Abstract
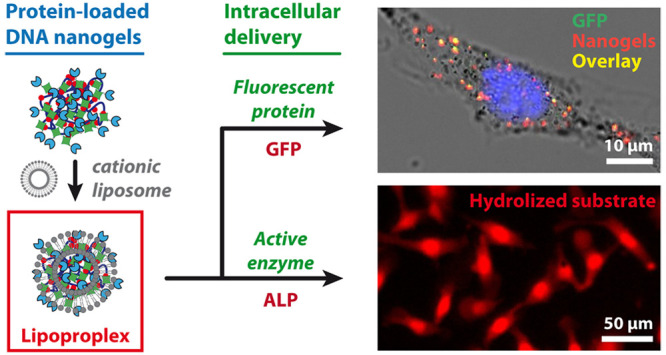
Using functional proteins for therapeutic purposes due to their high selectivity and/or catalytic properties can enable the control of various cellular processes; however, the transport of active proteins inside living cells remains a major challenge. In contrast, intracellular delivery of nucleic acids has become a routine method for a number of applications in gene therapy, genome editing, or immunization. Here we report a functionalizable platform constituting of DNA–protein nanogel carriers cross-linked through streptavidin–biotin or streptactin–biotin interactions and demonstrate its applicability for intracellular delivery of active proteins. We show that the nanogels can be loaded with proteins bearing either biotin, streptavidin, or strep-tag, and the resulting functionalized nanogels can be delivered into living cells after complexation with cationic lipid vectors. We use this approach for delivery of alkaline phosphatase enzyme, which is shown to keep its catalytic activity after internalization by mouse melanoma B16 cells, as demonstrated by the DDAO–phosphate assay. The resulting functionalized nanogels have dimensions on the order of 100 nm, contain around 100 enzyme molecules, and are shown to be transfectable at low lipid concentrations (charge ratio R± = 0.75). This ensures the low toxicity of our system, which in combination with high local enzyme concentration (∼100 μM) underlines potential interest of this nanoplatform for biomedical applications.
Introduction
Due to their highly specific biological activities and intrinsic biocompatibility, delivery of active proteins into cells represents great interest not only for fundamental research1 and intracellular sensing,2 but also for numerous therapeutic applications.3−5 However, despite their high potential, proteins suffer from chemical and thermal degradation, kidney clearance, and difficulty to penetrate through biological membranes.6 In particular, enzymes present a major limitation to be sensitive to both medium and the temperature, leading to the loss of their catalytic activity in inappropriate conditions thus limiting their applicability in both enzyme replacement therapy7,8 and enzyme prodrug therapy.9,10 Another aspect requiring consideration is the localization of proteins. Indeed, while most therapeutic proteins currently on the market have an extracellular localization, the intracellular transport of active proteins aiming cytosolic targets could strongly expand their exploitation for biomedical purposes.11,12 Yet the transport to the cytosol is still a major challenge, mainly due to physical impediment of the cell membrane and possible protein inactivation due to endocytosis.13−15
Various vectorization systems have thus been proposed for protein protection and intracellular delivery, such as PEGylation, conjugation with cell-penetrating (CPP) or specifically engineered peptides, and formulations with liposomes or polymeric nanoparticles.5,16−18 DNA represents another promising protein carrier in nanomedicine,19,20 due to its intrinsic biocompatibility, programmability and transfectability. In fact, nucleic acids can be readily delivered into cells by using various nonviral cationic transfection agents, among which, cationic lipids represent the most involved vectors in clinical studies.21 In this view, recently, DNA has been bound to β-galactosidase and glucose oxidase in order to both protect and deliver them into the cells using lipofectamine as a transfection agent.2,22 In this case, individual enzyme molecules were covalently conjugated to several single-stranded DNA oligonucleotides. In another example, three luciferase molecules inside tubular DNA origami nanostructures were delivered into the cells via polyethylenimine transfection reagent.23 A modular protein delivery system was recently proposed by Ryu et al., where 3 different proteins could be bound to 3 extremities of Y-shaped DNA nanostructure via DNA-binding zinc-finger protein tags.24 This system was successfully applied for cytosolic delivery of therapeutic endogenous tumor suppressor PTEN (phosphatase and tensin homolog).
In order to expand DNA-based protein delivery to strong local concentrations of delivered protein, in this study we propose to incorporate a large number of active protein molecules into a three-dimensional DNA nanogel network. We have recently reported a new type of DNA nanogels where the intramolecular cross-linking was achieved not by hybridization or covalent DNA cross-linking, but by noncovalent specific interactions between a multibiotinylated DNA (bt-DNA) and streptavidin.25,26 Here, we show different strategies for functionalization of these nanogels with two model cargo proteins: green fluorescent protein (GFP), and alkaline phosphatase (ALP), which has been proposed for replacement therapy against the hypophosphatasia disorder.27 We then investigate the delivery of the resulting functionalized nanogels, containing up to 100 μM local protein concentration, into cells by using lipidic nucleic acid transfection agents, known for promoting endosomal escape after internalization of lipoplexes by disrupting the membrane integrity.28
Results
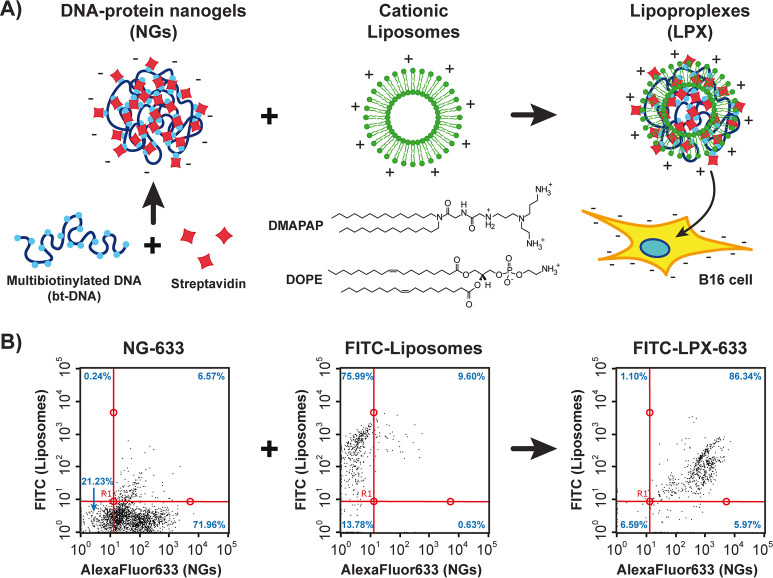
Formation of Complexes between DNA–Protein Nanogels and Cationic Vesicles: Lipoproplexes
