Abstract
We obtained exonuclease III (exoIII) footprints for a series of RNA polymerase II transcription complexes stalled between positions +20 to +51. Downstream advance of the exoIII footprint is normally tightly coordinated with RNA synthesis. However, arrested RNA polymerases slide back along the template, as indicated by exoIII footprints in which the last transcribed base is abnormally close to the downstream edge of the footprint. None of the polymerase II complexes stalled between +20 and +51 were arrested. Nevertheless, the exoIII footprints of complexes with 20-, 23-, or 25-nucleotide RNAs resembled those of arrested complexes, with the last transcribed base very close to the footprint’s front edge. The exoIII footprint of the +27 complex was displaced downstream by 17 bp compared to the footprint of the +25 complex. Many complexes between +27 and +42 also showed evidence of sliding back along the template. We compared the effects of template sequence and transcript length by constructing a new template in which the initial transcribed sequence was duplicated beginning at +98. The exoIII footprints of transcription complexes stalled between +122 to +130 on this DNA did not resemble those of arrested complexes, in contrast to the footprints of analogous complexes stalled over the same DNA sequences early in transcription. Our results indicate that the RNA polymerase II transcription complex passes through a major, sequence-independent structural transition about 25 bases downstream of the starting point of transcription. The fully mature form of the elongation complex may not appear until more than 40 bonds have been made.
It is increasingly evident that a significant number of eukaryotic genes are regulated during the transcript elongation process (for a recent review, see reference 29). One important control point is in the promoter-proximal region, where regulation occurs during the transition from initiation to elongation. For example, in the well-studied Drosophila hsp70 gene, RNA polymerases can access the promoter at normal temperatures but the newly initiated polymerases pause 21 to 35 bases downstream (reviewed in reference 17). Upon heat shock, these paused polymerases are released into productive elongation. The promoter-proximal attenuation mode of regulation has been observed in a number of other genes as well (see, for example, references 14, 21, and 25; see also the review in reference 29).
RNA polymerases paused in the promoter-proximal region resemble arrested RNA polymerases to some extent. Arrest, which can occur with Escherichia coli RNA polymerase as well as RNA polymerase II (29), results in stable ternary transcription complexes which cannot continue transcript elongation (9). Resumption of transcription by arrested polymerases requires cleavage of a segment of the 3′ end of nascent RNA, a reaction which is greatly stimulated by the transcript elongation factor TFIIS (also referred to as SII) for RNA polymerase II (9, 23) or by GreB for bacterial RNA polymerase (1).
A model to explain arrest which is now widely accepted postulates that RNA polymerases in ternary transcription complexes can translocate upstream instead of forming the next phosphodiester bond. These complexes remain stably clamped to the template during the sliding process (12, 13, 16, 19, 22; see also reference 6). Most importantly, since the transcription bubble and the RNA-DNA hybrid also translocate upstream, the 3′ end of the nascent RNA will be removed from the polymerase’s catalytic site. Thus, unless the translocation process is reversed, cleavage of the transcript at the new, upstream location of the catalytic center is necessary to realign the 3′ end of the RNA with the catalytic site and to allow RNA synthesis to continue (20, 26). Normally, upstream translocation is very unlikely in comparison with bond formation (3, 4, 11). However, at template locations which encode long runs of U residues in the transcript, the RNA-DNA hybrid is exceptionally unstable and upstream translocation may be favored (19, 22).
The location of the RNA polymerase along the template can be visualized by exonuclease III (exoIII) footprinting. During normal transcript elongation, the template segment protected by the transcription complex appears to translocate along the DNA in rough synchrony with transcript elongation (18, 27). However, at arrest sites, the exoIII footprint either remains stationary or shifts upstream relative to the footprints of earlier complexes (3, 12, 13, 19, 27, 30). It is important to stress that transcription complexes may partition between upstream and downstream conformers. We have described several RNA polymerase II elongation complexes which show evidence of both conformations, either by exoIII footprinting (27) or by the presence of large (7 to 17 nucleotides [nt]) transcript cleavage products in the presence of SII (11). In some, but not all, of these cases, a fraction of the transcription complexes appeared arrested since they failed to resume RNA synthesis immediately upon readdition of nucleoside triphosphates (NTPs). Thus, the tendency of a transcription complex to show measurable amounts of an upstream-translocated form by exoIII footprinting is an indication of the potential for arrest. In this context, it is particularly interesting that E. coli RNA polymerase tends to slide back along the template during the initial phases of transcription, until the addition of roughly the 28th base to the nascent RNA (12, 18; see also reference 15). As noted above, pausing by RNA polymerase II at similar promoter-proximal locations is a significant regulatory event in vivo. Thus, it is important to determine if RNA polymerase II is also prone to upstream translocation early in transcript elongation.
In the studies reported here, we examined the transition from initiation to elongation for RNA polymerase II by using a series of transcription complexes stalled because of NTP limitation between positions +20 and +51. None of these complexes were overtly arrested, since all were able to elongate their transcripts in 5-min chase reactions. However, the exoIII footprints of complexes with 20- to 25-nt RNAs were identical and resembled those of arrested complexes, while the footprint of a +27 complex was displaced forward by 17 bp. In contrast, transcription of a template in which the initial transcribed sequence was duplicated beginning at +98 showed coordinate advance of the polymerase footprint with RNA synthesis during transcription from +122 to +130. Thus, the RNA polymerase II transcription complex, like E. coli RNA polymerase, passes through a major, apparently sequence-independent structural transition about 25 bases downstream of the starting point of transcription. We also showed that footprint translocation and transcription do not become fully synchronized for RNA polymerase II until more than 40 bonds have been made.
MATERIALS AND METHODS
Reagents.
Ultrapure (fast-performance liquid chromatography purified) NTPs, deoxynucleoside triphosphates (dNTPs), and dideoxynucleoside triphosphates were obtained from Pharmacia LKB Biotechnology, and 32P-labeled NTPs and dNTPs were obtained from NEN. Bio-Gel A-1.5m was acquired from Bio-Rad. Exonuclease III, placental ribonuclease inhibitor, Taq polymerase, and restriction enzymes were purchased from BRL Life Technologies, Inc.
Plasmids.
All plasmids used in this study were based on pML20-23, which contains the adenovirus 2 major late promoter cloned into pUC18 (27). Plasmids pML20-42, -45, -46, and -47 differ from pML20-23 in two ways. First, a C residue at position +17 on the template strand was changed to G, and second, the DNA from +20 to the StuI site (which is located at +149 in pML20-23) was replaced with a much shorter DNA segment. The sequences of pML20-42, -45, -46, and -47, beginning at +1, are shown in Fig. 1.
FIG. 1.
Sequence of relevant portions of the nontemplate strand of the DNAs used in these experiments. The arrows indicate the transcription start site (+1). The first 20 nt of the initially transcribed regions from pML20-42, -45, -46, and -47 are designated by dashed underlining. The same sequence is also duplicated in pML20-49 starting at position +98. Gray boxes represent T-free segments, each of which has a StuI site (solid underline) at its downstream end. Note that the sequences downstream of the StuI sites are identical in all constructs. Note also that the sequence of the pML20-49 template starting from +98 is identical to that of pML20-42 starting at +1.
Construction of pML20-49 was a two-step process. First, the pML20-42 plasmid was modified by inserting a second copy of the sequence from +1 to +20 at position +20. From this intermediate construct we assembled pML20-49 by inserting, again at +20, a segment of the G-free cassette from pC2AT (28) (kindly provided by R. G. Roeder). The fragment of pC2AT was generated by PCR and inserted so that there were no G residues on the nontemplate strand of pML20-49 between +1 and the StuI site, which begins at +120. The nontemplate strand of pML20-49 starting at +98 is shown in Fig. 1. All of the pML20-40 series templates were verified by sequencing.
