Abstract
While in much of the Western world snakes are feared, in the small, rural, mountainous town of Cocullo, in the middle of central Italy, snakes are annually collected and celebrated in a sacro-profane ritual. Every 1st of May, Serpari (snake catchers) capture and showcase dozens of non-venomous snakes to celebrate the ritual of San Domenico. In order to detect potential zoonotic pathogens within this unique epidemiological context, parasites and microorganisms of snakes harvested for the “festa dei serpari” ritual were investigated. Snakes (n = 112) were examined and ectoparasites collected, as well as blood and feces sampled. Ectoparasites were identified morpho-molecularly, and coprological examination conducted through direct smear and flotation. Molecular screenings were performed to identify parasites and microorganisms in collected samples (i.e., Mesostigmata mites, Anaplasma/Ehrlichia spp., Rickettsia spp., Borrelia burgdorferi sensu lato, Coxiella burnetii, Babesia/Theileria spp., Cryptosporidium spp., Giardia spp., Leishmania spp. and helminths). Overall, 28.5% (32/112) of snakes were molecularly positive for at least one parasite and/or microorganism. Endosymbiont Wolbachia bacteria were identified from Macronyssidae mites and zoonotic vector-borne pathogens (e.g., Rickettsia, Leishmania), as well as orally transmitted pathogens (i.e., Cryptosporidium, Giardia, Proteus vulgaris, Pseudomonas), were detected from blood and feces. Thus, given the central role of the snakes in the tradition of Cocullo, surveys of their parasitic fauna and associated zoonotic pathogens may aid to generate conservation policies to benefit the human-snake interactions, whilst preserving the cultural patrimony of this event.
Author summary
The “festa dei serpari” is a unique sacro-profane ritual held each 1st of May in the small town of Cocullo, Central Italy. In this ceremony, dozens of non-venomous free-ranging snakes are captured by serpari (snake catchers) and showcased to thousands of pilgrims and tourists. Therefore, we aimed to assess the parasites and microorganisms of snakes within this unique epidemiological context to identify potential zoonotic pathogens. Snakes were examined and ectoparasites, blood and feces were collected, and morpho-molecular studies were performed. Overall, 28.5% (32/112) of snakes were positive for at least one parasite and/or microorganism. We identified new records of Mesostigmata mites as well as Leishmania tarentolae for the first time in Italian snakes. Importantly, we detected zoonotic microorganisms such as Rickettsia sp. in Aesculapian snake, as well as orally transmitted pathogens (i.e., Cryptosporidium, Giardia, Pseudomonas sp., Proteus vulgaris) from blood and feces of four species of snakes. Thus, snakes handled in this tradition may play a role in the zoonotic transmission of pathogens, given the contact with humans during this unique event.
Introduction
Snakes’ (Serpentes: Squamata) perception and interaction with human societies can be contrasting, generating fear and negative feelings (e.g., disgust, repulsion; [1,2]), being merely tolerated or even used for food or economic sources (i.e., snake charming) [3], or considered as new companion animals [4,5]. One of the major threats on snake conservation is the anthropogenic pressure, directly implicated in the decline of snake populations [6,7]. Similarly, habitat and biodiversity loss and climate change represent main threats on snake populations [8–11]. Important threats are also represented by the predation by domestic carnivores (i.e., dogs and cats) and the high density of some wild ungulates, such as wild boars [12–14], as well as the emergence of pathogens (e.g., Ophidiomyces ophidiicola) within wild populations of snakes [15]. Both factors above are connected, as many pathogens are transmitted by predation, with snakes being intermediate or definitive hosts of parasites, some of which are zoonotic [16].
The relationships and uses by human communities of reptiles is also known as ethnoherpetology [17], which studies the importance of reptiles in different ecological, economic, and cultural contexts [2]. Further investigations were conducted toward integrating One-Health parasitological approaches with ethnoherpetology (i.e., ethnoherpetoparasitology), which allowed to identify the microorganisms and parasites that these animals harbor, as well as the potential risk of zoonotic transmission to snake charmers and vendors in the souks of Marrakech, Morocco [3]. All of the above was assessed in a place where snakes are feared but highly tolerated, given their cultural and economic importance.
In Italy, these slithery animals are part of socio-cultural and religious aspects of the country’s history. One of the most ancient and iconic ethnoherpetological rituals across Europe, known as the “festa dei serpari” (also called the ritual of San Domenico), is performed in the small mountainous town of Cocullo, central Apennine (Abruzzo, central Italy) [6,18,19]. This mixed Catholic and pagan ceremony has been performed for centuries during the first days of May, with little to no alterations, consisting on placing four-lined snakes (i.e., E. quatuorlineata) on top of the statue of San Domenico [20]. Soon after, the snake-adorned statue is taken through the small town, in a religious procession with thousands of onlookers in attendance. During the main event, other species of snakes (e.g., western whip snake—Hierophis viridiflavus, Aesculapian snake—Zamenis longissimus, juvenile specimens of E. quatuorlineata) are handled by “serpari” (i.e., people that capture and handle the snakes) for thousands of pilgrims and tourists to photograph or interact with them [18,21]. In order to have a good number of snakes for the festival, the “serpari” are formally authorized by relevant authorities to capture snakes alive in the surroundings of the Cocullo municipality from the 19th of March till the 30th of April, after which they are obliged to release them in the same capture sites, within three days following the main event.
In Italy, most parasitological studies on snakes have been focused on Cryptosporidium, helminths and ectoparasites in exotic/pet snakes [22–24], with few investigations focused on wild species [25]. In the same context, the introduction of exotic parasites (e.g., Renifer aniarum) in grass snakes (Natrix natrix) and of snake fungal disease (SFD) caused by O. ophidiicola in dice snakes (Natrix tessellata) stresses the importance of monitoring the health status of wild populations of animals by accurate risk assessment [26–28]. In addition, aside from salmonellosis [29,30], zoonotic parasites of reptiles [16,31], including Reptile Vector-Borne diseases (RVBDs; [32]) have gained interest of the scientific community. Indeed, wild snakes are sentinels for zoonotic agents as they are reservoirs of a plethora of pathogens, playing a role in the life cycle of helminths (i.e., cestodes, nematodes and trematodes), pentastomids and vector-borne pathogens [16]. Ticks such as Ixodes ricinus have been collected from wild four-lined snakes (Elaphe quatuorlineata) from southern Italy, that tested positive for Mediterranean spotted-fever (Rickettsia helvetica), but not for Lyme disease (Borrelia burgdorferi sensu lato [33–35]). Other zoonotic parasites have been identified in free-ranging snakes, such as Spirometra erinaceieuropaei, Mesocestoides and Raillietiella [25,36]. Moreover, efforts have been carried out to assess possible emerging pathogens that could be a threat to the snake populations, such as bacteria [37], and the devastating keratinophilic fungus O. ophidiicola [27]. Considering all the above, the present study aimed to investigate parasites and microorganisms associated to snakes collected for the “festa dei serpari” ritual, as well as to identify potential zoonotic pathogens that these animals may harbor.
Methods
Ethics statement
The study was conducted in accordance with all applicable international, national, and/or institutional guidelines for the care and use of animals. One of the major goals was to monitor for threats that could negatively impact the health of snakes due to their handling and exposure to humans within the ritual, whilst continuing to preserve the cultural patrimony of this event. Protocols of snake sampling, handling, and capture by Serpari, as well as by scientific committee, is allowed under the National authorizations (National law DPR 357/97). The permit was granted by the Italian Ministry of Environment (n. 16271/2023 PNM and 79052/2023 PNM).
Animal examination and sampling
Given its cultural, religious, and historical importance, the ritual is accredited by local authorities, which in recent years have worked on increasing the awareness of the importance of conservation of the snake populations to perpetuate the traditional ritual. Since 2010, all “serpari” must declare captured snakes to a scientific committee established specifically for the purpose of the event by the major of the town (EF; GM). The scientific committee is in charge of the operations to record all captured animals along with their biometric data (i.e., weight, snout-vent length measures), and other details, such as sex, age class (juvenile, subadult, adult), and site of capture. The sex of snakes was determined by probing, using round-ended metallic probes, or palpating the hemipenes. Additionally, animals are marked with a subcutaneous Passive Integrated Transponder (PIT) tag, which allows to recognize animals in case of recaptures. Moreover, the scientific committee includes a veterinarian that performs clinical exams on all animals. This initiative has improved the welfare conditions of the collected snakes, as well as educated the “serpari” on animal husbandry. The added value of the presence of the scientific committee has permitted scientists to improve the knowledge of the distribution of different snake species at local and regional scale and to assess the snake population dynamics, accounting for more than 1300 animals examined to date [27].
Snakes captured for the “Festa dei serpari” event in Cocullo, Abruzzo (Fig 1) were examined the 29th and 30th of April 2023, as part of the annual monitoring program performed by the local authorities and a scientific committee (EF, GM). Animals were examined according to the protocols of the monitoring program described elsewhere [27]. After these procedures, animals were clinically assessed and examined for ectoparasites and when found they were removed by scarification methods and stored in 70% ethanol (Fig 2). Blood samples (~100 μl to 1 ml; Fig 3A) were drawn from all animals using the ventral coccygeal vein. Blood was divided between Whatman FTA Cards and 1.5 ml Eppendorf tubes which were later stored at -20°C. Blood smears were performed from all animals and then assessed for the presence of hemoparasites [38] using Diff-Quik stain [39], and later evaluated using an optical microscope (LEICA DM LB2, Germany). Cloacal swabs (Fig 3B) were performed from all animals and stored at -20°C. Whenever possible, fecal samples were collected and stored in 1.5 ml Eppendorf tubes at 4°C.
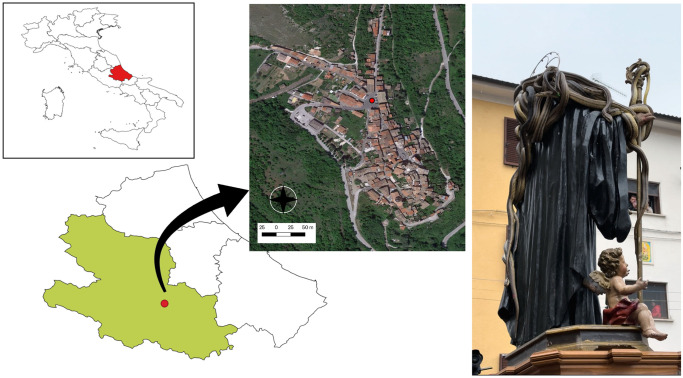
Fig 1. Map of the town of Cocullo, Abruzzo, Central Italy, where the ritual of “festa dei serpari” is celebrated.
