Abstract
By binding to serine-phosphorylated proteins, 14-3-3 proteins function as effectors of serine phosphorylation. The exact mechanism of their action is, however, still largely unknown. Here we demonstrate a requirement for 14-3-3 for Raf-1 kinase activity and phosphorylation. Expression of dominant negative forms of 14-3-3 resulted in the loss of a critical Raf-1 phosphorylation, while overexpression of 14-3-3 resulted in enhanced phosphorylation of this site. 14-3-3 levels, therefore, regulate the stoichiometry of Raf-1 phosphorylation and its potential activity in the cell. Phosphorylation of Raf-1, however, was insufficient by itself for kinase activity. Removal of 14-3-3 from phosphorylated Raf abrogated kinase activity, whereas addition of 14-3-3 restored it. This supports a paradigm in which the effects of phosphorylation on serine as well as tyrosine residues are mediated by inducible protein-protein interactions.
Members of the 14-3-3 family of proteins are highly conserved proteins which are expressed in all eukaryotic cells (reviewed in reference 1). They are thought to play important roles in a variety of signal transduction pathways, including those involved in cell cycle regulation and cell survival. Because 14-3-3 proteins bind to specific phosphoserine-containing sequences (44), they are likely to have an important role in signaling pathways mediated by serine/threonine protein kinases.
Many proteins are known to bind to 14-3-3 proteins, and the list of proteins continues to expand. In a few cases, the function of 14-3-3 binding is known. For example, 14-3-3 binding to the proapoptotic protein BAD blocks the interaction of BAD with the antiapoptotic protein BCL2 (65). In another case, 14-3-3 binding to the Cdc25C phosphatase prevents it from activating the Cdc2 kinase (49). In most cases, however, the effect of 14-3-3 binding is unknown. In this study, we have focused on the contribution made by 14-3-3 binding to Raf-1 kinase activity.
The Raf-1 kinase plays a central role in the signal transduction pathway induced by growth factors (reviewed in reference 3). In the cell, activation of Ras recruits Raf-1 to the plasma membrane, where it becomes activated (32, 57). Raf-1 in turn phosphorylates the kinase MEK, which in turn phosphorylates mitogen-activated protein (MAP) kinase. Although heavily studied over the last decade, the exact mechanism of Raf-1 activation is not known. An important clue to the mechanism of Raf-1 activation is the structure of constitutively active, oncogenic forms of Raf-1. These forms of Raf-1 lack the amino-terminal half of the molecule, suggesting that the amino-terminal domain suppresses Raf-1 kinase activity. One potential mechanism of activation is that lipids or proteins interacting with Raf-1 help to facilitate a change in conformation in which the inhibitory influence of the amino-terminal domain is removed. However, other models propose that phosphorylation of Raf-1 is required for its activation (42).
Recently, the 14-3-3 proteins have been implicated in the activation of Raf-1. 14-3-3 was first identified as a Raf-associated protein by several groups using both biochemical and genetic approaches (12, 14, 15, 23, 64). Although a positive role for 14-3-3 in Raf activation is supported by genetic experiments with yeast and Drosophila, in which 14-3-3 is required for Raf-1 activation (7, 23, 28), in vitro biochemical studies have been unable to support a role for 14-3-3 in regulating Raf-1 activity. Although it was initially reported that 14-3-3 can activate Raf-1 (14, 15, 23, 33), others have reported that 14-3-3 binding is not required for Raf-1 activity (39, 58).
One possible explanation for these inconsistencies is the fact that Raf-1 contains at least three potential 14-3-3 binding sites, one surrounding serine 259 (S259) a second surrounding S621 (43, 44), and a third, recently identified site in the cysteine-rich domain, between amino acids 136 and 187 (8). Mutagenesis studies suggest that phosphorylation of S259 has an effect contrasting to that of S621 phosphorylation on Raf-1 kinase activity. Mutation of S259 results in a constitutively active kinase, suggesting that phosphorylation of S259 is inhibitory (8, 39). In contrast, mutation of S621 results in an inactive kinase, suggesting that S621 phosphorylation is required for Raf-1 kinase activity (39, 43). 14-3-3 binding to these sites may potentiate the functional effects of phosphorylation at these two sites.
To selectively examine the positive role of 14-3-3 binding on Raf-1 kinase activity, we used a truncated, constitutively active form of Raf-1 that contains only one 14-3-3 binding site, the equivalent of S621. Using a phosphospecific S621 antibody, we analyzed the effect of dominant negative 14-3-3 expression as well as 14-3-3 overexpression on S621 phosphorylation and Raf-1 kinase activity. Our results demonstrate that S621 is an autophosphorylation site whose stability is directly regulated by 14-3-3 expression levels. These results clearly demonstrate a requirement for 14-3-3 for Raf-1 kinase activity and suggest that changes in 14-3-3 expression levels have a significant impact upon the magnitude of Raf-1 kinase signaling.
MATERIALS AND METHODS
Plasmid construction.
The glutathione S-transferase–14-3-3η fusion construct was created by generating BamHI and EcoRI restriction sites at the 5′ and 3′ ends, respectively, of the 14-3-3η cDNA by PCR. The cDNA product was subcloned into the vector pGEX-KT (20). For expression of wild-type and mutated 14-3-3η in eukaryotic cells, EcoRI and XhoI restriction sites were introduced by PCR at the 5′ and 3′ ends for subsequent cloning into a modified pcDNA3.1 vector (Invitrogen) which adds an N-terminal Myc epitope tag (11) to the expression product. CT-Raf was generated by PCR and encodes residues 326 to 648 of Raf-1. It was cloned into the XhoI and EcoRI sites of a modified pBluescript vector which adds an N-terminal Myc tag to the expressed product (50). The resulting construct was digested with EcoRI and subcloned into a modified pcDNA3.1 vector which adds an N-terminal FLAG tag to the expressed product. Site-directed mutagenesis to generate point or deletion mutations was done by inverse PCR (56). All introduced mutations were verified by DNA sequencing. The reporter plasmid pB4XCAT (21) was kindly provided A. Prendergast. The constitutively active MEK expression construct R4F Mek (36), was kindly provided by N. Ahn.
Recombinant protein expression and purification.
Recombinant GST fusion proteins were expressed and purified from Escherichia coli as described previously (20). Protein was quantitated by comparison to bovine serum albumin (BSA) standards on Coomassie blue-stained sodium dodecyl sulfate (SDS)-polyacrylamide gels.
Peptides.
Peptides were synthesized, purified, and analyzed as previously described (45). All peptides were shown to consist of a single species of the correct molecular weight by mass spectrometry (Washington University Mass Spectrometry Facility).
SPR studies.
Surface plasmon resonance (SPR) studies were performed with a BiaCore 2000 (Pharmacia). The substrate peptide and assay conditions have been described previously (44). Briefly, 5 μl of a biotinylated Raf phosphopeptide (1 nM; corresponding to amino acids 251 to 265 of human Raf-1) was immobilized on streptavidin-coated sensor chips (SA5; Pharmacia) at a flow rate of 5 μl/min at 25°C. This generally resulted in a resonance unit value of 80 to 100 U. To compare the relative affinities of the mutated 14-3-3η proteins, GST fusion proteins (1 μM) were allowed to flow over the immobilized peptide at a rate of 5 μl/min for 5 min. The resulting increase in resonance units remaining after two wash steps was used to calculate the percentage of binding of mutated proteins relative to that of the wild-type protein. To test the inhibitory capacity of the mutated peptides, 1 μM GST–14-3-3 was first incubated with 50 μM peptide in duplicate. Peptides that could not inhibit greater than 50% of binding were not further analyzed. The remaining peptides were then tested at concentrations of between 1 and 50 μM to determine 50% inhibitory concentrations.
Transfections and CAT assays.
293 cells were plated at a density of 3 × 105 cells/well in six-well tissue culture dishes and allowed to adhere for 4 to 6 h. Cells were transfected with SuperFect reagent (Qiagen, Chatsworth, Calif.) according to the manufacturer’s directions, using a total of 3 μg of DNA for each transfection. Transfected cells were cultured in medium without serum for 24 h after transfection and then harvested by lysis in 120 μl of 20 mM Tris-Cl (pH 7.5)–2 mM MgCl2–0.1% Nonidet P-40 (NP-40) and cleared by centrifugation. Chloramphenicol acetyltransferase (CAT) activity was measured by the method of Seed and Sheen (52). For expression in HeLa cells, the vaccinia virus-T7 expression system was used as described previously (16, 17).
Recombinant baculovirus production.
For the production of recombinant baculoviruses, cDNAs were first subcloned into the vector pFastBac-HT (Life Technologies, Gaithersburg, Md.). The wild-type and mutated forms of CT-Raf were ligated into the EcoRI site, and the mouse MEK-1 was ligated into the BamHI and KpnI sites. The GST constructs were generated by introducing 5′ NcoI and 3′ EcoRI sites into coding sequence from pGEX-KT constructs by PCR. Recombinant baculoviruses were then generated by using the Bac-to-Bac HT Baculovirus Expression System (Life Technologies) according to the manufacturer’s instructions.
In vitro kinase assays.
Cells were lysed on ice in a buffer containing 25 mM Tris (pH 8.0), 100 mM NaCl, 25 mM NaF, 5 mM EDTA, 100 μM vanadate, 1% NP-40, and 1% β-octylglucoside with protease inhibitors. After clearing, immunoprecipitates were prepared with antibodies to Raf-1 (C-12; Santa Cruz) and protein A-Sepharose. Immunoprecipitates were washed three times with lysis buffer and then once with 2× kinase buffer (40 mM Tris [pH 7.4], 200 mM NaCl, 10 mM MgCl2, 100 μM vanadate). Kinase reactions were performed in two steps. First, immunoprecipitates were incubated with 10 μl of 1× kinase buffer containing 100 μM cold ATP, 1 mM dithiothreitol, and 0.5 μg of purified MEK (Santa Cruz) for 10 min at room temperature. Next, 40 μl of 1× kinase buffer containing 3 μg of kinase-inactive MAP kinase (30) and 20 μCi of [γ-32P]ATP was added and incubated with constant agitation for 30 min at room temperature. Phosphorylated proteins were analyzed by SDS-polyacrylamide gel electrophoresis (SDS-PAGE), and the radioactivity in the dried gels was quantitated with a phosphorimager.
