SUMMARY
Protein structure, function, and evolution depend on local and collective epistatic interactions between amino acids. A powerful approach to defining these interactions is to construct models of couplings between amino acids that reproduce the empirical statistics (frequencies and correlations) observed in sequences comprising a protein family. The top couplings are then interpreted. Here, we show that as currently implemented, this inference unequally represents epistatic interactions, a problem that fundamentally arises from limited sampling of sequences in the context of distinct scales at which epistasis occurs in proteins. We show that these issues explain the ability of current approaches to predict tertiary contacts between amino acids and the inability to obviously expose larger networks of functionally relevant, collectively evolving residues called sectors. This work provides a necessary foundation for more deeply understanding and improving evolution-based models of proteins.
In brief
A current approach for understanding and designing proteins is to make models of epistatic interactions between amino acids from available sequence data comprising a protein family. This work shows that as currently implemented, these models unequally represent the pattern of these interactions. These insights provide a basis for improving next-generation models.
INTRODUCTION
The basic characteristics of natural proteins are the ability to fold into compact three-dimensional structures, to carry out chemical reactions, and to adapt as conditions of selection fluctuate. To understand how these properties are encoded in the amino acid sequence, a powerful approach is statistical inference from datasets of homologous sequences—the study of evolutionary constraints on and between amino acids. In different implementations, this approach has led to the successful prediction of protein tertiary structure contacts,1–4 protein-protein interactions,5–7 mutational effects,8–11 and even the design of synthetic proteins that fold and function in a manner indistinguishable from their natural counterparts.12–14 A major result from these studies is the sufficiency of pairwise correlations in multiple sequence alignments (MSAs) to specify many key aspects of proteins. This result motivates the search for statistical models of protein sequences that capture these correlations as a route to understanding and designing proteins.
What characteristics underlie a “good” statistical model of protein sequences? The native state of a protein represents a fine balance of large opposing forces between atoms that operate with strong distance dependence to produce marginally stable structures. Thus, many complex and non-intuitive patterns of interdependence between amino acids (epistasis) are possible, all consistent with the compact, well-packed character of tertiary structures. Indeed, many studies show that amino acids act heterogeneously and cooperatively within proteins, producing epistasis between amino acids on vastly different scales. At one level, there are local, pairwise interactions that define direct contacts in the tertiary structure. But, at another level, there are collectively acting networks of amino acids that mediate folding13,15,16 and central aspects of protein function—binding,17 catalysis,18 and allosteric communication.19 Past work show that both scales are represented in the pattern of empirical correlations in MSAs20,21 resulting in different sequence-based methods for understanding protein structure22 and function.23 Thus, a basic requirement for statistical models of protein sequences is to account for both local and collective amino acid epistasis.
A fundamental problem in making such models is the lack of a ground truth for validating all features of the inference process. For example, local epistasis can be verified by direct contacts in atomic structures of members of a protein family,1–4 but a similar benchmark for global collective actions of amino acids is not broadly available. Indeed, the inspiration for building statistical models from evolutionary data is, in part, to provide hypotheses for the collective behaviors of amino acids as a route to understanding protein function. How then can we better understand the inference process itself? In this work, we take the approach of using “toy models”24–26 in which we (1) specify a pattern of amino acid couplings for a hypothetical protein, (2) generate synthetic sequences that satisfy those constraints, and (3) examine the ability of statistical inference methods to learn these patterns (Figures 1A and 1B). This analysis shows that in practical contexts, model inference is systematically skewed, given limited sampling of sequences. The consequence is that features of different size and strength are unevenly inferred with current methods. These findings are confirmed in a real protein model system in which experimental data allow us to verify both structural contacts and functional amino acid networks. This work clarifies apparent inconsistencies in the current interpretation of coevolution in proteins and opens a path toward new methods for more completely inferring the information content of protein sequences.
Figure 1. Inference in a toy model of proteins.
The model assumes a sequence of length with possible amino acids at each position.
(A) The pattern of input couplings between sequence positions, in Frobenius norm form. There are three types of features: three isolated pairwise couplings (“contacts,” 2–5, 4–7, and 6–9), a small collective group (“small sector,” all possible couplings within positions 11–14), and a large collective unit (“large sector,” all possible couplings within positions 15–20). All non-zero couplings have the same magnitude, see text.
(B) The strategy used in this work, in which we make the input model (step 1), sample sequences from a Boltzmann distribution defined by the input and compute the empirical first and second order statistics and (step 2), and use the DCA approach to infer back the input couplings from the sampled sequences (step 3).
(C) Frobenius norm of the empirical correlation matrix computed from the sampled sequences, showing that the collective groups are most strongly correlated.
(D) The inferred couplings with usual settings in DCA (regularization ). As described in the text, (A) and (D) show normalized couplings in the zero-sum gauge .
RESULTS
Inference from toy models
A generative statistical model of protein sequences is provided by the direct coupling analysis (DCA),3,22 or more generally a Markov random field. This method starts with a MSA of a protein family comprised of sequences by positions, and makes the assumption that each sequence is a sample from a Boltzmann distribution of a Potts model:
| (Equation 1) |
where represents the intrinsic propensity of each amino acid to occur at each position (the “fields”), represents the constraints between amino acids , at pairs of positions , (the “couplings”), and is the probability of sequence .
The parameters are inferred by maximum likelihood and are related to the frequencies and joint frequencies of amino acids at positions , by the consistency equations
| (Equation 2) |
where when and zero otherwise. The probability distribution can also be viewed as the maximum entropy model that reproduces the empirical frequencies and of the protein family.3 In practice, exact inference of the parameters is computationally intractable because the number of terms in Equation 2 is excessively large, but effective approximations exist. In this work, we use pseudo-likelihood maximization (plmDCA),27 but we show in the STAR Methods that our results are not approximation dependent.
A critical fact is that in nearly every practical situation, the inference is carried out with very limited sampling. Typically, MSAs may contain on the order of N = 103 — 105 sequences, which often reduces to an even lower effective diversity due to phylogenetic relationships (Neff, see STAR Methods). This number of sequences is usually not enough to provide sufficient sampling of the many possible pairwise statistical observations . This undersampling necessitates the use of statistical regularization during the inference process to avoid overfitting. A standard approach is the so-called L2 regularization, meaning that the log-likelihood function is penalized by a term proportional to the L2 norm of the parameters. The larger the regularization, the more constrained the parameters. If the fields and the couplings are regularized separately, this changes the consistency equations to
| (Equation 3) |
where and are the regularization parameters. How does one choose these parameters? Because the inference is unsupervised and cross-validation strategies cannot be applied, the standard approach is to empirically set them by their ability to predict protein properties of interest.9,28
One strategy to assess inference methods is to make use of artificial data generated by a model for which the parameters are known. For example, one can specify an initial model inferred from real data, generate novel sequences from the model, and attempt to re-infer the model from the sampled sequences.29 However, the use of a generative model as a benchmark cannot address features of the real data that were mis-represented or even omitted by choices made in the initial inference process.
To more formally study the influence of sample size and regularization on the inference process, we made a toy model of a hypothetical protein obeying Equation 1 with input parameters , and asked whether these parameters can in fact be inferred from sequences sampled from the model (Figure 1B). The model comprises positions and possible amino acids and has the following characteristics: all fields are set to zero , and couplings have the pattern shown in Figure 1A. There are three isolated pairwise couplings at pairs of positions (2,5), (4,7), and (6,9), a medium-sized interconnected group containing all possible couplings between positions (11–14), and a larger-sized interconnected group containing all possible couplings within positions (15–20). The isolated pairwise couplings mirror the concept of coevolving contacts in protein structures, whereas the interconnected groups of couplings represent the concept of a cooperatively evolving group of positions (sectors). All non-zero couplings have the same strength . We made the choice of setting fields to zero for simplicity but show in the STAR Methods that adding fields leads to a lower effective alphabet per position but does not alter the general conclusions of this work regarding the effects of undersampling. Note that is a four-dimensional array, but for presentation, Figure 1A (and all such panels below) shows the Frobenius norm (see STAR Methods). We also normalize inferred parameters by the input Frobenius value in the zero-sum gauge , so that perfect inference corresponds to for all non-zero couplings.
