Abstract
Mammalian cells have established mechanisms to reduce the abundance of mRNAs that harbor a nonsense codon and prematurely terminate translation. In the case of the human triosephosphate isomerase (TPI gene), nonsense codons located less than 50 to 55 bp upstream of intron 6, the 3′-most intron, fail to mediate mRNA decay. With the aim of understanding the feature(s) of TPI intron 6 that confer function in positioning the boundary between nonsense codons that do and do not mediate decay, the effects of deleting or duplicating introns have been assessed. The results demonstrate that TPI intron 6 functions to position the boundary because it is the 3′-most intron. Since decay takes place after pre-mRNA splicing, it is conceivable that removal of the 3′-most intron from pre-mRNA “marks” the 3′-most exon-exon junction of product mRNA so that only nonsense codons located more than 50 to 55 nucleotides upstream of the “mark” mediate mRNA decay. Decay may be elicited by the failure of translating ribosomes to translate sufficiently close to the mark or, more likely, the scanning or looping out of some component(s) of the translation termination complex to the mark. In support of scanning, a nonsense codon does not elicit decay if some of the introns that normally reside downstream of the nonsense codon are deleted so the nonsense codon is located (i) too far away from a downstream intron, suggesting that all exon-exon junctions may be marked, and (ii) too far away from a downstream failsafe sequence that appears to function on behalf of intron 6, i.e., when intron 6 fails to leave a mark. Notably, the proposed scanning complex may have a greater unwinding capability than the complex that scans for a translation initiation codon since a hairpin structure strong enough to block translation initiation when inserted into the 5′ untranslated region does not block nonsense-mediated decay when inserted into exon 6 between a nonsense codon residing in exon 6 and intron 6.
For all organisms that have been studied, the abundance of mRNAs harboring a nonsense codon generated by either a frameshift or a nonsense mutation is generally no more than 20 to 25% of normal (for reviews, see references 22, 23, 30, and 36). In the case of mammalian cells, exceptions to this generalization arise when nonsense codon recognition is prevented by inhibitors of translation such as (i) a suppressor tRNA (3, 21), (ii) ribosome-binding drugs, including anisomysin, cycloheximide, emetine, puromycin, or pactamycin (8, 26, 34); (iii) a secondary structure within the 5′ untranslated region that blocks translation initiation (3); or (iv) polio virus infection, which inactivates cap-dependent translation (8). Exceptions also arise for nonsense codons followed by an in-frame translation reinitiation site (47) or residing within the distal end of the translational reading frame (reviewed in references 22 and 23).
For mRNA encoding human triosephosphate isomerase (TPI), the boundary between distal nonsense codons that do and do not reduce mRNA abundance resides between codons 192 and 195 of exon 6, i.e., between 52 and 43 nucleotides (nt) upstream of the 3′-most exon-exon junction, and appears to be determined by the position of the 3′-most intron, intron 6, within pre-mRNA (10). Nonsense codons upstream of the boundary result in the decay of nucleus-associated mRNA so that the levels of nuclear mRNA and, as a consequence, cytoplasmic mRNA are 20 to 25% of normal; nonsense codons downstream of the boundary fail to elicit mRNA decay (4, 12). Moving intron 6 of the TPI gene downstream by 33 bp results in a corresponding movement of the boundary within mRNA (10). Furthermore, a nonsense codon residing 4 bp upstream of intron 6 can be converted from the type that has no effect on mRNA abundance to the type that reduces mRNA abundance with an insertion that increases the distance between the nonsense codon and the intron to 61 bp; an insertion that increases the distance to only 42 bp is of no consequence to mRNA abundance (10). Similar data have recently been obtained for β-globin RNA (48).
It has been difficult to determine if TPI intron 6 positions the boundary between the two types of nonsense codons in a mechanism that depends on intron 6 splicing. The finding that deletion of the intron 6 splice sites has little effect on decay elicited by a nonsense codon at position 189 of exon 6 (12) indicates that either the splice sites are dispensable for intron 6-mediated decay or another cis-acting sequence is capable of effectively mediating decay in the absence of the splice sites. In fact, evidence that another sequence mediates decay, albeit not as efficiently as a spliceable intron 6, does exist: when all of intron 6 is deleted, a boundary is still evident in roughly the same position as when intron 6 is present, although the difference in the extent to which nonsense codons on either side of the boundary affect mRNA abundance is decreased (10). A comparable result exists for β-globin RNA: the 3′-most intron of β-globin RNA normally positions the boundary between nonsense codons that do and do not reduce mRNA abundance, but the intron can be deleted without significant consequence to either the position of the boundary or the extent to which mRNA abundance is decreased (48). Most simply interpreted, these results indicate that a β-globin RNA sequence other than the new 3′-most intron positions the boundary in the absence of the 3′-most intron. This sequence, which resides, at least in part, within the penultimate exon of β-globin mRNA may provide a failsafe mechanism in the case that the 3′-most intron fails to function properly in the nonsense-mediated decay mechanism (48). Nevertheless, we cannot rule out the possibility for TPI RNA that deletion of the 3′-most intron generates a new 3′-most intron that functions differently than the usual 3′-most intron in nonsense-mediated decay. Data indicate that TPI intron 2, when positioned with or without splice sites in the place of TPI intron 6, functions indistinguishably from intron 6 in the extent to which nonsense codons mediate decay (10). The penultimate intron of the mouse major urinary protein (MUP) gene also appears to function in the place of TPI intron 6 in nonsense-mediated decay (49). While it may be that any intron can functionally substitute for intron 6 as the boundary determinant, it is also possible that the cis-acting sequence known to function on behalf of intron 6 in a failsafe capacity is functioning as the boundary determinant.
The importance of an exon-exon junction downstream of a nonsense codon for the reduction in mRNA abundance is corroborated by studies of the mouse T-cell receptor β (TCR-β) gene (9). These studies demonstrated that deleting the three introns that normally reside downstream of a nonsense codon within exon 3 almost completely abrogates the nonsense-mediated reduction in mRNA abundance. In the absence of the three introns, the boundary between nonsense codons that do and do not effectively reduce mRNA abundance resides between 2 and 10 bp upstream of the new 3′-most intron, intron 2. It was not determined if the new 3′-most intron positions the boundary, and it is unclear why the boundary is characterized by a different distance from the 3′-most intron than the boundary for either the TPI gene or the β-globin gene (48). The nonsense codon residing 2 bp upstream of TCR-β intron 2 could be converted to the type that effectively reduces mRNA abundance by reinserting all three TCR-β introns downstream of the nonsense codon. Which of the three introns function(s) to reduce mRNA abundance was not determined. Insertion of the same three introns downstream of the normal termination codon also resulted in a reduction in mRNA abundance, indicating that the normal termination codon does not normally mediate a reduction in mRNA abundance because it resides within the final exon. In contrast to results with the TPI gene, deleting the splice sites of the 3′-most TCR-β intron almost completely abrogates the reduction in mRNA abundance mediated by a nonsense codon within the penultimate exon (9). Therefore, a cis-acting sequence comparable to that of TPI RNA that functions in the absence of a spliceable 3′-most intron either does not exist or does not function as effectively for TCR-β RNA.
Consistent with our studies demonstrating that a nonsense codon within TPI and β-globin mRNAs mediates decay only if located more than 50 to 55 nt upstream of the 3′-most exon-exon junction, a normal termination codon need not reside within the final exon in order to preclude its functioning in nonsense mRNA decay. This is exemplified by the MUP gene (5). Normally, MUP mRNA translation terminates within the penultimate exon, exon 6. However, the normal termination codon does not elicit nonsense-mediated decay since the boundary between nonsense codons that do and do not reduce mRNA abundance resides within exon 5 (5). Notably, a different scenario would be predicted from data for the TCR-β gene (9): the boundary would reside within MUP exon 6, and the normal termination codon would mediate a reduction in MUP mRNA abundance. Discrepancies between predictions made from studies of either the TPI or β-globin gene and the TCR-β gene illustrate the need to assess other exon-intron arrangements for the location of the boundary and the identification of cis-acting sequences that position the boundary.
With the goals of understanding (i) what determines whether or not an intron functions as a boundary determinant and (ii) the positional requirements of a nonsense codon, including a normal termination codon, that make it ineffective in mediating a reduction in mRNA abundance, different nonsense codons within TPI alleles having various exon-intron arrangements were assayed for an effect on mRNA production. The assays were designed so as not to be confounded by the cis-acting sequence that can function on behalf of intron 6. Data indicate that at least one intron is required for nonsense-mediated mRNA decay. Furthermore, TPI intron 6 normally functions to position the boundary between nonsense codons that do and do not reduce TPI mRNA abundance solely because it is the 3′-most intron. When TPI intron 6 is deleted (10), a sequence other than the new 3′-most exon-exon junction most likely positions the boundary, possibly as a failsafe mechanism in case the 3′-most intron fails to function in this capacity, as has been shown for β-globin RNA (48). Data suggest that there must be either an intron or the failsafe sequence residing at least 50 to 55 nt but not too far downstream of a nonsense codon or the nonsense codon will fail to elicit decay. These data indicate that a complex may assemble as a consequence of translation termination, begin scanning 50 to 55 nt downstream of the termination site, and scan only a limited distance for a downstream exon-exon junction or failsafe sequence. Evidence for scanning a limited distance downstream of a translation termination event has also been reported for S. cerevisiae (37). Our finding of what appears to be a limit to scanning for mammalian cells suggests that every exon-exon junction, not just the 3′-most junction, has the potential to function in nonsense-mediated decay. Models for nonsense-mediated mRNA decay are presented in view of these data and the finding that a hairpin structure of sufficient strength to block translation initiation when inserted into the 5′ TPI untranslated region does not block decay when inserted into exon 6, between a nonsense codon within exon 6 and intron 6.
