Abstract
Background
GBM is an aggressive grade 4 primary brain tumor (BT), with a 5%–13% 5-year survival. Most human GBMs manifest as immunologically “cold” tumors or “immune deserts,” yet the promoting or suppressive roles of specific lymphocytes within the GBM tumor microenvironment (TME) is of considerable debate.
Methods
We used meticulous multiparametric flow cytometry (FC) to determine the lymphocytic frequencies in 102 GBMs, lower-grade gliomas, brain metastases, and nontumorous brain specimen. FC-attained frequencies were compared with frequencies estimated by “digital cytometry.” The FC-derived data were combined with the patients’ demographic, clinical, molecular, histopathological, radiological, and survival data.
Results
Comparison of FC-derived data to CIBERSORT-estimated data revealed the poor capacity of digital cytometry to estimate cell frequencies below 0.2%, the frequency range of most immune cells in BTs. Isocitrate dehydrogenase (IDH) mutation status was found to affect TME composition more than the gliomas’ pathological grade. Combining FC and survival data disclosed that unlike other cancer types, the frequency of helper T cells (Th) and cytotoxic T lymphocytes (CTL) correlated negatively with glioma survival. In contrast, the frequencies of γδ-T cells and CD56bright natural killer cells correlated positively with survival. A composite parameter combining the frequencies of these 4 tumoral lymphocytes separated the survival curves of GBM patients with a median difference of 10 months (FC-derived data; P < .0001, discovery cohort), or 4.1 months (CIBERSORT-estimated data; P = .01, validation cohort).
Conclusions
The frequencies of 4 TME lymphocytes strongly correlate with the survival of patients with GBM, a tumor considered an immune desert.
Keywords: Cancer, Immunotherapy, Glioblastoma, Flow cytometry, digital cytometry
Key Points.
Flow cytometry is irreplaceable by digital cytometry assessing TME lymphocytes.
Paradoxically, CTL and helper T frequencies negatively correlate with GBM survival.
Frequencies of 4 tumoral lymphocytes predict the survival of newly diagnosed GBM.
Importance of the Study.
While gliomas are considered immunologically “cold” tumors, with lymphocytes usually accounting for <1%–2% of the tumor, this lymphocytic infiltrate strongly correlates with survival. We found that direct measurement of cell frequencies by FC is irreplaceable by frequency estimation by digital cytometry, failing to estimate rare cells. Using FC-quantified discovery cohort and TCGA validation cohort, we defined a novel parameter composed of the frequencies of 4 lymphocytic cells strongly predicting the survival of newly diagnosed GBM patients, with 10 months difference in median survival (FC data), or 4.1 months difference (TCGA CIBERSORTed data). We provide a survival prediction nomogram and an online tool utilizing this parameter and other significant prognostic factors as age, extent of resection, and postoperative damage. Gliomas challenge the dogma of “hot” and “cold” tumors, with IDH mutation decreasing lymphocyte infiltration but correlating with extended survival, and with CTL and Th frequencies correlating with poorer survival.
Glioblastoma (isocitrate dehydrogenase-wild-type (IDHWT) grade 4 astrocytoma- GBM), and IDH-mutated (IDHmut) grade 4 astrocytomas are primary brain tumors (BTs), which account for the majority of newly diagnosed primary BTs in adults.1 Despite intensive research, 5-year survival remains only 5%–13% under optimal approved therapies.2 Lower-grade gliomas (LGG; grades 2–3 [G2-3]), which include astrocytomas and oligodendrogliomas, have better prognosis but still pose a major health burden.1
The immunological tumor microenvironment (TME) in cancer has major effects on disease pathogenesis and progression.3 Various immune factors serve as either tumor-promoting or tumor-suppressing agents.4 Immunologically “hot” tumors, which recruit specific lymphocyte subsets, such as T cells, more favorably respond to immunotherapy and generally have better prognosis.5 Nonetheless, the brain is considered an immune-privileged organ, and BTs are considered “cold” tumors, or “immune deserts.”3 Current literature on the association between lymphocytic infiltrate and BT survival is highly controversial (Supplementary Table S1).
Flow cytometry (FC) is the gold standard for identification and quantification of immune cells. While the identification of immune cells in blood has rapidly evolved, many technical challenges impede effective monitoring of immune cell frequencies within tissues or tumors.6 These technical challenges together with the increasing abundance of transcriptomic data have prompted the field of “digital cytometry.” Algorithms, such as CIBERSORT/X7,8, deconvolute bulk-tumor gene expression data to estimate frequencies of different cell types within the tumor. The performance of those algorithms was established on synthetic data, hematological tumors, and peripheral blood samples, and not by using ground truth FC-based data from solid tumors.7,8
Since microscopic histopathology provides only partial assessment of disease subtype and prognosis9 the 2016 and, more recently, the 2021 WHO classification of central nervous system (CNS) tumors included various genomic features as integral part of glioma classification, with IDH mutations being the most prominent.10,11 Although considered oncogenic,12,13 IDH-mutated gliomas are associated with a better prognosis.10 They were demonstrated to have immunosuppressive qualities.14 The utility of TP53 and ATRX for prognosis was demonstrated for LGGs,11 and the methyl guanine methyl transferase (MGMT) gene methylation status was shown to be predictive for gliomas’ response to chemotherapy15; all these parameters are used in the current WHO guidelines.11 Knowledge of the interaction between the status of these genes and the immune infiltrate is scarce.16,17
Multiple studies aimed to provide survival prediction models for GBM.18 These have commonly utilized clinical factors, and those that have incorporated TME features were limited. FC-acquired lymphocyte frequencies have not been incorporated into any validated model.
Here, we introduce a single-center BT dataset, which incorporates FC-acquired frequencies of prominent TME lymphocytes, patient survival data, and detailed demographic, radiologic, clinical, molecular, and histopathological features of the patient cohort. We demonstrate the irreplaceable role of FC, rather than digital cytometry, in acquiring TME frequencies. We identify the immune-constraining effect of IDH mutation on most lymphocyte frequencies in the TME and the association of some of these lymphocytes with survival. Lastly, we provide and validate a prognostic model that combines TME frequencies of four lymphocytic subsets together with clinical parameters for survival prediction of newly diagnosed (IDHwt) GBM.
Materials and Methods
Human Subjects
Brain tumor tissue and blood samples were obtained from 102 patients who underwent surgery in the Neurosurgery Department at the Tel-Aviv Sourasky Medical Center between 2011 and 2020. All patients have signed an informed consent (IRB approval 0408-10-TLV). The tumors were pathologically classified by the institute’s neuropathologists. Samples were obtained from 59 GBMs, 4 IDH-mut G4 astrocytomas, 22 G2-3 LGGs, 10 brain metastases from solid malignancies (Met.), and 7 nontumorous brain samples (Table 1).
Table 1.
Patients’ Characteristics.
| GBM | IDH-mut G4 | LGG * | Metastasis** | Brain | P-value | |
|---|---|---|---|---|---|---|
| N | 59 | 4 | 22 | 10 | 7 | |
| Demographics | ||||||
| AGE (mean (SD)) | 61.4 (12.3) | 44 (26.9) | 41.4 (13.6) | 66.2 (8.2) | <.001 | |
| OS (median [95% CI]) | 422 [371,502] | 1027 [225, NA] | 6267 [3176, NA] | <.001 | ||
| Event = dead (%) | 54 (93.1) | 4 (100.0) | 7 (35.0) | <.001 | ||
| Molecular markers (N (%) of tested) | ||||||
| P53 overexpression (%) | 39 (64.3) | 3 (75) | 15 (65) | 0.91 | ||
| ATRX preserved (%) | 40 (88.9) | 1 (50.0) | 2 (40) | 0.003 | ||
| IDH-WT (%) | 59 (100) | 0 (0) | 3 (15) | |||
| MGMT hypermethylation (%) | 8 (42) | |||||
| Lymphocyte frequencies (log 10 (mean (SD))) | ||||||
| Th | −2.41 (0.69) | −3.15 (0.52) | −3.79 (0.96) | −2.27 (1.01) | −4.28 (0.28) | <.001 |
| CTL | −2.76 (0.98) | −2.85 (0.40) | −3.62 (0.78) | −2.61 (1.35) | −4.41 (0.80) | 0.001 |
| NK56 | −4.72 (1.02) | −5.15 (0.76) | −5.36 (0.80) | −4.65 (1.30) | −5.34 (0.80) | 0.14 |
| gdTc | −3.42 (0.87) | −4.05 (0.51) | −4.31 (1.02) | −3.40 (0.60) | −5.00 (0.75) | <.001 |
| BCs | −3.54 (0.90) | −3.81 (NA) | −4.23 (0.57) | −3.17 (1.32) | NaN (NA) | 0.48 |
Lymphocyte frequencies are log10 transformed. Reported numbers refer to patients for whom data were available. P-values refer to ANOVA test where multiple group continuous variables were compared, and Chi-squared test where categorical variables were compared. *Grades 2–3\lower-grade gliomas (LGGs) included 5 oligodendrogliomas, 4 astrocytomas, 4 oligoastrocytomas, 4 anaplastic astrocytomas, 2 anaplastic oligodendrogliomas, 1 anaplastic oligoastrocytoma, and 2 cases of LGG NOS. **Primary cancers included 3 breast carcinomas, 2 lung carcinomas, 2 melanomas and 3 unknown primary cancers. ATRX, alpha-thalassemia/mental retardation X-linked; IDH, isocitrate dehydrogenase; MGMT, O6 methylguanine-DNA methyl transferase; LGG, low grade glioma; NOS, not otherwise specified.
