Abstract
In this paper the design and use of a semi-quantitative real-time polymerase chain reaction assay (RT-PCR) for feline leukaemia virus (FeLV) provirus is described. Its performance is evaluated against established methods of FeLV diagnosis, including virus isolation and enzyme-linked immunoassay (ELISA) in a population of naturally infected cats. The RT-PCR assay is found to have both a high sensitivity (0.92) and specificity (0.99) when examined by expectation maximisation methods and is also able to detect a large number of cats with low FeLV proviral loads that were negative by other conventional test methods.
Feline leukaemia virus (FeLV) is a retrovirus of cats that is an important cause of disease and mortality. Following infection, there are a variety of outcomes that occur. Some cats mount an effective immune response, appear to recover from the infection, and then become immune to future re-infection. Others become persistently viraemic and although some of these cats remain disease free for long periods, the vast majority succumb to a variety of serious and fatal disease syndromes such as lymphoma or leukaemia, within a few years (Hoover et al 1975, Hardy et al 1976, Rojko and Kociba 1991). These diseased cats act as sources of infection, primarily from virus excreted in the mouth, to susceptible cats with which they come into contact.
The detection of virus is therefore important both in the diagnosis of FeLV-related disease and also for the identification of infected shedder cats in the control of the infection. Several methods are available to diagnose FeLV infection, including rapid tests for FeLV antigen in the plasma, virus isolation (VI) to detect infectious virus in the plasma, immunofluorescence (IF) which detects viral antigen in the blood leukocytes and the polymerase chain reaction (PCR) assays which detect FeLV proviral DNA in the blood cells. The tests most widely used in veterinary practice employ enzyme-linked immunosorbent assay (ELISA) and rapid immunomigration methods for detecting FeLV antigen. VI and IF are principally used as confirmatory tests for viraemia, whilst PCR, a technique developed more recently, is used to detect proviral DNA in the circulation.
In this study we describe the use of a real-time, semi-quantitative polymerase chain reaction assay (RT-PCR) to detect FeLV proviral DNA. We compared RT-PCR test results with results from both ELISA antigen tests and VI tests from a large number of cats from which samples had been submitted for diagnostic screening.
Methods
Detection of FeLV provirus by RT-PCR
Primers and probes
GenBank was searched for long terminal repeat (LTR) sequences belonging to both endogenous and exogenous FeLV genomes. Five were retrieved for use in the development of the RT-PCR assay. Three were chosen to represent each known subgroup of exogenous FeLV; Accession number M18247 for FeLV subgroup A, V01172 for FeLV subgroup B and M14331 for FeLV subgroup C. Two endogenous FeLV pseudogenes were chosen; M21479 for CFE-6 and M21480 for CFE-16. Omiga 2 software (Oxford Molecular, Oxford, UK) was used to perform a Clustal W alignment (Thompson et al 1994) of these sequences. Primers and a probe specific for the exogenous FeLV sequences were designed using Primer3 (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi) in the U3 region of the LTR (Table 1). Primers were ordered from Invitrogen (Invitrogen, Paisley, UK) and probes were synthesised by Cruachem (Cruachem, Glasgow, UK). The reporter dye was indodicarbocyanine (Cy5) and black hole quencher 3 (BHQ3) was the quencher. The primer pair was designed to amplify a 101 base pair PCR product from template of all three exogenous FeLV subgroups, but not the endogenous FeLV sequences. Previously designed primers and probes to the feline 28S rDNA gene were also included in the RT-PCR to act as an internal control (Table 1) (Dean et al 2005).
Table 1.
