Abstract
Monoclonal antibodies (mAbs) have transformed therapeutic strategies for various diseases. Their high specificity to target antigens makes them ideal therapeutic agents for certain diseases. However, a challenge to their application in clinical practice is their potential risk to induce unwanted immune response, termed immunogenicity. This challenge drives the continued efforts to deimmunize these protein therapeutics while maintaining their pharmacokinetic properties and therapeutic efficacy. Because mAbs hold a central position in therapeutic strategies against an array of diseases, the importance of conducting comprehensive immunogenicity risk assessment during the drug development process cannot be overstated. Such assessment necessitates the employment of in silico, in vitro, and in vivo strategies to evaluate the immunogenicity risk of mAbs. Understanding the intricacies of the mechanisms that drive mAb immunogenicity is crucial to improving their therapeutic efficacy and safety and developing the most effective strategies to determine and mitigate their immunogenic risk. This review highlights recent advances in immunogenicity prediction strategies, with a focus on protein engineering strategies used throughout development to reduce immunogenicity.
Key Points
| Antidrug antibody (ADA) rates vary among protein therapeutics, and immunogenicity remains a major challenge in the development of mAb therapies. |
| Multiple factors related to the therapeutic protein and to the patient may lead to ADA generation. |
| Early assessment of immunogenicity risk can contribute to the development of more effective and selective protein therapeutics with low ADAs, ultimately improving patient outcomes. |
| Multiple strategies exist to limit and mitigate immunogenicity risk during therapeutic protein design or drug development. |
Introduction
As of 2022, the number of biopharmaceutical agents with active licenses in the USA is greater than 620, with monoclonal antibodies (mAbs) accounting for more than 50% of all new approvals in the past few years [1, 2]. Initially, murine mAbs derived from mouse B cell hybridomas entered clinical studies. However, the success of those mAbs was very limited because of the human immune system’s high recognition of those murine mAbs as foreign protein [3]. To reduce this risk, mAbs were then generated by fusing the variable domain (Fab) encoded by B cells from immunized mice with the constant domain (Fc) of human IgG. These chimeric antibodies retain the antigen-binding specificity of the original murine antibody and interact with human effector cells and proteins due to their human constant regions. Although these mouse–human chimeric antibodies were successfully applied for diagnostic purposes, their application as treatments was limited due to the generation of a robust antidrug antibody (ADA) response in patients, leading to increased drug clearance and even fatal adverse events in some cases [4]. Nevertheless, the seminal work with chimeric antibodies laid the groundwork for future developments in antibody engineering and for the development of humanized and fully human antibodies.
Humanization is a widely utilized technique to deimmunize mAbs by replacing nonhuman components with human ones. The process may involve retaining only the complementarity-determining regions (CDRs)—the portions of the mAb that bind to the target antigen—from the nonhuman (typically mouse) antibody and replacing the rest of the antibody structure with human sequences [5, 6]. Such a more “human-like” antibody is less likely to be recognized as foreign by the patient’s immune system, thus reducing the risk of an immune response toward the antibody. Compared with chimeric mAbs, humanization reduces, but does not eliminate, immunogenicity. Indeed, even fully human antibodies with both the CDRs and frameworks derived from human immunoglobulin gene repertoires may potentially provoke an unwanted immune response [7]. Therefore, ADA rates vary substantially among mAbs (Table 1); accordingly, immunogenicity remains a major challenge in the development of mAb therapies. In this review, we provide a comprehensive overview of the strategies currently employed to mitigate immunogenicity risk of these protein therapeutics. Although the focus of this review is on ADAs directed against mAbs, the principles often apply to other types of protein therapeutics.
Table 1.
Overview of FDA approved fully human or humanized mAbs and their reported ADA rates
| mAb | Target | Isotype | Type | ADA (range) (%) | USA approval |
|---|---|---|---|---|---|
| Adalimumab | TNF-α | IgG1 | Human | 3–61 | 2002 |
| Aducanumab | amyloid-β | IgG1 | Human | 0.6 | 2021 |
| Alemtuzumab | CD52 | IgG1 | Humanized | 29–83 | 2001 |
| Alirocumab | PCSK9 | IgG1 | Human | 5.5 | 2015 |
| Anifrolumab | IFNAR1 | IgG1 | Human | 1.7 | 2021 |
| Atezolizumab | PD-L1 | IgG1 | Humanized | 13–48 | 2016 |
| Avelumab | PD-L1 | IgG1 | Human | 8.5–19.1 | 2017 |
| Belimumab | BLyS | IgG1 | Humanized | 0.6–4.8 | 2011 |
| Benralizumab | IL-5R | IgG1 | Human | 13 | 2017 |
| Bevacizumab | VEGF-A | IgG1 | Humanized | 0.2–0.6 | 2004 |
| Bezlotoxumab | C. difficile toxin B | IgG1 | Human | 0 | 2016 |
| Bimekizumab | IL-17A,IL-17F | IgG1 | Humanized | 31–45 | 2023 |
| Brodalumab | IL17RA | IgG2 | Human | 3 | 2017 |
| Brolucizumab | VEGF-A | scFv | Human | 53–76 | 2019 |
| Burosumab | FGF23 | IgG1 | Human | 0–19 | 2018 |
| Canakinumab | IL-1β | IgG1 | Human | 1.5–3.1 | 2009 |
| Cemiplimab | PD-1 | IgG4 | Human | 2.2 | 2018 |
| Certolizumab | TNF-α | IgG1 | Humanized | 8–28 | 2008 |
| Crizanlizumab | P-selectin | IgG2 | Humanized | 0–1.6 | 2019 |
| Daclizumab | CD25 | IgG1 | Humanized | 12–19 | 2016 |
| Daratumumab | CD38 | IgG1 | Human | 0 | 2015 |
| Denosumab | RANKL | IgG2 | Human | 0.67 | 2010 |
| Dostarlimab | PD-1 | IgG4 | Humanized | 2.5 | 2021 |
| Dupilumab | IL‐4Rα | IgG4 | Human | 1–16 | 2017 |
| Durvalumab | PD-L1 | IgG1 | Human | 0.8–3 | 2017 |
| Eculizumab | C5 | IgG2/4 | Humanized | 0–3 | 2007 |
| Efalizumab | CD11a | IgG1 | Humanized | 6.3 | 2003 |
| Elotuzumab | SLAMF7 | IgG1 | Humanized | 18.5–36 | 2015 |
| Epcoritamab | CD20, CD3 | IgG1 | Humanized | 2.6 | 2023 |
| Emapalumab | IFNG | IgG1 | Human | 3–5 | 2018 |
| Eptinezumab | CGRP | IgG1 | Humanized | 18–20.6 | 2020 |
| Erenumab | CGRPR | IgG2 | Human | 2.6–6.2 | 2018 |
| Evinacumab | ANGPTL3 | IgG4 | Human | 0 | 2021 |
| Evolocumab | PCSK9 | IgG2 | Human | 0.3 | 2015 |
| Faricimab | VEGF-A/Ang-2 | IgG1 | Humanized | 8.4–10.4 | 2022 |
| Fremanezumab | CGRP | IgG2a | Human | 0.4–1.6 | 2018 |
| Galcanezumab | CGRP | IgG4 | Human | 4.8–12.5 | 2018 |
| Glofitamab | CD20, CD3e | IgG1 | Humanized | 1.1 | 2023 |
| Golimumab | TNFα | IgG1 | Human | 19–31 | 2009 |
| Guselkumab | IL-23 | IgG1 | Human | 2–9 | 2017 |
| Ibalizumab | CD4 | IgG4 | Humanized | 0.6 | 2018 |
| Idarucizumab | dabigatran | Fab | Humanized | 2–4 | 2015 |
| Ipilimumab | CTLA-4 | IgG1 | Human | 1.1–36.7 | 2011 |
| Ixekizumab | IL-17A | IgG4 | Humanized | 5.2–22 | 2016 |
| Lanadelumab | pKal | IgG1 | Human | 5–12 | 2018 |
| Lecanemab | Aβ | IgG1 | Humanized | 40.9 | 2023 |
| Loncastuximab tesirine | CD19 | IgG1 | Humanized | 0 | 2021 |
| Mepolizumab | IL-5 | IgG1 | Humanized | < 2–6 | 2015 |
| Mosunetuzumab | CD20, CD3 | IgG1 | Humanized | 0 | 2022 |
| Mogamulizumab | CCR4 | IgG1 | Humanized | 14.1 | 2018 |
| Natalizumab | α4-integrin | IgG4 | Humanized | 9–10 | 2004 |
| Naxitamab | GD2 | IgG1 | Humanized | 8–23 | 2020 |
| Necitumumab | EGFR | IgG1 | Human | 4.1 | 2015 |
| Nivolumab | PD-1 | IgG4 | Human | 11 | 2014 |
| Obinutuzumab | CD20 | IgG1 | Humanized | 0.27 | 2013 |
| Ocrelizumab | CD20 | IgG1 | Humanized | ~ 1 | 2017 |
| Ofatumumab | CD20 | IgG1 | Human | < 1 | 2009 |
| Olaratumab | PDGFRα | IgG1 | Human | 3.5 | 2016 |
| Omalizumab | IgE | IgG1 | Humanized | < 0.1 | 2003 |
| Palivizumab | RSV F protein | IgG1 | Humanized | 1.1–1.5 | 1998 |
| Panitumumab | EGFR | IgG2 | Human | 0.5–5.3 | 2006 |
| Pembrolizumab | PD-1 | IgG4 | Humanized | 2.1 | 2014 |
| Pertuzumab | HER2 | IgG1 | Humanized | 0.3–3 | 2012 |
| Polatuzumab vedotin | CD79b | IgG1 | Humanized | 1.4–6 | 2019 |
| Ramucirumab | VEGFR2 | IgG1 | Human | 3 | 2014 |
| Ranibizumab | VEGF-A | IgG1 | Humanized | 1–9 | 2022 |
| Ravulizumab | C5 | IgG2/4 | Humanized | 0.5–1.4 | 2018 |
| Reslizumab | IL-5 | IgG4 | Humanized | 4.8–5.4 | 2016 |
| Relatlimab and Nivolumab | LAG-3 | IgG4 | Human | 3.8–5.6 | 2022 |
| Retifanlimab | PD-1 | IgG4 | Humanized | 2.9 | 2023 |
| Risankizumab | IL-23 | IgG1 | Humanized | 12.1–24 | 2019 |
| Romosozumab | sclerostin | IgG2 | Humanized | 18.1 | 2019 |
| Rozanolixizumab | FcRn | IgG4 | Humanized | 37 | 2023 |
| Sarilumab | IL-6R | IgG1 | Human | 9.2 | 2017 |
| Satralizumab | IL-6R | IgG2 | Humanized | 38–73 | 2020 |
| Secukinumab | IL-17A | IgG1 | Human | < 1 | 2015 |
| Spesolimab | IL-36R | IgG1 | Humanized | 46 | 2022 |
| Sutimlimab | C1s | IgG4 | Humanized | 0 | 2022 |
| Teclistamab | BCMA, CD3 | IgG4 | Humanized | 0.5 | 2022 |
| Teplizumab | CD3 | IgG1 | Humanized | 57 | 2022 |
| Teprotumumab | IGF-1R | IgG1 | Human | 0 | 2020 |
| Tezepelumab | TSLP | IgG2 | Human | 5 | 2021 |
| Tildrakizumab | IL-23 | IgG1 | Humanized | 6.5 | 2018 |
| Tocilizumab | IL-6R | IgG2 | Humanized | 2 | 2010 |
| Tralokinumab | IL-13 | IgG4 | Human | 1.4–4.6 | 2021 |
| Trastuzumab | HER2 | IgG1 | Humanized | 10 | 1998 |
| Tremelimumab | CTLA-4 | IgG2a | Human | 11 | 2022 |
| Ustekinumab | IL-12, IL-23 | IgG1 | Human | 6–12.4 | 2009 |
| Vedolizumab | α4β7 integrin | IgG1 | Humanized | 4–13 | 2014 |
The Clinical Consequences Associated with Immunogenicity of mAbs
ADA formation results in different clinical consequences, ranging from no effect to severe toxicity. Relevant adverse effects of ADAs include: (1) impairment of treatment efficacy by altering the bioavailability, pharmacodynamics, or pharmacokinetics of mAbs, (2) interference with the function of endogenous proteins through crossreactivity, or (3) immune modulatory effects [8–10].
ADAs can be categorized into neutralizing and nonneutralizing antibodies, each with implications for mAb or other protein therapeutic treatment. Neutralizing ADAs (NAbs) directly inhibit the biological activity of the protein therapeutic by binding to its active site or to regions critical for its function [11], thereby reducing the efficacy of the therapy. Whereas non-neutralizing ADAs do not inhibit the biological activity of the protein therapeutic directly, such ADA responses can still impact the pharmacokinetics and pharmacodynamics of the drug [8]. Both types of ADAs can modify the clearance of the protein therapeutic from circulation or alter its distribution and, thus, reduce efficacy [12].
Severe adverse effects of ADA often involve immune complex formation. These complexes can induce complement activation, triggering an inflammatory cascade that can produce severe infusion reactions [13, 14]. For example, the immunogenicity of brolucizumab, an antibody used for the treatment of neovascular age-related macular degeneration, resulted in ADAs that were associated with the development of retinal vasculitis/retinal vascular occlusion (RV/RO) in some patients. Formation of immune complexes with brolucizumab is a proposed mechanism for RV/RO induced in some patients through cellular responses such as enhanced antigen presentation, platelet aggregation, endothelial cell activation, and cytokine release [15].