Phosphate labeling.
For 32P labeling of recombinant proteins, 3 × 106 Sf9 cells were infected with the appropriate recombinant baculovirus at a multiplicity of infection of 10. Six hours after infection, the culture medium was replaced with phosphate-free Dulbecco’s modified Eagle’s medium, adjusted to pH 6.2 with HCl, containing 1% dialyzed fetal bovine serum. One hour later, 1 mCi of [32P]orthophosphoric acid (NEN, Boston, Mass.) was added to each culture and incubated for 12 h. Cells were washed once with ice-cold phosphate-buffered saline (PBS) and then with 1 ml of NP-40 lysis buffer (25 mM HEPES [pH 7.4], 150 mM NaCl, 25 mM NaF, 10% glycerol, 1% NP-40, 100 μM sodium vanadate, 100 μM microcystin LR, 10 μg of aprotinin per ml, 10 μg of leupeptin per ml, 1 μg of pepstatin A per ml, 20 μM phenylmethylsulfonyl fluoride, and 5 mM benzamidine). After the lysates were cleared, immunoprecipitates were prepared with Raf-1 antibody (Santa Cruz Biotechnology) and protein A-Sepharose and washed six times in NP-40 lysis buffer containing 0.1% SDS. The Raf-1 immunoprecipitates were resolved by SDS-PAGE and transferred to nitrocellulose, and the phosphoproteins were visualized by phosphorimaging.
Phospho-S621 antibody.
A rabbit polyclonal antibody to a phosphorylated peptide consisting of residues 615 to 625 of Raf-1 was generated by Quality Controlled Biochemicals (Hopkinton, Mass.). The antibody was affinity purified and tested by enzyme-linked immunosorbent assay to confirm its specificity for the phosphorylated peptide. Purified GST–CT-Raf as well as GST–full-length Raf was tested in vitro for the ability to be phosphorylated by C-TAK1 purified as a histidine-tagged protein from bacteria. After tryptic cleavage, peptides were separated by high-pressure liquid chromatography (HPLC) essentially as described by Morrison et al. (43).
To test the specificity of the antibody, we generated both phosphorylated and unphosphorylated GST–CT-Raf. This was done by transforming bacteria with a plasmid encoding GST–CT-Raf alone (for the unphosphorylated form) or cotransforming bacteria with plasmids encoding GST–CT-Raf and C-TAK1, followed by selection in both ampicillin and kanamycin to produce phosphorylated GST–CT-Raf. To generate the C-TAK1-encoding plasmid, we ligated an NcoI-HpaI DNA fragment encoding a hexahistidine-tagged form of the C-TAK1 kinase domain (residues 1 to 412) into pBB131 (10a). Whole-cell lysates were prepared and immunoblotted with antibodies to Raf and pS621.
Western blotting.
Antiserum against 14-3-3 proteins was prepared in rabbits by immunization with a mixture of GST-14-3-3 proteins (beta, eta, tau, and sigma). Polyclonal rabbit antibody preparations against Raf-1 (C-12) and MEK (C-18) were purchased from Santa Cruz. Proteins separated by SDS-PAGE were transferred to nitrocellulose membranes. The membranes were blocked by incubation in 5% BSA or a mixture of 3% BSA and 1% nonfat dry milk, incubated for 1 to 16 h in a dilution of the appropriate primary antibody-antiserum preparation, and then visualized by chemiluminescence (Pierce).
RESULTS
Mutations of 14-3-3 which impair ligand binding result in a dominant negative phenotype.
While searching for suppressors of constitutively active Ras in the sevenless signaling pathway, Chang and Rubin isolated dominant negative forms of 14-3-3 (7). Although the exact mechanism of the dominant negative effect is unknown, it seemed likely that the effect is related to a reduction in the affinity of 14-3-3 for its ligands. To test this hypothesis directly, our strategy was to generate other 14-3-3 molecules with impaired ligand binding and then test whether they could also function as dominant negative proteins.
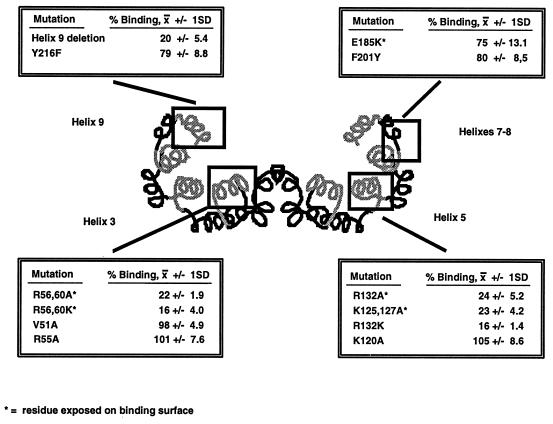
We generated mutated forms of 14-3-3, guided by the recently described crystal structure (35, 62) and the high degree of sequence conservation between all known forms of 14-3-3. The crystal structures of 14-3-3 demonstrate that the molecules form dimers, with each monomer composed of nine alpha helices. The 14-3-3 dimers form a cup-like shape with an inner concave surface composed of helices 3, 5, 7, and 9. Because this inner surface forms the ligand binding site (63), it seemed likely that conserved residues exposed on this surface would be important for ligand binding (66). Mutations were therefore introduced into a variety of conserved residues in helices 3, 5, 7, and 9 of the 14-3-3η fusion protein. The abilities of the mutated fusion proteins to bind to a serine-phosphorylated Raf-1 peptide at a concentration of 1 μM were then compared (Fig. 1) by SPR.
FIG. 1.
Binding of mutated 14-3-3η proteins to a serine-phosphorylated Raf peptide. A schematic representation of the helices comprised by a 14-3-3 dimer, with the relative locations of amino acid substitutions introduced into 14-3-3η by site-directed mutagenesis, is shown. Each mutant was expressed as a GST fusion protein, and binding of the proteins at 1 μM to a serine-phosphorylated Raf peptide by SPR was compared at equilibrium. Shown is the average percent binding ± one standard deviation (SD) determined in two separate experiments for each mutated protein relative to the binding obtained with the wild-type 14-3-3η fusion protein at an equal concentration. Helices shown in gray are those expected to contain amino acids lining the proposed binding surface.
Substitutions of alanine for the conserved arginines at positions 56 and 60 in helix 3, the arginine at position 132, or the lysines at positions 125 and 127 in helix 5 all resulted in significantly impaired binding. Affinity measurements performed by fluorescence polarization and SPR demonstrated in most cases greater-than-10-fold-weaker binding (data not shown). Even mutation of arginines 56 and 60 to like-charged lysine residues significantly impaired binding, as did a complete deletion of helix 9 (amino acids 212 to 248), which had not previously been implicated in ligand binding (63). In contrast, mutation of nonconserved residues or residues not exposed on the inner surface of the binding groove (e.g., V51A, R55A, or K120A) had little or no effect on the binding of 14-3-3 to the phosphoserine-containing peptide. Thus, conserved residues from the inner concave surface of the 14-3-3 molecule are required for ligand binding.
We confirmed that the dominant negative mutations described by Chang and Rubin (7) did impair ligand binding by introducing the mutations, E185K, F201Y, and Y216F, into the human 14-3-3η cDNA. GST fusions of the mutated proteins were produced, and each was tested for the ability to bind to a serine-phosphorylated Raf-1 peptide. Each of these mutated forms of 14-3-3 displayed an approximately 15 to 25% decrease in binding compared to the wild-type protein (Fig. 1), confirming that these mutations do impair ligand binding. Scatchard analysis, performed by fluorescence polarization and SPR, suggested that the change in affinity of these mutated proteins is small; they are less than twofold weaker than the wild type (data not shown).
The mutated forms of 14-3-3 were then tested for the ability to inhibit a constitutively active form of Raf-1. Because Raf-1 has two 14-3-3 binding sites and because 14-3-3 may mediate both positive and negative effects on Raf-1 kinase activity (42, 44), we decided to use an amino-truncated, constitutively active form of Raf-1 (CT-Raf). This would allow us to focus exclusively on the positive role of 14-3-3 in Raf-1 activity. This form of Raf-1, comprising amino acids 326 through 648 of the full-length molecule, lacks the amino-terminal domains and contains only one potential 14-3-3 binding site, the equivalent of S621 in Raf-1 (44). S621 is known to be constitutively phosphorylated in cells and required for Raf kinase activity (43).
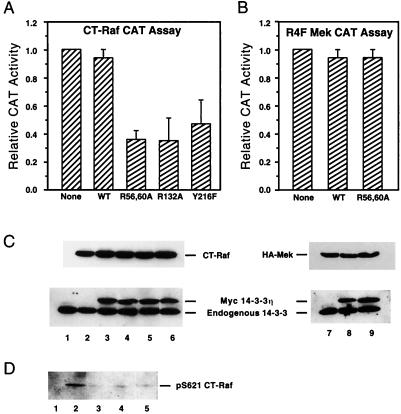
CT-Raf was coexpressed with either the wild-type, R56,60A, R132A, or Y216F form of 14-3-3η in 293 cells. To assess CT-Raf activity, we used a reporter construct (pB4XCAT) which is stimulated upon activation of the MAP kinase cascade (21). Twenty-four hours after transfection, cell lysates were analyzed for CAT activity. Overexpression of mutated forms of 14-3-3η but not wild-type 14-3-3η inhibited CT-Raf-induced CAT activity (Fig. 2A). This inhibition was specific for Raf-1, because the ability of constitutively active MEK to transactivate the reporter construct was not affected by coexpression with mutated forms of 14-3-3 (36) (Fig. 2). Immunoblotting of the cell lysates with antibodies to 14-3-3 and Raf-1 demonstrated similar levels of expression (Fig. 2C). These results suggest that one mechanism for the dominant negative effect of 14-3-3 may be related to decreased ligand binding affinity.