We used a Markov chain Monte Carlo sampling procedure to draw an MSA of N = 300 independent sequences from the model (Figure 1B, step 2), a number that mirrors the undersampling observed in natural protein families. Figure 1C shows the position by position magnitudes of correlations between amino acids in the sampled sequences. The pattern is heterogeneous, with stronger correlations within the larger interconnected groups of positions. This is because the larger the group, the more constraints exist on it to conform to the motif. In this context, how does DCA work to infer the input couplings from the empirical statistics? With standard settings for3 regularization , DCA emphasizes the isolated pairwise couplings, whereas the collective features are hardly discernible relative to noise (Figure 1D).
Inference as a function of sample size
Why does DCA selectively emphasize the isolated couplings and under-represent those that make up larger collective features? The answer lies in examining the dependence of the inferred couplings on the degree of sampling in the MSA (Figures 2A and 2B). The data in the weakly regularized Figure 2B indicate that inferred couplings show three properties as a function of MSA size: (1) they exhibit a property where the value of sharply peaks at a characteristic MSA size, (2) they peak at different characteristic MSA sizes depending on the size of the group they belong to (non-interacting pairs, isolated pairs, small collective, and large collective units), and (3) they only approach their correct values ( for non-zero couplings) at the limit of very large sampling (large sampling limit shown in Figure S1I). This sharp peak is mitigated by regularization, but persists even at realistic values (Figure 2A). At the MSA size chosen in our example (N = 300), the isolated couplings dominate the inference, with all collective features lower in magnitude. Figure 2B also shows that if the MSA contained more sequences, we could suppress the isolated pairwise couplings and instead emphasize the collective features.
Figure 2. Inference of model features as a function of MSA size.
(A) Normalized magnitude of inferred couplings as a function of MSA size, averaged for positions comprising the different sized features in the input model (Figure 1A). The inference is carried out with the same regularization as in Figure 1D. Sharp increase in the smallest scale can be visible for low sampling sizes. For this value of regularization, even full sampling will not reproduce the input value ( for all interacting position pairs and 0 otherwise).
(B) Same as (A) for a very small value of regularization , to demonstrate the unmitigated effect even more clearly. The data show that features in the amino acid sequence display a sharp peak at characteristic levels of sampling in order of their effective size. Because we use this low value of regularization, interactions of any size can reach their input values, but only at the limit of infinite sampling.
(C) Inferred couplings for an even simpler model of just positions and amino acids either without (black, ) or with (red) input interactions. The traces show cases of deterministic (solid) or random (dashed) sampling of sequences. As described in the text, this model provides a simple mechanistic understanding of the origin of the peak property.
What is the mechanism of the peaking of inferred couplings as a function of MSA size? To study this, we made an even simpler model of just two positions, each with possible amino acids and with no fields or couplings; that is, with no constraints at all. With infinite sampling, all correlations between amino acids at the two positions must be zero and the inference will return the correct result that all fields and couplings are zero. With finite number of sequences, however, the inferred parameters are generally non-zero. For example, consider the situation in which we deterministically draw amino acid pairs uniquely and without repetition to form an MSA of size N while keeping amino acid frequencies at both sites uniform. If , some amino acid pairs will be observed and the rest will be absent, requiring inferred couplings in the Potts model to be infinite to account for the absences. The point of regularization is to prevent such an outcome, constraining the difference between the largest and smallest couplings for the case of this simplified model to satisfy , where is the regularization parameter. It is then easy to show that the magnitude of couplings over all amino acid pairs will be unimodal, with a maximum at the point where the sampling produces the same number of observed and missing pairs—that is, when (Figure 2C, solid black curve, and see STAR Methods for derivation). The true value of the interaction is only reached with complete sampling . This shows the basic mechanism of the peaking—a sampling-dependent maximization of inferred couplings with a magnitude that is simply set by the strength of regularization.
Generalizing to include a non-zero input coupling (red curve in Figure 2C) has the effect of displacing the peak curve to the right and shifting the inferred coupling at large sampling to the correct input value (Figure 2C, compare black and red curves). This makes sense: with stronger coupling, more sampling is generally necessary to draw all possible amino acid states. Thus, as shown in Figure 2B, the position of a peak is a function of the effective size and magnitude of the input interaction. Pure sampling noise in uncoupled positions peaks at the lowest MSA size, followed in sequence by isolated pairwise couplings and collective features of increasing size. Relaxing the model to use random, rather than deterministic sampling of amino acid pairs just further increases the sampling required for inferring couplings, either without (Figure 2C, black dashed curve) or with (Figure 2C, red dashed curve) true interactions.
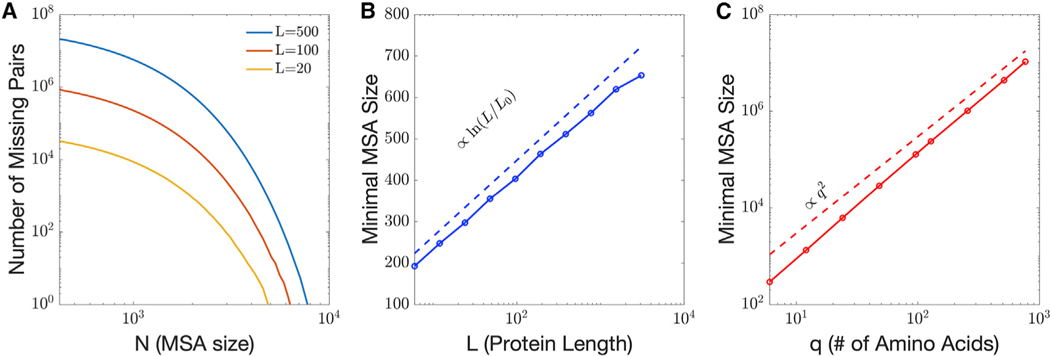
How many sequences are required to avoid the undersampling regime? The minimal MSA size will in general depend on the pattern and strength of constraints. However, a lower bound can be estimated by considering a fully unconstrained model—a model with no input interactions at all (Figure 3). For comparative purposes, one indicator of a lower bound is the number of sequences required to observe every possible pair of amino acids in every pair of positions at least once. Using this measure, we find that the lower bound MSA size scales with sequence length as (Figure 3B) and scales with amino acid alphabet size as (Figure 3C); these scalings are verified for the case of no interactions by analytical calculation (see STAR Methods). With constraints, the MSA size needed to observe all pairs can be orders of magnitude larger, depending on the strength and structure of constraints (see Figures 2 and S4 as examples). For real proteins with a sequence length of 100 or greater, tens of thousands of effective sequences are required just to overcome the lowest possible scale (pure noise associated with non-interacting positions), and many more sequences may be required to fully sample various scales of true interactions (Figure S4). Based on these considerations, we expect that nearly all cases of model inference operate in the undersampled regime.
Figure 3. The undersampled regime for unconstrained models.
(A) Number of missing pairs as a function of MSA size for and various sequence lengths , averaged over 30 realizations, showing that complete sampling for a totally random MSA requires on the order of 104 sequences.
(B) Average minimal MSA size required to avoid the critical undersampling regime in unconstrained models as a function of the sequence length , for . We observe a scaling in ln .
(C) Average minimal MSA size to avoid the critical undersampling regime in unconstrained models as a function of the alphabet size of possible amino acids, for L = 10. We observe a scaling that tends to q2.
The toy model provides another insight into the contact prediction process. A common practice in DCA is to apply an average product correction (APC), which removes a background value from inferred couplings.30 This approach has been justified by its role in mitigating the effects of phylogenetic bias. However, APC also improves the inference of isolated contacts in our toy model, which includes no notion of phylogeny (Figure S5). Our work, therefore, suggests a more general explanation for APC: it works by suppressing the spurious couplings between non-interacting positions that arise due to undersampling in the data. Because, in this limit, the non-interacting couplings are comparable in magnitude to the smallest scale of true couplings between positions (Figure 2), APC helps to separate signal from noise and improve contact prediction in protein structures (Figure S5).
