Abstract
We have isolated a human RNA polymerase II complex that contains chromatin structure remodeling activity and histone acetyltransferase activity. This complex contains the Srb proteins, the Swi-Snf complex, and the histone acetyltransferases CBP and PCAF in addition to RNA polymerase II. Notably, the general transcription factors are absent from this complex. The complex was purified by two different methods: conventional chromatography and affinity chromatography using antibodies directed against CDK8, the human homolog of the yeast Srb10 protein. Protein interaction studies demonstrate a direct interaction between RNA polymerase II and the histone acetyltransferases p300 and PCAF. Importantly, p300 interacts specifically with the nonphosphorylated, initiation-competent form of RNA polymerase II. In contrast, PCAF interacts with the elongation-competent, phosphorylated form of RNA polymerase II.
Transcription is a key step at which gene expression is regulated. The first step of transcription involves the recognition of promoter DNA sequences and the formation of a transcription initiation complex (54). DNA in the cell is associated with histone proteins to form nucleosomal arrays that are compacted into a highly ordered protein-DNA structure known as chromatin (32). Chromatin has been shown to be a barrier to transcription, as well as to most processes requiring access of proteins to DNA (30, 76). Increasing evidence suggests that the accessibility of factors to DNA packed into chromatin is an important event in the regulation of transcription. Activities that increase the accessibility of proteins to DNA packaged into chromatin have been characterized (70). The Swi-Snf complex can remodel nucleosome structure to facilitate the access of DNA-binding proteins to DNA (34, 58). Other nucleosome-remodeling factors have also been isolated from yeast (RSC [5]), Drosophila (NURF [69], CHRAC [72], and ACF [24]), and human (34, 74) cell extracts. The conservation of these machineries emphasizes the importance of their function.
Direct covalent modification of core histones has also been implicated in the activation of transcription. An increase in histone acetylation correlates with increased levels of transcription (18, 22). The finding that well-studied coactivators, such as Gcn5 (4), p300/CBP (3, 53), and TAFII250 (44), are histone acetyltransferases is consistent with a role for histone acetylation in transcriptional activation. Conversely, the finding that histone deacetylases are components of a Sin3 repressor complex (1, 21, 23, 26, 46, 80) also agrees with the observation that histone acetylation plays a key role in regulating transcription. However, whether histones are the actual and only targets of these acetyltransferases and deacetylases remains to be determined.
Histone acetyltransferases and deacetylases have been demonstrated to interact with sequence-specific transcription factors (1, 13, 21, 23, 26, 46, 73, 80) and with components of the general transcription machinery (25, 73). This suggests a model whereby these activities can be recruited to promoters to alter the acetylation state of promoter-bound histones. This, in turn, would increase or decrease promoter accessibility (18).
RNA polymerase II (RNAPII) is a 12-subunit complex in which the largest subunit contains a carboxy-terminal domain (CTD) composed of a heptapeptide repeat sequence (YSPTSPS) ich in amino acids that can be phosphorylated (78). The phosphorylation of the CTD is highly regulated and can modulate the association of proteins with RNAPII (50, 65). The RNAPII that contains an unphosphorylated CTD (IIA form) is able to form transcription initiation complexes (10, 14, 39). The elongating RNAPII contains a highly phosphorylated CTD (IIO form) (14, 52, 57).
The CTD of RNAPII is essential for viability (2, 79). In yeast, the cold-sensitive growth phenotype associated with partial truncation of the CTD led to the isolation of the SRB (suppressor of RNA polymerase B) family of genes (51). The Srb proteins are associated with RNAPII and a subset of general transcription factors forming an RNAP holoenzyme complex (31). A similar complex, referred to as the mediator, differing slightly in polypeptide composition has also been isolated by using a biochemical approach to search for activities that mediate the response to transcriptional activators (29). This complex not only mediates activated transcription but also increases basal transcription and allows increased phosphorylation of the CTD. Several independent studies have also suggested negative roles for the Srb-mediator complex in transcription (7, 20, 33, 66).
Complexes similar to the yeast RNAPII holoenzyme have been isolated from mammalian cells (9, 40, 56). However, the mammalian complexes show greater heterogeneity than the ones derived from yeast. The mammalian complexes contain a variety of different proteins in addition to Srb homologs and general transcription factors (GTFs) (9, 40, 56), such as proteins involved in recombination and DNA double-strand break repair (40), transcription-coupled repair (71), and RNA processing (such as polyadenylation and splicing factors) (43); elongation factors (56); coactivators mediating the response to Gal4-VP16 (40, 56), Gal4-SP1 (56), and the human immunodeficiency virus type 1 transactivator Tat (12, 68); coactivators that contain histone acetyltransferase activity such as CBP (47); the breast cancer tumor suppressor gene product BRCA1 (62); and RNA helicase A (RHA) (48). As it became clear that chromatin remodeling is an important step in the regulation of transcription, the existence of RNAPII complexes containing activities capable of modifying chromatin was predicted. Indeed, a yeast RNAPII complex associated with the Swi-Snf chromatin remodeling complex has been reported (75). However, the existence of this complex has been controversial and a similar complex has not been detected in mammals.
Here we report the isolation of an RNAPII complex that contains human Srb proteins and factors that modify chromatin structure, including the Swi-Snf complex and the histone acetyltransferases CBP and PCAF.
MATERIALS AND METHODS
Purification of RNAPII complexes.
RNAPII complexes were purified as follows. HeLa cell nuclear extracts (2.3 g) were fractionated on a 500-ml phosphocellulose (Sigma) column which was step eluted with buffer C (20 mM Tris-HCl [pH 7.85], 0.2 mM EDTA, 10 mM β-mercaptoethanol, 10% [vol/vol] glycerol, 0.2 mM phenylmethylsulfonyl fluoride [PMSF]) containing 0.1 M KCl (BC100) (0.8 g of protein), 0.3 M KCl (0.5 g of protein), 0.5 M KCl (0.3 g of protein), and 1.0 M KCl (0.1 g of protein). The 0.5 M KCl eluate was dialyzed against BC100, and proteins were loaded onto a 50-ml DEAE-cellulose (Whatman) column equilibrated with BC100. The flowthrough fraction (also referred to as the 0.1 M KCl eluate; 0.1 g of protein) and the 0.35 M KCl eluates were collected. The 0.35 M KCl fraction (0.2 g of protein) was dialyzed against BC100 and loaded onto a 40-ml S-Sepharose (Sigma) column equilibrated with BC100. This column was developed with 20 column volumes of a linear gradient from 0.1 to 0.8 M KCl in buffer C. Fractions containing RNAPII were pooled and dialyzed against buffer C containing 500 mM KCl, 0.01% (vol/vol) Triton X-100, 0.05% (vol/vol) Nonidet P-40 (NP40), and 20% (vol/vol) glycerol. One-sixth of the pool was loaded onto a CL-4B Sepharose (Pharmacia) gel filtration column (350 ml; 2.5 by 75 cm) equilibrated in the same buffer. Fractions of 1.2 ml were collected and analyzed by Western blot and transcription assays. Blue dextran (Sigma), thyroglobulin (Sigma), and core RNAPII (41) were loaded onto the same column as molecular weight markers. Fractions containing Swi-Snf subunits were pooled, dialyzed against BC100, and further chromatographed on a Mono S fast protein liquid chromatography 5/5 column (Pharmacia). The column was developed with a 10-column-volume linear gradient from 0.1 to 1.0 M KCl in buffer C. Every other fraction was analyzed by Western blotting and for histone acetyltransferase activity.
Western blot analysis.
Blots were incubated with 3% (wt/vol) nonfat dry milk for 1 h at room temperature with shaking. Following three washes with TTBS (10 mM Tris-HCl buffer [pH 7.5], 0.2 M NaCl, 0.05% [vol/vol] Tween 20), blots were incubated with primary antibodies in 0.1% (wt/vol) bovine serum albumin containing TTBS for 2 h at room temperature with gentle agitation. Blots were washed again with TTBS and incubated with secondary antibodies conjugated to alkaline phosphatase (Promega) or horseradish peroxidase (Bio-Rad) for 30 min at room temperature. Blots were washed with TTBS and developed according to the manufacturer’s protocol (Bio-Rad or Boehringer Mannheim).
Immunoprecipitation.
