Abstract
The aim of this study was to determine the prevalence and risk factors for Mycoplasma haemofelis (Mhf) and ‘Candidatus Mycoplasma haemominutum’ (Mhm) infections in domestic cats tested for feline immunodeficiency virus (FIV) and feline leukemia virus (FeLV) with a commercial enzyme-linked immunosorbent assay (ELISA) kit. Based on serological testing, cats were grouped as i) FIV-positive (n=25); ii) FeLV-positive (n=39); iii) FIV/FeLV-positive (n=8); and iv) FIV/FeLV-negative (n=77). Complete blood counts were followed by DNA extraction, species-specific polymerase chain reaction (16S rRNA gene) for Mhf and Mhm and Southern blotting for all animals. Mhf DNA was found in 4.0, 2.6, 12.5 and 7.8% of the cats from groups i, ii, iii and iv, respectively, while 32, 5.1, 50 and 5.2% of these animals had an Mhm infection. Cats with FIV (OR=4.25, P=0.009) and both FIV and FeLV (OR=7.56, P=0.014) were at greater risk of being hemoplasma infected than retroviral-negative cats, mainly due to Mhm infection (OR=8.59, P=0.001 and OR=18.25, P=0.001, respectively). Among pure-breed cats, FIV-positive status was associated with hemoplasma infection (OR 45.0, P=0.001).
Mycoplasma haemofelis and ‘Candidatus Mycoplasma haemominutum’, commonly referred to as feline hemoplasmas, are pleomorphic bacteria that parasitize erythrocytes of domestic cats (Neimark et al 2001, Messick 2004). They are rod, spherical or ring shaped Gram-negative bacteria which can be found individually or in chains across the red cell surface (Messick 2004). The hemoplasmas are simple parasites with a small genome that replicate by binary fission. To date, these organisms have not been cultivated outside their natural hosts (Harvey 2006). The feline hemoplasmas were formerly classified as rickettsial organisms (and named Haemobartonella felis) but based on sequence analysis of the 16S rRNA gene, two different parasites have been identified in cats with similarity scores supporting their new classification as members of the genus Mycoplasma (Neimark et al 2001, 2002, Foley and Pedersen 2001). Recently, a third feline hemoplasma species, ‘Candidatus M turicensis’ was identified in Switzerland (Willi et al 2005) and also found in United Kingdom, South Africa, Australia and USA, suggesting a worldwide distribution (Willi et al 2006a, Terry et al 2006). In Brazil, this parasite has thus far been only described in zoo-maintained wild felids (Willi et al 2007).
Cats experimentally infected with ‘Candidatus M haemominutum’ or M haemofelis exhibit different signs. While in the first case there are minimal clinical signs of infection and anemia is mild or absent, cats infected with M haemofelis often have a severe hemolytic anemia (Berent et al 1998, Foley et al 1998). Mycoplasma haemofelis may cause primary disease in cats and is also commonly recognized as a pathogen in conjunction with retroviruses, including feline immunodeficiency virus (FIV) and feline leukemia virus (FeLV) and other debilitating diseases (Kociba et al 1983, Bobade et al 1988, Grindem et al 1990, Lappin 1995, George et al 2002, Harrus et al 2002). It has been suggested that cats having a pre-existing retroviral infection may be at a greater risk for becoming infected with feline hemoplasmas (Nash and Bobade 1986, Grindem et al 1990, Lappin 1995, Harrus et al 2002, George et al 2002, Inokuma et al 2004, Luria et al 2004). Further, cats that were experimentally infected with retroviruses and ‘Candidatus M haemominutum’ developed more severe anemia than cats infected with the parasite alone (George et al 2002). Studies also indicate that M haemofelis and ‘Candidatus M haemominutum’ may act as cofactors, accelerating the rate of progression of feline retrovirus related diseases. Thus, cats that are co-infected with hemoplasmas and retroviruses may be at greater risk for the development of lymphoma, leukemia, and immunodeficiency syndrome (Priester and Hayes 1973, Cotter et al 1975, Bobade et al 1988, George et al 2002, Harrus et al 2002).
Based on the above information, we hypothesized that cats having retroviral infections are at greater risk than non-retroviral infected animals for having a hemoplasma infection. Thus, the aim of this study was to determine the prevalence of hemoplasma infections in cats selected on the basis of having pre-existing retroviral infections (FIV and/or FeLV) and compare these findings to a reference group. The risk factors for hemoplasma infection in cats with pre-existing retroviral-positive status were also evaluated. To date, there are no studies in Brazil critically examining whether or not hemoplasma infections are associated with retroviral infection in cat. The use of a pre-selected population of retroviral-positive cats was also unique to our study.
Materials and methods
Animals/group division
We prospectively collected samples from 149 cats admitted to a feline-only veterinary clinic in Rio de Janeiro, Brazil from February 2005 to February 2006; this included all cats tested for retroviral exposure/infection during that time period. Samples were collected once and sequential admissions were not accessed in our study. Most tests were performed by the attending clinician based on clinical findings, and some for prophylactic purposes (eg, introducing a new animal in the household). Animals were tested for the retroviruses with a commercially available, in-house ELISA test kit (Snap FIV Antibody/FeLV Antigen Combo; IDEXX Laboratories, Westbrook, ME, USA) using serum or plasma, according to the manufacturer's instructions. This test detects FeLV p27 antigen and specific antibodies to FIV. The owner's written consent was obtained for inclusion of the cat in the study. Based on serological testing, cats were grouped for statistical analyses as i) FIV-positive (n=25); ii) FeLV-positive (n=39); iii) FeLV and FIV-positive (n=8); and iv) FeLV and FIV negative, also defined as the base comparison or reference group (n=77).
