Abstract
Introduction
Image-based machine learning holds great promise for facilitating clinical care; however, the datasets often used for model training differ from the interventional clinical trial-based findings frequently used to inform treatment guidelines. Here, we draw on longitudinal imaging of psoriasis patients undergoing treatment in the Ultima 2 clinical trial (NCT02684357), including 2,700 body images with psoriasis area severity index (PASI) annotations by uniformly trained dermatologists.
Methods
An image-processing workflow integrating clinical photos of multiple body regions into one model pipeline was developed, which we refer to as the “One-Step PASI” framework due to its simultaneous body detection, lesion detection, and lesion severity classification. Group-stratified cross-validation was performed with 145 deep convolutional neural network models combined in an ensemble learning architecture.
Results
The highest-performing model demonstrated a mean absolute error of 3.3, Lin’s concordance correlation coefficient of 0.86, and Pearson correlation coefficient of 0.90 across a wide range of PASI scores comprising disease classifications of clear skin, mild, and moderate-to-severe disease. Within-person, time-series analysis of model performance demonstrated that PASI predictions closely tracked the trajectory of physician scores from severe to clear skin without systematically over- or underestimating PASI scores or percent changes from baseline.
Conclusion
This study demonstrates the potential of image processing and deep learning to translate otherwise inaccessible clinical trial data into accurate, extensible machine learning models to assess therapeutic efficacy.
Keywords: Deep learning, Psoriasis assessment, Clinical trial imaging, Accurate PASI prediction, Deep convolutional neural network
Introduction
Psoriasis is a chronic inflammatory disease that damages the skin and impacts several other organ systems [1]. Psoriasis is driven by a confluence of genetic, immunologic, and behavioral factors and affects between 2 and 4% of the population worldwide with overall prevalence across demographic groups [2]. While effective systemic and targeted therapies have been developed for psoriasis, non-treatment and undertreatment remain significant problems, with over 50% of patients reporting dissatisfaction with their treatment [3].
Consensus treatment guidelines for psoriasis use the psoriasis area severity index (PASI) clinical assessment to measure disease severity and provide target values for treatment efficacy [4]. The PASI assessment is performed by a trained physician evaluating the overall body surface area (BSA) or involvement that is affected by psoriatic plaques and assessing the severity of plaques in the categories of erythema (redness), induration (thickness), and desquamation (scaling) to generate a composite score on a scale of 0–72 [5]. The PASI score and thresholds for percent change upon treatment (i.e., PASI75, PASI100) are routinely used for eligibility criteria and primary endpoints of therapeutic efficacy in interventional clinical trials.
Image-based machine learning workflows to generate PASI scores have potential future use to augment and uniformize PASI assessment in clinical trials, clinical care, and remote monitoring applications [6]. Prior studies have employed image-based machine learning models to perform PASI assessments [7–10]. However, these efforts have not trained their machine learning models on interventional studies collecting longitudinal, standardized imaging data. As a result, machine learning-assisted PASI scoring may not accurately measure individual’s change upon treatment, potentially hindering deep learning adoption to assess therapeutic efficacy in clinical trials and routine clinical practice settings. Here, we have used interventional clinical imaging data to train a deep learning workflow, called the “one-step PASI” framework, to integrate clinical photos of multiple body regions into one model pipeline for simultaneous body detection, lesion detection, and lesion severity classification to generate PASI scores (without head and neck).
Methods
Ethics
This study utilizes data from the UltIMMA-2 trial, which was done in accordance with the study protocol, International Council for Harmonisation of Technical Requirements for Pharmaceuticals for Human Use (ICH) guidelines, applicable local regulations, and Good Clinical Practice guidelines governing clinical study conduct, and ethical principles outlined in the Declaration of Helsinki. All study-related documents (including the study protocols) were approved by an Institutional Review Board or Independent Ethics Committee at each study site, and all patients provided written informed consent before participation.
Data Pre-Processing and Tiling
The imaging dataset utilized for this study comprised 60 psoriasis patients who underwent five or six site visits, resulting in a total of 338 visits among all participants. For each subject visit, a total of 8 raw images (online suppl. Fig. 1; for all online suppl. material, see https://doi.org/10.1159/000536499) were collected as follows: four images were taken of the upper extremities, capturing the front left, front right, back left, and back right orientations; two images were captured of the lower extremities, covering the front and back orientations; and two images were obtained of the trunk, representing the front and back orientations. This resulted in a total of 2,700 images, with only two subjects having six images available. Images were resized from the raw 4,928 × 3,264 pixels to 128 × 128 pixels (see supplementary materials for further discussion). Despite pixel reduction, the resized images maintained original ratios to prevent distortion of relative involvement scores used for PASI scoring.
