Summary
Human induced pluripotent stem cell (hiPSC)-derived macrophages provide a valuable tool for disease modeling and drug discovery. Here, we present a protocol to generate functional macrophages from hiPSCs using a feeder-free hematopoietic differentiation technique. We describe steps for preparing hiPSCs, mesodermal differentiation, hematopoietic commitment, and macrophage differentiation and expansion. We then detail assays to characterize their phenotype, polarization, and phagocytic functions. The functional macrophages generated here could be used to generate organoids for disease modeling and drug discovery studies.
For complete details on the use and execution of this protocol, please refer to Jeong et al.1 and Heo et al.2
Subject areas: Cell Biology, Cell culture, Cell isolation, Stem Cells, Cell Differentiation
Graphical abstract
Highlights
-
•
7-stage stepwise differentiation of hiPSC-derived macrophages
-
•
Steps for mesoderm induction with high concentration of BMP4 treatment
-
•
Generation, expansion, and characterization of functional macrophages
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Human induced pluripotent stem cell (hiPSC)-derived macrophages provide a valuable tool for disease modeling and drug discovery. Here, we present a protocol to generate functional macrophages from hiPSCs using a feeder-free hematopoietic differentiation technique. We describe steps for preparing hiPSCs, mesodermal differentiation, hematopoietic commitment, and macrophage differentiation and expansion. We then detail assays to characterize their phenotype, polarization, and phagocytic functions. The functional macrophages generated here could be used to generate organoids for disease modeling and drug discovery studies.
Before you begin
The protocol below was initially optimized using the CMC (CMC-003/009/011) and iPS-NT4-S1 human induced pluripotent stem cell (hiPSC) lines.3,4,5,6 However, it has also been used to generate hematopoietic progenitors and macrophages from multiple hiPSC lines. hiPSC lines may display variable capacity to differentiate into hematopoietic lineages during the induction protocol, but we have outlined key points and possible modifications that can be made to minimize this effect.
Culture of human induced pluripotent stem cells (hiPSCs)
Timing: 2 weeks before the start of differentiation
-
1.Thawing of hiPSCs.
-
a.Prepare a Vitronectin XF–coated plate or 35-mm dish.Note: When maintaining undifferentiated hiPSCs by mechanical passaging, we recommend the use of 35-mm dishes for easy handling.
-
i.Thaw Vitronectin XF at room temperature and dilute in Cell Adhere Dilution Buffer (final concentration of 10 μg/mL).
-
ii.Add the diluted Vitronectin XF to each well of a plate and incubate for at least 1 h at room temperature before use.Note: If not used immediately, the plate must be sealed with Parafilm to prevent evaporation of the Vitronectin XF solution. The coated plate can be stored at 2°C–8°C and will remain stable for 1 week.Note: Cover the cultureware surface with the recommended volume of diluted Vitronectin XF according to the manufacturer’s instructions.
Cultureware Volume of diluted Vitronectin XF 6-well plate 1 mL/well 100 mm dish 6 mL/dish T-25 cm2 flask 3 mL/flask T-75 cm2 flask 8 mL/flask -
iii.Rinse each well with 1 mL of Cell Adhere Dilution Buffer.
-
iv.Add 1 mL of TeSR-E8 medium supplemented with 1 μL of 10 mM Rho-associated protein kinase (ROCK) inhibitor (Y-27632).Note: When handling Y-27632, protect it from light. Ensure it is protected from light when handling dishes with added Y-27632.
-
v.Incubate the plate in a humidified incubator (37°C, 5% CO2) for at least 60 min.
-
i.
-
b.Thaw frozen cryovials containing hiPSCs in a 37°C water bath until only a small amount of ice is visible in the vial.Note: hiPSC cultures were cryopreserved as aggregates harvested from one well of a 6-well plate at the time they would normally be ready for passaging. Each cryovial contains approximately 1–2×106 cells per vial (can vary depending on the confluency of the cultures). Plate the equivalent of one cryovial of cells into 1 or 2 wells of a coated 6-well plate.
-
c.Wipe the outsides of the cryovials with 70% (v/v) ethanol to disinfect them, then dry them and place them in a biosafety cabinet.
-
d.Using a serological pipette, quickly and gently transfer the cells into a sterile 15-mL conical tube.
-
e.Slowly add 3 mL of E8 medium dropwise to the cells in the 15-mL tube. While adding the medium, gently rock the tube back and forth to mix the hiPSCs.
-
f.Centrifuge the cells at 600 g for 2 min.
-
g.Aspirate the supernatant and resuspend the cell pellet in 1 mL of E8 medium.
-
h.Transfer the cell solution to the prepared plate and gently rock the plate side to side and back and forth to distribute the aggregates across the bottom of the well.
-
i.Incubate cells overnight to allow colonies to attach.
-
a.
-
2.Culture of hiPSCs.
-
a.Feed the hiPSCs by removing approximately 95% of the medium from the wells to avoid drying out the cells.
-
b.Add 2 mL of fresh prewarmed E8 medium to each well of the plate and incubate cells at 37°C in a humidified incubator with 5% CO2.
-
a.
Note: It is highly recommended to monitor the hiPSCs daily under phase contrast microscopy to verify their morphology and confluency, the presence of differentiated cells, and the absence of contamination.
-
3.Mechanical passaging of hiPSCs.Note: Before passaging, it may be necessary to manually remove spontaneously differentiating whole colonies using a drawn-out glass Pasteur pipette, hypodermic syringe, or 10- to 20-μL pipette tip under a dissecting microscope (Figure 1).
-
a.Cells should be passaged once they reach 80%–90% confluency.
-
b.Prepare the Vitronectin XF–coated plate 1 day before passaging.
-
c.Aspirate spent medium from the cultureware and wash the cells with KnockOut Dulbecco’s Modified Eagle Medium (DMEM) once.
-
d.Add 1 mL of prewarmed E8 medium to each well and dissect colonies into multiple pieces under a dissecting microscope using a 10- to 20-μL pipette tip or syringe.
-
e.Gently scrape the dissected colonies with a cell scraper and transfer the detached small clumps to the prepared Vitronectin XF–coated plate containing prewarmed E8 medium.Note: To avoid cell culture contamination, manual passaging must be performed aseptically under a laminar flow hood using sterile materials. Adjust split ratios depending on the number of cell clusters and variations in rates of growth and attachment among the cell lines.
-
f.Carefully place the cultureware in the incubator, and gently swirl the plate back and forth and from side to side to evenly distribute the cells.
-
g.Incubate cells overnight to allow colonies to attach.
 CRITICAL: After thawing, hiPSCs should be passaged at least once before starting the differentiation protocol. We recommend starting the hematopoietic differentiation after 2–3 passages to remove as many unwanted differentiated colonies as possible, which is critical to achieving efficient and reproducible hematopoietic induction.
CRITICAL: After thawing, hiPSCs should be passaged at least once before starting the differentiation protocol. We recommend starting the hematopoietic differentiation after 2–3 passages to remove as many unwanted differentiated colonies as possible, which is critical to achieving efficient and reproducible hematopoietic induction.