Affinity-purified antibodies (approximately 2 μg) were incubated with 15 μl of protein A-agarose beads (Repligen) for 30 min at room temperature. After a washing with buffer E (20 mM Tris-HCl [pH 7.85], 0.2 mM EDTA, 1 mM dithiothreitol [DTT], 10% [vol/vol] glycerol, 0.2 mM PMSF) containing 0.1 M KCl and 0.05% (vol/vol) NP-40, 100 μl of the DEAE-cellulose-bound fraction was incubated with each antibody-bead complex for 5 h. Immunoprecipitates were collected by centrifugation and washed four times each with 1 ml of buffer E containing 0.4 M KCl and 0.05% (vol/vol) NP-40. Protein complexes were eluted from the beads with 20 μl of Laemmli loading buffer (2% [wt/vol] sodium dodecyl sulfate [SDS], 100 mM DTT, 60 mM Tris-HCl [pH 6.8], 0.001% [wt/vol] bromophenol blue, 10% [vol/vol] glycerol) for Western blot analysis.
Affinity purification of CDK8-containing complexes.
Affinity purification of CDK8-containing complexes was performed with antibodies against CDK8. Affinity-purified anti-CDK8 antibodies were covalently immobilized onto protein A-agarose beads (≈1 mg/ml). Following equilibration in buffer E containing 0.1 M KCl and 0.1% (vol/vol) NP-40, antibody-cross-linked beads (0.5 ml) were incubated with the DEAE-cellulose-bound fraction (0.35 M KCl eluate; 4 mg of protein) at 4°C with rotation. After incubation for 2 h, the beads were washed extensively with buffer E containing 0.7 M KCl and 0.1% (vol/vol) NP-40. Protein complexes were eluted from the column with 2.5 ml of 0.2 M glycine, pH 2.5, in five fractions. Forty microliters of the glycine eluate was used in the silver staining analysis. The beads were also used directly in functional assays.
Supercoiling reduction assay.
Plasmid chromatin was assembled and purified as described previously (55). Each reaction mixture (60 μl) contained chromatin (40 ng; assembled as described in reference 55), topoisomerase I, and 4 mM ATP in buffer (4 mM MgCl2, 2 mM HEPES buffer [pH 7.5], 1 mM DTT, 20 mM KCl). The fraction used in the assay was the affinity-purified CDK8 complex (20 μl of beads), control antibody beads (20 μl), or highly purified Swi-Snf complex (Superose 12 fraction; 10 μl, 0.4 μg). Reaction mixtures were incubated for 30 min at 30°C, and reactions were terminated by the addition of 50 μl of stop mixture (20 mM EDTA [pH 8.0], 0.2 M NaCl, 1% SDS, 0.25 mg of glycogen per ml). Following treatment with proteinase K for 30 min at 37°C, the DNA was extracted with phenol-chloroform-isoamyl alcohol (25:24:1), ethanol precipitated, and resolved by agarose gel electrophoresis. The agarose gel was transferred to a nitrocellulose membrane for Southern hybridization using a 32P-end-labeled probe that hybridizes to the β-galactosidase gene in the backbone plasmid (55). The DNA topoisomers were visualized by autoradiography.
Histone acetyltransferase assay.
The assay was performed in a 50-μl reaction volume containing 100 mM Tris-HCl buffer (pH 8.0), 8% (vol/vol) glycerol, 1 mM DTT, 1 mM PMSF, 10 μM butyric acid, and 0.1 mM EDTA with 130 nCi of [3H]acetyl-coenzyme A and 50 μg of calf thymus core histones and incubated for 30 min at 30°C. Reaction products were spotted onto Whatman P-81 paper, air dried, extensively washed in 50 mM sodium carbonate buffer (pH 9.1), and dried. The amount of [3H]acetate incorporated was determined by liquid scintillation counting.
Transcription assays.
Transcription reaction mixtures were reconstituted as described previously (11). Transcription factors were isolated as described in reference 41.
Protein interaction studies.
M2 affinity beads (Kodak; 10 μl) were incubated for 1 h at 4°C with SF9 cell extracts containing baculovirus-expressed wild-type or truncated PCAF (77) or p300 (53) polypeptides as indicated. The beads were then washed extensively with 1 M KCl and 0.1% (vol/vol) NP-40 in buffer E. Prior to incubation with RNAPII, beads were equilibrated with buffer E containing 0.1 M KCl and 0.1% (vol/vol) NP-40. An RNAPII fraction (1.2 μg; Fig. 5A and B) containing approximately equal amounts of the IIA and IIO forms was mixed with the beads and incubated at 4°C for 1 h with rotation. The beads were washed extensively with buffer E containing 0.1 M KCl and 0.1% (vol/vol) NP-40. The immunoprecipitates were electrophoresed on a 5-to-20%-gradient SDS-polyacrylamide gel and analyzed by Western blotting.
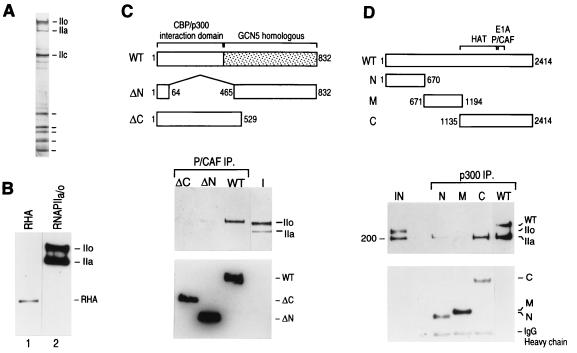
FIG. 5.
(A) Silver-stained gel of core RNAPII containing the phosphorylated (IIo) and nonphosphorylated (IIa) forms of the large subunit that was used in the interaction experiments shown in panels C and D. (B) Western blot analysis reveals the absence of RHA in the RNAPII preparation used in the interaction experiments. Lane 1 shows a Western blot of a crude fraction containing RHA, and lane 2 shows a Western blot of the RNAPII (50% of input) preparation used in the interaction studies. Blots were analyzed for RHA and subsequently reprobed for the largest subunit of RNAPII (IIa and IIo). (C) PCAF directly interacts with the phosphorylated form of RNAPII. Different deletion mutant polypeptides of PCAF (77) were immobilized on beads through their N-terminal FLAG tags and were incubated with a mixture of phosphorylated and nonphosphorylated forms of RNAPII (I; 10% of input). Immunoprecipitates (IP) were extensively washed as described in Materials and Methods and were analyzed by Western blotting using anti-CTD antibodies (top gel) and anti-FLAG antibodies (bottom gel). The diagram above the blots shows the different deletion PCAF proteins that were used in the assay. WT, wild type. (D) p300 directly interacts with the nonphosphorylated form of RNAPII through its C-terminal domain. Experiments were performed as described for panel C. The diagram at the top illustrates the different truncated p300 proteins (53) used. The top blot shows the results of Western blotting with anti-CTD and anti-FLAG antibodies; the bottom blot shows the results of Western blotting with anti-FLAG antibodies. IgG, immunoglobulin G.
RESULTS
Resolution of two RNAPII complexes.
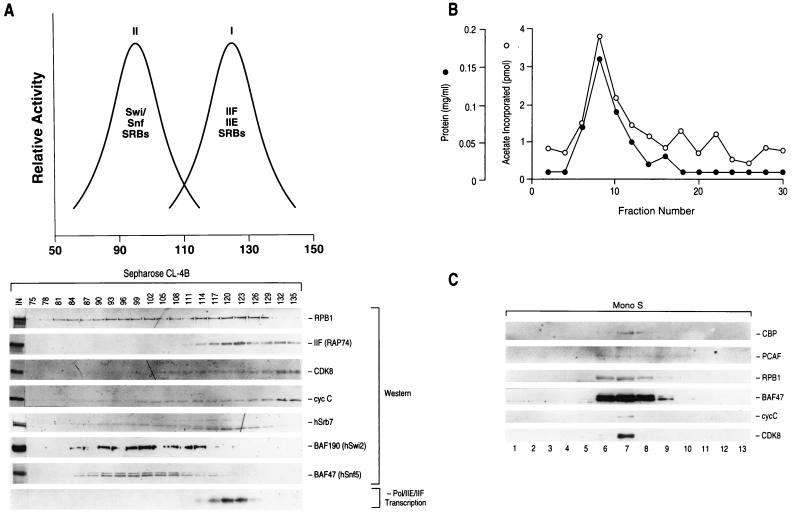
In HeLa cell nuclear extracts, RNAPII exists in large-molecular-weight complexes (40, 56). We previously reported the purification of an RNAPII complex that contains stoichiometric amounts of transcription factors IIE and IIF, substoichiometric amounts of human Srb (hSrb) homologs (Srb7, Srb10, and Srb11), DNA repair proteins, and limiting amounts of TFIIH (11, 40). This complex can efficiently initiate transcription upon the addition of TBP, TFIIB, and TFIIH. Transcription reconstituted with TBP, TFIIB, TFIIH, and fractions derived from a Sepharose CL-4B gel filtration column revealed the presence of an RNAPII complex with a native molecular mass of approximately 1.5 MDa (11) (Fig. 1A). Interestingly, Western blot analysis using antibodies raised against the CTD of RNAPII revealed that a substantial amount of RNAPII eluted with a native molecular mass greater than 2 MDa (Fig. 1A). This RNAPII complex may be similar to the larger RNAPII complex detected by glycerol gradient sedimentation analysis in our previous study (40).
