Abstract
The PKR protein kinase is a critical component of the cellular antiviral and antiproliferative responses induced by interferons. Recent evidence indicates that the nonstructural 5A (NS5A) protein of hepatitis C virus (HCV) can repress PKR function in vivo, possibly allowing HCV to escape the antiviral effects of interferon. NS5A presents a unique tool by which to study the molecular mechanisms of PKR regulation in that mutations within a region of NS5A, termed the interferon sensitivity-determining region (ISDR), are associated with sensitivity of HCV to the antiviral effects of interferon. In this study, we investigated the mechanisms of NS5A-mediated PKR regulation and the effect of ISDR mutations on this regulatory process. We observed that the NS5A ISDR, though necessary, was not sufficient for PKR interactions; we found that an additional 26 amino acids (aa) carboxyl to the ISDR were required for NS5A-PKR complex formation. Conversely, we localized NS5A binding to within PKR aa 244 to 296, recently recognized as a PKR dimerization domain. Consistent with this observation, we found that NS5A from interferon-resistant HCV genotype 1b disrupted kinase dimerization in vivo. NS5A-mediated disruption of PKR dimerization resulted in repression of PKR function and inhibition of PKR-mediated eIF-2α phosphorylation. Introduction of multiple ISDR mutations abrogated the ability of NS5A to bind to PKR in mammalian cells and to inhibit PKR in a yeast functional assay. These results indicate that mutations within the PKR-binding region of NS5A, including those within the ISDR, can disrupt the NS5A-PKR interaction, possibly rendering HCV sensitive to the antiviral effects of interferon. We propose a model of PKR regulation by NS5A which may have implications for therapeutic strategies against HCV.
The interferon (IFN)-induced double-stranded RNA (dsRNA)-activated protein kinase, PKR (52), is a key component of the antiviral and antiproliferative effects of interferon (reviewed in reference 13). As a member of the IFN-induced gene family, PKR is transcriptionally activated from a low level of expression upon cellular exposure to IFN (52). Activation of PKR catalytic function proceeds through a process of dsRNA binding, dimerization, and autophosphorylation (reviewed by Clemens and Elia [13]). Tight regulation of PKR is essential for controlling the function of PKR substrates, the best characterized of which is the protein synthesis initiation factor 2, alpha subunit (eIF-2α). PKR phosphorylates serine 51 of eIF-2α, leading to limitations in functional eIF-2, a concomitant inhibition of mRNA translation initiation, and repression of cell growth (13, 51). Another PKR substrate is IκB, the inhibitor of nuclear factor kappa B (NF-κB) (43). By phosphorylating IκB, PKR functions within dsRNA- and IFN-signaling pathways to induce NF-κB-dependent transcription (44; reviewed in reference 77). PKR may also be required for IFN-γ signaling processes (44) and is a key mediator of stress-induced apoptosis (18). Constitutive repression of PKR induces malignant transformation of mammalian cells (3, 42), thus identifying PKR as a potential tumor suppressor (4). Tumor suppressor function has been attributed to PKR-dependent eIF-2α phosphorylation (54), though the other roles played by PKR may contribute to cell growth regulation as well (59).
PKR is best understood for its role in the IFN-induced cellular antiviral response (for reviews of the IFN response, see references 68 and 69). Within the IFN response, PKR-mediated phosphorylation of eIF-2α provides a key antiviral function by phosphorylating eIF-2α to block protein synthesis and thereby inhibit viral replication (reviewed in reference 37). To facilitate replication and avoid the antiviral effects of IFN, eukaryotic viruses have evolved a diverse repertoire of mechanisms to repress PKR function during infection (24). We have recently determined that hepatitis C virus (HCV), a member of the Flaviviridae (31, 70), encodes a mechanism to repress PKR. The ability of HCV to regulate PKR lies within the viral nonstructural 5A (NS5A) protein, which binds to a distinct region of PKR to repress kinase function (27). HCV is of particular interest due to the emergence of a global HCV epidemic comprising approximately 2% of the world population.
To date, type I IFN remains the only approved anti-HCV therapeutic agent, but it is effective in only 20% of HCV-infected individuals (1, 23, 49). HCV infection is characterized by progressive liver pathology, often developing into a chronic disease state, perhaps due in part to the high number of IFN-resistant viral quasispecies within the infected population (17, 71). Problematically, chronic HCV infection has been epidemiologically linked to the development of hepatocellular carcinoma and is currently the leading indicator for adult liver transplants in the United States (20). A goal of the present study was to define the molecular mechanism which underlies the ability of HCV to evade the antiviral effects of IFN and induce disease.
Most relevant are the observations that sequence variation from the prototypic IFN-resistant HCV J strain (36) within the NS5A protein of the HCV polyprotein cleavage product has been associated with sensitivity to IFN in Japanese HCV genotype 1b (HCV-1b) subtypes (21, 22, 45). Viral isolates with multiple amino acid substitutions within a region of NS5A, termed the IFN sensitivity-determining region (ISDR; amino acids [aa] 2209 to 2248), were eliminated from HCV-infected patients during IFN therapy, while those exhibiting the prototypic ISDR sequence were IFN resistant, persisting at therapy cessation. We have recently demonstrated that NS5A from IFN-resistant strains of HCV-1a and -1b can physically bind PKR by an ISDR-dependent mechanism to inhibit kinase function, implicating NS5A as a mediator of the IFN-resistant HCV phenotype (27). We hypothesized that mutations within the ISDR may similarly disrupt NS5A function to render HCV sensitive to the PKR-mediated antiviral effects of IFN. In this study we conducted a detailed molecular analysis of PKR regulation by ISDR sequence variants of NS5A previously described for IFN-resistant and sensitive clinical isolates of HCV-1b (21). We show that NS5A from IFN-resistant HCV disrupts a critical step of PKR activation, resulting in repression of PKR function and a block in eIF-2α phosphorylation. Mutations in the PKR-binding domain of NS5A, localized to within the ISDR, abrogated the PKR-regulatory function of NS5A. Taken together, these results suggest a molecular mechanism for IFN sensitivity of HCV that is defined, at least in part, by the sequence of the PKR-binding domain of NS5A.
MATERIALS AND METHODS
Plasmid construction and site-directed mutagenesis.
Plasmids pGBT10 and pGAD425 encode the GAL4 DNA-binding domain (BD) and transcription activation domain (AD), respectively (25). pAD-PKR K296R, pAD-PKR 244-551, and pAD-PKR 244-296 encode the indicated AD-PKR fusion constructs and have been described previously (25). All NS5A 1b expression constructs were generated from pAD-NS5A, which harbors wild-type (wt) NS5A from a clinical isolate of IFN-resistant HCV-1b (NS5A 1b-wt) (27). To facilitate yeast two-hybrid protein interaction analysis, pAD-NS5A was cleaved with restriction enzymes NdeI and BamHI and the 1.4-kb insert encoding full-length NS5A (aa 1973 to 2419) was cloned into the corresponding sites of pGBT10 to yield pBD-NS5A 1b-wt. pBD-NS5A 1973-2361 encodes a BD fusion of NS5A aa 1973 to 2361 and was generated by subcloning the NdeI/SalI insert from pBD-NS5A 1b-wt into the corresponding sites of pGBT10. pBD-NS5A 2120-2274 encodes a BD fusion of NS5A aa 2120 to 2274 constructed by ligating the internal 462-bp EcoRI/BstYI fragment from pBD-NS5A 1b-wt into the EcoRI/BamHI sites of pGBT10. pBD-NS5A 1973-2208, pBD-NS5A 2209-2274, and pBD-NS5A 2180-2251 encode BD fusions of NS5A aa 1973 to 2208, 2209 to 2274, and 2180 to 2251, respectively, and were generated by PCR amplification of the corresponding pBD-NS5A 1b-wt coding region, using the restriction site-linked oligonucleotide primer pairs shown in Table 1. PCR products were directly cloned into pCR2.1 (Invitrogen) as described by the plasmid manufacturer. PCR products encoding NS5A aa 1973 to 2208 and 2209 to 2274 were released from pCR2.1 by digestion with restriction enzymes NdeI/SalI and EcoRI/SalI, respectively. The resultant insert DNA was ligated into the appropriate sites of pGBT10 and pGBT9 (Clontech) to yield pBD-NS5A 1973-2208 and pBD2209-2274. The PCR product encoding NS5A aa 2180 to 2251 was released from pCR2.1 by NcoI/BamHI digestion, and the resultant 213-bp fragment was ligated into the identical sites of pAS2-1 (Clontech) to yield pBD-NS5A 2180-2251. pBD-ΔISDR encodes an ISDR deletion mutant of NS5A from IFN-resistant HCV-1a (27).
TABLE 1.
Sequences of PCR NS5A oligonucleotidesa
| Construct | Senseb | Antisense | nt |
|---|---|---|---|
| NS5A 1b-wt | 5′ TAAGCTTATGGGC | 5′ CAAGCTTGGATCC | 6260–7598 |
| 1973-2419 | TCCGGCTCGTGGCT | TTAGGACATTGAGC | |
| NS5A | 5′ CATATGGGCTCCGG | 5′ GTCGACCGCAGACA | 6260–6965 |
| 1973-2208 | CTCGTGGCTA | ACTGGCTAGCTGA | |
| NS5A | 5′ GAATTCCCTTCCTT | 5′ GTCGACCTCCGCCG | 6966–7165 |
| 2209-2274 | GAAGGCAACATGC | GAACGGATAC | |
| NS5A | 5′ CCTTCCATGGCCCA | 5′ ATCGGATCCTTATA | 6877–7094 |
| 2180-2251 | CATTACAGCAGAGACG | CCACCTTATTCTCTGA |
Amino acid and nucleotide (nt) positions correspond to the PCR-amplified region. Numbering is based on the prototypic HCV-J sequence (36).
Cloning restriction sites (described in text) are underlined.
We used site-directed mutagenesis (Chameleon double-stranded site-directed mutagenesis kit; Stratagene) to introduce ISDR mutations corresponding to IFN-sensitive strains of HCV-1b into pBD-NS5A 1b-wt. Mutagenesis reactions were carried out as described by the manufacturer, using the mutagenic primers shown in Table 2. Template DNA was denatured by incubation at 100°C for 5 min, followed by annealing of the indicated mutagenic primer and the ScaI-to-StuI selection primer 5′ GTGACTGGTGAGGCCTCAACCAAGTC (StuI restriction site underlined). T7 DNA polymerase-primer extension products were ligated and selected for the primer-encoded mutation(s) by digestion with restriction enzyme ScaI and subsequent transformation into Escherichia coli XlmutS. By this method, we constructed a set of isogenic NS5A constructs, identical to NS5A 1b-wt except for the defined mutations introduced into the ISDR (Table 3). pBD-NS5A 1b-2 and pBD-NS5A 1b-4 were generated directly from pBD-NS5A 1b-wt and encode a single (A2224V) or multiple (P2209L, T2214A, and T2217G) ISDR amino acid mutations, respectively (Table 3). pBD-NS5A-5, encoding the ISDR mutations P2209L, T2214A, T2217G, and A2224V, was produced by introducing an A2224V mutation into pBD-NS5A-4.