See Methods Appendix for information on lesion grouping and classification terminology.
Tissue Processing, Flow Cytometry, and Calculating the Frequencies of Peripheral Blood Mononuclear Cells (PBMC) and Tumoral Lymphocytes
Fresh brain lesions were dissociated using gentle enzymatic dissociation using our enzymatic protocol.19 PBMC from patients and healthy donors were purified from blood samples using Ficoll (GE Healthcare, Waukesha, WI). PBMC and dissociated tumor/tissue (at least 2 × 106 cells) were stained with ViViD amine viability dye (Invitrogen, Eugene, OR) and incubated with an antibody panel including CD3, CD4, CD45, CD56 (eBioscience, San-Diego, CA), CD8, CD66b, TCRγδ (BD Biosciences, Franklin Lakes, NJ), and CD19, CD14 (Biolegend, San-Diego, CA). CD14 and CD66b shared excitation and emission as ViViD, excluding dead and unwarranted cells on a single “dump” channel. The samples were FC-analyzed on 3-laser FACS CANTO II (BD).
All brain lesions were analyzed together with their cognate PBMC, serving to guide correct gate placement and to provide positive control for rare or missing tumoral immune subsets. Single cells were isolated on light scatter properties and RBCs were removed on CD45-ViViddimmest. The nucleated cells were gated on CD45 for tumoral leukocytes, and then for viability (ViViDlow) and for nonexpression of unwarranted lymphocytic markers (Dump channel - CD14: monocytes, macrophages, and microglia; CD66b: granulocytes). T cells were gated as CD4 (Th) or CD8 (CTL) positive CD3 + cells. γδ-T cells were gated on their γδ-TCR from the CD3 + 4-8− cells. CD3 + CD56 + natural killer T (NK-T) were found in negligible numbers. The CD3- population was then gated for CD56bright NK cells (NK56: a less-mature, less cytotoxic NK population), and for CD19 + B cells (BCs). FC data were analyzed on FlowJoX software. Live lymphocyte frequencies were calculated from total nucleated cells. Frequencies were displayed on a log-10 scale as cells per million (CPM), and added 1 × 10-6 to avoid taking the log of zero. For tissue preparation protocol and additional information see Methods Appendix).
Acquisition of Molecular Parameters
Molecular parameters were documented according to standard practice for all glioma patients. This included institutional molecular tests for the IDH1 R132H mutation, ATRX preservation, and P53 overexpression. The testing method for MGMT gene methylation, performed for some patients, is provided in the Methods Appendix.
Acquisition of Demographic and Clinical Parameters
Demographic and clinical parameters were collected for 45 GBM patients who underwent full lymphocytic TME assessment. This included patient age, tumor location, Eastern Cooperative Oncology Group (ECOG) performance score at diagnosis (ECOG PS), major postoperative complication (PO comp), and major postoperative neurological deficits (PO neuroDef) which have demonstrated prognostic value in glioma.20 The administered therapies included standard adjuvant chemoradiation (Stupp) protocol,21 anti-angiogenic bevacizumab use,22 and other therapeutic modalities. GBM cohort parameters are summarized in Table 2.
Table 2.
Newly Diagnosed GBM Cohort Characteristics.
| Feature | Level | Overall | Chemoradiation | No chemoradiation | P |
|---|---|---|---|---|---|
| N | 45 | 36 | 9 | ||
| Demographics | |||||
| Age > 60 (%) | (60.0) 27 | (58.3) 21 | (66.7) 6 | 0.65 | |
| ECOG > 1 (%) | (13.3) 6 | (11.1) 4 | (22.2) 2 | 0.38 | |
| Overall survival (median [95% CI]) | 426 (374, 540) | 492 (405, 712) | 273 (152, NA) | ||
| Event = dead (%) | 40\45 | 33\36 | 7\9 | 0.24 | |
| Molecular markers (positive\tested) | |||||
| Methylated MGMT | 4\14 | 4\11 | 0\3 | 0.6 | |
| P53 overexpression | 28\42 | 21\29 | 7\13 | 0.24 | |
| ATRX preserved | 30\33 | 20\23 | 10\10 | 0.59 | |
| Therapy | |||||
| Bevacizumab Tx = yes(%) | (57.8) 26 | (63.9) 23 | (33.3) 3 | 0.1 | |
| Other Tx modalities* = Yes(%) | (37.8) 17 | (44.4) 16 | (11.1) 1 | 0.065 | |
| Clinical features | |||||
| Hemisphere = R (%) | (51.1) 23 | (44.4) 16 | (77.8) 7 | 0.073 | |
| Location (%) | Frontal | (24.4) 11 | 8 (22.2) | (33.3) 3 | 0.97 |
| Temporal | (28.9) 13 | (30.5) 11 | (22.2) 2 | ||
| Parietal | (17.8) 8 | 7 (19.4) | (11.1) 1 | ||
| FT | (13.3) 6 | (13.9) 5 | (11.1) 1 | ||
| other | (15.6) 7 | (13.9) 5 | (22.2) 2 | ||
| Post-op neurological deficit** = yes (%) | (15.5) 7 | (11.1) 4 | (33.3) 3 | 0.26 | |
| Post-op complication***= yes (%) | (11.1) 5 | (13.9) 5 | (0.0) 0 | 0.55 | |
| Radiological features | |||||
| Pre-op tumor vol. (mean (SD)) | (26.2) 37.15 | (21.86) 33.10 | (37.53) 57.38 | 0.023 | |
| Post-op tumor vol. (mean (SD)) | (3.74) 2.70 | (4.01) 2.72 | (2.15) 2.60 | 0.936 | |
| % Tumor resected (mean (SD)) | (13.46) 91.33 | (14.38) 90.79 | (7.46) 94.02 | 0.568 | |
| Lymphocytic TME scores | |||||
| PC1 (mean (SD)) | (1.52) 0.00 | (1.53) 0.17 | −(1.15) 0.92 | 0.081 | |
| PC2 (mean (SD)) | (1.02) 0.00 | −(0.92) 0.09 | (1.41) 0.47 | 0.188 |
Reported numbers refer to patients for whom data were available. P-values refer to t-test for comparison of continuous variables’ means and Chi-squared test for comparison of categorical variables. *Other therapeutic modalities included: gliadel© wafers implantation, CCNU protocol, carboplatin salvage therapy, stereotactic radiosurgery, autologous tumor vaccines, virotherapy, checkpoint inhibition (anti-PD1, anti-PDL-1, anti-CTLA4). **Postoperative neurological deficit included: persistent changes in alertness and confusion, memory and cognitive disorders, major aphasia or dysphasia, limb paresis or plaegia, and general functional decline. Most patients suffered from more than one deficit. ***Major postoperative complication included: brain abscess, postoperative aseptic meningitis, tumor bed bleeding requiring surgical intervention, surgical-site infection requiring surgical revision, new cardiac disease, ICU referral (N = 1 for each). ECOG—Eastern Cooperative Oncology Group, OS, overall survival; MGMT, O6–methyl guanine DNA methyltransferase; ATRX, alpha-thalassemia/mental retardation X-linked; Tx, treatment; R, right; FT, fronto-temporal; pre-op, preoperative; post-op, postoperative; vol., volume, PD1, programmed death receptor 1; PDL-1, programmed death ligand 1; CTLA4, cytotoxic T-lymphocyte associated antigen 4; ICU, intensive care unit.
Volumetric Assessment of Surgical Intervention
Forty-five GBM patients underwent preoperative and immediate postoperative MRI assessment (Table 2). Supplementary Figure M1 shows a histogram of the distribution of percent resected tumor.23 For further information on the volumetric assessment see Methods Appendix.