FIG. 2.
Binding-impaired 14-3-3 proteins inhibit CT-Raf kinase activity. (A) Activity of CT-Raf coexpressed with wild-type (WT) or mutated 14-3-3η proteins. 293 cells were transfected with a Ras-responsive reporter plasmid (pB4XCAT) and expression plasmids encoding CT-Raf and the indicated 14-3-3η construct. Cell lysates were analyzed 24 h later for CAT activity. Values were normalized to the amount of activity obtained from CT-Raf alone. Results shown are averages and standard deviations from four experiments. (B) Binding-impaired 14-3-3 proteins have no effect on the activity of constitutively active MEK. 293 cells were transfected as for panel A, using the constitutively active MEK instead of CT-Raf. CAT assays were performed 24 h after transfection, and the results are expressed as described for panel A. (C) Immunoblots of lysates from panel A or B. Equal aliquots of each lysate were separated on 10% gels by SDS-PAGE and transferred to nitrocellulose. Immunoblots were then developed with anti-Raf, anti-14-3-3, or antihemagglutinin (anti-HA) antibodies as indicated. Lane 1, untransfected; lane 2, CT-Raf only; lanes 3 to 6, CT-Raf with wild-type 14-3-3η (lane 3) or the R56,60A (lane 4), R132A (lane 5), or Y216F (lane 6) mutant; lane 7, HA-R4F Mek alone; lanes 8 and 9, HA-R4F Mek with wild-type 14-3-3η (lane 8) or R56,60A 14-3-3η (lane 9). (D) Coexpression of binding-impaired 14-3-3 proteins with CT-Raf destabilizes the phosphorylation of CT-Raf at S621. Equal aliquots of lysates from panel A were separated by SDS-PAGE and transferred to nitrocellulose. The blots were developed with the phosphospecific anti-p621 antibody. Lane 1, untransfected; lanes 2 to 5, CT-Raf with wild-type 14-3-3η (lane 2) or the R56,60A (lane 3), R132A (lane 4), or Y216F (lane 5) mutant.
Overexpression of dominant negative forms of 14-3-3 results in decreased phosphorylation of S621 in Raf.
Previous studies demonstrated that the phosphorylation of S621 is required for Raf-1 kinase activity, because mutation of this site results in an inactive kinase (43). We therefore wanted to confirm that the S621 site in Raf-1 was, in fact, phosphorylated in cells expressing wild-type or mutated forms of 14-3-3.
For these experiments, we generated an antibody that specifically recognizes the phosphorylated S621 site (anti-p621). Use of a phosphospecific antibody has several advantages over phosphate labeling with 32P. Because phosphate labeling is dependent on phospate turnover, it may not reflect the true in vivo phosphorylation status of a particular residue. Also, radiation is known to stimulate Raf-1 activity and may affect the phosphorylation of Raf-1 (26).
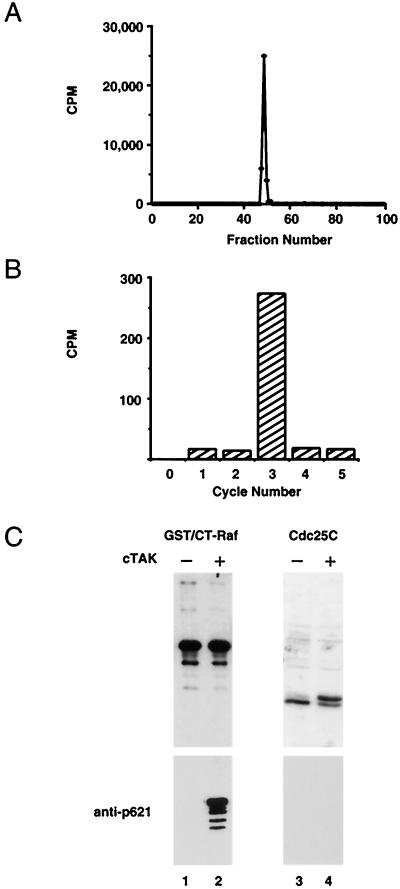
To test the specificity of the antibody, a Cdc25C kinase, C-TAK1 (46, 49a), was used to phosphorylate CT-Raf specifically at S621. The site on CT-Raf phosphorylated by C-TAK1 was determined by digesting in vitro-phosphorylated CT-Raf with trypsin and subjecting the tryptic peptides to phosphoamino acid analysis and also to reverse-phase HPLC. Phosphoamino acid analysis revealed only phosphoserine, demonstrating that C-TAK1 phosphorylated CT-Raf on one or more serine residues (data not shown). HPLC analysis revealed a single phosphopeptide eluting in fractions 49 to 51 (Fig. 3A). Edman degradation of the phosphopeptides derived after proteolysis with trypsin or endoproteinase Lys-C confirmed that the phosphorylated serine corresponds to S621 (data not shown). Immunoblotting of extracts from bacteria expressing truncated Raf-1 with or without C-TAK1 demonstrated that the antibody recognized only the phosphorylated form of Raf-1 (Fig. 3B, bars 1 and 2). Recognition was specific for the S621 site, as the antibody did not recognize the related phosphorylation site S216 of Cdc25C (Fig. 3B, bars 3 and 4) (49), nor did it recognize a form of full-length Raf-1 lacking the S621 site but containing the S259 site (data not shown).
FIG. 3.
Specificity of the anti-Raf phosphoserine 621 antibody. (A) Tryptic peptide analysis of CT-Raf phosphorylated by C-TAK1. CT-Raf was expressed in bacteria as a GST fusion protein. After purification, it was labeled in vitro with purified C-TAK1 and [γ-32P]ATP and then digested with trypsin. Peptides were resolved by HPLC. Radioactivity associated with each fraction was measured by scintillation counting. (B) Manual Edman degradation of tryptic fraction 51. Fraction 51 from panel A was subjected to manual Edman degradation. Bars represent radioactivity released from the membrane. The starting radioactivity associated with the membrane was 376 cpm. (C) Anti-phospho-S621 immunoblotting. CT-Raf or histidine-tagged Cdc25C was either expressed alone (lanes 1 and 3) or coexpressed with C-TAK1 (lanes 2 and 4) in bacteria. Lanes 1 and 2, cell extracts were analyzed by SDS-PAGE, transferred to nitrocellulose, and immunoblotted with antibodies to Raf-1 (top panel) or with anti-p621 (lower panel). Lanes 3 and 4, His-tagged Cdc25C was purified by nickel chromatography and Coomassie blue stained (upper panel) or blotted with anti-p621 (lower panel).
Lysates from cells coexpressing CT-Raf with mutated or wild-type forms of 14-3-3η were immunoblotted with the anti-p621 Raf-1 antibody. The antibody recognized a 36-kDa protein from cells coexpressing CT-Raf with wild-type 14-3-3η (Fig. 2D, lane 2), but little if any S621-phosphorylated CT-Raf was detected from cells expressing the dominant negative forms of 14-3-3 (Fig. 2D, lanes 3 to 5). Immunoblotting demonstrated equivalent levels of CT-Raf as well as 14-3-3 in all lysates (data not shown). This result suggests that the dominant negative forms of 14-3-3 mediate their inhibitory effects by causing the loss of serine phosphorylation on target proteins. One important function of 14-3-3 binding may be to protect such sites from phosphatases, as proposed by Dent et al. (10).
Raf-1 proteins which are unable to bind 14-3-3 are not phosphorylated.
To confirm the hypothesis that 14-3-3 binding is required for maintaining S621-phosphorylated Raf-1 in vivo, our strategy was to generate mutated forms of CT-Raf that retained the phosphorylation site but were unable to bind 14-3-3. We reasoned that forms of CT-Raf that cannot bind 14-3-3 should not maintain phosphorylation on S621. Although we had previously defined a favored sequence motif for 14-3-3 binding (RSxpSxP) (44), our studies had also demonstrated that this motif was not absolute.
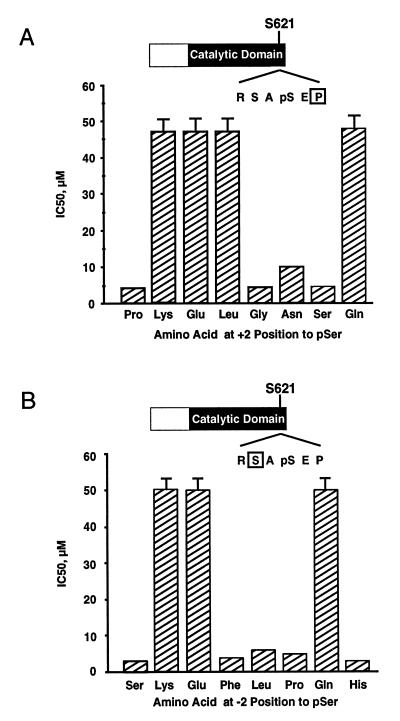
A series of serine-phosphorylated Raf-1 peptides containing a range of amino acid substitutions at either the +2 or −2 position relative to the phosphoserine were generated. Each peptide was analyzed for its ability to inhibit the binding of GST–14-3-3 to the wild-type Raf-1 phosphopeptide by using SPR. As shown in Fig. 4A, substitution of lysine, glutamic acid, leucine, or glutamine for the proline in the +2 position essentially eliminated the ability of the phosphopeptide to competitively inhibit 14-3-3 binding, with 50% inhibitory concentrations of greater than 50 μM. Peptides containing these residues in the +2 position therefore cannot bind 14-3-3. Similarly, peptides with a substitution of lysine, glutamic acid, or glutamine for the serine in the −2 position also did not compete, demonstrating that peptides containing these sequences are unable to bind 14-3-3. In contrast, a number of substitutions, such as glycine or serine in the +2 position and phenylalanine or leucine in the −2 position, were able to inhibit 14-3-3 binding similarly to the wild-type peptide, demonstrating that these sequences are able to bind 14-3-3. These finding are consistent with those recently reported by Yaffe et al. (63).
