Abstract
The giant protein titin (TTN) is a sarcomeric protein that forms the myofibrillar backbone for the components of the contractile machinery which plays a crucial role in muscle disorders and cardiomyopathies. Diagnosing TTN pathogenic variants has important implications for patient management and genetic counseling. Genetic testing for TTN variants can help identify individuals at risk for developing cardiomyopathies, allowing for early intervention and personalized treatment strategies. Furthermore, identifying TTN variants can inform prognosis and guide therapeutic decisions. Deciphering the intricate genotype–phenotype correlations between TTN variants and their pathologic traits in cardiomyopathies is imperative for gene-based diagnosis, risk assessment, and personalized clinical management. With the increasing use of next-generation sequencing (NGS), a high number of variants in the TTN gene have been detected in patients with cardiomyopathies. However, not all TTN variants detected in cardiomyopathy cohorts can be assumed to be disease-causing. The interpretation of TTN variants remains challenging due to high background population variation. This narrative review aimed to comprehensively summarize current evidence on TTN variants identified in published cardiomyopathy studies and determine which specific variants are likely pathogenic contributors to cardiomyopathy development.
Keywords: TTN, Titin, Cardiomyopathy, Variant, Genetic
Subject terms: Genetics, Cardiology, Medical research, Molecular medicine
Introduction
Cardiomyopathies refer to a diverse range of complex diseases affecting heart muscle, which can lead to abnormalities in the structure and function of the myocardium. These abnormalities occur in the absence of other conditions like coronary artery disease, hypertension, or valvular heart disease1,2. The American Heart Association (AHA) has categorized cardiomyopathies into genetic, acquired or mixed forms like virally induced post-myocarditis cardiomyopathy. The European Society of Cardiology Organization (ESCO) proposed an alternative classification system dividing cardiomyopathies into two subgroups—familial/genetic cardiomyopathies and non-familial/non-genetic cardiomyopathies3,4. Based on morpho-functional phenotypes5, cardiomyopathies are classified into hypertrophic cardiomyopathy (HCM), dilated cardiomyopathy (DCM), restrictive cardiomyopathy (RCM), and arrhythmogenic right ventricular (ARVC) which each one has their specific features6. The hallmark features of cardiomyopathies are genetic and clinical heterogeneity, variable expressivity, and incomplete penetrance. Numerous genes and mutations have been identified that can cause the various types of cardiomyopathies. The majority of known mutations are linked to DCM and HCM, while fewer are associated with RCM and ARVC. Cardiomyopathies demonstrate considerable genetic heterogeneity—mutations in various different genes can lead to cardiomyopathy. There is also phenotypic heterogeneity, where mutations in the same gene can result in diverse types and degrees of severity of cardiomyopathy7. Cardiomyopathy following myocarditis is probably the result of an interaction interplay between the viral infection and a person's inherent susceptibility. Certain subgroups induced by viral infection may be influenced, at least partially, by genetic factors, suggesting that the elimination of the virus and the immune response could be genetically predetermined8.
Among the genes involved in cardiomyopathies, the TTN gene plays a central role which is attributable to its key structural properties and mechanical function within the striated muscle sarcomeres9. TTN is a major human muscle disease-related gene that encodes the largest human protein, Titin, which is a fundamental structural and functional unit of striated muscles10,11. Due to the size and complexity of this gene, its sequencing was difficult to study the mutations and variants. The initial family studies were performed with primer pairs searching on the exons contained in a 280 kb genomics 2q31 region. This indeed led to the identification of titin mutations causing DCM by Gramlich et al.12 Subsequently, the introduction of NGS has allowed for the exploration of larger cohorts and various clinical entities.
Following the development of next-generation sequencing (NGS), as a potent tool for sequencing large and complex genes, TTN gene sequencing which was previously impossible to conduct a comprehensive analysis, has been performed. This improvement in study tools has led to identifying more than 60,000 TTN missense variants reported in the 1000 Genomes Project13,14. Determining which TTN variants actually cause disease versus which are benign is challenging. The goal of this review is to discuss the current state of understanding regarding the challenges in establishing clear associations between particular TTN mutations and specific cardiomyopathy subtypes in a clinical context.
Method and materials
Systematic search, selection criteria and data collection
The study systematically collected TTN variants associated with cardiomyopathy from the Human Gene Mutation Database (HGMD) and public archive of interpretations of clinically relevant variants (ClinVar). In prioritizing data reliability, only variants with documented reference articles were included, while those lacking reference articles were excluded. The search strategy, extending until February 2023, employed key parameters such as Position on Chromosome, Human Genome Variation Society (HGVS) DNA, HGVS Protein, exon or intron number, and dbSNP identifiers.
Variant annotation and pathogenicity assessment
The annotation of TTN variants involved a comprehensive pathogenicity assessment using multiple tools. This included the application of the American College of Medical Genetics and Genomics (ACMG) guidelines, consultation of ClinVar for variant interpretation, insights from Mutation Taster regarding potential pathogenicity, the use of the Combined Annotation Dependent Depletion (CADD) scoring system for deleteriousness prediction, and evaluation through Genomic Evolutionary Rate Profiling (GERP) to assess evolutionary conservation which are explain more in the following.
We determineded the ACMG score for each variant using franklin, an online database (https://franklin.genoox.com/clinical-db). After adding the name in this website, varints ACMG score anongside with other features are provided.
ACMG score
The American College of Medical Genetics and Genomics (ACMG) previously established guidelines for interpreting sequence variants. With the rapid advancements in sequencing technology over the past decade, this report suggests the adoption of standardized terms such as “pathogenic,” “likely pathogenic,” “uncertain significance,” “likely benign,” and “benign” to characterize variants found in genes associated with Mendelian disorders. Additionally, the recommendation outlines a systematic approach for classifying variants into these categories, relying on various types of evidence, including population data (Population, disease-specific, and sequence databases), computational data (using silico tools for missense prediction, splice site prediction and nucleotide conservation prediction), functional data, and segregation data15,16.
In this classification a variant is considered pathogenic if it meets the requirement of having a very strong criterion (PVS1) along with at least one strong criterion (PS1-PS4), or alternatively, two or more moderate criteria (PM1-PM6), or a combination of one moderate criterion and one supporting criterion (PP1-PP5). Another condition is that a variant can be classified as pathogenic if it satisfies the condition of having at least two strong criteria (PS1-PS4). Additionally, a variant can be considered pathogenic if it meets the criteria of having one strong criterion (PS1-PS4) and either three moderate criteria (PM1-PM6), two moderate criteria and at least two supporting criteria (PP1-PP5), or one moderate criterion and at least four supporting criteria (PP1-PP5)16.
A variant is considered likely pathogenic if it satisfies the condition of having one very strong criterion (PVS1) in combination with one moderate criterion (PM1-PM6). Alternatively, a likely pathogenic variant may exhibit one strong criterion (PS1-PS4) along with one to two moderate criteria (PM1-PM6). Another criterion designates a variant as likely pathogenic if it possesses one strong criterion (PS1-PS4) and at least two supporting criteria (PP1-PP5). Furthermore, likely pathogenic variants may be identified if they meet the requirement of having three or more moderate criteria (PM1-PM6). Additionally, a variant is classified as likely pathogenic if it has two moderate criteria (PM1-PM6) and at least two supporting criteria (PP1-PP5), or if it exhibits one moderate criterion (PM1-PM6) along with at least four supporting criteria (PP1-PP5)16. More information is provided in “Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology”16.
The ACMG score for each variant is determined using Franklin, an online database available at https://franklin.genoox.com/clinical-db. Upon entering the variant's name on this website, the ACMG score, along with other relevant features, is provided.
CADD score
CADD, or Combined Annotation Dependent Depletion, serves as a tool for evaluating the deleteriousness of various genetic variants, including single nucleotide changes, multi-nucleotide substitutions, and insertion/deletion variants within the human genome. In contrast to many other annotation tools that often rely on a singular type of information or have limited applicability, CADD offers a versatile metric that objectively combines diverse annotations. The framework integrates multiple annotations into a unified metric by comparing variants that have undergone natural selection with simulated mutations. It incorporates information from more than 60 genomic features to assess single nucleotide variants and short insertions and deletions across the reference assembly. The C-scores generated by CADD demonstrate robust correlations with allelic diversity, pathogenicity of coding and non-coding variants, and experimentally measured regulatory effects. Notably, C-scores of variants associated with complex traits in genome-wide association studies (GWAS) are significantly higher than matched controls, showing correlation with study sample size, indicative of improved accuracy in larger GWAS. CADD employs a machine learning model that distinguishes between simulated de novo variants, potentially encompassing neutral or harmful alleles, and variants persisting in human populations since the split from chimpanzees.
CADD's capability to quantitatively prioritize functional, deleterious, and disease-causing variants spans a wide range of functional categories, effect sizes, and genetic architectures. This tool enhances the scoring of coding variants through features derived from the ESM-1v protein language model and improves the scoring of regulatory variants using features from a convolutional neural network trained on open chromatin regions. For more information CADD has been detailed in four publications17–20.
MutationTaster
MutationTaster is a web-based application designed to assess the disease-causing potential of DNA sequence variants. It employs in silico tests to estimate the impact of a variant on the gene product or protein, conducting assessments at both the protein and DNA levels. Unlike tools limited to single amino acid substitutions, MutationTaster can handle a variety of variants, including synonymous and intronic ones21. The software, written in Perl programming language and utilizes integrated databases (Ensembl, UniProt, ClinVar, ExAC, 1000 Genomes Project, phyloP and phastCons) to filter out known harmless polymorphisms. Various tests, such as amino acid substitution, conservation, domain functionality, splicing effects, and regulatory element abrogation, are performed on the remaining single-nucleotide polymorphisms (SNPs). The results are evaluated by a Naive Bayes classifier, and the output indicates whether the alteration is known or predicted to be harmless or disease-causing, providing detailed information about the mutation. While the tool demonstrates a raw accuracy of approximately 90%, considering knowledge about common polymorphisms and known disease mutations significantly improves the rate of correct classifications. However, it is important to note that predictions of clinical effects suffer from a lack of specificity, a common constraint across various prediction methods22,23.
GERP
Comparative genomic approaches have historically identified mutation sites under purifying selection by examining conserved sequences across distantly related species. Additionally, the performance of such approaches may be limited for short-lived functional elements that don't exhibit sequence conservation across numerous species. Genomic Evolutionary Rate Profiling (GERP) score is associated with the strength of selection (Nes). Results indicate that the GERP score is linked to the intensity of purifying selection. Nevertheless, variations in selection coefficients or turnover of functional elements over time can significantly impact the GERP distribution, leading to unexpected relationships between GERP and Nes24. The GERP score is characterized as the decrease in the count of substitutions in the multi-species sequence alignment in comparison to the neutral expectation. GERP++ scores span from − 12.3 to 6.17, with elevated scores signifying a greater level of evolutionary constraint.
Data integration
Data integration encompassed the consolidation of relevant information, including Position on Chromosome, HGVS DNA, HGVS Protein, exon or intron number, and dbSNP identifiers. Rigorous quality control measures were then applied to ensure the accuracy and consistency of data extraction and annotation.
Statistical analysis
Descriptive statistics were employed for a comprehensive analysis, summarizing the distribution of TTN variants in terms of positions, types, and associated pathogenicity.
Ethical considerations
Ethical considerations are considered in the study, with a commitment to adhering to Data reliability and responsible data handling. In the present study, it is important to note that no human subjects were involved, as this investigation is a comprehensive review rather than an experimental study. The research focused on analyzing reported variants available on PubMed, and ethical approval or consent from human participants was not applicable.
Results
The molecular structure of titin
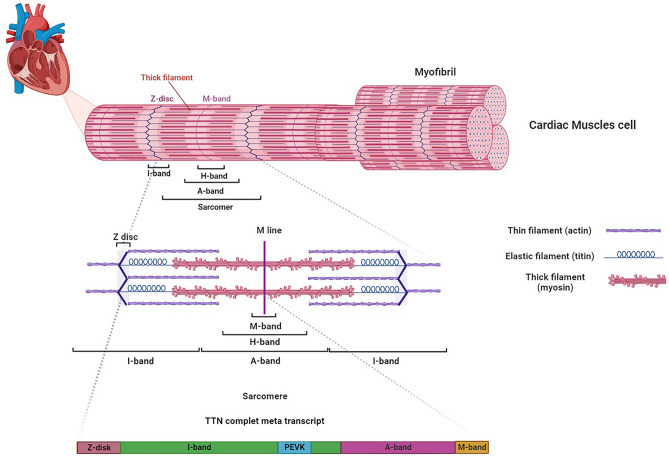
The TTN gene located on the second human chromosome in the 2q31 area. This gene contains 364 exons, which their translation produces a 4200-kDa protein with ~ 38,000 amino acid residues, the largest polypeptide found in the human body. The Titin giant protein, also known as connectin, is the third most abundant protein found in striated muscle among the vertebrates, after myosin and actin. The Titin is a flexible filament that is more than 1 µm long and 3–4 nm wide and spans half of the sarcomere as the repeating contractile unit that gives striated muscle characteristic striped appearance25.
Titin has a complex multidomain structure which is composed of four main structural and functional regions: the N-terminal Z-line acts as an anchor for the sarcomeric Z-disk; the I-band provides elastic properties; the A-band stabilizes the thick filament; and the C-terminal M-line extremity overlaps in an antiparallel orientation with another titin molecule's C-terminus, allowing for modulation of titin expression and turnover via the tyrosine kinase domain26.
The N-terminus contains immunoglobulin (Ig) domains, fibronectin (FN) domains, and a Z-disk region27. The rest of the titin molecule includes an elastic I-band region, a spring-like Pro-Glu-Val-Lys (PEVK) domain, three unique sequences called Novex 1, 2, and 3, cardiac-specific N2B and N2A domains, a thick A-band region, and an M-band region where the C-terminus is embedded.
Extensive alternative splicing in the 364 exons of TTN leads to forming various molecular isoforms. Previous studies have shown three main titin isoforms expressed in cardiomyocytes: the adult N2B isoform, the adult N2BA isoform, and the fetal cardiac titin (FCT) isoform. The distinct characteristics of each titin isoform arise from differences in their I-band sequences, while the Z-disk, A-band, and M-line regions are highly conserved across all isoforms28. Due to the longer extensible I-band region, the N2BA isoform is more compliant than N2B. The N2BA isoform contains additional spring-like elements in the PEVK and tandem Ig regions, leading to lower passive tension in cardiomyocytes compared to other isoforms29–31.
Molecular structure of sarcomere and the interaction of Titin with thin and thick filaments is demonstrated in Fig. 1.
Figure 1.
Molecular structure of sarcomere and the interaction of Titin with thin and thick filaments.
Z-disk
The Z-disk region spans 826 amino acids horizontally across the structure and contains seven Ig domains separated by Z-insertion sequences. As the site of numerous structural and functional interactions with myofibrillar and sarcolemmal proteins, the Z-disk is critical for myofibril assembly, stability, and signaling. Z-disks anchor essential proteins like titin-Tcap (telethonin), which enables key Z-disk functions including mechanosensing. Mechanosensing involves recruiting other interacting and signaling partners to the Z-disk in response to mechanical stimuli. Overall, Z-disks play indispensable roles in anchoring titin and enabling vital structural and sensory functions32–34.
The Z-disk interacts with small ankyrin proteins, spectrin, desmin, and obscurin, connecting it to other cytoskeletal structures. Filamin C links the Z-disk to costameres via integrins and sarcoglycans, participating in mechanosensory pathways. Additionally, the Z-disk binds nebulin, which helps stabilize Z-disk anchorage through interactions with actin, desmin, CapZ and myopalladin. α-Actinin binding also enhances Z-disk mechanical stability. Overall, the Z-disk forms critical protein interactions that provide structural support and sensory functions35–40.
I-band
The I-band region of titin displays extensive alternative splicing, generating diverse isoforms that confer tissue-specific mechanical properties in cardiac and skeletal muscles. Through alternative splicing mechanisms, a spectrum of isoforms emerges, tailoring titin's mechanical functions to meet the needs of different muscle types. The I-band thus acts as a central adapter, converting titin into specialized molecular springs via splicing variability. This interactive segment contains a meta-transcript with principal cardiac and skeletal isoforms. Key components include immunoglobulin folds, the cardiac N2B zone, and the skeletal N2A zone containing nonrepetitive sequences and immunoglobulin domains. The proline-glutamate-valine-lysine (PEVK) domain follows, acting as a spring-like element. Together, the I-band components enable the elasticity of titin38,41.
The I-band region has distinct proximal and distal segments with specialized roles. The proximal I-band maintains sarcomere integrity, while the medial/distal I-band acts as a bidirectional molecular ruler setting resting length and passive tension42. The I-band also functions as a biochemical stress sensor through interactions with αβ-crystallin, a chaperone that stabilizes I-band immunoglobulin domains. Additionally, metabolic enzymes like DRAL, FHL1, and FHL2 associate with I-band sarcomere regions via the Gαq-MAPK pathway37,43,44. Indeed, though I-band interactions with the Ca+2-dependent proteases Calpain-1 and Calpain-3, I-band not only contributes to a sarcomeric quality control pathway but also serves as a reservoir for inactive forms of Calpain-345,46.
A-band
The A-band spans the sarcomere from M-line to M-line, containing thick filaments of myosin. Within the A-band, titin forms a network that maintains the structural integrity of the thick filaments and regulates their length. The A-band exclusively contains fibronectin type III (FnIII) motifs. Immunoglobulin (Ig) and FnIII motifs are arranged in two super-repeats bisected by Ig folds. Unlike the elastic I-band, the A-band is inextensible, providing myosin binding sites that function as stable anchors. A-band super-repeat domains interact with and position sarcomeric myosin binding protein C (MyBP-C). The A-band also contains binding sites for muscle ring finger proteins MURF1 and MURF2. MURF1 likely facilitates quality control and protein turnover at the sarcomere center, while MURF2 interactions aid formation of mature A-band structures36,38.
M-band
The M-band integrates structural, signaling, metabolic and protein quality control functions. It contains a putative serine/threonine kinase domain and immunoglobulin cross-hatched rectangle (CII) domains interspersed with M-insertion sequences47. While its kinase activity is debated, the M-band kinase domain likely participates in stress sensing through Ca2+-calmodulin-regulated mechanochemical signaling38,48. During sarcomerogenesis, myomesin constructs an M-band scaffold linking titin to myosin thick filaments, establishing the myomesin-titin-myosin stability axis49. The M-band also senses metabolic stress via ligands DRAL/FHL2 that tether metabolic enzymes, and enables ubiquitin-mediated turnover through interactions with nbr1, p62, MURF1 and MURF250. MURF2 binding facilitates M-band's role in cardiac development51. Additionally, the extreme C-terminal TTN/calpain-3/p94 interaction participates in M-band-associated protein turnover37,52.
The molecular function of titin
Since the discovery of titin, the complexity and diverse functional roles of titin in health and disease continue to emerge. As the third filament system of the sarcomere alongside actin and myosin, titin forms a unique filament network in cardiomyocytes that engages in mechanical and signaling roles10. During muscle development, titin likely controls the assembly of actin and myosin contractile proteins, regulating sarcomere size and thick filament structure. In mature muscle, titin contributes to elasticity mechanisms affecting sarcomere resting lengths and tension-related processes25.
The enormity and intricate three-dimensional structure of titin provides structural support to maintain sarcomere integrity during contraction while generating passive tension during stretching. Additionally, the numerous titin-binding proteins arranged in signaling hotspots allow titin to participate in mechanosensing and signal transduction26,53. Thus, titin has multifaceted roles beyond viscoelastic force generation: (a) centering thick filaments for optimal active force; (b) assembling sarcomeres; (c) mechanochemical signaling through binding partners; and (d) potentially enabling length-dependent activation underlying the Frank-Starling law54.
Comparative analysis of TTN variants
In this study we found 611 distant TTN variant which were not benign and they were pathogenic, likely pathogenic or variant of uncertain significance (VUS).
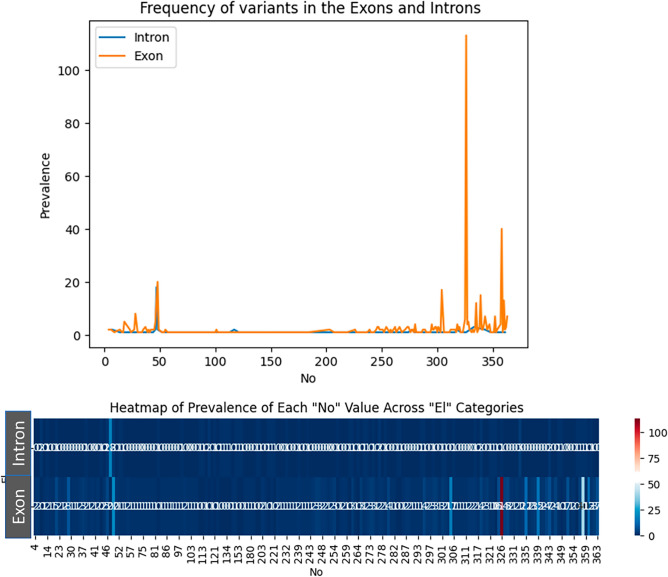
85% of the variants were reported in exon fragments, while 15% were reported in intron fragments. In ACMG classification, 69.6% of the variants were classified as Pathogenic, 21.6% as Likely Pathogenic, and 8.8% as Variants of Uncertain Significance (VUS). Substitution accounted for 57.25% of the variants, deletion for 29.62%, duplication for 7.36%, and insertion for 5.72%.
The majority of variants occurred in the interval from exon 200 to the end of the molecule, with the hotspot regions identified at exon 326 and 358 being the most common points for variations (Fig. 2).
Figure 2.
Prevalence of variants in different exons and introns in TTN.
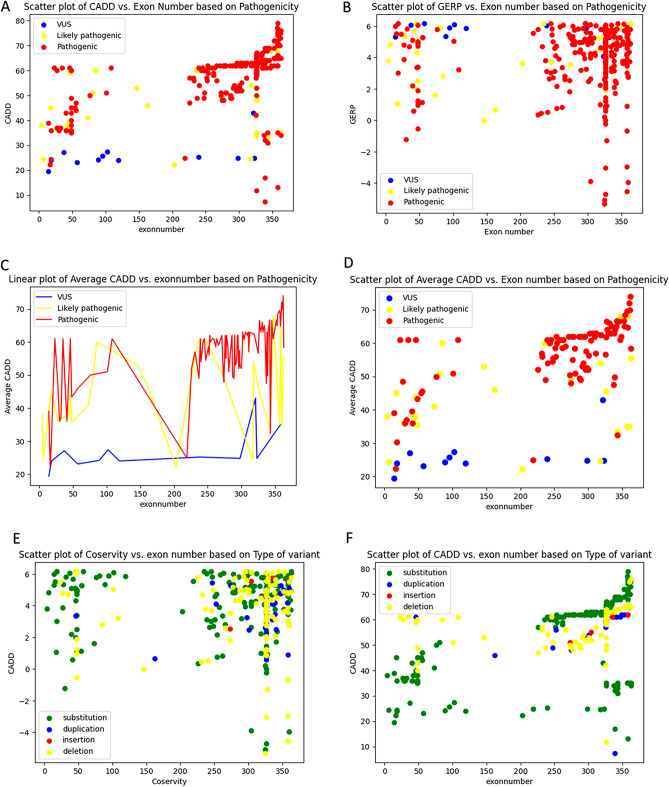
Most pathogenic variants are located after the exon 326 to the end of the molecule which has higher CADD number compared to others (Fig. 3A). The Genomic Evolutionary Rate Profiling (GERP) score is used to compare the gene nucleotides among the species in the TTN gene24. It is supposable that the nucleotides and exons which are conserved in the evolution, can be considered a vital element for survive and loss of function of these components are associated with death and the prevention of its inheritance. In the comparison of the conservity of the gene nucleotides, it can be concluded that most the variants have a notable GERP score which indicates their conservity (Fig. 3B).
Figure 3.
Comparative analysis of TTN variants with their pathogenicity, type of alternation, and conservity.
In comparing the average CADD score of various exons, it can be concluded that exons with higher CADD scores are located in the end of the gene and the middle part of the gene, the average CADD score is not notable. The first few exons of the gene have a higher CADD score but in the last exons, the CADD score is increased considerably especially in the last 50 exons. VUS variants have less CADD score and likely pathogenic variants also have lesser scores compared to pathogenic variants (Fig. 3C,D).
In the comparing type of genetic alternation in variants, it can be concluded that the most common alternations are substitution and deletions. Most of the deletions have high score numbers while substitutions have various CADD scores. Most of the insertion and duplications also have notable CADD score because of frameshift events while in the substitutions we can observe some lesser CADD score which is not exists in other types of alternations. As demonstrated, most of the pathogenic variants in the first parts of the gene are deletions but the most pathogenic variants in the last parts of the gene have substitutions (Fig. 3E,F).
The biogenesis pathways of TTN
Role of alternative splicing
TTN gene consists of 364 exons as translatable parts according to NCBI55 and is estimated to code 34,350 amino acid residues according to UniProt56. TTN can be spliced in different ways to produce different transcript forms. Since alternative splicing of TTN, the protein has various sizes. The I-band, M-line, and Z-disc areas of Titin are the most variable parts, which lead to various isoforms with a wide range of elasticity. Due to variations in the I-band area, different muscle types have varying degrees of elasticity. The Titin gene's I-band encoding region is the site of many splicing processes resulting in isoforms with various spring compositions. This process even can discriminate cardiac Titin with skeletal muscle Titin.
All cardiac Titin isoforms have exon 49, which contains the N2B sequence; however, skeletal muscle does not57. The cardiac isoform known as N2B Titin is a small 2970-kDa weight protein produced by splicing exons 49/50. Deletion of Titin N2B region causes diastolic dysfunction and cardiac atrophy58. Another isoform is N2BA which is made up of exons 102 to 109, which code for the N2A element. A specific property of this isoform is that it contains more PEVK segments and is longer with more Ig domains58.
I-band and its isoforms in cardiac compliance and DCM
Protein composition patterns can change among different populations and even in various stages of human life. The isoform transforming of sarcomeric proteins in the troponin complex, Myosine heavy chain (MHC), Myosine light chain (MLC) and Titin from fetal to adult through transcriptional changes or alternative splicing is the essential element of myofibril maturation59.
A study by Lahmers et al.59 revealed that fetal titin isoforms are expressed in neonates, containing additional spring elements in the tandem Ig and PEVK regions. This leads to lower stiffness compared to adults, explained by the unique spring composition of fetal cardiac titin in neonates. Changes in titin expression during development likely impact functional transitions and diastolic filling as the heart matures. The fetal cardiac titin isoform, with its extra Ig and PEVK spring elements, gradually disappears postnatally in a species-dependent manner.
In the human heart, the ratio of titin isoform expression is established based on passive tension. There is a high correlation between titin-based passive tension and I-band region size, with lower tension associated with a larger, more elastic I-band. In healthy adult hearts, the N2BA and N2B titin isoforms express at 30–40% and 60–70% respectively. The relative levels of these two isoforms are a key determinant of cardiomyocyte stiffness60. Titin plays a central role in the passive ventricular tension. Animal studies have proved that the N2BA isoform is present in the near-term fetus 6 days before birth but after birth disappears and is replaced by a smaller N2B isoform, which predominates in 1-week-old neonate and adults. Adult cardiomyocytes have 15 times more passive tension compared to fetal cardiomyocytes which is confirmed by immunofluorescence microscopy. This transformation is compatible with the heart's function in each stage of life which after birth needs more passive tension to pump the blood effectively through the vessels61.
Alternative splicing of the TTN gene plays significant roles in cardiac diseases like dilated cardiomyopathy (DCM). In DCM, the more compliant N2BA isoform is upregulated, decreasing passive stiffness and increasing chamber compliance. Overall, variable expression and splicing of titin isoforms critically influence myocardial passive tension and compliance30,31,62,63.
Hidalgo et al.64 conducted sophisticated experiments to identify the mechanisms influencing myocardial passive stiffness by modifying the phosphorylation state of titin. The study revealed that titin serves as a substrate not only for protein kinase A but also for protein kinase G and protein kinase C α (PKCα). The researchers pinpointed the PEVK region of titin as the primary site for PKCα phosphorylation, demonstrating that phosphorylation at this site enhances passive tension in the myocardium.