Primer and probe sequences included in the FeLV and 28S rDNA RT-PCR assay
| FeLV 226 for | 5′-TCCCCAGTTGACCAGAGTTC-3′ |
| FeLV 326 rev | 5′-GATGGCTCGTTTTATAGCAGAAAG-3′ |
| FeLV 272 probe | 5′-Cy5-AATCCCCATGCCTCTCGCTTCTGTA-BHQ3-3′ |
| Cat 28S rDNA 521 for | 5′-AGCAGGAGGTGTTGGAAGAG-3′ |
| Cat 28S rDNA 620 rev | 5′-AGGGAGAGCCTAAATCAAAGG-3′ |
| Cat 28S rDNA 557 Taqman | 5′-Texas Red-TGGCTTGTGGCAGCCAAGTGT-BHQ2-3′ |
DNA was extracted from 100 μl of EDTA blood using the DNeasy blood Kit (Qiagen, Crawley, UK) in accordance with the manufacturer's instructions. RT-PCR was performed using an iCycler IQ (Bio-Rad Laboratories, Hemel Hempstead, UK). The RT-PCR reaction consisted of 12.5 μl of Hotstartaq Master mix (Qiagen), 100 nM FeLV forward and reverse primers, 100 nM FeLV Taqman probe, 200 nM 28S rDNA forward and reverse primers, 200 nM 28S rDNA Taqman probe, a final MgCl2 concentration of 6 mM, 5 μl genomic DNA (approx. 100 ng) and water to 25 μl. After an initial incubation at 95°C for 15 min, 45 cycles of 95°C for 10 s and 60°C for 30 s were carried out. Fluorescence was detected at 620 nm and 680 nm at each annealing step (60°C). EDTA blood samples from five cats known to be FeLV positive, based on a positive ELISA (PetChek FeLV, IDEXX Laboratories, Wőrrstadt, Germany) and positive FeLV isolation (Jarrett and Ganiere 1996), and from six specific pathogen-free derived cats known to be FeLV negative were used as controls. Threshold values were then calculated using the iCycler software ver3.0.
RT-PCR efficiency and linearity
Serial 10-fold dilutions of genomic DNA isolated from an FeLV positive blood sample showed the efficiency of RT-PCR amplification to be 95.4% with a slope of −3.436 and linearity (r=0.995) over a 105 range of starting templates.
Test evaluation
The RT-PCR was then evaluated by using 465 samples of EDTA blood submitted by veterinarians to Langford Veterinary Diagnostics Laboratories for FeLV testing between September 2002 and June 2004. These were consecutive samples for which there was sufficient sample volume to perform all tests. Samples were collected and tested by ELISA, FeLV VI and RT-PCR for FeLV provirus. ELISA testing was carried out as samples were submitted whilst samples for RT-PCR and VI were frozen at −70°C. The samples examined by RT-PCR were analysed as one group at the end of the study. The samples examined by VI were grouped and tested weekly. ELISA tests (PetChek FeLV; IDEXX Laboratories, Worrstadt, Germany) were performed and results interpreted according to the manufacturer's instructions. Virus isolation was carried out according to the method described by Jarrett and Ganiere (1996).
Evaluation of discrepant samples
Samples that were negative by conventional tests but positive by RT-PCR and for which adequate sample volume remained were examined further for antibodies to the feline oncornavirus-associated tumour cell-membrane antigen (FOCMA) and FeLV neutralising antibodies according to the techniques described by Madewell and Jarrett (1983) and Jarrett and Ganiere (1996), respectively. Sixteen RT-PCR positive samples and 16 control samples (negative by all tests) were tested for anti-FOCMA antibodies, and seven RT-PCR positive samples and three control samples were tested for FeLV neutralising antibodies.
Statistical analyses
Data were collated and then analysed using WinEpisope 2.0 (CLIVE Project, Royal (Dick) School of Veterinary Studies, Edinburgh) and SPSS 11.0. (SPSS Inc Chicago, Illinois, USA). Threshold cycle values (CT) were converted to relative copy number (RCN) using a technique described by Dean et al (2005). Samples were grouped according to test results, ie, positive by all tests, positive by RT-PCR and ELISA only and positive by RT-PCR alone and examined for differences in RCN by a Kruskal–Wallis test with Dunn's post test with a null hypothesis of no difference in RCN between groups. Receiver operator characteristic (ROC) analysis was performed to establish RT-PCR threshold cycle cut off values to optimise test sensitivity and specificity estimations. Further calculations of test sensitivity and specificity were performed by using expectation maximisation methods (Pouillot et al 2002) (TAGS, http://www.epi.ucdavis.edu/diagnostictests/QUERY.HTM).