Data collection
Information regarding animal history and risk factors for hemoplasma and feline retroviruses' infections were obtained by the clinician (from the owner and clinical examination) at the time of presentation or from clinics' files later. Unavailable information regarding any risk factor was considered to be a ‘missing variable’ for statistical analysis. The following information was collected: sex, age, breed, neuter status, fight/bites history with other cats, blood transfusion history, presence of fleas, outdoor-roaming status, multiple cats' household, and vaccination status (feline herpesvirus 1, feline calicivirus, feline panleukopenia virus and/or rabies). Cats were also clinically evaluated for anorexia, lethargy/depression, pale mucous membranes, icterus, tachycardia (>220 beats per minute), tachypnea (>60 breaths per minute), and fever (temperature ≥39.2°C).
Complete blood count
Blood was collected in EDTA tubes for complete blood count (CBC). Manual blood counts and evaluation of blood smear preparations were performed by a well-trained veterinarian. The CBC results, including packed cell volume (PCV) (reference range: 24–45%; 0.24–0.45 SI), and white blood cell count (WBC) (reference range: 5000–19,500 cells/mm3; 5.0×109/l–19.5×109/l SI), were evaluated as risk factors for hemoplasma infections in this study. EDTA-blood was stored at 4°C until DNA extraction.
DNA extraction
DNA was extracted within 1 week of collection with a commercial kit (Generation Capture Column, Gentra Systems, Minneapolis, MN, USA) according to manufacturer's instructions. Extracted DNA was stored at −20°C until PCR testing was performed.
Polymerase chain reaction
All DNA samples were tested for the presence of M haemofelis and ‘Candidatus M haemominutum’ 16S rRNA gene. To amplify a 393 base-pair (bp) partial sequence of M haemofelis 16S rRNA gene, the PCR was performed using 5 μl of DNA template per 25 μl reaction containing a final concentration of 50 mM KCl, 10 mM Tris–HCl, 0.1% Triton X-100, 2.5 mM MgCl2, 0.2 mM each dNTP, 0.2 mM each primer, 1.0 U of Taq DNA polymerase (Promega, Madison, WI, USA), and sterile water up to the final volume. The cycling conditions consisted of an initial denaturation step of 10 min at 94°C, followed by 43 cycles of 94°C for 45 s, 54°C for 45 s, 72°C for 1 min and a final elongation step at 72°C for 7 min (Berent et al 1998). For ‘Candidatus M haemominutum’, the PCR was performed, using 5 μl of DNA template per reaction containing a final concentration of 50 mM KCl, 10 mM Tris–HCl, 0.1% Triton X-100, 2.5 mM MgCl2, 0.2 mM each dNTP, 0.2 mM each primer, 1.0 U of Taq DNA polymerase (Promega, Madison, WI, USA), completing 25 μl with sterile water. The cycling conditions consisted of an initial denaturation at 94°C for 4 min, followed by 35 cycles of 94°C for 30 s, 53°C for 1 min, and 70°C for 45 s followed by a 5 min final elongation step at 70°C, resulting in a 130 bp product (Foley et al 1998). Samples were cooled to 4°C after the final elongation step in both PCR procedures. For all amplifications, previously known positive and negative samples for M haemofelis or ‘Candidatus M haemominutum’ DNA were used as controls. The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene was used as a housekeeping target in all samples to verify amplifiable DNA as previously described (Birkenheuer et al 2003).
The amplification products were analyzed by electrophoresis using 1.5% agarose gels in 1% Tris–Acetate-EDTA buffer (40 mM Tris–acetate and 1 mM EDTA) solution containing 0.5 mg/ml of ethidium bromide (Invitrogen, Carlsbad, CA, USA). Results were visualized and documented under UV transillumination (Kodak Gel Logic 100 Imaging System, Eastman Kodak Corporation, Rochester, NY, USA). Standard precautions to avoid cross contamination were observed.
Southern blot hybridization
Following electrophoresis, the gels were denatured and PCR products were transferred to a nylon membrane using the Turboblotter Rapid Downward Transfer Systems (Whatman Schleicher and Schuell, Keene, NH, USA) (Chomczynski 1992). Specific M haemofelis and ‘Candidatus M haemominutum’ DNA probes were synthesized replacing regular dNTPs in the conventional PCR performed herein by the PCR DIG labeling mix, resulting in a 393 and 130 bp chemically-labeled sequences for both hemoplasmas, respectively. These steps were performed according to the manufacturer's instructions. Membranes were prehybridized for 2 h at 45°C using the DIG Easy Hyb solution followed by overnight hybridization at 45°C with specific probes. Washing was first performed in 2×saline–sodium citrate (SSC)/0.1% sodium dodecyl sulfate (SDS) solution at room temperature (RT) three times for 10 min each and then in 0.2×SSC/0.1% SDS buffer at 68°C for 30 min. Membrane was blocked for 4 h with 2×blocking/1×maleic acid buffer solution/RT. This was followed by equilibration in 1×washing solution for 5 min/RT and antibody binding with anti-Dig-alkaline phosphatase conjugate in 1×blocking/1×maleic acid solution (1:20,000). Membranes were equilibrated with 1×detection solution for 5 min/RT and then soaked with CDP-Star, ready-to-use chemiluminescent substrate for alkaline phosphatase during an additional 5 min incubation. Finally, membranes were sealed and autoradiographed. Except for SDS and SSC (Promega, Madison, WI, USA), all hybridization assays, including probes' synthesis, were done using DIG System reagents for non-radioactive nucleic acid labeling and detection (Roche Diagnostics Corporation, Indianapolis, IN, USA).