-
a.
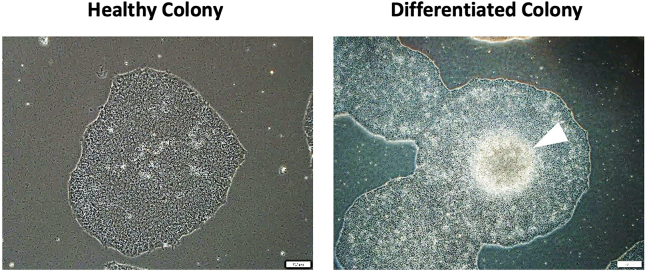
Figure 1.
Example images of healthy and differentiated colonies that are maintained in culture prior to the onset of differentiation
Healthy colonies are densely composed of homogeneous single cells and appear as a single layer (left image). In differentiated colonies, a cluster of brightly glowing cells is produced from the center of the colony (right image, white arrowhead). These colonies will increase in size over time, and if there are differentiated colonies present, they will be less efficient at differentiating the desired hematopoietic progenitor cells. Scale bars are 200 μm.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| PE mouse anti-human CD31 (1:500) | BD Biosciences | 555446 |
| FITC mouse anti-human CD34 (1:500) | BD Biosciences | 555821 |
| APC mouse anti-human CD45 (1:500) | BD Biosciences | 555485 |
| APC mouse anti-human CD43 (1:500) | BD Biosciences | 560198 |
| 7-Amino-actinomycin D (7-AAD) (1:500) | BD Biosciences | 559925 |
| APC anti-mouse/human CD11b antibody (1:500) | BioLegend | 101212 |
| APC anti-human CD14 antibody (1:500) | BioLegend | 325608 |
| FITC anti-human CD40 antibody (1:500) | BioLegend | 334306 |
| APC anti-human CD80 antibody (1:500) | BioLegend | 305220 |
| PE anti-human CD163 antibody (1:500) | BioLegend | 333606 |
| APC anti-human CD206 (MMR) antibody (1:500) | BioLegend | 321110 |
| Chemicals, peptides, and recombinant proteins | ||
| Vitronectin XF | STEMCELL Technologies | 07180 |
| Cell Adhere dilution buffer | STEMCELL Technologies | 07183 |
| TeSR-E8 | STEMCELL Technologies | 05990 |
| STEMdiff APEL 2 medium | STEMCELL Technologies | 28995 |
| KnockOut DMEM | Gibco | 10829018 |
| RPMI 1640 medium | Gibco | 22400089 |
| Fetal bovine serum (FBS) | HyClone | SH30071 |
| Y-27632 dihydrochloride | Tocris | 1254 |
| 100X Insulin-Transferrin-Selenium-X (ITS-X) supplement | Gibco | 51500056 |
| Recombinant human BMP4 protein | R&D | 314-BP |
| Recombinant human VEGF 165 protein | R&D | 293-VE |
| Recombinant human SCF protein | R&D | 255-SC |
| Recombinant human Thrombopoietin (NS0-expressed) protein | R&D | 288-TPN |
| Recombinant human IL-3 protein | R&D | 203-IL |
| Recombinant human IL-6 protein | R&D | 206-IL |
| Recombinant human Flt-3 ligand/FLT3L protein | R&D | 308-FK |
| Recombinant human M-CSF | PeproTech | 300-25 |
| Lipopolysaccharides | Sigma | L2630 |
| Human IFN-γ | Miltenyi Biotec | 130-096-872 |
| Human IL-4 | Miltenyi Biotec | 130-095-373 |
| Human IL-13 | Miltenyi Biotec | 130-112-409 |
| Latex beads, carboxylate-modified polystyrene, fluorescent yellow-green | Sigma-Aldrich | L4530-1ML |
| Experimental models: Cell lines | ||
| CMC hiPSC lines | Korea National Stem Cell Bank | CMC-003/009/011 |
| iPS-NT4-S1 hiPSC lines | CHA University | N/A |
| Oligonucleotides | ||
| Primer: GAPDH forward | Bioneer | TGCACCACCAACTGCTTAGC |
| Primer: GAPDH reverse | Bioneer | GGCATGGACTGTGGTCATGAG |
| Primer: T forward | Bioneer | ATGAGCCTCGAATCCACATAGT- |
| Primer: T reverse | Bioneer | TCCTCGTTCTGATAAGCAGTCA |
| Primer: MIXL1 forward | Bioneer | GGATCCAGGTATGGTTCCAG |
| Primer: MIXL1 reverse | Bioneer | GGAGCACAGTGGTTGAGGAT |
| Software and algorithms | ||
| GraphPad Prism 10.0 | GraphPad Software | https://www.graphpad.com/ |
| FlowJo software | Tree Star | https://www.flowjo.com/ |
| Other | ||
| 35 mm cell culture dish | SPL | 11035 |
| 6-well clear multiple well plates | Corning | 3516 |
| 70 μm cell strainer | SPL | 93070 |
| Dissection microscope | Olympus | CKX53 |
Materials and equipment
Basal 1 Medium (Stage 2–3)
| Reagent | Final concentration | Amount |
|---|---|---|
| STEMdiff APEL medium | N/A | 495 mL |
| Insulin-Transferrin-Selenium-X (ITS-X) (100×) | 1× | 5 mL |
| Total | N/A | 500 mL |
Store in aliquots at −20°C for a maximum of 7 months. Defrost at 4°C before use.
Basal 2 Medium (Stage 4)
| Reagent | Final concentration | Amount |
|---|---|---|
| RPMI 1640 Medium | N/A | 450 mL |
| Fetal Bovine Serum (FBS) | 10% | 50 mL |
| Total | N/A | 500 mL |
Store at 4°C and keep for a maximum of 3–4 weeks.
Stage 1 Cytokines
| Reagent | Final concentration | Reconstitution |
|---|---|---|
| BMP4 | 50 ng/mL | 50 μg in 1 mL (50 ng/μL) |
Aliquot 50 μL into a 1.5 mL microcentrifuge tube and store at −80°C for a maximum of 7 months. Limit freezing and thawing cycles.
Stage 2 Cytokines
| Reagent | Final concentration | Reconstitution |
|---|---|---|
| VEGF | 40 ng/mL | 10 μg in 0.5 mL (20 ng/μL) |
| SCF | 50 ng/mL | 50 μg in 2 mL (25 ng/μL) |
Aliquot 50 μL into a 1.5 mL microcentrifuge tube and store at −80°C for a maximum of 7 months. Limit freezing and thawing cycles.
Stage 3 Cytokines
| Reagent | Final concentration | Reconstitution |
|---|---|---|
| SCF | 50 ng/mL | 50 μg in 2 mL (25 ng/μL) |
| TPO | 10 ng/mL | 25 μg in 2.5 mL (10 ng/μL) |
| IL-3 | 50 ng/mL | 50 μg in 2 mL (25 ng/μL) |
| IL-6 | 50 ng/mL | 50 μg in 2 mL (25 ng/μL) |
| Flt-3L | 50 ng/mL | 25 μg in 1 mL (25 ng/μL) |
Aliquot 50 μL into a 1.5 mL microcentrifuge tube and store at −80°C for a maximum of 7 months. Limit freezing and thawing cycles.
