Abstract
Genetic studies have suggested that chromatin structure is involved in repression of the silent mating type loci in Saccharomyces cerevisiae. Chromatin mapping at nucleotide resolution of the transcriptionally silent HMLα and the active MATα shows that unique organized chromatin structure characterizes the silent state of HMLα. Precisely positioned nucleosomes abutting the silencers extend over the α1 and α2 coding regions. The HO endonuclease recognition site, nuclease hypersensitive at MATα, is protected at HMLα. Although two precisely positioned nucleosomes incorporate transcription start sites at HMLα, the promoter region of the α1 and α2 genes is nucleosome free and more nuclease sensitive in the repressed than in the transcribed locus. Mutations in genes essential for HML silencing disrupt the nucleosome array near HML-I but not in the vicinity of HML-E, which is closer to the telomere of chromosome III. At the promoter and the HO site, the structure of HMLα in Sir protein and histone H4 N-terminal deletion mutants is identical to that of the transcriptionally active MATα. The discontinuous chromatin structure of HMLα contrasts with the continuous array of nucleosomes found at repressed a-cell-specific genes and the recombination enhancer. Punctuation at HMLα may be necessary for higher-order structure or karyoskeleton interactions. The unique chromatin architecture of HMLα may relate to the combined requirements of transcriptional repression and recombinational competence.
Transcriptional repression of the silent-mating-type loci is fundamental for the haploid yeast life cycle. The a or α mating type is determined by expression of master regulatory genes of the active MAT locus near the centromere of chromosome III. Identical genes present at the HM (haploid mating) loci near the telomeres of the same chromosome, HML carrying α information and HMR bearing a information, are not transcribed, thus preserving the unique mating type. The HM loci serve as donors during the gene interconversion event that allows a homothallic haploid cell to switch mating type, ensuring a diploid population in the wild. In addition to transcriptional repression, the DNA of the silenced HM loci is protected from HO endonuclease, which makes a double-strand break at the MAT HO site to initiate mating-type switching (40, 46, 55, 75).
Silencing of the HM-mating-type loci in Saccharomyces cerevisiae is remarkably similar to long-term, epigenetic inactivation of specific genomic domains in complex eukaryotes. X-chromosome inactivation (33) and gene imprinting (82) in mammalian cells, telomeric silencing (22) in yeast, and position effect variegation (reviewed by Henikoff [28]) in Drosophila melanogaster are examples of such parallel situations. Epigenetic states are thought to be achieved by chromosomal condensation into heterochromatin. The molecular events leading to such position-dependent, gene-independent transcriptional repression of a chromosomal region are not well understood. Silencing mechanisms in yeast are likely to involve a repressive chromatin structure equivalent to that of heterochromatin (87). The genetics of yeast silencing have been intensively studied (for a review, see reference 45), and a large number of cis-DNA elements and trans-acting proteins involved in the establishment and maintenance of repression at the silent-mating-type loci have been identified. Studies of histone mutations and modifications are consistent with the involvement of chromatin organization in gene silencing (34, 35, 50, 58). However, chromatin structure at the silent mating type loci has not been analyzed in detail.
Two cis-acting elements are necessary for repression at the silent-mating-type loci. The E and I silencers, which flank both HMR and HML are essential or important for silencing (1, 7, 47, 68). Each silencer consists of either a Rap1p and/or an Abf1p binding site (9, 71) and a binding site for the origin recognition complex (ORC), termed an autonomously replicating sequence consensus site (ACS) (4). The silencers of both the HMR and HML loci are functionally similar, yet their efficiencies in conferring gene silencing are different (69), and functional cooperativity between two distant silencers can enhance repression (6). Transcriptional repression of the genes located between the E and I silencers is independent of their sequence, chromosomal origin, and orientation. Transplacement of the mating-type genes outside the silent locus causes activation of their transcription, while heterologous RNA polymerase II or III genes inserted into the HM loci become silenced (7, 29, 68).
Among the trans-acting factors, the four Sir (silent information regulator) proteins, initially identified by genetic screens for loss of repression at the HM loci (25, 39, 63, 64), function without directly binding to DNA. Null mutations of sir2, sir3, and sir4 result in complete derepression of the HM loci, whereas only partial derepression was observed in the absence of Sir1p (31, 63). Sir1p binds to the Orc1p subunit of ORC, which binds the ACS of the silencers. A role for Sir1p in the establishment of silencing via binding to ORC was suggested (12, 89). While passage through S phase is required for the establishment of silencing, the role of ORC is independent of replication initiation at the silencers (18). Sir3p and Sir4p have been shown to form homo- and heterodimers in vivo and also to interact with the carboxy-terminal region of Rap1p in vitro (23, 51, 52). ORC and at least one of Rap1p or Abf1p bind to the silencer (9, 15). Subsequently, Sir3p and Sir4p can be tethered to the silencers by virtue of their interactions with the initiation complex and establish a matrix (24) which could support a repressive chromatin structure across the entire locus. H3 and H4 N-terminal tail interactions with Sir3p and Sir4p (26, 35) are consistent with this hypothesis. In addition, the histone H4 amino-terminal regions are indispensable for HM silencing (35, 58, 88). Overexpressed Sir3p has been shown by immunoprecipitation to physically spread in a histone H4-dependent manner as far as silencing extends both at the subtelomeric regions and at the HM loci (27). The extent of silencing can be correlated to the level of overexpression of Sir3p. Although Sir4p and Rap1p are required for that effect at telomeres (62, 78), their relative contributions to the postulated repressive structure at HMLα or whether they spread in association with Sir3p remains unclear.
Mutations of N-terminal tails of histone H3 alone have little effect on repression at the HM loci but appear to increase the severity of other mutations that affect silencing. These amino-terminal regions of the core histones are known to be sites for posttranslational modification by histone acetyltransferases, and nucleosomes of silent regions are hypoacetylated, similar to the histones in inactive chromatin from metazoan organisms (11, 91). SIR2, a protein involved in HM silencing, promotes deacetylation of histones, an activity which is characteristic of repressed chromatin (8). Other genes, such as NAT1, ARD1, SAS2, and SAS3, with predicted protein products bearing similarities to acetyltransferases also contribute to HM silencing (54, 60, 94). NAT1 and ARD1 are N-terminal acetylases with a different function from histone acetyltransferases. Their role in silencing is likely to be indirect; nearly 20% of yeast proteins have altered isoelectric points in a nat1 background, suggesting that many diverse effects could arise from mutations in these genes.
Derepression of the silent mating type loci leads to transcription of the a1 and a2 or α1 and α2 genes from HMRa or HMLα, respectively, as well as cleavage by HO endonuclease at its site located near the 3′ end of the a1 and α1 coding sequences. Taken together, these studies suggest a correlation between a specific chromatin configuration and this transcriptional repression at HMLα and HMRa. The only previous study to address this suggestion at HMLα showed that the HO endonuclease recognition site of the HML locus was more accessible to DNase I cleavage in sir mutants than in wild type, as might have been expected from its differential accessibility to the HO endonuclease in the two genetic backgrounds (55). To directly examine the role of chromatin structure in silencing, we have performed a high-resolution analysis of the chromatin organization of ∼4 kb of yeast chromosome III, which includes the silenced HMLα region. In addition, we compare this structure with that of the active MATα locus. Finally, we focus on modifications of chromatin structure at HMLα consequent to various null mutants of the SIR protein genes and an H4 amino-terminal-region deletion.
MATERIALS AND METHODS
S. cerevisiae strains, generously provided by J. E. Haber, were all derivatives of DBY745 (S288C): tNR (HMLα matΔ::LEU2 hmrΔ::ADE1 lys5 leu2 ura3 trp1) (“HMLα only”); JKM115 (hmlΔ::ADE1 MATα hmrΔ::ADE1 lys5 leu2 ura3 trp1) (“MATα only”); K30 (ho MATα leu2 trp1 his4 ura3); and 23-Δ2 (ho mat Δ200 bp [a-like] leu2 trp1 his4 ura3). The mat deletion is between his mutations αx109 αx52 and was originally constructed by K. Tatchell (86) and by S. Y. Roth: PKY913 (MATα Δhhf1::HIS3 Δhhf2::LEU2, pUK613[hhf2 del(4-23)], lys2 leu2 ura3 trp1 ade2 arg4 his3 thr4 tyr) (35). The strains constructed during this study were YKW01 (tNR sir1::URA3), YKW03 (tNR sir3::URA3), and YKW04 (tNR sir4::URA3). They were propagated in yeast extract-peptone-dextrose (YEPD) complete medium. Mating ability was tested by mating tester strains, MATa or MATα ura2, with the strain to be assessed on YEPD plates for 16 h, and subsequently selecting for growth on minimal medium (SD) plates.
The SIR1, SIR3, and SIR4 genes of the HMLα-only strain (tNR) were replaced by standard methods (65) with the URA3 gene by transformation with the linearized disruption plasmids D1528 (77), pSR-sir4 (61), and pCTC73 (12), which were generously provided by D. Shore and S. Reimer. Transformation was assessed by uracil prototrophy. sir3 and sir4 mutants were screened for their ability to mate with a cells. The wild-type tNR cells mate with α cells. sir1 mutants were screened for their ability to mate with both cell types. The disruption of SIR1, SIR3, and SIR4 was verified by PCR analysis.
