Abstract
The retinoblastoma (pRB) family of proteins includes three proteins known to suppress growth of mammalian cells. Previously we had found that growth suppression by two of these proteins, p107 and p130, could result from the inhibition of associated cyclin-dependent kinases (cdks). One important unresolved issue, however, is the mechanism through which inhibition occurs. Here we present in vivo and in vitro evidence to suggest that p107 is a bona fide inhibitor of both cyclin A-cdk2 and cyclin E-cdk2 that exhibits an inhibitory constant (Ki) comparable to that of the cdk inhibitor p21/WAF1. In contrast, pRB is unable to inhibit cdks. Further reminiscent of p21, a second cyclin-binding site was mapped to the amino-terminal portions of p107 and p130. This amino-terminal domain is capable of inhibiting cyclin-cdk2 complexes, although it is not a potent substrate for these kinases. In contrast, a carboxy-terminal fragment of p107 that contains the previously identified cyclin-binding domain serves as an excellent kinase substrate although it is unable to inhibit either kinase. Clustered point mutations suggest that the amino-terminal domain is functionally important for cyclin binding and growth suppression. Moreover, peptides spanning the cyclin-binding region are capable of interfering with p107 binding to cyclin-cdk2 complexes and kinase inhibition. Our ability to distinguish between p107 and p130 as inhibitors rather than simple substrates suggests that these proteins may represent true inhibitors of cdks.
Orderly progression through the cell cycle requires the orchestration of growth-promoting and -restraining signals. The cyclin-dependent kinases (cdks) are believed to constitute some of the most important factors driving proliferation of eukaryotic cells (40). These proteins function by phosphorylating substrates required to effect each transition through the cycle. The growth-promoting influences of the cdks are counterbalanced by two groups of cdk inhibitors (CKIs), which include at least seven members. One group includes p21 (also known as WAF1/Cip1), p27 (Kip1), and p57 (Kip2), while the second group is comprised of p15, p16, p18, and p19 (also known as INK4 proteins; reviewed in reference 36). These low-molecular-weight inhibitors were classified based on sequence homology and the kinases inhibited by each. The first group is thought to bind primarily to those kinases involved in G1 and S phase progression (i.e., kinases associated with cyclins A, D, and E), while the second group exclusively inhibits kinases associated with cyclin D.
The retinoblastoma tumor suppressor (pRB) and the related proteins p107 and p130 comprise another class of proteins involved in limiting cell cycle progression. pRB is thought to control entry into S phase in part by repressing the activity of E2F, a transcription factor known to promote proliferation (40). Another member of the pRB family, p107, regulates cell cycle progression by at least two distinct mechanisms (38, 42). p107 can also inhibit the activity of the E2F transcription factor (34, 45). In addition, p107 can interact with the cdks cyclin A-cdk2 and cyclin E-cdk2 through a second domain independent of the one required for E2F binding (42). p107 forms stoichiometric complexes with these kinases and E2F in a temporally defined manner, with the p107-cyclin E-cdk2 complex appearing in late G1 phase and p107-cyclin A-cdk2 appearing later in S phase (3, 29, 37).
Biochemical and structural studies have identified an amino acid sequence in the spacer region of p107 required for binding cyclins (43), and related sequences have been found in other cyclin-binding proteins. This short sequence motif, termed the LFG motif for the residues important for the interaction, was initially identified in the p21-p27-p57 family of mammalian and Drosophila CKIs (9, 27), and structural studies have established the importance of this motif in p27-cyclin A interactions (33). A similar sequence was noted in the spacer region of p130, and a related, but nonidentical, sequence was identified in the E2F family of transcription factors (1, 25, 26). Interestingly, this E2F sequence is necessary and sufficient for conferring cyclin A-cdk2 binding to certain members (E2F-1, -2, and -3) of this transcription factor family but not others, resulting in their phosphorylation and inhibition of activity (14).
Previously, we showed that in vitro reconstitution of stoichiometric complexes containing either p107 or p130 and cyclin A-cdk2 or cyclin E-cdk2 resulted in the loss of kinase activity directed toward an exogenous substrate, histone H1 (41). Interestingly, endogenous p130-kinase complexes isolated from human cells exhibited similar properties, and we could distinguish two cellular p130-cyclin-cdk2 complexes that lacked and contained associated E2F activity.
In this study, we have begun to address the mechanism by which p107 regulates the activity of associated cdk2 in vivo and in vitro. We have surveyed cells lacking p107 and the related p130 protein and found that the total kinase activity associated with cdk2 increases in these cells, and in complementary experiments modest increases in p107 expression in human cells significantly decreased endogenous cdk2 activity. By several biochemical criteria, we show that p107 can act as a bona fide CKI with an inhibitory constant (Ki) similar to that of p21/WAF1. Although p107 is a strong substrate for cyclin A-cdk2, cyclin D-cdk4, and cyclin E-cdk2, the ability to dissect regions of the protein that function as efficient substrates but not inhibitors suggests that inhibition does not occur simply by a preferred-substrate mechanism. In distinguishing between cyclin-cdk2 substrates and inhibitors, our experiments also point to a major difference between p107 and its relative pRB: while the former is an effective inhibitor in vitro, the latter is not. Through systematic mutagenesis of p107, we define a previously uncharacterized portion of p107 that can inhibit both cyclin A-cdk2 and cyclin E-cdk2. Interestingly, this region of p107 contains a sequence related to other cyclin-binding domains, and we show that in some settings, it is required in vivo for growth suppression. These findings prompt a comparison with the CKI p21, which also inhibits cdk activity through dual cyclin-binding sites.
MATERIALS AND METHODS
Cell lines and transfections.
The human osteosarcoma cell line Saos-2 and cervical carcinoma C33A were obtained from E. Harlow. Transfections, nocodazole treatment, and fluorescence-activated cell sorter (FACS) analysis were performed essentially as described previously (42). Briefly, for C33A transfections, cells were transfected with 2 μg of cytomegalovirus (CMV)-CD20 and 23 μg of each p107 or p130 expression plasmid (except for the p130AA and p130AAA mutants, in which case 11.5 μg of DNA was used), unless noted otherwise, by standard calcium phosphate methods. After 12 to 14 h, precipitates were removed and cells were washed twice with phosphate-buffered saline and allowed to incubate further for 24 h. Cells were then treated with 40 ng of nocodazole per ml for an additional 12 h and harvested for FACS and Western blot analyses. CD20-positive cells were analyzed with a FACScan (Becton Dickinson) equipped with CellQuest and ModFit software. Data are presented as averages from at least three separate transfection experiments.
Plasmids and peptides.
Deletion and point mutageneses were carried out by PCR. Construction of the p107ΔSA mutant involved digestion of CMV-p107 with SphI and AccI, creation of blunt ends with T4 polymerase, and ligation. To create an expression vector for Δ10N protein containing the first 409 amino acids of p107, a BamHI-DraIII fragment was excised from CMV-p107Δ10, T4 DNA polymerase blunted, and subcloned into BamHI-EcoRI-digested, Klenow polymerase-blunted pGEX-KG. Δ10NAAA was made by replacing an EagI-BsmI fragment from pGEX-KG-Δ10N with the corresponding one from CMV-p107AAA. All mutations were confirmed by DNA sequencing. Oligonucleotide sequence information and details for plasmid construction are available upon request. The glutathione S-transferase (GST)–p53 expression plasmid was a gift of H. Lu.
The following p107 peptides were synthesized: p107N (ACRKSIIPTV), p107N-mut (AAAASIIPTV), and p107S (SAKRRLFGED). All peptides were synthesized by Research Genetics, Inc.
Antibodies.
Polyclonal antibodies against p107 (C-18), cyclin A (H-432), cdk2 (M2), and p130 (C-20) were obtained from Santa Cruz Biotechnology, and anti-influenza virus hemagglutinin (HA) antibody 12CA5 was obtained from Berkeley Antibody Co. Monoclonal antibodies against cyclin E (HE12) and p107 (a mixture of SD2, -4, -6, and -9 used for immunoprecipitations or SD9 used for Western blotting) were provided by N. Dyson and E. Harlow. For immunoprecipitations that were followed by Western blotting, antibodies were first coupled to protein A-Sepharose by standard methods (20).
Recombinant protein production.
p107, N385, and pRB were purified to near homogeneity by chromatography over an affinity column bearing amino acid residues 20 to 29 of human papillomavirus E7 as described previously (13) except that the column was washed with an additional step of 1.0 HMGNB (25 mM HEPES [pH 7.6], 1 M NaCl, 10% glycerol, 0.1% Nonidet P-40 [NP-40], 5 mM β-mercaptoethanol, and 0.2 mM phenylmethylsulfonyl fluoride [PMSF]). Eluates from the E7 column were subsequently dialyzed against 0.1 HEMGNDP buffer (100 mM KCl, 25 mM HEPES [pH 7.6], 0.1 mM EDTA, 10 mM MgCl2, 10% glycerol, 1 mM dithiothreitol [DTT], 0.01% NP-40, and 0.2 mM PMSF). In experiments involving cyclin D-cdk4, buffer D (150 mM NaCl, 50 mM HEPES [pH 7.6], 1 mM EDTA, 2.5 mM EGTA, 10% glycerol, 0.1% Tween 20, 5 mM β-mercaptoethanol, and 0.2 mM PMSF) was used instead of 0.1 HMGNB. p21 was produced in bacteria and purified as described previously (43).
Purification of GST-tagged proteins.