T Cell-Dependent or T Cell-Independent Pathways to ADA Development
ADAs can arise through either T cell-dependent or T cell-independent pathways. The T cell-dependent pathway requires the internalization of the mAb, or other protein therapeutics, and processing by antigen-presenting cells (APCs), leading to the presentation of peptides derived from the therapeutic agent by human leukocyte antigen (HLA) class II (HLA-II) molecules and the recognition of the peptide-HLA-II complexes by T cells with cognate T cell receptors (TCRs). The immune response to recognition depends on the surrounding cytokines [16]. ADAs are produced when T helper cells (Th) interact with B cells leading to B cell differentiation into plasma cells that can release ADAs.
The T cell-independent pathway involves mAbs or protein therapeutics that bind B cell receptors (BCRs) directly, stimulating B cells to differentiate into plasma cells that produce ADAs [17–19].
Factors Intrinsic to a Protein Therapeutic that Contribute to Immunogenicity
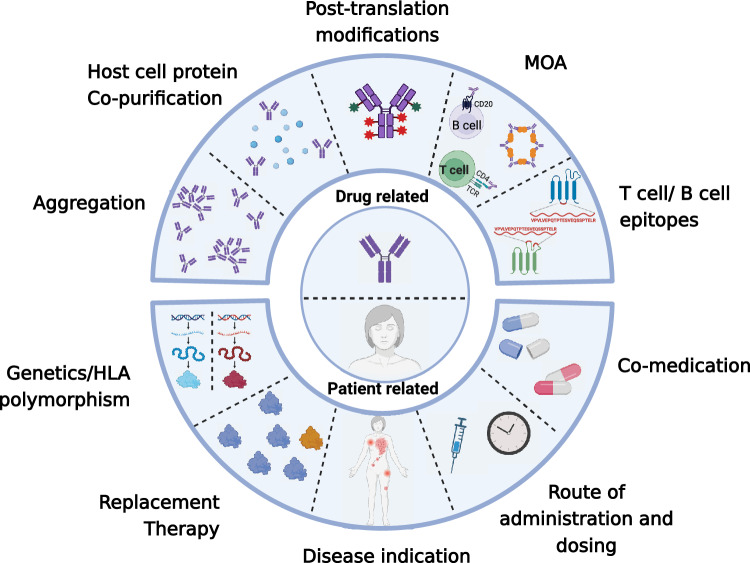
Complex, multifactorial elements contribute to ADA generation, including properties intrinsic to the protein therapeutic, properties of the drug preparation, and patient characteristics (Fig. 1). It is still unclear which factors have the most substantial influence and how these multiple factors interact with each other. Attributes of the protein therapeutic itself, such as amino acid sequence, three-dimensional structure, and posttranslational modifications (PTMs), can all factor into ADA development in a patient. Additionally, manufacturing-related impurities, dosing regimen or administration route, and even the mechanism of action (MOA) are additional elements that can impact ADA responses.
Fig. 1.
Factors that can influence protein therapeutics immunogenicity. The center of the diagram represents drug related factors (top) and patient related factors (bottom). Surrounding the inner circle are illustrations depicting the drug related factors and patient related factors on the top and bottom, respectively. MOA mechanism of action
Preparation-Related Factors: Host Cell Proteins
Protein therapeutics are primarily produced in and purified from genetically engineered mammalian expression systems. Although rigorously manufactured in controlled environments, process-related impurities, such as host cell proteins (HCPs), often, but typically at trace amount, copurify with protein therapeutics. Like other proteins, HCPs have the potential to be immunogenic themselves or act as adjuvants. Additionally, specific HCPs may impact the efficacy of the therapeutic protein or produce toxicity in patients when their levels in the product are increased [20]. An exemplary case of this phenomenon is lebrikizumab, a humanized IgG4 antibody used for the treatment of severe asthma. A preparation of lebrikizumab, produced and purified from Chinese hamster ovary (CHO) cells, contained the phospholipase B-like 2 (PLBL2) protein, an HCP that was a process-related impurity. More than ∼90% of subjects during the phase II clinical study developed an immune response to PLBL2. These findings emphasized the need for rigorous monitoring and control of HCPs in protein therapeutics to ensure patient safety and optimize treatment outcomes [20].
Aggregation
Aggregates can range in size from dimers of the protein therapeutic protein to visible particles with formation occurring at various stages of production, transport, and storage, even after delivery to the patient [21, 22]). The presence of aggregates, mainly those with a high molecular weight rather than dimers, may augment the immune response toward protein therapeutics by triggering both T cell-dependent and T cell-independent pathways [23–26]. Activation of the innate immune response by aggregates can lead to enhanced dendritic cell maturation and antigen presentation [27], thereby stimulating the T cell-mediated pathway to ADA. Aggregate-mediated crosslinking of the BCR may directly trigger the formation of antibodies targeting both the aggregates and the monomeric form of the protein therapeutic.
Several studies showed a correlation between the presence of aggregation and unwanted immune responses [21]. For example, high degree of aggregation in the product likely caused the increased immunogenicity and Nab production observed following the administration of recombinant erythropoietin or human growth hormone (hGH) [28, 29]. Thus, drug manufacturing practices should be optimized to limit the formation of protein aggregates.
Isoelectric Point of mAbs
Specific properties of some mAbs present a greater risk for ADA development. mAbs with a high isoelectric point (pI), which corresponds to a high amount of positive charge, are more immunogenic than mAbs with those with a low pI [12]. This is likely due to the interaction of positively charged mAbs with negatively charged cell surfaces and proteins, which can lead to receptor mediated uptake and facilitates the internalization and presentation of mAb-derived peptides by APCs. Positively charged mAbs can also exhibit nonspecific binding to off-target proteins, leading to immune complexes and faster mAb clearance [30, 31].
Posttranslational Modifications
Posttranslational modifications of antibodies may lead to an immunogenic response [32, 33]. Antibodies have diverse numbers and positions of conserved N-linked glycosylation sites, which are present on both the antigen-binding fragment (Fab) and the common portion (Fc) [34]. IgG molecules are the most abundant antibodies in serum and most mAb therapeutics of IgGs. IgG molecules are N-glycosylated at the conserved asparagine position, Asn297, in the Fc region, a modification that is essential for Fc effector functions. Changes to the N-linked glycosylation pattern of the Fc domain can alter the safety and efficacy of an mAb [35–37].
Glycan patterns are highly variable and depend on the host glycosylation machinery. Thus, the glycoform of a protein therapeutic largely depends on the host cells. For mAbs, variations in glycosylation can have a substantial influence on function in patients [38, 39], whereas mAbs produced in CHO cells have glycosylation patterns similar to human antibodies. mAbs produced in murine myeloma cells have glycosylation patterns that differ from produced by human cells, which may lead to an immunogenic response when administered to patients [40]. For example, mouse glycosylation with galactose-α-1,3-galactose on the Fab portion of cetuximab, a chimeric mouse–human IgG1 mAb against epidermal growth factor receptor (EGFR), resulted in hypersensitivity reactions in many patients mediated by preexisting IgE antibodies against this oligosaccharide on cetuximab [41–43]. Cetuximab lacking these galactose modifications exhibited less immunogenicity, demonstrating the impact of glycosylation patterns on mAb immunogenicity [41]. Thus, many mAbs are produced using expression systems that confer glycosylation patterns like those found on endogenous human proteins [44–46].
Glycosylation is not the only type of posttranslational modification that is important. Other posttranslational modifications affect the quality of protein therapeutics. These include PTMs such as oxidation, deamidation, and isomerization, all of which increase immune activation potential toward the mAbs [47]. Thus, quality control measures for mAbs and other protein therapeutics should include an analysis of the propensity to undergo chemical alteration events.
T Cell and B Cell Epitopes
The presence of T cell and B cell epitopes serves as an additional factor intrinsic to protein therapeutics, which contributes to their immunogenicity [48, 49]. Epitopes recognized by T cells are typically linear fragments of the amino acid sequence of a protein therapeutic. Such peptides are presented by HLA-II and can be recognized and bound by TCRs to initiate CD4+ T cells.
Epitopes recognized by B cells can be either linear or conformational. Like the linear peptide antigens bound by HLA-II and recognized by TCRs, linear epitopes recognized by BCRs consist of a contiguous sequence of amino acids. In contrast, conformational epitopes are formed by amino acids that are close in the three-dimensional structure but not necessarily contiguous in the primary sequence [50]. Since most B cell epitopes are conformational, they are more challenging to predict than T cell epitopes. ADA production can arise through direct B cell activation by protein therapeutic epitopes or through activation by T cells. Therefore, minimizing both T cell and B cell epitopes during mAb design can reduce potential immunogenicity.
Mechanism of Action of mAbs
Immunogenicity of an mAb can be influenced by the MOA of the drug. In some cases, especially for cancer therapy, the goal of the mAb is to trigger an immune response against the target antigen, which may affect the immunogenic response toward the protein therapeutic itself. Another factor influencing immunogenicity of mAbs is whether the target is a cell-surface molecule or a soluble molecule. Those targeting cell-surface antigens tend to produce a higher immune response than those targeting soluble antigens. The higher response could be due to enhanced uptake of the cell-surface antigen–mAb complexes by APCs, thus inciting a stronger immune response [51]. Additionally, immune complexes formed between mAbs and soluble antigens are typically smaller and less immunogenic than those formed with cell-surface antigens. The immune complexes formed with soluble antigens are typically cleared faster, reducing the chance of stimulating an immune response [52].
The MOA of mAb-targeting antigens present on malignant and normal hematological cells may also influence potential immunogenicity. B cell-depleting mAbs have a low risk of immunogenicity. The use of anti-CD19 (such as tafasitamab and loncastuximab) and anti-CD20 mAbs (such as ofatumumab, obinutuzumab, and rituximab) for the treatment of acute lymphocytic leukemia (ALL) or B-cell chronic lymphocytic leukemia (CLL) resulted in very low ADA rates with no ADA-related clinical consequences [53]. In contrast, antibodies targeting T cell functions or APCs may have a higher likelihood of immunogenicity. For example, administration of the immune checkpoint inhibitor (ICI) ipilimumab caused a flare of an underlying autoimmune disease [54], and the development of ADAs was observed in 30% of patients (range 13–54%) following atezolizumab treatment. However, in most patients, the incidences of ADA for most ICIs have been relatively low, and their presence has not shown significant effects on safety, pharmacokinetics, or efficacy in large patient populations [53].
Patient-Related Factors that Contribute to Immunogenicity
Within a patient, genetic-based differences in the immune system, additional medications, disease indication, the presence of diseases or disorders (comorbidities) other than the one targeted by the protein therapeutic, and the route of administration or dosing regimen can all impact ADA development and clinical consequences thereof. The interplay between the protein therapeutic and the patient’s unique characteristics can impact the safety and effectiveness of such therapy. Thus, customizing protein therapeutics to patients is a key aspect of personalized medicine that is important to study for improving therapeutic outcomes.
Genetics and HLA-II Polymorphism
Genetic variability, particularly within genes encoding proteins responsible for controlling immune responses and HLA-II presentation, can predispose individuals to the formation of ADAs [55]. Three distinct loci—HLA-DR, HLA-DP, and HLA-DQ—encode the HLA-II proteins. These genes are highly polymorphic [56]. HLA-II proteins bind a diversity of peptides 13–25 amino acids in length [57]. The binding affinity between a peptide and HLA-II depends on both the peptide’s amino acid at the anchor residue position (the backbone of the peptide directed into the pocket of the HLA-II) and the specific HLA-II variant [57–59]. Each patient’s HLA-II repertoire determines whether peptides from the protein therapeutic bind and are presented by APCs to T cells. Thus, the HLA-II repertoire dictates the risk of a patient developing ADAs. For example, patients with HLA-DQ-03, HLA-DQ-05, and HLA-DRβ-11 were suggested to have a higher risk for ADA formation in response to treatment with mAb-targeting tumor necrosis factor (TNF) [60].
Replacement therapy is typically the administration of an exogenous protein as a protein therapeutic to patients who fail to naturally produce the fully functional protein or who fail to produce enough of the protein. Such patients tend to have an increased immunogenicity risk, because central or peripheral tolerance toward the sequence of the protein was not developed [61]. Hemophilia patients who lack a functional clotting factor, such as factor VIII (FVIII) in hemophilia A or factor IX in hemophilia B, are at risk of an ADA that impairs the effectiveness of the treatment or causes adverse effects [62]. For patients receiving FVIII, the choice of protein therapeutic is critical because there are six variants of the encoding gene. The clinically used FVIII products match only two of the variants, the H1 and H2 haplotypes. However, patients with the H3 or H4 haplotypes, which are found only in African Americans, had a higher incidence of ADAs than those with the H1 or H2 haplotypes, suggesting that the mismatch of the FVIII amino acid sequence in the recombinant FVIII product and that encoded by the H3 or H4 haplotypes caused the high incidence of ADA [63]. Therefore, demographic factors, such as race and ethnicity, can have significant effects on the immunogenic potential of protein therapeutics. Genetic screening and matching the patient with the appropriate recombinant protein is one approach to reducing ADA-targeting protein-based replacement therapies.
Variation in Drug Target Abundance
The amount of the target of an mAb can influence the immunogenicity of the therapeutic agent. One example is the interaction between TNFα and the mAb adalimumab [64]. Patients with lower baseline levels of TNFα prior to adalimumab treatment had a higher frequency of ADAs against adalimumab than patients with higher amounts of TNFα at baseline [65]. One proposed mechanism for this difference is the presence of a surplus of unbound adalimumab triggering an immune response in patients with low amounts of TNFα; whereas, those with higher initial TNFα levels bound more molecules of adalimumab, lessening the chance of triggering an immune response and ADA production [66].
