Abstract
An antibody that specifically recognized phosphothreonine 72 in ets-2 was used to determine the phosphorylation status of endogenous ets-2 in response to colony-stimulating factor 1 (CSF-1)/c-fms signaling. Phosphorylation of ets-2 was detected in primary macrophages, cells that normally express c-fms, and in fibroblasts engineered to express human c-fms. In the former cells, ets-2 was a CSF-1 immediate-early response gene, and phosphorylated ets-2 was detected after 2 to 4 h, coincident with expression of ets-2 protein. In fibroblasts, ets-2 was constitutively expressed and rapidly became phosphorylated in response to CSF-1. In both cell systems, ets-2 phosphorylation was persistent, with maximal phosphorylation detected 8 to 24 h after CSF-1 stimulation, and was correlated with activation of the CSF-1 target urokinase plasminogen activator (uPA) gene. Kinase assays that used recombinant ets-2 protein as a substrate demonstrated that mitogen-activated protein (MAP) kinases p42 and p44 were constitutively activated in both cell types in response to CSF-1. Immune depletion experiments and the use of the MAP kinase kinase inhibitor PD98059 indicate that these two MAP kinases are the major ets-2 kinases activated in response to CSF-1/c-fms signaling. In the macrophage cell line RAW264, conditional expression of raf kinase induced ets-2 expression and phosphorylation, as well as uPA mRNA expression. Transient assays mapped ets/AP-1 response elements as critical for basal and CSF-1-stimulated uPA reporter gene activity. These results indicate that persistent activation of the raf/MAP kinase pathway by CSF-1 is necessary for both ets-2 expression and posttranslational activation in macrophages.
Macrophage colony-stimulating factor 1 (CSF-1) controls the proliferation and differentiation of cells of the mononuclear phagocyte cell lineage. The actions of CSF-1 are mediated through an integral membrane receptor tyrosine kinase, the product of the c-fms proto-oncogene (22). As with other tyrosine kinase receptors, ligand binding leads to c-fms autophosphorylation, assembly of phosphotyrosine-dependent signaling complexes, and the subsequent activation of signal transduction pathways (25). Pathways controlled by CSF-1/c-fms include phosphatidylinositol 3-kinase (23), JAK-STATs (19), c-src-related kinases (6), and the ras pathway (3, 7). The latter two pathways have been demonstrated to be critical for the mitogenic action of CSF-1 (3, 6, 15).
CSF-1 stimulation results in the stable, persistent expression of specific genes, for example, the urokinase plasminogen activator (uPA) gene, in mature macrophages or in fibroblasts engineered to express c-fms (3, 14, 27). The uPA gene encodes an extracellular protease involved in cellular migration in many cell types, including metastatic tumor cells (2) and macrophages (5, 27).
The uPA promoter contains regions conserved across species up to 8.2 kb 5′ to the transcription start site (1, 5, 9). Within these regions of homology, two compound ets/AP-1 growth factor- and oncogene-responsive elements have been identified at −2.4 and −6.9 kb upstream of the transcription initiation site (1, 9, 27). In transient transfections, oncoprotein ras collaborates with either ets-1 or ets-2 to superactivate the uPA promoter via the compound ets/AP-1 enhancer located at −2.6 kb relative to the transcription initiation site (34). Collaboration between ras and exogenously expressed ets factors depends on ras-dependent phosphorylation at threonine residues Thr 38 and Thr 72 in ets-1 and ets-2, respectively (34). The Thr 38 residue of ets-1 has been shown to be phosphorylated in a CSF-1-dependent manner in NIH 3T3 cells that exogenously express both ets-1 and c-fms (21). The phosphorylation sites are contained in a 100-amino-acid domain that is conserved between ets-1 and ets-2 and also in the Drosophila melanogaster protein pointed P2 (4, 20). The conserved N-terminal domain of the ets factor pointed P2 has been shown to be a nuclear target for ras signaling pathways critical for differentiation of the R7 photoreceptor cell in Drosophila, and thus defines a target for ras signaling pathways that is conserved through evolution from flies to humans (4, 20).
One well-characterized effector pathway activated by the ras-GTP complex is the raf/MEK-1/mitogen-activated protein (MAP) kinase pathway (16, 31). However, the exact identity of the ras effector pathways that CSF-1/c-fms engage to persistently activate the uPA promoter have not been defined. The ras/MAP kinase pathway has been shown to activate TCF/elk-1 ets family transcription factors, but these events occur early after growth factor stimulation and result in regulation of immediate-early genes such as c-fos (reviewed in reference 31). In PC-12 cells, activation of trkA has been shown to sustain activation of ras and MAP kinases p42 and p44 over several hours, leading to the proposition that the duration and strength of the ras signal are the critical variables that distinguish how cells interpret ras/MAP kinase signals generated by different environmental stimuli (reviewed in reference 16).
In the present study, we present evidence for a signaling cascade initiated by CSF-1/c-fms in either macrophages or heterologous cells that ectopically express c-fms. This pathway involves stimulation of the ras pathway, resulting in continuous activation of MAP kinases p42 and p44 and stable phosphorylation of ets-2 at threonine 72, events that are correlated with the induction of uPA transcription by CSF-1.
MATERIALS AND METHODS
Cell culture and RNA analysis.
NIH 3T3 cells containing genes expressing c-fms protein were grown in Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal calf serum (FCS). Prior to stimulation with CSF-1 (1 U = 0.01 ng) or platelet-derived growth factor (PDGF, BB isoform; Upstate Biotechnology, Inc.), cells were grown in DMEM with 0.1% FCS for 16 to 24 h. Cells were stimulated with 104 U of CSF-1 per ml (27). RAW264 cells were maintained in RPMI media containing heat-inactivated 5% FCS. RAW264 cells expressing the estrogen receptor-active raf kinase fusion protein were obtained from Julie Hambleton and Tony DeFranco and were stimulated with β-estradiol (10−7 M) as described previously (11). The isolation of bone marrow-derived macrophages (BMMs) has been previously described (27), and the cells were grown in RPMI media containing 10% FCS and 104 U of CSF-1 per ml. For the experiments described here, BMMs were deprived of CSF-1 for 16 to 24 h and then restimulated with 104 U of CSF-1 per ml as indicated in the figure legends.
For experiments with the specific MEK-1 inhibitor PD98059, serum-starved cells were treated with the drug at a final concentration of 50 μM (10) for 15 min prior to addition of CSF-1. The drug was dissolved in dimethyl sulfoxide, and control cells were treated with vehicle alone.
RNA isolation and Northern blotting were performed as previously described (14, 27).
Production of anti-phosphopeptide T72 ets-2 antibody and Western blotting.
The peptides LPLL(p-T)PCS and LPLLTPCSKA corresponding to amino acids 68 to 77 of human ets-2 were synthesized. The position of the phosphate at threonine 72 was confirmed by nuclear magnetic resonance. The phosphopeptide was coupled to keyhole limpet hemocyanin and used to immunize two New Zealand White rabbits. Collected serum was pooled and passed over a column to which the nonphosphopeptide was coupled, and material that did not bind to this column was collected and passed over a phosphopeptide affinity column. Bound material was eluted from this second column with glycine (pH 2) buffer, dialyzed against phosphate-buffered saline and stored at −70°C before use. Polyclonal, phosphorylation-independent ets-2 antibodies were produced by immunization of rabbits with a recombinant ets-2 protein corresponding to amino acid residues 60 to 167 [ets-2(60–167)].
Western blotting was performed with nitrocellulose membranes and the ECG detection system (Amersham) as previously described (17, 34).
In-gel and MAP kinase assays.