FIG. 1.
(A) Resolution of two different RNAPII complexes by Sepharose CL-4B gel filtration chromatography. The upper panel summarizes the Western blot results shown below. The lower panel shows Western blots of fractions of the Sepharose CL-4B gel filtration column. Blots were probed with antibodies as indicated at the side of each blot. IIF (RAP74), the RAP74 subunit of TFIIF cycC, cyclin C. Input (1%; IN) and fraction numbers are indicated on the top of the lower panel. The last row shows the transcription activity of the column fractions obtained by using the adenovirus major late promoter in a system reconstituted with TBP, TFIIB, and TFIIH. (B) The amount of acetate incorporated into core histones and the protein concentration of each Mono S (see Materials and Methods) column fraction. (C) Coelution of CBP, PCAF, RNAPII (RPB1), BAF47, cyclin C, and CDK8 by Mono S ion exchange chromatography. Fractions from a similar Mono S column were analyzed by Western blotting using antibodies against CBP, PCAF, RPB1, BAF47, cyclin C, and CDK8.
To further characterize this large (>2-MDa)-molecular-mass RNAPII complex, Western blot analyses using antibodies against various proteins were performed (Fig. 1A). This complex was devoid of GTFs but contained the hSrb homologs CDK8 (hSrb10), cyclin C (hSrb11), and hSrb7. In yeast, the Swi-Snf chromatin remodeling complex has been reported to be a component of an RNAPII holoenzyme complex (75). Therefore, we analyzed whether subunits of the human Swi-Snf (74) complex coeluted with RNAPII during gel filtration chromatography. The previously reported 1.5-MDa RNAPII complex containing GTFs did not contain human Swi-Snf components (fractions 117 to 129). However, the Swi-Snf components BAF190 (Swi2-Snf2) and BAF47 (Snf5) coeluted with the higher-molecular-weight RNAPII complex (fractions 87 to 108), suggesting the possibility that the Swi-Snf complex and RNAPII are associated. This Swi-Snf complex and RNAPII appeared to elute with a larger apparent molecular mass than that reported for core RNAPII (0.6 MDa) and for the Swi-Snf complex (2 MDa), suggesting that the Swi-Snf complex we have purified is in association with RNAPII and/or other proteins. These protein fractions were pooled and further fractionated on a Mono-S column. We observed cofractionation of RNAPII, BAF47, and hSrb polypeptides (Fig. 1C; also, see below).
Coimmunoprecipitation of the Swi-Snf complex.
The presence of the Swi-Snf complex in an RNAPII complex has been controversial, identified by one laboratory (75) but not by another (45). Therefore, we further investigated the putative association of this chromatin remodeling factor with the RNAPII holoenzyme by coimmunoprecipitation studies. Immunoprecipitation was performed under highly stringent conditions (0.4 M KCl, 0.05% NP-40) with antibodies against two hSrb proteins. Antibodies against hSrb10 (CDK8) and hSrb7 coimmunoprecipitated RNAPII, as expected (Fig. 2A). Importantly, these antibodies also immunoprecipitated the Swi-Snf complex as detected in a Western blot analysis using antibodies against the BAF47 subunit of the remodeling complex (Fig. 2A). The immunoprecipitation observed was specific, as control antibodies were not capable of coimmunoprecipitating any of the components of the RNAPII complex. This result is consistent with the coelution of RNAPII, hSrbs, and the Swi-Snf complex on multiple chromatographic steps as shown above. Although the stoichiometry of the Swi-Snf complex within the RNAPII complex has not been carefully analyzed, we believe, based on multiple chromatographic steps, that only a subpopulation of the Swi-Snf complex is associated with the RNAPII complex.
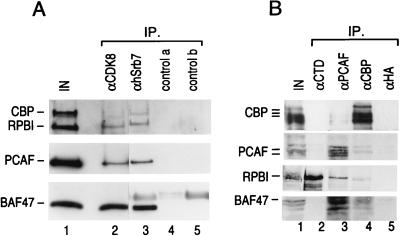
FIG. 2.
(A) Western blot analysis of the immunoprecipitates using antibodies against CDK8 and hSrb7. Immunoprecipitates (IP) of anti-CDK8 (lane 2), anti-Srb7 (lane 3), and control (lanes 4 and 5) antibodies and 10% of the input protein fraction (lane 1) were analyzed by SDS–5 to 12% PAGE. The Western blots were probed for CBP/p300 and RNAPII (top panel), PCAF (middle panel), and Swi-Snf component BAF47 (bottom panel). (B) Western blot analysis of the immunoprecipitates using antibodies against histone acetyltransferases PCAF and CBP. Immunoprecipitates of anti-CTD (lane 2), anti-PCAF (lane 3), anti-CBP (lane 4), and control (lane 5) antibodies and 10% of the input protein fraction (lane 1) were analyzed by SDS–5 to 12% PAGE and Western blot analysis. The Western blots were probed for CBP/p300 (top panel), PCAF (second panel), RNAPII (third panel), and Swi-Snf component BAF47 (bottom panel).
Affinity purification of CDK8 (hSrb10)-cyclin C (hSrb11) containing the Swi-Snf complex.
Because it is difficult to maintain the integrity of large-molecular-weight complexes during multiple chromatographic steps, we devised an affinity purification scheme to isolate the large RNAPII complex. Since our goal was to analyze whether the Swi-Snf complex was associated with the RNAPII complex by an approach different from that described above, antibodies to one of the Srb polypeptides, CDK8 (hSrb10), were used in the affinity purification procedure. The rationale behind choosing human Srb10 for the affinity purification step was based on the copurification and coimmunoprecipitation studies described above, as well as the regulation of the yeast SUC2 gene, which revealed a genetic interaction between Srb10 (and other components of the RNAPII holoenzyme complex) and the Swi-Snf complex (7, 66).
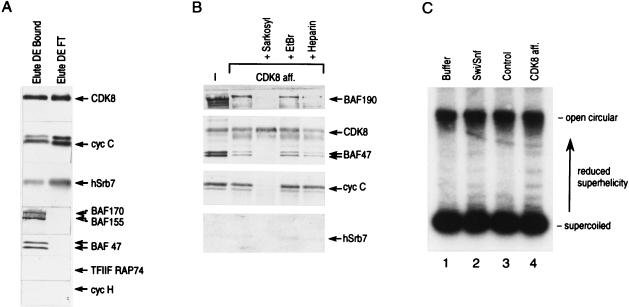
Interestingly, we observed that CDK8 (hSrb10) exists in multiple complexes in HeLa cell nuclear extracts (60). Upon fractionation of HeLa cell nuclear extracts on a phosphocellulose column, the majority of CDK8 and its regulatory subunit, cyclin C (hSrb11), eluted in the 0.1 to 0.3 and 0.3 to 0.5 M KCl fractions (Fig. 3A). The 0.3 to 0.5 M KCl eluate contains most of the GTFs as well as RNAPII and the Swi-Snf complex (Fig. 3A). Therefore, this material was further fractionated on a DEAE-cellulose column. Most of the RNAPII, GTFs, and the Swi-Snf complex bound to this column (Fig. 3A), whereas approximately half of the CDK8-cyclin C complex was found in the flowthrough fraction (see Materials and Methods and the legend to Fig. 3A). The DEAE-cellulose-bound fraction was used as the input sample for affinity chromatography. The affinity purification procedure isolated a CDK8 complex containing Swi-Snf subunits (BAF190, BAF170, BAF155, BAF60, and BAF47), cyclin C, hSrb7, and limiting amounts of RNAPII, as judged by Western blot analysis (Fig. 3B). This complex was specifically associated with CDK8, since none of these polypeptides were retained by the control antibody column (Fig. 3B, lane 4). This affinity-purified complex lacks GTFs (Fig. 3B and data not shown), reminiscent of the larger RNAPII complex purified by conventional chromatography. This was the case even under conditions where lower-concentration salt washes were used in the affinity purification step and RNAPII was more abundant in the complex (data not shown).