TABLE 2.
Sequences of mutagenic NS5A oligonucleotides
| Construct | Mutagenic primera | Mutation(s) |
|---|---|---|
| NS5A 1b-2 | 5′ GACTCCCCAGATGTTGACCTCATC | A2224V |
| NS5A 1b-5b | ||
| NS5A 1b-4 | 5′ TTGTCTGCGCTTTCCTTGAAGGCAGCATGCACTGGCCGTCACGAC | P2209L, T2214A, T2217G |
Underlined codons correspond to the indicated amino acid mutations.
See Materials and Methods for details on the construction of this construct.
TABLE 3.
Sequences of the PKR-binding region and ISDR, with the corresponding IFN sensitivities of NS5A expression constructs
| Namea | PKR-binding region (aa 2209–2274)b | IFN responsec | Reference(s) |
|---|---|---|---|
| 1b-pt | PSLKATCTTHHDSPDADLIEANLLWRQEMGGNITRVESENKVVMLDSFDPLRAEEDEREVSVAAEI | R | 21 |
| 1b-wt | ---------R-----------------------------------------H-----K----P--- | R | 14 |
| 1b-2 | ---------R-----V-----------------------------------H-----K----P--- | R/S | 21, 78 |
| 1b-4 | L----A- GR-----------------------------------------H-----K----P--- | Sd | |
| 1b-5 | L----A- GR-----V-----------------------------------H-----K----P--- | S | 21 |
| 1a-wt | --------AN------E----------------------------V-----V-------I--P--- | R | 27 |
pt, HCV-J prototype reference sequence (GenBank accession no. D90208); wt, wild-type parental HCV-1b clone (GenBank accession no. AF034151 [NS5A coding region only]).
The ISDR sequence is in boldface.
R, IFN resistant; S, IFN sensitive; R/S, independently reported as IFN resistant or IFN sensitive in separate studies.
For expression of NS5A in Saccharomyces cerevisiae, the entire 1.4-kb insert of pBD-NS5A 1b-wt was amplified by PCR using the restriction enzyme-linked oligonucleotides shown in Table 1. PCR products were cloned into the SrfI site of pCR-Script (Stratagene) and released from the resultant plasmid by HindIII digestion. The gel-purified insert DNA was cloned into the HindIII site of pYES2 (Invitrogen) to yield pYES-NS5A 1b-wt expressing NS5A under control of the galactose-inducible GAL1 promoter. Construction of pYES-NS5A 1b-2, pYES-NS5A 1b-4 and pYES-NS5A 1b-5 was facilitated by replacing the internal 1.1-kb SacII/SalI fragment of pYES-NS5A 1b-wt with the internal SacII/SalI fragment from the corresponding pBD constructs. pYex-PKRΔ295-300 was generously provided by P. Romano (Small Molecule Therapeutics, Inc.).
For expression of NS5A in mammalian cells, the entire 1.4-kb NS5A coding region of pYES-NS5A 1b-wt and pYES-NS5A 1b-5 was released by HindIII digestion and cloned into the HindIII site of pFLAG-CMV2 (Eastman Kodak Co.). The resulting plasmids, pFlagNS5A 1b-wt and pFlagNS5A 1b-5, respectively, encode full-length wt and mutant NS5A fused at the N terminus to the 8-aa FLAG epitope tag sequence (FLAG-NS5A) under control of the cytomegalovirus immediate-early promoter. pNeo-NS5A 1a-wt was constructed by cloning the HindIII/XbaI insert of pYES2-NS5A (27) into the corresponding site of pcDNA1Neo. pNeo-PKR K296R encodes the full-length inactive human PKR K296R mutant (5). For construction of pGST-NS5A 1b-wt, the 1.4-kb NcoI/XhoI insert DNA from pAD-NS5A (27) was isolated and the 3′-recessed termini were filled in with Klenow polymerase (66). The resulting blunt-ended DNA was cloned into the SmaI site of pGEX-2TK (Pharmacia Biotech), fusing the NS5A coding region in frame to the plasmid-encoded glutathione S-transferase (GST) protein. pGST-K3L encodes a GST fusion of the 88-aa vaccinia virus K3L protein (a very kind gift from E. Beattie, University of Washington). pGST-NS1 encodes a GST fusion of the influenza virus NS1 protein and was a generous gift from R. Krug (Sloan-Kettering). The fusion between the N-terminal 132 aa of the phage λ cI repressor and the catalytically inactive PKR K296R was constructed in pcI168 to yield pcI-PKR K296R as recently described (73). To avoid the cellular toxicity that is associated with wt PKR expression (5, 63), we used the full-length inactive PKR K296R mutant (5) for all PKR-protein interaction analyses. The nucleotide sequence of each new construct was verified by dideoxy nucleotide sequence analysis (Applied Biosystems).
Cell culture and transfection.
Cos-1 cells (American Type Culture Collection) were grown in Dulbecco’s modified Eagle medium (DMEM) containing 10% fetal bovine serum as described previously (74). For transient transfections, expression plasmid combinations were introduced into Cos-1 cells by the DEAE-dextran–chloroquine method exactly as described previously (74) or by a procedure using the Superfect transfection reagent as described by the manufacturer (Qiagen). Each set of transfections consisted of subconfluent 25-cm2 cultures of approximately 6 × 105 cells cotransfected with 5 μg of each expression plasmid in the following combinations: pcDNA1Neo-pNeoPKR K296R and pNeoNS5A 1a-wt–pNeoPKR K296R, or pFlag-pNeoPKR K296R, pFlagNS5A 1b-wt–pNeoPKR K296R, or pFlagNS5A 1b-5–pNeoPKR K296R. Cells were harvested 48 h posttransfection, and extracts were processed for immunoprecipitation or immunoblot analyses as described previously (26).
Protein analysis.
To prepare yeast extracts, cells from 20-ml liquid cultures were collected, washed once with ice-cold water, resuspended in ice-cold yeast lysis buffer (40 mM PIPES [pH 6.6], 100 mM NaCl, 1 mM dithiothreitol, 50 mM NaF, 37 mM β-glycerolphosphate, 120 mM ammonium sulfate, 10 mM 2-aminopurine, 15 mM EDTA, 0.2 mM phenylmethylsulfonyl fluoride), and lysed by the glass bead method as described previously (25). Cos-1 cell extracts were prepared in buffer I (50 mM KCl, 50 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol, 20% glycerol, 0.5% Triton X-100, 100 U of aprotinin per ml 1 mM phenylmethylsulfonyl fluoride, 20 mM Tris [pH 7.5]) exactly as described previously (74). Extracts were clarified by 4°C centrifugation at 12,000 × g; supernatants were collected and stored at −70°C. Cell extract protein concentration was determined using the Bio-Rad Bradford assay as described by the manufacturer.
Determination of protein expression was carried out by immunoblot analyses of 25 to 50 μg of total protein from cell extracts as previously described (26). Proteins were separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and transferred to nitrocellulose membranes. Bound proteins were detected by probing the membranes with primary monoclonal antibodies specific to NS5A (anti-NS5A; a generous gift from T. Imagawa, Osaka University), human PKR (anti-PKR [47]; generously provided by A. Hovanessian, Pasteur Institute), FLAG epitope (anti-FLAG; Eastman Kodak), and GAL4 AD and GAL4 BD, (anti-AD and anti-BD, respectively; Clontech). Proteins were visualized by enhanced chemiluminescence and autoradiography. To control for potential errors in protein loading, blots were also probed with an actin-specific monoclonal antibody (anti-actin; ICN).
Immunoprecipitations were carried out with extracts representing 2 × 106 transfected Cos-1 cells as previously described (26). Extracts (150 μl) were thawed on ice and precleared by a 1-h incubation with protein G-agarose beads (Boehringer Mannheim) at 4°C. Supernatants were recovered by 4°C centrifugation (12,000 × g) and mixed with anti-NS5A (1:500) or anti-FLAG (Eastman Kodak) M2 affinity gel in a final volume of 600 μl of buffer I and incubated at 4°C for 2 or 16 h, respectively. Anti-NS5A immunocomplexes were recovered by an additional incubation with protein G-agarose beads equilibrated in buffer I. Immunocomplexes were washed five times with 1 ml each of ice-cold buffer I. Anti-FLAG M2 affinity gel immunocomplexes were further washed three times with cold Tris-buffered saline (50 mM Tris [pH 7.5]) and eluted by the addition of competitor FLAG peptide as described by the manufacturer (Eastman Kodak). Immunocomplexes were recovered by centrifugation, diluted in SDS sample buffer, and incubated at 100°C for 5 min. Immunoprecipitation products were resolved by electrophoresis on SDS–12% acrylamide gels and processed for immunoblot analysis as described above.
For isoelectric focusing (IEF) of eIF-2α, yeast strains were grown 16 h in uracil-deficient synthetic defined medium containing 2% dextrose (SD medium), diluted to an optical density at 600 nm of 0.4 in uracil-deficient synthetic defined medium containing 2% raffinose and 10% galactose (SGAL medium), and grown for an additional 4 to 9 h at 30°C. Yeast extracts were prepared as described for immunoblot analysis. Proteins (16 μg) were separated by vertical IEF (19) and blotted to nitrocellulose membranes. eIF-2α was detected by immunoblot analysis using a rabbit polyclonal antiserum specific to yeast eIF-2α (a generous gift from T. Dever, National Institutes of Health). In these experiments, an increase in the level of the less acidic, basally phosphorylated form of eIF-2α indicates a concomitant decrease in the level of hyperphosphorylated eIF-2α, which is phosphorylated by PKR on serine 51 (19, 63).
Yeast methods.
Details of the yeast two-hybrid assay have been extensively described elsewhere (25, 27). This assay utilizes specific induction of a HIS reporter gene to support growth of S. cerevisiae Hf7c (Clontech) on histidine-deficient medium as an indicator of a two-hybrid protein interaction. S. cerevisiae Hf7c [MATa ura 3-52 his 3-200 lys2-801 ade2-101 trp1-901 leu2-3,112 gal4-542 gal80-538 LYS2::GAL1-HIS3 URA3::(GAL4 17-mers)3-CYC1-lacZ] was transformed with the indicated 2μm TRP1 and LEU2 expression plasmids harboring the corresponding GAL4 AD and BD fusion constructs. Transformed strains were plated onto SD medium lacking tryptophan and leucine (+His medium). After 3 days at 30°C, strains were streaked onto SD medium lacking tryptophan, leucine, and histidine (−His medium) and incubated for 3 to 6 days at 30°C. The resultant histidine-depleted colonies were replica streaked onto +His and −His media and incubated for 3 to 5 days at 30°C. Specific growth on −His medium was scored as positive for a two-hybrid protein interaction.
