Abstract
Estimating total infection levels, including unreported and asymptomatic infections, is important for understanding community disease transmission. Wastewater can provide a pooled community sample to estimate total infections that is independent of case reporting biases toward individuals with moderate to severe symptoms and by test-seeking behavior and access. We derive three mechanistic models for estimating community infection levels from wastewater measurements based on a description of the processes that generate SARS-CoV-2 RNA signals in wastewater and accounting for the fecal strength of wastewater through endogenous microbial markers, daily flow, and per-capita wastewater generation estimates. The models are illustrated through two case studies of wastewater data collected during 2020–2021 in Virginia Beach, VA, and Santa Clara County, CA. Median simulated infection levels generally were higher than reported cases, but at times, were lower, suggesting a discrepancy between the reported cases and wastewater data, or inaccurate modeling results. Daily simulated infection estimates showed large ranges, in part due to dependence on highly variable clinical viral fecal shedding data. Overall, the wastewater-based mechanistic models are useful for normalization of wastewater measurements and for understanding wastewater-based surveillance data for public health decision-making but are currently limited by lack of robust SARS-CoV-2 fecal shedding data.
Keywords: COVID-19, infectious disease surveillance, mechanistic modeling, SARS-CoV-2, wastewater
Graphical Abstract:
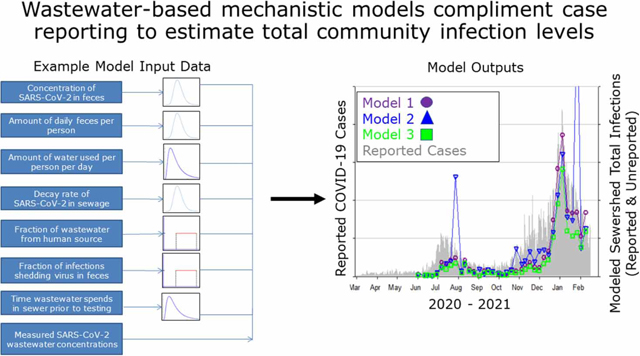
INTRODUCTION
SARS-CoV-2, the virus that causes COVID-19 infections, primarily causes respiratory illness (CDC 2021a). However, the RNA from this virus is also present in feces of infected symptomatic, pre-symptomatic, post-symptomatic, and asymptomatic individuals (Cheung et al. 2020; Foladori et al. 2020; WHO 2020a; Wolfel et al. 2020; Zhang et al. 2020; Zheng et al. 2020). Environmental surveillance through the testing of wastewater for evidence of pathogens has a long history of use in public health, particularly for poliovirus and more recently antimicrobial resistance (Asghar et al. 2014; WHO 2020b, 2020c). SARS-CoV-2 RNA has been reported in untreated wastewater and settled solids (e.g., sludge) in a number of countries (WHO 2020a), and wastewater SARS-CoV-2 RNA monitoring data have proven useful as an indicator of community illness in conjunction with traditional case reporting and surveillance methods (Medema et al. 2020; Peccia et al. 2020; D’Aoust et al. 2021; Fernandez-Cassi et al. 2021; Saguti et al. 2021; Weidhaas et al. 2021; Hewitt et al. 2022). Because of this, wastewater surveillance systems are being implemented for the COVID-19 response to provide data on overall infection trends and variant tracking within specific populations as a complement to clinical- and individual-based surveillance data for public health decision-making (Bivins et al. 2020; Medema et al. 2020; Peccia et al. 2020; Betancourt et al. 2021; Graham et al. 2021; Zhu et al. 2021; Kirby et al. 2022).
In addition to community infection trends, wastewater-based surveillance data have been proposed as a tool for estimating absolute community-level COVID-19 infections (Ahmed et al. 2020; Bivins et al. 2020; Hart & Halden 2020; Gerrity et al. 2021; Wurtzer et al. 2021). Estimating absolute infection levels (reported and unreported) is important for understanding disease transmission and designing effective mitigation strategies but has proven difficult to achieve using traditional surveillance indicators due to the large and variable relative burden of unreported cases (Bivins et al. 2020; Medema et al. 2020). To date, methods for estimating wastewater-based SARS-CoV-2 infections have included mechanistic-, statistical-, and epidemiological-based numerical modeling approaches (Baud et al. 2020; Ceylan 2020; Medema et al. 2020; Paul et al. 2020; Turk et al. 2020; Gerrity et al. 2021; Huisman et al. 2022; Kaplan et al. 2022; Weidhaas et al. 2021). For a mechanistic example, Ahmed et al. (2020) proposed an approach for COVID-19 wastewater surveillance in Australia based on the concentration of SARS-CoV-2 RNA in wastewater, the volume wastewater generated daily in a catchment, the number of SARS-CoV-2 RNA copies shed in stool by an infected individual each day, and the concentration of SARS-CoV-2 RNA in feces, each of which is represented by a combination of point estimate values and statistical distribution estimates in a Monte Carlo-based numerical simulation. Kaplan et al. (2022) proposed an epidemiologically- and statistically-calibrated scaling model to estimate incidence in a community from sludge RNA, a transmission dynamics model that aligns lagged epidemic indicators, and a site-specific scaling factor.
Mechanistic wastewater models are based on the processes that generate SARS-CoV-2 RNA signals in wastewater without additional statistical calibration against reported cases. Although wastewater-based disease research is rapidly advancing, the previously proposed mechanistic models fail to account for salient attributes of the wastewater processes, such as target decay within the system, and to account for variability in available data to estimate human contribution to the wastewater stream. In addition, the proposed statistical and epidemiological wastewater-infection-based models are either site-specific, using statistical correction factors dependent on reported cases, or are dependent on other clinical surveillance data; both limit the ability of wastewater to provide estimates independent of the variability in these clinical surveillance data (Li et al. 2020; Wu et al. 2020; Fernandez-Cassi et al. 2021). Moreover, epidemiologic compartmental models anchored to hospitalization or death rates are limited in their ability to provide near real-time infection estimates due to delayed nature of these indicators (Kaplan et al. 2021).
In this study, we derive three mechanistic models for estimating community infection levels from wastewater measurements that are agnostic to location. Each model uses a distinct approach to account for the human fecal content of wastewater, including endogenous microbial markers, wastewater flow measurements, and population-level estimates of per-capita wastewater generation. A numerical method is used to simulate the models, which are illustrated through two case studies of wastewater data and COVID-19 case data collected over several months in 2020–2021 (Virginia Beach, VA, and Santa Clara County, CA). We also propose a mathematical approach for connecting wastewater-based infection estimates to clinical surveillance data for public health interpretation. Finally, we describe the uncertainty associated with each modeling approach and the critical data needed to effectively use wastewater pathogen measurements to estimate community infections for public health decision-making.
METHODS
Our model formulations were developed to estimate the fraction of infected individuals in a community that could be shedding SARS-CoV-2 RNA either through respiratory or fecal secretions, or both, regardless of symptoms (referred to hereafter as ‘infected’). Some of these individuals may not be considered ‘infected’ from a clinical perspective because the fecal shedding of viral RNA does not necessarily correspond with respiratory symptoms or shedding (Wolfel et al. 2020; Zheng et al. 2020). We use a stochastic, Monte Carlo numerical simulation-based mechanistic modeling approach (Soller & Eisenberg 2008) encompassing the following three related formulations: (1) a flow generation formulation based on per-capita domestic potable water use adjusted for wastewater generation from non-domestic sources; (2) a fecal strength formulation that is a modified flow generation formulation to include an endogenous human fecal control as a proxy for dilution and loss of human feces and associated pathogens during the sewer transport and testing processes; and (3) a flow receipt formulation based on collection site daily flow measurements and wastewater utility provided sewershed population estimates. The inputs of the models are microbial wastewater influent concentrations (i.e., SARS-CoV-2 RNA and endogenous control when applicable), where influent is defined as the untreated wastewater entering a treatment plant containing both solids and liquids.
The three formulations are summarized below, and detailed mathematical specifications are provided in Supplementary Material (SI) Section A. The model formulations, as applicable, incorporate the following: variability in each model parameter using data from the peer reviewed scientific literature; differences between disease presentation that exhibit gastrointestinal (GI) symptoms and those that do not (i.e., account for differences in total fecal matter shed for those with and without GI symptoms); per-capita water use; the decay of viral RNA during transport within the wastewater collection system, and finally, the fraction of wastewater that results from human fecal waste (versus commercial, industrial, run-off, etc.) (refer to Supplementary Material, Table A1 for a comparison of the three formulations). All three modeling approaches are fundamentally dependent on the levels of SARS-CoV-2 RNA in the feces of infected individuals and are related as shown mathematically in Supplementary Material, Section A.
