Abstract
Translocation of preproteins across the mitochondrial outer membrane is mediated by the TOM complex. This complex consists of receptor components for the initial contact with preproteins at the mitochondrial surface and membrane-embedded proteins which promote transport and form the translocation pore. In order to understand the interplay between the translocating preprotein and the constituents of the TOM complex, we analyzed the dynamics of the TOM complex of Neurospora crassa and Saccharomyces cerevisiae mitochondria by following the structural alterations of the essential pore component Tom40 during the translocation of preproteins. Tom40 exists in a homo-oligomeric assembly and dynamically interacts with Tom6. The Tom40 assembly is influenced by a block of negatively charged amino acid residues in the cytosolic domain of Tom22, indicating a cross-talk between preprotein receptors and the translocation pore. Preprotein binding to specific sites on either side of the outer membrane (cis and trans sites) induces distinct structural alterations of Tom40. To a large extent, these changes are mediated by interaction with the mitochondrial targeting sequence. We propose that such targeting sequence-induced adaptations are a critical feature of translocases in order to facilitate the movement of preproteins across cellular membranes.
The import of proteins into mitochondria is mediated by multisubunit translocases in the outer (TOM complex) and inner (TIM complex) membranes of the organelles (23, 28, 33). The TOM complex contains components which expose domains to the cytosol and act as preprotein receptors. The major import receptors are Tom20 and Tom22, which are essential for the specific recognition, unfolding, and translocation of the majority of preproteins (22). Both components interact with preproteins and cooperate in the formation of a presequence binding site termed the cis site (3, 20, 25, 26, 34). Another binding site for a more restricted set of preproteins, especially for members of the mitochondrial carrier family, is Tom70 (13, 35, 36), which acts in conjunction with Tom37 (12). From this binding site, preproteins are transferred to Tom20-Tom22 before entering the translocation pore (19).
Other components of the TOM complex (Tom40, Tom5, Tom6, and Tom7) are deeply embedded in the outer membrane and are believed to form the translocation pore. Tom40 is an essential protein and was found in the vicinity of polypeptide chains in transit (31, 37, 39). The protein was suggested to be a central element of the preprotein-conducting pore of the mitochondrial outer membrane. The small members of the TOM complex are not essential by themselves, but combined deletion of their genes and those of other components of the translocase is lethal (1, 6, 15). Studies on the function of the small TOM complex proteins suggest that they play distinct roles. Tom6 and Tom7 were found to influence the stability of the TOM complex (1, 15). For Tom5 a function in facilitating preprotein transfer from the receptors into the translocation pore was reported (6).
Much information has been recently obtained on how mitochondrial preproteins are recognized by the receptor components and how preproteins move across the outer membrane (reviewed in reference 23). Comparatively little is known, however, about structural rearrangements occurring within the TOM complex in response to preprotein binding, insertion, and membrane translocation. Such dynamic alterations of the TOM complex might be a crucial feature of the translocation process, as they might be linked to the stepwise and progressive movement of the polypeptide chain across the membrane. Therefore, knowledge of changes in the spatial arrangement of various members of the translocase are important for a comprehensive description of the molecular events leading to preprotein transfer across the outer membrane.
To investigate the dynamic behavior of the TOM complex during preprotein transfer, we have chosen to analyze the molecular environment of a key component of the TOM complex, Tom40, at various stages of translocation across the outer membrane. Deeper insights into the structure of Tom40, its interaction with other TOM complex components, and the dynamic cross-talk between Tom40 and preproteins in transit should provide information about the translocation process at the molecular level.
Our findings show that Tom40 undergoes multiple conformational changes during the various stages of preprotein translocation. The alterations affect both the structure of the Tom40 oligomer and its interaction with other members of the TOM complex. These structural rearrangements are triggered, at least to a large extent, by interaction with the mitochondrial targeting sequence. Our data suggest that such targeting sequence-induced adaptations of the translocase are crucial for the movement of preproteins across the mitochondrial outer membrane.
MATERIALS AND METHODS
General biochemical procedures.
Isolation of mitochondria or mitochondrial outer membrane vesicles (OMV) from Neurospora crassa and the yeast Saccharomyces cerevisiae was performed as described elsewhere (5, 24). The TOM complex was purified from OMV isolated from N. crassa GR-107 carrying a hexahistidinyl-tagged tom22 gene instead of the wild-type copy. OMV were solubilized in buffer A (50 mM KCl, 10 mM MOPS-KOH [pH 7.0]) containing 1% digitonin. Samples were centrifuged for 30 min at 226,000 × g, and the supernatant was applied to a Ni-nitrilotriacetic acid (NTA) agarose affinity matrix. The column was washed with buffer A containing 0.5% digitonin, and bound protein was eluted with an imidazole gradient (0 to 300 mM). The TOM complex was recovered in one main peak. The enrichment of TOM complex proteins over the major outer membrane protein porin was at least 1,000-fold (21). Antibodies against N. crassa Tom6 were raised in rabbits by injecting a peptide corresponding to the 12 N-terminal residues. The peptide was coupled to keyhole limpet hemocyanine (Pierce). A chemiluminescence kit (ECL Kit; Amersham) and goat anti-rabbit antibodies conjugated to horseradish peroxidase were used for immunostaining.
Yeast strains.
The yeast strain SEY6210 was used for cross-linking experiments (17). A yeast strain with an N-terminal hexahistidinyl-tagged version of Tom40 was constructed by transforming strain W303 MATa with the vector pVT102U (38) carrying the tom40his6 gene.
Translocation of precursor proteins.