Results
The results of testing the 465 blood samples submitted by veterinarians to Langford Veterinary Diagnostics to evaluate the FeLV RT-PCR are shown in Table 2. Of these samples, 101 tested positive by RT-PCR; of these, 42 were positive by both VI and ELISA, two were positive only by VI and 12 were positive only by ELISA. Forty-five were found to be positive by RT-PCR only. Of the 364 samples that were negative by RT-PCR, one was positive by both VI and ELISA, two were positive by ELISA only, and five were negative by VI but gave equivocal results by ELISA. The remaining 356 samples were negative by both ELISA and VI as well as RT-PCR.
Table 2.
Results of the blood samples examined by FeLV RT-PCR, FeLV VI and FeLV ELISA
| VI + | VI − | |||
|---|---|---|---|---|
| ELISA + | ELISA − | ELISA + | ELISA − | |
| RT-PCR + | 42 | 2 | 12 | 45 |
| RT-PCR − | 1 | 0 | 2 | 356 |
| 5∗ | ||||
Equivocal results by ELISA.
Threshold cycle (CT) values were recorded for each RT-PCR positive result. The lower the CT value, the higher the copy number of proviral DNA copies in the blood cell DNA. All 18 samples with a CT under 20 were also positive by ELISA and VI. Of the 30 samples with CT values between 20 and 25, most (71%, n=21) were ELISA and VI positive; 26% (n=8) of samples were ELISA positive but VI negative, and one sample with a CT value of 24.3 was negative by ELISA and VI. Of the 53 samples with CT over 25, only three were ELISA and VI positive (CT 26.1, 30, 30.2), two were ELISA negative and VI positive (CT 34.8, 38.6), and four were ELISA positive and VI negative (CT 25.1, 26.1, 31.7, 36.6). These results are shown in Table 3.
Table 3.
Number of blood samples testing positive for FeLV by VI and ELISA at different RT-PCR CT values
| RT-PCR CT values | VI + | VI − | ||
|---|---|---|---|---|
| ELISA + | ELISA − | ELISA + | ELISA − | |
| <20 | 18 | 0 | 0 | 0 |
| 20–25 | 21 | 0 | 8 | 1 |
| >25 | 3 | 2 | 4 | 44 |
There was a highly significant difference between RCN values from samples positive by all tests and samples positive by RT-PCR alone (P<0.001) when examined by a Kurskal–Wallis test with Dunn's post test. There was also a highly significant difference between RCN values from samples positive by both RT-PCR and ELISA and samples positive by RT-PCR alone (P=0.01). No difference was found (P>0.05) between RCN values from samples positive by all tests and the RCN values from samples that were positive by RT-PCR and ELISA alone.