Novex variants and tiny titin results alternative splicing
The whole sequence of the human TTN gene contains three isoform-specific mutually exclusive exons named novel exons (novex), which encode for the I-band sequence. Novex1 is presented in exon 45, novex-2 is located in exon 46, and novex-3 is placed in exon 48. The novex-1 and novex-2 Titin isoforms are encoded by transcripts that either include the novex-1 or novex-2 exons. Early stop-gain codon in the novex-3 transcript produces a remarkably tiny isoform (700 kDa) known as novex-3 Titin. The 'tiny Titin' isoform, expressed in all striated muscles, stretches from the Z-disc to the novex-3 domain (C-terminus). Therefore, stress-induced sarcomeric rearrangement may be mediated by novex-3 Titin because of its regulatory involvement in calcium level and GTPase-associated myofibrillar pathways65. Furthermore, novexes 2 and 3 may be linked to DCM or ARVC based on the expression levels of novex variations in human cardiac tissues affected by cardiomyopathies. Previous research suggests that novex variations may be attributable to cardiomyopathy66.
Splicing regulation of alternative splicing
Encoding Titin by a single gene into various forms is the result of different mRNA splice pathways which leads to Titin isoform classes57. The titin gene contains 409 introns, enabling generation of 57 distinct mRNA transcripts through extensive alternative splicing. These include 29 unspliced forms and 28 spliced isoforms. Additional diversity arises from 5 alternative promoters, 9 non-overlapping final exons, and 9 verified polyadenylation sites. The resulting mRNAs vary in: 3’ end truncations, 5’ end truncations, presence/absence of 173 cassette exons, overlapping exons with different borders, and splicing versus retention of 3 introns67.
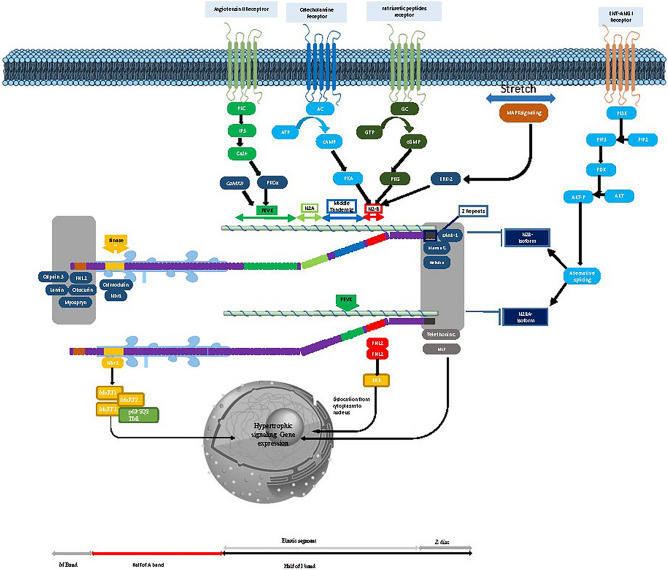
RBM20 regulates a subset of genes involved in developing the heart's muscles by modulating their mRNA alternative splicing. Titin, known to undergo extremely complex alternative splicing, is one of the RBM20’s targets. RBM20 specifically manipulates alternative splicing within the I-band of TTN pre-mRNA, which possesses the highest frequency of the alternative splicing process. It has been demonstrated that some alterations in the protein can produce pathogenic TTN isoforms, which are believed to lead to DCM68. Surprisingly, Khan et al.69, detected 80 distinct circRNAs among nearly a thousand from human hearts, indicating that the I-band of Titin is a hotspot region of circRNAs. Remarkably, the introns on each side of the back-spliced junctions were enriched in RBM20 binding sites, and the introns related to the TTN circRNAs had a five-fold higher frequency of RBM20 binding sites compared to a control set of introns. Studies on the RBP20 knock-out animals, and a cardiac sample of heterozygous RBM20 mutation carrier with substantially compromised synthesis of TTN circRNAs, both provided evidence that RBP20 is involved in the biogenesis of these TTN circRNAs69. Furthermore, the most recent study by Czubak et al.70, also found that Type 1 diabetes patients' human skeletal muscles included a significant amount of circRNAs primarily derived from the I-band of Titin. Titin has considerable interaction with other functional and structural proteins of sarcomeres. So, it is assumable that it has numerous binding sites for muscle-associated proteins and serves as an adhesion template for contractile machinery assembly in cardiac cells. So, it should be considered a dynamic and transformable molecule.
The role of TTN variants in cardiomyopathies
Heterozygous mutations in TTN are commonly associated with cardiomyopathies and TTN has been reported as the most common gene involved in cardiomyopathies71. The mutations can be broadly classified into two categories, which are truncating or missense mutations. Truncating mutations lead to premature termination of Titin protein synthesis, resulting in either an altered protein or the loss of functional domains. In contrast, missense mutations result in the replacement of amino acids, potentially causing interference with the typical operation of the Titin protein36.
The ongoing inquiry into the exact molecular mechanisms by which TTN mutations lead to cardiomyopathies illuminates the intricate relationship between TTN mutations and various forms of cardiomyopathies. The haploinsufficiency model is a notable mechanism that proposes the presence of truncating mutations in one allele of the TTN gene results in a reduction in Titin expression, consequently inducing a functional deficit of Titin protein. The phenomenon mentioned above possesses the capability to disrupt the sarcomere assembly process, alter the mechanical properties of cardiac muscle cells, and prevent the heart's contractile function, leading to the manifestation of cardiomyopathy. Another proposed mechanism which even can be manifest in dominant pattern is missense mutations. This occurrence takes place when the mutated form of the Titin protein impairs the normal functioning of the unaltered Titin protein, leading to compromised assembly and operation of the sarcomere.
Moreover, it is plausible that TTN mutations may trigger aberrant splicing occurrences, leading to the production of deficient or abnormal Titin isoforms, thus playing a role in the pathogenesis of cardiomyopathy c. The bioinformatics analysis of reported variants in TTN related to cardiomyopathies has been shown in Table 1.
Table 1.
Bioinformatics analysis of Pathogenic, Likely pathogenic, Unknown Significance reported variants in TTN related to cardiomyopathies.
| No | Position on Chr. 2 | HGVS DNA | HGVS Protein | Exon/Intron | dbSNP | ACMG | ClinVar | Mutation Taster | CADD | GERP | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 179391826 | c.107889del | p.Lys35963AsnfsTer9 | E.363 | rs281864930 | P | P | DC | 76 | 5.79 | 195 |
| 2 | 179391848 | c.107867T > C | p.Leu35956Pro | E.363 | rs267607156 | LP | LP | DC | 35 | 6.17 | 196 |
| 3 | 179391875 | c.107840T > A | p.Ile35947Asn | E.363 | rs281864928 | P | VUS | DC | 34 | 4.9 | 196 |
| 4 | 179391915 | c.107800G > T | p.Gly35934Ter | E.363 | rs368277535 | LP | VUS | DC | 76 | 6.05 | 197 |
| 5 | 179391925 | c.107780-107790delinsTGAAAGAAAAA | p.Glu35927-Trp35930delinsValLysGluLys | E.363 | rs281864927 | P | P | DC | 65 | 4.87 | 198 |
| 6 | 179391925 | c.107780-107781insTGAAAGAAAAA | p.Glu35927AspfsTer6 | E.363 | NA | LP | NA | PO | 65 | 4.52 | 196 |
| 7 | 179391972 | c.107743A > C | p.Thr35915Pro | E.363 | NA | LP | NA | DC | 32 | 6.06 | 196 |
| 8 | 179392207 | c.107646del | p.Ser35883GlnfsTer10 | E.362 | NA | LP | NA | DC | 75 | 5.76 | 199 |
| 9 | 179392218 | c.107635C > T | p.Gln35879Ter | E.362 | rs757082154 | P | VUS | DC | 75 | 4.87 | 196 |
| 10 | 179392275 | c.107578C > T | p.Gln35860Ter | E.362 | rs1009131948 | P | LP/P | DC | 73 | 3.75 | 200 |
| 11 | 179393000 | c.107377 + 1G > A | – | I.361 | rs112188483 | P | P/LP | NA | NA | 4.96 | 201 |
| 12 | 179393027 | c.107351del | p.Ser35784Ter | E.361 | rs778765016 | P | NA | DC | 81 | 4.97 | 202 |
| 13 | 179393094 | c.107284C > T | p.Arg35762Ter | E.361 | rs1477669354 | P | LP | DC | 70 | 4.36 | 203 |
| 14 | 179393272 | c.107208del | p.Phe35736LeufsTer15 | E.360 | NA | P | NA | DC | 75 | 5.17 | 204 |
| 15 | 179393329 | c.107149C > T | p.Gln35717Ter | E.360 | rs369157062 | P | NA | DC | 81 | 5.56 | 202 |
| 16 | 179393480 | c.106998dup | p.Ala35667SerfsTer6 | E.360 | rs1031891465 | LP | NA | NA | 65 | 4.56 | 202 |
| 17 | 179393500 | c.106978C > T | p.Gln35660Ter | E.360 | rs1687693219 | P | NA | DC | 81 | 5.56 | 205 |
| 18 | 179393519 | c.106959T > A | p.Tyr35653Ter | E.360 | rs369450212 | LP | NA | DC | 41 | − 7.15 | 202,206 |
| 19 | 179393524 | c.106954C > T | p.Arg35652Ter | E.360 | rs565675340 | P | P | DC | 70 | − 3.94 | 207 |
| 20 | 179393564 | c.106914G > C | p.Trp35638Cys | E.360 | rs758497512 | LP | VUS | DC | 35 | 5.55 | 205 |
| 21 | 179393709 | c.106768dup | p.His35590ProfsTer2 | E.360 | NA | P | LP | NA | 65 | 5.10 | 202 |
| 22 | 179393738 | c.106740del | p.Ala35581GlnfsTer36 | E.360 | NA | P | LP | DC | 75 | 5.53 | 202 |
| 23 | 179393845 | c.106668del | p.Lys35556AsnfsTer6 | E.360 | rs587776772 | P | P | DC | 75 | 2.76 | 208 |
| 24 | 178529118 | c.106632-106633del | p.Leu35545LysfsTer3 | E.360 | NA | P | NA | DC | 9.91 | 1.97 | 204 |
| 25 | 179393849 | c.106629del | p.Ala35544ProfsTer2 | E.360 | rs869312069 | P | LP | DC | 75 | 2.82 | 202 |
| 26 | 179393907 | c.106571del | p.Lys35524ArgfsTer22 | E.360 | rs199469666 | P | NA | DC | 73 | 3.39 | 208 |
| 27 | 179394686 | c.106531 + 1G > A | – | I.359 | rs760915007 | P | P | NA | NA | 5.61 | 209 |
| 28 | 179394796 | c.106422del | p.Phe35475SerfsTer3 | E.359 | NA | LP | NA | DC | 72 | − 0.80 | 206 |
| 29 | 179394967 | c.106374 + 1del | – | I.358 | rs763404256| | LP | VUS | NA | NA | 5.13 | 202 |
| 30 | 179395292 | c.106050del | p.Glu35351AsnfsTer54 | E.358 | NA | LP | NA | DC | 74 | − 10.5 | 206 |
| 31 | 179395323 | c.106019del | p.Gly35340ValfsTer65 | E.358 | rs727504482 | P | NA | DC | 74 | 5.23 | 210 |
| 32 | 179395428 | c.105910-105914del | p.Thr35304CysfsTer3 | E.358 | NA | P | NA | DC | 73 | 3.24 | 206 |
| 33 | 179395510 | c.105832C > T | p.Gln35278Ter | E.358 | NA | LP | NA | DC | 11.95 | 2.7 | 211 |
| 34 | 179395528 | c.105814del | p.Thr35272HisfsTer21 | E.358 | rs759645441 | LP | NA | DC | 66 | 0.59 | 202 |
| 35 | 179395600 | c.105739-105742dup | p.Lys35248SerfsTer2 | E.358 | rs866421715 | LP | NA | NA | 62 | 0.88 | 202 |
| 36 | 179395807 | c.105528-105535del | p.Gln35176HisfsTer9 | E.358 | rs199469665 | P | LP | DC | 66 | 3.57 | 212 |
| 37 | 179395811 | c.105523-105531del | p.His35175-Val35177del | E.358 | NA | VUS | NA | PO | 53 | 3.49 | 199 |
| 38 | 179395856 | c.105486del | p.Trp35162CysfsTer8 | E.358 | rs1553485330 | P | P | DC | 66 | 4.78 | 213 |
| 39 | 179395919 | c.105423C > A | p.Tyr35141Ter | E.358 | NA | LP | NA | DC | 64 | − 4.25 | 214 |
| 40 | 179396571 | c.104771C > A | p.Ser34924Ter | E.358 | rs1559003939 | P | LP | DC | 75 | 5.56 | 215 |
| 41 | 179396675 | c.104666-104667del | p.Pro34889ArgfsTer3 | E.358 | NA | P | LP | DC | 66 | − 0.66 | 216 |
| 42 | 179396929 | c.104413C > T | p.Arg34805Ter | E.358 | rs750519430 | P | LP/P | DC | 71 | 4.59 | 217 |
| 43 | 179397250 | c.104092C > T | p.Arg34698Ter | E.358 | rs727504184 | P | LP | DC | 79 | 4.19 | 202,218 |
| 44 | 179397397 | c.103945C > T | p.Arg34649Ter | E.358 | rs995029896 | P | LP | DC | 74 | 3.46 | 219 |
| 45 | 179397492 | c.103850-103851insAAC | p.Lys34618AspfsTer2 | E.358 | NA | VUS | NA | PO | 62 | 0.00 | 210 |
| 46 | 179397546 | c.103796G > A | p.Arg34599Lys | E.358 | rs1362778188 | LP | NA | DC | 35 | 5.80 | 205 |
| 47 | 179397637 | c.103705A > T | p.Lys34569Ter | E.358 | rs1553490574 | P | LP | DC | 75 | 5.94 | 202 |
| 48 | 179397824 | c.103518del | p.Ala34507LeufsTer8 | E.358 | rs1553491220 | P | LP | DC | 66 | − 4.55 | 209 |
| 49 | 179397934 | c.103408G > T | p.Glu34470Ter | E.358 | rs769023413 | LP | VUS | DC | 68 | 5.78 | 202 |
| 50 | 179397982 | c.103360del | p.Glu34454AsnfsTer3 | E.358 | rs760768093 | P | P | DC | 66 | 3.87 | 220 |
| 51 | 179398245 | c.103096-103097insSVAelement | – | E.358 | rs1575266261 | NA | LP | NA | NA | NA | 202 |
| 52 | 179398266 | c.103073-103076dup | p.Ser34359ArgfsTer2 | E.358 | NA | P | LP | NA | 62 | 0.89 | 202 |
| 53 | 179398340 | c.103002-103003insA | p.Ala34335SerfsTer7 | E.358 | NA | P | NA | DC | 62 | 3.24 | 205 |
| 54 | 179398393 | c.102949C > T | p.Gln34317Ter | E.358 | rs397517787 | P | LP | DC | 75 | 5.5 | 80 |
| 55 | 179398396 | c.102946del | p.Tyr34316ThrfsTer3 | E.358 | NA | P | NA | DC | 66 | 4.28 | 205 |
| 56 | 179398410 | c.102932C > G | p.Ser34311Ter | E.358 | NA | LP | NA | DC | 72 | 5.6 | 221 |
| 57 | 179398712 | c.102630del | p.Val34211Ter | E.358 | rs869312101 | p | VUS | DC | 66 | 4.82 | 202 |
| 58 | 179398819 | c.102523C > T | p.Arg34175Ter | E.358 | rs752697861 | P | P | DC | 13.12 | 4.23 | 221 |
| 59 | 179398833 | c.102509G > A | p.Trp34170Ter | E.358 | NA | P | NA | DC | 73 | 5.38 | 205 |
| 60 | 179399071 | c.102271C > T | p.Arg34091Trp | E.358 | rs140319117 | P | VUS | DC | 35 | 4.82 | 205 |
| 61 | 179399128 | c.102214T > A | p.Trp34072Arg | E.358 | NA | LP | NA | DC | 34 | 5.88 | 204 |
| 62 | 179399285 | c.102057del | p.Asn34020ThrfsTer9 | E.358 | NA | P | LP | DC | 66 | − 2.96 | 204 |
| 63 | 179400115 | c.101227C > T | p.Arg33743Ter | E.358 | rs794729305 | P | LP | DC | 76 | 4.63 | 222 |
| 64 | 179400229 | c.101113del | p.Ser33705LeufsTer4 | E.358 | NA | P | NA | DC | 65 | 4.4 | 213 |
| 65 | 179400244 | c.101098-101099insT | p.Asp33700ValfsTer13 | E.358 | rs869312122 | P | LP | DC | 62 | 5.59 | 202 |
| 66 | 179400320 | c.101021-101022del | p.Arg33674IlefsTer4 | E.358 | rs869312087 | P | LP | DC | 65 | 3.01 | 202 |
| 67 | 179400405 | c.100936-100937del | p.Val33646HisfsTer26 | E.358 | NA | LP | NA | DC | 65 | 4.08 | 205 |
| 68 | 179400516 | c.100826G > A | p.Arg33609Gln | E.358 | rs771243505 | VUS | VUS | DC | 35 | 5.3 | 223 |
| 69 | 179400517 | c.100825C > T | p.Arg33609Ter | E.358 | rs1057518195 | P | LP/P | DC | 72 | 5.3 | 224 |
| 70 | 179400577 | c.100766-1G > T | – | I.357 | rs185589320 | LP | NA | NA | NA | 5.3 | 202 |
| 71 | 179400887 | c.100587G > A | p.Trp33529Ter | E.357 | rs1064793560 | P | LP | DC | 70 | 5.76 | 225 |
| 72 | 179400913 | c.100558-100561dup | p.Gly33521AspfsTer25 | E.357 | rs1553501572 | P | LP | NA | 62 | 4.18 | 213 |
| 73 | 179401029 | c.100445C > A | p.Ser33482Ter | E.357 | rs869312086 | P | LP | DC | 77 | 5.76 | 202 |
| 74 | 179401230 | c.100244C > T | p.Pro33415Leu | E.357 | rs72648282 | LP | VUS | DC | 35 | 5.76 | 226 |
| 75 | 179402067 | c.99865 + 2T > C | – | I.355 | rs1453570860 | P | NA | NA | NA | 5.53 | 199 |
| 76 | 179403522 | c.99034A > T | p.Lys33012Ter | E.354 | rs771511344 | P | LP | DC | 72 | 5.71 | 199 |
| 77 | 179403562 | c.98994del | p.Lys32998AsnfsTer63 | E.354 | rs727504535 | P | P | DC | 65 | 3.68 | 222 |
| 78 | 179403888 | c.98774del | p.Gly32925ValfsTer56 | E.353 | NA | P | LP | DC | 65 | 6.15 | 210 |
| 79 | 179404189 | c.98603del | p.Phe32868SerfsTer11 | E.352 | NA | P | NA | DC | 65 | 3.44 | 201 |
| 80 | 179404241 | c.98551C > T | p.Arg32851Ter | E.352 | rs553821887 | P | VUS | DC | 69 | 3.78 | 202 |
| 81 | 179404286 | c.98506C > T | p.Arg32836Ter | E.352 | rs869312085 | P | LP | DC | 72 | 4.88 | 202 |
| 82 | 179404492 | c.98299-98300del | p.Arg32767GlyfsTer2 | E.352 | rs397517776 | P | P | DC | 65 | 4.91 | 202 |
| 83 | 179404493 | c.98299del | p.Arg32767GlyfsTer26 | E.352 | rs772061676 | P | LP | DC | 65 | 3.65 | 202 |
| 84 | 179404524 | c.98265-98268dup | p.His32757AsnfsTer4 | E.352 | rs869312067 | P | LP | NA | 62 | 5.02 | 202 |
| 85 | 179404687 | c.98105del | p.Pro32702LeufsTer15 | E.352 | NA | P | NA | DC | 65 | 6.17 | 213 |
| 86 | 179405030 | c.97863G > A | p.Trp32621Ter | E.351 | NA | LP | NA | DC | 68 | 5.96 | 201 |
| 87 | 179406990 | c.97492 + 1G > A | – | I.349 | rs727505319 | P | NA | NA | NA | 6.17 | 227 |
| 88 | 179407385 | c.97192 + 4A > G | – | I.348 | rs370069759 | VUS | VUS | NA | NA | 4.4 | 202 |
| 89 | 179407531 | c.97050dup | p.Glu32351ArgfsTer6 | E.348 | rs794729365 | P | P | NA | 62 | 5.27 | 228 |
| 90 | 179407808 | c.96892C > T | p.Gln32298Ter | E.347 | rs201108270 | LP | VUS | DC | 68 | 5.91 | 202 |
| 91 | 179408200 | c.96500-96501insAGAATTC | p.Gly32168GlufsTer27 | E.347 | NA | P | NA | DC | 61 | 6.03 | 205 |
| 92 | 179408240 | c.96460dup | p.Thr32154AsnfsTer39 | E.347 | rs869312084 | P | LP | NA | 61 | 4.75 | 202 |
| 93 | 179408364 | c.96336-96337insC | p.Lys32113GlnfsTer3 | E.347 | NA | P | NA | DC | 61 | 5.32 | 80 |
| 94 | 179408990 | c.95966del | p.Asn31989ThrfsTer2 | E.345 | rs72648265 | P | LP | DC | 64 | 6.17 | 199 |
| 95 | 179409084 | c.95872C > T | p.Arg31958Ter | E.345 | NA | P | LP | DC | 69 | 5.23 | 229 |
| 96 | 179410544 | c.95416 + 3–95416 + 4insCCT | – | I.343 | NA | LP | NA | NA | NA | 3.31 | 199 |
| 97 | 179410545 | c.95415–95416 + 2del | – | I.343 | rs769407533 | P | LP | NA | NA | 5.82 | 202 |
| 98 | 179410592 | c.95371G > C | p.Gly31791Arg | E.343 | NA | P | VUS | DC | 31 | 5.82 | 230 |
| 99 | 179410605 | c.95358C > G | p.Asn31786Lys | E.343 | rs869320743 | P | P | DC | 31 | 4.95 | 231 |
| 100 | 179410622 | c.95341C > T | p.Arg31781Ter | E.343 | NA | P | NA | DC | 69 | 2.95 | 205 |
| 101 | 179410768 | c.95195C > T | p.Pro31732Leu | E.343 | rs753334568 | P | LP/P | DC | 35 | 5.82 | 231 |
| 102 | 179410778 | c.95185T > C | p.Trp31729Arg | E.343 | rs869320741 | LP | P | DC | 34 | 5.82 | 231 |
| 103 | 179410799 | c.95164C > T | p.Gln31722Ter | E.343 | NA | P | NA | DC | 66 | 4.95 | 199 |
| 104 | 179410829 | c.95134T > C | p.Cys31712Arg | E.343 | rs869320740 | LP | P | DC | 33 | 5.82 | 231 |
| 105 | 179411050 | c.95008C > T | p.Arg31670Ter | E.342 | rs1322596650 | P | P | DC | 68 | 4.78 | 232 |
| 106 | 179411199 | c.94859T > G | p.Leu31620Ter | E.342 | rs561946873 | LP | NA | DC | 70 | 6.03 | 207 |
| 107 | 179411200 | c.94852-94858del | p.Ala31618TyrfsTer37 | E.342 | rs869312066 | P | LP | DC | 64 | 4.51 | 202 |
| 108 | 179411203 | c.94855C > T | p.Arg31619Ter | E.342 | rs869312121 | P | LP | DC | 68 | 2.36 | 202 |
| 109 | 179411339 | c.94816C > T | p.Arg31606Ter | E.341 | rs1060500435 | P | LP | DC | 69 | 1.72 | 233 |
| 110 | 179411593 | c.94562dup | p.Thr31522AsnfsTer12 | E.341 | rs869312083 | P | LP | NA | 61 | 2.50 | 202 |
| 111 | 179411905 | c.94344-94347del | p.Lys31448AsnfsTer8 | E.340 | rs727503546 | P | P | DC | 64 | 5.67 | 234 |
| 112 | 179411967 | c.94285T > A | p.Trp31429Arg | E.340 | NA | LP | NA | DC | 35 | 6.03 | 196 |
| 113 | 179412186 | c.94167del | p.Phe31389LeufsTer7 | E.339 | rs747837187 | LP | NA | DC | 64 | 5.26 | 202 |
| 114 | 179412199 | c.94154C > G | p.Ser31385Ter | E.339 | rs548010682 | LP | NA | DC | 72 | 6.03 | 207 |
| 115 | 179412246 | c.94103-94107del | p.Ile31368SerfsTer34 | E.339 | rs769488730 | P | P | DC | 64 | 5.33 | 199 |
| 116 | 179412456 | c.93897del | p.Phe31299LeufsTer14 | E.339 | rs397517758 | P | P | DC | 64 | 3.15 | 80 |
| 117 | 179412902 | c.93451G > T | p.Glu31151Ter | E.339 | NA | P | NA | DC | 67 | 5.65 | 199 |
| 118 | 179413151 | c.93202G > T | p.Glu31068Ter | E.339 | NA | P | NA | DC | 68 | 5.65 | 205 |
| 119 | 179413187 | c.93166C > T | p.Arg31056Ter | E.339 | rs72648250 | P | LP/P | DC | 69 | 5.65 | 202 |
| 120 | 179413477 | c.92876G > A | p.Trp30959Ter | E.339 | rs72648249 | P | NA | DC | 67 | 5.22 | 199 |
| 121 | 179413670 | c.92683C > T | p.Asp30885SerfsTer30895Ter | E.339 | rs869312065 | P | LP | DC | 16.84 | 5.3 | 202 |
| 122 | 179413694 | c.92652-92659del | p.Asp30885SerfsTer3 | E.339 | rs1559175090 | P | LP | DC | 63 | 2.1 | 224 |
| 123 | 178549148 | c.92478dup | p.Val30827SerfsTer22 | E.339 | NA | P | LP | NA | 7.36 | 3.45 | 235 |
| 124 | 179414036 | c.92317C > T | p.Arg30773Ter | E.339 | rs794729301 | P | LP/P | DC | 68 | 3.79 | 225 |
| 125 | 179414065 | c.92284-92288dup | p.Ser30763ArgfsTer7 | E.339 | rs756367933 | P | VUS | NA | 64 | 4.17 | 202 |
| 126 | 179414119 | c.92234C > A | p.Ser30745Ter | E.339 | NA | P | NA | DC | 67 | 5.74 | 205 |
| 127 | 179414186 | c.92167C > T | p.Pro30723Ser | E.339 | rs758537709 | P | VUS | DC | 32 | 5.73 | 213 |
| 128 | 179414303 | c.92146C > T | p.Gln30716Ter | E.338 | NA | P | NA | DC | 70 | 5.73 | 205 |
| 129 | 179414366 | c.92083T > C | p.Ser30695Pro | E.338 | rs768267695 | LP | NA | DC | 31 | 5.74 | 236 |
| 130 | 179414574 | c.91875del | p.Pro30626GlnfsTer2 | E.338 | rs757451467 | P | P | DC | 63 | 4.82 | 205 |
| 131 | 179414812 | c.91753T > G | p.Phe30585Val | E.337 | rs1060500507 | P | VUS | DC | 34 | 5.74 | 237 |
| 132 | 179414850 | c.91715dup | p.Asn30572LysfsTer16 | E.337 | rs779129892 | P | VUS | NA | 61 | 4.08 | 202 |
| 133 | 179415706 | c.91551-91552del | p.Asp30519Ter | E.336 | NA | P | NA | DC | 63 | 3.18 | 205 |
| 134 | 179416527 | c.91097-91100dup | p.Asn30367LysfsTer3 | E.335 | NA | P | NA | NA | 61 | 5.07 | 79 |
| 135 | 179416849 | c.90778dup | p.Tyr30260LeufsTer12 | E.335 | rs397517750 | P | LP | NA | 61 | 4.10 | 199 |
| 136 | 179416870 | c.90757G > A | p.Gly30253Arg | E.335 | – | P | NA | DC | 35 | 5.9 | 205 |