Template preparation for in vitro transcription reactions.
Plasmid DNAs were prepared by double banding in CsCl. Plasmids were linearized by digestion with either SstI for nontemplate strand labeling or PstI for labeling of the template strand. Linearized DNA was treated with calf intestine phosphatase and labeled with [γ-32P]ATP to a typical specific activity of 3 × 106 to 6 × 106 cpm/μg with T4 polynucleotide kinase (New England Biolabs). DNA labeled at the SstI end was subsequently digested with EcoRI, which gave two fragments: a uniquely single-end-labeled DNA containing the promoter and the transcribed sequence (approximately 2.8 kb) and a small (12 bp) end-labeled EcoRI-SstI fragment. Similarly, DNA labeled at the PstI end was digested with HindIII, which also gave a uniquely single-end-labeled DNA and a short (25 bp) fragment. Template DNAs were purified by phenol-chloroform extraction and ethanol precipitation before use in transcription.
Assembly, purification, and analysis of ternary transcription complexes.
Ternary complexes stalled at specific positions on the DNA were generated essentially as previously described (27). Briefly, preinitiation complexes were assembled on end-labeled templates by incubation with HeLa cell nuclear extract. In a typical experiment, the reaction volume was 200 μl, and the total template DNA concentration in these reactions was 20 to 35 μg/ml. Residual NTPs were removed by gel filtration on Bio-Gel A-1.5m. On all templates, complexes were advanced to position +20 (U20 complexes) by incubation with 2 mM ApC, 10 μM dATP, 20 μM UTP, and 1 μM [α-32P]CTP at 30°C for 5 min followed by another 5-min incubation after the addition of CTP to 20 μM. The stalled ternary complexes were further purified by Sarkosyl rinsing: Sarkosyl was added to 1% and incubated for 5 min at 30°C, followed by Bio-Gel A-1.5m gel filtration. A complete description of Sarkosyl rinsing is given in reference 7. Sarkosyl-rinsed U20 complexes were incubated at 37°C with 8 mM MgCl2 and either 20 μM ATP, GTP, and CTP for 5 min (for all templates except pML20-49) or 20 μM ATP, UTP, and CTP for 10 min (for pML20-49) followed by another round of gel filtration. The last step was omitted for production of the U20, A23, G25, and C27 complexes on the pML20-42 template.
After the Sarkosyl rinsing-gel filtration step, all complexes assembled on the SstI end-labeled template were incubated with restriction enzymes StuI and HindIII (0.2 U/μl each) for 10 min at 37°C. Analogous complexes assembled on templates labeled at the PstI site were treated with StuI and EcoRI (0.2 U/μl each) for 10 min at 37°C. Placental ribonuclease inhibitor was added to the reaction mixture at this and all subsequent steps to a final concentration of 50 U/ml. After the restriction enzyme digestion, complexes were either immediately treated with exoIII or incubated with an appropriate subset of NTPs and then with exoIII, as indicated later in the text. The exoIII digestion was done for 6 min at 37°C at either a 2- or a 5-U/μl final concentration. The reactions were stopped by adding EDTA to a final concentration of 20 mM. In the indicated cases, half of the sample underwent additional treatment with RNase A (50 μg/ml for 30 min at 37°C). DNA and RNA were purified by proteinase K digestion (0.2 mg/ml for 10 min at 37°C), phenol-chloroform extraction and ethanol precipitation. Samples were resolved on denaturing polyacrylamide gels and visualized by autoradiography (Kodak X-AR or Kodak Biomax) and with a PhosphorImager (Molecular Dynamics).
Markers.
Exact DNA length markers were generated by primer extension with the same DNA template employed in the experiment, dNTP mixes with a single dideoxynucleoside triphosphate, [α-32P]dCTP, Taq polymerase, and synthesized primers. Primers were phosphorylated before addition to the reaction.
RESULTS
Experimental design.
In order to study transcription complexes paused early in the elongation process, we constructed a set of four templates, pML20-42, -45, -46 and -47, which allowed us to stall RNA polymerase II at many locations from 20 to 51 bases downstream of the transcription start site. We use the term “stall” to designate RNA polymerases which have stopped transcription because the next NTP required for chain elongation is missing from the reaction mixture. Stalled polymerases are not arrested; that is, they resume transcription when the necessary NTPs are supplied. The initial transcribed regions of the pML20-40 series DNAs are shown in Fig. 1. Each promoter contains the adenovirus 2 major late TATA element and initiator. The prototypical template, pML20-42, consists of a polypyrimidine segment from +2 to +20 on the nontemplate strand followed by a cleavage site for the restriction enzyme StuI. Downstream of the StuI site is a triplet repeat segment ( … TTTGGGAAACCC … ) and vector DNA. All of the pML20-40 constructs are based on the pML20-23 plasmid which we used in our previous study of RNA polymerase II elongation complexes (27). In pML20-23, the StuI site begins at +147 (Fig. 1). To construct pML20-42, the DNA between +22 and +147 in pML20-23 was deleted; for pML20-45, -46 and -47, an additional 5, 10, or 15 bases upstream of the StuI site were added back into pML20-42. Note that while the original sequence upstream of the StuI site in pML20-23 was generally preserved in pML20-45, -46 and -47 (Fig. 1), some base changes were necessary to allow convenient assembly of transcription complexes at desired template locations. RNA polymerases stalled at or downstream of the StuI site on the pML20-40 series templates should for the most part contact the same DNA sequences as polymerases stalled at comparable locations on pML20-23. This is significant because exoIII footprints of RNA polymerase II advanced in synchrony with transcription as the polymerase transcribed the triplet repeat segment in pML20-23 (27). Thus, the pML20-40 series plasmids should not contain DNA sequences at or downstream of the site of RNA polymerase stalling which provide an intrinsic block to the advance of the RNA polymerase or which tend to provoke upstream translocation by stalled RNA polymerases. The most upstream DNA contacts for some transcription complexes on the pML20-40 series plasmids will differ from those on pML20-23, because the initial transcribed region has replaced the original upstream DNA. We will return to this point below.
The generation of RNA polymerase II complexes halted at defined sites on the template has been described in detail previously (7, 27). Briefly, preinitiation complexes were formed by incubation of DNA with HeLa cell nuclear extract and purified by gel filtration. Incubation of these complexes with ApC, GTP, UTP, and [α-32P]CTP resulted in the production of stable U20 complexes (Fig. 1). These complexes were purified by the addition of 1% Sarkosyl followed by gel filtration (Sarkosyl rinsing), which removes the detergent, the NTPs, and most proteins, including free transcript elongation factors. The Sarkosyl-rinsed U20 complexes were advanced to the StuI site, digested with StuI, and then gel filtered again to remove NTPs. Treatment with StuI was necessary because only about 1% of the templates are occupied by RNA polymerase II; unless the background created by incomplete exoIII digestion of these nontranscribed DNAs is eliminated, it is not possible to detect the exoIII protection conferred by the transcription complexes (27). All of the complexes we analyzed stopped transcription within 12 bases in either direction of a StuI site in order to guarantee that the restriction enzyme site would be protected. After StuI treatment, the RNA polymerases were advanced to various downstream stalling sites with subsets of the NTPs and treated with exoIII.
Elongation complexes stalled in the initially transcribed region are transcriptionally competent.