Flow generation formulation
The flow generation formulation is based on a mechanistic conceptualization of daily SARS-CoV-2 RNA input into the wastewater stream resulting from the fraction of SARS-CoV-2-infected individuals fecally shedding the virus , both with and without GI symptoms , over the course of an individual’s daily water use . The formulation then accounts for time within the sewer system and resulting decay over that period to estimate the total daily SARS-CoV-2 RNA concentration at the wastewater treatment plant influent (or settled solids) collection point (Equation (1)). Settled solids concentrations are converted to estimated wastewater influent concentrations via the use of a viral-specific solid-to-liquid partitioning coefficient following methods described in Kim et al. (2022) and Graham et al. (2021). Of note, does not include time spent in the sedimentation basin for settled solids measurements, which in some systems could be longer than time spent in the sewer system.
| (1) |
where is the fraction of infected individuals that shed viral RNA in feces (unitless); is the fraction of infected individuals with diarrhea (unitless); is the SARS-CoV-2 RNA concentration in feces among infected individuals with diarrhea (viral copies/mL); is the volume of feces per person per day among infected individuals with diarrhea (mL/day); is the SARS-CoV-2 RNA concentration in feces among infected individuals without diarrhea (viral copies/g); is the mass of feces per person per day among infected individuals without diarrhea (g/day); is the wastewater generated per person per day (L/person day); is the pseudo-first-order decay coefficient for SARS-CoV-2 RNA in sewage (1/day); is the time wastewater spends in the sewer prior to reaching the wastewater treatment plant (days); is the volume fraction of wastewater coming from a source that could potentially contain SARS-CoV-2 RNA (unitless).
By rearranging the equation, the fraction of the population with an active infection can be estimated for a specified median level of SARS-CoV-2 in the wastewater influent (Equation (2)).
| (2) |
The defining attribute of this formulation is a variable that defines the fraction of daily municipal wastewater that could be contributing SARS-CoV-2 RNA into the waste stream represents a variable, but time-invariant, estimate of the ‘human fecal strength’ (FS) of the wastewater. The salient assumption for this formulation is that it is feasible (e.g., for a wastewater treatment plant manager at the site monitored) to estimate with reasonable accuracy.
Fecal strength formulation
The flow generation formulation is limited when cannot be estimated accurately or when temporal variation of fecal strength is important. Such temporal variation could occur because of increased flows due to wet weather or variable wastewater composition due to dynamic water usage and human behavioral patterns (e.g., governmental restricted water usage or communities with large population influxes such as tourist locations). To address this, we reconfigured the flow generation model to define FS-based on an endogenous microbial control – a wastewater constituent that is present in relatively stable, high, and measurable concentrations in human excreta – rather than an expert judgement point estimate. We used Pepper Mild Mottle Virus (PMMoV) as the example human fecal-specific endogenous control (Zhang et al. 2006; Hamza et al. 2011).
This alternative approach for parameterization of fecal strength is the ratio of the concentration of endogenous control measured in wastewater, adjusted for decay in the sewer, to the expected wastewater endogenous control concentration if only human wastewater sources were contributing (e.g., sources of wastewater that could potentially contain SARS-CoV-2 RNA). A theoretical fecal strength of 1 would mean that human-associated wastewater is undiluted by other sources (e.g., industrial or run-off). The notation for the variables in the fecal strength formulation and the flow receipt formulation follow those presented above for the flow generation formulation where refers to the concentration in feces and refers to the mass of feces, with subscripts providing details about the specific parameters. FS is defined as follows, noting that this quotient is a unitless fraction:
| (3) |
where is the pseudo-first-order decay coefficient for endogenous control in sewage (1/day); is the endogenous control concentration in stool (gc/g); and is the stool mass generated per person per day (g/person day).
To demonstrate the mechanistic relatedness of the flow generation and fecal strength formulations, can be substituted for in Equation (2) of the fecal strength formulation as follows:
| (4a) |
Comparison of Equations (2) and (4a) indicates that two additional pieces of information are needed for this formulation compared to the flow generation formulation: (1) the concentration of the endogenous control in stool , which does not distinguish concentrations between diarrheal and non-diarrheal stool and (2) the corresponding decay coefficient .
Assuming that the concentration in stool is ~1.0 g/mL (Penn et al. 2018) and that the decay rate constants of the endogenous control and SARS-CoV-2 RNA are negligible or similar (which is most plausible for an RNA viral control such as PMMoV), the fecal strength formulation simplifies as follows (details provided in Supplementary Material, Section A):
| (4b) |
This fecal strength formulation (Equation (4a) and (4b)) has both advantages and disadvantages compared to the flow generation formulation (Equation (2)). The advantages are that the endogenous control (e.g., PMMoV RNA) measurements can theoretically serve as a normalizing control in that they can reflect the strength of human feces in the wastewater, the viral losses and dilution that occur as stool is transported from excreta through the sewage collection system to the sampling location, and the viral losses that occur during laboratory processing and testing. The primary disadvantage of this formulation is that it relies on measurements of the endogenous control concentrations in human feces, of which data are limited, and that endogenous control measurements must be made in each sample. This disadvantage is further complicated by the fact that wastewater processing and testing methods are not standardized, meaning that absolute concentrations generally cannot be compared across laboratories (without first conducting inter-lab comparisons).
Flow receipt formulation
Like the flow generation formulation, the flow receipt formulation relies on flow rate measurements to estimate the number of active SARS-CoV-2 infections within a sewershed () (Equation (5d)). However, in this formulation, the flow estimates are measured at the sampling site, often at the influent to a wastewater treatment plant over the course of one day, which enables flow to vary from sample-to-sample (or day to day). To put flow on a per-capita basis, this formulation also requires an explicit sewershed population estimate and estimate of the number of infected individuals in a population (). This formulation is derived from a mass balance equation on the number of RNA copies in wastewater:
| (5a) |
where
| (5b) |
Thus,
| (5c) |
or
| (5d) |
Similar to the derivations provided above, this flow receipt formulation is equivalent to the fecal strength formulation under specific, realistic conditions (mathematical details are provided in the Supplementary Material, refer to formulas A3.6–A3.11). This formulation also assumes that the SARS-CoV-2 RNA wastewater measurement multiplied by the average daily flow rate reasonably represents the average SARS-CoV-2 RNA load over one day. A primary advantage of this formulation is that, for samples collected at treatment plants, flow measurements and population estimates are commonly available. Therefore, the flow receipt formulation reduces the requisite number of laboratory measurements compared to the other two formulations. Moreover, flow measurements and population estimates may have lower relative error and/or variability than microbial endogenous control measurements, which vary on a log-scale basis. The disadvantages of the flow receipt formulation are that it does not reflect SARS-CoV-2 RNA losses that occur during laboratory processing or unpredictable SARS-CoV-2 RNA losses in the sewage system that are attributable to processes other than the modeled gene target decay. This formulation also requires a static wastewater contributing population estimate, total daily flow measurements, and an aggregated average value for (used to account for variability in influent over the course of a day into a treatment facility).
Wastewater modeling case studies
The characteristics of the wastewater case studies are summarized in Table 1, including information on geographic area and population represented, sampling and testing methodologies, and clinical case data. For both case studies, viral wastewater concentration data (SARS-CoV-2 and PMMoV RNA) were measured by reverse transcriptase quantitative polymerase chain reaction (RT-qPCR) using digital droplet PCR technology (ddPCR). Note that the SARS-CoV-2 RNA and PMMoV RNA data were collected from primary settled solids rather than wastewater influent for the San Jose, CA case study (SJ). For the purposes of numerical modeling, the observed settled solids concentrations of SARS-CoV-2 RNA and PMMoV RNA were converted to estimated wastewater influent concentrations using a solid-to-liquid partitioning coefficient following methods described in Kim et al. (2022) and Graham et al. (2021). Use of a fixed assumes the viral RNA remains at a consistent level of equilibrium between water and solids through the sample collection period.
Table 1.