Chemical amounts of pSu9(1-69)-DHFR with a hexahistidinyl tag at the C terminus [pSu9(1-69)-DHFRhis6] were purified by Ni-NTA affinity chromatography from extracts of the Escherichia coli strain DH5α carrying the pQE60-pSu9(1-69)-DHFRhis6 overexpression vector (25). OMV were suspended in import buffer (bovine serum albumin [0.25 mg/ml], 20 mM KCl, 2.5 mM MgCl2, 10 mM MOPS (morpholine propanesulfonic acid)-KOH [pH 7.2]) in the absence or presence of 1 mM NADPH and 1 μM methotrexate (MTX). In experiments using mitochondria, the import buffer was supplemented with 220 mM sucrose and with 30 μM carbonyl cyanide m-chlorophenylhydrazone to dissipate the membrane potential across the inner membrane. pSu9-DHFRhis6 was then added and incubated with OMV or mitochondria for the desired times at various temperatures. Samples were diluted with high- or low-salt buffer (10 mM MOPS-KOH and 1 mM EDTA [pH 7.2] plus 120 or 20 mM KCl, respectively) containing 220 mM sucrose for experiments with mitochondria. Finally, OMV or mitochondria were reisolated by centrifugation for 20 min at 125,000 × g or 10 min at 12,000 × g, respectively.
Cross-linking.
For cross-linking experiments, intact mitochondria, OMV, or the purified TOM complex was suspended in SEMK buffer (220 mM sucrose, 1 mM EDTA, 10 mM MOPS [pH 7.2], and 20 mM KCl) and incubated with various cross-linking reagents (all from Pierce) for 30 min at 25°C. The concentration of the cross-linkers was 440 μM for disuccinimidyl glutarate (DSG), 300 μM for dithiobis(succinimidylpropionate) (DSP), 250 μM for 1,4-di-[3′-(2′-pyridyldithio)propionamido]butane (DPDPB), and 1 mM for 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride (EDC). Excess cross-linker was quenched by the addition of 80 mM glycine (pH 8.0) and incubation for 10 min at 25°C. Aliquots were removed before and after the addition of the cross-linking reagents; proteins were precipitated with trichloroacetic acid and analyzed by immunostaining.
Gel filtration analysis.
Purified OMV (900 μg) were solubilized in buffer G (30 mM KCl, 6% glycerol, 10 mM MOPS–KOH, 2% digitonin [pH 7.2]). After a clarifying spin (20 min at 125,000 × g), the supernatant was applied on a Superose 6 gel filtration column (25 ml column volume; Pharmacia) and chromatographed in buffer G at a flow rate of 0.2 ml/min. Fractions (0.5 ml) were collected and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunostaining with antibodies against Tom40 and other TOM components. Calibration standards used were as follows: S. cerevisiae alcohol dehydrogenase (150 kDa), apoferritin (440 kDa), and β-thyroglobulin (660 kDa).
RESULTS
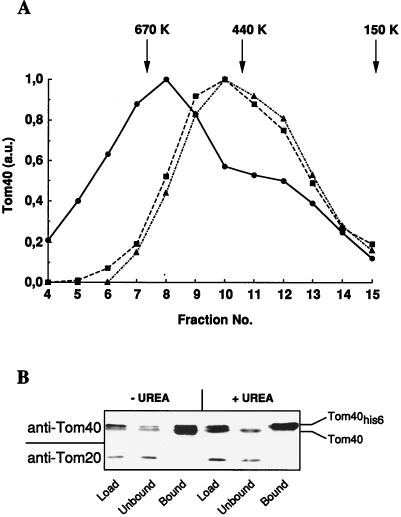
To investigate the oligomeric state of Tom40 in the TOM complex, OMV were isolated from N. crassa mitochondria (24), solubilized in buffer containing 1% digitonin, and subjected to gel filtration. Tom40 was found in an assembly with a molecular mass of 600 kDa (Fig. 1A). In addition, this complex contained the receptors Tom20, Tom22, and Tom70, as verified by immunostaining (not shown). Treatment of the OMV with trypsin before or after the solubilization resulted in a shift of the Tom40-containing complex to an apparent molecular mass of 470 kDa. Trypsin degrades the receptors of the TOM machinery but leaves Tom40 intact (19, 24). These data suggest an oligomeric structure of Tom40 in the mitochondrial outer membrane.
FIG. 1.
Tom40 forms an oligomeric structure in the outer membrane of N. crassa and yeast mitochondria. (A) Untreated OMV (circles) and trypsin-treated OMV (squares) were solubilized in buffer G. A third sample was solubilized before treatment with trypsin (triangles). All samples were applied to a Superose 6 column and chromatographed and fractions were collected as described in Materials and Methods. Tom40 was detected by immunostaining and quantitated by densitometry. The peak of elution of various marker proteins of the indicated molecular masses is marked by arrows. a.u., arbitrary units. (B) Mitochondria were isolated from a yeast strain expressing a hexahistidinyl-tagged version of Tom40 (Tom40his6). The organelles were solubilized in buffer B (50 mM Tris-HCl [pH 7.4], 200 mM KCl, 10 mM imidazole, and 0.5% Triton X-100) containing 7 M urea where indicated. The extract (Load) was applied to a Ni-NTA affinity resin. Bound and unbound material was analyzed by immunostaining using antibodies against Tom40 and Tom20. Wild-type Tom40 does not bind to the Ni-NTA affinity resin (not shown).
Further evidence for the existence of Tom40 oligomers was obtained by employing yeast cells that express a hexahistidinyl-tagged version of Tom40 (termed Tom40his6) in addition to the wild-type protein. Mitochondria were isolated from these cells and used to purify Tom40his6 by affinity chromatography. Wild-type Tom40 could be copurified with the tagged protein, even under conditions that disrupted the TOM complex, in particular the interaction between Tom40 and the receptors, e.g., Tom20 (Fig. 1B) and Tom70 (not shown). No such copurification of wild-type Tom40 with Tom40his6 was observed when solubilization was performed under denaturing conditions by the addition of urea. These results indicate a tight interaction between the subunits of the Tom40 oligomer, which appears to be more stable than the TOM complex.