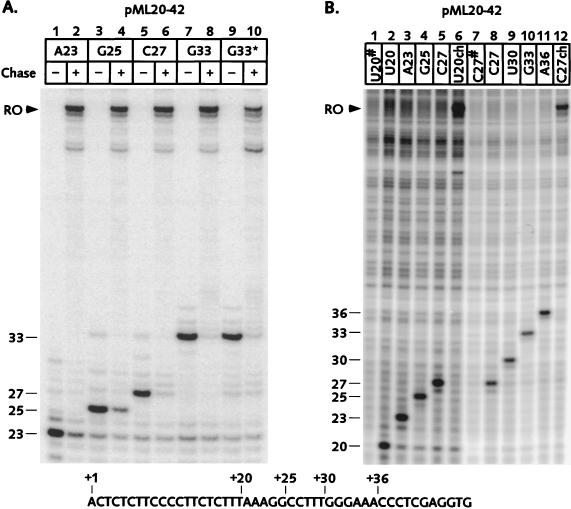
Although the transcribed segments of the pML20-40 series plasmids did not contain any sequences which resembled known arrest sites, we wanted to verify that all complexes involved in the study were transcriptionally competent. Transcripts generated on the pML20-42 template are shown in Fig. 2A. In this experiment, in which only the RNA was labeled, complexes were initially brought to position +23, gel filtered, and then advanced to +25, +27, or +33 by incubation with the appropriate subset of NTPs. All of these complexes could chase to the end of the template (Fig. 2) when all four NTPs were supplied. Thus, complexes A23, G25, C27, and G33 were genuinely stalled and not arrested. We always observed a small proportion (5 to 10%) of complexes that failed to restart whenever RNA polymerase II was halted in chain elongation at any template position. The amount of residual RNA seen in Fig. 2A is typical for stalled complexes (27). We performed similar experiments for all of the transcription complexes presented in this paper. In all cases only a very small proportion of the nascent RNA failed to chase (data not shown).
FIG. 2.
RNA polymerase II elongation complexes stalled in the initially transcribed region remain transcriptionally competent. Stalled transcription complexes were generated on the pML20-42 template as described in Materials and Methods except that in panel A the complexes were initially advanced to +23 and Sarkosyl rinsed at this location, not at +20. The sequence of the template strand of pML20-42 starting at +1 is shown at the bottom of the figure. For both panels, RNAs were resolved on 12% polyacrylamide gels. (A) In this experiment only the RNA was labeled. Complexes were advanced from +23 to the indicated locations by incubation with a subset of the NTPs, except for the G33* complex (lane 9) which was generated by incubating C27 complex with UTP. Portions of each complex were chased to the end of the template by adding missing NTPs (even numbered lanes). The arrowhead indicates runoff (RO) transcript. (B) U20 complexes were supplied with subsets of the NTPs to form A23, G25, or C27 complexes, or chased to runoff (RO, lane 6). In a separate experiment, C27 complexes formed from U20 complexes were gel filtered and further advanced to form U30, G33, or A36 complexes, or chased to runoff. Portions of the U20 and C27 complexes were treated with RNase A (#; lanes 1 and 7) to distinguish between labeled RNA and background DNA bands.
The exoIII protection patterns generated by RNA polymerase II elongation complexes stalled at sequential pausing sites in the initially transcribed region.
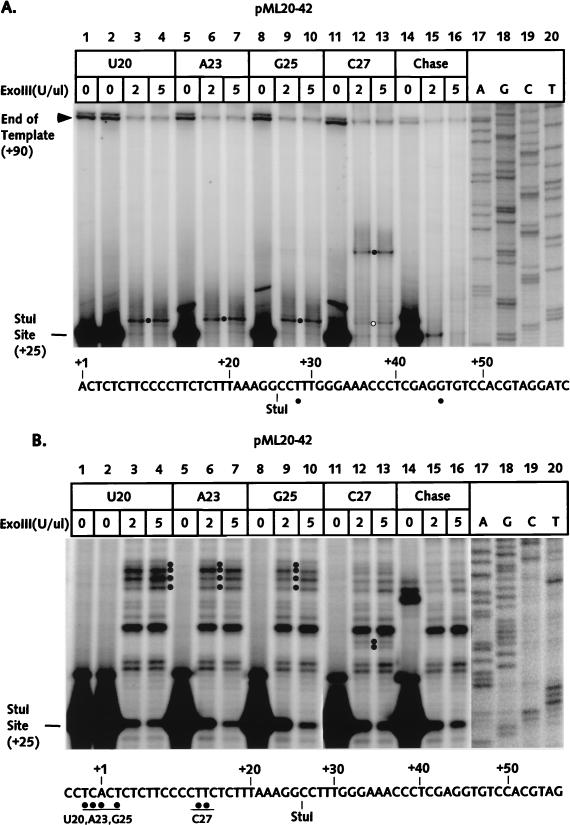
Our initial exoIII studies were performed on stalled complexes at positions +20, +23, +25, and +27 on the pML20-42 template. The RNAs for each complex are shown in Fig. 2B, lanes 2 to 5. In this case both template and transcript were labeled; to distinguish between RNA and a background of bands from the DNA, a portion of the U20 sample was treated with RNase A (lane 1). The ability to chase the U20 RNA to the end of the template (lane 6) indicated that the U20 complex was fully transcriptionally competent. The downstream, or front-edge, boundaries of template protection for the U20, A23, G25, and C27 complexes were determined by exoIII digestion after StuI cleavage, as described above. The results are shown in Fig. 3A. Most of the template DNA was cleaved by StuI (lanes 1 and 2), consistent with transcription of only a few percent of the templates by RNA polymerase II. When U20 complexes were treated with exoIII, the StuI-resistant DNA was truncated to a band whose downstream edge maps to +28 (band marked by dot, lanes 3 and 4). This band was absent when the complexes were chased before exoIII digestion (lanes 15 and 16), consistent with our assignment of the band as the front edge of the U20 transcription complex. The location of the U20 complex front edge, only 7 nt downstream of the last transcribed base, was surprising. This configuration is typical for arrested complexes, in which the RNA polymerase has translocated upstream (27; see also reference 3), even though the U20 complex showed no evidence of arrest. Furthermore, the front edges of the A23 and G25 complexes were identical to the U20 front edge (Fig. 3A, lanes 6, 7, 9, and 10), while the leading edge of the C27 complex was displaced 17 bases down the template, to +45 (lanes 12 and 13).
FIG. 3.
The exoIII footprints of U20, A23, G25, and C27 complexes on the pML20-42 template. A segment of nontemplate DNA strand sequence is shown at the bottom of each panel. (A) The front-edge boundaries of RNA polymerase II elongation complexes were determined with DNA labeled at the 5′ end of the nontemplate strand as described in Materials and Methods. DNAs were resolved on a 6% polyacrylamide gel. ExoIII reactions on U20 complexes chased to the end of the template with all four NTPs are shown in lanes 14 to 16. Solid dots mark the position of the major downstream protection boundary for each complex; the residual protection at +28 in lanes 12 and 13 is indicated by the open dot. The numbers in the left margin indicate the positions of bands relative to the transcription start site. The actual lengths of the DNA fragments equal the number indicated plus the 70-bp upstream fragment. Exact DNA length markers (lanes 17 to 20) were generated by primer extension from the same DNA template used for transcription. (B) The rear-edge boundaries of RNA polymerase elongation complexes (indicated by solid dots) were obtained as described for panel A, except that DNA was labeled at the 5′ end of the template strand. DNAs were resolved on a 10% polyacrylamide gel. The actual length of the StuI-cut DNA fragment is 75 nt.
As with the leading edges, the upstream boundaries of the U20, A23, and G25 complexes were essentially identical, while the C27 boundary was displaced well downstream (Fig. 3B). The rear-edge boundary was reproducibly more diffuse for several of the complexes (compare with Fig. 3A); we also observed this effect with the upstream boundaries of a number of the complexes we studied previously (27). The bands corresponding to the upstream boundaries were absent when the complexes were chased to runoff before exoIII digestion (Fig. 3B, lanes 15 and 16).