MATERIALS AND METHODS
DNA mutagenesis and plasmid constructions.
All products of PCR and oligonucleotide-directed mutagenesis were characterized by DNA sequencing prior to expression in mammalian cells.
(i) Deleting TPI introns 1 through 6 or 1 through 5.
pmCMV-TPINorm, Δ(introns1–6) and pmCMV-TPI189Ter, Δ(introns1–6) were generated by replacing the intron-containing 3.0-kbp NcoI-NcoI fragment that extends from exon 1 into exon 7 with the corresponding intronless 1.13-kbp NcoI-NcoI fragment from pMT-TPINorm, Δ(introns1–6) and pMT-TPI189Ter, Δ(introns1–6) (28, 29). Analogously, pmCMV-TPINorm, Δ(introns1–5) and pmCMV-TPI189Ter, Δ(introns1–5) were generated by replacing the 3.0-kbp fragment with the corresponding 2.01-kbp NcoI-NcoI fragments of pMT-TPINorm, Δ(introns1–5) (29) and pMT-TPI189Ter, Δ(introns1–5), respectively. pMT-TPI189Ter, Δ(introns1–5) was constructed by substituting the 674-bp EagI-MscI fragment of pMT-TPINorm, Δ(introns1–5) that extends from exon 1 into exon 7 with the corresponding mutagenized fragment. pmCMV-TPI23Ter, Δ(introns1–5) was generated by ligating the 3.44-kbp NcoI-PstI fragment from pmCMV-TPINorm, Δ(introns1–5), which extends from 3′-flanking DNA through the pUC13 vector and mCMV promoter to the initiation codon of TPI exon 1; the 105-bp NcoI-EagI fragment from pMT-TPI23Ter, which extends from the initiation codon to the EagI site of exon 1 and harbors codon 23; and the 2.0-kbp EagI-PstI fragment from pmCMV-TPINorm, Δ(introns1–5).
(ii) Inserting an extra copy of TPI intron 6 into the Klenow-filled EcoRI site of TPI exon 7.
PCR-produced TPI intron 6 was prepared as described earlier (10) and inserted in both orientations into the Klenow-filled EcoRI site of the 2.78-kbp XhoI-NcoI fragment that extends from TPI intron 1 into TPI exon 7. This fragment had been cloned into the XhoI and Klenow-filled BamHI sites of pGEM7-Zf(+). Derivatives of pMT-TPINorm, pMT-TPI189Ter, pMT-TPI195Ter, pMT-TPI208Ter, and pMT-TPI214Ter were generated by substituting the 372-bp MscI-AvrII fragment that extends from exon 6 into exon 7 with the corresponding fragment that harbors the PCR-produced intron. In order to construct derivatives of pMT-TPI237Ter, the 2.09-kbp PstI-PstI fragment from the pMT-TPI construct harboring the extra copy of TPI intron 6 was inserted into the PstI site of pBluescriptKS(−). After introducing the 237Ter mutation in vitro by using the antisense oligonucleotide 5′-AATTCGGGCTAGAGGGAAGCAC-3′ (nucleotides corresponding to the mutagenized codon are underlined, and the mutagenic nucleotide is italicized), the pMT-TPI237 derivative was generated by substituting the 1.96-kbp PstI-PstI fragment of pMT-TPINorm with the corresponding fragment that harbors the PCR-produced intron.
(iii) Inserting MUP intron 5 into the filled EcoRI site of TPI exon 7.
PCR-produced MUP intron 5 was generated by using the sense oligonucleotide 5′-TTGACCTATCCAACTGCAGTAATCAGG-3′ and the antisense oligonucleotide 5′-CCTGGAGGCAGCCAGCTGTAGTGTGAGAAC-3′. The PCR product was digested with PstI and PvuII, each of which cleaves within an underlined sequence of the oligonucleotide. The PstI end was made blunt with Klenow, and the resulting full-length intron (159 bp) was inserted in both orientations into the Klenow-filled EcoRI site of TPI exon 7 as described above for the TPI intron 6 insertions. Derivatives of pMT-TPINorm, pMT-TPI189Ter, pMT-TPI195Ter, pMT-TPI208Ter, pMT-TPI214Ter, and pMT-TPI237Ter were generated analogously to the way corresponding derivatives harboring an extra copy of TPI intron 6 were generated.
(iv) Moving TPI intron 2 downstream of its normal position in the absence of introns 3 through 6.
pMT-TPI23Ter, Δ(introns3–6), which consists of a TPI allele harboring a nonsense codon at position 23 and lacking introns 3 to 6, was generated by replacing the 2.6-kbp EagI-MscI fragment of pMT-TPI23Ter with the 1.8-kbp EagI-MscI fragment of pMT-TPINorm, Δ(introns3–6). pMT-TPI70–71Ter, Δ(introns3–6), which consists of a TPI allele harboring a nonsense codon spanning codons 70 and 71 and lacking introns 3 through 6, was generated by Klenow-filling in the EagI site that spans codons 21 through 23 within exon 1 of pMT-TPINorm, Δ(introns3–6) (28) to create a 4-bp insertion (5′-GGCC-3′ on the sense strand). Subsequently, intron 2 was deleted and an Ecl136II site was generated within codon 94 of exon 3 of both nonsense-free and nonsense-containing plasmids. To this end, pBluescriptKS(−) harboring the 2.34-kbp KpnI-KpnI fragment extending from TPI intron 1 into 3′ flanking sequences of each plasmid was mutagenized by using the antisense oligonucleotide 5′-CATGCCAGGGΔCTGATCTCC-3′, which deletes intron 2, together with the antisense oligonucleotide 5′-CTGAGAGCTCCAGG-3′, which generates the Ecl136II site. PCR-produced intron 2 (10) was then inserted into the created Ecl136II site so as to position the intron 46 bp downstream of its normal position, which is 70 bp downstream of the nonsense codon. Subsequently, the 2.24-kbp KpnI-KpnI fragment that extends from intron 1 into 3′ flanking DNA and that harbors the inserted intron was used to replace the corresponding fragment of pMT-TPINorm, Δ(introns3–6) and pMT-TPI70–71Ter, Δ(introns3–6).
(v) Inserting a hairpin structure into codon 209 of the TPI gene.
The 1.96-kbp PstI-PstI fragment from pMT-TPINorm that harbored an in vitro-generated insertion of the 6-bp HpaI site within codon 209 (10) was subcloned into the PstI site of pBluescriptKS(−). The 52-bp HindIII-BamHI fragment containing on each strand a hairpin structure (hp) having an 18-bp stem (ΔG = −61 kcal/mol) was excised from pSP64 · hp7 (20) and inserted into the HpaI site. pmCMV-TPINorm, hp@codon209 was generated by substituting the 889-bp BstEII-AvrII fragment that extends from intron 5 into exon 7 with the corresponding fragment from the pBluescript subclone. pmCMV-TPI189Ter, hp@codon209 was generated similarly, except the fragment from the pBluescript subclone harbored 189Ter, which was introduced by oligonucleotide-directed mutagenesis (15).
Cell culture, cell transfection, and RNA purification.
Mouse Ltk− cells and NIH 3T3 cells were propagated in minimal essential medium containing 10% fetal calf serum and 5% bovine calf serum. L cells were transiently transfected with DEAE dextran (4), and NIH 3T3 cells were transiently transfected with calcium phosphate (2). Total-cell, nuclear, and cytoplasmic RNAs were purified as previously described (46).
RNA blot hybridization.
Total, nuclear, or cytoplasmic RNA (25 μg) was denatured, electrophoresed in a 1.5% agarose gel, and transferred to a nylon (Zeta-bind) membrane (12). Blot hybridization was performed by using two DNA fragments that had been 32P labeled by random priming (12). MT-TPI or mCMV-TPI RNA was detected by using a 299-bp NcoI-NdeI fragment that derived from the 3′ untranslated region of human TPI cDNA. MT-Gl or mCMV-Gl RNA was detected by using a 170-bp BalI-DraI fragment that included 158 bp of exon 3 and 3′ flanking sequences from the mouse β-globin gene. Hybridization and wash conditions allowed for the detection of human but not mouse TPI RNA (12).
RESULTS
Deletion of all TPI introns eliminates the nonsense-mediated decay of TPI mRNA.
