alone is interfered by cirrhosis, the combination of CCL20 and AFP can overcome this defect and greatly improve the diagnostic efficiency of AFP. It is expected that multi-center, large-sample tests will be carried out in the future to further confirm the value of the combination of CCL20 and AFP in the diagnosis of HCC.
Availability of data and materials
The data associated with the study has been included in article/supp. material/referenced in article. The datasets analyzed in the current study are available in the TCGA repository. (https://gdc.cancer.gov/). Relevant data or materials can also be obtained from the corresponding author upon reasonable request.
Funding
This study was in part supported by the National Natural Science Foundation of China (81872276, 61973295), the Scientific Research Project of Anhui Provincial Health Commission (AHWJ2021b142), the Hefei municipal Natural Science Foundation (2022050), the Anhui Province's Key Research and Development Project (201904a07020092), and the CASHIPS Director's Fund (YZJJ2022QN48).
Ethics approval
Approval of the research protocol by an Institutional Reviewer Board: this study was approved by the Ethics Review Board of the Hefei Institutes of Physical Science, Chineses Academy of Sciences (Approval No. Y-2018-19).
Informed consent
This study was conducted in accordance with the Declaration of Helsinki, and all participants signed informed consent for their serum samples to be used for research at the time of admission.
Registry and the Registration No. of the study/trial: N/A.
Animal studies
N/A.
CRediT authorship contribution statement
Qingmei Deng: Data curation, Formal analysis, Methodology, Resources, Validation, Writing – original draft. Xinhui Zhang: Data curation, Formal analysis, Methodology, Resources, Software, Validation, Writing – original draft. Xiaofeng Wan: Data curation, Resources, Validation. Xin Zheng: Methodology, Resources, Validation. Hongzhi Wang: Data curation, Formal analysis, Investigation, Methodology, Resources, Validation. Jingyu Zhao: Methodology, Resources, Validation. Hong-Qiang Wang: Investigation, Methodology, Validation, Visualization. Wulin Yang: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Writing – original draft, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We are grateful to the TCGA repository for providing a convenient platform for data sharing, and to data contributors for generously sharing their important and meaningful datasets.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e26774.
Contributor Information
Hongzhi Wang, Email: wanghz@hfcas.ac.cn.
Wulin Yang, Email: yangw@cmpt.ac.cn.
Abbreviations
- ACC
Adrenocortical carcinoma
- BLCA
Bladder Urothelial Carcinoma
- BRCA
Breast invasive carcinoma
- CESC
Cervical squamous cell carcinoma and endocervical adenocarcinoma
- CHOL
Cholangio carcinoma
- COAD
Colon adenocarcinoma
- DLBC
Lymphoid Neoplasm Diffuse Large B-cell Lymphoma
- ESCA
Esophageal carcinoma
- GBM
Glioblastoma multiforme
- HNSC
Head and Neck squamous cell carcinoma
- KICH
Kidney Chromophobe
- KIRC
Kidney renal clear cell carcinoma
- KIRP
Kidney renal papillary cell carcinoma
- LAML
Acute Myeloid Leukemia
- LGG
Brain Lower Grade Glioma
- LIHC
Liver hepatocellular carcinoma
- LUAD
Lung adenocarcinoma
- LUSC
Lung squamous cell carcinoma
- MESO
Mesothelioma
- OV
Ovarian serous cystadenocarcinoma
- PAAD
Pancreatic adenocarcinoma
- PCPG
Pheochromocytoma and Paraganglioma
- PRAD
Prostate adenocarcinoma
- READ
Rectum adenocarcinoma
- SARC
Sarcoma
- SKCM
Skin Cutaneous Melanoma
- STAD
Stomach adenocarcinoma
- TGCT