FIG. 4.
Analysis of the effect of amino acid substitutions at the +2 or −2 position relative to pS621 in Raf on 14-3-3 binding. (A) Raf phosphopeptides containing the indicated amino acid substitution at the +2 position relative to S621 were assessed for the ability to inhibit the binding of a 14-3-3η fusion protein to immobilized, wild-type Raf phosphopeptide. For each mutated peptide, the concentration required to achieve 50% inhibition (IC50) of 14-3-3 binding is shown. Essentially identical results were obtained with 14-3-3β, 14-3-3τ, and 14-3-3ζ (data not shown). (B) Raf phosphopeptides containing the indicated amino acid substitution at the −2 position relative to S621 were assessed as for panel A. Shown are the peptide concentrations required to achieve 50% inhibition of 14-3-3 binding to the wild-type peptide. Essentially identical results were obtained with 14-3-3β, 14-3-3τ, and 14-3-3ζ (data not shown).
Two mutations expected to impair 14-3-3 binding and one mutation expected to support 14-3-3 binding were chosen for further analysis. Recombinant baculoviruses encoding CT-Raf proteins containing each of these mutations were generated. These viruses encoded glycine or leucine at the +2 position and lysine at the −2 position. Phosphorylation of S621 was determined by analyzing Raf-1 immunoprecipitates from [32P]orthophosphate-labeled SF9 cells infected with each of the mutated CT-Raf baculoviruses (Fig. 5). The anti-p621 antibody would not be used because it did not recognize any of the mutated CT-Raf proteins (data not shown).
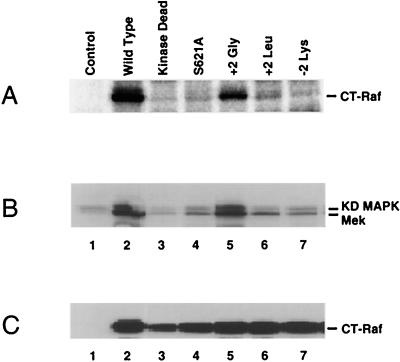
FIG. 5.
S621 phosphorylation and kinase activity of wild-type or mutated CT-Raf proteins. (A) Phosphate labeling of mutated CT-Raf proteins. Sf9 cells infected with recombinant baculoviruses encoding the indicated CT-Raf constructs or uninfected cells (control) were cultured in the presence of [32P]orthophosphoric acid for 12 h. Cells were lysed in NP-40 lysis buffer, and Raf-1 immunoprecipitates were prepared. After resolution by SDS-PAGE, the labeled proteins were visualized by phosphorimaging. Equivalent expression of each construct was verified separately by immunoblot analysis (data not shown). The data shown are representative of three separate experiments. (B) In vitro kinase activity of wild-type or mutated CT-Raf proteins. Sf9 cells were infected with a recombinant baculovirus encoding MEK-1 alone (control) or coinfected with the MEK-1 virus and a second recombinant baculovirus encoding the indicated CT-Raf construct. At 48 h postinfection, cell lysates were prepared. Anti-MEK-1 immunoprecipitations from each lysate were analyzed for MEK-1 kinase activity by using recombinant, kinase-deficient (KD) MAPK as a substrate. Lane 1, cells infected with MEK-1 alone. Lanes 2 to 7, cells infected with MEK-1 and the following CT-Raf constructs: lane 2, wild type; lane 3, K375M (kinase dead); lane 4, S621A; lane 5, +2 Gly (P623G); lane 6, +2 Leu (P623L); lane 7, −2 Lys (S619K). Data shown are representative of four separate experiments. (C) Expression levels of CT-Raf constructs used for panel B. Equal aliquots of lysates used for panel B were resolved by SDS-PAGE, transferred to nitrocellulose, and developed with an anti-Raf antibody. Lane contents are identical to those in panel B.
Phosphorylated CT-Raf was detected only in cells expressing either the wild-type protein or the mutant containing glycine in the +2 position (+2 Gly) (Fig. 5A, lanes 2 and 5). As expected, no phosphorylated CT-Raf was detected from the S621A mutant (lane 4). In addition, phosphorylation was not detected on the +2 Leu or the −2 Lys form of CT-Raf, both of which should be impaired for binding to 14-3-3. Interestingly, no S621 phosphorylation was detected on the kinase-dead form of CT-Raf (Fig. 5A, lane 3; see also Fig. 6B). This is consistent with a previous report demonstrating that unphosphorylated, kinase-competent Raf is able to autophosphorylate in vitro at S621 (40). These findings are consistent with the hypothesis that the ability to detect S621 phosphorylation is linked to 14-3-3 binding. Examination of Raf kinase activity demonstrated that the kinase activity correlated with S621 phosphorylation (Fig. 5B). Only the wild-type and the +2 Gly forms of CT-Raf demonstrated kinase activity (Fig. 5B, lanes 2 and 5). None of the other mutated forms of CT-Raf demonstrated any detectable kinase activity above the background level. These findings support the idea that 14-3-3 binding is required for Raf-1 kinase activity, at least in part, because it stabilizes the S621 phosphorylation. However, it is also possible that the +2 Leu and −2 Lys mutations of CT-Raf disrupt the recognition sequence required for autokinase activity, resulting in a failure to autophosphorylate at S621.
FIG. 6.
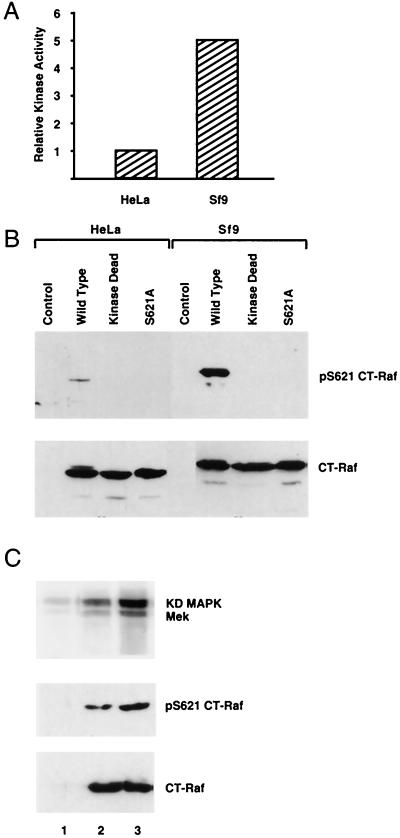
Kinase activity and S621 phosphorylation of CT-Raf expressed in HeLa or Sf9 cells. (A) Relative in vitro kinase activities of HeLa-expressed CT-Raf and Sf9-expressed CT-Raf. Anti-Raf immunoprecipitates from lysates of HeLa cells expressing FLAG–CT-Raf or Sf9 cells expressing baculovirus-encoded CT-Raf were analyzed for in vitro kinase activity by using a linked assay as described in Materials and Methods. Kinase activity from dried gels was quantitated by using a phosphorimager. CT-Raf protein expression levels from each cell type were quantitated by densitometric analysis of immunoblots prepared with equal aliquots of the lysates. The kinase activity observed from each cell type was then adjusted to reflect equivalent protein levels. (B) Analysis of S621 phosphorylation of CT-Raf constructs expressed in HeLa cells and Sf9 cells. Lysates of HeLa cells or Sf9 cells expressing the indicated CT-Raf construct were adjusted to contain equivalent amounts of CT-Raf protein as judged by anti-Raf immunoblotting (lower panel). Aliquots of each lysate were then immunoblotted with the anti-phospho-S621 antibody (upper panel). (C) Coexpression of 14-3-3 with CT-Raf in HeLa cells augments CT-Raf S621 phosphorylation and kinase activity. HeLa cells were either transfected with CT-Raf alone (lane 2) or CT-Raf plus 14-3-3β (lane 3) or left untransfected (lane 1). Raf immunoprecipitates were prepared and analyzed for in vitro kinase activity by using a linked assay (upper panel) as described in Materials and Methods. Equal aliquots of each lysate were analyzed by immunoblotting with the anti-p621 antibody (middle panel) or an anti-Raf antibody (lower panel). KD, kinase dead.
Increased 14-3-3 expression levels enhance Raf-1 phosphorylation and activity.
Our results suggested that variation in 14-3-3 expression levels might affect the stoichiometry of S621 phosphorylation and therefore the potential level of Raf-1 activity in the cell. In our preliminary experiments, we noted that CT-Raf was much less active in HeLa cells than when it was expressed in Sf9 cells (Fig. 6A). Immunoblotting with the anti-p621 antibody confirmed that S621 phosphorylation of CT Raf was much lower in HeLa cells than in Sf9 cells (Fig. 6B, compare wild-type lanes). Western blot analysis demonstrated that endogenous 14-3-3 levels in these two cell types are roughly equivalent on a per-cell basis (data not shown). However, the procedure used to transfect the HeLa cells is much less efficient than infection of Sf9 cells with baculovirus. Thus, when the relative efficiencies of transfection and infection are accounted for, the degree of overexpression of CT-Raf by HeLa cells is much greater than that by Sf9 cells on a per-cell basis, resulting in a lower ratio of 14-3-3 to CT-Raf in HeLa cells. To test whether lower levels of phosphorylation and kinase activity were due to a lower ratio of 14-3-3 to CT-Raf in HeLa cells, we coexpressed 14-3-3β with CT-Raf in HeLa cells (Fig. 6C). Coexpression of 14-3-3β with CT-Raf enhanced both the S621 phosphorylation of CT-Raf and its kinase activity. This suggests that the stoichiometry of Raf-1 phosphorylation on S621 could vary between cells and can, therefore, be directly modulated by 14-3-3 expression levels.