Experimental procedure for preparation, vectorization and delivery of DNA-streptavidin nanogels (NGs) is schematically shown in the Figure 1A. The scaffold of the NGs, a multibiotinylated 3480 base pairs (bp) DNA (bt-DNA), was prepared by PCR as previously described.26 Cross-linking of the resulting bt-DNA was performed by adding streptavidin labeled with AlexaFluor633 fluorophore to give fluorescent NGs (NG-633) enabling their tracking and analysis by fluorescence. The resulting NGs had an apparent diameter of 114 ± 36 nm, as measured by atomic force microscopy (AFM) (Figure S1A). For vectorization of the NGs, cationic liposomes constituted of equimolar amounts of zwitterionic lipid DOPE (1,2-dioleoyl-sn-glycero-3-phosphoethanolamine) and the cationic lipid DMAPAP (2-{3-[bis(3-amino-propyl)-amino]-propylamino}-N-ditetradecyl carbamoyl methyl-acetamide) were used.29 In analogy with the formation of DNA-lipid complexes (lipoplexes), in our case the complexation of DNA–protein nanogel with lipids would give complexes containing DNA, lipids, and proteins, which we named lipoproplexes (LPX). We thus investigated the interaction between NG-633 and DMAPAP/DOPE liposomes containing 5% DOPE labeled with fluorescein (FITC). First, the colloidal stability was assessed for the LPX-633 obtained at different charge ratios (R±) by fluorescence microscopy (Figure S2). We considered the charge of the nanogels to be dependent only on the DNA component, as streptavidin was shown not to affect the zeta-potential of bt-DNA upon its folding into nanogels,26 and the charge concentration of the liposomes was calculated by considering the DMAPAP to have 3 positive groups at physiological pH. It was found that aggregation of the LPX occurred at R± higher than 1.5. We thus fixed the ratio R± = 1.5 and analyzed the resulting LPX by flow cytometry, a method which enables codetection of different fluorophores within individual particles of nanometric size.30 The resulting dot-plots for NGs-633 only, FITC-labeled liposomes only, and FITC-LPX-633 are given in the Figure 1B. For NGs and liposomes, the dot plots demonstrate a higher fluorescence signal for AlexaFluor633 (71.96%) and FITC (75.99%) signals, respectively. However, when NGs and liposomes were mixed together, a large population of particles demonstrating the signal from both fluorophores (86.34%) was detected, indicating successful complexation. The observation that the mean FITC fluorescence in the LPX decreased, while the mean AlexaFluor633 fluorescence increased can be explained either by FRET energy transfer between the fluorophores, or by change of their fluorescent properties in lipidic microenvironment,31,32 both further confirming binding between DNA–protein nanogels and cationic lipids leading to the efficient formation of the lipoproplexes.
Figure 1.
Preparation and characterization of lipoproplexes. (A) Scheme of lipoproplex preparation by electrostatic binding of DNA–protein nanogels with cationic liposomes and their consequent delivery into B16 cells. (B) Flow cytometry analysis of DNA nanogels labeled with AlexaFluor633 (NG-633, left), liposomes labeled with FITC (middle), and the resulting lipoproplex complex of the fluorescent nanogels with fluorescent liposomes (FITC-LPX-633, right). The dot-plots represent the fluorescence intensity of the liposomes (FITC: λEx = 488 nm; λEm = 525/30 nm) versus the fluorescence intensity of the NGs (AlexaFluor633, λEx = 640 nm; λEm = 661/19 nm). Red lines separate the dot plots into 4 quadrants, and blue numbers show the percentage of detected events within each quadrant. [NG-633] = 2 μM (in DNA charge); [liposomes] = 3 μM (in DMAPAP charge). All measurements are performed in 10 mM phosphate buffer pH 7.4, [NaCl] = 75 mM.
Delivery of Lipoproplexes into Cells
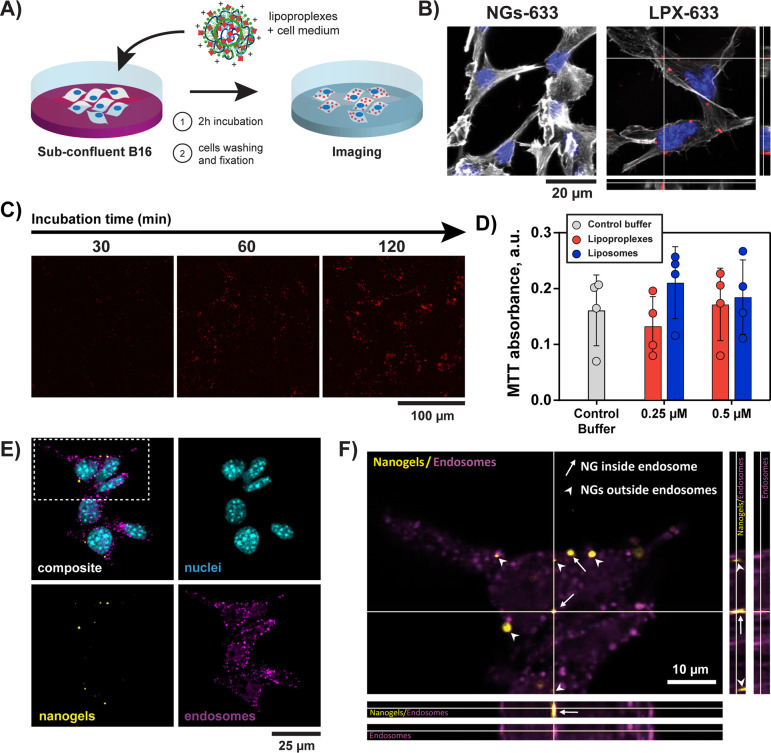
The possibility of delivery of the NGs-633 into living cells by their vectorization with cationic liposomes was then investigated with B16 mouse melanoma cells. The subconfluent cells were incubated with LPX-633 (or control solution containing NGs-633 in absence of liposomes) in the presence of DMEM medium without serum for 2 h (Figure 2A), then fixed and stained for subsequent microscopy observations. No NGs were observed within the cells after treating them with NGs in the absence of liposomes (Figure 2B, left). In contrast, when the cells were treated with LPX-633, fluorescent signal of AlexaFluor633 was clearly observed as fluorescent spots within the cells. By performing confocal microscopy acquisition at different Z-planes, the presence of the NGs-633 in the same focal planes as cytoskeleton and nucleus was confirmed thus proving that the NGs could be internalized by the cells (Figure 2B, right). Liposomes were thus necessary to deliver the nanogels into the cells. These observations were also confirmed by wide field fluorescence microscopy observations (Figure S3), where all tested lipoproplexes demonstrated corresponding fluorescence inside B16 cells, regardless of the fluorescent labeling. In contrast, only poor fluorescent signal was detected in B16 cells in the control samples where cells were treated with NG-633 in the absence of liposomes or with AlexaFluor633-streptavidin protein in the presence of liposomes. Contrary to several previous reports on intracellular delivery of DNA-based nanomaterials,33−35 we did not detect any internalization of our nanogels in cells in absence of the transfection agent. This difference in internalization could be due to a difference in rigidity between the highly structured tetrahedral DNA nanostructures having edges and corners34 and our soft nanogels, as it has been demonstrated that the penetration through the membrane depends on the rigidity of the particle.36 On the other hand, it has already been shown that the use of a transfection agent could increase the internalization of a DNA-based nanocages in cells.35 Moreover, we propose that this agent can allow the nanogel to escape degradation in lysosomes. Indeed, Liang et al. showed that bare DNA tetrahedron, after endocytosis, ends up trapped in lysosomes,34 whereas transfection agents, such as cationic lipids, are known to allow the exit of the endosomes.28
Figure 2.