Inference as a function of regularization strength
As explained above, the magnitude of inferred couplings in the undersampled regime is basically set by the strength of the regularization parameter . For example, with typical small , . But how do features of different sizes respond to regularization in the context of undersampling? To understand this, we carried out model inference for a fixed size MSA (N = 300) drawn from the toy model while varying the regularization strength (Figure 4A). The data show that for small regularization, the isolated pairwise couplings dominate (black), and collective features (blue and red) are inferred at or below the level of non-interacting pairs (green). As regularization is increased, different features take prominence, until ultimately features are inferred with magnitudes that are in order of their effective size—large collective > small collective > isolated pairs (Figure 4A). In this strong regularization regime, all inferred couplings decay like and resemble the empirical correlations (see STAR Methods for details). Remembering that the true input couplings are equal for all features and have normalized value , we can conclude that there is no single choice of a regularization parameter that can correctly infer the true pattern of couplings whenever sampling of sequences is limited (compare Figures 4B–4E with Figure 1A).
Figure 4. Inference of couplings as a function of the regularization parameter .
(A) The normalized magnitude of inferred couplings averaged over position pairs comprising isolated pairwise couplings (black), the small sized collective group (blue), and the large-sized collective group (red). Inferred couplings for position pairs with zero input couplings are pure undersampling noise and are shown in green.
(B–E) Values of corresponding to (B)–(E) are marked, and the true value for non-zero couplings is indicated. Note that the sharpness of the peak is influenced by regularization, but is nevertheless present at typical values. (B–E) For comparison with Figure 1A, the matrix inferred at increasing levels of regularization . The data show how features of different effective size dominate the inference as regularization is adjusted from small to large values. Note that DCA is traditionally carried out at small regularization strengths .
An even simpler model with just two features and two parameters provides an intuitive geometrical illustration of the problem (Figure 5). This model comprises sequences with positions and amino acids with a pattern of input interactions shown in Figure 5A. There is one isolated pairwise coupling between positions 1 and 2, and one collective group of couplings between positions 3–6 (Figure 5A), all with the same magnitude . The value of the coupling is chosen simply to be largely above random fluctuations. This makes the number of parameters to be inferred just two—(;)—enabling us to visualize the inference results on a 2D plane (Figure 5B). For an undersampled case (here, N = 4), the contours of the log-likelihood function being optimized (solid blue contours) show that the inference process has no finite maximum; without regularization, inferred values of couplings ; will diverge to infinity. This is consistent with the intuition that couplings must be infinity to account for unobserved amino acid configurations.
Figure 5. A geometrical explanation of regularized inference.
(A) The input coupling matrix for a toy model with positions and amino acids and with no fields h. The model has two parameters, one representing the isolated pairwise coupling (, positions 1 and 2) and the other the couplings in the collective set (, positions 3–6). The input values are .
(B) Inferred values of and from an N = 4 undersampled set of sequences for the toy model as a function of regularization . The solid contours show the landscape of the log-likelihood function being optimized, and the dashed contours show values of that are consistent with different strengths of regularization.
How does regularization correct this problem? The dashed line contours in Figure 5B show the curves along which the magnitude of (that is, ) is a constant for various regularization strengths. This defines the solutions to inference with regularization—the points (black filled circles, Figure 5B) where the solid contours are tangent to the dashed contours. Thus, the inferred solution is set by the regularization used, and there is no regularization at which the inferred solution matches the true solution . Also, note that at this level of undersampling, is always larger than . An analytical solution relating the regularization parameter and inferred values of shows how the ratio of these parameters depends on the relative size of the pairwise and collective units, and on the level of sampling (see STAR Methods).
Application to real problems
These findings have direct impact for model inference in real proteins. The pattern of empirical correlations between pairs of positions in MSAs of protein families reveals a hierarchy of correlation scales, both in terms of magnitude and size of the correlated unit. For example, in an MSA of 1,258 members of the AroQ family of chorismate mutase (CM) enzymes, a subset of more conserved positions display a pattern of strong interconnected correlations and the remainder of less conserved positions show weaker and more dispersed correlations14 (Figure 6A). This pattern is reminiscent of Figure 1C, the correlation matrix resulting from a toy model with features of different effective size. Positions in Figure 6A are ordered by their sensitivity to regularization (see STAR Methods), suggesting that with the undersampling that characterizes practical MSAs, the inference of couplings in Potts models will inevitably treat these groups unequally. Indeed, for the AroQ family inferred with standard weak regularization (, Figure 6B) highlights interactions between mostly unconserved positions with weak correlations, whereas inference with strong regularization (, Figure 6C) highlights interactions between the conserved, more collectively evolving positions (see Figure S6 for intermediate values of regularization). Thus, inference in the context of undersampling selectively represents the information content of protein sequences, with the emphasis of inferred couplings set by the regularization used (see color scale, Figures 6B and 6C).
Figure 6. Inference of positional couplings for the AroQ family of chorismate mutase enzymes.

(A) Positional conservation (by Kullback-Leibler relative entropy,23 bar graph) and the matrix of positional correlations for an MSA of 1,258 CM homologs. The positions are ordered by “sensitivity to regularization” (see text and STAR Methods).
(B and C) The coupling matrix for the CM family inferred with standard small regularization (, B) or strong regularization (, C), both ordered as in (A).
(D) AroQ CMs are dimers with two symmetric active sites formed by elements from both protomers (blue and silver); active site residues are highlighted in yellow stick bonds and a bound substrate analog in magenta. Shown is the structure of the E. coli CM domain (EcCM, PDB: 1ECM).
(E) Spatial organization of positions comprising the top 20 couplings inferred with weak (, blue spheres) or strong (, orange spheres) regularization.
How do these findings influence our understanding of protein structure and function? AroQ CMs occur in bacteria, archaea, plants, and fungi and catalyze the conversion of the intermediary metabolite chorismate to prephenate, a reaction essential for biosynthesis of the aromatic amino acids tyrosine and phenylalanine. Structurally, these enzymes form a compact domain-swapped dimer of relatively small protomers with two active sites (Figure 6D). The top terms in inferred with weak regularization () mainly correspond to direct contacts between amino acids in the three-dimensional structure (Figure S7), but are exclusively located within surface-exposed residues (Figure 6E, blue spheres). In contrast, top couplings inferred with strong regularization represent interactions between buried positions built around the enzyme active site (Figure 6E, orange spheres). The couplings inferred with strong regularization still include many direct tertiary structure contacts, but also comprise indirect, longer-range or substrate-mediated interactions (Figure S8). The key result is that regularization gradually shifts the pattern of inferred couplings from direct contacts at surface sites to a mixture of direct and indirect interactions within the protein core.
What is the functional meaning of these findings? To comprehensively evaluate this, we carried out a saturation single mutation screen (a “deep mutational scan [DMS]”) of the AroQ CM domain from E. coli (EcCM), following the effect on catalytic activity. This work is enabled by a quantitative select-seq assay for CM activity, reported recently.14 Briefly, a library comprising all single mutations was made by oligonucleotide-directed NNS-codon mutagenesis, expressed in a CM-deficient E. coli host strain (KA12/pKIMP-UAUC, see STAR Methods), and grown together as a single population under selective conditions. Deep sequencing of the populations before and after selection provides a log relative enrichment score for each mutant relative to wild type, which quantitatively reports the effect on catalytic power.14 This information is displayed as a heatmap in Figure 7A—a global survey of mutational effects in EcCM.
Figure 7. Functional analysis of positions in the E. coli CM domain.
(A) A deep mutational scan (DMS), showing the effect of every single mutation on the catalytic power relative to wild type (see STAR Methods). Blue shades indicate loss of function, red indicates gain of function, and white is neutral. The illustration above indicates the secondary structure.
(B and C) The distribution of mutational effects displayed for all amino acid substitutions (B) or for the average effect of mutations at each position (C). The data are fit to a Gaussian mixture model with two components (red curve).
(D) The average effect of mutations is shown as a heatmap and circles below mark the positions comprising the deleterious mode in (C) (black), positions comprising the top 20 couplings inferred with weak (, blue) or strong (, yellow) regularization (as in Figure 6), and positions comprising the sector as defined by the SCA method (red).