Overview of case studies including information on wastewater service area, sampling, testing, and clinical case data
| Characteristic | Virginia Beach, VA | San Jose, CA |
|---|---|---|
|
| ||
| Geographic area | Atlantic Treatment Plant, Hampton Roads region, VA, USA | San Jose wastewater treatment plant, Santa Clara County, CA, USA |
| Sewershed population | 343,016 | 1,458,000 |
| Wastewater sampling dates | 6/2/20 to 2/9/21 | 3/18/20 to 3/31/21 |
| Sampling frequency | 1 per week | Weekly from 3/18/20 to 7/14/20, then 5 × /week |
| Wastewater matrix | Primary influent | Primary settled solids |
| Number wastewater samples | 37 | 195 |
| Sample collection | 24-h flow-weighted composite | 24-h composite solids sample |
| Sample concentration | Electronegative membrane filtration with MgCl2 addition and acidification followed by extraction via NucliSENS easyMag. Solids were not removed or allowed to settle prior to filtration. | Dewatering by centrifugation at 24,000 g at 4 °C for 30 min and decanting supernatant. Extraction of 1.8 – 2.8 g of sample using the RNeasy PowerSoil total RNA kit with further purification using Zymo OneStep PCR inhibitor removal columns. |
| SARS-CoV-2 RNA RT-PCR target | N1, N2a | N |
| Detection technology | 1-step RT-ddPCR | 1-step RT-ddPCR |
| Case study reference | Gonzalez et al. (2020, 2021) | Graham et al. (2021); Kim et al. (2022); Wolfe et al. (2021) |
| Case data geographic area | The sewershed area determined from the treatment plant service area polygon and address-level case data | The sewershed service area of the treatment plant comprises approximately 75% of the cumulative cases for the entire county |
| Case data dates | Earliest available data hierarchy: (1) illness onset date; (2) specimen collection date of earliest associated lab; (3) date of diagnosis; (4) earliest date received by local/ county health department, date received by state, date of report, investigations start date, confirmation date, investigation create date | 1/27/2020 to 11/4/2021 reported by Santa Clara County Health District. |
PCR, polymerase chain reaction; ddPCR, digital droplet PCR.
In 2021, this changed to using a single N target, a slight modification of the N2 target published by Gonzalez et al. (2020, 2021).
Model parameterization and implementation
To parameterize the model for SARS-CoV-2, we used SARS-CoV-2 data from the scientific literature, other surrogate pathogen data when not available for SARS-CoV-2, and professional judgement as needed – details are provided in Supplementary Material, Section B. The model variables, corresponding statistical distributions, and distribution parameters used in the model formulations are shown in Table 2.
Table 2.
Model variables, distributions, and distribution parameters
| Model parameter | Description | Distribution | Distribution parameters |
|---|---|---|---|
|
| |||
| Ddiar | SARS-CoV-2 density in feces among infected individuals with diarrhea (log10 virus copies/mL) | Normal | (5.1, 0.76) |
| Vdiar | volume of feces per person per day among infected individuals with diarrhea (mL/day) | Normal | (1000, 100) |
| Fdiar | Fraction of infections with diarrhea (unitless) | Uniform | (0.2, 0.3) |
| Dnodiar | SARS-CoV-2 density in feces among infected individuals without diarrhea (log10 virus copies/g) | Normal | (4.067, 1.591) |
| Mnodiar | Mass of feces per person per day for infected individuals without diarrhea (g/day) | Lognormal | (4.84, 0.4) |
| Vww | Wastewater generated per person per day (L/day) | Lognormal | (5.397, 0.1595) |
| Tsewer | time wastewater spends in the sewer prior to reaching the wastewater treatment plant (h) | Lognormal | (1.2, 0.85) |
| Fshed | Fraction of infections resulting in shedding of viral RNA in feces (unitless) | Uniform | (0.6, 0.8) |
| Fcont | Volume fraction of wastewater coming from a source that could potentially be contributing to the observed reported influent SARS-CoV-2 concentration (unitless) | Uniform | (0.10, 0.85) |
| kvirus | Pseudo-first-order decay coefficient estimate for SARS-CoV-2 RNA target (day−1) | Point Estimate | 0.29 |
| kpMMoV | Pseudo-first-order decay coefficient estimate for PMMoV target (day−1) | Point Estimate | 0.29 |
| k_dSARS_CoV2 | SARS-CoV-2 Partitioning coefficient for conversion of settled solids density to wastewater influent density (unitless) | Point Estimate - Median | 900 |
| Point Estimate (lower, upper) | (280, 10,000) | ||
| k_d PMMoV | PMMoV Partitioning coefficient for conversion of settled solids density to wastewater influent density (unitless) | Point Estimate - Median | 2000 |
| Point Estimate (lower, upper) | (1000, 30,000) | ||
| DWW_endog | PMMoV density in feces (log10 virus copies/g) | Uniform from min-median; | (5.58, 8.28) |
| Uniform from median-max | (8.28, 9.99) | ||
For each formulation, we ran simulations using Eqns 2, 4a and 5 (for the flow generation, fecal strength, and flow receipt formulations, respectively) using case study SARS-CoV-2 influent concentration data to estimate . For the flow generation and fecal strength formulations, the measured daily average SARS-CoV-2 RNA concentration (via 24-h composite sampling) was converted to an estimated median SARS-CoV-2 RNA concentration. This conversion is necessary so that subsequent calculations are not biased by the measured SARS-CoV-2 RNA concentrations, which are inherently mean values and right skewed rather than the median values that are required for the subsequent calculations. For each observation, this conversion was accomplished by running an initial set of simulations to solve for an .value in which the simulated mean concentration equaled the observed mean. The median value of the simulated distribution was then used in the subsequent calculations as indicated above. For the flow receipt formulation, we computed an average value of through simulation for use in Eqn 5c for consistency with the conceptual model of flow receipt at a treatment plant rather than variation in from individuals. During each iteration of the numerical simulation, values for the parameters were drawn from their respective statistical distributions to estimate , and each Monte Carlo simulation comprised of 100,000 iterations for each case study and model formulation.
Model performance evaluations
We conducted a series of in-depth analyses to determine the extent to which selected parameter value choices in the three model formulations affected the output (see Supplementary Material, Section D for further detail). The parameters evaluated include the following: (1) the concentration of SARS-CoV-2 RNA in feces of infected individuals, (2) the duration of SARS-CoV-2 RNA shedding in feces, and (3) the partitioning coefficients for SARS-CoV-2 and PMMoV (for the San Jose, CA case study where primary sludge was sampled). Not all model parameters were evaluated in this way and other potentially important considerations related to sensitivity include, but are not limited to (1) the gene target used to quantify SARS-CoV-2 RNA in sewage; (2) uncertainty or variability of sewage concentration measurements including the approach to handling SARS-CoV-2 RNA values reported below quantifiable limits; and (3) relative contributions of non-fecal (mucosal) SARS-CoV-2 RNA shedding to the wastewater stream. Consideration of those additional factors could serve as a guide for future study.
RESULTS
Daily confirmed cases, cumulative confirmed cases, observed wastewater SARS-CoV-2 RNA and PMMoV RNA concentrations from Virginia Beach, VA (VB) are presented in Figure 1(a). These case data represent the sewershed area determined from the Atlantic wastewater treatment plant service area polygons and address-level cases as reported in Gonzalez et al. (2020, 2021). The daily confirmed cases, cumulative confirmed cases, and computed influent SARS-CoV-2 RNA and PMMoV RNA data for the San Jose, CA (SJ) case study are presented below in Figure 1(b). The SJ case data represent the City of San Jose treatment plant service area within Santa Clara County, which comprises approximately 75% of the population and 75% of the cumulative cases for the county.
Figure 1 |.