Internalization of DNA–protein nanogels mediated by cationic liposomes. (A) Scheme of experimental procedure for delivery of lipoproplexes into cells. (B) Confocal microscopy observation of B16 cells treated with AlexaFluor633-labeled nanogels in the absence (NG-633) and in the presence (LPX-633) of cationic lipids after 2h of incubation. Gray color corresponds to actin labeled with phalloidin (λEx= 488 nm and λEm = 495–550 nm), blue to the nucleus stained with DAPI (λEx= 405 nm and λEm = 415–450 nm), and red to nanogels containing AlexaFluor633-labeled streptavidin (λEx= 638 nm and λEm = 645–700 nm). Orthogonal views are given for the cells treated with LPX-633 (z-depth = 3 μm). C) Widefield fluorescence microscopy images of B16 cells incubated with LPX-633 during different times (λEx= 578 ± 11 nm and λEm = 641 ± 38 nm). D) MTT cell viability assay. Formazan absorbance for B16 cells after 2 h of incubation with different concentrations of lipoproplexes or liposomes. Circles show data points, and bars show the mean values ± sd. Four independent tests, statistic Kruskal–Wallis multiple comparison test: n.s. (E, F) Confocal microscopy analysis of B16 cell with EEA1 immunolabeling of endosomes after LPX-555 delivery. (E) Composite image and the separate channels of nanogels labeled with AlexaFluor555 (nanogels, colored in yellow λEx = 555 nm and λEm = 645–700 nm), EEA1 immunolabeling (endosomes, colored in magenta, λEx= 638 nm and λEm = 645–700 nm) and DAPI staining (nuclei, colored in blue, λEx= 405 nm and λEm = 415–450 nm). (F) Zoom on the selected region from (E) and orthogonal views along the white lines (z-depth = 2.29 μm). Arrows: nanogels colocalized with endosomes, arrowheads: nanogel not colocalized/close to endosomes.
The kinetics of lipoproplex internalization by the cells was then analyzed. Lipoproplex samples (or controls corresponding to NGs in absence of liposomes) were incubated with cells for 30, 60, or 120 min, before cell fixation and widefield fluorescence imaging (Figures 2C and Figure S4). We observed that nanogel internalization progressively increased upon incubation and reached a maximum after 2 h of incubation (Figure S5). The toxicity of the lipoproplexes after internalization in B16 cells was then assessed with MTT assay (Figure 2D).37 In this test, formazan absorbance at 570 nm is proportional to the number of viable cells, and no significant difference was observed between lipoproplexes, cationic liposomes, and the control sample (buffer alone), indicating no toxicity for the B16 cells (n = 4, Kruskall-Wallis multiple comparison test). Moreover, the detection by confocal microscopy of the nanogels inside the cells 26 h after their internalization (Figure S6), further confirmed their biocompatibility and stability in the cellular microenvironment.
We then investigated the fate of the nanogels after their internalization by the cells. First, we studied colocalization of FITC-labeled lipids with NG-633 at different times after their internalization by the cells. We found that 30 min after delivery, fluorescent signal corresponding to FITC-lipid could be colocalized with the NG-633 fluorescent signal, suggesting that NG-633 and FITC-lipid were still associated. After longer period (5 and 26 h post incubation with FITC-LPX-633), only fluorescent signal corresponding to NG-633 was observed inside cells (Figure S6). This suggests the dissociation of lipids from nanogels followed by the loss of fluorescent signal of FITC-liposome probably due to “dilution” of lipids in the cell lipidic environment following a probable lipid exchange occurring during FITC-LPX-633 cell internalization. Indeed, during internalization of nucleic acid vectorized by cationic liposomes, it is known that a lipid exchange occurs with endosomal membrane inducing disruption of endosomal membrane and delivery of nucleic acid in cytoplasm.28 In order to provide evidence of endosomal escape of our DNA–protein NGs inside cells, we performed immunolabeling of endosomes and lysosomes after the delivery of lipoproplexes. For this purpose, anti-EEA1 and anti-LAMP1 antibodies were used for the immunostaining of early endosomes and lysosomes, respectively. After 2 h of incubation of cells with fluorescent LPX (in this experiment, we used AlexaFluor555 streptavidin for nanogel formation, resulting in LPX-555 to allow multichannel observation of nanogels, cell nuclei and immunolabeling), we observed only rare events of colocalization of NG with EEA1 (Figure 2E and F). Interestingly, in most cases, the nanogels were found close to endosomal compartments. We thus hypothesize that this observation was performed after the endosomal escape. Concerning lysosomes, we observed no colocalization of NG with LAMP1 labeled compartments (Figure S7). The endosomal-lysosomal system is a dynamic process where intracellular membranous compartment interconvert from early endosomes, that maturate to late endosomes and then fused into lysosomes.38 As the lysosome appears to be the “last” step of this pathway, we can assume that if NGs are rarely observed inside early endosomes, it is expected that NGs inside lysosome may be more rare or even impossible to observe.
Intracellular Delivery of GFP-Loaded Lipoproplexes
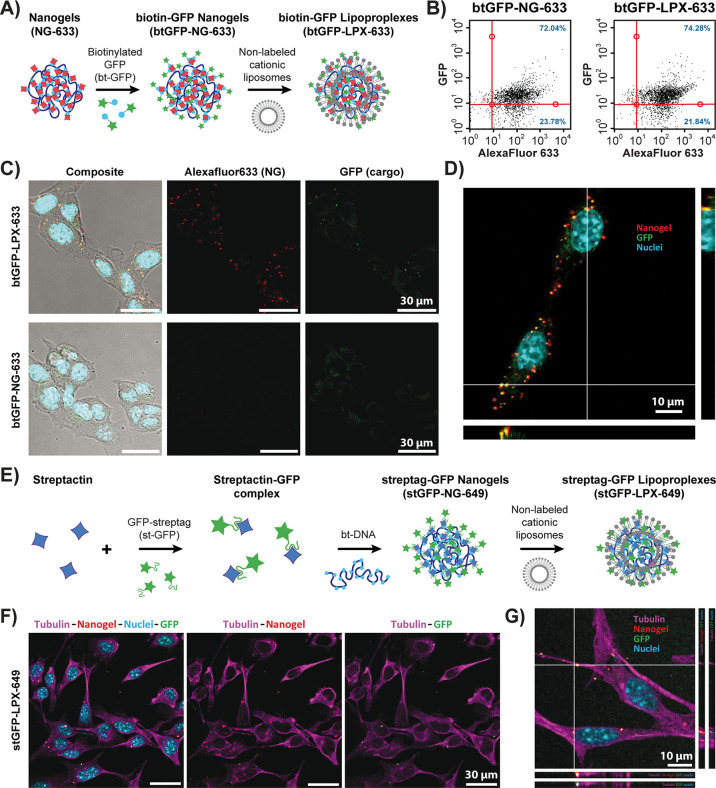
We then explored the possibility of loading biotinylated cargo proteins into already formed NGs. Biotinylated green fluorescent protein (btGFP) was used as a model protein due to its fluorescent properties. The preparation of btGFP-loaded nanogels (btGFP-NGs) was conducted in two steps: (i) DNA–protein nanogels were first formed with AlexaFluor633-functionalized streptavidin in the conditions described above, and (ii) btGFP was then added and bound to remaining available biotin-binding streptavidin sites on the nanogels (Figure 3A). By AFM, the size of the resulting GFP-functionalized nanogels was measured to be 130 ± 51 nm (Figure S1B). The NGs were then characterized by flow cytometry (Figures 3B and Figure S8A). Colocalization of the btGFP with AlexaFluor633 fluorescence in the NGs demonstrates successful formation of GFP-functionalized NGs (btGFP-NGs-633), as 72.04% of fluorescent events were observed in the upper-right quadrant of the dot plot. The NGs were then complexed with nonfluorescent cationic liposomes in order to form GFP-functionalized lipoproplexes, btGFP-LPX-633. Flow cytometry (Figure 3B right and Figure S8B) demonstrated that the formation of the lipoproplexes did not interfere with GFP functionality, since the signal of functional GFP protein was still codetected with AlexaFluor633 from the nanogels after adding the liposomes, and the percentage of colocalized fluorescence events in the upper-right quadrant of the dot plot stayed quite unaffected (74.28%).