Yeast were grown in YEPD at 30°C to mid-log phase (optical density at 600 nm of ∼1). Nuclei were isolated and then digested with micrococcal nuclease or DNase I (Worthington); DNA was purified as described previously (67, 85) with modifications as detailed by Weiss and Simpson (93). Protein-free DNA controls were obtained by either digesting purified, previously undigested DNA with a 50-fold-lower concentration of enzyme or by digesting a PCR product. Portions (4.4 kb) of the sequences including HMLα were amplified with oligonucleotides p108 and q152 (see below) as primers. PCR product (∼100 ng) was digested with 1.0 U of MNase or 0.05 U of DNase I per ml at 37°C for 3 min in the presence of 36 μg of carrier DNA (calf thymus). After ethanol precipitation, DNA was resuspended in 50 μl of 0.1× TE buffer.
MNase and DNase I cleavage sites were located by primer extension assay with Taq polymerase as described previously (70) with minor modifications (93). Oligonucleotides used as primers included (coordinates are base pair positions in the published sequence of S. cerevisiae chromosome III (56): p108, 10830–10855; p111, 11107–11134; p129, 12874–12895; p134, 13362–13386; p136, 13654–13673; p140, 13984–14013; q152, 15213–15187; q140, 14043–14017; q134, 13386–13362; q123, 12306–12283; q120, 12030–12008; q113, 11388–11367; and q1999, 199946–199916.
RESULTS
Unique organized chromatin domain at HMLα.
A high-resolution map of the chromatin structure of an ∼4-kb domain spanning HMLα was established by using primer extension analysis of micrococcal nuclease digests of isolated nuclei. Nuclease cutting patterns of the silent HMLα locus were compared to the pattern of identical sequences of the transcribed MATα locus in regions where they overlap as well as to nuclease digests of protein-free DNA. Nucleosome positions are inferred from areas of nuclease protection extending about 150 bp which are flanked by nuclease-sensitive cleavage sites. Due to the sequence identity of portions of the three mating-type loci, strains with deletions of MAT and HMR (i.e., the “HMLα only” strain) or HML and HMR (i.e., the “MATα only” strain) were used to create unique primer extension sites at HML and MAT, respectively. The inferred chromatin structure of HMLα in a wild-type background is summarized in Fig. 1.
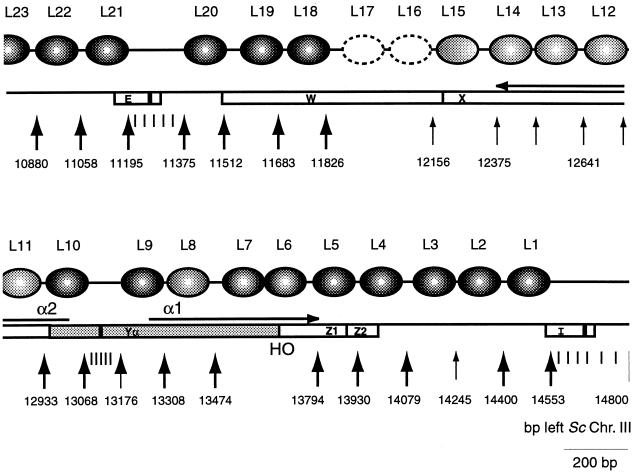
FIG. 1.
Schematic representation of the chromatin map of the entire HMLα locus. Map units correspond to base-pair positions of the published sequence of chromosome III (56). White boxes labeled E and I identify the silencer sequences; boxes labeled W, X, Yα, Z1, and Z2 identify the mating-type-locus regions. Black arrowheads identify sites that are hypersensitive to micrococcal nuclease; tick marks correspond to regions generally sensitive to nuclease cleavage and detailed in other figures. The black rectangles indicate the Rap1p binding sites. Dark-shaded ellipses indicate precisely positioned nucleosomes. Light-gray ellipses indicate more loosely positioned nucleosomes, and dashed ellipses indicate less-defined chromatin structure of the W region. The α1 and α2 coding regions are identified by arrows.
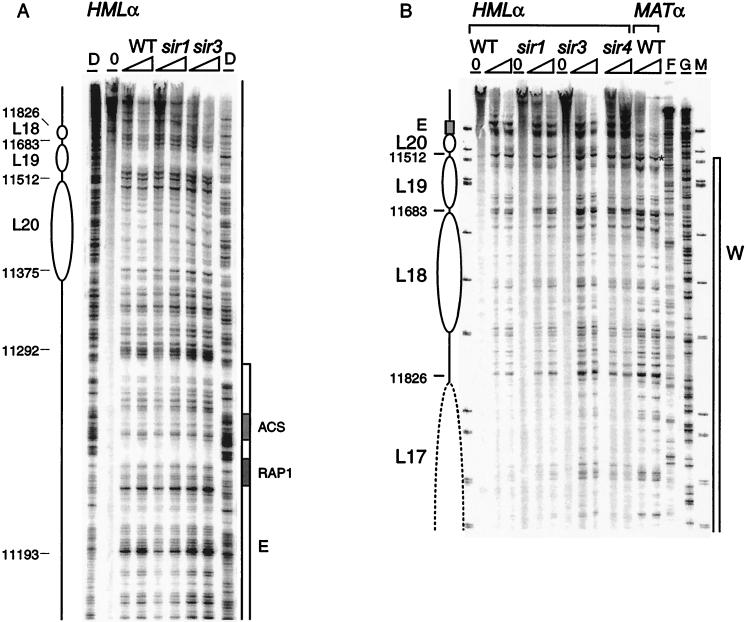
The entire chromatin domain between the E and I silencers, at positions 11352 to 14553, is organized into 20 nucleosomes, most of which are precisely defined in their location (Fig. 1). The critical cis-acting DNA elements that flank HMLα have a distinctive digestion pattern comprised of hypersensitive regions and protected regions. The protected regions correspond to the binding site for Rap1p and the ACS at the E silencer (see Fig. 9) and the binding site for Abf1p and the ACS at the I silencer when mapped at high resolution (data not shown).
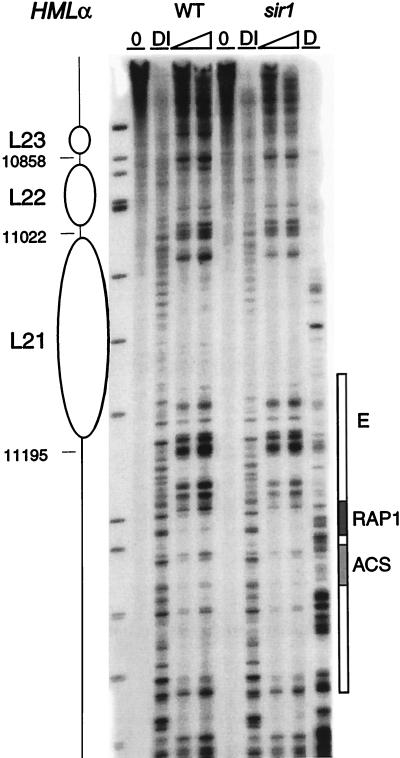
FIG. 9.
Chromatin structure near the E silencer outside HMLα in wild type and the sir1 mutant. The chromatin structure of the Watson strand was mapped by primer extension analysis of micrococcal nuclease cleavage sites with primer q113. Wild-type (WT) and sir1 mutant HMLα are as indicated. Symbols are as detailed in the legend to Fig. 2. Column DI shows extensions of DNase I-digested chromatin. Shaded boxes identify the Rap1p binding site and the ACS of the E silencer.
Internal to the I silencer, nine precisely located nucleosomes are present, extending through the α1 coding region and thus including the HO endonuclease recognition site at 13689 (Fig. 2 to 5). Immediately adjacent to the I element, the nucleosome array appears to be tightly packed (Fig. 2 and 3). Two nucleosomes (L4 and L5) protect the Z1 and Z2 regions (Fig. 3), and three more nucleosomes (L1 to L3) are accommodated between Z2 and the I silencer (Fig. 2). Precision of organization of this region is equivalent to that seen at other highly organized yeast chromatin domains, such as the recombination enhancer (93) and repressed a-cell specific genes (59, 90).
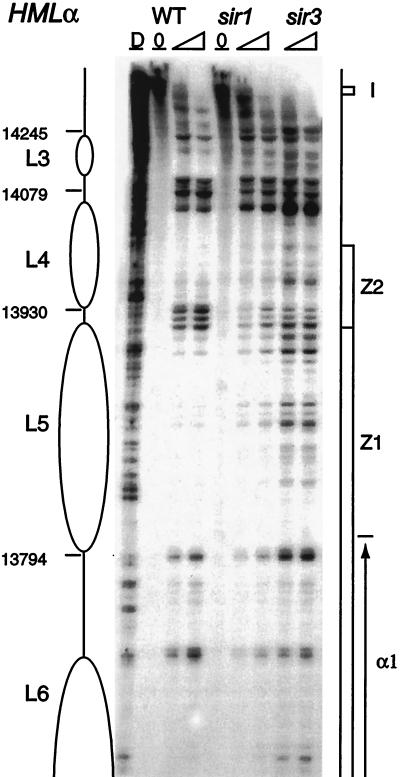
FIG. 2.
HMLα chromatin near the I silencer. The chromatin structure of the Crick strand was mapped by primer extension analysis of micrococcal nuclease cleavage sites with primer p140. Wild-type (WT) and sir1 and sir3 mutant cells are as indicated. Extensions of undigested (0) and micrococcal-nuclease-digested chromatin are also presented. The D columns indicate protein-free DNA digests as a control for micrococcal-nuclease sequence specificity. The C column shows a dideoxycytosine-terminated sequencing reaction. Coordinates are positions in the published sequence of S. cerevisiae chromosome III. The silencer is represented by a shaded rectangle. Ellipses correspond to inferred positions of nucleosomes.