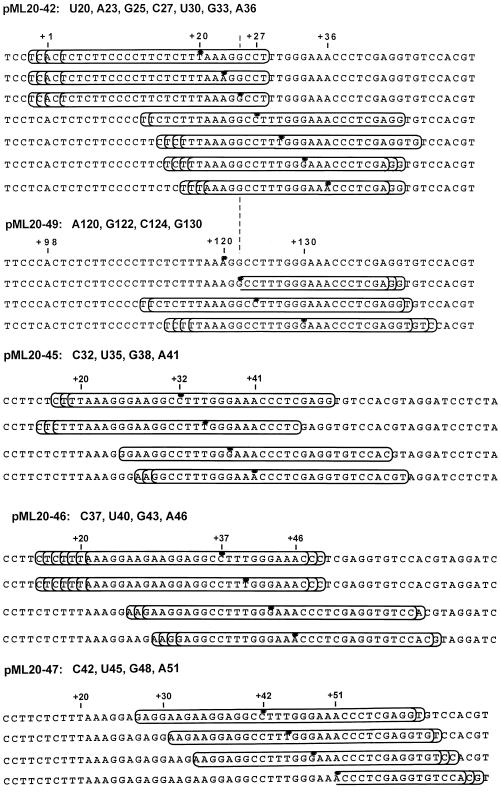
The results from Fig. 3 are summarized schematically in Fig. 7. The elongation complexes are represented by boxes and the positions of the last transcribed bases are designated by dots. For the sake of simplicity, only the nontemplate strand of DNA is shown, even though the rear edges were mapped on the template strand. The overall dimensions of the complexes, as judged by the length of template protected, did not change substantially among the four stalled complexes, consistent with the results obtained in earlier work (27). The strongly discontinuous advance of the exoIII footprint in complexes U20, A23, G25, and C27 is in sharp contrast to our previous results with a series of complexes stalled over analogous DNA sequences (but much further downstream of the transcription start site) on the pML20-23 template (27). However, the results in Fig. 3 are very similar to those reported for a series of E. coli RNA polymerase complexes stalled at positions +25 to +30 (18). It is particularly striking that the exoIII footprint of the bacterial polymerase advanced by nine bases as a result of adding only three more bases to the transcript of a complex at +27 (18). Apparent discontinuous movement of the RNA polymerase footprint was also reported in a comparison of the DNase I protection patterns obtained with E. coli RNA polymerase complexes stalled between +11 and +35 (15), although the differences between footprints were more complex than those seen with exoIII. Thus, a major structural transition may occur for both bacterial RNA polymerase and mammalian RNA polymerase II during transcription about 25 bases downstream of the transcription start site. We will return to this possibility in the Discussion.
FIG. 7.
Schematic summary of the exoIII protection experiments. Only the nontemplate strand of DNA is shown. The elongation complexes are represented by the boxes. The positions of transcript 3′ ends are designated by dots. Note that only the more downstream conformation is shown for complexes U30, G33, and A36. The pML20-42 and pML20-49 sequences are aligned to facilitate comparison of the footprints of pairs of analogous complexes, that is, complexes for which the transcript-template sequences are identical in the region near the 3′ end of the nascent RNA.
The unexpected results with the exoIII footprints of the U20, A23, G25, and C27 complexes prompted us to ask whether RNA polymerase II complexes stalled somewhat further downstream of +1 would also show anomalous footprints. The design of the pML20-42 template allowed us to generate complexes paused at positions +30, +33, and +36 (U30, G33, and A36 complexes, respectively). RNAs from each of these complexes are shown in Fig. 2B, lanes 8 to 11. Note that the C27 complex was fully active when chased (lane 12).
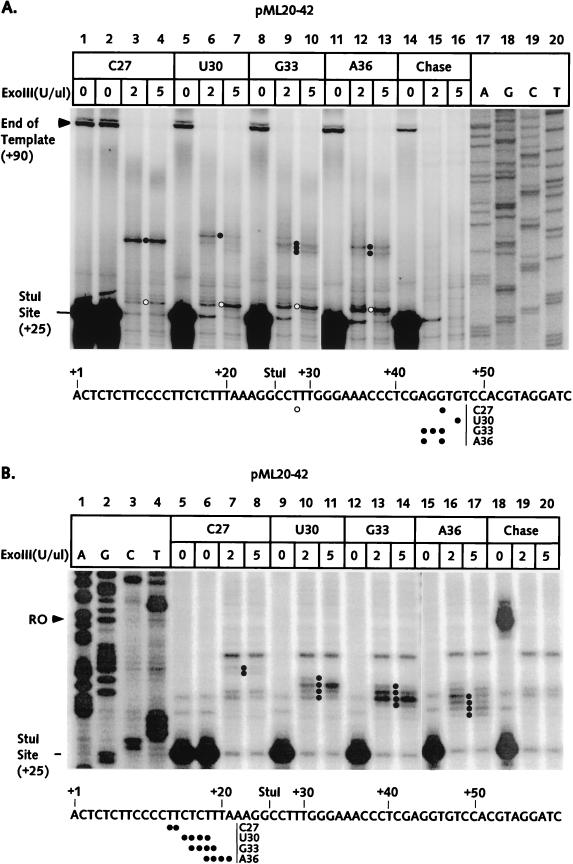
The front-edge boundaries of the C27, U30, G33, and A36 complexes were determined essentially as described above. The results, shown in Fig. 4A, were unanticipated in two respects. First, the front edges for the U30, G33, and A36 complexes were not substantially displaced downstream in comparison with the C27 complex boundary. Also, the boundaries of the U30, G33, and A36 complexes were clearly partitioned between a set of bands at +44 to +49 and a band at +28; this latter band appeared as a minor part of the C27 front-edge pattern as well (lanes 3 and 4, Fig. 4A; see also lane 13 of Fig. 3A). Both the +44 to +49 and the +28 bands were absent in the chase control (lanes 15 to 16). Thus, there appeared to be two populations of RNA polymerases within each of the stalled complexes. The more upstream conformation was the more abundant complex for G33 and A36, even though the last transcribed base is downstream of +28 for each of these complexes. In our earlier studies, the presence of two exoIII footprints for a single transcription complex correlated with a subset of arrested complexes (27). However, the U30, G33, and A36 complexes (Fig. 2B, lanes 8 to 11) did not show a significant population which failed to chase. The upstream exoIII boundaries of the C27, U30, G33, and A36 complexes are shown in Fig. 4B. Unlike the front-edge boundaries, the rear edges were displaced downstream with RNA synthesis, although the extent of this displacement did not match the increase in transcript length. Surprisingly, we failed to detect partitioning between two distinct conformations as we did for the front edges.
FIG. 4.
The exoIII footprints of RNA polymerase II elongation complexes between positions +27 and +36 on the pML20-42 template. The sequence of the nontemplate DNA strand is shown at the bottom of each panel. Numbers in the left margins of both panels indicate distances downstream of the transcription start site. (A) U20 complexes were supplied with ATP, GTP, and CTP to stall polymerase at position +27; the C27 complexes were then gel filtered, treated with StuI, and advanced to the indicated positions by incubating with subset of NTPs. Front-edge boundaries were obtained as described in Materials and Methods. DNAs were resolved on a 6% polyacrylamide gel. The exoIII reactions on C27 complexes chased to the end of the template with all four NTPs are shown in lanes 14 to 16. The front edges of the more downstream conformations (see text) are marked by solid dots and the front edges of the more upstream conformations are indicated by open dots. Exact DNA markers are shown in lanes 17 to 20. (B) The rear-edge boundaries of RNA polymerase elongation complexes were obtained as just described, except that the DNA was labeled at the 5′ end of the template strand. DNAs were resolved on a 10% polyacrylamide gel.