L’Aquila province is evidenced in light green. Red circles indicate the municipality of Cocullo, as well as the town square where the statue of San Domenico is covered in snakes. Map prepared using QGIS software—Buenos Aires version (link of the XYZ tile: https://gdg.sc.egov.usda.gov/GDGOrder.aspx).
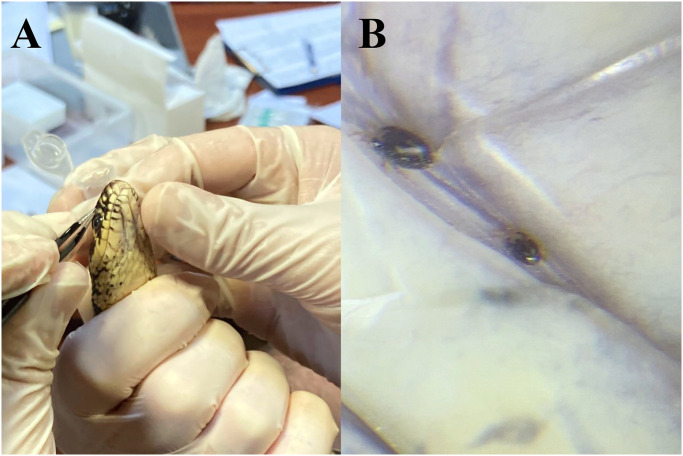
Fig 2. Ectoparasite collection from snakes.
a) mite collection from the gular region from a western whip-tail snake (H. viridiflavus); b) Macronyssidae mites in the gular area of a whip-tail snake (H. viridiflavus).
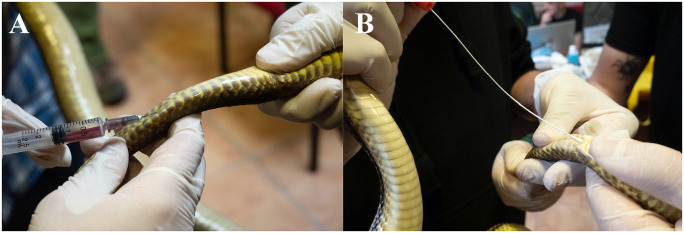
Fig 3. Blood and fecal sampling of snakes.
a) blood draw from the ventral coccygeal vein from a four-lined snake (E. quatuorlineata); b) cloacal swab from a four-lined snake (E. quatuorlineata).
Ectoparasite processing and identification
Ectoparasites were slide-mounted in Hoyer’s medium [40] and identified using dichotomous keys [41–43], as well as original species descriptions [42,44] were used for morphological identification of Mesostigmata mites. To assess the parasitic load of mites, descriptive statistics was calculated using Quantitative Parasitology software, version 3.0 [45]. Prevalence, mean abundance (i.e., number of mites per total number of hosts) and mean intensity (i.e., number of mites per number of infested hosts) were calculated.
Coprological studies
Fecal samples were stored at 4°C and analyzed within 48 hours. Due to the low volume obtained per individual snake (~50 μl), all samples were only analyzed microscopically through direct smear (using saline solution) to observe motile protozoa, helminths, acanthocephalans, and pentastomids, as well as a flotation test with a low-density solution was performed (saturated ZnCl solution, specific gravity 1350) [23].
Molecular screening
DNA of mites was extracted via lysis using the guanidine isothiocyanate protocol (GT) [46]. This protocol was adapted to avoid mite destruction, which allowed the preservation of a voucher for morphological evaluation [33]. Extractions were performed from individual mites. DNA was extracted from individual blood samples (n = 112), feces (n = 38), and cloacal swabs (n = 101) using commercial kits (QIAamp DNA Mini Kit and DNeasy PowerSoil kit, Qiagen, Hilden, Germany), according to the manufacturer’s instructions.
PCRs of the mites were performed to confirm species identity using two molecular markers: Cytochrome Oxidase subunit 1 (cox1; primers Cox1 LCO1490 and HCO2198), that amplifies 680 bp fragment [47], and primers for the 18S rRNA gene (18S+ and 18S−, respectively), which amplify a fragment of 480 bp of the V4 region [48]. Cycling conditions for both PCRs were initial denaturation at 94°C for 1 min, then 30 cycles of 20 s at 94°C, 50°C for 30 s and 72°C for 1 min and 30 s, with a final extension of 72°C for 7 min.
DNA extracted from mites, blood, feces, and cloacal swabs was analyzed for the detection of different microorganisms and parasites through cPCR and qPCR protocols (Table 1). All cPCR products were examined on 2% agarose gel stained with GelRed (VWR International PBI, Milan, Italy) and visualized on a GelLogic 100 gel documentation system (Kodak, New York, USA). Amplicons were then purified and sequenced in both directions using the same PCR primers, by the Big Dye Terminator version 3.1 chemistry in a 3130 Genetic Analyzer (Applied Bio-systems, Foster City, CA, USA). Sequences were edited and analyzed using Geneious Prime software version 9.0 (Biomatters Ltd., Auckland, New Zealand) [49] and compared with those available in the GenBank database by the Basic Local Alignment Search Tool (BLAST; http://blast.ncbi.nlm.nih.gov/Blast.cgi) for species identification.
Table 1. Pathogens screened in this study by conventional (c) and quantitative (q) PCR, with target genes, primers, probes nucleotide sequences and fragment length.
| Pathogens | Target gene | Primers | Sequence (5′−3′) | Fragment length (bp) | References | |
|---|---|---|---|---|---|---|
| cPCR | Anaplasma/Ehrlichia spp. | 16S rRNA | EHR-16SD | GGTACCYACAGAAGAAGTCC | 345 | [50] |
| EHR-16SR | TAGCACTCATCGTTTACAGC | |||||
| Borrelia burgdorferi sensu lato | Flagellin | FLA1 | AGAGCAACTTACAGACGAAATTAAT | 482 | [51] | |
| FLA2 | CAAGTCTATTTTGGAAAGCACCTAA | |||||
| Rickettsia spp. | gltA | CS-78F | GCAAGTATCGGTGAGGATGTAAT | 401 | [52] | |
| CS-323R | GCTTCCTTAAAATTCAATAAATCAGGAT | |||||
| Spotted Fever Group Rickettsiae | ompA | Rr190.70F | ATGGCGAATATTTCTCCAAAA | 632 | [53] | |
| Rr190.701R | GTTCCGTTAATGGCAGCATCT | |||||
| Coxiella burnetii | IS1111a | Trans-1 | TATGTATCCACCGTAGCCAGT | 687 | [54] | |
| Trans-2 | CCCAACAACACCTCCTTATTC | |||||
| Babesia/Theileria spp. | 18S rRNA | RLB-F | GAGGTAGTGACAAGAAATAACAATA | 460–520 | [55] | |
| RLB-R | TCTTCGATCCCCTAACTTTC | |||||
| Cestodes/Nematodes | cox1 | JB3 | TTTTTTGGGCATCCTGAGGTTTAT | 400 | [56] | |
| JB4.5 | TAAAGAAAGAACATAATGAAAATG | |||||
| Leishmania spp. | ITS1 | L5.8S | TGATACCACTTATCGCACTT | 320 | [57] | |
| LITSR | CTGGATCATTT-TCCGATG | |||||
| Trypanosomatidae | 18S rRNA | 18SN1F | GGATAACAAAGG AGCAGCCTCTA | 332 | [58] | |
| 18SN1R | CTCCACACT TTG GTTCTTGATTGA | |||||
| qPCR | Leishmania spp. | kinetoplast | LEISH-1 | AACTTTTCTGGTCCTCCGGGTAG | 120 | [59] |
| LEISH-2 | ACCCCCAGTTTCCCGCC | |||||
| Probe | 6-FAM-AAAAATGGGTGCAGAAAT-MGB | |||||
| Duplex Leishmania | ITS1 | L.i.t. -ITS1-F | GCAGTAAAAAAAAGGCCG | 150 | [60] | |
| L.i.t. -ITS1-R | CGGCTCACATAACGTGTCGCG | |||||
| Probe L.t. | 6-FAM-CACGCCGCGTATACAAAAACAC-MGB | |||||
| Probe L.i. | VIC-TAACGCACCGCCTATACAAAAGCA-MGB | |||||
| Giardia duodenalis | SSU | Giardia-80F | GACGGCTCAGGACAACGGTT | 62 | [61] | |
| Giardia-127R | TTGCCAGCGGTGTCCG | |||||
| Giardi-105 | Fam-5′- CCCGCGGCGGTCCCTGCTAG-3′-Tamra |
Additionally, fecal samples and cloacal swabs were tested using a multiplex (5plex) qPCR for Blastocystis hominis, Cryptosporidium spp., Dientamoeba fragilis, Entamoeba histolytica, and Giardia duodenalis assemblages A and B [62]. In order to have sequences of the positive samples, nested PCRs were performed for Giardia spp. and Cryptosporidium spp. as follows. For Giardia, a nested PCR amplifying a partial sequence of the triosephosphate isomerase (tpi) gene (532 bp) was used that detects all known assemblages [63, 64]. For Cryptosporidium spp., a nPCR targeting a fragment of the 18S rRNA gene was run [65].
Phylogenetic analyses
Mite 18S rRNA and Rickettsial gltA, were separately aligned against those closely related species available from GenBank database using the ClustalW application within MEGA7 software [66]. The Akaike Information Criterion (AIC) option in MEGA7 was used to establish the best nucleotide substitution model adapted to each sequence alignment. Tamura 3-parameter model with invariant sites (I) [66] was used to generate the gltA trees and Tamura 3-parameter model with invariant sites (I) and discrete Gamma distribution (G) for 18S rRNA of mites, and Kimura 2-parameter model with invariant sites (I) and discrete Gamma distribution (G) for 18S rRNA of Leishmania. Maximum likelihood (ML) phylogenetic inference was used with 2000 bootstrap replicates to generate the phylogenetic tree in MEGA7. Homologous sequences of 18S rRNA for Ixodes ricinus tick (Z74479) were used as outgroup to root the trees, as well as for Rickettsia including the gltA sequences from Rickettsia belli and Rickettsia canadensis (AB297809), and the 18S rRNA sequence of sequence of Trypanosoma brucei (XR_002989995).
Results
Overall, 112 snakes were examined and screened representing two families and five species (Table 2). Only two animals were not marked with PIT tags because of their small body size (one Coronella girondica, one Zamenis longissimus). Most individuals were apparently healthy, with three animals having some type of external lesion (Table 2).