Figure 3.
Delivery of lipoproplexes functionalized with GFP. (A) Scheme of postfunctionalization of NG-633 with biotinylated GFP (btGFP) and preparation of the corresponding lipoproplexes (btGFP-LPX-633). (B) Flow cytometry dot plots for btGFP-NG-633 (left) and btGFP-LPX-633 (right). The dot plots represent intensity of GFP fluorescence (λEx = 488 nm; λEm = 525/30 nm) versus the fluorescence intensity of the NG-633 (AlexaFluor633, λEx = 640 nm; λEm = 661/19 nm). Red lines separate the dot plots into 4 quadrants. Blue numbers give the percentage of observed events in the upper right and lower right quadrants. (C) Transmission and confocal microscopy images of B16 cells incubated either with btGFP-NG-633 (bottom) or with btGFP-LPX-633 (top). Cells are imaged in transmission light and overlaid with fluorescence signal from confocal observations. Green color corresponds to GFP cargo protein (λEx= 488 nm and λEm = 495–550 nm), cyan to the nucleus stained with DAPI (λEx= 405 nm and λEm = 415–450 nm), and red to nanogels containing AlexaFluor633-labeled streptavidin (λEx= 638 nm and λEm = 645–700 nm). (D) Higher magnification confocal microscopy observation of the sample from (C) with orthogonal views (z-depth = 5.79 μm). (E) Scheme of preparation of GFP-functionalized streptactin–DNA nanogels (stGFP-NG-649) based on streptag–streptactin binding and preparation of the corresponding lipoproplexes (stGFP-LPX-649). (F) Confocal microscopy observation of B16 cells incubated with stGFP-LPX-649. Green color corresponds to GFP cargo protein (λEx= 488 nm and λEm = 495–550 nm), cyan to the nucleus stained with DAPI (λEx= 405 nm and λEm = 415–450 nm), magenta to the immunolabeled tubulin with AlexaFluor555-labeled antibody (λEx = 552 nm and λEm = 584–613 nm), and red to nanogels containing DY649-labeled streptavidin (λEx = 638 nm and λEm = 645–700 nm). (G) Higher magnification confocal microscopy observation of the sample from (F) with orthogonal views (z-depth = 2.65 μm).
B16 cells were then incubated with the btGFP-LPX-633 and observed by confocal microscopy. Figure 3C shows the presence of fluorescent nanogels in the cells only when they have been complexed with the liposomes. Even though the fluorescence signal of the GFP was weaker, at least 76 ± 8% (for over 400 events) of the nanogels demonstrated colocalization of the AlexaFluor633 red fluorescent signal from streptavidin with the green fluorescence from the GFP (see example of composite image and orthogonal views in Figure 3D), in agreement with the flow cytometry analysis. We typically observed 7.7 ± 4.5 internalized particles per cell (for 140 cells) in a single confocal plane, and around 50 internalized nanogels per cell have been observed in one particular z-stack of 7 confocal planes shown in the Figure S9. Biotinylated GFP protein can thus be successfully delivered into the cells within DNA-streptavidin nanogels complexed with cationic liposome vector.
To extend the applicability of our method, we devised another functionalization approach by using GFP fused to the twin-streptag oligopeptide sequence (stGFP).39,40 Twin-streptag is a 28 amino acid tag having 2 binding sequences for the biotin-binding pocket of streptavidin, with a strong affinity (in the range of pM) for the engineered variant of streptavidin, streptactin. We first found that streptactin labeled with DY649 dye was also able to cross-link the bt-DNA, but at higher protein concentrations (above 300 nM) than with the streptavidin–biotin system (Figure S10A). To functionalize the nanogels, we first obtained a stoichiometric complex of stGFP with streptactin and then used it for cross-linking of bt-DNA (Figure 3E and Figure S10B). We found that even more stGFP–streptactin complex was necessary in this case to produce compact nanogels (around 1000 nM). We then formed the lipoproplexes with the stGFP-NG-649, incubated them with B16 cells, and imaged them by confocal microscopy (Figure 3F). Here again, fluorescent spots with colocalized fluorescence from streptactin (red) and GFP fluorescence (green) were found inside the cells when they were treated with stGFP-LPX-649 (Figure 3G). These results expand the functionalizability of DNA-streptavidin (or DNA-streptactin) nanogels and demonstrate that nanogel-based approach for intracellular protein delivery can also be applicable for cloned proteins bearing not biotin but specific oligopeptide tags.
