Abstract
Introduction
Coronavirus disease 2019 caused by coronavirus-2 (SARS-CoV-2) has emerged as an aggressive viral pandemic. Health care providers confront a challenging task for rapid development of effective strategies to combat this and its long-term after effects. Virus entry into host cells involves interaction between receptor-binding domain (RBD) of spike (S) protein S1 subunit with angiotensin converting enzyme present on host cells. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) is a moonlighting enzyme involved in cellular glycolytic energy metabolism and micronutrient homeostasis. It is deployed in various cellular compartments and the extra cellular milieu. Though it is known to moonlight as a component of mammalian innate immune defense machinery, till date its role in viral restriction remains unknown.
Method
Recombinant S protein, the RBD, and human GAPDH protein were used for solid phase binding assays and biolayer interferometry. Pseudovirus particles expressing four different strain variants of S protein all harboring ZsGreen gene as marker of infection were used for flow cytometry-based infectivity assays.
Results
Pseudovirus entry into target cells in culture was significantly inhibited by addition of human GAPDH into the extracellular medium. Binding assays demonstrated that human GAPDH binds to S protein and RBD of SARS-CoV-2 with nanomolar affinity.
Conclusions
Our investigations suggest that this interaction of GAPDH interferes in the viral docking with hACE2 receptors, thereby affecting viral ingress into mammalian cells.
Keywords: Severe acute respiratory syndrome coronavirus-2, GAPDH, ACE2, Inhibition, Innate immune defense
Introduction
COVID-19 (coronavirus disease 2019) is caused by the coronavirus SARS-CoV-2 (severe acute respiratory syndrome coronavirus-2), which spread rapidly, infecting over 550 million individuals across 220 countries (WHO Coronavirus [COVID-19] Dashboard, https://covid19.who.int/). The primary mechanism of the viral transmission among humans is usually via exposure to contaminated respiratory droplets [1]. Viral transmission occurs through fomites, larger respiratory droplets (>5 μm), aerosolized droplets (<5 μm), and droplet transmission [2, 3]. The symptoms of SARS-CoV-2 infection are milder than severe acute respiratory syndrome and Middle East respiratory syndrome; however, the human-to-human transmission is much faster. The mortality rate of SARS-CoV-2 is 3.4%, while that of SARS-CoV is 9.6% and Middle East respiratory syndrome is 35% [4]. Although the overall mortality rate is relatively low, people who have chronic diseases like diabetes mellitus, hypertension, and asthma tend to present with more severe disease outcomes [5, 6]. Apart from vaccine development, researchers have focused on the development of protective and therapeutic options. Some of the options explored are SARS-CoV-2 entry inhibition into the host cells, inhibition of virus replication within host cell, inflammatory response suppression, and antibody cocktails-based inhibition [7]. SARS-CoV-2 entry into host cells requires the cleavage of S protein at S1/S2 and S2′ location by host cellular proteases. This allows fusion of viral and cellular membranes driven by the S2 subunit. Spike protein binds to angiotensin-converting enzyme 2 (ACE2) [8] and utilizes host serine protease TMPRSS2 for S protein cleavage and providing further access to host cellular machinery [9]. SARS-CoV and SARS-CoV-2 protein sequences have 76% amino acid identity. Human ACE2 (hACE2) degrades Ang II to angiotensin 1–7 (Ang 1–7), which has actions opposite to that of Ang II. Enhanced Ang II levels upregulate ACE2 enzymatic activity. In ACE2-deficient mice, Ang II levels were double to that of wild-type mice, while Ang 1–7 levels were not detectable [10, 11]. Apart from functioning as a portal of entry into cells for SARS-CoV-2, host ACE2 receptors are exploited by corona viruses HCoV-NL63 and SARS-CoV [12]. SARS-CoV-2 is known to employ furin proteases for priming of S protein [13]. Recently, it has also been reported that neuropilin-1 (NRP1), which binds furin-cleaved substrates, enhances SARS-CoV-2 infectivity [14]. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH, EC 1.2.1.12) has been long recognized as a multifunctional glycolytic enzyme, whose classical involvement in energy production and glucose metabolism has been well defined [15]. Ubiquitously expressed in all prokaryotes and eukaryotes, it is a homotetrameric molecule assembled from 334 amino acids with a highly conserved sequence, each monomeric unit having a molecular weight of 36 kDa [16]. Expression mapping studies have revealed a significant disparity in GAPDH mRNA with abundant expression in muscle and heart tissues and a relatively lower expression in liver, brain, and kidney cells [17]. GAPDH is a moonlighting protein with numerous functions apart from glycolysis [18–20]. Investigations have revealed that GAPDH is also involved in a wide range of diverse activities such as membrane transport, cytoskeleton assembly, nuclear material export, protein phosphotransferase activity, membrane fusion, DNA replication and repair [21–23]. Apart from its normal physiological functions, several groups have identified the role of GAPDH in various pathological conditions such as infection biology, apoptosis, autophagy, efferocytosis, and neuronal degenerative disorders [16, 21, 24–28]. It is also identified to play a role in the mammalian innate immune defense machinery [29].
Multi-functionality of GAPDH is not limited to eukaryotes; it also exhibits moonlighting in prokaryotes. Investigators have reported that apart from a cytosolic localization, GAPDH exists on the surface of eukaryotic as well as prokaryotic cells and plays a vital role in microbial pathogenesis [30]. Bacterial cell surface GAPDH has the capability to interact with several mammalian proteins such as lysosome, laminin, fibronectin, plasminogen, plasmin, actin and mediates several processes for successful survival within the host tissue [31–35]. Apart from these diverse multiple functions, it also plays a crucial role in viral infection. GAPDH is known to interact with positive- and negative-strand 3′-end RNAs of bamboo mosaic virus and is involved in the transcription initiation from positive- and negative-strand RNA templates of the virus [36]. Another study also reported similar findings in case of Japanese encephalitis virus and tomato bushy stunt virus (TBSV), where GAPDH was observed to bind both positive and negative strand RNAs of viruses (with a preference for +ve strand), resulting in inhibition of viral replication [37]. Mammalian GAPDH possesses a well-characterized phospholipid membrane fusogenic potential [38]. GAPDH levels were observed to be enhanced after Vaccinia virus infection in human monocytes at mRNA and protein levels [39] and GAPDH of eukaryotic origin played a progressive role in cell-to-cell transmission of red clover necrotic mosaic virus [40].
