Abstract
Inefficient splicing of human immunodeficiency virus type 1 (HIV-1) RNA is necessary to preserve unspliced and singly spliced viral RNAs for transport to the cytoplasm by the Rev-dependent pathway. Signals within the HIV-1 genome that control the rate of splicing include weak 3′ splice sites, exon splicing enhancers (ESE), and exon splicing silencers (ESS). We have previously shown that an ESS present within tat exon 2 (ESS2) and a suboptimal 3′ splice site together act to inhibit splicing at the 3′ splice site flanking tat exon 2. This occurs at an early step in spliceosome assembly. Splicing at the 3′ splice site flanking tat exon 3 is regulated by a bipartite element composed of an ESE and an ESS (ESS3). Here we show that ESS3 is composed of two smaller elements (AGAUCC and UUAG) that can inhibit splicing independently. We also show that ESS3 is more active in the context of a heterologous suboptimal splice site than of an optimal 3′ splice site. ESS3 inhibits splicing by blocking the formation of a functional spliceosome at an early step, since A complexes are not detected in the presence of ESS3. Competitor RNAs containing either ESS2 or ESS3 relieve inhibition of splicing of substrates containing ESS3 or ESS2. This suggests that a common cellular factor(s) may be required for the inhibition of tat mRNA splicing mediated by ESS2 and ESS3.
Human immunodeficiency virus type 1 (HIV-1) is a complex retrovirus whose RNA splicing is dependent on the host cell splicing machinery (12, 50). HIV-1 pre-mRNA contains five 5′ splice sites and nine 3′ splice sites which are used to generate more than 30 different mRNA species by alternative splicing. In addition, a pool of unspliced RNA must be maintained to serve as genomic RNA and as mRNA for the gag and pol gene products (18, 21, 32, 34, 35, 38). To achieve the balance between spliced and unspliced RNAs, splicing of HIV-1 RNA is inefficient, a feature which is shared with all other retroviruses (4, 7, 11, 20, 22, 45–47, 52). In contrast to the major HIV-1 5′ splice sites, which are strong, the 3′ splice sites are weak (33). These are characterized by the presence of nonconsensus polypyrimidine tracts and branch point sites other than the eukaryotic consensus adenosine residue (2, 13, 17, 33, 43). In addition, exon sequences downstream of the 3′ splice sites enhance or inhibit splicing at the 3′ splice sites (2, 3, 39, 42, 51). Furthermore, the virus-encoded protein Rev plays an essential role in facilitating the transport of unspliced and singly spliced mRNAs, thus preventing the viral RNA from splicing to completion (19, 37; for a review, see reference 12).
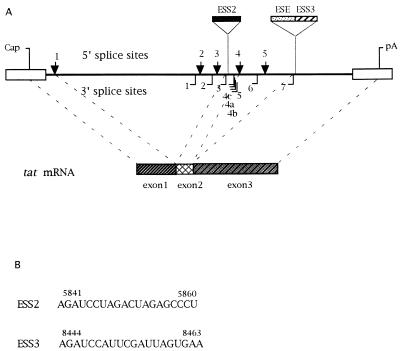
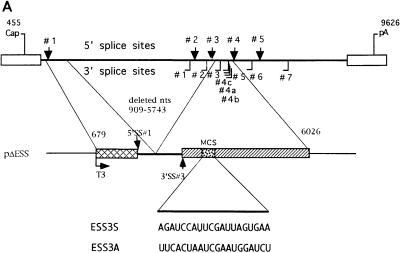
HIV-1 tat mRNA is formed by the splicing of two coding exons (exon 2 and exon 3) and one or more upstream noncoding exons. Exon 3 is also joined to rev exon 2 and is therefore referred to as tat/rev exon 3 (34). The 3′ splice sites flanking exon 2 and exon 3 (3′ splice sites 3 and 7, respectively [Fig. 1]) contain suboptimal polypyrimidine tracts, and the 3′ splice site flanking exon 3 has been reported to contain a nonconsensus branch point site (17, 33, 39, 43). In addition, we and others have demonstrated that tat/rev exon 3 splicing is regulated by an exon splicing enhancer (ESE) and a juxtaposed exon splicing silencer (ESS) within this exon (ESS3) (Fig. 1) (3, 42). An ESS is also present within tat exon 2 (ESS2), which has been localized to the RNA sequence CUAGACUAGA in the region of vpr encoding the C terminus and in the region of tat encoding the N terminus. It acts at an early step in the splicing pathway to selectively inhibit splicing at the upstream 3′ splice site flanking this tat exon (3). Addition of competitor RNA containing ESS2 to splicing reaction mixtures with HIV-1 RNA substrates containing ESS2 causes an increase in splicing at the upstream 3′ splice site (3′ splice site 3). Based on these data, we hypothesized that the inhibition by ESS2 is mediated by a negative cellular factor(s) which binds to the ESS (3). In this study, we have delineated the ESS3 element in tat/rev exon 3 and have investigated the mechanism by which it inhibits splicing. The data suggest that the inhibition by ESS2 and ESS3 involves binding of a common factor(s).
FIG. 1.
(A) Schematic diagram of HIV-1 splice sites and structure of tat mRNA. ESS2 and ESS3 are ESSs located within tat exon 2 and tat/rev exon 3, respectively. 5′ splice sites are indicated above the line by arrows; 3′ splice sites are denoted below the line as solid lines. “Cap” indicates the RNA initiation site, and “pA” represents the poly(A) site. The ESE is juxtaposed to the ESS3 within tat/rev exon 3. The regions which are spliced to form the tat mRNA are indicated by dashed lines. (B) Sequences of ESS2 and ESS3.
MATERIALS AND METHODS
Plasmid constructions.
Mutations were made either by PCR-mediated site-directed mutagenesis or by subcloning of annealed oligonucleotide pairs (36). Mutations were confirmed by sequencing. pΔESS has been described previously (2, 39) and was used as the parent plasmid for all mutant HIV-1 ESS3 minigene constructs shown in Fig. 3A. pHS1-X and pΔESS10 have been described previously (2). pHS1-X+ESS3 was constructed by standard cloning techniques by replacing the 20-nucleotide (nt) ESS2 sequence with a 20-nt ESS3 sequence. All plasmids were linearized with HindIII at nt 6026 of pNL4-3 (GenBank accession no. M19921) and used as templates to synthesize RNA substrates by in vitro transcription with T3 RNA polymerase.
FIG. 3.