| 137 | 179417040 | c.90587del | p.Lys30196ArgfsTer94 | E.335 | rs397517749 | P | LP | DC | 63 | 6.06 | 238 |
| 138 | 179417257 | c.90370G > T | p.Glu30124Ter | E.335 | rs1553539995 | P | LP | DC | 67 | 5.76 | 239 |
| 139 | 179417305 | c.90322-90323insT | p.Glu30108ValfsTer6 | E.335 | rs869312082 | P | LP | DC | 61 | 5.76 | 202 |
| 140 | 178552691 | c.90208-90209insSVAelement | – | E.335 | NA | NA | LP | NA | NA | NA | 202 |
| 141 | 179417539 | c.90087-90088del | p.Glu30029AspfsTer7 | E.335 | rs869312064 | P | LP | DC | 63 | 3.32 | 202 |
| 142 | 179417542 | c.90085del | p.Glu30029LysfsTer11 | E.335 | NA | P | NA | DC | 63 | 5.76 | 238 |
| 143 | 179417543 | c.90084del | p.Glu30029LysfsTer11 | E.335 | NA | LP | NA | DC | 63 | -9.19 | 199 |
| 144 | 179417724 | c.89900-89903del | p.Asn29967MetfsTer27 | E.335 | rs869312081 | P | LP | DC | 63 | 4.36 | 202 |
| 145 | 179417877 | c.89750dup | p.Val29918SerfsTer3 | E.335 | rs869312063 | P | LP | NA | 63 | 3.12 | 202 |
| 146 | 179418418 | c.89314G > T | p.Glu29772Ter | E.334 | NA | P | P | DC | 64 | 4.71 | 240 |
| 147 | 179418468 | c.89265G > A | p.Trp29755Ter | E.334 | rs1179247052 | P | LP | DC | 66 | 5.6 | 225 |
| 148 | 179418639 | c.89197 + 2T > G | – | I.333 | rs1575536935 | P | LP | DC | NA | 5.61 | 241 |
| 149 | 179418639 | c.89197–89197 + 2del | – | I.333 | rs397517741 | P | LP | NA | NA | 4.10 | 80 |
| 150 | 179418640 | c.89197 + 1G > C | – | I.333 | rs1131691873 | P | LP | DC | NA | 5.61 | 225 |
| 151 | 179418877 | c.88961G > A | p.Trp29654Ter | E.333 | NA | P | NA | DC | 66 | 5.61 | 205 |
| 152 | 179419329 | c.88745C > T | p.Ser29582Phe | E.332 | NA | LP | NA | DC | 35 | 5.66 | 237 |
| 153 | 179419370 | c.88703-88704del | p.His29568LeufsTer7 | E.332 | rs794729360 | P | P | DC | 63 | 5.29 | 242 |
| 154 | 179419765 | c.88421G > A | p.Trp29474Ter | E.331 | rs869025546 | P | LP | DC | 66 | 5.66 | 243 |
| 155 | 179422099 | c.87887-87890del | p.His29296ProfsTer104 | E.329 | rs869312120 | P | LP | DC | 63 | 5.77 | 202 |
| 156 | 179422273 | c.87716del | p.Gly29239AspfsTer32 | E.329 | rs869312028 | P | VUS | DC | 63 | 5.56 | 202 |
| 157 | 179422457 | c.87624C > A | p.Tyr29208Ter | E.328 | rs772121356 | P | LP | DC | 66 | 0.93 | 202 |
| 158 | 179422552 | c.87529A > T | p.Lys29177Ter | E.328 | NA | LP | NA | DC | 33 | 4.44 | 201 |
| 159 | 179422565 | c.87516del | p.Tyr29173ThrfsTer24 | E.328 | rs727503552 | P | LP | DC | 63 | -1.28 | 199 |
| 160 | 179422726 | c.87355del | p.Ala29119LeufsTer17 | E.328 | rs794729356 | P | P | DC | 63 | 5.63 | 244 |
| 161 | 179422902 | c.87179C > A | p.Ser29060Ter | E.328 | NA | P | NA | DC | 67 | 5.69 | 205 |
| 162 | 179423093 | c.87093del | p.Pro29032LeufsTer8 | E.327 | NA | P | NA | DC | 63 | 4.57 | 205 |
| 163 | 179423146 | c.87040C > T | p.Arg29014Ter | E.327 | rs776065839 | P | P | DC | 67 | 4.77 | 209 |
| 164 | 179423220 | c.86967G > A | p.Trp28989Ter | E.327 | rs869312062 | P | LP | DC | 66 | 5.76 | 202 |
| 165 | 179423314 | c.86872dup | p.Ser28958LysfsTer10 | E.327 | NA | P | NA | NA | 61 | 0.07 | 79 |
| 166 | 179424036 | c.86821 + 2T > A | – | I.326 | rs397517735 | P | P | DC | NA | 5.61 | 199 |
| 167 | 179424057 | c.86799-86802del | p.Gly28936Ter | E.326 | rs727504856 | P | P | DC | 63 | 1.24 | 228 |
| 168 | 179424114 | c.86742-86745del | p.Tyr28915ThrfsTer22 | E.326 | rs1415420768 | P | LP | DC | 63 | 0.88 | 225 |
| 169 | 179424219 | c.86640C > A | p.Tyr28880Ter | E.326 | NA | P | LP | DC | 65 | 3.92 | 202 |
| 170 | 179424219 | c.86640delC | p.His28881ThrfsX2 | E.326 | rs794729298 | P | LP | DC | 63 | 3.92 | 202 |
| 171 | 179424399 | c.86459-86460del | p.Ser28820TrpfsTer50 | E.326 | rs869312080 | P | LP | DC | 63 | 2.79 | 202 |
| 172 | 179424496 | c.86363G > A | p.Trp28788Ter | E.326 | rs1064793814 | P | P | DC | 66 | 5.87 | 199 |
| 173 | 179424743 | c.86116C > T | p.Arg28706Ter | E.326 | rs794729384 | P | P | DC | 64 | 2.05 | 232 |
| 174 | 179424783 | c.86076dup | p.Ser28693IlefsTer2 | E.326 | rs1285329277 | P | P | NA | 61 | 0.61 | 245 |
| 175 | 179424844 | c.86015G > A | p.Trp28672Ter | E.326 | NA | P | NA | DC | 66 | 5.87 | 223 |
| 176 | 179424968 | c.85891del | p.Ala28631LeufsTer3 | E.326 | rs1575610911 | P | LP | DC | 63 | 6.08 | 246 |
| 177 | 179425091 | c.85768C > T | p.Arg28590Ter | E.326 | rs748689777 | P | P | DC | 65 | 2.95 | 227 |
| 178 | 179425207 | c.85640-85652del | p.Pro28547GlnfsTer12 | E.326 | rs762286447 | P | LP | DC | 63 | 4.12 | 205 |
| 179 | 179425598 | c.85261-85262insAlu | – | E.326 | NA | P | LP | NA | NA | NA | 247 |
| 180 | 179425708 | c.85151G > A | p.Arg28384Gln | E.326 | rs1465916943 | LP | NA | DC | 34 | 5.09 | 223 |
| 181 | 179425709 | c.85150C > T | p.Arg28384Ter | E.326 | NA | P | LP | DC | 65 | 3.09 | 205 |
| 182 | 179425748 | c.85109-85111del | p.Lys28370-Ala28371delinsThr | E.326 | NA | P | NA | DC | 50 | 4.99 | 223 |
| 183 | 179425769 | c.85090C > T | p.Arg28364Ter | E.326 | rs770038577 | P | LP/P | DC | 66 | 5.09 | 202 |
| 184 | 179425848 | c.85008-85011del | p.Glu28338HisfsTer9 | E.326 | rs869312100 | P | VUS | DC | 62 | 0.90 | 202 |
| 185 | 179426041 | c.84819G > A | p.Trp28273Ter | E.326 | rs72648222 | P | P | DC | 66 | 5.78 | 199 |
| 186 | 179426302 | c.84557dup | p.Ile28187AsnfsTer6 | E.326 | rs1553564589 | P | LP | NA | 61 | 2.28 | 80 |
| 187 | 179426383 | c.84476del | p.Gly28159ValfsTer15 | E.326 | rs1553564694 | P | LP | DC | 62 | 5.56 | 80 |
| 188 | 179426471 | c.84388del | p.Cys28130ValfsTer44 | E.326 | NA | P | NA | DC | 62 | − 0.12 | 205 |
| 189 | 179426483 | c.84376C > T | p.Gln28126Ter | E.326 | rs869312119 | P | LP | DC | 66 | 5.22 | 202 |
| 190 | 179426940 | c.83919del | p.Asn27973LysfsTer2 | E.326 | NA | LP | NA | DC | 62 | − 1.64 | 223 |
| 191 | 179427344 | c.83515C > T | p.Arg27839Ter | E.326 | rs869312118 | P | P | DC | 67 | 5.76 | 202 |
| 192 | 179427362 | c.83497G > T | p.Gly27833Ter | E.326 | NA | P | P | DC | 66 | 4.87 | 199 |
| 193 | 179428087 | c.82772G > A | p.Trp27591Ter | E.326 | NA | P | NA | DC | 66 | 5.85 | 205 |
| 194 | 179428202 | c.82657G > T | p.Gly27553Ter | E.326 | rs869178171 | P | P | DC | 65 | 4.96 | 248 |
| 195 | 179428256 | c.82603A > G | p.Thr27535Ala | E.326 | rs775733174 | P | NA | DC | 24 | 4.8 | 236 |
| 196 | 179428346 | c.82513del | p.Ile27505PhefsTer20 | E.326 | rs869312060 | P | LP | DC | 62 | 0.86 | 202 |
| 197 | 179428522 | c.82337C > T | p.Ala27446Val | E.326 | rs780558473 | LP | NA | DC | 34 | 5.97 | 249 |
| 198 | 179428586 | c.82273C > T | p.Gln27425Ter | E.326 | rs371332011 | P | LP | DC | 65 | 5.97 | 202 |
| 199 | 179428871 | c.81988C > T | p.Gln27330Ter | E.326 | rs72648222 | P | NA | DC | 65 | 6.07 | 199 |
| 200 | 179428916 | c.81943G > T | p.Glu27315Ter | E.326 | rs373533040 | P | LP | DC | 66 | 6.07 | 202 |
| 201 | 179428920 | c.81942del | p.Glu27315AsnfsTer35 | E.326 | NA | LP | NA | DC | 62 | 0.49 | 223 |
| 202 | 179428980 | c.81878-81879del | p.Phe27293CysfsTer3 | E.326 | rs727504660 | P | P | DC | 62 | 3.73 | 199 |
| 203 | 179429341 | c.81518del | p.Pro27173HisfsTer17 | E.326 | rs869312079 | P | LP | DC | 62 | 4.63 | 202 |
| 204 | 179429515 | c.81340-81344del | p.Lys27114GlnfsTer9 | E.326 | rs886038928 | P | LP | DC | 62 | 4.1 | 250 |
| 205 | 179429538 | c.81321C > G | p.Tyr27107Ter | E.326 | rs557312035 | P | P | DC | 64 | 4.22 | 202 |
| 206 | 179429590 | c.81262-81269del | p.Gln27088CysfsTer5 | E.326 | rs869312059 | P | LP | DC | 62 | 4.82 | 202 |
| 207 | 179429862 | c.80997-81012del | p.Tyr26999Ter | E.326 | rs727503559 | P | LP | DC | 64 | 2.08 | 251 |
| 208 | 179430143 | c.80716C > T | p.Arg26906Ter | E.326 | rs727505284 | P | P | DC | 64 | 3.71 | 252 |
| 209 | 179430224 | c.80635C > T | p.Gln26879Ter | E.326 | rs79926414 | LP | VUS | DC | 65 | 5.49 | 202 |
| 210 | 179430320 | c.80539C > T | p.Gln26847Ter | E.326 | rs561152891 | P | NA | DC | 65 | 4.59 | 243 |
| 211 | 179430345 | c.80514del | p.Val26839LeufsTer5 | E.326 | NA | P | P | DC | 62 | 1.22 | 199 |
| 212 | 179430692 | c.80167C > T | p.Arg26723Cys | E.326 | rs1412497882 | LP | VUS | DC | 35 | 4.92 | 223 |
| 213 | 179430807 | c.80052del | p.Gly26685AspfsTer11 | E.326 | NA | LP | NA | DC | 62 | 3.32 | 223 |
| 214 | 179431048 | c.79809-79811del | p.Val26604del | E.326 | rs776591304 | VUS | NA | PO | 48 | 0.24 | 223 |
| 215 | 179431175 | c.79684C > T | p.Arg26562Ter | E.326 | rs869025545 | P | LP | DC | 65 | 4.03 | 253 |
| 216 | 179431293 | c.79566T > A | p.Tyr26522Ter | E.326 | NA | LP | NA | DC | 62 | − 2.28 | 205 |
| 217 | 179431416 | c.79443del | p.Cys26482ValfsTer16 | E.326 | NA | P | NA | DC | 62 | 2.22 | 243 |
| 218 | 179431868 | c.78991C > T | p.Arg26331Ter | E.326 | rs779996703 | P | P | DC | 65 | 1.45 | 254 |
| 219 | 179431880 | c.78979C > T | p.Arg26327Ter | E.326 | rs1419374180 | P | LP | DC | 65 | 0.75 | 232 |
| 220 | 179432352 | c.78507del | p.Gly26170ValfsTer3 | E.326 | rs869312058 | P | LP | DC | 62 | 3.06 | 202 |
| 221 | 179432357 | c.78502G > A | p.Ala26168Thr | E.326 | NA | LP | NA | DC | 28.7 | 5.75 | 199 |
| 222 | 179432675 | c.78184G > T | p.Glu26062Ter | E.326 | rs869312057 | P | LP | DC | 64 | 5.58 | 202 |
| 223 | 179432681 | c.78178G > T | p.Glu26060Ter | E.326 | rs794729289 | P | P | DC | 64 | 5.58 | 225 |
| 224 | 179432761 | c.78095-78098del | p.Arg26032ThrfsTer41 | E.326 | rs869312117 | P | LP | DC | 62 | 4.37 | 202 |
| 225 | 179433095 | c.77764C > T | p.Gln25922Ter | E.326 | rs794729288 | P | VUS | DC | 65 | 5 | 210 |
| 226 | 179433197 | c.77646-77662delinsAGA | p.Ile25883AspfsTer3 | E.326 | rs794729345 | P | LP | DC | 11.72 | 3.33 | 199 |
| 227 | 179433210 | c.77647-77649del | p.Ile25883del | E.326 | NA | LP | P | DC | 48 | 1.91 | 199 |
| 228 | 179433274 | c.77585del | p.Lys25862ArgfsTer25 | E.326 | NA | P | NA | DC | 62 | 6.03 | 205 |
| 229 | 179433407 | c.77452G > T | p.Glu25818Ter | E.326 | NA | P | P | DC | 63 | 6.03 | 205 |
| 230 | 179433438 | c.77421dup | p.Ser25808GlnfsTer19 | E.326 | rs730880343 | P | LP | NA | 61 | 3.64 | 80 |
| 231 | 179433632 | c.77227G > T | p.Glu25743Ter | E.326 | rs765997807 | P | LP | DC | 64 | 5.74 | 223 |
| 232 | 179433630 | c.77226-77229del | p.Ser25742ArgfsTer9 | E.326 | NA | P | NA | DC | 61 | 3.86 | 196 |
| 233 | 179433665 | c.77194C > T | p.Gln25732Ter | E.326 | NA | P | NA | DC | 64 | 5.74 | 243 |
| 234 | 179433714 | c.77145dup | p.Ser25716LeufsTer8 | E.326 | rs1205409465 | P | LP | NA | 60 | 3.91 | 225 |
| 235 | 179433758 | c.77101-77102insT | p.Pro25701LeufsTer9 | E.326 | NA | P | NA | DC | 60 | 5.83 | 199 |
| 236 | 179433759 | c.77100dup | p.Pro25701ThrfsTer9 | E.326 | rs794729343 | P | P | NA | 60 | 3.71 | 255 |
| 237 | 179433781 | c.77077-77078delATinsGA | p.Ile25693Asp | E.326 | NA | LP | NA | DC | 60 | 2.62 | 256 |
| 238 | 179434010 | c.76849-76850insGT | p.Ser25617CysfsTer18 | E.326 | NA | P | NA | DC | 60 | 3.76 | 243 |
| 239 | 179434060 | c.76790-76799del | p.Arg25597ThrfsTer9 | E.326 | NA | P | NA | DC | 61 | 4.08 | 79 |
| 240 | 179434161 | c.76697-76698del | p.Leu25566ArgfsTer3 | E.326 | NA | P | NA | DC | 61 | 2.12 | 199 |
| 241 | 179434463 | c.76393-76396del | p.Asn25465Ter | E.326 | rs727504483 | P | LP | DC | 59 | 2.75 | 210 |
| 242 | 179434473 | c.76383-76386del | p.Asn25462LysfsTer4 | E.326 | rs869312078 | P | LP | DC | 61 | 3.78 | 202 |
| 243 | 179434486 | c.76373del | p.Pro25458GlnfsTer9 | E.326 | rs869025553 | P | P | DC | 60 | 5.02 | 243 |
| 244 | 179434743 | c.76116-76117insA | p.His25373ThrfsTer4 | E.326 | rs869312077 | P | LP | DC | 61 | 3.03 | 202 |
| 245 | 179435035 | c.75824A > G | p.Tyr25275Cys | E.326 | NA | LP | NA | DC | 34 | 5.87 | 249 |
| 246 | 179435223 | c.75633-75636dup | p.Val25213CysfsTer25 | E.326 | rs1553603036 | P | LP | NA | 60 | 4.42 | 224 |
| 247 | 179435390 | c.75469C > T | p.Arg25157Ter | E.326 | rs1553603394 | P | P | DC | 64 | 0.01 | 220 |
| 248 | 179435609 | c.75250C > T | p.Arg25084Ter | E.326 | rs794729286 | P | P | DC | 64 | 4.74 | 257 |
| 249 | 179435628 | c.75231T > A | p.Tyr25077Ter | E.326 | NA | P | LP | DC | 63 | − 4.72 | 227 |
| 250 | 179435718 | c.75138-75141del | p.Lys25046AsnfsTer8 | E.326 | rs794729340 | P | P | DC | 60 | 4.16 | 258 |
| 251 | 179435736 | c.75123T > A | p.Tyr25041Ter | E.326 | rs753526510 | P | VUS | DC | 62 | − 0.24 | 202 |
| 252 | 179435976 | c.74880-74883dup | p.Pro24962AsnfsTer9 | E.326 | rs869312116 | P | LP | NA | 60 | 3.48 | 202 |
| 253 | 179436177 | c.74682C > A | p.Tyr24894Ter | E.326 | NA | P | NA | DC | 63 | 1.11 | 223 |
| 254 | 179436456 | c.74403del | p.Asn24802MetfsTer20 | E.326 | NA | P | NA | DC | 60 | 3.495 | 227 |
| 255 | 179436521 | c.74338C > T | p.Arg24780Ter | E.326 | rs794729285 | P | P | DC | 64 | 5.09 | 202 |
| 256 | 179436553 | c.74306dup | p.Asn24769LysfsTer2 | E.326 | rs869312056 | P | LP | NA | 59 | 4.02 | 202 |
| 257 | 179437013 | c.73846C > T | p.Arg24616Ter | E.326 | rs794729284 | P | P | DC | 64 | 3.98 | 259 |
| 258 | 179437291 | c.73568del | p.Pro24523HisfsTer4 | E.326 | rs1559415567 | P | P | DC | 59 | 3.58 | 260 |
| 259 | 179437750 | c.73109G > A | p.Trp24370Ter | E.326 | rs869312115 | P | LP | DC | 63 | 5.19 | 202 |
| 260 | 179438060 | c.72799C > T | p.Gln24267Ter | E.326 | NA | P | P | DC | 63 | 4.17 | 205 |
| 261 | 179438190 | c.72669del | p.Asp24224IlefsTer8 | E.326 | rs727504531 | P | P | DC | 59 | − 3.04 | 260 |
| 262 | 179438873 | c.71980-71986delGCATATGinsTA | p.Ala23994Ter | E.326 | rs794729338 | P | P | DC | 58 | 4.05 | 199 |
| 263 | 179439257 | c.71602C > T | p.Arg23868Ter | E.326 | rs397517689 | P | P | DC | 64 | 2.44 | 227 |
| 264 | 179439359 | c.71500C > T | p.Gln23834Ter | E.326 | rs730880242 | P | LP | DC | 63 | 5.7 | 243 |
| 265 | 179439438 | c.71421T > A | p.Tyr23807Ter | E.326 | NA | LP | NA | DC | 61 | − 4.4 | 205 |
| 266 | 179439506 | c.71353A > G | p.Thr23785Ala | E.326 | rs765937279 | P | NA | DC | 26.9 | 5.6 | 223 |
| 267 | 179439852 | c.71007dup | p.Gly23670ArgfsTer6 | E.326 | NA | P | NA | NA | 59 | 3.79 | 243 |
| 268 | 179439881 | c.70978C > T | p.Arg23660Ter | E.326 | rs1553612386 | P | P | DC | 63 | 5.51 | 243 |
| 269 | 179439924 | c.70935del | p.Ala23647LeufsTer19 | E.326 | NA | P | NA | DC | 59 | 5.06 | 205 |
| 270 | 179439980 | c.70879C > T | p.Gln23627Ter | E.326 | rs1575799625 | P | LP | DC | 64 | 4.71 | 203 |
| 271 | 179440068 | c.70791del | p.Gly23598GlufsTer8 | E.326 | rs869312076 | P | LP | DC | 58 | 5.02 | 202 |
| 272 | 179440084 | c.70775del | p.Val23592GlyfsTer4 | E.326 | rs1216966174 | LP | NA | DC | 59 | 3.16 | 202 |
| 273 | 179440168 | c.70690-70691dup | p.Thr23565SerfsTer5 | E.326 | NA | P | NA | NA | 59 | 4.66 | 199 |
| 274 | 179440565 | c.70294G > C | p.Val23432Leu | E.326 | NA | VUS | NA | DC | 32 | 5.76 | 237 |
| 275 | 179440697 | c.70162C > T | p.Arg23388Ter | E.326 | rs781540455 | P | P | DC | 63 | 2.78 | 261 |
| 276 | 179440982 | c.69877G > T | p.Gly23293Ter | E.326 | rs869312114 | P | LP | DC | 62 | 5.87 | 202 |
| 277 | 179440999 | c.69860G > A | p.Trp23287Ter | E.326 | NA | P | LP | DC | 63 | 5.87 | 248 |
| 278 | 179441016 | c.69843del | p.Val23282Ter | E.326 | rs869312075 | P | LP | DC | 53 | 3.28 | 202 |
| 279 | 179441101 | c.69758C > T | p.Thr23253Ile | E.326 | NA | LP | NA | DC | 31 | 5.74 | 236 |
| 280 | 179441300 | c.69671del | p.Pro23224HisfsTer10 | E.325 | NA | P | NA | DC | 54 | 4.37 | 205 |
| 281 | 179441341 | c.69630C > A | p.Tyr23210Ter | E.325 | rs777602537 | P | LP | DC | 62 | − 5.08 | 205 |
| 282 | 179441449 | c.69522T > G | p.Tyr23174Ter | E.325 | NA | P | P | DC | 63 | 0.22 | 199 |
| 283 | 179441479 | c.69491-69492del | p.Val23164GlyfsTer2 | E.325 | rs869312113 | P | LP | DC | 42 | − 5.32 | 202 |
| 284 | 179441510 | c.69458-69461dup | p.Asn23154LysfsTer14 | E.325 | rs397517679 | P | LP | NA | 57 | 2.62 | 80 |
| 285 | 179441550 | c.69421-69422insAAAAG | p.Gly23141GlufsTer38 | E.325 | rs1247353236 | P | LP | PO | 59 | 4.64 | 225 |
| 286 | 179441649 | c.69412 + 1G > A | – | I.324 | rs869312074 | P | LP | DC | NA | 5.72 | 202 |
| 287 | 179442329 | c.68824 + 5G > C | – | I.323 | rs749639627 | VUS | VUS | DC | NA | 5.79 | 199 |
| 288 | 179442329 | c.68824G > A | p.Glu22942Lys | E.323 | rs199506676 | VUS | VUS | DC | 24.8 | 4.08 | 202 |
| 289 | 179443336 | c.68329 + 2–68329 + 3insTT | – | I.321 | rs536078303 | LP | VUS | NA | NA | 5.39 | 246 |
| 290 | 179443339 | c.68328A > G | p.Thr22776 = | E.321 | rs1553619783 | VUS | VUS | DC | 43 | 5.78 | 199 |
| 291 | 179443889 | c.67868T > C | p.Ile22623Thr | E.320 | NA | LP | NA | DC | 31 | 5.98 | 262 |
| 292 | 179444012 | c.67745del | p.Val22582AlafsTer10 | E.320 | NA | P | NA | DC | 57 | 5.68 | 199 |
| 293 | 179444052 | c.67705-67706insLINE1 | – | E.320-I.319 | NA | P | LP | NA | NA | NA | 219 |
| 294 | 179444405 | c.67519C > T | p.Gln22507Ter | E.319 | rs1559490694 | P | LP | DC | 62 | 5.78 | 196 |
| 295 | 179444429 | c.67495C > T | p.Arg22499Ter | E.319 | rs574660186 | P | P | DC | 63 | 4.63 | 202 |
| 296 | 179444577 | c.67349-2A > C | – | I.318 | rs753948675 | P | P | DC | NA | 5.10 | 263 |
| 297 | 179444661 | c.67348 + 5G > A | – | I.318 | rs765587170 | VUS | VUS | PO | NA | 3.7 | 199 |
| 298 | 179444666 | c.67348C > T | p.Gln22450Ter | E.318 | NA | P | P | DC | 62 | 2.24 | 264 |
| 299 | 179444735 | c.67279C > T | p.Arg22427Ter | E.318 | rs1200988060 | P | LP | DC | 63 | 0.99 | 265 |
| 300 | 179444855 | c.67159del | p.Ile22387Ter | E.318 | rs869312092 | LP | VUS | DC | 54 | 4.48 | 202 |
| 301 | 179444925 | c.67089del | p.Lys22364ArgfsTer24 | E.318 | NA | P | NA | DC | 56 | 1.07 | 213 |
| 302 | 179445166 | c.66940G > T | p.Asp22314Tyr | E.317 | rs768380109 | LP | VUS | DC | 24.6 | 5.25 | 236 |
| 303 | 179446219 | c.66769 + 3–66769 + 7delAAGTAinsT | – | I.316 | NA | LP | NA | NA | NA | 4.29 | 266 |
| 304 | 179446300 | c.66695T > A | p.Val22232Glu | E.316 | NA | LP | NA | DC | 31 | 5.41 | 204 |
| 305 | 179446471 | c.66523-66524del | p.Leu22175IlefsTer8 | E.316 | rs866120156 | P | NA | DC | 52 | 2.96 | 202 |
| 306 | 179447667 | c.65860-65863dup | p.Asp21955ValfsTer3 | E313 –I.313 | NA | P | NA | NA | 57 | 3.88 | 229 |
| 307 | 179447693 | c.65837C > G | p.Ser21946Ter | E.313 | rs775504996 | P | NA | DC | 63 | 5.02 | 267 |
| 308 | 179448411 | c.65498G > C | p.Arg21833Thr | E.312 | NA | VUS | NA | DC | 24.7 | 5.14 | 205 |
| 309 | 179448433 | c.65476G > T | p.Glu21826Ter | E.312 | rs763824247 | P | LP | DC | 63 | 6.02 | 202 |
| 310 | 179449208 | c.65070del | p.Ile21691LeufsTer5 | E.311 | NA | P | NA | DC | 57 | 4.15 | 199 |
| 311 | 179449453 | c.64915C > T | p.Arg21639Ter | E.310 | rs1432889079 | P | LP | DC | 63 | 4.3 | 242 |
| 312 | 179450018 | c.64453C > T | p.Arg21485Ter | E.309 | rs768345594 | P | LP | DC | 62 | 5.25 | 202 |
| 313 | 179451443 | c.64185del | p.Ala21396LeufsTer26 | E.308 | NA | LP | NA | DC | 56 | − 10 | 205 |
| 314 | 179452145 | c.63794-1G > A | – | I.306 | rs2049262622 | P | LP | DC | NA | 5.98 | 268 |
| 315 | 179452435 | c.63601C > T | p.Arg21201Ter | E.306 | rs764243269 | P | P | DC | 63 | 4.92 | 202 |
| 316 | 179453427 | c.63025C > T | p.Arg21009Ter | E.304 | rs368452607 | P | LP | DC | 62 | 5.27 | 202 |
| 317 | 179453720 | c.62733G > A | p.Trp20911Ter | E.304 | NA | P | NA | DC | 63 | 6.07 | 243 |
| 318 | 179453730 | c.62722C > T | p.Arg20908Ter | E.304 | rs543860009 | P | P | DC | 62 | − 3.88 | 224 |
| 319 | 179453946 | c.62506C > T | p.Arg20836Ter | E.304 | rs757231565 | P | VUS | DC | 63 | 4.14 | 202 |
| 320 | 179454235 | c.62217T > A | p.Tyr20739Ter | E.304 | rs727503586 | P | P | DC | 62 | 2.63 | 199 |
| 321 | 179454531 | c.61921C > T | p.Arg20641Ter | E.304 | rs878854324 | P | P | DC | 63 | 5.2 | 268 |
| 322 | 179454576 | c.61876C > T | p.Arg20626Ter | E.304 | rs72646846 | P | P | DC | 62 | 5.17 | 242 |
| 323 | 179454770 | c.61682C > G | p.Ser20561Ter | E.304 | rs1114167324 | P | LP | DC | 62 | 4.21 | 244 |
| 324 | 179454784 | c.61668del | p.His20557MetfsTer20 | E.304 | NA | LP | NA | DC | 54 | − 0.84 | 223 |
| 325 | 179454957 | c.61495C > T | p.Arg20499Ter | E.304 | rs869312112 | P | LP | DC | 62 | 3.97 | 224 |
| 326 | 179455112 | c.61339del | p.Ile20447Ter | E.304 | rs1576086839 | P | LP | DC | 52 | 6.11 | 243 |
| 327 | 179455162 | c.61290T > A | p.Cys20430Ter | E.304 | NA | P | NA | DC | 63 | 6.11 | 199 |