Cloning and sequencing of PCR products
In order to confirm the identity of PCR products, a positive sample was selected from each gel run which showed specific amplicons to be cloned and sequenced. Prior to cloning of the PCR products, the 393 base-pair (bp) fragment of M haemofelis and 130 bp fragment of ‘Candidatus M haemominutum’ were purified with Zymoclean gel DNA recovery kit (Zymo Research Corp, Orange, CA, USA). Purified fragments were then cloned into pGEM T-easy vector system II (Promega, Madison, WI, USA). Recombinants were selected on the basis of the blue-white color of colonies. Plasmids containing proper size inserts were prepped and purified for sequencing using the QIAprep miniprep kit (Qiagen, Valencia, CA, USA). The above steps were performed according to the manufacturer's instructions. All clones were sequenced in both sense and antisense directions by a dideoxy terminator method using the Applied Biosystems Automated 3730 DNA analyzer at Cornell University, Ithaca, NY, USA. Sequencing data were analyzed with Blast 2.0.10 software (http://www.ncbi.nml.nih.gov/blast/).
Statistic analysis
Comparison of results between PCR and Southern blot (SB) results was undertaken using Fisher's exact test. Prevalence estimates and 95% exact confidence intervals were calculated for hemoplasmas for each group and reported as the percentage of cats with a positive test result (number of cats tested positive divided by the total number of cats tested×100). A logistic regression model was run for each of the three outcomes (hemoplasma, ‘Candidatus M haemominutum’ and M haemofelis) including only group (FIV-positive, FeLV-positive, FIV/FeLV-positive, and retroviral-negative cats) as a predictor. The odds ratios, 95% profile-likelihood confidence intervals, and P-values from likelihood ratio χ2 tests are reported. In addition, a logistic regression model was performed in order to evaluate the relationship between risk factors and retroviruses on presence of hemoplasma DNA. Thus for each possible risk factor, a logistic regression model was run, using the presence of that risk factor, retroviral status (FIV-positive, FIV/FeLV-negative), and the two-way interaction to predict the presence of hemoplasma DNA. Risk factors could only be evaluated individually due to the low number of hemoplasma-positive cats. If the interaction were found significant then the model was broken down by the risk factor to calculate odds ratios and 95% CI. P-values <0.05 were considered statistically significant for all tests in this study. Statistical evaluation was performed using the software SAS 9.1 (SAS Institute Inc, Cary, NC, USA).
Results
Hemoplasma PCR/Southern blot
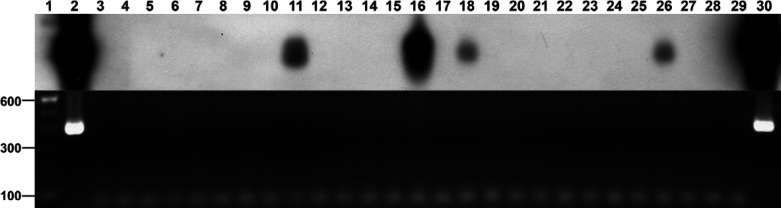
PCR were performed for all 149 samples, followed by SB. Eighteen of 149 samples tested were positive by PCR for at least one hemoplasma. Based on criteria described earlier, eight samples (44.4% of all PCR positive products) were selected for sequencing. Sequence analysis confirmed the identity was 100% for both M haemofelis and ‘Candidatus M haemominutum’ when compared to previously published sequences in the genbank database (data not shown). Further, there was a strong association between PCR and SB results (P<0.001). SB showed that 24/149 samples had a positive result, confirming the 18 PCR positive samples (Fig 1) and revealing six additional positive cats (Fig 2) (P<0.001). As SB was used to enhance the detection of any hemoplasma product made during PCR and is known to have a higher sensitivity, all statistical analyses were based on SB results. As determined by amplification of glyceraldehyde phosphate dehydrogenase (GAPDH), the presence of extracted DNA was confirmed for all 149 samples (data not shown).
Fig 1.
PCR (bottom) and Southern blot (top) results for ‘Candidatus Mycoplasma haemominutum’ 16S rRNA gene among cats having a positive FIV status. Lane 1: 100 bp molecular weight marker; Lane 2: Positive DNA control; Lane 3: Negative water control; Lane 4: Negative DNA control; Lanes 5 to 30: Clinical samples tested for ‘Candidatus M haemominutum’ DNA among FIV cats.
Fig 2.
PCR (bottom) and Southern blot (top) results for Mycoplasma haemofelis 16S rRNA gene among cats negative for both FIV and FeLV. Lane 1: 100 bp molecular weight marker; Lane 2: Positive DNA control; Lane 3: Negative water control; Lane 4: Negative DNA control; Lanes 5 to 29: Clinical samples tested for M haemofelis DNA among both FIV- and FeLV-negative cats; Lane 30: Previously known positive clinical sample.