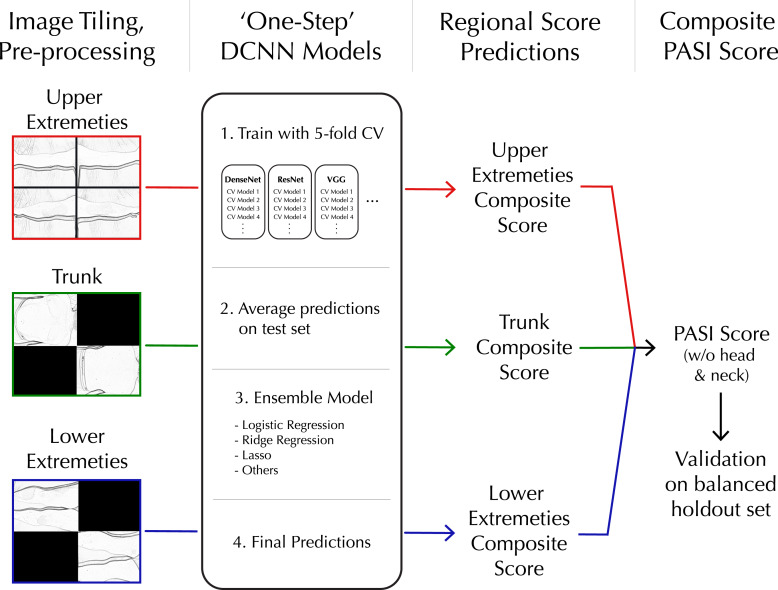
In order to establish a standardized workflow capable of processing a variable number of images from the three distinct body regions, the resized images from the same region within a patient visit were seamlessly stitched together in a 2 × 2 grid pattern, creating a single composite image (Fig. 1). Solid black squares filled in missing regions of the 2 × 2 composite image grids generated for the trunk and lower extremity regions because they each had two images. The four upper extremity images completed the 2 × 2 composite. Black squares contained no features for deep convolutional neural network (DCNN) models to detect and therefore did not interfere with learning. Sub-images comprising the 2 × 2 composite were randomly rotated, and their positions within the grid were shuffled to avoid position-detection bias in the trained PASI scoring model. The pre-processing stage yielded a collection of three 2 × 2 composite images, representing each body region, for every clinical visit. In total, there were 1,013 composite images that served as training samples for the DCNN models.
Fig. 1.
One-step workflow for processing whole-body imaging, training DCNN, and evaluation of composite PASI scores.
Images were also pre-processed with randomized brightness and contrast adjustments. Images used for training and testing were in full color but were converted to black and white for demonstration purposes in this manuscript to protect subject privacy.
Group-Stratified Train/Test Splitting
A group-stratified train-test splitting strategy was applied based on a greedy algorithm. Subject records were randomly shuffled into groups in a stepwise fashion, where train or test group assignments were made one subject at a time. This minimized scoring imbalances by assessing score distributions after each round of sorting. The scaling severity score was used to guide data splitting.
Scaling (desquamation) was the chosen guiding metric for data splitting because it has the highest correlation with other severity dimensions. Our imbalance measure is the average of absolute train-test differences in the adjusted percentage out of the total number of records for a given desquamation severity label over five labels, where the adjustment accounts for the different sizes of the cohorts. To generate the most representative dataset split, random shuffling was done multiple times with different random seeds.
PASI Scoring Algorithm
PASI scores are generated by (1) scoring the disease severity of multiple body regions and (2) combining all regional severity scores into a weighted composite PASI score for the whole body. Disease severity is determined by evaluating body regions for the degree of erythema, induration, scaling (i.e., desquamation), and lesion BSA (i.e., involvement). A regional composite score is calculated as seen in Equation 1, where Eregion, Iregion, and Sregion are the erythema, induration, and scaling severity, and Bregion is the BSA score for psoriasis lesions.
| (1) |
Next, a PASI score is calculated based on a linear combination of the regional composite scores of all four individual regions [5]. Due to subject privacy protection, there are no head and neck photos in the dataset used in this paper. A PASI sub-score without head and neck disease severity is derived using Equation 2. Cupper, Ctrunk, and Clower are the regional composite score of upper extremities, trunks, and lower extremities specifically.
| (2) |
In this study, the severity scores (Eregion, Iregion, and Sregion) and lesion BSA (Bregion) were evaluated by trained dermatologists and collected during the UltlMMA-2 trial. The regional composite score is derived based on Equation 1 and used as ground truth to train the DCNN models.
