Abstract
Using the DNA-binding domain of androgen receptor (AR) as a bait in a yeast two-hybrid screening, we have identified a small nuclear RING finger protein, termed SNURF, that interacts with AR in a hormone-dependent fashion in both yeast and mammalian cells. Physical interaction between AR and SNURF was demonstrated by coimmunoprecipitation from cell extracts and by protein-protein affinity chromatography. Rat SNURF is a highly hydrophilic protein consisting of 194 amino acid residues and comprising a consensus C3HC4 zinc finger (RING) structure in the C-terminal region and a bipartite nuclear localization signal near the N terminus. Immunohistochemical experiments indicated that SNURF is a nuclear protein. SNURF mRNA is expressed in a variety of human and rat tissues. Overexpression of SNURF in cultured mammalian cells enhanced not only androgen, glucocorticoid, and progesterone receptor-dependent transactivation but also basal transcription from steroid-regulated promoters. Mutation of two of the potential Zn2+ coordinating cysteines to serines in the RING finger completely abolished the ability of SNURF to enhance basal transcription, whereas its ability to activate steroid receptor-dependent transcription was maintained, suggesting that there are separate domains in SNURF that mediate interactions with different regulatory factors. SNURF is capable of interacting in vitro with the TATA-binding protein, and the RING finger domain is needed for this interaction. Collectively, we have identified and characterized a ubiquitously expressed RING finger protein, SNURF, that may function as a bridging factor and regulate steroid receptor-dependent transcription by a mechanism different from those of previously identified coactivator or integrator proteins.
Androgen receptor (AR) that mediates the biological actions of physiological androgens is a member of the superfamily of ligand-inducible transcription factors (59). Like other nuclear receptors, AR contains three major structural and interchangeable domains: the N-terminal transactivation domain, the central DNA-binding domain (DBD) that associates with specific androgen response elements (AREs) in target genes, and the C-terminal ligand binding domain (LBD) that binds physiological and synthetic androgens. Upon ligand binding, AR acquires a new conformational state (39), which enables the receptor to interact with AREs and converts the protein to a transcriptional activator. Molecular analyses have shown that the N-terminal half of AR, similar to that of other steroid receptors, contains sequences responsible for the activation function AF-1 (11, 34, 37, 47, 54, 59, 65). In addition to AF-1, another activation function (AF-2) has been identified in the LBDs of various nuclear receptors, including AR (11, 51). Besides encompassing nuclear localization signal (NLS) (38, 65, 75), the functional role of the hinge region residing between the DBDs and LBDs of steroid receptors has remained elusive.
Nuclear receptors have been shown to contact the basal transcription machinery, and it has been postulated that the DNA-bound transcription factors, including steroid receptors, stimulate the efficiency of transcription by affecting directly or indirectly the assembly of the preinitiation complex, which results in an increase in the rate of transcription initiation (12, 32, 48, 67). In addition to direct contacts between nuclear receptors and basal transcription factors such as TFIIB (14, 25, 35) and TFIIF (49), interactions with TATA-binding protein (TBP) or TBP-associated factors, TAFIIs, have been described (36, 50, 63, 64). TAFIIs are suggested to function as mediators between enhancer-bound receptor and basal transcription factors (36, 50, 63, 64). Functionally distinct TFIID complexes, comprising common and unique TAF subunits, may be responsible for specific interactions between TFIID and different activators of transcription (19, 20, 61). Additionally, protein-protein interaction screenings have provided evidence for several new coactivator proteins that can convey the effects of nuclear receptors to the basal transcription machinery. These coactivators are usually defined as limiting factors that enhance transcriptional activity of an upstream activator without being required for basal transcription. Examples of the factors include Ada2 (29), RIP-140 and RIP-160 (16, 17), ERAP140 (26), TIF1 (45), TRIP1/SUG1 (46, 71), ARA70 (74), and SRC-1 (53), together with a number of its variants or related proteins, such as GRIP1, TIF2 and ERAP160/p160 (27, 31, 42, 70). Many of these proteins are reported to function through a direct interaction with the AF-2s of nuclear receptors (32). CBP/p300 proteins that act as cointegrators of several signaling pathways have also been demonstrated to coordinate transcriptional signals from nuclear receptors (1, 18, 27, 42, 73); these proteins have inherent histone acetylase activity and are also capable of recruiting another histone acetyltransferase, PCAF (8, 52, 72).
Even though the DBDs of steroid receptors appear to be mainly involved in DNA binding and homodimerization of receptor monomers, there is evidence that this domain also serves as an interaction interface for other proteins (11, 41, 56, 64). To extend our understanding of the role of the DBD in androgen action, we conducted a yeast two-hybrid screening (23, 30) with this domain of AR as a bait. Here, we report cloning and characterization of a small nuclear RING finger protein termed SNURF, which interacts not only with AR DBD but also with the DBDs of the estrogen and progesterone receptors (ER and PR, respectively). This novel protein is capable of enhancing both steroid receptor-dependent and basal transcription. We suggest that SNURF acts as a bridging factor between steroid receptors and other transcription factors.
MATERIALS AND METHODS
Materials.
pPB(−285/+32)-LUC is a reporter that contains nucleotides −285 to +32 of the rat probasin promoter (56) in pGL3-Basic vector (Promega). pGRE2-E1b-CAT (pARE2-E1b-CAT in this report) contains two copies of rat tyrosine aminotransferase glucocorticoid-progesterone-androgen response element (GRE-PRE-ARE) inserted upstream of the adenovirus E1b TATA sequence (a gift from John A. Cidlowski, NIEHS, Research Triangle Park, N.C.) (3). pARE2-tk-LUC contains two copies of the same GRE-PRE-ARE inserted upstream of the thymidine kinase (tk) promoter (56). pSG5-hPR1 and pHG0 encoding human PR1 and GR, respectively, were gifts from Pierre Chambon (INSERM, Illkirch, France). pSG5-hGR was created by inserting hGR coding sequence from pGH0 as a BamHI fragment into the BamHI site of pSG5 (Stratagene). pCB6-WT18A (WT1) encoding the Wilms’ tumor gene product was from Frank J. Rauscher III (Wistar Institute, Philadelphia, Pa.). pXJ41-BTF1H, pXJ41-TAF30, pXJ42-TAF70β2, and pXJ42-TAF100 were gifts from Laszlo Tora (INSERM). pMOR encoding mouse ER was a gift from Malcolm G. Parker (Imperial Cancer Research Fund, London, England). pG5-CAT contains five Gal4-binding sites in front of the adenovirus E1b minimal promoter driving the CAT gene (Clontech). The β-galactosidase expression plasmid pCMVβ was purchased from Clontech. The following mammalian two-hybrid system vectors were used (all from Clontech): pM for expressing DBD of the Saccharomyces cerevisiae Gal4 protein (amino acid residues 1 to 147), pVP16 for expressing the transcriptional activation domain (VP16 AD) of the herpes simplex virus VP16 protein (amino acid residues 411 to 456), pM-VP16 encoding Gal4 DBD-VP16 AD fusion, and pVP16-CP for expressing a fusion of the VP16 AD to the polyomavirus coat protein. Testosterone was purchased from Makor Chemicals, and progesterone and dexamethasone were obtained from Sigma. Restriction endonucleases and DNA-modifying enzymes were purchased from Pharmacia Biotech. 3H-labeled acetyl coenzyme A was purchased from Dupont-New England Nuclear. Luciferase assay reagent and the TNT-coupled reticulocyte lysate system were purchased from Promega. The yeast two-hybrid system vectors, pVP16, and pLex-a and pLexN-a based on pBTM116 (10) and encoding bacterial LexA and LexA fused to a SV40 large T-antigen nuclear localization signal N terminal to the polylinker were, respectively, kind gifts from Stanley M. Hollenberg (Vollum Institute, Oregon Health Sciences University, Portland, Oreg.).
Isolation of partial cDNAs for AR-interacting proteins by using yeast two-hybrid system.
A yeast two-hybrid screening from mouse embryo E10.5 cDNA library (a gift from S. M. Hollenberg) was used to identify clones that interacted with hAR DBD as described by Hollenberg et al. (30). LexA fusion expression vector pLex-DBD (residues 554 to 644) was generated by inserting PCR-amplified cDNA fragment encoding the indicated amino acids of hAR in frame into the BamHI/SalI site of pLex-a and used as a bait. The library with randomly primed size-selected cDNA inserts (average size, 500 bp) in pVP16 vector used VP16 AD as a transcriptional activator. The yeast strain L40 (MATa trp1-901 leu2-3,112 LYS::(lexAop)4-HIS3 URA3::(lexAop)8-LacZ) was used in the assay. Approximately 2.5 × 107 transformants were screened for interaction in the presence of 0.5 mM 3-aminotriazole. All clones potentially interacting with AR were cured of the bait plasmid and tested against the negative control plasmids pLex-a, pLex-lamin, and pLex-WT1-ZF by a mating strategy with AMR70 [MATa his3D200 lys2-801am trp1-901 leu2-3,112 URA3::(lexAop)8-LacZ] (30). Of the 28 positive clones, 6 contained 400- to 500-bp inserts encoding overlapping fragments of SNURF.
cDNA cloning and characterization.
A rat testis λZapII cDNA library (Stratagene) was screened with 32P-labeled SNURF cDNA corresponding to amino acid residues 20 to 177 by using standard hybridization methods and washings under high-stringency conditions (6). Positive clones were converted in vivo into pBluescript plasmids according to the manufacturer’s instructions. The BLAST program (4) was used to search for DNA and protein sequence homologies in the databases at the National Center for Biotechnology Information, National Institutes of Health.
Plasmid construction.