GST-tagged recombinant Δ10N and p53 were overproduced and purified as follows. After a 1-h induction with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) at 37°C, bacteria were pelleted, washed once in phosphate-buffered saline, and sonicated in 0.1 HEMGNDP buffer containing 5 μg of leupeptin per ml and 5 μg of aprotinin per ml. Following sonication, lysates were precleared by centrifugation, and the resulting supernatants were incubated with glutathione-agarose for 1 h at 4°C. After extensive washing with 0.1 HEMGNDP, the proteins were eluted with elution buffer (100 mM Tris [pH 7.9], 120 mM NaCl, 7 mg of glutathione per ml, 1 mM DTT, and 0.2 mM PMSF). Proteins eluted from the glutathione-agarose beads were subsequently dialyzed against 0.1 HEMGNDP. GST-cyclin A-cdk2, GST-cyclin E-cdk2, and GST-cyclin D-cdk4 complexes were produced in insect cells by previously described methods (13, 14, 41). These complexes were purified as for all other GST-tagged proteins. Baculoviruses encoding HA-tagged cdk2 and GST-cyclins were generously provided by D. Morgan, W. Harper, and H. Piwinica-Worms.
In vitro kinase assays.
For kinase inhibition assays, purified p107, Δ10N, Δ10N-AAA, N385, or p21 was preincubated at room temperature for 30 min in kinase buffer (50 mM HEPES [pH 7], 10 mM MgCl2, 5 mM MnCl2, 1 mM DTT, 0.2 mg of bovine serum albumin per ml, 10 mM NaF, and 0.2 mM PMSF) with ∼0.8 ng of purified cyclin A-cdk2 or cyclin E-cdk2. Where noted, 1 μM ATP was used to phosphorylate p107 during the preincubation. Kinase buffer with 1 μM ATP, 2.5 μCi of [γ-32P]ATP (3,000 Ci/mmol), and 100 ng of the indicated substrate (or 1.25 μg in the case of histone H1) was then added, and the reactions were allowed to proceed for 15 min at 37°C. For kinase assays involving peptide competition, reactions were carried out by adding the indicated peptide at 15 μM during the preincubation. Phosphorylation levels were quantified with a PhosphorImager, and the data were plotted as percentages, with 100% representing the value for the reactions without any inhibitor. The values reported represent the averages and standard errors for three independent experiments.
Peptide competition assays.
Peptide competition assays were carried out by incubation of 40 ng of p107 with 40 ng of cyclin A-cdk2 or cyclin E-cdk2 for 1 h at 4°C in 20 μl of kinase buffer in the presence or absence of the indicated amount of peptide followed by a 1-h incubation with 500 μl of 0.1 HEMGNDP and 5 μl of a 50% slurry of glutathione-agarose beads. After extensive washing with 0.1 HEMGNDP, the precipitated proteins were resolved on a sodium dodecyl sulfate (SDS)–10% polyacrylamide gel and visualized by Western blot analysis as follows. After electrophoresis, the gels were transferred to a polyvinylidene difluoride membrane (Millipore), and the membranes were blocked for 1 h with 5% nonfat milk in Tris-buffered saline at pH 8.0 with 0.2% Tween 20, washed, and incubated with anti-p107, anti-cyclin A, or anti-cyclin E antibodies. The blots were then washed and developed by using an enhanced chemiluminescence detection system (NEN).
p107 and pRB binding assays.
Phosphorylation-binding experiments were carried out by incubation of 100 ng of p107 or 100 ng of pRB with 40 ng of cyclin A-cdk2 and 2.5 μCi of [γ-32P]ATP (3,000 Ci/mmol) for 30 min at 37°C followed by a 1-h incubation with 500 μl of 0.1 HEMGNDP and 5 μl of a 50% slurry of glutathione-agarose beads. After extensive washing with 0.1 HEMGNDP, the beads were resuspended in loading dye, boiled, loaded on an SDS–10% polyacrylamide gel, and visualized by autoradiography. Coprecipitation of cyclin A by p107 or Δ10N was carried out by incubating 50 ng of thrombin-cleaved GST-cyclin A or GST-cyclin E (thrombin was used to remove the GST moiety) with 100 ng of p107, N385, Δ10N, or Δ10N-AAA for 1 h at 4°C. Complexes were collected either by GST precipitation as described above or, in the case of p107 and N385, with 5 μl of E7 beads substituted for the GST beads.
Immunoprecipitations and deoxycholate release.
Extracts were generally made by cell lysis on ice for 30 min in E1A lysis buffer (50 mM HEPES [pH 7], 5 mM EDTA, 250 mM NaCl, 0.1% NP-40, 1 mM DTT, 0.2 mM PMSF, 2 μg of aprotinin per ml, 2 μg of leupeptin per ml, 10 mM NaF, and 50 mM β-glycerophosphate). For the analysis of p107 and p130 expression and cyclin association, approximately 25 to 200 μg of total protein was immunoprecipitated. Release of E2F associated with p107 by deoxycholate treatment and E2F gel mobility shift assays have been described previously (42).
The tetracycline-repressible p107 cell line (44) (a gift of L. Zhu and E. Harlow) was grown in Dulbecco modified Eagle medium containing 10% fetal calf serum, 500 μg of G418 per ml, and 1 μg of tetracycline per ml. After the cells were grown to 70% confluence, p107 was induced by removal of tetracycline. The cells were grown for 48 h after removal of tetracycline, and extracts of these cells were compared with those obtained from cells that were grown in the presence of tetracycline. The extracts were made in E1A lysis buffer as described above, and 300 μg of total protein was used for the immunoprecipitation with anti-cdk2 antibody as described above. Immunoprecipitates were incubated in kinase reactions as described above.
RESULTS
p107 and p130 are physiological inhibitors of cdk2 activity.
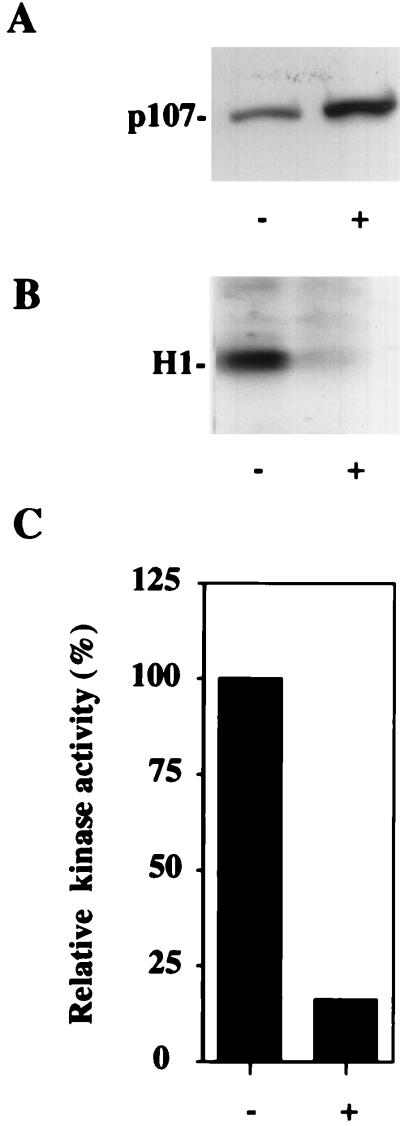
To test directly the notion that p107 is a physiological inhibitor of cdk2, we induced small increases in p107 expression in human Saos-2 osteosarcoma cells. In these cells, expression of p107 is controlled in a tetracycline-repressible manner (18, 44) (Fig. 1A). Here, removal of tetracycline led to a modest increase in p107 levels with a concomitant significant decrease in cdk2 kinase activity (Fig. 1B and C). These differences could not be ascribed to changes in the amounts of cyclin A, cyclin E, or cdk2 protein, since their levels were not altered upon induction of p107 (data not shown). Thus, we conclude that p107 is an inhibitor of cdk2 in vivo.
FIG. 1.
p107 and p130 are inhibitors of cdk2 activity in vivo. (A) p107 expression is modestly induced in a tetracycline-repressible cell line by removal of tetracycline. +, p107 induction; −, expression was not induced. (B) Induction of p107 levels shown in panel A causes significant reduction in histone H1 kinase activity. Results of a representative experiment are shown. (C) Quantitation of kinase activity shown in panel B.
In further experiments, when we immunoprecipitated cdk2 from lysates derived from wild-type, p107−/−, and p107−/− p130−/− mouse embryo fibroblasts (MEFs) (6, 24, 28), we noticed that cdk2-associated histone H1 kinase activity was elevated in either mutant cell type relative to that in wild-type cells. We reproducibly observed a 2-fold increase in kinase activity in p107−/− cells and a 2- to 2.5-fold increase in p107−/− p130−/− cells (data not shown). While these differences are modest, they are in keeping with the fact that the mutant cells have retained a host of biochemically redundant CKIs, namely, p21/WAF1, p27, and p57.
Unphosphorylated p107 and phosphorylated p107 inhibit cdks with an apparent Ki similar to that of p21/WAF1.