FIG. 5.
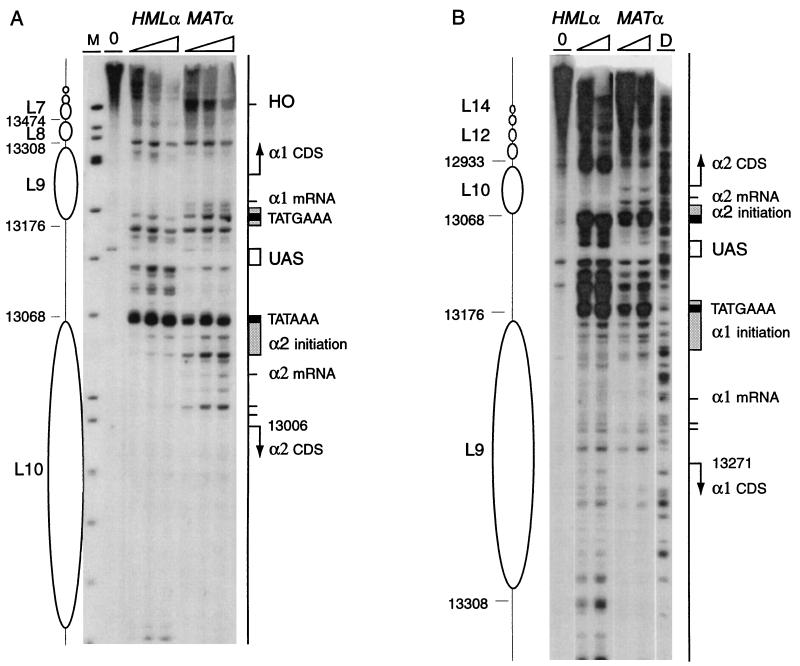
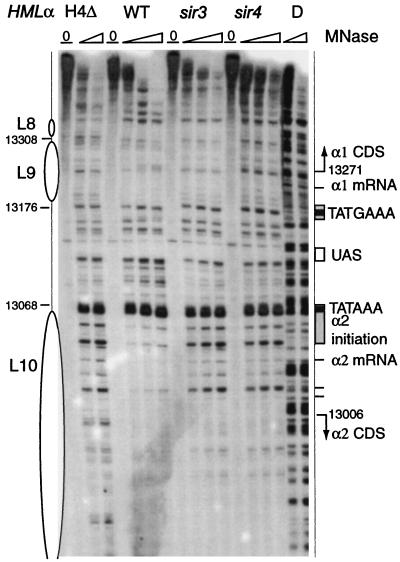
Chromatin structure of the α1 and α2 promoter region in HMLα and MATα. The chromatin structure was mapped by primer extension analysis of micrococcal nuclease cleavage sites with primer p129 (Crick strand) (A) and q134 (Watson strand) (B). Wild-type MATα and HMLα cells are as indicated. Symbols are as detailed in the legend to Fig. 2. The M column shows ΦX174/HinfI-digested DNA fragments for size indication. Sequence numbers shown correspond to those of HMLα and differ by 185,899 from the corresponding nucleotide in MATα. Shaded rectangles locate sequences necessary for transcription initiation; black rectangles indicate TATA elements, and the white box shows the shared UAS. Tick marks indicate points of transcription initiation of the α1 and α2 genes, and arrows show their coding sequences (CDS).
FIG. 3.
HMLα chromatin of the region between Z1 and the I silencer. The chromatin structure of the Crick strand was mapped by primer extension analysis of micrococcal nuclease cleavage sites with primer p136. Wild-type (WT) and sir1 and sir3 mutant cells are as indicated. Symbols are as detailed in the legend to Fig. 2. Rectangular boxes indicate the locations of mating-type-locus regions. The 3′ end of the α1 gene is indicated by an arrow.
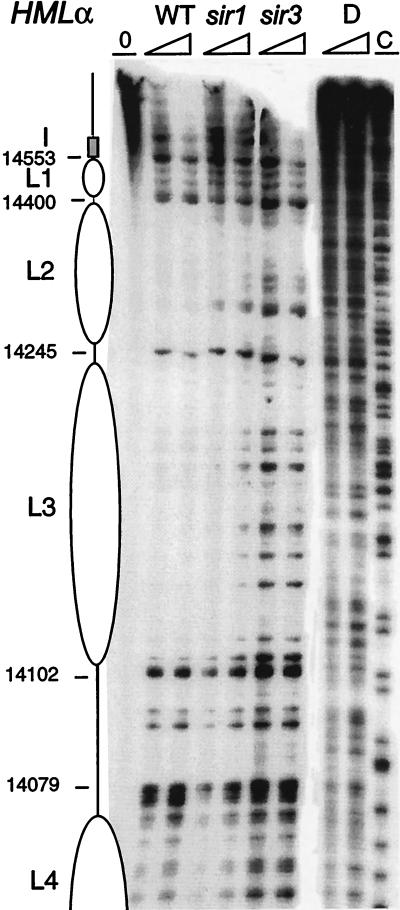
The creation of a double-strand break at the active MAT locus by the HO endonuclease initiates recombination of MAT with one of the HM loci during mating-type switching (43). Protection of the HO endonuclease recognition sequence at the Yα-Z1 border at HMLα (80) is essential for survival of the cell. Indeed, at the Yα-Z1 border two nuclease-hypersensitive sites exist at the active MATα (Fig. 4). Several sites flanking the HO site are also rather sensitive to nuclease cleavage. This region comprising the HO site is largely protected from nuclease cleavage at the silent HMLα. The area of protection around the HO site spans 316 bp from positions 13474 to 13789. While this could reflect binding of another, unknown trans-acting factor or protein complex that blocks access of HO to its cognate site in the silenced locus, it could equally well result from two closely packed nucleosomes (L6 and L7). The level of protection against micrococcal nuclease cutting in this region is not as striking as that observed for the first five nucleosomes (L1 to L5). Positioned nucleosomes L8 and L9 (Fig. 5B) return the precision of organization and protection against nuclease cutting to the level observed adjacent to the I element in nucleosomes L1 to L5.
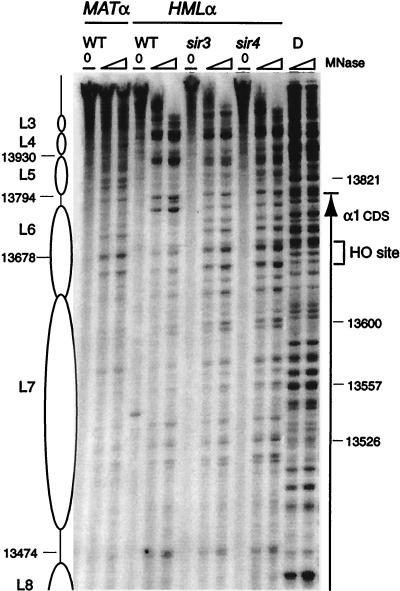
FIG. 4.
HMLα, wild type, sir3, and sir4, and MATα chromatin near the HO site and Yα-Z1 border. The chromatin structure of the Crick strand was mapped by primer extension analysis of micrococcal nuclease cleavage sites with primer p134. Wild-type (WT) MATα and wild-type and sir3 and sir4 mutant HMLα cells are as indicated. Symbols are as detailed in the legend to Fig. 2. The coding region of the α1 gene is indicated by an arrow.
Chromatin structure of the active MATα locus differs from that of the silenced locus in the region of the α1 gene (Fig. 4). In addition to the hypersensitivity observed at the HO site, numerous nuclease cleavage sites are present over parts of the α1 coding region. The Z1 and Z2 regions are more nuclease accessible at MATα in the region occupied by nucleosomes L4 and L5 in HMLα. The pattern at MATα is not identical to the nuclease digests of protein-free DNA. In particular, some sites of strong cleavage in protein-free DNA appear to be protected, and certain sites are more readily accessible. Accessibility of the active locus in the region protected by nucleosomes L6 and L7 at HMLα can be clearly detected in the chromatin digests of the sir3 and sir4 mutants of HMLα. Nuclease cutting patterns of the sir mutants in this region are reproducibly identical to the chromatin digests of MATα (which is somewhat underdigested in the experiment shown in Fig. 4). Disruption of L6 and L7 in MATα versus HMLα is seen to a similar extent on the other strand (see Fig. 10). The structure at MATα cannot be totally random chromatin.
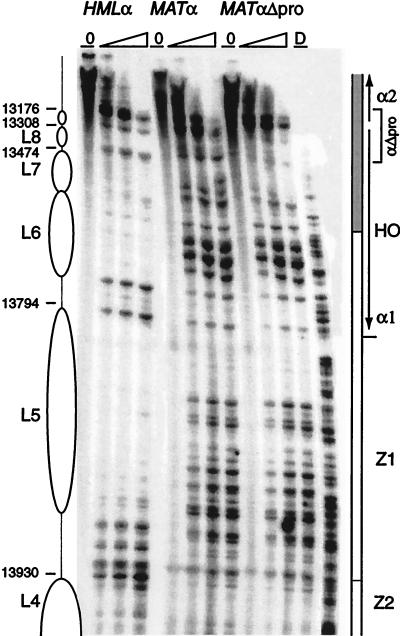
FIG. 10.