Survival Times and Events
Overall survival times (OS) were documented for all 85 glioma patients in this study. Fifty-four of the 59 GBM patients and all 4 IDH-mutated G4 Astrocytomas had died, and 2 patients were alive at the end of follow-up (May 2021). The remaining 3 patients were lost to follow-up. Seven of the 22 LGG patients had died and 15 were alive at the end of follow-up time (Table 1).
TCGA G4 Glioma Subjects and Corresponding CIBERSORT Immune Infiltrate Data
Immune infiltrate to GBM samples were obtained from Thorsson et al.12 who used the CIBERSORT algorithm to estimate leukocyte frequencies across multiple cancer types in The Cancer Genome Atlas (TCGA). Additional clinical and molecular data were obtained from correspondent TCGA Research Network resources24 (for further information see Methods appendix).
Statistical Analysis
FC-acquired cell frequencies were log10-scaled and tested for normal distribution by means of the Shapiro–Wilk normality test and QQ-plots. Correlation coefficients were evaluated with Pearson’s product moment. The statistical significance of correlations was evaluated using t-test. t-test was also used to compare the means of 2 normally distributed groups. ANOVA test was used to compare multiple normally distributed groups, while Tukey’s intervals method was applied for post hoc comparisons. Statistical hypothesis tests determined significance at P-value < .05. All statistical analyses were performed using R software for statistical computing.25
Dimensionality Reduction Methods
We used principal component analysis (PCA) on the R “base” package25 to incorporate 4 lymphocyte frequencies (Th, CTL NK56, and γδ-T) into one composite parameter with minimal loss of data. For the PCA we used only samples with full clinical data.
Survival Analysis and Visualization
Median survival with 95% CIs using the Kaplan–Meier method were used to report survival. Log-rank tests were used to evaluate survival differences between 2 groups. In cases where a continuous variable was used to predict survival, maximally selected log-rank statistic method was applied to set optimal cutpoints (further information in the Methods Appendix). Log-rank tests and Kaplan–Meier (KM) survival curves were generated with “survival”26 and “survminer”27 R packages. Survival difference was reported in median survival months. Cox proportional hazards (CPH) regression models were fitted to evaluate categorical and continuous survival-predicting variables using “survival” package. Backward-step selection algorithm was used to construct the multivariate CPH model. Nomogram was plotted using the “rms” package.28 An online dynamic nomogram tool was generated using the “DynNom” package.29 All plots were generated with R software.25 For specific plots and packages see Methods Appendix.
Data Availability Statement
The data presented in this study are available upon reasonable request from the corresponding authors.
Results
Lymphocyte Frequencies Differ Across Brain Lesions
We applied flow cytometry (FC) to evaluate lymphocyte frequencies in brain tumors and brain lesions (Figure 1A and B). Table 1 numerically lists the frequencies of Th, CTL, γδ-T, NK56, and BCs among 102 brain lesions in G4 gliomas, LGG, metastases, and brain lesions. The table lists cohorts’ molecular classification and demographic parameters.
Figure 1.
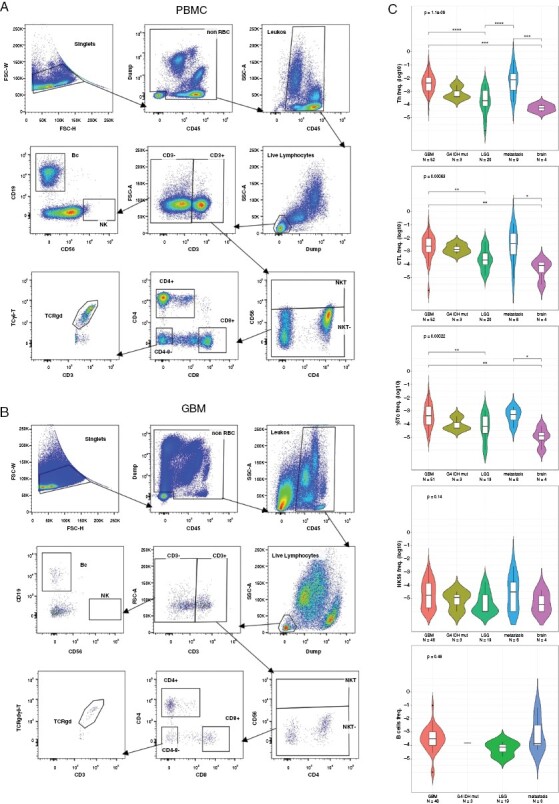
FC gating strategy and differential immune cell composition among different brain lesions. A representative flow cytometric analysis of the gating strategy of (A) PBMC and cognate, (B) freshly dissociated GBM sample of the same individual. (C) FC lymphocytic frequencies attained from 5 types of resected brain lesion samples: GBM (N = 48–52 per different cell types); grade 4 IDH-mutated astrocytoma (N = 3); LGG (N = 19–20); metastasis (N = 8–9); and nontumorous brain tissue (brain, N = 4). The frequency of 5 lymphocytic populations was determined from total nucleated cells: T-helper cells (Th), Cytotoxic T-lymphocyte cells (CTL), gamma-delta T cells (γδ-T), CD56-bright natural killer cells (NK56), and B-cells (Bc). Top labels report ANOVA P-value; brackets report post hoc Tukey’s interval significance (*<.05, **<.01, ***<.001, ****<.0001). FC, flow cytometry; PBMC, peripheral blood mononuclear cells; LGG, low grade glioma; Met—metastases.
Supplementary Figure M2 demonstrates the strong impact of use, or lack of use, of different components of our FC panel on the accuracy of frequency detection (discussed in depth in the Methods Appendix). Figure 1C depicts lymphocyte distribution among different lesions. Th, CTL, and γδ-T cells demonstrated similar trends across all malignant lesions, with the highest frequencies observed in GBM and Mets (interquartile range [IQR] ~10−2 to 10−3). Approximately one-fold lower frequencies were found in LGG and G4 IDH-mut astrocytoma, (IQR ~10−3 to 10−4), and additional one-fold lower frequencies were recorded in nontumorous brain lesions (Br) lesions (IQR~10−4 to 10−5). The infiltration of γδ-T cells was higher in GBM and Mets compared to LGG, G4 IDH-mut Br, although their frequencies were approximately 10-fold lower than those of CTL or Th (IQR ~10−3 to 10−4). NK56 and B cells showed relatively constant frequencies among different tumor types ranging from 10−3 to 10−5.
Comparison Between CIBERSORT, A Digital Cytometry Algorithm, and FC Using Real-World GBM Data
TME lymphocyte frequencies correlate with cancer prognosis.5,12 Recently, several digital cytometry tools were developed to estimate immune cell frequencies using bulk-tumor RNA-Seq data. CIBERSORT/X are the most used digital cytometry tools, yet their performance was only partially compared to solid tumor FC data.7,8
We compared CIBERSORT’s performance with that of FC by examining the distributions of TME lymphocytes across GBMs (Figure 2A). CIBERSORT could not “detect” cell populations with frequencies lower than ~0.2%, while our FC data showed that frequencies of approximately half of the more common lymphocytes (CTL, Th), and even higher fractions of the less common lymphocytes (NK56, Bc, γδ-Tc) are found below 0.2%. Notably, while γδ-T cells were evident in all our FC-analyzed specimens, they were lacking in 87% of the CIBERSORT samples. Moreover, all CIBERSORT approximated means significantly differed from the matching FC population (t-test P < .001 all comparisons,), except for γδ-T.
Figure 2.