Table 2. Species of snakes (scientific and common names) sampled, sex and clinical observations.
| Family | Species | Common name | n | Sex | Observations |
|---|---|---|---|---|---|
| Colubridae | Coronella girondica | Southern smooth snake | 1 | Female (1) | |
| Elaphe quatuorlineata | Four-lined snake | 66 | Male (47) Female (19) |
Abscesses of masses in their dorsal subcutaneous area (2) | |
| Hierophis viridiflavus | Western whip snake | 28 | Male (20) Female (8) |
Skin lesions compatible scale loses due to trauma (1) | |
| Zamenis longissimus | Aesculapian snake | 15 | Male (20) Female (8) |
||
| Natricidae | Natrix helvetica | Grass snake | 2 | Female (2) | |
| Total | 112 | Male (87) Female (38) |
Of the 101 blood smears performed and examined, none of them had visible hemoparasites. Additionally, ectoparasites were collected from 10.7% of snakes (12/112) being all of them mites (n = 46) of the Mesostigmata order. Species of mites, their sex and snake hosts, as well as infestation rates are summarized in Table 3. Namely, mites were identified as Hemilaelaps piger (Berlese, 1918) (Ixodorhynchidae: Mesostigmata) in two H. viridiflavus, one of them also co-infested with the second species identified, Ophionyssus sp. (Macronissydae: Mesostigmata). The latter mite species was identified in the remaining 11 infested snakes (Table 3). Mites were found mainly in the gular area (Fig 2B).
Table 3. Mite species, snake host, biological stage (nymph = N; male = M; female = F), infestation rates and sequence accession numbers (AN).
| Mites Species | Snake species | N snakes | n and sex of mites | Prevalence | Mean Intensity | Mean Abundance | Sequence AN |
|---|---|---|---|---|---|---|---|
| Hemilaelaps piger | Hierophis viridiflavus | 2* | 5F | 1.8% (2/112) | 2.5 (95% CI: 1–2.5) | 0.37 (95% CI: 0.16–0.79), | 18S rRNA: OR771478, OR771479 |
| Ophionyssus sp. | Elaphe quatuorlineata | 3 | 2M; 1F | 9.8% (11/112) | 3.73 (95% CI: 2–6.55) | 0.04 (95% CI: 0.0–0.16) | 18S rRNA: OR771465 |
| Hierophis viridiflavus | 8* | 1N; 3M; 32F | 18S rRNA: OR771463 cox1: OR763078 |
||||
| Zamenis longissimus | 1 | 2F | 18S rRNA OR771469 |
||||
| Total | 12 | 1N; 5M;35F (46) |
*one snake co-infested
Hemilaelaps piger were all females (n = 5) and some of them bearing eggs (Fig 4A). Morphologically, mites displayed features such as three pairs of sternal setae (Fig 4B) and one pair of metasternal setae. The species identified in this study belongs to the piger group, given that all coxae I bear two strong bifid spurs (Fig 4C), and coxae II and III have a single bifid spur and a simple seta. Observed females had dorsal shield bearing 34–36 setae. The sclerotized part of the sternal shield was short, arched and very broad, the anal shield is within a cribrum (Fig 4D), and the anal opening is in the anterior part of the anal shield. Both 18S gene sequences obtained herein had nucleotide identity of 99.2% with sequences of Hemilaelaps triangulus from captive snakes of Mexico (i.e., MT163322, MT163324, MT163324).
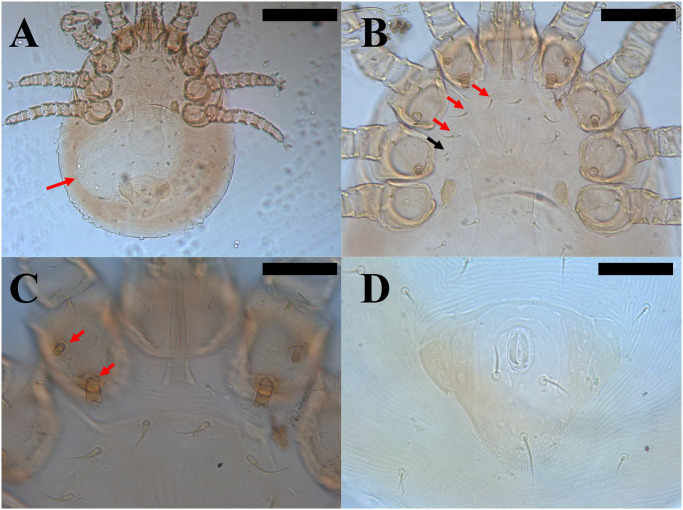
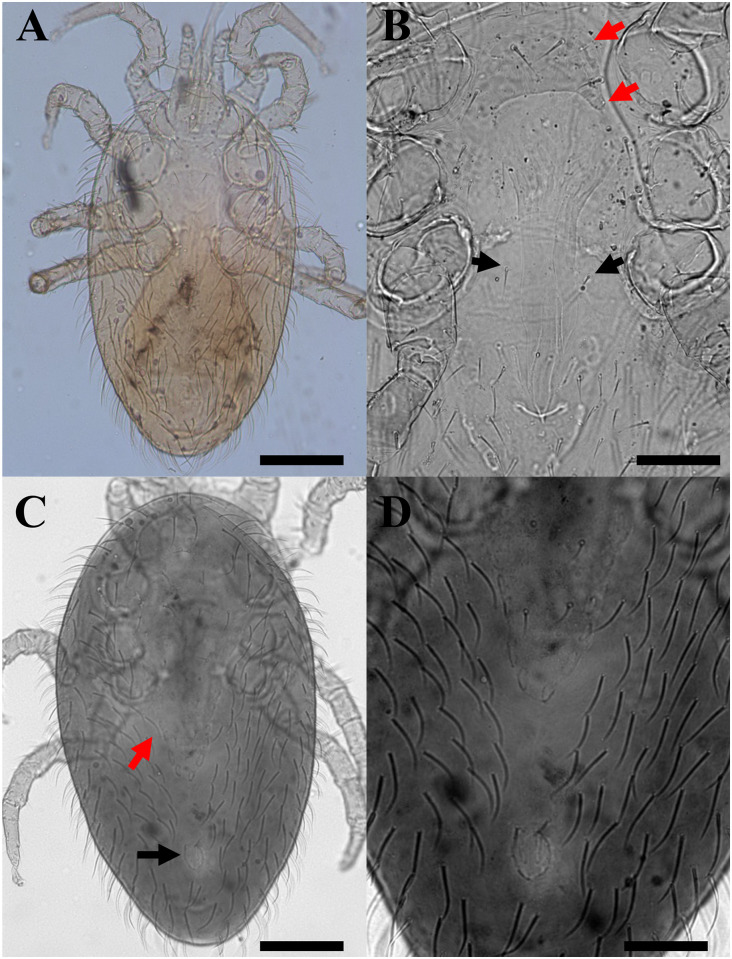
Fig 4. Morphological features of Hemilaelaps piger females.
a) egg in the idiosoma of a female (red arrow); b) Three pairs of sternal setae (red arrows) and one pair of metasternal setae (black arrow); c) coxae I bear two strong bifid spurs (red arrows) typical of the piger group; d) the anal shield within a cribrum. Scale bars: 200μm (a); 100μm (b); 50μm (c,d).
The remaining mites (n = 41) were all identified as Ophionyssus sp. (Fig 5A), most of them females (n = 32), characterized by having less than three pore pairs on sternal shield (Fig 5B). Also, the Genu III have 10 setae, and the epigynal shield is surrounded by the genital setae inserted on the integument (Fig 5B). Moreover, females had a dorsal shield divided in a large anterior and a small pygidial shield (Fig 5C) but differed in not having mesonotal scutae (Fig 5D) and having 9 pair of setae in the podonotal shield. Additionally, sequences obtained from 18S (n = 14) had high nucleotide similarities (99.7%) with Ophionyssus natricis from captive snakes of Italy (OP752167). On the other hand, cox1 sequences (n = 15) had low homology (85.8%) with those of O. natricis from captive snakes in Mexico (i.e., MT154424, MT154425). The 18S rRNA ML tree clustered the 13 generated sequences of Ophionyssus sp. with the available sequences of Ophionyssus natricis, with high bootstrap values (99%). On the other hand, the two sequences of H. piger clustered together with sequences of H. triangulus and Ixodorhynchus leptodeirae (Fig 6). Representative sequences herein generated were deposited in GenBank (Table 3).
Fig 5. Morphological features of Ophionyssus sp.
a) dorsal view of female Ophionyssus sp; b) pore pairs on sternal shield (black arrows); c) dorsal shield divided in a large anterior podonotal shield (red arrow) and a small pygidial shield (black arrow); d) absence of mesonotal scutae in between the podonotal and pygidial shield. Scale bars: 200μm (a,c); 50μm (b, d).
Fig 6. Maximum-likelihood phylogenetic trees of 18S rRNA genes of Mesostigmata mites.
Bootstrap values (>40%) are shown near the nodes. Ixodes ricinus was used as outgroup. Scale bar indicates nucleotide substitution per site. Sequences of this study are in bold.
Furthermore, of the 38 fecal samples collected from different snake species (i.e., 16 H. viridiflavus, 10 E. quatuorlineata, 2 Natrix helvetica, and 10 Z. longissimus), 39.4% (15/38) scored positive for parasites, with 26.3% (10/38) positive at direct fecal examination, and 13.1% (5/38) through flotation test, being only two positive for both tests (i.e., two H. viridiflavus; Table 4). Briefly, ciliates (i.e., Nyctotherus sp.) were observed in two H. viridiflavus and Coccidia (Fig 7A) were detected in two H. viridiflavus. Helminth eggs (i.e., Capillarid, Strongyle, Oxyurid, Trematoda, Cestoda; Fig 7B and 7C) were detected in 2 H. viridiflavus and one E. quatuorlineata. One H. viridiflavus had a Trombiculidae mite larva (Table 4; Fig 7D).
Table 4. Parasitic forms observed through direct and/or flotation fecal tests with species of snake hosts.
| Snake ID | Species | Sex | Direct fecal test | Flotation fecal test |
|---|---|---|---|---|
| CS007 | Zamenis longissimus | M | Flagellates | - |
| CS011 | Hierophis viridiflavus | M | Coccidia Capillarid eggs Kalicephalus-type eggs Trematode egg Acanthocephalan egg |
Coccidia |
| CS021 | Zamenis longissimus | M | Flagellates | - |
| CS025 | Elaphe quatuorlineata | F | Flagellates | - |
| CS043 | Hierophis viridiflavus | M | Flagellates | - |
| CS047 | Hierophis viridiflavus | M | Flagellates | - |
| CS051 | Elaphe quatuorlineata | F | Flagellates | - |
| CS078 | Hierophis viridiflavus | M | - | Nyctotherus sp. |
| CS097 | Hierophis viridiflavus | F | Flagellates | - |
| CS098 | Hierophis viridiflavus | M | Capillarid eggs Trombiculidae mite larva |
Capillarid eggs Cestode eggs |
| CS100 | Hierophis viridiflavus | F | - | Sporulated Eimeria |
| CS102 | Elaphe quatuorlineata | M | - | Oxyurid eggs |
| CS104 | Hierophis viridiflavus | M | Nyctotherus sp. | - |
Fig 7. Parasitic forms found in coprological exams H. viridiflavus.
a) sporulated Eimeria sp.; b) Capillarid egg; c) Taeniid egg; d) pseudoparasite Trombiculidae mite larva from. Scale bars: 100μm (a); 200μm (b,d); 20μm (c).