(E) The sector forms a physically contiguous network within the core of the CM enzyme linking the two active sites across the dimer interface.
(F) Inferred couplings as a function of regularization in the CM protein family. The graph shows the magnitude of inferred couplings averaged over couplings in experimentally functional positions as defined in the figure (red), direct contacts (black), and all other position pairs (light blue). Positions comprising the three groups are defined in the STAR Methods. In analogy with inference for toy models (Figure 4A), these data show that features of different effective size (here, pairwise contacts and interactions in mutationally sensitive positions) differentially dominate the inference as regularization is adjusted from small to large values.
The distribution of mutational effects is bimodal (Figures 7B and 7C), with one mode representing neutral variation and the other representing deleterious effects (black circles, Figure 7D). The comparison with positions inferred in the top couplings of is clear—the top couplings inferred with standard weak regularization occur almost exclusively at mutationally tolerant positions, whereas those inferred with strong regularization occur at functionally important positions (Figure 7D, , Fisher’s exact test) (Figures S7 and S8). Consistent with this, the top couplings inferred with strong regularization significantly overlap with the network of conserved, coevolving positions (the sector) defined by the statistical coupling analysis (SCA) method23 (, Fisher’s exact test) (Figures 7D and 7E).
A systematic analysis of the effect of regularization on inference of positional couplings is shown in Figure 7F. The data show that contacts and functional positions are differentially emphasized, with contacts acting similar to isolated pairwise couplings and functional sites acting similar to a more epistatic collective unit.
Discussion
The inference of coevolution between amino acids has been valuable, providing new hypotheses for protein mechanisms and global rules for design. One approach is based on Potts models, in which empirical frequencies and correlations of amino acids in a MSA are used to define a probability distribution for the protein family over all sequences.22 The Potts model has been demonstrated to reveal pairwise tertiary contacts between amino acids,3,22 opening the path to sequence-based structure prediction.1,4 In this regard, the apparent inability of Potts models to obviously describe collective interactions of amino acids has been puzzling.31 The collective interactions have been shown to specify native-state foldability,13 biochemical activities,10,12,17,18,32,33 allosteric communication,19,34,35 and evolvability,36 defining features of proteins that are essential for their biological function.
The work presented here explains the nature of this problem. With limited sampling of sequences in practically available MSAs, features of different effective size and conservation are differentially emphasized as a function of MSA size and regularization. With weak regularization, the inference focuses on small-scale, relatively unconserved, local interactions. The DMS in CM show that these tend to be functionally less important, local structural contacts. With strong regularization, inference emphasizes larger-scale, conserved features, which in CM are in functionally essential positions. A key point is that there is no single setting of regularization at which all features are correctly represented. In future work, it may be valuable to extend the experimental studies to comprehensive double mutagenesis, an approach that can directly probe the collective action of larger-scale statistical features in proteins.10
One consequence of biased inference is evident in the use of Potts models for protein design. Recent work shows that sequences drawn from a Potts models of chorismate mutate enzymes are indeed true synthetic homologs of the protein family, displaying function both in vitro and in vivo that recapitulates the activity of the natural counterparts.14 However, this result required sampling from the model at computational “temperatures” less than unity, a process that is meant to shift the energy scale to correct for regularization and to enforce under-estimated but functionally essential couplings. This procedure recovers protein function, but does so at the expense of dramatic reduction in sequence diversity of designed proteins compared with natural ones.14 In light of the work presented here, we can now understand this problem as a non-optimal solution to compensating the unequal inference of features by globally depressing the energy scale.
Can we then “correct” the inference process to more uniformly and accurately represent the biologically relevant patterns of amino acid interactions? Given that practical MSAs are usually grossly undersampled, the main parameter we can control is regularization. However, although no single regularization parameter can provide a proper inference for all scales of interactions, it seems clear that what is needed in the Potts model framework is a strategy for inhomogeneous regularization, where parameters in the model are inferred according to the level of sampling noise that acts on them. If done correctly, such a process should lead to a model that unifies the inference of both local and collective features and enables design of artificial proteins that recapitulate the sequence diversity of natural members of a protein family. With insights from the toy models presented here, the availability of powerful experimental systems such as the CMs may provide the foundation for this next advancement in sequence based models for proteins.
STAR★METHODS
RESOURCE AVAILABILITY
Lead Contact
Information and requests for resources and reagents should be directed to the lead contact. Rama Ranganathan (ranganathanr@uchicago.edu).
Materials Availability
This study did not generate new materials.
Data and code availability
Deep mutational scan data have been deposited at the Dryad database and are publicly available as of the data of publication. DOIs are listed in the key resources table.
All original code has been deposited at Zenodo and is publicly available as of the date of publication at. DOIs are listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Bacterial and virus strains | ||
|
| ||
| KA12 strain, Escherichia coli | Kast et al.37 | N/A |
|
| ||
| Biological samples | ||
|
| ||
| Plasmid pKTCTET-0 | Roderer et al.38 | N/A |
| Plasmid pKIMP-UAUC | Kast et al.37 | N/A |
|
| ||
| Deposited data | ||
|
| ||
| Chorismate Mutase Deep Mutational Scan | This work | https://datadryad.org/stash/share/3jzBqewGiS5_SrMrI92C8Dc1FVDmjdtHQ1Qx2Si_trY |
|
| ||
| Software and algorithms | ||
| plmDCA | Ekeberg et al.27 | https://github.com/magnusekeberg/plmDCA |
| ExactDCA and Markov-chain Monte Carlo for sequence generation | This work | https://doi.org/10.5281/zenodo.5919205 |
| bmDCA | Figliuzzi et al.16 | https://github.com/ranganathanlab/bmDCA |
METHOD DETAILS
Toy models and simulated data
Simulated data are generated from input Potts models, with couplings and fields . The inferred couplings and fields are denoted . The input model described in Figures 1, 2, and 3 involves zero fields and couplings with non-zero interactions set to equal strength . This choice makes the pattern of couplings favorable for and to have identical amino acids, excluding frustration. Sequences are generated from input models through a Markov-Chain Monte Carlo process using the Metropolis-Hastings algorithm. Each sample is obtained after Monte Carlo iterations starting from independent random sequences, a value sufficient to reproduce the true input couplings with complete sampling (Figure S1). All codes for creating the MSAs were written in house using MATLAB (Mathworks Inc.) and are available in a dedicated Zenodo repository release.
Inference and Gauge
Exact calculations were used for model inference in the small systems described in Figures 2C, 5, and S1H. The process involves numerical minimization of the negative log likelihood function, with a regularization term
| (Equation 4) |
where is the partition function, with running over the entire space of sequences.
For all other cases involving larger systems, we used the pseudo-likelihood maximization method plmDCA27,39 for approximate inference, with L2 regularization on both fields , and on couplings . The value of is set consistent with past work to be and the values of as indicated in the main text. For inference of Chorismate Mutase MSA, a standard sequence weighing step was added3 to reduce phylogenetic bias. In this step, for each sequence a number of similar (defined as having a Hamming distance less than ) sequences in MSA is calculated, including self. The statistical contribution of that sequence are then reduced by a factor that is the inverse of that number. The sum of these factors for all sequences defines the effective number of sequences, Neff. This value represents the diversity in the MSA, and the evaluation of sufficient sampling, in a more meaningful way than MSA size. For sequences of length and number of amino acids , the couplings and correlations comprise four dimensional arrays, and to represent them in as two-dimensional matrices, we take the Frobenius norm over amino acids, defined by
| (Equation 5) |
For couplings, this projection is gauge dependent and we implement it in the zero-sum (or Ising) gauge, such that
| (Equation 6) |
This gauge minimizes the Frobenius norm over all gauges. Note however that the inferred model is independent of the choice of the gauge. For comparison with input values , the inferred values are represented as which is inferred as 1 for all non-zero couplings, when the inference is well sampled (Figure S1I). For real data treatment, the Frobenius norm sum does not include the “gap” residue, since those are artifacts of the alignment process, however the general results of this paper do not depend on this particular choice.