Model Architecture
The one-step workflow trained a DCNN module with an ensemble learning architecture using the prespecified training dataset. This included base learners with 5-fold cross-validation (CV) (see online supplementary for details). 29 pre-trained DCNN models were imported from PyTorch packages for use as base learners [11]. The complete list of base learner models is available in online supplementary Table S2 [12–24]. For each of the 29 base learner DCNN architectures, 5 DCNN copies were created. Each of the DCNN copies was fine-tuned with 4 folds (out of 5 total folds) from the training dataset and subsequently predicted the last fold (1 out of 5 folds). 145 DCNNs were used across the 29 base learners. Predictions for each sample in the training dataset were generated by base learners and concatenated into one vector input to train the ensemble model. Four different logistic regression models were used for the ensemble model. These included logistic regression (meta_learner), logistic regression with intercept term (meta_learner_w_intercept), logistic regression with ridge penalty (meta_learner_ridge), and ridge penalty (meta_learner_ridge_w_intercept). The models are specifically trained to directly predict regional composite scores (Psub) in Equation 1. Consequently, the severity scores (Eregion, Iregion, and Sregion) are implicitly estimated by the DCNN models and cannot be directly obtained.
The one-step workflow tested the trained model by having each of the 5 DCNN models within the 29 base learners make predictions for the full testing dataset. For each image, the mean of the 5 predictions generated by each base learner was calculated. The 29 mean predictions generated for each image were concatenated into a vector input to the ensemble model. Finally, the ensemble model predicted the PASI score results for the testing dataset.
Model Implementation
A Dell Precision 7,550 laptop with an Intel i7-10850H CPU and Nvidia Quadro TRX 4000 graphic card was used for this workflow. The pre-trained DCNNs were imported from PyTorch (v.1.9.0) [11], with classification layers being updated for regression tasks. The training and testing pipeline was implemented with PyTorch and PyTorch Lightning (v1.4.0) [25]. Mean squared error (MSE) was the loss criterion, and the optimizer was stochastic gradient descent with momentum set to 0.9. The learning rate was initialized at 0.01 and reduced by 0.1 at the plateau. The plateau threshold was set to 0.0001, and the patience was 10. The training batch size was 16. Early stopping was applied during the training procedure with patience of 50 epochs.
Results
Image Dataset
To develop a deep learning workflow for prediction of PASI scores from imaging data, we utilized 2,700 standardized photos captured during UltIMMa-2, a randomized, placebo-controlled phase 3 interventional trial (NCT02684357) investigating the efficacy of Risankizumab to reduce the severity of psoriasis over the course of 16 weeks [26, 27]. 74.8% of study participants achieved 90% reduction in PASI score (PASI90) during the first 16 weeks, providing a large dynamic range of PASI scores within individuals. The demographic information of patients is available in online supplementary Table S1.
