Abstract
Background
Necroptosis could regulate immunity in cancers, and stratification of colorectal cancer (CRC) subtypes based on key genes related to necroptosis might be a novel strategy for CRC treatment.
Method
The RNA-sequencing data of CRC and other 31 types of cancers were obtained from The Cancer Genome Atlas (TCGA) database. Consensus clustering was performed based on protein-coding genes (PCGs) related to necroptosis score calculated by single sample gene set enrichment analysis (ssGSEA). Module genes showing a significant positive correlation with the necroptosis score were identified by weighted correlation network analysis (WGCNA) and further used to develop a risk stratification model applying least absolute shrinkage and selection operator (LASSO) and Cox regression analysis. The risks score for each sample in CRC cohorts, immunotherapy cohorts and pan-cancer study cohorts was calculated.
Result
Two subgroups (C1 cluster and C2 cluster) of CRC were identified based on the necroptosis score. Compared with C1 cluster, the survival possibility of C2 cluster was greatly reduced, the levels of necroptosis score, immune cell infiltration, immune score and expression of immune checkpoint molecules were significantly increased and immunotherapy response was less active. Low-risk patients defined by the risk model had a significant survival advantage than high-risk counterparts in both CRC and the other 31 cancer types. Furthermore, the risk model was also more efficient than the Tumor Immune Dysfunction and Exclusion (TIDE) tool in predicting OS and immunotherapy response for the samples in the immunotherapy cohort.
Conclusion
CRC patients were classified by necroptosis score-related PCGs, and a risk model was designed to evaluate the immunotherapy and prognosis of patients with CRC. The current molecular subtype and prognostic model could help stratify patients with different risks and predict their prognosis and immunotherapy sensitivity.
Keywords: Necroptosis, Colorectal cancer, Molecular subtypes, Risk stratification tools, Immunotherapy
1. Introduction
Colorectal cancer (CRC) is now the most frequent malignant tumor of the digestive tract and the second largest cause of cancer-related death worldwide [1,2]. It is estimated that by 2030, there will be over 2.2 million new cases of CRC and over 1.1 million deaths [3]. Accumulation of genetic susceptibility and epigenetic factors, eating habits, changes in intestinal microbiota and lack of physical activity are all associated with the complex pathogenesis of CRC [4]. Most CRC originates from precursors such as adenomas and transforms into adenocarcinomas [5]. The standard treatment for CRC is surgery, chemotherapy and radiotherapy [[6], [7], [8]], but there is limited variation between the effectiveness of these treatments and long-term survival, particularly for patients with metastatic CRC, where approximately 70%–75% of patients survive more than one year and 30%–35% of patients survive more than three years, compared to less than five years after diagnosis [9,10]. The molecular landscape of CRC on histological and clinical information of each patients differs greatly [[11], [12], [13]]. Therapeutic decisions based on molecular subtypes might be a promising strategy for treating CRC, and development of new drugs in a way that targets CRC subtypes has the potential to transform drug discovery research [14].
Necroptosis is involved in regulating a variety of pathological mechanisms of tumors, including tumorigenesis, metastasis, and immunity [15,16]. Necroptosis plays a dual regulatory role in tumor progression, as it not only promotes anti-tumor immunity, but also participates in the promotion of myeloid cell-induced adaptive immunosuppression to stimulate tumourigenesis [17]. Increasing evidence suggests that human tumors appear to gain an advantage from the down-regulation of key components in the molecular mechanism of necroptosis, which points to the potential of necroptosis as a novel cancer therapy [18,19]. Hence, many studies now define subtypes for cancer patients according to the key components involved in the necroptosis molecular mechanism and predict cancer prognosis and treatment response from a multi-molecular perspective. A recent study characterized two subtypes of renal clear cell carcinoma based on the expression patterns of necroptosis-related genes and established a gene signature as a prognostic predictor [20]. Similarly, tumor subtyping and prognostic multi-molecular signature according to necroptosis-related gene expression patterns have also been studied in bladder cancer [21], pancreatic cancer [22], hepatocellular carcinoma [23], and ovarian cancer [24] but not in CRC.
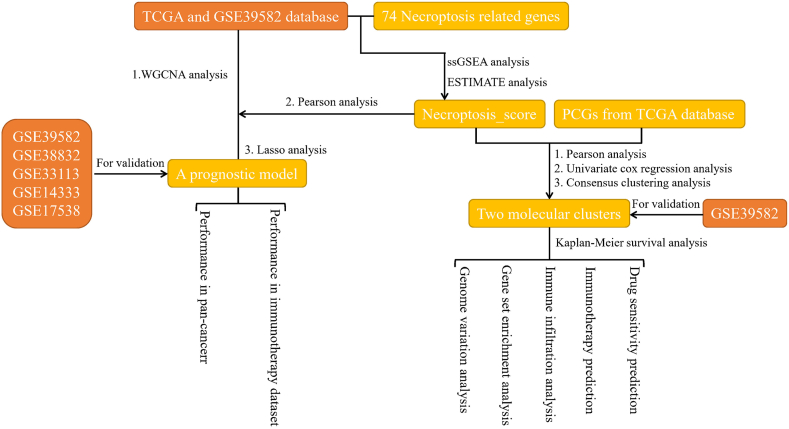
The work flow of the work could be seen in Fig. 1. The current study first calculated necroptosis score for CRC samples based on 74 necroptosis-related genes using ssGSEA. The molecular subtypes of CRC were classified according to the expression of prognostic necroptosis score-related genes in CRC, and the differences in survival, tumor microenvironment (TME) and response to immunotherapy and targeted/chemotherapeutic drugs were analyzed by multi-group analysis. Finally, we developed a necroptosis-related co-expression network and a prognostic risk model for CRC, aiming to improve the current risk stratification of CRC and to guide clinical decision-making process.
Fig. 1.
The work flow of the study.