Comparison between methods of inference
As noted in the main text, the inference of Potts models is computationally intractable for all but the smallest of systems for which exact calculations are possible (exactDCA). The calculations involve the estimation of the marginal , as a function of the model parameters and an exact estimation requires a sum over the space of all possible sequences (see Equation 3 of the main text). Several approximations have been proposed to address this computational problem,22 including the plmDCA method and Botlzmann machine learning (bmDCA).16 In the main text, we use an exact calculation for Figures 2C and 5 and the plmDCA method otherwise. Figure S1 shows comparisons of inference with these various approaches for the same input as in Figure 1(or an equivalent one with ; for exactDCA), demonstrating robustness of the claims in this work to the chosen method of inference. Finally, Figure S1I shows the plmDCA calculation for an MSA size that approaches convergence to full sampling with N = 107, for a small value of regularization, , such that input coupling can be recovered in full. This plot shows that plmDCA and that our Markov Chain Monte Carlo generation process are sufficient to recover the true input constraints with complete sampling.
Choices of interaction strength and structure
The input interactions were chosen to represent patterns of pairwise couplings, medium cooperative interaction units and large cooperative interaction units. The interactions between position pairs are chosen to be “ferromagnetic”, meaning that the occurrence of the same amino acid in both positions is favored. This choice does not limit the generality of the results however, because choosing any other favorable amino acid pair combinations in the absence of frustration would produce the same outcome. One useful consequence of the ferromagnetic case is that the number of amino acid motifs in every interaction regardless of size can be made equal to the number of amino acids . In real proteins, not all interactions will necessarily involve just favorable pairings or will avoid frustration. However, we note that the biases in inference reported here due to undersampling are only enhanced by having less than favorable amino acid pairs (see also next section below); thus, the ferromagnetic case is a conservative choice to illustrate the effects of heterogeneous undersampling noise. A second issue is the strength of couplings in the input model. The most justifiable way to demonstrate the uneven treatment of these three classes of interactions is to have each interaction between residues involved to be of the same magnitude , regardless of the size of unit to which they belong. In this work, we chose a value for interaction strength that simply keeps the smallest-scale feature, pairwise interactions, separable from pure noise. For example, Figure S2 shows that choosing too small a value for will make isolated pairwise interactions invisible to the inference, a choice to be avoided. Above some minimal limit, however, the specific value chosen is not critical for the results of this work.
Effects of positional conservation in the toy model
In this work, we keep the number of favored configurations (the motifs) equal to the number of amino acids , such that all ferromagnetic outcomes occur. But in real proteins, selective pressure on positions (whether due to first- or higher-order constraints) drives the conservation of certain motifs such that some configurations of amino acids are typically not observed within such motifs in MSAs. So, what happens if the number of favorable ferromagnetic pair configurations in our toy model is less than (homogeneously for all interacting position pairs)? Figures S3A–S3C show the case of inference with N = 300 and (that is, same as for Figure 1D), but with input patterns that have varying numbers of favorable motifs in the interactions (8,5,3). The plots show that the smaller the number of motifs the stronger is the effect that we are describing, where inference of true interactions of all scales are close to or below the level of pure noise. The reason is that the relative importance of positions and amino acids, which are not favored increases (because the effective alphabet size of interacting positions decreases; more below) when there are fewer motifs. These findings suggest that the tendency for conservation within larger collectively evolving networks should exacerbate the “invisibility” of of such features in standard inference approaches.
Another key feature of our model is that first-order constraints on positions taken independently (fields ) are set to zero, a simplification to exclusively focus on the inference of pairwise interactions . But then what is the effect of adding first-order constraints on the inference process? We compared a model with coupled pairs not subject to a field to one in which coupled pairs are subject to a non-zero field to out of the possible amino acids. All other parameter choices of the model are otherwise the same as in Figure 1 (inference shown in Figure S3D). The results show that inference in the presence of first-order conservation causes the inferred couplings to be interpreted as weaker. This is due to the Frobenius norm metric being extensive in the alphabet size; thus, with a smaller effective alphabet (Figure S3E), the conserved positions sum to a smaller Frobenius norm; as a consequence the magnitude of inferred couplings are smaller. Another consequence of the smaller alphabet is that it leads to a lesser degree of undersampling; hence the peak of inferred couplings shifts to small MSA size (Figure S3F). In real proteins, available data suggest that the larger collective features are selectively more conserved than the isolated pairwise interactions (see Figure 6A). The findings described here suggest two conclusions: (1) the conservation of larger-scale features serves to exacerbate the inability of Potts model inference to discern these larger-scale features (this contribution is entirely independent of undersampling), and (2) conservation alleviates sampling needs and so any undersampling phenomenon are driven by the strength of epistatic interactions, rather than first-order constraints.
Validity for realistic proteins
Finally, while we perform the calculations in the main paper on a small toy model, with an alphabet of only amino acids and only positions (to be able to observe full sampling for the stronger scales), the validity of the effect extends to larger systems. As an illustration, we performed a similar calculation, for an system with amino acids, where 50 pairwise couplings are chosen at random within the first 80 positions, in addition to cooperative units of size 5 and 8 positions – all with . The results of the inference as a function of MSA size are given in Figure S4 for two values of regularization, showing the peak in the inferred isolated couplings dominating over the other couplings. In these larger proteins, unlike in the smaller toy model, a coupling strength can be chosen even lower than the value we used in the toy models, while still discerning isolated couplings at the standard regularization, such that the non-interacting scale is more substantial and dominates over the rest. In practical cases, the effective alphabet in most interacting positions is lower than . As a result and as mentioned above, we may expect an additional reduction of the various inferred coupling with respect to the undersampling spurious signal in non-interacting positions and at the smaller scales.
Lower bound for minimal sampling
The effects that we describe in the main text arise from undersampling, such that certain combinations of pairs of amino acids are not represented in the data. The typical number of samples needed to overcome this problem depends on the specifics of the generating models; for example, models with stronger constraints, meaning larger couplings and/or larger collective units will require a larger number of samples. This phenomenon lies at the heart of the heterogeneity in sampling noise experienced by features of different effective size in practical multiple sequence alignments. To obtain an analytical expression for the lower bound on sampling, we consider here the least constrained model, which is a null model with no fields or couplings at all, and estimate the mean number of samples needed to observe all possible pairs of amino acids at least once, as a function of the number L of positions and the number of possible amino acids.
The numerical results (Figure 3 of the main text) suggest a scaling with as q2 and a scaling with L as ln L. These scaling relationships can be understood by a rough calculation that treats the combinations of amino acids independently. Starting with just one pair of positions (L = 2), a particular combination (a, b) of amino acids has probability of occurring in any particular sample, and the probability that it is not observed in samples is, therefore, . Treating all combinations of amino acids independently, the probability that one of the combinations is not observed is then . The necessary number of samples needed to observe all combinations of amino acids thus scales with as q2.
Extending the argument to positions under the same simplifying assumption that combinations of amino acids can be treated independently, the total number of combinations becomes and , from which it follows that the required number of sequences scales with L as ln .
As an example, with the model of Figure 2A, where L = 20 ad , the unconstrained model is predicted to require as a lower limit, which is beyond the peak corresponding to pairwise couplings but before the peak corresponding to couplings involved in collective units. For a length and amino acids, this limit corresponds to sequences. This is, however, only a lower bound that assumes a model with no constraints. Constraints can increase the required number of sequence for proper sampling by orders of magnitudes. Figure S4 shows the inference as a function of sample size, the equivalent of Figures 2A and 2B, for an and system. The low regularization peak is close to the predicted lower bound for the unconstrained positions, and slightly higher for the pairwise interactions. The small collective unit is nowhere near its peak even at MSA size N = 105, and so is still deeply undersampled. The more regularized result in Figure S4A shows the effect mitigated, but not removed, preserving artificial skew in favor of the pairwise interactions.