This imaging dataset included 60 psoriasis patients with images captured for five or six site visits, totaling 338 visits across all participants. At each visit, site staff used a Nikon D5100 camera to capture 4,928 × 3,264 pixel photos of three key body regions: four photos of the upper extremities, two of the trunk, and two of the lower extremities (online suppl. Fig. 1). Head and neck photos were not collected to preserve subject privacy. The three regions were scored in person in a clinical setting by uniformly trained dermatologists for region involvement, erythema, induration, and scaling severity. The images underwent pre-processing using the method outlined in the above section prior to being utilized for model training.
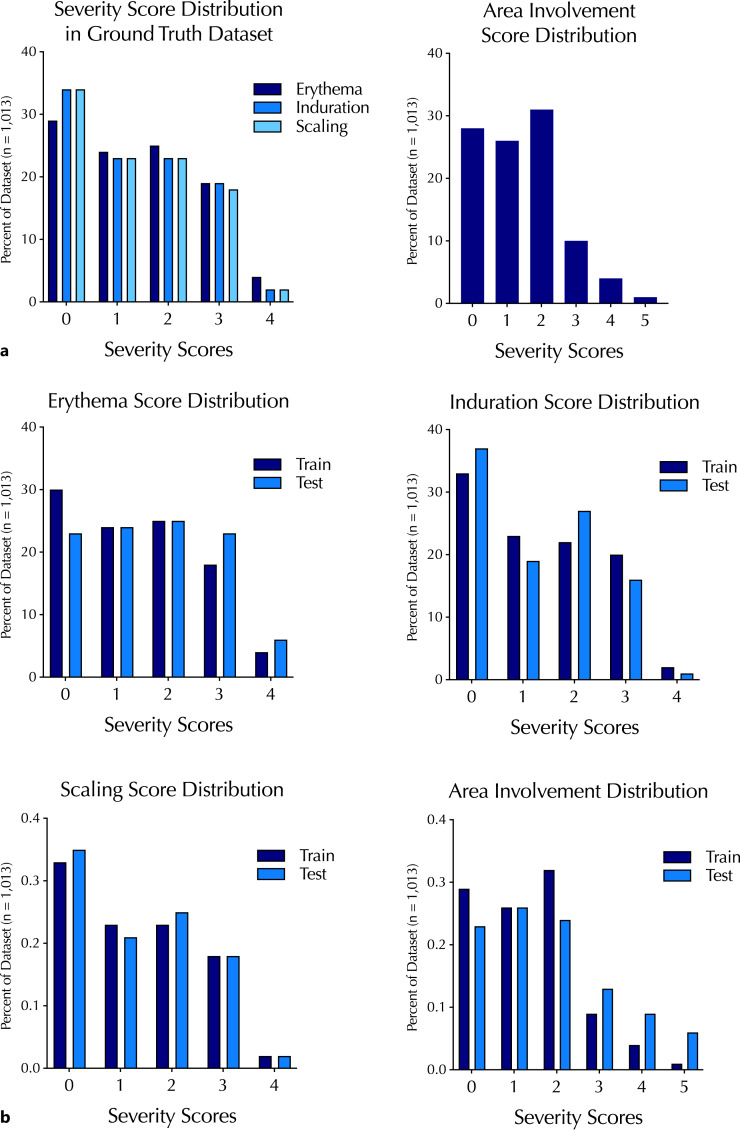
To ensure a balanced distribution of training and testing datasets, the images were sorted into training and testing datasets using the group-stratified train-test splitting strategy mentioned earlier. The ratio of 90% training and 10% testing was employed. Images associated with an individual subject were all assigned to either the training or testing set, ruling out potential within-subject information leakage between training and test sets. The training and testing datasets contained images with a similar distribution of erythema, induration, scaling, and involvement (Fig. 2). The numerical information on the severity score distribution is also available in online supplementary Table S3.
Fig. 2.
Distribution of psoriasis severity scores and involvement (or BSA) scores in the ground-truth annotated dataset overall (a) and their distribution between training and test sets (b).
Model Training and Performance
Both base and meta-learners were validated for PASI scoring accuracy using the test dataset. The mean predictions from the five DCNNs in each base learner were used for testing. Learning models were evaluated using mean absolute error (MAE), MSE, Lin’s concordance correlation coefficient (CCC) score, and Pearson correlation coefficient (PCC) (Table 1).
Table 1.
Top 10 models based on MAE performance
| Model | MAE | PCC | MSE | CCC |
|---|---|---|---|---|
| resnet34 | 3.336 | 0.900 | 27.191 | 0.830 |
| shufflenet_v2_x0_5 | 3.479 | 0.896 | 23.616 | 0.864 |
| densenet121 | 3.507 | 0.892 | 27.475 | 0.835 |
| resnet152 | 3.539 | 0.885 | 28.499 | 0.825 |
| resnet101 | 3.639 | 0.866 | 28.821 | 0.835 |
| vgg19_bn | 3.676 | 0.870 | 30.606 | 0.826 |
| resnext50_32 × 4d | 3.709 | 0.883 | 30.273 | 0.812 |
| meta_learner_ridge | 3.781 | 0.879 | 30.583 | 0.812 |
| mobilenet_v2 | 3.786 | 0.886 | 29.025 | 0.823 |
| vgg11_bn | 3.788 | 0.874 | 31.482 | 0.806 |
MAE, mean absolute error; PCC, Pearson correlation coefficient; MSE, mean squared error; CCC, Lin’s concordance correlation coefficient.
The resnet34 model [12] demonstrated the best performance as measured by MAE, MSE, and PCC in DCNN validation testing. The base learner models largely outperformed the four meta-learning architectures. The best meta-learner was the logistical regression model with ridge regularization, but it was ranked 8th among all learners.