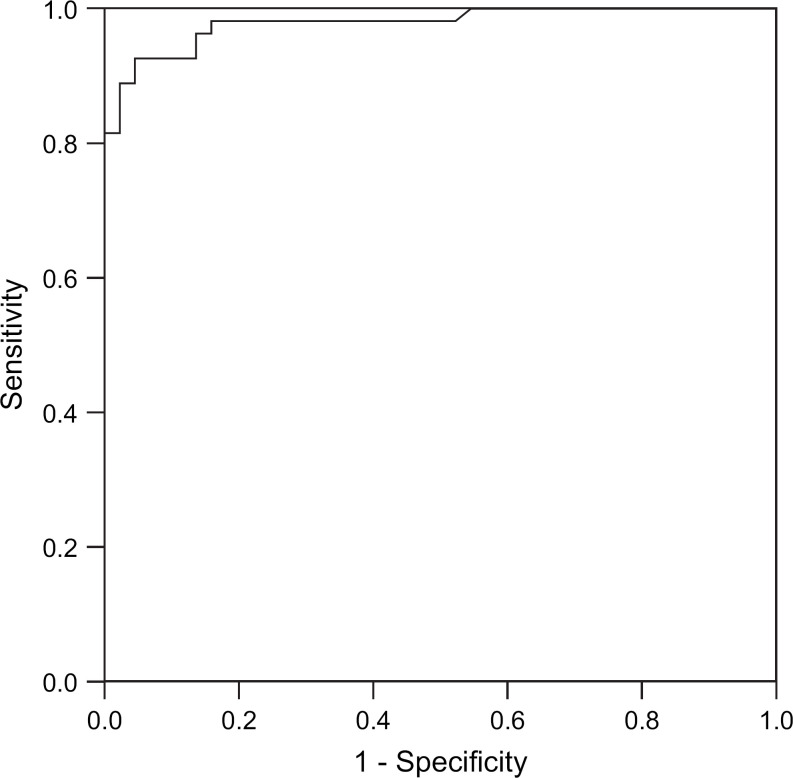
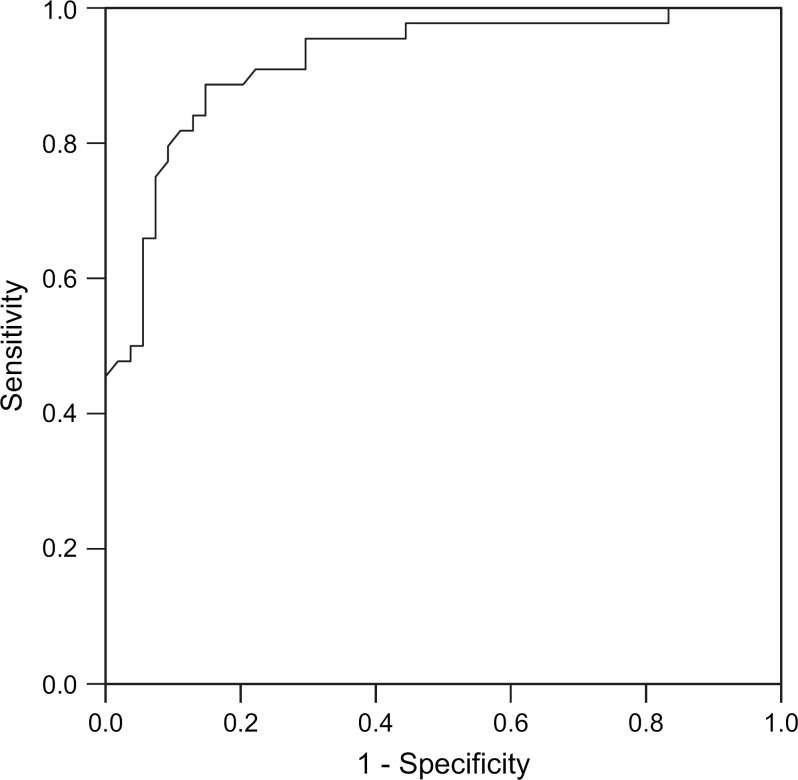
ROC curves for RT-PCR CT – ELISA and RT-PCR CT – VI are shown in Figs 1 and 2. The areas under the curves were 0.979 (standard error 0.012) and 0.920 (standard error 0.28), respectively. A CT value of <20 has 100% specificity and a CT <25 maximises test sensitivity for minimum loss in specificity. Diagnostic sensitivity was 0.9288 (95% CI 0.8003–0.9770) and specificity 0.9950 (95% CI 0.9803–0.9988) when calculated using a CT value of <25 as a cut off point for infection.
Fig 1.
ROC curve RT-PCR CT – ELISA.
Fig 2.
ROC curve RT-PCR CT – VI.