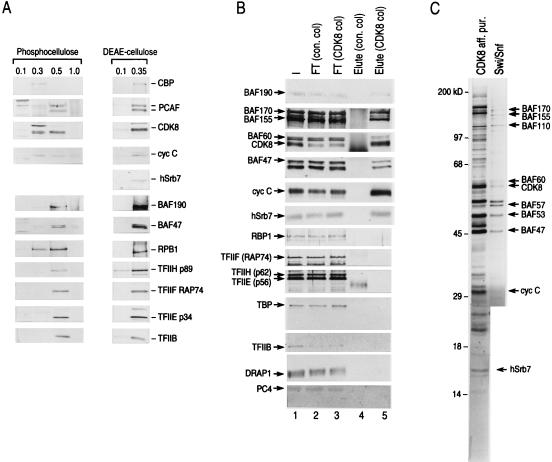
FIG. 3.
(A) Western blot analysis of the fractions derived from phosphocellulose and DEAE-cellulose columns. Western blots of eluates of 0.1 (0.18 mg of protein per ml), 0.3 (0.12 mg of protein per ml), 0.5 (0.08 mg of protein per ml), and 1.0 M KCl (0.16 mg of protein per ml) from a phosphocellulose column (left panel) and 0.1 (0.03 mg of protein per ml) and 0.35 M KCl (0.18 mg of protein per ml) eluates of the DEAE-cellulose column (right panel) derived from the phosphocellulose 0.5 M KCl fraction are shown (see Materials and Methods). Each eluate (20 μl) was loaded on an SDS–5 to 20% polyacrylamide gel for Western blot analysis. Each blot was analyzed for CDK8, cyclin C (cyc C), hSrb7, BAF190, BAF47, the largest subunit of RNAPII (RPB1), the 89-kDa subunit of TFIIH (ERCC3) (TFIIH p89), the large subunit of TFIIF (RAP74), the small subunit of TFIIE (p34), TFIIB, and the histone acetyltransferases CBP and PCAF. (B) Affinity-purified CDK8 complex contains hSrb proteins, Swi-Snf subunits (BAFs), and substoichiometric amounts of RNAPII but not GTFs, the corepressor DRAP1, or the coactivator PC4. Western blots of input (I), flowthrough (FT), and eluate (Elute) of anti-CDK8 (CDK8 col) and control antibody (con. col) columns are shown. Blots were probed for BAF190, BAF170, BAF155, BAF60, BAF47, CDK8, cyclin C, hSrb7, RPB1, TFIIF (RAP74), TFIIH (p62), TFIIE (p56), TBP, TFIIB, DRAP1, and PC4. (C) A silver-stained SDS-polyacrylamide gel containing the affinity-purified CDK8 complex and the Swi-Snf complex shows multiple common polypeptides. A silver-stained gel containing affinity-purified CDK8 complex (left) and Swi-Snf complex (right) was aligned to show common polypeptides (>29 kDa). Another silver-stained gel containing bands from 29 to 10 kDa showing polypeptides derived from the affinity-purified CDK8 complex is at the bottom of the left lane. Polypeptides were identified by Western blot analysis as indicated. Other polypeptides in this fraction which are common to the polypeptides present in the DEAE-cellulose flowthrough fraction-derived affinity-purified CDK8 complex were identified by microsequencing (67a).
Silver staining of the affinity-purified complex revealed the presence of multiple polypeptides, some of which were identified as CDK8, cyclin C, and hSrb7 (Fig. 3C). Importantly, when the migration of the polypeptides in the affinity-purified complex was compared to that of a purified human Swi-Snf complex by SDS-polyacrylamide gel electrophoresis (SDS-PAGE), multiple common polypeptides were observed (Fig. 3C). This result is in agreement with the Western blot analysis demonstrating the presence of Swi-Snf polypeptides in the CDK8 affinity-purified complex (Fig. 3B). The presence of the Swi-Snf polypeptides in this complex appears to be specific and solely mediated through protein-protein interactions because of the following experimental evidence. (i) Fractionation of the DEAE-cellulose flowthrough fraction by CDK8 affinity chromatography resulted in the purification of a complex of approximately 20 polypeptides, most of which were common to the complex described above, yet the Swi-Snf subunits were absent (Fig. 4A and data not shown). (ii) Treatment of the CDK8 affinity-purified complex with high concentrations of ethidium bromide (400 μg/ml) (35) did not affect the association of the Swi-Snf subunits (Fig. 4B, lane + EtBr). (iii) Concentrations of heparin (16 μg/ml) that abolish nonspecific protein-DNA interactions did not affect the integrity of the complex (Fig. 4B, lane + Heparin) lane 5). (iv) However, treatment with 0.1% Sarkosyl disrupted all interactions between CDK8 and other components of the complex without affecting the antigen-antibody interaction (Fig. 4B, lane + Sarkosyl; silver stain, data not shown). This implies that Swi-Snf, cyclin C, and hSrb7 are not bound to the affinity matrix nonspecifically but rather are tethered to the matrix through interactions with CDK8 itself or components of the CDK8 complex. Thus, using three different approaches, conventional chromatography (Fig. 1), coimmunoprecipitation (Fig. 2), and affinity chromatography (Fig. 3), we have demonstrated an association of the Swi-Snf complex with polypeptides known to be subunits of the RNAPII holoenzyme, namely hSrb7, hSrb10, and hSrb11.
FIG. 4.
(A) Comparison of Western blots of affinity-purified CDK8 complexes isolated from the DEAE-cellulose flow-through (Elute DE FT) and DEAE-cellulose-bound (Elute DE Bound) fractions. (B) Heparin or ethidium bromide does not disrupt the interaction of the components of the complex. Western blots of CDK8 complex affinity purified in the absence (second lane from left) or the presence of ethidium bromide (lane + EtBr), heparin (lane + Heparin), and Sarkosyl (lane + Sarkosyl) and input (lane 1; 1%) are shown. Either ethidium bromide (400 μg/ml), heparin (16 μg/ml), or sarkosyl (0.1%) was added as indicated to the DEAE-cellulose-bound fraction during the incubation with the antibody column as well as during the washes. (C) The affinity-purified CDK8 complex alters the linking number of reconstituted plasmid chromatin. Buffer (lane 1), highly purified Swi-Snf (Superose 6 column fraction; lane 2), the control antibody fraction (lane 3), and the affinity-purified CDK8 complex (lane 4) were incubated with plasmid reconstituted into chromatin in vitro (see Materials and Methods). Plasmid DNA was extracted and analyzed for a change in linking number.
The affinity-purified CDK8 complex remodels chromatin structure.
We next analyzed whether the Swi-Snf complex present in the Srb10-containing complex was functional by a plasmid linking number reduction assay. This assay measures the reduction in plasmid linking number in the presence of topoisomerase I that results from a loss of nucleosomes or an alteration of nucleosome structure. The addition of a purified Swi-Snf complex to a chromatinized plasmid resulted in a gradual reduction in the superhelicity of the plasmid compared to that of a plasmid incubated with buffer alone (Fig. 4C, compare lanes 1 and 2). When the CDK8 affinity-purified complex was added to the assay mixture, a pattern similar to that observed with purified Swi-Snf complex was observed. The amounts of Swi-Snf added to the reaction mixtures were similar and were normalized by using BAF47 Western blot units. The supercoiling reduction activity observed was specific to the CDK8-containing complex, as the control antibodies (lane 3) treated in the same manner did not contain such activity. This result demonstrates that the CDK8-containing complex is active in chromatin remodeling.
The RNAPII complex contains histone acetyltransferase activity.
Another activity that can increase chromatin accessibility is the modification of histones by acetylation. Previous studies have reported the presence of CBP within an RNAPII complex (47). Since the complex characterized above contains at least one nucleosome-remodeling activity, we investigated whether histone acetyltransferase activity was also present in the complex. Histone acetyltransferase activity coeluted with the large RNAPII complex in both the gel filtration (data not shown) and Mono-S (Fig. 1B) columns described above. We used Western blot analysis to identify the putative acetyltransferase polypeptides. We found that the histone acetyltransferases p300/CBP (the antibodies used in the Western blot analysis do not distinguish between CBP and p300) and PCAF (77) coeluted with RNAPII, Srb proteins, and the Swi-Snf complex (Fig. 1C). Another histone acetyltransferase, human Gcn5 (6, 77), that contains extensive homology with PCAF in the catalytic domain but that lacks the p300/CBP binding domain (77), was separated in earlier chromatographic steps and, therefore, was not present in the RNAPII complex (data not shown).