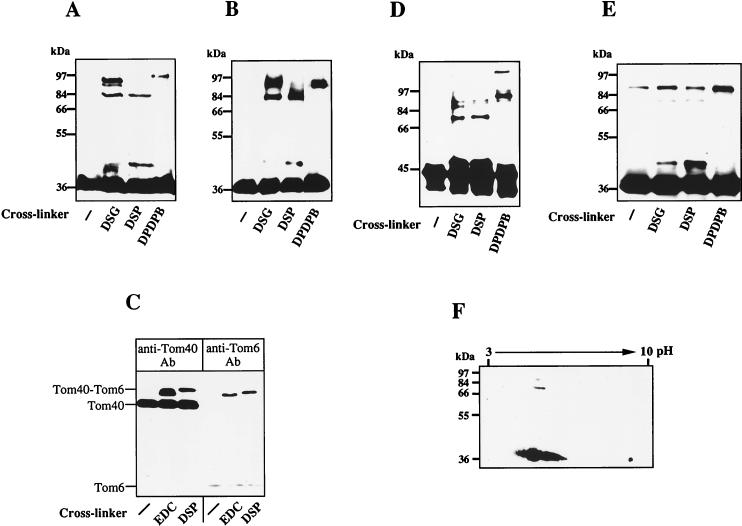
The Tom40 assembly was further studied by chemical cross-linking. Intact mitochondria or OMV isolated from N. crassa were treated with the cross-linking reagents DSG, DSP, or DPDPB. Tom40-containing cross-linking products were analyzed by nonreducing SDS-PAGE and immunostaining. Major bands corresponding to apparent molecular masses between 76 and 93 kDa were detected after treatment of both intact mitochondria and OMV with either of the three cross-linkers (Fig. 2A and B). These bands were not recognized by antibodies against porin (not shown), the most abundant protein in the mitochondrial outer membrane. The 76- to 93-kDa cross-linking products containing Tom40 represent various homodimers which exhibit different electrophoretic mobilities (see below). Another Tom40-specific cross-link with an apparent molecular mass of 45 kDa was formed by using DSP (Fig. 2A and B). Using a specific antibody, the cross-linking product was shown to contain Tom6, one of the small components of the N. crassa TOM complex (Fig. 2C) (24, 37). The adduct between Tom40 and Tom6 was also generated by cross-linking with the zero-length cross-linker EDC, indicating that the two proteins are in intimate contact with each other. Formation of the Tom40 cross-linking products was observed also with yeast mitochondria (Fig. 2D). The pattern of cross-linking products was similar to that observed with N. crassa mitochondria.
FIG. 2.
Tom40 forms homo-oligomers and interacts with Tom6. The indicated cross-linking reagents (see Materials and Methods) were added to intact mitochondria (A) or OMV (B). Samples were incubated for 30 min at 25°C before the cross-linkers were quenched. Proteins were analyzed by SDS-PAGE under nonreducing conditions and immunostaining with antibodies against Tom40. (C) The Tom40-containing 45-kDa band is a cross-linking adduct of Tom40 and Tom6. OMV were incubated with the cross-linker EDC or DSP for 30 min. Aliquots of each sample were analyzed by immunostaining with antibodies (Ab) against Tom40 and Tom6. Tom6 is only weakly stained due to its poor blotting efficiency. (D) Tom40 cross-linking products in yeast mitochondria. Isolated yeast mitochondria were treated with the indicated cross-linkers and analyzed by immunostaining for Tom40 as described for panel A. (E) The Tom40 cross-linking products are formed by using purified N. crassa TOM complex (21). As described for panel A, cross-linkers were added to the purified TOM complex and samples were incubated for 90 min at 0°C. Further analysis was performed as described for panel A. (F) The isoelectric point of the Tom40 cross-linking products is identical to that of the Tom40 monomer. Cross-linking with DSG was performed as described for panel B, using OMV. The sample was separated in the first dimension by isoelectric focusing and in the second dimension by SDS-PAGE (2). The pI values and the molecular masses of marker proteins are indicated.
To identify the cross-linking bands between 76 and 93 kDa as homodimers of Tom40, we utilized the purified TOM complex of N. crassa. This complex contains only a single protein in the size range of 25 to 65 kDa, namely Tom40 (21). Addition of the cross-linkers DSG, DSP, or DPDPB to this purified complex resulted in the formation of cross-links at 45, 76, and 93 kDa (Fig. 2E). The differences in the intensities of the various bands compared to the cross-linking pattern of mitochondria and OMV may be due to slight changes of the TOM complex conformation upon detergent solubilization. The isoelectric points of the cross-linking products of Tom40 were found by two-dimensional gel electrophoresis to be the same as that of the monomeric form of Tom40 (Fig. 2F). Together, these results strongly suggest that the cross-linking products with molecular masses between 76 and 93 kDa correspond to isoforms of homodimers of Tom40 cross-linked at different sites.