Intracellular Delivery of Enzyme-Loaded Lipoproplexes
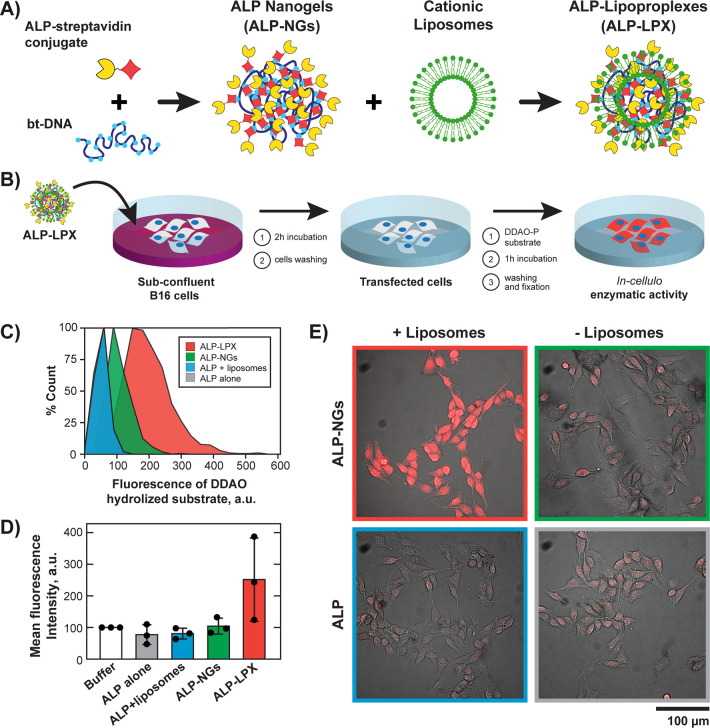
We finally investigated whether the lipoproplexes could be used as carriers for an active enzyme and if it would keep its catalytic activity after delivery into the cells. For this purpose, bt-DNA was directly cross-linked in one step with commercially available streptavidin conjugated to alkaline phosphatase (ALP) enzyme (Figure 4A)26 to give functionalized nanogels (ALP-NGs). The formation of ALP-NGs was characterized by AFM imaging (Figures S11), and the concentration of streptavidin-ALP conjugate of 15 nM (for 1 μM of bt-DNA in phosphate groups) was selected for further experiments, giving an apparent diameter of 133 ± 41 nm (Figure S1C). This ratio of streptavidin-ALP to bt-DNA corresponds to up to 100 enzymes per nanogel. The complexation of nonfluorescent ALP-NGs with FITC-labeled liposomes was then confirmed by flow cytometry (Figure S12). We observed that complexation with functionalized nanogels led to an increase of mean fluorescence signal distribution of liposomes from the charge ratio R± = 0.75 (i.e., 0.5 μM liposomes for 2 μM bt-DNA). Such a low R± ratio was thus sufficient for the formation of ALP-loaded lipoproplexes (ALP-LPX).
Figure 4.
Delivery of ALP-functionalized lipoproplexes and enzymatic activity assay in the cells. (A) Preparation of ALP-loaded lipoproplexes (ALP-LPX). (B) Schematic representation of the experimental procedure for ALP delivery and measurement of its activity inside the cells. (C) Flow cytometry distribution of hydrolyzed DDAO substrate inside B16 cells treated with ALP-LPX and in control samples (λEx = 640 nm; λEm = 661/19 nm). The distribution for ALP alone control is not visible, as it has practically the same shape as the one for ALP+liposomes control. (D) Normalized intracellular fluorescence intensities of hydrolyzed DDAO (mean ± s.d.) detected by fluorescence microscopy. White bar shows intrinsic ALP activity (normalized to 100%) for cells incubated with the phosphate buffer (in the absence of ALP). Black circles show the data points. (n = 3 independent experiments, Kruskal–Wallis Test and Dunn’s multiple comparison test, p = 0.0412 ALP-LPX vs ALP+liposomes and p = 0.0689 for ALP-LPX vs ALP alone). (E) Example of combined transmission and fluorescence microscopy images of hydrolyzed DDAO substrate after treatment of B16 cells with enzyme alone (ALP) or enzyme loaded into DNA nanogels (ALP-NGs) in the presence and absence of liposomes.
We then checked if the ALP enzyme kept its catalytic activity within the lipoproplexes in vitro. For the assay, 9H-(1,3-dichloro-9,9-dimethylacridin-2-one-7-yl) phosphate, diammonium salt (DDAO-P) fluorogenic substrate was used to follow the hydrolysis of its phosphate group by alkaline phosphatase via appearance of far-red fluorescence emission of the resulting DDAO. We found that incorporation of the ALP into the nanogels only slightly decreased the catalytic activity of the enzyme to 88% (Figure S13). Adding liposomes to the ALP alone did not affect its activity, whereas within ALP-loaded lipoproplexes, the enzyme conserved 79% of its initial catalytic activity, as compared to the activity of the native ALP-streptavidin conjugate. The ALP enzyme thus stayed available to the DDAO-P substrate and catalytically active even after loading into the DNA nanogels and their complexation with cationic lipids.
Finally, the delivery of ALP-functionalized lipoproplexes (ALP-LPX) into B16 cells was investigated (Figure 4B). The cells were treated with ALP-LPX for 2 h and washed to remove any remaining nanogels. The efficiency of delivery could not be assessed for the nonfluorescent NGs, instead we directly characterized the enzymatic activity of ALP inside the cells. For this purpose, DDAO-P substrate was added into the cell medium, and after 40 min of incubation the substrate was removed from the medium, cells were rinsed, and intracellular fluorescence of the hydrolyzed DDAO was analyzed either by flow cytometry after detaching (Trypsin-EDTA) and fixing the cells (Figure 4C), or by fluorescence microscopy after rinsing and fixing the cells onto culture plate (Figure 4D, E and Figures S14 and S15). We found that even in absence of added ALP, the B16 cells were able to slightly hydrolyze the DDAO-P substrate giving a basal level of alkaline phosphatase activity.41 Flow cytometry showed no effect of adding enzyme alone and enzyme in the presence of liposomes on the basal fluorescence of the cells. The distribution of fluorescence of hydrolyzed substrate slightly increased when cells were incubated with ALP-functionalized nanogels (ALP-NG) without liposomes, but the increase in fluorescence was much stronger when the cells were incubated with ALP-LPX (Figure 4E). Figure S15 shows fluorescence distributions for 3 independent experiments, and Figure 4D gives the average fluorescence value of the triplicate ALP delivery experiments normalized by the basal level of ALP activity inside B16 cells treated with phosphate buffer only. We systematically observed that fluorescence intensity corresponding to the hydrolyzed substrate inside the cells treated with ALP-LPX, was higher than for the controls containing phosphate buffer only, free enzyme (free ALP), ALP with liposomes or ALP-functionalized nanogels without liposomes, with significant difference for ALP-LPX vs ALP+liposomes and ALP-LPX vs ALP alone (Kruskal–Wallis Test and Dunn’s multiple comparison test). In an additional control, we compared ALP enzymatic activity inside cells incubated with functionalized and nonfunctionalized LPX. Figure S16 demonstrates that no enhancement of ALP enzymatic activity after delivery of the enzyme-free LPX was observed, indicating that the increased enzymatic activity in ALP-LPX was due to the catalytic activity of delivered enzyme and not a result of increasing the basal expression of alkaline phosphatase due to the interaction of lipoproplexes with cellular environment. All these results show that the ALP has been efficiently delivered inside the cells in its active state via functionalized lipoproplexes. To the best of our knowledge, it is a first example of efficient delivery of a large local number of active enzymes.