Overall, 28.5% (32/112) of snakes were molecularly positive for at least one microorganism or parasite (Table 5). Three Ophionyssus sp. of three different snake hosts (i.e., E. quatuorlineata, H. viridiflavus, Z. longissimus) were positive for the endosymbiont Wolbachia bacteria, similar to that detected in Dermanyssus gallinae from Japan (LC710644–97.62%) and from Spinturnix mites collected in bats from Thailand (KP165044–98.88%). In addition, one Z. longissimus snake was positive for Rickettsia (gltA gene) in blood, with high nucleotide identity (100%) to Rickettsia aeschlimannii detected in human blood from Kenya (RQB050057). Phylogenetic inference clustered the sequence of Rickettsia sp. generated in this study with Rickettsia aeschlimannii available in Genbank (Fig 8). On the other hand, Leishmania tarentolae was detected in cloacal swabs from two snakes (i.e., E. quatuorlineata—Ct 31,8; H. viridiflavus—Ct 31,65), whereas two Leishmania sp. 18S rRNA sequences were retrieved from the blood of snakes (i.e., E. quatuorlineata, H. viridiflavus) different from those positive in cloacal swabs. Phylogenetic inference, despite being discretely informative, clustered both sequences from this study with those of Sauroleishmania species, as well as clustering all the Leishmania subclade ones apart (Fig 9).
Table 5. Molecular identification of vector-borne and fecal pathogens detected in snakes.
| Species of snake (Infected/total) |
Vector-Borne Pathogen | Fecal pathogen | |||||
|---|---|---|---|---|---|---|---|
| cPCR (16S rRNA) | cPCR (gltA) | cPCR (18S rRNA) | dqPCR (ITS1) | cPCR (cox1) | cPCR—nPCR (18S rRNA) | 5plex-qPCR | |
| Coronella girondica (0/1) | |||||||
| Elaphe quatuorlineata (11/66) | (1)** Wolbachia sp.—LC710644 (97.62%) | (1)* Leishmania tarentolae - KC205986 (100%) |
(1)*** Leishmania tarentolae (Ct 31,8) |
(1)*** Oswaldocruzia filiformis - OQ346357 (87.96%) |
(1)*** Alveolate FJ410512 (95.82%) (1)*** Citrobacter freundii CP026235 (99.23%) (1)*** Pseudomonas sp. CP043060 (93.75%) |
(1)***
Blastocystis (2)*** Cryptosporidium (1)*** Giardia assemblage B |
|
| Hierophis viridiflavus (12/28) | (1)** Wolbachia sp.—LC710644 (97.40%) | (1)* Leishmania tarentolae - KC205986 (100%) |
(1)*** Leishmania tarentolae (Ct 31,65) |
(1)*** Achromobacter xylosoxidans HE798385 (97.77%) (1)*** Coccidia sp. MH590231 (88.7%) (1)*** Heteromita globosa - LC764482 (99.58%) (1)**** Pseudomonas brenneri LT629800 (98.65%) |
(2)***
Blastocystis (2)*** Cryptosporidium (1)**** Entamoeba |
||
| Natrix helvetica (1/2) | (1)**** Proteus vulgaris—CP054157 (99.18%) |
||||||
| Zamenis longissimus (8/15) | (1)** Wolbachia sp.—KP165044 (98.88%) | (1)* Rickettsia aeschlimannii—RQB050057 (100%) | (1)*** Rhabdias kafunata—OP605735 (89.46%) |
(1)**** Pseudomonas brenneri LT629800 (97.32%) (1)*** Stenotrophomonas sp. CP109812 (99.77%) (1)*** Citrobacter braakii CP113163 (98.08%) |
(2)*** Giardia assemblage B |
||
| Total (32/112) | (3/112) | (1/112) | (2/112) | (2/112) | (3/112) | (10/112) | (11/112) |
*Blood;
**Mite;
***Swab;
****Feces
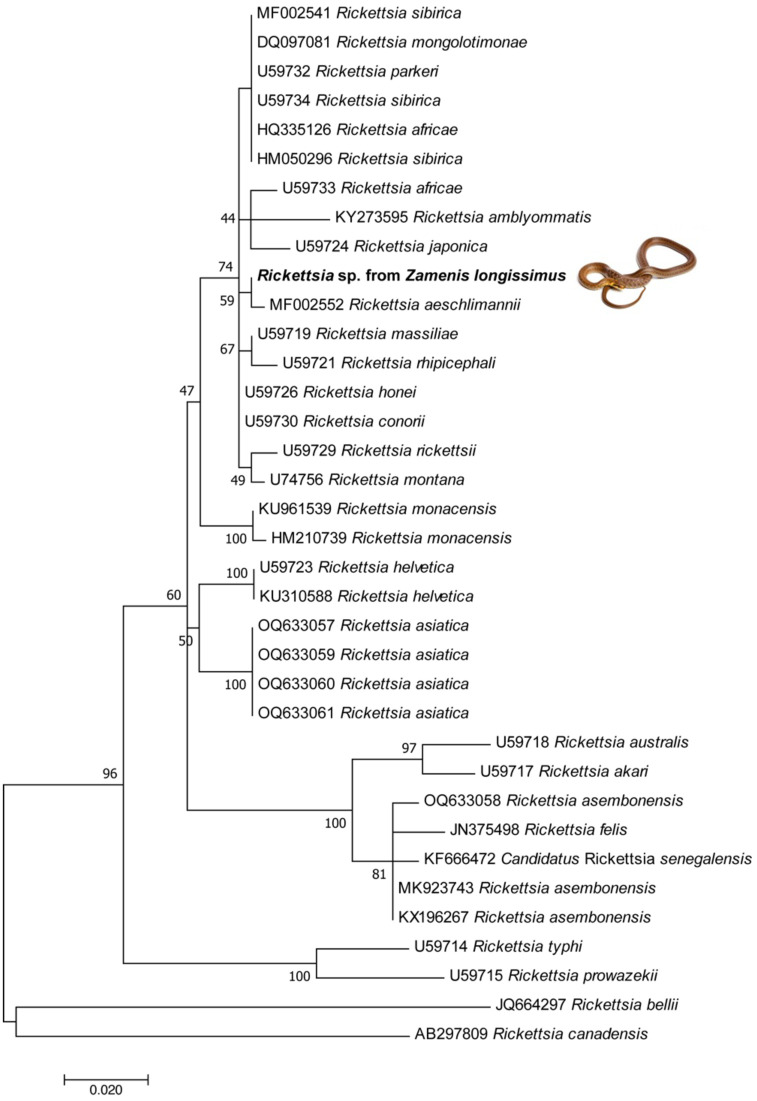
Fig 8. Maximum-likelihood phylogenetic trees of gltA genes of Rickettsia spp.
Bootstrap values (>40%) are shown near the nodes. Rickettsia belli, Rickettsia canadensis were used as outgroups. Scale bar indicates nucleotide substitution per site. Sequences of this study are in bold.
Fig 9. Maximum-likelihood phylogenetic trees of 18S rRNA genes of Leishmania spp.
Bootstrap values (>40%) are shown near the nodes. Trypanosoma brucei and Trypanosoma evansi were used as outgroups. Scale bar indicates nucleotide substitution per site. Sequences of this study are in bold.
Furthermore, various fecal pathogens of zoonotic potential were detected by the 5plex qPCR, with high CT values (Table 5). In brief, Blastocystis spp. and Cryptosporidium spp. were detected in two species of snakes (i.e., E. quatuorlineata, H. viridiflavus), whilst Entamoeba spp. were detected in one H. viridiflavus, and Giardia assemblage B in two other species of snakes (i.e., E. quatuorlineata, Z. longissimus). The nPCRs for Cryptosporidium spp. and Giardia spp. confirmed the two Cryptosporidum spp., but did not give specific bands for Giardia spp. However, several sequences of bacteria were obtained from feces and cloacal swabs by sequencing un-specific bands of the two nPCRs (i.e., Achromobacter xylosoxidans, Citrobacter braakii, Citrobacter freundii, Pseudomonas sp., Pseudomonas brenneri, Stenotrophomonas) and protozoa (i.e., Alveolate, Coccidia, Heteromita globosa), one of the positive snakes was also positive to protozoa in the coprological tests (i.e., CS011—H. viridiflavus; Table 3). Additionally, cox1 sequences from the cloacal swab were obtained for two nematodes, one from an E. quatuorlineata similar to Oswaldocruzia filiformis from Russia (OQ346357–87.96%), and one from Z. longissimus similar to Rhabdias kafunata from China (OP605735–89.46). Moreover, a sequence was retrieved from a N. natrix with high homology to Proteus vulgaris from wastewater facilities in Canada (CP054157–99.18%; Table 5). Representative sequences herein generated were deposited in GenBank (accession number OR753376 for gltA; OR755903 to OQ630505 for 16S rRNA; OQ632771 to OQ632773, OR771463 to OR771475 and OR771476-OR771477 for 18S rRNA; OQ672452, OR758867, OR761977, OR761978 and for OR763078 cox1).
Discussion
Ecto- and endoparasites were identified using a morpho-molecular approach from four of the five screened species of snakes collected for the “festa dei serpari” ritual. While most of the identified parasites are specific of reptiles and non-pathogenic (i.e., mites, helminths and protozoa), others transmitted by ticks (e.g., Rickettsia), as well as through fecal-oral transmission (i.e., Cryptosporidium spp., Giardia; Pseudomonas, Proteus vulgaris) have a zoonotic potential. The surveillance performed in this study with the local authorities, allowed to evaluate the parasitic fauna of free-ranging native snakes, which has been until now scarcely investigated or tackled in the Italian ophidic fauna.