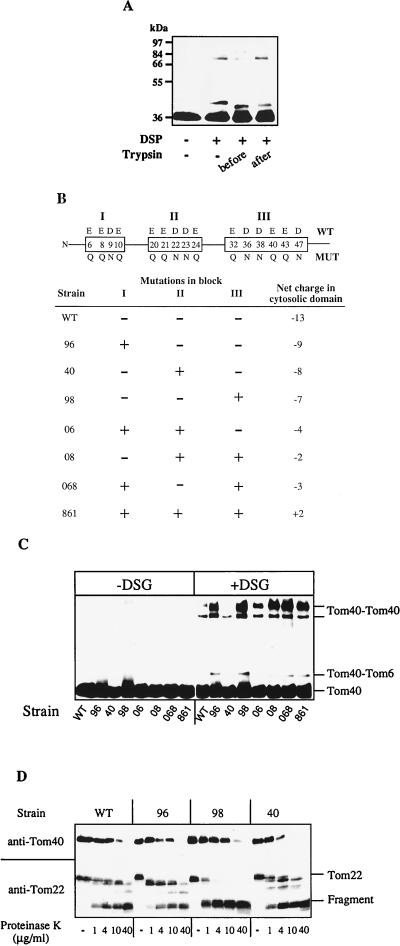
We next analyzed the potential influence of the cytosolic domains of the surface receptors on the oligomeric state of Tom40. OMV were treated with trypsin to degrade the receptors, and cross-linking was performed with DSP. The cross-linking pattern of the Tom40 homodimers changed markedly upon the proteolytic removal of the surface receptors. The 76-kDa band decreased in intensity, whereas the intensity of the 93-kDa band increased (Fig. 3A). No effect on the cross-linking pattern was observed when the trypsin treatment was performed after the cross-linking reaction. Thus, although the protease-sensitive receptors of the TOM complex are not essential for the oligomeric state of Tom40, they appear to influence the structural arrangement of the Tom40 oligomer. The intensity of cross-linking to Tom6, on the other hand, remained unchanged (Fig. 3A). The Tom40-Tom6 cross-linking product was slightly smaller because of the trypsin sensitivity of Tom6 (not shown). This observation suggests that the cytosolic domain of Tom6 is not required for the interaction between Tom40 and Tom6.
FIG. 3.
The cytosolic domains of the surface receptors modulate the structure of the Tom40 oligomer. (A) Cross-linking with DSP was performed as described in the legend to Fig. 2A by using untreated OMV (−) or OMV that were treated with trypsin (60 μg/ml) for 15 min on ice (before). With one sample, trypsin treatment was performed after the cross-linking reaction (after). Further analysis by immunostaining of Tom40 was performed as described in the legend to Fig. 2A. (B) Mutations introduced into the cytosolic domain of N. crassa Tom22. A schematic representation of the cytosolic domain of Tom22 is shown and the three blocks of negative charges (I, II, and III) are boxed. Above and below the boxes, the wild-type (WT) and mutant (MUT) residues, respectively, are given for the positions indicated inside the box. The lower panel presents the N. crassa strains expressing Tom22 mutant proteins with various combinations of the three mutated blocks. The net negative charge of the cytosolic domain is given on the right. (C) Mutations in the cytosolic domain of Tom22 alter the oligomeric structure of Tom40. OMV isolated from the various Tom22 strains were incubated without (−) or with (+) DSG. Further analysis of Tom40-containing cross-linking products was performed as described in the legend to Fig. 2A. (D) The mutations in the cytosolic domain of Tom22 affect the sensitivity of Tom22 and Tom40 to proteolytic attack. OMV isolated from the indicated Tom22 strains were treated with different concentrations of proteinase K (15 min at 0°C). After the addition of phenylmethylsulfonyl fluoride, further analysis and immunostaining for Tom40 and Tom22 were performed as described in the legend to Fig. 2A. The positions of Tom22 and its C-terminal 12-kDa fragment are indicated (18). WT, wild-type strain.
To investigate the cross-talk between the protease-sensitive receptors and Tom40 in detail, we made use of a series of N. crassa tom22 mutant strains in which negatively charged residues in the cytosolic domain of Tom22 were changed to neutral residues by in vitro mutagenesis (Fig. 3B) (27). Three blocks of negative charges can be distinguished in this domain. They were mutated either alone or in combination. OMV were isolated from these mutant tom22 strains, cross-linking with DSG was performed, and Tom40 was detected by immunostaining. The levels of Tom40-specific cross-linking in the various mutant OMV differed strongly from that in wild-type membranes (Fig. 3C). In those Tom22 mutant strains in which the third block (residues 32 to 47) was neutralized, Tom40 dimer formation was drastically increased compared to wild-type OMV. In strains carrying mutations in the other two blocks (residues 6 to 10 and 20 to 24), the Tom40-specific cross-links were hardly changed. In addition, we noted a striking correlation between the cross-linking efficiency and the protease sensitivity of both Tom40 and the mutant Tom22 proteins. In strains exhibiting largely increased cross-linking (strains 98, 08, 068, and 861), Tom40 was less susceptible to digestion by proteinase K while mutant Tom22 was more sensitive to this treatment than the wild-type protein (Fig. 3D and data not shown). On the contrary, the Tom40 cross-linking efficiency was only slightly or not affected when the sensitivity of Tom22 and Tom40 to proteinase K was comparable to that observed in wild-type OMV (strains 96, 40, and 06). We conclude that there is a direct influence of the cytosolic domain of Tom22 on the structural arrangement of Tom40 in the translocation pore. This effect is almost exclusively mediated by the third block of negatively charged amino acid residues in Tom22 and may indicate a modulating role of Tom22 on the Tom40 assembly. Apparently, changes on the surface of the TOM complex are transmitted to Tom40 and influence the structure of the translocation pore.