2. Materials and methods
2.1. Acquisition of CRC expression datasets
The expression profile and clinicopathological data of colon adenocarcinoma (COAD) samples were obtained from TCGA database (https://tcga-data.nci.nih.gov/tcga/), which stores data for a wide range of tumor types, including survival data for a total of 437 cases. The gene expressions of more CRC samples with survival characteristics were collected from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/, accession numbers: GSE39582, GSE38832, GSE33113, GSE14333 and GSE17538). Additionally, IMvigor210 and GSE135222 cohorts were utilized for immunotherapy evaluation of the risk model.
2.2. Identification of necroptosis score-related genes
Xin et al. integrated a necroptosis gene set containing 74 necroptosis-related genes [20]. Specific genetic information can be found in Table S1. Here, we utilized ssGSEA applying to calculate the enrichment score of the necroptosis gene set above using the “GSVA” package [25]. The PCGs exhibiting significant correlation with the necroptosis score were screened from TCGA-COAD by Pearson correlation analysis under the thresholds of |R | > 0.4 and P < 0.05.
2.3. Consensus clustering for CRC samples
The prognosis-related PCGs (exhibiting significant correlation with the necroptosis score) obtained above were screened by univariate cox regression analysis. Then, PCGs related to both CRC prognosis and necroptosis score were subjected to consensus clustering analysis adopting ConsensusClusterPlus package [26]. When running the program, the Pam unsupervised clustering algorithm was selected for 500 iterations, with each iteration randomly containing 80% of the samples, and the cluster number as in the range of 2–10.
2.4. Genome variation analysis
Mutation data of CRC samples were obtained from TCGA. The total number of mutations in the samples was measured, and genes with mutation number >3 were screened. Differences in mutation frequency between subtypes was identified by Fisher test and visualized by “maftools” [27]. Based on a previously published research [28], the DNA damage score and copy number burden score for each sample were calculated, including homologous recombination defect (HRD), fraction altered, number of segments and tumor mutation burden, and compared among clusters.
2.5. Gene set enrichment analysis (GSEA)
The c2.cp.kegg.v7.0.symbols.gmt gene set obtained from molecular signature database (http://www.gsea-msigdb.org/gsea/downloads.jsp) was used as the reference gene set, and pathway analysis of molecular subtypes in TCGA-COAD cohort and GSE39582 cohort was carried out employing GSEA [29]. The input file included the expression profile of CRC and the molecular subtype of the sample. Pathways with p < 0.05 and FDR<0.1 were considered as statistically different between subtypes.
2.6. Immune infiltration analysis
Microenvironment cell populations-counter (MCP-counter) [30] includes information about immunocytes and stromal cells. The abundance of immune cells and stromal cells was calculated here using MCP-Counter. SsGSEA quantified the immune cell fraction of each case based on the signature of 28 immune cells. ESTIMATE [31] evaluated the proportion of infiltrating stromal and immune components by calculating stromal and immune scores, and gave the ESTIMATE score used to evaluate TME. In addition to comparing the infiltration abundance of immune cell, the expression of 21 immune checkpoints from HisgAtlas database [32] in each molecular subtype was evaluated.
2.7. Immunotherapy response and drug sensitivity predictions
Drug resistance of immune checkpoint blockade (ICB) was evaluated by calculating T cell dysfunction signature, T cell exclusion signature and TIDE score using TIDE algorithm [33]. Six chemotherapy drugs including Cisplatin, Sunitinib, Saracatinib, Cyclopamine, Imatinib and Dasatinib were used for drug sensitivity prediction. Half-maximal inhibitory concentration (IC50) value was calculated by ridge regression algorithm in “pRRophetic” package [34].
2.7.1. Weighted gene correlation network analysis (WGCNA)
WGCNA was used to cluster genes with similar expression patterns into the same gene module for analyzing the relationship between gene networks and phenotypes [35]. Firstly, the Pearson correlation coefficients of all gene pairs were calculated to develop an adjacency matrix, and then the adjacency matrix with weighted coefficients was used to assess the network connections in the topological overlap matrix (TOM). Hierarchical clustering analysis based on TOM formed a cluster tree structure to identify gene co-expression modules. Dynamic hybrid-cutting algorithm was used to cluster genes into module, the minimum number of genes in each cluster was 300 and similar modules were merged together. The correlation of each coexpression modules with necroptosis score, immune score and molecular subtypes was analyzed by WGCNA. The coexpression modules with the highest correlation with necroptosis score from the TCGA-COAD cohort and GSE39582 cohort were selected, and overlapping genes between the two coexpression modules were further analyzed for developing a risk model.
2.8. Design and evaluation of a risk model
Of the 437 CRC samples in TCGA, 7/10 were used as the training cohort and 3/10 were used as the validation cohort to design the risk model. Firstly, univariate Cox regression analysis was performed to determine the prognostic correlation of necroptosis score-related genes screened by WGCNA in the training set. LASSO cox regression analysis using “glmnet” package [36] was performed to reduce the number of the candidate genes. Variables were selected according to the penalty parameters that met the minimum criteria in the 10-fold cross-validation, and multivariate Cox regression analysis was used to filter the variables and calculate the coefficient. The risk model was as follows: .where βi refers to the coefficient of the first variable, and Expi refers to the expression value of the variable. The grouping threshold for the risk score in each queue was determined by “survminer” [37]. According to the risk score and survival data of each sample, survival difference between risk groups in each cohort was estimated and compared by plotting Kaplan-Meier curve and performing log-rank test.
2.9. Statistical analysis
All the results of statistical analysis were produced by R program. The Kaplan-Meier curve and ROC analysis was completed in “survminer” package and the “survivalROC” package [38], respectively. The T-test and Kruskal-Wallis test were used to compare the continuous and ordered categorical variables between the two groups. P < 0.05 was indicated a statistical significance.