The substrate for both in-gel kinase and MAP kinase assays was the portion of human ets-2 corresponding to amino acid residues 60 to 167. This protein was expressed as a six-histidine-tagged recombinant protein in Escherichia coli K-12 by using the pET15b expression vector system and was purified to >95% purity by nickel-Sepharose affinity chromatography. Versions of the protein with either threonine or alanine at position 72 were produced and purified.
The in-gel and MAP kinase assays have been described in detail elsewhere (17). For in-gel kinase assays, 100 μg of total protein derived from BMMs or RAW264 cells was subjected to electrophoresis through a 12.5% acrylamide gel which had been copolymerized with 500 μg of either the threonine 72 or alanine 72 versions of ets-2 protein per ml. After electrophoresis, the gel was subjected to a denaturation-renaturation procedure, the in-gel kinase reaction was performed, and the gel was subjected to autoradiography for 24 h (17).
An anti-MAP kinase antibody coupled to Sepharose beads was used for the immune kinase experiments (Santa Cruz Biochemicals, Santa Cruz, Calif.). The immune complex obtained from incubation of cell extracts with this antibody was suspended in 30 μl of kinase buffer (20 mM HEPES [pH 7.2], 10 mM MgCl2, 1 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride, 10 μg of leupeptin per ml, 10 μg of aprotinin per ml, 1 mM orthovanadate, 1 mM EGTA, 10 mM sodium fluoride, 1 mM tetrasodium pyrophosphate, 0.1 mM β-glycerophosphate) that contained 1 μg of the purified His-tagged ets-2 protein corresponding to amino acids 60 to 167 and 5 μCi of [γ-32P]ATP. After incubation at room temperature for 30 min, the reaction was terminated by addition of sodium dodecyl sulfate (SDS) sample buffer and boiling. The supernatant was run on denaturing gels and analyzed by autoradiography. In some experiments, as indicated in the figure legends, cold ATP was included in the kinase reaction. For these experiments, phosphorylation of threonine 72 in the ets-2 substrate was determined by Western blotting with the antiphosphopeptide ets-2 antibody described above.
The jun kinase assays were performed with an influenza virus hemagglutinin (HA)-tagged version of p55-JNK2 and glutathione S-transferase (GST)–N-terminal c-jun recombinant proteins (residues 1 to 79) as previously described (13). Both wild-type (S63/S73) and mutated (S63/A73) versions of c-jun were employed (13).
Plasmids and transient transfections.
The parent plasmid for the murine uPA promoter-luciferase reporters was pGL2-B (Promega). The construction of the −8.2, −6.6, −6.6 Δets/AP-1, and −114 luciferase reporter plasmids has been previously described (1, 27). The −4.2, −2.6, and −2.2 reporters were derived from the −6.6 plasmid by digestion with HindIII, EcoRV, or BglII restriction sites located at these positions in the −6.6 plasmid. The mouse c-fms cDNA (3.7 kb) was placed into the simian virus 40 expression vector pECE (27). The fms-pECE plasmid was obtained from Changmin Chen (Centre for Molecular and Cellular Biology, University of Queensland, Queensland, Australia). Electroporation of RAW264 cells and determination of luciferase activity following transfection were performed as previously described (27).
RESULTS
Recombinant ets-2 protein is a specific substrate for purified MAP kinases p42 and p44 in vitro.
In work that depended on transient transfection systems, we demonstrated that threonine 72 of ets-2 was phosphorylated in a ras-dependent fashion (34). In an attempt to define kinases that catalyze phosphorylation of this site in ets-2 in vitro, an N-terminal region of ets-2 corresponding to the region conserved in the Drosophila pointed P2 protein (amino acids 60 to 167) was overexpressed as a six-histidine-tagged fusion protein in bacteria. The fusion protein was subsequently purified and used as a substrate for in vitro kinase reactions. Purified, recombinant MAP kinase p44 could utilize this portion of ets-2 as a substrate (Fig. 1A, lane 1). A recombinant ets-2 protein containing the Ala 72 substitution was not used as a substrate by MAP kinase p44 (Fig. 1A, lane 2). Furthermore, phosphoamino acid analysis of the 32P-labeled threonine 72 ets-2(60–167) protein revealed exclusive phosphorylation at threonine (data not shown). Identical results were obtained with purified MAP kinase p42 (data not shown).
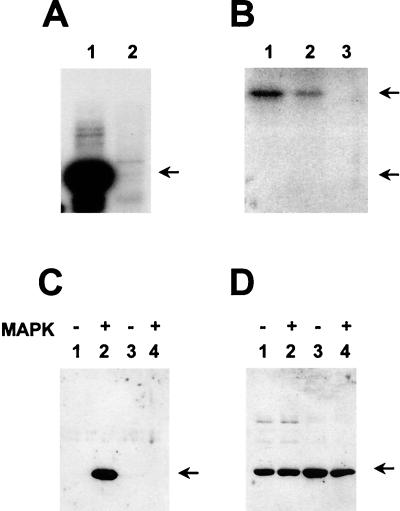
FIG. 1.
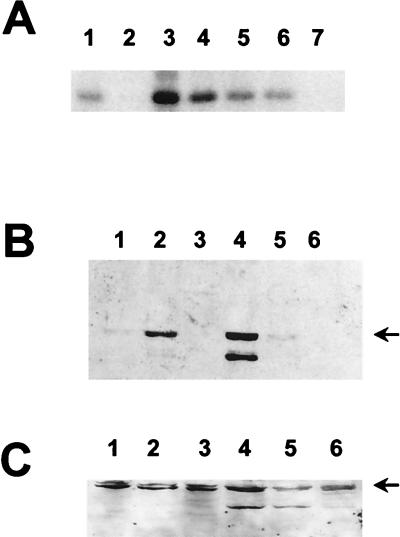
MAP kinases p42 and p44 phosphorylate position threonine 72 of ets-2 in vitro. (A) Recombinant ets-2 proteins corresponding to amino acids 60 to 167 that contained either threonine 72 or alanine 72 (200 ng of protein, lanes 1 and 2, respectively) were incubated with [γ-32P]ATP and purified, activated MAP kinase p44 for 30 min and separated on an SDS-15% polyacrylamide gel. Autoradiography was performed for 12 h. (B) In a parallel experiment, HA-tagged JUNK2 was expressed in NIH 3T3 cells, immunoprecipitated, and used in immune kinase assays with an N-terminal jun-GST fusion protein (amino acids 1 to 79), a jun-GST protein with residues 63 and 73 mutated from serine to alanine, or the threonine 72 version of ets-2(60–167) (200 ng of each protein, lanes 1 to 3, respectively). Autoradiography was performed for 12 h. (C and D) Western blot of recombinant human ets-2 proteins (amino acids 60 to 167) with threonine at position 72 (lanes 1 and 2) or alanine at position 72 (lanes 3 and 4). The proteins (200 ng) were incubated with nonradioactive ATP in the absence (lanes 1 and 3) or presence (lanes 2 and 4) of activated MAP kinase p42 (Upstate Biotech, Inc.) for 30 min. The blot was probed with affinity-purified anti-phosphopeptide T72 antibody (C [see Materials and Methods]). The same blot was stripped and probed with polyclonal antibody directed against ets-2 residues 60 to 167 (D).
In contrast, the MAP kinase family member p55-JNK2 (13) did not utilize the ets-2 recombinant protein (Fig. 1B lane 3) as a substrate under conditions where the N-terminal region of c-jun (residues 1 to 79) could be phosphorylated in vitro (Fig. 1B, lanes 1 and 2). In these experiments, the ets-2 protein was a poorer substrate than even the c-jun substrate that lacked the major phosphorylation sites at positions S63 and S73 (Fig. 1B, lane 2), which was still detectably phosphorylated at minor sites by JNK2 as previously reported (13). Additionally, both MAP kinase p38 (32) and the proline-directed kinase cdk4 were unable to catalyze phosphorylation of the ets-2 threonine 72 substrate in vitro (data not shown).