Displacement of 14-3-3 from S621 results in the loss of Raf kinase activity.
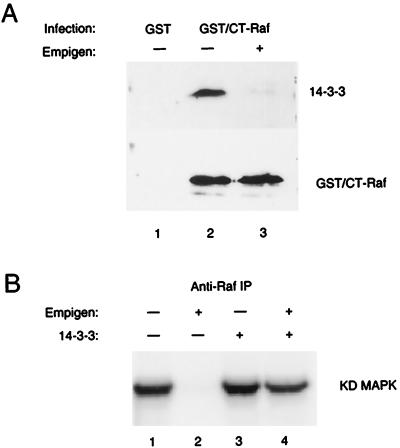
Is the binding of 14-3-3 to S621 required only to preserve S621 phosphorylation, or does 14-3-3 binding confer additional properties? To distinguish between these two possibilities, we used a zwitterionic detergent, Empigen-BB, to remove 14-3-3 from CT-Raf. This detergent was previously shown to efficiently disrupt the binding of 14-3-3 to simple epithelial keratin polypeptides (34). The ability of Empigen to displace 14-3-3 was verified by expressing a GST–CT-Raf fusion protein in SF9 cells and isolating the protein by using glutathione beads. The immobilized CT-Raf complexes were washed either with 1% NP-40 or with 1% Empigen (Fig. 7A). Immunoblotting with 14-3-3 antibodies demonstrated association of 14-3-3 with GST–CT-Raf but not with GST alone when complexes were washed with 1% NP-40 (Fig. 7A, lanes 1 and 2). As expected, incubation with 1% Empigen prior to immunoblotting eliminated all detectable 14-3-3 associated with CT-Raf (Fig. 7A, lane 3).
FIG. 7.
14-3-3 is required for CT-Raf kinase activity. (A) 14-3-3 can be removed from CT-Raf in vitro with the detergent Empigen-BB. Sf9 cells were infected with recombinant baculoviruses encoding either GST or a GST–CT-Raf fusion protein. Forty-eight hours after infection, the GST or GST–CT-Raf proteins were isolated from NP-40 lysates by using glutathione-agarose beads. The bead-bound complexes were then washed with either NP-40 lysis buffer (lanes 1 and 2) or NP-40 lysis buffer containing 1% Empigen-BB (lane 3), resolved by SDS-PAGE, transferred to nitrocellulose, and developed with an antibody to either 14-3-3 (upper panel) or Raf (lower panel). (B) Kinase activity of CT-Raf immunoprecipitates washed with Empigen BB, with or without addition of recombinant 14-3-3. Anti-Raf immunoprecipitates (IP) were prepared from lysates of Sf9 expressing CT-Raf and washed with NP-40 lysis buffer (lanes 2 and 4) or with 1% Empigen-BB (lanes 1 and 3). Following this wash step, purified, recombinant 14-3-3 protein was added as indicated (lanes 3 and 4). The in vitro kinase activity of the immunoprecipitates was then assessed by using a linked assay as described in Materials and Methods. Shown are results from one representative experiment in which recombinant 14-3-3τ was used. Other experiments were performed with recombinant 14-3-3β, with similar results. Quantitation of the 32P-labeled substrate in each lane was performed by volume analysis of the phosphorimaged data. Results for each lane normalized to lane 1: lane 1, 100%; lane 2, 0%; lane 3, 115%; lane 4, 75%. KD, kinase dead.
Immunoprecipitates were then tested for Raf kinase activity by using a linked kinase assay with MEK and MAP kinase (Fig. 7B) (30), and the results were quantitated by phosphorimage analysis. CT-Raf immunoprecipitates prepared with 1% Empigen were unable to activate MEK kinase activity as measured by the phosphorylation of kinase-inactive MAP kinase (Fig. 7B, lane 2), but the addition of purified, recombinant 14-3-3β or 14-3-3τ was able to restore approximately 75% of CT-Raf kinase activity obtained in the absence of Empigen. This demonstrates that the loss of kinase activity was due to displacement of 14-3-3 and not to other effects of the detergent. Exogenous 14-3-3 could, in some cases, moderately enhance the activity of CT-Raf in immunoprecipitates washed with 1% NP-40 (Fig. 7B, lane 3) (115% of the control value). This was a specific effect, as incubation with a mutated form of 14-3-3 (R56,60A) could not reconstitute kinase activity (data not shown). Thus, 14-3-3 binding not only is required for the stability of S621 phosphorylation but also is directly required for Raf-1 kinase activity.
DISCUSSION
To better understand the function of the 14-3-3 proteins, we examined the requirement for 14-3-3 in Raf kinase activity. Using a constitutively active, truncated form of Raf-1 which contains only a single 14-3-3 binding site, the equivalent of S621, we found that overexpression of dominant negative forms of 14-3-3 could inhibit the ability of constitutively active Raf to signal. Examination of the mechanism of this effect demonstrated that 14-3-3 binding both functions to maintain a critical phosphorylation site, S621, and is required for Raf kinase activity. This suggests that 14-3-3 levels regulate the stoichiometry of S621 phosphorylation. Consistent with this, we found that the stoichiometry of S621 phosphorylation of CT-Raf expressed in HeLa cells was low but could be enhanced by overexpression of 14-3-3. As S621 phosphorylation is required for Raf-1 kinase activity, changes in 14-3-3 expression can therefore have a significant impact on the magnitude of potential Raf-1 kinase activity.
The mechanism of action of the dominant negative forms of 14-3-3 is probably related to changes in the affinity of 14-3-3 for its binding partners. Because 14-3-3 proteins form dimers and because heterodimers can form between different isoforms (25), it seems likely that overexpression of our mutated 14-3-3 molecules inhibited wild-type 14-3-3 by forming mixed dimers with wild-type protein. Although our affinity measurements demonstrated that the dominant negative forms of 14-3-3 reported by Chang and Rubin (7) were only mildly impaired in their binding to a serine-phosphorylated peptide, this is not unexpected. More severe impairments of 14-3-3 binding might be predicted to be lethal, given that loss of one of the 14-3-3 isoforms is lethal (28). As suggested by Chang and Rubin (7), the dominant negative effect implies that normal 14-3-3 function requires both of the binding sites in the dimer. This could be related to a function for 14-3-3 in simultaneously recruiting two phosphorylated ligands (5) or may be related to enhancing overall affinity (63).
Since its discovery over 40 years ago, serine phosphorylation has been recognized as a major regulator of enzyme function in the cell. But exactly how serine phosphorylation achieves its effects is largely unknown. In most cases, it is assumed that phosphorylation is sufficient to change the function of a protein by causing a conformational change. Recently, however, it has become clear that phosphorylation sites can also serve as binding sites for other proteins, suggesting that phosphorylation regulates protein-protein interactions. The best-characterized example of this is the role of phosphotyrosine as a binding site for SH2 domains (48, 51). When a tyrosine kinase is activated, most of the functional effects are mediated by the recruitment of SH2 domain-containing proteins to phosphorylated tyrosine residues. Although the SH2-phosphotyrosine paradigm is well established, the role of phosphoserine binding proteins was, until recently, relatively uncharacterized.
14-3-3 proteins were among the first proteins demonstrated to have phosphoserine binding activity (44). The functional effects of this binding are just beginning to be understood. Examples of this include interactions of 14-3-3 with BAD and Cdc25C (49, 65). Both of these proteins are inhibited by serine phosphorylation and 14-3-3 binding. In both cases, however, the mechanism is indirect; 14-3-3 binding prevents interactions with their critical targets, bcl-2 and Cdc-2, respectively. Here we show that 14-3-3 can play other important roles. It is required for the serine phosphorylation and the activity of the serine kinase Raf-1.
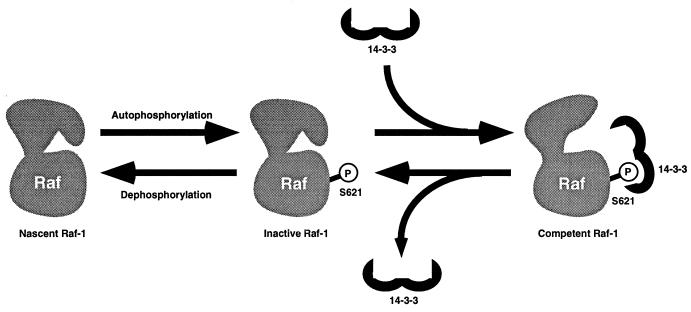
Our results suggest that 14-3-3 is critically involved in processing Raf-1 into a conformation that is competent for activation (Fig. 8). The inability to detect S621 phosphorylation of Raf-1 when 14-3-3 function is inhibited suggests that Raf-1 molecules are in a dynamic equilibrium between phosphorylated and unphosphorylated forms. S621 is phosphorylated by autophosphorylation and is rapidly dephosphorylated by an unknown phosphatase. Preliminary experiments demonstrate that this is self- or cis phosphorylation; we could not detect S621 phosphorylation of kinase-dead Raf-1 even when both wild-type and kinase-dead forms were coexpressed together (58a). By binding to phosphorylated S621, 14-3-3 protects this site from dephosphorylation and allows the kinase to become competent for activation. Our model can potentially explain how okadaic acid treatment results in Raf-1 activation (27a, 61a). Phosphatase inhibition might result in enhanced S621 phosphorylation. We are currently testing whether okadaic acid enhances S621 phosphorylation.