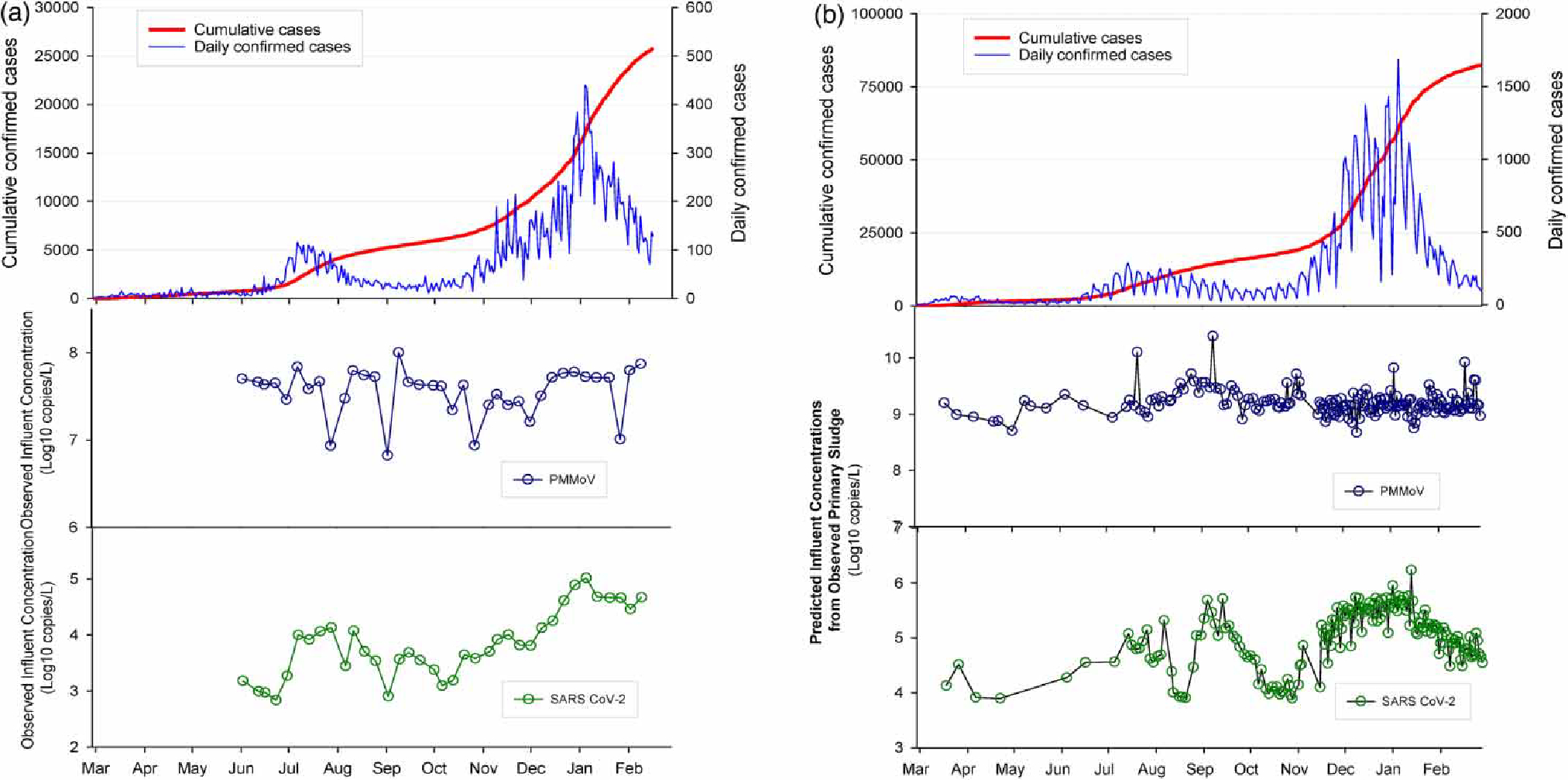
March 2020–February 2021 daily confirmed cases, cumulative confirmed cases, SARS-CoV-2 RNA and PMMoV RNA concentrations for (a) Virginia Beach, VA and (b) San Jose, CA.
Median values from the case study simulations are presented in Figure 2 alongside the reported daily case numbers (sewershed level for Virginia Beach and county level for San Jose). Note that the ratio of to the confirmed daily cases varies between the two case studies as shown on Figure 2(a) and 2(b). In general, the median values from all three formulations followed the reported case data trends well. However, daily simulated infection estimates exhibited large ranges (two orders of magnitude), in part due to the strong model dependence on highly variable viral fecal shedding data (Supplementary Material, Figure D1). Given the assumed uncertainty in the limited fecal SARS-CoV-2 RNA shedding data (further detail in Supplementary Material, Section C), the precision and accuracy of the estimated median values is unknown within the range of simulated values.
Figure 2 |.
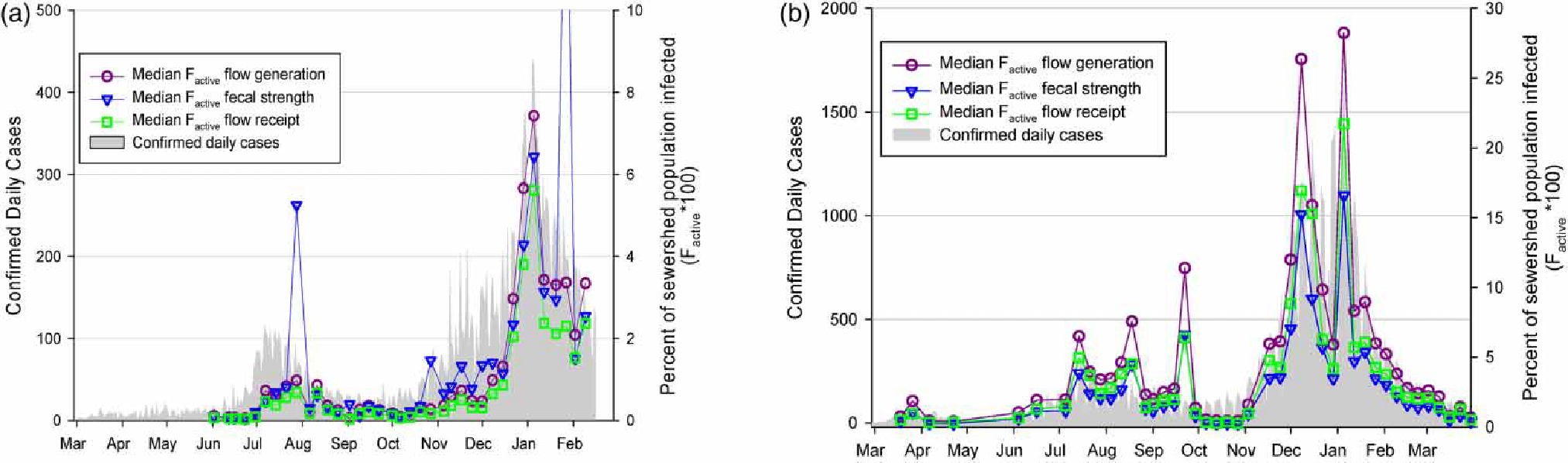
Estimated percent of population actively infected each day monitored for the three model formulations compared to reported cases for (a) Virginia Beach, VA and (b) San Jose, CA beginning March 2020. Note that the ratio of the left and right y-scales differs between plots (a) and (b).
To evaluate the simulated results within the context of the reported number of cases at each study location, we relied on published data indicating that infected individuals shed detectable SARS-CoV-2 RNA in their feces for a median of 21 days (Weiss et al. 2020; Zhou et al. 2020). Because the model formulations treat fecal shedding as a time-invariant parameter, we computed the cumulative number of confirmed cases over 21 days and plotted those against multiplied by the wastewater utility estimated sewershed contributing population (Figure 3). The slopes of the lines shown represent the average ‘ascertainment ratio (AR)’ (i.e., ratio of predicted to reported cases) over the period evaluated. As shown in Figure 3, at times the median simulated infection levels were lower than reported case burden. This observation suggests that there could be a discrepancy between the dates of reported cases and wastewater data (refer to Discussion), temporally varying nature of the true AR, and/or inaccuracies in the modeling results.
Figure 3 |.
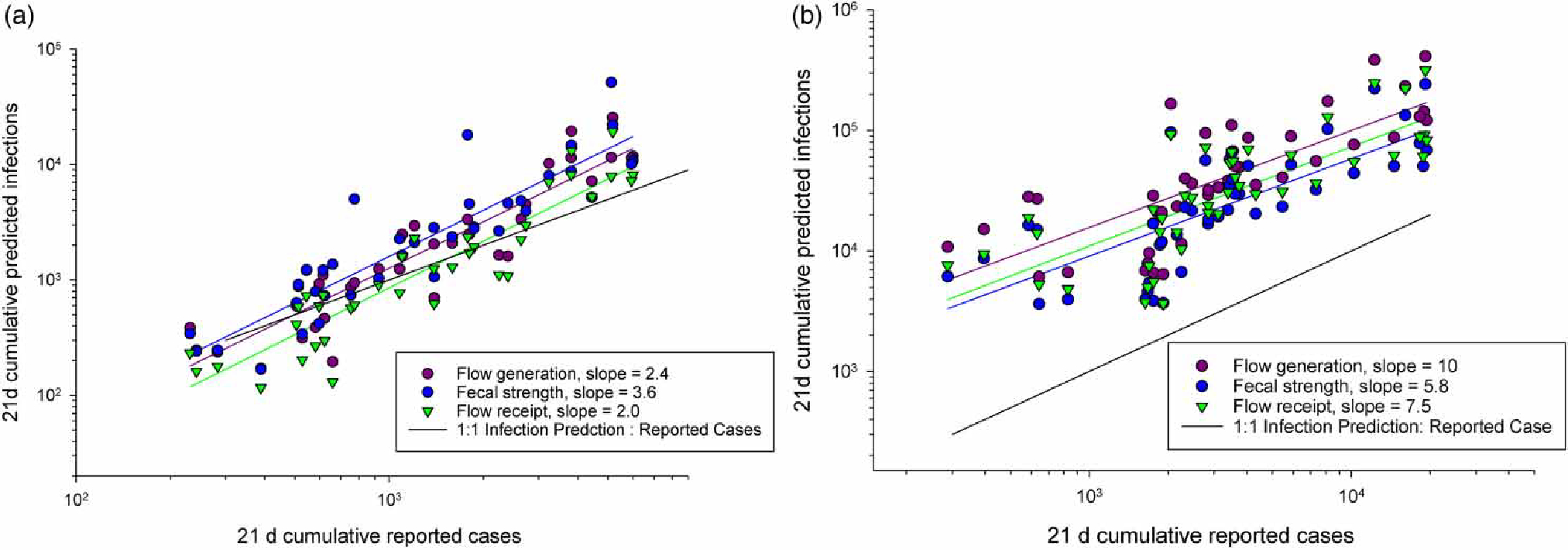
21-day reported cases versus predicted cases for the median outcome value of three model formulation simulations (a) Virginia Beach, VA and (b) San Jose, CA beginning March 2020. Reported slopes, representing ARs over surveyed period, are derived from linear regressions of linear scaled reported and estimated cases using the three model formulations.