3. Results
3.1. Immune correlation of necroptosis score
Immune relevance of necroptosis was explored using correlation analysis between necrosis scores (using ssGSEA for a total of 74 necrosis-related genes) and TME scores. In the TCGA-COAD cohort, necroptosis score was positively correlated with immune score, stromal score and ESTIMATE score, with a correlation coefficient of 0.775, 0.642 and 0.74, respectively (Fig. S1A). The necroptosis score in GSE39582 also showed a significant positive correlation with ESTIMATE score, immune score, and stromal score, respectively (Fig. S1B). Also a significantly positive correlation between necroptosis score and immune cells were also observed (Fig. S1C). Thus, necroptosis score may be a positive factor affecting immunity in CRC.
3.2. Two clusters of CRC defined based on the necroptosis score-related genes
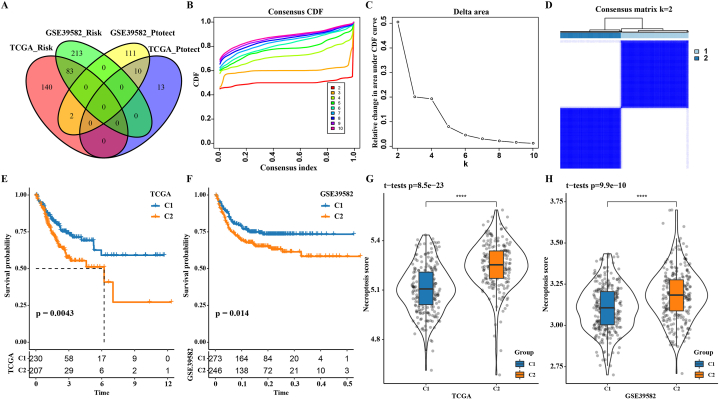
By analyzing the correlation between all PCGs and necroptosis score in TCGA-COAD, 1906 genes positively correlated with necroptosis score were screened. Univariate Cox regression analysis identified a total of 83 risk genes (hazard ratio (HR) > 1 and p < 0.05) and 10 protective genes (HR < 1 and p < 0.05) as the overlapping genes between TCGA-COAD and GSE39582 cohorts (Fig. 2A). These 93 prognostic necroptosis score-related genes were adopted to classify CRC. CRC was divided into two subgroups based on the results of cumulative distribution function (CDF) curve and consensus clustering delta area curve and Consensus matrix (Fig. 2B–D). Analyses on overall survival [31] of the two subgroups showed the same survival outcomes in both the TCGA-COAD cohort and GSE39582 cohort, specifically, the OS of C1 was always significantly longer than C2 (Fig. 2E and F). Necroptosis score also exhibited significantly different levels in the two subgroups of the two cohorts, with C2 having a significantly higher necroptosis score than C1 (Fig. 2G and H).
Fig. 2.
The necroptosis score related gene defined two clusters of CRC. A: The intersection of prognostic necroptosis score-related genes in TCGA-COAD and GSE39582 cohorts. B: Cumulative distribution function (CDF) curve with k in the range of 2–10. C: Consensus clustering delta area curve with k in the range of 2–10. D: Consensus matrix for k = 2. E: Prognostic Kaplan-Meier curve of two subgroups in the TCGA-COAD cohort. F: Kaplan-Meier curve involving OS in two subgroups of the GSE39582 cohort. G: Necroptosis score difference between C1 and C2 in the TCGA-COAD cohort. H: Necroptosis score difference between two subgroups in the GSE39582 cohort.
3.3. Genome variation and clinical characteristics of the two molecular subtypes
The genome variation and clinical characteristics of the two subtypes in the TCGA-COAD cohort were analyzed. It was found that there was no difference in the distributions of T stage, M stage, N stage, tumor stage, gender, or age between C1 and C2 (Fig. S2A). Except that the mutation rate of APC in C1 was higher than that in C2, almost all the other genes with a high mutation frequency showed higher mutation rate in C2 than in C1 (Fig. S2B). In terms of DNA damage score and copy number burden score, C2 had significantly higher number of segments than C1 (Fig. S2C).
3.4. GSEA and immune-related analysis revealed different immune states in the two molecular subtypes
Differences of biological pathways affected by C1 and C2 were analyzed in TCGA-COAD cohort and GSE39582 cohort. In the TCGA-COAD cohort, we observed that tumor-related pathways, such as JAK_ STAT_ Signaling_ PATHWAY, PATHWAYS_ IN_ CANCER, TGF_BETA_SIGNALING_PATHWAY, and WNT_ Signaling, were more enriched in the C2 cluster. Also, immunomodulatory pathways including T_CELL_RECEPTOR_SIGNALING_PATHWAY, CHEMOKINE_SIGNALING_PATHWAY, and B_CELL_RECEPTOR_SIGNALING_PATHWAY were more enriched in C2 cluster (Fig. 3A). In the GSE39582 cohort, the pathway in cancer and JAK-STAT signaling pathway and TGF- β signaling pathway of C2 were also more activated than in C1 (Fig. 3B). Therefore, we speculated that C2 cluster was more closely related to tumor progression and immunological characteristics. In order to explore the immune status of C1 and C2 from different aspects, the differences of immune cell infiltration and immune molecules between the two clusters were analyzed. All the 10 types of immune cells quantified by MCP-Counter showed significantly increased abundance in C2 compared with C1 (Fig. 4A). Moreover, the scores of all 28 immune cells in C1 were significantly higher than in C2 (Fig. 4C). Compared with C1, C2 also manifested a significant increase in stromal score and immune score and ESTIMATE score (Fig. 4B). Among the 21 immune checkpoint molecules analyzed, 19 showed significantly higher expressions in C2, including LAG3, ICOS, CD276, CD244, BTLA, CD80, CTLA4, CD47, CD160, ADORA2A, TNFSF4, PDCD1 (PD-1), HAVCR2, IDO1, VTCN1, CD27, CD274 (PD-L1), VISTA, GEM (Fig. 4D). Therefore, the two subtypes defined by necroptosis score-related genes played an important role in shaping the different TME characteristics of CRC.
Fig. 3.
GSEA of C1 and C2 in the TCGA-COAD cohort (A) and the GSE39582 cohort (B).
Fig. 4.