We investigated whether cats that were positive for FeLV proviral DNA by RT-PCR but negative by ELISA and VI had other evidence of having been infected with FeLV. Thus, of 16 samples (positive by RT-PCR alone) that were tested, 13 had anti-FOCMA antibodies, and of seven samples tested for virus neutralising antibodies three were positive. Of the 16 control samples (negative by all tests) also tested, none had detectable anti-FOCMA antibodies and none of the sera from the three samples tested for FeLV virus neutralising antibodies were positive.
Discussion
In this paper the development of a real-time semi-quantitative polymerase chain reaction assay for FeLV proviral DNA and its use in the diagnosis of FeLV infection in cats is reported. CT values of less than 25 showed a very high correlation with positive results for FeLV antigen (by ELISA) and circulating virus (by VI). Further, a CT of less than 20 had a specificity that approached 100% for FeLV viraemia. This indicates that the RT-PCR assay can be used to reliably confirm FeLV viraemia in cats that test positive by traditional methods.
There was agreement between the results of ELISA, VI and RT-PCR in 96% of the samples. However, some discrepancies were recognised between the tests. Fourteen samples were found to be ELISA positive and VI negative and were regarded as discordant. Such discordancy is well recognised (Jarrett et al 1982b, Lutz et al 1983) and has been reported to account for 10–30% of ELISA positive results (Jarrett et al 1982a, Jarrett et al 1991, Weijer and van Herwijnen 1995). In the present study, 24.6% (n=14) of ELISA positive results were discordant with VI. There are a variety of possible explanations, both biological and methodological, that have been proposed for such discordance. Possible biological explanations include expression of cross-reacting antigens from endogenous FeLV genes (Jarrett et al 1982b); localised infection with selective release of antigen but not virus, as has been shown for the mammary gland (Pacitti et al 1986); or recovery from viraemia but failure to clear antigenaemia in cats with transient infections after the immune response has been activated. The latter explanation is unlikely to explain the discrepancies in most cases, as given the sampling methods used in this study, it is improbable that cats would be sampled during this short period. Methodological concerns include cross reactivity in the ELISA with some other antigen (Lopez et al 1989), or a lack of sensitivity of the VI technique due to limited viability of virus during transport; although Jarrett et al (1982b) suggest that virus deterioration is not a concern for FeLV VI.
The concurrent use of PCR can help to clarify some of the discordant results, as shown by Miyazawa and Jarrett (1997) who detected proviral DNA in the blood cells of 13 of 39 discordant cats. In this present study, of the 14 discordant samples 12 (85%) were found to be positive by RT-PCR. CT values for these samples ranged from 20.4 to 36.6 with eight samples having high levels of provirus (CT value of <25). The RT-PCR was designed not to cross react with endogenous FeLV genes, therefore, the most likely explanation is that these cats had localised FeLV infections. Such localised infections have been well recognised and such cats have been shown to carry a risk, thought to be very low, of becoming persistently viraemic and potentially infectious to other cats (Jarrett et al 1982b, Lutz et al 1983, Pacitti et al 1986). Unfortunately, follow-up samples were not available from these cats to determine the eventual outcome in these cases.
The two remaining discordant samples which tested ELISA positive and VI negative were both RT-PCR negative. Possible explanations are that these results were falsely positive by ELISA, reflecting either the presence of an anti-mouse antibody or some other cross-reacting antigen in the sample serum (Lopez et al 1989), or that these two discordant results reflect a combined false negative VI and RT-PCR result.
A further sample was also considered to be potentially false negative by RT-PCR, as this sample was positive by both ELISA and VI. Although RT-PCR, when properly designed, is generally recognised to be a highly sensitive method, there are a variety of factors that can inhibit PCR assays. These include the presence of inhibitors such as free haemoglobin within the sample, or the failure of primers or probes to amplify or detect the PCR product because of sequence heterogeneity between different viral strains. In this study, no evidence for the presence of any inhibitors was found in any of the samples, as the internal control PCR assay for 28S feline DNA were always positive, indicating that DNA amplication had occurred. Sequence heterogeneity remains a possible explanation, as although the RT-PCR is designed to detect DNA from the LTR, a region of the FeLV genome thought to be highly conserved across FeLV strains, the possibility that heterogeneous, and as yet unsequenced, FeLV strains exist in the field cannot be ruled out.