The prevalence of hemoplasmas among FIV-positive cats was 36% (9/25), mainly due to the presence of ‘Candidatus M haemominutum’ (8/25, 32%). A low prevalence of hemoplasma DNA was observed among FeLV cats (2/39, 5.1%), whereas the highest rate of hemoplasma infection was found among both FIV- and FeLV-positive cats (50%). Among the cats in the base comparison group, 11.7% were hemoplasma infected. Due to the low number of double hemoplasma-positive cats (3/149; 2.0%) and for statistical purposes, these animals were included among ones positive only for ‘Candidatus M haemominutum’ or M haemofelis. The complete descriptive results for prevalence estimates and 95% CI for all hemoplasmas, “Candidatus M. haemominutum” and M. haemofelis among cats tested for FIV and FeLV are shown in Table 1.
Table 1.
Prevalence estimates by Southern blot (SB+) and 95% confidence intervals for all hemoplasmas, ‘Candidatus Mycoplasma haemominutum’ and Mycoplasma haemofelis among all cats tested for FIV and FeLV in a feline-only clinic in Rio de Janeiro, Brazil, from February 2005 to February 2006
| Retroviral status | Feline hemoplasma | |
|---|---|---|
| SB+ (%) | 95% Confidence intervals * | |
| FIV-positive (n= 25) | ||
| Hemoplasma † | 9/25 (36.0) | [18.0, 57.5] |
| Mhm ‡ | 8/25 (32.0) | [15.0, 53.5] |
| Mhf § | 1/25 (4.0) | [0.1, 20.4] |
| FIV and FeLV-positive (n=8) | ||
| Hemoplasma † | 4/8 (50.0) | [15.7, 84.3] |
| Mhm ‡ | 4/8 (50.0) | [15.7, 84.3] |
| Mhf § | 1/8 (12.5) | [0.3, 52.7] |
| FeLV-positive (n=39) | ||
| Hemoplasma † | 2/39 (5.1) | [0.6,17.3] |
| Mhm ‡ | 2/39 (5.1) | [0.6,17.3] |
| Mhf § | 1/39 (2.6) | [0.1,13.5] |
| FIV- and FeLV-negative (n=77) | ||
| Hemoplasma † | 9/77 (11.7) | [5.5, 21.0] |
| Mhm ‡ | 4/77 (5.2) | [1.4, 12.8] |
| Mhf § | 6/77 (7.8) | [2.9,16.2] |
Exact 95% confidence intervals for Binomial Proportion.
Positive for at least one hemoplasma DNA, double hemoplasma positive counted only once.
‘Candidatus Mycoplasma haemominutum’.
Mycoplasma haemofelis.
Risk factors
Significant results were found when comparing the FIV-positive group and both FIV- and FeLV-positive cats to the base comparison group regarding the presence of any hemoplasma and ‘Candidatus M haemominutum’ infection. Cats with FIV-positive status were at greater risk of having hemoplasma (OR=4.25, P=0.009) and ‘Candidatus M haemominutum’ infections (OR=8.59, P=0.001) than both FIV- and FeLV-negative cats. In addition, cats having both FIV- and FeLV-positive status were also at a greater risk of being hemoplasma (OR=7.56, P=0.014) and ‘Candidatus M haemominutum’ infected (OR=18.25, P=0.001) than cats without any retrovirus. FeLV cats did not show any increased risk for ‘Candidatus M haemominutum’ infection when compared to the reference group (P>0.05). In addition, no increased risk or significant associations were found regarding M haemofelis infection (outcome) in any group compared to cats with a negative FIV and FeLV status (P>0.05). The OR of hemoplasma infections (outcome) associated with the presence of a pre-existing positive ELISA status for FIV and/or FeLV and the 95% CI, compared to the reference (retroviral-negative status) group are shown in Table 2.
Table 2.
Risk of hemoplasma infections (outcome) and 95% confidence intervals (CI) associated with the presence of a pre-existing positive ELISA (FIV and/or FeLV-positive), compared to cats testing negative for both retroviruses (reference group)
| Outcome | Exposure (versus reference cats) | Odds ratio | 95% CI * | P-value * |
|---|---|---|---|---|
| Hemoplasma | FIV | 4.25 | [1.45, 12.66] | 0.009 |
| FeLV | 0.41 | [0.06, 1.69] | 0.232 | |
| FIV/FeLV | 7.56 | [1.55, 37.56] | 0.014 | |
| Mhm † | FIV | 8.59 | [2.42, 35.39] | 0.001 |
| FeLV | 0.99 | [0.13, 5.30] | 0.988 | |
| FIV/FeLV | 18.25 | [3.32, 110.85] | 0.001 | |
| Mhf ‡ | FIV | 0.496 | [0.01, 4.42] ** | 0.902 ** |
| FeLV | 0.314 | [0.01, 2.74] ** | 0.500 ** | |
| FIV/FeLV | 1.678 | [0.03, 17.49] ** | >0.999 ** |
Profile-likelihood confidence intervals reported and P-values obtained from likelihood ratio test.
P-values and confidence intervals reported from exact test due to small sample sizes.
‘Candidatus Mycoplasma haemominutum’.
Mycoplasma haemofelis.