Medical Condition
The coexistence of autoimmune disease can profoundly enhance the immunogenicity of mAbs. A prominent factor is the underlying inflammatory milieu that accompanies autoimmune diseases. This environment is rich in activated immune cells and cytokines that can boost the immune response to the mAb, leading to the generation of ADAs [67]. For example, rituximab, a chimeric anti-CD20 monoclonal antibody, demonstrated a low incidence of ADAs in patients with malignant tumors. However, it resulted in a higher emergence of treatment-related ADAs in a distinct patient cohort undergoing treatment for autoimmune diseases, such as rheumatoid arthritis or systemic lupus erythematosus [68]. Additionally, in autoimmune patients, antibodies against rituximab decreased the drug’s therapeutic efficacy and increased adverse reactions [69].
B cells can further enhance the diversity of their ADA repertoire using a process of affinity maturation. This results in ADAs being generated against additional regions of the therapeutic, a phenomenon called “epitope spreading,” in which the immune system inappropriately recognizes endogenous proteins [70, 71]. Thus, these patients face the risk of an autoimmune flare-up or development of a new autoimmune condition due to the mAb therapy.
Comedication
Individual variation in the susceptibility to ADAs also relates to other medications a patient is taking. Coadministration of drugs that suppress the immune system have been reported to reduce the development of ADAs. A study assessing adalimumab immunogenicity in juvenile idiopathic arthritis patients found that concomitant use of methotrexate, a commonly prescribed immunosuppressant, was associated with lower immunogenicity toward adalimumab than in patients who were receiving adalimumab as a monotherapy [72]. Additionally, a low-dose of methotrexate given at the beginning of a treatment with an enzyme replacement therapy of recombinant human acid α-glucosidase (rhGAA) was shown to attenuate the immune response and reduce the formation of anti-rhGAA in Pompe disease [73]. Similarly, combinational treatment with anti-CD20 antibodies can reduce B cell responses and ADA generation. Lower ADAs toward utomilumab, an anti-CD137 mAb, were observed in patients with non-Hodgkin lymphoma who received this mAb in combination treatment with rituximab, anti-CD20 mAb, compared with those receiving utomilumab as a single-agent treatment [74].
In contrast, treatments that promote immune responses can enhance the development of ADAs. In particular, combination treatment with ICI mAbs nivolumab (targeting PD-1) and ipilimumab (targeting CTLA-4) increased ADAs rates against nivolumab, whereas a lower incidence of ADAs occurred with nivolumab monotherapy [53].
In Silico Approaches to Reduce the Immunogenicity Risk of Protein Therapeutics
Rigorous preclinical and clinical assessments of immunogenicity risk are critical to ensure the safety and efficacy of protein therapeutics (Fig. 2). Comprehensive analysis of immunogenicity risk before clinical trials contributes to the development of more effective protein therapeutics and can guide clinical candidate selection and optimization for therapeutic candidates. Additionally, it provides valuable insights for decisions related to dosage and administration of the protein therapeutics, ultimately enhancing patient outcomes. Consequently, evaluation of immunogenicity risk prior to initiation of clinical trials is highly recommended by regulatory agencies such as the FDA. A recent survey spanning 5 years showed an increased use of in silico algorithms, human immune cell-based assays, and proteomics-based studies to improve protein-engineering strategies, including selection of lead molecules, amino acid sequence optimization, and deimmunization [75].
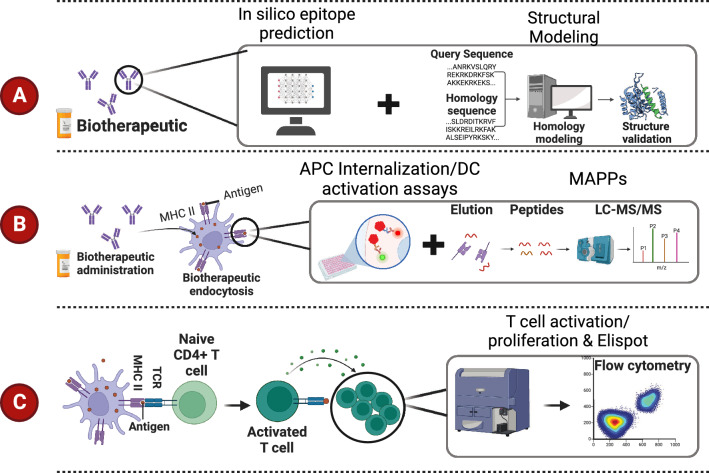
Fig. 2.
Selected tools employed for immunogenicity risk assessment. A In silico approaches are available for computational prediction of mAb immunogenicity risk. In silico tools are often employed for lead candidate selection, followed by further optimization of the drug candidate. B Some tools are specific to characterizing mAb internalization, presentation, and dendritic cell (DC) activation, such as antigen presenting cell (APC) internalization and DC activation assays as well as MHC-associated peptide proteomics (MAPPs) assays. C Some tools are specific to identify T cell response. The tools listed include enzyme-linked immunospot (ELISPOT) and flow cytometry-based methods that measure T cell activation and proliferation
Prior to performing in vitro or in vivo studies, various bioinformatic-based or computational approaches, collectively referred to here as “in silico” approaches, are valuable methods employed to limit immunogenicity of a candidate protein therapeutic and to optimize antibody sequences (Fig. 2A). Application of such strategies can also reduce effort and cost by limiting experiments requiring cell culture or animal studies.
These in silico strategies leverage an array of algorithms and databases. Two of the most used are the Immune Epitope Database (IEDB) [76] and Basic Local Alignment Search Tool (BLAST), the sequence alignment tool from the National Library of Medicine [72, 74]. The IEDB is used to predict potential immunogenic threats within a protein’s sequence, thereby enabling early interventions and design amendments in the drug development pipeline [78]. The IEDB contains a vast library of known T cell and B cell epitopes and provides access to many of the tools and algorithms and includes access to prediction tools for both T cell epitopes and B cell epitopes (http://tools.iedb.org/main/). Using alignments from BLAST searches, sequences of chimeric mAbs, for example, can be assessed for divergence from the human antibody sequence. Together, these bioinformatic tools inform many aspects of protein therapeutic or mAb development, such as selection of lead candidate protein therapeutics, optimization and engineering of the protein therapeutic, and prediction of the binding affinity of a peptide to the HLA-II pocket.
T Cell Epitope Prediction
A key strategy in immunogenicity prediction is the identification of linear sequences, typically around 15 amino acids, that serve as peptides presented by HLA-II to T cells [58, 59]. However, the sole use of the presence of T cell epitopes results in overpredicting clinical immunogenicity.
Algorithms that predict such antigens have been trained on extensive datasets of peptide–HLA-II binding affinities and/or mass spectrometry–HLA-II eluted-ligand data to predict peptide interaction or presentation on specific HLA-II alleles [79]. Tools that integrate peptide elution data and apply various machine-learning approaches have improved performance over predictors based solely on peptide-HLA-II binding algorithms [80–84]. One of the common algorithms available in the IEDB is the NetMHCIIpan 4.0. This algorithm employs machine-learning strategies to integrate both binding affinity and mass spectrometry-eluted ligand presentation data, thereby identifying peptides that are presented by multiple HLA-II [81].
Recently the same group has developed the NetMHCIIpan 4.2 [79, 85], which shows a superior performance of the HLA-DQ peptide by training and integrating immunoinformatic data mining models with high-quality HLA-DQ-specific mass spectrometry immunopeptidomic data into their model [86].
Multiple studies have demonstrated the use of in silico tools to assess the immunogenicity risk of protein therapeutics. NetMHCIIpan 3.2, a binding in silico algorithm and ISPRI, a commercial in silico option developed by Epivax, were both used to find the cause for the immunogenicity of a bispecific antibody (bsAb) that exhibited high immunogenicity in both preclinical and clinical studies [87]. Using in silico analysis, the monospecific and bispecific variants of the antibody were compared, revealing mechanisms for the higher immunogenicity of the bispecific antibodies. Based on the in silico analysis, it was suggested that a higher epitope count and a larger population were at risk for ADA development following the bispecific antibody compared with the monospecific variants [87]. Another study showed the ability of the NetMHCIIpan algorithm to successfully predicted peptide-HLA-DR presentation that can lead to T cell immunogenicity for infliximab and rituximab [88]. Lastly, the application of NetMHCIIpan-4.0 and graph-pMHC to assess the immunogenicity risk of mAbs was assessed. In this study, both algorithms showed to be effective in their ability to separate 107 mAbs into those with high immunogenicity and those with low immunogenicity [89]. Such tools have the potential to improve mAb sequence design and lead candidate selection; however, they also carry the risk of overprediction, thereby eliminating good candidates. The peptide-HLA interaction is one step in activating an immune response to a potential antigen. In vivo, proteolytic processing generates the peptides that are loaded onto HLA proteins for presentation [90]; in silico analysis typically accounts for all possible peptides that can be formed from a protein (not all computationally predicted peptides may be produced). Additionally, not all therapeutics will be taken up similarly by APCs and not all HLA-peptide complexes will be recognized by the TCR and, hence, will not activate T cells.
B Cell Epitope Prediction
B cell epitope prediction is another important step in immunogenicity assessment. However, the conformation-dependent nature of most B cell epitopes makes them challenging to predict due to the complexities associated with protein structure [50]. B cell prediction tools apply various machine-learning algorithms and structural bioinformatics methods to predict B cell epitopes based on the characteristics of known epitopes, such as hydrophilicity, flexibility, accessibility, and propensity to form turns or other secondary structures. Although promising, the performance of current computational tools for B cell epitope prediction remains suboptimal. There is a lack of high-quality, experimentally validated epitope data, especially for conformational epitopes. Consequently, the training data available for machine-learning algorithms are limited, which currently limits the accuracy of these tools.
Thus, the performance of B cell epitope-predicting algorithms is lower than for predicting potential T cell epitopes. Improving structure modeling will continue to enhance our ability to define B cell epitopes.
Structural Modeling
Homology modeling is a computational structure prediction method that generates three-dimensional protein structures when experimental data are not available [91]. The main principle of homology modeling relies on the evolutionary conservation of protein structures, where structurally similar proteins usually share similar functions [92]. The constant regions of mAbs are highly conserved and easily modeled from available experimental data, whereas the variable regions, which confer specificity to the mAb, can be modeled based on sequence similarities with known structures. More specifically, the three CDRs of the light chain, as well as CDR1 and CDR2 of the heavy chain in the variable region, contain conserved folds that can be predicted effectively by sequence similarity. However, due to its high diversity, both in sequence and length, the CDR3 of the heavy chain is the most difficult to model [93].
Several structural modeling tools are available, such as AlphaFold, which can predict protein structures even when a similar structure is unknown [94], ABodyBuilder2, which was trained to predict the structure of antibodies [95], SWISS-MODEL, which provides a user-friendly interface for homology modeling with fully automated workflows [96, 97], and Phyre2, which performs homology modeling and predicts various structural features without the requirement for a template [98]. For all homology models, the accuracy depends on the level of sequence identity between the target and the template: low sequence identity may result in less reliable models [92].
In Vitro Approaches to Predicting Immunogenicity
In vitro methods can be used to evaluate immunogenicity at different stages of immune activation and are commonly used to assess the immunogenicity risk of protein therapeutics (Fig. 2B–C). Such methods may help provide early indications for deimmunization planning or ranking of candidate drugs early in the selection process, leading to the identification of lower-risk clinical leads for further development and testing in clinical trials [75]. Standardization of in vitro assay procedures, readouts, and data interpretation would benefit the data interpretation by providing a consistent set of validated approaches and control molecules to evaluate immunogenicity risk at different stages of drug development.
Monitoring T Cell Activation and Proliferation as a Measure of Immunogenicity
In vivo, CD4 T cells are necessary to induce B cells to produce high-affinity specific antibodies. Thus, CD4 T cells isolated from peripheral blood mononuclear cells (PBMCs) of healthy donors are often used in assays as a surrogate to assess the potential for ADA development. Assays include detection of the release of specific immunomodulatory cytokines, induction of specific cell surface markers of activation, and stimulation of proliferation.
The enzyme-linked immunospot (ELISPOT) assay quantifies cytokines, such as interferon γ (IFN-γ) and interleukin-2 (IL-2), which are produced by activated CD4 T cells [99, 100]. The ELISPOT assay can detect T cells activated by coculturing with dendritic cells (DCs) preincubated with an mAb. If the mAb is immunogenic, the DCs activate the CD4 T cells leading to the production and release of IFN-γ, which is detected by ELISPOT [101]. Flow cytometric measurement of CD4 T cell activation surface markers, CD134 and CD137, accurately assessed the immunogenicity of multiple mAbs, although this assay was limited in predicting the immunogenicity risk of mAbs that inhibit TNFα, which itself is an immune-activating cytokine [102].
Various methods for evaluating the stimulation of proliferation of CD4 T cells in response to protein therapeutics are available. Historically, incorporation of radioactively labeled [3H] thymidine into the DNA of dividing cells was the main method for monitoring cell proliferation, and this approach has been applied to detect proliferation in cultures of PBMCs [103].