Following Wolfe et al. (2021), we plotted the observed daily cases as a function of the influent concentrations of SARS-CoV-2 RNA for both case studies, and influent concentrations of SARS-CoV-2 RNA normalized by observed PMMoV RNA concentrations to evaluate and compare across case studies the association between wastewater concentrations of SARS-CoV-2 RNA and reported cases (Figure 4). As shown, the datasets from the two case studies are more comparable when normalized by PMMoV highlighting a potentially important implementation consideration.
Figure 4 |.
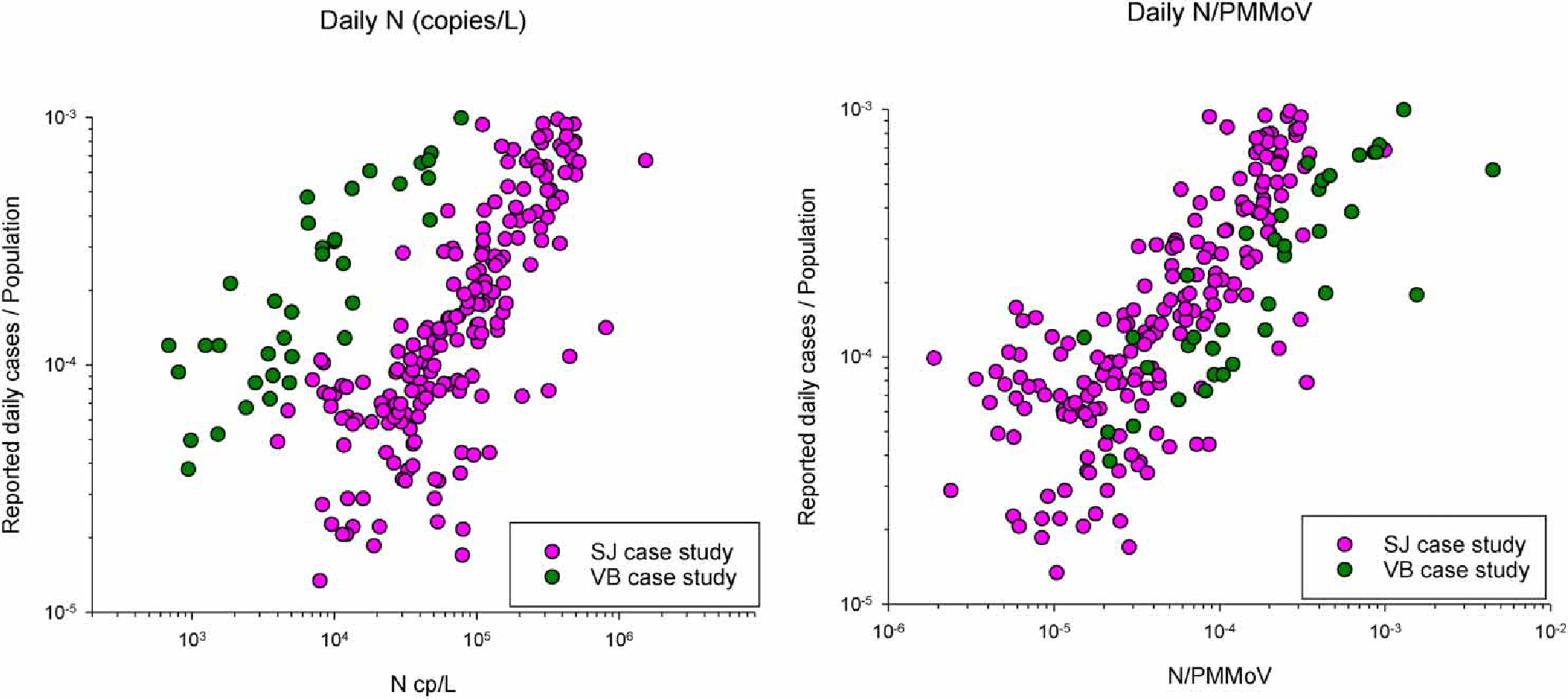
Reported daily cases versus observed influent concentrations of SARS-CoV-2 (left) and SARS-CoV-2/PMMoV (right) for Virginia Beach, VA and San Jose, CA case studies.
Model performance evaluations
Detailed results from the sensitivity evaluations are presented in Supplementary Material, Sections C and D. Section C includes the fecal shedding distribution evaluation, and Section D includes evaluation of the uncertainty in the simulation results (D1), variability/uncertainty in computed influent concentrations for the San Jose case study based on the range of partitioning coefficients (D2), fecal shedding duration (D3), and model formulation sensitivity from inter-site variability (D4). The sensitivity evaluations indicate that: (1) there is substantial variability/uncertainty in the model output from individual simulations (Supplementary Material, Figure D1). These results are likely driven primarily by the highly uncertain fecal shedding data (Supplementary Material, Section C); (2) computed influent concentrations are strongly influenced by the selected values for the partitioning coefficients for conditions using sludge data as input (Supplementary Material, Figure D2); and (3) the general trends observed within any given site were not dependent on the specific fecal shedding duration used (Supplementary Material, Figure D3). Further details are presented in Supplementary Material, Sections C and D.
DISCUSSION
This study describes a modeling framework for estimating community-level COVID-19 infection based on wastewater monitoring data that are independent of individual and clinical surveillance data. However, the case studies presented highlight the limited available data to parameterize key model inputs, resulting in high outcome variability and presumed uncertainty, in turn, limiting model application to support public health decision-making for the COVID-19 response. The mechanistic modeling framework described encompasses three complementary formulations, each of which requires a unique set of inputs for estimating human contribution to wastewater (referred to as fecal strength) and size of population represented by a wastewater sample. A principal benefit of the mechanistic modeling approach is that it provides the conversion of wastewater (or sludge) levels into estimated total cases for direct comparison and interpretation against clinical surveillance data for public health decision-making. For example, this wastewater modeling approach can aid in understanding the variable effective reproductive number through the epidemic by estimating a total community infection burden that includes unreported and sub-clinical infections (Huisman et al. 2022). An additional benefit of the mechanistic modeling approach is that it can be updated to aid in understanding relative differences in community infection as the underlying drivers of transmission change temporally, such as changing variants and increasing vaccination levels. It can also be useful to provide insights about relative infection levels during time periods where community-level testing is changing (increasing or decreasing) or reporting of testing results is delayed or aggregated (such as holidays or some weekends).
Available wastewater-related data, and public health decision-making needs will determine which of these three wastewater-based infection model formulations would be most suitable for a community. For example, if it is reasonable to estimate the fraction of wastewater that derives from industrial and/or other sources that are unlikely to contain viral contributions and to assume those estimates are stable over the modeled period; or if endogenous control data are not available or feasible to collect, the flow generation formulation may be most applicable. Conversely, if a community is generating paired SARS-CoV-2 RNA and endogenous control wastewater surveillance data, the fecal strength formulation may be desirable over the flow generation formulation to provide enhanced potential for inter-site comparability. Moreover, Figure 4 suggests that, when applying the model across lab methods, sample types, and locations, the FS model maybe more applicable because the fecal normalization incorporated into that model likely makes the ratio of SARS-CoV-2 to reported cases more consistent across sites and sample types.