Testicular Germ Cell Tumors
- THCA
Thyroid carcinoma
- THYM
Thymoma
- UCEC
Uterine Corpus Endometrial Carcinoma
- UCS
Uterine Carcinosarcoma
- UVM
Uveal Melanoma
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Llovet J.M., Kelley R.K., Villanueva A., Singal A.G., Pikarsky E., Roayaie S., Lencioni R., Koike K., Zucman-Rossi J., Finn R.S. Hepatocellular carcinoma. Nat. Rev. Dis. Prim. 2021;7:6. doi: 10.1038/s41572-020-00240-3. [DOI] [PubMed] [Google Scholar]
- 2.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global cancer statistics 2020: GLOBOCAN Estimates of incidence and mortality worldwide for 36 cancers in 185 Countries. CA Cancer J Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 3.Hepatocellular carcinoma. Nat. Rev. Dis. Prim. 2021;7:7. doi: 10.1038/s41572-021-00245-6. [DOI] [PubMed] [Google Scholar]
- 4.Yang J.D., Heimbach J.K. New advances in the diagnosis and management of hepatocellular carcinoma. BMJ. 2020;371:m3544. doi: 10.1136/bmj.m3544. [DOI] [PubMed] [Google Scholar]
- 5.Tsuchiya N., Sawada Y., Endo I., Saito K., Uemura Y., Nakatsura T. Biomarkers for the early diagnosis of hepatocellular carcinoma. World J. Gastroenterol. 2015;21:10573–10583. doi: 10.3748/wjg.v21.i37.10573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pinero F., Dirchwolf M., Pessoa M.G. Biomarkers in Hepatocellular Carcinoma: Diagnosis, Prognosis and Treatment Response Assessment. 2020 doi: 10.3390/cells9061370. Cells 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Galle P.R., Foerster F., Kudo M., Chan S.L., Llovet J.M., Qin S., Schelman W.R., Chintharlapalli S., Abada P.B., Sherman M., Zhu A.X. Biology and significance of alpha-fetoprotein in hepatocellular carcinoma. Liver Int. 2019;39:2214–2229. doi: 10.1111/liv.14223. [DOI] [PubMed] [Google Scholar]
- 8.Han K.Q., Han H., He X.Q., Wang L., Guo X.D., Zhang X.M., Chen J., Zhu Q.G., Nian H., Zhai X.F., Jiang M.W. Chemokine CXCL1 may serve as a potential molecular target for hepatocellular carcinoma. Cancer Med. 2016;5:2861–2871. doi: 10.1002/cam4.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li B., Su H., Cao J., Zhang L. CXCL13 rather than IL-31 is a potential indicator in patients with hepatocellular carcinoma. Cytokine. 2017;89:91–97. doi: 10.1016/j.cyto.2016.08.016. [DOI] [PubMed] [Google Scholar]
- 10.Sadeghi M., Lahdou I., Oweira H., Daniel V., Terness P., Schmidt J., Weiss K.H., Longerich T., Schemmer P., Opelz G., Mehrabi A. Serum levels of chemokines CCL4 and CCL5 in cirrhotic patients indicate the presence of hepatocellular carcinoma. Br. J. Cancer. 2015;113:756–762. doi: 10.1038/bjc.2015.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Korbecki J., Grochans S., Gutowska I., Barczak K., Baranowska-Bosiacka I. CC chemokines in a tumor: a review of pro-cancer and Anti-cancer properties of receptors CCR5, CCR6, CCR7, CCR8, CCR9, and CCR10 ligands. Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21207619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leake I. Alcoholic hepatitis: potential role of cytokine CCL20 in alcoholic hepatitis. Nat. Rev. Gastroenterol. Hepatol. 2014;11:76. doi: 10.1038/nrgastro.2014.9. [DOI] [PubMed] [Google Scholar]
- 13.Liu W., Wang W., Wang X., Xu C., Zhang N., Di W. Cisplatin-stimulated macrophages promote ovarian cancer migration via the CCL20-CCR6 axis. Cancer Lett. 2020;472:59–69. doi: 10.1016/j.canlet.2019.12.024. [DOI] [PubMed] [Google Scholar]