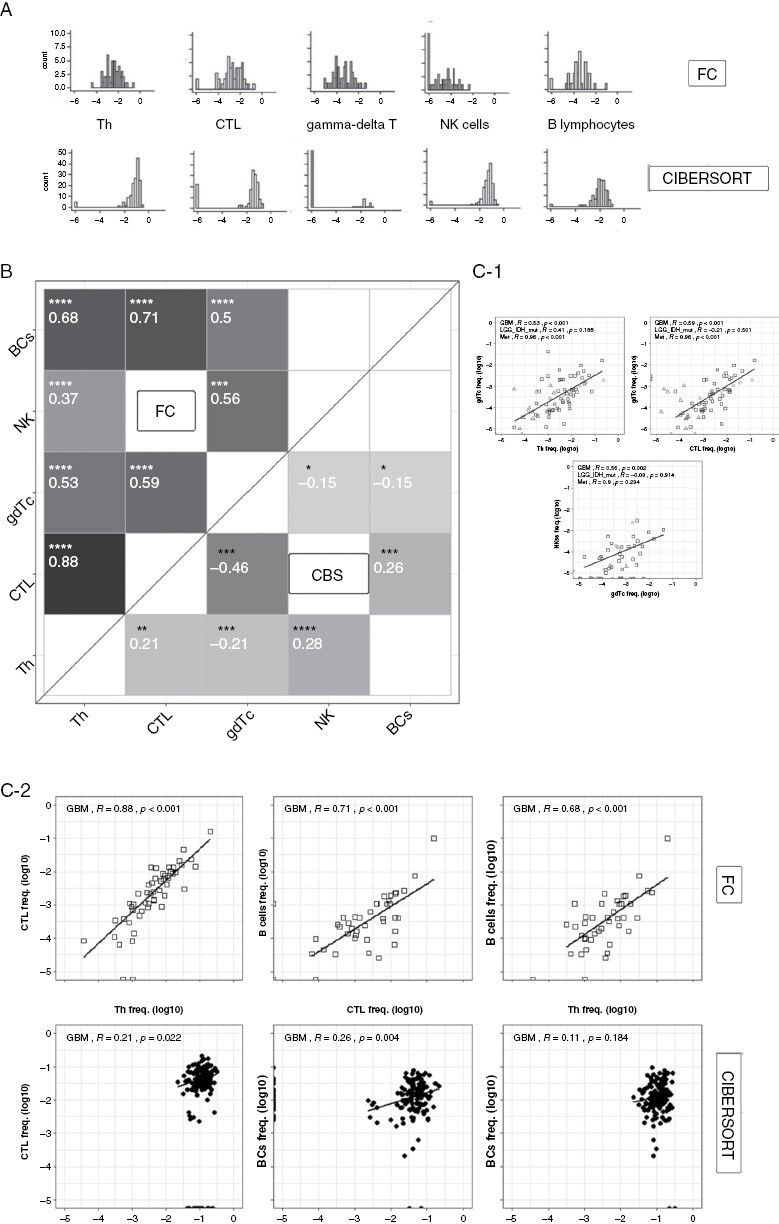
Comparison of FC- and CIBERSORT-acquired lymphocytic TME distributions and correlations. (A) Histograms depicting frequency distribution of GBM lymphocytes acquired by FC (top) or CIBERSORT (bottom). (B) Correlogram showing statistically significant lymphocytic correlations using GBM study data (top triangle) and CIBERSORT TCGA GBM data (bottom triangle). Tiles’ tone corresponds to correlation strength. (C-1) Scatter plots of select immune infiltrating cell populations indifferent brain lesions (GBM\LGG\metastasis) acquired by FC. Regression lines are plotted only for GBM samples. (C-2) Comparison of GBM immune-infiltrate scatter plots acquired by FC (top) and CIBERSORT (bottom) with regression lines. For C-1 and C-2, “R” indicates regression coefficient. All frequencies are displayed on the log-10 scale. (*<.1, **<.05, ***<.01, ****<.001). FC, flow cytometry; CBS-CIBERSORT TME, tumor microenvironment; GBM, glioblastoma multiforme; LGG, low grade glioma.
Figure 2B shows a correlogram that compares the correlations between all cell types in GBMs as calculated by FC and by CIBERSORT. FC-determined correlations were both strong and statistically significant, with the strongest correlations found between Th, CTL, and BCs with Rpearson ranging from + 0.68 to + 0.88. In contrast, CIBERSORT correlations between these cells ranged from significant + 0.26 for CTL-BCs to nonsignificant + 0.11 for Th-BCs. Moreover, several significant positive FC correlations as those between γδ-T and BCs or CTL (+0.5 and + 0.59, respectively) turned significantly negative in CIBERSORT (−0.15 and −0.46, respectively). Figure 2C-1 shows scatterplots of several cell types found in GBM, LGG, and metastasis using our FC data; Figure 2C-2 compares 3 representative correlations acquired using FC versus CIBERSORT on GBM data. The plots demonstrate the strong correlation found using FC between many different lymphocytic cell types in all brain tumors across all frequency ranges. In contrast, CIBERSORT demonstrated weaker correlations, and in considerably more limited frequency ranges. In summary, digital cytometry for lymphocyte-poor GBM constitutes a weak surrogate for FC enumeration of TME cellular frequencies.
P53 Overexpression, ATRX Preservation, and MGMT Methylation do not Affect Lymphocyte Infiltration to Gliomas
Alterations in TP53 expression, ATRX preservation, and MGMT methylation are major alterations affecting gliomas’ clinical behavior and thereby essential in their classification.10,11 Their effect on the lymphocytic TME in gliomas has been scarcely studied[17. Supplementary Figure S1 shows that alterations in these 3 genes had no significant effects on the lymphocytic infiltration to gliomas.
IDH Mutation Affects the Glioma Immunophenotype More Than the Pathological Tumor Grade
IDH1/2 mutations play a pathogenetic role in various malignancies13 and were shown to suppress CTL accumulation in gliomas [14] yet, their effects on other infiltrating lymphocytes are unknown. Grouping all gliomas together irrespective of grade,30 the lymphocytic infiltrate levels in IDHmut gliomas were found to be significantly decreased compared to their IDHwt counterparts (all P < 0.05, t-test, not shown). When adjusting for tumor grade, differences persisted for Th and γδ-T in the G4 gliomas (P < .001, Figure 3A).
Figure 3.
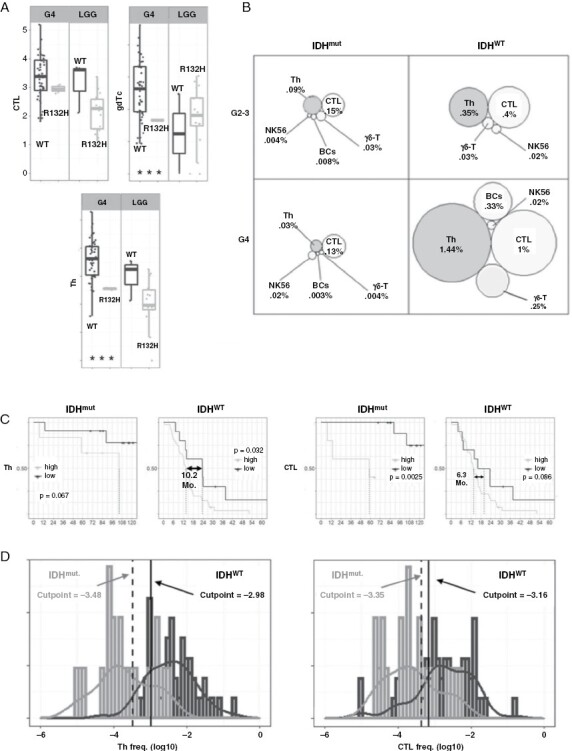
IDH mutation status affects glioma lymphocytic infiltrate and patient survival. Seventy glioma specimen were divided according to pathological tumor grade and IDH mutation status (IDHmut: G4 N = 4, G2-3 N = 17; IDHWT: GBM N = 48, G2-3 N = 3). (A) Boxplots showing IDH status effect on lymphocytic infiltrate, stratified for tumor grade. (B) Visual comparison of IDH status and tumor grade combined effect on lymphocytic TME. (C) Optimal Th and CTL frequency cutpoints for survival were obtained for 51 IDH-WT and 21 IDH-mutated gliomas. Kaplan–Meier (KM) survival curves display median survival differences (in months) for high versus low Th/CTL infiltrates. (D) Optimal, independently calculated, Th and CTL cutpoints (high vs. low) for IDH-mut (gray)/IDH-WT (black) populations. Histograms and estimated distributions are displayed for both populations. Exact cutpoints for survival prediction: Th: 10−3.48 to 10−2.98 [~330–1050 CPM], CTL: 10−3.35 to 10−3.16 [~445–690 CPM]. R132H, IDH gene R132H mutation; G2-3, grades 2–3; G4, grade 4; IDHmut/WT, mutated/wild-type IDH; Mo., months; CPM, cells per million; Th, helper T cells, CTL; cytotoxic T lymphocytes; NK56, CD56 + natural killer cells; γδ-T, gamma-delta T cells; BCs, B cells.
We assessed the influence of both tumor grade and IDH status on lymphocytic infiltrate finding that while IDH status significantly modified glioma infiltration of all lymphocytes (Th, CTL: P < .001; γδ-T: P = .001; NK56: P = .013, 2-way ANOVA), tumor grade did not significantly modify infiltration (P > .05 for all). Visual comparison of the 4 different subgroups (Figure 3B) underscores the closer proximity of lymphocytic infiltration stratified by IDH mutation compared to tumor grade.