FIG. 8.
Potential role for 14-3-3 in Raf-1 maturation. 14-3-3 may play a critical role in Raf-1 maturation. Raf-1 molecules may exist in dynamic equilibrium between phosphorylated and unphosphorylated forms. Raf-1 can autophosphorylate itself at S621, but in the absence of 14-3-3, this phosphorylation is rapidly lost in the cell, presumably via the action of a phosphatase. The binding of 14-3-3 to this site protects the phosphorylation from phosphatase activity and is proposed to stabilize a kinase-competent conformation in Raf. This 14-3-3-bound form of CT-Raf possesses constitutive activity, requiring no additional activation events. In the context of the full-length molecule, the binding of 14-3-3 to the pS621 site is proposed to result in a preactivated molecule whose kinase activity is repressed due to interactions with the amino-terminal domains. This form of the kinase would be competent to bind Ras and become activated, or derepressed, at the plasma membrane.
It is interesting that both protein kinase A (PKA) and PKC have similar C-terminal serine phosphorylation sites that are required for kinase activity (27, 55). In the case of PKC, this site (S660) is a cis-autophosphorylation site and is required in the maturation of PKC, to release PKC from the cytoskeleton into the cytoplasm. It will be interesting to determine whether S621 phosphorylation plays a similar role in Raf-1 maturation.
What is the function of 14-3-3 binding to Raf-1? One possibility is that it helps to maintain a conformation required for substrate recognition or catalytic activity. The location of the 14-3-3 binding site, S621, about 20 residues from the end of the kinase domain, suggests that it could interact with and inhibit the active site. Phosphorylation of this site and 14-3-3 binding might function to remove this inhibitory segment from the active site in a manner analogous to the role of calmodulin binding to CAM kinase I (19).
Another possibility is that 14-3-3 facilitates Raf-1 interactions with MEK (24, 41). This is important because Raf-1 is extremely selective about its substrates (13). Until the discovery that MEK-1 was the substrate for Raf-1 (9, 29), it was difficult to measure Raf-1 kinase activity in vitro. If 14-3-3 binding to Raf-1 facilitates substrate binding, loss of 14-3-3 binding would result in an inability to detect Raf-1 kinase activity towards MEK. 14-3-3 could function as a scaffold to hold MEK and Raf-1 together (5). Alternatively, 14-3-3 binding to Raf-1 might promote a conformation of Raf-1 that allows MEK binding.
Interestingly, although S621 and the sequence surrounding this serine residue are highly conserved throughout evolution, at least two members of the Raf family have been reported to have amino acids other than serine at the position equivalent to 621 of Raf-1. The oncogenic component of the murine sarcoma virus 3611, known as v-Raf, is reported to encode proline at the position equivalent to S621 (36a), while the Xenopus laevis Raf-1 cDNA is reported to encode leucine at this position (32a). Based on our current knowledge, these amino acid substitutions would not be expected to support 14-3-3 binding, contradicting our model. To address these apparent discrepancies, we mutated S621 of CT-Raf to proline and found that this mutation results in an inactive CT-Raf molecule (58a). In addition, DNA sequencing of six separate Xenopus Raf-1 cDNA clones demonstrates that this form of Raf-1 cDNA encodes a serine at the codon equivalent to that for S621 of human Raf-1 (data not shown). As the reported codons for v-Raf and Xenopus Raf (CCT and TTG) differ from the codons encoding serine (TCX) by only a single nucleotide, we suspect that these discrepancies may be due to sequencing errors. These findings again underscore the significance of S621 in Raf-1 activity.
These ideas also have implications regarding Raf kinase regulation. Although the S621 site has long been considered a candidate positive regulatory site, the finding that the site is constitutively phosphorylated has lessened its appeal as a regulatory site (43). Our results demonstrate that the site is constitutively phosphorylated because it is an autophosphorylation site. However, the stability of S621 phosphorylation requires 14-3-3 binding. Thus, 14-3-3 expression levels will regulate the stoichiometry of S621 phosphorylation and therefore the magnitude of potential Raf-1 activity. Furthermore, our data suggest that the primary mechanism of Raf-1 activation is relief from inhibition by the amino-terminal domains. Bound to 14-3-3, the kinase is preactivated and competent to phosphorylate substrates. Additional phosphorylations of Raf-1 as well as lipid binding may function mainly to relieve inhibition rather than to activate enzyme activity.
As bacterially expressed CT-Raf is inactive and not phosphorylated at S621, we have tested whether S621 phosphorylation of CT-Raf and 14-3-3 binding might be sufficient to generate an active form of CT-Raf. In several experiments, S621-phosphorylated CT-Raf in the presence or absence of 14-3-3 was completely inactive. Although trivial explanations like improper protein folding can explain our results, it is also possible that other factors or posttranslational modifications are required to generate an enzymatically active Raf-1 kinase.
As levels of 14-3-3 are likely to vary between different cells, tissues, and conditions, 14-3-3 may play an important secondary role in Raf-1 kinase regulation. Recently, it was reported that different intensities of Raf-1 activation can account for the ability of Raf-1 to induce either cell cycle arrest or cell proliferation (53). High-intensity Raf-1 activation leads to p21cip1 expression and cell cycle arrest, whereas low- to medium-intensity Raf-1 activation leads to cell proliferation. One possibility is that changes in 14-3-3 expression regulate changes in the intensity of Raf-1 kinase activity.
Our data suggest novel ways of thinking about the function of 14-3-3 in the cell. For example, they suggest that levels of 14-3-3 expression are critical. Because 14-3-3 is a highly abundant protein in the brain (4), it has been assumed that 14-3-3 is equally abundant and nonlimiting in other cells. However, 14-3-3 levels can be modulated, and changes in levels can affect cellular responses. The fact that 14-3-3ς overexpression can cause cell cycle arrest (22) and our finding that 14-3-3β overexpression can enhance Raf-1 activity both support the idea that 14-3-3 levels, at least in some cells, will be important. 14-3-3 expression levels are known to have tissue-specific patterns and are dynamically regulated throughout development (6, 37, 60, 61). In addition, 14-3-3 expression can be induced by certain growth factors, and levels are elevated in some skin and lung cancers (2, 31, 47, 54, 59). It will be important to determine whether it is the total 14-3-3 expression level that is important or whether specific 14-3-3 isoforms mediate these specific effects.
It seems likely that 14-3-3 expression levels in the cell play an important role in regulating the strength and duration of signaling responses. High 14-3-3 levels would potentiate signaling responses by having a large capacity to bind to and affect the function of serine-phosphorylated substrates. On the other hand, signaling reactions that occur with lower levels of 14-3-3 would be quenched quickly because kinase activation would generate more 14-3-3 binding sites than 14-3-3 molecules could protect, resulting in rapid turnover of phosphoserine.
Our work demonstrates that serine phosphorylation can effect specific responses by inducing binding sites for 14-3-3 proteins on target proteins. 14-3-3 binding protects the phosphate from dephosphorylation, and in the case of Raf-1, this binding is required for Raf-1 kinase activity. In this light, it is interesting to note the many similarities between the phosphoserine–14-3-3 system and the calcium-calmodulin system. Both involve the binding of ubiquitous alpha-helical, symmetrical molecules that are broadly involved in signaling processes (38). Both also have the capacity to bind a large number of potential targets during a signaling event and function to both inhibit and activate enzymes (18). This similarity underscores the importance of protein-protein interactions in mediating the effects of second messengers induced by signal transduction.
ACKNOWLEDGMENTS
We thank Jun Li and Mary Stephenson for invaluable technical assistance; N. Ahn, M. Cobb, and A.-M. Pendergast for providing reagents; Rich Thoma for performing the peptide mapping studies; and Andy Chan, Tony Muslin, and Steve Zheng for critical review of the manuscript and helpful discussion.
This work was supported by grants from the NIH (AI54094 to A.S.S.) and the Howard Hughes Medical Institute (GM47017 to H.P.-W. and GM18428 to P.R.G.).