- 14.Kwantwi L.B., Wang S., Sheng Y., Wu Q. Multifaceted roles of CCL20 (C-C motif chemokine ligand 20): mechanisms and communication networks in breast cancer progression. Bioengineered. 2021;12:6923–6934. doi: 10.1080/21655979.2021.1974765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kwantwi L.B., Wang S., Zhang W., Peng W., Cai Z., Sheng Y., Xiao H., Wang X., Wu Q. Tumor-associated neutrophils activated by tumor-derived CCL20 (C-C motif chemokine ligand 20) promote T cell immunosuppression via programmed death-ligand 1 (PD-L1) in breast cancer. Bioengineered. 2021;12:6996–7006. doi: 10.1080/21655979.2021.1977102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang D., Yuan W., Wang Y., Wu Q., Yang L., Li F., Chen X., Zhang Z., Yu W., Maimela N.R., et al. Correction to: serum CCL20 combined with IL-17A as early diagnostic and prognostic biomarkers for human colorectal cancer. J. Transl. Med. 2021;19:435. doi: 10.1186/s12967-021-03107-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ueda S., Goto M., Hashimoto K., Hasegawa S., Imazawa M., Takahashi M., Oh-Iwa I., Shimozato K., Nagao T., Nomoto S. Salivary CCL20 level as a biomarker for oral squamous cell carcinoma. Cancer Genomics Proteomics. 2021;18:103–112. doi: 10.21873/cgp.20245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shen Q., Polom K., Williams C., de Oliveira F.M.S., Guergova-Kuras M., Lisacek F., Karlsson N.G., Roviello F., Kamali-Moghaddam M. A targeted proteomics approach reveals a serum protein signature as diagnostic biomarker for resectable gastric cancer. EBioMedicine. 2019;44:322–333. doi: 10.1016/j.ebiom.2019.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klemm C., Dommisch H., Goke F., Kreppel M., Jepsen S., Rolf F., Dommisch K., Perner S., Standop J. Expression profiles for 14-3-3 zeta and CCL20 in pancreatic cancer and chronic pancreatitis. Pathol. Res. Pract. 2014;210:335–341. doi: 10.1016/j.prp.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 20.Benkheil M., Van Haele M., Roskams T., Laporte M., Noppen S., Abbasi K., Delang L., Neyts J., Liekens S. CCL20, a direct-acting pro-angiogenic chemokine induced by hepatitis C virus (HCV): potential role in HCV-related liver cancer. Exp. Cell Res. 2018;372:168–177. doi: 10.1016/j.yexcr.2018.09.023. [DOI] [PubMed] [Google Scholar]
- 21.Du L., Wang M., Li H., Li N., Wang F. Identification of CCL20 and LCN2 as efficient serological tools for detection of hepatocellular carcinoma. Dis. Markers. 2022;2022 doi: 10.1155/2022/7758735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cancer Genome Atlas Research N., Weinstein J.N., Collisson E.A., Mills G.B., Shaw K.R., Ozenberger B.A., Ellrott K., Shmulevich I., Sander C., Stuart J.M. The cancer genome atlas pan-cancer analysis project. Nat. Genet. 2013;45:1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dennis G., Jr., Sherman B.T., Hosack D.A., Yang J., Gao W., Lane H.C., Lempicki R.A. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 24.Tang Z., Kang B., Li C., Chen T., Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47:W556–W560. doi: 10.1093/nar/gkz430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li C., Tang Z., Zhang W., Ye Z., Liu F. GEPIA2021: integrating multiple deconvolution-based analysis into GEPIA. Nucleic Acids Res. 2021;49:W242–W246. doi: 10.1093/nar/gkab418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fu J., Wang H. Precision diagnosis and treatment of liver cancer in China. Cancer Lett. 2018;412:283–288. doi: 10.1016/j.canlet.2017.10.008. [DOI] [PubMed] [Google Scholar]
- 27.Chen J.G., Zhang S.W. Liver cancer epidemic in China: past, present and future. Semin. Cancer Biol. 2011;21:59–69. doi: 10.1016/j.semcancer.2010.11.002. [DOI] [PubMed] [Google Scholar]
