Abstract
The immune response to pathogens is regulated by a delicate balance of cytokines. The dysregulation of cytokine gene expression, including interleukin-12, tumor necrosis factor alpha, and gamma interferon (IFN-γ), following human retrovirus infection is well documented. One process by which such gene expression may be modulated is altered DNA methylation. In subsets of T-helper cells, the expression of IFN-γ, a cytokine important to the immune response to viral infection, is regulated in part by DNA methylation such that mRNA expression inversely correlates with the methylation status of the promoter. Of the many possible genes whose methylation status could be affected by viral infection, we examined the IFN-γ gene as a candidate. We show here that acute infection of cells with human immunodeficiency virus type 1 (HIV-1) results in (i) increased DNA methyltransferase expression and activity, (ii) an overall increase in methylation of DNA in infected cells, and (iii) the de novo methylation of a CpG dinucleotide in the IFN-γ gene promoter, resulting in the subsequent downregulation of expression of this cytokine. The introduction of an antisense methyltransferase construct into lymphoid cells resulted in markedly decreased methyltransferase expression, hypomethylation throughout the IFN-γ gene, and increased IFN-γ production, demonstrating a direct link between methyltransferase and IFN-γ gene expression. The ability of increased DNA methyltransferase activity to downregulate the expression of genes like the IFN-γ gene may be one of the mechanisms for dysfunction of T cells in HIV-1-infected individuals.
The molecular mechanisms underlying cytokine dysregulation following human retroviral infection are not well understood. Considerable data demonstrating aberrant cytokine production from both human T-cell leukemia virus type 1 (HTLV-1)- and human immunodeficiency virus type 1 (HIV-1)-infected cells have been obtained (37, 38, 67). Both viruses infect CD4+ T cells and monocytes/macrophages, cells which orchestrate the immune response primarily by the elaboration of cytokines. Therefore, elucidating the molecular mechanisms of cytokine dysregulation in HTLV-1- and HIV-1-infected cells is critical to understanding the mechanisms of pathogenesis of these viruses.
Methylation is an epigenetic mechanism for modulation of gene expression in mammalian cells (81, 83). Studies with knockout mice have demonstrated the absolute requirement for DNA methyltransferase (MTase), as homozygous mutant embryos die at mid-gestation (50). Faithful propagation of the methylation state (maintenance methylation) occurs directly after DNA replication. The process is mediated by an enzymatic methyl transfer reaction at cytosine located 5′ to guanosine (CpG dinucleotide) residues in the unmethylated nascent DNA strand across from methylated CpG dinucleotides. Acquisition of DNA methylation at a previously unmethylated site cannot be accomplished by maintenance methylation and requires de novo methylation (48). To date, only one DNA (cytosine-5) MTase has been identified in mammalian cells (7, 45, 82). While this DNA MTase prefers a hemimethylated substrate, it shows both maintenance and de novo activity in vitro and in vivo (48).
A substantial body of work implicates a role for altered DNA methylation patterns and regulation in the pathogenesis of cancer (3, 42, 46). Changes in the pattern of DNA methylation are often seen in human tumors (2, 12, 22, 27, 29). One of these changes is the aberrant methylation of normally unmethylated CpG islands in gene promoter regions and an associated decrease in expression of tumor suppressor genes such as the von Hippel-Lindau (35), p16 (36, 54), and Rb (63) genes. While it is unclear whether the observed changes in DNA methylation play a direct role in oncogenesis or whether they are the result of the transformation process, the substantial correlative data and a recent study using a combination of genetics and pharmacology to decrease levels of DNA MTase in mice (46) strongly support a causal role for aberrant methylation in the pathogenesis of some cancers.
In other studies, it has been demonstrated that overexpression of murine DNA MTase is transforming for NIH 3T3 cells (79) and that levels of DNA MTase are increased in neoplastic cells (17, 44), with incremental increases at the different stages of colon carcinoma progression (5, 40). Some studies have found more modest (two- to fourfold) increases in MTase upregulation, which were correlated with increases in cellular proliferation, making it difficult to determine whether this increase was biologically significant or merely secondary to cell proliferation (47, 71). Nonetheless, recent studies illustrate that chronically increased cellular levels of DNA MTase can result in aberrant CpG island hypermethylation in simian virus 40-transformed human fibroblasts (74, 75). While it is unclear at present how such increased expression of DNA MTase results in increased tumorigenesis, it is known that methylated cytosine residues are susceptible to deamination, which changes methylcytosine to thymine, ultimately resulting in a permanent genetic alteration. 5-Methylcytosine is then a potential endogenous mutagen (68, 73). Alternatively, an increase in DNA methylation might enhance pathogenesis via an epigenetic mechanism by inhibiting the expression of tumor suppressor or cell cycle genes as discussed above.
While there is no published information concerning the role of DNA MTase in the pathogenesis of human retroviruses, it is known that the DNA of both endogenous and exogenous retroviruses can be highly methylated in the genome of the host (31, 52, 86) and that increased DNA methylation can occur elsewhere in the genome of infected cells (15, 41). In the case of the human retroviruses HIV-1 and HTLV-1, several studies have shown increased methylation of the viral long terminal repeats and throughout the viral genome, suggesting methylation as a mechanism of suppression of viral expression (4, 69) and latency (58, 59, 70).
The present study was initiated to determine whether infection by HIV-1 modulates expression of DNA MTase. Modulation of this enzyme could result in widespread aberrant methylation of genes, resulting in alteration of gene expression and ultimately contributing to the pathogenic effects of these viruses. Since it had been previously shown that methylation of the gamma interferon (IFN-γ) gene promoter could silence the expression of this gene in primary T cells and T-cell lines (20, 21, 24, 32, 53, 64, 66, 84, 85), we examined the effect of HIV infection on IFN-γ gene expression. We report here that acute infection of lymphoid cells by HIV-1 results in increased DNA MTase expression, an overall increase in methylation of DNA in infected cells, and the de novo methylation of a CpG dinucleotide in the IFN-γ gene promoter, resulting in the subsequent downregulation of expression of this cytokine. Antisense DNA MTase experiments demonstrated that this enzyme regulates the expression of IFN-γ in lymphoid cells. The use of IFN-γ as a target gene for aberrant DNA methylation provides support for a potential mechanism for some pathologic consequences of human retroviral infection involving altered methylation of genes.
MATERIALS AND METHODS
Cell culture and separation of T-cell subsets.
All cell lines, with the exception of NK 3.3, used in this study were maintained in RPMI 1640 supplemented with 10% fetal calf serum, penicillin (100 μg/ml), streptomycin (100 μg/ml), and l-glutamine (300 μg/ml) (complete RPMI). NK 3.3, whose growth is cytokine dependent, was grown in complete medium plus interleukin-2 (IL-2) (80). The JMO and CS-3 CD4+ cell lines were developed in our laboratory by HTLV-1 transformation of peripheral blood mononuclear cells (PBMCs). Clonal populations of JMO were prepared by standard limiting dilution methods. T lymphocytes were obtained from leukopaks isolated from healthy donors. Mononuclear cells were separated by Ficoll-Hypaque gradient centrifugation followed by centrifugal elutriation to separate monocyte and lymphocyte populations as previously described (58). CD4+ and CD8+ T cells were obtained at >95% purity by incubating with specific monoclonal antibodies (MAbs) linked to magnetic beads and passed through a magnetic column (Milteny Biotec, Auburn, Calif.). Purity of populations was determined by flow cytometry analysis of purified populations. Cells were activated with phytohemagglutinin (PHA; 1 μg/ml) for 72 h and exposed to HIV-1 as previously described (58).
Cell cycle analysis of HIV-1-infected CD4+ T cells.
Cell cycle analysis by flow cytometry was performed as follows. A total of 2 × 106 cells were removed from infected and uninfected cultures at specified time points. Cells were washed twice with phosphate-buffered saline (PBS) and fixed in ice-cold ethanol for 1 h. Fixed cells were pelleted, resuspended in 0.5 ml of PBS containing propidium iodide (30 μg/ml) and RNase A (1 mg/ml; Sigma, St. Louis, Mo.), and analyzed for DNA content by flow cytometry. Cell cycle histograms were then analyzed with a multicycle computer program.
Plasmid constructs.
Expression vectors for human DNA MTase were prepared as previously described (75). Clones were selected in the sense (HMT) and antisense (TMH) orientations. pCMV-Neo-Bam was used as a vector control in all stable transfections. For stable transfections (2 × 106), cells were electroporated in complete RPMI at 250 V and 600 μF with 10 μg of plasmid. G418-resistant cells were isolated in selective medium (0.3 mg of G418 per ml; Life Technologies Inc., Gaithersburg, Md.). Clonal populations of G418-resistant cell lines were prepared by standard limiting dilution methods.
RPA.
A 364-bp (bp 4774 to 5138) fragment of the 3′ end of MTase subcloned into pBluescript (Stratagene, La Jolla, Calif.) was linearized with HindIII and in vitro transcribed, using a Promega (Madison, Wis.) RNA synthesis kit in the presence of [α-32P]UTP and T3 polymerase. The sense message (to detect a 280-bp antisense fragment) could be made by linearizing the same construct with DraI and using T7 polymerase in the transcription reaction. RNase protection assays (RPAs) were performed with an RPA II kit (Ambion) according to the manufacturer’s instructions. A human 18S rRNA probe (116 bp) and human ptriactin RNA probe (225 bp), or ptricyclophilin (Ambion) in some experiments, were included in each reaction mixture as an internal loading standard. The multiprobe template human cytokine-1 (Pharmingen) was transcribed and used according to the RPA II (Ambion) protocol. Briefly, total RNA (20 μg) was hybridized at 50°C with the mixed radioactive probes. RNA was digested with RNase A and RNase T1 at 37°C for 30 min, and protected fragments were precipitated and electrophoresed on a 6% sequencing gel. Gels were dried and scanned on a PhosphorImager (Molecular Dynamics, Palo Alto, Calif.), using Imagequant and Microsoft Excel software. Dried gels were also autoradiographed on XAR-5 film at −70°C.
Southern analysis.
Five micrograms of DNA, isolated from uninfected and infected cells, was digested with a restriction enzyme (BamHI, SnaBI, HpaII, or MspI) overnight under appropriate conditions as specified by the manufacturer (Boehringer Mannheim or New England Biolabs). Restriction digests were ethanol precipitated analyzed by electrophoresis in 0.8% agarose gels as described elsewhere (9). Membranes were hybridized with 32P-labeled full-length IFN-γ cDNA. The cDNA was labeled to a specific activity of 1 × 108 to 2 × 108 cpm/μg by random priming (Prime-it II; Stratagene) and transferred to a nylon membrane (Magna Graph; Micron Separations Inc., Westborough, Mass.). After hybridization, membranes were washed in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% sodium dodecyl sulfate at room temperature for 20 min followed by a wash in 0.2× SSC–0.1% sodium dodecyl sulfate at 65°C for 20 min. Blots were then exposed to Kodak X-Omat X-ray film at −70°C overnight. Images were quantitated with a PhosphorImager using Imagequant (Molecular Dynamics). Blots were stripped and reprobed with mitochondrial DNA to control for complete digestion.
Derivation and maintenance of antigen-specific human CD4+ T-cell clones.