The experimental results from Fig. 4 are summarized in Fig. 7. For clarity, only the more downstream of the U30, G33, and A36 front edges are shown. It seems very unlikely that the transcription complex could adopt a conformation in which it protects less than 10 base pairs of template (which would be the case for the A36 complex with a leading edge at +27 and a trailing edge at +17 to +21). In this context, it is important to note that our footprinting experiment involves 6 min of exposure to the exonuclease. If a particular stalled complex is in true equilibrium between two template locations, progressive exoIII digestion will tend to emphasize the more upstream of the possible front edges and the more downstream of the possible rear edges, thus leading to an apparently shortened footprint. This model does not indicate why we failed to detect a second, more upstream conformation in the rear-edge determination (Fig. 4B). At present, we do not have a good explanation for this discrepancy, but it is worth noting that the upstream exoIII boundaries for the U30, G33, and A36 complexes are quite diffuse. It would be very difficult to detect a second boundary if the signal from this edge were also spread out over many bands.
DNA protection patterns generated by RNA polymerase II elongation complexes stalled between positions +32 and +51.
To generate complexes stalled downstream of position +36 we used the pML20-45, -46 and -47 templates (Fig. 1). We determined the front and back edges of exoIII protection for C32, U35, G38, and A41 complexes on the pML20-45 template and for C37, U40, G43, and A46 complexes on the pML20-46 template. The results of these experiments are summarized in Fig. 7 (primary data are not shown). Most of these complexes showed the predicted exoIII footprint for stalled, transcriptionally competent complexes; that is, 30 to 35 bp of DNA was protected with the last transcribed base located just upstream of the center of the footprint (27). However, the U35 complex on pML20-45 and the U40 complex on pML20-46 did not conform to this pattern. The U35 complex footprint was displaced upstream slightly relative to the footprint of the C32 complex on the same template. Both of the U35 and U40 complexes have three U residues at the 3′ ends of their nascent RNAs. Thus, tight linkage of footprint translocation with RNA synthesis was not achieved even with complexes containing RNAs as long as 40 nt.
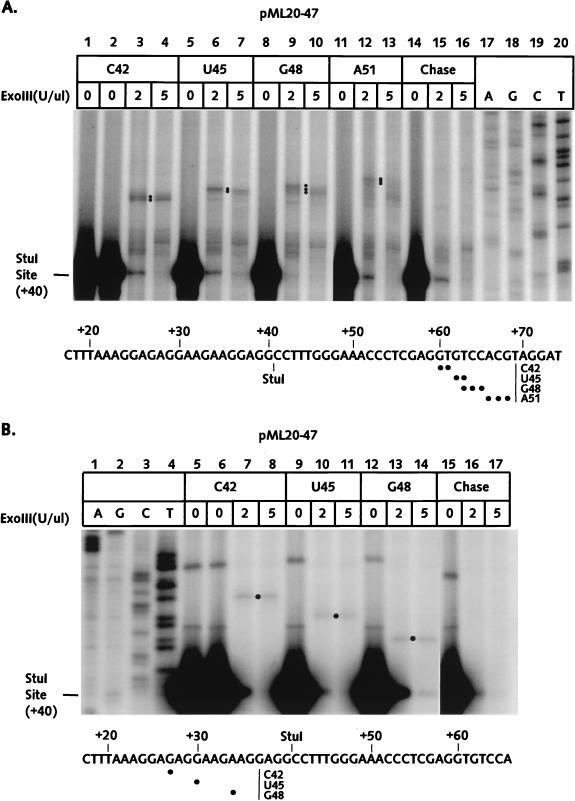
The pML20-47 template was used to produce C42, U45, G48, and A51 complexes. Results of mapping of the front and back edges of these complexes with exoIII are shown in Fig. 5A and B and summarized in Fig. 7. Although we were not able to determine unambiguously the back-edge boundary for the A51 complex, it seems clear that synchrony between RNA synthesis and footprint translocation was observed with this set of four complexes. It is particularly interesting that the U45 complex, unlike complexes U35 and U40, did not show any evidence of upstream translocation.
FIG. 5.
The exoIII footprints of RNA polymerase II complexes stalled between +42 and +51 on the pML20-47 template. The sequence of the nontemplate DNA strand is shown at the bottom of each panel. Numbers in the left margins of both panels indicate distances downstream of the starting point of transcription. (A) Front-edge boundaries of RNA polymerase elongation complexes were obtained as described in Materials and Methods. DNAs were resolved on a 6% polyacrylamide gel. Dots indicate the major boundaries. ExoIII reactions on C42 complexes chased to the end of the template are shown in lanes 14 to 16; exact DNA length markers are shown in lanes 17 to 20. (B) Rear-edge boundaries of RNA polymerase elongation complexes were obtained as just described, except that DNA was labeled at the 5′ end of the template strand. DNAs were resolved on a 10% polyacrylamide gel.
The exoIII protection patterns for RNA polymerase II complexes stalled far downstream of the transcription start site.
Footprint translocation and transcription were tightly linked during far-downstream transcription on pML20-23 (27), but this linkage was lost during transcription of similar DNA sequences in a promoter-proximal location (see Fig. 7). This difference could have resulted from the difference in transcript length between the two sets of complexes. However, the otherwise analogous complexes on the pML20-23 and pML20-40 series templates did not have identical upstream DNA contacts, because of the initial G-free cassette which is present in the pML20-40 series plasmids. In order to assess the role of transcript length without the complication of differences in template sequence, we constructed the pML20-49 template. In this DNA, the initially transcribed segment of pML20-42 was duplicated beginning at +98 in pML20-49. The duplicated regions are separated by a G-free cassette. The sequence of pML20-49 from +98 downstream is shown in Fig. 1.
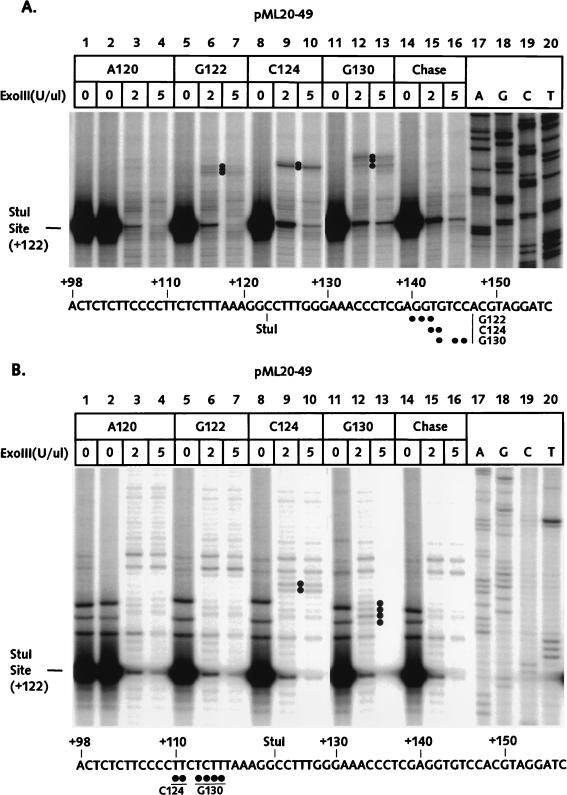
We assembled U20 complexes on the pML20-49 template, Sarkosyl rinsed the complexes, and advanced the RNA polymerases through the G-free cassette to +120 with ATP, UTP, and CTP. After a second round of gel filtration, some of the A120 complexes were walked to G122, C124, or G130. The front- and rear-edge boundaries of these complexes as determined with exoIII are shown in Fig. 6A and B and summarized in Fig. 7. Despite considerable effort, we failed to detect upstream or downstream edges for the A120 complex, although this complex was stable and fully transcriptionally competent (data not shown). Note that the numerous faint bands which appeared in the A120 footprinting reactions (lanes 3 and 4) were also present in the chase control (lanes 15 and 16). The C124 complex has the same underlying template sequence as the C27 complex (Fig. 7), and the front-edge footprints of both complexes were essentially identical. However, there is a dramatic difference in the footprints of the analogous pair of G25 and G122 complexes. The G25 footprint is translocated far upstream, such that the front edge is only 3 bp from the last transcribed base, while the front edge of the G122 complex is 18 to 20 bp downstream from the last transcribed base, as is typical of transcriptionally competent complexes. Finally, the G130 complex also has a typical stalled-complex footprint, while the analogous G33 complex has an abnormally short footprint shifted upstream relative to the last transcribed base. Most significantly, the G130 footprint showed no evidence for a second, far-upstream conformation, in contrast to the G33 complex. Since the template and transcript sequences are identical for at least 26 bases upstream of the last transcribed base for the G25-G122, C27-C124, and G33-G130 complex pairs, we conclude that the upstream translocation of the exoIII footprint for the G25 and G33 complexes cannot be attributed to the sequence context and therefore most probably results from proximity to the transcription start site.