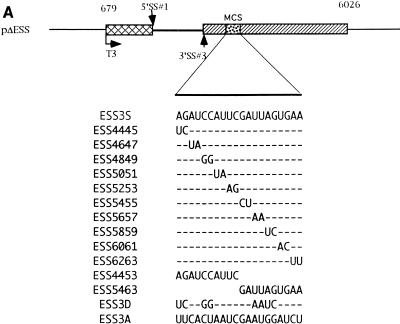
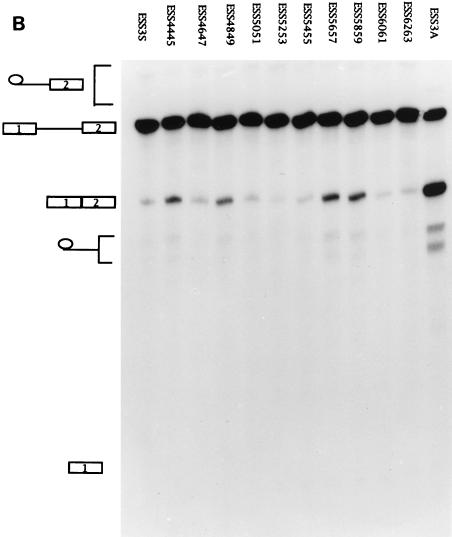
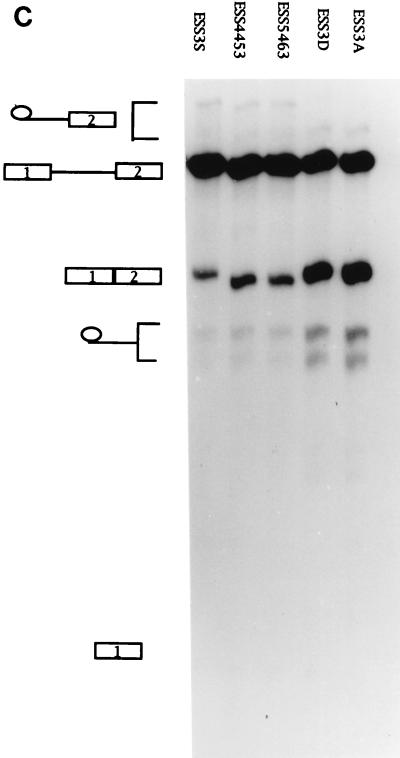
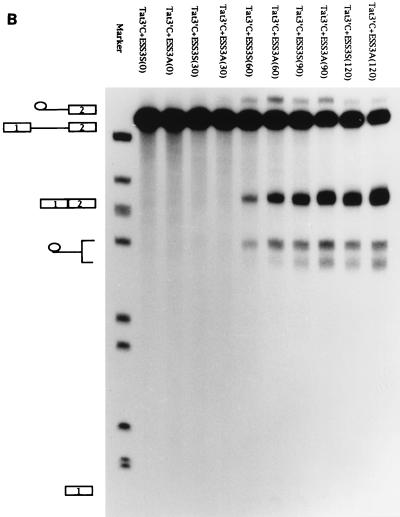
ESS3 is composed of two subelements. (A) Schematic diagram of the ESS3 mutants derived from pΔESS (see Materials and Methods). Abbreviations are defined in the legend for Fig. 2A. (B) Effects of single 2-nt substitution mutations in the ESS3 sequence on splicing efficiency. (C) Effects of the two regions of ESS3 and combination mutation on the splicing of tat exon 2. Splicing assay procedures and the identities of the band are as described in the legend for Fig. 2.
pAdML was a generous gift from Dr. R. Reed (Department of Cell Biology, Harvard University) (29). pAdML+ESS3S and pAdML+ESS3A were created by subcloning the AccI-AccI fragments containing the ESS from pESS3S and pESS3A into the unique AccI site downstream from the AdML 3′ splice site (see Fig. 4A). To prepare DNA templates for T7 RNA polymerase transcription, pAdML was linearized with BamHI and pAdML+ESS3S and pAdML+ESS3A were digested with MfeI.
FIG. 4.
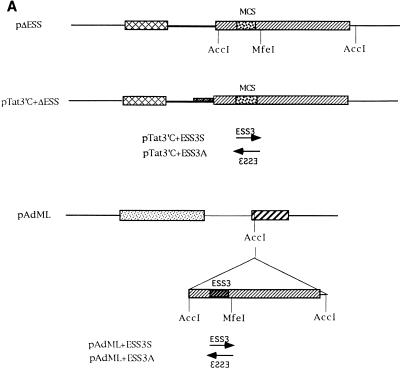
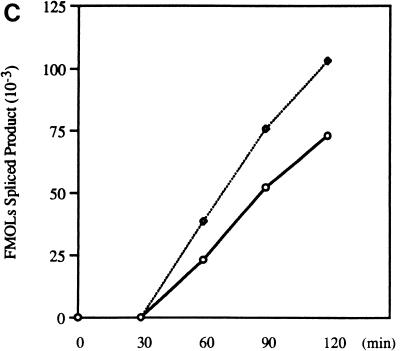
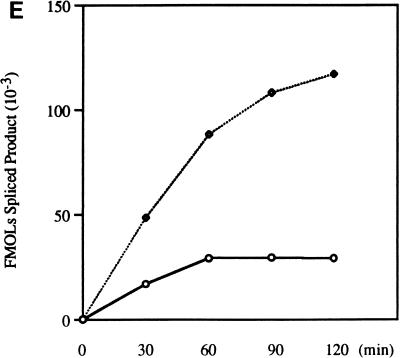
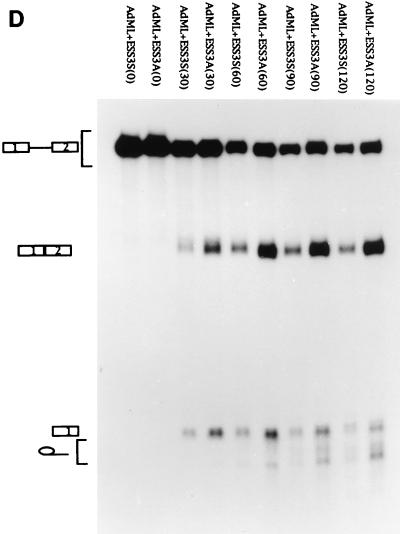
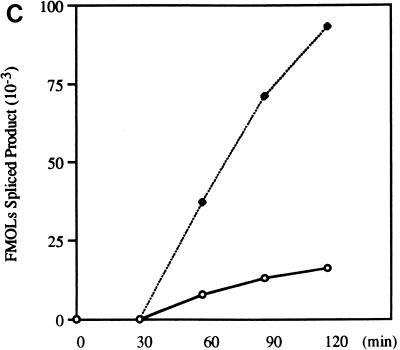
Effect of optimal 3′ splice sites on the function of ESS3. (A) Organization of DNA constructs (see Materials and Methods). pTat3′C+ΔESS is derived from pΔESS and has an optimized polypyrimidine tract and a consensus 3′ splice site (39). ESS3 was inserted into pTat3′C+ΔESS in both the sense (pTat3′C+ESS3S) and antisense (pTat3′C+ESS3A) orientations downstream from the 3′ splice site. MCS, multiple cloning site. (B) Time course of splicing for Tat3′C+ESS3S and Tat3′C+ESS3A RNA substrates. The corresponding RNA substrates were spliced and analyzed and the identities of the bands (left) are as described in the legend for Fig. 2. (C) Quantitation of the spliced products from the RNA substrates, Tat3′C+ESS3S (open symbols) and Tat3′C+ESS3A (filled symbols), shown in panel B. (D) ESS3 was inserted downstream of the 3′ splice site of pAdML in both the sense (pAdML+ESS3S) and antisense (pAdML+ESS3A) orientations. Plasmid linearization and transcription were performed as described in Materials and Methods. RNAs were spliced in vitro for various times as described in the legend for Fig. 2 with Mg2+ at a final concentration of 1 mM. (E) Quantitation of the spliced products from the RNA substrates, AdML+ESS3S (open symbols) and AdML+ESS3A (filled symbols), shown in panel D.