Purified protein derivative (PPD)-, tetanus toxoid (TTx)-, and Dermatophagoides pteronyssinus antigen (DP)-specific as well as staphylococcal enterotoxin B (SEB)-reactive T-cell clones were generated according to previously described techniques (23). PPD and TTx were purchased from Connaught, Inc. (Swiftwater, Pa.), SEB was purchased from Sigma, and DP was purchased from Miles, Inc. (Spokane, Wash.). Briefly, PBMCs at a concentration of 5 × 105 cells/ml in clone medium (EHAA [CLICK’s] medium supplemented with l-glutamine, 2-mercaptoethanol, 2% human AB serum, 10% fetal calf serum, penicillin-streptomycin, nonessential amino acids, and sodium pyruvate) were stimulated with either PPD (1 μg/ml), TTx (10 μg/ml), DP (10 IAU/ml), or SEB (0.1 μg/ml) in 24-well flat-bottomed plates for 7 days. Many but not all of the TH1 and TH2 clones used in these studies were derived in cultures supplemented with the T-helper cell-selective cytokines IL-4 and IL-12. For the generation of TH1 clones, these cultures were supplemented with recombinant human IFN-γ (rhIFN-γ; 10 U/ml), rhIL-12 (50 pg/ml), and anti-IL-4 MAb (10 μg/ml) over the culture period. For TH2 clones, bulk cultures were supplemented with rhIL-4 (200 U/ml) and anti-IFN-γ MAb (10 μg/ml). Forty-eight hours after the initiation of the cultures, rhIL-2 (10 U/ml) was added to each well. After 7 days of incubation, the cultures were harvested, extensively washed, replated in fresh clone medium supplemented with additional IL-2 (10 U/ml), and incubated for an additional 7 to 10 days. Viable T cells were then plated in limiting dilution cultures (0.5 cells/well) in 16 flat-bottomed 96-well plates containing 2 × 105 irradiated (1,200 rads) syngeneic PBMC feeder cells, specific antigen, and IL-2 (10 U/ml) in a final volume of 200 μl. The growing cultures were examined daily and supplemented with the TH1- and TH2-selecting cytokines (as described above) and at 10-day intervals with feeder cells and IL-2. Individual clones were isolated, characterized for lymphokine production by enzyme-linked immunosorbent assay (ELISA) and reverse transcription-PCR (RT-PCR) analysis and for the ability to respond to specific antigen in combination with syngeneic irradiated (1,200 rads) feeder cells. In maintaining T-helper clones, every 14 to 21 days, cells were restimulated with specific antigen in the presence of autologous PBMCs treated with mitomycin C at 25 μg/ml to prevent outgrowth of feeder cells and IL-2 (20 U/ml). After 96 h of antigenic stimulation, cells were subjected to two successive Ficoll-Hypaque centrifugations to remove dead cells. All clones were tested for their cytokine profile by ELISA and RT-PCR after stimulation with phorbol myristate acetate and monoclonal anti-CD3 as well as with antigen and antigen-presenting cells to phenotype each clone.
HIV infection of T cells and T-cell clones.
PHA-activated T cells, CD4+ T cells 3 days after activation, and antigen-stimulated clones (106 cells) 4 to 7 days postactivation (>95% viable by trypan blue exclusion) were inoculated with cell-free viral isolates at a multiplicity of infection (MOI) of 1 for 2 h before complete aspiration of medium, washing with PBS, and addition of fresh growth medium containing IL-2 (10 U/ml). Cells were aliquoted at 106 cells/ml in 24-well plates or tissue culture flasks. Cell lines were infected with cloned isolates or purified concentrated isolates at an MOI of 1. Three laboratory strains of HIV (BP-1 [59], ADA [26], and MN) as well as the defined clones NL43 and HXB-2 (stocks purified 1,000-fold; kindly provided by the AIDS Vaccine Program, Frederick, Md.) were used.
Cytokine mRNA expression of T-helper clones.
RT-PCR was also used to pedigree cell clones for cytokine expression. Primer pairs for IFN-γ and IL-4 (Clontech) and HIV SK38/39 were used to amplify cDNA prepared by Superscript II reverse transcription of 5 μg of RNA (GIBCO-BRL) according to the manufacturer’s instructions. Amplification was carried out by using [α-32P]dCTP as described previously (16); products were electrophoresed on 8% polyacrylamide gels and then subjected to autoradiography.
Quantitation of cytokine and p24 production by T-cell clones.
Quantitative determinations for each of the lymphokines from the 48-h supernatant of antigen-stimulated T-cell clones were determined by human IL-2, IL-4, IL-5, IL-10, and IFN-γ ELISAs (Quantikine; R&D Systems) as instructed by the manufacturer. The results are expressed in either nanograms/milliliter or units/milliliter based on a standard curve using recombinant cytokine within the ELISAs. Cytokine analyses after HIV infection were performed with cell-free supernatant, and the results were quantitated by ELISA for IL-4, IL-5 (R&D Systems), and IFN-γ (Medigenix) with sensitivities of 3 pg/ml, 1 pg/ml, and 1 IU/ml, respectively. Viral p24 antigen was determined by ELISA (Cellular Products, Buffalo, N.Y.) with a sensitivity of 10 pg/ml.
PCR analysis of IFN-γ promoter methylation status.
Primer pairs were chosen such that they flanked the methyl-sensitive SnaBI site at −55 of the IFN promoter. The upstream (US) SnaBI sense primer 31-48 with the antisense (AS) primer 870-890 yielded a product of ∼850 bp. An internal control primer, the downstream sense primer (DS) 319-342 with the AS 870-890 primer yielded a product of ∼470 bp and was used as an internal control. DNA aliquots of 200 ng to 1 μg were digested with BamHI and SnaBI overnight to ensure complete digestion; 20 ng of DNA was then amplified in a 50-μl PCR mixture containing 0.25 μl of [32P]dCTP as described above. Cycling conditions were 1 cycle of 94°C for 45 s, 60°C for 60 s, and 72°C for 45 s and 30 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s, followed by extension for 7 min at 72°C. PCR products were electrophoresed on 8% polyacrylamide gels, dried, autoradiographed, and quantitated on a PhosphorImager using Imagequant (Molecular Dynamics). Alternatively, 100 ng of DNA was amplified in a 50-μl reaction mixture with unlabeled deoxynucleoside triphosphates, and products were visualized by ethidium bromide staining following polyacrylamide gel electrophoresis. Quantitation was performed by densitometric analysis of gels photographed with Polaroid type 55 Pos/Neg film.
DNA methyltransferase enzymatic activity assay.
Cell lysates were prepared as previously described (40). Briefly, cells were lysed by sonication at 0°C in 5 volumes of 20 mM Tris-HCl (pH 7.4)–0.4 M NaCl–25% glycerol–5 mM EDTA–0.1% Nonidet P-40–1 mM dithiothreitol (DTT)–0.2 mM phenylmethylsulfonyl fluoride–100 μg of aprotinin per ml. After sonication, 1 volume of a 50% Chelex-100 resin was added, and the mixture was incubated for 10 min to remove nucleic acids from the lysate. The clarified lysate (∼200 mg of protein in 20 μl) was added to 200 μl of a solution containing 20 mM Tris-HCl (pH 7.4), 5 mM EDTA, 25% glycerol, 5 μCi of S-adenosyl-l-[methyl-3H]methionine (12 Ci/mmol; New England Nuclear), 4 μg of poly(dI-dC) · poly(dI-dC), 1 mM DTT, and bovine serum albumin (200 μg/ml). The assay mixture was then incubated at 37°C for 2 h and extracted twice with phenol-chloroform. The aqueous phase was made 0.1 M in NaOH and incubated at 50°C for 2 h. The solution was neutralized with HCl, and the radioactivity incorporated into DNA was measured by scintillation counting following trichloroacetic acid precipitation.
DNA methylation level.
A modified methyl accepting assay (79) was used to determine the methylation status of DNA isolated from uninfected and infected primary cell culture cultures, chronically infected cell lines, and the stably transfected transformed cell line JMO. DNA (200 ng) was incubated with 4 U of M · SssI CpG methylase (New England Biolabs) in the presence of 1.5 μM S-adenosyl-l-[methyl-3H]methionine (60 to 85 Ci/mmol; Amersham product no. TRK 581) and 1.5 μM nonradioactive S-adenosylmethionine (New England Biolabs). The reaction mixtures (20 μl) were incubated at 37°C for 4 h in a buffer containing 10 mM Tris-HCl (pH 7.9), 50 mM NaCl, 10 mM MgCl2, and 1 mM DTT. The reactions were stopped by the addition of 5 μl of 2.5 mM nonradioactive S-adenosylmethionine and spotted on GF/C 2.4-cm2 Whatman filter discs, which were air dried for 15 min and washed with 6 ml of 5% (wt/vol) trichloroacetic acid–70% (vol/vol) ethanol. Discs were counted in Econofluor in a Beckman liquid scintillation counter. Control reactions without DNA or enzyme added were included as background, and counts in the samples never exceeded 5% of those in the test samples. All samples were tested in triplicate, and values were obtained as disintegrations/minute per nanogram of DNA.
ELISPOT for IFN-γ expression.
The frequency of specific IFN-γ-producing cells was determined by ELISA spot assay (ELISPOT). Briefly 96-well nitrocellulose-bottomed plates (Millititer HA; Millipore Inc., Bedford, Mass.) were coated with specific mouse anti-human IFN-γ antibody, clone 1-D1k (Chromigenix, Montal, Sweden) by overnight incubation at 4°C. Cells to be analyzed were washed twice in appropriate incubation media. A known number of cells was then plated to the top row of the previously IFN-γ MAb-coated plate, serially twofold diluted down the plate, and incubated at 37°C for 16 h. Cells were then washed off, and IFN-γ-secreting cells were detected with a biotinylated second IFN-γ-specific antibody, clone 7-B6 (Chromigenix). Plates were next incubated with horseradish peroxidase-conjugated avidin and developed with TruBlu peroxidase substrate (Kirkegaard & Perry, Gaithersburg, Md.) to produce an insoluble precipitate that formed distinct spots that could be easily counted by light microscopy.
RESULTS
HIV-1 infection of lymphoid cells increases cellular capacity to methylate DNA by increasing DNA MTase expression.
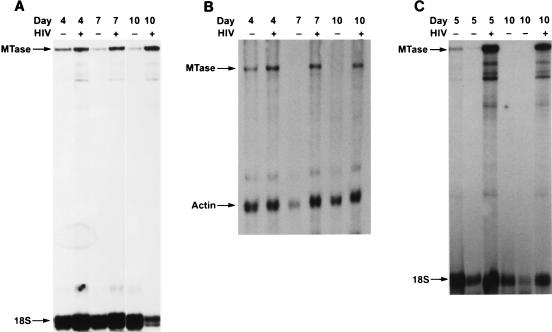
To determine whether there is a relationship between retroviral infection and the overall cellular capacity to methylate DNA, we first examined the time-dependent effect of acute HIV-1 infection of primary T cells and T-cell subsets on DNA MTase mRNA expression. As shown by RPA (Fig. 1A), a significant increase in DNA MTase mRNA was observed in PHA-activated, HIV-1-infected T cells compared to their PHA-activated, uninfected counterparts on days 4, 7, and 10 postinfection. Similarly, when CD4+ T cells were isolated, activated with PHA, and infected with HIV-1, levels of DNA MTase mRNA expression were higher in infected cells than in uninfected cells throughout a similar 10-day incubation period (Fig. 1B). This increase in DNA MTase RNA was seen with several T-cell-tropic HIV-1 isolates (BP-1, IIIB, and a clinical isolate) (data not shown). Similar results were obtained when CD4+ Hut 78 cells were infected with the molecularly cloned proviruses NL43 and HXB-2 (Table 1). Increases of 3- to 20-fold in MTase RNA could be seen as early as 2 days postinfection in both primary T cells and the T-cell line Hut 78.