Different immune states were detected in the two molecular subtypes. A: Abundances of 10 immune cell infiltration in two different subtypes of the TCGA-COAD cohort. B: The stromal score, immune score and ESTIMATE score of C2 compared to C1. C: The scores of 28 immune cells in two different subtypes of TCGA-COAD cohort. D: The expression of 21 immune checkpoint molecules in two different subtypes of TCGA-COAD cohort.
3.5. Immune escape, sensitivity to chemotherapy, and targeted therapy of the two CRC subtypes
C2 was significantly more immunoreactive than C1 but C2 had a significantly worse prognosis than C1, which could be explained by the immune escape in C2. To prove this hypothesis, immune escape in two molecular subtypes was detected estimated TIDE score, T cell dysfunction score and T cell exclusion score. In the TCGA-COAD cohort, C2 showed significantly higher TIDE score, T cell dysfunction score and T cell exclusion score than C1, indicating that immune escape was more likely to occur in C2 than C1. Responses of the two molecular subtypes to ICB therapy were also evaluated using TIDE. C1 had a considerably higher response rate to ICB therapy than C2 (0.65 vs 0.19). (Fig. 5A). Similarly, in the GSE39582 cohort, C2 had a significantly lower response rate to ICB therapy than C, and C2 had a significantly higher TIDE score, T cell dysfunction score, and T cell exclusion score than C1 (0.70 vs 0.22) (Fig. 5C).
Fig. 5.
Immune escape and sensitivity to chemotherapy and targeted therapy for two CRC subtypes. A: In the TCGA-COAD cohort, the TIDE score, T cell dysfunction score, T cell exclusion score and the response rate to ICB treatment of C1 and C2. B: The IC50 values of Cisplatin, Sunitinib, Saracatinib, Cyclopamine, Imatinib and Dasatinib in the two subtypes of the TCGA-COAD cohort. C: TIDE score, T cell dysfunction score, T cell exclusion score and response rates to ICB therapy for the two subtypes in the GSE39582 cohort. D: Sensitivity differences in of the two subtypes in the TCGA-COAD cohort to Cisplatin, Sunitinib, Saracatinib, Cyclopamine, Imatinib, and Dasatinib.
The drug sensitivity prediction results showed that C2 cluster was more sensitive to Cisplatin, Sunitinib, Saracatinib, Cyclopamine, Imatinib and Dasatinib than C1 cluster in both TCGA-COAD and GSE39582 cohorts (Fig. 5B and D).
3.6. Overlapping necroptosis score-related module genes between TCGA-COAD and GSE39582 cohorts
The optimal soft threshold power for a scale-free network (R2 = 0.85) was 7 when performing WGCNA using the data in TCGA-COAD (Fig. 6A). Eight gene modules of TCGA-COAD were obtained by hierarchical cluster analysis (Fig. 6B). In the GSE39582 cohort, the minimum soft threshold for a scale-free topology fitting index was 5 (Fig. S3A), and six modules were sectioned according to the clustering of the dynamic cutting method (Fig. S3B). The Pearson correlation coefficients of the gene modules in each cohort with immune score and stromal score, necroptosis score were calculated, and we found that the black module in the TCGA-COAD cohort had the highest correlation with necroptosis score (Fig. 6C). Among the gene modules in the GSE39582 cohort, the green module showed the highest correlation with necroptosis score (Fig. 6D). These two modules in two cohorts also had a significant positive correlation with both immune score and stromal score and shared a total of 674 overlapping genes (Fig. 6E).
Fig. 6.
Shared necroptosis score-related module genes in TCGA-COAD and GSE39582 cohorts. A: Network topology analysis for each soft-thresholding power value in TCGA-COAD cohort. B: Clustering tree of genes in TCGA-COAD cohort. C, D: The heatmap shows the correlation between modules in TCGA-COAD chohort and GSE39582 cohort and necroptosis score, immune score, and stromal score. E: The intersection of genes in the modules with the highest correlation with necroptosis score in the TCGA-COAD cohort and the GSE39582 cohort.
3.7. Development of a risk model and its prediction performance in various cohorts
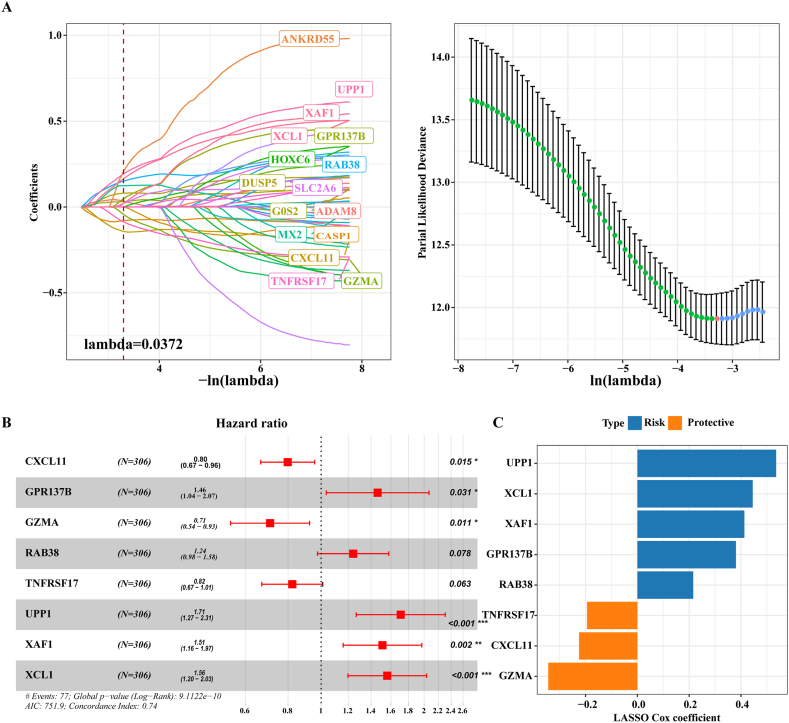
Univariate Cox regression analysis was performed using the above 674 genes in the training cohort, and 45 of these genes showed a p value within the threshold. LASSO Cox regression analysis reduced these 45 genes to 16 (Fig. 7A). Multivariate Cox regression analysis identified 8 genes with the most prognostic significance, namely, CXCL11, GPR137B, GZMA, RAB38, TNFRSF17, UPP1, XAF1, XCL1 (Fig. 7B). The expression of these 8 genes was weighted by LASSO regression coefficient, and the risk model was obtained: Risk score = (−0.225 × CXCL11) + 0.38 × GPR137B + (−0.344 × GZMA) + 0.216 × RAB38 + (−0.194 × TNFRSF17) +0.536 × UPP1 + 0.413 × XAF1+0.445 × XCL1 (Fig. 7C). Risk score was calculated and OS was analyzed in TCGA-COAD training cohort, verification cohort and full cohort, GSE39582, GSE38832, GSE33113, GSE14333 and GSE17538 cohorts, and significant survival advantage was found in the low-risk group compared with the high-risk group (Fig. 8A–H).