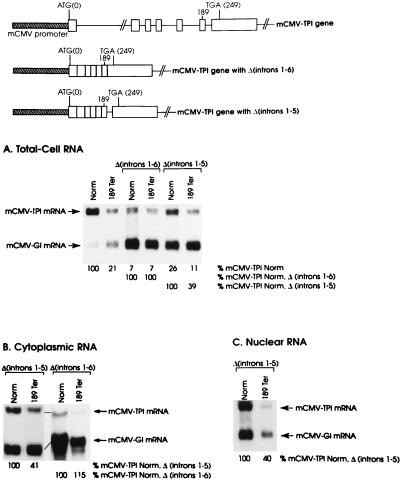
TPI intron 6 clearly plays an important role in positioning the boundary between nonsense codons that do and do not mediate TPI mRNA decay. However, deletion of the intron splice sites or deletion of the entire intron does not eliminate either the boundary or decay, presumably because of the function of another cis-acting sequence. In order to extend these results, the effect on nonsense-mediated decay of deleting all six introns was examined. Introns 1 through 6 were deleted in the context of alleles that were either nonsense-free (Norm) or harbored a nonsense codon at position 189 (189Ter) within exon 6. Each of the TPI alleles was transiently introduced into mouse NIH 3T3 cells in the form of a pmCMV-TPI plasmid, from which TPI gene transcription is driven by the mouse cytomegalovirus (mCMV) promoter (see Fig. 1). In order to control for variations in the efficiencies of cell transfection and RNA recovery, pmCMV-Gl, which contains a β-globin gene similarly driven by the mCMV promoter, was simultaneously introduced. Cells were harvested after 48 h, and the amounts of mCMV-TPI and mCMV-Gl mRNAs in total RNA were quantitated by using Northern blot hybridization. For each transfection, the amount of mCMV-TPI mRNA was normalized to the amount of mCMV-Gl mRNA and presented as a percentage of the normalized amount of the corresponding nonsense-free mRNA, which was considered to be 100.
FIG. 1.
The nonsense-mediated reduction in TPI mRNA abundance is eliminated by deleting all six TPI introns and is restored by reinserting TPI intron 6. NIH 3T3 cells were transfected with the reference pmCMV-Gl construct and the pmCMV-TPI construct specified above each lane, where Ter indicates a nonsense codon and Δ specifies deletion of the designated introns. RNA was purified from total-cell (A), cytoplasmic (B), or nuclear (C) fractions with guanidine isothiocyanate and by cesium chloride centrifugation (12, 46), and 25 μg was analyzed by blot hybridization to DNA fragments 32P labeled by random priming. TPI RNA was detected with a 299-bp NdeI-NcoI fragment that derives from the 3′ untranslated region of human TPI cDNA, and β-globin RNA was detected with a 170-bp BalI-DraI fragment that includes 158 bp of exon 3 from the mouse βmajor-globin gene (11). Hybridization was quantitated with a PhosphorImager. The level of mRNA from each mCMV-TPI allele was normalized to the level of mCMV-Gl mRNA in order to control for variations in the efficiencies of cell transfection and RNA recovery. Normalized values for mCMV-TPI mRNAs that derived from constructs harboring a nonsense codon at position 189 and a deletion of either introns 1 through 6 or introns 1 through 5 [189Ter, Δ(introns 1–6), or 189Ter, Δ(introns 1–5), respectively] were then calculated as a percentage of the corresponding construct harboring a nonsense-free sequence [Norm, Δ(introns 1–6), or Norm, Δ(introns 1–5), respectively]. Values shown are an average of the values obtained from two independently performed transfections, which did not differ by more than 7%.
Consistent with previous demonstrations that used other promoters to drive TPI gene expression in cells other than NIH 3T3 cells (see, e.g., references 3 and 10), a nonsense codon at position 189 (189Ter) within exon 6 of a construct harboring the usual intron configuration reduced the abundance of mCMV-TPI mRNA in total-cell RNA to 21% of normal (Fig. 1A). Deletion of all TPI gene introns [Δ(introns1–6)] reduced the level of nonsense-free mRNA to 7% of normal (Fig. 1A), a finding consistent with previous indications that introns are required for efficient formation of TPI RNA 3′ ends (28, 29), if not other processes. Deletion of all TPI gene introns in cis to 189Ter also resulted in an mRNA level that was 7% of normal (Fig. 1A), indicating that the nonsense codon was ineffective in mediating a reduction in mRNA abundance. Notably, the failure of 189Ter to reduce mRNA abundance is not attributable to a complete block in mRNA export to the cytoplasm (Fig. 1B), where nonsense codon recognition is known to take place. Possibly, the small amount of mRNA that is exported to the cytoplasm may be metabolized in such a way that it bypasses the nonsense-mediated pathway. As one example, the mRNA may not be translated. We deem it unlikely that the deletion of all introns reduces mRNA abundance to a level that precludes our detecting decay. Reinsertion of intron 6 into the intron-less constructs [Δ(introns1–5)] increased the level of nonsense-free mRNA in total-cell RNA to 26% of normal (Fig. 1A) and restored most, but not all, of the 189Ter-mediated reduction in mRNA abundance that characterizes total-cell RNA (Fig. 1A), cytoplasmic RNA (Fig. 1B), and nuclear RNA (Fig. 1C). These data indicate that a minimum of one intron is required for nonsense-mediated decay. At least when this intron is intron 6, decay is less effective than when all introns are present.
Inserting a copy of the 3′-most TPI intron, intron 6, into TPI exon 7 moves the boundary between nonsense codons that do and do not reduce TPI mRNA abundance from within exon 6 to within the new penultimate exon.
The boundary between nonsense codons that do and do not reduce TPI mRNA abundance normally resides within TPI exon 6 and appears to be a fixed distance upstream of the 3′-most exon-exon junction (10). For example, moving intron 6 to a position that is 33 bp downstream of its usual position within the TPI gene results in a comparable movement of the boundary (10). Notably, experiments of this type require that a series of nonsense codons be generated and analyzed in cis to the variously positioned intron. When the present studies were initiated, this type of experiment had been done only for the TPI gene and only by repositioning or deleting intron 6.
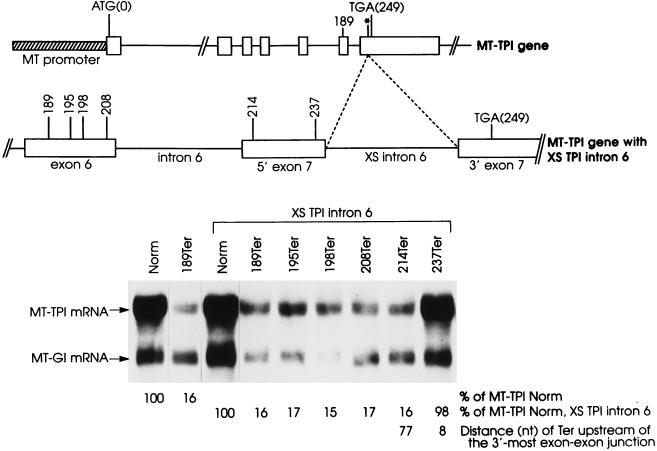
In order to learn more about how TPI intron 6 influences the boundary, an extra copy of intron 6 (128 bp) was generated by PCR and inserted into the EcoRI site of exon 7, which is located 91 bp downstream of the normally positioned intron 6 (Fig. 2). The intron was inserted into an allele that was nonsense-free or harbored a nonsense codon upstream of the insertion site at position 189, 195, 198, or 208 within exon 6 or position 214 or 237 within exon 7. In theory, one or both copies of intron 6 could function to position the boundary.
FIG. 2.
Inserting an extra copy of TPI intron 6 into TPI exon 7 moves the boundary between nonsense codons that do and do not reduce TPI mRNA abundance to the new penultimate exon, indicating that the intron functions in a context-dependent manner to position the boundary. NIH 3T3 cells were transfected, and total-cell RNA was purified and analyzed as described in the legend to Fig. 1. Transfecting plasmids consisted of the reference pMT-Gl construct and the pMT-TPI construct specified above each lane. Constructs with an extra (XS) copy of TPI intron 6 harbored the XS intron at the EcoRI site of TPI exon 7, which is designated with an asterisk in the diagram of the MT-TPI gene. The normalized value for MT-TPI mRNA harboring 189Ter and the usual intron arrangement was then calculated as a percentage of MT-TPINorm mRNA (Norm), which was considered to be 100. Similarly, normalized values for MT-TPI mRNAs that derived from constructs harboring two copies of intron 6 were calculated as a percentage of MT-TPINorm, XS TPI intron 6 mRNA (Norm, XS TPI intron 6). Values are representative of three independently performed experiments and did not differ by more than 6%. Notably, both copies of intron 6 were efficiently and accurately spliced from pre-mRNA as indicated by sequencing products of reverse transcriptase-PCR products that extended from exon 6 through exon 7 (data not shown).