Discussion
We reported a new vector-carrier-cargo platform called lipoproplex, schematically represented in Figure S17, and demonstrated its ability to transport active proteins through the cell membrane. For this purpose, we used DNA nanogels cross-linked via streptavidin–biotin or streptactin-biotin interactions. This platform enables loading of the protein cargo into DNA carrier in several ways (i.e., using streptavidinated, biotinylated or strep-tagged cargo proteins) thus opening the possibility of large number of functionalities that can be potentially introduced into a single nanogel. We used cationic liposomes for the vectorization of DNA–protein NGs and named the resulting complex as “lipoproplex”, which evokes the complex between DNA and liposomes (i.e., lipoplex) but is intended to emphasize the importance of the protein component in our DNA–protein nanogels. Three-dimensional structure of the nanogels demands lower concentration of cationic liposomes for complexation (down to R± = 0.75), which in combination with intrinsic biocompatibility of DNA nanoscaffold results in nontoxic protein delivery system as demonstrated by the MTT assay. For a proof of concept, here we investigated delivery of two proteins: biotinylated (or strep-tagged) fluorescent protein GFP and the enzyme alkaline phosphatase conjugated to streptavidin. In both cases, the complexation with cationic liposomes and subsequent internalization by the cells did not affect the respective properties of the proteins: GFP retained its fluorescence emission, whereas the ALP kept its efficiency to hydrolyze the DDAO-P substrate. Both strategies thus proved to be adapted for the nanogels functionalization.
By considering that all streptavidin-ALP conjugates enter the nanogels and each NG consists of one DNA scaffold molecule, under our conditions (15 nM of strep-ALP for 1 μM of bt-DNA in phosphates) each nanogel can contain up to 100 loaded enzyme molecules. By assuming the nanogels to have a spherical morphology and 130 nm diameter, the local concentration of the loaded protein can be evaluated to around 130 μM. High number of potentially loadable proteins can be useful for delivery into cells of multifunctional nanogels containing a large number of different proteins, and the high local concentration of the loaded proteins is promising for biomedical applications. This approach can thus be of particular interest for codelivery of several active enzymes at controlled stoichiometry. Cotransfection of genes coding for different enzymes was suggested for cancer therapy42,43 and treatment of metabolic diseases.44 But it was shown in some cases to result in the expression of only one or the other enzyme rather than the desired dual expression of both enzymes,45 because of the competition for the use of intracellular resources available for gene expression.46 Direct delivery of NGs containing defined enzymatic composition avoids the coexpression step and can thus overcome this issue. Moreover, high local enzyme concentrations within the nanogels can be of particular interest for creating or restoring complex metabolic pathways requiring the close spatial positioning of several catalytic processes within an enzymatic cascade.
Conclusions
In conclusion, we present a new DNA–protein nanogel platform for the transport of active loaded proteins into living cells. The loaded proteins have been efficiently internalized by the cells while keeping their specific properties, which underlines potential therapeutic interest of lipoproplexes for delivery and codelivery into the cells of active enzymes. We anticipate that DNA–protein nanogels could be employed as nanoscaffold for the transport of other biological and chemical entities, not only by incorporating them into the nanogel by streptavidin/streptactin-biotin binding, but also by orthogonal binding into DNA double helix (i.e., intercalative drugs, such as doxorubicin).24 Moreover, the applicability of our systems to biotinylated, streptavidinated, or strep-tagged recombinant proteins, nanoparticles, aptameric sequences, or antibodies opens up the possibility of using nanogels as universal scaffolds with a high degree of functionalizability that not only could encapsulate a number of desired features in a single nanosized material but also could protect the incorporated entities from the biological microenvironment. All of these results underline the great potential of using DNA transfection agents for soft DNA nanomaterials delivery.
Acknowledgments
This work was supported by Agence Nationale de la Recherche (ANR) grants ANR-22-CE06-0009 and ANR-18-CE07-0001 (S.R.), ANR-18-CE18-0005-01 (C.H.), ANR-18-CE06-0019 and ANR-21-CE18-0051 (D.B.). We thank the Plateforme d’Imagerie Cellulaire et Moléculaire, US25 Inserm, UAR3612 CNRS, Université Paris Cité, Faculté de Pharmacie, Paris, France for access to confocal microscopy. We also thank Anne-Marie Lachagès from UTCBS for her technical assistance and, in particular, for preparation of cationic liposomes.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jacs.3c08000.
Materials and methods, AFM images and frequency size distribution for different nanogels (Figure S1); fluorescent microscopy analysis of lipoproplex aggregation (Figure S2); wide-field microscopy observation of B16 cells treated with different lipoproplexes and controls (Figure S3); kinetics of lipoproplexes internalization and its analysis (Figures S4–S5); confocal microscopy images of B16 cells at different times after incubation with FITC-LPX-633 (Figure S6); LAMP1 immunolabeling on B16 cells after LPX-633 incubation (Figure S7); flow cytometry analysis of GFP-functionalized nanogels and lipoproplexes (Figure S8); z-stack projection of confocal planes for B16 cells treated with GFP-functionalized lipoproplexes (Figure S9); AFM observation of DNA–streptactin nanogels and their functionalization with st-GFP (Figure S10), AFM observation of formation of nanogels functionalized with streptavidin-ALP conjugate (Figure S11); flow cytometry analysis of formation of ALP-functionalized lipoproplexes (Figure S12); effect of incorporation into nanogels and lipoproplexes on the acellular enzymatic activity of ALP (Figure S13); fluorescence microscopy detection of intracellular catalytic activity of ALP after intracellular delivery of ALP-LPX (Figure S14); box plot fluorescence distributions for 3 individual experiments of ALP-LPX delivery (Figure S15); comparison of the ALP enzymatic activity of B16 treated with ALP-functionalized and nonfunctionalized lipoproplexes (Figure S16); and schematic representation of lipoproplex-based proteins delivery platform (Figure S17) (PDF)
Author Present Address
‡ ClinSearch, 110 Avenue Pierre Brossolette, 92240 Malakoff, France
Author Contributions
⊥ S.R. and C.H. contributed equally to this paper.
The authors declare no competing financial interest.