- 28.Maki H., Hasegawa K. Advances in the surgical treatment of liver cancer. Biosci Trends. 2022;16:178–188. doi: 10.5582/bst.2022.01245. [DOI] [PubMed] [Google Scholar]
- 29.Gao Y.X., Yang T.W., Yin J.M., Yang P.X., Kou B.X., Chai M.Y., Liu X.N., Chen D.X. Progress and prospects of biomarkers in primary liver cancer (Review) Int. J. Oncol. 2020;57:54–66. doi: 10.3892/ijo.2020.5035. [DOI] [PubMed] [Google Scholar]
- 30.Sangro B., Sarobe P., Hervas-Stubbs S., Melero I. Advances in immunotherapy for hepatocellular carcinoma. Nat. Rev. Gastroenterol. Hepatol. 2021;18:525–543. doi: 10.1038/s41575-021-00438-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang H.Y., Chen J., Xia C.C., Cao L.K., Duan T., Song B. Noninvasive imaging of hepatocellular carcinoma: from diagnosis to prognosis. World J. Gastroenterol. 2018;24:2348–2362. doi: 10.3748/wjg.v24.i22.2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cassinotto C., Aube C., Dohan A. Diagnosis of hepatocellular carcinoma: an update on international guidelines. Diagn Interv Imaging. 2017;98:379–391. doi: 10.1016/j.diii.2017.01.014. [DOI] [PubMed] [Google Scholar]
- 33.European Association for the Study of the Liver. Electronic address, e.e.e., and European Association for the Study of the, L. EASL Clinical Practice Guidelines: Management of hepatocellular carcinoma. J Hepatol. 2018;69:182–236. doi: 10.1016/j.jhep.2018.03.019. [DOI] [PubMed] [Google Scholar]
- 34.Johnson P., Zhou Q., Dao D.Y., Lo Y.M.D. Circulating biomarkers in the diagnosis and management of hepatocellular carcinoma. Nat. Rev. Gastroenterol. Hepatol. 2022 doi: 10.1038/s41575-022-00620-y. [DOI] [PubMed] [Google Scholar]
- 35.Pinto Marques H., Gomes da Silva S., De Martin E., Agopian V.G., Martins P.N. Emerging biomarkers in HCC patients: current status. Int. J. Surg. 2020;82S:70–76. doi: 10.1016/j.ijsu.2020.04.043. [DOI] [PubMed] [Google Scholar]
- 36.Williams F.M. Biomarkers: in combination they may do better. Arthritis Res. Ther. 2009;11:130. doi: 10.1186/ar2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fan T., Li S., Xiao C., Tian H., Zheng Y., Liu Y., Li C., He J. CCL20 promotes lung adenocarcinoma progression by driving epithelial-mesenchymal transition. Int. J. Biol. Sci. 2022;18:4275–4288. doi: 10.7150/ijbs.73275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mo M., Tong S., Huang W., Cai Y., Zu X., Hu X. High serum CCL20 is associated with tumor progression in penile cancer. J. Cancer. 2020;11:6812–6822. doi: 10.7150/jca.48939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kadomoto S., Izumi K., Mizokami A. The CCL20-CCR6 Axis in cancer progression. Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21155186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hanif H., Ali M.J., Susheela A.T., Khan I.W., Luna-Cuadros M.A., Khan M.M., Lau D.T. Update on the applications and limitations of alpha-fetoprotein for hepatocellular carcinoma. World J. Gastroenterol. 2022;28:216–229. doi: 10.3748/wjg.v28.i2.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hu J., Wang N., Yang Y., Ma L., Han R., Zhang W., Yan C., Zheng Y., Wang X. Diagnostic value of alpha-fetoprotein combined with neutrophil-to-lymphocyte ratio for hepatocellular carcinoma. BMC Gastroenterol. 2018;18:186. doi: 10.1186/s12876-018-0908-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data associated with the study has been included in article/supp. material/referenced in article. The datasets analyzed in the current study are available in the TCGA repository. (https://gdc.cancer.gov/). Relevant data or materials can also be obtained from the corresponding author upon reasonable request.