Differences between wastewater characteristics in communities may also influence which formulation is most appropriate. For example, in communities with substantial agricultural contributions to the community wastewater, the use of PMMoV RNA or another endogenous control may be more complicated as PMMoV may not be human fecal-specific in those settings (Zhang et al. 2006), and thus, make the flow generation or flow receipt formulation more appropriate. Another consideration is the population served by a treatment facility and whether the population is relatively static (i.e., are there large changes over short periods of time or does a large fraction of the population live in one sewershed and work in another). The FS formulation may be more representative of the true population contributing feces to wastewater, but it is also likely more vulnerable to outliers, as seen in Figure 2, because the formulation is dependent on two microbial measurements that can easily vary across orders of magnitude. Interpretation of the modeling results is complicated in cases where the population is more dynamic, and in such cases, careful consideration of these important factors is warranted.
Average modeled AR values ranged from 2 to 10 across the two case studies for the period of March 2020 to March 2021. While this range appears plausible given the reported levels of asymptomatic COVID-19 infections and estimates of unreported cases (Ma et al. 2021; Sah et al. 2021), the range of model results and sensitivity evaluations limit confidence in the model output for any specific data point. In particular, fecal shedding data for SARS-CoV-2 RNA and PMMoV RNA are presently limited, constraining the utility of the model. Moreover, multiple wastewater testing method evaluation studies have shown that absolute SARS-CoV-2 RNA wastewater measurements are not necessarily comparable across laboratory methods or wastewater sample types (Deng et al. 2021; Pecson et al. 2021; Foster et al. 2022; Kim et al. 2022). Therefore, it is unknown whether the differences in AR values between the two case studies reported here (mean AR values of 2–4 in VB compared to 6–10 in SJ) represent true differences in community infection levels captured by case reporting over the modeled period or are a function of differences in virus recovery, testing methods, and/or sample types (i.e., influent versus sludge). However, one interesting perspective from this modeling approach is the evaluation of the temporally varying nature of AR, which can vary due to many factors including community mitigation behavior (e.g., mask wearing, physical distancing), testing availability, circulating variants, and vaccination status. Few other analytical approaches have the ability to evaluate this characteristic on a time frame relevant for decision-making (Fernandez-Cassi et al. 2021; Huisman et al. 2022).
Although we conducted an extensive analysis of available data related to fecal shedding of SARS-CoV-2 RNA, there are limitations and uncertainties associated with those data. To mitigate potential bias associated with high levels of shedding, we selected best fitting distributions with an emphasis on the upper tails (refer to Supplementary Material, Section C for evaluation of this assumption on model estimates). However, future work is critically needed to characterize fecal shedding of SARS-CoV-2 RNA and human-specific microbial fecal targets, such as PMMoV RNA, to enhance the utility of this modeling framework. The model sensitivity evaluations showed that the mechanistic model was most sensitive to the fecal shedding parameter, which due to lack of available data was treated stochastically rather than temporally varying over the estimated shedding period. Temporal characterization of SARS-CoV-2 RNA fecal shedding over the full duration of infection, from exposure to recovery could allow for a time dependent model and in turn may reduce the uncertainty in community SARS-CoV-2 infection level estimates from wastewater. Similarly, inter-laboratory method evaluations that identify critical methodological impacts on measurement comparability would allow for model output comparability across sites. Finally, while shedding contributions via other bodily fluid types (e.g., mucous, urine, etc.) was found to be negligible compared to fecal contributions (Supplementary Material, Appendix B), these other shedding routes should be considered as variants emerge with potentially different clinical presentations.
Until more well characterized shedding data are available, site-specific relative ascertainment ratios (RAR) (relative to specific times of known infection and transmission profiles as described in the Supplementary Material, Section A for methods and Supplementary Material, Figures D3 and D4 for examples) are a present use for the described SARS-CoV-2 infection modeling framework given the uncertainties in the available input data. Relative AR values could provide insight into coverage of testing resources in relation to trends in total community cases relative to reported cases (Supplementary Material, Section D). This may become particularly useful to continue monitoring SARS-CoV-2 infection levels as mass screening of asymptomatic patients is no longer conducted due to elevated costs and increased vaccination coverage. Another possible approach that could help to minimize bias associated with the proposed mechanistic model uncertainty is the use of multiple formulations. This general approach has proven to be useful in other unrelated applications that need to account for substantial uncertainty in the modeling (i.e., long-range weather forecasting).
The described mechanistic modeling framework can be used for future wastewater surveillance targets, such as those diseases currently endemic in the United States, like norovirus, or other pathogens where sufficient high-quality input data are available. One potential complicating factor is the presence of pathogens that are also shed from non-human sources (i.e., parasitic protozoans such as Cryptosporidium spp.) as those other sources to the waste stream are not accounted for in the present framework. Antibiotic resistance genes (ARGs) are another promising target for wastewater surveillance, but in terms of modeling infection burden, ARGs have the added complication of being carried by multiple health relevant microorganisms. However, the current framework could be used for emerging pathogens whose characteristics are similar to currently well characterized pathogens – new SARS variants may fall into this category in the near term. In this case, the framework can be useful for complementing traditional epidemiological approaches (i.e., case counts, etc.) for broadly understanding pathogen spread within a community while the AR is unknown or changing.
Because of the public health utility of wastewater surveillance, the Centers for Disease Control and Prevention in the United States (CDC) launched the National Wastewater Surveillance System (NWSS) (CDC 2021b). NWSS provides novel national wastewater-based disease surveillance infrastructure to support public health action during and after the COVID-19 pandemic by providing coordination, laboratory and epidemiology capacity, and technical guidance for wastewater surveillance programs implemented by state, tribal, local, and territorial health departments (Kirby et al. 2021). NWSS runs a centralized data system that standardizes wastewater data submission, analysis, visualization, and sharing between jurisdictions and with the public. If the uncertainty in wastewater-infection modeling is sufficiently reduced, NWSS is positioned to rapidly integrate this analytic capacity into the data dashboard for public health use, which may become more relevant as CDC adds new targets, including emerging pathogens and antimicrobial resistance genes (CDC 2021b).
The interpretation of wastewater SARS-CoV-2 RNA data will continue to be multi-faceted and complicated as vaccination and variant emergence evolve. Understanding the extent to which these emerging factors will affect SARS-CoV-2 RNA fecal shedding will be critical to understand the utility of this modeling framework. Nevertheless, interpretation of wastewater SARS-CoV-2 RNA data will continue to be important to complement traditional surveillance data and the modeling framework presented can be useful for researchers and public health decision-makers.
Supplementary Material
HIGHLIGHTS.
Reported COVID-19 cases do not capture total infections which is important for understanding community disease transmission.
We present three wastewater mechanistic simulation models to estimate community infection levels and demonstrate them through two case studies.
Wastewater-based mechanistic models presented are useful for public health decisions but currently limited by limited SARS-CoV-2 fecal shedding data.
ACKNOWLEDGEMENTS
We thank the participating wastewater utilities, Hampton Roads Sanitation District and San José-Santa Clara Regional Wastewater Facility, for the use of their data and dedication to protecting public health during the COVID-19 response. We also thank Santa Clara County and Virginia Beach Health Department for gathering and publicly sharing their data used in this study, as well as their dedication to the COVID-19 response. Finally, we acknowledge Daniel Gerrity, Nasa Armstrong, and Zach Marsh for their intellectual feedback and discussion regarding model development. Use of trade names and commercial sources is for identification only and does not imply endorsement by the Centers for Disease Control and Prevention (CDC). The findings and conclusions in this report are those of the authors and do not necessarily represent those of the CDC.
Footnotes
CONFLICT OF INTEREST
The authors declare there is no conflict.
DATA AVAILABILITY STATEMENT
All relevant data are included in the paper or its Supplementary Information.