For determination of PKR and NS5A function in vivo, wt or mutant NS5A URA3 expression plasmids were transformed in S. cerevisiae RY1-1 [MATa ura3-52 leu2-3 leu2-112 gcn2Δ trp1-Δ63 LEU2::(GAL-CYC1-PKR)2] (63). This strain lacks the yeast eIF-2α kinase GCN2 and harbors two copies of wt human PKR integrated into the LEU2 locus under control of the galactose-inducible GAL-CYC1 hybrid promoter (10). When grown on SGAL medium, PKR is expressed and phosphorylates endogenous eIF-2α on serine 51, resulting in inhibition of mRNA translation and growth suppression (19, 63). Conversely, coexpression of wt NS5A represses these toxic effects associated with PKR function in this system, allowing strains coexpressing functional NS5A to grow on SGAL medium (27). RY1-1 strains harboring the indicated NS5A expression constructs were plated onto noninducing uracil-deficient SD medium and incubated at 30°C for 3 days. Single colonies were picked and cultured for 16 h at 30°C in uracil-deficient liquid SD medium. Aliquots of each culture were normalized to an optical density at 600 nm of 0.2 and serially diluted in 10-fold increments with sterile H2O. Then 2 μl of each dilution was applied in replicate onto uracil-deficient SD and SGAL media and incubated for 3 to 6 days at 30°C. Strains were scored for high-dilution growth on SGAL medium, which is indicative of NS5A-mediated repression of PKR (27).
Dimerization disruption assay.
The assay for dimerization disruption has been previously described (32, 33, 73). This assay utilizes sensitivity to phage λ-mediated cell lysis as an indicator of the dimerization state of a cI-PKR K296R fusion protein expressed from pcI-PKR K296R in E. coli. pcI-PKR K296R replicates under control of the p15A origin of replication and is thus a low-copy replicon compatible with plasmids that contain the ColE1 origin of replication, including the pGEX series of vectors (72). E. coli AG1688 (33) coexpressing cI-PKR K296R and the indicated GST fusion protein was assessed for resistance to cell lysis mediated by the phage λ cI deletion mutant λKH54 (33). E. coli AG1688 was grown to mid-log phase in liquid cultures consisting of Luria broth supplemented with 10 mM MgSO4 and 0.2% maltose. Bacteria (2.5-μl aliquots) were mixed with an equal volume of serial 10-fold dilutions of λKH54 containing 102 to 106 PFU each. The bacterium-phage mixture was applied to antibiotic-agar plates containing 0.1 mM isopropylthio-β-d-galactoside to induce expression of the plasmid-encoded fusion proteins. Plates were air dried, incubated 16 h at 37°C, and scored for resistance to λKH54-mediated cell lysis, which is an indicator for in vivo formation of functional cI-PKR K296R homodimers. The expression of each plasmid-encoded fusion protein was verified by immunoblot analysis (data not shown).
RESULTS
Mechanism of PKR regulation by NS5A: disruption of protein kinase dimerization.
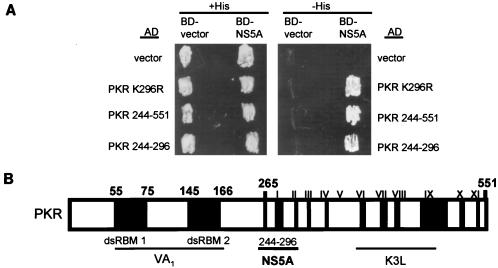
We previously determined that NS5A from IFN-resistant strains of HCV binds to PKR to repress kinase function (27). NS5A binding was mapped to within a broad region of the PKR catalytic domain defined by PKR aa 244 to 366. To better understand the molecular mechanism(s) of NS5A-mediated regulation of PKR, we identified a minimal NS5A-binding domain on the protein kinase. We used yeast two-hybrid analysis to examine the interaction between wt NS5A, isolated from an IFN-resistant strain of HCV-1b (27) fused to the GAL4 BD (BD-NS5A 1b-wt), and deletion mutants of PKR fused to the GAL4 AD. A two-hybrid protein interaction was confirmed by growth on −His medium (which is due to activation of the Hf7c HIS reporter [6]). We determined that each construct was efficiently expressed in the corresponding strains (data not shown). Hf7c yeast strains coexpressing BD-NS5A 1b-wt with AD-PKR (PKR K296R) or the AD-PKR deletion construct PKR 244-551 or PKR 244-296 all exhibited growth on −His medium, demonstrating a two-hybrid protein interaction within these strains (Fig. 1). These results define an NS5A-binding domain in PKR to within the 52-aa sequence defined by PKR aa 244 to 296. However, we cannot rule out the possibility that NS5A targets other regions of PKR. Importantly, the sequence defined by PKR aa 244 to 296 has recently been identified as a critical PKR dimerization domain (73).
FIG. 1.
NS5A-binding domain of PKR. (A) Hf7c yeast strains harboring the indicated AD and BD expression constructs were replica printed onto +His (left) and −His (right) media, incubated at 30°C for 4 days, and assayed for growth. Expression of AD-PKR and BD-NS5A 1b-wt constructs was confirmed by immunoblot analysis (not shown). Strains which grew on −His medium were scored positive for a two-hybrid protein interaction. (B) Structural representation of PKR. The positions of the two dsRNA-binding motifs (dsRBM 1 and 2) and the 11 protein kinase catalytic domain conservation regions (roman numerals) (28) are indicated in black. The regions of PKR which mediate interaction with the virus-encoded inhibitors adenovirus VA1 RNA (38), vaccinia virus K3L (16, 25), and HCV NS5A proteins (reference 27 and this study) are underlined.
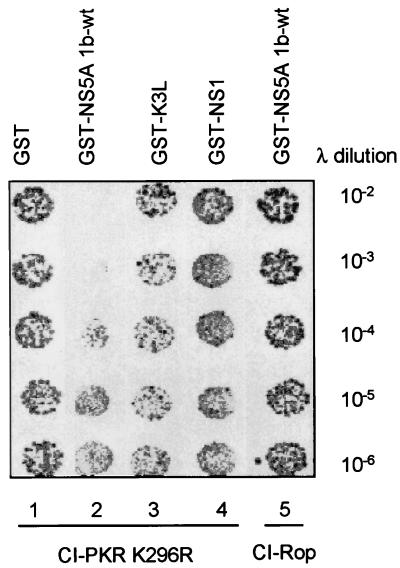
We reasoned that NS5A may inhibit PKR by disrupting the PKR dimerization process. We therefore tested the ability of NS5A 1b-wt to interfere with PKR dimerization in vivo, using a phage λ-based genetic assay in E. coli. In this assay, dimerization proteins, which are fused in frame to the DNA-binding domain of the phage λ cI repressor, mediate dimerization of the cI DNA-binding domain, which is required for binding to the λ promoter (32). When expressed in E. coli, the hybrid cI repressor mediates resistance to cell lysis induced by a cI deletion mutant of λ phage (λKH54 [33]) by dimerizing and binding to the λ promoter. This results in repression of phage gene expression within λKH52-infected E. coli that express the hybrid cI repressor. Conversely, coexpression of a dimerization inhibitor in this system releases λ gene repression through the disruption of cI dimers, resulting in E. coli lysis. Expression of full-length inactive PKR K296R fused at the N terminus to the cI DNA-binding domain (cI-PKR) was sufficient to repress λ gene expression upon λKH54 infection in E. coli (Fig. 2). Coexpression of GST had no effect on cI-PKR-mediated λ gene repression, as resistance to cell lysis was observed even after exposure to high concentrations of phage. Thus, the PKR component of cI-PKR facilitates protein dimerization and λ gene repression in vivo. Resistance to λKH54-mediated cell lysis was reduced approximately 1,000-fold in E. coli coexpressing cI-PKR with GST-NS5A (compare lanes 1 and 2 in Fig. 2), indicating that NS5A was disrupting the cI-PKR dimerization process. This effect of GST-NS5A 1b-wt was specific to cI-PKR, as cells coexpressing GST-NS5A 1b-wt with the dimerization control fusion protein, cI-Rop, repressed λ gene expression and λKH54-mediated cell lysis even after exposure to high phage concentrations (lane 5). Importantly, GST constructs encoding other viral inhibitors of PKR, including vaccinia virus K3L (GST-K3L; lane 3) and influenza virus NS1 (GST-NS1; lane 4), which target the PKR-substrate (16, 25) and PKR-dsRNA (38) interactions, respectively, did not affect the ability of E. coli to resist λKH54-mediated cell lysis when coexpressed with cI-PKR. Thus, NS5A specifically disrupts the PKR dimerization process in vivo.
FIG. 2.
NS5A disrupts PKR dimerization. E. coli AG1688 cells were cotransformed with expression plasmid combinations encoding cI-PKR K296R and GST (column 1), GST-NS5A 1b-wt (column 2), GST-K3L (column 3), or GST-NS1 (column 4), mixed with the indicated dilution of λKH54 phage, and spotted onto plates containing agar medium. Plates were incubated and visually scored for colony formation (dark spots) as described in Materials and Methods. Column 5 contains E. coli harboring cI-Rop and GST-NS5A 1b-wt expression plasmids (control). cI fusion protein dimerization is indicated by colony formation. Shown is a photograph of a plate from a representative experiment.
Identification of the PKR-interacting domain of NS5A: the ISDR is necessary but not sufficient for NS5A-PKR complex formation.