Rat AR expression vector pSG5-rAR and hAR expression vector pSG5-hAR were constructed as previously described (2, 54). pSG5-rARR590Q/Y603C and pSG5-rAR28.1 (38) were constructed by PCR. Fusion vectors containing indicated (as indicated in parentheses) amino acids of AR were constructed as follows: LexA fusion expression vectors pLex-DBD-s (residues 554 to 623) and pLex-HLBD (residues 624 to 919) were generated by inserting PCR-amplified cDNA fragments of hAR in frame into the BamHI/SalI site of pLex-a (pLexN-a for DBD-s). pLexA-R607/Y620C and pLexA-28.1 were generated by the same PCR protocol, except for the mutated template. pLex-rAR and pM-rAR encoding full-length rAR-LexA fusions were constructed by transferring the BamHI/PstI fragment of rAR from pGEM-3Z-rAR (51) into pLex-a and pM. pM-HLBD and pVP16-rAR-(5–538) have been previously described (34, 51). pLex-WT1-ZF (zinc finger, residues 312 to 419) was created by inserting a PCR-amplified cDNA fragment into the BamHI/SalI site of pLex-a. pLex-SNURF(20–177) was constructed by inserting a PCR-amplified cDNA fragment into BamHI/SalI site of pLex-a. pVP16-SNURF(1–194) was generated by transferring PCR amplified cDNA into the BamHI/NotI site of pVP16. pLex-hPR-DBD and pLex-mER-DBD containing hPR DBD and mER DBD (residues 562 to 652 and 184 to 274, respectively) were constructed by inserting PCR-generated cDNA fragments into the BamHI/SalI site of pLex-a.
pcDNA-SNURF was constructed by replacing the 5′-UTR plus the entire open reading frame of SNURF in pBluescript-SNURF with a PCR-generated SNURF with only nine nucleotides of the 5′-untranslated sequence. The cDNA insert encoding full-length SNURF was subsequently transferred into NotI/ApaI site of pcDNA3.1(+) expression vector (Invitrogen). pcDNA-SNURF(C→S), containing cysteines 136 and 139 mutated to serines, was constructed by replacing a KpnI fragment of pcDNA-SNURF corresponding to amino acids 1 to 189 with the corresponding cDNA fragment containing the indicated point mutations generated by the overlapping PCR mutagenesis strategy (6). pFLAG-SNURF was made by cloning a PCR-generated full-length SNURF cDNA fragment into the HindIII/BamHI site of pFLAG-CMV-2 expression vector (Kodak). pFLAG-SNURFΔID/(C→S) was constructed by replacing an XbaI/KpnI fragment of pFLAG-SNURF that corresponds to residues 18 to 194 with a PCR-generated cDNA fragment encoding amino acids 122 to 194. The pGEX-SNURF was obtained by inserting a PCR-generated cDNA fragment corresponding to SNURF(29–129) into the EcoRI/SalI site of pGEX-5X-1 vector (Pharmacia Biotech). The mammalian two-hybrid expression vector for VP16-SNURF (residues 20 to 177) was created by subcloning one of the cDNA inserts obtained in the two-hybrid screening into the EagI site of pET21c(+) (Novagen) and thereafter transferring the BamHI/XhoI fragment into pVP16. pM-SNURF coding for SNURF-Gal4 DBD fusion was constructed by transferring a BamHI fragment encoding full-length SNURF from pcDNA-SNURF into pM.
pTATA-LUC was constructed by inserting the 19-bp self-complementary oligonucleotide 5′-aAGCTTAGGGTATATAATGAagctt-3′ corresponding to adenovirus E1b TATA sequence into HindIII-digested pGL3-Basic. This plasmid was subsequently used as a recipient for insertion of other elements. An oligonucleotide duplex 5′-cATAGTACGTGATGTTCTAGGCCTAGTACGTGATGTTCTCgagct-3′ containing two AREs of the C3(1) gene flanked by SacI ends (43) was inserted into the SacI site, creating pARE2-TATA-LUC. To construct pSp1-TATA-LUC, an oligonucleotide duplex 5′-ggtaccCGGATCGGGGCGGGGCgagctc-3′ containing an Sp1-binding site with KpnI and SacI overhangs was inserted into the KpnI/SacI site in pTATA-LUC. For pSp12-TATA-LUC, a double-stranded oligonucleotide 5′-CGGATCGGGGCGGGGC-3′ was cloned into the SmaI site of pSp1-TATA-LUC; to create pSp1-AP1-TATA-LUC, a double-stranded oligonucleotide 5′-CGCTTGATGAGTCAGCCGGAA-3′ was annealed into the SmaI site of pSp1-TATA-LUC. The correctness of all constructs was verified by DNA sequencing by using the ALFexpress system (Pharmacia Biotech).
RNA preparation and Northern blot analysis.
RNA was extracted from different adult rat tissues by the LiCl-urea precipitation method (5) and enriched for poly(A)+ RNA by oligo(dT)-cellulose chromatography (7). Polyadenylated RNA samples (5 μg/lane) were fractionated on 1.3% agarose gels containing 2.2 M formaldehyde, then transferred to Hybond-N nylon membrane (Amersham), and finally immobilized onto the membrane by exposure to UV light (Stratalinker). Membrane was hybridized to 32P-labeled SNURF cDNA, washed at high stringency (0.2× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 0.1% sodium dodecyl sulfate [SDS], 52°C), and subjected to autoradiography. A human multiple tissue Northern blot (Clontech no. 7759-1) was hybridized to 32P-labeled SNURF cDNA according to the manufacturer’s protocol, washed at high stringency (0.1× SSC, 0.1% SDS, 50°C), and subjected to autoradiography.
Cell culture and transfections.
All mammalian cell lines were obtained from the American Type Culture Collection (ATCC). CV-1, COS-1, and HeLa cells were maintained in Dulbecco’s minimal essential medium containing penicillin (25 U/ml), streptomycin (25 U/ml), and 10% (vol/vol) fetal bovine serum (FBS). Chinese hamster ovary (CHO) cells were maintained in the same medium but also containing nonessential amino acids. Human prostate carcinoma PC-3 cells were cultured in nutrient mixture F-12 (HAM; Gibco BRL) in the presence of antibiotics and FBS as described above. Cells were transfected by the calcium phosphate precipitation method as described previously (6, 33, 54). The cells (1.5 × 106) were plated on a 10-cm dish 24 h before the precipitate with the indicated amounts of expression and reporter vectors was added. The β-galactosidase expression plasmid, pCMVβ (2 μg/10-cm plate), was used as an internal control for transfection efficiency. For preparation of whole-cell extracts, the indicated amounts of expression vectors/10-cm dish were transfected by electroporation into COS-1 cells as described earlier (33). For protein-protein interaction studies (2.3 × 105 cells/35-mm dish), 1.5 μg of chimeric expression vectors and 3 μg of pG5CAT reporter vector were transfected into CV-1 cells by using DOTAP transfection reagent according to manufacturer’s instructions (Boehringer Mannheim) (34). At 18 h after transfection, the medium was changed to one containing charcoal-stripped 2% (vol/vol) FBS in the presence or absence of testosterone as depicted in the figure legends. CAT and β-galactosidase activities were assayed as previously described (22, 54, 60). Protein concentration was determined by using Bio-Rad protein assay reagents. Luciferase activity was determined with reagents obtained from Promega with a Luminoskan RT reader (Labsystems) (55).
Antibodies, immunoblotting, and immunoprecipitation.
Bacterially expressed GST-SNURF(29–129) was purified by affinity chromatography on glutathione- Sepharose 4B (Pharmacia Biotech) essentially as described earlier (41), except that the fusion proteins were eluted in a buffer containing phosphate-buffered saline (PBS; 140 mM NaCl, 20 mM sodium phosphate [pH 7.4]), 10% (vol/vol) glycerol, and 10 mM reduced glutathione. Polyclonal antisera were raised against purified GST-SNURF(29–129) in rabbits by using 50 μg of protein at each immunization.
Whole-cell extracts from electroporated COS-1 cells were prepared as previously described (33) except that the extraction buffer contained 350 mM KCl, 50 mM NaF, 2 mM dithiothreitol, 15% glycerol, 20 mM sodium phosphate (pH 7.4), 20 mM β-glycerophosphate, 0.5 mM phenylmethylsulfonyl fluoride (PMSF), leupeptin (10 μg/ml), aprotinin (6 μg/ml), pepstatin A (2 μg/ml), and 0.5 μM okadaic acid. To assay the levels of different LexA-AR fusion proteins in yeast cells, cells from 50-ml cultures (grown and harvested as for the β-galactosidase assays) were resuspended in lysis buffer containing 50 mM Tris-HCl (pH 7.4), 140 mM NaCl, 1 mM EDTA, 0.5% Nonidet P-40, 0.1% SDS, 0.5% deoxycholate, 0.5 mM PMSF, aprotinin (10 μg/ml), and pepstatin A (5 μg/ml) and lysed by vortexing six times with glass beads at 4°C for 30 s. Lysates were centrifuged as described above. For immunoblot analysis, 15 μg of cell extracts were fractionated by electrophoresis on polyacrylamide gels under denaturing conditions (44) and electroblotted onto Immobilon-P membrane (Millipore) or Hybond ECL membrane (Amersham). FLAG-tagged proteins were detected with the M2 monoclonal antibody (10 μg/ml; Kodak) according to the manufacturer’s instructions, and immunocomplexes were visualized by using horseradish peroxidase-conjugated goat anti-mouse immunoglobulin G (IgG) antibody (Zymed) and the DAB peroxidase detection system (Sigma). ECL Western blotting detection reagents from Amersham were used with horseradish peroxidase-conjugated goat anti-rabbit IgG antibody (Zymed) to detect SNURF-rabbit IgG complexes. Immunoprecipitation from COS-1 cells was performed essentially as previously described (1) with 15 μg of pFLAG-SNURF (or pFLAG-CMV-2) and 15 μg of pSG5-rAR (or pSG5) per 10-cm plate.
Production of GST-SNURF in insect cells.
Baculovirus transfer plasmid pAcG3X-SNURF was constructed by cloning a PCR-generated full-length SNURF cDNA fragment into SmaI/BamHI site of pAcG3X (Pharmingen, San Diego, Calif.). Recombinant transfer plasmid was cotransfected into Spodoptera frugiperda Sf9 cells with a modified linear baculovirus DNA by using the BaculoGold transfection system (Pharmingen). Sf9 cells were maintained and infected either as monolayers (20 × 106 cells/150-cm2 flask at a multiplicity of infection of 10) or in suspension culture (1.5 × 106 to 2 × 106 cells/ml at a multiplicity of infection of 1) with TNM-FH medium containing 10% FBS, gentamicin (50 μg/ml), and amphotericin B (2.5 μg/ml) (40). The cells infected with recombinant baculovirus DNA were harvested at 65 h postinfection, and the soluble proteins were extracted by homogenization in a buffer containing 50 mM Tris-HCl (pH 7.8), 2 mM EDTA, 50 μM ZnCl2, 10% (vol/vol) glycerol, 300 mM NaCl, 1% (vol/vol) Triton X-100, 0.1% (vol/vol) Nonidet P-40, 0.5 mM PMSF, 5 μM leupeptin, 5 μM pepstatin A, and aprotinin (10 μg/ml), purified by affinity chromatography on glutathione-Sepharose, and eluted in a buffer containing 100 mM Tris-HCl (pH 8.0), 150 mM NaCl, 20 mM reduced glutathione, 15% (vol/vol) glycerol, and 0.5 mM PMSF.
Protein-protein interaction in vitro.
Protein-protein affinity chromatography with purified GST-DBD (41), GST-NTERM, GST-HLBD (56), GST-SNURF, or GST alone bound to glutathione-Sepharose (5 μg of protein/40 μl of resin) and 10 μl of [35S]methionine-labeled in vitro-translated protein was done in a buffer containing 50 mM Tris-HCl (pH 7.8), 150 mM KCl, 0.1% (vol/vol) Nonidet P-40, 0.1% (vol/vol) Triton X-100, 5 mM MgCl2, 0.5 mM EDTA, 10% (vol/vol) glycerol, 50 μM ZnCl2, 0.1 mM sodium orthovanadate, 0.5 mM PMSF, leupeptin (5 μg/ml), aprotinin (10 μg/ml), and pepstatin A (5 μg/ml) in a total volume of 500 μl at 4°C for overnight. The resin was subsequently washed four times with 1 ml of binding buffer. Bound proteins were released in an SDS-polyacrylamide gel electrophoresis (PAGE) sample buffer (44) and then analyzed by electrophoresis under denaturing conditions and fluorography (39).