Fig. 7.
Design of risk model. A: Parameter adjustment of variable selection and 10-fold cross-validation in LASSO model. B: Multivariate Cox regression forest map of the 8 most prognostic genes. C: The LASSO regression coefficients of the 8 most prognostic genes.
Fig. 8.
Survival prediction of CRC by risk model in various cohorts, including (A) training cohort, (B) validation cohort, and (C) complete cohort of TCGA-COAD, (D) GSE39582, (E) GSE38832, (F) GSE33113, (G) GSE14333, (H) GSE17538 cohorts.
3.8. Prognostic analysis of pan-cancer based on risk model
Each patient with different types of cancers in TCGA was also assigned with a risk score and grouped by the risk model to evaluate the prediction performance of the risk model in pan-cancer. In the cohort of all cancer types, patients in the high-risk group normally had an unfavorable prognosis (Fig. 9A–F).
Fig. 9.
Prognostic analysis of pan-cancer based on risk model.
3.9. The value of risk model in predicting immunotherapy response and prognosis in immunotherapy cohorts
The predictive effects of risk model and TIDE on prognosis and immunotherapy response were compared in two cohorts treated with PD-L1 blockade and anti-PD-1, respectively. In the IMvigor210 cohort with PD-L1 blocking therapy, the OS of samples with a high-risk score was significantly shorter than those with a low-risk score. The TIDE score was also significantly correlated with OS, but the accuracy for predicting OS for 0.5 year (0.62 vs 0.54), 1 year (0.64 vs 0.57) and 1.5 years (0.66 vs 0.57) was significantly lower than the risk model (Fig. 10A and B). The area under the ROC curve [11] of risk score for predicting immune treatment response was 0.61, which was higher than the AUC of TIDE (Fig. 10C). In the GSE135222 cohort treated with anti-PD-1, risk score also showed high predictive value, with AUCs of 0.85 and 0.83 at 0.5 and 1 year, respectively, and high-risk score was also significantly associated with a poor prognosis (Fig. 10D). However, the OS predicted by TIDE did not differ significantly between samples, although its AUC for predicting 1-year prognosis reached 0.81 (Fig. 10E). The risk score also performed significantly better than TIDE in predicting immunotherapy response (AUC: 0.81 vs 0.75) (Fig. 10F).
Fig. 10.
The value of risk model in predicting prognosis and immunotherapy response in immunotherapy cohorts. A: In the IMvigor210 cohort that receiving PD-L1 blocking therapy, the sample survival curve and ROC curve obtained by risk score were used. B: In the IMvigor210 cohort receiving PD-L1 blocking therapy, the sample survival curve and ROC curve obtained by TIDE were used. C: Risk score and TIDE predict the area under the ROC curve of immunotherapy response in the IMvigor210 cohort receiving PD-L1 blocking therapy. D: The Kaplan-Meier curve and ROC curves predicted by risk score to OS in the GSE135222 cohort receiving anti-PD1 therapy. E: TIDE predicts survival Kaplan-Meier curve and ROC curves in the GSE135222 cohort receiving anti-PD-1 therapy. F: Risk score and TIDE predicted the ROC curve of immunotherapy response in the GSE135222 cohort treated with PD-1.
4. Discussion
Clinical and molecular data all suggest the presence of heterogeneity in the treatment response of CRC patients and their prognostic outcomes, but currently the complex molecular heterogeneity of CRC is not fully understood [11,39]. Genomic analysis defines subsets of CRC patients to facilitate personalize treatment decisions [40]. Necroptosis apoptosis is a regulatory form of molecular-defined necrosis [15]. In this study, two subgroups of CRC were defined based on the expression patterns of necroptosis score-related prognostic genes. As the molecular characteristics of CRC and detection of specific mutations can be used to evaluate patients’ response to treatment and prognostic outcomes, these biomarkers can also serve as key tools for personalized treatment [41]. This study found that the necroptosis score was significantly positively correlated with immune score and stromal score, which was consistent with the results of a previous pan-cancer study [42]. The current results indicated that targeting necroptosis to induce anti-tumor immunity may be a feasible strategy, especially when apoptosis is blocked.
The necroptosis score calculated by a multi-group study varies significantly with tumor subtypes [43]. This study divided the two subgroups according to the expression pattern of prognosis necroptosis score-related genes, and the necroptosis score of C2 was significantly higher and closely correlated with the poor prognosis of CRC. Necroptosis participates in immune infiltration and immune regulation of cancers [44]. Here, immunomodulatory pathways, including B cell receptor signaling pathway, chemokine signaling pathway, FC gamma mediated phagocytosis, and T cell receptor signaling pathway, showed differential enrichment between the two molecular subtypes. More importantly, immune cell infiltration, immunescore and immune checkpoint molecule expression were significantly higher in C2 than C1, which demonstrated a strong immune activity of C2 and revealed the effect of necroptosis on immune heterogeneity of subtypes. Immunological activity of C2 was lower than C1 as the TIDE score, T cell dysfunction score, and T cell exclusion score were all significantly higher in C2 than C1, suggesting the possibility of immune escape in C2. We speculated that immune escape may be an important reason to explain the less active response of C2 to ICB therapy.