Given the extreme promiscuity exhibited by GAPDH for intermolecular interactions, we decided to test for any effect that GAPDH may have via spike protein on the corona of SARS-CoV-2. Utilizing pseudovirus expressing S protein from various strains of the virus and GAPDH protein, our current study indicates that GAPDH from three divergent organisms (human, bacteria, and rodent) could inhibit SARS-CoV-2 pseudovirus infection by binding to the receptor-binding domain (RBD) region of SARS-CoV-2 S protein and thus hindering the process of SARS-CoV-2 pseudovirus entry into host cells. These results were confirmed with the SARS-CoV-2 (Wuhan) strain of the virus in a cell culture model of viral infectivity. Our findings indicate an evolutionarily conserved protective role in innate host immune defense against viral incursion for GAPDH.
Materials and Methods
Cell Lines, Cell Culture, Plasmids, and Reagents
HEK 293T (ATCC, CRL-3216) and Vero E6 (ATCC, CCL-1586) cells were maintained in high glucose DMEM (SIGMA) supplemented with 10% FBS (GIBCO), penicillin (100 IU/mL), streptomycin (100 μg/mL), and HEPES (20 mm) in a 5% CO2 environment at 37°C and passaged every 2–3 days. SARS-Related Coronavirus 2, Wuhan-Hu-1 Spike-Pseudotyped Lentiviral Kit, NR-52948, was procured from BEI resources. It comprises five plasmids, namely, viral entry protein, spike glycoprotein (NR-52514), lentiviral backbone Luc2; ZsGreen (NR-52516), helper plasmid Gag; pol (NR-52517), helper plasmid Tat1b (NR-52518), helper plasmid Rev1b (NR-52519). SARS-CoV-2 B.1.617.2 (Delta strain) plasmid Addgene (#172320), SARS-CoV-2 B.1.617.1 (Kappa strain) plasmid Addgene (#172319), B.1.1.529/BA.1 (Omicron strain) (InvivoGen) (#plv-spike-v11) were used for variant pseudovirus production. Polyethylenimine, high molecular weight, water free, from Sigma Aldrich (# 408727) was used for plasmid transfection. Rabbit muscle GAPDH (RM GAPDH) was procured from Sigma Aldrich (#G2267); recombinant human GAPDH and Salmonella GAPDH [rH and rS GAPDH]) were cloned and purified as described in online supplementary materials and methods and Figure S1 (for all online suppl. material, see https://doi.org/10.1159/000535634). Anti-GAPDH antibody was from Calbiochem (# CB1001); antibody against bovine serum albumin, from Invitrogen (#MA5-15238), anti-rabbit IgG-HRP, and anti-mouse IgG were obtained from Sigma (# A9044 & #A0545). Recombinant spike, SARS-CoV-2 stabilized protein (#NR-52724), and recombinant spike glycoprotein RBD from SARS-CoV-2, novel coronavirus (#NR-52366), were procured from BEI resources.
Interaction Studies Using Solid Phase Binding Assay
The ability of pseudoviral particles and recombinant viral protein to bind recombinant human GAPDH protein was screened by solid phase binding assay. Briefly, ELISA grade polystyrene plates were coated for 12 h at 4°C with either 100 ng S protein or RBD peptide. As negative control, wells were coated with BSA. After blocking with 2% casein (prepared in PBST) for 2 h at RT, the immobilized proteins were allowed to interact with rH GAPDH in concentrations from 10,000 ng to 0.305 ng per well in triplicates. All incubations were carried out in PBST + 0.2% casein for 2 h at room temperature. Wells loaded with only buffer (no protein coating) were considered as control wells for background subtraction. Unbound GAPDH/BSA was removed by extensive rinsing with PBST. Captured protein was detected using 1 μg/mL of primary rabbit anti-GAPDH/1 μg/mL of primary mouse anti-BSA antibody. Secondary antibody used was anti-rabbit IgG-HRP (1:32,000 dilution)/anti-mouse IgG-HRP (1:32,000). TMB-H2O2 ELISA substrate (Sigma) was used to quantitate the bound secondary antibody by measuring OD450 using a plate reader and data were analyzed as described previously [41].
Binding Studies Using BLI
Quantification of the interaction between recombinant human GAPDH and COVID-19 partner (S protein/RBD peptide) was measured using an Octet K2 system (Pall Life Sciences, Fremont, CA, USA). COVID-19 proteins were immobilized onto aminopropylsilane optic sensor chips as ligand and evaluated for their binding affinity with the target. Briefly, 25 nm of S protein or RBD was immobilized to saturate the sensor for 600 s cycle followed by a 60 s baseline step using neutral buffer (150 mm NaCl, 4.96 mm KCl, 0.99 mm CaCl2, 2.1 mm MgCl2, 22.73 mm HEPES, pH 7.4) to remove unbound molecules. Cycles of 300 s each for association and dissociation steps were performed for binding with GAPDH and its removal in the working diluent. The assay was repeated with the reference biosensors to eliminate the possibility for non-specific interactions.
Pseudoviral Particles and SARS-CoV-2 Virus
HEK 293T cells were cultured to 65–70% confluence in complete DMEM media in standard tissue culture conditions and transfected with plasmids. A combination of five plasmids were utilized to produce the severe acute respiratory syndrome pseudovirus particles whose co-expression in HEK293T cells produces SARS-CoV-2 spike protein which decorates onto budding lentiviral particle envelopes containing reporter ZsGreen. Reporter protein enables fluorescence-based analysis of spike-mediated entry into host cells. Briefly, plasmids and PEI transfection reagent (1 mg/mL) were incubated in DMEM (SFM) for 5 min at 37°C in separate tubes. PEI and plasmid DNA were mixed at 3:1 ratio (w/w) and pre-incubated at 37°C for 30 min. For transfection, cells were first incubated in serum-free DMEM for 30 min (5% CO2) at 37°C. Subsequently, the SFM was replaced and cells incubated for a further 30 min at which time PEI: DNA mixture was added gently over the cells with gentle swirling for uniform distribution. Transfection was allowed to continue over the next 6–8 h in CO2 at which time the incubation mixture was replaced with complete DMEM. Sixty hours post-transfection, the culture supernatant was harvested from the plates and spun at 800 g to remove cellular debris and then filtered using 0.45 μm syringe filters. Pseudovirus particles using S gene of different variants, i.e., Wuhan (original), Delta, Kappa, and Omicron, were quantified as described in online supplementary materials and methods, Figure S1, and Table S1. SARS-CoV-2 virus stock (originally prepared from nasal swab of RT-PCR confirmed individual) was obtained from the MTCC and Gene Bank, CSIR-IMTECH (Chandigarh, India). Virus propagation was done in Vero E6 cell line maintained in 10% FBS containing DMEM and 1% L-glutamine at standard tissue culture conditions maintained in biosafety level-3 conditions. Culture supernatant was collected, centrifuged and aliquots stored at −70°C and diluted 1:104 in medium before use. The viral particles were cytopathic and its genome sequencing had revealed close relatedness with SARS-CoV-2 originating in Wuhan, China.