FIG. 6.
ExoIII footprints of RNA polymerase II elongation complexes stalled far from the transcription start site on the pML20-49 template. The sequence of the nontemplate DNA strand is shown at the bottom of each panel. Numbers in the left margins of both panels indicate distances downstream of the transcription start site. (A) Front-edge boundaries were determined as described in Materials and Methods. DNAs were resolved on a 6% polyacrylamide gel. The exoIII reactions on A120 complexes chased to the end of the template with all four NTPs are shown in lanes 14 to 16. Dots mark the positions of the major boundaries; note that no boundary could be detected above the chase background for the A120 complex. Exact DNA length markers are shown in lanes 17 to 20. (B) Rear-edge boundaries of RNA polymerase elongation complexes were obtained as just described, except that the DNA was labeled at the 5′ end of the template strand. DNAs were resolved on a 10% polyacrylamide gel. Boundaries were detected only for the C124 and G130 complexes.
DISCUSSION
We have investigated the movement of RNA polymerase II during early elongation by using a series of complexes stalled between positions +20 and +51. To summarize our central observation, we found that polymerase II transcription complexes pass through a major transition about 25 bases downstream of the transcription start site. In the case of several of the complexes we studied, for example those with 20-, 23-, or 25-nt nascent RNAs, the predominant template location as judged by exoIII footprinting was far upstream of the expected position. Upstream displacement of the footprint is characteristic of arrested transcription complexes (3, 12, 13, 19, 27, 30). Although RNA polymerase II complexes with upstream-displaced footprints are not always arrested (as, for example, we showed in our earlier work [27]; see also Rice et al. [24]), it is nevertheless striking that all of the promoter-proximal complexes we tested were able to elongate their nascent RNAs in 5-min chase reactions with excess NTPs.
In order to put these findings into the context of our current understanding of the mechanism of transcript elongation by RNA polymerase, it is useful to briefly review recent work in this area. Models have been put forward, based primarily on studies of E. coli RNA polymerase, to explain the apparent lack of synchrony of transcription and translocation of the polymerase footprint in certain transcription complexes. In the “sliding clamp” model (reviewed in reference 16), it is envisioned that RNA polymerase is able to translocate backwards along the DNA template as an alternative to making additional phosphodiester bonds. This upstream movement carries the transcription bubble, the RNA-DNA hybrid, and the active site away from the 3′ end of nascent RNA, resulting in a ternary complex which cannot continue transcription (12, 13, 19). The primary driving force for this upstream movement is thought to be the presence of a relatively weak RNA-DNA hybrid at the 3′ end of the transcript, which is replaced by a stronger hybrid via upstream translocation (19, 22). In particular, those complexes with the weakest possible hybrid at the 3′ end (U-A) should be especially prone to upstream translocation. Thus, the sliding clamp model suggests a simple explanation for the nonsynchrony of transcription and footprint translocation in arrested complexes: at arrest sites, RNA polymerase slides upstream to a stable, more energetically favored position. It is this stable upstream location, and not the transient downstream location that immediately preceded arrest, which is detected by the relatively slow footprinting procedure (12, 13). The sliding clamp model treats arrest as a dynamic phenomenon. There is no mechanistic or structural difference between stalled and arrested complexes (such as compression of the arrested polymerase along the template [2, 18]). Arrested complexes cannot restart transcription only because of the particularly unfavorable sequence context near the 3′ end of the RNA (19, 22).
This model envisions arrested complexes as stably occupying upstream template locations. It also predicts the existence of elongation-competent complexes in equilibrium between two or more conformers, with only the most downstream conformation capable of productive elongation (12). Many of our findings support this latter aspect of the sliding clamp model. Partitioning of the RNA polymerase II elongation complex between two locations is clearly illustrated by the complexes stalled at positions 27, 30, 33, and 36 on the pML20-42 template (Fig. 4A). In the C27 complex, the predominant footprint was that expected for a transcriptionally competent polymerase (27), with the transcript 3′ end centrally located. However, as transcription progressed to +30, +33, and +36, two distinct footprints became evident. The more upstream of these had the same front edge as complexes U20, A23, and G25. Thus, we suppose that complexes U30, G33, and A36 are in equilibrium between a relatively stable upstream conformation and a transcriptionally competent downstream conformation. During the 5-min chase period, essentially 100% of the complexes must occupy the downstream conformation for at least the brief time required to make an additional bond, since all of the complexes could resume transcription in 5 min when supplied with excess NTPs. Based on the relative proportion of the two footprints, the upstream conformer represents the predominant structure for the G33 and A36 complexes.
The most dramatic example of the existence of multiple conformers in a transcriptionally competent complex was provided by the A120 complex on the pML20-49 template. Despite repeated attempts, we could not detect any exoIII protection by either the upstream or the downstream edge of this complex. However, the complex was fully active in transcription; indeed, the footprintable G122, C124, and G130 complexes were produced by walking the A120 complex forward with subsets of the NTPs. When the A120 complex was chased with 3′ dGTP to generate a G121 complex, a weak front-edge footprint was observed (data not shown). We interpret the failure to detect any A120 footprint as evidence for many conformations which were relatively stable during the course of the experiment. With the footprint signal spread out over many bands, we presume that it became undetectable above the background.
Other aspects of our results are not easily explained by the sliding clamp model as described above. First, while we clearly observed upstream conformers for many of our complexes, upstream translocation in most of these cases could not have been driven by weak RNA-DNA hybrids. Complexes A23 and G25, for example, both show a single, apparently very stable template location well upstream of the expected position, but the 3′ ends of the RNAs in these complexes are not U rich. What drives upstream translocation for these complexes, and why are they not arrested?
The predominance of the upstream conformation in complexes U20, A23, and G25 must be primarily due to the length of the transcript and not to the template or transcript sequence. This is demonstrated by comparison with analogous complexes assembled on the pML20-49 template. Note that the RNA-DNA hybrids are identical in the G25-G122 and G33-G130 pairs (Fig. 7). However, the complexes stalled on the pML20-49 template did not show any upstream-displaced conformers. Since both sets of complexes were prepared in an identical manner, including Sarkosyl rinsing at position +20, the only difference is the lengths of the nascent RNAs. It is not immediately obvious why promoter-proximal stalling should strongly favor sliding back along the template, an effect which was also observed with E. coli RNA polymerase (18). It is, however, interesting that the first complex to show a stable downstream conformer was C27. The length of RNA which early RNA polymerase II transcription complexes will protect against attack by ribonuclease is about 25 bases (5, 17a). Thus, it is tempting to speculate that filling an RNA binding site or channel is necessary to begin to lock the RNA polymerase into a stable elongation configuration. If this idea is correct, the association of more than 25 bases of RNA with the polymerase must be required to complete the conversion to the elongation-committed form, since a tight linkage between transcript elongation and downstream translocation of the exoIII footprint is not achieved until the nascent RNA is more than 40 bases long (Fig. 7).