Flow cytometric methods using incorporation of the synthetic nucleoside analog of thymidine bromodeoxyuridine (BrdU) during the DNA synthesis phase of cell division, staining for the proliferation marker Ki67, or carboxyfluorescein diacetate succinimidyl ester (CFSE) staining provide less hazardous and more selective approaches to evaluate T cell proliferation. By combining proliferation markers with detection of the presence of CD4+-selective markers, such flow cytometry-based methods provide single-cell resolution, making them more suitable for studying heterogeneous cell populations [102, 104, 105]. CFSE is a cell-permeable dye that distributes evenly amongst all cells, and its concentration reduces by half with each cell division, enabling detection of up to ten rounds of cell division [105]. Measuring CD4 T cell proliferation by CFSE readout was shown to be valuable for assessing the immunogenicity risk of mAbs. To achieve better sensitivity, this assay required the enrichment of CD4 in the culture and the removal of CD8 T cells [106, 107]. Yet, measuring CD4 T cell proliferation with CFSE was a partially effective method for predicting immunogenicity risk of mAbs in PBMCs [107].
APC Internalization Assays
Internalization of a protein therapeutic by APCs initiates the presentation of protein therapeutic-derived peptides to T cells, which may trigger an immune response. Therefore, the propensity of mAbs to be internalized in vitro by APCs, particularly DCs, which are the most potent APCs, is a valuable assay of potential immunogenicity [108]. Two strategies have been used: either direct conjugation of a fluorescent dye to the protein therapeutic or indirect detection with a secondary, often fluorescently labeled, molecule [109]. Each has advantages and disadvantages. Indirect detection has the advantage of avoiding alteration of the therapeutic molecule; however, care is required to identify internalization of the protein therapeutic from binding to the surface of the APC. Direct conjugation of the protein therapeutic to the fluorescent dye runs the risk of altering the properties of the protein therapeutic and, for most fluorescent dyes, still requires a careful distinction between the signal that is a result of membrane binding versus that of internalization [110, 111]. The use of pHrodo dye as the fluorescent signal, which fluoresces only in acidic environments [111], enables detection only upon internalization into the lysosome or endosome compartments. A high-throughput DC internalization assay, which uses fluorescence resonance energy transfer (FRET), was developed that requires not only internalization but also antigen processing to produce a positive signal [112]. This FRET-based assay uses TAMRA and QSY7 as fluorophore and quencher, respectively. When conjugated to a folded protein molecule, this pair produces minimal signal, but when the conjugated protein is degraded, a bright fluorescent signal is observed as a result of separation of the fluorophore and quencher. Thus, when the fluorophore and quencher are conjugated to a mAb of interest, this assay provides a robust signal for DC internalization and processing of the mAb.
To avoid the variability associated with donor cells, as well as the limitations in cell numbers associated with using donor cells, the cell line THP1 has been explored as surrogate cells to study APC internalization and innate immune activation. THP1 cells can replicate the innate immune responses of monocyte-derived DCs (MoDCs) and primary CD14+ monocytes when exposed to therapeutic antibodies, enabling the development of a high-throughput internalization assay to assess immunogenic risk at the prelead candidate stage of mAb development [113].
These DC-based internalization assays have demonstrated a correlation between clinical immunogenicity and the rate of internalization of mAbs [112]. However, internalization and antigen processing are only the initial events in eliciting an immunogenic response; peptide binding to HLA and a T cell response are also needed to lead to ADAs against the protein therapeutics [87]. Thus, internalization assays should be combined with other in vitro or in silico studies, or both, to provide a more complete picture of the potential for immunogenicity.
MHC-Associated Peptide Proteomics (MAPPs) Assay
Proteomics provide a comprehensive strategy for detecting peptides presented by HLA proteins. This approach is referred to as MHC-associated peptide proteomics (MAPPs) for MHC-associated peptide proteomics. MAPPs involves an initial step of culturing the APCs, typically MoDC, with the protein therapeutic of interest, followed by an elution process to isolate HLA-II-peptide complexes from the APCs and, lastly, identification of the bound peptides by high-resolution mass spectrometry. The sequences of these peptides presented by HLA-II are ascertained by searching through the proteomic data with algorithms like Sequest, Mascot, or PEAKS [114–117]. These algorithms compare the proteomic data against public protein databases specifically filtered for human taxonomy and supplemented with protein sequences from the protein therapeutic under investigation. The identification of the peptides that are presented on the HLA-II molecules not only provides information about the immunogenic potential of a protein therapeutic but also provides key information about the antigenic portion of the protein therapeutic that may trigger the immune response [118].
MAPPs assays have been used in multiple contexts to evaluate the immunogenicity of mAbs and identify potential antigenic sequences. MAPPs was utilized to characterize HLA-DR-associated peptides from DCs loaded with inflixumab and rituximab, and the identified peptides were experimentally documented or found to align with known amino acid sequences that activate CD4+ T cells [119]. Similarly, a comparative study employing MAPPs explored the immunogenicity potential and T cell epitopes of two anti-IL17A antibodies, secukinumab and ixekizumab [120]. The frequency of ADAs observed clinically for these compounds aligned with the quantity of T cell-reactive peptides detected by MAPPs [120].
The primary advantages of MAPPs over other in vitro assays are that the data can be leveraged to identify the specific epitope(s) causing T cell activation, thereby providing key information for deimmunization of a protein therapeutic. However, similarly to the in silico analysis data, not all presented peptides will be recognized by the TCR and trigger T cell response, thus a careful interpretation of the MAPPs data should be conducted. Additionally, broad application of MAPPs is presently limited by its low-throughput capacity and limitation of the antibody used for immunoprecipitation prior to MAPPs [75].
Epitope Mapping
Epitope mapping is a powerful immunological technique employed to identify the specific epitopes of the protein therapeutic that are recognized by the ADA generated against the therapeutic. This technique is typically applied once ADAs or Nabs are found in patients and is used to help recognize the potential regions within the protein therapeutics that are recognized by the patient’s ADAs. Different methods, including peptide array, phage display, hydrogen–deuterium exchange mass spectrometry (HDX-MS), and various types of imaging technologies are employed to determine these antigen–antibody interactions. These methods may be combined with in silico prediction approaches to aid in the identification of the potential linear or conformational epitopes and map the epitopes onto the protein therapeutic.
Although epitope mapping to determine ADA binding is challenging due to the polyclonal nature of ADAs, knowledge of potential B cell epitopes can be used to reduce the immunogenicity of future protein therapeutics. For example, a short amino acid sequence within brolucizumab similar to that of a bacterial protein antigen was identified through a peptide-scanning approach [15]. This suggests that the ADA response toward brolucizumab was due to preexisting antibodies against this bacterial epitope. This result emphasized the need for considering sequence similarity to bacterial proteins in the design of protein therapeutics [15, 121]. Additionally, it demonstrated the importance of epitope mapping for a better understanding of ADA targets and for better design of mAbs for therapy.
Mitigation and Deimmunization Strategies
Based on the known factors that may trigger an unwanted immune response toward mAbs, multiple strategies have been proposed to limit the immunogenicity risk and mitigate it prior to or during treatment (Fig. 3). These include the preferential administration of fully human or humanized mAbs for therapy. However, these antibody-based therapies still have a risk of ADA production. Additionally, the implementation of a risk-based approach to mitigate patient-related factors, such as modification of the dosing regimen and administration schedules or cessation or addition of comedications, may also reduce the risk of ADA development.
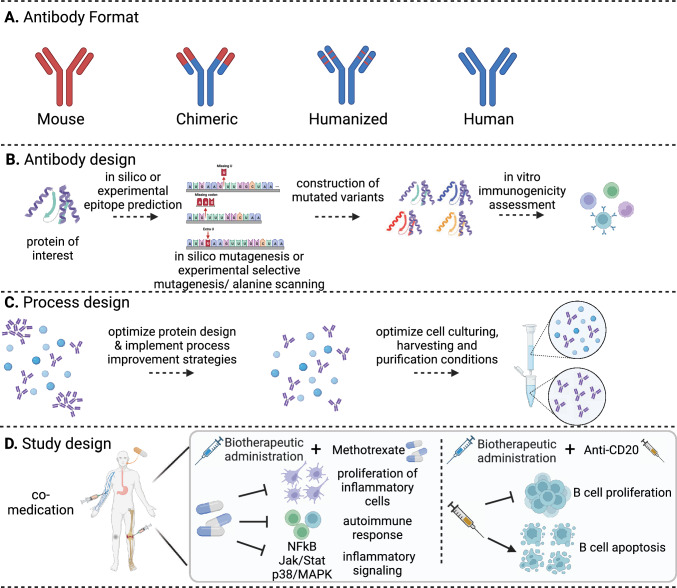
Fig. 3.
Design approaches for deimmunization of monoclonal antibodies (mAbs). Based on the source that leads to high immunogenicity, various design approaches can be applied for deimmunizing mAbs. A These approaches include the humanization process, which removes nonhuman segments within the mAbs. B Antibody design: a comprehensive strategy that includes screening of multiple variants with in silico and in vitro immunogenicity risk prediction tools and selection of a lead molecule with low immunogenicity risk. C Process design: a necessary step to reduce immunogenic factors such as host cell proteins (HCPs) and aggregates. D Study design: mitigation strategy that does not require alteration of the drug. Attenuation of the immunogenic response and reduction of anti-drug antibodies (ADA) toward mAbs is done by combininig the mAb treatment with comedication with immunosuppressant activity
Other methods of deimmunization by addressing the drug-related factors that lead to mAb immunogenicity include improving the quality of the therapeutic by design. Design-based strategies include removing predicted or known T cell and B cell epitopes.
Antibody Design, Development, and Selection of mAbs with Reduced Immunogenicity
Two approaches to eliminating potential or known T cell and B cell epitopes and designing mAbs with reduced immunogenicity risk are rational design and directed evolution. Rational design, as the name implies, is a process that employs a systematic, knowledge-based approach to design proteins with specific traits. It leverages an understanding of protein structure, function, and interaction with other molecules (such as the immune system components) to guide the precise modification of the protein. Directed evolution is an iterative process simulating natural evolution in a laboratory setting. This method generates a large library of mAb variants through random mutagenesis, recombination, or both, and then uses a high-throughput screening or selection process to identify variants with desired properties, such as reduced immunogenicity [34, 122].
In the context of mAbs, rational design could involve identifying and modifying immunodominant epitopes to reduce the risk of an immune response while preserving therapeutic functionality [123]. A rational design approach was used to deimmunize moxetumomab pasudotox (MP), calcitonin, Epo, human growth hormone (hGH), emicizumab, and IFN-β [124, 125]. Identification of immunogenic T cell epitopes in these protein therapeutics was performed experimentally. For example, MP peptides containing overlapping sequences, spanning the entire length of the antibody sequence, were generated and used to stimulate healthy donor PBMCs. Epitopes demonstrating immunogenic potential were then selectively mutated by alanine scanning or systematic substitutions with other viable amino acids. Peptides were then rescreened to identify mutagenic peptides that conferred reduced immunogenicity [126]. This strategy, which uses primarily experimental methods for T cell epitope identification, has been widely applied. However, this approach is expensive and labor intensive due to the need to generate dozens, and possibly hundreds, of overlapping peptides that expand the entire length of the protein therapeutic [127, 128]. With the advancement of in silico tools to identify T cell epitopes, this “trial and error mutagenic approach” spanning the entire sequence of the protein therapeutic can be avoided. Currently, the deimmunization process often involves in silico methods to predict and analyze potential epitopes, followed by site-directed mutagenesis to modify these epitopes (Fig. 3). The mAb variants generated are then tested for binding affinity, specificity, stability, and reduced immunogenicity [129].
In silico methods used to rationally design antibodies have also been applied successfully to reduce the immunogenicity of a recombinant activated human factor VII (rFVIIa) variant called vatreptacog alfa (VA). This recombinant protein was designed to provide increased procoagulant activity for hemophilia patients with inhibitors to factor VIII or factor IX [130]. Clinical trials revealed that the bioengineered VA variant led to the development of ADAs in some patients, leading to discontinuation of the product. To deimmunize the rFVIIa analog, an in silico prediction strategy, named rational immunogenicity determination (RID), was developed and applied to identify amino acid substitutions that reduced T cell epitopes without modifying conserved positions, which are likely functionally important. In vitro immunogenicity assays and thrombin generation assays showed that the redesigned protein therapeutics had reduced T cell responses without compromising the biological activity [130]. This proof-of-principle experimental validation of the deimmunization strategy provided a potential approach to mitigate immunogenicity risks associated with bioengineered proteins.
Epitope Shielding
Epitope shielding methods are used to disguise parts of the antigenic fragment from the immune system when removal of T cell or B cell epitopes could impair the functionality of the protein therapeutic. Although glycosylation is critical to some functions of antibodies, such as regulating ADCC and CDC activity, glycosylation can also alter protein therapeutic immunogenicity. mAb can bear N-glycosylation or O-glycosylation in the Fc region, typically at asparagine occupying position 297, and in the Fab region of IgG [131]. It has been previously shown that mannan (a polymer of mannose) glycosylation can increase the immunogenicity of antigens, thus removal of mannose content in the protein therapeutic may reduce its immunogenicity [38]. PEGylation involves attaching polyethylene glycol (PEG) polymers to mAbs to shield the surface epitopes from the immune system [132]. Compared with unmodified mAbs, the PEGylated versions have a longer circulatory half-life, improved water solubility, and reduced immunogenicity [133]. However, potential drawbacks include decreased biological activity, altered properties, PEG accumulation in organs, and the creation or presence of preexisting anti-PEG antibodies. PASylation and XTENylation are alternative shielding methods. PASylation involves the fusion of a long, unstructured polypeptide (proline–alanine–serine sequence) to the protein therapeutic protein, and XTENylation involves the attachment or association with XTEN (a proprietary unstructured hydrophilic, biodegradable protein polymer) [134, 135].
These shielding methods may be used to mitigate both product- and patient-related factors that contribute to immunogenicity. The engineered product may escape immune surveillance, enabling enhanced therapeutic efficacy and a longer half-life, thus reducing the need for frequent dosing [136]. However, these approaches need to be carefully evaluated for each protein therapeutic because they have the potential to introduce new epitopes or reduce efficacy of the final product.