Methods such as flavor reduction,40 which effectively decrease , or pseudo-counting,3 which effectively increase , will alleviate undersampling-induced spurious signals but come with biases of their own. Note also that pseudo-counts are formally equivalent to L2 regularization in the context of Gaussian models.41
Deterministic minimal model for the peak in the undersampled regime
The peak observed in Figure 2B arises in an undersampling regime where the log-likelihood has no extremum, and where the results are entirely determined by the regularization. This is illustrated by the equation shown in the main text, where we consider a minimal model of just L = 2 positions with possible amino acids and no constraint. In order to focus on the desired effect and isolate it from the contribution of sampling stochasticity, we analyze sets of sequences that have a uniform number of each of the amino acids, for any a, b at each position . We further assume that each combination (a, b) is either present or absent in a single sequence for each pair , . This defines a “deterministic” sampling procedure.
In this scenario, the inferred couplings can only take two values, depending on whether the combination of (, ) is observed or not at (, ). Using Equation 3 in the main text, this difference is given by
| (Equation 7) |
Assuming that is small, an expansion for large leads to equation in the beginning of this section and in the main text.
In the Ising gauge, the inferred couplings for the occurring and missing combinations of amino acids are, respectively, and . The Frobenius norm of the couplings is, therefore, maximal when there is the same number of missing and occurring pairs, at . The exact position of the peak is gauge and representation dependent, but the mechanism that leads to a non-monotonic dependence in sampling size is general.
The dependence of the peak on the input coupling can be studied by adding a strong ferromagnetic coupling between the two positions of the model. Most generated sequences then involve the beneficial ferromagnetic combinations of amino acids, which increases the number of samples needed to observe all combinations, and therefore shifts the position of the peak to larger values of sampling size, as seen in Figure 2C (red).
Strong regularization limit
In the strong regularization limit where , we have where is the correlation obtained from the inferred model . Using from Equation 3, we have, therefore,
| (Equation 8) |
and
| (Equation 9) |
In the strong regularization limit, the inferred couplings are, thus, proportional to the correlations, up to the addition of a rank-two correction. This correction is controlled by , the regularization parameter for the fields, and is negligible whenever the model reproduces the first order statistics, i.e., for all i, a.
Two-parameter minimal model
An even simpler model with just two features and two parameters provides an intuitive geometrical illustration of the problem (Figure 5). This model comprises sequences with positions and amino acids with a pattern of input interactions shown in Figure 5A. There is one isolated pairwise coupling between positions 1 and 2, and one collective group of couplings between positions 3–6 (Figure 5A), all with the same magnitude . This makes the number of parameters to be inferred just two, , enabling us to visualize the inference results on a 2D plane (Figure 5B). For a maximally undersampled case (here, N = 4, specifically chosen such that both amino acids are equally represented at every position), the contours of the log-likelihood function being optimized (solid blue contours) show that the inference process has no finite maximum; without regularization, inferred values of couplings , will diverge to infinity. This is consistent with the intuition that couplings must be infinity to account for unobserved amino acid configurations.
How does regularization correct this problem? The dashed line contours in Figure 5B show the curves along which the magnitude of (that is, ) is a constant for various regularization strengths. This defines the solutions to inference with regularization – the points (black filled circles, Figure 5B) where the solid contours are tangent to the dashed contours. Thus, the inferred solution is set by the regularization used, and there is no regularization at which the inferred solution matches the true solution . Also, note that at this level of undersampling, is always larger than . An analytical solution relating the regularization parameter and inferred values of which shows how the ratio of these parameters depends on the relative size of the pairwise and collective units, and on the level of sampling, is derived as follows. below.
We generate a very small data-set of N = 4 sequences with input couplings . We also assert that amino acids are uniformly represented at each position to focus on the inference of the couplings - this is not usually a problem in larger system, but here needs to be chosen as a condition. We typically obtain a data-set where every sequence has the maximal fitness of . To estimate and , consider more generally inferring a common coupling between positions based on the knowledge of a mean fitness given by . The partition function in the low-temperature limit is and for and the regularized log-likelihood function is . Differentiating with respect to and keeping again only leading term in , we thus obtain and for . Applied to our minimal model, this gives
| (Equation 10) |
which directly indicates an inequality and . The ratio can be roughly estimated to be in the limit where goes to zero. Figure 5B indeed indicates an asymptotically linear relationship with .
Average product correction
An average product correction (APC) is routinely used to predict contacts from the inferred couplings ,30 where pairs of positions are not scored by the Frobenius norm but by
| (Equation 11) |
The correction is aimed at removing a background value shared by positions , . The comparison of Figure S5 with Figure 4 shows that APC is indeed effective in enhancing the identification of isolated coupled pairs in the low-regularization limit. On the other hand, APC may not be necessarily useful for identifying large collective units: for instance, Figures S5B and S5C show that APC seems to highlight smaller scale patterns at the expense of larger scale patterns. As described in the main text, our work suggests that APC mainly works by removing spurious signals that arise by the smallest scale features in an alignment, the non-interacting positions. Since isolated positions scale closest to this random signal (Figures 2A and 2B), the APC is primarily effective at contacts prediction.
Multiple sequence alignment
Sequences of the AroQ family were acquired by three rounds of PSI-BLAST42 using residues 1–95 of EcCM (the chorismate mutase (CM) domain of the E.coli CM-prephenate dehrdratase) as the intial query (e-score cutoff 10−4). For alignment, we created a position-specific amino acid profile from 3D alignment of four CM atomic structures (PDB IDs 1ECM, 2D8E, 3NVT, and 1YBZ) and iteratively aligned nearest neighbor sequences from the PSI-BLAST using MUSCLE,43 each time updating the profile. The resulting multiple alignment was subject to minor hand adjustment using standard rules and trimmed sequentially (1) to retain positions present in EcCM, (2) to remove positions with more than 20% gaps, (3) to remove sequences with more than 30% gaps, and to remove excess sequences with more than 90% identity to each other. The final alignment contains 1258 sequences and 89 positions and is available in a dedicated ranganathanlab github repository.
Inference from real data
Figures 6 and 7F present results obtained by inferring a Potts model from a multiple sequence alignment of chorismate mutases (CMs) previously described in,14 using a standard sequence weighting parameter to reduce proximal phylogenetic effects. The inference is performed with plmDCA for different values of while keeping fixed. This value, though important, does not influence the results significantly as long as it is kept sufficiently low.
Coupling matrices with positions ordered along the primary sequences are represented in the top row of Figure S6. In Figures 6B and 6C and the bottom row of Figure S6, the positions are re-ordered to visually emphasize the differences arising from different choices of . The new order is based on a sensitivity to regularization measure, defined by
| (Equation 12) |
where indicates the coupling inferred with and where we compare here two extreme values of . The result in the ordered case shows how the protein positions seemingly decompose into two parts that are analogous to Figure 4, where the collective unit and the isolated pairwise couplings switch their relative importance. Note that if pairwise coupling represented the only signal in the data, we would expect a different picture: as regularization increases, the spurious signal would decrease and separate from true signals but the largest couplings would remain the same as when inferred with low regularization.
The dramatic switching of couplings between different groups of positions strongly argues for the presence of a heterogeneity of scales in real proteins.
Interpretation of top couplings
The top L/2 couplings inferred at low regularization typically represent contacts in three-dimensional structures.3 We examined the top 20 pairs with largest for the CM MSA, excluding pairs that are less than four positions apart along the linear sequence . Figures S7 and S8 represent the positions that contribute to the top 20 pairs with either weak regularization or strong regularization. The data show that inference with weak regularization identifies mainly direct contacts in the tertiary structure of E.coli CM (EcCM, PDB 1ecm) but that inference with strong regularization identifies also indirect or substrate-mediated interactions that are sensitive to mutations. Here a contact is defined as two residues with at least one pair of atoms approaching within 5Å in the crystal structure of EcCM. To examine the relationship of couplings to function, we define “experimentally significant” couplings as those including the 34 positions involved in the deleterious mode in Figure 7C, defined by fitting the data to a Gaussian mixture model. A position is said to be sensitive to mutations based on the findings presented in Figure 7D. This allows us to examine how top pairs found with different levels of regularization are related to CM function. The data show that top pairs obtained in the low-regularization limit correspond in large part to contacts (44%) and not at all to experimentally significant pairs (2%) (Figure S9A; see below for results with an average product correction that leads to a greater number of top pairs that are contacts, as well as an increase in the experimentally functional network). In contrast, 98% of top pairs obtained in the high-regularization limit are experimentally significant couplings (most of which even include both positions as mutationally sensitive 76%, and only 36% are contacts (Figure S9C). These contacts are distinct from those obtained with weak regularization and overlap with sector pairs (Figures 6E and 7E).