Lower Frequencies of Th and CTL Correlate with Extended Survival in IDHWT and IDHmut Glioma Patients
Given that lymphocytic infiltrate in gliomas is more strongly correlated with IDH status than with tumor grade, we segregated all gliomas to IDHmut and IDHWT and examined whether the lymphocytic infiltrate affected patient survival. Using the maximally selected log-rank statistics, we found in IDHWT (N = 51) gliomas, a significant survival benefit for lower Th infiltrates (a 10.2-month difference, P = .032) and a near-significant survival benefit for lower CTL infiltrates (a 6.3-month difference, P = .086; Figure 3C). Similar results were found in the IDHmut gliomas (N = 21) where significant survival extension was found for lower CTL infiltrates (P = .0025) and near-significant for lower Th infiltrates (P = 0.067). Median survival was not reached for IDHmut gliomas with lower Th or CTL infiltrates at the time of writing. Strengthening the validity of our finding, we saw that the independently calculated cutpoints for Th and CTL frequencies for either IDHWT or IDHmut distributions were adjacently located, and found near the intersection between the 2 histograms (Figure 3D). The significant survival benefit persisted when excluding G4 astrocytomas from the IDHmut, and G2-3 LGGs from the IDHWT analyses (removed N = 3 for either). Taken together, CTL or Th frequencies lower than approximately 600–700 CPM correlate with extended survival, independently of gliomas’ IDH status.
Lymphocytic Composition is a Powerful Independent Survival Prognostic Biomarker for Newly Diagnosed GBM
To evaluate the effect of TME lymphocytes on survival, we analyzed a set of 45 newly diagnosed GBM patients who underwent full lymphocytic assessment (Table 2). ECOG performance status (PS) scores were divided between (0–1) and (>1) and age was divided at 60 years. Univariate CPH regression models found the following parameters as significant hazard predictors: ECOG PS > 1 (HR = 4.83; P = .003), Postoperative (PO) neuroDef (HR = 6.72; P < .001), high resected tumor proportion (HR = 0.98, P = .047) and treatment using standard chemoradiation (HR = 0.05; P = .003; Supplementary Table 2A).31–33 no significant association was found between any single lymphocyte frequency and survival (not shown).
For a more comprehensive evaluation of GBM lymphocytic TME infiltrate we used principal component analysis (PCA). Principal Component (PC) 1, explaining 57.4% of variance, correlated positively with overall extent of infiltration to the tumor of all different lymphocytes (Supplementary Figure S2A–B). PC2, explaining 24% of variance, correlated positively with Th and CTL frequencies, and negatively with γδ-T cells and NK56, thus corresponding to the composition of the lymphocytic infiltrate. PC2 score is given by the formula: [0.28 × (Th)] + [0.59 × (CTL)] + [−0.36× (γδ-T)] + [−0.67 × (NK56)] (frequencies are log10-transformed, and statistically centered and scaled). In univariate CPH models, PC1 did not significantly predict survival (HR = 1.14, P = .312). Contrastingly, continuous PC2 score was marginally significant (HR = 1.46, P = .054), and PC2 undergoing median dichotomization was found a significant hazard-predicting factor (HR = 2.17, P = 0.031; Supplementary Table 2A). KM curves were used to illustrate PC2 strong impact on survival, showing a significant 10-month survival benefit for PC2low (Thlow CTLlow, γδ-Thigh NK56high) over PC2high tumors (below vs. above median, respectively, log-rank test: P < .001, Fig. 4A). Next, we used backward-step selection procedure to fit a multivariate CPH model for newly diagnosed GBM patients who received standard chemoradiation therapy (N = 36). This subgroup model found that PC2 score, age > 6033 PO neuroDef, and the extent of resection were significant hazard modifiers (Supplementary Table 2B). We composed a nomogram, which allowed a calculation of survival probability (Figure 4C) for newly diagnosed chemoradiation-treated GBM patients. All significant parameters included in the univariate models were also significant or marginally significant (P = .069, extent of resection) in the multivariate CPH.34 Multivariate nonsignificant ECOG PS > 1 was excluded from this model due to its considerable collinearity with statistically significant PO neuroDef (3/4 patients with ECOG PS > 1 had PO neuroDef). Based on this model, PC2 score had the strongest effect on survival probability, slightly stronger than PO neuroDef (found only in 4/36 of the patients). PC2 also showed similar effects in the non chemoradiation-treated group yet the smaller sample size did not enable reaching significance (CPH: HR = 1.7, P = .25, N = 9). Although acquired for 29% of patients, MGMT methylation was not associated with PC2 parameterization (Fisher’s exact, P = .19). An online dynamic nomogram tool to predict survival using this model is available at ?Impact-insert-start?> https://rg-medical-apps.shinyapps.io/GBM_prgonostic_calculator/.
Figure 4.

GBM survival prognostication using study’s principal component 2 (PC2), in the discovery and validation cohorts. Following principal component analysis (PCA) of 45 newly diagnosed GBM lymphocytic TME samples, PC2 was positively influenced by Th and CTL, and negatively by γδ-T and NK56. (A) KM survival curve shows survival extension for PC2low (below median) versus PC2high (above median) samples (P < .0001). PC2 scores were then calculated for TCGA GBM samples and optimal cutpoints were determined. (B) PC2 splits the survival curves of two CIBERSORT-derived TCGA GBM cohorts: all TCGA GBM patients (top) and TCGA GBM patients labeled as “treated” (bottom). (C) Nomogram for prognostication of newly diagnosed GBM patients who received chemoradiation therapy (N = 36), based on multivariate CPH model. GBM, glioblastoma multiforme, OS, overall survival, PC2, principal component 2, Mo., months, ECOG PS, Eastern Cooperative Oncology Group Performance Score. TME, tumor micro environment, Th, helper T cells, CTL, cytotoxic T lymphocytes, NK56, CD56 + natural killer cells, γδ-T, gamma-delta T cells, KM, Kaplan–Meier; TCGA, the cancer genome atlas; CPH, Cox proportional hazard.
Validation of the PC2 Prognosticator Using an Independent Cohort
To validate our model, we implemented a similar analysis scheme on the CIBERSORT deconvoluted TCGA GBM dataset from which all IDHmut GBM samples were removed. PC2 scores were calculated for each CIBERSORT data sample by means of our PCA formula. Correlations of PC2 with CTLs, γδ-T, and NK56 were at the same direction and at similar proportions as those for the FC data; Th correlated oppositely with PC2 at the CIBERSORT dataset than the FC dataset (Supplementary Figure S2–C). All available epidemiologic/demographic factors were obtained from the TCGA resources including patient age, sex, ethnicity, and treatment.24 As previously demonstrated (Figure 2), the capacity of CIBERSORT to estimate lymphocyte frequencies in GBM is limited. Nevertheless, with the larger TCGA cohort, KM curves, and log-rank tests demonstrated a significant 4.1–4.3 month extension of survival for PC2low patients among all 146 GBM patients as well as among the 110 patients labeled as “treated,” respectively (Figure 4B). Similarly to the FC data, univariate CPH models found the PC2 score (HR = 1.16, P = .021), any treatment (HR = 0.2, P < .001), and the age at index (HR = 1.02 P = .008) as significant hazard factors. Here, too, no single lymphocyte frequency showed a significant association with survival. (Supplementary Table 3A). A multivariate CPH model incorporating significant univariate hazard factors found PC2 and treatment as significant hazard-predicting factors (Supplementary Table 3B). The lack of TCGA-reported ECOG (or similar performance scores), extent of resection, and postoperative complications precluded the construction of a TCGA nomogram using a similar approach.
Discussion
Our study enriches the current understanding of the complexity of gliomas’ TME by connecting accurate immunological data to multiple clinical parameters. Our results demonstrate the unmatched capacity of FC to quantify the frequencies of lymphocytes within solid tumors and delineate technical approaches critical for correct cell quantification by FC. The data also demonstrate how such accurate quantification could uncover non-trivial connections between lymphocytic infiltration and patient survival.