The species composition of snake population (i.e., E. quatuorlineata, H. viridiflavus, Z. longissimus) is typical of the surroundings of Cocullo municipality, as already observed in previous screenings [27]. On the other hand, the absence of ticks in the snake population from this study may be due to the collection of the snakes during the early spring, where ophidians are less exposed to larvae and nymphs of I. ricinus [67]. Indeed, this tick species is most prevalent in woody areas of central Italy and was previously recorded in four-lined snakes from southern Italy [33]. In addition, immature stages of I. ricinus may prefer other reptile species that are more abundant and fossorial, such as Podarcis or Lacerta lizards [68,69]. The Ixodorhynchidae mites identified herein (H. piger) were described from an unknown snake collected in Florence (central Italy) [44]. Probably, H. viridiflavus is new host for this mite species, as well as Cocullo a new locality. The Ophionyssus species herein identified differs from the common snake mite, O. natricis, which is usually found in captive snakes around the world and may feed also on humans [70]. Moreover, the identified mite specimens morphologically differed from the 16 known species of Ophionyssus [42,43], with low homology in the cox1 nucleotide sequences compared to those of O. natricis deposited in Genbank. Given all the above, further studies are required to elucidate if the Ophionyssus species found is in fact a new species. Nonetheless, H. viridiflavus and E. quatuorlineata represent new hosts for Ophionyssus mites, as well as the Abruzzo region a new locality for Macronyssidae mites of snakes. Although all of the tested mites were negative for pathogens, the molecular detection of Wolbachia sp. in Ophionyssus sp. supports a previous finding in O. natricis from captive Boa constrictor [24], as well as in Ornithonyssus bursa [71]. The finding of Wolbachia in female mites, but not males, may be due to the role this endosymbiont bacterium displays in the reproduction of mites, through male-killing, feminization, and parthenogenesis [72,73].
Furthermore, the diversity and prevalence of the endoparasitic fauna of snakes recorded in this study (39.4%) were higher than that reported for captive snakes from Poland (i.e., 13.7%; [74]) and Italy (i.e., 10.5%; [23]), and lower from another survey conducted also in Italy (56.8%; [75]). Nevertheless, this study represents the first coprological survey of free-raging snakes in the Italian peninsula, without the need of euthanizing or working with recently dead/killed snakes [25,76,77]. Coccidia identified were morphologically similar with Eimeria [78], of mild pathogenicity, commonly observed in wild snakes. In addition, snakes were found infected with helminth eggs similar to Kalicephalus, Rhabdias and Strongyloides [79], as well as oxyurids and Capillaria eggs [80], with the latter being generally found in healthy animals. Overall, the findings of the above parasites may derive from the predation attitude of snakes [81] under different ecological contexts, therefore representing spurious parasites [82], rather than host specific ones. The possibility of having spurious parasites is higher in wild snakes than in those kept in captivity (e.g., 1.4%, 4/283; [82]), given that they actively feed on small preys. Given all the above, future studies are warranted to better distinguish real parasitic fauna of free-ranging snakes from spurious or free-living parasites. Importantly, molecular screening allowed to identify potentially zoonotic parasites and microorganisms, highlighting the need of an integrative approach using morpho-molecular techniques to assess wild animal populations under a One-Health perspective. For the zoonotic protozoa identified with the 5-plex qPCR, confirmatory sequences could only be retrieved for Cryptosporidium, but they were too short for reliable species identification. Zoonotic species, such as Cryptosporidium muris, Cryptosporidium parvum and Cryptosporidium tyzzeri were already identified in captive snakes [83]. Indeed, zoonotic Cryptosporidium species are spurious parasites in snakes, where resistant oocysts may contaminate the environment. Given that the diet of both species of snakes (i.e., E. quatuorlineata, H. viridiflavus) found positive for Cryptosporidium may include also rodents [84,85], further attempts should establish whether the captured snakes of Cocullo harbor zoonotic Cryptosporidium species or Cryptosporidium serpentis. The latter species may cause asymptomatic or chronic infections, being highly pathogenic, infectious, and irresponsive to therapeutic treatment [86]. The 5-plex qPCR may have low specificity and limited discriminatory power for the highly diverse protozoa species in snakes’ feces [62,87]. Moreover, mixed infections with related species of protozoa result in close melting curves hindering the detection of targeted species. Nonetheless, specific investigations on zoonotic pathogens in snakes are warranted considering that G. duodenalis is an important food and waterborne pathogen [88] as well as E. histolytica. The latter has never been reported in snakes [89], which typically host a highly pathogenic reptilian protozoan, Entamoeba invadens, causing necrotic enteritis and hepatitis [90], as well as the less pathogenic species Entamoeba ranarum [90]. Certainly, the integrative approach using morphological and molecular tools further permitted the identification of nematodes belonging to the genera Oswaldocruzia and Rhabdias from cloacal swabs, when fecal samples were not collected [91,92]. These genera of nematodes are potentially pathogenic to snakes and should be actively surveyed to assess their deleterious effects on the free-raging snake populations, also considering the zoonotic potential and emergence of snake-associated human parasites, such as Ophidascaris robertsi neural larva migrans [93].
While it was not possible to perform a definitive identification of zoonotic parasites, the sequences obtained from fecal and cloacal swabs allowed the detection of zoonotic bacteria (i.e., A. xylosoxidans, C. freundii, P. vulgaris, Pseudomonas), which were already detected in snakes, that may act both as reservoirs and spreaders [94,95]. As the bacteria above may be opportunistic pathogens of humans and multi-drug resistant strains have been identified [96], correct biosafety measures should be applied during and after the “festa dei serpari” event. This is mainly due to the fact that, when handled, free-ranging snakes defecate as a defense mechanism [97,98], therefore increasing the risks of contamination with pathogens such as Salmonella [29,99].
Regarding zoonotic vector-borne pathogens, the detection of R. aeschlimannii in Z. longissimus blood suggests exposure to tick bites, further corroborating the potential role of reptiles as reservoirs for Rickettisa spp. [100,101]. Again, molecular positivity for species of the spotted fever group (i.e., Rickettsia monacensis, Rickettsia helvetica) was reported in lizards from Italy [35] and snakes (i.e., Rickettsia asiatica) from Morocco [3]. As R. aeschlimannii and other rickettsiae (i.e., R. monacensis, Rickettsia massiliae) are considered as emerging human pathogens [102], further studies should be conducted to verify the occurrence of this species of Rickettsia, previously detected in Algeria from Hyalomma aegyptium ticks collected from tortoises [103].
The retrieval of the reptile-associated L. tarentolae from snakes in Italy represents new hosts (i.e., E. quatuorlineata, H. viridiflavus) and broadens its geographical distribution northwards, near the Lazio region where it was previously detected in human blood donors and sand flies [104]. Prior surveys from Italy yielded positive molecular results in species of lizards and geckos for L. infantum and L. tarentolae, in urban, peri-urban areas and dog shelters [105]. In addition, L. tarentolae was detected for the first time from cloacal swabs, however, attempts to isolate Leishmania spp. from snakes are warranted given that, thus far L. tarentolae has been isolated only from geckos of the Mediterranean basin [106,107]. To further address the epidemiological picture of Leishmania, entomological surveys are pivotal to describe the species composition of sand flies and address if there is also a sympatric occurrence of both L. tarentolae and L. infantum in the surrounding of the Cocullo municipality, given that L. tarentolae can potentially infect mammals (i.e., humans and dogs) [106].
Given all of the above, the population of snakes around Cocullo may be in part under a “positive” human-snake relationship, where traditional beliefs impact directly on reptile conservation. This, effect has been already studied in other cultural contexts for reptiles such as water monitor lizards in the surroundings of a small village in northern India [108]. As in Cocullo, villagers from the small town of Chak Manik have many beliefs (i.e., protecting the marshlands for their Gods and for the village to thrive), that indirectly have a positive effect on the vulnerable population of reptiles, being mutually beneficial for both the villagers and the reptile species [108]. Although highly subjective, overall health status and condition of the screened snakes was established as “apparently healthy”. Compared to the previous study, where 23 animals had some type of dermal abnormality [27], the three snakes that were herein reported as having skin lesions, were all probably signs of healed trauma or infection. However, given that it was not possible to discard fungal granuloma, annual screening of the collected snake population of Cocullo, assessing fungal, bacterial, and parasitic infections, should be encouraged to have a well-established and consistent surveillance program that will allow for rapid detection of harmful and zoonotic pathogens. Accordingly, results from this study will aid to create strategies to prevent zoonotic transmission of pathogens. Indeed, alongside the established snakes’ population monitoring efforts, pathogens surveillance using a multi-sectorial approach should be also performed annually to assess zoonotic pathogen emergence and their dynamics within the capture population of snakes, that are after released in the environment [109]. Data generated from the present study will be useful for local and national authorities to formulate proper prevention policies specific for the serpari as well as for tourist and pilgrims that participate in the event. For example, education and training of serpari on proper husbandry and protective measures when capturing and handling snakes will aid to reduce the risk of transmission of pathogens to this group of people that are at higher risk, as they are in contact with snakes for over a month [110]. Indeed, educating serpari on proper husbandry and procedures such as quarantine of snakes and the proper use of personal protective equipment (PPE), may reduce the risk of transmission from snake to snake, as well to humans [111]. On the other hand, coordinated policies with local authorities during the event are important to minimize the risk of oral-fecal transmission of pathogens from snakes to tourists. These prevention strategies should be focused on providing hand hygienization/disinfection places, as well as information on why and how to wash their hand after handling a wild animal [112]. Using a One-Health approach to monitor the snake population and reduce the risk of zoonotic transmission will thereby contribute to the conservation of the snakes and the perpetuation of the tradition.
Conclusion
Data presented here demonstrate that using an ethnoherpetoparasitological framework to assess the collected ophidian population prior to the annual celebration of the “festa dei serpari” ritual in Cocullo, is a useful approach that allowed for the assessment of the health of the handled snakes, as well as the risks of transmission of zoonotic pathogens present in wild populations. Although snake collected for the ritual harbored reptile-specific and non-pathogenic mites, helminths, and protozoa, the presence of zoonotic pathogens should not be disregarded. This is the case with vector-borne pathogens (e.g., Rickettsia, Leishmania), as well as opportunistic zoonotic pathogens (i.e., Cryptosporidium, Giardia, A. xylosoxidans, C. freundii, P. vulgaris, Pseudomonas) present in the feces of these scaley animals. Thus, snakes collected and showcased in the “festa dei serpari” are optimal sentinels and bioindicators of environmental and ophidian population health, as well as reservoirs of microorganisms that should be controlled through proper biosafety measures when handled by serpari to avoid the risk of zoonotic transmission. Effective public health policies within this unique epidemiological context are advocated, while promoting targeted conservation initiatives, education, and biosafety measures.