EMSA.
Electrophoretic mobility shift assays (EMSAs) with purified recombinant proteins or whole-cell extracts from COS-1 cells were carried out as previously described (33, 56).
Immunocytofluorescence.
CV-1 cells seeded on glass slips on 10-cm plastic plates were transfected with DOTAP transfection reagent with 1 μg of pFLAG-SNURF and 10 μg of pBSIISK (Stratagene) as the carrier DNA. Cells were fixed in 4% (wt/vol) paraformaldehyde and permeabilized, and the SNURF protein was then visualized by using anti-FLAG M2 monoclonal antibody (1:50 dilution) or anti-SNURF antiserum (1:500 dilution) and fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse or anti-rabbit secondary antibody, respectively (1:200 dilution; Jackson Immunoresearch Laboratories). AR was visualized with a polyclonal rabbit antiserum raised against full-length rAR and FITC-conjugated goat anti-rabbit secondary antibody as described elsewhere (43).
Immunohistochemistry.
Rat prostate specimens were frozen on dry ice and stored at −80°C. Cryostat sections (7 μm) from Tissue-Tek (Miles, Inc.)-embedded tissues were air dried and fixed in acetone at −20°C or with 4% (wt/vol) paraformaldehyde and dehydrated in a 70 to 100% (vol/vol) ethanol series at room temperature. Sections were stored at −20°C and rehydrated in PBS. After an extensive washing with PBS, sections were incubated with 0.03% (vol/vol) hydrogen peroxide in methanol for 30 min at room temperature to reduce endogenous peroxidase activity. Nonspecific binding was blocked by incubating the tissue sections for 30 min in 10% normal rabbit serum in PBS or in 1% blocking reagent (DIG DNA labeling and detection kit; Boehringer Mannheim) in 100 mM Tris-HCl and 150 mM NaCl (pH 7.9). Polyclonal SNURF antiserum was used in a 1:1,000 dilution and incubated at room temperature for 1 h or at 4°C for 14 to 16 h. After being washed with PBS, biotin-labeled anti-rabbit IgG and AB complex from Vectastain Elite-Kit (Vector Laboratories) were applied according to the manufacturer’s instructions. A peroxidase reaction was carried out with 0.02% (wt/vol) 3-amino-9-ethylcarbazole in 50 mM sodium acetate (pH 5.0) for 20 min at room temperature. Some sections were counterstained with Mayer hematoxylin (Merck) to visualize nuclei.
RESULTS
Isolation of a partial cDNA sequence for SNURF.
The yeast two-hybrid system was used to identify proteins that would interact with the DBD of AR. A fusion between LexA protein and hAR DBD, including part of the hinge region (amino acids 554 to 644), was used as a bait to screen a size-selected 10.5-day-old mouse embryo cDNA library (ca. 500-bp cDNA inserts) fused to the VP16 activation domain (VP16 AD) (30). Approximately 2.5 × 107 yeast transformants were screened, and about 400 candidates were obtained. Nonspecific interactions were eliminated by excluding clones that interacted with the LexA protein or with LexA fused to lamin or the zinc finger region of the Wilms’ tumor gene product, after which 28 colonies that interacted reproducibly with AR DBD were sequenced. Nucleotide sequence analysis revealed that six of these clones corresponded to fragments of a previously uncharacterized protein, here given the name SNURF (for small nuclear RING finger protein).
Structural characteristics of SNURF.
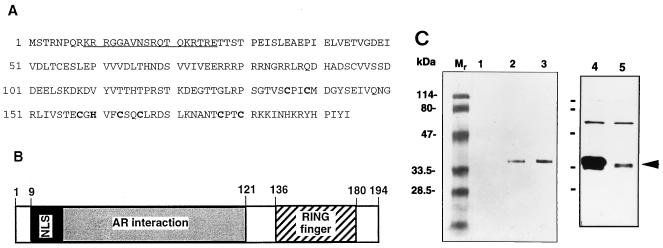
Sequence analysis of a 1.6-kb SNURF cDNA isolated from a rat testis cDNA library revealed an open reading frame for 194 amino acids, with a calculated molecular mass of 22 kDa and an isoelectric point of 8.0 (Fig. 1A). Comparison of the SNURF sequence with those available in current data resources showed a 91% amino acid (derived from cDNA) identity with an uncharacterized human zinc finger sequence recently deposited in the GenBank database (accession number 1843401), which is likely to represent the human homolog of SNURF. Further sequence analysis revealed a bipartite NLS consensus sequence near the N terminus of the protein, other clusters of basic amino acids that might also serve as NLSs, and a C3HC4-type zinc finger (a RING finger motif) in the C-terminal region (residues 136 to 180) (Fig. 1A and B). The latter motif is present in a number of regulatory proteins and has been suggested to mediate protein-protein interactions (62). Other regions of SNURF exhibited no obvious homology to proteins in the existing data resources. On the basis of the six SNURF cDNAs identified in the yeast screening, the region encompassing amino acids 20 to 121 is considered to be sufficient for a SNURF-AR interaction. This region comprises two acidic stretches separated by a sequence rich in arginine and contains seven potential casein kinase II (CKII) phosphorylation sites. The high local concentration of acidic residues is characteristic of many nuclear proteins involved in transcriptional activation.
FIG. 1.
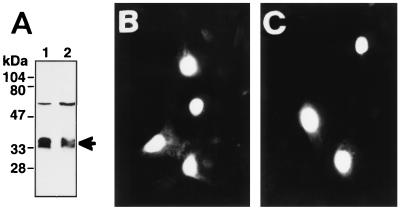
Characteristics of SNURF. (A) Predicted amino acid sequence of rat SNURF (GenBank accession no. AF022081). Cysteine and histidine residues in the consensus RING finger motif (C3HC4-type) presumably involved in zinc complexing are high-lighted, and the potential bipartite NLS is underlined. (B) Schematic structure of SNURF showing regions for bipartite NLS, AR interaction domain, and the RING finger structure. (C) Expression of SNURF protein in COS-1 cells. COS-1 cells were transfected by electroporation with pFLAG-SNURF (30 μg) (lanes 2 and 3), empty pFLAG-CMV-2 expression vector (lane 1), pcDNA-SNURF (15 μg) (lane 4), or empty pcDNA3.1 (lane 5). Cells were harvested 48 h after the transfection and then solubilized and analyzed by immunoblotting with the M2 monoclonal antibody against the FLAG epitope (lanes 1 to 3) as described in Materials and Methods or with an antiserum raised against SNURF in rabbits (1:2,500 dilution) and ECL detection reagents (Amersham) (lanes 4 and 5). The arrowhead depicts the mobility of the SNURF protein.
Expression of a plasmid encoding FLAG-tagged SNURF in COS-1 cells and analysis of the cell extracts by immunoblotting detected a protein with an apparent molecular mass of 35 kDa (Fig. 1C). An antiserum raised against recombinant SNURF in rabbits revealed a protein of similar size also in mock-transfected cells. A comparable Mr value was obtained for SNURF produced by translation in vitro with the reticulocyte lysate system (see Fig. 5C). The SNURF protein is highly hydrophilic and is particularly rich in Ser and Thr (19% of the residues), and about 30% of its residues are charged. Its aberrant migration on SDS-PAGE is, therefore, probably due to this asymmetric charge distribution.
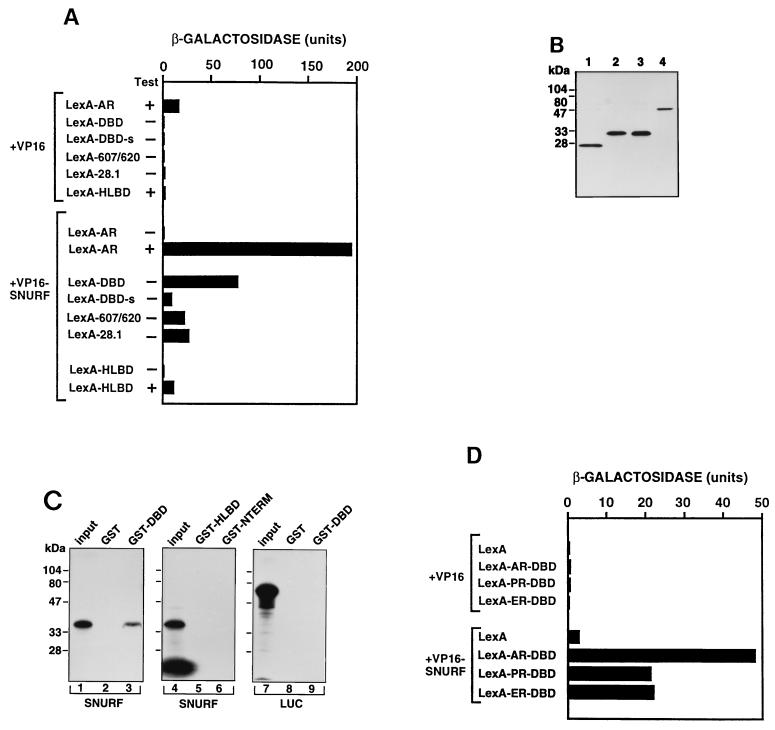
FIG. 5.
Characterization of SNURF-AR interactions in yeast cells and in vitro. (A) Sequences in both DBD and the hinge region of AR are needed for interaction with SNURF. Plasmids expressing LexA, LexA-AR, LexA-DBD, LexA-R607Q/Y620C, LexA-28.1, LexA-DBD-s, and LexA-HLBD were introduced into S. cerevisiae L40 together with expression plasmids for VP16 AD and VP16 AD fused to SNURF (VP16-SNURF, residues 20 to 177 of SNURF). Transformants were grown in the presence (+) or absence (−) of 50 nM testosterone (Test). β-Galactosidase activity in cell extracts of liquid yeast cultures was assayed according to the Clontech two-hybrid system manual. Each bar depicts the average of three independent yeast transformants. (B) Immunoblot analysis of LexA-AR fusion proteins expressed in S. cerevisiae L40. Extracts of yeast cells expressing LexA (lane 1), LexA-DBD (lane 2), LexA-DBD-s (lane 3), and LexA-HLBD (lane 4) were prepared as described in Materials and Methods. All of the samples originated from the same experiment and were processed simultaneously. Immunoblots were developed with a monoclonal LexA antibody from Clontech (1:2,000 dilution) and ECL Western blotting detection reagents. (C) Specific interaction of SNURF and AR DBD in vitro. 35S-labeled full-length SNURF was synthesized by translation in vitro and incubated with GST alone (lane 2), GST-DBD (lane 3), GST-HLBD (lane 5), or GST-NTERM (lane 6) adsorbed to glutathione-Sepharose, after which the matrix was washed and bound proteins were analyzed as described in Materials and Methods. Lanes 8 and 9 show the results of identical experiments in which 35S-labeled luciferase was incubated with GST alone (lane 8) or GST-DBD (lane 9). Lanes 1, 4, and 7 represent 15% of the amount of labeled proteins incubated with matrices. The labeled band migrating at the bottom of lane 4 represents free [35S]methionine. (D) Interaction of estrogen receptor (mouse ER, residues 184 to 274) and progesterone receptor (human PR, residues 562 to 652) DBDs with SNURF. Plasmids expressing LexA or LexA fused to the DBDs of AR, PR, or ER also including part of their hinge regions were introduced into L40 together with expression plasmids for VP16 AD and VP16-SNURF. Each bar depicts the mean of β-galactosidase activities in three independent yeast transformations.