Breed was the unique risk factor which appeared to contribute to a hemoplasma infection among retrovirus-positive cats compared to the animals that tested negative for both FIV and FeLV. In the logistic regression model predicting hemoplasma infection according to the breed (pure or mixed), retrovirus status (FIV-positive, FIV/FeLV-negative), and the interaction; the interaction term was statistically significant (P=0.019). Among pure-breed cats, FIV-positive animals were at greater risk to be infected with at least one hemoplasma (OR 45.0, 95% CI 4.6–1168, P=0.001). Among mixed-breed cats, FIV-positive status was not associated with hemoplasma infection (P=0.759). No association was found when evaluating the breed and the ‘Candidatus M haemominutum’ outcome. All the other evaluated risk factors showed no influence on the outcome (hemoplasma or ‘Candidatus M haemominutum’ infection) in the retrovirus-positive groups (data not shown). Due to the low number of positive M haemofelis outcomes, risk factor analysis regarding this parasite could not be performed. In addition, there were not enough positive results for either of the hemoplasmas to run the logistic model for risk factors in FeLV cats.
Discussion
This is the first in-depth study using molecular techniques to evaluate the relationship between retroviral-positive status and hemoplasma infection in domestic cats in Brazil. We found the prevalence of hemoplasmas among cats with antibodies to FIV was 36%, mainly due to the presence of ‘Candidatus M haemominutum’. Another study in Brazil using the same PCR protocols described herein, found hemoplasma infections in 56.7% of domestic cats that were anemic (Baumann et al 2006). The authors of this study found a higher occurrence of M haemofelis when compared to ‘Candidatus M haemominutum’ (37.8 and 10.8%, respectively). As our study population was not defined by the presence of anemia, and due to the fact we also used SB analysis in addition to PCR to identify the presence of hemoplasma DNA, direct comparisons cannot be made.
Our study corroborates, in part, the findings of Luria et al (2004) suggesting that cats with FIV were more likely to be ‘Candidatus M haemominutum’ infected. Their results were based on PCR, which may explain why the findings were similar. However, the presence of FIV antibodies was also associated with the presence of M haemofelis in this study. In contrast, we found no relationship -between FIV and/or FeLV status and M haemofelis infection nor was there any relationship between FeLV-positive cats and ‘Candidatus M haemominutum’ infection. Our study population was domestic rather than feral cats, which might explain this difference. A previous Brazilian study was unable to establish any relationship between hemoplasma and retroviral infections (Mendes-de-Almeida et al 2004). However, the population of cats sampled and method used for hemoplasma detection were different than those used in our study. Their study population was urban stray cats, whereas our study population was cats with pre-existing retroviral infections. Although none was infected with FeLV, antibodies to FIV were detected in 21% and a hemoplasma infection was detected in 38% of these stray cats. The latter findings were based on microscopy evaluation of stained blood films. Nevertheless, other authors have suggested a significant association between FeLV and hemoplasma infection using microscopic evaluation of blood smears as a diagnostic method for hemoplasma infection (Nash and Bobade 1986, Bobade et al 1988, Grindem et al 1990, Harrus et al 2002).
Several other studies indicate anemia or decreased PCV is a risk factor for hemoplasma infection in domestic cats (Grindem et al 1990, Jensen et al 2001, Harrus et al 2002, Tasker et al 2003, Kewish et al 2004). Whether looking at the anemic status or PCV (continuous variable) in the various groups, there was no relationship to the presence of hemoplasma infection in our logistic regression model. These findings were somewhat surprising and were in agreement with a study undertaken in Switzerland which also found no association between hemoplasma infection with anemia or PCV variations (Willi et al 2006b). Perhaps our results reflect a selection bias, as the presence of FIV and/or FeLV are well-known causes of anemia. In the aforementioned study, anemia was found when double hemoplasma or retroviral+hemoplasma infections were concurrent, but there was no statistical difference regarding hemoplasma status in retroviral infected and non-infected cats. Anemia, however, was not a risk factor for retroviral-positive status of cats in our study, even though a positive status was associated with a lower PCV. It might improve our ability to detect an association with anemia, if the number of hemoplasma infected cats in each group were higher. Willi et al (2006b) suggested that the progress of retroviral disease is necessary for the manifestation of the hemoplasma-induced anemia. We agree but also hypothesize that differences in the host–parasite relationship caused by varying geographical/seasonal conditions in these studies and/or stage of infection could be plausible explanations for failing to identify anemia as a risk factor for hemoplasma infection.
Logistic regression also identified pure-breed status as a risk factor for at least one hemoplasma infection among FIV animals, compared to cats negative for FIV and FeLV. This finding contrasts with other studies reporting mixed-breed or non-pedigree cats were more likely to have a hemoplasma infection (Hayes and Priester 1973, Tasker et al 2004). One possibility for these findings is that mixed-breed cats, even those with antibodies to FIV, are more resistant to hemoplasma infection. On the other hand, intensive breeding of pedigree cats, as with kennel raised dogs, might lead to increased prevalence of hemoplasma infections in this population (Kemming et al 2004).
Recently, a third species of hemoplasma – ‘Candidatus M turicensis’ – was described (Willi et al 2005) and found in Brazil among native zoo-maintained wild felids (Willi et al 2007). We were not equipped to run a test for identifying this parasite and are currently setting up a real-time PCR protocol in order to test samples for this newly described species.