Consistent with previous demonstrations with L cells (4, 10), a TPI gene driven by the mouse metallothionein (MT) promoter and harboring 189Ter in the context of the usual intron configuration reduced the abundance of MT-TPI mRNA in mouse NIH 3T3 cells to 16% of normal (Fig. 2). This is because the boundary between nonsense codons that do and do not reduce TPI mRNA abundance normally resides between codons 192 and 195, i.e., between 52 and 43 nt upstream of the usual 3′-most exon-exon junction (10). Inserting an extra copy of intron 6 into exon 7 moved the boundary from within exon 6 to within the new penultimate exon, i.e., between codons 214 and 237, which reside 77 and 8 nt upstream of the new 3′-most exon-exon junction (Fig. 2, XS TPI intron 6). These results demonstrate that the boundary is determined by the inserted copy and not the usual copy of intron 6, proving that it is the context of an intron within pre-mRNA rather than some feature of the intron per se that engenders intron function as a boundary determinant.
Inserting a copy of MUP intron 5 into TPI exon 7 also moves the boundary between nonsense codons that do and do not reduce TPI mRNA abundance from within exon 6 to within the new penultimate exon.
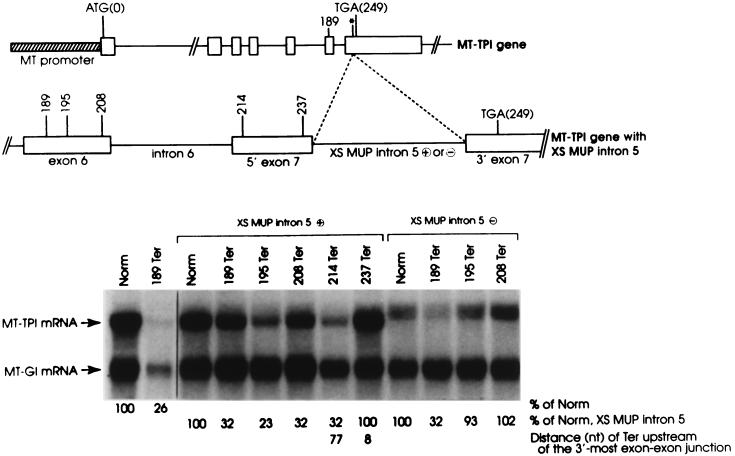
If any intron could function as a boundary determinant provided it were appropriately located within pre-mRNA, then an internal intron from another gene would function in place of the extra copy of TPI intron 6 to position the boundary within the new penultimate exon. Notably, this approach of testing for intron function is far superior to the previous approach of substituting intron 6 with the intron in question (12), since this approach is not confounded by contributions that derive from sequences that function as a putative failsafe determinant in the absence of intron 6. To test if an internal intron can function as a determinant, the penultimate intron, intron 5, of the mouse MUP gene was inserted in either orientation into the filled EcoRI site within TPI exon 7 of nonsense-free or nonsense-containing MT-TPI alleles (Fig. 3). The nonsense codons tested reside at position 189, 195, 208, 214, or 237.
FIG. 3.
Inserting a copy of MUP intron 5 into TPI intron 6 moves the boundary between nonsense codons that do and do not reduce TPI mRNA abundance to the new penultimate exon. L-cell transfections and the purification and analysis of total-cell RNA were as described in the legend to Fig. 1. XS MUP intron 5 denotes the insertion of MUP intron 5 into the EcoRI site of TPI intron 7, where plus (⊕) and minus (⊝) signs specify proper and improper orientations of the insertion, respectively. Values shown are an average of the values obtained from two independently performed experiments, which did not differ by more than 7%.
MUP intron 5 inserted in the proper orientation functioned just like the extra copy of TPI intron 6 (Fig. 2) by moving the boundary between nonsense codons that do and do not reduce TPI mRNA abundance to between codons 214 and 237 (Fig. 3, XS MUP intron 5 ⊕; sequence analysis of PCR-amplified MT-TPI cDNA [data not shown]). In contrast, MUP intron 5 inserted in the opposite orientation failed to move the boundary from its usual position as indicated by the failure of 195Ter to mediate a reduction in mRNA abundance (Fig. 3, XS MUP intron 5 ⊝). Notably, the intron inserted in the opposite orientation was not removed by splicing as indicated by the production of mRNA that includes the 159-nt inverted intron. Considering these results, any intron from any gene might function as a boundary determinant when properly positioned within TPI RNA.
In the absence of TPI introns 3 through 6, a nonsense codon within exon 2 located less than 50 to 55 bp upstream of intron 2 fails to reduce TPI mRNA abundance.
An obvious contextual feature in common to the boundary determinants of the normal TPI gene and the derivative TPI genes harboring an inserted copy of either TPI intron 6 or MUP intron 5 is residence as the 3′-most intron. However, the idea that the boundary within TPI mRNA is always predicted by the position of the 3′-most intron is confounded by the finding that deletion of TPI intron 6 does not appreciably affect the boundary (10). This result suggests that a sequence comparable to the failsafe exonic sequence within β-globin mRNA (48), rather than intron 5, i.e., the 3′-most intron in the absence of intron 6, positions the boundary in the absence of intron 6.
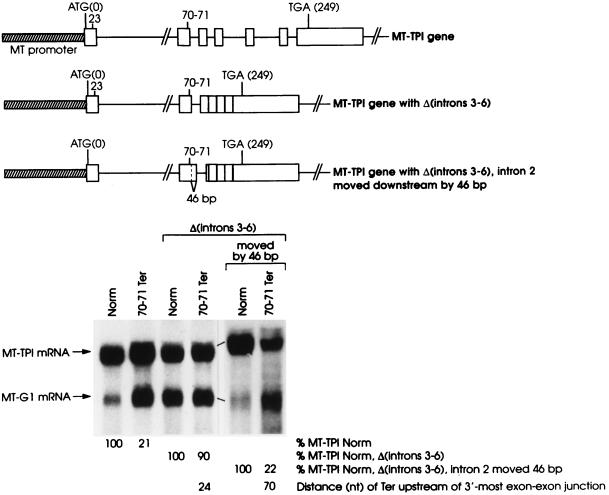
In order to determine if the deletion of intron 6 generally precludes the 3′-most intron from positioning the boundary, TPI intron 2 was made the 3′-most intron by deleting TPI introns 3 through 6. Introns 3 through 6 were deleted in the context of a nonsense-free allele and an allele that harbors a nonsense codon within exon 2. The nonsense codon, generated by a frameshift mutation within exon 1, spans codons 70 and 71 and resides within exon 2, 24 bp upstream of intron 2.
In the context of the full-length TPI gene, 70–71Ter reduced the abundance of TPI mRNA to 21% of normal (Fig. 4), which is expected given the presence of the downstream introns. However, 70–71Ter had essentially no effect on mRNA abundance when introns 3 through 6 were deleted (Fig. 4). Considering that a nonsense codon must reside more than 43 nt upstream of the usual 3′-most exon-exon junction in order to reduce TPI mRNA abundance, 70–71Ter may not reduce mRNA abundance in the absence of introns 3 through 6 because it resides only 24 nt upstream of the new 3′-most exon-exon junction, i.e., the junction generated by the removal of intron 2. Alternatively, the new 3′-most exon-exon junction may not be appropriately located for function in nonsense-mediated decay. For example, it may reside too far from the RNA 3′ end, which could also explain why TPI intron 5 does not function as would be expected of a boundary determinant in the absence of TPI intron 6 (10). Interestingly, whatever cis-acting failsafe sequence localizes the boundary to within exon 6 when intron 6 is the sole intron deleted must not function when introns 3 through 6 are deleted. Otherwise, 70–71Ter would reduce mRNA abundance in the absence of introns 3 through 6.
FIG. 4.
70–71Ter within exon 2 fails to mediate a reduction in mRNA abundance in the absence of introns 3 through 6 unless intron 2, the new 3′-most intron, is moved from residing 24 bp downstream of the nonsense codon to residing 70 bp downstream of the nonsense codon. NIH 3T3 cell transfections and the purification and analysis of total-cell RNA were as described in the legend to Fig. 1. Values shown are an average of the values obtained from two independently performed experiments, which did not differ by more than 8%.
In order to determine if 70–71Ter fails to elicit a reduction in mRNA abundance in the absence of introns 3 through 6 because it resides too close to intron 2, intron 2 was moved downstream by 46 bp in the context of the nonsense-free and 70–71Ter alleles. In so doing, the intron was moved to reside 70 bp downstream of codons 70 and 71, which is a distance that would be predicted to allow for nonsense-mediated decay provided intron 2 were to function comparably to intron 6 as a boundary determinant in the absence of introns 3 through 6. Results demonstrated that 70–71Ter did effectively reduce mRNA abundance when intron 2 was moved (Fig. 4). These findings, taken together with data shown in the earlier figures and data demonstrating that deletion of TPI intron 6 fails to move the boundary between nonsense codons that do and do not mediate mRNA decay, are consistent with the concept that a nonsense codon located less than 50 to 55 nt upstream of any 3′-most exon-exon junction of TPI mRNA will fail to mediate mRNA decay provided the nonsense codon resides a sufficient distance upstream of the failsafe sequence that functions on behalf of intron 6.
Indications that an intron must reside a minimum distance downstream of a nonsense codon or the nonsense codon will not mediate a reduction in mRNA abundance.