Expression and localization of SNURF.
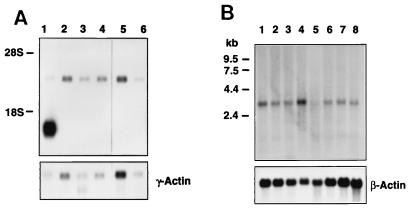
Northern blot analysis revealed the presence of a 3-kb SNURF mRNA in various rat tissues; in addition, a smaller SNURF mRNA species (ca. 1.6 kb) was expressed to a very high level in the testis (Fig. 2A). It is currently unknown whether this smaller mRNA species originates from the utilization of another polyadenylation signal in the 3′-untranslated region, alternative splicing of the primary transcript, or the use of a testis-specific promoter in the SNURF gene. SNURF mRNA is also widely expressed in human cells, as the 3-kb mRNA species was detected in a variety of tissues (Fig. 2B).
FIG. 2.
Expression of SNURF mRNA in rat and human tissues. (A) Northern blot of various rat tissues. Polyadenylated RNA samples (5 μg/lane) from testis (lane 1), prostate (lane 2), seminal vesicles (lane 3), brain (lane 4), spleen (lane 5), and kidney (lane 6). (B) Northern blot of poly(A)+ RNA (2 μg/lane) from various human tissues. Human multiple tissue Northern blot II (Clontech) contains RNA from spleen (lane 1), thymus (lane 2), prostate (lane 3), testis (lane 4), ovary (lane 5), small intestine (lane 6), colon (lane 7), and peripheral blood leukocytes (lane 8). Both blots were probed with 32P-labeled cDNA fragments of SNURF as described in Materials and Methods. To check the integrity of the RNA samples, the rat blot was reprobed with rat γ-actin, and the human blot was reprobed with human β-actin cDNA.
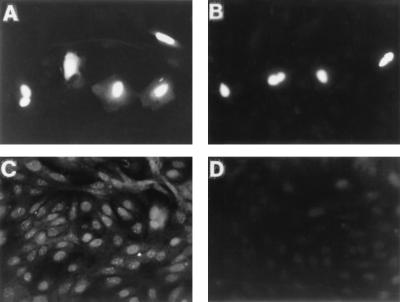
Subcellular localization of SNURF was initially determined by indirect immunofluorescence. Consistent with the presence of NLSs in the SNURF sequence, transiently expressed SNURF appeared to reside exclusively in CV-1 cell nuclei (Fig. 3A). Analysis of subcellular localization by using anti-FLAG monoclonal antibody confirmed the nuclear localization (Fig. 3B). Interestingly, subnuclear localization of endogenous SNURF displayed a fine punctate pattern (Fig. 3C). No staining was detected in controls incubated with the antiserum neutralized with GST-SNURF (Fig. 3D), whereas neutralization with GST alone did not influence the staining pattern (data not shown).
FIG. 3.
Immunocytochemical localization of SNURF in CV-1 cells. CV-1 cells seeded on glass slips on 10-cm plastic plates were transfected by using DOTAP transfection reagent and 1 μg of FLAG-SNURF expression vector as described in Materials and Methods. Cells were fixed in 4% (wt/vol) paraformaldehyde and permeabilized, and the SNURF protein was visualized with rabbit antiserum raised against GST-SNURF (A) or anti-FLAG M2 antibody (B) as described in Materials and Methods. (C) The distribution of endogenous SNURF in CV-1 cells as shown by using anti-SNURF antiserum. (D) Immunofluorescence of CV-1 cells with anti-SNURF antiserum neutralized with purified GST-SNURF fusion protein.
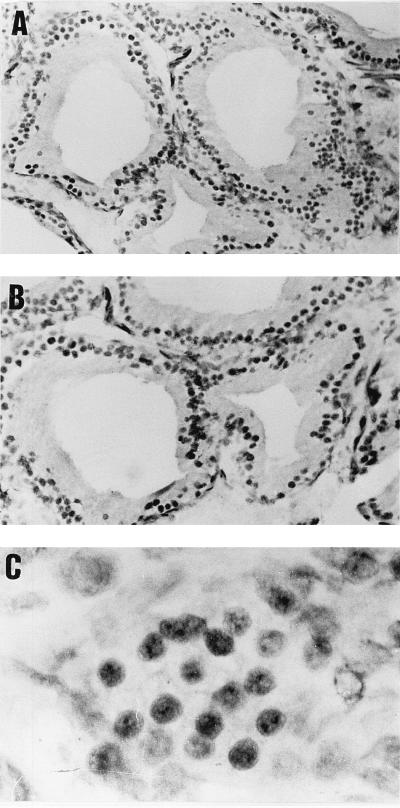
In rat prostate, SNURF antigen was detected solely in the nuclei of secretory epithelial cells (Fig. 4A) and an identical pattern of staining was seen with an antibody specific for AR (Fig. 4B). Negative controls with normal rabbit serum or without the primary antiserum showed no staining (data not shown). The nuclear staining exhibited a granular pattern with a few SNURF-positive granules per nucleus (Fig. 4C). It is also worth noting that prostatic epithelial cells exhibited heterogeneity in the amount of SNURF antigen, a finding similar to that previously reported for immunoreactive AR in these cells (58) and which was confirmed in the present study (Fig. 4B). The staining heterogeneity can be explained by the presence of different cell types in the epithelium of rat prostate and by the possibility that the SNURF gene is expressed in these cells in a cell cycle-dependent fashion.
FIG. 4.
Cellular distribution of SNURF and AR in rat prostate. The immunoperoxidase technique was applied to visualize SNURF and AR by using polyclonal antisera as described in Materials and Methods. (A) SNURF is localized in the nuclei of epithelial cells. (B) Immunoreactive AR protein shows the same pattern of distribution as for SNURF. (C) A higher magnification shows granular clusters of SNURF immunoreactivity in epithelial cell nuclei. Magnifications: ×550 (A), ×700 (B), ×2,800 (C).
Characterization of SNURF-AR interactions in yeast cells and in vitro.
To characterize the interaction between SNURF and AR in more detail, three derivatives of the original bait construct (R607Q/Y620C containing substitution of Arg-607 and Tyr-620 to Gln and to Cys, respectively; DBD-s lacking the hinge residues 624 to 644; and 28.1 devoid of residues 629 to 634), an LBD-hinge region construct (residues 624 to 919, designated HLBD), and full-length rAR were fused to LexA. The resulting hybrid proteins (LexA-R607Q/Y620C, LexA-DBD-s, LexA-28.1, LexA-HLBD, and LexA-AR), along with the original bait construct LexA-DBD, were expressed in the yeast strain L40 with either VP16 AD alone (VP16) or with VP16-AD fused to amino acids 20 to 177 of SNURF (VP16-SNURF). Activation of β-galactosidase reporter was measured in the presence or absence of 50 nM testosterone. Strong reporter gene activation was observed when LexA-AR was coexpressed with VP16-SNURF in the presence but not in the absence of androgen, indicating that AR-SNURF interaction is hormone-dependent in the context of full-length AR (Fig. 5A). The lack of interaction of full-length apo-AR is probably due to its association with heat-shock proteins and other chaperones or to steric hindrance presented by the ligand-free LBD for the interaction between AR DBD and SNURF. Interestingly, two amino acid changes in the C-terminal zinc finger of DBD (R607Q/Y620C) reduced the interaction to one-fourth. Deletion of the entire hinge region, or only six hinge region residues (629 to 634), from the original bait construct attenuated the activation brought about by VP16-SNURF to one-tenth and one-third, respectively. Coexpression of LexA-HLBD with VP16-SNURF did not induce reporter gene activity in the absence of steroid, and the addition of 50 nM testosterone elicited a weak activation (Fig. 5A). The differences among the LexA constructs could not be explained by their dissimilar expression levels, as immunoblot analyses of yeast extracts showed that the amounts of expressed proteins did not differ markedly from each other (Fig. 5B and data not shown). Furthermore, our previous ligand-binding assays indicated that LexA-AR and LexA-HLBD proteins were expressed at similar levels in these yeast cells (51). Taken together, these results indicate that both the DBD and residues in the proximal hinge region of AR are critical for an efficient interaction of SNURF with AR but do not rule out the possibility that other regions of AR may also contribute to SNURF-AR interaction.
Physical interaction between SNURF and AR proteins was assessed by GST pull-down experiments. 35S-labeled SNURF produced by translation in vitro was allowed to bind to AR DBD (residues 554 to 644), HLBD (residues 624 to 919), or AF-1 (residues 141 to 547) fused to GST and immobilized onto a glutathione-Sepharose matrix, after which the bound proteins were analyzed by SDS-PAGE. SNURF associated clearly with AR DBD in vitro, and as much as 10% of the input protein was recovered as complexes with DBD (Fig. 5C), whereas only negligible amounts of SNURF were bound to matrices containing either the HLBD or the AF-1 region of AR. The interaction was specific, as SNURF did not adhere to GST resin devoid of AR DBD and no binding of a control protein, 35S-labeled luciferase, was observed under identical conditions. Thus, there is a physical interaction between SNURF and the AR DBD.
SNURF is also capable of interacting with DBDs of other members of the steroid receptor family. Coexpression of LexA fusions of mER DBD (residues 184 to 274) or hPR DBD (residues 562 to 652) with VP16-SNURF increased LexA-dependent reporter gene activity in yeast cells, albeit to a lesser extent than AR DBD (Fig. 5D).
SNURF interacts with AR in mammalian cells.