REFERENCES
- 1.Aitken A. 14-3-3 proteins on the MAP. Trends Biochem Sci. 1995;20:95–97. doi: 10.1016/s0968-0004(00)88971-9. [DOI] [PubMed] [Google Scholar]
- 2.Autieri M V, Haines D S, Romanic A M, Ohlstein E H. Expression of 14-3-3 gamma in injured arteries and growth factor- and cytokine-stimulated human vascular smooth muscle cells. Cell Growth Differ. 1996;7:1453–1460. [PubMed] [Google Scholar]
- 3.Avruch J, Zhang X F, Kyriakis J M. Raf meets Ras: completing the framework of a signal transduction pathway. Trends Biochem Sci. 1994;19:279–283. doi: 10.1016/0968-0004(94)90005-1. [DOI] [PubMed] [Google Scholar]
- 4.Boston P, Jackson P, Kynoch P, Thompson R. Purification, properties and immunohistochemical localisation of human brain 14-3-3 protein. J Neurochem. 1982;38:1466–1474. doi: 10.1111/j.1471-4159.1982.tb07927.x. [DOI] [PubMed] [Google Scholar]
- 5.Braselmann S, McCormick F. BCR and RAF form a complex in vivo via 14-3-3 proteins. EMBO J. 1995;14:4839–4848. doi: 10.1002/j.1460-2075.1995.tb00165.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Byrjalsen I, Larsen P M, Fey S J, Christiansen C. Human endometrial proteins with cyclic changes in the expression during the normal menstrual cycle: characterization by protein sequence analysis. Hum Reprod. 1995;10:2760–2766. doi: 10.1093/oxfordjournals.humrep.a135788. [DOI] [PubMed] [Google Scholar]
- 7.Chang H C, Rubin G M. 14-3-3 epsilon positively regulates Ras-mediated signaling in Drosophila. Genes Dev. 1997;11:1132–1139. doi: 10.1101/gad.11.9.1132. [DOI] [PubMed] [Google Scholar]
- 8.Clark G J, Drugan J K, Rossman K L, Carpenter J W, Rogers G K, Fu H, Der C J, Campbell S L. 14-3-3 zeta negatively regulates raf-1 activity by interactions with the Raf-1 cysteine-rich domain. J Biol Chem. 1997;272:20990–20993. doi: 10.1074/jbc.272.34.20990. [DOI] [PubMed] [Google Scholar]
- 9.Dent P, Haser W, Haystead T A, Vincent L A, Roberts T M, Sturgill T W. Activation of mitogen-activated protein kinase kinase by v-Raf in NIH 3T3 cells and in vitro. Science. 1992;257:1404–1407. doi: 10.1126/science.1326789. [DOI] [PubMed] [Google Scholar]
- 10.Dent P, Jelinek T, Morrison D K, Weber M J, Sturgill T W. Reversal of Raf-1 activation by purified and membrane-associated protein phosphatases. Science. 1995;268:1902–1906. doi: 10.1126/science.7604263. [DOI] [PubMed] [Google Scholar]
- 10a.Duronio R J, Jackson-Machelski E, Heuckeroth R O, Olins P O, Devine C S, Yonemoto W, Slice L W, Taylor S S, Gordon J I. Protein N-myristoylation in Escherichia coli: reconstitution of a eukaryotic protein modification in bacteria. Proc Natl Acad Sci USA. 1990;87:1506–1510. doi: 10.1073/pnas.87.4.1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Evan G I, Lewis G K, Ramsay G, Bishop J M. Isolation of monoclonal antibodies specific for the human c-myc proto-oncogene product. Mol Cell Biol. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fantl W J, Muslin A J, Kikuchi A, Martin J A, MacNicol A M, Gross R W, Williams L T. Activation of Raf-1 by 14-3-3 proteins. Nature. 1994;371:612–614. doi: 10.1038/371612a0. [DOI] [PubMed] [Google Scholar]
- 13.Force T, Bonventre J V, Heidecker G, Rapp U, Avruch J, Kyriakis J M. Enzymatic characteristics of the c-Raf-1 protein kinase. Proc Natl Acad Sci USA. 1994;91:1270–1274. doi: 10.1073/pnas.91.4.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Freed E, Symons M, Macdonald S G, McCormick F, Ruggieri R. Binding of 14-3-3 proteins to the protein kinase Raf and effects on its activation. Science. 1994;265:1713–1716. doi: 10.1126/science.8085158. [DOI] [PubMed] [Google Scholar]
- 15.Fu H, Xia K, Pallas D C, Cui C, Conroy K, Narsimhan R P, Mamon H, Collier R J, Roberts T M. Interaction of the protein kinase Raf-1 with 14-3-3 proteins. Science. 1994;266:126–129. doi: 10.1126/science.7939632. [DOI] [PubMed] [Google Scholar]
- 16.Fuerst T R, Niles E G, Studier F W, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gauen L T, Kong A N, Samelson L E, Shaw A S. p59fyn associates with multiple T-cell receptor subunits through its unique amino-terminal domain. Mol Cell Biol. 1992;12:5438–5446. doi: 10.1128/mcb.12.12.5438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghosh A, Greenberg M E. Calcium signaling in neurons: molecular mechanisms and cellular consequences. Science. 1995;268:239–247. doi: 10.1126/science.7716515. [DOI] [PubMed] [Google Scholar]
- 19.Goldberg J, Nairn A C, Kuriyan J. Structural basis for the autoinhibition of calcium/calmodulin-dependent protein kinase I. Cell. 1996;84:875–887. doi: 10.1016/s0092-8674(00)81066-1. [DOI] [PubMed] [Google Scholar]
- 20.Guan K L, Dixon J E. Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal Biochem. 1991;192:262–267. doi: 10.1016/0003-2697(91)90534-z. [DOI] [PubMed] [Google Scholar]
- 21.Hauser C A, Westwick J K, Quilliam L A. Ras-mediated transcription activation: analysis by transient cotransfection assays. Methods Enzymol. 1995;255:412–426. doi: 10.1016/s0076-6879(95)55043-7. [DOI] [PubMed] [Google Scholar]
- 22.Hermeking H, Lengauer C, Polyak K, He T-C, Zhang L, Thiagalingam S, Kinzler K, Vogelstein B. 14-3-3ς is a p53-regulated inhibitor of G2/M progression. Mol Cell. 1997;1:3–11. doi: 10.1016/s1097-2765(00)80002-7. [DOI] [PubMed] [Google Scholar]
- 23.Irie K, Gotoh Y, Yashar B M, Errede B, Nishida E, Matsumoto K. Stimulatory effects of yeast and mammalian 14-3-3 proteins on the Raf protein kinase. Science. 1994;265:1716–1719. doi: 10.1126/science.8085159. [DOI] [PubMed] [Google Scholar]
- 24.Jelinek T, Catling A D, Reuter C W, Moodie S A, Wolfman A, Weber M J. RAS and RAF-1 form a signalling complex with MEK-1 but not MEK-2. Mol Cell Biol. 1994;14:8212–8218. doi: 10.1128/mcb.14.12.8212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones D H, Ley S, Aitken A. Isoforms of 14-3-3 protein can form homo- and heterodimers in vivo and in vitro: implications for function as adapter proteins. FEBS Lett. 1995;368:55–58. doi: 10.1016/0014-5793(95)00598-4. [DOI] [PubMed] [Google Scholar]
- 26.Kasid U, Suy S, Dent P, Ray S, Whiteside T L, Sturgill T W. Activation of Raf by ionizing radiation. Nature. 1996;382:813–816. doi: 10.1038/382813a0. [DOI] [PubMed] [Google Scholar]
- 27.Keranen L M, Dutil E M, Newton A C. Protein kinase C is regulated in vivo by three functionally distinct phosphorylations. Curr Biol. 1995;5:1394–1403. doi: 10.1016/s0960-9822(95)00277-6. [DOI] [PubMed] [Google Scholar]
- 27a.Kharbanda S, Saleem A, Emoto Y, Stone R, Rapp U, Kufe D. Activation of raf-1 and mitogen-activated protein kinases during monocytic differentiation of human myeloid leukemia cells. J Biol Chem. 1994;269:872–878. [PubMed] [Google Scholar]
- 28.Kockel L, Vorbruggen G, Jackle H, Mlodzik M, Bohmann D. Requirement for Drosophila 14-3-3 zeta in Raf-dependent photoreceptor development. Genes Dev. 1997;11:1140–1147. doi: 10.1101/gad.11.9.1140. [DOI] [PubMed] [Google Scholar]
- 29.Kyriakis J M, App H, Zhang X F, Banerjee P, Brautigan D L, Rapp U R, Avruch J. Raf-1 activates MAP kinase-kinase. Nature. 1992;358:417–421. doi: 10.1038/358417a0. [DOI] [PubMed] [Google Scholar]
- 30.Lange-Carter C, Johnson G. Assay of MEK kinases. Methods Enzymol. 1995;255:290–301. doi: 10.1016/s0076-6879(95)55032-1. [DOI] [PubMed] [Google Scholar]
- 31.Lee N H, Weinstock K G, Kirkness E F, Earle H J, Fuldner R A, Marmaros S, Glodek A, Gocayne J D, Adams M D, Kerlavage A R, et al. Comparative expressed-sequence-tag analysis of differential gene expression profiles in PC-12 cells before and after nerve growth factor treatment. Proc Natl Acad Sci USA. 1995;92:8303–8307. doi: 10.1073/pnas.92.18.8303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leevers S J, Paterson H F, Marshall C J. Requirement for Ras in Raf activation is overcome by targeting Raf to the plasma membrane. Nature. 1994;369:411–414. doi: 10.1038/369411a0. [DOI] [PubMed] [Google Scholar]
- 32a.Le Guellec R, Couturier A, Le Guellec K, Paris J, Le Fur N, Philippe M. Xenopus c-raf proto-oncogene: cloning and expression during oogenesis and early development. Biol Cell. 1991;72:39–45. doi: 10.1016/0248-4900(91)90076-y. [DOI] [PubMed] [Google Scholar]
- 33.Li S, Janosch P, Tanji M, Rosenfeld G C, Waymire J C, Mischak H, Kolch W, Sedivy J M. Regulation of Raf-1 kinase activity by the 14-3-3 family of proteins. EMBO J. 1995;14:685–696. doi: 10.1002/j.1460-2075.1995.tb07047.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liao J, Omary M B. 14-3-3 proteins associate with phosphorylated simple epithelial keratins during cell cycle progression and act as a solubility cofactor. J Cell Biol. 1996;133:345–357. doi: 10.1083/jcb.133.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu D, Bienkowska J, Petosa C, Collier R J, Fu H, Liddington R. Crystal structure of the zeta isoform of the 14-3-3 protein. Nature. 1995;376:191–194. doi: 10.1038/376191a0. [DOI] [PubMed] [Google Scholar]