Supplementary Material
References
- Kusmierz C. D.; Bujold K. E.; Callmann C. E.; Mirkin C. A. Defining the Design Parameters for in Vivo Enzyme Delivery Through Protein Spherical Nucleic Acids. ACS Cent. Sci. 2020, 6 (5), 815–822. 10.1021/acscentsci.0c00313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samanta D.; Ebrahimi S. B.; Kusmierz C. D.; Cheng H. F.; Mirkin C. A. Protein Spherical Nucleic Acids for Live-Cell Chemical Analysis. J. Am. Chem. Soc. 2020, 142 (31), 13350–13355. 10.1021/jacs.0c06866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kintzing J. R.; Filsinger Interrante M. V.; Cochran J. R. Emerging Strategies for Developing Next-Generation Protein Therapeutics for Cancer Treatment. Trends Pharmacol. Sci. 2016, 37 (12), 993–1008. 10.1016/j.tips.2016.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrov D. S. Therapeutic Proteins. Methods Mol. Biol. 2012, 899, 1–26. 10.1007/978-1-61779-921-1_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean S. N.; Turner K. B.; Medintz I. L.; Walper S. A. Targeting and Delivery of Therapeutic Enzymes. Ther. Delivery 2017, 8 (7), 577–595. 10.4155/tde-2017-0020. [DOI] [PubMed] [Google Scholar]
- Maximov V.; Reukov V.; Vertegel A. A. Targeted Delivery of Therapeutic Enzymes. J. Drug Delivery Sci. Technol. 2009, 19 (5), 311–320. 10.1016/S1773-2247(09)50066-4. [DOI] [Google Scholar]
- Desnick R. J.; Schuchman E. H. Enzyme Replacement Therapy for Lysosomal Diseases: Lessons from 20 Years of Experience and Remaining Challenges. Annu. Rev. Genomics Hum. Genet. 2012, 13 (1), 307–335. 10.1146/annurev-genom-090711-163739. [DOI] [PubMed] [Google Scholar]
- Concolino D.; Deodato F.; Parini R. Enzyme Replacement Therapy: Efficacy and Limitations. Ital. J. Pediatr. 2018, 44 (S2), 120. 10.1186/s13052-018-0562-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penet M.-F.; Chen Z.; Li C.; Winnard P. T.; Bhujwalla Z. M. Prodrug Enzymes and Their Applications in Image-Guided Therapy of Cancer: Tracking Prodrug Enzymes to Minimize Collateral Damage. Drug Delivery Transl. Res. 2012, 2 (1), 22–30. 10.1007/s13346-011-0052-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yata V. K.; Banerjee S.; Ghosh S. S. Folic Acid Conjugated-Bio Polymeric Nanocarriers: Synthesis, Characterization and In Vitro Delivery of Prodrug Converting Enzyme. Adv. Sci. Eng. Med. 2014, 6 (4), 388–392. 10.1166/asem.2014.1508. [DOI] [Google Scholar]
- Raman V.; Van Dessel N.; Hall C. L.; Wetherby V. E.; Whitney S. A.; Kolewe E. L.; Bloom S. M. K.; Sharma A.; Hardy J. A.; Bollen M.; Van Eynde A.; Forbes N. S. Intracellular Delivery of Protein Drugs with an Autonomously Lysing Bacterial System Reduces Tumor Growth and Metastases. Nat. Commun. 2021, 12 (1), 6116. 10.1038/s41467-021-26367-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y.; Tirrell M. V.; LaBelle J. L. Harnessing the Therapeutic Potential of Biomacromolecules through Intracellular Delivery of Nucleic Acids, Peptides, and Proteins. Adv. Healthc. Mater. 2022, 11 (12), 2102600. 10.1002/adhm.202102600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goswami R.; Jeon T.; Nagaraj H.; Zhai S.; Rotello V. M. Accessing Intracellular Targets through Nanocarrier-Mediated Cytosolic Protein Delivery. Trends Pharmacol. Sci. 2020, 41 (10), 743–754. 10.1016/j.tips.2020.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miersch S.; Sidhu S. S. Intracellular Targeting with Engineered Proteins. F1000Research 2016, 5, 1947. 10.12688/f1000research.8915.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu A.; Tang R.; Hardie J.; Farkas M. E.; Rotello V. M. Promises and Pitfalls of Intracellular Delivery of Proteins. Bioconjugate Chem. 2014, 25 (9), 1602–1608. 10.1021/bc500320j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisal D. S.; Kosloski M. P.; Balu-Iyer S. V. Delivery of Therapeutic Proteins. J. Pharm. Sci. 2010, 99 (6), 2557–2575. 10.1002/jps.22054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oba M.; Tanaka M. Intracellular Internalization Mechanism of Protein Transfection Reagents. Biol. Pharm. Bull. 2012, 35 (7), 1064–1068. 10.1248/bpb.b12-00001. [DOI] [PubMed] [Google Scholar]
- Dhankher A.; Lv W.; Studstill W. T.; Champion J. A. Coiled Coil Exposure and Histidine Tags Drive Function of an Intracellular Protein Drug Carrier. J. Controlled Release 2021, 339 (March), 248–258. 10.1016/j.jconrel.2021.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.; Champion J. A. Self-Assembling Nanocarriers from Engineered Proteins: Design, Functionalization, and Application for Drug Delivery. Adv. Drug Delivery Rev. 2022, 189, 114462. 10.1016/j.addr.2022.114462. [DOI] [PubMed] [Google Scholar]
- He L.; Mu J.; Gang O.; Chen X. Rationally Programming Nanomaterials with DNA for Biomedical Applications. Adv. Sci. 2021, 8 (8), 2003775. 10.1002/advs.202003775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendonça M. C. P.; Kont A.; Kowalski P. S.; O’Driscoll C. M. Design of Lipid-Based Nanoparticles for Delivery of Therapeutic Nucleic Acids. Drug Discovery Today 2023, 28 (3), 103505. 10.1016/j.drudis.2023.103505. [DOI] [PubMed] [Google Scholar]
- Brodin J. D.; Sprangers A. J.; McMillan J. R.; Mirkin C. A. DNA-Mediated Cellular Delivery of Functional Enzymes. J. Am. Chem. Soc. 2015, 137 (47), 14838–14841. 10.1021/jacs.5b09711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ora A.; Järvihaavisto E.; Zhang H.; Auvinen H.; Santos H. A.; Kostiainen M. A.; Linko V. Cellular Delivery of Enzyme-Loaded DNA Origami. Chem. Commun. 2016, 52 (98), 14161–14164. 10.1039/C6CC08197E. [DOI] [PubMed] [Google Scholar]
- Ryu Y.; Hong C. A.; Song Y.; Beak J.; Seo B. A.; Lee J.; Kim H.-S. Modular Protein-DNA Hybrid Nanostructures as a Drug Delivery Platform. Nanoscale 2020, 12 (8), 4975–4981. 10.1039/C9NR08519J. [DOI] [PubMed] [Google Scholar]
- Zhou L.; Morel M.; Rudiuk S.; Baigl D. Intramolecularly Protein-Crosslinked DNA Gels: New Biohybrid Nanomaterials with Controllable Size and Catalytic Activity. Small 2017, 13 (28), 1700706. 10.1002/smll.201700706. [DOI] [PubMed] [Google Scholar]
- Mariconti M.; Morel M.; Baigl D.; Rudiuk S. Enzymatically Active DNA-Protein Nanogels with Tunable Cross-Linking Density. Biomacromolecules 2021, 22 (8), 3431–3439. 10.1021/acs.biomac.1c00501. [DOI] [PubMed] [Google Scholar]
- Bowden S. A.; Foster B. L.. Alkaline Phosphatase Replacement Therapy for Hypophosphatasia in Development and Practice. In Therapeutic Enzymes: Function and Clinical Implications. Advances in Experimental Medicine and Biology; Springer, 2019; Vol. 1148, pp 279–322. 10.1007/978-981-13-7709-9_13 [DOI] [PubMed] [Google Scholar]
- Mui B.; Ahkong Q.; Chow L.; Hope M. Membrane Perturbation and the Mechanism of Lipid-Mediated Transfer of DNA into Cells. Biochim. Biophys. Acta - Biomembr. 2000, 1467 (2), 281–292. 10.1016/S0005-2736(00)00226-1. [DOI] [PubMed] [Google Scholar]
- Byk G.; Scherman D.; Schwartz B.; Dubertret C.. Lipopolyamines as Transfection Agents and Pharmaceutical Uses Thereof. U.S. Patent 6 171 612 B1, 2001.