Chromatin structure of the Z2-Z1-α1 region in a nontranscribed MAT compared to HMLα and MATα. The chromatin structure of the Watson strand was mapped by primer extension analysis of micrococcal nuclease cleavage sites with primers q140 for HMLα and q1999 for MAT. Wild-type HMLα and MATα are as indicated. MATαΔpro designates the strain that has a 200-bp deletion of the promoter sequences of the α1 and α2 genes at MATα. The location of the deletion is indicated by a bracket (αΔpro). Symbols are as defined in the legend to Fig. 2. Sequence numbers shown correspond to that of HMLα and differ by 185,899 from the corresponding nucleotide at MAT.
Striking differences in the nuclease cutting patterns of the active versus silent state are observed at the promoter region of the divergently transcribed α1 and α2 genes (Fig. 5). Transcription initiation and regulatory elements of the intergenic region have been determined by Siliciano and Tatchell (72). At HMLα, the precisely positioned nucleosomes L10 and L9 are present over the transcriptional initiation and mRNA start sites of the α2 (Fig. 5B) and the α1 genes (Fig. 5A and B), respectively. These regions are more nuclease sensitive at MATα, although the effect is more pronounced for the α2 gene than for the α1 gene. Several sites, including transcription start sites, in particular between site 13006 and the α2 TATA (Fig. 5A), are readily cut. The cleavage pattern is distinct from that in protein-free DNA (Fig. 6). In surprising contrast, promoter sequences between the two TATA boxes are generally more nuclease sensitive at HMLα than at MATα (Fig. 5). Both TATA elements are hypersensitive to cleavage at the transcribed and repressed loci. The TATA element of α2, but not of α1, is also strongly cut in protein-free DNA (Fig. 5 and 6). Possibly, transient binding of TATA-binding protein (TBP) does not allow footprinting of the transcription initiation complex in nuclei at this promoter. A series of strong cleavage sites in the region between the upstream activation sequence (UAS) and the initiation elements of Matα2 at HMLα are protected in MATα. Sequences of the shared UAS, which is situated 40 bp from α2 TATAAA and 54 bp from α1 TATGAA, are not subject to nuclease cleavage. However, the Rap1 binding site is immediately flanked by nuclease-hypersensitive AT-rich sequences. These sites have remarkably different susceptibilities to cleavage in chromatin than in protein-free DNA (Fig. 6). The protection in chromatin could result from an association of Rap1p with its binding site. Overall, chromatin at the promoter sequences is more accessible to micrococcal nuclease at HMLα than at MATα.
FIG. 6.
Impact of the sir mutations on the chromatin structure of the α1 and α2 promoter region. The chromatin structure of the Crick strand was mapped by primer extension analysis of micrococcal nuclease cleavage sites with primer p129. Wild-type (WT) and sir3, sir4, and histone H4 N-terminal deletion (ΔH4) mutant HMLα cells are as indicated. Symbols are as detailed in the legend to Fig. 5.
At HMLα less precise nucleosome organization characterizes the remainder of the α2 coding region, i.e., nucleosomes L11 to L14 (data not shown). At micrococcal nuclease concentrations sufficient to see a clear nucleosomal positioning at adjacent regions, this pattern is not observed in this region. But at a 10-fold-higher concentration the cleavage pattern seen is suggestive of precise nucleosome positioning. Two pairs of closely spaced nucleosomes cover the X region and thereby the coding region of the α2 gene in a continuous array from the nucleosome placed near the promoter. The fact that the linkers of positioned nucleosomes are subject to nuclease cleavage only at high enzyme concentrations could reflect the presence of a heterochromatic state or sequestering of the silenced region.
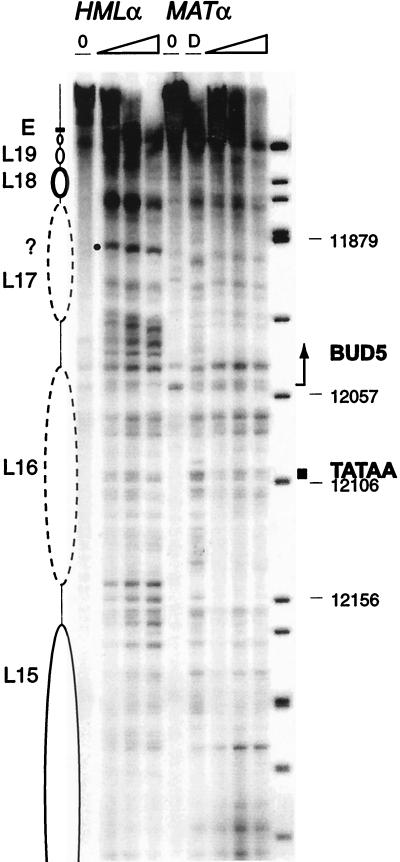
The chromatin structure of most of the W region of HMLα appears less organized (Fig. 7) but significantly different from the one at MATα. In fact, the promoter region and transcription initiation of the BUD5 gene, a GTPase required for bud site selection (10, 32), lies about 250 bp from the X region inside the W region. Its open reading frame extends 1.6 kb at MATα, thus including 500 bp of the W region at the 5′ part of the gene. At HML, where the truncated BUD5 is unlikely to be transcribed, two nucleosomes, L16 and L17, are present. However, some internal cutting in L16 and the existence of a mysterious band inside L17 indicate that their positioning is not extremely precise.
FIG. 7.
Chromatin structure of the W region in HMLα and MATα. The chromatin structure of the Watson strand was mapped by primer extension analysis of micrococcal nuclease cleavage sites with primer q123. HMLα and MATα are as indicated. Symbols are as detailed in the legend to Fig. 2. The dark box identifies the TATAA box, and the arrow shows the beginning of the BUD5 coding sequence. Sequence numbers shown correspond to those of HMLα, differ by 185,899 from the corresponding nucleotide in MATα, and are derived from the size of the ΦX174/HinfI-digested DNA fragments.
In contrast, like the region at the other end of the HMLα locus, nucleosomes are precisely positioned adjacent to the E silencer (Fig. 8). Three nucleosomes, L18 to L20, flank the E silencer from sites 11352 to 11826 inside the W region, where the chromatin appears remarkably similar for HMLα and MATα (Fig. 8 and data not shown). They are separated from the edge of the E element by 60 bp of DNA which has a nuclease cutting pattern resembling the one for the protein-free DNA control. Deletion of this D-region and either the Rap1 or ORC binding site was previously reported to lead to full derepression of HMLα (48). This sequence, which has no obvious protein binding motif, might have a role in spacing during the formation of the repressive chromatin organization near HML-E. In addition, at least two nucleosomes, L21 and L22, are positioned flanking the E silencer distally towards the telomere (Fig. 9), covering the 3′ end of the YCL069w open reading frame. YCL069w could code for a putative protein with homology to bacterial-drug-resistance factors (42). Its functionality has not been ascertained in yeast cells, but it is nonessential because a strain where HML was ligated to MAT is viable (81).
FIG. 8.
Chromatin structure near the E silencer. Chromatin structure was mapped by primer extension analysis of micrococcal nuclease cleavage sites with primer q120 (A) for the Watson strand between the W region and the silencer in the HMLα wild type (WT) and sir mutants and with p111 (B) for the Crick strand of the E silencer and adjacent region inside the HMLα in wild type and sir mutants. Wild-type MATα and wild-type and sir1, sir3, and sir4 mutant HMLα cells are as indicated. Symbols are as detailed in the legend to Fig. 2. Column G shows a dideoxyguanosine-terminated sequencing reaction. Column M shows ΦX174/HinfI-digested DNA fragments for size indication. Sequence numbers shown correspond to those of HMLα and differ by 185,899 from the corresponding nucleotide in MATα. Shaded boxes identify the Rap1p binding site and the ACS of the E silencer in panel A. MATα sequences are different from HMLα outside the W region (downstream [∗] in panel B).
In contrast to the organized chromatin outside HMLα at the E silencer, the centromere proximal region outside the I silencer exhibits random chromatin structure in both a and α cells (data not shown).
Impact of sir mutations on HMLα chromatin organization.
SIR1, SIR3, and SIR4 have been shown to be required for the maintenance of the repressed state of HMLα (45). Null mutants of these genes were created by replacing the promoter and part of the coding region with the URA3 gene (65). The HMLα-only strain used mates such as an a cell (79). In contrast, sir3 or sir4 mutants mate as α cells due to lack of silencing of HMLα, resulting in transcription of the α1 and α2 genes. The sir1 mutants mate with both a and α cells because the derepression of HMLα is only partial and the resulting mating phenotype is mixed. The Sir proteins are required for establishing and maintaining a repressive chromatin structure at HMLα (24, 55). Once the regions of HMLα where a particular chromatin organization characterizes the silent state of the locus were identified, the impact of the sir mutations on those structures was evaluated. Micrococcal nuclease cleavage sites of sir mutant HMLα were compared to the wild-type HMLα and MATα, as well as to HMLα in an H4 amino-terminal region (amino acids 4 to 23) deletion (ΔH4) mutant. In the ΔH4 mutant, HMLα is totally derepressed (26).