REFERENCES
- Ahmed W, Angel N, Edson J, Bibby K, Bivins A, O’Brien JW, Choi PM, Kitajima M, Simpson SL, Li J, Tscharke B, Verhagen R, Smith WJM, Zaugg J, Dierens L, Hugenholtz P, Thomas KV & Mueller JF 2020. First confirmed detection of sars-cov-2 in. untreated wastewater in australia: A proof of concept for the wastewater surveillance of covid-19 in.the community. The Science of the Total Environment 728, 138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asghar H, Diop OM, Weldegebriel G, Malik F, Shetty S, El Bassioni L, Akande AO, Al Maamoun E, Zaidi S, Adeniji AJ, Burns CC, Deshpande J, Oberste MS & Lowther SA 2014. Environmental surveillance for polioviruses in the global polio eradication initiative. J Infect Dis 210 Suppl 1, S294–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud D, Qi X, Nielsen-Saines K, Musso D, Pomar L & Favre G 2020. Real estimates of mortality following covid-19 infection. The Lancet Infectious diseases 20(1), 773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancourt WQ, Schmitz BW, Innes GK, Prasek SM, Pogreba Brown KM, Stark ER, Foster AR, Sprissler RS, Harris DT, Sherchan SP, Gerba CP & Pepper IL. 2021. Covid-19 containment on a college campus via wastewater-based epidemiology, targeted clinical testing and an intervention. Sci Total Environ 779, 146408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bivins A, North D, Ahmad A, Ahmed W, Alm E, Been F, Bhattacharya P, Bijlsma L, Boehm AB, Brown J, Buttiglieri G, Calabro V, Carducci A, Castiglioni S, Cetecioglu Gurol Z, Chakraborty S, Costa F, Curcio S, de Los Reyes FL 3rd, Vela J Delgado, Farkas K, Fernandez-Casi X, Gerba C, Gerrity D, Girones R, Gonzalez R, Haramoto E, Harris A, Holden PA, Islam MT, Jones DL, Kasprzyk-Hordern B, Kitajima M, Kotlarz N, Kumar M, Kuroda K, La Rosa G, Malpei F, Mautus M, McLellan SL, Medema G, Meschke JS, Mueller J, Newton RJ, Nilsson D, Noble RT, van Nuijs A, Peccia J, Perkins TA, Pickering AJ, Rose J, Sanchez G, Smith A, Stadler L, Stauber C, Thomas K, van der Voorn T, Wigginton K, Zhu K & Bibby K 2020. Wastewater-based epidemiology: Global collaborative to maximize contributions in the fight against covid-19. Environ Sci Technol 54, 7754–7757. [DOI] [PubMed] [Google Scholar]
- CDC 2021a. Covid-19, https://www.Cdc.Gov/coronavirus/2019-ncov/index.Html. Available: https://www.cdc.gov/coronavirus/2019-ncov/cases-updates/geographic-seroprevalence-surveys.html [accessed May 24, 2021 2021].
- CDC 2021b. National wastewater surveillance system (nwss), https://www.Cdc.Gov/coronavirus/2019-ncov/cases-updates/wastewater-surveillance.Html. [accessed May 24, 2021 2021].
- Ceylan Z 2020. Estimation of covid-19 prevalence in italy, spain, and france. Science of The Total Environment 729, 138817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung KS, Hung IF, Chan PP, Lung KC, Tso E, Liu R, Ng YY, Chu MY, Chung TW, Tam AR, Yip CC, Leung KH, Yim-Fong Fung A, Zhang RR, Lin Y, Cheng HM, Zhang AJ, To KK, Chan KH, Yuen KY & Leung WK 2020. Gastrointestinal manifestations of sars-cov-2 infection and virus load in fecal samples from the hong kong cohort and systematic review and meta-analysis. Gastroenterology 159(1), 81–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Aoust PM, Graber TE, Mercier E, Montpetit D, Alexandrov I, Neault N, Baig AT, Mayne J, Zhang X, Alain T, Servos MR, Srikanthan N, MacKenzie M, Figeys D, Manuel D, Juni P, MacKenzie AE & Delatolla R 2021. Catching a resurgence: Increase in sars-cov-2 viral rna identified in wastewater 48 h before covid-19 clinical tests and 96 h before hospitalizations. Sci Total Environ 770, 145319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng K, Uhlig S, Ip HS, Lea Killian M, Goodman LB, Nemser S, Ulaszek J, Pickens S, Newkirk R, Kmet M, Frost K, Hettwer K, Colson B, Nichani K, Schlierf A, Tkachenko A, Reddy R & Reimschuessel R 2021. Interlaboratory comparison of sars-cov2 molecular detection assays in use by u.S. Veterinary diagnostic laboratories. Journal of veterinary diagnostic investigation: official publication of the American Association of Veterinary Laboratory Diagnosticians, Inc 33, 1039–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Cassi X, Scheidegger A, Bänziger C, Cariti F, Tuñas Corzon A, Ganesanandamoorthy P, Lemaitre JC, Ort C, Julian TR & Kohn T 2021. Wastewater monitoring outperforms case numbers as a tool to track covid-19 incidence dynamics when test positivity rates are high. Water Research 200, 117252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foladori P, Cutrupi F, Segata N, Manara S, Pinto F, Malpei F, Bruni L & La Rosa G 2020. Sars-cov-2 from faeces to wastewater treatment: What do we know? A review. Sci Total Environ 743, 140444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster CSP, Stelzer-Braid S, Deveson IW, Bull RA, Yeang M, Au J-P, Ruiz Silva M, van Hal SJ, Rockett RJ, Sintchenko V, Kim KW & Rawlinson WD 2022. Assessment of inter-laboratory differences in sars-cov-2 consensus genome assemblies between public health laboratories in australia. Viruses 14, 185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerrity D, Papp K, Stoker M, Sims A & Frehner W 2021. Early-pandemic wastewater surveillance of sars-cov-2 in.southern nevada: Methodology, occurrence, and incidence/prevalence considerations. Water research X 10, 100086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez R, Curtis K, Bivins A, Bibby K, Weir MH, Yetka K, Thompson H, Keeling D, Mitchell J & Gonzalez D 2020. Covid-19 surveillance in southeastern virginia using wastewater-based epidemiology. Water Res 186, 116296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez R, Larson A, Thompson H, Carter E & Cassi XF 2021. Redesigning sars-cov-2 clinical rt-qpcr assays for wastewater rt-ddpcr. medRxiv:DOI 10.1101/2021.1103.1102.21252754. [DOI] [Google Scholar]
- Graham KE, Loeb SK, Wolfe MK, Catoe D, Sinnott-Armstrong N, Kim S, Yamahara KM, Sassoubre LM, Mendoza Grijalva LM, Roldan-Hernandez L, Langenfeld K, Wigginton KR & Boehm AB 2021. Sars-cov-2 rna in wastewater settled solids is associated with covid-19 cases in a large urban sewershed. Environ Sci Technol 55, 488–498. [DOI] [PubMed] [Google Scholar]
- Hamza IA, Jurzik L, Uberla K & Wilhelm M 2011. Evaluation of pepper mild mottle virus, human picobirnavirus and torque teno virus as indicators of fecal contamination in river water. Water Res 45, 1358–1368. [DOI] [PubMed] [Google Scholar]
- Hart OE & Halden RU 2020. Computational analysis of sars-cov-2/covid-19 surveillance by wastewater-based epidemiology locally and globally: Feasibility, economy, opportunities and challenges. Sci Total Environ 730, 138875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt J, Trowsdale S, Armstrong BA, Chapman JR, Carter KM, Croucher DM, Trent CR, Sim RE & Gilpin BJ 2022. Sensitivity of wastewater-based epidemiology for detection of sars-cov-2 rna in a low prevalence setting. Water Res 211, 118032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisman J, Scire J, Caduff L, Fernandez-Cassi X, Ganesanandamoorthy P, Kull A, Scheidegger A, Stachler E, Boehm A, Hughes B, Knudson A, Topol A, Wigginton K, Wolfe M, Kohn T, Ort C, Stadler T & Julian T 2022. Wastewater-based estimation of the effective reproductive number of sars-cov-2. Environ Health Perspect 130, 057011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan EH, Wang D, Wang M, Malik AA, Zulli A & Peccia J 2021. Aligning sars-cov-2 indicators via an epidemic model: Application to hospital admissions and rna detection in sewage sludge. Health care management science 24, 320–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan EH, Zulli A, Sanchez M & Peccia J 2022. Scaling sars-cov-2 wastewater concentrations to population estimates of infection. Scientific Reports 12, 3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Kennedy LC, Wolfe MK, Criddle CS, Duong DH, Topol A, White BJ, Kantor RS, Nelson KL, Steele JA, Langlois K, Griffith JF, Zimmer-Faust AG, McLellan SL, Schussman MK, Ammerman M, Wigginton KR, Bakker KM & Boehm AB 2022. Sars-cov-2 rna is enriched by orders of magnitude in primary settled solids relative to liquid wastewater at publicly owned treatment works. Environmental