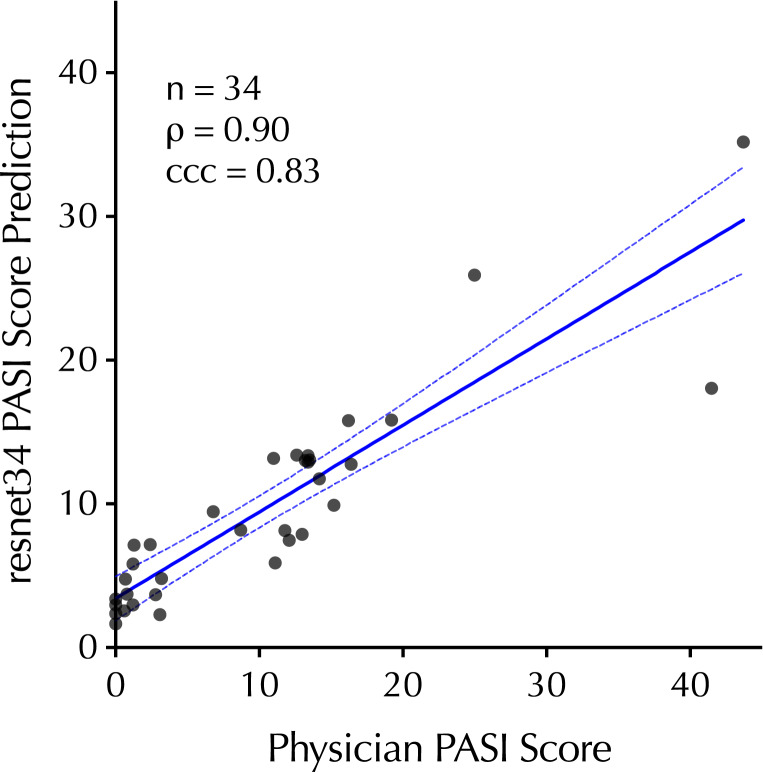
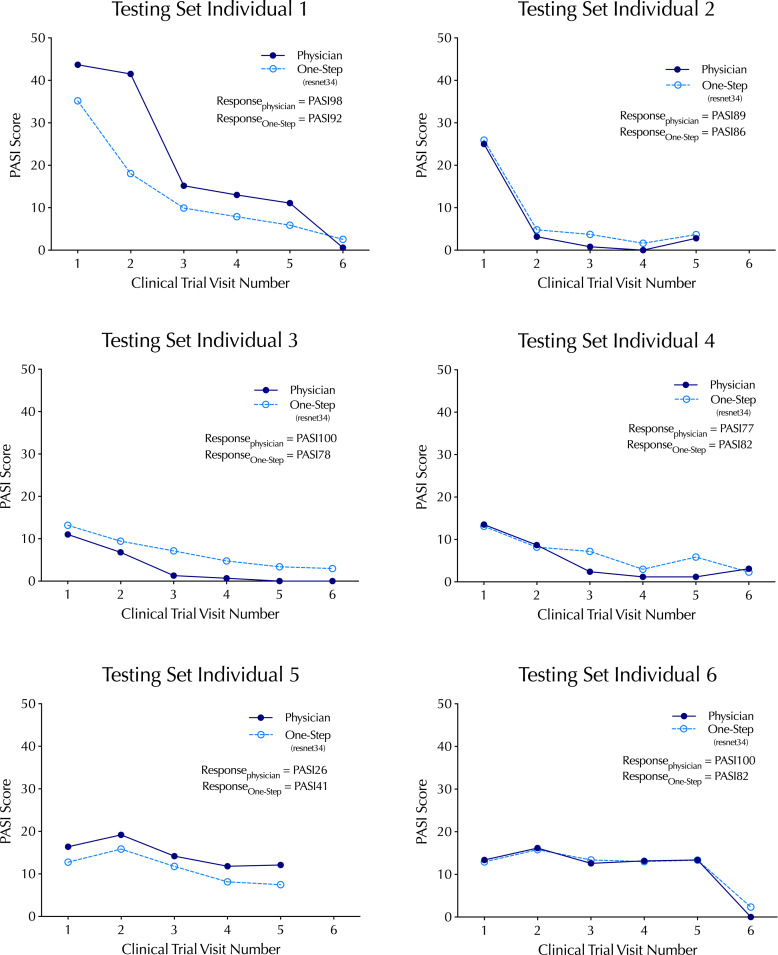
To explore the model’s performance in the test dataset, ground-truth physician PASI scores were plotted with the resnet34 PASI score predictions comprising 34 clinical trial visits across 6 subjects (Fig. 3). There was a strong correlation (p = 0.90) across the range of ground-truth PASI scores between 0 and 43.7, a range that covers overall psoriasis severity classifications of clear, mild, and moderate-to-severe disease [28]. To explore the one-step workflow for its capacity to track changes in an individual’s psoriasis severity over time, time-series analysis of individuals over the course of the study was performed, revealing that one-step PASI estimations closely tracked the trajectory of the physician-based PASI scores upon interventional treatment. The best-performing model did not systematically over- or underestimate PASI scores or the interventional response variable that denotes percent change from baseline (i.e., PASI75) (Fig. 4).
Fig. 3.
Correlation between the top-performing PASI estimation model (resnet34) predictions on the test dataset relative to in-clinic physician PASI scores (without head and neck). 95% CI is shown as dashed-blue lines.
Fig. 4.
Time-series of top-performing one-step PASI estimation model (resnet34) predictions relative to in-clinic physician PASI scores (without head and neck). The clinical trial endpoint metric, percent change from baseline in the PASI score (e.g., PASI98, PASI92), is listed for the model and physician.