In agreement with other recent reports of FeLV RT-PCR we found proviral DNA within a significant proportion of cats that had undetectable antigenaemia (Hofmann-Lehmann et al 2001, Flynn et al 2002, Torres et al 2005). In the present study 9.3% (n=47) of samples submitted for diagnostic screening tested positive by RT-PCR but negative by ELISA. These findings are similar to those reported by Hoffmann-Lehmann et al (2001) who found a similar discrepancy in 10% of Swiss cats. We also found that there was significantly less provirus (higher CT and lower RCN) in cats that were positive by RT-PCR alone than those that were positive by all tests. Given our calculated reaction efficiency of 96%, the differences in CT values represent an average 30,000-fold difference in proviral load between the two groups.
Our demonstration of antibodies to FeLV in these cats strongly supports the assumption that these discrepant results do indeed arise from cats that have been previously exposed to FeLV but have developed a sufficient immunological response to suppress viraemia and antigen production, but have yet to clear all provirus. Such an outcome following infection has been demonstrated recently by Torres et al 2005 in vaccinated cats.
The long-term consequences of a low-level FeLV proviral infection are currently unknown. However, the fate of pet cats exposed to FeLV in multi-cat households was examined by Hardy et al (1976). Over a follow-up period of 3.5 years, 85% of the viraemic cats died, while only 17% of cats that had recovered and remained free of virus, as indicated by negative results in an IF test, died. This was a life expectancy identical to that of cats for which there had been no known exposure to the virus. Therefore, it would appear that recovered cats are at a very low risk of developing FeLV-related disease, although as shown here and by others (Hoffman-Lehman et al 2001, Torres et al 2005), many cats may show persistence of FeLV proviral DNA following recovery. This study, for the first time, demonstrates that a high proportion of these cats have anti-FOCMA antibodies. As these antibodies are associated with protection against tumour development (Rojko and Kociba et al 1991), it would appear unlikely that such cats would develop FeLV-related lymphoid neoplasms. Whether or not such cats might develop other FeLV-related diseases in the longer term would require a longitudinal study.
This paper demonstrates that RT-PCR for FeLV provirus is a highly accurate test for the diagnosis of FeLV and can potentially be used to monitor proviral levels in cats persistently infected with FeLV. However, its use in investigating the role of latent FeLV infections in other disorders such as immunosuppressive or neoplastic disease remains to be fully explored. Hence, the significance of cats that have detectable proviral loads yet test negative by other tests requires clarification. Further, of the available diagnostic tests for FeLV infection, a negative provirus status, as determined by RT-PCR, is the most reliable for ruling out FeLV infection.
Acknowledgements
M.D. Pinches is sponsored by Axiom Veterinary Laboratories.