| 328 | 179455521 | c.60931C > T | p.Arg20311Ter | E.304 | rs869312055 | P | LP | DC | 62 | 5.23 | 202 |
| 329 | 179455598 | c.60854-60855insG | p.Asn20286LysfsTer13 | E.304 | NA | P | LP | DC | 55 | 5.535 | 205 |
| 330 | 179455719 | c.60733C > T | p.Arg20245Ter | E.304 | rs1057522256 | P | P | DC | 62 | 4.26 | 205 |
| 331 | 179455726 | c.60726T > A | p.Tyr20242Ter | E.304 | rs145423907 | LP | NA | DC | 61 | − 1.83 | 202 |
| 332 | 179455780 | c.60672del | p.Gly20225GlufsTer7 | E.304 | NA | P | NA | DC | 55 | 0.045 | 205 |
| 333 | 179456553 | c.59993G > A | p.Trp19998Ter | E.303 | NA | P | NA | DC | 62 | 6.16 | 79 |
| 334 | 179456704 | c.59926 + 1G > A | – | I.302 | rs553526525 | P | P | DC | NA | 6.16 | 269 |
| 335 | 179456766 | c.59865-59866insA | p.Gln19956ThrfsTer9 | E.302 | NA | P | NA | DC | 45 | 4.98 | 205 |
| 336 | 179456783 | c.59848C > T | p.Arg19950Ter | E.302 | rs1559598775 | P | LP | DC | 63 | 5.16 | 253 |
| 337 | 179457005 | c.59627-1G > A | – | I.301 | rs869312073 | P | LP | DC | NA | 6.03 | 202 |
| 338 | 179457273 | c.59460G > A | p.Trp19820Ter | E.301 | rs1250461669 | P | LP | DC | 62 | 6.03 | 225 |
| 339 | 179457321 | c.59411dup | p.Arg19805LysfsTer3 | E.301 | rs755261062 | P | LP | NA | 54 | 2.46 | 202 |
| 340 | 179457380 | c.59352del | p.Glu19785SerfsTer2 | E.301 | rs869312111 | P | LP | DC | 53 | 5.01 | 202 |
| 341 | 179457644 | c.59201-59202del | p.Pro19734ArgfsTer5 | E.300 | rs752948913 | P | LP | DC | 52 | 4.85 | 257 |
| 342 | 179457977 | c.58958G > C | p.Arg19653Pro | E.299 | NA | LP | NA | DC | 32 | 6.16 | 205 |
| 343 | 179458080 | c.58855del | p.Glu19619LysfsTer27 | E.299 | NA | LP | NA | DC | 52 | 6.16 | 199 |
| 344 | 179458083 | c.58852dup | p.Arg19618LysfsTer6 | E.299 | NA | LP | NA | NA | 54 | 1.43 | 205 |
| 345 | 179458293 | c.58732 + 2T > C | – | I.298 | rs869312054 | P | LP | DC | NA | 6.02 | 202 |
| 346 | 179458407 | c.58620del | p.Val19541PhefsTer22 | E.298 | rs1576147786 | P | LP | DC | 52 | 5.63 | 210 |
| 347 | 179458459 | c.58567-58568dup | p.Lys19524ValfsTer8 | E.298 | rs1553650442 | P | P | NA | 53 | 3.26 | 234 |
| 348 | 179458477 | c.58550T > C | p.Ile19517Thr | E.298 | rs72646838 | VUS | VUS | DC | 24.8 | 5.86 | 226 |
| 349 | 179458850 | c.58270G > T | p.Glu19424Ter | E.297 | rs72646837 | P | P | DC | 63 | 6.17 | 199 |
| 350 | 179458948 | c.58172del | p.Asp19391AlafsTer45 | E.297 | rs869312072 | P | LP | DC | 52 | 5.03 | 202 |
| 351 | 179459155 | c.58066dup | p.Glu19356GlyfsTer27 | E.296 | NA | LP | NA | NA | 54 | 4.11 | 199 |
| 352 | 179459226 | c.57995del | p.His19332ProfsTer18 | E.296 | rs397517633 | P | LP | DC | 52 | 6.17 | 80 |
| 353 | 179460233 | c.57847 + 1G > A | – | I.295 | rs397517631 | LP | VUS | DC | NA | 6.07 | 80 |
| 354 | 179460312 | c.57769C > T | p.Arg19257Ter | E.295 | rs794729275 | P | LP | DC | 62 | 5.08 | 270 |
| 355 | 179460320 | c.57761A > G | p.Tyr19254Cys | E.295 | NA | VUS | NA | DC | 33 | 5.98 | 10 |
| 356 | 179460363 | c.57718C > T | p.Arg19240Ter | E.295 | rs2051361827 | P | LP | DC | 62 | 3.94 | 79 |
| 357 | 179460478 | c.57603C > A | p.Cys19201Ter | E.295 | rs1418030810 | P | LP | DC | 62 | 5.17 | 225 |
| 358 | 179462264 | c.57544 + 1G > A | – | I.294 | rs2052045274 | P | LP | DC | NA | 6.06 | 202 |
| 359 | 179462478 | c.57331C > T | p.Arg19111Ter | E.294 | rs72646831 | P | P | DC | 62 | 4.23 | 228 |
| 360 | 179462682 | c.57215del | p.Gly19072GlufsTer12 | E.293 | rs397517628 | P | LP | DC | 54 | 5.87 | 80 |
| 361 | 179463603 | c.56834del | p.Gly18945ValfsTer6 | E.291 | rs869312110 | P | LP | DC | 53 | 4.97 | 202 |
| 362 | 179463948 | c.56572C > T | p.Arg18858Ter | E.290 | rs745376275 | P | LP | DC | 62 | 3.19 | 271 |
| 363 | 179464342 | c.56286T > A | p.Tyr18762Ter | E.289 | NA | P | NA | DC | 62 | − 1.01 | 205 |
| 364 | 179464422 | c.56206del | p.Thr18736ProfsTer8 | E.289 | rs869312109 | P | LP | DC | 49 | 4.5 | 202 |
| 365 | 179466193 | c.55525-55531del | p.Asp18509SerfsTer29 | E.287 | rs869312052 | P | LP | DC | 50 | 4.37 | 202 |
| 366 | 179466263 | c.55460-55461del | p.Lys18487SerfsTer3 | E.287 | rs1064796230 | P | P | DC | 49 | 4.96 | 272 |
| 367 | 179466466 | c.55351C > T | p.Arg18451Ter | E.286 | rs1440093502 | P | P | DC | 62 | 5.83 | 205 |
| 368 | 179466515 | c.55303-1G > A | – | I.285 | rs748369265 | P | VUS | DC | NA | 6.07 | 202 |
| 369 | 179466726 | c.55269 + 3A > G | – | I. 284 | rs72646820 | P | NA | NA | NA | 4.92 | 199 |
| 370 | 179468833 | c.54581G > T | p.Gly18194Val | E.282 | NA | LP | NA | DC | 26.8 | 6.16 | 205 |
| 371 | 179469477 | c.54339del | p.Glu18113AspfsTer10 | E.281 | rs796122911 | P | LP | DC | 51 | 4.33 | 205 |
| 372 | 179469738 | c.54166C > T | p.Arg18056Ter | E.280 | rs768431507 | P | LP | DC | 62 | 5.05 | 272 |
| 373 | 179469837 | c.54067C > T | p.Arg18023Ter | E.280 | rs1553682168 | P | P | DC | 62 | 4.83 | 273 |
| 374 | 179469882 | c.54022G > A | p.Glu18008Lys | E.280 | NA | P | NA | DC | 23.9 | 5.74 | 237 |
| 375 | 179469986 | c.53918del | p.Gly17973GlufsTer18 | E.280 | rs1486129583 | P | P | DC | 51 | 5.74 | 199 |
| 376 | 179470140 | c.53881 + 1G > T | – | I.279 | rs869312051 | P | LP | DC | NA | 5.63 | 202 |
| 377 | 179470359 | c.53656-53663del | p.Pro17886Ter | E.279 | NA | P | NA | DC | 49 | 3.10 | 205 |
| 378 | 179471841 | c.53488G > T | p.Gly17830Ter | E.278 | rs759231562 | P | LP | DC | 62 | 5.35 | 202 |
| 379 | 179471975 | c.53355G > A | p.Trp17785Ter | E.278 | rs794729273 | P | P | DC | 62 | 5.99 | 274 |
| 380 | 179472042 | c.53288-1G > C | – | I.277 | rs1553685927 | P | LP | DC | NA | 5.99 | 199 |
| 381 | 179472127 | c.53287 + 1G > T | – | I.277 | rs1064794266 | P | VUS | DC | NA | 5.99 | 199 |
| 382 | 179472156 | c.53259del | p.Lys17753AsnfsTer7 | E.277 | rs1389777522 | P | LP | DC | 48 | 5.19 | 205 |
| 383 | 179472209 | c.53206C > T | p.Arg17736Ter | E.277 | rs571702144 | P | LP | DC | 62 | 4.84 | 275 |
| 384 | 179472611 | c.52903C > T | p.Arg17635Ter | E.276 | NA | P | LP | DC | 62 | 5.16 | 276 |
| 385 | 179473206 | c.52406-2A > C | – | I.274 | rs753798236 | P | LP | DC | NA | 5.72 | 199 |
| 386 | 179473427 | c.52311-52312insTTGA | p.Gly17438LeufsTer12 | E.274 | NA | P | NA | DC | 46 | 4.90 | 205 |
| 387 | 179473511 | c.52223-52227dup | p.Asp17410ArgfsTer25 | E.274 | rs869312050 | P | LP | NA | 48 | 5.29 | 202 |
| 388 | 179473610 | c.52128del | p.Phe17376LeufsTer27 | E.274 | rs869312095 | LP | VUS | DC | 49 | 3.55 | 202 |
| 389 | 179474002 | c.52035-52036insTT | p.Leu17346PhefsTer4 | E.273 | rs869312049 | P | LP | DC | 51 | 2.55 | 202 |
| 390 | 179474121 | c.51913-51916del | p.Lys17305ValfsTer13 | E.273 | rs747513278 | P | LP | DC | 50 | 1.81 | 79 |
| 391 | 179474220 | c.51817G > T | p.Gly17273Ter | E.273 | NA | P | NA | DC | 61 | 5.85 | 205 |
| 392 | 179474816 | c.51436 + 1G > A | – | I.271 | rs761807131 | P | P | DC | NA | 5.48 | 244 |
| 393 | 179474817 | c.51436C > T | p.Gln17146Ter | E.271 | rs906494713 | P | P | DC | 62 | 5.48 | 224 |
| 394 | 179474936 | c.51317G > A | p.Trp17106Ter | E.271 | NA | P | NA | DC | 61 | 5.48 | 199 |
| 395 | 179476484 | c.50551 + 1G > A | – | I.268 | rs188050862 | LP | NA | DC | NA | 5.18 | 202 |
| 396 | 179476569 | c.50467C > T | p.Gln16823Ter | E.268 | NA | P | NA | DC | 62 | 5.08 | 205 |
| 397 | 179477005 | c.50247del | p.Phe16749LeufsTer15 | E.266 | rs869312071 | P | LP | DC | 56 | 2.6 | 202 |
| 398 | 179477082 | c.50170C > T | p.Arg16724Ter | E.266 | rs794729265 | P | P | DC | 62 | 2.83 | 202 |
| 399 | 179477226 | c.50026G > T | p.Glu16676Ter | E.266 | NA | P | NA | DC | 62 | 5.71 | 196 |
| 400 | 179477886 | c.49648 + 2del | – | I.264 | rs727504851 | P | P | NA | NA | 5.95 | 199 |
| 401 | 179478553 | c.49458G > A | p.Trp16486Ter | E.263 | rs869312108 | P | LP | DC | 61 | 6.07 | 202 |
| 402 | 179478665 | c.49346-1G > A | – | I.262 | rs869312070 | P | P | DC | NA | 6.07 | 202 |
| 403 | 179478861 | c.49263C > A | p.Tyr16421Ter | E.262 | NA | P | P | DC | 58 | 0.84 | 205 |
| 404 | 179478865 | c.49259del | p.Glu16420GlyfsTer23 | E.262 | NA | LP | NA | DC | 55 | 6.07 | 199 |
| 405 | 179478953 | c.49171C > T | p.Arg16391Ter | E.262 | rs570046043 | P | LP | DC | 59 | 3.38 | 277 |
| 406 | 179479481 | c.48761-1G > C | – | I.260 | rs876657665 | P | LP | DC | NA | 5.63 | 80 |
| 407 | 179480145 | c.48527G > A | p.Trp16176Ter | E.259 | rs869312048 | P | LP | DC | 61 | 5.96 | 202 |
| 408 | 179480423 | c.48405T > A | p.Cys16135Ter | E.258 | rs371722903 | LP | NA | DC | 61 | 4.62 | 202 |
| 409 | 179480446 | c.48382-48383insT | p.Lys16128IlefsTer6 | E.258 | rs771146720 | LP | NA | DC | 49 | 5.76 | 202 |
| 410 | 179481235 | c.48283C > T | p.Arg16095Ter | E.257 | rs374140736 | P | P | DC | 61 | 3.9 | 202 |
| 411 | 179481846 | c.47875 + 1G > A | – | I.255 | rs869312047 | P | LP | DC | NA | 5.76 | 202 |
| 412 | 179482115 | c.47697C > A | p.Cys15899Ter | E.254 | rs373040154 | P | LP | DC | 59 | 2.18 | 202 |
| 413 | 179482120 | c.47692C > T | p.Arg15898Ter | E.254 | rs775186117 | P | LP | DC | 61 | 0.77 | 202 |
| 414 | 179482230 | c.47582G > A | p.Ser15861Asn | E.254 | NA | VUS | NA | DC | 28.2 | 6.08 | 236 |
| 415 | 179482584 | c.47494C > T | p.Arg15832Ter | E.253 | rs751746401 | P | P | DC | 62 | 4.74 | 232 |
| 416 | 179482662 | c.47416del | p.Asp15806IlefsTer4 | E.253 | NA | P | NA | DC | 53 | 5.63 | 205 |
| 417 | 179483042 | c.47142-47143dup | p.Glu15715ValfsTer19 | E.252 | rs869312107 | P | LP | NA | 56 | 4.12 | 202 |
| 418 | 179483495 | c.46782C > A | p.Tyr15594Ter | E.251 | rs397517587 | P | LP | DC | 60 | 4.5999 | 202 |
| 419 | 179483504 | c.46773T > A | p.Tyr15591Ter | E.251 | rs397517586 | P | LP | DC | 57 | 3.1199 | 80 |
| 420 | 179485012 | c.46236C > A | p.Cys15412Ter | E.248 | rs368200299 | P | LP | DC | 60 | 2.73 | 202 |
| 421 | 179485178 | c.46069-46070del | p.Met15357ValfsTer4 | E.248 | rs397517584 | P | LP | DC | 51 | 5.0099 | 80 |
| 422 | 179485525 | c.45812T > G | p.Leu15271Ter | E.247 | rs869312046 | P | LP | DC | 60 | 5.83 | 202 |
| 423 | 179485581 | c.45756dup | p.Tyr15253IlefsTer15 | E.247 | rs869312045 | P | LP | NA | 49 | 5.44 | 202 |
| 424 | 179485589 | c.45732-45748del | p.Glu15245PhefsTer17 | E.247 | NA | P | NA | DC | 60 | 4.23 | 205 |
| 425 | 179485878 | c.45567C > A | p.Tyr15189Ter | E.246 | NA | LP | NA | DC | 48 | -9.56 | 278 |
| 426 | 179485878 | c.45566dup | p.Tyr15189Ter | E.246 | NA | P | NA | DC | 48 | 0.73 | 218 |
| 427 | 179486054 | c.45391delA | p.Ile15131TyrfsTer46 | E.246 | rs869312091 | LP | VUS | DC | 61 | 3.71 | 202 |
| 428 | 179486229 | c.45322C > T | p.Arg15108Ter | E.245 | rs1060500405 | P | VUS | DC | 61 | 6.17 | 243 |
| 429 | 179486244 | c.45307C > T | p.Arg15103Ter | E.245 | rs397517580 | P | VUS | DC | 61 | 3.01 | 80 |
| 430 | 179487411 | c.44899C > T | p.Arg14967Ter | E.243 | rs727505350 | P | VUS | DC | 60 | 2.65 | 205 |
| 431 | 179487495 | c.44816-1G > A | – | I.242 | rs749705939 | P | VUS | DC | NA | 5.54 | 210 |
| 432 | 179489209 | c.44798G > A | p.Cys14933Tyr | E.242 | NA | LP | NA | DC | 28 | 5.72 | 279 |
| 433 | 179490056 | c.44492G > C | p.Gly14831Ala | E.241 | NA | LP | NA | DC | 27.3 | 5.95 | 279 |
| 434 | 179494088 | c.44364del | p.Tyr14789ThrfsTer15 | E.240 | rs397517576 | P | VUS | DC | 54 | 0.52 | 205 |
| 435 | 179494967 | c.44281 + 1G > A | – | I.239 | rs771562210 | P | VUS | DC | NA | 6.04 | 202 |
| 436 | 179494968 | c.44281C > T | p.Pro14761Ser | E.239 | rs192766485 | VUS | VUS | DC | 25.2 | 6.04 | 202 |
| 437 | 179494977 | c.44272C > T | p.Arg14758Ter | E.239 | rs140743001 | P | VUS | DC | 61 | 3.14 | 202 |
| 438 | 179495983 | c.43792del | p.Val14598Ter | E.237 | rs869312044 | P | LP | DC | 49 | 3.81 | 202 |
| 439 | 179497082 | c.43539-43540insA | p.Ala14514SerfsTer10 | E.236 | NA | P | NA | DC | 45 | 5.51 | 199 |
| 440 | 179497414 | c.43319G > A | p.Trp14440Ter | E.235 | rs372663057 | LP | VUS | DC | 60 | 6.16 | 202 |
| 441 | 179498055 | c.42947-2A > G | – | I.232 | rs1553741357 | P | VUS | DC | NA | 6.17 | 199 |
| 442 | 179498176 | c.42909-42910del | p.Cys14303TrpfsTer12 | E.232 | rs1114167333 | P | LP | DC | 56 | 4.69 | 244 |
| 443 | 179498592 | c.42636del | p.Ala14213LeufsTer6 | E.231 | rs869312106 | P | LP | DC | 57 | 0.47 | 202 |
| 444 | 179500295 | c.41756A > G | p.Asp13919Gly | E.227 | NA | VUS | NA | DC | 24.4 | 6.05 | 237 |
| 445 | 179500825 | c.41473C > T | p.Arg13825Ter | E.226 | rs869312043 | P | VUS | DC | 57 | 0.36 | 202 |
| 446 | 179500851 | c.41447del | p.Gly13816AlafsTer18 | E.226 | rs869312042 | P | LP | DC | 47 | 5.8 | 202 |
| 447 | 179505267 | c.40723 + 1G > T | – | I.221 | rs371770198 | LP | VUS | DC | NA | 0.36 | 202 |
| 448 | 179506963 | c.40558 + 1G > A | – | I.219 | rs368219776 | LP | VUS | DC | NA | 5.55 | 199 |
| 449 | 179506964 | c.40558G > C | p.Val13520Leu | E.219 | rs587780488 | P | VUS | DC | 24.9 | 5.55 | 199 |
| 450 | 179514543 | c.39895 + 1G > T | – | I.211 | 179514543 | LP | VUS | DC | NA | 5.58 | 202 |
| 451 | 179516234 | c.39492dup | p.Glu13165Ter | E.207 | NA | P | NA | DC | 55 | 5.22 | 213 |
| 452 | 179516991 | c.39211G > T | p.Val13071Phe | E.203 | rs1334646153 | LP | VUS | DC | 22.2 | 3.64 | 199 |
| 453 | 179516996 | c.39204-39206dup | p.Thr13069dup | E.203 | NA | VUS | NA | NA | 60 | 0.72 | 210 |
| 454 | 179517379 | c.39043 + 1G > T | – | I.201 | rs373516134 | LP | NA | DC | NA | 5.64 | 202 |
| 455 | 179517464 | c.38960–3-38960-1del | – | I.200 | rs773282707 | LP | NA | DC | NA | 5.64 | 202 |
| 456 | 179523240 | c.37579-37582del | p.Lys12527HisfsTer419 | E.184 | NA | P | NA | DC | 43 | 2.07 | 266 |
| 457 | 179526509 | c.37262del | p.Lys12421SerfsTer526 | E.180 | rs867008501 | LP | NA | DC | 60 | 2.65 | 202 |
| 458 | 179532021 | c.35739dup | p.Pro11914SerfsTer7 | E.162 | rs968544783 | LP | VUS | NA | 46 | 0.67 | 202 |
| 459 | 179532190 | c.35692A > T | p.Arg11898Ter | E.161 | rs188568710 | LP | NA | DC | 52 | 4.84 | 202 |
| 460 | 179535816 | c.35308 + 1G > T | – | I.156 | rs1423135750 | P | VUS | DC | NA | 5.92 | 199 |
| 461 | 179537361 | c.34855 + 1G > A | – | I.153 | rs377319699 | LP | VUS | DC | NA | 5.25 | 202 |
| 462 | 179542346 | c.34291 + 2T > C | – | I.146 | rs186084940 | LP | NA | DC | NA | 6.17 | 202 |
| 463 | 179542507 | c.34132del | p.Leu11378TyrfsTer90 | E.146 | rs869025551 | LP | VUS | DC | 53 | -0.01 | 243 |
| 464 | 179544666 | c.33535del | p.Glu11179SerfsTer3 | E.140 | rs757135518 | LP | NA | DC | 47 | 3.86 | 202 |
| 465 | 179544980 | c.33418 + 1G > A | – | I.139 | rs746588865 | LP | VUS | DC | NA | 4.98 | 202 |
| 466 | 179547631 | c.32888-1del | – | I.134 | rs869312041 | P | VUS | DC | NA | 5.18 | 202 |
| 467 | 179549632 | c.32554 + 1G > C | – | I.130 | rs376018437 | LP | VUS | DC | NA | 5.81 | 202 |
| 468 | 179549717 | c.32471-1G > A | – | I.129 | rs371725574 | P | VUS | DC | NA | 5.81 | 202 |
| 469 | 179554062 | c.31966A > T | p.Lys10656Ter | E.124 | rs368775510 | LP | NA | DC | 40 | 2.7 | 202 |
| 470 | 179554624 | c.31763-1G > A | – | I.121 | rs202234172 | P | VUS | DC | NA | 5.29 | 227 |
| 471 | 179558336 | c.31594G > T | p.Val10532Phe | E.119 | rs763955552 | VUS | VUS | DC | 24 | 5.85 | 202 |
| 472 | 179558736 | c.31427-1G > A | – | I.117 | NA | P | NA | DC | NA | 6.16 | 199 |
| 473 | 179559325 | c.31426 + 1G > C | – | I.117 | rs6749719 | LP | VUS | DC | NA | 6.07 | 202 |
| 474 | 179559557 | c.31347del | p.Val10450TyrfsTer25 | E.116 | NA | P | NA | DC | 61 | 5.26 | 205 |
| 475 | 179560998 | c.30803-2A > G | – | I.113 | rs869312089 | LP | VUS | DC | NA | 5.5 | 202 |
| 476 | 179563643 | c.30683-2del | – | I.111 | rs1553868981 | LP | VUS | DC | NA | 5.57 | 202 |
| 477 | 179566913 | c.30484-30493del | p.Thr10162CysfsTer3 | E.108 | rs727504452 | P | LP | DC | 61 | 3.22 | 199 |
| 478 | 179567322 | c.30292G > T | p.Glu10098Ter | E.107 | NA | P | NA | DC | 58 | 5.72 | 227 |
| 479 | 179569962 | c.29543G > A | p.Arg9848Gln | E.103 | rs773444238 | VUS | NA | DC | 24.4 | 5.82 | 280 |
| 480 | 179571370 | c.29231G > A | p.Arg9744His | E.102 | rs760305440 | VUS | VUS | DC | 27.4 | 6.1 | 280 |
| 481 | 179571652 | c.29071A > T | p.Lys9691Ter | E.101 | rs376189903 | LP | NA | DC | 60 | 6.14 | 202 |
| 482 | 179571661 | c.29062del | p.Ala9688GlnfsTer7 | E.101 | rs869312040 | P | LP | DC | 51 | 5.05 | 202 |
| 483 | 179571683 | c.29042-2A > C | – | I.100 | rs6716782 | P | VUS | DC | NA | 6.16 | 202 |
| 484 | 179572327 | c.28967dup | p.Asp9656GlufsTer8 | E.100 | NA | LP | NA | NA | 42 | 3.43 | 202 |
| 485 | 179575947 | c.28016dup | p.Pro9340AlafsTer23 | E.97 | rs954237155 | LP | NA | NA | 54 | 2.52 | 202 |
| 486 | 179577042 | c.27607G > A | p.Glu9203Lys | E.95 | rs769097909 | VUS | VUS | DC | 25.7 | 5.88 | 202 |
| 488 | 179580418 | c.25723G > A | p.Gly8575Arg | E.89 | rs397517517 | VUS | VUS | DC | 24.2 | 5.33 | 205 |
| 489 | 179582078 | c.25383del | p.Lys8461AsnfsTer5 | E.88 | rs1452206214 | LP | NA | DC | 61 | -1.14 | 202 |
| 500 | 179582856 | c.24863-24877del | p.Asp8288-Ile8293delinsVal | E.86 | NA | LP | NA | DC | 60 | 3.55 | 210 |
| 490 | 179583072 | c.24749-24761del | p.Gly8250ValfsTer8 | E.85 | NA | LP | VUS | DC | 60 | 2.80 | 199 |
| 491 | 179583429 | c.24498dup | p.Val8167CysfsTer13 | E.84 | rs1282574211 | P | NA | NA | 41 | 3.99 | 243 |
| 492 | 179583702 | c.24227-2A > G | – | I.83 | rs373060681 | P | NA | DC | NA | 5.71 | 202 |
| 493 | 179584983 | c.23386C > T | p.Arg7796Ter | E.81 | rs748111134 | LP | VUS | DC | 51 | 5.91 | 202 |
| 495 | 179585718 | c.23014-2328del | p.Ser7672-Ser7676del | E.79 | NA | NA | DC | 61 | 2.29 | 210 | |
| 496 | 179586600 | c.22788-22790delCATinsG | p.Asp7596GlufsTer16 | E.78 | NA | LP | NA | DC | 60 | 5.08 | 266 |
| 497 | 179587599 | c.22027C > T | p.Gln7343Ter | E.76 | rs886043434 | P | VUS | DC | 50 | 5.8 | 196 |
| 498 | 179587773 | c.21961G > A | p.Glu7321Lys | E.75 | NA | P | NA | DC | 23.2 | 5.95 | 210 |
| 499 | 179588844 | c.21142C > T | p.Arg7048Ter | E73 | rs770579313 | LP | VUS | DC | 41 | 1.61 | 202 |
| 501 | 179590572 | c.20477C > A | p.Ser6826Ter | E.70 | NA | P | NA | DC | 42 | 4.85 | 214 |
| 502 | 179591958 | c.20134del | p.Asp6712IlefsTer5 | E.69 | NA | P | NA | DC | 51 | 6.17 | 199 |
| 503 | 179596801 | c.16895T > C | p.Ile5632Thr | E.57 | rs727504971 | VUS | VUS | DC | 23.1 | 6.17 | 262 |
| 504 | 179597846 | c.16057C > T | p.Arg5353Ter | E.55 | rs267599069 | P | VUS | DC | 47 | 5.29 | 281 |
| 505 | 179597615 | c.16288C > T | p.Arg5430Ter | E.55 | rs772235481 | P | VUS | DC | 44 | 5.29 | 282 |
| 506 | 179598224 | c.15796C > T | p.Arg5266Ter | E.54 | rs372277017 | P | VUS | DC | 45 | 1.13 | 202 |
| 507 | 179598245 | c.15776-1G > T | – | I.53 | rs869312094 | LP | VUS | DC | NA | 5.86 | 202 |
| 509 | 179598437 | c.15679del | p.Ile5227SerfsTer29 | E.53 | NA | LP | NA | DC | 59 | 0.07 | 202 |
| 510 | 179599054 | c.15496 + 1G > A | – | I.52 | rs397517481 | P | VUS | DC | NA | 5.86 | 210 |
| 511 | 179599091 | c.15460G > A | p.Gly5154Ser | E.52 | rs772907723 | VUS | NA | DC | 25.5 | 5.86 | 205 |
| 512 | 179602835 | c.14344-14345delAGinsGA | p.Ser4782Asp | E.49 | NA | VUS | NA | DC | 61 | 5.8 | 97 |
| 513 | 179602866 | c.14314T > C | p.Cys4772Arg | E.49 | NA | VUS | NA | DC | 26.6 | 5.8 | 205 |
| 514 | 179603088 | c.14093-1G > A | – | I.48 | rs869312099 | P | VUS | DC | NA | 5.37 | 202 |
| 515 | 179603867 | c.14092 + 1G > T | – | I.48 | NA | P | NA | DC | NA | 5.62 | 205 |
| 516 | 179603904 | c.14056del | p.Thr4686GlnfsTer9 | E.48 | rs869312104 | P | LP | DC | 49 | 0.97 | 202 |
| 517 | 179604264 | c.13696C > T | p.Gln4566Ter | E.48 | rs775072385 | LP | VUS | DC | 36 | 4.95 | 199 |
| 518 | 179604345 | c.13615-13616insT | p.Asn4539IlefsTer5 | E.48 | NA | LP | NA | DC | 61 | − 5.64 | 205 |
| 519 | 179604363 | c.13597del | p.Glu4533LysfsTer38 | E.48 | NA | LP | NA | DC | 55 | 3.84 | 205 |
| 520 | 179604368 | c.13592C > G | p.Ser4531Ter | E.48 | NA | P | P | DC | 36 | 4.91 | 199 |
| 521 | 179604528 | c.13432-13433insA | p.Cys4478Ter | E.48 | NA | LP | NA | DC | 56 | 4.66 | 230 |
| 522 | 179604852 | c.13108C > T | p.Gln4370Ter | E.48 | rs267607158 | P | P | DC | 35 | 5.02 | 97 |
| 523 | 179604950 | c.13010del | p.Lys4337SerfsTer14 | E.48 | NA | P | NA | DC | 49 | 3.46 | 199 |
| 524 | 179598224 | c.15796C > T | p.Arg5266Ter | E.48 | rs372277017 | P | VUS | DC | 45 | 1.13 | 202 |
| 525 | 179605203 | c.12757C > T | p.Gln4253Ter | E.48 | rs869312039 | P | LP | DC | 38 | 4.91 | 202 |