Production and characterization of antiserum specific for ets-2 phosphothreonine 72.
In order to directly measure the phosphorylation of endogenous ets-2 at amino acid position threonine 72, an antibody that was specific for the phosphorylated threonine residue was developed. For this purpose, the peptide PLL-pT-PCSKA (corresponding to amino acids 69 to 77 of ets-2) was synthesized and used to produce polyclonal rabbit serum. Following affinity purification (see Materials and Methods), the specificity of the antibody for detecting phosphothreonine 72 ets-2 in Western blotting experiments was tested with recombinant ets-2 proteins corresponding to amino acids 60 to 167.
For these experiments, the ets-2 region was incubated in vitro with nonradioactive ATP and activated MAP kinase p44 (Fig. 1C). These experiments showed that only wild-type threonine 72 protein which had been incubated with the MAP kinase preparation was recognized by the antibody (lane two versus lane 1 in Fig. 1C). Proteins containing the A72 substitution were not recognized by the antibody whether MAP kinase was present or absent (lanes 3 and 4). A second nondiscriminating (i.e., phosphorylation independent) antiserum directed against the ets-2 N-terminal region (amino acids 60 to 167) was also produced. When the blot shown in Fig. 1C was stripped and reprobed with the second antibody, equal loading of the recombinant proteins could be demonstrated (Fig. 1D). The phosphopeptide-specific anti-peptide antibody did not react with blots that contained 10 μg of recombinant unphosphorylated threonine 72 protein per lane, nor did it react with other phosphoproteins, for example, the N-terminal region of c-jun (data not shown).
Persistent phosphorylation of ets-2 in response to CSF-1/c-fms signaling in NIH 3T3 cells and in primary macrophages.
The anti-phosphopeptide T72 ets-2 antibody was used to determine the phosphorylation status of endogenous ets-2 in NIH 3T3 cells that express the human c-fms receptor tyrosine kinase (23, 24). Activation of c-fms tyrosine kinase activity by CSF-1 results in the activation of ras signaling pathways and the persistent activation of ras-responsive genes (3, 14, 27). For example, the activation of uPA mRNA expression in these cells following CSF-1 stimulation was demonstrated in Fig. 2A. In this experiment, Northern analysis reveals that uPA expression was stimulated within 30 min following growth factor treatment and that the expression of this mRNA persisted after 24 h of stimulation. In contrast, stimulation of the same NIH 3T3 cell line with PDGF resulted in only transient stimulation of uPA mRNA, with maximal stimulation after 30 min of PDGF treatment (Fig. 2A). We have previously reported this difference in the ability of CSF-1 and PDGF to stimulate the expression of ras-responsive genes in NIH 3T3 cells (3). Thus, if ets-2 is involved in growth factor-induced activation of uPA expression, it should be rapidly phosphorylated in response to CSF-1 or PDGF stimulation, but in addition it should remain phosphorylated for extended periods following CSF-1 treatment.
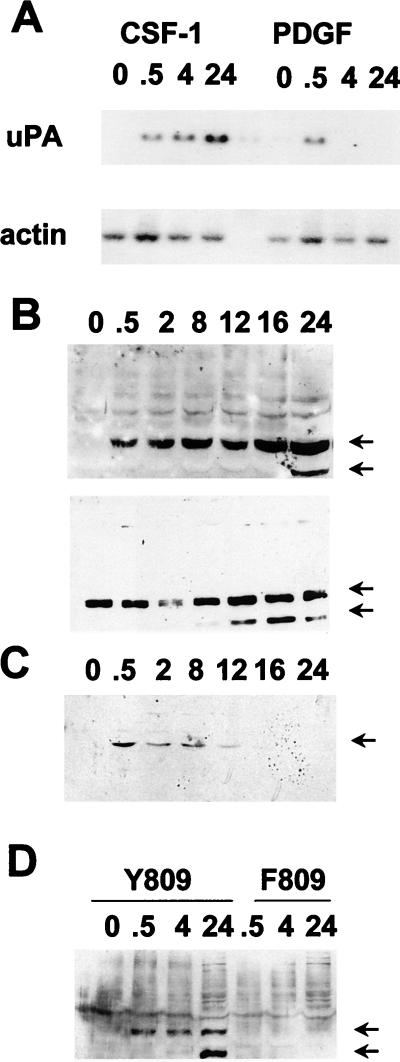
FIG. 2.
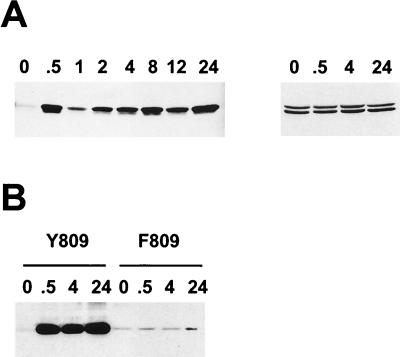
Kinetics of uPA mRNA induction and phosphorylation of ets-2 at threonine 72 in response to CSF-1/c-fms signaling in NIH 3T3 cells. (A) Northern blot of total RNA isolated from NIH 3T3 cells expressing c-fms following stimulation with CSF-1 and PDGF (2 ng/ml) for the times indicated (in hours) and probed with a mouse uPA probe (upper panel) or a γ-actin probe (lower panel). (B) Nuclear protein extracts were prepared from NIH 3T3 cells expressing c-fms following stimulation with CSF-1 for the times indicated (in hours). Extracts were run on a 10% SDS gel and Western blotted with the anti-pT72 ets-2 antibody (upper panel). The top arrow indicates the predicted location of ets-2 (54 kDa), and the lower arrow indicates a second ets-2-related band (45 kDa). The same samples in panel B were run on a second gel and probed with a nondiscriminating ets-2 antibody directed against amino acids 60 to 167 (bottom panel). (C) Nuclear protein extracts prepared from NIH 3T3 cells stimulated with 2 ng of PDGF (BB isoform) per ml for the times indicated (in hours) were analyzed by Western analysis with the anti-pT72 antibody, as described above. Only the 54-kDa form of ets-2 was detected (arrow). (D) NIH 3T3 cells expressing wild-type c-fms Y809 (lanes 1 to 4) or the c-fms F809 protein (lanes 5 to 7) were stimulated for increasing periods of time with CSF-1 as indicated (in hours). Nuclear protein extracts were prepared and analyzed by SDS-polyacrylamide gel electrophoresis (10% polyacrylamide gel) followed by Western blotting with the anti-phosphopeptide T72 ets-2 antibody.
Nuclear extracts were prepared from the NIH 3T3 cells expressing c-fms following CSF-1 treatment for increasing periods of time. These samples were analyzed by Western blotting with the anti-phosphopeptide T72 affinity-purified antibody (Fig. 2B). Specific binding of the antibody was not seen in the extracts prepared from serum-starved cells. However, the antibody detected a major band of 54 kDa, the predicted size for ets-2, within 30 min of CSF-1 treatment (Fig. 2B, upper panel, upper arrow). This band was observed throughout the CSF-1 time course up to 24 h after CSF-1 treatment. If antibody incubations were performed in the presence of the phosphopeptide used for immunization, the 54-kDa band was not detected (data not shown). The nuclear samples were analyzed with a nonphosphopeptide-selective ets-2 antibody raised against a recombinant ets-2 protein comprising amino acids 60 to 167 (Fig. 2B, lower panel). This antibody detected the same 54-kDa major species in all samples, including the serum-starved samples (Fig. 2B, lower panel, upper arrow). The 54-kDa band was not detected in cytoplasmic extracts (data not shown).