FIG. 1.
Expression of DNA MTase in HIV-infected PHA-activated T cells and purified T-cell subsets detected by RPA as described in Materials and Methods. Data shown are representative of four normal donors and five TH1 cell clones. Each experiment was done with a different healthy donor. (A) Elutriated PHA-activated lymphocytes; (B) purified PHA-activated CD4+ T cells; (C) TTx-specific TH1 clone H1.15. Days after infection are as indicated.
TABLE 1.
Upregulation of DNA MTase RNA in Hut 78 cells during acute HIV infection
| Infection | MTase RNA (relative units, 103)a | Proliferation (% of Hut 78)b |
|---|---|---|
| Hut 78 | 22–48 | |
| Mock | 18–63 | 94–96 |
| NL43 | 132–425 | 83–90 |
| HXB-2 | 262–380 | 85–88 |
Determined 48 h postinfection with a PhosphorImager (Molecular Dynamics) by comparing integrated volumes of MTase RPA signals on gels to an internal standard, actin and/or cyclophilin. Data shown are ranges of three separate experiments.
Measured 48 h postinfection by MTT assay (Promega) according to the manufacturer’s protocol. Data shown are ranges of three separate experiments.
As CD4+ helper T cells can be further subdivided into subsets based on their cytokine profiles (human TH1 cells express IFN-γ but not IL-4 and IL-5, while TH2 cells express IL-4 and IL-5 but not IFN-γ), we next examined the effect of HIV-1 on DNA MTase mRNA expression in antigen-specific human TH1 and TH2 clones following infection by HIV-1. In five antigen-specific TH1 clones and three antigen-specific TH2 clones, DNA MTase mRNA expression was 8- to 20-fold higher in the infected TH1 and TH2 clones than in the uninfected controls. A representative experiment is shown in Fig. 1C. DNA MTase mRNA expression was greatly upregulated at both day 5 and day 10 following HIV-1 infection in a TH1 clone compared to the expression of DNA MTase mRNA in the uninfected clone (Fig. 1C). These findings show that acute infection of CD4+ T cells, including both TH1 and TH2 T-cell subsets, with HIV-1 markedly upregulates DNA MTase mRNA expression.
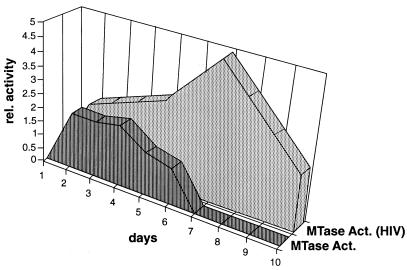
We next examined whether DNA MTase mRNA levels following acute HIV-1 infection are associated with increased cellular DNA MTase activity. As expected, DNA MTase activity was upregulated in uninfected cells following PHA stimulation of T-cell proliferation. Limiting amounts of exogenous IL-2 (10 U/ml) were added to CD4+ T cells after HIV-1 infection, which occurred 3 days after PHA activation. As a result, detectable DNA MTase activity in uninfected cells declined as proliferation declined, and no MTase activity was found at later time points (Fig. 2). In contrast, by day 4 following infection, DNA MTase activity in HIV-1-infected cells further increased above the level in uninfected control cells. By day 5, HIV-1-infected cells displayed peak levels of HIV-1 p24 production (data not shown) and levels of DNA MTase activity threefold higher than in uninfected cells. Under these culture conditions, the T cells cease proliferating between 7 and 10 days postinfection (10 to 13 days after PHA activation), as measured by cell counts and tritiated thymidine incorporation. In fact, a significant proportion of the primary cell culture undergoes HIV-1-induced apoptosis at day 7 and later time points following infection. It is not known if high levels of MTase activity play a role in this cell death. However, it was previously noted that increases in MTase expression above a certain threshold are toxic (75). Despite the lack of cell proliferation, a substantial level of MTase activity was present in the infected cells at day 10, while no increased MTase activity was detectable in the uninfected CD4+ T-cell control (Fig. 2).
FIG. 2.
Schematic depiction of DNA MTase activity in primary T cells with time following HIV-1 infection. DNA MTase activity was determined in infected cell lines maintained in log-phase growth and primary T cells following HIV-1 infection using S-adenosyl-l-[methyl-3H]methionine as the methyl donor and poly(dI-dC) · poly(dI-dC), which acts similarly to hemimethylated DNA, as the substrate as described in Materials and Methods. Primary CD4+ T cells from the same donor were activated with PHA for 3 days, and one set was infected with HIV-1 (day 1 on the graph). Parallel cultures of infected and uninfected cells from the same donor were harvested at daily intervals following infection. Relative values are given as the fold increase above that seen in day 3 activated T cells, which is set to equal zero. Data shown are the average of three separate experiments with a standard error of the mean of <20%.
Since DNA MTase activity is tightly linked to cell proliferation, it is important to determine whether this upregulation of DNA MTase activity by HIV-1 infection is secondary to increased cellular proliferation, as has been suggested for tumorigenesis models (47, 71). There is no compelling evidence that HIV-1 infection stimulates T-cell proliferation. Indeed, several studies show that it induces apoptosis (30, 56) and/or, through the vpr gene encoded by HIV-1, induces cell cycle arrest in the G2 phase of the cell cycle (1, 33). Also, infection of Hut 78, a continuously proliferating T-cell line, with NL43 and HXB-2 (Table 1) resulted in rates of proliferation 10 to 15% lower in the infected cells than in the mock-infected cells. Nonetheless, to further rule out that the differences in DNA MTase expression seen were due to the effects of the virus on cell proliferation, cell cycle analysis by propidium iodide staining and flow cytometry was performed on both the uninfected and infected cultures throughout the time course of the infection. No significant differences were seen in the cell cycle progression between the HIV-1-infected and uninfected CD4+ T cells up to 10 days postinfection (Table 2).
TABLE 2.
Cell cycle analysis of HIV-1-infected and uninfected CD4+ T cellsa
| Time (days) p.i.b | HIV-1 status | % of cells in:
|
|||
|---|---|---|---|---|---|
| G1 | G2/M | S | Apoptosis | ||
| 0 | − | 71 | 11 | 17 | 1 |
| 4 | − | 82 | 6.4 | 6.2 | 5.2 |
| + | 81 | 7.2 | 6.0 | 5.4 | |
| 7 | − | 70.8 | 4.4 | 2.3 | 22.9 |
| + | 66.6 | 4.8 | 2.6 | 26 | |
| 10 | − | 66.4 | 4.2 | 2.6 | 26.8 |
| + | 59 | 3.5 | 2.0 | 35.5 | |
Cell cycle analyses based on DNA content per cell were performed as described in Materials and Methods; 20,000 cells were analyzed. Data shown are representative of three separate experiments from individual normal donors.
Days postinfection (p.i.) of PHA-activated CD4+ T cells purified, activated, and infected as described in Materials and Methods. Day 0, time of infection, 3 days after PHA activation.
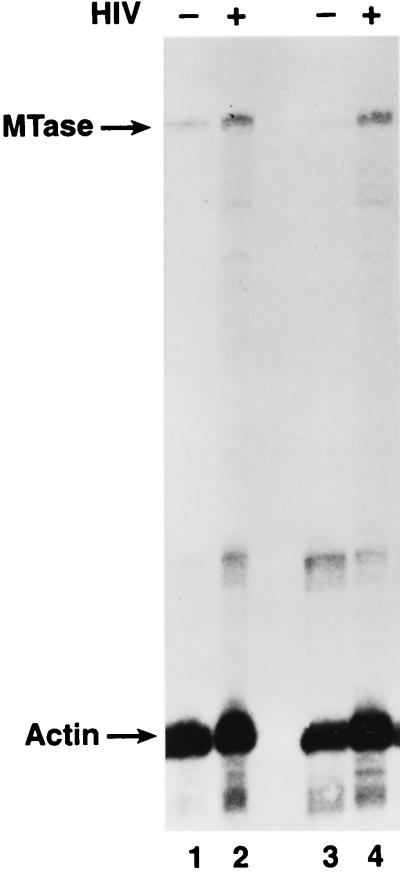
In addition, because the HIV-1 accessory gene vpr is also required for efficient replication of the virus in primary macrophages associated with its function in facilitating nuclear localization in these nondividing cells (14), we examined MTase mRNA levels in primary nondividing monocyte-derived macrophages (MDM) from normal donors. In MDM from two different donors expressing similar levels of HIV-1 p24 at 14 days after infection with the HIV-1 Bal isolate, the MTase mRNA levels were three- and fivefold higher in the infected culture than in the uninfected matched culture (Fig. 3). At that time, these cultures did not incorporate tritiated thymidine (data not shown). This finding provides further evidence that viral infection leads to upregulation of the MTase at the cellular level. Thus, it is likely that HIV-1 infection maintains increased levels of DNA MTase expression independent of proliferation. The mechanism of how HIV-1 infection maintains this upregulation of MTase activity is not clear. Expression of single gene products of HIV, including Nef, Tat, and Rev, using transient and stable transfection of human T-cell lines has not shown any clear ability to upregulate MTase mRNA expression (data not shown).
FIG. 3.
Expression of DNA MTase in 14-day MDM detected by RPA as described in Materials and Methods. Data shown indicate purified MDM (>94) isolated from two different healthy donors. Infection with HIV-1 monocytotropic strain Bal was performed 3 days following plastic adherence of monocytes. Positive infection was determined by p24gag ELISA on cell supernatants. Total RNA was isolated at day 14 postinfection (18 days of culture).
Increased genomic methylation in primary cells and cell lines following acute infection with HIV-1.
Having established that HIV-1 infection can increase the cellular capacity to methylate DNA, we examined whether this was reflected in an overall increase in genomic methylation of the infected cell population. This was assessed two ways. First, genomic DNA was digested with methyl-insensitive and -sensitive restriction enzymes (MspI and HpaII) followed by Southern blot analysis with an Alu-specific probe. Increased overall methylation was demonstrated after HIV infection, as mainly high-molecular-weight DNA was seen with the methyl-sensitive HpaII while the methyl-insensitive MspI gave low- as well as high-molecular-weight bands (data not shown). Second, a modified methyl accepting assay (79) was used to more quantitatively assess the methylation status of DNA isolated from uninfected and infected primary cell cultures as well as following acute infection of human cell lines. As shown in Table 3, an increase in overall genomic methylation was seen as early as 2 days after acute HIV-1 infection of both primary PHA-activated CD4+ T cells and the T-cell line Hut 78 compared to uninfected control cultures. The lack of cell death at day 2 postinfection indicates that HIV-1 infection does not select for a subpopulation of cells expressing higher levels of DNA MTase. By day 7, methylation levels of DNA in the HIV-1-infected cultures were as high as 152% of the level of uninfected controls. Daily cell proliferation measurements postinfection by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay showed that the proliferation rates of infected cultures were similar to or slightly less than those for the uninfected cultures (data not shown).
TABLE 3.