Stage 4 Cytokines
| Reagent | Final concentration | Reconstitution |
|---|---|---|
| M-CSF | 100 ng/mL | 100 μg in 1 mL (100 ng/μL) |
Aliquot 50 μL into a 1.5 mL microcentrifuge tube and store at −80°C for a maximum of 7 months. Limit freezing and thawing cycles.
Stage 4-1 Cytokines
| Reagent | Final concentration | Reconstitution |
|---|---|---|
| LPS | 100 ng/mL | 10 mg in 10 mL (1 mg/mL) |
| IFN-γ | 20 ng/mL | 20 μg in 1 mL (20 ng/μL) |
| IL-4 | 20 ng/mL | 20 μg in 1 mL (20 ng/μL) |
| IL-13 | 20 ng/mL | 20 μg in 1 mL (20 ng/μL) |
Aliquot 50 μL into a 1.5 mL microcentrifuge tube and store at −80°C for a maximum of 7 months. Limit freezing and thawing cycles.
Note: Cytokines were diluted with 10% FBS-PBS.
Step-by-step method details
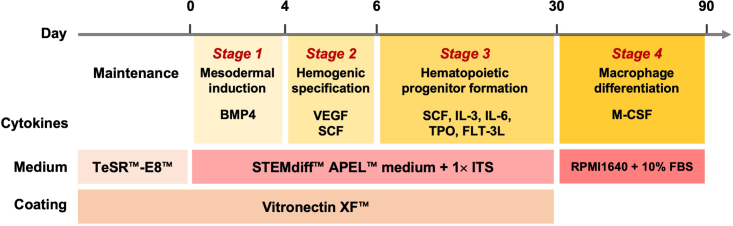
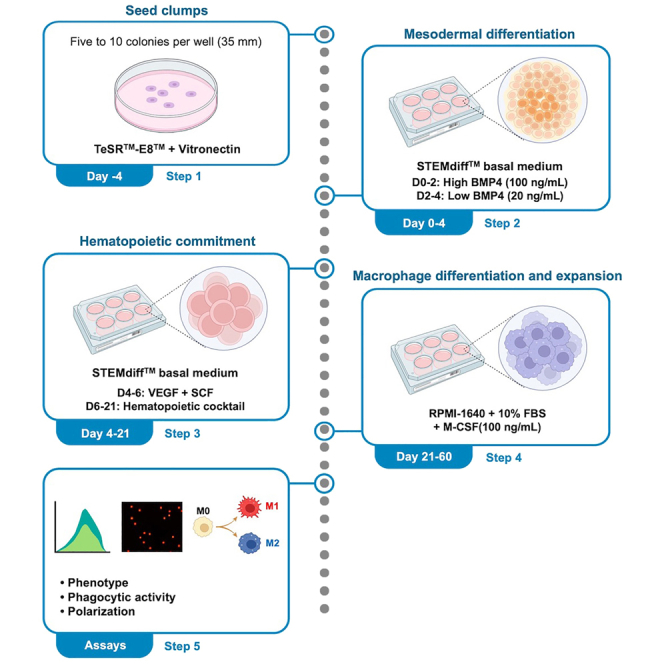
To mimic the ontogeny of the embryonic hematopoietic system as closely as possible in vitro, this protocol induces hiPSCs to functional macrophages in a stepwise manner: 1) mesodermal induction, 2) hemogenic specification, 3) hematopoietic progenitor formation, and 4) macrophage differentiation. This stepwise differentiation process will ensure the quality of efficient macrophage differentiation from multipotent hematopoietic progenitor hiPSCs. Each step consists of media and cytokines that sequentially induce hiPSCs into the hematopoietic lineage. All steps in this protocol should be performed within a biosafety cabinet and under aseptic conditions. A schematic overview of all steps is provided in Figure 2.
Figure 2.
Schematic overview of all steps of the protocol
Schematic overview of the four-step protocol for the induction of hematopoietic cells and macrophages from hiPSCs.
The yields of hematopoietic progenitors and macrophages can vary depending on hiPSC lines and culture methods. Thus, this method can be scaled up by selecting hiPSC lines that have a strong propensity to differentiate into the mesodermal lineage and adjusting the seeding number of hiPSC colonies.
Preparation of hiPSCs
Timing: 4–5 days, 30 min per day
-
1.
Prepare a Vitronectin XF–coated 6-well plate 1 day prior to passaging hiPSCs as described above.
-
2.Split hiPSCs into each well of a 6-well plate when cells reach 80–90% confluency.
-
a.Aspirate spent medium from the plates and wash cells with KnockOut DMEM once.
-
b.Add 1 mL of prewarmed E8 medium to each well and dissect colonies into multiple pieces under a dissecting microscope using a 10- to 20-μL pipette tip or syringe.
-
c.Gently transfer the detached small clumps to a prepared Vitronectin XF–coated plate containing prewarmed E8 medium.
-
d.Place the plate back into the incubator and gently move the plate back and forth and side to side to evenly distribute any clumps.
-
e.Incubate overnight in a humidified incubator (37°C, 5% CO2) to allow the clumps to attach and grow.
-
f.Culture the hiPSCs by changing the E8 medium every day until the colonies grow to approximately 500 μm in diameter.
-
a.
CRITICAL: In this step, the number, size, and distribution of colonies prior to the start of the mesodermal induction are important for ensuring high-yield production of hematopoietic progenitors and macrophages from hiPSCs. Based on our experience in the CMC and iPS-NT4-S1 hiPSC lines, achieving an even distribution of 5–7 colonies with diameters of approximately 500 μm in each well of a 6-well plate is a key step in ensuring the quality and number of hematopoietic progenitors and macrophages produced in later stages. However, these conditions may need optimization depending on the proliferative capacity and differentiation propensity of the hiPSC lines being used.
Mesodermal induction (stage 1)
Timing: days 0–4, 30 min per day
This is the stage at which the proper formation of the mesoderm is induced. Temporal regulation of bone morphogenetic protein 4 (BMP4) promotes early mesodermal differentiation of hiPSCs in a differentiation stage-specific manner. This is a successful strategy for increasing the efficiency of induction of the next step, hemogenic specification. μ.
CRITICAL: The colony sizes at the time of differentiation should be approximately 500 μm in diameter. Colony sizes that are too small or too large may affect the efficiency of hematopoietic differentiation.
Note: If too many clumps adhere to the plate or spontaneously differentiated cells are present before the start of the differentiation process, remove them mechanically from plate the using a 10- to 20-μL pipette tip under a dissection microscope.
Note: This day is defined as Day 0 of the differentiation process.
-
3.Prepare Hematopoietic Differentiation Medium (HDM). The HDM medium will continue to be used until Stage 3.
-
a.Add 1× Insulin-Transferrin-Selenium-X (ITS-X) supplement to STEMdiff APEL medium.
-
a.
-
4.Day 0–2: Prepare aliquots of HDM and prewarm in a 37°C water bath.