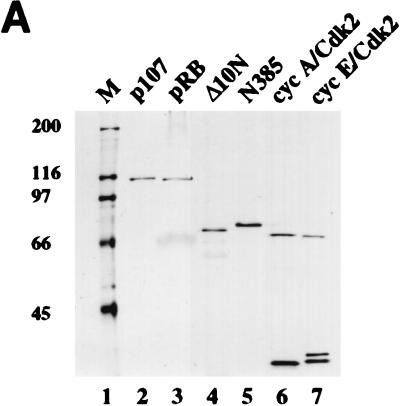
Previously we had shown that stoichiometric cyclin-cdk2 complexes containing either p107 or p130 exhibited greatly diminished kinase activity toward an exogenous substrate, histone H1, relative to free kinase complexes (41). However, given that both pRB-related proteins bound tightly to cyclin A-cdk2 and cyclin E-cdk2 and were potent substrates of these kinases, we were unable to distinguish between two potential mechanisms for kinase inhibition. In one scenario, p107 and p130 could inhibit each kinase in a manner similar to that of the p21-p27-p57 family of CKIs. Alternatively, p107 and p130 could act as preferred substrates to inhibit phosphorylation of exogenous substrates by simple substrate competition. Although the experiments described above render this latter possibility unlikely, we designed experiments using highly purified recombinant proteins (Fig. 2A) to test each possibility.
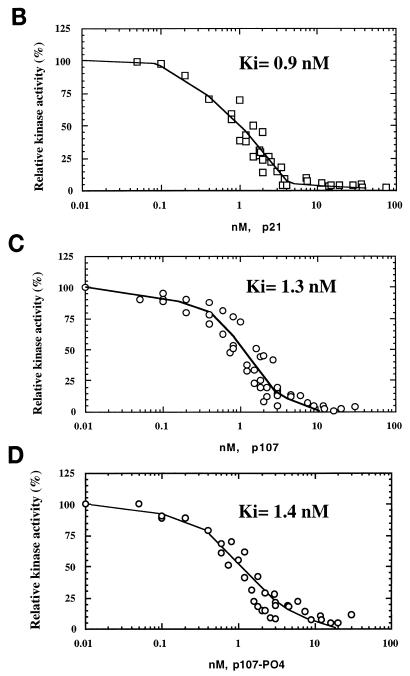
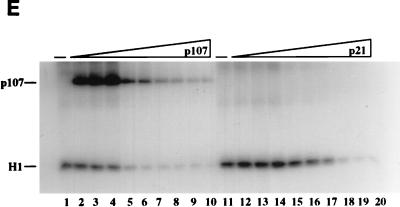
FIG. 2.
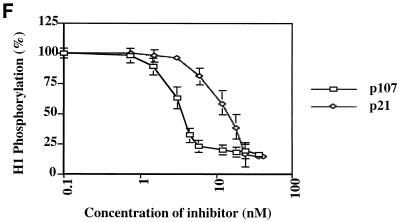
Inhibition of cyclin A (cyc A)- and cyclin E-cdk2 by p107 and p21 in vitro. (A) Recombinant proteins used in the in vitro assays. p107, pRB, N385, cyclin A-cdk2, and cyclin E-cdk2 were purified from insect cells infected with recombinant baculoviruses. GST-tagged Δ10N was purified from bacteria as described in Materials and Methods. Equal amounts of the indicated proteins were electrophoresed and visualized by silver staining. The sizes of molecular mass markers (lane M) (in kilodaltons) are indicated on the left. (B to D) Histone H1 kinase assays were performed with purified p21 from Escherichia coli or purified p107 (see Materials and Methods). Increasing concentrations of p21 (0.05 to 74 nM), p107 (0.01 to 30 nM), or prephosphorylated p107 (0.01 to 30 nM) were used as indicated. The relative levels of phosphorylated histone H1 were determined with a PhosphorImager. Histone H1 phosphorylation by cyclin A-cdk2 in the absence of an inhibitor was given a value of 100. Calculated average Ki values are indicated. (E) Histone H1 kinase assays comparing p21 and p107 as inhibitors of cyclin E-cdk2. Increasing concentrations of p107 (0.7 to 42 nM) (lanes 2 to 10) or p21 (0.7 to 42 nM) (lanes 12 to 20) were incubated with 1 ng of cyclin E-cdk2. For each set of reactions, a kinase-alone control (lanes −) was included. (F) The relative levels of phosphorylated histone H1 were determined as for panels B to D. Each value represents the mean and standard error of the mean for four independent experiments.
First, we compared the apparent inhibition constants (Kis) for cyclin A-cdk2 and cyclin E-cdk2 by using p107 and p21/WAF1, a known CKI. We performed histone H1 kinase assays in which we titrated increasing amounts of each inhibitor into the reaction mixtures. Remarkably, p107 and p21 exhibited similar apparent Kis for inhibition of cyclin A-cdk2, and we calculated Kis of 1.3 and 0.9 nM for p107 and p21, respectively (Fig. 2B to D). This estimate for the inhibitory constant for p21 is in agreement with ones made previously (0.5 nM [21]). However, p107 and p21 were markedly less potent as inhibitors of cyclin E-cdk2 (Fig. 2E and F). Moreover, p107 and p21 differentially inhibited cyclin E-cdk2, with Kis of 3.3 and 13.6 nM, respectively. This estimate for inhibition of cyclin E-cdk2 by p21 differs somewhat from the previously calculated value (3.7 nM [21]), perhaps due to differences in reaction conditions. We have also noted that purified, recombinant cyclin D-cdk4 robustly phosphorylates p107, consistent with a previous report (2), but cyclin D-cdk4 kinase activity is not significantly affected by amounts of p107 that abolish cyclin A-cdk2 activity (data not shown), suggesting that kinase inhibition by p107 is specific for cdk2-associated kinases.
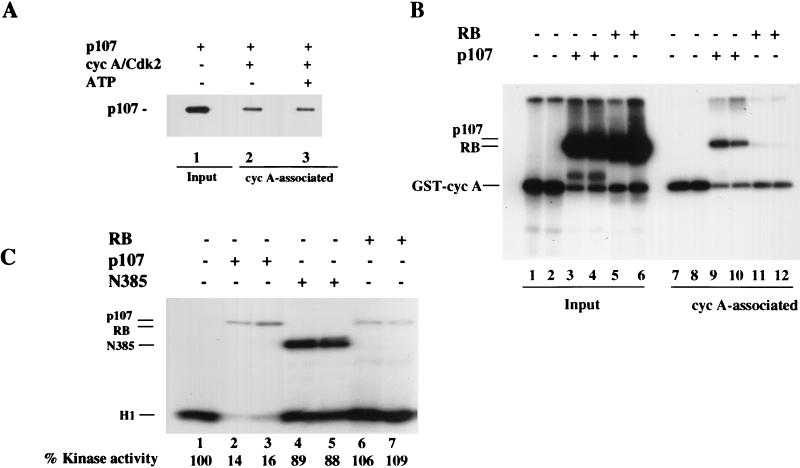
As expected, p107 was phosphorylated during the course of the kinase reaction. In principle, then, enhanced p107 phosphorylation could account for the decreased phosphorylation of histone H1. To rule out this possibility, we determined whether phosphorylated p107 was equally capable of inhibiting the phosphorylation of histone H1 by cyclin A-cdk2. Here, p107 was phosphorylated with unlabeled ATP and subsequently incubated with labeled ATP and substrate. In this experiment, further phosphorylation of p107 was not apparent, attesting to the fact that prephosphorylation of p107 approached completion (data not shown). Interestingly, phosphorylated p107 had a Ki similar to that of unphosphorylated p107 and p21 (Fig. 2B to D). The results of this experiment (and those presented below) argue further against a substrate competition model for kinase inhibition by p107 and instead support the idea that p107 is a direct and potent inhibitor of cyclin-cdk2 complexes. Since phosphorylated p107 retained the ability to inhibit cyclin A-cdk2, we determined whether the phosphorylated protein would be capable of stable binding to the kinase. Both forms of p107 bound cyclin A-cdk2 to similar extents (see Fig. 4A and B), in agreement with their comparable Ki values.
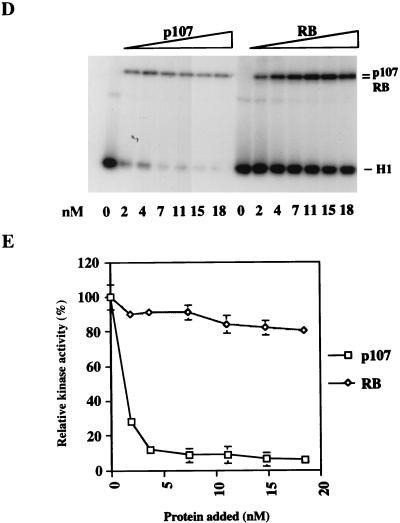
FIG. 4.
p107 and pRB as inhibitor and substrate, respectively, of cyclin A (cyc A)-cdk2. (A) p107 was allowed to bind cyclin A-cdk2 in the absence or presence of 1 μM ATP for 30 min at 37°C. Glutathione-agarose precipitation of GST-cyclin A-cdk2 was carried out as described in Materials and Methods, followed by immunoblot analysis of p107. The input amount of p107 is shown in lane 1. (B) p107 (lanes 3, 4, 9, and 10) and pRB (lanes 5, 6, 11, and 12) were incubated in a kinase reaction without histone H1 and with [γ-32P]ATP. After 30 min at 37°C, half of the reaction mixture (lanes 1 to 6) was taken and boiled in SDS sample buffer, while the other half (lanes 7 to 12) was incubated with glutathione-agarose beads and precipitated as described in Materials and Methods. (C) Cyclin A-cdk2 kinase reactions in the presence of 20 ng of p107 (lanes 2 and 3), 60 ng of N385 (lanes 4 and 5), or 20 ng of pRB (lanes 6 and 7). (D) An extended titration comparing the effects of equal amounts of pRB and p107 on cyclin A-cdk2 kinase activity. The amount of either protein is indicated below the autoradiogram. (E) Quantitation of the data shown in panel D. The relative percentage of kinase activity was determined in each case by the amount of 32P incorporated into histone H1, as measured with a PhosphorImager.
p107 inhibits phosphorylation of several cyclin A-cdk2 substrates.