Infectivity Assays
Infectivity of pseudoviral particles was evaluated by inoculating Vero cells with viral particles and then measuring the percentage cells expressing marker gene (ZsGreen) after 72 h as described below. The degree of infection by SARS-CoV-2 virus was quantified by evaluating infected cells for (1) expression of E gene by RT-PCR and (2) expression of S protein using Western blot as described below.
Vero E6 cells were seeded in 24 well plates (1 × 104 cell/well). After 24 h, the medium was removed and the cells incubated in fresh medium (untreated control) or fresh medium supplemented with GAPDH or BSA (non-specific protein control). To test for inhibitory effect of GAPDH on viral/pseudoviral infection, cells were infected with viral particles in presence of GAPDH (100 μg/mL) and compared with infectivity of cells in absence of the protein. As a negative control, BSA was utilized to replace GAPDH. Dose dependency of GAPDH on viral infectivity was determined by varying the concentration of GAPDH used. For this, culture medium was supplemented with GAPDH at different dilutions (100–0.001 μg/mL). Contact time for GAPDH and viral particles was 1 h in tube, then 24 h over the cells. After rinsing cells with fresh medium, any viral particles that had succeeded to infect the cells were allowed to replicate in situ for 48 h. A non-specific protein control was run in parallel where GAPDH was replaced by BSA (100 μg/mL). To check if GAPDH protein can maintain its stability over an extended period under culture conditions and continue to inhibit infection by virus, cells were pre-incubated with GAPDH for 12 h at which time any residual protein was rinsed away by three washes with PBS before processing the cells for infectivity assay as above. Briefly, cells were incubated with 300 μL/well of pseudoviral/viral particle suspension (MOI 2:1) made up of DMEM + 5% FBS. After inoculation for 24 h, the medium was replaced with complete DMEM and the cells were incubated for a further 72 h. After harvesting with PBS-EDTA, the cells were washed with PBS and analyzed for expression of ZsGreen using a BD FACS verse flow cytometer. Infectivity was determined as the percentage of cells expressing ZsGreen. From each sample, 104 cells were analyzed, all experiments repeated at least three times. For evaluation of cellular infection with SARS-CoV-2 virus, 104 cells/well, plated in 96 well plates were infected for 1 h using 100 μL of viral suspension. Subsequently, the supernatant was removed and replenished with fresh media and the cells incubated for 72 h at 37°C, 5% CO2. After 72 h, the cells were collected and processed for RNA isolation using TRIzol™ Reagent (#:15596026). The RNA was converted into cDNA using RevertAid First Strand cDNA Synthesis Kit, #K1621, Thermo Scientific. RT-PCR was set up in an Applied Biosystem 7300 Fast Real-Time PCR System using SYBR Green chemistry procured from Thermo Scientific Maxima SYBR Green/ROX qPCR Master Mix (2X) (Cat No. K0221). Expression of CoV E gene was evaluated using ACAGGTACGTTAATAGTTAATAGCGT and ATATTGCAGCAGTACGCACACA as forward and reverse primers, respectively. The expression of the target gene was normalized to the expression of the β-actin gene (forward primer CACCATTGGCAATGAGCGGTTC and reverse primer AGGTCTTTGCGGATGTCCACGT). For Western blot experiments to evaluate S protein expression, Vero E6 cells (104 per well of 96 well plate) were infected with SARS-CoV-2 viral supernatant as above. After 72 h, cells were harvested using RIPA lysis buffer. Lysate from 6 wells was pooled for each concentration of GAPDH or control used. Subsequently, the lysate was centrifuged at 12,000 g for 10 min at 4°C. Protein in supernatant was quantified using BCA method and 40 μg/well was loaded onto 10% SDS-PAGE gel. The resolved proteins were transferred to 0.22 μm nitrocellulose membrane and probed with anti-S antibody (RD Systems, #MAB105403). Pure S protein (100 ng) was used as positive control. Uninfected Vero E6 lysate served as negative control. GAPDH antibody (Calbiochem, #CD1001) was used to calculate fold change of S protein expression in Vero E6 lysate. ImageJ software was used to calculate fold change of S protein in GAPDH-treated lysate in comparison to control infected lysate.
Molecular Docking
Docking for human GAPDH and SARS-CoV-2 S protein was carried out using ClusPro 2.0 Web server [42–45]. Human GAPDH (PDB code 1ZNQ) was used as receptor and SARS-CoV-2 S protein (PDB code 6VXX) as ligand. Amino acids located at protein interface were obtained using PDB ePISA Web server [46]. The residues whose bond length was below 4 A° were taken into consideration. The pictorial representations for residues involved in interactions were shown by PyMOL server [47, 48].
Statistical Analysis
Each set was performed in duplicates, repeated at least three times. Data are represented as mean ± SD. Data were plotted using GraphPad Prism software 5.0 (GraphPad Software Inc., La Jolla, CA, USA). Statistical significance was calculated using unpaired t test.
Results
Interaction Studies Using Solid Phase Binding Assay and BLI
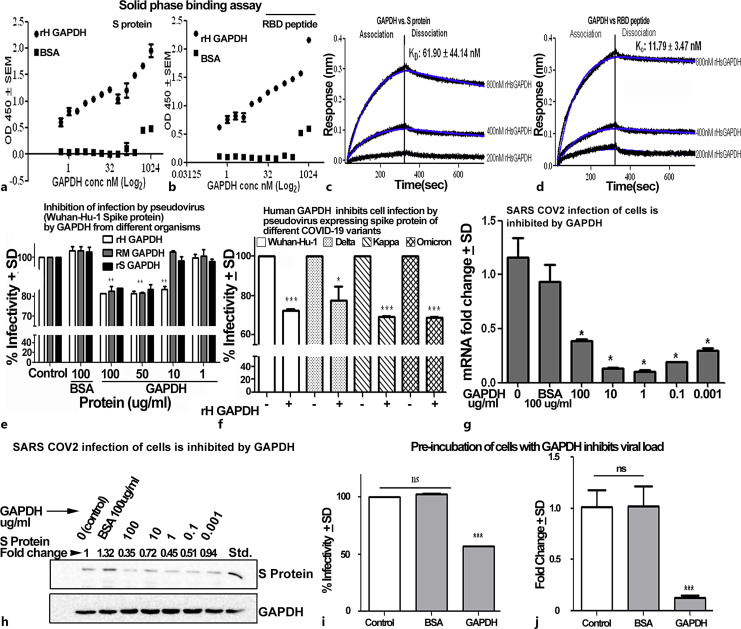
There are few reports where a viral protein has been shown to interact with host GAPDH and influence the host responses. One recent study has suggested that the non-structural 3 protein of dengue virus interacts with human GAPDH [49]. With this background, we explored if SARS-CoV-2 viral receptors protein could interact with human GAPDH. We observed that both S protein and SARS-CoV-2 RBD of S protein demonstrate significant capture of rH GAPDH in the solid phase assay (Fig. 1a, b). BLI confirmed the interaction between S protein and GAPDH with nanomolar affinity. For interaction between S protein and GAPDH, the KD was determined to be 61.90 ± 44.14 nm (Fig. 1c) while in case of RBD KD was 11.79 ± 3.47 nm (Fig. 1d).