According to the supplier, the ELISA detection kit used in this study has a sensitivity of 97.6 and 100% with a specificity of 99.1 and 99.5% for FIV and FeLV, respectively. However, the reliability of test results also depends on the prevalence of the disease (Hartmann et al 2001). A low prevalence of disease may lead to a low positive predictive-value of the test result; thus, the probability is low that the retrovirus is actually present in the cat when the test result is positive (Hardy 1991, Jacobson 1991). A previous study of all cats presenting to the feline-only veterinary clinic in Rio de Janeiro, Brazil reported a prevalence of 17.46% for FeLV and 16.66% for FIV (de Souza et al 2002). Based on these prevalence results and assuming ELISA sensitivity and specificity for FIV and FeLV as previously stated, the reliability of a positive ELISA result of 95% was predicted (Jacobson 1991). Factors which might lead to a false-positive ELISA result for FIV antibody detection are believed to be unlikely in the current study. Maternally derived antibodies to FIV in kittens younger than 6 months of age are reported to confound interpretation of positive test results (Richards 2003, Levy et al 2003). In addition, vaccination for FIV results in rapid production of antibodies in vaccinates that persist at least 1 year inducing a false-positive ELISA result (Levy et al 2004). However, there is no FIV vaccine licensed and being used in Brazil to date and all cats tested positive for FIV antibodies were adults. The prevalence among cats in our study might actually be higher than general population, as most were tested because of illness.
The use of Western blot (Lutz et al 1988, Egberink et al 1992) and direct fluorescence antibody (Hardy et al 1973) tests as the gold standards to confirm feline retroviral infections is restricted to only a few laboratories in Brazil, mostly for research purposes. These tests are expensive and do not provide an immediate result, sometimes needed for deciding clinical procedures. A recent report, comparing ELISA and PCR results for detection of retroviral infections in cats, confirmed the reliability of the commercial ELISA test (Arjona et al 2007). Thus, we believe that our results provide a ‘real world’ and accurate reflection of hemotrophic mycoplasma infections in cats with retroviral infections. The increasing number of cats as pets and the great number of infectious diseases found among domestic animals in Brazil and other tropical areas will provide a rich source of materials for future studies.
Acknowledgments
We would like to thank the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) for the financial support for this study and for Dr Macieira's personal research fellowship; the College of Veterinary Medicine of Cornell University, USA and the School of Veterinary Medicine of Purdue University, USA for the laboratory support; Dr George Moore, from the Department of Comparative Pathobiology of the School of Veterinary Medicine – Purdue University for the statistical and epidemiological assistance to evaluate our data; Dr Heloisa Justen de Souza, from Clínica Veterinária Gatos e Gatos and all practitioners for the great logistic support given to this study. Last but not least, we must acknowledge all owners who allowed their animals to be included in this study.
References
- Arjona A., Barquero N., Domenech A., Tejerizo G., Collado V.M., Toural C., Martin D., Gomez-Lucia E. Evaluation of a novel nested PCR for the routine diagnosis of feline leukaemia virus (FeLV) and feline immunodeficiency virus (FIV), Journal of Feline Medicine and Surgery 9, 2007, 14–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann A., Guimarães A.M.S., Silva C.C., Yamaguti M., Kozemjakim D.A., Messick J.B., Biondo A.W., Timenetsky J. Mycoplasma haemofelis and ‘Candidatus Mycoplasma haemominutum’ detection by PCR in anemic domestic cats (Felis catus) from Curitiba, Brazil: a preliminary study, Veterinary Clinical Pathology Supplement 35, 2006, 370. [Google Scholar]
- Berent L.M., Messick J.B., Cooper S.K. Detection of Haemobartonella felis in cats with experimentally induced acute and chronic infections, using a polymerase chain reaction assay, American Journal of Veterinary Research 59, 1998, 1215–1220. [PubMed] [Google Scholar]
- Birkenheuer A.J., Levy M.G., Breitschwerdt E.B. Development and evaluation of a seminested PCR for detection and differentiation of Babesia gibsoni (Asian genotype) and B canis DNA in canine blood samples, Journal of Clinical Microbiology 41, 2003, 4172–4177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobade P.A., Nash A.S., Rogerson P. Feline haemobartonellosis: clinical, haematological and pathological studies in natural infections and the relationship to infection with feline leukaemia virus, Veterinary Record 122, 1988, 32–36. [DOI] [PubMed] [Google Scholar]
- Chomczynski P. One-hour downward alkaline capillary transfer for blotting of DNA and RNA, Analytical Biochemistry 201, 1992, 134–139. [DOI] [PubMed] [Google Scholar]