- 36.Mansour S J, Matten W T, Hermann A S, Candia J M, Rong S, Fukasawa K, Vande W G, Ahn N G. Transformation of mammalian cells by constitutively active MAP kinase kinase. Science. 1994;265:966–970. doi: 10.1126/science.8052857. [DOI] [PubMed] [Google Scholar]
- 36a.Mark G E, Rapp U. Primary structure of v-raf: relatedness to the src family of oncogenes. Science. 1984;224:285–289. doi: 10.1126/science.6324342. [DOI] [PubMed] [Google Scholar]
- 37.McConnell J E, Armstrong J F, Hodges P E, Bard J B. The mouse 14-3-3 epsilon isoform, a kinase regulator whose expression pattern is modulated in mesenchyme and neuronal differentiation. Dev Biol. 1995;169:218–228. doi: 10.1006/dbio.1995.1139. [DOI] [PubMed] [Google Scholar]
- 38.Meador W E, Means A R, Quiocho F A. Modulation of calmodulin plasticity in molecular recognition on the basis of x-ray structures. Science. 1993;262:1718–1721. doi: 10.1126/science.8259515. [DOI] [PubMed] [Google Scholar]
- 39.Michaud N R, Fabian J R, Mathes K D, Morrison D K. 14-3-3 is not essential for Raf-1 function: identification of Raf-1 proteins that are biologically activated in a 14-3-3- and Ras-independent manner. Mol Cell Biol. 1995;15:3390–3397. doi: 10.1128/mcb.15.6.3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mischak H, Seitz T, Janosch P, Eulitz M, Steen H, Schellerer M, Philipp A, Kolch W. Negative regulation of Raf-1 by phosphorylation of serine 621. Mol Cell Biol. 1996;16:5409–5418. doi: 10.1128/mcb.16.10.5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moodie S A, Willumsen B M, Weber M J, Wolfman A. Complexes of Ras.GTP with Raf-1 and mitogen-activated protein kinase kinase. Science. 1993;260:1658–1661. doi: 10.1126/science.8503013. [DOI] [PubMed] [Google Scholar]
- 42.Morrison D K, Cutler R E. The complexity of Raf-1 regulation. Curr Opin Cell Biol. 1997;9:174–179. doi: 10.1016/s0955-0674(97)80060-9. [DOI] [PubMed] [Google Scholar]
- 43.Morrison D K, Heidecker G, Rapp U R, Copeland T D. Identification of the major phosphorylation sites of the Raf-1 kinase. J Biol Chem. 1993;268:17309–17316. [PubMed] [Google Scholar]
- 44.Muslin A J, Tanner J W, Allen P M, Shaw A S. Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell. 1996;84:889–897. doi: 10.1016/s0092-8674(00)81067-3. [DOI] [PubMed] [Google Scholar]
- 45.Muslin A J, Williams L T. Well-defined growth factors promote cardiac development in axolotl mesodermal explants. Development. 1991;112:1095–1101. doi: 10.1242/dev.112.4.1095. [DOI] [PubMed] [Google Scholar]
- 46.Ogg S, Gabrielli B, Piwnica W H. Purification of a serine kinase that associates with and phosphorylates human Cdc25C on serine 216. J Biol Chem. 1994;269:30461–30469. [PubMed] [Google Scholar]
- 47.Olsen E, Rasmussen H H, Celis J E. Identification of proteins that are abnormally regulated in differentiated cultured human keratinocytes. Electrophoresis. 1995;16:2241–2248. doi: 10.1002/elps.11501601356. [DOI] [PubMed] [Google Scholar]
- 48.Pawson T, Gish G D. SH2 and SH3 domains: from structure to function. Cell. 1992;71:359–362. doi: 10.1016/0092-8674(92)90504-6. [DOI] [PubMed] [Google Scholar]
- 49.Peng C Y, Graves P R, Thoma R S, Wu Z, Shaw A S, Piwnica-Worms H. Mitotic and G2 checkpoint control: regulation of 14-3-3 protein binding by phosphorylation of Cdc25C on serine-216. Science. 1997;277:1501–1505. doi: 10.1126/science.277.5331.1501. [DOI] [PubMed] [Google Scholar]
- 49a.Peng C-Y, Graves P R, Ogg S, Thoma R S, Byrnes M J, Wu Z, Stephenson M, Piwnica-Worms H. C-TAK1 protein kinase phosphorylates human Cdc25C on serine 216 and promotes 14-3-3 binding. Cell Growth Differ. 1998;9:197–208. [PubMed] [Google Scholar]
- 50.Richard S, Yu D, Blumer K J, Hausladen D, Olszowy M W, Connelly P A, Shaw A S. Association of p62, a multifunctional SH2- and SH3-domain-binding protein, with Src family tyrosine kinases, Grb2, and phospholipase C gamma-1. Mol Cell Biol. 1995;15:186–197. doi: 10.1128/mcb.15.1.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schlessinger J. SH2/SH3 signaling proteins. Curr Opin Genet Dev. 1994;4:25–30. doi: 10.1016/0959-437x(94)90087-6. [DOI] [PubMed] [Google Scholar]
- 52.Seed B, Sheen J. A simple phase-extraction assay for chloramphenicol acyltransferase activity. Gene. 1988;67:271–277. doi: 10.1016/0378-1119(88)90403-9. [DOI] [PubMed] [Google Scholar]
- 53.Sewing A, Wiseman B, Lloyd A, Land H. High intensity Raf signal causes cell cycle arrest mediated by p21(CIP1) Mol Cell Biol. 1997;17:5588–5597. doi: 10.1128/mcb.17.9.5588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shoji M, Kawamoto S, Setoguchi Y, Mochizuki K, Honjoh T, Kato M, Hashizume S, Hanagiri T, Yoshimatsu T, Nakanishi K, et al. The 14-3-3 protein as the antigen for lung cancer-associated human monoclonal antibody AE6F4. Hum Antibodies Hybridomas. 1994;5:123–130. [PubMed] [Google Scholar]
- 55.Shoji S, Titani K, Demaille J, Fischer E. Sequence of two phosphorylated sites in the catalytic subunit of bovine cardiac muscle adenosine 3′:5′-monophosphate-dependent protein kinase. J Biol Chem. 1979;254:6211–6225. [PubMed] [Google Scholar]
- 56.Spee J H, de Vos W M, Kuipers O P. Efficient random mutagenesis method with adjustable mutation frequency by use of PCR and dITP. Nucleic Acids Res. 1993;21:777–778. doi: 10.1093/nar/21.3.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stokoe D, Macdonald S G, Cadwallader K, Symons M, Hancock J F. Activation of Raf as a result of recruitment to the plasma membrane. Science. 1994;264:1463–1467. doi: 10.1126/science.7811320. [DOI] [PubMed] [Google Scholar]
- 58.Suen K L, Bustelo X R, Barbacid M. Lack of evidence for the activation of the Ras/Raf mitogenic pathway by 14-3-3 proteins in mammalian cells. Oncogene. 1995;11:825–831. [PubMed] [Google Scholar]
- 58a.Thorson, J. A., and A. S. Shaw. Unpublished data.
- 59.Vellucci V F, Germino F J, Reiss M. Cloning of putative growth regulatory genes from primary human keratinocytes by subtractive hybridization. Gene. 1995;166:213–220. doi: 10.1016/0378-1119(95)00543-9. [DOI] [PubMed] [Google Scholar]
- 60.Wang W, Shakes D C. Expression patterns and transcript processing of ftt-1 and ftt-2, two C. elegans 14-3-3 homologues. J Mol Biol. 1997;268:619–630. doi: 10.1006/jmbi.1997.1002. [DOI] [PubMed] [Google Scholar]
- 61.Watanabe M, Isobe T, Ichimura T, Kuwano R, Takahashi Y, Kondo H, Inoue Y. Molecular cloning of rat cDNAs for the zeta and theta subtypes of 14-3-3 protein and differential distributions of their mRNAs in the brain. Mol Brain Res. 1994;25:113–21. doi: 10.1016/0169-328x(94)90285-2. [DOI] [PubMed] [Google Scholar]
- 61a.Whitehurst C E, Owaki H, Bruder J T, Rapp U R, Geppert T D. The MEK kinase activity of the catalytic domain of RAF-1 is regulated independently of ras binding in T cells. J Biol Chem. 1995;270:5594–5599. doi: 10.1074/jbc.270.10.5594. [DOI] [PubMed] [Google Scholar]
- 62.Xiao B, Smerdon S J, Jones D H, Dodson G G, Soneji Y, Aitken A, Gamblin S J. Structure of a 14-3-3 protein and implications for coordination of multiple signaling pathways. Nature. 1995;376:188–191. doi: 10.1038/376188a0. [DOI] [PubMed] [Google Scholar]
- 63.Yaffe M, Rittinger K, Volinia S, Caron P, Aitken A, Leffers H, Gamblin S, Smerdon S, Cantley L. The structural basis for 14-3-3:phosphopeptide binding specificity. Cell. 1997;91:961–971. doi: 10.1016/s0092-8674(00)80487-0. [DOI] [PubMed] [Google Scholar]
- 64.Yamamori B, Kuroda S, Shimizu K, Fukui K, Ohtsuka T, Takai Y. Purification of a Ras-dependent mitogen-activated protein kinase kinase kinase from bovine brain cytosol and its identification as a complex of B-Raf and 14-3-3 proteins. J Biol Chem. 1995;270:11723–11726. doi: 10.1074/jbc.270.20.11723. [DOI] [PubMed] [Google Scholar]
- 65.Zha J, Harada H, Yang E, Jockel J, Korsmeyer S J. Serine phosphorylation of death agonist BAD in response to survival factor results in binding to 14-3-3 not BCL-X(L) Cell. 1996;87:619–628. doi: 10.1016/s0092-8674(00)81382-3. [DOI] [PubMed] [Google Scholar]
- 66.Zhang L, Wang H, Liu D, Liddington R, Fu H. Raf-1 kinase and exoenzyme S interact with 14-3-3 zeta through a common site involving lysine 49. J Biol Chem. 1997;272:13717–13724. doi: 10.1074/jbc.272.21.13717. [DOI] [PubMed] [Google Scholar]