The mechanism by which nonsense codons reduce the abundance of nucleus-associated mRNA is best characterized for TPI mRNA. All data indicate that nonsense codons function in cis to reduce TPI mRNA abundance by reducing the half-life of newly synthesized mRNA before it is released from an association with nuclei into the cytoplasm (4). mRNA decay could take place either during or just after mRNA transit across the nuclear pore complex (reviewed in references 22 to 24). Once released into the cytoplasm, TPI mRNA becomes associated with polysomes but is immune to further nonsense-mediated decay (12, 38). Data also indicate that decay is triggered by nonsense codon recognition after splicing (4, 46) in a mechanism that is indistinguishable from cytoplasmic translation (3, 47).
How, then, could an intron that is removed during the process of mRNA formation influence the metabolism of the fully spliced product? Conceivably, the presence of an intron within pre-mRNA could affect mRNP structure, i.e., one or more “marks” may be deposited on the mRNA at the position where the intron resided within pre-mRNA. In one possible scenario, once translation initiates on a nucleus-associated mRNA, if translating ribosomes fail to approximate the 3′-most exon-exon junction, then the mRNA is degraded (Fig. 5, model 1). Notably, decay is not elicited merely by the failure of translating ribosomes to approximate the junction, since a block in translation initiation abrogates decay (reviewed in references 22 and 23). As another possible scenario, after translation initiates and subsequently terminates, some component(s) of the termination complex may loop out or scan the mRNA downstream of the termination site. If the 3′-most exon-exon junction resides a sufficient distance (i.e., ∼50 nt or more) downstream of the termination site, then the mRNA is degraded when either the looped-out complex or the “scanner” reaches the junction (Fig. 5, model 2). Evidence for a scanner derives from the finding that reinitiation downstream of a nonsense codon abrogates decay (47), which indicates that something involved in the decay process senses what resides downstream of the nonsense codon. Evidence for a scanner having limits to the distance that can be scanned derives from the finding that 70–71Ter fails to elicit nonsense-mediated mRNA decay in the absence of introns 3 through 6. This finding suggests that something involved in the decay process recognizes neither the new 3′-most exon-exon junction, because the junction is too close to the nonsense codon, nor the failsafe sequence that functions on behalf of intron 6, because the sequence is too far from the nonsense codon.
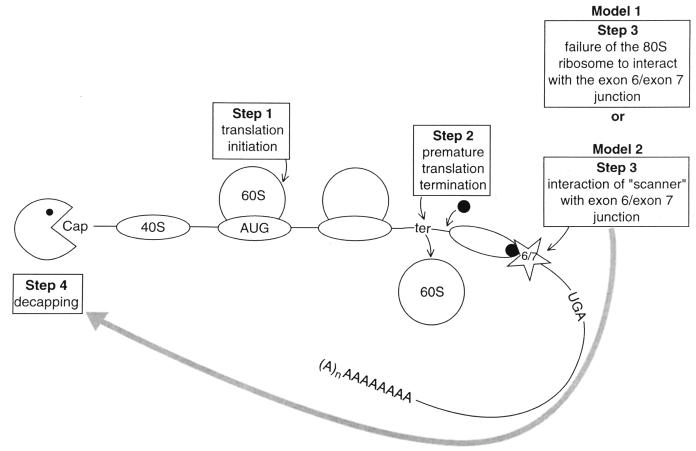
FIG. 5.
Possible mechanisms by which nonsense codons mediate the decay of TPI mRNA. Each model pertains to newly synthesized, nucleus-associated mRNA, the species that undergoes nonsense-mediated decay. The two models differ at Step 3. According to model 1, once translation initiates on TPI mRNA, if translating ribosomes fail to approximate the junction of the last two exons, then the mRNA is degraded. According to model 2, which seems more likely than model 1, once translation initiates and subsequently terminates ∼50 nt or more upstream of the junction of the last two exons, then a complex that could involve a 40S ribosome, eIF-2-GTP-Met-tRNAiMet, and/or, possibly, other component(s) of the translation termination complex, such as the mammalian homolog to S. cerevisiae Upflp (shown as the filled circle), could scan the mRNA downstream of the termination site. If the scanner reaches the junction of the last two exons or, possibly, any downstream exon-exon junction or the failsafe sequence that functions on behalf of intron 6, then the mRNA is degraded. Translation elongation, i.e., peptide bond formation, is not required for nonsense-mediated decay (47). AUG, initiation codon; ter, premature termination codon; UGA, normal termination codon; 40S and 60S, ribosome subunits. We believe the models for TPI mRNA can be generalized to other mRNAs. To do so, the exon 6–exon 7 junction would be substituted with the 3′-most exon-exon junction and UGA would be substituted with the normal termination codon. Furthermore, the consideration that all exon-exon junctions would be marked and the existence of a failsafe sequence would also pertain.
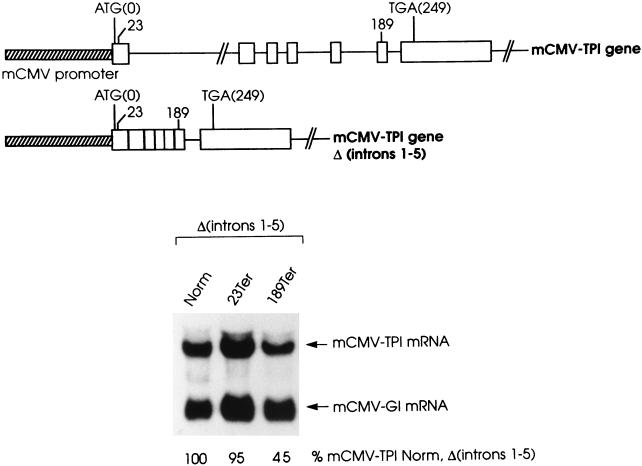
Additional evidence for a scanner would derive from finding that a nonsense codon requires an intron located a minimum distance downstream in order to elicit a reduction in mRNA abundance. To test for such evidence, the effects on TPI mRNA abundance of 23Ter within exon 1 and 189Ter within exon 6 were determined in the absence of introns 1 through 5 (Fig. 6). In the context of a full-length allele, each nonsense codon reduces mRNA abundance to ∼25% of normal (see, e.g., reference 47; Fig. 1 for 189Ter). In the absence of introns 1 through 5, 23Ter resides 559 bp upstream of the sole intron, intron 6, while 189Ter remains 61 bp upstream of intron 6. In the absence of the introns, the level of mRNA harboring 23Ter was 95% the level of mRNA that derives from the corresponding nonsense-free allele (Fig. 6), indicating that the nonsense codon was ineffective in mediating mRNA decay. In contrast, the level of mRNA harboring 189Ter was 45% the level of mRNA that derives from the corresponding nonsense-free allele (Fig. 6; see also Fig. 1), indicating that the nonsense codon still functions, albeit incompletely, in nonsense-mediated mRNA decay. These results reveal that the nonsense-mediated decay of TPI mRNA is optimal when (i) the mRNA derives from an allele harboring one or more introns in addition to the intron that positions the boundary and (ii) an intron resides a minimum distance downstream of the nonsense codon. We conclude that the need for either an intron or a failsafe sequence to be located a minimum distance downstream of the nonsense codon supports model 2 (Fig. 5), in which a complex scans downstream of the nonsense codon in order to elicit nonsense-mediated mRNA decay. It also indicates that exon-exon junctions in addition to the 3′-most junction may be “marked.”
FIG. 6.
Deleting TPI introns 1 through 5 eliminates the reduction in mRNA abundance brought about by 23Ter but not by 189Ter. NIH 3T3 cell transfections and the purification and analysis of total-cell RNA were as described in the legend to Fig. 1. Values shown are an average of the values obtained from two independently performed experiments, which did not differ by more than 5%.
A hairpin structure between codon 189 and intron 6 of the TPI gene is inconsequential to the reduction in mRNA abundance mediated by 189Ter.
One candidate for the scanner would be the complex of eIF-2-GTP-Met-tRNAiMet and the 40S ribosome, which is known to scan mRNAs of Saccharomyces cerevisiae for at least 400 nt downstream of a translation termination event in search of a reinitiation site (16–18). Since reinitiation downstream of a nonsense codon within exon 1 has been demonstrated for TPI mRNA (47), it is possible that a comparable complex also scans TPI mRNA downstream of a nonsense codon within exons 2, 3, 4, 5, or 6.