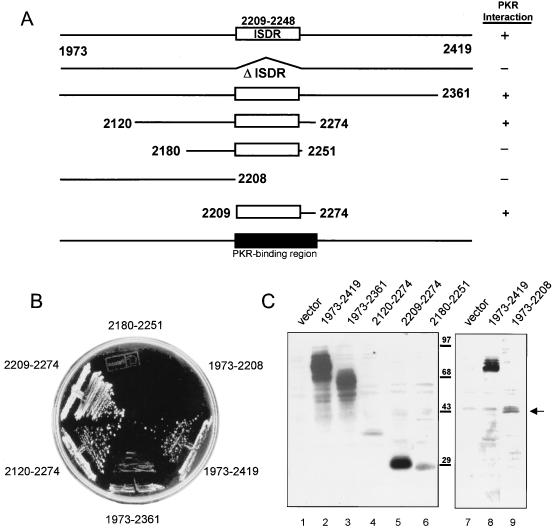
We previously demonstrated that the ISDR of NS5A was necessary for both interaction with and repression of PKR (27). We therefore conducted a detailed structural analysis of the NS5A-PKR interaction, using the yeast two-hybrid assay to determine the role of the ISDR in this interaction. TRP1 plasmids encoding full-length or deletion mutants of BD-NS5A 1b-wt (Fig. 3A; Table 3) were introduced into yeast strain Hf7c harboring a LEU2 plasmid encoding AD-PKR. All constructs were expressed in cotransformed strains (Fig. 3C). Strains harboring each BD-NS5A construct and AD-PKR or AD vector (interaction-negative control [not shown]) all grew on this medium. Interaction-negative control strains failed to grow on −His medium, demonstrating specificity for the described interactions (data not shown). Using an identical assay, we previously determined that the NS5A ISDR was required for interaction with PKR (27). Importantly, we found that an ISDR-inclusive 66-aa region of NS5A was required for complex formation with PKR in vivo (Fig. 3). The NS5A N-terminal region alone (aa 1973 to 2208) was not sufficient to interact with AD-PKR, as determined by the inability of strains harboring this construct to grow on −His medium (Fig. 3B). Moreover, we found that the ISDR-inclusive construct encoding NS5A aa 2180 to 2251 did not support growth on −His medium when expressed with AD-PKR. It is important to note that this construct was expressed to levels equal to or higher than those of the overlapping construct, BD-NS5A 2120-2274, which scored positive for PKR interaction in our assay (Fig. 3B and C). Those strains harboring BD-NS5A construct 1973-2419 or 1973-2361 exhibited growth on −His medium, implying a two-hybrid protein interaction. By these analyses, we determined that the PKR-binding region of NS5A mapped to within a 66-aa region comprising the ISDR and the adjacent C-terminal 26 aa (Fig. 3A). Thus, the ISDR was necessary but not sufficient for the NS5A-PKR interaction. Examination of the amino acid sequence within the PKR-binding domain of NS5A revealed that this region is highly conserved between our NS5A 1b-wt construct and the protoypic HCV-J sequence (Table 3). However, the NS5A-PKR interaction appears to tolerate nonconservative amino acid substitutions within this region (27).
FIG. 3.
PKR-binding domain of NS5A. (A) Structural representation of BD-NS5A fusion constructs. Deletion mutants were prepared from NS5A 1b-wt, except for the ΔISDR construct, which was prepared from NS5A 1a-wt (27). The ISDR and the PKR-binding region are shown as white and black rectangles, respectively. Terminal amino acid positions are indicated, with numbering based on the prototypic HCV-J polyprotein sequence (36). The PKR interaction of each construct (scored in panel B) is indicated at the right. (B) Yeast two-hybrid assay. Hf7c yeast strains harboring AD-PKR K296R were cotransformed with the indicated BD-NS5A deletion constructs. Strains were propagated on +His medium for 3 days (not shown), after which single colonies were streaked onto −His medium and assayed for growth. Shown is a −His plate incubated for 3 days at 30°C. Growth on −His medium is indicative of a two-hybrid protein interaction. In parallel experiments, we determined that the indicated BD-NS5A constructs did not interact with a construct encoding the GAL4 AD alone (not shown). (C) Immunoblot analysis. Extracts prepared from the yeast strains shown in panel B were separated by SDS-PAGE and subjected to immunoblot analysis using anti-NS5A (lanes 1 to 6) or anti-BD (lanes 7 to 9) monoclonal antibody. Lanes 1 and 7 contain extracts prepared from strains harboring the pGBT9 BD vector (control). Extracts are identified by the construct designation shown above the corresponding lane. BD-NS5A construct 1973-2419 was included as a positive control for the blot shown at the right. Positions of protein standards are indicated in kilodaltons. Arrow points to the protein expressed by the 1973-2208 construct.
The NS5A-PKR interaction is dependent on the sequence of the PKR-binding domain of NS5A and is disrupted by ISDR mutations.
ISDR mutations which correlate with IFN sensitivity of HCV localize to within the PKR-binding domain of NS5A (Table 3 and references 20 and 21). We used the yeast two-hybrid assay to examine the effects, if any, that defined ISDR mutations had on in vivo complex formation between NS5A and PKR. We first prepared a series of NS5A expression plasmids encoding wt and ISDR variants of NS5A corresponding to IFN-resistant and -sensitive strains of HCV, respectively. Rather than randomly assigning ISDR mutations, we used site-directed mutagenesis to construct ISDR variants of NS5A based on defined mutations previously identified within clinical isolates of IFN-sensitive strains of HCV (21). These mutations (Table 3) were introduced into the ISDR of NS5A 1b-wt, previously isolated from IFN-resistant HCV (27). We thus generated the isogenic NS5A constructs 1b-2, 1b-4, and 1b-5, which contained two, four, and five, respectively, amino acid changes from the prototype ISDR sequence from IFN-resistant HCV. These constructs were identical to NS5A 1b-wt except for defined mutations within the ISDR and thus allowed us to determine the effects of specific ISDR mutations on HCV-1b NS5A function. Table 3 compares the ISDR sequence and IFN response of the prototype IFN-resistant HCV J strain (36) with the ISDR sequence and response to IFN determined for the corresponding viral isolate from each wt and mutant NS5A construct. The IFN sensitivity corresponding to the ISDR sequence of construct 1b-4 has not been described, although based on previous work (21), we propose that such a sequence may correlate with an IFN-sensitive phenotype.
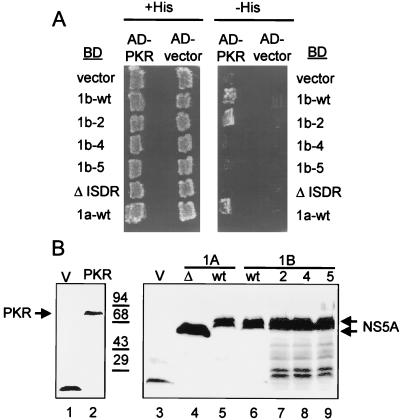
BD-NS5A constructs encoding full-length 1b-wt NS5A and isogenic ISDR variants (Table 3) were transformed into Hf7c yeast cells harboring plasmid-encoded AD-PKR or the AD control vector. As additional controls, we included a parallel assessment of strains harboring plasmids encoding AD-PKR and the HCV-1a NS5A construct BD-1a-wt (positive control) or BD-ΔISDR (negative control), the latter lacking the complete ISDR of the corresponding 1a-wt construct (27). The resulting yeast strains were replica printed onto +His and −His medium. All strains grew on +His medium (Fig. 4A). However, only those strains expressing BD-NS5A construct 1b-wt, 1b-2, or the 1a-wt control exhibited growth on −His medium, indicating that these constructs could bind PKR in vivo. Similar to the ΔISDR control, ISDR variants of BD-NS5A, 1b-4, and 1b-5 failed to interact with AD-PKR, as strains containing these constructs failed to grow on −His medium (Fig. 4A). Immunoblot analyses demonstrated that all BD-NS5A constructs and the AD-PKR construct were efficiently expressed in cotransformed yeast (Fig. 4B). Thus, isogenic NS5A constructs differing only in the ISDR sequence differentially interacted with PKR in vivo. Introduction of a single ISDR point mutation (NS5A 1b-2) was not sufficient to abolish the NS5A-PKR interaction, while multiple ISDR mutations did abolish complex formation with PKR (Fig. 4A). The inability of NS5A to bind PKR can therefore be attributed to multiple mutations within the ISDR which, importantly, have been associated with IFN-sensitive HCV quasispecies.
FIG. 4.
Effects of ISDR mutations on the NS5A-PKR interaction. (A) Yeast two-hybrid assay. Hf7c yeast strains harboring pGAD425 encoding AD-PKR K296R (AD-PKR) or the AD alone (AD-vector) were cotransformed with pGBT9 encoding the BD alone (vector), BD-NS5A 1a-wt, BD-NS5A-ΔISDR, or an isogenic set of BD-NS5A 1b-wt constructs possessing the ISDR sequence shown in Table 3. Strains were replica printed onto +His (left) and −His (right) media and incubated for 3 days at 30°C. Growth on −His medium is indicative of a two-hybrid protein interaction. (B) Immunoblot analysis. Extracts were prepared from the strains shown in panel A and subjected to immunoblot analysis using anti-AD (left) or anti-BD (right) monoclonal antibody. Lanes 1 and 2 show expression of the AD vector (V; control) and AD-PKR (PKR; arrow), respectively. Lanes 3 to 9 show expression of the BD vector (V; lane 3), BD-NS5A-ΔISDR (Δ; lane 4), BD-NS5A 1a-wt (wt; lane 5), BD-NS5A 1b-wt (wt; lane 6), BD-NS5A 1b-2 (2; lane 7), BD-NS5A 1b-4 (4; lane 8), and BD-NS5A 1b-5 (5; lane 9). Arrows at the right indicate positions of the ΔISDR and full-length BD-NS5A constructs. Positions of protein standards are shown in kilodaltons.
ISDR mutations abolish NS5A function.
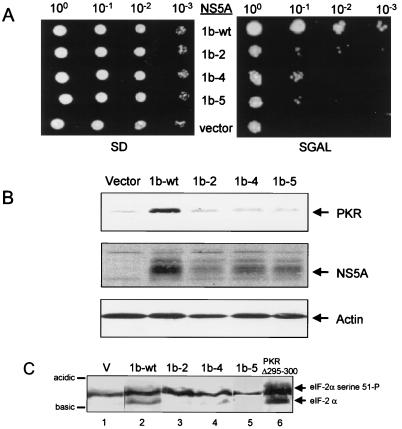
NS5A represses PKR function through a direct interaction with the protein kinase (27). Multiple ISDR mutations disrupt NS5A-PKR complex formation (Fig. 4). It was therefore essential to compare the abilities of wt and ISDR variants of NS5A to regulate PKR function in vivo. Expression plasmids encoding NS5A constructs 1b-wt, 1b-2, 1b-4, and 1b-5 under control of the GAL promoter were introduced into the gcn2Δ S. cerevisiae strain RY1-1 (63). This strain lacks the endogenous GCN2 protein kinase and harbors two integrated copies of wt human PKR under control of a galactose-inducible GAL-CYC hybrid promoter (10). When placed on galactose medium, mammalian PKR is expressed and phosphorylates serine 51 on the endogenous yeast eIF-2α, resulting in suppression of cell growth (63). We have demonstrated that NS5A 1a-wt can repress PKR function when coexpressed in strain RY1-1, resulting in reduced levels of eIF-2α phosphorylation, enhanced protein expression, and restoration of cell growth on galactose medium (27). Strains harboring an NS5A expression plasmid or the expression plasmid alone (vector; control) grew equally well when cell equivalents were serially plated onto noninducing medium containing dextrose as the sole carbon source (Fig. 5A, left). In contrast, only the strain coexpressing NS5A 1b-wt exhibited significant growth on inducing medium (Fig. 5A, right). By this method, we determined that strains coexpressing NS5A 1b-wt with PKR exhibited greater than a 100-fold increase in plating efficiency when grown on galactose medium compared to those strains coexpressing NS5A 1b-4 or 1b-5 and PKR. Thus, multiple ISDR mutations, corresponding to IFN-sensitive HCV, resulted in loss of the growth restoration phenotype associated with NS5A 1b-wt. In contrast, we observed only a 10-fold reduction in the plating efficiency of strains coexpressing PKR and NS5A 1b-2 (Fig. 5A). This may be due to a reduction in the PKR-binding affinity imposed by the A2224V point mutation within the NS5A 1b-2 ISDR, which does not completely abolish interaction with PKR (Fig. 4).