Most FC-assisted GBM studies to date did not utilize FC to broadly quantify lymphocytic infiltration into these tumors. Mohme et al. and Woroniecka et al. immunophenotyped peripheral blood (PB) and tumor-infiltrating lymphocytes (TILs) from newly diagnosed GBM patients.35,36 Davidson et al. compared exhaustion markers from GBM patients’ PD1+ TILs with PD1+ PB lymphocytes.37 While those studies characterized specific aspects of GBM TILs, they did not determine lymphocytes’ relative frequencies in the tumor or their association with survival. Several other studies immunophenotyped PB lymphocytes and not tumor samples of GBM patients.38,39
Digital Cytometry is a Poor Surrogate for FC in Quantifying Lymphocytic Infiltrates into Brain Tumors
Digital cytometry methods are widely regarded as an alternative to direct measurements of cellular frequencies by FC. The performance of those methods has been only partially tested in comparison to real-world FC data from solid tumors. Deconvolution algorithms were established using synthetic data and pre-mixed samples, and validated by using FC data on PB leukocytes or hematological malignancies rather than using data from solid tumors.8 Deconvolution algorithms rely on signature matrices per each cell subset, assuming uniform expression of genes. Yet leukocyte gene expression profiles dramatically change depending on tissue and various clinical factors.40 Our study compares FC-derived frequencies to digital cytometry using FC data derived from solid, nonhematological malignancies. Although different datasets were used, both FC and TCGA sets consisted of large sample numbers enabling a suitable assessment of lymphocytes’ distribution. Large differences were evident in both the frequency ranges and correlations found between cell types. Significant correlations in FC, turned weak, were lost, and even reversed using CIBERSORT. While most lymphocyte frequencies (measured by FC) were below 0.2% almost no cells were found under this frequency using CIBERSORT. Relying on a PBMC-derived gene expression reference profiles was shown as insufficient to identify tumor-infiltrating immune cells.41 Various deconvolution methods have demonstrated high minimal detection rates, tended to report the existence of lymphocytes when they are absent42 and performed poorly on clinical samples.43 Although considerably poorer in resolution than FC, CIBERSORT could validate our PC2 composite biomarker identified in the FC discovery cohort. Unlike CTL, NK56, and γδ-T, the Th subset correlated in opposite directions in FC and CIBERSORT. This might be attributable to limitations discussed above or to the fact that the minimal detection rate for deconvolution methods including CIBERSORT for Th is particularly high compared to CTLs or NK.42 The agreement of 3 out of 4 PC2 parameters may explain the smaller survival difference provided by PC2 using CIBERSORT-estimated data (4.2 months) versus FC-measured data (10.2 months). Thus, our data suggests that the poorer resolution of deconvoluted data can only be partially mitigated by larger patient cohorts.
Another approach to determining immune frequencies is using a DNA-methylomic-based approach, as methylCIBERSORT. In a large study on pediatric brain tumors, 6 broad immune subsets were estimated using this approach. However, when compared with FC, methylCIBERSORT missed flow-determined lymphocyte frequencies by > 100% or < 50% in the majority of evaluated PBMC samples. Concurrent with our findings, the study also found that high Th infiltration was associated with poorer survival in pediatric H3-G34 GBM.44 Another study used DNA-methylomics to study archived blood samples from patients with IDH/1p19q/TERT-wild-type gliomas; they found that among patients under 58, high CD4 + blood counts were associated with enhanced survival.45
The Broad Effects of IDH Mutation on the Lymphocytic Infiltrate to Gliomas
We found that most of the frequent glioma genetic alterations show no association with lymphocytic infiltration. In contrast, the R132H mutation in the IDH1 gene, accounting for ~85% of IDH1 mutations,13 was associated with a 10- to 100-fold decrease in lymphocytes’ infiltration to gliomas. Previous studies demonstrated lower T-cell infiltration to IDHmut gliomas,14,16,46 However, these studies did not study non-T-cell lymphocytes, nor did they examine association of the infiltrate with survival. In our study, infiltration was more strongly stratified according to IDH mutation than it was according to the WHO-2016 pathological grade,10 providing an immunological endorsement to the definition of grade 4 IDHmut astrocytomas as a clinically distinct entity from GBM, according to the 2021 WHO classification.11
The relationship between IDH mutations, lowered lymphocytic infiltration, and enhanced survival is not fully understood. IDH-mutated gliomas display altered cellular metabolism, resulting in the accumulation of 2-hydroxy-glutarate (2-HG), an onco-metabolite with immunosuppressive properties.14 Bunse et al. demonstrated suppression of T-cell activity by tumor-derived 2-HG, and improved antitumor immunity following inhibition of mutated IDH enzymatic function.47 Several clinical trials showed some promise for IDH inhibition in gliomas.48 Additionally, IDH1 R132H mutation generates an immunogenic epitope49 that could induce a significant immune reaction in mouse models.50
While above examples view the IDH mutation as a tumor-supportive mechanism, an opposite approach aims to exploit mutated IDH enzyme accumulation to modify DNA repair pathways, thereby upregulating glioma cell sensitivity to PARP inhibitors.51 Paradoxically, both IDH potentiation and inhibition are currently evaluated as therapeutic avenues in gliomas.47 In this study, we showed that the IDH R132H mutation drives a significant decrease in the lymphocytic infiltrate to gliomas. Notwithstanding, even within the vastly different ranges of lymphocytic infiltration in IDH-mutated versus wild-type-gliomas, higher Th and CTL frequencies were independently associated with poorer survival.
The Composition of the Lymphocytic Infiltrate into Gliomas Challenges the Immunological Hot/Cold Dogma
Immunologically “hot” tumors contain high T-cell infiltrate and a proinflammatory milieu. Those tumors more favorably respond to immunotherapy than “colder” tumors (immune-excluded or “desert”) and generally have a better prognosis.52 The hot/cold distinction was established on tumors such as melanoma and NSCLC. Scant evidence challenges this concept: high Treg infiltration is associated with a better prognosis in colorectal cancer, while high CTL infiltration is associated with a worse prognosis in some renal cell carcinomas.53 Numerous mechanisms underlie GBM as being a “cold” tumor.3 We show here that specific variations within the “cold range” of GBM are associated with survival, possibly in a counterintuitive manner. Our data question the applicability of the hot/cold concept in gliomas: First, independently of tumor grade, patients with IDHmut tumors with lower T-lymphocytic infiltrate survive longer than patients with IDHWT gliomas. Second, we show that the infiltration of the more frequent lymphocytes—Th and CTLs correlate with poorer survival, while the infiltration of the less frequent lymphocytes—γδ-T and NK56 correlate with enhanced survival.
Evidence on the clinical effects of lymphocytic infiltration on GBM and other gliomas survival are highly contradictory.53Supplementary Table S1 provides a review of studies utilizing various methodologies, aiming to link between TILs abundancy and glioma survival. Prior evidence exists for both positive and negative correlation with survival for all lymphocytes analyzed in our study. Among identified studies, those utilizing FC reached conclusions similar to ours. However, critical FC-related tools included in our panel (Supplementary Figure M2) were frequently not utilized. Other studies estimated glioma lymphocytic infiltration using deconvolution methods, the limitations of which are discussed above. Studies employing IHC generally use a single marker to identify cells; yet those usually cannot unequivocally identify a single cell type: e.g. CD4 is expressed by T-helpers, monocytes/macrophages, microglia, NK-T cells, and others. Additionally, IHC studies tumors in 2-dimensional (2D) slices and not in 3D pieces, potentially limiting the accuracy of evaluated frequencies in GBM, a tumor with high intra-tumoral heterogeneity.54 These limitations may explain the contradicting results found between different IHC studies (Supplementary Table S1).
The biological mechanisms underlying the correlation between high Th and CTL infiltration and poor survival, or high γδTc and NK infiltration and extended survival in GBM cannot be extracted from our data. We show that none of the tested major prognosticators is responsible for confounding this effect. Several studies showed that most Th and CTLs in the GBM TME express an exhausted phenotype failing to produce effective antitumor immunity.34–36,55 Many studies have pointed to the pro-tumoral roles of several Th subsets (eg, Treg or Th17), yet CTLs are usually linked with extended survival.53 It may be that increased inflammation in a more malignant tumor both shortens the survival and attracts more Th and CTLs, Further research is required to shed light on this unexpected correlation. Total numbers of γδTc and NKs were generally low, yet they both positively correlated with prognosis. This may stem from an unknown confounder both enhancing survival and increasing these cells’ frequencies. Alternatively direct antitumoral effects of these cells may be envisioned relating to their activation by GBM-related antigens such as MIC-A/B, ULBP, and DMAM1.56,57
One limitation of this study is that it focused on lymphocytic TME and not on the myeloid compartment. The myeloid compartment was studied by others and was shown to impact survival.58,59
A New Tool for GBM Prognostication Utilizing Tumoral Lymphocytic Infiltrate
Gorlia et al. published the first attempt to construct nomograms for survival prediction of newly diagnosed GBM patients.18 Multiple studies tried to outperform it, but most models used demographic or clinical factors similar to the original manuscript.31–33 Ferguson et al. included also radiographic assessments and PD-L1 expression as candidates for their prognostic model, yet did not include them in the final multivariate model.60 To our knowledge, Qin et al. is the only group who published immune-assisted prognostic score model for glioblastoma.61 Their model however was not stratified for IDH mutation status, which is strongly associated with both immune infiltration and survival.