Fig. 1.
a–d Human GAPDH binds with S protein and its RBD peptide segment. A solid phase binding assay was performed using MaxiSorp Nunc plates coated with 100 ng of either S protein (a) or RBD fragment (b). Subsequently, wells were incubated with increasing concentrations of recombinant human GAPDH. After extensive washing, the captured GAPDH was detected with monoclonal anti-GAPDH. Assay was performed multiple times in triplicates. Error bars indicate the standard deviation from triplicate values for each concentration tested. Affinity of interaction was determined by BLI. BLI sensorgrams indicate the binding of recombinant GAPDH with full-length S protein (c) and RBD (d). Data are plotted as response shift in nm versus time and the global equilibrium dissociation constant values (KD) were calculated. e GAPDH from diverse organisms is capable of inhibiting pseudoviral infectivity. Transduction of Vero cells by Wuhan strain S protein pseudovirus is inhibited by GAPDH from evolutionarily diverse sources in human infectivity assay using GAPDH from three organisms, each was tested at different concentrations. BSA was used as a non-specific protein control. All experiments were repeated three times, **p < 0.01. f Human GAPDH can inhibit infection by pseudovirus-expressing spike protein of different COVID-19 variants. Transduction of Vero cells by pseudoviral particles expressing S protein from four different variants is significantly inhibited by human GAPDH, ***p < 0.001, *p < 0.05, n = 3. g, h GAPDH inhibits infection of Vero E6 cells by SARS-CoV-2 virus. When Vero E6 cells were infected with SARS-CoV-2 viral particles in presence of human GAPDH, there was inhibition of viral RNA in supernatant of Vero E6. Viral load was evaluated by RT-PCR of SARS-CoV-2 E gene using total cellular cDNA, *p < 0.05, n = 3 (g) and Western blot of cell lysate to evaluate expression of S protein (h). i, j GAPDH can inhibit SARS-CoV-2 virus infection of Vero E6 host cells in culture up to 12 h post-application. Vero E6 cells were pre-incubated with 100 μg/mL GAPDH or BSA for 12 h at which time cells were rinsed and infected with either pseudoviral particles (i) or SARS-CoV-2 virus (j). After 72 h, the pseudovirus infected cells were evaluated for GFP expression and the % infectivity determined as compared to cells where GAPDH pre-incubation was omitted. In comparison to cells that had been left untreated or pre-treated with BSA, the GAPDH coated cells demonstrate a significantly lower infectivity, ns, not significant, ***p < 0.001, n = 3. SARS-CoV-2 virus infected cells were maintained in culture for 48 h at which time the cellular viral RNA expression was evaluated by RT-PCR for CoV E gene expression. In comparison to control cells, the cells pre-incubated with GAPDH demonstrate significantly lower viral load. ns, not significant, ***p < 0.001, n = 3.
GAPDH Inhibits Pseudovirus Variant’s Entry into the Host Cells
Previous studies have revealed that interaction between the spike protein and host cell receptors is vital for infection. To explore for any conserved role of GAPDH in modulating the cellular entry by SARS-CoV-2, we performed transduction assays of SARS-CoV-2 pseudovirus in the presence of GAPDH from evolutionary distant organisms. We tested GAPDH obtained from rabbit muscle, human, and bacteria (salmonella). Our studies revealed that GAPDH from all three sources could significantly reduce infection of Vero E6 cells with rH GAPDH being most effective (Fig. 1e). When pseudovirus particles decorated with S protein of different variants (Wuhan, Delta, Kappa, and Omicron) were used for transduction assays, we noted that in all variants the transduction was significantly decreased in host cells in presence of rH GAPDH (Fig. 1f). Overall, 100 μg/mL of GAPDH concentration was used for these assays. Error bars represent the standard deviation observed in technical triplicate values. The treated samples were compared to control, which was taken as 100% in each triplicate.
GAPDH Inhibits SARS-CoV-2 Virus Entry into the Host Cells
Finally, we tested if infection of cells with SARS-CoV-2 virus was inhibited by GAPDH. Incubation of Vero cells with SARS-CoV-2 virus in presence of varying concentration of rH GAPDH revealed a significant reduction in the viral titer as evaluated by RT-PCR-based assay of E gene expression in infected cells. Viral titer was significantly inhibited in all the concentrations of rH GAPDH used (7 pm–700 nm) (Fig. 1g). A Western blot of cellular S protein expression confirmed this result of lower viral load (Fig. 1h). Entry of fewer viral particles in the presence of GAPDH in the extracellular milieu leads to multiplication of a smaller viral load over the 72 h incubation period. This leads to fewer viral particles inside the cell as well as lower amount of virus released into the culture medium.
Pre-Treatment of Cells with GAPDH Provides Protection from Viral Infection
To investigate if the blocking effect of GAPDH in prevention of viral infection is only a transient phenomena, we first allowed cell surface to be treated with GAPDH and then allowed for infection with pseudovirus as well as SARS-CoV-2. In both cases, we found that pre-treatment of cells with GAPDH results in a significant decrease of cellular infection even after 12 h (Fig. 1i, j).