In contrast to the results obtained with CSF-1, treatment of the same NIH 3T3 cell line with PDGF resulted in a transient stimulation of ets-2 phosphorylation (Fig. 2D). The phosphorylation of ets-2 was maximal within 30 min following PDGF treatment and was undetectable after 8 to 12 h of treatment. The steady-state levels of ets-2 protein, determined by using the nondiscriminating antibody, were not affected by PDGF treatment (data not shown). Thus, ets-2 was phosphorylated at position threonine 72 in a manner consistent with the kinetics of activation of uPA mRNA by either CSF-1 or PDGF.
A mutation at an autophosphorylation site at tyrosine residue 809 in the human c-fms (809Y→809F) protein selectively abrogates the ability of ligand-activated receptor to stimulate mitogenic growth of NIH 3T3 cells and to stimulate expression of ras-responsive genes like that coding for uPA (14, 27). The F809 receptor also failed to stimulate the conversion of ras to the GTP complex, a conversion the wild-type Y809 receptor efficiently carries out (20a). This receptor failed to activate phosphorylation of ets-2 (Fig. 2D), providing an additional correlation between ets-2 phosphorylation and receptor-dependent activation of persistent gene expression.
In addition to the major band migrating at the predicted size for ets-2, a smaller, 45-kDa band was detected by both antibodies used (lower arrow in Fig. 2B and D). This band was especially prominent after 12 to 24 h of CSF-1 stimulation of cells. The identity of this cross-reacting species is under investigation, but preliminary data indicate that it may be derived by proteolysis of the 54-kDa protein species (data not shown).
The c-fms gene is usually predominantly expressed in monocytes and macrophages in adult mammals (22), and we have previously implicated ets-2 in uPA activation in this cell type (27). In BMMs deprived of CSF-1, the kinetics of uPA induction were delayed following restimulation with CSF-1 with respect to the situation observed in the artificial NIH 3T3 system. In BMMs, uPA expression was induced after 4 h and was maximal by 8 h following stimulation (Fig. 3A). When nuclear extracts from BMMs stimulated with CSF-1 were analyzed, the kinetics of ets-2 phosphorylation were also found to be delayed relative to those in the NIH 3T3 system, with little of the phosphothreonine 72 form of ets-2 seen until 4 to 8 h of CSF-1 treatment (Fig. 3B, upper panel, upper arrow). The phosphothreonine 72 form of ets-2 was observed following 12 to 24 h of growth factor stimulation and in cells continuously grown in the presence of CSF-1 (data not shown). When these samples were analyzed with nondiscriminating antibody [anti-ets-2(60–167) antibody], it was observed that in contrast to NIH 3T3 cells, ets-2 was not expressed in CSF-1-deprived BMMs, but was detectable after 4 h of CSF-1 treatment (Fig. 3B, lower panel, upper arrow).
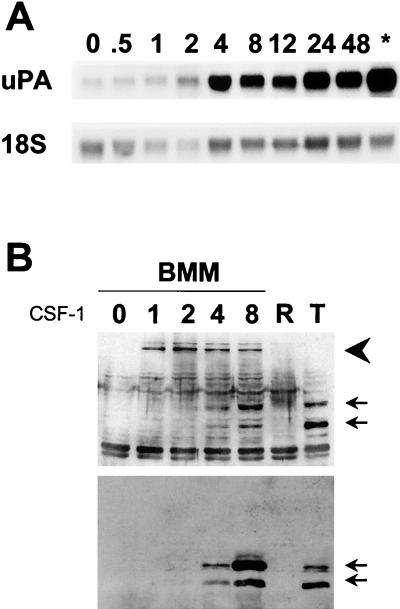
FIG. 3.
Kinetics of uPA mRNA induction and phosphorylation of ets-2 at threonine 72 in response to CSF-1/c-fms signaling in primary macrophages. (A) Northern blot of total RNA isolated from BMMs following stimulation with CSF-1 (time in hours as indicated; ∗, cells grown continually in CSF-1 without starvation) and probed with mouse uPA (upper panel) or an 18S RNA probe (lower panel). (B) Nuclear protein extracts were prepared from BMMs following stimulation with CSF-1 for the times indicated. Extracts were run on a 10% SDS gel and Western blotted with the anti-phosphopeptide T72 ets-2 antibody (upper panel). The two arrows indicate the predicted location of ets-2 (54 kDa) and a second related 45-kDa band, as described above, while the large arrowhead indicates the position of a 100-kDa cross-reacting protein species. The same samples as those used in the upper panel were analyzed in parallel with a nondiscriminating ets-2 antibody directed against amino acids 60 to 167 (bottom panel). Lanes R and T contained extracts prepared from RAW264 cells (negative control) or NIH 3T3 cells that express c-fms (positive control), respectively, both treated with CSF-1 for 24 h prior to extract preparation. Arrowheads indicate the predicted position of ets-2 p54 and the related 45-kDa protein, as described above.
As in the NIH 3T3 cells, the faster-migrating 45-kDa cross-reactive protein was again detected (Fig. 3B, lower arrows). In addition, the anti-phosphopeptide T72 antibody specifically cross-reacted with a protein with an apparent molecular mass of 100 kDa (Fig. 3B, arrowhead). This 100-kDa species was induced within 1 h following CSF-1 stimulation of BMMs and was detected at 8 h after stimulation as well. However, the 100-kDa protein was not recognized by the non-phosphopeptide ets-2 antibody (Fig. 3B, lower panel). The nature of the 100-kDa species is currently under investigation.
The expression and phosphorylation of ets-2 in the macrophage cell line RAW264 were also analyzed. CSF-1 does not stimulate uPA transcription in these cells (27), and ets-2 expression was not detected with either phosphopeptide ets-2 or nondiscriminating ets-2 antibodies, regardless of whether CSF-1 was added to the cell culture medium (Fig. 3B, lanes R). The anti-phosphopeptide Thr 72 cross-reacting 100-kDa band was also absent in RAW264 cells.
MAP kinase p42 and p44 activity correlates with ets-2 phosphorylation in BMMs and in NIH 3T3 cells.
Previous work has indicated that CSF-1 stimulation of MAP kinase p42 and p44 activity in macrophages is transient, with a peak of activity within 10 to 15 min following growth factor stimulation (12), suggesting that these kinases may not be the ets-2 kinase present in these cells. In order to identify kinases capable of phosphorylating ets-2 in primary macrophages, in-gel kinase assays were performed with the ets-2(60–167) recombinant protein as a substrate (Fig. 4A, left panel, lanes 1 to 6). In extracts derived from BMMs, the two major specific protein species detected by this assay migrated with mobilities of 42 and 44 kDa. Within 1 min of addition of CSF-1, the activity of these two kinases was induced. In addition, the 42-kDa kinase remained active following 4 h of CSF-1 stimulation, indicating that activation of this kinase was persistent in BMMs. In RAW264 cells treated with CSF-1, there was five- to sevenfold-less induction of this ets-2 kinase activity (Fig. 4A, lanes 7 to 12). When the Ala 72 form of the ets-2 pointed P2 domain was used as a substrate in this analysis, the p42 and p44 bands were not detected (Fig. 4A, right panel). In addition to the two specific bands, a number of constitutive phosphorylated bands were detected. All of these were also observed with the Ala 72 mutant ets-2 pointed domain (Fig. 4A, right panel) and probably represent autophosphorylation of resident kinases.
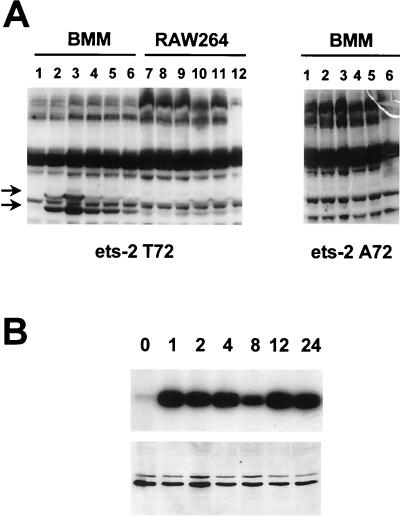
FIG. 4.