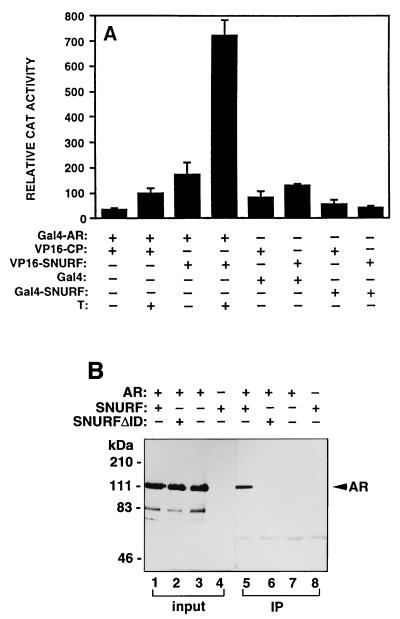
To confirm that SNURF interacts with AR also in mammalian cells, two-hybrid protein-protein interaction assays were carried out in cultured cells. When CV-1 cells were transiently transfected with a vector encoding rAR residues 3 to 902 fused to Gal4 DBD (Gal4-AR) and a vector encoding VP16 AD or VP16 AD fused to polyomavirus coat protein (VP16-CP), only minimal transcriptional activity was observed in the presence of testosterone (Fig. 6A and data not shown). Cotransfection of Gal4-AR with a plasmid encoding SNURF(20–177) as a VP16 AD fusion protein (VP16-SNURF) produced an increased reporter activity which was further augmented by the presence of testosterone, indicating hormone-enhanced recruitment of VP16-SNURF by Gal4-AR in CV-1 cells. The reason for the finding that there is some interaction of SNURF with apo-AR in the mammalian but not in the yeast two-hybrid system is not currently known. Like yeast cells, HLBD alone was not capable of efficiently associating with SNURF in mammalian cells (data not shown). Moreover, the N-terminal region encompassing the AF-1 did not interact with SNURF. Full-length SNURF tethered to Gal4 DBD (Gal4-SNURF) did not activate a minimal reporter controlled by Gal4 binding sites, suggesting that SNURF does not possess a strong intrinsic transcription activation function (Fig. 6A).
FIG. 6.
Interaction between AR and SNURF in mammalian cells. (A) The ability of rAR (residues 3 to 902) fused to the DBD of Gal4 (Gal4-AR) to interact with VP16 AD fused to SNURF residues 20 to 177 (VP16-SNURF) or to polyomavirus coat protein (VP16-CP) was examined in CV-1 cells by assaying chloramphenicol acetyltransferase (CAT) activity from the reporter plasmid pG5CAT. Cells (2.3 × 105 cells/35-mm dish) were transfected with 1.5 μg of each chimeric expression vector and 3 μg of pG5CAT reporter by using DOTAP transfection reagent. At 18 h after transfection, the medium was changed to one containing charcoal-stripped 2% (vol/vol) FBS in the presence (+) or absence (−) of 100 nM testosterone (T), and the cells were incubated for an additional 30 h. Interaction between VP16-SNURF and Gal4-SNURF was also examined. Transcriptional activity is expressed as the relative CAT activity corrected for protein concentration. The mean ± standard error values for at least three separate experiments are shown. The CAT activity derived from the interaction between hormone-bound Gal4-AR and VP16-SNURF corresponds to ca. 25% of that of a positive control; between p53 and large T antigen, tethered to Gal4 and VP16, respectively (34). (B) AR and SNURF are physically associated in COS-1 cells. COS-1 cells were transfected by electroporation with pFLAG-SNURF or pFLAG-SNURFΔID and pSG5-rAR as indicated. After a 30-h culture in the presence of 100 nM testosterone, whole-cell extracts were prepared and subjected to immunoprecipitation (IP) with mouse monoclonal anti-FLAG antibody. Immunoprecipitated proteins were analyzed by immunoblotting with a rabbit anti-AR antibody. Lanes 1 to 4 (input) represent portions of the cell extracts (5%) that were subjected to immunoblotting without prior immunoprecipitation.
To examine whether SNURF and AR are physically associated in intact cells, COS-1 cells were transfected with AR and FLAG-tagged SNURF expression vectors. Protein complexes associated with SNURF were first immunoprecipitated with the anti-FLAG monoclonal antibody, and the bound proteins were subsequently analyzed by immunoblotting with an AR-specific antibody. AR protein was clearly demonstrated in immunoprecipitates from cells transfected with both SNURF and AR but not from those expressing AR in the presence of SNURFΔID devoid of AR interaction domain (ID; residues 18 to 121) or only AR in the absence of SNURF, confirming that AR and SNURF are found as complexes in vivo (Fig. 6B).
Influence of SNURF overexpression on AR-dependent transcription.
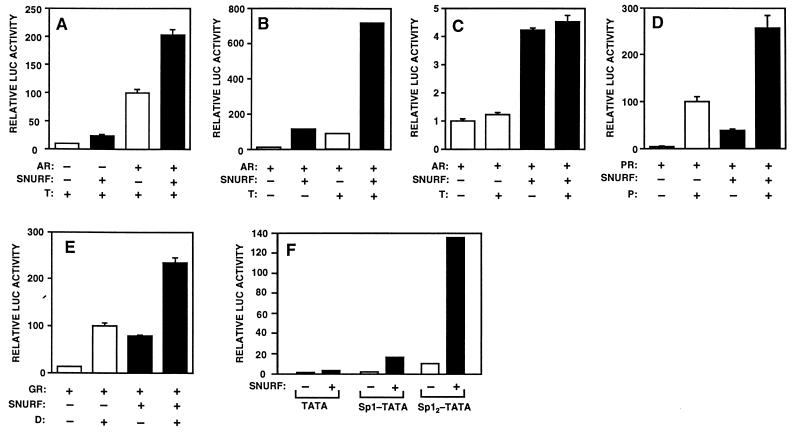
Transient transfection assays were performed in mammalian cell lines to investigate the influence of SNURF overexpression on AR-dependent transcription. COS-1 cells were transfected with expression vectors for SNURF and rAR along with the probasin promoter that is regulated by androgen. As shown in Fig. 7A, AR activated the reporter gene 10-fold in the presence of testosterone, and coexpressed SNURF increased further this AR-dependent transactivation. SNURF also activated transcription in the absence of AR, suggesting that it influences the function of other transcription factors governing probasin promoter activity (Fig. 7A). In the absence of cotransfected AR, testosterone did not influence reporter gene activity in the presence or absence of SNURF (data not shown). The above findings could not be explained by an increased cellular concentration of the receptor protein, as immunoblotting analysis of cell extracts showed that coexpressed SNURF does not alter AR concentration (data not shown). Similar results were obtained when other cell lines (e.g., CV-1, HeLa, CHO, and PC-3) were employed.
FIG. 7.
Influence of SNURF overexpression on AR-dependent transactivation. (A) SNURF enhances AR-dependent and basal transcription from the rat probasin promoter. COS-1 cells were transfected by the calcium phosphate method with 5 μg of pPB(−285/+32)-LUC reporter plasmid along with 1 μg of pSG5-rAR or empty pSG5 and 5 μg of SNURF expression vector (pcDNA-SNURF) or empty expression vector (pcDNA-3.1+) in the presence 25 nM testosterone (T) as depicted. β-Galactosidase expression plasmid, pCMVβ (2 μg), was used as a control for transfection efficiency. (B) CV-1 cells were transfected with 1 μg of pSG5-rAR, 5 μg of pARE2-TATA-LUC reporter, 2 μg of pCMVβ, and 5 μg of pcDNA-SNURF or empty expression vector in the presence or absence of testosterone (T) as depicted. (C) The experimental conditions were as in panel B, except that 5 μg of pTATA-LUC (devoid of AREs) was used as a reporter. Reporter gene activities are expressed relative to that achieved with pSG5-rAR in the presence of testosterone (100 in panels A and B; 1 in panel C), and the mean ± standard error values of at least three independent experiments are given. (D and E) SNURF activates PR- and GR-dependent transcription. (D) Effect of SNURF on the transcriptional activity of PR. CV-1 cells were transfected with 5 μg of pARE2-tk-LUC reporter containing two copies of the GRE-PRE-ARE element of the rat TAT gene upstream of the thymidine kinase promoter along with 1 μg of pSG5-hPR1, 5 μg of pcDNA-SNURF or empty expression vector pcDNA-3.1+, and 2 μg of pCMVβ in the presence or absence of 100 nM progesterone (P). (E) CV-1 cells were transfected as for panel D but with 1 μg of pSG5-hGR instead of pSG5-hPR1 in the presence or absence of 100 nM dexamethasone (D). Luciferase (LUC) activities are expressed relative to those achieved with pSG5-hPR1 and pSG5-hGR in the presence of progesterone and dexamethasone, respectively (those values being equal to 100), and the mean ± standard error values of at least three independent experiments are shown. (F) Effect of SNURF overexpression on Sp1 activity. CV-1 cells were transiently transfected with 5 μg of pTATA-LUC, pSp1-TATA-LUC, or pSp12-TATA-LUC reporters along with 5 μg of SNURF expression vector (pFLAG-SNURF) or empty expression vector (pFLAG-CMV-2) and also 2 μg of pCMVβ. Transcriptional activities are expressed as relative luciferase (LUC) activity normalized by using the β-galactosidase activity.
Next, we examined the effect of SNURF overexpression on the transcription from a minimal reporter gene construct regulated by two AREs in front of the E1b TATA sequence (pARE2-TATA-LUC). As was the case with the more complex probasin promoter, both basal and androgen-dependent transcription were markedly activated by coexpressed SNURF with pARE2-TATA-LUC as the reporter (Fig. 7B). The amount of SNURF expression plasmid used (5 μg of DNA/10-cm plate) brought about a maximal effect, and higher concentrations (10 to 15 μg of DNA/plate) resulted in a 35 to 50% decrease from this maximal effect. If AREs in the promoter region were deleted or replaced by estrogen response elements, SNURF-mediated enhancement of AR-dependent transactivation was not observed (Fig. 7C and data not shown). In agreement with the yeast two-hybrid interaction data (Fig. 5), mutations corresponding to R607Q/Y620C and 28.1 in the context of rAR expression vector attenuated the response to coexpressed SNURF by 60 ± 11% and 49 ± 16%, respectively.
The above results along with those depicted in Fig. 5D suggested that SNURF is not specific for AR. It was therefore pertinent to study the effects of SNURF on other steroid receptors and transcription factors. Similar to AR, PR- and GR-dependent transactivation from a reporter containing two steroid response elements in front of the thymidine kinase promoter was significantly augmented by overexpressed SNURF (Fig. 7D and E). We also compared activities of some simple promoter constructs containing binding sites for TBP, Sp1, and AP1 in the absence or presence of coexpressed SNURF. As shown in Fig. 7F, SNURF was capable of activating a weak promoter construct that comprises adenovirus E1b TATA as the sole transcription regulatory element. When the Sp1 binding site(s) was inserted in front of the TATA sequence, the effect of SNURF was more pronounced and resulted in a 8- to 14-fold activation of the reporter gene (Fig. 7F). Overexpression of SNURF also activated markedly (≥15-fold) transcription from a minimal promoter containing an AP1 site located between the Sp1 site and the TATA sequence (see Fig. 8D).
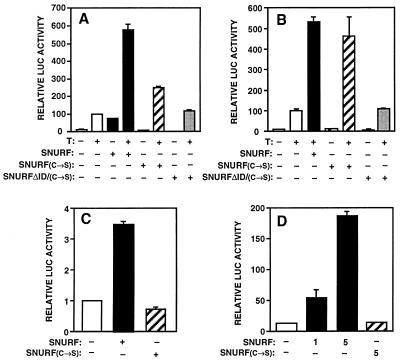
FIG. 8.