- Cotter S.M., Hardy W.D., Jr., Essex M. Association of feline leukemia virus with lymphosarcoma and other disorders in the cat, Journal of the American Veterinary Medical Association 166, 1975, 449–454. [PubMed] [Google Scholar]
- Egberink H.F., Keldermans C.E., Koolen M.J., Horzinek M.C. Humoral immune response to feline immunodeficiency virus in cats with experimentally induced and naturally acquired infections, American Journal of Veterinary Research 53, 1992, 1133–1138. [PubMed] [Google Scholar]
- Foley J.E., Harrus S., Poland A., Chomel B., Pedersen N.C. Molecular, clinical, and pathologic comparison of two distinct strains of Haemobartonella felis in domestic cats, American Journal of Veterinary Research 59, 1998, 1581–1588. [PubMed] [Google Scholar]
- Foley J.E., Pedersen N.C. ‘Candidatus Mycoplasma haemominutum’, a low-virulence epierythrocytic parasite of cats, International Journal of Systematic and Evolutionary Microbiology 51, 2001, 815–817. [DOI] [PubMed] [Google Scholar]
- George J.W., Rideout B.A., Griffey S.M., Pedersen N.C. Effect of preexisting FeLV infection or FeLV and feline immunodeficiency virus coinfection on pathogenicity of the small variant of Haemobartonella felis in cats, American Journal of Veterinary Research 63, 2002, 1172–1178. [DOI] [PubMed] [Google Scholar]
- Grindem C.B., Corbett W.T., Tomkins M.T. Risk factors for Haemobartonella felis infection in cats, Journal of the American Veterinary Medical Association 196, 1990, 96–99. [PubMed] [Google Scholar]
- Hardy W.D., Jr. General principles of retrovirus immunodetection tests, Journal of the American Veterinary Medical Association 199, 1991, 1282–1287. [PubMed] [Google Scholar]
- Hardy W.D., Jr., Hirshaut Y., Hess P. Detection of the feline leukemia virus and other mammalian oncornaviruses by immunofluorescence. Unifying concepts of leukemia, Bibliotheca Haematologica 39, 1973, 778–799. [DOI] [PubMed] [Google Scholar]
- Harrus S., Klement E., Aroch I., Stein T., Bark H., Lavy E., Mazaki-Tovi M., Baneth G. Retrospective study of 46 cases of feline haemobartonellosis in Israel and their relationships with FeLV and FIV infections, Veterinary Record 151, 2002, 82–85. [DOI] [PubMed] [Google Scholar]
- Hartmann K., Werner R.M., Egberink H., Jarrett O. Comparison of six in-house tests for the rapid diagnosis of feline immunodeficiency and feline leukaemia virus infections, Veterinary Record 149, 2001, 317–320. [DOI] [PubMed] [Google Scholar]
- Harvey J.W. Hemotrophic mycoplasmosis (Hemobartonellosis). Greene C.E. Infectious Diseases of the Dog and Cat, 3rd edn, 2006, Elsevier Inc.: Saint Louis, 252–260. [Google Scholar]
- Hayes H.M., Priester W.A. Feline infectious anaemia. Risk by age, sex and breed; prior disease; seasonal occurrence; mortality, Journal of Small Animal Practice 14, 1973, 797–804. [DOI] [PubMed] [Google Scholar]
- Inokuma H., Taroura S., Okuda M., Hisasue M., Itamoto K., Une S., Nakaichi M., Taura Y. Molecular survey of Mycoplasma haemofelis and ‘Candidatus Mycoplasma haemominutum’ infection in cats in Yamaguchi and surrounding areas, Journal of Veterinary Medical Science 66, 2004, 1017–1020. [DOI] [PubMed] [Google Scholar]
- Jacobson R.H. How well do serodiagnostic tests predict the infection or disease status of cats?, Journal of the American Veterinary Medical Association 199, 1991, 1343–1347. [PubMed] [Google Scholar]
- Jensen W.A., Lappin M.R., Kamkar S., Reagan W.J. Use of a polymerase chain reaction assay to detect and differentiate two strains of Haemobartonella felis in naturally infected cats, American Journal of Veterinary Research 62, 2001, 604–608. [DOI] [PubMed] [Google Scholar]
- Kemming G.I., Messick J.B., Enders G., Boros M., Lorenz B., Muenzing S., Kisch-Wedel H., Mueller W., Hahmann-Mueller A., Messmer K., Thein E. Mycoplasma haemocanis infection—a kennel disease?, Comparative Medicine 54, 2004, 404–409. [PubMed] [Google Scholar]
- Kewish K.E., Appleyard G.D., Myers S.L., Kidney B.A., Jackson M.L. Mycoplasma haemofelis and Mycoplasma haemominutum detection by polymerase chain reaction in cats from Saskatchewan and Alberta, Canadian Veterinary Journal 45, 2004, 749–752. [PMC free article] [PubMed] [Google Scholar]
- Kociba G.J., Weiser M.G., Olsen R.G. Enhanced susceptibility to feline leukemia virus in cats with Haemobartonella felis infection, Leukaemia Reviews International 1, 1983, 88–89. [Google Scholar]
- Lappin M.R. Opportunistic infections associated with retroviral infections in cats, Seminars in Veterinary Medicine and Surgery (Small Animal) 10, 1995, 244–250. [PubMed] [Google Scholar]
- Levy J., Richards J., Edwards D., Elston T., Hartmann K., Rodan I., Thayer V., Tompkins M., Wolf A., American Association of Feline Practitioners, Academy of Feline Medicine Advisory Panel 2001 Report of the American Association of Feline Practitioners and Academy of Feline Medicine Advisory Panel on feline retrovirus testing and management, Journal of Feline Medicine and Surgery 5, 2003, 3–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy J.K., Crawford P.C., Slater M.R. Effect of vaccination against feline immunodeficiency virus on results of serologic testing in cats, Journal of the American Veterinary Medical Association 225, 2004, 1558–1561. [DOI] [PubMed] [Google Scholar]