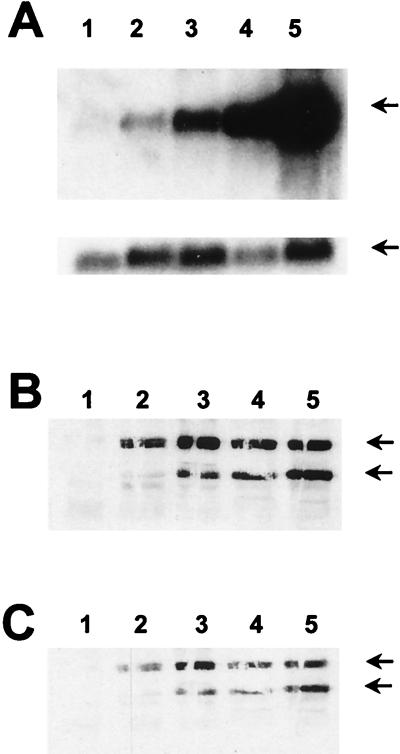
Kinetics of MAP kinase activation in response to CSF-1 stimulation in primary macrophages. (A) In-gel kinase assays with the ets-2(60–167) threonine 72 (left panel), or alanine 72 (right panel) protein as the substrate. BMMs were starved of CSF-1 for 16 h and then stimulated with CSF-1 for 1, 5, 30, 60, and 240 min (lanes 1 to 6). RAW264 cells were stimulated with CSF-1 for the same times (lanes 7 to 12). For these experiments, the 12.5% acrylamide gel was copolymerized with 500 μg of either the threonine 72 or alanine 72 versions of ets-2 protein per ml as indicated. Arrows indicate the migration of the major kinase bands identified in the BMMs at 42 and 44 kDa, when threonine 72 protein is used as a substrate. These bands are not seen in the gel containing the alanine 72 version of the protein. (B) Immunoprecipitation kinase assays were performed with BMMs that had been starved of CSF-1 for 16 h and then stimulated with CSF-1 for the times indicated (in hours). MAP kinases p42 and p44 were immunoprecipitated from 25 μg of whole-cell protein, and incorporation of 32P into the ets-2 threonine 72 protein substrate was measured in one-half of the sample following electrophoresis in a 15% SDS gel (top panel). The other half of the immune kinase assay was analyzed on a 10% SDS gel, and MAP kinase p42 and p44 were detected by Western blotting with a specific polyclonal antibody (bottom panel).
In order to confirm that the specific protein species detected in the in-gel kinase assay were MAP kinases p42 and p44, immune kinase assays were performed with BMM cell lysates with antibody that specifically recognizes these two MAP kinase species. Once again, the ets-2 recombinant protein was utilized as a substrate. These data were consistent with those of the in-gel kinase assays, demonstrating that MAP kinase was detected within 30 min of CSF-1 stimulation and that activity persisted up to 24 h following stimulation (Fig. 4B, upper panel). Analysis of these samples by Western blotting with an anti-MAPK kinase antibody revealed that the level of MAP kinase did not change during the CSF-1 time course (Fig. 4B, lower panel). Consistent with the in-gel kinase results, significant MAP kinase activity was not seen when such an assay was performed with CSF-1-treated RAW264 cells (data not shown).
The immune kinase assays as described above were repeated with lysates prepared from NIH 3T3 cells that express c-fms (Fig. 5A). Little MAP kinase activity was detected in CSF-1-starved cells, but persistent activation of MAP kinase activity was seen from 30 min up to 24 h following CSF-1 treatment (Fig. 5A, left panel). MAP kinase expression remains constant over this time course, as revealed by Western analysis with the same MAP kinase-specific antibody as that used above (Fig. 5A, right panel). Stimulation of F809–c-fms/NIH 3T3 cells with CSF-1 does not lead to increased MAP kinase activity (Fig. 5B), although Western analysis indicated that MAP kinase levels in these cells were comparable to those seen in the Y809–c-fms/NIH 3T3 cells (data not shown).
FIG. 5.
Kinetics of MAP kinase activation in response to CSF-1 stimulation in c-fms/NIH 3T3 cells. (A) Immunoprecipitation kinase assays were performed with extracts from NIH 3T3 cells expressing c-fms and stimulated with CSF-1 for various amounts of time, as indicated (in hours). MAP kinases were immunoprecipitated from 25 μg of whole-cell lysates and incubated with recombinant ets-2 protein (amino acids 60 to 167) and cold ATP. One-half of the kinase assays were analyzed by SDS-polyacrylamide gel electrophoresis (SDS-15% polyacrylamide gel) followed by Western blotting with the anti-phosphopeptide T72 ets-2 antibody (left panel). For select samples (as indicated), one-half of the kinase assay was analyzed with an SDS-10% polyacrylamide gel, and MAP kinase p42 and p44 was detected by Western analysis (right panel). (B) Immunoprecipitation-kinase assays were performed and analyzed as described above with extracts from NIH 3T3 cells expressing either wild-type c-fms (lanes 1 to 4) or the c-fms F809 mutant receptor (lanes 5 to 8). Samples were analyzed at the time points (in hours) indicated.
MAP kinases p42 and p44 are the major ets kinases detected in c-fms/NIH 3T3 cells.
To determine if kinases other than MAP kinases p42 and p44 could be detected following CSF-1 stimulation, MAP kinase p42 and p44 activity was depleted by five successive rounds of immune precipitation from lysates prepared from CSF-1-stimulated c-fms/NIH 3T3 cell lysates (Fig. 6A). This analysis revealed that after four rounds of antibody treatment, >95% of MAP kinase activity was removed from the cell lysates (lanes 3 to 7). At the same time, while ets kinase activity could be detected in the supernatant recovered after the second round of immunoprecipitation, no activity could be detected in the supernatant that was recovered after the fourth round (lane 1 versus lane 2).
FIG. 6.
MAP kinases p42 and p44 are the major ets-2 kinase in c-fms/NIH 3T3 cells. (A) Immunodepletion of MAP kinase p42 and p44 activity in c-fms/NIH 3T3 cells stimulated for 24 h with CSF-1. Five successive rounds of immunoprecipitation were performed to completely deplete MAP kinase activity, and the precipitated kinase activity was measured by 32P incorporation into the ets-2(60–167) substrate detected after SDS-PAGE (15% gel, lanes 3–7, respectively). The kinase activities remaining in the supernatant following the second and fourth rounds of immunoprecipitation (lanes 1 and 2, respectively) were also measured by 32P incorporation into the ets-2(60–167) substrate. (B) Inhibition of ets-2 phosphorylation by the MEK-1 inhibitor PD98059. NIH 3T3 cells that expressed c-fms were serum starved for 24 h (lane 1) and then treated with CSF-1 and dimethyl sulfoxide (the carrier used for PD98059) for 4 or 24 h (lanes 2 and 4, respectively) or with the combination of CSF-1 and PD98059 for 4 of 24 h (lanes 3 and 5, respectively). Cells were also treated with PD98059 alone for 24 h (lane 6). Nuclear extracts were prepared, and one-half of the extract was analyzed by Western blotting with the anti-phosphopeptide ets-2 antibody. (C) The second half of the samples described above was analyzed in parallel with the nondiscriminating ets-2 antibody. Lanes 1 to 6 contained the samples in the same order described above.