pTAT118 has been described previously (3) and was used to make RNA competitors containing ESS2. pTAT90S was constructed by subcloning the AccI-MfeI fragment from pESS3S between the AccI and EcoRI sites of pBluescript SK (Stratagene, La Jolla, Calif.). pEC4× was constructed according to methods previously described (27); it contains four tandem copies of the ESS2 sequence. Both plasmids were linearized with BamHI for transcription with T7 RNA polymerase. pHRR80 (obtained from Ambion, Austin, Tex.) contains an 80-nt fragment of human 18S rRNA (nt 715 to 794 the sequence under GenBank accession no. M10098), inserted downstream from the T7 promoter sequence of pUC18 in the antisense orientation. The construct was linearized with HindIII and transcribed with T7 RNA polymerase to generate a 116-nt RNA which was used as the control RNA in RNA competition assays.
In vitro RNA splicing and RNA competition assays.
RNA substrates of pΔESS and its derivatives were synthesized with T3 RNA polymerase and labeled with [32P]UTP. Uncapped RNA competitors were synthesized with T7 RNA polymerase with a MegaShortscript kit (Ambion). After DNase I treatment, the reaction mixtures were extracted twice with phenol-chloroform and passed through 1-ml Sephadex G-50 columns to remove the unincorporated nucleotides. The RNA was precipitated with ethanol in the presence of 0.3 M sodium acetate and dried under vacuum. The amount of RNA was quantitated spectrophotometrically, and the RNA was shown to be intact by agarose gel electrophoresis. HeLa cell nuclear extracts were prepared and splicing reactions were performed as previously described (2, 16, 25). For RNA competition assays, the substrates were spliced for 2 h at 30°C after the nuclear extracts were preincubated for 15 min with various amounts of nonradioactive competitor RNAs at the same temperature. The products of the splicing reactions were separated on 7 M urea–4% polyacrylamide gels. Gels were scanned for radioactivity and quantitated with an Instantimager analysis system (Packard, Meriden, Conn.). The amounts of spliced product were calculated based on the specific activity of the [32P]UTP used for labeling the precursor RNAs and were expressed as femtomoles of spliced product.
Analysis of spliceosome complexes.
Complex E formation and analysis were carried out as previously described (29). In brief, [32P]UTP-labeled RNA substrates were transcribed from linearized pESS3S and pESS3A DNA, labeled RNA was incubated under splicing conditions for 30 min in the absence of both ATP and MgCl2, and the complexes were separated on Sephacryl S500 columns (1.5 by 100 cm). Complex A assembly was analyzed by nondenaturing gel electrophoresis as previously described (23). The RNA substrates, ESS3S and ESS3A, were incubated at 30°C for various times in the presence of ATP (0.5 mM) and MgCl2 (3 mM). Complex formation was determined by electrophoresis on 4% native polyacrylamide gels with an acrylamide-to-N,N′-methylenebisacrylamide ratio of 80:1.
RESULTS
ESS3 is functional when inserted downstream of a suboptimal heterologous 3′ splice site.
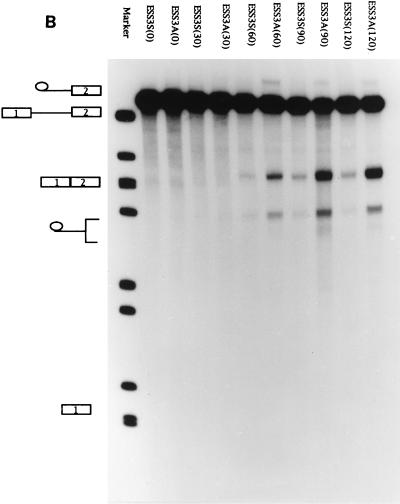
We showed previously that the 3′ splice site of tat exon 2 contains a nonconsensus polypyrimidine tract and that the negative effect of ESS2 is orientation dependent (2, 39). We have also shown that the ESS2 in tat exon 2 is active when inserted downstream of a suboptimal heterologous src splice site (2). We examined whether ESS3 was also active when placed downstream of a suboptimal heterologous 3′ splice site. For this experiment, we used construct pΔESS in which the ESS2 sequence had been deleted and the 20-nt ESS3 had been inserted in the sense and antisense orientations within tat exon 2 to create pESS3S and pESS3A, respectively (Fig. 2A). These constructs were used as templates to synthesize RNA substrates for in vitro splicing assays which were carried out for various times (Fig. 2B). As shown in Fig. 2C, the rate of splicing of substrate ESS3S was approximately 25% that of ESS3A. ESS3A, on the other hand, was spliced with an efficiency similar to that of ΔESS (data not shown). These results indicate that ESS3 behaves similarly to ESS2 in that it inhibits the efficiency of splicing when placed downstream of a suboptimal heterologous 3′ splice site.
FIG. 2.
ESS3 is functional when inserted downstream of a suboptimal heterologous 3′ splice site. (A) In pΔESS the region containing ESS2 has been deleted from pHS1-X and replaced with a multiple cloning site (MCS). ESS3 was inserted into pΔESS in both the sense (pESS3S) and antisense (pESS3A) orientations downstream of tat exon 2. [32P]UTP-labeled RNA substrates were synthesized as described in Materials and Methods and incubated in HeLa cell nuclear extracts for 2 h at 30°C under standard splicing conditions with 3 mM Mg2+ (25). T3, phage T3 RNA polymerase promoter; SS, splice site. Cap and pA, same as in Fig. 1. (B) Time course of splicing for ESS3S and ESS3A RNA substrates. Products of in vitro splicing were analyzed with denaturing polyacrylamide gels. The substrate for each lane is specified at the top, and the time (in minutes) of the reaction is shown in parentheses. The identities of bands are illustrated on the left side of the autoradiogram (top to bottom: lariat exon, pre-mRNA, spliced product, lariat, and 5′ exon). (C) Quantitation of the spliced products from RNA substrates ESS3S (open symbols) and ESS3A (filled symbols) shown in panel B.
ESS3 is composed of two subelements, and each element can inhibit splicing independently.
We have previously defined the ESS3 as a 20-nt sequence (3). We next determined whether the inhibitory activity of ESS3 was confined to certain sequences within ESS3. To this end, 2-nt substitution mutations were created to examine the significance of individual bases (Fig. 3A). Four mutant substrates (ESS4445, ESS4849, ESS5657, and ESS5859) showed significantly higher levels of splicing than the other mutant substrates, indicating that these nucleotides are important for the activity of ESS3 (Fig. 3B). However, the two groups of essential nucleotides in ESS3 (AGAUCC and UUAG) were separated by 6 nt instead of being in one contiguous region, as in ESS2. Since all of the mutant substrates were spliced with efficiencies lower than that of ESS3A, we determined the effect of combining mutations at positions 4445, 4849, 5657, and 5859 (ESS3D) (Fig. 3C). The level of splicing of this mutant was increased to comparable to that of ESS3A. This suggested that the two regions of ESS3 act additively to inhibit splicing.