We also predicted patients’ drug sensitivity to chemotherapy and observed that C2 cluster was more sensitive to these common chemotherapy drugs. Cisplatin and Imatinib (STI571) has also been used for CRC treatment [[45], [46], [47]]. In vitro study, we found that Sunitinib drug eluting beads and Dasatinib could effectively inhibit the growth of human CRC cells [48,49]. Saracatinib is the first inhibitor of Src family kinase used for treating human tumors [50] and Cyclopamine is an inhibitor for Hedgehog signaling [51]. Hence, as C2 cluster may not benefit from immunotherapy, these selected drugs with specific targets of action might be alternatives for C2 cluster treatment.
The risk model in this study was established using 8 genes (CXCL11, GPR137B, GZMA, RAB38, TNFRSF17, UPP1, XAF1 and XCL1), each of which has been previously studied in cancers. The relationship between CXCL11 and CRC is controversial. According to Cao et al., CXCL11 is positively correlated with cytotoxic genes including GZMA and immunosuppressive molecules, which can promote anti-tumor immunity and benefit patients with CRC [52]. A research of Liu et al. showed that in colon cancer, CXCL11 is secreted by overexpressed RBP-J κ to upregulate the expression of TGF-β 1 in tumor-associated macrophages, thus promoting the metastasis of colon cancer cells [53]. Additionally, chemotherapy drug Oxaliplatin suppresses CRC development through inhibiting CXCL11 secreted by tumor-associated fibroblasts and CXCR3/PI3K/AKT pathway. This study proposed that the opposite functions on CXCL11 may be resulted from CRC heterogeneity. Both GPR137 and RAB38 can promote the malignant progression of bladder cancer and pancreatic cancer [[54], [55], [56], [57]]. High expression of TNFRSF17 could prolong the OS time of patients with CC and this also indicates a higher sensitivity to immunotherapy [58]. According to previous studies, UPP1 is a tumor suppressor gene of CRC and the direct target of piR-1245 [59]. The role of XAF1 in gastrointestinal cancer has been widely studied, and it mediates apoptosis of colon cancer cells through ERK1/2 pathway [60]. In metastatic CRC, XAF1 could facilitate metastasis through VCP-RNF114-JUP axis [61]. As a chemokine, the transcript of XCL1 in tumor is related to a higher OS rate [62]. These studies highlight the potential of these genes as therapeutic targets for cancer. However, a better evaluation of CRC prognosis demands an overall consideration of all these genes from a “multi-molecular” point of view. In this study, the risk stratification model developed by integrating the 8 genes could predict patients’ prognosis in multiple independent CRC cohorts and had a significant effect on the OS prediction in 31 types of cancers. Moreover, the 8-gene signature also showed a significantly better effect on evaluating prognosis and immunotherapy response than TIDE in two ICB treatment cohorts.
Limitations in this study should be equally noticed. Firstly, the retrospective nature of our research demands further validation of the established molecular subtype and the prognostic model in prospective cohorts. Secondly, the possible functions of the hub necroptosis-related genes requires basic and clinical experiments to explore their actions in CRC development.
5. Conclusion
Using the prognostic necroptosis score-related genes, we defined molecular subtypes of CRC patients with different prognosis, immune activity and sensitivity to immunotherapy and chemotherapy/targeted drugs. Also, a risk stratification model showing a strong predictive efficiency for estimating prognostic outcomes and immunotherapy benefit was developed. Collectively, the current molecular subtype and prognostic model for CRC could be used to assist clinicians in deciding clinical intervention strategies and precise therapy for patients suffering from CRC.
Ethics declarations
Informed consent was not required for this study.
Funding
This research was supported by Technology Planning Program of Health Commission of Jiangxi Province, China (202312275).
Data availability statement
The datasets generated and/or analyzed during the current study are available in the [GSE39582] repository, [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc= GSE39582], [GSE38832] repository, [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc= GSE38832], [GSE33113] repository, [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc= GSE33113], [GSE14333] repository, [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc= GSE14333], [GSE15222] repository [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc= GSE15222], and [GSE17538] repository, [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc= GSE17538].
CRediT authorship contribution statement
Mengling Li: Writing – review & editing, Writing – original draft, Visualization, Validation, Software, Project administration, Methodology, Funding acquisition, Formal analysis, Conceptualization. Ming Lu: Writing – original draft, Validation, Resources, Project administration, Formal analysis. Jun Li: Writing – review & editing, Software, Methodology. Qingqing Gui: Validation, Software, Methodology, Investigation, Conceptualization. Yibin Xia: Writing – original draft, Visualization, Software, Formal analysis. Chao Lu: Visualization, Validation, Software, Project administration, Formal analysis. Hongchun Shu: Writing – review & editing, Validation, Project administration, Formal analysis.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
None.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e26781.