Discussion
In this study, we developed a “one-step” deep learning framework for multi-image processing and deep convolutional neural network (DCNN) training to predict PASI scores (without head and neck). This framework utilized 2,700 images of psoriasis patients’ skin while undergoing treatment in a controlled clinical setting to train and test model performance. The best-performing model trained by the framework (resnet34) produced predictions demonstrating a strong correlation (Pearson’s r = 0.9) with in-person physician scoring and a MAE of 3.3 on the scale of 0–54 (without head and neck). Importantly, the sub-score estimations for individuals over time tracked the trajectory of physician scoring without systemic over- or underestimates at an individual visit or the estimated percent change over time that is typically used in clinical trial settings (e.g., PASI75).
A key component of models from the one-step PASI framework is their training on highly standardized, interventional clinical trial data. In contrast to deep learning workflows trained with a large volume of cross-sectional images capturing a snapshot in time from a given patient, the UltIMMa-2 trial images allow the model to learn in the setting of within-person disease changes from baseline over time. In this dataset, individuals often progressed from severe disease to clear skin. Thus, in addition to evaluating single-point diagnostic accuracy, we investigated one-step models for their capacity to accurately assess therapeutic efficacy.
PASI scoring requires three different tasks: body detection from the background, disease lesion segmentation from healthy skin, and severity classification for detected lesions. Other methods use separate DCNNs to achieve each of the three tasks. For example, previous work from Li et al. [10] used 86,000 images labeled with lesion location for the segmentation. The one-step PASI framework implicitly integrates all three tasks into one DCNN model by pre-processing images from multiple body regions into a standard input. This allows the model to generalize learning from a relatively small sample size by connecting information across multiple body regions. Compared to the benchmark model ResNet-50 [10], the resnet34 model trained by the one-step PASI framework demonstrated better MAE (3.34 vs. 3.50) while also training on fewer images (2,700 vs. 5,205).
Despite the strong concordance between the one-step PASI predictions and ground-truth PASI scores, there are limitations to this work. The dataset contained no head and neck region photos. While there are regional severity scores for other body regions, the accuracy of facial, head, and neck region scoring is unknown, and this may bias the accuracy of the overall PASI score for patients whose psoriatic lesions are found primarily in this region (e.g., scalp psoriasis). The imaging to generate the dataset was highly standardized, so it is unknown how these models would perform with non-clinical image capture methods such as a smartphone.
There was a relatively small sample size used to train the DCNN model. Access to a larger dataset of clinical trial imaging would enable more robust DCNN training and allow for assessment of how well these models generalize to a larger population. This is particularly important for clinical applications in dermatology, where imbalances in skin tone representation in the training set may impart systematic biases in digital tools [29, 30]. We envision this method being employed on larger datasets in the future to evaluate this important limitation.
This study performed CV to tune hyperparameters and maximize the use of limited clinical image data. Five copies of each model architecture were trained with CV, so each image could be learned by four out of the five models, resulting in 145 DCNNs across 29 base learners. Then, these models were used to predict the last fold of images (not used to train the specific model), and the predictions were concatenated to train the ensemble model to avoid potential information leakages. While repeated CV could provide more comprehensive statistical measures, it would require significant time and resources, so it is left for future exploration. The current approach maximizes the efficient use of clinical image data. It can be applied to other datasets with limited samples, paving the way for more efficient and effective deep learning in medicine.
As mentioned earlier, the inclusion of head and neck images in our model development was limited due to privacy concerns. However, it is important to note that the full PASI scores, encompassing the head and neck regions, were estimated and collected on-site for the current dataset by trained dermatologists. Exploring potential correlations between the severity scores of the head and neck region and other body regions presents an intriguing avenue for future research.
This work uses AI/ML techniques to improve an existing, well-established clinical outcome assessment (COA), PASI. While not a digital measure in its original conception but consistent with the V3 framework developed by DiMe [31], PASI measures a meaningful aspect of psoriasis, including measurement concepts of discoloration, thickness, scaling, and skin coverage. Although PASI is a well-established COA, it has been criticized for its complex, manual, and subjective interpretations in the assessment. The proposed AI/ML-trained algorithms can potentially eliminate the lengthy, complex, manual scoring processes, easing the burden for physicians. In addition, by removing manual and subjective interpretation processes, the algorithms may reduce inter-rater variability and improve assessment accuracy across clinical sites.
In summary, the one-step PASI deep learning framework trains models that can accurately estimate PASI scores from clinical imaging. While further work with larger datasets will be needed to generalize this approach, this study demonstrates the potential of image processing and deep learning to translate otherwise inaccessible clinical trial data into accurate, extensible machine learning models with the potential to assess therapeutic efficacy.