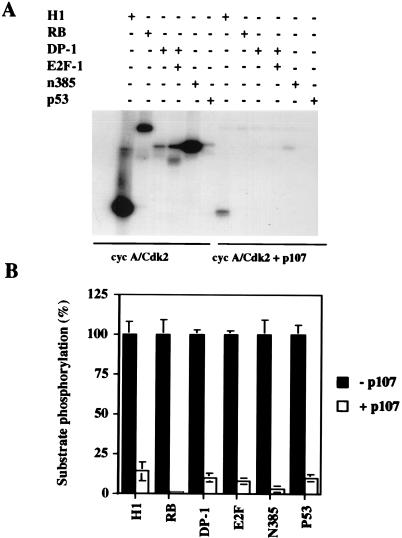
Earlier experiments did not address the possibility that p107 could preferentially inhibit the phosphorylation of some substrates but not others, in effect redirecting substrate usage by cdks. We reasoned that if p107 was truly an inhibitor of cyclin-cdk2 complexes, phosphorylation of all substrates should be diminished in the presence of this protein. We therefore tested the phosphorylation of several known cyclin A-cdk2 substrates. Only a handful of substrates have been identified thus far, and these include pRB, E2F-1 and its heterodimeric partner DP-1, and p53 (reviewed in reference 12). When equivalent amounts of these proteins (as judged by silver staining) were added to kinase reaction mixtures, phosphorylation of each substrate was considerably reduced, and in every case, the level of inhibition was also comparable (Fig. 3).
FIG. 3.
Inhibition of cyclin A (cyc A)-cdk2 substrates. (A) Kinase assays were performed in the absence (left) or presence (right) of 20 ng of p107 (previously phosphorylated). One hundred nanograms of the indicated purified proteins was added to the reaction mixture, with the exception of histone H1, for which 1.25 μg was used. (B) The amount of phosphorylation of each protein was quantified with a PhosphorImager, and the level of phosphorylation of each protein in the absence of p107 was set to 100%. The values represent the means and standard errors of the means for four different experiments.
pRB does not inhibit cyclin A-cdk2.
The p107 protein displays significant similarity with pRB, particularly in the carboxy-terminal half of the protein. To determine whether pRB was likewise able to inhibit cyclin A-cdk2, we performed comparisons between pRB and p107 under identical assay conditions. Although pRB is phosphorylated by the kinase to the same degree as p107, unlike p107, it cannot stably bind cyclin A-cdk2 after phosphorylation (Fig. 4B). More importantly, when we titrated equal amounts of p107 and pRB into kinase reaction mixtures (Fig. 4C) containing cyclin A-cdk2, we noted a striking difference in histone H1 phosphorylation. Here, p107, but not pRB, was able to inhibit cyclin A-cdk2. In further titration experiments (Fig. 4D and E), even a fivefold excess of pRB (relative to an amount of p107 that produced complete inhibition) did not significantly inhibit this kinase (80% of histone H1 kinase activity was retained). pRB was nevertheless heavily phosphorylated in these experiments (Fig. 4D). We conclude that pRB is a potent substrate for, but not an inhibitor of, cyclin A-cdk2.
An amino-terminal domain of p107 inhibits cdks.
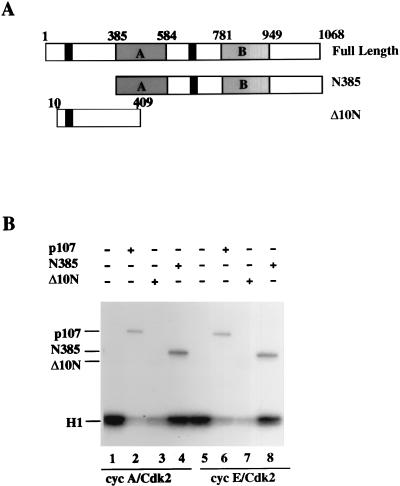
In earlier work, we had demonstrated that p107 required an amino-terminal domain to inhibit associated cyclin-cdk2 complexes completely (41). Consistent with this observation, when we titrated a purified carboxy-terminal fragment of p107 lacking 385 amino-terminal residues (termed N385) into kinase reaction mixtures, phosphorylation of histone H1 was not significantly altered, although the N385 protein was an excellent substrate (Fig. 4C, compare lanes 1 to 5). Notably, when equal amounts of protein were compared, N385 was phosphorylated to a greater extent than full-length p107 (data not shown and see Fig. 10E), confirming the idea that an inhibitory domain had been removed.
FIG. 10.
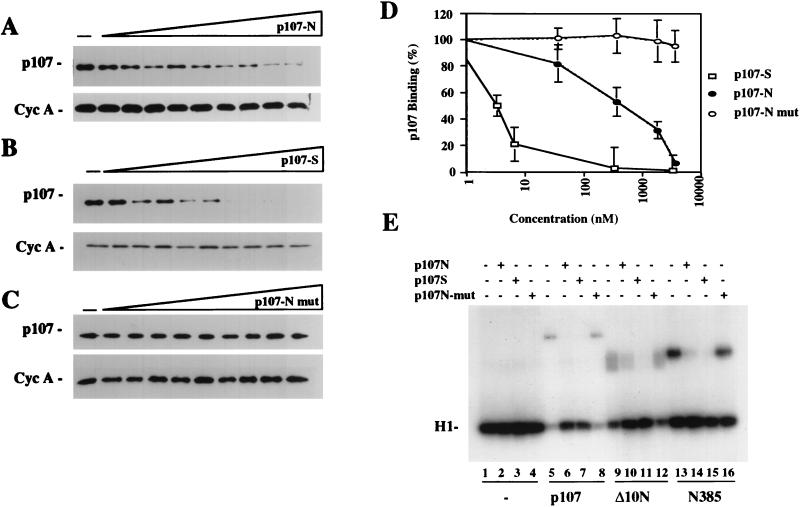
Peptide binding competition of p107 to cyclin A (Cyc A)-cdk2. (A to C) Western blot analysis carried out after GST precipitation of GST-cyclin A-cdk2 shows the capacity of peptides p107S and p107N, but not p107N-mut, to compete the binding of p107 to cyclin A-cdk2. The concentrations of peptides range from 2 nM to 5 μM. The amount of cyclin A retained was not affected by addition of peptide and was included as a control. (D) Western blots were densitometrically scanned and quantified by utilizing NIH Image 1.61 software. The values indicate the means and standard errors of the means for three independent experiments. (E) Cyclin A-cdk2 kinase reaction. The peptides p107S and p107N, but not p107N-mut, were able to partially reverse the inhibition of cyclin A-cdk2 by p107 or Δ10N. The peptides had no effect on the kinase reaction by themselves (compare lane 1 to lanes 2 to 4).
Since our experiments had identified a region in the amino-terminal one-third of p107 important for kinase inhibition, we examined the ability of a purified amino-terminal fragment to inhibit both cyclin A-cdk2 and cyclin E-cdk2. Figure 5A shows a representation of several p107 proteins that were tested. Interestingly, the GST-tagged amino-terminal fragment (called Δ10N) was a potent inhibitor of both kinases, although complete inhibition required a fivefold molar excess relative to full-length p107 (Fig. 5B). As a control, we could show that excessive (10-fold-larger) amounts of purified GST protein lacking additional residues had no effect on kinase reactions (data not shown). In contrast with the N385 protein, Δ10N is not significantly phosphorylated (data not shown and see Fig. 10E). Thus, we have identified both a carboxy-terminal region in p107 (N385) that is a potent substrate of cdks but which does not inhibit kinase activity and an amino-terminal fragment that is independently able to inhibit cyclin A-cdk2.
FIG. 5.
Inhibition of cyclin A (cyc A)- and cyclin E-cdk2 by the amino terminus of p107. (A) Schematic diagram of p107, N385, and Δ10N proteins. Shaded rectangles with A and B denote subdomains of the carboxy-terminal region conserved with pRB and p130, and black rectangles indicate potential amino-terminal and spacer region cyclin-binding domains. (B) Kinase reactions comparing equal amounts (approximately 20 ng) of p107 (lanes 2 and 6), Δ10N (lanes 3 and 7), and N385 (lanes 4 and 8). All of the reaction mixtures contained 1.25 μg of histone H1 as a substrate for cyclin A-cdk2 (lanes 1 to 4) or cyclin E-cdk2 (lanes 5 to 8), and each had been preincubated at room temperature in the presence of unlabeled 1 μM ATP. The positions of phosphorylated histone H1 and p107 proteins are indicated at the left.
The amino-terminal region of p107 is critical for growth suppression of C33A cells.
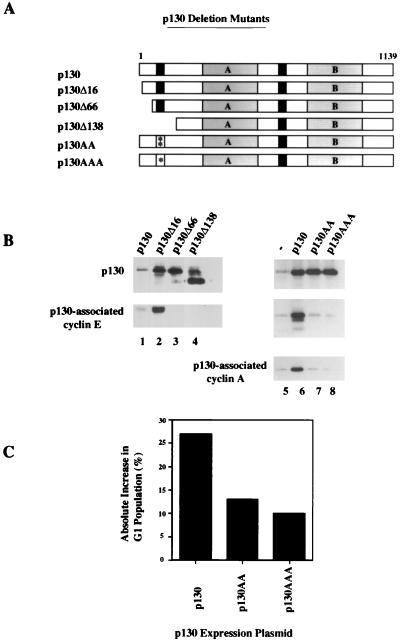
Previous studies had indicated that residues within the first 110 amino acids of p107 were important for growth suppression and complete inhibition of associated kinases (41). These data and those described above led us to examine this region of p107 and p130 in greater detail to delineate the location of a region potentially important for both cyclin binding and growth suppression. In parallel studies, we tested the effects of deletions and point mutations on cyclin binding and kinase inhibition.