Acute HIV-1 infection increases overall genomic DNA methylation
| Cells | Time (days) p.i.a | Mean DNA methylation level (% of control) ± SEM |
|---|---|---|
| Hut 78 | 3 | 114 ± 4 |
| 7 | 140 ± 5 | |
| CD4+ T | 1 | 96 ± 2.7 |
| 2 | 124 ± 6.3 | |
| 6 | 152 ± 7.4 |
Days postinfection (p.i.) of lymphoid cell lines and PHA-activated CD4+ T cells purified and activated as described in Materials and Methods. Cell lines were infected with cloned isolates (NL43 and HXB-2) as well as the primary isolate BP-1 at an MOI of 1.
Levels are expressed as percent increase over the methylation level of uninfected matched control cultures at each time point. Data shown are representative of three separate infections.
De novo methylation of the IFN-γ promoter occurs in TH1 cells following acute infection with HIV-1.
A recent study has shown that the cellular DNA MTase can directly affect the de novo methylation of CpG islands if enzyme levels are increased ninefold (75). As we consistently saw high levels of DNA MTase activity in TH1 clones following acute infection with HIV-1, we next sought to identify genomic targets of this increased methylation, particularly sites which may become de novo methylated. In this regard, of the many genes that would be potential candidates for aberrant methylation during HIV infection, we chose IFN-γ, since previous studies have demonstrated that the IFN-γ promoter contains a methyl-sensitive endonuclease SnaBI site at position 52 of the transcription start site (22, 65, 84) in a region of the promoter critical for promoter activity (22, 65). Further, the methylation status of the promoter at this site correlates with IFN-γ gene expression, as human and murine TH1 cells, large granular lymphocyte cells, and peripheral blood memory T cells, which express IFN-γ, show hypomethylation of the IFN-γ core promoter at this site (57, 84). Cells that do not express IFN-γ, such as TH2 cells and naive T cells, are hypermethylated at this site (22, 57, 84). Given the important role of IFN-γ both in the immune response to viral infections and in the development of TH1 cells (60), and for the reasons outlined above, we hypothesized that the IFN-γ gene represented a potential target of the increased DNA MTase activity demonstrated in HIV-1-infected T cells.
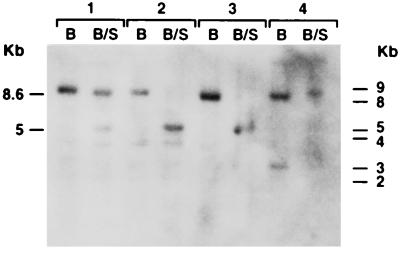
We first examined by Southern analysis the methylation status of the IFN-γ promoter in cell lines chronically infected with HIV-1. As a control for these studies, we also used an NK cell line which had been chronically infected by HTLV-1, as it had previously been shown that the IFN-γ promoter in this uninfected cell line was completely hypomethylated. The band detected at 5 kb in uninfected cell lines (lymphoid NK 3.3 and monocytoid THP-1) digested with BamHI and SnaBI is consistent with hypomethylation of this site (Fig. 4, groups 2 and 3). Interestingly, in both of the human retrovirally infected cell lines, the 5-kb band was greatly diminished or nonexistent and specific hybridization with the full-length probe now occurred at 8.6 kb (Fig. 4, groups 1 and 4, respectively; Fig. 5B). These data demonstrate that hypermethylation of this methyl-sensitive site can occur during HTLV-1 and HIV-1 infection in cell lines. Thus, infection with both known pathogenic human retroviruses correlates with altered methylation patterns of IFN-γ in the infected cells.
FIG. 4.
Southern analysis of the methylation status of the IFN-γ promoter at the SnaBI site in cell lines with or without HTLV-1 and HIV-1 infection. Digestions and analysis were performed as described in Materials and Methods. B, BamHI digest alone; B/S, BamHI and SnaBI digest of DNA. Group 1, HTLV-1-infected NK 3.3; group 2, uninfected NK 3.3; group 3, THP-1; group 4, HIV-ADA-infected THP-1.
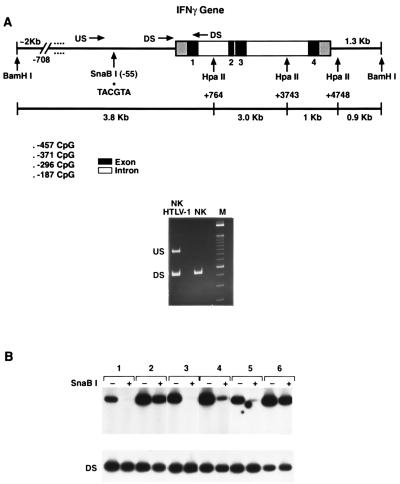
FIG. 5.
PCR analysis of IFN-γ promoter methylation status in T-cell clones and lymphoid cell lines. (A) (Top) Schematic showing the PCR primer locations (US, DS, and AS), the HpaII sites, the SnaBI site, and the CpG promoter sites in the IFN-γ gene. Solid boxes represent exons, and open boxes represent introns. (Bottom) When DNA was digested with SnaBI, PCR with the US primer pair plus the AS primer, which flank the SnaBI site, yields a product if the SnaBI site is methylated (HTLV-1-infected NK cells) but no product if the site is unmethylated (NK cells). As a DNA loading control, PCR was performed with the DS-AS primer pair, which does not flank the SnaBI site and should amplify independent of methylation status. Identical PCR products are generated from both the infected and the uninfected cells. (B) PCR with the US-AS primers in DNA, from various cells, digested (+) and not digested (−) with SnaBI. Group 1, uninfected human TH1 cell clone; group 2, uninfected human TH2 cell clone; group 3, uninfected NK 3.3 (NK cell line); group 4, NK 3.3 14 days after HTLV-1 infection; group 5, uninfected CS-3 (T cells); group 6, CS-3 7 days after HIV-1 infection.
To analyze the methylation status of the IFN-γ promoter in samples where few cells were available (e.g., HIV-1-infected TH1 clones and patient samples), we developed a PCR-based analysis of the methyl-sensitive SnaBI site. The premise of this method is that DNA cut by SnaBI will not yield a product with a PCR primer pair which spans the SnaBI site (Fig. 5A). The downstream primer pair which does not contain a SnaBI site and should have a PCR product was always included as a control for a poor PCR. The method was validated by the presence of a band in TH2 clones (Fig. 5C, group 2), which do not express the IFN-γ gene and are methylated at the SnaBI site as shown by Southern analysis (84) and by the lack of a PCR product in TH1 clones (Fig. 5C, group 1), which express the IFN-γ gene and are not methylated as determined by Southern analysis. Acute infection of susceptible lymphoid cell lines with either HTLV-1 or HIV-1 resulted in hypermethylation of the SnaBI site, as demonstrated by detection of increased PCR products (Fig. 5B; Fig. 5C, groups 3 versus 4 and 5 versus 6).
We next examined whether increased DNA MTase expression correlated with increased methylation of the IFN-γ promoter and decreased IFN-γ mRNA expression and production following acute infection by HTLV-1 and HIV-1 in primary cells. Since acute infection of primary TH1 clones by HIV-1 stimulated a marked increase in DNA MTase expression (Fig. 1C), the methylation status of the IFN-γ promoter in these HIV-1-infected TH1 clones was analyzed by PCR (Fig. 6A) following acute infection. A marked increase in the methylation of the IFN-γ promoter occurred in HIV-1-infected TH1 clones (Fig. 6A; Table 4). As shown by RT-PCR, IFN-γ mRNA expression was not detectable at day 10 following HIV infection of the TH1 clones (Fig. 6B). In contrast, detectable levels of IFN-γ mRNA were observed in the uninfected clones. Similarly, acute HTLV-1 infection of NK 3.3, which produces IFN-γ after stimulation with IL-2 (80), resulted in increased DNA MTase expression and concomitant downregulation of IFN-γ expression (data not shown). In five individual TH1 clones, IFN-γ expression and production correlated with the methylation status of the promoter during HIV-1 infection of TH1 clones in vitro (Table 4).
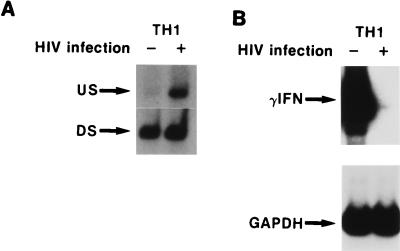
FIG. 6.
Correlation of HIV-1 infection and IFN-γ mRNA expression with methylation state of the IFN-γ promoter in TH1 clones. RNA or DNA was isolated at day 7 following infection. (A) IFN-γ promoter methylation status of the SnaBI site in SnaBI-digested DNA from an HIV-infected and uninfected TH1 clone as detected by the PCR procedure outlined for Fig. 4. (B) RT-PCR analysis of IFN-γ mRNA in the same TH1 clone used for panel A. Film was overexposed to demonstrate lack of IFN-γ mRNA expression 7 days following HIV-1 infection.
TABLE 4.
HIV-1 infection of human TH1 clones downregulates IFN-γ expression
| Clonea | Antigen | Mean HIV p24 (pg/ml) ± SEM (103)b | IFN-γ promoter methylation (fold increase)c | IFN-γ RNA expression (fold decrease)d |
|---|---|---|---|---|
| 1 (C01.D6) | SEB | 23,000 ± 1 | 10 | 14 |
| 2 (H1.15) | TTx | 31,500 ± 2.4 | 75 | >100 |
| 3 (H1.12) | TTx | 37,000 ± 2.9 | 15 | 20 |
| 4 (H1.11) | TTx | 43,500 ± 5.5 | 4 | 5 |
| 5 (H1.18) | TTx | 29,000 ± 3.3 | 60 | >100 |
T-cell clones were generated, maintained, and activated as described in Materials and Methods. Clones 2 to 5 were from same donor.
Antigen-activated clones were infected with either HIV-1 strain BP-1 (clones 2 to 5) or ADA (clone 1) as described in Materials and Methods. At day 7, medium was removed, stored, and used for HIV p24 assays (data are for triplicates of two experiments).
Determined with a PhosphorImager (Molecular Dynamics) by scanning films of PCR analysis of the methylation status of the IFN-γ promoter SnaBI site. The fold increase was calculated from the integrated volume of the product of the US-AS primer pair normalized to the DS-AS control (see Fig. 4) in the infected clone compared to the paired uninfected clone.
Determined by scanning with a PhosphorImager (Molecular Dynamics) films of RT-PCR analysis of IFN-γ expression; calculated from the integrated volume of the product normalized to the GAPDH internal control product in the infected clone compared to the paired uninfected clone.
To further strengthen the correlation between acute HIV infection, increased MTase activity and downregulation of IFN-γ, an ELISPOT for IFN-γ was developed. This assay assesses production of the cytokine at the level of the individual cell and allows determination of the number of cells producing IFN-γ in replicate cultures following HIV infection. Using three different TH1 clones (H1.12, H1.15, and H1.18) whose IFN-γ promoter is methylated during HIV-1 infection (Table 4), we found 67, 70, and 80% reductions, respectively, in the number of IFN-γ-producing cells 2 days after infection as shown by ELISPOT. All five TH1 clones showed greater than a fivefold increase in methylation of the IFN-γ promoter. Taken together, these data indicate that the IFN-γ gene is a target for de novo methylation as a result of the high levels of DNA MTase activity following acute HIV infection of primary lymphocytes.