Molecular Docking Studies Indicate the GAPDH and S Protein for a Structurally Stable Complex
Human liver GAPDH was used as receptor and S protein as ligand. S protein consists of three chains (A, B, C) whereas GAPDH has four chains (O, P, Q, R). Chain O (green), P (red), and R (yellow) of GAPDH were found to interact with chain B (cyan) of S protein via hydrogen bonds as well as salt bridges (online suppl. Fig. S1I). Bond lengths below 4 A° were considered during docking of amino acid residues involved in interaction. The interaction between full length S and GAPDH was determined using 30 representative structures as generated by docking software. All structures were of balanced models with least center energy value. This minimal energy shown in structures corroborates with results obtained from solid phase assay and BLI assays, showing higher interaction of GAPDH with S protein. The amino acid residues of human GAPDH involved in the surface contact with full length S protein are Lys 194, Lys 215, Lys 219, and Asn 225 from chain O, Asp 256, Asp 257, Gln 280, Ala 299, and Asp 305 from chain P, Arg 80 from chain R (Table 1). The other amino acids were not masked when GAPDH interacts with S protein as evident from docking study (online suppl. Fig. S1J). The interactions between two large protein molecules are usually attributed to a combinatorial effect of multiple amino acid interactions. Future studies could explore in depth the role of key amino acid residues involved in stabilizing these interactions using multiple single/double/triple amino acid mutants of GAPDH.
Table 1.
Amino acid residues of human GAPDH involved in interaction with S protein of SARS-CoV-2 fetched from PDBePISA are enlisted here
| S. No | GAPDH protein | GAPDH chain | S protein | S chain |
|---|---|---|---|---|
| 1 | Lys 194 | O | Asn 81, Asp 138 | B |
| 2 | Lys 215 | O | Trp 64 | B |
| 3 | Lys 219 | O | Tyr 28 | B |
| 4 | Asn 225 | O | Tyr 28 | B |
| 5 | Asp 256 | P | Lys 97 | B |
| 6 | Asp 257 | P | Lys 187 | B |
| 7 | Gln 280 | P | His 69 | B |
| 8 | Ala 299 | P | His 69 | B |
| 9 | Asp 305 | P | Arg 214 | B |
| 10 | Arg 80 | R | Cys 136 | B |
These residues involved hydrogen bond among them with bond length below 4 A°.
Discussion
The innate immune system in higher organisms has evolved to deploy multiple cellular and molecular defense mechanisms, critical for the recognition and restriction of virus entry into host cells. Cells secrete multiple effector molecules, which can play a decisive role in modulating the entry of pathogens. For example, the human surfactant protein D and lactoferrin are potent molecules whose presence at mucosal surfaces contributes toward defense against a wide variety of pathogens including inhibiting entry of SARS-CoV-2 virus due to interactions with the spike protein [50–52].
GAPDH is an abundantly expressed multifunctional protein that is secreted by cells and along with lactoferrin has been known to play a key role in the innate immune defenses [28, 41, 53]. These protective effects are manifested through close interaction between host molecules and host/microbial components that are vital for pathogen entry and survival. Earlier studies have confirmed that GAPDH is expressed on the surface of cells and also secreted into the extracellular fluid [54–57] especially during conditions of cellular stress such as iron depletion and hypoxic stress [41, 58, 59]. We first looked for evidence of any interaction between virus entry machinery and GAPDH. Our studies revealed that GAPDH binds to RBD of SARS-CoV-2 S protein. A wide range in the affinity of interaction between ACE2 receptor and RBD (ranging from 16.2 nm to 325.8 nm) has been documented earlier [60]. Our experiments revealed a relatively high binding affinity of ∼62 and 12 nm for binding of GAPDH with S protein and RBD fragment, respectively. This suggests that presence of extracellular GAPDH could affect the interaction between virus and its receptor(s) on host cells.
To explore if GAPDH could cause inhibition of viral entry into cells, we carried out infectivity assays and found that GAPDH could significantly inhibit the entry of pseudoviral particles as well as SARS-CoV-2 virus into the Vero E6 cells. Numerous studies have reported on the importance of viral spike protein-hACE2 receptor interaction leading to entry into host cells. Our investigations revealed that presence of GAPDH significantly reduces the quantum of viral entry into cells. As we observed that GAPDH interacts with spike protein, we postulate that this lowered viral entry could be due to GAPDH interfering in the interaction between spike protein and hACE2. The Coronaviridae family encompasses positive-strand RNA viruses infecting amphibians, birds, and mammals. Genetic evolution data reveal SARS-CoV origin in bats was by sequential recombination of bat SARSr-CoVs. Virus residing in civets and mammal underwent rapid mutations in S and orf genes. Strains further underwent mutations in S gene leading to the epidemic [61]. As the pandemic has spread, the virus has undergone numerous mutations leading to the emergence of newer variants. In several cases, these variants exhibit significantly altered properties in terms of infectivity, host fatality, and prognosis. To understand if the interaction of GAPDH with the S protein is variant dependent, we compared the inhibitory effect of GAPDH on viral infection using pseudoviral particles presenting with S protein of Delta, Kappa, and Omicron variants along with the original Wuhan variant. Interestingly, we found that GAPDH was able to significantly inhibit infection by all four variants tested. This suggests that the interaction of GAPDH and spike protein is independent of the evolutionary pressure driving other genetic alterations.
The COVID-19-induced global pandemic has been ongoing for over 2 years and resulted in >350 million infections worldwide. Vaccination at the global scale has narrowed the severe COVID-19-based complications, yet mortality rate remains high. While COVID-19 vaccines educate immune systems to identify a target protein on the virus (S protein), however, this could mutate to escape the host immune response. Indiscriminate use of anti-viral medication is also known to lead to accumulation of mutations in the pathogen. As entry of SARS-CoV-2 into cells can occur through several receptor molecules present on the surface of mammalian cells [62, 63], globally there are several attempts to supplement currently available therapeutic options with simple solutions to block viral entry into cells via application of any barrier/inhibitory molecules on target surfaces. Antiviral nasal sprays are believed to have a promising role in this regard. Any studies regarding the virus’s interactions with competing agents (antibodies, peptides, drugs, etc.) have relevance in terms of development of therapeutic options [64]. As SARS-CoV-2 is a virus requiring a biosafety level-3 containment, one way to simplify the safe conduct of such assays is to utilize spike protein expressing pseudoviral particles, which can easily be handled under biosafety level-1/2 containment.
Our studies have revealed that human GAPDH can interact with viral spike protein and inhibit viral entry into cells. We also observed that prior surface exposure of target cells to GAPDH can result in significant inhibition of viral entry upon subsequent exposure to virus. These results suggest that GAPDH (a host protein) could harbor the potential for use as a therapeutic agent to inhibit the viral entry for up to 12 h post-application. In future, this could possibly provide leads for the further development of peptide/protein-based blockers of viral infections, which could be deployed by way of upper-respiratory tracts sprays [65, 66]. In silico molecular docking analysis indicated that primarily basic amino acid residues of GAPDH interact with different amino acids in the B chain of the viral S protein. Intermolecular interactions between two large protein molecules are usually attributed to a combinatorial effect involving multiple amino acid interactions. Future investigations on deciphering their contributions could be carried out using multiple single/double/triple amino acid mutants of GAPDH.