FIG. 5.
ISDR mutations abolish NS5A function. (A) Yeast growth assay. Cell equivalents of RY1-1 yeast strains harboring the galactose-inducible URA3 expression plasmid pYES-NS5A 1b-wt (1b-wt), pYES-NS5A 1b-2 (1b-2), pYES-NS5A 1b-4 (1b-4), or pYES-NS5A 1b-5 (1b-5) or the pYES control (vector) were serially diluted and spotted onto SD or SGAL medium. Panels show colony formation after 5 days growth at 30°C. (B) Immunoblot analysis of protein extracts prepared from the yeast strains shown in panel A, probed sequentially with anti-PKR, anti-NS5A, and anti-actin (control) monoclonal antibodies. Arrows at the right denote positions of PKR, NS5A, and actin. Each lane represents 50 μg of total protein. (C) eIF-2α phosphorylation. Extracts prepared from the yeast strains shown in panel A were separated by single-dimension IEF and blotted onto a nitrocellulose membrane. Detection of eIF-2α was facilitated by probing the blot with anti-yeast eIF-2α serum. Each lane represents 20 μg of protein prepared from RY1-1 cells harboring pYES (V; lane 1), pYES-NS5A 1b-wt (1b-wt; lane 2), pYES-NS5A 1b-2 (1b-2; lane 3), pYES-NS5A 1b-4 (1b-4; lane 4), pYES-NS5A 1b-5 (1b-5; lane 5), or pYex-PKRΔ295-300 (PKR Δ295-300 [control]; lane 6). Arrows at the right show positions of hypophosphorylated eIF-2α (lower) and hyperphosphorylated eIF-2α, which is phosphorylated by PKR on serine 51. Bars at the left identify the acidic and basic ends of the blot.
It is well established that inhibition of PKR results in stimulation of mRNA translation and higher levels of plasmid-encoded protein expression within yeast and mammalian cells (27, 39, 74). Such a relationship was confirmed in assays using the RY1-1 strains described above, which coexpressed PKR and ISDR variants of NS5A. Immunoblot analysis of extracts prepared from these strains revealed that PKR and the respective NS5A constructs were expressed in each strain (Fig. 5B). Importantly, this analysis revealed that the relative levels of PKR and NS5A 1b-wt were significantly increased in the corresponding strain, while levels remained unchanged among the strains harboring the comparatively nonfunctional NS5A 1b-2, 1b-4, or 1b-5. Consistent with previous results (25, 26), the steady-state levels of actin, which are not limiting under these experimental conditions, did not change. Importantly, we determined that all NS5A constructs were expressed to equal levels in a yeast isogenic control strain which lacks PKR (data not shown). The relative expression patterns of the NS5A constructs in RY1-1 reflected the growth properties of strains on galactose medium (compare Fig. 5A and B). These results indicate that NS5A 1b-wt repressed the translational regulatory properties of PKR in vivo, resulting in restoration of cell growth and stimulation of protein synthesis. The loss of function associated with the NS5A ISDR variants suggest that the PKR-regulatory properties of NS5A were disrupted by the introduction of mutations within the ISDR.
To directly determine the effects of ISDR mutations on the PKR-regulatory function of NS5A, we analyzed the endogenous in vivo phosphorylation state of the PKR substrate, eIF-2α. Repression of PKR function in RY1-1 yeast strains results in a reduction in the level of the hyperphosphorylated form of eIF-2α, phosphorylated exclusively by PKR on serine 51 (63). By using single-dimension IEF, the hyperphosphorylated form of eIF-2α can be electrophoretically separated from the less acidic, hypophosphorylated form, which lacks serine 51 phosphorylation (19). Figure 5C shows an immunoblot from an IEF gel of extracts prepared from the RY1-1 strains represented in Fig. 5, which was probed with antisera specific to yeast eIF-2α. As a control, we included extracts from strains harboring either the expression plasmid devoid of insert or the transdominant-negative PKR mutant, PKR Δ295-300 (Fig. 5, lane 1 or 6, respectively). PKR Δ295-300 inhibits wt PKR when coexpressed in yeast, resulting in reduced levels of serine 51 phosphorylation and restoration of cell growth when plated on galactose medium (63). As seen in Fig. 5C, strains expressing NS5A 1b-wt or PKR Δ295-300 exhibited a significant reduction in the level of hyperphosphorylated eIF-2α, as demonstrated by a concomitant increase in the abundance of the hypophosphorylated eIF-2α isoform relative to the vector control strain (compare lanes 1 and 2). Strains expressing the isogenic NS5A variant 1b-2, 1b-4, or 1b-5 possessed predominantly the hyperphosphorylated isoform of eIF-2α, similar to the vector control strain (Fig. 5C; compare lanes 3 to 5 with lane 1). The respective level of serine 51-phosphorylated eIF-2α corresponded with the growth phenotype of each strain on galactose medium (Fig. 5A), where expression of NS5A 1b-wt facilitated growth on galactose and a reduction in serine 51-phosphorylation. This phenotype was reversed by the introduction of ISDR mutations into NS5A. Our results, taken together, demonstrate that ISDR mutations which correspond to IFN-sensitive HCV (Table 3) can disrupt the PKR-regulatory properties of NS5A.
The NS5A-PKR interaction in mammalian cells is disrupted by ISDR mutations.
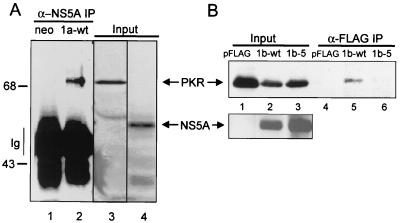
NS5A from IFN-resistant HCV represses the translational regulatory properties of PKR when expressed in mammalian cells (27). As suggested by the results from our yeast two-hybrid (Fig. 4) and growth (Fig. 5) assays, we predicted that the NS5A-directed repression of PKR occurring in mammalian cells was similarly mediated through a direct NS5A-PKR interaction. We therefore sought to determine if NS5A and PKR could form a stable complex in mammalian cells and what effect, if any, ISDR mutations had on complex formation. We carried out coimmunoprecipitation analyses from Cos-1 cells transiently cotransfected with plasmids encoding wt or ISDR variants of NS5A and full-length inactive human PKR. In these analyses, we used plasmids encoding NS5A from IFN-resistant HCV-1a (1a-wt), 1b-wt, and the 1b isogenic variant, 1b-5; the latter corresponding to IFN-sensitive HCV (Table 3). Immunoblot analysis of extracts prepared from cotransfected cells demonstrated that all constructs were efficiently expressed within 48 h of transfection (Fig. 6). PKR was recovered from anti-NS5A immunoprecipitates prepared from cells cotransfected with NS5A 1a-wt and from anti-FLAG immunoprecipitates prepared from cells harboring FLAG-tagged NS5A 1b-wt (Fig. 6A and B, respectively). In comparison, no PKR was detected in immunoprecipitates of FLAG-tagged NS5A 1b-5 (compare lanes 5 and 6 in Fig. 6B). Recovery of PKR was dependent on the presence of NS5A in the extract, as we failed to detect PKR in NS5A-specific immunoprecipitates prepared from extracts lacking wt NS5A (Fig. 6A and B, lanes 1 and 4, respectively). These results demonstrate that NS5A from IFN-resistant strains of HCV-1a and HCV-1b can form a stable and specific complex with PKR when expressed in mammalian cells. Consistent with our yeast two-hybrid results (Fig. 4), mutations within the ISDR which correspond to IFN-sensitive HCV (Table 3) disrupted the NS5A-PKR interaction within mammalian cells. The consistency between these results and those observed in our yeast studies (Fig. 4 and 5) validates the yeast system as a viable model for the study of NS5A-PKR interaction and the effects of this interaction on PKR function.
FIG. 6.
ISDR mutations disrupt the NS5A-PKR association in mammalian cells. Cos-1 cells were cotransfected with cytomegalovirus expression plasmids encoding PKR K296R and NS5A or with PKR K296R and the vector control. Extracts were prepared and mixed with anti-NS5A monoclonal antibody (A) or anti-FLAG resin (B). (A) Anti-NS5A immunocomplexes prepared from extracts harboring PKR K296R with vector control (neo; lane 1) or NS5A 1a-wt (1a-wt; lane 2) and input extract (Input; lanes 3 and 4) were separated by SDS-PAGE and subjected to immunoblot analysis using anti-PKR (lanes 1 to 3) or anti-NS5A (lane 4) monoclonal antibody. Lanes 3 and 4 represent the starting material from the immunoprecipitation (IP) reaction shown in lane 2. The vertical line at left indicates the broad band corresponding to the immunoglobulin (Ig) heavy chain. Positions of protein standards are indicated in kilodaltons. (B) Immunoblot analysis of input protein (lanes 1 to 3) or protein complexes (lanes 4 to 6) recovered by mixing extracts harboring PKR K296R with vector alone (pFLAG; lanes 1 and 4), FLAG-NS5A 1b-wt (1b-wt; lanes 2 and 5), or FLAG-NS5A 1b-5 (1b-5; lanes 3 and 6) with anti-FLAG resin. Blots were probed with a monoclonal antibody specific to human PKR (top) or NS5A (bottom). Arrows point to PKR and NS5A.
DISCUSSION
Mechanism of PKR regulation: disruption of PKR dimerization by NS5A.
Molecular studies of HCV have been hampered by the lack of a suitable tissue culture system in which to support viral replication in vitro, though HCV has been successfully passaged in a chimpanzee model (41). To overcome this deficiency, we have developed a series of reliable yeast and mammalian cell systems to study NS5A function and PKR regulation in vivo (27) and used them to determine the contribution of the ISDR in NS5A-mediated regulation of PKR.