Pharmaceutical Quality by Design
The Quality by Design (QbD) strategy is a systematic approach that aims to ensure product quality, safety, and efficacy by understanding the impact of critical quality attributes (CQAs) related to the product and production process on adverse immune effects QbD optimizes mAb production, purification, and downstream processes throughout the development program to ensure batch-to-batch comparability. Two of the major issues addressed by this strategy are aggregation and HCP contamination. Defining a limit for the acceptable levels of both product- and process-related impurities, supported by published safety limits and data gathered during clinical trials, is required [137].
With regard to aggregation, regulatory authorities have varying expectations regarding the acceptable level of aggregation propensity. Regardless of the regulatory requirements, efforts should be taken to both monitor and minimize aggregate formation, thereby both minimizing immunogenicity risk and maximizing the effective amount of the protein therapeutic in the preparation. According to the suggested FDA guidelines, minimization should be done as early as feasible in product development. Implementing manufacturing practices that minimize aggregate formation, incorporating purification steps that remove aggregates, selecting an appropriate cell system and formulation buffer, and determining the appropriate containers to minimize aggregation during storage are strategies to deimmunize a protein therapeutic due to aggregation [1]. Importantly, based on the FDA guidance, unless shown to be unnecessary, each final lot of an mAb preparation should be analyzed for aggregate content during lot release and at the end of its shelf life.
To address the issue of HCP copurification leading to increased immunogenicity risk, several methods have been explored at different stages of mAb manufacturing. One approach involves cell line engineering using CRISPR–Cas9-based gene editing to reduce or eliminate expression of genes encoding problematic HCPs [138]. By targeting specific genes responsible for HCP production, such as lipoprotein lipase (LPL), CHO cell lines were engineered to produce mAbs with substantially reduced amounts of this problematic HCP [139, 140]. CRISPR–Cas9 technology, which has high target selectivity and is cost effective, is a promising technique for eliminating problematic HCPs that are not essential to the growth, survival, or cellular production of the protein therapeutic. Other techniques focus on downstream processing and purification steps. Affinity precipitation, activated carbon membranes, flocculation reagents, and various types of peptide resins are being explored as alternatives or improvements to current purification methods [141].
Although there is no clear health authority guidance for the amount of HCPs that is safe within protein therapeutic preparations, levels below 100 ppm (< 100 ng/mg mAb protein) are generally recognized as acceptable. However, the final determination of the safe and acceptable level of HCP for any mAb product must be based on a risk assessment and depends on dose and frequency of administration [137].
Comedication
Various strategies can be employed to minimize the immunogenicity of monoclonal antibodies (mAbs) during clinical-stage treatment. These approaches can be implemented without requiring modification or reformulation of the protein therapeutic. One specific approach involves coadministering mAbs with an additional immunosuppressive agent, which reduces the overall immune response and may minimize the generation of ADAs. This immunosuppressive agent could be a drug targeting costimulatory signals crucial for T cell activation and function, thereby inhibiting T cell responses to mAbs and diminishing the probability of an adverse immune reaction [142]. In a typical immune response, T cell activation requires three signals: one delivered through the TCR upon recognition of an antigen presented on HLA, a second “costimulatory” signal provided by interactions between additional molecules present on the surface of T cells and APCs, and a third signal delivered through cytokines from an APC to a T cell [143, 144]. This costimulatory signal is crucial for a full immune response. Without this second signal, T cells become anergic, or nonreactive, rather than activated. This concept is exploited with costimulatory blockade, in which the aim is to prevent this second signal and inhibit T cell activation. One strategy for costimulatory blockade involves a fusion protein called CTLA-4-Ig (for example, abatacept and belatacept), which binds to the CD80 and CD86 proteins on APCs [142, 145]. CTLA-4-Ig prevents these proteins from interacting with CD28, a costimulatory molecule on T cells, effectively blocking the second signal and reducing T cell responses to therapeutic mAb [142]. Moreover, synthetic vaccine particles containing rapamycin (SVP-rapamycin) have shown efficacy in inducing immunogenic tolerance and reducing the formation of ADAs when coadministered with various protein therapeutics [146]. For example, the administration of SVP-rapamycin, alongside adalimumab, successfully prevented the formation of ADAs in transgenic mice expressing human TNFα [146].
Costimulation blockade strategies have been successful in reducing immunogenicity and increasing the efficacy of mAbs in various settings, particularly in transplantation and autoimmune diseases. However, they also carry risks, such as the potential for general immunosuppression and increased susceptibility to infections and malignancies [147]. As mentioned above, administration of the mAb with comedication of immune suppressors, such as methotrexate or CD20 blockade, can attenuate the immune response and may prevent ADA development toward the primary mAb protein therapeutic. Like with any therapeutic approach, balancing the benefits of reduced immunogenicity with potential side effects is a key concern when combining different treatments together.
Challenges, Unmet Needs, and Future Research Directions
ADAs are a complex group of analytes with variable isotypes, binding regions, and affinities, leading to different clinical consequences among patients. Due to the diverse consequences and possible adverse immunological responses caused by ADA development, clinical immunogenicity must be monitored during clinical trials. Analytical detection methods, such as the enzyme-linked immunosorbent assay (ELISA), meso scale discovery immunoassays (MSD), radioimmunoprecipitation (RIP) assay, and electrochemiluminescence (ECL), are commonly used to detect ADAs in the clinical setting. These immunoassays are not considered quantitative because standardized human polyclonal ADA reference materials are unavailable [148]. Additionally, the observed incidence of clinical ADAs is influenced by multiple factors, such as assay sensitivity, sample handling, and sampling collection time, which hinder the ability to compare ADA rates between protein therapeutics [149, 150]. The lack of standardization in the terminologies and methodologies for collecting, analyzing, and reporting immunogenicity results adds to the complexity of harmonizing clinical ADA data. Additionally, the incidence of ADA in patient populations varies substantially depending on the disease and specific therapeutic regimen, as well as patient-related factors, such as genetics, immune status, and comorbidities [151, 152]. Consequently, mining ADA data and clinical consequences across studies to enable data-based decisions regarding immunogenicity risk of mAbs before reaching the clinic are not yet possible, and ADA assessment of protein therapeutics at clinical stages requires empirical testing.
As the complexity and volume of biological data grow, in silico strategies to predict immunogenicity risk are increasingly employed. Although these methods offer an efficient preliminary screening of potential immunogenic threats, these tools are not definitive with one of the main challenges still being the accurate prediction of immunogenicity at the steps of BCR or TCR recognition and immune cell activation, not only peptide–HLA-II binding or presentation. Because the in silico strategies rely on machine-learning algorithms trained on existing data, they may fail to accurately predict the immunogenicity of novel sequences. Currently available algorithms for immunogenicity prediction often lack precision, because they depend primarily on the identification of linear sequence-based T cell epitopes, neglecting factors such as the influence of tertiary structure, posttranslational modifications, and individual patient factors on immunogenicity [153].
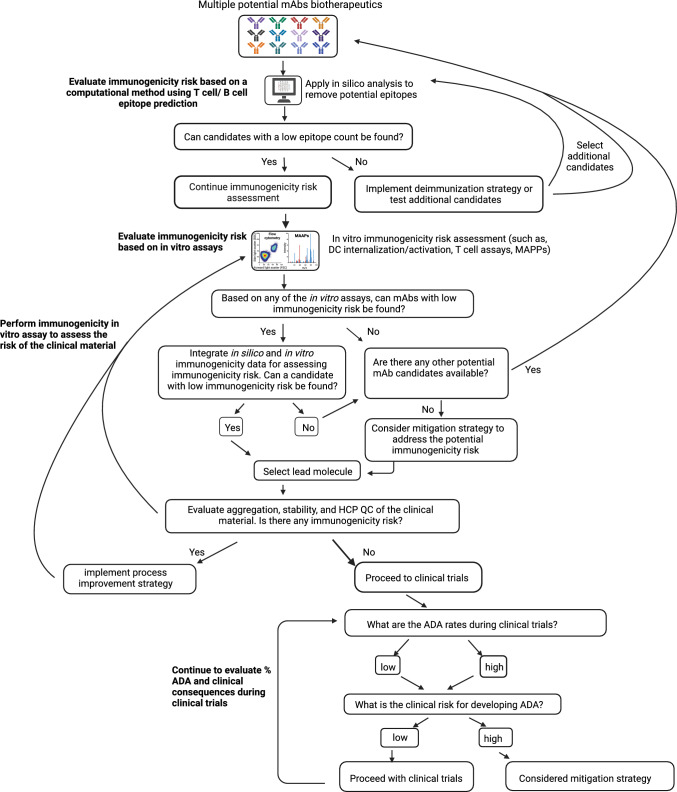
Several health authorities recommend performing immunogenicity risk assessment by in silico tools or in vitro assays before clinical trials and submitting an integrated immunogenicity risk assessment summary (ISI) that can evolve through the product’s lifecycle [154]. Therefore, a strategy that integrates multiple methods to assess immunogenicity risk of protein therapeutics, including a step-based evaluation of the immunogenicity risk at different stages of the drug development and offering a tailored deimmunization or mitigation plan to address the risk, will increase the pace of the development of safe and effective protein therapeutics (Fig. 4). As a first step, computational methods can help screen multiple candidates and assist with the selection of candidates with a potential low epitope count. These methods can also be applied for antibody design and removal of potential epitopes. As a next step, in vitro immunogenicity assessment assays can be conducted, and an integrated approach that combines different immunogenicity risk output can facilitate the selection of lead molecule selection based on its immunogenicity risk. Other factors will also determine the selection of a lead molecule, such as PK, affinity, and safety. Presence of aggregation, sequence variants, and HCPs are some of the factors that can influence immunogenicity and should be evaluated during production. Once the product reaches clinical trials, immunogenicity monitoring by ADA assays should be conducted with clinical samples. Those results will determine the true value of ADAs. The clinical consequences of development of ADAs, such as any potential safety risk to the patients, influence on PK/PD and efficacy should be strictly watched and may trigger the need for mitigation of immunogenicity (Fig. 4). Additional conversation may be required with health authorities to discuss the risk before or during clinical trials and review the mitigation plan.
Fig. 4.
Decision tree of the strategies to assess immunogenicity risk of mAbs. A suggested step-based flow chart guide for lead mAb therapeutic selection focuses on immunogenicity risk assessment at different stages of drug development. When data suggest a higher immunogenicity risk, deimmunization/mitigating strategies should be considered. ADA antidrug antibody, DC dendritic cell, HCP host cell protein, mAbs monoclonal antibodies, MAPPs MHC-associated peptide proteomics, QC quality control
Future research should focus on improving the accuracy and interpretability of machine-learning models and in vitro assays. Because the treatment regimen and the patient’s genetic makeup and immune status influence immunogenicity risk, integrating key genetic data and clinical details is important to minimize ADA development at the individual patient level. Fostering collaborations between computational scientists, experimental biologists, and clinicians will ensure the translational potential of these innovative strategies.
Acknowledgements
The authors thank Patricia Y. Siguenza and Steven Swanson for their support of the project; additionally, we also thank Nancy R. Gough (BioSerendipity, LLC) for editorial assistance. Figures were created with BioRender.com.
Declarations
Funding
This project was fully funded by Genentech Inc.
Disclosure of potential conflicts of interest
All authors are employees and may own stocks or stock options in Genentech Inc. The authors have no other conflicts of interest or subject matter of materials discussed in this manuscript.
Ethics approval
No ethics approval was required.
Patient consent to participate/publish
Not applicable.
Data availability
All data represented are available in the public domain.
Code availability
Not applicable.
Author contributions
C.H. and S.C. contributed equally to the planning, literature search, writing, and final approval of the manuscript.
References
- 1.FDA. (2014). Immunogenicity assessment for therapeutic protein products.