GAPDH is a highly conserved multifunctional molecule that has maintained its functions across organisms. Apart from chemical synthesis and recombinant human protein, GAPDH can also easily be purified from several GRASS (generally accepted as safe) sources [67]. We tested and confirmed that GAPDH from at least two other evolutionarily diverse organisms can inhibit pseudoviral transfection of Vero cells. Airway submucosal glands contribute to innate immunity and protect the lungs by secreting mucus. GAPDH has been identified as one of the numerous components of human mucus [68]. It is possible that one of the conserved innate immune functions of GAPDH could be to provide protection from viral infections. We speculate that fluctuations in the levels of extracellular GAPDH may contribute to success or failure of viral infection of cells. GAPDH expression on the exterior of the plasma membrane of mammalian as well as microbial cells is known to be regulated by cellular iron levels and a recent report from our laboratory has demonstrated a linkage between cellular iron levels and infection by SARS-CoV-2 virus [69].
A limitation of the current study is that the work did not involve animal model studies; however, in future before these are taken up it may be worthwhile to focus on deciphering the minimal conserved structural motif present in GAPDH responsible for this activity and seek to utilize it for prophylactic usage. As SARS-CoV-2 becomes endemic, it is expected that new virus variants will continue to emerge in future. There is likely a continuing need for effective therapeutics, especially as immunity wanes, vaccine hesitancy continues, and new virus variants emerge that escape natural and vaccine-induced antibodies.
Acknowledgments
Mr. Anil Theophilus and Mr. Randeep Sharma are thanked for technical assistance (IMTECH communication No. 01/2023).
Statement of Ethics
As the research work utilized only recombinant molecules and immortalized cell lines obtained from cell repositories, ethical approval was not required as per national guidelines. As the work did not utilize any tissues or fluid from human subjects, the need for consent for participation did not arise.
Conflict of Interest Statement
The authors have no conflicts of interest to declare.
Funding Sources
Financial support of DST, DBT, CSIR, ICMR, and UGC by way of research grants and fellowships is acknowledged.
Author Contributions
Chaaya Iyengar Raje and Manoj Raje conceptualized and planned the overall research work and finalized the manuscript. Rahul Dilawari and Gauav Kumar Chaubey planned most of the experiments, collected the data, performed analysis, and wrote the initial draft of paper. Asmita Dhiman carried out some of the experiments with SARS-CoV-2 virus. Sharmila Talukdar performed some of the RT-PCR experiments and collected and performed analysis of the data. Radheshyam Modanwal and Asmita Dhiman maintained the cell lines and assisted in tissue culture and microscopy experiments. Ajay Kumar cloned and purified human GAPDH along with Chaaya Iyengar Raje. All authors have seen the manuscript and consent to its publication.
Funding Statement
Financial support of DST, DBT, CSIR, ICMR, and UGC by way of research grants and fellowships is acknowledged.
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its online supplementary material files. Further inquiries can be directed to the corresponding author.
Supplementary Material
References
- 1. Singhal T. A review of coronavirus disease-2019 (COVID-19). Indian J Pediatr. 2020;87(4):281–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kutter JS, de Meulder D, Bestebroer TM, Lexmond P, Mulders A, Richard M, et al. SARS-CoV and SARS-CoV-2 are transmitted through the air between ferrets over more than one meter distance. Nat Commun. 2021;12(1):1653–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Peng X, Xu X, Li Y, Cheng L, Zhou X, Ren B. Transmission routes of 2019-nCoV and controls in dental practice. Int J Oral Sci. 2020;12(1):9–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Guo YR, Cao QD, Hong ZS, Tan YY, Chen SD, Jin HJ, et al. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak–an update on the status. Mil Med Res. 2020;7(1):11–0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Petrakis D, Margină D, Tsarouhas K, Tekos F, Stan M, Nikitovic D, et al. Obesity - a risk factor for increased COVID-19 prevalence, severity and lethality (Review). Mol Med Rep. 2020;22(1):9–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tadic M, Cuspidi C, Mancia G, Dell’Oro R, Grassi G. COVID-19, hypertension and cardiovascular diseases: should we change the therapy? Pharmacol Res. 2020;158:104906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Weinreich DM, Sivapalasingam S, Norton T, Ali S, Gao H, Bhore R, et al. REGN-COV2, a neutralizing antibody cocktail, in outpatients with Covid-19. N Engl J Med. 2021;384(3):238–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Belouzard S, Millet JK, Licitra BN, Whittaker GR. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses. 2012;4(6):1011–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–80. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Santos R, Ferreira AJ, Verano-Braga T, Bader M. Angiotensin-converting enzyme 2, angiotensin-(1-7) and Mas: new players of the renin-angiotensin system. J Endocrinol. 2013;216(2):R1–17. [DOI] [PubMed] [Google Scholar]
- 11. Wysocki J, Ye M, Rodriguez E, González-Pacheco FR, Barrios C, Evora K, et al. Targeting the degradation of angiotensin II with recombinant angiotensin-converting enzyme 2: prevention of angiotensin II–dependent hypertension. Hypertension. 2010;55(1):90–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liu DX, Liang JQ, Fung TS. Human coronavirus-229E,-OC43,-NL63, and-HKU1. In: Reference module in life sciences. 2020. p. B978-0-12-809633-8.21501-X. [Google Scholar]
- 13. Coutard B, Valle C, de Lamballerie X, Canard B, Seidah NG, Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antivir Res. 2020;176:104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cantuti-Castelvetri L, Ojha R, Pedro LD, Djannatian M, Franz J, Kuivanen S, et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science. 2020;370(6518):856–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Benham F, Povey S. Members of the human glyceraldehyde-3-phosphate dehydrogenase-related gene family map to dispersed chromosomal locations. Genomics. 1989;5(2):209–14. [DOI] [PubMed] [Google Scholar]