Statement of Ethics
This study utilizes data from the UltIMMA-2 trial, which was done in accordance with the study protocol, International Council for Harmonisation of Technical Requirements for Pharmaceuticals for Human Use (ICH) guidelines, applicable local regulations, and Good Clinical Practice guidelines governing clinical study conduct, and ethical principles outlined in the Declaration of Helsinki. All study-related documents (including the study protocols) were approved by an Institutional Review Board or Independent Ethics Committee at each study site, and all patients provided written informed consent before participation. More details can be found on ClinicalTrials.gov with identifier NCT02684357.
Conflict of Interest Statement
All authors are employees of AbbVie Inc. and may own AbbVie stock.
Funding Sources
This manuscript was sponsored by AbbVie. AbbVie contributed to the design, research, and interpretation of the data, writing, reviewing, and approving the publication.
Author Contributions
Y.X., S.Z., and L.W. designed this study and wrote the first manuscript draft. Y.X., S.Z., and D.W. performed the analyses. Y.X., S.Z., S.A., F.R., D.W., M.C., and L.W. contributed to the writing, interpretation of the content, and editing of the manuscript, revising it critically for important intellectual content. All authors take accountability for ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Funding Statement
This manuscript was sponsored by AbbVie. AbbVie contributed to the design, research, and interpretation of the data, writing, reviewing, and approving the publication.
Data Availability Statement
All image and annotation data used in this work is prohibited from broader distribution due to the data privacy outlined in the informed consent obtained from clinical trial subjects. Further inquiries can be directed to the corresponding author.
Supplementary Material
References
- 1. Lowes MA, Bowcock AM, Krueger JG. Pathogenesis and therapy of psoriasis. Nature. 2007;445(7130):866–73. [DOI] [PubMed] [Google Scholar]
- 2. Griffiths CEM, Van Der Walt JM, Ashcroft DM, Flohr C, Naldi L, Nijsten T, et al. The global state of psoriasis disease epidemiology: a workshop report [Internet]. UK: Blackwell Publishing Ltd Oxford; 2017. [cited 2024 January 8]. Available from: https://academic.oup.com/bjd/article-abstract/177/1/e4/6601929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Armstrong AW, Robertson AD, Wu J, Schupp C, Lebwohl MG. Undertreatment, treatment trends, and treatment dissatisfaction among patients with psoriasis and psoriatic arthritis in the United States: findings from the National Psoriasis Foundation surveys, 2003-2011. JAMA Dermatol. 2013;149(10):1180–5. [DOI] [PubMed] [Google Scholar]
- 4. Mrowietz U, Kragballe K, Reich K, Spuls P, Griffiths CEM, Nast A, et al. Definition of treatment goals for moderate to severe psoriasis: a European consensus. Arch Dermatol Res. 2011;303(1):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Langley RG, Ellis CN. Evaluating psoriasis with psoriasis area and severity index, psoriasis global assessment, and lattice system physician’s global assessment. J Am Acad Dermatol. 2004;51(4):563–9. [DOI] [PubMed] [Google Scholar]
- 6. Esteva A, Chou K, Yeung S, Naik N, Madani A, Mottaghi A, et al. Deep learning-enabled medical computer vision. NPJ Digit Med. 2021;4(1):5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fink C, Alt C, Uhlmann L, Klose C, Enk A, Haenssle HA. Precision and reproducibility of automated computer-guided Psoriasis Area and Severity Index measurements in comparison with trained physicians. Br J Dermatol. 2019;180(2):390–6. [DOI] [PubMed] [Google Scholar]
- 8. Wu D, Lu X, Nakamura M, Sekhon S, Jeon C, Bhutani T, et al. A Pilot Study to assess the reliability of digital image-based PASI scores across patient skin tones and provider training levels. Dermatol Ther. 2022;12(7):1685–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schaap MJ, Cardozo NJ, Patel A, De Jong E, Van Ginneken B, Seyger MMB. Image-based automated psoriasis area severity index scoring by convolutional neural networks. J Eur Acad Dermatol Venereol. 2022;36(1):68–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li Y, Wu Z, Zhao S, Wu X, Kuang Y, Yan Y, et al. PSENet: psoriasis severity evaluation network. Proceedings of the AAAI conference on artificial intelligence; 2020. p. 800. [Google Scholar]
- 11. Paszke A, Gross S, Massa F, Lerer A, Bradbury J, Chanan G, et al. Pytorch: an imperative style, high-performance deep learning library. Adv Neural Inf Process Syst. 2019;32. [Google Scholar]
- 12. He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. Proceedings of the IEEE conference on computer vision and pattern recognition; 2016. p. 770–8. [Google Scholar]
- 13. Howard A, Sandler M, Chu G, Chen LC, Chen B, Tan M, et al. Searching for mobilenetv3. Proceedings of the IEEE/CVF international conference on computer vision; 2019. p. 1314–24. [Google Scholar]
- 14. Huang G, Liu Z, Van Der Maaten L, Weinberger KQ. Densely connected convolutional networks. Proceedings of the IEEE conference on computer vision and pattern recognition; 2017. p. 4700–8. [Google Scholar]
- 15. Iandola FN, Han S, Moskewicz MW, Ashraf K, Dally WJ, Keutzer K. SqueezeNet: AlexNet-level accuracy with 50x fewer parameters and< 0.5 MB model size. 2016. arXiv preprint arXiv:160207360.