- Luria B.J., Levy J.K., Lappin M.R., Breitschwerdt E.B., Legendre A.M., Hernandez J.A., Gorman S.P., Lee I.T. Prevalence of infectious diseases in feral cats in Northern Florida, Journal of Feline Medicine and Surgery 6, 2004, 287–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz H., Arnold P., Hubscher U., Egberink H., Pedersen N., Horzinek M.C. Specificity assessment of feline T-lymphotropic lentivirus serology, Zentralblatt für Veterinärmedizin. Reihe B, Journal of Veterinary Medicine. Series B 35, 1988, 773–788. [DOI] [PubMed] [Google Scholar]
- Mendes-de-Almeida F., Faria M.C., Branco A.S., Serrão M.L., Souza A.M., Almosny N., Charme M., Labarthe N. Sanitary conditions of a colony of urban feral cats (Felis catus Linnaeus, 1758) in a zoological garden of Rio de Janeiro, Brazil, Revista do Instituto de Medicina Tropical de São Paulo 46, 2004, 269–274. [DOI] [PubMed] [Google Scholar]
- Messick J.B. Hemotrophic mycoplasmas (hemoplasmas): a review and new insights into pathogenic potential, Veterinary Clinical Pathology 33, 2004, 2–13. [DOI] [PubMed] [Google Scholar]
- Nash A.S., Bobade P.A. Haemobartonella felis infection in cats from the Glasgow area, Veterinary Record 119, 1986, 373–375. [DOI] [PubMed] [Google Scholar]
- Neimark H., Johansson K.E., Rikihisa Y., Tully J.G. Proposal to transfer some members of the genera Haemobartonella and Eperythrozoon to the genus Mycoplasma with descriptions of ‘Candidatus Mycoplasma haemofelis’, ‘Candidatus Mycoplasma haemomuris’, ‘Candidatus Mycoplasma haemosuis’ and ‘Candidatus Mycoplasma wenyonii’, International Journal of Systematic and Evolutionary Microbiology 51, 2001, 891–899. [DOI] [PubMed] [Google Scholar]
- Neimark H., Johansson K.E., Rikihisa Y., Tully J.G. Revision of haemotrophic Mycoplasma species names, International Journal of Systematic and Evolutionary Microbiology 52, 2002, 683. [DOI] [PubMed] [Google Scholar]
- Priester W.A., Hayes H.M. Feline leukemia after feline infectious anemia, Journal of the National Cancer Institute 51, 1973, 289–291. [DOI] [PubMed] [Google Scholar]
- Richards J. Retrovirus testing: the mainstay remains, Journal of Feline Medicine and Surgery 5, 2003, 1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza H.J.M., Teixeira C.H.R., Graça R.F.S. Epidemiological study of feline leukaemia virus and feline immunodeficiency virus infections in domestic cats in the city of Rio de Janeiro, Clínica-Veterinária 7, 2002, 14–21. [Google Scholar]
- Tasker S., Binns S.H., Day M.J., Gruffydd-Jones T.J., Harbour D.A., Helps C.R., Jensen W.A., Olver C.S., Lappin M.R. Use of a PCR assay to assess the prevalence and risk factors for Mycoplasma haemofelis and ‘Candidatus Mycoplasma haemominutum’ in cats in the United Kingdom, Veterinary Record 152, 2003, 193–198. [DOI] [PubMed] [Google Scholar]
- Tasker S., Braddock J.A., Baral R., Helps C.R., Day M.J., Gruffydd-Jones T.J., Malik R. Diagnosis of feline haemoplasma infection in Australian cats using a real-time PCR assay, Journal of Feline Medicine and Surgery 6, 2004, 345–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry J., Owens S., Sykes J.E. Epidemiology of hemoplasma infections in anemic cats using conventional and real-time PCR, Proceedings of the Merck/Merial National Veterinary Scholar Symposium, 2006, Louisiana State University: Baton Rouge, Louisiana. [Google Scholar]
- Willi B., Boretti F.S., Cattori V., Tasker S., Meli M.L., Reusch C., Lutz H., Hofmann-Lehmann R. Identification, molecular characterization, and experimental transmission of a new hemoplasma isolate from a cat with hemolytic anemia in Switzerland, Journal of Clinical Microbiology 43, 2005, 2581–2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willi B., Tasker S., Boretti F.S., Doherr M.G., Cattori V., Meli M.L., Lobetti R.G., Malik R., Reusch C.E., Lutz H., Hofmann-Lehmann R. Phylogenetic analysis of ‘Candidatus Mycoplasma turicensis’ isolates from pet cats in the United Kingdom, Australia, and South Africa, with analysis of risk factors for infection, Journal of Clinical Microbiology 44, 2006a, 4430–4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willi B., Boretti F.S., Baumgartner C., Tasker S., Wenger B., Cattori V., Meli M.L., Reusch C.E., Lutz H., Hofmann-Lehmann R. Prevalence, risk factor analysis, and follow-up of infections caused by three feline hemoplasma species in cats in Switzerland, Journal of Clinical Microbiology 44, 2006b, 961–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willi B., Filoni C., Catao-Dias J.L., Cattori V., Meli M.L., Vargas A., Martinez F., Roelke M.E., Ryser-Degiorgis M.P., Leutenegger C.M., Lutz H., Hofmann-Lehmann R. Worldwide occurrence of feline hemoplasma infections in wild felid species, Journal of Clinical Microbiology 45, 2007, 1159–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]