- 16. Sirover MA. New insights into an old protein: the functional diversity of mammalian glyceraldehyde-3-phosphate dehydrogenase. Biochim Biophys Acta. 1999;1432(2):159–84. [DOI] [PubMed] [Google Scholar]
- 17. Seidler NW. GAPDH: biological properties and diversity; 2012. [Google Scholar]
- 18. Nicholls C, Li H, Liu JP. GAPDH: a common enzyme with uncommon functions. Clin Exp Pharmacol Physiol. 2012;39(8):674–9. [DOI] [PubMed] [Google Scholar]
- 19. Boradia VM, Raje M, Raje CI. Protein moonlighting in iron metabolism: glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Biochem Soc Trans. 2014;42(6):1796–801. [DOI] [PubMed] [Google Scholar]
- 20. Boradia VM, Malhotra H, Thakkar JS, Tillu VA, Vuppala B, Patil P, et al. Mycobacterium tuberculosis acquires iron by cell-surface sequestration and internalization of human holo-transferrin. Nat Commun. 2014;5(1):4730–13. [DOI] [PubMed] [Google Scholar]
- 21. Sirover MA. Role of the glycolytic protein, glyceraldehyde-3-phosphate dehydrogenase, in normal cell function and in cell pathology. J Cell Biochem. 1997;66(2):133–40. [PubMed] [Google Scholar]
- 22. Sirover MA. On the functional diversity of glyceraldehyde-3-phosphate dehydrogenase: biochemical mechanisms and regulatory control. Biochim Biophys Acta. 2011;1810(8):741–51. [DOI] [PubMed] [Google Scholar]
- 23. Hara MR, Agrawal N, Kim SF, Cascio MB, Fujimuro M, Ozeki Y, et al. S-nitrosylated GAPDH initiates apoptotic cell death by nuclear translocation following Siah1 binding. Nat Cell Biol. 2005;7(7):665–74. [DOI] [PubMed] [Google Scholar]
- 24. Sirover MA. New nuclear functions of the glycolytic protein, glyceraldehyde-3-phosphate dehydrogenase, in mammalian cells. J Cell Biochem. 2005;95(1):45–52. [DOI] [PubMed] [Google Scholar]
- 25. Chaudhary S, Dhiman A, Dilawari R, Chaubey GK, Talukdar S, Modanwal R, et al. Glyceraldehyde-3-Phosphate dehydrogenase facilitates macroautophagic degradation of mutant huntingtin protein aggregates. Mol Neurobiol. 2021;58(11):5790–8. [DOI] [PubMed] [Google Scholar]
- 26. Chaudhary S, Dhiman A, Patidar A, Malhotra H, Talukdar S, Dilawari R, et al. Moonlighting glyceraldehyde-3-phosphate dehydrogenase (GAPDH) modulates protein aggregation. Biochim Biophys Acta, Mol Basis Dis. 2021;1867(10):166202. [DOI] [PubMed] [Google Scholar]
- 27. Chaudhary S, Patidar A, Dhiman A, Chaubey GK, Dilawari R, Talukdar S, et al. Exposure of a specific pleioform of multifunctional glyceraldehyde 3-phosphate dehydrogenase initiates CD14-dependent clearance of apoptotic cells. Cell Death Dis. 2021;12(10):892–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Patidar A, Malhotra H, Chaudhary S, Kumar M, Dilawari R, Chaubey GK, et al. Host glyceraldehyde-3-phosphate dehydrogenase-mediated iron acquisition is hijacked by intraphagosomal Mycobacterium tuberculosis. Cell Mol Life Sci. 2022;79(1):62–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dhiman A, Talukdar S, Chaubey GK, Dilawari R, Modanwal R, Chaudhary S, et al. Regulation of macrophage cell surface GAPDH alters LL-37 internalization and downstream effects in the cell. J Innate Immun. 2023;15:581–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mazzola JL, Sirover MA. Subcellular localization of human glyceraldehyde-3-phosphate dehydrogenase is independent of its glycolytic function. Biochim Biophys Acta. 2003;1622(1):50–6. [DOI] [PubMed] [Google Scholar]
- 31. Gozalbo D, Gil-Navarro I, Azorín I, Renau-Piqueras J, Martínez JP, Gil ML. The cell wall-associated glyceraldehyde-3-phosphate dehydrogenase of Candida albicans is also a fibronectin and laminin binding protein. Infect Immun. 1998;66(5):2052–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pancholi V, Fischetti VA. A major surface protein on group A streptococci is a glyceraldehyde-3-phosphate-dehydrogenase with multiple binding activity. J Exp Med. 1992;176(2):415–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Winram SB, Lottenberg R. The plasmin-binding protein Plr of group A streptococci is identified as glyceraldehyde-3-phosphate dehydrogenase. Microbiology. 1996;142(Pt 8)(8):2311–20. [DOI] [PubMed] [Google Scholar]
- 34. Gani Z, Boradia VM, Kumar A, Patidar A, Talukdar S, Choudhary E, et al. Mycobacterium tuberculosis glyceraldehyde-3-phosphate dehydrogenase plays a dual role–as an adhesin and as a receptor for plasmin (ogen). Cell Microbiol. 2021;23(5):e13311. [DOI] [PubMed] [Google Scholar]
- 35. Malhotra H, Patidar A, Boradia VM, Kumar R, Nimbalkar RD, Kumar A, et al. Mycobacterium tuberculosis glyceraldehyde-3-phosphate dehydrogenase (GAPDH) functions as a receptor for human lactoferrin. Front Cell Infect Microbiol. 2017;7:245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Prasanth KR, Huang YW, Liou MR, Wang RYL, Hu CC, Tsai CH, et al. Glyceraldehyde 3-phosphate dehydrogenase negatively regulates the replication of Bamboo mosaic virus and its associated satellite RNA. J Virol. 2011;85(17):8829–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yang SH, Liu ML, Tien CF, Chou SJ, Chang RY. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) interaction with 3'ends of Japanese encephalitis virus RNA and colocalization with the viral NS5 protein. J Biomed Sci. 2009;16(1):40–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. López Vinals AE, Farias RN, Morero RD. Characterization of the fusogenic properties of glyceraldehyde-3-phosphate dehydrogenase: fusion of phospholipid vesicles. Biochem Biophys Res Commun. 1987;143(1):403–9. [DOI] [PubMed] [Google Scholar]
- 39. Nahlik KW, Mleczko AK, Gawlik MK, Rokita HB. Modulation of GAPDH expression and cellular localization after vaccinia virus infection of human adherent monocytes. Acta Biochim Pol. 2003;50(3):667–76. [PubMed] [Google Scholar]
- 40. Kaido M, Abe K, Mine A, Hyodo K, Taniguchi T, Taniguchi H, et al. GAPDH-A recruits a plant virus movement protein to cortical virus replication complexes to facilitate viral cell-to-cell movement. PloS Pathog. 2014;10(11):e1004505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chauhan AS, Kumar M, Chaudhary S, Patidar A, Dhiman A, Sheokand N, et al. Moonlighting glycolytic protein glyceraldehyde-3-phosphate dehydrogenase (GAPDH): an evolutionarily conserved plasminogen receptor on mammalian cells. FASEB J. 2017;31(6):2638–48. [DOI] [PubMed] [Google Scholar]