Science: Water Research & Technology 8, 757–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby AE, Walters MS, Jennings WC, Fugitt R, LaCross N, Mattioli M, Marsh ZA, Roberts VA, Mercante JW, Yoder J & Hill VR 2021. Using wastewater surveillance data to support the covid-19 response - united states, 2020–2021. MMWR Morb Mortal Wkly Rep 70, 1242–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby AE, Welsh RM, Marsh ZA, Yu AT, Vugia DJ, Boehm AB, Wolfe MK, White BJ, Matzinger SR, Wheeler A, Bankers L, Andresen K, Salatas C New York City Department of Environmental P, Gregory DA., Johnson MC., Trujillo M., Kannoly S., Smyth DS., Dennehy JJ., Sapoval N., Ensor., Treangen T., Stadler LB. & Hopkins L. 2022. Notes from the field: Early evidence of the sars-cov-2 b.1.1.529 (omicron) variant in community wastewater - united states, november-december 2021. MMWR Morb Mortal Wkly Rep 71, 103–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Pei S, Chen B, Song Y, Zhang T, Yang W & Shaman J 2020. Substantial undocumented infection facilitates the rapid dissemination of novel coronavirus (sars-cov-2). Science 368, 489–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q, Liu J, Liu Q, Kang L, Liu R, Jing W, Wu Y & Liu M 2021. Global percentage of asymptomatic sars-cov-2 infections among the tested population and individuals with confirmed covid-19 diagnosis: A systematic review and meta-analysis. JAMA Network Open 4, e2137257–e2137257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema G, Heijnen L, Elsinga G, Italiaander R & Brouwer A 2020. Presence of sars-coronavirus-2 rna in sewage and correlation with reported covid-19 prevalence in the early stage of the epidemic in the netherlands. Environmental Science & Technology Letters 7, 511–516. [DOI] [PubMed] [Google Scholar]
- Paul R, Arif AA, Adeyemi O, Ghosh S & Han D 2020. Progression of covid-19 from urban to rural areas in the united states: A spatiotemporal analysis of prevalence rates. The Journal of Rural Health 36, 591–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peccia J, Zulli A, Brackney DE, Grubaugh ND, Kaplan EH, Casanovas-Massana A, Ko AI, Malik AA, Wang D, Wang M, Warren JL, Weinberger DM, Arnold W & Omer SB 2020. Measurement of sars-cov-2 rna in wastewater tracks community infection dynamics. Nature biotechnology 38, 1164–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecson BM, Darby E, Haas CN, Amha YM, Bartolo M, Danielson R, Dearborn Y, Di Giovanni G, Ferguson C, Fevig S, Gaddis E, Gray D, Lukasik G, Mull B, Olivas L, Olivieri A, Qu Y & Consortium SA-C-I. 2021. Reproducibility and sensitivity of 36 methods to quantify the sars-cov-2 genetic signal in raw wastewater: Findings from an interlaboratory methods evaluation in the u.S. Environmental Science: Water Research & Technology 7, 504–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penn R, Ward BJ, Strande L & Maurer M 2018. Review of synthetic human faeces and faecal sludge for sanitation and wastewater research. Water Res 132, 222–240. [DOI] [PubMed] [Google Scholar]
- Saguti F, Magnil E, Enache L, Churqui MP, Johansson A, Lumley D, Davidsson F, Dotevall L, Mattsson A, Trybala E, Lagging M, Lindh M, Gisslen M, Brezicka T, Nystrom K & Norder H 2021. Surveillance of wastewater revealed peaks of sars-cov-2 preceding those of hospitalized patients with covid-19. Water Res 189, 116620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sah P, Fitzpatrick MC, Zimmer CF, Abdollahi E, Juden-Kelly L, Moghadas SM, Singer BH & Galvani AP 2021. Asymptomatic sars-cov-2 infection: A systematic review and meta-analysis. Proceedings of the National Academy of Sciences 118, e2109229118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soller JA & Eisenberg JNS. 2008. An evaluation of parsimony for microbial risk assessment models. Environmetrics 19, 61–78. [Google Scholar]
- Turk PJ, Chou S-H, Kowalkowski MA, Palmer PP., Priem JS, Spencer MD., Taylor YJ. & McWilliams AD. 2020. Modeling covid-19 latent prevalence to assess a public health intervention at a state and regional scale: Retrospective cohort study. JMIR Public Health Surveill 6, e19353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidhaas J, Aanderud ZT, Roper DK, VanDerslice J, Gaddis EB, Ostermiller J, Hoffman K, Jamal R, Heck P, Zhang Y, Torgersen K, Laan JV & LaCross N 2021. Correlation of sars-cov-2 rna in wastewater with covid-19 disease burden in sewersheds. Sci Total Environ 775, 145790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss A, Jellingso M & Sommer MOA 2020. Spatial and temporal dynamics of sars-cov-2 in.covid-19 patients: A systematic review and meta-analysis. EBioMedicine 58, 102916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO 2020a. Antimicrobial resistance surveillance system (glass). Available from: Https://www.Who.Int/glass/en/. Accessed 19 august 2020.
- WHO 2020b. Guidelines for environmental surveillance of poliovirus circulation. Available from https://apps.Who.Int/iris/handle/10665/67854?Locale=en&mode=full. Accessed 19 august 2020.
- WHO 2020c. Status of environmental surveillance for sars-cov-2 virus, scientific brief, available from: Https://apps.Who.Int/iris/bitstream/handle/10665/333670/who-2019-ncov-sci_brief-environmentalsampling-2020.1-eng.Pdf. Accessed 19 august 2020.
- Wolfe MK, Topol A, Knudson A, Simpson A, White B, Vugia DJ, Yu AT, Li L, Balliet M, Stoddard P, Han GS, Wigginton KR & Boehm AB. 2021. High-frequency, high-throughput quantification of sars-cov-2 rna in wastewater settled solids at eight publicly owned treatment works in northern california shows strong association with covid-19 incidence. mSystems 6, e0082921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, Muller MA, Niemeyer D, Jones TC, Vollmar P, Rothe C, Hoelscher M, Bleicker T, Brunink S, Schneider J, Ehmann R, Zwirglmaier K, Drosten C & Wendtner C 2020. Virological assessment of hospitalized patients with covid-2019. Nature 581, 465–469. [DOI] [PubMed] [Google Scholar]
- Wu SL, Mertens AN, Crider YS, Nguyen A, Pokpongkiat NN, Djajadi S, Seth A, Hsiang MS, Colford J. M., Jr Jr, Reingold A, Arnold BF, Hubbard A & Benjamin-Chung J 2020. Substantial underestimation of sars-cov-2 infection in the united states. Nature communications 11, 4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtzer S, Waldman P, Ferrier-Rembert A, Frenois-Veyrat G, Mouchel JM, Boni M, Maday Y, consortium O, Marechal V & Moulin L 2021. Several forms of sars-cov-2 rna can be detected in wastewaters: Implication for wastewater-based epidemiology and risk assessment. Water Res 198, 117183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Breitbart M, Lee WH, Run JQ, Wei CL, Soh SW, Hibberd ML, Liu ET, Rohwer F & Ruan Y 2006. Rna viral community in human feces: Prevalence of plant pathogenic viruses. PLoS biology 4, e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Du R-H, Li B., Zheng X-S, Yang X-L, Hu B., Wang Y-Y, Xiao G-F, Yan B., Shi Z-L & Zhou P. 2020. Molecular and serological investigation of 2019-ncov infected patients: Implication of multiple shedding routes. Emerging Microbes & Infections 9, 386–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S, Fan J, Yu F, Feng B, Lou B, Zou Q, Xie G, Lin S, Wang R, Yang X, Chen W, Wang Q, Zhang D, Liu Y, Gong R, Ma Z, Lu S, Xiao Y, Gu Y, Zhang J, Yao H, Xu K, Lu X, Wei G, Zhou J, Fang Q, Cai H, Qiu Y, Sheng J, Chen Y & Liang T 2020. Viral load dynamics and disease severity in patients infected with sars-cov-2 in.zhejiang province, china, january-march 2020: Retrospective cohort study. BMJ 369, m1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, Xiang J, Wang Y, Song B, Gu X, Guan L, Wei Y, Li H, Wu X, Xu J, Tu S, Zhang Y, Chen H & Cao B 2020. Clinical course and risk factors for mortality of adult inpatients with covid-19 in.wuhan, china: A retrospective cohort study. The Lancet 395, 1054–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Oishi W, Maruo C, Saito M, Chen R, Kitajima M & Sano D 2021. Early warning of covid-19 via wastewater-based epidemiology: Potential and bottlenecks. Sci Total Environ 767, 145124. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data are included in the paper or its Supplementary Information.