Supporting information
Every 1st of may for more than 500 hundred years, the statue of San Domenico is covered with four-lined snakes (Elaphe quatuorlineata), followed by a procession through the small town of Cocullo, Italy.
(MP4)
Acknowledgments
Authors thank Giada Annoscia (UniBa) for her technical work in molecular biology in the laboratories of de Department of Veterinary Medicine of the university of Bari. Authors would like to thank the Serpari, the local community, authorities, and the veterinarian Pasqualino Piro for supporting and rendering possible this study. Authors would like to also thank, Rifcon GmbH for sponsoring since 2016 the professional terraria to house captured reptiles. Authors would like to also acknowledge the Major and the local community of Cocullo for supporting and allowing this annual survey.
Data Availability
Representative sequences were deposited in GenBank (https://www.ncbi.nlm.nih.gov/genbank) (accession number OR753376 for gltA; OR755903 to OQ630505 for 16S rRNA; OQ632771 to OQ632773, OR771463 to OR771475 and OR771476-OR771477 for 18S rRNA; OQ672452, OR758867, OR761977, OR761978 and for OR763078 cox1).
Funding Statement
D.O. and J.A.M.R. were partially supported by EU funding within the NextGenerationEU-MUR PNRR Extended Partnership initiative on Emerging Infectious Diseases (Project no. PE00000007, INF-ACT). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Alves R, Silva VN, Trovão DM, Oliveira JV, Mourão JS, Dias TL, et al. Students’ attitudes toward and knowledge about snakes in the semiarid region of Northeastern Brazil. J EthnobiolEthnom. 2014;10:1–8. doi: 10.1186/1746-4269-10-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pandey DP, Subedi Pandey G, Devkota K, Goode M. Public perceptions of snakes and snakebite management: implications for conservation and human health in southern Nepal. J Ethnobiol Ethnom. 2016;12:1–25. doi: 10.1186/s13002-016-0092-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mendoza-Roldan JA, Noll Louzada-Flores V, Lekouch N, Khouchfi I, Annoscia G, Zatelli A, et al. Snakes and Souks: Zoonotic pathogens associated to reptiles in the Marrakech markets. Morocco. PLOS Negl Trop Dis. 2023;17:e0011431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alves RRDN, Filho GAP. Commercialization and use of snakes in North and Northeastern Brazil: implications for conservation and management. Biod Conservat. 2007;16:969–985. [Google Scholar]
- 5.Kusrini M, Palesa SP, Masy’ud B. Snake pet ownership in the city: A case study in Greater Jakarta. Indonesia. Biodiv J Biol Div. 2021;22:1790–1798. [Google Scholar]
- 6.Filippi E, Luiselli L. Status of the Italian snake fauna and assessment of conservation threats. Biol Conserv. 2000;93:219–225. [Google Scholar]
- 7.Reading CJ, Luiselli LM, Akani GC, Bonnet X, Amori G, Ballouard JM. Are snake populations in widespread decline? Biol Let. 2010;6:777–780. doi: 10.1098/rsbl.2010.0373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Filippi E. The effects of timbering on the community structure of snakes at a Mediterranean area of central Italy. Amph Rep. 2003;24:75–79. [Google Scholar]
- 9.Filippi E. Effects of restoration habitat on snake species of Dghoumes National Park (Tunisia). Biod J. 2019;10:213–220. [Google Scholar]
- 10.Lourenço-de-Moraes R, Lansac-Toha FM, Schwind LTF, Arrieira RL, Rosa RR, Terribile LC, et al. Climate change will decrease the range size of snake species under negligible protection in the Brazilian Atlantic Forest hotspot. Sci Rep. 2019;9:8523. doi: 10.1038/s41598-019-44732-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zipkin E, DiRenzo G, Ray J, Rossman S, Lips K. Tropical snake diversity collapses after widespread amphibian loss. Sci. 2020;367:814–816. doi: 10.1126/science.aay5733 [DOI] [PubMed] [Google Scholar]
- 12.Filippi E, Luiselli L. Negative effect of the wild boar (Sus scrofa) on the populations of snakes at a protected mountainous forest in central Italy. Ecol Medit. 2002;28:93–98. [Google Scholar]
- 13.Mendoza Roldan JA, Otranto D. Zoonotic parasites associated with predation by dogs and cats. Parasites Vectors. 2023;16:55. doi: 10.1186/s13071-023-05670-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sogliani D, Mori E, Lovari S, Lazzeri L, Longoni A, Tabarelli De Fatis K. Citizen science and diet analysis shed light on dog-wildlife interactions in Italy. Biod Conserv. 2023:1–19. [Google Scholar]
- 15.Lorch JM, Knowles S, Lankton JS, Michell K, Edwards JL, Kapfer JM. Snake fungal disease: an emerging threat to wild snakes. Phil Trans Royal Soc B Biol Sci. 2016;371:20150457. doi: 10.1098/rstb.2015.0457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mendoza-Roldan JA, Modry D, Otranto D. Zoonotic parasites of reptiles: a crawling threat. Trends Parasitol. 2020;36:677–687. doi: 10.1016/j.pt.2020.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alves RRN, Vieira KS, Santana GG, Vieira WLS, Almeida WO, Souto WMS, et al. A review on human attitudes towards reptiles in Brazil. Environ Monit Asses. 2012;184:6877–6901. doi: 10.1007/s10661-011-2465-0 [DOI] [PubMed] [Google Scholar]
- 18.Filippi E, Luiselli L. Delayed reproduction in snakes subjected to human traditional rituals in central Italy. Vie et Milieu/Life Envir. 2003;53:111–118. [Google Scholar]
- 19.Achille G. Snakes of Italy: Herpetological Treatise on the Biology and Iconography of Italian Ophidians. 1st ed. Springer: Cham; 2015. [Google Scholar]
- 20.Pellegrini M, Di Francesco N, Di Tizio L, Di Toro F, D’Amico M, Cameli A, et al. Action Plan per la conservazion di Elaphe quatuorlineata (Lacépède, 1789) in Abruzzo. In: Atti XI Congresso Nazionale Societas Herpetologica Italica. 2017, pp. 273–279. Menegon M, Rodriguez-Prieto A, Deflorian AM, Eds. Edizioni Ianieri. Pescara. [Google Scholar]
- 21.Ten SF. Years of Geo-Archeo-Mythological Studies in the Abruzzo Region—Central Italy: An Updated Review. J Archaeol Anthropol. 2020;2:1–13. [Google Scholar]
- 22.Rinaldi L, Capasso M, Mihalca AD, Cirillo R, Cringoli G, Cacciò S. Prevalence and molecular identification of Cryptosporidium isolates from pet lizards and snakes in Italy. Paras. 2012;19:437–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cervone M, Fichi G, Lami A, Lanza A, Damiani GM, Perrucci S. Internal and external parasitic infections of pet reptiles in Italy. J Herpetol Med Surg. 2016;26:122–130. [Google Scholar]
- 24.Manoj RRS, Latrofa MS, Mendoza-Roldan JA, Otranto D. Molecular detection of Wolbachia endosymbiont in reptiles and their ectoparasites. Parasitol Res. 2021;120:3255–3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Santoro M, Aznar FJ, Mattiucci S, Kinsella JM, Pellegrino F, Cipriani P, et al. Parasite assemblages in the Western whip snake Hierophis viridiflavus carbonarius (Colubridae) from southern Italy. J Helminthol. 2013;87:277–285. [DOI] [PubMed] [Google Scholar]
- 26.Santoro M, Tkach VV, Mattiucci S, Kinsella JM, Nascetti G. Renifer aniarum (Digenea: Reniferidae), an introduced North American parasite in grass snakes Natrix natrix in Calabria, southern Italy. Dis Aquat Organ. 2011;95:233–240. [DOI] [PubMed] [Google Scholar]
- 27.Marini D, Filippi E, Montinaro G, Origgi FC. Screening of Ophidiomyces ophidiicola in the free-ranging snake community annually harvested for the popular ritual of San Domenico e dei Serpari (Cocullo, AQ, Italy). Acta Herpetol. 2023;18:45–52. [Google Scholar]
- 28.Marini D, Di Nicola MR, Crocchianti V, Notomista T, Iversen D, Coppari L. Pilot survey reveals ophidiomycosis in dice snakes Natrix tessellata from Lake Garda. Italy. Vet Res Commun. 2023;47:1707–1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Corrente M, Totaro M, Martella V, Campolo M, Lorusso A, Ricci M. Reptile-associated salmonellosis in man. Italy. Emerg Infec Dis. 2006;12:358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Corrente M, Sangiorgio G, Grandolfo E, Bodnar L, Catella C, Trotta A. Risk for zoonotic Salmonella transmission from pet reptiles: A survey on knowledge, attitudes and practices of reptile-owners related to reptile husbandry. Prev Vet Med. 2017;146:73–78. [DOI] [PubMed] [Google Scholar]
- 31.Pantchev N, Tappe D. Pentastomiasis and other parasitic zoonoses from reptiles and amphibians. Berl. Munch. Tierarztl. Wochenschr. 2011;124:528–535. [PubMed] [Google Scholar]
- 32.Mendoza-Roldan JA, Otranto D. Reptile vector-borne diseases of zoonotic concern. International J Parasitol Paras Wildl. 2021;15:132–142. doi: 10.1016/j.ijppaw.2021.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mendoza-Roldan JA, Colella V, Lia RP, Nguyen VL, Barros-Battesti DM, Iatta R, et al. Borrelia burgdorferi (sensu lato) in ectoparasites and reptiles in southern Italy. Parasites Vectors. 2019;12:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mendoza-Roldan JA, Colella V. Ixodes ricinus infesting snakes: insights on a new tick-host association in a Borrelia burgdorferi sensu lato endemic area. Acta Trop. 2019;193:35–37. [DOI] [PubMed] [Google Scholar]
- 35.Mendoza-Roldan JA, Manoj R, Latrofa MS, Iatta R, Annoscia G, Lovreglio P, et al. Role of reptiles and associated arthropods in the epidemiology of rickettsioses: A one health paradigm. PLoS Negl Trop Dis. 2021;15:e0009090. doi: 10.1371/journal.pntd.0009090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuchta R, Kołodziej-Sobocińska M, Brabec J, Młocicki D, Sałamatin R, Scholz T. Sparganosis (Spirometra) in Europe in the molecular era. Clin Infect Dis. 2021;72:882–890. [DOI] [PubMed] [Google Scholar]
- 37.Filippi E D Alterio GL, Brozzi AB, Micci M, Politi P, Mantero D. Note on the intestinal bacterial populations of free-living snakes in Italy. Herpetol Notes. 2010;3:263–265. [Google Scholar]
- 38.Telford SR. Hemoparasites of the Reptilia: Color atlas and text (No. 21482). 1st ed. CRC Press; Taylor & Francis; 2009. [Google Scholar]
- 39.Skipper R, DeStephano DB. A rapid stain for Campylobacter pylori in gastrointestinal tissue sections using Diff-Quik. J Histotechnol. 1989;12:303–304. [Google Scholar]
- 40.Krantz GW. A manual of acarology. 2nd ed. Oregon: st Univ Bookstores; 1978. [Google Scholar]