To further confirm that the raf/MEK-1/MAP kinase p42 and p44 pathway was the major CSF-1-induced ets-2 kinase pathway, phosphorylation of ets-2 in the presence of the specific MEK-1 inhibitor PD98059 (10) was determined with the anti-phosphopeptide T72 ets-2 antibody (Fig. 6B). This experiment demonstrated that in fibroblasts expressing c-fms, PD98059 blocked ets-2 phosphorylation at both early (Fig. 6B, lane 2 versus lane 3) and late (Fig. 6B, lane 4 versus lane 5) times following growth factor stimulation and also blocked CSF-1 stimulation of uPA mRNA expression (data not shown). At the same time, ets-2 expression, as detected with the non-phosphopeptide-specific antibodies, was not affected by drug treatment (Fig. 6C). The same experiments were attempted with primary mouse BMMs, but ets-2 expression could not be detected following treatment of cells with both CSF-1 and PD98059 (data not shown). Within hours of exposure to the drug PD98059, BMMs lose viability, as determined by trypan blue dye exclusion, making it difficult to determine if the lack of ets-2 expression was a specific or general effect of the drug.
Conditional expression of activated raf kinase in RAW264 cells induces ets-2 expression and phosphorylation and uPA mRNA.
CSF-1 is not able to stimulate ets-2 expression or phosphorylation in RAW264 cells. In order to determine where the defect in CSF-1 signaling occurs, RAW264 cells that express an estrogen-inducible form of raf kinase were used to activate MAP kinases in a CSF-1-independent fashion (11). We reasoned that such an analysis would reveal whether the signaling defect lies downstream or upstream of raf kinase.
Following estrogen treatment, analysis of uPA mRNA levels indicated that this gene was induced within 2 h and maximally stimulated 8 to 16 h after addition of estrogen (Fig. 7A, upper panel), after correction for RNA sample loading (lower panel, γ-actin rehybridization control). Northern analysis also demonstrated that ets-2 mRNA was induced in these cells following stimulation of raf activity (data not shown).
FIG. 7.
Conditional expression of activated raf kinase in RAW264 cells induces ets-2 expression and phosphorylation. RAW264 cells that express the estrogen receptor-raf fusion protein (11) were treated with estrogen (10−7 M) for 2, 4, 8, or 16 h (lanes 2 to 5, respectively) and compared to untreated cells (lane 1). (A) RNA was prepared from the untreated and stimulated cells and analyzed by Northern blotting with the uPA-specific probe (upper panel; the arrow indicates the position of uPA mRNA). The blot was rehybridized with an actin-specific probe to control for RNA loading (lower panel). (B and C) Protein extracts were prepared from the untreated and stimulated cells and analyzed by Western blotting with the anti-ets-2 phosphopeptide T72 antibody (B) or the nondiscriminating ets-2 antibody (C). Arrows indicate the positions of the p54 and p45 ets-2 proteins, respectively.
The expression and phosphorylation status of ets-2 protein were studied by Western blotting (Fig. 7B and C). Use of the anti-phosphopeptide T72 ets-2 antibody demonstrated that ets-2 was expressed and phosphorylated following activation of raf kinase activity (Fig. 7B). As with CSF-1 treatment of signaling-competent cells, both the p54 and p45 forms of ets-2 could be detected (see above). Use of the nondiscriminating ets-2 antibody again demonstrated that both ets-2 bands were detected. The kinetics of ets-2 expression and phosphorylation paralleled expression of uPA mRNA.
In control experiments, β-estradiol had no effect on ets-2 or uPA expression in either normal RAW264 cells or cells that contained the estrogen receptor vector lacking the raf kinase coding sequences (reference 11 and data not shown).
cis element requirements for CSF-1 induction of the uPA in RAW264 cells.
Given the results presented above, a possible explanation for the failure of CSF-1 signaling to induce uPA transcription in RAW264 cells is that the steady-state level of receptor is insufficient to provide the signal required for sustained activation of MAP kinases and subsequent phosphorylation of ets-2. Consistent with this hypothesis, previous studies have shown that the level of CSF-1 binding sites per cell and the level of c-fms mRNA are lower in RAW264 cells than in BMMs (35) and that the relative transcription of c-fms in run-on transcription assays is lower (27).
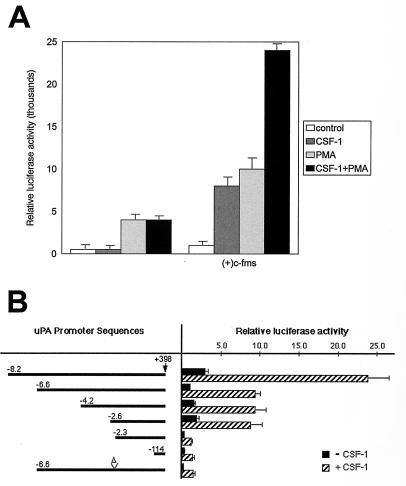
To determine if transient overexpression of c-fms from a heterologous promoter would rescue CSF-1 induction of uPA promoter activity, RAW264 cells were cotransfected with a c-fms expression plasmid and a uPA reporter plasmid (Fig. 8A). Both CSF-1 and phorbol ester were able to stimulate the uPA reporter, and the effects of the two agents were approximately additive (Fig. 8A). Cells transfected with control expression plasmid lacking the c-fms cDNA and a uPA reporter responded only to phorbol ester, and there was no interaction between CSF-1 and phorbol ester (Fig. 8A).
FIG. 8.
The ets/AP-1 enhancer is necessary for CSF-1 induction of uPA reporter gene activity in RAW264 cells. (A) RAW264 cells were transiently transfected with pGL2-uPA-6.6 with either the c-fms expression plasmid or the empty parent plasmid, pECE. Where indicated, 104 U of CSF-1 per ml was added immediately after electroporation, while 10−7 M phorbol myristate acetate (PMA) was added after 6 h, and cells were harvested after 24 h. (B) RAW264 cells were transiently transfected with a series of uPA promoter and enhancer mutations as represented in the first column. The Δ indicates the deletion of the ets/AP-1 element from the −6.6-kb promoter. A c-fms expression plasmid was included with the various uPA reporters in the transient transfection, cells were treated with or without 104 U of CSF-1 per ml for 24 h, and the cell lysates were assayed for luciferase activity. Luciferase activity is presented as relative light units per microgram of protein. The results of four experiments (A) or three experiments (B), each performed in duplicate, are presented with bars representing the standard error.
The response of a series of uPA reporter plasmids to CSF-1/c-fms activation was studied in order to determine the cis requirements for cytokine activation (Fig. 8B). A construct containing 8.2 kb of information upstream of the mRNA initiation site had maximal basal activity and was activated eightfold by CSF-1 in cells cotransfected with c-fms. The deletion from −8.2 to −6.6 kb, which eliminates the distal ets/AP-1 site at −6.9 kb (9), reduced both basal and CSF-1/c-fms-induced promoter activities approximately twofold. Further deletions to −4.2 and −2.6 had no significant effect on either basal or CSF-1-induced activity of the uPA promoter. However, a deletion to −2.3, which eliminates a second ets/AP-1 element located at −2.4 kb (27), resulted in a further eightfold reduction in both basal and CSF-1-induced promoter activity. Deletion of the uPA upstream region to −114 bp had no additional effect on either basal activity or the CSF-1/c-fms response. However, site-directed deletion of the conserved ets/AP-1 enhancer region located at −2.4 kb also reduced both basal and CSF-1-stimulated activity seven- to eightfold (Fig. 8B, −6.6 Δ). Thus, the ets/AP-1 site located at −2.4 kb distal to the uPA transcription initiation site is necessary, although not sufficient, for maximal CSF-1 induction of uPA promoter activity.