To test whether the two groups of essential nucleotides represented two subelements within ESS3, we constructed templates containing either the first or second 10 nt of ESS3 (pESS4453 or pESS5463, respectively). RNA substrates synthesized from these templates were analyzed by in vitro splicing (Fig. 3C). Splicing of both ESS4453 and ESS5463 mutant substrates was inhibited but to a lesser extent than that of the substrate containing the entire ESS sequence (ESS3S). These results implied that there are two subelements within ESS3 that act independently and additively to inhibit splicing.
ESS3 is less active in the context of optimal 3′ splice sites.
Results previously acquired at our laboratory have indicated that ESS2 activity is strongly influenced by the strength of the upstream 3′ splice site which is inhibited (39). Staffa and Cochrane, who used in vivo transfection studies, came to similar conclusions regarding ESS3 (42). However, the DNA constructs used in their study contained only the upstream ESS3 subelement (42). To determine if ESS3 inhibits splicing in the context of an optimal 3′ splice site, the entire 20-nt ESS3 was inserted into pTat3′C+ΔESS in either the sense or the antisense orientation (Fig. 4A). RNA substrates synthesized from these constructs have optimal polypyrimidine tracts and consensus 3′ splice sites flanking tat exon 2 (39). The effects of the improved 3′ splice sites on the function of ESS3 were tested by in vitro splicing assays carried out for the various times shown in Fig. 4B. The rate of splicing of substrate Tat3′C+ESS3S was approximately 75% that of Tat3′C+ESS3A. Comparison of the data in Fig. 2C and 4C indicated that the effect of ESS3 was significantly weakened in the presence of an optimal 3′ splice site.
To further examine the effect of ESS3 in the context of an unrelated RNA substrate that is not subject to regulation, we placed ESS3 in both orientations within exon 2 of an adenovirus major late gene construct (pAdML) (Fig. 4A). RNA substrates synthesized with this construct have been widely used to investigate basic splicing mechanisms, since these substrates contain consensus polypyrimidine tracts and branch point sequences (5, 9, 10, 30, 44). As shown in Fig. 4D and E, when the splicing assays were carried out with 1 mM Mg2+ for various times, the rate of splicing of substrate AdML+ESS3S was found to be approximately 25% that of substrate AdML+ESS3A. These data support the notion that ESS3 is active in the context of a heterologous RNA substrate.
Spliceosome assembly is inhibited prior to the formation of a complex (complex A) with an RNA substrate containing ESS3.
Previous results indicated that splicing is inhibited at an early step of spliceosome formation by ESS2 (3). Splicing of ESS3S appeared to be inhibited before the first step of the splicing reaction, since no lariat-exon intermediate accumulated even at the 2-h point (Fig. 2B). To further define the mechanism by which ESS3 acts to inhibit splicing, we investigated the step at which spliceosome assembly is blocked by ESS3. Functional spliceosomes are assembled in a stepwise manner through a series of intermediate complexes, termed complexes E, A, B, and C (24, 29). Formation of complex E, or the commitment complex, appears not to require the presence of ATP (6, 29). This is followed by a series of ATP-dependent steps leading to formation of complexes A, B, and C (26, 31).
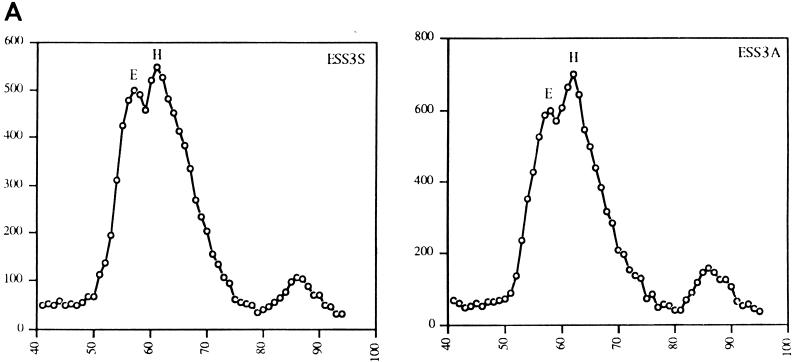
Two RNA substrates, ESS3S and ESS3A, in which ESS3 was inserted in the sense and antisense orientations, respectively, were used to study formation of spliceosome complexes. Both substrates were incubated with HeLa cell nuclear extracts under standard splicing conditions in the absence of ATP. The reaction mixtures were then subjected to gel filtration analysis under conditions previously shown to separate complex E and a nonspecific RNA-protein complex (complex H) (29). Incubation of control RNA lacking splice sites resulted in formation of only complex H, whereas the control substrate AdML formed both complexes H and E (data not shown) as described by Michaud and Reed (29). The elution profiles of the RNA substrates ESS3S and ESS3A are shown in Fig. 5A. These results indicated that two distinct peaks were present when either RNA substrate was used. One peak was eluted at a position corresponding to complex H, and the other was eluted at the position expected for complex E. These results suggested that the step of splicing inhibited by ESS3 occurred subsequent to formation of complex E.
FIG. 5.
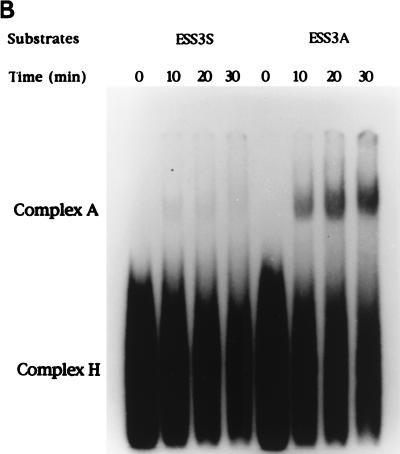
Inhibition of spliceosome complex formation by the HIV-1 ESS3. (A) RNA substrates (ESS3S and ESS3A) were incubated with HeLa cell nuclear extracts. The extracts were preincubated at room temperature to deplete endogenous ATP levels. Incubation was carried out in the absence of ATP and Mg2+ at 30°C for 25 min. The reaction mixtures were applied to a Sephacryl S500 column under conditions previously described (29). The positions of complexes E and H are shown. Numbers on the abscissa and ordinate indicate the fraction number and the amount of radioactivity in the samples (counts per minute), respectively. (B) RNA substrates were incubated with HeLa cell nuclear extracts in the presence of ATP and Mg2+ for the indicated times, and the complexes were analyzed on a 4% nondenaturing polyacrylamide gel.
To further test the effect of ESS3 on spliceosome assembly, we determined whether complex A was assembled with either or both substrates. After incubation of RNAs with nuclear extracts in the presence of ATP for various times, the reaction mixtures were applied to native polyacrylamide gels to examine the assembly of complexes (Fig. 5B). Complex A formation was detected for substrate ESS3A after 10 min. Substrate ESS3S, on the other hand, did not form a significant amount of complex A, even after a 30-min incubation. It appears, therefore, that assembly of both substrates can proceed to complex E formation but that the ESS3s substrate cannot form complex A. Thus, the inhibition of splicing caused by ESS3 takes place prior to the formation of complex A.