RING finger mutated SNURF is capable of enhancing AR-dependent transactivation but not basal transcription. (A) Influence of SNURF and SNURF(C→S) on transcription from the AR-dependent probasin promoter. CV-1 cells were transfected with 5 μg of pPB(−285/+32)-LUC reporter, 1 μg of pSG5-rAR, 2 μg of pCMVβ, and 5 μg of expression vectors of SNURF, SNURF(C→S), or SNURFΔID/(C→S) or the empty expression vector (pcDNA-3.1+) in the presence or absence of 100 nM testosterone (T) as indicated. (B) Transcription from a minimal AR-dependent promoter. Experimental conditions were as described for panel A, except that 5 μg of pARE2-TATA-LUC was used as the reporter. (C) Effect of SNURF and SNURF(C→S) on transcription from a minimal TATA-LUC promoter. Experimental conditions were as described for panel A, except that 5 μg of pTATA-LUC in the absence of pSG5-rAR was used. (D) Influence of SNURF and SNURF(C→S) on transcription from a simple Sp1-AP1-TATA promoter. CV-1 cells were transfected as described above but in the absence of pSG5-rAR, and 5 μg of pSp1-AP1-TATA-LUC containing a Sp1 and an AP1 binding site upstream of TATA sequence was used as the reporter. The amounts of pcDNA-SNURF and pcDNA-SNURF(C→S) are given in micrograms. Luciferase (LUC) activities were normalized for transfection efficiency by using the β-galactosidase activity.
Different regions of SNURF mediate activation of steroid receptor-dependent and basal transcription.
The RING finger motif has been proposed to be involved in protein-protein interactions (62). In the case of PML (15) and equine herpes virus IE110 protein (9), the two zinc atoms of the RING finger are ligated tetrahedrally by the conserved cysteine and histidine residues of the consensus sequence. To characterize the importance of the RING finger structure in the SNURF-mediated activation of transcription, two point mutations converting cysteines 136 and 139 to serines were introduced. The influence of C→S mutations on transcription from the natural AR-inducible probasin promoter was first investigated. As with COS-1 cells (Fig. 7A), overexpression of wild-type SNURF activated both basal and AR-dependent transcription from the probasin promoter in CV-1 cells (Fig. 8A). SNURF(C→S) variant was totally incapable of stimulating basal transcription but maintained ca. 50% of the wild-type SNURF’s ability to activate AR-dependent transcription (Fig. 8A). Furthermore, SNURF(C→S) protein was also able to efficiently activate AR-dependent pARE2-TATA-LUC reporter but did not influence basal transcription (Fig. 8B) or transcription from pTATA-LUC reporter in the presence of AR and testosterone (data not shown). Deletion of the receptor interaction region from SNURF(C→S) rendered it inactive in AR-regulated transcription, strengthening the notion that SNURF activates AR through direct protein-protein interaction (Fig. 8A and B). Together, these findings imply that the RING finger structure is not critically involved in the activation of steroid receptor function but does serve as an interaction interface for other regulatory proteins, such as basal transcription factors.
To test further the latter possibility, the effect of SNURF(C→S) on transcription from a minimal TATA promoter was analyzed. In contrast to wild-type SNURF, SNURF(C→S) was unable to enhance the activity of the promoter (Fig. 8C). In addition, increasing amounts of SNURF activated markedly an Sp1-AP1-TATA promoter, whereas SNURF(C→S) did not do so (Fig. 8D). These results lend further support to the conclusion that integrity of the RING finger region is indeed required for the SNURF-dependent activation of basal transcription. Immunoblot analyses of CV-1 cell extracts and immunofluorescence studies showed that the inability of SNURF(C→S) or SNURFΔID/(C→S) to enhance basal transcription was not due to its significantly lower expression level or altered subcellular localization (Fig. 9 and data not shown).
FIG. 9.
Comparison of SNURF and SNURF(C→S) proteins levels and subcellular localization in CV-1 cells. (A) CV-1 cells were transfected with 15 μg of expression vectors for SNURF (lane 1) or SNURF(C→S) (lane 2) by the calcium phosphate precipitation method. Cells were harvested 48 h later and then solubilized and analyzed by immunoblotting with anti-SNURF antiserum raised in rabbits (1:2,500 dilution) and ECL detection reagents as described in Materials and Methods. The arrow depicts the position of the SNURF protein. (B) Immunocytochemical localization of SNURF(C→S) (B) and SNURF (C) in the same experiment. Conditions for transfection and immunological analyses were as described for Fig. 3.
SNURF interacts with TBP in vitro.
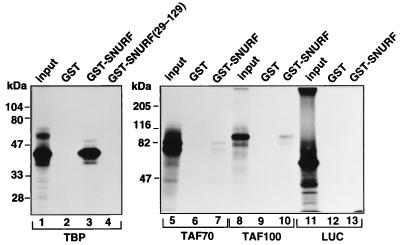
A 16-kDa multiprotein bridging factor 1 (MBF1) interacts with the DBD of D. melanogaster nuclear receptor FTZ-F1 (66), a region corresponding to that of AR employed in the interaction with SNURF. The finding that MBF1 binds to TBP prompted us investigate whether there is also a physical interaction between SNURF and TBP. As shown in Fig. 10, as much as one-third of the input TBP protein was recovered as complexes with full-length SNURF in a GST pull-down assay, whereas only negligible amounts of TBP were bound to matrices containing only GST or GST-SNURF(29–129) that encompasses the AR interaction domain but not the RING finger region. Under identical conditions, very low amounts of some TAFs (TAFII70, TAFII100, or TAFII30) and luciferase were bound to SNURF (Fig. 10 and data not shown), attesting to the specificity of SNURF-TBP interaction.
FIG. 10.
Specific interaction of SNURF and TATA-binding protein in vitro. 35S-labeled TBP, TAFII70, TAFII100, and luciferase were synthesized by translation in vitro and incubated with GST alone (lanes 2, 6, 9, and 12), GST-SNURF (lane 3, 7, 10, and 13), or GST-SNURF(29–129) (lane 4) adsorbed to glutathione-Sepharose, after which the matrices were washed, and the bound proteins were analyzed as described in Materials and Methods. Lanes 1, 5, 8, and 11 correspond to 20% of the input amounts of 35S-labeled proteins.
DISCUSSION
During the last few years, several putative coregulatory proteins have been identified for nuclear receptors. Most of them interact in an agonist-dependent or agonist-enhanced fashion with LBDs of a number of nuclear receptors and possibly convey the transcription activation function AF-2 residing in LBDs to the basal transcription machinery (32). Recently, a short sequence motif LXXLL (where X is any amino acid) in the coregulatory proteins has been identified to serve as a signature sequence that facilitates the interaction of these proteins with AF-2s of nuclear receptors (28, 68). Even though the physiological role of nuclear receptor coactivators remains to be elucidated, it is becoming clear that these ligand-inducible transcription factors are capable of associating, probably simultaneously, with multiple target proteins. Identification of an increasing number of proteins with the potential to enhance or repress steroid receptor action indicates that the activation process is complex and is controlled, at least in part, by the cellular ratio of coregulatory proteins.
In this study we have identified a novel nuclear receptor coactivator protein, SNURF, that does not contain LXXLL motifs but that does display significant activation potential in cotransfection experiments. The exact molecular mechanism underlying the recognition of AR by SNURF is currently unclear. The region encompassing residues 20 to 121 of SNURF, which does not contain the RING finger domain, is essential for the interaction. This region contains several potential CKII phosphorylation sites, and SNURF is indeed phosphorylated as efficiently as the model substrate β-casein by CKII in vitro (our unpublished observations), suggesting that the interactions mediated through this region of SNURF are modulated by its phosphorylation status. In AR, besides the DBD, residues 624 to 644 of the hinge region contributed to the AR-SNURF interaction, as the DBD alone was not sufficient to achieve strong interaction. Binding of SNURF to AR and SNURF-mediated transcriptional enhancement were compromised by mutations both in the DBD and the hinge region, mutations that do not affect DNA or hormone binding. The residues 617 to 633 in the proximal hinge region encompass the NLS (75). However, SNURF does not appear to be involved in the nuclear import of AR, as coexpression of SNURF with AR failed to alter the amount of nuclear AR in CV-1 cells (our unpublished results).
SNURF was able to activate transcription in an efficient fashion from both minimal and complex AR-dependent promoters; however, several features of the SNURF-mediated activation are different from those of the nuclear receptor coactivators characterized so far. First, AR DBD and the N-terminal part of hinge region, which are sufficient for interaction with SNURF, do not appear to possess an autonomous transcription activation function (this work and our unpublished results). In view of this, it is interesting that Schwerk et al. (64) have reported that there is a weak transactivation domain within the DBD of PR which mediates the interaction between hPR and TAFII110 in vitro. These investigators also suggested that this interaction is an important step in the PR-mediated transactivation process. Second, when tethered to enhancer elements by fusion to Gal4 DBD, SNURF did not exhibit clear autonomous transcriptional activation potential in mammalian or yeast cells. Third, AR binding to AREs was not influenced by SNURF in vitro (our unpublished results). It is, therefore, unlikely that activation of AR-dependent transcription is simply due to increased binding of the receptor to its cognate response elements. Moreover, recombinant SNURF does not possess significant binding affinity for double- or single-stranded DNA.
In addition to steroid receptors, SNURF also increased markedly the activities of other transcription factors, such as AP1 and Sp1, acting through their cognate response elements. Our data thus imply that SNURF functions as a more general coregulator, possibly as a bridging factor, participating in the coordination of the activities of multiple transcriptional signals from upstream factors. Besides a growing family of interacting proteins for nuclear receptor LBDs, there appears to be a group of proteins capable of recognizing the DBDs. Takemaru et al. (66) recently described another small cofactor, MBF1, which interacts with the C-terminal region of DBD of Drosophila nuclear receptor FTZ-F1, a region similar to the AR’s interaction domain with SNURF. Like SNURF, MBF1 does not bind to DNA but interconnects FTZ-Z1, MBF2, and TBP, which is essential for the FTZ-F1-induced transcription. Intriguingly, SNURF is also capable of binding to TBP in vitro. TLS/FUS has recently been demonstrated to act as a high-affinity binding protein for the DBDs of retinoid, glucocorticoid, and thyroid hormone receptors (57). TLS bears significant sequence homology to hTAFII68 and is also present in a subset of TFIID complexes (13). It is of considerable interest that when the zinc finger DNA-binding domain of another transcription factor, GATA-1, was used as a bait in the yeast two-hybrid screen, a novel zinc finger protein, FOG, was identified as a coactivator of GATA-1 (69), indicating that cofactors recognizing the zinc finger domains of transcription factors are not restricted to the C4-type fingers of nuclear receptors. Like SNURF, FOG Gal4 DBD fusions failed to stimulate transcription from promoters containing Gal4-binding sites, implying that its transcriptional activity is also context-dependent and probably involves other protein-protein or protein-DNA interactions. Point mutations in the RING finger domain of SNURF abolished selectively the activation of basal transcription and signals from Sp1 and AP1 but retained the capacity of SNURF to stimulate steroid receptor-dependent transcription. This suggests that the interaction with different regulators is mediated by distinct SNURF domains. The notion that RING finger motifs, present both in nuclear and cytoplasmic proteins with diverse regulatory functions, are mediating specific protein-protein interactions (15, 62) is thus supported by our findings. Moreover, the region encompassing the RING finger appeared to be mandatory for SNURF-TBP interaction in vitro.