A limitation of our study is the highly selected discovery cohort (newly diagnosed, IDHWT, chemoradiation-treated GBM). This cohort yielded a relatively small sample size (N = 36). Nonetheless, our PC2 biomarker was validated on a larger, independent cohort (N = 146).
Low PC2 scores (ie, Thlow CTLlow, γδ-Thigh NK56high) were significantly associated with extended median survival, ranging between 4.1 and 10 months survival difference in the validation and discovery cohort, respectively. To our knowledge, our model is the first to provide an independent immune-assisted prognostic biomarker for newly diagnosed, chemoradiation-treated GBM prognosis.
Taken together, our study provides a new standard to accurately quantify lymphocytic infiltration into brain tumors of all grades. It demonstrates how the quality of the utilized monitoring tool translates into a stronger predictive tool. Future studies should aim to understand the mechanisms underlying the association between specific infiltrating lymphocytes and disease survival. Such understanding could be channeled to designing more effective treatments for GBM.
Supplementary material
Supplementary material is available online at Neuro-Oncology (https://academic.oup.com/neuro-oncology).
Contributor Information
Rotem Gershon, The Cancer Immunotherapy Laboratory, The Tel-Aviv Sourasky Medical Center, Tel-Aviv, Israel.
Antonina Polevikov, The Cancer Immunotherapy Laboratory, The Tel-Aviv Sourasky Medical Center, Tel-Aviv, Israel.
Yevgeny Karepov, Neurosurgery Department, The Tel-Aviv Sourasky Medical Center, Tel-Aviv, Israel.
Anatoly Shenkar, The Cancer Immunotherapy Laboratory, The Tel-Aviv Sourasky Medical Center, Tel-Aviv, Israel.
Idan Ben-Horin, The Cancer Immunotherapy Laboratory, The Tel-Aviv Sourasky Medical Center, Tel-Aviv, Israel; Oncology Department, The Tel-Aviv Sourasky Medical Center, Tel-Aviv, Israel.
Tal Alter Regev, The Cancer Immunotherapy Laboratory, The Tel-Aviv Sourasky Medical Center, Tel-Aviv, Israel.
Meytal Dror-Levinsky, The Cancer Immunotherapy Laboratory, The Tel-Aviv Sourasky Medical Center, Tel-Aviv, Israel.
Kelly Lipczyc, The Cancer Immunotherapy Laboratory, The Tel-Aviv Sourasky Medical Center, Tel-Aviv, Israel.
Lital Gasri-Plotnitsky, The Cancer Immunotherapy Laboratory, The Tel-Aviv Sourasky Medical Center, Tel-Aviv, Israel.
Gil Diamant, The Cancer Immunotherapy Laboratory, The Tel-Aviv Sourasky Medical Center, Tel-Aviv, Israel; Neurosurgery Department, The Tel-Aviv Sourasky Medical Center, Tel-Aviv, Israel.
Nati Shapira, The Cancer Immunotherapy Laboratory, The Tel-Aviv Sourasky Medical Center, Tel-Aviv, Israel; Neurosurgery Department, The Tel-Aviv Sourasky Medical Center, Tel-Aviv, Israel.
Barak Bensimhon, The Cancer Immunotherapy Laboratory, The Tel-Aviv Sourasky Medical Center, Tel-Aviv, Israel.
Aharon Hagai, The Cancer Immunotherapy Laboratory, The Tel-Aviv Sourasky Medical Center, Tel-Aviv, Israel.
Tal Shahar, Neurosurgery Department, The Tel-Aviv Sourasky Medical Center, Tel-Aviv, Israel.
Rachel Grossman, Neurosurgery Department, The Tel-Aviv Sourasky Medical Center, Tel-Aviv, Israel.
Zvi Ram, Neurosurgery Department, The Tel-Aviv Sourasky Medical Center, Tel-Aviv, Israel.
Ilan Volovitz, The Cancer Immunotherapy Laboratory, The Tel-Aviv Sourasky Medical Center, Tel-Aviv, Israel; Neurosurgery Department, The Tel-Aviv Sourasky Medical Center, Tel-Aviv, Israel.
Funding
Israeli ministry of health—Ezvonot (to IV, ZR).
Conflict of interest statement
None of the authors declared any conflict of interests.
Authorship statement
conceptualization, methodology: I.V. and R.G. Formal analysis, software, visualization, original draft: R.G. Supervision: I.V., R.G., Z.R. Review and editing: I.V., G.D., T.S. Data collection and curation, validation, investigation: A.P., K.L., Y.K., B.B., N.S., A.S., A.H., I.B.-H., T.A.R, G.D., M.D.-L., L.G.-P. Funding acquisition: I.V. and Z.R.
Data Availability
The data will be made available upon reasonable request from the corresponding author
References
- 1. Ostrom QT, Gittleman H, Liao P, et al. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2010–2014. Neuro-oncology. 2017;19(suppl_5):v1–v88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Stupp R, Taillibert S, Kanner A, et al. Effect of tumor-treating fields plus maintenance temozolomide vs maintenance temozolomide alone on survival in patients with glioblastoma: a randomized clinical trial. JAMA. 2017. ;318(23):2306–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tomaszewski W, Sanchez-Perez L, Gajewski TF, Sampson JH.. Brain tumor microenvironment and host state: implications for immunotherapy. Clin Cancer Res.. 2019 Jul 15;25(14):4202–4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Riley RS, June CH, Langer R, Mitchell MJ.. Delivery technologies for cancer immunotherapy. Nat Rev Drug Discovery. 2019;18(3):175–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gajewski TF, Corrales L, Williams J, et al. Cancer immunotherapy targets based on understanding the T cell-inflamed versus non-T cell-inflamed tumor microenvironment. Adv Exp Med Biol. 2017;1036:19–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cherian S, Hedley BD, Keeney M.. Common flow cytometry pitfalls in diagnostic hematopathology. Cytometry Part B. 2019;96(6):449–463. [DOI] [PubMed] [Google Scholar]
- 7. Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12(5):453–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Newman AM, Steen CB, Liu CL, et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat Biotechnol. 2019;37(7):773–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cancer Genome Atlas Research Network. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med. 2015;372(26):2481–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. van den Bent MJ, Weller M, Wen PY, et al. A clinical perspective on the 2016 WHO brain tumor classification and routine molecular diagnostics. Neuro-oncology. 2017;19(5):614–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Louis DN, Perry A, Wesseling P, et al. 2021 WHO classification of tumors of the central nervous system: a summary. Neuro-oncology. 2021;23(8):1231–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Thorsson V, Gibbs DL, Brown SD, et al. The immune landscape of cancer. Immunity. 2018;48(4):812–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cohen AL, Holmen SL, Colman H.. IDH1 and IDH2 mutations in gliomas. Curr Neurol Neurosci Rep. 2013;13(5):345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kohanbash G, Carrera DA, Shrivastav S, et al. Isocitrate dehydrogenase mutations suppress STAT1 and CD8+ T cell accumulation in gliomas. J Clin Invest. 2017;127(4):1425–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Weller M, Stupp R, Hegi ME, et al. Personalized care in neuro-oncology coming of age: why we need MGMT and 1p/19q testing for malignant glioma patients in clinical practice. Neuro-oncology. 2012;14(Suppl 4):iv100–iv108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Berghoff AS, Kiesel B, Widhalm G, et al. Correlation of immune phenotype with IDH mutation in diffuse glioma. Neuro-oncology. 2017;19(11):1460–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chaurasia A, Park SH, Seo JW, Park CK.. Immunohistochemical analysis of ATRX, IDH1 and p53 in glioblastoma and their correlations with patient survival. J Korean Med Sci. 2016;31(8):1208–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gorlia T, van den Bent MJ, Hegi ME, et al. Nomograms for predicting survival of patients with newly diagnosed glioblastoma: prognostic factor analysis of EORTC and NCIC trial 26981-22981/CE 3. Lancet Oncol. 2008;9(1):29–38. [DOI] [PubMed] [Google Scholar]
- 19. Volovitz I, Shapira N, Ezer H, et al. A non-aggressive, highly efficient, enzymatic method for dissociation of human brain-tumors and brain-tissues to viable single-cells. BMC Neurosci. 2016;17(1):30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nayak L, DeAngelis LM, Brandes AA, et al. The Neurologic Assessment in Neuro-Oncology (NANO) scale: a tool to assess neurologic function for integration into the Response Assessment in Neuro-Oncology (RANO) criteria. Neuro-oncology. 2017;19(5):625–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Stupp R, Mason WP, Van Den Bent MJ, et al. ; European Organisation for Research and Treatment of Cancer Brain Tumor and Radiotherapy Groups. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. [DOI] [PubMed] [Google Scholar]