The series of five positioned nucleosomes, L1 to L5, between the Z1 region and the I silencer is disrupted in all the sir mutants (Fig. 2 and 3). The general pattern of hypersensitive sites which flanked positioned nucleosomes in the wild-type cells is maintained, but there is increased nuclease cleavage in the formerly protected regions. This disruption of organized chromatin structure is more pronounced for the sir3 strain than for the sir1 mutant (compare, for example, nucleosomes L2 and L3 [Fig. 2] and L4 [Fig. 3]) in the two strains. The pattern of MNase cleavage in the sir4 mutant strain resembled that for the sir3 mutant (data not shown). The sir3 and sir4 strains also show disruption of the chromatin organization around the HO endonuclease site in the region occupied by nucleosomes L6 and L7 in the wild type (Fig. 4). Susceptibilities to micrococcal nuclease cleavage in the sir3 and sir4 mutants are closely similar to those observed for this region at the active MATα locus. The generally more moderate effect of a sir1 mutation on chromatin may reflect the population effect, where HMLα is transcriptionally derepressed in only a fraction of cells.
A particular cleavage pattern at the promoter of α1 and α2 is the signature of transcriptional activity (Fig. 5 and 6). Transcription of α1 and α2 at HMLα in the sir mutants and the ΔH4 strain correlates with chromatin structure at the promoter, being essentially identical to that observed at MATα (Fig. 6). The transcription initiation and start sites, normally protected by nucleosomes L9 and L10, become nuclease accessible. For all mutant strains, the transcription start sites of Matα2, which are protected by nucleosome L10 in the silenced HML locus, are readily accessible. Curiously, the disruptive effect at the transcription initiation sites of Matα1 is less severe in general but in particular in the sir3 strain. As expected, the protection of the promoter region characteristic of the active MATα locus is mirrored at HMLα when the genes are derepressed by mutations in H4 (Fig. 6), sir3 and sir4 (Fig. 6), and sir1 (data not shown). The effect of the H4 mutation is more pronounced than that of the sir mutations, a finding which is consistent with the observation that sensitivity to micrococcal nuclease is generally increased in the chromatin of a strain deleted for the H4 amino-terminal tail (35). In the absence of the Sir proteins at HML and also at the transcribed MAT the nucleosomes are likely to still be present near the promoter region, but they will be in a more random position than at the silent HML.
The two nucleosome pairs, mapped at elevated nuclease concentrations, organizing the remainder of the α2 gene at HMLα are also disrupted when SIR3 and SIR4 are mutated (data not shown). The chromatin structure of the W region is similar in the sir mutants and in MATα (data not shown), as might be expected since this structure is already less organized than the remainder of the locus. Surprisingly, the nucleosomes flanking the E silencer, both inside (L18 to L20; Fig. 8) and outside (L21 and L22; Fig. 9) of HML, are still present in all examined sir mutants. Thus, in contrast to the significant alterations that occur in chromatin structure at the promoter and at the right-hand half of the silent locus, no distinctive differences between transcribed (sir) and silent (wild-type) loci are detectable in this left portion of the locus.
Inactivation of transcription at MATα is not sufficient to establish the HMLα specific nucleosomal organization.
We questioned whether the disorganized chromatin structure observed for most of the active MATα was the consequence of active nucleosome disruption caused by transcription. A strain constructed by combining two isolated XhoI linker mutations at MAT abolishing α1 and α2 transcription (86), thus creating an a-like strain, was used to compare the chromatin of the Z1-Z2-α1 region of HMLα with the active and transcription-blocked MATα. HMLα- and MATα-specific primers lying immediately outside the Z2 region were used. Positioned nucleosomes L5 to L8 are clearly seen in the Z1 and Z2 region and extending into the α1 coding region, blocking the HO endonuclease site, at HMLα in the wild-type strains (Fig. 10). These nucleosomes are disrupted at the active MATα, with extensive nuclease cutting across the mapped region. At the mutated, nontranscribed MAT locus, identical disruption of the nucleosomes is observed (Fig. 10). The highly organized chromatin structure of the HMLα locus is not present at the MAT locus irrespective of whether it is being actively transcribed or not. Particular features of the silent locus are necessary for establishing nucleosome positioning in the distinctive, silenced chromatin structure.
DISCUSSION
A central role for chromatin in the repression of genes in S. cerevisiae has been postulated for a number of loci. In contrast to genes where local, promoter-specific, chromatin structures have been observed, such as genes SUC2 (19), PHO5 (83), and ADH2 (92), larger domains of organized chromatin have been found at subtelomeric regions (44), at the recombination enhancer (93), and for a-cell-specific genes (74). Where examined in detail, these domains have consisted of continuous arrays of precisely positioned nucleosomes, delimited by the Matα2p-Mcm1p binding site and the 3′ end of the transcription unit for the a-specific genes (59, 90) or by two transcribed gene promoters flanking the recombination enhancer (93). Based on currently available evidence, particularly the results of histone H4 amino-terminal tail mutations (35, 88) and interactions of proteins known to be necessary for HM silencing with histones (26), the ∼3-kb silent-mating-type loci also represent regions of transcriptional repression where chromatin structure is important for regulation. In striking contrast to the continuous chromatin organization of other domains, chromatin at HML is discontinuous. While arrays of nucleosomes abut the E and I silencers, the arrays are punctuated by a 120-bp nucleosome-free region that encompasses the promoter of the divergent α1 and α2 genes (Fig. 1).
Adjacent to the silencers and flanking the promoter region, precisely positioned nucleosomes are located at HML. Each of these regions contains a binding site for Rap1p, the E and I silencers also have an ACS binding site for the ORC complex, and the I silencer contains an Abf1p binding site (6, 9, 30). Several of these proteins interact with proteins of the Sir group, and Sir3p and Sir4p interact with the amino-terminal regions of histones H3 and H4. The proposal has been made that Rap1p and/or the ORC complex bind to specific DNA sequences, recruit the Sir group, and then organize chromatin structure by interactions with histones. This scenario bears striking similarities to repression of a-cell-specific genes, where Matα2p and Mcm1p bind to specific DNA sequences, recruit the Ssn6p-Tup1p complex (which interacts with the amino-terminal regions of H3 and H4), and presumably organize chromatin structure (14, 17, 36, 41, 76). Defining the similarities between these two systems that both appear to produce organized chromatin should advance our understanding of how repressive nucleoprotein structures are established in eukaryotic cells.
In agreement with a current model for silencing in which one or more Sir proteins physically spread from the silencer over the silenced locus (26), the chromatin between HML-I and the promoter is disrupted in sir mutants. In contrast, the chromatin organization in the region near HML-E is not altered by any sir mutation. Organized chromatin near E does not depend on the presence of any individual Sir protein, and its establishment is not only nucleated towards the repressed locus, since positioned nucleosomes can be found flanking E on both sides. It seems likely that some of these features may arise from the proximity of E to the silenced telomere that is separated from HML by only 10 kb of untranscribed DNA (21). In the absence of transcribed genes, organized chromatin could be propagated from the telomere of chromosome III, where it is established in a Sir-dependent manner, to the vicinity of HML. This nucleosomal organization is likely to be independent of Sir proteins, since Sir3 was shown to only spread about 3 kb on a different telomere (62, 78). It has been shown that placing either HMLα or HML-E and/or HML-I near heterologous genes on chromosome III or on a plasmid alters the level of silencing (3, 49, 69) and that silencing is generally greater in the proximity of silenced regions such as telomeres. The proximity of a transcriptionally active chromosomal region to HML-I could increase the severity of single sir mutations, reflecting a context-dependent Sir protein role in the maintenance of highly organized chromatin.
In summary, in their native context, HML-E and HML-I seem functionally different, despite being equally competent at maintaining repression individually (47). HML-I has binding sites for both Abf1 and Rap1, while HML-E only has a Rap1 site. Abf1 and Rap1 can act as transcriptional activators when present at a promoter site (9, 15, 16, 71); possibly the Sir proteins prevent their activating function when recruited to the silencer. Destabilization of the silencing complex at a silencer due to the absence of one of the Sir proteins may consequently be more severe if two activators rather than a single one are present. While comparison of E and I at HML suggests differences in the role of Sir proteins in the establishment of organized chromatin, more in-depth indications of functional differences among the silencers should result from an ongoing characterization of chromatin near the HMR-E element that can silence this locus independently (59a).
In contrast to the parallel pathways for HML- and Matα2p-mediated chromatin assembly suggested above, the precise architecture around promoter elements differs strikingly for the two situations. At a-cell-specific promoters for STE6 and BAR1, a positioned nucleosome places the TATA box near the pseudodyad of the nucleosome core (59, 66, 90); inaccessibility of this critical element to the transcription machinery has been proposed as one mechanism that could lead to repression (70, 73, 74). Surprisingly, at HMLα, much of the 200-bp intergenic region between the divergently transcribed α1 and α2 genes, including the single shared UAS, is highly accessible to micrococcal nuclease digestion. No repressor binding site (other than that for Rap1p, which also serves as an activator) has been identified in the intergenic region, and both activators and the transcriptional machinery are readily available to transcribe both genes from an identical promoter at MATα. Hence, transcriptional repression at HMLα seems likely to be regulated structurally.
Several possibilities arise for such structural regulation. First, the transcription initiation sites for both genes are located in positioned nucleosomes. Although the TATA boxes are not blocked by histone-DNA interactions, assembly of the basal transcription machinery requires significantly greater lengths of DNA than that contacted directly by the TBP (72, 84), and sequences that would be involved in such interactions are sequestered in the positioned nucleosomes. At MATα, the entire region between the two TATA boxes is relatively protected, but the transcription initiation sites are susceptible to micrococcal nuclease cleavage, possibly reflecting TBP and associated factor binding and formation of the transcription initiation complex.