SNURF amino acid sequence is highly conserved between human and rat cells, and the expression of SNURF mRNA is not restricted to classical androgen target tissues. In keeping with the putative role of SNURF in transcriptional regulation, endogenous SNURF protein was detected in the nuclei of cultured cells and rat tissues. Even though SNURF shares no obvious homology, except for the consensus RING finger structure present also in TIF1 (45), with previously characterized coactivator proteins, subnuclear localization of many of these coregulators, such as RIP140, TIF1, and TIF2, shows a similar punctuate pattern (17, 45, 70). The composition and functional role of these nuclear structures, generally referred to as nuclear bodies, are poorly known, but several recent studies have suggested that these structures play a significant role in the regulation of gene expression (21, 24).
Collectively, the present study reports the identification and characterization of a novel nuclear regulatory protein which may interact by utilizing selective domains with steroid receptors and other transcription factors and mediate their synergistic function at the level of gene transcription. While a better understanding of the mechanisms of SNURF action will require identification of other interaction partners, the current results could be explained by SNURF functioning as a bridging factor between sequence-specific upstream factors and RNA polymerase II-basal transcription machinery.
ACKNOWLEDGMENTS
We thank Ms. Leena Pietilä, Pirjo Kilpiö, and Seija Mäki for excellent technical assistance; A. O. Brinkmann, P. Chambon, J. A. Cidlowski, M. G. Parker, F. J. Rauscher III, L. Tora, L. Seikku, and H. Santti for plasmids; and S. M. Hollenberg for providing the materials for the yeast two-hybrid system.
This work was supported by grants from the Medical Research Council of the Academy of Finland, the Finnish Foundation for Cancer Research, the Jalmari and Rauha Ahokas Foundation, and the University of Helsinki.
REFERENCES
- 1.Aarnisalo P, Palvimo J J, Jänne O A. CREB-binding protein in androgen receptor-mediated signaling. Proc Natl Acad Sci USA. 1998;95:2122–2127. doi: 10.1073/pnas.95.5.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adeyemo O, Kallio P J, Palvimo J J, Kontula K, Jänne O A. A single-base substitution in exon 6 of the androgen receptor gene causing complete androgen insensitivity: the mutated receptor fails to transactivate but binds to DNA in vitro. Hum Mol Genet. 1993;2:1809–1812. doi: 10.1093/hmg/2.11.1809. [DOI] [PubMed] [Google Scholar]
- 3.Allgood V E, Oakley R H, Cidlowski J A. Modulation by vitamin B6 of glucocorticoid receptor-mediated gene expression requires transcription factors in addition to the glucocorticoid receptor. J Biol Chem. 1993;268:20870–20876. [PubMed] [Google Scholar]
- 4.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 5.Auffray C, Rougeon F. Purification of mouse immunoglobulin heavy-chain messenger RNAs from total myeloma tumor RNA. Eur J Biochem. 1980;107:303–314. doi: 10.1111/j.1432-1033.1980.tb06030.x. [DOI] [PubMed] [Google Scholar]
- 6.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1997. [Google Scholar]
- 7.Aviv H, Leder P. Purification of biologically active globin messenger RNA by chromatography on oligothymidylic acid-cellulose. Proc Natl Acad Sci USA. 1972;69:1408–1412. doi: 10.1073/pnas.69.6.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bannister A J, Kouzarides T. The CBP coactivator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 9.Barlow P N, Luisi B, Milner A, Elliot M, Everett R. Structure of the C3HC4 domain by 1H-nuclear magnetic resonance spectroscopy. A new structural class of zinc-finger. J Mol Biol. 1994;237:201–211. doi: 10.1006/jmbi.1994.1222. [DOI] [PubMed] [Google Scholar]
- 10.Bartel P L, Chien C T, Sternglanz R, Fields S. Using the two-hybrid system to detect protein-protein interactions. In: Hartley D A, editor. Cellular interactions in development: a practical approach. Oxford, England: IRL Press; 1993. pp. 153–179. [Google Scholar]
- 11.Beato M, Herrlich P, Schütz G. Steroid hormone receptors: many actors in search for a plot. Cell. 1995;83:851–857. doi: 10.1016/0092-8674(95)90201-5. [DOI] [PubMed] [Google Scholar]
- 12.Beato M, Sanchez-Pacheco A. Interaction of steroid hormone receptors with the transcription initiation complex. Endocr Rev. 1996;17:587–609. doi: 10.1210/edrv-17-6-587. [DOI] [PubMed] [Google Scholar]
- 13.Bertolotti A, Lutz Y, Heard D J, Chambon P, Tora L. hTAFII68, a novel RNA/ssDNA-binding protein with homology to the pro-oncoproteins TLS/FUS and EWS is associated with both TFIID and RNA polymerase II. EMBO J. 1996;15:5022–5031. [PMC free article] [PubMed] [Google Scholar]
- 14.Blanco J C G, Wang I-M, Tsai S Y, Tsai M-J, O’Malley B W, Jurutka P W, Haussler M R, Ozato K. Transcription factor TFIIB and vitamin D receptor cooperatively activate ligand-dependent transcription. Proc Natl Acad Sci USA. 1995;92:1535–1539. doi: 10.1073/pnas.92.5.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Borden L B K, Boddy M N, Lally J, O’Reilley N J, Martin S, Howe K, Solomon E, Freemont P S. The solution structure of the RING finger domain from the acute promyelocytic leukaemia proto-oncoprotein PML. EMBO J. 1995;14:1532–1541. doi: 10.1002/j.1460-2075.1995.tb07139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cavaillés V, Dauvois S, Danielian P S, Parker M G. Interaction of proteins with transcriptionally active estrogen receptors. Proc Natl Acad Sci USA. 1994;91:10009–10013. doi: 10.1073/pnas.91.21.10009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cavaillés V, Dauvois S, L’horset F, Lopez G, Hoare S, Kushner P J, Parker M G. Nuclear factor RIP140 modulates transcriptional activation by the estrogen receptor. EMBO J. 1995;14:3741–3751. doi: 10.1002/j.1460-2075.1995.tb00044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chakravarti D, LaMorte V J, Nelson M C, Nakajima T, Juguilon H, Montminy M, Evans R M. Role of CBP/p300 in nuclear receptor signalling. Nature. 1996;383:99–103. doi: 10.1038/383099a0. [DOI] [PubMed] [Google Scholar]
- 19.Chen J-L, Attardi D L, Verrijzer C P, Yokomori K, Tjian R. Assembly of recombinant TFIID reveals differential coactivator requirements for distinct transcriptional activators. Cell. 1994;79:93–105. doi: 10.1016/0092-8674(94)90403-0. [DOI] [PubMed] [Google Scholar]
- 20.Chiang C-M, Roeder R G. Cloning of an intrinsic human TFIID subunit that interacts with multiple transcriptional activators. Science. 1995;267:531–536. doi: 10.1126/science.7824954. [DOI] [PubMed] [Google Scholar]
- 21.Dyck J A, Maul G D, Miller W H, Chen J D, Kakizuka A, Evans R M. A novel macromolecular structure is a target of the promyelocyte-retinoic acid receptor oncoprotein. Cell. 1994;76:333–343. doi: 10.1016/0092-8674(94)90340-9. [DOI] [PubMed] [Google Scholar]
- 22.Eastman A. An improvement to the novel rapid assay for chloramphenicol acetyltransferase gene expression. BioTechniques. 1987;5:73. [Google Scholar]
- 23.Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 24.Guiochon-Mantel A, Savouret J F, Quignon F, Delabre K, Milgrom E, De The H. Effect of PML and PML-RAR on the transactivation and subcellular distribution of steroid hormone receptors. Mol Endocrinol. 1995;9:1791–1803. doi: 10.1210/mend.9.12.8614415. [DOI] [PubMed] [Google Scholar]
- 25.Hadzig E, Desai-Yajnik V, Helmer E, Guo S, Wu S, Koudinova N, Casanova J, Raaka B M, Samuels H H. A 10-amino acid sequence in the N-terminal A/B domain of thyroid hormone receptor a is essential for transcriptional activation and interaction with the general transcription factor TFIIB. Mol Cell Biol. 1995;15:4507–4517. doi: 10.1128/mcb.15.8.4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Halachmi S, Marden E, Martin G, MacKay H, Abbondanza C, Brown M. Estrogen receptor-associated proteins—possible mediators of hormone-induced transcription. Science. 1994;264:1455–1458. doi: 10.1126/science.8197458. [DOI] [PubMed] [Google Scholar]
- 27.Hanstein B, Eckner R, DiRenzo J, Halachmi S, Liu H, Searcy B, Kurokawa R, Brown M. p300 is a component of an estrogen receptor coactivator complex. Proc Natl Acad Sci USA. 1996;93:11540–11545. doi: 10.1073/pnas.93.21.11540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heery D M, Kalkhoven E, Hoare S, Parker M G. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 29.Henriksson A, Almlöf T, Ford J, McEwan I J, Gustafsson J-Å, Wright A P. Role of the Ada adaptor complex in gene activation by the glucocorticoid hormone. Mol Cell Biol. 1997;17:3065–3973. doi: 10.1128/mcb.17.6.3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hollenberg S M, Sternglanz R, Cheng P F, Weintraub H. Identification of a new family of tissue-specific basic helix-loop-helix proteins with a two-hybrid system. Mol Cell Biol. 1995;15:3813–3822. doi: 10.1128/mcb.15.7.3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hong H, Kohli K, Trivedi A, Johnson D L, Stallcup M R. GRIP1, a novel mouse protein that serves as a transcriptional coactivator in yeast for the hormone binding domains of steroid receptors. Proc Natl Acad Sci USA. 1996;93:4948–4952. doi: 10.1073/pnas.93.10.4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Horwitz K B, Jackson T A, Bain D L, Richard J K, Takimoto G S, Tung L. Nuclear receptor coactivators and corepressors. Mol Endocrinol. 1996;10:1167–1177. doi: 10.1210/mend.10.10.9121485. [DOI] [PubMed] [Google Scholar]
- 33.Ikonen T, Palvimo J J, Kallio P J, Reinikainen P, Jänne O A. Stimulation of androgen-regulated transactivation by modulators of protein phosphorylation. Endocrinology. 1994;135:1359–1366. doi: 10.1210/endo.135.4.7925097. [DOI] [PubMed] [Google Scholar]
- 34.Ikonen T, Palvimo J J, Jänne O A. Interaction between amino- and carboxyl-terminal regions of rat androgen receptor modulates transcriptional activity and is influenced by nuclear receptor coactivators. J Biol Chem. 1997;272:29821–29828. doi: 10.1074/jbc.272.47.29821. [DOI] [PubMed] [Google Scholar]