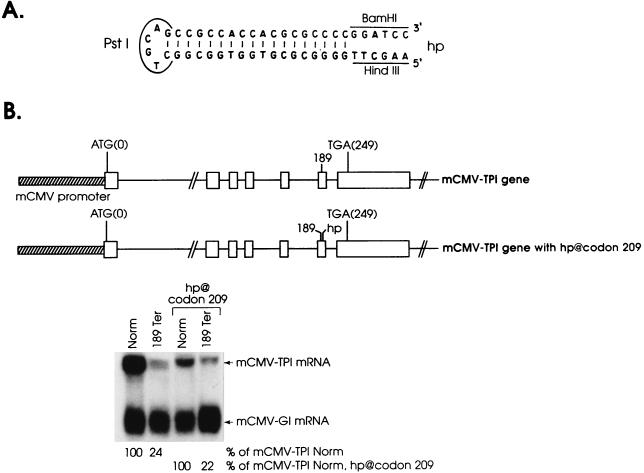
If model 2 (Fig. 5) is true, and the scanner consists only of the complex that scans for the initiation codon, then nonsense-mediated decay would be blocked by a secondary RNA structure of sufficient strength to block the scanner from reaching the junction of a downstream exon-exon junction. One such RNA structure is the hairpin (ΔG = −61 kcal/mol) that has been shown to be of sufficient strength to block translation initiation when inserted into the 5′ untranslated region of the TPI gene (3). To test the nature of the putative scanner, the hairpin (Fig. 7A) was inserted into codon 209 of exon 6 within an allele that was either nonsense-free or harbored 189Ter so the stem of the hairpin resides 11 bp upstream of intron 6 (Fig. 7B). The insertion did not preclude the reduction in mCMV-TPI mRNA abundance mediated by 189Ter (Fig. 7B). This result may indicate that any scanner involved in nonsense-mediated decay has an unwinding capability larger than that of the complex that scans for the initiation codon. Alternatively, the mark at the exon 6-exon 7 junction may encompass the hairpin so as to elicit decay regardless of the hairpin. Attempts to differentiate between these possibilities by (i) inserting the hairpin within exon 1, downstream of a nonsense codon within exon 1, or (ii) inserting a stronger hairpin (ΔG = −72 kcal/mol) within codon 209 of exon 6, downstream of 189Ter, failed because each hairpin precluded the production of a detectable level of mRNA, even when the construct was nonsense-free (38a).
FIG. 7.
Inserting a hairpin structure into exon 6, between 189Ter and intron 6, does not block the nonsense-mediated decay of TPI mRNA. NIH 3T3 cell transfections and the purification and analysis of total-cell RNA were as described in the legend to Fig. 1. The hairpin (hp) (A) was inserted as a Klenow-filled BamHI-HindIII fragment into codon 209, as indicated (B). Values shown are an average of the values obtained from two independently performed experiments, which did not differ by more than 5%.
DISCUSSION
Generally, a nonsense codon reduces the abundance of TPI mRNA provided TPI pre-mRNA harbors an intron located at least ∼50 nt but less than 550 nt downstream of the nonsense codon.
All cells appear to have mechanisms to reduce the abundance of mRNAs that prematurely terminate translation, undoubtedly because the translation of these mRNAs could result in the production of truncated proteins that are deleterious to cell viability (reviewed in references 22, 23, and 36). We show here that the mechanism for human TPI mRNA requires the presence of at least one intron within TPI pre-mRNA (Fig. 1). In theory, an intron may be required for the mRNA (i) to acquire an mRNP structure that is conducive to nonsense-mediated decay (see below), (ii) to be properly compartmentalized within the cell so as to be accessible to decay, (iii) to be produced in sufficient quantity so as to be targeted for decay, or (iv) in any combination of these possibilities. Since the nonsense-mediated decay of TPI mRNA is nucleus associated (4, 12), accessibility to decay may reflect how nuclear mRNA is exported to the cytoplasm. For example, it may be that TPI mRNA produced from an intron-less gene is immune to nonsense-mediated decay because it is not translated, either during export or ever. Countering the idea that mRNA from an intron-less gene is never translated, however, is the existence of commercially available cDNA expression vectors (e.g., pFlag-CMV-2 from Kodak) that are intron-less and successful in directing protein synthesis, at least in the case of some cDNAs. The idea that mRNA may require a particular nuclear history in order to be subject to nonsense-mediated decay is corroborated by the intron requirement and the finding that the presumed failsafe sequence that functions on behalf of intron 6 does not function in the absence of all introns, i.e., does not function with 189Ter to mediate decay (Fig. 1).
Introns are removed by the process of splicing, and it is reasonable to assume that the intron requirement of nonsense-mediated decay may reflect a splicing requirement. In fact, the splice sites of the 3′-most intron of the TCR-β gene have been shown to be critical for the reduction in mRNA abundance brought about by a nonsense codon in the penultimate exon (9). The situation is more complicated in the case of the TPI gene, where deletion of the splice sites of intron 6, the 3′-most intron, is of little consequence to nonsense-mediated decay (10). We propose that the failsafe sequence that functions in decay in the absence of the intron 6 splice sites (10) is exonic, as has been recently shown for β-globin RNA (48).
When an extra copy of TPI intron 6 or a copy of MUP intron 5 is inserted into TPI exon 7, each inserted copy functions to position the boundary between nonsense codons that do and do not reduce mRNA abundance (Fig. 2 and 3). Since MUP intron 5 is normally a penultimate intron, this result indicates that context engenders the function of the boundary determinant to the intron. Therefore, there is apparently nothing inherently different between a boundary-determining intron and other introns aside from being 3′-most and residing 3′ of a failsafe sequence.
Results described here and elsewhere (10) for the TPI gene demonstrate for several different introns at several different positions that mRNA harboring a nonsense codon less than ∼52 nt upstream of the 3′-most exon-exon junction fails to be subject to nonsense-mediated decay. The same conclusion can be drawn from our recent studies of the β-globin gene (48). As would be predicted from this conclusion, the normal termination codon of MUP mRNA, which resides within the penultimate exon, 20 nt upstream of the 3′-most exon-exon junction, does not mediate decay since the boundary between nonsense codons that do and do not reduce MUP mRNA abundance resides within the third-to-last exon (5).
Since the reduction in mRNA abundance mediated by two cis-residing nonsense codons is the same as that mediated by a single nonsense codon (11a; see, e.g., Fig. 4, in which 70–71Ter is in frame with other nonsense codons because it is the consequence of a frameshift mutation) and since the vast majority of intron-containing genes that have been examined are subject to nonsense-mediated decay (reviewed in references 22 and 23), the normal termination codon of the majority of intron-containing genes must not trigger nonsense-mediated decay. Nature seems to have ensured this for most genes by positioning the termination codon within the last exon, so that there are no downstream introns (19). For those genes producing mRNAs that normally terminate translation within an exon other than the last exon, results obtained for TPI, β-globin, and MUP genes indicate that the distance between the normal termination codon and the 3′-most exon-exon junction must be short enough to obviate nonsense-mediated decay. A striking indication that this hypothesis is valid and can generally be applied to genes as a way to predict which termination codons reduce mRNA abundance derives from a search of available gene sequences. A survey of 1,500 genes from species as diverse as fungi, plants, insects, and mammals demonstrated that the normal termination codon of product mRNAs from all but two (i.e., 98%) of the 101 genes found to have one or more 3′ untranslated exons resides less than 50 nt upstream of the 3′-most exon-exon junction (27). As with every generalization, there will likely be exceptions. Possibly, the TCR-β gene is an exception (27) since a nonsense codon located only 8 nt upstream of a 3′-most exon-exon junction within TCR-β mRNA has been reported to mediate a reduction in mRNA abundance (9). Notably, a nonsense codon fails to mediate decay if the closest downstream exon-exon junction is too far away (Fig. 6; see below).
Models for how an intron within pre-mRNA can influence the metabolism of nonsense-containing mRNA: components of the nuclear splicing complex may be exported to the cytoplasm on spliced mRNA and may interact with components of the cytoplasmic translation complex.
All data indicate that the nonsense-mediated decay of TPI mRNA involves nonsense codon recognition after splicing (3, 46) by a mechanism that is indistinguishable from cytoplasmic translation (3, 47). For example, nonsense codons interrupted by an intron within TPI pre-mRNA are capable of effectively mediating decay (46), a finding that has also been demonstrated for immunoglobulin μ mRNA (14) and TCR-β RNA (9). Since a suppressor tRNA abrogates the nonsense-mediated reduction in TPI mRNA abundance (3), as well as TCR-β mRNA abundance (21), a nonsense codon interrupted by an intron would probably not be recognized until after intron removal (46). Consistent with this idea, when the splicing pathway of TPI pre-mRNA is altered so that exon skipping takes place part of the time, only alternatively spliced mRNA that maintains the nonsense-containing exon after splicing is subject to nonsense-mediated decay. Also consistent with nonsense codon recognition after splicing, the nucleus-associated species that undergoes decay is fully spliced (4).
Since decay is restricted to newly synthesized mRNA that copurifies with nuclei, we have proposed that nonsense codon recognition takes place in the cytoplasm, on an mRNA that has yet to be released from an association with nuclei (reviewed in references 22 to 24). Recognition and decay may take place either while the mRNA is in transit across the nuclear pore or after transit but prior to release from an association with the cytoplasmic side of the pore or nuclear envelope. Nevertheless, we are left with the intriguing dilemma of understanding how intron 6, which is removed during the process of splicing, could position the boundary between nonsense codons that do and do not reduce TPI mRNA half-life, i.e., could affect the metabolism of the fully spliced product.