Repeating the analysis of Figure S9 with the APC, we verify that contact prediction is significantly improved (Figure S10). In particular, at low regularization, 80% of the top L/2 pairs are contacts (Figure S10A). In contrast, at high regularization, contact prediction is slightly improved , when compared to no APC, but the inference of experimentally significant pairs drops from 98% to 90% (Figure S10C). Note, however, that at high regularization, with or without APC, over 96% of top pairs can be interpreted as contacts or experimentally significant pairs.
In Figure 7F, we define 17 “statistically significant contacts” as top pairs obtained in the low-regularization limit with APC that are in the contact map, but are not part of the functional network positions presented in Figure 7D . Also, we define 32 “statistically significant experimental couplings” as top pairs obtained with strong regularization without APC that are in the functional network, but are not contacts.
Deep mutation library
A saturation single site mutational library for EcCM was constructed using oligonucleotide-directed NNS codon mutagenesis. To mutate each position, two mutageneic oliognucleotides (one sense, one antisense) were synthesized (IDT) that contain sequences complementary to ~ 15 base pairs (bp) on either side of the target position and an NNS codon at the target site (N is a mixture of A,T,C,G bases and S is a mixture of G and C). One round of PCR was carried out with either the sense or antisense oligonucleotide and a flanking antisense or sense primer. A second round amplification with first round products and both flanking primers produced the full-length double-stranded product, which was purified on agarose gel and quantitated using Picogreen (Invitrogen). All first round products were pooled in equimolar ratios, purified, digested with NdeI and XhoI, and ligated into correspondingly digested plasmid pKTCTET-0.38 For selection, the library was transformed into electrocompetent NEB 10-beta cells (NEB) to yield over 1000x transformants per gene, cultured overnight in 500 ml LB supplemented with 100 μg/ml ampicillin (Amp), and subject to plasmid purification. The library was diluted to 1 ng/ml to minimize multiple transformation and transformed into the CM-deficient strain KA12 containing the auxiliary plasmid pKIMP-UAUC37 to yield >1000x transformants per gene. The mixture was then recultured in 500 ml LB containing 100 μg/ml Amp and 30 μg/ml chloramphenicol (Cam) overnight, supplemented with 16% glycerol, and frozen at − 80°C.
Chorismate mutase selection assay
The selection assay followed a recently reported protocol.38 Briefly, glycerol stocks of KA12/pKIMP-UAUC carrying the saturation mutation library in pKTCTET-0 were cultured overnight at 30°C in LB supplemented with 100 μg/ml Amp and 30 μg/ml Cam. The culture was diluted to OD600 of 0.045 in M9c minimal medium38 supplemented with 100 μg/ml Amp, 30 μg/ml Cam, and 20 μg/ml each of L-phenylalanine (F) and L-tyrosine (Y) (M9cFY, non-selective conditions), grown at 30°C to OD600 ~ 0.2, and washed in M9c (no FY). An aliquot of the washed culture was used to inoculate 2 ml LB with 100 μg/ml Amp, and grown overnight at 37°C and harvested for plasmid purification (the pre-selected, or input sample). For selection, another aliquot of the washed culture was diluted to a calculated starting OD600 = 10−4 into 500 ml M9c supplemented with 100 μg/ml Amp, 30 μg/ml Cam, 3 ng/ml doxycycline (to induce CM gene expression from the Ptet promoter) and grown at 30°C for 24h to a final OD600 < 0.1. Fifty ml of the culture was harvested, re-suspended in 2 ml LB with 100 μg/ml Amp, grown overnight at 37°C, and harvested for plasmid purification (the selected sample). Input and selected samples were amplified using two rounds of PCR with KOD polymerase (EMD Millpore) to add adapters and indices for Illumina sequencing. Amplification in the first round included 6–9 random bases to aid initial focusing and part of the i5 or i7 Illumina adapters. The remaining adapter sequenes and TruSeq indicies were added in the second round. PCR was limited to 16 cycles and included high initial tempate concentration to minimize amplification bias. Final products were gel purified (Zymo Research), quantified by Qubit (ThermoFisher), and sequenced on an Illumina MiSeq system with a paired-end 250 cycle kit. Paired-end reads were joined using FLASH, trimmed to the NdeI and XhoI cloning sites and translated. Only exact matches to library variants were counted. Relative enrichments (r.e.) were calculated according to the equation where and represent the frequencies of each allele x in either selected (s) or input i pools and and represent those values for EcCM, the wild-type reference.
Supplementary Material
Highlights.
Direct coupling analysis models unequally represent epistatic patterns within proteins
Model inference is typically done in the limit of extreme undersampling of input data
Show why epistatic features of different sizes and strengths are unequally inferred
Findings are recapitulated in experimental data
ACKNOWLEDGMENTS
We thank M. Weigt, R. Monasson, S. Cocco, F. Zamponi, A.F. Bitbol, Y. Meir, N.S. Wingreen, and members of the Ranganathan and Rivoire laboratories for discussions. This work was supported by grant FRM AJE20160635870 (O.R.), grant ANR 17-CE30-0021-02 (O.R.), NIH grant RO1GM131697 (R.R.), a Data Science Discovery Award from the University of Chicago (R.R.) and a collaboration grant from the France-Chicago Center (R.R. and O.R.).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.cels.2022.12.013.
DECLARATION OF INTERESTS
R.R. is a founder and shareholder of Evozyne Inc. and a member of its corporate board. R.R. is also a member of the scientific advisory board of this journal.
REFERENCES
- 1.Marks DS, Colwell LJ, Sheridan R, Hopf TA, Pagnani A, Zecchina R, and Sander C. (December 2011). Protein 3D structure computed from evolutionary sequence variation. PLoS One 6, e28766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marks DS, Hopf TA, and Sander C. (November 2012). Protein structure prediction from sequence variation. Nat. Biotechnol. 30, 1072–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morcos F, Pagnani A, Lunt B, Bertolino A, Marks DS, Sander C, Zecchina R, Onuchic JN, Hwa T, and Weigt M. (December 2011). Direct-coupling analysis of residue coevolution captures native contacts across many protein families. Proc. Natl. Acad. Sci. USA 108, E1293–E1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ovchinnikov S, Park H, Varghese N, Huang P-S, Pavlopoulos GA, Kim DE, Kamisetty H, Kyrpides NC, and Baker D. (January 2017). Protein structure determination using metagenome sequence data. Science 355, 294–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bitbol A-F, Dwyer RS, Colwell LJ, and Wingreen NS (October 2016). Inferring interaction partners from protein sequences. Proc. Natl. Acad. Sci. USA 113, 12180–12185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gueudré T, Baldassi C, Zamparo M, Weigt M, and Pagnani A. (October 2016). Simultaneous identification of specifically interacting paralogs and interprotein contacts by direct coupling analysis. Proc. Natl. Acad. Sci. USA 113, 12186–12191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cong Q, Anishchenko I, Ovchinnikov S, and Baker D. (July 2019). Protein interaction networks revealed by proteome coevolution. Science 365, 185–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Figliuzzi M, Jacquier H, Schug A, Tenaillon O, and Weigt M. (January 2016). Coevolutionary landscape inference and the context-dependence of mutations in beta-lactamase TEM-1. Mol. Biol. Evol. 33, 268–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hopf TA, Ingraham JB, Poelwijk FJ, Schärfe CPI, Springer M, Sander C, and Marks DS (2017). Mutation effects predicted from sequence co-variation. Nat. Biotechnol. 35, 128–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salinas VH, and Ranganathan R. (July 2018). Coevolution-based inference of amino acid interactions underlying protein function. eLife 7, e34300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng RR, Nordesjö O, Hayes RL, Levine H, Flores SC, Onuchic JN, and Morcos F. (2016). Connecting the sequence-space of bacterial signaling proteins to phenotypes using coevolutionary landscapes. Mol. Biol. Evol 33, 3054–3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Russ WP, Lowery DM, Mishra P, Yaffe MB, and Ranganathan R. (September 2005). Natural-like function in artificial WW domains. Nature 437, 579–583. [DOI] [PubMed] [Google Scholar]