DISCUSSION
The data presented support a model in which CSF-1/c-fms receptor ligation leads to prolonged activation of MAP kinases p42 and p44, to sustained phosphorylation of ets-2 on residue threonine 72, and to stable induction of uPA transcription. These results emphasize a basic difference between macrophages and the fibroblast model for c-fms signaling and reinforce the importance of studying c-fms action in the biologically relevant monocyte/macrophage background (15). In macrophages, the expression of ets-2 is part of the CSF-1 immediate-early response, and both expression of this gene and activation of the factor by phosphorylation are mediated by the raf/MAPK kinase signaling pathway. Future studies will be directed toward understanding the regulation of the ets-2 gene in macrophages. Activation of immediate-early MAP kinase targets such as elk-1 (31, 32) or inactivation of repressors such as the ets factor erf-1 (26) may be involved in the induction of the ets-2 promoter.
The transient transfection studies with RAW264 cells support the hypothesis that ets-2 is necessary for CSF-1 induction of the uPA promoter. The ets/AP-1 element at −2.4 kb relative to the transcription initiation site has been shown previously to mediate the response to phorbol esters and oncoprotein ras in these cells (27, 34). Mutation of this site, along with the deletion of the region containing a fibroblast growth factor-responsive ets/AP-1 site located at −6.9 kb (9), resulted in a 90 to 95% decrease in both basal and CSF-1-induced uPA promoter activity. Transcription of another CSF-1-responsive gene in macrophages, the scavenger receptor, has also been shown to be dependent on redundant promoter and enhancer elements that include ets/AP-1-type elements (18, 33), indicating that common mechanisms control the expression of CSF-1 target genes.
The dependence of the basal activity upon the ets/AP-1 elements was unexpected given that the ets-2 protein is not detected in RAW264 cells. A potential explanation of the data lies in the observation that RAW264 cells and primary macrophages are able to respond to unmethylated CpG residues contained in the plasmid DNA used for the transfections (28). The unmethylated DNA response is similar to the response stimulated by bacterial lipopolysaccharide (LPS) (28, 29), and both LPS and CpG DNA can mimic the ability of CSF-1 to induce ets-2 expression and phosphorylation (29). Hence, the basal activity of the uPA promoter in transfected RAW264 cells may be interpreted as a response to plasmid DNA, which likely acts through the same ets/AP-1 elements.
The persistent activation of MAP kinases by CSF-1 in BMMs contrasts with previous work where kinase activity was reported to decline rapidly after an initial peak at 5 to 15 min following CSF-1 treatment (12). The ets-2 substrate employed in the present studies may be a more efficient monitor of specific MAP kinase activity than other substrates commonly used. The use of the ets-2 pointed P2 domain as a substrate has the additional advantage that it distinguishes activation of p42 and p44 from the activation of other proline-directed kinases, which were not able to phosphorylate this substrate. This is in contrast to the ets factor elk-1 and related proteins which are phosphorylated by multiple MAP kinase family members, including MAP kinase p42/p44, JNK, and p38 (31, 32).
How is the persistent signal in response to CSF-1 maintained? The results obtained with RAW264 cells suggest an answer to this question. In RAW264 cells, c-fms expression is likely below a threshold that allows persistent engagement of the ras/MAP kinase pathway and activation of the uPA promoter, and transient overexpression of c-fms rescues the signaling defect. In primary macrophages, CSF-1 and its receptor are rapidly internalized and degraded following CSF-1 binding so that there is a cycle of receptor-mediated ligand degradation (reviewed in reference 22). Thus, the steady-state level of c-fms could determine whether a transient or persistent signal is engaged in response to CSF-1. Similarly, the failure of PDGF to stimulate persistent phosphorylation of ets-2 in NIH 3T3 cells may reflect rapid turnover of the PDGF receptor with respect to c-fms following growth factor treatment. Such arguments are consistent with results obtained in other systems, for example, increasing steady-state levels of the epidermal growth factor or insulin receptors in PC-12 cells results in prolonged activation of the ras/MAP kinase pathway and neuronal differentiation of this cell type (8, 30).
In conclusion, the present studies indicate that CSF-1 action involves the continual activation of MAP kinase activity, leading to transcription factor phosphorylation and selective activation of gene transcription. Studies of CSF-1 signaling provide one paradigm for understanding how tyrosine kinase receptors trigger persistent signaling pathways as well as the biological consequences of such prolonged signaling events.
ACKNOWLEDGMENTS
L.F.F. and M.L.M. contributed equally to this work.
We thank Michael Karin for the HA-JUNK2 expression vector and GST vectors to express c-jun N-terminal peptides and Julie Hambleton and Tony DeFranco for providing the estrogen receptor-raf/RAW264 cells.
This work was supported by NIH grant CA-53271 (M.C.O.) and by the National Health and Medical Research Council of Australia (D.A.H.). The DNAX Research Institute is supported by the Schering Plough Corporation.
REFERENCES
- 1.Besser D, Presta M, Nagamine Y. Elucidation of a signaling pathway induced by FGF-2 leading to uPA gene expression in NIH 3T3 fibroblasts. Cell Growth Differ. 1995;6:1009–1017. [PubMed] [Google Scholar]
- 2.Blasi F. Molecular mechanisms of protease-mediated tumor invasiveness. J Surg Oncol. 1993;3:21–24. [PubMed] [Google Scholar]
- 3.Bortner D M, Ulivi M, Roussel M F, Ostrowski M C. The carboxy-terminal catalytic domain of the GTPase-activating protein inhibits nuclear signal transduction and morphological transformation mediated by the CSF-1 receptor. Genes Dev. 1991;5:1777–1785. doi: 10.1101/gad.5.10.1777. [DOI] [PubMed] [Google Scholar]
- 4.Brunner D, Ducker K, Oellers N, Hafen E, Scholz H, Klambt C. The Ets domain protein pointed-P2 is a target of MAP kinase in the sevenless signal transduction pathway. Nature. 1994;370:386–389. doi: 10.1038/370386a0. [DOI] [PubMed] [Google Scholar]
- 5.Cassady A I, Stacey K J, Nimmo K A, Murphy K M, von der Ahe D, Pearson D, Botteri F M, Nagamine Y, Hume D A. Constitutive expression of the urokinase plasminogen activator gene in murine RAW264 macrophages involves distal and 5′ non-coding sequences that are conserved between mouse and pig. Nucleic Acids Res. 1991;19:6839–6847. doi: 10.1093/nar/19.24.6839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Courtneidge S A, Dhand R, Pilat D, Twamley G M, Waterfield M D, Roussel M F. Activation of Src family kinases by colony stimulating factor-1, and their association with its receptor. EMBO J. 1993;12:943–950. doi: 10.1002/j.1460-2075.1993.tb05735.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.den Hertog J, Hunter T. Tight association of GRB2 with receptor protein-tyrosine phosphatase alpha is mediated by the SH2 and C-terminal SH3 domains. EMBO J. 1996;15:3016–3027. [PMC free article] [PubMed] [Google Scholar]
- 8.Dikic I, Schlessinger J, Lax I. PC12 cells overexpressing the insulin receptor undergo insulin-dependent neuronal differentiation. Curr Biol. 1994;4:702–708. doi: 10.1016/s0960-9822(00)00155-x. [DOI] [PubMed] [Google Scholar]