-
a.Each well of a 6-well plate requires 2 mL of HDM.
-
a.
Stage 1. Day 0–2 medium
| Reagent | Final concentration | Amount |
|---|---|---|
| Hematopoietic Differentiation Medium (HDM) | N/A | 998 μL |
| BMP4 (50 ng/μL) | 100 ng/mL | 2 μL |
| Total | N/A | 1 mL |
-
5.
Gently remove the E8 medium and replace it with 2 mL of the Stage 1 Days 0–2 medium per well of the plate.
-
6.
Incubate the cells for 2 days (48 h) in a humidified incubator (37°C, 5% CO2).
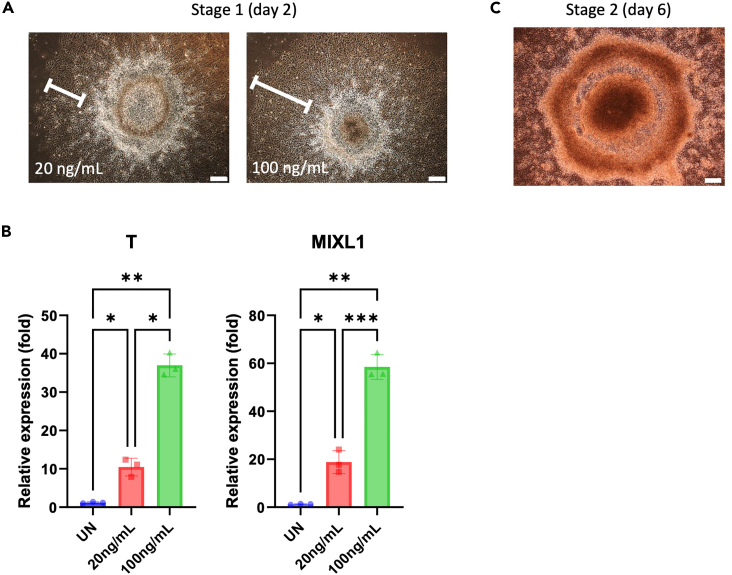
Note: On days 0–2, a brief induction with a high concentration of BMP4 (100 ng/mL) promotes the frequency and yield of primitive hematopoietic progenitors (CD34+CD45+ or CD34+CD43+) and mature blood cells (CD34-CD45+ or CD34-CD43+). Short-term BMP4 treatment can lead to morphological changes characterized by flattened and enlarged cells, which initially may appear on the edges of the colonies. Colonies treated with a high concentration of BMP4 show broader flattened areas and exhibit higher expression levels of mesodermal genes, such as Brachyury (T) and mix paired-like homeobox (MIXL1) (Figures 3A and 3B).
-
7.Day 2–4: Prepare aliquots of HDM and prewarm them in a 37°C water bath.
-
a.One well of a 6-well plate requires 2 mL.
-
a.
Stage 1. Day 2–4 medium
| Reagent | Final concentration | Amount |
|---|---|---|
| Hematopoietic Differentiation Medium (HDM) | N/A | 999.6 μL |
| BMP4 (50 ng/μL) | 20 ng/mL | 0.4 μL |
| Total | N/A | 1 mL |
-
8.
Gently remove the previous medium and replace it with the Stage 1 day 2–4 medium (2 mL per well of the plate).
-
9.
Incubate the cells for 2 days (48 h) in a humidified incubator (37°C, 5% CO2).
Note: Mesodermal differentiation efficiency can be assessed using qPCR (Figure 3B).
Figure 3.
Effect of BMP4 treatment concentration on early mesoderm differentiation
(A) Representative bright-field images of colonies in which the early mesoderm was induced with low (20 ng/mL) and high (100 ng/mL) concentrations of BMP4 in Stage 1. Scale bars are 200 μm.
(B) Differentiation days 0–2. The differences in mesoderm differentiation markers (T and MIXL1) between low (20 ng/mL) and high (100 ng/mL) doses of BMP4 were confirmed by quantitative reverse transcriptase polymerase chain reaction (qRT–PCR). The bars indicate the mean ± SD, n = 3, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. UN, undifferentiated hiPSCs.
(C) Representative bright-field images of colonies during the hemogenic specification differentiation of hiPSCs. Scale bar is 200 μm.
Hemogenic specification (stage 2)
Timing: days 4–6, 30 min per day
This is the hemogenic specification phase, which is characterized by the emergence of bipotent hemogenic precursors that can give rise to the hematopoietic and vascular lineages.
-
10.Days 4–6: Prepare aliquots of HDM and prewarm them in a 37°C water bath.
-
a.One well of a 6-well plate requires 3 mL of HDM.
-
a.
Stage 2. Day 4–6 medium
| Reagent | Final concentration | Amount |
|---|---|---|
| Hematopoietic Differentiation Medium (HDM) | N/A | 997 μL |
| VEGF (20 ng/μL) | 40 ng/mL | 2 μL |
| SCF (50 ng/μL) | 50 ng/mL | 1 μL |
| Total | N/A | 1 mL |
-
11.
Gently remove the previous medium and replace it with the Stage 2 days 4–6 medium (3 mL per well of the plate).
-
12.
Incubate the cells for 2 days (48 h) in a humidified incubator (37°C, 5% CO2).
Note: Colonies exhibit a plateau-like central area and a flat peripheral area during the hemogenic specification phase. The morphology then changes, forming beaded-like structures along the edges of the plateau area (Figure 3C).
Hematopoietic progenitor formation (stage 3)
Timing: days 6 up to 30 days, 30 min per day
This is the hematopoietic commitment phase, which is characterized by the emergence of multi-lineage hematopoietic progenitor cells (HPCs) and mature blood cells.
-
13.Days 6–30: Prepare aliquots of HDM and prewarm them in a 37°C water bath.
-
a.Each well of a 6-well plate requires 4 mL of HDM.
-
a.
Stage 3. Day 6–30 medium
| Reagent | Final concentration | Amount |
|---|---|---|
| Hematopoietic Differentiation Medium (HDM) | N/A | 995 μL |
| SCF (50 ng/μL) | 50 ng/mL | 1 μL |
| TPO (10 ng/μL) | 10 ng/mL | 1 μL |
| IL-3 (50 ng/μL) | 50 ng/mL | 1 μL |
| IL-6 (50 ng/μL) | 50 ng/mL | 1 μL |
| Flt-3L (50 ng/μL) | 50 ng/mL | 1 μL |
| Total | N/A | 1 mL |
-
14.
Gently remove the previous medium and replace it with the Stage 3 medium (4 mL per well of the plate).
-
15.
Incubate the cells in a humidified incubator (37°C, 5% CO2).
-
16.
Change the Stage 3 medium every 3 days until floating HPCs are no longer present.
CRITICAL: From day 12 of differentiation, floating HPCs are generated. After each medium change, the floating HPCs can be collected in a conical tube and spun down in a centrifuge for macrophage differentiation (Refer to Stage 4 of the protocol).