Second, the geometry of chromatin at and around the intergenic region at HMLα could preclude formation of the transcription initiation complex. The two TATA sites are separated by 105 bp, exactly 10 helical turns of DNA in solution. Since TBP creates an ∼80° bend when it binds to DNA and an 18-Å lateral displacement between upstream and downstream DNA when it binds to the TATA box (37, 38), the two nucleosomes which flank the intergenic region have the potential to be involved in a steric clash if TBP is bound to both TATA boxes. Rap1p binding to DNA also bends DNA by more than 50° (20, 53), so it is likely to affect this possible interaction. If Rap1p serves to anchor chromatin to a karyoskeletal element, the system becomes too complex to make mechanistic predictions based on known structures of proteins and the DNA involved.
Third, a higher-order structure which precludes transcription could be formed by the chromatin at HMLα. Looping of DNA from the HM loci has been shown to occur readily in vitro; loops between E and I silencers and between the silencers and the promoter region were observed and were shown to require Rap1p (30). Rap1 was initially isolated from a karyoskeletal fraction, and HML-E and HML-I were found to be associated with a “nuclear scaffold” fraction (30). While probably reflecting telomere location and therefore only indirectly the location of the nearby HM loci, immunofluorescence studies show colocalization of Rap1p, Sir3p, and Sir4p with telomeric DNA in discrete foci around the nuclear periphery (13, 57). Proximity of silenced loci to telomeres has been shown to be necessary for effective silencing (49). A recent study with topological measurements on circles containing all or parts of HMLα excised in vivo (5) showed a linking-number difference of ∼2 between samples from a wild type versus a sir3 background; the wild type had two more negative supercoils than did the mutant. While a number of reasons could lead to the linking-number deficit in the mutant strain, loss of a double loop of DNA, looped from E to UAS and from UAS to I, in the mutant strains is certainly consistent with this experimental result. Targeting of a LexA-Sir4p chimera to a plasmid by inclusion of LexA binding sequences led to partitioning of the plasmid on cell division, suggesting interaction of the plasmid-bound protein with a nuclear element that partitions equally between mother and daughter cells (2). Interestingly, partitioning was dependent on Rap1p, suggesting that this protein might form the anchor on the nuclear skeletal element which held the Sir4p-bound plasmid. One can envision Rap1p anchoring HMLα to a karyoskeletal element at three sites, interacting with a Sir protein complex that somehow organizes chromatin and thereby creating a substrate refractory to transcription initiation as well as sequestering the locus to a potentially repressive nuclear location. Differences in effects on chromatin structure along the length of the locus of the sir mutations, greatest at I and at the promoter and less near E, suggest that the structure is not homogeneous from end to end.
While the chromatin organization of HMLα seems intimately connected with transcriptional silencing, the locus is fully capable of participation in recombination. This is also true of loci involved in mammalian immunoglobulin gene recombination. Resolving the apparent paradox of transcriptional silencing coexisting with recombinational competence provides a healthy experimental challenge.
ACKNOWLEDGMENTS
We thank J. E. Haber, S. K. Reimer, D. Shore, and M. Grunstein for generous gifts of strains and plasmids; J. E. Haber, H. G. Patterton, and P. A. Grant for review of the manuscript; and members of the Simpson and Workman lab for their criticism and technical advice.
This study was supported by NIH grant GM52311.
REFERENCES
- 1.Abraham J A, Nasmyth K A, Strathern J N, Klar A J S, Hicks J B. Regulation of mating-type information in yeast. J Mol Biol. 1984;176:307–331. doi: 10.1016/0022-2836(84)90492-3. [DOI] [PubMed] [Google Scholar]
- 2.Ansari A, Gartenberg M R. The yeast silent information regulator Sir4p anchors and partitions plasmids. Mol Cell Biol. 1997;17:7061–7068. doi: 10.1128/mcb.17.12.7061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aparicio O M, Gottschling D E. Overcoming telomeric silencing: a trans-activator competes to establish gene expression in a cell cycle-dependent way. Genes Dev. 1994;8:1133–1146. doi: 10.1101/gad.8.10.1133. [DOI] [PubMed] [Google Scholar]
- 4.Bell S P, Stillman B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature. 1992;357:128–134. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- 5.Bi X, Broach J R. DNA in transcriptionally silent chromatin assumes a distinct topology that is sensitive to cell cycle progression. Mol Cell Biol. 1997;17:7077–7087. doi: 10.1128/mcb.17.12.7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boscheron C, Maillet L, Marcand S, Tsai-Pflugfelder M, Gasser S M, Gilson E. Cooperation at a distance between silencers and proto-silencers at the yeast HML locus. EMBO J. 1996;15:2184–2195. [PMC free article] [PubMed] [Google Scholar]
- 7.Brand A H, Breeden L, Abraham J, Sternglanz R, Nasmyth K. Characterization of a “silencer” in yeast: a DNA sequence with properties opposite to those of a transcriptional enhancer. Cell. 1985;41:41–48. doi: 10.1016/0092-8674(85)90059-5. [DOI] [PubMed] [Google Scholar]
- 8.Braunstein M, Rose A B, Holmes S G, Allis C D, Broach J R. Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes Dev. 1993;7:592–604. doi: 10.1101/gad.7.4.592. [DOI] [PubMed] [Google Scholar]
- 9.Buchman A R, Kimmerly W J, Rine J, Kornberg R D. Two DNA-binding factors recognize specific sequences at silencers, upstream activating sequences, autonomously replicating sequences, and telomeres in Saccharomyces cerevisiae. Mol Cell Biol. 1988;8:210–225. doi: 10.1128/mcb.8.1.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chant J, Corrado K, Pringle J R, Herskowitz I. Yeast BUD5, encoding a putative GDP-GTP exchange factor, is necessary for bud site selection and interacts with bud formation gene BEM1. Cell. 1991;65:1213–1224. doi: 10.1016/0092-8674(91)90016-r. [DOI] [PubMed] [Google Scholar]
- 11.ChenCleland T A, Smith M M, Le S Y, Sternglanz R, Allfrey V G. Nucleosome structural changes during derepression of silent mating-type loci in yeast. J Biol Chem. 1993;268:1118–1124. [PubMed] [Google Scholar]
- 12.Chien C T, Buck S, Sternglanz R, Shore D. Targeting of SIR1 protein establishes transcriptional silencing at HM loci and telomeres in yeast. Cell. 1993;75:531–541. doi: 10.1016/0092-8674(93)90387-6. [DOI] [PubMed] [Google Scholar]
- 13.Cockell M, Palladino F, Laroche T, Kyrion G, Liu C, Lustig A J, Gasser S M. The carboxy termini of Sir4 and Rap1 affect Sir3 localization: evidence for a multicomponent complex required for yeast telomeric silencing. J Cell Biol. 1995;129:909–924. doi: 10.1083/jcb.129.4.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cooper J P, Roth S Y, Simpson R T. The global transcriptional regulators, SSN6 and TUP1, play distinct roles in the establishment of a repressive chromatin structure. Genes Dev. 1994;8:1400–1410. doi: 10.1101/gad.8.12.1400. [DOI] [PubMed] [Google Scholar]
- 15.Diffley J, Stillman B. Transcriptional silencing and lamins. Nature. 1989;342:24. doi: 10.1038/342024a0. [DOI] [PubMed] [Google Scholar]
- 16.Diffley J F X, Stillman B. Purification of a yeast protein that binds to origins of DNA replication and a transcriptional silencer. Proc Natl Acad Sci USA. 1988;85:21220–21124. doi: 10.1073/pnas.85.7.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Edmondson D G, Smith M M, Roth S Y. Repression domain of the yeast global repressor Tup1 interacts directly with histones H3 and H4. Genes Dev. 1996;10:1247–1259. doi: 10.1101/gad.10.10.1247. [DOI] [PubMed] [Google Scholar]
- 18.Fox C A, Ehrenhofer-Murray A E, Loo S, Rine J. The origin recognition complex, SIR1, and the S phase requirement for silencing. Science. 1997;276:1547–1551. doi: 10.1126/science.276.5318.1547. [DOI] [PubMed] [Google Scholar]
- 19.Gavin I M, Simpson R T. Interplay of yeast global transcriptional regulators Ssn6p-Tup1 and Swi-Snf and their effect on chromatin structure. EMBO J. 1997;16:6263–6271. doi: 10.1093/emboj/16.20.6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gilson E, Roberge M, Giraldo R, Rhodes D, Gasser S M. Distortion of the DNA double helix by RAP1 at silencers and multiple telomeric binding sites. J Mol Biol. 1993;231:293–310. doi: 10.1006/jmbi.1993.1283. [DOI] [PubMed] [Google Scholar]
- 21.Gottschling D E. Telomere-proximal DNA in Saccharomyces cerevisiae is refractory to methyltransferase activity in vivo. Proc Natl Acad Sci USA. 1992;89:4062–4065. doi: 10.1073/pnas.89.9.4062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gottschling D E, Aparico O M, Billington B L, Zakian V A. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell. 1990;63:751–762. doi: 10.1016/0092-8674(90)90141-z. [DOI] [PubMed] [Google Scholar]