- 41.Fain A. Les acariens mesostigmatiques ectoparasites des serpents. Bull Inst Roy Sci Natur Belg. 1962;38:1–149. [Google Scholar]
- 42.Moraza ML, Irwin NR, Godinho R, Baird SJ, Bellocq JG. A new species of Ophionyssus Mégnin (Acari: Mesostigmata: Macronyssidae) parasitic on Lacerta schreiberi Bedriaga (Reptilia: Lacertidae) from the Iberian Peninsula, and a world key to species. Zootaxa. 2009;2007:58–68. [Google Scholar]
- 43.Gomes-Almeida BK, Pepato AR. A new genus and new species of macronyssid mite (Mesostigmata: Gamasina: Macronyssidae) from Brazilian caves including molecular data and key for genera occurring in Brazil. Acarol. 2021;61:501–526. [Google Scholar]
- 44.Fain A, Bannert B. Two new species of Ophionyssus Mégnin (Acari: Macronyssidae) parasitic on lizards of the genus Gallotia Boulenger (Reptilia: Lacertidae) from the Canary Islands. Inter J Acarol. 2000;26:41–50. [Google Scholar]
- 45.Rózsa L, Reiczigel J, Majoros G. Quantifying parasites in samples of hosts. J Parasitol. 2000;86:228–232. doi: 10.1645/0022-3395(2000)086[0228:QPISOH]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- 46.Chomczynski P. A reagent for the single-step simultaneous isolation of RNA, DNA and proteins from cell and tissue samples. Biotechn. 1993;15:532–534. [PubMed] [Google Scholar]
- 47.Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek AR. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol. 1994;3:294–299. [PubMed] [Google Scholar]
- 48.Otto JC, Wilson K. Assessment of the usefulness of ribosomal 18S and mitochondrial COI sequences in Prostigmata phylogeny. In Acarology: proceedings of the 10th international congress 2001, Vol. 100, No. 9. Melbourne: Csiro Publishing. [Google Scholar]
- 49.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinform. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martin AR, Brown GK, Dunstan RH, Roberts TK. Anaplasma platys: an improved PCR for its detection in dogs. Exp Parasitol. 2005;109:176–180. [DOI] [PubMed] [Google Scholar]
- 51.Wójcik-Fatla A, Szymanska J, Wdowiak L, Buczek A, Dutkiewicz J. Coincidence of three pathogens [Borrelia burgdorferi sensu lato, Anaplasma phagocytophilum and Babesia microti] in Ixodes ricinus ticks in the Lublin macroregion. Annals Agric Environ Med. 2009;16:151–158. [PubMed] [Google Scholar]
- 52.Labruna MB, Whitworth T, Horta MC, Bouyer DH, McBride JW, Pinter A, et al. Rickettsia species infecting Amblyomma cooperi ticks from an area in the state of Sao Paulo, Brazil, where Brazilian spotted fever is endemic. J Clin Microbiol. 2004;42:90–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Regnery RL, Spruill CL, Plikaytis B. Genotypic identification of rickettsiae and estimation of intraspecies sequence divergence for portions of two rickettsial genes. J Bacteriol. 1991;173:1576–1589. doi: 10.1128/jb.173.5.1576-1589.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Berri M, Laroucau K, Rodolakis A. The detection of Coxiella burnetii from ovine genital swabs, milk and fecal samples by the use of a single touchdown polymerase chain reaction. Vet Microbiol. 2000;72:285–293. [DOI] [PubMed] [Google Scholar]
- 55.Gubbels JM, De Vos AP, Van der Weide M, Viseras J, Schouls LM, De Vries E, et al. Simultaneous detection of bovine Theileria and Babesia species by reverse line blot hybridization. J Clin Microbiol. 1999;37:1782–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bowles J, Blair D, McManus DP. Genetic variants within the genus Echinococcus identified by mitochondrial DNA sequencing. Mol Bioch Parasitol. 1992;54:165–173. [DOI] [PubMed] [Google Scholar]
- 57.El Tai NO, El Fari M, Mauricio I, Miles MA, Oskam L, El Safi SH, et al. Leishmania donovani: intraspecific polymorphisms of Sudanese isolates revealed by PCR-based analyses and DNA sequencing. Exp Parasitol. 2001;97:35–44. [DOI] [PubMed] [Google Scholar]
- 58.Sadlova J, Bacikova D, Becvar T, Vojtkova B, England M, Shaw J, et al. Porcisia transmission by prediuresis of sand flies. Front Cell Inf Microbiol. 2022;12:981071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Francino O, Altet L, Sánchez-Robert E, Rodriguez A, Solano-Gallego L, Alberola J, et al. Advantages of real-time PCR assay for diagnosis and monitoring of canine leishmaniosis. Vet Parasitol. 2006;137:214–221. doi: 10.1016/j.vetpar.2006.01.011 [DOI] [PubMed] [Google Scholar]
- 60.Latrofa M, Mendoza-Roldan JA, Dantas-Torres F, Otranto D. A duplex real-time PCR assay for the detection and differentiation of Leishmania infantum and Leishmania tarentolae in vectors and potential reservoir hosts. Entomol Gen. 2021;41:543–551. [Google Scholar]
- 61.Nazeer JT, El Sayed KK, von Thien H, El-Sibaei MM, Abdel-Hamid MY, Tawfik RA. Use of multiplex real-time PCR for detection of common diarrhea causing protozoan parasites in Egypt. Parasitol Res. 2013;112:595. doi: 10.1007/s00436-012-3171-8 [DOI] [PubMed] [Google Scholar]
- 62.Lamien-Meda A, Schneider R, Walochnik J, Auer H, Wiedermann U, Leitsch D. A novel 5-Plex qPCR-HRM assay detecting human diarrheal parasites. Gut Pathog. 2020;12:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hopkins RM, Meloni BP, Groth DM, Wetherall JD, Reynoldson JA, Thompson RA. Ribosomal RNA sequencing reveals differences between the genotypes of Giardia isolates recovered from humans and dogs living in the same locality. J Parasitol. 1997;44–51. [PubMed] [Google Scholar]
- 64.Sulaiman IM, Fayer R, Bern C, Gilman RH, Trout JM, Schantz PM, et al. Triosephosphate isomerase gene characterization and potential zoonotic transmission of Giardia duodenalis. Emerg Infect Dis. 2003;9:1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ryan U, Xiao L, Read C, Zhou L, Lal AA, Pavlasek I. Identification of novel Cryptosporidium genotypes from the Czech Republic. Applied Environ Microbiol. 2003;69:4302–4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dantas-Torres F, Otranto D. Seasonal dynamics of Ixodes ricinus on ground level and higher vegetation in a preserved wooded area in southern Europe. Vet Parasitol. 2013;192:253–258. [DOI] [PubMed] [Google Scholar]
- 68.Amore G, Tomassone L, Grego E, Ragagli C, Bertolotti L, Nebbia P, et al. Borrelia lusitaniae in immature Ixodes ricinus (Acari: Ixodidae) feeding on common wall lizards in Tuscany, central Italy. J Med Entomol. 2007;44:303–307. [DOI] [PubMed] [Google Scholar]
- 69.Tomassone L, Ceballos LA, Ragagli C, Martello E, De Sousa R, Stella MC, et al. Importance of common wall lizards in the transmission dynamics of tick-borne pathogens in the northern Apennine Mountains. Italy. Microb Ecol. 2017;74:961–968. [DOI] [PubMed] [Google Scholar]
- 70.Mendoza-Roldan J, Napoli E, Perles L, Marino M, Spadola F, Berny P, et al. Afoxolaner (NexGard) in pet snakes for the treatment and control of Ophionyssus natricis (Mesostigmata: Macronyssidae). Parasites Vectors. 2023;16:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lareschi M, Cicuttin GL, Salvo MND, Ibañez L, Montalti D. The tropical fowl mite Ornithonyssus bursa (Acari: Mesostigmata: Macronyssidae) parasitizing the European starling Sturnus vulgaris (Aves: Passeriformes: Sturnidae), an invasive bird in central Argentina. An approach to the bacterial fauna of this mite. Rev Mex Biod. 2017;88:454–458. [Google Scholar]
- 72.Reeves WK, Dowling AP, Dasch GA. Rickettsial agents from parasitic dermanyssoidea (Acari: Mesostigmata). Experim Applied Acarol. 2006;38:181–188. doi: 10.1007/s10493-006-0007-1 [DOI] [PubMed] [Google Scholar]
- 73.Chaisiri K, McGarry JW, Morand S, Makepeace BL. Symbiosis in an overlooked microcosm: a systematic review of the bacterial flora of mites. Parasitol. 2015;142:1152–1162. [DOI] [PubMed] [Google Scholar]
- 74.Okulewicz A, Kaźmierczak M, Zdrzalik K. Endoparasites of exotic snakes (Ophidia). Helmint. 2014;51:31–36. [Google Scholar]
- 75.Papini R, Manetti C, Mancianti F. Coprological survey in pet reptiles in Italy. Vet Rec. 2011;169:207–207. doi: 10.1136/vr.d4398 [DOI] [PubMed] [Google Scholar]
- 76.Yildirimhan HS, Bursey CR, Goldberg SR. Helminth parasites of the grass snake, Natrix natrix, and the dice snake, Natrix tessellata (Serpentes: Colubridae), from Turkey. Comp Parasitol. 2007;74:343–354. [Google Scholar]
- 77.Carbonara M, Mendoza-Roldan JA, Lia RP, Annoscia G, Iatta R, Varcasia A, et al. Squamata reptiles as a potential source of helminth infections when preyed on by companion animals. Parasites Vectors. 2023;16:233. doi: 10.1186/s13071-023-05852-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Šlapeta JR, Modrý D, Ashe J, Koudela B. Description of Eimeria arabukosokokensis sp. n. (Apicomplexa: Eimeriidae) from Telescopus semiannulatus (Serpentes: Colubridae) with notes on eimerian coccidia from snakes of Eastern Kenya. Folia Parasitol. 2003;50:23–30. [DOI] [PubMed] [Google Scholar]
- 79.Hallinger MJ, Taubert A, Hermosilla C. Occurrence of Kalicephalus, Strongyloides, and Rhabdias nematodes as most common gastrointestinal parasites in captive snakes of German households and zoological gardens. Parasitol Res. 2020;119:947–956. [DOI] [PubMed] [Google Scholar]
- 80.Rataj AV, Lindtner-Knific R, Vlahović K, Mavri U, Dovč A. Parasites in pet reptiles. Acta Vet Scandin. 2011;53:1–21. doi: 10.1186/1751-0147-53-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wolf D, Vrhovec MG, Failing K, Rossier C, Hermosilla C, Pantchev N. Diagnosis of gastrointestinal parasites in reptiles: comparison of two coprological methods. Acta Vet Scand. 2014;56:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ellerd R, Saleh MN, Luksovsky JL, Verocai GG. Endoparasites of pet reptiles and amphibians from exotic pet shows in Texas, United States. Vet Parasitol Reg Stud Reports. 2022;27:100671. doi: 10.1016/j.vprsr.2021.100671 [DOI] [PubMed] [Google Scholar]
- 83.Díaz P, Rota S, Marchesi B, López C, Panadero R, Fernández G, et al. Cryptosporidium in pet snakes from Italy: molecular characterization and zoonotic implications. Vet Parasitol. 2013;197:68–73. [DOI] [PubMed] [Google Scholar]
- 84.Capula M, Filippi E. Elaphe quatuorlineata. In: Fauna d’Italia Vol. XLV Reptilia. Corti C, Capula M, Luiselli L, Mazzetti E, Sindaco R, Eds Calderini, Milano. 2011,pp- 489–493.