- 2.Walsh G, Walsh E. Biopharmaceutical benchmarks 2022. Nat Biotechnol. 2022;40(12):1722–1760. doi: 10.1038/s41587-022-01582-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reichert JM. Monoclonal antibodies in the clinic. Nat Biotechnol. 2001;19(9):819–822. doi: 10.1038/nbt0901-819. [DOI] [PubMed] [Google Scholar]
- 4.Hwang WYK, Foote J. Immunogenicity of engineered antibodies. Methods. 2005;36(1):3–10. doi: 10.1016/j.ymeth.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 5.Masson GR, Burke JE, Ahn NG, Anand GS, Borchers C, Brier S, et al. Recommendations for performing, interpreting and reporting hydrogen deuterium exchange mass spectrometry (HDX-MS) experiments. Nat Methods. 2019;16(7):595–602. doi: 10.1038/s41592-019-0459-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Riechmann L, Clark M, Waldmann H, Winter G. Reshaping human antibodies for therapy. Nature. 1988;332(6162):323–327. doi: 10.1038/332323a0. [DOI] [PubMed] [Google Scholar]
- 7.Wang Y-MC, Wang J, Hon YY, Zhou L, Fang L, Ahn HY. Evaluating and reporting the immunogenicity impacts for biological products—a clinical pharmacology perspective. APPS J. 2016;18(2):395–403. doi: 10.1208/s12248-015-9857-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krieckaert CLM, Bartelds GM, Lems WF, Wolbink GJ. The effect of immunomodulators on the immunogenicity of TNF-blocking therapeutic monoclonal antibodies: a review. Arthritis Res Ther. 2010;12(5):217. doi: 10.1186/ar3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Atzeni F, Talotta R, Salaffi F, Cassinotti A, Varisco V, Battellino M, et al. Immunogenicity and autoimmunity during anti-TNF therapy. Autoimmun Rev. 2013;12(7):703–708. doi: 10.1016/j.autrev.2012.10.021. [DOI] [PubMed] [Google Scholar]
- 10.Moots RJ, Xavier RM, Mok CC, Rahman MU, Tsai W-C, Al-Maini MH, et al. The impact of anti-drug antibodies on drug concentrations and clinical outcomes in rheumatoid arthritis patients treated with adalimumab, etanercept, or infliximab: Results from a multinational, real-world clinical practice, non-interventional study. PLoS ONE. 2017;12(4):e0175207. doi: 10.1371/journal.pone.0175207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shankar G, Arkin S, Cocea L, Devanarayan V, Kirshner S, Kromminga A, Quarmby V, et al. Assessment and reporting of the clinical immunogenicity of therapeutic proteins and peptides—harmonized terminology and tactical recommendations. AAPS J. 2014;16(4):658–673. doi: 10.1208/s12248-014-9599-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu L. Pharmacokinetics of monoclonal antibodies and Fc-fusion proteins. Protein Cell. 2018;9(1):15–32. doi: 10.1007/s13238-017-0408-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Korswagen LA, Bartelds GM, Krieckaert CLM, Turkstra F, Nurmohamed MT, van Schaardenburg D, et al. Venous and arterial thromboembolic events in adalimumab-treated patients with antiadalimumab antibodies: a case series and cohort study. Arthritis Rheumat. 2011;63(4):877–883. doi: 10.1002/art.30209. [DOI] [PubMed] [Google Scholar]
- 14.Petitpain N, Gambier N, Wahl D, Chary-Valckenaere I, Loeuille D, Gillet P, et al. Arterial and venous thromboembolic events during anti-TNF therapy: a study of 85 spontaneous reports in the period 2000–2006. Biomed Mater Eng. 2009;19(4–5):355–364. doi: 10.3233/bme-2009-0600. [DOI] [PubMed] [Google Scholar]
- 15.Kearns JD, Wassmann P, Olgac U, Fichter M, Christen B, Rubic-Schneider T, et al. A root cause analysis to identify the mechanistic drivers of immunogenicity against the anti-VEGF biotherapeutic brolucizumab. Sci Transl Med. 2023;15(681):eabq5068. doi: 10.1126/scitranslmed.abq5068. [DOI] [PubMed] [Google Scholar]
- 16.Talotta R, Rucci F, Canti G, Scaglione F. Pros and cons of the immunogenicity of monoclonal antibodies in cancer treatment: a lesson from autoimmune diseases. Immunother. 2019;11(3):241–254. doi: 10.2217/imt-2018-0081. [DOI] [PubMed] [Google Scholar]
- 17.Shikh MEME, Sayed RME, Szakal AK, Tew JG. T-independent antibody responses to t-dependent antigens: a novel follicular dendritic cell-dependent activity. J Immunol. 2009;182(6):3482–3491. doi: 10.4049/jimmunol.0802317. [DOI] [PubMed] [Google Scholar]
- 18.Obukhanych TV, Nussenzweig MC. T-independent type II immune responses generate memory B cells. J Exper Med. 2006;203(2):305–310. doi: 10.1084/jem.20052036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vos Q, Lees A, Wu ZQ, Snapper CM, Mond JJ. B-cell activation by T-cell-independent type 2 antigens as an integral part of the humoral immune response to pathogenic microorganisms. Immunol Rev. 2000;176(1):154–170. doi: 10.1034/j.1600-065x.2000.00607.x. [DOI] [PubMed] [Google Scholar]
- 20.Fischer SK, Cheu M, Peng K, Lowe J, Araujo J, Murray E, et al. Specific immune response to phospholipase b-like 2 protein, a host cell impurity in lebrikizumab clinical material. AAPS J. 2017;19(1):254–263. doi: 10.1208/s12248-016-9998-7. [DOI] [PubMed] [Google Scholar]
- 21.Moussa EM, Panchal JP, Moorthy BS, Blum JS, Joubert MK, Narhi LO, Topp EM. Immunogenicity of therapeutic protein aggregates. J Pharm Sci. 2016;105(2):417–430. doi: 10.1016/j.xphs.2015.11.002. [DOI] [PubMed] [Google Scholar]
- 22.Ratanji KD, Derrick JP, Dearman RJ, Kimber I. Immunogenicity of therapeutic proteins: influence of aggregation. J Immunotoxicol. 2014;11(2):99–109. doi: 10.3109/1547691x.2013.821564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rosenberg AS. Effects of protein aggregates: an immunologic perspective. AAPS J. 2006;8(3):E501–E507. doi: 10.1208/aapsj080359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shire SJ. Formulation and manufacturability of biologics. Curr Opin Biotechnol. 2009;20(6):708–714. doi: 10.1016/j.copbio.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 25.Chackerian B, Lenz P, Lowy DR, Schiller JT. Determinants of autoantibody induction by conjugated papillomavirus virus-like particles. J Immunol. 2002;169(11):6120–6126. doi: 10.4049/jimmunol.169.11.6120. [DOI] [PubMed] [Google Scholar]
- 26.Vaisman-Mentesh A, Gutierrez-Gonzalez M, DeKosky BJ, Wine Y. The molecular mechanisms that underlie the immune biology of anti-drug antibody formation following treatment with monoclonal antibodies. Front Immunol. 2020;11:1951. doi: 10.3389/fimmu.2020.01951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lundahl MLE, Fogli S, Colavita PE, Scanlan EM. Aggregation of protein therapeutics enhances their immunogenicity: causes and mitigation strategies. RSC Chem Biol. 2021;2(4):1004–1020. doi: 10.1039/d1cb00067e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seidl A, Hainzl O, Richter M, Fischer R, Böhm S, Deutel B, et al. Tungsten-induced denaturation and aggregation of epoetin alfa during primary packaging as a cause of immunogenicity. Pharm Res. 2012;29(6):1454–1467. doi: 10.1007/s11095-011-0621-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moore WV, Leppert P. Role of aggregated human growth hormone (hGH) in development of antibodies to hGH*. J Clin Endocrinol Metab. 1980;51(4):691–697. doi: 10.1210/jcem-51-4-691. [DOI] [PubMed] [Google Scholar]
- 30.Hermeling S, Crommelin DJA, Schellekens H, Jiskoot W. Structure-immunogenicity relationships of therapeutic proteins. Pharm Res. 2004;21(6):897–903. doi: 10.1023/b:pham.0000029275.41323.a6. [DOI] [PubMed] [Google Scholar]
- 31.Hu S, D’Argenio DZ. Predicting monoclonal antibody pharmacokinetics following subcutaneous administration via whole-body physiologically-based modeling. J Pharmacokinet Pharmacodyn. 2020;47(5):385–409. doi: 10.1007/s10928-020-09691-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Doyle HA, Gee RJ, Mamula MJ. Altered immunogenicity of isoaspartate containing proteins. Autoimmunity. 2007;40(2):131–137. doi: 10.1080/08916930601165180. [DOI] [PubMed] [Google Scholar]
- 33.Eggleton P, Haigh R, Winyard PG. Consequence of neo-antigenicity of the ‘altered self’. Rheumatol. 2008;47(5):567–571. doi: 10.1093/rheumatology/ken014. [DOI] [PubMed] [Google Scholar]
- 34.Arnold JN, Wormald MR, Sim RB, Rudd PM, Dwek RA. The impact of glycosylation on the biological function and structure of human immunoglobulins. Ann Rev Immunol. 2007;25(1):21–50. doi: 10.1146/annurev.immunol.25.022106.141702. [DOI] [PubMed] [Google Scholar]
- 35.Mimura Y, Katoh T, Saldova R, O’Flaherty R, Izumi T, Mimura-Kimura Y, et al. Glycosylation engineering of therapeutic IgG antibodies: challenges for the safety, functionality and efficacy. Protein Cell. 2018;9(1):47–62. doi: 10.1007/s13238-017-0433-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu L. antibody glycosylation and its impact on the pharmacokinetics and pharmacodynamics of monoclonal antibodies and fc-fusion proteins. J Pharm Sci. 2015;104(6):1866–1884. doi: 10.1002/jps.24444. [DOI] [PubMed] [Google Scholar]
- 37.Li W, Zhu Z, Chen W, Feng Y, Dimitrov DS. Crystallizable fragment glycoengineering for therapeutic antibodies development. Front Immunol. 2017;8:1554. doi: 10.3389/fimmu.2017.01554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boune S, Hu P, Epstein AL, Khawli LA. Principles of N-linked glycosylation variations of igg-based therapeutics: pharmacokinetic and functional considerations. Antibodies. 2020;9(2):22. doi: 10.3390/antib9020022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beck A, Wagner-Rousset E, Bussat M-C, Lokteff M, Klinguer-Hamour C, Haeuw J-F, et al. Trends in glycosylation, glycoanalysis and glycoengineering of therapeutic antibodies and Fc-fusion proteins. Curr Pharm Biotechnol. 2008;9(6):482–501. doi: 10.2174/138920108786786411. [DOI] [PubMed] [Google Scholar]
- 40.Barbosa MDFS. Immunogenicity of biotherapeutics in the context of developing biosimilars and biobetters. Drug Discov Today. 2011;16(7–8):345–353. doi: 10.1016/j.drudis.2011.01.011. [DOI] [PubMed] [Google Scholar]
- 41.Chung CH, Mirakhur B, Chan E, Le Q-T, Berlin J, Morse M, et al. Cetuximab-induced anaphylaxis and IgE specific for galactose-alpha-1,3-galactose. N Engl J Med. 2008;358(11):1109–1117. doi: 10.1056/nejmoa074943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lungulescu CV, Ungureanu BS, Turcu-Stiolica A, Ghimpau V, Artene SA, Cazacu IM, et al. The role of IgE specific for galactose-α-1,3-galactose in predicting cetuximab induced hypersensitivity reaction: a systematic review and a diagnostic meta-analysis. Sci Rep. 2020;10(1):21355. doi: 10.1038/s41598-020-78497-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Berg EA, Platts-Mills TAE, Commins SP. Drug allergens and food—the cetuximab and galactose-α-1,3-galactose story. Annal Allergy Asthma Immunol. 2014;112(2):97–101. doi: 10.1016/j.anai.2013.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang L, Biolsi S, Bales KR, Kuchibhotla U. Impact of variable domain glycosylation on antibody clearance: an LC/MS characterization. Anal Biochem. 2006;349(2):197–207. doi: 10.1016/j.ab.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 45.Almagro JC, Daniels-Wells TR, Perez-Tapia SM, Penichet ML. Progress and challenges in the design and clinical development of antibodies for cancer therapy. Front Immunol. 2018;8:1751. doi: 10.3389/fimmu.2017.01751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hötzel I, Theil F-P, Bernstein LJ, Prabhu S, Deng R, Quintana L, et al. A strategy for risk mitigation of antibodies with fast clearance. MAbs. 2012;4(6):753–760. doi: 10.4161/mabs.22189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zeunik R, Ryuzoji AF, Peariso A, Wang X, Lannan M, Spindler LJ, et al. Investigation of immune responses to oxidation, deamidation, and isomerization in therapeutic antibodies using preclinical immunogenicity risk assessment assays. J Pharm Sci. 2022;111(8):2217–2229. doi: 10.1016/j.xphs.2022.05.005. [DOI] [PubMed] [Google Scholar]
- 48.Chirino AJ, Ary ML, Marshall SA. Minimizing the immunogenicity of protein therapeutics. Drug Discov Today. 2004;9(2):82–90. doi: 10.1016/s1359-6446(03)02953-2. [DOI] [PubMed] [Google Scholar]
- 49.Schellekens H. How to predict and prevent the immunogenicity of therapeutic proteins. Biotechnol Ann Rev. 2008;14:191–202. doi: 10.1016/s1387-2656(08)00007-0. [DOI] [PubMed] [Google Scholar]
- 50.Potocnakova L, Bhide M, Pulzova LB. An introduction to b-cell epitope mapping and in silico epitope prediction. J Immunol Res. 2016;2016:6760830. doi: 10.1155/2016/6760830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Harding FA, Stickler MM, Razo J, DuBridge RB. The immunogenicity of humanized and fully human antibodies: residual immunogenicity resides in the CDR regions. MAbs. 2010;2(3):256–265. doi: 10.4161/mabs.2.3.11641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gogolák P, Réthi B, Hajas G, Rajnavölgyi É. Targeting dendritic cells for priming cellular immune responses. J Mol Recognit. 2003;16(5):299–317. doi: 10.1002/jmr.650. [DOI] [PubMed] [Google Scholar]