It is important to note that while transcript length is an overriding feature in the positioning of early elongation complexes on the template, transcript-template sequence also plays a role. For example, the G38 complex on the pML20-45 template was stalled at almost the same distance downstream of +1 as the C37 complex on the pML20-46 template, but the DNA protection pattern was quite different between these two complexes (Fig. 7). The U35 complex on the pML20-45 template and the U40 complex on the pML20-46 template both had nascent RNAs which end in three U residues, so it is not surprising that the footprints for these complexes were displaced upstream. However, complexes U30 on the pML20-42 template and U45 on the pML20-47 template, which also had three U residues at the end of their RNAs, gave footprints which were not displaced upstream. In this context, the observations of Rice et al. should be noted (24). In that study, DNase I footprints were generated of two elongation-competent RNA polymerase II complexes stalled at G135 and U138. The footprint of the more downstream U138 complex, whose nascent RNA ended with … UUU3′, was slightly upstream of the footprint for the G125 complex. Thus, the exact sequence context must be crucial in determining whether upstream displacement will occur.
Complexes such as G33 and A36, which showed partitioning between upstream and downstream conformers in footprinting, were entirely transcriptionally active when chased. This presumably reflects a very high probability that all of these complexes can occupy the downstream, transcriptionally competent state for at least the time required to make another bond (probably about 0.2 s; see reference 8) during the 5 min of incubation with high levels of NTPs. It is more difficult to understand the transcriptional activity of complexes such as U20, A23, and G25. The only detectable footprints for these complexes show a conformation in which the polymerase has translocated far upstream. This is the same pattern we (27) and others (3, 13, 19) have observed with arrested complexes. If the G25 complex, for example, can occupy a downstream conformation for a sufficient time to resume transcription during a 5-min chase, why do arrested complexes fail to resume elongation under the same conditions? This difference presumably reflects the importance in the arrest process of a U-rich 3′ end on the nascent RNA. We suppose that all complexes which show only upstream conformers by exoIII analysis have some probability of sliding downstream towards the transcriptionally competent configuration. However, the active site in arrested complexes may be prevented from reaching the 3′ end of the RNA because this segment of RNA is U rich in arrested complexes. The weak U-A hybrid would destabilize the complex to an increasing extent as downstream translocation continued, making it very unlikely that the active site would actually reach the 3′ end. In complexes such as G25, this barrier would not exist. Thus, two complexes which both show footprints translocated far upstream of the expected location might nevertheless have very different abilities to resume transcript elongation.
Arrested elongation complexes may cleave RNA as far upstream as 17 bases from the 3′ end when exposed to SII or pyrophosphate (11, 26). The size of the cleavage products correlates with the extent of upstream translocation of the elongation complex, because the catalytic site of RNA polymerase itself is the cleaving agent (26). The increment of SII-mediated cleavage in complexes stalled early in elongation has been addressed in a limited way by earlier work in this laboratory (9, 10). In particular, SII-mediated cleavage in a 20-mer complex (with a sequence very similar to the U20 complex tested here) liberated primarily dinucleotides and a much lower level of large cleavage fragments (10). This result appears to contrast with the footprint of the U20 complex determined in the present study, which shows that complex exclusively in the upstream conformation. To explain this apparent contradiction, we would note that during upstream translocation RNA polymerase should pass through intermediate steps between the most downstream and upstream conformers. If the residence time of RNA polymerase at these intermediate positions is usually sufficient for cleavage to occur, one would generally observe processive cleavage of the transcript from the 3′ end with the release of dinucleotide fragments. As noted above, the weak RNA-DNA hybrids in arrested complexes might cause the most downstream of the translocation intermediates in such complexes to be extremely short lived. Cleavage would therefore be improbable until RNA polymerase reached more upstream locations. Translocation kinetics will not affect the equilibrium distribution between upstream and downstream configurations. Thus, the experimentally observed footprints would look similar for truly arrested complexes and upstream-translocated, but elongation-competent, stalled complexes.
As a final comment, it is important to acknowledge certain technical limitations of our current study. First, all of our exoIII footprinting experiments (those described in both reference 27 and the present work) used RNA polymerases initiated at an adenovirus 2 major late promoter with a G-free initially transcribed region. Thus, we cannot address the role that different promoter elements (such as the TATA box or the initiator) might play in the structural transition from initiation to elongation. Second, it should also be emphasized that we probably removed all of the transcription initiation factors from our ternary complexes by the Sarkosyl rinsing procedure, and thus we cannot know what effect the retention of these factors would have on the results of the footprinting assay. We have not determined the protein content of our Sarkosyl-rinsed complexes directly since the amount of RNA polymerase, for example, would be much too low to detect by staining methods. However, we can infer the absence of initiation factors from a number of considerations. Based on the results of Zawel et al. (31), one would expect that TFIIB and TFIIE, which leave the transcription complex before the formation of the tenth bond, would be missing from all of the complexes we assayed. We can also assume that TFIID is not present in our complexes, since exoIII could not have digested through DNA in the promoter region to give upstream transcription complex boundaries in the vicinity of +1 (Fig. 3B) if the TATA box and surrounding DNA had been tightly complexed with protein. Since TFIIF strongly stimulates elongation by Sarkosyl-rinsed complexes (8), it seems unlikely that such complexes retain significant amounts of TFIIF. Perhaps the most interesting question is whether TFIIH remains in our rinsed complexes; this is a formal possibility, since Zawel et al. (31) showed that (in the absence of Sarkosyl rinsing) TFIIH does not leave the ternary complex until at least 30 bonds are made. One could imagine that the helicase activities in this factor might affect translocation along the template by the RNA polymerase. If this were true, the presence or absence of ATP in the walking-forward procedure after Sarkosyl rinsing should have a significant effect on the footprints. This was not what we observed; for example, complexes U20 and A23 had identical footprints (Fig. 7). Thus, neither the exoIII footprints nor the functional properties of the Sarkosyl-rinsed complexes make us suspect that residual transcription initiation factors were present in these complexes.
In summary, RNA polymerase II elongation complexes stalled early in elongation adopt a stable template location far upstream of the expected position for typical transcriptionally competent complexes. Since upstream translocation is almost certainly a requirement for arrest to occur, complexes which have already taken this step may be more sensitive to other exogenic factors which themselves are not sufficient to force normally elongating RNA polymerase into arrest. There are a number of candidates which might interact with RNA polymerase II in the promoter proximal region and modulate arrest. Further structural studies on the transcription complex will be needed to demonstrate the physiological relevance of these interactions for arrest.
ACKNOWLEDGMENTS
We thank David Setzer, Pieter de Haseth, Richard Gronostajski, Donna Driscoll, and Richard Padgett for advice and encouragement during the course of these studies and Richard Keene for comments on the manuscript.
This research was supported by grant GM 29487 from the National Institutes of Health.