- 35.Ing N H, Beekman J M, Tsai S Y, Tsai M-J, O’Malley B W. Members of the steroid hormone receptor superfamily interact with TFIIB (S300-II) J Biol Chem. 1992;267:17617–17623. [PubMed] [Google Scholar]
- 36.Jacq X, Brou C, Lutz Y, Davidson I, Chambon P, Tora L. Human TAFII30 is present in a distinct TFIID complex and is required for transcriptional activation by the estrogen receptor. Cell. 1994;79:107–117. doi: 10.1016/0092-8674(94)90404-9. [DOI] [PubMed] [Google Scholar]
- 37.Jenster G, van der Korput H A G M, van Vroonhoven C, van der Kwest T H, Trapman J, Brinkmann A O. Domains of the human androgen receptor involved in steroid binding, transcriptional activation and subcellular localization. Mol Endocrinol. 1991;5:1396–1404. doi: 10.1210/mend-5-10-1396. [DOI] [PubMed] [Google Scholar]
- 38.Jenster G, Trapman J, Brinkmann A O. Nuclear import of the human androgen receptor. Biochem J. 1993;293:761–768. doi: 10.1042/bj2930761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kallio P J, Jänne O A, Palvimo J J. Agonists, but not antagonists, alter the conformation of the hormone-binding domain of androgen receptor. Endocrinology. 1994;134:998–1001. doi: 10.1210/endo.134.2.8299593. [DOI] [PubMed] [Google Scholar]
- 40.Kallio P J, Palvimo J J, Mehto M, Jänne O A. Analysis of androgen receptor-DNA interactions with receptor proteins produced in insect cells. J Biol Chem. 1994;269:11514–11522. [PubMed] [Google Scholar]
- 41.Kallio P J, Poukka H, Moilanen A, Jänne O A, Palvimo J J. Androgen receptor mediated transcriptional regulation in the absence of direct interaction with a specific DNA element. Mol Endocrinol. 1995;9:1017–1028. doi: 10.1210/mend.9.8.7476976. [DOI] [PubMed] [Google Scholar]
- 42.Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, Lin C-S, Heyman R A, Rose D W, Glass C K, Rosenfeld M G. A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- 43.Karvonen U, Kallio P J, Jänne O A, Palvimo J J. Interaction of androgen receptors with androgen response elements in intact cells: roles of amino- and carboxyl-terminal regions and the ligand. J Biol Chem. 1997;272:15973–15979. doi: 10.1074/jbc.272.25.15973. [DOI] [PubMed] [Google Scholar]
- 44.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;22:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 45.Le Douarin B, Zechel C, Garnier J-M, Lutz Y, Tora L, Pierrat P, Heery D, Gronemeyer H, Chambon P, Losson R. The N-terminal part of TIF1, a putative mediator of the ligand-dependent activation function (AF-2) of nuclear receptors, is fused to B-raf in the oncogenic protein T18. EMBO J. 1995;14:2020–2033. doi: 10.1002/j.1460-2075.1995.tb07194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee J W, Ryan F, Swaffield S A, Johnston S A, Moore D D. Interaction of thyroid hormone receptor with a conserved transcriptional mediator. Nature. 1995;374:91–94. doi: 10.1038/374091a0. [DOI] [PubMed] [Google Scholar]
- 47.Mangelsdorf D J, Thummel C, Beato M, Herrlich P, Schütz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, Evans R M. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McEwan I J, Wright A P, Gustafsson J-Å. Mechanism of gene expression by the glucocorticoid receptor: role of protein-protein interactions. Bioessays. 1997;19:153–160. doi: 10.1002/bies.950190210. [DOI] [PubMed] [Google Scholar]
- 49.McEwan I J, Gustafsson J-Å. Interaction of the human androgen receptor transactivation function with the general transcription factor TFIIF. Proc Natl Acad Sci USA. 1997;94:8485–8490. doi: 10.1073/pnas.94.16.8485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mengus G, May M, Carré L, Chambon P, Davidson I. Human TAFII135 potentiates transcriptional activation by the AF-2s of the retinoic acid, vitamin D3, and thyroid hormone receptors in mammalian cells. Genes Dev. 1997;11:1381–1395. doi: 10.1101/gad.11.11.1381. [DOI] [PubMed] [Google Scholar]
- 51.Moilanen A, Rouleau N, Ikonen T, Palvimo J J, Jänne O A. The presence of a transcription activation function in the hormone-binding domain of androgen receptor is revealed by studies in yeast cells. FEBS Lett. 1997;412:355–358. doi: 10.1016/s0014-5793(97)00791-6. [DOI] [PubMed] [Google Scholar]
- 52.Ogryzko V V, Schilz R L, Russanove V, Howard B H, Nakatani Y. Transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 53.Oñate S A, Tsai S Y, Tsai M-J, O’Malley B W. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science. 1995;270:1354–1357. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- 54.Palvimo J J, Kallio P J, Ikonen T, Mehto M, Jänne O A. Dominant negative regulation of trans-activation by the rat androgen receptor: roles of the N-terminal domain and heterodimer formation. Mol Endocrinol. 1993;7:1399–1028. doi: 10.1210/mend.7.11.8114755. [DOI] [PubMed] [Google Scholar]
- 55.Palvimo J J, Partanen M, Jänne O A. Characterization of a cell specific modulatory element in the murine ornithine decarboxylase promoter. Biochem J. 1996;316:993–998. doi: 10.1042/bj3160993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Palvimo J J, Reinikainen P, Ikonen T, Kallio P J, Moilanen A, Jänne O A. Mutual transcriptional interference between RelA and androgen receptor. J Biol Chem. 1996;271:24151–24156. doi: 10.1074/jbc.271.39.24151. [DOI] [PubMed] [Google Scholar]
- 57.Powers C A, Mathur M, Raaka B M, Ron D, Samuels H H. TLS (translocated-in-liposarcoma) is a high affinity interactor for steroid, thyroid, and retinoid receptors. Mol Endocrinol. 1998;12:4–18. doi: 10.1210/mend.12.1.0043. [DOI] [PubMed] [Google Scholar]
- 58.Prins G S, Birch L, Greene G L. Androgen receptor localization in different cell types of the adult rat prostate. Endocrinology. 1991;129:3187–3199. doi: 10.1210/endo-129-6-3187. [DOI] [PubMed] [Google Scholar]
- 59.Quigley C A, DeBellis A, Marschke K B, El-Awady M K, Wilson E M, French F S. Androgen receptor defects: historical, clinical, and molecular perspectives. Endocr Rev. 1995;16:271–321. doi: 10.1210/edrv-16-3-271. [DOI] [PubMed] [Google Scholar]
- 60.Rosenthal N. Identification of regulatory elements of cloned genes with functional assays. Methods Enzymol. 1987;152:704–720. doi: 10.1016/0076-6879(87)52075-4. [DOI] [PubMed] [Google Scholar]
- 61.Sauer F, Hansen S K, Tjian R. Multiple TAFIIs directing synergistic activation of transcription. Science. 1995;270:1783–1788. doi: 10.1126/science.270.5243.1783. [DOI] [PubMed] [Google Scholar]
- 62.Saurin A J, Borden K L B, Boddy M N, Freemont P S. Does this have a familiar RING? Trends Biochem Sci. 1996;21:208–214. [PubMed] [Google Scholar]
- 63.Schulman I G, Chakravarti D, Juguilon H, Romo A, Evans R M. Interactions between the retinoid X receptor and a conserved region of the TATA-binding protein mediate hormone-dependent transactivation. Proc Natl Acad Sci USA. 1995;92:8288–8292. doi: 10.1073/pnas.92.18.8288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schwerk C, Klotzbucher M, Sachs M, Ulber V, Klein-Hitpass L. Identification of a transactivation function in the progesterone receptor that interacts with the TAFII110 subunit of the TFIID complex. J Biol Chem. 1995;270:21331–21338. doi: 10.1074/jbc.270.36.21331. [DOI] [PubMed] [Google Scholar]
- 65.Simental J A, Sar M, Lane M V, French F S, Wilson E M. Transcriptional activation and nuclear targeting signals of the human androgen receptor. J Biol Chem. 1991;266:510–518. [PubMed] [Google Scholar]
- 66.Takemaru K-I, Li F-Q, Ueda H, Hirose S. Multiprotein bridging factor 1 (MBF1) is an evolutionary conserved transcriptional coactivator that connects a regulatory factor and TATA element binding protein. Proc Natl Acad Sci USA. 1997;94:7251–7256. doi: 10.1073/pnas.94.14.7251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tjian R, Maniatis T. Transcriptional activation: a complex puzzle with few easy pieces. Cell. 1994;77:5–8. doi: 10.1016/0092-8674(94)90227-5. [DOI] [PubMed] [Google Scholar]
- 68.Torchia J, Rose D W, Inostroza J, Kamei Y, Westin S, Glass C K, Rosenfeld M G. The transcriptional co-activator p/CIP binds CBP and mediates nuclear receptor function. Nature. 1997;387:677–684. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- 69.Tsang A P, Visvader J E, Turner C A, Fujiwara Y, Yu A, Weiss M J, Crossley M, Orkin S H. FOG, a multitype zinc finger protein, acts as a cofactor for transcription factor GATA-1 in erythroid and megakaryotic differentiation. Cell. 1997;90:109–119. doi: 10.1016/s0092-8674(00)80318-9. [DOI] [PubMed] [Google Scholar]
- 70.Voegel J J, Hiene M J S, Zechel C, Chambon P, Gronemeyer H. TIF2, a 160 kDa transcriptional mediator for the ligand-dependent activation function AF-2 of nuclear receptors. EMBO J. 1996;15:3667–3675. [PMC free article] [PubMed] [Google Scholar]
- 71.vom Baur E, Zechel C, Heery D, Heine M J S, Garnier J M, Vivat V, Le Douarin B, Gronemeyer H, Chambon P, Losson R. Differential ligand-dependent interactions between the AF-2 activating domain of nuclear receptors and putative transcriptional intermediary factors mSUG1 and TIF1. EMBO J. 1996;15:110–124. [PMC free article] [PubMed] [Google Scholar]
- 72.Yang X-J, Ogryzko V V, Nishikawa J, Howard B H, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 73.Yao T-P, Ku G, Zhou N, Scully R, Livingston D M. The nuclear hormone receptor coactivator SRC-1 is a specific target of p300. Proc Natl Acad Sci USA. 1996;93:10626–10631. doi: 10.1073/pnas.93.20.10626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yeh S, Chang C. Cloning and characterization of a specific coactivator, ARA70, for the androgen receptor in human prostate cells. Proc Natl Acad Sci USA. 1996;93:5517–5521. doi: 10.1073/pnas.93.11.5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhou Z-X, Sar M, Simental J A, Lane M V, Wilson E M. A ligand-dependent bipartite nuclear targeting signal in the human androgen receptor. J Biol Chem. 1994;269:13115–13123. [PubMed] [Google Scholar]