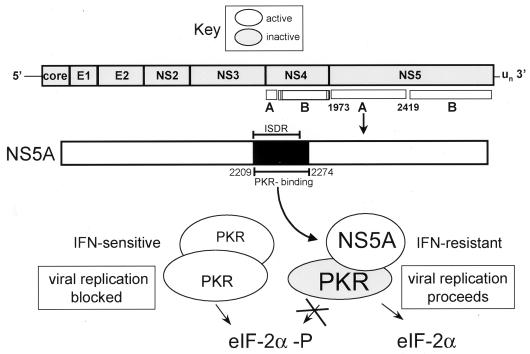
Activation of PKR is considered to be dependent on the ability of the kinase to dimerize and autophosphorylate in trans (2, 56, 57). This is supported by previous work which determined that active PKR resides as a dimer within mammalian cell extracts (46) and that the kinase forms a stable homodimeric complex in solution (9). Recent results indicate that PKR dimerization occurs via a mutually dependent two-step process involving dsRNA-dependent and -independent mechanisms (15, 55, 57), the latter mediated through PKR aa 244 to 296 (73). We propose a model for PKR regulation during HCV infection in which NS5A targets the PKR dimerization process by binding to within aa 244 to 296 of PKR (Fig. 1) by an ISDR-dependent mechanism (Fig. 7). Within an HCV-infected cell, viral quasispecies containing an ISDR sequence similar to the IFN-resistant HCV-J prototype (36) can persist during the course of IFN therapy through NS5A-mediated repression of PKR. In this case, the virus-encoded NS5A polyprotein cleavage product binds to PKR, targeting a kinase dimerization domain defined by PKR aa 244 to 296. Through sequences encoded within a 62-aa region spanning the ISDR and the adjacent C-terminal 26 aa (Fig. 3), NS5A disrupts the critical PKR dimerization process which is required for catalytic activity. Our data indicate that disruption of PKR dimerization results in repression of PKR function and a block in PKR-mediated eIF-2α phosphorylation within the host cell. NS5A-mediated repression of PKR thereby removes the PKR-imposed block on mRNA translation and viral replication induced by cellular exposure to IFN, thus allowing HCV to resist the antiviral effects of IFN (Fig. 7, lower right).
FIG. 7.
Role of NS5A in PKR regulation during HCV infection. HCV sensitivity to IFN is determined, at least in part, by the structure of the PKR-binding domain (dark region) within the NS5A cleavage product of the HCV polyprotein. During HCV infection NS5A from wt, IFN-resistant strains of HCV binds PKR, disrupting the critical PKR dimerization process. Resulting PKR monomers are unable to phosphorylate eIF-2α, and thus viral replication proceeds unobstructed (lower right). Mutations within the 66-aa PKR-binding region of NS5A, including the ISDR (indicated by bars), abolish the PKR-regulatory properties of HCV, rendering the virus sensitive to the antiviral actions of IFN. In this case, PKR remains active in a dimeric state and phosphorylates eIF-2α to inhibit mRNA translation and viral replication (lower left).
Conversely, those viral quasispecies exhibiting ISDR sequence divergence from the prototypic HCV J strain lack the ability to disrupt the PKR dimerization process and to repress PKR function within the HCV-infected host cell (Fig. 7, lower left). ISDR variants of HCV lacking the ability to bind PKR, and/or to disrupt PKR dimerization, are thereby rendered sensitive to the antiviral effects of IFN mediated through the translational regulatory and growth-suppressive properties of PKR (Fig. 7, lower left). Moreover, NS5A function may be controlled in part by posttranslational modifications which occur within the infected host cell. NS5A resides within the cell as a phosphoprotein, present in a variety of hypo- and hyperphosphorylated states (35, 75). Our results and those of others suggest that phosphorylation of NS5A occurs by a PKR-independent process (27) and may be mediated by a CMGC-like protein kinase or a cyclic AMP-dependent protein kinase activity (34, 62). Factors which modulate such activities may thus lead to regulation of NS5A function, possibly including NS5A-mediated repression of PKR.
NS5A defines a novel class of PKR inhibitors.
PKR plays a central role within the cellular response to IFN by limiting mRNA translation and transducing IFN-mediated signals which are required for establishment of the comprehensive IFN-induced antiviral state (44, 53; reviewed in reference 67). To avoid the IFN response, many viruses encode mechanisms to disrupt PKR function which target distinct steps within the PKR maturation, regulatory, and catalytic processes. The mechanisms by which virus-directed inhibitors disrupt PKR function can be classified into five broad categories: those which (i) interfere with dsRNA-mediated PKR activation, (ii) block PKR-substrate interactions, (iii) modulate the physical levels of PKR, (iv) dephosphorylate eIF-2α and/or modulate events downstream of eIF-2α, or (v) disrupt PKR dimerization (reviewed in reference 24). Our results indicate that NS5A belongs to this latter group of PKR inhibitors, which also includes the cellular oncoprotein P58IPK (25, 73). Indeed, both of these inhibitors bind to sites within the same dimerization domain of PKR (Fig. 1), resulting in repression of PKR function (26, 27). Disruption of PKR dimerization was specific to NS5A and was not observed in parallel analyses of other viral inhibitors of PKR which target distinct regions of the kinase (Fig. 2). Our results support previous studies indicating that PKR dimerization is a requisite step for catalytic function (15, 57) and identify the PKR dimerization process as a key element in the regulation of PKR function.
NS5A, PKR regulation, and IFN sensitivity: redefining the ISDR.
HCV infection is currently treated by parenteral administration of type I IFN, the only therapeutic approved for this disease (23). While high IFN response rates are associated with HCV genotypes 2 to 4, a significantly lower rate of response is observed within those individuals infected with genotype 1, suggesting that HCV encodes an IFN resistance mechanism(s) which is genotype 1 specific (76). Recent molecular epidemiological studies from Japan have identified the ISDR as a conserved region within the HCV genome of some IFN-resistant strains of HCV-1b; within Japanese patients, mutations within the ISDR of NS5A have been associated with increased HCV sensitivity to IFN (reviewed in reference 29). We previously determined that NS5A from IFN-resistant HCV-1a and -1b strains could bind to PKR to repress kinase function in vivo, a process that was dependent on the NS5A ISDR (27). Using a series of NS5A ISDR variants isogenic to NS5A 1b-wt, we have now demonstrated that mutations within the ISDR can abrogate the PKR-regulatory properties of NS5A in vivo, which, interestingly, may be dependent on NS5A sequences both within and proximal to the ISDR. The use of isogenic ISDR variants of NS5A in these studies allows us to attribute loss of NS5A function to mutations within the ISDR. However, due to the quasispecies nature of HCV, the possibility remains that mutations in other regions of NS5A outside the ISDR also contribute to loss of NS5A function.
It has been suggested that the sequence of the ISDR in HCV isolated from patient serum may be of predictive value in predetermining the therapeutic efficacy of IFN for specific clinical cases (12, 21). Recent controversy surrounds these observations due to a lack of correlation between ISDR sequence and IFN sensitivity of HCV isolates from western Europe and the United States (30, 40, 78, 58). These difference may reflect distinct geographical features of the HCV isolates or may be due simply to variations in study parameters, IFN-dosing regimens, and/or viral genotypes examined (29). Although the present study provides strong molecular evidence to support a role for NS5A in IFN resistance, it is possible that genetic differences outside the ISDR or NS5A contribute to IFN resistance in some HCV isolates (58). Our data emphasize the need to examine additional sequences within NS5A in addition to the ISDR as originally defined. Our results argue for redefining the ISDR to include, at least, the entire PKR-binding region of NS5A (NS5A aa 2209 to 2274). Determination of sequence variations in this redefined ISDR within liver-replicating viral quasispecies may allow for the further identification of critical amino acids residues within NS5A that are required for the repression of PKR function and resistance to the antiviral effects of IFN.
PKR regulation and HCV pathogenesis.
In addition to its antiviral function, PKR has been identified as a critical component in dsRNA and IFN-induced signaling processes and in transcriptional regulation and as an effector of apoptosis (reviewed by Williams [77]). Moreover, several independent studies have implicated PKR as a tumor suppressor, a property that is dependent on its ability to phosphorylate eIF-2α (3, 4, 7, 42, 54). Thus, NS5A-directed repression of PKR not only has implications for how HCV responds to IFN therapy but also suggests that NS5A may deregulate other PKR-dependent cellular processes. Recent results from our laboratory suggest that constitutive repression of PKR by NS5A disrupts PKR-dependent apoptosis and cell growth control (unpublished observations). Determining the long-term cellular effects from NS5A-mediated PKR repression will further our understanding of HCV pathogenesis during chronic infection.
ACKNOWLEDGMENTS
We thank Pat McGifford and the University of Washington photography service for excellent data photos. We are grateful to Dagma Daniel for excellent administrative support. We thank T. Dever (National Institute of Child Health and Human Development) and T. Imagawa (Osaka University) for antibodies to yeast eIF-2α and NS5A, respectively.
This work was supported in part by National Institutes of Health grants AI22646, RR00166, and AI41629 (M.G.K.) and AI41320-02 and AI39049-02 (D.R.G.) and by grants from the University of Washington Royalty Research Fund, Schering Plough, and Ribogene Corporation, Hayward, Calif. M.D. is a Howard Hughes undergraduate research fellow. M.G. is supported by the Helen Hay Whitney Foundation.