It is conceivable that nonsense codon recognition within TPI mRNA may be linked to the position of the exon 6–exon 7 junction by an interaction in the cytoplasm between components of the translation complex and components of the splicing complex that remain associated with the spliced product. Our finding that a nonsense codon located immediately downstream of the initiation codon is capable of mediating TPI mRNA decay indicates that peptide bond formation is not required for decay (47). Therefore, at least one intron, translation initiation, and translation termination are required for decay, but translation elongation is not. Nonsense-mediated decay may be elicited by the failure of translating ribosomes to reach the exon 6–exon 7 junction (Fig. 5, model 1). Alternatively, nonsense-mediated decay may be elicited when one or more component(s) of the translation termination complex, as a so-called “scanner,” reach the exon 6–exon 7 junction (Fig. 5, model 2) or, possibly, any downstream exon-exon junction or the failsafe sequence that functions on behalf of intron 6. In support of the scanning model, model 2, a newly discovered type of stabilizing element inserted between a termination codon and a downstream destabilizing element can prevent nonsense-mediated decay in S. cerevisiae (37), as can the presence of a translation reinitiation site in S. cerevisiae as well as in mammals (35, 37, 47). Also, the RNA binding and ATPase-helicase activities of S. cerevisiae Upflp, the human homolog of which also functions in nonsense-mediated decay in mammalian cells (1, 31, 39), are not needed in S. cerevisiae for enhancing translation termination but are needed to degrade nonsense-containing transcripts (42, 43). Also in support of scanning, we have found that a nonsense codon is ineffective in eliciting decay if the next downstream intron is located 559 bp away, in contrast to a nonsense codon that is followed by an intron located only 61 bp away (Fig. 6). Also in support of scanning, a nonsense codon is ineffective in eliciting decay if located less than 50 to 55 nt from the only downstream intron and too far upstream of the proposed failsafe sequence (Fig. 4). These findings indicate that for mammalian cells (i) there is a limit to the distance that can be scanned, i.e., there is a limit to the distance between a termination codon and a downstream exon-exon junction or failsafe sequence for nonsense-mediated decay, much as there is an ∼200-nt limit to the distance between a termination codon and a downstream destabilizing element in order for nonsense-mediated decay in S. cerevisiae (37) and (ii) all exon-exon junctions within an mRNA that derives from a multi-intron gene may be “marked” with a remnant(s) of the splicing machine that functions in nonsense-mediated decay. If all exon-exon junctions are marked, then nonsense codons located less than ∼50 nt upstream of the 3′-most exon-exon junction may fail to elicit decay because scanning begins downstream of the only downstream mark. Since there is no correlation between the efficiency with which a nonsense codon mediates decay and the number of exon-exon junctions that reside downstream of the nonsense codon (reviewed in references 22 and 23), we imagine that the interaction of the scanner with a properly positioned downstream junction must be efficient.
Data indicate that a hairpin structure of sufficient strength to block translation initiation (i.e., strong enough to block scanning by eIF-2-GTP-Met-tRNAiMet bound to a 40S ribosome for an initiation codon) does not block nonsense-mediated decay when inserted into exon 6, between a nonsense codon within exon 6 and the exon 6–exon 7 junction (Fig. 7). Provided that scanning takes place and that the hairpin is accessible to the scanner, this finding indicates that any scanner must have a greater RNA unwinding capability than the complex that scans for the initiation codon. A greater unwinding capability may be conferred by one or more factors that associate as a consequence of translation termination (Fig. 5, filled circle). A candidate for such a factor could be the mammalian homolog to S. cerevisiae Upf1p, a factor involved in nonsense-mediated decay in S. cerevisiae that is characterized by RNA binding and helicase activities (44).
While this study was undergoing review, Ruiz-Echevarria et al. (37) reported that the termination of translation at uORF4 of S. cerevisiae GCN4, which blocks ribosomes from reinitiating at downstream initiation sites, does not prevent nonsense-mediated decay. Since decay in S. cerevisiae is dependent on a destabilizing element that resides downstream of the nonsense codon and appears to function analogously to an exon-exon junction or the failsafe sequence of TPI RNA, the scanner may be distinct from the complex that reinitiates translation. Alternatively, the scanner could be the complex that reinitiates translation but either does not have to be capable of reinitiation or does not have to reinitiate efficiently in order to function in nonsense-mediated decay.
Since data implicate nuclear splicing as an effector of cytoplasmic translation, it is notable that certain splicing factors, including hnRNPA1 and SRp20, shuttle between the nucleus and the cytoplasm of mammalian cells (7, 32). Both proteins function in splice-site selection in vitro and in vivo (6 [and references therein], 13, 25, 45), and hnRNPA1 has been found in association with cytoplasmic mRNA (32, 33). Although there is no direct evidence for the presence of any splicing factor on exported mRNP, indirect evidence is provided by the finding that Ct-hrp36, a relative of human hnRNPA and -B proteins found in the insect Chironomus tentans, is incorporated into nascent pre-mRNP complexes, subsequently transported as a complex with mRNA through the nuclear pore to the cytoplasm, and found in polysomes (41). Therefore, Ct-hrp36 is an example of a factor that loads onto pre-mRNA, possibly as a component of the splicing complex, and could interact with the translation complex, as a component of cytoplasmic messenger ribonucleoprotein (mRNP).
Future studies that aim to define the role of trans-acting factors, including components of mRNP, will be required to resolve the mechanism by which introns within pre-mRNA function in the nonsense-mediated decay of mRNA.
ACKNOWLEDGMENTS
This work was supported by Public Health Service research grant DK33938 from the National Institutes of Health. J.P.L. was supported in part by a Bertha Scott Endowed Fellowship from the State University of New York at Buffalo.
We thank Marie Costa for technical assistance, Jack Gauldie for the mCMV immediate-early promoter, John Yates and Eszter Nagy for helpful discussions, and Eszter Nagy for comments on the manuscript.
REFERENCES
- 1.Applequist S E, Selg M, Raman C, Jäck H-M. Cloning and characterization of HUPF1, a human homolog of the Saccharomyces cerevisiae nonsense mRNA-reducing UPF1 protein. Nucleic Acids Res. 1997;25:814–821. doi: 10.1093/nar/25.4.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. Vol. 1. Brooklyn, N.Y: John Wiley & Sons, Inc.; 1995. Introduction of DNA into mammalian cells; pp. 9.1.1–9.1.3. [Google Scholar]
- 3.Belgrader P, Cheng J, Maquat L E. Evidence to implicate translation by ribosomes in the mechanism by which nonsense codons reduce the nuclear level of human triosephosphate isomerase mRNA. Proc Natl Acad Sci USA. 1993;90:482–486. doi: 10.1073/pnas.90.2.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belgrader P, Cheng J, Zhou X, Stephenson L, Maquat L E. Mammalian nonsense codons can be cis effectors of nuclear mRNA half-life. Mol Cell Biol. 1994;14:8219–8228. doi: 10.1128/mcb.14.12.8219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belgrader P, Maquat L E. Nonsense but not missense mutations can decrease the abundance of nuclear mRNA for the mouse major urinary protein, while both types of mutations can facilitate exon skipping. Mol Cell Biol. 1994;14:6326–6336. doi: 10.1128/mcb.14.9.6326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cáceres J F, Stamm S, Helfman D M, Krainer A R. Regulation of alternative splicing in vivo by overexpression of antagonistic splicing factors. Science. 1994;265:1706–1709. doi: 10.1126/science.8085156. [DOI] [PubMed] [Google Scholar]
- 7.Cáceres J F, Scredaton G R, Krainer A R. A specific subset of SR proteins shuttle continuously between the nucleus and the cytoplasm. Genes Dev. 1998;12:55–66. doi: 10.1101/gad.12.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carter M S, Doskow J, Morris P, Li S, Nhim R P, Sandstedt S, Wilkinson M F. A regulatory mechanism that detects premature nonsense codons in T-cell receptor transcripts in vivo is reversed by protein synthesis inhibitors in vitro. J Biol Chem. 1995;270:28995–29003. doi: 10.1074/jbc.270.48.28995. [DOI] [PubMed] [Google Scholar]
- 9.Carter M S, Li S, Wilkinson M F. A splicing-dependent regulatory mechanism that detects translation signals. EMBO J. 1996;15:5965–5975. [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng J, Belgrader P, Zhou X, Maquat L E. Introns are cis-effectors of the nonsense codon-mediated reduction in nuclear mRNA abundance. Mol Cell Biol. 1994;14:6317–6325. doi: 10.1128/mcb.14.9.6317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng J, Fogel-Petrovic M, Maquat L E. Translation to near the distal end of the penultimate exon is required for normal levels of spliced triosephosphate isomerase mRNA. Mol Cell Biol. 1990;10:5215–5225. doi: 10.1128/mcb.10.10.5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11a.Cheng, J., and L. E. Maquat. Unpublished data.