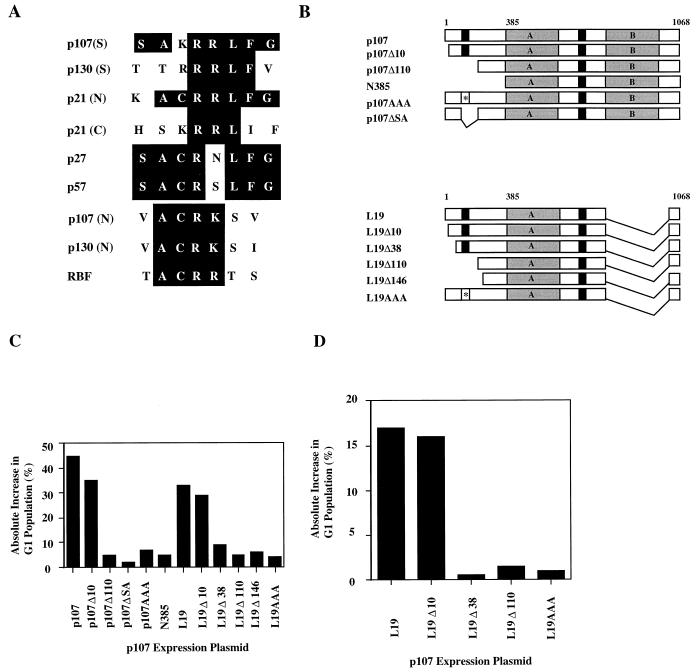
Recent biochemical and structural studies have identified a novel motif that confers tight binding to cyclins (Fig. 6A). This LFG motif is critical for interactions with several families of proteins, including the p21-p27-p57 group of CKIs, E2Fs, and the spacer regions of p107 and p130 (1, 5, 31, 33, 43). Interestingly, a second region with limited homology to the LFG motif was identified near the amino terminus of p107 and spanned residues 66 to 69 (Fig. 6A). Although this region of the protein is not highly conserved in pRB, it is strongly conserved in p130 and the Drosophila pRB-related factor, RBF (11, 19, 30, 45).
FIG. 6.
An amino-terminal region in p107 is important for growth suppression. (A) Schematic alignment of cyclin-binding domains found in the p21-p27-p57 and p107-p130 families of proteins. Previously identified sequences found in the spacer regions (S) of p107 and p130 are indicated. Homologous amino-terminal sequences (N) found in p107, p130, and the Drosophila pRB-related factor, RBF, are shown. (B) Diagram of p107 constructs used in transfection assays. Symbols used are as indicated in the legend to Fig. 5. Deletions and triple alanine point mutations (asterisks) are indicated. (C) Growth arrest of C33A cells mediated by p107 as determined by FACS analysis (see Materials and Methods). The ordinate shows the absolute difference in percentage of cells in the G1 phase of the cell cycle. Values represent averages for at least three independent experiments. (D) Growth arrest of Saos-2 cells by p107 L19 derivatives.
To begin examining the relevance of this region to cyclin binding, we took advantage of a growth suppression assay in which C33A human cervical carcinoma cells were cotransfected with various p107 test plasmids and a plasmid expressing the CD20 cell surface marker (45). CD20-positive cells were scored for cell cycle distribution by flow cytometry. To define a region important for growth suppression, we initially produced p107 deletions that removed 10, 24, 38, 76, and 110 amino-terminal residues, as well as an internal deletion of residues 68 to 132 (p107ΔSA) (Fig. 6B). Although we succeeded in expressing three deletions, p107Δ10, p107Δ110, and N385, we were unable to express many of these proteins at levels equivalent to that of full-length p107 owing to their instability in cells (data not shown).
As expected, expression of p107 provoked a potent G1 arrest, as evidenced by a large increase in this population, while the mutants truncated by 110 and 385 amino-terminal residues had little effect on cell cycle progression (Fig. 6B and C). On the other hand, p107Δ10 was nearly as potent a growth suppressor as the wild-type protein. This suggested that the region of interest is located between residues 10 and 110. Furthermore, since the protein encoded by p107ΔSA was unable to prevent cell proliferation, residues between 68 and 110 were clearly important for growth suppression. We also transfected a p107 construct termed L19 that lacks an E2F-binding domain but which retains the ability to suppress cell growth (Fig. 6A) (42). Starting with L19, we derived additional, stable amino-terminal deletion mutants of L19 that behaved in a manner similar to that of the corresponding mutants able to bind E2F described above (Fig. 6C).
To rule out the possibility that the effects described above were specific to C33A cells, we also transfected the human osteosarcoma cell line Saos-2 with L19 and mutant derivatives thereof. Use of the p107 L19 mutant bypasses growth suppression through E2F that occurs in this, but not the C33A, cell line (42). As shown in Fig. 6D, mutation of a putative amino-terminal domain dramatically compromised the growth-suppressive properties of p107 to an extent similar to that seen with C33A cells.
Identification of a second region in p107 important for cyclin binding.
Having identified a functionally important region between residues 10 and 110 that contains sequence similarity to other cyclin interaction sites, we made several additional deletions that specifically target this region of p107. Significantly, one p107 mutant, p107ΔSA, has two residues (arginine and lysine) of the putative amino-terminal cyclin-binding motif deleted and is unable to suppress growth of C33A cells (Fig. 6A and C). To confirm the idea that this region was critical for growth suppression, we generated clustered point mutations in p107 that converted the cysteine, arginine, and lysine residues to alanines (mutant p107AAA). Although expressed at levels equivalent to that of the wild-type protein, this mutant was unable to suppress growth of C33A cells (Fig. 6C). Likewise, the corresponding L19 mutant (L19AAA) was completely impaired in its ability to arrest both C33A and Saos-2 cells (Fig. 6C and 6D).
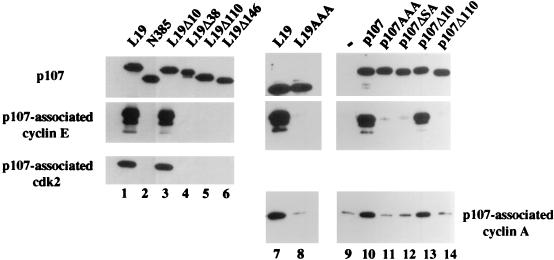
Next, we tested the ability of each p107 mutant to bind endogenous cyclins after expression in C33A or Saos-2 cells. As shown in Fig. 7, antibodies recognizing either p107 or an influenza virus HA tag on L19, or the Δ10 derivative of each, coprecipitated abundant amounts of cyclin A, cyclin E, and cdk2 (lanes 1, 3, 7, 10, and 13). In marked contrast, each of the larger amino-terminal deletions and the p107ΔSA deletion completely eliminated cyclin binding. Notably, the p107AAA and L19AAA mutants no longer associated with either cyclin (lanes 8 and 11). Thus, there was a direct correlation between the abilities of p107 to bind cyclins and suppress cell growth. Immunoblotting of identical samples of cell extracts indicated that the overall levels of cyclins A and E had not been altered by overexpression of p107 derivatives (data not shown). Although the deletion of 38 amino-terminal residues unexpectedly abrogated both cyclin binding and growth suppression (Fig. 6C and D and 7), we surmise that this indirectly results from the proximity of the deletion to the cyclin-binding region, which could destabilize its ability to bind cyclins. Alternatively, additional residues amino terminal to the putative cyclin-binding region could be important in stabilizing the association between cyclins and p107.
FIG. 7.
Mutations in p107 abolish binding to cyclins A and E. Each of the indicated proteins was expressed in C33A cells by transfection, and whole-cell extracts were immunoprecipitated with antibodies against p107 or the HA tag (which recognize the carboxy-terminal tag on p107 constructs). Immunoprecipitates were electrophoresed and subjected to immunoblotting with the indicated antibodies. A negative control transfection with empty expression vector (lane −) is indicated.
p130 mutants lacking the putative cyclin-binding domain are partially compromised in their growth-suppressive activity.
We also tested the effect of mutations of the ACRK region of p130 on growth suppression by using assays identical to those described for p107. Using sequence alignments, we produced deletion mutants of p130 that were similar to those generated for p107. These deletions removed 16, 66, and 138 amino-terminal residues and have deletion endpoints that correspond to the p107 Δ10, Δ38, and Δ110 mutants (Fig. 8A). In addition, we constructed clustered point mutations of the ACRK region of p130, converting the cysteine and arginine residues to alanines (p130AA mutant) or converting all three residues to alanine (p130AAA, which corresponds to the p107AAA mutant). We observed comparable levels of expression of p130 and each of the p130AA and p130AAA mutants and robust expression of the deletion derivatives (Fig. 8B). Each of these proteins was then immunoprecipitated from C33A cells after transfection, and coprecipitation of cyclin E and cyclin A was tested by immunoblotting. The pattern of cyclin association with each of the mutants recapitulated that seen with the corresponding p107 mutant (Fig. 7). Significantly, mutation of either the cysteine and arginine or all three residues resulted in a level of cyclin binding comparable to that of the negative control lacking exogenous p130 expression (Fig. 8B, compare lanes 1, 3, and 4).