- 9.D’Orazio D, Besser D, Marksitzer R, Kunz C, Hume D A, Kiefer B, Nagamine Y. Cooperation of two PEA3/AP1 sites in uPA gene induction by TPA and FGF-2. Gene. 1998;201:179–187. doi: 10.1016/s0378-1119(97)00445-9. [DOI] [PubMed] [Google Scholar]
- 10.Dudley D T, Pang L, Decker S J, Bridges A J, Saltiel A R. A synthetic inhibitor of the mitogen-activated protein kinase cascade. Proc Natl Acad Sci USA. 1995;92:7686–7689. doi: 10.1073/pnas.92.17.7686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hambleton J, McMahon M, DeFranco A L. Activation of Raf-1 and mitogen-activated protein kinase in murine macrophages partially mimics lipopolysaccharide-induced signaling events. J Exp Med. 1995;182:147–154. doi: 10.1084/jem.182.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jaworowski A, Christy E, Yusoff P, Byrne R, Hamilton J A. Differences in the kinetics of activation of protein kinases and extracellular signal-related protein kinase 1 in colony-stimulating factor 1-stimulated and lipopolysaccharide-stimulated macrophages. Biochem J. 1996;320:1011–1016. doi: 10.1042/bj3201011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kallunki T, Su B, Tsigelny I, Sluss H K, Derijard B, Moore G, Davis R, Karin M. JNK2 contains a specificity-determining region responsible for efficient c-Jun binding and phosphorylation. Genes Dev. 1994;8:2996–3007. doi: 10.1101/gad.8.24.2996. [DOI] [PubMed] [Google Scholar]
- 14.Langer S J, Bortner D M, Roussel M F, Sherr C J, Ostrowski M C. Mitogenic signaling by colony-stimulating factor 1 and ras is suppressed by the ets-2 DNA-binding domain and restored by myc overexpression. Mol Cell Biol. 1992;12:5355–5362. doi: 10.1128/mcb.12.12.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lioubin M N, Myles G M, Carlberg K, Bowtell D, Rohrschneider L R. Shc, Grb2, Sos1, and a 150-kilodalton tyrosine-phosphorylated protein form complexes with Fms in hematopoietic cells. Mol Cell Biol. 1995;14:5682–5691. doi: 10.1128/mcb.14.9.5682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marshall C J. Specificity of receptor tyrosine kinase signaling: transient versus sustained extracellular signal-regulated kinase activation. Cell. 1995;80:179–185. doi: 10.1016/0092-8674(95)90401-8. [DOI] [PubMed] [Google Scholar]
- 17.McCarthy S A, Chen D, Yang B-S, Garcia Ramirez J J, Cherwinski H, Chen X-R, Klagsbrun M, Hauser C A, Ostrowski M C, McMahon M. Rapid phosphorylation of Ets-2 accompanies mitogen-activated protein kinase activation and the induction of heparin-binding epidermal growth factor gene expression by oncogenic Raf-1. Mol Cell Biol. 1997;17:2401–2412. doi: 10.1128/mcb.17.5.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moulton K S, Semple K, Wu H, Glass C K. Cell-specific expression of the macrophage scavenger receptor gene is dependent on PU.1 and a composite AP-1/ets motif. Mol Cell Biol. 1994;14:4408–4418. doi: 10.1128/mcb.14.7.4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Novak U, Nice E, Hamilton J A, Paradiso L. Requirement for Y706 of the murine (or Y708 of the human) CSF-1 receptor for STAT1 activation in response to CSF-1. Oncogene. 1996;13:2607–2613. [PubMed] [Google Scholar]
- 20.O’Neill E M, Rebay I, Tjian R, Rubin G M. The activities of two Ets-related transcription factors required for Drosophila eye development are modulated by the Ras/MAPK pathway. Cell. 1994;78:137–147. doi: 10.1016/0092-8674(94)90580-0. [DOI] [PubMed] [Google Scholar]
- 20a.Ostrowski, M. C. Unpublished results.
- 21.Rabault B, Roussel M F, Quang C T, Ghysdael J. Phosphorylation of Ets1 regulates the complementation of a CSF-1 receptor impaired in mitogenesis. Oncogene. 1996;13:877–881. [PubMed] [Google Scholar]
- 22.Roth P, Stanley E R. The biology of CSF-1 and its receptor. Curr Top Microbiol Immunol. 1992;181:141–167. doi: 10.1007/978-3-642-77377-8_5. [DOI] [PubMed] [Google Scholar]
- 23.Roussel M F, Shurtleff S A, Downing J R, Sherr C J. A point mutation at tyrosine-809 in the human colony-stimulating factor 1 receptor impairs mitogenesis without abrogating tyrosine kinase activity, association with phosphatidylinositol 3-kinase, or induction of c-fos and junB genes. Proc Natl Acad Sci USA. 1990;87:6738–6742. doi: 10.1073/pnas.87.17.6738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roussel M F, Cleveland J L, Shurtleff S A, Sherr C J. Myc rescue of a mutant CSF-1 receptor impaired in mitogenic signalling. Nature. 1991;353:361–363. doi: 10.1038/353361a0. [DOI] [PubMed] [Google Scholar]
- 25.Roussel M F. Regulation of cell cycle entry and G1 progression by CSF-1. Mol Rep Dev. 1997;46:11–18. doi: 10.1002/(SICI)1098-2795(199701)46:1<11::AID-MRD3>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 26.Sgouras D N, Athanasiou M A, Beal G J, Jr, Fisher R J, Blair D G, Mavrothalassitis G J. ERF: an ETS domain protein with strong transcriptional repressor activity, can suppress ets-associated tumorigenesis and is regulated by phosphorylation during cell cycle and mitogenic stimulation. EMBO J. 1995;14:4781–4793. doi: 10.1002/j.1460-2075.1995.tb00160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stacey K J, Fowles L F, Colman M S, Ostrowski M C, Hume D A. Regulation of urokinase-type plasminogen activator gene transcription by macrophage colony-stimulating factor. Mol Cell Biol. 1995;15:3430–3441. doi: 10.1128/mcb.15.6.3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stacey K J, Sweet M J, Hume D A. Macrophages ingest and are activated by bacterial DNA. J Immunol. 1996;157:2116–2122. [PubMed] [Google Scholar]
- 29.Sweet M J, Stacey K J, Ross I L, Ostrowski M C, Hume D A. The involvement of Ets, rel and Sp1-like proteins in the lipopolysaccharide-mediated activation of the HIV-1-LTR in macrophages. J Inflamm. 1998;48:67–83. [PubMed] [Google Scholar]
- 30.Traverse S, Seedorf K, Paterson H, Marshall C J, Cohen P, Ullrich A. EGF triggers neuronal differentiation of PC12 cells that overexpress the EGF receptor. Curr Biol. 1994;4:694–701. doi: 10.1016/s0960-9822(00)00154-8. [DOI] [PubMed] [Google Scholar]
- 31.Treisman R. Regulation of transcription by MAP kinase cascades. Curr Opin Cell Biol. 1996;8:205–215. doi: 10.1016/s0955-0674(96)80067-6. [DOI] [PubMed] [Google Scholar]
- 32.Whitmarsh A J, Shore P, Sharrocks A D, Davis R J. Integration of MAP kinase signal transduction pathways at the serum response element. Science. 1995;269:403–407. doi: 10.1126/science.7618106. [DOI] [PubMed] [Google Scholar]
- 33.Wu H, Moulton K, Horvai A, Parik S, Glass C K. Combinatorial interactions between AP-1 and ets domain proteins contribute to the developmental regulation of the macrophage scavenger receptor gene. Mol Cell Biol. 1994;14:2129–2139. doi: 10.1128/mcb.14.3.2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang B-S, Hauser C A, Henkel G, Colman M S, Van Beveren C, Stacey K J, Hume D A, Maki R A, Ostrowski M C. Ras-mediated phosphorylation of a conserved threonine residue enhances the transactivation activities of c-Ets1 and c-Ets2. Mol Cell Biol. 1996;16:538–547. doi: 10.1128/mcb.16.2.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yue X, von Gavel S, Cassady A I, Stacey K J, Dunn T L, Hume D A. The resistance of macrophage-like tumour cell lines to growth inhibition by lipopolysaccharide and pertussis toxin. Br J Haematol. 1993;84:392–401. doi: 10.1111/j.1365-2141.1993.tb03092.x. [DOI] [PubMed] [Google Scholar]