REFERENCES
- 1.Alter M. Epidemiology of hepatitis C in the west. Semin Liver Dis. 1995;15:5–14. doi: 10.1055/s-2007-1007259. [DOI] [PubMed] [Google Scholar]
- 2.Barber G N, Jagus R, Meurs E F, Hovanessian A G, Katze M G. Molecular mechanisms responsible for malignant transformation by regulatory and catalytic domain variants of the interferon-induced enzyme RNA-dependent protein kinase. J Biol Chem. 1995;270:17423–17428. doi: 10.1074/jbc.270.29.17423. [DOI] [PubMed] [Google Scholar]
- 3.Barber G N, Thompson S, Lee T G, Strom T, Jagus R, Darveau A, Katze M G. The 58-kilodalton inhibitor of the interferon-induced double-stranded RNA-activated protein kinase is a tetratricopeptide repeat protein with oncogenic properties. Proc Natl Acad Sci USA. 1994;91:4278–4282. doi: 10.1073/pnas.91.10.4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barber G N, Wambach M, Thompson S, Jagus R, Katze M G. Mutants of the RNA-dependent protein kinase (PKR) lacking double-stranded RNA binding domain I can act as transdominant inhibitors and induce malignant transformation. Mol Cell Biol. 1995;15:3138–3146. doi: 10.1128/mcb.15.6.3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barber G N, Wambach M, Wong M-L, Dever T E, Hinnebusch A G, Katze M G. Translational regulation by the interferon-induced double-stranded-RNA-activated 68-kDa protein kinase. Proc Natl Acad Sci USA. 1993;90:4621–4625. doi: 10.1073/pnas.90.10.4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartel P L, Chien C-T, Sternglanz R, Fields S. Using the two-hybrid system to detect protein-protein interactions. In: Hartley D A, editor. Cellular interactions in development: a practical approach. Oxford, England: Oxford University Press; 1993. pp. 153–179. [Google Scholar]
- 7.Benkirane M, Neuveut C, Chun R F, Smith S M, Samuel C E, Gatignol A, Jeang K-T. Oncogenic potential of TAR RNA binding protein TRBP and its regulatory interaction with RNA-dependent protein kinase PKR. EMBO J. 1997;16:611–624. doi: 10.1093/emboj/16.3.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borowski P, Oehlmann K, Heiland M, Laufs R. Nonstructural protein 3 of hepatitis C virus blocks the distribution of free catalytic subunit of cyclic AMP-dependent protein kinase. J Virol. 1997;71:2838–2843. doi: 10.1128/jvi.71.4.2838-2843.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carpick B W, Graziano V, Schneider D, Maitra R K, Lee X, Williams B R G. Characterization of the solution structure between the interferon-induced double-stranded RNA-activated protein kinase and HIV-1 transactivating region RNA. J Biol Chem. 1997;272:9510–9516. doi: 10.1074/jbc.272.14.9510. [DOI] [PubMed] [Google Scholar]
- 10.Cesareni G, Murray J A H. Plasmid vectors carrying the replication origin of filamentous single-stranded phages. In: Setlow J K, editor. Genetic engineering: principles and methods. New York, N.Y: Plenum Press; 1987. pp. 135–154. [Google Scholar]
- 11.Chang J, Yang S-H, Cho Y-G, Hwang S B, Hahn Y S, Sung Y C. Hepatitis C virus core from two different genotypes has an oncogenic potential but is not sufficient for transforming primary rat embryo fibroblasts in cooperation with the H-ras oncogene. J Virol. 1998;72:3060–3065. doi: 10.1128/jvi.72.4.3060-3065.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chayama K, Tsubota A, Kobayashi M, Okamoto K, Hashimoto M, Miyano Y, Koike H, Koida I, Arase Y, Saitoh S, Suzuki Y, Murashima N, Ikeda K, Kumada H. Pretreatment virus load and multiple amino acid substitutions in the interferon sensitivity-determining region predict the outcome of interferon treatment in patients with chronic genotype 1b hepatitis C virus infection. Hepatology. 1997;25:745–749. doi: 10.1002/hep.510250342. [DOI] [PubMed] [Google Scholar]
- 13.Clemens M J, Elia A. The double-stranded RNA-dependent protein kinase PKR: structure and function. J Interferon Cytokine Res. 1997;17:503–524. doi: 10.1089/jir.1997.17.503. [DOI] [PubMed] [Google Scholar]
- 14.Clements J E, Zink M C. Molecular biology and pathogenesis of animal lentivirus infections. Clin Microbiol Rev. 1996;9:100–117. doi: 10.1128/cmr.9.1.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cosentino G P, Venkatesan S, Serluca F C, Green S R, Mathews M B, Sonenberg N. Double-stranded-RNA-dependent protein kinase and TAR RNA-binding protein form homo- and heterodimers in vivo. Proc Natl Acad Sci USA. 1995;92:9445–9449. doi: 10.1073/pnas.92.21.9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Craig A W, Cosentino G P, Donzé O, Sonenberg N. The kinase insert domain of interferon-induced protein kinase PKR is required for activity but not for interaction with the pseudosubstrate K3L. J Biol Chem. 1996;271:24526–24533. doi: 10.1074/jbc.271.40.24526. [DOI] [PubMed] [Google Scholar]
- 17.Cuthbert J A. Hepatitis C: progress and problems. Clin Microbiol Rev. 1994;7:505–532. doi: 10.1128/cmr.7.4.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Der S D, Yang Y-L, Weissman C, Williams B R G. A double-stranded RNA-activated protein kinase-dependent pathway mediating stress-induced apoptosis. Proc Natl Acad Sci USA. 1997;94:3279–3283. doi: 10.1073/pnas.94.7.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dever T E, Chen J-J, Barber G N, Cigan A M, Feng L, Donahue T F, London I M, Katze M G, Hinnebusch A G. Mammalian eukaryotic initiation factor eIF2α kinases functionally substitute for GCN2 protein kinase in the GCN4 translational control mechanism of yeast. Proc Natl Acad Sci USA. 1993;90:4616–4620. doi: 10.1073/pnas.90.10.4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Di Bisceglie A M. Hepatitis C and hepatocellular carcinoma. Semin Liver Dis. 1995;15:64–69. doi: 10.1055/s-2007-1007263. [DOI] [PubMed] [Google Scholar]
- 21.Enomoto N, Sakuma I, Asahina Y, Kurosaki M, Murakami T, Yamamoto C, Ogura Y, Izumi N, Maruno F, Sato C. Mutations in the nonstructural protein 5A gene and response to interferon in patients with chronic hepatitis C virus 1b infection. N Engl J Med. 1996;334:77–81. doi: 10.1056/NEJM199601113340203. [DOI] [PubMed] [Google Scholar]
- 22.Enomoto N, Sakuma I, Asahina Y, Kurosaki M, Murankami T, Yamamoto C, Izumi N, Marumo F, Sato C. Comparison of full-length sequences of interferon-sensitive and resistant hepatitis C virus 1b. J Clin Investig. 1995;96:224–230. doi: 10.1172/JCI118025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fried M, Hoofnagle J. Therapy of hepatitis C. Semin Liver Dis. 1995;15:82–91. doi: 10.1055/s-2007-1007265. [DOI] [PubMed] [Google Scholar]
- 24.Gale M, Jr, Katze M G. Molecular mechanisms of interferon resistance mediated by viral-directed inhibition of PKR, the interferon-induced protein kinase. Pharmacol Ther. 1998;78:29–46. doi: 10.1016/s0163-7258(97)00165-4. [DOI] [PubMed] [Google Scholar]
- 25.Gale M, Jr, Tan S-L, Wambach M, Katze M G. Interaction of the interferon-induced PKR protein kinase with inhibitory proteins P58IPK and vaccinia virus K3L is mediated by unique domains: implications for kinase regulation. Mol Cell Biol. 1996;16:4172–4181. doi: 10.1128/mcb.16.8.4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gale M, Jr, Blakely C M, Hopkins D A, Melville M W, Wambach M, Romano P R, Katze M G. Regulation of interferon-induced protein kinase PKR: modulation of P58IPK inhibitory function by a novel protein, P52rIPK. Mol Cell Biol. 1998;18:859–871. doi: 10.1128/mcb.18.2.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gale M, Jr, Korth M J, Tang N M, Tan S L, Hopkins D A, Dever T E, Polyak S J, Gretch D R, Katze M G. Evidence that hepatitis C virus resistance to interferon is mediated through repression of the PKR protein kinase by the nonstructural 5A protein. Virology. 1997;230:217–227. doi: 10.1006/viro.1997.8493. [DOI] [PubMed] [Google Scholar]
- 28.Hanks S K, Quinn A M, Hunter T. The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science. 1988;241:42–52. doi: 10.1126/science.3291115. [DOI] [PubMed] [Google Scholar]
- 29.Herion D, Hoofnagle J. The interferon sensitivity determining region: all hepatitis C virus isolates are not the same. Hepatology. 1997;25:769–771. doi: 10.1002/hep.510250346. [DOI] [PubMed] [Google Scholar]
- 30.Hofgärtner W T, Polyak S J, Sullivan D, Carithers R L, Jr, Gretch D R. Mutations in the NS5A gene of hepatitis C virus in North American patients infected with HCV genotype 1a or 1b. J Med Virol. 1997;53:118–126. doi: 10.1002/(sici)1096-9071(199710)53:2<118::aid-jmv3>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 31.Houghton M, Weiner A, Han J, Kuo G, Choo Q-L. Molecular biology of the hepatitis C viruses: implications for diagnosis, development and control of viral disease. Hepatology. 1991;14:381–388. [PubMed] [Google Scholar]
- 32.Hu J C. Repressor fusions as a tool to study protein-protein interactions. Structure. 1995;3:431–433. doi: 10.1016/s0969-2126(01)00176-9. [DOI] [PubMed] [Google Scholar]
- 33.Hu J C, O’Shea E K, Kim P S, Saur R T. Sequence requirements for coiled-coils: analysis with lambda repressor-GCN4 leucine zipper fusions. Science. 1990;250:1400–1403. doi: 10.1126/science.2147779. [DOI] [PubMed] [Google Scholar]
- 34.Ide Y, Tanimoto A, Sasaguri Y, Padmanabhan R. Hepatitis C virus NS5A protein is phosphorylated in vitro by a stably bound protein kinase from HeLa cells and by cAMP-dependent protein kinase A-alpha catalytic subunit. Gene. 1997;201:151–158. doi: 10.1016/s0378-1119(97)00440-x. [DOI] [PubMed] [Google Scholar]
- 35.Kaneko T, Tanji Y, Satoh S, Hijikata M, Asabe S, Kimura K, Shimotohno K. Production of two phosphoproteins from the NS5A region of the hepatitis C viral genome. Biochem Biophys Res Commun. 1994;205:320–326. doi: 10.1006/bbrc.1994.2667. [DOI] [PubMed] [Google Scholar]
- 36.Kato N, Hijikata M, Ootsuyama Y, Nakagawa M, Ohkoshi S, Sugimura T, Shimotohno K. Molecular cloning of the human hepatitis C virus genome from Japanese patients with non-A, non-B hepatitis. Proc Natl Acad Sci USA. 1990;87:9524–9528. doi: 10.1073/pnas.87.24.9524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Katze M G. The war against the interferon-induced dsRNA-activated protein kinase: can viruses win? J Interferon Res. 1992;12:241–248. doi: 10.1089/jir.1992.12.241. [DOI] [PubMed] [Google Scholar]