- 53.Davda J, Declerck P, Hu-Lieskovan S, Hickling TP, Jacobs IA, Chou J, et al. Immunogenicity of immunomodulatory, antibody-based, oncology therapeutics. J Immunother Cancer. 2019;7(1):105. doi: 10.1186/s40425-019-0586-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Callahan MK, Yang A, Tandon S, Xu Y, Subudhi SK, Roman RA, et al. Evaluation of serum IL-17 levels during ipilimumab therapy: correlation with colitis. J Clin Oncol. 2011;29:2505–2505. doi: 10.1200/jco.2011.29.15_suppl.2505. [DOI] [Google Scholar]
- 55.Kuriakose A, Chirmule N, Nair P. Immunogenicity of biotherapeutics: causes and association with posttranslational modifications. J Immunol Res. 2016;2016:1298473. doi: 10.1155/2016/1298473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Robinson J, Barker DJ, Georgiou X, Cooper MA, Flicek P, Marsh SGE. IPD-IMGT/HLA database. Nucl Acids Res. 2019;48(D1):D948–D955. doi: 10.1093/nar/gkz950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chicz RM, Urban RG, Lane WS, Gorga JC, Stern LJ, Vignali DAA, Strominger JL. Predominant naturally processed peptides bound to HLA-DR1 are derived from MHC-related molecules and are heterogeneous in size. Nature. 1992;358(6389):764–768. doi: 10.1038/358764a0. [DOI] [PubMed] [Google Scholar]
- 58.Mommen GPM, Frese CK, Meiring HD, Brink J van G. den, Jong APJM, de Els CACM, van Heck AJR (2014). Expanding the detectable HLA peptide repertoire using electron-transfer/higher-energy collision dissociation (EThcD). Proc Natl Acad Sci USA. 111(12):4507–4512. 10.1073/pnas.1321458111. [DOI] [PMC free article] [PubMed]
- 59.Peters B, Nielsen M, Sette A. T cell epitope predictions. Ann Rev Immunol. 2020;38(1):1–23. doi: 10.1146/annurev-immunol-082119-124838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Benucci M, Damiani A, Gobbi FL, Bandinelli F, Infantino M, Grossi V, et al. Correlation between HLA haplotypes and the development of antidrug antibodies in a cohort of patients with rheumatic diseases. Biologics. 2018;12:37–41. doi: 10.2147/btt.s145941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schurgers E, Wraith DC. Induction of tolerance to therapeutic proteins with antigen-processing independent T cell epitopes: controlling immune responses to biologics. Front Immunol. 2021;12:742695.. doi: 10.3389/fimmu.2021.742695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pandey GS, Yanover C, Howard TE, Sauna ZE. Polymorphisms in the F8 gene and MHC-II variants as risk factors for the development of inhibitory anti-factor viii antibodies during the treatment of hemophilia a: a computational assessment. PLoS Comput Biol. 2013;9(5):e1003066. doi: 10.1371/journal.pcbi.1003066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Viel K, Afshin A, Abshire TC, Iyer RV, Watts RG, Lutcher C, et al. Inhibitors of factor VIII in black patients with hemophilia. N Engl J Med. 2009;360(16):1618–1627. doi: 10.1056/nejmoa075760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vincent FB, Morand EF, Murphy K, Mackay F, Mariette X, Marcelli C. Antidrug antibodies (ADAb) to tumour necrosis factor (TNF)-specific neutralising agents in chronic inflammatory diseases: a real issue, a clinical perspective. Ann Rheum Dis. 2013;72(2):165. doi: 10.1136/annrheumdis-2012-202545. [DOI] [PubMed] [Google Scholar]
- 65.Bartelds GM, Krieckaert CLM, Nurmohamed MT, van Schouwenburg PA, Lems WF, Twisk JWR, et al. Development of antidrug antibodies against adalimumab and association with disease activity and treatment failure during long-term follow-up. JAMA. 2011;305(14):1460–1468. doi: 10.1001/jama.2011.406. [DOI] [PubMed] [Google Scholar]
- 66.Berkhout LC, Lami MJ, Ruwaard J, Hart MH, Heer PO, Bloem K, et al. Dynamics of circulating TNF during adalimumab treatment using a drug-tolerant TNF assay. Sci Transl Med. 2019 doi: 10.1126/scitranslmed.aat3356. [DOI] [PubMed] [Google Scholar]
- 67.Baker M, Reynolds HM, Lumicisi B, Bryson CJ. Immunogenicity of protein therapeutics: the key causes, consequences and challenges. Self/Nonself. 2010;1(4):314–322. doi: 10.4161/self.1.4.13904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Saffari F, Jafarzadeh A. Development of anti-rituximab antibodies in rituximab-treated patients: related parameters & consequences. Indian J Med Res. 2022;155(3–4):335–346. doi: 10.4103/ijmr.ijmr_312_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Leandro MJ, Cambridge G, Ehrenstein MR, Edwards JCW. Reconstitution of peripheral blood B cells after depletion with rituximab in patients with rheumatoid arthritis. Arthritis Rheumat. 2006;54(2):613–620. doi: 10.1002/art.21617. [DOI] [PubMed] [Google Scholar]
- 70.Salant DJ. Does epitope spreading influence responsiveness to rituximab in PLA2R-associated membranous nephropathy? Clin J Am Soc Nephrol. 2019;14:1122–4. doi: 10.2215/CJN.07300619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Disis ML, Goodell V, Schiffman K, Knutson KL. Humoral epitope-spreading following immunization with a HER-2/neu peptide based vaccine in cancer patients. J Clin Immunol. 2004;24:571–8. doi: 10.1023/B:JOCI.0000040928.67495.5. [DOI] [PubMed] [Google Scholar]
- 72.Ruperto N, Bazso A, Ravelli A, Malattia C, Filocamo G, Pistorio A, et al. The paediatric rheumatology international trials organization (PRINTO) Lupus. 2007;16(8):670–676. doi: 10.1177/0961203307079556. [DOI] [PubMed] [Google Scholar]
- 73.Kazi ZB, Desai AK, Troxler RB, Kronn D, Packman S, Sabbadini M, et al. An immune tolerance approach using transient low-dose methotrexate in the ERT-naïve setting of patients treated with a therapeutic protein: experience in infantile-onset Pompe disease. Genet Med. 2019;21(4):887–895. doi: 10.1038/s41436-018-0270-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gopal AK, Kahl BS, Flowers CR, Martin P, Ansell SM, Abella-Dominicis E, et al. Idelalisib is effective in patients with high-risk follicular lymphoma and early relapse after initial chemoimmunotherapy. Blood. 2017;129(22):3037–3039. doi: 10.1182/blood-2016-12-757740. [DOI] [PubMed] [Google Scholar]
- 75.Gokemeijer J, Wen Y, Jawa V, Mitra-Kaushik S, Chung S, Goggins A, et al. Survey outcome on immunogenicity risk assessment tools for biotherapeutics: an insight into consensus on methods, application, and utility in drug development. AAPS J. 2023;25(4):55. doi: 10.1208/s12248-023-00820-7. [DOI] [PubMed] [Google Scholar]
- 76.Immune epitope database & tools. https://www.iedb.org/. Accessed 12 Aug 2023.
- 77.Basic local alignment search tool. https://blast.ncbi.nlm.nih.gov/Blast.cgi. Accessed 12 Aug 2023.
- 78.Vita R, Mahajan S, Overton JA, Dhanda SK, Martini S, Cantrell JR, et al. The Immune Epitope Database (IEDB): 2018 update. Nucl Acids Res. 2019;47(Database issue):D339–D343. doi: 10.1093/nar/gky1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lundegaard C, Lamberth K, Harndahl M, Buus S, Lund O, Nielsen M. NetMHC-30: accurate web accessible predictions of human, mouse and monkey MHC class I affinities for peptides of length 8–11. Nucl Acids Res. 2008;36(Web Server issue):W509–W512. doi: 10.1093/nar/gkn202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Alvarez B, Reynisson B, Barra C, Buus S, Ternette N, Connelley T, et al. NNAlign_MA; mhc peptidome deconvolution for accurate mhc binding motif characterization and improved t-cell epitope predictions. Mol Cell Proteom. 2019;18(12):2459–2477. doi: 10.1074/mcp.tir119.001658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Reynisson B, Alvarez B, Paul S, Peters B, Nielsen M. NetMHCpan-4.1 and NetMHCIIpan-4.0: improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data. Nucl Acids Res. 2020;48(W1):gkaa379. doi: 10.1093/nar/gkaa379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Abelin JG, Harjanto D, Malloy M, Suri P, Colson T, Goulding SP, et al. Defining HLA-ii ligand processing and binding rules with mass spectrometry enhances cancer epitope prediction. Immunity. 2019;51(4):766–779.e17. doi: 10.1016/j.immuni.2019.08.012. [DOI] [PubMed] [Google Scholar]
- 83.Racle J, Michaux J, Rockinger GA, Arnaud M, Bobisse S, Chong C, et al. Robust prediction of HLA class II epitopes by deep motif deconvolution of immunopeptidomes. Nat Biotechnol. 2019;37(11):1283–1286. doi: 10.1038/s41587-019-0289-6. [DOI] [PubMed] [Google Scholar]
- 84.Chen B, Khodadoust MS, Olsson N, Wagar LE, Fast E, Liu CL, et al. Predicting HLA class II antigen presentation through integrated deep learning. Nat Biotechnol. 2019;37(11):1332–1343. doi: 10.1038/s41587-019-0280-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.NetMHCIIpan 4.2. https://services.healthtech.dtu.dk/services/NetMHCIIpan-4.0/. Accessed 12 Aug 2023.
- 86.Nilsson JB, Kaabinejadian S, Yari H, Peters B, Barra C, Gragert L, et al. Machine learning reveals limited contribution of trans-only encoded variants to the HLA-DQ immunopeptidome. Commun Biol. 2023;6(1):442. doi: 10.1038/s42003-023-04749-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cohen S, Chung S, Spiess C, Lundin V, Stefanich E, Laing ST, et al. An integrated approach for characterizing immunogenic responses toward a bispecific antibody. MAbs. 2021;13(1):1944017. doi: 10.1080/19420862.2021.1944017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Barra C, Ackaert C, Reynisson B, Schockaert J, Jessen LE, Watson M, et al. Immunopeptidomic data integration to artificial neural networks enhances protein-drug immunogenicity prediction. Front Immunol. 2020;11:1304. doi: 10.3389/fimmu.2020.01304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Thrift WJ, Perera J, Cohen S, Lounsbury NW, Gurung H, Rose C, et al. Graph-pMHC: graph neural network approach to mhc class II peptide presentation and antibody immunogenicity. bioRxiv. 2023 doi: 10.1101/2023.01.19.524779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Holland CJ, Cole DK, Godkin A. Re-directing CD4+ T cell responses with the flanking residues of MHC class II-bound peptides: the core is not enough. Front Immunol. 2013;4:172. doi: 10.3389/fimmu.2013.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Muhammed MT, Aki-Yalcin E. Homology modeling in drug discovery: overview, current applications, and future perspectives. Chem Biol Drug Des. 2019;93(1):12–20. doi: 10.1111/cbdd.13388. [DOI] [PubMed] [Google Scholar]
- 92.Schwede T. Protein modeling: what happened to the “protein structure gap”? Structure. 2013;21(9):1531–1540. doi: 10.1016/j.str.2013.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ruffolo JA, Chu L-S, Mahajan SP, Gray JJ. Fast, accurate antibody structure prediction from deep learning on massive set of natural antibodies. Nat Commun. 2023;14(1):2389. doi: 10.1038/s41467-023-38063-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596(7873):583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Abanades B, Wong WK, Boyles F, Georges G, Bujotzek A, Deane CM. ImmuneBuilder: Deep-Learning models for predicting the structures of immune proteins. Commun Biol. 2023;6(1):575. doi: 10.1038/s42003-023-04927-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucl Acids Res. 2018;46(Web Sever Issue):W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: an automated protein homology-modeling server. Nucl Acids Res. 2003;31(13):3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10(6):845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Klinman D. ELISPOT assay to detect cytokine-secreting murine and human cells. Curr Prot Immunol. 2008;83(1):6.19.1–6.19.9. doi: 10.1002/0471142735.im0619s83. [DOI] [PubMed] [Google Scholar]
- 100.Slota M, Lim J-B, Dang Y, Disis ML. ELISpot for measuring human immune responses to vaccines. Expert Rev Vacc. 2011;10(3):299–306. doi: 10.1586/erv.10.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Siegel M, Steiner G, Franssen LC, Carratu F, Herron J, Hartman K, et al. Validation of a dendritic cell and CD4+ T cell restimulation assay contributing to the immunogenicity risk evaluation of biotherapeutics. Pharmaceutics. 2022;14(12):2672. doi: 10.3390/pharmaceutics14122672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cohen S, Myneni S, Batt A, Guerrero J, Brumm J, Chung S. Immunogenicity risk assessment for biotherapeutics through in vitro detection of CD134 and CD137 on T helper cells. MAbs. 2021;13(1):1898831. doi: 10.1080/19420862.2021.1898831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Schultz HS, Reedtz-Runge SL, Bäckström BT, Lamberth K, Pedersen CR, Kvarnhammar AM, consortium A Quantitative analysis of the CD4+ T cell response to therapeutic antibodies in healthy donors using a novel T cell:PBMC assay. PLoS ONE. 2017;12(5):e0178544. doi: 10.1371/journal.pone.0178544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ito S, Ikuno T, Mishima M, Yano M, Hara T, Kuramochi T, et al. In vitro human helper T-cell assay to screen antibody drug candidates for immunogenicity. J Immunotoxicol. 2019;16(1):125–132. doi: 10.1080/1547691x.2019.1604586. [DOI] [PubMed] [Google Scholar]
- 105.Lyons AB. Analysing cell division in vivo and in vitro using flow cytometric measurement of CFSE dye dilution. J Immunol Methods. 2000;243(1–2):147–154. doi: 10.1016/s0022-1759(00)00231-3. [DOI] [PubMed] [Google Scholar]