Abbreviations
CRCcolorectal cancer
TCGAThe Cancer Genome Atlas
GEOGene Expression Omnibus
PCGprotein coding gene
ICBimmune checkpoint blockade
TMEtumor microenvironment
LASSOleast absolute shrinkage and selection operator
COADcolon adenocarcinoma
SsGSEAsingle sample Gene Set Enrichment Analysis
HRDhomologous recombination defect
MCP-countermicroenvironment cell populations-counter
TIDETumor Immune Dysfunction and Exclusion
IC50half-maximal inhibitory concentration
WGCNAweighted gene correlation network analysis
TOMtopological overlap matrix
CDFcumulative distribution function
OSoverall survival
Appendix A. Supplementary data
The following is/are the supplementary data to this article:
References
- 1.Siegel R.L., et al. Cancer statistics, 2022. CA A Cancer J. Clin. 2022;72(1):7–33. doi: 10.3322/caac.21708. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R.L., et al. Colorectal cancer statistics, 2020. CA A Cancer J. Clin. 2020;70(3):145–164. doi: 10.3322/caac.21601. [DOI] [PubMed] [Google Scholar]
- 3.Arnold M., et al. Global patterns and trends in colorectal cancer incidence and mortality. Gut. 2017;66(4):683–691. doi: 10.1136/gutjnl-2015-310912. [DOI] [PubMed] [Google Scholar]
- 4.Sieminska I., Baran J. Myeloid-derived suppressor cells in colorectal cancer. Front. Immunol. 2020;11:1526. doi: 10.3389/fimmu.2020.01526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harada S., Morlote D. Molecular pathology of colorectal cancer. Adv. Anat. Pathol. 2020;27(1):20–26. doi: 10.1097/PAP.0000000000000247. [DOI] [PubMed] [Google Scholar]
- 6.Johdi N.A., Sukor N.F. Colorectal cancer immunotherapy: options and strategies. Front. Immunol. 2020;11:1624. doi: 10.3389/fimmu.2020.01624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Poureau P.-G., et al. Tolerance and efficacy of regorafenib according to UGT pharmacogenetical status in the treatment of metastatic refractory colorectal cancer. Oncologie. 2021;23(2):195–202. [Google Scholar]
- 8.Zhang H., et al. Progression of exosome-mediated chemotherapy resistance in cancer. Oncologie. 2022;24(2):247–259. [Google Scholar]
- 9.Kuipers E.J., et al. Colorectal cancer. Nat. Rev. Dis. Prim. 2015;1 doi: 10.1038/nrdp.2015.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Biller L.H., Schrag D. Diagnosis and treatment of metastatic colorectal cancer: a review. JAMA. 2021;325(7):669–685. doi: 10.1001/jama.2021.0106. [DOI] [PubMed] [Google Scholar]
- 11.Martini G., et al. Present and future of metastatic colorectal cancer treatment: a review of new candidate targets. World J. Gastroenterol. 2017;23(26):4675–4688. doi: 10.3748/wjg.v23.i26.4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Priyamvada P., Ramaiah S. Potential signature therapeutic biomarkers TOP2A, MAD2L1, and CDK1 in colorectal cancer: a systems biomedicine-based approach. Biochem. Genet. 2023 doi: 10.1007/s10528-023-10544-0. [DOI] [PubMed] [Google Scholar]
- 13.Ma S., et al. RCN3 expression indicates prognosis in colorectal cancers. Oncologie. 2022;24(4):823–833. [Google Scholar]
- 14.Linnekamp J.F., et al. Colorectal cancer heterogeneity and targeted therapy: a case for molecular disease subtypes. Cancer Res. 2015;75(2):245–249. doi: 10.1158/0008-5472.CAN-14-2240. [DOI] [PubMed] [Google Scholar]
- 15.Sprooten J., et al. Necroptosis in immuno-oncology and cancer immunotherapy. Cells. 2020;9(8) doi: 10.3390/cells9081823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gong Y., et al. The role of necroptosis in cancer biology and therapy. Mol. Cancer. 2019;18(1):100. doi: 10.1186/s12943-019-1029-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu F., et al. Complex roles of necroptosis in cancer. J. Zhejiang Univ. - Sci. B. 2019;20(5):399–413. doi: 10.1631/jzus.B1900160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galluzzi L., et al. Necroptosis: mechanisms and relevance to disease. Annu. Rev. Pathol. 2017;12:103–130. doi: 10.1146/annurev-pathol-052016-100247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yan J., et al. Necroptosis and tumor progression. Trends Cancer. 2022;8(1):21–27. doi: 10.1016/j.trecan.2021.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xin S., et al. Identification and quantification of necroptosis landscape on therapy and prognosis in kidney renal clear cell carcinoma. Front. Genet. 2022;13 doi: 10.3389/fgene.2022.832046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nie S., et al. Identification of bladder cancer subtypes based on necroptosis-related genes, construction of a prognostic model. Front. Surg. 2022;9 doi: 10.3389/fsurg.2022.860857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fang K., et al. Comprehensive analysis of necroptosis in pancreatic cancer for appealing its implications in prognosis, immunotherapy, and chemotherapy responses. Front. Pharmacol. 2022;13 doi: 10.3389/fphar.2022.862502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen J., et al. A necroptosis-related gene signature for predicting prognosis, immune landscape, and drug sensitivity in hepatocellular carcinoma. Cancer Med. 2022;11(24):5079–5096. doi: 10.1002/cam4.4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Z., et al. Identification and verification of necroptosis-related gene signature with prognosis and tumor immune microenvironment in ovarian cancer. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.894718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hänzelmann S., Castelo R., Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinf. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilkerson M.D., Hayes D.N. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26(12):1572–1573. doi: 10.1093/bioinformatics/btq170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mayakonda A., et al. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018;28(11):1747–1756. doi: 10.1101/gr.239244.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thorsson V., et al. The immune landscape of cancer. Immunity. 2018;48(4):812–830.e14. doi: 10.1016/j.immuni.2018.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Subramanian A., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Becht E., et al. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol. 2016;17(1):218. doi: 10.1186/s13059-016-1070-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoshihara K., et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Y., et al. HisgAtlas 1.0: a Human Immunosuppression Gene Database. Database. 2017;2017 doi: 10.1093/database/bax094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang P., et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat. Med. 2018;24(10):1550–1558. doi: 10.1038/s41591-018-0136-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Geeleher P., Cox N., Huang R.S. pRRophetic: an R package for prediction of clinical chemotherapeutic response from tumor gene expression levels. PLoS One. 2014;9(9) doi: 10.1371/journal.pone.0107468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Langfelder P., Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinf. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Simon N., et al. Regularization paths for cox's proportional hazards model via coordinate descent. J. Stat. Software. 2011;39(5):1–13. doi: 10.18637/jss.v039.i05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Therneau T.M., Lumley T. Package ‘survival’. R. Top. Doc. 2015;128(10):28–33. [Google Scholar]