- 23.Graham I R, Chambers A. A Reb1p-binding site is required for efficient activation of the yeast RAP1 gene, but multiple binding sites for Rap1p are not essential. Mol Microbiol. 1994;12:931–940. doi: 10.1111/j.1365-2958.1994.tb01081.x. [DOI] [PubMed] [Google Scholar]
- 24.Grunstein M, Hecht A, Fisher-Adams G, Wan J, Mann R K, Strahl-Bolsinger S, Laroche T, Gasser S. The regulation of euchromatin and heterochromatin by histones in yeast. J Cell Sci. 1995;19:29–36. doi: 10.1242/jcs.1995.supplement_19.4. [DOI] [PubMed] [Google Scholar]
- 25.Haber J E, George J P. A mutation that permits the expression of normally silent copies of mating type information in Saccharomyces cerevisiae. Genetics. 1979;93:13–35. doi: 10.1093/genetics/93.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hecht A, Laroche T, StrahlBolsinger S, Gasser S M, Grunstein M. Histone H3 and H4 N-termini interact with SIR3 and SIR4 proteins: a molecular model for the formation of heterochromatin in yeast. Cell. 1995;80:583–592. doi: 10.1016/0092-8674(95)90512-x. [DOI] [PubMed] [Google Scholar]
- 27.Hecht A, StrahlBolsinger S, Grunstein M. Spreading of transcriptional repressor SIR3 from telomeric heterochromatin. Nature. 1996;383:92–96. doi: 10.1038/383092a0. [DOI] [PubMed] [Google Scholar]
- 28.Henikoff S. Position-effect variegation after 60 years. Trends Genet. 1990;120:422–426. doi: 10.1016/0168-9525(90)90304-o. [DOI] [PubMed] [Google Scholar]
- 29.Hicks J, Strathern J N, Klar A J S. Transposable mating type genes in Saccharomyces cerevisiae. Nature. 1979;282:473–478. doi: 10.1038/282478a0. [DOI] [PubMed] [Google Scholar]
- 30.Hofmann J F X, Laroche T, Brand A H, Gasser S M. RAP-1 factor is necessary for DNA loop formation in vitro at the silent mating type locus HML. Cell. 1989;57:725–737. doi: 10.1016/0092-8674(89)90788-5. [DOI] [PubMed] [Google Scholar]
- 31.Ivy J M, Klar A J S, Hicks J B. Cloning and characterization of four SIR genes of Saccharomyces cerevisiae. Mol Cell Biol. 1986;6:688–702. doi: 10.1128/mcb.6.2.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jacquet M, Buhler J M, Iborra F, Francingues-Gaillard M C, Soustelle C. The MAT locus revisited within a 9.8 kb fragment of chromosome III containing BUD5 and two new open reading frames. Yeast. 1991;7:881–888. doi: 10.1002/yea.320070815. [DOI] [PubMed] [Google Scholar]
- 33.Jamieson R V, Tam P P, Gardiner-Garden M. X-chromosome activity: impact of imprinting and chromatin structure. Int J Dev Biol. 1996;40:1065–1080. [PubMed] [Google Scholar]
- 34.Johnson L M, Kayne P S, Kahn E S, Grunstein M. Genetic evidence for an interaction between SIR3 and histone H4 in the repression of silent mating type loci in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1990;87:6286–6290. doi: 10.1073/pnas.87.16.6286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kayne P S, Kim U, Han M, Mullen J R, Yoshizaki F, Grunstein M. Extremely conserved histone H4 N-terminus is dispensable for growth but essential for repressing the silent mating type loci in yeast. Cell. 1988;55:27–39. doi: 10.1016/0092-8674(88)90006-2. [DOI] [PubMed] [Google Scholar]
- 36.Keleher C A, Redd M J, Schultz J, Carlson M, Johnson A D. Ssn6-Tup1 is a general repressor of transcription in yeast. Cell. 1992;68:709–719. doi: 10.1016/0092-8674(92)90146-4. [DOI] [PubMed] [Google Scholar]
- 37.Kim J L, Nikolov D B, Burley S K. Co-crystal structure of TBP recognizing the minor groove of a TATA element. Nature. 1993;365:520–527. doi: 10.1038/365520a0. [DOI] [PubMed] [Google Scholar]
- 38.Kim Y, Geiger J H, Hahn S, Sigler P B. Crystal structure of a yeast TBP/TATA-box complex. Nature. 1993;365:512–520. doi: 10.1038/365512a0. [DOI] [PubMed] [Google Scholar]
- 39.Klar A J S, Fogel S, Radin D N. Switching of mating-type a mutant allele in budding yeast Saccharomyces cerevisiae. Genetics. 1979;92:756–776. doi: 10.1093/genetics/92.3.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klar A J S, Strathern J N, Abraham J A. Involvement of double-strand chromosomal breaks for mating-type switching in Saccharomyces cerevisiae. Cold Spring Harbor Symp Quant Biol. 1984;49:77–88. doi: 10.1101/sqb.1984.049.01.011. [DOI] [PubMed] [Google Scholar]
- 41.Komachi K, Redd M J, Johnson A D. The WD repeats of Tup1 interact with the homeo domain protein alpha 2. Genes Dev. 1994;8:2857–2867. doi: 10.1101/gad.8.23.2857. [DOI] [PubMed] [Google Scholar]
- 42.Koonin E V, Bork P, Sander C. Yeast chromosome III: new gene functions. EMBO J. 1994;13:493–503. doi: 10.1002/j.1460-2075.1994.tb06287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kostriken R, Strathern J N, Klar A J S, Hicks J B, Heffron F. A site-specific endonuclease essential for mating-type switching in Saccharomyces cerevisiae. Cell. 1983;35:167–174. doi: 10.1016/0092-8674(83)90219-2. [DOI] [PubMed] [Google Scholar]
- 44.Kyrion G, Liu K, Liu C, Lustig A J. RAP1 and telomere structure regulate telomere position effects in Saccharomyces cerevisiae. Genes Dev. 1993;7:1146–1159. doi: 10.1101/gad.7.7a.1146. [DOI] [PubMed] [Google Scholar]
- 45.Laurenson P, Rine J. Silencers, silencing, and heritable transcriptional states. Microbiol Rev. 1992;56:543–560. doi: 10.1128/mr.56.4.543-560.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Loo S, Rine J. Silencers and domains of generalized repression. Science. 1994;264:1768–1771. doi: 10.1126/science.8209257. [DOI] [PubMed] [Google Scholar]
- 47.Mahoney D J, Broach J R. The HML mating-type cassette of Saccharomyces cerevisiae is regulated by two separate but functionally equivalent silencers. Mol Cell Biol. 1989;9:4621–4630. doi: 10.1128/mcb.9.11.4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mahoney D J, Marquardt R, Shei G, Rose A B, Broach J R. Mutations in the HML-E silencer of Saccharomyces cerevisiae yield metastable inheritance of transcriptional repression. Genes Dev. 1991;5:605–615. doi: 10.1101/gad.5.4.605. [DOI] [PubMed] [Google Scholar]
- 49.Maillet L, Boscheron C, Gotta M, Marcand S, Gilson E, Gasser S M. Evidence for silencing compartments within the yeast nucleus: a role for telomere proximity and Sir protein concentration in silencer-mediated repression. Genes Dev. 1996;10:1796–1811. doi: 10.1101/gad.10.14.1796. [DOI] [PubMed] [Google Scholar]
- 50.Megee P C, Morgan B A, Mittman B A, Smith M M. Genetic analysis of histone H4: essential role of lysines subject to reversible acetylation. Science. 1990;247:841–845. doi: 10.1126/science.2106160. [DOI] [PubMed] [Google Scholar]
- 51.Moazed D, Johnson A D. A deubiquinating enzyme interacts with SIR4 and regulates silencing in S. cerevisiae. Cell. 1996;86:667–677. doi: 10.1016/s0092-8674(00)80139-7. [DOI] [PubMed] [Google Scholar]
- 52.Moretti P, Freeman K, Coodly L, Shore D. Evidence that a complex of SIR proteins interacts with the silencer and telomere binding protein RAP1. Genes Dev. 1994;8:2257–2269. doi: 10.1101/gad.8.19.2257. [DOI] [PubMed] [Google Scholar]
- 53.Mueller T, Gilson E, Schmidt R, Giraldo R, Sogo J, Gross H, Gasser S M. Imaging the asymmetrical DNA bend induced by repressor activator protein 1 with scanning tunneling microscopy. J Struct Biol. 1994;113:1–12. doi: 10.1006/jsbi.1994.1027. [DOI] [PubMed] [Google Scholar]
- 54.Mullen J R, Kayne P S, Moerschell R P, Tsunasawa S, Gribskov M, Colavito-Shepanski M, Grunstein M, Sherman F, Sternglanz R. Identification and characterization of genes and mutants for an N-terminal acetyltransferase from yeast. EMBO J. 1989;8:2067–2075. doi: 10.1002/j.1460-2075.1989.tb03615.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nasmyth K A. The regulation of yeast mating-type chromatin structure by SIR: an action at a distance affecting both transcription and transposition. Cell. 1982;30:567–578. doi: 10.1016/0092-8674(82)90253-7. [DOI] [PubMed] [Google Scholar]
- 56.Oliver S G, et al. The complete DNA sequence of yeast chromosome III. Nature. 1992;357:38–46. doi: 10.1038/357038a0. [DOI] [PubMed] [Google Scholar]
- 57.Palladino F, Laroche T, Gilson E, Axelrod A, Pillus L, Gasser S M. SIR3 and SIR4 proteins are required for the positioning and integrity of yeast telomeres. Cell. 1993;75:543–555. doi: 10.1016/0092-8674(93)90388-7. [DOI] [PubMed] [Google Scholar]
- 58.Park E C, Szostak J W. Point mutations in the yeast histone H4 gene prevent silencing of the silent mating type locus HML. Mol Cell Biol. 1990;10:4932–4934. doi: 10.1128/mcb.10.9.4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Patterton H G, Simpson R T. Nucleosomal location of the STE6 TATA box and Mat alpha 2p-mediated repression. Mol Cell Biol. 1994;14:4002–4010. doi: 10.1128/mcb.14.6.4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59a.Ravindra, A., K. Weiss, and R. T. Simpson. Unpublished observations.