| 526 | 179605317 | c.12643C > T | p.Gln4215Ter | E.48 | rs368329612 | LP | VUS | DC | 35 | 2.51 | 202 |
| 527 | 179605373 | c.12587C > A | p.Ser4196Ter | E.48 | rs370912401 | P | VUS | DC | 35 | 3.39 | 202 |
| 528 | 179605482 | c.12478del | p.Thr4160ProfsTer8 | E.48 | NA | P | VUS | DC | 40 | 1.23 | 205 |
| 529 | 179605512 | c.12438-12448del | p.Ser4147ThrfsTer20 | E.48 | rs1553939749 | P | LP | DC | 43 | − 0.52 | 244 |
| 530 | 179605752 | c.12208G > T | p.Glu4070Ter | E.48 | rs397517830 | P | LP | DC | 36 | 4.73 | 283 |
| 531 | 179606008 | c.11952C > A | p.Tyr3984Ter | E.48 | NA | P | NA | DC | 38 | 0.85 | 205 |
| 532 | 179606286 | c.11674T > C | p.Cys3892Arg | E.48 | NA | VUS | NA | DC | 21.9 | 6.08 | 205 |
| 533 | 179606303 | c.11657del | p.Asp3886ValfsTer22 | E.48 | rs397517826 | P | VUS | DC | 60 | 6.08 | 80 |
| 534 | 179606362 | c.11598C > A | p.Tyr3866Ter | E.48 | NA | P | NA | DC | 37 | 6.08 | 205 |
| 535 | 179606445 | c.11497-11515del | p.Met3833CysfsTer3 | E.48 | NA | P | VUS | DC | 60 | 1.90 | 205 |
| 536 | 179612657 | c.11311 + 5184_11311 + 5194dup | – | I.47 | rs869312088 | VUS | VUS | NA | NA | 0.83 | 202 |
| 537 | 179611822 | c.11312-5174del | – | I.47 | rs869312097 | VUS | VUS | DC | NA | 3.71 | 202 |
| 538 | 179611814 | c.11312-5166C > T | – | I.47 | rs376396183 | VUS | NA | DC | NA | 1.64 | 202 |
| 539 | 179610598 | c.11312-3950del | – | I.47 | rs774991940 | VUS | VUS | DC | NA | 3.58 | 202 |
| 540 | 179612712 | c.11311 + 5139del | – | I.47 | rs750893661 | VUS | NA | DC | NA | 1.18 | 202 |
| 541 | 179616552 | c.11311 + 1299T > A | – | I.47 | rs1561044021 | P | NA | DC | NA | 3.52 | 205 |
| 542 | 179616684 | c.11311 + 1167del | – | I.47 | rs869312096 | VUS | VUS | DC | NA | 2.36 | 202 |
| 543 | 179613467 | c.11311 + 4384dup | – | I.47 | rs771985828 | VUS | VUS | DC | NA | 1.80 | 202 |
| 544 | 179613188 | c.11311 + 4663del | – | I.47 | rs781363456 | VUS | VUS | DC | NA | 3.13 | 202 |
| 545 | 179613422 | c.11311 + 4429G > T | – | I.47 | rs372994805| | VUS | VUS | DC | NA | 4.68 | 202 |
| 546 | 179613717 | c.11311 + 4134dup | – | I.47 | rs768458450 | VUS | VUS | NA | NA | 3.61 | 202 |
| 547 | 179610611 | c.11312-3963G > T | – | I.47 | rs148430495 | VUS | VUS | DC | NA | 5.94 | 202 |
| 548 | 179614105 | c.11311 + 3746C > G | – | I.47 | rs763408700 | LP | VUS | DC | NA | 5.25 | 202 |
| 549 | 179610383 | c.11312-3735G > T | – | I.47 | rs143376837 | VUS | VUS | DC | NA | 6.17 | 202 |
| 550 | 179614541 | c.11311 + 3310G > T | – | I.47 | rs372772094 | VUS | NA | DC | NA | 4.95 | 202 |
| 551 | 179615375 | c.11311 + 2476G > T | – | I.47 | rs373480236 | VUS | NA | DC | NA | 4.66 | 202 |
| 552 | 179616345 | c.11311 + 1506del | – | I.47 | rs777963995 | VUS | NA | DC | NA | 2.31 | 202 |
| 553 | 179620948 | c.11254 + 1G > C | – | I.46 | rs192945689 | LP | NA | DC | NA | 2.21 | 202 |
| 554 | 179620947 | c.11254 + 2T > C | – | I.46 | rs199565715 | LP | VUS | DC | NA | 0.77 | 202 |
| 555 | 179621013 | c.11190C > G | p.Tyr3730Ter | E.46 | rs373667402 | LP | VUS | DC | 38 | 1.99 | 202 |
| 556 | 179621020 | c.11183dup | p.Leu3729ThrfsTer9 | E.46 | rs778172350 | P | VUS | NA | 61 | 3.38 | 202 |
| 557 | 179621090 | c.11113del | p.Arg3705AspfsTer2 | E.46 | rs746386040 | LP | VUS | DC | 59 | 6.16 | 202 |
| 558 | 179621351 | c.10852C > T | p.Gln3618Ter | E46 | rs779064556 | LP | VUS | DC | 39 | 4.31 | 202 |
| 559 | 179621404 | c.10799C > A | p.Ser3600Ter | E.46 | rs374300381 | LP | VUS | DC | 40 | 6.17 | 202 |
| 560 | 179622355 | c.10592C > G | p.Ser3531Ter | E.45 | rs767420661 | LP | VUS | DC | 38 | 5.12 | 202 |
| 561 | 179622472 | c.10475-10476insAGAC | p.Lys3493AspfsTer10 | E.45 | NA | LP | NA | DC | 60 | 5.57 | 210 |
| 562 | 179623709 | c.10303 + 2T > C | – | I.44 | rs371596417 | P | VUS | DC | NA | 6.03 | 202 |
| 563 | 179629492 | c.9749-9750del | p.Val3250AlafsTer40 | E.42 | rs1445295628 | LP | NA | DC | 60 | 1.06 | 202 |
| 564 | 179629515 | c.9727C > T | p.Gln3243Ter | E.42 | rs869312093 | LP | VUS | DC | 38 | 5.69 | 202 |
| 565 | 179631234 | c.9577C > T | p.Arg3193Ter | E.41 | rs746115846 | P | VUS | DC | 36 | 0.59 | 211 |
| 566 | 179632509 | c.9448C > T | p.Arg3150Ter | E.40 | rs146572907 | P | VUS | DC | 43 | 5.11 | 202 |
| 567 | 179632576 | c.9381C > A | p.Tyr3127Ter | E.40 | NA | P | LP/P | DC | 36 | 2.19 | 205 |
| 568 | 179632841 | c.9205del | p.Val3069TyrfsTer23 | E.39 | NA | P | NA | DC | 61 | 2.956 | 243 |
| 569 | 179632884 | c.9164-2A > T | – | I.38 | rs777369921 | LP | VUS | DC | NA | 5.73 | 202 |
| 570 | 179633403 | c.9160G > C | p.Glu3054Gln | E.38 | – | VUS | NA | DC | 23.2 | 5.81 | 249 |
| 571 | 179633431 | c.9132del | p.Ala3045GlnfsTer14 | E.38 | rs36059692 | LP | NA | DC | 52 | 3.46 | 202 |
| 572 | 179634417 | c.8891-8892insC | p.Thr2965AspfsTer17 | E.37 | NA | P | NA | DC | 59 | 4.49 | 205 |
| 573 | 179634544 | c.8764G > T | p.Glu2922Ter | E.37 | NA | P | NA | DC | 38 | 5.93 | 205 |
| 574 | 179634621 | c.8687C > T | p.Thr2896Ile | E.37 | rs72647884 | VUS | VUS | DC | 27.1 | 6.06 | 226 |
| 575 | 179635166 | c.8353G > T | p.Gly2785Ter | E.35 | NA | P | NA | DC | 36 | 5.19 | 243 |
| 576 | 179635211 | c.8307-8308del | p.Ala2770HisfsTer4 | E.35 | rs869312037 | P | VUS | DC | 61 | 4.71 | 202 |
| 577 | 179636183 | c.7871dup | p.Pro2625AlafsTer9 | E.34 | rs1553997502 | LP | NA | NA | 43 | 3.86 | 202 |
| 578 | 179638333 | c.7450C > T | p.Gln2484Ter | E.32 | NA | P | VUS | DC | 37 | 5.82 | 199 |
| 579 | 179639171 | c.6820C > T | p.Gln2274Ter | E.30 | rs145649088 | P | VUS | DC | 36 | − 1.22 | 202 |
| 580 | 179640236 | c.6355G > T | p.Glu2119Ter | E.28 | rs869312098 | LP | VUS | DC | 36 | 5.33 | 202 |
| 581 | 179640343 | c.6248del | p.Arg2083LysfsTer56 | E.28 | rs72647879 | P | NA | DC | 61 | 3.54 | 236 |
| 582 | 179640343 | c.6248G > T | p.Arg2083Ile | E.28 | rs781676050 | VUS | NA | DC | 21.9 | 3.54 | 236 |
| 583 | 179640344 | c.6247del | p.Arg2083GlufsTer56 | E.28 | NA | P | NA | DC | 57 | 3.68 | 199 |
| 584 | 179640468 | c.6123G > A | p.Trp2041Ter | E.28 | 179640468 | LP | NA | DC | 36 | 5.19 | 202 |
| 585 | 179640970 | c.5622G > A | p.Trp1874Ter | E.28 | rs777078420 | P | NA | DC | 38 | 5.09 | 197 |
| 586 | 179641014 | c.5577G > C | p.Arg1859Ser | E.28 | NA | VUS | NA | B | 23.6 | 1.41 | 284 |
| 587 | 179641524 | c.5067G > A | p.Trp1689Ter | E.28 | rs375648277 | P | NA | DC | 37 | 5.33 | 202 |
| 588 | 179641962 | c.4724-4728del | p.Met1575SerfsTer6 | E.27 | rs756433029 | P | VUS | DC | 60 | 4.81 | 202 |
| 589 | 179641976 | c.4714C > T | p.Arg1572Ter | E.27 | rs1554008881 | P | VUS | DC | 37 | 3.89 | 203 |
| 590 | 179643691 | c.4118C > A | p.Ala1373Glu | E.24 | NA | VUS | NA | DC | 25 | 5.91 | 205 |
| 591 | 179644006 | c.3913G > A | p.Gly1305Arg | E.23 | NA | VUS | NA | DC | 25.9 | 5.72 | 205 |
| 592 | 179644174 | c.3742-3745del | p.Ser1248ProfsTer14 | E.23 | NA | P | LP | DC | 61 | 5.48 | 205 |
| 593 | 179647331 | c.3101-2A > T | – | I.18 | rs1060500467 | P | VUS | DC | NA | 5.54 | 199 |
| 594 | 179647533 | c.3100G > A | p.Val1034Met | E18 | rs142951505 | VUS | VUS | DC | 24 | 6.17 | 202 |
| 595 | 179647588 | c.3045C > G | p.Cys1015Trp | E.18 | NA | VUS | NA | DC | 25 | 4.09 | 223 |
| 596 | 179647599 | c.3034C > T | p.Arg1012Ter | E.18 | rs397517547 | P | VUS | DC | 36 | 3.03 | 80 |
| 597 | 179647707 | c.2926T > C | p.Trp976Arg | E.18 | rs267607155 | P | LP | DC | 24.5 | 6.17 | 285 |
| 598 | 179647707 | c.2926T > A | p.Trp976Arg | E.18 | rs267607155 | P | NA | DC | 24.5 | 6.17 | 10 |
| 599 | 179648447 | c.2841G > T | p.Ser947 = | E.17 | rs774074192 | LP | VUS | DC | 45 | 1.07 | 202 |
| 600 | 179649078 | c.2494G > T | p.Ala832Ser | E.16 | rs376133574 | P | VUS | DC | 22.3 | 5.52 | 202 |
| 601 | 179650574 | c.2370 + 1G > T | – | I.14 | rs375796806 | LP | NA | DC | NA | 4.99 | 202 |
| 602 | 179650717 | c.2228C > T | p.Ala743Val | E.14 | rs267607157 | VUS | P | PO | 19.42 | 5.3 | 97 |
| 603 | 179650808 | c.2137C > T | p.Arg713Ter | E.14 | rs727505277 | P | VUS | DC | 39 | 5.99 | 205 |
| 604 | 179658212 | c.1455dup | p.Ala486SerfsTer26 | E.9 | rs758662735 | P | NA | NA | 60 | 3.36 | 243 |
| 605 | 179659281 | c.1246-3del | – | I.7 | NA | VUS | NA | DC | NA | 1.44 | 199 |
| 606 | 179659646 | c.1245 + 3A > G | – | I.7 | rs757221300 | LP | VUS | DC | NA | 5.87 | 202 |
| 607 | 179613717 | c.11311 + 4134dup | – | I.47 | rs768458450 | VUS | VUS | NA | NA | 3.61 | 202 |
| 608 | 179664231 | c.897-898insT | p.Thr300TyrfsTer23 | E.6 | NA | P | NA | DC | 58 | − 2.83 | 205 |
| 609 | 179664293 | c.835C > T | p.Arg279Trp | E.6 | rs138060032 | LP | VUS | DC | 24.5 | 4.82 | 191 |
| 610 | 179665172 | c.533C > A | p.Ala178Asp | E.4 | NA | LP | NA | DC | 23.4 | 5.16 | 286 |
| 611 | 179665380 | c.325C > T | p.Arg109Ter | E.4 | rs150954246 | LP | VUS | DC | 38 | 3.8 | 202 |
Dilated cardiomyopathy
Idiopathic factors are just as significant in the pathophysiology of DCM as acquired variables (such as infections, poisons, or autoimmune diseases). Individuals harboring TTN mutations exhibit a higher susceptibility to developing DCM compared to other forms of the disease36,72–74. Idiopathic DCM, including familial and sporadic instances, has a genetic etiology, according to a vast number of studies75,76.
A review study by Chauveau et al.26 reported that Among the TTN mutations linked to DCM, 29 are categorized as nonsense mutations, with three of them occurring in the I-band, while the remaining 26 are located in the A-band. Additionally, 17 frameshift mutations are reported, with three in the I-band and 14 in the A-band. Furthermore, 18 mutations are predicted to affect TTN splicing TTN mutations, particularly truncating variants (TTNtv) in the A-band region and in exons that are highly utilized across the range of titin isoforms, have been shown in a number of studies to be strongly associated with the occurrence of DCM and its severity, accounting for the majority of cases77–80.
Although fewer TTNtv have been identified in pediatrics, a study by Fatkin et al.81 on the young population showed that the prevalence between adolescents and adults is similar, indicating that they need to have multiple clinical and genetic risk factors other than a single TTNtv to present with CDM. TTNtv accounts for 25% of familial cases and 18% of sporadic cases of idiopathic dilated cardiomyopathy82. The aforementioned TTNtv have demonstrated a remarkably low prevalence within the broader populace.
According to Fatkin et al. the prevalence of TTNtv is 20% among individuals with DCM, whereas only 0.5% of the general population carries this type of mutation83,84. The aforementioned data aligns with the results of Fang et al.85 survey, which indicated an overall prevalence rate of 17%. The survey also revealed that 23% of cases were familial, while 16% were sporadic. For example, mutations in the A-band are implicated as predominant genetic causes of DCM86–88.
An important question is how minor TTNtv carrier populations can avoid presenting with DCM. A convincing explanation comes from a study by Roberts et al.77 showing that the two major adult cardiac titin isoforms, N2BA and N2B, are responsible. These abundant full-length isoforms predominantly contain distal A-band exons, where most DCM-causing TTNtvs are located. However, mutations in proximal exons not present in all TTN transcripts do not cause DCM.
Hypertrophic cardiomyopathy
HCM is the most common inherited cardiomyopathy, frequently arising from sarcomere gene defects. Characterized by arrhythmias and heart failure symptoms due to left ventricular outflow obstruction, diastolic dysfunction, ischemia, or mitral regurgitation, HCM displays autosomal dominant inheritance. Mutations, predominantly missense, in one or more sarcomere genes underlie most cases of HCM. To date, over 1400 mutations have been identified in genes encoding primarily sarcomeric proteins89.
Due to the involvement of a vast range of mutations with distinctive penetrance, a comprehensive understanding of the pathophysiological mechanisms underlying the development of HCM in the presence of sarcomere-related gene mutations is still unfulfilling90. In a study conducted by Ingles et al.91 on 33 genes reported to have an association with HCM, only 8 genes (MYBPC3, MYH7, TNNT2, TNNI3, TPM1, ACTC1, MYL2, and MYL3) were shown to have a definitive impact on occurring HCM. It is estimated that around 30% of HCM patients have unidentified genes responsible for the condition.
The gene MYBPC3, which codes for cardiac myosin-binding protein C, is the most important gene in this process accounting for up to half of the mutations identified92–94. In the second place, MYH7, which is responsible for encoding the beta-myosin heavy chain, is present in approximately 15–25% of patients diagnosed with HCM92,95.
In comparison to other plausible etiologies of HCM, the presence of the TTN gene mutations exhibits a relatively low ranking. Several studies reported four TTN variants resulting in gain-of-function effects in HCM patients. Satoh et al.96 found a Z-line mutation (c.2219G > T, p.Arg740Leu) which increases alpha-actinin binding affinity. Two studies, similarly, reported a mutation in cardiac-specific N2B exon 49 [c.12347C > A, p.Ser4116Tyr] resulting in increased TTN binding to DRAL/FHL297,98. The TTN/T-CARP interaction is reinforced by the presence of two mutations located in exons 103 and 104-N2A, c.29231G > A, p.Arg9744 (initially reported as p.Arg8500His) and c.29543G > A, p.Arg9848Gln (initially reported as p.Arg8604Gln), as reported by Arimura et al.99. Lopes et al., in a different study, reported 219 TTN variants in a population of unrelated HCM patients. Of those 87% coexisted with mutations in HCM-related sarcomere gene defects and only 13% were found isolated26,100. However, in a study on 90 HCM patients and their close relatives, the mutation screening revealed no clue of the TTN gene being involved in their pathogenesis101. Similarly, Martijn Bos et al.102 detected no TTN mutation in a group of 389 HCM patients.
Restrictive cardiomyopathy
Restrictive cardiomyopathy is a diverse collection of disorders that primarily affect the myocardium, with a lesser impact on the endocardium and sub-endocardium. It is characterized by increased stiffness of the ventricular walls leading to restricted ventricular filling, which consequently results in significant diastolic dysfunction, elevated end-diastolic pressure, and reduced ejection fraction in the advanced stages103,104.
The epidemiology of this disease is not well understood in the literature due to classification and etiology reporting difficulties, but RCM is surely the least common form of cardiomyopathies, representing 2% to 5% of cases2,105. There are a variety of diseases that can cause it, including infiltrative disorders like amyloidosis and sarcoidosis, non-infiltrative disorders like diabetes and scleroderma, storage disease, endomyocardial disease, and cardiotoxicity brought on by chemotherapy or radiotherapy2.
Numerous genes that encode non-sarcomeric, sarcomeric, and sarcomere-associated proteins have been shown to play a role in RCM occurrence and inheritance. Examples include the TTR gene variants (V122I; I68L; L111M; T60A; S23N; P24S; W41L; V30M; V20I) and APOA1 gene in Amyloidosis; GLA gene in Fabry disease; GBA gene in Gaucher disease; HAMP, HFE, HFE2, HJV, PNPLA3, SLC40A1, TfR2 genes in Hereditary hemochromatosis; NPC1, NPC2 and SMPD1 genes in Niemann-Pick disease; AG3, CRYAB, DES, DNAJB6, FHL1, FLNC, LDB3, and MYOT genes in Myofibrillar myopathies; ABCC6 gene in Pseudoxanthoma elasticum; ACTC, MHC, TNNT2, TNNI3, TNNC1, DES, MYH, MYL3, and CRYAB genes in Sarcomeric protein disorders; WRN gene in Werner's syndrome; and BMP5, BMP7 and TAZ genes in Endocardial fibroelastosis1,2,106,107.
The role of TTN variants in RCM is relatively unknown and more investigations are needed to illustrate this fact. In 2013, for the first time, Peled et al. discovered a novel missense mutation (c.50057A > G, p.Tyr16686Cys) in the intersection of the A and I regions of Titin (IA junction). This mutation was found to play a role in early-onset familial RCM, which affected six members of a family. They asserted that Titin determines the sarcomere's resting tension, and their study offers genetic proof of its critical significance in diastolic function.36,108,109. In another study, Kizawa et al.110 found another novel TTN missense mutation (c.22769C > A, p.P7590Q) in a young boy with neurofibromatosis type 1, which is thought to be responsible for RCM co-occurrence. This de novo mutation is also located at the IA junction.
Arrhythmogenic right ventricular cardiomyopathy (ARVC)
Arrhythmogenic cardiomyopathy (ACM), is a rare and potentially life-threatening heart muscle disease with a prevalence of approximately 1:1000 to 1:5000111–113. Although asymptomatic in most instances upon diagnosis, it is characterized by palpitations, atypical chest pain, and syncope caused by cardiac arrhythmia, mostly in the right ventricle, which leads to the term “arrhythmogenic right ventricular cardiomyopathy (ARVC)”114–116. This condition is characterized by the progressive replacement of the myocardium with fibrofatty tissue, a process that begins at the epicardium, turns into a regional wall motion abnormality, and eventually spreads throughout the myocardium, resulting in the development of ventricular dilation and multiple aneurysms117–119.
The primary etiology of ACM is attributed to mutations in genes that encode desmosomal proteins, mainly with an autosomal dominant pattern of inheritance and over 30 percent of cases being familial. JUP, DSP, PKP2, DSG2, and DSC2 genes are the most probable to be involved. LMNA and TMEM43 are two additional genes that have been linked to the nuclear envelope, and there are genes that are shared with other cardiomyopathies (such as DES, PLN, TGFB3, TTN, and SCN5A)112,120–123.
Several studies have been conducted on the role of TTN variations in the pathogenesis of ARVC. In one study by Taylor et al.121, eight novel TTN variants (c.C29453T, p.Thr2896Ile; c.A97341G, p.Tyr8031Cys; c.C106734T, p.His8848Tyr; c.T215598C, p.Ile16949Thr; c.G221380A, p.Ala18579Thr; c.G226177T, p.Ala19309Ser; c.C272848T, p.Pro30847Leu; c.T281801C, p.Met33291Thr) were identified in seven unrelated families with well-established ARVC. They claimed the most prominent variant was Thr2896Ile, showing strong segregation evidence. In another investigation on the phenotype-genotype relationship of ARVC in 39 families, Brun et al.123 found 13% of their studied population, had rare TTN variants (c.29453C > T, p.Thr2896Ile; 281801T > C, Ala18579Thr; c.221380AG > T, p.Met33291Thr; c.226177G > T, p.Ala19309Ser; c.97341G > A, p.Tyr8031Cys; c.272848C > T, p.Pro30847Leu). In the investigation of the levels of Novex variant expression in human hearts with cardiomyopathies, Chen et al.124 came to the conclusion that this factor was altered in cardiomyopathies such as DCM and ARVC.
Other muscle disorders
Beyond cardiomyopathies, TTN mutations are implicated in numerous non-cardiac muscle disorders. According to Chauveau et al.26, 39 TTN mutations have been identified so far in four pure skeletal muscle myopathies: limb girdle muscular dystrophy type 2J (LGMD2J), late-onset autosomal dominant tibial muscular dystrophy (TMD), hereditary myopathy with early respiratory failure (HMERF), and congenital centronuclear myopathy (CNM). Additional conditions associated with TTN variants include early adult onset recessive distal titinopathy, early-onset myopathy with fatal cardiomyopathy, multi-minicore disease with heart disease, childhood-juvenile Emery-Dreifuss-like phenotype without cardiomyopathy, and adult-onset recessive proximal muscular dystrophy125.
Frequent TTN-related molecules in cardiomyopathies
There are several molecules which play a considerable role in the signaling and function of Titin. In the present study, we evaluated their interaction with Titin and consider their interaction with Titin in the pathogenesis of cardiomyopathies (Fig. 4).
Figure 4.
Illustration of the intricate signaling pathway implicated in the development of cardiomyopathy associated with Titin and other related proteins.