FIG. 8.
Mutations in p130 similar to those introduced into p107 also abolish cyclin binding and diminish growth-suppressive activity. (A) Schematic of p130 mutations. Symbols are described in legend to Fig. 6. (B) Cyclin-binding activity of transfected p130 constructs. Endogenous cyclins were coimmunoprecipitated with p130 by using anti-p130 antibodies as described in the legend to Fig. 7. (C) Growth arrest of C33A cells as determined in Fig. 6C. The transfected p130 plasmids are indicated below the graph, and results represent averages for several independent experiments.
Intriguingly, although the p130AA and p130AAA mutants had apparently lost the ability to bind cyclins in vivo and appeared to have reduced growth-suppressive activity, the extent of this defect differed from that of the corresponding p107AAA mutant (compare Fig. 8C with 6C). While the basis for this residual inhibitory activity is presently unknown, it is possible that the growth-suppressive properties of p130 could arise in part from additional mechanisms not utilized by p107.
Taking these observations and our observations on p107 together, we conclude that p107 and p130 require two binding sites for potent in vivo interactions with cyclins and growth suppression activity.
The mutant p107 and p130 proteins retain E2F-binding activity.
Given the rather severe effects on cyclin binding caused by deleting the amino-terminal domains of p107 and p130, it was important to determine whether the mutations globally disrupted the structures of these proteins. Since both of these proteins are able to bind cellular E2F, we relied on an assay in which endogenous E2F activity associated with p107 or p130 is released by the detergent deoxycholate. E2F released by this treatment can be assayed for DNA-binding activity by using a gel mobility shift assay with a labeled oligonucleotide containing an E2F-binding site. Each of the amino-terminal mutants retained the ability to bind E2F (data not shown), suggesting that the loss of cyclin binding was not due to simple protein unfolding.
Mutations in the amino-terminal cyclin-binding domain abrogate kinase inhibition.
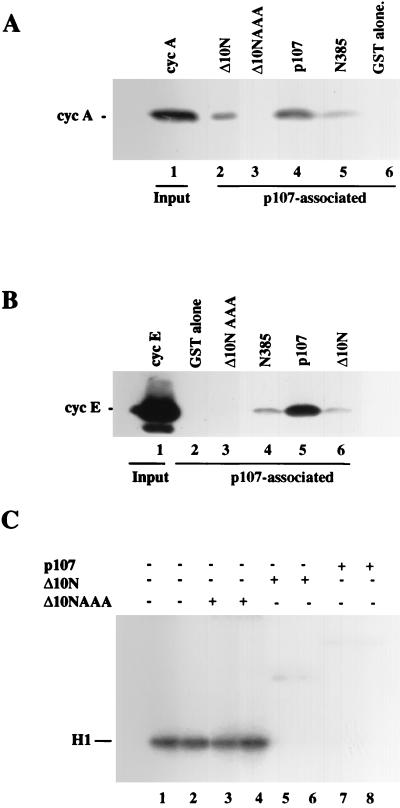
Having determined that the amino-terminal region of p107 was important for growth suppression and cyclin binding in transfection experiments, we then tested the role of the amino-terminal cyclin-binding site in cyclin-cdk2 binding and inhibition in vitro. First, we tested the ability of the Δ10N protein to bind cyclin A and cyclin E. Purified cyclins interacted with both full-length p107 and Δ10N, although binding of Δ10N to both cyclins, especially cyclin E, was significantly reduced (Fig. 9A). In addition, the N385 protein (which contains the spacer region), like Δ10N, was able to bind both cyclins, although it did so with lower efficiency than the full-length protein (Fig. 9A and B). Notably, however, only the amino-terminal Δ10N fragment was capable of inhibiting cdks. We conclude that p107 utilizes a second region near the amino terminus, in addition to the LFG domain residing in the spacer region, to bind and inhibit cyclin-cdk2 complexes.
FIG. 9.
Mutation of the p107 amino-terminal cyclin-binding domain abolishes kinase inhibition and binding. Cyclin A (cyc A) (A) and cyclin E (B) bind Δ10N and N385, albeit less efficiently than full-length p107, but do not bind the mutant Δ10N-AAA. (A) Western blot analysis of cyclin A bound to p107 derivatives. Thrombin-cleaved cyclin A, devoid of a GST tag, was tested for binding to GST alone (lane 2), Δ10N (lane 3), Δ10N-AAA (lane 4), p107 (lane 5), and N385 (lane 6). The input amount of cyclin A is shown in lane 1. (B) Western blot analysis of cyclin E bound to p107 derivatives. Thrombin-cleaved cyclin E, devoid of a GST tag, was tested for binding to GST alone (lane 2), Δ10NAAA (lane 3), N385 (lane 4), p107 (lane 5), and Δ10N (lane 6). The cyclin E input is indicated in lane 1. In each case, precipitation of N385 and p107 was performed with a human papillomavirus E7 peptide linked to Sepharose beads, and precipitation of Δ10N and Δ10N-AAA was performed with glutathione-agarose beads as described in Materials and Methods. (C) Kinase reactions comparing equal amounts of Δ10N-AAA (lanes 3 and 4), Δ10N (lanes 5 and 6), or p107 (lanes 7 and 8).
We also constructed, purified, and tested a GST-tagged amino-terminal fragment identical to Δ10N (described above) in which the three alanine mutations present in p107AAA have replaced the wild-type residues. We titrated equal amounts of this protein, termed Δ10N-AAA, and Δ10N into kinase reaction mixtures containing cyclin A-cdk2. Δ10N and full-length p107 were potent inhibitors, as expected, while the mutant version of Δ10N failed to inhibit kinase activity (Fig. 9C, compare lanes 3 to 8). Moreover, relative to that with Δ10N, introduction of the triple alanine mutation severely impaired binding to both cyclin A and cyclin E (Fig. 9A and B).
Peptides spanning several cyclin-binding domains specifically compete p107 binding to cyclin-cdk2 and reverse inhibition of kinases by p107.
As a final in vitro test of the specificity of interactions between cyclins and the amino-terminal cyclin-binding domain of p107, we performed competition experiments using peptides spanning the regions depicted in Fig. 6A as well as a peptide corresponding to the triple alanine point mutant of p107. Thus, peptides corresponding to the cyclin-binding motifs in the amino-terminal region of p107 (termed p107N) and the spacer region (termed p107S) were synthesized. In addition, a mutated peptide corresponding to p107N (p107N-mut) in which the cysteine, arginine, and lysine residues were converted to alanines was synthesized.
First, we determined whether coincubation of each peptide with p107 and cyclin A-cdk2 could effectively compete p107-kinase interactions. Figure 10 shows that peptide p107S effectively competed the binding of p107 to cyclin A-cdk2. Importantly, peptide p107N was also capable of competing this interaction, albeit more weakly, and the p107N-mut peptide was completely without effect at the highest concentration tested. As a control, we showed that the peptides did not interfere with retention of the cyclin A-cdk2 complex (lower panels in Fig. 10A to C). Experiments performed in parallel with cyclin E-cdk2 indicated that both the p107N and p107S peptides could compete the binding of this complex to p107, although each one was somewhat less effective at competing with p107 binding to this complex relative to the case for cyclin A-cdk2 (data not shown).
A similar profile was obtained when we titrated each of these peptides into kinase reaction mixtures in which cyclin A-cdk2 was inhibited by p107 (Fig. 10E). Both p107N and p107S were able to alleviate p107-mediated kinase inhibition (compare lanes 5 to 7). In contrast, the p107N-mut peptide did not alter inhibition by p107 at the highest concentration tested, nor did any of the peptides influence histone H1 kinase activity in the absence of p107 (Fig. 10E, lanes 1 to 4 and 8). Furthermore, we could show that peptides p107N and p107S, but not p107N-mut, could partially reverse kinase inhibition by the purified Δ10N fragment (lanes 8 to 12). As expected, the peptides had no effect on the activity of N385, which did not inhibit kinase activity (lanes 13 to 16). From these data, we conclude that the amino-terminal ACRK sequence found in p107 is important for mediating cyclin binding and kinase inhibition.
DISCUSSION
p107 was originally described as a pRB-related component of complexes that contained not only E2F but stoichiometric amounts of cyclin A-cdk2 and cyclin E-cdk2 as well (3, 15, 16, 37), and subsequent experiments suggested that p107 was a tightly binding substrate of these kinases. Our observations suggest that p107 not only is able to bind both kinases but also is capable of inhibiting the complexes to the same degree as most characterized CKIs (21). The inhibitory potential of p107 is not significantly affected by phosphorylation by the cyclin-cdk2 complex, not unlike certain CKIs, such as p27/Kip1, which are themselves phosphorylated during the process of inactivating these kinase complexes (35).
Furthermore, if p107 is a true kinase inhibitor, it should prevent the phosphorylation of known substrates. Indeed, this is the case. p107 can prevent the phosphorylation of several known cyclin A-cdk2 substrates, including pRB, E2F-1, DP-1, p53, and its own carboxy-terminal region, which contains most of the phosphorylation sites on p107, suggesting that p107 is capable of inhibiting the associated kinase rather than altering its substrate specificity, as suggested previously (22). In the previous study (22), it was shown that immunoprecipitates of p107 and p130 from cell extracts contained cyclin-cdk2 but showed little histone H1 kinase activity. However, the same immunoprecipitates nevertheless retained the ability to phosphorylate GST-pRB family protein fusions. Although the basis for this difference is currently unknown, it is important to note that our work has dealt with complexes reconstituted with highly purified proteins, while the work of Hauser et al. (22) has relied on immunoprecipitates of endogenous proteins, and the possibility that a contaminating kinase(s) activity was coprecipitated was not excluded.