Antisense constructs to DNA MTase prevent IFN-γ gene methylation and increase IFN-γ production.
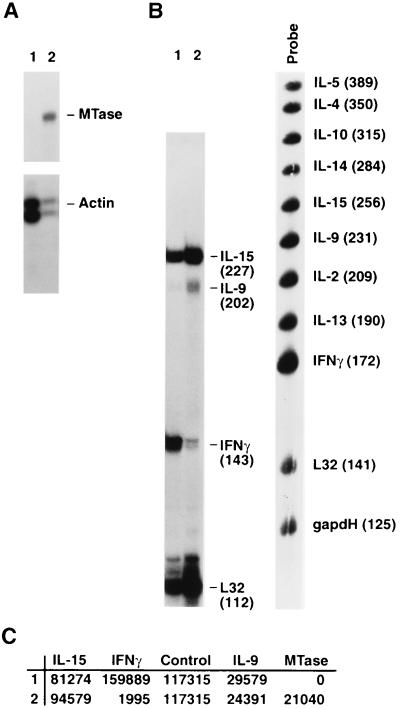
All of the above data predict that in HIV-1- and HTLV-1-infected lymphocytes, decreasing DNA MTase activity and the cellular capacity to methylate DNA will increase the expression of the IFN-γ gene. To test this directly, we used an antisense gene insertion approach to modulate DNA MTase expression. We achieved stable expression of a DNA MTase antisense construct in a lymphoid cell line, designated JMO (CD3− CD4+ CD2− CD8− CD19− CD14− CD25+ CD16− CD56+), developed in one of our laboratories. The parental JMO cell line and all single-cell clones of this cell line were shown to constitutively express detectable but variable levels of DNA MTase and IFN-γ as measured by RPA and ELISPOT, respectively (data not shown). To verify that HIV-1-induced DNA MTase expression could modulate IFN-γ expression in this cell line, JMO clone D8, whose basal MTase expression was lower (compare lanes 1 in Fig. 7A and 8A) and IFN-γ expression was higher (Table 5) than that of the parental JMO cell line, was infected, and total RNA was prepared 7 days postinfection. The RNA was analyzed by RPA as described in Materials and Methods. As shown in Fig. 7A, HIV-1 infection upregulated DNA MTase in JMO. Figure 7B shows a concomitant downregulation of IFN-γ mRNA in a multiprobe RPA to detect human cytokines (Pharmingen). Interestingly, the expression of IL-15 and IL-9 remained unchanged (Fig. 7B and C), demonstrating that there is not a general downmodulation of cytokine expression in the HIV-1-infected cells.
FIG. 7.
HIV-1 infection regulates MTase and IFN-γ expression in a lymphoid cell line. The T-cell line JMO clone D8 was infected with HIV-1. At 7 days postinfection, MTase and IFN-γ expression was measured, using 5 μg of total RNA in each RPA as described in Materials and Methods. (A) MTase RPA. Lane 1, uninfected JMO clone D8; lane 2, HIV-infected JMO clone D8. (B) Cytokine RPA. Lane 1, uninfected JMO clone D8; lane 2, HIV-infected JMO clone D8. Probe, human cytokine-1 multiprobe (Pharmingen). (C) PhosphorImager (Molecular Dynamics) quantitation of results in panels A and B, normalized to the actin and L32 controls, respectively.
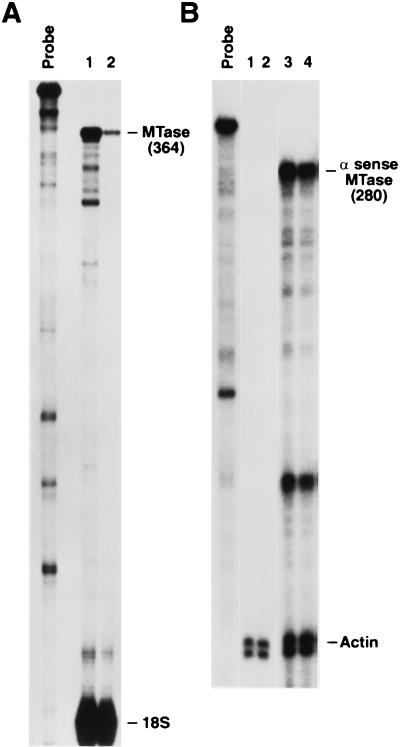
FIG. 8.
DNA MTase expression in stably transfected lymphoid cells. As described in Materials and Methods, the parental cell line JMO was stably transfected with an antisense DNA MTase expression vector to generate JMO-TMH cells and with the vector alone to derive the JMO-neo control cells. RPA of MTase sense and antisense expression was determined in the lymphoid cells. (A) Probe alone. Lane 1, JMO-neo; lane 2, JMO-TMH. (B) Probe alone. Lanes 1 and 2, clones of JMO-neo; lanes 3 and 4 clones of JMO-TMH.
TABLE 5.
Increased IFN-γ production in cells transfected with an antisense MTase vector
| Cell line (clone) | No. of IFN-γ-expressing cells/100 cellsb | IFN-γc (IU/well) |
|---|---|---|
| Single-cell clones | ||
| JMO (parental) | 22 ± 5 | 2 |
| JMO (D10) | 34 ± 6 | 1.7 |
| JMO (G9) | 27 ± 5 | 2 |
| JMO-TMH | 105 ± 6 | 26 |
| JMO-TMH (F7) | 98 ± 4 | 23 |
| JMO-TMH (D8) | 103 ± 6 | 20 |
| JMO-TMH (C7) | 93 ± 6 | 3 |
| Clone D8 | ||
| JMO (D8) | 36 ± 5 | 1.7 |
| HIV-infected JMO (D8), day 7d | 20 ± 4 | ND |
| HIV-infected JMO (D8)-TMHe | 36 ± 4 | ND |
JMO clones were generated and maintained as described in Materials and Methods.
Determined by ELISPOT as described in Materials and Methods.
Determined as ELISA international units per well (100 cells) of supernatant from the same wells of ELISPOT to give the quantity of IFN-γ per cell. ND, not determined.
ELISPOT analysis was performed 7 days after HIV infection of the cells.
After 7 days of HIV-1 infection, JMO clone D8 was transfected with an antisense MTase expression vector by using Superfect (Qiagen). After 48 h, the transfected cells were subjected to ELISPOT analysis.
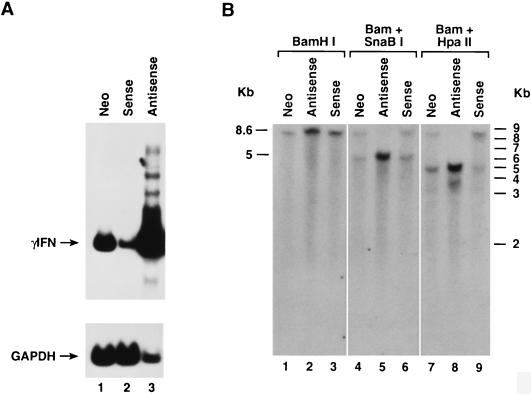
Furthermore, when parental JMO cells were stably transfected with the antisense MTase vector, the resulting cell line, JMO-TMH, showed a >90% reduction in DNA MTase expression (Fig. 8A) along with expression of the antisense mRNA (Fig. 8B) as determined by RPA. When IFN-γ mRNA expression from these cell lines was measured by RT-PCR, we found that IFN-γ mRNA was markedly higher in the antisense-expressing line (Fig. 9A, lane 3) than in the control cell lines stably transfected with neomycin (JMO-neo) and sense (JMO-HMT) constructs (Fig. 9A, lanes 1 and 2, respectively). The reduction in the IFN-γ mRNA in the sense transfectants was threefold when quantitated and normalized against the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) control. As previously described (75), the overexpression of sense MTase was not well tolerated by the cells. These transfected cells either lost overexpression of MTase or died within 10 days and were not further analyzed.
FIG. 9.
DNA MTase expression regulates IFN-γ expression in lymphoid cells. As described in Materials and Methods, the parental cell line JMO was stably transfected with an expression vector containing the full-length DNA MTase cDNA, in the antisense orientation, to generate JMO-TMH, with the vector alone to generate JMO-neo control, and with the sense-orientation DNA MTase cDNA to derive JMO-HMT as an additional control. (A) RT-PCR analysis of the expression of IFN-γ mRNA in these lymphoid cell lines. Lane 1, JMO-neo; lane 2, JMO-HMT (sense); lane 3, JMO-TMH (antisense). GAPDH mRNA is also shown as internal loading control for semiquantitative PCR. (B) Southern analysis of the methylation status of the IFN-γ region SnaBI site and first intronic region HpaII site. BamHI, BamHI-SnaBI, and BamHI-HpaII digests were performed as described in the text. Lanes 1, 4, and 7, JMO-neo control; lanes 2, 5, and 8, JMO-TMH, DNA MTase cDNA in the antisense orientation; lanes 3, 6, and 9, JMO-HMT, DNA MTase cDNA in the sense orientation.
Southern analysis showed partial methylation of the IFN-γ promoter in the control JMO-neo cell line (Fig. 9B, lanes 4 and 7), while JMO-TMH was completely hypomethylated not only at the SnaBI site (Fig. 9B, lane 5) but also at the HpaII site in the first intron, near the intronic enhancer elements (Fig. 5A; Fig. 9B, lane 8), as evidenced by the generation of 4.9- and 3.8-kb bands following BamHI-HpaII digestion. Expression of an antisense MTase in the JMO cell line shows a direct effect of DNA MTase activity on IFN-γ production. One hundred percent of cells carrying the antisense gene produced IFN-γ, compared to 22% of parental JMO as measured by ELISPOT (Table 5). On a per-cell basis, the antisense containing cells produced 5- to 10-fold more IFN-γ. These observations were similar for both the bulk culture and single-cell clones of JMO-TMH, indicating the results were not due to cell-to-cell variability (Table 5), even though there is heterogeneity in basal expression of MTase and IFN-γ among individual clones within the population of parental JMO cells. Thus, MTase expression and IFN-γ gene expression are directly linked in these cells. The ability to modulate MTase and IFN-γ by expression of antisense MTase allowed us to directly test the correlation between HIV-1 and MTase modulation of IFN-γ expression. The D8 clone of JMO, expressing IFN-γ in 36% of the population as determined by ELISPOT (Table 5), was infected for 7 days with HIV-1. At this time, ELISPOT analysis detected only 20% of the cells expressing IFN-γ. These cells were then transiently transfected with the antisense construct by using Superfect according to the manufacturer’s protocol. At 48 h, the cells were harvested and ELISPOT was performed. The HIV-infected JMO population which had 20% expressing cells now had 36% of the cells expressing IFN-γ compared to the same cells transfected with a control construct, which was unchanged at 20% expressing cells (Table 5). These data indicate a causal link between HIV-1 infection, MTase expression, and the downregulation of IFN-γ expression.