- 106.Wang S-Y, Racila E, Taylor RP, Weiner GJ. NK-cell activation and antibody-dependent cellular cytotoxicity induced by rituximab-coated target cells is inhibited by the C3b component of complement. Blood. 2008;111(3):1456–1463. doi: 10.1182/blood-2007-02-074716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Walsh RE, Lannan M, Wen Y, Wang X, Moreland CA, Willency J, et al. Post-hoc assessment of the immunogenicity of three antibodies reveals distinct immune stimulatory mechanisms. MAbs. 2020;12(1):1764829. doi: 10.1080/19420862.2020.1764829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Roche PA, Furuta K. The ins and outs of MHC class II-mediated antigen processing and presentation. Nat Rev Immunol. 2015;15(4):203–216. doi: 10.1038/nri3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Melendez R, Ordonia B, Guerrero J, Hassanzadeh A, Tran P, Low J, et al. Introducing dendritic cell antibody internalization as an immunogenicity risk assessment tool. Bioanalysis. 2022;14(10):703–713. doi: 10.4155/bio-2022-0024. [DOI] [PubMed] [Google Scholar]
- 110.Xue L, Hickling T, Song R, Nowak J, Rup B. Contribution of enhanced engagement of antigen presentation machinery to the clinical immunogenicity of a human interleukin (IL)-21 receptor-blocking therapeutic antibody. Clin Exper Immunol. 2016;183(1):102–113. doi: 10.1111/cei.12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Deora A, Hegde S, Lee J, Choi C-H, Chang Q, Lee C, et al. Transmembrane TNF-dependent uptake of anti-TNF antibodies. MAbs. 2017;9(4):680–695. doi: 10.1080/19420862.2017.1304869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wen Y, Cahya S, Zeng W, Lin J, Wang X, Liu L, et al. Development of a FRET-based assay for analysis of mabs internalization and processing by dendritic cells in preclinical immunogenicity risk assessment. AAPS J. 2020;22(3):68. doi: 10.1208/s12248-020-00444-1. [DOI] [PubMed] [Google Scholar]
- 113.Wen Y, Jawa V. The impact of product and process related critical quality attributes on immunogenicity and adverse immunological effects of biotherapeutics. J Pharm Sci. 2021;110(3):1025–1041. doi: 10.1016/j.xphs.2020.12.003. [DOI] [PubMed] [Google Scholar]
- 114.Quarmby V, Phung QT, Lill JR. MAPPs for the identification of immunogenic hotspots of biotherapeutics; an overview of the technology and its application to the biopharmaceutical arena. Expert Rev Proteom. 2018;15(9):733–748. doi: 10.1080/14789450.2018.1521279. [DOI] [PubMed] [Google Scholar]
- 115.Eng JK, Fischer B, Grossmann J, MacCoss MJ. A fast SEQUEST cross correlation algorithm. J Proteome Res. 2008;7(10):4598–4602. doi: 10.1021/pr800420s. [DOI] [PubMed] [Google Scholar]
- 116.Perkins DN, Pappin DJC, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20(18):3551–3567. doi: 10.1002/(sici)1522-2683(19991201)20:18<3551::aid-elps3551>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 117.Ma B, Zhang K, Hendrie C, Liang C, Li M, Doherty-Kirby A, Lajoie G. PEAKS: powerful software for peptide de novo sequencing by tandem mass spectrometry. Rapid Commun Mass Spectrom. 2003;17(20):2337–2342. doi: 10.1002/rcm.1196. [DOI] [PubMed] [Google Scholar]
- 118.Mommen GPM, Marino F, Meiring HD, Poelen MCM, Brink JAM, van GD, Mohammed S, et al. sampling from the proteome to the human leukocyte antigen-dr (hla-dr) ligandome proceeds via high specificity*. Mol Cell Proteom. 2016;15(4):1412–1423. doi: 10.1074/mcp.m115.055780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hamze M, Meunier S, Karle A, Gdoura A, Goudet A, Szely N, et al. Characterization of CD4 T cell epitopes of infliximab and rituximab identified from healthy donors. Front Immunol. 2017;8:500. doi: 10.3389/fimmu.2017.00500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Spindeldreher S, Karle A, Correia E, Tenon M, Gottlieb S, Huber T, et al. T cell epitope mapping of secukinumab and ixekizumab in healthy donors. MAbs. 2020;12(1):1707418. doi: 10.1080/19420862.2019.1707418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Karle AC, Wrobel MB, Koepke S, Gutknecht M, Gottlieb S, Christen B, et al. Anti-brolucizumab immune response as one prerequisite for rare retinal vasculitis/retinal vascular occlusion adverse events. Sci Transl Med. 2023;15(eabq681):5241. doi: 10.1126/scitranslmed.abq5241. [DOI] [PubMed] [Google Scholar]
- 122.Arnold FH. Innovation by evolution: bringing new chemistry to life (nobel lecture) Angew Chem Int Ed. 2019;58(41):14420–14426. doi: 10.1002/anie.201907729. [DOI] [PubMed] [Google Scholar]
- 123.Griswold KE, Bailey-Kellogg C. Design and engineering of deimmunized biotherapeutics. Curr Opin Struct Biol. 2016;39:79–88. doi: 10.1016/j.sbi.2016.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Mazor R, Eberle JA, Hu X, Vassall AN, Onda M, Beers R, et al. Recombinant immunotoxin for cancer treatment with low immunogenicity by identification and silencing of human T-cell epitopes. Proc Natl Acad Sci USA. 2014;111(23):8571–8576. doi: 10.1073/pnas.1405153111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Tangri S, Mothé BR, Eisenbraun J, Sidney J, Southwood S, Briggs K, et al. Rationally engineered therapeutic proteins with reduced immunogenicity. J Immunol. 2005;174(6):3187–3196. doi: 10.4049/jimmunol.174.6.3187. [DOI] [PubMed] [Google Scholar]
- 126.Mazor R, Vassall AN, Eberle JA, Beers R, Weldon JE, Venzon DJ, et al. Identification and elimination of an immunodominant T-cell epitope in recombinant immunotoxins based on Pseudomonas exotoxin A. Proc Natl Acad Sci USA. 2012;109(51):E3597–E3603. doi: 10.1073/pnas.1218138109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Cizeau J, Grenkow DM, Brown JG, Entwistle J, MacDonald GC. Engineering and biological characterization of VB6-845, an anti-EpCAM immunotoxin containing a T-cell epitope-depleted variant of the plant toxin bouganin. J Immunother. 2009;32(6):574–584. doi: 10.1097/cji.0b013e3181a6981c. [DOI] [PubMed] [Google Scholar]
- 128.Harding FA, Liu AD, Stickler M, Razo OJ, Chin R, Faravashi N, et al. A β-lactamase with reduced immunogenicity for the targeted delivery of chemotherapeutics using antibody-directed enzyme prodrug therapy. Mol Cancer Therapeut. 2005;4(11):1791–1800. doi: 10.1158/1535-7163.mct-05-0189. [DOI] [PubMed] [Google Scholar]
- 129.Arslan M, Karadağ D, Kalyoncu S. Protein engineering approaches for antibody fragments: directed evolution and rational design approaches. Turk J Biol. 2019;43(1):1–12. doi: 10.3906/biy-1809-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Jankowski W, McGill J, Lagassé HAD, Surov S, Bembridge G, Bunce C, et al. Mitigation of T-cell dependent immunogenicity by reengineering factor VIIa analogue. Blood Adv. 2019;3(17):2668–2678. doi: 10.1182/bloodadvances.2019000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Jefferis R. Glycosylation as a strategy to improve antibody-based therapeutics. Nat Rev Drug Discov. 2009;8(3):226–234. doi: 10.1038/nrd2804. [DOI] [PubMed] [Google Scholar]
- 132.Veronese FM. Peptide and protein PEGylation a review of problems and solutions. Biomaterials. 2001;22(5):405–417. doi: 10.1016/s0142-9612(00)00193-9. [DOI] [PubMed] [Google Scholar]
- 133.Dozier JK, Distefano MD. Site-specific PEGylation of therapeutic proteins. Int J Mol Sci. 2015;16(10):25831–25864. doi: 10.3390/ijms161025831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Schlapschy M, Binder U, Börger C, Theobald I, Wachinger K, Kisling S, et al. PASylation: a biological alternative to PEGylation for extending the plasma half-life of pharmaceutically active proteins. Protein Eng Des Sel. 2013;26(8):489–501. doi: 10.1093/protein/gzt023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Schellenberger V, Wang C, Geething NC, Spink BJ, Campbell A, To W, et al. A recombinant polypeptide extends the in vivo half-life of peptides and proteins in a tunable manner. Nat Biotechnol. 2009;27(12):1186–1190. doi: 10.1038/nbt.1588. [DOI] [PubMed] [Google Scholar]
- 136.Zinsli LV, Stierlin N, Loessner MJ, Schmelcher M. Deimmunization of protein therapeutics – Recent advances in experimental and computational epitope prediction and deletion. Comput Struct Biotechnol J. 2021;19:315–329. doi: 10.1016/j.csbj.2020.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.WHO. Guideline for the production and quality control of monoclonal antibodies and related products intended for medicinal use. WHO; 2022.
- 138.Cost GJ, Freyvert Y, Vafiadis A, Santiago Y, Miller JC, Rebar E, et al. BAK and BAX deletion using zinc-finger nucleases yields apoptosis-resistant CHO cells. Biotechnol Bioeng. 2010;105(2):330–340. doi: 10.1002/bit.22541. [DOI] [PubMed] [Google Scholar]
- 139.Sun T, Li C, Han L, Jiang H, Xie Y, Zhang B, et al. Functional knockout of FUT8 in Chinese hamster ovary cells using CRISPR/Cas9 to produce a defucosylated antibody. Eng Life Sci. 2015;15(6):660–666. doi: 10.1002/elsc.201400218. [DOI] [Google Scholar]
- 140.Ronda C, Pedersen LE, Hansen HG, Kallehauge TB, Betenbaugh MJ, Nielsen AT, Kildegaard HF. Accelerating genome editing in CHO cells using CRISPR Cas9 and CRISPy, a web-based target finding tool. Biotechnol Bioeng. 2014;111(8):1604–1616. doi: 10.1002/bit.25233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Tuameh A, Harding SE, Darton NJ. Methods for addressing host cell protein impurities in biopharmaceutical product development. Biotechnol J. 2023;18(3):2200115. doi: 10.1002/biot.202200115. [DOI] [PubMed] [Google Scholar]
- 142.Kinnear G, Jones ND, Wood KJ. Costimulation blockade: current perspectives and implications for therapy. Transplantation. 2012;95(4):527–535. doi: 10.1097/tp.0b013e31826d4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Mueller DL, Jenkins MK, Schwartz RH. Clonal expansion versus functional clonal inactivation: a costimulatory signalling pathway determines the outcome of T cell antigen receptor occupancy. Ann Rev Immunol. 1989;7(1):445–480. doi: 10.1146/annurev.iy.07.040189.002305. [DOI] [PubMed] [Google Scholar]
- 144.Trinchieri G. Interleukin-12 and the regulation of innate resistance and adaptive immunity. Nat Rev Immunol. 2003;3(2):133–146. doi: 10.1038/nri1001. [DOI] [PubMed] [Google Scholar]
- 145.Larsen CP, Pearson TC, Adams AB, Tso P, Shirasugi N, StrobertM E, et al. Rational development of LEA29Y (belatacept), a high-affinity variant of CTLA4-Ig with potent immunosuppressive properties. Am J Transpl. 2005;5(3):443–453. doi: 10.1111/j.1600-6143.2005.00749.x. [DOI] [PubMed] [Google Scholar]
- 146.Kishimoto TK, Ferrari JD, LaMothe RA, Kolte PN, Griset AP, O'Neil C, et al. Improving the efficacy and safety of biologic drugs with tolerogenic nanoparticles. Nat Nanotechnol. 2016;11:890–899. doi: 10.1038/nnano.2016.135. [DOI] [PubMed] [Google Scholar]
- 147.Riella LV, Paterson AM, Sharpe AH, Chandraker A. Role of the PD-1 pathway in the immune response. Am J Transpl. 2012;12(10):2575–2587. doi: 10.1111/j.1600-6143.2012.04224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Findlay JWA, Smith WC, Lee JW, Nordblom GD, Das I, DeSilva BS, et al. Validation of immunoassays for bioanalysis: a pharmaceutical industry perspective. J Pharm Biomed Anal. 2000;21(6):1249–1273. doi: 10.1016/s0731-7085(99)00244-7. [DOI] [PubMed] [Google Scholar]
- 149.Koren E, Smith HW, Shores E, Shankar G, Finco-Kent D, Rup B, et al. Recommendations on risk-based strategies for detection and characterization of antibodies against biotechnology products. J Immunol Methods. 2008;333(1–2):1–9. doi: 10.1016/j.jim.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 150.Mire-Sluis AR, Barrett YC, Devanarayan V, Koren E, Liu H, Maia M, et al. Recommendations for the design and optimization of immunoassays used in the detection of host antibodies against biotechnology products. J Immunol Methods. 2004;289(1–2):1–16. doi: 10.1016/j.jim.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 151.Jahn E-M, Schneider CK. How to systematically evaluate immunogenicity of therapeutic proteins – regulatory considerations. New Biotechnol. 2009;25(5):280–286. doi: 10.1016/j.nbt.2009.03.012. [DOI] [PubMed] [Google Scholar]
- 152.Schellekens H. Immunogenicity of therapeutic proteins: clinical implications and future prospects. Clin Therapeut. 2002;24(11):1720–1740. doi: 10.1016/s0149-2918(02)80075-3. [DOI] [PubMed] [Google Scholar]
- 153.Jawa V, Cousens LP, Awwad M, Wakshull E, Kropshofer H, Groot ASD. T-cell dependent immunogenicity of protein therapeutics: preclinical assessment and mitigation. Clin Immunol. 2013;149(3):534–555. doi: 10.1016/j.clim.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 154.Guideline on Immunogenicity assessment of therapeutic proteins. www.ema.europa.eu/en/documents/scientific-guideline/guideline-immunogenicity-assessment-therapeutic-proteins-revision-1_en.pdf. Accessed 12 Aug 2023.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data represented are available in the public domain.