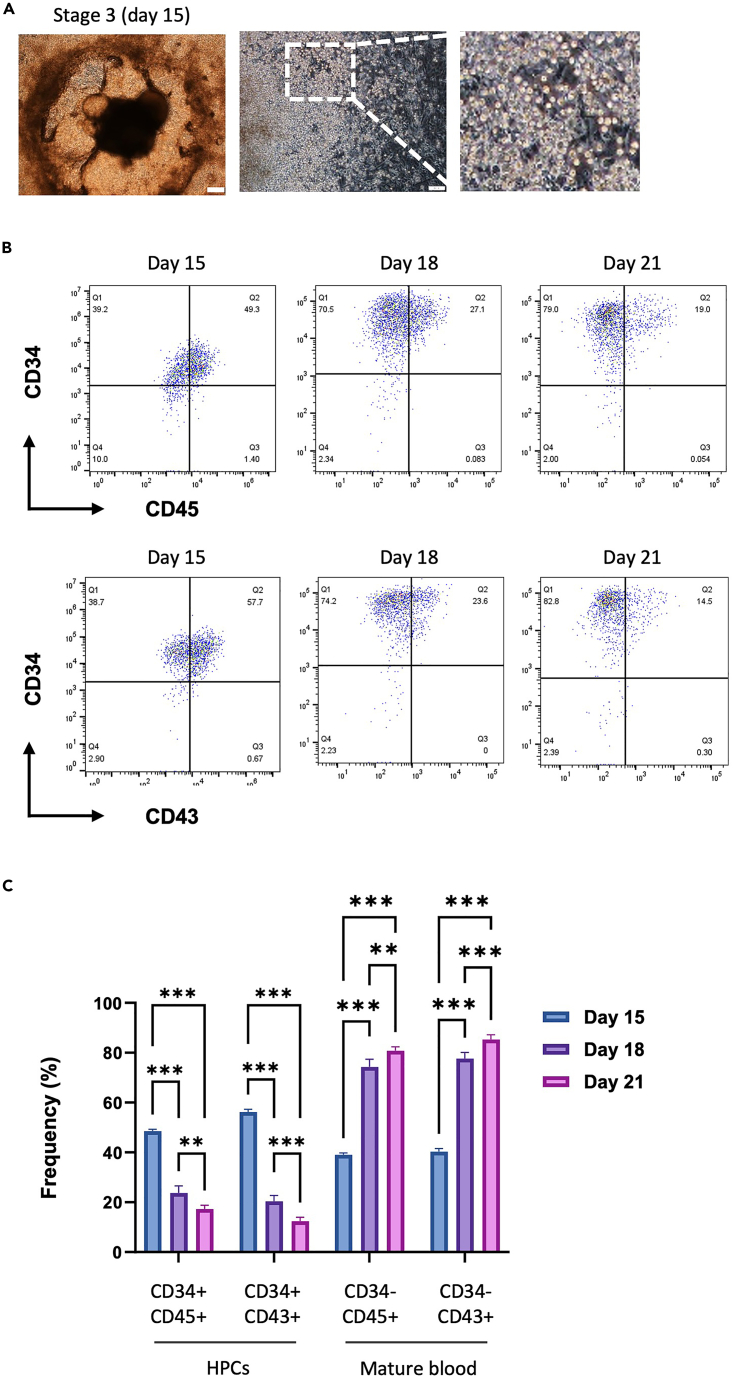
Note: As the commitment phase progresses, floating hematopoietic cells and vascular-like structures are observed on days 12–15, gradually increasing through day 21. There are variations depending on the cell lines, but floating HPCs can be produced in as few as 24 days and after as many as 30 days (Figure 4A).
Note: HPCs from days 12–18 of differentiation express the phenotype of primitive hematopoietic progenitors (CD34+CD45+ or CD34+CD43+). After 18 days, HPCs gradually adopt the phenotype of mature blood cells (CD34-CD45+ or CD34-CD43+) (Figures 4B and 4C).
Figure 4.
Changes in the morphology and phenotype of HPCs during the hematopoietic commitment phase
(A) Representative bright-field images of colonies (upper) and floating HPCs (lower) during the hematopoietic commitment phase of hiPSCs. Scale bars are 200 μm (upper) and 100 μm (lower).
(B) Representative FACS dot plots of CD34, CD43, and CD45 expression in generated HPCs.
(C) The generated HPCs were analyzed by flow cytometry for the phenotypes of primitive hematopoietic progenitors (CD34+CD45+ or CD34+CD43+) and mature blood cells (CD34-CD45+ or CD34-CD43+) during days 15–21. The bars indicate the mean ± SD, ∗∗p < 0.01, ∗∗∗p < 0.001.
Macrophage differentiation, expansion, and characterization (stage 4)
Macrophage differentiation and expansion
Timing: days 1 up to 60 days, 30 min per day
In this step, floating HPCs are harvested and replated to induce them into functional macrophages. High-density seeding of HPCs and macrophage-colony stimulating factor (M-CSF) supplementation are crucial to achieving a high-quality yield of functional macrophages that have both plasticity and phagocytic abilities.
Note: Regardless of the day on which the previous step (stage 1–3) was completed, we define day 1 of Stage 4 as the day on which we harvested the HPCs and proceeded to differentiate them into macrophages.
-
17.
Days 1–60: Prepare aliquots of macrophage differentiation and expansion medium (MDEM) and prewarm them in a 37°C water bath.
Stage 4. Macrophage Differentiation and Expansion medium (MDEM)
| Reagent | Final concentration | Amount |
|---|---|---|
| Basal 2 Medium (RPMI 1640 Medium+10% FBS) | N/A | 999 μL |
| M-CSF (100 ng/μL) | 100 ng/mL | 1 μL |
| Total | N/A | 1 mL |
-
18.
Collect the floating HPCs from the previous stage (Hematopoietic progenitor formation, stage 3) in a 15-mL tube and centrifuge them at 600 g for 2 min.
-
19.
Gently remove the supernatant medium and resuspend the cells in MDEM.
CRITICAL: Adjust the resuspension medium volume as appropriate for seeding cells depending on the surface area of the vessels being used. In our hands, HPCs need to be seeded at high density (at least 8×104 cells/cm2) for successful macrophage differentiation and subsequent expansion (see Table 1).
-
20.
Seed at the appropriate cell density into the desired culture plates and incubate the cells in a humidified incubator (37°C, 5% CO2) (Table 2).
-
21.
Change the MDEM every 3 days.
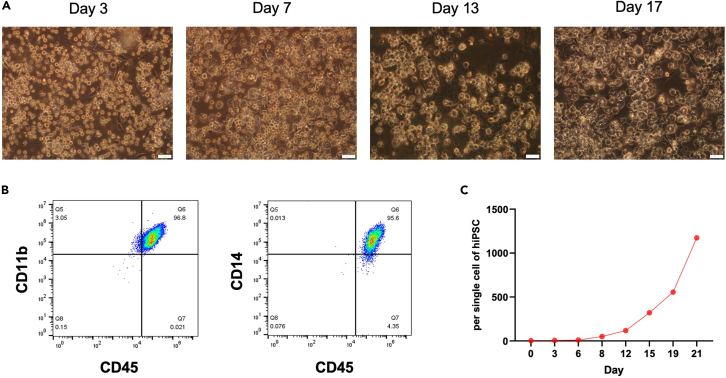
Note: For terminal differentiation of HPCs into macrophages, HPCs were cultured for 7–10 days in MDEM. Upon their maturation into macrophages, the small, round-shaped HPCs become larger and mostly remain roundish in shape, with no or little cytoplasmic extension (Figure 5A).