Calpain
Calpain, a family of Ca2+-dependent cytosolic cysteine proteases, plays a role in various cellular processes, including cell death and tissue remodeling126. It has been implicated in several cardiac conditions, including dilated cardiomyopathy, alcohol-related cardiomyopathy, chemotherapy-induced cardiomyopathy, arrhythmogenic cardiomyopathy, and diabetic cardiomyopathy127–131. Sustained over-expression of calpain-2, specifically in cardiomyocytes, induced age-dependent dilated cardiomyopathy in mice127.
MuRF1/2
Muscle ring finger (MuRF) proteins are muscle-specific ubiquitin E3 ligases that regulate the ubiquitin–proteasome system and modulate cardiac mass and function132. A study by Su et al.133 showed a higher prevalence of rare MuRF1 and MuRF2 variants in hypertrophic cardiomyopathy (HCM) patients compared to controls. HCM patients with these rare MuRF1/2 variants were younger and had greater maximum left ventricular wall thickness than those without the variants133.
ERK
ERK (Extracellular signal-regulated kinase) plays a central role in cardiac physiology and hypertrophy134–136. ERK signaling is implicated in various forms of cardiac hypertrophy and progression to heart failure135. Altered ERK activity has been linked to HCM134. ERKs are considered key regulators of cardiac hypertrophy since they are activated by most, if not all, stress stimuli known to induce hypertrophic growth137. Studies show that concurrently eliminating ERK1 and ERK2 in the heart leads to eccentric hypertrophy with chamber dilatation and cardiomyocyte elongation136.
NFAT
Nuclear factor of activated T-cells (NFAT) transcription factors are implicated in developing cardiac hypertrophy and heart failure138. Activation of NFAT signaling induces pathological remodeling of cardiomyocytes139. Inhibition of NFAT prevents maladaptive cardiac growth in response to stress stimuli140. Targeting NFAT signaling pathways may be therapeutic for specific cardiomyopathies141,142.
FHL1/2
Mutations in the four-and-a-half LIM domain proteins 1 and 2 (FHL1 and FHL2) are associated with reducing body myopathy and hypertrophic cardiomyopathy143. FHL1/2 are involved in sarcomere assembly and signaling and highly expressed in skeletal and cardiac muscle144,145. Abnormal FHL proteins cause structural defects in sarcomeres and impaired muscle contraction146. FHL1 mutations account for 8–10% of familial reducing body myopathy cases which can include cardiomyopathy147,148. Chu et al.145 reported FHL1 upregulation in Cardiac ventricles of two mouse models with cardiac hypertrophy and dilated cardiomyopathy.
MARP
Muscle ankyrin repeat proteins (MARPs), including CARP, Ankrd1/2, and DARP, are a family of ankyrin repeat proteins expressed in striated muscle that are induced by stress. MARPs play regulatory roles in the muscle stress response and hypertrophy pathogenesis149. Overexpression of CARP is linked to dilated cardiomyopathy in animal models150. In addition, Patients with hypertrophic, dilated, ischemic, and arrhythmogenic right ventricular cardiomyopathy are more likely to develop CARP upregulation62,149,151,152. Missense mutations in the Ankrd1 gene have recently been identified as the cause of dilated and hypertrophic cardiomyopathy in humans99,149,153,154. CARP modulation of gene expression may contribute to adverse ventricular remodeling in cardiomyopathies155.
Nbr1
Neighbor of BRCA1 gene 1 (Nbr1) is a cardiac-expressed protein involved in autophagy, protein degradation and sarcomere organization156. Several studies suggested role of Nbr1 overexpression in developing dilated cardiomyopathy157–159.
SRF
Serum response factor (SRF) is a transcription factor regulating cardiac gene expression important for adaptation to stress160. SRF inactivation in animal models causes dilated cardiomyopathy160. SRF likely controls genes involved in maintaining normal cardiac structure and function161. Alterations in SRF-dependent gene regulation may underlie some cardiomyopathies162.
MLP
Muscle LIM protein (MLP) is involved in mechanosensing and stretch response in cardiomyocytes163. MLP knockout mice develop dilated cardiomyopathy164. Loss of MLP leads to impaired myocyte stretch signaling and contraction165. MLP deficiency is implicated in some forms of familial dilated cardiomyopathy166.
MyBP-C
Myosin binding protein C (MyBP-C) is important for maintaining sarcomere structure and regulating muscle contraction167. Mutations in cardiac MyBP-C are the most common cause of hypertrophic cardiomyopathy168. Abnormal MyBP-C disrupts sarcomere function leading to reduced contractility and development of hypertrophy169.
Myomesin
Myomesin is a major component of the sarcomeric M-band involved in thick filament organization170. Myomesin mutations have been associated with hypertrophic and dilated cardiomyopathy in some patients171. Altered myomesin disrupts myofilament integrity and crosstalk resulting in cardiomyocyte damage172.
Sh2 domain
Src homology 2 (SH2) domains mediate protein–protein interactions in cell signaling cascades173. Mutations affecting SH2 domains of ZASP/Cypher proteins are linked to dilated cardiomyopathy174. Disruption of ZASP protein interactions likely impairs structural organization and signaling processes in cardiac muscle175.
Ras
Ras family small GTPases regulate growth and survival signaling176. Constitutively active mutant Ras expressed in mouse hearts causes dilated cardiomyopathy phenotype177. Hyperactive Ras leads to increased cell growth, altered metabolism and myocardial dysfunction178.
Raf
Raf kinases act downstream of Ras to activate MEK/ERK signaling involved in cell proliferation and differentiation179. Cardiac-specific expression of activated Raf in transgenic mice induces dilated cardiomyopathy180.
Alpha actinin
Alpha-actinin-2 (ACTN2) is the sole muscle isoform of α-actinin expressed in cardiac muscle181. Previous studies have shown that novel ACTN2 variants are associated with familial HCM182. Previous studies have shown that novel ACTN2 variants are associated with181. Mutations in ACTN2 have been linked to mild to moderate forms of HCM181. Disease modeling of an ACTN2 mutation has guided clinical therapy in HCM183. Genome-wide analyses have also demonstrated that ACTN2 mutations can cause HCM184.
Filamin C
In striated muscle, different forms of the Ank3 gene product (ankyrins-G) are produced due to tissue-specific alternative splicing. These ankyrins-G have a shared segment called the Obscurin/Titin-Binding-related Domain (OTBD), which is consistent across ankyrin genes and links obscurin and Titin to Ank1 gene products. Previously, it was suggested that the OTBD segment in ankyrins plays a unique role in muscle protein interactions. In recent studies, muscle proteins that can bind to the ankyrin-G OTBD were identified as plectin and filamin C, both crucial for muscle development and structure. These three proteins (ankyrin-G, plectin, and filamin C) are found together in skeletal muscle and are observed in the same regions (costameres) of adult muscle fibers185. Filamin C (FLNC) is an actin-binding cytoskeletal protein encoded by the FLNC gene, instrumental in maintaining sarcomeric integrity. While first identified as causative in myofibrillar myopathy, recent evidence reveals a key role for FLNC in cardiomyopathy pathogenesis. Truncated FLNC variants predominate in DCM and ARVC, while non-truncated forms are more common in hypertrophic cardiomyopathy and restrictive cardiomyopathy. The primary mechanisms underlying FLNC-associated cardiomyopathies are protein aggregation from non-truncating mutations and haploinsufficiency resulting from filamin C truncation186.
Nebulin
Members of the nebulin protein family, which includes nebulin, nebulette, LASP-1, LASP-2, and N-RAP, are diverse in size, expression pattern, and function, but they all bind to actin. While nebulin's presence in the heart is minimal, nebulette stands out for its heart-specific expression. Crucially, mutations in the nebulette gene have been linked to DCM. Transgenic mice with these mutations display symptoms that mirror this human heart condition187.
Mechanosensory signaling mechanism of titin
Titin plays a crucial role in mechanosensing, which is the ability of cells to sense mechanical forces. When muscles undergo stretch or contraction, Titin is subjected to mechanical stress and strain. This mechanical deformation of Titin can trigger mechanotransduction pathways, converting mechanical signals into biochemical signals. These pathways involve the activation of various signaling molecules, including kinases, phosphatases, and transcription factors, leading to cellular responses such as gene expression changes, protein synthesis, and remodeling of the contractile apparatus188 (Fig. 4).
Z disk region
The Z-disc region of Titin consists of Z-repeats and Ig-domains Z1 and Z2, forming the very NH2-terminal end. Telethonin connects two Titin molecules from one sarcomere, which is essential for sarcomere integrity. Cardiac telethonin undergoes phosphorylation by various kinases and mutations in telethonin are linked to various cardiac cardiomyopthies. Some mutations might disrupt its phosphorylation and, thus, its function. Telethonin interacts with the muscle LIM protein (MLP), together with actinin, MLP, Titin, and telethonin might form a complex that senses mechanical stretch50.
N2-B region
Cardiac-specific N2-B region which made up of Ig-domains can bind to two isoforms of the LIM domain protein, FHL-1 and FHL-2 which respond strongly to biomechanical stress, and can move to the nucleus to work as transcriptional co-activators. FHL-2's activity could suppress calcineurin, inhibiting pathological cardiac growth while FHL1 might connect to the MAPK signaling cascade. Under non-stimulating conditions, MEK1/2 anchors ERK in the cytoplasm, but after activation, it shifts ERK to the nucleus, activating specific transcription factors.
ERK2 has been seen to phosphorylate Titin's N2-Bus sequence, potentially affecting myofilament stiffness. Knocking down FHL1 in mice changed myofibrillar responsiveness and reduced hypertrophic signaling. Hence, the N2-B/FHL-1/MAPK complex might be a key biomechanical stress sensor in cardiomyocytes44,58,137,189,190.
M-band region
The M-band region of Titin, particularly the Titin kinase (TK) domain, is a significant area for hypertrophic signaling. TK's conformational changes, suggesting its role as a biomechanical stress sensor, might be biomechanically induced. When activated, TK interacts with Nbr1, forming a complex with p62/SQSTM1 and muscle-specific ubiquitin E3 ligases MuRF1, MuRF2, and MuRF3.
The TK signaling complex with the zinc-finger protein nbr1 is involved in mechanically-activated signaling. Nbr1 directs the ubiquitin-binding protein p62/SQSTM1 to sarcomeres where it interacts with the muscle-specific E3 ligase MuRF2, linked to the transactivation domain of serum response factor (SRF). Mechanical inactivity triggers MuRF2 nuclear migration, decreasing nuclear SRF and suppressing transcription. Mutations in the TK domain disrupt this mechanism, resulting in hereditary muscle disorders50,191.
Of course, it should be considered that subsequent investigations have proposed that TK functions as an inactive pseudokinase, utilizing its kinase scaffold to recruit MuRF1 for biomechanically regulated autophagy pathways192,193.
The hotspot region for TTN variants
In a quantitative analysis of variants, it was revealed that the most common hotspot region for variants is the exon number 326 which is located in the A band as the Fibronectin type III domain194 and has a more considerable number of variants compared to other parts which are followed by exon 358 (containing Ig-like domain and Fibronectin type III domain)194 and exon 48. Among the introns, intron 47 can be considered as the hotspot point for variants compared to other introns194 (Fig. 2).
Discussion
This study identified 611 distant TTN variants, classified as pathogenic, likely pathogenic, or variants of uncertain significance (VUS). These variants predominantly occurred in exon fragments (85%), with 69.6% classified as pathogenic, 21.6% as likely pathogenic, and 8.8% as VUS in ACMG classification. Substitutions accounted for 57.25% of the variants, deletions for 29.62%, duplications for 7.36%, and insertions for 5.72%. The majority of pathogenic variants were located after exon 326, exhibiting higher CADD scores. GERP scores indicated conservity among gene nucleotides, with most variants having notable GERP scores. Exons at the end of the gene displayed higher average CADD scores. VUS variants had lower CADD scores.
TTN, a functionally and structurally essential component of striated muscles, is the largest human protein10,11. It consists of four functional regions including N-terminal, I-band, A-band, and C-terminal26. The N-terminal is an anchor for Z-disk, which not only plays a crucial role in myofibril assembly and stability but also in sensory functions, protein interactions, and signaling pathways32–40. Owing to alternative splicing, I-band is the central adopter specializing titin for specific tissues. The elasticity of the titin is mostly attributable to the I-band unit38,41. On the contrary to the I-band, the A-band is not extensible and is a stable anchor for myosin fibers. It also interacts with various proteins contributing to protein turnover at the sarcomeric center38,41. The M-band constitutes the myomesin-titin-myosin and also senses and responds to the metabolic stress50.
The passive tension of the human heart is determined by the pattern of expression of titin isoforms. Expression of more elastic and larger I-band isoforms is associated with lower titin passive tension. The ratio of N2BA and N2B isoform expression determines the stiffness of cardiomyocytes60. If the balance between N2BA and N2B is disrupted and N2BA isoform upregulates, the decrease in passive stiffness of the heart brings about DCM30,31,62,63. Mutations in the TTN gene are speculated to bring about cardiomyopathies through disruption in sarcomere assembly or contractility, or triggering aberrant splicing30,31,62,63.
In accordance with our study, another study demonstrated that most TTN variants associated with TTN are located in the A-band unit followed by the I-band26. Truncating TTN variants located in the A-band region are the predominant TTN mutations associated with the DCM77–80,86–88. The N2BA and N2B isoforms contain distal exons of the A-band. Therefore, variants affecting the A-band and its distal regions are more frequently reported to manifest with DCM, while, the N-terminal mutations are less likely to bring about DCM, considering they are not expressed in N2BA and N2B isoforms77.
TTN mutations are not as prominent in HCM compared to DCM. HCM is speculated to arise from mutations in sarcomere-related genes; nonetheless, the exact pathophysiology of HCM is yet to be found90. Mutations in Sarcomeric, non-sarcomeric, and sarcomere-associated proteins are proposed to contribute to the development and inheritance of RCM1,2,106,107. Although the role of TTN variants in the pathogenesis and inheritance of RCM is not fully understood, it is known that titin is the key determinant of sarcomere resting tension and diastolic function36,108,109. Similarly, the impact of TTN mutations in ARVC is not yet determined. However, rare TTN variants have been reported in probands and family members of ARVC patients121,123.
The most common hotspot for mutations is exon 326 of the TTN gene which is located in the A-band region. Notably, the exon containing the most TTN variants is 358, also in the A-band. As presented, the TTN variants were primarily located in a small number of exons which are mostly situated at A- and I-bands. This localization of TTN variants might stem from the higher fatality of mutations in other locations, or conversely, these mutations do not exhibit clinical symptoms to prompt genetic evaluation.
The conservatory TTN exons seem to be associated with the pathogenicity of the variants This might be explained, at least in part, by the theory that more conserved nucleotides could be essential, and mutations affecting this nucleotide could be more pathogenic.
Acknowledgements
We appreciate the support from Cardiogenetic Research Center, Rajaie Cardiovascular Medical and Research Center, Iran University of Medical Sciences, Tehran, Iran.
Author contributions
A.G., E.K. and S.K. wrote the initial manuscript. S.K., M.M., A.F., and M.H. contributed to the research design. S.K. made a comprehensive revise. S.G.H., M.H., M.H.M., and N.N. contributed to the collection of data. All the authors read and approved the final manuscript.
Funding
This research received no specific grant from any funding agency, commercial or not-for-profit sectors.
Data availability
The datasets generated and/or analyzed during the current study are available in the the public archive of interpretations of clinically relevant variants (ClinVar) repository, (https://www.ncbi.nlm.nih.gov/clinvar/?term=TTN%5Bgene%5D&redir=gene).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ormerod, J.O. and A. Yavari, Cardiomyopathies. Medicine, 2022.
- 2.Ciarambino T, et al. Cardiomyopathies: An Overview. Int. J. Mol. Sci. 2021;22(14):7722. doi: 10.3390/ijms22147722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maron BJ, et al. Contemporary definitions and classification of the cardiomyopathies: An American heart association scientific statement from the council on clinical cardiology, heart failure and transplantation committee; quality of care and outcomes research and functional genomics and translational biology interdisciplinary working groups; and council on epidemiology and prevention. Circulation. 2006;113(14):1807–1816. doi: 10.1161/CIRCULATIONAHA.106.174287. [DOI] [PubMed] [Google Scholar]
- 4.Elliott P, et al. Classification of the cardiomyopathies: A position statement from the European society of cardiology working group on myocardial and pericardial diseases. Eur. Heart J. 2008;29(2):270–276. doi: 10.1093/eurheartj/ehm342. [DOI] [PubMed] [Google Scholar]
- 5.Maurizia Grasso B, Favalli V, Riccardo Bellazzi M. The MOGE (S) classification of cardiomyopathy for clinicians. J. Am. College Cardiol. 2014;64(3):304–318. doi: 10.1016/j.jacc.2014.05.027. [DOI] [PubMed] [Google Scholar]
- 6.Brandenburg R. Report of the WHO/ISFC task force on the definition and classification of cardiomyopathies. Br. Heart J. 1980;44:672–673. doi: 10.1136/hrt.44.6.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Czepluch FS, Wollnik B, Hasenfuß G, (2018) Genetic determinants of heart failure: facts and numbers, Wiley Online Library. Cham. 211-217 [DOI] [PMC free article] [PubMed]
- 8.Cannata A, et al. Myocarditis evolving in cardiomyopathy: When genetics and offending causes work together. Eur. Heart J. Suppl. 2019;21(Suppl B):B90–b95. doi: 10.1093/eurheartj/suz033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.The Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447(7145):661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gerull B. The rapidly evolving role of titin in cardiac physiology and cardiomyopathy. Can. J. Cardiol. 2015;31(11):1351–1359. doi: 10.1016/j.cjca.2015.08.016. [DOI] [PubMed] [Google Scholar]
- 11.Tabish AM, et al. Genetic epidemiology of titin-truncating variants in the etiology of dilated cardiomyopathy. Biophys. Rev. 2017;9(3):207–223. doi: 10.1007/s12551-017-0265-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gerull B, et al. Mutations of TTN, encoding the giant muscle filament titin, cause familial dilated cardiomyopathy. Nat. Genet. 2002;30(2):201–204. doi: 10.1038/ng815. [DOI] [PubMed] [Google Scholar]
- 13.Arbustini E, et al. Desmin accumulation restrictive cardiomyopathy and atrioventricular block associated with desmin gene defects. Eur. J. Heart Fail. 2006;8(5):477–483. doi: 10.1016/j.ejheart.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 14.Merlo M, et al. Poor prognosis of rare sarcomeric gene variants in patients with dilated cardiomyopathy. Clin. Transl. Sci. 2013;6(6):424–428. doi: 10.1111/cts.12116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Richards S, et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American college of medical genetics and genomics and the association for molecular pathology. Genet. Med. 2015;17(5):405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Richards S, et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American college of medical genetics and genomics and the association for molecular pathology. Genet. Med. 2015;17(5):405–423. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kircher M, et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46(3):310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rentzsch P, et al. CADD: Predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2019;47(D1):D886–d894. doi: 10.1093/nar/gky1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rentzsch P, et al. CADD-Splice-improving genome-wide variant effect prediction using deep learning-derived splice scores. Genome Med. 2021;13(1):31. doi: 10.1186/s13073-021-00835-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schubach M, et al. CADD v1.7: Using protein language models, regulatory CNNs and other nucleotide-level scores to improve genome-wide variant predictions. Nucleic Acids Res. 2024;52(D1):D1143–d1154. doi: 10.1093/nar/gkad989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schwarz JM, et al. MutationTaster evaluates disease-causing potential of sequence alterations. Nat. Methods. 2010;7(8):575–576. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- 22.Steinhaus R, et al. MutationTaster2021. Nucleic Acids Res. 2021;49(W1):W446–w451. doi: 10.1093/nar/gkab266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Šimčíková D, Heneberg P. Refinement of evolutionary medicine predictions based on clinical evidence for the manifestations of Mendelian diseases. Sci. Rep. 2019;9(1):18577. doi: 10.1038/s41598-019-54976-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huber CD, Kim BY, Lohmueller KE. Population genetic models of GERP scores suggest pervasive turnover of constrained sites across mammalian evolution. PLoS Genet. 2020;16(5):e1008827. doi: 10.1371/journal.pgen.1008827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tskhovrebova L, Trinick J. Titin: Properties and family relationships. Nat. Rev. Mol. Cell Biol. 2003;4(9):679–689. doi: 10.1038/nrm1198. [DOI] [PubMed] [Google Scholar]
- 26.Chauveau C, Rowell J, Ferreiro A. A rising titan: TTN review and mutation update. Hum. Mutat. 2014;35(9):1046–1059. doi: 10.1002/humu.22611. [DOI] [PubMed] [Google Scholar]
- 27.Tskhovrebova L, et al. Elasticity and unfolding of single molecules of the giant muscle protein titin. Nature. 1997;387(6630):308–312. doi: 10.1038/387308a0. [DOI] [PubMed] [Google Scholar]
- 28.LeWinter MM, Granzier H. Cardiac titin: A multifunctional giant. Circulation. 2010;121(19):2137–2145. doi: 10.1161/CIRCULATIONAHA.109.860171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lahmers S, et al. Developmental control of titin isoform expression and passive stiffness in fetal and neonatal myocardium. Circ. Res. 2004;94(4):505–513. doi: 10.1161/01.RES.0000115522.52554.86. [DOI] [PubMed] [Google Scholar]
- 30.Cazorla O, et al. Differential expression of cardiac titin isoforms and modulation of cellular stiffness. Circ. Res. 2000;86(1):59–67. doi: 10.1161/01.RES.86.1.59. [DOI] [PubMed] [Google Scholar]
- 31.Neagoe C, et al. Gigantic variety: expression patterns of titin isoforms in striated muscles and consequences for myofibrillar passive stiffness. J. Muscle Res. Cell Motil. 2003;24(2):175–189. doi: 10.1023/A:1026053530766. [DOI] [PubMed] [Google Scholar]
- 32.Knoll R, Buyandelger B, Lab M. The sarcomeric Z-disc and Z-discopathies. J. Biomed. Biotechnol. 2011;2011:569628. doi: 10.1155/2011/569628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Knoll R, Hoshijima M, Chien KR. Muscle LIM protein in heart failure. Exp. Clin. Cardiol. 2002;7(2–3):104–105. [PMC free article] [PubMed] [Google Scholar]
- 34.Knöll R, et al. A common MLP (muscle LIM protein) variant is associated with cardiomyopathy. Circ. Res. 2010;106(4):695–704. doi: 10.1161/CIRCRESAHA.109.206243. [DOI] [PubMed] [Google Scholar]
- 35.Clark KA, et al. Striated muscle cytoarchitecture: An intricate web of form and function. Ann. Rev. Cell Dev. Biol. 2002;18(1):637–706. doi: 10.1146/annurev.cellbio.18.012502.105840. [DOI] [PubMed] [Google Scholar]
- 36.Gigli M, et al. A review of the giant protein titin in clinical molecular diagnostics of cardiomyopathies. Front Cardiovasc. Med. 2016;3:21. doi: 10.3389/fcvm.2016.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Granzier HL, Labeit S. Titin and its associated proteins: The third myofilament system of the sarcomere. Adv. Protein Che. 2005;71:89–119. doi: 10.1016/S0065-3233(04)71003-7. [DOI] [PubMed] [Google Scholar]
- 38.Kontrogianni-Konstantopoulos A, et al. Muscle giants: Molecular scaffolds in sarcomerogenesis. Physiol. Rev. 2009;89(4):1217–1267. doi: 10.1152/physrev.00017.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Linke WA. Sense and stretchability: The role of titin and titin-associated proteins in myocardial stress-sensing and mechanical dysfunction. Cardiovasc. Res. 2008;77(4):637–648. doi: 10.1016/j.cardiores.2007.03.029. [DOI] [PubMed] [Google Scholar]
- 40.Miller MK, et al. The muscle ankyrin repeat proteins: CARP, ankrd2/Arpp and DARP as a family of titin filament-based stress response molecules. J. Mol. Biol. 2003;333(5):951–964. doi: 10.1016/j.jmb.2003.09.012. [DOI] [PubMed] [Google Scholar]
- 41.Trombitas K, et al. Titin extensibility in situ: Entropic elasticity of permanently folded and permanently unfolded molecular segments. J. Cell Biol. 1998;140(4):853–859. doi: 10.1083/jcb.140.4.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Helmes M, Trombitas K, Granzier H. Titin develops restoring force in rat cardiac myocytes. Circ. Res. 1996;79(3):619–626. doi: 10.1161/01.RES.79.3.619. [DOI] [PubMed] [Google Scholar]
- 43.Hojayev B, et al. FHL2 binds calcineurin and represses pathological cardiac growth. Mol. Cell. Biol. 2012;32(19):4025–4034. doi: 10.1128/MCB.05948-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hojayev B, et al. FHL2 binds calcineurin and represses pathological cardiac growth. Mol. Cell Biol. 2012;32(19):4025–4034. doi: 10.1128/MCB.05948-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Granzier HL, Labeit S. The giant protein titin: A major player in myocardial mechanics, signaling, and disease. Circ. Res. 2004;94(3):284–295. doi: 10.1161/01.RES.0000117769.88862.F8. [DOI] [PubMed] [Google Scholar]
- 46.Witt CC, et al. Induction and myofibrillar targeting of CARP, and suppression of the Nkx2.5 pathway in the MDM mouse with impaired titin-based signaling. J. Mol. Biol. 2004;336(1):145–54. doi: 10.1016/j.jmb.2003.12.021. [DOI] [PubMed] [Google Scholar]
- 47.Obermann WM, et al. Molecular structure of the sarcomeric M band: Mapping of titin and myosin binding domains in myomesin and the identification of a potential regulatory phosphorylation site in myomesin. EMBO J. 1997;16(2):211–220. doi: 10.1093/emboj/16.2.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gautel M, et al. A calmodulin-binding sequence in the C-terminus of human cardiac titin kinase. Eur. J. Biochem. 1995;230(2):752–759. [PubMed] [Google Scholar]
- 49.Musa H, et al. Targeted homozygous deletion of M-band titin in cardiomyocytes prevents sarcomere formation. J. Cell Sci. 2006;119(20):4322–4331. doi: 10.1242/jcs.03198. [DOI] [PubMed] [Google Scholar]
- 50.Kotter S, Andresen C, Kruger M. Titin: central player of hypertrophic signaling and sarcomeric protein quality control. Biol. Chem. 2014;395(11):1341–1352. doi: 10.1515/hsz-2014-0178. [DOI] [PubMed] [Google Scholar]
- 51.McElhinny AS, et al. Muscle-specific RING finger-2 (MURF-2) is important for microtubule, intermediate filament and sarcomeric M-line maintenance in striated muscle development. J. Cell Sci. 2004;117(15):3175–3188. doi: 10.1242/jcs.01158. [DOI] [PubMed] [Google Scholar]
- 52.Beckmann JS, Spencer M. Calpain 3, the “gatekeeper” of proper sarcomere assembly, turnover and maintenance. Neuromusc. Disord. 2008;18(12):913–921. doi: 10.1016/j.nmd.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Granzier H, et al. Titin: Physiological function and role in cardiomyopathy and failure. Heart Fail. Rev. 2005;10(3):211–223. doi: 10.1007/s10741-005-5251-7. [DOI] [PubMed] [Google Scholar]
- 54.Tskhovrebova, L. and J. Trinick, Roles of titin in the structure and elasticity of the sarcomere. Journal of Biomedicine and Biotechnology, 2010 (2010). [DOI] [PMC free article] [PubMed]
- 55.Medicine, N.L.o. TTN titin [ Homo sapiens (human) ]. 2024; Available from: https://www.ncbi.nlm.nih.gov/gene/7273.
- 56.Resource, T.U.P. Q8WZ42 · TITIN_HUMAN. Available from: https://www.uniprot.org/uniprotkb/Q8WZ42/entry.