- 85.Vanni S, Zuffi MA. Hierophis viridiflavus. In: Fauna d’Italia Vol. XLV Reptilia. Corti C, Capula M, Luiselli L, Mazzetti E, Sindaco R, Eds Calderini, Milano, 2011, pp. 489–493
- 86.Bogan JE Jr, Hoffman M, Dickerson F, Mitchell MA, Garner MM, Childress A, et al. Evaluation of paromomycin treatment for Cryptosporidium serpentis infection in eastern indigo snakes (Drymarchon couperi). J Herpetol Med Surg. 2021;31:307–314. [Google Scholar]
- 87.Robertson LJ, Clark CG, Debenham JJ, Dubey JP, Kváč M, Li J, et al. Are molecular tools clarifying or confusing our understanding of the public health threat from zoonotic enteric protozoa in wildlife? Int J Parasitol Paras Wildl. 2019;9:323–341. doi: 10.1016/j.ijppaw.2019.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Júlio C, Sá C, Ferreira I, Martins S, Oleastro M, Ângelo H, et al. Waterborne transmission of Giardia and Cryptosporidium at river beaches in Southern Europe (Portugal). J Water Heal. 2012;10:484–496. doi: 10.2166/wh.2012.030 [DOI] [PubMed] [Google Scholar]
- 89.Brewer LA, Denver MC, Whitney M, Eichinger DJ. Analysis of commercial Entamoeba histolytica ELISA kits for the detection of Entamoeba invadens in reptiles. J Zoo Wildl Med. 2008;39:493–495. [DOI] [PubMed] [Google Scholar]
- 90.McFarland A, Conley KJ, Seimon TA, Sykes JM IV. A retrospective analysis of amoebiasis in reptiles in a zoological institution. J Zoo Wildl Med. 2021;52:232–240. doi: 10.1638/2020-0148 [DOI] [PubMed] [Google Scholar]
- 91.Kirillova NY, Kirillov AA, Shchenkov SV, Chikhlyaev IV. Oswaldocruzia filiformis sensu lato (Nematoda: Molineidae) from amphibians and reptiles in European Russia: Morphological and molecular data. Nat Conserv Res. 2020;5:41–56. [Google Scholar]
- 92.Mihalca AD, Micluş V, Lefkaditis M. Pulmonary lesions caused by the nematode Rhabdias fuscovenosa in a grass snake. Natrix natrix. J Wildl Dis. 2010;46:678–681. [DOI] [PubMed] [Google Scholar]
- 93.Hossain ME, Kennedy KJ, Wilson HL, Spratt D, Koehler A, Gasser RB, et al. Human Neural Larva Migrans Caused by Ophidascaris robertsi Ascarid. Emerg Infect Dis. 2023;29:1900–1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Schmidt V, Mock R, Burgkhardt E, Junghanns A, Ortlieb F, Szabo I, et al. Cloacal aerobic bacterial flora and absence of viruses in free-living slow worms (Anguis fragilis), grass snakes (Natrix natrix) and European Adders (Vipera berus) from Germany. Eco Health. 2014;11:571–580. [DOI] [PubMed] [Google Scholar]
- 95.Padhi L, Panda SK, Mohapatra PP, Sahoo G. Antibiotic susceptibility of cultivable aerobic microbiota from the oral cavity of Echis carinatus from Odisha (India). Microb Pathog. 2020;143:104–121. doi: 10.1016/j.micpath.2020.104121 [DOI] [PubMed] [Google Scholar]
- 96.Morrison BJ, Rubin JE. Detection of multidrug-resistant Gram-negative bacteria from imported reptile and amphibian meats. J Applied Microb. 2020;129:1053–1061. doi: 10.1111/jam.14658 [DOI] [PubMed] [Google Scholar]
- 97.Gehlbach FR. Death-feigning and erratic behavior in leptotyphlopid, colubrid, and elapid snakes. Herpetol. 1970;26:24–34. [Google Scholar]
- 98.Claunch NM, Lind C, Lutterschmidt DI, Moore IT, Neuman-Lee L, Stahlschmidt Z, et al. Stress Ecology in Snakes. In: editor Penning D. Snakes. Nova Science Publishers, Inc. 2023; pp. 415–478. [Google Scholar]
- 99.Bertrand S, Rimhanen-Finne R, Weill FX, Rabsch W, Thornton L, Perevoščikovs J, et al. Salmonella infections associated with reptiles: the current situation in Europe. Eurosurveillance. 2008;13:18902. [PubMed] [Google Scholar]
- 100.Sánchez-Montes S, Isaak-Delgado AB, Guzmán-Cornejo C, Rendón-Franco E, Muñoz-García CI, Bermúdez S, et al. Rickettsia species in ticks that parasitize amphibians and reptiles: Novel report from Mexico and review of the worldwide record. Ticks Tick-borne Dis. 2019;10:987–994. [DOI] [PubMed] [Google Scholar]
- 101.Mendoza-Roldan JA, Ribeiro SR, Castilho-Onofrio V, Marcili A, Simonato BB, Latrofa MS, et al. Molecular detection of vector-borne agents in ectoparasites and reptiles from Brazil. Ticks Tick-borne Dis. 2021;12:101585. doi: 10.1016/j.ttbdis.2020.101585 [DOI] [PubMed] [Google Scholar]
- 102.Kernif T, Leulmi H, Raoult D, Parola P. Emerging tick-borne bacterial pathogens. Microbiol. Spectrum. 2016;4:1110–1128. doi: 10.1128/microbiolspec.EI10-0012-2016 [DOI] [PubMed] [Google Scholar]
- 103.Bitam I, Kernif T, Harrat Z, Parola P, Raoult D. First detection of Rickettsia aeschlimannii in Hyalomma aegyptium from Algeria. Clin Microbiol Inf. 2009;15:253–254. [DOI] [PubMed] [Google Scholar]
- 104.Pombi M, Giacomi A, Barlozzari G, Mendoza-Roldan J, Macrì G, Otranto D, et al. Molecular detection of Leishmania (Sauroleishmania) tarentolae in human blood and Leishmania (Leishmania) infantum in Sergentomyia minuta: unexpected host-parasite contacts. Med Vet Entomol. 2020;34:470–475. [DOI] [PubMed] [Google Scholar]
- 105.Mendoza-Roldan JA, Latrofa MS, Tarallo VD, Manoj RR, Bezerra-Santos MA, Annoscia G, et al. Leishmania spp. in Squamata reptiles from the Mediterranean basin. Transb Emerg Dis. 2022;69:2856–2866. [DOI] [PubMed] [Google Scholar]
- 106.Mendoza-Roldan JA, Votýpka J, Bandi C, Epis S, Modrý D, Tichá L, et al. Leishmania tarentolae: A new frontier in the epidemiology and control of the leishmaniases. Transb Emerg Dis. 2022;69:e1326–e1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mendoza-Roldan JA, Zatelli A, Latrofa MS, Iatta R, Bezerra-Santos MA, Annoscia G, et al. Leishmania (Sauroleishmania) tarentolae isolation and sympatric occurrence with Leishmania (Leishmania) infantum in geckoes, dogs and sand flies. PLoS Negl Trop Dis. 2022;16:e0010650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bhattacharya S, Koch A. Effects of traditional beliefs leading to conservation of Water Monitor Lizards (Varanus Salvator) and threatened marshlands in West Bengal. India. Herpetol Conservat Biol. 2018;3:408–414. [Google Scholar]
- 109.Rahman MT, Sobur MA, Islam MS, Ievy S, Hossain MJ, El Zowalaty ME, et al. Zoonotic Diseases: Etiology, Impact, and Control. Microorganisms. 2020;8:1405. doi: 10.3390/microorganisms8091405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bosch SA, Musgrave K, Wong D. Zoonotic disease risk and prevention practices among biologists and other wildlife workers-results from a national survey, US National Park Service, 2009. J Wildl Dis. 2013;49:475–585. doi: 10.7589/2012-06-173 [DOI] [PubMed] [Google Scholar]
- 111.Yeo LLizo S. Prevention is Better than Cure: An Overview of Disease Outbreak Management in Herptiles. Vet Clin North Am Exot Anim Pract. 2021;24:647–659. doi: 10.1016/j.cvex.2021.05.002 [DOI] [PubMed] [Google Scholar]
- 112.Li H, Chen Y, Machalaba CC, Tang H, Chmura AA, Fielder MD, et al. Wild animal and zoonotic disease risk management and regulation in China: Examining gaps and One Health opportunities in scope, mandates, and monitoring systems. One Health. 2021;13:100301. doi: 10.1016/j.onehlt.2021.100301 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Every 1st of may for more than 500 hundred years, the statue of San Domenico is covered with four-lined snakes (Elaphe quatuorlineata), followed by a procession through the small town of Cocullo, Italy.
(MP4)
Data Availability Statement
Representative sequences were deposited in GenBank (https://www.ncbi.nlm.nih.gov/genbank) (accession number OR753376 for gltA; OR755903 to OQ630505 for 16S rRNA; OQ632771 to OQ632773, OR771463 to OR771475 and OR771476-OR771477 for 18S rRNA; OQ672452, OR758867, OR761977, OR761978 and for OR763078 cox1).