-
22.
Subculture differentiated macrophages when their confluency reaches 90%. After 2–3 subcultures, macrophages rapidly proliferate and can be expanded for up to 60 days. At every subculture step, cells should be counted and replated at high density (approximately 8×104 cells/cm2) for successful expansion.
Note: The first step in subculturing is to detach the cells from the surface of the plate or dish. The differentiating HPCs or macrophages can be easily lifted by gentle pipetting. If the cells do not detach easily, you can use a scraper to gently peel them from the plate.
Table 1.
Number of cells seeding per plate type
| Plates | Surface area (cm2) | Seeding density (×105) |
|---|---|---|
| 6-well | 9.6 | 7.7 |
| 12-well | 3.5 | 2.8 |
| 24-well | 1.9 | 1.5 |
| 96-well | 0.32 | 0.26 |
Table 2.
The volume of medium per plate type
| Plates | Volume of medium (mL) |
|---|---|
| 6-well | 2 |
| 12-well | 1 |
| 24-well | 0.5 |
| 96-well | 0.2 |
Figure 5.
Morphological changes in HPCs following macrophage differentiation
(A) Representative bright-field images of HPC from days 3–17 as they differentiate into macrophages. Scale bars are 50 μm.
(B) Macrophage phenotype markers (CD45+CD11b+, CD45+CD14+) were measured by flow cytometry, and data were analyzed using FlowJo software (Tree Star).
(C) The graph shows the number of hiPSC-derived macrophages over the three weeks. The proliferation rate per 1 well was calculated by counting the number of macrophage cells.
Macrophage characterization: Phenotype analysis
Timing: 3–4 h
At 10 days after differentiation, macrophages are distinguished by their phenotype (CD45+CD11b+, CD45+CD14+). The phenotypes of the differentiated macrophages according to their polarity are then analyzed by flow cytometry (Figure 5B).
-
23.
Collect the cells in a 15-mL tube and centrifuge them at 600 g for 2 min.
-
24.
Aspirate the supernatant and resuspend the cells in 1 mL of 1% fetal bovine serum in phosphate-buffered saline (FBS–PBS).
-
25.
Filter the cells through a sterile 70-μm cell strainer.
-
26.
Add single cells in 100 μL of 1% FBS–PBS to each fluorescence-activated cell sorting (FACS) tube.
-
27.
Incubate the cells with antibodies at 4°C for 60 min.
-
28.
Add 3 mL of 1% FBS–PBS and centrifuge the cells at 600 g for 2 min to wash out any unbound antibodies.
-
29.
Aspirate the supernatant and add 200 μL of 1% FBS–PBS and 2 μL of 7-actinomycin D (7AAD).
Note: 7AAD is a dye that distinguishes between living and dead cells. 7AAD staining will exclude false signals from dead cells, ensuring analysis of only real signals expressed by living cells.
-
30.
Analyze the cells by flow cytometry.
Macrophage characterization: Phagocytosis
Timing: 1–2 h
Phagocytosis is the primary function of macrophages. Perform a phagocytosis assay using green fluorescent protein (GFP)-labeled carboxylate-modified polystyrene latex beads followed by phenotypic marker staining to confirm macrophage phagocytosis.
-
31.
Dilute the GFP-labeled carboxylate-modified polystyrene latex beads 10-fold in PBS or MDM.
-
32.
For a cell count of 1×106/mL, add 200 μL of the diluted beads to a cell culture dish.
-
33.
Incubate the cells for 30 min in a humidified incubator (37°C, 5% CO2).
-
34.
Collect the cells in a 15-mL tube, add 5 mL of 1% FBS–PBS, and centrifuge the cells at 600 g for 2 min to wash out any unassociated beads.
-
35.
Analyze cells by flow cytometry.
Note: For negative controls of macrophage phagocytosis, either HPCs that do not phagocytize or the same undifferentiated iPSCs used for differentiation can be used. Additional methods for analyzing phagocytosis include using unexposed macrophages with latex beads as a negative control7 and treatment with the phagocytosis inhibitor cytochalasin D (CyD).8,9
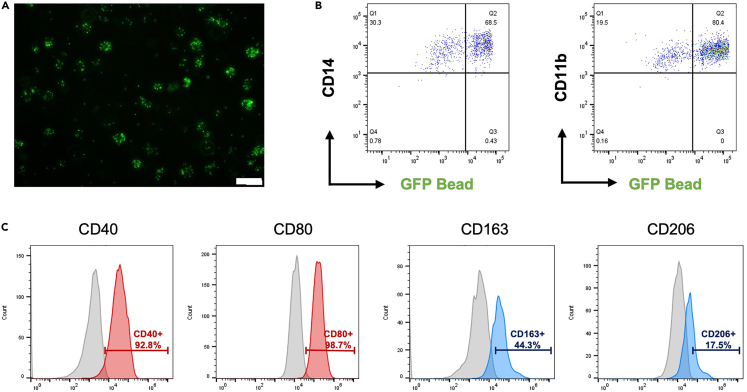
Note: The phagocytosis assay requires double staining with antibodies that can distinguish phenotypes after bead uptake to obtain accurate results (Figures 6A and 6B) (Refer to the macrophage characterization: phenotype analysis section).
Figure 6.
Analysis of the phagocytosis activity and polarization of macrophages
(A) Representative fluorescence image of the macrophage uptake of GFP-labeled carboxylate-modified polystyrene latex beads. Scale bar is 50 μm.
(B) Macrophage phagocytosis was analyzed by flow cytometry using GFP-labeled carboxylate-modified polystyrene latex beads. Frequencies for macrophage markers (CD14 and CD11b) and GFP-labeled carboxylate-modified polystyrene latex beads were measured by flow cytometry.
(C) The phenotypes of M1 (CD40, CD80) and M2 (CD163, CD206) polarized macrophages were measured by flow cytometry. The data were analyzed using FlowJo software (Tree Star).
Macrophage characterization: Polarization
Timing: day 2
In general, macrophage polarization is induced by internal and external stimuli, and cells of each polarity are functionally heterogeneous. Macrophage polarities are divided into two types, M1 and M2, which are separated by different stimulus signals.
CRITICAL: Macrophages are able to polarize after 10 days of differentiation. Importantly, the cytokines used during polarization induction have different reactivities depending on the cell density.10,11,12 Polarization induction of macrophages is typically performed at a density of 1×106 cells/well in a 6-well plate.
-
36.
Prepare aliquots of macrophage polarization medium (M1 or M2) and prewarm them in a 37°C water bath.