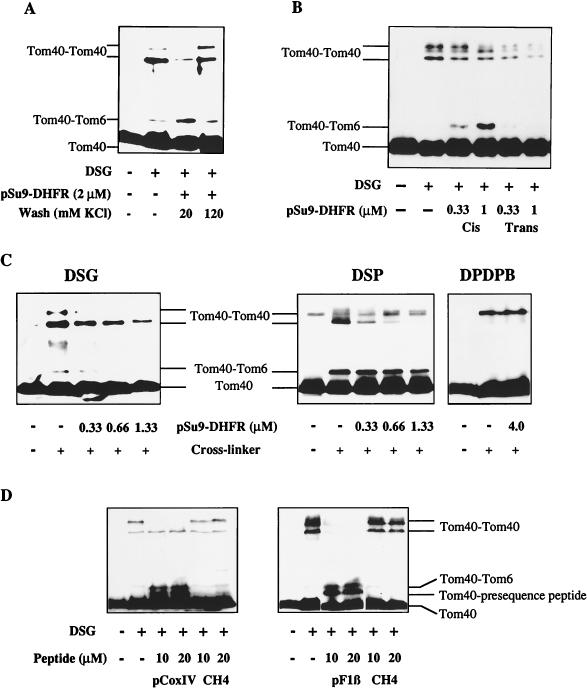
Do the structural alterations within the TOM complex play a role during preprotein binding to the receptors and transport across the outer membrane? To address this problem, we used pSu9-DHFR, a chimeric preprotein consisting of the first 69 amino acids of subunit 9 of the mitochondrial F0-ATPase fused to mouse dihydrofolate reductase (DHFR). Chemical amounts of pSu9-DHFR were added to OMV in the presence or absence of MTX, a specific ligand of DHFR which prevents the import of pSu9-DHFR into mitochondria (9). Addition of MTX results in selective binding of pSu9-DHFR to the cis site, which is formed mainly by the surface receptors Tom20 and Tom22 (25). In the absence of MTX, DHFR can unfold as the presequence translocates across the outer membrane and specifically associates with the trans site. This specific preprotein binding site is exposed to the intermembrane space and, at least in part, is formed by Tom40 (26, 30). After addition of pSu9-DHFR, cross-linking was performed with DSG. Upon preprotein binding to the cis site, the efficiency of Tom40 dimer formation decreased, while the cross-link between Tom40 and Tom6 increased in intensity (Fig. 4A). Hence, the interaction of the preprotein with surface receptors caused a major structural reorganization of the Tom40 assembly. These conformational changes were fully reversed when bound pSu9-DHFR was released from the cis site by treatment of the OMV with buffers of a higher ionic strength (Fig. 4A) (25, 30). Essentially the same observations were made with pSu9-DHFR bound to the surface of deenergized mitochondria (not shown; see reference 30).
FIG. 4.
Structural alterations in the Tom40 oligomer upon binding and movement of preproteins from the cis to the trans side of the outer membrane. (A) OMV were incubated with (+) or without (−) pSu9-DHFR in the presence of NADPH and MTX for 10 min at 0°C. The samples containing pSu9-DHFR were treated with low- or high-salt buffer (20 or 120 mM KCl, respectively). After reisolation of the OMV by centrifugation and resuspension in SEMK buffer containing NADPH and MTX, DSG was added. One of the samples lacking pSu9-DHFR was also treated with DSG, while the other one remained untreated. Further analysis and immunostaining against Tom40 were performed as described in the legend to Fig. 2A. (B) Preprotein binding to cis and trans sites alters the cross-linking pattern of Tom40. The indicated concentrations of pSu9-DHFR were added to OMV under conditions leading to specific binding to the cis or trans sites (see references 30 and 31). Cross-linking was performed with DSG, and samples were analyzed by immunostaining for Tom40 as described in the legend to Fig. 2A. (C) Binding of preproteins does not cause dissociation of the Tom40 oligomer. OMV were incubated for 20 min at 0°C with various concentrations of pSu9-DHFR. The cross-linkers DSG, DSP, and DPDPB were added as indicated for 30 min at 25°C. Further treatment and analysis by immunostaining for Tom40 was performed as described in the legend to Fig. 2A. (D) Presequence peptides alter the structure of Tom40. Isolated mitochondria were incubated with the indicated concentrations of the presequence peptides pCoxIV (residues 1 to 22 of the precursor of subunit IV of yeast cytochrome oxidase [11]) and pF1β (residues 1 to 32 of the precursor of the β subunit of yeast F1-ATPase) or with the control peptide CH4 (N-terminal 25 residues of N. crassa cytochrome c heme lyase [7]) for 15 min at 0°C. DSG was added, and further treatment and analysis by immunostaining of Tom40 were performed as described in the legend to Fig. 2A.
A strong decrease in the formation of the Tom40 dimers was observed upon association of pSu9-DHFR with the trans site (Fig. 4B); this was much stronger than that observed for preprotein binding to the cis site. Preprotein binding to the trans site did not result in increased formation of the Tom40-Tom6 cross-linking adduct (Fig. 4B), in marked contrast to what was observed for preprotein binding to the cis site (Fig. 4A and B). Thus, there is a differential effect on the Tom40 structure, resulting from preprotein binding to either the cis or trans sites. This is also evident from the cross-link between Tom40 and Tom6. Taken together, the structure of the TOM complex is dynamically altered during preprotein translocation across the outer membrane. Two stages can be distinguished; a first stage representing preprotein binding to the mitochondrial surface and a second one after movement of the preprotein from the cis to the trans site.
Does preprotein binding to the outer membrane cause a complete dissociation of the TOM complex, or does the altered cross-linking efficiency result from conformational changes? The influence of trans site-bound preprotein on cross-linking of Tom40 was tested by employing the cross-linkers DSG, DSP, and DPDPB, which differ in the length of their spacer arms (7.6, 12, and 20Å, respectively). With DSG the Tom40 dimer bands decreased upon increasing the concentration of added preprotein (Fig. 4C, left). Using DSP, the intensity of the Tom40 dimer with a lower apparent molecular mass was gradually reduced, while the larger product increased in intensity (Fig. 4C, middle). Formation of the Tom40 dimer was not influenced by preprotein binding using DPDPB with the long spacer arm, even when rather high concentrations of preprotein were used (Fig. 4C, right). The occurrence of the Tom40-Tom6 cross-linking product was also dependent on the cross-linker used. With DSG the extent of the Tom40-Tom6 cross-link decreased with increasing concentrations of preprotein, whereas the adduct was virtually unchanged upon preprotein binding using DSP (Fig. 4C). These data indicate that the Tom40 subunits do not dissociate upon preprotein binding but rather change their spatial arrangement.