Evidence that ESS2 and ESS3 share a cellular factor(s) necessary for the inhibition of tat mRNA splicing.
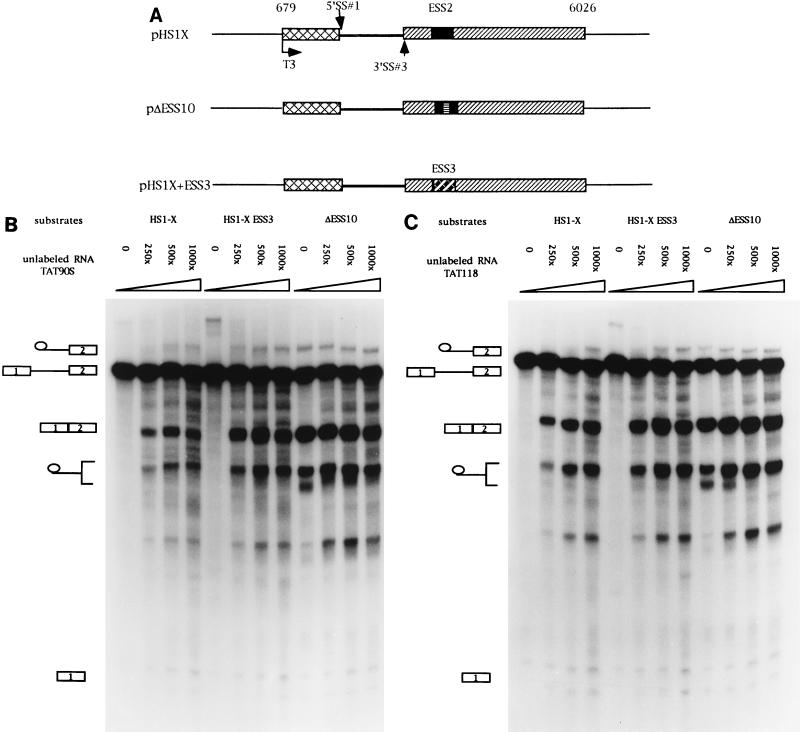
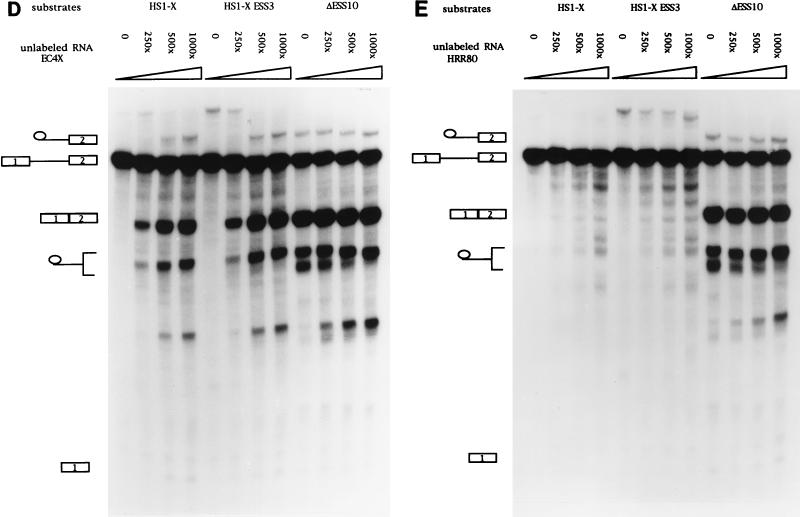
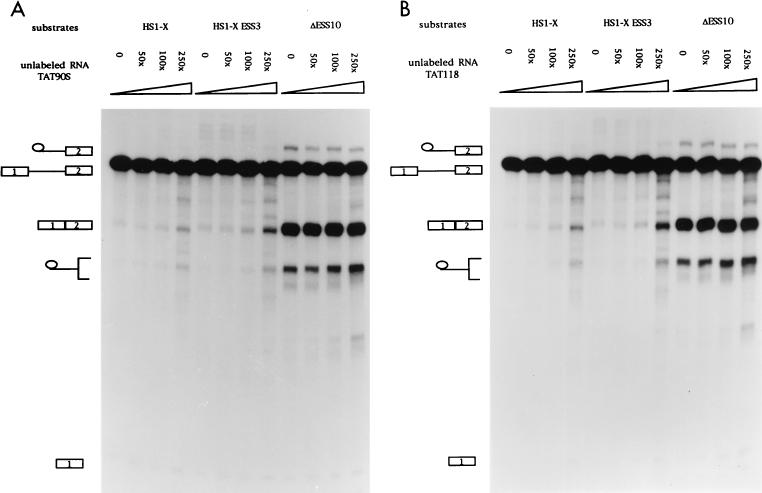
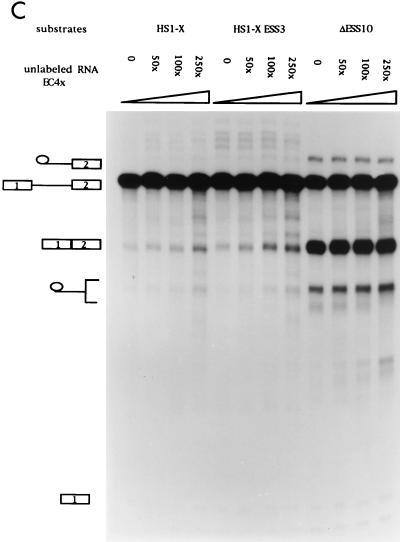
Previous assays with unlabeled competitor RNAs containing ESS2 suggested that the inhibition is mediated by a cellular factor(s) which binds to ESS2 (3). We next tested whether the inhibition of ESS3 was also mediated by a cellular factor and, if so, whether ESS3 and ESS2 bound to the same factor. RNA competition assays were carried out with three different RNA substrates (Fig. 6A). Substrates HS1-X and ΔESS10 had been previously used to study the function of ESS2 (2, 3). HS1-X contains the wild-type ESS2 sequence, whereas ΔESS10 contains a 10-nt heterologous sequence inserted in place of ESS2. The former substrate is spliced inefficiently whereas the latter substrate, because of the mutated ESS2 sequence, is spliced efficiently. The third RNA substrate was synthesized from HS1-X+ESS3, in which the 20-nt ESS3 sequence of HS1-X had been substituted for the 20-nt ESS2 sequence. As shown in Fig. 6B, splicing of HS1-X+ESS3 was very inefficient. Addition of competitor RNA containing ESS3 (TAT90S) derepressed splicing of HS1-X+ESS3 but had little effect on the efficiency of splicing of the ΔESS10 substrate. This suggested that a cellular factor specifically binds to ESS3 to mediate the inhibition of splicing. Interestingly, addition of TAT90S RNA also derepressed splicing of substrate HS1-X, which contains the ESS2 sequence. This suggests that the two ESS sequences bind to a common factor or factors. Comparison of the amounts of spliced product at the different molar excesses of competitor shown in Fig. 6B indicated that TAT90S competed approximately twofold more efficiently with the ESS3 substrate than with the ESS2 substrate. Splicing assays carried out at lower competitor concentrations (from 50- to 250-fold molar excesses) confirmed this small difference in efficiency (Fig. 7A). This suggests that ESS2 may have a slightly higher affinity for the putative common factor than does ESS3. A second RNA competitor containing the ESS2 sequence (TAT118) was tested against the same set of RNA substrates. As shown previously (3), this RNA specifically derepressed splicing of substrate HS1-X but had no effect on the splicing of ΔESS10, containing a mutated ESS2 (Fig. 6C). Addition of the ESS2 competitor also resulted in an increase in the splicing efficiency of substrate HS1-X+ESS3. Like the ESS3 competitor, TAT118 RNA competed more efficiently with the ESS3 substrate than with the ESS2 substrate. Splicing assays carried out at lower competitor concentrations confirmed this small difference (Fig. 7B).