- 13.Socolich M, Lockless SW, Russ WP, Lee H, Gardner KH, and Ranganathan R. (2005). Evolutionary information for specifying a protein fold. Nature 437, 512–518. [DOI] [PubMed] [Google Scholar]
- 14.Russ WP, Figliuzzi M, Stocker C, Barrat-Charlaix P, Socolich M, Kast P, Hilvert D, Monasson R, Cocco S, Weigt M, et al. (July 2020). An evolution-based model for designing chorismate mutase enzymes. Science 369, 440–445. [DOI] [PubMed] [Google Scholar]
- 15.Tian P, Louis JM, Baber JL, Aniana A, and Best RB (2018). Co-evolutionary fitness landscapes for sequence design. Angew. Chem. Int. Ed. Engl 57, 5674–5678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Figliuzzi M, Barrat-Charlaix P, and Weigt M. (2018). How pairwise coevolutionary models capture the collective residue variability in proteins? Mol. Biol. Evol 35, 1018–1027. [DOI] [PubMed] [Google Scholar]
- 17.McLaughlin RN, Poelwijk FJ, Raman A, Gosal WS, and Ranganathan R. (November 2012). The spatial architecture of protein function and adaptation. Nature 491, 138–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Halabi N, Rivoire O, Leibler S, and Ranganathan R. (August 2009). Protein sectors: evolutionary units of three-dimensional structure. Cell 138, 774–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reynolds KA, McLaughlin RN, and Ranganathan R. (December 2011). Hot spots for allosteric regulation on protein surfaces. Cell 147, 1564–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rivoire O. (April 2013). Elements of coevolution in biological sequences. Phys. Rev. Lett 110, 178102. [DOI] [PubMed] [Google Scholar]
- 21.Cocco S, Monasson R, and Weigt M. (August 2013). From principal component to direct coupling analysis of coevolution in proteins: low-eigenvalue modes are needed for structure prediction. PLoS Comp. Biol 9, e1003176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cocco S, Feinauer C, Figliuzzi M, Monasson R, and Weigt M. (January 2018). Inverse statistical physics of protein sequences: a key issues review. Rep. Prog. Phys 81, 032601. [DOI] [PubMed] [Google Scholar]
- 23.Rivoire O, Reynolds KA, and Ranganathan R. (June 2016). Evolution-based functional decomposition of proteins. PLoS Comp. Biol 12, e1004817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jacquin H, Gilson A, Shakhnovich E, Cocco S, and Monasson R. (2016). Benchmarking inverse statistical approaches for protein structure and design with exactly solvable models. PLOS Comp. Biol 12, e1004889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rivoire O. (2019). Parsimonious evolutionary scenario for the origin of allostery and coevolution patterns in proteins. Phys. Rev. E 100, 032411. [DOI] [PubMed] [Google Scholar]
- 26.Bravi B, Ravasio R, Brito C, and Wyart M. (2020). Direct coupling analysis of epistasis in allosteric materials. PLoS Comp. Biol 16, e1007630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ekeberg M, Hartonen T, and Aurell E. (2014). Fast pseudolikelihood maximization for direct-coupling analysis of protein structure from many homologous amino-acid sequences. J. Comp. Phys 276, 341–356. [Google Scholar]
- 28.Ekeberg M, Lövkvist C, Lan Yueheng, Weigt M, and Aurell E. (2013). Improved contact prediction in proteins: using pseudolikelihoods to infer potts models. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 87, 012707. [DOI] [PubMed] [Google Scholar]
- 29.Haldane A, and Levy RM (Mar 2019). Influence of multiple-sequence-alignment depth on potts statistical models of protein covariation. Phys. Rev. E 99, 032405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dunn SD, Wahl LM, and Gloor GB (2008). Mutual information without the influence of phylogeny or entropy dramatically improves residue contact prediction. Bioinformatics 24, 333–340. [DOI] [PubMed] [Google Scholar]
- 31.Anishchenko I, Ovchinnikov S, Kamisetty H, and Baker D. (August 2017). Origins of coevolution between residues distant in protein 3D structures. Proc. Natl. Acad. Sci. USA 114, 9122–9127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Narayanan C, Gagné D, Reynolds KA, and Doucet N. (2017). Conserved amino acid networks modulate discrete functional properties in an enzyme superfamily. Sci. Rep 7, 3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walker AS, Russ WP, Ranganathan R, and Schepartz A. (2020). Rna sectors and allosteric function within the ribosome. Proc. Natl. Acad. Sci. USA 117, 19879–19887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Süel GM, Lockless SW, Wall MA, and Ranganathan R. (2003). Evolutionarily conserved networks of residues mediate allosteric communication in proteins. Nat. Struct. Biol 10, 59–69. [DOI] [PubMed] [Google Scholar]
- 35.Novinec M, Korenč M, Caflisch A, Ranganathan R, Lenarčič B, and Baici A. (2014). A novel allosteric mechanism in the cysteine peptidase cathepsin K discovered by computational methods. Nat. Commun 5, 3287. [DOI] [PubMed] [Google Scholar]
- 36.Raman AS, White KI, and Ranganathan R. (2016). Origins of allostery and evolvability in proteins: a case study. Cell 166, 468–480. [DOI] [PubMed] [Google Scholar]
- 37.Kast P, Asif-Ullah M, Jiang N, and Hilvert D. (May 1996). Exploring the active site of chorismate mutase by combinatorial mutagenesis and selection: the importance of electrostatic catalysis. Proc. Natl. Acad. Sci. USA 93, 5043–5048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roderer K, Neuenschwander M, Codoni G, Sasso S, Gamper M, and Kast P. (December 2014). Functional mapping of protein-protein interactions in an enzyme complex by directed evolution. PLoS One 9, e116234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Balakrishnan S, Kamisetty H, Carbonell JG, Lee SI, and Langmead CJ (2011). Learning generative models for protein fold families. Proteins 79, 1061–1078. [DOI] [PubMed] [Google Scholar]
- 40.Rizzato F, Coucke A, de Leonardis E, Barton JP, Tubiana J, Monasson R, and Cocco S. (2020). Inference of compressed potts graphical models. Phys. Rev. E 101, 012309. [DOI] [PubMed] [Google Scholar]
- 41.Baldassi C, Zamparo M, Feinauer C, Procaccini A, Zecchina R, Weigt M, and Pagnani A. (2014). Fast and accurate multivariate gaussian modeling of protein families: predicting residue contacts and protein-interaction partners. PLoS One 9, e92721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, and Lipman DJ (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 Eprint. https://academic.oup.com/nar/article-pdf/25/17/3389/3639509/25-17-3389.pdf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Edgar RC (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 Eprint. https://academic.oup.com/nar/article-pdf/32/5/1792/7055030/gkh340.pdf. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Deep mutational scan data have been deposited at the Dryad database and are publicly available as of the data of publication. DOIs are listed in the key resources table.
All original code has been deposited at Zenodo and is publicly available as of the date of publication at. DOIs are listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Bacterial and virus strains | ||
|
| ||
| KA12 strain, Escherichia coli | Kast et al.37 | N/A |
|
| ||
| Biological samples | ||
|
| ||
| Plasmid pKTCTET-0 | Roderer et al.38 | N/A |
| Plasmid pKIMP-UAUC | Kast et al.37 | N/A |
|
| ||
| Deposited data | ||
|
| ||
| Chorismate Mutase Deep Mutational Scan | This work | https://datadryad.org/stash/share/3jzBqewGiS5_SrMrI92C8Dc1FVDmjdtHQ1Qx2Si_trY |
|
| ||
| Software and algorithms | ||
| plmDCA | Ekeberg et al.27 | https://github.com/magnusekeberg/plmDCA |
| ExactDCA and Markov-chain Monte Carlo for sequence generation | This work | https://doi.org/10.5281/zenodo.5919205 |
| bmDCA | Figliuzzi et al.16 | https://github.com/ranganathanlab/bmDCA |