We finally investigated whether it is the presequence part of the preprotein that induces the structural alterations within the TOM complex. Peptides corresponding to the presequences of subunit IV of cytochrome oxidase (pCoxIV) and of the β subunit of yeast F1-ATPase (pF1β) were incubated with isolated mitochondria, and cross-linking was performed using DSG. Addition of both peptides caused changes in the formation of the Tom40 dimers comparable to those observed upon addition of pSu9-DHFR (Fig. 4C and D). In addition, the presequence peptides induced formation of the cross-link between Tom40 and Tom6. In contrast, a control peptide not related to mitochondrial presequences (CH4) did not affect the cross-linking pattern. Thus, mitochondrial targeting sequences can induce structural changes within the TOM complex which are similar to those observed during preprotein translocation across the membrane. The influence of presequence peptides on the TOM complex is mediated most likely through their direct interaction with Tom40, as cross-linking of the presequence peptides to this protein occurred with high efficiency (Fig. 4D) (10, 31).
DISCUSSION
We have analyzed the molecular organization of Tom40, an essential component of the TOM complex. Our data suggest that Tom40 forms a homooligomeric assembly in the mitochondrial outer membrane and changes its structure during various stages of preprotein translocation across the outer membrane. The Tom40 oligomer appears to be relatively stable as it persists under conditions that lead to the dissociation of the receptor components from Tom40. Our cross-linking studies and the observation that hexahistidinyl-tagged Tom40 copurifies with the wild-type protein as well as gel filtration analysis suggest that Tom40 is organized as a dimer that forms a larger structural assembly of about 450 kDa.
After removal of the receptors by protease treatment, the TOM complex is able to translocate preproteins at a low but significant level (the so-called “bypass” translocation [29]). Thus, the Tom40 assembly contains the information to decipher the targeting signal in preproteins and likely represents the structural unit forming the translocation pore. In that function, Tom40 appears to be similar to Toc75, a component of the protein translocase of the chloroplast envelope membrane (14). Toc75 was reported to be a voltage-gated ion channel which presumably forms the central pore of the protein import machinery.
The Tom40 homo-oligomer can undergo various dynamic alterations that are important features of its function in preprotein translocation. Even though a precise molecular explanation of the observed rearrangements is not possible, a minimal model can be proposed, in which three conformational states can be distinguished. In the first state without bound preproteins, one molecule of Tom40 can be cross-linked to another Tom40 protein and to Tom6. Cross-linking of Tom40 is affected by removal of the cytosolic domains of the surface receptors, indicating a communication between receptors and Tom40. In particular, the lack of a negatively charged sequence in the cytosolic domain of Tom22 caused a major rearrangement of Tom40. This changed the structure of the Tom40 monomer, as indicated by its altered sensitivity to proteolytic attack and the relative vicinity to interacting proteins such as Tom6. These observations demonstrate that the cytosolic domain of Tom22 influences the conformation of the Tom40 oligomer. Since the negative charges on Tom22 are not essential for preprotein binding to OMV (27), it is conceivable that some of the negative charges of Tom22 are involved in the structural modulation of Tom40. The functional significance of the cross-talk between Tom40 and Tom22 might be related to the possible involvement of Tom22 in preprotein transfer into the translocation pore (19).
The other two conformational states depend on the interaction with a preprotein. In state two, a structural alteration in the Tom40 assembly is induced by preprotein binding to surface receptors at the cis site. These changes are triggered by the presequence and are fully reversed upon dissociation of the preprotein from the receptors. This indicates that the occupancy of the receptors by preproteins is sensed and transmitted from the mitochondrial surface to the Tom40 assembly, which participates in later stages of the translocation process. We propose that the observed changes reflect an opening of the translocation pore to facilitate the entry of the presequence into the membrane. Such a model would readily explain how the cytosolic domains of the receptors increase the efficiency of preprotein entry into the translocation pore, even though they are not obligatory for this process.
In state three, further changes in the chemical environment of Tom40 occur when the preprotein enters the outer membrane and binds in a stable fashion to the trans site (26, 30). These structural alterations are also triggered to a large extent by the N-terminal presequence. In the trans site, the preprotein was shown recently to be in close contact with Tom40 and tightly bound to the translocation machinery by interaction through both the presequence and the mature parts (30, 31). The intimate contact with the translocon maintains the preprotein in a translocation-competent state (26); i.e., it performs a chaperone-like function by preventing the unfolded polypeptide chain from aggregation.
Structural rearrangements of the translocation machinery during preprotein transfer might be a common feature of many translocases. The most striking alteration was reported for the translocation of ATPase SecA, a peripheral component of the bacterial plasma membrane (for a review, see reference 8). During translocation of a preprotein, SecA inserts a large domain into the membrane. This major rearrangement within SecA is accompanied by mutual changes occurring in SecG, a membrane-embedded component of the bacterial preprotein translocase. These dynamic alterations are accompanied by translocation of segments of the preprotein and thus appear to be hallmarks of the mechanism of bacterial protein translocation. Similarly, the translocon of the endoplasmic reticulum appears to undergo mechanistically important changes during preprotein movement across the membrane (reviewed in reference 32). As with the TOM complex, the signal sequence appears to represent the major trigger for these alterations. The N-terminal signal sequence, possibly through its interaction with a second signal binding site, opens the gated translocon on the lumenal side of the membrane (4, 16). Even though direct cross-linking data have not been reported, it seems likely that these changes are accompanied by conformational changes similar to those reported here for the TOM complex.
The present study documents important insights into the structural dynamics of the TOM complex during preprotein translocation. We have defined several alterations of the vicinity of Tom40 in response to the binding, membrane entry and translocation of a preprotein. Further refinement of our views on how the dynamic alterations within the translocase result in the directed transport of a preprotein across the lipid bilayer will depend on information about the structure of the membrane-embedded components of the TOM complex and their spatial arrangement in the membrane (21).