- Arruda D. C.; Lachagès A.-M.; Demory H.; Escriou G.; Lai-Kuen R.; Dugas P.-Y.; Hoffmann C.; Bessoles S.; Sarrabayrouse G.; Malachias A.; Finet S.; Gastelois P. L.; de Almeida Macedo W. A.; da Silva Cunha A.; Bigey P.; Escriou V. Spheroplexes: Hybrid PLGA-Cationic Lipid Nanoparticles, for in Vitro and Oral Delivery of SiRNA. J. Controlled Release 2022, 350, 228–243. 10.1016/j.jconrel.2022.08.030. [DOI] [PubMed] [Google Scholar]
- Hughes L. D.; Rawle R. J.; Boxer S. G. Choose Your Label Wisely: Water-Soluble Fluorophores Often Interact with Lipid Bilayers. PLoS One 2014, 9 (2), e87649. 10.1371/journal.pone.0087649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z.; Yomo D.; Gradinaru C. Choosing the Right Fluorophore for Single-Molecule Fluorescence Studies in a Lipid Environment. Biochim. Biophys. Acta - Biomembr. 2017, 1859 (7), 1242–1253. 10.1016/j.bbamem.2017.04.001. [DOI] [PubMed] [Google Scholar]
- Li J.; Pei H.; Zhu B.; Liang L.; Wei M.; He Y.; Chen N.; Li D.; Huang Q.; Fan C. Self-Assembled Multivalent DNA Nanostructures for Noninvasive Intracellular Delivery of Immunostimulatory CpG Oligonucleotides. ACS Nano 2011, 5 (11), 8783–8789. 10.1021/nn202774x. [DOI] [PubMed] [Google Scholar]
- Liang L.; Li J.; Li Q.; Huang Q.; Shi J.; Yan H.; Fan C. Single-Particle Tracking and Modulation of Cell Entry Pathways of a Tetrahedral DNA Nanostructure in Live Cells. Angew. Chemie Int. Ed. 2014, 53 (30), 7745–7750. 10.1002/anie.201403236. [DOI] [PubMed] [Google Scholar]
- Walsh A. S.; Yin H.; Erben C. M.; Wood M. J. A.; Turberfield A. J. DNA Cage Delivery to Mammalian Cells. ACS Nano 2011, 5 (7), 5427–5432. 10.1021/nn2005574. [DOI] [PubMed] [Google Scholar]
- Wang S.; Guo H.; Li Y.; Li X. Penetration of Nanoparticles across a Lipid Bilayer: Effects of Particle Stiffness and Surface Hydrophobicity. Nanoscale 2019, 11 (9), 4025–4034. 10.1039/C8NR09381D. [DOI] [PubMed] [Google Scholar]
- Mosmann T. Rapid Colorimetric Assay for Cellular Growth and Survival: Application to Proliferation and Cytotoxicity Assays. J. Immunol. Methods 1983, 65 (1–2), 55–63. 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- Hu Y.-B.; Dammer E. B.; Ren R.-J.; Wang G. The Endosomal-Lysosomal System: From Acidification and Cargo Sorting to Neurodegeneration. Transl. Neurodegener. 2015, 4 (1), 18. 10.1186/s40035-015-0041-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt T. G. M.; Koepke J.; Frank R.; Skerra A. Molecular Interaction Between the Strep-Tag Affinity Peptide and Its Cognate Target, Streptavidin. J. Mol. Biol. 1996, 255 (5), 753–766. 10.1006/jmbi.1996.0061. [DOI] [PubMed] [Google Scholar]
- Skerra A.; Schmidt T. G. Applications of a Peptide Ligand for Streptavidin: The Strep-Tag. Biomol. Eng. 1999, 16 (1–4), 79–86. 10.1016/S1050-3862(99)00033-9. [DOI] [PubMed] [Google Scholar]
- Burghoff S.; Gong X.; Viethen C.; Jacoby C.; Flögel U.; Bongardt S.; Schorr A.; Hippe A.; Homey B.; Schrader J. Growth and Metastasis of B16-F10 Melanoma Cells Is Not Critically Dependent on Host CD73 Expression in Mice. BMC Cancer 2014, 14 (1), 898. 10.1186/1471-2407-14-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.; Wang H.; Zhao J.; Ma J.; Wei L.; Wu S.; Xie T.; Shen F.; Trojan J.; Habib N.; Anthony D. D.; Wu M.; Guo Y. Enhancement of Immunogenicity of Tumor Cells by Cotransfection with Genes Encoding Antisense Insulin-like Growth Factor-1 and B7.1 Molecules. Cancer Gene Ther. 2000, 7 (3), 456–465. 10.1038/sj.cgt.7700137. [DOI] [PubMed] [Google Scholar]
- Uckert W.; Kammertöns T.; Haack K.; Qin Z.; Gebert J.; Schendel D. J.; Blankenstein T. Double Suicide Gene (Cytosine Deaminase and Herpes Simplex Virus Thymidine Kinase) but Not Single Gene Transfer Allows Reliable Elimination of Tumor Cells In Vivo. Hum. Gene Ther. 1998, 9 (6), 855–865. 10.1089/hum.1998.9.6-855. [DOI] [PubMed] [Google Scholar]
- De Sabbata G.; Boisgerault F.; Guarnaccia C.; Iaconcig A.; Bortolussi G.; Collaud F.; Ronzitti G.; Sola M. S.; Vidal P.; Rouillon J.; Charles S.; Nicastro E.; D’Antiga L.; Ilyinskii P.; Mingozzi F.; Kishimoto T. K.; Muro A. F. Long-Term Correction of Ornithine Transcarbamylase Deficiency in Spf-Ash Mice with a Translationally Optimized AAV Vector. Mol. Ther. - Methods Clin. Dev. 2021, 20, 169–180. 10.1016/j.omtm.2020.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S.; Shah B.; Fung C. S.; Chan P. K.; Wakefield D. L.; Kuhns S.; Goudar C. T.; Piret J. M. Engineering Protein Glycosylation in CHO Cells to Be Highly Similar to Murine Host Cells. Front. Bioeng. Biotechnol. 2023, 11, 1113994. 10.3389/fbioe.2023.1113994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Blasi R.; Marbiah M. M.; Siciliano V.; Polizzi K.; Ceroni F. A Call for Caution in Analysing Mammalian Co-Transfection Experiments and Implications of Resource Competition in Data Misinterpretation. Nat. Commun. 2021, 12 (1), 2545. 10.1038/s41467-021-22795-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.