To further analyze the association of p300/CBP and PCAF with the RNAPII complex, we performed coimmunoprecipitation experiments using different antibodies. Antibodies against hSrb7 and hSrb10 (CDK8) specifically immunoprecipitated p300/CBP and PCAF (Fig. 2A). Using antibodies against the two histone acetyltransferases detected above, we observed a coimmunoprecipitation of RNAPII and Swi-Snf (Fig. 2B). The immunoprecipitation was specific because the control antibodies failed to immunoprecipitate any polypeptides. More importantly, antibodies against the CTD of the largest subunit of RNAPII immunoprecipitated RNAPII but abolished the coimmunoprecipitation of p300/CBP, PCAF, and Swi-Snf. This result suggested that these proteins are tethered, directly or indirectly, to the RNAPII complex through interactions involving the CTD which are disrupted (competed) by the anti-CTD antibodies.
p300 and PCAF directly interact with different forms of RNAPII.
Previous studies have indicated that CBP is a component of the human RNAPII complex and that its association with RNAPII is mediated by RHA (48). We sought to expand these studies to analyze the specificity and directness of the interaction between RNAPII and the p300/CBP-associated factor, PCAF, by coimmunoprecipitation experiments. We analyzed whether truncated and full-length PCAF proteins could interact with core RNAPII. More importantly, we sought to determine whether histone acetyltransferases p300 and PCAF could interact with both the initiation-competent nonphosphorylated and elongation-competent phosphorylated forms of RNAPII. Antibodies against the tag that is fused to the N terminus of each of the recombinant proteins were used in immunoprecipitation experiments with different forms of RNAPII. The silver staining of an SDS-polyacrylamide gel demonstrates the purity of the core RNAPII used in these experiments (Fig. 5A). Western blot analyses showed that only full-length PCAF can efficiently interact with RNAPII (Fig. 5C). Significantly, PCAF interacts with the phosphorylated, elongating form of RNAPII (Fig. 5C). In contrast to data published by others, we observed a direct interaction between RNAPII and p300 (Fig. 5D). The C terminus of p300 was sufficient for this direct interaction. Moreover, p300 directly and specifically immunoprecipitated the nonphosphorylated form of RNAPII. No interaction with the phosphorylated form of RNAPII could be detected (Fig. 5D). The interaction between p300 and RNAPII was not mediated by RHA, since the RNAPII used in these experiments was free of contaminating RHA as determined by silver staining (Fig. 5A) and Western blot analysis (Fig. 5B). However, our studies do not rule out the possibility that RHA can enhance the interaction between CBP and RNAPII. Our results demonstrate that PCAF and p300 can interact directly with RNAPII. Although PCAF does not interact with the initiating form of RNAPII, it is possible that PCAF is brought into the preinitiation complex through an interaction with p300.
DISCUSSION
We have isolated an RNAPII complex containing factors that modify chromatin structure. Analysis of a gel filtration column reveals the existence of at least two different populations of RNAPII. The first elutes at approximately 1.5 MDa and contains GTFs and Srb homologs, while the second elutes at approximately 4 MDa and contains the Swi-Snf complex, Srb homologs, and histone acetyltransferases PCAF and p300/CBP. Whether these two RNAPII complexes are functionally distinct and carry out the transcription of different subsets of genes, whether they are species engaged in different steps of transcription, or whether they are subassemblies of a larger complex remains to be answered.
Three different RNAPII complexes have been isolated from yeast (7). Koleske and Young isolated the so-called RNAPII holoenzyme complex by using antibodies against different Srb polypeptides and Western blot analyses (31). This RNAPII complex contains the entire set of Srbs (Srb2 and Srb4 to Srb11) and most of the GTFs, except TBP and TFIIE. By contrast, Kim and coworkers (29) isolated the mediator complex by using a functional transcription assay analyzing for factors that mediate the response to transcription activation. The mediator is free of RNAPII and contains Srb2, Srb4 to Srb7, Rox3 (19), Gal11, Sin4, Rgr1 (37), and other polypeptides referred to as Meds (36, 45). This complex is devoid of Srb8 to Srb11. Interestingly, disruption of the SIN4 gene or a truncation of the RGR1 gene allowed the resolution of the mediator into two subcomplexes. One subcomplex is composed of Gal11, Sin4, Rgr1, and Med3 (37, 45), whereas the other subcomplex functionally resembles the mediator but lacks Gal11, Sin4, Rgr1, and Med3 (7). A third RNAPII complex has been described by Shi and coworkers (63). This complex appears to be functionally different from those described by Young and Kornberg as it lacks Srb and Med polypeptides but contains Paf1, Cdc73, Ccr1, and Hpr1 (8, 63).
The kinase-cyclin pair CDK8-cyclin C is the human homolog of the yeast Srb10-Srb11 complex (38). Previous studies have demonstrated the existence of different CDK8-cyclin C complexes in human cells (60). In agreement with these previous observations, we have chromatographically separated different CDK8-containing complexes. Immunoaffinity purification using antibodies against CDK8 demonstrated a tight association of the CDK8-cyclin C complex with the Swi-Snf complex as well as with hSrb7 and other polypeptides. This affinity-purified complex contains a substoichiometric amount of RNAPII. While this represents the first report of such a complex in mammalian cells, Wilson and coworkers demonstrated a physical and functional association between the Swi-Snf complex and the yeast RNAPII holoenzyme complex (75). However, apparently contradictory studies have been reported by Myers and coworkers, who found that the Swi-Snf complex is absent from a purified mediator complex (45). The studies of Carlson and coworkers analyzing the regulation of the SUC2 gene, together with our findings reported here may provide an explanation to the apparent paradox between the studies of Young and Kornberg. Neigeborn and Carlson identified the SNF family of genes based on their requirement for SUC2 expression (49). A subset of the SNF genes encode subunits of the Swi-Snf complex (SNF2, SNF5, SNF6, and SNF11), whereas SNF1 encodes a protein kinase. Components of the RNAPII holoenzyme complex were isolated as suppressors of snf1 mutations (SSN) (33, 66). The SSN genes encoding components of the RNAPII complex include SSN2 (SRB9), SSN3 (SRB10), SSN4 (SIN4), SSN5 (SRB8), SSN7 (ROX3), and SSN8 (SRB11) and are negative regulators of transcription (7, 20). Kornberg and colleagues isolated the mediator complex as an activity that mediates the response to activators by using a functional reconstituted transcription system. Therefore, this may have resulted in the purification of a complex lacking the negative regulator Srb8 to Srb11. The RNAPII complex isolated by Young and coworkers was purified by using antibodies against different Srb polypeptides in Western blot analyses. The findings presented here suggest that the Swi-Snf complex is tethered to the RNAPII complex through interactions mediated by the CDK8-cyclin C-containing complex. Thus, it is possible that the RNAPII complex is a dynamic entity composed of subcomplexes. One subcomplex is the mediator, which is involved in positive regulation; another subcomplex is composed of Sin4, Rgr1, Gal11, and Med3 (37), and another subcomplex includes Srb8 to Srb11 (7). Our studies suggest that the Swi-Snf complex is tethered to the RNAPII complex via an Srb10-Srb11-containing complex. Importantly, we have recently isolated a CDK8-containing complex similar to the one described in these studies but devoid of the Swi-Snf polypeptides. This complex was derived from the DEAE-cellulose flowthrough fraction described in the legend for Fig. 3A and functions to inhibit the response to transcriptional activators (67a). However, a large population of Swi-Snf complexes appears to be devoid of CDK8, judged by coimmunoprecipitation and chromatographic studies.
An important point that must be addressed is the relative abundance of the different RNAPII complexes in the cell. This is difficult to address experimentally, because upon cell disruption the majority of RNAPII (>90%) is found in the insoluble nuclear pellet (41). Extraction of RNAPII from this fraction requires sonication in high-ionic-strength buffer, which disrupts any large RNAPII complexes. Indeed, we and others have previously purified the core RNAPII from this insoluble protein fraction (39, 41, 61). By contrast, we find that the small proportion of RNAPII that remains in the soluble protein fraction upon cell disruption (the nuclear extract) is associated with other proteins in high-molecular-weight complexes (40). We have thus far purified two such RNAPII complexes, one containing GTFs and the other lacking GTFs, but containing the Swi-Snf complex. Based on yields from purification, we estimate that the former complex is more abundant. We also note that the complex we have purified by immunoaffinity chromatography using CDK8 antibodies contains only a small amount of RNAPII. This observation is in agreement with those of Kornberg and colleagues, who isolated a mediator complex containing a limiting amount of RNAPII (15, 27, 28, 45).