ACKNOWLEDGMENTS
We thank M. Brunner, D. A. Court, and T. Langer for helpful discussions and P. Heckmeyer and M. Braun for excellent technical assistance. We acknowledge the gift of presequence peptide pCoxIV by M. Cumsky.
Our work was supported by grants of the Sonderforschungsbereich 184 of the Deutsche Forschungsgemeinschaft, the Fonds der Chemischen Industrie, and the Medical Research Council of Canada and by a fellowship of the European Molecular Biology Organisation (to D.R.).
REFERENCES
- 1.Alconada A, Kübrich M, Moczko M, Hönlinger A, Pfanner N. The mitochondrial receptor complex: the small subunit Mom8b/Isp6 supports association of receptors with the general insertion pore and transfer of preproteins. Mol Cell Biol. 1995;15:6196–6205. doi: 10.1128/mcb.15.11.6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bjellqvist B, Sanchez J C, Pasquali C, Ravier F, Paquet N, Frutiger S, Hughes G J, Hochstrasser D. Micropreparative two-dimensional electrophoresis allowing the separation of samples containing milligram amounts of proteins. Electrophoresis. 1993;14:1375–1378. doi: 10.1002/elps.11501401212. [DOI] [PubMed] [Google Scholar]
- 3.Brix J, Dietmeier K, Pfanner N. Differential recognition of preproteins by the purified cytosolic domains of the mitochondrial import receptors Tom20, Tom22, and Tom70. J Biol Chem. 1997;272:20730–20735. doi: 10.1074/jbc.272.33.20730. [DOI] [PubMed] [Google Scholar]
- 4.Crowley K S, Liao S, Worrell V E, Reinhart G D, Johnson A E. Secretory proteins move through the endoplasmic reticulum membrane via an aqueous, gated pore. Cell. 1994;78:461–471. doi: 10.1016/0092-8674(94)90424-3. [DOI] [PubMed] [Google Scholar]
- 5.Daum G, Böhni P C, Schatz G. Import of proteins into mitochondria: cytochrome b2 and cytochrome c peroxidase are located in the intermembrane space of yeast mitochondria. J Biol Chem. 1982;257:13028–13033. [PubMed] [Google Scholar]
- 6.Dietmeier K, Hönlinger A, Bomer U, Dekker P J, Eckerskorn C, Lottspeich F, Kübrich M, Pfanner N. Tom5 functionally links mitochondrial preprotein receptors to the general import pore. Nature. 1997;388:195–200. doi: 10.1038/40663. [DOI] [PubMed] [Google Scholar]
- 7.Drygas M E, Lambowitz A M, Nargang F E. Cloning and analysis of the Neurospora crassa gene for cytochrome c heme lyase. J Biol Chem. 1989;264:17897–17907. [PubMed] [Google Scholar]
- 8.Duong F, Eichler J, Price A, Leonard M R, Wickner W. Biogenesis of the gram-negative bacterial envelope. Cell. 1997;91:567–573. doi: 10.1016/s0092-8674(00)80444-4. [DOI] [PubMed] [Google Scholar]
- 9.Eilers M, Schatz G. Binding of a specific ligand inhibits import of a purified precursor protein into mitochondria. Nature. 1986;322:228–232. doi: 10.1038/322228a0. [DOI] [PubMed] [Google Scholar]
- 10.Gaikwad A S, Cumsky M G. The use of chemical crosslinking to identify proteins that interact with a mitochondrial presequence. J Biol Chem. 1994;269:6437–6443. [PubMed] [Google Scholar]
- 11.Glaser S M, Cumsky M G. A synthetic presequence reversibly inhibits protein import into yeast mitochondria. J Biol Chem. 1990;265:8808–8816. [PubMed] [Google Scholar]
- 12.Gratzer S, Lithgow T, Bauer R E, Lamping E, Paltauf F, Kohlwein S D, Haucke V, Junne T, Schatz G, Horst M. Mas37p, a novel receptor subunit for protein import into mitochondria. J Cell Biol. 1995;129:25–34. doi: 10.1083/jcb.129.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hines V, Schatz G. Precursor binding to yeast mitochondria. J Biol Chem. 1993;268:449–454. [PubMed] [Google Scholar]
- 14.Hinnah S C, Hill K, Wagner R, Schlicher T, Soll J. Reconstitution of a chloroplast protein import channel. EMBO J. 1997;16:7351–7360. doi: 10.1093/emboj/16.24.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hönlinger A, Bömer U, Alconada A, Eckerskorn C, Lottspeich F, Dietmeier K, Pfanner N. Tom7 modulates the dynamics of the mitochondrial outer membrane translocase and plays a pathway-related role in protein import. EMBO J. 1996;15:2125–2137. [PMC free article] [PubMed] [Google Scholar]
- 16.Jungnickel B, Rapoport T A. A posttargeting signal sequence recognition event in the endoplasmic reticulum membrane. Cell. 1995;82:261–270. doi: 10.1016/0092-8674(95)90313-5. [DOI] [PubMed] [Google Scholar]
- 17.Kassenbrock C K, Cao W, Douglas M G. Genetic and biochemical characterization of ISP6, a small mitochondrial outer membrane protein associated with the protein translocation complex. EMBO J. 1993;12:3023–3034. doi: 10.1002/j.1460-2075.1993.tb05971.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keil P, Pfanner N. Insertion of MOM22 into the mitochondrial outer membrane strictly depends on surface receptors. FEBS Lett. 1993;321:197–200. doi: 10.1016/0014-5793(93)80107-6. [DOI] [PubMed] [Google Scholar]