Our results therefore suggest an important functional distinction between different members of the pRB family of proteins. All members of this family are thought to restrain cell growth as a consequence of transcriptional repression of E2F activity, and each is an excellent substrate for cyclin-cdk2 complexes. However, our work has distinguished at least two differences between pRB and p107 or p130. First, although pRB shows sequence similarity to p107 and p130, especially in the well-defined carboxy-terminal region (termed the pocket domain), it displays little similarity to these proteins in the amino-terminal region. In agreement with our identification of an amino-terminal region of p107 involved in kinase inhibition, we have shown here that pRB does not inhibit cyclin A-cdk2. Second, whereas pRB is not stably bound to this kinase before (10) or after (Fig. 4) phosphorylation, p107 remains bound to the kinase and inhibits its activity toward other substrates. Taken together, these experiments could provide an explanation for how pRB might function as a kinase substrate, while p107 and p130, with dedicated cyclin-binding domains, could function as specific inhibitors.
We have defined a second region of p107 and p130, spanning residues 67 to 69, that is required for efficient cyclin binding in addition to the LFG motif in the spacer region (43), and mutation of these amino acids results in the loss of inhibitory potential of the p107 amino-terminal domain. Furthermore, these p107 mutants were no longer bound by either cyclin-cdk2 complex in vivo, implying that binding of this portion of the molecule is important for both kinase and growth inhibition. Moreover, we showed that peptides corresponding to amino- and carboxy-terminal cyclin-binding sequences, but not a mutant version thereof, could specifically compete for the binding of cyclin A-cdk2 and cyclin E-cdk2 to p107 as well as restore histone H1 kinase activity inhibited by p107.
p107 displays an inhibitory spectrum similar to that of p21 and p27 with regard to the level of inhibition of cyclin A- and cyclin E-cdk2, although p107 may be more restricted than p21 and p27 in its specificity, since p107 does not appreciably inhibit cyclin D-cdk4 (data not shown). Interestingly, in our experiments, p107 inhibits cyclin E-cdk2 with a sixfold lower Ki than p21, suggesting that p107 may in some situations be a physiologically relevant inhibitor of cyclin E-cdk2. In addition to the ability to inhibit both cyclin A- and cyclin E-associated kinases, p107 and the CKI p21/WAF1 share another property, namely, the presence of dual cyclin-binding sites in their amino- and carboxy-terminal regions. It had been shown previously that p21/WAF1 utilizes two cyclin-binding domains, an amino-terminal one with an LFG motif known to be of critical importance in the binding of cyclin A to p27 (4, 17, 31, 33) and a second, related sequence near the carboxy terminus (Fig. 6A) (5).
Why is it necessary for p21 and p107 to engage cyclin-cdk2 complexes by using two binding sites? While we have not addressed the molecular architecture of cyclin-p107 or cyclin-p21 interactions here, we speculate that both cyclin-binding sites are essential for high-affinity interactions and potent inhibition. Indeed, although we could demonstrate detectable interactions between either binding site alone and cyclins in vitro, binding to full-length p107 was noticeably more efficient in vitro and in vivo, and both p107 binding sites were required in vivo to achieve efficient binding of cyclins and growth inhibition of C33A cells.
Although we have determined the existence of two cyclin-binding sites in p107, our studies point to a unique role for the amino-terminal region in kinase inhibition and growth suppression. The amino-terminal region of p107 is solely able to inhibit cyclin-cdk2 complexes in vitro, while equal or excessive amounts of the carboxy-terminal portion are unable to do so, although this portion is an efficient substrate for these kinases. A more detailed understanding of how these two regions of p107 differentially interact with cyclin-cdk2 will require a structural analysis of ternary p107-cyclin-cdk2 complexes.
It is intriguing to speculate that p107 and p130 may share overlapping functions as a CKI in the cell. p130 may share several properties of p107 both as an inhibitor and as a transcriptional regulator of E2F activity, as illustrated by numerous biochemical studies of these proteins and by recent mouse knockout experiments (6, 7, 23, 28, 32, 39). In the latter studies, it was shown that mice lacking not one but both proteins suffer lethal developmental abnormalities. Moreover, MEFs deficient in p107 and p130, but not either alone, showed strongly derepressed expression of specific genes, again suggesting that the two proteins have overlapping functions (24). Indeed, the two proteins also share a number of properties in our biochemical assays. For example, both appear to inhibit associated kinase activity (41), and both have sequences related to the LFG motif (8, 26). Thus, it is tempting to speculate that p130 may function in a manner analogous to p107. However, in our transfection assays, deletion of the amino-terminal cyclin-binding motif largely abolished the growth-suppressive activity of p107, while the growth arrest imposed by p130 was not altered as drastically by similar mutations (Fig. 6 and 8). This could suggest that p130 uses an additional mechanism(s) for restraining growth other than those used by p107. This possibility is currently under investigation.
The compensatory nature of p107 and p130 function has made it virtually impossible to study the normal role of each protein in vivo, and the function of different E2F complexes with p107 or p130 and cyclin-cdk2 remains obscure. In addition to studies related to kinase inhibition by these proteins, experiments are under way to dissect the transcriptional regulatory mechanisms through which each multiprotein complex functions in order to understand the normal role of p107 and p130 in cell cycle progression.
ACKNOWLEDGMENTS
We thank S. Woo for excellent technical help and I. Sanchez, J. Ross, and K. Cai for comments on the manuscript and helpful discussions. We thank N. Dyson, E. Harlow, T. Jacks, and B. Weinberg for wild-type and nullizygous MEFs. We are grateful to E. Harlow and L. Zhu for providing plasmids and cell lines and to D. Morgan, H. Lu, W. Harper, and H. Piwinica-Worms for plasmids and baculoviruses.
This work was supported in part by a Research Project Grant (RPG-98-074-01-GMC) from the American Cancer Society and the Department of Defense (U.S. Army award no. DAMD17-96-1-6092). B.D.D. is also most grateful to E. and K. Langone and to the Damon Runyon-Walter Winchell Cancer Fund for the generous donation of a Damon Runyon Scholar Award (DRS-01).
REFERENCES
- 1.Adams P D, Sellers W R, Sharma S K, Wu A D, Nalin C M, Kaelin W G. Identification of a cyclin-cdk2 recognition motif present in substrates and p21-like cyclin-dependent kinase inhibitors. Mol Cell Biol. 1996;16:6623–6633. doi: 10.1128/mcb.16.12.6623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beijersbergen R L, Carlee L, Verkhoven R M, Bernards R. Regulation of the retinoblastoma protein-related p107 by G1 cyclin complexes. Genes Dev. 1995;9:1340–1353. doi: 10.1101/gad.9.11.1340. [DOI] [PubMed] [Google Scholar]
- 3.Cao L, Faha B, Dembski M, Tsai L-H, Harlow E, Dyson N. Independent binding of the retinoblastoma protein and p107 to the transcription factor E2F. Nature. 1992;355:176–179. doi: 10.1038/355176a0. [DOI] [PubMed] [Google Scholar]
- 4.Chen I-T, Akamatsu M, Smith M L, Lung F-D T, Duba D, Roller P P, Fornace A J, O’Connor P M. Characterization of p21cip1/waf1 peptide domains required for cyclin E/cdk2 and PCNA interactions. Oncogene. 1996;12:595–607. [PubMed] [Google Scholar]
- 5.Chen J, Saha P, Kornbluth S, Dynlacht B D, Dutta A. Cyclin-binding motifs are essential for the function of p21CIP1. Mol Cell Biol. 1996;16:4673–4682. doi: 10.1128/mcb.16.9.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cobrinik D, Lee M-H, Hannon G, Mulligan G, Bronson R T, Dyson N, Harlow E, Beach D, Weinberg R A, Jacks T. Shared role of the pRB-related p130 and p107 proteins in limb development. Genes Dev. 1996;10:1633–1644. doi: 10.1101/gad.10.13.1633. [DOI] [PubMed] [Google Scholar]
- 7.Cobrinik D, Whyte P, Peeper D S, Jacks T, Weinberg R A. Cell cycle-specific association of E2F with the p130 E1A-binding domain. Genes Dev. 1993;7:2392–2404. doi: 10.1101/gad.7.12a.2392. [DOI] [PubMed] [Google Scholar]