Stage 4-1. Macrophage polarization medium (M1)
| Reagent | Final concentration | Amount |
|---|---|---|
| Basal 2 Medium (RPMI 1640 Medium+10% FBS) | N/A | 997 μL |
| M-CSF (100 ng/μL) | 100 ng/mL | 1 μL |
| LPS (1 mg/mL) | 100 ng/mL | 0.1 μL |
| IFN-γ (10 ng/μL) | 20 ng/mL | 2 μL |
| Total | N/A | 1 mL |
Stage 4-1. Macrophage polarization medium (M2)
| Reagent | Final concentration | Amount |
|---|---|---|
| Basal 2 Medium (RPMI 1640 Medium+10% FBS) | N/A | 995 μL |
| M-CSF (100 ng/μL) | 100 ng/mL | 1 μL |
| IL-4 (10 ng/μL) | 20 ng/mL | 2 μL |
| IL-13 (10 ng/μL) | 20 ng/mL | 2 μL |
| Total | N/A | 1 mL |
-
37.
Gently remove the previous medium and replace it with the macrophage polarization medium (M1 or M2) (2 mL per well of the plate).
-
38.
Incubate the cells for 48 h in a humidified incubator (37°C, 5% CO2).
Note: Macrophage phenotypes are different depending on their polarity (M0 macrophages serve as a negative control for polarized macrophages). M1 macrophages express the surface markers CD40 and CD80, and M2 macrophages express CD163 and CD206 (Figure 6C).
Expected outcomes
This protocol provides an efficient method to generate macrophages via an optimized hematopoietic differentiation process. We employed a robust serum-, feeder-, and xeno-free clinical-grade protocol for the stepwise induction of hematopoietic cells from hiPSCs for therapeutic purposes. In previous studies, we have shown that we can reliably induce the differentiation of various hiPSC lines into hematopoietic cells.1
In our protocol, we have established not only the production of macrophages but also assays to determine their phenotypes and evaluate the functional properties of the generated macrophages, allowing for consistent, high-quality differentiation. From approximately day 40 of the protocol (10 days after Stage 4 differentiation and culture), HPCs exhibit the phenotype of macrophages (CD11b, CD14) with mature blood characteristics (CD45) (Figure 5B). These properties are expressed as long as the macrophages proliferate and are maintained. Long-term cultivation of hiPSC-derived macrophages in the presence of M-CSF resulted in continuous expansion of these cells for more than 3 weeks. The cell yield in our protocol is approximately 1000 cells per single cell of hiPSCs (Figure 5C). Based on our experience, we have observed that macrophages can proliferate in media containing M-CSF for up to 8 weeks or more.
Phagocytosis is the most important functional aspect of macrophages. The macrophages that we can produce using this protocol are superior in terms of function. When establishing this protocol, using GFP-labeled carboxylate-modified polystyrene latex beads, we demonstrated that approximately 80% of macrophages were capable of phagocytosis (Figures 6A and 6B).
Macrophages become polarized in various diseases. Polarized macrophages differ in not only phenotype but also signaling pathways and intracellular functional properties. These attributes are divided into two groups, represented by the classical activation (M1) and alternative activation (M2) pathways.13 Macrophage polarity is controlled by varying the cytokines with which the generated macrophages are treated. The induced M1 macrophages upregulate surface markers such as CD40 and CD80, and the M2 macrophages upregulate CD163 and CD206 (Figure 6C).
Regulation of macrophage polarization is key to the prevention and treatment of various diseases and is a critical factor in infectious, tumor, inflammatory, and fibrotic diseases.14 Macrophages are a crucial immune component for improving the understanding of the pathological mechanisms of organoid systems (lung, intestine, brain, liver and blood vessel) in vitro. We previously published a study on the generation of macrophage-containing alveolar organoids from hiPSCs, which was successfully demonstrated for disease modeling and drug efficacy testing.2 Currently, we are further investigating the therapeutic efficacy and mechanisms of macrophages by modulating macrophage polarity for applications in disease models both in vitro and in vivo. We also expect that the generation of various forms of multicellular organoids, including these functional macrophages, will contribute to disease modeling and drug discovery endeavors.
Limitations
We aimed for homogeneous differentiation efficiency regardless of the hiPSC line. On average, this protocol can yield 40–60% HPCs at day 15 of differentiation. Nevertheless, the genetic background and source of the donor of the hiPSCs may increase or decrease the yield of differentiating HPCs. If differentiation into a particular cell line continues to fail, the yield of HPCs should be compared with those of different cell lines. In our experience, most cell lines differentiate with high efficiency based on the supplied protocols.
In addition, traditional mechanical passaging is subject to a large number of parameters compared to enzymatic passaging. Different experimenters have different levels of proficiency with mechanical passaging, which can result in differences in the size and number of hiPSC colonies seeded upon differentiation. For consistent differentiation rates, enzyme passaging or passage culture of equal amounts of single cells is efficient. Hematopoietic cell differentiation in single-cell cultures has been studied mostly via embryoid body (EB) formation; direct hematopoietic cell differentiation in single-cell cultures has only recently been reported.7,15 Therefore, research and further experiments on the direct differentiation of hematopoietic stem cells using more advanced single-cell cultures of equal cell numbers are needed to achieve consistent and efficient differentiation rates.
Troubleshooting
Problem 1
The number of colonies that are seeded before the start of the differentiation induction is too high (hiPSC preparation stage).
Potential solution
-
•
Control the seed number of colonies by manually removing colonies under a dissecting microscope using a glass Pasteur pipette, hypodermic syringe, or 10- to 20-μL pipette tip, as described in the mechanical passaging method.
Problem 2
Cell lines that have previously differentiated without any problems fail to differentiate.
Potential solution
-
•
The quality of the differentiation medium (STEMdiff APEL medium) is important, as is the quality of the cytokine stocks employed. Aliquots of differentiation media and cytokine stocks are stored at −80°C and thawed at 4°C prior to use. It is recommended that differentiation media and cytokine stocks be used in small, single-use batches whenever feasible. Avoid their frequent freeze/thaw cycles. Differentiation media and cytokines stored at 4°C for a long time after they are opened become less active, which will decrease the differentiation potential of the cells. It is recommended to store differentiation media and cytokines at 4°C for up to 1 week prior to use.
Problem 3
When collecting floating HPCs, a large amount of differentiated debris is present (Hematopoietic progenitor formation stage).
Potential solution
-
•
For the collection of floating HPCs, we use a 40-μm cell strainer to first filter out larger differentiated debris.
-
•
If a lot of debris is still present, wash the cells 2–3 times at low G-force (300–400 g).
-
•
Furthermore, CD34+ hematopoietic progenitors isolated using magnetic-activated cell sorting (MACS) with CD34 microbeads can be used to induce macrophage differentiation from uniform population of HPCs.
Problem 4
Macrophages do not detach from the bottoms of the plate wells when pipetting during subculture (macrophage differentiation and expansion stage).
Potential solution
-
•
If the cells do not detach easily, a scraper can be used to gently peel them from the plate. Macrophages in their differentiated naïve state are not adherent cells and therefore do not dissociate with enzymes such as trypsin or TrypLE.