References
- Dean R., Harley R., Helps C., Caney S., Gruffydd-Jones T. Use of quantitative real-time PCR to monitor the response of Chlamydophila felis infection to doxycycline treatment, Journal of Clinical Microbiology 43 (4), 2005, 1858–1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn J.N., Dunham S.P., Watson V., Jarrett O. Longitudinal analysis of feline leukemia virus-specific cytotoxic T lymphocytes: correlation with recovery from infection, Journal of Virology 76 (5), 2002, 2306–2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy W.D., Jr., McClelland A.J., Zuckerman E.E., Hess P.W., Essex M., Cotter S.M., MacEwen E.G., Hayes A.A. Prevention of the contagious spread of feline leukaemia virus and the development of leukaemia in pet cats, Nature 263 (5575), Sep 23, 1976, 326–328. [DOI] [PubMed] [Google Scholar]
- Hofmann-Lehmann R., Huder J.B., Gruber S., Boretti F., Sigrist B., Lutz H. Feline leukaemia provirus load during the course of experimental infection and in naturally infected cats, Journal of General Virology 82 (Pt 7), 2001, 1589–1596. [DOI] [PubMed] [Google Scholar]
- Hoover E.A., Olsen R.G., Hardy W.D., Jr., Schaller J.P., Mathes L.E., Cockerell G.L. Biologic and immunologic response of cats to experimental infection with feline leukaemia virus, Bibliotheca Haematologica 43, 1975, 180–183. [DOI] [PubMed] [Google Scholar]
- Jarrett O., Ganiere J.P. Comparative studies of the efficacy of a recombinant feline leukaemia virus vaccine, Veterinary Record 138 (1), 1996, 7–11. [DOI] [PubMed] [Google Scholar]
- Jarrett O., Golder M.C., Stewart M.F. Detection of transient and persistent feline leukaemia virus infections, Veterinary Record 110 (10), 1982a, 225–228. [DOI] [PubMed] [Google Scholar]
- Jarrett O., Golder M.C., Weijer K. Comparison of three methods of feline leukaemia virus diagnosis, Veterinary Record 110 (14), 1982b, 325–328. [DOI] [PubMed] [Google Scholar]
- Jarrett O., Pacitti A.M., Hosie M.J., Reid G. Comparison of diagnostic methods for feline leukemia virus and feline immunodeficiency virus, Journal of the American Veterinary Medical Association 199 (10), 1991, 1362–1364. [PubMed] [Google Scholar]
- Lopez N.A., Jacobson R.H., Scarlett J.M., Center S.A., Randolph J.F., Scott F.W. Sensitivity and specificity of blood test kits for feline leukemia virus antigen, Journal of the American Veterinary Medical Association 195 (6), Sep 15, 1989, 747–751. [PubMed] [Google Scholar]
- Lutz H., Pedersen N.C., Theilen G.H. Course of feline leukemia virus infection and its detection by enzyme-linked immunosorbent assay and monoclonal antibodies, American Journal of Veterinary Research 44 (11), 1983, 2054–2059. [PubMed] [Google Scholar]
- Madewell B.R., Jarrett O. Recovery of feline leukaemia virus from non-viraemic cats, Veterinary Record 112 (15), 1983, 339–342. [DOI] [PubMed] [Google Scholar]
- Miyazawa T., Jarrett O. Feline leukaemia virus proviral DNA detected by polymerase chain reaction in antigenaemic but non-viraemic (‘discordant’) cats, Archives of Virology 142 (2), 1997, 323–332. [DOI] [PubMed] [Google Scholar]
- Pacitti A.M., Jarrett O., Hay D. Transmission of feline leukaemia virus in the milk of a non-viraemic cat, Veterinary Record 118 (14), 1986, 381–384. [DOI] [PubMed] [Google Scholar]
- Pouillot R., Gerbier G., Gardner I.A. ‘TAGS’, a program for the evaluation of test accuracy in the absence of a gold standard, Preventive Veterinary Medicine 53 (1–2), 2002, 67–81. [DOI] [PubMed] [Google Scholar]
- Rojko J.L., Kociba G.L. Pathogenesis of infection by feline leukaemia virus, Journal of the American Veterinary Medical Association 199, 1991, 1305–1310. [PubMed] [Google Scholar]
- Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice, Nucleic Acids Research 22 (22), 1994, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres A.N., Mathiason C.K., Hoover E.A. Re-examination of feline leukemia virus: host relationships using real-time PCR, Virology 332 (1), 2005, 272–283. [DOI] [PubMed] [Google Scholar]
- Weijer K., van Herwijnen R. Detection of FeLV antigen, Veterinary Record 137 (5), 1995, 127. [DOI] [PubMed] [Google Scholar]