- 22. Gilbert MR, Dignam JJ, Armstrong TS, et al. A randomized trial of bevacizumab for newly diagnosed glioblastoma. N Engl J Med. 2014;370(8):699–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bloch O, Han SJ, Cha S, et al. Impact of extent of resection for recurrent glioblastoma on overall survival. J Neurosurg. 2012;117(6):1032–1038. [DOI] [PubMed] [Google Scholar]
- 24. The results shown here are in whole or part based upon data generated by the TCGA Research Network. https://www.cancer.gov/tcga
- 25. R Core Team. R: A language and environment for statistical computing. R Foundation fro Statistical Computing, Vienna, Austria; 2020. https://www.R-project.org/ [Google Scholar]
- 26. Therneau T. A Package for Survival Analysis in R. R package version 3.2-7; 2020. URL: https://CRAN.R-project.org/package=survival [Google Scholar]
- 27. Alboukadel K, Marcin K, and Przemyslaw B .Survminer: Drawing Survival Curves Using “ggplot2”. R Package Version 0.4.8; 2020. https://CRAN.Rproject.org/package=survminer [Google Scholar]
- 28. Frank EH. RMS: Regression Modeling Strategies. R Package Version 6.1-0; 2020. https://CRAN.R-project.org/package=rms [Google Scholar]
- 29. Amirhossein J, Davood R, Alberto A-I and John N.. DynNom: Visualising Statistical Models using Dynamic Nomograms. R Package Version 5.0.2; 2022. https://CRAN.R-project.org/package=DynNom [Google Scholar]
- 30. Klemm F, Maas RR, Bowman RL, et al. Interrogation of the microenvironmental landscape in brain tumors reveals disease-specific alterations of immune cells. Cell. 2020;181(7):1643–1660.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Senders JT, Staples P, Mehrtash A, et al. An online calculator for the prediction of survival in glioblastoma patients using classical statistics and machine learning. Neurosurgery. 2020;86(2):E184–E192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gittleman H, Lim D, Kattan MW, et al. An independently validated nomogram for individualized estimation of survival among patients with newly diagnosed glioblastoma: NRG oncology RTOG 0525 and 0825. Neuro-oncology. 2017;19(5):669–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liu ZY, Feng SS, Zhang YH, et al. Competing risk model to determine the prognostic factors and treatment strategies for elderly patients with glioblastoma. Sci Rep. 2021;11(1):1–0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Stuart WG., Graeme LH, Stuart JH, Statistical primer: multivariable regression considerations and pitfalls. Euro J Cardio-Thoracic Surg. 2019;55(2):179–185. [DOI] [PubMed] [Google Scholar]
- 35. Mohme M, Schliffke S, Maire CL, et al. Immunophenotyping of newly diagnosed and recurrent glioblastoma defines distinct immune exhaustion profiles in peripheral and tumor-infiltrating lymphocytes immunophenotyping of T cells in GBM. Clin Cancer Res. 2018;24(17):4187–4200. [DOI] [PubMed] [Google Scholar]
- 36. Woroniecka K, Chongsathidkiet P, Rhodin K, et al. T-cell exhaustion signatures vary with tumor type and are severe in glioblastomat-cell exhaustion signatures in glioblastoma. Clin Cancer Res. 2018;24(17):4175–4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Davidson TB, Lee A, Hsu M, et al. Expression of PD-1 by T cells in malignant glioma patients reflects exhaustion and activation immune landscape of PD-1 in glioma-infiltrating T cells. Clin Cancer Res. 2019;25(6):1913–1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mostafa H, Pala A, Högel J, et al. Immune phenotypes predict survival in patients with glioblastoma multiforme. J Hematol Oncol. 2016;9(1):1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chitadze G, Flüh C, Quabius ES, et al. In-depth immunophenotyping of patients with glioblastoma multiforme: Impact of steroid treatment. Oncoimmunology. 2017;6(11):e1358839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Monaco G, Lee B, Xu W, et al. RNA-seq signatures normalized by mRNA abundance allow absolute deconvolution of human immune cell types. Cell Rep. 2019;26(6):1627–1640.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schelker M, Feau S, Du J, et al. Estimation of immune cell content in tumour tissue using single-cell RNA-seq data. Nat Commun. 2017;8(1):2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sturm G, Finotello F, Petitprez F, et al. Comprehensive evaluation of transcriptome-based cell-type quantification methods for immuno-oncology. Bioinformatics. 2019;35(14):i436–i445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nadel BB, Oliva M, Shou BL, et al. Systematic evaluation of transcriptomics-based deconvolution methods and references using thousands of clinical samples. Brief Bioinform. 2021;22(6):bbab265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Grabovska Y, Mackay A, O’Hare P, et al. Pediatric pan-central nervous system tumor analysis of immune-cell infiltration identifies correlates of antitumor immunity. Nat Commun. 2020;11(1):4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Annette MM, John KW, Gayathri W, et al. Interactions of age and blood immune factors and noninvasive prediction of glioma survival. J Natl Cancer Inst 2022;114(3):446–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhang L, Sorensen MD, Kristensen BW, et al. D-2-hydroxyglutarate is an intercellular mediator in IDH-mutant gliomas inhibiting complement and T cells. Clin Cancer Res. 2018;24(21):5381–5391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bunse L, Pusch S, Bunse T, et al. Suppression of antitumor T cell immunity by the oncometabolite (R)-2-hydroxyglutarate. Nat Med. 2018;24(8):1192–1203. [DOI] [PubMed] [Google Scholar]
- 48. Gatto L, Franceschi E, Tosoni A, et al. IDH inhibitors and beyond: the cornerstone of targeted glioma treatment. Molecular Diagnosis & Therapy. 2021;25(4):457–473. [DOI] [PubMed] [Google Scholar]
- 49. Schumacher T, Bunse L, Pusch S, et al. A vaccine targeting mutant IDH1 induces antitumour immunity. Nature. 2014;512(7514):324–327. [DOI] [PubMed] [Google Scholar]
- 50. Pellegatta S, Valletta L, Corbetta C, et al. Effective immuno-targeting of the IDH1 mutation R132H in a murine model of intracranial glioma. Acta Neuropathol Commun. 2015;3(1):1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sulkowski PL, Corso CD, Robinson ND, et al. 2-Hydroxyglutarate produced by neomorphic IDH mutations suppresses homologous recombination and induces PARP inhibitor sensitivity. Sci Transl Med. 2017;9(375):eaal2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Liu YT, Sun ZJ.. Turning cold tumors into hot tumors by improving T-cell infiltration. Theranostics. 2021;11(11):5365–5386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Fridman WH, Zitvogel L, Sautès–Fridman C, Kroemer G.. The immune contexture in cancer prognosis and treatment. Nat Rev Clin Oncol. 2017;14(12):717–734. [DOI] [PubMed] [Google Scholar]
- 54. Hu LS, Hawkins-Daarud A, Wang L, Li J, Swanson KR.. Imaging of intratumoral heterogeneity in high-grade glioma. Cancer Lett. 2020;477:97–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Gieryng A, Pszczolkowska D, Walentynowicz KA, Rajan WD, Kaminska B.. Immune microenvironment of gliomas. Lab Investigat J Techn Method Pathol. 2017;97(5):498–518. [DOI] [PubMed] [Google Scholar]
- 56. Friese MA, Platten M, Lutz SZ, et al. MICA/NKG2D-mediated immunogene therapy of experimental gliomas. Cancer Res. 2003;63(24):8996–9006. [PubMed] [Google Scholar]
- 57. Choi H, Lee Y, Park SA, et al. Human allogenic γδ T cells kill patient-derived glioblastoma cells expressing high levels of DNAM-1 ligands. Oncoimmunology. 2022;11(1):2138152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Annovazzi L, Mellai M, Bovio E, et al. Microglia immunophenotyping in gliomas. Oncol Lett. 2018;15(1):998–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wu M, Shi Y, Zhu L, et al. Macrophages in glioblastoma development and therapy: a double-edged sword. Life (Basel). 2022;12(8):1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ferguson SD, Hodges TR, Majd NK, et al. A validated integrated clinical and molecular glioblastoma long-term survival-predictive nomogram. Neuro-oncol Adv. 2021;3(1):vdaa146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Qin Z, Zhang X, Chen Z, Liu N.. Establishment and validation of an immune-based prognostic score model in glioblastoma. Int Immunopharmacol. 2020;85:106636. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are available upon reasonable request from the corresponding authors.
The data will be made available upon reasonable request from the corresponding author