- 16. Krizhevsky A. One weird trick for parallelizing convolutional neural networks. 2014. arXiv preprint arXiv:14045997. [Google Scholar]
- 17. Krizhevsky A, Sutskever I, Hinton GE. Imagenet classification with deep convolutional neural networks. Commun ACM. 2017;60(6):84–90. [Google Scholar]
- 18. Ma N, Zhang X, Zheng HT, Sun J. Shufflenet v2: practical guidelines for efficient cnn architecture design. Proceedings of the European conference on computer vision ECCV; 2018. p. 116–31. [Google Scholar]
- 19. Sandler M, Howard A, Zhu M, Zhmoginov A, Chen LC. Mobilenetv2: inverted residuals and linear bottlenecks. Proceedings of the IEEE conference on computer vision and pattern recognition; 2018. p. 4510–20. [Google Scholar]
- 20. Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. 2014. rXiv preprint arXiv:14091556. [Google Scholar]
- 21. Szegedy C, Liu W, Jia Y, Sermanet P, Reed S, Anguelov D, et al. Going deeper with convolutions. Proceedings of the IEEE conference on computer vision and pattern recognition; 2015. p. 1–9. [Google Scholar]
- 22. Tan M, Chen B, Pang R, Vasudevan V, Sandler M, Howard A, et al. Mnasnet: platform-aware neural architecture search for mobile. Proceedings of the IEEE/CVF conference on computer vision and pattern recognition; 2019. p. 2820–8. [Google Scholar]
- 23. Xie S, Girshick R, Dollár P, Tu Z, He K. Aggregated residual transformations for deep neural networks. Proceedings of the IEEE conference on computer vision and pattern recognition; 2017. p. 1492–500. [Google Scholar]
- 24. Zagoruyko S, Komodakis N. Wide residual networks. 2016. arXiv preprint arXiv:160507146. [Google Scholar]
- 25. Falcon W. PyTorch lightning. GitHub Note: https://github.com/PyTorchLightning/pytorch-lightning [Internet]. 2019 March 30. Available from: https://www.pytorchlightning.ai.
- 26. Gordon KB, Strober B, Lebwohl M, Augustin M, Blauvelt A, Poulin Y, et al. Efficacy and safety of risankizumab in moderate-to-severe plaque psoriasis (UltIMMa-1 and UltIMMa-2): results from two double-blind, randomised, placebo-controlled and ustekinumab-controlled phase 3 trials. Lancet. 2018;392(10148):650–61. [DOI] [PubMed] [Google Scholar]
- 27. Augustin M, Lambert J, Zema C, Thompson EH, Yang M, Wu EQ, et al. Effect of risankizumab on patient-reported outcomes in moderate to severe psoriasis: the UltIMMa-1 and UltIMMa-2 randomized clinical trials. JAMA Dermatol. 2020;156(12):1344–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Finlay AY. Current severe psoriasis and the rule of tens. Br J Dermatol. 2005;152(5):861–7. [DOI] [PubMed] [Google Scholar]
- 29. Wen D, Khan SM, Ji Xu A, Ibrahim H, Smith L, Caballero J, et al. Characteristics of publicly available skin cancer image datasets: a systematic review. Lancet Digit Health. 2022;4(1):e64–74. [DOI] [PubMed] [Google Scholar]
- 30. Nelson BW, Low CA, Jacobson N, Areán P, Torous J, Allen NB. Guidelines for wrist-worn consumer wearable assessment of heart rate in biobehavioral research. NPJ Digit Med. 2020;3(1):90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Goldsack JC, Dowling AV, Samuelson D, Patrick-Lake B, Clay I. Evaluation, acceptance, and qualification of digital measures: from proof of concept to endpoint. Digit Biomark. 2021;5(1):53–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All image and annotation data used in this work is prohibited from broader distribution due to the data privacy outlined in the informed consent obtained from clinical trial subjects. Further inquiries can be directed to the corresponding author.