- 38.Katze M G, Wambach M, Wong M-L, Garfinkel M S, Meurs E, Chong K L, Williams B R G, Hovanessian A G, Barber G N. Functional expression of interferon-induced, double-stranded RNA-activated 68,000-Mr protein kinase in a cell-free system. Mol Cell Biol. 1991;11:5497–5505. doi: 10.1128/mcb.11.11.5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kawagishi-Kobayashi M, Silverman J B, Ung T L, Dever T E. Regulation of the protein kinase PKR by the vaccinia virus pseudosubstrate inhibitor K3L is dependent on residues conserved between the K3L protein and the PKR substrate eIF-2α. Mol Cell Biol. 1997;17:4146–4158. doi: 10.1128/mcb.17.7.4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Khorsi H, Castelain S, Wyseur A, Izopet J, Canva V, Rombout A, Capron D, Capron J-P, Lunel F, Stuyver L, Duverlie G. Mutations of hepatitis C virus 1b NS5A 2209-2248 amino acid sequence do not predict the response to recombinant interferon-alpha therapy in French patients. J Hepatol. 1997;27:72–77. doi: 10.1016/s0168-8278(97)80282-6. [DOI] [PubMed] [Google Scholar]
- 41.Kolykhalov A A, Agapov E V, Blight K J, Mihalik K, Feinstone S M, Rice C M. Transmission of hepatitis C by intrahepatic inoculation with transcribed RNA. Science. 1997;277:570–574. doi: 10.1126/science.277.5325.570. [DOI] [PubMed] [Google Scholar]
- 42.Koromilas A E, Roy S, Barber G N, Katze M G, Sonenberg N. Malignant transformation by a mutant of the IFN-inducible dsRNA-dependent protein kinase. Science. 1992;257:1685–1689. doi: 10.1126/science.1382315. [DOI] [PubMed] [Google Scholar]
- 43.Kumar A, Haque J, Lacoste J, Hiscott J, Williams B R G. Double-stranded RNA-dependent protein kinase activates transcription factor NF-κB by phosphorylating IκB. Proc Natl Acad Sci USA. 1994;91:6288–6292. doi: 10.1073/pnas.91.14.6288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kumar A, Yang Y-L, Flati V, Der S, Kadereit S, Deb A, Haque J, Reis L, Weissmann C, Williams B R G. Deficient cytokine signaling in mouse embryo fibroblasts with a targeted deletion in the PKR gene: role of IRF-1 and NF-κB. EMBO J. 1997;16:406–416. doi: 10.1093/emboj/16.2.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kurosaki M, Enomoto N, Murakami T, Sakuma I, Asahina Y, Yamamoto C, Ikeda T, Tozuka S, Izumi N, Marumo F, Sato C. Analysis of genotypes and amino acid residues 2209 to 2248 of the NS5A region of hepatitis C virus in relation to the response to interferon-β therapy. Hepatology. 1997;25:750–753. doi: 10.1002/hep.510250343. [DOI] [PubMed] [Google Scholar]
- 46.Langland J O, Jacobs B L. Cytosolic double-stranded RNA-dependent protein kinase is likely a dimer of partially phosphorylated Mr 66,000 subunits. J Biol Chem. 1992;267:10729–10736. [PubMed] [Google Scholar]
- 47.Laurent A G, Krust B, Galabru J, Svab J, Hovanessian A G. Monoclonal antibodies to interferon induced 68,000 Mr protein and their use for the detection of double-stranded RNA dependent protein kinase in human cells. Proc Natl Acad Sci USA. 1985;82:4341–4345. doi: 10.1073/pnas.82.13.4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee T G, Tang N, Thompson S, Miller J, Katze M G. The 58,000-Dalton cellular inhibitor of the interferon-induced double-stranded RNA-activated protein kinase (PKR) is a member of the tetratricopeptide repeat family of proteins. Mol Cell Biol. 1994;14:2331–2342. doi: 10.1128/mcb.14.4.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mansell C, Locarnini S. Epidemiology of hepatitis C in the east. Semin Liver Dis. 1995;15:15–32. doi: 10.1055/s-2007-1007260. [DOI] [PubMed] [Google Scholar]
- 50.Matsumoto M, Hsieh T-Y, Zhu N, VanArsdale T, Hwang S B, Gorbalenya A E, Lo S-Y, Ou J-H, Ware C F, Lai M C. Hepatitis C virus core protein interacts with the cytoplasmic tail of the lymphotoxin-β receptor. J Virol. 1997;71:1301–1309. doi: 10.1128/jvi.71.2.1301-1309.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Merrick W C, Hershey J W B. The pathway and mechanism of eukaryotic protein synthesis. In: Hershey J, Mathews M, Sonenberg N, editors. Translational control. Cold Spring Harbor, New York, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 31–70. [Google Scholar]
- 52.Meurs E, Chong K L, Galabru J, Thomas N, Kerr I, Williams B R G, Hovanessian A G. Molecular cloning and characterization of the human double-stranded RNA-activated protein kinase induced by interferon. Cell. 1990;62:379–390. doi: 10.1016/0092-8674(90)90374-n. [DOI] [PubMed] [Google Scholar]
- 53.Meurs E, Watanabe Y, Barber G N, Katze M G, Chong K L, Williams B R G, Hovanessian A G. Constitutive expression of human double-stranded RNA-activated p68 kinase in murine cells mediates phosphorylation of eukaryotic initiation factor 2 and partial resistance to encephalomyocarditis virus growth. J Virol. 1992;66:5805–5814. doi: 10.1128/jvi.66.10.5805-5814.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meurs E F, Galabru J, Barber G N, Katze M G, Hovanessian A G. Tumor suppressor function of the interferon-induced double-stranded RNA-activated protein kinase. Proc Natl Acad Sci USA. 1993;90:232–236. doi: 10.1073/pnas.90.1.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ortega L G, McCotter M D, Henry G L, McCormack S J, Thomis D C, Samuel C E. Mechanism of interferon action. Biochemical and genetic evidence for the intermolecular association of the RNA-dependent protein kinase PKR from human cells. Virology. 1996;215:31–39. doi: 10.1006/viro.1996.0004. [DOI] [PubMed] [Google Scholar]
- 56.Patel R C, Stanton P, Sen G C. Role of the amino-terminal residues of the interferon-induced protein kinase in its activation by double-stranded RNA and heparin. J Biol Chem. 1994;269:18593–18598. [PubMed] [Google Scholar]
- 57.Patel R C, Stanton P, Sen G C. Specific mutations near the amino terminus of double-stranded RNA-dependent protein kinase (PKR) differentially affect its double-stranded RNA binding and dimerization properties. J Biol Chem. 1996;271:25657–25663. doi: 10.1074/jbc.271.41.25657. [DOI] [PubMed] [Google Scholar]
- 58.Polyak S J, McCardle S, Lui S-L, Sullivan D, Chung M, Hofgärtner W T, Carithers R, McMahon B J, Mullins J I, Corey L, Gretch D R. Evolution of hepatitis C virus quasispecies in hypervariable region 1 and the putative interferon sensitivity-determining region during interferon therapy and natural infection. J Virol. 1998;72:4288–4296. doi: 10.1128/jvi.72.5.4288-4296.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Proud C G. PKR: a new name and new roles. Trends Biochem Sci. 1995;20:241–246. doi: 10.1016/s0968-0004(00)89025-8. [DOI] [PubMed] [Google Scholar]
- 60.Ray R B, Meyer K, Ray R. Suppression of apoptotic cell death by hepatitis C virus core protein. Virology. 1996;226:176–182. doi: 10.1006/viro.1996.0644. [DOI] [PubMed] [Google Scholar]
- 61.Ray R B, Lagging L M, Meyer K, Ray R. Hepatitis C virus core protein cooperates with ras and transforms primary rat embryo fibroblasts to tumorigenic phenotype. J Virol. 1996;70:4438–4443. doi: 10.1128/jvi.70.7.4438-4443.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Reed K E, Xu J, Rice C M. Phosphorylation of the hepatitis C virus NS5A protein in vitro and in vivo: properties of the NS5A-associated kinase. J Virol. 1997;71:7187–7197. doi: 10.1128/jvi.71.10.7187-7197.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Romano P R, Green S R, Barber G N, Mathews M B, Hinnebusch A G. Structural requirements for double-stranded RNA binding, dimerization, and activation of the human eIF-2α kinase DAI in Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:365–378. doi: 10.1128/mcb.15.1.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ruggieri A, Harada T, Matsuura Y, Miyamura T. Sensitization to Fas-mediated apoptosis by hepatitis C virus core protein. Virology. 1997;229:68–76. doi: 10.1006/viro.1996.8420. [DOI] [PubMed] [Google Scholar]
- 65.Sakamuro D, Furukawa T, Takegami T. Hepatitis C virus nonstructural protein NS3 transforms NIH 3T3 cells. J Virol. 1995;69:3893–3896. doi: 10.1128/jvi.69.6.3893-3896.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, New York, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 67.Samuel C E. Antiviral actions of interferon: interferon-regulated cellular proteins and their surprisingly selective antiviral activities. Virology. 1991;183:1–11. doi: 10.1016/0042-6822(91)90112-o. [DOI] [PubMed] [Google Scholar]
- 68.Sen G C, Lengyel P. The interferon system: a bird’s eye view of its biochemistry. J Biol Chem. 1992;267:5017–5020. [PubMed] [Google Scholar]
- 69.Sen G C, Ransohoff R M. Interferon-induced antiviral actions and their regulation. Adv Virus Res. 1993;42:57–102. doi: 10.1016/s0065-3527(08)60083-4. [DOI] [PubMed] [Google Scholar]
- 70.Shukla D D, Hoyne P A, Ward C W. Evaluation of complete genome sequences and sequences of individual gene products for the classification of hepatitis C viruses. Arch Virol. 1995;140:1747–1761. doi: 10.1007/BF01384339. [DOI] [PubMed] [Google Scholar]
- 71.Simmonds P. Variability of hepatitis C virus. Hepatology. 1995;21:570–583. doi: 10.1002/hep.1840210243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Smith D B, Johnson K S. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione-S-transferase. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- 73.Tan S-L, Gale M, Jr, Katze M G. Double-stranded RNA-independent dimerization of interferon-induced protein kinase PKR and inhibition of dimerization by the cellular P58IPK inhibitor. Mol Cell Biol. 1998;18:2431–2443. doi: 10.1128/mcb.18.5.2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tang N M, Ho C Y, Katze M G. The 58-kDa cellular inhibitor of the double stranded RNA-dependent protein kinase requires the tetratricopeptide repeat 6 and DnaJ motifs to stimulate protein synthesis in vivo. J Biol Chem. 1996;271:28660–28666. doi: 10.1074/jbc.271.45.28660. [DOI] [PubMed] [Google Scholar]
- 75.Tanji Y, Kaneko T, Satoh S, Shimotohno K. Phosphorylation of hepatitis C virus-encoded nonstructural protein NS5A. J Virol. 1995;69:3980–3986. doi: 10.1128/jvi.69.7.3980-3986.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tsubota A, Chayama K, Ikeda K, Yasuji A, Koida I, Saitoh S, Hashimoto M, Iwasaki S, Kobayashi M, Hiromitsu K. Factors predictive of response to interferon-α therapy in hepatitis C virus infection. Hepatology. 1994;19:1088–1094. [PubMed] [Google Scholar]
- 77.Williams B R G. The role of the dsRNA-activated kinase, PKR, in signal transduction. Semin Virol. 1995;6:191–202. [Google Scholar]
- 78.Zeuzem S, Lee J-H, Roth W K. Mutations in the nonstructural 5A gene of European hepatitis C virus isolates and response to interferon alfa. Hepatology. 1997;25:740–744. doi: 10.1002/hep.510250341. [DOI] [PubMed] [Google Scholar]
- 79.Zhu N, Khoshan A, Schneider R, Matsumoto M, Dennert G, Ware C F, Lai M C. Hepatitis C virus core protein binds to the cytoplasmic domain of tumor necrosis factor (TNF) receptor 1 and enhances TNF-induced apoptosis. J Virol. 1998;72:3691–3697. doi: 10.1128/jvi.72.5.3691-3697.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]