FIG. 6.
ESS2 and ESS3 share a cellular factor(s) necessary for the inhibition of tat mRNA splicing. (A) Diagrams of constructs used to synthesize RNA substrates for competition assays. SS, splice site. (B to E) In vitro splicing assays were carried out in the presence of increasing amounts of unlabeled RNA (250, 500, and 1,000× specify the molar excesses relative to the substrate concentrations). The identities of bands are as described in the legend for Fig. 2B. (B) TAT90S RNA (ESS3 plus the same flanking sequence as ESS2). (C) TAT118 RNA (ESS2 plus the flanking sequence). (D) EC4× RNA (four tandem copies of ESS2 without the flanking sequence). (E) HRR80 RNA (control). This RNA contains an 80-nt fragment of human 18S rRNA sequence but does not contain either ESS sequence.
FIG. 7.
Competition assays against the same set of RNA substrates as shown in Fig. 6A but with lower molar excesses of competitor RNAs. 50, 100, and 250× specify the molar excesses relative to the substrate concentrations. The identities of the bands are as described in the legend for Fig. 2B.
Because the ESS2 in TAT118 and the ESS3 in TAT90S share flanking sequences, we tested a second RNA competitor (EC4×), which contains four tandem copies of the ESS2 sequence with no flanking sequence. As shown in Fig. 6D and 7C, this competitor produced derepression of splicing of HS1-X and HS1-X+ESS3 similar to that by TAT118, indicating that no sequence except that of ESS2 is required for the effect. Surprisingly, the concentration dependence of derepression by EC4×, with four tandem copies of the ESS2 sequence, was not significantly different from that of TAT118, with one copy (compare Fig. 6D with Fig. 6C and Fig. 7C with Fig. 7B). This suggests that some flanking sequence may be necessary for binding the putative factor(s).
To further test the specificity of the competitor RNAs, we used an irrelevant RNA, HRR80, which contains an 80-nt sequence derived from human 18S rRNA. This RNA did not significantly affect the splicing efficiencies of the three RNA substrates (Fig. 6E). This indicated that the derepression by the ESS2 and ESS3 competitor RNAs was specific. These data strongly suggest that the inhibitory effect of ESS3 is mediated by a cellular factor(s) and that ESS3 and ESS2 may bind to a common cellular factor.
DISCUSSION
Our results, based on in vitro splicing assays, have indicated that the ESS present in the second tat coding exon of HIV-1 (ESS3) is comprised of two smaller elements and that each of these smaller elements can inhibit splicing when placed in the exon downstream of a heterologous weak 3′ splice site. We have shown that the core sequences necessary for ESS3 activity are AGAUCC and UUAG and that the core sequence necessary for ESS2 activity is CUAGACUAGA (39). The sequences necessary for ESS activity appear to have some common sequence features. In the first part of the sequence AGAPyPy is common, and in the downstream regions PyUAG is common. It is possible that two binding sites on the RNA are necessary for full activity of an ESS. ESS2 has two repeats of the sequence CUAGA, and this may also indicate the presence of two binding sites.
The sequences of several other potential ESS elements are known, and it is of interest to compare them to those of ESS2 and ESS3. HIV-1 cryptic exon 6D is a small exon present in the env gene. The inclusion of this exon results in the production of a chimeric Tat-Env-Rev fusion protein called Tev (28). Recently it has been shown that this exon contains a potential ESS which may act to inhibit splicing at the upstream cryptic 3′ splice site. The sequence of this ESS is CAAUAGUAGUAG (51). The human fibronectin alternative EDA exon contains a cis splicing element comprised of an ESE (GAAGAAGA) and an ESS (CAAG) (8). The K-SAM alternative exon of the human fibroblast growth factor receptor 2 gene has been reported to contain an ESS element with the sequence UAGGGCAGGC. Further experiments suggested that the functional portion of this element is limited to the sequence UAGG and that the dinucleotide AG appears to be of particular importance to its activity (14, 15). Although the sequence homology among these reported ESS elements is minimal, most of them contain UAG, and all elements contain at least one AG.
Data obtained by our laboratory have suggested that both ESS2 and ESS3 act to inhibit splicing by blocking the formation of functional spliceosomes at an early step (3). We showed in Fig. 5A that a substrate containing a functional ESS3 forms a complex that is eluted at the position expected for complex E. However, since the transition from complex E to complex A may involve several intermediate steps (9), we cannot determine whether the complexes shown in Fig. 5A are functional or aberrant complexes E, and further characterization of these complexes will be required to answer this question.
RNA competition assays suggested that a cellular factor or factors bind to the ESS sequence to mediate inhibition of splicing. There are a number of examples of RNA-binding proteins that inhibit splicing at an early step. In Saccharomyces cerevisiae, the ribosomal protein L32 binds to the first exon of its own pre-mRNA to prevent the formation of functional complex A (49). In Drosophila, Sxl protein promotes the female-specific splicing of tra pre-mRNA by blocking the utilization of the default tra 3′ splice site. Sxl regulation requires a poly(U) sequence located in the polypyrimidine tract of the default 3′ splice site. Thus, it may inhibit splicing at an early step by competing with U2AF for the binding to the polypyrimidine tract (48). Splicing of the Drosophila P-element third intron is repressed in the soma at an early step by blocking of U1 snRNP binding to the actual 5′ splice site and stabilization of U1 snRNP binding to an inactive pseudo-5′ splice site. This process is regulated by an alternative splicing factor, PSI (P-element somatic inhibitor), and a general splicing factor, hrp48 (homologous to mammalian hnRNPA1) (1, 40, 41).
Our data suggest that inhibition of splicing by ESS2 and ESS3 may involve the action of a common factor or factor(s). We showed that ESS2 RNA competitors are able to abrogate the inhibition by ESS3 and vice versa. This is not surprising, since the sequences necessary for ESS2 and ESS3 activities have some common features. The small but reproducible difference between the efficiencies of derepression of the two competitor RNAs could be explained by different binding affinities resulting from the sequence difference between ESS2 and ESS3. Alternatively, it is possible that inhibition by ESS2 and ESS3 may require the formation of a complex by a number of proteins. Thus, a unique factor may bind to each of ESS2 and ESS3, but the complexes may share one or more limiting factors.