- 19.Kiebler M, Keil P, Schneider H, van der Klei I, Pfanner N, Neupert W. The mitochondrial receptor complex: a central role of MOM22 in mediating transfer of preproteins from receptors to the general insertion pore. Cell. 1993;74:483–492. doi: 10.1016/0092-8674(93)80050-o. [DOI] [PubMed] [Google Scholar]
- 20.Komiya T, Rospert S, Schatz G, Mihara K. Binding of mitochondrial precursor proteins to the cytoplasmic domains of the import receptors Tom70 and Tom20 is determined by cytoplasmic chaperones. EMBO J. 1997;16:4267–4275. doi: 10.1093/emboj/16.14.4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Künkele K P, Heins S, Dembowski M, Nargang F E, Benz R, Thieffry M, Walz J, Lill R, Nussberger S, Neupert W. The preprotein translocation channel of the outer membrane of mitochondria. Cell. 1998;93:1009–1019. doi: 10.1016/s0092-8674(00)81206-4. [DOI] [PubMed] [Google Scholar]
- 22.Lill R, Nargang F E, Neupert W. Biogenesis of mitochondrial proteins. Curr Opin Cell Biol. 1996;8:505–512. doi: 10.1016/s0955-0674(96)80028-7. [DOI] [PubMed] [Google Scholar]
- 23.Lill R, Neupert W. Mechanisms of protein import across the mitochondrial outer membrane. Trends Cell Biol. 1996;6:56–61. doi: 10.1016/0962-8924(96)81015-4. [DOI] [PubMed] [Google Scholar]
- 24.Mayer A, Lill R, Neupert W. Translocation and insertion of precursor proteins into isolated outer membranes of mitochondria. J Cell Biol. 1993;121:1233–1243. doi: 10.1083/jcb.121.6.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mayer A, Nargang F E, Neupert W, Lill R. MOM22 is a receptor for mitochondrial targeting sequences and cooperates with MOM19. EMBO J. 1995;14:4204–4211. doi: 10.1002/j.1460-2075.1995.tb00094.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mayer A, Neupert W, Lill R. Mitochondrial protein import: reversible binding of the presequence at the trans side of the outer membrane drives partial translocation and unfolding. Cell. 1995;80:127–137. doi: 10.1016/0092-8674(95)90457-3. [DOI] [PubMed] [Google Scholar]
- 27.Nargang F E, Rapaport D, Ritzel R G, Neupert W, Lill R. Role of the negative charges in the cytosolic domain of TOM22 in the import of precursor proteins into mitochondria. Mol Cell Biol. 1998;18:3173–3181. doi: 10.1128/mcb.18.6.3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neupert W. Protein import into mitochondria. Annu Rev Biochem. 1997;66:861–915. doi: 10.1146/annurev.biochem.66.1.863. [DOI] [PubMed] [Google Scholar]
- 29.Pfaller R, Pfanner N, Neupert W. Mitochondrial protein import: bypass of proteinaceous surface receptors can occur with low specificity and efficiency. J Biol Chem. 1989;264:34–39. [PubMed] [Google Scholar]
- 30.Rapaport D, Mayer A, Neupert W, Lill R. Cis and trans sites of the TOM complex in unfolding and translocation of preproteins across the outer membrane of mitochondria. J Biol Chem. 1998;273:8806–8813. doi: 10.1074/jbc.273.15.8806. [DOI] [PubMed] [Google Scholar]
- 31.Rapaport D, Neupert W, Lill R. Mitochondrial protein import: Tom40 plays a major role in targeting and translocation of preproteins by forming a specific binding site for the presequence. J Biol Chem. 1997;272:18725–18731. doi: 10.1074/jbc.272.30.18725. [DOI] [PubMed] [Google Scholar]
- 32.Rapoport T A, Rolls M M, Jungnickel B. Approaching the mechanism of protein transport across the ER membrane. Curr Opin Cell Biol. 1996;8:499–504. doi: 10.1016/s0955-0674(96)80027-5. [DOI] [PubMed] [Google Scholar]
- 33.Schatz G. The protein import system of mitochondria. J Biol Chem. 1996;271:31763–31766. doi: 10.1074/jbc.271.50.31763. [DOI] [PubMed] [Google Scholar]
- 34.Schleiff E, Shore G C, Goping I S. Interactions of the human mitochondrial protein import receptor, hTom20, with precursor proteins in vitro reveal pleiotropic specificities and different receptor domain requirements. J Biol Chem. 1997;272:17784–17789. doi: 10.1074/jbc.272.28.17784. [DOI] [PubMed] [Google Scholar]
- 35.Schlossmann J, Dietmeier K, Pfanner N, Neupert W. Specific recognition of mitochondrial preproteins by the cytosolic domain of the import receptor MOM72. J Biol Chem. 1994;269:11893–11901. [PubMed] [Google Scholar]
- 36.Söllner T, Pfaller R, Griffiths G, Pfanner N, Neupert W. A mitochondrial import receptor for the ADP/ATP carrier. Cell. 1990;62:107–115. doi: 10.1016/0092-8674(90)90244-9. [DOI] [PubMed] [Google Scholar]
- 37.Söllner T, Rassow J, Wiedmann M, Schlossmann J, Keil P, Neupert W, Pfanner N. Mapping of the protein import machinery in the mitochondrial outer membrane by crosslinking of translocation intermediates. Nature. 1992;355:84–87. doi: 10.1038/355084a0. [DOI] [PubMed] [Google Scholar]
- 38.Vernet T, Dignard D, Thomas D Y. A family of yeast expression vectors containing the phage f1 intergenic region. Gene. 1987;52:225–233. doi: 10.1016/0378-1119(87)90049-7. [DOI] [PubMed] [Google Scholar]
- 39.Vestweber D, Brunner K P, Baker A, Schatz G. A 42K outer membrane protein is a component of the yeast mitochondrial import site. Nature. 1989;341:205–209. doi: 10.1038/341205a0. [DOI] [PubMed] [Google Scholar]