- 8.De Luca A, MacLachlan T K, Bagella L, Dean C, Howard C M, Claudio P P, Baldi A, Khalil K, Giordano A. Unique domain of pRb2/p130 acts as an inhibitor of Cdk2 kinase activity. J Biol Chem. 1997;272:20971–20974. doi: 10.1074/jbc.272.34.20971. [DOI] [PubMed] [Google Scholar]
- 9.de Nooij J C, Letendre M A, Hariharan I K. A cyclin-dependent kinase inhibitor, Dacapo, is necessary for timely exit from the cell cycle during Drosophila embryogenesis. Cell. 1996;87:1237–1247. doi: 10.1016/s0092-8674(00)81819-x. [DOI] [PubMed] [Google Scholar]
- 10.Dowdy S F, Hinds P W, Louie K, Reed S I, Arnold A, Weinberg R A. Physical interaction of the retinoblastoma protein with human D cyclins. Cell. 1993;73:499–511. doi: 10.1016/0092-8674(93)90137-f. [DOI] [PubMed] [Google Scholar]
- 11.Du W, Vidal M, Xie J-E, Dyson N. A homologue of the retinoblastoma family of proteins regulates E2F activity in Drosophila. Genes Dev. 1996;10:1206–1218. doi: 10.1101/gad.10.10.1206. [DOI] [PubMed] [Google Scholar]
- 12.Dynlacht B. Regulation of transcription by proteins that control the cell cycle. Nature. 1997;389:149–152. doi: 10.1038/38225. [DOI] [PubMed] [Google Scholar]
- 13.Dynlacht B D, Flores O, Lees J A, Harlow E. Differential regulation of E2Ftrans-activation by cyclin-cdk2 complexes. Genes Dev. 1994;8:1772–1786. doi: 10.1101/gad.8.15.1772. [DOI] [PubMed] [Google Scholar]
- 14.Dynlacht B D, Moberg K, Lees J A, Harlow E, Zhu L. Specific regulation of E2F family members by cyclin-dependent kinases. Mol Cell Biol. 1997;17:3867–3875. doi: 10.1128/mcb.17.7.3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ewen M, Faha B, Harlow E, Livingston D. Interaction of p107 with cyclin A independent of complex formation with viral oncoproteins. Science. 1992;255:85–87. doi: 10.1126/science.1532457. [DOI] [PubMed] [Google Scholar]
- 16.Faha B, Ewen M, Tsai L-H, Livingston D, Harlow E. Interaction between human cyclin A and adenovirus E1A-associated p107 protein. Science. 1992;255:87–90. doi: 10.1126/science.1532458. [DOI] [PubMed] [Google Scholar]
- 17.Fotedar R, Fitzgerald P, Rouselle T, Cannella D, Doree M, Messier H, Fotedar A. p21 contains independent binding sites for cyclin and cdk2: both sites are required to inhibit cdk2 kinase activity. Oncogene. 1996;12:2155–2164. [PubMed] [Google Scholar]
- 18.Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hannon G J, Demetrick D, Beach D. Isolation of the Rb-related p130 through its interaction with CDK2 and cyclins. Genes Dev. 1993;7:2378–2391. doi: 10.1101/gad.7.12a.2378. [DOI] [PubMed] [Google Scholar]
- 20.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 21.Harper J W, Elledge S J, Keyomarsi K, Dynlacht B, Tsai L-H, Zhang P, Dobrowolski S, Bai C, Connell-Crowley L, Swindell E, Fox M P, Wei N. Inhibition of cyclin-dependent kinases by p21. Mol Biol Cell. 1995;6:387–400. doi: 10.1091/mbc.6.4.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hauser P J, Agrawal D, Chu B, Pledger W J. p107 and p130 associated cyclin A has altered substrate specificity. J Biol Chem. 1997;272:22954–22959. doi: 10.1074/jbc.272.36.22954. [DOI] [PubMed] [Google Scholar]
- 23.Hijmans E M, Voorhoeve P M, Beijersbergen R L, Veer L J, Bernards R. E2F-5, a new E2F family member that interacts with p130 in vivo. Mol Cell Biol. 1995;15:3082–3089. doi: 10.1128/mcb.15.6.3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hurford R K, Cobrinik D, Lee M-H, Dyson N. pRB and p107/p130 are required for the regulated expression of different sets of E2F responsive genes. Genes Dev. 1997;11:1447–1463. doi: 10.1101/gad.11.11.1447. [DOI] [PubMed] [Google Scholar]
- 25.Krek W, Ewen M E, Shirodkar S Z, Arany Z, Kaelin W G, Livingston D M. Negative regulation of the growth-promoting transcription factor E2F-1 by a stably bound cyclin A-dependent protein kinase. Cell. 1994;78:161–172. doi: 10.1016/0092-8674(94)90582-7. [DOI] [PubMed] [Google Scholar]
- 26.Lacy S, Whyte P. Identification of a p130 domain mediating interactions with cyclin A/cdk2 and cyclin E/cdk2 complexes. Oncogene. 1997;14:2395–2406. doi: 10.1038/sj.onc.1201085. [DOI] [PubMed] [Google Scholar]
- 27.Lane M E, Sauer K, Wallace K, Jan Y N, Lehner C F, Vaessin H. Dacapo, a cyclin-dependent kinase inhibitor, stops cell proliferation during Drosophila development. Cell. 1996;87:1225–1235. doi: 10.1016/s0092-8674(00)81818-8. [DOI] [PubMed] [Google Scholar]
- 28.Lee M-H, Williams B O, Mulligan G, Mukai S, Bronson R T, Dyson N, Harlow E, Jacks T. Targeted disruption of p107: functional overlap between p107 and Rb. Genes Dev. 1996;10:1621–1632. doi: 10.1101/gad.10.13.1621. [DOI] [PubMed] [Google Scholar]
- 29.Lees E, Faha B F, Dulic V, Reed S I, Harlow E. Cyclin E/cdk2 and cyclin A/cdk2 kinases associate with p107 and E2F in a temporally distinct manner. Genes Dev. 1992;6:1874–1885. doi: 10.1101/gad.6.10.1874. [DOI] [PubMed] [Google Scholar]
- 30.Li Y, Graham C, Lacy S, Duncan A M V, Whyte P. The adenovirus E1A-associated 130-kD protein is encoded by a member of the retinoblastoma gene family and physically interacts with cyclins A and E. Genes Dev. 1993;7:2366–2377. doi: 10.1101/gad.7.12a.2366. [DOI] [PubMed] [Google Scholar]
- 31.Lin J, Reichner C, Wu X, Levine A. Analysis of wild-type and mutant p21WAF activities. Mol Cell Biol. 1996;16:1786–1793. doi: 10.1128/mcb.16.4.1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mayol X, Grana X, Baldi A, Sang N, Hu Q, Giordano A. Cloning of a new member of the retinoblastoma gene family (pRb2) which binds to the E1A transforming domain. Oncogene. 1993;8:2561–2566. [PubMed] [Google Scholar]
- 33.Russo A R, Jeffrey P D, Patten A K, Massague J, Pavletich N P. Crystal structure of the p27Kip1 cyclin-dependent-kinase inhibitor bound to the cyclin A-Cdk2 complex. Nature. 1996;382:325–331. doi: 10.1038/382325a0. [DOI] [PubMed] [Google Scholar]
- 34.Schwarz J K, Devoto S H, Smith E J, Chellappan S P, Jakoi L, Nevins J R. Interactions of the p107 and Rb proteins with E2F during the cell proliferation response. EMBO J. 1993;12:1013–1020. doi: 10.1002/j.1460-2075.1993.tb05742.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sheaff R J, Groudine M, Gordon M, Roberts J M, Clurman B E. Cyclin E-CDK2 is a regulator of p27Kip1. Genes Dev. 1997;11:1464–1478. doi: 10.1101/gad.11.11.1464. [DOI] [PubMed] [Google Scholar]
- 36.Sherr C, Roberts J. Inhibitors of mammalian G1 cyclin-dependent kinases. Genes Dev. 1995;9:1149–1153. doi: 10.1101/gad.9.10.1149. [DOI] [PubMed] [Google Scholar]
- 37.Shirodkar S, Ewen M, DeCaprio J A, Morgan D, Livingston D, Chittenden T. The transcription factor E2F interacts with the retinoblastoma product and a p107-cyclin A complex in a cell cycle-regulated manner. Cell. 1992;68:157–166. doi: 10.1016/0092-8674(92)90214-w. [DOI] [PubMed] [Google Scholar]
- 38.Smith E, Nevins J R. The Rb-related p107 protein can suppress E2F function independently of binding to cyclin A/cdk2. Mol Cell Biol. 1995;15:338–344. doi: 10.1128/mcb.15.1.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vairo G, Livingston D M, Ginsberg D. Functional interaction between E2F-4 and p130: evidence for distinct mechanisms underlying growth suppression by different retinoblastoma protein family members. Genes Dev. 1995;9:869–881. doi: 10.1101/gad.9.7.869. [DOI] [PubMed] [Google Scholar]
- 40.Weinberg R A. The retinoblastoma protein and cell cycle control. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 41.Woo M S-A, Sanchez I, Dynlacht B D. p130 and p107 use a conserved domain to regulate cellular cyclin-dependent kinase activity. Mol Cell Biol. 1997;17:3566–3579. doi: 10.1128/mcb.17.7.3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu L, Enders G, Lees J A, Beijersbergen R L, Bernards R, Harlow E. The pRB-related protein p107 contains two growth suppression domains: independent interactions with E2F and cyclin/cdk complexes. EMBO J. 1995;14:1904–1913. doi: 10.1002/j.1460-2075.1995.tb07182.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu L, Harlow E, Dynlacht B D. p107 uses a p21CIP1-related domain to bind cyclin/cdk2 and regulate interactions with E2F. Genes Dev. 1995;9:1740–1752. doi: 10.1101/gad.9.14.1740. [DOI] [PubMed] [Google Scholar]
- 44.Zhu L, Enders G H, Wu C-L, Starz M A, Moberg K H, Lees J A, Dyson N, Harlow E. Growth suppression by members of the retinoblastoma protein family. Cold Spring Harbor Symp Quant Biol. 1994;59:75–84. doi: 10.1101/sqb.1994.059.01.011. [DOI] [PubMed] [Google Scholar]
- 45.Zhu L, van den Heuvel S, Helin K, Fattaey A, Ewen M, Livingston D, Dyson N, Harlow E. Inhibition of cell proliferation by p107, a relative of the retinoblastoma protein. Genes Dev. 1993;7:1111–1125. doi: 10.1101/gad.7.7a.1111. [DOI] [PubMed] [Google Scholar]