The large RNAPII complex was also found to contain two or three histone acetyltransferases, p300 and/or CBP and PCAF. CBP and PCAF have been previously shown to interact with multiple transcriptional activators in various coactivator complexes. During multiple chromatographic procedures, we observed a large population of CBP and PCAF in other complexes. Whether these complexes contain RNAPII or other components of the RNAPII complex has not been determined. Interestingly, thus far no yeast RNAPII complex has been shown to contain histone acetyltransferase activity. In yeast, GCN5, a component of a coactivator complex (adapter), contains histone acetyltransferase activity. GCN5 exists in multimeric complexes termed ADA (17, 42) and SAGA (Spt-Ada-Gcn5 acetyltransferase) (17), which have been separated from the RNAPII complex. Two human Gcn5 homologs, hGcn5 and PCAF, have been isolated. Human Gcn5 was found not to cofractionate with the human RNAPII complexes. There are other nuclear histone acetyltransferases in yeast (59), and it is therefore possible that some may exist in association with the RNAPII holoenzyme complex. In support of this idea are recent experiments demonstrating that recruitment of the RNAPII holoenzyme complex to the PHO5 promoter is sufficient for the displacement of four positioned nucleosomes (16). Importantly, expression of the PHO5 promoter is independent of the Swi-Snf chromatin remodeling complex (16). Therefore, displacement of the four nucleosomes is unlikely to be mediated by the Swi-Snf complex, and it is possible that the RNAPII complex includes histone acetyltransferases or other nucleosome remodeling activities. The actual mechanism remains to be elucidated. These experiments demonstrate, however, that recruitment of the RNAPII holoenzyme complex is sufficient to remove four positioned nucleosomes at the PHO5 promoter in vivo and reveals a functional association of the RNAPII complex with chromatin remodeling activities.
In these studies we have also analyzed the nature of the interaction between RNAPII and the histone acetyltransferases. Our studies revealed that p300 and PCAF interact directly with RNAPII. Using a recombinant glutathione S-transferase–CBP truncated polypeptide, others observed a requirement for RHA for the interaction between CBP and RNAPII (48). The discrepancy between the two studies may be due to the different truncated proteins used. The studies of others used a smaller version of CBP, and it is possible that the RNAPII-interacting domain was removed. However, in support of the studies of Nakajimo and coworkers (48), we have found RHA in our RNAPII complexes (data not shown).
Importantly, we found that each of the acetyltransferases interacts with a different form of RNAPII. p300 was found to interact with the nonphosphorylated form of RNAPII, which is the initiation-competent form of the enzyme. By contrast, PCAF was found to interact with the phosphorylated, elongation-competent form of RNAPII. This suggests that the RNAPII complex may be loaded onto the promoter through interactions mediated by p300 or CBP. These two polypeptides have been shown to interact, in addition to RNAPII, with GTFs, a large number of different sequence-specific DNA-binding proteins, and coactivators (25, 64, 73). p300 or CBP or both are common components of different signal transduction pathways (25). It is possible that upon “activation” of p300/CBP, this large polypeptide interacts with the RNAPII complex. This newly formed complex would contain sequence-specific DNA-binding proteins (those interacting with p300/CBP) and would recruit chromatin remodeling activity and histone acetyltransferase activity to specific genes thereby facilitating DNA binding. Once the full preinitiation complex is formed, RNAPII can initiate transcription. Upon phosphorylation of the CTD, the interactions between RNAPII and factors are disrupted. Only interactions that tolerate this transition or that interact specifically with the phosphorylated form of RNAPII will remain associated with the elongating polymerase (67). This transition may allow PCAF to be transferred from p300/CBP to RNAPII during elongation where it can acetylate nucleosomal histones along the body of the gene.
ACKNOWLEDGMENTS
We thank Edio Maldonado for thoughtful comments and advice during early steps of the work. We also thank G. LeRoy for purifying the Swi-Snf complex, W. Wang and G. Crabtree for human Swi-Snf subunit antibodies, M. Montminy for the CBP antibodies, J. Hurwitz for the RNA antibodies, R. Young for the hSRB7 antibodies, and members of the Reinberg laboratory for helpful suggestions. We also thank Mike Hampsey for helpful comments on the manuscript.
This work was supported by a grant from the National Institutes of Health (GM-37120) and from the Howard Hughes Medical Institute to D.R.
ADDENDUM
Results similar to those reported here were reported by Neish et al. (49a) following submission of the manuscript.
REFERENCES
- 1.Alland L, Muhle R, Hou H, Potes J, Chin L, Schreiber-Agus N, DePinho R. Role for N-CoR and histone deacetylase in Sin3-mediated transcriptional repression. Nature (London) 1997;387:49–55. doi: 10.1038/387049a0. [DOI] [PubMed] [Google Scholar]
- 2.Allison L, Wong J, Fitzpatrick V, Moyle M, Ingles J. The C-terminal domain of the largest subunit of RNA polymerase II of Saccharomyces cerevisiae, Drosophila melanogaster, and mammals: a conserved structure with an essential function. Mol Cell Biol. 1988;8:321–329. doi: 10.1128/mcb.8.1.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bannister A, Kouzarides T. The CBP-co-activator is a histone acetyltransferase. Nature (London) 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 4.Brownell J, Zhou J, Ranalli T, Kobayashi R, Edmondson D, Roth S, Allis D. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 5.Cairns B, Lorch Y, Zhang M, Lacomis L, Erdjument-Bromage H, Tempst P, Du J, Laurent B, Kornberg R. RSC, an essential, abundant chromatin-remodelling complex. Cell. 1996;87:1249–1260. doi: 10.1016/s0092-8674(00)81820-6. [DOI] [PubMed] [Google Scholar]
- 6.Candau R, Moore P, Wang L, Barlev N, Ying C, Rosen C, Berger S. Identification of human proteins functionally conserved with the yeast putative adaptors ADA2 and GCN5. Mol Cell Biol. 1996;16:593–602. doi: 10.1128/mcb.16.2.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carlson M. Genetics of transcriptional regulation in yeast: connections to the RNA polymerase II CTD. Annu Rev Cell Dev Biol. 1997;13:1–23. doi: 10.1146/annurev.cellbio.13.1.1. [DOI] [PubMed] [Google Scholar]
- 8.Chang M, Jaehning J. A multiplicity of mediators: alternative forms of transcription complexes communicate with transcriptional regulators. Nucleic Acids Res. 1997;25:4861–4865. doi: 10.1093/nar/25.24.4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chao D, Gadbois E, Murray P, Anderson S, Sonu M, Parvin J, Young R. A mammalian SRB protein associated with an RNA polymerase II holoenzyme. Nature (London) 1996;380:82–85. doi: 10.1038/380082a0. [DOI] [PubMed] [Google Scholar]
- 10.Chestnut J, Stephens J, Dahmus M. The interaction of RNA polymerase II with the adenovirus-2 major late promoter is precluded by phosphorylation of the C-terminal domain of subunit IIa. J Biol Chem. 1992;267:10500–10506. [PubMed] [Google Scholar]
- 11.Cho H, Maldonado E, Reinberg D. Affinity purification of a human RNA polymerase II complex using monoclonal antibodies against transcription factor IIF. J Biol Chem. 1997;272:11495–11502. doi: 10.1074/jbc.272.17.11495. [DOI] [PubMed] [Google Scholar]
- 12.Cujec T P, Cho H, Maldonado E, Meyer J, Reinberg D, Peterlin B M. The human immunodeficiency virus transactivator Tat interacts with the RNA polymerase II holoenzyme. Mol Cell Biol. 1997;17:1817–1823. doi: 10.1128/mcb.17.4.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Currie R. NF-Y is associated with the histone acetyltransferases GCN5 and P/CAF. J Biol Chem. 1998;273:1430–1434. doi: 10.1074/jbc.273.3.1430. [DOI] [PubMed] [Google Scholar]