DISCUSSION
HTLV-1 and HIV-1 cause an overlapping set of pathogenic events including tumor development, immunodeficiency, and neurological symptoms (37, 49). Both human retroviruses preferentially infect CD4+ T lymphocytes, monocytes, and macrophages. It is the interplay of these cell types that orchestrates an immune response, primarily through the elaboration of cytokines. In the past decade, many reports have documented dysregulation of cytokines in vivo and in vitro following infection with these viruses (37, 38, 67). However, little is known about the molecular mechanism(s) involved in this cytokine dysregulation. In this report, we have shown that infection by HTLV-1 and HIV-1 can upregulate DNA MTase expression and increase the cellular capacity to methylate genes. Increased DNA MTase expression is seen not only in acutely infected cell lines of T-cell, NK cell, and monocyte/macrophage lineages but also following acute HIV infection of primary T cells and monocytes. We have shown herein that this increase in the cellular capacity to methylate genes not only results in an overall increase in DNA methylation in the infected cells but also has the capacity for de novo methylation. Among many possible candidate genes, we have identified one, the IFN-γ gene, and demonstrated de novo methylation at a site which had previously been shown to be critical in the transcriptional regulation of this gene.
Young et al. have reported (84, 85) that methylation of a CpG dinucleotide at −55 of the transcriptional start site of the IFN-γ could negatively regulate the interaction of nuclear DNA binding proteins with this site. They further demonstrated that this site is differentially methylated in the TH1 and TH2 helper T-cell subsets. TH1 cells, which produce IFN-γ, are hypomethylated at this site, while TH2 cells, which do not produce IFN-γ, are hypermethylated at this site. Previous studies showed that 5-azacytidine, a hypomethylating agent, stimulates IFN-γ production in cytotoxic T cells as well as in TH2 cells. These studies also demonstrated the specificity of this methylation, as expression other cytokines is not altered in these systems (21, 32, 84). One of the mechanisms by which DNA methylation has been shown to inhibit transcription is by inhibiting the recognition of the sequence motif by a transcription factor (39). Young et al. (84, 85) have also shown decreased levels of specific DNA binding protein complexes in TH1 cell nuclear extracts following in vitro methylation of this SnaBI site. It is of note that this site is not part of a CpG island, as there are only five CpG dinucleotides in the first 780 bp of the human IFN-γ promoter (Fig. 5A). Furthermore, none of the other CpG sites have been demonstrated to directly interact with DNA binding proteins. Here we detect marked increases in IFN-γ promoter methylation in lymphocytes following infection by HIV-1 or HTLV-1. Furthermore, we show that acute HIV-1 infection of TH1 clones in vitro results in increased DNA MTase activity, methylation of the SnaBI site in the promoter of the IFN-γ gene and subsequent downregulation of IFN-γ mRNA, and protein production from these infected cells. These results demonstrate a strong correlation during viral infection between increased DNA MTase levels, methylation of the IFN-γ promoter, and decreased IFN-γ production. Using an MTase antisense strategy, we observed a direct relationship between DNA MTase and IFN-γ gene expression through regulation of the methylation of the IFN-γ promoter.
The mechanism of this upregulation of DNA MTase expression during retroviral infection is not clear. Since DNA MTase activity is tightly linked to cellular proliferation, and some argue that in carcinogenesis, MTase upregulation is secondary to cell growth (47), it is possible that the upregulation of MTase activity by HIV-1 is secondary due to increases in cell proliferation. However, no difference in cell number, thymidine incorporation, or cell cycle status was seen between infected and uninfected primary CD4+ T cells. Moreover, infection of Hut 78, a continually proliferating T-cell line, with NL43 and HXB-2 resulted in rates of proliferation lower in the infected cells than in the mock-infected cells. Further, in 18-day monocyte cultures under conditions where labeling indices for tritiated thymidine were reported to be 1 positive cell per 1,000 whether or not the cells were HIV infected (77), high DNA MTase RNA expression is seen only in infected cultures. Whether or not proliferation is needed for initiation of HIV-1 infection, it is clear from the results presented here that HIV-1 infection maintains increased levels of DNA MTase expression in the absence of proliferation. On the other hand, since high levels of DNA MTase can be toxic to cells (75, 78), this MTase upregulation could be a nonspecific response to MTase-induced apoptotic signals. However, this is unlikely since approximately 20 to 30% of the cultured uninfected T cells were apoptotic by 10 days postinfection, due to a lack of IL-2, and no DNA MTase activity was detected in these cultures. Furthermore, the rapid increase in DNA MTase expression, overall genomic methylation, and decrease in IFN-γ production at the single-cell level by day 2 postinfection, in the absence of detectable cell death, indicates that the selection of a rare cell with these characteristics is not occurring. In addition, using a one-step assay where zidovudine inhibits viral replication, Fan et al. (20) also showed downregulation of IFN-γ mRNA in HIV-infected Hut 78 cells. Zidovudine blocked this downregulation, suggesting that the downregulation was a direct effect of viral replication.
These results provide a potential mechanism for dysregulation of expression of many genes following human retroviral infection. The overall increase in genomic methylation seen after HIV infection could have a toxic effect on the cells. In fact, a significant proportion of the primary cell cultures undergo HIV-1-induced apoptosis whether infected in vivo or in vitro (57). It is not known if the high level of MTase activity plays a role in this cell death; however, it should be noted that increases in MTase expression above a certain threshold are toxic (75). In addition, our finding that by increasing the cellular capacity to methylate genes, there is one gene that HIV-1 can downregulate, the IFN-γ gene, a key element in the host defense against viral infection, is of particular interest. The role of IFN-γ in differentiation of precursors of TH1 cells, downregulation of the production of TH2 cells, activation of monocytes to resist foreign invaders, and prevention of cellular spread of viruses has been well documented (60). Compromising any one of these functions could have an effect on some of the pathological consequences of HIV infection. Shearer and colleagues (13, 72) have hypothesized that there is an induction of a TH2 bias during HIV-1 infection and progression to AIDS. Our present data support this hypothesis in two ways. First, since IFN-γ expression is a requirement for designating a cell TH1, methylation of the IFN-γ promoter could lead to a low estimate of the number of TH1 cells in a population. Second, decreased IFN-γ expression could result in a decrease in production of TH1 cells (8) as well as a decrease in monocyte functions such as IL-12 production, which is observed in AIDS patients (11, 25). The presence of IFN-γ is needed for the long-term production of IL-12 in monocytes (51). Previous studies have suggested that the progression to AIDS is, at least in part, dependent on the ability to generate a TH1-like immune response (72). Specifically, aberrant methylation of the IFN-γ promoter could play a role in the gradual loss of type 1 response seen in AIDS patients.
Though beyond the scope of this study, these results indicate the need to define which, if any, of these potential pathogenic effects resulting from the aberrant methylation of the IFN-γ gene or any other gene are important in AIDS. The role of IFN-γ in the development of AIDS is not clear. Consistent with a decrease in TH1 function, several studies show a decrease in IFN-γ production (23, 38, 61, 87). In contrast, increased IFN-γ production has been reported (28, 62, 76). Since IFN-γ is produced by the CD4+ target cells of HIV-1 as well as cells reactive to HIV-1 infection (e.g., large granular lymphocyte and CD8+ T cells), these discrepancies in IFN-γ production could result from the fact that most studies have quantitated systemic IFN-γ in vivo or after short stimulation in vitro of bulk PBMC cultures from AIDS patients, potentially masking differential effects on target cells. Indeed, recent studies have shown that most IFN-γ is produced by CD8+ cells either in the periphery (28) or in the lymph nodes (19), where most viral replication occurs during the asymptomatic phase of AIDS (18). Single-cell analysis of IFN-γ production in T cells from HIV-1-infected individuals showed decreased numbers of CD4+ IFN-γ-producing cells but preserved numbers of IL-4-producing cells (55, 56). No effect was seen on the numbers of CD8+-producing cells. The effects of this decrease in CD4+ IFN-γ-producing cells on the cellular immune response and control of viral spread, particularly during acute infection, need to be determined. In this regard, recent evidence suggests that controlling viral load and spread is critical in determining the extent and progression of AIDS (6, 10, 43). If this variable course of lentiviral infection and long-term outcome is related to controlling viral spread, the number of CD4+ IFN-γ-producing cells present during HIV-1 infection could be a determinant of this control. We are now studying this question in a simian immunodeficiency virus primate model.
Our current findings may have implications for virus-induced tumorigenesis. Previous studies have shown that the promoter region of the calcitonin gene becomes densely methylated following cell infection with multiple tumorigenic viruses, including simian virus 40, Epstein-Barr virus, and HTLV-1 (15). Our results suggest that the underlying mechanism for this change could involve the early increases that we now define for DNA MTase activity following HIV and HTLV-1 infection of cells. While this study has focused on the de novo methylation of only one gene, the increases in overall genomic methylation demonstrated herein suggest that many genes may be altered by this mechanism during viral infection. The fact that multiple tumor suppressor genes are now known to be inactivated in association with aberrant promoter region methylation (34–36, 54) suggests that altered methylation of genes could be studied as a candidate early step in the tumorigenic activity of a range of viruses.
ACKNOWLEDGMENTS
We thank Kathleen Wieman and Jason Troxell for excellent technical assistance.
Portions of the work in the laboratory of S.B.B. were supported by NIH grant CA43318 and in part with funds from the NCI, NIH, under contract no. NOI-CO-56000.