Problem 5
Macrophages proliferate slowly, or the cells clump together to form spheres (macrophage differentiation and expansion stage)
Potential solution
-
•
Cell density in culture is of paramount importance for macrophages. If macrophages are proliferating more slowly than before, it is likely because the cells are less dense. In addition, if the cells clump together to form spheres, it is likely because the density of the cells in the culture is too high. They should be maintained at the appropriate cell density suggested in the protocol to ensure that they can be maintained and proliferated as single macrophages.
Problem 6
It is difficult to observe macrophage phagocytosis with the GFP-labeled beads (macrophage characterization: phagocytosis)
Potential solution
-
•
Most of the hiPSC-derived macrophages will have completed the phagocytosis of the beads within 1–2 h. In our protocol, we set the bead uptake time to 30 min. Due to differences in active macrophage uptake time depending on the experimental environment, it is recommended that an assay time point be set within 30 min to 2 h. In FACS analysis, phagocytosis still takes place during antibody staining; therefore, antibody staining should be performed at 4°C to delay any phagocytic activity.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Seok-Ho Hong (shhong@kangwon.ac.kr).
Technical contact
Questions about the technical specifics of performing the protocol should be directed to and will be answered by the technical contact, Suji Jeong (suji1566@naver.com).
Materials availability
This study did not generate new unique reagents.
Data and code availability
This study did not generate datasets or codes. Raw data for qPCR analysis is available from the corresponding authors on request.
Acknowledgments
This research was supported by a grant from the Korean Fund for Regenerative Medicine (KFRM) funded by the Korean government (the Ministry of Science and ICT and the Ministry of Health and Welfare) (22A0304L1-01) as well as the Bio & Medical Technology Development Program and Basic Science Research Program of the National Research Foundation (NRF) funded by the Korean government (MSIT) (2022M3A9E4016936) and Ministry of Education (2022R1A6A3A130637).
Author contributions
Conceptualization, S.J. and S.-H.H.; methodology, S.J.; writing – original draft, S.J. and S.-H.H.; writing – review and editing, S.-H.H.; funding acquisition, S.J. and S.-H.H.; supervision, S.-H.H.
Declaration of interests
The mesodermal induction with high dose of BMP4 and subsequent hematopoietic and macrophage differentiation are protected by published patents (KOR/10-2106710), and these intellectual property rights belong to KW-Bio Co., Ltd.
References
- 1.Jeong S., An B., Kim J.H., Han H.W., Kim J.H., Heo H.R., Ha K.S., Han E.T., Park W.S., Hong S.H. BMP4 and perivascular cells promote hematopoietic differentiation of human pluripotent stem cells in a differentiation stage-specific manner. Exp. Mol. Med. 2020;52:56–65. doi: 10.1038/s12276-019-0357-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Heo H.R., Hong S.H. Generation of macrophage containing alveolar organoids derived from human pluripotent stem cells for pulmonary fibrosis modeling and drug efficacy testing. Cell Biosci. 2021;11:216. doi: 10.1186/s13578-021-00721-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heo H.R., Song H., Kim H.R., Lee J.E., Chung Y.G., Kim W.J., Yang S.R., Kim K.S., Chun T., Lee D.R., Hong S.H. Reprogramming mechanisms influence the maturation of hematopoietic progenitors from human pluripotent stem cells. Cell Death Dis. 2018;9:1090. doi: 10.1038/s41419-018-1124-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim J.H., Jo H.Y., Ha H.Y., Kim Y.O. Korea National Stem Cell Bank. Stem Cell Res. 2021;53 doi: 10.1016/j.scr.2021.102270. [DOI] [PubMed] [Google Scholar]
- 5.Lim J.J., Kim H.J., Rhie B.H., Lee M.R., Choi M.J., Hong S.H., Kim K.S. Maintenance of hPSCs under Xeno-Free and Chemically Defined Culture Conditions. Int. J. Stem Cells. 2019;12:484–496. doi: 10.15283/ijsc19090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rim Y.A., Park N., Nam Y., Ham D.S., Kim J.W., Ha H.Y., Jung J.W., Jung S.M., Baek I.C., Kim S.Y., et al. Recent progress of national banking project on homozygous HLA-typed induced pluripotent stem cells in South Korea. J. Tissue Eng. Regen. Med. 2018;12:e1531–e1536. doi: 10.1002/term.2578. [DOI] [PubMed] [Google Scholar]
- 7.Chang Y., Syahirah R., Wang X., Jin G., Torregrosa-Allen S., Elzey B.D., Hummel S.N., Wang T., Li C., Lian X., et al. Engineering chimeric antigen receptor neutrophils from human pluripotent stem cells for targeted cancer immunotherapy. Cell Rep. 2022;40 doi: 10.1016/j.celrep.2022.111128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang R., Kobayashi Y., Kazumura K., Tsuchiya H., Morishita N., Inagawa H., Soma G.I. Development of an Evaluation Device for Phagocytic Activity of New Phagocytes Using Simple and pH-sensitive Particles that Do Not Require Pre-treatment. Anticancer Res. 2016;36:3613–3618. [PubMed] [Google Scholar]
- 9.Zhang R., Inagawa H., Takahashi M., Kawanishi H., Kazumura K., Tsuchiya H., Morishita N., Kobayashi Y., Masaki T., Kobara H., Soma G.I. Measurement of the Phagocytic Activity of Human Peripheral Blood Using a Highly Sensitive Fluorometric Detection Device Without Hemolysis. Anticancer Res. 2017;37:3897–3903. doi: 10.21873/anticanres.11771. [DOI] [PubMed] [Google Scholar]
- 10.Lee C.M., Hu J. Cell density during differentiation can alter the phenotype of bone marrow-derived macrophages. Cell Biosci. 2013;3:30. doi: 10.1186/2045-3701-3-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vaughan-Jackson A., Stodolak S., Ebrahimi K.H., Johnson E., Reardon P.K., Dupont M., Zhang S., McCullagh J.S.O., James W.S. Density dependent regulation of inflammatory responses in macrophages. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.895488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ruder A.V., Temmerman L., van Dommelen J.M.A., Nagenborg J., Lu C., Sluimer J.C., Goossens P., Biessen E.A.L. Culture density influences the functional phenotype of human macrophages. Front. Immunol. 2023;14 doi: 10.3389/fimmu.2023.1078591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao C., Mirando A.C., Sové R.J., Medeiros T.X., Annex B.H., Popel A.S. A mechanistic integrative computational model of macrophage polarization: Implications in human pathophysiology. PLoS Comput. Biol. 2019;15 doi: 10.1371/journal.pcbi.1007468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kadomoto S., Izumi K., Mizokami A. Macrophage Polarity and Disease Control. Int. J. Mol. Sci. 2021;23 doi: 10.3390/ijms23010144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jung J., Chang Y., Jin G., Deng Q., Lian X.L., Bao X. Chemically defined generation of human definitive hematopoietic stem and progenitor cells. STAR Protoc. 2023;4 doi: 10.1016/j.xpro.2022.101953. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This study did not generate datasets or codes. Raw data for qPCR analysis is available from the corresponding authors on request.