- 38.Heagerty P.J., Lumley T., Pepe M.S. Time-dependent ROC curves for censored survival data and a diagnostic marker. Biometrics. 2000;56(2):337–344. doi: 10.1111/j.0006-341x.2000.00337.x. [DOI] [PubMed] [Google Scholar]
- 39.Shen W., et al. Sangerbox: a comprehensive, interaction‐friendly clinical bioinformatics analysis platform. iMeta. 2022;1(3):e36. doi: 10.1002/imt2.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mody K., Bekaii-Saab T. Clinical trials and progress in metastatic colon cancer. Surg. Oncol. Clin. 2018;27(2):349–365. doi: 10.1016/j.soc.2017.11.008. [DOI] [PubMed] [Google Scholar]
- 41.Lieu C.H., Corcoran R.B., Overman M.J. Integrating biomarkers and targeted therapy into colorectal cancer management. Am. Soc. Clin. Oncol. Educ. Book. 2019;39:207–215. doi: 10.1200/EDBK_240839. [DOI] [PubMed] [Google Scholar]
- 42.Liu A., et al. Pan-cancer analysis of a novel indicator of necroptosis with its application in human cancer. Aging. 2022;14(18):7587–7616. doi: 10.18632/aging.204307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li G., et al. Multi-omics analysis reveals the panoramic picture of necroptosis-related regulators in pan-cancer. Aging. 2022;14(12):5034–5058. doi: 10.18632/aging.204124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu J., et al. Programmed cell death tunes tumor immunity. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.847345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li W., et al. PEGylated cisplatin nanoparticles for treating colorectal cancer in a pH-responsive manner. J. Immunol. Res. 2022;2022 doi: 10.1155/2022/8023915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tan L., et al. The interferon regulatory factor 6 promotes cisplatin sensitivity in colorectal cancer. Bioengineered. 2022;13(4):10504–10517. doi: 10.1080/21655979.2022.2062103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gondal M.N., et al. A personalized therapeutics approach using an in silico Drosophila patient model reveals optimal chemo- and targeted therapy combinations for colorectal cancer. Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.692592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lahti S., et al. In vitro biologic efficacy of sunitinib drug-eluting beads on human colorectal and hepatocellular carcinoma-A pilot study. PLoS One. 2017;12(4) doi: 10.1371/journal.pone.0174539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scott A.J., et al. Evaluation of the efficacy of dasatinib, a Src/Abl inhibitor, in colorectal cancer cell lines and explant mouse model. PLoS One. 2017;12(11) doi: 10.1371/journal.pone.0187173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ahangari F., et al. Saracatinib, a selective Src kinase inhibitor, blocks fibrotic responses in preclinical models of pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 2022;206(12):1463–1479. doi: 10.1164/rccm.202010-3832OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chianese G., et al. Hydroxylated cyclopamine analogues from Veratrum californicum and their hedgehog pathway inhibiting activity. Bioorg. Med. Chem. 2023;84 doi: 10.1016/j.bmc.2023.117265. [DOI] [PubMed] [Google Scholar]
- 52.Cao Y., et al. CXCL11 correlates with antitumor immunity and an improved prognosis in colon cancer. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.646252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu M., et al. Colon cancer cells secreted CXCL11 via RBP-Jkappa to facilitated tumour-associated macrophage-induced cancer metastasis. J. Cell Mol. Med. 2021;25(22):10575–10590. doi: 10.1111/jcmm.16989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Du Y., et al. G-protein-coupled receptor 137 accelerates proliferation of urinary bladder cancer cells in vitro. Biotechnol. Appl. Biochem. 2015;62(6):855–860. doi: 10.1002/bab.1329. [DOI] [PubMed] [Google Scholar]
- 55.Tian D.W., et al. RAB38 promotes bladder cancer growth by promoting cell proliferation and motility. World J. Urol. 2019;37(9):1889–1897. doi: 10.1007/s00345-018-2596-9. [DOI] [PubMed] [Google Scholar]
- 56.Cui X., et al. Knockdown of GPR137 by RNAi inhibits pancreatic cancer cell growth and induces apoptosis. Biotechnol. Appl. Biochem. 2015;62(6):861–867. doi: 10.1002/bab.1326. [DOI] [PubMed] [Google Scholar]
- 57.Li B.Y., et al. High expression of RAB38 promotes malignant progression of pancreatic cancer. Mol. Med. Rep. 2019;19(2):909–918. doi: 10.3892/mmr.2018.9732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Song Y., et al. CD8+ T cell-associated genes MS4A1 and TNFRSF17 are prognostic markers and inhibit the progression of colon cancer. Front. Oncol. 2022;12 doi: 10.3389/fonc.2022.941208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Weng W., et al. Novel evidence for a PIWI-interacting RNA (piRNA) as an oncogenic mediator of disease progression, and a potential prognostic biomarker in colorectal cancer. Mol. Cancer. 2018;17(1):16. doi: 10.1186/s12943-018-0767-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Moon J.R., et al. TGF-beta1 protects colon tumor cells from apoptosis through XAF1 suppression. Int. J. Oncol. 2019;54(6):2117–2126. doi: 10.3892/ijo.2019.4776. [DOI] [PubMed] [Google Scholar]
- 61.Xia J., et al. XAF1 promotes colorectal cancer metastasis via VCP-RNF114-JUP axis. J. Cell Biol. 2024;223(2) doi: 10.1083/jcb.202303015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bottcher J.P., et al. NK cells stimulate recruitment of cDC1 into the tumor microenvironment promoting cancer immune control. Cell. 2018;172(5):1022–1037 e14. doi: 10.1016/j.cell.2018.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analyzed during the current study are available in the [GSE39582] repository, [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc= GSE39582], [GSE38832] repository, [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc= GSE38832], [GSE33113] repository, [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc= GSE33113], [GSE14333] repository, [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc= GSE14333], [GSE15222] repository [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc= GSE15222], and [GSE17538] repository, [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc= GSE17538].