- 42. Desta IT, Porter KA, Xia B, Kozakov D, Vajda S. Performance and its limits in rigid body protein-protein docking. Structure. 2020;28(9):1071–81.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kozakov D, Beglov D, Bohnuud T, Mottarella SE, Xia B, Hall DR, et al. How good is automated protein docking? Proteins. 2013;81(12):2159–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kozakov D, Hall DR, Xia B, Porter KA, Padhorny D, Yueh C, et al. The ClusPro web server for protein–protein docking. Nat Protoc. 2017;12(2):255–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Vajda S, Yueh C, Beglov D, Bohnuud T, Mottarella SE, Xia B, et al. New additions to the C lus P ro server motivated by CAPRI. Proteins. 2017;85(3):435–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372(3):774–97. [DOI] [PubMed] [Google Scholar]
- 47. DeLano WL. Pymol: an open-source molecular graphics tool. CCP4 Newsl Protein Crystallogr. 2002;40(1):82–92. [Google Scholar]
- 48. DeLano WL. The PyMOL molecular graphics system; 2002. Available from: http://www.pymol.org. [Google Scholar]
- 49. Silva EM, Conde JN, Allonso D, Ventura GT, Coelho DR, Carneiro PH, et al. Dengue virus nonstructural 3 protein interacts directly with human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and reduces its glycolytic activity. Sci Rep. 2019;9(1):2651–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hsieh MH, Beirag N, Murugaiah V, Chou YC, Kuo WS, Kao HF, et al. Human surfactant protein D binds spike protein and acts as an entry inhibitor of SARS-CoV-2 pseudotyped viral particles. Front Immunol. 2021;12:1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Madan T, Biswas B, Varghese PM, Subedi R, Pandit H, Idicula-Thomas S, et al. A recombinant fragment of human surfactant protein D binds spike protein and inhibits infectivity and replication of SARS-CoV-2 in clinical samples. Am J Respir Cell Mol Biol. 2021;65(1):41–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lang J, Yang N, Deng J, Liu K, Yang P, Zhang G, et al. Inhibition of SARS pseudovirus cell entry by lactoferrin binding to heparan sulfate proteoglycans. PLoS One. 2011;6(8):e23710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Takaoka Y, Goto S, Nakano T, Tseng HP, Yang SM, Kawamoto S, et al. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) prevents lipopolysaccharide (LPS)-induced, sepsis-related severe acute lung injury in mice. Sci Rep. 2014;4(1):5204–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chauhan AS, Rawat P, Malhotra H, Sheokand N, Kumar M, Patidar A, et al. Secreted multifunctional Glyceraldehyde-3-phosphate dehydrogenase sequesters lactoferrin and iron into cells via a non-canonical pathway. Sci Rep. 2015;5(1):18465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sheokand N, Kumar S, Malhotra H, Tillu V, Raje CI, Raje M. Secreted glyceraldehye-3-phosphate dehydrogenase is a multifunctional autocrine transferrin receptor for cellular iron acquisition. Biochim Biophys Acta. 2013;1830(6):3816–27. [DOI] [PubMed] [Google Scholar]
- 56. Kumar S, Sheokand N, Mhadeshwar MA, Raje CI, Raje M. Characterization of glyceraldehyde-3-phosphate dehydrogenase as a novel transferrin receptor. Int J Biochem Cell Biol. 2012;44(1):189–99. [DOI] [PubMed] [Google Scholar]
- 57. Yamaji R, Chatani E, Harada N, Sugimoto K, Inui H, Nakano Y. Glyceraldehyde-3-phosphate dehydrogenase in the extracellular space inhibits cell spreading. Biochim Biophys Acta. 2005;1726(3):261–71. [DOI] [PubMed] [Google Scholar]
- 58. Malhotra H, Kumar M, Chauhan AS, Dhiman A, Chaudhary S, Patidar A, et al. Moonlighting protein glyceraldehyde-3-phosphate dehydrogenase: a cellular rapid-response molecule for maintenance of iron homeostasis in hypoxia. Cell Physiol Biochem. 2019;52(3):517–31. [DOI] [PubMed] [Google Scholar]
- 59. Chauhan AS, Kumar M, Chaudhary S, Dhiman A, Patidar A, Jakhar P, et al. Trafficking of a multifunctional protein by endosomal microautophagy: linking two independent unconventional secretory pathways. FASEB J. 2019;33(4):5626–40. [DOI] [PubMed] [Google Scholar]
- 60. Shang J, Ye G, Shi K, Wan Y, Luo C, Aihara H, et al. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581(7807):221–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Cui J, Li F, Shi Z-L. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019;17(3):181–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Nassar A, Ibrahim IM, Amin FG, Magdy M, Elgharib AM, Azzam EB, et al. A review of human coronaviruses’ receptors: the host-cell targets for the crown bearing viruses. Molecules. 2021;26(21):6455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Liu J, Lu F, Chen Y, Plow E, Qin J. Integrin mediates cell entry of the SARS-CoV-2 virus independent of cellular receptor ACE2. J Biol Chem. 2022;298(3):101710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zhang L, Narayanan KK, Cooper L, Chan KK, Skeeters SS, Devlin CA, et al. An ACE2 decoy can be administered by inhalation and potently targets omicron variants of SARS-CoV-2. EMBO Mol Med. 2022;14(11):e16109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Chonira V, Kwon YD, Gorman J, Case JB, Ku Z, Simeon R, et al. A potent and broad neutralization of SARS-CoV-2 variants of concern by DARPins. Nat Chem Biol. 2022;19(3):284–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Lin Y, Sun J, Cao X, Wang X, Chen X, Xu H, et al. Non-adjuvanted interferon-armed RBD protein nasal drops protect airway infection from SARS-CoV-2. Cell Discov. 2022;8(1):43–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Gani Z, Boradia VM, Raghu Ram J, Suryavanshi PM, Patil P, Kumar S, et al. Purification and characterization of glyceraldehyde-3-phosphate-dehydrogenase (GAPDH) from pea seeds. Protein Expr Purif. 2016;127:22–7. [DOI] [PubMed] [Google Scholar]
- 68. Joo NS, Evans IAT, Cho HJ, Park IH, Engelhardt JF, Wine JJ. Proteomic analysis of pure human airway gland mucus reveals a large component of protective proteins. PLoS One. 2015;10(2):e0116756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Chaubey GK, Dilawari R, Modanwal R, Talukdar S, Dhiman A, Raje CI, et al. Excess iron aggravates the severity of COVID-19 infection. Free Radic Biol Med. 2023;208:186–93. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its online supplementary material files. Further inquiries can be directed to the corresponding author.