- 14.Dahmus M. Reversible phosphorylation of the C-terminal domain of RNA polymerase II. J Biol Chem. 1996;271:19009–19012. doi: 10.1074/jbc.271.32.19009. [DOI] [PubMed] [Google Scholar]
- 15.Flanagan P, Kelleher R, Sayre M, Tshochner H, Kornberg R. A mediator required for activation of RNA polymerase II transcription in vitro. Nature (London) 1991;350:436–438. doi: 10.1038/350436a0. [DOI] [PubMed] [Google Scholar]
- 16.Gaudreau L, Schmid A, Blaschke D, Ptashne M, Horz W. RNA polymerase II holoenzyme recruitment is sufficient to remodel chromatin at the yeast PHO5 promoter. Cell. 1997;89:55–62. doi: 10.1016/s0092-8674(00)80182-8. [DOI] [PubMed] [Google Scholar]
- 17.Grant P, Duggan L, Cote J, Roberts S, Brownell J, Candau R, Ohba R, Owen-Hughes T, Allis D, Winston F, Berger S, Workman J. Yeast Gcn5 functions in two multiprotein complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- 18.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature (London) 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 19.Gustafsson C, Myers L, Li Y, Redd M, Lui M, Erdjument-Bromage H, Tempst P, Kornberg R. Identification of Rox3 as a component of mediator and RNA polymerase II holoenzyme. J Biol Chem. 1997;272:48–50. doi: 10.1074/jbc.272.1.48. [DOI] [PubMed] [Google Scholar]
- 20.Hampsey M. Molecular genetics of the RNA polymerase II general transcriptional machinery. Microbiol Mol Biol Rev. 1998;62:465–503. doi: 10.1128/mmbr.62.2.465-503.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hassig C, Fleischer T, Billin A, Schreiber S, Ayer D. Histone deacetylase activity is required for full transcriptional repression by mSin3A. Cell. 1997;89:341–347. doi: 10.1016/s0092-8674(00)80214-7. [DOI] [PubMed] [Google Scholar]
- 22.Hebbes T, Clayton A, Thorne A, Crane-Robinson C. A direct link between core histone acetylation and transcriptionally active chromatin. EMBO J. 1994;7:1395–1402. doi: 10.1002/j.1460-2075.1988.tb02956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heinzel T, Lavinsky R, Mullen T, Soderstrom M, Laherty C, Torchia J, Yang W-M, Brard G, Ngo S, Davie J, Seto E, Eisenman R, Rose D, Glass C, Rosenfeld M. A complex containing N-CoR, mSin3A and histone deacetylase mediates transcriptional repression. Nature (London) 1997;387:43–48. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- 24.Ito T, Bulger M, Pazin M, Kobayashi R, Kadonaga J. ACF, an ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell. 1997;90:145–155. doi: 10.1016/s0092-8674(00)80321-9. [DOI] [PubMed] [Google Scholar]
- 25.Janknecht R, Hunter T. Transcription. A growing coactivator network. Nature (London) 1996;383:22–23. doi: 10.1038/383022a0. [DOI] [PubMed] [Google Scholar]
- 26.Kadosh D, Struhl K. Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor Rpd3 histone deacetylase to target promoters. Cell. 1997;89:365–371. doi: 10.1016/s0092-8674(00)80217-2. [DOI] [PubMed] [Google Scholar]
- 27.Kelleher R, Flanagan P, Kornberg R. A novel mediator between activator proteins and the RNA polymerase II transcription apparatus. Cell. 1990;61:1209–1215. doi: 10.1016/0092-8674(90)90685-8. [DOI] [PubMed] [Google Scholar]
- 28.Kelleher R, Flanagan P, Chasman D, Ponticelli A, Struhl K, Kornberg R. Yeast and human TFIIDs are interchangeable for the response to acidic transcriptional activators in vitro. Genes Dev. 1992;6:296–304. doi: 10.1101/gad.6.2.296. [DOI] [PubMed] [Google Scholar]
- 29.Kim Y, Bjorklund S, Li Y, Sayre M, Kornberg R. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- 30.Kingston R, Bunker C, Imbalzano A. Repression and activation by multiprotein complexes that alter chromatin structure. Genes Dev. 1996;10:905–920. doi: 10.1101/gad.10.8.905. [DOI] [PubMed] [Google Scholar]
- 31.Koleske A, Young R. An RNA polymerase II holoenzyme responsive to activators. Nature (London) 1994;368:466–469. doi: 10.1038/368466a0. [DOI] [PubMed] [Google Scholar]
- 32.Kornberg R. Structure of chromatin. Annu Rev Biochem. 1977;46:931–954. doi: 10.1146/annurev.bi.46.070177.004435. [DOI] [PubMed] [Google Scholar]
- 33.Kuchin S, Yeghiayan P, Carlson M. Cyclin-dependent protein kinase and cyclin homologs SSN3 and SSN8 contribute to transcriptional control in yeast. Proc Natl Acad Sci USA. 1995;92:4006–4010. doi: 10.1073/pnas.92.9.4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kwon H, Imbalzano A, Khavari P, Kingston R, Green M. Nucleosome disruption and enhancement of activator binding by a human SWI/SNF complex. Nature (London) 1994;370:477–481. doi: 10.1038/370477a0. [DOI] [PubMed] [Google Scholar]
- 35.Lai J-S, Herr W. Ethidium bromide provides a simple tool for identifying genuine DNA-independent protein associations. Proc Natl Acad Sci USA. 1992;89:6958–6962. doi: 10.1073/pnas.89.15.6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee Y C, Min S, Gim B S, Kim Y-J. A transcriptional mediator protein that is required for activation of many RNA polymerase II promoters and is conserved from yeast to humans. Mol Cell Biol. 1997;17:4622–4632. doi: 10.1128/mcb.17.8.4622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Y, Bjorklund S, Jiang Y, Kim Y-J, Lane W, Stillman D, Kornberg R. Yeast global transcriptional regulators Sin4 and Rgr1 are components of mediator complex/RNA polymerase II holoenzyme. Proc Natl Acad Sci USA. 1995;92:10864–10868. doi: 10.1073/pnas.92.24.10864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liao S, Zhang D, Jefferey D, Koleske A, Thompson C, Chao D, Viljoen M, van Vuure H, Young R. A kinase-cyclin pair in the RNA polymerase II holoenzyme. Nature (London) 1995;374:193–196. doi: 10.1038/374193a0. [DOI] [PubMed] [Google Scholar]
- 39.Lu H, Flores O, Weinman R, Reinberg D. The nonphosphorylated form of RNA polymerase II preferentially associates with the preinitiation complex. Proc Natl Acad Sci USA. 1991;88:10004–10008. doi: 10.1073/pnas.88.22.10004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maldonado E, Shiekhattar R, Sheldon M, Cho H, Drapkin R, Rickert P, Lees E, Anderson C, Linn S, Reinberg D. A human RNA polymerase II complex associated with SRB and DNA-repair proteins. Nature (London) 1996;381:86–89. doi: 10.1038/381086a0. [DOI] [PubMed] [Google Scholar]
- 41.Maldonado E, Drapkin R, Reinberg D. Purification of human RNA polymerase II and general transcription factors. Methods Enzymol. 1996;174:72–100. doi: 10.1016/s0076-6879(96)74009-0. [DOI] [PubMed] [Google Scholar]
- 42.Marcus G, Silverman N, Berger S, Horiuchi J, Guarente L. Functional similarities and physical association between Gcn5 and Ada2-putative transcriptional adaptors. EMBO J. 1994;13:4807–4815. doi: 10.1002/j.1460-2075.1994.tb06806.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McCracken S, Fong N, Yankulov K, Ballantyne S, Pan G, Greenblatt J, Patterson S, Wickens M, Bentley D. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature (London) 1997;385:357–361. doi: 10.1038/385357a0. [DOI] [PubMed] [Google Scholar]
- 44.Mizzen C, Yang X-J, Kokubo T, Brownell J, Bannister A, Owen H, Workman J, Wang L, Berger S, Kouzarides T, Nakatani Y, Allis D. The TAFII250 subunit of TFIID has histone acetyltransferase activity. Cell. 1996;87:1261–1270. doi: 10.1016/s0092-8674(00)81821-8. [DOI] [PubMed] [Google Scholar]
- 45.Myers L, Gustafsson C, Bushnell D, Lui M, Erdjument-Bromage H, Tempst P, Kornberg R. The Med proteins of yeast and their function through the RNA polymerase II carboxy-terminal domain. Genes Dev. 1998;12:45–54. doi: 10.1101/gad.12.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nagy L, Kao H-Y, Chakravarti D, Lin R, Hassig C, Ayer D, Schreiber S, Evans R. Nuclear receptor repression is mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell. 1997;89:373–380. doi: 10.1016/s0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]
- 47.Nakajima T, Uchida C, Anderson S, Parvin J, Montminy M. Analysis of a cAMP-responsive activator reveals a two-component mechanism for transcription induction via signal-dependent factors. Genes Dev. 1997;11:738–747. doi: 10.1101/gad.11.6.738. [DOI] [PubMed] [Google Scholar]