- 57.Bang ML, et al. The complete gene sequence of titin, expression of an unusual approximately 700-kDa titin isoform, and its interaction with obscurin identify a novel Z-line to I-band linking system. Circ. Res. 2001;89(11):1065–1072. doi: 10.1161/hh2301.100981. [DOI] [PubMed] [Google Scholar]
- 58.Radke MH, et al. Targeted deletion of titin N2B region leads to diastolic dysfunction and cardiac atrophy. Proc. Natl. Acad. Sci. USA. 2007;104(9):3444–3449. doi: 10.1073/pnas.0608543104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lahmers S, Wu Y, Call DR, Labeit S, Granzier H. Developmental control of titin isoform expression and passive stiffness in fetal and neonatal myocardium. Circ. Res. 2004;94:505–513. doi: 10.1161/01.RES.0000115522.52554.86. [DOI] [PubMed] [Google Scholar]
- 60.Hidalgo C, Granzier H. Tuning the molecular giant titin through phosphorylation: Role in health and disease. Trends Cardiovasc. Med. 2013;23(5):165–171. doi: 10.1016/j.tcm.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Opitz CA, et al. Developmentally regulated switching of titin size alters myofibrillar stiffness in the perinatal heart. Circ. Res. 2004;94(7):967–975. doi: 10.1161/01.RES.0000124301.48193.E1. [DOI] [PubMed] [Google Scholar]
- 62.Nagueh SF, et al. Altered titin expression, myocardial stiffness, and left ventricular function in patients with dilated cardiomyopathy. Circulation. 2004;110(2):155–162. doi: 10.1161/01.CIR.0000135591.37759.AF. [DOI] [PubMed] [Google Scholar]
- 63.Opitz CA, et al. Developmentally regulated switching of titin size alters myofibrillar stiffness in the perinatal heart. Circ. Res. 2004;94(7):967–975. doi: 10.1161/01.RES.0000124301.48193.E1. [DOI] [PubMed] [Google Scholar]
- 64.Ahmed SH, Lindsey ML. Titin phosphorylation: Myocardial passive stiffness regulated by the intracellular giant. Circ. Res. 2009;105(7):611–613. doi: 10.1161/CIRCRESAHA.109.206912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kellermayer D, Smith JE, 3rd, Granzier H. Novex-3, the tiny titin of muscle. Biophys. Rev. 2017;9(3):201–206. doi: 10.1007/s12551-017-0261-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen Z, et al. Characterization of TTN novex splicing variants across species and the role of RBM20 in novex-specific exon splicing. Genes (Basel) 2018;9(2):86. doi: 10.3390/genes9020086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.View, A. Homo sapiens complex locus CCDC141andTTN, encoding titin and coiled-coil domain containing 141. . 2010; Available from: https://www.ncbi.nlm.nih.gov/ieb/research/acembly/av.cgi?db=human&term=ttn&submit=Go.
- 68.Legnini I, et al. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol. Cell. 2017;66(1):22–37. doi: 10.1016/j.molcel.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Khan MA, et al. RBM20 regulates circular RNA production from the titin gene. Circ. Res. 2016;119(9):996–1003. doi: 10.1161/CIRCRESAHA.116.309568. [DOI] [PubMed] [Google Scholar]
- 70.Czubak K, et al. Global increase in circular RNA levels in myotonic dystrophy. Front. Genet. 2019;10:649. doi: 10.3389/fgene.2019.00649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ellepola CD, et al. Genetic testing in pediatric cardiomyopathy. Pediatr. Cardiol. 2018;39(3):491–500. doi: 10.1007/s00246-017-1779-2. [DOI] [PubMed] [Google Scholar]
- 72.Waldmuller S, et al. Targeted 46-gene and clinical exome sequencing for mutations causing cardiomyopathies. Mol. Cell Probes. 2015;29(5):308–314. doi: 10.1016/j.mcp.2015.05.004. [DOI] [PubMed] [Google Scholar]
- 73.Marian AJ, Asatryan B, Wehrens XHT. Genetic basis and molecular biology of cardiac arrhythmias in cardiomyopathies. Cardiovasc. Res. 2020;116(9):1600–1619. doi: 10.1093/cvr/cvaa116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Monserrat L, et al. Genetics of cardiomyopathies: Novel perspectives with next generation sequencing. Curr. Pharm. Des. 2015;21(4):418–430. doi: 10.2174/138161282104141204123748. [DOI] [PubMed] [Google Scholar]
- 75.Schultheiss HP, et al. Dilated cardiomyopathy. Nat. Rev. Dis. Primers. 2019;5(1):32. doi: 10.1038/s41572-019-0084-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Weintraub RG, Semsarian C, Macdonald P. Dilated cardiomyopathy. The Lancet. 2017;390(10092):400–414. doi: 10.1016/S0140-6736(16)31713-5. [DOI] [PubMed] [Google Scholar]
- 77.Roberts AM, et al. Integrated allelic, transcriptional, and phenomic dissection of the cardiac effects of titin truncations in health and disease. Sci. Transl. Med. 2015;7(270):270ra6. doi: 10.1126/scitranslmed.3010134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Akinrinade O, et al. Genetics and genotype-phenotype correlations in Finnish patients with dilated cardiomyopathy. Eur. Heart J. 2015;36(34):2327–2337. doi: 10.1093/eurheartj/ehv253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.van Spaendonck-Zwarts KY, et al. Titin gene mutations are common in families with both peripartum cardiomyopathy and dilated cardiomyopathy. Eur. Heart J. 2014;35(32):2165–2173. doi: 10.1093/eurheartj/ehu050. [DOI] [PubMed] [Google Scholar]
- 80.Pugh TJ, et al. The landscape of genetic variation in dilated cardiomyopathy as surveyed by clinical DNA sequencing. Genet. Med. 2014;16(8):601–608. doi: 10.1038/gim.2013.204. [DOI] [PubMed] [Google Scholar]
- 81.Fatkin D, et al. Titin truncating mutations: A rare cause of dilated cardiomyopathy in the young. Progress Pediatr. Cardiol. 2016;40:41–45. doi: 10.1016/j.ppedcard.2016.01.003. [DOI] [Google Scholar]
- 82.Herman DS, et al. Truncations of titin causing dilated cardiomyopathy. N. Engl. J. Med. 2012;366(7):619–628. doi: 10.1056/NEJMoa1110186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fatkin D, Huttner IG. Titin-truncating mutations in dilated cardiomyopathy: The long and short of it. Curr. Opin. Cardiol. 2017;32(3):232–238. doi: 10.1097/HCO.0000000000000382. [DOI] [PubMed] [Google Scholar]
- 84.Akinrinade O, Koskenvuo JW, Alastalo TP. Prevalence of titin truncating variants in general population. PLoS One. 2015;10(12):e0145284. doi: 10.1371/journal.pone.0145284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fang HJ, Liu BP. Prevalence of TTN mutations in patients with dilated cardiomyopathy: A meta-analysis. Herz. 2020;45(Suppl 1):29–36. doi: 10.1007/s00059-019-4825-4. [DOI] [PubMed] [Google Scholar]
- 86.Akinrinade O, Alastalo TP, Koskenvuo JW. Relevance of truncating titin mutations in dilated cardiomyopathy. Clin. Genet. 2016;90(1):49–54. doi: 10.1111/cge.12741. [DOI] [PubMed] [Google Scholar]
- 87.Schafer S, et al. Titin-truncating variants affect heart function in disease cohorts and the general population. Nat. Genet. 2017;49(1):46–53. doi: 10.1038/ng.3719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yoskovitz G, et al. A novel titin mutation in adult-onset familial dilated cardiomyopathy. Am. J. Cardiol. 2012;109(11):1644–1650. doi: 10.1016/j.amjcard.2012.01.392. [DOI] [PubMed] [Google Scholar]
- 89.Herrero-Galán, E., et al., Conserved cysteines in titin sustain the mechanical function of cardiomyocytes. bioRxiv, (2020).
- 90.Marian AJ, Braunwald E. Hypertrophic cardiomyopathy: Genetics, pathogenesis, clinical manifestations, diagnosis, and therapy. Circ. Res. 2017;121(7):749–770. doi: 10.1161/CIRCRESAHA.117.311059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ingles J, et al. Evaluating the clinical validity of hypertrophic cardiomyopathy genes. Circ. Genom. Precis. Med. 2019;12(2):e002460. doi: 10.1161/CIRCGEN.119.002460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Van Driest, S.L., et al. Sarcomeric genotyping in hypertrophic cardiomyopathy. in Mayo Clinic Proceedings. 2005. Elsevier. [DOI] [PubMed]
- 93.Van Driest SL, et al. Myosin binding protein C mutations and compound heterozygosity in hypertrophic cardiomyopathy. J. Am. Coll. Cardiol. 2004;44(9):1903–1910. doi: 10.1016/j.jacc.2004.07.045. [DOI] [PubMed] [Google Scholar]
- 94.Richard P, et al. Hypertrophic cardiomyopathy: Distribution of disease genes, spectrum of mutations, and implications for a molecular diagnosis strategy. Circulation. 2003;107(17):2227–2232. doi: 10.1161/01.CIR.0000066323.15244.54. [DOI] [PubMed] [Google Scholar]
- 95.Van Driest SL, et al. Comprehensive analysis of the beta-myosin heavy chain gene in 389 unrelated patients with hypertrophic cardiomyopathy. J. Am. Coll. Cardiol. 2004;44(3):602–610. doi: 10.1016/j.jacc.2004.04.039. [DOI] [PubMed] [Google Scholar]
- 96.Satoh M, et al. Structural analysis of the titin gene in hypertrophic cardiomyopathy: Identification of a novel disease gene. Biochem. Biophys. Res. Commun. 1999;262(2):411–417. doi: 10.1006/bbrc.1999.1221. [DOI] [PubMed] [Google Scholar]
- 97.Itoh-Satoh M, et al. Titin mutations as the molecular basis for dilated cardiomyopathy. Biochem. Biophys. Res. Commun. 2002;291(2):385–393. doi: 10.1006/bbrc.2002.6448. [DOI] [PubMed] [Google Scholar]
- 98.Matsumoto Y, et al. Functional analysis of titin/connectin N2-B mutations found in cardiomyopathy. J. Muscle Res. Cell Motil. 2005;26(6–8):367. doi: 10.1007/s10974-005-9018-5. [DOI] [PubMed] [Google Scholar]
- 99.Arimura T, et al. Cardiac ankyrin repeat protein gene (ANKRD1) mutations in hypertrophic cardiomyopathy. J. Am. Coll. Cardiol. 2009;54(4):334–342. doi: 10.1016/j.jacc.2008.12.082. [DOI] [PubMed] [Google Scholar]
- 100.Lopes LR, et al. Genetic complexity in hypertrophic cardiomyopathy revealed by high-throughput sequencing. J. Med. Genet. 2013;50(4):228–239. doi: 10.1136/jmedgenet-2012-101270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Andersen PS, et al. Diagnostic yield, interpretation, and clinical utility of mutation screening of sarcomere encoding genes in Danish hypertrophic cardiomyopathy patients and relatives. Hum. Mutat. 2009;30(3):363–370. doi: 10.1002/humu.20862. [DOI] [PubMed] [Google Scholar]
- 102.Bos JM, et al. Genotype-phenotype relationships involving hypertrophic cardiomyopathy-associated mutations in titin, muscle LIM protein, and telethonin. Mol. Genet. Metab. 2006;88(1):78–85. doi: 10.1016/j.ymgme.2005.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lewis AB. Clinical profile and outcome of restrictive cardiomyopathy in children. Am. Heart J. 1992;123(6):1589–1593. doi: 10.1016/0002-8703(92)90814-C. [DOI] [PubMed] [Google Scholar]
- 104.Muchtar E, Blauwet LA, Gertz MA. Restrictive cardiomyopathy: Genetics, pathogenesis, clinical manifestations, diagnosis, and therapy. Circ. Res. 2017;121(7):819–837. doi: 10.1161/CIRCRESAHA.117.310982. [DOI] [PubMed] [Google Scholar]
- 105.Sayegh ALC, et al. Cardiac and peripheral autonomic control in restrictive cardiomyopathy. ESC Heart Fail. 2017;4(3):341–350. doi: 10.1002/ehf2.12142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yang Z, et al. Genotype-phenotype associations with restrictive cardiomyopathy induced by pathogenic genetic mutations. Rev. Cardiovasc. Med. 2022;23(6):185. doi: 10.31083/j.rcm2306185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Pereira NL, Grogan M, Dec GW. Spectrum of restrictive and infiltrative cardiomyopathies: Part 1 of a 2-part series. J. Am. College Cardiol. 2018;71(10):1130–1148. doi: 10.1016/j.jacc.2018.01.016. [DOI] [PubMed] [Google Scholar]
- 108.Peled Y, et al. Titin mutation in familial restrictive cardiomyopathy. Int. J. Cardiol. 2014;171(1):24–30. doi: 10.1016/j.ijcard.2013.11.037. [DOI] [PubMed] [Google Scholar]
- 109.Neiva-Sousa M, et al. Titin mutations: The fall of Goliath. Heart Fail. Rev. 2015;20(5):579–588. doi: 10.1007/s10741-015-9495-6. [DOI] [PubMed] [Google Scholar]
- 110.Kizawa M, et al. Identification of a novel titin variant underlying myocardial involvement in neurofibromatosis type 1. Can J Cardiol. 2018;34(10):1369 e5–1369 e7. doi: 10.1016/j.cjca.2018.07.473. [DOI] [PubMed] [Google Scholar]
- 111.Groeneweg JA, et al. Clinical presentation, long-term follow-up, and outcomes of 1001 arrhythmogenic right ventricular dysplasia/cardiomyopathy patients and family members. Circ. Cardiovasc. Genet. 2015;8(3):437–446. doi: 10.1161/CIRCGENETICS.114.001003. [DOI] [PubMed] [Google Scholar]
- 112.Corrado D, Thiene G. Arrhythmogenic right ventricular cardiomyopathy/dysplasia: Clinical impact of molecular genetic studies. Circulation. 2006;113(13):1634–1637. doi: 10.1161/CIRCULATIONAHA.105.616490. [DOI] [PubMed] [Google Scholar]
- 113.McKenna WJ, Judge DP. Epidemiology of the inherited cardiomyopathies. Nat. Rev. Cardiol. 2021;18(1):22–36. doi: 10.1038/s41569-020-0428-2. [DOI] [PubMed] [Google Scholar]
- 114.Marrone, D., et al., History of the discovery of Arrhythmogenic Cardiomyopathy: The history of arrhythmogenic cardiomyopathy (AC) is a paradigm in the progress of Cardiovascular Medicine knowledge, from nosology to diagnosis, treatment, and prevention. In this review, we focus on the discovery of this heart muscle disease at the beginning of Modern Medicine, something you cannot find on the Internet or PubMed. 2019, Oxford University Press.
- 115.Basso C, et al. Arrhythmogenic right ventricular cardiomyopathy. Lancet. 2009;373(9671):1289–1300. doi: 10.1016/S0140-6736(09)60256-7. [DOI] [PubMed] [Google Scholar]
- 116.Sen-Chowdhry S, et al. Arrhythmogenic right ventricular cardiomyopathy: Clinical presentation, diagnosis, and management. Am. J. Med. 2004;117(9):685–695. doi: 10.1016/j.amjmed.2004.04.028. [DOI] [PubMed] [Google Scholar]
- 117.Corrado D, et al. Spectrum of clinicopathologic manifestations of arrhythmogenic right ventricular cardiomyopathy/dysplasia: a multicenter study. J. Am. Coll. Cardiol. 1997;30(6):1512–1520. doi: 10.1016/S0735-1097(97)00332-X. [DOI] [PubMed] [Google Scholar]
- 118.Towbin JA, et al. 2019 HRS expert consensus statement on evaluation, risk stratification, and management of arrhythmogenic cardiomyopathy. Heart Rhythm. 2019;16(11):e301–e372. doi: 10.1016/j.hrthm.2019.05.007. [DOI] [PubMed] [Google Scholar]
- 119.List, P.G. and D. CTNNA, Arrhythmogenic Right Ventricular Cardiomyopathy Panel. 2021.
- 120.Calabrese F, et al. Arrhythmogenic right ventricular cardiomyopathy/dysplasia: Is there a role for viruses? Cardiovasc. Pathol. 2006;15(1):11–17. doi: 10.1016/j.carpath.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 121.Taylor M, et al. Genetic variation in titin in arrhythmogenic right ventricular cardiomyopathy-overlap syndromes. Circulation. 2011;124(8):876–885. doi: 10.1161/CIRCULATIONAHA.110.005405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sen-Chowdhry S, Syrris P, McKenna WJ. Role of genetic analysis in the management of patients with arrhythmogenic right ventricular dysplasia/cardiomyopathy. J. Am. Coll. Cardiol. 2007;50(19):1813–1821. doi: 10.1016/j.jacc.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 123.Brun F, et al. Titin and desmosomal genes in the natural history of arrhythmogenic right ventricular cardiomyopathy. J. Med. Genet. 2014;51(10):669–676. doi: 10.1136/jmedgenet-2014-102591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Chen Z, et al. Characterization of TTN novex splicing variants across species and the role of RBM20 in novex-specific exon splicing. Genes. 2018;9(2):86. doi: 10.3390/genes9020086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Savarese M, et al. Increasing role of titin mutations in neuromuscular disorders. J. Neuromuscul. Dis. 2016;3(3):293–308. doi: 10.3233/JND-160158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Martinez JA, et al. Calpain and caspase processing of caspase-12 contribute to the ER stress-induced cell death pathway in differentiated PC12 cells. Apoptosis. 2010;15(12):1480–1493. doi: 10.1007/s10495-010-0526-4. [DOI] [PubMed] [Google Scholar]
- 127.Ji XY, et al. Sustained over-expression of calpain-2 induces age-dependent dilated cardiomyopathy in mice through aberrant autophagy. Acta Pharmacol. Sin. 2022;43(11):2873–2884. doi: 10.1038/s41401-022-00965-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kartkaya K, et al. Protective effect of calpain inhibitor N-acetyl-L-leucyl-L-leucyl-L-norleucinal on acute alcohol consumption related cardiomyopathy. Mol. Biol. Rep. 2014;41(10):6743–6753. doi: 10.1007/s11033-014-3560-4. [DOI] [PubMed] [Google Scholar]
- 129.Ng, R., et al., Patient mutations linked to arrhythmogenic cardiomyopathy enhance calpain-mediated desmoplakin degradation. JCI Insight, 2019. 5(14). [DOI] [PMC free article] [PubMed]
- 130.Ni R, et al. Mitochondrial calpain-1 disrupts ATP synthase and induces superoxide generation in type 1 diabetic hearts: A novel mechanism contributing to diabetic cardiomyopathy. Diabetes. 2016;65(1):255–268. doi: 10.2337/db15-0963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Zuo S, et al. CRTH2 promotes endoplasmic reticulum stress-induced cardiomyocyte apoptosis through m-calpain. EMBO Mol. Med. 2018;10(3):e8237. doi: 10.15252/emmm.201708237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Willis MS, et al. Muscle ring finger 1 and muscle ring finger 2 are necessary but functionally redundant during developmental cardiac growth and regulate E2F1-mediated gene expression in vivo. Cell Biochem. Funct. 2014;32(1):39–50. doi: 10.1002/cbf.2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Su M, et al. Rare variants in genes encoding MuRF1 and MuRF2 are modifiers of hypertrophic cardiomyopathy. Int. J. Mol. Sci. 2014;15(6):9302–9313. doi: 10.3390/ijms15069302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Gilbert CJ, Longenecker JZ, Accornero F. ERK1/2: An integrator of signals that alters cardiac homeostasis and growth. Biology (Basel) 2021;10(4):346. doi: 10.3390/biology10040346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Gallo S, et al. ERK: A key player in the pathophysiology of cardiac hypertrophy. Int. J. Mol. Sci. 2019;20(9):2164. doi: 10.3390/ijms20092164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Mutlak M, Kehat I. Extracellular signal-regulated kinases 1/2 as regulators of cardiac hypertrophy. Front. Pharmacol. 2015;6:149. doi: 10.3389/fphar.2015.00149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Bueno OF, et al. The MEK1-ERK1/2 signaling pathway promotes compensated cardiac hypertrophy in transgenic mice. EMBO J. 2000;19(23):6341–6350. doi: 10.1093/emboj/19.23.6341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Heineke J, Molkentin JD. Regulation of cardiac hypertrophy by intracellular signalling pathways. Nat. Rev. Mol. Cell Biol. 2006;7(8):589–600. doi: 10.1038/nrm1983. [DOI] [PubMed] [Google Scholar]
- 139.Wilkins BJ, Molkentin JD. Calcium-calcineurin signaling in the regulation of cardiac hypertrophy. Biochem. Biophys. Res. Commun. 2004;322(4):1178–1191. doi: 10.1016/j.bbrc.2004.07.121. [DOI] [PubMed] [Google Scholar]
- 140.Bourajjaj M, et al. NFATc2 is a necessary mediator of calcineurin-dependent cardiac hypertrophy and heart failure. J. Biol. Chem. 2008;283(32):22295–22303. doi: 10.1074/jbc.M801296200. [DOI] [PubMed] [Google Scholar]
- 141.He X, et al. Cardiac CIP protein regulates dystrophic cardiomyopathy. Mol. Ther. 2022;30(2):898–914. doi: 10.1016/j.ymthe.2021.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kura B, et al. Oxidative Stress-Responsive MicroRNAs in Heart Injury. Int J Mol Sci. 2020;21(1):358. doi: 10.3390/ijms21010358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Windpassinger C, et al. An X-linked myopathy with postural muscle atrophy and generalized hypertrophy, termed XMPMA, is caused by mutations in FHL1. Am. J. Hum. Genet. 2008;82(1):88–99. doi: 10.1016/j.ajhg.2007.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Liang Y, et al. Four and a half LIM domain protein signaling and cardiomyopathy. Biophys. Rev. 2018;10(4):1073–1085. doi: 10.1007/s12551-018-0434-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Chu PH, et al. Expression patterns of FHL/SLIM family members suggest important functional roles in skeletal muscle and cardiovascular system. Mech. Dev. 2000;95(1–2):259–265. doi: 10.1016/S0925-4773(00)00341-5. [DOI] [PubMed] [Google Scholar]
- 146.Schessl J, et al. Clinical, histological and genetic characterization of reducing body myopathy caused by mutations in FHL1. Brain. 2009;132(Pt 2):452–464. doi: 10.1093/brain/awn325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Selcen D, et al. Reducing bodies and myofibrillar myopathy features in FHL1 muscular dystrophy. Neurology. 2011;77(22):1951–1959. doi: 10.1212/WNL.0b013e31823a0ebe. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.San Roman I, et al. Unclassifiable arrhythmic cardiomyopathy associated with Emery-Dreifuss caused by a mutation in FHL1. Clin. Genet. 2016;90(2):171–176. doi: 10.1111/cge.12760. [DOI] [PubMed] [Google Scholar]
- 149.Bang ML, et al. The muscle ankyrin repeat proteins CARP, Ankrd2, and DARP are not essential for normal cardiac development and function at basal conditions and in response to pressure overload. PLoS One. 2014;9(4):e93638. doi: 10.1371/journal.pone.0093638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Aihara Y, et al. Doxorubicin represses CARP gene transcription through the generation of oxidative stress in neonatal rat cardiac myocytes: Possible role of serine/threonine kinase-dependent pathways. J. Mol. Cell Cardiol. 2000;32(8):1401–1414. doi: 10.1006/jmcc.2000.1173. [DOI] [PubMed] [Google Scholar]
- 151.Wei YJ, et al. Upregulated expression of cardiac ankyrin repeat protein in human failing hearts due to arrhythmogenic right ventricular cardiomyopathy. Eur. J. Heart Fail. 2009;11(6):559–566. doi: 10.1093/eurjhf/hfp049. [DOI] [PubMed] [Google Scholar]
- 152.Zolk O, et al. Cardiac ankyrin repeat protein, a negative regulator of cardiac gene expression, is augmented in human heart failure. Biochem. Biophys. Res. Commun. 2002;293(5):1377–1382. doi: 10.1016/S0006-291X(02)00387-X. [DOI] [PubMed] [Google Scholar]
- 153.Duboscq-Bidot L, et al. Mutations in the ANKRD1 gene encoding CARP are responsible for human dilated cardiomyopathy. Eur. Heart J. 2009;30(17):2128–2136. doi: 10.1093/eurheartj/ehp225. [DOI] [PubMed] [Google Scholar]
- 154.Moulik M, et al. ANKRD1, the gene encoding cardiac ankyrin repeat protein, is a novel dilated cardiomyopathy gene. J. Am. Coll. Cardiol. 2009;54(4):325–333. doi: 10.1016/j.jacc.2009.02.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Kuo H, et al. Control of segmental expression of the cardiac-restricted ankyrin repeat protein gene by distinct regulatory pathways in murine cardiogenesis. Development. 1999;126(19):4223–4234. doi: 10.1242/dev.126.19.4223. [DOI] [PubMed] [Google Scholar]
- 156.Marsh T, Debnath J. Autophagy suppresses breast cancer metastasis by degrading NBR1. Autophagy. 2020;16(6):1164–1165. doi: 10.1080/15548627.2020.1753001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Bogomolovas J, et al. Induction of Ankrd1 in dilated cardiomyopathy correlates with the heart failure progression. Biomed. Res. Int. 2015;2015:273936. doi: 10.1155/2015/273936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Harris MP, et al. Perinatal versus adult loss of ULK1 and ULK2 distinctly influences cardiac autophagy and function. Autophagy. 2022;18(9):2161–2177. doi: 10.1080/15548627.2021.2022289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Radke MH, et al. Deleting full length titin versus the Titin M-band region leads to differential mechanosignaling and cardiac phenotypes. Circulation. 2019;139(15):1813–1827. doi: 10.1161/CIRCULATIONAHA.118.037588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Parlakian A, et al. Temporally controlled onset of dilated cardiomyopathy through disruption of the SRF gene in adult heart. Circulation. 2005;112(19):2930–2939. doi: 10.1161/CIRCULATIONAHA.105.533778. [DOI] [PubMed] [Google Scholar]
- 161.Miano JM. Serum response factor: Toggling between disparate programs of gene expression. J. Mol. Cell. Cardiol. 2003;35(6):577–593. doi: 10.1016/S0022-2828(03)00110-X. [DOI] [PubMed] [Google Scholar]
- 162.Kuwahara K, et al. Myocardin-related transcription factor A is a common mediator of mechanical stress-and neurohumoral stimulation-induced cardiac hypertrophic signaling leading to activation of brain natriuretic peptide gene expression. Mol. Cell. Biol. 2010;30(17):4134–4148. doi: 10.1128/MCB.00154-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Knoll R, Hoshijima M, Chien K. Cardiac mechanotransduction and implications for heart disease. J. Mol. Med. (Berl) 2003;81(12):750–756. doi: 10.1007/s00109-003-0488-x. [DOI] [PubMed] [Google Scholar]
- 164.Arber S, et al. MLP-deficient mice exhibit a disruption of cardiac cytoarchitectural organization, dilated cardiomyopathy, and heart failure. Cell. 1997;88(3):393–403. doi: 10.1016/S0092-8674(00)81878-4. [DOI] [PubMed] [Google Scholar]
- 165.Ehler E, Perriard JC. Cardiomyocyte cytoskeleton and myofibrillogenesis in healthy and diseased heart. Heart Fail. Rev. 2000;5(3):259–269. doi: 10.1023/A:1009861504264. [DOI] [PubMed] [Google Scholar]
- 166.Geier C, et al. Beyond the sarcomere: CSRP3 mutations cause hypertrophic cardiomyopathy. Hum. Mol. Genet. 2008;17(18):2753–2765. doi: 10.1093/hmg/ddn160. [DOI] [PubMed] [Google Scholar]
- 167.Karsai A, Kellermayer MS, Harris SP. Mechanical unfolding of cardiac myosin binding protein-C by atomic force microscopy. Biophys. J. 2011;101(8):1968–1977. doi: 10.1016/j.bpj.2011.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Maron BJ, Maron MS, Semsarian C. Genetics of hypertrophic cardiomyopathy after 20 years: Clinical perspectives. J. Am. Coll. Cardiol. 2012;60(8):705–715. doi: 10.1016/j.jacc.2012.02.068. [DOI] [PubMed] [Google Scholar]
- 169.Harris SP, et al. Hypertrophic cardiomyopathy in cardiac myosin binding protein-C knockout mice. Circ. Res. 2002;90(5):594–601. doi: 10.1161/01.RES.0000012222.70819.64. [DOI] [PubMed] [Google Scholar]
- 170.Agarkova I, Perriard J-C. The M-band: An elastic web that crosslinks thick filaments in the center of the sarcomere. Trends Cell Biol. 2005;15(9):477–485. doi: 10.1016/j.tcb.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 171.Forleo C, et al. Targeted next-generation sequencing detects novel gene-phenotype associations and expands the mutational spectrum in cardiomyopathies. PLoS One. 2017;12(7):e0181842. doi: 10.1371/journal.pone.0181842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Lange S, et al. Obscurin and KCTD6 regulate cullin-dependent small ankyrin-1 (sAnk1.5) protein turnover. Mol. Biol. Cell. 2012;23(13):2490–504. doi: 10.1091/mbc.e12-01-0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Krishnamoorthy S, et al. A novel phosphopeptide microarray based interactome map in breast cancer cells reveals phosphoprotein-GRB2 cell signaling networks. PLoS One. 2013;8(6):e67634. doi: 10.1371/journal.pone.0067634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Vatta M, et al. Mutations in Cypher/ZASP in patients with dilated cardiomyopathy and left ventricular non-compaction. J. Am. Coll. Cardiol. 2003;42(11):2014–2027. doi: 10.1016/j.jacc.2003.10.021. [DOI] [PubMed] [Google Scholar]
- 175.Hoshijima M. Mechanical stress-strain sensors embedded in cardiac cytoskeleton: Z disk, titin, and associated structures. Am. J. Physiol. Heart Circ. Physiol. 2006;290(4):H1313–H1325. doi: 10.1152/ajpheart.00816.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Pylayeva-Gupta Y, Grabocka E, Bar-Sagi D. RAS oncogenes: Weaving a tumorigenic web. Nat. Rev. Cancer. 2011;11(11):761–774. doi: 10.1038/nrc3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Sussman MA, et al. Altered focal adhesion regulation correlates with cardiomyopathy in mice expressing constitutively active rac1. J. Clin. Invest. 2000;105(7):875–886. doi: 10.1172/JCI8497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Yamaguchi K, et al. A spin correction procedure for unrestricted Hartree-Fock and Møller-Plesset wavefunctions for singlet diradicals and polyradicals. Chem. Phys. Lett. 1988;149(5–6):537–542. doi: 10.1016/0009-2614(88)80378-6. [DOI] [Google Scholar]
- 179.Wellbrock C, Hurlstone A. BRAF as therapeutic target in melanoma. Biochem. Pharmacol. 2010;80(5):561–567. doi: 10.1016/j.bcp.2010.03.019. [DOI] [PubMed] [Google Scholar]
- 180.Harris IS, et al. Raf-1 kinase is required for cardiac hypertrophy and cardiomyocyte survival in response to pressure overload. Circulation. 2004;110(6):718–723. doi: 10.1161/01.CIR.0000138190.50127.6A. [DOI] [PubMed] [Google Scholar]
- 181.Haywood NJ, et al. Hypertrophic cardiomyopathy mutations in the calponin-homology domain of ACTN2 affect actin binding and cardiomyocyte Z-disc incorporation. Biochem. J. 2016;473(16):2485–2493. doi: 10.1042/BCJ20160421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Girolami F, et al. Novel alpha-actinin 2 variant associated with familial hypertrophic cardiomyopathy and juvenile atrial arrhythmias: A massively parallel sequencing study. Circ. Cardiovasc. Genet. 2014;7(6):741–750. doi: 10.1161/CIRCGENETICS.113.000486. [DOI] [PubMed] [Google Scholar]
- 183.Prondzynski M, et al. Disease modeling of a mutation in alpha-actinin 2 guides clinical therapy in hypertrophic cardiomyopathy. EMBO Mol. Med. 2019;11(12):e11115. doi: 10.15252/emmm.201911115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Chiu C, et al. Mutations in alpha-actinin-2 cause hypertrophic cardiomyopathy: A genome-wide analysis. J. Am. Coll. Cardiol. 2010;55(11):1127–1135. doi: 10.1016/j.jacc.2009.11.016. [DOI] [PubMed] [Google Scholar]
- 185.Maiweilidan Y, Klauza I, Kordeli E. Novel interactions of ankyrins-G at the costameres: The muscle-specific Obscurin/Titin-Binding-related Domain (OTBD) binds plectin and filamin C. Exp. Cell Res. 2011;317(6):724–736. doi: 10.1016/j.yexcr.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 186.Song S, et al. Filamin C in cardiomyopathy: From physiological roles to DNA variants. Heart Fail. Rev. 2022;27(4):1373–1385. doi: 10.1007/s10741-021-10172-z. [DOI] [PubMed] [Google Scholar]
- 187.Bang ML, Chen J. Roles of nebulin family members in the heart. Circ. J. 2015;79(10):2081–2087. doi: 10.1253/circj.CJ-15-0854. [DOI] [PubMed] [Google Scholar]
- 188.Voelkel T, Linke WA. Conformation-regulated mechanosensory control via titin domains in cardiac muscle. Pflugers Arch. 2011;462(1):143–154. doi: 10.1007/s00424-011-0938-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Sheikh F, et al. An FHL1-containing complex within the cardiomyocyte sarcomere mediates hypertrophic biomechanical stress responses in mice. J. Clin. Invest. 2008;118(12):3870–3880. doi: 10.1172/JCI34472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.El-Bizri N, et al. Abstract 13402: FHL-1 contributes to and colocalizes with titin in cardiac hypertrophy. Circulation. 2020;142(Suppl_3):A13402–A13402. doi: 10.1161/circ.142.suppl_3.13402. [DOI] [Google Scholar]
- 191.Lange S, et al. The kinase domain of titin controls muscle gene expression and protein turnover. Science. 2005;308(5728):1599–1603. doi: 10.1126/science.1110463. [DOI] [PubMed] [Google Scholar]
- 192.Bogomolovas J, et al. Titin kinase ubiquitination aligns autophagy receptors with mechanical signals in the sarcomere. EMBO Rep. 2021;22(10):e48018. doi: 10.15252/embr.201948018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Bogomolovas J, et al. Titin kinase is an inactive pseudokinase scaffold that supports MuRF1 recruitment to the sarcomeric M-line. Open Biol. 2014;4(5):140041. doi: 10.1098/rsob.140041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Yang, K., et al., Clinical efficacy and safety of atorvastatin for chronic subdural hematoma: A randomized controlled trial.