- 12.Cheng J, Maquat L E. Nonsense codons can reduce the abundance of nuclear mRNA without affecting the abundance of pre-mRNA or the half-life of cytoplasmic mRNA. Mol Cell Biol. 1993;13:1892–1902. doi: 10.1128/mcb.13.3.1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chiara M D, Gozani O, Bennett M, Champion-Arnaud P, Palandjian L, Reed R. Identification of proteins that interact with exon sequences, splice sites, and the branchpoint sequence during each stage of spliceosome assembly. Mol Cell Biol. 1996;16:3317–3326. doi: 10.1128/mcb.16.7.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Connor A, Wiersma E, Schulman M J. On the linkage between RNA processing and RNA translatability. J Biol Chem. 1994;269:25178–25194. [PubMed] [Google Scholar]
- 15.Daar I O, Maquat L E. Premature translation termination mediates triosephosphate isomerase mRNA degradation. Mol Cell Biol. 1988;8:802–813. doi: 10.1128/mcb.8.2.802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dever T E, Yang W, Åström S, Byström A S, Hinnebusch A G. Modulation of tRNAiMet, eIF-2, and eIF-2B expression shows that GCN4 translation is inversely coupled to the level of eIF-2 · GTP · Met-tRNAiMet ternary complexes. Mol Cell Biol. 1995;15:6351–6363. doi: 10.1128/mcb.15.11.6351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garcia-Barrio M T, Naranda T, Vazquez de Aldana C R, Cuesta R, Hinnebusch A G, Hershey J W, Tamame M. GCD10, a translational repressor of GCN4, is the RNA-binding subunit of eukaryotic translation initiation factor-3. Genes Dev. 1995;9:1781–1796. doi: 10.1101/gad.9.14.1781. [DOI] [PubMed] [Google Scholar]
- 18.Grant C M, Miller P F, Hinnebusch A G. Requirements for intercistronic distance and level of eukaryotic initiation factor 2 activity in reinitiation on GCN4 mRNA vary with the downstream cistron. Mol Cell Biol. 1994;14:2616–2628. doi: 10.1128/mcb.14.4.2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hawkins J D. A survey on intron and exon lengths. Nucleic Acids Res. 1988;16:9893–9908. doi: 10.1093/nar/16.21.9893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kozak M. Circumstances and mechanisms of inhibition of translation by secondary structure in eucaryotic mRNAs. Mol Cell Biol. 1989;9:5134–5142. doi: 10.1128/mcb.9.11.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li S L, Leonard D, Wilkinson M F. T cell receptor (TCR) mini-gene mRNA expression regulated by nonsense codons: a nuclear-associated translation-like mechanism. J Exp Med. 1997;185:985–992. doi: 10.1084/jem.185.6.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maquat L E. When cells stop making sense: effects of nonsense codons on RNA metabolism in vertebrate cells. RNA. 1995;1:453–465. [PMC free article] [PubMed] [Google Scholar]
- 23.Maquat L E. Defects in RNA splicing and the consequence of shortened translational reading frames. Am J Hum Genet. 1996;59:279–286. [PMC free article] [PubMed] [Google Scholar]
- 24.Maquat L E. RNA export from the nucleus. In: Harford J B, Morris D K, editors. mRNA metabolism and post-transcriptional gene regulation. New York, N.Y: John Wiley & Sons, Inc.; 1997. pp. 107–125. [Google Scholar]
- 25.Mayeda A, Krainer A R. Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. Cell. 1992;68:365–375. doi: 10.1016/0092-8674(92)90477-t. [DOI] [PubMed] [Google Scholar]
- 26.Menon K P, Neufeld E F. Evidence for degradation of mRNA encoding alpha-l-iduronidase in Hurler fibroblasts with premature termination alleles. Cell Mol Biol. 1994;40:999–1005. [PubMed] [Google Scholar]
- 27.Nagy E, Maquat L E. A rule for termination codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem Sci. 1998;23:189–199. doi: 10.1016/s0968-0004(98)01208-0. [DOI] [PubMed] [Google Scholar]
- 28.Nesic D, Cheng J, Maquat L E. Sequences within the last intron function in RNA 3′ end formation in cultured cells. Mol Cell Biol. 1993;13:3359–3369. doi: 10.1128/mcb.13.6.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nesic D, Maquat L E. Upstream introns influence the efficiency of final intron removal and RNA 3′ end formation. Genes Dev. 1994;8:363–375. doi: 10.1101/gad.8.3.363. [DOI] [PubMed] [Google Scholar]
- 30.Peltz S W, Feng H, Welch E, Jacobson A. Nonsense-mediated decay in yeast. Prog Nucleic Acid Res Mol Biol. 1994;47:271–298. doi: 10.1016/s0079-6603(08)60254-8. [DOI] [PubMed] [Google Scholar]
- 31.Perlick H A, Medghalchi S M, Spencer F A, Kendzior R J, Dietz H C. Mammalian orthologues of a yeast regulatory of nonsense transcript stability. Proc Natl Acad Sci USA. 1996;93:10928–10932. doi: 10.1073/pnas.93.20.10928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Piñol-Roma S, Dreyfuss G. Shuttling of pre-mRNA binding proteins between nucleus and cytoplasm. Nature. 1992;355:730–732. doi: 10.1038/355730a0. [DOI] [PubMed] [Google Scholar]
- 33.Piñol-Roma S, Dreyfuss G. mRNA proteins: localization and transport between nucleus and cytoplasm. Trends Cell Biol. 1993;3:151–155. doi: 10.1016/0962-8924(93)90135-n. [DOI] [PubMed] [Google Scholar]
- 34.Qian L, Theodor L, Carter M, Vu M N, Sasaki A W, Wilkinson M F. T cell receptor-β mRNA splicing: regulation of unusual splicing intermediates. Mol Cell Biol. 1993;13:1686–1696. doi: 10.1128/mcb.13.3.1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ruiz-Echevarria M J, Peltz S W. Utilizing the GCN4 leader region to investigate the role of the sequence determinants in nonsense-mediated mRNA decay. EMBO J. 1996;15:2810–2819. [PMC free article] [PubMed] [Google Scholar]
- 36.Ruiz-Echevarria M J, Czaplinski K, Peltz S W. Making sense of nonsense in yeast. Trends Biochem Sci. 1996;21:433–438. doi: 10.1016/s0968-0004(96)10055-4. [DOI] [PubMed] [Google Scholar]
- 37.Ruiz-Echevarria M J, Gonzalez C I, Peltz S W. Identifying the right stop: determining how the surveillance complex recognizes and degrades an aberrant mRNA. EMBO J. 1998;17:575–589. doi: 10.1093/emboj/17.2.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stephenson L S, Maquat L E. Cytoplasmic mRNA for human triosephosphate isomerase is immune to nonsense-mediated decay despite forming polysomes. Biochimie. 1996;78:1043–1048. doi: 10.1016/s0300-9084(97)86728-4. [DOI] [PubMed] [Google Scholar]
- 38a.Sun, X., and L. E. Maquat. Unpublished data.
- 39.Sun, X., H. A. Perlick, H. C. Dietz, and L. E. Maquat. The mutated human homologue of yeast Upf1 protein has a dominant-negative effect on the decay of nonsense-containing mRNAs in mammalian cells. Proc. Natl. Acad. Sci. USA, in press. [DOI] [PMC free article] [PubMed]
- 40.Urlaub G, Mitchell P J, Ciudad C J, Chasin L A. Nonsense mutations in the dihydrofolate reductase gene affect RNA processing. Mol Cell Biol. 1989;9:2868–2880. doi: 10.1128/mcb.9.7.2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Visa N, Alzhanova-Ericsson A T, Sun X, Kiseleva E, Bjorkroth B, Wurtz T, Daneholt B. A pre-mRNA-binding protein accompanies the RNA from the gene through the nuclear pores and into polysomes. Cell. 1996;84:253–264. doi: 10.1016/s0092-8674(00)80980-0. [DOI] [PubMed] [Google Scholar]
- 42.Weng Y, Czaplinski K, Peltz S W. Identification and characterization of mutations in the UPF1 gene that affect nonsense suppression and the formation of the Upf protein complex but not mRNA turnover. Mol Cell Biol. 1996;16:5491–5506. doi: 10.1128/mcb.16.10.5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weng Y, Czaplinski K, Peltz S W. Genetic and biochemical characterization of the mutations in the ATPase and helicase regions of the Upf1 protein. Mol Cell Biol. 1996;16:5447–5490. doi: 10.1128/mcb.16.10.5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weng Y, Ruiz-Echevarria M J, Zhang S, Czaplinski K, Dinman J D, Peltz S W. Characterization of the nonsense-mediated mRNA decay pathway and its effect on modulating translation termination and programmed frameshifting. Mod Cell Biol. 1997;17:241–263. [Google Scholar]
- 45.Yang X, Bani M R, Lu S J, Rowan S, Ben-David Y, Chabot B. The A1 and A1B proteins of heterogeneous nuclear ribonucleoparticles modulate 5′ splice site selection in vivo. Proc Natl Acad Sci USA. 1994;91:6924–6928. doi: 10.1073/pnas.91.15.6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang J, Maquat L E. Evidence that the decay of nucleus-associated nonsense mRNA for human triosephosphate isomerase involves nonsense codon recognition after splicing. RNA. 1996;2:235–243. [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang J, Maquat L E. Evidence that translation reinitiation abrogates nonsense-mediated mRNA decay in mammalian cells. EMBO J. 1997;16:826–833. doi: 10.1093/emboj/16.4.826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang J, Sun X, Qian Y, Maquat L E. Intron function in the nonsense-mediated decay of β-globin mRNA: indications that pre-mRNA splicing in the nucleus can influence mRNA translation in the cytoplasm. RNA. 1998;4:801–815. doi: 10.1017/s1355838298971849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang, J., Y. Qian, and L. E. Maquat. Unpublished data.