REFERENCES
- 1.Borukhov S, Sagitov V, Goldfarb A. Transcript cleavage factors from E. coli. Cell. 1993;72:459–466. doi: 10.1016/0092-8674(93)90121-6. [DOI] [PubMed] [Google Scholar]
- 2.Chamberlin M J. The Harvey lectures, series 88. New York, N.Y: Wiley-Liss, Inc.; 1995. New models for the mechanism of transcription elongation and its regulation; pp. 1–21. [PubMed] [Google Scholar]
- 3.Gu W, Powell W, Mote J, Jr, Reines D. Nascent RNA cleavage by arrested RNA polymerase II does not require upstream translocation of the elongation complex on DNA. J Biol Chem. 1993;268:25604–25616. [PMC free article] [PubMed] [Google Scholar]
- 4.Gu W, Reines D. Identification of a decay in transcription potential that results in elongation factor dependence of RNA polymerase II. J Biol Chem. 1995;270:11238–11244. doi: 10.1074/jbc.270.19.11238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gu W G, Wind M, Reines D. Increased accommodation of nascent RNA in a product site on RNA polymerase II during arrest. Proc Natl Acad Sci USA. 1996;93:6935–6940. doi: 10.1073/pnas.93.14.6935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guajardo R, Sousa R. A model for the mechanism of polymerase translocation. J Mol Biol. 1997;265:8–19. doi: 10.1006/jmbi.1996.0707. [DOI] [PubMed] [Google Scholar]
- 7.Izban M G, Luse D S. Transcription on nucleosomal templates by RNA polymerase II in vitro: inhibition of elongation with enhancement of sequence-specific pausing. Genes Dev. 1991;5:683–696. doi: 10.1101/gad.5.4.683. [DOI] [PubMed] [Google Scholar]
- 8.Izban M G, Luse D S. Factor-stimulated RNA polymerase II transcribes at physiological elongation rates on naked DNA but very poorly on chromatin templates. J Biol Chem. 1992;267:13647–13655. [PubMed] [Google Scholar]
- 9.Izban M G, Luse D S. The RNA polymerase II ternary complex cleaves the nascent transcript in a 3′→5′ direction in the presence of elongation factor SII. Genes Dev. 1992;6:1342–1356. doi: 10.1101/gad.6.7.1342. [DOI] [PubMed] [Google Scholar]
- 10.Izban M G, Luse D S. SII-facilitated transcript cleavage in RNA polymerase II complexes stalled early after initiation occurs in primarily dinucleotide increments. J Biol Chem. 1993;268:12864–12873. [PubMed] [Google Scholar]
- 11.Izban M G, Luse D S. The increment of SII-facilitated transcript cleavage varies dramatically between elongation competent and incompetent RNA polymerase II ternary complexes. J Biol Chem. 1993;268:12874–12885. [PubMed] [Google Scholar]
- 12.Komissarova N, Kashlev M. RNA polymerase switches between inactivated and activated states by translocating back and forth along the DNA and the RNA. J Biol Chem. 1997;272:15329–15338. doi: 10.1074/jbc.272.24.15329. [DOI] [PubMed] [Google Scholar]
- 13.Komissarova N, Kashlev M. Transcriptional arrest: Escherichia coli RNA polymerase translocates backward, leaving the 3′ end of the RNA intact and extruded. Proc Natl Acad Sci USA. 1997;94:1755–1760. doi: 10.1073/pnas.94.5.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krumm A, Meulia T, Brunvand M, Groudine M. The block to transcriptional elongation within the human c-myc gene is determined in the promoter-proximal region. Genes Dev. 1992;6:2201–2213. doi: 10.1101/gad.6.11.2201. [DOI] [PubMed] [Google Scholar]
- 15.Krummel B, Chamberlin M J. Structural analysis of ternary complexes of Escherichia coli RNA polymerase-deoxyribonuclease I footprinting of defined complexes. J Mol Biol. 1992;225:239–250. doi: 10.1016/0022-2836(92)90918-a. [DOI] [PubMed] [Google Scholar]
- 16.Landick R. RNA polymerase slides home: pause and termination site recognition. Cell. 1997;88:741–744. doi: 10.1016/s0092-8674(00)81919-4. [DOI] [PubMed] [Google Scholar]
- 17.Lis J, Wu C. Protein traffic on the heat shock promoter: parking, stalling, and trucking along. Cell. 1993;74:1–4. doi: 10.1016/0092-8674(93)90286-y. [DOI] [PubMed] [Google Scholar]
- 17a.McKean, D., and D. Luse. Unpublished observations.
- 18.Nudler E, Goldfarb A, Kashlev M. Discontinuous mechanism of transcription elongation. Science. 1994;265:793–796. doi: 10.1126/science.8047884. [DOI] [PubMed] [Google Scholar]
- 19.Nudler E, Mustaev A, Lukhtanov E, Goldfarb A. The RNA-DNA hybrid maintains the register of transcription by preventing backtracking of RNA polymerase. Cell. 1997;89:33–41. doi: 10.1016/s0092-8674(00)80180-4. [DOI] [PubMed] [Google Scholar]
- 20.Orlova M, Newlands J, Das A, Goldfarb A, Borukhov S. Intrinsic transcript cleavage activity of RNA polymerase. Proc Natl Acad Sci USA. 1995;92:4596–4600. doi: 10.1073/pnas.92.10.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rasmussen E B, Lis J T. Short transcripts of the ternary complex provide insight into RNA polymerase II elongational pausing. J Mol Biol. 1995;252:522–535. doi: 10.1006/jmbi.1995.0517. [DOI] [PubMed] [Google Scholar]
- 22.Reeder T C, Hawley D K. Promoter proximal sequences modulate RNA polymerase II elongation by a novel mechanism. Cell. 1996;87:767–777. doi: 10.1016/s0092-8674(00)81395-1. [DOI] [PubMed] [Google Scholar]
- 23.Reines D, Ghanouni P, Li Q Q, Mote J. The RNA polymerase II elongation complex. Factor-dependent transcription elongation involves nascent RNA cleavage. J Biol Chem. 1992;267:15516–15522. [PMC free article] [PubMed] [Google Scholar]
- 24.Rice G A, Chamberlin M J, Kane C M. Contacts between mammalian RNA polymerase II and the template DNA in a ternary elongation complex. Nucleic Acids Res. 1993;21:113–118. doi: 10.1093/nar/21.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rougvie A E, Lis J T. Postinitiation transcriptional control in Drosophila melanogaster. Mol Cell Biol. 1990;10:6041–6045. doi: 10.1128/mcb.10.11.6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rudd M D, Izban M G, Luse D S. The active site of RNA polymerase II participates in transcript cleavage within arrested ternary complexes. Proc Natl Acad Sci USA. 1994;91:8057–8061. doi: 10.1073/pnas.91.17.8057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Samkurashvili I, Luse D S. Translocation and transcriptional arrest during transcript elongation by RNA polymerase II. J Biol Chem. 1996;271:23495–23505. doi: 10.1074/jbc.271.38.23495. [DOI] [PubMed] [Google Scholar]
- 28.Sawadogo M, Roeder R G. Factors involved in specific transcription by human RNA polymerase II: analysis by a rapid and quantitative in vitro assay. Proc Natl Acad Sci USA. 1985;82:4394–4398. doi: 10.1073/pnas.82.13.4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Uptain S M, Kane C M, Chamberlin M J. Basic mechanisms of transcript elongation and its regulation. Annu Rev Biochem. 1997;66:117–172. doi: 10.1146/annurev.biochem.66.1.117. [DOI] [PubMed] [Google Scholar]
- 30.Wang D, Meier T I, Chan C L, Feng G, Lee D N, Landick R. Discontinuous movements of DNA and RNA in RNA polymerase accompany formation of a paused transcription complex. Cell. 1995;81:341–350. doi: 10.1016/0092-8674(95)90387-9. [DOI] [PubMed] [Google Scholar]
- 31.Zawel L, Kumar K P, Reinberg D. Recycling of the general transcription factors during RNA polymerase II transcription. Genes Dev. 1995;9:1479–1490. doi: 10.1101/gad.9.12.1479. [DOI] [PubMed] [Google Scholar]