- 60.Reifsnyder C, Lowell J, Clarke A, Pillus L. Yeast SAS silencing genes and human genes associated with AML and HIV-1 Tat interactions are homologous with acetyltransferases. Nat Genet. 1996;14:42–49. doi: 10.1038/ng0996-42. [DOI] [PubMed] [Google Scholar]
- 61.Reimer S K, Buchman A R. Yeast silencers create domains of nuclease-resistant chromatin in an SIR4-dependent manner. Chromosoma. 1997;106:136–148. doi: 10.1007/s004120050233. [DOI] [PubMed] [Google Scholar]
- 62.Renauld H, Aparicio O M, Zierath P D, Billington B L, Chablani S K, Gottschling D E. Silent domains are assembled continuously from the telomere and are defined by promoter distance and strength, and by SIR3 dosage. Genes Dev. 1993;7:1133–1145. doi: 10.1101/gad.7.7a.1133. [DOI] [PubMed] [Google Scholar]
- 63.Rine J, Herskowitz I. Four genes responsible for a position effect on expression from HML and HMR in Saccharomyces cerevisiae. Genetics. 1987;116:9–22. doi: 10.1093/genetics/116.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rine J, Strathern J N, Hicks J B, Herskowitz I. A suppressor of mating-type locus mutations in Saccharomyces cerevisiae: evidence for and identification of cryptic mating-type loci. Genetics. 1979;93:877–901. doi: 10.1093/genetics/93.4.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rose M D, Winston F, Hieter P. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1990. [Google Scholar]
- 66.Roth S Y, Dean A, Simpson R T. Yeast α2 repressor positions nucleosomes in TRP1/ARS1 chromatin. Mol Cell Biol. 1990;10:2247–2260. doi: 10.1128/mcb.10.5.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Roth S Y, Shimizu M, Johnson L, Grunstein M, Simpson R T. Stable nucleosome positioning and complete repression by the yeast α2 repressor are disrupted by amino-terminal mutations in histone H4. Genes Dev. 1992;6:411–425. doi: 10.1101/gad.6.3.411. [DOI] [PubMed] [Google Scholar]
- 68.Schnell R, Rine J. A position effect on the expression of a tRNA gene mediated by the SIR genes in Saccharomyces cerevisiae. Mol Cell Biol. 1986;6:494–501. doi: 10.1128/mcb.6.2.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shei G J, Broach J R. Yeast silencers can act as orientation-dependent gene inactivation centers that respond to environmental signals. Mol Cell Biol. 1995;15:3496–3506. doi: 10.1128/mcb.15.7.3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shimizu M, Roth S Y, Szent-Gyorgyi C, Simpson R T. Nucleosomes are positioned with base pair precision adjacent to the α2 operator in Saccharomyces cerevisiae. EMBO J. 1991;10:3033–3041. doi: 10.1002/j.1460-2075.1991.tb07854.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shore D, Nasmyth K. Purification and cloning of a DNA binding protein from yeast that binds to both silencer and activator elements. Cell. 1987;51:721–732. doi: 10.1016/0092-8674(87)90095-x. [DOI] [PubMed] [Google Scholar]
- 72.Siliciano P G, Tatchell K. Transcription and regulatory signals at the mating type locus in yeast. Cell. 1984;37:969–978. doi: 10.1016/0092-8674(84)90431-8. [DOI] [PubMed] [Google Scholar]
- 73.Simpson R T. Nucleosome positioning can affect the function of a cis-acting DNA element in vivo. Nature. 1990;343:387–389. doi: 10.1038/343387a0. [DOI] [PubMed] [Google Scholar]
- 74.Simpson R T, Roth S Y, Morse R H, Patterton H-G, Cooper J P, Murphy M R, Kladde M P, Shimizu M. Nucleosome positioning and transcription. Cold Spring Harbor Symp Quant Biol. 1993;58:237–245. doi: 10.1101/sqb.1993.058.01.028. [DOI] [PubMed] [Google Scholar]
- 75.Singh J, Klar A J S. Active genes in budding yeast display enhanced in vivo accessibility to foreign DNA methylases: a novel in vivo probe for chromatin structure of yeast. Genes Dev. 1992;6:186–196. doi: 10.1101/gad.6.2.186. [DOI] [PubMed] [Google Scholar]
- 76.Smith D L, Desai A B, Johnson A D. DNA bending by the a1 and alpha 2 homeodomain proteins from yeast. Nucleic Acids Res. 1995;23:1239–1243. doi: 10.1093/nar/23.7.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Stone E M, Swanson M J, Romeo A M, Hicks J B, Sternglanz R. The SIR1 gene of Saccharomyces cerevisiae and its role as an extragenic suppressor of several mating-defective mutants. Mol Cell Biol. 1991;11:2253–2262. doi: 10.1128/mcb.11.4.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.StrahlBolsinger S, Hecht A, Luo K H, Grunstein M. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 1997;11:83–93. doi: 10.1101/gad.11.1.83. [DOI] [PubMed] [Google Scholar]
- 79.Strathern J N, Hicks J, Herskowitz I. Control of cell type in yeast by the mating type locus. The alpha 1-alpha 2 hypothesis. J Mol Biol. 1981;147:357–372. doi: 10.1016/0022-2836(81)90488-5. [DOI] [PubMed] [Google Scholar]
- 80.Strathern J N, Klar A J S, Hicks J B, Abraham J A, Ivy J M, Nasmyth K, Gill C. Homothallic switching of yeast mating-type is initiated by a double-stranded cut in the MAT locus. Cell. 1982;31:183–192. doi: 10.1016/0092-8674(82)90418-4. [DOI] [PubMed] [Google Scholar]
- 81.Strathern J N, Newlon C S, Herskowitz I, Hicks J B. Isolation of a circular derivative of yeast chromosome III: implication for the mechanism of mating type interconversion. Cell. 1979;18:309–319. doi: 10.1016/0092-8674(79)90050-3. [DOI] [PubMed] [Google Scholar]
- 82.Surani M A. Genomic imprinting: control of gene expression by epigenetic inheritance. Curr Opin Cell Biol. 1994;6:390–395. doi: 10.1016/0955-0674(94)90031-0. [DOI] [PubMed] [Google Scholar]
- 83.Svaren J, Hörz W. Transcription factors vs nucleosomes: regulation of the PH05 promoter in yeast. Trends Biochem Sci. 1997;22:93–97. doi: 10.1016/s0968-0004(97)01001-3. [DOI] [PubMed] [Google Scholar]
- 84.Sypes M A, Gilmour D S. Protein/DNA crosslinking of TFIID complex reveals novel interactions downstream of the transcription start. Nucleic Acids Res. 1994;22:807–814. doi: 10.1093/nar/22.5.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Szent-Gyorgyi C, Isenberg I. The organization of oligonucleosomes in yeast. Nucleic Acids Res. 1983;11:3717–3736. doi: 10.1093/nar/11.11.3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tatchell K, Nasmyth K A, Hall B D, Astell C, Smith M. In vitro mutation analysis of the mating-type locus in yeast. Cell. 1981;27:25–35. doi: 10.1016/0092-8674(81)90357-3. [DOI] [PubMed] [Google Scholar]
- 87.Thompson J S, Hecht A, Grunstein M. Histones and the regulation of heterochromatin in yeast. Cold Spring Harbor Symp Quant Biol. 1993;58:247–256. doi: 10.1101/sqb.1993.058.01.029. [DOI] [PubMed] [Google Scholar]
- 88.Thompson J S, Johnson L M, Grunstein M. Specific repression of the yeast silent mating locus HMR by an adjacent telomere. Mol Cell Biol. 1994;14:446–455. doi: 10.1128/mcb.14.1.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Triolo T, Sternglanz R. Role of interactions between the origin recognition complex and SIR1 in transcriptional silencing. Nature. 1996;381:251–253. doi: 10.1038/381251a0. [DOI] [PubMed] [Google Scholar]
- 90.Tsukagoshi, Y. T., and R. T. Simpson. Unpublished data.
- 91.Turner B M. Histone H4, the cell cycle and a question of integrity. Bioessays. 1995;17:1013–1015. doi: 10.1002/bies.950171204. [DOI] [PubMed] [Google Scholar]
- 92.Verdone L, Camilloni G, Di Mauro E, Caserta M. Chromatin remodelling during Saccharomyces cerevisiae ADH2 gene activation. Mol Cell Biol. 1996;16:1978–1988. doi: 10.1128/mcb.16.5.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Weiss K, Simpson R T. Cell type-specific chromatin organization of the region that governs directionality of yeast mating type switching. EMBO J. 1997;16:4352–4360. doi: 10.1093/emboj/16.14.4352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Whiteway M, Freedman R, Van Arsdell S, Szostak J W, Thorner J. The yeast ARD1 gene product is required for repression of cryptic mating-type information at the HML locus. Mol Cell Biol. 1987;7:3713–3722. doi: 10.1128/mcb.7.10.3713. [DOI] [PMC free article] [PubMed] [Google Scholar]