- 195.Campuzano O, et al. Rare titin (TTN) variants in diseases associated with sudden cardiac death. Int. J. Mol. Sci. 2015;16(10):25773–25787. doi: 10.3390/ijms161025773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Evila A, et al. Targeted next-generation sequencing reveals novel TTN mutations causing recessive distal titinopathy. Mol. Neurobiol. 2017;54(9):7212–7223. doi: 10.1007/s12035-016-0242-3. [DOI] [PubMed] [Google Scholar]
- 197.Lahrouchi N, et al. Utility of post-mortem genetic testing in cases of sudden arrhythmic death syndrome. J. Am. Coll. Cardiol. 2017;69(17):2134–2145. doi: 10.1016/j.jacc.2017.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Hackman P, et al. Tibial muscular dystrophy is a titinopathy caused by mutations in TTN, the gene encoding the giant skeletal-muscle protein titin. Am. J. Hum. Genet. 2002;71(3):492–500. doi: 10.1086/342380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Herman DS, et al. Truncations of titin causing dilated cardiomyopathy. New Engl. J. Med. 2012;366(7):619–628. doi: 10.1056/NEJMoa1110186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Krenn M, et al. Genotype-guided diagnostic reassessment after exome sequencing in neuromuscular disorders: Experiences with a two-step approach. Eur. J. Neurol. 2020;27(1):51–61. doi: 10.1111/ene.14033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Harris E, et al. A ‘second truncation’in TTN causes early onset recessive muscular dystrophy. Neuromusc. Disord. 2017;27(11):1009–1017. doi: 10.1016/j.nmd.2017.06.013. [DOI] [PubMed] [Google Scholar]
- 202.Roberts AM, et al. Integrated allelic, transcriptional, and phenomic dissection of the cardiac effects of titin truncations in health and disease. Sci. Transl. Med. 2015;7(270):270ra6. doi: 10.1126/scitranslmed.3010134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Schultze-Berndt A, et al. Reduced systolic function and not genetic variants determine outcome in pediatric and adult left ventricular noncompaction cardiomyopathy. Front. Pediatr. 2021;9:722926. doi: 10.3389/fped.2021.722926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Chauveau C, et al. Recessive TTN truncating mutations define novel forms of core myopathy with heart disease. Hum. Mol. Genet. 2014;23(4):980–991. doi: 10.1093/hmg/ddt494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Haas J, et al. Atlas of the clinical genetics of human dilated cardiomyopathy. Eur. Heart J. 2015;36(18):1123–1135. doi: 10.1093/eurheartj/ehu301. [DOI] [PubMed] [Google Scholar]
- 206.De Cid R, et al. A new titinopathy: Childhood-juvenile onset Emery-Dreifuss–like phenotype without cardiomyopathy. Neurology. 2015;85(24):2126–2135. doi: 10.1212/WNL.0000000000002200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Deo RC. Alternative splicing, internal promoter, nonsense-mediated decay, or all three: explaining the distribution of truncation variants in titin. Circ. Cardiovasc. Genet. 2016;9(5):419–425. doi: 10.1161/CIRCGENETICS.116.001513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Mukhtar, M.M. and M.A. Salih, C-Terminal Titin Deletions Cause a Novel Early-Onset Myopathy with Fatal Cardiomyopathy. 2007, University of Khartoum. [DOI] [PubMed]
- 209.Savarese M, et al. Genotype-phenotype correlations in recessive titinopathies. Genet. Med. 2020;22(12):2029–2040. doi: 10.1038/s41436-020-0914-2. [DOI] [PubMed] [Google Scholar]
- 210.Ceyhan-Birsoy O, et al. Recessive truncating titin gene, TTN, mutations presenting as centronuclear myopathy. Neurology. 2013;81(14):1205–1214. doi: 10.1212/WNL.0b013e3182a6ca62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Fattori F, et al. Centronuclear myopathies: genotype-phenotype correlation and frequency of defined genetic forms in an Italian cohort. J. Neurol. 2015;262(7):1728–1740. doi: 10.1007/s00415-015-7757-9. [DOI] [PubMed] [Google Scholar]
- 212.Hackman, P., et al., Salih myopathy. 2019.
- 213.Evila A, et al. Atypical phenotypes in titinopathies explained by second titin mutations. Ann. Neurol. 2014;75(2):230–240. doi: 10.1002/ana.24102. [DOI] [PubMed] [Google Scholar]
- 214.Witting, N., et al., Phenotypes, genotypes, and prevalence of congenital myopathies older than 5 years in Denmark. Neurology Genetics, 2017. 3(2). [DOI] [PMC free article] [PubMed]
- 215.Rich KA, et al. Novel heterozygous truncating titin variants affecting the A-band are associated with cardiomyopathy and myopathy/muscular dystrophy. Mol. Genet. Genomic. Med. 2020;8(10):e1460. doi: 10.1002/mgg3.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Hackman P, et al. Truncating mutations in C-terminal titin may cause more severe tibial muscular dystrophy (TMD) Neuromuscul. Disord. 2008;18(12):922–928. doi: 10.1016/j.nmd.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 217.Marschall C, Moscu-Gregor A, Klein HG. Variant panorama in 1,385 index patients and sensitivity of expanded next-generation sequencing panels in arrhythmogenic disorders. Cardiovasc. Diagn. Ther. 2019;9(Suppl 2):S292–S298. doi: 10.21037/cdt.2019.06.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Laquerriere A, et al. Mutations in CNTNAP1 and ADCY6 are responsible for severe arthrogryposis multiplex congenita with axoglial defects. Hum. Mol. Genet. 2014;23(9):2279–2289. doi: 10.1093/hmg/ddt618. [DOI] [PubMed] [Google Scholar]
- 219.Enriquez A, et al. Substrate characterization and outcomes of ventricular tachycardia ablation in TTN (Titin) cardiomyopathy: A multicenter study. Circ. Arrhythm. Electrophysiol. 2021;14(9):e010006. doi: 10.1161/CIRCEP.121.010006. [DOI] [PubMed] [Google Scholar]
- 220.Perić S, et al. A novel recessive TTN founder variant is a common cause of distal myopathy in the Serbian population. Eur. J. Hum. Genet. 2017;25(5):572–581. doi: 10.1038/ejhg.2017.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Haskell GT, et al. Whole exome sequencing identifies truncating variants in nuclear envelope genes in patients with cardiovascular disease. Circ. Cardiovasc. Genet. 2017;10(3):e001443. doi: 10.1161/CIRCGENETICS.116.001443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Nykamp K, et al. Sherloc: A comprehensive refinement of the ACMG-AMP variant classification criteria. Genet. Med. 2017;19(10):1105–1117. doi: 10.1038/gim.2017.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Dalin MG, et al. Massive parallel sequencing questions the pathogenic role of missense variants in dilated cardiomyopathy. Int. J. Cardiol. 2017;228:742–748. doi: 10.1016/j.ijcard.2016.11.066. [DOI] [PubMed] [Google Scholar]
- 224.Walsh R, et al. Reassessment of Mendelian gene pathogenicity using 7,855 cardiomyopathy cases and 60,706 reference samples. Genet. Med. 2017;19(2):192–203. doi: 10.1038/gim.2016.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Choi SH, et al. Association between titin loss-of-function variants and early-onset atrial fibrillation. Jama. 2018;320(22):2354–2364. doi: 10.1001/jama.2018.18179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Taylor M, et al. Genetic variation in titin in arrhythmogenic right ventricular cardiomyopathy–overlap syndromes. Circulation. 2011;124(8):876–885. doi: 10.1161/CIRCULATIONAHA.110.005405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Ware JS, et al. Shared genetic predisposition in peripartum and dilated cardiomyopathies. N. Engl. J. Med. 2016;374(3):233–241. doi: 10.1056/NEJMoa1505517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Gigli M, et al. Genetic risk of arrhythmic phenotypes in patients with dilated cardiomyopathy. J. Am. Coll. Cardiol. 2019;74(11):1480–1490. doi: 10.1016/j.jacc.2019.06.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Goli R, et al. Genetic and phenotypic landscape of peripartum cardiomyopathy. Circulation. 2021;143(19):1852–1862. doi: 10.1161/CIRCULATIONAHA.120.052395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Sevy A, et al. Improving molecular diagnosis of distal myopathies by targeted next-generation sequencing. J. Neurol. Neurosurg. Psychiatry. 2016;87(3):340–342. doi: 10.1136/jnnp-2014-309663. [DOI] [PubMed] [Google Scholar]
- 231.Pfeffer, G. and P.F. Chinnery, Hereditary myopathy with early respiratory failure. GeneReviews®[Internet], 2020.
- 232.Augusto JB, et al. Dilated cardiomyopathy and arrhythmogenic left ventricular cardiomyopathy: A comprehensive genotype-imaging phenotype study. Eur. Heart J. Cardiovasc. Imaging. 2020;21(3):326–336. doi: 10.1093/ehjci/jez188. [DOI] [PubMed] [Google Scholar]
- 233.van Waning JI, et al. Genetics, clinical features, and long-term outcome of noncompaction cardiomyopathy. J. Am. Coll. Cardiol. 2018;71(7):711–722. doi: 10.1016/j.jacc.2017.12.019. [DOI] [PubMed] [Google Scholar]
- 234.Norton N, et al. Exome sequencing and genome-wide linkage analysis in 17 families illustrate the complex contribution of TTN truncating variants to dilated cardiomyopathy. Circ. Cardiovasc. Genet. 2013;6(2):144–153. doi: 10.1161/CIRCGENETICS.111.000062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Cowan JR, et al. SOS1 gain-of-function variants in dilated cardiomyopathy. Circ. Genom. Precis. Med. 2020;13(4):e002892. doi: 10.1161/CIRCGEN.119.002892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Kostareva A, et al. Genetic spectrum of idiopathic restrictive cardiomyopathy uncovered by next-generation sequencing. PLoS One. 2016;11(9):e0163362. doi: 10.1371/journal.pone.0163362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Song JS, et al. Identification of pathogenic variants in genes related to channelopathy and cardiomyopathy in Korean sudden cardiac arrest survivors. J. Hum. Genet. 2017;62(6):615–620. doi: 10.1038/jhg.2017.8. [DOI] [PubMed] [Google Scholar]
- 238.Gerull B, et al. Identification of a novel frameshift mutation in the giant muscle filament titin in a large Australian family with dilated cardiomyopathy. J. Mol. Med. (Berl) 2006;84(6):478–483. doi: 10.1007/s00109-006-0060-6. [DOI] [PubMed] [Google Scholar]
- 239.Verdonschot JAJ, et al. Implications of genetic testing in dilated cardiomyopathy. Circ. Genom. Precis. Med. 2020;13(5):476–487. doi: 10.1161/CIRCGEN.120.003031. [DOI] [PubMed] [Google Scholar]
- 240.Herkert, J.C., Paediatric cardiomyopathies: an evolving landscape of genetic aetiology and diagnostic applications. 2019.
- 241.Vissing CR, et al. Dilated cardiomyopathy caused by truncating titin variants: Long-term outcomes, arrhythmias, response to treatment and sex differences. J. Med. Genet. 2021;58(12):832–841. doi: 10.1136/jmedgenet-2020-107178. [DOI] [PubMed] [Google Scholar]
- 242.Morales A, et al. Variant interpretation for dilated cardiomyopathy: Refinement of the american college of medical genetics and genomics/clingen guidelines for the DCM precision medicine study. Circ. Genom. Precis. Med. 2020;13(2):e002480. doi: 10.1161/CIRCGEN.119.002480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Akinrinade O, et al. Genetics and genotype–phenotype correlations in Finnish patients with dilated cardiomyopathy. Eur. Heart J. 2015;36(34):2327–2337. doi: 10.1093/eurheartj/ehv253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Klauke B, et al. High proportion of genetic cases in patients with advanced cardiomyopathy including a novel homozygous Plakophilin 2-gene mutation. PLoS One. 2017;12(12):e0189489. doi: 10.1371/journal.pone.0189489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Franaszczyk M, et al. Titin truncating variants in dilated cardiomyopathy–prevalence and genotype-phenotype correlations. PLoS One. 2017;12(1):e0169007. doi: 10.1371/journal.pone.0169007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Kuhnisch J, et al. Targeted panel sequencing in pediatric primary cardiomyopathy supports a critical role of TNNI3. Clin. Genet. 2019;96(6):549–559. doi: 10.1111/cge.13645. [DOI] [PubMed] [Google Scholar]
- 247.Hancks DC, Kazazian HH., Jr Active human retrotransposons: Variation and disease. Curr. Opin. Genet. Dev. 2012;22(3):191–203. doi: 10.1016/j.gde.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Chami N, et al. Nonsense mutations in BAG3 are associated with early-onset dilated cardiomyopathy in French Canadians. Can. J. Cardiol. 2014;30(12):1655–1661. doi: 10.1016/j.cjca.2014.09.030. [DOI] [PubMed] [Google Scholar]
- 249.Al-Shamsi A, et al. Whole exome sequencing diagnosis of inborn errors of metabolism and other disorders in United Arab Emirates. Orphanet. J. Rare Dis. 2016;11(1):94. doi: 10.1186/s13023-016-0474-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.LaDuca H, et al. Exome sequencing covers >98% of mutations identified on targeted next generation sequencing panels. PLoS One. 2017;12(2):e0170843. doi: 10.1371/journal.pone.0170843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Anderson JL, et al. Discovery of TITIN Gene truncating variant mutations and 5-Year outcomes in patients with nonischemic dilated cardiomyopathy. Am. J. Cardiol. 2020;137:97–102. doi: 10.1016/j.amjcard.2020.09.026. [DOI] [PubMed] [Google Scholar]
- 252.Cuenca S, et al. Genetic basis of familial dilated cardiomyopathy patients undergoing heart transplantation. J. Heart Lung Transplant. 2016;35(5):625–635. doi: 10.1016/j.healun.2015.12.014. [DOI] [PubMed] [Google Scholar]
- 253.Kolokotronis K, et al. New insights on genetic diagnostics in cardiomyopathy and arrhythmia patients gained by stepwise exome data analysis. J. Clin. Med. 2020;9(7):2168. doi: 10.3390/jcm9072168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Minoche AE, et al. Genome sequencing as a first-line genetic test in familial dilated cardiomyopathy. Genet. Med. 2019;21(3):650–662. doi: 10.1038/s41436-018-0084-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Fokstuen S, et al. Experience of a multidisciplinary task force with exome sequencing for Mendelian disorders. Hum. Genomics. 2016;10(1):24. doi: 10.1186/s40246-016-0080-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Campuzano Larrea O, et al. Post-mortem genetic analysis in juvenile cases of sudden cardiac death. Forensic Sci. Int. 2014;245:30–37. doi: 10.1016/j.forsciint.2014.10.004. [DOI] [PubMed] [Google Scholar]
- 257.van Lint FHM, et al. Large next-generation sequencing gene panels in genetic heart disease: Yield of pathogenic variants and variants of unknown significance. Neth. Heart J. 2019;27(6):304–309. doi: 10.1007/s12471-019-1250-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Yang Y, et al. Molecular findings among patients referred for clinical whole-exome sequencing. Jama. 2014;312(18):1870–1879. doi: 10.1001/jama.2014.14601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Mazzarotto F, et al. Reevaluating the genetic contribution of monogenic dilated cardiomyopathy. Circulation. 2020;141(5):387–398. doi: 10.1161/CIRCULATIONAHA.119.037661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Cowan JR, et al. Multigenic disease and bilineal inheritance in dilated cardiomyopathy is illustrated in nonsegregating LMNA pedigrees. Circ. Genomic Precis. Med. 2018;11(7):e002038. doi: 10.1161/CIRCGEN.117.002038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Brown EE, et al. Genetic dilated cardiomyopathy due to TTN variants without known familial disease. Circ. Genomic Precis. Med. 2020;13(6):e003082. doi: 10.1161/CIRCGEN.120.003082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Ceyhan-Birsoy O, et al. Next generation sequencing-based copy number analysis reveals low prevalence of deletions and duplications in 46 genes associated with genetic cardiomyopathies. Mol. Genet. Genomic Med. 2016;4(2):143–151. doi: 10.1002/mgg3.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Peat RA, et al. Diagnosis and etiology of congenital muscular dystrophy. Neurology. 2008;71(5):312–321. doi: 10.1212/01.wnl.0000284605.27654.5a. [DOI] [PubMed] [Google Scholar]
- 264.Lu C, et al. Molecular analysis of inherited cardiomyopathy using next generation semiconductor sequencing technologies. J. Transl. Med. 2018;16(1):241. doi: 10.1186/s12967-018-1605-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Bryen SJ, et al. Recurrent TTN metatranscript-only c.39974–11T>G splice variant associated with autosomal recessive arthrogryposis multiplex congenita and myopathy. Hum. Mutat. 2020;41(2):403–411. doi: 10.1002/humu.23938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Cummings BB, et al. Improving genetic diagnosis in Mendelian disease with transcriptome sequencing. Sci. Transl. Med. 2017;9(386):eaal5209. doi: 10.1126/scitranslmed.aal5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Hershberger, R.E., Exome Sequencing and Genome-Wide Linkage Analysis in 17 Families Illustrates the Complex Contribution of TTN Truncating Variants to Dilated Cardiomyopathy. [DOI] [PMC free article] [PubMed]
- 268.Hazebroek MR, et al. Prevalence of pathogenic gene mutations and prognosis do not differ in isolated left ventricular dysfunction compared with dilated cardiomyopathy. Circ. Heart Fail. 2018;11(3):e004682. doi: 10.1161/CIRCHEARTFAILURE.117.004682. [DOI] [PubMed] [Google Scholar]
- 269.Hoorntje ET, et al. The first titin (c.59926 + 1G > A) founder mutation associated with dilated cardiomyopathy. Eur. J. Heart Fail. 2018;20(4):803–806. doi: 10.1002/ejhf.1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Miszalski-Jamka K, et al. Novel genetic triggers and genotype-phenotype correlations in patients with left ventricular noncompaction. Circ. Cardiovasc. Genet. 2017;10(4):e001763. doi: 10.1161/CIRCGENETICS.117.001763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Fan LL, et al. Whole-exome sequencing reveals doubly novel heterozygous Myosin Binding Protein C and Titin mutations in a Chinese patient with severe dilated cardiomyopathy. Cardiol. Young. 2018;28(12):1410–1414. doi: 10.1017/S1047951118001403. [DOI] [PubMed] [Google Scholar]
- 272.Jansweijer JA, et al. Truncating titin mutations are associated with a mild and treatable form of dilated cardiomyopathy. Eur. J. Heart Fail. 2017;19(4):512–521. doi: 10.1002/ejhf.673. [DOI] [PubMed] [Google Scholar]
- 273.Wu L, et al. Next-generation sequencing to diagnose muscular dystrophy, rhabdomyolysis, and HyperCKemia. Can. J. Neurol. Sci. 2018;45(3):262–268. doi: 10.1017/cjn.2017.286. [DOI] [PubMed] [Google Scholar]
- 274.Yavarna T, et al. High diagnostic yield of clinical exome sequencing in Middle Eastern patients with Mendelian disorders. Hum. Genet. 2015;134(9):967–980. doi: 10.1007/s00439-015-1575-0. [DOI] [PubMed] [Google Scholar]
- 275.Tester DJ, et al. Cardiac genetic predisposition in sudden infant death syndrome. J. Am. Coll. Cardiol. 2018;71(11):1217–1227. doi: 10.1016/j.jacc.2018.01.030. [DOI] [PubMed] [Google Scholar]
- 276.Carnevale A, et al. Genomic study of dilated cardiomyopathy in a group of Mexican patients using site-directed next generation sequencing. Mol. Genet. Genomic. Med. 2020;8(11):e1504. doi: 10.1002/mgg3.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Cambon-Viala M, et al. Phenotype/genotype relationship in left ventricular noncompaction: Ion channel gene mutations are associated with preserved left ventricular systolic function and biventricular noncompaction: Phenotype/genotype of noncompaction. J. Card. Fail. 2021;27(6):677–681. doi: 10.1016/j.cardfail.2021.01.007. [DOI] [PubMed] [Google Scholar]
- 278.Bell CJ, et al. Carrier testing for severe childhood recessive diseases by next-generation sequencing. Sci. Transl. Med. 2011;3(65):65ra4. doi: 10.1126/scitranslmed.3001756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Willsey AJ, et al. De Novo coding variants are strongly associated with tourette disorder. Neuron. 2017;94(3):486–499 e9. doi: 10.1016/j.neuron.2017.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Arimura T, et al. Cardiac ankyrin repeat protein gene (ANKRD1) mutations in hypertrophic cardiomyopathy. J. Am. College Cardiol. 2009;54(4):334–342. doi: 10.1016/j.jacc.2008.12.082. [DOI] [PubMed] [Google Scholar]
- 281.Liu JS, et al. Whole-exome sequencing identifies two novel TTN mutations in Chinese families with dilated cardiomyopathy. Cardiology. 2017;136(1):10–14. doi: 10.1159/000447422. [DOI] [PubMed] [Google Scholar]
- 282.Posey JE, et al. Resolution of disease phenotypes resulting from multilocus genomic variation. N. Engl. J. Med. 2017;376(1):21–31. doi: 10.1056/NEJMoa1516767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Golbus JR, et al. Targeted analysis of whole genome sequence data to diagnose genetic cardiomyopathy. Circ. Cardiovasc. Genet. 2014;7(6):751–759. doi: 10.1161/CIRCGENETICS.113.000578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Campuzano O, et al. Post-mortem genetic analysis in juvenile cases of sudden cardiac death. Forensic Sci. Int. 2014;245:30–37. doi: 10.1016/j.forsciint.2014.10.004. [DOI] [PubMed] [Google Scholar]
- 285.Gerull B, et al. Mutations of TTN, encoding the giant muscle filament titin, cause familial dilated cardiomyopathy. Nat. Genet. 2002;30(2):201–204. doi: 10.1038/ng815. [DOI] [PubMed] [Google Scholar]
- 286.Hastings R, et al. Combination of whole genome sequencing, linkage, and functional studies implicates a missense mutation in titin as a cause of autosomal dominant cardiomyopathy with features of left ventricular noncompaction. Circ. Cardiovasc. Genet. 2016;9(5):426–435. doi: 10.1161/CIRCGENETICS.116.001431. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and/or analyzed during the current study are available in the the public archive of interpretations of clinically relevant variants (ClinVar) repository, (https://www.ncbi.nlm.nih.gov/clinvar/?term=TTN%5Bgene%5D&redir=gene).