- 48.Nakajima T, Uchida C, Anderson S, Lee C-G, Hurwitz J, Parvin J, Montminy M. RNA helicase A mediates association of CBP with RNA polymerase II. Cell. 1997;90:1107–1112. doi: 10.1016/s0092-8674(00)80376-1. [DOI] [PubMed] [Google Scholar]
- 49.Neigeborn L, Carlson M. Genes affecting the regulation of SUC2 gene expression by glucose repression in Saccharomyces cerevisiae. Genetics. 1984;108:845–858. doi: 10.1093/genetics/108.4.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49a.Neish A, Anderson S, Wei B S W, Parvin J. Factors associated with the mammalian RNA polymerase II holoenzyme. Nucleic Acids Res. 1998;26:847–853. doi: 10.1093/nar/26.3.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Neugebauer K, Roth M. Transcription units as RNA processing units. Genes Dev. 1997;11:3279–3285. doi: 10.1101/gad.11.24.3279. [DOI] [PubMed] [Google Scholar]
- 51.Nonet M, Young R. Intragenic and extragenic suppressors of mutations in the heptapeptide repeat domain of Saccharomyces cerevisiae RNA polymerase II. Genetics. 1989;123:715–724. doi: 10.1093/genetics/123.4.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.O’Brien T, Hardin S, Greenleaf A, Lis J. Phosphorylation of RNA polymerase II C-terminal domain and transcriptional elongation. Nature (London) 1994;370:75–77. doi: 10.1038/370075a0. [DOI] [PubMed] [Google Scholar]
- 53.Ogryzko V, Schiltz L, Russanova V, Howard B, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 54.Orphanides G, Lagrange T, Reinberg D. The general transcription factors of RNA polymerase II. Genes Dev. 1996;10:2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- 55.Orphanides G, LeRoy G, Chang C, Luse D, Reinberg D. FACT, a factor that facilitates transcript elongation through nucleosomes. Cell. 1998;92:105–116. doi: 10.1016/s0092-8674(00)80903-4. [DOI] [PubMed] [Google Scholar]
- 56.Pan G, Aso T, Greenblatt J. Interaction of elongation factors TFIIS and Elongin A with a human RNA polymerase II holoenzyme capable of promoter-specific initiation and responsive to transcriptional activators. J Biol Chem. 1997;272:24563–24571. doi: 10.1074/jbc.272.39.24563. [DOI] [PubMed] [Google Scholar]
- 57.Payne J, Laybourn P, Dahmus M. The transition of RNA polymerase II from initiation to elongation is associated with phosphorylation of the carboxyl-terminal domain of subunit IIa. J Biol Chem. 1989;264:19621–19629. [PubMed] [Google Scholar]
- 58.Peterson C, Tamkun J. The SWI-SNF complex: a chromatin remodeling machine? Trends Biochem Sci. 1995;20:143–146. doi: 10.1016/s0968-0004(00)88990-2. [DOI] [PubMed] [Google Scholar]
- 59.Reifsnyder C, Lowell J, Clarke A, Pillus L. Yeast SAS silencing genes and human genes associated with AML and HIV-1 Tat interactions are homologous with acetyltransferases. Nat Genet. 1996;14:42–49. doi: 10.1038/ng0996-42. [DOI] [PubMed] [Google Scholar]
- 60.Rickert P, Seghezzi W, Shanahan F, Cho H, Lees E. Cyclin C/CDK8 is a novel CTD kinase associated with RNA polymerase II. Oncogene. 1996;12:2631–2640. [PubMed] [Google Scholar]
- 61.Sawadogo M, Roeder R. Factors involved in specific transcription by human RNA polymerase II: analysis by a rapid and quantitative in vitro assay. Proc Natl Acad Sci USA. 1985;82:4394–4398. doi: 10.1073/pnas.82.13.4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Scully R, Anderson S, Chao D, Wanjiang W, Liyan Y, Young R, Livingston D, Parvin J. BRCA1 is a component of the RNA polymerase II holoenzyme. Proc Natl Acad Sci USA. 1997;94:5605–5610. doi: 10.1073/pnas.94.11.5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shi X, Chang M, Wolf A, Chang C, Frazer-Abel A, Wade P, Burton Z, Jaehning J. Cdc73p and Paf1p are found in a novel RNA polymerase II-containing complex from the Srbp-containing holoenzyme. Mol Cell Biol. 1997;17:1160–1169. doi: 10.1128/mcb.17.3.1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shikama N, Lyon J, LaThangue N. The p300/CBP family: integrating signals with transcription factors and chromatin. Trends Cell Biol. 1997;7:232–236. doi: 10.1016/S0962-8924(97)01048-9. [DOI] [PubMed] [Google Scholar]
- 65.Shuman S. Origins of mRNA identity: capping enzymes bind to the phosphorylated C-terminal domain of the RNA polymerase II. Proc Natl Acad Sci USA. 1997;94:12758–12760. doi: 10.1073/pnas.94.24.12758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Song W, Treich I, Qian N, Kuchin S, Carlson M. SSN genes that affect transcriptional repression in Saccharomyces cerevisiae encode SIN4, ROX3, and SRB proteins associated with RNA polymerase II. Mol Cell Biol. 1996;16:115–120. doi: 10.1128/mcb.16.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sqvestrup J, Li Y, Fellows J, Gnatt A, Bjorklund S, Kornberg R. Evidence for a mediator cycle at the initiation of transcription. Proc Natl Acad Sci USA. 1997;94:6075–6078. doi: 10.1073/pnas.94.12.6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67a.Sun, X., Y. Zhang, H. Cho, P. Rickert, E. Lees, W. Lane, and D. Reinberg. NAT, a human complex Srb polypeptides that functions as a negative regulator of activated transcription. Mol. Cell, in press. [DOI] [PubMed]
- 68.Sune C, Hayashi T, Liu Y, Lane W, Young R, Garcia-Blanco M. CA150, a nuclear protein associated with the RNA polymerase II holoenzyme, is involved in Tat-activated human immunodeficiency virus type 1 transcription. Mol Cell Biol. 1997;17:6029–6039. doi: 10.1128/mcb.17.10.6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tsukiyama T, Wu C. Purification and properties of an ATP-dependent nucleosome remodeling factor. Cell. 1995;83:1011–1020. doi: 10.1016/0092-8674(95)90216-3. [DOI] [PubMed] [Google Scholar]
- 70.Tsukiyama T, Wu C. Chromatin remodelling and transcription. Curr Opin Genet Dev. 1997;7:182–191. doi: 10.1016/s0959-437x(97)80127-x. [DOI] [PubMed] [Google Scholar]
- 71.van Gool A, Citterio E, Rademakers S, Os R, Vermeulen W, Constantinou A, Egly J-M, Bootsma D, Hoejmakers H. The Cockayne syndrome B protein, involved in transcription-coupled DNA repair, resides in an RNA polymerase II-containing complex. EMBO J. 1997;16:5955–5965. doi: 10.1093/emboj/16.19.5955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Varga-Weisz P, Wilm M, Bonte E, Dumas K, Mann M, Becker P. Chromatin-remodelling factor CHRAC contains the ATPases ISWI and topoisomerase II. Nature (London) 1997;388:598–602. doi: 10.1038/41587. [DOI] [PubMed] [Google Scholar]
- 73.Wade P, Pruss D, Wolffe A. Histone acetylation: chromatin in action. Trends Biochem Sci. 1997;22:128–132. doi: 10.1016/s0968-0004(97)01016-5. [DOI] [PubMed] [Google Scholar]
- 74.Wang W, Cote J, Xue Y, Khavari P, Biggar S, Muchardt C, Kalpana G, Goff S, Yaniv M, Workman J, Crabtree G. Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. EMBO J. 1996;15:5370–5382. [PMC free article] [PubMed] [Google Scholar]
- 75.Wilson C, Chao D, Imbalzano A, Schnitzler G, Kingston R, Young R. RNA polymerase II holoenzyme contains SWI/SNF regulators involved in chromatin remodelling. Cell. 1996;84:235–244. doi: 10.1016/s0092-8674(00)80978-2. [DOI] [PubMed] [Google Scholar]
- 76.Workman J, Abmayr S, Cromlish W, Roeder R. Transcriptional regulation by the immediate early promoter of pseudorabies virus during in vitro nucleosome assembly. Cell. 1988;55:211–219. doi: 10.1016/0092-8674(88)90044-x. [DOI] [PubMed] [Google Scholar]
- 77.Yang X-J, Ogryzko V, Nishikawa J, Howard B, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature (London) 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 78.Young R. RNA polymerase II. Annu Rev Biochem. 1991;60:689–715. doi: 10.1146/annurev.bi.60.070191.003353. [DOI] [PubMed] [Google Scholar]
- 79.Zehring W, Lee J, Weeks J, Jokerst R, Greenleaf A. The C-terminal repeat domain of RNA polymerase II largest subunit is essential in vivo but is not required for accurate transcription in vitro. Proc Natl Acad Sci USA. 1988;85:3698–3702. doi: 10.1073/pnas.85.11.3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang Y, Iratni R, Erdjument-Bromage H, Tempst P, Reinberg D. Histone deacetylases and SAP18, a novel polypeptide, are components of a human Sin3 complex. Cell. 1997;89:357–364. doi: 10.1016/s0092-8674(00)80216-0. [DOI] [PubMed] [Google Scholar]