The accumulation of unspliced and partially spliced HIV-1 RNA is necessary for HIV-1 replication. It is of interest that splicing at both tat 3′ splice sites appears to be inhibited at an early step by ESS elements by similar mechanisms and with mediation by a common cellular factor(s). This would allow a coordinated response of the two splice sites to the cellular environment, with the result being the preservation of the unspliced and singly spliced RNAs necessary for Rev-dependent transport to the cytoplasm.
ACKNOWLEDGMENTS
We thank Stanley Perlman, Richard Roller, and Brad Amendt for critical reading of the manuscript.
This research was supported by PHS grant AI36073 from the National Institute of Allergy and Infectious Diseases.
REFERENCES
- 1.Adams M D, Tarng R S, Rio D C. The alternative splicing factor PSI regulates P-element third intron splicing in vivo. Genes Dev. 1997;11:129–138. doi: 10.1101/gad.11.1.129. [DOI] [PubMed] [Google Scholar]
- 2.Amendt B A, Hesslein D, Chang L-J, Stoltzfus C M. Presence of negative and positive cis-acting RNA splicing elements within and flanking the first tat coding exon of human immunodeficiency virus type 1. Mol Cell Biol. 1994;14:3960–3970. doi: 10.1128/mcb.14.6.3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amendt B A, Si Z-H, Stoltzfus C M. Presence of exon splicing silencers within human immunodeficiency virus type 1 tat exon 2 and tat-rev exon 3: evidence for inhibition mediated by cellular factors. Mol Cell Biol. 1995;15:4606–4615. doi: 10.1128/mcb.15.8.4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arrigo S, Beemon K. Regulation of Rous sarcoma virus RNA splicing and stability. Mol Cell Biol. 1988;8:4858–4867. doi: 10.1128/mcb.8.11.4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bennett M, Michaud M S, Kingston J, Reed R. Protein components specifically associated with pre-spliceosome and spliceosome complexes. Genes Dev. 1992;6:1986–2000. doi: 10.1101/gad.6.10.1986. [DOI] [PubMed] [Google Scholar]
- 6.Bennett M, Piñol-Roma S, Staknis D, Dreyfuss G, Reed R. Differential binding of heterogeneous nuclear ribonucleoproteins to mRNA precursors prior to spliceosome assembly in vitro. Mol Cell Biol. 1992;12:3165–3175. doi: 10.1128/mcb.12.7.3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bouck J, Fu X-D, Skalka A M, Katz R A. Genetic selection for balanced retroviral splicing: novel regulation involving the second step can be mediated by transitions in the polypyrimidine tract. Mol Cell Biol. 1996;15:2663–2671. doi: 10.1128/mcb.15.5.2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caputi M, Casari G, Guenzi S, Tagliabue R, Sidoli A, Melo C A, Baralle F E. A novel bipartite splicing enhancer modulates the differential processing of the human fibronectin EDA exon. Nucleic Acids Res. 1994;22:1018–1022. doi: 10.1093/nar/22.6.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Champion-Arnaud P, Gozani O, Palandjian L, Reed R. Accumulation of a novel spliceosomal complex on pre-mRNAs containing branch site mutations. Mol Cell Biol. 1995;15:5750–5756. doi: 10.1128/mcb.15.10.5750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chiara M D, Gozani O, Bennett M, Champion-Arnaud P, Palandjian L, Reed R. Identification of proteins that interact with exon sequences, splice sites, and the branchpoint sequence during each stage of spliceosome assembly. Mol Cell Biol. 1996;16:3317–3326. doi: 10.1128/mcb.16.7.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coffin J M. Retroviridae and their replication. In: Fields B N, Knipe D M, editors. Fields virology. New York, N.Y: Raven Press; 1990. pp. 1437–1500. [Google Scholar]
- 12.Cullen B R. Regulation of human immunodeficiency virus replication. Annu Rev Microbiol. 1991;45:219–250. doi: 10.1146/annurev.mi.45.100191.001251. [DOI] [PubMed] [Google Scholar]
- 13.Damier L, Domenjoud L, Branlant C. The D1-A2 and D2-A2 pairs of splice sites from HIV-1 are highly efficient in vitro, in spite of an unusual branch site. Biochem Biophys Res Commun. 1997;237:182–187. doi: 10.1006/bbrc.1997.7091. [DOI] [PubMed] [Google Scholar]
- 14.Del Gatto F, Breathnach R. Exon and intron sequences, respectively, repress and activate splicing of a fibroblast growth factor receptor 2 alternative exon. Mol Cell Biol. 1995;15:4825–4834. doi: 10.1128/mcb.15.9.4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Del Gatto F, Gesnel M-C, Breathnach R. The exon sequence TAGG can inhibit splicing. Nucleic Acids Res. 1996;24:2017–2021. doi: 10.1093/nar/24.11.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dignam J D, Lebovitz R M, Roeder R G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dyhr-Mikkelsen H, Kjems J. Inefficient spliceosome assembly and abnormal branch site selection in splicing of an HIV-1 transcript in vitro. J Biol Chem. 1995;270:24060–24066. doi: 10.1074/jbc.270.41.24060. [DOI] [PubMed] [Google Scholar]
- 18.Felber B K, Drysdale C M, Pavlakis G N. Feedback regulation of human immunodeficiency virus type 1 expression by the Rev protein. J Virol. 1990;64:3734–3741. doi: 10.1128/jvi.64.8.3734-3741.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fischer U, Meyer S, Teufel M, Heckel C, Luhrmann R, Rautmann G. Evidence that HIV-1 Rev directly promotes the nuclear export of unspliced RNA. EMBO J. 1994;13:4105–4112. doi: 10.1002/j.1460-2075.1994.tb06728.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fu X-D, Katz R A, Skalka A M, Maniatis T. The role of branchpoint and 3′-exon sequences in the control of balanced splicing of avian retrovirus RNA. Genes Dev. 1991;5:211–220. doi: 10.1101/gad.5.2.211. [DOI] [PubMed] [Google Scholar]
- 21.Guatelli J C, Gingeras T R, Richman D D. Alternative splice acceptor utilization during human immunodeficiency virus type 1 infection of cultured cells. J Virol. 1990;64:4093–4098. doi: 10.1128/jvi.64.9.4093-4098.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katz R A, Skalka A M. Control of retroviral RNA splicing through maintenance of suboptimal processing signals. Mol Cell Biol. 1990;10:696–704. doi: 10.1128/mcb.10.2.696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Konarska M M. Analysis of splicing complexes and small ribonucleoprotein particles by native gel electrophoresis. Methods Enzymol. 1989;180:442–453. doi: 10.1016/0076-6879(89)80116-8. [DOI] [PubMed] [Google Scholar]
- 24.Konarska M M, Sharp P A. Electrophoretic separation of complexes involved in the splicing of precursors to mRNA. Cell. 1986;46:845–855. doi: 10.1016/0092-8674(86)90066-8. [DOI] [PubMed] [Google Scholar]