REFERENCES
- 1.Bartz S R, Rogel M E, Emerman M. Human immunodeficiency virus type 1 cell cycle control: Vpr is cytostatic and mediates G2 accumulation by a mechanism which differs from DNA damage checkpoint control. J Virol. 1996;70:2324–2331. doi: 10.1128/jvi.70.4.2324-2331.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baylin S B. Abnormal regional hypermethylation in cancer cells. AIDS Res Hum Retroviruses. 1992;8:811–820. [PubMed] [Google Scholar]
- 3.Baylin S B, Makos M, Wu J J, Yen R W, deBustros A, Vertino P, Nelkin B D. Abnormal patterns of DNA methylation in human neoplasia: potential consequences for tumor progression. Cancer Cells. 1991;3:383–390. [PubMed] [Google Scholar]
- 4.Bednarik D P, Cook J A, Pitha P M. Inactivation of HIV LTR by DNA CpG methylation. EMBO J. 1989;1:1157–1164. doi: 10.1002/j.1460-2075.1990.tb08222.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belinsky S L, Nikula K J, Baylin S B, Issa J-P J. Increased cytosine DNA-methyltransferase activity is target-cell-specific and an early event in lung cancer. Proc Natl Acad Sci USA. 1996;93:4045–4050. doi: 10.1073/pnas.93.9.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benveniste O, Vaslin B, Le grand R, Cheret A, Matheux F, Theodoro F, Cranage M P, Dormont D. Comparative interleukin (IL)-2/interferon (IFN)-γ and IL-4/IL-10 responses during acute infection of macaques inoculated with attenuated nef-truncated or pathogenic SIVmac251 virus. Proc Natl Acad Sci USA. 1996;93:3658–3663. doi: 10.1073/pnas.93.8.3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bestor T, Laudana A, Mattaliano R, Ingram V. Cloning and sequencing of a cDNA encoding DNA methyltransferase of mouse cells: the carboxyl-terminal domain of the mammalian enzymes is related to bacterial restriction methyltransferases. J Mol Biol. 1988;203:971–983. doi: 10.1016/0022-2836(88)90122-2. [DOI] [PubMed] [Google Scholar]
- 8.Bradley L M, Dalton K D, Croft M. A direct role for IFN-γ in regulation of TH1 cell development. J Immunol. 1996;157:1350–1358. [PubMed] [Google Scholar]
- 9.Brown T. Analysis of DNA sequences by blotting and hybridization. In: Ausubel F A, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Greene Publishing and Wiley Interscience; 1994. pp. 2.91–2.10.16. [Google Scholar]
- 10.Chakrabarti L, Cumont M-C, Montagnier L, Hurtrel B. Variable course of primary simian immunodeficiency virus infection in lymph nodes: relation to disease progression. J Virol. 1994;68:6634–6642. doi: 10.1128/jvi.68.10.6634-6643.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chehimi J, Starr S E, Frank I, D’Andrea A, Ma X, Macgregor R R, Sennelier J, Trinchieri G. Impaired IL-12 production in human immunodeficiency virus infected patients. J Exp Med. 1994;179:1361–1367. doi: 10.1084/jem.179.4.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clark S J, Harrison J, Frommer M. CpNpG methylation in mammalian cells. Nat Genet. 1995;10:20–26. doi: 10.1038/ng0595-20. [DOI] [PubMed] [Google Scholar]
- 13.Clerici M, Hakim F T, Venzon D J, Blatt S, Hendrix C W, Wynn T A, Shearer G M. Changes in interleukin-2 and interleukin-4 production in asymptomatic, human immunodeficiency virus-seropositive individuals. J Clin Invest. 1993;91:759–765. doi: 10.1172/JCI116294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Conner R I, Chen B K, Choe S, Landau N R. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology. 1995;206:935–944. doi: 10.1006/viro.1995.1016. [DOI] [PubMed] [Google Scholar]
- 15.de Bustros A, Nelkin B D, Silverman A, Ehrlich G, Poiesz B, Baylin S B. The short arm of chromosome 11 is a “hot spot” for hypermethylation in human neoplasia. Proc Natl Acad Sci USA. 1988;85:5693–5697. doi: 10.1073/pnas.85.15.5693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Derse D D, Mikovits J A, Polianova M, Felber B K, Ruscetti F W. Virions released from cells transfected with a molecular clone of human T-cell leukemia virus type 1 give rise to primary and secondary infections of T cells. J Virol. 1995;69:1907–1912. doi: 10.1128/jvi.69.3.1907-1912.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.El-Deiry W S, Nelkin B D, Celano P, Yen R W, Falco J P, Hamilton S R, Baylin S B. High expression of the DNA methyltransferase gene characterizes human neoplastic cells and progression stages of colon cancer. Proc Natl Acad Sci USA. 1991;88:3470–3474. doi: 10.1073/pnas.88.8.3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Embretson J, Zupancic M, Beneke J, Till M, Wolinsky S, Ribas J L, Burke A, Haase A T. Analysis of human immunodeficiency virus-infected tissues by amplification and in situ hybridization reveals latent and permissive infections at single-cell resolution. Proc Natl Acad Sci USA. 1993;90:357–361. doi: 10.1073/pnas.90.1.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Emilie D, Peuchmaur M, Maillot M C, Crevon M C, Brousse N, Delfraissy J F, Dormont J, Galanaud P. Production of interleukins in human immunodeficiency virus-1-replicating lymph nodes. J Clin Invest. 1990;86:148–159. doi: 10.1172/JCI114678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fan J, Li P, Kok T-W, Burrell C J. AZT blocks down-regulation of IL-2 and IFN-γ gene expression in HIV acutely infected cells. Arch Virol. 1997;142:1035–1043. doi: 10.1007/s007050050139. [DOI] [PubMed] [Google Scholar]
- 21.Farrar W L, Ruscetti F W, Young H A. 5-Azacytidine treatment of a murine cytotoxic T cell line alters gamma interferon gene induction by interleukin 2. J Immunol. 1985;138:1551–1554. [PubMed] [Google Scholar]
- 22.Feinberg A P, Gehrke C W, Kuo K C, Ehrlich M. Reduced genomic 5-methylcytosine content in human colonic neoplasia. Cancer Res. 1988;48:1159–1161. [PubMed] [Google Scholar]
- 23.Fitch F, Gajewski T. Production of T-cell clones. In: Coligan J, Kruisbeck A, Marguiles D, Shevach E, Stober W, editors. Current protocols in immunology. J. New York, N.Y: Wiley and Sons; 1991. pp. 3.13.1–3.13.9. [Google Scholar]
- 24.Fitzpatrick, D. R., D. Macaranas, K. Shirley, and A. Kelso. 1997. Clonal analysis of demethylation patterns in the interferon-gamma promoter following CD8+ T cell activation. J. Interferon Cytokine Res. 17(Suppl. 2):S82.
- 25.Gazzinelli R T, Bala S, Stevens R, Baseler M, Kubin M, Trinchieri G, Sher A. HIV infection suppresses type 1 lymphokine and IL-12 responses to Toxoplasma gondii but fails to inhibit the synthesis of other parasite-induced monokines. J Immunol. 1995;155:1565–1571. [PubMed] [Google Scholar]
- 26.Gendelman H, Orenstein J, Martin M, Ferrua C, Mitra R, Phipps T, Wahl L, Lane H C, Fauci A. Efficient isolation and propagation of human immunodeficiency virus on colony stimulating factor 1-treated monocytes. J Exp Med. 1988;167:1428–1441. doi: 10.1084/jem.167.4.1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goelz S E, Vogelstein B, Hamilton S R, Feinberg A P. Hypomethylation of DNA from benign and malignant human colon neoplasms. Science. 1985;228:187–190. doi: 10.1126/science.2579435. [DOI] [PubMed] [Google Scholar]
- 28.Graziosi C, Gantt K R, Vaccarezza M, Demarest J F, Daucher M, Saag M S, Shaw G M, Quinn T C, Cohen O J, Welbon C C, Pantaleo G, Fauci A S. Kinetics of cytokine expression during primary human immunodeficiency virus type 1 infection. Proc Natl Acad Sci USA. 1996;93:4386–4391. doi: 10.1073/pnas.93.9.4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Greger V, Passarge E, Hopping W, Messmer E, Horsthemki B. Epigenetic changes may contribute to the formation and spontaneous regression of retinoblastoma. Hum Genet. 1989;83:155–158. doi: 10.1007/BF00286709. [DOI] [PubMed] [Google Scholar]
- 30.Groux H, Torpier G, Monte D, Mouton Y, Capron A, Ameisen J C. Activation-induced death by apoptosis in CD4+ T cells from human immunodeficiency virus-infected asymptomatic individuals. J Exp Med. 1992;175:331–336. doi: 10.1084/jem.175.2.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gunthert U, Schweiger M, Stupp M, Doerfler W. DNA methylation in adenovirus, adenovirus-transformed cells, and host cells. Proc Natl Acad Sci USA. 1976;73:3923–3927. doi: 10.1073/pnas.73.11.3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hardy K J, Peterlin B M, Atchison R E, Stobo J D. Regulation of expression of the human interferon gene. Proc Natl Acad Sci USA. 1985;82:8173–8177. doi: 10.1073/pnas.82.23.8173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.He J, Choe S, Walker R, Di Marzio P, Morgan D O, Landau N R. Human immunodeficiency virus type 1 viral protein R (Vpr) arrests cells in the G2 phase of the cell cycle by inhibiting p34cdc2 activity. J Virol. 1995;69:6705–6711. doi: 10.1128/jvi.69.11.6705-6711.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Herman J G, Jen J, Merlo A, Baylin S B. Hypermethylation-associated inactivation indicates a tumor suppressor role for p15INK4B1. Cancer Res. 1996;56:722–727. [PubMed] [Google Scholar]
- 35.Herman J G, Latif F, Weng Y, Lerman M I, Zbar B, Liu S, Samid D, Duan D-H, Gnarra J R, Linehan W M, Baylin S B. Silencing of the VHL tumor-suppressor gene by DNA methylation in renal cell carcinoma. Proc Natl Acad Sci USA. 1994;91:9700–9704. doi: 10.1073/pnas.91.21.9700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Herman J G, Merlo A, Mao L, Lapidus R G, Issa J-P J, Davidson N E, Sidransky D, Baylin S B. Inactivation of the CDKN2/p16/MTS1 gene is frequently associated with aberrant DNA methylation in all common human cancers. Cancer Res. 1995;55:4525–4530. [PubMed] [Google Scholar]
- 37.Hollsberg P, Hafler D A. Pathogenesis of diseases induced by human lymphotropic virus type 1 infection. N Engl J Med. 1993;328:1173–1182. doi: 10.1056/NEJM199304223281608. [DOI] [PubMed] [Google Scholar]
- 38.Hyjek E, Lischner H W, Hyslop T, Bartkowiak J, Kubin M, Trinchieri G, Kozbor D. Cytokine patterns during progression to AIDS in children with perinatal HIV infection. J Immunol. 1995;155:4060–4067. [PubMed] [Google Scholar]
- 39.Iguchi-Ariga S M M, Schaffner W. CpG methylation of the cAMP responsive enhancer/promoter TGACGTCA abolishes specific factor binding as well as transcriptional activation. Genes Dev. 1989;3:612–619. doi: 10.1101/gad.3.5.612. [DOI] [PubMed] [Google Scholar]
- 40.Issa J P, Vertino P M, Wu J, Sazawal S, Celano P, Nelkin B D, Hamilton S R, Baylin S B. Increased cytosine DNA-methyltransferase activity during colon cancer progression. J Natl Cancer Inst. 1993;85:1235–1240. doi: 10.1093/jnci/85.15.1235. [DOI] [PubMed] [Google Scholar]
- 41.Jahner D, Jaenisch R. Retrovirus-induced de novo methylation of flanking host sequences correlates with gene inactivity. Nature (London) 1985;315:594–597. doi: 10.1038/315594a0. [DOI] [PubMed] [Google Scholar]
- 42.Jones P A, Buckley J D. The role of DNA methylation in cancer. Adv Cancer Res. 1990;54:1–23. doi: 10.1016/s0065-230x(08)60806-4. [DOI] [PubMed] [Google Scholar]
- 43.Jurriaans S, Van Gemen B, Weverling G J, Van Strijp D, Nara P, Coutinho R, Koot M, Schuitemaker H, Goudsmit J. The natural history of HIV-1 infection: virus load and virus phenotype independent determinants of clinical course? Virology. 1994;204:223–233. doi: 10.1006/viro.1994.1526. [DOI] [PubMed] [Google Scholar]