- 25.Krainer A R, Maniatis T, Ruskin B, Green M R. Normal and mutant human beta-globin pre-mRNAs are faithfully and efficiently spliced in vitro. Cell. 1984;36:993–1005. doi: 10.1016/0092-8674(84)90049-7. [DOI] [PubMed] [Google Scholar]
- 26.Kramer A. The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu Rev Biochem. 1996;65:367–409. doi: 10.1146/annurev.bi.65.070196.002055. [DOI] [PubMed] [Google Scholar]
- 27.Liaw G. Improved protocol for directional multimerization of a DNA fragment. BioTechniques. 1994;17:668–670. [PubMed] [Google Scholar]
- 28.Luciw P A. Human immunodeficiency viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. Philadephia, Pa: Lippincott-Raven Publishers; 1996. pp. 1881–1952. [Google Scholar]
- 29.Michaud S, Reed R. An ATP-independent complex commits pre-mRNA to the mammalian spliceosome assembly pathway. Genes Dev. 1991;5:2534–2546. doi: 10.1101/gad.5.12b.2534. [DOI] [PubMed] [Google Scholar]
- 30.Michaud S, Reed R. A functional association between the 5′ and 3′ splice site is established in the earliest prespliceosome (E) complex in mammals. Genes Dev. 1993;7:1008–10020. doi: 10.1101/gad.7.6.1008. [DOI] [PubMed] [Google Scholar]
- 31.Moore M J, Query C C, Sharp P A. Splicing of precursors to mRNAs by the spliceosome. In: Gesteland R F, Atkins J R, editors. The RNA world. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1993. pp. 303–357. [Google Scholar]
- 32.Muesing M A, Smith D H, Cabradilla C D, Benton C V, Lasky L A, Capon D J. Nucleic acid structure and expression of the human AIDS/lymphadenopathy retrovirus. Nature. 1985;313:450–458. doi: 10.1038/313450a0. [DOI] [PubMed] [Google Scholar]
- 33.O’Reilly M M, McNally M T, Beemon K L. Two strong 5′ splice sites and competing suboptimal 3′ splice sites involved in alternative splicing of human immunodeficiency virus type 1 RNA. Virology. 1995;213:373–385. doi: 10.1006/viro.1995.0010. [DOI] [PubMed] [Google Scholar]
- 34.Purcell D F J, Martin M A. Alternative splicing of human immunodeficiency virus type 1 mRNA modulates viral protein expression, replication, and infectivity. J Virol. 1993;67:6365–6378. doi: 10.1128/jvi.67.11.6365-6378.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robert-Guroff M, Popovic M, Gartner S, Markham P, Gallo R C, Reitz M S. Structure and expression of tat-, rev-, and nef-specific transcripts of human immunodeficiency virus type 1 in infected lymphocytes and macrophages. J Virol. 1990;64:3391–3398. doi: 10.1128/jvi.64.7.3391-3398.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 37.Schwartz S, Felber B, Pavlakis G N. Expression of human immunodeficiency virus type 1 vif and vpr mRNAs is rev-dependent and regulated by splicing. Virology. 1991;183:677–686. doi: 10.1016/0042-6822(91)90996-o. [DOI] [PubMed] [Google Scholar]
- 38.Schwartz S, Felber B K, Benko D M, Fenyö E-M, Pavlakis G N. Cloning and functional analysis of multiply spliced mRNA species of human immunodeficiency virus type 1. J Virol. 1990;64:2519–2529. doi: 10.1128/jvi.64.6.2519-2529.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Si Z, Amendt B A, Stoltzfus C M. Splicing efficiency of HIV-1 tat RNA is determined by both a suboptimal 3′ splice site and a 10 nucleotide exon splicing silencer element located within tat exon 2. Nucleic Acids Res. 1997;25:861–867. doi: 10.1093/nar/25.4.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Siebel C W, Admon A, Rio D C. Soma-specific expression and cloning of PSI, a negative regulator of P element pre-mRNA splicing. Genes Dev. 1995;9:269–283. doi: 10.1101/gad.9.3.269. [DOI] [PubMed] [Google Scholar]
- 41.Siebel C W, Kanaar R, Rio D C. Regulation of tissue-specific P-element pre-mRNA splicing requires the RNA-binding protein PSI. Genes Dev. 1994;8:1713–1725. doi: 10.1101/gad.8.14.1713. [DOI] [PubMed] [Google Scholar]
- 42.Staffa A, Cochrane A. Identification of positive and negative splicing regulatory elements within the terminal tat-rev exon of human immunodeficiency virus type 1. Mol Cell Biol. 1995;15:4597–4605. doi: 10.1128/mcb.15.8.4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Staffa A, Cochrane A. The tat/rev intron of human immunodeficiency virus type 1 is inefficiently spliced because of suboptimal signals in the 3′ splice site. J Virol. 1994;68:3071–3079. doi: 10.1128/jvi.68.5.3071-3079.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Staknis D, Reed R. Direct interactions between pre-mRNA and six U2 small nuclear ribonucleoproteins during spliceosome assembly. Mol Cell Biol. 1994;14:2994–3005. doi: 10.1128/mcb.14.5.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stoltzfus C M. Synthesis and processing of avian sarcoma retrovirus RNA. Adv Virus Res. 1988;35:1–37. doi: 10.1016/s0065-3527(08)60707-1. [DOI] [PubMed] [Google Scholar]
- 46.Stoltzfus C M, Fogarty S J. Multiple regions in the Rous sarcoma virus src gene intron act in cis to affect the accumulation of unspliced RNA. J Virol. 1989;63:1669–1676. doi: 10.1128/jvi.63.4.1669-1676.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Touchman J W, D’Souza I, Heckman C A, Zhou R, Biggart N W, Murphy E C., Jr Branchpoint and polypyrimidine tract mutations mediating the loss and partial recovery of the Moloney murine sarcoma virus MuSVts110 thermosensitive splicing phenotype. J Virol. 1995;69:7724–7733. doi: 10.1128/jvi.69.12.7724-7733.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Valcarcel J, Singh R, Zamore P D, Green M R. The protein Sex-lethal antagonizes the splicing factor U2AF to regulate alternative splicing of transformer pre-mRNA. Nature. 1993;362:171–175. doi: 10.1038/362171a0. [DOI] [PubMed] [Google Scholar]
- 49.Vilardell J, Warner J R. Regulation of splicing at an intermediate step in the formation of the spliceosome. Genes Dev. 1994;8:211–220. doi: 10.1101/gad.8.2.211. [DOI] [PubMed] [Google Scholar]
- 50.Vogt P K. Historical introduction to the general properties of retroviruses. In: Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 1–26. [PubMed] [Google Scholar]
- 51.Wentz M P, Moore B E, Cloyd M W, Berget S M, Donehower L A. A naturally arising mutation of a potential silencer of exon splicing in human immunodeficiency virus type 1 induces dominant aberrant splicing and arrests virus production. J Virol. 1997;71:8542–8551. doi: 10.1128/jvi.71.11.8542-8551.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang L, Stoltzfus C M. A suboptimal src 3′ splice site is necessary for efficient replication of Rous sarcoma virus. Virology. 1995;206:1099–1107. doi: 10.1006/viro.1995.1033. [DOI] [PubMed] [Google Scholar]