- 44.Kautiainen T L, Jones P A. DNA methyltransferase levels in tumorigenic and nontumorigenic cells in culture. J Biol Chem. 1986;261:1594–1598. [PubMed] [Google Scholar]
- 45.Kumar S, Cheng X, Klimasauskas S, Mi S, Posfai J, Roberts R J, Wilson G G. The DNA (cytosine-5) methyltransferases. Nucleic Acids Res. 1994;22:1–10. doi: 10.1093/nar/22.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Laird P W, Jaenisch R. DNA methylation and cancer. Hum Mol Genet. 1994;3:1487–1495. doi: 10.1093/hmg/3.suppl_1.1487. [DOI] [PubMed] [Google Scholar]
- 47.Lee P J, Washer L L, Law D J, Boland C R, Horon I L, Feinberg A P. Limited up-regulation of DNA methyltransferase in human colon cancer reflecting increased cell proliferation. Proc Natl Acad Sci USA. 1996;93:10366–10370. doi: 10.1073/pnas.93.19.10366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leonhardt H, Bestor T H. Structure, function and regulation of mammalian DNA methyltransferase. In: Jost J P, Saluz H P, editors. DNA methylation: molecular biology and biological significance. Basel, Switzerland: Birkhauser Verlag; 1993. pp. 109–119. [DOI] [PubMed] [Google Scholar]
- 49.Levy J. HIV pathogenesis and long term survival. AIDS. 1993;7:1401–1410. doi: 10.1097/00002030-199311000-00001. [DOI] [PubMed] [Google Scholar]
- 50.Li E, Bestor T H, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 51.Ma X, Chow J M, Gri G, Carra G, Gerosa F, Wolf S F, Dzialo R, Trinchieri G. The interleukin 12 p 40 gene promoter is primed by interferon γ in monocytic cells. J Exp Med. 1996;183:147–157. doi: 10.1084/jem.183.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Masucci M G, Salazzar-Contreras B, Ragnar E, Falk K, Minarovits J, Ernberg I, Klein G. 5-Azacytidine upregulates the expression of Epstein-Barr virus nuclear antigen 2 (EBNA-2) through EBNA-6 and latent membrane protein in Burkitt’s lymphoma line Real. J Virol. 1989;63:3135–3141. doi: 10.1128/jvi.63.7.3135-3141.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Melvin M J, McGurn M E, Bort S J, Gibson C, Lewis D B. Hypomethylation of the interferon gamma gene correlates with its expression by primary T-lineage cells. Eur J Immunol. 1995;25:426–430. doi: 10.1002/eji.1830250218. [DOI] [PubMed] [Google Scholar]
- 54.Merlo A, Herman J G, Mao L, Lee D J, Gabrielson E, Burger P C, Baylin S B, Sidransky D. 5′ CpG island methylation is associated with transcriptional silencing of the tumor suppressor p16/CDKN2/MTS1 in human cancers. Nat Med. 1995;1:686–692. doi: 10.1038/nm0795-686. [DOI] [PubMed] [Google Scholar]
- 55.Meyaard L, Hovenkamp E, Keet R P M, Hooibrink B, De Jong I H, Otto S A, Miedema F. Single-cell analysis of IL-4 and IFN-γ production by T cells from HIV-infected individuals. J Immunol. 1996;157:2712–2718. [PubMed] [Google Scholar]
- 56.Meyaard L, Otto S, Hooibrink B, Miedema F. Quantitative analysis of CD4+ T cell function in the course of human immunodeficiency virus infection. J Clin Invest. 1994;94:1947–1952. doi: 10.1172/JCI117545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Meyaard L, Otto S A, Jonker R R, Mijnster M J, Keet R P M, Miedema F. Programmed cell death of T cells in HIV-1 infection. Science. 1992;257:217–219. doi: 10.1126/science.1352911. [DOI] [PubMed] [Google Scholar]
- 58.Mikovits J A, Lohrey N C, Schuloff R, Courtless J, Ruscetti F W. Immune activation of HIV expression from latently infected monocytes from asymptomatic seropositive patients. J Clin Invest. 1992;90:1486–1491. doi: 10.1172/JCI116016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mikovits J, Raziuddin A, Gonda M, Ruta M, Lohrey N, Kung H-F, Ruscetti F W. Negative regulation of HIV replication in monocytes: distinctions between restricted and latent expression in THP-1 cells. J Exp Med. 1990;171:1705–1720. doi: 10.1084/jem.171.5.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mossman T, Coffman R. TH1 and TH2 cells: different patterns of lymphokine secretion lead to different functional properties. Annu Rev Immunol. 1989;7:145–173. doi: 10.1146/annurev.iy.07.040189.001045. [DOI] [PubMed] [Google Scholar]
- 61.Murray H W, Scavuzzo D A, Kelly C D, Rubin B Y, Roberts R B. T4+ cell production of IFN-γ and the clinical spectrum of patients at risk for and with acquired immunodeficiency syndrome. Arch Intern Med. 1988;148:1613–1620. [PubMed] [Google Scholar]
- 62.Navikas V, Link J, Wahren B, Persson C, Link H. Increased levels of interferon-gamma (IFN-gamma), IL-4 and transforming growth factor-β (TGF-β) mRNA expressing blood mononuclear cells in human HIV infection. Clin Exp Immunol. 1994;96:59–65. doi: 10.1111/j.1365-2249.1994.tb06230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ohtani-Fujita N, Fujita T, Aoike A, Osifchin N E, Robbins P D, Sakai T. CpG methylation inactivates the promoter activity of the human retinoblastoma tumor-suppressor gene. Oncogene. 1993;8:1063–1067. [PubMed] [Google Scholar]
- 64.Pang Y, Norihisa Y, Benjamin D, Kantor R R S, Young H A. Interferon-γ gene expression in human B-cell lines: induction by interleukin-2, protein kinase C activators, and possible effect of hypomethylation on gene regulation. Blood. 1992;80:724–732. [PubMed] [Google Scholar]
- 65.Penix L, Weaver W M, Pang Y, Young H A, Wilson C B. Two essential regulatory elements in the human IFN-γ promoter confer specific expression in T-cells. J Exp Med. 1993;178:1483–1496. doi: 10.1084/jem.178.5.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Penix L A, Sweetser M T, Weaver W M, Hoeffler J P, Kerppola T K, Wilson C B. The regulatory element of the interferon-γ promoter mediates selective expression in T cells. J Biol Chem. 1996;271:31964–31972. doi: 10.1074/jbc.271.50.31964. [DOI] [PubMed] [Google Scholar]
- 67.Poli P, Fauci A S. Role of cytokines in the pathogenesis of human immunodeficiency virus infection. In: Aggarwaral B, Puri R, editors. Human cytokines: their role in disease and therapy. Cambridge, Mass: Blackwell Science; 1995. pp. 421–450. [Google Scholar]
- 68.Rideout W M, III, Coetzee G A, Olumi A F, Jones P A. 5-Methylcytosine as an endogenous mutagen in the human LDL receptor and p53 genes. Science. 1990;249:1288–1290. doi: 10.1126/science.1697983. [DOI] [PubMed] [Google Scholar]
- 69.Saggioro D, Forino M, Chieco-Bianchi L. Transcriptional block of HTLV-1 LTR by sequence-specific methylation. Virology. 1991;182:68–75. doi: 10.1016/0042-6822(91)90649-v. [DOI] [PubMed] [Google Scholar]
- 70.Saggioro D, Panozzo M, Chieco-Bianchi L. Human T-lymphotropic virus type I transcriptional regulation by methylation. Cancer Res. 1990;50:4968–4973. [PubMed] [Google Scholar]
- 71.Schmutte C, Yang A S, Nguyen T T, Beart R W, Jones P A. Mechanisms for the involvement of DNA methylation in colon carcinogenesis. Cancer Res. 1996;56:2375–2381. [PubMed] [Google Scholar]
- 72.Shearer G M, Clerici M. Early T-helper cell defects in HIV infection. AIDS. 1991;5:245–253. doi: 10.1097/00002030-199103000-00001. [DOI] [PubMed] [Google Scholar]
- 73.Shen J C, Rideout W M, Jones P A. High frequency mutagenesis by a DNA methyltransferase. Cell. 1992;71:1073–1080. doi: 10.1016/s0092-8674(05)80057-1. [DOI] [PubMed] [Google Scholar]
- 74.Vertino P M, Issa J-P, Pereira-Smith O M, Baylin S B. Stabilization of DNA methyltransferase levels and CpG island hypermethylation precede SV40 induced immortalization of human fibroblasts. Cell Growth Differ. 1994;5:1395–1402. [PubMed] [Google Scholar]
- 75.Vertino P M, Yen R-W C, Gao J, Baylin S B. De novo methylation of CpG island sequences in human fibroblasts overexpressing DNA (cytosine-5-methyltransferase) Mol Cell Biol. 1996;16:4555–4565. doi: 10.1128/mcb.16.8.4555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vykarnam A, Matear P, Meager A, Kelly G, Stanley B, Weller I, Beverley P. Altered production of tumor necrosis factors alpha and beta and interferon-gamma by HIV-1 infected individuals. Clin Exp Immunol. 1991;84:109–115. doi: 10.1111/j.1365-2249.1991.tb08132.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Weinberg J B, Matthews T J, Cullen B R, Malim M H. Productive human immunodeficiency virus type 1 (HIV-1) infection of nonproliferating human monocytes. J Exp Med. 1991;174:1477–1482. doi: 10.1084/jem.174.6.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wu J, Herman J G, Wilson G, Lee R Y, Yen R-W C, Mabry M, de Bustros A, Nelkin B D, Baylin S B. Expression of prokaryotic Hha I DNA methyltransferase is lethal and transforming to eukaryotic cells. Cancer Res. 1996;56:616–622. [PubMed] [Google Scholar]
- 79.Wu J, Issa J-P, Herman J G, Bassett D E, Jr, Nelkin B D, Baylin S B. Expression of an exogenous eukaryotic DNA methyltransferase gene induces transformation of NIH 3T3 cells. Proc Natl Acad Sci USA. 1993;90:8891–8895. doi: 10.1073/pnas.90.19.8891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ye J, Ortaldo J, Conlon K, Winkler-Pickett R, Young H. Cellular and molecular mechanisms of IFN production induced by IL-2 and IL-12. J Leukocyte Biol. 1995;58:225–233. doi: 10.1002/jlb.58.2.225. [DOI] [PubMed] [Google Scholar]
- 81.Yeivin A, Razin A. Gene methylation patterns and expression. In: Host J P, Saluz H P, editors. DNA methylation: molecular biology and biological significance. Basel, Switzerland: Birkhauser Verlag; 1993. pp. 523–568. [Google Scholar]
- 82.Yen R-W C, Vertino P M, Nelkin B D, Yu J J, El-Deiry W, Cumaraswamy A, Lennon G G, Trask B J, Celano P, Baylin S B. Isolation and characterization of the cDNA encoding human DNA methyltransferase. Nucleic Acids Res. 1992;20:2287–2291. doi: 10.1093/nar/20.9.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yisraeli J, Szyf M. DNA methylation and gene expression. In: Razin A, Cedar H, Riggs A D, editors. DNA methylation: biochemistry and biological significance. New York, N.Y: Springer-Verlag; 1984. pp. 353–378. [Google Scholar]
- 84.Young H A, Ghosh P, Ye J, Lederer J, Lichtman A, Gerard J, Penix L, Wilson C B, Melvin A J, McGurn M E, Lewis D B, Taub D D. Differentiation of the T helper phenotypes by analysis of the methylation state of the IFN-γ gene in immune cell regulation. J Immunol. 1994;153:3603–3610. [PubMed] [Google Scholar]
- 85.Young H A, Ghosh P. Molecular regulation of cytokine gene expression: interferon-γ as a model system. Prog Nucleic Acid Res Mol Biol. 1997;56:109–127. doi: 10.1016/s0079-6603(08)61004-1. [DOI] [PubMed] [Google Scholar]
- 86.Youssoufian H, Hammer S M, Hirsch M S, Mulder C. Methylation of the viral genome in an in vitro model of herpes simplex virus latency. Proc Natl Acad Sci USA. 1982;79:2207–2210. doi: 10.1073/pnas.79.7.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang M, Gong J, Iyer D V, Jones B E, Modlin R L, Barnes P F. T cell cytokine responses in persons with tuberculosis and human immunodeficiency virus infection. J Clin Invest. 1994;94:2435–2441. doi: 10.1172/JCI117611. [DOI] [PMC free article] [PubMed] [Google Scholar]